Abstract
Objective
Sharing health research data is essential for accelerating the translation of research into actionable knowledge that can impact health care services and outcomes. Qualitative health research data are rarely shared due to the challenge of deidentifying text and the potential risks of participant reidentification. Here, we establish and evaluate a framework for deidentifying qualitative research data using automated computational techniques including removal of identifiers that are not considered HIPAA Safe Harbor (HSH) identifiers but are likely to be found in unstructured qualitative data.
Materials and Methods
We developed and validated a pipeline for deidentifying qualitative research data using automated computational techniques. An in-depth analysis and qualitative review of different types of qualitative health research data were conducted to inform and evaluate the development of a natural language processing (NLP) pipeline using named-entity recognition, pattern matching, dictionary, and regular expression methods to deidentify qualitative texts.
Results
We collected 2 datasets with 1.2 million words derived from over 400 qualitative research data documents. We created a gold-standard dataset with 280K words (70 files) to evaluate our deidentification pipeline. The majority of identifiers in qualitative data are non-HSH and not captured by existing systems. Our NLP deidentification pipeline had a consistent F1-score of ∼0.90 for both datasets.
Conclusion
The results of this study demonstrate that NLP methods can be used to identify both HSH identifiers and non-HSH identifiers. Automated tools to assist researchers with the deidentification of qualitative data will be increasingly important given the new National Institutes of Health (NIH) data-sharing mandate.
Keywords: qualitative research data, deidentification, natural language processing, data sharing
INTRODUCTION
Qualitative research methods generate unstructured non-numeric textual data from patients, physicians, and other individuals in the form of interviews, focus groups, or narrative descriptions of health-related experiences. Qualitative research provides unique insight into health behaviors, attitudes, motivations, and subjective experiences and is especially useful for exploring sensitive, private, or stigmatizing experiences.1,2 Sharing health data is essential for transforming research into actionable knowledge that can impact health care services and outcomes. Data sharing is required by major United States (US) funders and deidentified biomolecular data, as well as data derived from electronic health records (EHR), including clinical notes, are increasingly being shared in order to better understand and improve health.3 Notably, qualitative healthcare data, including data collected outside of the EHR, are rarely shared.4,5 A major barrier to sharing qualitative data is the challenge of deidentifying unstructured non-numeric text.5
A new National Institutes of Health (NIH) data sharing policy mandates data sharing beginning in 2023 and does not distinguish between types of data to be shared. The policy defines data broadly as “recorded factual material commonly accepted in the scientific community as necessary to validate and replicate research findings” including unpublished data. The policy requires all NIH-funded investigators to submit Data Management and Sharing Plans to “integrate data sharing into the routine conduct of research.”6 Lack of data sharing plans can lead to termination of an award or impact future funding decisions and is an allowable cost. Researchers will need to be prepared for broader data sharing going forward—including qualitative data—in light of this revised NIH policy.
A substantial volume of data is generated in the healthcare domain through qualitative methods, suggesting this is an untapped resource available to supplement numeric and structured health data. A search of PubMed for qualitative health research articles including interviews and/or focus groups in English yields 11 507 published articles in the last 5 years. A majority of research projects funded through the National Human Genome Research Institute’s “ethical, legal, and social implications” (ELSI) program use qualitative or mixed methods.7 Eighty-two percent of initial research projects funded by the Patient-Centered Outcomes Research Institute (PCORI) include qualitative methods which are deemed essential to PCORI’s mission of ensuring patients’ voices are represented in research.8 PCORI also mandates that qualitative methods are used to inform the development of all validated measures and requires funded researchers to share all data generated.9–12
Qualitative research is resource intensive requiring significant time for collection, processing, and analyses.5 The benefits of qualitative data sharing (QDS) include saving research resources, reducing the data collection burden on participants, enabling secondary analyses, facilitating student training, and enhancing transparency, openness, and the ability to verify findings.4,5 In concurrent work, we explored the attitudes of qualitative researchers, data curators, institutional review board (IRB) members, and qualitative research participants regarding the barriers and benefits of QDS.4,13 While attitudes toward QDS vary, there is broad willingness and support of QDS among stakeholder groups. The biggest barriers to QDS currently are lack of knowledge, resources, and algorithms and support software, to facilitate the deidentification of qualitative data so that it can be responsibly and ethically shared in a data repository.4,13
The Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule provides 2 methods for deidentification of data: the Safe Harbor Method and expert determination.14 The HIPAA Safe Harbor method requires the removal of 18 identifiers and is the most commonly used process for deidentification. HIPAA also permits expert statistical determination in place of removing the 18 HIPAA Safe Harbor identifiers.14 However, deidentifying qualitative data presents unique challenges. The qualitative text contains identifiers—not considered HIPAA Safe Harbor identifiers (hereafter called HSH identifiers)—which in combination with other details could lead to reidentification of an individual. For instance, common redaction algorithms would remove all 18 HSH identifiers including names and dates, but dates may appear as “Christmas Eve 2004” in narrative text and would not be recognized as one of the 18 HSH identifiers. Narrative text may contain an identifier such as “CEO of Purina since 2010” rather than an individual first and last name. While neither “Christmas Eve 2004” nor “CEO of Purina since 2010” are HSH identifiers, they could identify an individual nonetheless—the former indirectly when mentioned as a birthday, for example, the latter more directly than a first and last name, which are rarely unique.
In addition, current deidentification tools have focused and trained on unstructured data such as clinical notes in EHRs but are not sufficient for qualitative research data which are structured differently. Qualitative research data mostly contain well-formed sentences occurring within structured conversations between multiple speakers, whereas clinical notes contain sentence fragments and observations by a single provider often using complex medical terms. Additionally, qualitative data are often transcribed and contain stutters, pauses, and timestamps. Some identifiers which are important to deidentify or redact from clinical notes—like timestamps—are generally only present in qualitative text because of transcription practices (ie, timestamps indicate words that could not be transcribed) and should not be marked for deidentification in qualitative data as it will create too many false positives, illustrating the unique nature of qualitative data.
Importantly, when qualitative data contain HSH identifiers, they are primarily names and geographic locations while the vast majority of HSH identifiers such as device, vehicle, medical record, social security, or fax numbers, are not present in qualitative data. Qualitative data are more likely to contain non-HSH identifiers than the majority of HSH identifiers. The majority of existing text deidentification tools and services rely on HIPAA-defined standards for deidentification and focus on removing only the 18 HSH identifiers and were developed for clinical records or narratives.15–22 Select text deidentification tools and services include the following limited number of non-HSH identifiers that are commonly found in clinical notes: age, gender, organizations, timestamp, and professions,17,23–25 but no system comprehensively covers all of these non-HSH identifiers. Philter, Scrubber, and Physionet tools are among a few of the state-of-the-art research-based tools developed and used for deidentification of clinical notes, but all of these tools have been trained for medical corpora and do not include identifiers beyond the HSH identifiers.15,16,19,26 Qualitative data are closer in content (vocabulary) and style (grammatical structure, syntactic structure) to general news articles and conversational data than the medical corpora. In the field of clinical notes deidentification, researchers have also used and shown the effectiveness of advanced computational methods like machine and deep learning in identifying HSH identifiers.27,28 The Irish Qualitative Data Archive (IQDA) Qualitative Data Anonymizer is one of the few tools developed specifically for qualitative data deidentification and contains features such as name mapping management and highlighting, but only recognizes names. However, the tool’s ability to automatically identify text believed to be Protected Health Information (PHI) is based on a user-supplied dictionary of words and their replacements is not very effective or comprehensive.29
In this paper, we describe the development of an automated computational framework for identification and removal of both HSH identifiers and non-HSH identifiers present in unstructured qualitative texts collected in biomedical and health settings. Currently, there is no standard for determining when such qualitative narrative text is adequately deidentified given the data contain non-HSH identifiers. For the purposes of this project, we define data as adequately deidentified when no one except the researcher(s) who gathered the data and the participant who provided it can recognize the individuals discussed in the text.30 We assume this standard will enable deidentified data to be deposited in a repository with restricted access, and potentially open access. This leaves open the question of whether other individuals—who are not the primary researcher or participant—could identify someone based on the data. Future research is needed to determine what is an adequate standard of deidentification, and if fewer or more identifiers should be included in the software.
We do not refer to the anonymization of data since some have suggested anonymity is almost never possible with qualitative research given that primary researchers who collect qualitative data will likely always be able to identify individual participants.30 Kayaalp distinguishes deidentification from anonymization as follows: deidentification describes a specific process or method to minimize the risk of reidentification of an individual, while anonymization is a goal rather than a specific method.26 The current project aims to balance preventing reidentification of an individual while ensuring adequate contextual detail remains in the data to enable others to analyze and interpret them.
OBJECTIVE
The goal of this study is to develop and evaluate a framework for deidentifying qualitative research data collected during healthcare research using automated computational techniques. Our goal is to achieve the deidentification of qualitative text which goes beyond the removal of the 18 HSH identifiers. To the best of our knowledge, this is the first study that focuses on deidentification of qualitative health research data not contained in the EHR. We developed a natural language processing (NLP) pipeline to deidentify the qualitative research data and validated the pipeline using qualitative expert evaluations.
MATERIALS AND METHODS
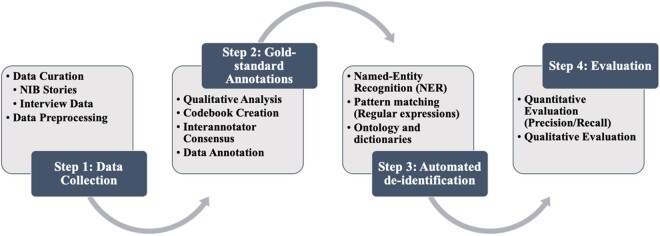
We performed an in-depth analysis and qualitative review of 2 different types of qualitative health research data, which resulted in the creation of gold-standard data containing HSH and non-HSH identifiers. Next, we used manual review to create a gold-standard dataset to develop and evaluate an NLP pipeline to automatically extract identifiers from qualitative texts and replace the identifiers with contextual replacement categories. Figure 1 provides a visual representation of our approach. We followed a multistep approach using NLP techniques, pattern matching, and dictionary-based identification.
Figure 1.
Overall approach for the qualitative research data review, development, and validation of the natural language processing (NLP)-based deidentification pipeline.
Step 1: Data collection and preprocessing
We used 2 types of qualitative research datasets for this study. The first dataset consisted of narrative texts in the form of stories written by patients, family caregivers, and healthcare providers published in the academic journal Narrative Inquiry in Bioethics (NIB). NIB publishes first-person narratives focused on a common healthcare topic or experience. The authors of NIB stories are typically not anonymous and often reveal sensitive and private information about themselves and others with their consent. The second dataset consisted of semistructured in-depth interviews with qualitative researchers, data curators, IRB members, and research participants regarding their attitudes toward QDS collected as part of this project. Interviews were audio-recorded and then transcribed verbatim by a professional transcription service.4,13
Both datasets used in this study provided unique challenges and opportunities for the deidentification analysis. The published NIB stories are reviewed by journal editors before publishing and are thus mostly grammatically well-structured narrative text. The NIB stories describe a wide variety of healthcare contexts including cancer care, end of life, vaccination preferences, and infertility. The second dataset from the interview study provides consistency in topic but created challenges due to the unstructured format of a transcript containing text of conversations between a researcher and participant, multiple transcription errors, and filler words such as “err” and “hmm.” The interview documents were preprocessed to remove any text containing words describing the actions of the interviewee such as [chuckles], [laughter], or [makes sputtering sounds]. We performed our deidentification analysis on both datasets, which provided considerable advantages over past attempts to develop a deidentification framework using qualitative research documents and not clinical notes and/or medical records.15–19,23–25
Step 2: Qualitative review and analysis
Qualitative research and ethics subject matter experts analyzed the collected NIB stories and interview transcripts with the following goals: (1) to understand and identify the kinds of identifiers most commonly found in qualitative texts; (2) to suggest additional categories other than the 18 HSH identifiers that can potentially reveal an individual’s identity; and (3) to create a gold-standard dataset that can be used in the next step for the deidentification modules of identification and substitution.
Members of the research team used the qualitative data analysis software Dedoose, which enables multiple users to code the same documents on the cloud and blinded to one another, to annotate identifiers in health-related stories.31 We used an established qualitative coding process of consensus coding.32,33 In the first stage, we created a codebook with operationalizations of the non-HSH identifiers we hoped to capture and the 18 HSH identifiers. When the first codebook was finalized, multiple independent coders annotated the 15 pilot datasets blinded to one another (4 coders annotated NIB stories, 3 coders annotated QDS interview files). Coding was manually checked for every instance of coder agreement and disagreement and each instance of disagreement was discussed by the entire team. Codebook development involved multiple meetings with the research team to operationalize, refine, and determine which additional non-HSH identifiers should be included or potentially rejected. The additional 55 files coded during the second phase involved a similar coding process with 2 independent coders, and using a refined codebook after the first round of coding.
Disagreement between coders generally fell into 2 categories. In some cases, disagreements were due to coder error where a coder simply missed an identifier that should have been coded according to the codebook. In these cases, after team discussion coding was corrected. The second area of disagreement occurred when definitions of variables were vague. In these cases, we refined definitions to improve operationalization.
Additionally, some variables we originally intended to flag were dropped. In initial phases of codebook development, we had hoped to include broad categories for things we deemed to be personally identifying and sensitive such as stigmatizing illnesses. Therefore, we initially tried to capture any health-related information under a code called “health status,” but soon recognized this was an enormous category that included any mention of a disease, treatment, disability, symptoms, medications, and more. Flagging every instance of any health status information removes essential contextual detail and is generally not personally identifying. There are many diseases, such as Alzheimer’s Disease, which may be stigmatized but are not rare enough to be personally identifying and provide critical contextual details about an individual. The team determined that capturing rare diseases only, as defined by a rare disease list, would enable us to capture potentially individually identifying information due to a rare disease, while leaving much health-related information present in the text.
Similarly, we considered including gender as this could be an identifier (ie, males with breast cancer) but flagging every instance of gender in a transcript also required removing all gendered words such as waitress, husband, wife, son, daughter, uncle, mother, he, or she, which are ubiquitous in transcripts. The team rejected including gender because it would adversely affect the readability of a transcript and because gender is an important piece of contextual information. Additionally, “sex as a biological variable” must be reported to NIH. We also included a category of “numbers” in our non-HSH identifiers as numbers often reveal unique traits (ie, being born with 3 limbs) or outlier values (ie, weight, height, or number of children) that can be individually identifying. We did, however, exclude the number “one” from our pipeline because this is both a number and a pronoun (eg, “one should always wear sunscreen”), and flagging every instance of “one” generates too many false positives.
We initially planned to include all instances of professions, but professions were present throughout transcripts suggesting they provide important context, and removing all professions adversely affects the readability of data. This category was also difficult to operationalize during coding because the category was overly broad and could include terms such as “mayor of,” “boss”, “CEO,” “researcher,” “faculty,” or “graduate students” that often were not individually identifying. By including the non-HSH organization/institution category in our pipeline, the team evaluated the typical risks and then determined that a profession could likely remain in the data without being individually identifying, for instance “CEO of Purina” would become “CEO of Inst/Org.”
Ultimately, the team determined that smaller, more narrowly defined categories of identifiers were preferable because they could be well operationalized for annotation, were more likely to be personally identifying, and also enable more granularity for users in potentially determining what types of identifier categories to look for within the data.
Our final codebook contained 2 broad categories of identifiers: 18 HSH identifiers and 8 additional categories of non-HSH identifiers (see Table 1) that must be removed, replaced with more general terms, or at least evaluated to achieve an adequate degree of deidentification of qualitative research data.
Table 1.
List of categories identified during qualitative analysis
| Category name | Identifier category and classification |
|---|---|
| Name | HSH: Names |
| Location |
|
| Date/time/age |
|
| Numbers |
|
| Web emails/URLs | HSH: Email addresses, web universal resource locators (URLs), internet protocol (IP) addresses. |
| Organization | Non-HSH: Institution or organization name: References to the name of an institution or organization that is not categorized as an HSH geographic region smaller than a state such as “Barnes-Jewish Hospital” or “Washington University in St Louis” which are not actual addresses and constitute multiple potential locations. Proper names of institutions or organizations would go here such as “Pfizer” or “World Health Organization”. |
| Rare diseases | Non-HSH: Commonly recognized rare diseases obtained from public databases. |
| Race, ethnicity | Non-HSH: References to NIH racial/ethnic categories, indigenous status, or nationality such as “Most patients were from Haiti” “A Hispanic nurse working on the psychiatric ward treated me”. |
| Sexual orientation | Non-HSH: Reference to sex, gender, or sexual orientation that is not heterosexual including LGBTQI. |
| Other | Non-HSH: Rare events and other rare references not captured under any existing category and that are unlikely to be captured by automation such as “He won the Olympic gold medal for swimming in Houston” or “Nobel laureate in 1995”. |
Note: Each category contained HSH and/or non-HSH identifiers. Identifier text was replaced by its corresponding category name in the deidentified text.
HSH: HIPAA Safe Harbor.
Step 3: Identification and substitution algorithm
We then developed an NLP pipeline for identifying different types of HSH and non-HSH identifiers in text data. In addition, we have utilized and customized some components from 2 existing toolkits Natural Language Tool Kit (NLTK) and Stanford Named Entity Recognizer34 in implementing our 18 HSH identifier modules including names, location, telephone numbers, and email addresses.
The identification of non-HSH identifiers was accomplished by using various NLP techniques. In order to detect various categories of identifiers including names, location, and organization, we used a set of pretrained models for Named Entity Recognition (NER). NER models are built to label sequences of words in textual data that represent an entity such as names of things, including person and company names, or location. One of the more common categorizations of this type of data is into person, location, and organization, which was used in the MUC-7 Named Entity task.35,36 These categories map very closely to the aspects in the text that need to be identified and suggested for the removal of identifiers. For this study, we used the publicly available 3- and 7-class NER classifiers from StanfordNER, which are pretrained models to identify person, location, and organization in newswire text,37 to suggest identifiers. NERs (including StanfordNER) typically utilize conditional random fields (CRFs) to identify different categories of entities and label sequential data such as text. A key feature of CRFs is that they are able to take into account the context surrounding words.
We also implemented some regular expressions-based pattern matching to extract identifiers such as time, date, age, and other numbers. Lastly, we used dictionaries for some of the identifier categories, including rare diseases and sexual orientation, and race/ethnicity for which we created a repository of possible words representing each category and extracted the identifiers based on their match. We also used the dictionary approach for certain special dates such as the holidays “Thanksgiving” or “Memorial Day.”
After developing the above methods for identifying HSH and non-HSH identifiers in qualitative text documents, the next step in the deidentification pipeline was to determine appropriate substitution texts for the identifiers to maintain the readability of deidentified text and minimize information loss. The category names defined in Table 1 were used to create unique substitution texts for each identifier.
Step 4: Performance evaluation
We empirically evaluated the accuracy of the NLP pipeline by comparing the tagged identifiers with the gold-standard annotations. Gold-standard annotations were produced by 4 team members who reviewed text independently and then met to achieve consensus or refine the operationalization of variables where discrepancies in rating existed. The metrics used for the evaluation are:
Precision (measure of exactness) P = Ncorrect/[Ncorrect + Nspurious]
Recall (measure of completeness) R = Ncorrect/Nexpected
F1-score (weighted average of precision and recall).
We implemented our deidentification and evaluation pipeline in the Python programming language and used shell scripting for integration of software modules. We reviewed the incorrect instances (false positives and false negatives) to understand the potential reasons for the incorrect identification.
RESULTS
We collected more than 400 qualitative research data documents (with 1.2 million words) and deidentified them using our NLP pipeline. These data documents were sourced from 2 distinct datasets. The first dataset contained 304 unique stories by patients, family caregivers, and healthcare providers published in the academic research journal NIB. The second dataset consisted of 120 semistructured interviews conducted with 4 unique participant groups. Table 2 provides the descriptive statistics for the NIB and interview datasets and the number of identifier tokens tagged by our NLP pipeline for each set.
Table 2.
Descriptive statistics of the 2 datasets used in the study
| Dataset 1 (NIB stories) | Dataset 2 (Interviews) | Total | |
|---|---|---|---|
| Number of files | 304 | 120 | 424 |
| Number of word tokens | 547 733 | 683 580 | 1 231 313 |
| Mean length of file (# word tokens) | 1801.75 | 5696.50 | 2904.04 |
NIB: Narrative Inquiry in Bioethics.
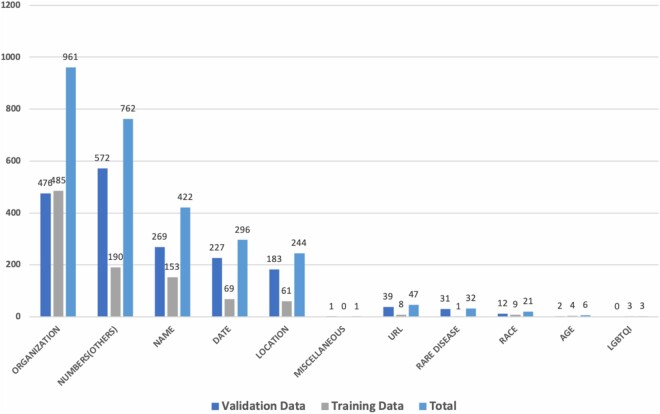
Figure 2 provides the distribution of different identifier categories in the 2 qualitative research datasets. We observe a difference in the distribution of the categories, owing to the nature of the datasets. Since the first dataset contained personal stories and experiences written by patients, caregivers, or healthcare providers, they contained many more identifiers in Name (HSH) and Organization (non-HSH) categories. In contrast, qualitative interviews contained far fewer identifiers in Names (HSH) but each qualitative transcript contained a median of 5 unique identifier categories (range 2–8/transcript), with a mean of 40 identifiers/transcript across all categories.
Figure 2.
Distribution of various identifier categories HIPAA Safe Harbor (HSH identifiers) and non-HSH in the 2 datasets.
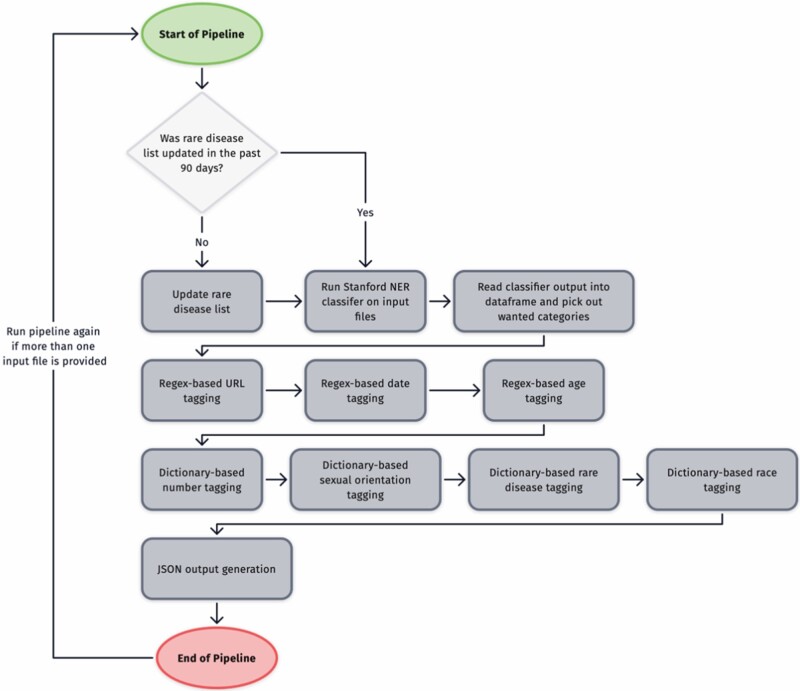
We empirically evaluated the accuracy of the NLP pipeline by comparing the tagged identifiers with the gold-standard annotations on 70 (15 pilot followed by 55 additional files) qualitative research documents. The 15 pilot files were used to iteratively develop the NLP pipeline described in Figure 3. Once developed, the pipeline was tested on 55 additional files. The metrics used for the evaluation were precision, recall, and F-score. Table 3 provides the descriptive statistics for the 2 datasets and the number of identifiers/tokens tagged by our NLP pipeline.
Figure 3.
The flowchart of how our NLP pipeline deidentifies the qualitative research documents.
Table 3.
Descriptive statistics of the 2 datasets used in the study and the number (%) of identifiers (HSH and non-HSH) extracted using the NLP pipeline from each set; and gold-standard evaluation of the NLP system
| Dataset name (number of files) | Token count | Identifier count (%) | Precision | Recall | F1 score | |
|---|---|---|---|---|---|---|
| Pilot files | NIB stories (6 files) | 12 620 | 389 (3 %) | 0.93 | 0.92 | 0.93 |
| QDS interviews (9 files)—Iteration 1 | 85 590 | 650 (1 %) | 0.98 | 0.83 | 0.90 | |
| QDS interviews (9 files)—Iteration 2a | 85 590 | 650 (1 %) | 0.98 | 0.90 | 0.94 | |
| Additional files | NIB stories (25 files) | 48 807 | 858 (2%) | 0.93 | 0.98 | 0.95 |
| QDS interviews (30 files)—Iteration 1 | 139 323 | 998 (1%) | 0.97 | 0.81 | 0.88 | |
| QDS interviews (30 files)—Iteration 2a | 139 323 | 998 (1%) | 0.97 | 0.95 | 0.96 | |
| Total | 70 | 286 340 | 2888 (1%) | 0.95 | 0.88 | 0.91 |
| Total—Iteration 2a | 70 | 286 340 | 2888 (1%) | 0.95 | 0.96 | 0.96 |
We performed an error analysis after Iteration 1 and observed that a single name of the organization which repeated as part of an interview question in every transcript of dataset 2, was being missed by our pipeline and driving the low recall. Iteration 2 results show the performance of the pipeline after the removal of one problematic organization name that was not recognized.
HSH: HIPAA Safe Harbor; NIB: Narrative Inquiry in Bioethics; QDS: qualitative data sharing.
Our results demonstrate that only a small percentage of word/word tokens in qualitative research data contains any form of HSH or non-HSH identifiers. The number of identifiers varied in our 2 datasets but ranged between 1% and 3% of the total word tokens (Table 3). These identifiers are located within large volumes of unstructured qualitative text, making manually locating them challenging. Our NLP pipeline has a consistent F1-score of ∼0.90 for both QDS and NIB datasets.
We performed an error analysis of the above results (Iteration 1), by analyzing the false-positive and false-negative identifiers. We observed that a single name of the organization which repeated as part of an interview question in every transcript of dataset 2, was being missed by our pipeline and driving the low recall. We identified that this organization name was missed due to both the unique format of the organization name, as well as the length and the structure of the whole sentence in which it occurred. The sentence contained 3 long-named organizations with few connecting words in the middle. Hence, we revised our pipeline analysis (Iteration 2) by adding the one organization name as a dictionary item to showcase the pipeline performance for dataset 2 (Table 3) after the removal of one problematic organization name. We thought that this was a reasonable approach as users of a deidentification system would likely have used the system in a similar manner. The majority of the other identifiers missed belonged to the Others non-HSH category (see “Discussion” section).
DISCUSSION
The goal of this study was to establish and evaluate a framework for deidentifying qualitative research data collected during healthcare research using automated computational techniques including removal of non-HSH identifiers commonly found in unstructured qualitative data. This study is uniquely focused solely on developing novel predictive deidentification algorithms with contextual substitutions to enhance the deidentification of qualitative health research data that includes non-HSH identifiers. We performed both qualitative and computational analysis of qualitative research data documents and obtained insights into the occurrence and nature of identifiers in these documents and the effectiveness of computational models in deidentification. We performed a gold-standard, annotation-based validation to evaluate the performance of the NLP pipeline in accurately deidentifying the qualitative research data. Our automated NLP pipeline significantly improves (consistent F1 score of ∼0.90) on prior attempts29,38 and is uniquely able to identify both HSH and 8 additional categories of non-HSH identifiers in unstructured qualitative text.
We found very few HSH and non-HSH identifiers in qualitative text (1–3% of all words). The low overall frequency is due to the large volume of unstructured qualitative text wherein identifiers are located; manually locating them would be extraordinarily time-consuming. Importantly, the majority of identifiers found were non-HSH identifiers, with the only exception being a large number of individual names contained in the first set of NIB stories. The presence of HSH names in NIB stories is unsurprising as they are published narratives where authors have frequently agreed to identify themselves and others and are required to obtain informed consent from others who might be identified in their detailed narrative. However, this finding suggests that some forms of qualitative data, such as ethnographies or detailed field notes taken by a researcher, may contain more identifiers and present greater challenges in terms of deidentification.
In contrast, qualitative interviews conducted between an individual and a researcher contained primarily non-HSH identifiers. Notably, qualitative interviews and focus groups are the most common methods used in qualitative health science research, and are likely representative of the majority of qualitative data collected. The only HSH identifiers contained in interview transcripts were names and a URL, but the remaining 16 HSH identifiers were not present. Here again, this finding is not unexpected given that many of the HSH identifiers relate to medical record, device, fax, telephone, or social security numbers, that we would not expect to be present in qualitative interviews where people describe experiences and attitudes. The majority of identifiers present in interviews were non-HSH. Each qualitative transcript contains a median of 5 unique identifier categories, with a mean of 40 identifiers/transcript across all categories. The most common identifier was organization (median 9/transcript), which is non-HSH and not currently captured by the majority of systems. While there are no standards to determine when qualitative texts are sufficiently deidentified, the presence of 5 unique identifier categories per transcript is sufficient to warrant removal as individual identity could be inferred from their combination.
Our findings indicate that non-HSH identifiers, such as organization, are particularly important for deidentification of qualitative data. For example, consider a participant who is a Hispanic, male nurse. Hispanic male nurses are not unique within the United States, but such a description may be highly unique at a particular hospital or research site, which indicates that research location is an important variable for deidentification especially when research is conducted at a single site. By not disclosing the study location or by conducting a study at multiple sites, qualitative researchers could reduce the likelihood that any individual could be reidentified and this may be a best practice going forward. Nevertheless, considering that most of the identifiers are only “indirect” (requiring more than one piece of information to infer an identity) and that the pipeline has a consistent F1 score of ∼0.90, the resulting data are likely highly deidentified.
Future research is needed to determine whether additional non-HSH identifiers should be included in our pipeline or whether some of our existing categories may be irrelevant going forward. For instance, we considered including professions as one of our non-HSH identifiers but determined that this detail may be contextually important and could likely remain in the data so long as other non-HSH identifiers such as location/organization are removed. However, our NLP pipeline may need to include additional categories of non-HSH identifiers, such as profession, going forward.
Some identifiers are simply not feasible to be extracted using automated computational techniques, a category we designate as “other” to indicate identifiers that are not captured under any existing category (Table 1). For instance, Olympic gold medalist swimmer would not be captured using our algorithm as either an HSH or non-HSH identifier. However, this piece of information could be as identifying as providing a first and last name if it is combined with other details such as the year of the games, or the location (eg, Houston). This indicates that tools for dedentification of qualitative data will always require human input to search for identifiers that will not be found automatically and confirm when data have been adequately deidentified.
We believe that automated computational methods can assist researchers who wish to share qualitative data in an efficient and ethically responsible manner, and will be especially important in light of the revised NIH data sharing policy. Given the complexity of identifiers present in qualitative research data, the goal of automatic deidentification should be to improve the efficiency and quality of the processes rather than to replace the need for careful attention from a highly trained human user. There are some identifiers that will only be found by a human user, such as those in the “other” category. The ultimate goal of our research is to develop a user-friendly cloud-based application that will enable users to deidentify qualitative text data, view the substitutions proposed and validate, customize, and download the final deidentified text. Given that there are no automated tools to assist qualitative researchers, who currently must manually deidentify data, our work represents a significant advance.
CONCLUSIONS
Identifiers in qualitative documents are more likely to be non-HSH and to our knowledge, there are no existing tools that capture both HSH and all 8 of the non-HSH identifiers our team has determined are present in qualitative healthcare data. Computational methods utilizing NLP methods can be effectively employed to deidentify qualitative research data. We propose to extend on our current work by utilizing machine-, deep learning, and ontology-based analyses to significantly enhance our deidentification capabilities and generate substitutions that avoid information loss. In addition, there is a need to conduct qualitative studies to determine whether we are capturing sufficient non-HSH identifiers in our system by gathering evidence directly from participants and researchers regarding the acceptability of deidentified qualitative data in masking individual identity while maintaining adequate contextual detail.
FUNDING
This project was supported by a grant from the National Human Genome Research Institute of the U.S. National Institutes of Health under award number, R01HG009351 and the National Center for Advancing Translational Sciences under award number UL1TR002345. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Human Genome Research Institute.
AUTHOR CONTRIBUTIONS
Study concept and design: J.D., A.L., A.G., and J.M.; Data collection: A.G., J.M., and H.W.; Analysis and interpretation of data: J.M.D., A.L., A.G., and J.M.; Draft of the manuscript: A.G., A.L., J.M., and X.M.; Review, revisions, and approval of final manuscript: A.G., J.M., A.L., and J.D. J.D. provided overall study supervision.
ACKNOWLEDGMENTS
The authors would like to thank Meredith Parsons, Kari Baldwin, Heidi Walsh, Elyssa Smith, Ruby Varghese, and Cynthia Hudson Vitale for their contributions to coding and annotations.
Contributor Information
Aditi Gupta, Institute for Informatics, Washington University, St. Louis, Missouri, USA .
Albert Lai, Institute for Informatics, Washington University, St. Louis, Missouri, USA .
Jessica Mozersky, Bioethics Research Center, Division of General Medical Sciences, Washington University, St. Louis, Missouri, USA.
Xiaoteng Ma, Institute for Informatics, Washington University, St. Louis, Missouri, USA .
Heidi Walsh, Bioethics Research Center, Division of General Medical Sciences, Washington University, St. Louis, Missouri, USA.
James M DuBois, Bioethics Research Center, Division of General Medical Sciences, Washington University, St. Louis, Missouri, USA.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The dataset rom the NIB stories is available through the Journal Narrative Inquiry in Bioethics, Johns Hopkins University Press. The second dataset of qualitative interviews underlying this article cannot be shared publicly due to the privacy of individuals that participated in the study. The deidentified version of this data will be made available at the ICPSR University of Michigan repository, and can be accessed with a website URL.
REFERENCES
- 1. Power R. The role of qualitative research in HIV/AIDS. AIDS 1998; 12 (7): 687–95. [DOI] [PubMed] [Google Scholar]
- 2. Al-Busaidi ZQ. Qualitative research and its uses in health care. Sultan Qaboos Univ Med J 2008; 8 (1): 11–9. [PMC free article] [PubMed] [Google Scholar]
- 3. National Institutes of Health. Draft NIH Policy for Data Management and Sharing. Bethesda, MD: Office of The Director, National Institutes of Health; 2019.
- 4. Mozersky J, Walsh H, Parsons M, McIntosh T, Baldwin K, DuBois JM. Are we ready to share qualitative research data? Knowledge and preparedness among qualitative researchers, IRB members, and data repository curators. IASSIST Q 2020; 43 (4): 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. DuBois JM, Strait M, Walsh H. Is it time to share qualitative research data? Qual Psychol 2018; 5 (3): 380–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. National Institutes of Health. Final NIH Policy for Data Management and Sharing. In: NIH, ed. NOT-OD-21-013. Vol NOT-OD-21-013. NIH Grants & Funding. Bethesda, MD: Office of The Director, National Institutes of Health; 2020.
- 7. National Institutes of Health (NIH). The Ethical, Legal and Social Implications (ELSI) Research Program. 2015. https://www.genome.gov/page.cfm?pageID=17515632#beginSearch. Accessed January 13, 2015.
- 8. Vandermause R, Barg FK, Esmail L, Edmundson L, Girard S, Perfetti AR. Qualitative methods in patient-centered outcomes research. Qual Health Res 2017; 27 (3): 434–42. [DOI] [PubMed] [Google Scholar]
- 9.Patient Centered Outcomes Research Institute (PCORI). PCORI Methodology Standards Report. Washington, DC; 2019. https://www.pcori.org/research-results/about-our-research/research-methodology/pcori-methodology-standards. Accessed August 11, 2021.
- 10. Patient Centered Outcomes Research Institute (PCORI). PCORI Policy for Data Management and Sharing. Washington, DC; 2018. https://www.pcori.org/about-us/governance/policy-data-management-and-data-sharing. Accessed August 11, 2021.
- 11. Bingham CO 3rd, Bartlett SJ, Merkel PA, et al. Using patient-reported outcomes and PROMIS in research and clinical applications: experiences from the PCORI pilot projects. Qual Life Res 2016; 25 (8): 2109–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. US Department of Health and Human Services. PROMIS (Patient Reported Outcomes Measurement Information System). National Institutes of Health. 2021. https://www.healthmeasures.net/explore-measurement-systems/promis. Accessed August 12, 2021.
- 13. Mozersky J, Parsons M, Walsh H, Baldwin K, McIntosh T, DuBois JM. Research participant views regarding qualitative data sharing. Ethics Hum Res 2020; 42 (2): 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Summary of the HIPAA Privacy Rule. Washington, DC: Department of Health and Human Services; 2003. https://www.hhs.gov/hipaa/for-professionals/privacy/index.html. Accessed August 11, 2021.
- 15. Norgeot B, Muenzen K, Peterson TA, et al. Protected Health Information filter (Philter): accurately and securely de-identifying free-text clinical notes. NPJ Digit Med 2020; 3 (1): 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.NLM-Scrubber. https://scrubber.nlm.nih.gov/. Accessed August 12, 2021.
- 17. Aberdeen J, Bayer S, Yeniterzi R, et al. The MITRE Identification Scrubber Toolkit: design, training, and assessment. Int J Med Inform 2010; 79 (12): 849–59. [DOI] [PubMed] [Google Scholar]
- 18. CliniDeID—Automatic clinical text de-identification. Clinacuity. https://www.clinacuity.com/clinideid/. Accessed August 12, 2021.
- 19. Neamatullah I, Douglass MM, Lehman L-WH, et al. Automated de-identification of free-text medical records. BMC Med Inform Decis Mak 2008; 8 (1): 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stubbs A, Uzuner Ö. Annotating longitudinal clinical narratives for de-identification: the 2014 i2b2/UTHealth corpus. J Biomed Inform 2015; 58 Suppl: S20–S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stubbs A, Kotfila C, Uzuner Ö. Automated systems for the de-identification of longitudinal clinical narratives: overview of 2014 i2b2/UTHealth shared task Track 1. J Biomed Inform 2015; 58 Suppl: S11–S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uzuner Ö, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc 2007; 14 (5): 550–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Amazon Comprehend Detect PHI. Amazon. https://docs.aws.amazon.com/comprehend/latest/dg/how-medical-phi.html. Accessed August 12, 2021.
- 24. Amazon Comprehend Medical. Amazon. https://aws.amazon.com/comprehend/medical/. Accessed August 12, 2021.
- 25. Google Cloud Healthcare API. Google. https://cloud.google.com/healthcare. Accessed August 12, 2021.
- 26. Kayaalp M. Modes of de-identification. AMIA Annu Symp Proc 2017; 2017: 1044–50. [PMC free article] [PubMed] [Google Scholar]
- 27. Liu Z, Tang B, Wang X, Chen Q. De-identification of clinical notes via recurrent neural network and conditional random field. J Biomed Inform 2017; 75S: S34–S42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dernoncourt F, Lee JY, Uzuner O, Szolovits P. De-identification of patient notes with recurrent neural networks. J Am Med Inform Assoc 2017; 24 (3): 596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. IQDA Qualitative Data Anonymizer. London, UK; 2011. https://www.lse.ac.uk/library/research-support/research-data-management/anonymisation-and-data-protection. Accessed August 11, 2021.
- 30. Saunders B, Kitzinger J, Kitzinger C. Anonymising interview data: challenges and compromise in practice. Qual Res 2015; 15 (5): 616–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dedoose. Version 8.0.35 web application for managing, analyzing, and presenting qualitative and mixed method research data,. 2018. Los Angeles, CA: SocioCultural Research Consultants, LLC. www.dedoose.com. Accessed August 12, 2021.
- 32. Roller MR, Lavrakas PJ. Applied Qualitative Research Design: A Total Quality Framework Approach. New York: Guilford Press; 2015. [Google Scholar]
- 33. Saldaña J. The Coding Manual for Qualitative Researchers. 3rd ed. Thousand Oaks, CA: Sage Publications Ltd.; 2016. [Google Scholar]
- 34. Finkel JR, Grenager T, Manning C. Incorporating non-local information into information extraction systems by Gibbs sampling. In: Proceedings of the 43rd Annual Meeting on Association for Computational Linguistics; 2005; Ann Arbor, MI.
- 35. Chinor N. MUC-7 Named Entity Task Definition. 1997. http://www.itl.nist.gov/iaui/894.02/related_projects/muc/proceedings/ne_task.html. Accessed November 4, 2016.
- 36. The Dryad Repository at North Carolina State University. DataDryad About. 2016. http://datadryad.org/. Accessed February 16, 2017.
- 37. Finkel JR, Grenager T, Manning C. Incorporating Non-local Information into Information Extraction Systems by Gibbs Sampling. In: Proceedings of the 43nd Annual Meeting of the Association for Computational Linguistics (ACL 2005); 2005; Ann Arbor, MI.
- 38. UK Data Archive. Managing and Sharing Data: Best Practices for Researchers. Vol. 3. Wivenhoe Park, Colchester, Essex: University of Essex; 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset rom the NIB stories is available through the Journal Narrative Inquiry in Bioethics, Johns Hopkins University Press. The second dataset of qualitative interviews underlying this article cannot be shared publicly due to the privacy of individuals that participated in the study. The deidentified version of this data will be made available at the ICPSR University of Michigan repository, and can be accessed with a website URL.