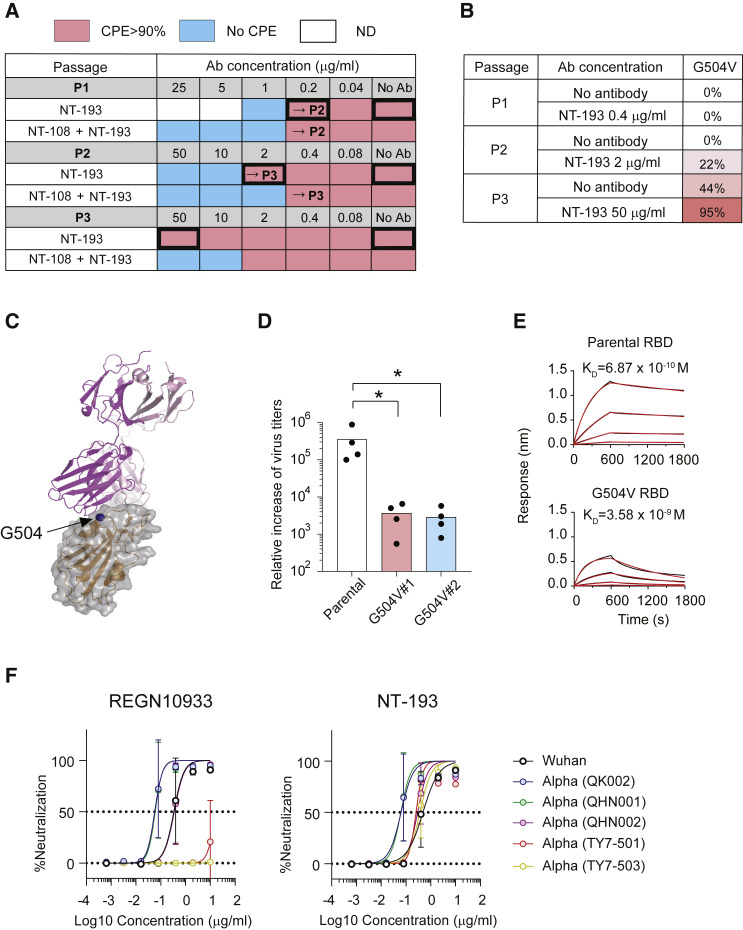
Figure 5.
NT-193 is resistant to escape mutations of SARS-CoV-2
(A) Passaging results of escape mutant screening are shown, with the qualitative percentage of CPE observed in each dilution (red, >90% CPE; blue, no CPE; white, not determined). Bold-bordered boxes indicate dilutions that were sequenced for identifying escape mutations.
(B) Identified escape mutations by deep sequencing of passaged virus. Viral RNAs were isolated at day 4 after infection from wells with the highest monoclonal antibody concentration among detectable CPE. RNA sequencing (RNA-seq) analysis was performed to identify changes relative to the input virus. Percentages of sequencing reads that contained the indicated mutant sequence are shown.
(C) Position of G504 is highlighted in dark blue.
(D) VeroE6 cells were infected with virus solution, including parental or G504 mutant viruses, and then the increase of virus titers from day 0 to day 3 was plotted. Each dot represents the number from the individual experiment, and the combined data from four independent experiments are shown. Statistical significance is indicated above the columns; ∗p < 0.05 (two-tailed Mann-Whitney test).
(E) Binding affinity between RBD (parental or G504 mutant) and ACE2 protein was determined by biolayer interferometry.
(F) Serially diluted REGN10933 and NT-193 were mixed with the indicated virus strains and then loaded onto VeroE6/TMPRSS2 cell lines. At day 3, the CPEs were visualized by crystal violet staining.
Values represent mean ± SD. The representative data from three independent experiments are shown (E and F). See also Figure S6.