Abstract
Ovarian cancer is one of the most lethal gynecologic malignancies for women. Due to the lack of efficient target therapy, the overall survival rate for patients with advanced ovarian cancer is still low. Illustrating the molecular mechanisms dictating ovarian cancer progression is critically important to develop novel therapeutic agents. Here, we found that RNA-binding motif protein 11 (RBM11) was highly elevated in ovarian cancer tissues compared with normal ovary, while RBM11 depletion in ovarian cancer cells resulted in impaired cell growth and invasion. Moreover, knockdown of RBM11 also retarded tumor growth in the A2780 ovarian cancer xenograft model. Mechanically, we found that RBM11 positively regulated Akt/mTOR signaling pathway activation in ovarian cancer cells. Thus, these results identify RBM11 is a novel oncogenic protein and prognostic biomarker for ovarian cancers.
1. Introduction
Ovarian cancer is one of the leading causes of death for gynecologic tumors among women, and more than 300,000 new ovarian cancer cases were diagnosed each year [1]. Although the rapid progression in cancer research studies, the five-year survival rate for ovarian cancer is still low (<47%) due to its fast progression and frequent recurrence [2, 3], which demands novel therapeutic regimens. Understanding the mechanism for ovarian progression and identifying novel molecular switch that dictate ovarian cancer malignancy are critically important to develop novel treatments for this deadly disease.
RNA-binding proteins (RBPs) are proteins that bind to RNA molecules and participate in posttranscriptional control of RNA functions [4–6]. RBPs play important roles in various cellular processes, such as RNA splicing, RNA stabilization, RNA transportation, RNA localization, and RNA modification [7, 8]. More than 2000 proteins have been identified to interact with RNA transcripts and are recognized as RBPs. RBPs are involved in various physiological and pathological processes including development, metabolism, proliferation, pluripotency, tumors, and immunity [9–11]. Recently, the functions of RBPs in cancers have been extensively studied [6]. RBPs could exert either oncogenic or tumor suppression roles in different human cancers [12]. For instance, RNA-binding protein Nova1 and NELFE promote cancer growth in hepatocellular carcinoma (HCC) cells [13, 14], whereas RBM47 and RBM43 have been shown to suppress HCC growth [15, 16].
RNA-binding motif protein 11 (RBM11) is a RNA splicing factor containing an RNA Recognition Motif (RRM) at the amino terminus (N-terminal), and its expression has been observed in multiple normal human tissues, including the brain, testis, and spleen [17]. Although the physiological function of RBM11 has not been clearly defined, it is putatively linked to Down syndrome due to its chromosome 21 location [17]. Recently, upregulation of RBM11 protein level has been observed in glioblastoma and RBM11 could promote glioblastoma cell proliferation and invasion in vitro and in vivo [18]. Nonetheless, the oncogenic roles of RBM11 in other cancer types beyond glioblastoma are needed to be further investigated. Here, we studied the functions of RBM11 in ovarian cancer and found that RBM11 was overexpressed in ovarian cancer tissues and exerts oncogenic roles in ovarian cancer through activating Akt/mTOR signaling. Thus, these results provide evidences that RBM11 is an oncogenic protein in cancers.
2. Materials and Methods
2.1. Cell Culture and Transfection
Human ovarian cancer cells A2780 and OVCAR-3 were obtained from the American Type Culture Collection (ATCC) and cultured in the Roswell Park Memorial Institute- (RPMI-) 1640 medium supplemented with 10% fetal bovine serum (FBS; Hyclone). Cells were tested for mycoplasma contamination every two months. Cells were transfected with plasmid DNA using lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instruction.
2.2. Plasmids and Antibodies
RBM11 and control shRNAs were purchased from Sigma-Aldrich (St. Louis, MO, USA), and their targeting sequences were as follows: RBM11 shRNA-1,5′-GTT CCG AAA GTC TAA GAA GAA-3′; RBM11 shRNA-2,5′-CCC AGC TCA TAT AAA TGG ACT-3′. The flag-RBM11 plasmid was purchased from Sino Biological Inc. (Beijing, China). Anti-RBM11 (17220-1-AP) antibody was purchased from Proteintech (Wuhan, China), and anti-pAkt S473 (#9271), anti-Akt (#4691), anti-pmTOR S2448 (#5536), and anti-mTOR (#2972) antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). Anti-β-actin (sc-47778) and anti-flag (sc-166355) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
2.3. Establishment of RBM11 Stable Knockdown Cells
RBM11 shRNA and lentiviral packaging vectors (psPAX2 and pCMV-VSV-G) were cotransfected into 293FT cells by lipofectamine 3000 transfection reagent. 72 hours after transfection, virus-containing medium supernatant was harvested by centrifugation. OVCAR-3 and A2780 cells were infected with lentivirus in the presence of 10 μg/ml of polybrene. After selection by puromycin, RBM11 knockdown efficacy was confirmed by western blot.
2.4. Colony Formation Assay
Cells expressing control or RBM11 shRNA were seeded in a 6-well plate at a density of 1 × 103 cells per well. Cells were grown for 10 days; then, the clones were fixed and stained with 0.5% crystal violet in 20% methanol followed by counting the number of colonies. Relative colony formation was expressed as a percentage normalized to cells expressing control shRNA.
2.5. Transwell Invasion Assay
The upper chambers of transwell inserts (Corning, NY, USA) were coated with matrigel (1 : 10 dilution) (Invitrogen, Carlsbad, CA, USA) at 37°C for 1 hour. Cells were starved for 24 hours before harvesting and suspending at 1 × 105/ml in a serum-free medium. Then, 200 μl of cell suspension was seeded into a matrigel-coated upper chamber of inserts. The bottom chamber of inserts was placed into a 24-well plate with a 500 μl medium containing 10% FBS. After culturing at 37°C for 24 h, the transwell inserted was removed, cells on the upper membrane were wiped using a cotton swap, and the cells on the bottom chamber were then stained with crystal violet in 20% methanol and counted.
2.6. Xenograft Tumor Growth
The cancer xenograft was established as previously described [19]. Briefly, 4-6 weeks of female nude mice were purchased from GemPharmatech (Nanjing, China). All the animal protocols were approved by the Animal Care and Use Committee of the First Affiliated Hospital of Xi'an Jiaotong University. A2780 xenograft was performed as previously reported [20]; briefly, 1 × 107 of A2780 cells expressing control or RBM11 shRNA were subcutaneously implanted into mouse flanks. Tumor growth was monitored, the tumor length (L) and width (W) were measured every five days, and tumor volume (V) was calculated by formula V = (W2 × L)/2.
2.7. Immunohistochemistry Assay
The ovarian cancer tissues and adjacent normal ovarian tissues were purchased from Alenabio Co., Ltd. (Xi'an, China). The tumor sections were firstly deparaffinized with 100% xylene, followed by rehydration using gradient ethanol (100%, 95%, 70%, 30%, and 0%). After inactivation of endogenous peroxidase by 3% H2O2 and heat-induced retrieval antigen, IHC staining was performed using the R.T.U Vectastain Kit (Vector Laboratories, Burlingame, CA) according to the manufacturer's instructions. The Ki67 (abcam, ab15580) antibody was diluted at 1 : 500. The percentage of Ki67-positive cells was determined from three separate fields.
2.8. Western Blot Assay
Western blot was performed as previously described. Briefly, proteins in cell lysates were resolved on SDS-PAGE and were transferred on PVDF membrane after electrophoresis. After blocking in 5% milk in PBS, the membrane was incubated with primary antibody and second antibody sequentially, following band detection by enhanced chemiluminescence (ECL) using Pierce™ ECL Western Blotting Substrate (GE Healthcare Bio-Sciences).
2.9. Statistical Analysis
A two-sided unpaired Student's t-test was used to calculate the statistical significance of differences. Results were considered statistically significant at p < 0.05. All data are presented as mean ± standard deviation (S.D.) from at least three replicates.
3. Results
3.1. RBM11 Is Overexpressed in Ovarian Cancer Tissues
RBM11 has been shown to exert oncogenic roles in glioblastoma cells [18]; to examine its function in ovarian cancers, we first examined RBM11 mRNA expression and its gene copy numbers in ovarian cancer tissues from the Cancer Genome Atlas (TCGA) database and found that although RBM11 copy number was not frequently amplified, RBM11 mRNA was highly elevated in ovarian cancer tissue (Figure 1(a) and Supplementary Figure 1). To further validate the RBM11 protein levels in ovarian cancer tissues, we performed Immunohistochemistry (IHC) staining and the result (Figures 1(b) and 1(c)) showed that RBM11 protein level was significantly overexpressed in ovarian cancer tissues compared with normal ovary. In addition, a high level of RBM11 was associated with poor survival in ovarian cancer patients (Figure 1(d)).
Figure 1.
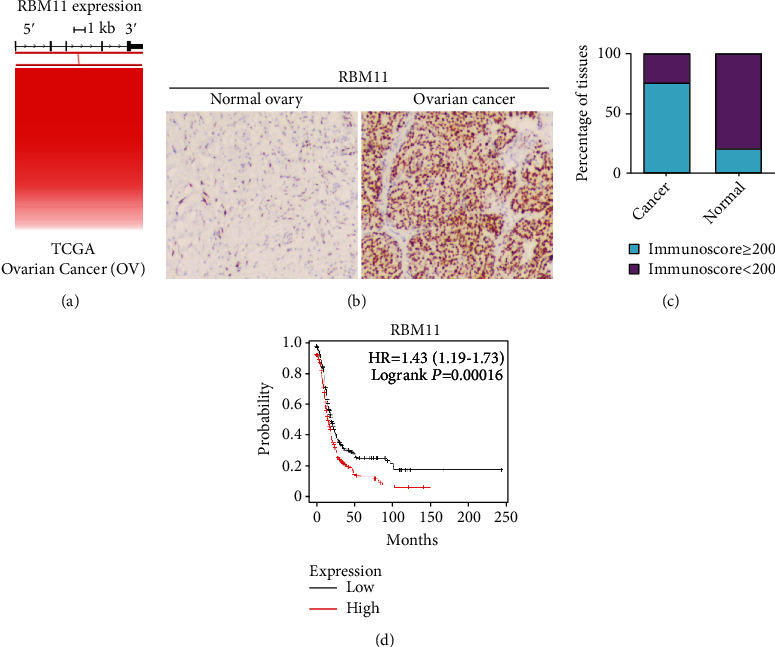
RBM11 is overexpressed in ovarian cancer tissues and is highly associated with poor survival in ovarian cancer patients. (a) RBM11 mRNA expression in ovarian cancer tissues was assessed with TCGA database. (b, c) RRM1 protein levels in normal ovary and ovarian cancer tissue were analyzed by Immunohistochemistry (IHC) staining. Representative image of staining (b) and quantitative analysis of immunoscore (c) were provided. (d) Overall survival assay in ovarian cancer patients with high RBM11 mRNA level and low RBM11 mRNA level was performed using the Kaplan-Meier plotter online server (https://www.kmplot.com/analysis).
3.2. RBM11 Promotes Ovarian Cancer Cell Growth
To test the role of RBM11 in ovarian cancer progression, we knocked down RBM11 expression in ovarian cancer cells including OVCAR-3 and A2780 using two distinct shRNA targeting the RBM11 coding region (Figure 2(a)). Interestingly, we found that silence of RBM11 significantly inhibited growth in both OVCAR-3 and A2780 cells (Figure 2(b)). Moreover, clonogenic formation assay also further revealed that depletion of RBM11 significantly reduced colony formation capacity in OVCAR-3 and A2780 cells (Figure 2(c)). These results demonstrated that RBM11 is required for ovarian cancer cell proliferation in vitro.
Figure 2.
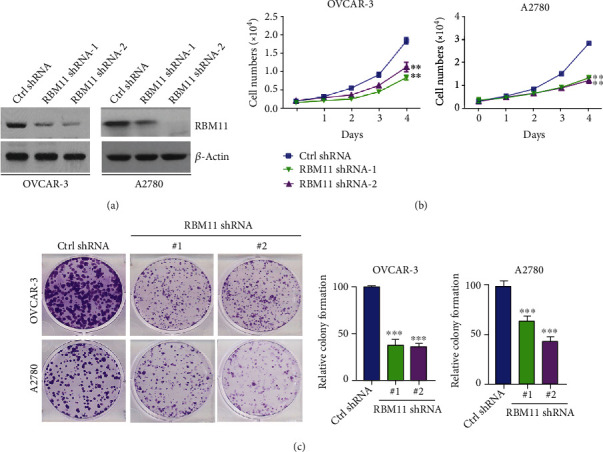
Knockdown of RBM11 inhibits ovarian cancer cell growth. (a) Western blot analysis of RBM11 protein level in OVCAR-3 and A2780 cells stably transfected with control (ctrl), RMB11 shRNA-1, or RBM11 shRNA-2. (b) Cell growth curve of OVCAR-3 and A2780 cells expressing ctrl shRNA, RBM11 shRNA-1, or RBM11 shRNA-2. (c) Colony formation assay of cells expressing ctrl shRNA or RBM11 shRNA. Left, the representative colony growth; right, the relative quantification of colony number. ∗∗p < 0.01 and ∗∗∗p < 0.001, by 2-tailed t-test, when compared with cells expressing ctrl shRNA.
3.3. RBM11 Enhances Ovarian Cancer Invasion
Beyond proliferation, the invasion capacity of cancer cells is also an important factor, affecting the prognosis of cancer patients. Therefore, we performed a transwell assay to evaluate the role of RBM11 in ovarian cancer invasion, as shown in Figure 3(a); the number of invaded OVCAR-3 and A2780 cells expressing RBM11 shRNA is remarkable less than that expressing control (ctrl) shRNA. To further confirm the effect of RBM11 on ovarian cell invasion, we ectopically expressed flag tagged RBM11 into OVCAR-3 and A2780 cells (Figure 3(b)). Similarly, overexpression of flag-RBM11 significantly enhanced cell invasion (Figure 3(c)). Thus, these results indicated that RBM11 is also essential for ovarian cancer invasion and metastasis.
Figure 3.
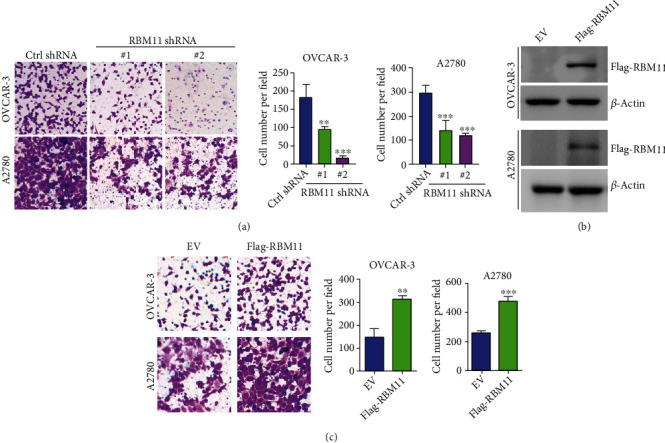
RBM11 promotes ovarian cancer invasion. (a) Transwell invasion analysis of OVCAR-3 and A2780 cells expressing ctrl or RBM11 shRNA. Left, representative staining of cells on the bottom membrane of the transwell insert was provided; right, cells on the bottom membrane per field were quantified. (b) Flag-tagged RBM11 was stably expressed in OVCAR-3 or A2780 cells; the expression of flag-RBM11 was examined by western blot. (c) Transwell analysis of OVCAR-3 or A2780 cells expressing flag-RBM11. Representative image (left) and quantitative analysis (right) were shown. ∗∗p < 0.01 and ∗∗∗p < 0.001, by 2-tailed t-test, when compared with cells expressing ctrl shRNA.
3.4. RBM11 Enhances Akt/mTOR Activation in Ovarian Cancer Cells
As one of the most frequently activated signaling pathways in ovarian cancer cells, Akt plays critical roles in ovarian cancer proliferation and invasion [21]. Akt is commonly activated by phosphatidylinositol 3 kinase (PI3K) and activates the downstream signal through mTOR to promote translation of target genes involved in cell proliferation and invasion [22]. To explore the molecular mechanism by which RBM11 exerts its oncogenic function in ovarian cancer cells, we tested whether RBM11 could affect Akt signaling. As shown in Figure 4(a), we detected a significant decrease of Akt phosphorylation and mTOR phosphorylation in cells expressing RBM11 shRNA, which suggests that RBM11 might positively regulate Akt signaling activation. To further confirm the role of RBM11 in Akt signaling, exogenous RBM11 was ectopically expressed in OVCAR-3 and A2780 cells (Figure 4(b)). Consistent with RBM11 knockdown, overexpression of RBM11 resulted in an increase of phosphorylation of Akt and phosphorylation of mTOR (Figure 4(c)). These results suggest that RBM11 promotes ovarian cancer progression through stimulating Akt/mTOR signaling pathways.
Figure 4.
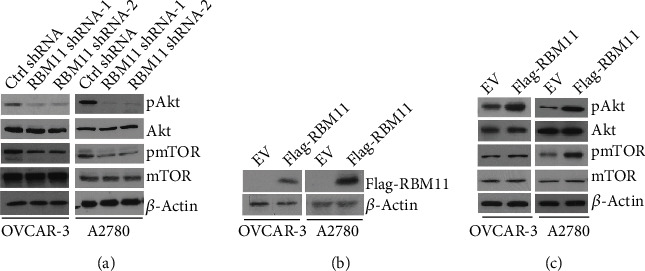
RBM11 promotes Akt/mTOR activation in ovarian cancer cells. (a) Western blot analysis of pAkt (S473), Akt, pmTOR (S2448), and mTOR levels in cells expressing control (ctrl) or RBM11 shRNA. (b) Flag-tagged RBM11 was transfected into OVCAR-3 and A2780 cells, and the expression of flag-RBM11 was validated by western blot assay. (c) Western blot analysis of indicated protein levels in cells transfected with empty vector (EV) or flag-RBM11.
3.5. RBM11 Knockdown Retards Ovarian Cancer Growth In Vivo
To validate the oncogenic roles of RBM11 in ovarian cancer in vivo, we constructed a xenograft model using A2780 cells expressing control (ctrl) or RBM11 shRNA. Consistent with in vitro results, we found that knockdown of RBM11 significantly inhibited tumor growth in the A2780 xenograft model (Figure 5(a)). Meanwhile, A2780 xenograft tumors with RBM11 knockdown contained significantly less Ki67 protein level, a well-defined proliferation marker, compared with tumors expressing control shRNA (Figure 5(b)). Thus, these results demonstrated that RBM11 promotes ovarian cancer growth in vivo.
Figure 5.
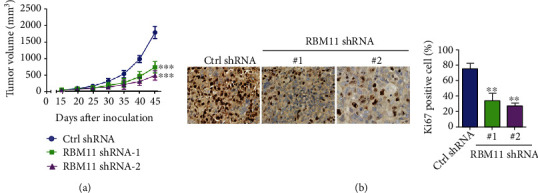
RBM11 silence retarded tumor growth in the A2780 xenograft model. (a) A2780 cells expressing control (ctrl) or RBM11 shRNA were implanted into nude mice, the tumor volumes were measured, and the tumor growth curve was shown. (b) Immunohistochemistry (IHC) analysis of Ki67 levels in tumor tissue. ∗p < 0.01 and ∗∗∗p < 0.001, by 2-tailed t-test, when compared with cells expressing ctrl shRNA.
4. Discussion
Ovarian cancer is a leading cause of death from gynecological malignancies. Although the prognosis is relatively favorable if diagnosed at an early stage, the 5-year survival rates are only 30-40% for most ovarian cancer patients when they are detected at an advanced stage [1]. Currently, surgery followed by traditional chemotherapy including carboplatin and paclitaxel is employed as the first-line treatment for patients with high-grade epithelial ovarian cancer [3]. Beyond chemotherapy, PARP inhibitors such as olaparib and rucaparib showed favorable outcomes during the maintenance therapy for patients with advanced ovarian cancer [23]; however, PARP inhibitors are only approved and effective for patients with BRCA1/2 mutation, which accounts for less than 40% of ovarian cancer cases [24]. Therefore, it is an urgent need to develop novel targeted therapies for patients with high-grade ovarian cancers. In the present study, we identified that RBM11 is especially expressed in ovarian cancer tissues, but not in normal ovary tissue, which demonstrates that RBM11 is a feasible target for drug design in future ovarian cancer treatment. However, the functional domain of RBM11 used as the drug interacting pocket needed to be further identified.
Regulation of RNA metabolism is important to maintain normal cellular and physiological circumstances. RNA-binding proteins (RBPs) are involved in posttranscriptional control of transcriptome via alternative splicing [25], RNA modification [26], and RNA stability [4]. Accumulating evidences indicate that dysregulation of RNA metabolism resulted from abnormal RBP function plays critical roles in development [27], immunity [9], and neural function [28], as well as cancer progression [9, 12, 23]. However, the function of this group of proteins in ovarian cancer has rarely been reported. In this study, we discovered a novel mechanism governing ovarian cancer progression through RBM11. RBM11 enhances ovarian cancer proliferation and invasion in vitro and in vivo, which highlighted the importance of RBPs in ovarian cancer progression.
Among the members of RNA-binding protein, the biochemical and physiological function of RBM11 has not been well defined. It is previously reported that RBM11 protein was upregulated in glioblastoma tissues and promoted glioblastoma cell progression [18]; however, the underlying mechanism for RBM11's oncogenic roles in cancer cells has not been defined. In our present study, we investigated the function of RBM11 in ovarian cancers. In agreement with the glioblastoma study, we also found that RBM11 was highly elevated in ovarian cancer tissues and positively regulated ovarian cancer growth and invasion. Furthermore, we demonstrated that RBM11 affected the Akt/mTOR signaling pathway, which provides evidence and explanation why RBM11 could promote cancer cell progression.
5. Conclusion
RBM11 is elevated in ovarian cancer tissues and promotes ovarian cancer growth and invasion through activating the Akt/mTOR signaling pathway. Our finding has identified a functional role of RBPs in ovarian cancer progression and provided a novel molecular target for ovarian cancer therapy.
Acknowledgments
This study was supported by basic research funding from the First Affiliated Hospital of Xi'an Jiaotong University.
Data Availability
All data used to support the findings of this study are included within the article and are available upon request from the corresponding author.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Authors' Contributions
C.F and Z.M conceived and designed the experiments. C.F, M.Y, G.L, X.Z, and W.C performed the experiments and analyzed the results. Z.M wrote the manuscript. J.S revised the manuscript. All authors read and approved the content of the final manuscript.
Supplementary Materials
Supplementary Figure S1: RBM11 copy number in TCGA ovarian cancer database was analyzed by UCSC Xena online software (https://xena.ucsc.edu/).
References
- 1.Stewart C., Ralyea C., Lockwood S. Ovarian cancer: an integrated review. Seminars in Oncology Nursing. 2019;35(2):151–156. doi: 10.1016/j.soncn.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Eisenhauer E. A. Real-world evidence in the treatment of ovarian cancer. Annals of Oncology. 2017;28(Supplement 8):viii61–viii65. doi: 10.1093/annonc/mdx443. [DOI] [PubMed] [Google Scholar]
- 3.Pujade-Lauraine E. New treatments in ovarian cancer. Annals of Oncology. 2017;28(Supplement 8):viii57–viii60. doi: 10.1093/annonc/mdx442. [DOI] [PubMed] [Google Scholar]
- 4.Gebauer F., Schwarzl T., Valcarcel J., Hentze M. W. Rna-binding proteins in human genetic disease. Nature Reviews. Genetics. 2021;22(3):185–198. doi: 10.1038/s41576-020-00302-y. [DOI] [PubMed] [Google Scholar]
- 5.Hentze M. W., Castello A., Schwarzl T., Preiss T. A brave new world of rna-binding proteins. Nature Reviews. Molecular Cell Biology. 2018;19(5):327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 6.Pereira B., Billaud M., Almeida R. Rna-binding proteins in cancer: old players and new actors. Trends Cancer. 2017;3(7):506–528. doi: 10.1016/j.trecan.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Guo L., Kim H. J., Wang H., et al. Nuclear-import receptors reverse aberrant phase transitions of rna-binding proteins with prion-like domains. Cell. 2018;173(3):677–692.e20. doi: 10.1016/j.cell.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ravanidis S., Doxakis E. Rna-binding proteins implicated in mitochondrial damage and mitophagy. Frontiers in Cell and Developmental Biology. 2020;8(372) doi: 10.3389/fcell.2020.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore M. J., Blachere N. E., Fak J. J., et al. Zfp36 rna-binding proteins restrain t cell activation and anti-viral immunity. Elife. 2018;7 doi: 10.7554/eLife.33057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang E., Lu S. X., Pastore A., et al. Targeting an rna-binding protein network in acute myeloid leukemia. Cancer Cell. 2019;35(3):369–384.e7. doi: 10.1016/j.ccell.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., Ping C., Tan P., et al. Hnrnpll controls pluripotency exit of embryonic stem cells by modulating alternative splicing of tbx3 and bptf. The EMBO Journal. 2021;40(4, article e104729) doi: 10.15252/embj.2020104729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang D., Lee Y., Lee J. S. Rna-binding proteins in cancer: functional and therapeutic perspectives. Cancers (Basel) 2020;12(9):p. 2699. doi: 10.3390/cancers12092699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang H., Takai A., Forgues M., et al. Oncogenic activation of the rna binding protein nelfe and myc signaling in hepatocellular carcinoma. Cancer Cell. 2017;32(1):101–114.e8. doi: 10.1016/j.ccell.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y. A., Liu H. N., Zhu J. M., Zhang D. Y., Shen X. Z., Liu T. T. Rna binding protein nova1 promotes tumor growth in vivo and its potential mechanism as an oncogene may due to its interaction with Gabaa Receptor-γ2. Journal of Biomedical Science. 2016;23(1):p. 71. doi: 10.1186/s12929-016-0288-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng H., Liu J., Qiu Y., et al. Rna-binding motif protein 43 (rbm43) suppresses hepatocellular carcinoma progression through modulation of cyclin b1 expression. Oncogene. 2020;39(33):5495–5506. doi: 10.1038/s41388-020-1380-7. [DOI] [PubMed] [Google Scholar]
- 16.Shen D. J., Jiang Y. H., Li J. Q., Xu L. W., Tao K. Y. The RNA-binding protein RBM47 inhibits non-small cell lung carcinoma metastasis through modulation of AXIN1 mRNA stability and Wnt/β-catentin signaling. Surgical Oncology. 2020;34:31–39. doi: 10.1016/j.suronc.2020.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Pedrotti S., Busa R., Compagnucci C., Sette C. The rna recognition motif protein rbm11 is a novel tissue-specific splicing regulator. Nucleic Acids Research. 2012;40(3):1021–1032. doi: 10.1093/nar/gkr819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavlyukov M. S., Yu H., Bastola S., et al. Apoptotic cell-derived extracellular vesicles promote malignancy of glioblastoma via intercellular transfer of splicing factors. Cancer Cell. 2018;34(1):119–135.e10. doi: 10.1016/j.ccell.2018.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shu Z., Li Z., Huang H., et al. Cell-cycle-dependent phosphorylation of rrm1 ensures efficient DNA replication and regulates cancer vulnerability to atr inhibition. Oncogene. 2020;39(35):5721–5733. doi: 10.1038/s41388-020-01403-y. [DOI] [PubMed] [Google Scholar]
- 20.Banerji U., Sain N., Sharp S. Y., et al. An in vitro and in vivo study of the combination of the heat shock protein inhibitor 17-allylamino-17-demethoxygeldanamycin and carboplatin in human ovarian cancer models. Cancer Chemotherapy and Pharmacology. 2008;62(5):769–778. doi: 10.1007/s00280-007-0662-x. [DOI] [PubMed] [Google Scholar]
- 21.Mundi P. S., Sachdev J., McCourt C., Kalinsky K. Akt in cancer: new molecular insights and advances in drug development. British Journal of Clinical Pharmacology. 2016;82(4):943–956. doi: 10.1111/bcp.13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Z., Han X., Ou D., et al. Targeting pi3k/akt/mtor-mediated autophagy for tumor therapy. Applied Microbiology and Biotechnology. 2020;104(2):575–587. doi: 10.1007/s00253-019-10257-8. [DOI] [PubMed] [Google Scholar]
- 23.Mirza M. R., Coleman R. L., Gonzalez-Martin A., et al. The forefront of ovarian cancer therapy: update on parp inhibitors. Annals of Oncology. 2020;31(9):1148–1159. doi: 10.1016/j.annonc.2020.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez Martin A. Progress in PARP inhibitors beyond BRCA mutant recurrent ovarian cancer? The Lancet Oncology. 2017;18(1):8–9. doi: 10.1016/S1470-2045(16)30621-0. [DOI] [PubMed] [Google Scholar]
- 25.Zhou C., Gao X., Hu S., et al. Rbm-5 modulates u2af large subunit-dependent alternative splicing in C. elegans. RNA Biology. 2018;15(10):1295–1308. doi: 10.1080/15476286.2018.1526540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y., Shi Y., Shen H., Xie W. m6A-binding proteins: the emerging crucial performers in epigenetics. Journal of Hematology & Oncology. 2020;13(1):p. 35. doi: 10.1186/s13045-020-00872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorkovic Z. J. Role of plant rna-binding proteins in development, stress response and genome organization. Trends in Plant Science. 2009;14(4):229–236. doi: 10.1016/j.tplants.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 28.Ghanbari M., Ohler U. Deep neural networks for interpreting rna-binding protein target preferences. Genome Research. 2020;30(2):214–226. doi: 10.1101/gr.247494.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: RBM11 copy number in TCGA ovarian cancer database was analyzed by UCSC Xena online software (https://xena.ucsc.edu/).
Data Availability Statement
All data used to support the findings of this study are included within the article and are available upon request from the corresponding author.


