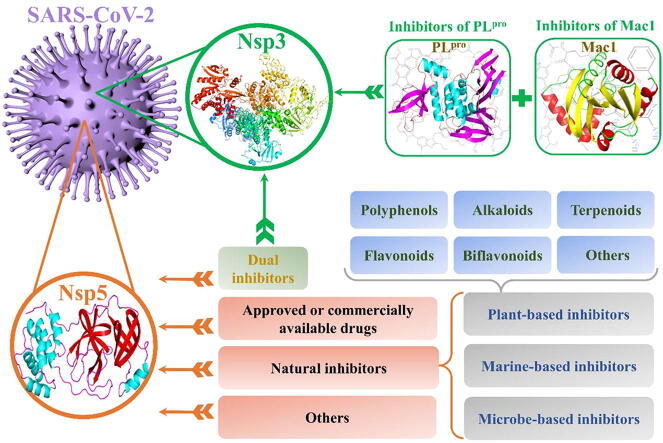
Graphical abstract
Potential inhibitors against nsp3 and nsp5 of SARS-CoV-2 obtained through an extensive literature retrieval.
Keywords: SARS-CoV-2, COVID-19, Non-structural protein 3 (nsp3), nsp5, Inhibitors
Abstract
There is an urgent need to develop effective treatments for coronavirus disease 2019 (COVID-19), which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The rapid spread of SARS-CoV-2 has resulted in a global pandemic that has not only affected the daily lives of individuals but also had a significant impact on the global economy and public health. Although extensive research has been conducted to identify inhibitors targeting SARS-CoV-2, there are still no effective treatment strategies to combat COVID-19. SARS-CoV-2 comprises two important proteolytic enzymes, namely, the papain-like proteinase, located within non-structural protein 3 (nsp3), and nsp5, both of which cleave large replicase polypeptides into multiple fragments that are required for viral replication. Moreover, a domain within nsp3, known as the macrodomain (Mac1), also plays an important role in viral replication. Inhibition of their functions should be able to significantly interfere with the replication cycle of the virus, and therefore these key proteins may serve as potential therapeutic targets. The functions of the above viral targets and their corresponding inhibitors have been summarized in the current review. This review provides comprehensive updates of nsp3 and nsp5 inhibitor development and would help advance the discovery of novel anti-viral therapeutics against SARS-CoV-2.
1. Introduction
Since the end of 2019, a novel respiratory disease that manifests as pneumonia has spread across many countries, and has garnered significant attention worldwide. Deep whole-genome sequencing of the patient samples revealed that the pathogen responsible for this respiratory disease is a novel coronavirus (nCoV), tentatively named 2019-nCoV by the World Health Organization (WHO). Similar to the severe acute respiratory syndrome coronavirus (SARS-CoV), 2019-nCoV belongs to the zoonotic β-coronaviruses family, and it shares a high degree of homology with SARS-CoV. On February 11, 2020, the International Committee on Taxonomy of Viruses (ICTV) renamed this coronavirus as SARS-CoV-2 [1]. The WHO termed the disease caused by SARS-CoV-2 as coronavirus disease 2019 (COVID-19) [2]. SARS-CoV-2 can achieve person-person transmission through a variety of ways, including prolonged close contact or via the inhalation of virus-containing aerosols [3], [4]. People who are infected with SARS-CoV-2 typically present a fever, cough and chest discomfort, and severe patients develop acute respiratory distress syndrome (ARDS), leading to death in some cases [5], [6]. SARS-CoV-2 has spread worldwide due to its high transmission rate, and the number of deaths caused by this disease continues to increase. The global COVID-19 pandemic not only brings panic or worry to people, but also poses a great threat to the global economy, public health and the daily lives of people all over the world [7], [8]. Considering the current grave situation, the development of reliable inhibitors to combat SARS-CoV-2 has become a critical task. Currently, extensive efforts have been made to identify reliable inhibitors against SARS-CoV-2, and some inhibitors have even entered the stage of clinical trials. The results of some clinical trials have been briefly summarized in Table S1. Through the evaluation of possible inhibitors, it was found that some inhibitors failed to achieve the expected efficacy or showed obvious negative effects in clinical trials, which brings great difficulties to the treatment of COVID-19.
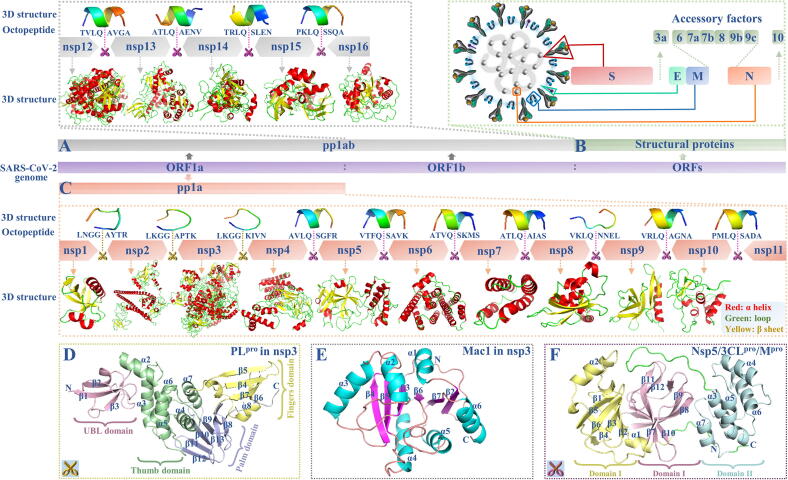
To identify effective viral inhibitors, it is necessary to have a comprehensive understanding of the potential therapeutic targets for SARS-CoV-2, and the study of the structural properties of SARS-CoV-2 can provide insights for the discovery of therapeutic targets and inhibitors. Recent developments in next-generation sequencing (NGS) technologies and related bioinformatics analysis methods can greatly assist in SARS-CoV-2 research. Within a month of the COVID-19 outbreak, the genome of SARS-CoV-2 was made available on public databases, such as the National Center for Biotechnology Information (NCBI) database [9]. SARS-CoV-2 has the largest genome among the known RNA viruses. The SARS-CoV-2 genome consists of 10 open reading frames (ORFs). Among them, ORF1a and ORF1b occupy approximately two-thirds of the whole SARS-CoV-2 genome, and they are used as templates to encode two large replicase polypeptides, pp1a and pp1ab, after the virus invades the host cell [10]. The pp1a and pp1ab can only initiate replication of their own genetic materials after they are cleaved into various fragments that perform different functions [11]; they can be separately cleaved into 11 non-structural proteins (nsps; nsp1-nsp11) and 16 nsps (nsp1-nsp16) by the virus-encoded 3-chymotrypsin-like proteinase (nsp5, also called 3CLpro or Mpro) and the papain-like proteinase (PLpro) that is present in nsp3 (Fig. 1A and C). Detailed information on the cleavage sites, cleavable octopeptides and the nsps of the replicase polypeptides in SARS-CoV-2 are shown in Fig. 1A and C, and they can be accurately predicted and included in our newly updated ZCURVE_CoV database (http://tubic.tju.edu.cn/CoVdb) [12]. According to the predicted results in Table S2, it was found that three nsps (nsp1-nsp3) were produced via the cleavage by PLpro, while the others were produced by 3CLpro. Three-dimensional (3D) models of these nsps are shown in Fig. 1A and Fig. 1C and can be downloaded directly from https://zhanglab.ccmb.med.umich.edu/C-I-TASSER/2019-nCoV [13]. Some of the models are also available in the Protein Data Bank (PDB) database (http://www.rcsb.org) [14]. The 3D models of the cleavable octopeptides were also predicted using a suitable PEP-FOLD3 software tool with a lower limit of 5 amino acids (http://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD3) [15], as shown in Fig. 1. As described by Chou et al., Gan et al. and Du el al., the cleavable octapeptide of SARS-CoV should be able to bind to the active site of 3CLpro, and therefore, modified octapeptides could be used as ideal SARS-CoV inhibitors [16], [17], [18]. Similarly, modified octapeptides of SARS-CoV-2 are also expected to become candidate inhibitors for SARS-CoV-2, but there is a lack of relevant research in this respect. Apart from ORF1a and ORF1b, the remaining ORFs are distributed in the last third of the SARS-CoV-2 genome, and they encode at least four structural proteins: spike (S), envelope (E), membrane (M) and nucleocapsid (N), as well as some accessory proteins, including 3a, 6, 7a, 7b, 8, 9b, 9c, and 10 (Fig. 1B) [19].
Fig. 1.
Structure of SARS-CoV-2: (A) replicase polypeptides pp1ab and corresponding cleavable octopeptide, (B) structural proteins and (C) replicase polypeptides pp1a and corresponding cleavable octopeptide. Structures of non-structural proteins: (D) PLpro in nsp3, (E) Mac1 in nsp3 and (F) nsp5.
S protein, as one of the four important structural proteins, plays a vital role in virus invasion and membrane fusion due to its two major regions (S1 and S2) [20]. S protein is regarded as an ideal target for vaccines and antibodies [20], [21], [22], and related studies have been well summarized by different groups [23], [24], [25], [26]. Among all non-structural proteins, two viral proteases (nsp5 and nsp3) appear to be particularly important. Nsp5 and PLpro in nsp3 are responsible for cleaving the large replicase polypeptides into fragments that can perform multiple functions, thereby assisting the replication of the virus. Meanwhile, PLpro and nsp5 contain highly conserved and well-defined druggable binding sites, whereas no binding sites are found in some nsps, such as nsp2, nsp7 and nsp8 [27]. In addition to PLpro, nsp3 also contains another important domain (macrodomain) related to viral replication and to the host’s innate immunity. Thus, nsp3 and nsp5 may potentially be good therapeutic targets for COVID-19. To promote a better understanding of the functions of nsp3 and nsp5 in SARS-CoV-2 infection and the current state of inhibitor discovery, we conducted a comprehensive review of the functions of these potential therapeutic targets and the development of corresponding inhibitors. This review is meant to provide a convenient resource of different viral targets and would assist with the development of specific inhibitors.
2. Structure, function and inhibitors of nsp3
SARS-CoV-2 nsp3 is a multi-domain enzyme with 1,945 amino acids, and it is the largest protein among all nsps. Nsp3 is remarkable because of the existence of two functional domains. One of the functional domains is a catalytically active PLpro domain. As shown in Fig. 1D, the PLpro can be divided into four distinct domains: the fingers (β4-β7), thumb (α2-α7), palm (β8-β13) and ubiquitin-like (UBL, β1-β3), and the first three domains constitute the catalytic core of the protein [28]. The druggable substrate-binding pocket is mainly determined by nine residues (R166, L185, L199, V202, E203, M206-M208 and K232) [29]. PLpro participates in the efficient cleavage of the N-terminal replicase polyprotein to produce functional proteins; this process of functional protein production plays an essential role in maintaining the basic cellular processes of SARS-CoV-2, including viral replication [30]. The other important functional domain of nsp3 is the ADP-ribose phosphatase (ADRP) domain, also known as macrodomain (Mac1) or Macro X domain. Mac1 mainly consists of seven β-sheets (β1-β7) and six α helices (α1-α6) (Fig. 1E). Among them, four β-sheets (β3 and β5-β7), α1, and two loops (β3-α2 and β6-α5) are involved in the formation of binding pocket [31]. Mac1 is a highly conserved and unique sequence in the genomes of many viruses and organisms, including human [32], [33], and inhibitors targeting this enzyme may have broad-spectrum antiviral activity. Notably, both PLpro and Mac1 can help the virus escape the antiviral immune response of the host [34]. Consequently, inhibiting the activity of PLpro and Mac1 will interfere with the replication cycle of the virus, maintain the innate immune pathways of the host and reduce the infection rate. In view of the importance of these two functional domains of nsp3, we summarized the inhibitor-related research that has been performed regarding these domains; moreover, we selected the best inhibitors from each study, as shown in Fig. 2. To gain insight into the binding modes of inhibitors, the binding mechanisms of these selected inhibitors to proteins are summarized, as shown in Table S3.
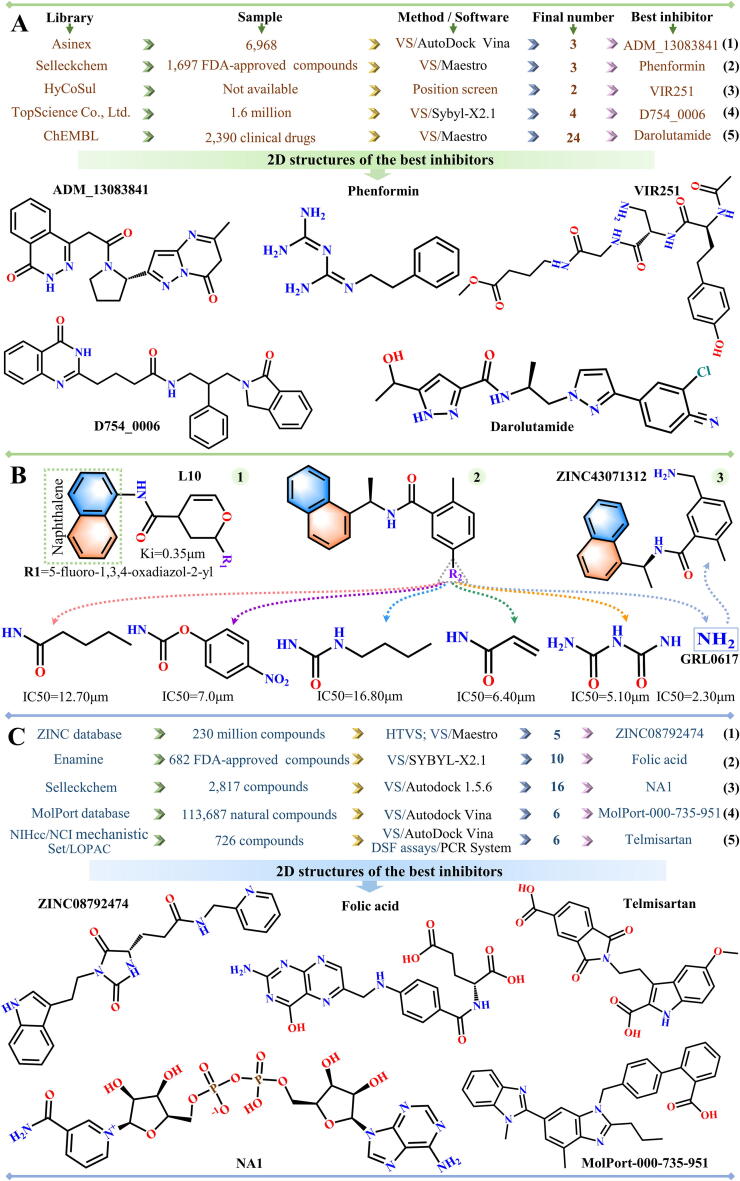
Fig. 2.
Flowchart illustrating the screening of inhibitors against (A-B) PLpro and (C) Mac1.
2.1. Inhibitors of SARS-CoV-2 PLpro
In April 2020, Osipiuk et al. first reported the crystal structure of SARS-CoV-2 PLpro, which is available in PDB (PDB ID: 6W9C). Based on this structure, studies have been performed to explore potential inhibitors of PLpro enzymatic activity. Alamri and colleagues conducted virtual screening (VS) of 6,968 protease inhibitors from Asinex library (https://www.asinex.com/protease) using AutoDock Vina software, and three compounds (ADM_13083841, LMG_15521745 and SYN_15517940) were identified as potential inhibitors of PLpro (Fig. 2A1) [35]. Kandeel et al. used the Maestro software to virtually screen 1,697 FDA-approved inhibitors from Selleckchem Inc. database (http://www.selleckchem.com) and obtained 26 compounds with lower negative docking scores (< -7 kcal/mol) [36]. After evaluating the binding free energies between compounds and PLpro, three compounds (phenformin, quercetin and ritonavir) were considered as potential inhibitors (Fig. 2A2). Rut et al. performed a comprehensive location screening of the LKGG motif based on the Hybrid Combinatorial Substrate Library (HyCoSul), and the results indicated that the tetrapeptides VIR250 and VIR251 are expected to behave as potent peptide inhibitors (Fig. 2A3) [37]. Using the structure of apo PLpro as a reference, they predicted the structures of VIR250 and VIR251 bound to PLpro (PDB IDs: 6WUU and 6WX4). Based on the interaction mode between VIR251 and SARS-CoV-2 PLpro, Pang et al. conducted virtual screening of 1.6 million compounds from TopScience Co., Ltd. (Shanghai, China) using the Sybyl-X2.1 software, verified their results through molecular dynamics (MD) simulations, and found that D754_0006 was the best among the four shortlisted inhibitors (F403_0159, F112_0109, G805_0497 and D754_0006) (Fig. 2A4) [38]. Based on the same molecular structure, a virtual screening study was carried out by Delre et al. to search SARS-CoV-2 PLpro inhibitors from 2,390 inhibitors used in clinical trials, as reported in the ChEMBL bank (https://www.ebi.ac.uk/chembl). Two covalent inhibitors (curcumin and afatinib) and several different types of non-covalent inhibitors (protein kinase inhibitors [dasatinib, pexidartinib and copanlisib], protease inhibitors [amprenavir, indinavir, anagliptin, boceprevir and semagacestat], adrenergic receptor modulators [vilanterol, arformoterol and atenolol], ACE inhibitors and direct oral anticoagulants [cilazaprilat, edoxaban and rivaroxaban] and inhibitors belonging to other groups [acotiamide, bentiromide, lymecycline, canagliflozin, darolutamide, lafutidine, vilazodone and methotrexate]) were obtained from this search [39]. Notably, covalent inhibitor binds irreversibly to the receptor through covalent bonds, while the binding of non-covalent inhibitor to the receptor is a reversible process. In general, the binding affinity of a covalent inhibitor to the target is stronger than that of the non-covalent inhibitor to the target [40], [41].
In addition, naphthalene-based inhibitors of SARS-CoV PLpro are highly effective in reducing the activity of SARS-CoV-2 PLpro enzyme [42]. Based on the naphthalene scaffold structure, Bhati and Osipiuk et al. analyzed several SARS-CoV-2 PLpro-related drugs through computational models and experiments, respectively. Among the 20 naphthalene-based compounds designed by Bhati, L10 (C18H14FN3O3) was considered to be the most potent anti-SARS-CoV-2 PLpro inhibitor (Fig. 2B1) [43], with a docking score of −8.81 kcal/mol. Osipiuk et al. tested the biochemical activity of their newly designed compounds, and six of them showed significant anti-PLpro activity in vitro, particularly compound 1 (GRL0617) (PDB ID: 7CMD) [44]. The structures and corresponding half maximal inhibitory concentration (IC50) values for these six inhibitors are shown in Fig. 2B2. In view of the strong inhibitory activity of GRL0617 against SARS-CoV-2 PLpro, a GRL0617-based virtual screening of chemical structures in the Binding database (BindingDB: http://www.bindingdb.org/bind/index.jsp) was conducted, five compounds (ZINC43063883, ZINC387735, ZINC78808978, ZINC43071312 and ZINC993539) were selected as potential inhibitors of SARS-CoV-2 PLpro [45], and ZINC43071312 showed the strongest binding ability to SARS-CoV-2 PLpro (Fig. 2B3).
2.2. Inhibitors of SARS-CoV-2 Mac1
The Mac1 of SARS-CoV-2 nsp3 has the characteristic of binding to ADP-ribose [46]; therefore, small molecules that can bind tightly to the binding sites of Mac1 and ADP-ribose are expected to be potential SARS-CoV-2 treatments. Based on an in-depth understanding of the interaction mechanism between Mac1 and ADP-ribose, investigations have been conducted on the inhibitors of this binding. Selvaraj et al. selected five inhibitors (ZINC08765069, ZINC08792474, ZINC08879336, ZINC08879971 and ZINC00897592) from 230 million compounds in the ZINC database (http://zinc.docking.org) via high-throughput virtual screening (HTVS), extra precision (XP) docking and quantum polarized ligand docking (QPLD) (Fig. 2C1) [46]. Based on the virtual screening method, 10 alternative inhibitors of Mac1 (folic acid, telmisartan, methotrexate, bosentan, lapatinib, gefitinib, ketoconazole, carvedilol, glyburide and avanafil) were identified from a screening of 682 FDA-approved compounds in the Enamine database by SYBYL-X2.1 (Fig. 2C2) [47], 16 inhibitors (ribostamycin sulfate, lactobionic acid, neohesperidin, lactitol, salvianolic acid, adenosine-5-diphosphoribose, folic acid, naringin, melibiose, maltose, rutin, nucleotide analogue 1 [NA1], NA2, NA3, nadide and citicholine) were identified from 2,682 FDA-approved compounds and 135 nucleoside analogues in the Selleckchem database by Autodock 1.5.6 (Fig. 2C3) [48], and 6 inhibitors (MolPort IDs: 000-735-951, 002-517-673, 021-745-738, 028-854-978, 028-856-111 and 035-700-887) were identified from 113,687 natural compounds in the MolPort database by Glide procedures (Fig. 2C4) [49]. After an in-depth assessment of the characteristics of inhibitors using various analyses, including MD analysis and absorption, distribution, metabolism, excretion and toxicity (ADMET) analysis, Folic acid, NA1 and MolPort-000-735-951 were determined to be the best among the inhibitors screened by the aforementioned research groups (Fig. 2C2-C4). Notably, the MD method applied by these researchers is a method commonly used for exploring the binding ability of ligands to receptors, which plays an important role in drug design [50], [51], [52]. Additionally, the Eppendorf Mastercycler ep Realplex Quantitative Realtime PCR System and AutoDock Vina software were separately used by Virdi et al. to conduct differential scanning fluorimetry (DSF) assays and virtual screening of 726 compounds, and steroids (estradiol valerate and flunisolide), β-lactams (cefaclor and cefatrizine) and benzimidazoles (rabeprazole and telmisartan) were identified as suitable compounds (Fig. 2C5) [53].
3. Structure, function and inhibitors of nsp5
Nsp5 is also described as the main protease (Mpro) or 3CLpro. It is a dimer structure composed of two monomers (residues 1–306) and each monomer has three domains (domains I, II and III), corresponding to residues 8–101, 102–184 and 201–303, respectively (Fig. 1F) [54]. As reported by Jin et al., residues located between domains I and II form the binding pocket of 3CLpro. Although the monomer state of 3CLpro is inactive, the homodimer state formed by the dimerization of two monomers is active. Indeed, monomers and dimers exist simultaneously in the solution, and their equilibrium is affected by many aspects, including the binding of inhibitors and protein concentration [55], [56], [57]. 3CLpro is essential for replication and transcription of the virus due to its function in cleaving replicase polypeptides (pp1a and pp1ab). Therefore, inhibiting the activity of this enzyme can significantly affect the replication and transcription of SARS-CoV-2. In addition, there are no homologues of 3CLpro in human [58], 3CLpro inhibitors are likely to cause fewer side effects on human. Therefore, 3CLpro is a potential drug target against COVID-19. A literature retrieval revealed that investigators are looking for inhibitors of 3CLpro from three main classes: natural compounds, approved or commercially available drugs and others.
3.1. Inhibitors of SARS-CoV-2 3CLpro screened from natural compounds
Natural compounds have the advantages of cost-effectiveness, high efficiency and low toxicity. Researchers are thus committed to mining inhibitors of COVID-19 from natural compounds. A literature retrieval revealed that the natural inhibitors currently under exploration are mainly derived from plants, marine organisms and microorganisms. The following is a summary of the natural inhibitors of SARS-CoV-2 3CLpro, and the binding mechanisms of these natural inhibitors to 3CLpro are listed in Table S4.
3.1.1. Plant-based natural inhibitors
3.1.1.1. Polyphenols
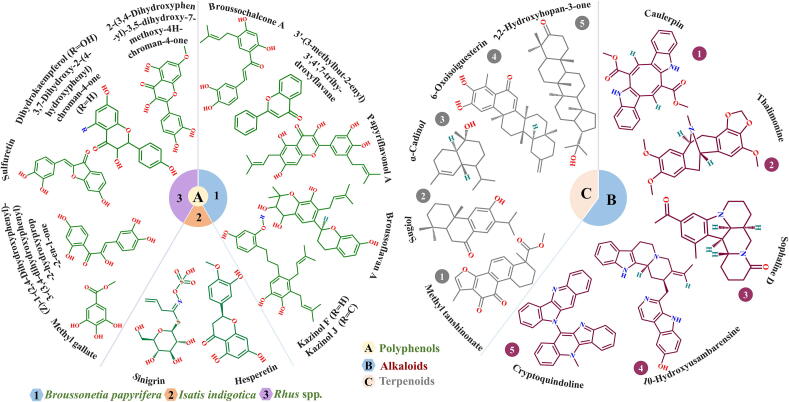
Polyphenols are a class of chemically diverse natural metabolites, and their structures are characterized by multiple phenol groups. Polyphenols have received attention in the pharmaceutical field owing to their important properties including pathogen defense, anti-oxidation and cancer prevention [59]. Studies have shown that some plant-derived polyphenols can inactivate SARS-CoV-2 by inhibiting the activity of SARS-CoV-2 3CLpro. Based on the structure of SARS-CoV-2 3CLpro, Ghosh et al. evaluated the inhibitory ability of 18 polyphenolic compounds extracted from Broussonetia papyrifera (10 compounds) and Isatis indigotica root (8 compounds) using computational approaches. Notably, all the selected compounds have known anti-SARS-CoV activity. After evaluating the binding affinity, structural stability and physicochemical properties, six possible polyphenolic inhibitors of SARS-CoV-2 3CLpro (broussochalcone A, papyriflavonol A, 3′-(3-methylbut-2-enyl)-3′,4′,7-trihy-droxyflavane, broussoflavan A, kazinol F and kazinol J) were screened from Broussonetia papyrifera and two inhibitors (sinigrin and hesperetin) were identified from Isatis indigotica (Fig. 3A1-A2) [60], [61]. Another study found that six polyphenolic compounds extracted from Rhus spp. could also potentially assist in the treatment of COVID-19, and the structures of these compounds are shown in Fig. 3A3 [62].
Fig. 3.
Structures of potential inhibitors targeting SARS-CoV-2 3CLpro: (A) polyphenolic inhibitors, (B) alkaloid inhibitors and (C) terpenoid inhibitors.
3.1.1.2. Alkaloids
Alkaloids are a class of natural compounds found mainly in plants, particularly flowering plants. Alkaloids are considered to be one of the most pharmacologically active substances found in plants, including those that assist in the defense against pathogens [63]. Recently, computational analyses were performed on 10 public bioactive compounds and 20 alkaloid compounds, all of which have known antiviral activity [64], [65]. After evaluating the compounds from various aspects including physiochemical properties, three alkaloid compounds (caulerpin, thalimonine and sophaline D) were identified as potential inhibitors of SARS-CoV-2 3CLpro (Fig. 3B1-B3). Similarly, a computational approach was applied by Gyebi et al. to explore inhibitors of SARS-CoV-2 3CLpro from 62 alkaloid compounds in African plants, which yielded two drug candidates, that is 10-hydroxyusambarensine and cryptoquindoline (Fig. 3B4-B5) [66].
3.1.1.3. Terpenoids and their derivatives
Structurally, terpenoids contain one or more isoprene units, and their general structure is (C5H8)n. Terpenoids can be classified into different types according to the number of isoprene unit, such as monoterpenes, sesquiterpenes and diterpenes [67]. They are involved in metabolic pathways of all living organisms and have a variety of pharmacological applications [68]. Importantly, previous studies have shown that some subtypes of terpenoids have strong antiviral activity against coronaviruses, such as CoV-229E and SARS-CoV [69], [70]. Thus, terpenoids and their derivatives may be helpful in the treatment of COVID-19.
In view of these findings, Diniz et al. reviewed 34 anti-coronavirus terpenoids from several studies, and identified three terpenoid compounds (methyl tanshinonate, sugiol and α-cadinol) that could bind tightly to SARS-CoV-2 3CLpro based on the binding free energies calculated using the molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) method (Fig. 3C1-C3) [71]. In addition, Gyebi et al. not only explored the antiviral properties of alkaloids, but also discovered two potential SARS-CoV-2 3CLpro inhibitors (6-oxoisoiguesterin and 22-hydroxyhopan-3-one) from a set of 100 terpenoids extracted from African plants (Fig. 3C4-C5) [66]. Therefore, the aforementioned studies also suggested that terpenoids and their derivatives can be potentially developed into drugs against SARS-CoV-2.
3.1.1.4. Flavonoids
Flavonoids are widely distributed in many parts of plants and are indispensable compounds for physiological processes in plants. Flavonoids, especially glycosylated flavonoids, are considered as potential inhibitors with the ability to inhibit the activities of multiple proteases, including SARS-CoV 3CLpro [72]. In addition, a study has shown that almost all the annotated flavonoids extracted from Salvadora persica could stably bind to SARS-CoV-2 3CLpro [73], suggesting that flavonoids may be suitable for the treatment of COVID-19.
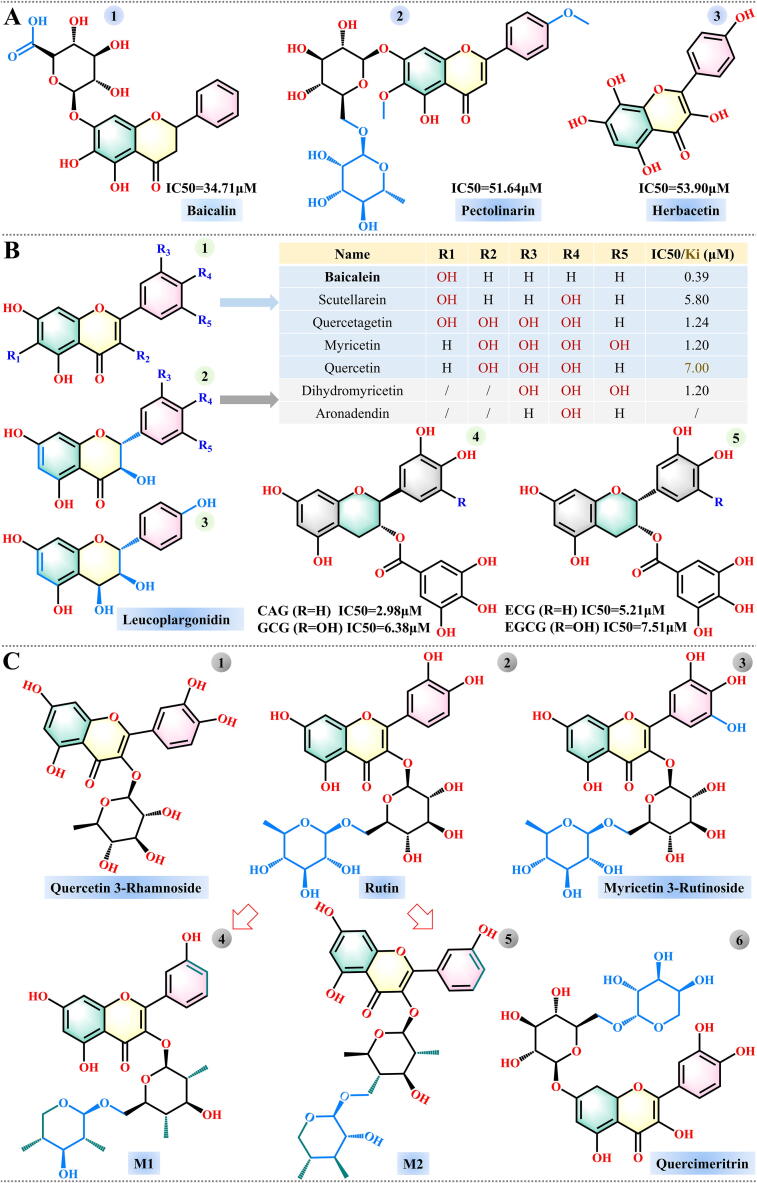
Taking into account the sequence similarity between SARS-CoV and SARS-CoV-2 3CLpro, Jo et al. mined the flavonoid library using a proteolytic method, and three compounds (baicalin, pectolinarin and herbacetin) exhibited better inhibitory effects, with baicalin showing the greatest effect [74]. The measured IC50 values for these three compounds were 34.71, 51.64 and 53.90 μM, respectively (Fig. 4A1-A3). However, baicalin, as one of the four main ingredients (baicalein, baicalin, wogonin, and wogonoside) of Scutellaria baicalensis, was not the compound with the strongest SARS-CoV-2 3CLpro inhibitory activity. Experimental investigation by Liu and colleagues found that the flavonoid baicalein was the most promising inhibitor of SARS-CoV-2 among the four, with a corresponding IC50 of 0.39 μM. They then tested 10 analogues of baicalein obtained from suppliers, and 4 flavonoid compounds (scutellarein, dihydromyricetin, quercetagetin and myricetin) had the highest inhibitory activity against SARS-CoV-2 3CLpro (Fig. 4B1 and 4B2) [75]. It is worth noting that the high binding affinity between myricetin and SARS-CoV-2 3CLpro has been confirmed by other computational study [76]. Quercetin is also an analogue of baicalein, and it was recognized as the most promising inhibitor of SARS-CoV-2 3CLpro among 150 compounds, with an inhibition constant (Ki) of 7 μM [77]. Other studies on SARS-CoV-2 3CLpro have also identified quercetin as potential inhibitor. Sen et al. performed molecular docking and dynamic studies on 1,040 natural compounds derived from an in-house database, and three (quercetin, aronadendin and leucopelargonidin) of the four identified inhibitors of SARS-CoV-2 3CLpro belong to the flavonoids group (Fig. 4B2 and B3) [78]. Furthermore, a large number of natural flavonoids obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov) were evaluated, and three compounds (quercetin 3-rhamnoside, rutin and myricetin 3-rutinoside) were selected as drug candidates (Fig. 4C1-C3). Among them, quercetin 3-rhamnoside had the lowest negative docking score (−9.7 kcal/mol), whereas the rutin-3CLpro complex was the most stable [79]. Another drug study on SARS-CoV-2 3CLpro also confirmed that rutin had the strongest inhibitory activity among 18 compounds extracted from Manilkara hexandra (Roxb.) [80]. Hence, rutin and its derivatives are expected to be potential natural inhibitors of SARS-CoV-2 3CLpro. Huynh et al. optimized the structure of rutin and developed two hydrophobic analogues of rutin (M1 and M2), as shown in Fig. 4C4-C5 [81]. The binding of compounds M1 and M2 to SARS-CoV-2 3CLpro was significantly enhanced compared with the binding of rutin.
Fig. 4.
Structures of flavonoid inhibitors of SARS-CoV-2 3CLpro.
Flavonoids can be divided into several sub-categories according to their chemical structure [82]. Anthocyanins represent one of the flavonoid groups, and have been extensively studied by Fakhar et al. [83]. They conducted a virtual screening of 3,435 anthocyanin-derived compounds from the PubChem database and selected two optimal drug candidates (compound IDs: 44256921 and 131751762) based on their docking score, physicochemical properties, structural stability and binding free energy. The basic structure of anthocyanins and these two compounds are shown in Fig. S1. Another class of flavonoids, flavan-3-ols, has also attracted attention. Zhu et al. performed docking simulations and experimental investigations on 10 main Flavan-3-ols compounds, and found that four of them (catechin gallate [CAG], epicatechin gallate [ECG], gallocatechin gallate [GCG] and epigallocatechin-3-gallate [EGCG]) had the potential to be utilized as inhibitors of SARS-CoV-2 3CLpro, with IC50 values of 2.98, 5.21, 6.38 and 7.51 μM, respectively (Fig. 4B4-B5) [84]. These four compounds are abundant in tea plants, particularly green tea [85], [86]. The ingredients in green tea may be helpful in fighting SARS-CoV-2, owing to their multi-faceted functions [87]. In fact, a study of eight compounds with known antiviral activity in green tea also found that EGCG, ECG and GCG are the best drug candidates for inhibiting the activity of SARS-CoV-2 3CLpro [88]. Other related studies have also confirmed the inhibitory effect of EGCG against SARS-CoV-2 3CLpro [89], [90], [91].
3.1.1.5. Biflavonoids
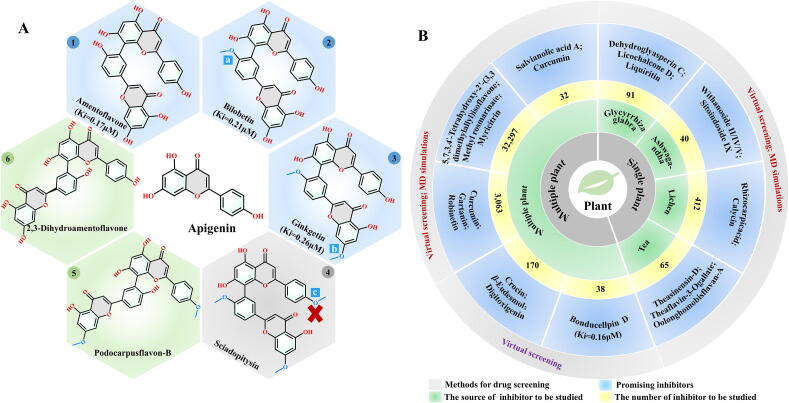
Previous experiments confirmed that four biflavonoids (amentoflavone, bilobetin, ginkgetin, sciadopitysin) and eight diterpenoids isolated from Torreya nucifera showed inhibitory activity against SARS-CoV [92]. Recently, the inhibitory effect of these compounds against SARS-CoV-2 was evaluated by Ghosh et al. [93]. Their computation results show that three of the four bioflavonoids: amentoflavone, bilobetin and ginkgetin, can bind stably to the binding sites of 3CLpro (PDB ID: 6LU7). The binding free energies of these three inhibitors to the protein calculated using the molecular mechanics generalized Born surface area (MM-GBSA) method were −59.57, −66.31 and −63.62 kcal/mol, respectively, and the corresponding Ki values were 0.17, 0.21 and 0.26 μM, respectively (Fig. 5A1-A3). As shown in Fig. 5A1-A3, the screened compounds share structural similarity and the differences among them originate from the different substitutions at positions a and b of the apigenin motif. Notably, biflavonoid sciadopitysin was also derived from the replacement of the apigenin motif at positions a, b, and c (Fig. 5A4), but it was revealed to be a toxic molecule that is not suitable for use as an antiviral drug. Based on this result, substitutions on the structure of apigenin can be attempted to identify inhibitors with more favorable physical and chemical properties.
Fig. 5.
Structures and sources of potential inhibitors targeting SARS-CoV-2 3CLpro: (A) structures of biflavonoid inhibitors of SARS-CoV-2 3CLpro and (B) potential inhibitors selected from different plants.
Flavonoids and biflavonoids are expected to behave as inhibitors of SARS-CoV-2 3CLpro. Bharadwaj et al. elucidated that four best inhibitors screened from 653 natural compounds in the NP-lib database are flavonoids (rutin, quercimeritrin 6′'-O-L-arabinopyranoside) (Fig. 4C2 and C6) and biflavonoids (2,3-dihydroamentoflavone and podocarpusflavon-B) (Fig. 5A5-A6) [94]. Therefore, there is an urgent need to design and optimize the structure of flavonoids and biflavonoids to develop powerful inhibitors of SARS-CoV-2.
3.1.1.6. Others
Based on the above studies, it can be inferred that some medicinal plants may be good sources of compounds that could potentially behave as inhibitors targeting SARS-CoV-2. Due to the lack of effective drugs, some medicinal plants have been studied under emergency situations, and a few studies have attempted to extract antiviral medicinal ingredients from a single plant [95], [96], [97], [98]. Potential inhibitors of SARS-CoV-2 3CLpro, including dehydroglyasperin C, licochalcone D and liquiritin, were found in Glycyrrhiza glabra, while other compounds including withanoside II, withanoside IV, withanoside V and sitoindoside IX were extracted from Withania somnifera (Ashwagandha). Moreover, calycin and rhizocarpic acid have been found in Lichen. Efforts have also been made to identify the most effective inhibitors by screening compounds from multiple plants [99], [100], [101], [102], [103]. For example, Mahmud et al. identified three potential inhibitors (curcumin, gartanin and robinetin) that can stably bind to SARS-CoV-2 3CLpro by screening 3,063 compounds from >200 plants. For ease of reference, all prospective SARS-CoV-2 3CLpro inhibitors and their corresponding sources are shown in Fig. 5B.
3.1.2. Natural inhibitors extracted from marine organisms
The marine ecosystem is rich in resources and comprises a wide variety of organisms. Recently, an increasing number of natural medicines for preventing or treating many diseases have been extracted from marine organisms. According to previous studies, some marine-derived natural compounds have been demonstrated to possess antiviral and antibacterial activities [104], [105]. Moreover, some marine-derived natural compounds have activities that are applied in treating nervous and immune systems, while others are used as diagnostic tools. Therefore, marine natural compounds represent a favorable source for the development of pharmaceuticals that may be used to alleviate COVID-19 pandemic. The marine natural product library (http://docking.umh.es/downloaddb) is a valuable database for mining marine drugs, which contains >10,000 compounds. Structure-based and ligand-based virtual screening studies were performed separately using this database, and 17 of 14,064 marine natural compounds were selected as putative inhibitors of SARS-CoV-2 3CLpro. Among these, heptafuhalol A has the lowest binding free energy [106]. Other computational studies demonstrated that four marine drugs (eribulin mesylate, plitidepsin, trabectedin and fostularin 3) also had excellent affinity for binding to SARS-CoV-2 3CLpro [107], [108].
3.1.3. Natural inhibitors of microbial origin
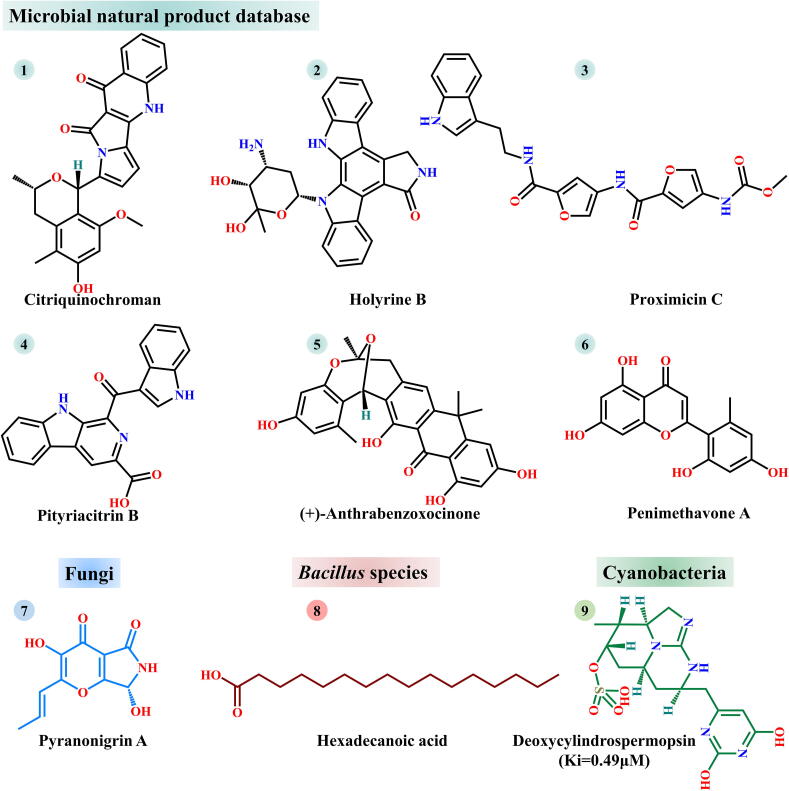
The earth is rich in microorganisms, and bioactive substances acquired from microorganisms are of great significance in the development of novel drugs. Therefore, natural inhibitors of microbial origin have attracted extensive attention of the pharmaceutical industry. Since the outbreak of SARS-CoV-2, researchers have searched for inhibitors of microbial origin. There is currently a large microbial natural product database (https://www.npatlas.org/joomla/index.php) containing >20,000 compounds from bacteria and fungi [109]. To obtain high-efficiency inhibitors that target SARS-CoV-2 3CLpro activity, one research group conducted a layer-by-layer screening of 24,581 compounds from this database, and six of them (citriquinochroman, holyrine B, proximicin C, pityriacitrin B, (+)-anthrabenzoxocinone and penimethavone A) were identified to be high potential for inhibiting SARS-CoV-2 3CLpro (Fig. 6[1]–[6]) [110]. Another study conducted computational screening of 100 fungal metabolites from PubChem using molecular docking and MD simulations. Among these 100 selected metabolites, pyranonigrin A was regarded as a potent inhibitor of SARS-CoV-2 3CLpro (Fig. 6[7]) [111]. Based on similar computational approaches, hexadecanoic acid and deoxycylindrospermopsin were selected from the metabolites of Bacillus species and cyanobacteria, respectively (Fig. 6[8]–[9]) [112], [113].
Fig. 6.
Microbe-derived natural inhibitors of SARS-CoV-2 3CLpro. Compounds 1–6, 7, 8 and 9 were selected from microbial natural product database, fungi, Bacillus species and cyanobacterial metabolites, respectively.
3.2. Inhibitors of SARS-CoV-2 3CLpro screened from approved or commercially available drugs
Given the current critical public health situation, i.e. lack of effective drugs to control COVID-19, the repurposing of already approved or commercially available drugs is a quick and desirable strategy to develop safe and effective treatments. Drugrepurposing is a process of re-screening existing drugs for their new applications using related techniques, and therefore, it is also regarded as drug recycling, drug repositioning, etc [114]. Previously approved drugs have many undeniable advantages over newly developed drugs. For example, they have known safety, pharmacokinetics and toxicity profiles, which not only save time and investments, but also reduce the possibility of negative effects on the human body [115]. Additionally, there are infrastructures available for the large-scale production of approved or commercially available drugs, which greatly improves the efficiency of drug production [116]. It is therefore a good strategy to screen inhibitors that can control the activity of SARS-CoV-2 3CLpro among the approved or commercially available drugs. Of course, this strategy will also encounter many challenges including patent application, investment and unexpected negative effects [117].
A literature retrieval revealed that the main sources of approved drugs are existing databases, including DrugBank (http://www.drugbank.ca), Druglib (http://www.druglib.com), eDrug3D (http://chemoinfo.ipmc.cnrs.fr/edrug3d), Reaxys (https://www.reaxys.com), Selleckchem Inc., Targetmol (https://www.targetmol.com), SuperDrugs2 (http://cheminfo.charite.de/superdrug2), ZINC, PubChem, in-house [118], Screen-Well ® FDA v. 2.0 Approved Drug, Drug Target Common (DTC) (https://drugtargetcommons.fimm.fi), Enamine and BindingDB databases, and the commonly used methods for drug screening are VS, HTVS, molecular docking and MD simulations. The likely potential inhibitors, selected via studies on approved or commercially available drugs from different sources, are summarized in Table 1. Among these, inhibitors identified in multiple studies are shown in bold, i.e. tipranavir, cobicistat, lopinavir, ritonavir, teniposide, bromocriptine, saquinavir and atazanavir.
Table 1.
Potential inhibitors of SARS-CoV-2 3CLpro selected from approved or commercially available drugs.
| Source | Numbera | Method | Potential inhibitor |
|---|---|---|---|
| SuperDrug2 | 4,600 clinically approved drugs [125] | VS; Molecular docking; MD simulations |
Binifibrate; Bamifylline |
| 3,987 FDA-approved drugs [126] | VS; Molecular docking; MD simulations; Experimental evaluation |
Ivermectin (IC50 = 21.53 μM); Tipranavir (IC50 = 27.66 μM); Boceprevir (IC50 = 31.36 μM); Micafungin (IC50 = 47.63 μM); Paritaprevir (IC50 = 73.38 μM); Ombitasvir (IC50 = 75.49 μM); |
|
| DrugBank | 2,100 FDA-approved drugs [127] | Cobicistat (IC50 = 6.7 μM) | |
| 2,454 FDA-approved drugs [128] | VS; Molecular docking; MD simulations |
Hyaluronic acid; Acarbose; Lopinavir | |
| 2,800 FDA-approved drugs [129] | Tipiracil; Aprepitant | ||
| Commercially available drugs [130] | Leuprolide; Nelfinavir; Ritonavir; Teniposide; Valrubicin | ||
| Druglib | 1,051 FDA-approved drugs [131] | R428; Teniposide; VS-5584; Setileuton | |
| ZINC | 1,615 FDA-approved drugs and available in the market [132] | Dihydroergotamine (Kd = 107.6 μM); Midostaurin (Kd = 43.5 μM); Ziprasidone | |
| 4,384 approved drugs [133] | Ergotamine; Bromocriptine; Meclocycline; Amrubicin | ||
| 5,903 approved clinical drugs [134] | Viomycin; Capastat; Carfilzomib; Saquinavir | ||
| PubChem | 77 FDA-approved drugs [135] | Lopinavir-Ritonavir; Tipranavir; Raltegravir | |
| 400 commercially available curcumin analogues [136] | Cyclohexanone | ||
| DrugBank; PubChem |
Approved drugs and bioactive compounds [137] | ChEMBL275592; Montelukast; ChEMBL288347; Bromocriptine; Saquinavir |
|
| Selleckchem | 3,809 conformations [138] | HTVS; Molecular docking; MD simulation |
Amikacin (Kd = 17.5 nM) |
| 487 FDA-approved drugs [139] | VS | Ribavirin; Telbivudine; Vitamin B12; Nicotinamide | |
| Selleckchem; Targetmol |
3,118 FDA-approved drugs [140] | Angiotensin II; GHRP-2; Indinavir; Polymyxin B; Fexofenadine; Atazanavir; Cobicistat; Caspofungin; Lopinavir | |
| e-Drug3D; Reaxys |
6,466 approved drugs [141] | Perampanel; Carprofen; Celecoxib; Alprazolam; Trovafloxacin; Sarafloxacin; Ethyl biscoumacetate | |
| In-house database | 2,000 approved drugs [142] | VS; Experimental evaluation |
Manidipine (IC50 = 4.8 μM); Boceprevir(IC50 = 5.4 μM); Lercanidipine(IC50 = 16.2 μM); Bedaquiline (IC50 = 18.7 μM); Efonidipine (IC50 = 38.5 μM) |
| Screen-Well | 774 FDA-approved drugs [143] | HTVS; Molecular docking |
Ethacrynic acid (IC50 = 1.11 μM); Naproxen (IC50 = 3.45 μM); Allopurinol (IC50 = 3.77 μM); Butenafine hydrochloride (IC50 = 5.40 μM); Raloxifene hydrochloride (IC50 = 5.61 μM); Tranylcypromine hydrochloride (IC50 = 8.64 μM); Saquinavir mesylate (IC50 = 9.92 μM) |
| DTC; BindingDB | 3,410 FDA-approved drugs [144] | Deep learning | Atazanavir (Kd = 94.94 nM); Remdesivir (Kd = 113.13 nM); Efavirenz (Kd = 199.17 nM); Ritonavir (Kd = 204.05 nM); Dolutegravir (Kd = 336.91 nM) |
| Literatures | 100 clinically approved drugs [145] | Experimental evaluation | Teicoplanin (IC50 = 1.5 μM) |
| PubChem | 22 FDA-approved glucocorticoids [146] | Molecular docking; MD simulations |
Ciclesonide; Dexamethasone; Betamethasone; Hydrocortisone; Fludrocortisone; Triamcinolone |
| 40 FDA-approved non-steroidal anti-inflammatory drugs [147] |
Sulfinpyrazone; Indomethacin; Auranofin | ||
| 9 FDA-approved angiotensin receptor blocker drugs [148] |
Olmesartan (IC50 = 1.808 μM) | ||
| Enamine | 8960 commercially available compounds [149] | HTVS; Molecular docking; MD simulation; Experimental evaluation |
Z1244904919 (IC50 = 0.73 μM); Z1759961356(IC50 = 0.69 μM) |
Note: 1. The inhibitors in bold indicate that they have been confirmed through different studies.
2. The binding mechanisms of all inhibitors to 3CLpro are listed in Table S5.
The number of all compounds used for drug screening.
3.3. Others
Based on the aforementioned literature review, it is clear that existing databases play an important role in mining for inhibitors. In addition to the databases mentioned in this review, other databases have also attracted widespread attention, including Existing drug database (https://www.pnas.org/content/suppl/2020/10/12/2010470117.DCSupplemental), Asinex antiviral database (https://www.asinex.com/antiviral), ChEMBL, SuperNatural II (http://bioinformatics.charite.de/supernatural), WithDrawn (http://cheminfo.charite.de/withdrawn), Compound Library (https://www.pharmchem1.uni-bonn.de/www-en/pharmchem1-en/mueller-laboratory/compound-library), Chemical Abstract Services (CAS) (https://www.cas.org/covid-19-antiviral-compounds-dataset), LASSBio (http://www.lassbio.icb.ufrj.br), PDB database, SwissSimilarity (http://www.swisssimilarity.ch), Life Chemicals database (https://lifechemicals.com) and Pharmit (http://pharmit.csb.pitt.edu). Following the sudden outbreak of SARS-CoV-2 infections, researchers have extensively screened the existing databases to investigate effective inhibitors of SARS-CoV-2. Some studies were performed on a single database, while others were performed on multiple databases. In addition, existing literature also provides sets of potential compounds eligible for drug screening. The corresponding studies are summarized in Table 2. Meanwhile, some groups have carried out the redesign, modification, and re-synthesis of drugs for SARS-CoV-2 3CLpro based on known inhibitor, key pharmacophore or active sites of Mpro [119], [120], [121], which may stimulate the discovery of novel drugs to alleviate the threat of COVID-19.
Table 2.
Potential inhibitors of SARS-CoV-2 3CLpro selected from existing databases or literature.
| Source | Number | Method | Potential inhibitor |
|---|---|---|---|
| ZINC | 2,000 [150] | VS | ZINC32960814/12006217/03231196/33173588 |
| 1,500a[151] | ZINC20291569/90403206/95480156 | ||
| 5,811b[152] | SCAR protocol | Telcagepant; Vidupiprant; Poziotinib; Fostamatinib. | |
| 606 million [153] | VS; MD simulations |
(−)-Taxifolin; Rhamnetin | |
| 1,000c[154] | ZINC000621278586/000621285995 | ||
| Asinex; ChEMBL | > 8,722 [155] | BBB_26580140; SCHEMBL12616233/18616095/20148701 | |
| SuperNatural II; In-house database; SuperDrug2; WithDrawn |
360,000 [156] | SN00017653 (SuperNatural II); Pseudostellarin C (in-house); Eledoisin (SuperDrug2); Naldemedine (SuperDrug2); Saralasin (WithDrawn); Saquinavir (WithDrawn) |
|
| Chemical Abstract Services | 35,000 [157] | SKS-01; SKS-02; SKS-03; SKS-04; | |
| Literatures; PubChem; Asinex |
> 10,584d[158] | pq8; pq9; pq10; A12 Note: Structures of these inhibitors are shown in Fig. S2. |
|
| Protein Data Bank | 2,692 [159] | PubChem IDs: 118098670; 104161460; 163632044 | |
| Literature | 49e[160] | Ethaselen (IC50 = 4.51 μM) | |
| PubChem | 10,433f[161] | 6-Deaminosinefungin; UNII-O9H5KY11SV | |
| Life Chemicals; Asinex | 21,207g[162] | F2679-0163(Life chemicals); F6355-0442 (Life chemicals); 8250 (Asinex) | |
| Pharmit | 213.5 million [163] | CSC057752019; PubChem-22029441/-11210821; MCULE-9349798441; MolPort-045-918-905 |
|
| DrugBank; Literatures; In-house database |
2,736h[164] | Acteoside; Chebulinic acid; Delphinidin-3,5-diglucoside; Saquinavir; Lithospermic acid B; 11m_32045235 |
|
| DrugBank | 10,036b[165] | DB02388; Cobicistat | |
| 10,246 [166] | Levothyroxine; Amobarbital; ABP-700 | ||
| 13,227 [167] | VS | DB02128/01871/04502/02378/03063/04378/03648/04353/04692/047https://doi.org/10/08732/11938/03395/04653/04758/07571/07934/12955;DBMET00084/01550/01549/01548 | |
| ChEMBL | 1,485,144 [168] | CHEMBL1559003/2237553/1511674/3260476/1170272/1335000/2235580/3264032/1447105/589899/1539803/2216842/427787/1339675/3126648/1302388/3234783/1807774/2087984/2387487/2113271/476947/399042/2000247/3236740/1447944/1760165/2087965 | |
| 33 protease inhibitors [169] | Paritaprevir; Ciluprevir; Simeprevir; Deldeprevir; Indinavir; Saquinavir; Faldaprevir; Brecanavir; Grazoprevir; Lopinavir |
||
| Protein Data Bank; SwissSimilarity |
133i[170] | Melatonin | |
| Existing drug database | > 2,500 [171] | VS; MD simulations; Experimental evaluation |
Dipyridamole (Ki = 0.04 μM); Hydroxychloroquine (Ki = 0.36 μM); Chloroquine (Ki = 0.56 μM) |
| LASSBio | 2,300 [172] | VS; Experimental evaluation |
LASSBio-1945 (IC50 = 15.97 μM) |
| DrugBank; PubChem | 5,016j[173] | HTVS; MD simulations | Isavuconazonium; α-KI; Pentagastrin |
| Compound Library | 23,000 [174] | HTVS | Azanitrile (Ki = 24.0 nM); Pyridyl ester(Ki = 10.0 nM) |
| In-house database; TargetMol; Selleckchem; Antivirus Drug Library |
~ 10,000 [54] | HTVS; VS; Experimental evaluation |
Ebselen (IC50 = 0.67 μM); Disulfiram (IC50 = 9.35 μM); Tideglusib (IC50 = 1.55 μM); Carmofur (IC50 = 1.82 μM); Shikonin (IC50 = 15.75 μM); PX-12 (IC50 = 21.39 μM) Note: These inhibitors have been further confirmed [175], [176], [177], [178], [179]. |
| AVPdb database | 88 [180] | Molecular docking; MD simulations |
P14, P39, P41, P74 Note: Sequences of these inhibitors are shown in Table S7. |
Note: The binding mechanisms of all inhibitors to 3CLpro are listed in Table S6.
These compounds were selected according to the pharmacophore features of inhibitor N3.
Approved or investigational drugs.
Protease-inhibitor-like compounds.
Analogs of Chloroquine.
Se-containing heterocyclic compounds.
The compounds that have > 90% structural similarity with the 10 high-affinity molecules previously reported (PubChem IDs: 5281605, 16394003, 19323586, 44137675, 118737648, 787400, 8386889, 34755, 65482 and 145998233).
12,485 anti-SARS-CoV-2 or antiviral compounds in the Life Chemicals database and 8,722 antiviral compounds in Asinex database.
2,454 FDA approved drugs from DrugBank, 144 coronavirus Mpro inhibitors from published literature and 138 natural compounds from an in-house database.
74 ligand-Mpro complexes and 59 analogues.
6 analogs of SARS-CoV-2 3CLpro inhibitor (13b) [58] in DrugBank and 5,010 analogs of 11 antiviral agents (Atazanavir, Darunavir, Fosamprenavir, Indinavir, Lopinavir, Ritonavir, Saquinavir, Tipranavir, Delavirdine, Nevirapine and Remdesivir) in PubChem.
4. Dual inhibitors
The inhibitors summarized above are mainly for a single target (nsp3 or nsp5) of SARS-CoV-2. Of course, the development of dual inhibitors for different targets of SARS-CoV-2 is also a more promising direction that cannot be ignored, which may greatly reduce the combination of drugs and improve the efficiency of treatment. To better understand the current development status of dual inhibitors, we have briefly summarized the studies on dual inhibitors related to nsp3 and nsp5, which are listed in Table 3.
Table 3.
Dual inhibitors against multiple SASR-CoV-2 targets including nsp3, nsp5 or both.
| Promising dual inhibitor | Source | Target |
|---|---|---|
| Naloxone; Fluoxetine; Dopal; Thiamine Phenylephrine; Epinephrine; Aspirin Pseudoephedrine; Benzenebutyrate; Nelfinavir; Tipranavir |
ZINC database [181], [182] | Nsp5; PLpro |
| Bemcentinib; MFCD00832476 | ZINC PubChem and NPASSa databases [183] |
|
| Ginkgolic acid (IC50 = 1.79; 16.30 μM); Anacardic acid (IC50 = 2.07; 17.08 μM) |
MedChemExpress [184] | |
| 2001083-68-5; 2001083-69-6; 833463-19-7 |
CAS Antiviral COVID19 database [185] | Nsp5; RdRpb |
| Nakinadine B; Amphimedoside C 20-hepacosenoic acid; |
Amphimedon sp[186] | |
| Hydroxymatairesinol | Sesame [187] | Nsp5; PLpro; RdRp |
| MCULE-3732245601–0 (IC50 = 0.45; 0.085; 0.29 μM) MCULE-7013373725–0 (IC50 = 0.11; 0.063; 0.29 μM) |
MCULE library [188] | Nsp5; PLpro; furin protease |
| 19 acetylsesterstatin 3 12β-acetoxy,16-epi-hyrtiolide |
Hyrtios erectus[189] | Nsp5; nsp15 |
| Cefiderocol; Plazomicin | PubChem database [190] | Nsp5; nsp16 |
| Vanganciclovir | Nsp5; N protein | |
| Rutin | DrugBank [191] | Nsp5; S protein |
| Solanine, Acetoside; Rutin; Withanone | PubChem database [192], [193] |
NPASS: Natural Product Activity and Species Source.
RdRp: RNA-dependent RNA polymerase.
5. Summary and outlook
The paucity of relevant information on therapeutic targets and development of inhibitor against SARS-CoV-2 has hindered the treatment of COVID-19. In this review, we have summarized the functions and the most likely potential inhibitors of viral targets (nsp3 and nsp5) to promote the development of drugs against SARS-CoV-2. During the literature retrieval for this review, we deem that existing databases play an important role in drug screening. Based on the different domains of nsp3, the inhibitors of nsp3 can be divided into two categories: PLpro inhibitors and Mac1 inhibitors. Inhibitors of nsp5 can be divided into three categories according to their sources: natural compounds, approved or commercially available inhibitors and others. Natural compounds are derived mainly from plants, marine organisms and microorganisms. Plant-derived compounds can be further subdivided into six categories: polyphenols, alkaloids, terpenoids, flavonoids, biflavonoids and others. Meanwhile, the binding mechanisms of the aforementioned promising inhibitors to the targets are also summarized. From the perspective of methods used in drug screening, VS, HTVS, molecular docking and MD simulations are commonly used. However, there is still a need to develop faster and more accurate methods for drug screening.
Although extensive effort has been made to explore and develop SARS-CoV-2 inhibitors, most of the current research is focused on in silico analysis, and there is a lack of relevant experimental confirmation or in vitro verification. Future research should be conducted with more in-depth experimental investigations and in vitro verification based on the available computational data. Moreover, studies of dual inhibitors for multiple targets of SARS-CoV-2 are relatively lacking. In addition to the development of single inhibitors against COVID-19, more attention should also be paid to the exploration of dual inhibitors in the future. Another issue that cannot be ignored is that the proteases of SARS-CoV-2 may undergo unpredictable mutations at any time, and some mutations may enhance the structural stability and drug resistance of the proteases [122], [123], which brings great challenge to the drug design targeting them. It is recommended to develop relevant technologies or software to predict high-risk or drug-resistant mutations, so as to find effective drugs to combat the mutant SARS-CoV-2 in advance. The last thing that needs to be pointed out is the importance of selective inhibitors, because some inhibitors of nsp3 and nsp5 may be related to the activity of the host protein, such as human cathepsins L and B [124].
CRediT authorship contribution statement
Fangfang Yan: Investigation, Software, Visualization, Writing – original draft. Feng Gao: Conceptualization, Project administration, Funding acquisition, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Key Research and Development Program of China [Grant number 2018YFA0903700]; and the National Natural Science Foundation of China [Grant numbers 21621004 and 31571358]. The authors would like to thank Prof. Chun-Ting Zhang for the invaluable assistance and inspiring discussions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.08.036.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Wu Y.T., Ho W.Z., Huang Y.W., Jin D.Y., Li S.Y., Liu S.L. SARS-CoV-2 is an appropriate name for the new coronavirus. Lancet. 2020;395:949–950. doi: 10.1016/S0140-6736(20)30557-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Organization WH. WHO Director-General’s remarks at the media briefing on 2019-nCoV on 11 February 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020 2020; Accessed 11 February 2020.
- 3.Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klompas M., Baker M.A., Rhee C. Airborne transmission of SARS-CoV-2: theoretical considerations and available evidence. JAMA. 2020;324:441–442. doi: 10.1001/jama.2020.12458. [DOI] [PubMed] [Google Scholar]
- 5.Helms J., Kremer S., Merdji H., Clere-Jehl R., Schenck M., Kummerlen C. Neurologic features in severe SARS-CoV-2 infection. New Engl J Med. 2020;382:2268–2270. doi: 10.1056/NEJMc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2020;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poland G.A. SARS-CoV-2: a time for clear and immediate action. Lancet Infect Dis. 2020;20:531–532. doi: 10.1016/S1473-3099(20)30250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanov D. Predicting the impacts of epidemic outbreaks on global supply chains: a simulation-based analysis on the coronavirus outbreak (COVID-19/SARS-CoV-2) case. Transport Res E-Log. 2020;136 doi: 10.1016/j.tre.2020.101922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sayers E.W., Agarwala R., Bolton E.E., Brister J.R., Canese K., Clark K. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2019;47:D23–D28. doi: 10.1093/nar/gky1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y., Liu Q.Y., Guo D.Y. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 12.Gao F., Ou H.Y., Chen L.L., Zheng W.X., Zhang C.T. Prediction of proteinase cleavage sites in polyproteins of coronaviruses and its applications in analyzing SARS-CoV genomes. FEBS Lett. 2003;553:451–456. doi: 10.1016/S0014-5793(03)01091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C.X., Zheng W., Huang X.Q., Bell E.W., Zhou X.G., Zhang Y. Protein structure and sequence reanalysis of 2019-nCoV genome refutes snakes as its intermediate host and the unique similarity between its Spike protein insertions and HIV-1. J Proteome Res. 2020;19:1351–1360. doi: 10.1021/acs.jproteome.0c00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sussman J.L., Lin D., Jiang J., Manning N.O., Prilusky J., Ritter O. Protein Data Bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallogr D Biol Crystallogr. 1998;54:1078–1084. doi: 10.1107/s0907444998009378. [DOI] [PubMed] [Google Scholar]
- 15.Lamiable A., Thévenet P., Rey J., Vavrusa M., Derreumaux P., Tufféry P. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016;44:W449–W454. doi: 10.1093/nar/gkw329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou K.C., Wei D.Q., Zhong W.Z. Binding mechanism of coronavirus main proteinase with ligands and its implication to drug design against SARS. Biochem Bioph Res Co. 2003;308:148–151. doi: 10.1016/S0006-291X(03)01342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gan Y.R., Huang H., Huang Y.D., Rao C.M., Zhao Y., Liu J.S. Synthesis and activity of an octapeptide inhibitor designed for SARS coronavirus main proteinase. Peptides. 2006;27:622–625. doi: 10.1016/j.peptides.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du Q.S., Wang S.Q., Zhu Y., Wei D.Q., Guo H., Sirois S. Polyprotein cleavage mechanism of SARS CoV Mpro and chemical modification of the octapeptide. Peptides. 2004;25:1857–1864. doi: 10.1016/j.peptides.2004.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu A.P., Peng Y.S., Huang B.Y., Ding X., Wang X.Y., Niu P.H. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sternberg A., Naujokat C. Structural features of coronavirus SARS-CoV-2 spike protein: targets for vaccination. Life Sci. 2020;257 doi: 10.1016/j.lfs.2020.118056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant O.C., Montgomery D., Ito K., Woods R.J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci Rep. 2020;30:1128. doi: 10.1038/s41598-020-71748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu F., Xiang R., Deng X., Wang L., Yu Z., Tian S. Receptor-binding domain-specific human neutralizing monoclonal antibodies against SARS-CoV and SARS-CoV-2. Signal Transduct Tar. 2020;5:1949–1960. doi: 10.1038/s41392-020-00318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niu L., Wittrock K.N., Clabaugh G.C., Srivastava V., Cho M.W. A structural landscape of neutralizing antibodies against SARS-CoV-2 receptor binding domain. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.647934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gavor E., Choong Y.K., Er S.Y., Sivaraman H., Sivaraman J. Structural basis of SARS-CoV-2 and SARS-CoV antibody interactions. Trends Immunol. 2020;41:1006–1022. doi: 10.1016/j.it.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang S., Zhang X., Du L. Therapeutic antibodies and fusion inhibitors targeting the spike protein of SARS-CoV-2. Expert Opin Ther Tar. 2021;25:415–421. doi: 10.1080/14728222.2020.1820482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu C.R., Liu Y., Yang Y.Y., Zhang P., Zhong W., Wang Y.L. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao X.P., Qin B., Chen P., Zhu K., Hou P., Wojdyla J.A. Crystal structure of SARS-CoV-2 papain-like protease. Acta pharm Sin B. 2021;11:237–245. doi: 10.1016/j.apsb.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavasotto C.N., Lamas M.S., Maggini J. Functional and druggability analysis of the SARS-CoV-2 proteome. Eur J Pharmacol. 2021;890 doi: 10.1016/j.ejphar.2020.173705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michalska K., Kim Y., Jedrzejczak R., Maltseva N.I., Stols L., Endres M. Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes. IUCrJ. 2020;7:814–824. doi: 10.1107/S2052252520009653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robson B. The use of knowledge management tools in viroinformatics. Example study of a highly conserved sequence motif in Nsp3 of SARS-CoV-2 as a therapeutic target. Comput Biol Med. 2020;125 doi: 10.1016/j.compbiomed.2020.103963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fehr A.R., Jankevicius G., Ahel I., Perlman S. Viral macrodomains: unique mediators of viral replication and pathogenesis. Trends Microbiol. 2018;26:598–610. doi: 10.1016/j.tim.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alamri M.A., ul M.T., Qamar, Mirza M.U., Alqahtani S.M., Froeyen M., Chen L.L. Discovery of human coronaviruses pan-papain-like protease inhibitors using computational approaches. J Pharm Anal. 2020;10:546–559. doi: 10.1016/j.jpha.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kandeel M., Abdelrahman A.H.M., Oh-Hashi K., Ibrahim A., Venugopala K.N., Morsy M.A. Repurposing of FDA-approved antivirals, antibiotics, anthelmintics, antioxidants, and cell protectives against SARS-CoV-2 papain-like protease. J Biomol Struct Dyn. 2021;39:5129–5136. doi: 10.1080/07391102.2020.1784291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: a framework for anti–COVID-19 drug design. Sci Adv. 2020;6:eabd4596. doi: 10.1126/sciadv.abd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pang J., Gao S., Sun Z.X., Yang G.S. Discovery of small molecule PLpro inhibitor against COVID-19 using structure-based virtual screening, molecular dynamics simulation, and molecular mechanics/Generalized Born surface area (MM/GBSA) calculation. Struct Chem. 2021;32:879–886. doi: 10.1007/s11224-020-01665-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delre P., Caporuscio F., Saviano M., Mangiatordi G.F. Repurposing known drugs as covalent and non-covalent inhibitors of the SARS-CoV-2 papain-like protease. Front Chem. 2020;8 doi: 10.3389/fchem.2020.594009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manandhar A., Blass B.E., Colussi D.J., Almi I., Abou-Gharbia M., Klein M.L. Targeting SARS-CoV-2 M3CLpro by HCV NS3/4a inhibitors: in silico modeling and in vitro screening. J Chem Inf Model. 2021;61:1020–1032. doi: 10.1021/acs.jcim.0c01457. [DOI] [PubMed] [Google Scholar]
- 41.Bauer R.A. Covalent inhibitors in drug discovery: from accidental discoveries to avoided liabilities and designed therapies. Drug Discov Today. 2015;20:1061–1073. doi: 10.1016/j.drudis.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Freitas B.T., Durie I.A., Murray J., Longo J.E., Miller H.C., Crich D. Characterization and noncovalent inhibition of the deubiquitinase and delSGylase activity of SARS-CoV-2 papain-like protease. ACS Infect Dis. 2020;6:2099–2109. doi: 10.1021/acsinfecdis.0c00168. [DOI] [PubMed] [Google Scholar]
- 43.Bhati S. Structure-based drug designing of naphthalene based SARS-CoV PLpro inhibitors for the treatment of COVID-19. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e05558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osipiuk J., Azizi S.-A., Dvorkin S., Endres M., Jedrzejczak R., Jones K.A. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat Commun. 2021;12:743. doi: 10.1038/s41467-021-21060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jamalan M., Barzegari E., Gholami-Borujeni F. Structure-based screening to discover new inhibitors for papain-like proteinase of SARS-CoV-2: An in silico study. J Proteome Res. 2021;20:1015–1026. doi: 10.1021/acs.jproteome.0c00836. [DOI] [PubMed] [Google Scholar]
- 46.Selvaraj C., Dinesh D.C., Panwar U., Boura E., Singh S.K. High-throughput screening and quantum mechanics for identifying potent inhibitors against Mac1 domain of SARS-CoV-2 Nsp3. IEEE ACM T Comput Bi. 2021;18:1262–1270. doi: 10.1109/TCBB.2020.3037136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung L.S., Gund T.M., Narayan M. Comparison of binding site of remdesivir and its metabolites with NSP12-NSP7-NSP8, and NSP3 of SARS CoV-2 virus and alternative potential drugs for COVID-19 treatment. Protein J. 2020;39:619–630. doi: 10.1007/s10930-020-09942-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh A.K., Kushwaha P.P., Prajapati K.S., Shuaib M., Gupta S., Kumar S. Identification of FDA approved drugs and nucleoside analogues as potential SARS-CoV-2 A1 pp domain inhibitor: an in silico study. Comput Biol Med. 2021;130 doi: 10.1016/j.compbiomed.2020.104185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Debnath P., Debnath B., Bhaumik S., Debnath S. In silico identification of potential inhibitors of ADP-Ribose phosphatase of SARS-CoV-2 nsp3 by combining E-pharmacophore- and receptor-based virtual screening of database. ChemistrySelect. 2020;5:9388–9398. doi: 10.1002/slct.202001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Vivo M., Masetti M., Bottegoni G., Cavalli A. Role of molecular dynamics and related methods in drug discovery. J Med Chem. 2016;59:4035–4061. doi: 10.1021/acs.jmedchem.5b01684. [DOI] [PubMed] [Google Scholar]
- 51.Yan F.F., Gao F. Comparison of the binding characteristics of SARS-CoV and SARS-CoV-2 RBDs to ACE2 at different temperatures by MD simulations. Brief Bioinform. 2021;22:1122–1136. doi: 10.1093/bib/bbab044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J.Z., Wang X.Y., Pang L.X., Zhang J.Z.H., Zhu T. Effect of mutations on binding of ligands to guanine riboswitch probed by free energy perturbation and molecular dynamics simulations. Nucleic Acids Res. 2019;47:6618–6631. doi: 10.1093/nar/gkz499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Virdi R.S., Bavisotto R.V., Hopper N.C., Vuksanovic N., Melkonian T.R., Silvaggi N.R. Discovery of drug-like ligands for the Mac1 domain of SARS-CoV-2 Nsp3. SLAS Discov. 2020;25:1162–1170. doi: 10.1177/2472555220960428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 55.Silvestrini L., Belhaj N., Comez L., Gerelli Y., Lauria A., Libera V. The dimer-monomer equilibrium of SARS-CoV-2 main protease is affected by small molecule inhibitors. Sci Rep. 2021;11:9283. doi: 10.1038/s41598-021-88630-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tekpinar M., Yildirim A. Impact of dimerization and N3 binding on molecular dynamics of SARS-CoV and SARS-CoV-2 main proteases. J Biomol Struct Dyn. 2021 doi: 10.1080/07391102.2021.1880481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fan K.Q., Wei P., Feng Q., Chen S.Q., Huang C.K., Ma L. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J Biol Chem. 2004;279:1637–1642. doi: 10.1074/jbc.M310875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang L.L., Lin D.Z., Sun X.Y.Y., Curth U., Drosten C., Sauerhering L. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petti S., Scully C. Polyphenols, oral health and disease: a review. J Dent. 2009;37:413–423. doi: 10.1016/j.jdent.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 60.Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Identification of polyphenols from Broussonetia papyrifera as SARS CoV-2 main protease inhibitors using in silico docking and molecular dynamics simulation approaches. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1802347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Depicting the inhibitory potential of polyphenols from Isatis indigotica root against the main protease of SARS CoV-2 using computational approaches. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1858164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sherif Y.E., Gabr S.A., Hosny N.M., Alghadir A.H., Alansari R. Phytochemicals of rhus spp. as potential inhibitors of the SARS-CoV-2 main protease: molecular docking and drug-likeness study. Evid-Based Compl Alt. 2021;2021:8814890. doi: 10.1155/2021/8814890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roy A. A review on the alkaloids an important therapeutic compound from plants. IJPB. 2017;3:1–9. [Google Scholar]
- 64.Garg S., Roy A. In silico analysis of selected alkaloids against main protease (Mpro) of SARS-CoV-2. Chem-Biol Interact. 2020;332 doi: 10.1016/j.cbi.2020.109309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abdelrheem D.A., Ahmed S.A., Abd El-Mageed H.R., Mohamed H.S., Rahman A.A., Elsayed K.N.M. The inhibitory effect of some natural bioactive compounds against SARS-CoV-2 main protease: insights from molecular docking analysis and molecular dynamic simulation. J Environ Sci Heal A. 2020;55:1373–1386. doi: 10.1080/10934529.2020.1826192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gyebi G.A., Ogunro O.B., Adegunloye A.P., Ogunyemi O.M., Afolabi S.O. Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CLpro): an in silico screening of alkaloids and terpenoids from African medicinal plants. J Biomol Struct Dyn. 2021;39:3396–3408. doi: 10.1080/07391102.2020.1764868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kennedy D.O., Wightman E.L. Herbal extracts and phytochemicals: plant secondary metabolites and the enhancement of human brain function. Adv Nutr. 2011;2:32–50. doi: 10.3945/an.110.000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bergman M.E., Davis B., Phillips M.A. Medically useful plant terpenoids: biosynthesis, occurrence, and mechanism of action. Molecules. 2019;24:3961. doi: 10.3390/molecules24213961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chang F.R., Yen C.T., Ei-Shazly M., Lin W.H., Yen M.H., Lin K.H. Anti-human coronavirus (anti-HCoV) triterpenoids from the leaves of euphorbia neriifolia. Nat Prod Commun. 2012;7:1415–1417. [PubMed] [Google Scholar]
- 70.Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Diniz L.R.L., Perez-Castillo Y., Elshabrawy H.A., de Sousa D.P. Bioactive terpenes and their derivatives as potential SARS-CoV-2 proteases inhibitors from molecular modeling studies. Biomolecules. 2021;11:74. doi: 10.3390/biom11010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jo S., Kim S., Shin D.H., Kim M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzym Inhib Med Ch. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Owis A.I., El-Hawary M.S., El Amir D., Aly O., Abdelmohsen U.R., Kamel M.S. Molecular docking reveals the potential of Salvadora persica flavonoids to inhibit COVID-19 virus main protease. RSC Adv. 2020;10:19570–19575. doi: 10.1039/d0ra03582c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jo S., Kim S., Kim D.Y., Kim M.S., Shin D.H. Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro. J Enzym Inhib Med Ch. 2020;35:1539–1544. doi: 10.1080/14756366.2020.1801672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu H.B., Ye F., Sun Q., Liang H., Li C.M., Li S.Y. Scutellaria baicalensis extract and baicalein inhibit replication of SARS-CoV-2 and its 3C-like protease in vitro. J Enzym Inhib Med Ch. 2021;36:497–503. doi: 10.1080/14756366.2021.1873977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mahmud S., Uddin M.A.R., Zaman M., Sujon K.M., Rahman M.E., Shehab M.N. Molecular docking and dynamics study of natural compound for potential inhibition of main protease of SARS-CoV-2. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1796808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abian O., Ortega-Alarcon D., Jimenez-Alesanco A., Ceballos-Laita L., Vega S., Reyburn H.T. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int J Biol Macromol. 2020;164:1693–1703. doi: 10.1016/j.ijbiomac.2020.07.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sen D., Debnath P., Debnath B., Bhaumik S., Debnath S. Identification of potential inhibitors of SARS-CoV-2 main protease and spike receptor from 10 important spices through structure-based virtual screening and molecular dynamic study. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1819883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cherrak S.A., Merzouk H., Mokhtari-Soulimane N. Potential bioactive glycosylated flavonoids as SARS-CoV-2 main protease inhibitors: a molecular docking and simulation studies. PLoS ONE. 2020;15 doi: 10.1371/journal.pone.0240653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abd El-Mordy F.M., El-Hamouly M.M., Ibrahim M.T., Abd El-Rheem G., Aly O.M., Abd El-kader A.M. Inhibition of SARS-CoV-2 main protease by phenolic compounds from Manilkara hexandra (Roxb.) Dubard assisted by metabolite profiling and in silico virtual screening. RSC Adv. 2020;10:32148–32155. doi: 10.1039/d0ra05679k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huynh T., Wang H., Luan B. Structure-based lead optimization of herbal medicine rutin for inhibiting SARS-CoV-2's main protease. Phys Chem Chem Phys. 2020;22:25335–25343. doi: 10.1039/d0cp03867a. [DOI] [PubMed] [Google Scholar]
- 82.Panche A.N., Diwan A.D., Chandra S.R. Flavonoids: an overview. J Nutr Sci. 2016;5 doi: 10.1017/jns.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fakhar Z., Faramarzi B., Pacifico S., Faramarzi S. Anthocyanin derivatives as potent inhibitors of SARS-CoV-2 main protease: an in-silico perspective of therapeutic targets against COVID-19 pandemic. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1801510. [DOI] [PubMed] [Google Scholar]
- 84.Zhu Y., Xie D.Y. Docking characterization and in vitro inhibitory activity of flavan-3-ols and dimeric proanthocyanidins against the main protease activity of SARS-Cov-2. Front Plant Sci. 2020;11 doi: 10.3389/fpls.2020.601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dai X.L., Liu Y.J., Zhuang J.H., Yao S.B., Liu L., Jiang X.L. Discovery and characterization of tannase genes in plants: roles in hydrolysis of tannins. New Phytol. 2020;226:1104–1116. doi: 10.1111/nph.16425. [DOI] [PubMed] [Google Scholar]
- 86.Wang P.Q., Liu Y.J., Zhang L.J., Wang W.Z., Hou H., Zhao Y. Functional demonstration of plant flavonoid carbocations proposed to be involved in the biosynthesis of proanthocyanidins. Plant J. 2020;101:18–36. doi: 10.1111/tpj.14515. [DOI] [PubMed] [Google Scholar]
- 87.Upadhyay S., Tripathi P.K., Singh M., Raghavendhar S., Bhardwaj M., Patel A.K. Evaluation of medicinal herbs as a potential therapeutic option against SARS-CoV-2 targeting its main protease. Phytother Res. 2020;34:3411–3419. doi: 10.1002/ptr.6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors – an in silico docking and molecular dynamics simulation study. J Biomol Struct Dyn. 2021;39:4362–4374. doi: 10.1080/07391102.2020.1779818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chiou W.C., Chen J.C., Chen Y.T., Yang J.M., Hwang L.H., Lyu Y.S. The inhibitory effects of PGG and EGCG against the SARS-CoV-2 3C-like protease. Biochem Bioph Res Co. 2021 doi: 10.1016/j.bbrc.2020.12.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Du A., Zheng R., Disoma C., Li S., Chen Z., Li S. Epigallocatechin-3-gallate, an active ingredient of traditional chinese medicines, inhibits the 3CLpro activity of SARS-CoV-2. Int J Biol Macromol. 2021;176:1–12. doi: 10.1016/j.ijbiomac.2021.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jang M., Park Y.I., Cha Y.E., Park R., Namkoong S., Lee J.I. Tea polyphenols EGCG and theaflavin inhibit the activity of SARS-CoV-2 3CL-Protease in vitro. Evid-Based Compl Alt. 2020;2020:5630838. doi: 10.1155/2020/5630838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ryu Y.B., Jeong H.J., Kim J.H., Kim Y.M., Park J.Y., Kim D. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorgan Med Chem. 2010;18:7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Computer aided identification of potential SARS CoV-2 main protease inhibitors from diterpenoids and biflavonoids of Torreya nucifera leaves. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1841680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bharadwaj S., Dubey A., Yadava U., Mishra S.K., Kang S.G., Dwivedi V.D. Exploration of natural compounds with anti-SARS-CoV-2 activity via inhibition of SARS-CoV-2 Mpro. Brief Bioinform. 2021;22:1361–1377. doi: 10.1093/bib/bbaa382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hejazi I.I., Beg M.A., Imam M.A., Athar F., Islam A. Glossary of phytoconstituents: can these be repurposed against SARS CoV-2? A quick in silico screening of various phytoconstituents from plant Glycyrrhiza glabra with SARS CoV-2 main protease. Food Chem Toxicol. 2021;150 doi: 10.1016/j.fct.2021.112057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tripathi M.K., Singh P., Sharma S., Singh T.P., Ethayathulla A.S., Kaur P. Identification of bioactive molecule from Withania somnifera (Ashwagandha) as SARS-CoV-2 main protease inhibitor. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1790425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Joshi T., Sharma P., Joshi T., Pundir H., Mathpal S., Chandra S. Structure-based screening of novel lichen compounds against SARS Coronavirus main protease (Mpro) as potentials inhibitors of COVID-19. Mol Divers. 2021;25:1665–1677. doi: 10.1007/s11030-020-10118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J Biomol Struct Dyn. 2021;39:3449–3458. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mahmud S., Uddin M.A.R., Paul G.K., Shimu M.S.S., Islam S., Rahman E. Virtual screening and molecular dynamics simulation study of plant-derived compounds to identify potential inhibitors of main protease from SARS-CoV-2. Brief Bioinform. 2021;22:1402–1414. doi: 10.1093/bib/bbaa428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aanouz I., Belhassan A., El-Khatabi K., Lakhlifi T., El-ldrissi M., Bouachrine M. Moroccan medicinal plants as inhibitors against SARS-CoV-2 main protease: computational investigations. J Biomol Struct Dyn. 2021;39:2971–2979. doi: 10.1080/07391102.2020.1758790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tahir, ul, Qamar, M,, Alqahtani, SM,, Alamri, MA,, Chen, LL. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharm Anal. 2020;10:313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gurung A.B., Ali M.A., Lee J., Farah M.A., Al-Anazi K.M. Unravelling lead antiviral phytochemicals for the inhibition of SARS-CoV-2 Mpro enzyme through in silico approach. Life Sci. 2020;255 doi: 10.1016/j.lfs.2020.117831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ibrahim M.A.A., Abdelrahman A.H.M., Hussien T.A., Badr E.A.A., Mohamed T.A., El-Seedi H.R. In silico drug discovery of major metabolites from spices as SARS-CoV-2 main protease inhibitors. Comput Biol Med. 2020;126 doi: 10.1016/j.compbiomed.2020.104046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lindequist U. Marine-derived pharmaceuticals - challenges and opportunities. Biomol Ther. 2016;24:561–571. doi: 10.4062/biomolther.2016.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mayer A.M.S., Rodríguez A.D., Berlinck R.G.S., Fusetani N. Marine pharmacology in 2007–8: Marine compounds with antibacterial, anticoagulant, antifungal, anti-inflammatory, antimalarial, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous system, and other miscellaneous mechanisms of action. Comp Biochem Phys C. 2011;153:191–222. doi: 10.1016/j.cbpc.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gentile D., Patamia V., Scala A., Sciortino M.T., Piperno A., Rescifina A. Putative inhibitors of SARS-CoV-2 main protease from a library of marine natural products: a virtual screening and molecular modeling study. Mar Drugs. 2020;18:225. doi: 10.3390/md18040225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kalhotra P., Chittepu V.C.S.R., Osorio-Revilla G., Gallardo-Velazquez T. Field-template, QSAR, ensemble molecular docking, and 3D-RISM solvation studies expose potential of FDA-approved marine drugs as SARS-CoVID-2 main protease inhibitors. Molecules. 2021;26:936. doi: 10.3390/molecules26040936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Khan M.T., Ali A., Wang Q., Irfan M., Khan A., Zeb M.T. Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2—a molecular dynamic study. J Biomol Struct Dyn. 2021;39:3627–3637. doi: 10.1080/07391102.2020.1769733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.van Santen J.A., Jacob G., Singh A.L., Aniebok V., Balunas M.J., Bunsko D. The natural products atlas: an open access knowledge base for microbial natural products discovery. ACS Central Sci. 2019;5:1824–1833. doi: 10.1021/acscentsci.9b00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sayed A.M., Alhadrami H.A., El-Gendy A.O., Shamikh Y.I., Belbahri L., Hassan H.M. Microbial natural products as potential inhibitors of SARS-CoV-2 main protease (Mpro) Microorganisms. 2020;8:970. doi: 10.3390/microorganisms8070970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rao P., Shukla A., Parmar P., Rawal R.M., Patel B., Saraf M. Reckoning a fungal metabolite, Pyranonigrin A as a potential Main protease (Mpro) inhibitor of novel SARS-CoV-2 virus identified using docking and molecular dynamics simulation. Biophys Chem. 2020;264 doi: 10.1016/j.bpc.2020.106425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Alam S., Sadiqi S., Sabir M., Nisa S., Ahmad S., Abbasi S.W. Bacillus species; a potential source of anti-SARS-CoV-2 main protease inhibitors. J Biomol Struct Dyn. 2021 doi: 10.1080/07391102.2021.1873188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Naidoo D., Roy A., Kar P., Mutanda T., Anandraj A. Cyanobacterial metabolites as promising drug leads against the Mpro and PLpro of SARS-CoV-2: an in silico analysis. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1794972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ashburn T.T., Thor K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 115.Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 116.Cha Y., Erez T., Reynolds I.J., Kumar D., Ross J., Koytiger G. Drug repurposing from the perspective of pharmaceutical companies. Brit J Pharmacol. 2018;175:168–180. doi: 10.1111/bph.13798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Novac N. Challenges and opportunities of drug repositioning. Trends Pharmacol Sci. 2013;34:267–272. doi: 10.1016/j.tips.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 118.Xu H.Y., Zhang Y.Q., Liu Z.M., Chen T., Lv C.Y., Tang S.H. ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 2019;47:D976–D982. doi: 10.1093/nar/gky987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang C.-H., Stone E.A., Deshmukh M., Ippolito J.A., Ghahremanpour M.M., Tirado-Rives J. Potent noncovalent inhibitors of the main protease of SARS-CoV-2 from molecular sculpting of the drug perampanel guided by free energy perturbation calculations. ACS Central Sci. 2021;7:467–475. doi: 10.1021/acscentsci.1c00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Forrestall K.L., Burley D.E., Cash M.K., Pottie I.R., Darvesh S. 2-Pyridone natural products as inhibitors of SARS-CoV-2 main protease. Chem-Biol Interact. 2021;335 doi: 10.1016/j.cbi.2020.109348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bzówka M., Mitusińska K., Raczyńska A., Samol A., Tuszyński J.A., Góra A. Structural and evolutionary analysis indicate that the SARS-CoV-2 Mpro is a challenging target for small-molecule inhibitor design. Int J Mol Sci. 2020;21:3099. doi: 10.3390/ijms21093099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Deshmukh M.G., Ippolito J.A., Zhang C.-H., Stone E.A., Reilly R.A., Miller S.J. Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease. Structure. 2021;29:823–833. doi: 10.1016/j.str.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Steuten K., Kim H., Widen J.C., Babin B.M., Onguka O., Lovell S. Challenges for targeting SARS-CoV-2 proteases as a therapeutic strategy for COVID-19. ACS Infect Dis. 2021;7:1457–1468. doi: 10.1021/acsinfecdis.0c00815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Arun K.G., Sharanya C.S., Abhithaj J., Francis D., Sadasivan C. Drug repurposing against SARS-CoV-2 using E-pharmacophore based virtual screening, molecular docking and molecular dynamics with main protease as the target. J Biomol Struct Dyn. 2021;39:4647–4658. doi: 10.1080/07391102.2020.1779819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mody V., Ho J., Wills S., Mawri A., Lawson L., Ebert M.C.C.J.C. Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents. Commun Biol. 2021;4:93. doi: 10.1038/s42003-020-01577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gupta A., Rani C., Pant P., Vijayan V., Vikram N., Kaur P. Structure-based virtual screening and biochemical validation to discover a potential inhibitor of the SARS-CoV-2 main protease. ACS Omega. 2020;5:33151–33161. doi: 10.1021/acsomega.0c04808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kumar P., Bhardwaj T., Kumar A., Gehi B.R., Kapuganti S.K., Garg N. Reprofiling of approved drugs against SARS-CoV-2 main protease: an in-silico study. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1845976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Baby K., Maity S., Mehta C.H., Suresh A., Nayak U.Y., Nayak Y. Targeting SARS-CoV-2 main protease: a computational drug repurposing study. Arch Med Res. 2021;52:38–47. doi: 10.1016/j.arcmed.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fiorucci D., Milletti E., Orofino F., Brizzi A., Mugnaini C., Corelli F. Computational drug repurposing for the identification of SARS-CoV-2 main protease inhibitors. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1796805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bharadwaj S., Azhar E.I., Kamal M.A., Bajrai L.H., Dubey A., Jha K. SARS-CoV-2 Mpro inhibitors: identification of anti-SARS-CoV-2 Mpro compounds from FDA approved drugs. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1842807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gupta A., Zhou H.X. Profiling SARS-CoV-2 main protease (MPRO) binding to repurposed drugs using molecular dynamics simulations in classical and neural network-trained force fields. ACS Comb Sci. 2020;22:826–832. doi: 10.1021/acscombsci.0c00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jiménez-Alberto A., Ribas-Aparicio R.M., Aparicio-Ozores G., Castelán-Vega J.A. Virtual screening of approved drugs as potential SARS-CoV-2 main protease inhibitors. Comput Biol Chem. 2020;88 doi: 10.1016/j.compbiolchem.2020.107325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang Q., Zhao Y., Chen X.J., Hong A. Virtual screening of approved clinic drugs with main protease (3CLpro) reveals potential inhibitory effects on SARS-CoV-2. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1817786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kumar Y., Singh H., Patel C.N. In silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing. J Infect Public Heal. 2020;13:1210–1223. doi: 10.1016/j.jiph.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Basu S., Veeraraghavan B., Ramaiah S., Anbarasu A. Novel cyclohexanone compound as a potential ligand against SARS-CoV-2 main-protease. Microb Pathogenesis. 2020;149 doi: 10.1016/j.micpath.2020.104546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Abu-Saleh A.A.-A.A., Awad I.E., Yadav A., Poirier R.A. Discovery of potent inhibitors for SARS-CoV-2's main protease by ligand-based/structure-based virtual screening, MD simulations, and binding energy calculations. Phys Chem Chem Phys. 2020;22:23099–23106. doi: 10.1039/d0cp04326e. [DOI] [PubMed] [Google Scholar]
- 138.Ahmed M.Z., Zia Q., Haque A., Alqahtani A.S., Almarfadi O.M., Banawas S. FDA-approved antiviral and anti-infection agents as potential inhibitors of SARS-CoV-2 main protease: an in silico drug repurposing study. J Infect Public Heal. 2021;14:611–619. doi: 10.1016/j.jiph.2021.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kandeel M., Al-Nazawi M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci. 2020;251 doi: 10.1016/j.lfs.2020.117627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Maffucci I., Contini A. In silico drug repurposing for SARS-CoV-2 main proteinase and spike proteins. J Proteome Res. 2020;19:4637–4648. doi: 10.1021/acs.jproteome.0c00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gimeno A., Mestres-Truyol J., Ojeda-Montes M.J., Macip G., Saldivar-Espinoza B., Cereto-Massagué A. Prediction of novel inhibitors of the main protease (M-pro) of SARS-CoV-2 through consensus docking and drug reposition. Int J Mol Sci. 2020;21:3793. doi: 10.3390/ijms21113793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ghahremanpour M.M., Tirado-Rives J., Deshmukh M., Ippolito J.A., Zhang C.H., Cabeza de Vaca I. Identification of 14 known drugs as inhibitors of the main protease of SARS-CoV-2. ACS Med Chem Lett. 2020;11:2526–2533. doi: 10.1021/acsmedchemlett.0c00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chiou W.C., Hsu M.S., Chen Y.T., Yang J.M., Tsay Y.G., Huang H.C. Repurposing existing drugs: identification of SARS-CoV-2 3C-like protease inhibitors. J Enzym Inhib Med Ch. 2021;36:147–153. doi: 10.1080/14756366.2020.1850710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Beck B.R., Shin B., Choi Y., Park S., Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput Struct Biotec. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tripathi P.K., Upadhyay S., Singh M., Raghavendhar S., Bhardwaj M., Sharma P. Screening and evaluation of approved drugs as inhibitors of main protease of SARS-CoV-2. Int J Biol Macromol. 2020;164:2622–2631. doi: 10.1016/j.ijbiomac.2020.08.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Elmaaty A.A., Alnajjar R., Hamed M.I., Khattab M., Khalifa M.M., Al-Karmalawy A.A. Revisiting activity of some glucocorticoids as a potential inhibitor of SARS-CoV-2 main protease: theoretical study. RSC Adv. 2021;11:10027–10042. doi: 10.1039/d0ra10674g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Abo Elmaaty A., Hamed M.I.A., Ismail M.I., Elkaeed E.B., Abulkhair H.S., Khattab M. Computational insights on the potential ofsSome NSAIDs for treating COVID-19: priority set and lead optimization. Molecules. 2021;26:3772. doi: 10.3390/molecules26123772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Alnajjar R., Mostafa A., Kandeil A., Al-Karmalawy A.A. Molecular docking, molecular dynamics, and in vitro studies reveal the potential of angiotensin II receptor blockers to inhibit the COVID-19 main protease. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e05641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yang J.Y., Lin X.Y., Xing N., Zhang Z., Zhang H.W., Wu H.B. Structure-based discovery of novel nonpeptide inhibitors targeting SARS-CoV-2 Mpro. J Chem Inf Model. 2021;61:3917–3926. doi: 10.1021/acs.jcim.1c00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Abdusalam A.A.A., Murugaiyah V. Identification of potential inhibitors of 3CL protease of SARS-CoV-2 from ZINC database by molecular docking-based virtual screening. Front Mol Biosci. 2020;7 doi: 10.3389/fmolb.2020.603037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Haider Z., Subhani M.M., Farooq M.A., Ishaq M., Khalid M., Akram M.N. In-silico pharmacophoric and molecular docking-based drug discovery against the Main Protease (M pro) of SARS-CoV-2, a causative agent COVID-19. Pak. J. Pharm. Sci. 2020;33:2697–2705. [PubMed] [Google Scholar]
- 152.Liu S., Zheng Q., Wang Z.Y. Potential covalent drugs targeting the main protease of the SARS-CoV-2 coronavirus. Bioinformatics. 2020;36:3295–3298. doi: 10.1093/bioinformatics/btaa224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fischer A., Sellner M., Neranjan S., Smieško M., Lill M.A. Potential inhibitors for novel coronavirus protease identified by virtual screening of 606 million compounds. Int J Mol Sci. 2020;21:3626. doi: 10.3390/ijms21103626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kavitha K., Sivakumar S., Ramesh B. 1,2,4 triazolo 1,5-a pyrimidin-7-ones as novel SARS-CoV-2 Main protease inhibitors: in silico screening and molecular dynamics simulation of potential COVID-19 drug candidates. Biophys Chem. 2020;267 doi: 10.1016/j.bpc.2020.106478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ahmad S., Waheed Y., Ismail S., Najmi M.H., Ansari J.K. Rational design of potent anti-COVID-19 main protease drugs: An extensive multi-spectrum in silico approach. J Mol Liq. 2021;330 doi: 10.1016/j.molliq.2021.115636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Abel R., Paredes Ramos M., Chen Q., Pérez-Sánchez H., Coluzzi F., Rocco M. Computational prediction of potential inhibitors of the main protease of SARS-CoV-2. Front Chem. 2020;8 doi: 10.3389/fchem.2020.590263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zia K., Khan S.A., Ashraf S., Nur-e-Alam M., Ahmed S., Ul-Haq Z. Probing CAS database as prospective antiviral agents against SARS-CoV-2 main protease. J Mol Struct. 2021;1231 doi: 10.1016/j.molstruc.2021.129953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Achutha A.S., Pushpa V.L., Suchitra S. Theoretical insights into the anti-SARS-CoV-2 activity of chloroquine and its analogs and in silico screening of main protease inhibitors. J Proteome Res. 2020;19:4706–4717. doi: 10.1021/acs.jproteome.0c00683. [DOI] [PubMed] [Google Scholar]
- 159.Havranek B., Islam S.M. An in silico approach for identification of novel inhibitors as potential therapeutics targeting COVID-19 main protease. J Biomol Struct Dyn. 2021;39:4304–4315. doi: 10.1080/07391102.2020.1776158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rakib A., Nain Z., Sami S.A., Mahmud S., Islam A., Ahmed S. A molecular modelling approach for identifying antiviral selenium-containing heterocyclic compounds that inhibit the main protease of SARS-CoV-2: an in silico investigation. Brief Bioinform. 2021;22:1476–1498. doi: 10.1093/bib/bbab045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mohammad T., Shamsi A., Anwar S., Umair M., Hussain A., Rehman M.T. Identification of high-affinity inhibitors of SARS-CoV-2 main protease: towards the development of effective COVID-19 therapy. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yadav R., Imran M., Dhamija P., Chaurasia D.K., Handu S. Virtual screening, ADMET prediction and dynamics simulation of potential compounds targeting the main protease of SARS-CoV-2. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1796812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Andrianov A.M., Kornoushenko Y.V., Karpenko A.D., Bosko I.P., Tuzikov A.V. Computational discovery of small drug-like compounds as potential inhibitors of SARS-CoV-2 main protease. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1792989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gahlawat A., Kumar N., Kumar R., Sandhu H., Singh I.P., Singh S. Structure-based virtual screening to discover potential lead molecules for the SARS-CoV-2 main protease. J Chem Inf Model. 2020;60:5781–5793. doi: 10.1021/acs.jcim.0c00546. [DOI] [PubMed] [Google Scholar]
- 165.Ibrahim M.A.A., Abdelrahman A.H.M., Hegazy M.-E.F. In-silico drug repurposing and molecular dynamics puzzled out potential SARS-CoV-2 main protease inhibitors. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1791958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tejera E., Munteanu C.R., López-Cortés A., Cabrera-Andrade A., Pérez-Castillo Y. Drugs repurposing using QSAR, docking and molecular dynamics for possible inhibitors of the SARS-CoV-2 Mpro protease. Molecules. 2020;25:5172. doi: 10.3390/molecules25215172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pinzi L., Tinivella A., Caporuscio F., Rastelli G. Drug repurposing and polypharmacology to fight SARS-CoV-2 through inhibition of the main protease. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.636989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tsuji M. Potential anti-SARS-CoV-2 drug candidates identified through virtual screening of the ChEMBL database for compounds that target the main coronavirus protease. FEBS Open Bio. 2020;10:995–1004. doi: 10.1002/2211-5463.12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hakmi M., Bouricha E.M., Kandoussi I., Harti J.E., Ibrahimi A. Repurposing of known anti-virals as potential inhibitors for SARS-CoV-2 main protease using molecular docking analysis. Bioinformation. 2020;16:301–306. doi: 10.6026/97320630016301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Feitosa E.L., Júnior F.T.D.S.S., Nery Neto J.A.D.O., Matos L.F.L., Moura M.H.D.S., Rosales T.O. COVID-19: rational discovery of the therapeutic potential of Melatonin as a SARS-CoV-2 main protease inhibitor. Int J Med Sci. 2020;17:2133–2146. doi: 10.7150/ijms.48053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Li Z., Li X., Huang Y.Y., Wu Y., Liu R., Zhou L. Identify potent SARS-CoV-2 main protease inhibitors via accelerated free energy perturbation-based virtual screening of existing drugs. P Natl Acad Sci. 2020;117:27381–27387. doi: 10.1073/pnas.2010470117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Franco L.S., Maia R.C., Barreiro E.J. Identification of LASSBio-1945 as an inhibitor of SARS-CoV-2 main protease (M-PRO) through in silico screening supported by molecular docking and a fragment-based pharmacophore model. RSC Med Chem. 2021;12:110–119. doi: 10.1039/d0md00282h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Achilonu I., Iwuchukwu E.A., Achilonu O.J., Fernandes M.A., Sayed Y. Targeting the SARS-CoV-2 main protease using FDA-approved Isavuconazonium, a P2–P3 α-ketoamide derivative and Pentagastrin: An in-silico drug discovery approach. J Mol Graph Model. 2020;101 doi: 10.1016/j.jmgm.2020.107730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Breidenbach J., Lemke C., Pillaiyar T., Schäkel L., Al Hamwi G., Diett M. Targeting the main protease of SARS-CoV-2: from the establishment of high throughput screening to the design of tailored inhibitors. Angew Chem Int Ed. 2021;60:2–9. doi: 10.1002/anie.202016961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jin Z.M., Zhao Y., Sun Y., Zhang B., Wang H.F., Wu Y. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat Struct Mol Biol. 2020;27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 176.Menéndez C.A., Byléhn F., Perez-Lemus G.R., Alvarado W., de Pablo J.J. Molecular characterization of ebselen binding activity to SARS-CoV-2 main protease. Sci Adv. 2020;6:eabd0345. doi: 10.1126/sciadv.abd0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ma C.L., Hu Y.M., Townsend J.A., Lagarias P.I., Marty M.T., Kolocouris A. Ebselen, disulfiram, carmofur, PX-12, tideglusib, and shikonin are nonspecific promiscuous SARS-CoV-2 main protease inhibitors. ACS Pharmacol Transl Sci. 2020;3:1265–1277. doi: 10.1021/acsptsci.0c00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Xu L., Tong J., Wu Y., Zhao S., Lin B.-L. A computational evaluation of targeted oxidation strategy (TOS) for potential inhibition of SARS-CoV-2 by disulfiram and analogues. Biophys Chem. 2021;276 doi: 10.1016/j.bpc.2021.106610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Li J., Zhou X., Zhang Y., Zhong F., Lin C., McCormick P.J. Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode. Sci Bull. 2021;66:661–663. doi: 10.1016/j.scib.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mahmud S., Paul G.K., Biswas S., Afrose S., Mita M.A., Hasan M.R. Prospective role of peptide-based antiviral therapy against the pain Protease of SARS-CoV-2. Front Mol Biosci. 2021;8 doi: 10.3389/fmolb.2021.628585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rajpoot S., Alagumuthu M., Baig M.S. Dual targeting of 3CLpro and PLpro of SARS-CoV-2: a novel structure-based design approach to treat COVID-19. Curr Res Struct Biol. 2021;3:9–18. doi: 10.1016/j.crstbi.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mitra K., Ghanta P., Acharya S., Chakrapani G., Ramaiah B., Doble M. Dual inhibitors of SARS-CoV-2 proteases: pharmacophore and molecular dynamics based drug repositioning and phytochemical leads. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1796802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jade D., Ayyamperumal S., Tallapaneni V., Joghee Nanjan C.M., Barge S., Mohan S. Virtual high throughput screening: Potential inhibitors for SARS-CoV-2 PLPRO and 3CLPRO proteases. Eur J Pharmacol. 2021;901 doi: 10.1016/j.ejphar.2021.174082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chen Z., Cui Q., Cooper L., Zhang P., Lee H., Chen Z. Ginkgolic acid and anacardic acid are specific covalent inhibitors of SARS-CoV-2 cysteine proteases. Cell Biosci. 2021;11:45. doi: 10.1186/s13578-021-00564-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Aouidate A., Ghaleb A., Chtita S., Aarjane M., Ousaa A., Maghat H. Identification of a novel dual-target scaffold for 3CLpro and RdRp proteins of SARS-CoV-2 using 3D-similarity search, molecular docking, molecular dynamics and ADMET evaluation. J Biomol Struct Dyn. 2021;39:4522–4535. doi: 10.1080/07391102.2020.1779130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shady N.H., Hayallah A.M., Mohamed M.F.A., Ghoneim M.M., Chilingaryan G., Al-Sanea M.M. Targeting 3CLpro and SARS-CoV-2 RdRp by amphimedon sp. metabolites: a computational study. Molecules. 2021;26:3775. doi: 10.3390/molecules26123775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Allam A.E., Amen Y., Ashour A., Assaf H.K., Hassan H.A., Abdel-Rahman I.M. In silico study of natural compounds from sesame against COVID-19 by targeting Mpro, Plpro and RdRp. RSC Adv. 2021;11:22398–22408. doi: 10.1039/d1ra03937g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Elseginy S.A., Fayed B., Hamdy R., Mahrous N., Mostafa A., Almehdi A.M. Promising anti-SARS-CoV-2 drugs by effective dual targeting against the viral and host proteases. Bioorg Med Chem Lett. 2021;43 doi: 10.1016/j.bmcl.2021.128099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Elhady S.S., Abdelhameed R.F.A., Malatani R.T., Alahdal A.M., Bogari H.A., Almalki A.J. Molecular docking and dynamics simulation study of hyrtios erectus isolated scalarane sesterterpenes as potential SARS-CoV-2 dual target inhibitors. Biology. 2021;10:389. doi: 10.3390/biology10050389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yadav R., Parihar R.D., Dhiman U., Dhamija P., Kumar S. Docking of fda approved drugs targeting nsp-16, n-protein and main protease of sars-cov-2 as dual inhibitors. Biointerface Res Appl Chem. 2020;11:9848–9861. [Google Scholar]
- 191.Kumari A., Rajput V.S., Nagpal P., Kukrety H., Grover S., Grover A. Dual inhibition of SARS-CoV-2 spike and main protease through a repurposed drug, rutin. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1864476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Teli D.M., Shah M.B., Chhabria M.T. In silico screening of natural compounds as potential inhibitors of SARS-CoV-2 main protease and Spike RBD: targets for COVID-19. Front Mol Biosci. 2021;7 doi: 10.3389/fmolb.2020.599079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Patil V.S., Hupparage V.B., Malgi A.P., Deshpande S.H., Patil S.A., Mallapur S.P. Dual inhibition of COVID-19 spike glycoprotein and main protease 3CLpro by Withanone from Withania somnifera. Chin Herb Med. 2021;13:359–369. doi: 10.1016/j.chmed.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.