Abstract
Aims/hypothesis
Type 2 diabetes increases the risk of cardiovascular and renal complications, but early risk prediction could lead to timely intervention and better outcomes. Genetic information can be used to enable early detection of risk.
Methods
We developed a multi-polygenic risk score (multiPRS) that combines ten weighted PRSs (10 wPRS) composed of 598 SNPs associated with main risk factors and outcomes of type 2 diabetes, derived from summary statistics data of genome-wide association studies. The 10 wPRS, first principal component of ethnicity, sex, age at onset and diabetes duration were included into one logistic regression model to predict micro- and macrovascular outcomes in 4098 participants in the ADVANCE study and 17,604 individuals with type 2 diabetes in the UK Biobank study.
Results
The model showed a similar predictive performance for cardiovascular and renal complications in different cohorts. It identified the top 30% of ADVANCE participants with a mean of 3.1-fold increased risk of major micro- and macrovascular events (p = 6.3 × 10−21 and p = 9.6 × 10−31, respectively) and a 4.4-fold (p = 6.8 × 10−33) higher risk of cardiovascular death. While in ADVANCE overall, combined intensive blood pressure and glucose control decreased cardiovascular death by 24%, the model identified a high-risk group in whom it decreased the mortality rate by 47%, and a low-risk group in whom it had no discernible effect. High-risk individuals had the greatest absolute risk reduction with a number needed to treat of 12 to prevent one cardiovascular death over 5 years.
Conclusions/interpretation
This novel multiPRS model stratified individuals with type 2 diabetes according to risk of complications and helped to target earlier those who would receive greater benefit from intensive therapy.
Graphical abstract
Supplementary Information
The online version contains peer-reviewed but unedited supplementary material available at 10.1007/s00125-021-05491-7.
Keywords: ADVANCE trial, Cardiovascular complications, Genetics, Polygenic risk score, Prediction, Renal complications, UK Biobank
Introduction
Diabetes increases the risk of serious and costly cardiovascular and renal complications [1, 2]. Prediction of risk prior to development of these complications is crucial to enable the targeting of individuals who could benefit from early intervention [3]. Genetic information, with which one is born, can be used to enable early detection of risk. Genome-wide association studies (GWAS) identified multiple common variants associated with type 2 diabetes [4–6], renal [7, 8] and cardiovascular diseases [9], and hypertension [10]. Individually, these genetic variants account for only a small effect size but the combination of hundreds of them into polygenic risk scores (PRSs) or genome-wide polygenic scores was recently introduced to predict risk of diseases [11–14] including type 2 diabetes [15]. Recent studies suggest that combining several PRSs of related traits into a joint model could optimise prediction [16–18]. Our aim was to develop a multi-polygenic risk score (multiPRS) prediction model that did not include any past risk/outcome data. Because of their common risk factors, overlap of its pathogenic mechanisms and correlations among them, a multiPRS composed of ten weighted PRSs (10 wPRS), gathering genomic variants associated with cardiovascular and renal complications and their key risk factors, was combined with sex and first principal component (PC1) of ethnicity, age at onset and diabetes duration into one logistic regression model, to classify or to predict micro- and macrovascular endpoints of type 2 diabetes [19–23]. The performance of the model was assessed by C-statistics in individuals of European descent with type 2 diabetes in the ADVANCE trial [24, 25] extended to its post-trial follow-up, ADVANCE-ON [26], for a total of nearly 10 years of observation. The multiPRS developed in ADVANCE was validated in participants with type 2 diabetes in the UK Biobank [27] and three smaller external cohorts. We also assessed whether this approach could help in early identification of individuals who could benefit most from the intensive therapy such as administered in ADVANCE [28, 29].
Methods
Ethics
The ADVANCE, CLINPRADIA, CanPath and post-MONICA studies were approved by the ethics commitees of their coordinating centres and by each participating centre. Only participants who provided written informed consent to the genetic sub-studies took part in the analysis. A material transfer agreement was signed with UK Biobank that covers Research Tissue Bank (RTB) under projects 49731 and 59642. The data analysis was approved by the ethics committee of the Centre hospitalier de l’Université de Montéal (CHUM).
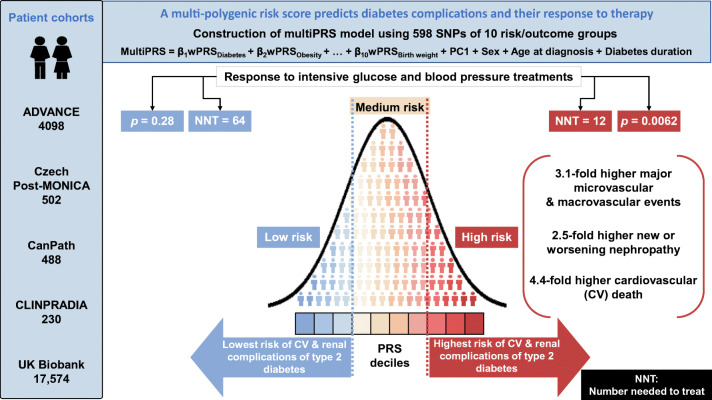
Patient cohorts
Five cohorts were studied here, details of which can be found in the electronic supplementary material (ESM) Methods. ADVANCE (Action in Diabetes and Vascular Disease: Preterax and Diamicron MR Controlled Evaluation) was used to construct the prediction model and to assess its clinical utility. It was a 2 × 2 factorial design, RCT of BP lowering (perindopril-indapamide vs placebo) and intensive glucose control (gliclazide-MR-based intensive intervention with a target of 6.5% [48 mmol/mol] HbA1c vs standard care) in individuals with type 2 diabetes (ClinicalTrials.gov registration no. NCT00145925). A total of 11,140 participants were recruited from 215 centres in 20 countries. Participants were older than 55 years and diagnosed with type 2 diabetes after the age of 30 years. ADVANCE-ON was a 5 year post-trial observational extension of ADVANCE conducted in 80% of participants [26]. Here, we studied a subset of 4098 genotyped individuals of European descent with type 2 diabetes whose clinical characteristics are summarised in ESM Table 1. Data from the UK Biobank [18] were used to validate the model developed in ADVANCE and construct an independent model in 17,604 participants of white British origin with type 2 diabetes (ESM Fig. 1, ESM Table 1). The model was validated in three independent cohorts with phenotypes available in each one. The Czech post-MONICA (the WHO Monitoring Trends and Determinants in Cardiovascular Disease) study was a cross-sectional survey investigating the determinants of cardiovascular risk factors in a 1% random sample of the general population in nine districts of the Czech Republic, stratified by age and sex [30]. Among the 502 individuals genotyped, 106 had albuminuria. CLINPRADIA (Management of Albuminuria in Hypertensive Diabetics) (ClinicalTrials.gov registration no. NCT 01907958) was a multicentre study to evaluate the management of microalbuminuria in hypertensive patients with type 2 diabetes in Canada. The study was performed in 230 individuals with type 2 diabetes (mean age 67 years). Forty per cent of participants had albuminuria at study entry. CanPath (Canadian Partnership for Tomorrow’s Health) brings together five Canadian regional cohorts: 488 (mean age 58 years) individuals of European origin with type 2 diabetes were analysed here.
Outcomes description
Macroalbuminuria is defined as urinary albumin/creatinine ratio (UACR) of >33.9 mg/mmol (300 mg/g). Low eGFR is defined as eGFR below 60 ml min−1 [1.73 m]−2. New or worsening nephropathy is defined as the development of macroalbuminuria, doubling of serum creatinine to a level of at least 200 mmol/l, or end-stage renal disease (ESRD). ESRD is defined as the need for dialysis or renal transplantation, or death due to renal disease. The outcome ‘major microvascular events’ is a composite of ESRD, defined as the requirement for renal replacement therapy, death from renal disease, requirement for retinal photocoagulation or diabetes-related blindness in either eye. The outcome ‘major macrovascular events’ is a composite of non-fatal myocardial infarction, non-fatal stroke or cardiovascular death. The outcome ‘combined major microvascular or macrovascular events’ is defined as death from CVD, non-fatal stroke or non-fatal myocardial infarction, and new or worsening renal or diabetic eye disease.
Creation of wPRS and multiPRS
We identified 26 factors and outcomes that we grouped into ten groups of risk/outcomes: diabetes, obesity, BP, albuminuria, GFR, biomarkers, lipids, stroke, CVDs and low birthweight, with SNPs obtained from 47 publications cited in ESM Table 2 of large-scale GWAS consortia conducted in hundreds of thousands of individuals of European descent. We used their summary statistics, included in the National Human Genome Research Institute GWAS Catalog and HuGE navigator (https://www.ebi.ac.uk/gwas/home), and extracted 598 SNPs, listed in ESM Table 3 together with their effect size (β). Descriptive summary statistics were computed, using frequencies (%) for categorical variables and means (±SD) for continuous variables. A binomial test was used to compare the two proportions of categorical variables. We constructed 10 wPRS for the ten risk groups, as different SNPs contribute with different weights to the PRS value, by summing the product of the number of risk alleles for each participant by the effect size of those SNPs, i.e., where is the allele frequency of ith subject in jth SNP for kth phenotype and β is the effect size attributed to the SNP for the same phenotype in the original GWAS (ESM Figs 2, 3 and ESM Table 3). The effect size attributed to each of the 598 SNPs was obtained from the same group of complications or risk factors. As an example, the effect size of a SNP associated with diabetes was used in the generation of the PRSdiabetes only. If the same SNP was also shown to be associated with albuminuria in meta-analyses, the effect size used in the other PRS was the one from the original meta-analysis of albuminuria and not the effect size derived from the meta-analysis of diabetes. Variants in five genes (TCF7L2, ADCY5, FTO, GCKR and HNF1A) were used in two PRSs. The number of SNPs or the unit used is not the same for the 26 predictors, so the wPRS had to be scaled by the sum of its effect coefficients and multiplied by the number of loci of that specific trait. With this scaling, each risk predictor will have an equivalent weight at an equivalent number of loci. The 10 wPRS, sex, PC1 of ethnicity, age at onset and diabetes duration were included as covariates in a logistic regression model, that we named multiPRS model, to classify prevalent or to predict new type 2 diabetes complications as illustrated in ESM Fig. 3. For clarity, we generated a PRS for each of the ten following traits: PRSdiabetes, PRSobesity, PRSblood pressure, PRSalbuminuria, PRSglomerular filtration rate, PRSbiomarkers, PRSlipids, PRSstroke, PRScardiovascular diseases and PRSlow birthweight. These 10 wPRS were constructed as described above and the effect sizes from the logistic regression model for each of these 10 wPRS are shown in ESM Fig. 3.
We developed the model using the ADVANCE cohort (referred to herein as the ADVANCE model) and compared the accuracy of the model using C-statistics with tenfold cross-validation with a 10–90 data split of ADVANCE, to avoid overfitting. We tested the model in the UK Biobank (used as an external validation dataset) and three other smaller cohorts of individuals with type 2 diabetes, using pROC package (https://cran.r-project.org/web/packages/pROC/pROC.pdf) in R. Given the size of the UK Biobank resource, an alternative (white British-specific) model was also fit to the UK Biobank samples, with tenfold cross-validation, to replicate the approach (referred to herein as the UK Biobank model).
Statistical analyses
MultiPRS thirds and treatment effects were examined through cumulative hazards curves with the use of Cox proportional hazard models. The logrank test was used to compare the cumulative hazards over the period of 9.5 years (ADVANCE and ADVANCE-ON) to examine trial and post-trial effects of the intensive BP-lowering and intensive glucose therapies on cardiovascular death, all-cause death and ESRD in the three genetic risk groups. The Hosmer–Lemeshow test is used to test for goodness of fit of multiPRS in logistic regression models. The test assesses whether the observed event rates match expected event rates in subgroups of the model population. The subgroups analysed are sex and ethnicity. Unless stated otherwise, a p value less than 0.05 is considered statistically significant. Additional details of statistical analyses, genotyping, imputation as well as the stepwise approach for selection of SNPs and creation of multiPRS are included in ESM Methods, ESM Figs 2, 3 and ESM Tables 2, 3.
Genotyping and imputation
ADVANCE participants were genotyped using the Affymetrix Genome-Wide Human SNP Arrays 5.0 or 6.0 or the Affymetrix UK Biobank Axiom Arrays (Affymetrix, Santa Clara, CA, USA). The genotype calling of participants from UK Biobank was performed by Affymetrix on two closely related purpose-designed arrays: ~50,000 participants were run on the UK BiLEVE Axiom array and the remaining ~450,000 were run on the UK Biobank Axiom array. The post-MONICA samples were genotyped with the Affymetrix Genome-Wide array 6.0 and those from CLINPRADIA with UK Biobank Axiom arrays. Participants in CanPath were genotyped using UK BioBank Axiom or GSA Illumina arrays. Quality control was performed on the final genotypes before imputation as described in ESM Methods [25]. A PC analysis using 34,570 independent SNPs on individuals of European descent in ADVANCE was done with EIGENSOFT 3.0 package (https://github.com/argriffing/eigensoft, version 3.0). It was used to adjust for genetic ethnicity and all individuals from other cohorts were projected onto these PCs (ESM Fig. 4). Only PC1 was used here, as it reflects best the genetic structure from East (Balto-Slavic) and West (Celtic) of Europe within these cohorts. Additional PCs could be needed when expanding to other geo-ethnic groups, such as southern European individuals for which population structure is reflected on second PC (PC2) (ESM Fig. 4).
Hierarchical clustering
We performed unsupervised hierarchical clustering (hclust complete method) as described by the R Core Team [31] on the Euclidean distance matrix of the predicted risk values of our models (myocardial infarction, stroke, heart failure, major macrovascular events, cardiovascular and all-cause death). Heatmaps were constructed using R heatmap.2 from gplots library [31, 32].
Prediction using clinical risk scores
For comparison between clinical and multiPRS-based scores, we calculated the ADVANCE clinical risk score that includes age at diagnosis, known duration of diabetes, sex, and baseline pulse pressure, treated hypertension, atrial fibrillation, retinopathy, HbA1c, UACR and non-HDL-cholesterol [33], and the widely used Framingham risk score (FRS) that includes age, sex, total cholesterol, HDL-cholesterol, smoking status, diabetes, systolic BP and BP treatment as predictors [34].
Results
Performance metrics of the multiPRS model as a classifier and predictor of cardiovascular and renal outcomes of type 2 diabetes
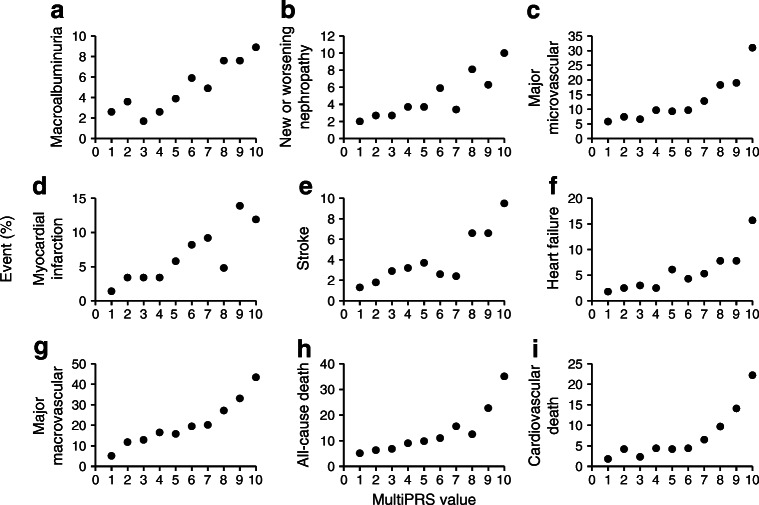
We developed a multiPRS model using the ADVANCE cohort, with tenfold cross-validation (referred to herein as the ADVANCE model). The baseline characteristics of 4098 individuals with type 2 diabetes in ADVANCE are shown in ESM Table 1. They were recruited in 14 European countries and in Australia, New Zealand and Canada allowing other European-descent cohorts to be projected appropriately onto principal components reflecting population structure (ESM Fig. 4). The percentages of ADVANCE participants who had an outcome including death during the 5 year follow-up increased exponentially according to multiPRS score rising sharply in the last 3 deciles of the distribution (Fig. 1). The ADVANCE model was validated in the UK Biobank and three smaller cohorts of individuals with type 2 diabetes (Table 1). The ADVANCE multiPRS model had similar area under the receiver operating characteristic (ROC) curves, AUCs (95% CI) for classification of prevalent stroke, myocardial infarction, low eGFR and albuminuria in ADVANCE and UK Biobank (Table 1). The AUCs were higher for stroke and myocardial infarction in the CanPath cohort while they were almost identical for albuminuria in ADVANCE, UK Biobank and CLINPRADIA. The multiPRS had a low but significant AUC for albuminuria in individuals with the metabolic syndrome from the general population of post-MONICA. Given the size of the UK Biobank resource (ESM Table 1), the multiPRS model was also fit to the UK Biobank samples, with tenfold cross-validation, to replicate the approach (referred to herein as the UK Biobank model). The UK Biobank model’s AUCs were similar to those of the ADVANCE model when tested in the UK Biobank (Table 1). These results show that the multiPRS model developed in ADVANCE had a similar discrimination power for cardiovascular and renal complications of type 2 diabetes in different patient cohorts.
Fig. 1.
(a–i) Percentage of events along multiPRS deciles. ADVANCE participants were stratified into equal deciles along multiPRS scoring, from lowest to highest score. Each point represents the percentage of event occurrence in the decile
Table 1.
Performance (AUC) of the multiPRS model for prevalent and incident cases of T2D complications in different cohorts
| T2D complication | Training/testing cohorts | Validation cohorts | ||||
|---|---|---|---|---|---|---|
| ADVANCE model n = 4098 |
UKBB model n = 17,574 |
UK Biobank n = 17,574 |
CanPath n = 488 |
CLINPRADIA n = 230 |
Czech post-MONICA n = 502 |
|
| Stroke | 0.61 (0.59, 0.64) | 0.61 (0.59, 0.63) | 0.59 (0.57, 0.60) | 0.80 (0.63, 0.97) | – | – |
| Myocardial infarction | 0.58 (0.56, 0.60) | 0.67 (0.66, 0.68) | 0.63 (0.62, 0.64) | 0.78 (0.68, 0.89) | – | – |
| Low eGFR | 0.72 (0.70, 0.74) | 0.70 (0.69, 0.72) | 0.67 (0.65, 0.69) | – | – | – |
| Macroalbuminuria | 0.63 (0.59, 0.68) | 0.65 (0.62, 0.69) | 0.63 (0.59, 0.66) | – | 0.62 (0.53, 0.71) | 0.56 (0.50, 0.62) |
| Incident stroke | 0.62 (0.58, 0.67) | 0.65 (0.62, 0.67) | 0.62 (0.59, 0.65) | – | – | – |
| Incident myocardial infarction | 0.64 (0.61, 0.68) | 0.65 (0.63, 0.67) | 0.61 (0.59, 0.64) | – | – | – |
Data expressed as AUC (95% CI)
30 participants with missing genotypes were excluded from the analysis of UK Biobank
The multiPRS model is composed of the 10 wPRS, PC1, sex, age at diagnosis and diabetes duration. The classification of cases vs non-cases of cardiovascular and renal complications of T2D by the multiPRS model was assessed in parallel in the ADVANCE (ADVANCE model) and the UK Biobank (UKBB model) sets. Incident cases are defined as having an outcome during the study (free of outcome at baseline), and control participants were those who did not have a specific outcome at any time during the study. AUCs and percentile-based CIs were estimated from ROC curves and calculated from the predicted risks derived from the regression models with tenfold cross-validation. The predictors of the ADVANCE model (dataset where the model was constructed) were also tested in the UK Biobank that was used as a validation dataset. The ADVANCE model was also assessed in three independent cohorts for complications available in each of them
T2D, type 2 diabetes
The primary outcome of the ADVANCE trial was the composite of major macrovascular and microvascular events as intention-to-treat by intensive control of BP [24, 26, 28, 29]. The AUCs of Table 2 represent the discrimination between incident cases, defined as having an outcome during the ADVANCE trial (free of outcome at baseline), and control participants who did not have a specific outcome at any time during the study. The AUCs were 0.67 (95% CI 0.65, 0.70) for combined micro- and macrovascular events, 0.67 (0.64, 0.70) for microvascular and 0.68 (0.66, 0.70) for macrovascular events (Table 2). The ADVANCE model predicted incident stroke with the same AUC of 0.62 in ADVANCE and UK Biobank (Table 1). It predicted incident myocardial infarction with AUC values of 0.64 (0.61, 0.68) in ADVANCE and 0.61 (0.59, 0.64) in UK Biobank; these AUC values were slightly lower than with the UK Biobank predictive model (AUC = 0.65 for both incident stroke and myocardial infarction) (Table 1).
Table 2.
Prediction performance and risk stratification thresholds of incident cases in ADVANCE
| Prediction of incident cases (n) | ADVANCE | High risk (30%) | High risk (10%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| PP % |
AUC 95% CI |
AUC1 95% CI |
OR | PPV % |
NPV % |
OR | PPV % |
NPV % |
|
| Combined micro- or macrovascular (844) | 40 | 0.67 (0.65, 0.70) | 0.71 (0.68, 0.74) | 3.1 | 68 | 54 | 3.5 | 80 | 60 |
| Major microvascular (334) | 13 | 0.67 (0.64, 0.70) | 0.68 (0.64, 0.72) | 3.1 | 28 | 86 | 3.7 | 41 | 81 |
| Major macrovascular (559) | 21 | 0.68 (0.66, 0.70) | 0.73 (0.70, 0.76) | 3.1 | 44 | 74 | 3.5 | 55 | 66 |
| Stroke (154) | 4 | 0.66 (0.62, 0.71) | 0.74 (0.70, 0.79) | 3.1 | 12 | 94 | 2.9 | 16 | 96 |
| Myocardial infarction (192) | 7 | 0.67 (0.63, 0.70) | 0.69 (0.65, 0.74) | 2.2 | 16 | 92 | 2.1 | 19 | 94 |
| Heart failure (225) | 6 | 0.68 (0.65, 0.72) | 0.74 (0.68, 0.78) | 3.1 | 15 | 93 | 3.9 | 29 | 90 |
| Macroalbuminuria (150) | 4 | 0.65 (0.60, 0.69) | 0.67 (0.62, 0.72) | 2.3 | 19 | 91 | 2.1 | 27 | 97 |
| Low eGFR (1009) | 41 | 0.64 (0.62, 0.66) | 0.69 (0.66, 0.72) | 4.0 | 59 | 55 | 5.1 | 74 | 46 |
| New/worsening nephropathy (198) | 5 | 0.64 (0.60, 0.68) | 0.66 (0.62, 0.70) | 2.5 | 27 | 94 | 2.5 | 21 | 95 |
| Cardiovascular death (283) | 7 | 0.72 (0.69, 0.75) | 0.78 (0.74, 0.81) | 4.4 | 20 | 91 | 4.7 | 35 | 87 |
| All-cause death (549) | 13 | 0.69 (0.67, 0.72) | 0.74 (0.72, 0.77) | 3.1 | 33 | 84 | 4.4 | 47 | 74 |
The multiPRS model is composed of the 10 wPRS, PC1, sex, age at diagnosis and diabetes duration. The number of cases as well as the period prevalence (PP) of each event during the 5 year follow-up of ADVANCE are indicated. AUC represents the discrimination between incident cases, defined as having an outcome during the 5 years of ADVANCE (free of outcome at baseline), and control participants who did not have a specific outcome at any time during the ADVANCE trial. AUC1 was calculated using a control group that includes normotensive participants only. PPV and NPV were adjusted for the prevalence of the specific outcome. OR: frequency of a specific outcome in high-risk group/frequency of the outcome in the remainder of the population
Outcomes are defined in the Methods section
Higher AUCs (AUC1 in Table 2) were observed for most outcomes when cases were compared with normotensive individuals who did not have a specific outcome at any time during the study with AUCs1 around 0.70 for most cardiovascular outcomes and death. Adjustment for treatment assignment did not modify the AUC values (ESM Table 4). The multiPRS model was well calibrated (expected vs observed event rates are similar) for cardiovascular death in the whole population (π = 0.67) with better fit (the closer the π value is to 1, the better the fit) for men (π = 0.66) than women (π = 0.48) and for Slavic (π = 0.77) than Celtic (π = 0.44) individuals. The best fit was observed for all-cause death, π-values exceeding 0.8 in both Celtic (West of Europe) and Slavic (East of Europe) individuals (ESM Fig. 5).
These results indicate that the multiPRS model can predict individual as well as combined cardiovascular and renal complications of type 2 diabetes before these complications appear.
Risk stratification
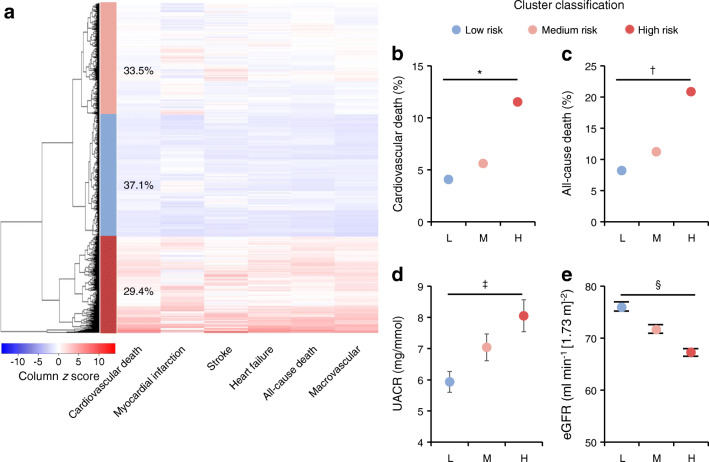
In order to determine the optimal risk thresholds, we explored two possible cut-offs: one of 10% that corresponds to the last decile of multiPRS score shown in Fig. 1 and a higher threshold of 30% that corresponds to the last 3 deciles. As expected, the positive predictive value (PPV) was higher for the top 10% than the 30% high-risk threshold (Table 2). However, when the frequency of the high risk is lower than the prevalence of the complication, as in the case of total death, low eGFR, micro- and macrovascular events and their combination that have prevalence higher than 10%, the PPV is high but the sensitivity (% of people who are at risk are detected) is low, and the 10% high-risk threshold will identify a lower percentage of individuals who will have an outcome. False negatives must be minimised and moderate PPV (with its greater proportion of false positives) is more acceptable as no harm is likely to be done in protecting patients against a diabetic complication even if that complication would not occur without it. The 30% threshold showed in general similar or even better negative predictive values (NPVs) than the 10% threshold. Furthermore, the ratio of events between the 10% and 30% thresholds and the remainder of the population demonstrated a clear enrichment of at-risk individuals with no clinically relevant differences between the two thresholds. For instance, the ADVANCE participants in the 30% high-risk threshold had a mean of 3.1-fold increased risk of major micro- and macrovascular events (p = 6.3 × 10−21 and p = 9.6 × 10−31, respectively), and a 4.4-fold (p = 6.8 × 10−33) increased risk of cardiovascular death than the remainder of participants. The pertinence of the 30% high-risk cut-off was confirmed by unsupervised hierarchical clustering analysis that identified three main clusters of individuals representing 37.1%, 33.5% and 29.4% of ADVANCE individuals having a low, medium or high risk of major macrovascular events including myocardial infarction, stroke, heart failure, and cardiovascular death (Fig. 2a). Twenty per cent of individuals in the high multiPRS stratum have died during the 5 years of the ADVANCE trial compared with 8% in the low multiPRS category (Fig. 2c). The difference between these two groups was also highly significant for renal events known to contribute to the high mortality rate in high-risk individuals (Fig. 2d,e).
Fig. 2.
(a–e) Clustering of combined macrovascular disease risk by multiPRS using unsupervised hierarchical clustering algorithm. This clustering method identified three main clusters of individuals with low (blue; L), medium (pink; M) or high (red; H) combined macrovascular risk representing 37.1%, 33.5% and 29.4%, respectively, of ADVANCE participants. (a) The multiPRS values for each participant and each outcome are represented by z score (blue colour: negative score, red colour: positive score) in the heat map. (b, c) The incidence (%) of cardiovascular (*p = 1.5 × 10−13) (b) and all-cause death (†p = 1.8 × 10−21) (c) were compared between the high and the low clusters. (d, e) Differences in UACR (‡p = 1 × 10−4) (d) and eGFR (§p = 2 × 10−44) (e) values were determined between the high and the low clusters. eGFR is based on CKD-EPI formula
Contribution of genomic and non-genomic factors to the risk prediction model
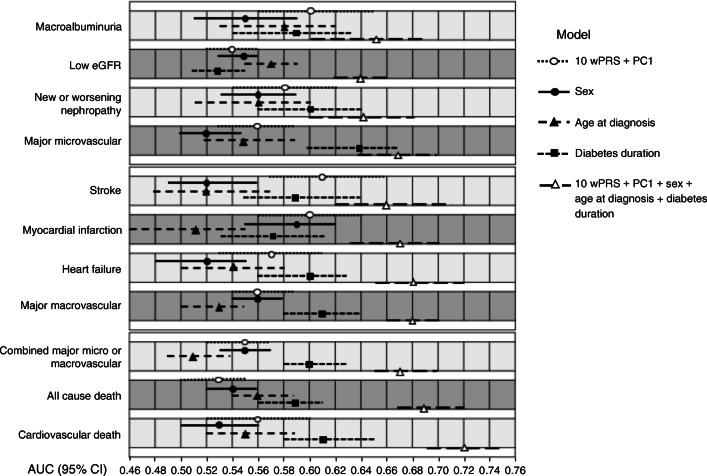
We conducted several complementary analyses to assess the contribution of the different factors to the prediction performance. For the outcomes for which we had a single wPRS developed specifically for the trait, we could determine that: (1) each one of the 10 wPRS contributed significantly to at least one of these traits (ESM Table 5); (2) the contribution of a trait-specific PRS was generally more important than the contribution of the other PRSs; (3) the outcomes were best predicted by more than one PRS; and finally, (4) the combination of 10 wPRS improved the AUCs (ESM Table 5). We previously reported significant differences in several of the diabetes outcomes between Europeans of Slavic and Celtic origins that can be stratified by PC1 of genetic diversity [25]. Here, we are showing that PC1 by itself had a variable impact on the AUC values, but its addition generally improved the prediction performance of the model. (ESM Table 5). Negligible changes were noted here by adding more PCs of ethnicity. The genomic component (10 wPRS and PC1) was generally more predictive than sex or age at onset of diabetes (Fig. 3). Diabetes duration had a higher AUC than the 10 wPRS particularly for heart failure, micro- and macrovascular events and death. The combination of genomic and non-genomic determinants yielded the model with the highest predictive performance (Fig. 3).
Fig. 3.
Contribution of genomic and non-genomic factors to the risk prediction model. AUCs (95% CI) are shown for incident complications (free of outcome at study entry). White circles indicate AUCs with genomic component 10 wPRS + PC1 only; black circles indicate AUCs with sex alone; black triangles represent AUCs with age at diagnosis alone; black squares are AUCs with diabetes duration alone; white triangles indicate AUCs of the full model. Upper section: microvascular and renal outcomes; middle section: macrovascular and cardiac outcomes; lower section: combined micro- and macrovascular outcomes and death
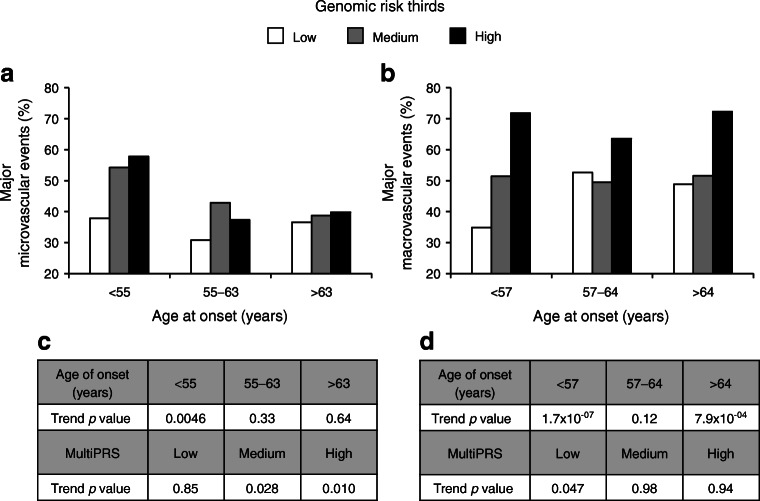
Figure 4a shows that the highest risk of microvascular events was seen in carriers of high genomic (10 wPRS and PC1) risk with OR High vs Low = 1.53 (95% CI 1.08, 2.17), p = 0.017 and younger age at onset of diabetes (OR Old vs Young = 0.61 [0.43, 0.87], p = 0.0057). This contrasts with macrovascular events, for which the highest risk was seen in the highest genomic risk group with OR High vs Low = 2.78 (2.02, 3.81), p = 2.6 × 10−10 independently of the age at onset of diabetes (Fig. 4b). It is noteworthy that the stratification capacity of the genomic component of the model was best in people with earlier onset of diabetes for both major micro- and macrovascular events, as shown by the ptrend values of 4.6 × 10−3 for major microvascular and 1.7 × 10−7 for major macrovascular events (Fig. 4c, d).
Fig. 4.
Frequency of major microvascular and macrovascular events by genomic (10wPRS + PC1) and age at onset of diabetes strata. ADVANCE participants were stratified into equal thirds of low, medium and high genomic risk strata and of <55, 55–63 and >63 years of age at diagnosis of diabetes. The control participants used are normotensive individuals with no major microvascular (a) and macrovascular (b) events at any time during the 4.5 year follow-up of the ADVANCE study. ORs were calculated between high and low genomic component of the multiPRS (OR 1.53 [95% CI 1.08, 2.17] p = 0.017) and between age at onset >63 years and <55 years (OR 0.61 [0.43, 0.87] p = 0.0057) for microvascular events. For macrovascular events, the ORs between high and low genomic component of the multiPRS were (OR 2.78 [2.02, 3.81] p = 2.6 × 10−10) and (OR 1.22 [0.91, 1.64] p = 0.19) between age at onset >64 years and <57 years. (c, d) The trend testing was done within formal regression analysis using parametric method separately for different age categories and genomic strata. Major macrovascular and major microvascular events are defined in the Methods
Clinical utility of the multiPRS model
The multiPRS model did not perform better than the ADVANCE clinical score, knowing that the latter must be considered ‘optimistic’ as our subset of patients was part of the population from which it was developed and included such clinical outcomes as atrial fibrillation, albuminuria and low eGFR (ESM Table 6). However, its AUC values were generally higher than those of the FRS. Even though our primary aim was not to develop a model that outperforms existing clinical scores but one that can predict before symptoms appear, it should be noted that the multiPRS improved the prediction of diabetes outcomes of the two clinical scores. For instance, the continuous net reclassification index (NRI) was 45% for myocardial infarction and 62% for cardiovascular death of people initially classified by the FRS, and was 41% for myocardial infarction and 26% for cardiovascular death of those initially classified with the ADVANCE clinical score (ESM Table 7).
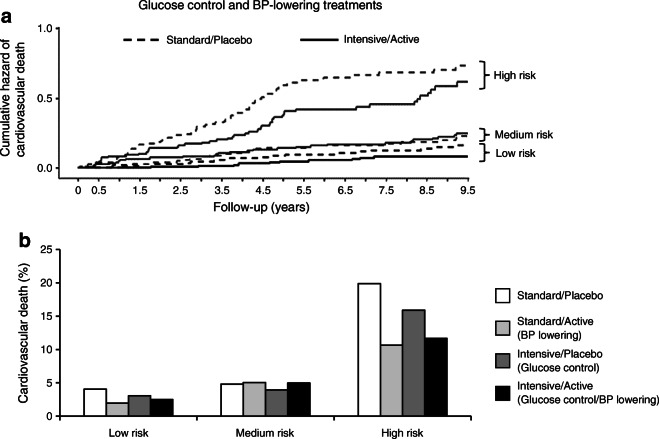
The cumulative incidence of death was significantly different (p < 0.0001) between individuals with low, medium and high predicted risk (ESM Fig. 6). We also noted that the intensive BP control achieved during ADVANCE led to a significant reduction of total death (HR 0.797, p = 0.046) and cardiovascular death (HR 0.677, p = 0.009) in individuals within the highest third of predicted risk, and these reductions remained significant during ADVANCE-ON (ESM Fig. 6, left panel). No such benefit was observed with intensive glycaemic control (ESM Fig. 6, right panel), while glucose control reduced ESRD in individuals carrying the highest predicted risk values (HR 0.345, p = 0.043 in ADVANCE), remaining significant at the end of ADVANCE-ON (HR 0.455, p = 0.026) (ESM Fig. 7). Fifty-nine per cent of ESRD cases occurred in the highest multiPRS third (ESM Fig. 7). The reduction of cardiovascular death by ADVANCE therapy occurred mainly in the high-risk group (HR 0.61 [95% CI 0.40, 0.93], p = 0.021) and remained significant during ADVANCE-ON (Fig. 5a) and the number needed to treat (NNT) to prevent one cardiovascular death could be reduced by as much as fivefold. For instance, NNT = 12 (p = 0.0062) in the high-risk third compared with NNT = 64 (not significant) in the low-risk third (Fig. 5b).
Fig. 5.
Cumulative hazard plots of cardiovascular death stratified by multiPRS strata and glucose and BP-lowering treatments. (a) Adjusted cumulative hazard curves for 9.5 year cardiovascular death by combined active BP and intensive glucose-lowering treatment arms in the high, medium and low multiPRS thirds. The control participants used are normotensive individuals. The effect of BP and glucose-lowering treatment was assessed using HRs and was significant for individuals included in the high-risk group (HR 0.61 [95% CI 0.40, 0.93], p = 0.021 at year 4.5 follow-up of ADVANCE trial, and HR 0.67 [0.47, 0.95], p = 0.023 at year 9.5 follow-up of ADVANCE-ON follow-up). (b) Frequency (%) of cardiovascular death in low, medium and high multiPRS risk strata and risk reduction by treatment arms in ADVANCE. The four treatment arms are Standard/Placebo, Standard/Active for BP lowering, Intensive/Placebo for glucose control and Intensive/Active for glucose control and BP lowering, respectively. The NNT is 64, p = 0.28 in the low multiPRS third compared with NNT 12, p = 0.0062 in the high-risk group for the combined therapy in ADVANCE
Discussion
Novel evidence suggests that for many adult-onset common diseases, a significant degree of the heritability could be captured with a large number of common SNPs identified through GWAS [35]. Diabetes is associated with microvascular and macrovascular outcomes and the line of demarcation between their pathogenetic mechanisms is blurred [19–21]. For instance, in ADVANCE, we previously reported that increases in UACR or decreases in eGFR in individuals with type 2 diabetes were independent predictors of cardiovascular events and death [22]. More recently, we showed that the combination of changes in both eGFR and UACR is a better predictor of major macrovascular events than when the two are assessed separately [23]. Other groups showed that a clinical score that captures different types of complications is more powerful in predicting mortality risk than a simple count of complications [36]. Furthermore, we reported a polygenic overlap between ischaemic stroke and kidney function [37], and a shared genetic architecture has been revealed between type 2 diabetes and BP regulation [10]. We included SNPs associated with micro- and macrovascular outcomes in addition to their common risk factors in our multiPRS and provide evidence that combining 10 wPRS of related traits with sex, PC1, age at onset and diabetes duration into a joint prediction model (named here multiPRS model) allows the prediction of both microvascular and macrovascular endpoints of type 2 diabetes. Our study provides the first evidence that combining 10 wPRS of related traits in a joint model could optimise the prediction of microvascular and macrovascular endpoints of type 2 diabetes. The accuracy and generalisation of prediction of cardiovascular and renal outcomes of diabetes with the multiPRS was shown in individuals with type 2 diabetes of five independent cohorts (Table 1).
Our multiPRS model yielded an AUC of 0.65 for incident stroke in UK Biobank. An AUC of 0.64 was recently reported with a metaGRS (defined as multiple GRSs combined into one meta-score) used to predict ischaemic stroke in the general white British population of the UK Biobank [18, 27]. Our prediction model was slightly superior to the genetic risk score derived from 204 variants associated with coronary risk published by ACCORD, a clinical trial with a design similar to that of ADVANCE, [38] suggesting that integration of 10 wPRS of related traits into a multiPRS model could improve the performance of the model.
The prediction model developed here outperformed the FRS, and improved the individual 5 year risk reclassification when added to the ADVANCE and FRS clinical scores but, more importantly, it can be used well before the apparition of outcomes. In ADVANCE, we observed that the penetrance of outcomes differs between macro- and microvascular complications [39]. Here, we are showing that, for macrovascular events, the greatest risk was in the oldest age groups with the longest duration of diabetes. By contrast, for microvascular events, the highest risk was also seen with the longest duration of diabetes, but in people with the youngest onset of diabetes (Fig. 4). Individuals in the high-risk group had a diagnosis of diabetes 3 years younger (57 years) than those of the low-risk group (60 years) for microvascular events, while for macrovascular events, the age at onset of diabetes was 62 years in the high-risk and 58 years in the low-risk groups. Finally, the efficacy of stratifying participants along a gradient of multiPRS was better seen in younger participants or in those with early onset of type 2 diabetes for both micro- and macrovascular complications, suggesting that the usefulness of a multiPRS is in primary prevention before target organ damage occurs.
Clinical utility of genetic risk scores emerged over the past few years with the demonstration that individuals in the highest genetic risk category had the largest clinical benefit from therapy [40]. The capacity to detect individuals with the best response to medication is one of the most important results of our study. Figure 5 illustrates three components of this study: (1) individuals classified into the low multiPRS category did not benefit from intensive therapies compared with participants of the higher multiPRS thirds; (2) the combination of intensive glucose and BP control showed the best reduction in risk, as reported in ADVANCE [29]; and finally, (3) the highest thirds of multiPRS had the lowest NNT with combined therapies of ADVANCE, i.e. PRS risk classification is clinically effective in reducing the burden of the disease. A limitation of our study is that ADVANCE participants were older than 65 years at entry or 55 years if they had a substantial elevated risk of cardiovascular disease. The possibility that all individuals with newly diagnosed type 2 diabetes could benefit from our multiPRS screening should be explored further. The UK Prospective Diabetes Study showed that at diagnosis, 20% of individuals already had diabetes-related complications. We are showing here that our prediction model could identify individuals with albuminuria in the prediabetes phase of the post-MONICA study suggesting that individuals at high risk for diabetes should also be screened using the multiPRS model.
We recently completed a cost–utility analysis to evaluate the practical implications of the prediction of pre-ESRD and death with the multiPRS vs usual albuminuria screening from a Canadian healthcare system and societal perspective. The analysis showed that, for a lifetime horizon, the polygenic risk scoring was less expensive and more efficacious in terms of quality adjusted life years than usual screening [41].
Our multiPRS scoring is generalisable to individuals of European descent, and our multi-ethnic study in preparation suggests a potential of applying our model to diverse populations. It is possible that, in the future, more SNPs will be added and that this, in addition to more complex machine learning models that would capture non-linear effects between the wPRS and existing clinical predictors, will further improve the predictive power [42]. The possibility that our multiPRS model could predict multimorbidity in non-diabetic individuals should also be explored, as recent evidence demonstrated that novel classes of glucose-lowering medication such as SGLT2 class, which improves heart failure in individuals with diabetes, are also effective in individuals without diabetes [43], suggesting common determinants. The polygenic prediction model developed here is based on common genomic variants that are present at birth, and a few reliable demographic variables that are routinely collected during clinical practice without requiring the presence of any clinical manifestations or initial outcomes, i.e. in the pre-symptomatic phase, suggesting its usefulness in primary prevention. It could be applied in a trial to help identify participants who could benefit from novel glucose-lowering treatments or high-risk individuals for cardiovascular outcomes trials for type 2 diabetes medications by reducing cost or time to obtain sufficient endpoints to allow a better estimate of risk.
Supplementary Information
(PDF 1997 kb)
Acknowledgements
We thank J.-L. Chiasson, Honorary Professor of Medicine at Université de Montréal for his insightful comments on the manuscript. We also thank M.-R. Guertin and P. Dumas (CRCHUM and Medpharmgene, Montreal, Québec, Canada) for their clinical and organisational assistance with the ADVANCE and CLINPRADIA studies.
Authors’ relationships and activities
JT and PH report grants from Servier and OPTITHERA during the conduct of the study and have a patent (number 62/685.642) applied for on 15 June 2018, N/Réf.: 019231-0002, licensed to OPTITHERA. JC reports grants and personal fees from Servier during the conduct of the study and grants from Idorsia outside the submitted work. MM reports personal fees from Servier, Novo Nordisk, Merck Sharp & Dohme outside the submitted work. MM is the president of a not-for-profit French Foundation entitled Fondation Francophone pour la Recherche sur le Diabète. He receives no personal fee for this function, but this foundation receives grants from the Association Française des Diabétiques and from Sanofi, Eli Lilly, Novo Nordisk, Merck Sharp & Dohme, Roche and Abbott. BW reports personal fees from Servier, Daiichi Sankyo, Boehringer Ingelheim and Novartis outside the submitted work. NP has received personal speaker fees from Servier, Takeda and Novo Nordisk, and advisory board activities from AstraZeneca and Novo Nordisk, and has received grants for his research group relating to diabetes mellitus from Diabetes UK, NIHR Efficacy and Mechanism Evaluation Programme (EME), Julius Clinical and the British Heart Foundation with a pending grant from Novo Nordisk. NP holds no stocks or shares in any such companies. SZ reports fees from Sanofi, AstraZeneca, Novo Nordisk and MSD Australia outside the submitted work. GM reports personal fees from Boehringer Ingelheim, Daiichi Sankyo, Ferrer, MEDTRONIC, Menarini International, Merck, Novartis, Recordati and Servier outside the submitted work. AR is one of the inventors on several patents filed by The George Institute for Global Health on Compositions for the Treatment of Hypertension. None of the inventors have a financial interest in these planned products. He is seconded to work for George Health Enterprises (the social enterprise arm of The George Institute for Global Health). He has received investment to develop fixed-dose combination products containing aspirin, statin and BP-lowering drugs. AR sits on a Data and Safety Monitoring Board for Idorsia. MW reports personal fees from Amgen and Kirin outside the submitted work. All other authors declare that there are no relationships or activities that might bias, or be perceived to bias, their work.
Abbreviations
- ESRD
End-stage renal disease
- FRS
Framingham risk score
- GWAS
Genome-wide association studies
- MultiPRS
Multi-polygenic risk score
- NNT
Number needed to treat
- NPV
Negative predictive value
- PC
Principal component
- PPV
Positive predictive value
- PRS
Polygenic risk score
- ROC
Receiver operating characteristic
- UACR
Urinary albumin/creatinine ratio
- wPRS
Weighted PRS
Contribution statement
JT and PH contributed equally to the study design, data collection, data analyses, figures, data interpretation, literature search and writing of the manuscript and performed the CLINPRADIA clinical study. They led the ADVANCE genetic sub-study committee which also includes JC, SH and MW. MHal, CL, CHiz, LS and RA contributed to the literature search, figures, data analyses, data interpretation and drafting of the manuscript. FH and F-CMB developed the database and contributed to the data analyses, interpretation and drafting of the manuscript. RT performed the statistical analyses that were reviewed by MW and JH and critically reviewed the manuscript. JC, MM, SH, DM, BW, NP, SZ, SC, GM, DEG, AR, LL and MW participated in the design and execution of the ADVANCE trial as members of the ADVANCE Management Committee and critically reviewed and commented on the manuscript. AK and RC collected the data for the post-MONICA study, contributed to the analyses and critically reviewed the manuscript. MA, VB, M-JF, MHar and PA contributed to the CanPath analysis, data interpretation and critically reviewed the manuscript. PS and CHis contributed to the genetic analyses, data interpretation and critically reviewed the manuscript. JH, RA and CHis contributed to the analysis of UK Biobank and critically reviewed the manuscript. All authors have approved the final version of the manuscript. PH is the guarantor of this work.
Funding
This work was supported by Genome Québec, Canadian Institutes of Health Research (CIHR), Ministère de l’Économie et de l’Innovation du Québec (MEIE), Consortium Québécois sur la Découverte du Médicament (CQDM), OPTITHERA, Servier and Canada Research Chair in Predictive Genomics to PH. JH is a Fonds la Recherche du Québec en Santé (FRQS) Junior 1 Research Scholar supported by the Institute for Data Valorization (IVADO) and Fondation de l’Institut de Cardiologie de Montréal (FICM). This research has been conducted using data from UK Biobank, a major biomedical database: www.ukbiobank.ac.uk. The study funders were not involved in the design of the study; the collection, analysis, and interpretation of data; writing the report; and did not impose any restrictions regarding the publication of the report.
Data availability
The list of SNPs used to generate the multiPRS model is included in ESM Table 3. The list of GWAS data is referenced in ESM Table 3. Other data are available on request from JT or PH. Parts of this work were presented in Late Breaking abstract form at the 78th Scientific Sessions of the American Diabetes Association, Orlando, FL, USA, 22–26 June 2018. A preliminary report of the work was made publicly available in MedRxiv on 19 November 2019.
Footnotes
The original online version of this article was revised: "The authors regret that they omitted to acknowledge the clinical and organisational assistance of M.-R. Guertin and P. Dumas (CRCHUM and Medpharmgene, Montreal, Québec, Canada) in the ADVANCE and CLINPRADIA studies.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Johanne Tremblay and Pavel Hamet contributed equally to this study.
Change history
8/27/2021
A Correction to this paper has been published: 10.1007/s00125-021-05544-x
Contributor Information
Johanne Tremblay, Email: johanne.tremblay@umontreal.ca.
Mark Woodward, Email: markw@georgeinstitute.org.au.
Pavel Hamet, Email: pavel.hamet@umontreal.ca.
References
- 1.The Emerging Risk Factors Collaboration Diabetes mellitus, fasting blood glucose concentration, and risk of vascular disease: a collaborative meta-analysis of 102 prospective studies. Lancet. 2010;375(9733):2215–2222. doi: 10.1016/S0140-6736(10)60484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacobs E, Hoyer A, Brinks R, Kuss O, Rathmann W. Burden of mortality attributable to diagnosed diabetes: a nationwide analysis based on claims data from 65 million people in Germany. Diabetes Care. 2017;40(12):1703–1709. doi: 10.2337/dc17-0954. [DOI] [PubMed] [Google Scholar]
- 3.American Diabetes Association 9. Cardiovascular disease and risk management: standards of medical care in diabetes—2018. Diabetes Care. 2018;41(Supplement 1):S86–S104. doi: 10.2337/dc18-S009. [DOI] [PubMed] [Google Scholar]
- 4.Abraham G, Inouye M. Genomic risk prediction of complex human disease and its clinical application. Curr Opin Genet Dev. 2015;33:10–16. doi: 10.1016/j.gde.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Läll K, Mägi R, Morris A, Metspalu A, Fischer K. Personalized risk prediction for type 2 diabetes: the potential of genetic risk scores. Genet Med. 2017;19(3):322–329. doi: 10.1038/gim.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lei X, Huang S. Enrichment of minor allele of SNPs and genetic prediction of type 2 diabetes risk in British population. PLoS One. 2017;12(11):e0187644. doi: 10.1371/journal.pone.0187644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravizza S, Huschto T, Adamov A, et al. Predicting the early risk of chronic kidney disease in patients with diabetes using real-world data. Nat Med. 2019;25(1):57–59. doi: 10.1038/s41591-018-0239-8. [DOI] [PubMed] [Google Scholar]
- 8.Wuttke M, Li Y, Li M, et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet. 2019;51(6):957–972. doi: 10.1038/s41588-019-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anand SS, Meyre D, Pare G, et al. Genetic information and the prediction of incident type 2 diabetes in a high-risk multiethnic population. Diabetes Care. 2013;36(9):2836–2842. doi: 10.2337/dc12-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evangelou E, Warren HR, Mosen-Ansorena D, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50(10):1412–1425. doi: 10.1038/s41588-018-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mihaescu R, Moonesinghe R, Khoury MJ, Janssens A. Predictive genetic testing for the identification of high-risk groups: a simulation study on the impact of predictive ability. Genome Med. 2011;3(7):51. doi: 10.1186/gm267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escott-Price V, Shoai M, Pither R, Williams J, Hardy J. Polygenic score prediction captures nearly all common genetic risk for Alzheimer’s disease. Neurobiol Aging. 2017;49:214.e7–214.e11. doi: 10.1016/j.neurobiolaging.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 13.Rosenberg NA, Edge MD, Pritchard JK, Feldman MW. Interpreting polygenic scores, polygenic adaptation, and human phenotypic differences. Evol Med Public Heal. 2019;2019(1):26–34. doi: 10.1093/emph/eoy036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson G. On the utilization of polygenic risk scores for therapeutic targeting. PLoS Genet. 2019;15(4):e1008060. doi: 10.1371/journal.pgen.1008060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khera AV, Chaffin M, Aragam KG, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turley P, Walters RK, Maghzian O, et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50(2):229–237. doi: 10.1038/s41588-017-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krapohl E, Patel H, Newhouse S, et al. Multi-polygenic score approach to trait prediction. Mol Psychiatry. 2018;23(5):1368–1374. doi: 10.1038/mp.2017.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abraham G, Malik R, Yonova-Doing E, et al. Genomic risk score offers predictive performance comparable to clinical risk factors for ischaemic stroke. Nat Commun. 2019;10(1):5819. doi: 10.1038/s41467-019-13848-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan CL, Currie CJ, Stott NCH, Smithers M, Butler CC, Peters JR. The prevalence of multiple diabetes-related complications. Diabet Med. 2000;17(2):146–151. doi: 10.1046/j.1464-5491.2000.00222.x. [DOI] [PubMed] [Google Scholar]
- 20.Nickerson HD, Dutta S. Diabetic complications: current challenges and opportunities. J Cardiovasc Transl Res. 2012;5(4):375–379. doi: 10.1007/s12265-012-9388-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avogaro A, Fadini GP. Microvascular complications in diabetes: a growing concern for cardiologists. Int J Cardiol. 2019;291:29–35. doi: 10.1016/j.ijcard.2019.02.030. [DOI] [PubMed] [Google Scholar]
- 22.Ninomiya T, Perkovic V, de Galan BE, et al. Albuminuria and kidney function independently predict cardiovascular and renal outcomes in diabetes. J Am Soc Nephrol. 2009;20(8):1813–1821. doi: 10.1681/ASN.2008121270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohkuma T, Jun M, Chalmers J, et al. Combination of changes in estimated GFR and albuminuria and the risk of major clinical outcomes. Clin J Am Soc Nephrol. 2019;14(6):862–872. doi: 10.2215/CJN.13391118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel A, ADVANCE Collaborative Group. MacMahon, et al. Effects of a fixed combination of perindopril and indapamide on macrovascular and microvascular outcomes in patients with type 2 diabetes mellitus (the ADVANCE trial): a randomised controlled trial. Lancet. 2007;370(9590):829–840. doi: 10.1016/S0140-6736(07)61303-8. [DOI] [PubMed] [Google Scholar]
- 25.Hamet P, Haloui M, Harvey F, et al. PROX1 gene CC genotype as a major determinant of early onset of type 2 diabetes in slavic study participants from action in diabetes and vascular disease. J Hypertens. 2017;35:S24–S32. doi: 10.1097/HJH.0000000000001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zoungas S, Chalmers J, Neal B, et al. Follow-up of blood-pressure lowering and glucose control in type 2 diabetes. N Engl J Med. 2014;371(15):1392–1406. doi: 10.1056/NEJMoa1407963. [DOI] [PubMed] [Google Scholar]
- 27.Sudlow C, Gallacher J, Allen N, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel A, MacMahon S, Chalmers J, et al. Intensive blood glucose control and vascular outcomes in patients with type 2 diabetes. N Engl J Med. 2008;358(24):2560–2572. doi: 10.1056/NEJMoa0802987. [DOI] [PubMed] [Google Scholar]
- 29.Zoungas S, de Galan BE, Ninomiya T, et al. Combined effects of routine blood pressure lowering and intensive glucose control on macrovascular and microvascular outcomes in patients with type 2 diabetes: new results from the ADVANCE trial. Diabetes Care. 2009;32(11):2068–2074. doi: 10.2337/dc09-0959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krajcoviechova A, Tremblay J, Wohlfahrt P, et al. The impact of blood pressure and visceral adiposity on the association of serum uric acid with albuminuria in adults without full metabolic syndrome. Am J Hypertens. 2016;29(12):1335–1342. doi: 10.1093/ajh/hpw098. [DOI] [PubMed] [Google Scholar]
- 31.R Core team . R: a language and environment for statistical computing. Vienna, Austria: R foundation for statistical computing; 2018. [Google Scholar]
- 32.Warnes GR, Bolker B, Bonebakker L et al (2016) gplots: various R programming tools for plotting data, R package version 3.0.1. R foundation for statistical computing, Vienna, Austria
- 33.Kengne AP, Patel A, Marre M, et al. Contemporary model for cardiovascular risk prediction in people with type 2 diabetes. Eur J Cardiovasc Prev Rehabil. 2011;18(3):393–398. doi: 10.1177/1741826710394270. [DOI] [PubMed] [Google Scholar]
- 34.D’Agostino RB, Vasan RS, Pencina MJ, et al. General cardiovascular risk profile for use in primary care. Circulation. 2008;117(6):743–753. doi: 10.1161/CIRCULATIONAHA.107.699579. [DOI] [PubMed] [Google Scholar]
- 35.Visscher PM, Wray NR, Zhang Q, et al. 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101(1):5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young BA, Lin E, Von Korff M, et al. Diabetes complications severity index and risk of mortality, hospitalization, and healthcare utilization. Am J Manag Care. 2008;14(1):15–23. [PMC free article] [PubMed] [Google Scholar]
- 37.Holliday EG, Traylor M, Malik R, et al. Polygenic overlap between kidney function and large artery atherosclerotic stroke. Stroke. 2014;45(12):3508–3513. doi: 10.1161/STROKEAHA.114.006609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morieri ML, Gao H, Pigeyre M, et al. Genetic tools for coronary risk assessment in type 2 diabetes: a cohort study from the ACCORD clinical trial. Diabetes Care. 2018;41(11):2404–2413. doi: 10.2337/dc18-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zoungas S, Woodward M, Li Q, et al. Impact of age, age at diagnosis and duration of diabetes on the risk of macrovascular and microvascular complications and death in type 2 diabetes. Diabetologia. 2014;57(12):2465–2474. doi: 10.1007/s00125-014-3369-7. [DOI] [PubMed] [Google Scholar]
- 40.Mega JL, Stitziel NO, Smith JG, et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 2015;385(9984):2264–2271. doi: 10.1016/S0140-6736(14)61730-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guinan K, Beauchemin C, Tremblay J, et al. Economic evaluation of a new polygenic risk score to predict nephropathy in adult patients with type 2 diabetes. Can J Diabetes. 2021;45(2):129–136. doi: 10.1016/j.jcjd.2020.06.010. [DOI] [PubMed] [Google Scholar]
- 42.Dias R, Torkamani A. Artificial intelligence in clinical and genomic diagnostics. Genome Med. 2019;11(1):70. doi: 10.1186/s13073-019-0689-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zannad F, Ferreira JP, Pocock SJ, et al. SGLT2 inhibitors in patients with heart failure with reduced ejection fraction: a meta-analysis of the EMPEROR-reduced and DAPA-HF trials. Lancet. 2020;396(10254):819–829. doi: 10.1016/S0140-6736(20)31824-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1997 kb)
Data Availability Statement
The list of SNPs used to generate the multiPRS model is included in ESM Table 3. The list of GWAS data is referenced in ESM Table 3. Other data are available on request from JT or PH. Parts of this work were presented in Late Breaking abstract form at the 78th Scientific Sessions of the American Diabetes Association, Orlando, FL, USA, 22–26 June 2018. A preliminary report of the work was made publicly available in MedRxiv on 19 November 2019.