Summary
The mesencephalic locomotor region (MLR) is a key midbrain center with roles in locomotion. Despite extensive studies and clinical trials aimed at therapy-resistant Parkinson’s disease (PD), debate on its function remains. Here, we reveal the existence of functionally diverse neuronal populations with distinct roles in control of body movements. We identify two spatially intermingled glutamatergic populations separable by axonal projections, mouse genetics, neuronal activity profiles, and motor functions. Most spinally projecting MLR neurons encoded the full-body behavior rearing. Loss- and gain-of-function optogenetic perturbation experiments establish a function for these neurons in controlling body extension. In contrast, Rbp4-transgene-positive MLR neurons project in an ascending direction to basal ganglia, preferentially encode the forelimb behaviors handling and grooming, and exhibit a role in modulating movement. Thus, the MLR contains glutamatergic neuronal subpopulations stratified by projection target exhibiting roles in action control not restricted to locomotion.
Keywords: Motor controls, locomotion, rearing, forelimb behaviors, brainstem, mesencephalic locomotor region, basal ganglia, substantia nigra, spinal cord, deep brain stimulation
Graphical abstract
Highlights
-
•
Glutamatergic mesencephalic locomotor region (MLR) neurons segregate by axonal target
-
•
MLR neurons projecting to basal ganglia output nuclei are labeled by Rbp4 transgene
-
•
Spinally projecting MLR neurons encode rearing and regulate body extension
-
•
Rbp4-MLR neurons are mostly tuned to forelimb movements and modulate behavior
The functionally diverse mesencephalic locomotor region (MLR) contains distinct subpopulations of glutamatergic neurons with different axonal targets, with activity correlated to specific behaviors beyond just locomotion.
Introduction
Locomotion is essential for survival across all species and is the terrestrial motor program translocating the entire body. It enables many forms of controlled interactions with the environment, including exploratory locomotion such as seeking food, as well as more urgent responses such as escaping from danger. Irrespective of the chosen locomotor form, its successful completion requires controlled postural adjustments of the entire body, the coordinated recruitment of limbs to translocate the body and the efficient suppression of other motor programs not compatible with locomotion. These behavioral observations raise the question of the underlying neuronal circuit mechanisms involved in the selection and regulation of locomotion and other forms of body movements.
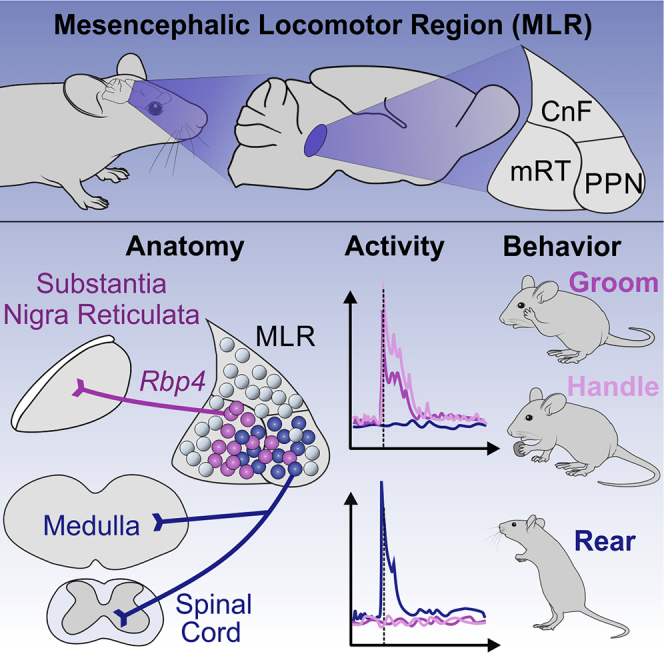
The mesencephalic locomotor region (MLR) is a midbrain area that combines various attributes central to the regulation of locomotion by integrating many inputs and projecting to both descending and ascending targets. The historic definition of the MLR has been functional and based on the fact that its electrical stimulation can elicit full-body locomotion, with speed and gaits scaling with the stimulation intensity (Noga et al., 1988; Shik et al., 1966). Several questions related to MLR function arose following these observations, driven by attempts to define its precise location and the neuronal identities responsible for the observed effects. Anatomically, the confines of the MLR by its original functional definition include a midbrain area comprising the pedunculopontine nucleus (PPN), the rostro-caudally contiguous area of the pre-cuneiform and cuneiform nuclei (pCnF and CnF), and the adjacent mesencephalic reticular formation (mRT). These areas contain intermingled glutamatergic and GABAergic neurons, and the PPN also contains cholinergic neurons (Martinez-Gonzalez et al., 2011; Mena-Segovia and Bolam, 2017; Tubert et al., 2019; Wang and Morales, 2009), necessitating a strategy to dissect and understand MLR function by location and neurotransmitter identity.
Mapping the broader MLR region by electrical stimulation and optogenetic techniques led to observations of functional diversity within the MLR. It is established that glutamatergic MLR (MLR-vGlut2) neurons constitute the neuroanatomical basis for the short-latency locomotion-promoting behavior observed upon MLR stimulation (Caggiano et al., 2018; Capelli et al., 2017; Josset et al., 2018; Lee et al., 2014; Roseberry et al., 2016). Using spatially more restricted dissection, studies agree that glutamatergic CnF (CnF-vGlut2) neurons elicit and control high-speed locomotion (Caggiano et al., 2018; Josset et al., 2018), resonating with other work (Jordan, 1998; Opris et al., 2019; Skinner and Garcia-Rill, 1984; Takakusaki et al., 2016). In contrast, evidence regarding the function of neurons in and around the PPN is more ambiguous. Electrical stimulation unraveled sites in the ventral PPN eliciting inhibition of muscle tone and sites in the dorsal PPN eliciting mixed responses (Takakusaki et al., 2016), while other studies identified locomotion-promoting sites in the PPN (Skinner and Garcia-Rill, 1984). Studies applying optogenetics to glutamatergic PPN (PPN-vGlut2) neurons did not provide definitive evidence on their function either, with reports supporting roles in low-speed exploratory locomotion (Caggiano et al., 2018), locomotion arrest (Josset et al., 2018), or both (Carvalho et al., 2020). Of note, recent studies were also mostly focused on limb dynamics during locomotion and did not take into account postural changes required for locomotion, as described before (Mori et al., 1992).
Studies on PPN are also of clinical importance. Application of PPN deep brain stimulation (DBS) (Lozano et al., 2017) to ameliorate parkinsonian gait and balance symptoms yields diverse findings (Nowacki et al., 2019; Thevathasan et al., 2018; Tubert et al., 2019). A recent review article stresses the fact that functional diversity in the PPN area is likely the key reason for the lack of consensus on applied strategies to ameliorate Parkinson’s disease (PD) symptoms, despite ongoing clinical work over many years (Garcia-Rill et al., 2019). Together, while it is clear that CnF-vGlut2 neurons can drive locomotion within an escape context, results on the function of the adjacent regions, including PPN, cannot be reconciled. These divergent observations underscore the need to better characterize the functional neuronal diversity within this midbrain region, also with respect to other motor behaviors, given that only a fraction of MLR neurons encode locomotion (Caggiano et al., 2018; Carvalho et al., 2020; Roseberry et al., 2016). Moreover, it is important to consider aspects other than speed regulation and limb coordination in locomotion, knowing that its successful execution also entails postural adjustments and suppression of other motor programs.
One still poorly explored dimension of the MLR is the diversity of output structures targeted by glutamatergic neurons in the PPN area. In addition to the descending projections to the medulla (Caggiano et al., 2018; Capelli et al., 2017; Noga et al., 1988), minor projections to the spinal cord (SC) also exist (Liang et al., 2012), which have not been studied functionally. PPN-vGlut2 neurons also have multiple ascending targets, including several basal ganglia components, basal forebrain, and thalamus (Martinez-Gonzalez et al., 2011). First reports begin to suggest that MLR-vGlut2 axons influence target structures differentially (Assous et al., 2019; Lee et al., 2014; Yoo et al., 2017). Yet, whether these effects represent differential responses of targets to a signal broadcasted by one population or reflect the action of distinct neuronal subpopulations remains unknown. The latter might explain the different findings in studies carrying out MLR neuron stimulations and DBS in PD patients.
Here, we identify and functionally dissect glutamatergic MLR subpopulations based on the premise that target connectivity might be linked to function. We found that MLR-vGlut2 neurons residing in spatial proximity segregate into separate neuronal populations based on axonal targets, transgenic marker expression, neuronal activity profiles, and roles in behavior. A spinally projecting population (MLR>SC) is distinct from an ascending population targeting basal ganglia output structures specifically marked by the Rbp4Cre transgene (MLR-Rbp4). While MLR>SC neurons are positively modulated during rearing, MLR-Rbp4 neurons are mostly recruited during the forelimb behaviors handling and grooming. Optogenetic perturbation experiments demonstrate a role for MLR>SC neurons in body extension while pointing to a function for MLR-Rbp4 neurons in modulation of various behaviors. We conclude that the proximity of functionally diverse MLR subpopulations likely explains the diverse results on glutamatergic MLR neurons and provides essential information for devising new strategies to ameliorate PD symptoms involving the PPN area.
Results
MLR-vGlut2 neurons divide into separate descending and ascending populations
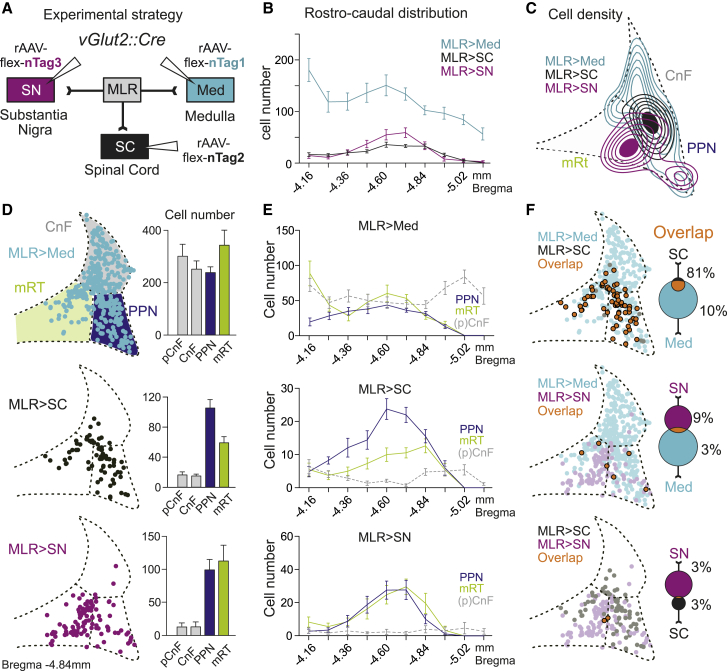
We first determined the precise location of glutamatergic MLR neurons with descending and/or ascending projections. We injected adeno-associated viruses (AAVs) with Cre-dependent conditional expression and retrograde neuronal targeting potential (rAAV) (Tervo et al., 2016) into select descending and ascending MLR projection targets. We delivered rAAVs expressing different marker proteins targeted to nuclei (nTag) to allow for detection of cell body location. We restricted our analysis to glutamatergic MLR neurons by injecting variants of rAAV-flex-nTag into vGlut2Cre mice (Figure 1A). To target MLR-vGlut2 neurons with descending projections, we injected rAAV-flex-nTag1 and -Tag2 into the medullary reticular formation (Med) and SC, respectively (see STAR Methods). As a major ascending target of MLR-vGlut2 neurons, we injected rAAV-flex-nTag3 centered into the substantia nigra (SN) reticulata (SNr), the main basal ganglia output structure in rodents (Oorschot, 1996; Smith et al., 1998) (Figure 1A).
Figure 1.
Glutamatergic MLR neurons segregate by projection target
(A) Strategy for retrograde labeling of glutamatergic MLR neurons from substantia nigra (SN), spinal cord (SC), and medulla (Med).
(B) Average number (±SEM) of labeled cells along the rostro-caudal axis (n = 9).
(C) Cell density from an example animal at bregma −4.84 mm containing PPN, mRT, and CnF subdivisions.
(D) Left: two-dimensional reconstruction of MLR neurons projecting to Med, SC, or SN at bregma −4.84 mm (n = 3). Right: quantification of labeled cell number in MLR subregions for each subpopulation (n = 9). Error bars represent SEM.
(E) Average number (±SEM) of labeled cells for the three retrograde injections in PPN, mRT, and (p)CnF along its rostro-caudal axis (n = 9).
(F) Pairwise comparison of the cellular overlap between MLR subpopulations. Two-dimensional distribution of single or double (orange) labeled cells at bregma −4.84 mm (left) and total percentage of overlapping cells for each subpopulation pair shown in Venn diagrams (right; n = 6 per pair). Percentage of double-labeled neurons (mean ± SEM) were Med+SC/SC, 81.3% ± 2.7%; Med+SC/Med, 10.1% ± 1.1%; Med+SN/SN, 8.8% ± 2.4%; Med+SN/Med, 2.7% ± 0.8%; SN+SC/SN, 2.8% ± 0.9%; SN+SC/SC, 3.1% ± 0.8%.
See also Figure S1.
We assessed the location of MLR>Med, MLR>SC and MLR>SN neurons. To get a measure of distributions for retrogradely labeled MLR-vGlut2 neurons, we reconstructed cell body position based on nTag labeling. We restricted our analysis to the rostro-caudal levels of the PPN, the perimeter of which is defined by the presence of cholinergic neurons (Martinez-Gonzalez et al., 2011; Mena-Segovia and Bolam, 2017), the rostro-caudally contiguous pCnF/CnF and the adjacent mRT (Franklin and Paxinos, 2007). We found that MLR>Med neurons were most numerous and widely distributed (Figures 1B–1E and S1), in agreement with previous work demonstrating that glutamatergic neurons in both PPN and pCnF/CnF subregions project to the caudal medulla (Caggiano et al., 2018; Capelli et al., 2017). In contrast, we observed a more restricted localization for MLR>SC and MLR>SN neurons (Figures 1B–1E and S1).
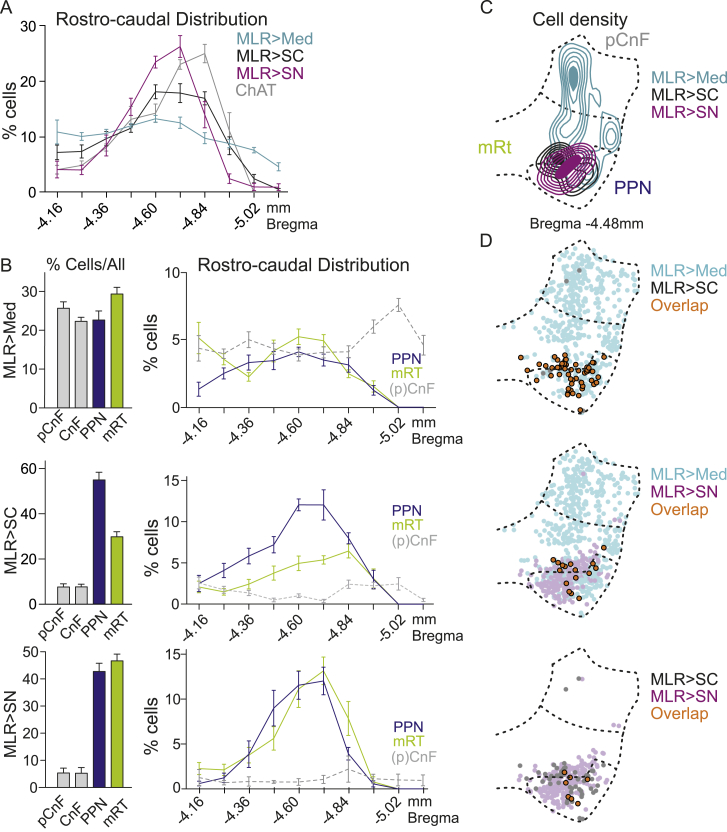
Figure S1.
Distribution of glutamatergic MLR neuron subpopulations, related to Figure 1
(A) Percentage of labeled cells (mean ± SEM) for each of the three MLR subpopulations (n = 9) or ChAT positive cells along the rostro-caudal axis (n = 5).
(B) Left, percentage of labeled cells for the 3 retrogradely labeled populations in each MLR subregion (n = 9; pCnF, CnF, PPN and mRT). Right, mean percentage of labeled cells in each MLR subregion along its rostro-caudal axis (n = 9; mean ± SEM; pCnF and CnF pooled for this analysis).
(C) Cell density from one example animal at Bregma −4.48 mm in a studied MLR section containing PPN, mRT and CnF subdivisions at this level.
(D) Pairwise comparison of the cellular overlap between MLR subpopulations by projection target in two-dimensional distribution of single or double (orange) labeled cells at Bregma −4.48 mm (n = 3).
Analysis of neuronal positioning revealed that MLR>Med neurons are scattered throughout the four MLR subdomains and along the rostro-caudal axis (Figures 1D, 1E, and S1). In contrast, MLR>SC and MLR>SN neurons were preferentially located within the PPN and mRT, with only rare residence in the pCnF/CnF domains (Figures 1D, 1E, and S1). In addition, MLR>SN neurons were more frequently located within the mRT immediately adjacent to PPN particularly in the caudal part, while MLR>SC neurons exhibited a slightly laterally shifted and more PPN-biased residence (Figures 1C, 1D, and S1). In summary, glutamatergic neurons within the PPN and immediately adjacent mRT subregion projecting to the three studied target sites Med, SC and SN are located in close spatial proximity and intermingled. The remainder of the MLR subregions, including the pCnF/CnF and large parts of the mRT, contained mostly neurons projecting to the Med.
Given the close spacing of glutamatergic neurons within the PPN and adjacent mRT with respect to different projection targets, we determined whether single neurons project to multiple targets. We found that most MLR>SC neurons (81.3% ± 2.7%) also project to the Med, indicating that the majority of spinally projecting neurons collateralize to medullary targets (Figures 1F and S1). In contrast, only few MLR-vGlut2 neurons with descending projections to the medulla (2.7% ± 0.8%) or SC (3.1% ± 0.8%) elaborate bifurcating axons projecting to the SN (Figures 1F and S1). We conclude that the assessed descending and ascending MLR-vGlut2 populations are largely separate entities. Due to the proximity of these neurons in and close to the PPN region, possibly divergent functions cannot be assessed purely by location.
Rbp4Cre transgene marks glutamatergic MLR neurons with SN projections
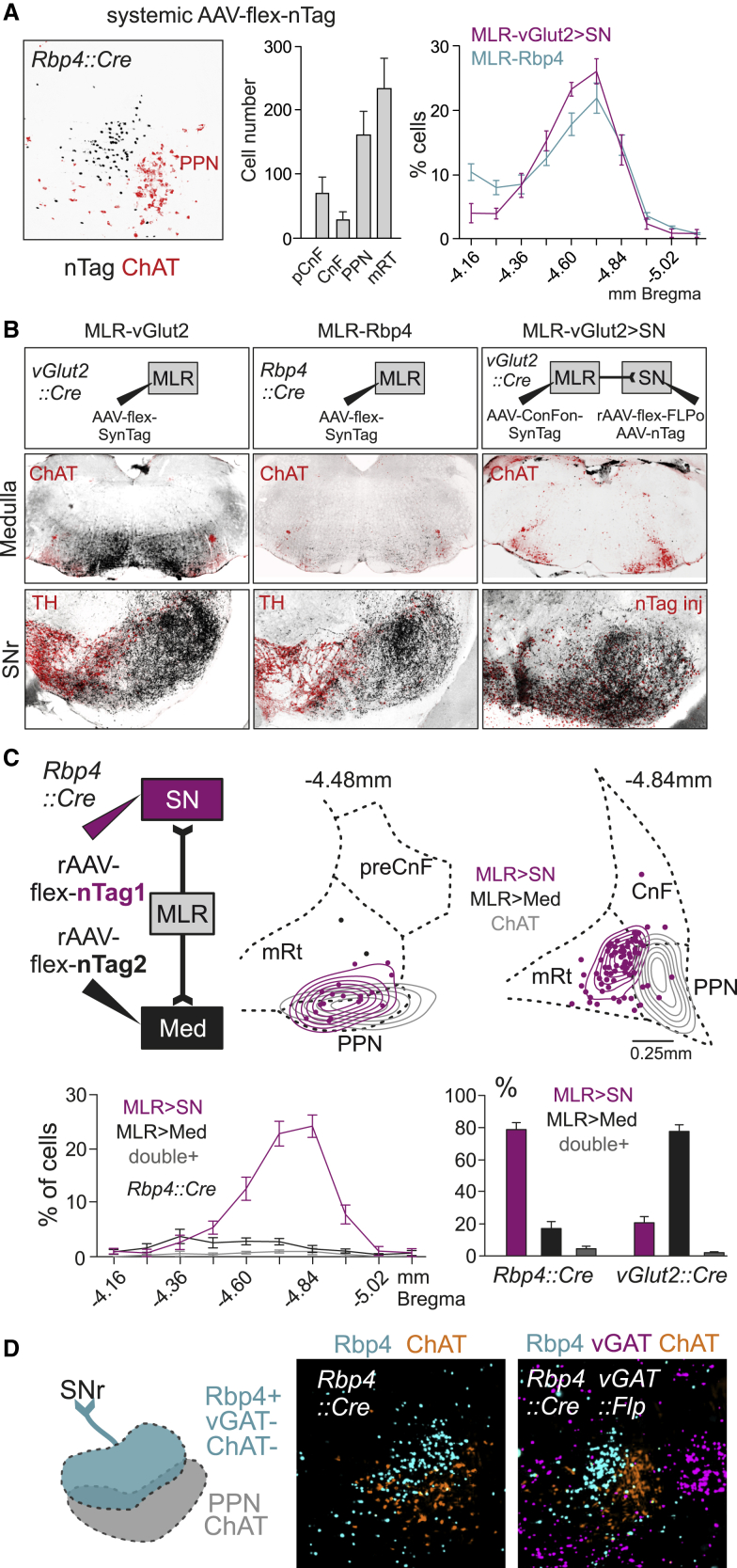
The cluster of glutamatergic SN-projecting neurons immediately adjacent to cholinergic PPN neurons prompted us to determine whether we can find genetic means to access these neurons. We used systemic injections of AAV-PHP.eB-flex-nTag viruses, an AAV variant efficiently transducing the central nervous system (Chan et al., 2017), into existing mouse lines expressing Cre recombinase. We found that the transgenic mouse line Rbp4Cre, widely used to target layer 5 pyramidal tract (PT) neurons in the cerebral cortex (Gerfen et al., 2013), also exhibits selective expression in a cluster of neurons immediately adjacent and partially intermingled with cholinergic PPN neurons (Figure 2A). Quantification of MLR-Rbp4 neurons revealed distribution profiles with more prominent occupancy of mRT and PPN subdivisions than pCnF and CnF, and the rostro-caudal distribution profile aligned with the one for glutamatergic MLR>SN neurons (Figure 2A).
Figure 2.
Rbp4 transgene marks basal ganglia output projecting MLR neurons
(A) Picture showing localization of MLR Rbp4-transgene-positive neurons (black) adjacent to cholinergic PPN neurons (red) (left). Average number of Rbp4-transgene-positive cells in each subregion (n = 5; middle; Error bars represent SEM). Comparison between the rostro-caudal distribution of Rbp4-transgene-positive neurons (n = 5) and glutamatergic neurons projecting to SN (n = 9; right; ± SEM).
(B) Scheme and images showing the distribution of descending (Med) versus ascending (SN) synaptic terminals arising from glutamatergic (vGlut2) (left), Rbp4 transgene positive (middle), or MLR>SN projecting glutamatergic (right) MLR neurons. Med sections are counterstained for ChAT, and SNr sections are counterstained for Tyrosine Hydroxylase (TH; left, middle) or AAV-nTag co-injected for injection specificity (right).
(C) Distribution of Rbp4-transgene-positive MLR neurons revealed by retrograde labeling in Rbp4::Cre mice. (top) Experimental scheme, cell density (lines), and distribution (dots) of MLR>SN and MLR>Med Rbp4 neurons compared to ChAT PPN neurons from one example mouse at bregma −4.48 and −4.84mm. (bottom) Rostro-caudal distribution of Rbp4-transgene-positive MLR subpopulations (n = 5) and percentage of glutamatergic (n = 6) or Rbp4-transgene-positive (n = 5) MLR>SN, MLR>Med, or MLR>SN/Med (double-positive) neurons (Error bars represent SEM).
(D) Neurotransmitter phenotype of MLR-Rbp4 neurons. Left: summary diagram of findings demonstrating that MLR-Rbp4 neurons do not express vesicular GABA transporter (vGAT) or ChAT (n = 3).
See also Figure S2.
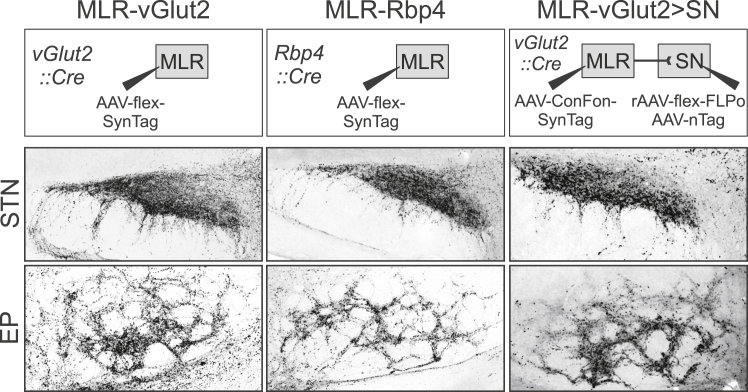
To visualize the synaptic targets of MLR-Rbp4 neurons, we injected a virus expressing a protein tag fused to synaptophysin (AAV2.9-flex-SynTag) into the MLR of Rbp4Cre mice (Figure 2B). We compared the synaptic distribution patterns to straight injections of the same tracer into vGlut2Cre mice and to injections targeting specifically glutamatergic MLR>SN neurons (Figure 2B) (Fenno et al., 2014). AAV2.9-flex-SynTag injections into vGlut2Cre mice revealed dense synaptic terminations in the Med and the SN (Figure 2B), other basal ganglia output structures (Figure S2), as well as thalamus and basal forebrain (data not shown). Much in contrast, the synaptic output of either MLR-Rbp4 or MLR>SN-vGlut2 neurons was strong in the ascending direction to the SN and other basal ganglia structures but minimal in the descending direction, with only sparse synapses in the Med (Figures 2B and S2).
Figure S2.
Ascending connectivity of MLR-vGlut2 neurons, related to Figure 2
Visualization of synaptic terminations in subthalamic nucleus (STN; top row) or entopeduncular nucleus (EP; bottom row) derived from glutamatergic (vGlut2) MLR neurons (left), Rbp4-transgene expressing MLR neurons (middle) or SN-projecting glutamatergic MLR neurons (right).
To quantify these findings at the neuronal level, we injected rAAV-flex-nTags into the SN and Med of Rbp4Cre mice (Figure 2C). We found that injection of rAAV-flex-nTag1 into the SN of Rbp4Cre mice marked a selective MLR neuron cluster, while injections of rAAV-flex-nTag2 into the Med led only to very sparse labeling (Figure 2C). We next analyzed the distribution of all MLR-Rbp4 neurons marked by the dual-injection experiment. We found that the large majority of marked neurons projects to the SN, while Med-projecting neurons were dominant for the analogous experiment carried out in vGlut2Cre mice (Figure 2C). We conclude that the Rbp4Cre transgene is expressed in MLR neurons with ascending projections to the SN and other basal ganglia structures but fails to express in MLR neurons with descending projections to the Med.
Although MLR-Rbp4 neurons occupy the same MLR subregion as MLR>SN glutamatergic neurons and have similar synaptic projection patterns, it is still possible that MLR-Rbp4 neurons are inhibitory or cholinergic, since these three neuronal subtypes are intermingled within the PPN (Martinez-Gonzalez et al., 2011; Mena-Segovia and Bolam, 2017; Wang and Morales, 2009). We found negligible overlap between MLR-Rbp4 neurons and the cholinergic marker choline acetyltransferase ChAT (0.36% ± 0.17%; n = 3; Figure 2D). Moreover, additionally using vGATFLP mice as tool to mark inhibitory neurons, we only found small overlap between MLR-vGAT and MLR-Rbp4 neurons (1.9% v 0.72%; n = 3; Figure 2D). By exclusion, the predominant neurotransmitter used by MLR-Rbp4 neurons is most likely glutamate (Figure 2D). Together, our findings reveal the existence of two anatomically and genetically separable but intermingled glutamatergic PPN/mRT populations.
Differential action tuning of glutamatergic MLR subpopulations
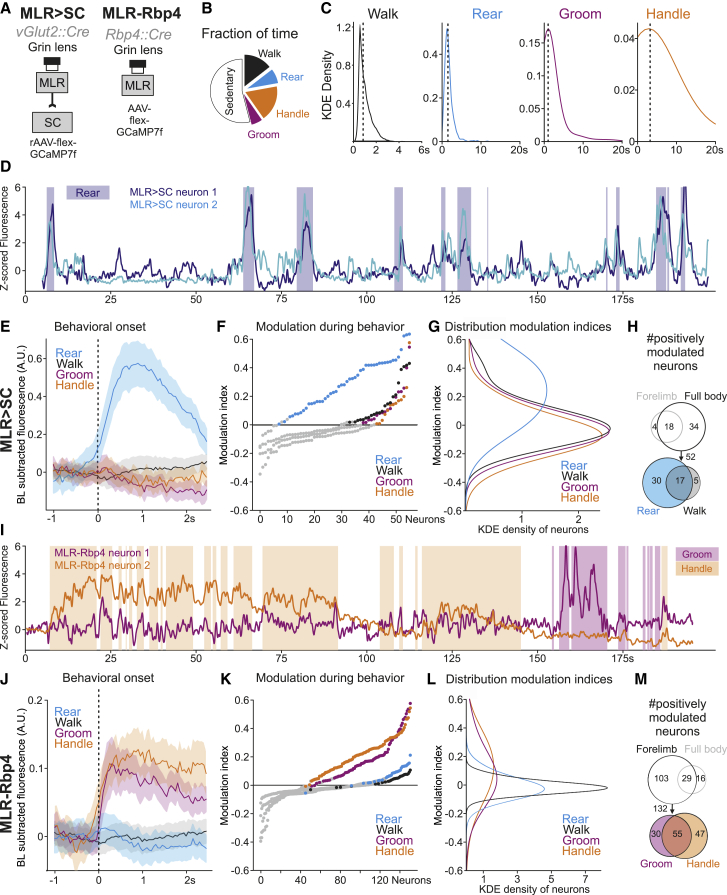
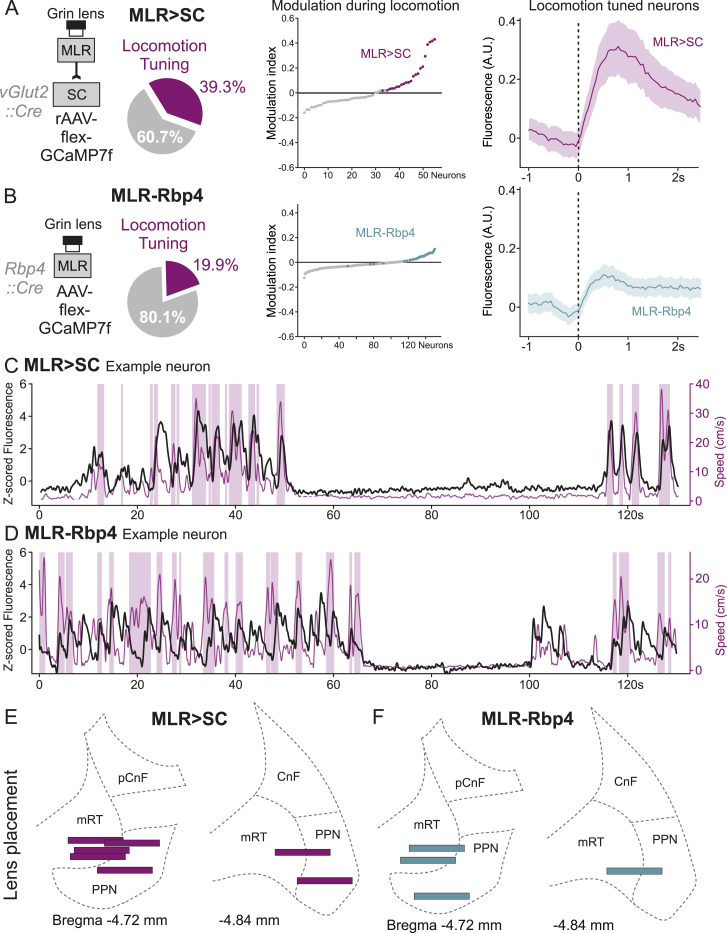
The possibility to target descending and ascending excitatory MLR neuronal populations allowed us to next determine recruitment profiles during self-motivated behavior in the open field by monitoring their activity through a gradient index (GRIN) lens coupled to a miniaturized fluorescent microscope (Figure 3A). Since previous work demonstrated that the activity of a fraction of MLR-vGlut2 neurons tracks locomotor state (Caggiano et al., 2018; Carvalho et al., 2020; Roseberry et al., 2016), we first assessed whether we also detect neurons preferentially active during locomotor bouts. To determine neuronal activity changes during behavior, we computed the mean fluorescence during the studied behavior and subtracted the mean fluorescence during frames for which no behavior was detected, resulting in a modulation index assigned to every neuron for a behavior compared to still episodes. We found that only a fraction of neurons is positively modulated during locomotion, a property more prominently associated with MLR>SC than MLR-Rbp4 neurons (MLR>SC: 39.3%; MLR-Rbp4: 19.9%; Figures S3A–S3E).
Figure 3.
Differential recruitment of MLR subpopulations during behavior
(A) Approach for in vivo calcium imaging of MLR>SC and MLR-Rbp4 neurons.
(B) Fraction of time mice spend sedentary, walking, rearing, handling or grooming in the open field (n = 11).
(C) Distribution for lengths of locomotor, rearing, grooming, handling episodes observed in open field (kernel density estimate [KDE] density; dashed lines: median of distribution).
(D) Z-scored fluorescence with overlaid rearing episodes from two rearing-tuned example neurons.
(E) Baseline (BL) subtracted average fluorescence (±SEM) of all MLR>SC neurons aligned to behavioral onset (56 neurons, n = 7 mice).
(F) Graphs depicting mean evoked fluorescence during rearing, walking, grooming, and handling of all MLR>SC neurons in rising order (colored dots, positively modulated neurons; gray dots, all others).
(G) KDE density of neurons from graphs shown in (F).
(H) Number of positively modulated MLR>SC neurons across behaviors (Venn diagrams). Comparison between forelimb and full-body behaviors (top) and between the full-body behaviors rear and walk (bottom).
(I) Z-scored fluorescence with overlaid grooming and handling episodes from two MLR-Rbp4 example neurons.
(J) Baseline (BL) subtracted average fluorescence (±SEM) of all MLR-Rbp4 neurons aligned to behavioral onset (152 neurons, n = 4 mice).
(K) Graphs depicting mean evoked fluorescence during rearing, walking, grooming, and handling of MLR-Rbp4 neurons in rising order (colored dots, positively modulated neurons; gray dots, all others).
(L) KDE density of neurons from the graphs shown in (K).
(M) Number of positively modulated MLR-Rbp4 neurons across different behaviors (Venn diagrams). Comparison between forelimb and full-body behaviors (top) and the forelimb behaviors groom and handle (bottom).
See also Figures S3 and S4.
Figure S3.
Subpopulation of MLR neurons recruited during locomotion, related to Figure 3
(A) Left: Experimental approach used for calcium imaging of spinally-projecting glutamatergic MLR neurons and fraction of MLR > SC neurons positively modulated by locomotion (39.3%; 56 neurons, n = 7 mice). Middle: Graphs depicting modulation indices during open field locomotion of MLR > SC neurons in rising order (neurons positively modulated by locomotion in magenta dots; all other neurons shown as gray dots). Right: Baseline subtracted average fluorescence (± SEM) of locomotion-tuned MLR > SC neurons, aligned to locomotion onset.
(B) Left: Experimental approach used for calcium imaging of Rbp4 transgene positive MLR neurons and fraction of MLR-Rbp4 neurons positively modulated by locomotion (19.9%; 152 neurons, n = 4 mice). Middle: Graphs depicting modulation indices during open field locomotion of MLR-Rbp4 neurons in rising order (positively modulated neurons are depicted in cyan; all other neurons shown as gray dots). Right: Baseline subtracted average fluorescence (± SEM) of locomotion-tuned MLR-Rbp4 neurons, aligned to locomotion onset.
(C, D) Two representative locomotion-tuned example neurons (C: MLR > SC; D: MLR-Rbp4) from our experimental dataset. Speed traces (magenta), locomotor bout time windows (transparent magenta boxes) and Z-scored fluorescence (black) are depicted. Note low fluorescence for both neurons in non-locomotion time windows in the center.
(E) Anatomical reconstruction of GRIN lens placements for MLR > SC (left) and MLR-Rbp4 (right) experiments shown on corresponding atlas sections (Bregma level indicated).
See also Figure S4.
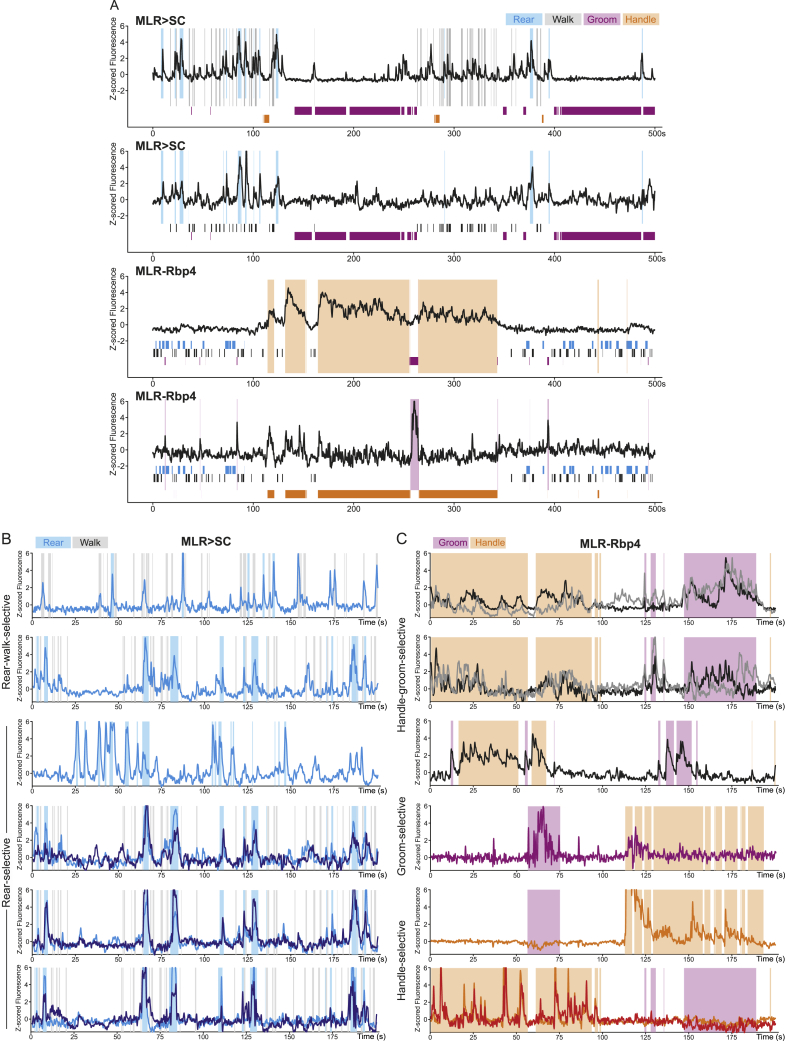
These findings raised the question of whether MLR>SC and MLR-Rbp4 neurons are recruited during other behaviors. Therefore, we tracked the occurrence of the other frequent spontaneous behaviors rearing, grooming, and handling of available food in the open field (Figure 3B). Behavioral episodes were identified using a supervised learning algorithm employing high-speed video and inertial sensor data (Figure 3C). Analyzing the responses of MLR>SC neurons (Figures 3D–3H), we found a prominent population increase in fluorescence associated with the onset of rearing (Figure 3E). In contrast, onsets of locomotion or the forelimb behaviors handling and grooming did not result in increased recruitment of the overall MLR>SC population (Figure 3E). We next determined the modulation indices of individual MLR>SC neurons and their distribution for the four analyzed behaviors. We found most striking recruitment of MLR>SC neurons during rearing, while only few neurons were strongly positively modulated during any of the other three behaviors (Figures 3F and 3G). Thus, also at the single neuron level, positive modulation during rearing was the most prominent effect, while the impact of modulation during other behaviors can be detected in some neurons but is much smaller (Figures 3E–3G). We also investigated the relationship between neuronal activity and rearing episodes by single-neuron and single-trial analysis (Figures 3D, S4A, and S4B). Individual MLR>SC neurons exhibited differential dynamics in timing and magnitude, as can be particularly well discerned in analyzing several MLR>SC neurons imaged in one mouse over the same behavioral time frames (Figures 3D, S4A, and S4B). Together, these findings demonstrate that MLR>SC neurons are preferentially tuned to rearing.
Figure S4.
Representative example neurons for MLR > SC and MLR-Rbp4, related to Figure 3
Representative MLR > SC and MLR-Rbp4 example neurons (A: as indicated; B: MLR > SC; C: MLR-Rbp4) from our experimental dataset with Z-scored fluorescence and behavioral time (transparent boxes overlaid to traces or shown as boxes below traces to indicate identity of behavior carried out; colors indicate different behaviors as depicted). Note that for some examples, two fluorescent traces in different colors are shown for space reasons. Examples illustrate differential dynamics of neurons at the single cell and single trial level.
MLR-Rbp4 neurons exhibited a very distinct recruitment profile from the one observed for MLR>SC neurons (Figures 3I–3M). Population analysis of all MLR-Rbp4 neurons showed strong recruitment at the onset of the forelimb behaviors handling and grooming but no overall recruitment during rearing and locomotion (Figure 3J). These findings were corroborated by analysis of modulation indices for individual neurons, for which we found that many MLR-Rbp4 neurons were strongly modulated during handling and/or grooming, while only few were modulated weakly during rearing and locomotion (Figures 4K and 4L). Determining the number of positively tuned neurons to the four different behaviors also confirmed the strong overrepresentation of tuning toward forelimb behaviors compared to full-body behaviors and revealed abundant overlap between the grooming and handling population (Figure 3M). The temporal structure of the observed neuronal activity patterns was complex, differed between grooming and handling, and did not simply reflect the general behavioral state of the mouse (Figures 3I, S4A, and S4C). While neuronal recruitment clearly occurred during the behavioral time windows, neurons were not constantly active but instead exhibited peaks within the behavioral time window. Furthermore, different neurons in one mouse tracked over the same behavioral time windows exhibited distinct dynamics (Figures 3I and S4C), suggesting the existence of fine neuronal differences with respect to precise behavioral engagement. Together, these findings demonstrate that MLR-Rbp4 neurons exhibit rich temporal coding for different aspects of forelimb movements.
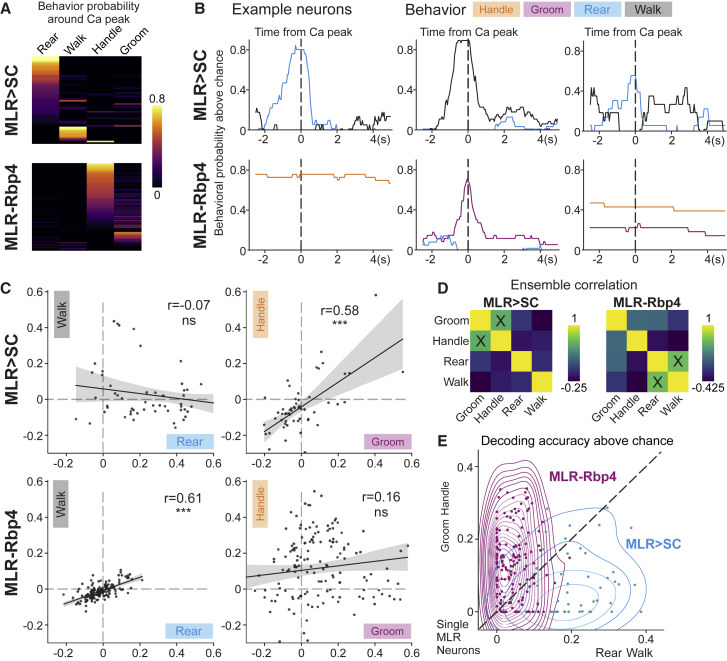
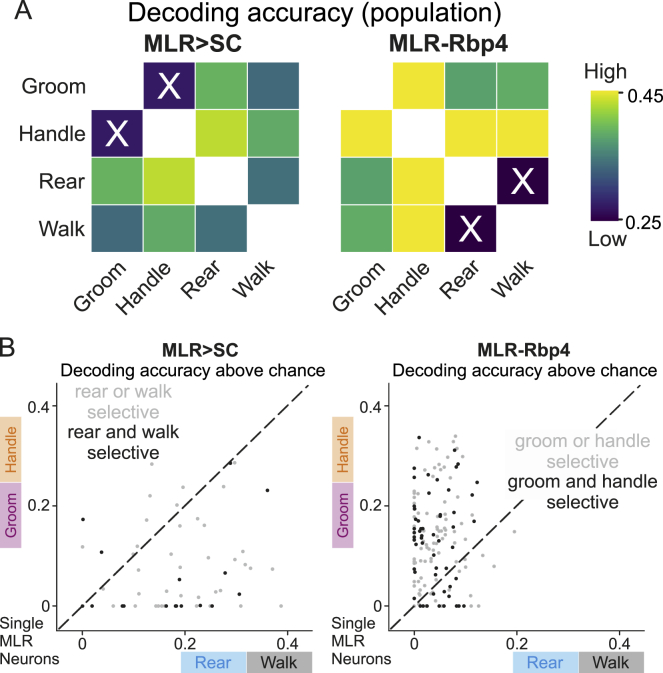
Figure 4.
Differential behavioral decoding from MLR>SC and MLR-Rbp4 neuron activity
(A) Behavioral probability above chance around peak calcium activity of all recorded MLR>SC and MLR-Rbp4 neurons for four behaviors (MLR>SC: 56 neurons from n = 7 mice; MLR-Rbp4: 152 neurons from n = 4 mice).
(B) Behavioral probability above chance of handle, groom, rear, and walk for six example neurons around time from calcium peak (T = 0; top: three MLR>SC neurons; bottom: three MLR-Rbp4 neurons).
(C) Correlation plots for behavioral modulation indices with respect to different behaviors for all studied MLR>SC (top) and MLR-Rbp4 (bottom) neurons and relative Spearman correlation coefficients (r). Shaded region represents 95% confidence intervals. ∗∗∗p ≤ 0.001; ns, not significant.
(D) Spearman correlation of neuronal modulation indices for all neurons for MLR>SC (left) and MLR-Rbp4 (right) neurons.
(E) Decoding accuracy of each neuron above chance for all MLR-Rbp4 and MLR>SC neurons (single neurons: dots, overlaid by KDE density isolines).
See also Figure S5.
Differential decoding of behaviors from MLR neuron populations
To further explore the relationship between neuronal activity profiles and the different behaviors, we took a reverse approach. We extracted the highest peaks of neuronal activity for all analyzed neurons and computed the probability of each behavior occurring at the time of peak (Figure 4A) or in a −2.5s to +5s time window for single neurons (Figure 4B). We found that MLR > SC neurons exhibited the highest probability of being strongly recruited during rearing, followed by walking, but the highest peaks were rarely found during handling and grooming (Figures 4A and 4B). In contrast, the highest activity peaks for MLR-Rbp4 neurons were most prominently associated with handling, followed by grooming, while walking and rearing were only poorly represented (Figures 4A and 4B). Together, these findings support the observation that the majority of MLR>SC neurons are recruited during full-body behaviors, and MLR-Rbp4 neurons exhibit the most striking recruitment during the forelimb behaviors handling and grooming.
To characterize the population-level representation of full-body and forelimb movements in MLR>SC and MLR-Rbp4 neurons, we performed a correlation analysis of modulation indices across the four different behaviors (Figures 4C and 4D). Analysis of MLR>SC neurons revealed no correlation between rearing and locomotion tuning, suggesting dissimilar overall recruitment profiles of MLR>SC ensembles during these two full-body behaviors. In contrast, modulation indices for the few handling- or grooming-tuned MLR>SC neurons were very small and exhibited a strong positive correlation to each other (Figures 4C and 4D). Correlation analysis of MLR-Rbp4 neurons across different behaviors revealed no significant correlation between grooming and handling modulation indices. In contrast, rearing and walking modulation indices were very small but highly correlated (Figures 4C and 4D). Together, these findings suggest that MLR>SC neuron activity may have low decoding accuracy to distinguish handling from grooming, while MLR-Rbp4 neurons may be poor predictors to distinguish rearing from walking. On the other hand, one may expect MLR>SC neurons to be good predictors of rearing and walking, while MLR-Rbp4 neurons may distinctly encode grooming and handling.
To test this hypothesis, we applied a generalized linear model. We used 80% of the recording time to train the models and 20% to test the accuracy of differentiating each pair of behaviors studied based on neuronal recording data (Figures 4E and S5A; with 100-fold cross validation). We found that MLR>SC neurons performed worse at distinguishing between forelimb behaviors compared to all other behavioral pairs, while MLR-Rbp4 neurons were worse at distinguishing the two full-body behaviors compared to all other behavioral pairs (Figures 4E and S5A). This statement was true not only for neurons tuned to a single behavior (i.e., handle or groom; rear or walk) but also for neurons tuned to two behaviors (i.e., handle and groom; rear and walk) (Figure S5B), suggesting that the fine details of behavioral recruitment at the neuronal level determine the neuronal fingerprint allowing decoding of behavior also for populations recruited during multiple behaviors. Together, these findings suggest that within the MLR>SC population, neuronal encoding is sufficiently rich to distinguish the full-body behaviors rearing and walking or distinguish these from forelimb behaviors. In contrast, MLR-Rbp4 neurons provide rich information about the forelimb behaviors of grooming and handling but do not carry information to differentiate the full-body behaviors rearing and walking.
Figure S5.
Differential behavioral decoding from MLR > SC and MLR-Rbp4 neuronal activity, related to Figure 4
(A) Average population decoding accuracy above chance across all mice analyzed for MLR > SC (n = 7) and MLR-Rbp4 (n = 4) populations. Note low decoding ability for MLR-Rbp4 neurons with respect to differentiating rearing from walking, and low decoding accuracy for MLR > SC neurons to distinguish handling from grooming consistent with results for single neurons as well.
(B) Same plot as shown in Figure 4E but differentiating neurons with bi-selective behavioral tuning (black) from neurons with tuning to a single behavior (gray). Note random distribution of the bi-selective neurons in their ability to distinguish distinct behaviors.
MLR > SC neurons regulate body extension
We next studied the role of MLR>SC and MLR-Rbp4 neurons in behavior through complementary loss- and gain-of-function experiments. Considering the observation that both MLR>SC and MLR-Rbp4 populations exhibit sophisticated tuning properties related to multiple behaviors, optogenetic perturbation of each entire population is predicted to test the impact that joint down- or upregulation of neuronal activity exhibits on behavioral output.
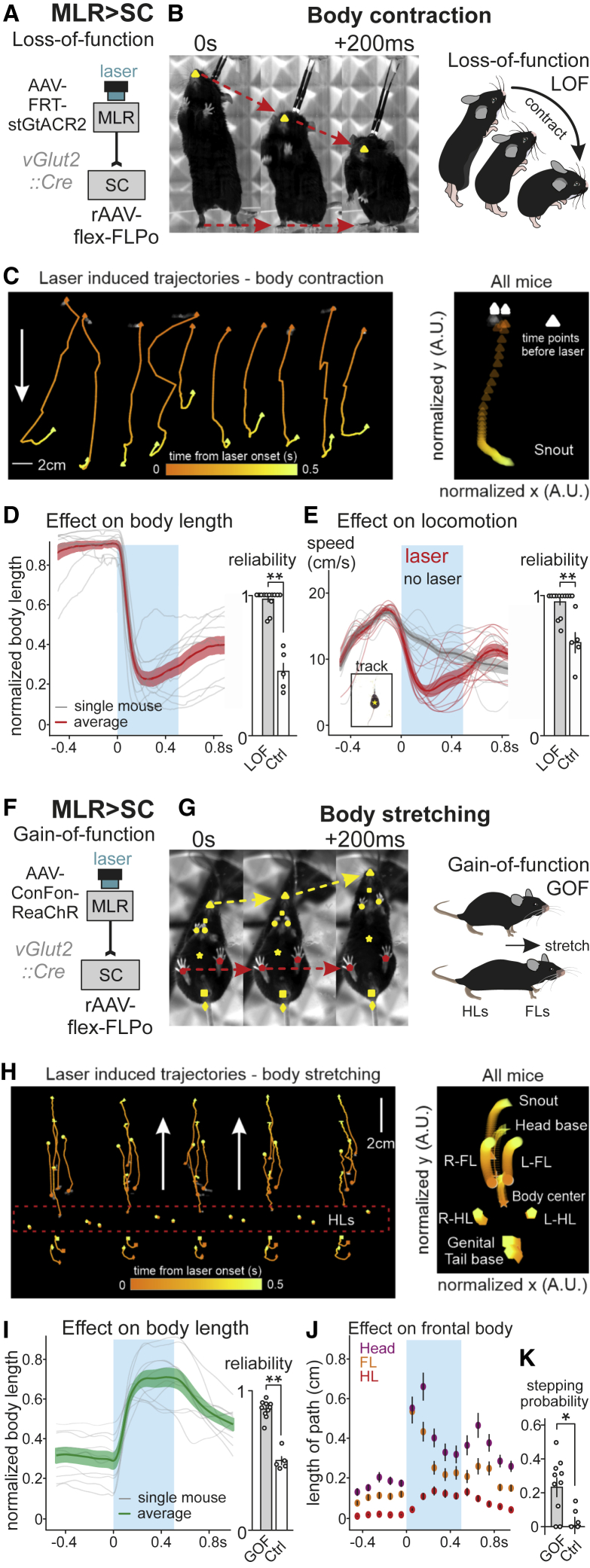
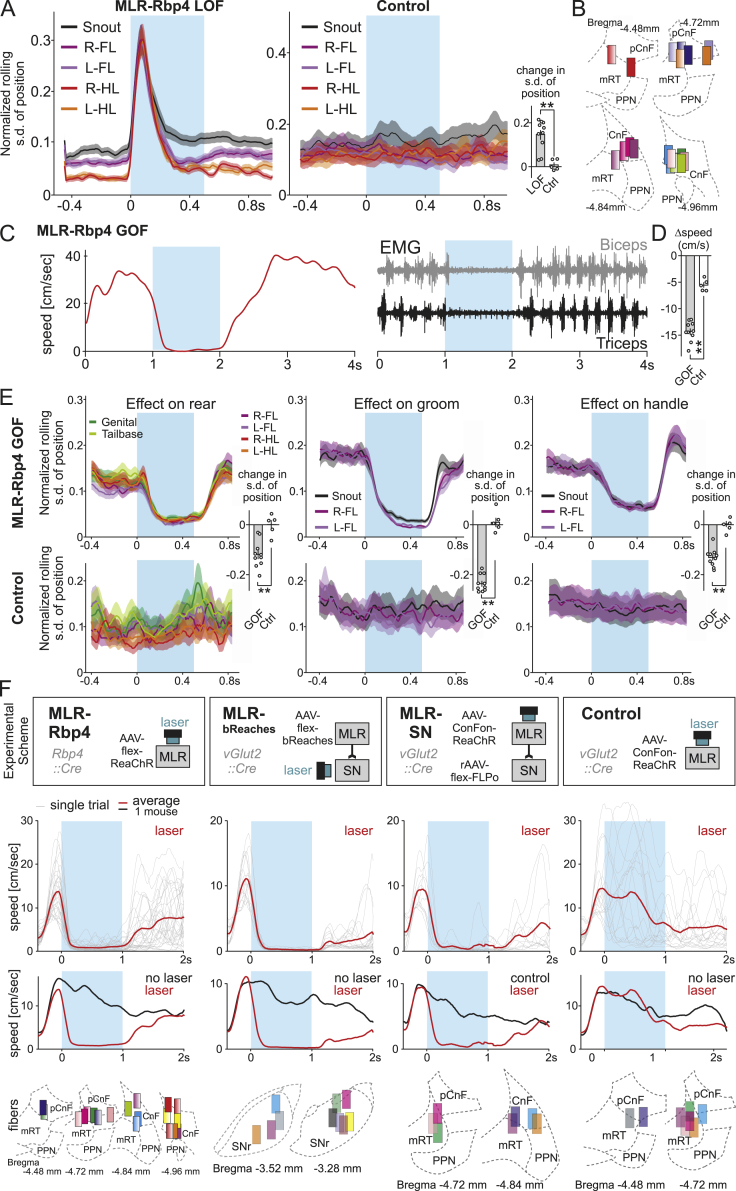
For loss-of-function experiments, we expressed the soma-targeted anion-conducting channelrhodopsin stGtACR2 (Mahn et al., 2018) in glutamatergic MLR>SC neurons (Figures 5A and S6A). Since many MLR>SC neurons exhibit modulation during rearing, we first performed bilateral optogenetic activation of stGtACR2 in MLR>SC neurons during rearing (Figures 5B and S6B). We observed that shortly after onset of optogenetic inhibition, mice terminated rearing, shortening their body to reach a stable position on the ground (Figure 5B; Video S1). We used the markerless pose estimation approach DeepLabCut (Mathis et al., 2018) to track the nose of mice during optogenetic inhibition and found that the termination of rearing was highly reproducible across trials and mice (Figures 5B–5D). Notably, within 200 ms after stimulation onset, the body of the mice shortened considerably, interrupting the ongoing rearing episode, an effect not observed in control mice (Figures 5D and S6B). Stimulation applied during ongoing locomotion resulted in reduced locomotor speed (Figures 5E and S6B; Video S1), which we interpret as interference with locomotion through the induced postural changes.
Figure 5.
MLR neurons with spinal projections regulate body extension
(A) Approach to target the optogenetic inhibitor stGtACR2 into MLR>SC neurons for loss-of-function (LOF) experiments (n = 13).
(B) Left: snapshots from video analysis of rearing mouse just before laser onset (0 s) to +200 ms after laser onset, with one intermediate frame (snout: yellow). Right: Cartoon representation of body contraction effect induced by optogenetic LOF of MLR>SC neurons during rearing.
(C) DeepLabCut tracking of snout position upon optogenetic inhibition of MLR>SC neurons (white points before stimulation; orange to yellow points from 0 to 500 ms of laser stimulation), showing single trials (left) and normalized snout trajectory over all trials and mice (right) (n = 13).
(D) Average (±SEM) of normalized body length (red) of rearing mice and single mouse averages (gray) upon optogenetic inhibition (blue window) of MLR>SC neurons and reliability of laser-induced decrease in average body length (0.5 being chance level) in mice expressing stGtACR2 (LOF, n = 13) or GFP (Ctrl, n = 5) in MLR > SC neurons.
(E) Average (±SEM) of locomotor speed upon closed loop optogenetic inhibition (blue window, red line) of MLR>SC neurons during locomotion and control trials with no laser stimulation (black line) with single-mouse averages of the two conditions and reliability of locomotor speed decrease (n = 13), compared to the reliability of occurrence of the same phenotype in control mice upon light application (n = 5).
(F) Approach used to target the optogenetic activator ReaChR into MLR>SC neurons for gain-of-function (GOF) experiments (n = 10 mice).
(G) Left: snapshots from video analysis of stationary mouse just before laser onset (0 s) to +200 ms after laser onset, with one intermediate frame. Snout, head base, forelimbs, hindlimbs, body center, genital, and tail base are marked through DeepLabCut analysis. Right: cartoon representation of the body extension effect induced by optogenetic GOF of MLR>SC neurons in stationary mice.
(H) DeepLabCut tracking of body part position upon optogenetic activation of MLR>SC neurons through laser application (white points before stimulation; orange to yellow points from 0 to 500 ms of laser stimulation), showing single trials (left) and normalized body part trajectories over all trials and mice (right) (n = 10; compared to n = 5 control mice).
(I) Average (±SEM) of normalized body length (green) of stationary mice and single-mouse averages (gray) upon optogenetic activation (blue window) of MLR>SC neurons and reliability of laser induced increase in body length (n = 10) compared to the probability of observing an increase in body length in control mice upon light application (n = 5).
(J) Binned average path length (±SEM) for head (average of snout and head base), forelimbs (FL; average of left and right forelimb), and hindlimbs (HL; average of left and right hindlimb) for stationary mice upon laser application (blue window) to MLR>SC neurons for optogenetic activation (n = 10).
(K) Graph depicting probability above baseline levels (baseline: application of a 0-mW laser with same closed-loop protocol) to initiate at least one cycle of four limb stepping after body stretching upon optogenetic activation of MLR>SC neurons (n = 10) and light application in control mice (n = 5). Stimulations were performed when mice were sedentary with all four paws on ground in an unrestrained open field environment.
See also Figure S6.
∗p ≤ 0.05 ∗∗p ≤ 0.01.
Figure S6.
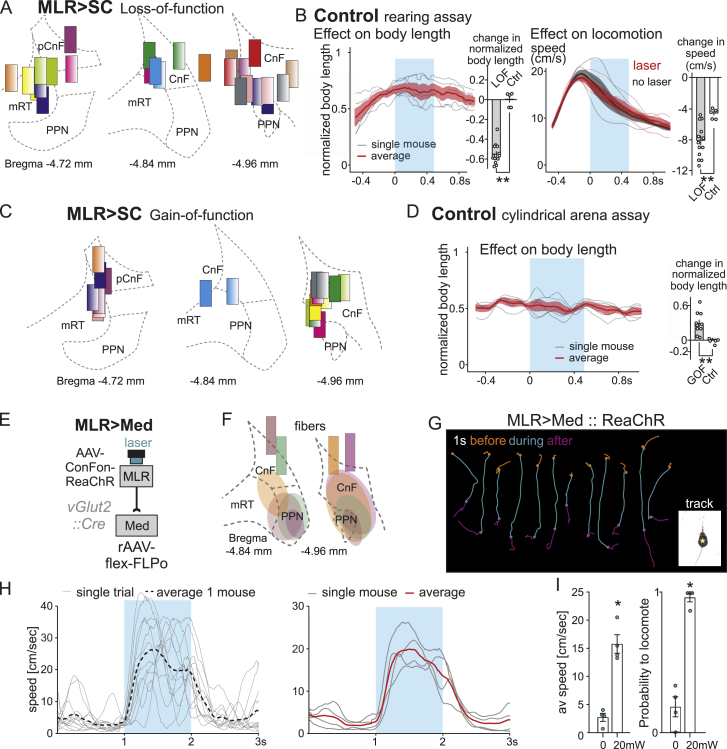
Optogenetic stimulation of medulla-projecting MLR neurons elicits locomotion, related to Figure 5
(A) Anatomical confirmation of fiber placements for MLR > SC loss-of-function (LOF) mice at corresponding rostro-caudal levels (distance from Bregma indicated).
(B) (left) Average (±SEM in red; single mouse averages in gray) of normalized body length upon light application (blue window) in control mice during rearing and bar plot comparing light-evoked change in normalized body length in MLR > SC LOF (n = 13) and control mice (n = 5) (right). Average (±SEM; single mouse averages) of locomotor speed upon light application (red) and in no laser trials (black) in control mice and bar plot comparing light-evoked change in speed in MLR > SC LOF (n = 13) and control mice (n = 5).
(C) Anatomical confirmation of fiber placements for MLR > SC gain-of-function (GOF) mice at corresponding rostro-caudal levels (distance from Bregma indicated).
(D) Average (±SEM in red; single mouse averages in gray) of normalized body length upon light application (blue window) in stationary control mice and bar plot comparing light-evoked change in normalized body length in MLR > SC GOF (n = 10) and control mice (n = 5).
(E) Experimental approach for optogenetic activation of medulla-projecting glutamatergic MLR neurons (MLR > Med).
(F) Anatomical confirmation of fiber placement and expression of optogenetic activator for MLR > Med GOF mice at corresponding rostro-caudal levels (distance from Bregma indicated).
(G) Single trajectories of center of body mass for 1 s before (orange), during (cyan) and after (magenta) laser application, visualizing induction of locomotion by stimulation of MLR > Med neurons (multiple trials from example mouse).
(H) Analysis of locomotor speed based on tracked center of body mass over time for single trials and average thereof (left) as well as single mice (n = 4) and average thereof (right), for optogenetic activation (blue window) of MLR > Med neurons.
(I) Quantification of average speed during the laser ON period and probability to locomote for optogenetic activation of MLR > Med neurons. (n = 4 mice; p < 0.05 calculated with the Mann-Whitney U test).
∗p ≤ 0.05 ∗∗p ≤ 0.01.
Video sequence demonstrating that optogenetic inhibition of MLR > SC neurons during rearing leads to body contraction, and that optogenetic activation of MLR > SC neurons leads to body stretching.
We next performed gain-of-function experiments by targeting the optogenetic activator Red-activatable channelrhodopsin (ReaChR; Lin et al., 2013) to MLR>SC neurons (Figures 5F and S6C). Bilateral stimulation of stationary mice resulted in consistent body stretching shortly after onset of optogenetic stimulation irrespective of body position (Figure 5G; Video S1). Using DeepLabCut (Mathis et al., 2018), we found that both hindlimbs remain essentially stable on the ground throughout the stimulation period, but optogenetically induced body extension had a striking impact on frontal body parts, resulting in a joint forward movement of head and forelimbs (Figures 5H, 5J, and S6D). Quantification of many trials over mice confirmed this finding and demonstrates that optogenetic activation of MLR>SC neurons elicits body stretching (Figures 5I and 5J). In contrast, optogenetic stimulation of vGlut2-expressing MLR neurons with projections to the medulla (MLR>Med neurons) elicited reliable locomotion (Figures S6E–S6I), in agreement with previous work showing that MLR-vGlut2 projections to the caudal medulla convey a descending locomotor signal (Capelli et al., 2017). Interestingly, when conditions favored transition to locomotion, i.e., with the mouse facing away from walls with all feet on the ground, optogenetic activation of MLR>SC neurons induced body stretching transitioning into at least one full stepping cycle in a fraction of trials (Figure 5K), suggesting that body stretching may facilitate the transition to locomotion. Notably, a fraction of rearing-tuned MLR>SC neurons are also recruited during locomotion (Figures 3H, S3A, and S3C). Neither body extension nor stepping could be elicited by light application in the sole presence of the fluorophore (Figures 5I–5K and S6D). Together, our findings support a model in which spinally projecting MLR neurons are required for postural body adjustments needed for full-body exploratory behaviors, while reliable locomotion-promoting effects of excitatory MLR neuron stimulation rely on interaction with caudal medullary circuits.
MLR-Rbp4 neurons modulate behavior through impacting basal ganglia
To determine the role of MLR-Rbp4 neurons in behavior, we performed loss-of-function experiments by expressing stGtACR2 in Rbp4 neurons (Figure 6A). We reasoned that acutely reducing neuronal activity in MLR-Rbp4 neurons might lead to generalized disinhibition of behaviors due to reduced excitatory drive onto behavioral inhibition-promoting basal ganglia output structures (Figures 2 and S2). Bilateral optogenetic inhibition of MLR-Rbp4 neurons indeed led to uncoordinated body movements (Figure 6B; Video S2). Optogenetically induced movements entailed all body parts with highly variable movement sequences across trials (Figures 6B, S7A, and S7B; Video S2), independent of the particular movement a mouse was engaged with at the time of stimulation (data not shown). We found that the aberrant optogenetically induced movements came to a halt while optogenetic stimulation was still ongoing, and no additional excessive movement was observed at or after laser offset (Figures 6C, S7A, and S7B; Video S2). Light application in control mice did not elicit speed changes of tracked body parts (Figure S7A). Together, these findings demonstrate that optogenetic inhibition of MLR-Rbp4 neurons leads to rapid misbalancing of motor output across all body parts, likely due to perturbation of the physiologically fine-tuned signaling between MLR-Rbp4 neurons and basal ganglia output structures.
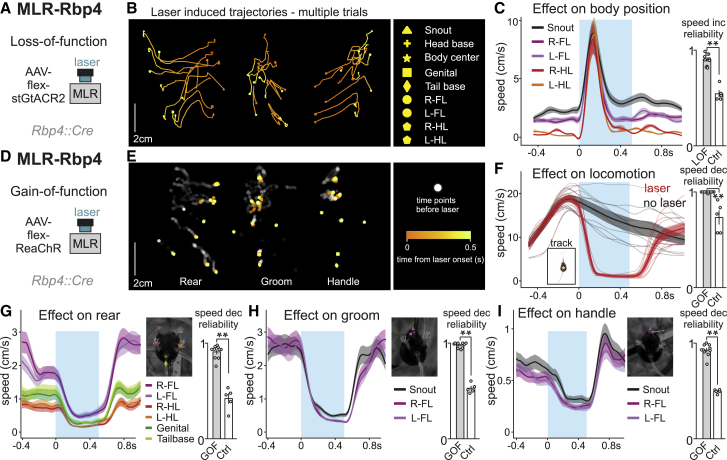
Figure 6.
Opposite perturbation of MLR-Rbp4 neurons elicits pro- and antikinetic modulation of behavior
(A) Approach to target the optogenetic inhibitor stGtACR2 into MLR-Rbp4 neurons for LOF experiments (n = 10).
(B) DeepLabCut tracking of body part position (list shown to the right) upon optogenetic inhibition of MLR-Rbp4 neurons through laser application (white points before stimulation; orange to yellow points from 0 to 500 ms of laser stimulation), showing three single trials from one mouse.
(C) Average speed of body parts (±SEM) upon optogenetic inhibition (blue window) of MLR-Rbp4 neurons and reliability of laser induced speed increase (n = 10) compared to the probability of increase in speed in control mice upon light application (n = 5).
(D) Approach to target the optogenetic activator ReaChR into MLR-Rbp4 neurons for GOF experiments (n = 10).
(E) DeepLabCut tracking of body part position (list shown in B) upon optogenetic activation of MLR-Rbp4 neurons through laser application (white points before stimulation; orange to yellow points from 0 to 500 ms of laser stimulation), showing three single trials during rearing, grooming, or handling.
(F) Average (±SEM) of center-of-body-mass speed upon closed loop optogenetic activation (blue window) of MLR-Rbp4 neurons during locomotion and control no laser trials (black) with single-mouse averages of the two conditions (n = 10). Graph to the right shows reliability in speed decrease upon optogenetic activation of MLR-Rbp4 neurons compared to the probability of observing a locomotor speed decrease in control mice expressing GFP upon light application (n = 5).
(G–I) Average (±SEM) of speed of body parts (as indicated: forelimbs, hindlimbs, genitals, tail base, and snout) for rearing (left), grooming (middle), and handling (right) trials (n = 10). Graphs to the right show reliability in speed decrease upon optogenetic activation of MLR-Rbp4 neurons compared to the probability of observing a decrease in speed in control mice upon light application (n = 5).
See also Figure S7.
∗∗p ≤ 0.01.
Figure S7.
Optogenetic perturbation of SN-projecting MLR neurons, related to Figure 6
(A) Effect of laser stimulation on normalized rolling s.d. of position (mean ± SEM) of body parts (forelimbs, hindlimbs, snout) upon laser application in MLR-Rbp4 loss-of-function (LOF) (n = 10; left) and control mice (n = 5, right) and bar plot comparing light-induced change in the normalized s.d. of position (mean ± SEM) in MLR-Rbp4 LOF (n = 10) and control mice (n = 5).
(B) Anatomical confirmation of fiber placements for MLR-Rbp4 LOF mice at corresponding rostro-caudal levels (Bregma indicated).
(C) Electromyography in the forelimb muscles biceps and triceps during ongoing locomotion and upon optogenetic stimulation of MLR-Rbp4 neurons in an example mouse (blue window: laser application; red line: speed trace). Note that behavioral arrest is accompanied by cessation of biceps and triceps alternating muscle contraction.
(D) Comparison of the change in speed (mean ± SEM) between the MLR-Rbp4 gain-of-function (GOF; n = 10) and control mice (n = 5) upon light application.
(E) Effect of light application on normalized rolling s.d. of position (mean ± SEM) for different body parts (as indicated: forelimbs, hindlimbs, genitals, tail base, snout) for rearing (left), grooming (middle) and handling (right) trials upon laser application in MLR-Rbp4 GOF (n = 10) and control mice (n = 5). The bar plots indicate the comparison of the light induced change in the normalized rolling s.d. of position (mean ± SEM) for each behavior between MLR-Rbp4 GOF (n = 10) and control mice (n = 5).
(F) Top: Experimental strategy for optogenetic activation of MLR-Rbp4 neurons (n = 10), axonal terminals of vGlut2-MLR neurons over SN (n = 10), vGlut2-MLR neurons with projections to SN (n = 7), compared to control mice (n = 8) not expressing optogenetic activators. Middle: Graphs from example mice showing speed versus time plots and depict single trials (in gray) and their averages for closed loop laser application (red), no laser application (black), as well as an overlay of these two experimental conditions. Bottom: Anatomical confirmation of fiber placements for analyzed mice at corresponding rostro-caudal levels (Bregma indicated; one fiber of one animal not shown due to unilateral dorsal fiber placement).
∗∗p ≤ 0.01.
Video sequence demonstrating that optogenetic inhibition of MLR-Rbp4 neurons leads to uncoordinated body movements, and that optogenetic activation of MLR-Rbp4 neurons leads to stalling of ongoing behaviors (grooming, handling, rearing, locomotion).
Based on these findings, one may predict that optogenetic activation of MLR-Rbp4 neurons has the opposite effect, i.e., would lead to stalling of body movements. To address this question, we targeted MLR-Rbp4 neurons with the optogenetic activator ReaChR (Figure 6D). We found that bilateral optogenetic activation of MLR-Rbp4 neurons during movement leads to rapid stalling of the body (Video S2). Body parts rapidly stopped moving shortly after laser onset as observed in analysis of single stimulation trials (Figure 6E). We found that stimulation during ongoing locomotion led to reliable stopping of locomotion compared to no laser trials or control mice (Figures 6F and S7D; Video S2). Locomotion stops were accompanied by cessation of regular limb muscle contractions during rhythmic stepping determined by electromyographic recordings (Figure S7C). Also the other three behaviors (rear, groom, and handle) were efficiently halted by application of optogenetic stimulation to MLR-Rbp4 neurons, exhibiting rapid speed decreases for moving body parts, not observed in control mice (Figures 6G–6I and S7E). Lastly, to determine whether the observed behavioral effects are not due to genetic targeting through a transgenic line, we applied optogenetic stimulation on excitatory MLR neurons retrogradely targeted from the SN or stimulated axons from MLR-vGlut2 neurons expressing an optogenetic activator (Rajasethupathy et al., 2015) in the SN during locomotion (Figure S7F). We found that all three approaches induced termination of locomotion during the period of laser application (Figure S7F). These findings demonstrate that loss- and gain-of-function perturbations have opposite impacts, and together suggest that excitatory MLR inputs to basal ganglia structures play a more holistic modulatory role to orchestrate body movements.
Discussion
Locomotion is a universal animal behavior engaging distributed neuronal circuits. Cumulative work on the MLR has elicited discussion with respect to understanding its function in natural locomotor behavior (Caggiano et al., 2018; Ferreira-Pinto et al., 2018; Josset et al., 2018), but it is also unclear with respect to application of DBS to treat therapy-resistant PD symptoms (Garcia-Rill et al., 2019). Here, we show that dedicated neuronal populations are recruited during different forms of body movement, notably not restricted to locomotion. We believe that these findings shed light on both ongoing debates and call for a radically updated view of neuronal function in this midbrain region. We will discuss the implications of our work for motor system function and design of future DBS interventions.
Functional separation of glutamatergic MLR neurons by projection target
Recent studies on the control of locomotion by the midbrain began to functionally dissect the broader MLR area. Studies agree on important roles of specifically glutamatergic MLR neurons in the regulation of locomotion. Most recordings from glutamatergic MLR neurons were focused on correlating their activity with positive locomotion attributes, yet many neurons remain uncharacterized (Caggiano et al., 2018; Carvalho et al., 2020; Roseberry et al., 2016). Our work reveals that anatomically identified subpopulations of excitatory MLR neurons are recruited during different body movements and that locomotion is but one type of movement involving excitatory MLR neurons. We describe two cleanly divided populations of ascending and descending glutamatergic neurons in the PPN region characterized by essentially opposite behavioral recruitment profiles. An ascending population with terminations in the basal ganglia structures SNr, entopeduncular nucleus, and subthalamic nucleus (STN) is positively modulated during the forelimb movements grooming and handling. In contrast, neurons with descending projections to the SC are recruited mostly during rearing. To what extent inhibitory and cholinergic PPN neurons share this organizational principle remains to be determined, but single-cell reconstructions of cholinergic PPN neurons revealed a high degree of collateralization, distinct from non-cholinergic PPN neurons (Mena-Segovia et al., 2008). Nevertheless, cholinergic PPN neurons are organized along the rostro-caudal axis according to preferential projection targets (Mena-Segovia and Bolam, 2017). In lamprey, medullary cholinergic projections were implicated in locomotion control via fast ionotropic mechanisms (Le Ray et al., 2003), and in cat, MLR neurons also recruit spinally projecting monoaminergic neurons to modulate locomotion (Noga et al., 2017).
What are possible functional implications for the regulation of motor behavior that might follow from our observations? MLR-Rbp4 neurons are glutamatergic and project to generally behavior-inhibitory basal ganglia structures, the recruitment of which by excitatory inputs is predicted to increase inhibitory drive to their output structures (Hikosaka, 2007). Through this connectivity loop, joint recruitment of MLR-Rbp4 neurons provides strong inhibition to neurons in the brainstem motor output pathways and motor thalamus. In agreement, optogenetic activation of MLR-Rbp4 neurons stalls all forms of body movements, likely due to the indirect negative impact on overall motor output pathways. In contrast, optogenetic inhibition of MLR-Rbp4 neurons leads to rapid, explosive body movements, which we interpret as a misbalancing of excitatory drive at the level of basal ganglia output structures, thereby leading to disinhibition of many body behaviors.
In light of the up-to-now studied function of MLR in locomotion, our findings on the prominent modulation of MLR-Rbp4 neurons during forelimb movements might be interpreted as keeping command pathways promoting locomotion repressed whenever forelimb movements are executed. An alternative possibility is that the MLR region might also actively contribute to non-locomotor behaviors. In such a model, the activity of MLR-Rbp4 neurons through their indirect action on SNr might help to modulate the choice or dynamics of specific forelimb and orofacial actions. Since different forelimb actions can be decoded from MLR-Rbp4 neurons, their impact on SNr neurons might contribute to the selection of appropriate downstream neurons. Their role might thus not be restricted to limiting recruitment of locomotion-promoting brainstem circuits. Testing these alternative models awaits the identification of entry points to separately target ascending projection neurons active selectively during specific forms or phases of body movement but leaving other subpopulations unaffected by the applied manipulation.
MLR>SC neurons control body extension
We also address the role of spinally projecting MLR neurons, which up to now have been neglected as a minor population with unknown function. Our loss- and gain-of-function experiments support a role for MLR>SC neurons in regulation of body extension. Our work agrees with models in which locomotion-promoting neurons, as opposed to body extension-controlling neurons, within the MLR area act on spinal circuits primarily indirectly through descending projections to intermediary neurons located in the caudal brainstem (Capelli et al., 2017; Garcia-Rill and Skinner, 1987; Shefchyk et al., 1984). We found that medulla-projecting glutamatergic neurons distribute broadly within all MLR subdivisions, whereas spinally projecting counterparts are largely intermingled with cholinergic PPN neurons and reside in the neighboring mRT. In rodents, glutamatergic MLR neurons implicated in the regulation of high-speed locomotion were recently demonstrated to reside within the pCnF/CnF (Caggiano et al., 2018), but these do not project to the SC. Moreover, high-speed locomotion depends on glutamatergic neurons within the medulla subdivision lateral paragiganto-cellular nucleus (LPGi) and ablation of these neurons attenuates the locomotor drive provided by glutamatergic MLR neurons (Capelli et al., 2017). Furthermore, in other species including the lamprey and the cat, normal or MLR-induced locomotion can modulate the activity of medullary reticulospinal neurons in nuanced ways (Brocard and Dubuc, 2003; Deliagina et al., 2000; Matsuyama and Drew, 2000; Perreault et al., 1993). Together, these data lend support to the idea that neurons in the midbrain charged with roles in complex regulatory parameters of limbed locomotion, including instructions on how to move limbs and at what speed, do not communicate with spinal circuits directly but engage at least the medulla as an intermediate processing step. This is conceptually similar to recent findings on neuronal circuits controlling the construction of forelimb movements (Ruder et al., 2021). In this work, neurons with direct spinal projections from the rostral lateral medulla can induce unilateral forelimb reaching movements, while digit involving forelimb movements can only be elicited by stimulation of neurons with targets in the caudal medulla (Ruder et al., 2021). In an analogous model, MLR>SC neurons may carry signals for postural adjustments to aid the body to engage in exploratory activities, including locomotion.
Implications of distinct MLR subpopulations for DBS interventions in PD patients
Our findings on functionally distinct excitatory MLR populations is also of value to the DBS field, where the PPN has been hotly discussed (Garcia-Rill et al., 2019; Nowacki et al., 2019; Tubert et al., 2019). Some PD patients exhibit postural instability and gait impairment that are resistant to dopamine replacement therapy or STN-DBS. PPN-DBS was tried as a possible intervention to ameliorate these resistant symptoms. Yet, results are not convincing, with patients reporting minimal benefits and many side effects. Decades of discussing about the best possible location within the PPN for stimulation did not lead to a solution.
Reviewing clinical and basic literature, it has been proposed that the PPN might be too complex a brain region for reliable direct DBS interventions (Tubert et al., 2019). Our results agree with this assessment. Even if DBS were to target primarily neuronal cell bodies, the intermingling of neurons within the PPN/mRT with distinct functions would make it impossible to reliably target one or the other separately to assess possible outcomes in patients cleanly. Our findings also reconcile the apparently conflicting published results in the field of basic research (Caggiano et al., 2018; Carvalho et al., 2020; Josset et al., 2018), since direct virus injections into the PPN/mRT area, inherently leads to co-infection of functionally mixed neurons. Only disentanglement of glutamatergic PPN/mRT neurons by projection target can reveal the behavioral output that stimulation of these neurons produces.
We conclude that while targeting the adequate neurons within the PPN/mRT region might indeed be beneficial for patients, current technology fails to produce reproducible benefits. Our work suggests that targeting spinally projecting MLR neurons might be beneficial for postural stabilization, whereas promoting limb stepping might need targeting of medulla-projecting populations. In contrast, targeting SN-projecting MLR population is likely going to be of limited value, since it encodes many behavioral parameters and its perturbation likely leads to non-controlled behavioral effects. Interestingly, the stalling of ongoing movements induced by stimulation of SNr-projecting MLR neurons resembles the freezing episodes of PD patients. This observation raises the question of how this population is affected in the parkinsonian state, opening new entry points into PD symptom research. At a clinical level, only once technologies for accessing functionally more uniform neuronal populations become available in humans can we expect to produce the needed better outcomes for PD patients.
Limitations of the study
Individual MLR-Rbp4 neurons change their firing profiles in highly varied and nuanced patterns during natural behavior and are never modulated as an entire population, like we artificially impose during optogenetic experiments. Therefore, the natural impact of MLR-Rbp4 neurons during behavior on SNr and other basal ganglia structures cannot be deduced from whole-population loss- or gain-of-function experiments. It most likely depends on the precise connectivity patterns between different populations of MLR neurons and basal ganglia recipient neurons, as well as their recruitment profiles during natural behavior regulated through their inputs. In line with this idea, different behaviors are encoded by specific neuronal ensembles in the striatum (Klaus et al., 2017), and SNr neurons divide into at least seven populations based on their projection targets (McElvain et al., 2021). Movement therefore entails the orchestrated activation and inhibition of these ensembles in adequate sequence and vigor. It is conceivable that the functionally diverse MLR-Rbp4 ensembles we describe here are part of a broader network, including the basal ganglia, and responsible for the selection of desired and the inhibition of nonselected motor programs. Further tool development for perturbation of neuronal circuits may help in reinforcing the results of our and related studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| chicken anti-GFP | Invitrogen | Cat# A10262; RRID: AB_2534023 |
| chicken anti-Myc | Invitrogen | Cat# A21281; RRID: AB_2535826 |
| chicken anti-TH | Neuromics | Cat# CH23006; RRID: AB_2201403 |
| goat anti-ChAT | Millipore | Cat# AB144P; RRID: AB_2079751 |
| mouse anti-Myc | ATCC | Cat CRL-1729; RRID: CVCL_G671 |
| mouse anti-NeuN | Millipore | Cat# MAB377; RRID: AB_2298772 |
| mouse anti-V5 | Invitrogen | Cat# R960CUS; RRID: AB_2792973 |
| rabbit anti-RFP | Rockland | Cat# 600-401-379; RRID: AB_2209751 |
| Donkey anti-rabbit Cy3 | Jackson ImmunoResearch | Cat#711-165-152; RRID: AB_2307443 |
| Donkey anti-goat Cy5 | Invitrogen | Cat# A-21447; RRID: AB_2535864 |
| Donkey anti-chicken 488 | Jackson ImmunoResearch | Cat#703-545-155; RRID: AB_2340375 |
| Donkey anti-chicken Cy5 | Jackson ImmunoResearch | Cat#703-605-155; RRID: AB_2340379 |
| Donkey anti-goat 488 | Invitrogen | Cat# A-11055; RRID: AB_2534102 |
| Donkey anti-mouse 647 | Invitrogen | Cat# A-31571; RRID: AB_162542 |
| Donkey anti-mouse Cy3 | Invitrogen | Cat# A-31570; RRID: AB_2536180 |
| Donkey anti-mouse DyL405 | Jackson ImmunoResearch | Cat# 715-475-150; RRID: AB_2340839 |
| Donkey anti-goat DyL405 | Jackson ImmunoResearch | Cat# 705-475-147; RRID: AB_2340427 |
| Virus strains | ||
| AAV-flex-SynGFP | Pivetta et al., 2014 | N/A |
| AAV-ConFon-SynGFP | This study | N/A |
| AAV-flex-ReaChR-YFP | Capelli et al., 2017 | N/A |
| AAV-flex-Flp-H2B-V5 | Capelli et al., 2017 | N/A |
| AAV-H2B-10xMyc | Capelli et al., 2017 | N/A |
| AAV-flex-TdTomato | Capelli et al., 2017 | N/A |
| AAV-flex-Flp-H2B-V5 | Ruder et al., 2021 | N/A |
| AAV-flex-H2B-GFP | Ruder et al., 2021 | N/A |
| AAV-flex-H2B-TdTomato | Ruder et al., 2021 | N/A |
| AAV-flex-H2B-V5 | Ruder et al., 2021 | N/A |
| AAV-Con-Fon-ReaChR-Citrine-YFP | Ruder et al., 2021 | N/A |
| AAV-frt-H2B-TdTomato | This study | N/A |
| AAV-flex-GCaMP7f | Dana et al., 2019 | N/A |
| AAV-flex-stGtACR2-FusionRed | Mahn et al., 2018 | N/A |
| AAV-frt-stGtACR2-FusionRed | This study | N/A |
| AAV-flex-bReaChEs | Rajasethupathy et al., 2015 | N/A |
| Deposited data | ||
| CNMF-E | Pnevmatikakis et al., 2016; Zhou et al., 2018 | https://github.com/zhoupc/CNMF_E |
| Experimental models: organisms/strains | ||
| Mouse: vGlut2Cre: Slc17a6tm2(cre)Lowl | Jackson Laboratory | JAX:028863 |
| Mouse: Rbp4Cre: Tg(Rbp4-cre)KL100Gsat/Mmucd | MMRRC | MMRRC_031125-UCD |
| Mouse: vGATFLP: Slc32a1tm1.1(flpo)Hze | Jackson Laboratory | JAX:029591 |
| Software and algorithms | ||
| MATLAB (v2017b) | Mathworks | https://www.mathworks.com/RRID:SCR_001622 |
| GraphPad PRISM (v7.0) | GraphPad PRISM | https://www.graphpad.com:443/RRID:SCR_002798 |
| Python (v3.7) | Python | https://www.python.org/RRID:SCR_008394 |
| Knime (v3.3.1) | Knime | https://www.knime.com/RRID:SCR_006164 |
| CorelDraw (vX6 to X9) | Corel | https://www.coreldraw.com/en/RRID:SCR_014235 |
| Inscopix Data Acquisition Software (v1.2.1 and 1.4.1) | Inscopix | https://www.inscopix.com |
| Bonsai (v2.3) | NeuroGEARS | https://bonsai-rx.org |
| CinePlexStudio (v3.7.1) | Plexon | https://plexon.com |
| DeepLabCut | Mathis Lab (Mathis et al., 2018) | http://mackenziemathislab.org/deeplabcut |
| Other | ||
| 200mm: MFC_200/230-0.48_3.5mm_ZF1.25_FLT Mono Fiberoptic Cannula | Doric | https://neuro.doriclenses.com/ |
| 200mm: MFC_200/230-0.48_6mm_ZF1.25_FLT Mono Fiberoptic Cannula | Doric | https://neuro.doriclenses.com/ |
| ProView Lens Probe 0.6 mm diameter, ~7.3 mm length | Inscopix | https://www.inscopix.com |
| Microendoscope (Inscopix nVista 3.0) | Inscopix | https://www.inscopix.com |
| Wired 9-axis motion sensor | Champalimaud Foundation’s Scientific Hardware Platform | https://www.cf-hw.org |
| Harp WEAR basestation | Champalimaud Foundation’s Scientific Hardware Platform | https://www.cf-hw.org |
| Clock synchronizer | Champalimaud Foundation’s Scientific Hardware Platform | https://www.cf-hw.org |
| PlexBright Optogenetic Stimulation System | Plexon | https://plexon.com |
| OmniPlex Neural Recording Data Acquisition System | Plexon | https://plexon.com |
| FV1000 confocal microscope | Olympus | http://www.olympusconfocal.com/products/fv1000/index.html |
| ZEISS Axio Imager 2 | Zeiss | https://www.zeiss.com/microscopy/int/products/light-microscopes/axio-imager-2-for-biology.html |
| CSU-W1 Confocal Scanner Unit | Yokogawa | https://www.yokogawa.com/solutions/products-platforms/life-science/spinning-disk-confocal/csu-w1-confocal-scanner-unit/ |
| OxyletPro System - Treadmill | Panlab | https://www.harvardapparatus.com/catalog/product/view/id/9001/s/oxyletpro-system-treadmill-with-indirect-calorimetry/category/448/ |
| Ace 2 Area Scan Cameras | Basler AG | a2A1920-160umBAS |
| Pike Cameras | Allied Vision | https://www.alliedvision.com/en/support/technical-documentation/pike-documentation.html |
| Cobolt 06-MLD; 473nm; 100mW | HÜBNER Photonics | https://hubner-photonics.com/products/lasers/diode-lasers/06-01-series/ |
| Model 2650 Micropositioner | Kopf | https://kopfinstruments.com/product/model-2650-micropositioner/ |
| Mouse schemes | Zenodo, scidraw.io |
https://doi.org/10.5281/zenodo.3925993 https://doi.org/10.5281/zenodo.3925913 |
Resource availability
Lead contact
Further information or requests for reagents and resources should be addressed to the lead contact, Silvia Arber (silvia.arber@unibas.ch).
Materials availability
All originally made constructs for AAV production described in this manuscript are available upon request by contacting the lead contact.
Data and code availability
All custom-made scripts and codes for analysis are available upon request by contacting the lead author.
Experimental model and subject details
Animals
We used adult male and female vGlut2Cre (RRID: IMSR_JAX:028863), Rbp4Cre (RRID: MMRRC_031125-UCD) and vGATFLP (RRID: IMSR_JAX:029591) mice, maintained on a mixed genetic background (129/C57B16). Experimental animals were 2- to 4-month-old heterozygous, backcrossed to C57Bl6. They originated from different litters, were randomly allocated to experimental groups and identified by earmarks. All procedures pertaining to housing, surgery, behavioral experiments and euthanasia were performed in compliance with the Swiss Veterinary Law guidelines.
Method details
Virus production, injections, and implantations
Most adeno-associated viruses (AAV) used in this work are based on a backbone derived from Allen Brain (AAV-CAG-flex-tdTomato-WPRE-bGH). Previously described viruses include: AAV-flex-SynGFP (Pivetta et al., 2014), referred to as AAV-flex-SynTag, as well as AAV-flex-ReaChR-eYFP, AAV-flex-Flp-H2B-V5, AAV-H2B-10xMyc, AAV-flex-TdTomato (Capelli et al., 2017), AAV-flex-Flp-H2B-V5, AAV-flex-H2B-GFP, AAV-flex-H2B-TdTomato, AAV-flex-H2B-V5 (last three referred to as AAV-flex-nTagX), AAV-Con-Fon-ReaChR-Citrine-YFP (Ruder et al., 2021). Not previously reported viral constructs were designed in analogy to above constructs: AAV-frt-H2B-TdTomato (referred to as AAV-frt-nTagX), AAV-flex-GCaMP7f (Dana et al., 2019), AAV-flex-stGtACR2-FusionRed and AAV-frt-stGtACR2-FusionRed (Mahn et al., 2018). The AAV-Con-Fon-SynGFP construct was designed following a published strategy (Fenno et al., 2014). The AAV-flex-bReaChEs construct (Rajasethupathy et al., 2015) was created using previously described strategies with an Ef1alpha promoter. To infect neuronal soma, a 2.9 serotype plasmid was used for production as in previous studies (Basaldella et al., 2015; Esposito et al., 2014; Pivetta et al., 2014; Ruder et al., 2021). For retrograde labeling of neurons by means of axonal infection, a rAAV2-retro capsid plasmid (Tervo et al., 2016) was used for coating as described previously (Capelli et al., 2017; Ruder et al., 2021). For systemic labeling of the central nervous system, a PHP.eB serotype was used (Chan et al., 2017) to produce AAV-PHP.eB-flex-nTag and AAV-PHP.eB-frt-nTag viruses. Genomic titers for AAVs used in this study were between 1-5x10e13 and produced following standard protocols. Viruses were delivered to the target brain regions via stereotaxic injection with high precision stereotaxic instruments (Kopf Instruments, Model 1900) under isoflurane anesthesia as previously described (Capelli et al., 2017; Esposito et al., 2014; Ruder et al., 2021). Viral injections in the spinal cord were targeted to the cervical spinal segments C1-C8. Injections in the medulla spanned the rostro-caudal extent of the gigantocellular reticular formation and its subdivisions (Gi, GiA, GiV) and the lateral paragigantocellular nucleus (LPGi), in agreement with anterograde tracing experiments, revealing that the most abundant synaptic output of glutamatergic MLR neurons is directed to this brainstem region (Figure 2B) (Caggiano et al., 2018; Capelli et al., 2017). The stereotaxic coordinates for brain injections are defined as antero-posterior (AP), medio-lateral (ML) and dorso-ventral (DV) (AP; ML; DV) in mm, taking lambda as a reference for the AP and ML axis for MLR and Med injections, while bregma was used as a reference point for the AP and ML axis for SNr; the reference for the DV axis was the dura mater surface at the site of the respective burr hole: MLR (−0.2; −1.19; −3,1); Med (−1.95; −0.7; −5,4); SNr (−3.1; −1.65; −4,6). For synaptic tracing experiments, we injected AAV-flex-SynTag and waited at least two weeks for expression before analysis. Triple or double rAAV injections in vGlut2Cre mice were performed for the combinations of spinal cord, Med and SNr. Before injection, the different rAAVs were diluted with saline solution to obtain the same titer for all viruses. We used the combination of AAV-flex-H2B-GFP, AAV-flex-H2B-TdTomato, and/or AAV-flex-H2B-V5 for most injections, but AAV-flex-TdTomato was used for some spinal cord injections. We added AAV2.9-H2B-10xMyc to the mix to label the injection site. Viruses were allowed to express for at least two weeks before analysis. Double rAAV injections in the Med and SNr of Rbp4Cre mice were performed using an analogous approach. Systemic labeling of the central nervous system was achieved with intravenous delivery of AAV-PHP.eB via retro-orbital injections under anesthesia (Challis et al., 2019), followed by tissue processing at least 4 weeks later. For optogenetic manipulation of projection-specific glutamatergic MLR subpopulations, we used the optogenetic activator ReaChR or inhibitor stGtACR2. ReaChR has been previously demonstrated to activate neurons in the brainstem and elicit behavior (Capelli et al., 2017; Lin et al., 2013; Ruder et al., 2021), and stGtACR2 has been used successfully to silence neurons, including excitatory glutamatergic neurons in subcortical structures (Karigo et al., 2021; Pamukcu et al., 2020). For targeting, rAAV-flex-Flp-H2B-V5 was injected in the cervical spinal cord, Med or SNr of vGlut2Cre mice, with AAV2.9-H2B-10xMyc added to the mix to visualize the injection site. AAV-Con-Fon-ReaChR-Citrine-YFP (for activation) or AAV-frt-stGtACR2-FusionRed (for inhibition) was subsequently injected in the MLR. To target the Rbp4-transgene expressing neurons in the MLR, AAV-flex-ReaChR-Citrine-YFP (for activation) and AAV-flex-stGtACR2-FusionRed (for inhibition) were injected in the MLR of Rbp4Cre mice. This strategy allowed us to restrict the expression of opsins to the identified MLR neuron subpopulations. To control for the effect of light application during optogenetic manipulations, AAV-flex-H2B-GFP was injected in the MLR of vGlut2Cre mice. After injections were completed, optic fibers were implanted bilaterally 200 μm above the injection site in the MLR (diameter: 200 μm: MFC_200/230-0.48_Xmm_ZF1.25_FLT Mono Fiberoptic Cannula; X refers to fiber length according to the stereotaxic coordinates; Doric lenses). For experiments concerning optogenetic stimulation of glutamatergic MLR terminals over SNr, AAV2.9-flex-bReaChEs was injected in the MLR bilaterally and an optic fiber was implanted in the SNr (−3.1; −1.65; −4,6) of vGlut2Cre mice. All optogenetic stimulation experiments were performed > 2 weeks after injection, to allow for adequate viral expression. For microendoscope calcium imaging experiments, the fluorescent calcium sensor flex-GCaMP7f was expressed in MLR neurons either by retrograde infection from the cervical spinal cord in vGlut2Cre mice (MLR > SC) using a rAAV construct, or direct injection of an AAV2.9 construct into the MLR in Rbp4Cre mice (MLR-Rbp4). After injection, a 0.7mm diameter needle was slowly lowered through the burr hole until a depth of 100 μm above the injection site, in order to create a path for the lens. Through this procedure, brain damage was kept minimal due to lateral pushing of tissue rather than removal. After the needle was retracted, a 0.6mm-diameter gradient index (GRIN) lens (ProView Lens Probe 0.6 mm diameter, ∼7.3mm length, Inscopix; small diameter to keep brain damage minimal) was implanted directly above the injection site. A Micropositioner (Kopf) was used to descend into the tissue with the needle first and GRIN lens after, at a speed of 10μm per second to minimize tissue damage. Mice were closely monitored after implantation and throughout the entire experimental period and no obvious behavioral consequences from lens implantation were observed. At least 4 weeks after virus injection and lens implantation, the microendoscope was connected and the field of view was inspected to determine the best focal plane. Subsequently, we mounted the baseplate.
After termination of experiments, injection sites were assessed by using choline acetyltransferase (ChAT) or tyrosine hydroxylase (TH) immunohistochemistry (see immunohistochemistry and microscopy section) to visualize the cholinergic clusters of the PPN, the motor nuclei in the brainstem and the dopaminergic neurons of the substantia nigra compacta (SNc) and VTA. For optogenetic and microendoscope imaging experiments, we also visualized the tip of the artifact left by the implant on the parenchyma to confirm correct placement. We employed a widely used mouse brain atlas as reference to assess the specificity of our injections and implantations (Franklin and Paxinos, 2007). Only mice with confirmed anatomical accuracy were included in the subsequent analysis (number of mice passing anatomical exclusion criteria: anatomical retrograde tracing experiments: n = 9 of 16 vGlut2Cre mice and n = 5 of 7 Rbp4Cre mice; systemic labeling of Rbp4-positive neurons in Rbp4Cre mice: n = 5 of 5; systemic labeling of Rbp4-positive and vGAT-positive neurons in Rbp4CrevGATFLP mice: n = 3 of 3; MLR > SC calcium imaging experiments: n = 7 of 12; Rbp4 calcium imaging experiments: n = 4 of 8; MLR > SC ReaChR experiments n = 10 of 10, MLR > SC stGtACR2 experiments n = 13 of 14, MLR-Rbp4 ReaChR n = 10 of 10, MLR-Rbp4 stGtACR2 n = 10 of 10; MLR > Med ReaChR experiments: n = 4 of 7; MLR > SN ReaChR experiments: n = 7 of 10; stimulation of MLR-vGlut2 axonal terminals over SN: n = 10 of 10).
Immunohistochemistry and microscopy
After termination of all experiments, mice were euthanized and brains and spinal cords were collected for processing, as previously described (Capelli et al., 2017). Briefly, animals were anaesthetized with a ketamine–xylazine solution and transcardially perfused with PBS, followed by a solution containing 4% paraformaldehyde (PFA) in PBS. The brain and spinal cord were dissected, post-fixed overnight in 4% PFA, and incubated in 30% sucrose (w/v) in PBS for at least two days before cryopreservation. Brain and spinal cord tissue were cut on a cryostat at 80 μm thickness (coronal sections for brain tissue and transverse sections for spinal cord tissue), with the exception of all the MLR spanning sections from triple or double rAAV injection experiments, which were cut at 40 μm thickness. Floating sections were collected in sequential order into individual wells and incubated for 1 hour in blocking solution (1% BSA, 0.2% Triton X-100, PBS). Primary antibodies were then applied in blocking solution and incubated for 1–3 days at 4°C. Fluorophore-coupled secondary antibodies (Jackson or Invitrogen) were applied to floating sections after extensive washing and incubated for 1 day at 4°C. Sections were then washed and mounted with anti-bleach preservative medium on slides in sequential rostro-caudal order. Primary antibodies used in this study were: chicken anti-GFP (Invitrogen), chicken anti-Myc (Invitrogen), chicken anti-TH (Neuromics), goat anti-ChAT (Millipore), mouse anti-Myc (ATCC), mouse anti-NeuN (Millipore), mouse anti-V5 (Invitrogen), rabbit anti-RFP (Rockland). For low-resolution overview imaging, slides were scanned with an Axioscan light microscope (Zeiss). For higher resolution imaging, we used a FV1000 confocal microscope (Olympus) or an Axio Imager M2 microscope (Zeiss) with a Yokogawa CSU W1 Dual camera T2 spinning disk confocal scanning unit.
Microendoscopic calcium imaging of MLR subpopulations
One-photon calcium imaging of MLR-Rbp4 and glutamatergic MLR > SC neurons was recorded in freely moving animals in a 35x35cm open field arena, using a microendoscope (Inscopix nVista 3.0) controlled with the Inscopix Data Acquisition Software (versions 1.2.1 and 1.4.1). The recording sessions started at least one week after baseplate implantation (see behavioral experiments). In the first imaging session, the focal setting was adjusted to select the focal distance that allowed for optimal visualization of the field of view. Data was acquired continuously at 20 Hz. Animal behavior during microendoscopy experiments was monitored with 2 video cameras (Pike, Allied Vision Inc.) acquiring at 100fps, controlled by the software Bonsai version 2.3 (NeuroGEARS Ltd.). Additionally, an Inertial Measurement Unit (IMU) (Wired 9-axis motion sensor, Champalimaud Foundation’s Scientific Hardware Platform) mounted on the microendoscope was used to measure the accelerometer, gyroscope and magnetometer data of the animal, sampled at 100Hz. All recording equipment was connected to a harp wear device and clock synchronizer (Champalimaud Foundation’s Scientific Hardware Platform) to synchronize the video, IMU measurements and calcium imaging time stamps. All behavioral movies and data synchronization information were saved with Bonsai.
Optogenetic perturbation experiments
Optogenetic stimulation was performed using a PlexBright Optogenetic Stimulation System (Plexon Inc.) in combination with a laser (Cobolt 06-MLD; 473nm; 100mW). Light was delivered via a patch cord (Doric Lenses), connected to the animal’s implant. Laser intensity was measured at the beginning of every session with an optical power meter (Thorlabs Inc.) on the tip of an optic fiber of the same length as the one implanted to ensure accurate stimulation strengths. Locomotion perturbation experiments were carried out in a 35x35cm open field arena and recorded with one camera (Pike, Allied Vision Inc.) from above at 100 fps. Laser timestamps and camera exposure timestamps were collected using Plexon Inc. software. We tracked center-of-body mass online using the CinePlexStudio tracking function (CinePlexStudio v3.7.1. Plexon Inc.) in order to be able to trigger the laser in a closed loop fashion (Figure 5, 6, and S7). For stimulation during locomotion, the laser was triggered when the speed crossed a threshold value for a given duration of time (either 12.5cm/s for 100ms or 5cm/s for 200ms). For stimulation during rest in the open field arena for the MLR > SC ReaChR mice, the laser was triggered when the speed of the animal was below 3cm/s for 500ms. Additionally, for both of the above, a set of control trials were collected with the same thresholds but with the laser power set to 0mW to enable comparison of the perturbation with natural locomotion (no laser controls). All other optogenetic perturbation experiments were carried out in a 20 cm diameter arena and recorded from below and side for pose estimation with Basler cameras (Ace 2 series). A minimum of 10 trials was used for each mouse for each experimental condition. Laser and camera exposure timestamps were collected using the Inscopix DAQ system (software version 1.4.1). Continuous laser stimulation was used for optogenetic experiments at laser powers as described below. For MLR > SC stGtACR2 experiments, we used bilateral stimulation powers of 5mW for 500ms. In the cylindrical arena, we encouraged rearing by attaching spaghetti to the walls of the cylinder, adjusted to the size of the mouse to allow it to rear comfortably and reach the spaghetti. The laser was triggered manually in a randomized fashion during rearing trials. For MLR > SC ReaChR experiments, we used powers of 5-10mW for 500ms, with trials from all powers pooled. Mice were bilaterally stimulated manually when they were stationary in the circular arena or in a closed-loop fashion in the open field arena (speed < 3cm/s for 500ms). For MLR-Rbp4 stGtACR2 experiments, we used powers of 1mW for 500ms. Mice were bilaterally stimulated manually when they were stationary or during spontaneous rearing, grooming or handling of spaghetti. For MLR-Rbp4 ReaChR experiments, we used 0.1mW laser power for 500ms. Mice were bilaterally stimulated manually during spontaneous rearing, grooming or handling of spaghetti. For optogenetic stimulation of MLR > Med neurons, SNr-projecting glutamatergic MLR neurons or glutamatergic MLR terminals over SNr, we used 1 s unilateral stimulation at 20mW laser power. To control for the effect of light, we performed the application of light under identical conditions in the control mice expressing GFP instead of the opsins for each of the above experiments, as described in the ‘Virus production, injections and implantations’ section.
Electromyography
For electromyographic (EMG) recordings during stimulation of MLR-Rbp4 neurons, injection and fiber implantation were conducted as described above (see virus production, injections and implantations). Cable preparation and EMG implantation of the biceps and triceps muscle of the forelimb were conducted as previously reported (Miri et al., 2017). Acquisition was carried out together with optogenetic stimulation (1 s continuous light, 20mW), during locomotion on a treadmill, set to 10 cm/s to encourage continuous locomotion. The signal was amplified and bandpass filtered (A-M systems 1700, gain 100, bandpass 100-1000 Hz) and acquired using a plexon recording system (Omniplex, Plexon Inc.) at 5000 Hz. Mean subtraction was applied to correct for the DC offset. Movies were recorded from the side with a video camera (Pike, Allied Vision Inc.) acquiring at 100 fps, controlled by the Cineplex Studio software (Plexon Inc.).
Quantification and statistical analysis
All data are presented as mean ± SEM unless otherwise stated, and significance levels are indicated as: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. No statistical methods were used to predetermine sample size. All plots, scripts and analysis were generated or performed in MATLAB v2017b (The Mathworks Inc.), GraphPadPrism7 (GraphPad Inc.), Python 3.7 or KNIME (v3.3.1). Figures were assembled using CorelDraw (versions X6 to X9; Corel Inc). All statistical tests used in this study and exact number of mice used in each experiment are spelled out in the corresponding Figure legends.
Anatomical reconstructions
To map the local distribution and cellular overlap between MLR subpopulations stratified by projections, viral injections and tissue processing were performed as described above. Images from 40 μm thick sections encompassing the full MLR area along its rostro-caudal axis were acquired using a FV1000 confocal microscope (Olympus) with a 20X objective (z-steps of 1 μm) and stitched offline. We used every third MLR image along the rostro-caudal axis for further analysis to best fit the anatomical properties depicted in the atlas (Franklin and Paxinos, 2007). Maximal projection intensity of z stack mosaics was used to detect MLR neurons using a custom-built workflow in KNIME (Dietz and Berthold, 2016). Virally expressed markers were detected automatically using KNIME cell segmentation nodes, or assigned manually in experiments where AAV-flex-TdTomato was used for injection. Cell segmentation parameters were adjusted to fit the results obtained by manual detection of one example image for each set of confocal acquisitions (sigma range used:4-5, threshold range used: 15-25, watershed threshold range used: 56000-65500). Automatic spot detection was visually validated on every section for all experiments. Detection of td-Tomato- and ChAT-expressing neurons was done manually in the same KNIME workflow. MLR subregions were drawn manually following the atlas (Franklin and Paxinos, 2007) using KNIME interactive annotator. The coordinates from the annotated MLR subregions were then used to split detected MLR neurons according to their location. Colocalizing neurons were detected using the feature calculator node from KNIME. We then extracted the number and x-y position of detected overlapping and single-positive neurons in each MLR subregion. Extracted x-y coordinates were used to plot the distribution of labeled neurons using custom-built MATLAB scripts. Density plots were generated using 2d-kernel density estimate, plotting 6 density lines of highest density using the MATLAB function kde2d.
Behavioral classification in the open field arena
Top-down view open field videos to track the locomotor state of the mouse were saved in .avi format and subsequently cropped to regions of interest and split into multiple shorter files using a MATLAB script. The machine learning software Ilastik (version 1.1.5) was used to track the position of mice in the open field. For every acquisition, a training session with refinement via machine learning was used to instruct the software to detect the mouse body and distinguish it from the background. The features used for this purpose were color/intensity (Gaussian smoothing), edge (Gaussian gradient magnitude, difference of Gaussians) and texture (structure tensor eigenvalues, hessian of Gaussian eigenvalues). For each of the features, the probability was calculated using a sigma of 0.3, 1, 3.5 and 10 pixels. With this training, a probability map with the positional information of the mouse for each video was created. From this, we obtained the center of body mass (COBM) of the mouse by extracting the centroid of the filled area corresponding to the animal tracking, with a custom-made MATLAB script. For open field video analysis, we used the x-y position of the COBM in each frame to calculate the instantaneous animal speed as pixel displacement per frame, which was converted into cm/s by incorporating the knowledge of the pixel size in cm and the frame rate (in fps) of each video.
Locomotion bouts were then excluded from the data for behavioral classification purposes. The rest of the video data was split in a training set and tracking set. The training set was manually annotated to identify grooming, handling and rearing during the open field session. Training data was used to train a one-dimensional convolutional neural network implemented in a custom-script in Python 3.7 to recognize behavior from IMU data. The IMU data was median filtered. The accelerometer data from the three axes was high pass filtered with a Butterworth filter (0.5Hz critical frequency). Sensor data was then z-scored. Briefly the network architecture consisted of two 1D convolutional layers (rectifier or “relu” activation) followed by a max pooling layer. This was followed by two more 1D convolutional layers (rectifier or “relu” activation), one global average pooling layer and a dropout layer before the output of the network composed of a dense layer composed of three softmax units (Groom, Handle and Rear). The trained network was used to predict the behavior for the training set and the output prediction was then manually curated to reach perfect annotation of behavioral episodes in the open field arena. To directly relate the behavior to the calcium imaging data, it was down sampled to 20Hz to match the microendoscopic image acquisition frequency. For each behavior, the distribution of the duration of behavioral episodes was studied by computing KDE density through Seaborn, a Python data visualization.
Detection and analysis of locomotor speed during optogenetic perturbation
To study the effects of perturbation on locomotion (Figures 5, 6, S6, and S7), mice were studied in the open field arena as described above. The speed was obtained using the Ilastik tracking. The speed is first median filtered and then a Savitzky–Golay filter was applied. We then temporally aligned the instantaneous speed information with laser onset time stamps, recorded by the Plexon system. The average speed for all trials for each mouse in laser on and control trials was plotted along with the s.e.m. and average across mice in a window of −0.5 s before laser stimulation onset and 1.0 s after stimulation onset. The probability of speed reduction of locomotion for MLR > SC stGtACR2 and MLR-Rbp4 ReaChR experiments was calculated as the proportion of trials in which the average speed in the laser stimulation window was below the average speed in the 500ms preceding the stimulation. For MLR > Med perturbation experiments, the probability to initiate a locomotor bout during the 1 s period of laser stimulation was quantified to compute reliability of the observed stimulation effect. To visualize the trajectory of the animals before, during and after laser-induced locomotion, we displayed the 2D trajectories of the COBM before, during and after the one-second laser-ON period (1 s each time window). For MLR-SC ReaChR experiments in the open field arena, we estimated the probability of the laser stimulation being followed by at least one full four limb stepping cycle in the trials where the animal was on all four limbs and not facing the walls of the arena. The trials which led to at least one complete step cycle in the laser on window were used to estimate the probability. In the same way, the probability was estimated for the control trials where the power was set to 0mW using the same parameters to trigger stimulation. This probability in the control trials was subtracted from the one with the laser on to obtain the control subtracted probability of stepping in Figure 5K.
Pose estimation and analysis for optogenetic perturbation experiments
The machine learning algorithm DeepLabCut (Mathis et al., 2018) was used in combination with high-speed videography (100fps, Basler Ace 2 cameras) to characterize behavioral phenotypes in optogenetic experiments. Video frames were synchronized with the laser pulses using the Inscopix DAQ system. The network for the MLR > SC ReaChR, MLR-Rbp4 stGtACR2 and MLR-Rbp4 ReaChR experiments was trained using at least 300 frames distributed equally over all the videos obtained from recording the mice from below. The training frames were annotated with the following body parts: nose, head base, forepaws, wrists, hands, body center, forelimb hands, hindlimb balls, genitals and tail base. For MLR > SC stGtACR2 experiments, the network was trained on at least 200 frames of videos of the mice recorded from the side distributed equally over all the corresponding videos. Here, the snout was annotated for quantification of the height of the mouse during rearing episodes. To ensure reliable tracking of all body parts, for all videos, trials in which any of the body parts in analysis were not tracked reliably (p < 0.4) for a period of over 200ms were excluded. For the others, in case of p falling below the threshold of 0.4, we linearly interpolated the trajectories. For MLR-Rbp4 stGtACR2 experiments, snout, forelimb hands and hindlimb balls were used for analysis because of their reliable tracking. For MLR-Rbp4 ReaChR experiments during grooming and handling, snout, forelimb hands and hindlimb balls were used for analysis because of their reliable tracking; for tracking during rearing, forelimb hands, hindlimb balls, genitals and tail base were used for analysis because of their reliable tracking.
For body length analysis in MLR > SC ReaChR experiments, the obtained trajectories were centered at the tail base position (onset of laser on time window) and rotated to have the snout vertically align with the origin. Trajectories were normalized between 0 and 1 for averaging across mice in order to account for differences in dimensions between animals. Body length was calculated as the sum of the distances of the nose to head base, head base to body center, body center to genital and genital to tail base. For body length analysis in MLR > SC stGtACR2 experiments, the body length was calculated as the height of the snout from a side camera. The obtained body length was then median filtered and Savitzky-Golay filtered for both MLR > SC ReaChR and MLR > SC stGtACR2 experiments. Each trial was normalized between 0 and 1 to account for different starting positions of the subjects and correct for differences in body size between mice. For path length analysis for MLR > SC ReaChR experiments, the absolute path traveled by each plotted body part was computed in 100ms time bins. For MLR-Rbp4 perturbation experiments, speed of all body parts was calculated from a differential of the coordinates. Rolling standard deviation of two-dimensional position (Figure S7A, E) was computed with a 100ms centered moving window for each coordinate axis and then averaged across them. All standard deviation measurements were then normalized on a trial by trial basis between 0 and 1.
For MLR > SC ReaChR experiments, the reliability of the stimulation inducing the behavioral phenotype was measured as the fraction of trials in which the maximum body length during the laser-on window was higher than the maximum body length during the 500ms window preceding the laser stimulation. For MLR > SC stGtACR2 experiments, the reliability was calculated as the fraction of trials in which the average body length during laser-on window was lower than the average body length during the window of 500ms preceding laser stimulation. For MLR-Rbp4 stGtACR2 experiments, reliability of the stimulation in inducing behavioral phenotypes was measured as the fraction of trials in which the max speed during the laser-on window was higher than the max speed during the 500ms before laser stimulation for all studied body parts. For MLR-Rbp4 ReaChR experiments, reliability was computed as the fraction of trials in which the average speed during laser application was lower than the average speed during the 500ms preceding laser stimulation for all studied body parts. All reliability computations were also done for the relative controls expressing GFP instead of the opsin. Note that for all light controls expressing GFP, the reliability of observing behavioral phenotypes is distributed around 0.5 as is expected by chance, indicating no effect of light application in the absence of optogenetic tools. Reliability of speed decrease is instead higher for locomotor speed even in control mice due to the natural tendency to slow down after a certain speed threshold is crossed (note the overlap between laser and no laser conditions in control mice (Figure S6B, right)). The reliability of the optogenetic experiments was compared to the controls using the Wilcoxon ranked sum test (MLR > SC stGtACR2, body length: p = 0.001; MLR > SC stGtACR2, locomotion: p = 0.003; MLR > SC ReaChR: p = 0.002; MLR-Rbp4 stGtACR2: p = 0.002; MLR-Rbp4 ReaChR, Grooming: p = 0.002; MLR-Rbp4 ReaChR, Handling: p = 0.002; MLR-Rbp4 ReaChR, Rearing: p = 0.002; MLR-Rbp4 ReaChR, Locomotion: p = 0.002).
To compare the effects of optogenetic manipulations and light application in control mice expressing GFP in MLR-vGlut2 neurons, for each variable considered, we calculated the light induced change as the difference between the mean of the variable during light application and the mean of the variable in the preceding 50ms. Specifically, for MLR > SC stGtacr2 experiments, we performed this computation on normalized body length for the rearing assay and locomotor speed for the stimulation during ongoing locomotion (Figure S6B); for MLR > SC ReaChR experiments, we performed this computation on normalized body length (Figure S6D); for MLR-Rbp4 stGtacr2 experiments, on normalized s.d. of pose for all body parts recorded (Figure S7A); for MLR-Rbp4 ReaChR experiments, we performed this computation on normalized s.d. of pose for all body parts recorded in each behavior (Figure S7E) and locomotor speed for the stimulation during ongoing locomotion (Figure S7D). The same was performed on the relative controls to allow for statistical comparison. The Wilcoxon ranked sum test was used to compare the effects of optogenetic perturbation and light application (MLR > SC stGtACR2, body length: p = 0.001; MLR > SC stGtACR2, locomotion speed: p = 0.007; MLR > SC ReaChR body length: p = 0.002; MLR-SC ReaChR no-laser subtracted probability of stepping: p = 0.028; MLR-Rbp4 stGtACR2, p = 0.007; MLR-Rbp4 ReaChR, Grooming: p = 0.002; MLR-Rbp4 ReaChR, Handling: p = 0.003; MLR-Rbp4 ReaChR, Rearing: p = 0.005; MLR-Rbp4 ReaChR, Locomotion speed: p = 0.002).
EMG data plotting
For EMG plotting, biceps and triceps signal was temporally aligned to animal speed of locomotion on the treadmill, tracked as described above (see behavioral tracking). For plotting, we used EMG data processed as described above and as published (Ruder et al., 2021) and the speed trace was smoothened using a moving average window (150ms).
Calcium image processing and analysis
All fluorescence movies were processed using a custom-made script. First, all frames were spatially binned by a factor of 4. To correct the movie for translational movements and rotations, motion correction was performed. Then, neuronal signals were extracted using the ‘constrained non-negative matrix factorization for endoscopic data’ (CNMF-E) framework (Pnevmatikakis et al., 2016; Zhou et al., 2018). ‘C_raw’ obtained from the CNMF-E was used for all further analysis of the fluorescent signal of the neurons. Calcium traces for each neuron were normalized through division by the 99th percentile for all analyses or z-scored for single-neuron examples plotting and peak detection analysis. The average fluorescence for all neurons of the MLR > SC or MLR-Rbp4 subpopulation was calculated centered around the onset for each behavior in a window from −1 s to 2.5 s. From this, we subtracted the average fluorescence during the baseline period defined from −1 s to −0.05 s with respect to behavioral onset, to obtain the baseline subtracted mean fluorescence as shown in Figures 3 and 4. For example neurons obtained from the same mouse, the traces in the same time window of a behavioral session were overlaid. In the case of correlation of neuronal activity with speed, speed obtained from COBM tracking with Ilastik was median filtered and then a Savitzky-Golay filter was applied. To quantitatively assess neuronal tuning to each of the behavioral categories, we computed a modulation index: the activity of a single neuron during each behavior was averaged and subtracted by the average activity of that neuron during frames not detected by any of the four behaviors. To assess the statistical significance of the obtained modulation index, we shuffled each neuronal time series 1000 times and computed the 99.9 percentile (p < 0.001) of the distribution of modulation indices for that neuron. Any value higher than the 99.9 percentile was considered significant. To study the general distribution of modulation indices for different behaviors in MLR neuronal populations, we plotted all neuron modulation indices and sorted them with ascending order for each behavior. We also performed a KDE analysis to display the distribution of modulation indices in each behavioral category using the Python library Seaborn. The baseline subtracted average fluorescence of the MLR > SC and MLR-Rbp4 neurons significantly tuned to locomotion was calculated as described above. Venn diagrams were plotted to compare the number of neurons positively modulated to each behavior. The “full-body” class comprised locomotion and rearing and “forelimb” class comprised grooming and handling. A linear regression analysis was performed between the modulation indices in the behaviors of Rear versus Walk and Groom versus Handle respectively for each of MLR > SC and MLR-Rbp4 (Figures 5 C, D). The modulation indices in these behaviors were plotted along with the linear fit and the 95% confidence interval for that regression indicated by the shaded region along the line. The Spearman’s correlation coefficient and p value were calculated for each of the pairs. For MLR > SC neurons, there was a significant positive correlation between the modulation indices for Groom and Handle with a Spearman’s r = 0.58 and p < 0.001 and a no significant correlation between the modulation indices for Rear versus Walk (r = −0.07, p = 0.606). For MLR-Rbp4 neurons, there was no significant correlation between the modulation indices of Groom versus Handle (r = 0.16, p = 0.053), while there was a significant positive correlation between the modulation indices for Rear versus Walk (r = 0.61, p < 0.001). The results for the Spearman’s correlation coefficient for the modulation indices for all pairs of behaviors are summarized in Figure 4D.
To identify peaks in neuronal calcium activity we used SciPy, a Python library for scientific computing. Specifically, we applied the “find_peaks” function using as parameters a height of 3 and a distance of 100. We computed the probability of each behavior occurring at the time of peak for each neuron. This probability was then corrected to be above chance, accounting for differences in time spent during each behavior during the imaging session (Figure 4A). Precisely, we computed the fraction of frames in each session assigned to each behavioral category and subtracted it from the obtained peak triggered probabilities for each neuron on a single mouse basis. The full dataset for each subpopulation was then plotted as a heatmap and sorted according to Handle, Groom, Rear, Walk for MLR-Rbp4 neurons and to Rear, Walk, Handle, Groom for MLR > SC neurons. For Figure 4B, the probability of behaviors above chance was plotted in time from −2.5 s to +5 s from time of Calcium peak for single representative example neurons.
For decoding analyses, we used scikit-learn, a Python library for statistical learning and glmnet (version 2.2.1), a Python wrapper for the fortran library used in the homonymous R package. Specifically, all decoding analyses were performed through the use of a Regularized Logistic Regression, LogitNet, with an elastic net penalty (alpha = 0.5, 0 for ridge, 1 for lasso) with 100-fold cross-validation and averaging of decoding accuracy. For each model fit, a shuffled version of the time series was used to evaluate chance decoding accuracy (for single neuron decoding, at the single neuron level; for population decoding, at the single mouse level). Final results were obtained by subtracting mean decoding accuracy with chance decoding accuracy and every negative value was set to 0. For population decoding in Figure S5A, the decoding accuracies obtained from each mouse were averaged and plotted. We applied models trained to distinguish between each behavioral pair separately.
Acknowledgments
We are grateful to M. Mielich, C. Pivetta, and K. Fidelin for experimental help; J. Hinz for precious advice and sharing his pipeline for extracting calcium imaging data; N. Whittle and G. Martins for help setting up miniscope calcium imaging experiments; P. Argast and P. Buchmann from the FMI mechanical workshop for building devices for behavioral experiments; R. Thierry for support with scripting for data analysis; K. Yamauchi for help setting up the high-speed video recording system; and P. Caroni for discussions and comments on the manuscript. M.J.F.P. was supported by the Portuguese Foundation for Science and Technology (PhD student fellowship PD/BD/105867/2014); H.K. by a Biozentrum PhD fellowship; and M.S.E. by funding from the Synapsis Foundation, the Brain and Behavior Foundation NARSAD Young Investigator Grant, and HFSP Career Development Award, CONICET, and PICT grant 2018-00607. All authors were supported by funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (Descent, grant agreement 692617), the Swiss National Science Foundation, the Kanton Basel-Stadt, the Novartis Research Foundation, and the Louis Jeantet Prize for Medicine.
Author contributions
All authors were involved in the design of experiments and data analysis. M.J.F.P. and M.S.E. carried out most anatomical characterization of neuronal subpopulations. M.J.F.P., H.K., and A.F. carried out neuronal recording experiments. H.K. and A.F. analyzed neuronal recording experiments. M.J.F.P., H.K., and A.F. performed and analyzed gain-of-function experiments. H.K. and A.F. performed and analyzed loss-of-function experiments. M.S. processed most tissues for post-experimental validation. S.A. initiated the project and wrote the manuscript. All authors discussed the experiments and commented on the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: July 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.07.002.
References
- Assous M., Dautan D., Tepper J.M., Mena-Segovia J. Pedunculopontine glutamatergic neurons provide a novel source of feedforward inhibition in the striatum by selectively targeting interneurons. J. Neurosci. 2019;39:4727–4737. doi: 10.1523/JNEUROSCI.2913-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basaldella E., Takeoka A., Sigrist M., Arber S. Multisensory Signaling Shapes Vestibulo-Motor Circuit Specificity. Cell. 2015;163:301–312. doi: 10.1016/j.cell.2015.09.023. [DOI] [PubMed] [Google Scholar]
- Brocard F., Dubuc R. Differential contribution of reticulospinal cells to the control of locomotion induced by the mesencephalic locomotor region. J. Neurophysiol. 2003;90:1714–1727. doi: 10.1152/jn.00202.2003. [DOI] [PubMed] [Google Scholar]
- Caggiano V., Leiras R., Goñi-Erro H., Masini D., Bellardita C., Bouvier J., Caldeira V., Fisone G., Kiehn O. Midbrain circuits that set locomotor speed and gait selection. Nature. 2018;553:455–460. doi: 10.1038/nature25448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelli P., Pivetta C., Soledad Esposito M., Arber S. Locomotor speed control circuits in the caudal brainstem. Nature. 2017;551:373–377. doi: 10.1038/nature24064. [DOI] [PubMed] [Google Scholar]
- Carvalho M.M., Tanke N., Kropff E., Witter M.P., Moser M.B., Moser E.I. A Brainstem Locomotor Circuit Drives the Activity of Speed Cells in the Medial Entorhinal Cortex. Cell Rep. 2020;32:108123. doi: 10.1016/j.celrep.2020.108123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis R.C., Ravindra Kumar S., Chan K.Y., Challis C., Beadle K., Jang M.J., Kim H.M., Rajendran P.S., Tompkins J.D., Shivkumar K. Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nat. Protoc. 2019;14:379–414. doi: 10.1038/s41596-018-0097-3. [DOI] [PubMed] [Google Scholar]
- Chan K.Y., Jang M.J., Yoo B.B., Greenbaum A., Ravi N., Wu W.L., Sánchez-Guardado L., Lois C., Mazmanian S.K., Deverman B.E., Gradinaru V. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 2017;20:1172–1179. doi: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H., Sun Y., Mohar B., Hulse B.K., Kerlin A.M., Hasseman J.P., Tsegaye G., Tsang A., Wong A., Patel R. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat. Methods. 2019;16:649–657. doi: 10.1038/s41592-019-0435-6. [DOI] [PubMed] [Google Scholar]
- Deliagina T.G., Zelenin P.V., Fagerstedt P., Grillner S., Orlovsky G.N. Activity of reticulospinal neurons during locomotion in the freely behaving lamprey. J. Neurophysiol. 2000;83:853–863. doi: 10.1152/jn.2000.83.2.853. [DOI] [PubMed] [Google Scholar]
- Dietz C., Berthold M.R. KNIME for Open-Source Bioimage Analysis: A Tutorial. Adv. Anat. Embryol. Cell Biol. 2016;219:179–197. doi: 10.1007/978-3-319-28549-8_7. [DOI] [PubMed] [Google Scholar]
- Esposito M.S., Capelli P., Arber S. Brainstem nucleus MdV mediates skilled forelimb motor tasks. Nature. 2014;508:351–356. doi: 10.1038/nature13023. [DOI] [PubMed] [Google Scholar]
- Fenno L.E., Mattis J., Ramakrishnan C., Hyun M., Lee S.Y., He M., Tucciarone J., Selimbeyoglu A., Berndt A., Grosenick L. Targeting cells with single vectors using multiple-feature Boolean logic. Nat. Methods. 2014;11:763–772. doi: 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Pinto M.J., Ruder L., Capelli P., Arber S. Connecting Circuits for Supraspinal Control of Locomotion. Neuron. 2018;100:361–374. doi: 10.1016/j.neuron.2018.09.015. [DOI] [PubMed] [Google Scholar]
- Franklin K.B., Paxinos G. Third Edition. Elsevier; San Diego: 2007. The Mouse Brain in Stereotaxic Coordinates. [Google Scholar]
- Garcia-Rill E., Skinner R.D. The mesencephalic locomotor region. I. Activation of a medullary projection site. Brain Res. 1987;411:1–12. doi: 10.1016/0006-8993(87)90675-5. [DOI] [PubMed] [Google Scholar]
- Garcia-Rill E., Saper C.B., Rye D.B., Kofler M., Nonnekes J., Lozano A., Valls-Solé J., Hallett M. Focus on the pedunculopontine nucleus. Consensus review from the May 2018 brainstem society meeting in Washington, DC, USA. Clin. Neurophysiol. 2019;130:925–940. doi: 10.1016/j.clinph.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen C.R., Paletzki R., Heintz N. GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron. 2013;80:1368–1383. doi: 10.1016/j.neuron.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikosaka O. GABAergic output of the basal ganglia. Prog. Brain Res. 2007;160:209–226. doi: 10.1016/S0079-6123(06)60012-5. [DOI] [PubMed] [Google Scholar]
- Jordan L.M. Initiation of locomotion in mammals. Ann. N Y Acad. Sci. 1998;860:83–93. doi: 10.1111/j.1749-6632.1998.tb09040.x. [DOI] [PubMed] [Google Scholar]
- Josset N., Roussel M., Lemieux M., Lafrance-Zoubga D., Rastqar A., Bretzner F. Distinct Contributions of Mesencephalic Locomotor Region Nuclei to Locomotor Control in the Freely Behaving Mouse. Curr. Biol. 2018;28:884–901.e883. doi: 10.1016/j.cub.2018.02.007. [DOI] [PubMed] [Google Scholar]
- Karigo T., Kennedy A., Yang B., Liu M., Tai D., Wahle I.A., Anderson D.J. Distinct hypothalamic control of same- and opposite-sex mounting behaviour in mice. Nature. 2021;589:258–263. doi: 10.1038/s41586-020-2995-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus A., Martins G.J., Paixao V.B., Zhou P., Paninski L., Costa R.M. The Spatiotemporal Organization of the Striatum Encodes Action Space. Neuron. 2017;96:949. doi: 10.1016/j.neuron.2017.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Ray D., Brocard F., Bourcier-Lucas C., Auclair F., Lafaille P., Dubuc R. Nicotinic activation of reticulospinal cells involved in the control of swimming in lampreys. Eur. J. Neurosci. 2003;17:137–148. doi: 10.1046/j.1460-9568.2003.02417.x. [DOI] [PubMed] [Google Scholar]
- Lee A.M., Hoy J.L., Bonci A., Wilbrecht L., Stryker M.P., Niell C.M. Identification of a brainstem circuit regulating visual cortical state in parallel with locomotion. Neuron. 2014;83:455–466. doi: 10.1016/j.neuron.2014.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Paxinos G., Watson C. Spinal projections from the presumptive midbrain locomotor region in the mouse. Brain Struct. Funct. 2012;217:211–219. doi: 10.1007/s00429-011-0337-6. [DOI] [PubMed] [Google Scholar]
- Lin J.Y., Knutsen P.M., Muller A., Kleinfeld D., Tsien R.Y. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat. Neurosci. 2013;16:1499–1508. doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano A.M., Hutchison W.D., Kalia S.K. What Have We Learned About Movement Disorders from Functional Neurosurgery? Annu. Rev. Neurosci. 2017;40:453–477. doi: 10.1146/annurev-neuro-070815-013906. [DOI] [PubMed] [Google Scholar]
- Mahn M., Gibor L., Patil P., Cohen-Kashi Malina K., Oring S., Printz Y., Levy R., Lampl I., Yizhar O. High-efficiency optogenetic silencing with soma-targeted anion-conducting channelrhodopsins. Nat. Commun. 2018;9:4125. doi: 10.1038/s41467-018-06511-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Gonzalez C., Bolam J.P., Mena-Segovia J. Topographical organization of the pedunculopontine nucleus. Front. Neuroanat. 2011;5:22. doi: 10.3389/fnana.2011.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis A., Mamidanna P., Cury K.M., Abe T., Murthy V.N., Mathis M.W., Bethge M. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 2018;21:1281–1289. doi: 10.1038/s41593-018-0209-y. [DOI] [PubMed] [Google Scholar]
- Matsuyama K., Drew T. Vestibulospinal and reticulospinal neuronal activity during locomotion in the intact cat. I. Walking on a level surface. J. Neurophysiol. 2000;84:2237–2256. doi: 10.1152/jn.2000.84.5.2237. [DOI] [PubMed] [Google Scholar]
- McElvain L.E., Chen Y., Moore J.D., Brigidi G.S., Bloodgood B.L., Lim B.K., Costa R.M., Kleinfeld D. Specific populations of basal ganglia output neurons target distinct brain stem areas while collateralizing throughout the diencephalon. Neuron. 2021;109:1721–1738 e1724. doi: 10.1016/j.neuron.2021.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena-Segovia J., Bolam J.P. Rethinking the Pedunculopontine Nucleus: From Cellular Organization to Function. Neuron. 2017;94:7–18. doi: 10.1016/j.neuron.2017.02.027. [DOI] [PubMed] [Google Scholar]
- Mena-Segovia J., Sims H.M., Magill P.J., Bolam J.P. Cholinergic brainstem neurons modulate cortical gamma activity during slow oscillations. J. Physiol. 2008;586:2947–2960. doi: 10.1113/jphysiol.2008.153874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miri A., Warriner C.L., Seely J.S., Elsayed G.F., Cunningham J.P., Churchland M.M., Jessell T.M. Behaviorally Selective Engagement of Short-Latency Effector Pathways by Motor Cortex. Neuron. 2017;95:683–696.e611. doi: 10.1016/j.neuron.2017.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S., Matsuyama K., Kohyama J., Kobayashi Y., Takakusaki K. Neuronal constituents of postural and locomotor control systems and their interactions in cats. Brain Dev. 1992;14(Suppl):S109–S120. [PubMed] [Google Scholar]
- Noga B.R., Kettler J., Jordan L.M. Locomotion produced in mesencephalic cats by injections of putative transmitter substances and antagonists into the medial reticular formation and the pontomedullary locomotor strip. J. Neurosci. 1988;8:2074–2086. doi: 10.1523/JNEUROSCI.08-06-02074.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noga B.R., Turkson R.P., Xie S., Taberner A., Pinzon A., Hentall I.D. Monoamine Release in the Cat Lumbar Spinal Cord during Fictive Locomotion Evoked by the Mesencephalic Locomotor Region. Front. Neural Circuits. 2017;11:59. doi: 10.3389/fncir.2017.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki A., Galati S., Ai-Schlaeppi J., Bassetti C., Kaelin A., Pollo C. Pedunculopontine nucleus: An integrative view with implications on Deep Brain Stimulation. Neurobiol. Dis. 2019;128:75–85. doi: 10.1016/j.nbd.2018.08.015. [DOI] [PubMed] [Google Scholar]
- Oorschot D.E. Total number of neurons in the neostriatal, pallidal, subthalamic, and substantia nigral nuclei of the rat basal ganglia: a stereological study using the cavalieri and optical disector methods. J. Comp. Neurol. 1996;366:580–599. doi: 10.1002/(SICI)1096-9861(19960318)366:4<580::AID-CNE3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Opris I., Dai X., Johnson D.M.G., Sanchez F.J., Villamil L.M., Xie S., Lee-Hauser C.R., Chang S., Jordan L.M., Noga B.R. Activation of Brainstem Neurons During Mesencephalic Locomotor Region-Evoked Locomotion in the Cat. Front. Syst. Neurosci. 2019;13:69. doi: 10.3389/fnsys.2019.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamukcu A., Cui Q., Xenias H.S., Berceau B.L., Augustine E.C., Fan I., Chalasani S., Hantman A.W., Lerner T.N., Boca S.M., Chan C.S. Parvalbumin+ and Npas1+ Pallidal Neurons Have Distinct Circuit Topology and Function. J. Neurosci. 2020;40:7855–7876. doi: 10.1523/JNEUROSCI.0361-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault M.C., Drew T., Rossignol S. Activity of medullary reticulospinal neurons during fictive locomotion. J. Neurophysiol. 1993;69:2232–2247. doi: 10.1152/jn.1993.69.6.2232. [DOI] [PubMed] [Google Scholar]
- Pivetta C., Esposito M.S., Sigrist M., Arber S. Motor-circuit communication matrix from spinal cord to brainstem neurons revealed by developmental origin. Cell. 2014;156:537–548. doi: 10.1016/j.cell.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Pnevmatikakis E.A., Soudry D., Gao Y., Machado T.A., Merel J., Pfau D., Reardon T., Mu Y., Lacefield C., Yang W. Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron. 2016;89:285–299. doi: 10.1016/j.neuron.2015.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P., Sankaran S., Marshel J.H., Kim C.K., Ferenczi E., Lee S.Y., Berndt A., Ramakrishnan C., Jaffe A., Lo M. Projections from neocortex mediate top-down control of memory retrieval. Nature. 2015;526:653–659. doi: 10.1038/nature15389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseberry T.K., Lee A.M., Lalive A.L., Wilbrecht L., Bonci A., Kreitzer A.C. Cell-Type-Specific Control of Brainstem Locomotor Circuits by Basal Ganglia. Cell. 2016;164:526–537. doi: 10.1016/j.cell.2015.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruder L., Schina R., Kanodia H., Valencia-Garcia S., Pivetta C., Arber S. A functional map for diverse forelimb actions within brainstem circuitry. Nature. 2021;590:445–450. doi: 10.1038/s41586-020-03080-z. [DOI] [PubMed] [Google Scholar]
- Shefchyk S.J., Jell R.M., Jordan L.M. Reversible cooling of the brainstem reveals areas required for mesencephalic locomotor region evoked treadmill locomotion. Exp. Brain Res. 1984;56:257–262. doi: 10.1007/BF00236281. [DOI] [PubMed] [Google Scholar]
- Shik M.L., Severin F.V., Orlovskiĭ G.N. [Control of walking and running by means of electric stimulation of the midbrain] Biofizika. 1966;11:659–666. [PubMed] [Google Scholar]
- Skinner R.D., Garcia-Rill E. The mesencephalic locomotor region (MLR) in the rat. Brain Res. 1984;323:385–389. doi: 10.1016/0006-8993(84)90319-6. [DOI] [PubMed] [Google Scholar]
- Smith Y., Bevan M.D., Shink E., Bolam J.P. Microcircuitry of the direct and indirect pathways of the basal ganglia. Neuroscience. 1998;86:353–387. doi: 10.1016/s0306-4522(98)00004-9. [DOI] [PubMed] [Google Scholar]
- Takakusaki K., Chiba R., Nozu T., Okumura T. Brainstem control of locomotion and muscle tone with special reference to the role of the mesopontine tegmentum and medullary reticulospinal systems. J. Neural Transm. (Vienna) 2016;123:695–729. doi: 10.1007/s00702-015-1475-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tervo D.G., Hwang B.Y., Viswanathan S., Gaj T., Lavzin M., Ritola K.D., Lindo S., Michael S., Kuleshova E., Ojala D. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron. 2016;92:372–382. doi: 10.1016/j.neuron.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevathasan W., Debu B., Aziz T., Bloem B.R., Blahak C., Butson C., Czernecki V., Foltynie T., Fraix V., Grabli D., Movement Disorders Society PPN DBS Working Groupin collaboration with the World Society for Stereotactic and Functional Neurosurgery Pedunculopontine nucleus deep brain stimulation in Parkinson’s disease: A clinical review. Mov. Disord. 2018;33:10–20. doi: 10.1002/mds.27098. [DOI] [PubMed] [Google Scholar]
- Tubert C., Galtieri D., Surmeier D.J. The pedunclopontine nucleus and Parkinson’s disease. Neurobiol. Dis. 2019;128:3–8. doi: 10.1016/j.nbd.2018.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.L., Morales M. Pedunculopontine and laterodorsal tegmental nuclei contain distinct populations of cholinergic, glutamatergic and GABAergic neurons in the rat. Eur. J. Neurosci. 2009;29:340–358. doi: 10.1111/j.1460-9568.2008.06576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J.H., Zell V., Wu J., Punta C., Ramajayam N., Shen X., Faget L., Lilascharoen V., Lim B.K., Hnasko T.S. Activation of Pedunculopontine Glutamate Neurons Is Reinforcing. J. Neurosci. 2017;37:38–46. doi: 10.1523/JNEUROSCI.3082-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Resendez S.L., Rodriguez-Romaguera J., Jimenez J.C., Neufeld S.Q., Giovannucci A., Friedrich J., Pnevmatikakis E.A., Stuber G.D., Hen R. Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. eLife. 2018;7:e28728. doi: 10.7554/eLife.28728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video sequence demonstrating that optogenetic inhibition of MLR > SC neurons during rearing leads to body contraction, and that optogenetic activation of MLR > SC neurons leads to body stretching.
Video sequence demonstrating that optogenetic inhibition of MLR-Rbp4 neurons leads to uncoordinated body movements, and that optogenetic activation of MLR-Rbp4 neurons leads to stalling of ongoing behaviors (grooming, handling, rearing, locomotion).
Data Availability Statement
All custom-made scripts and codes for analysis are available upon request by contacting the lead author.