Abstract
The process of multicellular organismal development hinges upon the specificity of developmental programs: for different parts of the organism to form unique features, processes must exist to specify each part. This specificity is thought to be hardwired into gene regulatory networks, which activate cohorts of genes in particular tissues at particular times during development. However, the evolution of gene regulatory networks sometimes occurs by mechanisms that sacrifice specificity. One such mechanism is network co-option, in which existing gene networks are re-deployed in new developmental contexts. While network co-option may offer an efficient mechanism for generating novel phenotypes, losses of tissue specificity at redeployed network genes could restrict the ability of the affected traits to evolve independently. At present, there has not been a detailed discussion regarding how tissue-specificity of network genes might be altered due to gene network co-option at its initiation, as well as how trait independence can be retained or restored after network co-option. A lack of clarity about network co-option makes it more difficult to speculate on the long-term evolutionary implications of this mechanism. In this review, we will discuss the possible initial outcomes of network co-option, outline the mechanisms by which networks may retain or subsequently regain specificity after network co-option, and comment on some of the possible evolutionary consequences of network co-option. We place special emphasis on the need to consider selectively-neutral outcomes of network co-option to improve our understanding of the role of this mechanism in trait evolution.
Keywords: Gene regulatory network, developmental evolution, modularity, pleiotropy
How does one part become different from other parts?
The biology of gene regulatory networks (GRNs) has played a key role in our understanding of how parts are differentiated during development (Davidson 2010). Each gene (or network “node”) within a GRN is deployed through the action of transcription factors which bind specifically to cis-regulatory elements (CREs) to activate its tissue-specific expression (Levine 2010; Farley, Olson, and Levine 2015). Phenotypic changes can often be traced to changes in GRN structure that have tissue-specific effects (Wray 2007; Carroll 2008; Stern and Orgogozo 2008), and thus understanding the mechanisms by which GRNs can be modified gives us insight into evolution.
One mechanism that has emerged as a potential player in the evolution of GRNs is the phenomenon of “gene network co-option” (For definition, see Box 1), particularly in the origins of novel phenotypes (True and Carroll 2002; Olson 2006; Shubin, Tabin, and Carroll 2009; Monteiro 2012; Peter and Davidson 2015). Changes to a single regulator (an “initiating trans change”) in an existing GRN could recruit many terminal effectors in just one or a few steps to produce a novel phenotype, rather than a slow accumulation of the necessary mutations in the CRE of each effector (Box Figure 1).
Box 1.
The term “co-option” has been applied to a range of phenomena. For instance, co-option is sometimes used to describe a case in which a gene network with a known ancestral function may evolve along a certain lineage to be employed instead for a novel derived function (e.g. (Hinman, Nguyen, and Davidson 2007; Suryamohan et al. 2016). Our usage here is somewhat more restricted. For our purposes, “gene network co-option” (or “network co-option”) is a specific way of modifying the developmental “program.” In network co-option, a regulatory factor is deployed in a new location or at a new time during development such that this factor interacts with already existing cis-regulatory elements (CREs) in the next developmental time step. These extant CREs were previously functional in the process of specifying some other trait, i.e. regulated nodes in an existing part (gene regulatory network or “GRN”) of the developmental program. Thus, the activation of these CREs may initiate a second instantiation of some or all of the subsequent time steps of that preexisting program (Box Figure 1). The regulatory machinery that defines the GRN of this other trait is therefore being re-used, recruited, or “co-opted” to a new location or at a novel point in time (True and Carroll 2002; Shubin, Tabin, and Carroll 2009). Our usage therefore defines co-option as a mechanism, not as an outcome per se. This distinction is important, as the deployment of an existing GRN in a novel location could occur by other mechanisms, such as de-novo construction of network connections or some combination of de-novo building and co-option.
“Co-option” of a terminal effector gene via changes to that gene’s locus is not conceptually distinct from what we describe here (e.g. Gompel et al. 2005), but our focus is specifically co-option of multiple interconnected elements in networks simultaneously. Other closely related and interesting phenomena that we do not discuss here are co-option of host gene expression by pathogens (e.g. Saeij et al. 2007; Faust et al. 2017) and the alternate developmental trajectories induced in cancer cells via co-option of extant network architecture (e.g. Shah et al. 2013; Minafra et al. 2014). It would be interesting to connect these areas of research to the concepts discussed here in the future.
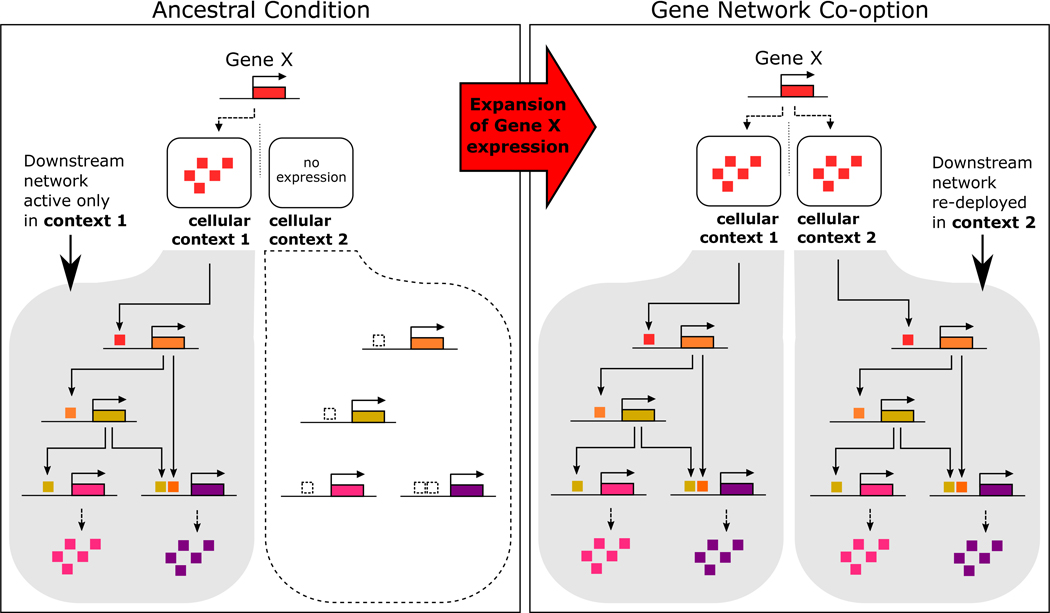
Box Figure 1. Redeployment of a gene regulatory network via gene network co-option.
(Left) The ancestral condition reflects that gene X directly regulates downstream targets only in cellular context 1 and not in context 2 because it is not expressed there. (Right) Co-option occurs when there is an expansion of the expression domain of gene X, such that it is now also expressed in cellular context 2 in the derived condition. The novel expression of gene X in cellular context 2 results in the redeployment of the downstream targets of gene X in cellular context 2, employing its existing cis-regulatory elements (CREs).
While co-option is a mechanism for rapidly establishing a complex network in a tissue, because the CREs of a co-opted GRN have their function expanded, network co-option is predicted to cause an immediate loss of the tissue-specificity for the reused CREs (Duboule and Wilkins 1998; Rebeiz, Patel, and Hinman 2015; Rice and Rebeiz 2019). If a large number of co-opted CREs are causally linked to an increased number of phenotypes (i.e. increased pleiotropy for every co-opted CRE), this lack of specificity could be detrimental over evolutionary time, as it may preclude the independent movement of affected traits towards fitness maxima, at least via changes to those CREs (Fisher 1930; Hansen 2003). That is, an excess of pleiotropic linkage between traits as a result of co-option could ultimately act as a relative constraint, impinging on the evolvability of traits. Since we generally do not observe such strong pleiotropic constraints (Wagner and Zhang 2011), we must either assume that network co-option is quite rare, or explain why repeated occurrences of network co-option do not hamper evolvability. The growing number of studies that implicate co-option suggest that this mechanism is common enough to warrant a discussion of the latter.
We presume that networks as a whole regain or maintain specificity after network co-option, as we observe some degree of modularity for GRNs that have thus far been characterized (Davidson and Erwin 2006; Wagner et al. 2008; Sabarís et al. 2019). What we would like to provide here is a thorough analysis of how the specificity of network nodes fluctuates over time after network co-option. We hope this will be informative for our understanding of co-option as a mechanism, and in particular, our understanding of how this mechanism might relate to evolvability. We will break down the phenomenon by first outlining the range of possible immediate outcomes for a given instance of network co-option. We will then describe the mechanisms by which network nodes may either retain or subsequently regain specificity of their cis-regulatory information. Finally, we will draw on our outline of this mechanism to discuss the potential role(s) of network co-option in the evolution of organismal parts.
1. Immediate outcomes of network co-option
Network co-option events can theoretically yield a wide range of outcomes. In most cases, there will exist many differences in the trans-regulatory landscape of the cells of the novel (differing in space or time) context and that of the context in which the network operated previously. Distinct regulatory information in the novel cellular context can intersect or interfere with the newly redeployed network at any point downstream of the initiating trans change, and thus individual cases of network co-option may differ in the number of network genes redeployed, as well as the identities of downstream targets. We can visualize the spectrum of possible outcomes at the initiation of network co-option by outlining four broad categories (Figure 1): Wholesale co-option, partial co-option, functionally divergent co-option, and aphenotypic co-option.
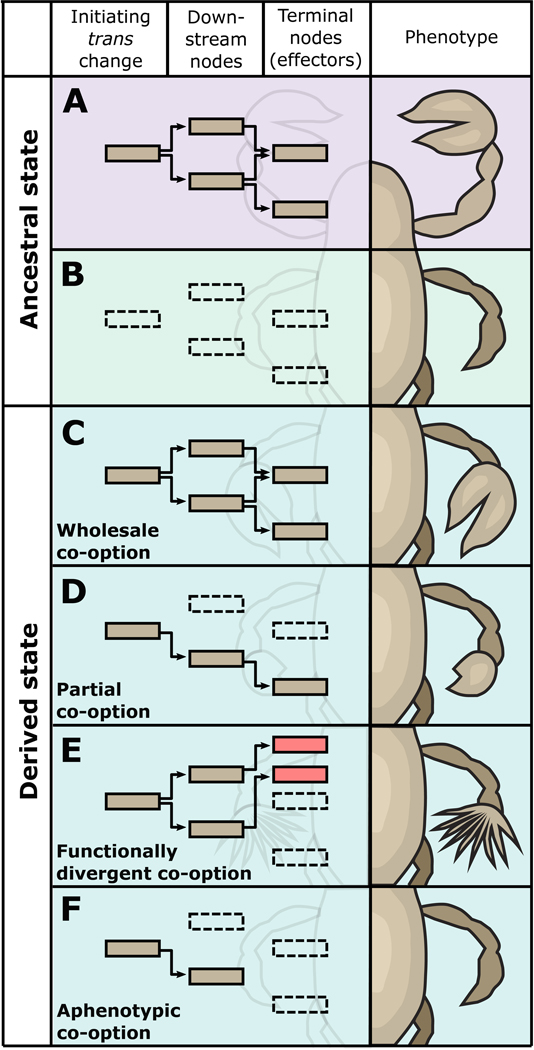
Figure 1. Network Co-option results in a range of possible outcomes.
A. Ancestral location and outcome of network deployment, showing the phenotype in that context. B. Prior to network co-option, the network is inactive in the second location. (C-F) Activation of the upstream “initiating trans factor” results in redeployment of some or all of this network in the second location. C. Wholesale co-option involves the redeployment of the entire network in the novel context, resulting in the recapitulation of the phenotype that appears in the ancestral context. D. Partial co-option, in which some of the downstream transcription factors and terminal effectors are not redeployed in the novel context. The phenotype in the novel context may share some features with the phenotype in the ancestral context. E. Functionally-divergent co-option is similar to D, except that in the novel context, some of the downstream targets of the redeployed network are distinct from the ancestral context. The phenotype is not necessarily recognizable as being associated with the phenotype in the ancestral context. F. Aphenotypic co-option, in which no terminal effectors are activated, although there are changes to the upstream developmental program. No phenotype is observed, apart from changes of expression that can be detected experimentally.
1.1. Wholesale co-option
One possibility is that the entire, or nearly the entire network downstream of the initiating trans change is redeployed in the novel tissue. We call such cases “wholesale co-option”. The result of wholesale co-option is that the same set of terminal effectors is activated in the novel location, and there will be a recapitulation or near-recapitulation in the novel location of the trait generated by the network downstream of the initiating trans change in the ancestral location (Figure 1C).
Gain-of-function homeotic transformations are an illustrative example of wholesale network re-use, where the initiating trans change may be the introduction of a Hox gene. For example, in Drosophila melanogaster the antennae can be transformed into legs by the overexpression of the homeobox gene Antennapedia (Schneuwly, Klemenz, and Gehring 1987). Here, the addition of a single upstream factor results in the deployment of an entire leg formation network in a different location, with the terminal result being easily recognizable as the trait for which the network is generally employed in wild type animals. Likewise, misexpression of the eyeless (ey) gene in Drosophila melanogaster is capable of generating ectopic eyes (Halder, Callaerts, and Gehring 1995). Similar kinds of homeotic transformations involving changes to single factors have also been observed in floral parts (Coen and Meyerowitz 1991; Álvarez-Buylla et al. 2010).
Wholesale co-option might also be common when repeated structures, such as neurons, muscles, epithelial appendages, and even serially-homologous body segments increase in number, as these networks have already been subject to recurrent reuse and therefore may possess nodes capable of “selector-like” (García-Bellido 1975; Mann and Carroll 2002) or “input-output” (Stern and Orgogozo 2009) function (i.e. largely sufficient to produce the phenotype). For example, Marcellini and Simpson (2006) showed that the expanded domain of a single enhancer of the gene scute was sufficient to explain the derived condition in which the number or dorsocentral bristles increased from two to four in the Drosophilid species Drosophila quadrilineata (Marcellini and Simpson 2006). The D. quadrilineata enhancer was able to recapitulate the derived condition when used to drive scute expression in D. melanogaster (ordinarily possessing only two dorsocentral bristles), demonstrating that the existing downstream regulatory logic was used in the construction of the novel pair of bristles, consistent with wholesale co-option.
We define wholesale co-option as an instance of network co-option in which the direct downstream consequences of the redeployment of the initiating trans factor are identical (or nearly identical) in the novel cellular context to those in the ancestral cellular context. This definition does not specify what qualifies as a recapitulated “trait.” A trait does not, for example, need be a discrete organ like a bristle, eye, or wing. It could instead be a characteristic such as pigmentation or a chemical signal, provided that the initiating trans change is sufficient to recapitulate the downstream effect. For instance, a possible example of wholesale co-option is in the exoskeletalization of the elytra (exoskeletalized forewings) of beetles. Experiments on Tribolium castaneum suggest that the derived state of elytral exoskeletalization may have evolved via redeployment of the entire exoskeletalization network of the body wall to the beetle forewing, involving novel upstream regulatory roles of existing wing-patterning elements (Tomoyasu et al. 2009). The recapitulated “trait” in this case is the exoskeletal fate of cells in the forewing tissue.
1.2. Partial network co-option and functionally-divergent network co-option
Wholesale co-option represents the most comprehensive case of network re-use. The striking differences between structures that we believe were built via co-option and the ancestral traits from which the networks were co-opted suggests that simple transplantation of entire networks will be rare. Rather, we anticipate that in the majority of co-option cases, only a portion of the network downstream of the initiating trans change will be redeployed (Erwin, this issue). We refer to cases wherein some subset, but not the entirety of a downstream network is re-deployed as “partial co-option” (Figure 1D). Network architecture can be highly context-dependent (Luscombe et al. 2004), and the fidelity of the network redeployment can range from substantial, in which case many features of the ancestral trait are identifiable, to quite minimal, such that the imported elements of the ancestral trait are unrecognizable, or nearly so, in the novel context. Many factors may come into play to prevent activation of some downstream nodes, including (but not be limited to): other tissue specific transcription factors, tissue-specific post-transcriptional modification (e.g. splicing, phosphorylation, protein cleavage), extrinsic signaling from adjacent tissues (Barolo and Posakony 2002), and boundary conditions set up by developmental timing or mechanical constraints (Davidson 2012; Green and Batterman 2017; Womack, Metz, and Hoke 2019).
A similar phenomenon occurs in a case of what we will call “functionally divergent” co-option (Figure 1E). In these cases, upstream network architecture is co-opted, but the terminal effectors activated by that upstream network differ in the novel context. The upstream nodes would be active in both ancestral and novel settings, but the CREs of the distinct downstream nodes would not be.
We sketch out the distinctions between wholesale, partial, and functionally divergent co-option largely for theoretical purposes here, to clearly outline the full range of possible implications for this mechanism. Empirically, these outcomes would only be definitively distinguishable from each other at the time the co-option was first initiated, and would require detailed knowledge of the network CREs, such that the activity of those CREs and the downstream targets of network genes could be compared across contexts. Any subsequent changes to these networks would obscure the distinction between categories of co-option.
Many well-known examples of network co-option referenced in the literature likely represent instances of partial co-option, functionally-divergent co-option, or some combination of both. For example, in contrast to the ectopic eyes generated by Pax6 mis-expression, a fascinating example of a possible partial network co-option of an eye network was found in a study of extinct dipterans. Two species in the genus Eohelea possessed a structure on their wings that bore a remarkable resemblance to the compound eyes of individuals of those same species, leading the authors to conclude that this structure was likely built through network re-use. However, it appears that in the case of this novel wing structure, the novelty consisted only of the cuticular part of the eye, and was not an entire ectopic eye (Dinwiddie, 2010). We note that in this case because these are not extant species, it is not possible to distinguish between a partial network co-option and a wholesale co-option of a single independent part of the eye network.
Functionally divergent co-option may often result from the co-option of signaling pathways, which are utilized throughout development and quite commonly implicated in the formation of novel traits (Loredo et al. 2001; Harris, Fallon, and Prum 2002; Cebra-Thomas et al. 2005; Harris et al. 2005; Wasik and Moczek 2011; Nakamura et al. 2015). For example, studies on butterfly eyespots suggest that the evolution of these novelties likely included functionally divergent co-option of a deeply conserved anterior-posterior boundary-forming network. The downstream consequences of this network in the novel context provides pattern information for the color phenotypes manifested in scales (Carroll et al. 1994; Keys et al. 1999). Thus, an important process was co-opted, that is, the formation of a particular transcription factor landscape pattern, yet the downstream targets differed in the novel location. We currently do not understand how these functionally divergent outcomes became connected to the anterior-posterior boundary network, and many hypotheses exist (Özsu and Monteiro 2017). A similar case has been suggested in plants, for which a abaxial-adaxial polarity gene network responsible for the flattening of organs such as leaves appears to have been co-opted to cause flattening of stamen filaments, a derived condition (Almeida et al. 2014).
A well-known example that appears to support partial co-option is the redeployment of the leg network in beetle head horns (Moczek and Nagy 2005). Not every gene of the canonical leg network is actually required for horn development, as evidenced by the lack of a phenotype when the gene dachshund was knocked down (Armin P. Moczek and Rose 2009). There is also evidence for partial co-option in the case of the posterior lobe (a genital structure) of fruit flies in the Drosophila melanogaster clade. The evolution of this novel structure appears to have involved co-option of part, but not all, of a network responsible for the development of an ancestral larval structure (Glassford et al. 2015). Additional examples include tree-hopper helmets, the evolution of which appears to have involved co-opted elements of the wing-patterning network (Prud’homme et al. 2011; Fisher, Wegrzyn, and Jockusch 2019), bilaterian appendages, which may have involved redeployment of an existing network for anteroposterior patterning (Lemons et al. 2010), and the use of Hox genes in the evolution of paired vertebrate limbs (Zakany and Duboule 2007). We imagine that these cases represent an amalgam of partial and functionally-divergent co-option, but we are still uncovering the full picture of how the ancestral networks were reused and rewired. Future work characterizing these networks more extensively will help us understand how and when nodes were lost and gained across contexts.
1.3. Co-option resulting in downstream regulatory expression only (aphenotypic co-option)
Finally, network co-option may involve the introduction of an upstream regulator that results in the deployment of some of the upstream network nodes in the novel context, but causes no phenotype (defined as measurable change in morphology/behavior); only transcription factor expression patterns are altered. Such cases of “aphenotypic” co-option could ensue if there is a total lack of activation, or inadequate activation, of required terminal effector genes (Figure 1F). Aphenotypic co-option is essentially an extreme case of partial network co-option. As the term “pleiotropy” is usually restricted to mean that changes to one locus can induce multiple phenotypes (Wagner et al. 2008), aphenotypic co-option generates no new pleiotropy, although changes to the developmental program have occurred.
Examples of aphenotypic co-option are lacking, but there are reasons to believe they exist, or at least we have evidence of the possibility in that we often observe expression patterns for which we can offer no functional explanation. For instance, RNAi screens sometimes find that knockdowns of some transcription factors expressed in the tissue of interest have no phenotype (e.g. Staller et al. 2013; Zattara et al. 2016). Similarly, a comparison of the expression of 20 genes in imaginal tissues of four very closely related species in the melanogaster clade uncovered striking differences in expression across species, many of which are not connected to any known phenotype (Rebeiz et al. 2011). Many CREs for genes exhibit expression patterns outside the focal tissue of a given study, these patterns having no known functional role (e.g. late anterior expression driven by the minimal even-skipped stripe 2 enhancer in Drosophila embryos (Janssens et al. 2006)). Besides the aforementioned examples, many more cases like these may suffer from the “file drawer problem” (Rosenthal 1979). Such results are usually ignored, or interpreted as evidence of robustness, but may sometimes represent cases of aphenotypic network redeployment or non-functional nodes of partially co-opted networks.
Aphenotypic co-option as an idea has generally received sparse attention, although it has been mentioned as a possibility in the past (True and Carroll 2002). While considering such an outcome on its own may seem irrelevant, when considered in the light of long evolutionary periods, this phenomenon could nonetheless have some quite interesting implications, as we will discuss in Sections 3 and 4.
2. How do GRNs maintain or recover specificity after network co-option?
Restoration of at least partial regulatory specificity of co-opted CREs is almost certain to be a pervasive phenomenon, considering the number of morphological novelties we have discussed here that arose through likely network co-options but now apparently evolve in a largely independent manner (e.g. treehopper helmets (Prud’homme et al. 2011), beetle horns (Emlen, Corley Lavine, and Ewen-Campen 2007; Moczek 2009), butterfly wing spots (Brunetti et al. 2001; Oliver et al. 2012), and feathers (Prum 1999; Prum and Brush 2002; Prum 2005)).
The process of re-establishing CRE tissue specificity could happen in two ways: In cis, via changes to the co-opted CREs themselves, or in trans, via changes outside the network that introduce tissue-specific regulators of the co-opted CREs. We discuss these possibilities below. We note that although our discussion is centered on co-opted CREs, multiple studies have noted pleiotropy in enhancer sequences in the absence of network co-option events (Nagy et al. 2018; Rebeiz et al. 2011; Preger-Ben Noon et al. 2018), and the mechanisms we describe below apply broadly to the evolution of regulatory specificity.
Changes in trans
Many genes outside of the co-opted GRN will likely have pre-existing roles in the tissue that predated the network co-option event. These genes may be available for genetic tinkering that yields tissue-specific modifications, or could contribute to an immediate plastic response that could modulate pleiotropy, and be genetically modified later (West-Eberhard 2005). Novel expression domains of such genes beyond the co-opted GRN could also arise subsequently, after co-option has occurred, and be exploited to achieve tissue independence at that future time. A modification of this type can be inferred from the striking instance of wholesale co-option demonstrated for the embryonic skeleton of sea urchins (a derived trait) which employs a GRN co-opted from adult skeletogenesis (Gao and Davidson 2008). In this case, while most of the genes in the co-opted network are shared across the two contexts, implicating wholesale co-option, the embryonic skeletogenesis network incorporates a small number of genes that are not part of the adult skeletogenesis network. One of these genes, tbrain (tbr), has known ancestral roles in endomesoderm specification in other echinoderms, leading the authors to suggest that the addition of this regulator was a modification to the embryonic skeletogenesis network that occurred after the initial co-option event.
Direct regulators of the initiating trans factor could be targets for modification, if these differ between the novel and ancestral contexts. For instance, in the example discussed above concerning the exoskeletalization of elytra in beetles, the authors showed that the gene apterous (ap), which is part of the ancestral wing network, has gained a novel role in redeploying the exoskeleton network in the elytra, whereas ap is not a direct regulator of the exoskeleton network in the mesonotum (a cuticular part of a thoracic segment). Consequently, when RNAi was performed targeting ap, defects were seen in exoskeletalization of the forewing, but not the mesonotum (Tomoyasu et al. 2009). This demonstrates that, in principle, exoskeletalization could be targeted independently in the elytra by modifying regulators further upstream of the co-opted network, even if the exoskeletalization network itself were pleiotropic.
Changes in cis
If tissue-specificity is to be regained via changes to CREs of nodes in the co-opted GRN, the first requirement is that the ancestral and novel tissues must have at least one qualitative or quantitative difference in cellular regulatory content (e.g. transcription factor identity, activity, or concentration) at the time the node in question is active. That is, there must exist some form of potentially exploitable tissue specificity, otherwise all changes to the CRE would necessarily affect both the ancestral and novel contexts. We suspect that this is usually the case, although in principle it is possible that the regulatory states of the ancestral and novel contexts would not differ at all after co-option (i.e. if the only difference between the two contexts prior to co-option was the presence/absence of the initiating trans factor itself). In such cases, multiple mutations would be required to regain tissue-specificity.
There are two primary mechanisms that can mitigate or eliminate the potential for pleiotropic constraint at a co-opted CRE: regulatory input diversification (Figure 2B) and context-specific redundant enhancers (Figure 2D). The result of these mechanisms would be either a facilitation of independent regulation of the node in the novel and ancestral contexts, or a modulation of the CRE in question in one context (for instance inactivating it in one context). A critical note is that while these features that restore CRE tissue-specificity may evolve subsequent to network co-option, they also may already be in place at the time of co-option. Indeed, the modifications to CREs we describe here would be causal explanations of why some nodes are not expressed in partial or aphenotypic co-option outcomes. Similarly, we note that although entirely novel mutations could be the causal changes in these mechanisms, novel genetic combinations of existing variants already segregating in the population could also alter pleiotropy or mask it through epistasis (Pavlicev et al. 2008). Given the pervasive nature of epistasis in natural populations (Phillips 2008; Mackay 2014) this may be a frequently employed path to recapturing lost specificity. It is also important to mention that tracing the process of how tissue specificity was restored may be very difficult when comparing species that have diverged for long periods of time. Once sufficient time has passed, the footprints of this process will have been erased. As such, evidence for this mechanism will likely be found in cases where at least some of the ancestral pleiotropic and redundant enhancers are still detectable.
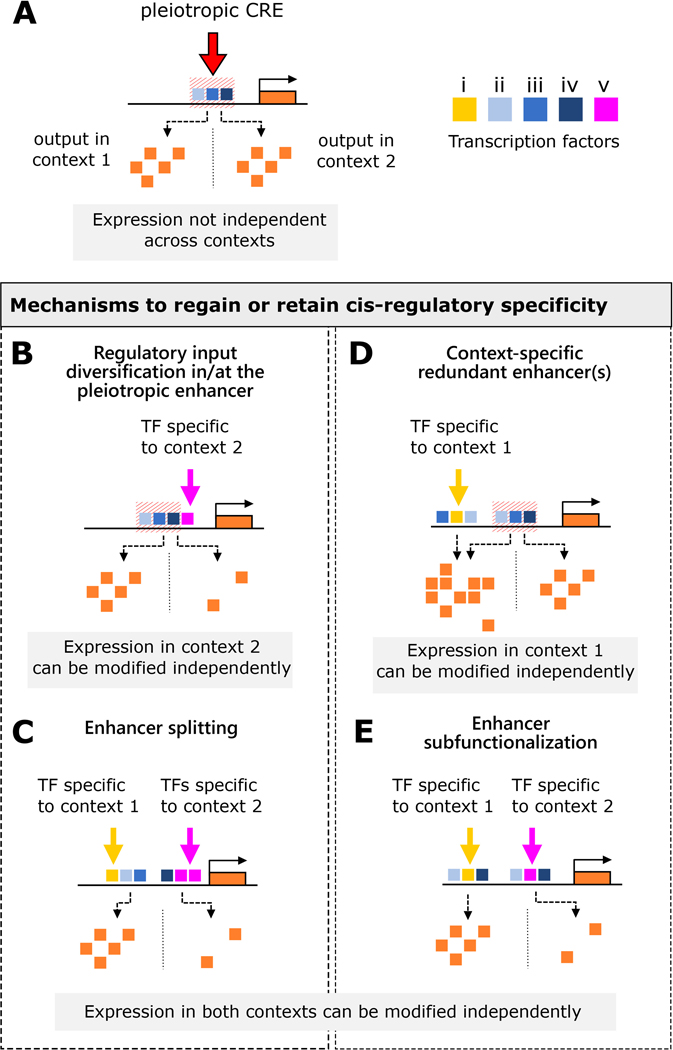
Figure 2. Mechanisms to retain or regain specificity of pleiotropic CREs.
A. A gene that is redeployed during network co-option possesses a pleiotropic cis-regulatory element that drives expression in contexts 1 and 2. The CRE is activated by the binding of transcription factors ii, iii, and iv, and the output of expression in both contexts is not independent. B. Regulatory input diversification: evolution of a binding site for transcription factor v, which acts as a repressor, only affects expression in context 2, as v is not present in context 1. Further modification can occur to achieve greater or full independence via enhancer splitting (C), in which a single enhancer fragments into two enhancers employing context-specific activators. D. Redundant enhancers: A second enhancer for the target gene affects expression in context 1 only, due to the fact that this redundant enhancer requires the binding of transcription factor i, which is not present in context 2. Further modification can occur via enhancer subfunctionalization (E), in which redundant enhancers that have partial or full overlap in their expression profiles gain or lose binding sites for context-specific factors to achieve complete independence.
Regulatory input diversification
The diversification of regulatory inputs (Figure 2B) mitigates pleiotropy via binding sites at the pleiotropic enhancer that affect the regulatory outcome of the enhancer differentially across tissues. For example, this might be the gain of a binding site for a repressor that is only present in one tissue. Enhancer splitting (Figure 2C) is an extension of the process above, and is related to the idea of enhancer sprawl (Rice and Rebeiz 2019), in which it is understood that enhancers sometimes expand and contract due to the addition and removal of binding sites via turnover. In this case, tissue-specific binding sites accumulate such that a single enhancer that has some tissue-specific and some pleiotropic binding sites may eventually split into two completely separate tissue-specific cis-regulatory elements in adjacent positions on the DNA.
Specificity conferred by redundant CREs
Redundant CREs (also called “shadow enhancers”) are defined here as at least two CREs driving expression of the same target gene in redundant or semi-redundant expression patterns (Hong, Hendrix, and Levine 2008; Barolo 2012). Redundant CREs are a route to at least partial recovery of modularity in cases of co-option because if the CREs employ a unique set of regulators, one or both of the redundant CREs may drive expression differently across the two tissues. We already have several empirical examples of redundant enhancer pairs that display different regulatory logic (Wunderlich et al. 2015; Vincent et al. 2018), lending credence to this potential route to specificity.
With redundant CREs, there are two possible conditions. First, a redundant CRE could drive expression in only one of the tissues (Figure 2D). This could in fact provide an immediate mode of retaining specificity, as a redundant CRE of this type could already exist in the cis-regulatory region of a GRN node at the time of co-option. A redundant CRE of this kind could also evolve later and restore specificity (Rebeiz and Tsiantis 2017). However, in the two cases just described, the redundant CRE from the co-opted network is still pleiotropic. CRE sub-functionalization (Figure 2E), in which two redundant CREs of a single gene (i.e. a redundant CRE pair) each evolve independent roles specific to one of their initial developmental contexts, would be required to erase all pleiotropic linkages between ancestral and novel contexts (e.g. the “cis-regulatory element duplication, degeneration, and complementation” model (Monteiro and Gupta 2016)). This may or may not be favorable, as robustness via redundant enhancers is also considered to be potentially beneficial (Perry et al. 2010; Frankel et al. 2010; Barolo 2012).
3. The action of selection on co-opted networks over time
Co-option is often viewed as a potential mechanism for facilitating the origins of morphological novelties. In other words, the appeal of this mechanism rests in its possible explanatory value with regard to evolution. However, while network co-option is often invoked in this way (True and Carroll 2002; Olson 2006; Shubin, Tabin, and Carroll 2009; Monteiro 2012; Peter and Davidson 2015), in order to appreciate the full evolutionary implications of this mechanism, we must carefully consider the manner in which we expect natural selection and neutral processes to operate on co-opted networks after they occur.
As with any mutation, in a simple sense there are three potential fitness effects of an initiating trans change (Figure 3, A): beneficial, neutral, or deleterious (Figure 3, B). When thinking about network co-option and evolution, the added element to consider is the concomitant reduction of tissue-specificity, which may have long term consequences (Figure 3, C). Below we discuss the evolutionary implications of pleiotropy at co-opted network CREs, given each of the three possible fitness consequences of the initiating mutation.
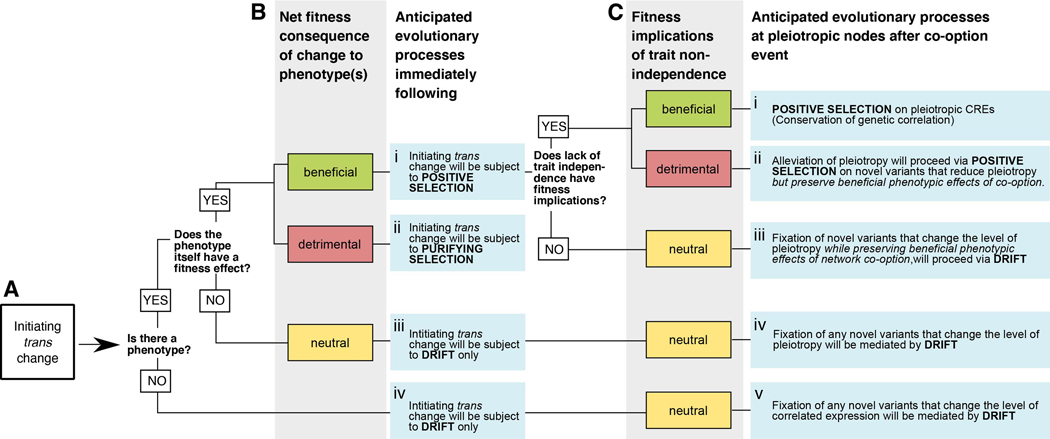
Figure 3. Fitness effects of network co-option and the influence of pleiotropy upon subsequent change.
Figure depicts a decision tree that partitions the evolutionary ramifications of GRN co-option and subsequent modification. Starting on the left side of the diagram with the initiating trans change (A), we may first make standard predictions about evolutionary response to that mutation based on its phenotypic effects (or lack thereof) (B). After network co-option, the fitness effects of decreased tissue-specificity are considered as the network is subject to additional modifications (C).
Initiating trans change with positive fitness effects
If network co-option generates a novel phenotype and the net effect on fitness is beneficial, we expect that the initiating trans change, would be under positive selection (Figure 3, B, i). If the beneficial phenotype is in the novel deployment context, this situation exemplifies what is imagined to be the major upshot of network co-option as a mechanism for evolutionary change that we discussed in the introduction: a novel, beneficial, phenotype produced in one or just a few evolutionary steps. Still, while the overall phenotype may be beneficial, the effects of pleiotropy resulting from the network co-option may be detrimental, either due to the lack of modularity between the ancestral and novel traits that limits adaptation, or because some nodes have negative pleiotropic effects (Figure 3, C, ii). In such cases, selection should favor mutations that maintain the expression of beneficial co-opted nodes in both tissues but restore specificity, for example, inactivation of particular nodes that have negative pleiotropic effects. In spite of the popularity of this view of network co-option, we do not have empirical examples that explicitly demonstrate this sequence of events.
In theory, it could be that when a co-option event occurs, the resulting pleiotropy is itself advantageous, in the sense that if the two traits usually experience selection together and in the same direction, the genetic correlation between them would allow selection to operate more efficiently. A simple example would be a trait that tracks environmental conditions, such as fur thickness or cryptic coloration, expanding via co-option to a new location on the body. If selection on the character in both contexts is uniform, the novel and ancestral traits would functionally amount to only one trait with respect to the co-opted network. In such cases, there could be selection against variants that broke up pleiotropy at CREs in the co-opted network, as trait independence would represent an unnecessary increase in complexity, possibly slowing the ability of the population to adapt overall (Orr 2000; Welch and Waxman 2003)(Figure 3, C, i). To our knowledge this particular outcome of network co-option does not currently have direct empirical support, although the idea of selectively maintaining beneficial pleiotropy has been suggested more generally (e.g. between interacting parts such as integrated skeletomuscular traits (Karasik and Kiel 2010)). A possible scenario of this sort could also occur in plants, where it is known that male and female floral parts (androecium and gynoecium) of some species share much of their developmental toolkit (Dornelas et al. 2011). A correlation of male and female floral structures could be favorable in some cases if it were required for efficient pollination.
Alternatively, the network co-option could confer a fitness benefit in the novel location, and the existence of pleiotropic roles of any given CRE could be neutral, or nearly neutral (Figure 3, C, iii). This could happen if there is only selection on the beneficial novel trait and the ancestral trait is either completely neutral (i.e. has no function), or if the majority of phenotypic changes to the ancestral trait via mutations at co-opted nodes would be neutral such that the pleiotropy is nearly neutral. Over time, evolution of the pleiotropic cis-regulatory regions could erode the genetic correlation by chance if changes arose to increase regulatory independence without negatively impacting the phenotype(s). Otherwise, because the CRE confers a functional benefit to the novel context, it may be conserved and the correlation could be maintained incidentally. A study of the genetic correlations among tetrapod limb developmental serial homologs suggests that covariance structures that result from reuse of networks (as is thought to be the case with hindlimbs and forelimbs (Sears, Capellini, and Diogo 2015)) can persist for long periods (Young and Hallgrímsson 2005). The authors found that the correlation between the lengths of hind limbs and forelimbs is only broken in cases of extreme functional necessity, such as is observed in the extremely divergent limb and digit proportions that enabled flight in the lineage leading to bats. However, we do not know whether the covariance structure in this case was maintained actively or passively, and the authors conclude that stabilizing selection on such correlations may often be an important factor beyond genetic constraints (Young and Hallgrímsson 2005; Hallgrímsson et al. 2009).
Another important possibility to consider is that the mutation confers a benefit in the ancestral context and initiates network co-option neutrally elsewhere as a byproduct. In such cases, the initiating mutation could be subject to selection irrespective of the co-option per se (this causal mechanism for selectively neutral traits is discussed in Lovejoy et al. 2002). A modeling study showed that the addition of genes to a network generally improved the “fit” of the model to its target data, which suggests that recruitment of genes to already functioning networks could be common (Spirov, Sabirov, and Holloway 2012), and might be a source of this type of “collateral” network co-option. This could be difficult to detect, as neutral phenotypes generated by network co-option could appear to be under selection if there is selection on the genetically correlated character (Lande and Arnold 1983). We will discuss the implications of this potential outcome more in the section on neutral fitness outcomes below.
Initiating trans change with detrimental fitness effects
If network co-option is deleterious, the initiating trans change should be lost due to purifying selection (Figure 3, B, ii) unless it is fixed by drift, which is more likely in small populations and when the fitness consequence is mild (Fisher 1930). It is also possible that the phenotypic consequences of a given co-option event on the novel tissue were initially beneficial or neutral, and only later became detrimental (e.g. accompanying a change in environment that alters selective regime or developmental plasticity, epistatic changes that reveal larger effects on phenotype, etc.). In such cases, the upstream mutation may have already been fixed in the population. In either of the above cases, the detrimental effects of network co-option could either be eliminated by another change at the upstream trans factor that reverts the co-option, or the effects could be reduced over time by evolving tissue specific repression of downstream network nodes individually. Any given case of this latter process would be indistinguishable from an initial state of partial or aphenotypic co-option, although in some cases a comparison across species or populations that diverged after the initiating trans change might reveal a history of modifications deactivating the co-opted network.
Initiating trans change with selectively neutral effects
One possible outcome of an initiating trans change that incurs network co-option is the generation of a phenotype which is completely neutral with respect to fitness (Figure 3, B, iii). Another neutral outcome, which we discussed in section one, is the possibility that no phenotype is generated in the novel tissue at all (e.g. aphenotypic co-option) (Figure 3, B, iv). In both of these cases, the genetic correlation generated between the tissues is also likely neutral, unless future mutations alter the neutrality of the phenotype or induce a new phenotype via the previously aphenotypic network. Otherwise, both the initiating trans change (Figure 3, B, iii, iv), and any future mutations that alter the genetic correlation generated by co-option would be fixed only by drift (Figure 3, C, iv, v). We have no reason at all to believe that fixation of a mutation of this type this would be more uncommon than the stochastic fixation of any other neutral mutation. This scenario is therefore especially important to consider in small populations that are more heavily influenced by drift. Modeling and analysis of changes to gene expression across species of Heliconius butterflies (Catalán, Briscoe, and Höhna 2019), fish (Whitehead and Crawford 2006), and primates (Khaitovich, Pääbo, and Weiss 2005; Chaix et al. 2008) all showed that the majority of changes to gene expression across species were consistent with neutral evolution, lending credibility to this possibility.
With respect to latent expression generated by co-opted networks specifically, we do not currently have examples. However, a study on the evolution of Onthophagus beetle horns suggested that exploitation of an existing expression pattern in the beetle anterodorsal head tissue was important to the evolution of the novel horn structures. A key member of this gene network, an ortholog of the Drosophila gene orthodenticle (otd), was also found to be expressed ancestrally in the anterodorsal head tissue of an outgroup species (Tribolium castaneum), which lacks horns. Interestingly, knockdown of otd in Tribolium does not induce detectable defects in the head, suggesting that this pattern is not functional in Tribolium (Zattara et al. 2016).
4. Network co-option and the origin and diversification of traits
Our breakdown of the phenomenon of network co-option in the previous sections now puts us in a position to offer a few discussion points on the relationship between network co-option and the evolvability of traits. This is in no way a comprehensive list. Our comments here will hopefully serve as a jumping-off point for further conversation.
First, as we discussed in Section 1, it is important to recognize that the comprehensive case, wholesale co-option, is likely not the most common outcome. We anticipate that many more instances of network co-option will be only partial, and therefore a degree of trait independence may be retained even at the onset of network co-option. It has been suggested that intermediate levels of pleiotropy maximize evolvability (Hansen 2003), and thus many cases of co-option may be well within the range of pleiotropic effects that do not cause serious problems for evolvability. Nevertheless, in such cases that the pleiotropy generated by network co-option acts as a constraint, many routes exist to modify CREs directly in cis or via their regulators to regain specificity, as we discussed in Section 2.
Second, we must keep in mind that the effects of pleiotropy are not always detrimental. Not all phenotypes generated by co-option will initially, or ever, affect fitness, and not all co-option events will have a phenotype. These neutral outcomes would still alter modularity in the strict sense that the co-opted CREs would have decreased potential to confer tissue-specificity, however there would be no immediate visibility of these events in terms of selection, and thus the evolvability of ancestral traits would not be affected, at least initially. Models that allow for neutral pleiotropic effects of co-option would improve our understanding in this area. One model predicting the degree to which pleiotropy would act as a developmental constraint revealed that the level of constraint was sensitive to changing the fitness effect of pleiotropy (Otto 2004). As has been pointed out previously in the case of gene pleiotropy (Stern and Orgogozo 2008), concerns about pleiotropic constraint resultant from network co-option may be mitigated by a clearer understanding of the forms pleiotropy can take as a result of this mechanism.
Beyond simply failing to obstruct the evolvability of traits, we should also keep in mind that neutral or nearly neutral outcomes of network co-option that are retained stochastically (phenotypes, expression patterns) could provide a reservoir of cryptic genetic variation. Such variation may have phenotypic and selective consequences later if subsequent mutations activate processes, such as additional network co-option events, downstream of these nodes. Initially aphenotypic outcomes of co-option might therefore contribute positively to trait evolvability (e.g. in cases of preadaptation). This possibility has been noted before (True and Carroll 2002), but we currently lack empirical examples to support this conjecture. However, there is a growing interest in understanding how cellular and morphological phenotypes may evolve neutrally (Ruths and Nakhleh, 2013; Zhang, 2018; Wideman et al, 2019). A recent study on cryptic genetic variation demonstrated that neutral mutations accumulated at the level of a single protein facilitated subsequent adaptation of that protein (Zheng, Payne, and Wagner 2019). This result might scale up to the level of networks. More examples such as these that examine multi-gene interactions, and especially comparative analyses of the network architecture of such cases will help us understand the role of network co-option in the generation of cryptic genetic variation.
The implications of the observations above are magnified when we consider that the simple version of co-option that begins with GRN deployment in one tissue and expands to deployment in two tissues is probably not realistic. More extensive effects across many tissues are likely to be common. As networks evolve downstream of newly redeployed nodes after network co-option, a complex collage forms rather than a pre-made template which is simply “copy-pasted” to a new location. Indeed, this view is supported by a mathematical modelling study, which demonstrated that the construction of a novel expression domain is facilitated by reuse of multiple but distinct existing modules that contribute to that domain elsewhere (Espinosa-Soto and Wagner 2010). Such a scenario, wherein CRE pleiotropy is spread out over multiple ancestral GRNs, might lend more flexibility to circumventing developmental constraints for both the ancestral and novel structures. To be sure, there are likely to be many cases where network co-option does result in constraint on some properties. For example, it has been suggested that limb outgrowth was constrained to have anterior-posterior polarity due to the fact that Hox genes were co-opted to initiate extension from the body wall (Tarchini, Duboule, and Kmita 2006).
We are still in the process of discovery in the area of network co-option, and there are many ways forward. With respect to modelling, it would be very enlightening to incorporate network co-option into dynamical models (Irons and Monk 2007; Alexander et al. 2009; Verd, Monk, and Jaeger 2019), which take spatio-temporal information into account when designating modules. Models incorporating some of the neutral outcomes of co-option that we discussed here would also be very useful. Beyond model development, many more empirical examples of network co-option are needed. In particular, to gain insight into the evolution of co-opted networks, we need examples wherein the structure of known or suspected co-opted networks is compared across species. One study investigated the expression of genes in the network co-opted to generate eyespots across 21 species of Nymphalid butterflies (Oliver et al. 2012). They showed that the expression of some network members was highly conserved, whereas repeated losses of others suggests that these nodes were more evolutionarily labile or possibly not necessary in the first place. More examples like this one would greatly improve our understanding of how co-opted networks are incorporated into existing networks and change over time. We hope the framework that we have outlined highlights the scope of possibilities that accompany network co-option and inspires a wide range of research questions into this intriguing mechanism of developmental evolution.
5. Concluding remarks
In writing this review, we were reminded of the way that general thinking has progressed with regard to genes. What began by attributing strict functional identities to individual genes (“a gene for function x”), eventually became more nuanced in light of empirical data that was inconsistent with a one-to-one view (Duboule and Wilkins 1998). Considering network reuse as a mechanism of altering development similarly complicates our concept of GRN identity. GRNs are not tidy, self-contained “programs” for specific traits (Nijhout 1990; DiFrisco and Jaeger 2019), but are instead highly context-dependent and may therefore yield different outcomes in different developmental circumstances. This suggests that we must caution ourselves against falling into a “GRN for function x” trap, and instead recognize that the GRN for any given trait will be a haphazard assembly of parts, often with a few spare odds and ends, drawn from existing GRNs over evolutionary time. Like all products of evolution, GRNs will be the result of evolutionary “tinkering” (Jacob 1977): functional, but messy.
Acknowledgements
We would like to acknowledge Aaron Novick, Gavin Rice, and Ben Vincent for their helpful comments on this manuscript. Our work on the specificity of co-opted networks is supported by the National Institutes of Health (GM112758 to MR).
Glossary
- Cis-regulatory element (CRE)
A stretch of non-coding DNA that is physically upstream, downstream, or in the intron, of a given gene, and which influences the expression of that gene at some time(s) and location(s) during the development or adult life of the organism
- Developmental context
A temporal and spatial domain in a tissue during which specific developmental events, such as the activation of a GRN or morphogenetic process occurs
- Epistasis
The condition in which the phenotypic effect or effects of a particular allele at a particular genetic locus are influenced by the presence of one or more alleles elsewhere in the genome
- Evolvability
Evolvability has many definitions (for discussion see Pigliucci 2008), but can be roughly defined as the ability of a system to change adaptively
- Gene regulatory network (GRN)
Semi-autonomous regulatory modules responsible for characters or phenotypes. This is by no means the only way to define GRNs, as we suggest in our discussion. However, a full discussion of gene regulatory network ontology is outside the scope of this work
- Initiating trans change
The mutation(s) that cause the deployment of a regulatory factor (e.g. transcription factor, signaling molecule) in a novel developmental context to activate its downstream network, initiating a co-option event
- Initiating trans factor
In network co-option, the regulatory factor whose activity in the novel context was triggered by the initiating trans change, and which is responsible for the co-option of downstream network
- Modularity
With respect to development, the phenomenon of partial independence of organismal parts during development (Gunter P. Wagner and Altenberg 1996 Bolker 2000)
- Phenotype
A measurable morphological or behavioral character, trait, behavior, or quality
- Pleiotropy
The condition in which a single mutation causally affects (alters) two or more traits
- Regulatory state
The total set of regulatory factors (transcription factors, co-factors, and signaling molecules) present, and their concentrations, at a given time and in a given cell or specified location during development (Peter and Davidson 2015)
- Terminal effector
A gene that contributes directly, via its participation in control of cellular proteins, to the mechanical behavior or physical phenotype of a cell or a group of neighboring cells (Jacquelyn Smith, Rebeiz, and Davidson 2018)
- Trans-landscape
See Regulatory State, above
References
- Alexander Roger P., Kim Philip M., Emonet Thierry, and Gerstein Mark B.. 2009. “Understanding Modularity in Molecular Networks Requires Dynamics.” Science Signaling 2 (81): pe44–pe44. 10.1126/scisignal.281pe44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida Ana Maria Rocha, Roxana Yockteng, James Schnable, Alvarez-Buylla Elena R., Freeling Michael, and Specht Chelsea D.. 2014. “Co-Option of the Polarity Gene Network Shapes Filament Morphology in Angiosperms.” Scientific Reports 4 (1): 1–9. 10.1038/srep06194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Buylla Elena R., Ambrose Barbara A., Flores-Sandoval Eduardo, Englund Marie, Adriana Garay-Arroyo, García-Ponce Berenice, Eduardo de la Torre-Bárcena, et al. 2010. “B-Function Expression in the Flower Center Underlies the Homeotic Phenotype of Lacandonia Schismatica (Triuridaceae).” The Plant Cell 22 (11): 3543–59. 10.1105/tpc.109.069153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo Scott. 2012. “Shadow Enhancers: Frequently Asked Questions about Distributed Cis-Regulatory Information and Enhancer Redundancy.” BioEssays : News and Reviews in Molecular, Cellular and Developmental Biology 34 (2): 135–41. 10.1002/bies.201100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo Scott, and Posakony James W.. 2002. “Three Habits of Highly Effective Signaling Pathways: Principles of Transcriptional Control by Developmental Cell Signaling.” Genes & Development 16 (10): 1167–81. 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- Bolker Jessica A. 2000. “Modularity in Development and Why It Matters to Evo-Devo.” Integrative and Comparative Biology 40 (5): 770–76. 10.1093/icb/40.5.770. [DOI] [Google Scholar]
- Brunetti CR, Selegue JE, Monteiro A, French V, Brakefield PM, and Carroll SB. 2001. “The Generation and Diversification of Butterfly Eyespot Color Patterns.” Current Biology: CB 11 (20): 1578–85. 10.1016/s0960-9822(01)00502-4. [DOI] [PubMed] [Google Scholar]
- Carroll SB, Gates J, Keys DN, Paddock SW, Panganiban GE, Selegue JE, and Williams JA. 1994. “Pattern Formation and Eyespot Determination in Butterfly Wings.” Science (New York, N.Y.) 265 (5168): 109–14. 10.1126/science.7912449. [DOI] [PubMed] [Google Scholar]
- Carroll Sean B. 2008. “Evo-Devo and an Expanding Evolutionary Synthesis: A Genetic Theory of Morphological Evolution.” Cell 134 (1): 25–36. 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Catalán Ana, Briscoe Adriana D., and Sebastian Höhna. 2019. “Drift and Directional Selection Are the Evolutionary Forces Driving Gene Expression Divergence in Eye and Brain Tissue of Heliconius Butterflies.” Genetics 213 (2): 581–94. 10.1534/genetics.119.302493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebra-Thomas Judith, Tan Fraser, Sistla Seeta, Estes Eileen, Bender Gunes, Kim Christine, Riccio Paul, and Gilbert Scott F.. 2005. “How the Turtle Forms Its Shell: A Paracrine Hypothesis of Carapace Formation.” Journal of Experimental Zoology Part B: Molecular and Developmental Evolution 304B (6): 558–69. 10.1002/jez.b.21059. [DOI] [PubMed] [Google Scholar]
- Chaix R, Somel M, Kreil DP, Khaitovich P, and Lunter GA. 2008. “Evolution of Primate Gene Expression: Drift and Corrective Sweeps?” Genetics 180 (3): 1379–89. 10.1534/genetics.108.089623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen Enrico S., and Meyerowitz Elliot M.. 1991. “The War of the Whorls: Genetic Interactions Controlling Flower Development.” Nature 353 (6339): 31–37. 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Davidson Eric H. 2010. The Regulatory Genome: Gene Regulatory Networks In Development And Evolution. Elsevier. [Google Scholar]
- Davidson Eric H., and Erwin Douglas H.. 2006. “Gene Regulatory Networks and the Evolution of Animal Body Plans.” Science 311 (5762): 796–800. [DOI] [PubMed] [Google Scholar]
- Davidson Lance A. 2012. “Epithelial Machines That Shape the Embryo.” Trends in Cell Biology 22 (2): 82–87. 10.1016/j.tcb.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFrisco James, and Jaeger Johannes. 2019. “Beyond Networks: Mechanism and Process in Evo-Devo.” Biology and Philosophy 34 (6): 54. 10.1007/s10539-019-9716-9. [DOI] [Google Scholar]
- Dornelas Marcelo C., Maistro Patreze Camila, Angenent Gerco C., and Immink Richard G. H.. 2011. “MADS: The Missing Link between Identity and Growth?” Trends in Plant Science 16 (2): 89–97. 10.1016/j.tplants.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Duboule Denis, and Wilkins Adam S. 1998. “The Evolution of `bricolage’.” Trends in Genetics 14 (2): 54–59. 10.1016/S0168-9525(97)01358-9. [DOI] [PubMed] [Google Scholar]
- Emlen Douglas J., Corley Lavine Laura, and Ewen-Campen Ben. 2007. “On the Origin and Evolutionary Diversification of Beetle Horns.” Proceedings of the National Academy of Sciences of the United States of America 104 Suppl 1 (May): 8661–68. 10.1073/pnas.0701209104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Soto Carlos, and Wagner Andreas. 2010. “Specialization Can Drive the Evolution of Modularity.” PLOS Computational Biology 6 (3): e1000719. 10.1371/journal.pcbi.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley Emma K., Olson Katrina M., and Levine Michael S.. 2015. “Regulatory Principles Governing Tissue Specificity of Developmental Enhancers.” Cold Spring Harbor Symposia on Quantitative Biology 80: 27–32. 10.1101/sqb.2015.80.027227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust Tyler B., Binning Jennifer M., Gross John D., and Frankel Alan D.. 2017. “Making Sense of Multifunctional Proteins: Human Immunodeficiency Virus Type 1 Accessory and Regulatory Proteins and Connections to Transcription.” Annual Review of Virology 4 (1): 241–60. 10.1146/annurev-virology-101416-041654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher Cera R., Wegrzyn Jill L., and Jockusch Elizabeth L.. 2019. “Co-Option of Wing-Patterning Genes Underlies the Evolution of the Treehopper Helmet.” Nature Ecology & Evolution, December, 1–11. 10.1038/s41559-019-1054-4. [DOI] [PubMed] [Google Scholar]
- Fisher Ronald A. 1930. The Genetical Theory of Natural Selection. Oxford: Clarendon Press. [Google Scholar]
- Frankel Nicolás, Davis Gregory K., Vargas Diego, Wang Shu, Payre François, and Stern David L.. 2010. “Phenotypic Robustness Conferred by Apparently Redundant Transcriptional Enhancers.” Nature 466 (7305): 490–93. 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Feng, and Davidson Eric H.. 2008. “Transfer of a Large Gene Regulatory Apparatus to a New Developmental Address in Echinoid Evolution.” Proceedings of the National Academy of Sciences 105 (16): 6091–96. 10.1073/pnas.0801201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Bellido A. 1975. “Genetic Control of Wing Disc Development in Drosophila.” Ciba Foundation Symposium 0 (29): 161–82. 10.1002/9780470720110.ch8. [DOI] [PubMed] [Google Scholar]
- Glassford William J., Johnson Winslow C., Dall Natalie R., Sarah Jacquelyn Smith Yang Liu, Boll Werner, Noll Markus, and Rebeiz Mark. 2015. “Co-Option of an Ancestral Hox-Regulated Network Underlies a Recently Evolved Morphological Novelty.” Developmental Cell 34 (5): 520–31. 10.1016/j.devcel.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel Nicolas, Prud’homme Benjamin, Wittkopp Patricia J., Kassner Victoria A., and Carroll Sean B.. 2005. “Chance Caught on the Wing: Cis -Regulatory Evolution and the Origin of Pigment Patterns in Drosophila.” Nature 433 (7025): 481–87. 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- Green Sara, and Batterman Robert. 2017. “Biology Meets Physics: Reductionism and Multi-Scale Modeling of Morphogenesis.” Studies in History and Philosophy of Science Part C: Studies in History and Philosophy of Biological and Biomedical Sciences 61 (February): 20–34. 10.1016/j.shpsc.2016.12.003. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, and Gehring WJ. 1995. “Induction of Ectopic Eyes by Targeted Expression of the Eyeless Gene in Drosophila.” Science 267 (5205): 1788–92. 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Hallgrímsson Benedikt, Jamniczky Heather, Young Nathan M., Rolian Campbell, Parsons Trish E., Boughner Julia C., and Marcucio Ralph S.. 2009. “Deciphering the Palimpsest: Studying the Relationship Between Morphological Integration and Phenotypic Covariation.” Evolutionary Biology 36 (4): 355–76. 10.1007/s11692-009-9076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen Thomas F. 2003. “Is Modularity Necessary for Evolvability?: Remarks on the Relationship between Pleiotropy and Evolvability.” Biosystems 69 (2): 83–94. 10.1016/S0303-2647(02)00132-6. [DOI] [PubMed] [Google Scholar]
- Harris Matthew P., Fallon John F., and Prum Richard O.. 2002. “Shh-Bmp2 Signaling Module and the Evolutionary Origin and Diversification of Feathers.” Journal of Experimental Zoology 294 (2): 160–76. 10.1002/jez.10157. [DOI] [PubMed] [Google Scholar]
- Harris Matthew P., Williamson Scott, Fallon John F., Meinhardt Hans, and Prum Richard O.. 2005. “Molecular Evidence for an Activator–Inhibitor Mechanism in Development of Embryonic Feather Branching.” Proceedings of the National Academy of Sciences 102 (33): 11734–39. 10.1073/pnas.0500781102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman Veronica F., Nguyen Albert, and Davidson Eric H.. 2007. “Caught in the Evolutionary Act: Precise Cis-Regulatory Basis of Difference in the Organization of Gene Networks of Sea Stars and Sea Urchins.” Developmental Biology 312 (2): 584–95. 10.1016/j.ydbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Hong Joung-Woo, Hendrix David A., and Levine Michael S.. 2008. “Shadow Enhancers as a Source of Evolutionary Novelty.” Science (New York, N.Y.) 321 (5894): 1314. 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons David J., and Nicholas AM Monk. 2007. “Identifying Dynamical Modules from Genetic Regulatory Systems: Applications to the Segment Polarity Network.” BMC Bioinformatics 8 (1): 413. 10.1186/1471-2105-8-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob François. 1977. “Evolution and Tinkering.” Science 196 (4295): 1161–66. [DOI] [PubMed] [Google Scholar]
- Smith Jacquelyn, Sarah Mark Rebeiz, and Davidson Lance. 2018. “From Pattern to Process: Studies at the Interface of Gene Regulatory Networks, Morphogenesis, and Evolution.” Current Opinion in Genetics & Development 51 (August): 103–10. 10.1016/j.gde.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens Hilde, Hou Shuling, Jaeger Johannes, Kim Ah-Ram, Myasnikova Ekaterina, Sharp David, and Reinitz John. 2006. “Quantitative and Predictive Model of Transcriptional Control of the Drosophila Melanogaster Even Skipped Gene.” Nature Genetics 38 (10): 1159–65. 10.1038/ng1886. [DOI] [PubMed] [Google Scholar]
- Karasik David, and Kiel Douglas P.. 2010. “Evidence for Pleiotropic Factors in Genetics of the Musculoskeletal System.” Bone 46 (5): 1226–37. 10.1016/j.bone.2010.01.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley Peter D., and William George Hill. 1990. “Variation Maintained in Quantitative Traits with Mutation–Selection Balance: Pleiotropic Side-Effects on Fitness Traits.” Proceedings of the Royal Society of London. Series B: Biological Sciences 242 (1304): 95–100. 10.1098/rspb.1990.0110. [DOI] [Google Scholar]
- Keys David N., Lewis David L., Selegue Jane E., Pearson Bret J., Goodrich Lisa V., Johnson Ronald L., Gates Julie, Scott Matthew P., and Carroll Sean B.. 1999. “Recruitment of a Hedgehog Regulatory Circuit in Butterfly Eyespot Evolution.” Science 283 (5401): 532–34. [DOI] [PubMed] [Google Scholar]
- Khaitovich Philipp, Svante Pääbo, and Gunter Weiss. 2005. “Toward a Neutral Evolutionary Model of Gene Expression.” Genetics 170 (2): 929–39. 10.1534/genetics.104.037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande Russell, and Arnold Stevan J.. 1983. “The Measurement of Selection on Correlated Characters.” Evolution 37 (6): 1210–26. 10.2307/2408842. [DOI] [PubMed] [Google Scholar]
- Lemons Derek, Fritzenwanker Jens H., Gerhart John, Lowe Christopher J., and William McGinnis. 2010. “Co-Option of an Anteroposterior Head Axis Patterning System for Proximodistal Patterning of Appendages in Early Bilaterian Evolution.” Developmental Biology 344 (1): 358–62. 10.1016/j.ydbio.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine Mike. 2010. “Transcriptional Enhancers in Animal Development and Evolution.” Current Biology : CB 20 (17): R754–63. 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loredo Grace A., Brukman Alla, Harris Matthew P., Kagle David, Leclair Elizabeth E., Gutman Rachel, Denney Erin, et al. 2001. “Development of an Evolutionarily Novel Structure: Fibroblast Growth Factor Expression in the Carapacial Ridge of Turtle Embryos.” Journal of Experimental Zoology 291 (3): 274–81. 10.1002/jez.1103. [DOI] [PubMed] [Google Scholar]
- Lovejoy C. Owen, Meindl Richard S., Ohman James C., Heiple Kingsbury G., and White Tim D.. 2002. “The Maka Femur and Its Bearing on the Antiquity of Human Walking: Applying Contemporary Concepts of Morphogenesis to the Human Fossil Record.” American Journal of Physical Anthropology 119 (2): 97–133. 10.1002/ajpa.10111. [DOI] [PubMed] [Google Scholar]
- Luscombe Nicholas M., Madan Babu M, Yu Haiyuan, Snyder Michael, Teichmann Sarah A., and Gerstein Mark. 2004. “Genomic Analysis of Regulatory Network Dynamics Reveals Large Topological Changes.” Nature 431 (7006): 308–12. 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- Mackay Trudy F. C. 2014. “Epistasis and Quantitative Traits: Using Model Organisms to Study Gene–Gene Interactions.” Nature Reviews Genetics 15 (1): 22–33. 10.1038/nrg3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann Richard S, and Carroll Sean B. 2002. “Molecular Mechanisms of Selector Gene Function and Evolution.” Current Opinion in Genetics & Development 12 (5): 592–600. 10.1016/S0959-437X(02)00344-1. [DOI] [PubMed] [Google Scholar]
- Marcellini Sylvain, and Simpson Pat. 2006. “Two or Four Bristles: Functional Evolution of an Enhancer of Scute in Drosophilidae.” PLoS Biology 4 (12): e386. 10.1371/journal.pbio.0040386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minafra Luigi, Valentina Bravatà, Giusi Irma Forte, Francesco Paolo Cammarata, Carla Gilardi Maria, and Messa Cristina. 2014. “Gene Expression Profiling of Epithelial–Mesenchymal Transition in Primary Breast Cancer Cell Culture.” Anticancer Research 34 (5): 2173–83. [PubMed] [Google Scholar]
- Moczek AP 2009. “On the Origins of Novelty and Diversity in Development and Evolution: A Case Study on Beetle Horns.” Cold Spring Harbor Symposia on Quantitative Biology 74: 289–96. 10.1101/sqb.2009.74.010. [DOI] [PubMed] [Google Scholar]
- Moczek Armin P., and Nagy Lisa M.. 2005. “Diverse Developmental Mechanisms Contribute to Different Levels of Diversity in Horned Beetles.” Evolution & Development 7 (3): 175–85. 10.1111/j.1525-142X.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- Moczek Armin P., and Rose Debra J.. 2009. “Differential Recruitment of Limb Patterning Genes during Development and Diversification of Beetle Horns.” Proceedings of the National Academy of Sciences of the United States of America 106 (22): 8992–97. 10.1073/pnas.0809668106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro A, and Gupta MD. 2016. “Identifying Coopted Networks and Causative Mutations in the Origin of Novel Complex Traits.” Current Topics in Developmental Biology 119: 205–26. 10.1016/bs.ctdb.2016.03.003. [DOI] [PubMed] [Google Scholar]
- Monteiro Antónia. 2012. “Gene Regulatory Networks Reused to Build Novel Traits.” BioEssays 34 (3): 181–86. 10.1002/bies.201100160. [DOI] [PubMed] [Google Scholar]
- Nagy Olga, Nuez Isabelle, Savisaar Rosina, Peluffo Alexandre E., Yassin Amir, Lang Michael, Stern David L., Matute Daniel R., David Jean R., and Courtier-Orgogozo Virginie. 2018. “Correlated Evolution of Two Copulatory Organs via a Single Cis-Regulatory Nucleotide Change.” Current Biology 28 (21): 3450–3457.e13. 10.1016/j.cub.2018.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Tetsuya, Klomp Jeff, Pieretti Joyce, Schneider Igor, Gehrke Andrew R., and Shubin Neil H.. 2015. “Molecular Mechanisms Underlying the Exceptional Adaptations of Batoid Fins.” Proceedings of the National Academy of Sciences 112 (52): 15940–45. 10.1073/pnas.1521818112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhout H F. 1990. “Metaphors and the Role of Genes in Development.” BioEssays : News and Reviews in Molecular, Cellular and Developmental Biology. [DOI] [PubMed] [Google Scholar]
- Oliver Jeffrey C., Tong Xiao-Ling, Gall Lawrence F., Piel William H., and Monteiro Antónia. 2012. “A Single Origin for Nymphalid Butterfly Eyespots Followed by Widespread Loss of Associated Gene Expression.” PLoS Genetics 8 (8): e1002893. 10.1371/journal.pgen.1002893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson Eric N. 2006. “Gene Regulatory Networks in the Evolution and Development of the Heart.” Science (New York, N.Y.) 313 (5795): 1922–27. 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA 2000. “Adaptation and the Cost of Complexity.” Evolution; International Journal of Organic Evolution 54 (1): 13–20. 10.1111/j.0014-3820.2000.tb00002.x. [DOI] [PubMed] [Google Scholar]
- Otto Sarah P. 2004. “Two Steps Forward, One Step Back: The Pleiotropic Effects of Favoured Alleles.” Proceedings of the Royal Society B: Biological Sciences 271 (1540): 705–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özsu Nesibe, and Monteiro Antónia. 2017. “Wound Healing, Calcium Signaling, and Other Novel Pathways Are Associated with the Formation of Butterfly Eyespots.” BMC Genomics 18 (October). 10.1186/s12864-017-4175-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlicev Mihaela, Kenney-Hunt Jane P., Norgard Elizabeth A., Roseman Charles C., Wolf Jason B., and Cheverud James M.. 2008. “Genetic Variation in Pleiotropy: Differential Epistasis as a Source of Variation in the Allometric Relationship Between Long Bone Lengths and Body Weight.” Evolution 62 (1): 199–213. 10.1111/j.1558-5646.2007.00255.x. [DOI] [PubMed] [Google Scholar]
- Perry Michael W., Boettiger Alistair N., Bothma Jacques P., and Levine Michael. 2010. “Shadow Enhancers Foster Robustness of Drosophila Gastrulation.” Current Biology: CB 20 (17): 1562–67. 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter Isabelle S., and Davidson Eric H.. 2015. Genomic Control Process: Development and Evolution. 1 edition. London, UK ; San Diego, CA, USA: Academic Press. [Google Scholar]
- Phillips Patrick C. 2008. “Epistasis--the Essential Role of Gene Interactions in the Structure and Evolution of Genetic Systems.” Nature Reviews. Genetics 9 (11): 855–67. 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigliucci Massimo. 2008. “Is Evolvability Evolvable?” Nature Reviews Genetics 9 (1): 75–82. 10.1038/nrg2278. [DOI] [PubMed] [Google Scholar]
- Noon Preger-Ben, Ella Gonzalo Sabarís, Ortiz Daniela M., Sager Jonathan, Liebowitz Anna, Stern David L., and Frankel Nicolás. 2018. “Comprehensive Analysis of a Cis-Regulatory Region Reveals Pleiotropy in Enhancer Function.” Cell Reports 22 (11): 3021–31. 10.1016/j.celrep.2018.02.073. [DOI] [PubMed] [Google Scholar]
- Prud’homme Benjamin, Minervino Caroline, Hocine Mélanie, Cande Jessica D., Aouane Aïcha, Dufour Héloïse D., Kassner Victoria A., and Gompel Nicolas. 2011. “Body Plan Innovation in Treehoppers through the Evolution of an Extra Wing-like Appendage.” Nature 473 (7345): 83–86. 10.1038/nature09977. [DOI] [PubMed] [Google Scholar]
- Prum RO 1999. “Development and Evolutionary Origin of Feathers.” The Journal of Experimental Zoology 285 (4): 291–306. [PubMed] [Google Scholar]
- Prum Richard O. 2005. “Evolution of the Morphological Innovations of Feathers.” Journal of Experimental Zoology. Part B, Molecular and Developmental Evolution 304 (6): 570–79. 10.1002/jez.b.21073. [DOI] [PubMed] [Google Scholar]
- Prum Richard O., and Brush Alan H.. 2002. “The Evolutionary Origin and Diversification of Feathers.” The Quarterly Review of Biology 77 (3): 261–95. 10.1086/341993. [DOI] [PubMed] [Google Scholar]
- Rebeiz Mark, Jikomes Nick, Kassner Victoria A., and Carroll Sean B.. 2011. “Evolutionary Origin of a Novel Gene Expression Pattern through Co-Option of the Latent Activities of Existing Regulatory Sequences.” Proceedings of the National Academy of Sciences of the United States of America 108 (25): 10036–43. 10.1073/pnas.1105937108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz Mark, Patel Nipam H., and Hinman Veronica F.. 2015. “Unraveling the Tangled Skein: The Evolution of Transcriptional Regulatory Networks in Development.” Annual Review of Genomics and Human Genetics 16 (1): 103–31. 10.1146/annurev-genom-091212-153423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz Mark, and Tsiantis Miltos. 2017. “Enhancer Evolution and the Origins of Morphological Novelty.” Current Opinion in Genetics & Development 45 (August): 115–23. 10.1016/j.gde.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Gavin, and Rebeiz Mark. 2019. “Evolution: How Many Phenotypes Do Regulatory Mutations Affect?” Current Biology 29 (1): R21–23. 10.1016/j.cub.2018.11.027. [DOI] [PubMed] [Google Scholar]
- Rosenthal Robert S. 1979. “The File Drawer Problem and Tolerance for Null Results.” In . 10.1037/0033-2909.86.3.638. [DOI] [Google Scholar]
- Sabarís Gonzalo, Laiker Ian, Preger-Ben Noon Ella, and Frankel Nicolás. 2019. “Actors with Multiple Roles: Pleiotropic Enhancers and the Paradigm of Enhancer Modularity.” Trends in Genetics 35 (6): 423–33. 10.1016/j.tig.2019.03.006. [DOI] [PubMed] [Google Scholar]
- Saeij JPJ, Coller S, Boyle JP, Jerome ME, White MW, and Boothroyd JC. 2007. “Toxoplasma Co-Opts Host Gene Expression by Injection of a Polymorphic Kinase Homologue.” Nature 445 (7125): 324–27. 10.1038/nature05395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneuwly Stephan, Klemenz Roman, and Gehring Walter J.. 1987. “Redesigning the Body Plan of Drosophila by Ectopic Expression of the Homoeotic Gene Antennapedia.” Nature 325 (6107): 816–18. 10.1038/325816a0. [DOI] [PubMed] [Google Scholar]
- Sears Karen E., Capellini Terence D., and Diogo Rui. 2015. “On the Serial Homology of the Pectoral and Pelvic Girdles of Tetrapods.” Evolution 69 (10): 2543–55. [DOI] [PubMed] [Google Scholar]
- Shah Sandeep N., Cope Leslie, Poh Weijie, Belton Amy, Roy Sujayita, Talbot C. Conover Jr, Sukumar Saraswati, Huso David L., and Resar Linda M. S.. 2013. “HMGA1: A Master Regulator of Tumor Progression in Triple-Negative Breast Cancer Cells.” PLOS ONE 8 (5): e63419. 10.1371/journal.pone.0063419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubin Neil, Tabin Cliff, and Carroll Sean. 2009. “Deep Homology and the Origins of Evolutionary Novelty.” Nature 457 (7231): 818–23. 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- Spirov Alexander V., Sabirov Marat A., and Holloway David M.. 2012. “In Silico Evolution of Gene Cooption in Pattern-Forming Gene Networks.” The Scientific World Journal 2012 (December). 10.1100/2012/560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller Max V., Yan Dong, Randklev Sakara, Bragdon Meghan D., Wunderlich Zeba B., Tao Rong, Perkins Lizabeth A., DePace Angela H., and Perrimon Norbert. 2013. “Depleting Gene Activities in Early Drosophila Embryos with the ‘Maternal-Gal4–ShRNA’ System.” Genetics 193 (1): 51–61. 10.1534/genetics.112.144915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern David L. 2000. “Perspective: Evolutionary Developmental Biology and the Problem of Variation.” Evolution 54 (4): 1079–91. 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- Stern David L, and Virginie Orgogozo. 2008. “The Loci of Evolution: How Predictable Is Genetic Evolution?” Evolution; International Journal of Organic Evolution 62 (9): 2155–77. 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern David L., and Orgogozo Virginie. 2009. “Is Genetic Evolution Predictable?” Science 323 (5915): 746–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryamohan Kushal, Hanson Casey, Andrews Emily, Sinha Saurabh, Molly Duman Scheel, and Halfon Marc S.. 2016. “Redeployment of a Conserved Gene Regulatory Network during Aedes Aegypti Development.” Developmental Biology 416 (2): 402–13. 10.1016/j.ydbio.2016.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarchini Basile, Duboule Denis, and Kmita Marie. 2006. “Regulatory Constraints in the Evolution of the Tetrapod Limb Anterior–Posterior Polarity.” Nature 443 (7114): 985–88. 10.1038/nature05247. [DOI] [PubMed] [Google Scholar]
- Tomoyasu Yoshinori, Arakane Yasuyuki, Kramer Karl J., and Denell Robin E.. 2009. “Repeated Co-Options of Exoskeleton Formation during Wing-to-Elytron Evolution in Beetles.” Current Biology: CB 19 (24): 2057–65. 10.1016/j.cub.2009.11.014. [DOI] [PubMed] [Google Scholar]
- True John R., and Carroll Sean B.. 2002. “Gene Co-Option in Physiological and Morphological Evolution.” Annual Review of Cell and Developmental Biology 18 (1): 53–80. 10.1146/annurev.cellbio.18.020402.140619. [DOI] [PubMed] [Google Scholar]
- Verd Berta, Nicholas AM Monk, and Johannes Jaeger. 2019. “Modularity, Criticality, and Evolvability of a Developmental Gene Regulatory Network.” Edited by Lee Altenberg and Patricia J Wittkopp. ELife 8 (June): e42832. 10.7554/eLife.42832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent Ben J., Staller Max V., Francheska Lopez-Rivera Meghan D. J. Bragdon, Pym Edward C. G., Biette Kelly M., Wunderlich Zeba, Harden Timothy T., Estrada Javier, and DePace Angela H.. 2018. “Hunchback Is Counter-Repressed to Regulate Even-Skipped Stripe 2 Expression in Drosophila Embryos.” PLoS Genetics 14 (9): e1007644. 10.1371/journal.pgen.1007644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner Gunter P., and Altenberg Lee. 1996. “Perspective: Complex Adaptations and the Evolution of Evolvability.” Evolution 50 (3): 967–76. 10.2307/2410639. [DOI] [PubMed] [Google Scholar]
- Wagner Günter P., Kenney-Hunt Jane P., Pavlicev Mihaela, Peck Joel R., Waxman David, and Cheverud James M.. 2008. “Pleiotropic Scaling of Gene Effects and the ‘Cost of Complexity.’” Nature 452 (7186): 470–72. 10.1038/nature06756. [DOI] [PubMed] [Google Scholar]
- Wagner Günter P., and Zhang Jianzhi. 2011. “The Pleiotropic Structure of the Genotype–Phenotype Map: The Evolvability of Complex Organisms.” Nature Reviews Genetics 12 (3): 204–13. 10.1038/nrg2949. [DOI] [PubMed] [Google Scholar]
- Wasik Bethany R., and Moczek Armin P.. 2011. “Decapentaplegic (Dpp) Regulates the Growth of a Morphological Novelty, Beetle Horns.” Development Genes and Evolution 221 (1): 17–27. 10.1007/s00427-011-0355-7. [DOI] [PubMed] [Google Scholar]
- Welch John J., and Waxman David. 2003. “Modularity and the Cost of Complexity.” Evolution 57 (8): 1723–34. 10.1111/j.0014-3820.2003.tb00581.x. [DOI] [PubMed] [Google Scholar]
- West-Eberhard Mary Jane. 2005. “Developmental Plasticity and the Origin of Species Differences.” Proceedings of the National Academy of Sciences of the United States of America 102 Suppl 1 (May): 6543–49. 10.1073/pnas.0501844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead Andrew, and Crawford Douglas L.. 2006. “Neutral and Adaptive Variation in Gene Expression.” Proceedings of the National Academy of Sciences 103 (14): 5425–30. 10.1073/pnas.0507648103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womack Molly C., Metz Marissa J., and Hoke Kim L.. 2019. “Larger Genomes Linked to Slower Development and Loss of Late-Developing Traits.” The American Naturalist, August, 000–000. 10.1086/705897. [DOI] [PubMed] [Google Scholar]
- Wray Gregory A. 2007. “The Evolutionary Significance of Cis-Regulatory Mutations,” 11. [DOI] [PubMed] [Google Scholar]
- Wunderlich Zeba, Bragdon Meghan D. J., Vincent Ben J., White Jonathan A., Estrada Javier, and DePace Angela H.. 2015. “Krüppel Expression Levels Are Maintained through Compensatory Evolution of Shadow Enhancers.” Cell Reports 12 (11): 1740–47. 10.1016/j.celrep.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young Nathan M., and Hallgrímsson Benedikt. 2005. “Serial Homology and the Evolution of Mammalian Limb Covariation Structure.” Evolution 59 (12): 2691–2704. 10.1111/j.0014-3820.2005.tb00980.x. [DOI] [PubMed] [Google Scholar]
- Zakany Jozsef, and Duboule Denis. 2007. “The Role of Hox Genes during Vertebrate Limb Development.” Current Opinion in Genetics & Development 17 (4): 359–66. 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Zattara Eduardo E., Busey Hannah A., Linz David M., Tomoyasu Yoshinori, and Moczek Armin P.. 2016. “Neofunctionalization of Embryonic Head Patterning Genes Facilitates the Positioning of Novel Traits on the Dorsal Head of Adult Beetles.” Proceedings of the Royal Society B: Biological Sciences 283 (1834). 10.1098/rspb.2016.0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Jia, Payne Joshua L., and Wagner Andreas. 2019. “Cryptic Genetic Variation Accelerates Evolution by Opening Access to Diverse Adaptive Peaks.” Science 365 (6451): 347–53. 10.1126/science.aax1837. [DOI] [PubMed] [Google Scholar]