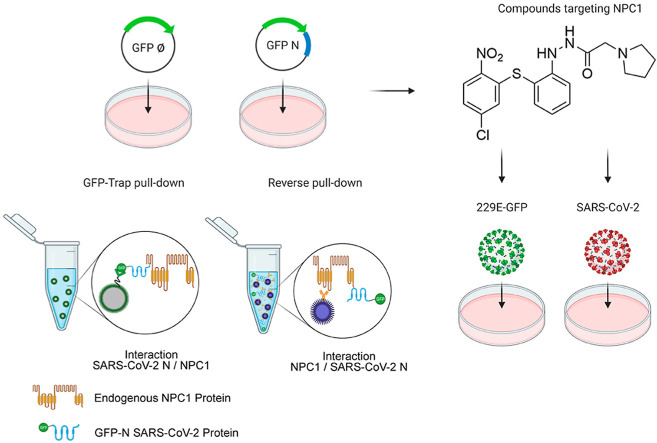
Abstract
Niemann-Pick type C1 (NPC1) receptor is an endosomal membrane protein that regulates intracellular cholesterol traffic. This protein has been shown to play an important role for several viruses. It has been reported that SARS-CoV-2 enters the cell through plasma membrane fusion and/or endosomal entry upon availability of proteases. However, the whole process is not fully understood yet and additional viral/host factors might be required for viral fusion and subsequent viral replication. Here, we report a novel interaction between the SARS-CoV-2 nucleoprotein (N) and the cholesterol transporter NPC1. Furthermore, we have found that some compounds reported to interact with NPC1, carbazole SC816 and sulfides SC198 and SC073, were able to reduce SARS-CoV-2 viral infection with a good selectivity index in human cell infection models. These findings suggest the importance of NPC1 for SARS-CoV-2 viral infection and a new possible potential therapeutic target to fight against COVID-19.
Keywords: Niemann-pick type C1 (NPC1), SARS-CoV-2, SARS-CoV-2 nucleoprotein, Potential therapeutic target, Endosomes
Graphical abstract
1. Introduction
The current coronavirus disease 19 (COVID-19) pandemic, caused by the emerging and pathogenic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has affected more than 170 millions of people worldwide and constitutes one of the greatest challenges in this century due to a great loss of lives, more than four millions up to date (WHO Coronavirus dashboard July 8, 2021) (WHO, 2021). These facts highlight the urgent need for developing efficient therapeutics against SARS-CoV-2 since there are only a few licensed treatments available (Indari et al., 2021). A better understanding of the biology of SARS-CoV-2 is vital in order to develop effective therapeutics.
A key step in the biology of SARS-CoV-2 is the cell entry mechanism upon interaction with the angiotensin-converting enzyme 2 (ACE2) receptor at the plasma membrane, which mediates fusion of membranes (Lan et al., 2020; Shang et al., 2020a, 2020b; Wang et al., 2008; Zhou et al., 2020). But, similarly to SARS-CoV-1, SARS-CoV-2 virus can also enter via endosomes after binding ACE2 receptor and subsequent cathepsin L processing (Inoue et al., 2007; Wang et al., 2008). In fact, the inhibition of both alternatives is necessary for full inhibition of SARS-CoV-2 entry (Hoffmann et al., 2020; Ou et al., 2020). Considering that SARS-CoV-2 may enter the cells by endocytosis, it would be partially acid pH-dependent and should exit endosomes by membrane fusion to start replication (Tharappel et al., 2020).
Several viruses interact with proteins related to the cholesterol transport. An example of this is the Hepatitis C virus (HCV), which uses the cellular Niemann-Pick C1-like 1 protein (NPC1L1), an homologous protein to NPC1 acting as a transmembrane-domain cell surface cholesterol-sensing receptor that is expressed on the apical surface of intestinal enterocytes and human hepatocytes (Sainz et al., 2012, Scott and Ioannou, 2004).
The Niemann-Pick type C1 (NPC1) is a late-endosomal membrane protein required for transport of cholesterol into the cells (Li et al., 2016). This protein localizes in late endosomes and it has 3 large loops (or domains A,C and I) that protrude into the endosome lumen (Scott and Ioannou, 2004).
NPC1 has been reported to be important for the infectivity of several viruses such as HCV (Stoeck et al., 2018), Ebola virus (EBOV) (Carette et al., 2011; Côté et al., 2011), HIV (Tang et al., 2009), Chikungunya virus (CHIKV) (Wichit et al., 2017), and several flaviviruses including Dengue (DENV) (Jupatanakul et al., 2014; Poh et al., 2012) and Zika virus (ZIKV) (Sabino et al., 2019), among others (Osuna-Ramos et al., 2018; Wichit et al., 2017).
For EBOV, two simultaneous publications first described NPC1 as an intracellular host receptor (Carette et al., 2011; Côté et al., 2011). For other viruses, like HCV, NPC1 has been described to contribute to viral replication; in this case, the pharmacological inhibition of NPC1 function results on the reduction of HCV replication (Stoeck et al., 2018). The FDA-approved drug, imipramine, which its known to interfere with NPC1 function, has been also reported to reduce CHIKV replication (Wichit et al., 2017) and HCV infection (Mingorance et al., 2014).
As SARS-CoV-2 can be internalized via clathrin- and non-clathrin-mediated endocytosis; several reports suggested a theoretical role for NPC1 in SARS-CoV-2 infection via an undetermined mechanism (Ballout et al., 2020; Daniloski et al., 2021; Sturley et al., 2020; Vial et al., 2021).
We hypothesized that NPC1 might play an important role in SARS-CoV-2 infection. To investigate this, we have first analyzed the potential interaction between SARS-CoV-2 proteins and NPC1 using an immunoprecipitation assay. Furthermore, in this study we found that specific drugs acting through NPC1 were able to reduce SARS-CoV-2 infection.
2. Materials and Methods
2.1. Cell culture and viruses
Human embryonic kidney cells 293T/17 (HEK 293T; ATCC-CRL-11268) and Vero E6 cells (ATCC CRL-1586) were cultured in complete Dulbecco modified Eagle medium (DMEM) (supplemented with 100 IU/ml penicillin, 100 μg/ml streptomycin, 1X GlutaMAX and 10% heat-inactivated fetal bovine serum) at 37 °C and 5% CO2 atmosphere. Huh-7 Lunet C3 cells, a gift from T. Pietschman (Twincore, Germany), were cultured at 37 °C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 100 IU/ml penicillin, 100 μg/ml streptomycin, 10 mM HEPES, 1X NEAA and 10% of heat-inactivated fetal bovine serum (FBS). A549 expressing human ACE2 (A549-ACE2) were kindly provided by Dr. Juan Ortín (CNB–CSIC, Spain) and maintained under 2.5 μg/ml blasticidin selection except for infection assays, and cultured at 37 °C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 100 IU/ml penicillin, 100 μg/ml streptomycin, 1% L-glutamine and 10% FBS.
For virus infections, we used common human cold coronavirus 229E (HCoV-229E), which expresses the green fluorescent protein (GFP) gene (HCoV-229E-GFP) (Cervantes-Barragan et al., 2010). This recombinant virus was kindly given by V. Thiel, at the University of Bern, in Switzerland. The HCoV-229E-GFP infection experiments were conducted at 33 °C and 5% CO2.
SARS-CoV-2 strain NL/2020 virus stock was kindly provided by Dr. Molenkamp from Erasmus University (Rotterdam, NL) through the EVAg virus repository. The virus was propagated by inoculating Vero E6 cells at MOI 0.001 and harvesting supernatants at 48 h post-inoculation. Virus titer was determined by determining TCID50 in Vero E6 cells and by endpoint dilution and immunofluorescence microscopy for A549-ACE2 cells. Recombinant Vesicular stomatitis virus (rVSV-luc) pseudotypes were generated as previously described (Whitt, 2010) and kindly provided by Dr. Rafael Delgado (Hospital 12 de Octubre, Spain). Briefly, BHK-21 cells were transfected to express VSV-G with Lipofectamine 3000 following the manufacturer's instructions (Invitrogen, Carlsbad, USA). After 24 h, transfected cells were inoculated with a replication-deficient rVSV-Luc pseudotype that contains firefly luciferase instead of the VSV-G open reading frame, rVSV ΔG-luciferase (G* ΔG-luciferase, Kerafast) during 1 h at 37 °C. The supernatants were harvested at 24 h, 48 h post-inoculation, centrifuged at 800×g for 10 min, and stored at −80 °C. Infectious titer was determined by limiting dilution of each rVSV-luc virus-containing supernatants on Vero E6 cells.
2.2. Design and construction of plasmid that express the SARS-CoV-2 N tag to EGFP
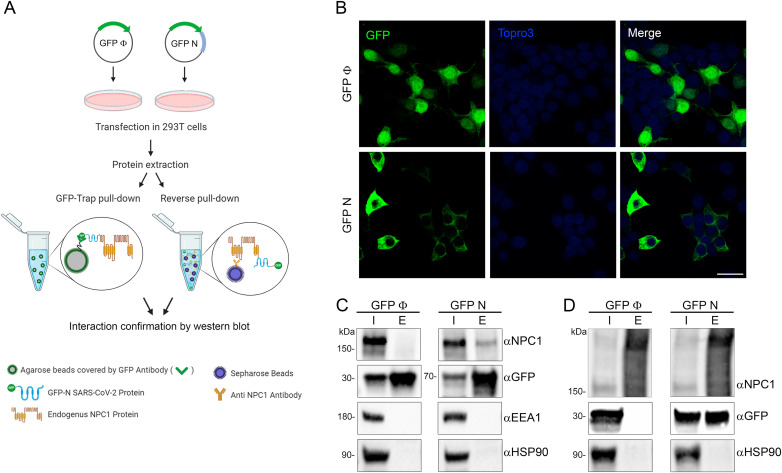
The methodology used in this part of the study (Fig. 1 A) was previously described (García-Dorival et al., 2016). To generate the SARS-CoV-2-N with N-terminal EGFP tag (EGFP-N), a codon optimized cDNA sequence for the ORF of SARS-CoV-2 N (NCBI reference sequence number: NC_045512) was cloned into the pEGFP-C1 (by GeneArt-Thermo Fisher Scientific). Once cloned, the sequence of the plasmid EGFP-N was confirmed by sequencing (Gene Art–Thermo Fisher Scientific).
Fig. 1.
Immunoprecipitation analysis of SARS-CoV-2 N protein with endogenous NPC1. (A) Schematic representation of the methodology used in this study. (B) Expression of SARS-CoV-2 N tag to EGFP (EGFP-N) and the control EGFP in HEK 293T cell confirmed by immunofluorescence. As expected, HEK 293T cells showed transient expression of EGFP (upper panel) and EGFP-N protein (lower panel) with a different distribution. EGFP, Topro3 and Merge are indicated in upper panels in colors. Scale bar 25 μm. (C) Detection of EGFP-N, EGFP and cellular proteins analyzed in the immunoprecipitation assay (where “I” refers to input sample and “E” refers to elution sample) by Western blot together with endogenous NPC1, EEA1, HSP90. EGFP-N and EGFP were both detected at the expected molecular weights (MW). (D) Endogenous NPC1, HSP90 and transfected EGFP-N and EGFP were detected at the expected MW from the co-immunoprecipitations (reverse pulldown). MW of the studied proteins: NPC1175kD, EEA1180kD, HSP9090kD, EGFP-N70kD, EGFP27kD. All immunoprecipitation assays experiments were repeated three times to ensure reproducibility (Fig. S2 and S3).
2.3. Expression of EGFP-N and EGFP in HEK 293T cells
To transfect HEK 293T cells, four 60 mm dishes were seeded with 2.5 × 106 cells 24 h prior to transfection in DMEM complete medium described above. Then, a transfection of 4 μg of plasmids EGFP or EGFP-N for each 60 mm dish was done using Lipofectamine 2000 (Thermo Fisher Scientific), following the instructions of the manufacturer. Twenty-four hours post transfection, cells were harvested, lysed and immunoprecipitated using a GFP-Trap kit (Chromotek).
2.4. Immunoprecipitations (IP)
EGFP-N and EGFP immunoprecipitations (IP) were done using a GFP-Trap®_A (Chromotek). To do the IPs, the cell pellet was resuspended in 200 μl of lysis buffer (10 mM Tris/Cl pH 7.5; 150 mM NaCl; 0.5 mM EDTA; 0.5% NP40) and then incubated for 30 min on ice. The lysate was then clarified by centrifugation at 14000×g and diluted five-fold with dilution buffer (10 mM Tris/Cl pH 7.5; 150 mM NaCl; 0.5 mM EDTA). The GFP-Trap agarose beads were equilibrated with ice-cold dilution buffer and then incubated with diluted cell lysate overnight at 4 °C on a rotator, followed by centrifugation at 2500×g for 2 min. The bead pellet was washed two times with wash buffer (10 mM Tris/Cl pH 7.5; 150 mM NaCl; 0.5 mM EDTA). After removal of wash buffer, the beads were resuspended in 100 μl of sample buffer Laemmli 2X Concentrate (Sigma Aldrich) and boiled at 95 °C for 10 min to elute the bound proteins. Buffers used for immunoprecipitations were all supplemented with Halt™ Protease Inhibitor Cocktail EDTA-Free (Thermo Fisher Scientific).
2.5. Co-immunoprecipitation (Co-IP)
Co-IP for NPC1 was performed using 50 μl of the Immobilized Recombinant Protein G Resin (Generon) and specific antibodies against NPC1 (Abcam, ab108921) as previously described (García-Dorival et al., 2016). The cell pellets were incubated for 30 min on ice with 200 μl lysis buffer. The lysate was then clarified by centrifugation and diluted five-fold with dilution buffer prior to adding 2 μg of the primary antibody and then incubated at 4 °C on a rotator for 2 h. The protein G resin (Generon) were equilibrated with ice-cold dilution buffer and then incubated at 4 °C on a rotator with diluted cell lysate containing the antibody overnight at 4 °C on a rotator, followed by centrifugation at 2500×g for 2 min to remove non-bounds fractions. The wash and elution steps were performed as described previously for GFP co-immunoprecipitation.
2.6. Western blot analysis
To confirm the expression of GFP and GFP-N proteins, an SDS-PAGE and a Western blot (WB) was done. For the SDS-PAGE, Mini-PROTEAN TGX gels were used (Bio-Rad 4561096), then, the gels were transferred to PVDF membranes using the Trans-Blot Turbo Transfect Pack (Bio-Rad 1704159) and the Trans-Blot Turbo system (Bio-Rad). Following this, the transferred membranes were blocked in 10% skimmed milk powder dissolved in TBS-0.1% Tween (TBS-T) (50 mM Tris-HCl (pH 8.3), 150 mM NaCl and 0.5% (v/v) Tween-20) buffer for 1 h at room temperature. Primary antibodies NPC1 (Abcam, ab108921), HSP90 (Enzo Life Sciences, ADI-SPA-835), EEA1 (BD Biosciences, 610457) and SARS spike protein (Novus, NB100–56578SS) were diluted in 5% skimmed milk powder dissolved in TBS-T at 1:1000 with the exception of GFP (B-2) (sc-9996) at 1:4000, and then incubated at 4 °C overnight. After three washes, blots were incubated with appropriate anti-horseradish peroxidase (HRP) secondary antibody diluted in 5% skimmed milk powder dissolved in TBS-T at 1:5000 for 1 h at room temperature. Blots then were developed using enhanced chemiluminescence reagent (Bio-Rad) and detected with ChemiDoc™ XRS Gel Imaging System using Image Lab™ software (Bio-Rad).
2.7. Production of SARS-CoV-2 N protein in the baculovirus system
The sequence of the N protein published in the NCBI database was selected (GenBank accession number: 43740575/NCBI reference sequence number: NC_045512). The codon usage of the N encoding gene was optimized for its expression in insect cells (OptimumGene™-Codon Optimization algorithm) and the coding sequence for this protein was synthesized by the company GenScript. The donor plasmid pFastBac 1 containing an expression cassette expressing the recombinant protein under the control of the polyhedrin promoter was obtained. The Bacmid for the generation of the baculovirus was prepared in E. coli DH10Bac bacterial cells containing the mini-Tn-7-replicon. Bacmids were transfected in Sf9 cells and a viral clone selection was made by two rounds of plaque cloning to obtain the working virus stock. The baculovirus genome region was sequenced to determine the integrity of the N gene in the recombinant baculovirus named rBacN.
2.8. SARS-CoV-2 N protein production in pupae
The production of SARS-CoV-2 N protein in insect pupae (Tricoplusia ni; T. ni) was performed as previously described (Escribano et al., 2020). Briefly, pupae were allocated in the inoculation robot that dispensed a maximum of 5 μl with the baculovirus titers protein in 5 days pupae incubation time in constant temperature and humidity chambers. After that period, pupae were collected and stored frozen, before downstream processing. T.ni pupae containing the recombinant protein were homogenized in extraction buffer. Then, subsequent steps of clarification, diafiltration and His-tag purification were carried, out in order to obtain purified SARS-CoV-2 N protein. Protein concentration, yield and level of purity were determined by SDS-PAGE analysis using 4–20% or 12% Mini-Protean TGX precast gels from Bio-Rad. Gels were stained with QC Colloidal stain (3 ng sensitivity) in the case of concentration and yield evaluation and with SYPRO Ruby (1 ng sensitivity) in the case of level purity analysis, both from Bio-Rad. Recombinant SARS-CoV-2 N protein produced in pupae was measured by band densitometry with the ChemiDoc™ XRS Gel Imaging System using Image Lab™ software (Bio-Rad). A BSA standard curve was used for quantification.
2.9. ELISA assays
High-binding 96-well ELISA plates (Nunc) were coated with 0.5 μg/well of purified SARS-CoV-2 N protein in carbonate/bicarbonate buffer 0.05 M pH 9.6 and allowed to bind over night at 4 °C. Then, endogenous human NPC1 and HSP90 were purified using Immobilized Recombinant Protein G Resin (Generon) and 4 μg of specific antibodies against NPC1 (Abcam, ab108921) or HSP90 (Enzo Life Sciences, ADI-SPA-835) respectively. This procedure was performed as described in Co-IP assays except the elution step, that in this case was done with glycine 200 mM pH 2.5. Serial dilutions of endogenous NPC1 and HSP90 were added to the plate and capture was allowed to proceed for 1 h at 37 °C. Subsequently, plates were washed with PBS-T (PBS 0.1% Tween 20) and the binding of NPC1 to SARS-CoV-2 N protein was detected with a rabbit anti-NPC1 antibody (1:2000), revealed with an anti-rabbit HRP (1:2000) using a colorimetric substrate (OPD) and finally, quantified by absorbance at 492 nm in the EnSight multimode plate reader of PerkinElmer.
2.10. Compounds studied
All the compounds tested in this work have a purity ≥95% by HPLC. SC compounds were synthesized at Centro de Investigaciones Biológicas Margaritas Salas (CIB–CSIC) following described procedures. All these molecules belong to the Medicinal and Biological Chemistry (MBC) library (Sebastián-Pérez et al., 2017) and some of them were previously characterized as potential inhibitors of the protein-protein interaction between NPC1 and EBOV glycoprotein (EBOV-GP) (Sebastián-Pérez et al., 2017; Lasala et al., 2021). The compounds tested in this study (Fig. 2 ) were resuspended at 50 mM in DMSO. Working concentrations of compounds were determined by cytotoxicity assays. Sulfides SC198 and SC073, and carbazole SC816, were used at working concentrations of 50, 50 and 5 μM respectively, for the HCoV-229E-GFP experiments in Huh-7 cells, and at concentrations of 25, 25 and 3.125 μM respectively for the SARS-CoV-2 assay in Vero E6 cells and A549-ACE2 cells. For the HCoV experiment in Huh-7 cells, benzothiazepine SC397, was used at a concentration of 75 μM. For the SARS-CoV-2 experiment in Vero E6 cells and A549-ACE2 cells, this compound was used at a concentration of 100 μM.
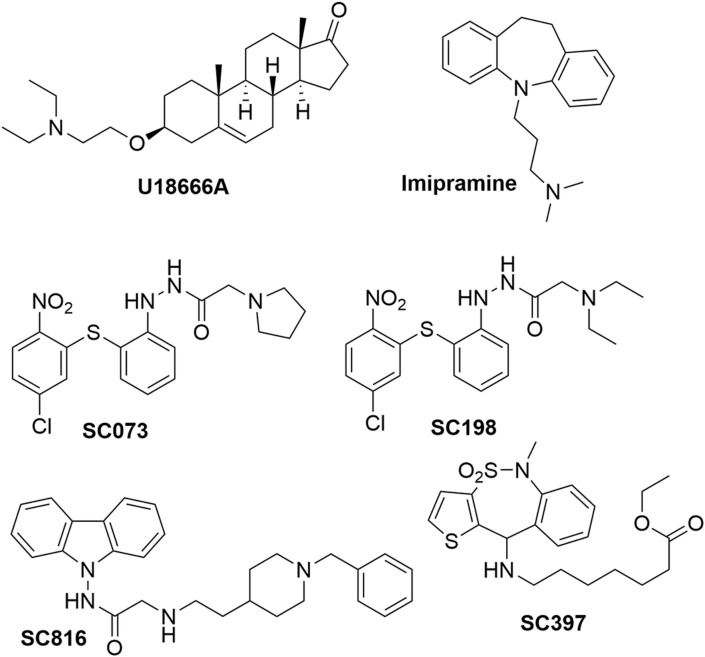
Fig. 2.
Chemical structure of small molecules used in this study. Figure of the drugs used in this study. Imipramine, a hydrophobic amine and FDA-approved antidepressant drug; the class II cationic amphiphilic compound U18666A; sulfides (SC198 and SC073), carbazole (SC816) and benzothiazepine (SC397).
Class II cationic amphiphilic compound U18666A, which is a drug that blocks cholesterol flux out of lysosomes, and also inhibits EBOV infection (Lu et al., 2015) was also tested in this study. U18666A was acquired from Sigma-Aldrich and used at 10 μM in Huh-7 cells or 12.5 μM in Vero E6 and A549-ACE2 cells. Imipramine, a hydrophobic amine and FDA-approved antidepressant drug, was also acquired from Sigma-Aldrich and used at 25 μM in Huh-7 cells or 12.5 μM in Vero E6 and A549-ACE2 cells (Herbert et al., 2015; Wichit et al., 2017).
2.11. Cytotoxicity assays
Huh-7, Vero E6 and A549-ACE2 cells were seeded in 96-well plates and incubated with DMEM containing each compound at concentrations ranging from 0 to 100 μM. After 24 h, cell viability was measured by Cell Titer 96 AQueous Non-Radioactive Cell Proliferation Assay (Promega) following the manufacturer's instructions. Absorbance was measured at 490 nm using an ELISA plate reader.
Cell viability was reported as the percentage of absorbance in treated cells relative to DMSO- treated cells. The 50% cytotoxic concentration (CC50) was calculated and non-toxic working concentrations (over 80% cell viability) were chosen to test the activities of these compounds on HCoV-229E-GFP and SARS-CoV-2 infected cells, respectively.
The estimation of the half maximal inhibitory concentration (IC50) was performed in those compounds with at least 50% efficacy and a dose-dependent inhibition of the infection. The values of the half maximal inhibitory concentration (IC50) of the infection table correspond to the mean of three independent experiments. The IC50 values and dose-response curves were estimated using GraphPad Prism v6.0 with a 95% confidence interval.
2.12. Evaluation of SARS-CoV-2 infection efficiency by immunofluorescence microscopy
Vero E6 and A549-ACE2 cells were seeded onto 96-well plates as described above and infected in the presence of the indicated compound doses, including 1 h pre-incubation period before virus inoculation. Multiplicity of infection (moi) was 0.01 for Vero E6 and 0.05 for A549-ACE2 cells. Twenty-four hours post infection, cultures were fixed for 20 min at RT with a 4% formaldehyde solution in PBS, washed twice with PBS and incubated with incubation buffer (3% BSA; 0.3% Triton X100 in PBS) for 1 h. A monoclonal antibody against N protein was diluted in incubation buffer (1:2000; Genetex HL344) and incubated with the cells for 1 h, time after which the cells were washed with PBS and subsequently incubated with a 1:500 dilution of a goat anti-rabbit conjugated to Alexa 488 (Invitrogen-Carlsbad, CA). Nuclei were stained with DAPI (Life Technologies) during the secondary antibody incubation using the manufacturer's recommendations. Cells were washed with PBS and imaged using an automated multimode reader (TECAN Spark Cyto; Austria). Overall infection efficiency was determined by determining the total fluorescence per well, using vehicle-treated cells (DMSO) and mock-infected cells as reference values.
2.13. Flow cytometry analysis
Detection of HCoV-229E-GFP infected cells was performed by flow cytometry. Huh-7 cells were pre-treated with compounds at the indicated concentrations in growth medium for 1 h at 33 °C, followed by infection with HCoV-229E-GFP at a multiplicity of infection (moi) of 1 pfu/cell for 24 h. Cells were washed twice with growth medium after 90 min of adsorption at 33 °C, and incubated with DMEM 10% for 24 h. Cells were then harvested with PBS-EDTA 5 mM, and diluted in PBS. Detection of HCoV-229E-GFP infected cells was performed by analyzing GFP expression. In order to determine the percentage of infected cells per condition, 15,000 cells/time point were scored using FACS Canto II flow cytometer (BD Sciences) and analyzed using the FlowJo software. Untreated control infected cultures yielded 75–90% of infected cells from the total cells examined. Infected cell percentages obtained after drug treatments were normalized to DMSO values.
2.14. Recombinant pseudotype rVSV-G infection assay
We used a replication-deficient recombinant Vesicular stomatitis virus luciferase (rVSV-G-luc) pseudotype that contains firefly luciferase instead of the VSV-G as described above. Vero E6 cells (1 × 104 cells/well) were seeded onto 96-well plates the day before. Compounds were diluted in 2% complete medium to achieve the final concentration. Compound dilutions were applied to the cell cultures 1 h before virus inoculation. Pretreatment was removed and fresh compound dilutions containing the rVSV-G-luc pseudotype were used to inoculate the cultures for 24 h. Twenty-four hours post-inoculation, cells were lysed for luciferase activity determination using Steady-Glo Luciferase Assay System (Promega, Madison, USA) and luminescence was quantified in the EnSight multimode plate reader of PerkinElmer (Waltham, USA). Relative infection values were determined by normalizing the data to the average relative light units detected in DMSO- treated cells.
2.15. Statistical analysis
The experimental data was analyzed by one-way ANOVA by Graph Pad Prism 6 software. For multiple comparisons, Bonferroni's correction was applied. Values were expressed in graph bars as mean ± SD of at least three independent experiments unless otherwise noted. A p value ˂0.05 was considered as statistically significant.
3. Results
3.1. Interaction of SARS-CoV-2 N protein with NPC1
Given a first attempt to detect interaction between SARS-CoV-2 spike glycoprotein (S), the outer receptor interacting protein, and NPC1 failed (Fig. S1), we further investigated the interaction of SARS-CoV-2 Nucleoprotein (N) with NPC1. SARS-CoV-2 N protein was expressed as an EGFP-fusion protein (EGFP-N) in HEK 293T cells in order to perform immunoprecipitations (IP) to study their potential as interacting partners (Fig. 1A). Similar approaches have been used to detect the protein interaction partners for other viral proteins such as the EBOV nucleoprotein (NP) or viral protein VP24 (García-Dorival et al., 2014, 2016). HEK 293T cells were selected for this study due to their high efficiency of transfection, being the line of choice for protein-protein interaction studies of several viruses including SARS-CoV-2 (Gordon et al., 2020).
Protein expression of EGFP-N and EGFP, which was used as a control, was confirmed by fluorescence and Western blot analysis (Fig. 1B and C). Then, proteins were extracted from lysed cells (input sample) and assayed for IP using a high affinity EGFP immunoprecipitation kit (GFP-Trap). After IP, both input and bound (or elution) samples were analyzed by Western blot; proteins corresponding to the molecular weight of EGFP-N (70 kDa) and the EGFP (27 kDa) were detected using an anti-EGFP antibody (Fig. 1C). NPC1, as an endogenous protein, was also detected in both input samples (EGFP-N and EGFP); but only in the bound fraction of EGFP-N sample (Fig. 1C).
3.2. Validation of SARS-CoV-2 N interaction with NPC1
To validate the specific interaction between EGFP-N and NPC1, two cellular proteins were selected as controls. In this case, HSP90 chaperone and endosomal protein EEA1 were used as load and negative controls respectively (Fig. 1C). To further validate the interaction between SARS-CoV-2 N and NPC1, co-immunoprecipitations (Co-IP or reverse pull downs) against NPC1 were performed using protein G-beads and specific monoclonal antibodies against NPC1 (Fig. 1D). Bound samples obtained from the Co-IP were then analyzed by Western blot, which confirmed the presence of SARS-CoV-2 N (Fig. 1D).
To further investigate the interaction between NPC1 and SARS-CoV-2 N protein, an ELISA assay was designed using an antibody against NPC1. A dose-dependent positive reaction was observed when we increased concentrations of NPC1 in plates coated with SARS-CoV-2 N protein, while control protein HSP90 yielded no signal (Fig. S4).
3.3. Functional assays
We further tested this interaction by using different small molecules, some of these compounds were able to target NPC1 in different regions. An example of these is imipramine, a Food and Drug Administration (FDA)-approved drug, which inhibits EBOV infection by targeting NPC1 (Rodriguez-Lafrasse et al., 1990). Also, as a reference drug, we used the U18666A compound (Fig. 2), a well-known compound that inhibits the cholesterol transport function of NPC1. This compound also has an effect on the entry of several infectious viruses including EBOV and African swine fever virus (ASFV) (Lu et al., 2015; Tang et al., 2009; Wichit et al., 2017; Cuesta-Geijo et al., 2016).
In addition to this, we studied whether other small molecules known to disrupt the interaction between NPC1 and other viral proteins (e.g. EBOV-GP), could affect the interaction between NPC1 and SARS-CoV-2. In order to do this, we assayed a set of antiviral compounds that were initially selected using a computational technique called virtual screening from the MBC chemical library and focused on the EBOV-GP/NPC1 interaction (Lasala et al., 2021). Noteworthy, sulfides SC073 and SC198 as well as carbazole SC816 (Fig. 2) are known to disturb the NPC1 domain C-GP interaction in an ELISA assay (Lasala et al., 2021), while benzothiazepines SC397 do not affect this interaction in the same assay. Based on these results, the three classes of small molecules were included in our study for comparative purposes.
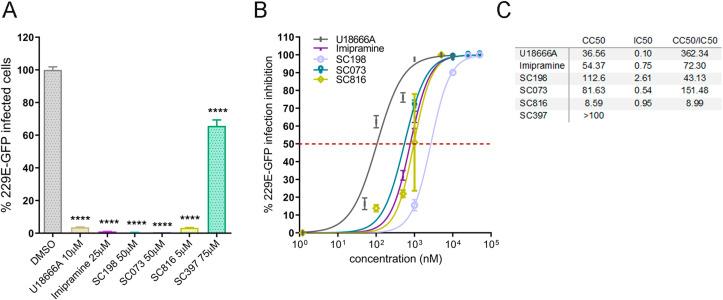
To functionally characterise this interaction, these selected small molecules were assayed as inhibitors of human coronavirus (HCoV) infections, specifically against HCoV-229E-GFP and SARS-CoV-2 infection. First, Huh-7 cells were pre-treated with indicated doses of compounds or DMSO and infected with 1 pfu/cell for 24 h at 33 °C as described in Materials and Methods. Then, cells were harvested and HCoV-229E infection scored by GFP expression in flow cytometry. Treatment with U18666A, imipramine, sulfides (SC073 and SC198) and carbazole SC816 yielded significant inhibition >96% of HCoV-229E-GFP infection at micromolar concentrations, while benzothiazepine SC397 showed weak inhibition at 75 μM (Fig. 3 A) and no remarkable effect at lower concentrations (Fig. S6A and S6B).
Fig. 3.
Activity of selected small-molecules against HCoV-229E-GFP. (A) Infectivity of HCoV-229E-GFP (229E-GFP) in Huh-7 cells at 24 hpi measured by GFP fluorescence percentages relative to the controls in cells pretreated 1 h before infection with the small molecules at the indicated concentrations or DMSO (****p < 0.0001). (B) Normalized dose-response sigmoidal curve displaying dose values at nM scale on X axis and % of infection inhibition on Y axis using GraphPad 6. (C) IC50 values (μM) were determined for those compounds with higher inhibitory effect. The curves for the determination of CC50 in Huh-7 cells are shown in Fig. S5A.
The estimation of the half maximal inhibitory concentration (IC50) was performed in those compounds with at least 50% efficacy. The dose-dependent inhibition curves of the infection (Fig. 3B) were analyzed in order to calculate the values of the IC50 (Fig. 3C). A good selectivity index (CC50/IC50) was obtained for all the inhibitory compounds. Carbazole SC816 and sulfides (SC198 and SC073) exhibited selectivity index values of 8.99, 43.13 and 151.48 respectively; being the most effective sulfide SC073. Dose-response effects for these chemicals are included in Fig. S6B.
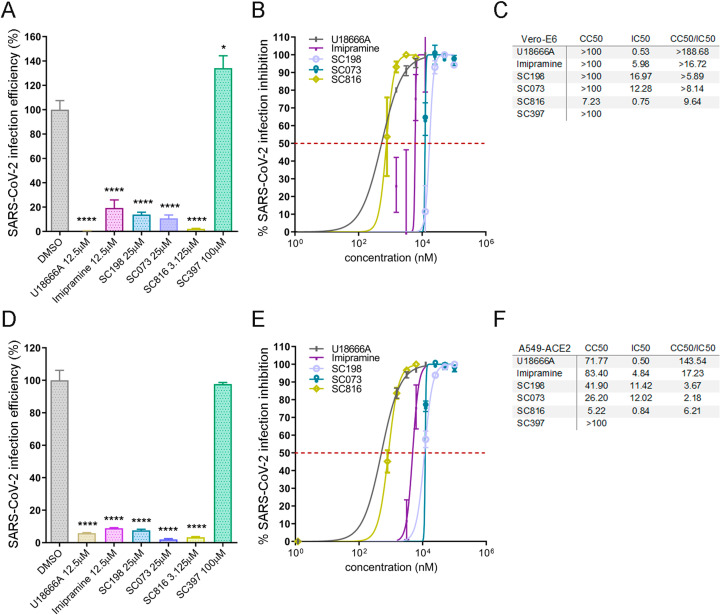
Interestingly, the antiviral activity against HCoV-229E-GFP of compounds U18666A, imipramine, SC198, SC073 and SC816 in Huh-7 cells, was confirmed using SARS-CoV-2 infection in Vero E6 (Fig. 4A and S7A) and A549-ACE2 cells (Fig. 4D and S7B).
Sulfides SC198 (25 μM) and SC073 (25 μM) decreased infectivity to 13% and 10% respectively, while carbazole SC816 (3.125 μM) to 2%. Benzothiazepine SC397 (100 μM) did not show any effect in SARS-CoV-2 infection efficiency in Vero E6 cells (Fig. S7A).
Dose inhibition curves and IC50 values were estimated for those compounds that decrease significantly the infection efficiency in Vero E6 cells (Fig. 4 B and C) finding potent antiviral compounds. To obtain CC50 and the selectivity index the values of the cytotoxicity assay were used. Cytotoxicity assay results are shown in Fig. S5B. In this case, the selectivity index values of these compounds ranged from 6 to 10 (Fig. 4C).
Fig. 4.
Activity of selected small molecules inhibitors of NPC1 and related compounds against SARS-CoV-2. Infectivity of SARS-CoV-2 in (A) Vero E6 and (D) A549-ACE2 cells at 24 hpi measured by immunofluorescence microscopy. The percentage of infection efficiency was determined by determining the total fluorescence per well, using vehicle-treated cells (DMSO) and mock-infected cells as reference values. Normalized dose-response sigmoidal curve displaying dose values at nM scale on X axis and % of infection inhibition on Y axis using GraphPad 6 in (B) Vero E6 and (E) A549-ACE2. IC50 values (μM) were determined for these compounds for (C) Vero E6 and (F) A549-ACE2. The curves for the determination of CC50 in Vero E6 and A549-ACE2 cells are shown in Fig. S5B and S5C respectively.
In order to verify the antiviral activity of these compounds in human cell lines, infection experiments were performed in human lung carcinoma A549 cells constitutively overexpressing ACE2 (A549-ACE2). These results revealed a robust antiviral activity for SC816, SC198 and SC073 in this infection model, as well as for U18666A and Imipramine as expected. Interestingly, benzothiazepine SC397 showed no antiviral activity in both HCoV independently of the cell type used. As SC397 do not affect NPC1 interaction, these results are in agreement with our hypothesis.
While effective compounds inhibited between 80 and 99% SARS-CoV-2 infection, in Vero-E6 we detected much less inhibition using the RNA virus VSV-G, that preferentially enters the cell at the level of the early endosome (Fig. S8).
4. Discussion
This study found a novel interaction between the SARS-CoV-2 N protein and NPC1. The SARS-CoV-2 N protein is a structural protein that binds to the viral RNA conferring stability by forming a helical ribonucleoprotein complex named nucleocapsid (Mariano et al., 2020; Ye et al., 2020). Even though the main function of the SARS-CoV-2 N protein is to bind to the RNA, other functions during the SARS-CoV-2 replication have been described for the N protein including acting as an antagonist of interferon signalling (Mu et al., 2020). Therefore, further studies need to be done in order to fully understand the role of the N protein in SARS-CoV-2.
NPC1, is a late-endosomal membrane protein required for transport of cholesterol into cells (Li et al., 2016, Scott and Ioannou, 2004). Mutations in the NPC1 gene result in the Niemann-Pick disease, which is a rare disease that cause an accumulation of cholesterol in the endosomal/lysosomal system (Carstea et al., 1997, Scott and Ioannou, 2004).
It has been reported that some viruses might use/need NPC1 at some stages of their infectious cycle. Therefore, the interaction of drugs with NPC1 protein can result in a severe reduction of viral infection.
EBOV is an example of an important pathogenic virus that uses NPC1, in this particular case, as a host receptor (Carette et al., 2011; Côté et al., 2011). EBOV entry is mediated by the viral glycoprotein (GP) which is organized in trimeric spikes at the viral surface (White et al., 2008). NPC1 binding requires the processing of viral GP. GP cleavage by endosomal cathepsins unmasks the binding site for NPC1 by removing heavily glycosylated C-terminal residues and the glycan cap to produce the cleaved form of the N-terminal receptor binding subunit GP1 (GPCL). Finally, GPCL-NPC1 binding within endosomes is required to mediate fusion and viral escape into the host cytoplasm (Carette et al., 2011; Côté et al., 2011). Thus, this protein is acting as a true intracellular receptor (Miller et al., 2012). Therefore, NPC1 could be used as an important druggable target for EBOV (Côté et al., 2011).
Another example is HCV, in this virus there is evidence that HCV RNA replication depends on NPC1 and its function as lipid transporter along the endosome-lysosomal pathway (Stoeck et al., 2018). Moreover, inhibition of NPC1 by CRISPR/Cas9 system was reported to reduced HCV viral replication (Stoeck et al., 2018).
It has been found as well, that compounds able to interfere with NPC1 can affect viral replication for several viruses. Compound U18666A, extensively used as a pharmacological tool to study the role of NPC1, blocks intracellular cholesterol efflux mediated by NPC1. It has been demonstrated that U18666A can affect DENV entry and trafficking (Poh et al., 2012). Besides, this compound its known to severely impair the infections caused by EBOV (Côté et al., 2011; Herbert et al., 2015; Lu et al., 2015), HIV (Tang et al., 2009), DENV (Jupatanakul et al., 2014; Poh et al., 2012), CHIKV (Wichit et al., 2017), and ZIKV (Sabino et al., 2019). In the present study, compound U18666A (10 or 12.5 μM) also induced a considerable reduction of HCoV infections including 229E-GFP (97% of inhibition, CC50 = 36.56) and 99% of SARS-CoV-2 inhibition in Vero E6 (CC50 > 100) and 94% of inhibition in A549-ACE2 cells (CC50 = 71.77).
In this work, we have identified NPC1 as cellular protein interactors of SARS-CoV-2-N protein, thus being a potential drug target for SARS-CoV-2 infection. Therefore, the inhibition of this interaction could impair SARS-CoV-2 infection, with the subsequent decrease of virus spread. Furthermore, we have also tested in this study some reported antiviral compounds that interact with NPC1. These compounds are known to have an effect on EBOV replication; potentially acting by disruption of the interaction between EBOV-GP and NPC1, such as sulfides SC073 and SC198 and carbazole SC816.
In this study, it was found that imipramine, sulfides (SC073, SC198) and carbazole SC816 inhibited over 95% the infection of SARS-CoV-2 in Vero-E6 and A549-ACE2 with an inhibitory concentration 50 (IC50) at the low micromolar range while benzothiazepine SC397 showed no significant inhibition (Fig. 4). Additionally, we have also included in our study benzothiazepine SC397 as a negative control because it showed no disruption of the above mentioned interaction (Galindo et al., 2021; Lasala et al., 2021). In fact, SC397, reported as unable to interact with NPC1, showed no inhibition of HCoV. This highlights the specificity of the interaction and the inhibitor drugs thereby. In addition to this, another assay testing the compounds was performed using VSV, which needs an early/less acidic endosomes as reported (Lay Mendoza et al., 2020; White and Whittaker, 2016). As expected, no severe impact was observed in the infection compared to HCoV-229E-GFP and SARS-CoV-2 (Fig. S8). In fact, the localisation of NPC1 in the late endosomes may explain why these compounds might be less relevant for VSV-G-driven entry, which occurs preferentially at early endosomes. VSV-G is also able to penetrate into the cytoplasm through the intraluminal vesicles (ILV) of endosomal transport intermediates, called multivesicular bodies (MVBs)(Le Blanc et al., 2005). This could explain why these compounds still have some impact in VSV-G infection.
It is not clear yet the specific role of NPC1 during SARS-CoV-2 infection, and therefore further studies need to be done. However, there are some preliminary studies that show that NPC1 can have a role during SARS-CoV-2 infection (Daniloski et al., 2021; Vial et al., 2021). An example of this is the report of Daniloski et al. (2021), were they used a genome -scale CRISPR loss of function for several proteins and they found that NPC1 was a host factor required for SARS-CoV-2 infection.
One possibility is that similarly to EBOV, SARS-CoV-2 could traffic the endocytic pathway inside the early and late endosomes (LE) to finally fuse with lysosomes, an essential stage for viral uncoating and fusion (Ballout et al., 2020). Therefore, viral infection can be abrogated by drugs interfering endosome acidification, as previously evidenced (Ou et al., 2020). Also, SARS-CoV-2 pseudovirions infection is inhibited by drugs targeting late endosome proteins, such as cathepsin L, two-pore channel 2 (TPC2), or PIKfyve. Inhibitors targeting these proteins dramatically reduce infection, indicating that those are crucial host factors for endocytosed SARS-CoV-2 entry (Ou et al., 2020; Galindo et al., 2021). LE/lysosomes are relevant organelles to develop therapeutic targets against infection (Daniloski et al., 2021; Sturley et al., 2020; Tang et al., 2020; Zhou et al., 2020).
In addition to this, for other coronavirus, such as Infectious bronchitis virus (IBV), N protein is involved in immune regulation but also plays key roles in viral replication such as binding to viral RNA (Fan et al., 2005), and unwinds double-stranded nucleic acid acting as an RNA chaperone (Zúñiga S, 2007). It is involved in the RNA packaging into new virions and assembly, thus, can be detected at replication sites. These replication sites are originated from endoplasmic reticulum (ER) membranes, budding from the ERGIC (ER-Golgi intermediate compartment) (Cong et al., 2020; Stertz et al., 2007) and are called double membrane vesicles (DMVs), partially opened by a molecular pore for RNA export (Wolff et al., 2020).
Importantly, the generation of these DMVs require massive reorganization of cholesterol enriched intracellular membranes, similar to HCV. HCV replication in ER-derived DMVs, depends on intact lipid transport from the endolysosomal compartment. Since ER is poor in cholesterol content (Holthuis and Menon, 2014), HCV usurps the cholesterol transporter NPC1, located at the late endosomal compartment, to collect cholesterol to the replication sites, ensuring its stability (Stoeck et al., 2018). A similar mechanism could be shared by SARS-CoV-2 in order to maintain the integrity of the replication vesicles via the N-NPC1 interaction as a cholesterol supplier to the viral replication complex.
A recent study discovered that SARS-CoV-2 non-structural protein 7 interacts with the late endosomes (LE)-associated GTPase Rab7a, and its depletion causes retention of ACE2 receptor inside endosomes (Gordon et al., 2020). All these studies coincided to highlight the importance of the endosomal pathway in SARS-CoV-2 infection. Furthermore, there are other reports supporting the relevance of several proteins involved in cholesterol regulation, including NPC1 in SARS-CoV-2 infection (Daniloski et al., 2021). These studies support our findings. Also, the cholesterol biosynthesis pathway is downregulated at infection and, drug treatments that regulate this pathway impact the infection (Hoagland et al., 2020). Finding a similar potential cellular target shared by different viruses can be very useful as one can use the same/similar compound to treat several diseases.
Due to recent evidence, and according to other authors suggesting the important role of NPC1 in SARS-CoV-2 infection (Ballout et al., 2020; Daniloski et al., 2021; Hoagland et al., 2020; Sturley et al., 2020; Vial et al., 2021), here we propose NPC1 as a potential therapeutic target for SARS-CoV-2 to combat COVID-19 pandemic. In this study, we have shown for first-time experimental evidence of the interaction of SARS-CoV-2-N and NPC1. These results were corroborated by using different chemical tools able to interact with NPC1 opening new avenues for antiviral development. In view of this important finding, future medical and therapeutic efforts will be directed to this target and to the regulation of cellular cholesterol in SARS-CoV-2 infection.
5. Conclusion
Here we reported for the first time the interaction between SARS-CoV-2 N protein and NPC1. This interaction unveiled a novel host-based target for antivirals and a potential host factor for SARS-CoV-2 infectivity. As in other viruses, this interaction could possibly regulate the entry from LE and/or modify cholesterol efflux from LE and alter the lipid composition of replicative membranes for its own benefit (Chernomordik and Kozlov, 2008).
These results are of utmost importance because finding a similar potential cellular target shared by different viruses can be very useful for the development of broad-spectrum antivirals.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are thankful to Dr. V. Thiel from the University of Bern, Switzerland for HCoV-229E-GFP, Dr. T. Pietschman, Twincore, Germany for Huh-7 Lunet C3 cells, Dr. R. Delgado (Hospital 12 de Octubre, Spain for VSV-G-luc, Prof. Juan Ortín (CNB–CSIC, Spain) for A549 cells and Dr. R. Molenkamp (Erasmus University Medical Center, Rotterdam, Netherlands; participant of the EVA-GLOBAL project) for the SARS-CoV-2 strain NL/2020 virus. EVA (European Virus Archive; grant agreement No 871029). BioRender.com was used to created icons in Figures. This research was partially supported through “La Caixa” Banking Foundation (LCF/PR/HR19/52160012), Instituto de Salud Carlos III (ISCIII–COV20/01007), the Spanish Ministry of Science and Innovation (RTI2018-097305-R-I00), CSIC (PIE-RD-COVID-19 refs. E201980E024 and E202020E079) and the European Union-NextGenerationEU.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.antiviral.2021.105167.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Ballout R.A., Sviridov D., Bukrinsky M.I., Remaley A.T. The lysosome: a potential juncture between SARS‐CoV‐2 infectivity and Niemann‐Pick disease type C, with therapeutic implications. Faseb. J. 2020;34(6):7253–7264. doi: 10.1096/fj.202000654R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J.E., Raaben M., Wong A.C., Herbert A.S., Obernosterer G., Mulherkar N., Kuehne A.I., Kranzusch P.J., Griffin A.M., Ruthel G., Dal Cin P. Ebola virus entry requires the cholesterol transporter Niemann–Pick C1. Nature. 2011;477(7364):340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstea E.D., Morris J.A., Coleman K.G., Loftus S.K., Zhang D., Cummings C., Gu J., Rosenfeld M.A., Pavan W.J., Krizman D.B., Nagle J. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 1997;277(5323):228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- Cervantes-Barragan L., Züst R., Maier R., Sierro S., Janda J., Levy F., Speiser D., Romero P., Rohrlich P.S., Ludewig B., Thiel V. Dendritic cell-specific antigen delivery by coronavirus vaccine vectors induces long-lasting protective antiviral and antitumor immunity. MBio. 2010;1(4):e00171. doi: 10.1128/mBio.00171-10. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik L.V., Kozlov M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008;15(7):675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Ulasli M., Schepers H., Mauthe M., V’kovski P., Kriegenburg F., Thiel V., de Haan C.A., Reggiori F. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J. Virol. 2020;94(4):e01925. doi: 10.1128/JVI.01925-19. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté M., Misasi J., Ren T., Bruchez A., Lee K., Filone C.M., Hensley L., Li Q., Ory D., Chandran K., Cunningham J. Small molecule inhibitors reveal Niemann–Pick C1 is essential for Ebola virus infection. Nature. 2011;477(7364):344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta-Geijo M.Á., Chiappi M., Galindo I., Barrado-Gil L., Muñoz-Moreno R., Carrascosa J.L., Alonso C. Cholesterol flux is required for endosomal progression of African swine fever virions during the initial establishment of infection. J. Virol. 2016;90(3):1534–1543. doi: 10.1128/JVI.02694-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z., Jordan T.X., Wessels H.H., Hoagland D.A., Kasela S., Legut M., Maniatis S., Mimitou E.P., Lu L., Geller E., Danziger O. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell. 2021;184(1):92–105. doi: 10.1016/j.cell.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano J.M., Cid M., Reytor E., Alvarado C., Nuñez M.C., Martínez-Pulgarín S., Dalton R.M. Chrysalises as natural production units for recombinant subunit vaccines. J. Biotechnol. 2020;X(6):100019. doi: 10.1016/j.btecx.2020.100019. [DOI] [PubMed] [Google Scholar]
- Fan H., Ooi A., Tan Y.W., Wang S., Fang S., Liu D.X., Lescar J. The nucleocapsid protein of coronavirus infectious bronchitis virus: crystal structure of its N-terminal domain and multimerization properties. Structure. 2005;13(12):1859–1868. doi: 10.1016/j.str.2005.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo I., Garaigorta U., Lasala F., Cuesta-Geijo M.A., Bueno P., Gil C., Delgado R., Gastaminza P., Alonso C. Antiviral drugs targeting endosomal membrane proteins inhibit distant animal and human pathogenic viruses. Antivir. Res. 2021;186:104990. doi: 10.1016/j.antiviral.2020.104990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Dorival I., Wu W., Armstrong S.D., Barr J.N., Carroll M.W., Hewson R., Hiscox J.A. Elucidation of the cellular interactome of Ebola virus nucleoprotein and identification of therapeutic targets. J. Proteome Res. 2016;15(12):4290–4303. doi: 10.1021/acs.jproteome.6b00337. [DOI] [PubMed] [Google Scholar]
- García-Dorival I., Wu W., Dowall S., Armstrong S., Touzelet O., Wastling J., Barr J.N., Matthews D., Carroll M., Hewson R., Hiscox J.A. Elucidation of the Ebola virus VP24 cellular interactome and disruption of virus biology through targeted inhibition of host-cell protein function. J. Proteome Res. 2014;13(11):5120–5135. doi: 10.1021/pr500556d. [DOI] [PubMed] [Google Scholar]
- Gordon D.E., Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S., Braberg H., Jureka A.S., Obernier K., Guo J.Z., Batra J., Kaake R.M. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370(6521) doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A.S., Davidson C., Kuehne A.I., Bakken R., Braigen S.Z., Gunn K.E., Whelan S.P., Brummelkamp T.R., Twenhafel N.A., Chandran K., Walkley S.U. Niemann-pick C1 is essential for ebolavirus replication and pathogenesis in vivo. MBio. 2015;6(3):e00565–15. doi: 10.1128/mBio.00565-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland D.A., Clarke D.J., Moeller R., Han Y., Yang L., Wojciechowicz M.L., Lachmann A., Oguntuyo K.Y., Stevens C., Lee B., Chen S. Modulating the transcriptional landscape of SARS-CoV-2 as an effective method for developing antiviral compounds. Biorxiv. 2020 [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis J.C., Menon A.K. Lipid landscapes and pipelines in membrane homeostasis. Nature. 2014;510(7503):48–57. doi: 10.1038/nature13474. [DOI] [PubMed] [Google Scholar]
- Indari O., Jakhmola S., Manivannan E., Jha H.C. An update on antiviral therapy against SARS-CoV-2: how far have we come? Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.632677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Tanaka N., Tanaka Y., Inoue S., Morita K., Zhuang M., Hattori T., Sugamura K. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 2007;81(16):8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupatanakul N., Sim S., Dimopoulos G. Aedes aegypti ML and Niemann-Pick type C family members are agonists of dengue virus infection. Dev. Comp. Immunol. 2014;43(1):1–9. doi: 10.1016/j.dci.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lasala F., García-Rubia A., Requena C., Galindo I., Cuesta-Geijo M.A., García-Dorival I., Bueno P., Labiod N., Luczkowiak J., Martinez A., Campillo N.E. Identification of potential inhibitors of protein-protein interaction useful to fight against Ebola and other highly pathogenic viruses. Antivir. Res. 2021;186:105011. doi: 10.1016/j.antiviral.2021.105011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay Mendoza M.F., Acciani M.D., Levit C.N., Santa Maria C., Brindley M.A. Monitoring viral entry in real-time using a luciferase recombinant vesicular stomatitis virus producing SARS-CoV-2, EBOV, LASV, CHIKV, and VSV glycoproteins. Viruses. 2020;12(12):1457. doi: 10.3390/v12121457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Blanc I., Luyet P.P., Pons V., Ferguson C., Emans N., Petiot A., Mayran N., Demaurex N., Fauré J., Sadoul R., Parton R.G. Endosome-to-cytosol transport of viral nucleocapsids. Nat. Cell Biol. 2005;7(7):653–664. doi: 10.1038/ncb1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang J., Coutavas E., Shi H., Hao Q., Blobel G. Structure of human niemann–pick C1 protein. Proc. Natl. Acad. Sci. Unit. States Am. 2016;113(29):8212–8217. doi: 10.1073/pnas.1607795113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F., Liang Q., Abi-Mosleh L., Das A., De Brabander J.K., Goldstein J.L., Brown M.S. Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. Elife. 2015;4 doi: 10.7554/eLife.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariano G., Farthing R.J., Lale-Farjat S.L., Bergeron J.R. Structural characterization of SARS-CoV-2: where we are, and where we need to be. Frontiers in molecular biosciences. 2020;7:344. doi: 10.3389/fmolb.2020.605236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E.H., Obernosterer G., Raaben M., Herbert A.S., Deffieu M.S., Krishnan A., Ndungo E., Sandesara R.G., Carette J.E., Kuehne A.I., Ruthel G. Ebola virus entry requires the host‐programmed recognition of an intracellular receptor. EMBO J. 2012;31(8):1947–1960. doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingorance L., Friesland M., Coto-Llerena M., Pérez-del-Pulgar S., Boix L., López-Oliva J.M., Bruix J., Forns X., Gastaminza P. Selective inhibition of hepatitis C virus infection by hydroxyzine and benztropine. Antimicrob. Agents Chemother. 2014;58(6):3451–3460. doi: 10.1128/AAC.02619-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J., Fang Y., Yang Q., Shu T., Wang A., Huang M., Jin L., Deng F., Qiu Y., Zhou X. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell discovery. 2020;6(1):1–4. doi: 10.1038/s41421-020-00208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna-Ramos J.F., Reyes-Ruiz J.M., Del Ángel R.M. The role of host cholesterol during flavivirus infection. Frontiers in cellular and infection microbiology. 2018;8:388. doi: 10.3389/fcimb.2018.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1):1–12. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh M.K., Shui G., Xie X., Shi P.Y., Wenk M.R., Gu F. U18666A, an intra-cellular cholesterol transport inhibitor, inhibits dengue virus entry and replication. Antivir. Res. 2012;93(1):191–198. doi: 10.1016/j.antiviral.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lafrasse C., Rousson R., Bonnet J., Pentchev P.G., Louisot P., Vanier M.T. Abnormal cholesterol metabolism in imipramine-treated fibroblast cultures. Similarities with Niemann-Pick type C disease. Biochim. Biophys. Acta Lipids Lipid. Metabol. 1990;1043(2):123–128. doi: 10.1016/0005-2760(90)90284-5. [DOI] [PubMed] [Google Scholar]
- Sabino C., Basic M., Bender D., Elgner F., Himmelsbach K., Hildt E. Bafilomycin A1 and U18666A efficiently impair ZIKV infection. Viruses. 2019;11(6):524. doi: 10.3390/v11060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz B., Barretto N., Martin D.N., Hiraga N., Imamura M., Hussain S., Marsh K.A., Yu X., Chayama K., Alrefai W.A., Uprichard S.L. Identification of the Niemann-Pick C1–like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat. Med. 2012;18(2):281–285. doi: 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C., Ioannou Y.A. The NPC1 protein: structure implies function. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2004;1685(1–3):8–13. doi: 10.1016/j.bbalip.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Sebastián-Pérez V., Roca C., Awale M., Reymond J.L., Martinez A., Gil C., Campillo N.E. Medicinal and biological chemistry (MBC) library: an efficient source of new hits. J. Chem. Inf. Model. 2017;57(9):2143–2151. doi: 10.1021/acs.jcim.7b00401. [DOI] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz S., Reichelt M., Spiegel M., Kuri T., Martínez-Sobrido L., García-Sastre A., Weber F., Kochs G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361(2):304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck I.K., Lee J.Y., Tabata K., Romero-Brey I., Paul D., Schult P., Lohmann V., Kaderali L., Bartenschlager R. Hepatitis C virus replication depends on endosomal cholesterol homeostasis. J. Virol. 2018;92(1):e01196. doi: 10.1128/JVI.01196-17. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturley S.L., Rajakumar T., Hammond N., Higaki K., Márka Z., Márka S., Munkacsi A.B. Potential COVID-19 therapeutics from a rare disease: weaponizing lipid dysregulation to combat viral infectivity. J. Lipid Res. 2020;61(7):972–982. doi: 10.1194/jlr.R120000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Leao I.C., Coleman E.M., Broughton R.S., Hildreth J.E. Deficiency of niemann-pick type C-1 protein impairs release of human immunodeficiency virus type 1 and results in Gag accumulation in late endosomal/lysosomal compartments. J. Virol. 2009;83(16):7982–7995. doi: 10.1128/JVI.00259-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharappel A.M., Samrat S.K., Li Z., Li H. Targeting crucial host factors of SARS-CoV-2. ACS Infect. Dis. 2020;6(11):2844–2865. doi: 10.1021/acsinfecdis.0c00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vial C., Calderón J.F., Klein A.D. NPC1 as a modulator of disease severity and viral entry of SARSCoV-2. Curr. Mol. Med. 2021;21(1):2–4. doi: 10.2174/1566524020666200713175426. [DOI] [PubMed] [Google Scholar]
- Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., Jiang C. SARS coronavirus entry into host cells through a novel clathrin-and caveolae-independent endocytic pathway. Cell Res. 2008;18(2):290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43(3):189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.M., Whittaker G.R. Fusion of enveloped viruses in endosomes. Traffic. 2016;17(6):593–614. doi: 10.1111/tra.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitt M.A. Generation of VSV pseudotypes using recombinant ΔG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J. Virol Methods. 2010;169(2):365–374. doi: 10.1016/j.jviromet.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2021. WHO Coronavirus Report.https://covid19.who.int December 2020. [Google Scholar]
- Wichit S., Hamel R., Bernard E., Talignani L., Diop F., Ferraris P., Liegeois F., Ekchariyawat P., Luplertlop N., Surasombatpattana P., Thomas F. Imipramine inhibits chikungunya virus replication in human skin fibroblasts through interference with intracellular cholesterol trafficking. Sci. Rep. 2017;7(1):1–12. doi: 10.1038/s41598-017-03316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G., Limpens R.W., Zevenhoven-Dobbe J.C., Laugks U., Zheng S., de Jong A.W., Koning R.I., Agard D.A., Grünewald K., Koster A.J., Snijder E.J. A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020;369(6509):1395–1398. doi: 10.1126/science.abd3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., West A.M., Silletti S., Corbett K.D. Architecture and self‐assembly of the SARS‐CoV‐2 nucleocapsid protein. Protein Sci. 2020;29(9):1890–1901. doi: 10.1002/pro.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Tsybovsky Y., Gorman J., Rapp M., Cerutti G., Chuang G.Y., Katsamba P.S., Sampson J.M., Schön A., Bimela J., Boyington J.C. Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe. 2020;28(6):867–879. doi: 10.1016/j.chom.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga S., Sola I., Moreno J.L., Sabella P., Plana-Durán J., Enjuanes L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology. 2007;357(2):215–227. doi: 10.1016/j.virol.2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.