Abstract
Aberrant inflammation in the central nervous system (CNS) has been implicated as a major player in the pathogenesis of human neurodegenerative disease. We developed a novel approach to derive microglia from human pluripotent stem cells (hPSCs) and built a defined hPSC-derived tri-culture system containing pure populations of hPSC-derived microglia, astrocytes, and neurons to dissect cellular crosstalk along the neuroinflammatory axis in vitro.
We used the tri-culture system to model neuroinflammation in Alzheimer’s Disease with hPSCs harboring the APPSWE+/+ mutation and their isogenic control. We found that complement C3, a protein that is increased under inflammatory conditions and implicated in synaptic loss, is potentiated in tri-culture and further enhanced in APPSWE+/+ tri-cultures due to microglia initiating reciprocal signaling with astrocytes to produce excess C3. Our study defines the major cellular players contributing to increased C3 in AD and presents a broadly applicable platform to study neuroinflammation in human disease.
Introduction
Over recent years, neuroinflammation has been increasingly implicated in the progression of various neurodegenerative disorders such as Alzheimer’s1, Parkinson’s2, Amyotrophic Lateral Sclerosis (ALS)3 as well as in aging4. Microglia are thought to be key players in triggering an inflammatory state in the brain that may be beneficial initially but can precipitate or exacerbate disease pathology when prolonged. Other glial cells such as astrocytes interact with microglia and may further contribute to aberrant inflammation and neurotoxicity4,5. However, parsing out cellular crosstalk in vivo and understanding species specific differences in the neuroinflammatory response6 remain major challenges in the field. Human pluripotent stem cell (hPSC) technology has the potential to overcome those challenges and to present a fully human, defined and scalable platform to study neuroinflammation. An essential requirement for such an hPSC-based model are differentiation strategies that can reproducibly generate pure populations of microglia, astrocytes, and neurons in a synchronized, robust, efficient, and timely manner. Protocols based on dual-SMAD inhibition7,8 allow for the efficient production of highly pure neural precursors and postmitotic neurons from hPSCs. Likewise, we have recently reported a strategy to rapidly derive pure populations of astrocytes from hPSCs9. In contrast, several protocols have been published for generating microglial-like cells from hPSCs10–16; but those approaches commonly rely on embryoid body formation and are poorly defined with respect to ontogeny10,13, require manipulations such as hypoxia for patterning11, or necessitate cell-sorting for purity12. Importantly, current microglial differentiation protocols are not specifically optimized towards primitive hematopoiesis as defined by early induction of KDR+CD235A+ hemangioblasts17. Recapitulating those early developmental steps in vitro is important as the microglial lineage entirely traces back to primitive hematopoiesis18.
Results
Narrow window for primitive hematopoietic patterning of hPSCs
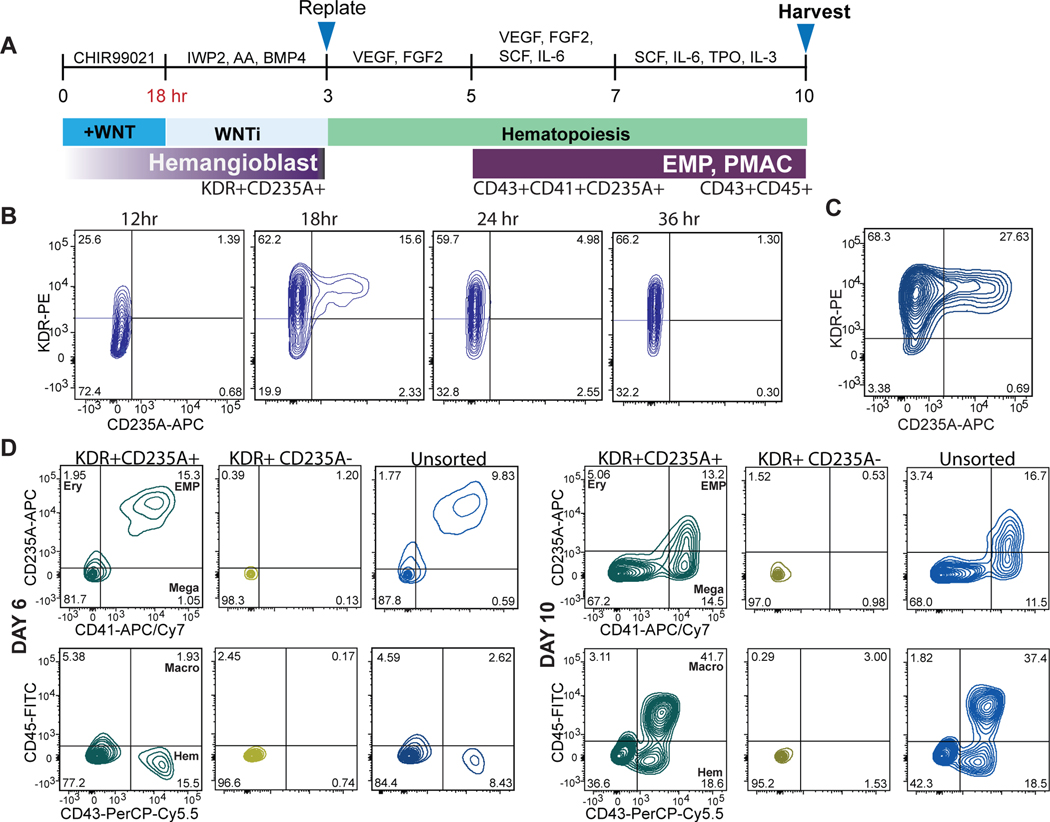
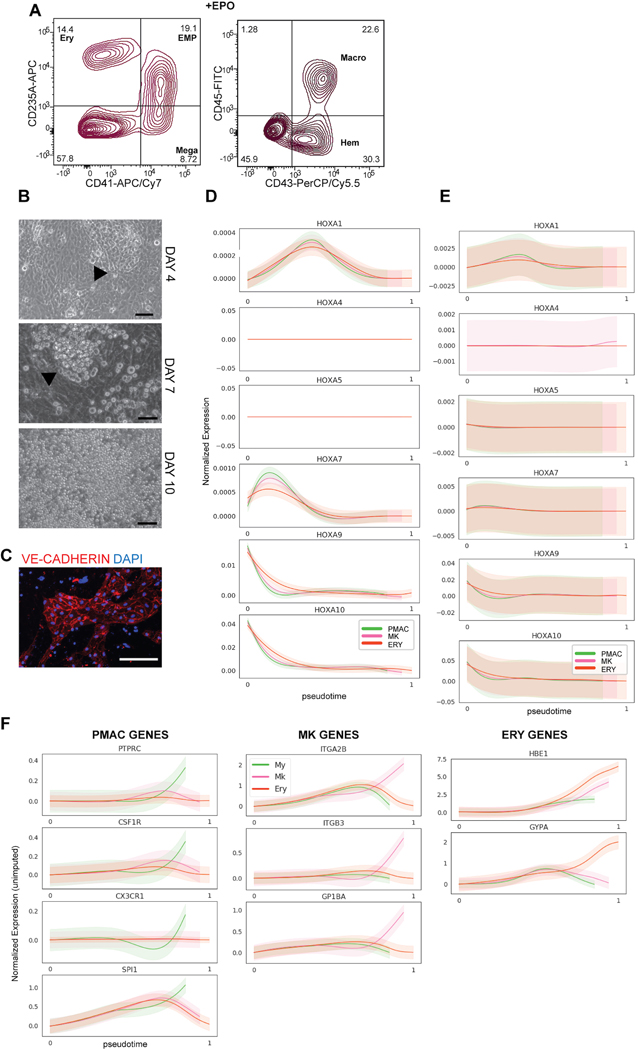
Here we present a novel strategy to direct the differentiation of hPSCs into microglial precursors via primitive hematopoiesis (Fig. 1A). First, we patterned hPSCs towards primitive streak mesoderm by activating WNT signaling via treatment with the GSK3ß inhibitor CHIR99021. We then inhibited WNT signaling by using the porcupine inhibitor IWP2 and concurrently activated Nodal signaling, mimicked by exposure to Activin A. Those conditions promoted the generation of KDR+CD235A+ primitive hematopoietic hemangioblasts versus KDR+CD235A- definitive hematopoietic precursors consistent with the paradigm proposed in Sturgeon et al17. Remarkably, we observed that WNT inhibition must occur within a very narrow developmental window, limited to 18–24 hours post WNT activation, to efficiently generate a KDR+CD235A+ population (Fig. 1B). After optimizing cell density and small molecule concentrations, WNT activation for 18 hours followed by WNT inhibition and Nodal activation for ~ 2 days (2 days + 6 hours) yields ~27% KDR+CD235A+ cells by day 3 of differentiation (Fig. 1C). We next asked whether KDR+CD235A+ hemangioblasts vs. the KDR+CD235A- definitive precursors can give rise to hematopoietic cells under those culture conditions. The KDR+CD235A+ hemangioblast population produced CD41+CD235A+CD43+ hematopoietic cells within 3 days after isolation by cell sorting (day 6 of hPSC differentiation) in the presence of minimal hematopoietic cytokines (Fig. 1D). In contrast, the KDR+CD235A- population failed to produce hematopoietic cells. Those data are consistent with reports suggesting that definitive precursors give rise to hematopoietic cells only at later stages of differentiations, under hypoxic conditions, or in the presence of additional hematopoietic cytokines19.
Figure 1. Patterning towards primitive hematopoiesis occurs during a narrow developmental window.
A) Schematic for differentiating primitive hematopoietic cells from hPSCs. WNT activation followed by WNT inhibition allows KDR+CD235A+ progenitors to emerge, which are re-plated in hematopoietic cytokines for 8 days to yield CD45+ macrophage precursors at day 10 of differentiation. B) FACS analysis timecourse shows WNT inhibition initiated at 18hr post WNT-activation induces the emergence of the KDR+CD235A+ hemangioblast population. WNT-inhibition initiated at 12hr, 24hr, or 36hr post WNT-activation does not yield significant numbers of KDR+CD235A+ hemangioblasts. C) FACS analysis of optimized WNT activation followed by inhibition shows efficient generation of ~30% KDR+CD235A+ hemangioblast population by day 3. Optimization included titrating the hPSC seeding density (60,000 cells/cm2 and concentrations of small molecule inhibitors and growth factors including CHIR99021 (3uM), IWP2 (2uM), Activin A (7.5–10ng/mL), BMP4 (30–40ng/mL), FGF2 (20ng/mL). D) FACS analysis of the differentiation at days 6 and 10 shows that only differentiations containing re-plated KDR+CD235A+ cells within the population (pure KDR+CD235A+, and unsorted) produce CD43+CD235A+CD41+ primitive EMPs by Day 6 and CD45+ macrophage precursors by Day 10 of differentiation. Markers for the differentiated hematopoietic populations are as follows: primitive EMP = CD43+CD41+CD235A+, Ery (erythrocyte) = CD43+CD235A+CD41-, Mega (megakaryocyte) = CD41+CD235A-, Macro (macrophage precursor) = CD45+, Hem (early hematopoietic committed) = CD43+. Gating was performed according to Supplementary Figure 2.
By day 7 after re-plating (day 10 of differentiation), the KDR+CD235A+ fraction yielded 41% CD45+ cells, the population that later gives rise to microglia. Other populations which arose include uncommitted CD41+CD235A+CD43+ primitive-erythromyeloid progenitors (primitive EMPs), CD41+ megakaryocytes, and CD235A+ erythrocytes (only when treated with erythropoietin, EPO) (Extended Data 1A). Primitive EMPs, megakaryocytes, and erythrocytes are all lineages produced during primitive hematopoiesis20. The definitive KDR+CD235A- population produced still no hematopoietic cells, demonstrating all hematopoietic cells produced by day 10 of differentiation are derived from KDR+CD235A+ hemangioblasts (Fig. 1D). Interestingly, unsorted samples containing both the KDR+CD235A- and KDR+CD235A+ populations produced hematopoietic cells at nearly the same efficiency as the sorted KDR+CD235A+ population, suggesting that KDR+CD235A+ cells do not need to be purified for the robust production of hematopoietic cells (Fig. 1D). This finding streamlines the differentiation process by eliminating the cell sorting step and instead allows for the simple re-plating of the mixed population at Day 3 in the presence of hematopoietic cytokines. Almost 60% of the cells at day 10 of differentiation are of hematopoietic identity, forming floating colonies in semi-suspension above a VE-CADHERIN+ hemogenic endothelium (Extended Data 1B, C).
Developmental stages of hPSC-derived microglia are similar to in vivo differentiation trajectory
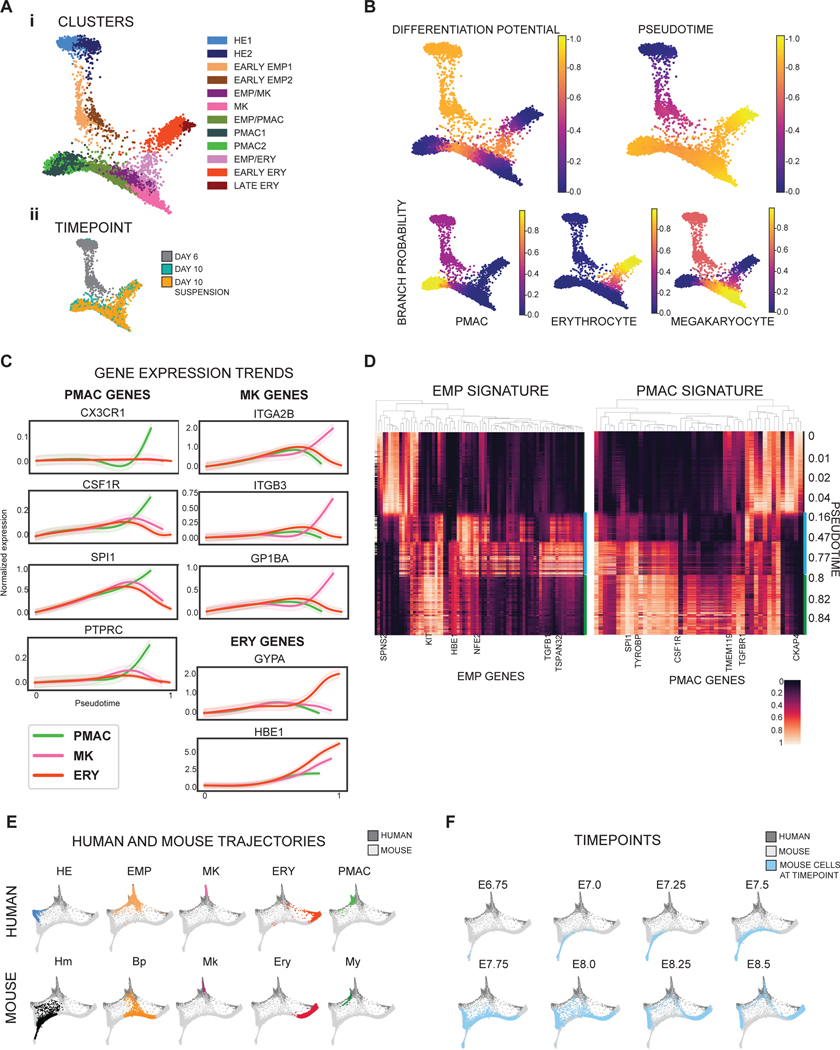
We next performed single cell RNA-sequencing (scRNA-seq) at day 6 and day 10 of differentiation (in the presence of EPO) to fully characterize the heterogeneity of the cells produced, and to identify the developmental trajectory of the transitioning cellular states given the paucity of data on human primitive hematopoiesis in current literature. Cells from pooled day 6 and day 10 scRNA-seq data were grouped into separate clusters using the PhenoGraph clustering algorithm21 and a force-directed graph layout of the cells was generated to visualize the data (Fig. 2A). The more mature hematopoietic populations of erythrocytes (ERY), megakaryocytes (MK), and macrophage precursors (PMAC) were identified by the trajectory analysis tool Palantir22 as three distinct arms stemming from a common primitive EMP population, mimicking the trajectory of primitive hematopoiesis. To investigate the differentiation status of individual cells in the trajectory, we used Palantir’s ‘differentiation potential’ calculation22, which views differentiation as steps through a Markov chain which can only be taken between similar cells and away from the starting point of differentiation. A cell with high differential potential can take a path of Markov steps to multiple arms and corresponds to an undifferentiated state, whereas low differentiation potential is represented by a lack of available Markov paths from a specific cell to other arms of differentiation. We found that the highest differentiation potential fell within the hemogenic endothelial clusters and the primitive EMP-like cells at the center of the map, and the lowest fell at the more mature hematopoietic clusters at the ends of the three arms (Fig. 2B). We further used Palantir to model the expression trends of key genes over pseudotime along the trajectory of each arm and found that the erythrocyte arm expressed the signature genes of embryonic hemoglobin (HBE1) and GYPA, the megakaryocyte arm expressed key megakaryocytic genes of ITGA2B, ITGB3, and GP1BA, and the macrophage precursor arm expressed CSF1R, PTPRC, SPI1, and CX3CR123 (Fig. 2C). In addition, we assessed expression of HOXA genes and CDX4 (markers of definitive hematopoiesis)24 along all three arms and found absent expression (CDX4 was undetectable) or very low expression, further indicating that our hematopoietic cells are of the primitive hematopoietic lineage (Extended Data 1D). Complementing this imputed gene trend modeling, we plotted the unimputed expression of those signature genes against pseudotime and obtained comparable results in which each arm enriched for the expression of its corresponding identity genes while showing near absent HOXA expression (Extended Data 1E, F). Thus, we ensured that the trends as inferred by Palantir from imputed data are also observed in non-imputed data.
Figure 2. Single cell RNA-sequencing validates the stages of microglial development within the in vitro differentiation.
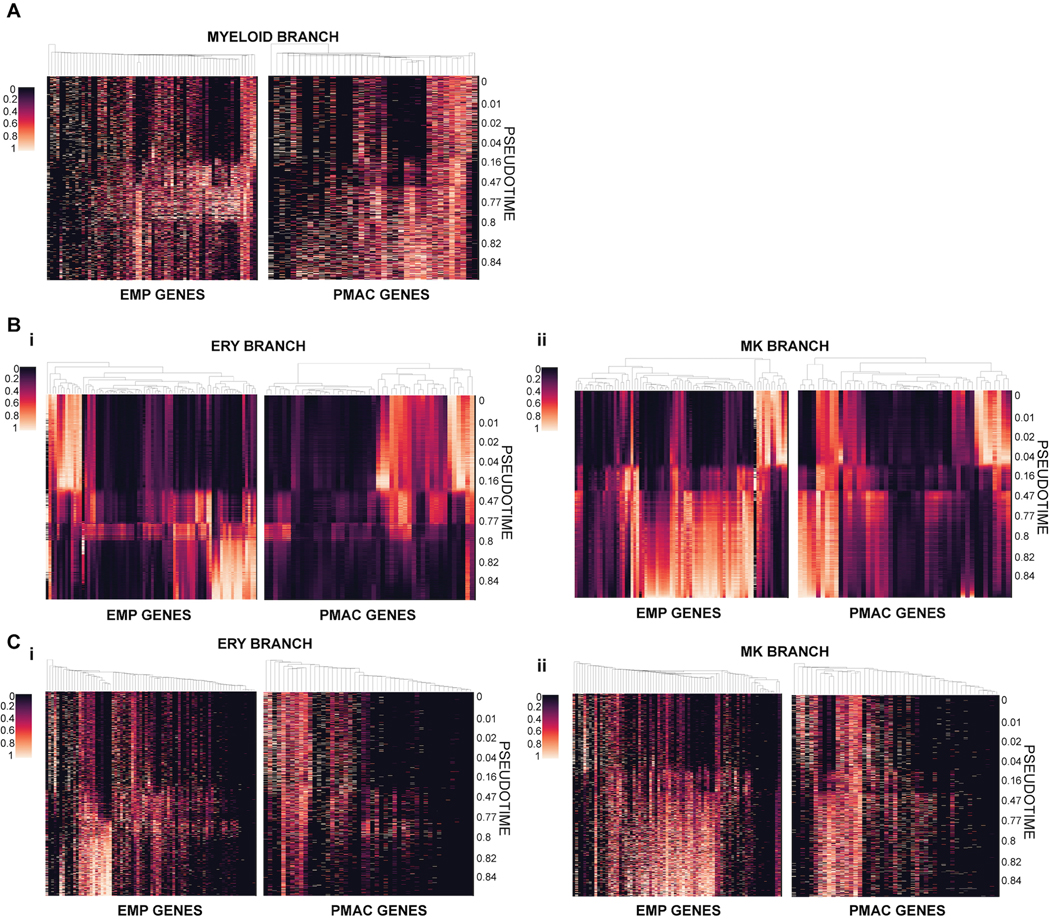
A) i) Force-directed graph layout of combined data from Day 6 and Day 10 cultures reveals hemogenic endothelium (HE), primitive erythromyeloid progenitor (EMP), erythrocyte (ERY), megakaryocyte (MK), and early macrophage (PMAC) clusters. ii) Breakdown of the samples used to generate the force-directed graph layout show that Day 6 of differentiation contains the HE and early EMP clusters and Day 10 of differentiation contains the later stage EMPs and ERY, MK, and PMAC clusters. B) Palantir analysis shows that HE clusters (set to be the start of the trajectory, with differentiation potential near 1.0 and pseudotime near 0.0), progress through a primitive EMP intermediate and branch into 3 separate differentiation trajectories towards erythrocytic (ERY), megakaryocytic (MK), and macrophage (PMAC) end populations (differentiation potential near 0.0 and pseudotime near 1.0). C) Imputed expression trends of key signature genes over pseudotime as calculated by Palantir. Separate differentiation arms (PMAC, MK, ERY) increasingly express key markers of ERY (GYPA, HBE1), MK (ITGA2B, ITGB3, GP1BA), and PMAC (CX3CR1, CSF1R, SPI1, PTPRC) identity over pseudotime. PMAC (PMAC arm, green), MK (megakaryocytic arm, pink), and ERY (erythrocyte arm, orange). n = 6743 cells. Error bars = SD. D) Heatmap of imputed gene expression data from cells along the PMAC trajectory ordered by pseudotime compared to mouse yolk sac primitive EMP and PMAC gene signatures25 shows higher expression of primitive EMP genes between pseudotimes 0.16 – 0.8 (marked by blue bar), corresponding to the in vitro human primitive EMP clusters, and higher expression of PMAC genes between pseudotimes 0.8 – 1 (green bar), corresponding to the in vitro human PMAC clusters. Gene expression was re-scaled per gene to fall between 0 and 1. E) Force-directed graph layout co-embedding of the in vitro human data (dark grey) with mouse gastrulation data (light grey) shows close mapping between the human PMAC cluster and the mouse My (myeloid) cluster. Mouse clusters are Hm (hemogenic endothelium), Bp (blood progenitor), Mk (megakaryocyte), Ery (erythrocyte), and My (myeloid). F) Mouse gastrulation data taken from distinct timepoints in mouse development (E6.75, E7.0, E7.25…) is highlighted in blue over the co-embedded force-directed graph layout of the in vitro human data (dark grey) and the mouse gastrulation data containing all timepoints (light grey). At E8.5, mouse gastrulation clusters are enriched (blue) in the area of the force-directed layout to which the in vitro human PMAC cluster most closely maps.
We compared the composition and trajectories of our differentiation to data from in vivo mouse development to verify whether we are indeed producing microglial precursors in a dish. We isolated the expression profiles of the cells along only the macrophage precursor arm and compared their gene expression to the gene signatures of primitive-EMPs and pre-macrophages (microglial precursors known as PMACS) derived from in vivo profiling of mouse microglial development25 (Fig. 2D). Indeed, at pseudotimes of 0.16–0.8 which correspond to the portion of the trajectory in which the early primitive-EMP and primitive-EMP/PMAC clusters fall (Fig. 2A, B), our cells show higher expression of the mouse primitive EMP signature. At pseudotimes of 0.8 and above which correspond to the mature PMAC1/2 clusters, our cells show higher expression of the mouse PMAC signature25. These trends were confirmed using unimputed data as well (Extended Data 2A). We note that of the 141 genes present in the mouse EMP and pMAC signatures, 130 genes were present in our scRNAseq dataset. A subset of these genes was separately expressed in the human primitive EMP or PMAC clusters with some genes expressed in both (Fig. 2D), likely representing human to mouse, species-specific differences. To demonstrate specificity, we isolated the expression profiles of cells along the megakaryocyte arm or the erythrocyte arm and compared those genes to the gene signatures of mouse primitive EMPs and PMACs. We noticed a slight increase in the expression of primitive EMP genes but no increased expression in PMAC genes, showing that the stage-specific enrichment of the primitive EMP and PMAC signatures along the macrophage trajectory is unique to this lineage (Extended Data 2B–C).
We also mapped our data onto whole mouse gastrulation scRNA-seq data26 by using the MNN batch correction algorithm27. We found that our PMAC cluster maps near the mouse gastrulation myeloid cluster (Fig. 2E) and maps most closely to populations which emerge at E8.5, 1 day before microglial precursor cells initially seed the mouse brain25 (Fig. 2F). Taken together, these data demonstrate the presence of primitive EMPs and EMP-derived PMACs in our cultures which closely correlate to mouse myeloid cells at the time of early microglial development, indicating that our in vitro hPSC differentiation strategy follows the postulated developmental roadmap for microglia.
Two methods to generate microglia from hPSCs
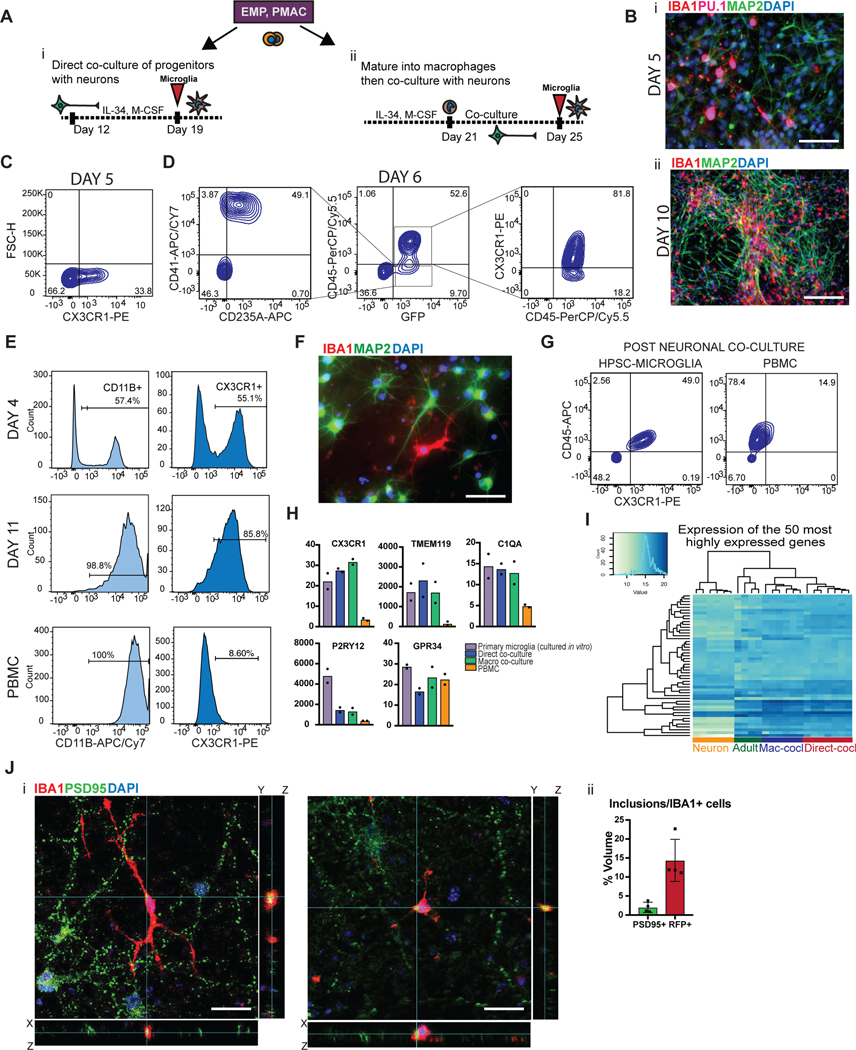
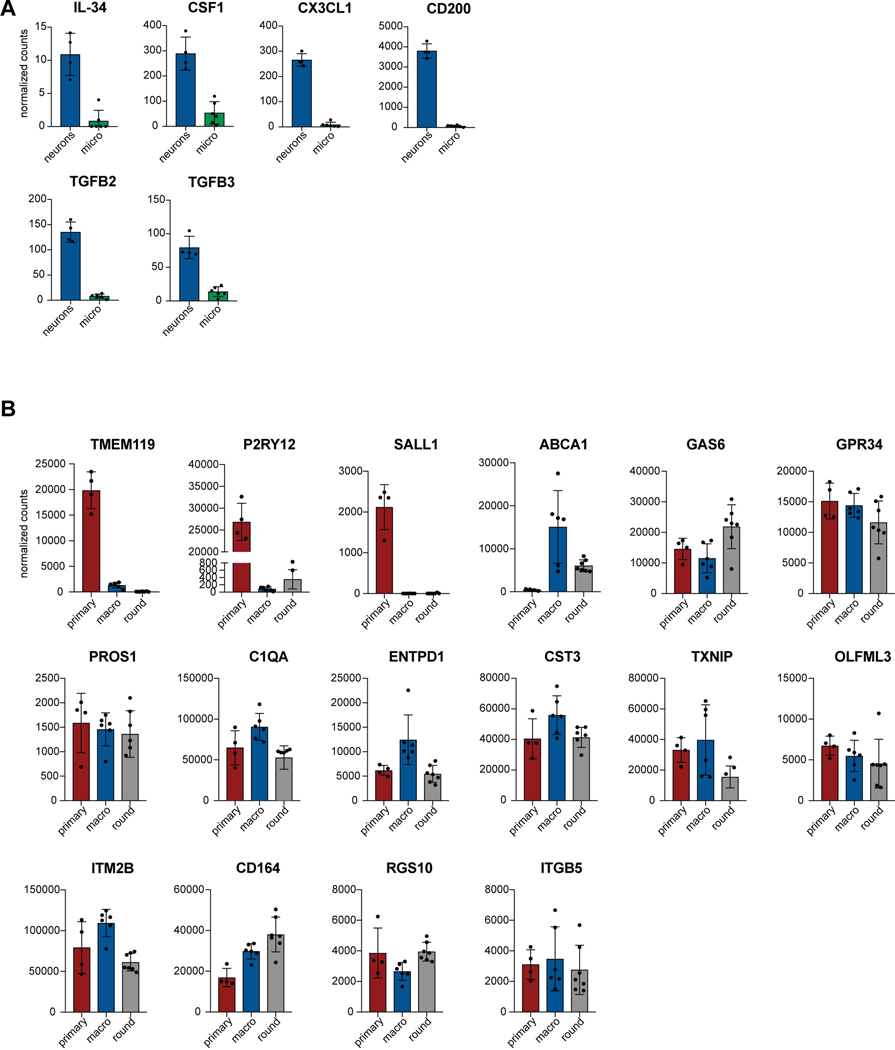
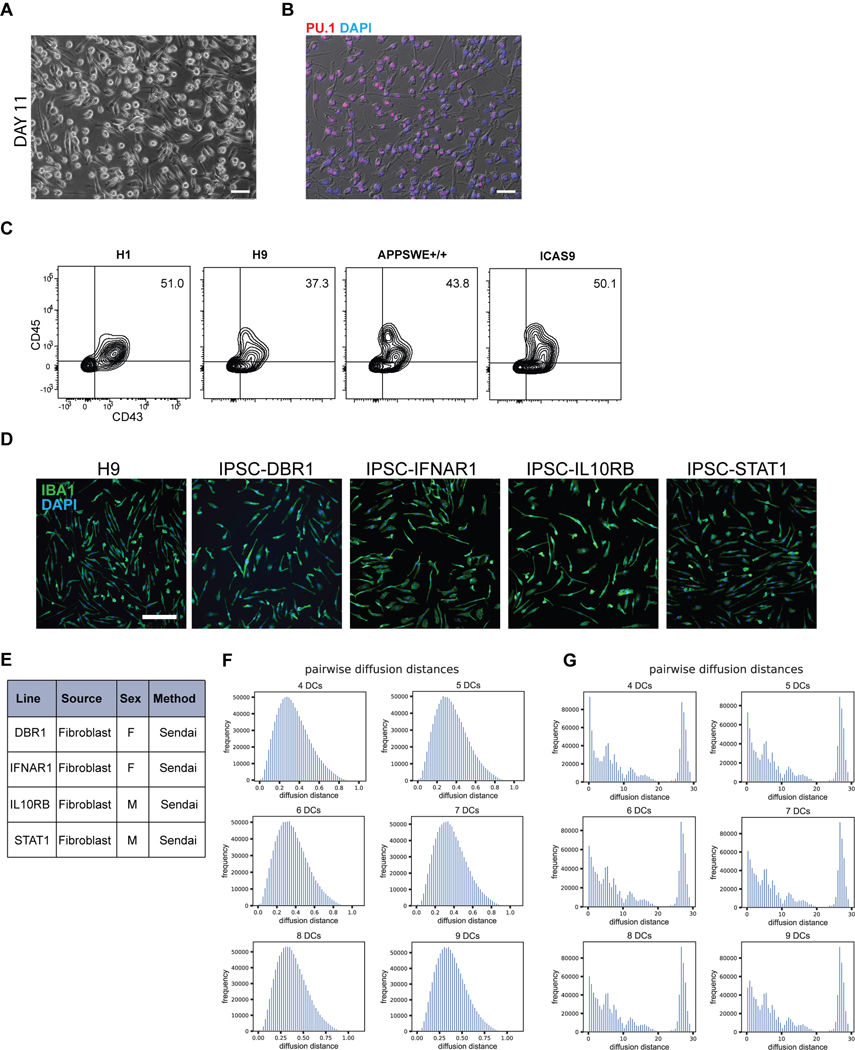
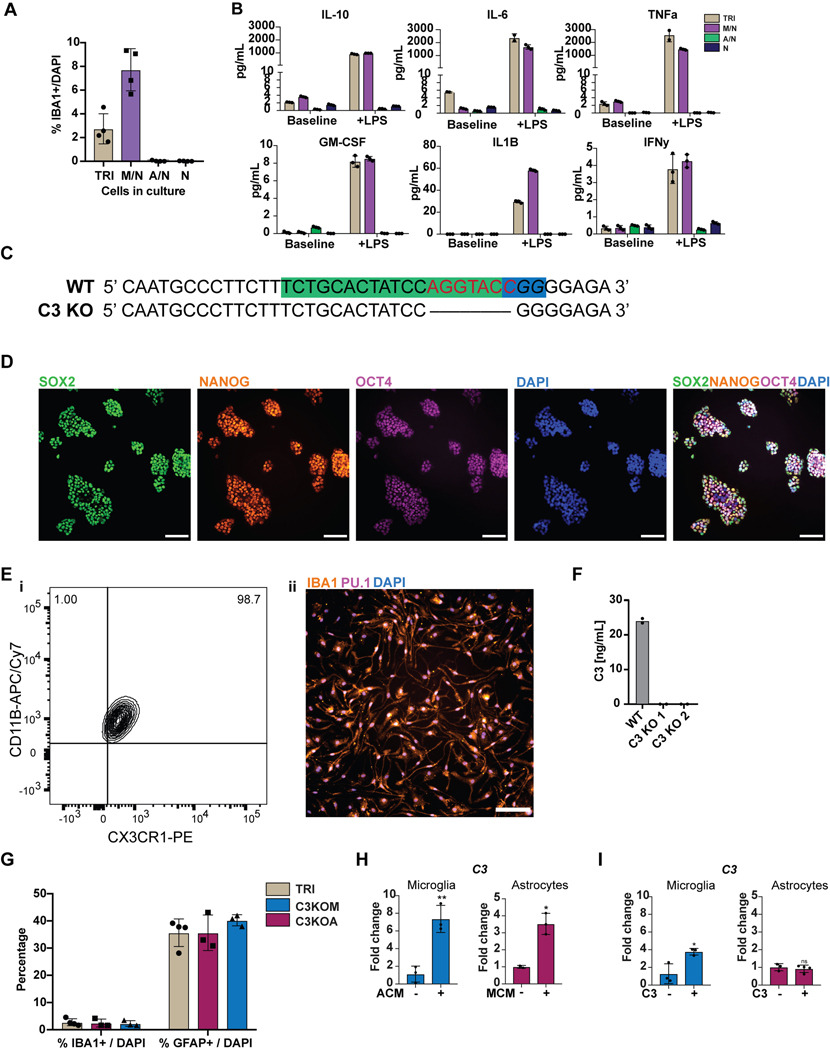
We next established two separate methods to functionally mature primitive EMPs or PMACs into microglia (Fig. 3A). The first method mimics the in vivo developmental trajectory during which microglial precursors seed the brain and develop into microglia within the neural environment25. To recapitulate this paradigm, we harvested cells in suspension at day 10 and directly co-cultured them with post-mitotic, day 30 hPSC-derived cortical neurons in the presence of IL-34 and M-CSF, cytokines important for microglial survival and maturation28 (Fig. 3Ai). Additionally, RNAseq of the hPSC-derived cortical neurons shows that they themselves express multiple cytokines which can support microglial maturation, including IL-34, CSF1, CX3CL1, CD200, and expression of TGFB2, which has been specifically identified as a growth factor critical for maintaining a homeostatic phenotype for microglia in culture29 (Extended Data 3A). 50,000 cells in suspension were seeded per 300,000 cortical neurons, yielding ~16% hematopoietic cells in co-culture at the time of plating. Remarkably, within 4 days of co-culture, adherent, ramified, IBA1+ and PU.1+ microglial-like cells emerged (Fig. 3Bi). By 10 days of co-culture, IBA1+ cells were evenly distributed throughout the neuronal culture (Fig. 3Bii). These cells were also CX3CR1+ and made up more than 30% of the co-cultures as early as day 5, indicating ongoing cell proliferation (Fig. 3C).
Figure 3. Two different methods to derive hPSC-microglia from the PMAC stage that are molecularly and functionally similar to primary microglia.
A) Schematic for two methods to derive microglia from PMACS. i) Day 10 microglial progenitors are co-cultured with postmitotic hPSC-derived neurons for 1 week in the presence of IL-34 and M-CSF. ii) Day 10 microglial progenitors are matured into primitive macrophages with IL-34 and M-CSF, then co-cultured with hPSC-derived neurons for 4 days. B)i. IF shows ramified IBA1+PU.1+ cells after day 5 of direct co-culture of day 10 progenitors with postmitotic hPSC-derived cortical neurons. Scale bar = 50 μM. ii. IF shows IBA1+ cells distributed evenly throughout the neuronal culture by day 10 of co-culture. Scale bar = 200uM. C) FACS analysis shows that over 30% of cells in co-culture at day 5 express CX3CR1+. D) FACS analysis shows that GFP+ day 10 progenitors co-cultured with hPSC-derived cortical neurons for 6 days are ~84% CD45+ indicating commitment to the microglial lineage. Of these over 80% are CX3CR1+, indicating maturation into early microglia. The ~15% GFP+ population that is CD45- is ~50% CD41+CD235A+ (primitive EMPs), and ~50% uncommitted. E) FACS analysis shows that maturing Day 10 progenitors in IL-34 and M-CSF without neurons yields a progressively pure population of primitive macrophages expressing CD11B (~99%) and CX3CR1 (>85%) by 11 days in culture. PBMCs matured in parallel express CD11B (100%) but not CX3CR1+ at day 11. F) IF shows ramified IBA1+ microglial-like cells after culturing primitive macrophages with hPSC-derived cortical neurons for 4 days. Scale bar = 50 μM. G) FACS analysis shows that co-cultured microglial cells maintain expression of CX3CR1 and have a lower expression of CD45 than PBMC-matured macrophages co-cultured with cortical neurons, which largely do not express CX3CR1 and have higher CD45 expression. H) qPCR of a panel of microglial-specific genes shows that hPSC-microglia generated from either method express these genes at similar levels to human primary microglial cDNA (Celprogen, commercially available), whereas PBMC-derived macrophages do not. Fold change is over day 10 progenitors for CX3CR1, TMEM119, C1QA, GPR34, and over hESCs for P2RY12. n=2 technical replicates of a representative qPCR. I) Bulk RNA-sequencing of microglia derived from 2 different methods (Mac-cocl = method ii, matured alone then co-cultured, Direct-cocl = method i, direct co-culture) show similarity to each other and to acutely isolated adult primary microglia from postmortem samples. hPSC-derived neurons sorted from co-culture grouped separately. n=3 samples per group. J) i) Confocal imaging shows microglia co-cultured with d70 hPSC-derived cortical neurons for 30 days contain inclusions of postsynaptic proteins (PSD95). Scale bar = 50 μM. ii) Quantification of inclusions show that inclusions containing general neuronal matter (tagged with RFP) are greater in volume than inclusions containing postsynaptic protein (PSD95). n=4 fields. Error bars = SD, center = mean.
To address whether other primitive hematopoietic lineages emerge in these co-cultures, we generated GFP+ primitive hematopoietic cells from a constitutively active nuclear GFP hPSC line (MEL1 GPI:H2B-GFP) (Supplementary Figure 1) and co-cultured these cells with hPSC-derived cortical neurons. At day 6 of co-culture, we found that the majority of GFP+ cells were CD45+ indicating that they differentiated along a microglial rather than megakaryocytic or erythrocytic trajectory. Of these, 82% expressed CX3CR1, indicating that most CD45+ cells had already transitioned towards microglia (Fig. 3D). Approximately 15% of GFP+ cells were not CD45+. Half of those cells were immature CD41+CD235A+ primitive EMPs, whereas the other half were negative for these markers, possibly indicating an even earlier hematopoietic-committed stage. These data demonstrate that co-culturing day 10 primitive EMPs and PMACs with cortical neurons yields a robust population of microglial cells within 4 days, though small populations of uncommitted hematopoietic cells persist.
To derive an even more pure and synchronized population of microglia, we developed a second strategy of maturing microglia from the progenitor stage without co-culture (Fig. 3Aii). We took the bulk population of hematopoietic cells in suspension at day 10 followed by exposure to either serum containing medium (RPMI + 10% serum with the addition of IL-34 and M-CSF) or by using defined, serum-free conditions (IMDM/F12 with the addition of IL-34 and M-CSF) for 7 – 11 days. At 4 days of culture, half of the cells had transitioned to the primitive macrophage stage, with 50–60% expressing CD11B (mature macrophage/microglial marker) and CX3CR1 (restricted to tissue-resident macrophages such as microglia)30. By 11 days of culture, close to 99% of the cells expressed CD11B and over 85% expressed CX3CR1 (Fig. 3E). At this stage, all cells were adherent, displayed an elongated morphology, and were PU.1+ (Extended Data 4A, B). In contrast, primary human peripheral blood mononuclear cells (PBMCs), matured in parallel under the same culture conditions, expressed CD11B but largely lacked CX3CR1 expression (Fig. 3E). We found this protocol to be highly reproducible in more than 10 different hPSC lines, showing high CD45+ efficiency at Day 10 of differentiation and resulting in a pure population of IBA1+ primitive macrophages by Day 21 (Extended Data 4C–E).
We co-cultured the resulting pure population of primitive macrophages with hPSC-derived cortical neurons to fully transition these cells to microglia. After 4 days of co-culture, the microglial cells appeared ramified and continued to be IBA1+ (Fig. 3F). When compared to matured PBMCs that were co-cultured with neurons, the microglial-like cells had lower levels of CD45 and maintained CX3CR1, whereas the PBMC-derived cells were CD45+ in the absence of CX3CR1 (Fig. 3G). Additionally, single-cell RNA sequencing on the sorted hPSC-derived microglial cells revealed that they represent a homogenous cell population devoid of undifferentiated precursors (Extended Data 4F,G). Pairwise distances calculated between cells in the microglial sample after diffusion map embedding fall in a clean unimodal distribution, indicating transcriptional variance among these cells is centered around one rather than multiple phenotypes (Extended Data 4F). In contrast, the distribution of pairwise diffusion distances calculated between cells at day 10, prior to co-culture, show multiple distant peaks indicating the heterogenous nature of these cells (Extended Data 4G).
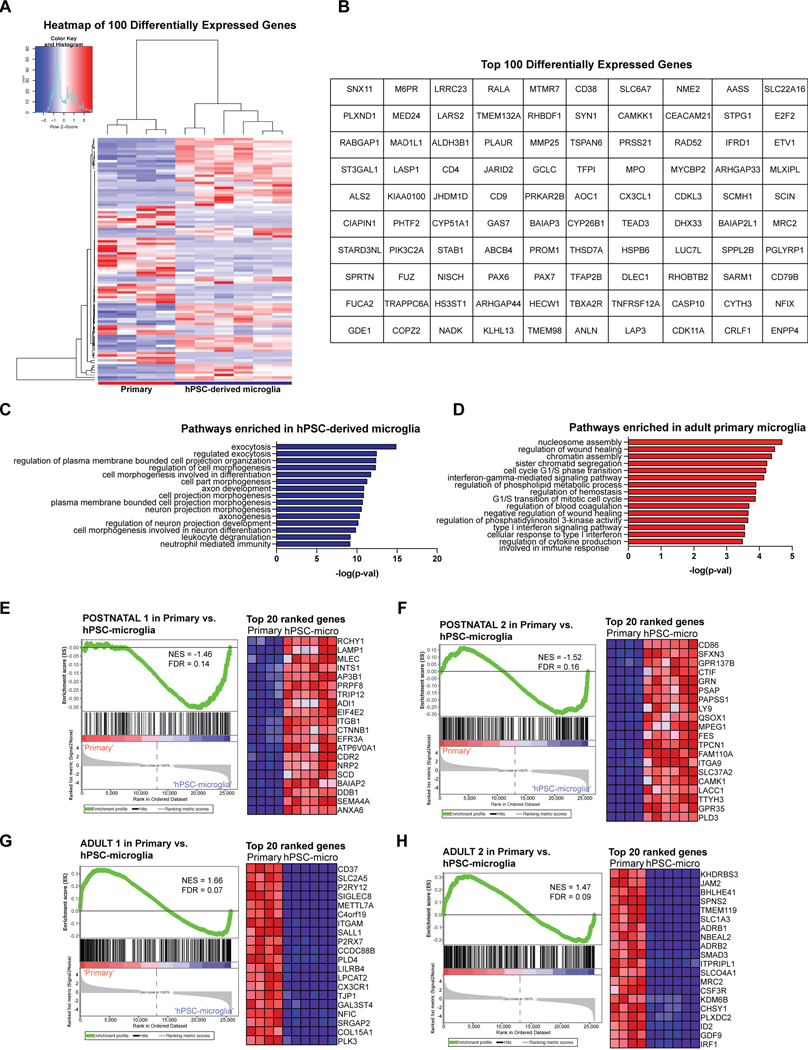
We next asked whether either of the two derivation methods yielded cells transcriptionally more similar to primary human microglia. We assessed the expression of signature microglial genes in the hPSC-derived microglial cells sorted from co-culture after 14 days using CD45+CX3CR1+ as compared to primary microglial cells cultured in vitro (cDNA from Celprogen)31,32 (Fig. 3H). hPSC-derived microglial cells derived from either method expressed the signature microglial genes of TMEM119, C1QA, CX3CR1, and GPR34 at similar levels to the primary microglia cultured in vitro, whereas PBMCs did not express these markers. We also compared hPSC-derived microglia to acutely isolated primary adult human microglia from postmortem tissue via bulk RNA sequencing. Following unsupervised hierarchical clustering, we found that both methods yielded microglial cells which cluster with primary human microglia obtained from postmortem cortical brain tissue (frontal and temporal, aged 60–77 years old) (Fig. 3I). However, neither method falls exactly in the same sub-branch as the acutely isolated adult primary human microglia. GO pathway analysis on the genes differentially expressed between the hPSC-derived microglia and acutely isolated adult primary microglia (Extended Data 5A,B) revealed that the adult microglia were enriched in cell cycle transition and immune activation pathways, potentially due to the fact that these cells were isolated from an in vivo, aged environment, whereas hPSC-derived microglia were enriched in cell differentiation and projection neuron developmental pathways, potentially due to contaminating hPSC-derived cortical neuronal cells sorted from co-culture (Extended Data 5C,D). We also performed GSEA using 7 stage-specific signatures of developing microglia33 to test whether the hPSC-derived microglial cells are at an earlier developmental stage than the acutely isolated adult primary microglia. Indeed, we found that the hPSC-derived microglia enriched for the postnatal microglial gene signatures, whereas the acutely isolated adult primary microglia enriched for the adult microglial gene signatures, further explaining why hPSC-derived microglia did not cluster exactly with acutely isolated adult primary microglia (Extended Data 5E–H). Finally, we also compared the expression of signature microglial genes in hPSC-derived microglia to those in the acutely isolated adult primary microglia using normalized counts from the RNA-sequencing data (Extended Data 3B). The majority of microglial-specific markers were expressed in the hPSC-derived microglia at similar levels to those in acutely isolated adult microglia (GAS6, GPR34, PROS1, C1QA, to name a few). However, TMEM119, P2RY12, and SALL1 were expressed at much lower levels in in the hPSC-derived microglia than the acutely isolated adult primary microglia. We postulate that this is due to the in vitro culture of the hPSC-derived microglial cells because these genes have been shown to be downregulated during in vitro culture34, and we have shown that our cells express more similar levels of these genes when compared to in vitro cultured primary microglia (Fig. 3H). We also postulate that the differences in TMEM119, P2RY12, and SALL1 expression come from the postnatal-like stage of the hPSC-derived microglia as compared to the acutely isolated adult microglia. In fact, TMEM119, P2RY12, and SALL1 are all found in the Adult 1/2 panel of gene signatures33 (Extended Data 5G,H).
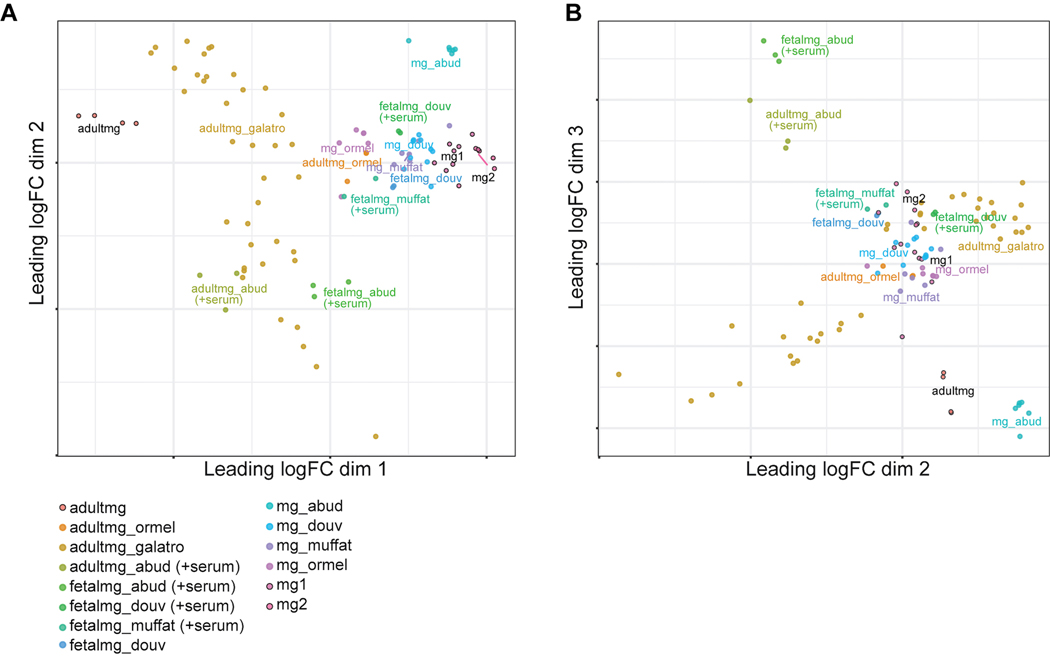
Finally, we compared the transcriptome of our hPSC-derived microglia to those of previously published microglial differentiations with publicly available RNA-seq datasets10–12,35 using multidimensional scaling (MDS) following the methodology in Grubman et al36 (Extended Data 6A,B). We also included the transcriptomes of the fetal and adult primary microglia used as comparisons in these studies, and we added the datasets of an additional study which specifically profiled acutely isolated adult primary microglia from human postmortem brain tissue37. In our analysis, dimension 1 vs. 2 separated the adult primary microglia from the fetal primary microglia/hPSC-derived microglia (Extended Data 6A), whereas dimension 2 vs. 3 separated out the serum-cultured fetal and adult primary microglia used in Abud et al11 (Extended Data 6B). We found that our hPSC-derived microglia (mg1 or mg2) clustered closely to the microglia differentiated from other protocols in both dimension 1 vs. 2 and 2 vs.3, and nearer to the fetal microglia as opposed to the adult microglia (Extended Data 6A). This suggests that our hPSC-derived microglia are highly similar to published differentiated microglia and are also more similar to fetal primary microglia than adult.
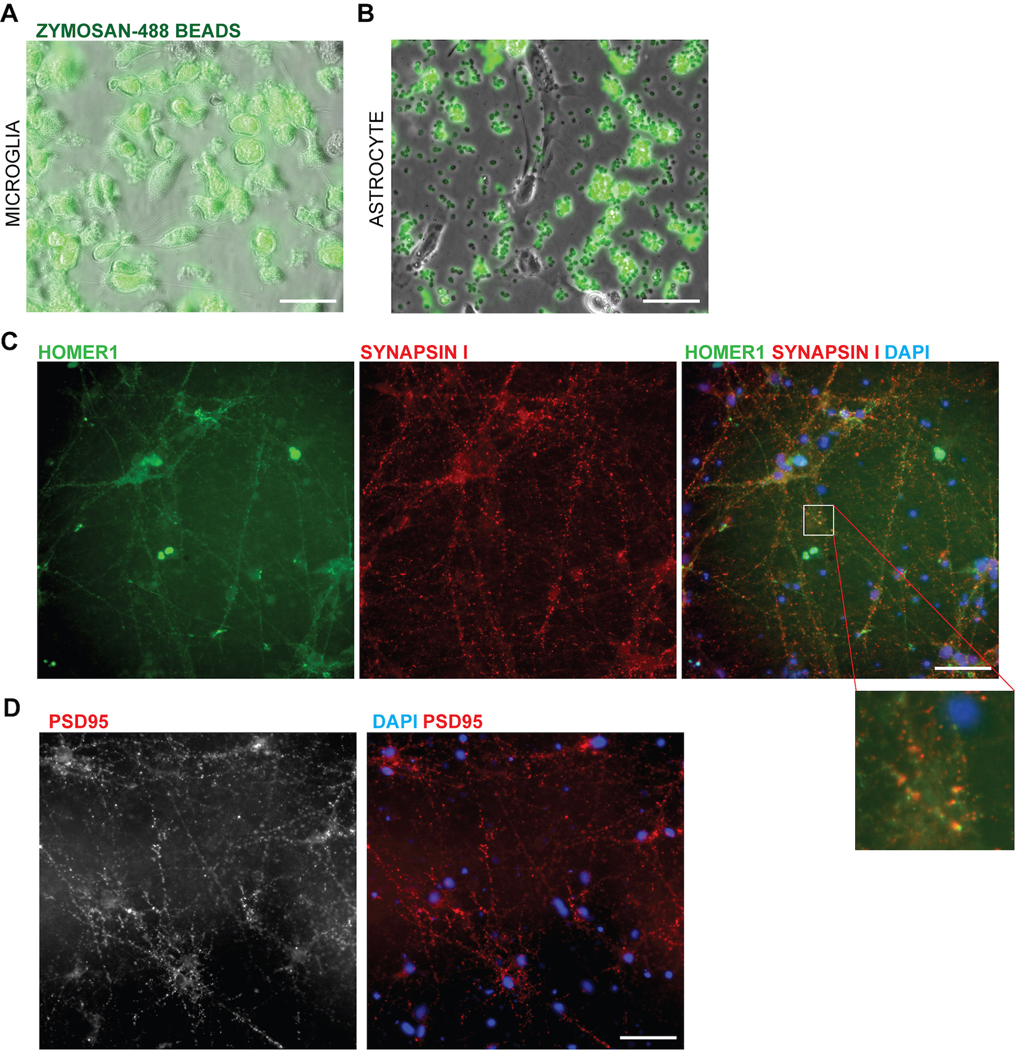
We next evaluated functional similarities between hPSC-derived microglial cells and primary microglia. We observed that hPSC-derived microglial cells in co-culture with neurons survey their environment, retracting and extending their processes to sample the surrounding neurons akin to homeostatic microglia in vivo38 (Supplementary Video 1, 2). When challenged with the yeast-antigen zymosan, the cells were also able to perform efficient phagocytosis39 as compared to an astrocyte control (Extended Data 7A–B, Supplementary Video 3). Finally, another role of microglial cells is to prune synapses in the developing brain40. When co-cultured with mature hPSC-derived neurons forming synapses (day 70 and older; (Extended Data 7C,D)), microglial cells showed inclusions containing synaptic material upon confocal imaging (Fig. 3Ji). While there were more inclusions that contained general neuronal material tagged with RFP, the number of inclusions specifically composed of synaptic materials was 1–2% (Fig. 3Jii), resembling the basal level of synaptic uptake reported for primary microglial cells during homeostasis41.
Tri-culture system models neuroinflammatory axis between microglia and astrocytes
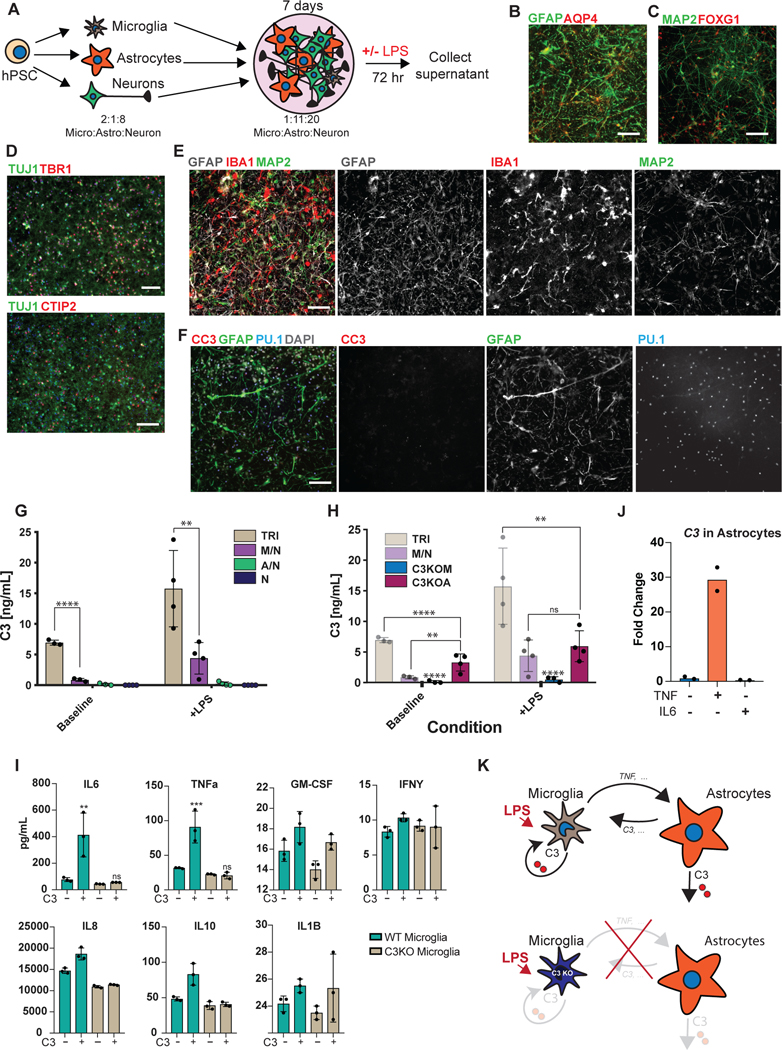
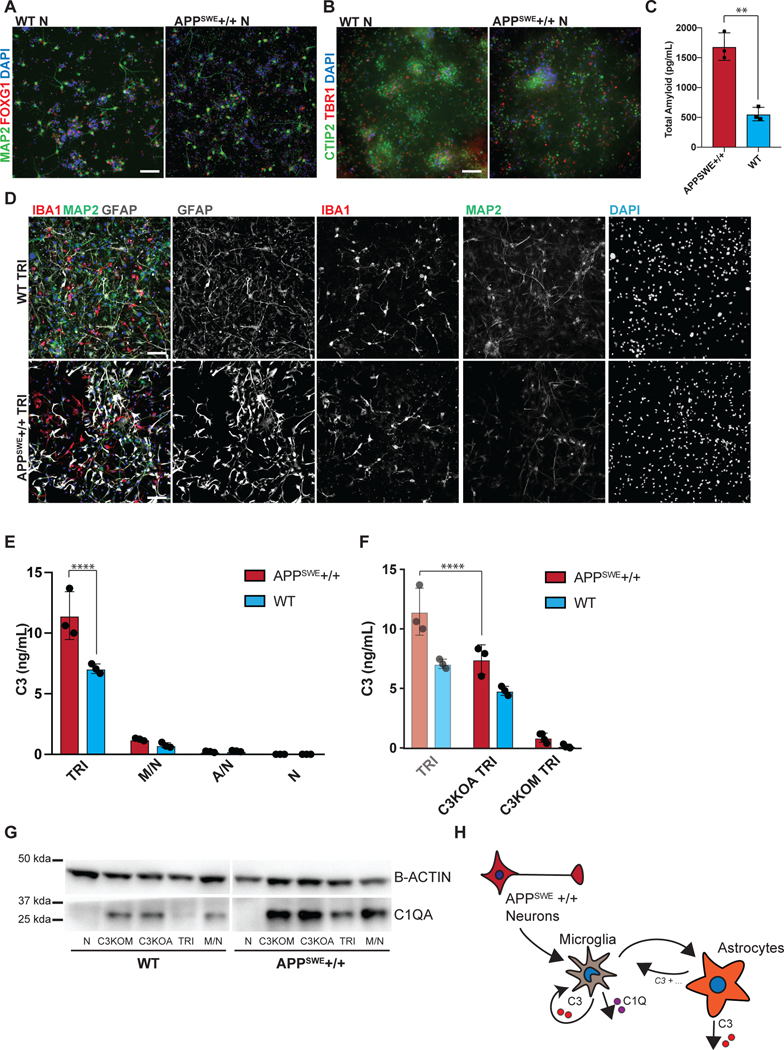
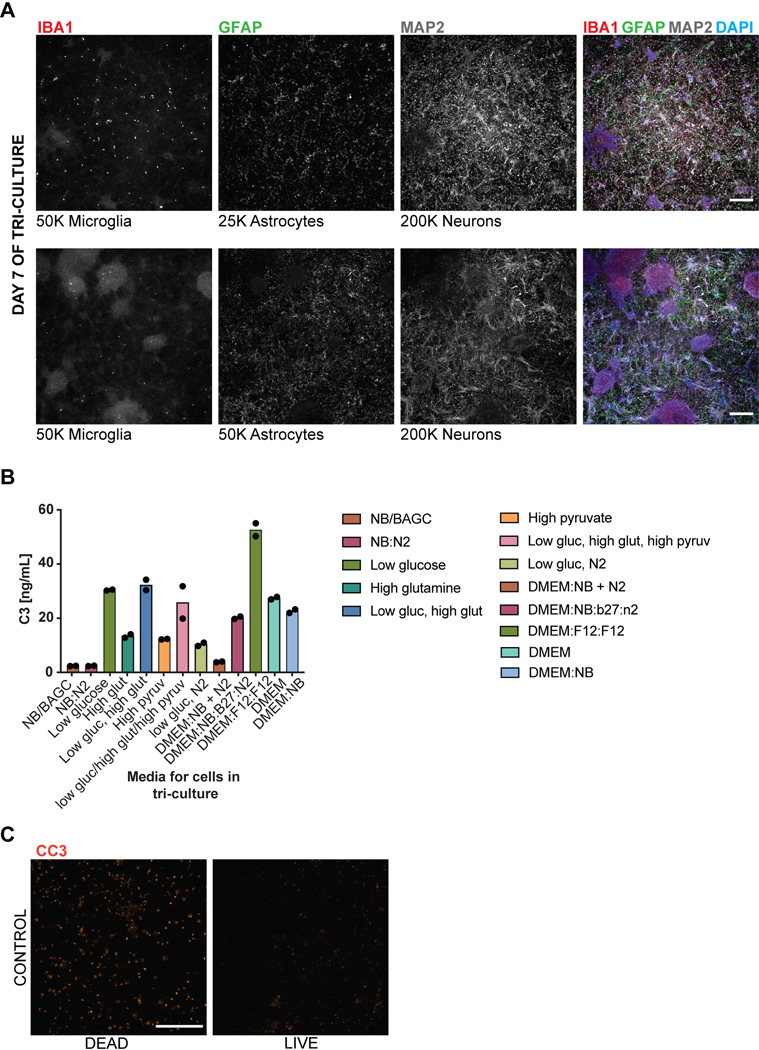
The ability to efficiently generate nearly pure microglia within 25 days of differentiation, with properties that are similar to their primary counterpart, set the stage to build a functional, fully hPSC-derived tri-culture platform composed of human microglia combined with similarly pure populations of human astrocytes9 and cortical neurons7,8 (Fig. 4A). The hPSC-derived astrocytes were generated as described recently9 and all expressed GFAP with a subset expressing both AQP4 and GFAP (Fig. 4B). Likewise, we generated pure hPSC-derived neurons that were of cortical neuronal identity expressing the telencephalic marker FOXG1 and the deep layer markers TBR1 and CTIP2 (Fig. 4C, D). For establishing a tri-culture platform, we identified the optimal ratio of each cell type at initial plating at 2:1:8, with 50,000 microglia:25,000 astrocytes: 200,000 neurons per cm2. Those conditions allowed for microglial cell attachment and survival in the presence of astrocytes, as increased astrocyte numbers interfered with the attachment of microglial cells (Extended Data 8A). After 1 week of tri-culture, cell ratios stabilized in which the microglial number decreased as not all plated cells attached, and the astrocyte number increased due to proliferation, resulting in a final ratio close to 1:11:20 of microglia:astrocytes:neurons. Base media culture conditions were optimized, in the presence of IL-34 and M-CSF, to reduce production of baseline inflammatory cytokines using complement C3 production as readout (Extended Data 8B). The NB/BAGC condition resulted in very low baseline C3 induction with excellent neuronal survival and maintenance8. After one week, tri-cultures showed ramified IBA1+ microglial cells and many GFAP+ astrocyte processes interacting with MAP2+ cortical neurons (Fig. 4E). The tri-cultures were largely devoid of any apoptotic, Cleaved Caspase 3 (CC3)+ cells confirming robust survival of all three cell types (Fig. 4F, Extended Data 8C).
Figure 4. hPSC-derived microglia cultured with hPSC-derived astrocytes and neurons builds a functional human tri-culture system that allows for the modeling of the neuroinflammatory axis in vivo.
A) Schematic of tri-culture differentiation. hPSC-derived microglia, astrocytes, and neurons are initially plated at a ratio of 2:1:8 which then stabilizes to a ratio of ~1:11:20 microglia (~2–3%), astrocytes (~35%), and neurons at the time of assay (1 week). B) IF shows that hPSC-derived astrocytes are GFAP+ and some are also AQP4+. Scale bar = 100uM. C) IF shows that postmitotic hPSC-derived neurons express FOXG1. Scale bar = 100uM. D) IF shows that day 50 hPSC-derived neurons express TBR1 and CTIP2. Scale bar = 40uM. E) IF of tri-culture shows IBA1+ microglia and GFAP+ astrocytes interacting with MAP2+ neurons. F) IF of tri-culture shows minimal CC3 staining. Scale bar = 100 μM in (E) – (F) G) ELISA shows that tri-cultures (TRI) have increased C3 as compared to microglia/neuron only (M/N) cultures, which is exacerbated upon LPS treatment. C3 is not present in astrocyte/neuron (A/N) and neuron only (N) cultures. n = 4 samples (distinct culture supernatants), 1-way ANOVA with Tukey’s post-hoc test, **** p < 0.0001, F = 53.10, df=5; ** p= 0.0039, F=16.41, df = 5. Error bars = SD, center = mean. H) ELISA shows that C3KOA tri-cultures have less C3 as compared to WT tri-cultures but more than M/N cultures. C3KOM cultures have minimal C3. Upon LPS treatment, C3KOA tri-cultures have less C3 release as compared to WT tri-cultures; C3KOM again have minimal C3 release. n=4 samples (distinct culture supernatants), 1-way ANOVA with Tukey’s post-hoc test, **** p< 0.0001, ** p=0.0032, F= 53.10, df=5 for baseline tests; ** p= 0.0028, F=16.41, df = 5. Translucent bars (TRI, M/N) represent data originally presented in (G). Error bars = SD, center = mean. I) C3 (1ug/mL) induces IL-6 and TNF-a in wildtype microglia but not C3KO microglia. GM-CSF, IFN-g, and IL-1B are in contrast not induced upon C3 addition. ** p= 0.0022; *** p=0.0005, n= 3 cell culture supernatants, 1-way ANOVA with Sidak’s post-hoc test, Error bars = SD, center = mean. J) TNF (100ng/mL) but not IL-6 (100ng/mL) added for 48 hr to astrocytes induces C3 expression normalized to untreated astrocytes. n=2 independent experiments. K) Neuroinflammatory loop in tri-culture is initiated by LPS-activated microglia signaling to astrocytes which signal back to microglia leading to increased C3 release. Mediators include TNF secreted by stimulated microglia inducing C3 in astrocytes, and C3 secreted by both astrocytes and microglia inducing C3 in microglia.
To test whether the triculture can recapitulate a neuroinflammatory axis between microglia, astrocytes, and neurons, we challenged our system with the inflammatory stimulus lipopolysaccharide (LPS) to pharmacologically model a neuroinflammatory state42. We focused on the production of complement C3 as a surrogate marker of neuroinflammation, because recent literature suggests that it is increased during several neuroinflammatory states including aging43 and neurodegenerative disorders such as AD44. In AD, C3 is directly implicated in causing aberrant synaptic pruning45,46. We first assessed C3 secretion by ELISA under various co-culture conditions using established plating ratios (Fig. 4A, materials & methods): tri-culture, microglia and neurons, astrocytes and neurons, and neurons only.
At baseline, C3 was only present in cultures that contained microglia while C3 levels were extremely low in astrocyte/neuron co-cultures and undetectable in neuron only cultures. Interestingly, in tri-cultures, the baseline C3 levels were dramatically higher than in microglia/neuron only cultures, suggesting a potentiation of C3 secretion via cellular cross talk in the presence of both microglia and astrocytes (Fig. 4G). After LPS treatment, C3 was increased in all cultures containing microglia, but again greatly potentiated under tri-culture conditions (Fig. 4G). To rule out the possibility that such potentiation could simply reflect an increase in microglial numbers, we quantified IBA+ cells by immunofluorescence using a high-content imaging microscope. IBA1+ positive cells were actually somewhat decreased in tri-cultures versus microglia/neuron co-cultures ruling out an increase in microglial numbers as the cause of higher C3 levels in tri-culture (Extended Data 9A). We further corroborated that LPS stimulation induces an inflammatory state beyond C3 secretion by observing increased levels of classical inflammatory cytokines by ELISA such as IL-6, TNF-a, IL-1B, IFN-g, and GM-CSF, of which IL-6 and TNF show potentiation in tri-culture as well (Extended Data 9B).
To define the key cellular players contributing to C3 potentiation in tri-culture, we next generated a C3 KO hPSC line using CRISPR/Cas9 (Extended Data 9C, D) which showed a complete lack of C3 production after differentiation to microglia (Extended Data 9E, F). We differentiated this line into C3 KO astrocytes and C3 KO microglia and generated tri-cultures that contained C3 KO astrocytes, wildtype microglia, and wildtype neurons (C3KOA) or wildtype astrocytes, C3 KO microglia, and wildtype neurons (C3KOM). The number of microglia scored by % IBA1/DAPI (3–5%) and the number of astrocytes scored by % GFAP/DAPI (>30%) were similar across the wildtype tri-culture, C3KOA, and C3KOM cultures (Extended Data 9G).
C3KOA cultures at baseline and upon LPS stimulation showed reduced C3 levels as compared to WT tri-cultures but higher levels than microglia/neuron only cultures, indicating that both astrocytic C3 and increased microglial C3 contribute to overall C3 potentiation in WT tri-culture (Fig. 4H). These data demonstrate reciprocal signaling between microglia and astrocytes in tri-culture, in which microglia activate astrocytes, and astrocytes re-activate microglia. We further corroborated evidence for such reciprocal signaling by testing whether conditioned medium from astrocyte/neuron or microglia/neuron co-cultures can induce C3 expression in microglia or astrocytes respectively. Indeed, microglia-conditioned medium (MCM) induced C3 expression in astrocytes, indicating the ability of microglial-secreted factors to activate astrocytes, and astrocyte-conditioned medium (ACM) induced C3 expression in microglia, indicating the ability of astrocyte-secreted factors to activate microglia (Extended Data 9H).
Interestingly, in C3KOM cultures at baseline or stimulated with LPS, C3 levels were very low, suggesting that microglia must express C3 in order to effectively induce astrocytes to secrete C3 in tri-culture (Fig. 4H). We asked whether this is due to C3 itself acting as an inductive factor for C3 expression. We added C3 to astrocyte/neuron cultures and found that it did not induce C3 expression in astrocytes, however, adding C3 to microglia/neuron cultures did increase C3 expression (Extended Data 9I). These data indicate that C3 is itself a feed-forward mediator of C3 expression in microglia, which is normally secreted by both microglia and astrocytes in tri-culture. We next asked whether the collapse of C3 in C3KOM tri-cultures could also be due to an altered state in C3KO microglia that is less primed to activation, such that C3KO microglia act as a “resistor” within the inflammatory loop. Indeed, when examining a panel of cytokines secreted by WT or C3KO microglia at baseline or upon the addition of inflammatory C3, we found that C3KO microglia were unable to induce secretion of two inflammatory cytokines, TNFa and IL-6 (Fig. 4I). Conversely, TNF-a but not IL-6 was able to directly induce C3 expression when added to astrocyte/neuron cultures, identifying it as one of the inducers of astrocytic C3 secreted by microglia upon activation (Fig. 4J).
These data characterize cellular crosstalk between microglia and astrocytes in an inflammatory loop present at baseline and exacerbated upon pharmacological induction of a neuroinflammatory state. In this loop, C3-producing microglia are the initiating cell which signal to astrocytes to produce C3, which in turn re-induce microglia to produce more C3. Among the milieu of mediators which facilitate this crosstalk, we identified C3 secreted from microglia and astrocytes as one such feed-forward inducer of C3 in microglia, as well as TNF-a secreted upon microglial activation as a C3 inducer in astrocytes (Fig. 4K). These studies demonstrate the power of our hPSC-derived tri-culture system to identify an emergent cellular property upon culture of all 3 cell types together (C3 potentiation), the major cellular contributions to this potentiation (astrocytic activation and microglial re-activation), as well as two key cytokine mediators that facilitate this potentiation (C3, TNF-a).
Increased complement in tri-culture model of Alzheimer’s Disease
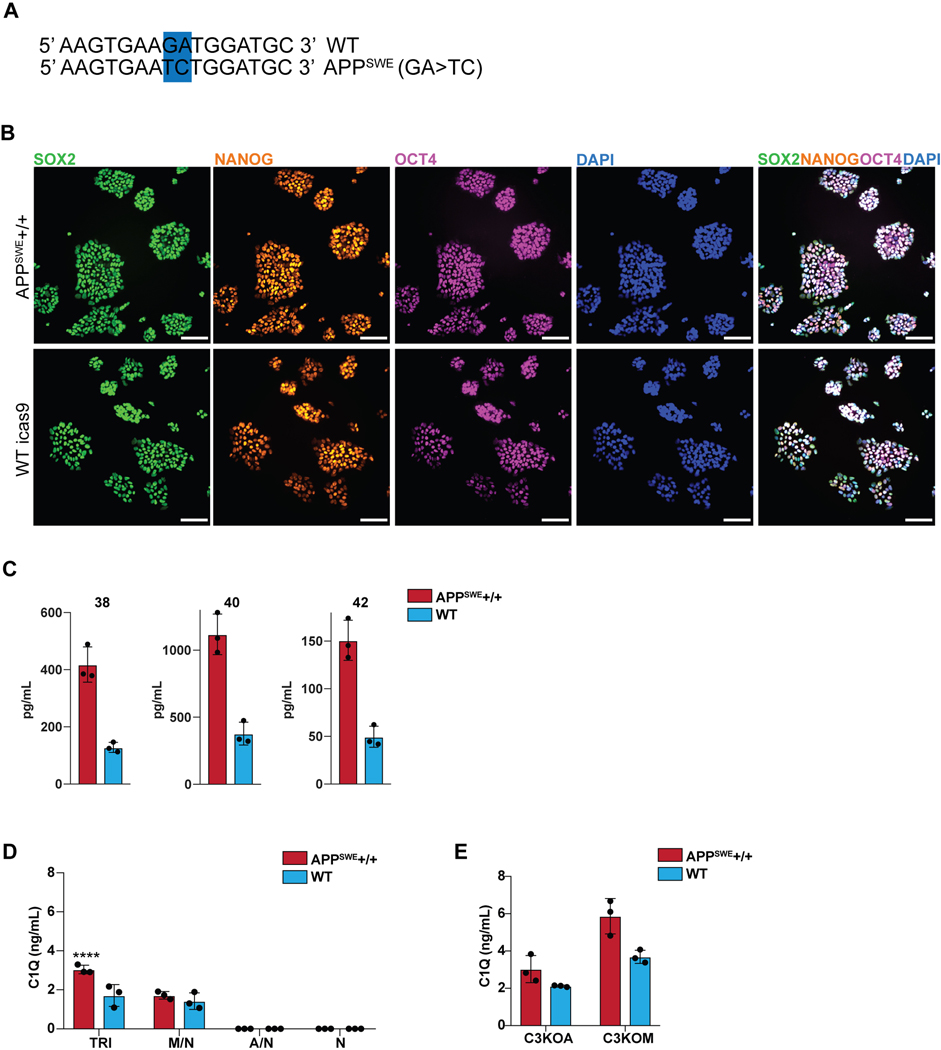
We next applied this platform to model neuroinflammation in a disease state, Alzheimer’s Disease (AD). We used a targeted human ESC line homozygous for the APPSWE mutation47 as well as its isogenic control (Extended Data 10A,B). Differentiated AD- and isogenic control hPSC-derived neurons were validated by FOXG1 and MAP2 expression as well as CTIP2 and TBR1 for cortical identity (Fig. 5A, B). The APPSWE+/+ neurons alone showed increased amyloid-beta production, a hallmark of the APPSWE+/+ model of Alzheimer’s Disease47 (Fig. 5C), with elevated levels of all three amyloid peptides (Extended Data 10C). We plated WT differentiated astrocytes (GFAP+) and WT differentiated microglia (IBA1+) in co-culture with matured APPSWE+/+ neurons (day 80) or isogenic control neurons to construct tri-cultures (Fig. 5D). After eight days, we measured the C3 levels by ELISA in tri-cultures with APPSWE+/+ neurons vs. those containing isogenic control neurons. Interestingly, C3 levels significantly increased in APPSWE +/+ neuron-containing tri-cultures (Fig. 5E). In contrast, C3 was not highly produced or increased in co-cultures of APPSWE+/+ neuron/WT astrocyte or in APPSWE+/+ neuron only cultures indicating that microglia must be present in the cultures for robust C3 production.
Figure 5. Tri-culture model of Alzheimer’s Disease shows increased C3 in AD tri-cultures caused by reciprocal signaling from microglia to astrocytes.
A) IF shows that the day 80 APPSWE+/+ neurons and isogenic control neurons express FOXG1 B) and the cortical layer markers CTIP2 and TBR1. Scale bar = 40 μM in (A) and (B). C) ELISA shows that day 50 APPSWE+/+ neurons secrete more total amyloid than isogenic control neurons. n=3 distinct culture supernatants, two-tailed t-test, ** p = 0.0016, Error bars = SD, center = mean. D) IF shows APPSWE+/+ tri-cultures with d80 APPSWE+/+ neurons (MAP2+), wildtype hPSC-derived microglia (IBA1+) and astrocytes (GFAP+), and isogenic control tri-cultures with d80 isogenic control neurons (MAP2+), wildtype hPSC-derived microglia (IBA1+) and wildtype astrocytes (GFAP+). Scale bar = 100 μM. E) ELISA shows that APPSWE+/+ tri-cultures secrete more C3 than isogenic control tri-cultures. n=3 (distinct culture supernatants), 2-way ANOVA with Bonferroni post-hoc test, **** p<0.0001. Error bars = SD, center = mean. F) ELISA shows that C3KOA tri-cultures with APPSWE+/+ neurons show lower C3 secretion as compared to APPSWE+/+ tri-cultures with wildtype astrocytes. C3KOM APPSWE+/+ tri-cultures secrete low levels of C3. n=3 (distinct culture supernatants), 2-way ANOVA with post-hoc Tukey’s test, **** p<0.0001, F=9.577, df=1. C3KOA tri-cultures with APPSWE+/+ neurons show higher levels of C3 secretion as compared to C3KOA tri-cultures with WT neurons. n=3 separate culture supernatants, 2-way ANOVA with Bonferroni post-hoc test, *** p=0.0007, F=182.9, df=5. Translucent bars (TRI) represent data originally presented in E). Error bars = SD, center = mean. G) Western blot (cropped) shows that APPSWE+/+ cultures which contain microglia show higher levels of C1QA as compared to isogenic control cultures (TRI, M/N, C3KOA, C3KOM). H) Neuroinflammatory loop schematic in an in vitro model of AD where APPSWE+/+ neurons activate microglia which activate reciprocal signaling to astrocytes leading to increased C3 release, as well as cause increased C1Q secretion and deposition.
To determine whether the source of the increased C3 in the APPSWE+/+ tri-cultures is due to astrocytes or microglia, we generated microglia and astrocytes from C3KO hPSCs and assembled tri-cultures containing C3 KO astrocytes, wildtype microglia, and APPSWE+/+ neurons or isogenic control neurons (C3KOA); or wildtype astrocytes, C3 KO microglia, and APPSWE+/+ neurons or isogenic control neurons (C3KOM). Remarkably, in C3KOA AD tri-cultures, we observed greatly reduced levels of C3 comparable to those of isogenic control tri-cultures, suggesting that some increased C3 in the AD tri-cultures is secreted by astrocytes activated by microglia. However, C3KOA AD tri-cultures still showed increased C3 levels compared to C3KOA isogenic tri-cultures as well as microglia/neuron cultures, suggesting that some increased C3 in the AD tri-cultures is secreted by microglia activated by astrocytes (Fig. 5F). In C3KOM cultures, we detected only low levels of C3 production, indicating that C3-expressing microglia must be present in order to induce robust C3 expression in APPSWE+/+ tri-cultures (Fig. 5F). These data suggest that the neuroinflammatory loop is exacerbated in tri-cultures containing AD neurons, which further trigger reciprocal signaling between microglia and astrocytes leading to the emergent property of increased C3 production when all 3 cell types are cultured together.
Upstream of C3 is C1Q, a complement protein which complexes with a cleavage product of complement C3 and tags synaptic material for clearance46. Strikingly, we found that there was an increase in C1Q protein deposition found in APPSWE+/+ cultures when compared to WT cultures (Fig. 5G), but only under co-culture conditions that include microglia. C1Q has been shown to accumulate in AD in vivo46,48, and we recapitulate this finding in our in vitro model of AD. We corroborate these data with C1Q ELISA results showing increased C1Q secretion in APPSWE+/+ tri-cultures when compared to WT tri-cultures, and the presence of C1Q secretion again only in cultures containing microglia (Extended Data 10D). These data suggest that microglia are the main source of C1Q49 in our system and that APPSWE+/+ tri-cultures have increased C1Q secretion and accumulation as compared to WT tri-cultures. C1Q has been identified as an inductive factor secreted from activated microglia that can activate astrocytes5, and in a previous study we have shown that in combination with other inflammatory cytokines, C1Q can activate hPSC-astrocytes9. Given the higher levels of C1Q secretion in APPSWE+/+ tri-cultures (Extended Data 10D), we postulate that C1Q may act as an inductive factor in AD tri-cultures (Fig. 5E) and possibly other inflammatory cytokines involved in microglia-astrocyte cellular crosstalk.
Interestingly, despite increased C1Q secretion in AD tri-cultures, the deposition of C1Q was lowest under tri-culture conditions when compared to microglia/neuron, C3KOA, and C3KOM cultures for both APPSWE+/+ and WT cultures (Fig. 5G). This seemingly contradictory finding suggests that there may be a homeostatic balance in minimizing C1Q deposition when microglia, astrocytes, and neurons are cultured together, such that microglia and astrocytes can optimally phagocytose C1Q-complexes and deplete C1Q accumulation (Fig. 5G). We speculate that if the system is perturbed, by either the addition of AD neurons, the subtraction of astrocytes, or the deletion of C3, homeostasis is disrupted leading to higher levels of C1Q accumulation. In C3KOM tri-cultures, there is increased C1Q deposition and secretion, suggesting an autocrine C3-C1q negative inhibition in microglial cells during homeostasis that is disinhibited upon C3 knockout (Extended Data 10E). In C3KOA tri-cultures, C1Q deposition is increased but not its secretion (Extended Data 10E), suggesting that there may be an altered state in C3KO astrocytes that hinders their ability to clear C1Q- complexes50, either directly or indirectly by an inability to signal to microglia to perform this function. It will be intriguing to use our culture system in future studies to further dissect the mechanisms responsible in both microglia and astrocytes for guiding the balance between C1Q secretion and deposition.
Based on the results from our novel in vitro tri-culture system, we built a model of the cellular contributions to neuroinflammation in Alzheimer’s Disease focused on complement C3 (Fig. 5H). We report increased C3 levels in AD versus isogenic control tri-cultures and reveal that this increase is due to astrocytic C3 induced by microglia as well as due to microglial C3 re-induced by astrocytes. The results point to an inflammatory loop involving APPSWE+/+ neurons which trigger microglia to induce reciprocal signaling with astrocytes. In addition, we find increased C1Q in AD cultures in the presence of microglia, as a candidate AD-specific modulator of the cellular crosstalk between microglia and astrocytes.
Discussion
These findings demonstrate that our tri-culture system enables the dissection of cellular crosstalk by genetic manipulation, and allowed us to study the mechanisms of increased complement C3 production in tri-culture upon LPS stimulation and in a model of AD. Furthermore, the technology could be readily adapted to study any other disease-relevant neuroinflammatory pathway in AD or other neurodegenerative disorders. The identification of the key cellular players contributing to the neuroinflammatory axis in humans should enable the development of directed, cell-type specific therapeutic strategies. In fact, the hPSC-derived tri-culture system could serve as a scalable platform for the screening of compounds that specifically target the crosstalk between microglia, astrocytes, and neurons as a novel therapeutic modality in the fight against Alzheimer’s Disease or other neurodegenerative disorders.
METHODS
Derivation of microglia from hPSCs
hPSCs maintained in Essential 8 media were dissociated by Accutase to obtain a single cell suspension. 60,000 cells/cm2 were plated in E8 medium containing Activin A (R&D 338-AC) (7.5ng/mL), BMP4 (R&D) (30ng/mL), CHIR 99021 (Tocris) (3uM), and ROCK inhibitor (Y-27632) (10uM) onto Matrigel-coated plates. After 18hr, medium was changed to Essential 6 medium containing Activin A (10ng/mL), BMP4 (40ng/mL), and IWP-2 (Selleck) (2uM). On day 2, medium was changed to Essential 6 medium containing Activin A (10ng/mL), BMP4 (40ng/mL), IWP-2 (2uM), and FGF2 (R&D) (20ng/mL). On day 3, cultures were dissociated with Accutase and replated at 60,000 cells/cm2 in Essential 6 medium containing VEGF (R&D) (15ng/mL), FGF2 (5ng/mL), and ROCK inhibitor (Y-27632) (10uM). On day 4, the ROCK inhibitor was removed and medium was changed to Essential 6 with VEGF (15ng/mL) and FGF2 (5ng/mL). On day 5 and 6, cultures were fed with Essential 6 medium containing VEGF (15ng/mL), FGF2 (5ng/mL), SCF (200ng/mL), and IL-6 (20ng/mL). On days 7 and 9, medium was changed to Essential 6 with SCF (100ng/mL), IL-6 (10ng/mL), TPO (30ng/mL), and IL-3 (30ng/mL). On day 10, the cells in suspension were collected and either 1) co-cultured with cortical neurons in neurobasal containing B27, L-glutamine, and BDNF, Ascorbic Acid, GDNF, cAMP and IL-34 (100ng/mL) and M-CSF (20ng/mL) for 5 days for direct transition to microglia, or 2) cultured in RPMI with 10% FBS, L-glutamine, and Penicillin/Streptavidin with IL-34 (100ng/mL) and M-CSF (10ng/mL) for 7–11 days until cells were adherent and elongated to transition to primitive macrophages. For serum-free culture, cells in suspension on day 10 were harvested and cultured in 75% IMDM, 25% F12 medium containing B27, L-glutamine, and IL-34 (100ng/mL) and M-CSF (20ng/mL) for 7–11 days. Transitioned macrophages were then co-cultured with cortical neurons with the addition of IL-34 and M-CSF for 7 days for upregulation of microglial-specific markers. IPSC lines used to test reproducibility of the microglial differentiation (DBR1, IFNAR1, IL10RB, STAT1) were each separately derived from patient fibroblasts and were reprogrammed using a nonintegrating Sendai viral vector in the Notarangelo lab at the NIH.
Cortical neuron protocol
hPSCs were dissociated with Accutase and plated at 200,000 cells/cm2 onto Matrigel-coated plates in Essential 8 medium with ROCK inhibitor (Y-27632) (10uM). Cells were treated with Essential 6 medium containing LDN193189 (100nM) and SB431542 (10uM) for 12 days, with the addition of XAV939 (2uM) for the first 4 days of differentiation. Cultures were fed with N2 medium with 1:1000 B27 supplement for an additional week for the development of neural progenitor cells (NPCs). NPCs were then dissociated and re-plated on Poly-ornithine/fibronectin/laminin coated plates and maintained in neurobasal, BDNF, Ascorbic Acid, GDNF, cAMP, L-glutamine, and B27 supplement for neuronal differentiation and maturation.
Astrocyte protocol
hPSCs were differentiated into astrocytes according to Tchieu et al. Briefly, cortical neural stem cells were pulsed with NFIA through an inducible lentiviral construct for 5 days, after which CD44+ progenitors are sorted and re-plated and maintained in astrocyte induction medium containing N2, HB-EGF (10ng/mL), and LIF (10ng/mL) for a minimum of 4 weeks.
FACS analysis
Cells were dissociated with Accutase for 20min and resuspended in FACS buffer containing 1% BSA, 2mM EDTA, 30ug/mL DNAse I and Normocin in PBS. Cells were washed and incubated in FACS buffer with antibody for 30 minutes on ice at 4 degrees in the dark. Cells were washed and resuspended in FACS buffer and strained through 40uM caps to eliminate cell clumps. Data collection was done on a BD-LSRII and FACSDiva v8. Gating (Fig. S2) and subsequent analysis was done using FlowJo software v10.5.3. FACS antibodies used include KDR-PE – R&D, FAB357P, Clone 89106 at 1:60, CD235A-APC – BD Biosciences at 1:100, 551336, Clone GA-R2, CD41-Apc/Cy7 – Biolegend, 303715, Clone HIP8, CD43-PerCP/Cy5.5 -BD Biosciences, 563521, Clone 1G10, CD45-FITC – BD Biosciences, 560976, Clone HI3O, CX3CR1-PE – Biolegend, 341604, 2A9–1, and Cd11b-APC/Cy7 – Biolegend, 301351, ICRF44 all at 5ul/100ul test.
Droplet-based scRNA-seq library preparation and sequencing
Four samples were prepared for single cell sequencing at different days of the microglial differentiation: ‘Day 6’ at day 6 of differentiation, ‘Day 10’ at day 10 of differentiation, ‘Day 10 suspension’ which only included cells in suspension at day 10 of differentiation, and ‘Microglia’ which included end-stage microglial cells cultured with neurons for 14 days. ‘Day 6’ and ‘Day 10’ samples were prepared by treating cultures with Accutase for 20 minutes to achieve a single cell suspension. ‘Day 10 suspension’ was prepared by collecting and straining cells in suspension through a 40uM filter to achieve a single cell suspension. ‘Microglia’ was prepared by sorting microglial-neuronal co-cultures for CX3CR1+. All samples were resuspended at 1000 cells/μL in FACS buffer before sequencing. Single cell sequencing was performed using 10X genomics Chromium Single Cell 3’ Library & Gel bead Kit V2 according to manufacturer’s protocol. An input of 8,700 cells was added to each 10x channel. Libraries were sequenced on an Illumina NovaSeq device.
scRNA-seq data preprocessing
scRNA-seq data was processed using the SEQC processing pipeline51. SEQC generates a cells-by-genes count matrix after read alignment, multi-mapping read resolution, cell barcode and UMI correction. SEQC included a first filtering step removing 1) putative empty droplets based on the cumulative distribution of molecule counts per barcode, 2) putative apoptotic cells based on a >20% of molecules derived from the mitochondria, and (3) removal of low-complexity cells identified as cells where the detected molecules are aligned to a small subset of genes. The number of cells per sample after SEQC processing was: 5253, 4320, 5555, 4961. Median library sizes were 19,195, 4039, 10,126 and 16,716 molecules per cell (Day 6, Microglia, Day 10, Day 10 suspension, respectively). Counts were normalized for library size by dividing each gene molecule count by the total number of molecules detected in the cell, then multiplying by 10,000 to convert the original counts to transcripts per 10,000. Data was then log transformed using natural log and pseudo-count 1.
Cell filtering
For each sample, cells were clustered using the PhenoGraph clustering algorithm21. Clusters of cells with low numbers of detected genes (~200) as putative empty droplets. Clusters with high mitochondrial RNA and a low number of detected genes were removed as putative dying cells. Four clusters not pertaining to hematopoietic differentiation were removed, including two early mesoderm clusters which expressed low levels of MESP1 and PDGFRA but not KDR, PECAM1, or CDH5, one cluster belonging to the cardiac lineage expressing NKX2.5 and ISL1, and one cluster belonging to mature endothelial cells.
Nearest neighbor graph construction
Principal components were used to calculate Euclidean distances between cells. An adaptive Gaussian kernel was used to convert Euclidean distances between cells’ k nearest neighbors into affinities, as described in 52. By using a Gaussian kernel, affinities between cells decrease exponentially with their distance, thereby increasing affinity to nearby cells and decreasing affinity to distant cells compared to the original Euclidean distances. Moreover, by using kernels with cell-adapted widths, differences in densities across regions of the data manifold are accounted for. Nearest neighbor graphs were used as a basis for force-directed graph layouts and diffusion map embeddings.
Clustering and force-directed graph layout
Data from Day 6, Day 10, and Day 10 suspension samples were pooled for trajectory modeling. Principal component analysis was performed on the data, and the first 20 principal components were selected for further analyses, to reduce noise due to the high degree of dropouts in scRNAseq53. Force-directed graph-layouts were calculated using the ForceAtlas2 algorithm54, based on the 30-nearest neighbors graph of the data that was constructed as described above. Clustering was done with PhenoGraph, using default parameter settings21.
Diffusion map embedding
To approximate the low-dimensional data manifold representing the differentiation trajectory, a diffusion map embedding was constructed using an adaptive Gaussian-kernel based nearest-neighbor graph (k=20, described above)52. Construction of a diffusion map is a non-linear method to recapitulate the low-dimensional structure underlying high-dimensional observations. The first four diffusion components of the diffusion map were selected for trajectory modeling. The diffusion distances between cells, i.e. the Euclidean distances between cells in the ‘diffusion map space’, were subsequently converted into pseudotime distances between individual cells as described in 55. Whereas distances in standard diffusion maps are related to a random Markov walk of length 1 along the edges of the ‘affinity graph’, diffusion distances in multi-scale space generalize over random walks of all lengths, thereby better capturing phenotypic similarities and differences between cells.55
Trajectory characterization
To further characterize the trajectory, we used Palantir22. Palantir is a tool that, using pseudotime distances, identifies trajectory endpoints (‘terminal cells’) in data of differentiating cells, and moreover measures entropy in cell phenotypes to measure their plasticity (‘differentiation potential’) and commitment to specific cell fates (‘branch probability’). As the input approximate start cell of the Palantir trajectory, a random cell from the CDH5-high, KDR-high, and PECAM1-high hemogenic endothelium clusters was used. The number of neighbors was set to k=20, the number of diffusion components was set to 4. For all other parameters, default settings were used.
Calculation of gene trends over pseudotime
To recover expression trends of individual genes over pseudotime, we first imputed our processed count matrix using MAGIC56. MAGIC is a method to denoise the cell count matrix and fill in zeros due to dropouts, by sharing information across similar cells via data diffusion. MAGIC was run with number of neighbors k = 40, random walk length t=6, and default further settings. Based on the imputed count matrix, gene trends were calculated using generalized additive models (GAMs) as described in 22. Spearman correlations between unimputed gene counts and pseudotime (Fig. S2B) were calculated per branch. For each branch, only those cells were included that had a branch probability equal to or higher than the branch probability at the start of the trajectory.
Diffusion distance distributions
To investigate whether cells in the Microglia sample were still in the process of differentiating, we inspected the distribution of pairwise diffusion distances among the cells. In accordance with single-cell studies of cell differentiation57,58, we assume that differentiation happens asynchronously: different cells are assumed to be at different stages of differentiation at one point in time. Therefore, differentiating cells’ transcriptomes are expected to lie on an elongated manifold. In contrast, cells with a homogenous phenotype are thought to be centered around one (multi-dimensional) mode, thus lying on an approximately sphere-shaped ‘manifold’. The distribution of pairwise distances between cells, calculated over multiple dimensions, is informative as to the shape of the manifold: if this distribution is unimodal, it suggests the cells lie on a spherical manifold.
A multi-scaled diffusion map embedding (described earlier) of the cells was used to calculate pairwise distances between the cells. Multi-scaled diffusion maps are thought to capture the shape of the manifold well, while reducing levels of noise as compared to the non-embedded data. Distances were calculated for different numbers (4 – 9) of diffusion components, to show robustness of the results to the number of components selected.
Comparison to mouse gene signatures
For comparison to previously published mouse signatures of EMPs and pMACs25, we translated signature mouse gene names that had a one-to-one mouse human orthologue (as defined by Ensembl BioMarts59) to human gene names. All other genes were excluded from the analysis. We furthermore excluded genes for which no transcripts were detected in our data. Heatmaps showing pseudotime expression trends of signature genes (Fig. 2D, 3I) were based on MAGIC56 imputed expression values. Imputed gene expression was normalized for each gene to range between 0 and 1. All cells that had a minimum myeloid branch probability of 0.1 were included in the heatmap. Cells were ordered by pseudotime, and genes were clustered using centroid clustering. For the macrophage gene signature, cells from the Microglia sample were appended to the trajectory cells and given an artificial pseudotime of 1.1.
Integration into single cell mouse embryogenesis atlas
For integration of trajectory data into a recently published single-cell transcriptomics atlas of mouse gastrulation and early organogenesis26, data of all cells annotated as haemato-endothelial lineage in the dataset was used (15875 cells). Only genes with a one-to-one mouse-human orthologue (as described earlier) were included in the analysis. To further restrict organism-related bias, the gene set was limited to genes that were highly variable in the reference mouse data. Highly variable genes were defined as described in 60. The final number of genes included was 1356. To perform further batch correction, a fast implementation of Mutual Nearest Neighbors batch correction61 was used. fastMNN (https://rdrr.io/github/LTLA/batchelor/man/fastMNN.html) performs batch correction on the principal component matrix instead of the gene expression matrix. The first 20 principal components of the pooled data were used for batch correction. Batch correction was first performed among samples from within the same time point, after which batch correction was performed between time points. The sample order used for fastMNN was: mouse data, late to early, then the human Day 10 and Day 10 suspension samples (pooled), and finally the Day 6 sample. Force-directed graph layout was calculated as described earlier. A graph of the clusters of mouse and human data was constructed using PAGA62. Only graph edges with a weight of 0.2 or higher were used for the force-directed layout54 of the graph.
Data analysis platform
The SCANPY platform (v1.4) was used for data analysis63.
Immunohistochemistry, live/dead assay, and high-content imaging
Cells were fixed in 4% PFA for 10 minutes at room temperature, permeabilized with 0.1% Triton for 5 minutes, washed with 0.2% Tween-20 in PBS for 5 minutes, and blocked with 5% donkey serum in 0.2% Tween-20 in PBS for 30 minutes. Primary antibodies were diluted in blocking solution and incubated with the sample overnight at 4 degrees. Secondary antibodies (Alexa 488, 555, and 647) were diluted in blocking solution and incubated with the sample at room temperature for 45 minutes. DAPI stain was used to identify cell nuclei. Live/dead assay was performed with CC3, with the control of hPSC-derived cortical neurons incubated with 70% methanol for 30 minutes. ImageExpress Micro Confocal High-Content Imaging System was used to quantify microglial cell numbers in culture. 9 fields were taken at 5x magnification to scan an entire 96-well culture well. Primary antibodies include: IBA1 – Wako #019–19741 1:500, MAP2 – Sigma-Aldrich #M1406 1:200, PU.1- Biolegend #658002 1:100, PSD95- Abcam #ab2723 1:200, GFAP – Biolegend #829401 1:1000, AQP4 – Santa Cruz Biotech #sc-9888 1:500, FOXG1 – Takara #M227 1:500, TBR1 – Abcam #ab183032 1:200, TUJ1 – Sigma-Aldrich #T2200 1:1000, CTIP2 – Abcam #ab18465 1:200, CC3 – Cell Signaling Tech #9664S 1:200, C1Q – Dako #A013602–1 1:1000. Secondary antibodies include donkey anti-mouse Alexa 488 – ThermoFisher #A-21202, donkey anti-chick Alexa 488 -JacksonImmunoResearch #AB_2340375, donkey anti-rabbit Alexa 555 – ThermoFisher #A-31572, donkey anti-mouse Alexa 555 – ThermoFisher #A-31570, donkey anti-mouse Alexa 647 - ThermoFisher, #A-31571, donkey anti-chick Alexa 647 - JacksonImmunoResearch, #AB_2340379 at 1:500.
Engulfment of synaptic proteins imaging
Microglia were co-cultured with day 70+ neurons on ibidi culture dishes for up to 30 days and stained with PSD95 and IBA1 (100 days total). Cultures were imaged on the Leica SP8 confocal microscope equipped with white light laser technology and standard argon lasers (458, 476, 488, 496 and 514 nm) at 40x magnification. Data was processed and analyzed with Imaris v 9.2: a surface volume mask was generated in the IBA1 channel, within which another mask was generated for the PSD95 channel to determine the volume of PSD95 inclusions/volume of IBA1 in a given Z-stack.
Phagocytosis assay and surveying assay
For the phagocytosis assay, microglial cells or astrocyte controls were incubated with Zymosan A bioparticles conjugated with Alexa fluor 488 for 5hr in an Olympus Vivaview fluorescent incubator microscope. For the surveying assay, microglial cells were infected with a lenti-viral construct expressing GFP and were co-cultured with d50 cortical neurons for 7 days, then incubated in an Olympus Vivaview fluorescent incubator microscope for 16hr. Time lapse imaging was compiled at 2min/frame.
RNA-sequencing
50,000 – 100,000 hPSC-derived microglia from 3 different hPSC lines, H1, H9, and the wildtype iPSC line SA241–1, were sorted from neuronal co-cultures by CX3CR1+. RNA was extracted using the Zymo RNA Micro Kit. RNA from primary human microglia was obtained after sorting postmortem tissue from the frontal and temporal lobes from patients aged 60–77 years. All samples were submitted to the MSKCC Integrated Genomics Core for paired end SMARTER-sequencing and 30–40 million reads. Analysis was done through a standard pipeline through the MSKCC Bioinformatics core: FASTQ files were mapped using the rnaSTAR aligner. Output SAM files are processed using PICARD tools. The mapped reads were then processed using HTSeq to compute a raw expression count matrix, which was then processed using DESeq from R/BioConductor to analyze differential expression between samples.
Primary human microglia
Brain tissue was provided by the Netherlands Brain Bank (NBB). Informed consent for brain autopsy, the use of tissue, and use of clinical information was obtained pre-mortem. The procedures of the NBB are in accordance with all national laws and the procedures have been approved by the ethics committee of the VU University Medical Center (Amsterdam, Netherlands). Brain tissues were collected from one male and three female donors without a history of a neurological or psychiatric disorder with an age range between 60 and 77 years. Microglia were isolated from the medial frontal gyrus and temporal superior gyrus as described before64. In short, 2–10 g of tissue was collected within 13 hours after death and stored in Hibernate medium on ice, and the isolation procedure was started within 2 to 24 h after autopsy. A single-cell suspension was generated by dissociating the tissue mechanically through a metal sieve, followed by an enzymatic digestion using collagenase Type I (3,700 U/ml; Worthington) and DNaseI (200 μg/ml; Roche). Microglia were further purified using a Percoll gradient and positive selection using CD11b-conjugated magnetic microbeads (Miltenyi Biotec). Microglia were lysed in RLT buffer (Qiagen), and RNA was extracted according to the protocol provided by the RNeasy Mini Kit (Qiagen).
GO Analysis and GSEA
GO analysis was performed using geneontology.org. The analysis type was PANTHER overrepresentation test, released 04/07/2020, the reference list was Homo Sapiens (all genes in database), the annotation data set was GO biological process complete, and the test type was Fisher with FDR correction. GSEA was performed using the GSEA software v4.0.3 from the Broad Institute, using the clusters defined in Matcovitch-Natan et al33 defining microglial developmental stages.
Comparison to publicly available microglial datasets
Publicly available datasets from the following studies were integrated according to the methodology in Grubman et al36 before multidimensional scaling. The following data sets were included: Muffat et al10, GSE85839, 8 samples; Douvaras et al12, GSE97744, 14 samples; Abud et al11, GSE89189, 12 samples; Ormel et al35, GSE102335, 9 samples; Galatro et al37, GSE99074, 38 samples. A total of 17 samples were integrated from our study (13 hPSC-derived microglia, 4 adult primary microglia). Briefly, count matrices were converted to log2 reads per kilobase million (log2-RPKM), and the differences between batches were removed using the removeBatchEffect from limma, specifying each dataset as a batch as well as specifying common groups as design (adultmg, hPSC-microglia, fetal microglia). MDS was performed using the plotMDS function from limma.
Tri-culture system, co-culture ratios, and LPS assay
Cortical neurons were differentiated from hPSCs and re-plated at 200,000 cells/cm2 on plates coated with Poly-ornithine/fibronectin/laminin and allowed to mature for 50–70 days in neurobasal with BDNF, Ascorbic Acid, GDNF, and cAMP (NB/BAGC). Astrocytes differentiated from hPSCs were dissociated with Accutase for 20–30min and then plated on top of the neurons at 25,000 cells/cm2 and were allowed to settle for 4 days in NB/BAGC. Microglia differentiated from hPSCs were then dissociated with Accutase for 10min and then plated on top of the astrocyte/neuron culture at 50,000 cells/cm2 in NB/BAGC with IL-34 (100ng/mL) and M-CSF (20ng/mL). Media was changed every other day with fresh addition of IL-34 and M-CSF. Other co-cultures included microglia and neurons (50,000 microglia/cm2, 200,000 neurons/cm2), astrocytes and neurons (25,000 astrocytes/cm2, 200,000 neurons/cm2), and neurons only (200,000 neurons/cm2). After the cultures were cultured for a minimum of 7 days, LPS was added to cultures at 1ug/mL for 72hr. Culture media was collected and spun down at 2000 rpm for 5 minutes, and the supernatant was frozen at −80 degrees Celsius until further analysis.
Cytokine ELISA
Following manufacturer’s protocols, culture supernatants were analyzed for C3 and C1Q using the Millipore Luminex Multiplex Kit on the FlexMap 3D system. Supernatants from the +/− LPS assay as well as the C3+/− assay on WT and C3KO microglia were also sent to Eve Technologies for multiplexed analysis of 14 inflammatory cytokines using the human high sensitivity T-cell discovery array 14-plex.
Conditioned medium assays
Medium from hPSC-derived microglia/neuron co-cultures was harvested after 48hr to represent microglial conditioned medium (MCM). MCM was added to astrocyte/neuron co-cultures for 48hr before the cells were harvested for C3 expression analysis by qPCR. Medium from hPSC-derived astrocyte/neuron co-cultures was harvested after 48hr to represent astrocyte conditioned medium (ACM). ACM was added to microglia-neuron co-cultures for 48hr before the cells were harvested for C3 expression analysis by qPCR.
Cytokine addition assays
Human C3 (Millipore) (1ug/mL) was added to microglia/neuron or astrocyte/neuron co-cultures for 48hr before cells were harvested for C3 expression analysis. Human TNF-a (R&D) (100ng/mL) and IL-6 (R&D) (100ng/mL) was added to astrocyte/neuron co-cultures for 48hr before cells were harvested for C3 expression analysis.
Cell line engineering
CRISPR/Cas9 KO of C3
The PX458 vector65 containing the guide 5’ TCTGCACTATCCAGGTA 3’ was nucleofected into H1 hESCs. Cells were sorted on the basis of GFP expression and cultured as single cell clones in E8 media with the cloneR supplement. Clones were picked onto replicate plates and genomic DNA was extracted using Bradley Lysis Buffer and Proteinase K treatment. A 450bp PCR product was amplified around the gRNA cut site and the clones were screened for indels by Sanger sequencing. Clones with indels were subsequently picked and expanded, karyotyped, and differentiated into microglia for further validation by ELISA for a lack of C3 protein secretion.
AD lines
APPSWE+/+ and WT iCas9 lines were generated using the gRNA and oligonucleotides from Paquet et al 47 in an H9 background.
Primers
Primers used for qPCR include: C1QA (QIAGEN QT00997745), C3 (F 5’ AAAAGGGGCGCAACAAGTTC 3’, R 5’ GATGCCTTCCGGGTTCTCAA 3’), CX3CR1 (F 5’ TGGGGCCTTCACCATGGAT 3’, R 5’ GCCAATGGCAAAGATGACGGAG 3’), TMEM119 (F 5’ CTTCCTGGATGGGATAGTGGAC 3’, R 5’ GCACAGACGATGAACATCAGC 3’), P2RY12 (F 5’ AAGAGCACTCAAGACTTTAC 3’, R 5’ GGGTTTGAATGTATCCAGTAAG 3’), GPR34 (F 5’ GAAGACAATGAGAAGTCATACC 3’, R 5’ TGTTGCTGAGAAGTTTTGTG 3’).
Statistics and reproducibility
All data presented in this study is representative of at least 3 independent experiments, including immunofluorescence panels, ELISA results, and FACS data. Western blots and gel electrophoresis were repeated at least twice. No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those reported in previous publications8,9. Cells were randomly assigned into experimental culture groups (TRI, M/N, A/N, N) after accounting for their respective genotypes (APPSWE+/+, WT, C3KO). Data collection and analysis were not performed blind to the conditions of the experiments because all quantified data were collected in an automated fashion (ELISA, FACS, ImageExpress) such that bias would not be introduced. Data distribution was assumed to be normal but this was not formally tested. Data are presented as the mean +/− SD. All statistical analyses were performed using GraphPad Prism v.8.0.1: 1-way ANOVA with Tukey’s post-hoc or Sidak’s tests to compare multiple groups, 2-way ANOVA with Tukey’s or Bonferroni post-hoc tests to compare multiple groups with two independent variables, and unpaired two-tailed Student’s t-test to compare two groups. Statistical differences were considered significant with p<0.05 as indicated in figure legends. Samples were excluded if data collection failed (machine clogs during FACS or ELISA collection – 1 sample in 4g), or after the outlier test – 2 samples in S9b. Four RNA samples from three postmortem human brain tissue samples were used for RNA-sequencing experiments, donors were aged 77, male, aged 60, female, and aged 84, female.
Reporting summary
Additional information on study design is available in the Life Sciences Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. Characterization of primitive hematopoiesis up to Day 10 of differentiation.
A) FACS analysis shows that addition of erythropoietin (EPO) from Day 6 to Day 10 of differentiation causes the emergence of CD235A+ erythrocytes at Day 10 of differentiation, as well as a reduction in the percentage of Macro (macrophage precursor) fated cells. B) Brightfield images over days 4–10 of the differentiation show that round hematopoietic cells progressively proliferate in semi-suspension through day 10, black arrows point to hematopoietic cells. Scale bar = 100 μM (Day 4, 10) and 50uM (Day 7). C) IF shows VE-cadherin+ hemogenic endothelium at Day 10 of differentiation. Scale bar = 100 μM. D) Imputed gene expression trends over pseudotime as calculated by Palantir of HOXA genes in the 3 differentiation trajectory arms show nearly absent HOXA1–7 expression, and minimal HOXA9 and 10 expression at low pseudotimes (õne log fold lower than genes in Fig. 2C). PMAC (PMAC arm, green), MK (megakaryocytic arm, pink), and ERY (erythrocyte arm, orange). E) Unimputed gene expression trends of HOXA1–10 genes show similar trends to imputed trends shown in A, nearly absent HOXA1–7 expression and minimal HOXA9–10 expression only at low pseudotimes in all differentiation trajectory arms. PMAC (PMAC arm, green), MK (megakaryocytic arm, pink), and ERY (erythrocyte arm, orange). F) Unimputed gene expression trends of key signature genes over pseudotime as calculated by Palantir show similar trends to imputed gene trends shown in Fig. 2C. Separate differentiation arms increasingly express their corresponding identity markers over pseudotime: ERY (GYPA, HBE1), MK (ITGA2B, ITGB3, GP1BA), and PMAC (CX3CR1, CSF1R, SPI1, PTPRC). MY (PMAC arm, green), MK (megakaryocytic arm, pink), and ERY (erythrocyte arm, orange). n = 6743 and error bars = SD for (A)-(C).
Extended Data Fig. 2. Heatmaps of EMP and PMAC signatures generated with unimputed data along PMAC, ERY, and MK arms.
A) Heatmaps showing unimputed counts of mouse yolk sac EMP PMAC signature genes25 along the PMAC arm ordered by pseudotime. There is increased expression of the EMP and PMAC signature genes over pseudotime along the PMAC trajectory, corresponding to the same increase in expression shown in Fig. 2D with imputed data. B) i) Heatmap of gene expression data from cells along the erythrocyte trajectory (ERY) ordered by pseudotime compared to mouse yolk sac EMP and PMAC gene signatures shows increased expression of EMP genes at pseudotimes corresponding to the in vitro human EMP/ERY clusters, but no increase in expression of the PMAC signature genes at any pseudotime. ii) Likewise, heatmap of imputed gene expression data from cells along the megakaryocytic trajectory (MK) ordered by pseudotime compared to mouse yolk sac EMP and PMAC gene signatures shows increased expression of EMP genes at pseudotimes corresponding to the human EMP/MK clusters, but no increase in expression of the PMAC signature at any pseudotime. C) Heatmaps generated from unimputed data show the same trends of increased expression of EMP signature genes at pseudotimes when EMP/ERY and EMP/MK clusters emerge, but no increased expression of the PMAC signature genes. Gene counts were individually scaled to range from 0 to 1 for all heatmaps.
Extended Data Fig. 3. Normalized counts of genes expressed by hPSC-derived microglia and hPSC-derived neurons in co-culture.
A) hPSC-derived neurons co-cultured with microglia express signature genes important for microglial maturation (IL-34, CSF1, CX3CL1, CD200, TGFB2, TGFB3), whereas hPSC-derived microglia express low levels or do not express these genes. neurons= hPSC-derived neurons, micro = hPSC-derived microglia from method ii of differentiation. n=4 samples for neurons and n=6 for microglia. Error bars = SD, center = mean. B) hPSC-derived microglia co-cultured with hPSC-derived neurons express a large panel of microglial genes at nearly the same levels as adult acutely isolated primary microglia, except TMEM119, P2RY12, and SALL1. primary = adult acutely isolated microglia, n=4, macro = method ii, matured alone then co-cultured, n=6, round = method i, direct co-culture, n=6. Error bars = SD, center = mean.
Extended Data Fig. 4. Characterization and reproducibility of maturing primitive EMPs/PMACs to homogenous hPSC-derived microglia without co-culture.
A) Brightfield image of differentiating primitive EMPs/PMACs shows that by 11 days of culture in IL-34 and M-CSF, the cells are adherent on TC-treated plastic and display an elongated morphology. B) IF shows that all cells at day 11 uniformly express the myeloid transcription factor PU.1. Scale bar = 50uM for (A) and (B). C) FACS analysis of day 10 differentiation cultures show a 37 – 51% induction of CD43+CD45+ macrophage precursors that is reproducible across 4 different hPSC lines. D) After 11 days of culture in IL-34 and M-CSF, a pure population of IBA1+ cells is reproducible in 5 hPSC lines (including 4 iPSC lines). Scale bar = 100μM. E) Each iPSC line used to test reproducibility of the microglia differentiation was derived from patient fibroblasts using a nonintegrating Sendai viral vector. F) Pairwise diffusion distances calculated between cells in the microglial sample after diffusion map embedding fall in a unimodal distribution. Pairwise distances calculated using different numbers of diffusion components are shown, with consistent results. G) In contrast, pairwise diffusion distances between cells in the heterogenous Day 10 sample have multiple peaks across different numbers of diffusion components.
Extended Data Fig. 5. GO pathway analysis and GSEA on differentially expressed genes between hPSC-derived microglia and acutely isolated adult primary microglia.
A) Heatmap of top 100 differentially expressed genes between hPSC-derived microglia derived from method ii and acutely isolated adult primary microglia using DESEQ. B) Table of top 100 differentially expressed genes between hPSC-derived microglia (method ii) and adult primary microglia using DESEQ. C) GO pathway analysis identifies neuronal developmental pathways as enriched in hPSC-derived microglia (method ii). D) GO pathways analysis identifies immune activation pathways as enriched in acutely isolated adult primary microglia. E-F) GSEA on 7 embryonic to adult microglial gene signatures33 (yolk sac, embryonic 1 and 2, postnatal 1 and 2, adult 1 and 2) reveals that hPSC-derived microglia (method ii) enrich for postnatal 1 and 2 signatures with NES = −1.46 and FDR = 0.14; NES = −1.52, FDR = 0.16. G-H) GSEA reveals that adult primary microglia enrich for adult 1 and 2 signatures with NES = −1.52 and FDR = 0.16; NES = −1.47, FDR = 0.09. Heatmaps show top 20 ranked genes from each gene signature in primary vs. hPSC-microglia (method ii).
Extended Data Fig. 6. Transcriptomic comparison of hPSC-derived microglia to microglia from previously published differentiation protocols.
A) MDS analysis using published datasets from 4 different microglial protocols10–12,35 and 1 study profiling acutely isolated adult primary microglia from postmortem human brain tissue37 reveals that hPSC-derived microglia from both method i (mg1) and method ii (mg2) cluster near the microglia differentiated from published protocols as well as near fetal microglia. adultmg = acutely isolated adult primary microglia sequenced in our study, mg1 = hPSC-derived microglia from method i, mg2 = hPSC-derived microglia from method ii of our study. adultmg_(ormel, galatro, abud) = adult microglia from Ormel et al35, Galatro et al37, Abud et al11, fetal_mg(abud, douv, muffat) = fetal microglia from Abud et al11, Douvaras et al12, Muffat et al10, mg_(abud, douv, muffat, ormel) = hPSC-microglia from Abud et al11, Douvaras et al12, Muffat et al10, and Ormel et al35. Dimension 1 vs. 2 separates the adult primary microglia from the fetal microglia/hPSC-derived microglia. B) Dimension 2 vs. 3 separates out the adult primary microglia and fetal microglia cultured in serum used in Abud et al11. Our hPSC-derived microglia (mg1, mg2) cluster near the other differentiated microglia (mg_ormel, mg_douv, mg_muffat) and fetal microglia (fetalmg_douv, fetalmg_douv+serum, fetalmg_muffat+serum).
Extended Data Fig. 7. Microglia perform efficient phagocytosis of zymosan-coated beads and mature hPSC-derived neurons form synapses in vitro.
A) Fluorescent microscopy shows that microglial cells contain zymosan-conjugated fluorescent beads as inclusions within 4 hours of incubation. Scale bar = 70uM. B) Astrocyte control does not contain fluorescent bead inclusions after 4hr of incubation. Scale bar = 280 μM. C) IF shows that hPSC-derived cortical neurons at day 70 express a punctate distribution of the pre-synaptic SYNI and post-synaptic HOMER1. Putative synapses are stained where both SYNI and HOMER1 are side-by-side (white arrow). D) IF shows that day 70 hPSC-derived cortical neurons also express the post-synaptic marker PSD95 in a punctate distribution. Scale bar = 50 μM (C) and (D).
Extended Data Fig. 8. Optimization of tri-culture ratio and culture media to maximize cell survival and minimize cell activation at baseline.
A) IF shows that tri-cultures with increased numbers of astrocytes plated (50K) show fewer microglial cells (IBA1+) attached at Day 7 than with tri-cultures containing fewer (25K) astrocytes. Scale bar = 200 μM. B) NB/BAGC and NB:N2 base media formulations show the lowest secretion of C3 in the tri-culture by ELISA. Addition of DMEM base, F12 supplement, low glucose, high glutamine, and high pyruvate all increase C3 levels in baseline tri-cultures. n=2 technical replicates of cell culture supernatant. C) Control staining for CC3. IF of hPSC-derived neurons killed by 70% methanol incubation for 30 minutes shows bright CC3+ whereas IF of fixed cells without the methanol treatment does not. Scale bar = 100 μM.
Extended Data Fig. 9. Characterization of C3KO hPSC line and C3KO-derived microglia and activation properties of conditioned medium and C3 addition to microglia and astrocytes.
A) Cell scoring by ImageExpress microscopy shows higher %IBA1+ / DAPI in microglia/neuron (M/N) cultures vs. tri-cultures at day 7. n=4 distinct cell culture wells. Error bars = SD, center = mean. B) Cytokine ELISA panel shows the secretion of inflammatory cytokines upon LPS stimulation only in cultures containing microglia, including IL-10, IL-6, TNFa, GM-CSF, IL1B, and IFNy. n=3 cell culture supernatants. Error bars = SD, center = mean. C) Sanger sequencing shows that the C3 KO hPSC line has a 7bp deletion (red) near the targeted PAM site (blue), guide is marked in green. D) IF shows that the C3KO hPSC line expresses the pluripotency markers of SOX2, NANOG, and OCT4. Scale bar = 100μM. E) i) FACS analysis shows that microglia differentiated from the C3KO line are a pure population expressing CD11B+ and CX3CR1+. ii) IF shows that these cells are all IBA1+ and PU.1+. Scale bar = 100μM. F) ELISA shows that C3KO microglia (densely cultured alone) do not secrete C3 protein as compared to WT microglia. n=2 cell culture supernatants. G) Cell scoring by ImageExpress shows similar numbers of IBA1+ microglia and GFAP+ astrocytes between the different tri-cultures (TRI, C3KOM, C3KOA). n= 4 distinct cell culture wells. Error bars = SD, center = mean. H) 48hrs of astrocyte-conditioned medium (ACM) treatment induces C3 expression in hPSC-derived microglia, normalized to untreated microglia. **p=0.0037, two-tailed t-test, n=3. 48 hrs of microglia-conditioned medium (MCM) treatment induces C3 in astrocytes, normalized to untreated astrocytes, n=3 independent experiments. *p=0.0122, two-tailed t-test. Error bars = SD, center = mean. I) C3 (1ug/mL) addition to microglia induces C3 expression after 48 hr, normalized to untreated microglia. *p=0.0217, two-tailed t-test, n=3 independent experiments. C3 addition to astrocytes does not induce C3 expression after 48hr of treatment, n=3. Error bars = SD, center = mean.
Extended Data Fig. 10. Characterization of the APPSWE+/+ and WT isogenic lines.
A) Sanger sequencing shows that the APPSWE+/+ line is homozygous for the GA>TC mutation as compared to the isogenic wildtype line. B) IF shows that both lines express the pluripotency markers of SOX2, NANOG, and OCT4. Scale bar = 100μM. C) Quantification of amyloid peptides 38, 40, and 42 in neuronal cultures shows that APPSWE+/+ neurons have higher levels of all peptides as compared to isogenic WT neurons. n=3 cell culture supernatants. D) Increased C1Q secretion by ELISA in APPSWE+/+ tri-cultures, ****p<0.0001, two-way ANOVA with Sidak’s post hoc test, n = 3 cell culture supernatants, error bars = SD, center = mean. E) Increased C1Q secretion in C3KOM but not C3KOA tri-cultures. n=3 cell culture supernatants, error bars = SD, center = mean.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the members the Studer lab for their helpful discussions and support for this project. Additionally, we would like to thank the MSKCC flow cytometry core for cell sorting, A. Viale at Integrated Genomics Operation Core (MSKCC) for the RNA-sequencing studies, P. Zumbo and D. Betel at the WCMC Applied Bioinformatics Core for MDS analysis, S. Ralph at the automated optical microscopy service with the Microscopy and Image Analysis Core (WCMC) for high-content imaging and analysis, V. Boyko at the Molecular Cytology Core (MSKCC) for help with confocal imaging and quantification, and the Molecular Cytogenetics core (MSKCC) for karyotyping analysis. We also thank the team of the Netherlands Brain Bank for providing us with post-mortem brain tissue, and the Notarangelo lab at NIH for reprogramming the IPSC lines used to test reproducibility of the microglial differentiation (DBR1, IFNAR1, IL-10RB, STAT1). S.R.G was supported by the Ruth L. Kirschstein Individual Predoctoral NRSA for MD/PhD Fellowship (1F30MH115616-01) and by a Medical Scientist Training Program grant from the NIGMS of the National Institutes of Health (NIH) under award number (T32GM007739) to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program. J.T. was supported by a Tri-I Starr Stem Cell Scholar fellowship. N.S. was supported by a Glenn/AFAR Postdoctoral Fellowship, R.W. was supported by an F32 Ruth Kirschstein Postdoctoral fellowship (MH116590), and G.C was supported by a EMBO long-term postdoctoral fellowship and a NYSTEM postdoctoral fellowship. The work was supported in part by R21 NS084334, 1R01AG056298 to L.S. and the core grant P30CA008748.
Footnotes
DATA AVAILABILITY
All RNA-sequencing data, bulk and scRNA-seq, have been deposited to GEO under GSE139549 and GSE139552. Publicly available RNA-seq datasets used include GSE8583910, GSE9774412, GSE8918911, GSE10233535, and GSE9907437. Other data and reagents in this study are available from the corresponding author upon reasonable request.
COMPETING FINANCIAL INTERESTS
L.ST. holds equity and is a scientific co-founder and paid consultant of BlueRock Therapeutics.
CITED REFERENCES
- 1.Keren-Shaul H. et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290.e1217, doi: 10.1016/j.cell.2017.05.018 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Lecours C. et al. Microglial Implication in Parkinson’s Disease: Loss of Beneficial Physiological Roles or Gain of Inflammatory Functions? Front Cell Neurosci 12, doi: 10.3389/fncel.2018.00282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geloso MC et al. The Dual Role of Microglia in ALS: Mechanisms and Therapeutic Approaches. Front Aging Neurosci 9, doi: 10.3389/fnagi.2017.00242 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke LE et al. Normal aging induces A1-like astrocyte reactivity. Proc Natl Acad Sci U S A 115, E1896–e1905, doi: 10.1073/pnas.1800165115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liddelow SA et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487, doi:doi: 10.1038/nature21029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith AM & Dragunow M. The human side of microglia. Trends Neurosci 37, 125–135, doi: 10.1016/j.tins.2013.12.001 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Chambers SM et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27, 275–280, doi: 10.1038/nbt.1529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi Y. et al. Combined small-molecule inhibition accelerates the derivation of functional cortical neurons from human pluripotent stem cells. Nat Biotechnol 35, 154–163, doi: 10.1038/nbt.3777 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tchieu J. et al. NFIA is a gliogenic switch enabling rapid derivation of functional human astrocytes from pluripotent stem cells. Nat Biotechnol 37, 267–275, doi: 10.1038/s41587-019-0035-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muffat J. et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med 22, 1358–1367, doi: 10.1038/nm.4189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abud EM et al. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron 94, 278–293.e279, doi: 10.1016/j.neuron.2017.03.042 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douvaras P. et al. Directed Differentiation of Human Pluripotent Stem Cells to Microglia. Stem Cell Reports 8, 1516–1524, doi: 10.1016/j.stemcr.2017.04.023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haenseler W. et al. A Highly Efficient Human Pluripotent Stem Cell Microglia Model Displays a Neuronal-Co-culture-Specific Expression Profile and Inflammatory Response. Stem Cell Reports 8, 1727–1742, doi: 10.1016/j.stemcr.2017.05.017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takata K. et al. Induced-Pluripotent-Stem-Cell-Derived Primitive Macrophages Provide a Platform for Modeling Tissue-Resident Macrophage Differentiation and Function. Immunity 47, 183–198.e186, doi: 10.1016/j.immuni.2017.06.017 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Pandya H. et al. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat Neurosci 20, 753–759, doi: 10.1038/nn.4534 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brownjohn PW et al. Functional Studies of Missense TREM2 Mutations in Human Stem Cell-Derived Microglia. Stem Cell Reports 10, 1294–1307, doi: 10.1016/j.stemcr.2018.03.003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sturgeon CM, Ditadi A, Awong G, Kennedy M. & Keller G. Wnt Signaling Controls the Specification of Definitive and Primitive Hematopoiesis From Human Pluripotent Stem Cells. Nat Biotechnol 32, 554–561, doi: 10.1038/nbt.2915 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginhoux F. et al. Fate Mapping Analysis Reveals That Adult Microglia Derive from Primitive Macrophages. Science 330, 841–845, doi: 10.1126/science.1194637 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ditadi A. & Sturgeon CM Directed differentiation of definitive hemogenic endothelium and hematopoietic progenitors from human pluripotent stem cells. Methods, 4–11, doi: 10.1016/j.ymeth.2015.10.001 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Palis J. Hematopoietic stem cell-independent hematopoiesis: emergence of erythroid, megakaryocyte, and myeloid potential in the mammalian embryo. FEBS Lett 590, 3965–3974, doi: 10.1002/1873-3468.12459 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Levine JH et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, 184–197, doi: 10.1016/j.cell.2015.05.047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Setty M. et al. Characterization of cell fate probabilities in single-cell data with Palantir. Nat Biotechnol 37, 451–460, doi: 10.1038/s41587-019-0068-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kierdorf K. et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci 16, 273–280, doi: 10.1038/nn.3318 (2013). [DOI] [PubMed] [Google Scholar]
- 24.ES N. et al. Differentiation of Human Embryonic Stem Cells to HOXA + Hemogenic Vasculature That Resembles the Aorta-Gonad-Mesonephros. Nature biotechnology 34, doi: 10.1038/nbt.3702 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Mass E. et al. Specification of tissue-resident macrophages during organogenesis. Science 353, doi: 10.1126/science.aaf4238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pijuan-Sala B. et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495, doi: 10.1038/s41586-019-0933-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haghverdi L, Lun ATL, Morgan MD & Marioni JC Batch effects in single-cell RNA sequencing data are corrected by matching mutual nearest neighbours. Nat Biotechnol 36, 421–427, doi: 10.1038/nbt.4091 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y. et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol 13, 753–760, doi: 10.1038/ni.2360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bohlen CJ et al. Diverse Requirements for Microglial Survival, Specification, and Function Revealed by Defined-Medium Cultures. Neuron 94, 759–773.e758, doi: 10.1016/j.neuron.2017.04.043 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greter M, Lelios I. & Croxford AL Microglia Versus Myeloid Cell Nomenclature during Brain Inflammation. Front Immunol 6, doi: 10.3389/fimmu.2015.00249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butovsky O. et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci 17, 131–143, doi: 10.1038/nn.3599 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennett ML et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A 113, E1738–1746, doi: 10.1073/pnas.1525528113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matcovitch-Natan O. et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science 353, aad8670, doi: 10.1126/science.aad8670 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Gosselin D. et al. An environment-dependent transcriptional network specifies human microglia identity. Science 356, doi: 10.1126/science.aal3222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ormel PR et al. Microglia innately develop within cerebral organoids. Nature Communications 9, 1–14, doi:doi: 10.1038/s41467-018-06684-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.G A et al. A CX3CR1 Reporter hESC Line Facilitates Integrative Analysis of In-Vitro-Derived Microglia and Improved Microglia Identity upon Neuron-Glia Co-culture. Stem cell reports 14, doi: 10.1016/j.stemcr.2020.04.007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.TF G. et al. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nature neuroscience 20, doi: 10.1038/nn.4597 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Nimmerjahn A, Kirchhoff F. & Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318, doi: 10.1126/science.1110647 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Wake H. & Fields RD Physiological Function of Microglia. Neuron Glia Biol 7, 1–3, doi: (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevens B. et al. The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178, doi: 10.1016/j.cell.2007.10.036 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Schafer DP et al. Microglia Sculpt Postnatal Neural Circuits in an Activity and Complement-Dependent Manner. Neuron 74, 691–705, doi: 10.1016/j.neuron.2012.03.026 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Z. et al. in J Neurosci Vol. 32 11706–11715 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi Q. et al. Complement C3-Deficient Mice Fail to Display Age-Related Hippocampal Decline. J Neurosci 35, 13029–13042, doi: 10.1523/jneurosci.1698-15.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu T. et al. Complement C3 Is Activated in Human AD Brain and Is Required for Neurodegeneration in Mouse Models of Amyloidosis and Tauopathy. Cell Rep 28, 2111–2123.e2116, doi: 10.1016/j.celrep.2019.07.060 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Shi Q. et al. Complement C3 deficiency protects against neurodegeneration in aged plaque-rich APP/PS1 mice. Sci Transl Med 9, doi: 10.1126/scitranslmed.aaf6295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hong S. et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716, doi: 10.1126/science.aad8373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paquet D. et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129, doi: 10.1038/nature17664 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Afagh A, Cummings BJ, Cribbs DH, Cotman CW & Tenner AJ Localization and cell association of C1q in Alzheimer’s disease brain. Exp Neurol 138, 22–32, doi: 10.1006/exnr.1996.0043 (1996). [DOI] [PubMed] [Google Scholar]
- 49.Fonseca MI et al. Cell-specific deletion of C1qa identifies microglia as the dominant source of C1q in mouse brain. J Neuroinflammation 14, 48, doi: 10.1186/s12974-017-0814-9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morizawa YM et al. Reactive astrocytes function as phagocytes after brain ischemia via ABCA1-mediated pathway. Nature Communications 8, 1–15, doi:doi: 10.1038/s41467-017-00037-1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS REFERENCES
- 51.Azizi E. et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 174, 1293–1308 e1236, doi: 10.1016/j.cell.2018.05.060 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haghverdi L, Buettner F. & Theis FJ Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics 31, 2989–2998, doi: 10.1093/bioinformatics/btv325 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Stegle O, Teichmann SA & Marioni JC Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet 16, 133–145, doi: 10.1038/nrg3833 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Jacomy M, Venturini T, Heymann S. & Bastian M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS One 9, e98679, doi: 10.1371/journal.pone.0098679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haghverdi L, Buttner M, Wolf FA, Buettner F. & Theis FJ Diffusion pseudotime robustly reconstructs lineage branching. Nat Methods 13, 845–848, doi: 10.1038/nmeth.3971 (2016). [DOI] [PubMed] [Google Scholar]
- 56.van Dijk D. et al. Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell 174, 716–729 e727, doi: 10.1016/j.cell.2018.05.061 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buenrostro JD et al. Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell 173, 1535–1548 e1516, doi: 10.1016/j.cell.2018.03.074 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Velten L. et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat Cell Biol 19, 271–281, doi: 10.1038/ncb3493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kinsella RJ et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011, bar030, doi: 10.1093/database/bar030 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Satija R, Farrell JA, Gennert D, Schier AF & Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33, 495–502, doi: 10.1038/nbt.3192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haghverdi L, Lun ATL, Morgan MD & Marioni JC Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 36, 421–427, doi: 10.1038/nbt.4091 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolf FA et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol 20, 59, doi: 10.1186/s13059-019-1663-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolf FA, Angerer P. & Theis FJ SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15, doi: 10.1186/s13059-017-1382-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sneeboer MAM et al. Microglia in post-mortem brain tissue of patients with bipolar disorder are not immune activated. Transl Psychiatry 9, 153, doi: 10.1038/s41398-019-0490-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ran FA et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308, doi: 10.1038/nprot.2013.143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.