Abstract
Automatic segmentation of brain anatomy has been a key processing step in quantitative neuroimaging analyses. An extensive body of literature has relied on Freesurfer segmentations. Yet, in recent years, the multi-atlas segmentation framework has consistently obtained results with superior accuracy in various evaluations. We compared brain anatomy segmentations from Freesurfer, which uses a single probabilistic atlas strategy, against segmentations from Multi-atlas region Segmentation utilizing Ensembles of registration algorithms and parameters and locally optimal atlas selection (MUSE), one of the leading ensemble-based methods that calculates a consensus segmentation through fusion of anatomical labels from multiple atlases and registrations. The focus of our evaluation was twofold. First, using manual ground-truth hippocampus segmentations, we found that Freesurfer segmentations showed a bias towards over-segmentation of larger hippocampi, and under-segmentation in older age. This bias was more pronounced in Freesurfer-v5.3, which has been used in multiple previous studies of aging, while the effect was mitigated in more recent Freesurfer-v6.0, albeit still present. Second, we evaluated inter-scanner segmentation stability using same day scan pairs from ADNI acquired on 1.5T and 3T scanners. We also found that MUSE obtains more consistent segmentations across scanners compared to Freesurfer, particularly in the deep structures.
Keywords: MRI, Segmentation, Freesurfer, MUSE, Brain, ROI
1. Introduction
Segmentation of brain anatomy has been a key image processing step in neuroimaging studies, as it enables assessment of regional brain volumes in a range of neurological diseases and conditions for image-based diagnosis, monitoring of disease progression, and tracking of neuro-developmental and aging-related brain changes (Giorgio 2013; Janowitz et al., 2014; Raz et al., 2010; Wierenga et al., 2014). Volumetric analyses of cortical structures provided markers of neuro-degeneration in various disorders including multiple sclerosis (MS), schizophrenia (SCZ) and Alzheimer’s disease (AD), as well as in normal aging (Bonilha et al., 2008; Brewer et al., 2009; Bakkour et al., 2013; Charil et al., 2007; Dicks et al., 2019). Previous studies also reported significant associations between volumes of sub-cortical deep brain structures, including the thalamus, caudate, putamen and amygdala, and neuropsychiatric and neuro-degenerative conditions such as AD and SCZ, suggesting that both cortical and subcortical structures are variably related to different neurodegenerative conditions (Ferreira et al., 2017; Janowitz et al., 2014; Apostolova et al., 2006; Goldstein et al., 1999; Satterthwaite et al., 2016).
Multiple algorithms and methods have been developed for segmenting anatomical regions of interest (ROIs) in a fully automated way. One of the most widely used automated segmentation methods is the publicly available FreeSurfer software package (Fischl et al., 2002; 2004). Freesurfer combines a surface-based stream for cortical segmentation with a volume-based stream for segmentation of subcortical structures. The surface-based stream first calculates an initial surface that delineates the white matter, and then refines this surface to calculate the pial surface. The volume-based stream uses a subject-independent probabilistic atlas, which is automatically derived from a training set consisting of multiple hand-labeled atlas images. After a high dimensional nonlinear volumetric alignment of the target image to a common atlas space, ROI labels are automatically assigned to each voxel by finding a segmentation that maximizes the probability of the input data given the prior probabilities from the training set. At present, an important body of literature and neuropsychiatry findings are based on Freesurfer segmentations (Kikinis et al., 2010; Rohrer2011; Messina et al., 2011; Sabuncu et al., 2011). Freesurfer has been widely tested for its accuracy, precision and repeatability. However, various evaluations have also revealed limitations of that approach (Mccarthy 2015; Keller et al., 2012; Mulder et al., 2014), which might be critical in aging and multi-site studies.
Instead of relying on a single atlas, the multi-atlas segmentation (MAS) framework utilizes multiple reference atlas images that are independently warped to the target image space, and their reference labels are fused together to derive a consensus segmentation. This process has important advantages, the most notably the robustness against individual registration errors by the virtue of the ensemble label fusion process. The MUSE algorithm (Doshi et al., 2016) extended the ensemble approach to multiple deformable registration algorithms applied at different regularizations, allowing a higher variation within the ensemble. Additionally, in the label fusion step, MUSE uses a spatially adaptive strategy to select atlases based on their local similarity to the target image. Effectively, atlases most similar to a target image being segmented have more influence on its segmentation. Even more importantly, this process is spatially-adaptive, i.e. atlases most suitable for someone’s hippocampus segmentation might not necessarily be best for thalamus segmentation. MUSE was the top-ranking method in the MICCAI-SATA challenge on deep brain segmentation (Asman et al., 2013), and has been used in several studies (Habes et al., 2016; Satterthwaite et al., 2016; Wee 2017; Tian et al., 2018).
Our primary motivation herein was to evaluate brain anatomy segmentations obtained using these two automated methods in multi-site brain aging studies. Importantly, our comparative evaluations included both versions v6.0 and v5.3 of Freesurfer, thus also providing a comparison between newer and older Freesurfer versions. This comparison may be informative for guiding processing pipeline updates in longitudinal studies that previously used older versions of Freesurfer for segmentation. Accurately measuring subtle brain aging changes, and particularly hippocampal volume change, is important for early identification of pathologic processes. Moreover, being able to perform multi-site studies has become critical, as large consortia of meta- and mega-analyses are being formed in order to achieve a sufficiently large sample size, in view of the heterogeneity and complexity of brain aging. Toward this goal, we utilized expert-based manual hippocampus segmentations provided by the EADC-ADNI Harmonized Protocol project for manual hippocampal segmentation (HarP) (Frisoni et al., 2015), and same-day scan/re-scan images from 1.5T and 3T MRI scans in ADNI.
2. Materials and methods
2.1. MR imaging data
We used two publicly available datasets for quantitative evaluations (Table 1). The first dataset (Dataset1) was provided by the EADC-ADNI Harmonized Protocol project for manual hippocampal segmentation (HarP) (Frisoni et al., 2015). The main aim of the HarP project was to harmonize existing protocols for manual segmentation of hippocampus in order to derive standardized ground truth labels that will be used as benchmark. The HarP dataset included T1-weighted scans of 135 ADNI subjects (Mean Age: 75.014, Slice Thickness: 1.2 mm,with 44 Controls, 45 MCI, 45 AD) and left and right hippocampal labels for these scans delineated by expert raters. 4 Due to mismatch related to image format conversion for one subject scan, our final sample included a total of 134 subjects.
Table 1.
General Characteristics of datasets that were used in validation experiments.
| CN | MCI | AD | Total | |
|---|---|---|---|---|
| Dataset1: EADC-ADNI | ||||
| Count | 44 | 45 | 45 | 134 |
| Age | 76.18 ± 7.45 | 74.44 ± 8.00 | 74.45 ± 8.10 | 75 ± 7.84 |
| Sex(M/F) | 22/22 | 26/19 | 21/24 | 69/65 |
| Dataset2: ADNI 1.5T, ADNI 3T * | ||||
| Count | 37 | 53 | 23 | 113 |
| Age | 75.72 ± 4.20 | 76.02 ± 7.90 | 75.03 ± 8.36 | 75.72 ± 6.97 |
| Sex(M/F) | 15/22 | 35/18 | 08/15 | 58/55 |
For each subject Dataset2 includes a pair of 1.5T and 3T scans acquired on same day.
Our Second dataset (Dataset2) consisted of T1-weighted scans of 113 ADNI-1 subjects who underwent 1.5T and 3T T1-weighted MR Imaging at the same day or within similar dates (12 scans within 1 month; 4 scans within 2 months; 1 scan within 3 months). All subjects belonged to the patient group with mean age of 75.7. High resolution structural scans were acquired using 1.5T (TR = 2400 ms,Flip Angle = 8°, with acquisition matrix of 256 × 256 × 166, yielding voxel size of 0.9 × 0.9 × 1.2 mm) and 3T (TR = 2300 ms, Flip Angle = 8°, with acquisition matrix of 256 × 256 × 170, yielding voxel size of 1.0 × 1.0 × 1.2) MR scanners from different vendors (GE, Philips and Siemens).
2.2. ROI segmentation methods
2.2.1. Freesurfer volumetric segmentation
FreeSurfer is a software package to analyze and visualize structural neuroimaging data (Fischl et al., 2002, 2004). A widely used functionality of Freesurfer is to perform cortical and subcortical segmentation. Freesurfer segmentation consists of surface and volume-based streams with multiple processing steps. After transformation to Tailarach space, the target image is corrected for intensity inhomogeneities (bias field) and non-brain tissues are removed automatically. A high dimensional nonlinear volumetric alignment to the atlas space is performed for transferring atlas label information to the target image. The alignment includes surface deformation to optimally place gray matter (GM) / white matter (WM) and GM / cerebro-spinal fluid (CSF) boundaries, and surface inflation and registration to spherical atlas to parcellate cerebral cortex into units based on gyral and sulcal structure. The final segmentation is based on both a subject-independent probabilistic atlas, which was built from a training set with manual labels, and subject-specific measured values. The final label assignment at each image voxel is achieved by finding the segmentation that maximizes the probability of input signal given the prior probabilities from the training set. Freesurfer segments the brain into 34 cortical ROIs per hemisphere, which were defined based on a parcellation scheme on an inflated representation of cortex (Desikan et al., 2006).
We applied Freesurfer v5.3 and v6.0 (released in April 2017) segmentation using the default parameters (recon-all -i T1image -s sub -sd SUBJECTS\_DIR -all) for 1.5T scans. For 3T scans, we run Freesurfer with the “−3 T”’ flag (recon-all -i T1image -s sub -sd SUBJECTS\_DIR −3 T -all). Freesurfer volume estimates were calculated using Desikan Killiany Atlas for all subjects using the command asegstats2table.
2.2.2. MUSE segmentation
The MUSE algorithm follows the multi-atlas image registration and label fusion framework. In this framework, multiple atlases with manually or semi-automatically drawn reference labels are independently registered to the target scan using deformable registration. Candidate labels from multiple registrations are fused together to calculate a consensus segmentation. Image preprocessing in MUSE included inhomogeneity correction with N4 (Tustison et al., 2010) and multi-atlas skull-stripping (Doshi et al., 2016). After skull-stripping, 11 atlas images are warped to the target image using 2 different deformable registration algorithms DRAMMS (Ou et al., 2011) and ANTS (Avants et al., 2014), and using two different regularization parameter values for these algorithms, resulting in a high variation within the ensemble, which is desirable to be able to better capture the inter-individual variability in target anatomy. The label fusion includes a local similarity term to select locally optimal atlases and an intensity term to refine the segmentation consistently with the target image’s intensity profile. MUSE reference atlases included 153 anatomical ROIs. We run MUSE using the default parameters.
2.3. Quality control of segmentations
Freesurfer and MUSE segmentations were subjected to a quality control (QC) procedure that aimed to detect and exclude cases with gross errors in the segmentations, which typically happen due to major failures of atlas to target image registrations. We should note that this is different from extensive QC on individual ROIs performed in studies with relatively modest sample sizes. In this analysis, we mainly focused on establishing an automated and reproducible processing procedure, in view of large scale datasets. Accordingly, cases were flagged for exclusion only if overall segmented ROI did not overlap with the actual brain boundaries. The QC procedure involved automatic ranking of scans based on a quality score derived from ROI volumes, followed by visual inspections of the segmentations guided by the ranking score. A quality score is automatically calculated for each segmentation mask by comparing ROI volumes extracted from this mask against the distributions obtained from the entire sample. Specifically, we used PCA to reduce data dimensionality by projecting ROI values to a lower dimensional space that optimally explains the variance of the data, and we calculated the Mahalanobis distance of each sample to the sample mean in the PCA space. We used an in-house visualization tool for visual inspections, which displayed boundaries of segmented ROIs on a subset of image slices in axial, sagittal and coronal views. A binary QC flag, i.e. accept or exclude, was assigned to each segmentation, based on the consensus of two independent raters.
2.4. Determination of common ROI labels
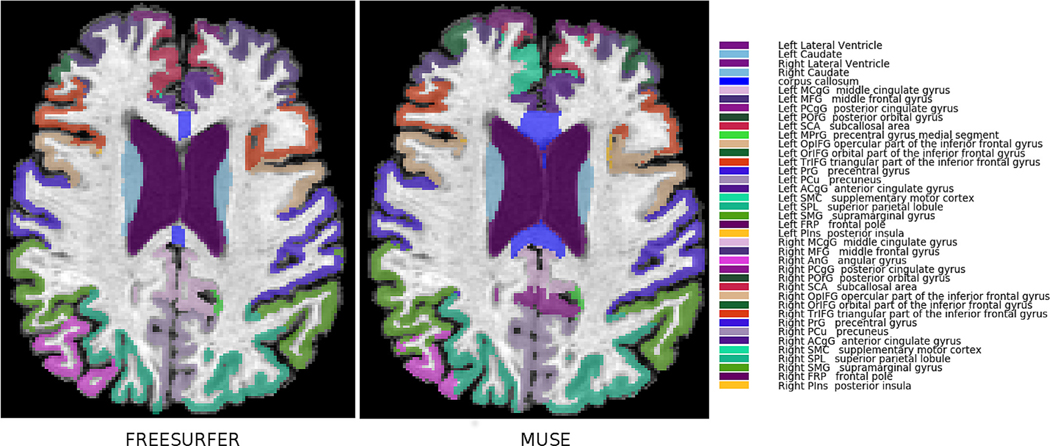
As the reference label definitions are different for Freesurfer and MUSE, a direct comparison of ROIs is not possible. Particularly, the cortical ROIs are substantially different both in terms of naming of anatomical regions and in the details of the delineation of their boundaries. Therefore, a direct comparison of the exact volumes produced by these two methods could not be applied, hence we focused on analyzing age trends and inter-scanner consistency. In order to facilitate this process, we applied a semi-automated process to identify approximate correspondences between the two atlas ROI definitions. We computed MUSE and Freesurfer segmentations of a single reference scan and calculated the percent overlap between each pair of ROIs from the two methods. MUSE and Freesurfer ROIs were matched to each other using a greedy matching algorithm guided by the maximum percent overlap between ROI pairs. The matching algorithm allowed grouping of multiple ROIs in one set to find the optimal coverage of a single ROI in the other set. Matched ROIs have been inspected by two manual raters (DS and MH) based on the ROI names and spatial correspondences between them. Revisions have been done after mutual agreement of the two raters. The final set of ROIs included 7 deep structure regions and 31 cortical regions. A visualization of the matched ROIs between MUSE and Freesurfer is presented in Fig. 1. A complete list of the matching between MUSE and Freesurfer ROIs is given in suppl. Fig. 1.
Fig. 1.
ROI atlas denoting common GM ROIs in MUSE and Freesurfer.
We also computed a set of composite ROIs in order to perform comparative analyses at a coarser level. Composite ROIs included 6 lobe level regions, the “cortex” ROI that is the combination of all cortical GM ROIs, and the “sub-cortex” ROI that combines all sub-cortical GM ROIs.
2.5. Statistical analyses
In Dataset1, for which manual hippocampus labels were available, the agreement between Freesurfer and MUSE against manual labels was measured by comparing the volumes of segmented hippocampus ROIs. Note that as the delineations of the hippocampus region were different in the reference atlases for each method and in manual segmentations, a direct calculation of the overlap between segmentations, e.g. by calculating the Dice score between them, was not suitable for comparisons.
We used Pearson pairwise correlation (r) and Lin’s Concordance Correlation Coefficient (CCC) to measure the reproducibility between manually segmented hippocampal labels and labels automatically segmented using MUSE and Freesurfer. Pearson correlation is a measure of linear associations of ROI volumes obtained using different segmentation methods. Concordance correlation coefficient measures the agreement between ROI volumes from two segmentation methods and penalizes differential mean shifts between them. The concordance correlation coefficient is defined as:
where μx and μy are the means and σx and σy are the variances of the two variables, and ρ is the correlation coefficient between them.
We computed Bland-Altman plots between gold standard (Manual) ROI volumes and the ROI volumes calculated using the three automatic segmentations. A Bland-Altman plot is a graphical method that is extensively used for comparing the agreements of two measurements of the same variable and for detecting the presence of systematic bias and amount of variation between the two.
We tested the significance of age bias of volume differences between automated and ground-truth segmentations using a linear regression model for each method separately. We fitted a linear model (estimated using OLS) in R4.0.0 to predict the delta (difference in volume) between ground truth segmentations and those obtained by each of the three methods, Freesurfer v6.0, Freesurfer v5.3 and MUSE, with Age as the predictor variable. The regression model was:
where k is the segmentation method for which the volume difference from manual segmentation is calculated and Agei represent ith subject’s age. P-values were corrected using FDR correction for multiple comparisons of same subject across different methods. In order to assess the significance of the correlation between volume difference and age for different diagnosis groups, we stratified subjects based on diagnosis (CN, MCI and AD) and ran separate linear regression models for each method and each diagnosis group (9 different models in total).
We used the Dataset2 for evaluating the robustness of each segmentation method to scanner variations between 1.5T and 3T scanners. For each method, we calculated Pearson pair-wise correlations (r) and concordance correlation coefficients between individual ROI volumes calculated from each pair of matching 1.5T and 3T scans. This analysis was performed for cortical and subcortical structures individually, as well as for each composite ROI.
To determine the significance of the differences between methods we used the Wilcoxon signed rank test between the ROI correlation values across scanners obtained by the two methods. We applied two independent tests, one by grouping together correlation values of all cortical ROIs, and the second by grouping correlation values of all subcortical ROIs. A non-parametric test was used to avoid any normality assumptions and the p-values were corrected for multiple comparisons.
3. Experimental results
All 132 subjects in Dataset1 were segmented using fully automated Freesurfer and MUSE pipelines. Average run time per scan for MUSE was approximately 50.4 min for individual registrations (running them in parallel) and 61.17 min for subsequent label fusion. The actual computation time depends on the number of atlases chosen for MUSE. Run time per scan for Freesurfer was approximately 9.22 h for the entire pipeline using a single thread.
In our visual inspection of segmentation results from Dataset1 and Dataset2, all MUSE segmentations passed the visual QC. For the Freesurfer segmentations, 2 cases from Dataset1 were flagged for exclusion due to gross errors (suppl. Fig. 2). The final sample included 132 subjects for Dataset1 and 113 subjects for Dataset2 after exclusion of the flagged cases.
3.1. Analysis of hippocampal volume differences
Mean volumes of right and left hippocampi calculated from automatic and manual segmentations are given in Table 2.
Table 2.
Left, Right & Total hippocampus volumes computed using manual ground-truth, Freesurfer and MUSE segmentations
| Method | Hippo. R | Hippo. L | Hippo. Total |
|---|---|---|---|
| Manual | 2769.165 ± 590.64 | 2664.277 ± 593.87 | 5433.442 ± 1154.88 |
| Freesurfer-v5.3 | 3158.656 ± 729.29 | 3071.671 ± 704.73 | 6230.328 ± 1378.62 |
| Freesurfer-v6.0 | 3288.765 ± 637.11 | 3177.69 ± 595.06 | 6466.45 ± 1197.69 |
| MUSE | 3362.186 ± 606.61 | 3085.091 ± 585.19 | 6447.28 ± 1162.57 |
| Manual – Freesurfer-v5.3(% vol diff) | 14.06 | 15.29 | 14.66 |
| Manual – Freesurfer-v6.0(% vol diff) | 18.76 | 19.27 | 19.01 |
| Manual – MUSE(% vol diff) | 21.41 | 15.79 | 18.65 |
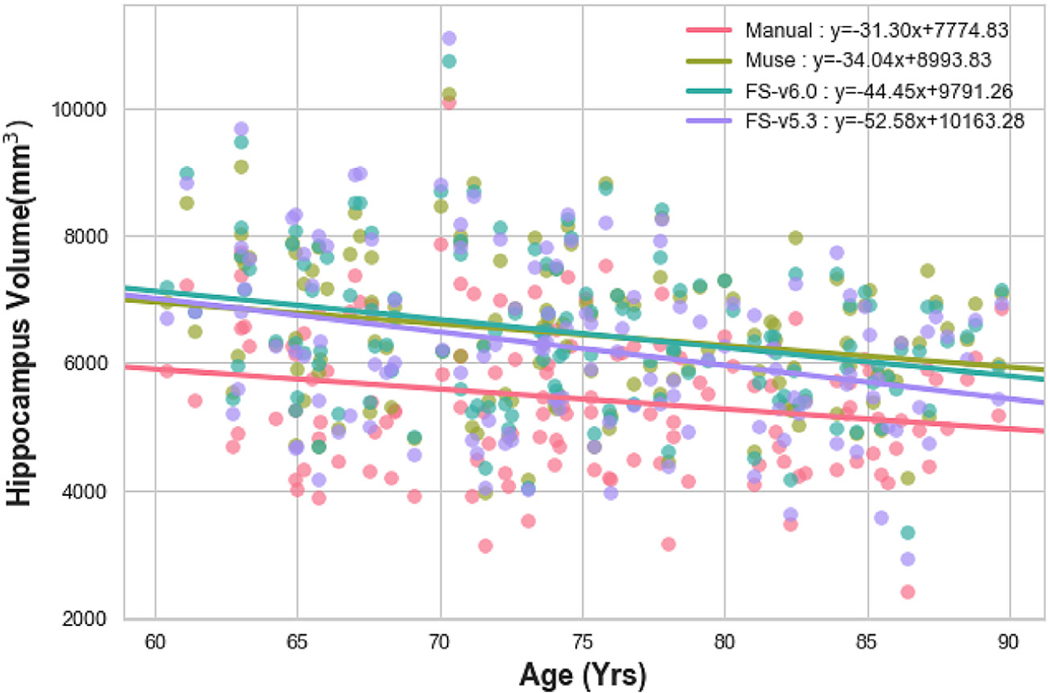
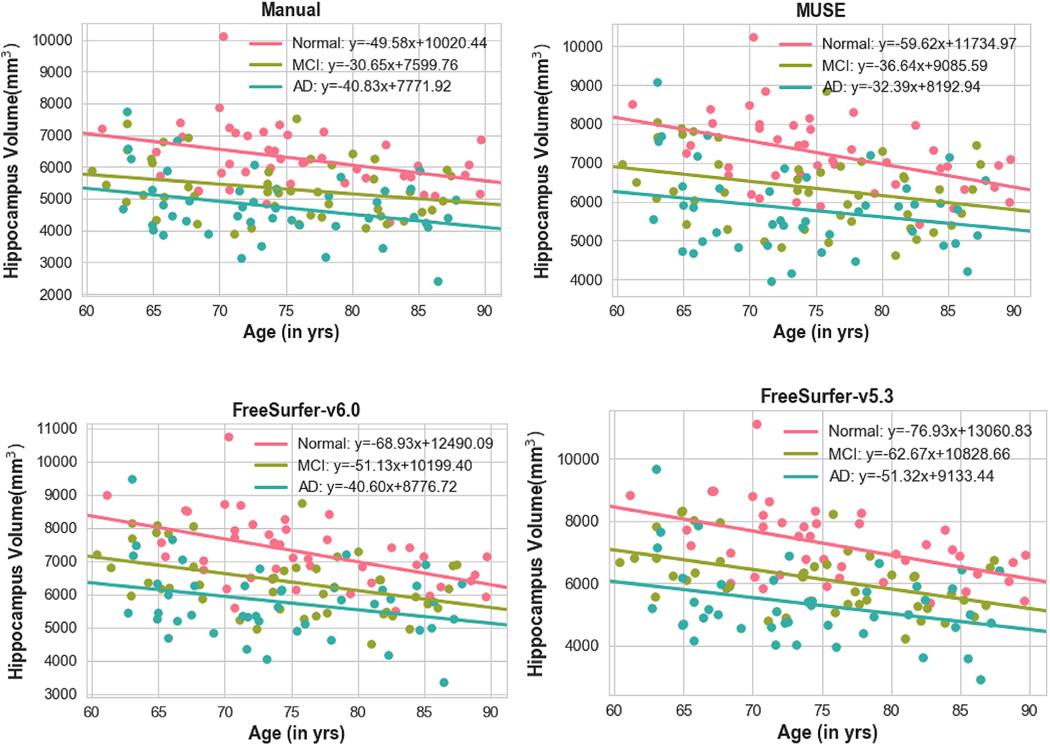
Fig. 2 shows the age trends of hippocampus volumes for manual and automatic segmentations. We found that MUSE and manual segmentations had a similar slope (s_MUSE = −34.04, s_Manual = −31.29), while Freesurfer segmentation had a significantly higher slope with age (s_Freesurfer-v5.3 = −52.57,s_Freesurfer-v6.0 = −44.44). Note that differences in intercepts between methods are expected and they are due to differences in ROI definitions. Age trends of hippocampal volumes for CN, MCI and AD subjects are shown in Fig. 3.
Fig. 2.
Hippocampal volumes calculated from MUSE (Green), Freesurfer-v6.0(Blue), Freesurfer-v5.3(Purple) and manual(Pink) segmentations, plotted against age at scan time.
Fig. 3.
Hippocampal volumes calculated from MUSE, and manual segmentation, plotted against age at scan time and grouped by disease category. Normal, MCI and AD subjects are shown with Pink, Green and Blue colors, respectively.
We verified possible bias in volume estimations using Bland Altman plots. Fig. 4 shows trends of volume differences of Freesurfer and MUSE segmentations from manual segmentations, plotted against mean hippocampus volumes.
Fig. 4.
Bland-Altman plots showing comparisons between hippocampal volumes calculated from Freesurfer-v6.0 and manual segmentations (left), Freesurfer-v5.3 and manual segmentations(middle) and MUSE and manual segmentations (right), against hippocampus volume. The black solid line represents the mean difference and dotted lines show upper and lower limits defined as 1.96 standard deviation from mean. X-axis and Y-axis denotes the mean values ((M1 + M2)/2) and difference (M1-M2) respectively. Regression lines have been fit to the data points for each automatic method to visualize potential bias.
The estimated regression line indicates a negative trend for Freesurfer, suggesting that larger hippocampus volumes lead to a larger difference in volume estimation in comparison to manual segmentations. For MUSE, regression line shows a constant difference trend for increasing hippocampus volumes. Bland Altman plots of volume differences against age are shown in Fig. 5. Similarly, both Freesurfer versions v6.0 and v5.3 has a negative slope, indicating a bias towards under-segmentation of hippocampus for older people, and hence introduction of spurious age trends. The age bias in Freesurfer segmentations was also present when the subjects were grouped by disease category (CN, MCI, AD) (suppl. Fig. 3).
Fig. 5.
Scatter plot of differences showing hippocampal volumes calculated from Freesurfer-v6.0 and manual segmentations (left), Freesurfer-v5.3 and manual segmentations (middle), and MUSE and manual segmentations (right), against subject’s age at scan time.
Linear regression models showed that the observed age bias was significant in both Freesurfer-v5.3 and Freesurfer-v6.0 (Table 3). The volume difference of Freesurfer-v5.3 from the ground-truth segmentation had a higher negative slope with higher age, with a difference of 21.28 mm3 per year of Age, 95% CI, p < 0.05, FDR corrected.
Table 3.
Age correlations of volume differences between automated and ground-truth segmentations.
| ΔMUSE |
ΔFS_v5.3 |
ΔFS_v6.0 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Predictors | Estimates | p | pcorr | Estimates | p | pcorr | Estimates | p | pcorr |
| (Intercept) | 1218.99 | <0.001*** | 2388.45 | <0.001*** | 2016.43 | <0.001*** | |||
| Age | −2.74 | 0.53265 | 0.53265 | −21.28 | 0.00105** | 0.00315** | −13.15 | 0.00324** | 0.00487** |
| Observations | 132 | 132 | 132 | ||||||
Results of regression models assessing the age bias for different segmentations across each diagnosis group are given in supplementary Table S1. We found that the age bias was predominantly driven by for the MCI subjects, both for Freesurfer-v5.3 and v6.0.
3.2. Reproducibility analysis across field strengths
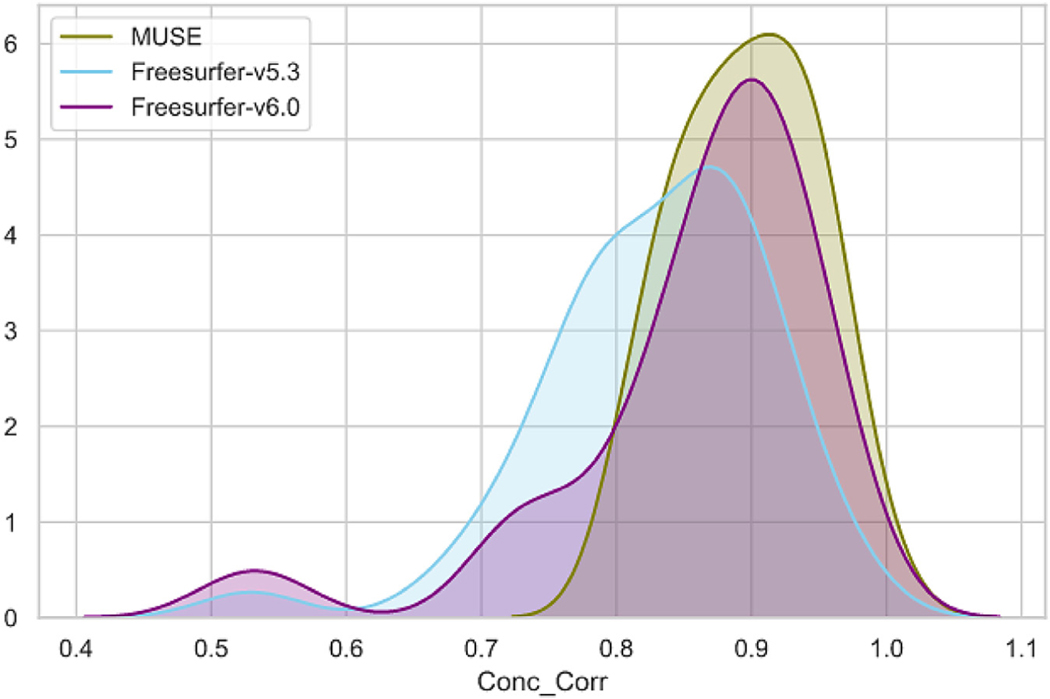
We compared volumes of matched ROIs for 1.5T and 3T same day scan pairs segmented using the automated methods. This comparison included all individual and composite ROIs. We calculated the Pearson and Concordance correlations between 1.5T and 3T volumes for each ROI. Concordance correlation values for all individual ROIs are shown in Fig. 6.
Fig. 6.
Concordance correlation between ROI volumes of 1.5T and 3T ADNI scan pairs obtained using Freesurfer-v6.0, Freesurfer-v5.3 and MUSE segmentations. The ROIs are grouped as cortical and subcortical, and sorted by MUSE correlation values in decreasing order, separately for each group, to better highlight the differences.
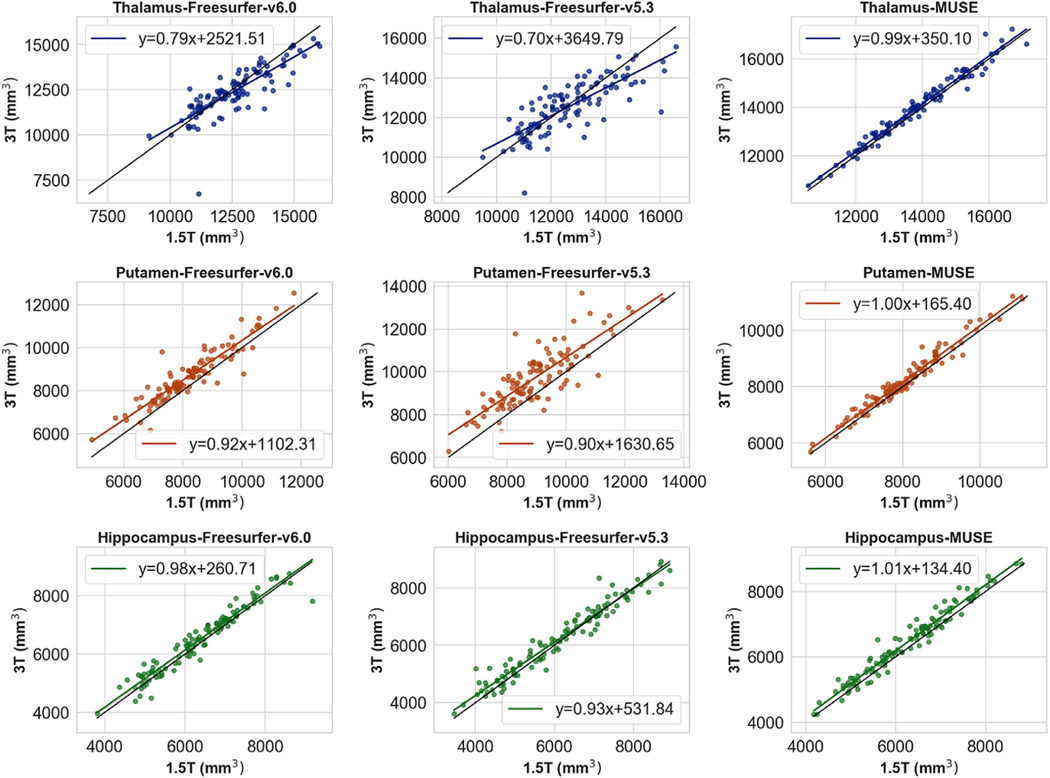
Freesurfer-v6.0 obtained higher correlations for all ROIs compared to Freesurfer-v5.3, suggesting that the major version update in Freesurfer resulted in considerable differences in the final segmentation, improving the overall accuracy. MUSE obtained consistently higher correlation compared to Freesurfer-v5.3, while in cortical ROIs Freesurfer-v6.0 and MUSE showed comparable performance. Importantly, the differences between Freesurfer and MUSE were higher in the segmentation of the deep structures, MUSE obtaining higher correlations in all deep structures except hippocampus. Scatter plots for the volumes of selected deep structures for each method against ground truth segmentation volumes are given in Fig. 7. The distribution of correlation coefficients from all ROIs for each method is presented in Fig. 8.
Fig. 7.
Scatter plots of ROI volumes for segmentations of 1.5T and 3T ADNI scan pairs. The plots show the volumes of 3 deep structures, thalamus, putamen and hippocampus, calculated using Freesurfer-v6.0(left), Freesurfer-v5.3(middle) and MUSE (right).
Fig. 8.
Histogram showing the distribution of Concordance Correlation between 1.5T and 3T ADNI scan pairs calculated using Freesurfer-v6.0(blue), Freesurfer-v5.3 (purple) and MUSE (green).
The results for composite ROIs were similar, with higher correlations for MUSE in subcortical regions, while in cortical regions MUSE and Freesurfer-v6.0 had comparable reproducibility at lobe level ROIs (suppl. Fig. 4). The Wilcoxon-tests comparing Freesurfer and MUSE segmentations indicated that correlations of ROI volumes across scanners were significantly different between MUSE and both Freesurfer versions for sub-cortical ROIs. For the deep structures, the differences were significant between MUSE and Freesurfer-v5.3, but not for Freesurfer-v6.0 (Table 4).
Table 4.
Mean correlation, concordance correlation and Wilcox p-values for segmentation of matching 1.5T and 3T ADNI scan pairs by each Freesurfer version and MUSE.
| Cortical |
Sub-cortical |
|||||
|---|---|---|---|---|---|---|
| Freesurfer-v5.3 | Freesurfer-v6.0 | MUSE | Freesurfer-v5.3 | Freesurfer-v6.0 | MUSE | |
| Mean Correlation | 0.8867 | 0.8982 | 0.9045 | 0.83992 | 0.88656 | 0.97425 |
| Mean Concordance Correlation | 0.83678 | 0.85236 | 0.87805 | 0.78113 | 0.85295 | 0.96042 |
| Wilcox signed rank p-val | 0.0029 | 0.5052 | 0.03125 | 0.01796 | ||
| Wilcox signed rank (FDR corr) | 0.0058 | 0.5167 | 0.03125 | 0.03125 | ||
4. Discussion
Freesurfer is one of the most widely used tools in neuroimaging research for segmenting brain anatomy. Although Freesurfer is extensively validated and it has been used in a large number of studies, various studies have also reported high rates of failures resulting in exclusion of large number of scans, inconsistencies in segmentations and age related bias (Wenger et al., 2014; Cherbuin et al., 2009).
In recent years, methods that use the multi atlas label fusion framework have obtained state-of-the-art accuracy, showing that consensus segmentation using multiple atlases may significantly improve the segmentation accuracy, while making it also more robust to sporadic registration errors/imperfections (Iglesias and Sabuncu, 2015; Warfield, 2017).
Herein we evaluated MUSE by comparing it to Freesurfer, which is the current standard for brain anatomy segmentation. While the accuracy, reliability and reproducibility of Freesurfer has been well tested across various studies and segmentation tools, it has not been compared with current multi-atlas methods.
Importantly, Freesurfer had a major revision update in 2017 (v6.0). Over 1600 neuroimaging publications used Freesurfer for quantification of brain volumes (as listed in pubmed.gov). Most of these publications used previous versions dating before the Freesurfer-v6.0 release. Considering that previous stable version of Freesurfer (v5.3) has been used by a large number of studies in the past, we performed comparisons using both versions of Freesurfer. Our hypothesis was that MUSE would overcome some of Freesurfer’s limitations. We specifically focused on two tasks, which are important in multi-site aging studies: bias of segmentation with age, and reproducibility across different field strengths. The latter was a test of inter-scanner reproducibility, an issue that is of rapidly rising significance with the emergence of large-scale meta/mega-analyses that pool data from multiple studies (Thompson et al., 2014; Davatzikos, 2018).
Automatic segmentation methods typically use a reference atlas with manually defined ROI labels and apply image registration for transferring these labels to target image space. Freesurfer is based on the registration of a single probabilistic atlas, and label assignment based on both aligned atlas probabilities and target image intensities. In contrast, multi-atlas techniques take advantage of the consensus labeling of multiple atlases. The advantage of the multi atlas approach is twofold: a) multiple atlases allow capturing a broader anatomical variation, e.g. by including atlases from subjects with different sex and age, thereby allowing the label fusion algorithm to select subject-appropriate atlases on a regional basis; and b) even when the image registration fails for one or more atlas images, the voting between multiple atlases presumably helps obtain a correct segmentation, unless there is a systematic registration error that affects a majority of the atlases. Specifically, in MUSE, two different registration algorithms were used to increase the variations within the ensemble, and a local similarity ranking strategy was used to give more weights to atlases locally more similar to the target scan in the fusion.
We found that hippocampus segmentations by both Freesurfer-v6.0 and Freesurfer-v5.3 showed a bias towards over-segmentation of larger hippocampi, and under-segmentation of hippocampi in smaller or older individuals, although this age bias was reduced with the newer version. On the other hand, MUSE segmentations were more consistent with ground-truth segmentations, without bias with subject age or hippocampus volume. This is an important finding, when evaluating age effects on brain volumes. Our results suggest that Freesurfer introduces spurious age effects, which can obviously lead to false biological interpretations. One reason could be that Freesurfer does not use the population-based specific template.
In our reproducibility analyses, we found that MUSE obtained consistently higher correlations between ROI volumes calculated across the two scanners in comparison to Freesurfer 5.3. Importantly, we also found that Freesurfer 6.0 segmentations showed consistent improvement in all ROIs compared to Freesurfer-v5.3. While Freesurfer 6.0 was comparable to MUSE for cortical ROIs, in deep structures MUSE obtained very consistent segmentations across subjects with higher correlations compared to both Freesurfer versions. This finding is consistent with previously reported state-of-the-art accuracy of MAS methods in segmentation of deep structures, and could be explained by the advantage of using a model based (i.e. a-priori defined ground-truth ROI labels) consensus labeling in these regions where the tissue contrast alone is not informative enough to guide the segmentation.
High number of Freesurfer exclusions based on detailed visual verification of individual ROIs was previously reported (Mccarthy 2015; Zandifar et al., 2017). Such an extensive QC was out of scope of our analysis that focused on mostly automated processing in view of largescale datasets. In our QC for detecting gross errors, MUSE outperformed Freesurfer in terms of overall failure rates. While ~1% of all segmentations (2 scans from 247 in total) were excluded due to Freesurfer failures based on our case by case visual QC of general segmentation quality, all MUSE segmentations have obtained a positive QC result. The robustness of MUSE was expected, as multiple atlases provide various representations of the anatomy, while the label fusion of multiple warped atlases allows the method to correct the effect of individual registrations that failed, unless the failure is not systematic to a majority of the atlases.
This work has also some limitations. A major problem for a systematic comparison is the limited availability of ground truth segmentations. Manual segmentation of anatomical ROIs is a difficult and time consuming task. For this reason, there are very few datasets that provide manually segmented ROI labels. Also, because Freesurfer and MUSE use their own atlas sets with different ROI label denotations, a direct comparison of the two methods is not possible. MUSE uses a set of 35 scans and their semi-automatically segmented ROI labels as reference atlases. We did not prefer to use these scans in our comparisons, as this would be biased towards MUSE, even with cross-validation. Also, our experiments were limited to comparisons between Freesurfer and the multi atlas segmentation approach. In recent years deep learning methods obtained state of the art accuracy in various problems in neuroimaging. While a comparison to more recent deep learning methods for segmentation would be very informative, this is out of the scope of this paper.
Our comparative evaluations have shown that MUSE, a multi-atlas ROI segmentation method, can help alleviate some of the limitations of Freesurfer and related methods, by virtue of leveraging multiple atlases, registration methods and parameters, thereby offering both the advantages of a consensus-based methods and of regional adaptivity of the atlases to the target anatomy. Critically, MUSE also displayed significantly higher inter-scanner consistency, thereby offering promise that multi-site, multi-study meta/mega-analyses can be performed more accurately. Given these favorable results and increasing availability of parallel and cloud computing capacities, multi-atlas segmentation has a great potential of becoming the standard approach for segmentation of brain images in population studies and in clinical applications.
Supplementary Material
Acknowledgments
This work was supported by the National Institute on Aging (grant number 1RF1AG054409), the National Institute of Mental Health (grant number 5R01MH112070), National Institutes of Health (grant number 1RF1AG059869) and National Institutes of Health (grant number 75N95019C00022).
Footnotes
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neuroimage.2020.117248.
References
- Apostolova LG, Dutton RA, Dinov ID, Hayashi KM, Toga AW, Cummings JL, Thompson PM, 2006. Conversion of mild cognitive impairment to Alzheimer disease predicted by hippocampal atrophy maps. Arch. Neurol. 63, 693–699. doi: 10.1001/archneur.63.5.693. [DOI] [PubMed] [Google Scholar]
- Asman A, Alireza Akhondi-Asl, Wang H, Tustison N, Avants B, Warfield SK, Landman B, 2013. Miccai 2013 segmentation algorithms, theory and applications (SATA) challenge results summary. Presented at the MICCAI Challenge Workshop on Segmentation: Algorithms, Theory and Applications (SATA). [Google Scholar]
- Avants BB, Tustison NJ, Stauffer M, Song G, Wu B, Gee JC, 2014. The In-sight ToolKit image registration framework. Front Neuroinform 8. doi: 10.3389/fn-inf.2014.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkour A, Morris JC, Wolk DA, Dickerson BC, 2013. The effects of aging and Alzheimer’s disease on cerebral cortical anatomy: specificity and differential relationships with cognition. Neuroimage 76, 332–344. doi: 10.1016/j.neuroimage.2013.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilha L, Molnar C, Horner MD, Anderson B, Forster L, George MS, Nahas Z, 2008. Neurocognitive deficits and prefrontal cortical atrophy in patients with schizophrenia. Schizophr Res 101, 142–151. doi: 10.1016/j.schres.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer JB, Magda S, Airriess C, Smith ME, 2009. Fully-automated quantification of regional brain volumes for improved detection of focal atrophy in Alzheimer disease. AJNR Am J Neuroradiol 30, 578–580. doi: 10.3174/ajnr.A1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charil A, Dagher A, Lerch JP, Zijdenbos AP, Worsley KJ, Evans AC, 2007. Focal cortical atrophy in multiple sclerosis: relation to lesion load and disability. Neuroimage 34, 509–517. doi: 10.1016/j.neuroimage.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Cherbuin N, Anstey KJ, Réglade-Meslin C, Sachdev PS, 2009. In vivo hippocampal measurement and memory: a comparison of manual tracing and automated segmentation in a large community-based sample. PLoS ONE 4, e5265. doi: 10.1371/journal.pone.0005265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C, 2018. BRAIN AGING HETEROGENEITY ELUCIDATED VIA MACHINE LEARNING: THE MULTI-SITE ISTAGING DIMENSIONAL NEUROIMAGING REFERENCE SYSTEM. Alzheimer’s & Dementia: The Journal of the Alzheimer’s Association 14, P1476–P1477. doi: 10.1016/j.jalz.2018.06.2505. [DOI] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ, 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31, 968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Dicks E, Vermunt L, van der Flier WM, Visser PJ, Barkhof F, Scheltens P, Tijms BM Alzheimer’s Disease Neuroimaging Initiative, 2019. Modeling grey matter atrophy as a function of time, aging or cognitive decline show different anatomical patterns in Alzheimer’s disease. Neuroimage Clin 22, 101786. doi: 10.1016/j.nicl.2019.101786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi J, Erus G, Ou Y, Resnick SM, Gur RC, Gur RE, Satterthwaite TD, Furth S, Davatzikos C Alzheimer’s Neuroimaging Initiative, 2016. MUSE: MUlti-atlas region Segmentation utilizing Ensembles of registration algorithms and parameters, and locally optimal atlas selection. Neuroimage 127, 186–195. doi: 10.1016/j.neuroimage.2015.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira D, Hansson O, Barroso J, Molina Y, Machado A, Hernández-Cabrera JA, Muehlboeck J-S, Stomrud E, Nägga K, Lindberg O, Ames D, Kalpouzos G, Fratiglioni L, Bäckman L, Graff C, Mecocci P, Vellas B, Tsolaki M, K ł oszewska I, Soininen H, Lovestone S, Ahlström H, Lind L, Larsson E-M, Wahlund L-O, Simmons A, Westman Ethe AddNeuroMed consortium, for the Alzheimer’s Disease Neuroimaging Initiative (ADNI), Australian Imaging Biomarkers and Lifestyle Study of Ageing (AIBL) research group, 2017. The interactive effect of demographic and clinical factors on hippocampal volume: A multicohort study on 1958 cognitively normal individuals. Hippocampus 27, 653–667. doi: 10.1002/hipo.22721. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM, 2002. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fischl B, van der Kouwe A, Destrieux C, Halgren E, Ségonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale AM, 2004. Automatically parcellating the human cerebral cortex. Cereb. Cortex 14, 11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Frisoni GB, Jack CR, Bocchetta M, Bauer C, Frederiksen KS, Liu Y, Preboske G, Swihart T, Blair M, Cavedo E, Grothe MJ, Lanfredi M, Martinez O, Nishikawa M, Portegies M, Stoub T, Ward C, Apostolova LG, Ganzola R, Wolf D, Barkhof F, Bartzokis G, DeCarli C, Csernansky JG, deToledoMorrell L, Geerlings MI, Kaye J, Killiany RJ, Lehéricy S, Matsuda H, O’Brien J, Silbert LC, Scheltens P, Soininen H, Teipel S, Waldemar G, Fellgiebel A, Barnes J, Firbank M, Gerritsen L, Henneman W, Malykhin N, Pruessner JC, Wang L, Watson C, Wolf H, deLeon M, Pantel J, Ferrari C, Bosco P, Pasqualetti P, Duchesne S, Duvernoy H, Boccardi M EADC-ADNI Working Group on The Harmonized Protocol for Manual Hippocampal Volumetry and for the Alzheimer’s Disease Neuroimaging Initiative, 2015. The EADC-ADNI Harmonized Protocol for manual hippocampal segmentation on magnetic resonance: evidence of validity. Alzheimers Dement 11, 111–125. doi: 10.1016/j.jalz.2014.05.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgio A, De Stefano N, 2013. Clinical use of brain volumetry. J Magn Reson Imaging 37, 1–14. doi: 10.1002/jmri.23671. [DOI] [PubMed] [Google Scholar]
- Goldstein JM, Goodman JM, Seidman LJ, Kennedy DN, Makris N, Lee H, Tourville J, Caviness VS, Faraone SV, Tsuang MT, 1999. Cortical abnormalities in schizophrenia identified by structural magnetic resonance imaging. Arch. Gen. Psychiatry 56, 537–547. doi: 10.1001/archpsyc.56.6.537. [DOI] [PubMed] [Google Scholar]
- Habes M, Toledo JB, Resnick SM, Doshi J, Van der Auwera S, Erus G, Janowitz D, Hegenscheid K, Homuth G, Völzke H, Hoffmann W, Grabe HJ, Davatzikos C, 2016. Relationship between APOE Genotype and Structural MRI Measures throughout Adulthood in the Study of Health in Pomerania Population-Based Cohort. AJNR Am J Neuroradiol 37, 1636–1642. doi: 10.3174/ajnr.A4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias JE, Sabuncu MR, 2015. Multi-atlas segmentation of biomedical images: A survey. Med Image Anal 24, 205–219. doi: 10.1016/j.media.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowitz D, Schwahn C, Borchardt U, Wittfeld K, Schulz A, Barnow S, Biffar R, Hoffmann W, Habes M, Homuth G, Nauck M, Hegenscheid K, Lotze M, Völzke H, Freyberger HJ, Debette S, Grabe HJ, 2014. Genetic, psychosocial and clinical factors associated with hippocampal volume in the general population.Transl Psychiatry 4, e465. doi: 10.1038/tp.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller SS, Gerdes JS, Mohammadi S, Kellinghaus C, Kugel H, Deppe K, Ringelstein EB, Evers S, Schwindt W, Deppe M, 2012. Volume estimation of the thalamus using freesurfer and stereology: consistency between methods. Neuroinformatics 10, 341–350. doi: 10.1007/s12021-012-9147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikinis Z, Fallon JH, Niznikiewicz M, Nestor P, Davidson C, Bobrow L, Pelavin PE, Fischl B, Yendiki A, McCarley RW, Kikinis R, Kubicki M, Shenton ME, 2010. Gray matter volume reduction in rostral middle frontal gyrus in patients with chronic schizophrenia. Schizophr. Res. 123, 153–159. doi: 10.1016/j.schres.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy CS, Ramprashad A, Thompson C, Botti J-A, Coman IL, Kates WR, 2015. A comparison of FreeSurfer-generated data with and without manual intervention. Front Neurosci 9, 379. doi: 10.3389/fnins.2015.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina D, Cerasa A, Condino F, Arabia G, Novellino F, Nicoletti G, Salsone M, Morelli M, Lanza PL, Quattrone A, 2011. Patterns of brain atrophy in Parkinson’s disease, progressive supranuclear palsy and multiple system atrophy. Parkinsonism Relat. Disord. 17, 172–176. doi: 10.1016/j.parkreldis.2010.12.010. [DOI] [PubMed] [Google Scholar]
- Mulder ER, de Jong RA, Knol DL, van Schijndel RA, Cover KS, Visser PJ, Barkhof F, Vrenken H Alzheimer’s Disease Neuroimaging Initiative, 2014. Hippocampal volume change measurement: quantitative assessment of the reproducibility of expert manual outlining and the automated methods FreeSurfer and FIRST. Neuroimage 92, 169–181. doi: 10.1016/j.neuroimage.2014.01.058. [DOI] [PubMed] [Google Scholar]
- Ou Y, Sotiras A, Paragios N, Davatzikos C, 2011. DRAMMS: Deformable registration via attribute matching and mutual-saliency weighting. Med Image Anal 15, 622–639. doi: 10.1016/j.media.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz N, Ghisletta P, Rodrigue KM, Kennedy KM, Lindenberger U, 2010. Trajectories of brain aging in middle-aged and older adults: regional and individual differences. Neuroimage 51, 501–511. doi: 10.1016/j.neuroimage.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer JD, Lashley T, Schott JM, Warren JE, Mead S, Isaacs AM, Beck J, Hardy J, de Silva R, Warrington E, Troakes C, Al-Sarraj S, King A, Borroni B, Clarkson MJ, Ourselin S, Holton JL, Fox NC, Revesz T, Rossor MN, Warren JD, 2011. Clinical and neuroanatomical signatures of tissue pathology in frontotemporal lobar degeneration. Brain 134, 2565–2581. doi: 10.1093/brain/awr198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabuncu MR, Desikan RS, Sepulcre J, Yeo BTT, Liu H, Schmansky NJ, Reuter M, Weiner MW, Buckner RL, Sperling RA, Fischl BAlzheimer’s Disease Neuroimaging Initiative, 2011. The dynamics of cortical and hippocampal atrophy in Alzheimer disease. Arch. Neurol. 68, 1040–1048. doi: 10.1001/archneurol.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Wolf DH, Calkins ME, Vandekar SN, Erus G, Ruparel K, Roalf DR, Linn KA, Elliott MA, Moore TM, Hakonarson H, Shinohara RT, Davatzikos C, Gur RC, Gur RE, 2016. Structural Brain Abnormalities in Youth With Psychosis Spectrum Symptoms. JAMA Psychiatry 73, 515–524. doi: 10.1001/jamapsychiatry.2015.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME, Toro R, Jahanshad N, Schumann G, Franke B, Wright MJ, Martin NG, Agartz I, Alda M, Alhusaini S, Almasy L, Almeida J, Alpert K, Andreasen NC, Andreassen OA, Apostolova LG, Appel K, Armstrong NJ, Aribisala B, Bastin ME, Bauer M, Bearden CE, Bergmann Ø, Binder EB, Blangero J, Bockholt HJ, Bøen E, Bois C, Boomsma DI, Booth T, Bowman IJ, Bralten J, Brouwer RM, Brunner HG, Brohawn DG, Buckner RL, Buitelaar J, Bulayeva K, Bustillo JR, Calhoun VD, Cannon DM, Cantor RM, Carless MA, Caseras X, Cavalleri GL, Chakravarty MM, Chang KD, Ching CRK, Christoforou A, Cichon S, Clark VP, Conrod P, Coppola G, Crespo-Facorro B, Curran JE, Czisch M, Deary IJ, de Geus EJC, den Braber A, Delvecchio G, Depondt C, de Haan L, de Zubicaray GI, Dima D, Dimitrova R, Djurovic S, Dong H, Donohoe G, Duggirala R, Dyer TD, Ehrlich S, Ekman CJ, Elvsåshagen T, Emsell L, Erk S, Espeseth T, Fagerness J, Fears S, Fedko I, Fernández G, Fisher SE, Foroud T, Fox PT, Francks C, Frangou S, Frey EM, Frodl T, Frouin V, Garavan H, Giddaluru S, Glahn DC, Godlewska B, Goldstein RZ, Gollub RL, Grabe HJ, Grimm O, Gruber O, Guadalupe T, Gur RE, Gur RC, Göring HHH, Hagenaars S, Hajek T, Hall GB, Hall J, Hardy J, Hartman CA, Hass J, Hatton SN, Haukvik UK, Hegenscheid K, Heinz A, Hickie IB, Ho B-C, Hoehn D, Hoekstra PJ, Hollinshead M, Holmes AJ, Homuth G, Hoogman M, Hong LE, Hosten N, Hottenga J-J, Hulshoff Pol HE, Hwang KS, Jack CR, Jenkinson M, Johnston C, Jönsson EG, Kahn RS, Kasperaviciute D, Kelly S, Kim S, Kochunov P, Koenders L, Krämer B, Kwok JBJ, Lagopoulos J, Laje G, Landen M, Landman BA, Lauriello J, Lawrie SM, Lee PH, Le Hellard S, Lemaître H, Leonardo CD, Li C, Liberg B, Liewald DC, Liu X, Lopez LM, Loth E, Lourdusamy A, Luciano M, Macciardi F, Machielsen MWJ, MacQueen GM, Malt UF, Mandl R, Manoach DS, Martinot J-L, Matarin M, Mather KA, Mattheisen M, Mattingsdal M, MeyerLindenberg A, McDonald C, McIntosh AM, McMahon FJ, McMahon KL, Meisenzahl E, Melle I, Milaneschi Y, Mohnke S, Montgomery GW, Morris DW, Moses EK, Mueller BA, Muñoz Maniega S, Mühleisen TW, Müller-Myhsok B, Mwangi B, Nauck M, Nho K, Nichols TE, Nilsson L-G, Nugent AC, Nyberg L, Olvera RL, Oosterlaan J, Ophoff RA, Pandolfo M, PapalampropoulouTsiridou M, Papmeyer M, Paus T, Pausova Z, Pearlson GD, Penninx BW, Peterson CP, Pfennig A, Phillips M, Pike GB, Poline J-B, Potkin SG, Pütz B, Ramasamy A, Rasmussen J, Rietschel M, Rijpkema M, Risacher SL, Roffman JL, Roiz-Santiañez R, Romanczuk-Seiferth N, Rose EJ, Royle NA, Rujescu D, Ryten M, Sachdev PS, Salami A, Satterthwaite TD, Savitz J, Saykin AJ, Scanlon C, Schmaal L, Schnack HG, Schork AJ, Schulz SC, Schür R, Seidman L, Shen L, Shoemaker JM, Simmons A, Sisodiya SM, Smith C, Smoller JW, Soares JC, Sponheim SR, Sprooten E, Starr JM, Steen VM, Strakowski S, Strike L, Sussmann J, Sämann PG, Teumer A, Toga AW, Tordesillas-Gutierrez D, Trabzuni D, Trost S, Turner J, Van den Heuvel M, van der Wee NJ, van Eijk K, van Erp TGM, van Haren NEM, van ‘t Ent D, van Tol M-J, Valdés Hernández MC, Veltman DJ, Versace A, Völzke H, Walker R, Walter H, Wang L, Wardlaw JM, Weale ME, Weiner MW, Wen W, Westlye LT, Whalley HC, Whelan CD, White T, Winkler AM, Wittfeld K, Woldehawariat G, Wolf C, Zilles D, Zwiers MP, Thalamuthu A, Schofield PR, Freimer NB, Lawrence NS, Drevets W, 2014. The ENIGMA Consortium: largescale collaborative analyses of neuroimaging and genetic data. Brain Imaging and Behavior. doi: 10.1007/s11682-013-9269-5. [DOI] [PMC free article] [PubMed]
- Tian Q, Bair W-N, Resnick SM, Bilgel M, Wong DF, Studenski SA, 2018. β-amyloid deposition is associated with gait variability in usual aging. Gait Posture 61, 346–352. doi: 10.1016/j.gaitpost.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC, 2010. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29, 1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee P, Wang Z, 2017. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers (Basel) 9. doi: 10.3390/cancers9050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger E, Mårtensson J, Noack H, Bodammer NC, Kühn S, Schaefer S, Heinze H-J, Düzel E, Bäckman L, Lindenberger U, Lövdén M, 2014. Comparing manual and automatic segmentation of hippocampal volumes: reliability and validity issues in younger and older brains. Hum Brain Mapp 35, 4236–4248. doi: 10.1002/hbm.22473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga L, Langen M, Ambrosino S, van Dijk S, Oranje B, Durston S, 2014. Typical development of basal ganglia, hippocampus, amygdala and cerebellum from age 7 to 24. Neuroimage 96, 67–72. doi: 10.1016/j.neuroimage.2014.03.072. [DOI] [PubMed] [Google Scholar]
- Zandifar A, Fonov V, Coupé P, Pruessner J, Collins DL Alzheimer’s Disease Neuroimaging Initiative, 2017. A comparison of accurate automatic hippocampal segmentation methods. Neuroimage 155, 383–393. doi: 10.1016/j.neuroimage.2017.04.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.