Abstract
Recently, automatic computer-aided detection (CAD) of COVID-19 using radiological images has received a great deal of attention from many researchers and medical practitioners, and consequently several CAD frameworks and methods have been presented in the literature to assist the radiologist physicians in performing diagnostic COVID-19 tests quickly, reliably and accurately. This paper presents an innovative framework for the automatic detection of COVID-19 from chest X-ray (CXR) images, in which a rich and effective representation of lung tissue patterns is generated from the gray level co-occurrence matrix (GLCM) based textural features. The input CXR image is first preprocessed by spatial filtering along with median filtering and contrast limited adaptive histogram equalization to improve the CXR image's poor quality and reduce image noise. Automatic thresholding by the optimized formula of Otsu's method is applied to find a proper threshold value to best segment lung regions of interest (ROIs) out from CXR images. Then, a concise set of GLCM-based texture features is extracted to accurately represent the segmented lung ROIs of each CXR image. Finally, the normalized features are fed into a trained discriminative latent-dynamic conditional random fields (LDCRFs) model for fine-grained classification to divide the cases into two categories: COVID-19 and non-COVID-19. The presented method has been experimentally tested and validated on a relatively large dataset of frontal CXR images, achieving an average accuracy, precision, recall, and F1-score of 95.88%, 96.17%, 94.45%, and 95.79%, respectively, which compare favorably with and occasionally exceed those previously reported in similar studies in the literature.
Keywords: Computer-aided detection, COVID-19, Radiological images, GLCM-Based texture features, Latent-dynamic conditional random fields, Cross-validation
1. Introduction
The coronavirus disease 2019 (so-called COVID-19) is a serious and highly contagious disease caused by infection with a newly discovered virus, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which has first emerged from Wuhan city, Hubei Province of China in December 2019, and since then it has rapidly spread to almost all countries and territories in the world, causing an unprecedented global pandemic in the history of mankind [1,2]. On January 30, 2020, the Director-General of the World Health Organization (WHO) has declared the Chinese outbreak of COVID-19 a public health emergency of international concern (PHEIC) posing a significant risk to countries with vulnerable fragile health systems, and subsequently the outbreak was officially recognized as a pandemic in March 2020 by the WHO [3]. As of March 2021, over 119 million global cases of COVID-19 have been reported worldwide, including above 2.6 million acknowledged deaths. Meanwhile, around 94 million people have recovered since the beginning of the epidemic, while there are almost 20 million active cases worldwide, and the pandemic is still escalating, with no signs of stopping or even throttling down. The United States, India, and Brazil are now among the countries facing the worst of the global outbreak of the pandemic.
According to WHO, the new COVID-19 virus is primarily transmitted among people in the same vicinity, especially through contact routes with small respiratory droplets containing the pathogen, expelled by an infected person during expiratory events (e.g., coughing, sneezing, laughing, breathing, and talking). The droplets that carry the virus can normally travel only short distances through the air, before they fall onto surrounding surfaces or the ground. Virologist have now found that the virus can spread not only via aerosol droplets expelled by the patient while speaking, sneezing, or coughing, but also by touching contaminated hands and inanimate surfaces or objects. For example, when an infected person sneezed or coughed, and a droplet landed on a surface, other people could become infected by touching that surface [4,5]. Importantly, based on currently available credible evidence emerged from an analysis of thousands of cases recorded in China, airborne spread has not yet been reported for COVID-19, and thus it does not appear to be a major driver of transmission. Mounting evidence has indicated that the virus could spread even if people didn't have symptoms. However, infected people are highly contagious during the first three days after they develop symptoms [6].
Fever is usually referred to as the most common symptom of COVID-19 infection, though not everyone develops it. Other possible signs or symptoms well observed by physicians, may include dry cough, shortness of breath, fatigue, body ache, loss of appetite, and distorted sense of smell or taste, which is often likely to indicate viral pneumonia [7]. According to the WHO, the intrinsic incubation period (i.e., the time from infection to the onset of symptoms) is currently believed to occur within 2–14 days [8], though symptoms typically appear within 4 or 5 days after exposure. The WHO has earlier recommended masks for healthy people, including those who don't exhibit COVID-19 symptoms, only if they take care of a person suspected of being infected or confirmed to be infected with COVID-19. Moreover, enough emphasis has been given by doctors, and health care experts are stressing on the importance of wearing face masks during the pandemic for protection from the airborne sneeze and cough. On December 11, 2020, the US Food and Drug Administration (FDA) has issued the first emergency use authorization (EUA) for emergency use of Pfizer/BioNTech COVID-19 vaccine for the prevention of COVID-19 for individuals 16 years of age and older. Currently, other vaccines, different from Pfizer-BioNTech's vaccine, have been authorized in the US and recommended to prevent COVID-19 such as Moderna's vaccine and, most recently, Johnson & Johnson's vaccine. According to the Centers for Disease Control (CDC), these vaccines are found to be not only safe and effective, but critical to protecting people from COVID-19.
As per the interim phase 3 trial data released on November 23, 2020 by the University of Oxford, the Oxford-AstraZeneca COVID-19 vaccine has been first approved for emergency use in the UK's vaccination programme, and the first vaccination outside of a trial has been administered on January 4, 2021. It is worth mentioning that the Pfizer-BioNTech's, Moderna's and Oxford-Astrozaneca's vaccines all have a dual dose requirement, spaced out a few weeks apart. In several regions of the world, there are multiple candidate vaccines against COVID-19 currently under investigation, with large-scale Phase 3 clinical trials. On February 27, 2021, large-scale clinical trials have been carried out for two COVID-19 vaccines (i.e., AstraZeneca and Novavax) in the U.S., demonstrating their consistent and robust high-level efficacy in preventing severe COVID-19 disease. Furthermore, Atrium Health as a part of the COVID-19 Prevention Network (CoVPN) is currently aiming at enrolling thousands of volunteers in large-scale clinical trials for testing a variety of investigational vaccines and monoclonal antibodies intended to protect people from COVID-19.
The primary laboratory diagnostic tool for SARS-CoV-2 (COVID-19) infection is the detection of the viral RNA (genetic material) in samples collected from nose or throat swabs. Today, real-time reverse-transcriptase polymerase chain reaction (RT-PCR), quantitative RT-PCR, and nested RT-PCR are the most widely used techniques employed for the diagnosis of COVID-19 [9]. In other words, the COVID-19 RT-PCR test is a most accurate nucleic acid test that looks for replication of the viral RNA in the patient's sample. Radiological imaging techniques such as digital CXR and thoracic computed tomography (CT) could potentially be used for early screening, diagnosis, and treatment of patients with suspected or confirmed COVID-19 infections [10,11].
Due to the low sensitivity of RT-PCR quantification, a highly-false negative rate is obtained. Therefore, it is difficult to confidently diagnose COVID-19 infection that affects the timely treatment and disposition of infected patients. On the other hand, radiological imaging (CXR and CT) are capable of playing a crucial role in the evaluation of the clinical course of COVID-19 and then in the selection of appropriate management of infected patients. Although the CT scan is gold standard for diagnosis, digital CXR systems are still particularly useful in emergency diagnosis and treatment, because they are fast, easy, relatively inexpensive, widely available, and able to deliver lower radiation doses [11,12]. The significantly large number of individuals in the U.S. who tested positive for Covid-19 makes the use of regular screening on a daily basis enormously challenging for physicians. Hence, on March 2020, the U.S. federal administration has encouraged health experts and researchers to embrace artificial intelligence (AI), machine learning (ML), and other emerging technologies to combat the COVID-19 global pandemic [10].
In this paper, our main contribution is twofold: on one hand, an automated detection framework is developed to detect COVID-19 precisely and quickly from CXR images. On the other hand, the presented CAD system is trained and optimized with five-fold validations using data from two different digital X-ray datasets, i.e., IEEE8023/Covid Chest X-Ray [13,14] and ChestX-ray8 [15]. In the proposed framework, a minimal set of GLCM based features is extracted to properly model the texture information in CXR images. The final reduced set of texture features is then fed into a discriminative LDCRF model to perform fine-grained classification for detection of COVID-19 from CXR images. The findings suggest that the use of a reduced set of features in the proposed framework not only significantly simplifies the work of the discriminative LDCRF classifier, but also could potentially improve the classification accuracy and the scalability of the approach. An additional advantage of diminishing the size of extracted features is that the time complexity of the system drops significantly. In addition, it is anticipated that findings of this study are likely to be of interest and hopefully assistance to other researchers who are contemplating carrying out similar studies on automated CAD of COVID-19 from radiological images. The rest of this paper is structured as follows: a brief overview of relevant literature is presented in Section 2. Section 3 describes the proposed automated CAD framework for COVID-19 from radiological images. Experimental results and performance evaluations are demonstrated in Section 4. Finally, in Section 5, conclusion and perspectives for future work are drawn.
2. Related work
Just after the emergence of COVID-19 epidemic in late 2019, a growing number of researchers in the fields of AI and medical image understanding have shown increasing interest in developing high performance automatic CAD systems for COVID-19 from chest radiographic images. Thanks to the efforts of these researchers, multiple AI studies using chest radiographic images have been emerging to aid physicians to manage patients with COVID-19 pneumonia [8,[16], [17], [18]]. In Ref. [19], a patch-based deep learning convolutional neural network (CNN) approach for COVID-19 diagnosis from CXR images, using a relatively small number of trainable parameters. In this approach, full lung regions are segmented from the CXR image, by using a fully connected DenseNet 103 (FC-DenseNet 103) semantic segmentation network model built from 103 convolutional layers. In regard to classification, multiple random patches (i.e., ROIs) are extracted from the segmented lung regions to be then fed as inputs to the multimodal ML classification model. The CXR image samples used in this study were collected from both healthy persons and patients diagnosed with tuberculosis, bacterial pneumonia, and viral pneumonia caused by COVID-19 infection. The diagnostic model performed quite well, achieving an overall accuracy 88.9% of and an F1-score of 84.40%.
In a similar vein, Ozturk et al. [11] have suggested the deep learning DarkCovidNet model to automatically detect and diagnose COVID-19 from digital CXR images. Their model consisted of a total of 17 convolutional layers, and was potentially capable of handling both binary (COVID-19 vs. non COVID) and multinomial classification (COVID-19 vs. bacterial pneumonia vs. non-COVID-19 viral pneumonia). Overall diagnostic accuracies of 98.08% and 87.02% were obtained for binary and multinomial classifications, respectively. Similarly, in. [20], Fan et al. presented a deep learning network (Inf-Net) for COVID-19 lung infection segmentation to identify suspicious regions indicative of COVID-19 on chest CT images, based on randomly selected propagation. In their approach, a parallel partial decoder is utilized to yield the global representation of the final segmented maps. Then, implicit reverse attention and explicit edge attention are used to model the boundaries and enhance the representations. The model archived quite satisfactory segmentation results, with a Dice of 0.74 and an alignment index of 0.89.
In May 2020, in Ref. [18], the authors introduced a deep learning network model (COVID-Net) to distinguish patients with confirmed COVID-19 infection from healthy individuals and those with pneumonia, based on CXR images. On the same CXR image dataset, the classification performance of the model was then compared with the those of VGG-19 and ResNet-50 models. The outcome of the comparison concluded that COVIDNet significantly outperformed both VGG-16 and ResNet-50 models, with positive predictive values (PPVs) of 0.90, 0.91, and 0.99 for healthy, pneumonia, and COVID-19, respectively. Moreover, in Ref. [21], a deep learning COVIDXNet model was proposed to differentiate patients with COVID-19 from healthy persons, using 50 CXR images. Seven well-established deep learning networks has successfully been employed as feature extractors. A concrete comparison, of the COVIDXNet modelling scheme to other two deep learning models (i.e., VGG-19 and DensNet201), has demonstrated that the COVIDXNet model results in better results, with a potential diagnostic performance rate of 90%.
Additionally, in Ref. [22], Ahuja et al. developed a three-stage deep learning model for detecting COVID-19 from CT scan images, by conducting a binary classification task. In this approach, data augmentation, abnormality localization, and transfer learning have been employed, with different backend deep networks (i.e., ResNet18, ResNet50, ResNet101, and SqueezeNet). Experiments showed that the pre-trained Resnet18 with the transfer learning strategy is able to give the best diagnostic performance, achieving 99.82%, 97.32%, and 99.40% accuracies on the training, validation, and testing sets, respectively. In their study [23], the authors presented five pre-trained CNN based models (i.e., ResNet50, ResNet101, ResNet152, InceptionV3, and Inception-ResNetV2) as a screening tool for the early detection of COVID-19 pneumonia infected patients using CXR radiographs. Three binary classifications with four classes (COVID-19, healthy, viral pneumonia, and bacterial pneumonia) were preformed over 5-fold cross-validation. Regarding the classification performance results obtained, the prediction capability of the three models to differentiate COVID-19 positive patients from those without COVID-19 was assessed, and interestingly they found that the pre-trained ResNet-50 has best performance as compared to the other models, achieving an overall accuracy of 98%.
Ardakani et al. [24] presented a rapid diagnostic AI approach for COVID-19 using 1020 chest CT slices from 108 patients with laboratory proven COVID-19 (COVID-19 set) and 86 patients with other atypical and viral pneumonia diseases (non-COVID-19 set). Ten CNNs were utilized to accurately discriminate COVID-19 patients from other diseases in CT images, i.e., AlexNet, SqueezeNet, VGG-16, VGG-19, MobileNet-V2, GoogleNet, ResNet-18, ResNet-50, ResNet-101, and Xception. The authors have reported that among all networks, ResNet-101 and Xception achieved the best diagnostic performance; ResNet-101 could distinguish patients with COVID-19 from non-COVID-19 patients, with an AUC of 0.99 (and 100%, 99.02% and 99.51% of sensitivity, specificity and accuracy, respectively), whereas Xception presented an AUC of 0.994 (with 98.04%, 100%, 99.02% of sensitivity, specificity and accuracy, respectively). However, this study revealed that the diagnostic performance of the radiologist was generally moderate in differentiating COVID-19 from other viral pneumonias on chest CT, with an AUC of 0.873 (and 89.21%, 83.33% and 86.27% of sensitivity, specificity and accuracy, respectively). Furthermore, it was concluded that not only ResNet-101 is best considered as a high sensitivity model to screen and diagnose COVID-19 cases, it can also best serve as an assistance diagnostic tool in radiology departments.
In [25], Hassanien et al. introduced a totally methodology for detecting COVID-19 cases using CXR images, where an assist vector gadget (i.e., a support vector machine (SVM)) is used to classify the COVID-19 affected CXR images from others, using deep features. This technique is intended as an auspicious computer-aided diagnostic tool to assist the clinical practitioners and radiologists in early detection of COVID-19 infected cases. The diagnostic system has shown promising results in terms of high accuracy in classification of the infected lung with COVID-19 infection, where average sensitivity, specificity and accuracy of 95.76%, 99.7%, and 97.48% have been achieved, respectively.
3. Proposed methodology
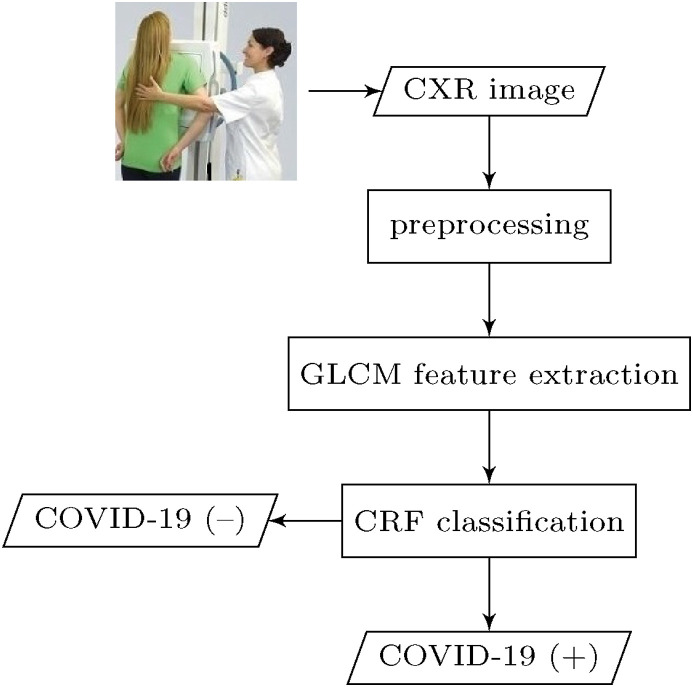
In this section, we present proposed methodology for automatic detection of COVID-19 pneumonia. The overall conceptual block diagram depicting the main steps of the proposed CAD system of COVID-19 is shown in Fig. 1 .
Fig. 1.
Functional block diagram of the main steps in the presented CAD system for COVID-19.
The general structure of the proposed CAD framework works as follows: as an initial step, each lung region suspected of having COVID-19 infection is first segmented by applying iterative automatic thresholding and morphological operations. Then, an optimized set of second-order statistical texture features is extracted from gray level co-occurrence matrix (GLCM) of both COVID-19 and normal healthy CXR image samples. A 1D vector representation is obtained from the extracted GLCM-based texture features and then fed into a CRF model for COVID-19 classification. In the following subsections, the workflow steps of the proposed methodology are explained in detail.
3.1. Image preprocessing
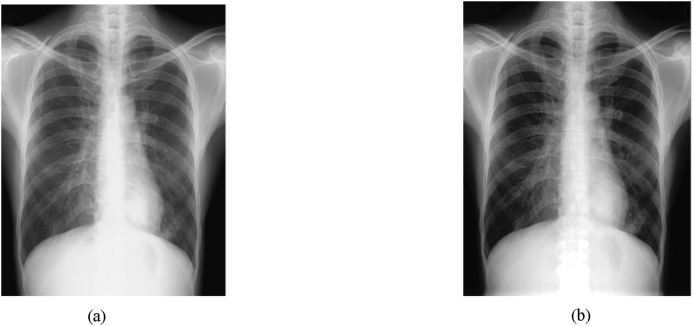
The image preprocessing step is basically responsible for detecting and reducing the amount of artifacts from the image. In CXR images, this step is necessary, since many CXR images contain some noise and unwanted artifacts such as patient cloths and wire that have to be removed to diagnose COVID-19 accurately. In the proposed methodology, the preprocessing step involves three main processes: (i) image resizing, (ii) noise removal by applying a simple 2D smoothing filter, (iii) image contrast enhancement [26] by pixel adjustment and histogram equalization (see Fig. 2 ).
Fig. 2.
Contrast-image enhancement:(a) original image and (b) enhanced image via pixel adjustment and histogram equalization.
The ultimate aim of lung segmentation is to isolate lung regions in CXR images and Computed Tomography (CT) scans. The automated lung segmentation is mostly performed as a pre-processing step of lung CXR image analysis and plays a pivotal role in all lung disease diagnosis. For the segmentation of lung regions in a CXR image, the proposed method involves iterative automatic thresholding and masking operations applied to contrast-enhanced CXR images. The lung segmentation procedure begins with applying automatic thresholding in each CXR image plane, based on Otsu discrimination criterion [27]. Binary masks are then crated and combined to generate a mask that achieves improved lung boundary segmentation accuracy. The segmented lung image may additionally include over-segmented areas (tiny blobs) which are too small to be part of lung tissues. To tackle this problem, a straightforward solution is to apply a morphological-area opening filter on the binary image. In addition, a finer segmented image which includes only the lung tissues can be obtained by smoothing the binary image with an iterative median filter of adjustable size.
In order to solve the problem of detecting extremely small non-lung tissues and to avoid confusion between isolated artifacts and lung tissues, an adaptive morphological open-close filter is iteratively applied to the resulting binary image to remove objects that are too small from the binary image, while maintaining larger objects in shape and size. This filter is best realized as a cascade of erosion and dilation operations using locally adaptive structuring elements. The morphological operations (opening and closing) for filling holes and boundary smoothing of the segmented lung regions are defined as follows:
| (1) |
| (2) |
where ϵ E and δ E denote the two basic morphological operations erosion and dilation, of an image f(x), respectively, which are defined using flat structuring elements (SEs) E(x) as follows:
| (3) |
| (4) |
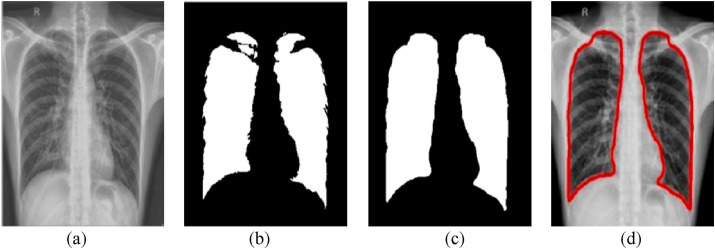
where ∧ and ∨ denotes the minimum and maximum operators, respectively. Finally, a modified canny edge detector [28] is applied to perform robust lung boundary detection. A sample of lung segmentation along with boundary detection can be seen in Fig. 3 .
Fig. 3.
Sample lung segmentation:(a) pre-processed image, (b) Otsu's thresholding, (c) a cascade of dilation, hole filling and erosion morphological operations, and (d) detected lung boundary.
3.2. Haralick feature extraction
Generally speaking, feature extraction is a key step in various image classification and pattern recognition applications, which aims to retrieve discriminative features from image data (or ROIs), and later these features are fed as input vectors to the machine learning models, from which a visual description, interpretation or understanding of image contents can be automatically provided by the machine. Specifically, in the proposed CAD system for COVID-19, an adaptive GLCM-based feature extraction technique is employed to obtain a compact set of discriminative texture features [29] (e.g., contrast, entropy, homogeneity, etc.) from chest X-ray images. Here, a second-order methodology is adopted to extract second-order statistical texture features, which considers the connectivity of cluster pixels in an input chest X-ray image I. To significantly facilitate the computation of GLCM, the gray level of the pixels of each preprocessed CXR image is quantized to a reduced number ℓ of gray levels (ℓ is experimentally set to 4). Moreover, this quantization was found to dramatically reduce the computational costs of GLCM, as it is computationally very costly to compute the GLCM form all 256 gray levels. The GLCM, can be constructed as a matrix of frequencies, by simply counting the number of times each pair of quantized gray-levels occurs as neighbors in the quantized image, . More formally, each element, P(i, j), in the GLCM can be readily computed as follows,
| (5) |
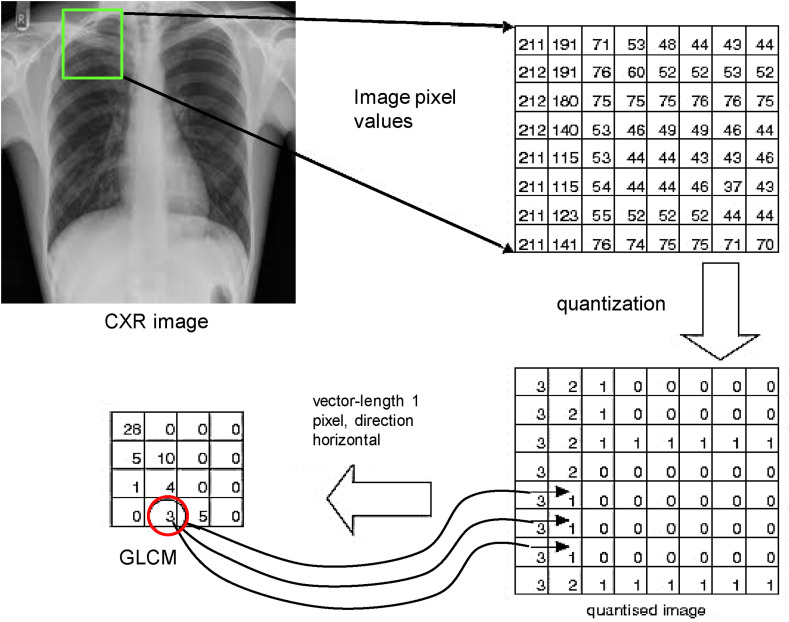
where δ = (Δx, Δy) is the displacement vector expressed in pixel units along x- and y-directions. Fig. 4 below shows an illustrative example of how the GLCM is created from the quantized input image.
Fig. 4.
An illustration example of the computation of the GLCM, where δ = (1, 0).
It should be noted that it is possible for a GLCM to be created with several displacement vectors, e.g.,
| (6) |
for the 8 immediate neighbors of a pixel. Then, a normalized GLCM, representing the estimated probability of the combinations of pairs of neighboring gray-levels in the image can be computed as follows,
| (7) |
Note that the above normalized GLCM can be thought of as a probability mass function of the gray-level pairs in the image, form which the following set of optimized texture features can be quantitatively calculated.
Contrast–measurement of the local variations: This feature measures the local variations in intensity present in an image (or ROIs). More specifically, the contrast measure returns the value of the intensity contrast between a pixel and its neighbor pixel over the entire image. Hence, it has been argued that a low-contrast image includes a smooth range of grays, whereas a high-contrast image contains more tones at either end of the spectrum (black and white). This implies that a low-contrast image is not characterized by low gray levels, but rather by low spatial frequencies. Therefore, the GLCM contrast is highly associated with spatial frequencies, which can be efficiently computed as follows:
| (8) |
where N represents the total number of gray levels used (i.e., the dimension of the GLCM).
Energy, also called uniformity or angular second moment (ASM), represents the degree of homogeneity of gray distribution and the thickness of texture, which always provides a stabilising measure of greyscale patterns; thus, for a lager value on the Energy feature, a more stable regulation is obtained. Formally speaking, Energy is quadratic sum of GLCM elements and measures the gray level concentration of intensity in GLCM, which is calculated from the GLCM using the expression given as:
| (9) |
Inverse difference moment (IDM): measures the local homogeneity of an image that reflects the intensity variation within the image. When the homogeneity value is increased, the variation intensity in image is decreased. Moreover, it is the inverse of contrast; thus, the higher the contrast, the lower the homogeneity and vice versa. More specifically, Homogeneity is a measure of the closeness of the distribution of GLCM elements to its diagonal, which defines the uniformity of a texture, and it is derived from the GLCM using the following formula:
| (10) |
Entropy– a standard measure of randomness is another important GLCM feature to distinguish an image texture, which is commonly classified as a first degree measure of the amount of disorder in an image. The GLCM derived entropy is inversely proportional to GLCM energy, and it can be easily calculated from the GLCM elements using the following formula:
| (11) |
where lg denotes the base 2 logarithm.
Mean: It appears to be the optimal GLCM texture measure, and thus it is much more useful than other GLCM textural features. The GLCM Mean is not simply the average of all the original pixel values in the image window; rather it is mathematically equivalent to GLCM dissimilarity, where each pixel value is weighted by the frequency of its occurrence and a specific neighbor pixel value. The GLCM Mean, μ = (μ i, μ j), is defined as:
| (12) |
where μ i and μ j are the GLCM means in the horizontal and vertical directions, respectively.
Standard Deviation, also called the square root of GLCM variance, is a measure of the variance of the gray levels in the GLCM and should not be confused with the marginal variance. GLCM variance in texture performs the same task as does the popular descriptive statistic called variance. However, the GLCM variance makes use of GLCM, thus it deals exclusively with the dispersion around the mean of combinations of reference and neighbor amplitudes, so that it is not the same as the variance of input amplitudes that can be derived by the ‘Volume Statistics’ attribute. Formally, the GLCM variance is computed as follows:
| (13) |
where σ i and σ j are the GLCM standard deviations in the horizontal and vertical directions, respectively.
Correlation: The correlation feature obtained from GLCM exhibits similar discriminative capability as the Contrast feature, and it gives a quantitative measure of how a pixel is closely connected or correlated with its neighbor over the image. In other words, Correlation figures out the linear gray-level dependency between the pixels at the specified positions relative to each other. The range of correlation property lies between [−1, 1]. The strength of the association between images is quantified by a correlation score, e.g., a correlation value close to +1 or −1 implies that two images are highly associated (positively or negatively, respectively). The correlation of the GLCM can be derived as follows,
| (14) |
where μ i, σ i, μ j, σ j denote the mean and standard deviation values for the normalized GLCM elements computed in horizontal and vertical directions, respectively.
RMS: This feature is statistically expressed as the square root of the arithmetic mean of the square of the ordinates of a given sample of n values. Formally, RMS can be stated as follows,
| (15) |
where Y 1, Y 2, …, Y 1 denote the ordinates of the sample.
Smoothness: The goal of image smoothing is to reduce image noise and reduce detail. Smoothing is a common statistical technique to handle image data by creating approximating functions that attempt to capture important patterns in the image data. The smoothness feature concerns the image texture which would be either smooth or coarse. In texture analysis, a smooth texture is typically characterized as having few pixels in a relatively constant gray-level run, and a coarse texture as having many pixels in such a run. Moreover, image contrast mainly reflects the smoothness degree of the image texture structure; if the GLCM has small off-diagonal elements, the image has low contrast, and then the texture is smooth. Formally, this provides a contribution to the GLCM as follows,
| (16) |
Skewness: This texture feature is a measure of asymmetry of a histogram distribution, which reflects gray level histogram properties of a given image (or ROI), with a value of zero for a normal distribution. For univariate data Y1, Y2, …, Y N, the skewness is given by the following formula:
| (17) |
where , σ, and N are the mean, the standard deviation, and the size of data points. Note that in Skewness computation, σ is calculated with N in the denominator rather than N − 1.
Kurtosis This feature is a statistically second-order parameter that quantifies the amount of histogram deviating from the characteristic bell shape of a normal distribution. More specifically, kurtosis can well describe the distribution tails. There are two types of kurtosis; a positive Kurtosis indicates a sharper distribution than the normal, whereas a negative Kurtosis indicates a flatter distribution than the normal. Note that a Gaussian distribution has a kurtosis of 0. Similar to Skewness, Kurtosis can be calculated, as follows,
| (18) |
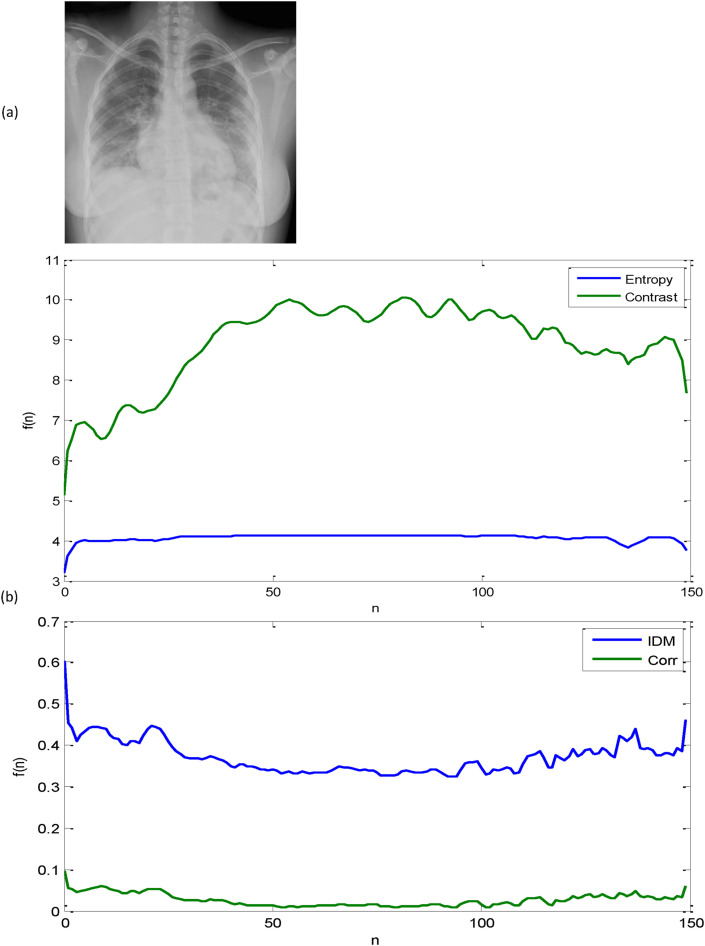
where , σ, and N are the mean, the standard deviation, and the size of data points, respectively. Fig. 5 displays a sample CXR image of a confirmed COVID-19 case along with plots of its GLCM-based texture features. At this point, it is pertinent to emphasize that before starting the feature concatenation process, each attribute of the GLCM based texture features is normalized into [0,1] to allow for equal weighting among each type of feature. The resulting features are then fed into a CRF model for finer-grained classification. Furthermore, it could be argued that the normalized GLCM based features have immense potential for more accurate and robust feature classification that in turn has a significantly positive influence on the performance of the proposed CAD system for COVID-19.
Fig. 5.
A sample of a CXR image of a confirmed COVID-19 case along with plots of its GLCM-based texture features: (a) Original image and (b) a sample of four GLCM-based texture features.
3.3. Feature classification
In this section, we give details of the feature classification module in our CAD system for distinguishing COVID-19 from non-COVID-19 cases. Broadly speaking, the key purpose of the classification module in the current CAD system is to classify each CXR image on the collected dataset into one of two diagnostic categories (COVID-19 and non-COVID-19), based on the extracted texture features. The classification module depends to a large extent on the availability of a set of clinical diagnostic cases. This set of previously diagnostic (or diagnostically labeled) cases is conventionally referred to as the “training set”, and thus the utilized learning strategy is termed as “supervised learning”. For the current task of COVID-19 classification, there are several classification techniques available in the existing literature, such as Artificial Neural Network (ANN), Support Vector Machines (SVMs), Bayesian Network (BN), k-Nearest Neighbor (k-NN), Conditional Random Fields (CRFs), etc. In the present work, we opt to adopt the latent-dynamic CRF (LDCRF) model for the feature classification task. Due to its inherent dependence on CRFs, the LDCRF model is characterized as a typical discriminative probabilistic latent variable model that could potentially describe the sub-structure of a class label and successfully and reliably learn dynamics between class labels. Additionally, it has been shown that the LDCRF model could perform significantly well in many large-scale object detection problems [30,31], and also it demonstrates superior performance in learning relevant context and integrating it with visual observations, when compared with other machine learning methods such as hidden Markov models, hidden Semi-Markov models, and Naive Bayes.
The LDCRF models have historically emerged as a potential extension to the original CRFs to learn the hidden interaction between features, and they could be interpreted as undirected probabilistic graphical models that provide powerful tools for segmenting and labeling sequential data. Consequently, they are directly applicable to sequential data, eliminating the need for windowing the signal. In this manner, each label (or state) suggests a specific diagnostic case. As LDCRF models have a class label for each observation, they could learn and accurately classify lung patterns in unsegmented CXR images. Moreover, the LDCRF models could perfectly infer the lung patterns in both training and testing stages.
Formally speaking, the primary task of the LDCRF model, as described by Morency et al. [32], aims to learn a direct mapping between a sequence of raw observations (or features) x = ⟨x 1, x 2, …, x m⟩ and a sequence of labels y = ⟨y 1, y 2, …, y m⟩, where each y j is a label for the j-th observation in a sequence, which represents a member of a set Y of possible class labels. A feature vector represents an image observation x j. For each sequence, suppose h = ⟨h 1, h 2, …, h m⟩ be a set of latent substructure variables, which are not observed in the training data and will therefore form a set of ‘hidden’ variables in the established model, as depicted in Fig. 6 below.
Fig. 6.
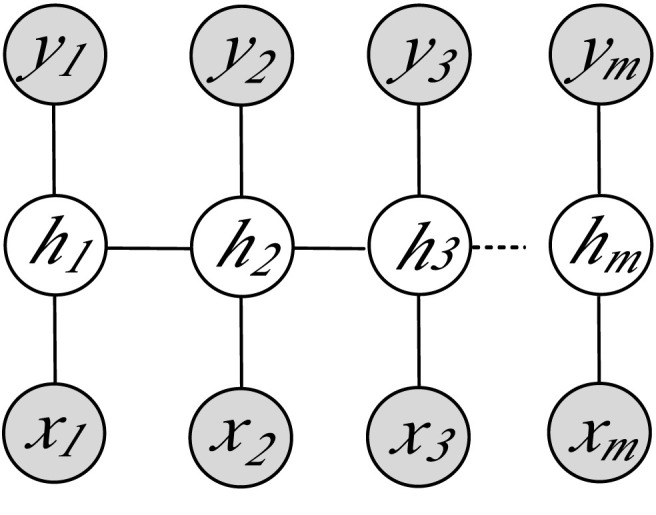
Graphical representation of LDCRF model, where hj denotes the hidden state assigned to the j-th observation xj, and yj is the class label of xj. The filled gray nodes correspond to the observed model variables.
Given the above definitions, we can thus formulate a latent-conditional model as follows:
| (19) |
where θ is a set representing the optimal model parameters. Now, given a collection of training examples, each labeled with its correct class value , the goal of the training procedure is to learn the optimal model parameters θ from the objective function [33] given by
| (20) |
where n denotes the number of training samples. It is worthwhile pointing out that in Eq. (20) above, there are two terms on the right-hand side: the first one is the log-likelihood of the training data, while the second one corresponds to the log of a Gaussian prior with variance σ 2, i.e.,
| (21) |
To estimate the optimal model parameters, we apply an iterative gradient ascent algorithm for maximizing the objective function:
| (22) |
Once the parameters θ* are learned, the trained model can then make predictions about unseen (test) data via inductive inference:
| (23) |
where y* denotes the predicted class label for an unseen sample x. For a more detailed discussion of LDCRF training and inference, the interested reader is encouraged to refer to Ref. [32].
4. Experimental results
In this section, we present and discuss in detail the results obtained from a series of extensive experiments conducted to corroborate the success of the proposed CAD model for automated identification of COVID-19 through demonstrating or confirming its superior performance characteristics.
4.1. Datasets
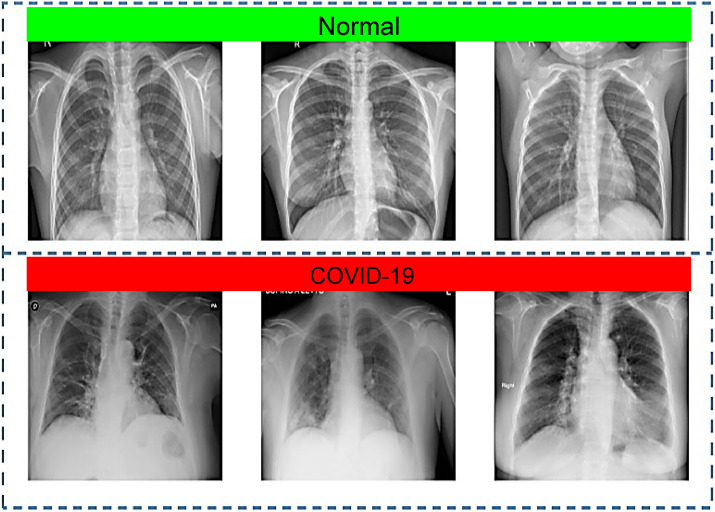
To evaluate the performance of the proposed CAD system, we created a mixed dataset generated by the fusion of multiple publicly available datasets. Firstly, a set of 500 CXR images representing confirmed COVID-19 cases was randomly selected from Github repository developed by Cohen et al. [34], where the CXR images were collected from different open sources and hospitals, and 83 female and 175 male were diagnosed or tested positive for COVID-19, with an average age of around 55 years among the infected patients. Though, the full metadata are not provided for all patients in this dataset. Additional 1800 representative images for COVID-19 positive cases were taken from various sources and repositories such as another Github source [35], SIRM database [36], TCIA [37], radiopaedia.org [38], and Mendeley [39]. Thus, the dataset used here consists of a total of 2,300 CXR images for COVID-19 positive cases. The normal (no finding) CXR images were obtained from the Chest X-ray8 database developed by Wang et al. [15] that comprises a total of 108,948 frontal view images from 32,717 patients. For this study, 2,300 normal CXR images were randomly selected to be used in the experiments. Hence, the prepared dataset used to train and evaluate the proposed CAD system has a total of 4,600 CXR images for COVID-19 positive cases and normal patients. A sample of the dataset images can be seen in Fig. 7 above.
Fig. 7.
A sample of CXR images from the used dataset: normal (Row 1) and COVID-19 cases (Row 2).
4.2. Performance evaluation and analysis
For computational efficiency, the dataset images are first converted from color to 8-bit grayscale format and down-sampled to a fixed resolution of 128 × 128 pixels, as a preprocessing step prior to the feature extraction phase. Due to the absence of independent dataset, a train-test split is randomly applied to the dataset images, with a ratio of 80% and 20% for training and test set, respectively. In order to obtain reliable and robust results independent from the training and test datasets, a k-fold cross-validation procedure (with k = 5) has been used throughout the evaluation experiments. More specifically, out of the entire dataset images, in each fold, 3,680 images are randomly assigned for training, and the remaining 920 images are used for testing. Thus, the classification performance measurements are averaged over k splits. To quantitatively assess the performance of the present CAD system, several evaluation metrics including accuracy, precision, recall, and F1-score are utilized, which can be conveniently expressed as follows.
Accuracy: Accuracy is one of the most intuitive and widely used performance measure, and it simply is the probability that a randomly chosen instance (positive or negative) will be correct. More specifically, it is the probability that the diagnostic test yields the correct determination, i.e.,
| (24) |
Precision: Precision concerns the ability to correctly detect positive classes from all predicted positive, which is then defined as a ratio of correctly predicted positive classes to the total predicted positive classes:
| (25) |
From the equation above, it is pretty obvious that high precision always relates to a low false positive rate.
Recall: Recall, also referred to as sensitivity, hit rate, or true positive rate, can be generally thought as a model's ability to identify all the positive cases, and it can thus be expressed as follows:
| (26) |
One should notice that the above equation suggests that high recall is certainly associated with a low false negative rate.
F1-score: Although this score is not as intuitive as accuracy, it is particularly advantageous in measuring how precise and robust the classifier is. F1 score, which serves as a single measure of test performance that takes into account both precision and recall, is commonly calculated as a weighted average of the precision and recall,
| (27) |
where TP (true positive) and TN (true negative) are correctly predicted positive and negative COVID-19 cases, respectively, while FP (false positive) and FN (false negative) are incorrectly predicted positive and negative COVID-19 cases, respectively. Table 1 presents the cross-classification table: standard-of-reference COVID-19/non-COVID-19 vs. model's prediction COVID-19/non-COVID-19.
Table 1.
Cross-classification: model's prediction COVID-19/non-COVID-19 cases.
| COVID-19 | non-COVID-19 | |
|---|---|---|
| Test (+) | 453 | 23 |
| Test (−) | 37 | 467 |
The figures in Table 1 suggest that the proposed CAD model has significant potential to deliver a competitive performance in terms of average accuracy, precision, recall and F1-score, achieving scores 95.88%, 96.17%, 94.45%, and 95.79% respectively, in light of the equations given above and based on 5-fold cross-validation. In regard to the confidence, the proposed diagnostic model has achieved an average ROC area under the curve (AUC) of 0.956 (95% confidence interval: 0.92–0.97). Moreover, the approximated 95% confidence intervals of the positive predictive value (PPV) and negative predictive value (NPV) of the diagnostic model are 96% (95% CI: 0.947 to 0.984) and 95% (95% CI: 0.937 to 0.972), respectively. In order to demonstrate the superior performance of the proposed CAD framework over existing state-of-the-art in terms of various assessment metrics, a comprehensive performance comparison with other similar works [40, 41, 42, 43, 44] in the literature has been performed. The results of the quantitative comparison are presented in Table 2 .
Table 2.
Quantitative performance comparison of the proposed CAD system with those presented in previous works.
| Method | Accuracy (%) | Precision (%) | Recall (%) | F1-score (%) |
|---|---|---|---|---|
| Our method | 95.88 | 96.17 | 94.46 | 95.79 |
| Podder et al. [40] | 94.06 | 94.00 | 94.00 | 94.00 |
| Echtioui et al. [41] | 94.14 | 96.00 | 86.00 | 91.07 |
| Shukla et al. [42] | 87.79 | 92.43 | 90.16 | 91.28 |
| Panwar et al. [43] | 88.10 | 97.62 | 82.00 | 89.13 |
| Konar et al. [44] | 93.1 | 89.0 | 83.5 | 82.6 |
The average computation time of the presented CAD system for COVID-19 is approximately 3.4 s from image pre-processing to the final stage of automatic CAD of COVID-19 pneumonia, so that it can operate sufficiently fast for real-time processing, due to its relatively low computational costs for lung and lung-lesion segmentation task, as well as the real-time GLCM based texture feature extraction and classification. The proposed automated CAD system has been designed and implemented for much of its framework in C++ under Microsoft Visual Studio 2016 using OpenCV Vision Library as an ideal implementation solution for real-time digital image processing and object detection written in optimized C/C++ and designed for multi-core processors. All experiments, including tests and evaluations, have been carried out on a desktop PC with an Intel(R) Core(TM) i7 CPU 2.60 GHz processor and 8 GB of RAM running Microsoft Windows 10 Professional x64 edition.
5. Conclusion
This paper has presented a CAD method for the identification of COVID-19 cases from CXR images, where a rich representation is constructed from an optimized set of GLCM based texture features to accurately represent the segmented lung tissue ROIs of each CXR image. The extracted features are normalized and then fed into a discriminative LDCRF model for final COVID-19 classification. On a large publicly available dataset of frontal CXR images, the method has been rigorously tested and validated, achieving an average accuracy of 95.88%, with precision, recall and F1-score of 96.17%, 94.45%, and 95.79%, respectively, using 5-fold cross-validation. These results provide evidence that the proposed CAD methodology can help guide radiologists and medical physicists in obtaining a robust diagnosis model to distinguish COVID-19 for non COVID-19 cases confidently and achieve rapid and accurate infection detection tests. As prospects for future work, our aim is twofold. On the one hand, we intend develop an improved approach for lung tissue feature extraction, based on the integration of extensive texture features (e.g., GLCM, HOG, LBP, etc.) to construct a hybrid feature descriptor. On the other hand, we plan to extend our experimentations to additional public CXR datasets from patients who are positive or suspected of COVID-19 or other viral and bacterial pneumonias.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Lu H., Stratton C., Tang Y.-W. Outbreak of pneumonia of unknown etiology in wuhan, China: the mystery and the miracle. J. Med. Virol. 2020;92:401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mustafa N. Research and statistics: coronavirus disease (covid-19) Int. J. Syst. Dynam. Appl. 2021;10:1–20. [Google Scholar]
- 3.Ji P., Song A., Xiong P., Yi P., Xu X., Li H. Egocentric-vision based hand posture control system for reconnaissance robots. J. Intell. Rob. Syst. 2017;87:583–599. [Google Scholar]
- 4.Wu F., Zhao S., Yu B., Chen Y., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E., Zhang Y.-Z. Author correction: a new coronavirus associated with human respiratory disease in China. Nature. 2020;580:1476–4687. doi: 10.1038/s41586-020-2202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Cao B. Clinical features of patients infected with 2019 novel coronavirus in wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holshue M.L., DeBolt C., et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singhal T. A review of coronavirus disease-2019 (covid-19) Indian J. Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. Fs12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanne J., Little B., Chung J., Elicker B., Ketai L. Essentials for radiologists on covid-19: an update—radiology scientific expert panel. Radiology. 2020;296:E113–E114. doi: 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miranda Pereira R., Bertolini D., Teixeira L., Silla C., Costa Y. Covid-19 identification in chest x-ray images on flat and hierarchical classification scenarios. Comput. Methods Progr. Biomed. 2020;194 doi: 10.1016/j.cmpb.2020.105532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alimadadi A., Aryal S., Manandhar I., Munroe P.B., Joe B., Cheng X. Artificial intelligence and machine learning to fight covid-19. Physiol. Genom. 2020;52(4):200–202. doi: 10.1152/physiolgenomics.00029.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of covid-19 cases using deep neural networks with x-ray images. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robson B. Computers and viral diseases. preliminary bioinformatics studies on the design of a synthetic vaccine and a preventative peptidomimetic antagonist against the sars-cov-2 (2019-ncov, covid-19) coronavirus. Comput. Biol. Med. 2020;119 doi: 10.1016/j.compbiomed.2020.103670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen J.P., Morrison P., Dao L. COVID-19 image data collection. https://ui.adsabs.harvard.edu/abs/2020arXiv200311597C arXiv e-prints. URL.
- 14.Chowdhury M., Rahman T., Khandakar A., Mazhar R., Kadir M., Mahbub Z., Islam K., Khan M.S., Iqbal A., Al-Emadi N., Reaz M.B.I., Islam M. Can ai help in screening viral and covid-19 pneumonia? IEEE Acc. 2020;8:132665–132676. doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- 15.Wang X., Peng Y., Lu L., Lu Z., Bagheri M., Summers R.M. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2017. Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases; pp. 3462–3471. [DOI] [Google Scholar]
- 16.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., jing Liu J. Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: relationship to negative RT-PCR testing. Radiology. 2020;296:E41–E45. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with covid-19 pneumonia in wuhan, China: a descriptive study, the Lancet. Infect. Dis. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.L. Wang, Z. Q. Lin, A. Wong, Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images, Sci. Rep. 10. [DOI] [PMC free article] [PubMed]
- 19.Oh Y., Park S., Ye J.C. Deep learning covid-19 features on cxr using limited training data sets. IEEE Trans. Med. Imag. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 20.Fan D.P., Zhou T., Ji G.P., Zhou Y., Chen G., Fu H., Shen J., Shao L. Inf-net: automatic covid-19 lung infection segmentation from ct images. IEEE Trans. Med. Imag. 2020;39(8):2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 21.E. E.-D. Hemdan, M. Shouman, M. Karar, Covidx-net: A Framework of Deep Learning Classifiers to Diagnose Covid-19 in X-Ray Images, ArXiv abs/2003.11055.
- 22.Ahuja S., Panigrahi B.K., Dey N., Rajinikanth V., Gandhi T. Deep transfer learning-based automated detection of covid-19 from lung ct scan slices. Appl. Intell. 2020:1–15. doi: 10.1007/s10489-020-01826-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.A. Narin, C. Kaya, Z. Pamuk, Automatic Detection of Coronavirus Disease (Covid-19) Using X-Ray Images and Deep Convolutional Neural Networks, ArXiv abs/2003.10849. [DOI] [PMC free article] [PubMed]
- 24.Ardakani A.A., Kanafi A.R., Acharya U., Khadem N., Mohammadi A. Application of deep learning technique to manage covid-19 in routine clinical practice using ct images: results of 10 convolutional neural networks. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103795. 103795 – 103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.A. E. Hassanien, L. N. Mahdy, K. A. Ezzat, H. H. Elmousalami, H. A. Ella, Automatic X-Ray Covid-19 Lung Image Classification System Based on Multi-Level Thresholding and Support Vector Machine, medRxiv, .
- 26.Cadena L., Zotin A., Cadena F. Lecture Notes in Engineering and Computer Science: Proceedings of the International MultiConference of Engineers and Computer Scientists. 2018. Enhancement of medical image using spatial optimized filters and openmp technology; pp. 324–329. [Google Scholar]
- 27.Otsu N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybernet. 1979;9(1):62–66. [Google Scholar]
- 28.Canny J. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986;(6):679–698. [PubMed] [Google Scholar]
- 29.Haralick R.M., Shanmugam K., Dinstein I. Textural features for image classification. IEEE Trans. Syst. Man Cybernet. SMC-3. 1973;(6):610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- 30.Deufemia V., Risi M., Tortora G. Sketched symbol recognition using latent-dynamic conditional random fields and distance-based clustering. Pattern Recogn. 2014;47(3):1159–1171. [Google Scholar]
- 31.Ramírez G.A., Fuentes O., Crites S.L., Jimenez M., Ordonez J. CVPR Workshops. 2014. Color analysis of facial skin: detection of emotional state; pp. 474–479. [Google Scholar]
- 32.Morency L.P., Quattoni A., Darrell T. CVPR ’07. 2007. Latent-dynamic discriminative models for continuous gesture recognition. [Google Scholar]
- 33.Lafferty J., McCallum A., Pereira F. Proceedings of the Eighteenth International Conference on Machine Learning. 2001. Conditional random fields: probabilistic models for segmenting and labeling sequence data; pp. 282–289. [Google Scholar]
- 34.Cohen J.P., Morrison P., Dao L., Roth K., Duong T.Q., Ghassemi M. 2006. Covid-19 Image Data Collection: Prospective Predictions Are the futurearXiv. [Google Scholar]
- 35.Covid 19 chest x-ray. https://github.com/agchung ([Online])
- 36.Covid-19 database | sirm. https://www.sirm.org/en/category/articles/covid-19-database/ ([Online])
- 37.Frederick Nat Lab The cancer imaging archive. 2018. https://www.cancerimagingarchive.net/about-the-cancer-imaging-archive-tcia/ the cancer imaging archive (tcia) [Online], 1.
- 38.Gaillard F., Radiopaedia.org . 2014. Radiopaedia.org, the Wiki-Based Collaborative Radiology Resource.http://radiopaedia.org/ [Online] [Google Scholar]
- 39.Alqudah A.M., Qazan S. vol. 4. 2020. (augmented Covid-19 X-Ray Images Dataset) [Google Scholar]
- 40.Podder P., Bharati S., Mondal M., Kose U. Application of machine learning for the diagnosis of covid-19. Data Sci. COVID-19. 2021;20:175–194. doi: 10.1016/B978-0-12-824536-1.00008-3. [DOI] [Google Scholar]
- 41.Echtioui A., Zouch W., Ghorbel M., Mhiri C., Hamam H. Detection methods of covid-19. SLAS Technol. Transl. Life Sci. Innovat. 2020;25:1–7. doi: 10.1177/2472630320962002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.P. Shukla, A. Verma, Abhishek, S. Verma, M. Kumar, Interpreting svm for medical images using quadtree, Multimed. Tool. Appl. 79. doi:10.1007/s11042-020-09431-2. [DOI] [PMC free article] [PubMed]
- 43.Panwar H., Gupta P., Siddiqui M.K., Morales-Menendez R., Singh V. Application of deep learning for fast detection of covid-19 in x-rays using ncovnet, Chaos. Solitons Fractals. 2020;138 doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konar D., Panigrahi B.K., Bhattacharyya S., Dey N., Jiang R. Auto-diagnosis of covid-19 using lung ct images with semi-supervised shallow learning network. IEEE Acc. 2021;9:28716–28728. doi: 10.1109/ACCESS.2021.3058854. [DOI] [Google Scholar]