Abstract
Loss of heterozygosity is a common mode of adaptation in asexual diploid populations. Because mitotic recombination frequently extends the full length of a chromosome arm, the selective benefit of loss of heterozygosity may be constrained by linked heterozygous mutations. In a previous laboratory evolution experiment with diploid yeast, we frequently observed homozygous mutations in the WHI2 gene on the right arm of Chromosome XV. However, when heterozygous mutations arose in the STE4 gene, another common target on Chromosome XV, loss of heterozygosity at WHI2 was not observed. Here, we show that mutations at WHI2 are partially dominant and that mutations at STE4 are overdominant. We test whether beneficial heterozygous mutations at these two loci interfere with one another by measuring loss of heterozygosity at WHI2 over 1,000 generations for ∼300 populations that differed initially only at STE4 and WHI2. We show that the presence of an overdominant mutation in STE4 reduces, but does not eliminate, loss of heterozygosity at WHI2. By sequencing 40 evolved clones, we show that populations with linked overdominant and partially dominant mutations show less parallelism at the gene level, more varied evolutionary outcomes, and increased rates of aneuploidy. Our results show that the degree of dominance and the phasing of heterozygous beneficial mutations can constrain loss of heterozygosity along a chromosome arm, and that conflicts between partially dominant and overdominant mutations can affect evolutionary outcomes.
Keywords: overdominance, loss of heterozygosity, experimental evolution, yeast, diploid
Significance
In diploid populations, it is beneficial for partially dominant beneficial mutations to lose heterozygosity, but it is deleterious for overdominant beneficial mutations to do so. Because loss-of-heterozygosity tracts often encompass entire chromosome arms, a conflict will arise when a partially dominant beneficial mutation and an overdominant beneficial mutation exist in close proximity. We demonstrate that this conflict occurs, and that it restricts loss of heterozygosity, resulting in more variable evolutionary outcomes.
Introduction
The pace of adaptation in asexual diploids is strongly dependent on the dominance of new beneficial mutations. Theoretically, the probability of a given beneficial mutation fixing in a population is the product of its coefficient of selection (s) and its degree of dominance (h), where h = 0 is fully recessive and h = 1 is fully dominant (Orr and Otto 1994). Beneficial alleles with a low degree of dominance (h ≈ 0) are unlikely to fix in asexual diploid populations, a phenomenon known as Haldane’s Sieve (Haldane 1924; Connallon and Hall 2018). Depending on the degree of dominance, some adaptive pathways that are open to haploids, will be improbable or inaccessible to diploids. Changes to the spectrum of beneficial mutations and to the genetic targets of selection between haploid and diploid populations provide experimental evidence of this constraint (Fisher et al. 2018; Marad et al. 2018).
Recessive beneficial mutations can be converted to beneficial homozygous mutations through loss-of-heterozygosity (LOH) events during asexual propagation of diploid yeast populations (Gerstein et al. 2014). For highly heterozygous populations, such as interspecific hybrids, LOH becomes the dominant mechanism of adaptation (Smukowski Heil et al. 2017; James et al. 2019) due to a high rate of LOH relative to point mutation (Barbera and Petes 2006; Lee et al. 2009; Dutta et al. 2021) and to a reservoir of beneficial mutations that are masked in the heterozygous state.
The ability of a given mutation to escape Haldane’s sieve by LOH will depend on its genomic location since the rate of LOH varies across the yeast genome (Lee et al. 2009). The rate of LOH increases with distance from the centromere (Peter et al. 2018) and several hotspots for LOH have been identified, most strikingly at the rDNA locus on Chromosome XII (Fisher et al. 2018; Marad et al. 2018). LOH can occur by gene conversion, which produces small (∼3 kb) interstitial conversion tracts, or by break-induced replication, which leads to LOH conversion tracts that extend through the telomere (Symington et al. 2014; Sui et al. 2020; Dutta et al. 2021). The latter occur at a lower rate but are more frequently observed in experimental evolution (Smukowski Heil et al. 2017; Marad et al. 2018; James et al. 2019).
Two special cases of dominance further constrain adaptation in diploids. Underdominance (h < 0) occurs when the heterozygote is less fit than either homozygous genotype. Underdominant mutations are unlikely to establish as heterozygotes and therefore underdominance impedes access to potentially adaptive homozygous genotypes. At the other extreme, overdominance (h > 0) occurs when the heterozygote is more fit than either homozygous genotype. Overdominant mutations should readily establish in populations and be maintained as heterozygous by selection against homozygous genotypes (Fisher 1928). Because little is known about the distribution of the degree of dominance of new mutations in diploids, the relative importance of underdominant and overdominant mutations in genome evolution is unclear. Theoretically, overdominant mutations are predicted to be a major contributor to the maintenance of genetic variation (Maruyama and Nei 1981), and a frequent, if perhaps only transient, outcome of diploid evolution (Manna et al. 2011; Sellis et al. 2011). Though genome-wide scans have turned up little evidence of overdominance (Szulkin et al. 2010; Hedrick 2012; Goudie et al. 2014), laboratory-evolution experiments demonstrate that overdominant mutations can contribute to short-term adaptation in diploid populations (Sellis et al. 2016; Leu et al. 2020).
If overdominant mutations are frequent in evolving asexual diploid populations, fitness conflicts may arise when overdominant (h > 0) and partially dominant (0 < h < 1) heterozygous beneficial mutations arise in close proximity to one another on a chromosome. This is because an LOH event would convert both mutations to the homozygous state resulting in a fitness loss due to the overdominant mutation and a fitness gain due to the partially dominant mutation. We demonstrate that this conflict arises in experimental evolution between an overdominant mutation in STE4 and a partially dominant mutation in WHI2, both of which are located on the right arm of Chromosome XV. We show experimentally that adaptive LOH at the WHI2 locus is slowed by the presence of the overdominant STE4 mutations.
Results
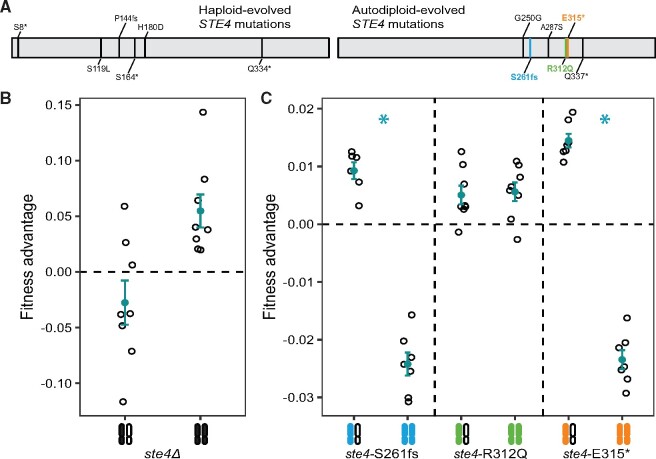
We previously identified 20 genes that are targets of selection in 46 laboratory-evolved populations yeast that were propagated asexually for 4,000 generations (Fisher et al. 2018). By Generation 1,000, all 46 populations had autodiploidized and therefore most adaptation occurred in the diploid state. Unlike true diploids (MATa/α), which do not mate, autodiploids (MATa/a or MATα/α) produce mating pheromones and will readily mate with cells of the opposite mating type. Therefore, autodiploids, like haploids (MATa or MATα), should benefit from mutations that inactivate the mating pathway (Lang et al. 2009). However, only one mating pathway gene, STE4, is identified as a common target of selection across the 46 autodiploid populations (Fisher et al. 2018). In contrast, mutations in STE4, STE5, STE11, and STE12 are overrepresented in 40 closely matched haploid populations (Lang et al. 2013). Though not identified in either experiment, STE7 is also a target of selection in haploids (Lang et al. 2009; Buskirk et al. 2017). We have demonstrated previously that mating pathway mutations are loss-of-function based on a combination of their mutational spectra, allele reconstruction, and gene deletion (Lang et al. 2009, 2013; Buskirk et al. 2017). However, STE4 mutations in autodiploids are inconsistent with simple loss-of-function: not only are they found in only one gene, but all six STE4 mutations are heterozygous and cluster in a small (260 bp) region of the coding sequence (X2(1, N = 6) = 18.76, P < 10−4, fig. 1A).
Fig. 1.
Adaptive STE4 mutations differ between haploid and autodiploid populations. (A) Mutations previously identified in the STE4 gene in haploid (Lang et al. 2013) and autodiploid (Fisher et al. 2018) populations. The positions of mutations across the coding sequence of STE4 in haploid populations did not deviate from random expectation (X2(1, N = 6)=0.74363, P = 0.39). Conversely, autodiploid mutations accumulated nonrandomly across the linear 1,276 bp sequence of the STE4 gene, (X2(1, N = 6)=18.76, P < 10−4). Bolded mutations are ones that were selected for reconstruction. (B) Deletion of STE4 is deleterious in autodiploids when heterozygous and beneficial when homozygous. (C) Three evolved autodiploid alleles were reconstructed in an ancestral background. ste4-S261fs and ste4-E315* are overdominant in the ancestral background. (B and C) A filled and open chromosome represents heterozygosity and two filled chromosomes represent homozygosity. Open points are fitness measures of eight biological replicates following MATa/a conversion. Filled points show mean fitness ± SE.
STE4 encodes the highly conserved beta subunit of the heterotrimeric G protein complex. We mapped the positions of the evolved mutations onto the homology-predicted structure of Ste4p (supplementary fig. S1, Supplementary Material online). All six autodiploid mutations impact residues near the C-terminal end of the protein, with three of these (a frameshift and two nonsense mutations) resulting in truncations of the final ∼100 amino acids. Two missense mutations both occur in a putative random coil that derives from a yeast-specific insertion (Sondek et al. 1996). One synonymous mutation also arose in this region.
Adaptive STE4 Mutations in Autodiploids Are Either Dominant or Overdominant
We hypothesized that the discrepancy between patterns of sequence evolution in haploids and autodiploids is because the beneficial mating pathway mutations in haploids are recessive. We first tested whether the fitness benefit of evolved ste4 mutations in autodiploids is phenocopied by gene deletion, as is the case with ste4 mutations arising in haploid populations. We generated STE4 deletion (ste4Δ) strains as haploids, as heterozygous autodiploids, and as homozygous autodiploids. As reported previously, we find that ste4Δ mutants are beneficial in a haploid background (supplementary fig. S2, Supplementary Material online). Similarly, homozygous ste4Δ/ste4Δ mutants are beneficial in MATa/a diploids (fig. 1B). Surprisingly, however, heterozygous STE4/ste4Δ mutants are underdominant: substantially less fit than the either homozygous genotype (fig. 1C).
The underdominance of STE4 deletions confirms that evolved autodiploid mutations are not purely loss-of-function, as these would be deleterious. We used CRISPR-Cas9 allele-swaps to reconstruct three autodiploid-evolved ste4 alleles (one frameshift, one nonsense, and one missense mutation, fig. 1A). For each allele, we then assayed the fitness of the haploid mutant and the homozygous and heterozygous autodiploid mutants. In a haploid background, the evolved frameshift and nonsense alleles have a fitness benefit (2.1 ± 0.3% and 2.2 ± 0.4%, respectively; mean ± SE, P < 10−4 both genotypes), whereas the missense allele is neutral (−0.7 ± 0.3%, P = 0.995, supplementary fig. S2, Supplementary Material online; Note that we also verified that the synonymous PAM site mutation is neutral, supplementary fig. S3, Supplementary Material online).
In the heterozygous autodiploids, like in the haploids, the ste4-S261fs and ste4-E315* mutations are beneficial (0.8 ± 0.2% and 1.2 ± 0.1%, P = 0.02 and P < 10−4, respectively), and the ste4-Arg312Gln mutation is neutral (0.2 ± 0.2%, P = 0.49, fig. 1C). The fitness effect of the heterozygous evolved alleles is ∼40–50% of the fitness advantage conferred in haploids. Given that 95% of mutations in the autodiploids are heterozygous after 4,000 generations, it is unsurprising that all six STE4 mutations are heterozygous, and we expected that the homozygous mutations would be equally (if not more) fit than the heterozygous STE4 mutations. However, the ste4-S261fs and ste4-E315* homozygous mutants have fitness defects of −2.1 ± 0.1% and −2.4 ± 0.1%, respectively (mean ± SE, P < 10−3 for both comparisons, fig. 1C). Therefore, rather than showing an additive fitness effects as predicted, two STE4 missense mutations are overdominant (h > 1).
Linkage to Overdominant STE4 Alleles Delays, but Does Not Prevent, Adaptive LOH
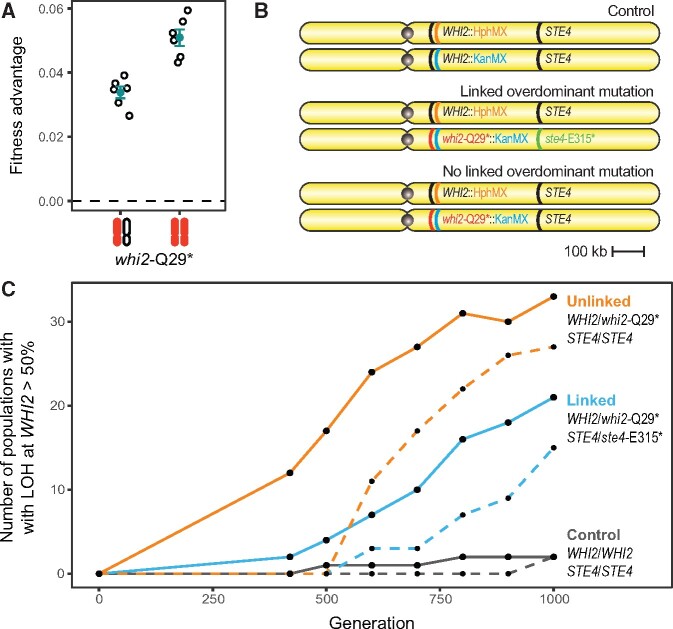
We next examined whether, in our populations, the presence of overdominant ste4 mutations constrains adaptive LOH at linked loci. Although most mutations in our evolved autodiploids are heterozygous, there are two large genomic regions that are prone to high rates of LOH; these regions are identifiable based on the clustering of homozygous mutations in evolved genomes (Fisher et al. 2018). One of these regions on the right arm of Chromosome XV contains the STE4 locus as well as three other common targets of selection in our experimental system: WHI2, SFL1, and PDR5. Unlike STE4, putative adaptive mutations in WHI2, SFL1, and PDR5 are commonly observed to be homozygous in evolved clones. We show that an evolved mutation in WHI2 (whi2-Q29*) is partially dominant, having a 3.4 ± 0.2% benefit when heterozygous and a 5.1 ± 0.3% benefit when homozygous (mean ± SE, fig. 2A).
Fig. 2.
The presence of a linked overdominant STE4 mutation restricts, but does not prevent, loss of heterozygosity at WHI2. (A) An evolved Q29* in the WHI2 gene, which is centromeric to STE4, reported in (Fisher et al. 2018) is partially dominant and most beneficial when homozygous. A filled and open chromosome represents heterozygosity and two filled chromosomes represent homozygosity. Open points are fitness measures of eight biological replicates following MATa/a conversion. Filled points show mean fitness ± SE. (B) Schematic of the three genotypes that were constructed for an evolution experiment. LOH in any genotype is determined by loss of double drug resistance and the direction of the LOH is determined by the single drug to which resistance is lost. (C) About 288 populations (96 whi2-Q29*/WHI2 STE4/STE4, 96 whi2-Q29*/WHI2 ste4-E315*/STE4, 95 wild-type control) were evolved for 1,000 generations. Solid lines show the number of populations with detected LOH over time. Dashed lines show the number of populations in which whi2-Q29*homozygous genotypes were fixed. Lines are colored by group (orange: ste4-E315* linked, blue: STE4 wild-type, black: control). LOH of whi2-Q29* was observed in a higher fraction of unlinked populations at Generation 420 (Fisher’s exact, P = 0.001), however, the difference becomes smaller over time and is nonsignificant at Generation 1,000 (Fisher’s exact, P = 0.08). At Generation 1,000, WHI2 LOH was detectable in 33 whi2-Q29* STE4 populations, 21 whi2-Q29* ste4-E315* populations, and two control populations.
WHI2, SFL1, and PDR5 are all centromere proximal to STE4, and since conversion tracts produced by mitotic recombination frequently extend from a medial breakpoint to the telomere (Sui et al. 2020; Dutta et al. 2021), adaptive LOH of any of these three loci is likely to extend through the STE4 locus. We examined evolved autodiploid genotypes for evidence of LOH events occurring after a STE4 mutation arises on the right arm of Chromosome XV and we find none. There are two populations with fixed homozygous Chromosome XV mutations, but each contain heterozygous STE4 alleles, indicating that the LOH event on Chromosome XV occurred before STE4 mutations in these populations. The inverse—unfixed Chromosome XV homozygous mutations on a fixed ste4 mutant background, which would indicate LOH on Chromosome XV after a mutation at STE4—is not observed in any of the three populations with fixed STE4 mutations (supplementary table S1, Supplementary Material online).
To explicitly test the hypothesis that overdominant STE4 alleles decrease the likelihood of adaptive LOH at linked loci, we performed an evolution experiment using three strains that differed only on the right arm of Chromosome XV (fig. 2B). One strain contained a heterozygous beneficial and partially dominant whi2-Q29* mutation and was wild-type at the STE4 locus (WHI2/whi2-Q29*, STE4/STE4). A second strain contained the same heterozygous whi2-Q29* mutation in cis with a heterozygous overdominant ste4-E315* mutation (WHI2/whi2-Q29*, STE4/ste4-E315*). A control strain was wild type at both loci.
We evolved 96 replicate populations of each strain (95 for control strain) for 1,000 generations. We tested for LOH events at the WHI2 locus every 100 generations starting at Generation 400. All three strains carried KanMX and HphMX drug-resistance cassettes tightly linked to the WHI2 loci on each chromosome. By assaying for loss of the ability to grow on double drug (G418+Hygromycin), our assay is sensitive to LOH events that reach frequency of 0.5 in the population (supplementary fig. S4, Supplementary Material online).
The presence of a heterozygous and partially dominant whi2-Q29* adaptive mutation drives a high rate of fixation of LOH events relative to the wild-type control populations (fig. 2C). However, among populations with a whi2-Q29* allele, the fraction of populations experiencing LOH is significantly lower in populations with a telomeric overdominant ste4-E315* mutation at Generation 420 (Fisher’s exact, P = 0.001, fig. 2C) and remains lower through all time points assayed, although this becomes nonsignificant at Generations 900 and 1,000 (Fisher’s exact, P = 0.066, 0.077). Although time-course LOH dynamics show an effect of linkage to an overdominant allele, the linkage did not prevent LOH at WHI2, as would be predicted by the additive fitness effects of both homozygous mutations (figs. 1C and 2B).
Although both the WHI2 and STE4 loci are on the right arm of Chromosome XV, they are 700 kb apart. We hypothesized that LOH events that occurred in populations with both WHI2/whi2-Q29* and STE4/ste4-E315* heterozygosities might involve short tracts of mitotic recombination that included the WHI2 locus but not the STE4 locus. We sequenced the STE4 locus in all 15 linked populations that had lost heterozygosity at the WHI2 locus. Only two populations remained heterozygous at STE4 locus. The other 13 populations were homozygous for both the partially dominant whi2-Q29* mutation and the overdominant ste4Q315* mutation (supplementary fig. S5, Supplementary Material online).
LOH of ste4Q315* Cannot Be Explained by Compensatory Mutations
Two possibilities could explain the observed LOH of ste4-E315*: either mutations at other loci on the right arm of Chromosome XV changed the net fitness effect of LOH or mutations elsewhere in the genome altered the fitness effect or the degree of dominance of either the ste4-E315* or the whi2-Q29* mutation. To test these possibilities we performed whole-genome sequencing on two clones each from 20 populations: seven that lost heterozygosity at both loci (whi2-Q29*/whi2-Q29*, ste4-E315*/ste4-E315*), two that lost heterozygosity at WHI2 but not STE4 in ste4-E315* linked populations (whi2-Q29*/whi2-Q29*, STE4/ste4-E315*), nine that lost heterozygosity in a STE4 wild-type background (whi2-Q29*/whi2-Q29*), and two control populations that are wild-type at both loci (supplementary fig. S5, Supplementary Material online). We identified 914 nuclear mutations normally distributed across the 40 clones (supplementary data set 1, Supplementary Material online, Shapiro–Wilk test, P = 0.36) with a mean of 30.7 mutations per clone. Twelve mutations were homozygous (not including WHI2 or STE4 alleles).
To identify putative de novo driver mutations on Chromosome XV that could explain the repeated occurrence of what should be a deleterious LOH event, we looked for homozygous nonsynonymous mutations on the right arm of Chromosome XV that are found in both clones (and thus were likely present before the LOH event). Of the 914 mutations, none meet these criteria, thus ruling out de novo evolution of linked beneficial mutations as an explanation for the repeated LOH of a the overdominant ste4-E315* mutation.
Next we looked for possible epistatic modifiers of STE4. Ninety-six genes either share Gene Ontology terms with STE4 or are known physical and/or genetic interactors with STE4 (supplementary table S2, Supplementary Material online). One of the of seven populations that lost heterozygosity of ste4-E315* acquired heterozygous missense mutations in two of these genes, STE7 and PTC2. None of the other sequenced populations contained mutations in any of these 96 genes. We also took an unbiased approach by searching for genes that were mutated in more than one of the populations that lost heterozygosity of ste4-E315*. However, of the 101 genes containing fixed (present in both sequenced clones) nonsynonymous mutations across the nine populations carrying an overdominant STE4 allele, none were mutated in more than one population. In contrast, five of 93 genes accruing nonsynonymous fixed mutations in unlinked populations are mutated in multiple populations. Unlinked populations are significantly enriched for multihit genes in this comparison (Fisher’s exact, P = 0.02).
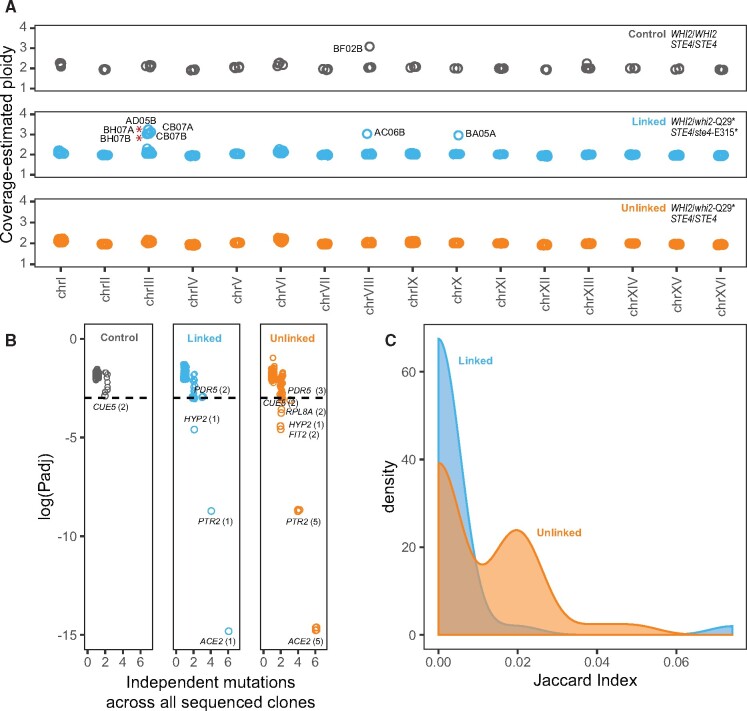
New point mutations cannot account for LOH in all populations. To explore other possible mechanisms of modifying or escaping the overdominance of ste4-E315* we looked for evidence of structural evolution in our sequenced populations. First, we verified that Chromosome XV read depth in all clones is consistent with genome-wide coverage (fig. 3A), indicating that LOH events were due to mitotic recombination and not chromosome loss. We next identified aneuploidies and copy number variants (CNVs) in each clone. We find three different chromosomal aneuploidies across six populations, five of which were initiated with a ste4-E315* mutation and one of which was a control population (fig. 3A). Among the ste4-E315* populations, trisomy-VIII and trisomy-X were found in individual clones of two populations that lost heterozygosity at both loci. Although Chromosome III aneuploidies were found in three ste4-E315* populations, only two of these populations lost heterozygosity at both whi2-Q29* and ste4-E315* and one lost heterozygosity at whi2-Q29* but retained heterozygosity at ste4-E315* (fig. 3A). We also identified two large CNVs on Chromosome III (supplementary table S3 and fig. S6, Supplementary Material online). An amplification of 93 kb on the right arm of Chromosome III, is found in both clones of a ste4-E315*-containing population in conjunction with a Chromosome III trisomy (fig. 3A). The other CNV, a 16-kb deletion on the right arm of Chromosome III, was detected in a wild-type STE4 population that lost heterozygosity at WHI2-Q29*.
Fig. 3.
Populations with linked partially dominant and overdominant mutations show less parallelism and more varied evolutionary outcomes. (A) Copy number of each chromosome based on median read depth in control populations (top, black), whi2-Q29* ste4-E315* populations (middle, blue), and whi2-Q29* STE4 populations (bottom, orange). Chromosome XV is diploid in all populations indicating that LOH is not due to chromosome loss. Trisomy of three different chromosomes was detected. Aneuploidy was observed in six total populations, five of which contained an overdominant STE4 mutation. Red asterisks indicate populations with Chromosome III trisomy that did not lose ste4-E315* heterozygosity. No aneuploidies were detected in genotypes with only whi2-Q29* mutations. (B) A probability of recurrence method (6) was used to identify genes receiving significantly more mutations than expected by chance. Points are jittered on both axes to show overlapping data. A threshold of P < 0.05 identifies seven genes as being targets of adaptive mutations in the 20 populations examined. Only five of the 21 total mutations in these genes occur in ste4-E315* linked populations. (C) Distribution of Jaccard Indices among pairwise comparisons of whi2-Q29* ste4-E315* and whi2-Q29* STE4 populations. The J distribution of linked populations is shifted significantly lower than the J distribution of unlinked populations (Wilcoxon rank-sum, Wlinked = 402, Wunlinked = 894, P < 0.001). (A–C) Points and lines are colored by genotype: (black: control, blue: whi2-Q29* ste4-E315*, orange: whi2-Q29* STE4).
There are no clear structural events shared by all populations that experienced LOH at an overdominant STE4 allele. Chromosome III is an apparent hotspot of structural evolution in this experiment—however, most populations that underwent LOH at an overdominant locus did not contain any Chromosome III copy number variations and one of the populations that did experience Chromosome III trisomy did not lose heterozygosity at ste4-E315*. Consequently, we cannot attribute LOH to Chromosome III copy number variation. Nonetheless, populations carrying both whi2-Q29* and ste4-E315* alleles were enriched for aneuploidies (Fisher’s exact, P = 0.03) relative to populations with only a whi2-Q29* allele, which were all found to be euploid and had no detectable CNVs.
Populations Carrying Linked Dominant and Overdominant Beneficial Mutations Show a Broad Range of Evolutionary Outcomes
Taken together, we find that ste4-E315* whi2-Q29* populations are significantly depleted for recurrently mutated genes and significantly enriched for aneuploidies, a standard signature of parallel evolution. This implies that populations seeded with an overdominant mutation telomeric to a partially dominant beneficial mutation may experience a greater range of possible evolutionary outcomes. To explore this further, we aggregated all 487 mutations to coding sequences (excluding synonymous mutations) across all populations to identify targets of adaptive mutations using a recurrence-based statistical method (supplementary data set 2, Supplementary Material online). We found that only five of 21 mutations to seven adaptive targets occurred in populations seeded with overdominant alleles. Since nine of the 20 sequenced populations carried an overdominant STE4 mutation, our null expectation would be that about half of driver mutations would be found in ste4-E315* populations. Instead, we find these populations to exhibit less parallelism at the gene level. We looked more closely at differences in parallelism in the set of 487 moderate to high effect coding sequence mutations by calculating the Jaccard Index (J) for all pairwise combinations of populations. The distribution of J is significantly left shifted in comparisons between ste4-E315*-whi2-Q29* populations relative to comparisons between STE4-whi2-Q29* populations (Wilcoxon rank-sum, Wlinked = 402, Wunlinked = 894, P < 0.001). Populations of both genotypes did not significantly differ in the total number of mutations accrued (t(15.734) = −1.68, P = 0.112, supplementary fig. S7, Supplementary Material online). However, because populations lacking an overdominant STE4 mutation tended to accrue slightly more mutations (49.4 12.8) than those carrying ste4-E315* (39.9 11.2), we used a multiple regression to show that starting genotype (P = 0.008), but not number of mutations (P = 0.211), is a significant predictor of J (F(2,69) = 3.89, P = 0.025). Therefore, we find genotypes with an overdominant STE4 allele to be evolving more divergently at the sequence level than genotypes that are otherwise identical but lack an overdominant STE4 allele.
Discussion
Most beneficial mutations that arise in asexually evolving diploid populations are heterozygous and are at least partially dominant. Here, we show that the degree of dominance can constrain adaptive evolution at linked loci. Mechanistically these constraints arise due to conflicting effects that LOH has on partially dominant and overdominant beneficial mutations. Given the fitness effects of LOH of the whi2-Q29* and the ste4-E315* alleles we expect the net effect of LOH over both of these loci to be deleterious. Suppression of LOH at linked loci could occur, however, if the net fitness effect is neutral (or even weakly beneficial) due to clonal interference from more-fit beneficial mutations.
Given that LOH conversion tracts frequently extend the full length of a chromosome arm (Barbera and Petes 2006; Lee et al. 2009; Dutta et al. 2021), the linked effects we observe will hold true for all chromosomes in asexually evolving diploid populations, particularly in the early stages of adaptation when overdominant beneficial mutations will be most frequent (Manna et al. 2011; Sellis et al. 2011). The strength of the linked effects, however, will vary depending on local rates of mitotic recombination and the length of repair tracts as well as the distribution of mutational effects on fitness—and the degree of dominance of those mutations—along a chromosome. Comprehensive analyses of gene deletions in yeast reveal few underdominant or overdominant deletions (Agrawal and Whitlock 2011), however, this may not be indicative of the distribution of dominance nonloss of function mutations. Indeed, in our evolution experiments we observe examples of overdominant, underdominant, and recessive beneficial mutations in STE4.
Most theory addressing mutational dominance and constraint focuses on the consequences of recessiveness, namely the constraints imposed by Haldane’s sieve (Charlesworth and Charlesworth 1999; Orr and Betancourt 2001) and the load imposed by recessive deleterious mutations (Charlesworth and Charlesworth 1999; Chasnov 2000). Underdominance is most frequently invoked as a cause of reproductive isolation (Barton and De Cara 2009), but our findings suggest an underappreciated role as an evolutionary constraint. For example, though a homozygous deletion of STE4 would be beneficial in a MATa/a strain, this mutation is underdominant and would be able to fix only in extremely small or fragmented populations (Newberry et al. 2016).
Two of the evolved STE4 mutations we identified demonstrate a strong degree of overdominance when engineered into an ancestral background. Recent theoretical examination of adaptation in diploids has renewed interest in the significance of overdominant mutations in adaptation and suggested overdominant polymorphisms may be a frequent mode of adaptation (Manna et al. 2011; Sellis et al. 2011). These models find that when selection on a trait is stabilizing, strong effect heterozygous mutations that overshoot the fitness optimum as homozygotes should be common. Experimental evolution provides a way to empirically test this prediction. The few examples of overdominance arising de novo in laboratory evolution include amplifications of glucose transporter genes in glucose-limited chemostat populations (Sellis et al. 2016). Overdominance of a CNV is well explained by an “overshoot” of an optimal gene copy number, and hexose transporter amplifications have been previously shown to exhibit sign epistasis with mutations that upregulate their expression (Kvitek and Sherlock 2011).
Mitotic recombination resulting in LOH is a common and important mechanism of adaptation in laboratory evolving diploid yeast (Gerstein et al. 2014; Smukowski Heil et al. 2017; Fisher et al. 2018; James et al. 2019). Most of these reported instances of LOH in asexual yeast adaptation involve a large conversion tract that runs from the break point to the telomere. This means that there is effective linkage between loci that are kilobases apart. Overdominant beneficial mutations will therefore impose constraint on mitotic recombination along the full length of a chromosome arm. We tested this using a partially dominant beneficial mutation at the WHI2 locus and an overdominant mutation at the STE4 locus, both on the right arm of Chromosome XV. Linked populations must either lose heterozygosity at the WHI2 locus while maintaining heterozygosity at the distal STE4 locus or suffer the fitness cost loss of LOH at the STE4 locus.
Although our results support the hypothesis that overdominant mutations restrict adaptive LOH at linked loci, after the first few hundred generations, LOH is repeatedly observed in populations bearing an overdominant STE4 allele. Although two of populations lost heterozygosity only at WHI2 (presumably through gene conversion rather than break-induced replication), 13 lost heterozygosity at both loci. One possible explanation is that de novo mutations arose that mitigated the deleterious effect of LOH at the STE4 locus. Whole-genome sequencing identified only one population that acquired mutations in known interactors of STE4. Therefore, while modifying mutations may occur, there are no obvious candidates to explain all of the instances of LOH at STE4. We do observe an enrichment for aneuploidies in the linked populations, though the specific changes we found (gains of additional copies of Chromosomes III and VIII) have been observed before (Fisher et al. 2018).
Analysis of genome sequence evolution shows that unlinked whi2-Q28* populations accrue more mutations in common targets of selection, including ACE2 and PTR2, which have been reported previously (Fisher et al. 2018). In contrast, the whi2-Q28*/ste4-Q315* linked populations show a wider range of evolutionary outcomes at the genome sequence level. The lack of known targets of selection suggests that the ste4-Q315* mutation is affecting the evolution of this population beyond constraining LOH at WHI2. That no clear pattern emerges from the genome-sequence evolution in the linked populations shows that there is not a single strong suppressor ste4-Q315* mutation. Rather, it suggests that there are multiple ways to (at least partially) compensate for the deleterious homozygous ste4-Q315* mutation. The difference in the modes of adaptation between two nearly identical genomes (differing only by a single heterozygous mutation), indicate that small changes in the genome can introduce constraints on genome evolution and influence evolutionary outcomes.
Materials and Methods
Analysis of Evolved Mutations in STE4
We previously identified STE4 mutations from whole-genome sequencing data reported for 40 haploid (Lang et al. 2013) and 46 autodiploid (Fisher et al. 2018) yeast populations. The mutational target window of STE4 (in bp) was calculated for both haploids and autodiploids. The probability of all mutations occurring in the observed window was calculated separately for haploids and autodiploids using a one-sample proportions test.
Homology modeling of Ste4p was performed automatically on the SWISS-MODEL web server to visualize the positions of mutated residues. The best scoring model was based on the structure of G protein subunit beta (Gnb1) from Rattus norvegicus (41.96% identity, 0.63 GMQE, −3.31 QMEAN, PDB ID: 6CMO). Visualization of the Ste4p model was done in PyMOL Molecular Graphics System, version 2.3.0. This structure does not contain a 33 amino acid yeast-specific insertion.
Construction of Evolved Mutation and STE4 Deletion Strains
Strains used in this study are described in supplementary table S4, Supplementary Material online. Reconstruction experiments were performed in the same W303 ancestral background (yGIL432; MATa, ura3Δ::pFUS1-yEVenus, ade2-1, his3-11,15, leu2-3,112, trp1-1, CAN1, bar1Δ::ADE2, hmlαΔ::LEU2, GPA1::NatMX). Briefly, deletion strains were generated by integrating the ste4Δ::KanMX locus from the deletion collection. Crosses of strains carrying null alleles were performed by first transforming with a STE4-expressing plasmid from the MoBY ORF plasmid collection to compliment ste4Δ. Three evolved STE4 alleles were selected for reconstruction, 81ΔT (S261fs), G943T (E315*), and G935A (R312Q). Alleles were reconstructed in yGIL432 using CRISPR-Cas9 alleles swaps. We constructed constitutive Cas9-expressing plasmids starting from pML104 (Addgene 67638) expressing a STE4-specific guide RNAs (5′-CTACCCCTAC TTATATGGCA-3′) and cotransformed the plasmid along with a 500-bp linear repair template (gBlock, IDT) encoding the one of three evolved alleles as well as a synonymous C954A PAM site substitution. A strain containing just the synonymous PAM site was also isolated to verify neutrality (supplementary fig. S3, Supplementary Material online). To minimize variation due to transformation and Cas9 activity, one successful transformant per allele was backcrossed twice and the resulting diploid was sporulated and tetrad dissected. For each allele, spores were genotyped at STE4 and intercrossed to generate heterozygous and homozygous mutants. Crosses of strains carrying evolved ste4 alleles were performed by first transforming with a plasmid from the MoBY ORF plasmid collection to compliment STE4. Mutants carrying an evolved whi2-C85T (Q29*) allele were generated in identical fashion with two exceptions. The evolved substitution is within the WHI2 gRNA used (5′-ACAGTACGAA GGTAACGAGG-3′), and therefore no synonymous mutation was introduced to eliminate Cas9 activity. A correct whi2-Q29* was backcrossed once and intercrossed to produce homozygotes and heterozygotes. All diploid genotypes were converted to MATa/a as described above. We also generated strains containing dominant drug cassettes tightly linked to the WHI2 locus using CRISPR. We inserted either HphMX or KanMX 220 bp downstream of WHI2 or whi2-Q29* by transforming with the same gRNA (5′-ATCCCCTTCT GCAAATAACG-3′) and Cas9-expressing plasmid and cotransforming with linear drug cassettes flanked by 40 bp of homology to the targeted region. Successful transformants were then backcrossed to either a wild-type background or a ste4-G943T (described above) mutant. Crosses were sporulated and spores were selected in which drug-marker tagged mutant and wild-type WHI2 alleles are present on the same chromosome as both mutant and wild-type STE4. Correct spores were crossed to generate three genotypes: WHI2::HphMX/WHI2::KanMx STE4/STE4, WHI2::HphMX/whi2-Q29*::KanMx STE4/ste4-E315*, and WHI2::HphMX/whi2-Q29*::KanMx ste4-E315*/STE4. All three genotypes were converted to MATa/a as described above. Eight replicate MATa/a colonies were picked for each mating-type conversion to be used for downstream analysis. Given the individual fitness effects of whi2-Q28* (3.4 ± 0.2% or 5.1 ± 0.3% when heterozygous or homozygous, respectively) and ste4-Q315* (1.2 ± 0.1% or −2.4 ± 0.1% when heterozygous or homozygous, respectively), we expect the net effect of LOH over both of these loci to be deleterious. This is a safe assumption since WHI2 and STE4 are not known to interact, nor is there evidence of a genetic interaction based on co-occurrence of mutations (Fisher et al. 2019).
Fitness Assays
We measured the effects of complete gene deletions and evolved STE4 mutations on fitness using competitive fitness assays as previously reported (Buskirk et al. 2017). Briefly, query cultures were mixed 1:1 with a ploidy and mating-type matched fluorescently labeled ancestral strain. There is no fitness effect of the fluorescent reporter within our error of detection (∼0.5%). Cocultures were propagated in a 96-well plate in an identical to the evolution experiment in in which the variants arose for 50 generations. Saturated cultures were sampled for flow cytometry at ten-generation intervals. Flow cytometry data were analyzed with FlowJo 10.3. Selective coefficients were calculated as the slope of the best-fit line of the natural log of the ratio between query and reference strains against time.
Two technical replicates each of eight biological replicates were averaged for analysis of all MATa/a genotypes and all deletion mutants. Evolved mutations in a haploid background were averaged from four technical replicates of a single clone. Fitness data for haploid and diploid genotypes were analyzed independently using a one-way analysis of variance ANOVA. Post hoc comparisons using the Tukey test were carried out to identify genotypes with significantly different fitness than wild-type controls.
Short-Term Evolution Experiment
We examined the effect of evolved ste4 alleles on likelihood of LOH at a linked locus, WHI2. We first validated the homozygous and heterozygous fitness benefits of an evolved allele, whi2-C85T (Q29*), via mutant reconstruction and fitness assays as described above. We then generated strains containing dominant drug cassettes tightly linked to the WHI2 locus to investigate the effect of STE4 linkage on LOH along the right arm of Chromosome XV (supplementary methods, Supplementary Material online).
Three strains (WHI2::HphMX/WHI2::KanMx STE4/STE4, WHI2::HphMX/whi2-Q29*::KanMx STE4/ste4-E315*, and WHI2::HphMX/whi2-Q29*::KanMx ste4-E315*/STE4) were grown in 10 ml overnight cultures in YPD with 0.4 mg/ml G418 and 0.6 mg/ml Hygromycin B. Saturated cultures were diluted 1:1,000 to initiate 287 128 μl cultures across three 96-well plates. Plates were incubated unshaken at 30 °C and propagated daily in an identical fashion to the original evolution experiment in which the mutations arose (Lang et al. 2013). After 500 generations heterozygosity was assayed by spotting 2 μl (∼5,000 cells) to double drug plates (YPD with 0.4 mg/ml G418, 0.6 mg/ml Hygromycin B) and to both single drug agar plates. Plates were inspected for speckled spots (indicating homozygous genotypes in ≥50% the population) and absence of growth (indicating LOH sweeps). We compared the number of populations with evidence of LOH polymorphism or sweeps between genotypes using a Fisher’s exact test with a Bonferroni correction for multiple comparisons.
Whole-Genome Sequencing and Analysis
We sequenced nine of 15 linked populations and nine of 27 unlinked populations in which a homozygous WHI2 allele genotype fixed. The nine populations for each group were chosen to be representative of the spectrum of dynamics observed in the evolution experiment (i.e., some populations that underwent LOH early in the experiment and some that underwent LOH late). Each population was struck to singles on YPD to obtain two clones for sequencing. Clones were grown overnight in 5 ml YPD and then frozen as cell pellets at −20 °C. Genomic DNA was isolated from frozen pellets via phenol–chloroform extraction and ethanol precipitation. Total genomic DNA was used in a Nextera library preparation as described previously (Buskirk et al. 2017). All individually barcoded clones were pooled and paired-end sequenced on NovaSeq 6000 sequencer at the Genomics Core Facility at the Lewis-Sigler Institute for Integrative Genomics, Princeton University.
Raw sequence data were concatenated and then demultiplexed using a custom python script from L. Parsons (Princeton University). Adapter sequences were removed using Trimmomatic (Bolger et al. 2014). Reads were then aligned to a customized W303 genome using BWA v. 0.7.7 (Li and Durbin 2009) and variants were called using FreeBayes v0.9.21-24-381. VCFtools was used to filter variants common to all genomes. Remaining mutations were annotated using a strain-background customized annotation file (Matheson et al. 2017). All putative evolved mutations were confirmed manually using IGV (Thorvaldsdóttir et al. 2013).
Each genome was independently examined for structural variants using Samtools depth (Li et al. 2009). Aneuploidies were detected by dividing median chromosome coverage by median genome-wide coverage for each chromosome. CNVs were similarly detected using a sliding 1-kb window across each chromosome. Putative CNVs identified were confirmed by visual inspection of chromosome coverage plots.
A list of 92 candidate genes for the modification of overdominance at STE4 was curated by concatenating a list of all known STE4 interactors and all genes annotated to the GO term “pheromone dependent signal transduction involved in conjugation with cellular fusion” and all genes annotated to children of this GO term. The above GO term was selected out of the 17 terms assigned to STE4 because changes to pheromone-induced signaling is thought to be the cellular basis of the fitness effect of STE4 mutations (Lang et al. 2009). These searches were performed in YeastMine (Balakrishnan et al. 2012).
Identification of Genic Targets of Selection and Quantification of Parallelism
To identify parallel targets of selection we first removed all nonprotein coding and synonymous mutations to improve our signal. We then identified parallel targets of selection as described previously (Fisher et al. 2018). Briefly, we calculate the expected number of mutations for each gene, σ, using the Poisson distribution weighted by the length (L) of the gene in base-pairs:
| (1) |
where M is the total number of coding sequence mutations in the data set. The probability of observing k mutations in gene σ is therefore:
| (2) |
We use expression (eq. 2) to calculate the P value for the observed number of mutations in each gene. We then applied a Benjamin–Hochberg post hoc adjustment to correct for multiple hypothesis testing.
Parallelism was quantified using the Jaccard Index (Bailey et al. 2015), which calculates the similarity between two sets of mutated genes by quantifying the overlap of their union. Again, nonprotein coding and synonymous mutations were excluded to increase the signal of adaptive parallelism. A value of J was calculated for pairwise combinations of populations. The distribution of J for all pairs of linked populations was compared with the distribution of J for all pairs of unlinked populations using a Wilcoxon rank-sum test with continuity correction.
Statistical Analyses
All statistical analyses reported were performed using tools in the R Stats package in R v.3.6.2. All plots were produced in R using the ggplot2 package (Wickham 2016) except supplementary figures S5 and S7, Supplementary Material online, which were produced using base R plotting.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank members of the Lang Lab for comments on this manuscript. This work was supported by the National Institutes of Health (Grant No. 1R01GM127420 to G.I.L.). K.J.F. is a Morgridge Metabolism Interdisciplinary Fellow of the Morgridge Institute for Research.
Author Contributions
K.J.F. and G.I.L. conceived of the project and designed experiments. K.J.F. and R.C.V. performed experiments. K.J.F. and R.C.V. analyzed the data. K.J.F. and G.I.L. wrote the manuscript.
Data Availability
The raw short-read sequencing data reported in this article have been deposited in the NCBI BioProject database (accession no. PRJNA634573).
Literature Cited
- Agrawal AF, Whitlock MC.. 2011. Inferences about the distribution of dominance drawn from yeast gene knockout data. Genetics 187(2):553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SF, Rodrigue N, Kassen R.. 2015. The effect of selection environment on the probability of parallel evolution. Mol Biol Evol. 32(6):1436–1448. [DOI] [PubMed] [Google Scholar]
- Balakrishnan R, et al. 2012. YeastMine—an integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit. Database (Oxford). 2012:bar062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera MA, Petes TD.. 2006. Selection and analysis of spontaneous reciprocal mitotic cross-overs in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 103(34):12819–12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH, De Cara MAR.. 2009. The evolution of strong reproductive isolation. Evolution 63(5):1171–1190. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskirk SW, Peace RE, Lang GI.. 2017. Hitchhiking and epistasis give rise to cohort dynamics in adapting populations. Proc Natl Acad Sci U S A. 114(31):8330–8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D.. 1999. The genetic basis of inbreeding depression. Genet Res. 74(3):329–340. [DOI] [PubMed] [Google Scholar]
- Chasnov JR.2000. Mutation-selection balance, dominance and the maintenance of sex. Genetics 156(3):1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, Hall MD.. 2018. Genetic constraints on adaptation: a theoretical primer for the genomics era. Ann N Y Acad Sci. 1422(1):65–87. [DOI] [PubMed] [Google Scholar]
- Dutta A, Dutreux F, Schacherer J.. 2021. Loss of heterozygosity results in rapid but variable genome homogenization across yeast genetic backgrounds. Elife 10:e70339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher KJ, Buskirk SW, Vignogna RC, Marad DA, Lang GI.. 2018. Adaptive genome duplication affects patterns of molecular evolution in Saccharomyces cerevisiae. PLoS Genet. 14(5):e1007396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher KJ, Kryazhimskiy S, Lang GI.. 2019. Detecting genetic interactions using parallel evolution in experimental populations. Philos Trans R Soc Lond B Biol Sci. 374(1777):20180237–20180237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA.1928. The possible modification of the response of the wild type to recurrent mutations. Am Nat. 62(679):115–126. [Google Scholar]
- Gerstein AC, Kuzmin A, Otto SP.. 2014. Loss-of-heterozygosity facilitates passage through Haldane’s sieve for Saccharomyces cerevisiae undergoing adaptation. Nat Commun. 5:1–9. [DOI] [PubMed] [Google Scholar]
- Goudie F, Allsopp MH, Oldroyd BP.. 2014. Selection on overdominant genes maintains heterozygosity along multiple chromosomes in a clonal lineage of honey bee. Evolution 68(1):125–136. [DOI] [PubMed] [Google Scholar]
- Haldane JBS.1924. A mathematical theory of natural and artificial selection. Part II: the influence of partial self-fertilisation, inbreeding, assortative mating, and selective fertilisation on the composition of Mendelian populations, and on natural selection. Biol Rev. 1(3):158–163. [Google Scholar]
- Hedrick PW.2012. What is the evidence for heterozygote advantage selection? Trends Ecol Evol. 27(12):698–704. [DOI] [PubMed] [Google Scholar]
- James TY, et al. 2019. Adaptation by loss of heterozygosity in Saccharomyces cerevisiae clones under divergent selection. Genetics 213(2):665–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitek DJ, Sherlock G.. 2011. Reciprocal sign epistasis between frequently experimentally evolved adaptive mutations causes a rugged fitness landscape. PLoS Genet. 7(4):e1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, et al. 2013. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500(7464):571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Murray AW, Botstein D.. 2009. The cost of gene expression underlies a fitness trade-off in yeast. Proc Natl Acad Sci U S A. 106(14):5755–5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, et al. 2009. A fine-structure map of spontaneous mitotic crossovers in the yeast Saccharomyces cerevisiae. PLoS Genet. 5(3):e1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu JY, Chang SL, Chao JC, Woods LC, McDonald MJ.. 2020. Sex alters molecular evolution in diploid experimental populations of S. cerevisiae. Nat Ecol Evol. 4(3):453–460. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna F, Martin G, Lenormand T.. 2011. Fitness landscapes: an alternative theory for the dominance of mutation. Genetics 189(3):923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marad DA, Buskirk SW, Lang GI.. 2018. Altered access to beneficial mutations slows adaptation and biases fixed mutations in diploids. Nat Ecol Evol. 2(5):882–889. [DOI] [PubMed] [Google Scholar]
- Maruyama T, Nei M.. 1981. Genetic variability maintained by mutation and overdominant selection in finite populations. Genetics 98(2):441–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson K, Parsons L, Gammie A.. 2017. Whole-genome sequence and variant analysis of W303, a widely-used strain of Saccharomyces cerevisiae. G3 (Bethesda) 7(7):2219–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberry MG, McCandlish DM, Plotkin JB.. 2016. Assortative mating can impede or facilitate fixation of underdominant alleles. Theor Popul Biol. 112:14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA, Betancourt AJ.. 2001. Haldane’s sieve and adaptation from the standing genetic variation. Genetics 157(2):875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA, Otto SP.. 1994. Does diploidy increase the rate of adaptation? Genetics 136(4):1475–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter J, et al. 2018. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556(7701):339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellis D, Callahan BJ, Petrov DA, Messer PW.. 2011. Heterozygote advantage as a natural consequence of adaptation in diploids. Proc Natl Acad Sci U S A. 108(51):20666–20671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellis D, Kvitek DJ, Dunn B, Sherlock G, Petrov DA.. 2016. Heterozygote advantage is a common outcome of adaptation in Saccharomyces cerevisiae. Genetics 203(3):1401–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smukowski Heil CS, et al. 2017. Loss of heterozygosity drives adaptation in hybrid yeast. Mol Biol Evol. 34(7):1596–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondek J, Bohm A, Lambright DG, Hamm HE, Sigler PB.. 1996. Crystal structure of a GA protein βγdimer at 2.1 Å resolution. Nature 379(6563):369–374. [DOI] [PubMed] [Google Scholar]
- Sui Y, et al. 2020. Genome-wide mapping of spontaneous genetic alterations in diploid yeast cells. Proc Natl Acad Sci U S A. 117(45):28191–28200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Rothstein R, Lisby M.. 2014. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 198(3):795–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulkin M, Bierne N, David P.. 2010. Heterozygosity-fitness correlations: a time for reappraisal. Evolution 64(5):1202–1217. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP.. 2013. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14(2):178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H.2016. ggplot2: elegant graphics for data analysis. New York: Springer. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw short-read sequencing data reported in this article have been deposited in the NCBI BioProject database (accession no. PRJNA634573).