Abstract
To improve laboratory safety we thermally treated naso-oropharyngeal samples before testing with the cobas SARS-CoV-2 assay. This study aimed to determine if thermal treatment significantly affects the qualitative detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the quantitative measurement of cobas SARS-CoV-2 ORF1a and E-gene target copy number using an in-house quantitative method. A collection of positive (n = 238) and negative samples (n = 196) was tested in parallel comparing thermal treatment (75 °C for 15 minutes) to room-temperature. There were no significant differences in the final qualitative outcomes for thermal treatment versus room-temperature (99.8% agreement) despite a statistically significant reduction (P < 0.05) in target copy number following thermal treatment. The median ORF1a and E-gene reduction in target copy number was 0.07 (1.6%) and 0.22 (4.2%) log10 copies/mL respectively. The standard curves for both ORF1a and E-gene targets were highly linear (r2 = 0.99). Good correlation was observed for ORF1a (r2 = 0.96) and E-gene (r2 = 0.98) comparing thermal treatment to room-temperature control.
Keywords: Thermal treatment, SARS-CoV-2, Cobas, Qualitative, Quantitative
1. Introduction
Since the start of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, we have seen shortages of laboratory reagents, consumables and personal protective equipment (PPE) [(Tang et al., 2020; World Health Organization (WHO) 2020)]. Our initial testing protocols included guanidine hydrochloride inactivation of SARS-CoV-2 to improve laboratory safety and avoid the use of additional PPE (eye protection, N95 mask, disposable gown) (Corman et al., 2020; Tang et al., 2020). When we transitioned SARS-CoV-2 testing to the cobas 6800 system (Roche Diagnostics, Basel, Switzerland) we were concerned about transferring un-capped samples to the cobas 6800 instrument so returned to using additional PPE as a precaution. We were also concerned about the pipetting workload associated with the addition of guanidine hydrochloride to samples before cobas testing, potential loss of assay sensitivity from sample dilution and assay nonspecific interference. The effects of temperature on the viability of SARS-CoV-2 and other coronaviruses have been reported (Chin et al., 2020; Pagat et al., 2007; Rabenau et al., 2005; Yunoki et al., 2004). Our initial investigations suggested thermal treatment of nasopharyngeal samples resulted in statistically significant higher cycle threshold (Ct) values for E-gene (P 0.040), suggesting a reduction in detectable virus RNA (Pryce et al., 2021). Despite marginally higher cobas Ct values for thermally treated samples, conflicting findings were observed for the qualitative detection of SARS-CoV-2, specifically detection of ORF1a and E-gene targets at the limit of detection. The chief aim of this study was to determine if thermal treatment of naso-oropharyngeal samples before cobas SARS-CoV-2 testing affects the qualitative detection of SARS-CoV-2 and the quantitative detection of RNA target copy number. In addition, we developed and present here a quantitative method for cobas SARS-CoV-2 ORF1a and E-gene targets, using a commercially available SARS-CoV-2 standard to assess the effects of thermal treatment on RNA target copy number. We also investigated quantitative and qualitative results of storage at room-temperature for up to 48 hours for thermally treated and non-thermally treated samples in case we encounter testing delays.
2. Material and methods
2.1. Patient samples
A combined nasopharyngeal and oropharyngeal swab from each patient was inoculated into 3 mL of either Copan UTM-RT media (Brescia, Italy), CITOSWAB (Citotest Scientific Jiangsu, People's Republic of China) or Virus Transport Media (VTM) prepared by PathWest Media (Centers for Disease Control and Prevention, Atlanta GA 2020). All samples were initially tested as part of routine testing using the cobas SARS-CoV-2 assay (Roche Diagnostics, Basel, Switzerland). This test is performed on either the cobas 6800 or cobas 8800 instrument (Roche) and is a walkaway sample-to-result assay. The cobas SARS-CoV-2 assay targets ORF1a (a nonstructural region that is unique to SARS-CoV-2) and E-gene (a structural protein envelope gene for pan-sarbecovirus detection). According to the manufacturer's instructions, a sample is SARS-CoV-2 positive if ORF1a is detected with or without E-gene detection. In the case of positivity with E-gene alone, the result should be reported as SARS-CoV-2 presumptive positive.
All SARS-CoV-2 positive samples were stored as aliquots at -80 °C. To prepare positive samples for this study (n = 238; positive sample group) a 0.2 mL aliquot from each sample was diluted with 1.8 mL of a nasopharyngeal/throat matrix (1:10 dilution). The matrix consisted of pooled cobas SARS-CoV-2 negative patient samples (oro-nasopharyngeal swabs) in VTM. The pooled matrix tested negative with cobas SARS-CoV-2 and Xpert Xpress SARS-CoV-2 (Cepheid, Sunnyvale, California, USA). All dilutions were prepared in cobas omni secondary tubes (Ref. 06438776001). Cobas SARS-CoV-2 negative samples were stored at 4 °C and were not diluted (n = 196; negative sample group). All samples were tested following protocols issued by the manufacturer (room-temperature control). Un-capped samples in cobas omni secondary tubes were transferred to the cobas 6800 system by laboratory personnel wearing additional PPE (eye protection, N95 mask, disposable gown). Samples were retrieved from the cobas 6800 following sample aspiration with additional PPE, capped and thermally treated for 75 °C for 15 minutes in a QBD4 dry block heater (Grant Instruments, Cambridge, United Kingdom), then retested within 2 hours (thermal treatment) without the use of additional PPE. The qualitative results and Ct values were recorded for ORF1a, E-gene and Internal Control (IC).
2.2. Quantitative standards, external control and analysis
Quantitative standards were prepared from a commercially available SARS-CoV-2 standard (Exact Diagnostics, Fort Worth, Texas). Exact Diagnostics SARS-CoV-2 standard contains E-gene and ORF1ab synthetic RNA transcripts quantitated to 200,000 copies/mL using Bio-Rad Digital Droplet PCR (Hercules, California). We pooled multiple vials and 10-fold serially diluted in molecular grade water (G-Biosciences, St. Louis, Missouri, USA) to prepare 6 standards over the range of 0.30 to 5.30 log10 copies/mL. Each standard was tested with cobas SARS-CoV-2 in duplicate (no thermal treatment) on a single run using cobas SARS-CoV-2 kit lot G18524. The mean Ct value at each concentration was used to calculate ORF1a and E-gene standard curves and regression. Both replicates at each dilution were required to be positive to be included in the standard curve and regression analysis. The regression formulas were used to calculate the ORF1a and E-gene copy number for all positive samples and controls over 3 consecutive runs. The testing for all positive samples and the quantitative standards was performed using the same cobas SARS-CoV-2 kit lot (G18524). As part of the NRL QConnect programme (NRL, Victoria, Australia) an external control (EQC) was also performed routinely to monitor reproducibility (Optitrol NAT SARS-CoV-2; DiaMex, Heidelberg, Germany). A single lot number of EQC (DM20119) was tested over 19 runs.
2.3. Effects over time
A low-titre positive patient sample in VTM (≈3.00 log10 copies/mL) was used to study effects over time. Two replicates were thermally treated then tested at 2, 4, 6, 10, 24 and 48-hour intervals (kept at room temperature). Another 2 replicates remained at room-temperature and were tested in parallel with the thermally treated samples. The Ct values for ORF1a, E-gene and IC was recorded for each time point and the ORF1a and E-gene copy number was calculated using the standard curves.
2.4. Statistical analysis
All results of ORF1a and E-gene (Ct values and log10 copies/mL) for thermal treatment and room-temperature were compared using the Wilcoxon signed-rank test (P < 0.05). The same statistical approach was applied for all IC Ct values. The median for each group was calculated and used to determine the percentage reduction or gain in Ct value or log10 target copy number/mL. Differences in the mean and standard deviation were also calculated for comparison. Correlation between comparing thermal treatment to room temperature for ORF1a-positive samples (n = 180) and E-gene-positive samples (n = 201) was also performed. The mean and standard deviation was calculated for the EQC. All statistical analyses were performed by Excel (Microsoft, Redmond, WA, USA) and MedCalc v15.4 (New York, NY, USA).
2.5. Ethics statement
Not applicable: the residual samples used in the study were de-identified and results were not used to clinically manage patients [National Statement on Ethical Conduct in Human Research 2007 (May 2015) by the National Health and Medical Research Council, Australian Research Council and Australian Vice-Chancellors’ Committee].
3. Results
3.1. Qualitative analysis
The qualitative results comparing thermal treatment to room-temperature for the positive sample group (n = 238) and negative sample group (n = 196) are shown in the Supplementary Material and are summarized in Table 1 . All samples in the negative sample group were negative (n = 196). Samples in the positive sample group were positive (n = 180; ORF1a positive, E-gene positive or negative), presumptive positive (n = 13; ORF1a negative, E-gene positive), or negative (n = 22; ORF1a and E-gene negative) for both thermal treatment and room-temperature. Discordant results of paired samples comparing thermal treatment and room-temperature were also observed in the positive sample group (n = 23). We expected negative results and discordant results for some samples in the positive sample group as they were diluted 10-fold from archival cobas SARS-CoV-2 positive material already at the lower limit of detection. These discordant results all demonstrate late Ct values as shown in the Supplementary Material.
Table 1.
Summary of cobas SARS-CoV-2 results for 434 samples.
| Result outcome | No. of samples | Sample group | cobas SARS-CoV-2 result for: | |
|---|---|---|---|---|
| Heat treatment | Room temperature control | |||
| 1 | 180 | Positive | Detected | Detected |
| 2 | 13 | Positive | Presumptive positive | Presumptive positive |
| 3 | 6 | Positive | Detected | Presumptive positive |
| 4 | 5 | Positive | Presumptive positive | Detected |
| 5 | 6 | Positive | Presumptive positive | Negative |
| 6 | 6 | Positive | Negative | Presumptive positive |
| 7 | 22 | Positive | Negative | Negative |
| 8 | 196 | Negative | Negative | Negative |
Detected indicates ORF1a positive, E-gene positive.
Presumptive positive indicates ORF1a negative, E-gene positive.
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
No significant differences in the qualitative outcomes were observed. The room-temperature group resulted in positive (n = 185), presumptive positive (n = 25) and negative (n = 224) outcomes compared to positive (n = 186), presumptive positive (n = 24) and negative (n = 224) for the thermal treatment group. No samples were inhibited (all had a positive IC).
3.2. Standard curves
The Ct values for each target concentration and standard curves for ORF1a and E-gene are shown in the Supplementary Material. The results for both targets were highly linear. ORF1a demonstrated R-squared value of 0.9988 over the range of 1.30 to 5.30 log10 copies/mL (5 standards). ORF1a detection at 0.30 log10 copies/mL was not reproducible and was omitted from the standard curve. E-gene demonstrated R-squared value of 0.9994 over the range of 2.30 to 5.30 log10 copies/mL (4 standards). E-gene detection at 1.30 log10 copies/mL was not reproducible and was omitted from the standard curve. Both replicates were negative at 0.30 log10 copies/mL for E-gene.
3.3. Statistical and quantitative analysis
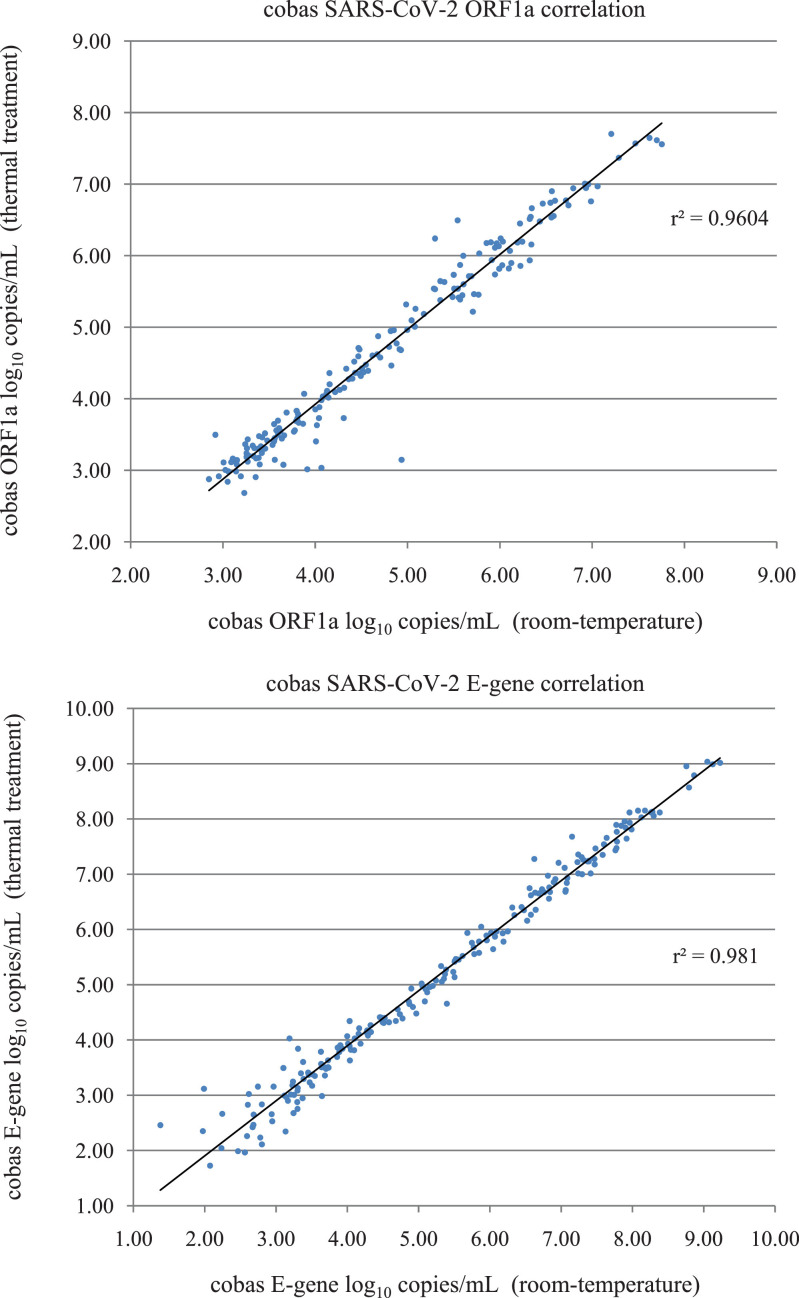
All Ct values for ORF1a, E-gene and internal control for all samples are shown in the Supplementary Material. The quantitative measurement of ORF1a and E-gene targets in copies/mL and log10 copies/mL are also shown, including the ORF1a and E-gene targets above and below the calculated standard curve range; the standard curve for each target was assumed to be linear above and below the calculated standard curve range to simplify statistical analysis. A summary of the Wilcoxon Signed-Ranks statistical analysis for ORF1a, E-gene and IC are summarized in Table 2 . The mean differences and standard deviation differences are also shown for comparison. For thermal treatment compared to room-temperature, we found a significant difference (P < 0.05) in median Ct and/or quantitative values for ORF1a, E-gene and IC for all samples. The median ORF1a and E-gene reduction in target copy number was 0.07 (1.6%) and 0.22 (4.2%) log10 copies/mL respectively. Good correlation was observed for ORF1a (r2 = 0.96) and E-gene (r2 = 0.98) comparing thermal treatment to room-temperature control (Fig. 1 ).
Table 2.
Summary of the Wilcoxon signed-rank test (P < 0.05) for thermally treated samples compared to room-temperature control.
| Target | Number compared | Median thermal treatment | Median room-temperature | Median difference | Mean difference | Standard deviation difference | P value |
|---|---|---|---|---|---|---|---|
| ORF1a Ct | 180 | 28.25 | 28.06 | +0.19 | +0.13 | 0.81 | 0.024 |
| ORF1a log10 copies/mL | 180 | 4.42 | 4.49 | -0.07 | -0.04 | 0.28 | 0.029 |
| E-gene Ct | 201 | 29.88 | 29.28 | +0.60 | +0.31 | 0.71 | < 0.01 |
| E-gene log10 copies/mL | 201 | 5.02 | 5.24 | -0.22 | -0.11 | 0.26 | < 0.01 |
| IC (SARS-CoV-2 positive) Ct | 180 | 33.57 | 34.49 | -0.92 | -0.44 | 0.92 | < 0.01 |
| IC (SARS-CoV-2 negative) Ct | 218 | 33.10 | 33.03 | +0.07 | +0.05 | 0.41 | < 0.01 |
| IC (all samples) Ct | 434 | 33.27 | 33.30 | -0.03 | -0.18 | 0.73 | < 0.01 |
IC indicates cobas SARS-CoV-2 internal control.
Ct indicates cycle threshold.
log10 copies/mL indicates the concentration of target quantified using the Exact Diagnostics SARS-CoV-2 standard curves.
Fig. 1.
Correlation in log10 copies/mL obtained by cobas SARS-CoV-2 following thermal treatment for ORF1a (target 1) and E-gene (target 2) compared to cobas SARS-CoV-2 room-temperature control. Linear regression was performed using samples positive for SARS-CoV-2 for both thermal treatment and room-temperature control for ORF1a (n = 180) and E-gene (n = 201). The r2 correlation is indicated.
The EQC was assessed over 19 runs as shown in the Supplementary Material. ORF1a demonstrated a mean of 3.95 ± 0.20 log10 copies/mL and E-gene 4.77 ± 0.23 log10 copies/mL across 12 reagent lot numbers. ORF1a demonstrated a mean of 3.88 ± 0.12 log10 copies/mL and E-gene 4.63 ± 0.21 log10 copies/mL for the positive sample group tested with the quantitative standards (lot number G18524).
3.4. Effects over time
The Ct values for ORF1a, E-gene and internal control and the quantitative results over time for the thermally treated and room-temperature replicates are shown in the Supplementary Material. We observed no evidence of increasing Ct values or decline in target copy number over 2, 4, 6, 10, 24 and 48-hour intervals for either thermally treated or room-temperature replicates. The Ct values and quantitative results remained stable over the 48-hour time period.
4. Discussion
We implemented thermal treatment of patient samples to improve laboratory safety and reduce additional PPE use (Pryce et al., 2021). Preliminary results with a limited number of positive samples (n = 34) showed increased Ct values for thermal treatment compared to room-temperature despite a potential improvement in the qualitative detection of SARS-CoV-2 for thermally treated samples. Our aim was to provide clarity using a greater number of positive samples (n = 238) and include more negative control samples to verify that thermal treatment does not cause nonspecific results. We sought to improve this assessment by measuring ORF1a and E-gene target copy number with a commercial quantitative standard. We also assessed thermal treatment compared to room temperature over time to obtain a better understanding of the stability of the ORF1a and E-gene targets with storage at ambient temperature.
A direct comparison of thermal treatment and room-temperature using undiluted original patient material would have been ideal. However, following initial routine testing and our SARS-CoV-2 surveillance testing, the residual sample volume was limited to conduct parallel re-testing with sufficient sample remaining for future research. To overcome this we diluted all positive samples in our collection with a pooled oro-nasopharyngeal matrix derived from SARS-CoV-2-negative patient samples to maximise the number of positive samples from different patients in this study. The closest representation to an original sample was maintained with this approach.
We found no significant differences in the qualitative outcomes (detected, presumptive or negative) for thermally treated samples compared to room-temperature samples in this study. In the previous study (Pryce et al., 2021) we observed additional positive results (n = 3) and presumptive results (n = 3) for thermal treatment compared to room-temperature (17.6%; 6/34 samples in the positive group), compared to only 1 additional positive result for thermal treatment (0.4%; 1/238 samples in the positive group) in this study. In the previous study we performed 10-fold serial dilutions (n = 34) for each patient sample (n = 8) compared to a single dilution for each patient this study (n = 238). Although assay sensitivity is best shown by testing replicates at the lower limit of detection, we were limited due to costs and reagents to perform similar 10-fold serial dilutions for all positive samples. Nevertheless, the current study included sufficient samples of low concentration where the qualitative results were not different overall. We conclude with this larger study that there are no significant qualitative differences between thermally treated and room-temperature samples. Although not validated by the manufacturer, there was no evidence that thermal treatment leads to non-specificity.
Other investigators using a digital droplet PCR method have demonstrated a median drop in SARS-CoV-2 copy number of 50% to 66% after heating samples (n = 63) at different SARS-CoV-2 concentrations for 80 °C for 20 minutes (Chen et al., 2020). Whilst digital droplet PCR methods are useful for the sensitive detection and quantification of SARS-CoV-2 (de Kock et al., 2021), they are not practical for routine diagnostic SARS-CoV-2 detection as digital droplet PCR machines lack the necessary throughput required for front-line testing (Vasudevan et al., 2021). The cobas 6800/8800 instrument is a sample-to-result platform widely used for molecular diagnostics and SARS-CoV-2 was added in response to the pandemic with recent reports of strong correlation with other assays and utility emerging (Poljak et al., 2020). The exceptional test performance of quantitative cobas 6800/8800 assays (using an internal quantitation standard) for blood-borne virus testing is well known (Roh et al., 2021; Tan et al., 2018; Yao et al., 2018). We sought to utilise the platform as a quantitative assay using an external standard as a reference. To our knowledge a quantitative cobas SARS-COV-2 method has not been published. To assess the loss of target copy number following thermal treatment we developed standard curves for the quantitation of ORF1a and E-gene copy number for cobas SARS-CoV-2 test. The ORF1a and E-gene standard curves were linear (r2 > 0.99) and the cobas SARS-CoV-2 test was shown to be a reproducible assay in our laboratory using a commercially available quality control. We demonstrated statistically significant reduction in the median target copy number for ORF1a (P 0.03) and E-gene (P < 0.01) after heating samples for 75 °C for 15 minutes. However in contrast, the median and mean difference was quantitatively small for both targets (<5%) and is unlikely to be clinically significant. We conclude the negligible loss in ORF1a and E-gene target copy number following thermal treatment is outweighed by a significant improvement of laboratory safety and handling of SARS-CoV-2, particularly with no evidence of detrimental qualitative outcomes.
There are conflicting reports of the effect of thermal treatment of SARS-CoV-2. Hemati et al. (2020) demonstrated that thermal inactivation of patient samples (60 °C for 30 minutes) results significantly lower Ct values for N and ORF1ab gene (P 0.009 and P 0.32 respectively) using an in-house PCR method (Hemati et al., 2021). However, the number of clinical samples tested by PCR (Ct values compared) was low (n = 7). Burton et al. (2021) showed thermal inactivation (56 °C and 60 °C) of a single strain of SARS-CoV-2 did not affect PCR sensitivity using the method of Corman et al. (2020), but showed a minimum increase of 3 Ct values when treated at 80 °C for 30 minutes (Burton et al., 2021). In comparison, we demonstrated mean Ct value increases of +0.13 for ORF1a and +0.31 for E-gene following thermal treatment at 75 °C for 15 minutes when tested with a commercial SARS-CoV-2 assay, using a large number of positive samples (key point of difference). We also assess the effects of thermal treatment of negative controls (nonspecific results were not observed).
We routinely thermally treat samples and leave the aliquots overnight at 4 °C to avoid delays in processing the next day and to maximise throughput. Investigations past 48 hours were not performed as our laboratory has an expected test-turnaround-time of <24 hours from collection. Initially we speculated that thermal treatment may inactivate nucleases and preserve the sample over extended periods of time. To investigate we performed quantitative measurement of ORF1a and E-gene targets comparing thermal treatment and room temperature over time intervals. No reduction in ORF1a or E-gene copy number was observed over a 48-hour duration. This is an important finding for remote collection where refrigeration may not be immediately available and transport to the testing laboratory may be delayed. Delays in testing may also occur due to overwhelming workload. We conclude that SARS-CoV-2 RNA targets remain stable in VTM over the 6-12 hour time delays that may be encountered due to workload. Investigations with other media and manufacturers are ongoing with similar results (data not shown). We recommend other laboratories conduct their own in-house evaluation of the media locally available for cobas or other SARS-CoV-2 testing.
Due to safety concerns at the time and suboptimal recovery of SARS-CoV-2 from culture, our laboratory did not confirm the inactivation efficacy of 75 °C for 15 minutes. However, we implemented a thermal treatment method that exceeds the temperature and time duration from a previously published method of 70 °C for 5 minutes for complete SARS-CoV-2 inactivation in virus transport medium (Chin et al., 2020). Subsequently, a report from the Institut Pasteur (France) has shown that SARS-CoV-2 is relatively sensitive to heat inactivation using a dry heating block and can be inactivated in less than 30 minutes, 15 minutes and 3 minutes at 56 °C, 65 °C and 95 °C respectively (Batéjat et al., 2021) In our laboratory, thermal treatment before cobas SARS-CoV-2 testing is a simple precautionary method to improve safety when transferring un-capped samples to the cobas 6800 instrument, which does not affect the qualitative detection SARS-CoV-2 with this assay. The use of additional PPE has also been reduced.
Acknowledgments
The authors are grateful to all laboratory personnel that work in the Department of Clinical Microbiology, for their dedication, diligence and perseverance.
Funding
This study did not receive funding support.
Declaration of competing Interest
The authors declare that they have no competing interests.
Authors’ contributions
Todd Pryce: Conceptualization, Methodology, Data analysis, Writing- Original draft preparation, Writing – Reviewing and Editing. Peter Boan: Data analysis, Writing – Reviewing and Editing. Ian Kay: Writing – Reviewing and Editing. James Flexman: Writing – Reviewing and Editing.
Supplementary materials
The authors have provided raw data in the supplementary file submitted.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.diagmicrobio.2021.115519.
Appendix. Supplementary materials
References
- Batéjat C, Grassin Q, Manuguerra JC, Leclercq I. Heat inactivation of the severe acute respiratory syndrome coronavirus 2. J Biosaf Biosecur. 2021;3:1–3. doi: 10.1016/j.jobb.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton J, Love H, Richards K, Burton C, Summers S, Pitman J, et al. The effect of heat-treatment on SARS-CoV-2 viability and detection. J Virol Methods. 2021;290 doi: 10.1016/j.jviromet.2021.114087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention, Atlanta GA. 2020. Preparation of viral transport medium (SOP#: DSR-052–03). [8th March 2021]. Available at: https://www.cdc.gov/coronavirus/2019-ncov/downloads/Viral-Transport-Medium.pdf. Accessed 8th March 2021
- Chen H, Wu R, Xing Y, Du Q, Xue Z, Xi Y, et al. Influence of different inactivation methods on severe acute respiratory syndrome coronavirus 2 RNA Copy Number. J Clin Microbiol. 2020;58:1–8. doi: 10.1128/jcm.00958-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin AWH, Chu JTS, Perera MRA, Hui KPY, Yen HL, Chan MCW, et al. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1:e10. doi: 10.1016/s2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.es.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kock R, Baselmans M, Scharnhorst V, Deiman B. Sensitive detection and quantification of SARS-CoV-2 by multiplex droplet digital RT-PCR. Eur J Clin Microbiol Infect Dis. 2021;40:807–813. doi: 10.1007/s10096-020-04076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemati M, Soosanabadi M, Ghorashi T, Ghaffari H, Vahedi A, Sabbaghian E, et al. Thermal inactivation of COVID-19 specimens improves RNA quality and quantity. J Cell Physiol. 2021;236:4966–4972. doi: 10.1002/jcp.30206. [DOI] [PubMed] [Google Scholar]
- Pagat AM, Seux-Goepfert R, Lutsch C, Lecouturier V, Saluzzo JF, Kusters IC. Evaluation of SARS-coronavirus decontamination procedures. Appl Biosaf. 2007;12:100–108. https://doi.org/10.1177%2F153567600701200206. [Google Scholar]
- Poljak M, Korva M, Knap Gašper N, Fujs Komloš K, Sagadin M, Uršič T, et al. Clinical evaluation of the cobas SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the midst of the COVID-19 pandemic. J Clin Microbiol. 2020;58:1–7. doi: 10.1128/jcm.00599-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryce TM, Boan PA, Kay ID, Flexman JP. Thermal treatment of nasopharyngeal samples before cobas SARS-CoV-2 testing. Clin Microbiol Infect. 2021;27:149–150. doi: 10.1016/j.cmi.2020.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabenau HF, Cinatl J, Morgenstern B, Bauer G, Preiser W, Doerr HW. Stability and inactivation of SARS coronavirus. Med Microbiol Immunol. 2005;194:1–6. doi: 10.1007/s00430-004-0219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh J, Kim S, Kwak E, Park J, Park Y. Performance evaluation of the Roche cobas 6800 system for quantifying cytomegalovirus DNA in plasma and urine samples. J Clin Virol. 2021;138 doi: 10.1016/j.jcv.2021.104816. [DOI] [PubMed] [Google Scholar]
- Tan NK, Carrington D, Pope CF. Verification of the Roche cobas® 6800 PCR 200 µl and 500 µl protocols for the quantification of HIV-1 RNA, HBV DNA and HCV RNA and evaluation with COBAS® Ampliprep/COBAS®TaqMan® assays. J Med Microbiol. 2018;67:1711–1717. doi: 10.1099/jmm.0.000838. [DOI] [PubMed] [Google Scholar]
- Tang YW, Schmitz JE, Persing DH, Stratton CW. Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol. 2020;58:e00512–e00520. doi: 10.1128/jcm.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan HN, Xu P, Servellita V, Miller S, Liu L, Gopez A, et al. Digital droplet PCR accurately quantifies SARS-CoV-2 viral load from crude lysate without nucleic acid purification. Sci Rep. 2021;11:780. doi: 10.1038/s41598-020-80715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO). Coronavirus disease (COVID-2019) situation reports. 2020. [8th March 2021]. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- Yao JD, Young S, Heilek GM, Marino E, Paxinos EE, Marins EG, Valsamakis A. Diagnosis and monitoring of HCV infection using the cobas® HCV test for use on the cobas® 6800/8800 systems. J Clin Virol. 2018;102:63–69. doi: 10.1016/j.jcv.2018.02.017. [DOI] [PubMed] [Google Scholar]
- Yunoki M, Urayama T, Yamamoto I, Abe S, Ikuta K. Heat sensitivity of a SARS-associated coronavirus introduced into plasma products. Vox Sang. 2004;87:302–303. doi: 10.1111/j.1423-0410.2004.00577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.