Figure S1.
Genome-scale CRISPRi fitness profiling in Mtb H37Rv, related to Figures 1 and 2, STAR Methods and Table S1
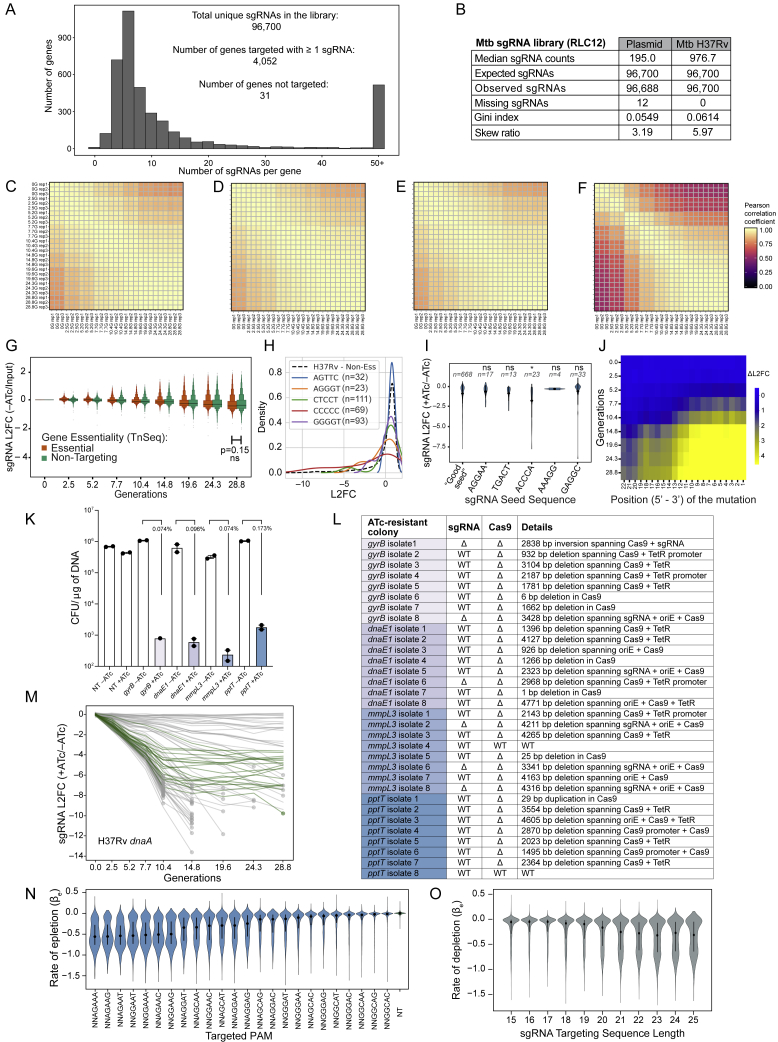
(A) Histogram depicting the number of sgRNAs per gene in the Mtb CRISPRi library (RLC12; Addgene #163954).
(B) Next generation sequencing quality-control metrics for the Mtb CRISPRi library. The “Plasmid” column depicts metrics for the RLC12 plasmid library following cloning and isolation from E. coli. The “H37Rv Mtb” column depicts library metrics following transformation and expansion in Mtb H37Rv. Skew ratio represents the ratio between top and bottom 10% of sgRNA counts.
(C-F) Correlation heatmap of the triplicate screens depicted in Figure 1A. Panel (C) depicts the correlation between non-targeting sgRNAs in the –ATc cultures; panel (D) depicts the correlation between non-targeting sgRNAs in the +ATc cultures; panel (E) depicts the correlation between TnSeq essential gene targeting sgRNAs in the –ATc cultures; panel (F) depicts the correlation between TnSeq essential gene targeting sgRNAs in the +ATc cultures. G, generation.
(G) Boxen plots comparing time-dependent changes in sgRNA L2FC values (mean ± quantiles) comparing –ATc to Input (i.e., generation 0). sgRNAs are grouped according to whether they target genes defined as Essential by TnSeq (n = 63,867 sgRNAs) or Non-Targeting sgRNAs (n = 1,658). ns, not significant.
(H) Density plot to detect potential new “bad-seed” sequences. The plot shows the L2FC (+ATc/–ATc at generation 24.3) of all sgRNAs targeting non-essential genes (dashed line), and sgRNAs targeting non-essential genes that contain the indicated sgRNA seed sequences (defined as the five PAM-proximal nucleotides of the sgRNA targeting sequence) displaying the strongest depletion from the library. See STAR Methods “Estimate of the “bad-seed” effect” for more detail.
(I) Violin plot showing the behavior of sgRNAs containing the strongest “bad-seed” sequences identified for SpydCas9 (Cui et al., 2018). Only sgRNAs targeting a CRISPRi non-essential gene were analyzed. sgRNAs with a PAM-proximal ‘ACCCA’ sequence (n = 24) show some evidence for target-independent depletion (i.e., "bad-seed" behavior). Dot and error bars represent mean and SD. ∗p = 0.021; ns, not significant.
(J) Heatmap showing the behavior of mismatched sgRNAs in the competitive fitness experiment depicted in Figure 1A. ΔL2FC represents the difference in depletion between essential gene-targeting sgRNAs with perfectly matching targeting sequences and the corresponding mismatched sgRNAs. Mismatched sgRNAs contain mismatches between the sgRNA targeting sequence and the gene target at the indicated position (x axis; 22 is the sgRNA nucleotide furthest from the PAM). Mismatched sgRNAs were not designed but were the result of errors during library synthesis or cloning.
(K) Frequency of ATc-resistant colonies that occur after transformation of four unique sgRNAs targeting the essential genes gyrB (ms0005), dnaE1 (ms3178), mmpL3 (ms0250), and pptT (ms2648) in Msmeg. Dots represent transformations performed in biological duplicate; error bars indicate median ± 95% CI. CFU, colony forming unit; NT, non-targeting.
(L) Table summarizing the mutations observed in the CRISPRi plasmid in independent ATc-resistant colonies. All but two isolates show unique deletions, duplications, or an inversion (all generically marked as Δ to indicate lack of CRISPRi functionality) within the sgRNA, Cas9, or both. WT, wild-type; TetR, Tet repressor protein; oriE, E. coli origin of replication.
(M) Line plot showing all sgRNAs targeting dnaA (rv0001) in the Mtb H37Rv CRISPRi fitness experiment. “Flatliner” sgRNAs of presumed CRISPRi-resistant subpopulations are indicated in green. See STAR Methods for details.
(N and O) Distribution of sgRNA depletion slopes (βe) for sgRNAs targeting essential genes (n = 63,867 sgRNAs) stratified by targeted PAM sequence (N) or sgRNA targeting sequence length (O). Black dots and lines show the median and 25%–75% percentiles. Dot and error bars represent mean and SD. NT, non-targeting.