Abstract
Background
Major depression is a substantial public health concern that can affect personal relationships, reduce people's ability to go to school or work, and lead to social isolation. Multi-gene pharmacogenomic testing that includes decision-support tools can help predict which depression medications and dosages are most likely to result in a strong response to treatment or to have the lowest risk of adverse events on the basis of people's genes.
We conducted a health technology assessment of multi-gene pharmacogenomic testing that includes decision-support tools for people with major depression. Our assessment evaluated effectiveness, safety, cost-effectiveness, the budget impact of publicly funding multi-gene pharmacogenomic testing, and patient preferences and values.
Methods
We performed a systematic literature search of the clinical evidence. We assessed the risk of bias of each included study using the Cochrane Risk of Bias Tool and the Risk of Bias Assessment Tool for Nonrandomized studies (RoBANS) and the quality of the body of evidence according to the Grading of Recommendations Assessment, Development, and Evaluation (GRADE) Working Group criteria.
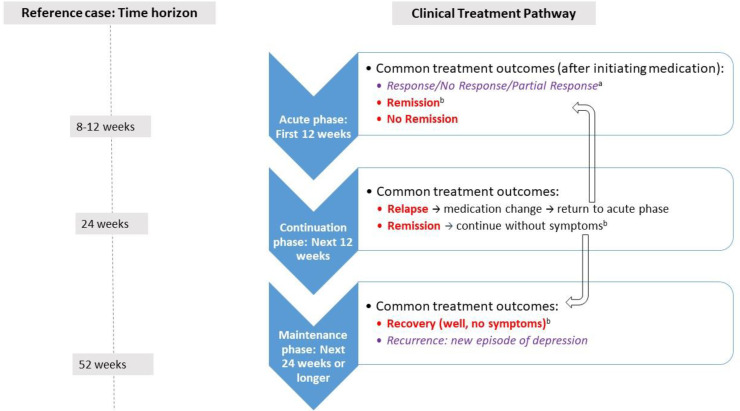
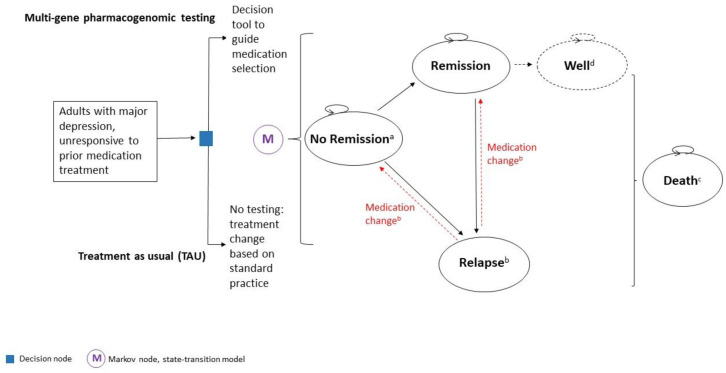
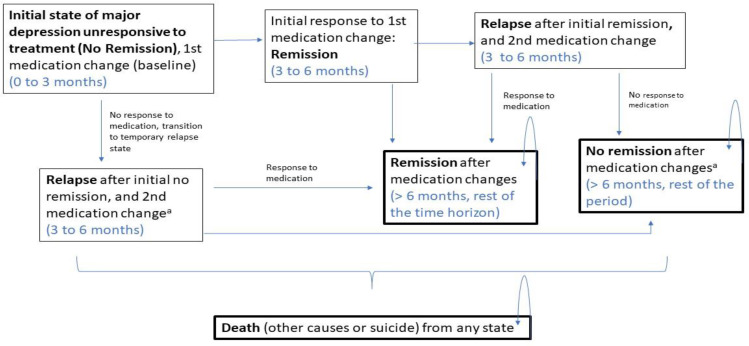
We performed a systematic literature search of the economic evidence to review published cost-effectiveness studies on multi-gene pharmacogenomic testing that includes a decision-support tool in people with major depression. We developed a state-transition model and conducted a probabilistic analysis to determine the incremental cost of multi-gene pharmacogenomic testing versus treatment as usual per quality-adjusted life-year (QALY) gained for people with major depression who had inadequate response to one or more antidepressant medications. In the reference case (with GeneSight-guided care), we considered a 1-year time horizon with an Ontario Ministry of Health perspective. We also estimated the 5-year budget impact of publicly funding multi-gene pharmacogenomic testing for people with major depression in Ontario.
To contextualize the potential value of multi-gene pharmacogenomic testing that includes decision-support tools, we spoke with people who have major depression and their families.
Results
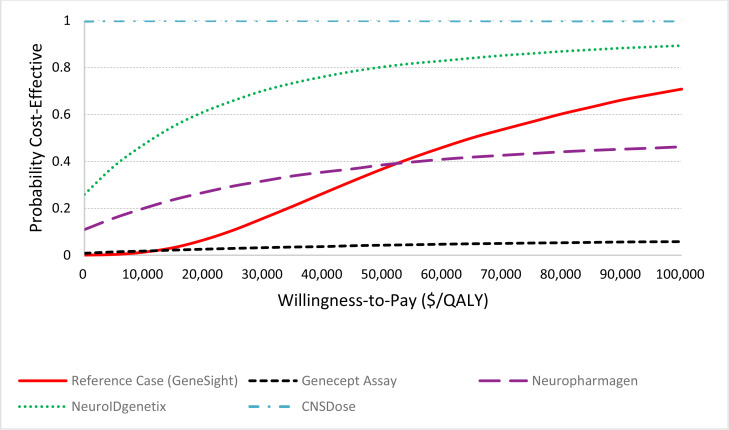
We included 14 studies in the clinical evidence review that evaluated six multi-gene pharmacogenomic tests. Although all tests included decision-support tools, they otherwise differed greatly, as did study design, populations included in studies, and outcomes reported. Little or no improvement was observed on change in HAM-D17 depression score compared with treatment as usual for any test evaluated (GRADE: Low–Very Low). GeneSight– and NeuroIDgenetix–guided medication selection led to statistically significant improvements in response (GRADE: Low–Very Low) and remission (GRADE: Low–Very Low), while treatment guided by CNSdose led to significant improvement in remission rates (GRADE: Low), but the study did not report on response. Results were inconsistent and uncertain for the impact of Neuropharmagen, and no significant improvement was observed for Genecept or another unspecified test for either response or remission (GRADE: Low–Very Low). Neuropharmagen may reduce adverse events and CNSDose may reduce intolerability to medication, while no difference was observed in adverse events with GeneSight, Genecept, or another unspecified test (GRADE: Moderate–Very Low). No studies reported data on suicide, treatment adherence, relapse, recovery, or recurrence of depression symptoms.
Our review included four model-based economic studies and found that multi-gene pharmacogenomic testing was associated with greater effectiveness and cost savings than treatment as usual, over long-term (i.e., 3-,5-year and lifetime) time horizons. Since none of the included studies was fully applicable to the Ontario health care system, we conducted a primary economic evaluation.
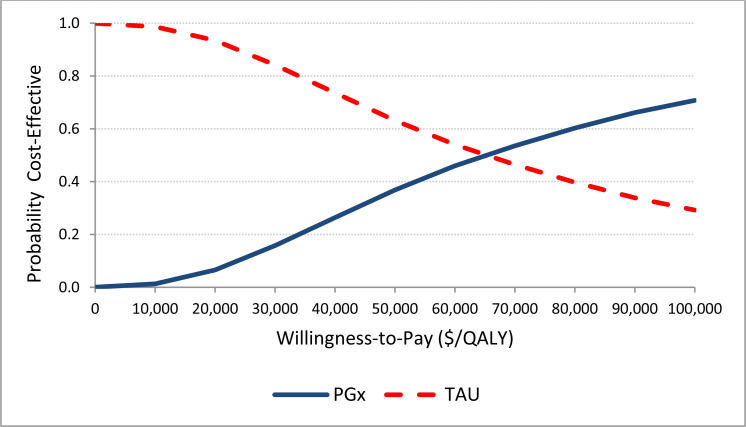
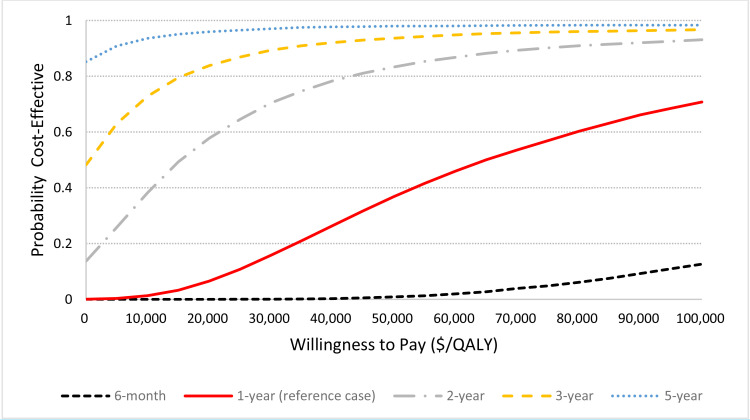
Our reference case analysis over the 1-year time horizon found that multi-gene pharmacogenomic testing (with GeneSight) was associated with additional QALYs (0.03, 95% credible interval [CrI]: 0.005; 0.072) and additional costs ($1,906, 95% Crl: $688; $3,360). An incremental cost-effectiveness ratio was $60,564 per QALY gained. The probability of the intervention being cost-effective (vs. treatment as usual) was 36.8% at a willingness-to-pay amount of $50,000 per QALY (i.e., moderately likely not to be cost-effective), rising to 70.7% at a willingness-to-pay amount of $100,000 per QALY (i.e., moderately likely to be cost-effective). Evidence informing economic modeling of the reference case with GeneSight and other multi-gene pharmacogenomic tests was of low to very low quality, implying considerable uncertainty or low confidence in the effectiveness estimates. The price of the test, efficacy of the intervention on remission, time horizon, and analytic perspective were major determinants of the cost-effectiveness results. If the test price were assumed to be $2,162 (compared with $2,500 in the reference case), the intervention would be cost-effective at a willingness-to-pay amount of $50,000 per QALY; moreover, if the price decreased to $595, the intervention would be cost saving (or dominant) compared with treatment as usual.
At an increasing uptake of 1% per year and a test price of $2,500, the annual budget impact of publicly funding multi-gene pharmacogenomic testing in Ontario over the next 5 years ranged from an additional $3.5 million in year 1 (at uptake of 1%) to $16.8 million in year 5. The 5-year budget impact was estimated at about $52 million.
People with major depression and caregivers generally supported multi-gene pharmacogenomic testing because they believed it could provide guidance that fit their values. They hoped such guidance would speed symptom relief, would reduce side effects and help inform their medication choices. Some patients expressed concerns over maintaining confidentiality of test results and the possibility that physicians would sacrifice patient-centred care to follow pharmacogenomic guidance.
Conclusions
Multi-gene pharmacogenomic testing that includes decision-support tools to guide medication selection for depression varies widely. Differences between individual tests must be considered, as clinical utility observed with one test might not apply to other tests. Overall, effectiveness was inconsistent among the six multi-gene pharmacogenomic tests we identified. Multi-gene pharmacogenomic tests may result in little or no difference in improvement in depression scores compared with treatment as usual, but some tests may improve response to treatment or remission from depression. The impact on adverse events is uncertain. The evidence, however, is uncertain, and therefore our confidence that these observed effects reflect the true effects is low to very low.
For the management of major depression in people who had inadequate response to at least one medication, some multi-gene pharmacogenomic tests that include decision support tools are associated with additional costs and QALYs over the 1-year time horizon, and maybe be cost-effective at the willingness-to-pay amount of $100,000 per QALY. Publicly funding multi-gene pharmacogenomic testing in Ontario would result in additional annual costs of between $3.5 million and $16.8 million, with a total budget impact of about $52 million over the next 5 years.
People with major depression and caregivers generally supported multi-gene pharmacogenomic testing because they believed it could provide guidance that fit their values. They hoped such guidance would speed symptom relief, would reduce side and help inform their medication choices. Some patients expressed concerns over maintaining confidentiality of test results and the possibility that physicians would sacrifice patient-centred care to follow pharmacogenomic guidance.
Objective
This health technology assessment evaluates the effectiveness, safety, and cost-effectiveness of multi-gene pharmacogenomic testing that includes decision-support tools to guide medication selection for people with major depression. It also evaluates the budget impact of publicly funding multi-gene pharmacogenomic testing to guide medication selection for people with major depression and the experiences, preferences, and values of people with major depression.
Background
Health Condition
Major depression, also known as major depressive disorder or clinical depression, is a mood disorder and a leading cause of disability in Ontario.1
Symptoms of depression are highly individual but are most often characterized by persistent feelings of sadness, hopelessness, excessive guilt, or despair accompanied by a loss of interest or pleasure in previously enjoyed life activities.2,3 Other symptoms can include decreased energy, fatigue, and inability to concentrate as well as changes in weight, appetite, or sleep patterns.3 Some people have thoughts about self-harm, death, or suicide.2,3 Symptoms of depression can lead to serious distress or inability to perform daily functions.3 Many affected people become unable to cope with simple aspects of everyday life, which can greatly affect quality of life, personal relationships, and people's ability to go to school or work, and can lead to social isolation.4
Major depression can range in degree from mild to severe. A clinical diagnosis of an episode of major depression is defined by the Diagnostic and Statistical Manual of Mental Disorders, fifth edition (DSM-5) as experiencing five or more clinically relevant symptoms during the same 2-week period, in addition to clinically relevant distress or impairment in important areas of daily function.5
There is no single cause of major depression. Various factors in combination are thought to contribute to the disorder, including genetics or a family history of depression, as well as biological, environmental, and social factors.1,2
Clinical Need and Target Population
Depression is the most prevalent mental illness for all ages.4 An estimated 11.3% of Canadian adults will have depression at some point during their lifetime.2 Data from Statistics Canada's 2012 Canadian Community Health Survey (CCHS) on Mental Health show 4.8% of people in Ontario aged 15 years and older reported symptoms for major depression in the previous 12 months.4
Depression affects people of all ages and cultures. In Ontario, people aged 15 to 24 years reported higher rates of depression (6.6%) than any other age group, followed by those aged 25 to 44 years (6.2%).4 Women report higher rates of depression than men (5.8% vs. 3.8%, respectively).4
Current Treatment Options
Pharmacotherapy for Depression
Pharmacotherapy (treatment with medication), alone or in combination with other therapies, is considered a first-line treatment for moderate-to-severe major depression.6 Antidepressants (drugs used to treat depression) are one of the most commonly used medications among Canadians, prescribed to an estimated 13.7% of women aged 25 to 79, 4.2% of men aged 25 to 44, and 8.2% of men aged 45 to 64.7 Available antidepressants commonly include selective serotonin reuptake inhibitors (SSRIs), serotonin and norepinephrine reuptake inhibitors, tricyclic antidepressants, and monoamine oxidase inhibitors.
Many people do not benefit from, or are unable to tolerate, their prescribed pharmacotherapy. Studies have found more than 50% of patients with major depression do not respond to their first medication, and an estimated 30% do not respond to two or more medications.8 Additionally, side effects often lead to a lack of adherence to medication.6,9 Poor adherence results in a lower probability of achieving remission and carries a large burden associated with medication side effects and medical costs.
Selecting pharmacotherapy for depression is often difficult given the many available medications. Canadian depression guidelines recommend more than 15 medications as first-line pharmacotherapy.6 People who do not respond to initial treatment could have a dose adjustment, could be prescribed an adjunct treatment, or could be switched to another medication.6,10 Various factors can help guide medication selection and dosage, such as clinical features, comorbid conditions, concomitant pharmacotherapy, potential side effects, and patient preferences.6 However, very little evidence shows that these factors alone improve rates of remission or reduce the number of adverse events. Selection is therefore often empirical, leading to a long trial-and-error process before an acceptable treatment response is achieved with minimal or no side effects.
Health Technology Under Review
Pharmacogenomics and Depression Pharmacotherapy
Genetic variation (i.e., differences in DNA sequence) among individuals is a potential contributor to differences in depression medication effect.11,12 Pharmacogenomics or pharmacogenetics is the study of how differences in genes (allelic variants) affect individual responses to various medications.13 The term “pharmacogenetics” is used to refer to how variation in a single gene affects a drug's response, whereas “pharmacogenomics” is a broader term used in the study of how all genes can affect drug responses.14 For the purposes of this report, we will use the term “pharmacogenomics” to refer to any drug–gene testing.
Genetic variation can affect the way drugs are absorbed, distributed, metabolized, or eliminated from the body (i.e., genes associated with pharmacokinetics) as well as the mechanism of action or effect of a drug (i.e., genes associated with pharmacodynamics).15 An estimated 40% of variance in response to antidepressants could result from common genetic variants, although information about specific genes and common variants identified is limited.16
The most studied example of the impact of genetic variations on pharmacokinetics of medications for depression involves the cytochrome P450 (CYP450) family of drug-metabolizing enzymes.11,12,17 Differences in the activity of these enzymes may lead to greater or lower exposure to medications that are metabolized by them. Knowledge of specific variations in the genes encoding these enzymes (i.e., an individual's genotype) can be used to try to predict how an individual will metabolize a medication (i.e., an individual's phenotype). Individuals with variants that are known to reduce an enzyme's function (e.g., intermediate or poor metabolizers) may not be able to break down and eliminate certain medications from the body, and therefore may be exposed to more drug than needed and have an increased risk of side effects or overdose compared with an extensive (normal) metabolizer. Individuals with variants that increase an enzyme's function (e.g., ultra-rapid metabolizers) may result in insufficient medication exposure, and subsequently and poor response.11,12 The opposite effects would be observed for medications that become pharmacologically active after metabolism (i.e., pro-drugs).
Two of the most studied CYP450 genes in the context of antidepressant medications are those for the CYP2D6 and CYP2C19 enzymes.11,18,19 These enzymes are extensively involved in metabolism of many SSRIs and tricyclic antidepressants.11,12 For CYP2D6 and CYP2C19, respectively, an estimated 1% to 20% and 2% to 5% of people are categorized as ultra-rapid metabolizers, 1% to 13% and 18% to 45% are intermediate metabolizers, and 1% to 10% and 2% to 15% are poor metabolizers.12 Approximately 2% to 30% of people have a CYP2C19 genotype that indicates a rapid metabolizer phenotype. While these phenotypes are based on average multiethnic frequencies, the distribution of these allelic variants and phenotypes vary substantially with ethnicity.11,12,18
Any drug, however, can be metabolized by multiple CYP450 enzymes and non-CYP450 enzymes, and therefore not all variations in one or more genes involved in a drug's metabolism will affect response to a specific medication. In addition, not every genetic variant would affect protein (i.e., enzymatic) function.
Few studies have examined how variants in pharmacodynamic genes change antidepressant effect. Some examples include genes that encode one of the serotonin receptors (e.g., HTR2A) or proteins involved in transport of serotonin (e.g., SLC6A4).20
Pharmacogenomic Testing to Guide Medication Selection
Pharmacogenomic testing for people with major depression involves assessing relevant genes to predict which psychotropic medications and dosages are most likely to result in a strong treatment response and have the lowest risk of causing an adverse event. Testing can be performed either before a new medication is started, or after response to one or more medications is considered inadequate (i.e., lack of clinical improvement, inability to tolerate treatment, or side effects develop). Pharmacogenomic testing is most-often non-invasive, requiring a painless cheek swab or a saliva sample to obtain a person's DNA. Samples are most often collected in a doctor's office or pharmacy, rather than a laboratory. Alternatively, a blood sample can also be used for DNA extraction. The turnaround time for testing depends on the specific test requested, but results among tests available in Canada usually take 2 to 40 days.21
Pharmacogenomic testing can be done in multiple ways. Single-gene testing can test for variants of an individual gene (e.g., CYP2C19) that might affect how a certain drug or class of drugs is prescribed. Multi-gene testing, or panel testing, can simultaneously test for variants of multiple genes known to be involved in the pharmacokinetics or pharmacodynamics of psychotropic drugs (e.g., testing for multiple CYP450 gene variations). Results from these tests provide clinicians with a person's genotype (e.g., the number of functional alleles), and might include their corresponding phenotype (e.g., ultra-rapid metabolizer), or a list of medications that are associated with each gene tested.
Several commercial multi-gene assays have been developed that provide pharmacogenomic-based decision-support tools to help guide medication dosage and selection for people with depression.22,23 These tools often use a proprietary algorithm, or combinatorial testing approach, to predict a combined phenotype for various medications. Test algorithms simultaneously assess the combined or relative effects of multiple gene variants for a given medication.24 Some tools combine genetic and non-genetic information (e.g., clinical characteristics, drug–drug interactions) to make treatment recommendations. Decision-support tools generally recommend medications likely to be safe and effective (i.e., no identified gene–drug interactions), identify those that could have some drug–gene interactions, or specify medications that should be avoided due to significant gene–drug interactions.23,24 Recommendations regarding treatment doses and monitoring are sometimes also provided.
Numerous pharmacogenomic tests are available; a recent review cites more than 30 commercial tests around the world that assess treatment outcomes in depression.25 It is unclear if all of these tests include a decision-support tool; however, each test assesses different genes, analyzes different variants of individual genes, includes different medications, and uses different methods to predict treatment outcomes and make recommendations.22 Additionally, reporting structures and level of detail in results and therapeutic implications vary widely across tests. Some focus on individual drug recommendations; others provide information about appropriate drug classes, and some focus on individual gene results.22
Regulatory Information
Pharmacogenomic testing is not subject to Health Canada approval unless the test is sold as a test kit, defined as a test that is sold to multiple laboratories.26 Tests that are offered as laboratory services, or laboratory-developed tests, are subject to licensing approval at the provincial level. Currently, no multi-gene pharmacogenomic tests used for selection of psychotropic medications are approved by Health Canada or have been licensed by the Ontario Ministry of Health. Tests that are ordered in Canada and performed in a laboratory outside of Canada do not require approval at the federal or provincial level (Laboratories and Genetics Branch, Ontario Ministry of Health, oral communication, January 2020).
Similar federal and laboratory approvals are required by the US Food and Drug Administration (FDA).26,27 In 2018, the FDA issued a statement warning consumers about the use of pharmacogenetic tests that claim to predict how a patient will respond to specific medications, noting these tests have not been evaluated by the FDA and might not be accurate or supported by scientific or clinical evidence.28 The FDA recommended caution in use of these tests and that only information in FDA-approved drug labelling should be used to determine medication treatment. The FDA further noted that the relationship between DNA variations and the effectiveness of antidepressant medication has not been established.28
Ontario and Canadian Context
Multi-gene pharmacogenomic testing for guiding medication selection among people with depression is currently not publicly funded in Ontario. Tests can be ordered through out-of-pocket payment or be covered by some private insurance plans29; reported prices range from $200 to $2,300.21
A recently published scan of pharmacogenetic testing options for psychiatry in Canada21 identified at least 13 pharmacogenomic tests available to people in Ontario.21 Tests are either targeted at people being treated for depression or other psychiatric disorders (e.g., GeneSight,30 Genecept,31 Neuropharmagen) or include recommendations for numerous drug classes but include a decision support for antidepressant drugs (e.g., Treatgxplus,32 PillCheck,33 myDNA,34 MatchMyMeds,35 RightMed,36 CEN4GEN). These tests are being sold
through private laboratories within Ontario, through laboratories in other provinces or countries, or online through direct-to-consumer sales. Some tests require clinician requisition and interpretation, several tests are sold at participating pharmacies across the province, and others can be obtained by consumers directly online or can be ordered and shipped to a physician's office. Some companies that offer tests direct to the consumer offer additional phone consultation or interpretation services with a geneticist, pharmacist, or physician.
From 2012 to 2018, combinatorial pharmacogenomic testing was made available to select primary care providers and psychiatrists in Ontario via the Centre for Addiction and Mental Health's (CAMH's) Individualized Medicine: Pharmacogenomic Assessment and Clinical Treatment (IMPACT) study. During this time, 11,200 people in Ontario taking or planning to take psychiatric medicines received pharmacogenomic testing,37 of which about 8,000 received the GeneSight test through partnership with Myriad Genetics, in an ongoing study.38
Pharmacogenomic Testing Guidelines and Recommendations
Several guidelines and health technology assessment agencies have provided recommendations relating to pharmacogenomic testing for people receiving treatment for depression (Table 1). Only groups recommending whom to test, rather than actions after testing, are included in Table 1.
Table 1:
Guidelines and Health Technology Assessment Recommendations on Use of Pharmacogenomic Testing for Guiding Treatment Among People With Depression
| Agency, Year | Recommendation or Statement |
|---|---|
| APA Task Force for Biomarkers and Novel Treatments, 201823 | “[T]here is insufficient evidence to support widespread use of combinatorial pharmacogenetic decision support tools at this point in time” |
| CANMAT, 20166 | “… CANMAT does not recommend routine use of pharmacogenetic testing. … Pharmacogenetic testing and/or TDM may be helpful in individual circumstances, including inability to tolerate minimum doses (i.e., to detect poor metabolizers), repeated failure to respond to high doses (i.e., to detect ultrarapid metabolizers), and to detect nonadherence” |
| Washington State HealthCare Authority, 201639 | “Based on these findings the committee voted to not cover pharmacogenomic testing for selected conditionsa” |
| EGAPP Working Group, 200727 | “[We] found insufficient evidence to support a recommendation for or against use of CYP450 testing in adults beginning SSRI treatment for non-psychotic depression. In the absence of supporting evidence, and with consideration of other contextual issues, EGAPP discourages use of CYP450 testing for patients beginning SSRI treatment until further clinical trials are completed” |
Abbreviations: APA, American Psychiatric Association; CANMAT, Canadian Network for Mood and Anxiety Treatments; CYP450, cytochrome P450; EGAPP, Evaluation of Genomic Applications in Practice and Prevention; SSRI, selective serotonin reuptake inhibitor; TDM, therapeutic drug monitoring.
Selected conditions include depression, mood disorders, psychosis, anxiety, ADHD (attention deficit hyperactivity disorder), and substance use disorder.
Expert Consultation
We engaged with experts in the specialty areas of psychiatry, family medicine, pharmacy, genetics, pharmacology, and ethics to help inform our understanding of aspects of the health technology and our methodologies and to contextualize the evidence.
PROSPERO Registration
This health technology assessment has been registered in PROSPERO, the international prospective register of systematic reviews (CRD #42020168084), available at: https://www.crd.york.ac.uk/PROSPERO.
Clinical Evidence
Research Question
What is the clinical utility of multi-gene pharmacogenomic testing that includes decision-support tools to guide medication selection compared with treatment as usual for people with major depression?
Methods
Clinical Literature Search
We performed a clinical literature search on January 24, 2020, to retrieve studies published from database inception until the search date. We used the Ovid interface in the following databases: MEDLINE, Embase, the Cochrane Central Register of Controlled Trials, the Cochrane Database of Systematic Reviews, the Health Technology Assessment database, and the National Health Service Economic Evaluation Database (NHS EED), and PsycINFO.
A medical librarian developed the search strategies using controlled vocabulary (e.g., Medical Subject Headings) and relevant keywords. The final search strategy was peer reviewed using the PRESS Checklist.40
We created database auto-alerts in MEDLINE, Embase, and PsycINFO, and monitored them for the duration of the assessment period. We also performed a targeted grey literature search of health technology assessment agency websites as well as clinical trial and systematic review registries. See Appendix 1 for our literature search strategies, including all search terms.
Eligibility Criteria
STUDIES
Inclusion Criteria
English-language full-text publications
Studies published from database inception until January 24, 2020
Randomized controlled trials, non-randomized studies, systematic reviews, and meta-analyses
Exclusion Criteria
Animal and in vitro studies
Non-systematic reviews, narrative reviews, abstracts, editorials, letters, case reports, and commentaries
Unpublished data, draft data, and manuscripts
Gene discovery, analytical validity, and clinical validity studies
Non-comparative studies (e.g., non-comparative before–after cohort studies)
PARTICIPANTS
Inclusion Criteria
-
Adults (aged 18 years and over) with a primary diagnosis of major depression requiring pharmacological treatment
-
○Studies with combined populations were included only if results for the depression subgroup could be extracted
-
○
-
Subpopulations
-
○Medication-naive (initiating pharmacological treatment)
-
○Inadequate response to one or more medications (i.e., lack of clinical improvement, unable to tolerate treatment, or developed side effects)
-
○
Exclusion Criteria
Bipolar depression
Children and adolescents
INTERVENTIONS
Inclusion Criteria
-
Multi-gene (two or more genes) pharmacogenomic tests that include a clinical decision-support tool to guide depression medication selection
-
○Decision-support tools defined as choice of medication or dosage recommendations or guidance
-
○
Exclusion Criteria
Single-gene tests
Tests that do not provide medication or dosage recommendations
COMPARATORS
Inclusion Criteria
No pharmacogenomic testing to guide depression medication selection or dose adjustment (treatment as usual)
Exclusion Criteria
Studies comparing different pharmacogenomic tests or genes
OUTCOME MEASURES
-
Change in depression outcomes
-
○Change in depression scores (e.g., HAM-D17); a minimally clinically important difference was defined as a score between 2 and 3 on the HAM-D41
-
○Response∗ (reduction in depression scores)
-
○Remission∗ (asymptomatic period [no clinically relevant symptoms])
-
○Relapse∗ (return of symptoms during remission)
-
○Recovery∗ (sustained remission)
-
○Recurrence∗ (return of symptoms after recovery)
-
○
Medication adherence
Suicide (thoughts, attempt, or completed)
Adverse events or side effects
Quality of life
-
Impact on therapeutic decisions
- ∗Definitions as specified in individual research articles.
Literature Screening
A single reviewer conducted an initial screening of titles and abstracts using Covidence42 and then obtained the full texts of studies that appeared eligible for review according to the inclusion criteria. This single reviewer then examined the full-text articles and selected studies eligible for inclusion. The reviewer also examined reference lists and consulted content experts for any additional relevant studies not identified through the search.
Data Extraction
A single reviewer extracted relevant data on study characteristics and risk-of-bias items using a data form to collect information on the following:
Source (e.g., citation information, study type)
Methods (e.g., study design, study duration and years, participant allocation, allocation sequence concealment, blinding, reporting of missing data, reporting of outcomes, whether the study compared two or more groups)
Outcomes (e.g., outcomes measured, number of participants for each outcome, number of participants missing for each outcome, outcome definition and source of information, unit of measurement, upper and lower limits [for scales], time points at which the outcomes were assessed)
In cases where multiple publications reported on the same study, we extracted data primarily from the primary study and referred to others to supplement results or methodological information, as necessary.
Where essential data were presented only in graphic form and clearly visible in figures, we approximated summary estimates (e.g., mean difference or percentage change) using WebPlotDigitizer software.43 This tool was used only to extract summary estimates for primary outcome measures and final follow-up periods. Given potential inaccuracies, data were not extracted for variance surrounding the effect estimate (e.g., interquartile ranges, standard errors, range) and were not incorporated into meta-analysis.
Statistical Analysis
Proportions and numbers of events were calculated from reported data where clear outcome definitions, numerators, and denominators were available.
We calculated risk ratios for dichotomous data and mean differences from baseline to follow-up or between follow-up measures for continuous data, along with 95% confidence intervals. Where studies adjusted analysis accounting for repeated measures for continuous outcome data, summary estimates were not calculated on the raw data and instead were reported as stated in the primary study.
Where data were available and it was appropriate given minimal methodological (e.g., study design), clinical diversity (e.g., study populations), or statistical heterogeneity, we generated pooled summary estimates using random effects models in Review Manager.42-44 In addition, risk differences were calculated to complement the relative effects for outcomes based on the HAM-D17 scale. Where preferred summary estimates could not be calculated or pooling of data was inappropriate, we present the data in figure or tabular form and provide narrative analysis. Owing to heterogeneity between individual tests, meta-analysis and narrative synthesis was performed among studies evaluating the same pharmacogenomic test.
Subgroup analyses were planned among individual tests based on prior medication use (treatment naive vs. inadequate response to one or more treatments) and treatment provider (psychiatrist vs. primary care provider). We were, however, unable to do these analyses because of the limited number of studies and lack of appropriate and relevant data. Some studies conducted their own subgroup analyses for other factors, which we reported as available but did not analyze further or critically appraise.
Critical Appraisal of Evidence
We assessed risk of bias using the Cochrane Risk of Bias Tool, Version 1.0 for RCTs44 and Risk of Bias tool for Non-randomized Studies (RoBANS) for non-randomized studies45 (Appendix 2).
We evaluated the quality of the body of evidence for each outcome according to the Grading of Recommendations Assessment, Development, and Evaluation (GRADE) Handbook.39 The body of evidence was assessed for the following considerations: risk of bias, inconsistency, indirectness, imprecision, and publication bias. The overall rating reflects our certainty in the evidence.
Results
Clinical Literature Search
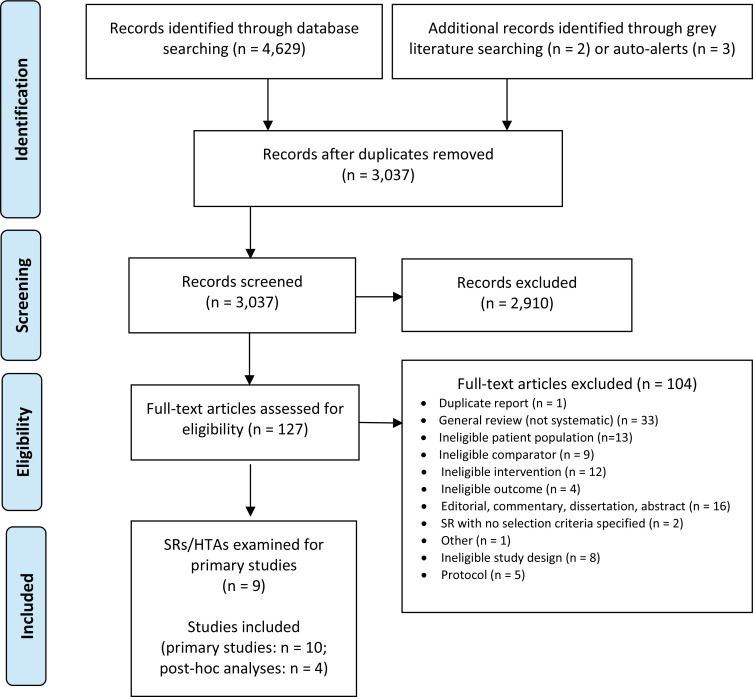
The database search of the clinical literature yielded 4,629 citations published between inception and January 24, 2020. We identified five additional studies from other sources. In total, we identified 14 studies (10 primary comparative studies and four post-hoc analyses of the primary studies) that met our inclusion criteria. We identified an additional nine systematic reviews and health technology assessments that met our selection criteria and were examined for additional primary studies. See Appendix 2 for studies excluded after full-text review. Figure 1 presents the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) flow diagram for the clinical literature search.
Figure 1: PRISMA Flow Diagram—Clinical Search Strategy.
Abbreviations: HTA, health technology assessment; PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-analyses; SR, systematic review.
Source: Adapted from Moher et al.46
Characteristics of Included Studies
SYSTEMATIC REVIEWS AND HEALTH TECHNOLOGY ASSESSMENTS EXAMINED
Nine systematic reviews and health technology assessments were identified that evaluated the use of multi-gene pharmacogenomic testing to guide medication selection among people with depression.39,47-54 Previous reviews were used for the purpose of cross-referencing and ensuring no relevant literature was missed. No additional primary studies were identified from these reviews, and no review included all studies or outcomes assessed in the present review. A summary of identified reviews is presented in Appendix 2, Table A1.
PRIMARY STUDIES
Table 2 summarizes study design and characteristics for the ten included primary studies and four post-hoc analyses. Eight of ten studies were RCTs, while two studies were non-randomized open-label studies.55,56 Length of follow-up ranged from 8 to 12 weeks. One RCT included 24-month follow-up data for the pharmacogenomic test-guided arm; however, results were not comparative and therefore not included in the review.57 The study by Bradley et al58 randomized a combined depression and/or anxiety population but was included as relevant outcomes were stratified separately for the depression (with or without anxiety) cohort. Outcomes that included only the combined population (depression or anxiety) were excluded. A corrigendum to the study by Han et al was published after completion of our systematic review, and all values are based on the corrected version of the originally published article.59
Table 2:
Summary of Study Design and Characteristics of Included Studies
| Author, Year Country | Study Design | N PGx/TAU | Setting and Provider Type | Inclusion Criteria | Exclusion Criteria | PGx Test, No. Genes | Length of FU, wk |
|---|---|---|---|---|---|---|---|
| GeneSight Studies | |||||||
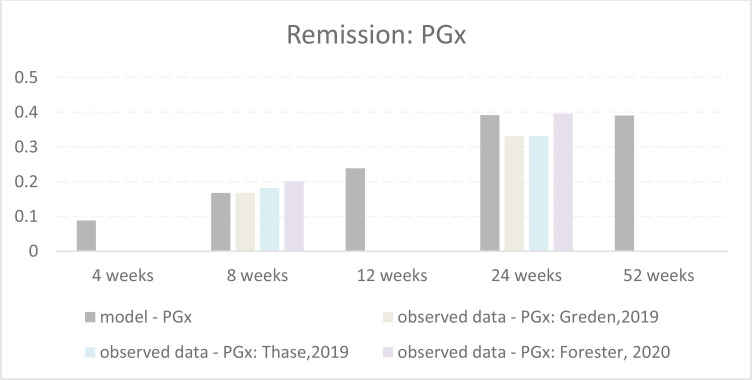
| GUIDED trial, Greden et al, 201957 United States Post hoc Analyses Thase et al, 201968; Dunlop et al, 201966; Forester et al, 202067 |
RCT | 760/781 | • Outpatients from 60 academic and community sites • Psychiatric and primary care providers |
• Age ≥ 18 • MDD (≥ 11 QIDS-C16 and QIDS-SR16 rating scale) • Inadequate response (no clinical improvement or intolerable SEs) to at least one treatment included in test report within current episode Forester et al, 2020, Subgroup • Aged 65 years or older Thase et al, 2019, Subgroup • Taking medications subject to gene-drug interactions at baselinea |
• Significant suicide risk • Severe co-occurring psychiatric or cognitive disordersb • Unstable or significant medical conditionsb • Inpatients Per Protocol Cohort • HAM-D17 < 14 at baseline • Protocol violations or clinician did not view test report |
GeneSight, 8 genes | 8 |
| Winner et al, 201365 United States |
RCT | 26/25 | • Outpatient clinics • Psychiatrists, psychiatric NPs |
• MDD or depressive disorder NOS • HAM-D17 ≥ 14 |
• Bipolar disorder, schizophrenia, schizoaffective disorders, active substance abuse or dependence • ECT • Depression requiring hospitalization |
GeneSight, 5 genes | 10 |
| Hall-Flavin et al, 201355 United States |
Prospective cohort | 114/113 | • Outpatient hospital clinic • Psychiatrists |
• Aged 18–72 y • Primary diagnosis of MDD or depressive disorder NOS (DSM-IV) • HAM-D17 ≥ 14 |
Bipolar disorder type I, schizophrenia, or schizoaffective disorder | GeneSight, 5 genes | 8 |
| Hall-Flavin et al, 201256 United States |
Prospective cohort | 25/26 | • Outpatient behavioural clinic • Psychiatrist |
• Aged 25–75 • Primary diagnosis of MDD based on DSM-IV (HAM-D17 ≥ 14) |
Bipolar disorder type I, schizophrenia, or schizoaffective disorder | GeneSight, 5 genes | 8 |
| Neuropharmagen Studies | |||||||
| Han et al, 201860 Korea |
RCT | 52/48 | • 2 university teaching hospitals • Psychiatrists |
• Aged ≥ 20 y • MDD (DSM-5) • ≥ 3 on CGI-I despite current treatment with proper dose and duration (≥ 6 wk) OR intolerance to current therapy |
• Not receiving antidepressant • Other psychiatric diagnosesb • Hospitalized within 8 wk• CBT or other psychotherapy • Clinical trial in past month • ECT within 8 wk • Pregnant or breastfeeding |
Neuro-pharmagen, 20 genes | 8 |
| Perez et al, 201762 Spain Post hoc Subgroups Menchon et al, 201969 |
RCT | 155/161 | • Outpatients and inpatients from 18 hospitals and mental health centres • Psychiatrists |
• Aged ≥ 18 y • MDD (DSM-5) • CGI-S ≥ 4 at screening and randomization • Required medication de novo or receiving treatment and required substitution or augmentation with antidepressant |
• Primary psychiatric diagnoses other than MDD • Pregnant or breastfeeding • Treatment with quinidine, cinacalcet, or terbinafine Per Protocol Analysis • Clinician prescribed against test recommendation |
Neuro-pharmagen, 30 genes | 12 |
| Other Pharmacogenomic Tests | |||||||
| Perlis et al, 202061 United States |
RCT | 151/153 | • 21 outpatient centres • Not specified |
• Aged 18–75 y • Primary diagnosis of nonpsychotic MDD (DSM-5, MINI 7.0) • SIGH-D17 score > 18 • Failure (inefficacy or intolerable AEs) of at least one prior adequate trial of standard antidepressant for current episode |
• Other psychiatric diagnosesb • History of suicidal behaviour within 12 mo or active suicidal thoughts with intent • 4 or more failed pharmacologic interventions in current episode • ECT, TMS, or psychotherapy (CBT or DBT) within 90 d • Current psychotherapy allowed if frequency is not increased • Unstable or active medical conditions (that could jeopardize safety or participation)c |
Genecept, 18 genes | 8 |
| Bradley et al, 201858 United States | RCT | 352/333 randomized (depresssion cohort: 237/213) | • 20 independent clinical sites in psychiatry, obstetrics and gynecology, internal medicine, family medicine | • Aged 19–87 y • Depression or anxiety (DSM-5 or site procedures and MINI Psychiatric interview)d • New to treatment (newly diagnosed or treated for < 6 wk) or inadequately controlled (lack of efficacy or discontinuation due to AE or intolerability) |
• Bipolar disorder, schizophrenia, personality disorder, traumatic physical injury • Significant risk for suicide or hospitalization • History of chronic renal dysfunction or chronic kidney disease, malabsorption, pregnancy, abnormal hepatic function |
NeuroID-genetix, 10 genes | 12 |
| Shan et al, 201963 China |
RCT | 31/43 | • Single-hospital outpatients and inpatients • Same psychiatrist treated both groups |
• Aged 18–51 y • MDD (DSM-5) • HAM-D17 ≥ 17, and depressive mood ≥ 2 • No psychotic symptoms • At least a junior high school education level • Han population in China • Treatment naive or interrupted medication for > 2 wk (4 wk for fluoxetine) |
• Any other diagnosis on DSM-5 • Physical illness (e.g., liver and kidney disease, CV diseases) • Any combination with other antipsychotic medications, including typical and atypical antipsychotic and mood stabilizer • Pregnancy |
Not specified, 5 genes | 8 |
| Singh et al, 201564 Australia |
RCT | 74/74 | • NR • Psychiatrist |
• Principal diagnosis of MDD (DSM-5) • HAM-D > 18 • Caucasian only |
• Other active psychiatric diagnosesb • Pregnant or breastfeeding • Hepatic or renal impairment • Co-prescribed CYP2D6, CYP2C19, ABCB1 inducers or inhibitors • Grapefruit juice drinker or smokers |
CNSDose, NR | 12 |
Abbreviations: AABCB1, ATP binding cassette subfamily B member 1; AE, adverse effect; C16, clinician rated; CBT, cognitive behavioural therapy; CGI, Clinical Global Impressions Scale I (improvement) or S (severity of illness); CV, cardiovascular; CYP, cytochrome P; DBT, dialectical behaviour therapy; DSM, Diagnostic and Statistical Manual of Mental Disorders; ECT, electroconvulsive therapy; FU, follow-up; HAM-D, Hamilton Depression Rating Scale; MDD, major depressive disorder; MINI 7.0, Mini International Neuropsychiatric Interview, Version 7.0; SIGH-D17, 17-item version of the Structured Interview Guide for the Hamilton Depression Rating Scale; NOS, not otherwise specified; NP, nurse practitioner; NR, not reported; PGx, pharmacogenomic testing group; QIDS, Quick Inventory of Depressive Symptomatology; RCT, randomized controlled trial; SE, side effect; TAU, treatment as usual; TMS, transcranial magnetic stimulation.
Patients in the “use with caution” and “use with increased caution and with more frequent monitoring” categories.
Full list of excluded conditions listed in supplementary methods for primary article.
In the opinion of the site investigator; list of examples provided in primary article.
Only data for the depression cohort were used in the present analysis (excluding those with anxiety alone).
All studies required a principal diagnosis of major depressive disorder for inclusion; however, most studies further limited the population to those with moderate or severe depression using different depression scale thresholds. Three studies limited their population to patients who had inadequate response (lack of efficacy or intolerable adverse events) to one or more medications at baseline,57,60,61 and three combined treatment-naive participants with participants who had inadequate response to prior medication.58,62,63 The remaining four studies55,56,64,65 did not specify current or previous pharmacotherapy trials as part of their selection criteria.
Among the included studies, six pharmacogenomic tests that include decision-support tools were evaluated: GeneSight (2 RCTs,57,65 3 post-hoc analyses,66-68 and 2 non-randomized studies55,56), Neuropharmagen (2 RCTs60,62 and 1 post-hoc analysis69), CNSDose (1 RCT64), Genecept (1 RCT61), NeuroIDgenetix (1 RCT58), and an unspecified test (1 RCT63). Specific details of each genetic test and its corresponding decision-support tool are shown in Appendix 6, Table A4. The CNSDose test used by Singh et al64 tests for variants in multiple genes and uses a proprietary combinatorial approach to develop an interpretive report; however, the publication provided no details about the genes and variants included, which therefore could not be summarized here. Among the other five tests, the number of included genes ranged from 5 to 30, with large variation in specific variants assessed and number of medications included in the report. Two versions of the GeneSight test were analyzed; three additional genes were added to the test used in the Greden et al57 study. Several tests used a proprietary combinatorial algorithm to classify medications, and most tests classified medications into risk categories based on the potential for gene-drug interactions. The studies evaluating the NeuroIDgenetix test58 and Neuropharmagen tests60,62 both noted additional non-gene factors were included within the test report, but it is unclear if these are linked to or combined with the genetic test recommendations.
The group receiving treatment as usual was poorly described in all studies, with no information regarding standard guidelines or prescribing practices followed. All studies swabbed patients' cheeks in the group receiving treatment as usual for pharmacogenomic testing but did not provide results to patients or clinicians until completion of the study's follow-up period.
Psychiatrists treated patients in all studies, while two studies also included primary care providers.57,58 Clinicians treated patients in both arms of the study, with one study including only a single psychiatrist for all patients.63 Similarly, the level of experience or training of clinicians was not adequately summarized.
Baseline Characteristics
Appendix 5, Table A3, summarizes baseline patient characteristics for each primary study.
Mean age ranged from 41 to 52 years across all studies, except for Shan et al63 who limited inclusion to a maximum of 51 years and observed a mean age of 26 to 29 years. Most participants were female in all studies. Seven study populations were predominately White or people of European ancestry (range: 63%–100%); one studied solely a Korean population60 and another only a Han Chinese population.63
The mean number of previous medication trails at baseline ranged from 1.7 to 4.7 across the six studies that reported this measure; in one study approximately 30% failed to benefit from two or three medications.61 Only three studies reported on the mean number of antidepressants being taken at baseline, ranging from 1.7 to 2.9.55,56,65
Only the largest study, by Greden et al57 (GeneSight), reported baseline pharmacogenomic test categories, noting only 18.3% of participants were taking a medication that was considered incongruent with the test among a population who had all failed to benefit from one or more treatment trials (i.e., categorized as use with increased caution and with more frequent monitoring). Fifteen percent of patients were also taking medications that were not included on the GeneSight report.
Studies provided limited information on which treatments patients started, their genetic congruency by treatment, or which treatments were subsequently switched to or augmented.
Risk of Bias in Included Studies
All included RCTs were at high risk of bias owing to various study design, analysis, or reporting issues (Appendix 7, Table A5). The primary concern was that all studies had clinicians who were not blinded to treatment. Most studies had outcome assessors blinded for some outcomes; however, several studies had clinician assessors who were not blinded for one or all outcomes. Shan et al63 did not blind clinicians or patients to treatment. Blinding is particularly important given the subjective nature of depression outcomes and potential for clinicians or assessors to influence perceived outcomes. Additionally, minimal information was provided on patient recruitment, with potential for selection bias, as clinicians were involved in both recruitment and treatment of patients.
Loss to follow-up was greater than a quarter to over a third of patients in each arm of three studies,57,60,63 with minimal information regarding reasons for such substantive losses.
Two studies were at high risk of bias due to selective outcome reporting, whereby only a proportion of results for patients were presented58 or important outcomes listed in the protocol were not reported in initial or post-hoc publications.57 Selective outcome reporting may have been present in other studies; however, detailed methods and study protocols were not available for all. The study by Han et al was at high risk of bias owing to serious errors in statistical analyses and accounting of patients, which was recently noted by a corrigendum.59,60
All but one study was funded by the pharmacogenomic test manufacturer. Most included authors and analysts employed by the manufacturer. While no study was downgraded for this reason alone, this factor could bias results in favour of the intervention, as has been noted in previous literature.70
Similarly, significant risk of bias was observed among the two non-randomized trials55,56 (Appendix 7, Table A6); their primary issues were lack of consideration of potential confounding variables, a lack of blinding of outcome assessors, and incomplete outcome data.
Change in Depression Score
Change in depression scores was most frequently measured using the HAM-D17 scale or the structured version of the scale, known as SIGH-D17, which we consider equivalent for the purposes of this review. Several studies also reported on changes in the 16-item Quick Inventory of Depressive Symptomatology (QIDS-C16), 9-item Patient Health Questionnaire (PHQ-9), Clinical Global Impressions Scale—Severity (CGI-S), and HAM-D6. Results are reported for the longest follow-up periods (8 to 12 weeks) in Tables 3 and 4, and for earlier follow-up periods in Appendix 8. A summary of the various depression scales is provided in Appendix 4, Table A2. For all scales, a higher score indicates worse depressive symptoms.
Table 3:
Change in HAM-D17 Depression Scores at Final Follow-Up
| Author, Year | No. PGx/TAU | Mean Score at Follow-up (SD) or Mean Δ From Baseline (SD) | Percent Decrease From Baseline to Follow-Up | P Valuea | ||
|---|---|---|---|---|---|---|
| PGx | TAU | PGx | TAU | |||
| GeneSight | ||||||
| Greden et al, 201957 | Allb: 621/678 PPb: 560/607 |
15.1c (NR) 15.4c (NR) |
15.7c (NR) 16.1c (NR) |
26.7 27.2 |
23.5 24.4 |
.07 .11 |
| Winner et al, 201365 | 25/24 | NR | NR | 30.8 | 20.7 | .29 |
| Hall-Flavin et al, 201355 | 72/93 | 12.3d (NE) | 15.4d (NE) | 46.9 | 29.9 | < .0001e |
| Hall-Flavin et al, 201256 | 22/22 | 14.1d (NR) | 17.5d (NR) | 30.8 | 18.2 | .04f |
| Neuropharmagen | ||||||
| Han et al, 201859,60h | 52/48 | Δ –16.1 (6.8) | Δ –12.1 (8.2) | NR | NR | MD: .04g |
| Perez et al, 201762 | 143/142 | Δ –8.01d (NE) | Δ –6.45d (NE) | NR | NR | MD: .08 |
| Genecept | ||||||
| Perlis et al, 202061 | 146/150 | 12.77 (6.65) | 11.90 (6.68) | 43.34 | 45.99 | .52 |
| Unspecified Test | ||||||
| Shan et al, 201963 | ITT: 31/40 PP: 21/27 |
8.10 (4.12) 6.76 (2.88) |
9.88 (5.49) 8.26 (4.84) |
60.86 NR |
52.38 NR |
.21 MD: .32 |
Abbreviations: Δ, change from baseline to follow-up; CI, confidence interval; ITT, intention to treat; MD, mean difference; NE, not estimated from graph; NR, not reported; PGx, pharmacogenomic-guided treatment selection; PP, per protocol; TAU, treatment as usual; SD, standard deviation.
P values reflect differences in percent decrease from baseline to follow-up between the PGx and TAU groups unless otherwise noted as MD.
“All” cohort included all patients who met eligibility criteria. Per protocol cohort excluded patients who had a score of < 14 on HAM-D17 at baseline as well as those with protocol violations or if clinician did not view pharmacogenomic report prior to baseline. Only patients who completed 8-week follow-up were included in both analyses.
Calculated from reported percentage decrease scores and baseline HAM-D17 scores for people who completed week 8.
Estimated using WebPlotDigitizer software based on graphic data provided in the publication. Not estimated from graph indicates standard errors and deviations were not extracted from figures.
Mixed model for repeated measures found P < .001.
Mixed model for repeated measures found P < .05.
Mean difference and confidence intervals not calculated from reported data, as reported P values are adjusted for various factors.
Data based on results from corrigendum.
Table 4:
Change in Depression Scores on Alternative Depression Scales at Final Follow-Up
| Scale Test | Author, Year | No. PGx/TAU | Mean at Follow-Up (SD) or Mean Δ From baseline (SD) | % Decrease From Baseline | P Valuea | ||
|---|---|---|---|---|---|---|---|
| PGx | TAU | PGx | TAU | ||||
| QIDS-C16 | |||||||
| GeneSight | Greden et al, 201957 | 621/678 | NR | NR | 35.1 | 32.9 | .19 |
| Winner et al, 201365 | 25/24 | NR | NR | 27.6 | 22.1 | NS | |
| Hall-Flavin et al, 201355 | 72/93 | 9.65b (NE) | 11.24b (NE) | 44.8 | 26.4 | < .0001c | |
| Hall-Flavin et al, 201256 | 22/22 | 10.92b (NR) | 13.91b (NR) | 31.2 | 7.2 | .002d | |
| Genecept | Perlis et al, 202061 | 146/150 | Δ –6.04 (5.4) | Δ –6.45 (5.1) | NR | NR | MD: 0.39 |
| 9-Item Patient Health Questionnaire | |||||||
| GeneSight | Greden et al, 201957 | 621/678 | NR | NR | 34.1 | 29.3 | .04 |
| Winner et al, 201365 | 25/24 | NR | NR | 35.4 | 21.3 | .18 | |
| Hall-Flavin et al, 201355 | 72/93 | 10.07b (NE) | 11.61b (NE) | 40.1 | 19.5 | < .0001e | |
| Neuropharmagen | Han et al, 201859,60 | 52/48 | Δ –13.6 (6.8) | Δ –9.8 (7.8) | NR | NR | .05f |
| HAM-D6 | |||||||
| GeneSight | Dunlop et al, 201966 (Greden et al, 201957) |
621/677 | NR | NR | 28.3 | 23.9 | .023 |
| Clinical Global Impression Scale-Severity | |||||||
| Neuropharmagen | Perez et al, 201762 | CR: 144/143 PR: unclear |
Δ –1.14 (1.13) Δ –1.09 (1.37) |
Δ –0.87 (1.13) Δ –0.87 (1.38) |
NR NR |
NR NR |
MD: .04 MD: .18 |
| Han et al, 201859,60 | CR: 52/48 | Δ –3.3 (1.4) | Δ –2.3 (1.8) | NR | NR | MD: .02f | |
| Genecept | Perlis et al, 202061 | 146/150 | Δ –1.74 (1.26) | Δ –1.65 (1.21) | NR | NR | MD: .56 |
Abbreviations: Δ, change from baseline to follow-up; CR, clinician-rated; HAM-D6, 6-item Hamilton Depression Rating Scale; ITT, intent-to-treat; MD, mean difference; NE, not estimated; NR, not reported; NS, not significant; PGx, pharmacogenomic-guided treatment selection; PR, patient rated; QIDS-C16, 16-item Quick Inventory of Depressive Symptomatology; TAU, treatment as usual; SD, standard deviation.
P values reflect differences in percent decrease from baseline to follow-up between PGx and TAU groups unless otherwise noted as MDs. Values are presented as reported in original article.
Estimated using WebPlotDigitizer software based on graphs in the publication. Standard errors and deviations were not extracted from figures.
Result based on endpoint analysis. Repeated measures analysis P < .001.
Result based on endpoint analysis. Repeated measures analysis P = .05.
Result based on endpoint analysis. Repeated measures analysis P = .002.
Mean differences and CIs not calculated from reported data, as reported P values were adjusted by authors.
Most studies reported on percentage changes from baseline to follow-up. While mean differences in scores were considered the most clinically relevant outcome, few studies directly reported the mean differences and variances between groups. Where provided or estimated, a mean change of 2 to 3 points was considered clinically relevant for HAM-D17 scores.41 No minimal clinically important differences were identified for other depression outcome measures in studies. Given reported P values for mean differences were obtained with methods accounting for repeated measures and often adjusted for additional factors, we did not calculate the unadjusted mean differences and variances between groups unless data were clearly presented.
17-ITEM HAMILTON DEPRESSION RATING SCALE
Results for studies reporting change in depression score based on the 17-item Hamilton Depression Rating Scale (HAM-D17 or HDRS) are grouped by specific test and summarized in Table 3 and Appendix 8, Table A23. Across all studies general improvements in depression scores were seen in both the pharmacogenomic-guided treatment groups and treatment as usual groups. Overall, results were either inconsistent within a specific test or found no statistically significant difference between groups (GRADE: Low to Very Low).
GeneSight
All four GeneSight studies reported the percentage change from baseline to follow-up on the HAM-D17 as the primary outcome measure of the study. Meta-analysis was not performed because mean change scores were not reported by any study.
The RCT evidence suggests little to no difference in the percentage decrease in HAM-D17 depression scores from baseline to final follow-up (range: 8–10 weeks) among those who received pharmacogenomic-guided medication selection compared with those who received treatment as usual. Results, however, are not statistically significant and are very uncertain (GRADE: Very Low; Appendix 7, Table A7). Mean HAM-D17 scores at follow-up were not reported by either study; however, based on reported baseline values and percent decreases, the raw data suggest no clinically meaningful differences between groups in the Greden et al57 trial (mean difference of 0.67 points) when using the predefined threshold of a 2- to 3-point decrease.
In contrast to the RCT data, the open-label studies by Hall-Flavin et al55,56 suggest pharmacogenomic-guided treatment selection results in a large percent decrease in HAM-D17 scores compared with treatment as usual at 8 weeks' follow-up; however, results were very uncertain (GRADE: Very Low; Appendix 7). The mean difference in scores based on graphs in the studies were clinically meaningful, ranging from a decrease of 3.1 to 3.4 points, although variance could not be assessed. Both studies noted similar results were observed when a mixed model for repeated measures analysis was performed, and Hall-Flavin et al (in 2013)55 observed similar results using post-hoc imputation methods accounting for missing data (data not shown). No significant differences were observed at 2 weeks in either study, or at 4 weeks in the study by Hall-Flavin et al (in 2012)56 (Appendix 8, Table A23).
Neuropharmagen
Both Neuropharmagen studies found pharmacogenomic-guided testing improved mean HAM-D17 depression scores from baseline to follow-up compared with treatment as usual (Table 3). However, the larger and higher-quality study by Perez et al62 did not find a statistically significant difference (P = .08), and the effect size was not a clinically meaningful difference based on unadjusted data (1.6 points). Han et al59,60 observed a statistically significant reduction in mean scores (P = .036), with a clinically meaningful decrease of 4 points. The GRADE for this body of evidence is assessed as Low (Appendix 7).
Genecept
Medication selection guided by the Genecept pharmacogenomic tool appears to result in no difference on the percent change in SIGH-D17 depression score compared with treatment as usual (P = .516) (GRADE: Low; Appendix 7). Using unadjusted data by the authors, we found the mean difference in scores was not clinically or statistically meaningful, with the point estimate favouring treatment as usual (mean difference 0.87, 95% CI −0.65 to 2.39). Depression scores improved from baseline in both arms and indicated mild depression at final follow-up (SIGH-D17 < 14). Similar results were observed at the 2-, 4-, and 6-week follow-up periods (Table A23, Appendix 8).
Another Unspecified Test
Depression medication selection guided by the pharmacogenomic test evaluated by Shan et al63 led to little or no improvement in change of HAM-D17 scores at follow-up compared with those who received treatment as usual; however, results were not statistically significant and very uncertain (Grade: Very Low; Appendix 7). Final scores were less than 10 in both arms at follow-up. Results were consistent with the per-protocol analysis as well as for earlier follow-up periods (Appendix 8).
OTHER DEPRESSION SCALES
Results for studies reporting change in depression score based on the QIDS-C16, PHQ-9, HAM-D6, and CGI-S depression scales are grouped by specific test and summarized below and in Table 4 and Appendix 8.
16-Item Quick Inventory of Depressive Symptomatology
Five studies reported on changes in QIDS-C16 scores (Table 4); all observed results similar to those for HAM-D17.
The two GeneSight RCTs57,65 indicated pharmacogenomic-guided treatment selection may have little to no effect on percent change scores, but the evidence is very uncertain (GRADE: Very Low; Appendix 7). Final scores for each group, however, were not provided. On the contrary, both non-randomized studies found GeneSight-guided treatment selection may improve depression scores based on percent decrease in QIDS-C16 from baseline. These results, however, were also highly uncertain (GRADE: Very Low; Appendix 7). Changes were not statistically significant until 4 weeks for the Hall-Flavin et al55 study (in 2013) nor at the final 8-week follow-up for Hall-Flavin et al56 (in 2012) (Appendix 8, Table A24).
The evidence suggests pharmacogenomic-guided treatment selection with Genecept may not reduce depression symptoms according to mean change from baseline QIDS-C16 scores, with the effect estimate favouring treatment as usual (mean difference 0.48, 95% CI −0.61 to 1.56; P = .39) (GRADE: Low; Appendix 7).
9-Item Patient Health Questionnaire
In contrast to the primary outcome of HAM-D17, which found no significant difference, the two GeneSight RCTs showed inconsistent results for percent change from baseline with the PHQ-9 score. The largest study by Greden et al57 observed a statistically significant improvement in PHQ-9 among people who received pharmacogenomics-guided treatment selection compared with treatment as usual (P = .04), while Winner et al65 observed no significant difference. Neither study provided final follow-up scores for assessment of mean differences, and we are uncertain about the effect observed (GRADE: Very Low; Appendix 7). The non-randomized trial observed a large percent decrease from baseline with pharmacogenomics-guided treatment selection at 8 weeks (multi-gene pharmacogenomic-guided testing: 40% vs. treatment as usual: 19.5%); however, the mean change in scores was 1.5 points and results were very uncertain (GRADE: Very Low; Appendix 7). No significant difference was observed at 2 or 4 weeks of follow-up (Appendix 8, Table A24).
Han et al59,60 found Neuropharmagen reduced PHQ-9 scores with a mean difference in change from baseline of 3.8 points; however, results were not statistically significant (P = .054) and were very uncertain (GRADE: Very Low; Appendix 7). No significant difference was observed when the 15-item PHQ score was used for evaluation (P = .239).
6-Item Hamilton Depression Rating Scale
A post-hoc analysis of the data provided by Greden et al57 was reassessed to calculate 6-item Hamilton Depression Rating Scale (HAM-D6) scores.66 Based on this reassessment of data, GeneSight led to a small but statistically significant reduction in percent change from baseline score compared with treatment as usual (28.3% vs. 23.9%; P = .02); however, no baseline scores were provided for assessment of mean differences in scores. Findings are very uncertain, as the GRADE for this body of evidence was assessed as Very Low, primarily owing to the post-hoc nature of this analysis (Appendix 7).
Clinical Global Impressions Scale
Three studies reported on the CGI-S as a measure of improvement in depression scores.60-62
Both Neuropharmagen studies found a statistically significant decrease in mean change of the clinician-rated CGI-S among people who received pharmacogenomic-guided testing compared with treatment as usual. The mean change from baseline based on unadjusted analyses, however, ranged greatly from −0.27 to −1.0. The GRADE for this body of evidence was assessed as Low (Appendix 7). No difference was observed when the patient-rated questionnaire was used (P = .184).
The evidence suggests Genecept-guided medication selection does not improve the mean change in depression scores based on CGI-S compared with treatment as usual (mean difference −0.08, 95%CI −0.33 to 0.18; P = .5612) (GRADE: Low; Appendix 7).
SUBPOPULATION ANALYSES
Planned subgroup analyses based on prior medication use (treatment naive vs. inadequate response to prior medications) or provider type could not be performed given a paucity of data for each test and few studies included for each test. However, subgroup analyses on these and other factors as reported by individual studies are summarized below and presented in Appendix 8.
Prior Medication Use
Only Perez et al62 reported on specific subgroups by prior depression medication use in post-hoc analysis (Appendix 8, Table A25). This study found a clinically (mean difference = 3 points) and statistically significant reduction in HAM-D17 depression scores among people who failed one to three medications at baseline and were randomized to pharmacogenomics-guided treatment selection with the Neuropharmagen decision-support tool compared with those who received treatment as usual (P = .008). The definition of treatment failure was not specified by the study. This study further noted no statistically significant differences between the two randomized groups among those who failed no medications at baseline or those who failed four or more medications (P values not reported). No comparison was made, however, between the three different subgroups and no evaluation of all patients who failed one or more medications was provided.
OTHER SUBGROUPS
A post-hoc analysis of the Perez et al Neuropharmagen trial by Menchon et al69 performed several sub-population analyses among the original dataset (Appendix 8, Table A25). This study found significant differences in change in HAM-D17 depression scores between those receiving pharmacogenomic-guided treatment selection compared with treatment as usual when HAM-D17 was greater than or equal to 18 at baseline, among those who were either less than or equal to 1 year since time of diagnosis or less than or equal to 5 years, as well as for those aged less than 60 years of age. No direct comparisons, however, were made between subgroups evaluated.
Similarly, Forester et al67 assessed a subgroup of patients from the Greden et al57 trial who were aged 65 years and older at baseline, and as with the original cohort analysis, found no statistically significant difference in depression scores between the pharmacogenomic-guided treatment selection group and treatment as usual.
Two of the original GeneSight studies and two post-hoc analyses of the Greden et al57 trial further analyzed results based on baseline classifications of patients' potential gene–drug interactions (Appendix 8, Table A26). These analyses excluded people for whom baseline medications were not listed on the GeneSight interpretive report (N = 228 in the Greden et al57 trials, N = 20 in the Hall-Flavin et al study, others not specified). The Winner et al65 RCT and post-hoc analyses68 of the Greden et al trial57 found statistically significant decreases in percent change in depression score among those in the pharmacogenomic-guided group who were on a “red or yellow bin” medication at baseline, where red bin is defined as “use with caution and more frequent monitoring” and yellow bin is “use with caution”. No difference was observed among those who were on one of these medications and subsequently switched medications, or for any individual grouping alone (green, yellow, or red bin). No data were provided to determine the mean change in scores from baseline and subsequent clinical significance of this data. Similarly, the open-label study observed a significant decrease only among those in the red bin at baseline.
Response
Response to treatment for depression, defined as an improvement of 50% or more in depression score from baseline, was reported by eight studies in addition to three post-hoc analyses of the GUIDED trial by Greden et al66-68 and a post-hoc analysis of the AB-Gen trial by Perez et al.62,69 Response to treatment was most often measured using the HAM-D17 or SIGH-D17 scales. Several studies also reported response using the QIDS-C16, PHQ-9, HAM-D6, and CGI-S.
17-ITEM HAMILTON DEPRESSION RATING SCALE
Results for studies reporting response based on the HAM-D17 depression scale (including the SIGH-D scale) are grouped by specific test and summarized in Figure 2 and Appendix 8. The overall rate of response in the pharmacogenomic-guided arm ranged greatly across studies, from 25% in the largest Genesight trial to 74% with the unspecified test by Shan et al.63 Overall, a statistically significant improvement in response was observed with the GeneSight (GRADE: Low–Very Low) and NeuroIDgenetix tests (Grade: Low), with other pharmacogenomic tests observing no statistically significant difference or inconsistent results (Grade: Low–Very Low).
Figure 2: Meta-Analysis for Relative Risk of Response with PGx Compared With TAU Based on HAM-D17 Scale.
Abbreviations: CI, confidence interval; df, degrees of freedom; HAM-D17, 17 item Hamilton Depression Rating Scale; M-H, Mantel-Haenzel test; PGx, pharmacogenomic-guided medication selection; TAU, treatment as usual.
a All studies are randomized controlled trials except where specified.
b Insufficient data were provided by Han et al for calculation of an effect estimate. Results for this study are shown in text and Appendix.
c Estimates for events and total numbers were calculated from data provided in study. Estimates can vary from publication because of variation in statistical analyses used or rounding differences.
GeneSight
Meta-analysis of the two GeneSight RCTs found a 34% relative improvement in response among people who received pharmacogenomic-guided treatment compared with treatment as usual (Figure 2); however, this finding was based on low-quality evidence as assessed by GRADE (Appendix 7). This relative improvement corresponds to an absolute rate of improvement of 7% (95% CI 2%–11%) and a number needed to treat of 15 (Appendix 8, Table A27). The total number of patients achieving response by the end of follow-up was less than 27% in either group.
The open-label study by Hall-Flavin et al (in 2013)55 also found pharmacogenomic-guided treatment may improve response based on the HAM-D17 scale compared with treatment as usual (RR 1.60; 95% CI 1.04–2.46; P = .03); however, results were very uncertain (GRADE: Very Low). The percent response among the pharmacogenomic-guided treatment arm was observed to be higher than in either randomized trial (43.1% vs. 26% and 36%; Table A27), which could reflect the lack of blinding in this study.
Neuropharmagen
Inconsistent and uncertain results were observed between the two Neuropharmagen trials on response rate. The larger and higher-quality trial by Perez et al observed no statistically significant difference in response (RR 1.13, 95% CI 0.86–1.48; P = .39) between groups at 12 weeks of follow-up (Figure 2). Han et al observed a statistically significant improvement (P = .014) with pharmacogenomic-guided treatment selection at 8 weeks, although the relative risk and variance could not be calculated from data provided (Table A27). Percent response was similar in the arms receiving treatment as usual between studies, however, was larger in the pharmacogenomic-guided arm in the Han et al59,60 trial (64.7% vs. 45.4% in Perez et al).
The evidence is very uncertain, as GRADE for this body of evidence was assessed as Very Low (Appendix 7).
NeuroIDgenetix
Bradley et al58 found people were 37% more likely to respond to treatment at 12 weeks with NeuroIDgenetix-guided medication selection relative to people receiving treatment as usual (GRADE: Low; Appendix 7). This represented an absolute increase of 17% (95% CI 5%–29%), and a number needed to treat of 6 (Appendix 8, Table A27). This analysis limited to those with moderate to severe depression at baseline, excluding the portion of the randomized patient population with mild depression at baseline. No data were provided for the full study population with depression.
Genecept
Perlis et al61 found that pharmacogenomic-guided medication selection with the Genecept decision-support tool does not improve response to depression treatment relative to standard care (Grade: Low; Appendix 7). Genecept-guided patients were 17% less likely respond to treatment relative to treatment as usual. Results were imprecise, however; confidence intervals included both reduction and improvement in benefit.
Unspecified Test
The evidence suggests that the unspecified test by Shan et al63 may improve the relative rate of response compared with treatment as usual; however, confidence intervals spanned both a large benefit and reduced effect (P = .14) (GRADE: Low; Appendix 7). In this trial response rates were much larger than those observed with any other study in both the intention-to-treat (74.2% and 57.5%) and per-protocol (90.5% and 70.4%) analyses for the pharmacogenomic-guided and treatment as usual groups, respectively (Appendix 8, Table A27).
OTHER SCALES MEASURING TREATMENT RESPONSE
Relative risk of response based on non–HAM-D17 depression scales are shown in Table 5.
Table 5:
Relative Risk of Response for Pharmacogenomic-Guided Medication Selection Compared With Treatment as Usual Based on Alternative Depression Scales
| Measure | Author, Year (Primary Study) | % Response (n/N) | RR (95% CI) | P Value | |
|---|---|---|---|---|---|
| PGx | TAU | ||||
| GeneSight | |||||
| QIDS-C16 | Greden et al, 201957 | 34.1 (212a/621) | 31.4 (213a/678) | 1.09 (0.93–1.27) | .29 |
| Hall-Flavin et al, 201355 | 44.4 (32/72) | 23.7 (22/93) | 1.88 (1.20–2.94) | .005 | |
| 9-Item Patient Health Questionnaire | Greden et al, 201957 | 39.7 (247a/621) | 31.6 (214a/678) | 1.26 (1.09–1.46) | .01 |
| Hall-Flavin et al, 201355 | 50.7 (36/72) | 31.2 (29/93) | 1.60 (1.10–2.35) | .01 | |
| HAM-D6 | Dunlop et al, 201966 (Greden et al, 201957) | 29.6 (184a/621) | 22.5 (152a/677) | 1.32 (1.10–1.59)a | .004 |
| Neuropharmagen | |||||
| PGI-I (score ≤ 2) | Perez et al, 201762 | 12 wk: 47.79 (NR) 8 wk 40.56 (NR) 4 wk: 28.47 (NR) |
12 wk: 36.11 (NR) 8 wk: 37.41 (NR) 4 wk: 31.97 (NR) |
OR 1.62 (1.0–2.61) NR NR |
.047 NS NS |
| PGI-I (sustained responseb) | 38.5 (NR) | 34.4 (NR) | NR | NS | |
| Genecept | |||||
| Clinical Global Impression–Improvement (≤ 3) | Perlis et al, 202061 | 87.7 (128/146) | 78.7 (118/150) | 1.11 (1.01–1.24) | .04 |
Abbreviations: CI, confidence interval; HAM-D6, 6-item Hamilton Depression Rating Scale; NR, not reported; NS, not significant; OR, odds ratio; PGI-I, Patient Global Impression of Improvement; PGx, pharmacogenomic-guided treatment; QIDS-C16, 16-item Quick Inventory of Depressive Symptomatology (clinician rated); RR, relative risk; TAU, treatment as usual.
Calculated from data provided in study. Estimates might vary from those in publication because of variation in statistical analyses used or rounding differences.
PGI-I ≤ 2 on at least two consecutive evaluations and maintained until final study visit.
9-Item Patient Health Questionnaire and 16-Item Quick Inventory of Depressive Symptomatology
The large GUIDED trial57 and the non-randomized GeneSight study55 evaluated response using the PHQ-9 and QIDS-C16 scales. With the PHQ-9 scale, both studies found statistically significant improvement in the relative rate of response, although the effect size was much smaller in the randomized trial (Table 5). The absolute risk of improvement was 8% (95% CI 3%–13%) in the Greden et al57 trial. Similarly, a large and significant improvement was observed by Hall-Flavin et al with the QIDS-C16 scale (RR 1.88; 95% CI 1.20–2.94); however, no statistically significant improvement was observed by Greden et al57 (RR 1.09; 95% CI 0.93–1.27). The evidence, however, is very uncertain about the effect of GeneSight-guided treatment selection on response rates for each of these outcome measures (GRADE: Very Low; Appendix 7).
Patient Global Impression of Improvement and Clinical Global Impression–Improvement
Using a PGI-I score of 2 or less as the primary measure of response, Perez et al found pharmacogenomic-guided treatment selection with Neuropharmagen may improve response at 12 weeks relative to treatment as usual (Table 5). However, the confidence interval included no effect (RR 1.62; 95% CI 1.0–2.61) (GRADE: Low; Appendix 7). The authors stated there was no statistically significant effect on sustained response, defined as PGI-I of 2 or less on two consecutive evaluations and maintained until final study visit (Table 5).
Han et al59,60 also evaluated the proportion of patients showing scores of 1 or 2 in the Clinical Global Impressions Scale—Improvement (CGI-I) at the end of treatment, finding no statistically significant difference in the proportion of patients achieving this outcome with Neuropharmagen-guided medication selection compared with treatment as usual (pharmacogenomic-guided treatment: 71.2% vs. treatment as usual: 58.3%; P = .197). This outcome was not considered a definition of response by the study and therefore not included within the GRADE analysis.
Similarly, Perlis et al61 found Genecept may improve relative response rate, defined as 3 or less on the CGI-I scale, compared with treatment as usual; however, confidence intervals ranged from very small or no difference to a modest improvement (RR 1.11; 95% CI 1.01–1.24) (GRADE: Low; Appendix 7).
HAM-D6
A post-hoc analysis of the Greden et al57 GUIDED trial by Dunlop et al66 reanalyzed the original study data with the HAM-D6 depression scale (an abbreviated version of the HAM-D17), finding a similar improvement in the relative rate of response with pharmacogenomic-guided care as the rate observed with the HAM-D17 scale and an absolute increase of 7% (Table 5). The results from this analysis, however, are uncertain (GRADE: Low; Appendix 7).
SUBPOPULATION ANALYSES
Given a paucity of data for each test, formal subgroup analyses we had planned to assess on prior medication use (treatment naive vs. inadequate response to prior medications) or provider type could not be performed. Subgroup analyses as performed by individual studies are presented below and summarized in Appendix 8, Table A28.
Prior Medication Use
Only three studies clearly limited their study enrollment to people who had inadequate response, with study results shown in the section above.57,60,61 Among studies with a combined population of treatment-naive participants and those with inadequate response, only two commented on differences in response rates between these groups.58,62
Bradley et al58 reported greater improvement in response for the experimental group compared with the group receiving treatment as usual when the population was limited to patients with treatment-resistant depression (62% vs. 44%; P = .01). This comparison, however, was included only as a post-hoc analysis within the discussion. Further description of this population was not provided.
Based on PGI-I, Perez et al62 found a greater rate of response with pharmacogenomic-guided testing than with treatment as usual when analysis limited to those individuals who failed one to three previous treatments (OR 2.39; 95%CI 1.28–4.44; P = .006). This post-hoc analysis excluded people who failed four or more treatments, and no direct comparison was made to people who had not failed prior treatment.
Other
Consistent results were observed in the Greden et al57 GeneSight trial when analysis was limited to patients aged 65 years and older,67 while subgroups of the Perez et al62 Neuropharmagen trial showed statistically significant improvement in response among those aged less than 60 years but not for those aged 60 years or older. Perez et al62 further found no significant difference in response based on HAM-D17 when limiting analysis to those people with baseline HAM-D17 scores of less than 18 or of 25 or greater. They did notice a significant improvement in people with scores of 18 and over. In contrast, Bradley et al found a greater improvement with NeuroIDGenetix in those with scores of 24 or greater but saw no improvement in those with mild depression. No clear trend was observed in relation to time since diagnosis in the subgroup analysis of Perez et al.62,69
Several studies stratified results for response based on genetic test results at baseline (i.e., people on genetically congruent and non-congruent medications). A post-hoc analysis68 of the Greden et al57 study found a statistically significant improvement in response between the GeneSight-guided treatment arm and treatment as usual when limiting to people receiving yellow or red bin medications at baseline. The absolute difference in response between the two groups was similar to that observed for the overall cohort in Greden et al57. A separate analysis directly comparing to these participants with those who were receiving genetically congruent medications at baseline was not provided. As seen with the overall cohort, Perlis et al61 found no significant difference in response with Genecept-guided care compared with treatment as usual when comparing people taking concordant versus discordant medications.
Remission
The impact of pharmacogenomic-guided treatment on remission from depression was reported by nine primary studies (eight RCTs and one non-randomized study) and three post-hoc publications of RCTs. Various depression scales were used to assess remission within individual studies. Remission was defined as a depression score at follow-up of 7 or less on the HAM-D17 scale, 5 or less on QIDS-C16, less than 5 on PHQ-9, and 4 or less on HAM-D6.
17-ITEM HAMILTON DEPRESSION RATING SCALE
Results for the eight studies reporting remission based on the HAM-D17 (or SIGH-D) are summarized in Figure 3 and Appendix 8. Rates of remission at follow-up ranged from 16.8% to 75% in the intervention arms of included trials, with a range of 9% to 51.8% in the treatment as usual arms. Overall, the evidence from three tests (GeneSight, NeuroIDgenetix, CNSDose) suggested statistically significant improvements in relative rates of remission (Figure 3). There was uncertainty in effect on remission among the remaining three pharmacogenomic tools (RR 0.78–1.36), none of which were statistically significant.
Figure 3: Meta-Analysis for Relative Risk of Remission With PGx Medication Selection Compared With TAU Based on HAM-D17.
Abbreviations: CI, confidence interval; df, degrees of freedom; HAM-D17, 17-Item Hamilton Depression Rating Scale; M-H, Mantel-Haenzel test; PGx, pharmacogenomic-guided treatment; RCT, randomized controlled trial; TAU, treatment as usual.
a All studies are RCTs except where specified.
b Han et al provided insufficient data for calculation of effect estimate. Results for this study are shown in text and Appendix 8.
c Estimates for events and total numbers were calculated from data provided in study. Estimates might vary from publication owing to variation in statistical analyses used or rounding differences.
Sources: Bradley et al, 2018,58 Greden et al, 2019,57 Hall-Flavin et al, 2013,55 Han et al, 2018,60 Perez et al, 2017,62 Perlis et al, 2020,61 Shan et al, 2019,63 Singh et al, 2015,64 Winner et al, 2013.65
GeneSight
Meta-analysis of the two GeneSight RCTs showed a 50% relative improvement in remission among people who received pharmacogenomic-guided treatment compared with treatment as usual (RR 1.50; 95% CI 1.14–1.96) (Figure 3; GRADE: Low, Appendix 7). This corresponds to an absolute increase in remission of 6% (95% CI 2%–9%) with pharmacogenomic-guided testing and a number needed to treat of 17 (Appendix 8).
In contrast to the combined RCT data, the open-label study55 did not find a statistically significant improvement in the relative risk of remission among people who received pharmacogenomic-guided treatment rather than treatment as usual (Figure 3; RR 1.42; 95% CI 0.84–2.39). Results for this outcome were very uncertain (GRADE: Very Low; Appendix 7). The proportion of people achieving remission in both arms of this study was larger than proportions in either of the RCTs.
Neuropharmagen
Meta-analysis of the two Neuropharmagen RCTs could not be performed given the lack of data from the Han et al59,60 trial and differences in study populations. Overall, the effect was very uncertain. The larger trial by Perez et al62 found little to no difference in relative risk of remission between the two groups (Figure 3), with data assessed only post hoc. Han et al59,60 found no statistically significant difference between groups (14.2% difference; P = .147) (Appendix 8, Table A29) (GRADE: Very Low; Appendix 7).
NeuroIDgenetix
One trial of NeuroIDgenetix58 reported remission among a small subset of randomized participants with severe depression at baseline (HAM-D17 ≥ 24). This was considered a post-hoc analysis as methods planned for results in all patients with HAM-D17 > 18. This study found pharmacogenomic-guided medication selection may result in a large increase in remission relative to treatment as usual (RR 2.65; 95% CI 1.18–5.95; Figure 3) (GRADE: Very Low; Appendix 7). This represented an absolute increase of 22% (95% CI 4%–39%), and a number needed to treat of 5 (Appendix 8, Table A29). No data were provided for participants with moderate depression (n = 168) who were included within other study outcome assessments. Authors noted that no significant improvements were observed among patients with mild depression, although no data were provided.
Genecept
The evidence from one study suggested pharmacogenomic-guided treatment selection with Genecept may result in a lower rate of remission relative to treatment as usual using the SIGH-D test (a standardized version of the HAM-D17); however, results did not reach statistical significance (RR 0.78; 95% CI 0.54–1.14). The GRADE for this outcome was assessed as Low (Appendix 7).
CNSDose
The evidence suggests CNSDose-guided medication selection may lead to a large improvement in remission relative to treatment as usual (RR 2.52, 95% CI 1.71–3.73) (GRADE: Low; Appendix 7). The absolute rate of improvement was 43 % (95% CI 29%–58%) with a number needed to treat of 3 (Appendix 8, Table A29).
Unspecified Test
Shan et al63 found an unspecified test may improve rate of remission; however, results were very uncertain with confidence intervals spanning both benefit and harm (P = .17) (GRADE: Very Low; Appendix 7). This study had a substantially higher percentage of patients achieving remission in both the intervention arm as well as the treatment as usual arm (76% and 51%, respectively) compared with other trials identified (multi-gene pharmacogenomic-guided testing range: 15%–72%; treatment as usual range: 8.3%–33%).
REMISSION WITH ALTERNATIVE DEPRESSION SCALES
In addition to HAM-D17, remission rates were reported using various alternative scales by two GeneSight studies55,66 (Table 6). Similar to results using HAM-D17, Greden et al57 observed the rate of remission may improve when evaluated using the QIDS-C16 scale (RR 1.34; 95% CI 1.05–1.69), (GRADE: Low; Appendix 7). However, the effect is very uncertain based on the HAM-D6 scale in a post-hoc publication (RR 1.28; 95% CI 1.02–1.61) (GRADE: Very Low; Appendix 7). When assessed with the PHQ-9 scale, a trend toward a benefit was also observed; however, confidence intervals cross no effect (RR 1.26; 95% CI 0.98–1.60). (GRADE: Very Low; Appendix 7). The absolute increase with each scale, however, was smaller than that observed with HAM-D17.
Table 6:
Relative Risk of Remission for PGx Compared With TAU Based on QIDS-C16, PHQ-9, and HAM-D6 Scales
| Measure | Author, Year (Primary Study) | % Remission (n/N) | RR (95% CI) | P Value | |
|---|---|---|---|---|---|
| PGx | TAU | ||||
| GeneSight | |||||
| QIDS-C16 | Greden et al, 201957 | 20.9 (130a/621) | 15.6 (106/678) | 1.34 (1.06–1.69)a | .01 |
| Hall-Flavin et al, 201355 | 26.4 (19/72) | 12.9 (12/93) | 2.05 (1.06–3.93)a | .028 | |
| PHQ-9 | Greden et al, 201957 | 18.6 (115a/621) | 14.8 (100a/678) | 1.26 (0.98–1.60)a | .066 |
| Hall-Flavin et al, 201355 | 25.4 (18a/72) | 16.1 (15a/93) | 1.73 (0.80–3.74)a | .14 | |
| HAM-D6 | Dunlop et al, 201966 (Greden et al, 201957) | 20.8 (129a/621) | 16.2 (110a/677) | 1.28 (1.02–1.61)a | .031 |
Abbreviations: CI, confidence interval; HAM-D6, 6-item Hamilton Depression Rating Scale; PGx, pharmacogenomic-guided medication selection; PHQ-9, Patient Health Questionnaire–9; QIDS-C16, 16-Item Quick Inventory of Depressive Symptomatology (clinician rated); RR, relative risk; TAU, treatment as usual.
Calculated from data provided in study. Estimates may vary from publication due to variation in statistical analyses used or rounding differences.
Findings from the open-label study by Hall-Flavin et al55 were very uncertain but similarly observed a 2-times increase in remission with pharmacogenomic-guided treatment selection relative to treatment as usual with the QIDS-C16 scale (GRADE: Very Low), but no statistically significant difference in the rate of remission using the PHQ-9 scale (P = .14) (Grade: Very Low; Appendix 7).
SUBPOPULATION ANALYSES
Given that no more than two studies were included for each test and lack of data, preplanned subgroup analyses by previous use of depression medications (i.e., treatment naive vs. inadequate response to treatment) and by provider type could not be performed. Results for subgroups as reported by individual studies are shown below and are summarized in Appendix 8, Table A30.
Inadequate Response to Medication
The three studies that limited the randomized patient population to participants who had inadequate response to one or more medications at baseline57,60,61 each used a different test and had conflicting results as shown above (Figure 3).
Bradley et al58 (IDGenetix) reported greater improvement in response when they limited to those patients with treatment-resistant depression (42% vs. 27%; P = .03). Yet this finding was included only as a post-hoc analysis within the discussion, and this population was not further defined. Additionally, researchers provided no data on the population without treatment-resistant depression.
Other
Several studies stratified or sub-grouped results by age, depression severity, or medication congruency (Appendix 8, Table A30).
Perez et al62 performed a post-hoc analysis limited to patients with a HAM-D17 depression score of 19 or greater; however, no statistically significant difference was observed between the Neuropharmagen-guided group and the group that received treatment as usual.
A post-hoc analysis of the Greden et al57 Genesight trial assessed remission among patients who were aged 65 years and older, noting a larger absolute improvement among those receiving pharmacogenomic-guided treatment compared with treatment as usual than was observed in the overall cohort; however, no direct comparisons were made to those who were aged less than 65 years.
Additionally, two studies stratified results from the Greden et al57 GeneSight trial based on medication congruency with test results at baseline (i.e., those receiving medications considered congruent and non-congruent based on the genetic testing results). Both were post-hoc analyses and found statistically significant improvements in those who were taking medications classified as “use with caution” (i.e., yellow bin) or “use with caution and more frequent monitoring” (i.e., red bin) at baseline and received pharmacogenomic-guided testing compared with those who received treatment as usual. No direct comparison was made, however, with people taking medications classified as “use as directed” (i.e., green bin) at baseline, and analyses excluded patients with medications that were not included on the GeneSight report at baseline. The original Greden et al57 paper classified congruency differently than the post-hoc analyses, including patients taking yellow bin medications as being congruent, given they required only minor clinical modifications (Appendix 8, Table 30).
Adverse Events and Side Effects
Six of the primary studies included treatment side effects or adverse reactions as outcomes, which were defined in the original articles (Table 7). Bradley et al58 reported data for a combined depression and anxiety cohort only, with no stratification among the depression cohort, and therefore results were not included.
Table 7:
Adverse Events for PGx Compared With TAU
| Author, Year | Measure | N PGx/N TAU | PGx | TAU | Summary Estimate (95% CI) | P Value |
|---|---|---|---|---|---|---|
| GeneSight | ||||||
| Greden et al, 201957 | Number of side effectsb Proportion of side effects |
560/607c | Mean 0.243 (SE 0.029) 15.6% |
Mean 0.237 (SE 0.028) 15.3% |
MD 0.01 (−;0.07 to 0.09)a RR 1.03 (0.78 to 1.34)a |
.855 .881 |
| Neuropharmagen | ||||||
| Han et al, 201860 | Change in FIBSER FIBSER frequency domain (≤ 2) FIBSER intensity domain (≤ 2) FIBSER burden domain(≤ 2) |
52/48 | Mean Δ –4.1 (SD 5.3) 96.2% 94.2% 92.3% |
Mean Δ –1.6 (SD 5.9) 83.3% 52.1% 50.0% |
NRd NRd NRd NRd |
.001 .035 < .001 < .001 |
| Perez et al, 201762 | FIBSER burden domain (≤ 2) for tolerability subpopulatione | 97/80 | 6 wk: 66.7% 12 wk: 68.5% |
6 wk: 50% 12 wk: 51.4% |
OR 2.0 (1.07 to 3.75) OR 2.06 (1.09 to 3.89) |
.029 .026 |
| Change in FIBSER frequency domain Change in FIBSER intensity domain Change in FIBSER burden domain |
143/143 | Mean Δ –0.68 (SD 2.35) Mean Δ –0.60 (SD 2.01) Mean Δ –0.57 (SD 2.00) |
Mean Δ –0.25 (SD 2.38) Mean Δ –0.09 (SD 1.92) Mean Δ –0.01 (SD 1.72) |
NRf NRf NRf |
.1280 .0303 .0125 |
|
| Genecept | ||||||
| Perlis et al, 202061 | Change in FIBSER frequency domaing Change in FIBSER intensity domaing Change in FIBSER burden domaing |
150/153 | Mean Δ –0.1 (SD 2.18) Mean Δ 0.0 (SD 1.86) Mean Δ –0.2 (SD 1.55) |
Mean Δ –0.2 (SD 2.18) Mean Δ 0.0 (SD 1.90) Mean Δ –0.2 (SD 1.59) |
MD 0.10 (−;0.39 to 0.59)a MD 0.00 (−;0.42 to 0.42)a MD 0.00 (−;0.35 to 0.35)a |
.69a 1.00a 1.00 a |
| CNSDose | ||||||
| Singh et al, 201564 | Intolerability rateh | 74/74 | 4% | 15% | RR 1.13 (1.01 to 1.25) | .027 |
| Unspecified Test | ||||||
| Shan et al, 201963 | Proportion of adverse reactionsi | 31/43j | 55.56% | 57.89% | NRe | NS |
Abbreviations: CI, confidence interval; FIBSER, Frequency, Intensity, and Burden of Side Effects Ratings; MD, mean difference; Mean Δ, mean change from baseline; NR, not reported; NS, not significant; OR, odds ratio, PGx, pharmacogenomic-guided treatment; PP, per protocol; RR, relative risk; SD, standard deviation, SE, standard error; TAU, treatment as usual.
Calculated using Review Manager on basis of data provided in article.
Measured at week 8. Defined as a probability of a causal link to the medication; side effects unrelated to medications not included.
Data provided only for per-protocol cohort and not overall cohort as with other outcomes.
Insufficient data provided to confirm denominator.
Calculated on basis of tolerability subpopulation (FIBSER > 0 at baseline).
Study data and P values assessed using ANOVA and therefore unadjusted values not calculated.
Based on symptoms over past week. Calculated as mean change from baseline to follow-up.
Patient needed to reduce dose or stop the antidepressant.
Assessed using Treatment Emergent Symptom Scale.
Number was included in full analysis protocol; however, unclear denominator was used for analyses.
Greden et al57 found little to no difference in the mean number of side effects (MD 0.01 [95% CI–0.07 to 0.09]) or the proportion of patients with a side effect (RR 1.03 [95% 0.78–1.34]) between the GeneSight-guided group and the treatment as usual group at week 8 (GRADE: Low, Appendix 7). The most common adverse events reported included dry mouth, nausea, headache, constipation, and fatigue. Similar results were observed in the subgroup analysis by Forester et al,67 which was limited to people aged 65 years and older (P = .435).
Several studies60-62 reported on the Frequency, Intensity, and Burden of Side Effects Ratings (FIBSER) scale, a 3-item scale used to assess side effects of antidepressant treatment over the previous week.71 The three questions cover the domains of frequency, intensity, and burden of side effects, with scores ranging from 0 to 6 for each question; higher scores indicate greater effect (Appendix 4, Table A2). Clinical relevance is considered a score of 3 or greater on the burden subscale, indicating the side effect should be addressed or treatment should be changed.
Overall, treatment selection guided by the Neuropharmagen test mayresult in either a greater reduction in the mean change from baseline FIBSER score or a higher proportion of patients achieving a score of 2 or less on all subscales at final follow-up (Table 7) (GRADE: Low; Appendix 7). Results were statistically significant for all outcomes in both studies, except for the mean change in FIBSER frequency score observed by Perez et al62 at week 12 (P = .128). Results were, however, statistically significant at the 6-week follow-up for all domains. When limited to participants reporting side effects related to burden at baseline (FIBSER > 0), the odds of achieving a Burden subscore of 2 or less were two times higher for the Neuropharmagen-guided group than for treatment as usual at both 6 and 12 weeks of follow-up (Table 7). Han et al60 reported the most common adverse events for pharmacogenomic-guided treatment were sleep disturbance, anxiety, and somnolence and for treatment as usual were headache, anxiety, and somnolence. Perez et al62 did not report on specific adverse events observed.
On the contrary, Perlis et al61 observed no statistically significant differences in the mean change in any FIBSER subscale from baseline to follow-up with Genecept-guided treatment compared with treatment as usual (GRADE: Moderate). Data were also reported for changes at 2- and 4-week follow-up; however, authors observed no meaningful differences at any time point (data not shown).
Singh et al64 observed a 13% relative reduction in the rate of intolerability to medication, defined as a requirement to reduce the dose or stop the antidepressant, when guided with CNSDose compared with treatment as usual (P = .027) (GRADE: Low; Appendix 7). The main reactions observed were considered mild: headache, dizziness, drowsiness, nausea, vomiting, dry mouth, constipation, diarrhea, decreased appetite, and tachycardia.
Shan et al,63 however, found no significant difference in adverse reactions between pharmacogenomic-guided treatment and treatment as usual when measured by the Treatment Emergent Symptom Scale. (GRADE: Very Low; Appendix 7).
Change in Treatment Prescribing Patterns
Three studies reported on how pharmacogenomic-guided testing affected clinician decision-making or medication selection (Table 8). Bradley et al58 reported data only for a combined depression and/or anxiety cohort, with no stratification among the depression cohort, and therefore results were not included.
Table 8:
Impact on Therapeutic Decision for PGx Compared With TAU
| Author, Year | Measure | PGx | TAU | P Value |
|---|---|---|---|---|
| GeneSight | ||||
| Hall-Flavin et al, 201355 | Switched, augmented, or dose-adjusted medication | 76.8% | 44.1% | < .001 |
| Winner et al, 201365 | Switched, augmented, or dose-adjusted medication | 53% | 58% | .66 |
| Mean no. of medications prescribed | Mean 1.9 | Mean 1.7 | .27 | |
| CNSDose | ||||
| Singh et al, 201565 | Medication dosing different from usual practicea | 65% | NA | NA |
Abbreviations: NA, not applicable, PGx, pharmacogenomic-guided testing; TAU, treatment as usual.
Definition not specified by study.
The randomized trial by Winner et al65 noted no difference in the proportion of patients that had their medication switched, augmented, or dose-adjusted with GeneSight-guided treatment selection compared with treatment as usual, with 53% and 58% experiencing a change in each group, respectively. On the contrary, the open-label study by Hall-Flavin et al55 found more patients had a change in their medication selection with GeneSight, with 77% having their medication switched, augmented, or dose-adjusted compared with only 44% in the treatment as usual arm (P < .001).
Singh et al64 noted 65% of medication dosing was different from usual practice with the CNSDose-guided treatment selection; however, it was unclear how the study assessed or defined this difference.
SUBGROUPS BY BASELINE MEDICATION CLASSIFICATION AND TEST RESULTS
Several studies reported on changes in medication selection or types of medications provided based on baseline genetic test results and congruency with baseline medications (Table 9). Studies showing the proportion of patients taking congruent medications at follow-up suggest prescribing patterns that differ between groups, but do not necessarily indicate that more changes occurred in either group, rather that clinicians were following the test results.
Table 9:
Treatment Selection Based on Genetic Test Classification for PGx Versus TAU
| Author, Year (Primary Study) | Measure | Proportion (%) | P Value | |
|---|---|---|---|---|
| PGx | TAU | |||
| GeneSight | ||||
| Greden et al, 201857 | Taking congruent medications at follow-up (green or yellow bin)a | Baseline: 79.4 Follow-up: 91.2 |
Baseline: 77.5 Follow-up: 76.3 |
NR |
| Forester et al, 201967 (Greden et al, 201857) | Distribution of medication's gene-drug interaction severity category at follow-up (aged ≥ 65 y) | Green bina: 59.7 Yellow bina: 26.0 Red bina: 14.3 |
Green bina: 25.0 Yellow bina: 50.0 Red bina: 25.0 |
Overall < .001 |
| Thase et al, 201968(Greden 201857) | Medication switch among those in yellow or red bin at baselinea,b,c | 65.8 | 52.3 | < .001 |
| Taking congruent medications at follow-up (green bin) among those in yellow or red bin at baselinea,b | 66.4 | 20 | NR | |
| Winner et al, 201365 | Switched, augmented, or dose-adjusted among those in red bin at baselinea,b | 100 | 50 | .02 |
| Hall-Flavin et al, 201355 | Switched, augmented, or dose-adjusted among those in red bin at baselinea,b | 93.8 | 55.6 | .01 |
| Taking greena bin medication at follow-up | 40 | 27.6 | NS | |
| Hall-Flavin et al, 201256 | Difference in prescribing patterns at follow-up | Green bina: NE Yellow bina: NE Red bina: 5.9 |
Green bina: NE Yellow bina: NE Red bina: 21.4 |
Overall .02 |
| Neuropharmagen | ||||
| Perez et al, 201762 | Prescribed medications in disagreement with test result | 17 participants | NA | NA |
| Unspecified Test | ||||
| Shan et al, 201963 | Prescribed medications in “Use as directed” category Prescribed medications in “Use with caution” category |
NR 3.2 |
37.5 40 |
NR NR |
| Prescribed medications in “Use with increased caution and more frequent monitoring” category | 0 | 22.5 | NR | |
Abbreviations: NA, not applicable; NE, not estimable; NR, not reported; NS, not significant; PGx, pharmacogenomic-guided treatment; TAU, treatment as usual.
Medications were categorized as green bin (use as directed), yellow bin (use with caution), or red bin (use with increased caution and more frequent monitoring).
Medication switch was defined as dropping a medication and adding a different medication during first 8 weeks.
Analysis considered only people taking at least one medication on test report.
GeneSight
All primary GeneSight studies evaluated various measures of therapeutic decisions based on baseline medication genetic classification or the proportion taking medications considered to be genetically congruent at follow-up. Greden et al57 found a similar proportion of people were taking medications considered to be genetically congruent with test results at baseline, but more people in the pharmacogenomic-guided group were taking a congruent medication at follow-up than in the treatment-as-usual group. Subgroup analyses from this trial found more patients switched if their medications were yellow or red bin at baseline (P < .001). Among people taking yellow or red bin medications at baseline, more were taking a green bin medication at follow-up if their treatment was guided by the GeneSight test (66.4% vs. 20%). Similarly, two GeneSight studies found more patients switched, augmented, or dose-adjusted treatment if their medications were considered red bin at baseline55,65; the third study noted differences in overall prescribing patterns at follow-up based on medication bin classification. However, Hall-Flavin et al (in 2013)55 found no statistically significant difference in the proportion taking a green bin medication at follow-up.
Neuropharmagen
Perez et al62 noted only 17 participants who received pharmacogenomic-guided treatment were taking medications that were in disagreement with the test results. However, no prescribing data were provided for participants receiving treatment as usual.
Unspecified Test
Shan et al63 found nearly all patients (97%) in the pharmacogenomic-guided group were prescribed medications in the “use as directed category” compared with only 37.5% in the treatment as usual group. More patients who received treatment as usual were likely to be given a medication in the “use with caution” category.
Suicide
No studies reported on suicide as an outcome of interest.
Relapse, Recovery, Recurrence
No studies reported on relapse, recovery, or recurrence of depression symptoms as an outcome of interest.
Quality of Life
No studies reported on quality of life as an outcome of interest.
Treatment Adherence
No studies reported on treatment adherence as an outcome of interest.
Ongoing Studies
We are aware of the following ongoing or recently completed (not yet published) studies that have potential relevance to this review.
Table 10:
Ongoing or Recently Completed Comparative Studies on Multi-gene Pharmacogenomic Testing
| Clinicaltrials.gov Identifier | Title | Genetic Test Evaluated |
|---|---|---|
| NCT02466477 | Pharmacogenomic Decision Support With GeneSight Psychotropic to Guide the Treatment of Major Depressive Disorder | GeneSight |
| NCT03749629 | Comparative Effectiveness of Pharmacogenomics for Treatment of Depression (CEPIO-D) | GeneSight |
| NCT03952494 | Individualizing Antidepressant Treatment Using Pharmacogenomics and EHR-driven Clinical Decision Support (MyGenes) | Genomind Professional PGx Express |
| NCT03591224 | Pharmacogenomic Testing to Optimize Antidepressant Drug Therapy | Pillcheck |
| NCT03468309 | Medication Optimization Using Pharmacogenetic Testing and the G-DIG to Reduce Polypharmacy in a Mental Health Population (MedOPT) | Genecept Assay and G-DIG decision tool |
Discussion
Major depressive disorder is a serious public health issue resulting in major personal, societal, and economic burdens.1,72 Multi-gene pharmacogenomic testing that includes decision-support tools for people with major depression is intended to predict which psychotropic medications and dosages are most likely to result in a treatment response and have the lowest risk of an adverse event based on a person's genetic profile.
Overall, we found inconsistent outcome reporting and inconsistent findings across the six multi-gene pharmacogenomic tests with decision-support tools identified. No improvement or little improvement across all depression outcomes was observed with Genecept-guided medication selection as well as with an unspecified pharmacogenomic test evaluated by Shan et al. The evidence found little to no difference on the impact of GeneSight-guided medication selection on depression scores, with inconsistent and uncertain results observed for Neuropharmagen. We found no evidence evaluating how NeuroIDgenetix or CNSDose effected change in depression scores.
We found GeneSight and NeuroIDgenetix led to statistically significant improvements in both response and remission, while CNSDose did not have evidence on response, but showed a statistically significant improvement in remission. The effect of Neuropharmagen on response and remission was inconsistent across studies evaluated. However, the evidence remains uncertain for all outcomes across all tests with a GRADE rating of low to very low for these outcomes, and therefore our confidence that these estimated effects reflect the true effect is low to very low.
Similarly, the impact of testing on adverse side effects from medication selection was inconsistent and uncertain, with little to no difference observed for some tests (i.e., GeneSight, Genecept, and an unspecified test), while the remaining tests reported some improvement.
No data were identified for any test that evaluated the impact of testing on important outcomes such as suicide, treatment adherence, or longer-term outcomes (relapse, recovery, or recurrence of depression symptoms). Similarly, no comparative outcomes were assessed beyond 12 weeks of follow-up.
Subgroup analyses (e.g., treatment naive vs. inadequate response to treatment) could not be specified to identify the populations most likely to benefit from pharmacogenomic-guided treatment. This was due to limited stratification of data and few studies evaluating each test.
On the whole, these findings are consistent with other published systematic reviews and health technology assessments (summarized in Appendix 3). The present review, however, is the most up to date, incorporating the most recent studies and a wider breadth of outcomes. Several previous reviews mathematically combined data across the various tests, leading to positive overall effect estimates for response or remission; however, we thought this was inappropriate given the variations in the tests themselves, as well as differences in patient cohorts included as described below.
Differences Across Included Tests and Considerations
Each pharmacogenomic test and decision-support tool included in this review uses a different combination of genes and variants, a different model to combine genes and provide a predicted phenotype, as well as a different format and level of detail of information presented within the decision-support tool. A recent study found both genotypic and phenotypic results varied across four of the tests included in the present review when assessed on the same five participants, with only modest concordance in medication recommendations.73
It is known that the level of evidence for individual gene–drug interactions ranges substantially, and numerous genetic variants for a single gene have various levels of evidence.11,12,74 Given that algorithms applied to predict outcomes are not disclosed by the companies, results do not tell us which key genetic variants are involved, the level of evidence behind each included gene, or which is the best way to combine them to predict clinical outcomes. Clinical utility evidence surrounding decision-support tools can therefore be considered only in the tools' entirety and not for the genes they consider. There is currently no consensus on the minimum set of genes required for a pharmacogenomic test, and various professional guidelines, regulatory bodies, and tests make different recommendations for the same medications. Consequently, careful consideration of the differences between individual tests is necessary, and any clinical utility observed with one test might not apply to a similar test on the market.
Few available pharmacogenomic tests that include decision-support tools incorporate important clinical and lifestyle characteristics (e.g., age, sex, weight, smoking status, liver or kidney function) into the decision algorithm or consider other concomitant inhibitors or inducers of drug-metabolizing enzymes that are known to have an impact on drug exposure as well as on therapeutic outcomes. For example, a person who is classified as a normal metabolizer for a specific enzyme might more closely resemble a poor metabolizer if the person is taking a medication known to inhibit the enzyme.
Last, results from included studies might not be generalizable to multi-ethnic populations like Ontario's. Most of the literature pertaining to gene–drug interactions has focused on testing in Caucasian or European populations, with only limited data and understanding of pharmacogenetic variation in non-Caucasian populations. It has been shown that the frequency of alleles among various cytochrome P450 genes commonly included within pharmacogenomic tests varies greatly between biogeographical groups; some alleles are very rare among Caucasians, but are common among people with other backgrounds.11,12,75,76 Therefore, a failure to include all relevant alleles within a specific
pharmacogenomic test can lead to false-negative phenotypes for certain ethnicities, and subsequent differences in medication selection and dosing recommendations. Further, studies included within the present review were primarily or entirely among people of European descent or Caucasian, with few exceptions (such as one study limited to a Chinese population and one to a Korean population). One example can be observed from the Neuropharmagen studies that came from two very different populations (over 90% Caucasian and 100% Korean), and could be a driver of differences in results observed.60,62
Generalizability to Ontario Context
In addition to considerations for how each test was developed, their applicability to the Ontario context needs to be considered.
Components of standard care provided across various studies might not reflect practice in Ontario. No study clearly established protocols or guidelines for the treatment as usual arm, and minimal information was provided by the studies regarding which medications were provided at baseline and how patients moved between medications (i.e., treatment switches, augmented treatment, dosage changes) or how often changes were made throughout a study period. There was also minimal information on the level of experience or training of prescribing clinicians, which could greatly influence the effect observed in the standard care arm and therefore might or might not represent the current standard of care in Ontario. The ability to accurately interpret each decision-support tool is a critical factor in successfully implementing pharmacogenomic data and recommendations.
Genetic test versions used in the included studies might not reflect the tests currently being marketed or could change with future iterations of tests. For instance, the GeneSight studies included in the review ranged from five genes in the earlier studies to eight genes in the most recent study, while the current version of the test includes an additional four genes (for a total of 12). These variations could affect both the validity and subsequent clinical utility of a test depending on the evidence surrounding these newer genes as well as the methods used to incorporate the new data into the combinatorial model.
According to a recent scan of available pharmacogenomic tests in psychiatry available in Canada,21 only three of the pharmacogenomic tests evaluated in this review are currently available for out-of-pocket purchase in Ontario (Genecept, GeneSight, and Neuropharmagen), and we found minimal information on current availability or plans to enter the Canadian market for IDGenetix or CNSDose. At least nine other tests are available in Ontario, for which we found no clinical utility data.
Last, in addition to a potential lack of generalizability of the included study populations to the multiethnic population of Ontario, the included studies were predominantly among women and people aged 40 to 50 years and therefore may not be applicable to the wider population in Ontario.
Limitations of the Review
The current review did not assess the analytical or clinical validity of pharmacogenomic testing. While a large body of literature has evaluated clinical validity data for pharmacogenomic testing, a previous health technology assessment by the Washington State Healthcare Authority found that the clinical validity data are limited to only associational evidence with small effect size for single genes or gene variants and selected outcomes. Most important, the data do not capture the unique algorithm-based phenotyping used by marketed tests.39 Researchers concluded that evaluating the many associational studies would not be practical and that clinical usefulness would be limited, particularly when considering the utility of a pharmacogenomic test. Additionally, clinical validity is often utilized as a proxy measure for clinical utility; therefore, a strong impact on patient outcomes relative to standard care would imply overall clinical validity of the test recommendations, although individual or combined genetic variants within the test might have different levels of evidence or might not be clinically valid. This uncertainty supports our focus on the clinical utility of pharmacogenomic tests with decision-support tools as a whole.
Some studies within our review attempted to evaluate groups who would most benefit from pharmacogenomic-guided testing, based on their baseline medications and congruency with test results. Specifically, GeneSight studies generally found that patients on medications in the “use with caution and more frequent monitoring” classification (i.e., red bin) or the combined red and “use with caution” category (i.e., yellow bin) observed the greatest benefit when treatment was guided by the pharmacogenomic test results. While these results help support the clinical validity of the test recommendations, it is impossible to identify this subgroup of patients without testing all patients at baseline, and therefore clinical utility must consider all patients who would require testing because of inadequate treatment response or before starting depression medication.
Conclusions
We identified ten primary studies and four post-hoc follow-up studies that evaluated six pharmacogenomic tests that include decision-support tools. The most-reported outcomes were change in depression score, response, and remission of depression; the HAM-D17 was the most frequently used depression scale to evaluate these outcomes. No data were identified for any test that evaluated the impact of testing on important outcomes such as suicide or treatment adherence, or on longer-term outcomes like relapse, recovery, or recurrence of depression symptoms.
Overall, we found the evidence of GRADE assessment Low to Very Low certainty, primarily owing to serious concerns over risk of bias. Given the heterogeneous evidence, we consider each test individually as follows.
Genesight
CHANGE IN DEPRESSION SCORE
Pharmacogenomic-guided treatment selection with GeneSight may have little to no effect on depression scores as measured by the HAM-D17, QIDS-C16, PHQ-9, or HAM-D6 scales, but the evidence is very uncertain (GRADE: Very Low)
RESPONSE
Pharmacogenomic-guided treatment selection with GeneSight may improve response to treatment relative to treatment as usual when assessed with the HAM-D17 or HAM-D6 scales (GRADE: Low), but we are very uncertain of the effect when assessed with the PHQ-9 or QIDS-C16 scales (GRADE: Very Low).
REMISSION
Pharmacogenomic-guided treatment selection with GeneSight may improve the rate of remission from depression compared with treatment as usual when assessed with HAM-D17 or QIDS-C16 (GRADE: Low), but we are very uncertain of the effect when assessed with PHQ-9 or HAM-D6 scales (GRADE: Very Low).
SIDE EFFECTS AND ADVERSE EVENTS
Pharmacogenomic-guided treatment selection with GeneSight appears to have little to no difference on the mean number of side effects or the proportion of patients with a side effect compared with treatment as usual at 8 weeks (GRADE: Low).
CHANGE IN TREATMENT
Study results were inconsistent on how GeneSight-guided treatment selection affected the proportion of patients who had their medication switched, augmented, or dose-adjusted compared with treatment as usual.
Neuropharmagen
CHANGE IN DEPRESSION SCORE
Results were inconsistent between studies and across scales when the effect of Neuropharmagen-guided treatment on depression scores was assessed using the HAM-D17, PHQ-9, or CGI-S scores. The evidence is uncertain (GRADE: Low–Very Low).
RESPONSE
The evidence was inconsistent and very uncertain about the effect of Neuropharmagen-guided treatment selection on the response rate when measured using the HAM-D17 scale (GRADE: Very Low) but may improve response relative to treatment as usual as defined using the PGI-I scale—although confidence intervals include no effect (GRADE: Low).
REMISSION
The evidence is very uncertain about the effect of pharmacogenomic-guided testing with Neuropharmagen on relative risk of remission, as assessed by the HAM-D17 scale, with no statistically significant differences observed between groups (GRADE: Very Low).
SIDE EFFECTS AND ADVERSE EVENTS
Pharmacogenomic-guided treatment selection with Neuropharmagen may reduce side effects compared with treatment as usual, based on the FIBSER scale (GRADE: Low).
CHANGE IN TREATMENT
We found no evidence evaluating how Neuropharmagen-guided testing affected changes in treatment decisions.
NeuroIDgenetix
CHANGE IN DEPRESSION SCORE
We found no evidence evaluating the effectiveness of NeuroIDgenetix-guided treatment on changes in depression scores.
RESPONSE
NeuroIDgenetix-guided medication selection may improve response to treatment compared with treatment as usual (GRADE: Low).
REMISSION
Pharmacogenomic-guided treatment selection with NeuroIDgenetix may result in a higher rate of remission from depression among people with severe depression at baseline, but we are very uncertain (HAM-D17 ≥ 24) (GRADE: Very Low)
SIDE EFFECTS/ADVERSE EVENTS
We found no evidence evaluating how NeuroIDgenetix-guided treatment affected side effects or adverse events.
CHANGE IN TREATMENT
We found no evidence evaluating how NeuroIDgenetix-guided treatment affected change in treatment decisions.
Genecept
CHANGE IN DEPRESSION SCORE
Evidence suggests that Genecept-guided medication selection does not reduce depression scores relative to treatment as usual when assessed with the HAM-D17, QIDS-C16, or CGI-S scales (GRADE: Low).
RESPONSE
Genecept-guided medications selection does not improve response to depression medication relative to treatment as usual when assessed using the HAM-D17 scale (Grade: Low), with little to no difference in response based on a score of 3 or less on the CGI-I scale (Grade: Low).
REMISSION
Pharmacogenomic-guided treatment selection with Genecept does not improve remission from depression relative to treatment as usual based on the SIGH-D scale (GRADE: Low).
SIDE EFFECTS AND ADVERSE EVENTS
Pharmacogenomic-guided treatment selection with Genecept likely results in little to no difference in side effects based on the FIBSER scale assessment (GRADE: Moderate).
CHANGE IN TREATMENT
We found no evidence evaluating how Neuropharmagen-guided testing affected change in treatment decisions.
CNSDose
CHANGE IN DEPRESSION SCORE
We found no evidence evaluating the impact of CNSDose-guided treatment on change in depression scores.
RESPONSE
We found no evidence evaluating the effectiveness of CNSDose-guided treatment on treatment response.
REMISSION
CNSDose-guided medication selection may lead to a large improvement in remission relative to treatment as usual, based on the HAM-D17 scale (GRADE: Low).
SIDE EFFECTS AND ADVERSE EVENTS
Evidence suggests pharmacogenomic-guided treatment selection with CNSDose may result in a reduction in the rate of medication intolerability compared with treatment as usual; however, results are uncertain (GRADE: Low).
CHANGE IN TREATMENT
Clinicians who guided treatment selection with CNSDose stated medication dosing was different from usual practice in 65% of cases.
Unspecified Decision-Support Tool
CHANGE IN DEPRESSION SCORE
Depression medication selection guided by the unspecified pharmacogenomic test with decision-support tool evaluated by Shan et al led to little or no improvement in depression scores based on the HAM-D17 scale compared with treatment as usual; however, results were not statistically significant and were very uncertain (Grade: Very Low).
RESPONSE
Shan et al found an unspecified pharmacogenomic test with decision-support tool may improve rate of response; however, results were observed to have confidence intervals spanning both a large benefit and reduced effect (GRADE: Low).
REMISSION
Shan et al found an unspecified pharmacogenomic test with decision-support tool may improve rate of remission as assessed by the HAM-D17 score; however, results were very uncertain with confidence intervals spanning both benefit and harm (GRADE: Very Low).
SIDE EFFECTS AND ADVERSE EVENTS
The unspecified pharmacogenomic test with decision-support tool evaluated by Shan et al may have no effect on adverse reactions compared with treatment as usual, but results are very uncertain (GRADE: Very Low).
CHANGE IN TREATMENT
We found no evidence evaluating the effect of the unspecified test by Shan et al on change in treatment decisions.
Economic Evidence
Research Question
What is the cost-effectiveness of multi-gene pharmacogenomic testing that includes decision-support tools to guide medication selection compared with treatment as usual for people with major depression?
Methods
Economic Literature Search
We performed an economic literature search on January 24, 2020, to retrieve studies published from database inception until the search date. To retrieve relevant studies, we used the clinical search strategy and applied an economic and costing filter.
We created database auto-alerts in MEDLINE, Embase, and PsycINFO and monitored them for the duration of the assessment period. We also performed a targeted grey literature search of health technology assessment agency websites, clinical trial and systematic review registries, and the Tufts Cost-Effectiveness Analysis Registry. See Appendix 1 for our literature search strategies, including all search terms.
Eligibility Criteria
STUDIES
Inclusion Criteria
English-language full-text individual-level or model-based comparative economic studies published from database inception until January 24, 2020, or later as identified via auto-alert search updates
Cost-effectiveness analyses, cost–utility analyses, cost–benefit analyses, or cost–consequence analyses
Exclusion Criteria
Narrative or systematic reviews, letters/editorials, commentaries, case reports, conference abstracts, study protocols, guidelines, and unpublished studies
Costing studies, feasibility analyses, or cost-of-illness studies
POPULATION
Inclusion Criteria
-
Adults (aged 18 years and older) with major depression requiring pharmacological treatment (i.e., medication naive or treated and inadequately responsive to one or more medications)
-
○Studies with combined populations were included only if results for the depression subgroup could be extracted
-
○
Exclusion Criteria
Bipolar depression
Children or adolescents
INTERVENTIONS
Inclusion Criteria
Multi-gene (2 or more genes) pharmacogenomic tests that include a clinical decision–support tool to guide depression medication selection
Exclusion Criteria
Single-gene tests or tests that provide no decision-support tool to guide treatment or dosage recommendations
COMPARATORS
Inclusion Criteria
No pharmacogenomic testing to guide depression medication selection or dose adjustment (i.e., treatment as usual)
Exclusion Criteria
Studies comparing various pharmacogenomic tests or genes
OUTCOME MEASURES
Costs
Health outcomes (e.g., response, remission, recovery, relapse, recurrence), quality-adjusted life-years (QALYs), or disability-adjusted life-years (DALYs)
Incremental costs
Incremental effectiveness
Incremental cost-effectiveness ratios (e.g., cost per incremental QALY gained) or incremental net benefit
Literature Screening
A single reviewer conducted an initial screening of titles and abstracts using Covidence42 and then obtained the full texts of studies that appeared eligible for review according to the inclusion criteria. This reviewer then examined the full-text articles and selected studies eligible for inclusion.
Data Extraction
We extracted relevant data on study characteristics and outcomes to collect information about the following:
Source (e.g., citation information, study type)
Methods (e.g., study design, analytic technique, perspective, time horizon, population, intervention[s], comparator[s])
Outcomes (e.g., health outcomes, costs, incremental cost-effectiveness ratios [ICERs])
Study Applicability and Limitations
We determined the usefulness of each identified study for decision-making by applying a modified quality appraisal checklist for economic evaluations originally developed by the National Institute for Health and Care Excellence (NICE) in the United Kingdom to inform the development of NICE's clinical guidelines.77 We modified the wording of the questions to remove references to guidelines and to make it specific to Ontario. Next, we separated the checklist into two sections. In the first section, we assessed the applicability of each study to the research question (directly, partially, or not applicable). In the second section, we assessed the limitations (minor, potentially serious, or very serious) of the studies we found to be directly or partially applicable.
Results
Economic Literature Search
The database search of the economic literature search yielded 471 citations published from database inception until January 24, 2020. We identified three additional studies from other sources, for a total of 363 after removing duplicates. These additional articles were identified through auto-alerts in MEDLINE or Embase or through additional search of the grey literature.
We excluded 319 articles on the basis of information in the title and abstract and obtained 44 potentially relevant articles for full-text assessment. Four studies met the inclusion criteria and were assessed to establish the applicability of their findings to the Ontario context. See Appendix 9 for a list of some studies excluded after full-text review. Figure 4 presents the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) flow diagram for the economic literature search.
Figure 4: PRISMA Flow Diagram—Economic Search Strategy.
Abbreviation: PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-analyses.
Source: Adapted from Moher et al.46
Overview of Included Economic Studies
Table 11 presents study designs, populations, outcomes, and results of the four included studies in detail. Below, we summarize these findings.
Table 11:
Results of Economic Literature Review—Summary
| Author, Year Country of Publication | Study Design, Analytic Technique, Perspective, Discounting, Time Horizon | Population | Intervention and Comparator | Results | ||
|---|---|---|---|---|---|---|
| Health Outcomes | Costsa | Cost-Effectiveness | ||||
| Tanner et al, 202078 Canada |
Study design: Model-based CEA Analytic technique: Markov cohort model Perspective: Canadian public health care system (i.e., public payer including both direct and indirect costs) Discounting: 3% Time horizon: 5 y |
Adults with moderate to severe major depression Mean age, y: 32 Female, %: NR |
Intervention: PGx-guided treatment Comparator: TAU (no PGx) |
Mean QALYs, intervention, and TAU: NR Mean difference, intervention vs. TAU: 0.168 |
Currency, y: 2018 CAD Total (direct, indirect, and testing) costs (mean), intervention, and TAU: NR Total costs, mean difference: –$2,431 PGx test cost (GeneSight): $2,500 |
Reference case: Compared with TAU, PGx-guided treatment is dominant (more effective and cost saving) Sensitivity analyses: Three more analyses address differences in effectiveness of intervention. In all analyses, intervention was dominant PSA: 94.5% to 96.7% chance of PGx-guided treatment being cost-effective over TAU at WTP of $50,000/QALY in the analyses One-way deterministic analyses of key drivers: rate of remission (assuming a 25% lower rate of remission led to incremental costs of the intervention, but ICER remained < $50,000/QALY) |
| Groessl et al, 201879 United States |
Study design: model-based CEA Analytic technique: Markov cohort model Perspective: societal Discounting: 3% Time horizon: 3 y |
Adults with moderate to severe major depression, treatment naive or with inadequately controlled disease Mean age, y: 48 Female, %: NR |
Intervention: PGx-guided treatment Comparator: TAU (no PGx) |
Mean QALYs for intervention and TAU: 2.07 and 1.97 Mean difference for intervention vs. TAU: 0.10 |
Currency, y: USD, 2016 Total (direct, indirect, and testing) costs (mean) for intervention and TAU: $44,697 and $47,295 Direct medical costsb (mean) for intervention and TAU: $29,990 and $32,908 Total costs, mean difference: –$2,598 Direct medical costs,b mean difference: –$2,918 PGx test cost (IDgenetix): $2,000 |
Reference case: compared with TAU, PGx-guided treatment is dominant (more effective and cost saving) Sensitivity analyses: PSA was not done One-way deterministic analyses: results were robust despite changes to response rate, costs, utilities |
| Najafzadeh et al, 201781 United States |
Study design: model-based CEA Analytic technique: discrete event simulation model Perspective: societal Discounting: 3% Time horizon: 3 y |
Adults with moderate or severe major depression or anxiety, receiving treatment level 1 as per the STAR∗D study (citalopram or equivalent therapy)8,88 Mean age, y: 48 (SD 14.5) Female, %: 73 |
Intervention: PGx-guided treatment Comparator: TAU (no PGx) |
Mean QALYs for intervention and TAU: 2.09 (95% CrI: 1.88 to 2.28) and 1.94 (95% CrI: 1.66 to 2.21) Mean difference for intervention vs. TAU: 0.15 (95% CrI: 0.04 to 0.28) |
Currency, y: USD, 2017 Total (direct, indirect, and testing) costs (mean) for intervention and TAU: $14,124 (95% CrI: $10,703 to $17,630) and $14,659 (95% CrI: $10,384 to $19,275) Direct medical costsb (mean) for intervention and TAU: $10,530 (95% CrI: $7,487 to $13,600) and $10,323 (95% CrI: $6,568 to $14,433) Total costs, mean difference: –$535 (95% CrI: –$2,902 to $1,692) Direct medical costs,b mean difference: $207 (95% CrI: considered?–$1,671 to $2,022) PGx test cost (IDgenetix): $2,000 |
Reference case: compared with TAU, PGx-guided treatment is dominant (more effective and cost saving) Sensitivity analyses: PSA: 98% chance of PGx-guided treatment being cost-effective over TAU at WTP of $50,000/QALY One-way deterministic analyses of key drivers time horizon (< 1 y: ICER > $50,000/QALY) and study perspective (if direct medical costs including testing were considered: ICER $1,394/QALY) |
| Hornberger et al, 201580 United States |
Study design: model-based CEA Analytic technique: Markov cohort model Perspective: societal Discounting: 3% Time horizon: lifetime |
Adults with major depression nonresponsive (did not benefit from) ≥ 1 course of antidepressant therapy Mean age, y: 44 Female, %: NR |
Intervention: PGx-guided treatment Comparator: TAU (no PGx) |
Mean QALYs for intervention and TAU: 13.624 and 13.308 Mean difference, intervention vs. TAU: 0.316 |
Currency, y: USD, 2013 Total (direct, indirect, and testing) costs (mean) for intervention and TAU: $272,751 and $276,515 Direct medical costsb (mean for intervention and TAU: $208,260 and $211,971 Total costs, mean difference: –$3,764 Direct medical costs,b mean difference: –$3,711 PGx test cost (GeneSight): $2,500 |
Reference case: compared with TAU, PGx-guided treatment is dominant (more effective and cost saving) Sensitivity analyses: PSA: 94.5% chance of PGx-guided treatment being cost-effective over TAU at WTP of $50,000/QALY in the analyses One-way deterministic analyses of key drivers: cost of PGx ($3,125 vs. $2,500) or duration of catch-up period (1 y vs. 3 y) led to incremental costs of the intervention, but the ICER remained < $50,000/QALY |
Abbreviations: CEA, cost-effectiveness analysis; ICER, incremental cost-effectiveness ratio; NA, not applicable; NR, not reported; PGx, multi-gene pharmacogenomic test; STAR∗D study, Sequenced Treatment Alternatives to Relieve Depression; PSA, probabilistic sensitivity analysis; QALY, quality-adjusted life-year; SD, standard deviation; TAU, treatment as usual; USD, United States dollars; WTP, willingness to pay.
Negative costs indicate savings.
Cost estimate did not include cost of testing.
STUDY METHODS
Study Design and Study Perspective
All four studies, published between 2015 and 2020, were model-based cost-effectiveness analyses.78-81 Three studies (Groessl et al,79 Najafzadeh et al,81 and Hornberger et al80) were conducted from a societal perspective of the United States that included direct medical costs related to health care utilization and indirect costs related to productivity lost, but that did not consider costs to health care for informal caregivers or all other costs to government (e.g., social services).79-81 The most recent study by Tanner et al78 considered the perspective of the Canadian public health care system that corresponded to the broader government perspective, as defined by guidelines by the Canadian Agency for Drugs and Technologies in Health.82 This analysis included both direct medical costs and some direct non-medical costs (i.e., social services), but omitted direct costs to informal caregivers, some rehabilitation and community-based services (e.g., home care), and a broader spectrum of indirect costs.
Time Horizon and Discounting
Two studies, by Groessl et al and Najafzadeh et al,79,81 described the natural and clinical course of major depression and modeled the costs and consequences over 3 years. Tanner et al78 considered a slightly longer time horizon of 5 years, while Hornberger et al80 considered a person's lifetime. In all studies (i.e., in the reference case analysis), the discount rate for both costs and QALYs was 3%.
Analytic (Modeling) Technique
Groessl et al79 and Hornberger et al80 used a Markov cohort analytic technique. Tanner et al78 modified the model developed by Hornberger et al, while Najafzadeh et al81 used an individual-level discrete event simulation (DES). None of the studies considered modeling of the diagnostic accuracy of multi-gene pharmacogenomic testing or adherence of providers or patients to the indicated treatment (based on results of testing).83 Analyses were inconsistent in the choice of major depression states or outcomes and included some of the following major depression stages either as health states or events: response,79-81 no response,79-81 remission,78,79 no remission,78 relapse,79,81 no relapse,81 and death (from suicide or other causes).78-81
Two analyses, with longer time horizons (5 years or lifetime), extrapolated the benefit of an initial treatment choice guided by multi-gene pharmacogenomic testing for about 3 years; after this period, the effectiveness of the initial treatment would start to decline and could catch up with the effectiveness of the control treatment or usual care.78,80 This assumption, about duration of the efficacy of multi-gene pharmacogenomic guided therapy, was based on a 2003 systematic review by Geddes et al,84 who suggested that the benefit of continual use of antidepressants persisted between 24 and 36 months after initial response or remission.84 Another analysis, based on a DES model,81 simulated individual-level health trajectories (dependent on baseline patient characteristics such as age, sex, disease severity, and treatment level) and considered various treatment outcomes (remission, response, or non-response), adverse events, relapse, and death (from suicide or other causes). Changes in medications (i.e., switch, augmentation, or dose change) after baseline were not modeled or presented in detail in any of the published economic studies. Simplified assumptions related to medication changes that occurred later, after a relapse, were made only in a DES modeling study by Najafzadeh et al.81
Study Population
Two studies included adults (mean ages 44 and 48 years) with major depression who did not benefit from at least one course of antidepressants.80,81 Only one study79 included a mixed sample of people (mean age 48 years), who never used antidepressant medications (treatment naive) or had not benefited from previous medications. This study did not report proportions of patients in treatment subgroups; nor did it assign various clinical pathways to present the course of the disease and treatment for these two patient subgroups. Tanner et al did not clearly define their study population with respect to current or prior use of antidepressants.78 Their study included adults with moderate-to-severe major depression, aged 32 years on average (i.e., median age of patients at the onset of major depression78). This suggests that the target population in the Tanner study included newly diagnosed cases for which antidepressants are indicated but treatment had not yet begun.
Interventions/Comparators
All studies examined the cost-effectiveness of multi-gene pharmacogenomic tests that include a decision-support tool aimed to guide depression medication selection (i.e., IDGenetix79,81 and GeneSight78,80). Effectiveness of the multi-gene pharmacogenomic testing to guide treatment was based on the results of manufacturer-supported randomized controlled clinical trials57,58 in the three modeling studies,78,79,81 or the meta-analyses of prospective studies and clinical trials (the GeneSight test solely) in the two modeling studies.78,80 Most participants in these clinical studies57,58 (which were used to inform the cost-effectiveness analyses) had not benefitted from two to three courses of antidepressants before the study began.
In all studies, the control was treatment as usual, which included medications, selected on the basis of standard practice and clinical pharmacologic guidelines.
Assessment of Health Outcomes
In all studies, the effectiveness of the intervention was estimated using QALYs. Differences between groups in other health outcomes, such as rates of suicides79-81 or remission,78,81 were reported.
Assessment of Costs
The cost of multi-gene pharmacogenomic testing was applied as a one-time cost per person, ranging from $2,000 to $2,500 USD in the US-based analyses. The cost was $2,500 CAD in the Canadian study.78 These prices were taken from manufacturers' websites or published sources; it is unclear whether prices were adjusted for mark-ups.
All studies used aggregate estimates for direct medical costs and indirect costs, as estimated in the literature. Direct costs in the US-based cost-effectiveness analyses79-81 were derived from US registries, claims data, and the literature, and were reported in aggregate. Direct medical costs included medications, outpatient clinical care, physician services, psychotherapy, and hospitalization. In one of these studies,81 the total cost estimate (an economic analysis by Greenberg et al85) included costs related to suicide, in addition to direct medical costs. Indirect costs were measured as the costs of productivity loss, costs of absenteeism and presentism, up to the retirement age of 65 years. This approach to costing suggests a limited societal perspective because some cost components recommended for a full societal perspective were omitted, such as costs to informal caregivers, employers (e.g., hiring), government (e.g., social services), and patients (e.g., out-of-pocket payments or premiums).86
The most recent Canadian study,78 which considered a broader federal government payer perspective,82 included the direct medical costs from an administrative database in Manitoba.87 This patient-level analysis of a cohort of patients with depression and controls without depression, matched for age, sex, and place of residence, estimated costs of health care utilization (e.g., hospital services, physician services, prescription drugs, long-term care services, psychotherapy). Total annual direct costs were $10,064 for people with depression and $2,832 for those without depression (including costs of prescription drugs: $1,441 and $557 [2018 CAD], respectively). The two cohorts were not matched by comorbidity status, as comorbidities were considered a study outcome and were present in 43% of patients with depression. The annual estimate of total non-medical costs related to social services use including rent assistance and employment income assistance (people with depression vs. people without depression) was $1,522 vs. $510 per year, respectively; however, other types of costs to government82 (such as rehabilitation, other social services, and informal caregiving) were not considered.
STUDY FINDINGS
All included economic analyses had consistent findings with respect to the cost-effectiveness of treatment guided by multi-gene pharmacogenomic tests versus treatment as usual (Table 11). In the reference case analyses, which considered the total costs (direct and indirect), treatment guided by pharmacogenomic tests was associated with cost savings and greater QALYs, dominating treatment as usual.
Sensitivity Analysis
Robustness of the cost-effectiveness estimates was explored through one-way deterministic sensitivity analyses, subgroup analyses, and probabilistic analysis (PA) (Table 11). One-way deterministic analyses examined the influence on the findings of variations in clinical and utility parameters (e.g., remission and response rates, health state utilities, starting age, disease severity, duration of benefits of the intervention), cost parameters (e.g., cost of care, cost of treatment guided by pharmacogenomic tests), study perspective, and time horizon. These analyses suggested the following parameters influenced the cost-effectiveness results:
Remission rate—Tanner et al78 found that a reduction of the remission rate of the intervention by 25% (reference case: 18.9%) would change the ICER from cost-saving to cost-effective but the estimate would remain below the willingness to pay amount of $50,000 per QALY. Reporting of this analysis is unclear, as the authors reported changes in costs only (e.g., $284) and not changes in QALYs
Duration of the beneficial effect of the intervention—Hornberger et al and Tanner et al78,80 assumed the beneficial effect of the treatment guided by a pharmacogenomic test would remain constant over 3 years. Hornberger et al found that a shorter 1-year beneficial effect of the intervention was associated with incremental costs.80 The authors reported that the ICER remained below $50,000 USD per QALY, but data related to changes in the QALYs, total costs, or ICER are unavailable (not published)
Time horizon—Najafzadeh et al81 showed that, if benefits and costs of treatment guided by a pharmacogenomic test were accrued over shorter periods (12 weeks or less than 1 year), which could correspond to a maximum follow-up of people receiving treatment as usual and multi-gene pharmacogenomic interventions in two major clinical trials,57,58,68 then the ICER would be well above $50,000 USD per QALY. The authors did not explain changes in the estimates of QALYs, costs, or the ICER; but one possible reason could be a lack of time to fail to benefit from treatment as usual (and enter relapse) and to accumulate downstream cost savings with the intervention (due to stable remission and recovery). Another possible explanation could be that costs associated with monitoring and follow-up might continue in people who achieved remission, thus obscuring cost savings of the intervention for several months
Study perspective—Najafzadeh et al81 also showed that the ICER changed as a function of payer perspective. Thus, when only direct medical costs were considered, pharmacogenomic-guided treatment versus treatment as usual became associated with incremental costs of $207 USD and incremental QALYs of 0.15, resulting in the ICER of $1,394 USD per QALY (i.e., the estimate is still below a commonly used willingness-to-pay amount of $50,000 USD/QALY)
In addition, two studies79,81 conducted subgroup analyses confirming similar findings of the original analyses. Groessl et al79 examined a subgroup of people with severe depression; compared with treatment as usual, treatment guided by pharmacogenomic tests resulted in greater cost savings and QALYs than the reference case analysis in a mix of people with moderate to severe depression (savings: $5,810 vs. $2,598 USD [reference case]); and 0.17 vs. 0.10 QALYs [reference case]). Najafzadeh et al81 examined a subgroup of people with anxiety only. The intervention remained cost-effective (incremental QALYs: 0.12 and incremental total (direct and indirect) costs: $4 USD; ICER: $35 USD/QALY, as reported in the original article).
Last, three studies (Table 11) conducted PA and showed that, compared with treatment as usual, treatment guided by multi-gene pharmacogenomic tests was highly likely to be cost-effective (probability of 0.94–0.98) at a willingness-to-pay amount of $50,000 per QALY.78,80,81 The probability of the intervention being dominant (cost saving and more effective) ranged from 0.6781 to 0.75.80
Applicability and Limitations of Included Studies
Appendix 10 presents the results of the quality appraisal checklist for economic evaluations applied to the included studies. All studies were partially applicable to our research question (Appendix 10, Table A31). In three studies,79-81 the estimates of costs were based on US data; the Canada-based cost-effectiveness analysis78 examined a broader perspective (of the federal government) and used a discount rate of 3%. Therefore, results of the included studies were not directly applicable to Ontario.
We assessed the methodological quality of the included studies and found that all studies had potentially serious limitations (Appendix 10, Table A32). Appropriate analytic modeling techniques were chosen. However, there is substantial heterogeneity in models that considered a variety of depression outcomes (e.g., response, remission, relapse).
Major depression is a chronic and episodic disease involving recurrent episodes of depression. Relapse is a short-term outcome in which acute depression symptoms reappear between 3 and 6 months from the start of the single episode and treatment.6,89-91 Recurrence is defined as a totally new episode of depression, occurring at least 6 to 9 months after recovery.6,89-91 Some models included relapse as a health state,78,79,81 but recurrence and recovery were not captured even when the authors followed participants over their lifetimes. Also, modeling of utilities and effectiveness of the intervention and treatment as usual strategies for a short-term relapse state was not transparent. For example, it was unclear whether and how utilities changed when a person transitioned to a relapse health state. Efficacy of pharmacogenomic-guided treatment on relapse was not reported in the primary studies57 but was calculated using various data sources,8,88,92 without exploring the impact of methodological quality or potential bias of the original sources on the cost-effectiveness results.
Given the lack of data, it is unclear if potentially favourable effectiveness of pharmacogenomic-guided treatment could be easily extrapolated over the long term. Authors of the included DES study81 showed that, when the effect of the intervention was extrapolated over a short term (for less than 1 year), the cost-effectiveness of pharmacogenomic-guided treatment versus treatment as usual was unfavourable (i.e., ICER > $50,000/QALY). Authors of another study80 modeled a decline of the benefit of the intervention, catching up with the level of benefit associated with usual care after 3 years. The 2003 review by Geddes et al,84 which supported assumptions related to duration of the effectiveness of the intervention, examined the probability of relapse in people who used relatively old types of antidepressants; thus, the duration of beneficial effects from new classes of antidepressants has not been corroborated in novel clinical studies that include multi-gene pharmacogenomic testing. Also, reporting on the modeling of costs of prescription drugs over time is limited. It is unclear whether cost savings associated with the intervention were overestimated because models did not allow for long-term use of drugs (i.e., during the maintenance phase of depression), as suggested by clinical practice guidelines6 for people with hard-to-treat depression.
In addition, the included studies partially examined decision, parameter, and structural model uncertainties using deterministic one-way sensitivity analyses to elucidate determinants of the cost-effectiveness of multi-gene pharmacogenomic-guided treatment.
Last, all studies had potential conflicts of interest because some of the authors were employees of or consultants to companies that developed the multi-gene pharmacogenomic tests with decision-support tools. Only one economic study81 did not receive funding from a manufacturer to conduct the study.
Discussion
Our review of the four model-based economic studies78-81 found that multi-gene pharmacogenomic testing combined with decision-support tools to guide medication selection in adults with major depression was associated with greater effectiveness and cost savings than treatment as usual. In general, the population of interest was people who previously did not benefit from treatment with antidepressants. None of the included studies were directly applicable to the Ontario health care system, and their results could not be generalized to Ontario.
Although all studies suggested robust cost-effectiveness benefits over the 3-year, 5-year, or lifetime time horizon, underlying assumptions related to extrapolating long-term effectiveness and costs were not completely substantiated by current evidence. For example, the duration of follow-up in clinical randomized controlled trials used to support the efficacy of pharmacogenomic-guided treatment is relatively short (8–24 weeks57,58,68); also, these studies did not capture the effectiveness of the intervention on relapse or recurrence over the long term. In a sensitivity analysis of one study,81 pharmacogenomic-guided treatment was not cost-effective at a willingness-to-pay amount of $50,000 per QALY if the time horizon was less than 1 year. Therefore, a long-term extrapolation of the effects of multi-gene pharmacogenomic testing and modeling of possible savings over the long term ought to be carefully conducted to prevent bias in estimates of the ICER.
In addition, the current 2016 guidelines from the Canadian Network for Mood and Anxiety Treatments (CANMAT)6 suggest pharmacogenomic testing for medication selection in people with major depression should be used carefully, resorting to this tool only if people have treatment-resistant disease. The evidence that supported these guidelines has not been updated with the most recent large clinical trials.57,58,68 Thus, guidance for appropriate use of multiple-gene pharmacogenomic testing that include a decision-support tool in Canada could change if the new evidence is included.
A possible limitation of our review is that we used no testing as the only comparator and did not consider single-gene or multiple-gene pharmacogenomic testing with or without a decision-support tool. Because our comparator was no testing, we could not evaluate diagnostic outcomes that assess the effectiveness of the test to detect relevant variants (e.g., sensitivity, specificity, positive predictive value, or negative predictive value). However, our systematic search was broad, and we did not identify any study that modeled diagnostic test accuracy and compared the cost-effectiveness of multiple-gene panels versus single-gene tests. Research findings suggested adequate analytical performance or the precision and accuracy of the multi-gene combinatorial pharmacogenomic test that included 12 genes associated with psychotropic medication metabolism, side effects, and mechanisms of action with regards to the individual gene components and genotype results (> 99.9% overall, and 99.4%–100% for individual gene components).93 Further, multi-gene pharmacogenomic testing had better discriminatory and predictive validity of patient-related outcomes than single-gene testing.94
Conclusions
Although the economic studies included in our review found that multi-gene pharmacogenomic testing used to guide medication selection in adults with major depression could be cost-saving and more beneficial than treatment as usual, long-term effectiveness of the intervention (1 year or longer) has not been investigated, making the conclusions uncertain. Moreover, none of the studies were done from the perspective of the Ontario Ministry of Health or were directly applicable to Ontario. Given these limitations, we undertook a primary economic evaluation to examine the cost-effectiveness and budget impact of publicly funding multi-gene pharmacogenomic testing that includes decision-support tools on medication selection in adults with major depression in Ontario.
Primary Economic Evaluation
Our review of the four model-based economic studies78-81 found that multi-gene pharmacogenomic testing that includes decision-support tools to guide medication selection in adults with major depression was associated with greater effectiveness and cost savings compared with treatment as usual. In general, the population of interest was people who previously did not benefit from treatment with antidepressants. Methodological quality of the reviewed studies is limited because the studies considered assumptions and data not supported by long-term clinical evidence. None of the included studies was fully applicable to the Ontario health care system. Therefore, we conducted a primary economic evaluation to inform policy- and decision-making about the cost-effectiveness of multi-gene pharmacogenomic-guided treatment of adults with major depression in Ontario.
Research Question
What is the incremental cost per quality-adjusted life-year (QALY) gained of multi-gene pharmacogenomic testing that includes a decision-support tool to guide medication selection compared with treatment as usual, for people with major depression who had inadequate response to at least one medication, from the perspective of the Ontario Ministry of Health?
Methods
The information presented in this report follows the reporting standards set out by the Consolidated Health Economic Evaluation Reporting Standards (CHEERS) statement.95
Type of Analysis
We conducted a cost–utility analysis to examine total and incremental mean costs and mean QALYs per person of multi-gene pharmacogenomic testing used to guide antidepressant selection compared with treatment as usual. We estimated incremental cost-effectiveness ratio (ICER), expressed as an incremental cost per QALY gained. The QALY outcome could be more appropriate for decision-making related to allocation of resources for various technologies across different conditions and is suggested by the Canadian guidelines for economic evaluation,82 among others.
We conducted a reference case analysis and sensitivity analyses. Our reference case analysis adhered to the Canadian Agency for Drugs and Technologies in Health (CADTH) guidelines82 when appropriate, and represented the analysis with the most likely set of input parameters and model assumptions. Our sensitivity analyses explored how the results were affected by varying input parameters and model assumptions. Scenario analyses were conducted to examine structural uncertainty.
Target Population
Our target population considered people aged 18 years or older with major depression requiring pharmacological treatment (mean age57,58 48 years). It considered people who had inadequate response (i.e., lack of clinical improvement, poor tolerance, or side effects) to one or more antidepressant medications. Adults with bipolar depression were not considered in our study.
The mean age and baseline characteristics of our target population closely resembled characteristics of patients in currently available clinical trials57,58 that also informed the cost-effectiveness models78-81 included in our economic evidence review and our clinical review. Multi-gene pharmacogenomic testing could be performed before initiation of a new medication; however, our clinical review found no studies that restricted the population solely to treatment-naive participants (although a few studies included mixed populations), and consequently, our economic analysis could not evaluate the cost-effectiveness of the intervention for this patient subgroup. Also, the 2016 Canadian clinical guidelines suggested that it is more appropriate to implement multi-gene pharmacogenomic-guided treatment when people have not benefited from multiple antidepressant medications.6
Perspective
We conducted this analysis from the perspective of the Ontario Ministry of Health. We explored a societal perspective in sensitivity analysis.
Intervention and Comparator
Table 12 summarizes the intervention and comparator evaluated in the reference case analysis.
Table 12:
Intervention and Comparator Evaluated in Primary Economic Model: Reference Case Analysis
| Intervention | Comparator | Population | Outcomes |
|---|---|---|---|
| Multi-gene (i.e., 2 or more genes) pharmacogenomic tests that include a clinical decision-support toola to guide depression medication selection or dose adjustment (e.g., GeneSight, Myriad) | TAU: Antidepressant therapy according to the current CANMAT guidelines6 without use of multi-gene pharmacogenomic testing | Adults with major depression who had inadequate response to at least one medication | Direct medical costs QALYs |
Abbreviations: CANMAT, Canadian Network for Mood and Anxiety Treatments; ICER, incremental cost-effectiveness ratio; QALY, quality-adjusted life years; TAU, treatment as usual.
Decision-support tools are defined as tools that guide choice of medication or dosage recommendations.
Multi-gene pharmacogenomic testing that include a decision-support tool represents the intervention strategy. These types of genetic tests represent companion diagnostics (i.e., a test that measures a person's protein or gene expression or detects genetic variation for the purpose of informing treatment decisions).96 Our clinical evidence review examined the effectiveness of several multi-gene pharmacogenomic tests that include decision-support tools: i.e., GeneSight (Myriad), Genecept Assay (Dynacare), Neuropharmagen (InSource Diagnostics), NeuroIDgenetix (AltheaDx), and CNSDose (cnsdose). Consequently, these interventions were further considered in our economic evaluation. Of note, three of these interventions (GeneSight, Genecept Assay, Neuropharmagen) were reviewed by Maruf et al in their analysis of the psychiatric pharmacogenomic tests available in Canada.21 As shown in our economic evidence review, cost-effectiveness was examined for some of these tests (GeneSight, NeuroIDgenetix).78-81 Last, we have not found relevant clinical evidence for people with solely major depression in the rest of the 10 interventions suggested by Maruf et al21 (e.g., Pillcheck [GeneYouIn Inc], TreatGxPlus [GenXysHealth Care Systems], myDNA Medication Test Kit [Multi by RxOME Pharmacogenomics Canada Inc]); therefore, we did not consider these interventions in our analysis.
Our clinical review (see Appendix 6, Table A4) and a review by Maruf et al21 indicated that multi-gene pharmacogenomic tests are considerably different in terms of gene panels21 and their availability in Ontario or Canada. Moreover, their recommendations for the medication choice can differ, and their effectiveness and costs vary (see clinical review, Results section, above, and Table 13).
Table 13:
Multi-gene Pharmacogenomic Tests Considered
| Name of Test (Company) | Available in Canada? | Effectiveness (Studies Included in our Clinical Review)a | Cost | Feasibility Studies in Ontario? |
|---|---|---|---|---|
| GeneSight (Myriad) | Yes, in Ontario | Greden et al, 201957 Winner et al, 201365 Hall-Flavin et al, 201355 |
$2,500 CAD78 | Yes97,b |
| Genecept Assay (Dynacare) | Yes | Perlis et al, 202061 | $495 CAD21 | Unclear |
| Neuropharmagen Core (InSource Diagnostics) | Yes | Han et al, 201860 Perez et al, 201762 |
$400 USD21 | Unclear |
| NeuroIDgenetix (AltheaDx)c | No, in United States | Bradley et al, 201858 | $2,000 USD81 | No |
| CNSDose (cnsdose) | No, in Australia and United States | Singh et al, 201564 | $299 AUD98 | No |
Effectiveness: Clinical studies included in our clinical review examined test efficacy in reference case population. See details on included genes/alleles in the clinical review, Appendix 6, Table A4.
IMPACT (Individualized Medicine: Pharmacogenetics Assessment and Clinical Treatment) study, partially supported by the Ontario Ministry of Research and Innovation grant of $7 million, and matching funds from the Centre for Addiction and Mental Health (CAMH), totalling about $19.5 million (available at http://impact.camhx.ca/en/clinicians-study).
Also known as IDgenetix in our economic evidence review.
Given this heterogeneity between multi-gene pharmacogenomic tests (Table 13), we decided to examine the GeneSight test (and its corresponding effectiveness and cost) in the reference case, and other tests (with their corresponding effectiveness and costs) in sensitivity analyses. The GeneSight test was elected because effectiveness data were based on randomized controlled clinical trials (RCTs) for the population of interest, the test cost is available for Ontario, and a feasibility study has been conducted in Ontario (i.e., the IMPACT [Individualized Medicine: Pharmacogenetics Assessment and Clinical Treatment] study,97 supported via private–public partnership, with partial funding from the Ontario government [available at http://impact.camhx.ca/en/clinicians-study]).
Treatment as usual represents pharmacotherapy with antidepressants for management of people with moderate-to-severe major depression, following current treatment clinical guidelines, without consideration of companion diagnostics (i.e., a trial-and-error sequential treatment).6 Various types of antidepressants are available for treatment of major depression: selective serotonin reuptake inhibitors (SSRIs), serotonin and noradrenaline reuptake inhibitors, tricyclic antidepressants, and monoamine oxidase inhibitors.6 Current 2016 CANMAT guidelines6 suggest different lines of therapy for the management of major depression including various medication classes. A physician could prescribe several drugs that are recommended or listed as first-line therapy.6 First-line medications could also be used either as initial, switch, or augmentation therapy (e.g., SSRIs: citalopram, 20–40 mg; sertraline, 50–200 mg), before considering second-line therapy (e.g., amitriptyline, various doses; quetiapine, 150–600 mg). In our analysis, we did not specify which line of therapy or class of medication was used in treatment as usual because medication options for our target population vary greatly; moreover, the current evidence related to pharmacogenomic-guided treatment lacks information on specific changes in medications after the testing and over the trial period (more details in Main Assumptions section).
Discounting and Time Horizon
We used a short-term time horizon of 52 weeks in our reference case analysis. This duration was proposed to capture the effectiveness of the intervention shown for response and remission outcomes in currently available evidence. Major depression has an episodic and chronic nature, and it is difficult to extrapolate the benefit of multi-gene pharmacogenomic-guided treatment over a long period because of the lack of observed data and because (1) in modeling the long-term course of this disease, we need to allow for the possibility that a major depressive episode might recur (e.g., after 9–12 months); and (2) we need reliable inputs on long-term effectiveness of our intervention versus treatment as usual on mitigating relapse and recurrence events over several years. Use of the short-term time horizon for the reference case was supported by experts.
In accordance with the CADTH guidelines,82 and given our time horizon of 1 year, we did not apply an annual discount rate of 1.5% in the reference case analysis. However, discounting was applied in a scenario analysis with longer follow-up. All costing estimates in our analyses were expressed in 2020 Canadian dollars.
Main Assumptions
The model's main assumptions are as follows:
Benefit of medication selected after the testing would be shown within the first 8 to 12 weeks
Given the lack of clinical evidence, we chose not to model the rate of adherence to prescribed therapy regimens (i.e., a simplifying assumption)
Cost of multi-gene pharmacogenomic testing would be incurred one time,99 at the beginning of model simulation
Medication changes after baseline: We were unable to model changes in medication dose, augmentation, and switches from one drug to another that are usually done in assessments of the cost-effectiveness of single-gene pharmacogenomic tests.100-104 Currently available multi-gene pharmacogenomic testing studies57,58 include multiple genes associated with metabolism of various antidepressant medications; however, researchers did not provide enough information about how specific types or classes of initial antidepressants were selected or changed over time. In these studies, after the testing, a decision support-tool (report) was provided, and all data related to changes in (unspecified) medication pathways were reported on aggregate levels (e.g., classified as congruent: “use as directed” and “use with caution” or as incongruent: “use with increased caution and with more frequent monitoring”). Last, these studies provided overall effectiveness estimates on aggregate level (not by the type of medication or for all patient subgroups; see our clinical review, Results section)
-
Medication change after relapse: We assumed that people who did not achieve remission and who experienced relapse within 6 months of the initial therapy would change their medications:
-
○Given poor documentation in the current studies of precise medication change algorithms, we simplified modeling and assumed that a person would start the next (step 2) treatment within 1 year
-
○We assumed a sequential medication pattern from the Sequenced Treatment Alternatives to Relieve Depression (STAR∗D) trial,8,88,92 with the corresponding risk ratios for remission ascertained between the sequential (step 1 and step 2) therapies (more details on inputs in the following sections)
-
○
The short-term time horizon was justified by lack of data on the long-term efficacy of multi-gene pharmacogenomic-guided treatment compared with treatment as usual and by lack of information on the prognostic value of the intervention
Model Structure
We developed a health state-transition (Markov) cohort model to evaluate the cost-effectiveness of multi-gene pharmacogenomic-guided treatment compared with treatment as usual. As explained above, we simplified a course of major depression to a short term that includes one major depression episode because we lack reliable data on the long-term effectiveness of multi-gene pharmacogenomic-guided treatment. Figure 5 presents a short-term clinical treatment pathway aligned with clinical practice guidelines,6,89,90,105 including the treatment phases and outcomes that are usually evaluated during one episode of major depression:
Figure 5: Clinical Treatment Pathway.
a Response: reduction of symptoms, ≥ 50% decrease from baseline depression scale scores.
b Remission: free of depressive symptoms, assessed at a single time point; recovery: no symptoms sustained for > 8 weeks.
Note: fonts distinguish treatment outcomes that were included or not included in our analysis. Purple (italic) outcomes (e.g., response/partial response or no response to treatment or recurrence) were not included in our model; red (bold) outcomes were considered in our models for the reference case (i.e., remission/no remission/relapse) and scenario (i.e., recovery) analysis. Blue block arrows indicate phases of treatment (acute, continuation, and maintenance). Outline arrow from relapse in the continuation phase suggests that a medication change (boldface) needs to be evaluated again for treatment response (i.e., going back to the acute treatment phase). Outline arrow from remission to recovery or to recurrence suggests that remission in the maintenance phase could lead to full recovery or to another, entirely new, episode of depression (i.e., recurrence).
In the first (acute) phase, therapy is initiated, and outcomes such as clinical improvement, response, and remission are usually measured over 8 to 12 weeks. Response represents alleviation in depression symptoms that corresponds to at least a 50% decrease in depression scores at trial endpoint (e.g., 8 or 12 weeks57,58) compared with baseline scores.6,91 Remission indicates that a person is free of depression symptoms (as measured by the depression scale score: e.g., the 17-item Hamilton Rating Scale for Depression score 7 or less).91 Progression of the disease and response to the initiated therapy is also evaluated by examining other outcomes such as partial response and presence of residual symptoms after achieving remission6,91; however, these outcomes were not assessed in the currently available clinical trials (see our clinical review)
After the acute phase, a person continues with the therapy for the next 12 weeks in the continuation phase. During this period, a person could experience relapse or could continue to improve social and physical function (i.e., remain stable and in remission). Relapse represents the reappearance of previous depression symptoms within 6 months of acute response,6,89,91 thus requiring a change in medication. After relapse, the process of assessing response to the new treatment begins again
In the maintenance treatment phase, a person continues the treatment for at least 6 months. During this period, long-term treatment outcomes such as recovery or recurrence are monitored. Recovery indicates that a treated person remains stable and in full remission for at least 2 or 3 months.89,91 Recurrence is a long-term outcome; it represents a full new episode of depression that occurs in the maintenance phase or later (in general, after 9 months of acute response)89,91
Ideally, all depression-related outcomes would be considered in a modeling exercise. As shown in Figure 5 and explained in detail below, we were unable to address all health outcomes.
Figure 6 represents a simplified Markov model schematic. A more detailed model description is provided in Appendix 11 (Figure A1). Our modeling approach follows the clinical treatment pathway presented in Figure 5. However, limited data meant we could not include all treatment outcomes. While clinical trials57,58 reported response and remission, these outcomes were measured at the end of trial follow-up (8 or 12 weeks). As a result, we could not infer all possible conditional probabilities for the modeling purpose (e.g., a proportion of people in remission, conditional on positive response to treatment or a proportion of people who responded to treatment but did not achieve remission). Thus, we have chosen to simplify the model, assuming that remission could be more clinically relevant than response. In addition, we did not model recurrence because we cannot know how effective the intervention would be over the long term.
Figure 6: Simplified Model Structure.
a This health state is “No remission—major depression continues to be unresponsive to treatment.”
b Relapse could occur only once during the time horizon (after no response to prescribed medication at baseline); another medication change was modeled after the occurrence of relapse.
c Death due to suicide or other causes.
d Well health state was included in a scenario analysis only.
The cohort's starting age was 48 years.57,58 The cohort included people with major depression unresponsive to at least one medication. In the current trials,57,58 the majority of participants had not benefited from an average of three medications and had untreated moderate-to-severe major depression.
In the reference case, the cohort's outcomes were accumulated over the time horizon of 52 weeks, using a cycle length of 1 month. At the beginning of the simulation, people could either receive the intervention (i.e., multi-gene pharmacogenomic testing that includes a decision-support tool to guide the medication choice) or treatment as usual (see Figure 6 and Appendix 11). The model included the following health states:
No remission – major depression unresponsive to treatment—A health state that represents major depression unresponsive to medication. People would enter this state at the beginning of simulation (at the time they start with either the intervention or treatment as usual) and would remain in it during the acute phase. From this state, people would transition to either remission or relapse, after a first medication change at baseline. Those whose symptoms do not respond to medication within the first 3 months would transition to the relapse health state, which requires another medication change (see Main Assumptions). People could transition back to the no remission state, after there is no response to subsequent therapy (initiated post-relapse). Their symptoms could stay in no remission until the end of the time horizon or death
Remission—A health state associated with no depression symptoms after treatment has begun. People would transition to this health state during the acute phase. Their symptoms could remain in remission after initial therapy or could relapse and transition to the relapse state. People could transition back to remission if their symptoms respond to a subsequent therapy initiated post-relapse. Their symptoms could stay in remission until the end of the time horizon or death
Relapse—A health state associated with reappearance of depressive symptoms from either no remission or remission after treatment initiation. This is a temporary health state (for about 2 months) during which patients receive a subsequent (step 2) treatment (i.e., treatment change) and are monitored for response. We assumed a possibility of one relapse given the time horizon. As described in the prior section (Main Assumptions), we had limited data and understanding of specific changes in the medication pathway after baseline by treatment outcomes; thus, it was difficult to ascertain which antidepressant would follow the medication initiated at the start of simulation. For simplicity, we modeled medication change in general and according to sequential medication pattern of the STAR∗D trial, without assessing specific outcomes of a single antidepressant or a medication class, and we used aggregate evidence on the effectiveness and cost of medications (see Main Assumptions).87,88,106 From this state, depending on the progress of their disease, people could transition back to more permanent states of no remission or remission (see Figure 6 and Figure A1)
Well (recovery)—A health state included in a scenario analysis only. It represents a natural course to recovery, where people have no depression symptoms for at least 2 months after the continuation phase (i.e., meaning that they were in remission for at least 6 months; see Figure 5); in the well state, people have stable, sustained remission and continue with medications
Death—During each cycle (month), based on the lifetime probabilities of Ontario's population,107 a person has a chance of dying from all causes, from any of the modelled health states. In addition, we modeled a possibility of death by suicide from all states87
Clinical Outcomes and Utility Parameters
We used several different input parameters to populate the model, informing the natural and clinical course of a major depression episode, effectiveness of the intervention, health state utilities, and costs.
NATURAL HISTORY
To model the natural history and clinical course of one episode of major depression, we informed input parameters from the literature sources (Table 14). In the arm receiving treatment as usual, the probability of initial remission (after medication change at baseline) and the probability of side effects of treatment were based on results of a blinded randomized-controlled clinical trial (RCT) by Greden et al, identified by our clinical review.57 The probability of relapse in the arm receiving treatment as usual was estimated from a systematic review by Sim et al.106 Sim et al meta-analyzed 45 RCTs to determine the efficacy of antidepressants within 12 months after initiating the treatment. We estimated remission rates with a subsequent treatment for both strategies from the results of the STAR∗D trial.88 The corresponding rate for remission with a next therapy (e.g., step 2) was compared with the initial therapy to obtain the risk ratio (e.g., an estimated remission rate ratio for step 2 vs. step 1 was 0.83: 0.366 [step 1]/0.306 [step 2]). This ratio was applied to an initial rate of remission (achieved after baseline) to estimate a remission rate with the subsequent therapy. Last, we accounted for age-dependent background mortality in Ontario,107 adjusted for an increased risk of death for people with unresolved depression, and the probability of suicide, based on Canadian data.87
Table 14:
Natural History Inputs for Treatment as Usual
| Model Parameter | Mean (SE)a,b | Distribution (Parameters)b | Reference |
|---|---|---|---|
| Probability of remission in TAU (initial treatment at baseline) • Estimated per monthc |
0.114 (0.012) • 0.059 (0.002) |
Beta (α: 77.178; β: 599.822) | Greden et al, 201957 |
| Probability of relapse in TAU • Estimated per monthc |
0.233 (0.14) • 0.043 (0.025) |
Beta (α: 1.891; β: 6.226) | Sim et al, 2015106 |
| Risk ratio for remission, next treatment vs. initial treatment in step 2d | 0.83 | NA (fixed) | Rush et al, 20068 |
| Probability of transitioning to the well state, in those with initial remission (maintenance treatment phase)e • Estimated per monthc,e |
0.66 (0.07) • 0.086 (0.006) |
Beta (α: 29.565; β: 15.231) | Williams et al, 2009108 |
| Probability of side effects due to treatment with antidepressants in TAU | 0.153 (0.015) | Beta (α: 92.718; β: 513. 282) | Greden et al, 201957 |
| Annual probability of all-cause mortality, starting at age 48 y • Estimated per monthc |
0.00198 • 1.65 × 10−4 |
NA (fixed, Life Table) | Ontario Life Tables 2016–2018, Statistics Canada, 2020107 |
| Risk ratio for all-cause mortality, no remission vs. remission | 1.7 (0.026) | Lognormal (mean: 0.531; SE: 0.008) | Tanner et al, 201987 |
| Annual probability of suicide, in people with no remission • Estimated per monthc |
0.0004 • 3.33 × 10−5 |
NA (fixed) | Tanner et al, 201987 |
| Annual probability of suicide, in people with remission • Estimated per monthc |
0.0001 • 8.33 × 10−6 |
NA (fixed) | Tanner et al, 201987 |
Abbreviations: NA, not applicable; SE, standard error; TAU, treatment as usual.
Standard errors were estimated whenever data were available; those associated with the probability of transitioning to the well state were assumed to be 10% of the mean. Input parameters presented as the point estimates (without SEs) were assumed to be fixed, given the limited data.
Beta distributions were assigned to probability estimates in probabilistic analysis where applicable. Standard error of the mean (SE) was estimated from 95% confidence intervals or from original data. Two parameters of the beta distribution (α, β) were derived from the mean and SE (stated for each model parameter). Formulas for these calculations, derived from the mean and SE, are: α = ([Mean2] × [1 – Mean])/([SE2] – Mean); β = ([{1 – Mean} × {1 – Mean}] × Mean)/([SE2] – 1). Lognormal distributions were assigned for risk ratio inputs (wherever possible), using two distribution parameters: μ (mean of logs) and σ (SE, standard deviation of logs). Distribution parameters values were based on original data; further adjustments and transformations to the model cycle length of 1 month were performed (see footnote c).
Markov model used a cycle length of 1 month and all rates and probabilities were adjusted appropriately.
When a person does not benefit from initial treatment and starts a second treatment, the probability of remission is decreased (by 0.83 times) as shown in the STARD∗D trial for the step 2 treatment: probability of remission with step 1 ÷ probability of remission with step 2: 0.366/0.306 = 0.83.8
Well health state was included in a scenario analysis only.
Given uncertainty with respect to inputs for relapse and remission, we conducted several sensitivity analyses to estimate the impact of these parameters on the cost-effectiveness results. In addition, we modeled the well health state in a scenario analysis. In this analysis, we modeled the probability of sustained remission within the maintenance treatment phase for those who have already achieved remission. This input was informed by a systematic review and a meta-analysis done by Williams et al, who evaluated data from 11 RCTs in 3,745 patients (mean age 49 years) with an average of three depressive episodes.108 The authors examined the continued efficacy of antidepressants over the maintenance phase (i.e., at least 6–12 months of treatment).108
IMPACT OF INTERVENTION ON NATURAL HISTORY
Our clinical review examined results of the published studies on the efficacy of multi-gene pharmacogenomic testing that includes decision-support tools to guide treatment of people with depression. Studies that were included in our clinical review were also used in our economic analyses. Our reference case analysis considered the results of a blinded RCT by Greden et al57 (Table 15). This was a large RCT with 1,167 participants that examined the effectiveness of the GeneSight test, and that was appraised as being at high risk of bias (see clinical review and Appendix 7, Table A5) and as offering low confidence in the evidence on the remission outcome (see clinical review, Appendix 6, and Appendix 7, Table A16). As discussed in the prior section (Intervention and Comparator), the effectiveness of other multi-gene pharmacogenomic-guided tests (i.e., Genecept Assay, Neuropharmagen, NeuroIDgenetix, and CNSDose) was determined from other studies.58,60-62,64 The evidence that informed modeling of the effectiveness of other multi-gene pharmacogenomic interventions was also of low to very low quality, consequently introducing substantial uncertainty. The cost-effectiveness of other multi-gene pharmacogenomic-guided tests was evaluated in sensitivity analyses.
Table 15:
Effectiveness of Multi-gene Pharmacogenomic-Guided Treatment
| Model Parameter | Mean (SE/95% CI)a,b | Distribution (Parameters)b | Reference |
|---|---|---|---|
| Risk ratio for remission (intervention vs. TAU) | 1.47 (1.12; 1.94) | Lognormal (mean: 0.385; SE: 0.140) | Greden et al, 201957 |
| Relative risk, relapse (intervention vs. TAU) | 0.39 (0.04) | Lognormal (mean: –0.942; SE: 0.0512) | Tanner et al, 202078 |
| Ratio for remission (next treatment vs. initial treatment: step 2d) | 0.83 | NA (fixed) | Rush, 20068 |
| Probability of transitioning to the well state, in those with initial remission (maintenance treatment phase)e • Estimated per monthc,e |
0.66 (0.07) • 0.086 (0.006) |
Beta (α: 29.565; β: 15.231) | Williams et al, 2009108 |
| Probability of side effects due to treatment with antidepressants | 0.156 (0.015) | Beta (α: 87.204; β: 471.796) | Greden et al, 201957 |
Abbreviations: CI, confidence interval, NA, not applicable; SE, standard error; TAU, treatment as usual.
Standard errors were estimated whenever data were available; those associated with relapse outcomes were assumed to be 10% of mean.
Beta and lognormal distributions were assigned for probabilities and risk ratio (relative risk), respectively, in probabilistic analysis. Two parameters of the beta distribution (α, β) were derived from the mean and SE (stated for each model parameter). Two parameters of the lognormal distribution were μ (mean of logs) and σ (SE, standard deviation of logs). Distribution parameter values were based on original data; further adjustments and transformations to model cycle length of 1 month were performed.
Markov model used a cycle length of 1 month and all rates and probabilities were adjusted appropriately.
When a person did not benefit from initial treatment assigned at baseline and started a second treatment, probability of remission was decreased (by 0.83 times) compared with baseline as shown in the STARD∗D trial for the step 2 treatment: probability of remission with step 1 ÷ probability of remission with step 2: 0.366/0.306 = 0.83.8
Well health state was included in a scenario analysis only.
All published RCTs followed participants over the short term (maximum 12 weeks for both arms); thus, the effectiveness of multi-gene pharmacogenomic-guided treatment on relapse or other long-term outcomes (recovery or recurrence) is uncertain. For the reference case, we assumed the relative risk of relapse from data provided in a Canada-based cost-effectiveness analysis by Tanner et al.78 The probability of relapse was reported at 9.1% based on 24-week follow-up data from an RCT by Greden et al,57 for people who continued with the intervention. Tanner et al78 further estimated the risk ratio of the multi-gene pharmacogenomic-guided intervention versus treatment as usual assuming the probability of relapse with treatment as usual of 23.3%, ascertained from a meta-analysis of RCTs by Sim et al.106 Given this considerable parameter uncertainty, we further extensively explored parameter inputs related to relapse and remission with the intervention in sensitivity analyses.
HEALTH STATE UTILITIES
We performed a targeted literature search in MEDLINE for health state utilities on February 3, 2020, to retrieve studies published from database inception until the search date. We based the search on the population and intervention of the clinical search strategy with a methodologic filter applied to limit retrieval to health state utilities. See Appendix 1 for our literature search strategies, including all search terms. This search did not identify any additional relevant studies.
Thus, we examined the inputs of the economic studies from our economic evidence review and identified health states utilities related to remission, no remission, relapse, and disutility of treatment with antidepressants (Table 16), all reported in a study by Mrazek et al.109 Disutility values due to medication-related side effects ranged from −0.01 (dry mouth or nausea) to −0.12 (nervousness or lightheadedness) and were further explored in sensitivity analysis.109,110
Table 16:
Utilities Used in Economic Model
| Health State Utility | Mean (SE)a | Distribution (Parameters)a,b | Reference |
|---|---|---|---|
| Remission | 0.826 (0.065) | Beta (α: 27.272; β: 5.745) | Mrazek et al, 2013109 |
| No remission | 0.552 (0.120) | Beta (α: 8.928; β: 7.246) | Mrazek et al, 2013109 |
| Relapse | 0.417 (0.126) | Beta (α: 5.969; β: 8.344) | Mrazek et al, 2013109 |
| Disutility associated with medication side effects | –0.055 (0.03) | Beta (α: 3.121; β: 53.629) | Mrazek et al, 2013109,110 Najafzadeh et al, 201781 |
| Well (recovery)b | 0.940 (0.03) | Beta (α: 57.967; β: 3.700) | Lenert et al, 2000111 |
Abbreviation: SE, standard error.
Beta distributions were assigned in probabilistic analysis. Two parameters of the beta distribution (α, β) will be derived from the mean and SE (stated for each model parameter). Distribution parameter values were based on original data; further adjustments and transformations to the model cycle length of 1 month were performed.
Well health state was included in a scenario analysis only.
For the scenario analysis including the well health state, in which people were assumed to be stable, we assigned the utilities found by Lenert et al,111 for people with major depression who achieved the state near normal health (Table 16).
Cost Parameters
Table 17 and Appendix 11 (Table A33) present cost parameters used in the economic model. Costs of multi-gene pharmacogenomic tests that include decision-support tools were based on the literature and published sources (see Table 13).21,78,81,98 In the reference case, we used data available for the GeneSight test, while the test cost was changed in sensitivity analyses to accommodate other tests (see the Analysis section).
Table 17:
Costs and Resource Use Inputs in Economic Model
| Variable | Total Costs, $ Mean (SE)a | Distribution (Parameters)b | Reference |
|---|---|---|---|
| Multi-gene Pharmacogenomic Testing (One-Time Cost) | |||
| Testing including sample transportation costs | 2,500 (625) | Gamma (α: 16; λ : 0.0064) | Tanner et al, 202078 |
| Physician costs (for 2 visits) | 135.5 (67.75/visit) | NA (fixed) | OHIP code K005113 |
| Direct Medical Costs c,d | |||
| Remission, total annual costs (2018 CAD) • Medication (prescription drug) costs, annual (2018 CAD)c ○ Medication costs, monthly (2020 CAD): First 6 months ÷ rest of follow-upd • Health care service resource use and hospitalization costs, annual (2018 CAD)c ○ Health care service resource use including hospitalization costs, monthly (2020 CAD)d • Physician costs, annual (2018 CAD)c ○ Physician costs, monthly (2020 CAD)d |
2,832 (STD: 7,601; SE: 12.36)c,f 527 (STD: 2,101; SE: 3.42)c 122.86 (0.58)/44.93 (0.29)d 1,701 (STD: 6,623; SE: 10.77)c 145.02 (0.92)d 605 (STD: 737; SE: 1.20)c 51.58 (0.10)d |
– – Gamma (α: 44,984.200; λ : 366.156)/Gamma (α: 23,793.824; λ : 529.571) – Gamma (α: 24,945.616; λ : 172.013) – Gamma (α: 254,841.929; λ : 4,940.672) |
Tanner et al, 201987; Tanner et al, 202078 – – – – – – |
| No remission (or relapse), total annual costs (2018 CAD) • Medication (prescription drug) costs, annual (2018 CAD)c ○ Medication costs, monthly (2020 CAD) |
10,064 (STD: 41,113; SE: 94.30)c,g 1,441 (STD: 2,962; SE: 6.79)c 122.86 (0.58)d |
– – Gamma (α: 44,984.200; λ : 366.156) |
Tanner et al, 201987; Tanner et al, 202078 – – |
| • Health care service resource use and hospitalization costs, monthly (2018 CAD) ○ Health care service resource use and hospitalization costs, monthly (2020 CAD) • Physician costs, annual (2018 CAD) ○ Physician costs, monthly (2020 CAD) |
7,192 (STD: 38,761; SE: 88.91)c 613.17 (7.58)d 1,431 (STD: 3,282; SE: 7.53)c 122.00 (0.64)d |
– Gamma (α: 6,543.522; λ : 10.672) – Gamma (α: 36,133.020; λ : 296.166) |
– – – – |
| Welle • Medication (prescription drug) costs, annual (2018 CAD)c ○ Medication costs, monthly (2020 CAD) • Physician costs, annual ○ Physician costs, monthly (2020 CAD) |
– 527 (STD: 2,101; SE: 3.42)c 44.93 (0.29)d – 47.70 |
– – Gamma (α: 23,793.824; λ : 529.571) – NA (fixed) |
– Tanner et al, 201987 – – OHIP code K033113 |
Abbreviations: NA, not applicable; OHIP, Ontario Health Insurance Plan; SE, standard error; STD, standard deviation.
Estimates of standard error were calculated from observed published data whenever possible; otherwise, SEs are assumed to be 25% of mean cost (e.g., cost of testing, SE=$625).
For inputs with calculated SEs, we assigned gamma distributions in probabilistic analysis. Two parameters of the gamma distribution (α, λ) are derived from the mean and SE. Formulas for these calculations are: α = (Mean2)/(SE2); λ = Mean/([Mean × SE]2).
Cost estimates are presented in this table as reported in the original paper (2018 CAD)87; SEs were calculated from the reported standard deviations and sample sizes (SE = STD/√(N), where N for the cohort of patients with depression was 190,065 and for the cohort of patients without depression was 378,177).87
To estimate cost per model cycle length of 1 month, we first inflated estimates from 2018 CAD to 2020 CAD using the Canadian Consumer Price Index (CPI).114 (137.4 [2020]/134.3 [2018]): for example, in no remission, annual cost of prescription drug was $1,441 in 2018 CAD, and was converted to $1,474 in 2020 CAD. Next, inflation-adjusted annual cost was transformed into the monthly estimate: $1474/12 = $123.
Well health state was included in a scenario analysis only.
Mean health care services utilization per year (a person without depression) was 8.5 (STD: 8.8) physician visits; 5.0 (STD: 5.2) family doctor visits; 3.5 (STD: 5.9) visits with a specialist; 0.1 (STD:0.5) sessions of psychotherapy; 0.1 (STD: 0.3) hospitalizations; 1.9 (STD: 8.3) days in hospital; 0.4 (STD: 3.5) days in intensive care unit; 0.1 (STD: 0.4) emergency department admissions; and 4.2 (29.5) days receiving long-term care (original article,87 Table 4).
Mean health care services utilization yearly (a person with depression) was 18.6 (STD: 27.8) physician visits; 11.0 (STD: 15.0) family doctor visits; 7.6 (STD: 19.4) visits with a specialist; 1.7 (STD: 4.7) sessions of psychotherapy; 0.5 (STD: 4.1) hospitalizations; 8.3 (STD: 40.5) days in hospital; 0.7 (STD: 0.5) days in intensive care unit; 0.4 (STD: 2.6) emergency department admissions; and 16.0 (61.2) days receiving long-term care (original article,87 Table 4).
The reference case cost of testing includes costs associated with the test, sample transportation, and reporting (i.e., decision-support report). Results are available to physicians within 36 hours.21 In addition to the cost of the test, two physician visits are required for the intervention: one to initiate the testing request and another to discuss the results (based on the decision-support report) and further treatment. In our analysis, the cost of testing (including physician visits) was applied as a one-time cost (Table 17). Our sensitivity analysis further explored the impact of the test price and additional visits with a health care provider required during the testing stage (i.e., visits with a family physician or a physician and pharmacist112 to order the test and discuss the test results varied from none to three).
For our analysis, the direct medical costs of major depression were based on a costing analysis by Tanner et al that used individual-level data from administrative, clinical, and social services databases available at the Manitoba Centre for Health Policy.87 It compared direct and indirect costs between a cohort of patients with depression (n = 190,065) and a cohort of patients without depression (n = 378,177). Direct medical costs included prescription drug costs and costs associated with health care utilization such as physician services, hospitalizations (initial and readmission), admissions to emergency departments, and outpatient costs, and psychotherapy. This study found that the annual direct costs of patients with depression were $10,064 compared with $2,832 for patients without depression (all in 2018 Canadian dollars). Patients with depression (average age 45 years [standard deviation (SD) 20]) had at least double the mean number of family physician (11.0 vs. 5.0) and specialist (7.6 vs. 3.5) visits yearly, compared with patients without depression. More specifically, based on the reported data (original article,87 Table 4), a person with depression required, on average, the following health care services per year: 18.6 (SD 27.8) physician visits; 11.0 (SD 15.0) family doctor visits; 7.6 (SD 19.4) visits with a specialist; 1.7 (SD 4.7) sessions of psychotherapy; 0.5 (SD 4.1) hospitalizations; 8.3 (SD 40.5) days in hospital; 0.7 (SD 0.5) days in intensive care unit; 0.4 (SD 2.6) emergency department admissions; and 16.0 (SD 61.2) days receiving long-term care. The corresponding utilization estimates for a person without depression were 8.5 (SD 8.8) physician visits; 5.0 (SD 5.2) family doctor visits; 3.5 (SD 5.9) visits with a specialist; 0.1 (SD 0.5) sessions of psychotherapy; 0.1 (SD 0.3) hospitalizations; 1.9 (SD 8.3) days in hospital; 0.4 (SD 3.5) days in intensive care unit; 0.1 (SD 0.4) emergency department admissions; and 4.2 (SD 29.5) days receiving long-term care (see original article,87 Table 4). Prescription drugs costs included the dispensing fees (as the total drug cost was calculated as a sum of drug ingredient cost and dispensing fee).87 The medication costs were based on pharmacy claims for formulary drugs dispensed to all Manitobans that are captured in the Drug Program Information Network (DPIN) database. This database includes all drug claims regardless of type of insurance coverage and payer; thus, the estimated prescription drug costs likely captured drugs covered by both public and private drug insurance plans. The drug claims included in this study covered the use of various types of prescribed antidepressants (e.g., norepinephrine reuptake inhibitors: maprotiline, bupropion; SSRIs: venlafaxine, duloxetine, desvenlafaxine, atomoxetine, fluoxetine, citalopram, paroxetine, sertraline, etc.; tricyclic antidepressants: imipramine, clomipramine, amitriptyline, etc.; and other antidepressants: mirtazapine, nefazodone, etc.; for more details see the original article,87 Supplemental Material, Table 4).
The study also included indirect costs to the federal government (i.e., social services: rent assist payments and employment and income assistance) of $1,522 and $510, respectively, for depressed and nondepressed patients. We considered these costs in a scenario analysis that addressed the broader government and societal perspectives (see Analysis section for more details).
The direct medical cost estimates, used for our model's health states (see Table 17 and Appendix 11, Table A33), are categorized into three cost components: the cost of medication, cost of physician services, and costs of other health care services including hospitalization, as reported in the study by Tanner et al.87 For the health states of no remission or relapse, the cost inputs by the cost category were calculated from the annual estimates reported for people with depression, and for the health state of remission, they were calculated from the annual estimates reported for people without depression.87 Similar assumptions about a costing approach for modeling various depression health states were made in previously published economic evaluations.78-81 We further adjusted the annual cost estimates for inflation and transformed them to our model cycle of 1 month. Given the 1-year time horizon, we assumed that people with depression adhered to the medication (chosen after baseline) through the whole state of remission. This assumption was based on the current clinical practice, which suggested a long-term use of antidepressants during and after the maintenance treatment phase before considering a drug holiday.6 The cost of medication for people achieving remission was modeled as time-dependent: in the first 6 months from baseline, the cost was assumed to be same between the remission and no remission states ($122.9/month); after 6 months (i.e., the start of the maintenance treatment phase [see Figure 5]), the medication cost continued to accrue but reflected the cost generated by people who attained remission ($44.9/month), as suggested in the prior economic evaluations.78,81
Since medication and health care services costs are important components of the cost-effectiveness analysis, we tested our input parameter assumptions in a sensitivity analysis. Last, in a scenario analysis that included the well health state, the cost of physician services slightly decreased to reflect a cost of monthly follow-up with a family physician, based on the Ontario Health Insurance Plan (OHIP) schedule.113
Internal Validation
Formal internal validation was conducted by a secondary health economist. This included testing the mathematical logic of the model and checking for errors and accuracy of parameter inputs and equations.
Model outputs were compared with all available observed data in relevant clinical trials.57,67,68 The model estimated an 8-week probability of remission of 0.168 and 0.112, respectively, in the intervention and treatment-as-usual arms; these estimates closely correspond to the observed data (Appendix 11, Figures A2 and A3). An estimated probability of remission at 6 months in the intervention arm was also within a close range of the reported estimates.57,67,68 External validation over long-term time horizons was not conducted owing to a lack of long-term studies and our incomplete understanding of possible target values for model calibration or validation over these periods.
Analysis
We calculated the reference case estimates through probabilistic analysis (PA) by running a Markov cohort of 10,000 patients (simulations). Types of distributions assigned to each input parameter used in the PA are presented in the input parameter tables (Tables 14 to 17). The PA simultaneously captured the uncertainty in all model parameters.
For each intervention, we calculated the mean costs and mean QALYs with their corresponding 95% credible intervals (CrIs). We also calculated the incremental mean costs and incremental mean QALYs (with the corresponding 95% CrIs) and the ICER, if applicable, for multi-gene pharmacogenomic-guided treatment compared with treatment as usual, expressed as incremental $ per QALY gained. The results of our reference case analysis were also presented in a scatter plot on the cost-effectiveness plane or a cost-effectiveness acceptability curve (CEAC). We presented uncertainty quantitatively as the probability that an intervention is cost-effective at specific willingness-to-pay values. We described uncertainty qualitatively (at the commonly used willingness-to-pay amounts of $50,000/QALY and $100,000/QALY), using one of five categories defined by the Ontario Decision Framework115: highly likely to be cost-effective (80%–100% probability of being cost-effective), moderately likely to be cost-effective (60%–79% probability), uncertain if cost-effective (40%–59% probability), moderately likely to not be cost-effective (20%–39% probability), or highly likely to not be cost-effective (0–19% probability).
SENSITIVITY ANALYIS AND SCENARIOS
As mentioned, we examined the cost-effectiveness of other multi-gene pharmacogenomic tests in our sensitivity analysis (see Appendix 12, Table A34). The robustness of our results on reference case cost-effectiveness given various parameter assumptions was also explored as follows (see Appendix 12, Table A35):
Effectiveness of the reference case intervention with respect to remission and relapse
Changes in the disutility value assigned in the reference case
Changes in the cost of the reference case test
Changes in the number of visits with a health care provider (i.e., physicians) during the stage of testing: none (no additional physician visits, assuming that ordering of the test and discussion of the test results were part of treatment as usual) to three additional visits (to account for longer transportation time or a service by other health care providers such as pharmacists112)
Changes in costs of prescription drugs and in costs of health care services
We examined structural uncertainty in the following scenarios (see Appendix 12, Table A36):
Changes in duration of the time horizon (e.g., 6 months, 2, 3, and 5 years), assuming that effectiveness of the intervention declines after the third year of treatment
Inclusion of the well health state to explore changes to the incremental costs and QALYs and uncertainty of decision-making if recovery is considered as an outcome
Inclusion of indirect costs to explore changes to the incremental costs and QALYs if the analytic perspective is broadened
We did not conduct a subgroup analysis of treatment-naive people with major depression who were about to start their first medication. Although a few studies have included mixed population of people with major depression, we lack clinical data on the effectiveness of multi-gene pharmacogenomic-guided treatment for treatment-naive people only that are necessary for modeling purposes.
All analyses were conducted using TreeAge Pro 2020.116 Where 2020 costs were unavailable, we used the Consumer Price Index to adjust to 2020 Canadian dollars.114,117
Results
Our economic evaluation estimated the cost-effectiveness of multi-gene pharmacogenomic-guided treatment compared with treatment as usual for adults with major depression who had inadequate response to one or more antidepressant medications. Table 18 presents the results of our reference case cost–utility analysis.
Table 18:
Cost–Utility Analysis: PGx Versus TAU
| Strategy | Mean Costs, $ (95% CrI) | Mean QALYs (95% CrI) | Mean Incremental Costs,a $ (95% CrI) | Incremental QALYsb Mean (95% CrI) | ICER: $/QALY Gainedc |
|---|---|---|---|---|---|
| TAU | 8,850.79 (8,493; 9,216) |
0.587 (0.421; 0.744) |
— | — | — |
| PGx | 10,757.28 (9,450; 12,231) |
0.6186 (0.468; 0.759) |
1,906.48 (688; 3,360) |
0.031 (0.005; 0.072) |
60,564 |
Abbreviations: CrI, credible interval; ICER, incremental cost-effectiveness ratio; PGx, multi-gene pharmacogenomic-guided treatment; QALY, quality-adjusted life-year; TAU, treatment as usual.
Incremental cost = mean cost (strategy PGx) – mean cost (strategy TAU).
Incremental effect = mean effect (strategy PGx) – mean effect (strategy TAU).
Results might appear incorrect owing to rounding.
Reference Case Analysis
Over a 1-year time horizon, multi-gene pharmacogenomic-guided treatment was not cost-effective at a willingness-to-pay amount of $50,000 per QALY gained, but it was cost-effective at a willingness-to-pay amount of $100,000 per QALY gained (Table 18). Compared with treatment as usual, the multi-gene pharmacogenomic-guided intervention was associated with additional 0.03 QALYs (95% credible interval [CrI]: 0.005; 0.072) and additional $1,906 (95% Crl: $688; $3,360), yielding an ICER of $60,564 per QALY gained.
Figures 7 and 8 represent the uncertainty around the estimated ICER. There was substantial uncertainty regarding the cost-effectiveness of multi-gene pharmacogenomic-guided treatment (Figure 7). Thus, in 6,259 of 10,000 simulations (62.6%), this intervention was more effective and more costly than treatment as usual, and above a willingness-to-pay amount of $50,000 per QALY. In 3,680 of 10,000 simulations (36.8%), the intervention was more effective and more costly, and below a willingness-to-pay amount of $50,000 per QALY. The intervention was inferior (less effective and more costly than treatment as usual) in 56 of 10,000 simulations.
Figure 7: Scatter Plot of Simulated Pairs of Incremental Costs and Effects in the Cost-Effectiveness Plane—Multi-gene Pharmacogenomic-Guided Treatment Versus Treatment as Usual.
Note: Effectiveness is expressed in quality-adjusted life years (QALYs). Negative QALYs indicate that intervention was associated with worse quality-adjusted survival. Diagonal dashed line that crosses the origin indicates a willingness-to-pay value of $50,000 per QALY gained.
Figure 8: Cost-Effectiveness Acceptability Curve for Reference Case—PGx Versus TAU.
Abbreviations: PGx, multi-gene pharmacogenomic-guided treatment; QALY, quality-adjusted life-year; TAU, treatment as usual.
Figure 8 further presents the probability of cost-effectiveness of multi-gene pharmacogenomic-guided treatment that includes a decision support tool versus treatment as usual across various willingness-to-pay values. Over the 1-year time horizon, this probability was 36.8% at a willingness-to-pay amount of $50,000 per QALY, reaching 70.7% at a willingness-to-pay amount of $100,000 per QALY. These findings suggest that the cost-effectiveness of multi-gene pharmacogenomic-guided treatment (at a test price of $2,500 ± 625) was uncertain but could be cost-effective if decision makers have higher willingness-to-pay amounts.115
Sensitivity Analysis
We conducted more than 40 analyses to examine parameter and structural uncertainty. The estimates of ICER and incremental net benefit (INB) are presented in Appendix 13, Tables A37 and A38.
PARAMETER UNCERTAINTY
Three input parameters substantially influenced the cost-effectiveness of multi-gene pharmacogenomic-guided intervention examined over the 1-year time horizon for the reference case: the effectiveness of the intervention on remission and on relapse, and the cost of testing.
Effectiveness of Intervention on Remission
Our analyses suggested that the cost-effectiveness of the reference case intervention would become more favourable (i.e., ICER < willingness-to-pay of $50,000/QALY) and more certain with an increase in the risk ratio (RR) associated with a positive effect of the intervention on remission (i.e., an increase of 25% or higher of the log odds ratio of the intervention with the corresponding shift of the distribution toward greater effectiveness of the intervention compared with the estimate used in the reference case; see details on the estimates in Appendix 12, Table A35, and results in Appendix 13, Table A37). As a reminder, the effectiveness of the reference case test on remission and relapse was assumed from an RCT by Greden et al (see Table 15).57 Thus, if we were to assume an RR of 1.81 (95% CI: 1.22; 2.26) for the remission outcome compared with the reference case RR of 1.47 (95% CI: 1.12; 1.94), given the same reduction of relapse (RR: 0.39; see Table 15), an ICER of multi-gene pharmacogenomic-guided treatment over treatment as usual would be $31,235 per QALY gained. The probability of cost-effectiveness of the intervention would range from 65% at a willingness-to-pay amount of $50,000 per QALY to 79% at a willingness-to-pay amount of $100,000 per QALY (compared with 37% and 71%, respectively, in the reference case). Further, if we were to assume an RR of 1.81 (95% CI: 1.22; 2.26) with no effectiveness of the intervention on the relapse outcome (RR = 1), the ICER would change to $40,396 per QALY (see Appendix 13, Table A37). The probability of cost-effectiveness of the intervention would be 54% at $50,000 per QALY and 79% at $100,000 per QALY.
Effectiveness of Intervention on Relapse
Changes in the RR associated with a reduction of relapse with the multi-gene pharmacogenomic-guided intervention considerably affected the ICER. If we assumed no reduction of relapse rates with the intervention (RR = 1 vs. RR = 0.39 in the reference case, while holding all other parameter estimates the same), the ICER increased to $81,165 per QALY (from $60,564/QALY in the reference case). The probability of cost-effectiveness of the intervention versus treatment as usual decreased to 23% at a willingness-to-pay amount of $50,000 per QALY and to 55% at a willingness-to-pay amount of $100,000 per QALY, suggesting high uncertainty.
Cost of Testing
Our threshold analysis of the price of the reference case test found that, at a cost of $2,161.70 or less (compared with the reference case price of $2,500), the multi-gene pharmacogenomic-guided intervention would be cost effective at a willingness-to-pay amount of $50,000 per QALY (see Appendix 13, Table A37). It would be cost saving if the test price decreased to $595.20. At a lower-end price of $450, suggested in the literature,21 the reference case intervention was cost saving with a high (93%) probability of cost-effectiveness at a willingness-to-pay amount of $50,000 per QALY.
Uncertainty Due to Other Input Parameters
Changes in values of the rest of inputs, such as number of physician visits during the testing stage, costs of medication or of health care services, and disutility of antidepressant therapy, did not substantially affect the ICER (see Appendix 13, Table A37). The estimates fluctuated within 10% of the reference case ICER (i.e., between $56,259/QALY and $66,296/QALY vs. $60,564/QALY, reference case), and remained above a willingness-to-pay amount of $50,000 per QALY.
TEST-SPECIFIC COST-EFFECTIVENESS
As previously mentioned, multi-gene pharmacogenomic-guided interventions represent a heterogeneous class of tests, different in their effectiveness and costs. In our sensitivity analyses, which were specific to each test, we showed considerable changes in the ICER and probability of cost-effectiveness of the intervention compared with intervention with the GeneSight test, used in the reference case (see Appendix 13, Table A37).
The most favourable cost-effectiveness was found with the NeuroIDgenetix and CNSDose interventions that showed a high probability of cost-effectiveness (more than 80%) at commonly used willingness-to-pay amounts (Figure 9). However, these tests are not currently available in Ontario, and the quality of studies used to inform the effectiveness model input was poor (see clinical review, Results section, and Appendices 7, Table A5, A16, A18, A20).
Figure 9: Cost-Effectiveness Acceptability Curves for Sensitivity Analyses of Various Multi-gene Pharmacogenomic-Guided Tests.
Abbreviation: QALY, quality-adjusted life-year.
Another two tests, Genecept Assay and Neuropharmagen, which are approved by Health Canada, fared much worse for cost-effectiveness when compared with the reference case test: the Genecept Assay was dominated by treatment as usual and the probability that the intervention would be cost-effective at commonly used willingness-to-pay values was less than 5%. The ICER of Neuropharmagen versus treatment as usual was $100,859 per QALY, and the probability that the intervention would be cost-effective at commonly used willingness-to-pay values was less than 46%. These findings could be explained by the lack of statistically significant improvement in remission with these interventions, despite their relatively low costs (about $500; see Appendix 12, Table A34). In addition, the clinical evidence that informed this modeling was of low to very low quality (see clinical review, Results section; and Appendix 7, Table A17 and A19).
SCENARIOS
Two structural assumptions affected the cost-effectiveness of the reference case for multi-gene pharmacogenomic-guided treatment in scenario analyses: duration of the time horizon and costs considered under the analytic perspective. Restructuring the model to include the well health state did not greatly affect cost-effectiveness of the intervention (see Appendix 13, Table A38).
Time Horizon
As the time horizon increased, the ICER decreased, and the certainty in the estimate or the probability of the intervention being cost-effective at commonly used willingness-to-pay amounts substantially changed (Figure 10 and Table A38). For example, the ICER of the reference case for multi-gene pharmacogenomic-guided treatment versus treatment as usual over 3 years was about $244 per QALY (compared with the reference case ICER of about $60,564 per QALY over 1 year or the ICER of about $185,990 over 6 months). If the time horizon were 5 years (and regardless of whether the intervention affected the reduction of relapse or not), multi-gene pharmacogenomic-guided treatment was more effective and less costly than treatment as usual. At a willingness-to-pay amount of $50,000 per QALY, the probability that the intervention would be cost-effective rose steeply from about 37% over 1 year (reference case) to 83% over 2 years, 97% over 3 years, and 98% over 5 years (see Figure 10). However, given the poor quality of evidence and lack of long-term data to support long-term cost-effectiveness modeling, these results must be treated with caution.
Figure 10: Cost Effectiveness Acceptability Curve for Multi-gene Pharmacogenomic-Guided Treatment Over Various Time Horizons.
Abbreviation: QALY, quality-adjusted life-year.
Note: Y-axis represents probability of cost-effectiveness of intervention vs. treatment as usual. X-axis represents willingness-to-pay amounts from 0 to $100,000/QALY.
Analytic Perspective
This scenario analysis showed that multi-gene pharmacogenomic-guided treatment became more cost-effective than treatment as usual at a willingness-to-pay amount of $50,000 per QALY over the 1-year time horizon if the perspective of the analysis changed and included direct non-medical costs such as costs related to social services, short- and long-term disability, and indirect costs (productivity loss of patients). These findings could be explained by a decrease in the estimate of incremental costs if we included additional costs to the government and society caused by disability claims and productivity loss. These costs would further reduce the ICER (e.g., in the reference case, expected incremental mean costs would be $1,906.48 and incremental mean effectiveness would be 0.031 QALYs; in the scenario analysis, mean costs would be $1,524 and incremental mean effectiveness would be 0.031 QALYs). When all costs to society were considered, the probability of the intervention being cost-effective rose to 48% at a willingness-to-pay amount of $50,000 per QALY and 77% at $100,000 per QALY (compared with direct medical costs alone in the reference case of 37% and 71%, respectively).
Discussion
We conducted a full economic evaluation to determine the cost-effectiveness of multi-gene pharmacogenomic testing that includes a decision-support tool to guide medication selection compared with treatment as usual for the management of major depression in Ontario.
Over the short-term horizon of 1 year and compared with treatment as usual, multi-gene pharmacogenomic-guided treatment was associated with increases in mean expected costs of $1,906 (95% CrI: $688; $3,360) and mean expected QALYs of 0.03 (95% CrI: 0.005; 0.072), yielding an ICER of $60,564 per QALY gained. The reference case intervention (at a test price of $2,500) was more likely to be cost-effective at a higher commonly used willingness-to-pay amount of $100,000 per QALY (71% compared with 37% at a willingness-to-pay amount of $50,000/QALY). Our modeling approach assumed a short time horizon because all currently available evidence examined effectiveness of this multi-gene pharmacogenomic intervention over the short term (8–24 weeks57,58,61,62,64,65; see clinical review); no effectiveness data on long-term outcomes such as recovery or recurrence are available.
As we lengthened the time horizon to 3 or 5 years (assuming constant effectiveness of the intervention over the first 2 years), the intervention became cost-effective or cost saving, reaching a relatively high probability of cost-effectiveness over treatment as usual of more than 80% at a lower commonly used willingness-to-pay amount of $50,000 per QALY. These findings can be explained by sustained slow accumulations of QALYs and savings in downstream expenditures over time; cost savings further balanced out the relatively high cost of the intervention (i.e., $2,500 for the testing plus two physician visits required during the testing stage at a total cost of about $135). However, our findings need to be treated with caution given the poor quality of evidence and lack of long-term data.
Our study population featured people who have been already treated with antidepressants because clinical evidence for treatment-naive people with major depression is very limited. Therefore, we could not determine the value of the intervention for people taking antidepressants for the first time or to prevent depression within a pre-emptive testing pathway.
We did not model adherence to prescribed therapeutic regimens because we lack published evidence on adherence or compliance outcomes (see clinical review) and because subsequent changes in clinical care pathways and in health outcomes are not documented for those who might drop out from the intervention or treatment-as-usual strategies. Consequently, we could have overestimated the benefits of the intervention over treatment as usual.
Future research should evaluate the short- and long-term impact of multi-gene pharmacogenomic-guided interventions on adherence so that the economic value of these novel interventions can be fully ascertained. Last, we were unable to address equity issues because the evidence on multi-gene pharmacogenomic-guided interventions is predominantly available for White populations (see clinical review, Discussion section).
Assuming the Ontario Ministry of Health perspective, we showed that the 1-year cost-effectiveness of the reference case depended mostly on the effectiveness of the intervention on remission and relapse, and on the cost of testing:
If future studies attain much higher effectiveness estimates of the intervention on remission compared with treatment as usual (e.g., a change in the RR from 1.47 [in the reference case] to 1.81; see Table A35, sensitivity analysis), the ICER of multi-gene pharmacogenomic-guided treatment would be much lower than a willingness-to-pay amount of $50,000 per QALY (Table A37). This estimate would hold even if the intervention had no large impact on the relapse outcome. Notably, some preliminary results from a recent clinical trial in Ontario suggested a relative increase of 88% with the multi-gene pharmacogenomic-guided intervention compared with treatment as usual118
The cost of the test would need to decrease by about $340 (i.e., from $2,500 to $2,161) for the reference case intervention to become cost-effective at a willingness to pay value of $50,000 per QALY. It would need to decrease by about $1,820 (i.e., from $2,500 to $595) for the intervention to become cost saving. These estimates of the threshold cost for the reference case test could make the multi-gene pharmacogenomic-guided intervention more appealing to decision-makers and policy makers. Recently, a lower price for the GeneSight test of about $1,569 USD (about $2,000 CAD) was approved by US Medicare (personal communication with the manufacturer119 and Bruce Quinn, MD,120 February 2021). With this new estimate, the ICER would be about $44,700 per QALY, and the probability of the reference case test being cost-effective at willingness-to-pay amounts of $50,000 per QALY and $100,000 per QALY would be about 52% and 82%, respectively
Given the heterogeneity between various multi-gene pharmacogenomic-guided interventions, we also examined the cost-effectiveness of each test identified by our clinical review (using the corresponding information for their costs and effectiveness). In addition to the GeneSight test that was used in the reference case analysis, two other interventions, Genecept Assay and Neuropharmagen, that are potentially available in Ontario, were examined in our sensitivity analysis 21 However, compared with treatment as usual, these tests were not cost-effective at commonly used willingness-to-pay amounts given low remission rates, despite their relatively low costs (see Appendix 12, Table A34, and Appendix 13, Table A37). Another two tests, NeuroIDgenetix and CNSDose, likely unavailable in Ontario, had favourable rates of cost-effectiveness, despite their quite variable price.
Our results partially agree with the findings of prior economic studies that showed cost savings with the GeneSight and NeuroIDgenetix pharmacogenomic-guided interventions over the long term (3–5 years), from the broader perspectives of society or government.78-81 Similar to our analysis, some of these studies concluded that the cost-effectiveness of the intervention depended on the time horizon,81 analytic perspective,81 and the effectiveness of the intervention on achieving remission.78,80,81
All our results need to be interpreted with caution given that the clinical evidence that informed our economic modeling—while not sparse or very limited—was of low to very low quality and therefore uncertain. In addition, caution needs to be exerted in comparing different multi-gene pharmacogenomic-guided interventions because these tests used various black box algorithms that do not specify how the commercial company weights or applies each gene in terms of specific medications.73,121 Further, most currently available multi-gene pharmacogenomic-guided interventions do not account for demographic factors (e.g., age, sex), clinical characteristics (e.g., body mass index, liver and renal function), and concomitant use of inhibitors or inducers of drug-metabolizing enzymes that have known effects on drug exposure and therapeutic outcomes.
Strengths and Limitations
Our modeling study has provided some new insights through a thorough, systematic investigation of conditions under which the intervention could become more or less cost-effective at commonly used willingness-to-pay amounts. Given the short-term effectiveness data, we used a 1-year time horizon in the reference case analysis and explored the long-term cost-effectiveness of multi-gene pharmacogenomic-guided interventions in sensitivity analysis.
As with any modeling study, our analyses are limited by parameter and structural assumptions:
We simplified the course of depression to use all relevant data within a specified time horizon. While we followed the clinical treatment outcome pathway, it is possible it would take a longer time to ascertain improvements in depression outcomes in current clinical practice, to decide whether medications should be changed or not, and to determine for how long one should be re-treated before starting with the next line of therapy
Given the lack of long-term data, our reference case model did not address all long-term outcomes such as recurrence. We partially addressed a recovery outcome in a scenario analysis by including the well health state. However, more research should be done to corroborate the duration of the effectiveness of multi-gene pharmacogenomic-guided interventions for a person who achieves a stable state of full remission. In addition, we simplified long-term models and did not account for multiple relapse outcomes or recurrences of the major depressive episode. As a result, the results of our and all other economic evaluations that modeled this intervention over the long term need to be interpreted with caution
Given currently published evidence, we were unable to model in detail changes after testing and after relapse in specific medication classes. We conducted several sensitivity analyses to address uncertainty in cost parameters. Nevertheless, our estimates of reduced costs in medication and health care services with the intervention need to be interpreted with caution
We also used data from a published Canadian study87 to inform costing inputs for the health states included in the model. Our approach likely introduced some inaccuracies in estimating health care utilization and associated costs with remission, relapse, or no remission. We tested our assumptions in sensitivity analyses: changes in health-state costs (associated with health care services including hospitalization, physician services, or medication costs) did not have a large impact on the cost-effectiveness results. Nevertheless, further studies should apply advanced methods122,123 to analyzing costs reported in administrative data to determine precisely resource utilization and corresponding phase-specific costs for the health states of remission, no remission, and relapse in people with major depression in Ontario or Canada
Our analysis considered various ways of implementing the intervention by considering various numbers of visits during the testing phase to accommodate different providers (e.g., family physicians, pharmacists) and variability in the tests' turnaround time. Although we found no substantial change in the cost-effectiveness of the intervention, our modeling is hypothetical and might not account for the complexity of implementing this intervention in actual clinical practice97,112,124-126
Generalizability
The findings of our economic analysis are generalizable to people with major depression who have had inadequate response to at least one medication. Current clinical evidence is available for predominantly White populations. Consequently, future studies should examine thoroughly the effectiveness and cost-effectiveness of multi-gene pharmacogenomic-guided interventions for people with newly diagnosed major depression (treatment naive) solely, and in all racial and ethnic groups to fill a gap in the literature with respect to equity and external validity.
Conclusions
Multi-gene pharmacogenomic testing that includes a decision-support tool represents a heterogeneous class of interventions that have different effectiveness, costs, and cost-effectiveness compared with treatment as usual (i.e., no genetic testing). The quality of the evidence informing our economic modeling is low to very low; therefore, our modelled effectiveness estimates are uncertain. Our analyses considering a 1-year time horizon found that some multi-gene pharmacogenomic interventions would be cost-effective at a willingness-to-pay amount of $100,000 per QALY, or lower, if they had similar or greater effectiveness on the remission outcome and were less costly than the reference case test.
Budget Impact Analysis
We estimated the potential budget impact of publicly funding multi-gene pharmacogenomic testing to guide medication selection for people with major depression in Ontario over the next 5 years. The analysis was done from the perspective of the Ontario Ministry of Health. All costs were reported in 2020 Canadian dollars.
Research Question
What is the potential 5-year budget impact for the Ontario Ministry of Health of publicly funding multi-gene pharmacogenomic testing that includes a decision-support tool to guide medication selection for people with major depression who have had inadequate response to at least one medication?
Methods
Analytic Framework
We estimated the budget impact of publicly funding multi-gene pharmacogenomic testing that includes a decision-support tool to guide medication selection using the cost difference between two scenarios: (1) current clinical practice without public funding for multi-gene pharmacogenomic testing (the current scenario) and (2) anticipated clinical practice with public funding for multi-gene pharmacogenomic testing (the new scenario). Figure 11 presents the budget impact model schematic.
Figure 11: Schematic Model of Budget Impact.
We conducted a reference case analysis and sensitivity analyses. Our reference case analysis represented the analysis with the most likely set of input parameters and model assumptions. Our sensitivity analyses explored how results were affected by varying input parameters and model assumptions.
Key Assumptions
The assumptions in this analysis are described in the primary economic evaluation. In addition, we considered the following:
Multi-gene pharmacogenomic testing is not publicly funded in Ontario; therefore, we assumed no use of this test in the current scenario
We assumed that all people who are offered this testing would accept it because we found no published data about test refusals in Ontario or elsewhere, and information obtained during patient engagement for this report indicated a preference for using multi-gene pharmacogenomic tests that include decision-support tools
An uptake rate of multi-gene pharmacogenomic testing of 1% per year, over the next 5 years (i.e., a maximum of 5% in year 5), is based on previous uptake of the intervention in the IMPACT (Individualized Medicine: Pharmacogenetics Assessment and Clinical Treatment) study (personal email communication with the manufacturer, May 4, 2020)97; in scenario analyses, we assumed higher uptake rates (3% or 5% per year) as suggested in other published studies.99,127 We further explored an even higher uptake of the intervention in younger populations, based on the current OHIP+ policy that covers medication costs in youth and young adults aged between 15 and 25 years128
Regardless of the number of times the test could be used over a person's lifetime to support medication selection (which could be more than once because of the changes in gene selection and algorithms included in the technology), only one-time costs associated with multi-gene pharmacogenomic testing will be incurred and reimbursed by the Ministry of Health (based on the manufacturers' policy99)
Target Population
Our study population included people with a primary diagnosis of major depression as defined by the Diagnostic and Statistical Manual of Mental Disorders, fifth edition (DSM-5), criteria.6,90,129 According to data from Statistics Canada's 2012 Canadian Community Health Survey (CCHS) on Mental Health, 4.8% of the Ontario population aged 15 years and older had reported symptoms of major depression in the previous 12 months.4 This estimate excludes people with bipolar depression.
About 50% of people with major depression do not respond to their first antidepressant medication, and an estimated 30% do not respond to two or more medications.8 In clinical trials that evaluated the efficacy of multi-gene pharmacogenomic testing, most study participants did not adequately respond to at least two antidepressants at the study entry.57,58 Therefore, for the target population in our reference case, we estimated that 30% of people with major depression would require multi-gene pharmacogenomic testing. However, we considered an expansion of this population in sensitivity analyses.
Using the most recent Ontario population projections from the Ontario Ministry of Finance, we estimated the total number of people in Ontario aged 15 years or older (from 2021 to 2025) (Table 19).130 Of these, we assumed about 4.8% are to would be diagnosed with major depression in year 1, and about 30% of this population would be eligible for multi-gene pharmacogenomic testing.57,58
Table 19:
Target Population for Multi-gene Pharmacogenomic Testing in Ontario
Thus, over the next 5 years, the number of people eligible for multi-gene pharmacogenomic testing would range from about 183,550 in year 1 to 194,110 in year 5. Notably, approximately 8,400 people were tested through the IMPACT study, most from Ontario. This study, which was partially supported by an Ontario Ministry of Research and Innovation grant (of $19.5 million: the Ontario Government provided $7 million, the Centre for Addiction and Mental Health (CAMH) invested $10.5 million, and $2 million was donated by a private donor; more information is available at http://impact.camhx.ca/en/clinicians-study).
Current Intervention Mix
As mentioned above (see Key Assumptions), we assumed no use of multi-gene pharmacogenomic testing for major depression in the current scenario.
Uptake of New Intervention and New Intervention Mix
In the reference case, we assumed that access to multi-gene pharmacogenomic testing would increase by 1% each year over the first 5 years (i.e., the maximum uptake of 5% in year 5). This relatively low uptake of the intervention in the reference case was based on our consultations and on findings from the literature with respect to barriers to implementation of multi-gene pharmacogenomic testing.97,112 For instance, Liu et al suggested that education of both providers and patients in the testing process is key to ensuring proper implementation of the information.97 Liu et al also implied that use of pharmacogenetic tests relies heavily on the attitudes of physicians who are the intersection among patients, pharmacists, and geneticists. They identified research that found that 90% of participants lacked confidence in their physician's ability to understand and use genomic information. Moreover, another study included in the review by Liu et al97 found that, after pharmacogenetic testing, about 60% of providers did not recommend using the test results at all, and about 40% suggested that test results should be filed for future use.131
Given an annual uptake of 1%, we estimated that about 1,835 eligible people with major depression would have access to multi-gene pharmacogenomic testing in year 1, rising to about 8,792 in year 5 (Table 20). Over the 5 years, a total of 27,063 persons would undergo testing. This assumption was conservative; higher annual uptake rates (including very high coverage in the subgroup of young adults) were examined in sensitivity analyses.
Table 20:
Volume After Accounting for Uptake of Multi-gene Pharmacogenomic Testing in Ontario During Years 1 to 5
| Year 1 | Year 2 | Year 3 | Year 4 | Year 5 | |
|---|---|---|---|---|---|
| No. of eligible people with major depression | 183,547 | 186,51 | 189,263 | 191,791 | 194,106 |
| Uptake rate | 0.01 | 0.02 | 0.03 | 0.04 | 0.05 |
| No. of people who continue TAU | 181,711 | 182,818 | 183,696 | 184,342 | 184,773 |
| No. of people to be assessed with multi-gene pharmacogenomic testinga | 1,835 | 3,694 | 5,512 | 7,230 | 8,792 |
Abbreviation: TAU, treatment as usual.
Uptake rate applied to approximate total of remaining people eligible for testing in specific year, reference case analysis: e.g., year 1: 183,547 × 0.01 = 1,835; year 2: (186,512 – 1,835) × 0.02 = 3,694. Those tested in prior years are subtracted from population in following years, as cost of test is applied only once over a person's lifetime.99
No mix of multi-gene pharmacogenomic testing interventions is expected in the future scenario (given the lack of data on commercially available and funded tests of a similar nature). However, medication replacement in a subset of people with major depression, guided by the results of multi-gene pharmacogenomic testing, could result in some cost savings over time because of potentially better compliance and better response to newly selected antidepressants.132,133
Resources and Costs
The proposed resource use and associated costs are described in the primary economic evaluation. As mentioned previously, the budget impact was analyzed from the perspective of the Ontario Ministry of Health, and all costs were reported in 2020 Canadian dollars.
Internal Validation
The secondary health economist conducted formal internal validation. This process included checking for errors and ensuring the accuracy of parameter inputs and equations in the budget impact analysis.
Analysis
We conducted a reference case analysis and sensitivity analysis, using the cost estimates calculated from our 1-year reference case cost-utility model. In the reference case analysis, we estimated the 5-year budget impact of publicly funding multi-gene pharmacogenomic testing that includes a decision-support tool to guide medication selection for people with major depression in Ontario.
We took a simplified, more conservative approach to calculate total budget impact. This decision was justified by our finding of substantial uncertainty in the expected effectiveness and cost savings with the intervention over long-term periods of 3 or 5 years. Therefore, we did not pursue a cumulative cohort approach that would accumulate potential cost savings due to medication and health care resource use reductions with the intervention because this approach would potentially overestimate downstream savings over 5 years. Our approach seems reasonable because the Ministry of Health ought to be advised on the imminent investment required for multi-gene pharmacogenomic testing if this technology is recommended for public funding. Whether the province would see large reductions in downstream costs should be corroborated in the implementation stage.
The sensitivity analysis considered several scenarios that could potentially affect the budget impact: uptake rate, expansion of target population in the reference case, cost of multi-gene pharmacogenomic testing, number of clinical visits associated with testing, and OHIP+ coverage of medication costs in youth and young adults.
SCENARIO 1: UPTAKE RATE
In this scenario we estimated how increases in use of multi-gene pharmacogenomic testing over time (i.e., increases in the uptake rate) affect the budget impact. We conducted two analyses:
One analysis with the uptake rate of 3% in year 1, increasing by 3% per year to 15% in year 5 (compared with the reference case analysis assuming an increase of 1% per year, and reaching 5% in year 5)
Another analysis assuming an increase in the uptake of 5% per year (with the rate of 25% in year 5). These uptake rate estimates99 were proposed for implementation of the GeneSight test in the United States
SCENARIO 2: EXPANSION OF REFERENCE CASE TARGET POPULATION
In this scenario analysis (Table 21), we explored a larger population of people with major depression who could be considered eligible for multi-gene pharmacogenomic testing:
Table 21:
Scenario 2: Expansion of Target Population
| Year 1 | Year 2 | Year 3 | Year 4 | Year 5 | |
|---|---|---|---|---|---|
| Estimated no. of people in Ontario | 12,746,315 | 12,952,196 | 13,143,292 | 13,318,835 | 13,479,594 |
| Treatment-Naive Subpopulation | |||||
| No. of people with new diagnosis of major depressiona | 191,195 | 194,283 | 197,149 | 199,783 | 202,194 |
| No. of people for whom first antidepressant therapy is indicateda | 63,094 | 64,113 | 65,059 | 65,928 | 66,724 |
| People Who Did Not Respond to at Least One Medication | |||||
| No. of people with major depressionb | 611,823 | 621,705 | 630,878 | 639,304 | 647,021 |
| No. of people who did not respond to at least one antidepressantb | 305,912 | 310,853 | 315,439 | 319,652 | 323,510 |
| Both groups, total | 369,006 | 374,966 | 380,498 | 385,580 | 390,234 |
Assuming annual incidence of major depression of 1.5% and that for 33% of this subgroup treatment with medication would be indicated.
Assuming prevalence of major depression would be 4.8% and that 50% of this subgroup would not respond to first medication.
Treatment-naive population—People with major depression in which antidepressants are indicated but never administered are treatment naive. Our clinical review did not identify any study that established the effectiveness of this intervention only for the subpopulation of people who are treatment naive; therefore, this scenario may be hypothetical. We used data from Lam et al90 to estimate the incidence of major depressive episodes in Canada (i.e., about 2.9% over 2 years or about 1.5% per year); about 33% of these people90 would be treated with antidepressants
People who did not respond to the first treatment—This group comprised people with major depression who did not respond to their first antidepressant medication (i.e., about 50% of those diagnosed with major depression8)
Both subgroups—The combined population comprised people with major depression requiring antidepressant therapy (i.e., those who were treatment naive plus those who did not respond to at least one medication)
Total number of people who would be tested over the 5 years at an uptake rate of 1% per year was estimated at 54,407 (including a total of 9,303 persons in the treatment-naive group).
SCENARIO 3: COST OF MULTI-GENE PHARMACOGENOMIC TESTING
Based on the literature, the price of multi-gene pharmacogenomic testing ranges from about $450 to $3,700.21,78 However, our clinical review did not identify any evidence for the majority of less costly tests that were examined in a study by Al Maruf et al.21 Therefore, for the reference case, we assumed that the price of testing in Canada would be around $2,500, as suggested in a Canadian cost-effectiveness analysis by Tanner et al.78
In this scenario analysis, we tested the change in the budget impact if the reference case price of multi-gene pharmacogenomic testing changed, assuming:
Price of the test at a lower end of $450
Price of the test increased or decreased by 25% and 50%
-
Price of the test same as the threshold value established in our cost-effectiveness analysis:
-
○For the intervention to be cost-effective at a willingness-to-pay amount of $50,000 per QALY, the price of the test had to be about $2,162
-
○For the intervention to be cost saving, the price of the test had to be about $595
-
○
SCENARIO 4: NUMBER OF CLINICAL VISITS ASSOCIATED WITH TESTING
In addition to the cost of the test during the stage of testing, additional physician visits are needed to order the test and discuss results. In the reference case, we accounted for two such clinical visits; in this scenario, we explored changes in the budget impact if we considered:
No additional clinical visit—Testing would be accounted for in a regular follow-up visit
One additional visit—In one visit, physicians and patients would discuss test results and subsequent action for medication selection
Three additional visits—Three visits would account for possible involvement of other health care providers (e.g., pharmacists) in the circle of care
SCENARIO 5: OHIP+ COVERAGE OF PHARMACOGENOMIC-GUIDED TREATMENT FOR YOUTH AND YOUNG ADULTS
This scenario reflected extension of the current OHIP+ policy that covers medication costs in youth and young adults aged between 15 and 25 years.128 Table 22 estimates the number of people eligible for pharmacogenomic testing by age groups, assuming the same prevalence of major depression as in the reference case.
Table 22:
Scenario 5: Extending Coverage to Young Adults (OHIP+ Scenario)
| Year 1 | Year 2 | Year 3 | Year 4 | Year 5 | |
|---|---|---|---|---|---|
| No. of people in Ontario aged 15 to 24 ya No. of people aged 15 to 24 y with major depressionb No. of people aged 15 to 24 eligible for multi-gene pharmacogenomic testingc |
1,798,491 86,328 25,898 |
1,823,917 87,548 26,264 |
1,847,143 88,663 26,599 |
1,867,725 89,651 26,895 |
1,879,433 90,213 27,064 |
| No. of people in Ontario aged ≥ 25 ya No. of people with major depression aged ≥ 25 yb No. of people eligible for multi-gene pharmacogenomic testing aged ≥ 25 yc |
10,785,699 517,714 155,314 |
10,962,097 526,181 157,854 |
11,126,282 534,062 160,218 |
11,282,015 541,537 162,461 |
11,431,084 548,692 164,608 |
Table 23 shows volumes estimated on the basis of differences in the uptake rates between age groups. We assumed a relatively high uptake (and public funding) of the intervention for people aged 15 to 24 years (starting with 20% in year 1 and increasing to 100% in year 5), and 1% uptake per year for those older than 25 years (i.e., the same uptake as in the reference case).
Table 23:
Scenario 5: OHIP+ Volume, Accounting for Uptake of Multi-gene Pharmacogenomic Testing in Ontario, Years 1 to 5
| Year 1 | Year 2 | Year 3 | Year 4 | Year 5 | |
|---|---|---|---|---|---|
| Eligible (with major depression): aged 15 to 24 y Uptake rate Assessed with a multi-gene pharmacogenomic test: aged 15 to 24 ya |
28,233 0.20 5,647 |
28,657 0.40 9,204 |
29,045 0.60 8,516 |
29,330 0.80 4,771 |
29,499 1.00 1,361 |
| Eligible (with major depression): aged ≥ 25 y Uptake rate Assessed with a multi-gene pharmacogenomic test: aged ≥ 25 ya |
155,314 0.01 1,553 |
157,854 0.02 3,126 |
160,218 0.03 4,666 |
162,461 0.04 6,125 |
164,608 0.05 7,457 |
| Total volume: pharmacogenomic testing (all age groups) | 7,200 | 12,330 | 13,182 | 10,896 | 8,818 |
Uptake rate applied to approximate total number remaining eligible for testing in the specific year: e.g., year 1 (aged 15–24): 28,233 × 0.20 = 5,647, or year 1 (aged ≥ 25): 155,314 × 0.01 = 1,553; year 2 (aged 15–24): (28,657 – 5,647) × 0.40 = 9,204 or year 2 (aged ≥ 25): (157,854 – 1,553) × 0.02 = 3,126; year 3 (aged 15—24): (29,045 – 5,647 – 9,204) × 0.60 = 8,516 or year 3 (aged ≥ 25): (160,218 – 1,553 – 3,126) × 0.03 = 4,666, etc. Those tested in prior years are subtracted from the population in the following years, as the cost of test is applied only once over a person's lifetime.99
Over the 5 years, the total number of assessed persons (aged 15–24 years) would be 29,499; the total number of assessed persons for the rest of the population would be 22,927. This accumulates to a total of 52,426 persons to be tested over the 5 years.
With respect to budget impact calculations in scenarios 2 to 4, we assumed the same rate of uptake for multi-gene pharmacogenomic testing (1% per year) as in the reference case. As shown in Table 23, the uptake rate varied by age group in Scenario 5. For scenarios 1,3, 4, and 5, we assumed the same prevalence of major depression as in the reference case.
All budget impact analyses were conducted using Microsoft Excel for Office 365.134
Results
Reference Case
Table 24 presents the budget impact of publicly funding multi-gene pharmacogenomic testing that includes a decision support tool to guide medication selection in people with major depression whose symptoms did not adequately respond to prior medication treatment. Adopting multi-gene pharmacogenomic testing in Ontario at an uptake rate of 1% in year 1 (increasing to 5% in year 5) would lead to additional costs of about $3.5 million in year 1 to about $16.8 million in year 5. The total 5-year budget impact would be about $51.6 million.
Table 24:
Budget Impact Analysis of Reference Case Results for Multi-gene Pharmacogenomic Testing in Ontario
| Budget Impact, $ Milliona | ||||||
|---|---|---|---|---|---|---|
| Year 1 | Year 2 | Year 3 | Year 4 | Year 5 | Totalb,c | |
| Current Scenario | ||||||
| Total costs | $1,624.20 | $1,650.44 | $1,674.79 | $1,697.16 | $1,717.64 | $8,364.22 |
| Cost of testing | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Medication cost | 248.29 | 252.30 | 256.02 | 259.44 | 262.57 | 1,278.63 |
| Other direct costsd | 1,375.91 | 1,398.14 | 1,418.76 | 1,437.71 | 1,455.07 | 7,085.59 |
| Future Scenario | ||||||
| Total costs | $1,627.70 | $1,657.47 | $1,685.29 | $1,710.93 | $1,734.39 | $8,415.77 |
| Cost of testing | 4.84 | 9.73 | 14.52 | 19.05 | 23.17 | 71.31 |
| Medication cost | 248.18 | 252.08 | 255.70 | 259.01 | 262.05 | 1,277.02 |
| Other direct costsd | 1,374.68 | 1,395.66 | 1,415.07 | 1,432.86 | 1,449.17 | 7,067.44 |
| BI | ||||||
| Total BI | 3.50 | 7.04 | 10.50 | 13.77 | 16.75 | 51.55 |
| BI: Cost of testing | 4.84 | 9.73 | 14.52 | 19.05 | 23.17 | 71.31 |
| BI: Medication costb | –0.11 | –0.22 | –0.33 | –0.43 | –0.52 | –1.61 |
| BI: Other direct costsd | –1.23 | –2.48 | –3.70 | –4.85 | –5.90 | –18.15 |
Abbreviation: BI, budget impact.
In 2020 Canadian dollars.
Negative costs indicate savings.
Results might appear incorrect due to rounding. The primary goal of our BIA was to estimate cost difference between two scenarios. Estimates of total costs for current and new scenarios are limited by data sources and methods used for this analysis, and they do not necessarily represent actual total costs of care.
Other direct medical costs include costs for physicians and health care services, excluding costs of medication.
The cost of testing accounted for most of the estimated budget impact, ranging from $4.8 million in year 1 to $23.2 million in year 5, yielding a total cost of $71.3 million over the next 5 years. Increases in costs with this intervention were counterbalanced by reduced medication and other direct medical costs for physician and health care services.
Sensitivity Analysis
In all scenario analyses, the total budget impact was affected by the cost of testing (Table 25). Consistent with the reference case, the intervention was associated with savings in medication and other health care services costs. Below we summarize the most important findings for each scenario:
Table 25:
Budget Impact in Sensitivity Analysis
| Scenarios Assessed | Budget Impact, $ Milliona | |||||
|---|---|---|---|---|---|---|
| Year 1 | Year 2 | Year 3 | Year 4 | Year 5 | Totalb,c | |
| Reference Case | ||||||
| Budget impact | 3.50 | 7.04 | 10.50 | 13.77 | 16.75 | 51.55 |
| Budget impact: cost of testing | 4.84 | 9.73 | 14.52 | 19.05 | 23.17 | 71.31 |
| Scenario 1a: Uptake of Multi-gene Pharmacogenomic Testing: Increment of 3% per Year (Year 1, 3%; Year 5, 15%) | ||||||
| Budget impact | 10.49 | 20.69 | 29.64 | 36.54 | 40.86 | 138.21 |
| Budget impact: cost of testing | 14.51 | 28.62 | 41.00 | 50.55 | 56.52 | 191.20 |
| Scenario 1b: Uptake of Multi-gene Pharmacogenomic Testing: Increment of 5% per Year (Year 1, 15%; Year 5, 35%) | ||||||
| Budget impact | 17.48 | 33.78 | 46.39 | 53.53 | 54.64 | 205.82 |
| Budget impact: cost of testing | 24.18 | 46.73 | 64.17 | 74.06 | 75.58 | 284.72 |
| Scenario 2a: Expansion of reference case target population to treatment-naive population only | ||||||
| Budget impact | 1.20 | 2.42 | 3.61 | 4.73 | 5.76 | 17.72 |
| Budget impact: cost of testing | 1.66 | 3.35 | 4.99 | 6.55 | 7.96 | 24.51 |
| Scenario 2b: Expansion of reference case target population to people who have not responded to at least one medication | ||||||
| Budget impact | 5.83 | 11.73 | 17.50 | 22.95 | 27.91 | 85.91 |
| Budget impact: cost of testing | 8.06 | 16.22 | 24.21 | 31.75 | 38.61 | 118.85 |
| Scenario 2c: Expansion of reference case target population to include both groups | ||||||
| Budget impact | 7.03 | 14.14 | 21.11 | 27.69 | 33.67 | 103.63 |
| Budget impact: cost of testing | 9.72 | 19.57 | 29.20 | 38.30 | 46.57 | 143.36 |
| Scenario 3a: Price of multi-gene pharmacogenomic test reduced to lower-end price of $450 | ||||||
| Budget impact | –0.27 | –0.53 | –0.80 | –1.05 | –1.27 | –3.92 |
| Budget impact: cost of testing | 1.07 | 2.16 | 3.23 | 4.23 | 5.15 | 15.85 |
| Scenario 3b: Price of multi-gene pharmacogenomic test reduced by 25% of reference case price (to $1,875) | ||||||
| Budget impact | 2.35 | 4.73 | 7.06 | 9.26 | 11.26 | 34.65 |
| Budget impact: cost of testing | 3.69 | 7.43 | 11.08 | 14.54 | 17.68 | 54.41 |
| Scenario 3c: Price of multi-gene pharmacogenomic test reduced by 50% of reference case price (to $1,250) | ||||||
| Budget impact | 1.20 | 2.42 | 3.61 | 4.74 | 5.76 | 37.50 |
| Budget impact: cost of testing | 2.54 | 5.12 | 7.64 | 10.02 | 12.18 | 37.50 |
| Scenario 3d: Price of multi-gene pharmacogenomic test ($2,161) if threshold for cost-effectiveness of intervention is achieved | ||||||
| Budget impact | 2.88 | 5.79 | 8.64 | 11.33 | 13.78 | 42.41 |
| Budget impact: cost of testing | 4.22 | 8.48 | 12.66 | 16.61 | 20.20 | 62.17 |
| Scenario 3e: Price of multi-gene pharmacogenomic test ($595) if cost saving threshold is achieved | ||||||
| Budget impact | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Budget impact: cost of testing | 1.34 | 2.70 | 4.03 | 5.28 | 6.42 | 19.77 |
| Scenario 4a: Clinical visits associated with testing: no additional visits are needed (testing incorporated in current care) | ||||||
| Budget impact | 3.25 | 6.54 | 9.76 | 12.80 | 15.56 | 47.90 |
| Budget impact: cost of testing | 4.59 | 9.23 | 13.78 | 18.08 | 21.98 | 67.66 |
| Scenario 4b: Clinical visits associated with testing: one additional visit is needed | ||||||
| Budget impact | 3.37 | 6.79 | 10.13 | 13.29 | 16.16 | 49.73 |
| Budget impact: cost of testing | 4.71 | 9.48 | 14.15 | 18.56 | 22.58 | 69.49 |
| Scenario 4c: Clinical visits associated with testing: 3 additional visits are needed | ||||||
| Budget impact | 3.62 | 7.29 | 10.88 | 14.27 | 17.35 | 53.40 |
| Budget impact: cost of testing | 4.96 | 9.98 | 14.90 | 19.54 | 23.77 | 73.16 |
| Scenario 5: Inclusion of OHIP+ pharmacogenomic-guided treatment coverage for youth and young adults | ||||||
| Budget impact | 13.72 | 23.49 | 25.12 | 20.76 | 16.80 | 99.89 |
| Budget impact: cost of testing | 18.98 | 32.50 | 34.74 | 28.72 | 23.24 | 138.17 |
All costs are in 2020 Canadian dollars.
Negative costs indicate savings.
Results might appear incorrect because of rounding.
Scenario 1: Change in uptake rate—With an increased rate of uptake of 3% and 5% per year (compared with 1% per year), respectively, the total 5-year budget impact would increase about 2.7 times and 4 times. A similar rate of increase is expected for the budget associated with testing
Scenario 2: Expansion of reference case target population—As expected, our analyses suggested that inclusion of treatment-naive people with depression would increase the total 5-year budget by about $18 million (the additional cost for testing alone would be $24.5 million); if all eligible people with major depression were considered for testing, the total budget would double (from $52 million for the reference case to about $104 million)
Scenario 3: Price of multi-gene pharmacogenomic testing—A decrease in the price of the test would substantially affect the total budget impact. If the test price decreased to a threshold estimate at which the intervention was cost-effective at a willingness-to-pay amount of $50,000 per QALY (i.e., a decrease from $2,500 to $2,161), the total 5-year budget and the test-related budget would be about 15% to 20% lower than the reference case. Moreover, if we assumed a cost-saving price point ($595), the total budget would be zero, as downstream cost savings would balance out the cost of the testing (at about $19.8 million)
Scenario 4: Number of clinical visits during testing—Assuming no additional visits with a physician would be needed during testing or three visits would be needed to accommodate a wider circle of care had a marginal impact. For instance, if no clinical visits were included in the care, the total budget would decrease by 7% compared with the reference case budget (which accounted for two clinical visits); if three visits were assumed, the total budget would increase by about 3.5%
Scenario 5: OHIP+ coverage for pharmacogenomic-guided treatment—The total budget impact of about $52 million in the reference case would change to about $99.9 million if full access were enabled for youth and young adults over the next 5 years. The additional cost of testing over 5 years would almost double, compared with the reference case ($138 million vs. $71 million)
Discussion
We conducted a model-based budget impact analysis to estimate the additional costs of publicly funding multi-gene pharmacogenomic testing in Ontario. This pharmacogenomic intervention includes a test with a decision support tool aimed to guide medication selection in people with major depression whose symptoms have not adequately responded to prior medication treatment.
At an uptake rate of 1% per year (a total of 27,063 persons tested over 5 years), the province would require an additional $3.5 million in year 1 to $16.8 million in year 5, or a 5-year total of $51.6 million. The cost of testing (at a test price of $2,500) accounted for about $4.8 million (in year 1) to $23.2 million (in year 5), with a total of $71.3 million over the next 5 years. The difference between the overall budget and the test-related budget suggests that the health care system could achieve some savings with this intervention (owing to reductions in medication and other health care costs). Our findings agree with results of several published studies that explored administrative data and showed reductions in the cost of medications and health care services after adoption of multi-gene pharmacogenomic-guided treatment.99,135-141 However, given uncertainty in the expected savings over a longer term and lack of long-term data on the effectiveness of the intervention, estimates of reductions in long-term downstream cost savings need to be confirmed in future phase 4 clinical studies.
As expected, our estimate of the budget impact was sensitive to the rate of test uptake over time, test eligibility, test use among people with depression (i.e., testing only the most vulnerable group with treatment-resistant disease vs. including newly diagnosed people as well), and test price.
If we were to use the test over the whole clinical pathway, so as to include all potentially eligible groups (such as treatment-naive people or those who have tried and failed to benefit from at least one medication), then the total number who would be eligible over 5 years would be about 54,407 persons (including 9,303 persons in the treatment-naive group). In this case, the total and test-related budget would be twice as high as the reference case budget.
-
An actual price of multi-gene pharmacogenomic testing is proprietary. It is determined during negotiations with the province if the technology is approved for public funding. The cost of the test is reimbursed once over a person's lifetime (even if the testing is repeated multiple times because the panel could expand to include new genes)99; however, the various multi-gene pharmacogenomic interventions used for management of major depression do differ. Although we included all these tests under the same umbrella, their costs and effectiveness and cost-effectiveness vary considerably (see Table 13; Figure 9; Appendix 12, Table A34; and Appendix 13, Table A37). Our analyses considered the most conservative cost estimate of the pharmacogenomic test available in Ontario. We showed that the price of the intervention is one of the most important drivers of the cost-effectiveness and budget impact results. Consequently, considerable savings to the province could be achieved by setting a lower price for the test:
-
○At a threshold price, for which the reference case intervention was shown to be cost-effective at a willingness-to-pay amount of $50,000 per QALY ($2,161 vs. $2,500 in the reference case), investment in this technology would decrease by about $9.1 million (from $71.3 to $62.2 million). With the most recently approved GeneSight price of $1,569 USD (about $2,000 CAD), the total budget impact would decrease from about $52 million to about $38 million, and the budget associated with this technology would decrease to about $58 million (from $71 million in the reference case)
-
○Moreover, if the price were $595, the province could expect no additional costs, except for the investment in the technology (about $19.8 million over 5 years)
-
○Last, we explored a scenario that reflects how the technology would be implemented in alignment with current OHIP+ policies for medication costs in youth and young adults.128 With OHIP+ coverage for pharmacogenomic-guided treatment, 29,499 persons aged 15 to 24 years would gain full access to the technology over 5 years; in addition, about 22,927 persons in other age groups would gain access at an uptake rate of 1% (a total of 52,426 persons to be tested). As expected, the total budget for testing would increase to $99.9 million from about $52 million in the reference case (estimated for 27,063 people). Also, the test-related budget would almost double (to $138 million from $71 million in the reference case). This estimate of the budget impact might be conservative because full adoption of the technology by young adults could be difficult to achieve.
-
○
Strengths and Limitations
Our analyses are restricted by our assumptions and uncertainty in parameter inputs that informed the model. Our estimate of the budget impact is conservative, and it depends on expected savings in downstream costs with the intervention, which are uncertain. We conducted several scenario analyses to examine factors that could affect changes in the overall budget, and in particular, the rates of uptake and the price of testing. However, further estimations of possibly larger downstream cost savings within Ontario's health system would be advisable during the implementation stages if this technology is recommended for public funding.
As mentioned above, tests for multi-gene pharmacogenomic identification are heterogeneous; they vary in cost, effectiveness, and availability in Ontario. Given established effectiveness data in several RCTs for the GeneSight test (see Clinical Evidence section) and large prior investments in the province through public–private partnerships,97 we considered the GeneSight test with the most conservative estimate of its price in the reference case. Consequently, we examined factors that could affect the reference case estimate, with specific attention to the test price. Also, some multi-gene pharmacogenomic tests proposed by Maruf el al21 were not included in our analyses because of limited clinical evidence. Last, the economic impact of various models of implementation is out of the scope of this study; however, if implementation were to occur through pharmacists, uptake rates could be substantially higher,142,143 resulting in much greater budget impact. In summary, when considering any multi-gene pharmacogenomic tests for public funding, it is of primary importance to review their efficacy data, and of secondary importance to review their cost.
With respect to the implementation of this technology in Ontario, if multi-gene pharmacogenomic pharmacogenetic testing were to be recommended for public funding, health care providers would be undertaking the difficult task of navigating the current labyrinth of pharmacogenetic testing options. For example, laboratories would need to provide the actual test results (i.e., results for each variant/single nucleotide polymorphism genotyped) and, when applicable, non-genetic factors (e.g., smoking status), which could be included in the tests' algorithms. Several studies suggest that both providers and people with major depression would require education to enable proper implementation.97,112 In terms of funding of these technologies (given financial risks associated with uncertain clinical effectiveness), the Ministry of Health might like to consider options such as outcomes-based agreements between the Ministry and manufacturers to spread the associated financial risks and uncertainties while more data are collected through research.
Conclusions
Our analysis examined publicly funding multi-gene pharmacogenomic testing that includes a decision support tool to guide medication selection in people with major depression in Ontario whose symptoms have not adequately responded to prior medication treatment. At an increasing uptake of 1% per year and a per-person test price of $2,500, adopting multi-gene pharmacogenomic-guided treatment would lead to additional costs of $3.5 million in year 1 to $16.8 million in year 5. The total additional costs over 5 years were estimated at about $52 million.
Preferences and Values Evidence
Objective
The objective of this analysis was to explore the underlying values, needs, and priorities of those who have lived experience with major depression, as well as the preferences and perceptions of both patients and caregivers relating to the use of multi-gene pharmacogenomic testing that includes decision-support tools to guide medication selection for major depression.
Background
Exploring patient preferences and values provides a unique source of information about people's experiences of a health condition and the health technologies or interventions used to manage or treat that health condition. It includes the impact of the condition and its treatment on people with the health condition, their family and other caregivers, and the person's personal environment. Engagement also provides insights into how a health condition is managed by the province's health system.
Information shared from lived experience can also identify gaps or limitations in published research (e.g., outcomes important to those with lived experience that are not reflected in the literature Additionally, lived experience can provide information and perspectives on the ethical and social values implications of health technologies or interventions.
Because the needs, preferences, priorities, and values of those with lived experience in Ontario are important to consider to understand the impact of the technology in people's lives, we sometimes speak directly with people who live with a given health condition, including those with experience of the technology or intervention we are exploring.
For this analysis, we examined perspectives and experiences of major depression among patients and families, some of whom sought multi-gene pharmacogenomic testing that includes decision-support tools to guide their medication selection. We conducted our examination in two ways:
A review by the Canadian Agency for Drugs and Technologies in Health (CADTH) of the published qualitative evidence
Direct engagement by Ontario Health with people diagnosed with major depression, and their families
Qualitative Evidence
Ontario Health collaborated with CADTH to conduct this health technology assessment. Because the literature on pharmacogenomic testing is limited, CADTH conducted a review of the qualitative literature on patient perspectives and experiences.144 We have summarized the key findings of this review below.
Key Findings
The rapid qualitative evidence synthesis included 13 primary studies that explored the views and understanding of patients and providers on pharmacogenomic testing
Patients and providers saw pharmacogenomic testing as beneficial. Although they sometimes wanted more information, most patients and providers said that pharmacogenomic testing helped them narrow their choices to the “best” medication so they could avoid adverse reactions
Patients and providers expressed worries about how pharmacogenomic testing would limit patient-centred care by limiting patients' choices of medications. They also raised concerns about having to select less effective or more expensive medications to avoid any potential adverse reactions flagged by the pharmacogenomic test results
Patients and providers raised concerns about the potential for genetic discrimination by insurers and employers, and about privacy and confidentiality. Limited access to pharmacogenomic test results was considered to be a key strategy for mitigating this risk
Pharmacogenomic test results can shape patient care over the life course. The potential for secondary findings from pharmacogenomic testing made patients worry about how results would affect them in the present and the future. The potential for pharmacogenomic test results to affect current and future family members also troubled patients and providers
The review found limited information about the use of and views on pharmacogenomic testing by disease or type of test. Findings point to the need for faster results from pharmacogenomic testing in life-limiting or rapidly progressing conditions. In areas such as mental health, pharmacogenomic testing was used less routinely, and generally applied when patients experienced adverse reactions or limited effectiveness. Providers and patients expected pharmacogenomic test results to be just one of several types of information they used for decision-making
Direct Patient Engagement
Methods
PARTNERSHIP PLAN
The partnership plan for this health technology assessment focused on consulting people diagnosed with major depressive disorder about their experiences with multi-gene pharmacogenomic tests that include decision-support tools to guide medication selection. We included the perspectives of families and caregivers of these people. We engaged people via one-on-one phone interviews.
We used a qualitative interview, as this method of engagement allowed us to explore central themes in the experiences of people who been diagnosed with major depressive disorder, as well as experiences of their families and caregivers.145 The sensitivity of exploring people's experiences with a health condition and their quality of life also supports our choice of interview methods.
PARTICIPANT OUTREACH
We used an approach called purposive sampling,146-149 which involves actively reaching out to people with direct experience of the health condition and health technology or intervention being reviewed. We approached a variety of clinicians, rehabilitation facilities, mental health facilities, community support groups, and partner organizations, including the Centre for Addictions and Mental Health, the Canadian Mental Health Association, and mental health units at various Local Health Integration Networks, to spread the word about this engagement activity and to contact people who have been assessed for major depressive disorder, and their family members and caregivers.
Inclusion Criteria
Adults (aged > 18 years) with major depression requiring pharmacological treatment.
-
Subpopulations included the following:
-
○Medication naive (initiating pharmacological treatment)
-
○Inadequate response with one or more medications (owing to lack of clinical improvement, intolerance, or side effects)
-
○
Exclusion Criteria
People younger than 18 years
People with a diagnosis of bipolar depression
Participants
For this project, we spoke with a total of 15 participants. Thirteen of the participants had received a diagnosis of major depression, five of whom had tried multi-gene pharmacogenomic-guided testing. Three participants we spoke to were caregivers, all of whom had a family member who had been diagnosed with major depression; one of these caregivers also had a diagnosis of major depression. Twelve participants were from the greater Toronto area; the remaining three lived in northern Ontario.
APPROACH
At the beginning of the interviews and surveys, we explained the role of Ontario Health, the purpose of this health technology assessment, the risks of participation, and how participants' personal health information would be protected. We gave this information to participants both verbally and in a letter of information (Appendix 14). We then obtained participants' verbal consent before starting the interview. With participants' consent, we audio-recorded and then transcribed interviews. All respondents who completed surveys remained anonymous.
Interviews lasted approximately 20 to 30 minutes. The interview was loosely structured as a series of open-ended questions. Questions were based on a list developed by the Health Technology Assessment International Interest Group on Patient and Citizen Involvement in Health Technology Assessment.150 Questions focused on how major depression affected participants' quality of life, their experiences with treatments to manage their depression, and their experiences (if any) with multi-gene pharmacogenomic-guided testing, and their perceptions of the benefits or limitations of using this testing. For family members and caregivers, questions focused on their perceptions of how the diagnosis of major depression affected their own and the patient's quality of life, and how management of the condition affected family members and caregivers themselves. See Appendix 15 for our interview and survey guide.
DATA EXTRACTION AND ANALYSIS
We used a modified grounded-theory method to analyze interview transcripts and survey results. The grounded-theory approach allowed us to organize and compare information on experiences across participants. This method consists of a repetitive process of obtaining information though interviews, documenting, and analyzing responses while simultaneously collecting, analyzing, and comparing information.157,158 We used the qualitative data analysis software program NVivo30 to identify and interpret patterns in the data. The patterns we identified allowed us to highlight the preferences and decision-making factors of those who received multi-gene pharmacogenetic testing to guide medication selection for their major depression.
Results
IMPACT OF MAJOR DEPRESSIVE DISORDER
Participants in this review reported having been diagnosed with major depressive disorder many years earlier. Causes of depression varied. Participants said it was either triggered by major life events or developed on its own and progressed over time. Symptoms greatly affected the quality of life of both participants and their families and caregivers.
Impact on People With Major Depressive Disorder
Participants faced a range of depression symptoms, which lasted both short and long periods. Some of the most common symptoms included anxiety, weariness, lethargy, fogginess, lack of will to do anything, and suicidal thoughts. Participants explained how many of these feelings also led to attempts at suicide. These symptoms greatly affected their lives as everyday things became a struggle:
When I'm depressed, I'm pathetic depressed. I have no ambition, no decision-making abilities, or anything.
I tend to feel sad and cry a lot, and it's affected my ability to care for my family. At one point I started gaining a lot of weight and just stopped caring.
There were times where I thought I could go onto the roof and just end this right now.
Impact on Families and Caregivers
Families and caregivers were also greatly affected. They expressed feelings of helplessness, worry, and projected depression. As well, they believed that their family member's lack of desire to do anything meant that caregivers had to increase caring and take over duties around the home. Caregivers tried to be understanding and be sources of support and comfort for their loved ones.
Dealing with my daughter's depression, I realized I was having similar problems. Seeing her that way gave me anxiety and depression. But I can't let her see that. That would make her symptoms worse.
My family is not happy. My husband would look after me till the end, no matter what shape I'm in, but I don't want him to live that.
My mom and dad were always stressed and worried about me. They had to financially help me out a bit because I couldn't do anything. They were very supportive.
World Suicide Prevention day is a big day in my family because of everything that's happened. My father actually said, “It was a hard journey with you, watching you struggle, and not be able to help you or kiss it better.”
MANAGING DEPRESSION SYMPTOMS
Medication Use for Symptom Relief
Participants tried a variety of medications over their lifetime to manage depression symptoms. They felt frustrated because they had to try many medications before finding one that worked while causing minimal to no side effects. Participants would typically try a medication for a while, and if it did not work or caused a lot of adverse effects, they would have to adjust the dose or begin weaning themselves off it. Participants said there was a lot of trial and error with medications to find one that worked. In some cases, the medication would work but only for a time before it had to be changed or the dose had to be adjusted. Few participants were successful at finding a medication that worked and had been using it for years. Participants described the process of trying to find the right medication as difficult and confusing, and said that it felt like a waiting game where they never knew what the outcome was going to be:
Trying to figure out what is best to do when you're trying to balance depression, headaches, insomnia (which are common side effects with depression medications), and one of the depression medications was making my depression worse—trying to figure out what to do was really hard. With the depression, your thinking and decision-making faculties aren't great. It's not a great time to be in a situation where you have to be objective and assess how you feel, to figure out if one medication is making you feel better than [you felt] 4 weeks ago on a different medication. It's difficult. And all the medications have different benefits and downsides. It's crazy.
There was a lot of trial and error. We would go through adjusting the dosages. And if that didn't work, we'd add some in or take some out. Without really knowing how it was going to affect me, it was kind of a guessing game. They really had no way of knowing which medication would have a negative or positive effect on me.
The process of finding the medication was painful. It was emotionally draining. My mood was all over the place.
Impact of Medication Use
Trying to find the right medication had a substantial impact on participants and their families. Participants had to face many different side effects with every new medication they tried. Though side effects were different for everyone, participants generally felt debilitated and unable to take care of themselves or their responsibilities and were emotionally unavailable. This affected their relationships as well:
The impact of the medications was a lot. I was a zombie for 2 or 3 years while it was impacting my personal life. I wasn't able to cope with my kids or family responsibilities.
It's impacted my relationship with my kids for sure. It's affected my bonding with them and communicating with them. They're so much closer to my husband than [to] me because of this.
And prior to all of this they were glued to me like there's no tomorrow. But I've lost all of that. It's very painful, and I feel a lot of regret.
When I first went on my antidepressant, my daughter was dealing with her own health issues. The antidepressants weren't doing anything and were causing me to be emotionally unstable. It got so bad that I couldn't cope with my child anymore and had to put her in a group home. I'm not stable emotionally to take on such a big challenge.
I got together with my boyfriend when I had just started a new medication and I just got sadder and sadder. I was very apathetic; I didn't care about anything. I was so agitated and irritable that I had to tell the guy to leave because I couldn't handle it any more.
Other Ways to Manage Depression Symptoms
Aside from medications, participants also mentioned attending therapy sessions with psychologists, psychotherapists, psychiatrists, social workers, or virtual mental health services. Other therapies participants had tried included Internet cognitive behavioural therapy, electroshock therapy, dialectical behaviour therapy, and repetitive transcranial magnetic stimulation. Many had also tried less conventional routes, including meditation and yoga. Some had successfully managed symptoms with these therapies, in some cases more successfully than with medication. However, most participants had gotten either short-term or no relief from depression symptoms:
I was seeing a counsellor and social worker for 4 years. I had a lot of other issues in my life previously that I had dealt with, and I guess all these things together is what caused this depression.
I'm in group therapy, and I've done some [dialectical behaviour therapy] and [cognitive behavioural therapy] treatments. Mindfulness stuff. And all this stuff has helped a lot, not necessarily the medication.
I've tried a number of different medications and talk therapy as well. It has not been completely successful. I've had recurrences throughout and sort of a constant level of some mood problem. I see a psychiatrist regularly and a therapist. I've had [repetitive transcranial magnetic stimulation] therapy, which I think was very helpful.
BARRIERS TO DEPRESSION SYMPTOM MANAGEMENT
Although a few participants mentioned finding symptom relief through either medications or other means, most were still looking for a solution. Participants relied primarily on medication for symptom relief; however, they were unable to do better than trial and error to find one that worked. Using trial and error was noted to be one of the main barriers to finding the right medication because it is time consuming, leads to many unwanted side effects, and leads to a lot of inconsistencies in medication use. Participants also felt uncomfortable taking medications or seeking treatment owing to the stigma surrounding depression and excessive medication use. A few participants mentioned having trouble accessing medications or other forms of symptom relief owing to geography. Some participants thought that a major barrier to getting the right treatment was not feeling heard by their clinicians. In all cases, participants faced many obstacles to find symptom relief.
Time-Consuming Trial and Error
Participants said it could take a few months to try a medication and figure out whether it would be the right one for them. Participants were frustrated and thought they were wasting a lot of time with the trial-and error process. They were also worried about uncertainty around medication use:
All of these medications take several weeks before you can understand what is happening. After 2 months the doctor suggests trying something different. It's like sitting on a fence waiting to see whether a medication will work or cause side affects. This was a huge worry. Especially for my daughter because she was prone to be suicidal and cut herself, and we didn't know if the medication was going to make it worse or better. It took so long.
We tried a bunch of medications then increased the dose, but it was so uncertain and it takes so long to know if it's working. I remember it was ridiculous how long I had to wait.
The fact that it took 4 years was a huge waste of my potential. And I was suicidal.
Unwanted Side Effects of Medication
Participants spoke of some of the unwanted side effects that resulted from trying different medications. They explained how, every time they tried a new medication, they were faced with an exhaustive list of side effects. This greatly reduced quality of life for both participants and families. In some cases, even if the medication was effective at reducing depression symptoms, the side effects were so unpleasant that participants stopped taking the medications:
At one point they switched my dose to about three times higher than my normal because it had stopped working. But that made me suicidal. As well, it impacted my heart and gave me diarrhea, problems with sleep, brain fogginess every day, blurred vision, weight loss, [and] headaches, and I spent a lot of time being bedridden.
One of the side effects I faced was sleepiness. That really knocked me out, but my doctor seemed to think it worked well. But I could not do anything; I basically just slept all day. And when I wasn't sleeping, then I was basically a zombie.
Some of the side effects I'd get were dry mouth, a certain fuzziness in thinking, and my movements were a little slower than usual.
After being on the Prozac for a couple of years, I started sleeping during the day and stayed wide awake but in a zombie-like state at night. I also faced sexual side effects.
Switching between medications was also an issue. Whether they were being slowly weaned or starting something new the next day, participants seemed to develop many adverse effects. They spoke of going through an ordeal each time they had to switch:
I realized my medication wasn't working properly for me, and when I started getting off it … I was getting all of these terrible reactions.
I was prescribed a new medication by my family doctor, but I ended up being hospitalized, so had to switch medications. But at the time, the switch was too quick, … too sudden. … It felt like I had just gotten electroshock therapy. And the third day it was so uncomfortable. It was very scary.
Changing medication is difficult. It takes such a long time and it causes side effects in itself. My psychiatrist feels it's best that I don't change medications when there are things going on in my life because we want to know what the cause of the changes in my mood [is]. I feel like I've ended up … taking medications that I may or may not still need to be taking, and sort of experimenting.
Inconsistent Use of Medication
Inconsistency in medication use was a common issue among participants trying various medications. Given the many unwanted and unpredictable side effects, participants were reluctant to take their medication or try new ones. They did not consider potential side effects worth the risk:
I was already taking a lot of drugs, and I wasn't really ready to throw another drug into the mix because I'm already having really bad side effects. So unless someone tells me that drug is going to work, I wouldn't want to risk getting something worse.
For the past 2 years, I asked for my medication to be stopped completely because of all of the side effects I was facing. I asked the doctor to stop it completely. And I did for a while because I had to get back on.
I have a heart condition, so I stopped taking my antidepressant medication for one or two days to see if it made a difference. I noticed a huge reduction in my symptoms. No more shaking, no more heart issues. But 6 months later I went through withdrawal.
Some became inconsistent with their medication regimen after having tried many in their lifetime with no success. Feelings of hopelessness made them hesitant to continue their regimen or trying anything new:
I stopped taking medications after 4 years of trying because I realized they weren't doing anything for me. My doctor said I was stable with the medications, but I wanted to get off them. I consulted with a pharmacist how to do it, then was able to stop altogether.
I have avoided or flat out refused some of my medications, because I wasn't sure it was going to help me.
I wish there were less back and forth [with the medications I had to take]. People get discouraged and inconsistent when they have to do so much back and forth.
Stigma
Many participants spoke of the stigma around depression and excessive medication use. They were ashamed and felt anxious about others knowing the amount of medications they had taken over their lifetime. They were not comfortable sharing with their family and friends that they had depression or how they were trying to manage it:
The stigma of it and the internalized stigma made it difficult. If people around me were more attuned to it and had taken it more seriously would have made it easier. But being alone with it was a barrier.
Many people are reluctant to seek help. There is still a lot of stigma around mental health. And if you say to people, “You have got to overcome your stigma or see a psychologist,” it might help, but that doesn't always happen.
FINANCIAL BURDEN
Both participants and their families described the financial burden of finding symptom relief. Most participants had some sort of insurance coverage for their medications. However, they reflected on how difficult it must be for people without coverage to be able to afford the number of medications they were taking.
Participants expressed interest in trying tests like multi-gene pharmacogenomic-guided testing to get more guidance on their medication selection. However, they felt held back because they could not afford to pay out of pocket for it. They admitted that costs of testing and treatment discouraged them from wanting to find something to relieve their symptoms or try things like pharmacogenomic-guided testing:
I had a psychiatrist several years ago suggest that I try pharmacogenomic testing. I have since learned more about it, and I wanted to try it but was too late to join the CAMH [Centre for Addiction and Mental Health] study. But I could not get in, and I can't afford to pay out of pocket for it.
Nobody has suggested the pharmacogenomic-guided test to me because of the expense. They know I can't afford it.
I fail to see how someone who is clinically depressed—and believe me, clinical depression is an absolutely horrifying state to be in—I don't see how they should be required to pay anything up front.
It's very expensive. I'm self employed, but I'm very fortunate to have a drug plan from when I was laid off from a past job. My medication alone costs from $2,000 to $3,000 a year, and that does not include the cost of therapy. So that's definitely a financial burden.
ACCESS ISSUES
Participants living in northern Ontario communities spoke of issues they had accessing appropriate treatment. Residents explained that these communities lack clinicians to prescribe the right medications. Also, getting appointments to see their clinicians or get access to different therapies often took longer than it would for those living in the greater Toronto area. Multi-gene pharmacogenomic-guided testing was not mentioned to most of these participants during their consultations with clinicians:
It was difficult to find someone to talk to and to listen to us. In the immediate area we looked around for almost 2 months before we found someone that would help us. We talked to social workers in between, but it was insufficient.
I was in Sudbury and thought that in southern Ontario there may have been better access to things and better knowledge about things there. Now that I live in Toronto, I see that. There were no good options available in Sudbury available for me.
One of the problems was lack of access to doctors [here in Sudbury]. We would not be able to see them while I was trying the medication. There was a really prolonged period of trying it out, which felt like a waste of time because I didn't know if it's going to work, and I had no one to talk to about it.
There's a lack of services in [northern communities]; they have one [cognitive behavioural therapy program for] anxiety and depression, but it takes 2 years to get in. Versus in Toronto it takes 2 weeks to get in. Here in [northern Ontario] it's like, “If you are going to die, then you're dead.”
LACK OF COMMUNICATION WITH CLINICIANS
Some participants thought they were unable to communicate well with their clinicians and were unable to take an active role in their own care. Participants did not feel heard by their clinicians and thought it led to being misdiagnosed or not being treated effectively for their depression symptoms. This delayed participants from getting the right treatment and finding symptom relief:
My psychologist still thought I had bipolar [personality disorder]. And we had conversations about me getting off the bipolar meds. I had the strong suspicion that I didn't actually have bipolar, but after clarifying with my doctor they said, “If it's going ok with your current meds, then lets just keep it going.” But I felt like it wasn't doing anything for my depression.
[I didn't feel] like I was the patient who would go in and be treated by the doctor. But I didn't feel like an active participant in it, and I felt that was a barrier to effective treatment.
I found a doctor and I would go in, and he would just say, “This is working, this isn't working; try these pills; you're fine; ok, goodbye.” At one point I had an opportunity to go to CAMH to try pharmacogenomic-guided testing, and I thought it would be great, so I asked the psychiatrist to make a referral to CAMH. I thought, “This is great; these guys are really involved at CAMH.” And when I asked for another referral to CAMH, he started yelling at me over the phone. His last words to me were “I don't give a shit.” So I called CAMH and got an appointment with a new doctor, and he started me on new medications.
PATIENT PERSPECTIVES ON MULTI-GENE PHARMACOGENOMIC TESTING
Experience with Multi-gene Pharmacogenomic Diagnostic Testing
A few participants had tried multi-gene pharmacogenomic testing to guide their medication selection. This was either done in a laboratory through their clinicians' recommendation, or through the IMPACT (Individualized Medicine: Pharmacogenetics Assessment and Clinical Treatment) study at CAMH. Those who tried it had mixed results. Some said that it provided incorrect guidance, given that that the recommended medications did not help to manage their symptoms. Others thought the guidance recommended medication that worked for a short period before they had to adjust their dosage or try something new. A few said that the test recommended medications that helped manage their depression symptoms and have continued to work years later:
My doctor told me to get this test done, so I went to a lab and got a blood test. It showed that I'd have bad side effects with the one I was on, so based on that we switched to a different one. It was helpful to know whether to increase the drug I was on and if it would make me better or not. But the testing showed it would make it worse. So that was helpful.
It was part of a study that was being done. I received a kit in the mail, and I sent back a swab and we got back a list telling me the different kind of medications whether it was red, green, or yellow.
Not all participants knew what multi-gene pharmacogenomic testing was, so we provided an overview to participants to get their perspectives on it. We explained that it was a non-invasive test that required taking a sample of DNA that is used to assess a person's genetic make-up. This information is used to predict which medications and dosages are most likely to work best to manage their depression and would result in the lowest risk of side effects. We also explained that this test is not 100% accurate, so it could recommend medications that do not work and do result in side effects. Given this information, we asked what their perspectives were on taking a test like this to help guide their medication selection.
Positive Perspectives on Multi-gene Pharmacogenomic Testing
Given all the facts and the personal experiences of participants, they felt positively about the use of multi-gene pharmacogenomic testing to guide medication selection. Participants who successfully controlled symptoms with the medications identified through pharmacogenomic-guided testing were especially supportive of the test after seeing firsthand what it could do. Participants who had tried the test but found the guidance to be incorrect in assessing their success with medications, still supported testing and found value in it. Those who had not tried pharmacogenomic testing also supported it and thought the benefits outweighed the potential lack of success.
Overall, participants thought that pharmacogenomic-guided testing would help narrow down the right medication and reduce the trial and error required to find something effective. They thought this would subsequently result in faster symptom relief, fewer adverse effects, and less wasted time, and would take them a step in the right direction given its basis on a patient's genetic information rather than random choice.
Faster Symptom Relief
Participants valued the pharmacogenomic-guided test because they felt it would help them find a medication faster than trial and error would. It took years for some participants to find treatment that worked, and in turn, a long time before they felt any symptom relief. They thought if they had guidance through something like the pharmacogenomic-guided test, that they would have been relieved of depression symptoms a lot faster:
Of course, it can't be done with 100% precision. Fair enough. But I think it's likely with testing and more sophistication that the drug regimen of a patient can be made much more precise a lot faster and there will be less back and forth between different medications.
The test could definitely be useful. It could reduce the amount of time it can take to get symptom relief—that's a big thing. That's a huge thing.
Reduced Adverse Effects
Participants thought using pharmacogenomic-guided testing would help reduce the adverse effects from medications. By narrowing down medications that cause the fewest or no side effects, participants believed they would be less likely to get unwanted side effects:
I would want to do it. If it reduced my side effects, that would be great.
If it helps you avoid the months that it takes to try something that is not going to work for you or going to have bad side effects for you, then it's great. Anything that can help make the process more efficient is fantastic.
I'm hoping that one day we can have tests like this readily available to help find the best medication solution. Just as an example, I have a bad reaction to some of my medications. But if this kind of test was more standard and could give us hints about what we should or shouldn't be taking to stop those reactions, that would be helpful.
Time Saved
Participants thought pharmacogenomic-guided testing would help save time spent trying to find an effective medication. They thought that using the test would help reduce time spent on trial and error with various medications. Participants valued this fact because they thought a lot of time was wasted on trial and error:
A lot of my life has been spent trying to figure out the right option. If … another option doesn't just require you to go through the process of waiting and seeing if it works, then you could get people back to living more effectively much quicker, and not just feeling incapacitated all the time.
There's a period when you start a new medication of figuring it out and getting it right. It can take a while. I feel like over time with this technology, finding a medication will become more effective. To me pharmacogenomic testing will be the future of this field.
The test could definitely be useful. It could save a lot of time; that is a big thing. That is a huge thing. My friend lost 10 to 15 years of her life just because she could not get the right drug. She was desperate at that time. She said if she didn't get it, she was probably going to end up at the bottom of Lake Ontario.
Step in the Right Direction
Many participants agreed that, even if the pharmacogenomic-guided test does not help to identify an effective medication, using the test would help point participants in the right direction. Many participants spent much time trying different medications with no knowledge of potential outcomes. Participants thought that this test at least provides some guidance as to what medications could work best and would not lead to participants going through random trial and error:
If we try it then we know more. Whether it works or does not work, it takes us in a good direction.
It takes such a long time to make any change. You have to increase or decrease so slowly. And you have to wait till you stabilize. And then see how you're doing before you can decide whether
the increase or decrease was the right move to make. It just takes such a long time that to have the testing to give us a hint in what direction to go was really useful.
I understand that it doesn't work 100% of the time; it doesn't work for everybody. But none of the current medications do, either. But it at least can give you a head start in the right direction. It may turn out not to be the right direction, but it doesn't put you further behind because you've got to start with something. And you're just as likely to start with a medication that may not work for you. The fact that the testing might not work for you isn't a reason not to do it. It might help. So might as well try it.
Trial and Error Was Not Random
Participants valued having a basis for the medications they tried. Many thought the process of trial and error was random, and that was likely the reason the medications did not work and caused a lot of adverse reactions. Participants believed using genetic information would provide a more medical basis for the medications used, which could result in a better chance of finding one that works and has fewer adverse effects:
Given that we've had to go through a lot of trial and error with different medications, and so far the only basis for diagnosis has been talking about it, I would be supportive of something that has more of a medical basis rather than just trying random ones.
If they [were] found to fit a particular genetic profile, then I think that's a splendid Idea for people who may need drugs and are prepared to take them.
It's really great to be able to get something on paper that you can show your doctors to help decide what to take. Having that makes it a lot easier for the trial-and-error process because there's no longer this guessing game. It's like having a guideline letting them know which ones to take or which ones to take with caution.
Negative Perspectives on Multi-gene Pharmacogenomic Testing
For the most part, participants who were directly engaged did not seem to have any concerns about pharmacogenomic-guided testing. Some had said that, owing to the cost of the testing, they did not get it done, because they could not afford it along with their other medication costs.
Impact on Patient-Centred Care
Participants had some concerns when it came to how the test results would affect their care. They worried that the guidance provided by the test would limit their medication options. They might want to opt for certain medications over others but were concerned that physicians would prefer following the test guidance over listening to patients' opinions. For instance, they could choose to use medications that the guidance says will be effective at reducing symptoms but might cause unwanted side effects, but clinicians might not prescribe it because they worry about the extent of these effects:
I had a psychiatrist several years ago suggest that I try pharmacogenomic testing, but it was so new at that point … they didn't know too much. But I did not want to come up with these test results and be forced onto medication that I was having reactions to.
Preferences and Values Evidence Discussion
Qualitative evidence from direct patient engagement illustrates a strong preference for multi-gene pharmacogenomic testing that includes decision-support tools to guide medication selection for people with major depressive disorder. These tests offer guidance on medications that are most likely to reduce depression symptoms and reduce the chance of adverse events. All sources of evidence indicate a strong inclination toward faster symptom relief, reduced adverse events, reduced time spent on medication trials, getting a potentially more effective medication, and having a medical basis for medications being tried.
People with major depression and families have some reservations about how the results of pharmacogenomic-guided tests are handled. Although Canada recently passed a law regarding prevention of genetic discrimination (Genetic Non-Discrimination Act, 2017: https://laws-lois.justice.gc.ca/eng/annualstatutes/2017_3/page-1.html), participants were still concerned about privacy and confidentiality of data gathered and the potential for genetic discrimination. Genetic discrimination is less of a concern in Canada than in other countries, given the policies for privacy of personal health information.
Another concern among people with major depression was how test results would affect patient care. This is valid in some cases, especially where patients' opinions are less valued or heard even before trying pharmacogenomic-guided testing, or in cases where clinicians have preconceived ideas of patients' health status and are considering some symptoms over others. Having the guidance available could further affect care, as clinicians could focus on pharmacogenomic test guidance over patients' preference.
Limitations
The direct patient engagement conducted for this analysis provided a good range of perspectives; however, some perspectives were missing. Although a portion of participants had tried pharmacogenomic testing, it was either offered through a clinical study (IMPACT study through CAMH) or was done at a laboratory when recommended by a clinician. Direct-to-consumer kits were not accessed by any of the participants in this review. This is a limitation, as people's experience with self-testing kits was not captured. These tests can be found online or at some pharmacies; however, the costs can be prohibitive for many, as we saw in our results.
Participants who were part of the IMPACT study had more knowledge of pharmacogenomic testing than those who had received testing from a laboratory or through their clinicians' recommendation. People with more knowledge about the test were able to share more perspectives about it. People who had not received pharmacogenomic testing did not know much about the test beyond the fact that it recommends which medication to use. They were unaware that testing provided evidence based on multiple genes or that it recommended which medications would be less likely to lead to adverse events, or even that the test would not be 100% accurate. However, when the test was described to them, these participants were very interested in it. Based on this description, they had many positive perspectives but also some negative perspectives about the test.
Qualitative Evidence Versus Direct Patient Engagement
The CADTH qualitative evidence was consistent with what we heard through direct patient engagement. Some key similarities were people's perspectives regarding multi-gene pharmacogenomic testing.
The agency's findings of people's positive perspectives showed that people with major depression believe getting pharmacogenomic testing would help them find symptom relief faster. They also believed that the test would allow them to select medications that avoided or reduced adverse reactions. A key finding from both CADTH and our patient engagement was that participants considered pharmacogenomic test results to take them a step in the right direction. Even if medications recommended through the guidance were ineffective, participants still believed they would be closer to finding an effective and safe treatment.
Comparable negative perspectives appeared through the two sources of evidence as well. Results showed participants were concerned about how pharmacogenomic testing would influence the care they would receive. Unlike the results of direct patient engagement, the CADTH results additionally indicated people's concerns over the privacy and confidentiality of their information. Participants thought that information gathered through the pharmacogenomic-guided test had the potential to be accessed and misused. Participants expressed particular concern about potential for genetic discrimination from employers and insurers.
Preferences and Values Evidence Conclusions
Although results among those who had tried pharmacogenomic-guided testing varied, participants' preferences and values generally supported having some guidance that speeds symptom relief by recommending a medication that works, with reduced side effects, and help inform their medication choices. People with major depression and caregivers alike valued multi-gene pharmacogenomic testing because they believed it could provide guidance that fit these values. There were some concerns that pharmacogenomic testing for medications would reduce patient-centred care insofar as people's preferences for pharmacotherapy treatment would not be included in treatment decisions.
Conclusions of the Health Technology Assessment
Multi-gene pharmacogenomic tests that include decision-support tools represent a heterogeneous class of interventions that have different effectiveness, costs, and cost-effectiveness compared with treatment as usual (i.e., no genetic testing).
We identified 10 primary studies and four post-hoc follow-up studies that evaluated six pharmacogenomic tests with decision-support tools. The most-reported outcomes were change in depression score, response, and remission of depression; the HAM-D17 was the most frequently used depression scale to evaluate these outcomes. No data were identified for any test that evaluated the impact of testing on important outcomes such as suicide or treatment adherence, or on longer-term outcomes like relapse, recovery, or recurrence of depression symptoms.
Overall, there was inconsistent effectiveness across the six multi-gene pharmacogenomic tests identified. Pharmacogenomic testing resulted in little to no difference in change in HAM-D17 scores as compared with treatment as usual, while some tests may improve response to treatment or remission from symptoms. The evidence was inconsistent with regard to the impact on side effects. The evidence, however, is uncertain, and therefore our confidence that these observed effects reflect the true effects is low to very low.
Although the economic studies included in our systematic review of the literature found that multi-gene pharmacogenomic testing used to guide medication selection in adults with major depression could be cost-saving and more beneficial than treatment as usual, long-term effectiveness of the intervention (1 year or longer) has not been investigated. Moreover, none of the studies used the perspective of the Ontario Ministry of Health or were directly applicable to Ontario. Given these limitations, we undertook a primary economic evaluation to examine the cost-effectiveness and budget impact of multi-gene pharmacogenomic testing that includes decision-support tools in adults with major depression in Ontario.
Our analyses in people who did not respond to prior medications found that some multi-gene pharmacogenomic interventions would be cost-effective at a willingness-to-pay amount of $100,000 per QALY or lower over a 1-year time horizon, if they had similar or greater effectiveness on the remission outcome and were less costly than the reference case test (i.e., GeneSight).
At an increasing uptake of 1% per year and a per-person test price of $2,500, adopting multi-gene pharmacogenomic-guided treatment would lead to additional costs of $3.5 million in year 1 to $16.8 million in year 5. The total additional costs over 5 years were estimated at about $52 million.
Although results among those who had tried pharmacogenomic-guided testing varied, participants' preferences and values generally supported having some guidance to find faster symptom relief by recommending a medication that works, with reduced side effects, and would help inform their medication choices. People with depression and caregivers alike valued multi-gene pharmacogenomic testing because they believed it could provide guidance that fit these values. There were some concerns that pharmacogenomic testing for medications would reduce patient-centred care if people's preferences for pharmacotherapy treatment were not considered in treatment decisions.
Acknowledgments
This report was developed by a multidisciplinary team from Ontario Health. The primary clinical epidemiologist was Milica Jokic, the secondary clinical epidemiologist was Stacey Vandersluis, the medical librarian was Caroline Higgins, the primary health economist was Olga Gajic-Veljanoski, the secondary health economist was Xuanqian Xie, and the patient and public partnership analyst was Aroma Akhund.
The medical editor was Elizabeth Betsch. Others involved in the development and production of this report were Jamie Turnbull, Claude Soulodre, Susan Harrison, Saleemeh Abdolzahraei, Elisabeth Smitko, Sarah McDowell, Vivian Ng, David Wells, Andrée Mitchell, Charles de Mestral, and Nancy Sikich.
We would like to thank the following people for lending their expertise to the development of this report:
Dr. June Carroll, Department of Family and Community Medicine, University of Toronto
Dr. James Kennedy, Tanenbaum Centre for Pharmacogenetics, CAMH, and Psychiatry and Medical Science, University of Toronto
Dr. Roger McIntyre, Psychiatry and Pharmacology, University of Toronto
Dr. Ute Schwarz, Departments of Medicine, Physiology and Pharmacology, Schulich School of Medicine and Dentistry, University of Western Ontario
Dr. Mina Tadrous, Women's College Hospital
Dr. Wendy Ungar, Technology Assessment at SickKids (TASK), Hospital for Sick Children Research Institute
We also thank our lived experience participants who generously gave their time to share their stories with us for this report.
The statements, conclusions, and views expressed in this report do not necessarily represent the views of those we consulted.
Abbreviations
- CADTH
Canadian Agency for Drugs and Technologies in Health
- CAMH
The Centre for Addiction and Mental Health
- CANMAT
Canadian Network for Mood and Anxiety Treatments
- CCHS
Canadian Community Health Survey
- CEAC
Cost-effectiveness acceptability curve
- CGI-I
Clinical Global Impressions Scale—Improvement
- CGI-S
Clinical Global Impressions Scale—Severity of illness
- CHEERS
Consolidated Health Economic Evaluation Reporting Standards
- CI
Confidence interval
- CrI
Credible interval
- CYP
Cytochrome P450
- DALY
Disability-adjusted life-year
- DES
Discrete event simulation
- DPIN
Drug Program Information Network
- DSM
Diagnostic and Statistical Manual of Mental Disorders
- FDA
US Food and Drug Administration
- FIBSER
Frequency, Intensity, Burden of Side Effects Rating
- GRADE
Grading of Recommendations Assessment, Development, and Evaluation
- HAM-D6
6-Item Hamilton Depression Rating Scale
- HAM-D17 or HDRS
17-Item Hamilton Depression Rating Scale
- ICER
Incremental cost-effectiveness ratio
- IMPACT
Individualized Medicine: Pharmacogenetic Assessment and Clinical Treatment
- NHS EED
National Health Service Economic Evaluation Database
- NICE
National Institute for Health and Care Excellence
- OHIP
Ontario Health Insurance Plan
- OR
Odds ratio
- PA
Probabilistic analysis
- PGI-I
Patient Global Impression of Improvement
- PHQ-9
9-Item Patient Health Questionnaire
- PRESS Checklist
Checklist assessing Peer Review of Electronic Search Strategies
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-analyses
- QALY
Quality-adjusted life-year
- QIDS-C16
Quick Inventory of Depressive Symptomatology (clinician rated)
- QIDS-SR16
Quick Inventory of Depressive Symptomatology (self-rated)
- RCT
Randomized controlled trial
- RD
Risk difference
- RoBANS
Risk of Bias Among Non-Randomized Studies
- RR
Relative risk
- SD
Standard deviation
- SE
Standard error
- SSRI
Selective serotonin reuptake inhibitor
Glossary
- Adverse event
An adverse event is an unexpected medical problem that happens during treatment for a health condition. Adverse events may be caused by something other than the treatment.
- Budget impact analysis
A budget impact analysis estimates the financial impact of adopting a new health care intervention on the current budget (i.e., the affordability of the new intervention). It is based on predictions of how changes in the intervention mix will impact the level of health care spending for a specific population. Budget impact analyses are typically conducted for a short-term period (e.g., 5 years). The budget impact, sometimes referred to as the net budget impact, is the estimated cost difference between the current scenario (i.e., the anticipated amount of spending for a specific population without using the new intervention) and the new scenario (i.e., the anticipated amount of spending for a specific population following the introduction of the new intervention).
- Cohort model
In economic evaluations, a cohort model is used to simulate what happens to a homogeneous cohort (group) of patients after receiving a specific health care intervention. The proportion of the cohort who experiences certain health outcomes or events is estimated, along with the relevant costs and benefits. In contrast, a microsimulation model follows the course of individual patients.
- Cost–benefit analysis
A cost–benefit analysis is a type of economic evaluation that expresses the effects of a health care intervention in terms of a monetary value so that these effects can be compared with costs. Results can be reported either as a ratio of costs to benefits or as a simple sum that represents the net benefit (or net loss) of one intervention over another. The monetary valuation of the different intervention effects is based on either prices that are revealed by markets or an individual or societal willingness-to-pay amount.
- Cost–consequence analysis
A cost–consequence analysis is a type of economic evaluation that estimates the costs and consequences (i.e., the health outcomes) of two or more health care interventions. In this type of analysis, the costs are presented separately from the consequences.
- Cost-effective
A health care intervention is considered cost-effective when it provides additional benefits, compared with relevant alternatives, at an additional cost that is acceptable to a decision-maker based on the maximum willingness-to-pay amount.
- Cost-effectiveness acceptability curve
In economic evaluations, a cost-effectiveness acceptability curve is a graphical representation of the results of a probabilistic sensitivity analysis. It illustrates the probability of health care interventions being cost-effective over a range of willingness-to-pay amounts. Willingness-to-pay amounts are plotted on the horizontal axis of the graph, and the probability of the intervention of interest and its comparator(s) being cost-effective at corresponding willingness-to-pay amounts is plotted on the vertical axis.
- Cost-effectiveness analysis
Used broadly, “cost-effectiveness analysis” may refer to an economic evaluation used to compare the benefits of two or more health care interventions with their costs. It may encompass several types of analysis (e.g., cost-effectiveness analysis, cost–utility analysis). Used more specifically, “cost-effectiveness analysis” may refer to a type of economic evaluation in which the main outcome measure is the incremental cost per natural unit of health (e.g., life-year, symptom-free day) gained.
- Cost-effectiveness plane
In economic evaluations, a cost-effectiveness plane is a graph used to show the differences in cost and effectiveness between a health care intervention and its comparator(s). Differences in effects are plotted on the horizontal axis, and differences in costs are plotted on the vertical axis.
- Cost–utility analysis
A cost–utility analysis is a type of economic evaluation used to compare the benefits of two or more health care interventions with their costs. The benefits are measured using quality-adjusted life-years, which capture both the quality and quantity of life. In a cost–utility analysis, the main outcome measure is the incremental cost per quality-adjusted life-year gained.
- Deterministic sensitivity analysis
Deterministic sensitivity analysis is an approach used to explore uncertainty in the results of an economic evaluation by varying parameter values to observe the potential impact on the cost-effectiveness of the health care intervention of interest. One-way sensitivity analysis accounts for uncertainty in parameter values one at a time, whereas multiway sensitivity analysis accounts for uncertainty in a combination of parameter values simultaneously.
- Disability-adjusted life-year (DALY)
The disability-adjusted life-year (DALY) is a health-related quality-of-life measure used to quantify the burden of disease from ill health, disability, or premature death. One disability-adjusted life-year represents the loss of one year of full health. Disability-adjusted life-years enable comparisons across different diseases, such that a disease that may cause premature death (e.g., measles) can be compared with a disease that may cause disability (e.g., cataracts).
- Discounting
Discounting is a method used in economic evaluations to adjust for the differential timing of the costs incurred and the benefits generated by a health care intervention over time. Discounting reflects the concept of positive time preference, whereby future costs and benefits are reduced to reflect their present value. The health technology assessments conducted by Ontario Health use an annual discount rate of 1.5% for both future costs and future benefits.
- Disutility
A disutility is a decrease in utility (i.e., a decrease in preference for a particular health outcome) typically resulting from a particular health condition (e.g., experiencing a symptom or complication).
- Dominant
A health care intervention is considered dominant when it is more effective and less costly than its comparator(s).
- Health-related quality of life
Health-related quality of life is a measure of the impact of a health care intervention on a person's health. It includes the dimensions of physiology, function, social life, cognition, emotions, sleep and rest, energy and vitality, health perception, and general life satisfaction.
- Health state
A health state is a particular status of health (e.g., sick, well, dead). A health state is associated with some amount of benefit and may be associated with specific costs. Benefit is captured through individual or societal preferences for the time spent in each health state and is expressed in quality-adjusted weights called utility values. In a Markov model, a finite number of mutually exclusive health states are used to represent discrete states of health.
- Incremental cost
Incremental cost is the additional cost, typically per person, of a health care intervention versus a comparator.
- Incremental cost-effectiveness ratio (ICER)
The incremental cost-effectiveness ratio (ICER) is a summary measure that indicates, for a given health care intervention, how much more a health care consumer must pay to get an additional unit of benefit relative to an alternative intervention. It is obtained by dividing the incremental cost by the incremental effectiveness. Incremental cost-effectiveness ratios are typically presented as the cost per life-year gained or the cost per quality-adjusted life-year gained.
- Incremental net benefit
Incremental net benefit is a summary measure of cost-effectiveness. It incorporates the differences in cost and effect between two health care interventions and the willingness-to-pay amount. Net health benefit is calculated as the difference in effect minus the difference in cost divided by the willingness-to-pay amount. Net monetary benefit is calculated as the willingness-to-pay amount multiplied by the difference in effect minus the difference in cost. An intervention can be considered cost-effective if either the net health or net monetary benefit is greater than zero.
- Markov model
A Markov model is a type of decision-analytic model used in economic evaluations to estimate the costs and health outcomes (e.g., quality-adjusted life-years gained) associated with using a particular health care intervention. Markov models are useful for clinical problems that involve events of interest that may recur over time (e.g., stroke). A Markov model consists of mutually exclusive, exhaustive health states. Patients remain in a given health state for a certain period of time before moving to another health state based on transition probabilities. The health states and events modelled may be associated with specific costs and health outcomes.
- Microsimulation model
In economic evaluations, a microsimulation model (e.g., an individual-level or patient-level model) is used to simulate the health outcomes for a heterogeneous group of patients (e.g., patients of different ages or with different sets of risk factors) after receiving a particular health care intervention. The health outcomes and health events of each patient are modelled, and the outcomes of several patients are combined to estimate the average costs and benefits accrued by a group of patients. In contrast, a cohort model follows a homogeneous cohort of patients (e.g., patients of the same age or with the same set of risk factors) through the model and estimates the proportion of the cohort who will experience specific health events.
- Ministry of Health perspective
The perspective adopted in economic evaluations determines the types of costs and health benefits to include. Ontario Health develops health technology assessment reports from the perspective of the Ontario Ministry of Health. This perspective includes all costs and health benefits attributable to the Ministry of Health, such as treatment costs (e.g., drugs, administration, monitoring, hospital stays) and costs associated with managing adverse events caused by treatments. This perspective does not include out-of-pocket costs incurred by patients related to obtaining care (e.g., transportation) or loss of productivity (e.g., absenteeism).
- Monte Carlo simulation
Monte Carlo simulation is an economic modeling method that derives parameter values from distributions rather than fixed values. The model is run several times, and in each iteration, parameter values are drawn from specified distributions. This method is used in microsimulation models and probabilistic sensitivity analysis.
- Multiway sensitivity analysis
A multiway sensitivity analysis is used to explore uncertainty in the results of an economic evaluation. It is done by varying a combination of model input (i.e., parameter) values simultaneously between plausible extremes to observe the potential impact on the cost-effectiveness of the health care intervention of interest.
- Natural history of a disease
The natural history of a disease is the progression of a disease over time in the absence of any health care intervention.
- OHIP+
Ontario Health Insurance Plan coverage of medication costs in youth and young adults aged between 15 and 25 years
- One-way sensitivity analysis
A one-way sensitivity analysis is used to explore uncertainty in the results of an economic evaluation. It is done by varying one model input (i.e., a parameter) at a time between its minimum and maximum values to observe the potential impact on the cost-effectiveness of the health care intervention of interest.
- Probabilistic sensitivity analysis
A probabilistic sensitivity analysis is used in economic models to explore uncertainty in several parameters simultaneously and is done using Monte Carlo simulation. Model inputs are defined as a distribution of possible values. In each iteration, model inputs are obtained by randomly sampling from each distribution, and a single estimate of cost and effectiveness is generated. This process is repeated many times (e.g., 10,000 times) to estimate the number of times (i.e., the probability) that the health care intervention of interest is cost-effective.
- Quality-adjusted life-year (QALY)
The quality-adjusted life-year (QALY) is a generic health outcome measure commonly used in cost–utility analyses to reflect the quantity and quality of life-years lived. The life-years lived are adjusted for quality of life using individual or societal preferences (i.e., utility values) for being in a particular health state. One year of perfect health is represented by one quality-adjusted life-year.
- Reference case
The reference case is a preferred set of methods and principles that provide the guidelines for economic evaluations. Its purpose is to standardize the approach of conducting and reporting economic evaluations, so that results can be compared across studies.
- Risk difference
Risk difference is the difference in the risk of an outcome occurring between one health care intervention and an alternative intervention.
- Scenario analysis
A scenario analysis is used to explore uncertainty in the results of an economic evaluation. It is done by observing the potential impact of different scenarios on the cost-effectiveness of a health care intervention. Scenario analyses include varying structural assumptions from the reference case.
- Sensitivity analysis
Every economic evaluation contains some degree of uncertainty, and results can vary depending on the values taken by key parameters and the assumptions made. Sensitivity analysis allows these factors to be varied and shows the impact of these variations on the results of the evaluation. There are various types of sensitivity analysis, including deterministic, probabilistic, and scenario.
- Societal perspective
The perspective adopted in an economic evaluation determines the types of costs and health benefits to include. The societal perspective reflects the broader economy and is the aggregation of all perspectives (e.g., health care payer and patient perspectives). It considers the full effect of a health condition on society, including all costs (regardless of who pays) and all benefits (regardless of who benefits).
- Switch therapy
Dropping a medication and adding a different medication during first 8 to 12 weeks
- Time horizon
In economic evaluations, the time horizon is the time frame over which costs and benefits are examined and calculated. The relevant time horizon is chosen based on the nature of the disease and health care intervention being assessed, as well as the purpose of the analysis. For instance, a lifetime horizon would be chosen to capture the long-term health and cost consequences over a patient's lifetime.
- Utility
A utility is a value that represents a person's preference for various health states. Typically, utility values are anchored at 0 (death) and 1 (perfect health). In some scoring systems, a negative utility value indicates a state of health valued as being worse than death. Utility values can be aggregated over time to derive quality-adjusted life-years, a common outcome measure in economic evaluations.
- Willingness-to-pay amount
A willingness-to-pay amount is the monetary value a health care consumer is willing to pay for added health benefits. When conducting a cost–utility analysis, the willingness-to-pay amount represents the cost a consumer is willing to pay for an additional quality-adjusted life-year. If the incremental cost-effectiveness ratio is less than the willingness-to-pay amount, the health care intervention of interest is considered cost-effective. If the incremental cost-effectiveness ratio is more than the willingness-to-pay amount, the intervention is considered not to be cost-effective.
Appendices
Appendix 1: Literature Search Strategies
Clinical Evidence Search
Search Date: January 24, 2020
Databases searched: MEDLINE, Embase, Cochrane Database of Systematic Reviews, Cochrane Central Register of Controlled Trials, CRD Health Technology Assessment Database, NHS Economic Evaluation Database, PsycINFO
Database: EBM Reviews – Cochrane Central Register of Controlled Trials <December 2019>, EBM Reviews – Cochrane Database of Systematic Reviews <2005 to January 21, 2020>, EBM Reviews – Health Technology Assessment <4th Quarter 2016>, EBM Reviews – NHS Economic Evaluation Database <1st Quarter 2016>, Embase <1980 to 2020 Week 03>, Ovid MEDLINE(R) ALL <1946 to January 23, 2020>, PsycINFO <1967 to January Week 3 2020>
Search Strategy:
-
1
Depressive Disorder/ (145792)
-
2
Depressive Disorder, Major/ (50224)
-
3
Depressive Disorder, Treatment-Resistant/ (2743)
-
4
((depress∗ adj2 (major or disorder∗ or resist#nt or refractory or chronic or recurrent or symptom∗)) or TRD or MDD or melancholia∗ or unipolar∗) .ti,ab,kf. (459505)
-
5
Mental Disorders/ge [Genetics] (4737)
-
6
Depression/ge [Genetics] (2789)
-
7
Antidepressive Agents/ (129544)
-
8
(((anti depressive or antidepressive) adj2 (drug∗ or agent∗ or medication∗)) or anti depressant∗ or antidepressant∗) .ti,ab,kf. (206981)
-
9
Psychotropic Drugs/ (77274)
-
10
(((psychoactive or psychiatric) adj2 (drug∗ or agent∗ or medication∗)) or psychopharmaceutical∗ or psycho pharmaceutical∗ or psychotropic∗) .ti,ab,kf. (76135)
-
11
Serotonin Uptake Inhibitors/ (65062)
-
12
(((serotonin or 5 ht or 5ht or 5 hydroxytryptamine or 5hydroxytryptamine) adj1 (update inhibitor∗ or reuptake inhibitor∗)) or SSRI or SSRIs).ti,ab,kf. (57640)
-
13
Antidepressive Agents, Tricyclic/ (42263)
-
14
tricyclic∗.ti,ab,kf. (43824)
-
15
Monoamine Oxidase Inhibitors/ (25999)
-
16
(monoamine oxidase or mao inhibitor∗) .ti,ab,kf. (34982)
-
17
“Serotonin and Noradrenaline Reuptake Inhibitors”/ (5813)
-
18
((serotonin adj2 noradrenaline) or SNRI or SNRIs).ti,ab,kf. (10160)
-
19
or/1-18 (944315)
-
20
Pharmacogenetics/ (31231)
-
21
Pharmacogenomic Testing/ (1521)
-
22
(pharmacogen∗ or PGx∗ or CPGx∗) .ti,ab,kf. (43128)
-
23
Precision Medicine/ (25023)
-
24
(precision adj1 medicine).ti,ab,kf. (20291)
-
25
or/20-24 (94116)
-
26
19 and 25 (5495)
-
27
animals/ not humans/ (5488343)
-
28
26 not 27 (5466)
-
29
Case Reports/ or Comment.pt. or Editorial.pt. or (Letter not (Letter and Randomized Controlled Trial)).pt. or Congress.pt. (5425981)
-
30
28 not 29 (5139)
-
31
limit 30 to english language [Limit not valid in CDSR; records were retained] (4710)
-
32
31 use medall,coch,cctr,clhta,cleed (1586)
-
33
major depression/ (180728)
-
34
chronic depression/ (215)
-
35
melancholia/ (200799)
-
36
treatment resistant depression/ (6664)
-
37
((depress∗ adj2 (major or disorder∗ or resist#nt or refractory or chronic or recurrent or symptom∗)) or TRD or MDD or melancholia∗ or unipolar∗) .tw,kw. (469881)
-
38
∗mental disease/ (86691)
-
39
∗depression/ (219247)
-
40
or/38-39 (304526)
-
41
genetics/ (742827)
-
42
genomics/ (108024)
-
43
or/41-42 (839177)
-
44
40 and 43 (1862)
-
45
antidepressant agent/ (92071)
-
46
(((anti depressive or antidepressive) adj2 (drug∗ or agent∗ or medication∗)) or anti depressant∗ or antidepressant∗) .tw,kw. (211545)
-
47
psychotropic agent/ (28814)
-
48
(((psychoactive or psychiatric) adj2 (drug∗ or agent∗ or medication∗)) or psychopharmaceutical∗ or psycho pharmaceutical∗ or psychotropic∗) .tw,kw. (77911)
-
49
serotonin uptake inhibitor/ (68676)
-
50
(((serotonin or 5 ht or 5ht or 5 hydroxytryptamine or 5hydroxytryptamine) adj1 (update inhibitor∗ or reuptake inhibitor∗)) or SSRI or SSRIs).tw,kw. (59572)
-
51
tricyclic antidepressant agent/ (31766)
-
52
tricyclic∗.tw,kw. (44710)
-
53
monoamine oxidase inhibitor/ (25999)
-
54
(monoamine oxidase or mao inhibitor∗) .tw,kw. (35217)
-
55
serotonin noradrenalin reuptake inhibitor/ (5462)
-
56
((serotonin adj2 noradrenaline) or SNRI or SNRIs).tw,kw. (10641)
-
57
or/33-37,44-56 (912502)
-
58
exp pharmacogenetics/ (41482)
-
59
pharmacogenetic testing/ (1489)
-
60
pharmacogenetic variant/ (1007)
-
61
(pharmacogen∗ or PGx∗ or CPGx∗) .tw,kw,dv. (47887)
-
62
∗personalized medicine/ (23776)
-
63
(precision adj1 medicine).tw,kw,dv. (20499)
-
64
or/58-63 (100223)
-
65
57 and 64 (5804)
-
66
(exp animal/ or nonhuman/) not exp human/ (10552538)
-
67
65 not 66 (5702)
-
68
Case Report/ or Comment/ or Editorial/ or (letter.pt. not (letter.pt. and randomized controlled trial/)) or conference abstract.pt. (11000834)
-
69
67 not 68 (4779)
-
70
limit 69 to english language [Limit not valid in CDSR; records were retained] (4352)
-
71
70 use emez (2467)
-
72
major depression/ (180728)
-
73
recurrent depression/ (814)
-
74
treatment resistant depression/ (6664)
-
75
((depress∗ adj2 (major or disorder∗ or resist#nt or refractory or chronic or recurrent or symptom∗)) or TRD or MDD or melancholia∗ or unipolar∗) .ti,ab,id. (465152)
-
76
mental disorders/ (300694)
-
77
genetics/ (742827)
-
78
genomics/ (108024)
-
79
or/77-78 (839177)
-
80
76 and 79 (2337)
-
81
antidepressant drugs/ (61980)
-
82
(((anti depressive or antidepressive) adj2 (drug∗ or agent∗ or medication∗)) or anti depressant∗ or antidepressant∗) .ti,ab,id. (206439)
-
83
(((psychoactive or psychiatric) adj2 (drug∗ or agent∗ or medication∗)) or psychopharmaceutical∗ or psycho pharmaceutical∗ or psychotropic∗) .ti,ab,id. (76530)
-
84
serotonin reuptake inhibitors/ (26961)
-
85
(((serotonin or 5 ht or 5ht or 5 hydroxytryptamine or 5hydroxytryptamine) adj1 (update inhibitor∗ or reuptake inhibitor∗)) or SSRI or SSRIs).ti,ab,id. (57689)
-
86
tricyclic antidepressant drugs/ (11811)
-
87
tricyclic∗.ti,ab,id. (43832)
-
88
monoamine oxidase inhibitors/ (25999)
-
89
(monoamine oxidase or mao inhibitor∗) .ti,ab,id. (34333)
-
90
serotonin norepinephrine reuptake inhibitors/ (396)
-
91
((serotonin adj2 noradrenaline) or SNRI or SNRIs).ti,ab,id. (10161)
-
92
or/72-75,80-91 (816925)
-
93
genetic testing/ (93965)
-
94
(pharmacogen∗ or PGx∗ or CPGx∗) .ti,ab,id. (41918)
-
95
precision medicine/ (25023)
-
96
(precision adj1 medicine).ti,ab,id. (18890)
-
97
or/93-96 (169919)
-
98
92 and 97 (4239)
-
99
case report/ or editorial.dt. or comment reply.dt. or letter.dt. (4618141)
-
100
98 not 99 (4009)
-
101
limit 100 to english language [Limit not valid in CDSR; records were retained] (3708)
-
102
101 use psyb (576)
-
103
32 or 71 or 102 (4629)
-
104
103 use medall (1402)
-
105
103 use emez (2467)
-
106
103 use coch (0)
-
107
103 use cctr (181)
-
108
103 use clhta (0)
-
109
103 use cleed (3)
-
110
103 use psyb (576)
-
111
remove duplicates from 103 (3075)
Economic Evidence Search
Search Date: 24 January 2020
Databases searched: MEDLINE, Embase, Cochrane Database of Systematic Reviews, Cochrane Central Register of Controlled Trials, CRD Health Technology Assessment Database, NHS Economic Evaluation Database, PsycINFO
Database: EBM Reviews – Cochrane Central Register of Controlled Trials <December 2019>, EBM Reviews – Cochrane Database of Systematic Reviews <2005 to January 21, 2020>, EBM Reviews – Health Technology Assessment <4th Quarter 2016>, EBM Reviews – NHS Economic Evaluation Database <1st Quarter 2016>, Embase <1980 to 2020 Week 03>, Ovid MEDLINE(R) ALL <1946 to January 23, 2020>, PsycINFO <1967 to January Week 3 2020>
Search Strategy:
-
1
Depressive Disorder/ (145792)
-
2
Depressive Disorder, Major/ (50224)
-
3
Depressive Disorder, Treatment-Resistant/ (2743)
-
4
((depress∗ adj2 (major or disorder∗ or resist#nt or refractory or chronic or recurrent or symptom∗)) or TRD or MDD or melancholia∗ or unipolar∗) .ti,ab,kf. (459505)
-
5
Mental Disorders/ge [Genetics] (4737)
-
6
Depression/ge [Genetics] (2789)
-
7
Antidepressive Agents/ (129544)
-
8
(((anti depressive or antidepressive) adj2 (drug∗ or agent∗ or medication∗)) or anti depressant∗ or antidepressant∗) .ti,ab,kf. (206981)
-
9
Psychotropic Drugs/ (77274)
-
10
(((psychoactive or psychiatric) adj2 (drug∗ or agent∗ or medication∗)) or psychopharmaceutical∗ or psycho pharmaceutical∗ or psychotropic∗) .ti,ab,kf. (76135)
-
11
Serotonin Uptake Inhibitors/ (65062)
-
12
(((serotonin or 5 ht or 5ht or 5 hydroxytryptamine or 5hydroxytryptamine) adj1 (update inhibitor∗ or reuptake inhibitor∗)) or SSRI or SSRIs).ti,ab,kf. (57640)
-
13
Antidepressive Agents, Tricyclic/ (42263)
-
14
tricyclic∗.ti,ab,kf. (43824)
-
15
Monoamine Oxidase Inhibitors/ (25999)
-
16
(monoamine oxidase or mao inhibitor∗) .ti,ab,kf. (34982)
-
17
“Serotonin and Noradrenaline Reuptake Inhibitors”/ (5813)
-
18
((serotonin adj2 noradrenaline) or SNRI or SNRIs).ti,ab,kf. (10160)
-
19
or/1-18 (944315)
-
20
Pharmacogenetics/ (31231)
-
21
Pharmacogenomic Testing/ (1521)
-
22
(pharmacogen∗ or PGx∗ or CPGx∗) .ti,ab,kf. (43128)
-
23
Precision Medicine/ (25023)
-
24
(precision adj1 medicine).ti,ab,kf. (20291)
-
25
or/20-24 (94116)
-
26
19 and 25 (5495)
-
27
economics/ (277073)
-
28
economics, medical/ or economics, pharmaceutical/ or exp economics, hospital/ or economics, nursing/ or economics, dental/ (856344)
-
29
economics.fs. (429280)
-
30
(econom∗ or price or prices or pricing or priced or discount∗ or expenditure∗ or budget∗ or pharmacoeconomic∗ or pharmaco-economic∗) .ti,ab,kf. (1079774)
-
31
exp “costs and cost analysis”/ (635087)
-
32
(cost or costs or costing or costly).ti. (286654)
-
33
cost effective∗.ti,ab,kf. (354761)
-
34
(cost∗ adj2 (util∗ or efficacy∗ or benefit∗ or minimi∗ or analy∗ or saving∗ or estimate∗ or allocation or control or sharing or instrument∗ or technolog∗)).ab,kf. (239214)
-
35
models, economic/ (13271)
-
36
markov chains/ or monte carlo method/ (86086)
-
37
(decision adj1 (tree∗ or analy∗ or model∗)).ti,ab,kf. (48441)
-
38
(markov or markow or monte carlo).ti,ab,kf. (142635)
-
39
quality-adjusted life years/ (41796)
-
40
(QOLY or QOLYs or HRQOL or HRQOLs or QALY or QALYs or QALE or QALEs).ti,ab,kf. (83606)
-
41
((adjusted adj1 (quality or life)) or (willing∗ adj2 pay) or sensitivity analys∗s).ti,ab,kf. (134216)
-
42
or/27-41 (2856034)
-
43
26 and 42 (505)
-
44
animals/ not humans/ (5488343)
-
45
43 not 44 (505)
-
46
Case Reports/ (2071976)
-
47
45 not 46 (505)
-
48
47 use medall,coch,cctr (140)
-
49
26 use cleed,clhta (3)
-
50
48 or 49 (143)
-
51
limit 50 to english language [Limit not valid in CDSR; records were retained] (129)
-
52
major depression/ (180728)
-
53
chronic depression/ (215)
-
54
melancholia/ (200799)
-
55
treatment resistant depression/ (6664)
-
56
((depress∗ adj2 (major or disorder∗ or resist#nt or refractory or chronic or recurrent or symptom∗)) or TRD or MDD or melancholia∗ or unipolar∗) .tw,kw. (469881)
-
57
∗mental disease/ (86691)
-
58
∗depression/ (219247)
-
59
or/57-58 (304526)
-
60
genetics/ (742827)
-
61
genomics/ (108024)
-
62
or/60-61 (839177)
-
63
59 and 62 (1862)
-
64
antidepressant agent/ (92071)
-
65
(((anti depressive or antidepressive) adj2 (drug∗ or agent∗ or medication∗)) or anti depressant∗ or antidepressant∗) .tw,kw. (211545)
-
66
psychotropic agent/ (28814)
-
67
(((psychoactive or psychiatric) adj2 (drug∗ or agent∗ or medication∗)) or psychopharmaceutical∗ or psycho pharmaceutical∗ or psychotropic∗) .tw,kw. (77911)
-
68
serotonin uptake inhibitor/ (68676)
-
69
(((serotonin or 5 ht or 5ht or 5 hydroxytryptamine or 5hydroxytryptamine) adj1 (update inhibitor∗ or reuptake inhibitor∗)) or SSRI or SSRIs).tw,kw. (59572)
-
70
tricyclic antidepressant agent/ (31766)
-
71
tricyclic∗.tw,kw. (44710)
-
72
monoamine oxidase inhibitor/ (25999)
-
73
(monoamine oxidase or mao inhibitor∗) .tw,kw. (35217)
-
74
serotonin noradrenalin reuptake inhibitor/ (5462)
-
75
((serotonin adj2 noradrenaline) or SNRI or SNRIs).tw,kw. (10641)
-
76
or/52-56,63-75 (912502)
-
77
exp pharmacogenetics/ (41482)
-
78
pharmacogenetic testing/ (1489)
-
79
pharmacogenetic variant/ (1007)
-
80
(pharmacogen∗ or PGx∗ or CPGx∗) .tw,kw,dv. (47887)
-
81
∗personalized medicine/ (23776)
-
82
(precision adj1 medicine).tw,kw,dv. (20499)
-
83
or/77-82 (100223)
-
84
76 and 83 (5804)
-
85
Economics/ (277073)
-
86
Health Economics/ or Pharmacoeconomics/ or Drug Cost/ or Drug Formulary/ (134083)
-
87
Economic Aspect/ or exp Economic Evaluation/ (465726)
-
88
(econom∗ or price or prices or pricing or priced or discount∗ or expenditure∗ or budget∗ or pharmacoeconomic∗ or pharmaco-economic∗) .tw,kw. (1111135)
-
89
exp “Cost”/ (594273)
-
90
(cost or costs or costing or costly).ti. (286654)
-
91
cost effective∗.tw,kw. (367733)
-
92
(cost∗ adj2 (util∗ or efficac∗ or benefit∗ or minimi∗ or analy∗ or saving∗ or estimate∗ or allocation or control or sharing or instrument∗ or technolog∗)).ab,kw. (250767)
-
93
Monte Carlo Method/ (67259)
-
94
(decision adj1 (tree∗ or analy∗ or model∗)).tw,kw. (52638)
-
95
(markov or markow or monte carlo).tw,kw. (148112)
-
96
Quality-Adjusted Life Years/ (41796)
-
97
(QOLY or QOLYs or HRQOL or HRQOLs or QALY or QALYs or QALE or QALEs).tw,kw. (87566)
-
98
((adjusted adj1 (quality or life)) or (willing∗ adj2 pay) or sensitivity analys∗s).tw,kw. (155292)
-
99
or/85-98 (2469037)
-
100
84 and 99 (477)
-
101
(exp animal/ or nonhuman/) not exp human/ (10552538)
-
102
100 not 101 (474)
-
103
Case Report/ (4430636)
-
104
102 not 103 (472)
-
105
104 use emez (329)
-
106
limit 105 to english language [Limit not valid in CDSR; records were retained] (316)
-
107
major depression/ (180728)
-
108
recurrent depression/ (814)
-
109
treatment resistant depression/ (6664)
-
110
((depress∗ adj2 (major or disorder∗ or resist#nt or refractory or chronic or recurrent or symptom∗)) or TRD or MDD or melancholia∗ or unipolar∗) .ti,ab,id. (465152)
-
111
mental disorders/ (300694)
-
112
genetics/ (742827)
-
113
genomics/ (108024)
-
114
or/112-113 (839177)
-
115
111 and 114 (2337)
-
116
antidepressant drugs/ (61980)
-
117
(((anti depressive or antidepressive) adj2 (drug∗ or agent∗ or medication∗)) or anti depressant∗ or antidepressant∗) .ti,ab,id. (206439)
-
118
(((psychoactive or psychiatric) adj2 (drug∗ or agent∗ or medication∗)) or psychopharmaceutical∗ or psycho pharmaceutical∗ or psychotropic∗) .ti,ab,id. (76530)
-
119
serotonin reuptake inhibitors/ (26961)
-
120
(((serotonin or 5 ht or 5ht or 5 hydroxytryptamine or 5hydroxytryptamine) adj1 (update inhibitor∗ or reuptake inhibitor∗)) or SSRI or SSRIs).ti,ab,id. (57689)
-
121
tricyclic antidepressant drugs/ (11811)
-
122
tricyclic∗.ti,ab,id. (43832)
-
123
monoamine oxidase inhibitors/ (25999)
-
124
(monoamine oxidase or mao inhibitor∗) .ti,ab,id. (34333)
-
125
serotonin norepinephrine reuptake inhibitors/ (396)
-
126
((serotonin adj2 noradrenaline) or SNRI or SNRIs).ti,ab,id. (10161)
-
127
or/107-110,115-126 (816925)
-
128
genetic testing/ (93965)
-
129
(pharmacogen∗ or PGx∗ or CPGx∗) .ti,ab,id. (41918)
-
130
precision medicine/ (25023)
-
131
(precision adj1 medicine).ti,ab,id. (18890)
-
132
or/128-131 (169919)
-
133
127 and 132 (4239)
-
134
economics/ or economy/ (376174)
-
135
pharmacoeconomics/ or health care economics/ (195126)
-
136
(econom∗ or price or prices or pricing or priced or discount∗ or expenditure∗ or budget∗ or pharmacoeconomic∗ or pharmaco-economic∗) .tw. (1083440)
-
137
exp “costs and cost analysis”/ (635087)
-
138
cost∗.ti. (308292)
-
139
cost effective∗.tw. (362017)
-
140
(cost∗ adj2 (util∗ or efficacy∗ or benefit∗ or minimi∗ or analy∗ or saving∗ or estimate∗ or allocation or control or sharing or instrument∗ or technolog∗)).ab. (236877)
-
141
markov chains/ (22392)
-
142
(decision adj1 (tree∗ or analy∗ or model∗)).tw. (51423)
-
143
(markov or markow or monte carlo).tw. (145117)
-
144
(QOLY or QOLYs or HRQOL or HRQOLs or QALY or QALYs or QALE or QALEs).tw. (86753)
-
145
((adjusted adj1 (quality or life)) or (willing∗ adj2 pay) or sensitivity analys∗s).tw. (152432)
-
146
or/134-145 (2388077)
-
147
133 and 146 (294)
-
148
case report/ (4430636)
-
149
147 not 148 (291)
-
150
149 use psyb (29)
-
151
limit 150 to english language [Limit not valid in CDSR; records were retained] (26)
-
152
51 or 106 or 151 (471)
-
153
152 use medall (118)
-
154
152 use emez (316)
-
155
152 use coch (0)
-
156
152 use cctr (8)
-
157
152 use clhta (0)
-
158
152 use cleed (3)
-
159
152 use psyb (26)
-
160
remove duplicates from 152 (364)
Search for Intervention-Related Health State Utilities
Search Date: February 3, 2020
Database searched: MEDLINE
Database: Ovid MEDLINE(R) ALL <1946 to January 31, 2020>
Search Strategy:
-
1
Depressive Disorder/ (71880)
-
2
Depressive Disorder, Major/ (29097)
-
3
Depressive Disorder, Treatment-Resistant/ (1169)
-
4
((depress∗ adj2 (major or disorder∗ or resist#nt or refractory or chronic or recurrent or symptom∗)) or TRD or MDD or melancholia∗ or unipolar∗) .ti,ab,kf. (133656)
-
5
Mental Disorders/ge [Genetics] (4741)
-
6
Depression/ge [Genetics] (2790)
-
7
Antidepressive Agents/ (42541)
-
8
(((anti depressive or antidepressive) adj2 (drug∗ or agent∗ or medication∗)) or anti depressant∗ or antidepressant∗) .ti,ab,kf. (66267)
-
9
Psychotropic Drugs/ (20731)
-
10
(((psychoactive or psychiatric) adj2 (drug∗ or agent∗ or medication∗)) or psychopharmaceutical∗ or psycho pharmaceutical∗ or psychotropic∗) .ti,ab,kf. (24218)
-
11
Serotonin Uptake Inhibitors/ (19196)
-
12
(((serotonin or 5 ht or 5ht or 5 hydroxytryptamine or 5hydroxytryptamine) adj1 (update inhibitor∗ or reuptake inhibitor∗)) or SSRI or SSRIs).ti,ab,kf. (17337)
-
13
Antidepressive Agents, Tricyclic/ (10289)
-
14
tricyclic∗.ti,ab,kf. (16257)
-
15
Monoamine Oxidase Inhibitors/ (10064)
-
16
(monoamine oxidase or mao inhibitor∗) .ti,ab,kf. (15940)
-
17
“Serotonin and Noradrenaline Reuptake Inhibitors”/ (312)
-
18
((serotonin adj2 noradrenaline) or SNRI or SNRIs).ti,ab,kf. (3391)
-
19
or/1-18 (304962)
-
20
Pharmacogenetics/ (11729)
-
21
Pharmacogenomic Testing/ (535)
-
22
(pharmacogen∗ or PGx∗ or CPGx∗) .ti,ab,kf. (17065)
-
23
Precision Medicine/ (16713)
-
24
(precision adj1 medicine).ti,ab,kf. (9053)
-
25
or/20-24 (42291)
-
26
19 and 25 (1713)
-
27
Quality-Adjusted Life Years/ (11791)
-
28
(quality adjusted or adjusted life year∗) .ti,ab,kf. (16207)
-
29
(qaly∗ or qald∗ or qale∗ or qtime∗) .ti,ab,kf. (10376)
-
30
(illness state$1 or health state$1).ti,ab,kf. (6292)
-
31
(hui or hui1 or hui2 or hui3).ti,ab,kf. (1463)
-
32
(multiattribute∗ or multi attribute∗) .ti,ab,kf. (869)
-
33
(utility adj3 (score$1 or valu∗ or health∗ or cost∗ or measure∗ or disease∗ or mean or gain or gains or index∗)).ti,ab,kf. (14143)
-
34
utilities.ti,ab,kf. (6903)
-
35
(eq-5d or eq5d or eq-5 or eq5 or euro qual or euroqual or euro qual5d or euroqual5d or euro qol or euroqol or euro qol5d or euroqol5d or euro quol or euroquol or euro quol5d or euroquol5d or eur qol or eurqol or eur qol5d or eurqol5d or euro?qul or eur?qul5d or euro∗ quality of life or European qol).ti,ab,kf. (10879)
-
36
(euro∗ adj3 (5 d or 5d or 5 dimension∗ or 5dimension∗ or 5 domain∗ or 5domain∗)).ti,ab,kf. (3833)
-
37
(sf36∗ or sf 36∗ or sf thirtysix or sf thirty six).ti,ab,kf. (21569)
-
38
(time trade off$1 or time tradeoff$1 or tto or timetradeoff$1).ti,ab,kf. (1854)
-
39
((qol or hrqol or quality of life).ti. or ∗quality of life/) and ((qol or hrqol∗ or quality of life) adj2 (increas∗ or decreas∗ or improve∗ or declin∗ or reduc∗ or high∗ or low∗ or effect or effects of worse or score or scores or change$1 or impact$1 or impacted or deteriorate$)).ab. (30759)
-
40
Cost-Benefit Analysis/ and (cost effectiveness ratio∗ and (perspective∗ or life expectanc∗)).ti,ab,kf. (3311)
-
41
∗quality of life/ and (quality of life or qol).ti. (52196)
-
42
quality of life/ and ((quality of life or qol) adj3 (improve∗ or chang∗)).ti,ab,kf. (23880)
-
43
quality of life/ and ((quality of life or qol) adj (score$1 or measure$1)).ti,ab,kf. (11347)
-
44
quality of life/ and health-related quality of life.ti,ab,kf. (30688)
-
45
quality of life/ and ec.fs. (9850)
-
46
quality of life/ and (health adj3 status).ti,ab,kf. (8720)
-
47
(quality of life or qol).ti,ab,kf. and cost-benefit analysis/ (12178)
-
48
models, economic/ (9822)
-
49
or/27-48 (155801)
-
50
26 and 49 (21)
-
51
limit 50 to english language (20)
Grey Literature Search
Search Dates: January 27–28, 2020
Websites searched:
Alberta Health Evidence Reviews, Alberta Health Services, BC Health Technology Assessments, Canadian Agency for Drugs and Technologies in Health (CADTH), Institut national d'excellence en santé et en services sociaux (INESSS), Institute of Health Economics (IHE), McGill University Health Centre Health Technology Assessment Unit, Centre Hospitalier de l'Universite de Quebec-Universite Laval, Health Technology Assessment Database, National Institute for Health and Care Excellence (NICE), Agency for Healthcare Research and Quality (AHRQ) Evidence-based Practice Centers, Australian Government Medical Services Advisory Committee, Council of Australian Governments Health Technologies, Centers for Medicare & Medicaid Services Technology Assessments, Institute for Clinical and Economic Review, Ireland Health Information and Quality Authority Health Technology Assessments, Washington State Health Care Authority Health Technology Reviews, Health Technology Wales, Oregon Health Authority Health Evidence Review Commission, Veterans Affairs Health Services Research and Development, Italian National Agency for Regional Health Services (AGENAS), Australian Safety and Efficacy Register of New Interventional Procedures −Surgical (ASERNIP-S), Belgian Health Care Knowledge Centre, Ludwig Boltzmann Institute for Health Technology Assessment, Ministry of Health Malaysia Health Technology Assessment Section, Swedish Agency for Health Technology Assessment and Assessment of Social Services, PROSPERO, EUnetHTA, ClinicalTrials.gov, Tufts Cost-Effectiveness Analysis Registry
Keywords used:
pharmacogenomic, pharmacogenomics, pharmacogenetic, pharmacogenetics, gene panel, gene panels, pgx, cpgx, precision medicine, depression, depressive, dépression, pharmacogénomique, pharmacogénétique, génétique, panel de gènes
Clinical results (included in PRISMA): 1
Economic results (included in PRISMA): 1
Ongoing Clinical Trials (ClinicalTrials.gov): 23
Ongoing HTAs (PROSPERO/EUnetHTA/MSAC): 1
Appendix 2: Selected Excluded Studies—Clinical Evidence
For transparency, we provide a list of studies that readers might have expected to see but that did not meet the inclusion criteria, along with the primary reason for exclusion.
| Citation | Primary Reason for Exclusion |
|---|---|
| Tanner JA, Davies PE, Voudouris NC, Shahmirian A, Herbert D, Braganza N, Guglia A, Dechairo BM, Kennedy JL. Combinatorial pharmacogenomics and improved patient outcomes in depression: treatment by primary care physicians or psychiatrists. J Psychiatr Res. 2018;104:157-62. | Comparator (not comparative) |
| Olson MC, Maciel A, Gariepy JF, Cullors A, Saldivar JS, Taylor D, Centeno J, Garces JA, Vaishnavi S. Clinical impact of pharmacogenetic-guided treatment for patients exhibiting neuropsychiatric disorders: a randomized controlled trial. Prim Care Companion CNS Disord. 2017;19(2). | Population (not limited to depression) |
| Espadaler J, Tuson M, Lopez-Ibor JM, Lopez-Ibor F, Lopez-Ibor MI. Pharmacogenetic testing for the guidance of psychiatric treatment: a multicenter retrospective analysis. CNS Spectr. 2016;22:315-24. | Population (not limited to depression) |
| Winner JG, Carhart JM, Altar CA, Goldfarb S, Allen JD, Lavezzari G, Parsons KK, Marshak AG, Garavaglia S, Dechairo BM. Combinatorial pharmacogenomic guidance for psychiatric medications reduces overall pharmacy costs in a 1 year prospective evaluation. Curr Med Res & Opin. 2015;31(9):1633-43. | Population (not limited to depression) |
| Breitenstein B, Scheuer S, Pfister H, Uhr M, Lucae S, Holsboer F, Ising M, Bruckl TM. The clinical application of ABCB1 genotyping in antidepressant treatment: a pilot study. CNS Spectrums. 2014;19:165-75. | Intervention (not a multi-gene decision-support tool) |
| Fagerness J, Fonseca E, Hess GP, et al. Pharmacogenetic-guided psychiatric intervention associated with increased adherence and cost savings. Am J Manag Care. 2014;20(5):e146-e156. | Population (not limited to depression) |
| Rundell JR, Harmandayan M, Staab JP. Pharmacogenomic testing and outcome among depressed patients in a tertiary care outpatient psychiatric consultation practice. Transl Psychiatry. 2011;1:e6. | Intervention (not limited to multi-gene decision-support tools) |
Appendix 3: Summary of Identified Systematic Reviews and Health Technology Assessments Meeting Study Selection Criteria
Table A1:
Characteristics of Identified Systematic Reviews
| Author, Year Literature Search End Date | Population | Intervention | Comparator(s) | Outcomes | Study Types Included |
|---|---|---|---|---|---|
| Brown et al, 202048 NS | • Adults (18 and older) • MDD |
GeneSight Psychotropic test | NS; control group required | • Depression symptom improvement • Response • Remission |
Open-label and RCTs |
| CADTH, 202050 December 2019 |
• Adults (18–60 y) • Depression |
Guided carea | Usual care (e.g., no testing) | •Response rate • Remission rate • Optimized dosing regimen • No. of changes in treatment choice • Harm (AEs, morbidity, mortality) |
HTAs, SRs, RCTs, non-randomized studies |
| Bousman et al, 201947 May 2018 |
• Adults • MDD |
Pharmacogenetic-guided decision-support tools for antidepressant treatment | Control group required | Remission | RCTs |
| Rosenblat et al, 201852 December 2017 |
MDD | Pharmacogenomic testing | Control group required | • Clinical outcomes (efficacy) • Response and remission primary outcomes |
Prospective cohort studies |
| Rosenblat et al, 201753 October 2015 |
• Adults (18–75 y) • MDD |
Pharmacogenomic testing | Not required | Clinical outcomes (efficiency) | NS |
| HQO, 201749 February 2016 |
• Age NS • Mood disorders, anxiety, schizophrenia |
Assurex GeneSight | Usual care (e.g., no testing) | • Suicide • Remission • Response • Depression score • Quality of life • Impact on therapeutic decisions • Patient and clinician satisfaction |
Excluded case reports |
| Washington State, 201639 November 2016 (last update; original was August 2016) |
• All ages • Prescribed medications for depression, mood disorder, psychosis, anxiety, ADHD, substance use disorder |
Guided carea with clinical lab tests for genetic variants in targeted genes or in panels of genes | Usual care (e.g., no testing) | • Physician and patient decision making on drug choice or dose • Adherence to treatment • Patient response • Adverse events • Cost and cost effectiveness |
RCTs, non-randomized trials, prospective cohort studies, case-control groups |
| Peterson et al, 201751,151 February 2017 |
• Adults • MDD |
Any pharmacogenomic testing platform | • Usual care (e.g., no testing) • Other types of risk prediction tools |
• Remission • Response • Quality of life • Functional capacity • Precision of effectiveness (time to remission, response, capacity, treatment switches) • Harms |
No restrictions |
| AHRQ/EGAPP, Thakur et al54 May 2006 |
• Age NS • Non-psychotic depression |
CYP450 genotypes for SSRIs | Usual care (e.g., no testing) | • Clinical outcomes (depression, quality of life, work absenteeism, harms of subsequent management) | NS |
Abbreviations: ADHD, attention-deficit hyperactivity disorder; AEs, adverse events; AHRQ, Agency for Healthcare Research and Quality; CYP, cytochrome P450; EGAPP, Evaluation of Genomic Applications in Practice and Prevention; HTAs, health technology assessments; HQO, Health Quality Ontario; MDD, major depressive disorder; NS, not specified; RCTs, randomized controlled trials; SRs, systematic reviews; SSRIs, selective serotonin reuptake inhibitors.
Guided care includes guiding drug selection and dose.
Appendix 4: Depression and Adverse Event Scales Used by Included Primary Studies
Table A2:
Scales and Scoring Used by Included Primary Studies
| Scale | Description | Number of Items and Scoring |
|---|---|---|
| HAM-D17152 | 17-item rating scale pertaining to symptoms of depression experienced over the past week | Total score ranging from 0–52; higher scores reflect greater severity of depression |
| HAM-D6153 | 6-item subscale of HAM-D17 (see above) | Total score ranging from 0–22; higher scores reflect greater severity of depression |
| SIGH-D17154 | Standardized manner of administration and scoring of the HAM-D17 scale (see above) | Total score ranging from 0–52; higher scores reflect greater severity of depression |
| QIDS-C16 or QIDS-SR16155 | 16 item scale pertaining to symptoms of depression over the last seven days | Total score ranging from 0–27; higher scores reflecting greater severity of depression |
| PHQ-9156 | 9-item scale pertaining to symptoms of depression over the last 2 weeks | Total score ranging from 0–27; higher scores reflect greater severity of depression |
| CGI-S and CGI-I157 | A two-component scale applicable to all psychiatric disorders, rated by the clinician: CGI-S rates illness severity as a single clinician question and CGI-I compares the patient's overall clinical condition to the week before medication initiation | Each question is scored from 1–7, ranging from 1 (normal) to 7 (among the most extremely ill patient) for CGI-S and 1 (very much improved since initiation of treatment) to 7 (very much worse since initiation of treatment) for CGI-I |
| FIBSER71 | A 3-question scale to assess patient side effects over the last week believed to be caused by depression medications. Three questions each correspond to the subscales of Frequency, Intensity, and Burden | Each question is rated from 0–6, ranging from 0 (least severe) to 6 (most severe). Clinical relevance is rated for each question: 0–2 indicates no changes needed, 3–4 suggests side effects should be addressed, and 5–6 indicates change in treatment |
Abbreviations: CGI-I, Clinical Global Impressions Scale I (improvement) or S (severity of illness); FIBSER, Frequency, Intensity and Burden of Side Effects; HAM-D6, 6-item Hamilton Depression Rating Scale; HAM-D17, 17-item Hamilton Depression Rating Scale; PHQ-9, 9-Item Patient Health Questionnaire; QIDS-C16, 16-Item Quick Inventory of Depressive Symptomatology (clinician-rated); QIDS-SR16, 16-Item Quick Inventory of Depressive Symptomatology (patient self-report); SIGH-D17, Structured Interview Guide for the HAM-D17.
Appendix 5: Additional Study Details
Table A3:
Primary Study Baseline Characteristics
| Author, Year Test | Mean Age (SD), y | Sex (% F) | Race/Ethnicity (%) | Psychiatric Comorbidities | Baseline HAM-D17 (SD) | Mean No. Previous Medication Trials (SD) | Baseline PGx Drug Category | |
|---|---|---|---|---|---|---|---|---|
| PGx | TAU | |||||||
| Greden et al, 2019a GeneSight |
PGx: 47.3 (14.6) TAU: 48 (14.4) |
PGx: 71.6 TAU: 68.5 |
White 80 Black 16.1 Asian 1.7 American Indian or Alaska Native 0.7 Native Hawaiian or Pacific Islander 0.1 Other/multi 1.4 |
White 82.6 Black 12.7 Asian 2.4 American Indian or Alaska Native 0.4 Native Hawaiian or Pacific Islander 0.1 Other/multi 1.8 |
PP cohort overall GAD 15.2 Panic or social phobia 15.2 PTSD 4.9 |
PGx: 20.5 (4.8) TAU: 20.7 (4.9) |
PGx: 3.4 (3.0) TAU: 3.5 (3.0) |
PP cohort cverall Use as directed 25.5% Use with caution 41.1% Use with increased caution 18.3% Not on report 15.0% |
| Winner et al, 201365 GeneSight |
PGx: 50.6 (14.6) TAU: 47.8 (13.9) |
PGx: 69 TAU: 92e |
Non-Hispanic White 96 African American 4 |
Non-Hispanic White 100 | NR | NR | PGx: 4.3 TAU: 4.5 |
NR |
| Hall-Flavin et al, 201355 GeneSight |
PGx: 41 (12.8) TAU: 44 (12.1) |
PGx: 69 TAU: 77 |
NR: almost exclusively identified as European ancestry | NR | PGx: 23 (5.1) TAU: 22.5 (5.4) |
PGx: 3.6 (3.5) TAU: 4.7 (3.5)e |
NR for all baseline categories Medication not on PGx report: TAU (16.6%), PGx (11.6%) |
|
| Hall-Flavin et al, 201256 GeneSight |
PGx: 42.1 (13.6) TAU: 42.6 (13.1) |
PGx: 54.5 TAU: 54.5 |
NR: almost exclusively identified as European ancestry | NR | NR | PGx: 1.7 (0.8) TAU: 2.2 (1.4) |
NR | |
| Bradley et al, 201858,b NeuroID-genetix |
PGxb: 47.8 (14.5) TAUb: 47.3 (15.2) |
PGxb: 73 TAUb: 72 |
Caucasianb 63 African Americanb 18 Hispanicb 16 Asianb 1 Otherb 2 |
Caucasianb 63 African Americanb 18 Hispanicb 17 Asianb 1 Otherb 1 |
PGX: depression + anxiety 43.9c TAU: depression + anxiety: 46.9c |
PGx: 20 (5.8)c TAU: 20 (5.6)c |
NR | NR |
| Han et al, 201860 Neuro-pharmagen |
PGx: 44.2 (16.1) TAU: 43.9 (13.8) |
PGx: 76.9 TAU: 72.9 |
Korean 100 | Korean 100 | NR | PGx: 24.5 (4.6) TAU: 23.1 (5.0) |
PGx: 2.5 (2.2) TAU: 2.1 (1.5) |
NR |
| Perez et al, 201762 Neuro-pharmagen |
PGx: 51.74 (12.0) TAU: 50.74 (13.1) |
PGx: 63.9 TAU: 63.4 |
Caucasian 93.5 Latin American 4.5 Other 2.0 |
Caucasian 91.3 Latin American 6.2 Other 2.5 |
Overall Anxiety 35.8 Substance abuse 12.6 |
PGx: 19.5 (6.0) TAU: 19.0 (5.7) |
PGx: 2.5 (2.3) TAU: 2.6 (2.1) |
NR |
| Perlis et al, 202061 Genecept |
PGx: 47.8 (12.4) TAU: 47.6 (12.1) |
PGx: 70.9 TAU: 72.5 |
Asian 0 American Indian or Alaskan Native 1.3 Black or African American 21.9 Native Hawaiian or other Pacific Islander 0.7 White 73.5 Other 2.6 |
Asian 0.7 American Indian or Alaskan Native 0.7 Black or African American 24.8 Native Hawaiian or other Pacific Islander 1.3 White 71.9 Other 0.7 |
NR | PGx: 22.5 (3.4) TAU: 22.1 (3.2) |
PGx 0–0.7 1–70.2 2 or 3–29.1 TAU 1–66 2 or 3–33.3 > 3–0.7 |
NR |
| Shan et al, 201963 Not specified |
PGx: 26.5 (7.9) TAU: 28.8 (8.9) |
PGx: 63 TAU: 65 |
Han Chinese 100 | Han Chinese 100 | Excluded other diagnoses | PGx: 21.0 (3.8) TAU: 20.9 (5.7) |
NR | NR |
| Singh et al, 201564 CNSDose |
PGx: 44.2 TAU: 44.3 |
PGx: 58 TAU: 61 |
Caucasian 100 | Caucasian 100 | Excluded other active diagnoses | PGx: 24.8 TAU: 24.7 |
NR | NR |
Abbreviations: GAD, generalized anxiety disorder; HAM-D17, 17-item Hamilton Depression Rating Scale; NR, not reported; PGx, pharmacogenomic-guided treatment; PP, per protocol; PTSD, post-traumatic stress disorder; SD, standard deviation; TAU, treatment as usual.
Results reported for full cohort (intention to treat) unless not reported, as specified. No substantial differences in demographics or disease between per protocol and full cohorts.
Data for depression cohort (N = 450) not reported separately. Estimates reflect combined anxiety or depression cohort (N = 685) and are not specific to those with depression alone. Depression cohort data alone were included in results.
Percentage of patients among patients in depression cohort (N = 450).
Estimated from graph using WebPlotDigitizer.
Statistically significant difference between PGx and TAU.
Appendix 6: Pharmacogenomic Tests and Decision-Support Tools Reviewed Table A4: Genetic Tests and Reporting By Included Studiesa
Table A4:
Genetic Tests and Reporting By Included Studiesa
| GeneSight | NeuroIDgenetix Bradley et al, 201858 | Neuropharmagen Han et al, 201860; Perez et al, 201762 | Genecept Perlis et al, 202061 | Unspecified Shan et al, 201963 | ||
|---|---|---|---|---|---|---|
| Greden et al, 201957 | Winner et al, 201365; Hall-Flavin et al, 201355 and 201256 | |||||
| Report classifications | Combinatorial algorithm with medications placed into coloured categories: •“Use as directed”/green bin • “Use with caution”/yellow bin • “Use with caution and with more frequent monitoring”/red bin |
Algorithm applied Medications classified into categories: •Use as directed • Use with caution or increased monitoring |
Algorithm applied Medications stratified into coloured categories: •No relevant genetic variants found/White • Increased likelihood of positive response or lower risk of ADR/Green • Need for drug dose monitoring or lower likelihood of positive response/Yellow • Increased risk of ADR/Red |
Gene-based results and impact on drug classes: •Use caution • Therapeutic options • No known gene-drug interactions. Includes drug interaction summary |
Combinatorial algorithm with 3 categories: •Use as directed • Use with caution • Use with increased caution and more frequent monitoring |
|
| No. of medications | 38 | 26 | “Over 40” | 50 (Perez et al)/59 (Han et al) | NR: ~68 current version | 16 |
| No. of genes | 8 | 5 | 10 | 30 | 18 | 5 |
| Non-gene considerations | NR | NR | Also screens for potential metabolic interactions between concomitant medications as well as for lifestyle factors | Pharmacological interactions and clinical conditions and lifestyle influences | NR | NR |
| Genes and Number of Alleles and Variants Assessed | ||||||
| CYP1A2 | 15 (−3860G>A, −2467T>delT, −739T>G, −729C>T, −163C>A, 125C>G, 558C>A, 2116G>A, 2473G>A, 2499A>T, 3497G>A, 3533G>A, 5090C>T, 5166G>A, 5347C>T) | 15 (−3860G>A, −2467T>delT, −739T>G, −729C>T, −163C>A, 125C>G, 558C>A, 2116G>A, 2473G>A, 2499A>T, 3497G>A, 3533G>A, 5090C>T, 5166G>A, 5347C>T) | 2 (NG_008431.2:g.28338G>A, NM_000761.4:c.-9-154C>A) | 2 (∗1, ∗1F) | Y | 13 (∗1, ∗1C, ∗1E, ∗1F, ∗1K, ∗3, ∗4, ∗6, ∗7, ∗8, ∗11, ∗15, ∗16) |
| CYP2B6 | 4 (∗1, ∗4, ∗6, ∗9) | 2 (∗1, ∗6) | Y | |||
| CYP2C9 | 6 (∗1, ∗2, ∗3, ∗4, ∗5, ∗6) | 6 (NM_000771.3:c.430C>T, NM_000771.3:c.1075A>C, NM_000771.3:c.1080C>G, NM_000771.3:c.817delA, M_000771.3:c.449G>A, NM_000771.3:c.1003C>T) | 6 (∗1, ∗2, ∗3, ∗6, ∗8, ∗27) | Y | ||
| CYP2C19 | 9 (∗1, ∗2, ∗3, ∗4, ∗5, ∗6, ∗7, ∗8, ∗17) | 8 (∗1, ∗2, ∗3, ∗4, ∗5, ∗6, ∗7, ∗8) | 3 (NM_000769.2:c.681G>A, NM_000769.2:c.636G>A, NM_000769.2:c.-806C>T) | 8 (∗1, ∗2, ∗3, ∗5, ∗7, ∗8, ∗17, ∗27) | Y | 10 (∗1, ∗2, ∗3, ∗4, ∗5, ∗6, ∗7, ∗8, ∗9, ∗17) |
| CYP2D6 | 18 (∗1, ∗2, ∗2A, ∗3, ∗4, ∗5 (deletion), ∗6, ∗7, ∗8, ∗9, ∗10, ∗11, ∗12, ∗14, ∗15, ∗17, ∗41, gene duplication) | 18 (∗1, ∗2, ∗2A, ∗3, ∗4, ∗5, ∗6, ∗7, ∗8, ∗9, ∗10, ∗11, ∗12, ∗14, ∗15, ∗17, ∗41, gene duplication) | 8 (NM_000106.5:c.886C>T, NM_001025161.2:c.1304G> C, NM_001025161.2:c.622delA, NG_008376.3:g.6047G>A, NC_000022.10:g.(42519196 42521970) (42531353 4253 4124)del (GRCh37), NM_001025161.2:c.688 690 delAAG, NM_000106.5:c.100C>T, NM_001025161.2:c.832+39G >A) | 26 (∗1, ∗2, ∗3, ∗4, ∗5, ∗6, ∗7, ∗8, ∗9, ∗10, ∗11, ∗12, ∗14, ∗15, ∗17, ∗19, ∗20, ∗29, ∗30, ∗35, ∗40, ∗41, ∗69, ∗1xN, ∗2xN, ∗35×2) | Y | 20 (∗1, ∗2, ∗2A, ∗3, ∗4, ∗5, ∗6, ∗7, ∗8, ∗9, ∗10, ∗11, ∗12, ∗14A, ∗14B, ∗15, ∗17, ∗36, ∗41, CNV) |
| CYP3A4 | 4 (∗1, ∗13, ∗15A, ∗22) | NM_017460.5:c.522-191C>T | 2 (∗1, ∗22) | Y – CYP3A4/5 | ||
| CYP3A5 | 2 (NM_001190484.2:c.219-237A>G, NM_001291830.1:c.594G>A) | Y – CYP3A4/5 | ||||
| SLC6A4 | 2 (L, S) | 2 (L, S) | 2 (5-HTTLPR, NM_001045.5:c.-1760C>T) | Y (2 variations) | L, S | |
| HTR2A | 1 (−1438 G>A) | NR | 2 (NM_000621.4:c.-998G>A, NM_000621.4:c.614-2211T>C) | rs7997012, A>G | ||
| COMT | NM_000754.3:c.472G>A | rs4680 | Y | |||
| BDNF | rs6265 | Y | ||||
| OPRM1 | Y (RS1799971) | Y | ||||
| MTHFR | 2 (NM_005957.3:c.665C>T, NM_005957.3:c.1286A>C) | Y | ||||
| Other: CACNA1C, ANK3, 5HT2C, MC4R, DRD2, ADRA2A, GRIK1 | Y | |||||
| Other: ABCB1, AKT2, CACNG2, CES1, CRHR1, DDIT4, DRD3, EPHX1, FCHSD1, GRIK2, GRIK4, HLA-A, HTR1A, HTR2A, HTR2C, LPHN3, NEFM, RGS4, RPTOR, SLC6A4, UGT2B15 | Y (variants provided in original article) | |||||
Abbreviations: ADR, adverse drug reaction; CYP, cytochrome P450; NR, not reported.
Insufficient data provided on CNSDose within published article by Singh et al64 as well as on online website for current version of test. Not included in summary table.
Appendix 7: Critical Appraisal of Clinical Evidence
Table A5:
Risk of BiasAmong Randomized Controlled Trials (Cochrane Risk-of-Bias Tool)
| Author, Year | Allocation Concealment | Blinding of Participants and Personnel | Incomplete Outcome Data | Selective Reporting | Other Bias |
|---|---|---|---|---|---|
| Greden et al, 201957 | Low | Highb | Unclear riskc | Highd,e | Highf |
| Winner et al, 201365 | Lowa | Highg | Low | Low | Highf,v |
| Perez et al, 201762 | Low | Highh | Low | Low/highi | Highf |
| Han et al, 201860 | Low | Highj | Highk | Low | Highf,w,u |
| Singh et al, 201564 | Lowa | Highl | Low | Low | Highf,m |
| Bradley et al, 201858 | Lowa | Highn | Lowo | Highp,q | Highf |
| Perlis et al, 202061 | Lowa | Highn | Low | Low | Highf |
| Shan et al, 201963 | Low | Highr | High risks | Low | Hight |
Allocation concealment was not described in detail. Assessment based on clear randomization process was described.
Treating clinicians were not blinded and were involved in recruitment of patients. Patients and raters were blinded.
In overall cohort, loss to follow-up by 8 weeks was considered high (31% pharmacogenic-guided treatment, 25% for treatment as usual). Reasons for discontinuation were not reported in detail; however, baseline characteristics were similar between groups among those who completed 8-week follow-up. No formal intention-to-treat analysis was performed to account for lost patients.
Protocol planned for all outcomes to also be analyzed at 12 weeks but none of these results were reported because of unplanned potential unblinding of clinicians before 12 weeks. Not all secondary outcomes were reported in manuscript (e.g., outcomes based on Clinical Global Impressions Scale).
Post-hoc, unplanned analyses conducted on the Greden et al data (Thase et al, 201968; Dunlop et al, 201966; Forester et al, 202067) were assessed at high risk of bias owing to selective reporting.
Study received funding from manufacturer or included authors working for manufacturer.
Treating clinicians were not blinded and were involved in recruitment of patients. Patients (exception of 4 patients receiving treatment as usual who did not have test) and raters were blinded.
Treating clinicians were not blinded and were the assessors for most outcomes; clinicians were also involved in recruitment of patients. Patients and telephone assessors for Patient Global Impression of Improvement; were blinded.
Several analyses were assessed to be at higher risk of bias based on post-hoc evaluations including response and remission with HAM-D17 scale and analysis of 1–3 failed medications. All other outcomes were assessed as low risk of bias for this domain. Results from Menchon et al were all post-hoc analyses and assessed at high risk of bias.
Treating clinician and assessors were not blinded. Only patients were blinded. Clinicians were also involved in recruitment of patients.
Loss to follow-up was high and not balanced between groups (25% pharmacogenic-guided treatment, 37.5 treatment as usual) with more losses from adverse events with treatment as usual.
Intention-to-treat analysis with last observation carried forward was performed; however, this might not account for potential risk of bias. Many patients were not included in original publication and subsequently reported in a corrigendum, increasing uncertainty about completeness of outcome data.
Treating clinicians were not blinded and were involved in recruitment of patients. Only patients and HAM-D assessors were blinded.
(Notes continued on the next page)
How patients were identified or recruited for study was unclear.
Treating clinicians were not blinded and were involved in recruitment of patients. Patients and all raters were blinded
Number of dropouts was not substantive, with similar numbers in each group, but no information on reasons for drop out or from which patient population (i.e., depression or anxiety) was supplied.
Data were presented only for a subset of the population. No data were reported on patients with mild depression, and only remission data were reported for severe depression. Definition of moderate depression varied from methods to results.
Protocol on clinicaltrials.gov reported change in HAM-D17 scores as an outcome but was not reported in publication.
Treating clinicians and patients were not blinded. Rater for assessment scales was blinded.
Loss to follow-up was high (37% for pharmacogenetic-guided testing, 32% for treatment as usual), and reasons for losses were not provided. Authors did do both per-protocol and intention-to-treat analyses; however, this might not address potential risk of bias.
A single psychiatrist treated all patients. It is unclear if this psychiatrist was originally treating the patients before enrollment.
Participantss were prohibited from using any combination of other new antidepressant, antipsychotic, mood stabilizer, or central nervous system stimulant and anti-addiction agents throughout study period. Discontinuation criteria were said to be established in protocol, but no details were provided.
Significantly more women were randomized to the treatment as usual arm than to the pharmacogenomic-guided treatment arm.
Corrigendum published 2 years after study completion identified substantial errors in original publication related to statistical analyses, inclusion of covariates, and missing patient data. It is unclear if version presented in corrigendum was peer reviewed.
Table A6:
Risk Of Bias Among Nonrandomized Studies (RoBANS)
| Author, Year | Selection of Participants | Confounding Variables | Measurement of Exposure | Blinding of Outcome Assessment | Incomplete Outcome Data | Selective Outcome Reporting |
|---|---|---|---|---|---|---|
| Hall-Flavin et al, 201355 | Low | Higha | Low | Highb | Highc | Low |
| Hall-Flavin et al, 201256 | Low | Higha | Low | Highb | Low | Lowd |
Abbreviation: RoBANS, Risk of Bias Assessment for Non-randomized Studies.
Major confounding variables were not considered during design stage, and no analysis to adjust for confounding factors was considered. Pharmacogenomic treatment group had fewer previously failed psychiatric medication trials than treatment as usual group. No information was provided about other treatments that might have been used.
No mention was made of blinding assessors, and no information was provided on who completed assessment.
Large and unbalanced numbers of dropouts in both groups, with larger number of dropouts by 8 weeks in guided group (36.8% in pharmacogenomic-guided treatment vs. 17.6% in treatment as usual). No differences were observed in measured baseline characteristics, and two methods of data imputation were applied to account for incomplete outcome data (although these were post-hoc imputations).
Authors noted raw changes in score as primary outcome, but focused on percent change in results.
Table A7:
GRADE Evidence Profile for Comparison of GeneSight-Guided Treatment Selection With Treatment as Usual—Change in Depression Score
| No. of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| 17-Item Hamilton Depression Rating Scale | |||||||
| 2 (RCTs) | Very serious limitations (−;2)a | No serious limitationsb | No serious limitationsc | Serious limitations (−;1)d,e | Undetected | None | ⊕ Very low |
| 2 (observational) | Serious limitations (−;1)a | No serious limitationsb | No serious limitationsc | Serious limitations (−;1)d,f | Undetected | None | ⊕ Very low |
| 16-item Quick Inventory of Depressive Symptomatology | |||||||
| 2 (RCTs) | Very serious limitations (−;2)a | No serious limitationsb | No serious limitationsc | Serious limitations (−;1)d | Undetected | None | ⊕ Very low |
| 2 (observational) | Serious limitations (−;1)a | No serious limitationsb | No serious limitationsc | Serious limitations (−;1)d,f | Undetected | None | ⊕ Very low |
| 9-Item Patient Health Questionnaire | |||||||
| 2 (RCTs) | Very serious limitations (−;2)b | No serious limitationsb | No serious limitationsc | Serious limitations (−;1)d | Undetected | None | ⊕ Very low |
| 1 (observational) | Serious limitations (−;1)a | Noneg | No serious limitationsc | Serious limitations (−;1)d,f | Undetected | None | ⊕ Very low |
| 6-Item Hamilton Depression Rating Scale | |||||||
| 1 (RCT) | Very serious limitations (−;2)a | Noneg | No serious limitationsc | Serious limitations (−;1)d | Undetected | None | ⊕ Very low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; RCT, randomized controlled trial.
See Risk of Bias Table A5 and Table A6. Observational studies begin at low quality GRADE and were not downgraded further owing to very serious risk of bias issues.
Insufficient data were available to judge consistency of data between studies.
Only percent changes from baseline were reported, which did not allow for assessment of clinically meaningful differences in mean scores.
No measures of variance were reported and therefore they could not be appropriately assessed.
Based on data from the larger RCT by Greden et al, estimated effect estimates did not meet the clinically meaningful threshold of a 2- to 3-point difference in mean HAM-D scores.
Study sample sizes were small and unlikely to meet optimal information size.
Not evaluable owing to single study.
Table A8:
GRADE Evidence Profile for Comparison of Neuropharmagen-Guided Treatment Selection and Treatment as Usual—Change in Depression Score
| No. of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| 17-Item Hamilton Depression Rating Scale | |||||||
| 2 (RCTs) | Serious limitations (−;1)a | No serious limitationsb | No serious limitations | Serious limitations (−;1)cd | Undetected | None | ⊕⊕ Low |
| 9-Item Patient Health Questionnaire | |||||||
| 1 (RCT) | Very serious limitations (−;2)a | Nonee | No serious limitations | Serious limitations (−;1)f | Undetected | None | ⊕ Very Low |
| Clinical Global Impression Scale–Severity | |||||||
| 2 (RCTs) | Serious limitations (−;1)a | No serious limitationsb | No serious limitations | Serious limitations (−;1)c | Undetected | None | ⊕⊕ Low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; RCT, randomized controlled trial.
See Risk of Bias Table A5. Han et al was considered to have very serious limitations related to risk of bias, but given the Perez et al study was much larger, we chose to downgrade only one level to reflect risk of bias in that study.
Insufficient data were available to judge consistency of data between studies, and findings were downgraded owing to uncertainty between study estimates.
Summary estimates or measures of variance between groups were not reported for the largest trial and therefore could not be appropriately assessed.
Based on unadjusted graphic values, the largest trial by Perez et al62 did not achieve statistical significance or a clinically meaningful threshold of a 2- to 3-point difference in mean scores for the Hamilton Depression Rating Scale.
Not evaluable owing to single study.
Small study which would not meet optimal information size. Summary estimate with confidence intervals could not be calculated given adjustments in data, and authors did not report variance around estimates to allow us to appropriately assess imprecision. Results were not statistically significant.
Table A9:
GRADE Evidence Profile for the Comparison of Genecept-Guided Treatment Selection and Treatment as Usual—Change in Depression Score
| No. of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| 17-Item Hamilton Depression Rating Scale | |||||||
| 1 (RCT) | Serious limitations (−;1)a | Noneb | No serious limitations | Serious limitations (−;1)c | Undetected | None | ⊕⊕ Low |
| 16-Item Quick Inventory of Depressive Symptomatology | |||||||
| 1 (RCTs) | Serious limitations (−;1)a | Noneb | No serious limitations | Serious limitations (−;1)d | Undetected | None | ⊕⊕ Low |
| Clinical Global Impression Scale–Severity | |||||||
| 1 (RCTs) | Serious limitations (−;1)a | Noneb | No serious limitations | Serious limitations (−;1)d | Undetected | None | ⊕⊕ Low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; RCT, randomized controlled trial.
See Risk of Bias Table A5.
Not evaluable owing to single study.
Mean difference was not clinically meaningful and ranged from potential harm to small benefit.
Mean differences crossed both potential benefit and harm.
Table A10:
GRADE Evidence Profile for Comparison of Treatment Guided by Unspecified Pharmacogenomic Test With Treatment as Usual—Change in Depression Score
| No. of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| 17-Item Hamilton Depression Rating Scale | |||||||
| 1 (RCT) | Very serious limitations (−;2)a | Noneb | No serious limitations | Serious limitations (−;1)c,d | Undetected | None | ⊕ Very Low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; RCT, randomized controlled trial.
See Risk of Bias Table A5.
Not evaluable owing to single study.
No measures of variance for adjusted analyses were reported, and therefore imprecision could not be appropriately assessed. Based on reported data, point estimates did not meet clinically meaningful threshold of a 2- to 3-point difference in mean scores from Hamilton Depression Rating Scale.
Study sample sizes were small and unlikely to meet optimal information size.
Table A11:
GRADE Evidence Profile for Comparison of GeneSight-Guided Treatment Selection and Treatment as Usual—Response
| No. of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| Response Based on HAM-D17 | |||||||
| 2 (RCTs) | Very serious limitations (−;2)a | No serious limitations | No serious limitations | No serious limitations | Undetected | None | ⊕⊕ Low |
| 1 (observational) | Serious limitations (−;1)a | No serious limitations | No serious limitations | Serious limitations (−;1)b | Undetected | None | ⊕ Very Low |
| Response Based on QIDS-C16 | |||||||
| 1 (RCTs) | Very serious limitations (−;2)a | No serious limitations | No serious limitations | Serious limitations (−;1)c | Undetected | None | ⊕ Very Low |
| 1 (observational) | Serious limitations (−;1)a | No serious limitations | No serious limitations | Serious limitations (−;1)b | Undetected | None | ⊕ Very Low |
| Response Based on 9-Item Patient Health Questionnaire | |||||||
| 1 (RCTs) | Very serious limitations (−;2)b | No serious limitations | No serious limitations | No serious limitations | Undetected | None | ⊕⊕ Low |
| 1 (observational) | Serious limitations (−;1)a | No serious limitations | No serious limitations | Serious limitations (−;1)b | Undetected | None | ⊕ Very Low |
| Response Based on HAM-D6 | |||||||
| 1 (RCT) | Very serious limitations (−;2)a | No serious limitations | No serious limitations | No serious limitations | Undetected | None | ⊕⊕ Low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; HAM-D6, 6-item Hamilton Depression Rating Scale; HAM-D17, 17-item Hamilton Depression Rating Scale; QIDS-C16, 16-Item Quick Inventory of Depressive Symptomatology (clinician-rated); RCT, randomized controlled trial.
See Risk of Bias Tables A5 and A6. Observational studies begin at low-quality GRADE and were not downgraded further owing to very serious risk of bias issues.
Study had a small sample size, and number included at follow-up did not meet sample size calculation.
Effect estimate crosses null effect including both benefit and harm in effect.
Table A12:
GRADE Evidence Profile for the Comparison of Neuropharmagen-Guided Treatment Selection and Treatment as Usual – Response
| No. of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| Response Based on 17-Item Hamilton Depression Rating Scale | |||||||
| 2 (RCTs) | Very serious limitations (−;2)a | No serious limitationsb | No serious limitations | Serious limitations (−;1)c | Undetected | None | ⊕ Very Low |
| Response Based on Patient Global Impression of Improvement | |||||||
| 1 (RCT) | Serious limitations (−;1)a | No serious limitations | No serious limitations | Serious limitations (−;1)d | Undetected | None | ⊕⊕ Low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; RCT, randomized controlled trial.
See Risk of Bias Table A5.
Insufficient data were provided by Han et al60 to assess effect size and confidence intervals.
Perez et al62 had wide confidence intervals surrounding effect estimate, including both benefit and harm with intervention. Only summary of effect and statistical significance was provided by Han et al.60
Confidence intervals are wide, spanning very large benefit to no effect.
Table A13:
GRADE Evidence Profile for the Comparison of NeuroIDgenetix-Guided Treatment Selection With Treatment as Usual—Response
| No. of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| Response Based on 17-Item Hamilton Depression Rating Scale | |||||||
| 1 (RCT) | Very serious limitations (−;2)a | No serious limitations | No serious limitations | No serious limitations | Undetected | None | ⊕⊕ Low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; RCT, randomized controlled trial.
See Risk of Bias Table A5.
Table A14:
GRADE Evidence Profile for Comparison of Genecept-Guided Treatment Selection With Treatment as Usual—Response
| No. of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| Response Based on 17-Item Hamilton Depression Rating Scale | |||||||
| 1 (RCT) | Serious limitations (−;1)a | No serious limitations | No serious limitations | Serious limitations (−;1)b | Undetected | None | ⊕⊕ Low |
| Response Based on Clinical Global Impression Scale—Improvement | |||||||
| 1 (RCT) | Serious limitations (−;1)a | No serious limitations | No serious limitations | Serious limitations (−;1)c | Undetected | None | ⊕⊕ Low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; RCT, randomized controlled trial.
See Risk of Bias Table A5.
Confidence intervals span both increased risk and small benefit.
Confidence intervals span both improvement and little to no difference.
Table A15:
GRADE Evidence Profile for Comparison of Unspecified Pharmacogenomic-Guided Treatment Selection and Treatment as Usual—Response
| No. of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| Response Based on 17-Item Hamilton Depression Rating Scale | |||||||
| 1 (RCT) | Serious limitations (−;1)a | No serious limitations | No serious limitations | Serious limitations (−;1)b | Undetected | None | ⊕⊕ Low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; HAM-D17, 17-item Hamilton Depression Rating Scale; RCT, randomized controlled trial.
See Risk of Bias Table A5.
Study had very small sample size and was unlikely to meet optimal information size. Confidence intervals span both large benefit and potential harm.
Table A16:
GRADE Evidence Profile for Comparison of GeneSight-Guided Treatment Selection With Treatment as Usual—Remission
| No. of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| Remission Based on HAM-D17 | |||||||
| 2 (RCT) | Very serious limitations (−;2)a | No serious limitations | No serious limitations | No serious limitations | Undetected | None | ⊕⊕ Low |
| 1 (observational) | Serious limitations (−;1)a | No serious limitations | No serious limitations | Serious limitations (−;1)b,c | Undetected | None | ⊕ Very Low |
| Remission Based on QIDS-C16 | |||||||
| 1 (RCT) | Very serious limitations (−;2)a | No serious limitations | No serious limitations | No serious limitations | Undetected | None | ⊕⊕ Low |
| 1 (observational) | Serious limitations (−;1)a | No serious limitations | No serious limitations | Serious limitations (−;1)b | Undetected | None | ⊕ Very Low |
| Remission Based on 9-Item Patient Health Questionnaire | |||||||
| 1 (RCT) | Very serious limitations (−;2)a | No serious limitations | No serious limitations | Serious limitations (−;1)c | Undetected | None | ⊕ Very Low |
| 1 (observational) | Serious limitations (−;1)a | No serious limitations | No serious limitations | Serious limitations (−;1)b,c | Undetected | None | ⊕ Very Low |
| Remission Based on HAM-D6 | |||||||
| 1 (RCT) | Very serious limitations (−;2)a | No serious limitations | No serious limitations | No serious limitations | Undetected | None | ⊕ Very Low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; HAM-D6, 6-item Hamilton Depression Rating Scale; HAM-D17, 17-item Hamilton Depression Rating Scale; QIDS-C16, 16-Item Quick Inventory of Depressive Symptomatology (clinician-rated); RCT, randomized controlled trial.
See Risk of Bias Table A5 and Table A6. Observational studies begin at low-quality GRADE and were not downgraded further owing to very serious risk of bias issues.
Study had small sample size, and number included at follow-up did not meet sample size calculation.
Effect estimate crosses null effect including both large benefit and no effect or harm.
Table A17:
GRADE Evidence Profile for Comparison of Neuropharmagen-Guided Treatment Selection With Treatment as Usual—Remission
| No. of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| Remission Based on 17-Item Hamilton Depression Rating Scale | |||||||
| 2 (RCTs) | Very serious limitations (−;2)a | No serious limitationsb | No serious limitations | Serious limitations (−;1)c | Undetected | None | ⊕ Very Low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; RCT, randomized controlled trial.
See Risk of Bias Table A5.
Insufficient data were provided by Han et al to assess effect size and confidence intervals.
Table A18:
GRADE Evidence Profile for Comparison of NeuroIDgenetix-Guided Treatment Selection With Treatment as Usual—Remission
| No. of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| Remission Based on 17-Item Hamilton Depression Rating Scale | |||||||
| 1 (RCT) | Very serious limitations (−;2)a | No serious limitations | No serious limitations | Serious limitations (−;1)b | Undetected | Nonec | ⊕ Very Low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; RCT, randomized controlled trial.
See Risk of Bias Table A5.
Sample size was small and unlikely to meet optimal information size for this outcome.
Effect size was large; however, we did not upgrade the evidence because data were from a single study with other serious limitations.
Table A19:
GRADE Evidence Profile for Comparison of Genecept-Guided Treatment Selection With Treatment as Usual—Remission
| No. of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| Remission Based on 17-Item Hamilton Depression Rating Scale | |||||||
| 1 (RCT) | Serious limitations (−;1)a | No serious limitations | No serious limitations | Serious limitations (−;1)b | Undetected | None | ⊕⊕ Low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; RCT, randomized controlled trial.
See Risk of Bias Table A5.
Confidence intervals span both increased risk and a small benefit.
Table A20:
GRADE Evidence Profile for Comparison of CNSDose-Guided Treatment Selection With Treatment as Usual—Remission
| No. of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| Remission Based on 17-Item Hamilton Depression Rating Scale | |||||||
| 1 (RCT) | Serious limitations (−;1)a | No serious limitations | No serious limitations | Serious limitations (−;1)b | Undetected | Nonec | ⊕⊕ Low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; RCT, randomized controlled trial.
See Risk of Bias Table A5.
Small sample size and no power calculation, although data were unlikely to meet optimal information size.
Effect size was large; however, we did not upgrade the evidence because data were from a single study with other serious limitations.
Table A21:
GRADE Evidence Profile for Comparison of Unspecified Pharmacogenomic-Guided Treatment Selection and Treatment as Usual—Remission
| No. of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| Response Based on Hamilton Depression Rating Scale | |||||||
| 1 (RCT) | Very serious limitations (−;2)a | No serious limitations | No serious limitations | Serious limitations (−;1)b | Undetected | None | ⊕ Very Low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; RCT, randomized controlled trial.
See Risk of Bias Table A5.
Effect estimate spans both large benefit and small harm. Sample size was small and unlikely to meet optimal information size.
Table A22:
GRADE Evidence Profile for Comparison of Pharmacogenomic-Guided Treatment Selection With Treatment as Usual—Side Effects and Adverse Events
| No. of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| GeneSight—Mean Side Effects | |||||||
| 1 (RCT) | Very serious limitations (−;2)a | No serious limitations | No serious limitations | No serious limitations | Undetected | None | ⊕⊕ Low |
| Neuropharmagen—FIBSER Scores | |||||||
| 2 (RCTs) | Serious limitations (1)a | No serious limitationsb | No serious limitations | Serious limitations (1)c | Undetected | None | ⊕⊕ Low |
| Genecept—FIBSER Scores | |||||||
| 1 (RCT) | Serious limitations (1)a | No serious limitations | No serious limitations | No serious limitations | Undetected | None | ⊕⊕⊕ Moderate |
| CNSDose—Intolerability Rate | |||||||
| 1 (RCT) | Serious limitations (1)a | No serious limitations | No serious limitations | Serious limitations (1)d | Undetected | None | ⊕⊕ Low |
| Unspecified Test—Adverse Reactions | |||||||
| 1 (RCT) | Very serious limitations (−;2)a | No serious limitations | No serious limitations | Serious limitations (1)e | Undetected | None | ⊕ Very Low |
Abbreviations: FIBSER, Frequency, Intensity and Burden of Side Effects Rating; GRADE, Grading of Recommendations Assessment, Development, and Evaluation; RCT, randomized controlled trial.
See risk of bias tables in Appendix 7.
Insufficient data were provided by studies to calculate summary estimates and confidence intervals. Overall, summary estimates appeared to be consistent between studies except for FIBSER Frequency score. Given small uncertainty with both inconsistency and imprecision, only imprecision was downgraded.
Insufficient data were provided to calculate summary estimate and variance around all scores. Studies also differed in their reported measures of FIBSER. Given small uncertainty with both inconsistency and imprecision, only imprecision was downgraded.
Study had small sample size and likely was underpowered. Confidence intervals ranged from very small difference to large effect.
No point estimate could be calculated from data provided, and only a non-significant result was provided.
Appendix 8: Additional Calculations and Subgroup Analyses
Table A23:
Results of Change in HAM-D17 Depression Scores With Less Than 8-Week Follow-Up
| Author, Year | N PGx/TAU | Mean at Follow-up (SD) | % Decrease from Baseline to Follow-Up | P Valuea | ||
|---|---|---|---|---|---|---|
| PGx | TAU | PGx | TAU | |||
| Genesight | ||||||
| Winner et al, 201365 | 6 wk: 25/24 4 wk: 25/24 |
NR NR |
NR NR |
35.4 28.3 |
18.5 19.8 |
.04 .27 |
| Hall-Flavin et al, 201355 | 4 wk: 72/93 2 wk: 72/93 |
NE NE |
NE NE |
NR NR |
NR NR |
.0002 NS |
| Hall-Flavin et al, 201256 | 4 wk: 22/22 2 wk: 22/22 |
NE NE |
NE NE |
NR NR |
NR NR |
NS NS |
| Neuropharmagen | ||||||
| Perez et al, 201762 | 6 wk: 146/146 | NE | NE | NR | NR | .036 |
| Other | ||||||
| Shan et al, 201963 | 4 wk: 31/40b 2 wk: 31/40b |
10.68 (4.17) 12.77 (4.67) |
11.03 (4.83) 13.33 (4.27) |
48.50 38.66 |
46.87 35.78 |
MD: 0.901 MD: 0.696 |
| Genecept | ||||||
| Perlis et al, 202061 | 6 wk: 146/150 4 wk: 146/150 2 wk: 146/150 |
13.93 (7.04) 15.43 (6.67) 17.39 (5.95) |
14.02 (7.17) 15.66 (6.42) 17.77 (5.77) |
38.05c 31.74c 22.76c |
35.34c 28.50c 19.60c |
.444c .306c .246c |
Abbreviations: MD, mean difference; NE, not estimated; NR, not reported; NS, not significant; PGx, pharmacogenomic-guided treatment selection; SD, standard deviation; TAU, treatment as usual.
P values reflect differences in percent decrease from baseline to follow-up unless otherwise noted.
Based on full analysis set (intention-to-treat analysis).
Values for mixed effects models with repeated measures.
Table A24:
Change in Depression Scores on Alternative Depression Scales (< 8-Week Follow-Up)
| Test | Author, Year | N Participants PGx/TAU | Mean at Follow-up (SD) or Mean Change (Δ) from Baseline to Follow-up (SD) | MD (95% CI)a | P for % Change or MDb | |
|---|---|---|---|---|---|---|
| PGx | TAU | |||||
| QIDS-C16 | ||||||
| Genesight | Hall-Flavin et al, 201355 | 4 wk: 86/98 2 wk: 97/105 |
NE NE |
NE NE |
NR NR |
%Δ: 0.0002 %Δ: NS |
| Hall-Flavin et al, 201256 | 4 wk: 22/22 2 wk: 22/22 |
NE NE |
NE NE |
NR NR |
%Δ: NS %Δ: NS |
|
| Genecept | Perlis et al, 202061 | 6 wk: 146/150 4 wk: 146/150 2 wk: 146/150 |
Δ –5.12 (5.17) Δ –4.48 4.63) Δ –3.27 (4.45) |
Δ –5.35 (5.36) Δ –4.03 (4.54) Δ –2.64 (3.91) |
0.41 (−;0.69, 1.50)c –0.17 (−;1.14, 0.81)c –0.56 (−;1.48, 0.37)c |
MD: 0.465c MD: 0.735c MD: 0.236c |
| 9-Item Patient Health Questionnaire | ||||||
| Genesight | Hall-Flavin, 201355 | 4 wk: 86/98 2 wk: 97/105 |
NE NE |
NE NE |
NR NR |
%Δ: NS %Δ: NS |
| CGI-S | ||||||
| Neuropharmag en | Perez, 201762 | CR 6 wk: 144/143 PR 6 wk: unclear |
Δ –0.67 (0.85) Δ –0.77 (1.09) |
Δ –0.53 (0.86) Δ –0.65 (1.16) |
NR NR |
MD: 0.1433 MD: 0.3595 |
| Genecept | Perlis, 202061 | 6 wk: 146/150 4 wk: 146/150 2 wk: 146/150 |
Δ –1.42 (1.18) Δ –1.04 (1.08) Δ –0.61 (0.938) |
Δ –1.33 (1.14) Δ –0.95 (0.975) Δ –0.52 (0.775) |
–0.08 (−;0.32, 0.16)c –0.09 (−;0.30, 0.12)c –0.08 (−;0.26, 0.10)c |
MD: 0.49c MD: 0.40c MD: 0.38c |
Abbreviations: CGI-S, Clinical Global Impressions Scale (severity of illness); CI, confidence interval; CR, clinician-rated; MD, mean difference; NE, data in graph but not estimated; NR, not reported; NS, not significant; PGx, pharmacogenomic-guided treatment; QIDS-C16, 16-Item Quick Inventory of Depressive Symptomatology (clinician-rated); PR, patient-rated; SD, standard deviation; TAU, treatment as usual.
Mean difference at follow-up, unless change from baseline data (Δ) reported by original article. Values only as reported in articles.
Percent changes from baseline estimates were not reported by any study and are therefore not shown. P values are presented as reported in original article.
Mean differences were based on mixed effect model with repeated measures. Change scores reported are not adjusted.
Table A25:
Change in HAM-D17 Scores—Sub-population Analyses
| Author, Year (Primary Study) | Subgroup (N PGx/TAU) | Mean Changea From Baseline to Follow-Up (SD) | P Value | |
|---|---|---|---|---|
| PGx | TAU | |||
| Severity of Depression at Baseline | ||||
| Menchon et al, 201969 (Perez et al62) | HAM-D17 < 18 (61/61)b HAM-D17 ≥ 18 (91/96)b HAM-D17 ≥ 25 (37/23)b |
–4.5 (5.4) 10.5 (8.2) –13.0 (8.8) |
–4.8 (5.7) –7.6 (7.8) –10.2 (8.0) |
.742 .020 .250 |
| Prior Depression Medication Use | ||||
| Perez et al, 201762 | 1–3 failed (90/83)b 0 failed (21/23)b 1 failed (27/28)b 2 failed (32/27)b 3 failed (31/28)b 4+ failed (30/33)b |
–8.86 (7.37) –7.67 (8.68) –9.19 (7.28) –8.88 (7.92) –8.55 (7.09) –5.87 (7.86) |
–5.86 (7.4) –7.57 (6.96) –5.79 (6.95) –5.93 (7.69) –5.86 (7.82) –7.24 (6.51) |
.0083 NS NS NS NS NS |
| Time Since Diagnosis | ||||
| Menchon et al, 201969 (Perez et al, 201762) | ≤ 1 y (79/73)b ≤ 5 y (113/111)b >5 y (42/50)b |
–9.5 (8.7) –8.8 (8.0) –6.0 (6.6) |
–6.6 (7.3) –6.2 (7.7) –7.0 (5.8) |
.035 .022 .518 |
| Age | ||||
| Forester et al, 202067 (Greden et al, 201957) | Age ≥ 65 (86/98) | % Δ 26.7 (3.6) | % Δ 18.7 (3.4) | .102 |
| Menchon et al, 201969 (Perez et al, 201762) | Age < 60 (111/122)b Age ≥ 60 (44/39)b |
–8.0 (7.8) –8.0 (7.5) |
–5.6 (7.2) –9.1 (6.4) |
.02 .522 |
Abbreviations: Δ, change from baseline; HAM-D17, 17-item Hamilton Depression Rating Scale; MD, mean difference; NR, not reported; NS, not significant; PR, patient-rated; SD, standard deviation.
Mean difference at follow-up, unless change from baseline data (Δ) reported by original article.
Number of participants at baseline; unclear which values used in follow-up data calculation. and therefore mean differences and confidence intervals were not calculated.
Table A26:
Change in Depression Scores—Sub-population Analyses by Genetic Test Interpretive Report Classification for Baseline Medications
| Author, Year (Primary Publication) | Interpretive Report Bin Classification at Baselined (N PGx/TAU) | Measure | % Decrease | P Value | |
|---|---|---|---|---|---|
| PGx | TAU | ||||
| GeneSight | |||||
| Thase et al, 201968 (Greden et al, 201957) | Reda or Yellowb (357/430)d Reda or Yellowb + switched medication (235/225)d |
HAM-D17 | 27.1 30.0 |
22.1 22.3 |
.029 .11 |
| Dunlop et al, 201966 (Greden et al, 201957) | Yellowb or Reda (357/429) | HAM-D6 | 28.6 | 21.3 | .004 |
| Winner et al, 201365 | Greenc (6/6) Yellowb (11/8) Reda (7/6) Yellowb or Reda (18/14) |
HAM-D17 | NE NE 33.1 NR |
NE NE 0.8 NR |
.79 .23 .06 .03 |
| Hall-Flavin et al, 201355 | Greenc,d (17/21) Yellowb,d (31/42) Reda,d (16/18) |
HAM-D17 | NE NE 42.5 |
NE NE 16.6 |
.05 .12 .01 |
| Greenc,d (17/21) Yellowb,d (31/42) Reda,d (16/18) |
QIDS-C16 | NE NE 41.9 |
NE NE 11 |
.09 .02 .004 |
|
Abbreviations: Δ, change from baseline; HAM-D17, 17-item Hamilton Depression Rating Scale; NE, not estimated from graph; NS, not significant; PGx, pharmacogenomic-guided treatment; QIDS-C16, 16-item Quick Inventory Depression Scale, clinician-rated; TAU, treatment as usual.
Red – Use with caution and more frequent monitoring.
Yellow – Use with caution
Green – Use as directed.
Patients who were taking more than one medication at baseline were classified based on their most severe classification. Analyses excluded patients who were taking medications that were not included on test report.
Table A27:
Risk Difference in Response for Pharmacogenomic-Guided Medication Selection Compared With Treatment as Usual Based on HAM-D17
| Author, Year | Follow-Up (wk) | (N PGx/TAU) | % Response | Risk Difference (95% CI) | P Valueb | |
|---|---|---|---|---|---|---|
| PGx | TAU | |||||
| GeneSight | ||||||
| Greden et al, 201957 | 8 | Alla: 621/678 PPa: 560/607 |
26.1 26 |
19.8 19.9 |
0.06 (0.02, 0.11)b 0.06 (0.01, 0.11)b |
.007b .01b |
| Winner et al, 201365 | 10 | 25/24 | 36 | 20.8 | 0.15 (−;0.10, 0.40)b | .23b |
| Hall-Flavin et al, 201355 | 8 | 72/93 | 43.1 | 26.9 | 0.16 (0.02, 0.31)b | .03b |
| Neuropharmagen | ||||||
| Han et al, 201860 Perez et al, 201762 |
8 12 |
NR 141/139 |
64.7 45.4 |
39.6 40.3 |
NR 0.05 (−;0.06, 0.17)b |
.014c .39b |
| Genecept | ||||||
| Perlis et al, 202061 | 8 | 146/150 | 39.7 | 48 | –0.08 (−;0.20, 0.03)b | .17 |
| NeuroIDgenetix | ||||||
| Bradley et al, 201858 | 12 8 |
140/121d 140/121d |
64 49 |
46 41 |
0.17 (0.05, 0.29)b,d 0.08 (−;0.4, 0.20)b,d |
.0045b .20b |
| Unspecified Test | ||||||
| Shan et al, 201963 | 8 | ITT: 31/40 PP: 21/27 |
74.2 90.5 |
57.5 70.4 |
0.17 (−;0.05, 0.38)b 0.20 (−;0.01, 0.41)b |
.144 .152 |
Abbreviations: CI, confidence interval; HAM-D17, 17-item Hamilton Depression Rating Scale; ITT, intent to treat; NR, not reported; PGx, pharmacogenomic-guided treatment; PP, per protocol; RR, relative risk, TAU, treatment as usual.
Full cohort included all patients who met eligibility criteria. Per-protocol cohort excluded patients with score of < 14 on HAM-D17 at baseline and patients with protocol violations or whose clinicians did not view pharmacogenomic report before baseline. Only patients who completed 8-week follow-up were included in both analyses.
Calculated from data provided in study. Estimates might vary from publication owing to variation in statistical analyses used or rounding differences.
P value is provided for difference in proportions and might not reflect risk difference.
Only patients with moderate and severe depression were included in analysis (excluded mild depression).
Table A28:
Response Rates for Pharmacogenomic-Guided Medication Selection Compared With Treatment as Usual—Post-Hoc Stratifications and Subgroup Analyses by Baseline Characteristics
| Author, Year (Primary Study) | Sub-Population | N PGx/TAU | % Responsea | Summary Estimate as Reported | P Value | |
|---|---|---|---|---|---|---|
| PGx | TAU | |||||
| Subgroup: Inadequate Response | ||||||
| Bradley et al, 201858 | Inadequately controlledb | NR | 62 (NR) | 44 (NR) | NR | .01 |
| Perez et al, 201762 | Failed 1–3 medications | 90/83 | PGI-I: 51.8 | PGI-I: 31 | OR 2.39 (95% CI 1.28–4.44) | .0058 |
| Subgroup: Age | ||||||
| Forester et al, 202067 (Greden et al, 201957) | Age ≥ 65 y | 86/98 | 29.6 | 16.1 | NR | .032 |
| Menchon et al, 201969 (Perez et al, 201762) | Age < 60 y | 111/122c | HAM-D: 45.2 PGI-I: 46 |
HAM-D: 28.6 PGI-I: 29.6 |
NR | .013 .015 |
| Age ≥ 60 y | 44/39c | HAM-D: 35.1 PGI-I: 52.8 |
HAM-D: 55.9 PGI-I: 55.6 |
NR | .079 .813 |
|
| Subgroup: Depression Severity | ||||||
| Perez et al, 201762 | HAM-D17 ≥ 19d | 79/71 | 49.4 | 31 | OR 2.17 (95% CI 1.11–4.24) | .02 |
| Menchon et al, 201969 (Perez et al, 201762) | HAM-D17 <18 | 61/61c | HAM-D: 34.5 PGI-I: 46.3 |
HAM-D: 45.6 PGI-I: 41.8 |
NR NR |
.223 .638 |
| HAM-D17 ≥ 18 | 91/96c | HAM-D: 48.2 PGI-I: 48.8 |
HAM-D: 28 PGI-I: 32.6 |
NR NR |
.008 .031 |
|
| HAM-D17 ≥ 25 | 37/23c | HAM-D: 51.4 PGI-I: 43.2 |
HAM-D: 28.6 PGI-I: 40 |
NR NR |
.094 .800 |
|
| Bradley et al, 201858 | 12 wk: HAM-D17 ≥ 24 | 40/53 | 73 | 36 | OR 4.72 (95% CI 1.93, 11.52) | .001 |
| 8 wk: HAM-D17 ≥ 24 | 40/53 | 55 | 28 | NR | NR | |
| Mild depression | NR | NR | NR | NR | NS | |
| Subgroup: Time Since Diagnosis | ||||||
| Menchon et al, 201969 (Perez et al, 201762) | ≤ 1 y | 79/73c | HAM-D: 52.1 PGI-I: 54.3 |
HAM-D: 39.7 PGI-I: 36.9 |
NR NR |
.149 .043 |
| ≤ 5 y | 113/111c | HAM-D: 49 PGI-I: 48.5 |
HAM-D: 34 PGI-I: 32 |
NR NR |
.034 .019 |
|
| 5 y | 42/50c | HAM-D: 25.6 PGI-I: 46.2 |
HAM-D: 37.8 PGI-I: 44.7 |
NR NR |
.207 .891 |
|
| Subgroup: Baseline Test Results Based on Medication Classification | ||||||
| Thase et al, 201968 (Greden et al, 201957) | Yellow/red bin at baselined | 357/430 | 27.0 | 19 | NR | .008 |
| Yellow/red bin at baselined and switchedf | 235/225 | 29.8 | 19.4 | NR | .011 | |
| Dunlop et al, 202066 (Greden et al, 201957) | Yellow/red bin at baselined | 357/429 | HAM-D6: 29.5 | HAM-D6: 19.5 | NR | .001 |
| Perlis et al, 202061 | Concordant vs. discordant with assay recommendation | NR | NR | NR | NR | NS |
Abbreviations: CI, confidence interval; HAM-D17, 17-item Hamilton Depression Rating Scale; NR, not reported; NS, not significant; OR, odds ratio, PGI-I, Patient Global Impression of Improvement; PGx, pharmacogenomic-guided treatment; PP, per protocol; TAU, treatment as usual.
Results are based on definition of 50% reduction in HAM-D17, unless otherwise specified.
Unclear to which risk population this applies.
Baseline values and follow-up numbers were not reported.
Medications were categorized as green bin (use as directed), yellow bin (use with caution), or red bin (use with increased caution and more frequent monitoring).
Provided in post-hoc analysis for comparison purposes.
“Switched” was defined as stopping 1 medication and adding 1 medication.
Table A29:
Risk Difference in Remission for Pharmacogenomic-Guided Medication Selection Compared With Treatment as Usual Based on HAM-D17
| Author, Year | FU wk | N PGX/TAU | % Remission | Risk Difference (95% CI) | P Valueb | |
|---|---|---|---|---|---|---|
| PGx | TAU | |||||
| GeneSight | ||||||
| Greden et al, 201957 Winner et al, 201365 Hall-Flavin et al, 201355 |
8 10 8 |
PP: 560/607 All: 621/104 25/24 72/93 |
15.3 16.8 20 30.6 |
10.1 11.4 8.3 21.5 |
0.05 (0.01 to 0.09)a 0.05 (0.02 to 0.09)a 0.12 (−;0.08 to 0.31)a 0.09 (−;0.04 to 0.23)a |
.007 .005 .23a .19 |
| Neuropharmagen | ||||||
| Han et al, 201860 Perez et al, 201762 |
8 12 |
NR 141a/139a |
39.2 34 |
25.0 33.1 |
NE 0.01 (−;0.10 to 0.12)a |
.147 .866 |
| Genecept | ||||||
| Perlis et al, 202061 | 8 | 146/150 | 24 | 30.7 | –0.07 (−;0.17 to 0.03)a | .23 |
| NeuroIDgenetix | ||||||
| Bradley et al, 201858 | 12 | HAM-D17 ≥ 24e: 40/53 Mild depression |
35 NR | 13 NR |
0.22 (0.04 to 0.39)a NR |
.01a NS |
| 8 | HAM-D17 ≥ 24e: 40/53 | 25 | 9 | 0.16 (0.00 to 0.31)a | .05a | |
| CNSDose | ||||||
| Singh et al, 201564 | 12 | 74/74 | 72 | 28 | 0.43 (0.29 to 0.58)a | <.0001 |
| Other | ||||||
| Shan et al, 201963 | 8 | ITT: 31/40 PP: 21/27 |
61.3 76.2 |
45 51.8 |
0.16 (−;0.07 to 0.39)a 0.24 (−;0.02 to 0.51)a |
.173 .133 |
Abbreviations: CI, confidence interval; FU, follow-up; HAM-D, Hamilton Depression Rating Scale; ITT, intention to treat; NE, not estimable; NR, not reported; NS, not significant; PGx, pharmacogenomic-guided medication selection; PP, per protocol; TAU, treatment as usual.
Calculated based on data provided in article. Estimates might vary from those in published studies owing to differences in analysis, adjustments, or rounding errors.
As reported in original study unless otherwise noted. No major differences were observed in P values with unadjusted analyses performed in current review.
Table A30:
Remission Rates for Pharmacogenomic-Guided Medication Selection Compared With Treatment as Usual—Post-Hoc Stratifications and Subgroup Analyses by Baseline Characteristics
| Author, Year (Primary Study) | Sub-population | N PGx/TAU | % Remissiona | Summary Estimate (95% CI) as Reported | P Value | |
|---|---|---|---|---|---|---|
| PGx | TAU | |||||
| Subgroup: Age | ||||||
| Forester et al, 202067 (Greden et al, 201957) | Age ≥ 65 y | 86/98 | 20.1 | 7.4 | NR | .014 |
| Subgroup: Depression Severity | ||||||
| Perez et al, 201762 | HAM-D17 ≥ 19b | 79/71 | 27.8 | 19.7 | OR 1.57 (0.73–3.37) | .244 |
| Subgroup: Inadequate Response to Medication or Treatment Resistance | ||||||
| Bradley et al, 201858 | Inadequately controlledc | NR | 42 | 27 | NR | .03 |
| Subgroup: Medication Congruency at Baseline | ||||||
| Thase et al, 201968 (Greden et al, 201957) | Yellow/red bind | 357/430 | 18.2 | 10.7 | NR | .003 |
| Yellow/red bind and switchede | 235/225 | 20.3 | 11.1 | NR | .008 | |
| Dunlop et al, 201966 (Greden et al, 201957) | Yellow/red bind at baseline (HAM-D6) | 357/429 | 22.2 | 14.3 | NR | .005 |
Abbreviations: CI, confidence interval; HAM-D, 6-item Hamilton Depression Rating Scale; HAM-D17, 17-item Hamilton Depression Rating Scale; NR, not reported; OR, odds ratio, PGx, pharmacogenomic-guided treatment; PP, per protocol; TAU, treatment as usual.
Results were based on HAM-D17 unless otherwise specified.
This post-hoc analysis was for comparison purposes only.
Inadequate control was not defined by article. Result was reported only in discussion post-hoc, which did not specify which cohort was used (moderate or severe + moderate depression).
Medications were categorized as green bin (use as directed), yellow bin (use with caution), or red bin (use with increased caution and more frequent monitoring).
Switched was defined as stopping one medication and adding one medication.
Appendix 9: Examples of Excluded Studies—Economic Evidence
For transparency, we provide a list of some studies that readers might have expected to see in the economic evidence review but that did not meet the inclusion criteria, along with the primary reason for exclusion.
| Citation | Primary Reason for Exclusion |
|---|---|
| Fabbri C, Kasper S, Zohar J, Souery D, Montgomery S, Albani D, et al. Cost-effectiveness of genetic and clinical predictors for choosing combined psychotherapy and pharmacotherapy in major depression. Journal of Affective Disorders 2021;279:722–9. | Intervention: does not match criteria of a PGx test that includes a decision-support tool |
| Jablonski MR, Lorenz R, Li J, Dechairo BM. Economic outcomes following combinatorial pharmacogenomic testing for elderly psychiatric outpatients. Journal of Geriatric Psychiatry and Neurology, 2019;33(6):324-32. | Study type: costing analysis, ICER not estimated Population: wider spectrum, all psychiatric patients |
| Sluiter RL, Janzing JGE, van der Wilt GJ, Kievit W, Teichert M. An economic model of the cost-utility of pre-emptive genetic testing to support pharmacotherapy in patients with major depression in primary care. Pharmacogenomics 2019;19(5):480-9. | Intervention: single-gene pharmacogenomic testing |
| Tanner JA, Brown LC, Yu K, Li J, Dechairo BM. Canadian medication cost savings associated with combinatorial pharmacogenomic guidance for psychiatric medications. Clinicoeconomics & Outcomes Research 2019;11:779-87. | Study type: costing analysis, ICER not estimated Population: wider spectrum, patients with bipolar disorder included |
| Gidding LG, Spigt M, Winkens B, Herijgers O, Dinant GJ. PsyScan e-tool to support diagnosis and management of psychological problems in general practice: a randomised controlled trial. British Journal of General Practice 2018;68(666):e18-e27. | Intervention Population |
| Brown LC, Lorenz RA, Li J, Dechairo BM. Economic utility: combinatorial pharmacogenomics and medication cost savings for mental health care in a primary care setting. Clinical Therapeutics 2017;39(3):592-602. | Study type: costing analysis, ICER not estimated Population: wider spectrum, all psychiatric patients |
| Serretti A, Olgiati P, Bajo E, Bigelli M, De Ronchi D. A model to incorporate genetic testing (5-HTTLPR) in pharmacological treatment of major depressive disorders. World J Biol Psychiatry 2011;12(7):501-15. | Intervention: single-gene pharmacogenomic testing |
Abbreviations: ICER, incremental cost-effectiveness ratio; PGx, multi-gene pharmacogenomic testing.
Appendix 10: Results of Applicability and Limitation Checklists for Studies Included in Economic Literature Review
Table A31:
Applicability of Studies Evaluating Cost-Effectiveness of Multi-gene Pharmacogenomic-Guided Treatment Versus Treatment as Usual in People With Major Depression
| Author, Year, Country of Publication | Is the study population similar to the question?a | Are the interventions similar to the question?a | Is the health care system studied sufficiently similar to Ontario?a | Were the perspectives clearly stated? If yes, what were they?a | Are all direct effects included? Are all other effects included where they are material?a | Are all future costs and outcomes discounted? If yes, at what rate?a | Is the value of health effects expressed in terms of quality-adjusted life-years?a | Are costs and outcomes from other sectors fully and appropriately measured and valued?a | Overall Judgmentb |
|---|---|---|---|---|---|---|---|---|---|
| Tanner et al, 2020, Canada78 | Partially | Partially | Yes | Unclear | Unclear | Yes, 3% | Yes | Partially | Partially applicable |
| Groessl et al, 2018, United States79 | Partially | Partially | No | Yes, societal | No | Yes, 3% | Yes | Partially | Partially applicable |
| Najafzadeh et al, 2017, United States81 | Partially | Partially | No | Yes, societal | Yes | Yes, 3% | Yes | Partially | Partially applicable |
| Hornberger et al, 2015, United States80 | Partially | Partially | No | Partially | No | Yes, 3% | Yes | Partially | Partially applicable |
Response options were “yes,” “partially,” “no,” “unclear,” and “NA” (not applicable).
Response options for overall judgment were “directly applicable,” “partially applicable,” or “not applicable.”
Table A32:
Limitations of Studies Evaluating Cost-Effectiveness of Multi-gene Pharmacogenomic-Guided Treatment Versus Treatment as Usual in People With Major Depression
| Author, Year, Country of Publication | Does the model structure adequately reflect the nature of the health condition under evaluation?a | Is the time horizon sufficiently long to reflect all important differences in costs and outcomes?a | Are all important and relevant health outcomes included?a | Are the clinical inputsb obtained from the best available sources?a | Do the clinical inputsb match the estimates contained in the clinical sources?a | Are all important and relevant (direct) costs included in the analysis?a | Are the estimates of resource use obtained from the best available sources?a | Are the unit costs of resources obtained from the best available sources?a | Is an appropriate incremental analysis presented, or can it be calculated from the reported data?a | Are all important and uncertain parameters subjected to appropriate sensitivity analysis?a | Is there a potential conflict of interest?a | Overall Judgmentc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tanner et al, 2020, Canada78 | Partially | Yes | Partially | Partially | Unclear | Unclear | Partially | Partially | Yes | No | Yes | Potentially serious limitations |
| Groessl et al, 2018, United States79 | Partially | No | Partially | Partially | Unclear | Yes | Unclear | Unclear | Yes | No | Yes | Potentially serious limitations |
| Najafzadeh et al, 2017, United States81 | Partially | Yes | Yes | Partially | Unclear | Yes | Unclear | Unclear | Yes | Yes | Yes | Potentially serious limitations |
| Hornberger et al, 2015, United States80 | Partially | Yes | No | Partially | Unclear | Yes | Unclear | Unclear | Yes | Partially | Yes | Potentially serious limitations |
Response options for all items were “yes,” “partially,” “no,” “unclear,” and “NA” (not applicable).
Clinical inputs include relative treatment effects, natural history, and utilities.
Response options for overall judgment were “minor limitations,” “potentially serious limitations,” or “very serious limitations.”
Appendix 11: Methods—Reference Case Model Structure, Model Inputs, and Validation
Reference Case Model
Figure A1: Markov Model Schematic—Reference Case.
Note: This figure depicts our 1-year Markov state-transition model for the reference case, which gives a possibility of having a sequence of two medication trials: a first (initial) medication change occurs at baseline; a second medication change is made after relapse. The model includes 7 health states, of which 4 are temporary, represented by a solid rectangle without an arrow. The other 3 health states (remission, no remission, or death) are permanent, represented by a dotted rectangle with an arrow. The temporary health states (initially no remission, initial remission, and relapse after no remission at baseline or after initial remission) indicate that a person remains in those states for a certain period, depicted by blue text on the figure (e.g., 3–6 months); after that period, one must transition toward one of the permanent health states.
a At the relapse state, the second change of medication is modeled; according to treatment pathway (see Figure 5), about 3 months are needed for evaluation of the response to medication, and subsequent transition to more permanent states of remission or no remission.
Estimation of Costs for Economic Model
Table A33:
Costs and Resource Use Inputs in Economic Model, Additional Information
| Variable | Unit Cost, $ Mean (SE)a | Frequency (Number)f,g | Total Costs, $ Mean (SE)a | Distribution (Parameters)b | Reference |
|---|---|---|---|---|---|
| Multi-gene Pharmacogenomic Testing (One-Time Cost) | |||||
| Testing including sample transportation costs | 2,500 (625) | 1 | 2,500 (625) | Gamma (α: 16; λ : 0.0064) | Tanner et al, 202078 |
| Physician costs (2 visits) | 67.75 | 2 | 135.5 | NA (fixed) | OHIP code K005113 |
| Direct Medical Costs c,d | |||||
| Remission, total annual costs (2018 CAD) • Medication (prescription drug) costs, annual (2018 CAD)c ○ Medication costs, monthly (2020 CAD): First 6 mo/Rest of follow-upd • Health care service resource use and hospitalization costs, annual (2018 CAD)c,f ○ Health care service resource use including hospitalization costs, monthly (2020 CAD) d,f |
2,832 (STD: 7,601; SE: 12.36)c,f 527 (STD: 2,101; SE: 3.42)c 122.86 (0.58)/44.93 (0.29)d 1,701 (STD: 6,623; SE: 10.77)c,f – – – 145.02 (0.92)d,f – |
NA 1 1 Hospitalizations: 0.1 (1.9 d in hospital and 0.4 d in ICU) ED admissions: 0.1 Long-term care, d: 4.2 – Hospitalizations: 0.1/12 ED admissions: 0.1/12 |
2,832 (STD: 7,601; SE: 12.36)c,f 527 (STD: 2,101; SE: 3.42)c 122.86 (0.58)/44.93 (0.29)d 1,701 (STD: 6,623; SE: 10.77)c – – – 1645.02 (0.92)d – |
NA NA Gamma (α: 44,984.200; λ : 366.156)/Gamma (α: 23,793.824; λ : 529.571) NA – – – Gamma (α: 24,945.616; λ : 172.013) – |
Tanner et al, 201987; Tanner et al, 202078 – – – – – – – – |
| • Physician costs, annual (2018 CAD)c,f ○ Physician costs, monthly (2020 CAD)d,f |
– – 605 (STD: 737; SE: 1.20)c,f 51.58 (0.10)d,f |
Long-term care, d: 4.2/12 – Physician visits: 8.5 (family doctor visits: 5.0; specialist visits: 3.5; psychotherapy sessions: 0.1) Physician visits: 8.5/12 |
– – 605 (STD: 737; SE: 1.20)c 51.58 (0.10) d |
– – NA Gamma (α: 254,841.929; λ : 4,940.672) |
– – – – |
| No remission (or relapse), total annual costs (2018 CAD) • Medication (prescription drug) costs, annual (2018 CAD)c,g ○ Medication costs, monthly (2020 CAD) • Health care service resource use and hospitalization costs, monthly (2018 CAD)c,g ○ Health care service resource use and hospitalization costs, monthly (2020 CAD)d,g |
10,064 (STD: 41,113; SE: 94.30)c,g 1,441 (STD: 2,962; SE: 6.79)c 122.86 (0.58)d 7,192 (STD: 38,761; SE: 88.91)c,g – – 613.17 (7.58)d,g – |
1 1 1 Hospitalizations: 0.5 (8.3 d in hospital; 0.7 d in ICU) ED admissions: 0.4 Long-term care, d: 16 Hospitalizations: 0.5/12 ED admissions: 0.4/12 |
10,064 (STD: 41,113; SE: 94.30)c,g 1,441 (STD: 2,962; SE: 6.79)c 122.86 (0.58)d 7,192 (STD: 38,761; SE: 88.91)c – – 613.17 (7.58)d – |
NA NA Gamma (α: 44,984.200; λ : 366.156) NA – – Gamma (α: 6,543.522; λ : 10.672) – |
Tanner et al, 201987; Tanner et al, 202078 – – – – – – – |
| • Physician costs, annual ○ Physician costs, monthly (2020 CAD) |
— 1,431 (STD: 3,282; SE: 7.53)c,g 122.00 (0.64)d,g |
Long-term care, d: 16/12 Physician visits: 18.6 (family doctor visits: 11.0; specialist visits: 7.6; psychotherapy sessions: 1.7) Physician visits: 18.6/12 |
— 1,431 (STD: 3,282; SE: 7.53)c 122.00 (0.64)d,g |
— — Gamma (α: 36,133.020; λ : 296.166) |
— — — |
| Welle • Medication (prescription drug) costs, annual (2018 CAD)c ○ Medication costs, per month (2020 CAD) • Physician costs ○ Physician costs, monthly (2020 CAD) |
— 527 (STD: 2,101; SE: 3.42)c 44.93 (0.29) d — 47.70 |
— 1 1 — 1 |
— 527 (STD: 2,101; SE: 3.42)c 44.93 (0.29)d — 47.70 |
— NA Gamma (α: 23,793.824; λ : 529.571) – NA (fixed) |
— Tanner et al, 201987 — — OHIP code K033113 |
Abbreviations: ED, emergency department; ICU, intensive care unit; NA, not applicable; OHIP, Ontario Health Insurance Plan; SE, standard error; STD, standard deviation.
Estimates of SEs were calculated from observed published data whenever possible; otherwise, SEs are assumed to be 25% of mean cost (e.g., cost of testing, SE = $625).
For inputs with calculated SEs, we assigned gamma distributions in probabilistic analysis. Two parameters of gamma distribution (α, λ) are derived from the mean and SE. Formulas for these calculations are: α = (Mean2)/(SE2); λ = Mean/([Mean × SE]2).
Cost estimates are presented in the table as reported in the original paper (2018 CAD)87; SEs were calculated from reported standard deviations and sample sizes (SE = STD/√) where n for the cohort of patients with depression was 190,065 and n for the cohort of patients without depression was 378,177).87
To estimate the cost for the 1-month model cycle, we first inflated the estimates from 2018 CAD to 2020 CAD using the Canadian Consumer Price Index114: (137.4 [2020]/134.3 [2018]): for example, in no remission, the annual cost of prescription drug was $1,441 in 2018 CAD and was converted to $1,474 in 2020 CAD. Next, the inflation-adjusted annual cost was transformed into the monthly estimate: $1,474/12 = $123.
Well health state was included in a scenario analysis only.
Mean health care services utilization yearly (for a person without depression) was 8.5 (STD: 8.8) physician visits; 5.0 (STD: 5.2) family doctor visits; 3.5 (STD: 5.9) visits with a specialist; 0.1 (STD: 0.5) sessions of psychotherapy; 0.1 (STD: 0.3) hospitalizations; 1.9 (STD: 8.3) days in hospital; 0.4 (STD: 3.5) days in ICU; 0.1 (STD: 0.4) ED admissions; and 4.2 (STD: 29.5) days receiving long-term care (original article,87 Table 4).
Mean health care services utilization yearly (for a person with depression) was 18.6 (STD: 27.8) physician visits; 11.0 (STD: 15.0) family doctor visits; 7.6 (STD: 19.4) visits with a specialist; 1.7 (STD: 4.7) sessions of psychotherapy; 0.5 (STD: 4.1) hospitalizations; 8.3 (STD: 40.5) days in hospital; 0.7 (STD: 0.5) days in ICU; 0.4 (STD: 2.6) ED admissions; and 16.0 (STD: 61.2) days receiving long-term care (original article, 87 Table 4).
External Validation, Reference Case Model
Figure A2: Probability of Remission in the PGx Arm, Model Estimates vs. Observed Data.
Abbreviation: PGx, multi-gene pharmacogenomic-guided treatment.
Note: Observed data in PGx arms are available for 8- and 24-week visits.
Sources: Forester et al, 202067; Greden et al, 201957; Thase et al, 2019.68
Figure A3: Probability of Remission in the TAU Arm, Model Estimates vs. Observed Data.
Abbreviation: TAU, treatment as usual.
Note: Observed data for TAU arms are available for 8-week visit only.
Sources: Forester et al, 202067; Greden et al, 201957; Thase et al, 2019.68
Appendix 12: Methods—Sensitivity and Scenario Analyses
Table A34:
Test-Specific Sensitivity Analyses (PAs)
| Test-Specific PAs: Parameter Uncertainty | Parameters: Reference Case Analysis | Source | Parameters: Sensitivity Analysis | Source | ||
|---|---|---|---|---|---|---|
| Genecept Assay | ||||||
| Parameters | Mean (SE/95% CI) a | Distribution a,b | Reference | Mean (SE/95% CI) a | Distribution a,b | Reference |
| Risk ratio for remission (intervention vs. TAU) | 1.47 (1.12; 1.94) | Lognormal | Greden et al, 201957 (GeneSight) | 0.78 (0.54; 1.14) | Lognormal | Perlis, 202061 on Genecept Assay |
| Probability of remission with TAU | 0.114 (0.012) | Beta | Greden et al, 201957 | 0.31 (0.04) | Beta | Perlis, 202061 |
| Relative risk of relapse (intervention vs. TAU) | 0.39 (0.04) | Lognormal | Tanner et al, 202078 | 1 | NA | Assumption owing to lack of data |
| Probability of relapse with TAU | 0.233 (0.14) | Beta | Sim et al, 2015106 | 0.233 (0.14) | Beta | Sim et al, 2015106 |
| Probability of side effects: • With intervention • With TAU |
0.156 (0.015) 0.153 (0.015) |
Beta | Greden et al, 201957 | 0.156 (0.015) 0.153 (0.015) |
Beta | Greden et al, 201957 |
| Cost of interventionc | $2,500 (625) | Gamma | Tanner et al, 202078 | $495 ($123.7) | Gamma | Maruf, 202021 |
| Neuropharmagen | ||||||
| Parameters | Mean (SE/95% CI) a | Distribution a,b | Reference | Mean (SE/95% CI) a | Distribution a,b | Reference |
| Risk ratio for remission (intervention vs. TAU) | 1.47 (1.12; 1.94) | Lognormal | Greden et al, 201957 (GeneSight) | 1.03 (0.74; 1.43) | Lognormal | Perez, 201762 on Neuropharmagen |
| Probability of remission with TAU | 0.114 (0.012) | Beta | Greden et al, 201957 | 0.33 (SE: 0.04) | Beta | Perez, 201762 |
| Relative risk of relapse (intervention vs. TAU) | 0.39 (0.04) | Lognormal | Tanner et al, 202078 | 1 | NA | Assumption owing to lack of data |
| Probability of relapse with TAU | 0.233 (0.14) | Beta | Sim et al, 2015106 | 0.233 (0.14) | Beta | Sim et al, 2015106 |
| Probability of side effects: • Intervention • TAU |
0.156 (0.015) 0.153 (0.015) |
Beta | Greden et al, 201957 | 0.315 (SE:0.05) 0.486 (SE: 0.06) |
Beta | Perez, 201762 |
| Cost for interventionc | $2,500 (625) | Gamma | Tanner et al, 202078 | $400 USD = $529.49 (SE: 132.37; 2020 CAD) | Gamma | Maruf, 202021 |
| NeuroIDgenetix | ||||||
| Parameters | Mean (SE/95% CI) a | Distribution a,b | Reference | Mean (SE/95% CI) a | Distribution a,b | Reference |
| Risk ratio for remission (intervention vs. TAU) | 1.47 (1.12; 1.94) | Lognormal | Greden et al, 201957 (GeneSight) | 2.65 (1.18; 5.95) | Lognormal | Bradley, 201858 on NeuroIDgenetix |
| Probability of remission with TAU | 0.114 (0.012) | Beta | Greden et al, 201957 | 0.13 (SE: 0.05) | Beta | Bradley, 201858 |
| Relative risk, relapse (intervention vs. TAU) | 0.39 (0.04) | Lognormal | Tanner et al, 202078 | 1 | NA | Assumption owing to lack of data |
| Probability of relapse, TAU | 0.233 (0.14) | Beta | Sim et al, 2015106 | 0.233 (0.14) | Beta | Sim et al, 2015106 |
| Probability of side effects: • Intervention • TAU |
0.156 (0.015) 0.153 (0.015) | Beta | Greden et al, 201957 | 0.156 (0.015) 0.153 (0.015) |
Beta | Assumed to be same as reference case (no statistically significant difference was found between groups, but data were not reported), Greden et al, 201957 |
| Cost of interventionc | $2,500 (625) | Gamma | Tanner et al, 202078 | $2,000 USD = $2,647.44 (SE:661.86; 2020 CAD) | Gamma | Najafzadeh, 201781 |
| CNSDose | ||||||
| Parameters | Mean (SE/95% CI) a | Distribution a,b | Reference | Mean (SE/95% CI) a | Distribution a,b | Reference |
| Risk ratio for remission (intervention vs. TAU) | 1.47 (1.12; 1.94) | Lognormal | Greden et al, 201957 (GeneSight) | 2.52 (1.71; 3.73) | Lognormal | Singh, 201564 on CNSDose |
| Probability of remission with TAU | 0.114 (0.012) | Beta | Greden et al, 201957 | 0.28 (SE: 0.05) | Beta | Singh, 201564 |
| Relative risk of relapse (intervention vs. TAU) | 0.39 (0.04) | Lognormal | Tanner et al, 202078 | 1 | NA | Assumption owing to lack of data |
| Probability of relapse with TAU | 0.233 (0.14) | Beta | Sim et al, 2015106 | 0.233 (0.14) | Beta | Sim et al, 2015106 |
| Probability of side effects: • Intervention • TAU |
0.156 (0.015) 0.153 (0.015) | Beta | Greden et al, 201957 | 0.04 (SE:0.02) 0.15 (SE: 0.04) |
Beta | Singh, 2015 (side effects based on reported intolerability rate)64 |
| Cost of interventionc | $2,500 (625) | Gamma | Tanner et al, 202078 | $299 AUD = $283.83 (SE: 70.96; 2020 CAD) | Gamma | CNSDose website98: $299 AUD = $283.83 (SE: 70.96) (2020 CAD) |
Abbreviations: CI, confidence interval; NA, not applicable; PA, probabilistic analysis; SE, standard error; TAU, treatment as usual.
Standard errors were estimated whenever data were available. The SE associated with the relative risk of relapse was assumed to be 10% of the mean, and SEs associated with price of tests were assumed to be 25% of the mean.
Beta distributions were assigned to probability estimates in probabilistic analysis where applicable. Standard error of the mean (SE) was estimated from 95% CIs or from original data. Two parameters of the beta distribution (α, β) were derived from the mean and SE (stated for each model parameter). Formulas for these calculations, derived from the mean and SE, are: α = ([Mean2] × [1 – Mean])/([SE2] – Mean); β = ([{1 – Mean} × {1 – Mean}] × Mean)/([SE2] – 1). Lognormal distributions were assigned for risk ratio inputs (wherever possible), using two distribution parameters: μ (mean of logs) and σ (SE, standard deviation of logs). Distribution parameters' values were based on original data; further adjustments and transformations to the model cycle of 1 month were performed. We assigned gamma distributions to cost input parameters. Two parameters of the gamma distribution (α, λ) are derived from the mean and SE. Formulas for these calculations are: α = (Mean2)/(SE2); λ = Mean/([Mean × SE]2).
Cost estimates, as reported in the original papers. Cost inputs were transferred to 2020 CAD using appropriate exchange currency converter (using a conversion rate published on August 12, 2020, and available at https://www.xe.com/currencyconverter/convert/).
Table A35:
Other Parameter-Specific Sensitivity Analyses: Remission, Relapse, Disutilities, and Costs
| Sensitivity Analysis: Parameter Uncertainty | Reference Case Analysis | Sensitivity Analysis | ||||
|---|---|---|---|---|---|---|
| Remission After Baseline | ||||||
| Parameter | Mean (95% CI/SE) a | Distribution a,b | Source | Mean (95% CI or SE) a | Distribution a,b | Source |
| Risk Ratio for Remission Based on MA | ||||||
| Risk ratio for remission (intervention vs. TAU) | 1.47 (1.12; 1.94) | Lognormal | Greden et al, 201957 (GeneSight) | 1.50 (1.14; 1.96) | Lognormal | Clinical review, meta-analysis (two GeneSight RCTs57,65) |
| % Change in Risk of Remission (± 25%; ± 50% on Reference Case Log Odds Ratio Distribution), With or Without Assumption on the RR of Relapse: 8 Pas | ||||||
| Risk ratio for remission (intervention vs. TAU) Relative risk, relapse (intervention vs. TAU) |
1.47 (1.12; 1.94) 0.39 (0.04) |
Lognormal (mean: 0.385; SE: 0.140) Lognormal (mean: –0.942; SE: 0.0512) |
Greden et al, 201957 (GeneSight) Tanner et al, 202078 |
1.64 (1.18; 2.26) 0.39 (0.04) |
Lognormal (mean: 0.385 + 0.25 × 0.385; SE: 0.14 + 0.14 × 0.25) Lognormal |
PA1: +25% on the log OR of remission, RR of relapse unchanged (RR = 0.39 [SE: 0.04])78 — |
| Risk ratio for remission (intervention vs. TAU) Relative risk, relapse (intervention vs. TAU) |
1.47 (1.12; 1.94) 0.39 (0.04) |
Lognormal (mean: 0.385; SE: 0.140) Lognormal (mean: –0.942; SE: 0.0512) |
Greden et al, 201957 (GeneSight) Tanner et al, 202078 |
1.64 (1.18; 2.26) 1 |
Lognormal (mean: 0.385 + 0.25 × 0.385; SE: 0.14 + 0.14 × 0.25) NA |
PA2: +25% on log odds ratio, RR of relapse = 1 — |
| Risk ratio for remission (intervention vs. TAU) Relative risk, relapse (intervention vs. TAU) |
1.47 (1.12; 1.94) 0.39 (0.04) |
Lognormal (mean: 0.385; SE: 0.140) Lognormal (mean: –0.942; SE: 0.0512) |
Greden et al, 201957 (GeneSight) Tanner et al, 202078 |
1.34 (1.10; 1.63) 0.39 (0.04) |
Lognormal (mean: 0.385 – 0.25 × 0.385; SE: 0.14 – 0.14 × 0.25) Lognormal |
PA3: –25% on log OR of remission, RR of relapse unchanged (RR = 0.39 [SE: 0.04])78 — |
| Risk ratio for remission (intervention vs. TAU) | 1.47 (1.12; 1.94) | Lognormal (mean: 0.385; SE: 0.140) | Greden et al, 201957 (GeneSight) | 1.34 (1.10; 1.63) | Lognormal (mean: 0.385 – 0.25 × 0.385; SE: 0.14 – 0.14 × 0.25) | PA4: –25% on log OR of remission, RR of relapse = 1 |
| Relative risk, relapse (intervention vs. TAU) | 0.39 (0.04) | Lognormal (mean: –0.942; SE: 0.0512) | Tanner et al, 202078 | 1 | NA | — |
| Risk ratio for remission (intervention vs. TAU) | 1.47 (1.12; 1.94) | Lognormal (mean: 0.385; SE: 0.140) | Greden et al, 201957 (GeneSight) | 1.81 (1.22; 2.66) | Lognormal (mean: 0.385 + 0.50 × 0.385; SE: 0.14 + 0.14 × 0.50) | PA5: +50% on the log OR of remission, RR of relapse unchanged (RR = 0.39 [SE: 0.04])78 |
| Relative risk, relapse (intervention vs. TAU) | 0.39 (0.04) | Lognormal (mean: –0.942; SE: 0.0512) | Tanner et al, 202078 | 0.39 (0.04) | Lognormal | — |
| Risk ratio for remission (intervention vs. TAU) | 1.47 (1.12; 1.94) | Lognormal (mean: 0.385; SE: 0.140) | Greden et al, 201957 (GeneSight) | 1.81 (1.22; 2.66) | Lognormal (mean: 0.385 + 0.50 × 0.385; SE: 0.14 + 0.14 × 0.50) | PA6: +50% on the log OR of remission, RR of relapse unchanged (RR = 0.39 [SE: 0.04])78 |
| Relative risk, relapse (intervention vs. TAU) | 0.39 (0.04) | Lognormal (mean: –0.942; SE: 0.0512) | Tanner et al, 202078 | 1 | NA | — |
| Risk ratio for remission (intervention vs. TAU) | 1.47 (1.12; 1.94) | Lognormal (mean: 0.385; SE: 0.140) | Greden et al, 201957 (GeneSight) | 1.21 (1.07; 1.39) | Lognormal (mean: 0.385 – 0.50 × 0.385; SE: 0.14 – 0.14 × 0.50) | PA7: –50% on log OR of remission, RR of relapse unchanged (RR = 0.39 [SE: 0.04])78 |
| Relative risk, relapse (intervention vs. TAU) | 0.39 (0.04) | Lognormal (mean: –0.942; SE: 0.0512) | Tanner et al, 202078 | 0.39 (0.04) | Lognormal | — |
| Risk ratio for remission (intervention vs. TAU) | 1.47 (1.12; 1.94) | Lognormal (mean: 0.385; SE: 0.140) | Greden et al, 201957 (GeneSight) | 1.21 (1.07; 1.39) | Lognormal (mean: 0.385 – 0.50 × 0.385; SE: 0.14 – 0.14 × 0.50) | PA8: –50% on log OR of remission, RR of relapse unchanged (RR = 0.39 [SE: 0.04])78 |
| Relative risk, relapse (intervention vs. TAU) | 0.39 (0.04) | Lognormal (mean: –0.942; SE: 0.0512) | Tanner et al, 202078 | 1 | NA | — |
| Threshold Analysis | ||||||
| Risk ratio for remission | 1.47 (1.12; 1.94) | Lognormal (mean: 0.385; SE: 0.140) | Greden et al, 201957 (GeneSight) | Threshold analysis | NA | DA1: Threshold analysis on risk ratio for remission, assuming RR of relapse unchanged (RR = 0.39 [SE: 0.04])78 |
| Relative risk, relapse | 0.39 (0.04) | Lognormal (mean: –0.942; SE: 0.0512) | Tanner et al, 202078 | 0.39 (0.04) | NA | — |
| Risk ratio for remission | 1.47 (1.12; 1.94) | Lognormal (mean: 0.385; SE: 0.140) | Greden et al, 201957 (GeneSight) | Threshold analysis | NA | DA2: Threshold analysis on risk ratio for remission, assuming RR of relapse = 1 |
| Relative risk, relapse | 0.39 (0.04) | Lognormal (mean: –0.942; SE: 0.0512) | Tanner et al, 202078 | 1 | NA | — |
| Relapse | ||||||
| Parameter | Mean (SE) a | Distribution a,b | Source | Mean (95% CI or SE) a | Distribution a,b | Source |
| Relative risk, relapse (intervention vs. TAU) | 0.39 (0.04) | Lognormal (mean: –0.942; SE: 0.0512) | Tanner et al, 202078 | 1 | NA | NA, assumption based on lack of comparative long-term clinical data |
| Medication-Related Disutility | ||||||
| Parameter | Mean (SE) a | Distribution a,b | Source | Mean (95% CI or SE) a | Distribution a,b | Source |
| Disutility associated with medication side effects | –0.055 (0.03) | Beta | Mrazek et al, 2013,109,110 Najafzadeh et al, 201781 |
–0.01 (0.03∗) | Beta | Revicki, 1998110 |
| Disutility associated with medication side effects | –0.055 (0.03) | Beta | Mrazek et al, 2013,109,110 Najafzadeh et al, 201781 |
–0.12 (0.03∗) | Beta | Revicki, 1998110 |
| Cost of Testing | ||||||
| Parameter | Mean (SE) a | Distribution a,b | Source | Mean (SE) a | Distribution a,b | Source |
| Cost of PGx: lower range | $2,500 (625)78 | Gamma | Tanner et al, 202078 | $450 (112.5) | Gamma | Assumption on range for this cost was based on Maruf et al, 2020,21 and Tanner et al78 |
| Cost of PGx: upper range | $2,500 (625)78 | Gamma | Tanner et al, 202078 | $3,750 (937.5) | Gamma | Assumption on range for this cost was based on Maruf et al, 2020,21 and Tanner et al78 |
| Cost of PGx: threshold analysis | $2,500 (625)78 | Gamma | Tanner et al, 202078 | NA | NA | Deterministic, one-way threshold analysis |
| No. of Required Visits With a Physician or a Health Care Provider During Testing | ||||||
| Parameter | Mean | Distribution a,b | Source | Mean | Distribution a,b | Source |
| Visits required during testing | 2 | NA | Expert consultation | 0, 1, and 3 | NA | Expert consultation, 3 visits were assumed to accommodate pharmacists as another provider in circle of care or a delay in receiving results of test |
| Costs of Prescription Drugs in Remission (2020 CAD) | ||||||
| Parameter | Mean (SE) a | Distribution a,b | Source | Mean (SE) a | Distribution a,b | Source |
| Medication costs, monthly (2020 CAD): first 6 mo/Rest of follow-up (see Table 6)c | $122.86 (0.58)/44.93 (0.29) | Gamma | Tanner et al, 201987; Tanner et al, 202078 | $122.86/mo | Gamma | Constant higher prescription drug costs over 1-year time horizon; source for cost inputs was Tanner et al, 201987 |
| Medication costs, monthly (2020 CAD): first 6 mo/Rest of follow-up (see Table 6)c | $122.86 (0.58)/$44.93 (0.29) | Gamma | Tanner et al, 201987; Tanner et al, 202078 | $44.93 (0.29) | Gamma | Constant lower prescription drug costs over 1-year time horizon; source for cost inputs was Tanner et al, 201987 |
| Costs Associated With Health Care Service Use and Hospitalization in Remission (2020 CAD) | ||||||
| Parameter | Mean (SE) a | Distribution a,b | Source | Mean (SE) a | Distribution a,b | Source |
| Health care service and hospitalization costs, monthly (see Table 6)c | $145.02 (0.92) | Gamma | Tanner et al, 201987; Tanner et al, 202078 | 0–6 mo: $613.17 (7.58) 7–12 mo: $145.02 (0.92) |
Gamma | Time-dependent decrease in health care spending (see Table 6); source for cost inputs was Tanner et al, 201987 |
| Health care service and hospitalization costs, monthly (see Table 6)c | $145.02 (0.92) | Gamma | Tanner et al, 201987; Tanner et al, 202078 | $290.04 (0.92) | Gamma | 2x higher spending in remission |
Abbreviations: CI, confidence interval; NA, not applicable; PGx, multi-gene pharmacogenomic-guided treatment that includes a decision support tool; SE, standard error; TAU, treatment as usual.
Standard errors were estimated whenever data were available; SEs associated with relative risk of relapse was assumed to be 10% of mean; SEs associated with price of tests were assumed to be 25% of the mean.
Beta distributions were assigned to probability estimates in probabilistic analysis where applicable. Standard error of the mean (SE) was estimated from 95% CIs or from original data. Two parameters of the beta distribution (α, β) were derived from mean and SE (stated for each model parameter). Formulas for these calculations, derived from the mean and SE, are: α = ([Mean2] × [1 – Mean])/([SE2] – Mean); β = ([{1 – Mean} × {1 – Mean}] × Mean)/([SE2] – 1). Lognormal distributions were assigned for risk ratio inputs (wherever possible), using two distribution parameters: μ (mean of logs) and σ (SE, standard deviation of logs). Distribution parameters' values were based on original data; further adjustments and transformations to model cycle of 1 month were performed. We assigned gamma distributions to cost input parameters. Two parameters of the gamma distribution (α, λ) are derived from the mean and SE. Formulas for these calculations are: α = (Mean2)/(SE2); λ = Mean/([Mean × SE] 2).
Cost estimates were adjusted per model cycle of 1 month; see Table 6 for more information.
Table A36:
Probabilistic Analyses, Scenarios
| Scenarios | Reference Case | Scenarios |
|---|---|---|
| Time horizon | Time horizon: 1 y Discounting: 0% |
Time horizons: 6 mo, 2 y, 3 y, and 5 y Discounting: 1.5% for time horizons > 1 y Effectiveness of PGx: Constant over first 2 years and declines to effectiveness of usual treatment from year 3 One relapse modeled over time horizon All parameter inputs were same as in reference case Additional analyses were performed for each time horizon (6 mo, 2 y, 3 y, and 5 y) with the RR of relapse (intervention) = 1 |
| Well health state | Not included | Well state included in these scenarios |
| Analytic perspective: inclusion of non-medical and indirect costsa | MOH perspective: solely direct medical costs | Analysis 1 : inclusion of social services: non-medical direct costs paid by Canadian government87: $1,522 (SD: $4,176) in no remission, and $510 (SD: $2,507) in remission, yearly (2018 CAD) Analysis 2: inclusion of social services costs paid by government (analysis 1)87 plus costs by private payer for disability claims, no remission159: annual short-term disability claimed costs of $6,263 (2011 CAD, N = 79) and $7,832 (2012 CAD, N = 86) and annual long-term disability claimed costs of $13,598 (2011 CAD, N = 80) and $13,927 (2012 CAD, N = 89)b Analysis 3: social perspective, inclusion of social services costs,87 costs of disability,159 and costs related to absenteeism and productivity loss85: $3,219 (SD:$6,587, N = 9,990, 2010 USD) in no remission and $1,191 (SD: $2,391, N = 9,990, 2010 USD) in remission |
Abbreviations: PGx, multi-gene pharmacogenomic testing; RR, risk ratio; SD, standard deviation; CAD, Canadian dollar; N; sample size.
Cost estimates were as reported in original papers. Cost inputs were transferred to 2020 CAD, using the Canadian Consumer Price Index (CPI).
Average per-person claimed costs were calculated (2011 CAD estimates were transformed into 2012 CAD using CPI); SEs were assumed to be 25% of mean.
Appendix 13: Results of Sensitivity and Scenario Analyses
Table A37:
Sensitivity Analyses for PGx Versus TAU
| Sensitivity Analyses | PGx vs. TAU: ICER ($/QALY)a,b; INB > or < 0 ($)a,b; Δ C ($); Δ E (QALY) |
|---|---|
| Reference Case Analysisc | |
| Time horizon: 1 y | ICER: 60,564; INB < 0; Δ C = $1,906; Δ E = 0.031 |
| Test-Specific Analyses c,d | |
| Genecept Assay | ICER: Dominated; INB < 0; Δ C = $1,194; Δ E = –0.024 |
| Neuropharmagen | ICER: 110,859; INB < 0; Δ C = $574; Δ E = 0.005 |
| NeuroIDgenetix | ICER: 9,735; INB > 0; Δ C = $811; Δ E = 0.083 |
| CNSDose | ICER: Dominant; INB > 0; Δ C = –$1,431; Δ E = 0.078 |
| Remission After Baselinec | |
| Meta-analysis of two GeneSight RCTs, RR of remission = 1.50, 95% CI: 1.14; 1.96 | ICER: 57,722; INB < 0; Δ C = $1,878; Δ E = 0.032 |
| +25% on log odds ratio, RR of remission = 1.64, 95% CI: 1.18; 2.26 (vs. 1.47, 95% CI: 1.12; 1.94, reference case); and RR of relapse = 0.39 +25% on log odds ratio, RR of remission = 1.64, 95% CI: 1.18; 2.26; and RR of relapse = 1 |
ICER: 42,290; INB > 0; Δ C = $1,715; Δ E = 0.039 ICER: 56,569; INB < 0; Δ C = $1,852; Δ E = 0.033 |
| –25% on log odds ratio, RR of remission = 1.34, 95% CI: 1.10; 1.63; and RR of relapse = 0.39 –25% on log odds ratio, RR of remission = 1.34, 95% CI: 1.10; 1.63; and RR of relapse = 1 |
ICER: 87,082; INB < 0; Δ C = $2,085; Δ E = 0.024 ICER: 122,580; INB < 0; Δ C = $2,204; Δ E = 0.018 |
| +50% on log odds ratio, RR of remission = 1.81, 95% CI: 1.22; 2.66; and RR of relapse = 0.39 +50% on log odds ratio, RR of remission = 1.81, 95% CI: 1.22; 2.66; and RR of relapse = 1 |
ICER: 31,235; INB > 0; Δ C = $1,510; Δ E = 0.048 ICER: 40,396; INB > 0; Δ C = $1,656; Δ E = 0.041 |
| –50% on log odds ratio, RR of remission= 1.21, 95% CI: 1.07; 1.39; and RR of relapse = 0.39 –50% on log odds ratio, RR of remission = 1.21, 95% CI: 1.07; 1.39; and RR of relapse = 1 |
ICER: 132,487; INB < 0; Δ C = $2,249; Δ E = 0.017 ICER: 206,053; INB < 0; Δ C = $2,360; Δ E = 0.011 |
| Threshold (one-way, deterministic) analysis, RR of relapse = 0.39 Threshold (one-way, deterministic) analysis, RR of relapse = 1 |
RR of remission = 1.507; ICER ≤ $50,000/QALY; INB ≥ 0 RR of remission = 1.691 ICER ≤ $50,000/QALY; INB ≥ 0 |
| Relapsec | |
| RR, relapse = 1 (vs. RR of relapse = 0.39, reference case) | ICER: 81,165; INB < 0; Δ C = $2,035; Δ E = 0.025 |
| Threshold (one-way, deterministic) analysis | No threshold value identified |
| Medication-Related Disutilityc | |
| Lower range: –0.01 (vs. –0.055, reference case) Upper range: –0.12 |
ICER: 60,535; INB < 0; Δ C = $1,906; Δ E = 0.031 ICER: 60,605; INB < 0; Δ C = $1,906; Δ E = 0.031 |
| Cost of Testingc | |
| Lower range: $450 (vs. $2,500, reference case) | ICER: Dominant; INB > 0; Δ C = –$143; Δ E = 0.031 |
| Upper range: $3,750 | ICER: 100,355; INB < 0; Δ C = $3,159; Δ E = 0.031 |
| Threshold (one-way, deterministic) analysis | Cost-effectiveness threshold: Test cost = $2,161.70; ICER ≤ $50,000/QALY; INB ≥ 0 Cost-saving threshold (break-even point): Test cost = $595.20; ICER: Dominant; INB > 0 |
| No. of Visits With a Health Care Provider Required During Testingc | |
| None (vs. 2 visits, reference case) | ICER: 56,259; INB < 0; Δ C = $1,771; Δ E = 0.031 |
| 1 visit | ICER: 58,411; INB < 0; Δ C = $1,839; Δ E = 0.031 |
| 3 visits | ICER: 62,716; INB < 0; Δ C = $1,974; Δ E = 0.031 |
| Costs of Prescription Drugs in Remissionc | |
| Lower cost: $44.90 (vs. time-dependent decrease, reference case) | ICER: 59,372; INB < 0; Δ C = $1,869; Δ E = 0.031 |
| Higher cost: $122 | ICER: 62,452; INB < 0; Δ C = $1,966; Δ E = 0.031 |
| Costs of Health Care Service Utilizationc | |
| Time-dependent: high to low, $613 to $145, after 6 mo (vs. constant $145, reference case) | ICER: 66,282; INB < 0; Δ C = $2,086; Δ E = 0.031 |
| Double costs in reference case: $290 | ICER: 66,296; INB < 0; Δ C = $2,087; Δ E = 0.031 |
Abbreviations: Δ E, incremental effects; Δ C, incremental costs; ICER, incremental cost-effectiveness ratio; INB, incremental net benefit; PGx, multi-gene pharmacogenomic-guided treatment that includes a decision support tool; QALY, quality-adjusted life-year; RCTs, randomised controlled trials; RR, risk ratio; TAU, treatment as usual.
All costs are in 2020 Canadian dollars.
ICER = Δ C ÷ Δ E and INB = Δ E × $50,000/QALY – Δ C; if INB($) > 0, then the strategy is cost-effective at a willingness-to-pay amount of $50,000/QALY gained; otherwise, the strategy (PGx) is not cost-effective. Dominant strategy means that PGx intervention is associated with lower costs and greater QALYs. Negative incremental costs indicate savings. If PGx was dominated, this means that TAU was associated with lower costs and greater effects. Changes in Δ C or Δ E might not be obvious owing to rounding.
Probabilistic analyses included 10,000 simulations.
Costs and effectiveness of specific PGx interventions were paired and RR of relapse was assumed to be 1, given the lack of data.
Table A38:
Scenario Analyses for PGx Versus TAU
| Scenario Analyses | PGx vs. TAU: ICER ($/QALY)a,b; INB > or < 0 ($)a,b; Δ C ($); Δ E (QALY) |
|---|---|
| Reference Case Analysis c | |
| Time horizon: 1 y | ICER: 60,564; INB < 0; Δ C = $1,906; Δ E = 0.031 |
| Time horizon c,d | |
| 6 mo (vs. 1 y in reference case); RR of relapse = 0.39 6 mo; RR of relapse = 1 |
ICER: 185,993; INB < 0; Δ C = $2,392; Δ E = 0.013 ICER: 221,284; INB < 0; Δ C = $2,421; Δ E = 0.011 |
| 2 y; RR of relapse = 0.39 2 y; RR of relapse = 1 |
ICER: 14,373; INB > 0; Δ C = $959; Δ E = 0.067 ICER: 23,800; INB > 0; Δ C = $1,273; Δ E = 0.053 |
| 3 y; RR of relapse = 0.39 3 y; RR of relapse = 1 |
ICER: 244; INB > 0; Δ C = $25; Δ E = 0.102 ICER: 6,375; INB > 0; Δ C = $521; Δ E = 0.082 |
| 5 y; RR of relapse = 0.39 5 y; RR of relapse = 1 |
ICER: Dominant; INB > 0; Δ C = –$1,788; Δ E = 0.171 ICER: Dominant; INB > 0; Δ C = –$937; Δ E = 0.137 |
| Well Health State c | |
| Addition of well state, time horizon = 1 y, RR of relapse = 0.39 | ICER: 59,329; INB < 0; Δ C = 1,898; Δ E = 0.032 |
| Addition of well state, time horizon = 1 y, RR of relapse = 1 | ICER: 79,811; INB < 0; Δ C = 2,029; Δ E = 0.025 |
| Analytic Perspective c | |
| Inclusion of direct non-medical costs to the government (vs. solely direct medical costs in reference case); time horizon = 1 y | ICER: 57,155; INB < 0; Δ C = 1,799; Δ E = 0.031 |
| Inclusion of disability-related costs in addition to direct non-medical costs | ICER: 56,230; INB < 0; Δ C = 1,770; Δ E = 0.031 |
| Societal perspective (all direct and indirect costs) | ICER: 48,424; INB < 0; Δ C = 1,524; Δ E = 0.031 |
Abbreviations: Δ E, incremental effects; Δ C, incremental costs; ICER, incremental cost-effectiveness ratio; INB, incremental net benefit; PGx, multi-gene pharmacogenomic-guided treatment that includes a decision support tool; QALY, quality-adjusted life-year; RCT, randomized controlled trial; RR, risk ratio; TAU, treatment as usual.
All costs are in 2020 Canadian dollars.
ICER = Δ C ÷ Δ E and INB = Δ E × $50,000/QALY – Δ C; if INB ($) > 0, then the strategy is cost-effective at a willingness-to-pay amount of $50,000/QALY gained; otherwise, the strategy (PGx) is not cost-effective. Dominant strategy means that PGx intervention is associated with lower costs and greater QALYs. Negative incremental costs indicate savings. If PGx was dominated, this means that TAU was associated with lower costs and greater effects. Changes in Δ C or Δ E might not be obvious owing to rounding.
Probabilistic analyses included 10,000 simulations.
Costs and effectiveness were not discounted at 1.5% in the reference case and short-term scenarios; but discounting was applied in long-term scenarios assuming time horizon > 1 y.
Appendix 14: Letter of Information
Appendix 15: Interview Guide
Author contributions
This report was developed by a multidisciplinary team from Ontario Health. The primary clinical epidemiologist was Milica Jokic, the secondary clinical epidemiologist was Stacey Vandersluis, the medical librarian was Caroline Higgins, the primary health economist was Olga Gajic-Veljanoski, the secondary health economist was Xuanqian Xie, and the patient and public partnership analyst was Aroma Akhund.
Key Messages
What Is This Health Technology Assessment About?
People with major depression often have persistent feelings of sadness and loss of interest or pleasure in activities they once enjoyed. Symptoms of major depression can harm personal relationships, reduce people's ability to go to school or work, and lead to social isolation.
Most people with major depression are treated with drugs. However, many people do not benefit from, or are unable to tolerate, their prescribed depression drugs. Genetic variation, or differences in people's DNA, can contribute to differences in response to depression medications. Multi-gene pharmacogenomic testing that includes a decision-support tool can help predict which depression medications and dosages are most likely to result in a strong treatment response or have the lowest risk of an adverse event based on a person's genes.
This health technology assessment looked at how safe, effective, and cost-effective multi-gene pharmacogenomic tests that include decision support tools are for people with major depression. It also looked at the budget impact of publicly funding multi-gene pharmacogenomic testing and at the experiences, preferences, and values of people with major depression.
What Did This Health Technology Assessment Find?
Multi-gene pharmacogenomic tests that include decision support tools are a heterogeneous group, and differences between tests need to be considered. Overall, the six multi-gene pharmacogenomic tests we identified showed inconsistent effectiveness. Pharmacogenomic testing resulted in little to no difference in change in scores on the 17-Item Hamilton Depression Rating Scale as compared with treatment as usual, while some tests may improve response to treatment or remission from symptoms. The evidence, however, is uncertain, and therefore our confidence that these observed effects reflect the true effects is low to very low.
There is substantial uncertainty in the cost-effectiveness of multi-gene pharmacogenomic tests that include decision-support tools for the management of major depression. Over the next 5 years, publicly funding multi-gene pharmacogenomic testing in Ontario would result in additional annual costs ranging from about $3.5 million in year 1 to about $16.8 million in year 5, for a total budget impact of about $52 million.
People with major depression and caregivers generally supported multi-gene pharmacogenomic testing because they believed it could provide guidance that fit their values. They hoped such guidance would speed symptom relief, would reduce side effects, and would help inform their medication choices. There were also concerns however about whether pharmacogenomic testing would reduce patient-centred care so that patients' preferences for treatment might not be included in treatment decisions.
Contributor Information
Ontario Health (Quality):
Milica Jokic, Stacey Vandersluis, Caroline Higgins, Olga Gajic-Veljanoski, Xuanqian Xie, and Aroma Akhund
About Us
Ontario Health is an agency of the Government of Ontario. Our mandate is to connect and coordinate our province's health care system in ways that have not been done before to help ensure that Ontarians receive the best possible care. We work to support better health outcomes, patient experiences, provider experiences, and value for money spent.
For more information about Ontario Health, visit ontariohealth.ca.
References
- (1).Ratnasingham SCJ, Rehm J, Manson H, Kurdyak PA. Opening eyes, opening minds: the Ontario burden of mental illness and addictions report. An ICES/PHO Report [Internet]. Toronto: Institute for Clinical Evaluative Sciences and Public Health Ontario; 2012. [Google Scholar]
- (2).Public Health Agency of Canada. What is depression? [Internet]. Ottawa: The Agency; 2016. [cited Nov 2019]. Available from: https://www.canada.ca/en/public-health/services/chronic-diseases/mental-illness/what-depression.html#R3 [Google Scholar]
- (3).Centre for Addiction and Mental Health. Depression [Internet]. Toronto: The Centre; 2019. [cited Nov 2019]. Available from: https://www.camh.ca/en/health-info/mental-illness-and-addiction-index/depression [Google Scholar]
- (4).Brien S, Grenier L, Kapral ME, Kurdyak P, Vigod S. Taking stock: a report on the quality of mental health and addictions services in Ontario. An HQO/ICES report [Internet]. Toronto: Health Quality Ontario and Institute for Clinical Evaluative Sciences; 2015. [Google Scholar]
- (5).Tolentino JC, Schmidt SL. DSM-5 criteria and depression severity: implications for clinical practice. Front Psychiatry. 2018;9:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kennedy SH, Lam RW, McIntyre RS, Tourjman SV, Bhat V, Blier P. Canadian Network for Mood and Anxiety Treatments (CANMAT) 2016 clinical guidelines for the management of adults with major depressive disorder: section 3. Pharmacological treatments. Can J Psychiatry. 2016;61(9):540–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Rotermann M, Sanmartin C, Hennessy D, Arthur M. Prescription medication use by Canadians aged 6 to 79. Ottawa: Statistics Canada; 2014. Contract No.: Catalogue No. 82-003-X. [PubMed] [Google Scholar]
- (8).Rush AJ, Trivedi MH, Wisniewski SR, Nierenberg AA, Stewart JW, Warden D. Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR∗D report. Am J Psychiatry. 2006;163(11):1905–17. [DOI] [PubMed] [Google Scholar]
- (9).Roberson AM, Castro VM, Cagan A, Perlis RH. Antidepressant nonadherence in routine clinical settings determined from discarded blood samples. J Clin Psychiatry. 2016;77(3):359–62. [DOI] [PubMed] [Google Scholar]
- (10).Health Quality Ontario. Major depression care for adults and adolescents. Quality Standard [Internet]. Toronto: Health Quality Ontario; 2016. Available from: https://www.hqontario.ca/portals/0/documents/evidence/quality-standards/qs-depression-clinical-guide-1609-en.pdf [Google Scholar]
- (11).Hicks JK, Bishop JR, Sangkuhl K, Muller DJ, Ji Y, Leckband SG. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98(2):127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Hicks JK, Sangkuhl K, Swen JJ, Ellingrod VL, Muller DJ, Shimoda K. Clinical Pharmacogenetics Implementation Consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102(1):37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Genetics Home Reference. What is pharmacogenomics [Internet]. Bethesda, MD: U.S. National Library of Medicine; 2019. [Available from: https://ghr.nlm.nih.gov/primer/genomicresearch/pharmacogenomics [Google Scholar]
- (14).Kalow W. Pharmacogenetics and pharmacogenomics: origin, status, and the hope for personalized medicine. Pharmacogenomic J. 2006;6:162–5. [DOI] [PubMed] [Google Scholar]
- (15).Licinio J, Wong ML. Pharmacogenomics of antidepressant treatment effects. Dialogues Clin Neurosci. 2011;13(1):63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Tansey KE, Guipponi M, Hu X, Domenici E, Lewis G, Malafosse A. Contribution of common genetic variants to antidepressant response. Biol Psychiatry. 2013;73(7):679–82. [DOI] [PubMed] [Google Scholar]
- (17).Matchar DB, Thakur ME, Grossman I, McCrory DC, Orlando LA, Steffens DC. Testing for cytochrome P450 polymorphisms in adults with non-psychotic depression treated with selective serotonin reuptake inhibitors (SSRIs). Evid Rep Technol Assess (Full Rep). 2007;Jan(146):1–77. [PMC free article] [PubMed]
- (18).Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3(2):229–43. [DOI] [PubMed] [Google Scholar]
- (19).Nassan M, Nicholson WT, Elliott MA, Rohrer Vitek CR, Black JL, Frye MA. Pharmacokinetic pharmacogenetic prescribing guidelines for antidepressants: a template for psychiatric precision medicine. Mayo Clin Proc. 2016;91(7):897–907. [DOI] [PubMed] [Google Scholar]
- (20).Fabbri C, Serretti A. Pharmacogenetics of major depressive disorder: top genes and pathways toward clinical applications. Curr Psychiatry Rep. 2015;17(7):50. [DOI] [PubMed] [Google Scholar]
- (21).Maruf AA, Fan M, Arnold PD, Muller DJ, Aitchison KJ, Bousman CA. Pharmacogenetic testing options relevant to psychiatry in Canada: Options de tests pharmacogénétiques pertinents en psychiatrie au Canada. Can J Psychiatry. 2020;65(8):521–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Bousman CA, Hopwood M. Commercial pharmacogenetic-based decision-support tools in psychiatry. Lancet Psychiatry. 2016;3(6):585–90. [DOI] [PubMed] [Google Scholar]
- (23).Zeier Z, Carpenter LL, Kalin NH, Rodriguez CI, McDonald WM, Widge AS. Clinical implementation of pharmacogenetic decision support tools for antidepressant drug prescribing. Am J Psychiatry. 2018;175(9):873–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Winner JG, Dechairo B. Combinatorial versus individual gene pharmacogenomic testing in mental health: a perspective on context and implications on clinical utility. Yale J Biol Med. 2015;88(4):375–82. [PMC free article] [PubMed] [Google Scholar]
- (25).Fabbri C, Zohar J, Serretti A. Pharmacogenetic tests to guide drug treatment in depression: comparison of the available testing kits and clinical trials. Prog Neuropsychopharmacol Biol Psychiatry. 2018;86:36–44. [DOI] [PubMed] [Google Scholar]
- (26).Joly Y, Ramos-Paque E. Approval of new pharmacogenomic tests: is the Canadian regulatory process adequate? Can J Law Tech. 2010;8(2):215–41. [Google Scholar]
- (27).Evaluation of Genomic Applications in Practice Prevention Working Group. Recommendations from the EGAPP Working Group: testing for cytochrome P450 polymorphisms in adults with nonpsychotic depression treated with selective serotonin reuptake inhibitors. Genet Med. 2007;9(12):819–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).U.S. Food and Drug Administration. The FDA warns against the use of many genetic tests with unapproved claims to predict patient response to specific medications: FDA safety communication [Internet]. 2018. [Cited Nov 2019]. Available from: https://www.fda.gov/medical-devices/safety-communications/fda-warns-against-use-many-genetic-tests-unapproved-claims-predict-patient-response-specific#actions
- (29).Benefits Canada. Pharmacogenetic testing a growing area as pilot projects, research get underway [Internet]. Montreal: Transcontinental Media G.P.; 2018. [cited 2020 Jan]. Available from: https://www.benefitscanada.com/news/pharmacogenetics-testing-offering-opportunities-in-disability-drug-cost-management-110047 [Google Scholar]
- (30).Myriad Neuroscience. The GeneSight® Psychotropic Test [Internet]. 2019. [Cited Dec 2019]. Available from: https://genesight.com/
- (31).Dynacare. Genecept Assay (National) [Internet]. [Cited Dec 2019]. Available from: https://www.dynacare.ca/specialpages/secondarynav/find-a-test/nat/genecept%C2%A0assay.aspx?sr=NAT&st=
- (32).LifeLabs Genetics. What is TreatGxPlus? [Internet]. 2019. [Cited Dec 2019]. Available from: https://www.lifelabsgenetics.com/product/treatgxplus-service/
- (33).Pillcheck. What is pillcheck? [Internet]. 2019. [Cited Dec 2019]. Available from: https://www.pillcheck.ca/
- (34).My DNA Life. myDNA. Don't guess. Ask your DNA. [Internet]. 2019. [cited Dec 2019]. Available from: https://www.mydna.life/en-ca/
- (35).DNA Labs Genetic Testing Solutions. Unlock the potential of your DNA with MatchMyMeds [Internet]. 2019. [cited 2020 Jan]. Available from: https://dnalabs.ca/matchmymeds/
- (36).OneOme. Personalized prescriptions with the RightMed Solution [Internet]. Minneapolis, MN: OneOme; 2019. [cited 2020 Jan]. Available from: https://oneome.com/home-international [Google Scholar]
- (37).Centre for Addiction and Mental Health. IMPACT: Individualized Medicine: Pharmacogenetic Assessment and Clinical Treatment [Internet]. Toronto: The Centre; 2017. [cited Nov 2019]. Available from: http://impact.camhx.ca/en/home.php [Google Scholar]
- (38).Tanner JA, Davies PE, Voudouris NC, Shahmirian A, Herbert D, Braganza N. Combinatorial pharmacogenomics and improved patient outcomes in depression: treatment by primary care physicians or psychiatrists. J Psychiatr Res. 2018;104:157–62. [DOI] [PubMed] [Google Scholar]
- (39).Washington State Health Care Authority. Pharmacogenomic testing for selected conditions. Final evidence report. [Internet]. Olympia, WA: The Authority; 2016. cited Nov 2019]. Available from: www.hca.wa.gov/about-hca/health-technology-assessmentshtap@hca.wa.gov [Google Scholar]
- (40).McGowan J, Sampson M, Salzwedel DM, Cogo E, Foerster V, Lefebvre C. PRESS peer review of electronic search strategies: 2015 guideline statement. J Clin Epidemiol. 2016;75:40–6. [DOI] [PubMed] [Google Scholar]
- (41).Canadian Agency for Drugs and Technologies in Health. Table 18: Validity and minimal clinically important difference of outcome measures. 2016. In: Aripiprazole (Abilify): depression, major depressive disorder (MDD) [Internet]. Ottawa, ON: The Agency. Available from: https://www.ncbi.nlm.nih.gov/books/NBK409740/table/T25/ [PubMed]
- (42).Covidence systematic review software [Internet]. Melbourne (Australia): Veritas Health Innovation; Undated [Available from: https://www.covidence.org/home. [Google Scholar]
- (43).Rohatgi A. WebPlotDigitizer [Computer program]. Version 4.3 [Internet]. Pacifica (CA): Ankit Rohatgi; 2020. [Available from: https://automeris.io/WebPlotDigitizer [Google Scholar]
- (44).Higgins J, Altman D, Sterne JA. Assessing risk of bias in included studies. 2011 March [cited Nov 2019]. In: Cochrane handbook for systematic reviews of interventions [Internet]. Cochrane. Version 5.1.0. [cited Nov 2019]. Available from: www.training.cochrane.org/handbook
- (45).Kim SY, Park JE, Lee YJ, Seo HJ, Sheen SS, Hahn S. Testing a tool for assessing the risk of bias for nonrandomized studies showed moderate reliability and promising validity. J Clin Epidemiol. 2013;66(4):408–14. [DOI] [PubMed] [Google Scholar]
- (46).Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol. 2009;62(10):1006–12. [DOI] [PubMed] [Google Scholar]
- (47).Bousman CA, Arandjelovic K, Mancuso SG, Eyre HA, Dunlop BW. Pharmacogenetic tests and depressive symptom remission: a meta-analysis of randomized controlled trials. Pharmacogenomics. 2019;20(1):37–47. [DOI] [PubMed] [Google Scholar]
- (48).Brown L, Vranjkovic O, Li J, Yu K, Al Habbab T, Johnson H. The clinical utility of combinatorial pharmacogenomic testing for patients with depression: a meta-analysis. Pharmacogenomics. 2020;21(8):559–69. [DOI] [PubMed] [Google Scholar]
- (49).Health Quality Ontario. Pharmacogenomic testing for psychotropic medication selection: a systematic review of the Assurex GeneSight Psychotropic test. Ont Health Technol Assess Ser [Internet]. 2017. Apr;7(4):1–39. Available from: http://www.hqontario.ca/Evidence-to-Improve-Care/Journal-Ontario-HealthTechnology-Assessment-Series. [PMC free article] [PubMed] [Google Scholar]
- (50).Li K, Loshak H. Pharmacogenomic testing in depression: a review of clinical effectiveness, cost-effectiveness, and guidelines [Internet]. Ottawa (ON): Candian Agency for Drugs and Technologies in Health; 2020. [PubMed] [Google Scholar]
- (51).Peterson K, Dieperink E, Ferguson L, Anderson J, Helfand M. Evidence brief: the comparative effectiveness, harms, and cost-effectiveness of pharmacogenomics-guided antidepressant treatment versus usual care for major depressive disorder. Washington (DC): Department of Veterans Affairs (US); 2016. Contract No.: Project #09-199. [PubMed] [Google Scholar]
- (52).Rosenblat JD, Lee Y, McIntyre RS. The effect of pharmacogenomic testing on response and remission rates in the acute treatment of major depressive disorder: a meta-analysis. J Affect Disord. 2018;241:484–91. [DOI] [PubMed] [Google Scholar]
- (53).Rosenblat J, Lee Y, McIntyre R. Does pharmacogenomic testing improve clinical outcomes for major depressive disorder? A systematic review of clinical trials and cost-effectiveness studies. J Clin Psychiatry. 2017;78(6):720–9. [DOI] [PubMed] [Google Scholar]
- (54).Thakur M, Grossman I, McCrory DC, Orlando LA, Steffens DC, Cline KE. Review of evidence for genetic testing for CYP450 polymorphisms in management of patients with nonpsychotic depression with selective serotonin reuptake inhibitors. Genet Med. 2007;9(12):826–35. [DOI] [PubMed] [Google Scholar]
- (55).Hall-Flavin DK, Winner JG, Allen JD, Carhart JM, Proctor B, Snyder KA. Utility of integrated pharmacogenomic testing to support the treatment of major depressive disorder in a psychiatric outpatient setting. Pharmacogenet Genomics. 2013;23(10):535–48. [DOI] [PubMed] [Google Scholar]
- (56).Hall-Flavin DK, Winner JG, Allen JD, Jordan JJ, Nesheim RS, Snyder KA. Using a pharmacogenomic algorithm to guide the treatment of depression. Transl Psychiatry. 2012;16(2):e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Greden JF, Parikh SV, Rothschild AJ, Thase ME, Dunlop BW, DeBattista C. Impact of pharmacogenomics on clinical outcomes in major depressive disorder in the GUIDED trial: a large, patient- and rater-blinded, randomized, controlled study. J Psychiatr Res. 2019;111:59–67. [DOI] [PubMed] [Google Scholar]
- (58).Bradley P, Shiekh M, Mehra V, Vrbicky K, Layle S, Olson MC. Improved efficacy with targeted pharmacogenetic-guided treatment of patients with depression and anxiety: a randomized clinical trial demonstrating clinical utility. J Psychiatr Res. 2018;96:100–7. [DOI] [PubMed] [Google Scholar]
- (59).Han C, Wang SM, Bahk WM, Lee SJ, Patkar AA, Masand PS. Corrigendum: A pharmacogenomic-based antidepresant treatment for patients with major depressive disorder: results from an 8-week, randomized, single-blinded clinical trial. Clin Psychopharmacol Neurosci. 2020;18(4):641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Han C, Wang SM, Bahk WM, Lee SJ, Patkar AA, Masand PS. A pharmacogenomic-based antidepressant treatment for patients with major depressive disorder: results from an 8-week, randomized, single-blinded clinical trial. Clin Psychopharmacol Neurosci. 2018;16(4):469–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Perlis RH, Dowd D, Fava M, Lencz T, Krause DS. Randomized, controlled, participant- and rater-blind trial of pharmacogenomic test-guided treatment versus treatment as usual for major depressive disorder. Depress Anxiety. 2020;37(9):834–41. [DOI] [PubMed] [Google Scholar]
- (62).Perez V, Salavert A, Espadaler J, Tuson M, Saiz-Ruiz J, Saez-Navarro C. Efficacy of prospective pharmacogenetic testing in the treatment of major depressive disorder: results of a randomized, double-blind clinical trial. BMC Psychiatry. 2017;17(1):250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Shan X, Zhao W, Qiu Y, Wu H, Chen J, Fang Y. Preliminary clinical investigation of combinatorial pharmacogenomic testing for the optimized treatment of depression: a randomized single-blind study. Front Neurosci. 2019;13(960):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Singh AB. Improved antidepressant remission in major depression via a pharmacokinetic pathway polygene pharmacogenetic report. Clin Psychopharmacol Neurosci. 2015;13(2):150–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Winner JG, Carhart JM, Altar CA, Allen JD, Dechairo BM. A prospective, randomized, double-blind study assessing the clinical impact of integrated pharmacogenomic testing for major depressive disorder. Discov Med. 2013;16(89):219–27. [PubMed] [Google Scholar]
- (66).Dunlop BW, Parikh SV, Rothschild AJ, Thase ME, DeBattista C, Conway CR. Comparing sensitivity to change using the 6-item versus the 17-item Hamilton depression rating scale in the GUIDED randomized controlled trial. BMC Psychiatry. 2019;19(1):420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Forester BP, Parikh SV, Weisenbach S, Ajilore O, Vahia I, Rothschild AJ. Combinatorial pharmacogenomic testing improves outcomes for older adults with depression. Am J Geriatr Psychiatry. 2020;28(9):933–45. [DOI] [PubMed] [Google Scholar]
- (68).Thase ME, Parikh SV, Rothschild AJ, Dunlop BW, DeBattista C, Conway CR. Impact of pharmacogenomics on clinical outcomes for patients taking medications with gene-drug interactions in a randomized controlled trial. J Clin Psychiatry. 2019;80(6):19m12910. [DOI] [PubMed] [Google Scholar]
- (69).Menchon JM, Espadaler J, Tuson M, Saiz-Ruiz J, Bobes J, Vieta E. Patient characteristics driving clinical utility in psychiatric pharmacogenetics: a reanalysis from the AB-GEN multicentric trial. J Neural Transm (Vienna). 2019;126(1):95–9. [DOI] [PubMed] [Google Scholar]
- (70).Lundh A, Lexchin J, Mintzes B, Schroll JB, Bero L. Industry sponsorship and research outcome: systematic review with meta-analysis. Intensive Care Med. 2018;44(10):1603–12. [DOI] [PubMed] [Google Scholar]
- (71).Wisniewski SR, Rush AJ, Balasubramani GK, Trivedi MH, Nierenberg AA, Investigators S. Self-rated global measure of the frequency, intensity, and burden of side effects. J Psychiatr Pract. 2006;12(2):71–9. [DOI] [PubMed] [Google Scholar]
- (72).Steensma C, Loukine L, Orpana H. Describing the population health burden of depression: health-adjusted life expectancy by depression status in Canada. Health Promot Chronic Dis Prev Can. 2016;36(10):205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Bousman CA, Dunlop BW. Genotype, phenotype, and medication recommendation agreement among commercial pharmacogenetic-based decision support tools. Pharmacogenomics J. 2018;18(5):613–22. [DOI] [PubMed] [Google Scholar]
- (74).PharmGKB. Clinical guideline annotations [Internet]. 2020. [cited 2020 Nov 2020]. Available from: https://www.pharmgkb.org/guidelineAnnotations
- (75).Hwang R, Shinkai T, De Luca V, Ni X, Potkin SG, Lieberman JA. Association study of four dopamine D1 receptor gene polymorphisms and clozapine treatment response. J Psychopharmacol. 2007;21(7):718–27. [DOI] [PubMed] [Google Scholar]
- (76).Owen RP, Sangkuhl K, Klein TE, Altman RB. Very important pharmacogene: CYP2D6 [Internet]. Stanford (CA): PharmGKB; c2001-2021. [cited 2020 Nov]. Available from: https://www.pharmgkb.org/vip/PA166170264 [Google Scholar]
- (77).National Institute for Health and Care Excellence. Appendix I: Quality appraisal checklist—economic evaluations. 2012. [cited 2016 Jan]. In: Methods for the development of NICE public health guidance, 3rd ed [Internet]. London: The Institute. Available from: https://www.nice.org.uk/process/pmg4/chapter/appendix-i-quality-appraisal-checklist-economic-evaluations [PubMed]
- (78).Tanner JA, Davies PE, Overall CC, Grima D, Nam J, Dechairo BM. Cost-effectiveness of combinatorial pharmacogenomic testing for depression from the Canadian public payer perspective. Pharmacogenomics. 2020;21(18):521–31. [DOI] [PubMed] [Google Scholar]
- (79).Groessl EJ, Tally SR, Hillery N, Maciel A, Garces JA. Cost-effectiveness of a pharmacogenetic test to guide treatment for major depressive disorder. Journal of Managed Care and Specialty Pharmacy. 2018;24(8):726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Hornberger J, Li Q, Quinn B. Cost-effectiveness of combinatorial pharmacogenomic testing for treatment-resistant major depressive disorder patients. Am J Managed Care 2015;21(6):e357–e65. [PubMed] [Google Scholar]
- (81).Najafzadeh M, Garces JA, Maciel A. Economic evaluation of implementing a novel pharmacogenomic test to guide treatment of patients with depression and/or anxiety. Pharmacoeconomics. 2017;35(12):1297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Canadian Agency for Drugs and Technologies in Health. Guidelines for the economic evaluation of health technologies: Canada—4th edition, version 1. CADTH ethods and guidelines [Internet]. 2017. [cited 2017 April 4]. Available from: https://www.cadth.ca/guidelines-economic-evaluation-health-technologies-canada-4th-edition
- (83).Canadian Agency for Drugs and Technologies in Health. Guidelines for the economic evaluation of health technologies: Canada. 4th ed. Appendix—Specific guidance for treatments with companion diagnostics. CADTH methods and guidelines [Internet]. 2019. [cited 2020 29 April]. Available from: https://www.cadth.ca/sites/default/files/pdf/cp0008-guidelines-for-economic-evaluation-of-health-technologies.pdf
- (84).Geddes JR, Carney SM, Davies C, Furukawa TA, Kupfer DJ, Frank E. Relapse prevention with antidepressant drug treatment in depressive disorders: a systematic review. Lancet. 2003;361(9358):653–61. [DOI] [PubMed] [Google Scholar]
- (85).Greenberg PE, Fournier AA, Sisitsky T, Pike CT, Kessler RC. The economic burden of adults with major depressive disorder in the United States (2005 and 2010). J Clin Psychiatry. 2015;76(2):155–62. [DOI] [PubMed] [Google Scholar]
- (86).Kim DD, Silver MC, Kunst N, Cohen JT, Ollendorf DA, Neumann PJ. Perspective and costing in cost-effectiveness analysis, 1974-2018. Pharmacoeconomics. 2020;38(10):1135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Tanner JA, Hensel J, Davies PE, Brown LC, Dechairo BM, Mulsant BH. Economic burden of depression and associated resource use in Manitoba, Canada. Can J Psychiatry. 2019;65(5):338–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Rush AJ, Trivedi MH, Wisniewski SR, Stewart JW, Nierenberg AA, Thase ME. Bupropion-SR, sertraline, or venlafaxine-XR after failure of SSRIs for depression. N Engl J Med. 2006;354(12):1231–42. [DOI] [PubMed] [Google Scholar]
- (89).Patten SB, Kennedy SH, Lam RW, O'Donovan C, Filteau MJ, Parikh SV. Canadian Network for Mood and Anxiety Treatments (CANMAT) clinical guidelines for the management of major depressive disorder in adults. I. Classification, burden and principles of management. J Affect Disord. 2009;117(Suppl 1):S5–S14. [DOI] [PubMed] [Google Scholar]
- (90).Lam RW, Kennedy SH, Parikh SV, MacQueen GM, Milev RV, Ravindran AV. Canadian Network for Mood and Anxiety Treatments (CANMAT) 2016 clinical guidelines for the management of adults with major depressive disorder: introduction and methods. Can J Psychiatry. 2016;61(9):506–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Nierenberg AA, DeCecco LM. Definitions of antidepressant treatment response, remission, nonresponse, partial response, and other relevant outcomes: a focus on treatment-resistant depression. J Clin Psychiatry. 2001;62(Suppl 16):5–9. [PubMed] [Google Scholar]
- (92).Rush AJ. STAR∗D: what have we learned? Am J Psychiatry. 2007;164(2):201–4. [DOI] [PubMed] [Google Scholar]
- (93).Jablonski MR, King N, Wang Y, Winner JG, Watterson LR, Gunselman S. Analytical validation of a psychiatric pharmacogenomic test. Per Med. 2018;15(3):189–97. [DOI] [PubMed] [Google Scholar]
- (94).Altar CA, Carhart JM, Allen JD, Hall-Flavin DK, Dechairo BM, Winner JG. Clinical validity: combinatorial pharmacogenomics predicts antidepressant responses and healthcare utilizations better than single gene phenotypes. Pharmacogenomics J. 2015;15(5):443–51. [DOI] [PubMed] [Google Scholar]
- (95).Husereau D, Drummond M, Petrou S, Carswell C, Moher D, Greenberg D. Consolidated Health Economic Evaluation Reporting Standards (CHEERS)—explanation and elaboration: a report of the ISPOR Health Economic Evaluation Publication Guidelines Good Reporting Practices Task Force. Value Health. 2013;16(2):231–50. [DOI] [PubMed] [Google Scholar]
- (96).Canadian Agency for Drugs and Technologies in Health. Guidelines for the economic evaluation of health technologies: Canada. 4th ed. Appendix—Specific guidance for treatments with companion diagnostics. CADTH Methods and Guidelines [Internet]. 2019. [cited 2020 November 19]. Available from: https://www.cadth.ca/sites/default/files/pdf/cp0008-guidelines-for-economic-evaluation-of-health-technologies.pdf
- (97).Liu JCW, Gorbovskaya I, Bousman C, Brown LC, Mueller DJ. Chapter 37. Opportunities and challenges of implementation models of pharmacogenomics in clinical practice. 2020. In: Personalized Psychiatry [Internet]. Cambridge (MA): Academic Press. Available from: 10.1016/B978-0-12-813176-3.00037-7 [DOI] [Google Scholar]
- (98).CNSDose [Internet]. Melbourne, Australia: Amplis (formerly CNSDose); 2020. [cited 2021 February]. Available from: https://cnsdose.com/faq [Google Scholar]
- (99).Maciel A, Cullors A, Alukowiak A, Garces J. Estimating cost savings of pharmacogenetic testing for depression in real-world clinical settings. Neuropsychiatric Disease and Treatment. 2018;14:225–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Olgiati P, Bajo E, Bigelli M, De Ronchi D, Serretti A. Should pharmacogenetics be incorporated in major depression treatment? Economic evaluation in high- and middle-income European countries. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2012;36(1):147–54. [DOI] [PubMed] [Google Scholar]
- (101).Olgiati P, Serretti A, Bigelli M, De Ronchi D, Bajo E. Is pharmacogenetic testing ready for antidepressant treatment? A cost-effectiveness simulation. European Neuropsychopharmacology; September; Paris, France; 2011. p. S389.
- (102).Perlis RH, Patrick A, Smoller JW, Wang PS. When is pharmacogenetic testing for antidepressant response ready for the clinic? A cost-effectiveness analysis based on data from the STAR∗D study. Neuropsychopharmacology. 2009;34(10):2227–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Serretti A, Olgiati P, Bajo E, Bigelli M, De Ronchi D. A model to incorporate genetic testing (5-HTTLPR) in pharmacological treatment of major depressive disorders. World J Biol Psychiatry. 2011;12(7):501–15. [DOI] [PubMed] [Google Scholar]
- (104).Sluiter RL, Janzing JGE, van der Wilt GJ, Kievit W, Teichert M. An economic model of the cost-utility of pre-emptive genetic testing to support pharmacotherapy in patients with major depression in primary care. Pharmacogenom J. 2019;19(5):480–9. [DOI] [PubMed] [Google Scholar]
- (105).Health Quality Ontario. Psychotherapy for major depressive disorder and generalized anxiety disorder: a health technology assessment. Ont Health Technol Assess Ser. 2017;17(15):1–167. [PMC free article] [PubMed] [Google Scholar]
- (106).Sim K, Lau WK, Sim J, Sum MY, Baldessarini RJ. Prevention of relapse and recurrence in adults with major depressive disorder: systematic review and meta-analyses of controlled trials. Int J Neuropsychopharmacol. 2015;19(2):pyv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Statistics Canada. Life tables, Canada, provinces and territories: 2016 to 2018, catalogue no. 84-537-X [Internet]. Ottawa (ON): Statistics Canada; 2020. [cited 2020 May]. Available from: https://www150.statcan.gc.ca/n1/pub/84-537-x/2019002/xls/2016-2018_Tbl-eng.xlsx [Google Scholar]
- (108).Williams N, Simpson AN, Simpson K, Nahas Z. Relapse rates with long-term antidepressant drug therapy: a meta-analysis. Hum Psychopharmacol. 2009;24(5):401–8. [DOI] [PubMed] [Google Scholar]
- (109).Mrazek DA, Hornberger JC, Altar CA, Degtiar I. A review of the clinical, economic, and societal burden of treatment-resistant depression: 1996-2013. Psychiatr Serv. 2014;65(8):977–87. [DOI] [PubMed] [Google Scholar]
- (110).Revicki DA, Wood M. Patient-assigned health state utilities for depression-related outcomes: differences by depression severity and antidepressant medications. J Affect Disord. 1998;48(1):25–36. [DOI] [PubMed] [Google Scholar]
- (111).Lenert LA, Sherbourne CD, Sugar C, Wells KB. Estimation of utilities for the effects of depression from the SF-12. Med Care. 2000;38(7):763–70. [DOI] [PubMed] [Google Scholar]
- (112).Weinstein S, Carroll JC, Jukic S, Mcgivney MS, Klatt P. Perspectives of a pharmacist-run pharmacogenomic service for depression in interdisciplinary family medicine practices. J Am Coll Clin Pharm. 2020;3(2):417–24. [Google Scholar]
- (113).Ontario Ministry of Health. Schedule of benefits: physician services under the Health Insurance Act [Internet]. Toronto (ON): Queen's Printer for Ontario; 2020. [cited 2020 May]. Available from: http://www.health.gov.on.ca/en/pro/programs/ohip/sob/physserv/sob_master20200429.pdf [Google Scholar]
- (114).Statistics Canada. Consumer price index, health and personal care, by province (Canada) [Internet]. Ottawa (ON)2016 [08 May 2020]. Available from: https://www150.statcan.gc.ca/t1/tbl1/en/tv.action?pid=1810000601
- (115).Krahn M, Miller F, Bayoumi A, Brooker AS, Wagner F, Winsor S. Development of the Ontario decision framework: a values based framework for health technology assessment. International journal of technology assessment in health care. 2018;34(3):290–9. [DOI] [PubMed] [Google Scholar]
- (116).TreeAge Pro decision analysis software. TreeAge Software, Inc. Williamstown (MA). Available at: http://www.treeage.com. [Google Scholar]
- (117).Canadian Agency for Drugs and Technologies in Health. Guidance document for the costing of health care resources in the Canadian setting [Internet]. Ottawa (ON): The Agency; 2016. [cited 2017 April 4]. Available from: https://www.cadth.ca/guidance-document-costing-health-care-resources-canadian-setting [Google Scholar]
- (118).Kennedy JL, Tiwari A, Clement Z, Mueller DJ, Davies P, Braganza N., editors. Clinical utility of combinatorial pharmacogenetic testing in depression: Canadian patient- and rater-blinded, randomized, controlled trial. Proceedings of the American College of Neuropsychopharmacology; 2020. Dec 6-9, 2020; Virtual.
- (119).Centers for Medicare and Medicaid Services. CMS releases proposed prices for clin lab fee schedule CY2021–Public comment open [Internet]. Baltimore (MD): The Centers; 2020. [cited 2021 March 9]. Available from: http://www.discoveriesinhealthpolicy.com/2020/09/cms-releases-proposed-prices-for-clin.html [Google Scholar]
- (120).Quinn B. Discoveries in health policy [Internet]. 2021. [cited 2021 9 Mar]. Available from: http://www.discoveriesinhealthpolicy.com/2020/09/cms-releases-proposed-prices-for-clin.html
- (121).Bousman CA, Eyre HA. “Black box” pharmacogenetic decision-support tools in psychiatry. Braz J Psychiatry. 2020;42(2):113–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Krahn MD, Bremner KE, Luo J, Alibhai SM. Health care costs for prostate cancer patients receiving androgen deprivation therapy: treatment and adverse events. Curr Oncol. 2014;21(3):e457–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Krahn MD, Bremner KE, Zagorski B, Alibhai SM, Chen W, Tomlinson G. Health care costs for state transition models in prostate cancer. Med Decis Making. 2014;34(3):366–78. [DOI] [PubMed] [Google Scholar]
- (124).Psychiatric Neurogenetic Section, Centre for Addiction and Mental Health. IMPACT Study [Internet]. Toronto (ON): The Centre; 2020. [Available from: http://impact.camhx.ca/en/clinicians-study [Google Scholar]
- (125).Bousman CA, Zierhut H, Muller DJ. Navigating the labyrinth of pharmacogenetic testing: a guide to test selection. Clin Pharmacol Ther. 2019;106(2):309–12. [DOI] [PubMed] [Google Scholar]
- (126).Anderson DW, Bousman CA, Chapman K. Engaging in building the educational support needed to deliver precision health in Canada. Healthc Manage Forum. 2020;33(3):135–9. [DOI] [PubMed] [Google Scholar]
- (127).Roe N, Passariello C, Brown L, Li J, Jablonski M, Dechairo BM. Prospective evaluation of the economic utility of combinatorial pharmacogenomics in generalized anxiety disorder and major depressive disorder. CNS Spectrums. 2018;23 (1):99–100. [Google Scholar]
- (128).Sousa C. 2018 Ontario budget: budget papers, a plan for care and opportunity [Internet]. Toronto (ON): Ministry of Finance, Goverment of Ontario; 2018. [cited 2020 19 May]. Available from: https://budget.ontario.ca/2018/budget2018-en.pdf [Google Scholar]
- (129).Parikh SV, Quilty LC, Ravitz P, Rosenbluth M, Pavlova B, Grigoriadis S. Canadian Network for Mood and Anxiety Treatments (CANMAT) 2016 clinical guidelines for the management of adults with major depressive disorder: section 2. Psychological treatments. Can J Psychiatry. 2016;61(9):524–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Ontario Ministry of Finance. Ontario population projections update, 2018-2046 [Internet]. Toronto (ON): Queen's Printer for Ontario; 2020. [cited 2020 19 May]. Available from: https://www.fin.gov.on.ca/en/economy/demographics/projections/ [Google Scholar]
- (131).Vassy JL, Davis JK, Kirby C, Richardson IJ, Green RC, McGuire AL. How primary care providers talk to patients about genome sequencing results: risk, rationale, and recommendation. J Gen Intern Med. 2018;33(6):877–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Winner JG, Carhart JM, Altar CA, Goldfarb S, Allen JD, Lavezzari G. Combinatorial pharmacogenomic guidance for psychiatric medications reduces overall pharmacy costs in a 1 year prospective evaluation. Curr Med Res Opinion. 2015;31(9):1633–43. [DOI] [PubMed] [Google Scholar]
- (133).Winner J, Allen JD, Altar CA, Spahic-Mihajlovic A. Psychiatric pharmacogenomics predicts health resource utilization of outpatients with anxiety and depression. Translational Psychiatry. 2013;3:e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Excel spreadsheet software. Microsoft Corporation. Redmond (WA). Available at: http://www.products.office.com/Microsoft/Excel.
- (135).Allen JD, Carhart JC, Marshak AG, Spivak AJ, Altar CA, Dechairo BM. GeneSight psychotropic decreases medication costs in a large, prospective case-control project. CNS Spectrums. 2015;20 (1):65. [Google Scholar]
- (136).Brown LC, Lorenz RA, Li J, Dechairo BM. Economic utility: combinatorial pharmacogenomics and medication cost savings for mental health care in a primary care setting. Clinical Therapeutics. 2017;39(3):592–602.e1. [DOI] [PubMed] [Google Scholar]
- (137).Edwards AM, Perlis RH, Krause DS. 128 factors associated with cost savings following use of a pharmacogenetic assay in individuals with mood and anxiety disorders. CNS Spectr. 2020;25(2):281–2. [Google Scholar]
- (138).Fagerness J, Fonseca E, Hess GP, Scott R, Gardner KR, Koffler M. Pharmacogenetic-guided psychiatric intervention associated with increased adherence and cost savings. Am J Manag Care. 2014;20(5):e146–56. [PubMed] [Google Scholar]
- (139).Jablonski MR, Lorenz R, Li J, Dechairo BM. Economic outcomes following combinatorial pharmacogenomic testing for elderly psychiatric patients. J Geriatric Psychiatry Neurol. 2019;33(6):324–32. [DOI] [PubMed] [Google Scholar]
- (140).Mayhew M, Jablonski M, Li J, Dechairo B, Healthson A. Combinatorial pharmacogenomics reduces polypharmacy and medication cost in elderly patients with anxiety and depression. Am J Geriatric Psychiatry. 2017;25 (3 Suppl 1):S143–S4. [Google Scholar]
- (141).Tanner JA, Brown LC, Yu K, Li J, Dechairo BM. Canadian medication cost savings associated with combinatorial pharmacogenomic guidance for psychiatric medications. ClinicoEcon. 2019;11:779–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (142).Breaux S, Desrosiers FAD, Neira M, Sinha S, Nislow C. Pharmacogenomics at the Point of Care: A Community Pharmacy Project in British Columbia. J Pers Med. 2020;11(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (143).Papastergiou J, Tolios P, Li W, Li J. The Innovative Canadian Pharmacogenomic Screening Initiative in Community Pharmacy (ICANPIC) study. J Am Pharm Assoc (2003). 2017;57(5):624–9. [DOI] [PubMed] [Google Scholar]
- (144).Smith A, Losak H. Pharmacogenomic testing for medication selection: a rapid qualitative review (CADTH rapid response report: summary with critical appraisal) [Internet]. Ottawa (ON): Canadian Agency for Drugs and Technologies in Health; 2020. [cited 2020 Dec 17]. Available from: https://www.cadth.ca/sites/default/files/rr/2020/RC1246%20PxGx%20Qualitative%20FINAL.pdf [PubMed] [Google Scholar]
- (145).Kvale S. Interviews: an introduction to qualitative research interviewing. Thousand Oaks (CA): Sage; 1996. [Google Scholar]
- (146).Kuzel AJ. Sampling in qualitative inquiry. In: Miller WL, Crabtree BF, editors. Doing qualitative research. Thousand Oaks (CA): Sage; 1999. p. 33–45. [Google Scholar]
- (147).Morse J. Emerging from the data: cognitive processes of analysis in qualitative research. In: Morse J, editor. Critical issues in qualitative research methods. Thousand Oaks (CA): Sage; 1994. p. 23–41. [Google Scholar]
- (148).Patton MQ. Qualitative research and evaluation methods. 3rd ed.Thousand Oaks (CA): Sage; 2002. [Google Scholar]
- (149).Strauss AL, Corbin JM. Basics of qualitative research: techniques and procedures of developing a grounded theory. 2nd ed.Thousand Oaks (CA): Sage; 1998. [Google Scholar]
- (150).Health Technology Assessment International. Introduction to health technology assessment [Internet]. Edmonton (AB): Health Technology Assessment International; 2015. [cited 2018 Apr 30]. Available from: http://www.htai.org/fileadmin/HTAi_Files/ISG/PatientInvolvement/v2_files/Resource/PCISG-Resource-Intro_to_HTA__KFacey__Jun13.pdf [Google Scholar]
- (151).Peterson K, Dieperink E, Anderson J, Boundy E, Ferguson L, Helfand M. Rapid evidence review of the comparative effectiveness, harms, and cost-effectiveness of pharmacogenomics-guided antidepressant treatment versus usual care for major depressive disorder. Psychopharmacology (Berl). 2017;234(11):1649–61. [DOI] [PubMed] [Google Scholar]
- (152).Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960;23:56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (153).Licht RW, Qvitzau S, Allerup P, Bech P. Validation of the Bech-Rafaelsen Melancholia Scale and the Hamilton Depression Scale in patients with major depression; is the total score a valid measure of illness severity? Acta Psychiatr Scand. 2005;111(2):144–9. [DOI] [PubMed] [Google Scholar]
- (154).Williams JB. A structured interview guide for the Hamilton Depression Rating Scale. Arch Gen Psychiatry. 1988;45(8):742–7. [DOI] [PubMed] [Google Scholar]
- (155).Rush AJ, Trivedi MH, Ibrahim HM, Carmody TJ, Arnow B, Klein DN. The 16-Item Quick Inventory of Depressive Symptomatology (QIDS), clinician rating (QIDS-C), and self-report (QIDS-SR): a psychometric evaluation in patients with chronic major depression. Biol Psychiatry. 2003;54(5):573–83. [DOI] [PubMed] [Google Scholar]
- (156).Spitzer R, Williams J, Kroenke K. Patient Health Questionnaire (PHQ-9) [Internet]. New York (NY): Pfizer Inc.; 1999. [cited 2020 April]. Available from: https://www2.gov.bc.ca/assets/gov/health/practitioner-pro/bc-guidelines/depression_patient_health_questionnaire.pdf [Google Scholar]
- (157).Busner J, Targum SD. The clinical global impressions scale: applying a research tool in clinical practice. Psychiatry (Edgmont). 2007;4(7):28–37. [PMC free article] [PubMed] [Google Scholar]
- (158).Dynacare, Genomind. White paper: the genes analyzed by Genecept Assay [Internet]. Brampton (ON): Dynacare; 2017. [cited 2020 Nov]. Available from: https://www.dynacare.ca/DYN/media/DYN/eng/Patients%20Ind/White-Paper_The-Genes-Analyzed-by-the-Genecept-Assay-2-0_final_2017JL25.pdf [Google Scholar]
- (159).Kellar J, Mittmann N, von Heymann C, Zingaro J, Kuriakose B, Li A. Costs of employees with treatment-resistant depression based on a Canadian private claims database. Value Health. 2014;17(7):A456–7. [DOI] [PubMed] [Google Scholar]