The styryl and ester substituents are displaced to opposite sides of the plane through the pyridine ring while the acetyl group is rotated well out of that plane. In the crystal, inversion-related C—H⋯O hydrogen bonds form chains extending parallel to the a-axis direction, which pack with partial intercalation of the styryl and ester substituents.
Keywords: crystal structure, pyridine, styryl, ester, hydrogen bond, Hirshfeld surface analysis
Abstract
In the title molecule, C21H20N2O3S, the styryl and ester substituents are displaced to opposite sides of the plane of the pyridine ring. In the crystal, C—H⋯O hydrogen bonds form chains extending parallel to the a-axis direction, which pack with partial intercalation of the styryl and ester substituents. A Hirshfeld surface analysis indicates that the most significant contributions to the crystal packing are from H⋯H (43.6%), C⋯H/H⋯C (15.6%), O⋯H/H⋯O (14.9%) and N⋯H/H⋯N (11.2%) contacts.
Chemical context
Numerous pyridine-containing natural products and synthetic organic compounds with various biophysio- and pharmacological activities have been reported (Gibson et al., 2007 ▸; Vidaillac et al., 2007 ▸). These scaffolds are also of widespread interest in supramolecular and coordination chemistry, as well as for materials science (Balasubramanian & Keay, 1996 ▸). The above findings promoted us to study the crystal structure of the title compound, C21H20N2O3S.
Structural commentary
The styryl substituent and the ester group are displaced to opposite sides of the plane of the pyridine ring (Fig. 1 ▸). The dihedral angle between the mean planes of the phenyl (C8–C13) and pyridine (N1/C1–C5) rings is 27.86 (3)°. The C1—C2—C14—C15 torsion angle of 68.1 (2)° indicates that the acetyl group is rotated well out of the plane of the pyridine ring, while the N1—C4—S1—C18 torsion angle of −5.66 (12)° shows that the link to the ester group is nearly coplanar with the pyridine ring.
Figure 1.
The title molecule with labelling scheme and displacement ellipsoids at the 50% probability level.
Supramolecular features
In the crystal, inversion dimers are formed by intermolecular C15—H15A⋯O2 hydrogen bonds between a methyl H atom of the acetyl group and the carbonyl O atom of the ester function. These dimers are further linked by inversion-related C18—H18B⋯O1 hydrogen bonds between a methylene H atom and the carbonyl O atom of the acetyl group (Table 1 ▸) to form ribbons of molecules extending parallel to the a-axis direction (Fig. 2 ▸). The chains pack with a partial intercalation of the styryl and ester substituents (Fig. 3 ▸).
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C15—H15A⋯O2i | 0.98 (2) | 2.56 (2) | 3.375 (2) | 139.9 (17) |
| C18—H18B⋯O1ii | 0.965 (17) | 2.493 (17) | 3.2989 (17) | 140.9 (13) |
Symmetry codes: (i) -x+2, -y+1, -z+1; (ii) -x+1, -y+1, -z+1.
Figure 2.
A portion of one hydrogen–bonded chain in a view along the c-axis direction. C—H⋯O hydrogen bonds are depicted by dashed lines.
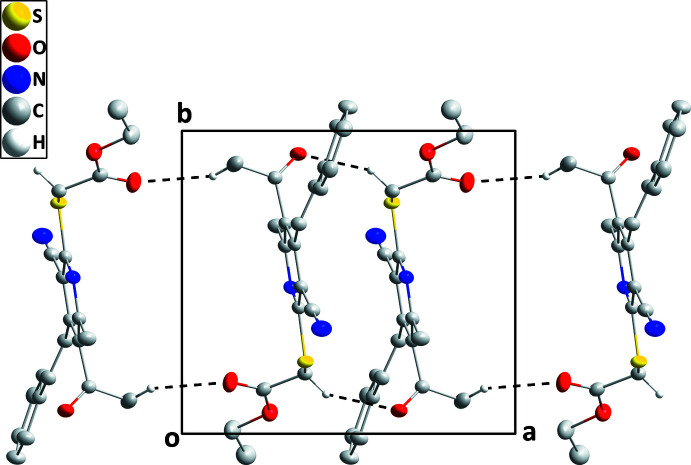
Figure 3.
Packing of the molecules in the title compound in a view along the b-axis direction. C—H⋯O hydrogen bonds are depicted by dashed lines.
Hirshfeld surface analysis
To quantify the intermolecular interactions in the title compound, a Hirshfeld surface analysis was performed and two-dimensional fingerprint plots were generated using Crystal Explorer (Turner et al., 2017 ▸). The Hirshfeld surface mapped over d norm in the range −0.1607 to +1.3888 arbitrary units is depicted in Fig. 4 ▸, where the red regions indicate apparent hydrogen bonds in this structure. The intensities of the red areas are greater for C15—H15A⋯O2 and C18—H18B⋯O1, indicating the strongest interactions as compared to other red spots on the Hirshfeld surface; Table 2 ▸ lists corresponding close intermolecular contacts. The two-dimensional fingerprint plots (Fig. 5 ▸) reveal that the largest contributions are from H⋯H (43.6%; Fig. 5 ▸ b), C⋯H/H⋯C (15.6%; Fig. 5 ▸ c), O⋯H/H⋯O (14.9%; Fig. 5 ▸ d) and N⋯H/H⋯N (11.2%; Fig. 5 ▸ e) interactions. Other interactions contributing less to the crystal packing are S⋯H/H⋯S (5.9%), C⋯C (4.4%), N⋯C/C⋯N (1.5%), S⋯O/O⋯S (1.1%), O⋯C/C⋯O (1.0%), O⋯O (0.3%), N⋯N (0.2%) and S⋯C/C⋯S (0.2%).
Figure 4.
A view of the three-dimensional Hirshfeld surface for the title compound, plotted over d norm in the range −0.1607 to +1.3888 a.u.
Table 2. Summary of short interatomic contacts (Å) in the title compound.
| Contact | Distance | Symmetry operation |
|---|---|---|
| H20B⋯H16B | 2.53 | x, 1 + y, z |
| H18B⋯H7 | 2.42 | 1 − x, 1 − y, 1 − z |
| O2⋯H10 | 2.613 | {3\over 2} − x, {1\over 2} + y, {1\over 2} − z |
| H15A⋯O2 | 2.56 | 2 − x, 1 − y, 1 − z |
| N2⋯H20A | 2.63 | −{1\over 2} + x, {3\over 2} − y, −{1\over 2} + z |
| H11⋯H11 | 2.31 | 1 − x, −y, −z |
| H12⋯H20A | 2.47 | −{1\over 2} + x, {1\over 2} − y, −{1\over 2} + z |
| H16A⋯H21B | 2.49 | {3\over 2} − x, −{1\over 2} + y, {3\over 2} − z |
Figure 5.
A view of the two-dimensional fingerprint plots for the title compound, showing (a) all interactions, and delineated into (b) H⋯H, (c) C⋯H/H⋯C, (d) O⋯H/H⋯O and (e) N⋯H/H⋯N interactions. The d i and d e values are the closest internal and external distances (in Å) from given points on the Hirshfeld surface.
Database survey
A search of the Cambridge Structural Database (version 5.42, update 1, Feb 2021; Groom et al., 2016 ▸) for related structures with the 2-sulfanylpyridine-3-carbonitrile moiety of the title compound gave the following matches: ethyl 4-methyl-2-phenyl-6-thioxo-1,6-dihydro-5-pyrimidinecarboxylate monohydrate (DEWCIS; Cunha et al., 2007 ▸), ethyl 4-(5-ethoxycarbonyl-6-methyl-2-phenyl-4-pyrimidinyldisulfanyl)-6-methyl-2-phenyl-5-pyrimidinecarboxylate (DEWCAK; Cunha et al., 2007 ▸), ethyl 4-{[(4-chlorophenyl)methyl]sulfanyl}-6-methyl-2-phenylpyrimidine-5-carboxylate (NILKOL; Stolarczyk et al., 2018 ▸), (4-{[(4-chlorophenyl)methyl]sulfanyl}-6-methyl-2-phenylpyrimidin-5-yl)methanol (NILKUR; Stolarczyk et al., 2018 ▸) and 4-{[(4-chlorophenyl)methyl]sulfanyl}-5,6-dimethyl-2-phenylpyrimidine (NILLAY; Stolarczyk et al., 2018 ▸).
Compound DEWCIS crystallizes in the space group P21/c with one molecule in the asymmetric unit. N—H⋯O, O—H⋯N and O—H⋯S interactions involving the water molecules, as well as π–π stacking interactions between the molecules along the b axis contribute to the formation of layers parallel to the bc plane. The stability of the molecular packing is achieved by van der Waals interactions between these layers. Compound DEWCAK crystallizes in the space group P
 with one molecule in the asymmetric unit. In the crystal structure of DEWCAK, there are no classical hydrogen bonds. The molecular packing is stabilized by C—H⋯π interactions and π–π stacking interactions. Compound NILKOL crystallizes in the space group P
with one molecule in the asymmetric unit. In the crystal structure of DEWCAK, there are no classical hydrogen bonds. The molecular packing is stabilized by C—H⋯π interactions and π–π stacking interactions. Compound NILKOL crystallizes in the space group P
 with one molecule in the asymmetric unit, whereas compounds NILKUR and NILLAY crystallize in the space group P21/c with two and one molecules, respectively, in their asymmetric units. The conformation of each molecule is best defined by the dihedral angles formed between the pyrimidine ring and the planes of the two aryl substituents attached at the 2- and 4-positions. The only structural difference between the three compounds is the substituent at the 5-position of the pyrimidine ring, but they present significantly different features in their hydrogen-bonding interactions. NILKOL displays a chain structure whereby the chains are further extended into a two-dimensional network. In NILKUR and NILLAY, the hydrogen-bonded chains have no further aggregation.
with one molecule in the asymmetric unit, whereas compounds NILKUR and NILLAY crystallize in the space group P21/c with two and one molecules, respectively, in their asymmetric units. The conformation of each molecule is best defined by the dihedral angles formed between the pyrimidine ring and the planes of the two aryl substituents attached at the 2- and 4-positions. The only structural difference between the three compounds is the substituent at the 5-position of the pyrimidine ring, but they present significantly different features in their hydrogen-bonding interactions. NILKOL displays a chain structure whereby the chains are further extended into a two-dimensional network. In NILKUR and NILLAY, the hydrogen-bonded chains have no further aggregation.
Synthesis and crystallization
A mixture of 5-acetyl-3-cyano-6-methyl-4-styrylpyridine-2(1H)-thione (3.24 g, 10 mmol), ethyl chloroacetate (1.1 ml, 10 mmol) and sodium acetate trihydrate (1.5 g, 11 mmol) in ethanol (40 ml) was heated under reflux for 30 min. The solid that formed after dilution with water (20 ml) was filtered off and recrystallized from methanol in the form of fine colourless crystals of the title compound, yield 85%; m.p. 343–344 K. Its IR spectrum showed characteristic absorption bands at 2219 cm−1 for (C≡N), at 1748 cm−1 for (C=O, non conjugated ester) and at 1724 cm−1 for (C=O, conjugated ester). The 1H NMR spectrum (400 MHz, DMSO-d 6) displayed a multiplet at δ = 6.60–7.63 ppm (7H: CH=CH and Ar-Hs), a multiplet at δ = 4.16–4.37 ppm (6H: two OCH2 and SCH2), a singlet at δ = 2.52 ppm (3H, CH3 at C-6, overlapped with solvent signal) and a multiplet at δ = 1.21–1.27 ppm (6H: two CH3 of ester groups).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. The C-bound H atoms were refined freely, while the H atoms of the C16 methyl group were placed geometrically (C—H = 0.98 Å) and refined as riding atoms with U iso(H) = 1.5U eq(C).
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | C21H20N2O3S |
| M r | 380.45 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 150 |
| a, b, c (Å) | 10.7365 (4), 9.7590 (3), 18.5600 (7) |
| β (°) | 90.066 (1) |
| V (Å3) | 1944.67 (12) |
| Z | 4 |
| Radiation type | Cu Kα |
| μ (mm−1) | 1.67 |
| Crystal size (mm) | 0.27 × 0.12 × 0.05 |
| Data collection | |
| Diffractometer | Bruker D8 VENTURE PHOTON 100 CMOS |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| Tmin, Tmax | 0.80, 0.92 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 14722, 3912, 3520 |
| R int | 0.034 |
| (sin θ/λ)max (Å−1) | 0.625 |
| Refinement | |
| R[F2 > 2σ(F 2)], wR(F 2), S | 0.033, 0.088, 1.04 |
| No. of reflections | 3912 |
| No. of parameters | 314 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.22, −0.29 |
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989021005600/wm5610sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021005600/wm5610Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989021005600/wm5610Isup3.cml
CCDC reference: 2087180
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
Author contributions are as follows: Conceptualization, EAB, MSA and SKM; methodology, JTM, EAB and MA; investigation, JTM, SKM, and EAB; writing (original draft), JTM, AM, SKM and EAB; writing (review and editing of the manuscript), MA and SKM; visualization, MA, SKM and JTM; funding acquisition, SAHA and SKM; resources, MA, JTM, EAB and SKM. AAA, VNK and FNN; supervision, SKM and MA.
supplementary crystallographic information
Crystal data
| C21H20N2O3S | F(000) = 800 |
| Mr = 380.45 | Dx = 1.299 Mg m−3 |
| Monoclinic, P21/n | Cu Kα radiation, λ = 1.54178 Å |
| a = 10.7365 (4) Å | Cell parameters from 9951 reflections |
| b = 9.7590 (3) Å | θ = 4.8–74.6° |
| c = 18.5600 (7) Å | µ = 1.67 mm−1 |
| β = 90.066 (1)° | T = 150 K |
| V = 1944.67 (12) Å3 | Column, colourless |
| Z = 4 | 0.27 × 0.12 × 0.05 mm |
Data collection
| Bruker D8 VENTURE PHOTON 100 CMOS diffractometer | 3912 independent reflections |
| Radiation source: INCOATEC IµS micro–focus source | 3520 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.034 |
| Detector resolution: 10.4167 pixels mm-1 | θmax = 74.6°, θmin = 4.8° |
| ω scans | h = −13→12 |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | k = −11→12 |
| Tmin = 0.80, Tmax = 0.92 | l = −21→22 |
| 14722 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.033 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.088 | w = 1/[σ2(Fo2) + (0.0426P)2 + 0.7692P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.04 | (Δ/σ)max = 0.001 |
| 3912 reflections | Δρmax = 0.22 e Å−3 |
| 314 parameters | Δρmin = −0.28 e Å−3 |
| 0 restraints | Extinction correction: SHELXL 2018/3 (Sheldrick, 2015b), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: dual | Extinction coefficient: 0.0041 (3) |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. Independent refinement of the hydrogen atoms attached to C16 led to an unreasonable geometry so these were included as riding contributions (C—H = 0.98 Å) with an AFIX 137 instruction. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.62964 (3) | 0.76240 (3) | 0.45327 (2) | 0.02319 (11) | |
| O1 | 0.65133 (9) | 0.07606 (9) | 0.47995 (6) | 0.0295 (2) | |
| O2 | 0.85513 (10) | 0.82982 (12) | 0.54910 (6) | 0.0367 (3) | |
| O3 | 0.73641 (9) | 0.93032 (11) | 0.63373 (5) | 0.0304 (2) | |
| N1 | 0.67271 (10) | 0.51703 (11) | 0.51422 (6) | 0.0222 (2) | |
| N2 | 0.58138 (13) | 0.65298 (13) | 0.27261 (7) | 0.0351 (3) | |
| C1 | 0.66330 (11) | 0.37425 (13) | 0.38230 (7) | 0.0203 (3) | |
| C2 | 0.69019 (12) | 0.30604 (13) | 0.44698 (7) | 0.0202 (3) | |
| C3 | 0.69103 (12) | 0.38044 (13) | 0.51167 (7) | 0.0215 (3) | |
| C4 | 0.65042 (12) | 0.58371 (13) | 0.45305 (7) | 0.0201 (3) | |
| C5 | 0.64281 (12) | 0.51610 (13) | 0.38647 (7) | 0.0208 (3) | |
| C6 | 0.64972 (13) | 0.30725 (14) | 0.31165 (7) | 0.0233 (3) | |
| H6 | 0.6751 (17) | 0.363 (2) | 0.2711 (10) | 0.038 (5)* | |
| C7 | 0.59846 (13) | 0.18469 (14) | 0.30086 (7) | 0.0239 (3) | |
| H7 | 0.5717 (16) | 0.1327 (18) | 0.3420 (10) | 0.033 (4)* | |
| C8 | 0.57238 (12) | 0.12114 (13) | 0.23083 (7) | 0.0222 (3) | |
| C9 | 0.58480 (13) | 0.19075 (14) | 0.16518 (7) | 0.0245 (3) | |
| H9 | 0.6136 (17) | 0.284 (2) | 0.1648 (9) | 0.034 (5)* | |
| C10 | 0.55575 (13) | 0.12655 (16) | 0.10077 (8) | 0.0276 (3) | |
| H10 | 0.5629 (17) | 0.178 (2) | 0.0567 (10) | 0.041 (5)* | |
| C11 | 0.51380 (14) | −0.00811 (16) | 0.10078 (8) | 0.0295 (3) | |
| H11 | 0.4913 (18) | −0.0529 (19) | 0.0555 (10) | 0.040 (5)* | |
| C12 | 0.50215 (15) | −0.07840 (15) | 0.16501 (8) | 0.0312 (3) | |
| H12 | 0.4716 (17) | −0.170 (2) | 0.1652 (10) | 0.037 (5)* | |
| C13 | 0.53114 (14) | −0.01448 (15) | 0.22967 (8) | 0.0279 (3) | |
| H13 | 0.5240 (17) | −0.0659 (18) | 0.2763 (10) | 0.036 (5)* | |
| C14 | 0.72087 (12) | 0.15479 (13) | 0.44955 (7) | 0.0220 (3) | |
| C15 | 0.84103 (15) | 0.10914 (17) | 0.41681 (10) | 0.0351 (3) | |
| H15A | 0.907 (2) | 0.149 (2) | 0.4470 (12) | 0.056 (6)* | |
| H15B | 0.846 (2) | 0.008 (2) | 0.4173 (12) | 0.058 (6)* | |
| H14C | 0.8516 (19) | 0.147 (2) | 0.3658 (12) | 0.051 (6)* | |
| C16 | 0.71490 (14) | 0.31393 (14) | 0.58331 (7) | 0.0282 (3) | |
| H16A | 0.723453 | 0.384753 | 0.620421 | 0.042* | |
| H16B | 0.791792 | 0.260036 | 0.580824 | 0.042* | |
| H16C | 0.645041 | 0.253596 | 0.595442 | 0.042* | |
| C17 | 0.60967 (13) | 0.59215 (14) | 0.32293 (7) | 0.0245 (3) | |
| C18 | 0.63109 (13) | 0.80140 (14) | 0.54760 (7) | 0.0231 (3) | |
| H18A | 0.6100 (17) | 0.717 (2) | 0.5740 (10) | 0.037 (5)* | |
| H18B | 0.5672 (16) | 0.8694 (18) | 0.5554 (9) | 0.027 (4)* | |
| C19 | 0.75457 (13) | 0.85366 (14) | 0.57467 (7) | 0.0244 (3) | |
| C20 | 0.84825 (15) | 0.98717 (19) | 0.66682 (9) | 0.0380 (4) | |
| H20A | 0.904 (2) | 0.910 (2) | 0.6807 (12) | 0.053 (6)* | |
| H20B | 0.896 (2) | 1.047 (2) | 0.6298 (12) | 0.056 (6)* | |
| C21 | 0.80775 (18) | 1.0701 (2) | 0.73071 (10) | 0.0410 (4) | |
| H21A | 0.883 (2) | 1.109 (2) | 0.7550 (13) | 0.065 (7)* | |
| H21B | 0.762 (2) | 1.014 (2) | 0.7639 (12) | 0.051 (6)* | |
| H21C | 0.751 (2) | 1.144 (2) | 0.7167 (11) | 0.049 (6)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.03027 (19) | 0.01732 (17) | 0.02199 (18) | 0.00121 (12) | −0.00303 (12) | −0.00091 (11) |
| O1 | 0.0316 (5) | 0.0202 (5) | 0.0367 (6) | 0.0000 (4) | 0.0043 (4) | 0.0046 (4) |
| O2 | 0.0248 (5) | 0.0464 (7) | 0.0389 (6) | 0.0060 (4) | −0.0025 (4) | −0.0121 (5) |
| O3 | 0.0289 (5) | 0.0356 (6) | 0.0266 (5) | 0.0020 (4) | −0.0059 (4) | −0.0092 (4) |
| N1 | 0.0257 (6) | 0.0202 (5) | 0.0208 (6) | −0.0021 (4) | −0.0033 (4) | −0.0004 (4) |
| N2 | 0.0473 (8) | 0.0295 (6) | 0.0285 (7) | −0.0007 (6) | −0.0097 (6) | 0.0023 (5) |
| C1 | 0.0199 (6) | 0.0200 (6) | 0.0209 (6) | −0.0030 (5) | −0.0009 (5) | −0.0009 (5) |
| C2 | 0.0203 (6) | 0.0180 (6) | 0.0224 (6) | −0.0021 (5) | −0.0018 (5) | 0.0007 (5) |
| C3 | 0.0224 (6) | 0.0201 (6) | 0.0220 (7) | −0.0025 (5) | −0.0032 (5) | 0.0008 (5) |
| C4 | 0.0199 (6) | 0.0192 (6) | 0.0211 (6) | −0.0021 (5) | −0.0023 (5) | −0.0002 (5) |
| C5 | 0.0223 (6) | 0.0203 (6) | 0.0199 (6) | −0.0022 (5) | −0.0033 (5) | 0.0012 (5) |
| C6 | 0.0283 (7) | 0.0222 (6) | 0.0195 (6) | −0.0010 (5) | −0.0012 (5) | −0.0010 (5) |
| C7 | 0.0281 (7) | 0.0245 (7) | 0.0193 (6) | −0.0024 (5) | 0.0000 (5) | −0.0010 (5) |
| C8 | 0.0233 (6) | 0.0230 (6) | 0.0201 (6) | −0.0001 (5) | −0.0010 (5) | −0.0028 (5) |
| C9 | 0.0271 (7) | 0.0233 (7) | 0.0232 (7) | −0.0021 (5) | −0.0012 (5) | 0.0003 (5) |
| C10 | 0.0290 (7) | 0.0329 (7) | 0.0209 (7) | 0.0010 (6) | −0.0022 (5) | 0.0007 (6) |
| C11 | 0.0337 (7) | 0.0323 (7) | 0.0226 (7) | 0.0009 (6) | −0.0057 (6) | −0.0064 (6) |
| C12 | 0.0418 (8) | 0.0230 (7) | 0.0287 (8) | −0.0048 (6) | −0.0045 (6) | −0.0046 (6) |
| C13 | 0.0368 (8) | 0.0240 (7) | 0.0228 (7) | −0.0035 (6) | −0.0018 (6) | −0.0006 (5) |
| C14 | 0.0244 (6) | 0.0202 (6) | 0.0214 (6) | −0.0002 (5) | −0.0042 (5) | −0.0005 (5) |
| C15 | 0.0311 (8) | 0.0294 (8) | 0.0448 (10) | 0.0052 (6) | 0.0068 (7) | 0.0029 (7) |
| C16 | 0.0393 (8) | 0.0239 (7) | 0.0214 (7) | −0.0016 (6) | −0.0065 (6) | 0.0022 (5) |
| C17 | 0.0293 (7) | 0.0211 (6) | 0.0231 (7) | −0.0022 (5) | −0.0044 (5) | −0.0016 (5) |
| C18 | 0.0257 (7) | 0.0207 (6) | 0.0230 (7) | 0.0004 (5) | 0.0007 (5) | −0.0027 (5) |
| C19 | 0.0277 (7) | 0.0223 (6) | 0.0231 (6) | 0.0038 (5) | −0.0038 (5) | −0.0008 (5) |
| C20 | 0.0329 (8) | 0.0429 (9) | 0.0383 (9) | 0.0026 (7) | −0.0140 (7) | −0.0119 (7) |
| C21 | 0.0458 (10) | 0.0446 (10) | 0.0324 (9) | −0.0011 (8) | −0.0100 (7) | −0.0095 (7) |
Geometric parameters (Å, º)
| S1—C4 | 1.7581 (13) | C9—H9 | 0.960 (19) |
| S1—C18 | 1.7918 (14) | C10—C11 | 1.389 (2) |
| O1—C14 | 1.2112 (16) | C10—H10 | 0.96 (2) |
| O2—C19 | 1.2026 (17) | C11—C12 | 1.381 (2) |
| O3—C19 | 1.3417 (17) | C11—H11 | 0.98 (2) |
| O3—C20 | 1.4578 (18) | C12—C13 | 1.388 (2) |
| N1—C4 | 1.3301 (17) | C12—H12 | 0.956 (19) |
| N1—C3 | 1.3482 (17) | C13—H13 | 1.003 (19) |
| N2—C17 | 1.1473 (19) | C14—C15 | 1.4946 (19) |
| C1—C2 | 1.4025 (18) | C15—H15A | 0.98 (2) |
| C1—C5 | 1.4038 (18) | C15—H15B | 0.99 (2) |
| C1—C6 | 1.4723 (18) | C15—H14C | 1.02 (2) |
| C2—C3 | 1.4032 (18) | C16—H16A | 0.9800 |
| C2—C14 | 1.5130 (17) | C16—H16B | 0.9800 |
| C3—C16 | 1.5013 (18) | C16—H16C | 0.9800 |
| C4—C5 | 1.4032 (18) | C18—C19 | 1.5061 (19) |
| C5—C17 | 1.4378 (18) | C18—H18A | 0.988 (19) |
| C6—C7 | 1.332 (2) | C18—H18B | 0.965 (17) |
| C6—H6 | 0.968 (19) | C20—C21 | 1.500 (2) |
| C7—C8 | 1.4669 (18) | C20—H20A | 1.00 (2) |
| C7—H7 | 0.961 (18) | C20—H20B | 1.04 (2) |
| C8—C13 | 1.3958 (19) | C21—H21A | 1.00 (2) |
| C8—C9 | 1.4016 (19) | C21—H21B | 0.96 (2) |
| C9—C10 | 1.385 (2) | C21—H21C | 0.97 (2) |
| C4—S1—C18 | 102.24 (6) | C12—C13—H13 | 120.2 (10) |
| C19—O3—C20 | 115.85 (11) | C8—C13—H13 | 119.0 (10) |
| C4—N1—C3 | 118.68 (11) | O1—C14—C15 | 122.21 (12) |
| C2—C1—C5 | 116.93 (11) | O1—C14—C2 | 119.95 (12) |
| C2—C1—C6 | 124.86 (12) | C15—C14—C2 | 117.77 (12) |
| C5—C1—C6 | 118.14 (11) | C14—C15—H15A | 105.7 (13) |
| C1—C2—C3 | 119.21 (12) | C14—C15—H15B | 109.9 (13) |
| C1—C2—C14 | 122.32 (11) | H15A—C15—H15B | 110.5 (18) |
| C3—C2—C14 | 118.47 (11) | C14—C15—H14C | 111.5 (12) |
| N1—C3—C2 | 122.74 (12) | H15A—C15—H14C | 107.7 (17) |
| N1—C3—C16 | 114.90 (11) | H15B—C15—H14C | 111.2 (17) |
| C2—C3—C16 | 122.34 (12) | C3—C16—H16A | 109.5 |
| N1—C4—C5 | 122.14 (12) | C3—C16—H16B | 109.5 |
| N1—C4—S1 | 120.40 (10) | H16A—C16—H16B | 109.5 |
| C5—C4—S1 | 117.46 (10) | C3—C16—H16C | 109.5 |
| C4—C5—C1 | 120.22 (12) | H16A—C16—H16C | 109.5 |
| C4—C5—C17 | 119.59 (12) | H16B—C16—H16C | 109.5 |
| C1—C5—C17 | 120.16 (12) | N2—C17—C5 | 178.94 (16) |
| C7—C6—C1 | 124.98 (12) | C19—C18—S1 | 113.86 (10) |
| C7—C6—H6 | 120.3 (11) | C19—C18—H18A | 108.7 (11) |
| C1—C6—H6 | 114.6 (11) | S1—C18—H18A | 107.8 (11) |
| C6—C7—C8 | 126.26 (13) | C19—C18—H18B | 110.0 (10) |
| C6—C7—H7 | 118.6 (11) | S1—C18—H18B | 106.7 (10) |
| C8—C7—H7 | 115.1 (11) | H18A—C18—H18B | 109.7 (14) |
| C13—C8—C9 | 118.47 (12) | O2—C19—O3 | 124.17 (13) |
| C13—C8—C7 | 118.34 (12) | O2—C19—C18 | 126.38 (13) |
| C9—C8—C7 | 123.18 (12) | O3—C19—C18 | 109.43 (11) |
| C10—C9—C8 | 120.64 (13) | O3—C20—C21 | 107.39 (14) |
| C10—C9—H9 | 119.7 (11) | O3—C20—H20A | 108.7 (13) |
| C8—C9—H9 | 119.7 (11) | C21—C20—H20A | 112.2 (13) |
| C9—C10—C11 | 120.03 (13) | O3—C20—H20B | 110.0 (12) |
| C9—C10—H10 | 118.6 (12) | C21—C20—H20B | 111.1 (12) |
| C11—C10—H10 | 121.4 (12) | H20A—C20—H20B | 107.4 (17) |
| C12—C11—C10 | 119.98 (13) | C20—C21—H21A | 109.1 (13) |
| C12—C11—H11 | 119.8 (11) | C20—C21—H21B | 110.5 (13) |
| C10—C11—H11 | 120.2 (11) | H21A—C21—H21B | 109.7 (18) |
| C11—C12—C13 | 120.18 (13) | C20—C21—H21C | 111.6 (12) |
| C11—C12—H12 | 120.1 (11) | H21A—C21—H21C | 110.1 (18) |
| C13—C12—H12 | 119.7 (11) | H21B—C21—H21C | 105.8 (18) |
| C12—C13—C8 | 120.69 (13) | ||
| C5—C1—C2—C3 | 2.15 (18) | C5—C1—C6—C7 | −140.67 (14) |
| C6—C1—C2—C3 | −175.01 (12) | C1—C6—C7—C8 | 173.53 (13) |
| C5—C1—C2—C14 | −176.70 (11) | C6—C7—C8—C13 | 172.91 (14) |
| C6—C1—C2—C14 | 6.15 (19) | C6—C7—C8—C9 | −8.2 (2) |
| C4—N1—C3—C2 | 1.39 (19) | C13—C8—C9—C10 | 0.5 (2) |
| C4—N1—C3—C16 | −179.73 (12) | C7—C8—C9—C10 | −178.32 (13) |
| C1—C2—C3—N1 | −3.20 (19) | C8—C9—C10—C11 | 0.0 (2) |
| C14—C2—C3—N1 | 175.69 (12) | C9—C10—C11—C12 | −0.6 (2) |
| C1—C2—C3—C16 | 177.99 (12) | C10—C11—C12—C13 | 0.6 (2) |
| C14—C2—C3—C16 | −3.12 (19) | C11—C12—C13—C8 | 0.0 (2) |
| C3—N1—C4—C5 | 1.39 (19) | C9—C8—C13—C12 | −0.5 (2) |
| C3—N1—C4—S1 | −178.47 (10) | C7—C8—C13—C12 | 178.38 (14) |
| C18—S1—C4—N1 | −5.66 (12) | C1—C2—C14—O1 | −114.75 (15) |
| C18—S1—C4—C5 | 174.48 (10) | C3—C2—C14—O1 | 66.40 (17) |
| N1—C4—C5—C1 | −2.31 (19) | C1—C2—C14—C15 | 68.12 (17) |
| S1—C4—C5—C1 | 177.55 (10) | C3—C2—C14—C15 | −110.73 (15) |
| N1—C4—C5—C17 | 175.55 (12) | C4—S1—C18—C19 | 98.96 (10) |
| S1—C4—C5—C17 | −4.59 (17) | C20—O3—C19—O2 | 0.6 (2) |
| C2—C1—C5—C4 | 0.44 (18) | C20—O3—C19—C18 | 179.32 (12) |
| C6—C1—C5—C4 | 177.79 (12) | S1—C18—C19—O2 | −26.34 (19) |
| C2—C1—C5—C17 | −177.42 (12) | S1—C18—C19—O3 | 154.93 (10) |
| C6—C1—C5—C17 | −0.06 (18) | C19—O3—C20—C21 | 179.23 (14) |
| C2—C1—C6—C7 | 36.5 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C15—H15A···O2i | 0.98 (2) | 2.56 (2) | 3.375 (2) | 139.9 (17) |
| C18—H18B···O1ii | 0.965 (17) | 2.493 (17) | 3.2989 (17) | 140.9 (13) |
Symmetry codes: (i) −x+2, −y+1, −z+1; (ii) −x+1, −y+1, −z+1.
Funding Statement
This work was funded by National Science Foundation grant 1228232; Tulane University.
References
- Balasubramanian, M. & Keay, J. G. (1996). In Comprehensive Heterocyclic Chemistry II, edited by A. R Katritzky, C. W. Rees & E. F. V. Scriven, pp. 245–300. Oxford: Pergamon.
- Brandenburg, K. & Putz, H. (2012). DIAMOND, Crystal Impact GbR, Bonn, Germany.
- Bruker (2016). APEX3 and SAINT. Bruker AXS, Inc., Madison, Wisconsin, USA.
- Cunha, S., Bastos, R. M., Silva, P. O., Nobre Costa, G. A., Vencato, I., Lariucci, C., Napolitano, H. B., de Oliveira, C. M. A., Kato, L., da Silva, C. C., Menezes, D. & Vannier-Santos, M. A. (2007). Monatsh. Chem. 138, 111–119.
- Gibson, V. C., Redshaw, C. & Solan, G. A. (2007). Chem. Rev. 107, 1745–1776. [DOI] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Stolarczyk, M., Bryndal, I., Matera-Witkiewicz, A., Lis, T., Królewska-Golińska, K., Cieślak, M., Kaźmierczak-Barańska, J. & Cieplik, J. (2018). Acta Cryst. C74, 1138–1145. [DOI] [PubMed]
- Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). Crystal Explorer 17. University of Western Australia. http://hirshfeldsurface.net
- Vidaillac, C., Guillon, J., Arpin, C., Forfar-Bares, I., Ba, B. B., Grellet, J., Moreau, S., Caignard, D. H., Jarry, C. & Quentin, C. (2007). Antimicrob. Agents Chemother. 51, 831–838. [DOI] [PMC free article] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989021005600/wm5610sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021005600/wm5610Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989021005600/wm5610Isup3.cml
CCDC reference: 2087180
Additional supporting information: crystallographic information; 3D view; checkCIF report