In the title compound, the benzimidazole ring system is inclined to the the benzene ring by 78.04 (10)°. The crystal structure features O—H⋯N and C—H⋯O hydrogen bonding and C—H⋯π and π–π interactions.
Keywords: crystal structure, benzimidazole, Tauc plot, Hirshfeld surface analysis
Abstract
In the title compound, C15H12N2O2, the benzimidazole ring system is inclined to the benzene ring by 78.04 (10)°. The crystal structure features O—H⋯N and C—H⋯O hydrogen bonding and C—H⋯π and π–π interactions, which were investigated using Hirshfeld surface analysis.
Chemical context
Benzimidazole is a naturally ocurring compound, being present in vitamin B12 (Crofts et al., 2014 ▸) and may also be synthesized from benzoic acid and o-phenylenediamine in presence of an excess of acid. Benzimidazole and its derivatives show biological activities such as antibacterial, antifungal (Yadav et al., 2015 ▸), antimicrobial (Shruthi et al., 2016 ▸), and anticancer (Kalalbandi et al., 2015 ▸). Cyanobenzyl compounds are used as intermediates in the synthesis of species that possess significant pharmaceutical properties. Compounds having carboxylic acid as a functional group have shown chelating properties and thus have potential applications in the field of biology. Such groups are also helpful in building metal–organic frameworks that usually form supramolecular networks due to extensive hydrogen bonding and weak interactions. For example, 4-[(1H-benzo[d]imidazol-1-yl)methyl]benzoic acid has been used to construct coordination polymers with different metal ions (Ahmad et al., 2013 ▸). Herein, we report the title compound, 2-[(1H-benzimidazol-1-yl)methyl]benzoic acid, which was synthesized by a condensation reaction of benzimidazole and 2-(bromomethyl) benzonitrile in acetonitrile followed by a hydrolysis process.
Structural commentary
The asymmetric unit of the title compound is illustrated in Fig. 1 ▸. The molecule is non-planar with a dihedral angle of 78.04 (10) between the benzimidazole ring system and the benzene ring. The N1—C8—C7 angle is 113.31° and the C9—N1—C8—C7 torsion angle is −116.8 (2)°,. The C10—C15 bond length [1.408 (3) Å] is comparable to that in a similar benzimidazole derivative (Faizi et al., 2017 ▸). The C—O bond lengths [C1—O1 = 1.319 (3) and C1—O2 = 1.216 (3) Å] are in the expected range (Kamaal et al., 2019 ▸).
Figure 1.
Asymmetric unit of title compound, with atom labelling and displacement ellipsoids are drawn at the 50% probability level.
Supramolecular features
In the crystal, the molecules are connected via O—H⋯N and C—H⋯O hydrogen bonds (Table 1 ▸), forming a 1D framework along the b-axis direction (Fig. 2 ▸). C—H⋯π and π–π interactions [centroid–centroid distance = 3.6166 (15) Å] between the N1/N2/C9/C10/C15 and C2–C7 rings also occur, leading to the formation of the supramolecular structure (Fig. 3 ▸).
Table 1. Hydrogen-bond geometry (Å, °).
Cg1, Cg2, Cg3 and Cg4 are the centroids of the N1/N2/C9/C10/C15, C2–C7, C10–C15 and N1/N2/C9–15 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯N2i | 0.88 (3) | 1.73 (3) | 2.592 (3) | 164 (4) |
| C8—H8A⋯O1ii | 0.99 (1) | 2.62 (1) | 3.374 (3) | 133 (1) |
| C4—H4⋯Cg1iii | 0.95 (1) | 2.99 (1) | 3.865 (3) | 155 (1) |
| C4—H4⋯Cg3iii | 0.95 (1) | 2.51 (1) | 3.408 (3) | 157 (1) |
| C4—H4⋯Cg4iii | 0.95 (1) | 2.51 (1) | 3.454 (3) | 170 (1) |
| C5—H5⋯Cg2iii | 0.95 (1) | 2.76 (1) | 3.554 (3) | 142 (1) |
Symmetry codes: (i) -x, y+{\script{1\over 2}}, -z+{\script{3\over 2}}; (ii) -x+1, y-{\script{1\over 2}}, -z+{\script{3\over 2}}; (iii) x+{\script{1\over 2}}, -y+{\script{3\over 2}}, -z+1.
Figure 2.
View of the crystal packing along the a axis, showing O—H⋯N and C—H⋯O hydrogen-bonding interactions forming a one-dimensional chain.
Figure 3.
The hydrogen bonding and C—H⋯π and π–π interactions form zigzag chains, giving a supramolecular structure along the bc plane.
Database survey
A search of the Cambridge Structural Database (CSD, Version 5.42, November 2020; Groom et al., 2016 ▸) found five examples of similar compounds: bis(pentafluorophenyl)-(μ-{1,1′-[1,2-phenylenebis(methylene)]bis(1H-benzimidazole)})digold(I) acetone solvate (WOPLIZ; Zheng et al., 2019 ▸), 3,3′-[1,2-phenylenebis(methylene)]bis(1-ethylbenzimidazolium) dibromide (LANHAL; Haque et al., 2012 ▸), 2-[(1H-benzimidazol-1-yl)methyl]benzonitrile (JONYUJ; Akkoç et al., 2017 ▸), 1-[(2-cyanophenyl)methyl]-3-[(2-methylphenyl)methyl]-1H-benzimidazol-3-ium (JONZAQ; Akkoç et al., 2017 ▸) and 1-(2-cyanobenzyl)-3-methyl-1H-3,1-benzimidazol-3-ium bromide (MOCWAE; Ghdhayeb et al., 2014 ▸).
Hirshfeld surface analysis
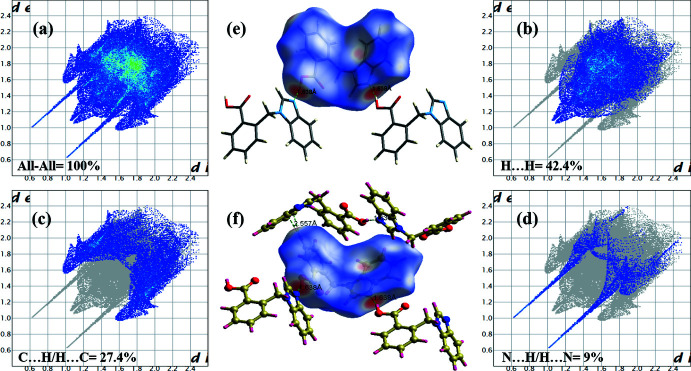
A Hirshfeld surface analysis was performed and the two-dimensional fingerprint plots generated (McKinnon et al., 2007 ▸; Spackman & Jayatilaka et al., 2009 ▸) using CrystalExplorer17 (Turner et al., 2017 ▸). The Hirshfeld surface mapped over d norm, colour-mapped from red (shorter distance than the sum of van der Waals radii) through white to blue (longer distance than the sum of the van der Waals radii). The principal weak interactions are clearly visible. The surface coverage corresponding to O—H⋯N and C—H⋯O interactions are 9% and 11.8%, respectively. The dark-red spot indicates significant hydrogen bonding.
The two-dimensional finger plots are given in Fig. 4 ▸. The principal contributions to the overall surface are from H⋯H (42.4%, Fig. 4 ▸ b), C⋯H/H⋯C (27.4%, Fig. 4 ▸ c) and N⋯H/H⋯N 9% (Fig. 4 ▸ d) interactions. The contributions of interactions such as C⋯C 4.8% are negligible.
Figure 4.
The Hirshfeld surface of the title compound mapped over d norm, in the range −0.722 to 1.183. (a) The overall two-dimensional finger plot of the title compound and those delineated into (b) H⋯H (42.4%), (c) C⋯H/ H⋯C (27.4%) and (d) N⋯H/H⋯N (9%) interactions, (e) significant hydrogen bonding and (f) extended supramolecular form.
Electronic transition analysis
Electro-conducting materials synthesized by conjugated organic compounds show promising electronic properties due to the availability of delocalized electrons, except for semiconducting materials such as TiO2, ZnO and other metal oxide nano-materials, which are electro-conducting in themselves (Odziomek et al., 2017 ▸). The electronic properties of organic compounds depend on the electronic transition between the highest occupied molecular orbital (HOMO) or valence band and lowest occupied molecular orbital (LUMO) or conduction band. In a simple method, the energy band gap (Eg) of organic molecule is determined by a Tauc plot from the absorption spectra (λmax = 245 nm, in this case). The band gap energy, Eg = 4.6 eV, of the title compound is very large (Fig. 5 ▸). This large band gap arises due to high π-conjugation or polarization in the title molecule system. The title molecule could be useful for developing or enhancing the organic electronic properties of conducting materials such as metal–organic frameworks.
Figure 5.
Energy band gap of the title molecule by Tauc plot from absorption spectra.
Synthesis and crystallization
In an equimolar ratio, benzimidazole (2 g, 16.9 mmol) and dry K2CO3 (4.66 g, 33.85 mmol) were mixed in a round-bottom flask in acetonitrile (MeCN, 60 ml) under an inert atmosphere. The mixture was then allowed to stirred for 60 min at 363 K then treated with 2-(bromomethyl) benzonitrile (3.31 g, 16.9 mmol), and the resulting solution refluxed for 24 h. After completion of this step, the solution was allowed to cool to room temperature and the mixture was poured slowly onto ice–water (100 ml) under constant stirring. A greenish muddy crystalline precipitate was obtained and it was left to stand at 293 K for two days. After two days, a crystalline powder of 2-[(1H-benzo[d]imidazol-1-yl)methyl]benzonitrile was obtained (Ahmad et al., 2013 ▸).
The title compound was synthesized by hydrolysis of 2-[(1H-benzo[d]imidazol-1-yl)methyl]benzonitrile, 2 g being mixed with 20 equimolar of potassium hydroxide (6.86 g, 8.58 mmol) in water. The solution was refluxed at 373 K for 36 h, the resultant solution was then allowed to cool at room temperature and then poured onto ice–water, and after that acidified using 6 N HCl for protonation. The protonated solution was kept for slow evaporation. After two weeks, pale-yellow cubic crystals were obtained in good yield, which were suitable for data collection. The reaction scheme is shown in Fig. 6 ▸.
Figure 6.
Reaction scheme.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C15H12N2O2 |
| M r | 252.28 |
| Crystal system, space group | Orthorhombic, P212121 |
| Temperature (K) | 100 |
| a, b, c (Å) | 6.5690 (8), 12.7956 (15), 14.1278 (16) |
| V (Å3) | 1187.5 (2) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.10 |
| Crystal size (mm) | 0.38 × 0.21 × 0.14 |
| Data collection | |
| Diffractometer | Bruker APEXII CCD |
| Absorption correction | Multi-scan (SADABS; Bruker, 2014 ▸) |
| No. of measured, independent and observed [I ≥ 2u(I)] reflections | 18798, 2095, 1759 |
| R int | 0.107 |
| (sin θ/λ)max (Å−1) | 0.596 |
| Refinement | |
| R[F2 > 2σ(F 2)], wR(F 2), S | 0.044, 0.091, 1.09 |
| No. of reflections | 2095 |
| No. of parameters | 176 |
| No. of restraints | 1 |
| H-atom treatment | All H-atom parameters refined |
| Δρmax, Δρmin (e Å−3) | 0.24, −0.28 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989021006435/ex2044sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021006435/ex2044Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989021006435/ex2044Isup3.cml
CCDC reference: 2091351
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors are grateful to the Department of Applied Chemistry, Aligarh Muslim University, for providing laboratory facilities. Author contribution are as follows. Conceptualization, MA and AM; methodology, AA and MS; investigation, MM, SK, SJ and JAK; writing (original draft), AA and ND; writing (review and editing of the manuscript), AAfall and ADA; visualization, MA, AA and AM; funding acquisition, AM; resources, ND and MA; supervision, MA and AM.
supplementary crystallographic information
Crystal data
| C15H12N2O2 | Dx = 1.411 Mg m−3 |
| Mr = 252.28 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, P212121 | Cell parameters from 1209 reflections |
| a = 6.5690 (8) Å | θ = 2.9–22.1° |
| b = 12.7956 (15) Å | µ = 0.10 mm−1 |
| c = 14.1278 (16) Å | T = 100 K |
| V = 1187.5 (2) Å3 | Block, colourless |
| Z = 4 | 0.38 × 0.21 × 0.14 mm |
| F(000) = 528.253 |
Data collection
| Bruker APEXII CCD diffractometer | 1759 reflections with I≥ 2u(I) |
| Detector resolution: X-ray pixels mm-1 | Rint = 0.107 |
| φ and ω scans | θmax = 25.1°, θmin = 3.2° |
| Absorption correction: multi-scan (SADABS; Bruker, 2014) | h = −8→8 |
| k = −17→17 | |
| 18798 measured reflections | l = −18→18 |
| 2095 independent reflections |
Refinement
| Refinement on F2 | 21 constraints |
| Least-squares matrix: full | Primary atom site location: iterative |
| R[F2 > 2σ(F2)] = 0.044 | All H-atom parameters refined |
| wR(F2) = 0.091 | w = 1/[σ2(Fo2) + (0.0284P)2 + 0.3178P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.09 | (Δ/σ)max = 0.004 |
| 2095 reflections | Δρmax = 0.24 e Å−3 |
| 176 parameters | Δρmin = −0.28 e Å−3 |
| 1 restraint |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.2861 (4) | 0.82904 (18) | 0.71973 (18) | 0.0170 (6) | |
| C2 | 0.4789 (4) | 0.81263 (19) | 0.66504 (18) | 0.0166 (6) | |
| C3 | 0.5267 (4) | 0.88501 (19) | 0.59451 (17) | 0.0186 (6) | |
| H3 | 0.4333 (4) | 0.93965 (19) | 0.58083 (17) | 0.0223 (8)* | |
| C4 | 0.7074 (4) | 0.8790 (2) | 0.54393 (18) | 0.0213 (6) | |
| H4 | 0.7359 (4) | 0.9282 (2) | 0.49537 (18) | 0.0256 (8)* | |
| C5 | 0.8447 (4) | 0.80132 (19) | 0.56462 (18) | 0.0204 (7) | |
| H5 | 0.9705 (4) | 0.79769 (19) | 0.53166 (18) | 0.0245 (8)* | |
| C6 | 0.7985 (4) | 0.7280 (2) | 0.63408 (18) | 0.0196 (6) | |
| H6 | 0.8941 (4) | 0.6743 (2) | 0.64778 (18) | 0.0235 (7)* | |
| C7 | 0.6164 (4) | 0.7311 (2) | 0.68391 (17) | 0.0163 (6) | |
| C8 | 0.5804 (4) | 0.6465 (2) | 0.75638 (17) | 0.0179 (6) | |
| H8a | 0.7114 (4) | 0.6115 (2) | 0.77035 (17) | 0.0215 (7)* | |
| H8b | 0.5313 (4) | 0.6790 (2) | 0.81572 (17) | 0.0215 (7)* | |
| C9 | 0.2503 (4) | 0.54778 (19) | 0.76721 (18) | 0.0197 (6) | |
| H9 | 0.2023 (4) | 0.58470 (19) | 0.82111 (18) | 0.0236 (7)* | |
| C10 | 0.2677 (4) | 0.43913 (19) | 0.65142 (17) | 0.0176 (6) | |
| C11 | 0.2311 (4) | 0.3622 (2) | 0.58413 (19) | 0.0229 (6) | |
| H11 | 0.1089 (4) | 0.3224 (2) | 0.58480 (19) | 0.0274 (8)* | |
| C12 | 0.3793 (4) | 0.3456 (2) | 0.51608 (19) | 0.0248 (7) | |
| H12 | 0.3580 (4) | 0.2940 (2) | 0.46874 (19) | 0.0297 (8)* | |
| C13 | 0.5601 (4) | 0.40366 (19) | 0.51595 (18) | 0.0223 (7) | |
| H13 | 0.6590 (4) | 0.39013 (19) | 0.46844 (18) | 0.0267 (8)* | |
| C14 | 0.5993 (4) | 0.4801 (2) | 0.58275 (18) | 0.0197 (6) | |
| H14 | 0.7226 (4) | 0.5188 (2) | 0.58240 (18) | 0.0236 (8)* | |
| C15 | 0.4494 (4) | 0.49744 (19) | 0.65048 (17) | 0.0156 (6) | |
| N1 | 0.4321 (3) | 0.56758 (15) | 0.72553 (14) | 0.0158 (5) | |
| N2 | 0.1467 (3) | 0.47270 (16) | 0.72598 (15) | 0.0195 (5) | |
| O1 | 0.1923 (3) | 0.91615 (15) | 0.69535 (13) | 0.0228 (5) | |
| O2 | 0.2246 (3) | 0.77045 (13) | 0.78116 (12) | 0.0227 (4) | |
| H1 | 0.078 (4) | 0.924 (3) | 0.727 (2) | 0.077 (13)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0165 (14) | 0.0131 (13) | 0.0214 (14) | −0.0029 (12) | −0.0021 (12) | −0.0043 (12) |
| C2 | 0.0186 (14) | 0.0126 (13) | 0.0184 (14) | −0.0064 (11) | −0.0017 (12) | −0.0040 (11) |
| C3 | 0.0246 (16) | 0.0118 (14) | 0.0193 (14) | 0.0002 (12) | −0.0039 (12) | −0.0027 (11) |
| C4 | 0.0257 (16) | 0.0207 (14) | 0.0175 (14) | −0.0068 (13) | 0.0054 (13) | −0.0014 (12) |
| C5 | 0.0217 (15) | 0.0172 (14) | 0.0223 (16) | −0.0060 (12) | 0.0068 (12) | −0.0042 (12) |
| C6 | 0.0187 (14) | 0.0166 (13) | 0.0235 (15) | 0.0024 (12) | −0.0007 (12) | −0.0050 (12) |
| C7 | 0.0166 (13) | 0.0158 (14) | 0.0164 (13) | −0.0037 (12) | −0.0020 (11) | −0.0056 (11) |
| C8 | 0.0166 (13) | 0.0181 (14) | 0.0190 (14) | −0.0010 (12) | −0.0033 (12) | −0.0009 (12) |
| C9 | 0.0241 (15) | 0.0163 (14) | 0.0186 (14) | 0.0038 (12) | 0.0022 (13) | 0.0027 (12) |
| C10 | 0.0210 (14) | 0.0149 (13) | 0.0170 (13) | 0.0010 (12) | −0.0025 (12) | 0.0032 (11) |
| C11 | 0.0235 (15) | 0.0183 (14) | 0.0268 (15) | −0.0006 (13) | −0.0034 (13) | 0.0007 (12) |
| C12 | 0.0339 (17) | 0.0159 (14) | 0.0244 (16) | 0.0026 (14) | −0.0028 (14) | −0.0030 (13) |
| C13 | 0.0295 (17) | 0.0185 (15) | 0.0188 (14) | 0.0077 (12) | 0.0031 (13) | 0.0007 (12) |
| C14 | 0.0201 (15) | 0.0178 (14) | 0.0212 (14) | −0.0002 (12) | 0.0010 (12) | 0.0031 (12) |
| C15 | 0.0198 (15) | 0.0117 (13) | 0.0153 (13) | 0.0008 (11) | −0.0027 (12) | 0.0017 (11) |
| N1 | 0.0160 (12) | 0.0144 (11) | 0.0170 (11) | −0.0010 (9) | −0.0006 (10) | 0.0000 (10) |
| N2 | 0.0222 (12) | 0.0136 (12) | 0.0226 (12) | −0.0009 (10) | 0.0014 (11) | 0.0032 (10) |
| O1 | 0.0170 (11) | 0.0199 (10) | 0.0316 (11) | 0.0030 (9) | 0.0012 (9) | 0.0042 (9) |
| O2 | 0.0250 (10) | 0.0171 (9) | 0.0259 (11) | 0.0012 (8) | 0.0075 (9) | 0.0031 (9) |
Geometric parameters (Å, º)
| C1—C2 | 1.499 (3) | C9—H9 | 0.950 (4) |
| C1—O1 | 1.319 (3) | C9—N1 | 1.356 (3) |
| C1—O2 | 1.216 (3) | C9—N2 | 1.313 (3) |
| C2—C3 | 1.396 (3) | C10—C11 | 1.390 (3) |
| C2—C7 | 1.405 (3) | C10—C15 | 1.408 (3) |
| C3—H3 | 0.950 (4) | C10—N2 | 1.388 (3) |
| C3—C4 | 1.388 (3) | C11—H11 | 0.9501 (4) |
| C4—H4 | 0.950 (4) | C11—C12 | 1.385 (4) |
| C4—C5 | 1.373 (4) | C12—H12 | 0.950 (4) |
| C5—H5 | 0.950 (4) | C12—C13 | 1.401 (4) |
| C5—C6 | 1.391 (4) | C13—H13 | 0.950 (4) |
| C6—H6 | 0.951 (4) | C13—C14 | 1.383 (3) |
| C6—C7 | 1.389 (4) | C14—H14 | 0.949 (4) |
| C7—C8 | 1.508 (3) | C14—C15 | 1.391 (4) |
| C8—H8a | 0.990 (4) | C15—N1 | 1.394 (3) |
| C8—H8b | 0.990 (3) | O1—H1 | 0.878 (18) |
| C8—N1 | 1.469 (3) | ||
| O1—C1—C2 | 112.3 (2) | N1—C9—H9 | 123.20 (14) |
| O2—C1—C2 | 124.2 (2) | N2—C9—H9 | 123.20 (15) |
| O2—C1—O1 | 123.5 (2) | N2—C9—N1 | 113.6 (2) |
| C3—C2—C1 | 117.7 (2) | C15—C10—C11 | 121.1 (2) |
| C7—C2—C1 | 123.3 (2) | N2—C10—C11 | 129.7 (2) |
| C7—C2—C3 | 118.9 (2) | N2—C10—C15 | 109.2 (2) |
| H3—C3—C2 | 119.23 (15) | H11—C11—C10 | 121.25 (15) |
| C4—C3—C2 | 121.5 (2) | C12—C11—C10 | 117.5 (3) |
| C4—C3—H3 | 119.23 (15) | C12—C11—H11 | 121.25 (16) |
| H4—C4—C3 | 120.26 (15) | H12—C12—C11 | 119.48 (16) |
| C5—C4—C3 | 119.5 (2) | C13—C12—C11 | 121.0 (2) |
| C5—C4—H4 | 120.26 (15) | C13—C12—H12 | 119.48 (15) |
| H5—C5—C4 | 120.16 (15) | H13—C13—C12 | 118.94 (15) |
| C6—C5—C4 | 119.7 (2) | C14—C13—C12 | 122.1 (2) |
| C6—C5—H5 | 120.16 (16) | C14—C13—H13 | 118.94 (16) |
| H6—C6—C5 | 119.13 (16) | H14—C14—C13 | 121.59 (16) |
| C7—C6—C5 | 121.7 (2) | C15—C14—C13 | 116.8 (3) |
| C7—C6—H6 | 119.13 (16) | C15—C14—H14 | 121.59 (16) |
| C6—C7—C2 | 118.6 (2) | C14—C15—C10 | 121.5 (2) |
| C8—C7—C2 | 124.1 (2) | N1—C15—C10 | 105.3 (2) |
| C8—C7—C6 | 117.3 (2) | N1—C15—C14 | 133.2 (2) |
| H8a—C8—C7 | 108.91 (14) | C9—N1—C8 | 125.7 (2) |
| H8b—C8—C7 | 108.91 (13) | C15—N1—C8 | 127.9 (2) |
| H8b—C8—H8a | 107.7 (3) | C15—N1—C9 | 106.4 (2) |
| N1—C8—C7 | 113.31 (19) | C10—N2—C9 | 105.4 (2) |
| N1—C8—H8a | 108.91 (12) | H1—O1—C1 | 111 (2) |
| N1—C8—H8b | 108.91 (12) | ||
| C1—C2—C3—C4 | 176.5 (2) | C8—N1—C9—N2 | −179.6 (2) |
| C1—C2—C7—C6 | −174.8 (2) | C8—N1—C15—C10 | 179.7 (3) |
| C1—C2—C7—C8 | 4.3 (3) | C8—N1—C15—C14 | −1.3 (3) |
| C2—C3—C4—C5 | −1.2 (3) | C9—N1—C15—C10 | 1.0 (2) |
| C2—C7—C6—C5 | −1.9 (3) | C9—N1—C15—C14 | 179.9 (2) |
| C2—C7—C8—N1 | 75.2 (3) | C9—N2—C10—C11 | −179.2 (2) |
| C3—C4—C5—C6 | 1.8 (3) | C9—N2—C10—C15 | 0.4 (2) |
| C4—C5—C6—C7 | −0.2 (3) | C10—C11—C12—C13 | 0.6 (3) |
| C5—C6—C7—C8 | 178.9 (2) | C10—C15—C14—C13 | 0.7 (3) |
| C6—C7—C8—N1 | −105.6 (2) | C11—C12—C13—C14 | −0.3 (3) |
| C7—C8—N1—C9 | −116.8 (2) | C12—C13—C14—C15 | −0.4 (3) |
| C7—C8—N1—C15 | 64.7 (3) | C13—C14—C15—N1 | −178.1 (2) |
Hydrogen-bond geometry (Å, º)
Cg1, Cg2, Cg3 and Cg4 are the centroids of the N1/N2/C9/C10/C15, C2–C7, C10–C15 and N1/N2/C9–15 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···N2i | 0.88 (3) | 1.73 (3) | 2.592 (3) | 164 (4) |
| C8—H8A···O1ii | 0.99 (1) | 2.62 (1) | 3.374 (3) | 133 (1) |
| C3—H3···O1 | 0.95 (1) | 2.28 (1) | 2.648 (3) | 102 (1) |
| C8—H8B···O2 | 0.99 (1) | 2.38 (1) | 2.846 (3) | 108 (1) |
| C9—H9···O2 | 0.95 (1) | 2.45 (1) | 2.861 (3) | 106 (1) |
| C4—H4···Cg1iii | 0.95 (1) | 2.99 (1) | 3.865 (3) | 155 (1) |
| C4—H4···Cg3iii | 0.95 (1) | 2.51 (1) | 3.408 (3) | 157 (1) |
| C4—H4···Cg4iii | 0.95 (1) | 2.51 (1) | 3.454 (3) | 170 (1) |
| C5—H5···Cg2iii | 0.95 (1) | 2.76 (1) | 3.554 (3) | 142 (1) |
Symmetry codes: (i) −x, y+1/2, −z+3/2; (ii) −x+1, y−1/2, −z+3/2; (iii) x+1/2, −y+3/2, −z+1.
Funding Statement
This work was funded by University Grants Commission; Department of Pharmacy, University of Science and Technology, Ibb Branch, Yemen.
References
- Ahmad, M. & Bharadwaj, P. K. (2013). Polyhedron, 52, 1145–1152.
- Akkoç, S., Kayser, V., İlhan, I. O., Hibbs, D. E., Gök, Y., Williams, P. A., Hawkins, B. & Lai, F. (2017). J. Organomet. Chem. 839, 98–107.
- Bourhis, L. J., Dolomanov, O. V., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2015). Acta Cryst. A71, 59–75. [DOI] [PMC free article] [PubMed]
- Bruker (2014). APEX2, SAINT and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Crofts, T. S., Men, Y., Alvarez-Cohen, L. & Taga, M. E. (2014). Front. Microbiol. 5, PMID 25431570. [DOI] [PMC free article] [PubMed]
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Faizi, M. S. H., Dege, N. & Malinkin, S. (2017). Acta Cryst. E73, 1180–1183. [DOI] [PMC free article] [PubMed]
- Ghdhayeb, M. Z., Haque, R. A. & Budagumpi, S. (2014). J. Organomet. Chem. 757, 42–50.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Haque, R. A., Iqbal, M. A., Budagumpi, S., Hemamalini, M. & Fun, H.-K. (2012). Acta Cryst. E68, o573. [DOI] [PMC free article] [PubMed]
- Kalalbandi, V. K. A. & Seetharamappa, J. (2015). Med. Chem. Commun. 6, 1942–1953.
- Kamaal, S., Faizi, M. S. H., Ali, A., Ahmad, M., Gupta, M., Dege, N. & Iskenderov, T. (2019). Acta Cryst. E75, 159–162. [DOI] [PMC free article] [PubMed]
- McKinnon, J. J., Jayatilaka, D. & Spackman, M. A. (2007). Chem. Commun. 3814–3816. [DOI] [PubMed]
- Odziomek, K., Ushizima, D., Oberbek, P., KurzydŁowski, K. J., Puzyn, T. & Haranczyk, M. (2017). J. Microsc. 265, 34–50. [DOI] [PubMed]
- Shruthi, N., Poojary, B., Kumar, V., Hussain, M. M., Rai, V. M., Pai, V. R., Bhat, M. & Revannasiddappa, B. C. (2016). RSC Adv. 6, 8303–8316.
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm, 11, 19–32.
- Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer17. University of Western Australia. http://hirshfeldsurface.net
- Yadav, G. & Ganguly, S. (2015). Eur. J. Med. Chem. 97, 419–443. [DOI] [PubMed]
- Zheng, Q., Borsley, S., Nichol, G. S., Duarte, F. & Cockroft, S. L. (2019). Angew. Chem. Int. Ed. 58, 12617–12623. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989021006435/ex2044sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021006435/ex2044Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989021006435/ex2044Isup3.cml
CCDC reference: 2091351
Additional supporting information: crystallographic information; 3D view; checkCIF report