Abstract
Oxidative stress can damage organs, tissues, and cells through reactive oxygen species (ROS) by oxidizing DNA, proteins, and lipids, thereby resulting in diseases. However, the underlying molecular mechanisms remain to be elucidated. In this study, employing scanning ion conductance microscopy (SICM), we explored the early responses of human embryonic kidney (HEK293H) cells to oxidative DNA damage induced by potassium chromate (K2CrO4). We found that the short term (1−2 h) exposure to a low concentration (10 μM) of K2CrO4 damaged the lipid membrane of HEK293H cells, resulting in structural defects and depolarization of the cell membrane and reducing cellular secretion activity shortly after the treatment. We further demonstrated that the K2CrO4 treatment decreased the expression of the cytoskeleton protein, β-actin, by inducing oxidative DNA damage in the exon 4 of the β-actin gene. These results suggest that K2CrO4 caused oxidative DNA damage in cytoskeleton genes such as β-actin and reduced their expression, thereby disrupting the organization of the cytoskeleton beneath the cell membrane and inducing cell membrane damages. Our study provides direct evidence that oxidative DNA damage disrupted human cell membrane integrity by deregulating cytoskeleton gene expression.
Keywords: oxidative stress, oxidative DNA damage, β-actin, scanning ion conductance microscopy, cell membrane, chromate
1. INTRODUCTION
Oxidative stress resulting from endogenously and environmentally induced reactive oxygen species (ROS) can damage organelles and biomolecules1–3 including mitochondria, endoplasmic reticulum, peroxisomes,4–6 DNA, RNA, proteins, and lipids,7 resulting in cell dysfunction. Also, oxidative stress can target cytoskeleton to disrupt the organization of cytoskeletons. It has been shown that oxidative stress mediated by H2O2 and other types of free radicals can alter cell membrane property and actin polymerization8 and disrupt microfilament cytoskeleton,9 thereby causing changes of cell morphology.10–12 The studies further suggest that oxidative stress leads to the oxidation of sulfhydryl groups of cysteine of the key cytoskeleton protein, actin, and increases the carbonylation of the protein causing abnormality of cell morphology such as the formation of blebs on cell membrane.9 Since ROS can damage DNA, proteins, and lipids, it is possible that ROS may disrupt cell membrane integrity through multiple channels. It has been proposed that the cytoskeleton may be the primary line of proteins that are attacked by ROS during oxidative stress. However, it remains to be elucidated how oxidative stress can affect the integrity of the cell membrane. Given the fact that cytoskeleton proteins are constitutively expressed in cells to maintain cell morphology, it is conceivable that cytoskeleton genes are susceptible to oxidative DNA damage. This notion is supported by recent studies showing that oxidative DNA damage is associated with actin cytoskeleton remodeling.13–15 We further hypothesize that oxidative DNA damage can alter the integrity of the cell membrane by inducing DNA damage in the cytoskeletal genes and modulate the expression of the genes.
To test this hypothesis, we studied the effect of short-term exposure to the oxidative DNA damaging agent, potassium chromate (K2CrO4) on the membrane of human embryonic kidney (HEK293H) cells. We initially examined the structural changes of the cell membrane after chromate treatment using potentiometric scanning ion conductance microscopy (P-SICM), as shown in Figure 1. SICM is a scanning probe microscope (SPM) technique that uses a glass or quartz nanopipette as the scanning probe and ionic current through the nanopipette as the feedback signal to acquire the surface topography image with nanoscale resolution under physiological conditions.16–20 SICM can be applied to reveal the nanoscale changes of the cell membrane during endocytosis,21 the early interaction with virus particles22 or protein,23 and the cytotoxicity of ZnO nanoparticles.24 In our previous studies, we have utilized SICM to map and reveal submicrometer size changes of the cell membrane induced by cell-penetration polymer nanoparticles.25 Based on SICM, a potential sensing probe can be added to qualitatively map the surface charge/potential distribution of the cell membrane (see Figure 1), first introduced by Baker et al.26 We have used P-SICM to reveal both the topography and surface potential/charge changes induced by positively charged polymer nanoparticles27 and elevated extracellular potassium concentration.28 Here, our P-SICM studies found that the exposure to low concentrations of K2CrO4 did not obviously affect surface charge/potential of the cell membrane, but resulted in a roughened cell membrane and the appearance of submicrometer- to micrometer-sized defects in the HEK293H cell membrane, indicating the disruption of the cell membrane structure. The cell membrane damage is also confirmed by the transmembrane potential measurements, showing a less negative transmembrane potential after treatment with a higher chromate concentration. Moreover, by monitoring the ionic current flux through the nanopipette probe, we found that the cellular secretion activity was suppressed after the K2CrO4 treatment. We further demonstrated that the K2CrO4 treatment induced single-strand DNA (ssDNA) breaks in the β-actin gene by a newly invented DNA damage profiling assay. Accordingly, the cellular level of β-actin protein is decreased, as revealed by immunofluorescence and immunoblotting assays. These results indicate that oxidative DNA damage accumulated in the β-actin gene disrupts its transcription, leading to a reduced amount of β-actin protein. Subsequently, this disrupts the cytoskeleton structure underneath the membrane of HEK293H cells, resulting in structural defects in the cell membrane.
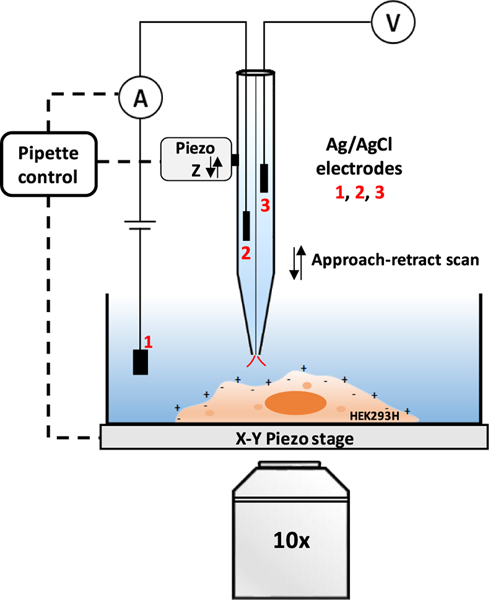
Figure 1.
Scheme of P-SICM on simultaneous recording of the topography and surface potential of cell membrane. The nanopipette is mounted on a Z-piezo actuator. The ion current that flows between the Ag/AgCl electrodes 1 and 2 is used as the feedback signal to control the distance between the nanopipette tip and the cell surface. Electrode 3 in another barrel is connected to a high-impedance differential amplifier to measure the extracellular potential near the pipette tip.
2. MATERIALS AND METHODS
2.1. HEK293H Cell Culture and Chromate Treatment.
The HEK293H cell line was obtained from ThermoFisher Scientific Invitrogen (Waltham, MA) and cultured with a DMEM/high glucose medium (GE Healthcare Life sciences, Piscataway) supplemented with fetal bovine serum 10% (ThermoFisher Scientific Gibco, Waltham, MA) in humidified incubator with 5% CO2 at 37 °C. Cells were seeded at a density of 1 × 105 per well. To record the transmembrane potential, cells were placed on a cover glass treated with poly-L-lysine 0.01% (Sigma-Aldrich). For the SICM imaging, cells were seeded on a polydimethylsiloxane (PDMS) substrate in a petri dish with 2 mm of thickness. The PDMS substrate was coated with collagen hydrogel matrix (5 μg/cm2) to improve the cell adhesion.29 If not mentioned otherwise, the HEK293H cells were treated with 10 μM K2CrO4 in extracellular solution for 90 min. For the fixed cell experiments by SICM, two batches of chromate treated cells were fixed with 4% paraformaldehyde for 10 min. Before patch-clamp or SICM experiments, the treated cells were rinsed two times with the extracellular solution.
2.2. Patch-Clamp Experiments.
The intracellular solution was prepared with the following composition: NaCl 8 mM, KCl 132.5 mM, MgCl2 2 mM, HEPES 10 mM, and 7.2 pH adjusted with NaOH. The extracellular or bath solution contained NaCl 130 mM, KCl 3 mM, CaCl2 2 mM, MgCl2 1.2 mM, HEPES 10 mM, and 7.4 pH adjusted with NaOH. The chemicals were purchased from Fisher Chemical and Sigma-Aldrich without further purification. All solutions were prepared using deionized water (∼18 MΩ) from a water purification system (Ultra Purelab system, ELGA/Siemens).
Borosilicate based glass micropipettes with filament (O.D. 1 mm, I.D. 0.58 mm, Sutter Instruments USA) were used after cleaning, and the micropipettes were prepared laser pipette puller (P-2000, Sutter Instruments, Novato, CA) with the following parameters: HEAT = 268, FIL = 4, VEL = 50, DEL = 225, and PUL = 100. The micropipettes were filled with the intracellular solution giving a final resistance between 4 and 5 MΩ.
Transmembrane potential recordings were performed at room temperature with the current-clamp configuration in whole-cell mode using the Axopatch 200B amplifier (Molecular Devices, San Jose, CA). Digidata 1440A controlled by the software Axoscope 10.5 (Molecular Devices) was used for the data acquisition.
2.3. Scanning Ion Conductance Microscopy.
2.3.1. P-SICM Setup and Nanopipettes.
The P-SICM (see Figure 1) is based on a commercial SPM (XE-Bio system, Park system Inc. Santa, CA) and mounted on the stage of the inverted microscope (Nikon Elipse Ti-U). Double-barrier quartz nanopipettes (O.D. 1.2 mm, I.D. 0.90 mm, Sutter Instruments USA) were prepared by the pipette puller with the following parameters: HEAT = 805, FIL = 3, VEL = 40, DEL = 220, and PUL = 165. The formed double-barrel nanopipettes with the diameter of each nanopore around 170−200 nm were filled with the extracellular solution and used as the probe for SICM imaging.27,30 It should be noted that the pore diameter is relatively bigger than the usual nanopore diameter below 100 nm size. The HEK293H cells show high secretion activities, which introduce a bigger noise in the measured ionic current during live cell imaging. We therefore used the nanopipettes with bigger nanopore diameter for all the measurements.
2.3.2. Imaging and Recordings.
As shown in Figure 1, P-SICM in the approach-retract scan (ARS) mode (also called hopping mode)19 was used to simultaneously acquire topography and surface membrane potential images. One barrel of the nanopipette was connected to a low noise current amplifier (DLPCA-200, FEMTO) with a 109 gain. The measured ionic current was used as the feedback to control the vertical position of the nanopipette tip. The second barrel was connected to a high impedance differential amplifier to measure the local potential change at the pipette tip. To improve the robustness of the potential image, we used the difference value ΔV to construct the potential image. The ΔV is defined as the potential difference between the potential values at the closest point and the farthest point from the cell surface in each ARS cycle.27
The topography image resolution was ∼80 to 100 nm/pixel with the typical setting here. The time to acquire an image of 128 × 128 pixels was about 25 min. The distance between the sample surface and tip of the pipette was controlled by a setpoint of a 1% decrease of the current. The tip approaching speed is about 75 μm/s, and the actual current change is about 8.2%. The current of the nanopipette (I), surface potential ΔV, and piezo Z displacement were recorded with an oscilloscope (Yokogama DL850 ScopeCorder) with a sampling rate of 3 kHz and a bandwidth of 500 or 400 Hz. Repeated SICM imaging was always conducted on different cells from different bathes to avoid cell-to-cell variations. All the measurements were conducted at room temperature in an HEPES bath solution.
2.3.3. Single-Point Measurement of the Ionic Current by SICM.
Nanopipettes were filled with extracellular solution and mounted on the SICM Z-piezo actuator and placed around 5 μm away from the membrane surface of HEK293H cells. The current was recorded with a constant sample bias 0.1 V applied in the bath solution.
2.4. Mapping of DNA Damage Profile in the Exon 4 of the β-Actin Gene Using DNA Damage Profiling Assay.
ssDNA breaks in the exon 4 of the β-actin gene were detected using DNA damage profiling assay recently invented by the Liu Laboratory. The technology was under the disclosure of patent application, and the details are reported somewhere else. Briefly, genomic DNA in untreated HEK293H cells and cells treated by 10 μM chromate for 120 min were isolated using a Wizard(R) Genomic DNA purification kit (Promega, Madison, WI). ssDNA breaks were subjected to 5′-phosphorylation at the 5′-end of the strand breaks and primer extension by vent DNA polymerase generating double-strand DNA fragments with varying sizes that resulted from the ssDNA breaks occurring at different nucleotides. DNA fragments were then ligated with a double-strand DNA barcode at 16 °C overnight. Subsequently, the ligated products were subjected to PCR amplifications with a primer labeled by 6-fluorescein (6-FAM). The PCR products were separated by capillary electrophoresis, and their sizes were determined by the GeneMapper software V.5.
2.5. Detection of β-Actin by Immunofluorescence.
HEK293H cells were seeded in an 8-well chamber slide (Ibidi, GmbH, Martinsried, Germany) at a density of 1 × 104 in 200 μL of culture medium. Cells were treated with 10 μM K2CrO4 for 120 min. Untreated cells were used as a negative control. After the chromate treatment, cells were washed with 1X PBS (pH 7.4) twice and fixed using 4% paraformaldehyde in 1X PBS at room temperature for 30 min. Cells were washed with 1X PBS for 5 min three times. Cells were then permeabilized with 0.1% Triton X-100 in PBS for 10 min and washed with 1X PBS for three times. Cells were then incubated with blocking buffer containing 1% BSA, 22.52 mg/mL glycine in PBST (PBS + 0.1%(v/v) Tween 20) for 30 min and were incubated with a mouse monoclonal anti-β-actin primary antibody at 1:1000 dilution (5 μg/mL)(Abcam, ab8226, Cambrige, MA) in PBST with 1% BSA overnight at 4 °C. Cells were subsequently washed three times with 1X PBS and incubated with an anti-mouse secondary antibody conjugated with Alexa-Fluor-594 (1:10000) (Abcam, ab150116, Cambridge, MA) in 1% BSA in PBS at room temperature for 1 h. Cells were washed with 1X PBS three times in the dark and incubated with 5 μg/mL DAPI in PBS for 1 min. Fluorescence images were acquired using a Nikon C1 confocal microscope equipped with a 60× objective and Nikon NIS Elements software.
2.6. Detection of β-Actin Using Immunoblotting.
For detection of β-actin in HEK293H cells using immunoblotting, 3 × 105 cells were seeded in a 2 mL culture medium in a 6-well plate overnight. Cells were treated with 10 μM K2CrO4 for 120 min. Untreated cells were used as a negative control. The cells were washed with 1X PBS and collected in PBS and pelleted by centrifugation at 3000 rpm for 5 min. The pellets were resuspended in ice-cold lysis buffer (0.1% v/v NP-40, 20% v/v glycerol, 1 mM EDTA, 200 mM KCl and 10 mM Tris-HCl, pH 7.8). The cell lysates were subjected to centrifugation at 12,000 rpm for 30 min. The supernatant of the lysates was collected. The protein concentrations of cell lysates were determined using Bradford assay. Cell lysates (30 μg protein) were mixed with 4× loading buffer and denatured at 95 °C for 5 min. After separation by SDS-PAGE, the proteins were transferred onto PVDF membranes. A SDS-PAGE gel was used as a loading control with the same amount of total proteins loaded. The gel was subject to Coomassie blue staining. The proteins in the lanes were used as loading controls. The PVDF membrane was subjected to blocking in TBST (20 mM Tris-base, 140 mM NaCl, 0.1% (v/v) Tween 20, pH 7.6) containing 1% (w/v) BSA and incubated with an anti-beta-actin primary antibodies (1:1000)(Abcam, ab8226, Cambridge, MA) at 4 °C overnight. The membrane was then washed with TBST three times and incubated with a HRP-labeled goat anti-mouse secondary antibody (1:10,000) (Abcam, ab6768, Cambridge, MA). The membrane was incubated with the ECL reagent (Pierce, Rockford, IL) for 5 min and exposed to an X-ray film (Fuji). The gel image was then developed by a film developer (SRX-101A, Konica Minolta, Wayne, NJ).
2.7. Data Analysis.
The SICM data analysis was accomplished with XEI (Park system Inc. Santa, CA), Gwyddion, Origin Pro (OriginLab Corp.) software and Matlab script (MathWorks) and LabVIEW programs. The potential difference images were constructed by LabVIEW programs; the 3D topography and enhanced color images were analyzed by XEI (Park Systems). The surface roughness was estimated by the root-mean square (RMS) of the whole topography image with a 5 × 5 μm size. Gaussian fit to the data was used to generate the mean value of ΔV. To avoid cell-to-cell variations, the experiments were always repeated several times on different cells and different batches. Results are reported as mean ± SD. The quantification of immunofluorescence imaging was performed using the Image J software. To compare the data groups, one-way ANOVA test and Mann Whitney test were employed and P < 0.05 indicates a statistically significant difference.
3. RESULTS AND DISCUSSIONS
3.1. Transmembrane Potential Change.
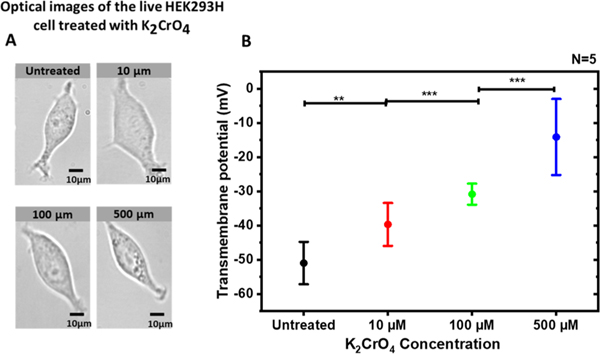
Transmembrane potential measurements were accomplished to assess the dose−response effects of K2CrO4 treatment on HEK293H cells. Cells were treated for 90 min with various K2CrO4 concentrations. As shown in Figure 2A, no apparent morphological changes of the live cells were observed in the bright-field microscope images after the chromate treatments with different concentrations. The footprint analysis of individual cells also reveals no noticeable differences (Figure S1). However, the measured transmembrane potentials become less negative with the increase of the extracellular potassium chromate concentration, as shown in Figure 2B. The depolarized cell membrane suggests that small damages likely occur in the cell membrane after the chromate treatment although they cannot be observed in conventional optical microscope images.
Figure 2.
Dose–response effect of potassium chromate on the transmembrane potential of the HEK293H cells. (A) Bright-field microscope images (40×) of the HEK293H cells after 90 min K2CrO4 treatment with various concentrations. (B) Transmembrane potential of the HEK293H cells after the treatment of K2CrO4 with different concentrations. The average transmembrane potentials are untreated (−50.9 ± 2.48 mV), K2CrO4 [10 μM] (−39.6 ± 2.5 mV), K2CrO4 [100 μM] (−30.8 ± 1.2 mV), and K2CrO4 [500 μM] (−14.1 ± 4.5 mV). For statistics with N = 5: untreated vs 10 μM, “**” P < 0.01; 10 μM vs 100 μM, “***” P < 0.001; 100 μM vs 500 μM, “***” P < 0.001. Both optical images and transmembrane potential measurements were taken about 90 min after the treatment.
To understand the cytotoxicity of chromate, we also conducted trypan blue exclusion tests (see Figure S2). After the 90 min treatment of 10 μM K2CrO4, most cells are alive without noticeable changes. With the increasing concentrations of K2CrO4, more cells exhibited cell death with a round shape and blue color under a bright-field microscope. However, the survived cells after chromate treatment with various dosages still proliferate in the following days, indicating that they recovered from the damage through self-repair after the shortterm exposure to K2CrO4. Because cell damages have been confirmed by the transmembrane potential measurement at the lowest dosage of 10 μM K2CrO4, we mainly studied the cells treated with this dosage in the following experiments if not mentioned otherwise.
3.2. Topography Studies of Fixed HEK293H Cells.
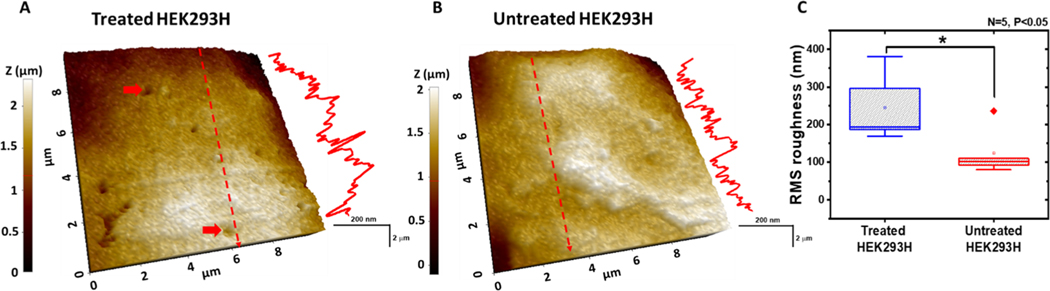
Because the bright-field optical microscope cannot resolve structural changes of cell membrane after chromate treatment, we explored the fine structural changes of the membrane of treated HEK293H cell by SICM. In the first step, we imaged fixed cells. The Figure 3A shows the 3D topography image of the membrane of treated cells. The height profile across the red dash line is shown at the right side of the topography image. For comparison, Figure 3B presented the typical topography data of the untreated cell. Additional topography images of treated and untreated cells are shown in Figure S3. Membrane defects, including micrometer- and submicrometer-sized pits and holes (some are indicated by the red arrow), are clearly observed in the topography image of the treated cell but not in the image of the untreated cell. In general, the treated cells appear rougher, which is also consistent with the height profile. As shown in Figure 3C, the statistical analysis of the treated cells shows an almost two-fold increase in the mean surface roughness (245.0 ± 40.6 nm) compared with the mean value of the untreated cells (124.0 ± 28.4 nm).
Figure 3.
SICM topography images of fixed HEK293H cells. (A, B) Topography of the cell membrane surface of the fixed HEK293H cell with (A) and without (B) 90 min treatment of 10 μM K2CrO4. The red arrows in (A) show damaged sites on the cell membrane. (C) RMS surface roughness of treated and untreated HEK293H cells. Treated vs untreated, “*” P < 0.05.
3.3. Topography and Surface Membrane Potential Studies of Live HEK293H Cells.
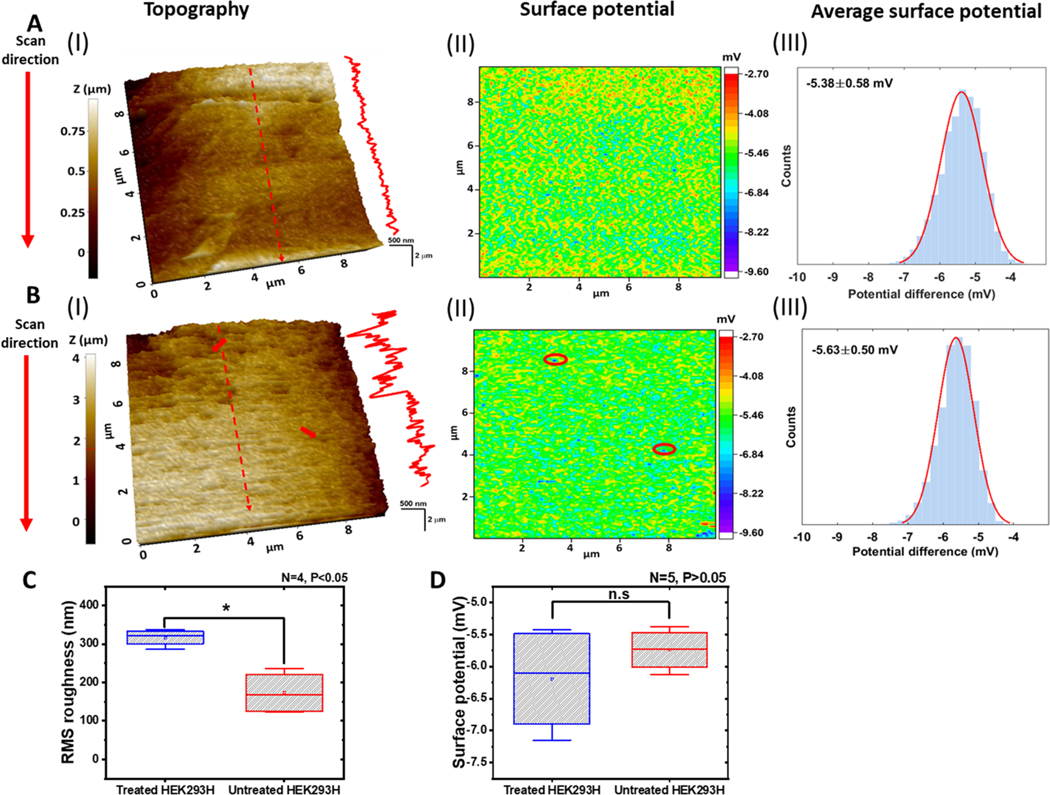
The cell membrane of live HEK293H cells was also imaged by P-SICM. More images are shown in Figure S4. Figure 4A (I) shows the typical topography image of the cell membrane of the untreated HEK293H live cell. A height profile across the red dash line is shown at the right side of the surface topography. The topography roughness was calculated to be 203 nm. Figure 4A (II) shows the corresponding potential ΔV image in the form of a heatmap with more negative potentials in blue color. The potential distribution over the cell membrane is relatively homogeneous. Figure 4A (III) shows the surface potential histogram, which was constructed by all the ΔV data points of the ΔV image in (II). The Gaussian fit (solid line) to the ΔV histogram gives a mean value of ΔV = −5.38 ± 0.58 mV.
Figure 4.
SICM images of live HEK293H cells. (A, B) Topography (I) and potential (II) images of the cell membrane surface of the untreated (A) and treated (B) HEK293H cells. The red arrows in (B (I)) point to two damage sites on the cell membrane. The same locations are circled in (B (II)). Height profiles are through the red dashed lines marked on the topography images. The potential distributions in (B (III)) with Gaussian fits (red solid lines) were generated from potential images (B (II)). (C) RMS surface roughness of treated and untreated cells. Treated vs untreated, “*” P < 0.05 with N = 4. (D) Mean surface potential of treated and untreated cells, Treated vs untreated, “n.s” P > 0.05 with N = 5. In all the SICM images, the scanning direction was from the top to the bottom.
A typical SICM image of treated live cells is shown in Figure 4B. The images were taken about 90 min after the treatment. As shown in Figure 4B (I), the surface of the treated cell appears rougher than that of the untreated cell. The increase of roughness is more obvious in the height line profile at the right side. The membrane topography presented a roughness of 381 nm, which is bigger than the untreated cells. The two red arrows show pore or pit like structures on the cell membrane, which appeared as blue color dots in the ΔV image in Figure 4B (II). In previous studies, we often observed the damaged sites of the cell membrane appear in blue color (more negative) in the potential image, which may be attributed to the more negative interior of the cell.27,28 In Figure 4B (III), the Gaussian fit to the ΔV histogram resulted in a mean value of −5.63 ± 0.50 mV. This value is still very close to that of the untreated cell in Figure 4A (III). Therefore, other than the defect sites, the overall surface charge was not greatly impacted by chromate treatment. In all the cases, the scanning was from the top to the bottom and the time to complete an image was about 25 min. The defects always appear less in the topography image at the later time of the imaging. We also notice that the defects in the topography images of the live cell membrane are less obvious as in the topography images of the fixed cell membrane. Therefore, the real time topography change of the live cell membrane during SICM imaging likely reveals the dynamic repair of the membrane by the cell after the damage.
As shown in Figure 4C, the RMS surface roughness value of treated cell was about two-fold rougher (316.0 ± 11.1 nm) than the value of untreated cells (173.0 ± 28.1 nm). In contrast, no significant differences were found in mean ΔV values between treated (−6.19 ± 0.84 mV) and untreated (−5.74 ± 0.33 mV) cells (Figure 4D). In light of these results, low-dose short term K2CrO4 treatment can impair the structure of the cell membrane, while the extracellular surface charge/potential of the membrane was not significantly modified.
3.4. Probing Cell Secretion Activities by Single-Point Time-Resolved Ionic Current Measurement.
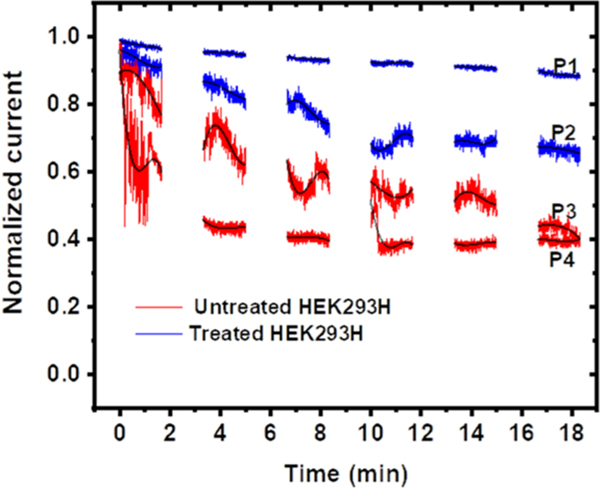
During SICM imaging of HEK293H live cells, we noticed that the magnitude of baseline ionic current gradually dropped over time. This non-random current change phenomenon has not been observed on several other cell lines we have imaged, including red blood cell, HeLa, and fibroblast cells. This is likely attributed to the high secretion activities of HEK293H cells.31 The secreted particles may attach to the glass surface of the nanopipette tip, leading to the partial blocking of ionic current through the nanoscale orifice of the nanopipette tip. We sought to use the current drop to indirectly measure the secretion activities of HEK293H cells. For all the measurements, the nanopipette tip was placed near the center of cells. Figure 5 shows the typical normalized ionic current changes over time on both untreated and treated HEK293H cells. The complete current−time traces are shown in Figure S5. Each current measurement was taken by a new nanopipette on a new batch of HEK293H cells. The measurements on treated cells were generally taken 90 min after the chromate treatment. On untreated cells, the ionic current exhibited a larger current drop over the same time span. In contrast, the ionic current change on treated cells is smaller. The current normally drops faster at the beginning along with more fluctuations and bigger RMS noise. Therefore, the adsorption of secreted particles to the glass nanopipette tip mainly occurred at the initial 5−10 min, and the current was gradually stabilized at the later time. This was attributed to the saturation of adsorption. The overall reduced current drop on treated cells suggests that less adsorption occurred at the nanopipette tip, which is attributed to a disruption of normal secretion activity by the chromate treatment. The chromate treatment often generates reactive oxidative species, such as hydroxyl radicals (OH·) inside the cells, suppressing the intracellular signaling.32,33 Therefore, the time-resolved ionic current measurements by the nanopipette near the cell membrane indicate the disrupted secretion activity of chromate-treated cells.
Figure 5.
Nanopipette single-point measurement near the cell surface. The time-dependent changes of normalized ionic current on two treated (blue color traces, recorded with nanopipettes P1 and P2) and untreated HEK293H cells (red color traces, recorded with nanopipettes P3 and P4).
3.5. K2CrO4 Induced DNA Damage in the Encoding Region of the β-Actin Gene.
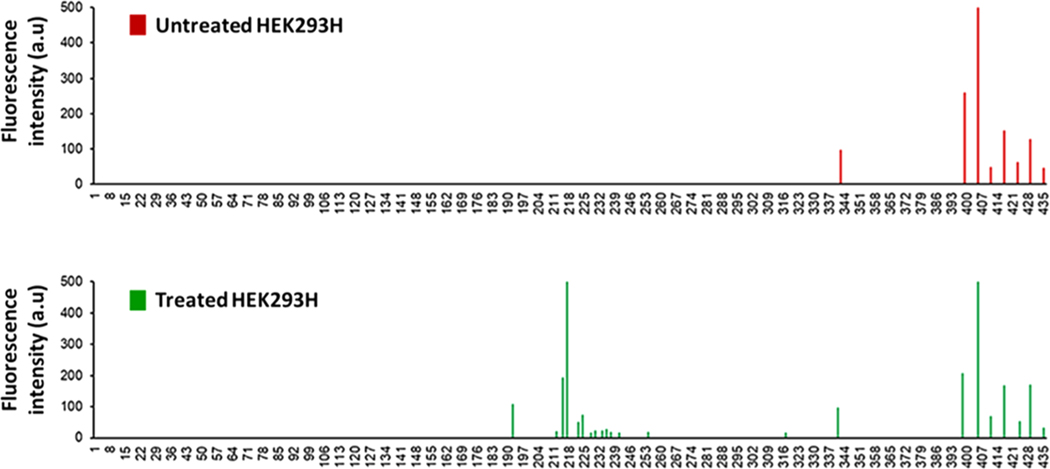
We then asked if K2CrO4 disrupted cell membrane integrity by inducing DNA damage in the β-actin gene that may in turn reduce the level of β-actin. We tested this hypothesis by detecting the profiles of ssDNA breaks in the exon 4 of the β-actin gene induced by 10 μM K2CrO4 using DNA damage profiling assay. We found that in untreated HEK239H cells, ssDNA breaks resulting from endogenous DNA damage that occurred at a few nucleotides located at the 3′-end of the exon 4 of the β-actin gene (Figure 6, panel on the top). Treatment of 10 μM K2CrO4 for 120 min resulted in the accumulation of ssDNA breaks at the nucleotides in the middle of the exon of the gene, as shown at bottom panel of Figure 6. The new features in the profile indicate that K2CrO4 treatment induced DNA damage in the encoding region of the β-actin gene. This further suggests that it is highly likely that oxidative DNA damage induced by chromate treatment results in downregulation of β-actin to disrupt the organization and structure of the cytoskeleton, thereby altering the integrity of cell membrane topology.
Figure 6.
Landscape profile of ssDNA breaks at the exon 4 of the β-actin gene in the untreated (top panel) and treated HEK293H cells (bottom panel). The peaks in the profiles represent ssDNA breaks located at a specific nucleotide at the exon 4 of the β-actin gene that are illustrated in the 5′ to 3′ direction counted from the initial nucleotide of the encoding sequence in the gene. The height of the peaks indicates the abundance of ssDNA breaks in the gene.
3.6. Chromate Treatment Reduced Cellular Level of β-Actin.
Since K2CrO4 treatment resulted in oxidative DNA damage in the encoding region of β-actin (Figure 6) and damages in the topography of the cell membrane surface (Figures 3 and 4), we reason that the chromate treatment may reduce the level of β-actin protein and thus disrupt the membrane structure of cells. To test this, we compared the level of β-actin protein in untreated and treated HEK293H cells using immunofluorescence.
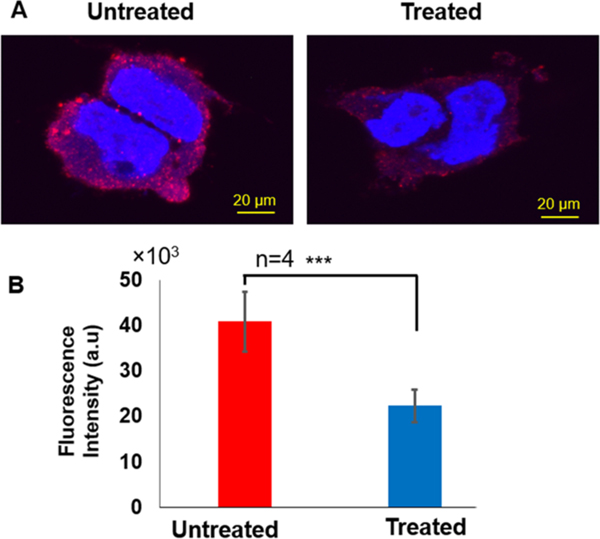
As shown in the fluorescence images in Figure 7A, the fluorescence from β-actin protein in the treated cell is much weaker. In Figure 7B, the mean fluorescence intensity of labeled β-actin protein obtained from individual untreated cells was about two-fold higher compared with the treated cells. We further measured the level of β-actin protein using immunoblotting. We found that the level of β-actin in both untreated and treated cells was similar (Figure S6A) and the relative amount of β-actin protein (vs control) in treated cells was only slightly lower than untreated cells (Figure S6B). The much smaller decrease of the β-actin protein level in immunoblotting result suggests that the total level of β-actin in cells was not significantly changed by the chromate treatment. Because the immunoblotting detected the β-actin level of the whole-cell extracts, the difference in the analysis of cellular β-actin between the two methods may suggest that the K2CrO4 treatment mainly reduced the level of β-actin protein underneath the cell membrane, leading to the disruption of cytoskeleton structure underneath cell membrane. Subsequently, the scarcity of β-actin underneath the cell membrane led to the formation of small pores in the cell membrane and increased permeability of cell membrane, which further decreased the transmembrane potential.
Figure 7.
Immunofluorescence staining of β-actin protein. (A) Immunofluorescence images for untreated (left) and treated (right) HEK293H cells. (B) The mean fluorescence intensity of β-actin protein in untreated (red bar graph) and treated (blue bar graph) HEK293H cells. There is a statistically significant difference between untreated and treated (“***” P < 0.001, n = 4). All experiments were performed in four biological replicates as described in Materials and Methods.
4. CONCLUSIONS
In summary, we used SICM to reveal the subtle structural damages on the HEK293H cell membrane upon short-term (90 min) exposure to the DNA damaging agent, K2CrO4. These changes were not observed in the bright-field microscope images, underscoring the capability of SICM imaging. The surface potential of the cell membrane was also probed by the P-SICM, and no detectable difference was observed between treated and untreated cells. We also showed that after the K2CrO4 treatment, the cells were depolarized with less negative transmembrane potential and cellular secretion activity was disrupted. We further explored the molecular mechanism underlying chromate-induced cell membrane damage. The immunofluorescence images and immunoblotting results revealed the decrease of β-actin proteins in the cell after the K2CrO4 treatment. Using DNA damage profiling assay, we further demonstrated that K2CrO4 treatment caused the accumulation of oxidative DNA damage that exhibited a unique profile in the encoding region of a key cytoskeletal protein, β-actin. Therefore, oxidative DNA damage reduced the level of β-actin protein especially underneath the cell membrane, thereby leading to the defects in the membrane, inducing cell membrane depolarization, and increasing permeability. It should be noted that the early responses induced by the short-term low concentration chromate treatment are not permanent. The real time SICM images of live cells reveal that the membrane damages are gradually repaired. This is also supported by the trypan blue staining experiments showing a high survival rate of cells in the following days. Our results provide new insights into the dynamic effects of oxidative stress on the cell membrane surface and the underlying molecular mechanisms by which oxidative DNA damage disrupts the structure and integrity of the cell membrane.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NSF (CBET1454544) to J.H and National Institutes of Health grant (R01ES023569) to Y.L. This work was also partially supported by the ERC program of NSF under NSF Cooperative Agreement no. EEC-1647837. P.S.T. was supported by Florida International University (FIU) Dissertation Year Fellowship. We acknowledge the use of facilities at the FIU Sequencing Core.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsabm.0c01461.
Footprint analysis of cell, trypan blue cell viability tests, additional SICM results of fixed and live cells, current time traces, and immunoblotting results (PDF)
AUTHOR INFORMATION
Complete contact information is available at: https://pubs.acs.org/10.1021/acsabm.0c01461
The authors declare no competing financial interest.
Contributor Information
Alberto S. Rubfiaro, Physics Department, Florida International University, Miami, Florida 33199, United States.
Pawlos S. Tsegay, Biochemistry Ph.D. Program, Florida International University, Miami, Florida 33199, United States.
Yanhao Lai, Department of Chemistry and Biochemistry, Florida International University, Miami, Florida 33199, United States.
Emmanuel Cabello, Physics Department, Florida International University, Miami, Florida 33199, United States.
Mohammad Shaver, Department of Biomedical Engineering, Florida International University, Miami, Florida 33199, United States.
Joshua Hutcheson, Department of Biomedical Engineering and Biomolecular Science Institute, Florida International University, Miami, Florida 33199, United States.
Yuan Liu, Biochemistry Ph.D. Program, Department of Chemistry and Biochemistry, and Biomolecular Science Institute, Florida International University, Miami, Florida 33199, United States.
Jin He, Physics Department and Biomolecular Science Institute, Florida International University, Miami, Florida 33199, United States.
REFERENCES
- (1).Kim J-H; Kang J-C Oxidative stress, neurotoxicity, and metallothionein (MT) gene expression in juvenile rock fish Sebastes schlegelii under the different levels of dietary chromium (Cr6+) exposure. Ecotoxicol. Environ. Saf. 2016, 125, 78–84. [DOI] [PubMed] [Google Scholar]
- (2).Winterbourn CC Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008, 4, 278–286. [DOI] [PubMed] [Google Scholar]
- (3).Redza-Dutordoir M; Averill-Bates DA Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta, Mol. Cell Res. 2016, 1863, 2977–2992. [DOI] [PubMed] [Google Scholar]
- (4).Poli G; Leonarduzzi G; Biasi F; Chiarpotto E Oxidative Stress and Cell Signalling. Curr. Med. Chem. 2004, 11, 1163–1182. [DOI] [PubMed] [Google Scholar]
- (5).Wise JTF; Wang L; Xu J; Zhang Z; Shi X Chapter 10 - Oxidative stress of Cr(III) and carcinogenesis. In The Nutritional Biochemistry of Chromium (III) (Second Edition); Vincent JB, Ed.; Elsevier: 2019; pp. 323–340. [Google Scholar]
- (6).Moldovan L; Moldovan NI Oxygen free radicals and redox biology of organelles. Histochem. Cell Biol. 2004, 122, 395–412. [DOI] [PubMed] [Google Scholar]
- (7).Ayala A; Muñoz MF; Argü S Lipid Peroxidation: Production, Metabolism, and Signaling Mechanisms of Malondialdehyde and 4-Hydroxy-2-Nonenal. Oxid. Med. Cell. Longevity 2014, 2014, 360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Zhu D; Tan KS; Zhang X; Sun AY; Sun GY; Lee JC-M Hydrogen peroxide alters membrane and cytoskeleton properties and increases intercellular connections in astrocytes. J. Cell Sci. 2005, 118, 3695–3703. [DOI] [PubMed] [Google Scholar]
- (9).Dalle-Donne I; Rossi R; Milzani A; Di Simplicio P; Colombo R The actin cytoskeleton response to oxidants: from small heat shock protein phosphorylation to changes in the redox state of actin itself. Free Radical Biol. Med. 2001, 31, 1624–1632. [DOI] [PubMed] [Google Scholar]
- (10).Mirabelli F; Salis A; Perotti M; Taddei F; Bellomo G; Orrenius S Alterations of surface morphology caused by the metabolism of menadione in mammalian cells are associated with the oxidation of critical sulfhydryl groups in cytoskeletal proteins. Biochem. Pharmacol. 1988, 37, 3423–3427. [DOI] [PubMed] [Google Scholar]
- (11).Jewell S; Bellomo G; Thor H; Orrenius S; Smith M Bleb formation in hepatocytes during drug metabolism is caused by disturbances in thiol and calcium ion homeostasis. Science 1982, 217, 1257. [DOI] [PubMed] [Google Scholar]
- (12).Lemasters JJ; DiGuiseppi J; Nieminen AL; Herman B Blebbing, free Ca2+ and mitochondrial membrane potential preceding cell death in hepatocytes. Nature 1987, 325, 78–81. [DOI] [PubMed] [Google Scholar]
- (13).Katsuno H; Toriyama M; Hosokawa Y; Mizuno K; Ikeda K; Sakumura Y; Inagaki N Actin Migration Driven by Directional Assembly and Disassembly of Membrane-Anchored Actin Filaments. Cell Rep. 2015, 12, 648–660. [DOI] [PubMed] [Google Scholar]
- (14).Wilson C; Gonzalez-Billault C´ Regulation of cytoskeletal dynamics by redox signaling and oxidative stress: implications for neuronal development and trafficking. Front. Cell. Neurosci. 2015, 9, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Meng F; Jiao X-F; Chen F; Zhang X-Y; Duan Z-Q; Ding Z-M; Wu D; Wang Y-S; Zhang S-X; Miao Y-L; Huo L-J Isobutylparaben Negatively Affects Porcine Oocyte Maturation Through Increasing Oxidative Stress and Cytoskeletal Abnormalities. Environ. Mol. Mutagen. 2020, 61, 433–444. [DOI] [PubMed] [Google Scholar]
- (16).Hansma P; Drake B; Marti O; Gould S; Prater C The scanning ion-conductance microscope. Science 1989, 243, 641. [DOI] [PubMed] [Google Scholar]
- (17).Ushiki T; Nakajima M; Choi M; Cho SJ; Iwata F Scanning ion conductance microscopy for imaging biological samples in liquid: a comparative study with atomic force microscopy and scanning electron microscopy. Micron 2012, 43, 1390–1398. [DOI] [PubMed] [Google Scholar]
- (18).Miragoli M; Moshkov A; Novak P; Shevchuk A; Nikolaev VO; El-Hamamsy I; Potter CMF; Wright P; Kadir SHSA; Lyon AR; Mitchell JA; Chester AH; Klenerman D; Lab MJ; Korchev YE; Harding SE; Gorelik J Scanning ion conductance microscopy: a convergent high-resolution technology for multiparametric analysis of living cardiovascular cells. J. R. Soc., Interface 2011, 8, 913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Novak P; Li C; Shevchuk AI; Stepanyan R; Caldwell M; Hughes S; Smart TG; Gorelik J; Ostanin VP; Lab MJ; Moss GW; Frolenkov GI; Klenerman D; Korchev YE Nanoscale live-cell imaging using hopping probe ion conductance microscopy. Nat. Methods 2009, 6, 279–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Korchev YE; Bashford CL; Milovanovic M; Vodyanoy I; Lab MJ Scanning ion conductance microscopy of living cells. Biophys. J. 1997, 73, 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Shin W; Gillis KD Measurement of Changes in Membrane Surface Morphology Associated with Exocytosis Using Scanning Ion Conductance Microscopy. Biophys. J. 2006, 91, L63–L65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Shevchuk AI; Hobson P; Lab MJ; Klenerman D; Krauzewicz N; Korchev YE Imaging Single Virus Particles on the Surface of Cell Membranes by High-Resolution Scanning Surface Confocal Microscopy. Biophys. J. 2008, 94, 4089–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Parres-Gold J; Chieng A; Wong Su S; Wang Y Real-Time Characterization of Cell Membrane Disruption by α-Synuclein Oligomers in Live SH-SY5Y Neuroblastoma Cells. ACS Chem. Neurosci. 2020, 11, 2528–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Yang X; Liu X; Lu H; Zhang X; Ma L; Gao R; Zhang Y Real-time investigation of acute toxicity of ZnO nanoparticles on human lung epithelia with hopping probe ion conductance microscopy. Chem. Res. Toxicol. 2012, 25, 297–304. [DOI] [PubMed] [Google Scholar]
- (25).Shan Y; Panday N; Myoung Y; Twomey M; Wang X; Li W; Celik E; Moy V; Wang H; Moon JH; He J Scanning Ion Conductance Microscopic Study for Cellular Uptake of Cationic Conjugated Polymer Nanoparticles. Macromol. Biosci. 2016, 16, 599–607. [DOI] [PubMed] [Google Scholar]
- (26).Chen C-C; Zhou Y; Morris CA; Hou J; Baker LA Scanning Ion Conductance Microscopy Measurement of Paracellular Channel Conductance in Tight Junctions. Anal. Chem. 2013, 85, 3621–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Chen F; Manandhar P; Ahmed MS; Chang S; Panday N; Zhang H; Moon JH; He J Extracellular Surface Potential Mapping by Scanning Ion Conductance Microscopy Revealed Transient Transmembrane Pore Formation Induced by Conjugated Polymer Nanoparticles. Macromol. Biosci. 2019, 19, 1800271. [DOI] [PubMed] [Google Scholar]
- (28).Chen F; Panday N; Li X; Ma T; Guo J; Wang X; Kos L; Ke H; Gu N; He J Simultaneous mapping of nanoscale topography and surface potential of charged surfaces by scanning ion conductance microscopy. Nanoscale 2020, 12, 20737–20748. [DOI] [PubMed] [Google Scholar]
- (29).Staunton JR; Doss BL; Lindsay S; Ros R Correlating confocal microscopy and atomic force indentation reveals metastatic cancer cells stiffen during invasion into collagen I matrices. Sci. Rep. 2016, 6, 19686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Nakajima M; Mizutani Y; Iwata F; Ushiki T Scanning ion conductance microscopy for visualizing the three-dimensional surface topography of cells and tissues. Semin. Cell Dev. Biol. 2018, 73, 125–131. [DOI] [PubMed] [Google Scholar]
- (31).Jeppesen DK; Hvam ML; Primdahl-Bengtson B; Boysen AT; Whitehead B; Dyrskjøt L; Ørntoft TF; Howard KA; Ostenfeld MS Comparative analysis of discrete exosome fractions obtained by differential centrifugation. J. Extracell. Vesicles 2014, 3, 25011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Benedikter BJ; Weseler AR; Wouters EFM; Savelkoul PHM; Rohde GGU; Stassen FRM Redox-dependent thiol modifications: implications for the release of extracellular vesicles. Cell. Mol. Life Sci. 2018, 75, 2321–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Krylatov AV; Maslov LN; Voronkov NS; Boshchenko AA; Popov SV; Gomez L; Wang H; Jaggi AS; Downey JM Reactive Oxygen Species as Intracellular Signaling Molecules in the Cardiovascular System. Curr. Cardiol. Rev. 2018, 14, 290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.