Abstract
The transmembrane α-helices of membrane proteins are in general highly hydrophobic, and they enter the lipid bilayer through a lateral gate in the Sec61 translocon. However, some transmembrane α-helices are less hydrophobic and form membrane channels or substrate-binding pockets. Insertion of these amphipathic transmembrane α-helices into the membrane requires the specific membrane-embedded insertase called the endoplasmic reticulum membrane complex (EMC), which is a multi-subunit chaperone distinct from the GET insertase complex. Four recent cryo-EM studies on the eukaryotic EMC have revealed their remarkable architectural conservation from yeast to human; a general consensus on the substrate transmembrane helix-binding pocket; and the evolutionary link to the prokaryotic insertases of the tail-anchored membrane proteins. These structures provide a solid framework for future mechanistic investigation.
Keywords: Cryo-EM, structural biology, membrane protein folding, insertase, membrane protein biogenesis
Graphical Abstract
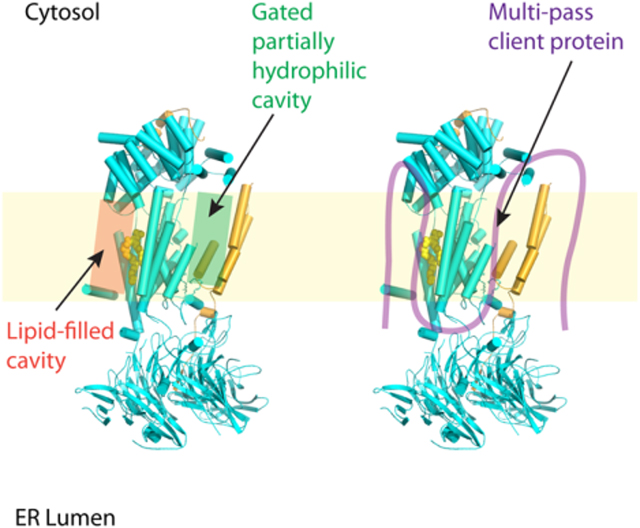
EMC has eight subunits in yeast and nine in human. EMC is a dual-function complex that inserts transmembrane domain of the tail-anchored proteins and chaperones biogenesis of multi-pass transmembrane proteins containing partially hydrophilic transmembrane helices. The EMC insertase and holdase activities are separable. The EMC structure contains two sizable cavities — a gated partially hydrophilic cavity and a lipid-filled hydrophobic cavity. Both cavities are involved in the EMC functions.
INTRODUCTION
Membrane proteins participate in essential cell activities such as signal transduction, molecular transport, and immune recognition [1]. About 20–30% of all proteins in eukaryotes are membrane proteins, which contain hydrophobic transmembrane domains (TMDs) that are embedded in the lipid bilayer. Membrane proteins are either co-translationally or post-translationally inserted into the endoplasmic reticulum (ER) membrane before reaching their final destinations [2]. In additional to the translocon, several conserved membrane protein chaperones are involved in the transmembrane domain insertion and/or folding processes, including the transmembrane recognition complex (TRC in mammalian, or Get [guided entry of tail-anchored proteins] in yeast), the prokaryotic YidC [3, 4], the eukaryotic EMC [5–10], the signal recognition particle (SRP)-independent targeting proteins Snd1–3 [11], the TMCO1 complex [12], and the PAT complex [13].
The GET/TRC pathway inserts about half of the tail-anchored (TA) membrane proteins encoded in the human genome. TA proteins contain a single TMD at their C-terminal [5–7, 14]. In this pathway, TA proteins are first captured by Sgt2 (SGTA in mammals) and then passed on to cytoplasmic Get3 (TRC40 in mammals) via the Get4/5 scaffold and are subsequently targeted to the ER membrane with the help of the Get1/Get2 insertase (WRB/CAML in mammals). However, how the other half of the TA proteins are inserted into ER membrane has been unclear for a long time. The EMC is a conserved TMD insertase identified in recent years [15, 16]. It mediates the insertion of a number of TA proteins [9], and more importantly, chaperones the biogenesis of many multi-pass membrane proteins [8, 10]. Like EMC, YidC also functions in both TA insertion and membrane protein folding [3, 4].
The EMC was originally discovered by the Jonathan Weissman lab in 2009 as an ER multi-protein complex that influences membrane protein quality control [17]. Subsequent studies suggested that the EMC is involved in a diverse array of cellular functions including biogenesis, quality control, and the trafficking of many membrane proteins [18–25]. In 2018, the Manu Hegde lab was the first to identify the exact function of EMC as a TMD insertase [9, 10]. Using functional studies with in vivo knockout strains and in vitro reconstituted liposomes, they elegantly demonstrated that EMC is a TMD insertase for both TA and multi-pass membrane proteins, particularly those containing less hydrophobic and partially hydrophilic TMDs. The mammalian EMC is a nine-subunit complex (EMC1–7, either EMC8 or EMC9, and EMC10) [22]. The Saccharomyces cerevisiae EMC was first reported to have six subunits, Emc1–6. Two additional proteins, Sop4p and Ydr056cp, were co-purified with Emc1–6 and were noted to be homologous to the mammalian EMC7 and EMC10 by bioinformatic analysis [17, 26]. A previous bioinformatic analysis suggested that Emc3 is a member of the evolutionarily conserved Oxa1/Alb3/YidC family, which includes the eukaryotic insertase Get1 and the prokaryotic insertase YidC [27]. Therefore, Emc3 is likely the catalytic subunit of the EMC. The exact functions of other EMC component proteins are unclear. Most recently, four labs reported high-resolution cryo-EM structures of the yeast and human EMCs, providing the first structural insights into their molecular mechanisms (Fig. 1A–C, Table 1) [28–31]. In this review, we describe structural features of and a plausible TMD insertion mechanism for the EMC.
Figure 1. Comparison of the cryo-EM structures of yeast and human EMCs.
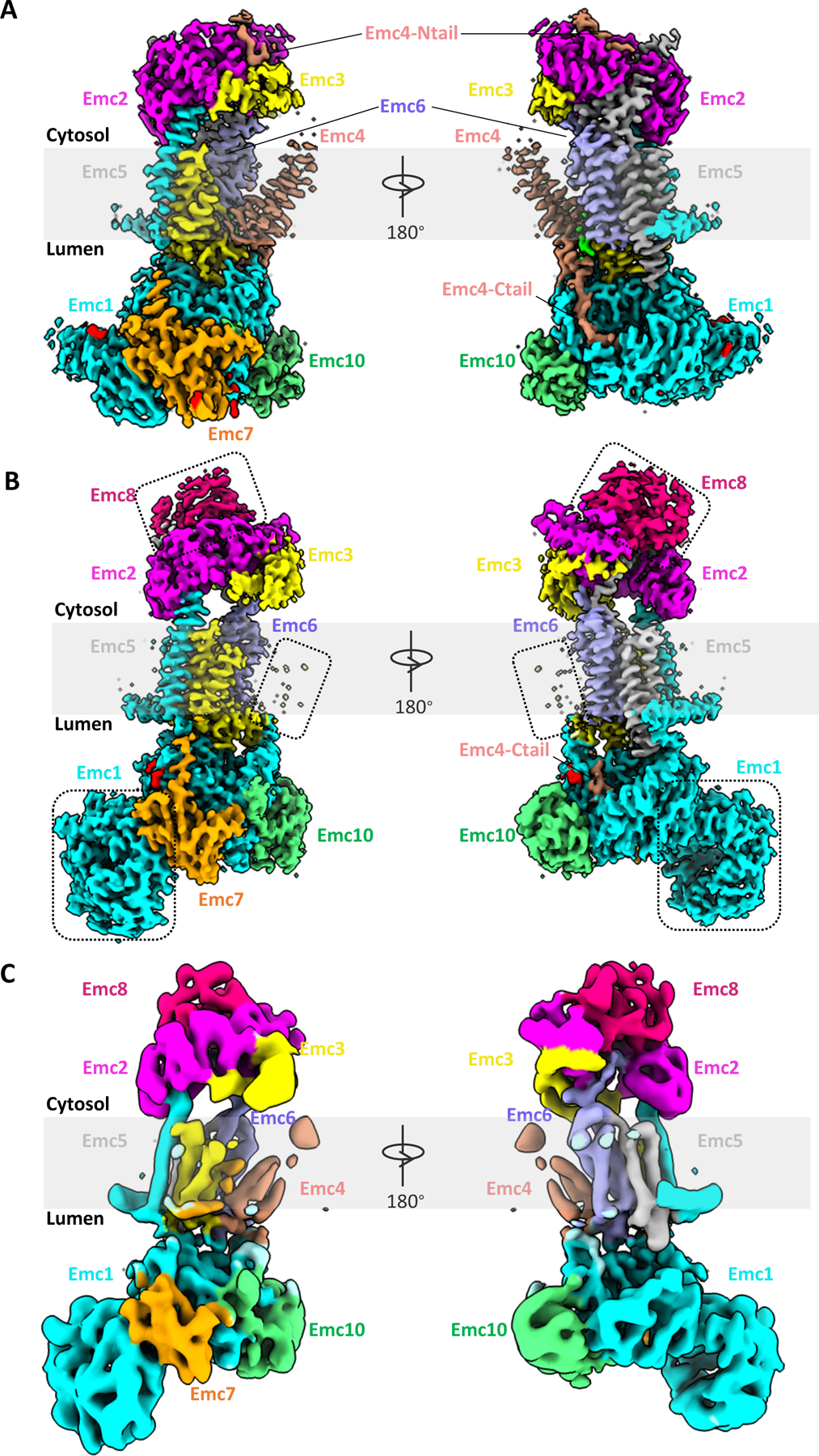
(A) Cryo-EM 3D map of the yeast EMC at 3.0-Å overall resolution in front and back views, as viewed from within membrane plane (EMD-21587). (B) Cryo-EM 3D map of the human EMC at 3.4-Å overall resolution (EMD-21929). Dotted rectangles mark unique features in the human EMC: metazoan-specific EMC8 (top), disordered TMD of EMC4 (middle), and NTD1 of EMC1 (bottom). (C) Cryo-EM 3D map of the human EMC at 6.4-Å overall resolution (EMD-11058).
Table 1.
List of recent reported structures of the yeast and human EMC complexes.
| Structure | Organism | Detergent/Nanodisk | Resolution (Å) | EMDB ID | PDB ID | References |
|---|---|---|---|---|---|---|
| 1 | Human | Nanodisk | 3.4 | EMD-21929 | 6WW7 | [30] |
| 2 | Yeast (S. cerevisiae) | Digitonin | 3.0 | EMD-21587 | 6WB9 | [29] |
| 3 | Human | LMNG | 6.4 | EMD-11058 | 6Z3W | [28] |
| 4 | Yeast (S. cerevisiae) | Nanodisk | 3.2 | EMD-23003 | 7KRA | [31] |
| Yeast (S. cerevisiae) | DDM | 4.3 | EMD-23033 | 7KTX | ||
| Human | Nanodisk | 3.4 | EMD-11732 | 7ADO | ||
| Human | GDN | 3.6 | EMD-11733 | 7ADP |
The eukaryotic EMC structure is highly conserved from yeast to humans
The cryo-EM structure of the S. cerevisiae EMC at resolution of 3.0 Å was derived from an endogenous sample purified using the detergent digitonin (Fig. 1A) [29]. In contrast, the human EMC complex sample was purified from a stable cell line expressing one tagged EMC subunit. The cryo-EM structures of the human EMC were determined by two groups at a resolution of 3.4 Å reconstituted in lipid nanodisks and at 6.4 Å in the detergent lauryl maltose neopentyl glycol (LMNG) (Fig. 1B–C) [28, 30]. The structures of the yeast and human EMCs are largely similar (Fig. 2A–B). Therefore, we will mainly focus on the yeast EMC structure, which had the highest resolution, but we will also describe unique features of the human EMC. During the preparation of this review, another group reported two cryo-EM structures of the yeast EMC at 3.2-Å resolution when reconstituted in lipid nanodisks and at 4.3-Å in DDM, as well as two structures of human EMC at 3.4-Å resolution reconstituted in lipid nanodisks and at 3.6-Å in the detergent glyco-diosgenin (GDN) [31] (Table 1). These structures are mostly identical to the earlier reported structures.
Figure 2. The atomic models of yeast and human EMCs shown in cartoons.
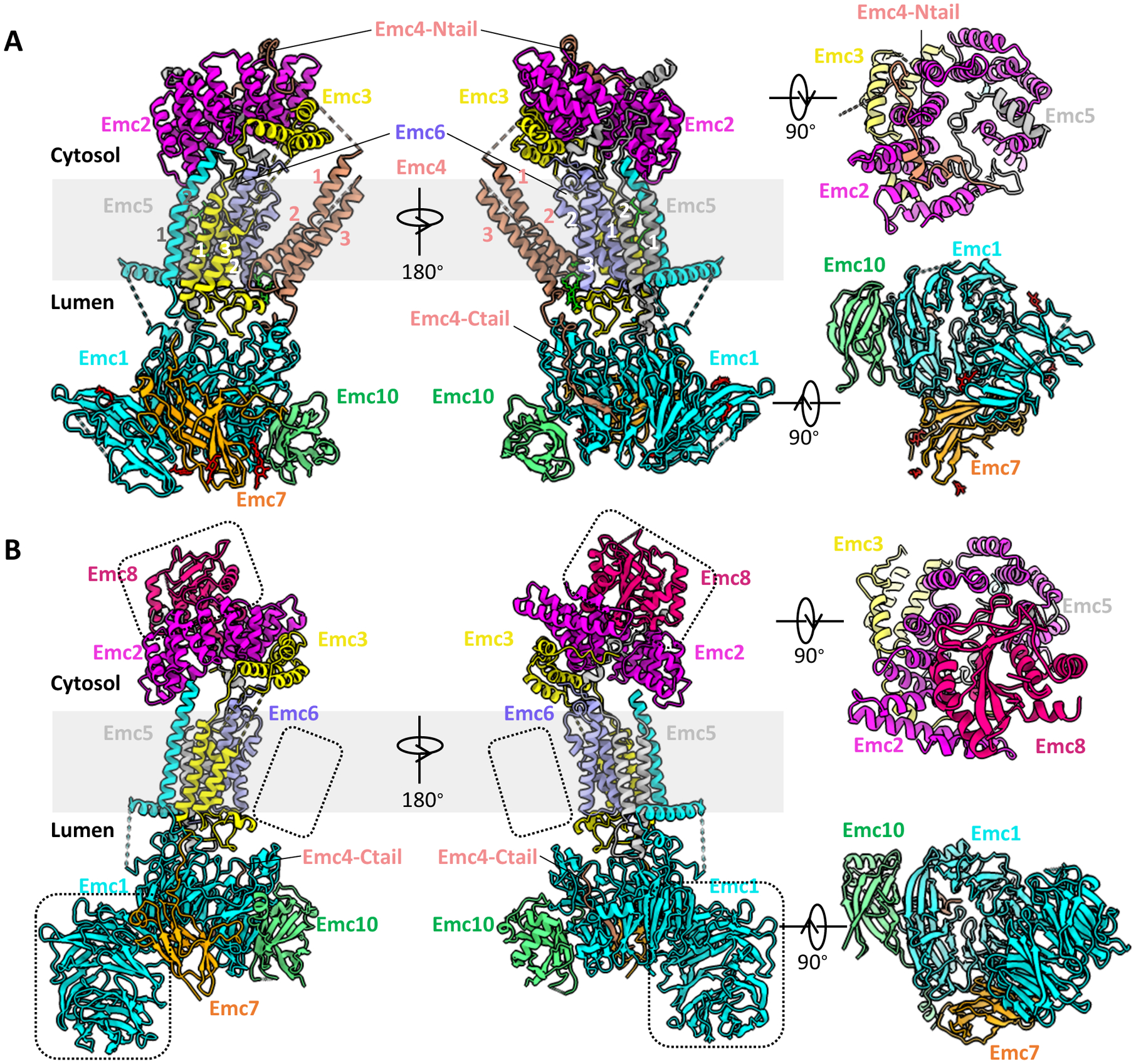
(A) Structure of the yeast EMC (PDB ID 6WB9). (B) Structure of human EMC (PDB ID 6WW7). In each panel, a front (left), a side (middle), a top (top right), and a bottom view (bottom right) are provided. Dotted black squares in panel B highlight unique features of the human EMC: EMC8 (top), disordered TMD of EMC4 (middle), and NTD1 of EMC1 (bottom).
In the yeast structure, most regions of all eight EMC subunits (Emc1–7 and Emc10) are unambiguously resolved (Figs. 1A, 2A). The overall structure is approximately 160 Å tall and 80 Å wide. It has a three-tiered architecture consisting of a transmembrane region, a large lumenal region, and a smaller cytosolic region. Among them, five subunits (Emc1 and Emc3–6) are transmembrane proteins with a total of 12 TMHs, and three subunits (Emc2, Emc7, and Emc10) are aqueous proteins (Fig. 2A).
In the transmembrane region, the putative catalytic subunit Emc3 is centrally located and is surrounded by other subunits (Fig. 2A). The TMHs of Emc1, Emc5 and Emc6 pack tightly against Emc3. A horizontal helix of Emc1 is half embedded in the lumenal side of ER membrane and may stabilize the transmembrane region of EMC. In contrast, the TMHs of Emc4 tilt away, creating a large cavity that is open to both cytosol and lipid bilayer. This cavity is surrounded by Emc3, Emc4, and Emc6, and it is probably the substrate binding site. Notably, the Emc4 TMHs appear to have a partial swinging movement like that of a pendulum, suggesting that the cavity size is expandable to accommodate a substrate TMD. In the human structures, the region corresponding to the Emc4 TMHs is more flexible (Figs. 1B–C, 2B), leading to some uncertainty in assignment of the three TMHs, as discussed below.
The lumenal region of the yeast EMC is mainly formed by Emc1, Emc7, Emc10, and a C-terminal loop of Emc4 (Fig. 2A). The large soluble domain of Emc1 can be divided into two subdomains, N-terminal domain 1 (NTD1) and NTD2. The NTD2 is an eight-bladed β-propeller, a typical tryptophan–aspartic acid (WD) repeat structure, which usually functions as a hub to mediate protein–protein or protein–substrate interactions. It locates in the center of the EMC lumenal region and organizes Emc1-NTD1, Emc7, Emc10, and the C-terminal loop of Emc4 around it. Interestingly, the yeast Emc1 NTD1 is a three-stranded β-sheet, but the human EMC1 is an eight-bladed β-propeller (Fig. 2B).
The cytosolic region of the yeast EMC is composed of Emc2 and the cytosolic domain of Emc3, a N-terminal loop of Emc4, and a C-terminal loop of Emc5 (Fig. 2A). Emc2 forms a shallow, right-handed spiral structure with seven tetratricopeptide repeats (TPRs), and it scaffolds other cytosolic components of the EMC. Emc2 is tilted 30° away from the ER membrane and partially caps the large cavity in the transmembrane region. Such architecture allows a large substrate to access the cavity in the transmembrane region from the cytosol. An aqueous subunit, either EMC8 or 9, is unique to human EMC and is attached on the top of EMC2, thus occupying the analogous binding site of the N-terminal loop found in yeast Emc4 (Fig. 2B). The crystal structure of the EMC2–EMC9 heterodimer (PDB: 6Y4L) further confirms this mode of interaction [28]. Therefore, unlike the yeast Emc4, whose N-terminal loop interacts with and is stabilized by Emc2, the N-terminal loop of human EMC4 is disordered.
Does Emc4 contain one or three transmembrane helices?
As mentioned above, Emc4 was modeled with three TMHs in the yeast EMC structure (Figs. 1A, 2A, 3A–D) [29]. The modeling is in agreement with the structure based on the 6.4-Å 3D map of the human EMC (Fig. 1C) [28]. The three TMHs were tentatively suggested to be the single TMH of EMC4, EMC7, and EMC10 in the cryo-EM analysis of human EMC at 3.4-Å average resolution, but they were not modeled in the structure due to the low resolution in this region (Figs. 1B, 2B) [30]. This region was also not modeled in the deposited structures of the latest publication [31]. Therefore, whether Emc4/EMC4 is a 3-TMH or a 1-TMH protein is unresolved.
Figure 3. Modeling of the yeast Emc4 subunit.
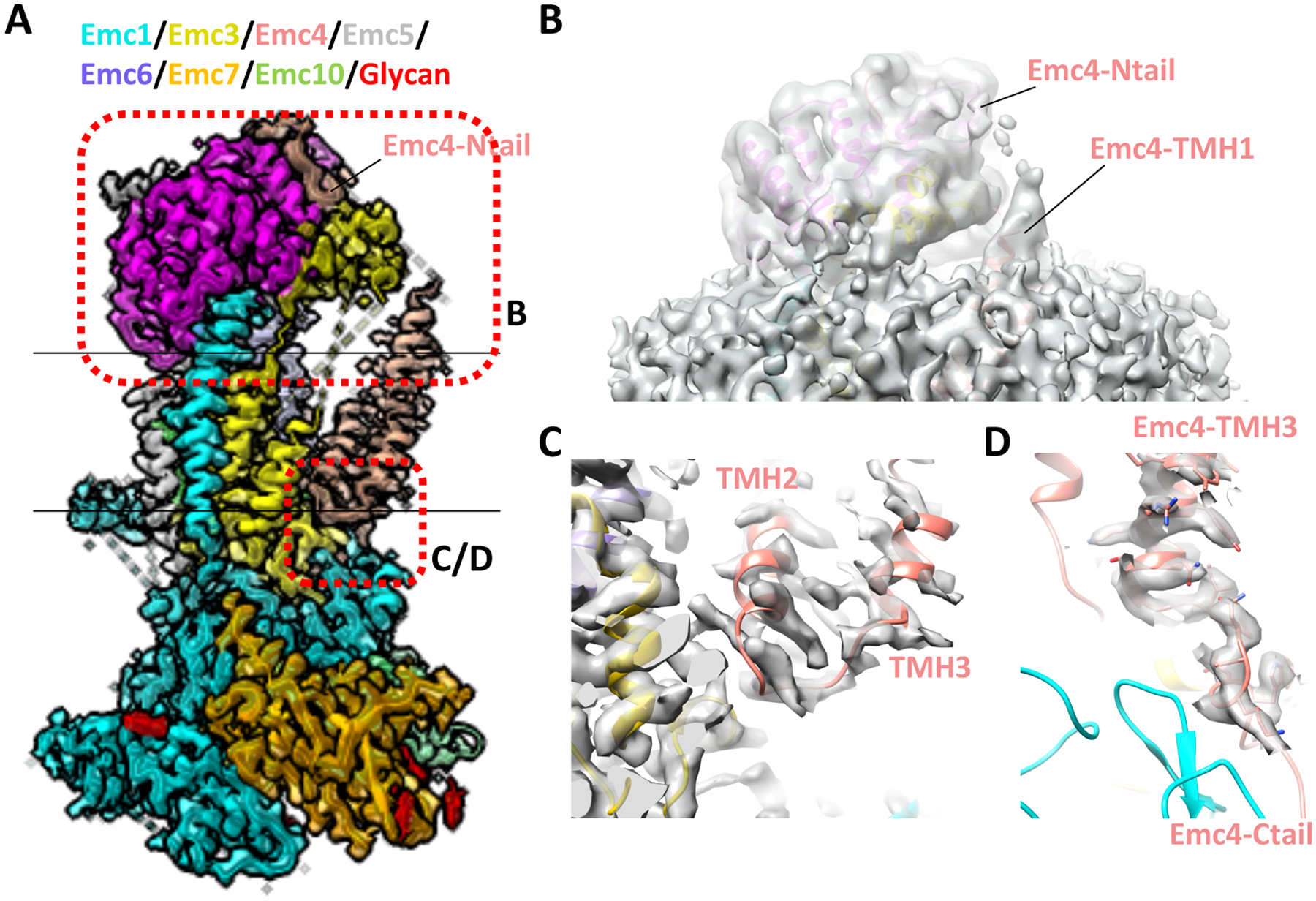
(A) Cryo-EM map of the yeast EMC superposed with the structure shown in cartoon (EMD-21587, PDB ID 6WB9). (B) Linker peptide density between the Emc4 N-tail and TMH1 is shown at a lower threshold. (C) Linker peptide density between Emc4 TMH2 and TMH3. (C) Linker peptide density between Emc4 TMH3 and the C-tail.
The yeast EMC map had the best density quality in the Emc4 region. Both the N-terminal and C-terminal loops of Emc4 were unambiguously resolved, residing in the cytosolic and lumenal sides of the membrane, respectively (Figs. 1A, 2A, 3A). This observation indicates that the yeast Emc4 contains either one or three, but not two, TMHs. Furthermore, the peptide loop density connecting the last TMH and the C-terminal loop of Emc4 is continuous and of good quality (Fig. 3D), so the assignment of this helical density to the last TMH of Emc4 is unambiguous.
The question then was the identity of the first two TMHs. They were assigned to Emc4 rather than to Emc7 and Emc10, for the following reasons. 1) These two TMH densities are connected by a continuous density in the yeast 3D map, suggesting that they belong to one protein (Emc4), not two (Emc7 and Emc10) (Fig. 3C). 2) In the low-pass filtered yeast EMC 3D map, one of these two TMH densities extends over to the cytosol and connects to the N-terminal loop of Emc4 (Fig. 3B), suggesting that this TMH is TMH1 of Emc4 and hence the other is TMH2 of Emc4. These structural features are also present in the latest cryo-EM structures of yeast EMC [31]. 3) Although both Emc7 and Emc10 have long disordered C-terminal regions that may contain TMH, the modeled C-terminus of Emc10 is far from the two TMHs densities (Figs. 1A, 2A). Furthermore, unlike the human EMC10, the yeast Emc10 is not predicted to have a TMH, based on topology algorithms (TMHMM online server) [32]. Therefore, modeling three TMHs for the yeast Emc4 is based on solid density features, and the yeast EMC model is consistent with bioinformatic analysis.
The next question was whether the architecture of human EMC4 is the same as that of yeast Emc4. Among the two independent cryo-EM structures of the human EMC, the density of the EMC4 region in the 6.4-Å map was surprisingly better defined than in the 3.4-Å map, in which the EMC4 region is barely visible (although other regions are much better resolved) (Figs. 1B–C, 2B). In the 6.4-Å human EMC map [28], EMC4 was modeled to have three TMHs, just as the yeast Emc4. Modeling three TMHs for human EMC4 is supported by three lines of evidence. 1) The EMC4 density in the human 3D map highly resembles the corresponding region of the yeast EMC map. 2) EMC4 is predicted to have three TMHs by trRosetta [33], a de novo structure prediction program that accurately predicted the TMHs of other EMC subunits. 3) A protease protection-based assay and an AbK-mediated crosslinking study were consistent with the three-TMH model of the human EMC4. Thus, it seems likely that the human EMC4 has the same three-TMH topology as yeast Emc4 and that the three TMHs are folded just as in the yeast Emc4. However, further studies are required to fully address this question.
Putative substrate-binding pocket
As an insertase, EMC is expected to have a TMH-binding pocket in the transmembrane region to facilitate the insertion of a client TMD into the ER membrane. There are two large cavities in the transmembrane domain (Figs. 1A–C, 4A–B). The first cavity (red arrowhead) surrounded by Emc3, Emc4, and Emc6 in the transmembrane region is accessible from either the membrane or the cytosol. The second cavity (blue arrow) in back of the first cavity, is filled with lipid molecules, and is accessible only from the membrane. It is surrounded by Emc5 in the membrane region, the horizontal helix (HH) of Emc1 from the cytosolic side, and Emc7 from the lumen. The first cavity appears to be gated and is expected to be the substrate-binding pocket [29–31]. That expectation was supported by several structural and functional considerations. Emc3 is predicted to be the catalytic subunit of EMC, based on bioinformatic analysis identifying Emc3 as a member of the evolutionarily conserved Oxa1/Alb3/YidC insertase family [27]. Indeed, EMC3 is the primary substrate interaction partner [28].
Figure 4. The transmembrane region of the EMC contains a client-binding pocket.
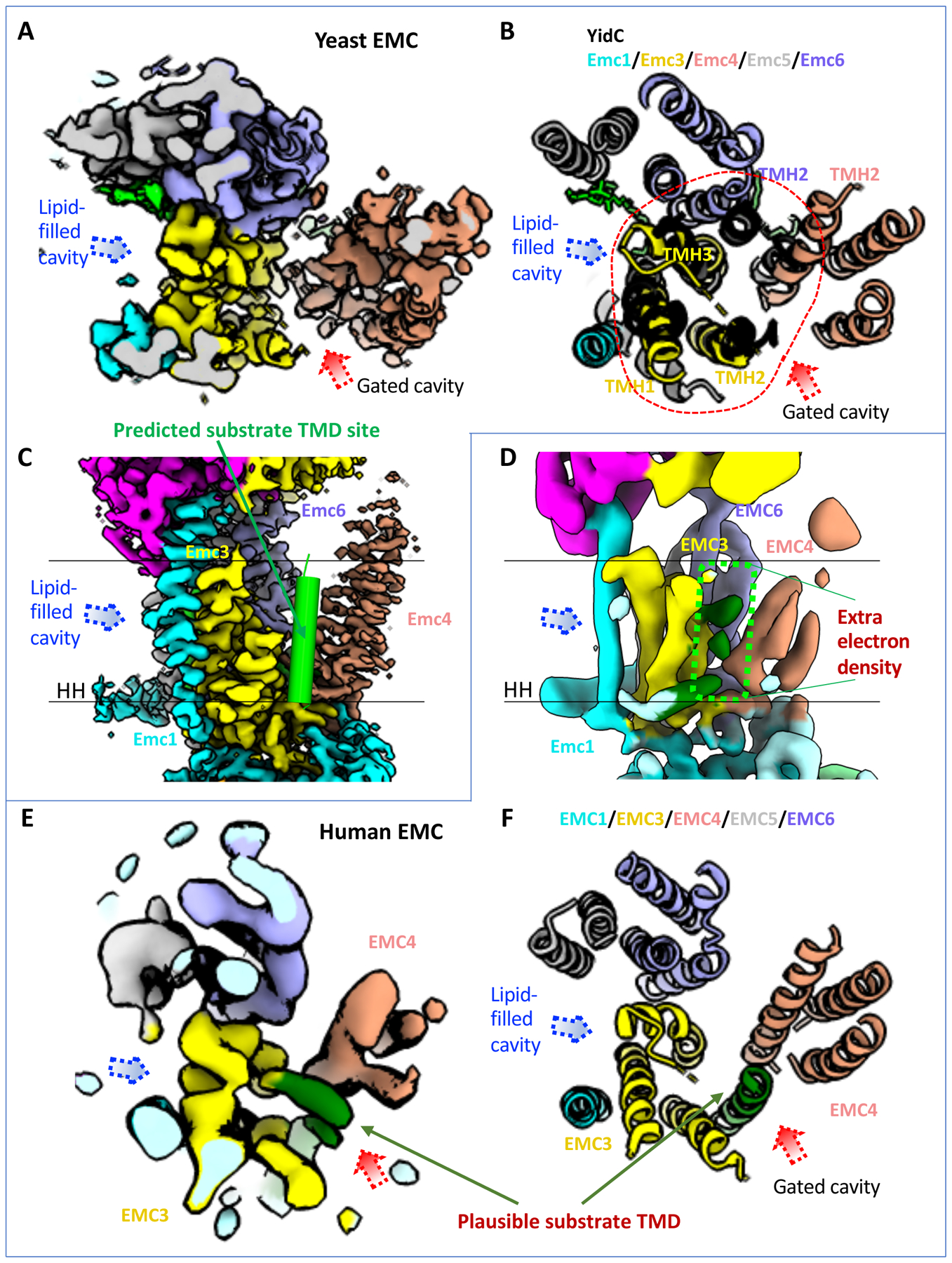
(A) Cryo-EM map of transmembrane domain of the yeast EMC in a top (cytosolic) view (EMD-21587). Dotted red and blue arrowheads mark two large, potential client TMD-binding cavities. (B) Superposition of the YidC structure (PDB ID 5Y83) in black cartoon and the transmembrane domain of yeast EMC (PDB ID 6WB9) in a top (cytosolic) view. Red dashes encircle the five EMC α-helices aligned with YidC. The putative client TMD position in EMC is shown by the red arrowhead, which is based on previously reported EM structure of a YidC–ribosome complex [4]. (C) A front view of the yeast EMC transmembrane region (EMD-21587). The green cylinder represents a client TMD located between TMH2 of Emc3 and TMH2 of Emc4 in the putative client-binding pocket. (D) A front view of the human EMC transmembrane region (EMD-11058). Two parallel black lines mark the lipid bilayer position. The dashed green rectangle highlights several density peaks in the putative client-binding pocket. The label “HH” in (C-D) refers to the amphipathic horizontal helix of EMC1. (E-F) Top view of the transmembrane region of the human EMC 3D map (EMD-11058, panel E) and atomic model (PDB ID 6Z3W, panel F), showing it in the same orientation and region as in the yeast EMC in (panels A-B). The rod-like weak density was originally assigned to the TMD of EMC7. This density is located at the position of the predicted substrate TMD and is likely from a co-purified endogenous substrate.
The substrate pocket is formed by five TMHs: the three TMHs of Emc3, TMH2 of Emc4, and TMH2 of Emc6, and they assemble into a YidC-like fold (Fig. 4B). YidC is a well-known prokaryotic insertase that binds a TMH substrate in a pocket between its TM3 and TM5 [4, 34, 35]. Based on the striking structural conservation between EMC and YidC, it is likely that this cavity is EMC’s client-binding site and that the substrate binds in the groove between TMH2 of Emc3 and TMH2 of Emc4, which correspond to the substrate-binding TM3 and TM5 of YidC (Fig. 4B–C). The groove between TMH2 of Emc3 and TMH2 of Emc4 is partially hydrophilic, featuring numerous polar residues, including K26, N122, S125, T130, N137, N188, Q129, and Q199 of yeast Emc3 and Q99 and T105 of yeast Emc4. The human EMC includes R31, N114, R180, Ser184, Q189, and D190 of EMC3 and R108 and Q111 of EMC4. The partial hydrophilicity resembles the substrate-binding pocket found in other protein-transporting channels such as YidC, Sec61 and the Hrd complex [36–38]. The substrate TMH binding by YidC relies on positively charged residues, such as R72 in Bacillus halodurans YidC [36], R260 in Thermotoga maritima YidC [39], and R366 in E. coli YidC [35, 40]. Thus, the EMC contains a structurally equivalent residue, K26 of yeast Emc3 or R31 of human EMC3, that is essential for EMC’s function [29]. In the human EMC, a second positively charged residue (EMC3 R180) is also important for function [30, 31]. Thus, the cavity surrounded by Emc3, Emc4, and Emc6 is the likely client-binding site.
Interestingly, there are several weak densities in the space between TMH2 of EMC3 and TMH2 of EMC4—the putative substrate binding pocket—in the 6.4-Å resolution structure of human EMC (Fig. 4D–E); these densities are absent in the higher resolution map (Fig. 1B). Although originally suggested to be a part of the EMC [28], these weak densities likely belong to a co-purified endogenous TMD substrate, based on the multiple structures now available. Relevant to the possibility, in a recent structural work, an endogenous TMD substrate was unexpectedly co-purified with Spf1, which is a TMD dislocase [41].
The insertase and holdase activities of EMC are separable
The Weissman lab developed a dual fluorescent EMC client reporter assay for a comprehensive structure-based functional interrogation of EMC [31]. Beside the previously identified C-terminal insertase activity for tail-anchored proteins and N-terminal insertase activity for some multi-pass membrane proteins, EMC was also proposed to have the holdase chaperone activity for some multi-pass membrane proteins, whose biogenesis requires the EMC but does not rely on its terminal insertase activity. The authors monitored the biogenesis of three types of client: the transmembrane domain of squalene synthase (SQS) whose C-terminal single TMH is inserted into membrane by EMC, the beta-1 adrenergic receptor (B1AR), a seven-TMH G protein coupled receptor whose N-terminal TMD is inserted by EMC, and the four-TMH transmembrane protein 97 (TMEM97) whose membrane incorporation requires the holdase activity of EMC. By probing about 50 EMC mutations in EMC1, EMC2, EMC3, and EMC5, they found that multiple mutations decreased TMEM97 biogenesis, increased SQS production, and largely sustained the B1AR level. This unexpected observation led to the conclusion that EMC is a multi-functional enzyme and that the insertase activity is separable from the holdase chaperone activity.
Taking advantage of the EMC subunit knockout cell lines and the fluorescent client reporter assay, the authors probed the functional relevance of the lipid filled membrane cavity in the back of the substrate TMD-binding cavity (Fig. 4A–F). They found that mutations around the cavity destabilized all three substrates, and they concluded that this cavity may hold a regulatory function in client biogenesis. We suggest that both cavities may bind a multi-pass client simultaneously, with the lipid-filled cavity holding a hydrophobic TMH, and the gated cavity interacting with a partially hydrophilic TMH.
Perspective
The yeast and human EMC structures reveal the architecture of the complex as well as a remarkable evolutionary conservation of the TMH insertion mechanism in the prokaryotic insertases (Fig. 5, top panel). However, many questions remain to be answered. For example, what is the detailed molecular mechanism of TMD insertion facilitated by EMC? How does the EMC specifically recognize its substrate? How does EMC cooperate with the translocon and the ribosome in co-translational TMD insertion? And how do the soluble domains of EMC contribute to TMH insertion?
Figure 5. Models for tail-anchored protein and multi-pass membrane protein biogenesis.
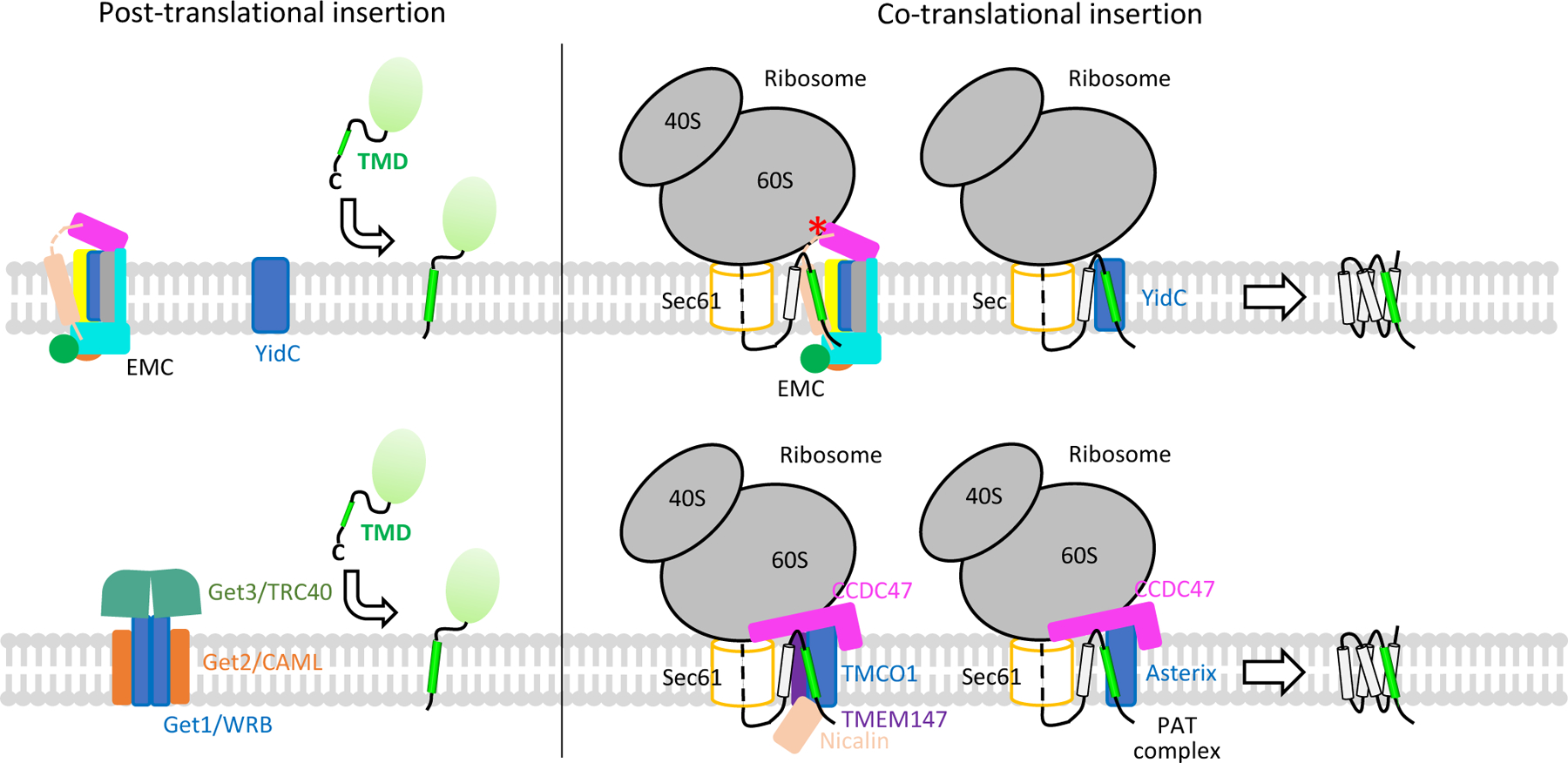
The top panel shows that YidC and EMC are able to facilitate membrane insertion of both tail-anchored proteins and multi-pass membrane proteins. The red asterisk indicates the collision point between the EMC cytosolic region and the ribosome, which could prevent the EMC from physically contacting the translocon. The bottom panel presents the GET complex that facilitates tail-anchored protein biogenesis, the TMCO1 complex, and the PAT complex that facilitate multi-pass membrane protein folding. The GET complex and ribosome–TMCO1 complex are sketched based on published cryo-EM maps (GET: EMD-10266 and EMD-11607; TMCO1: EMD-21426 and EMD-21427) [12, 14].
Recently, two additional ER-resident TMD chaperones have been reported, the PAT complex and the ribosome-associated TMCO1 complex (Fig. 5, bottom panel) [12, 13]. PAT is a small complex of 60 kDa, composed of CCDC47 and Asterix [13]. The TMCO1 complex also contains CCDC47, along with nicalin, TMCO1, and TMEM147 [12]. A cryo-EM structure of the ribosome–TMCO1–translocon complex indicates that TMCO1 cooperates with, but does not bind to, the translocon to facilitate multi-pass membrane protein biogenesis [12]. It is not clear how PAT and EMC cooperate with the translocon and whether they have a similar architecture. However, it was noted that the large cytosolic domain of EMC may collide with the ribosome, preventing EMC from physically associating with the translocon [28]. It is also unclear whether TMCO1, PAT, and EMC have distinct membrane protein substrates or whether they chaperone a subset of shared substrates, and if so, what is the spatial and temporal relationship of these membrane chaperones with respect to the translocon? We look forward to the answers to these questions in coming years.
Acknowledgements.
The authors are indebted to the members of the Li lab for their contributions and valuable discussions on the topic of this review, and to David Nadziejka for technical editing of the manuscript. This work was supported by the U.S. National Institutes of Health (CA231466 to H.L.) and the Van Andel Institute (to H.L).
Abbreviations:
- Cryo-EM
cryo-electron microscopy
- EMC
ER membrane complex
- ER
endoplasmic reticulum
- GDN
detergent glyco-diosgenin
- GET
guided entry of tail-anchored proteins
- HH
horizontal helix
- LMNG
lauryl maltose neopentyl glycol
- TA
tail-anchored
- TMD
transmembrane domain
- TMH
transmembrane helix
- TPR
tetratricopeptide repeat
- TRC
transmembrane recognition complex
- WD repeat
tryptophan–aspartic acid repeat
Footnotes
Conflicts of interests: The authors declare that they have no competing interests associated with the manuscript.
REFERENCES
- 1.Anson L (2009) Membrane protein biophysics, Nature. 459, 343. [DOI] [PubMed] [Google Scholar]
- 2.Shao S & Hegde RS (2011) A flip turn for membrane protein insertion, Cell. 146, 13–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiefer D & Kuhn A (2007) YidC as an essential and multifunctional component in membrane protein assembly, Int Rev Cytol. 259, 113–38. [DOI] [PubMed] [Google Scholar]
- 4.Kedrov A, Wickles S, Crevenna AH, van der Sluis EO, Buschauer R, Berninghausen O, Lamb DC & Beckmann R (2016) Structural Dynamics of the YidC:Ribosome Complex during Membrane Protein Biogenesis, Cell Rep. 17, 2943–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borgese N, Coy-Vergara J, Colombo SF & Schwappach B (2019) The Ways of Tails: the GET Pathway and more, Protein J. 38, 289–305. [DOI] [PubMed] [Google Scholar]
- 6.Mateja A & Keenan RJ (2018) A structural perspective on tail-anchored protein biogenesis by the GET pathway, Curr Opin Struct Biol. 51, 195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shan SO (2019) Guiding tail-anchored membrane proteins to the endoplasmic reticulum in a chaperone cascade, J Biol Chem. 294, 16577–16586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shurtleff MJ, Itzhak DN, Hussmann JA, Schirle Oakdale NT, Costa EA, Jonikas M, Weibezahn J, Popova KD, Jan CH, Sinitcyn P, Vembar SS, Hernandez H, Cox J, Burlingame AL, Brodsky JL, Frost A, Borner GH & Weissman JS (2018) The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins, Elife. 7, e37018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guna A, Volkmar N, Christianson JC & Hegde RS (2018) The ER membrane protein complex is a transmembrane domain insertase, Science. 359, 470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chitwood PJ, Juszkiewicz S, Guna A, Shao S & Hegde RS (2018) EMC Is Required to Initiate Accurate Membrane Protein Topogenesis, Cell. 175, 1507–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aviram N, Ast T, Costa EA, Arakel EC, Chuartzman SG, Jan CH, Hassdenteufel S, Dudek J, Jung M, Schorr S, Zimmermann R, Schwappach B, Weissman JS & Schuldiner M (2016) The SND proteins constitute an alternative targeting route to the endoplasmic reticulum, Nature. 540, 134–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGilvray PT, Anghel SA, Sundaram A, Zhong F, Trnka MJ, Fuller JR, Hu H, Burlingame AL & Keenan RJ (2020) An ER translocon for multi-pass membrane protein biogenesis, Elife. 9, e56889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chitwood PJ & Hegde RS (2020) An intramembrane chaperone complex facilitates membrane protein biogenesis, Nature. 584, 630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDowell MA, Heimes M, Fiorentino F, Mehmood S, Farkas A, Coy-Vergara J, Wu D, Bolla JR, Schmid V, Heinze R, Wild K, Flemming D, Pfeffer S, Schwappach B, Robinson CV & Sinning I (2020) Structural Basis of Tail-Anchored Membrane Protein Biogenesis by the GET Insertase Complex, Mol Cell. 80, 72–86 e7. [DOI] [PubMed] [Google Scholar]
- 15.Volkmar N & Christianson JC (2020) Squaring the EMC - how promoting membrane protein biogenesis impacts cellular functions and organismal homeostasis, J Cell Sci. 133, jcs243519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alvira S, Corey RA, Collinson I & Romisch K (2020) Membrane protein biogenesis by the EMC, EMBO J, e107407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS & Schuldiner M (2009) Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum, Science. 323, 1693–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satoh T, Ohba A, Liu Z, Inagaki T & Satoh AK (2015) dPob/EMC is essential for biosynthesis of rhodopsin and other multi-pass membrane proteins in Drosophila photoreceptors, Elife. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lahiri S, Chao JT, Tavassoli S, Wong AK, Choudhary V, Young BP, Loewen CJ & Prinz WA (2014) A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria, PLoS Biol. 12, e1001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richard M, Boulin T, Robert VJ, Richmond JE & Bessereau JL (2013) Biosynthesis of ionotropic acetylcholine receptors requires the evolutionarily conserved ER membrane complex, Proc Natl Acad Sci U S A. 110, E1055–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Louie RJ, Guo J, Rodgers JW, White R, Shah N, Pagant S, Kim P, Livstone M, Dolinski K, McKinney BA, Hong J, Sorscher EJ, Bryan J, Miller EA & Hartman J. L. t. (2012) A yeast phenomic model for the gene interaction network modulating CFTR-DeltaF508 protein biogenesis, Genome Med. 4, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christianson JC, Olzmann JA, Shaler TA, Sowa ME, Bennett EJ, Richter CM, Tyler RE, Greenblatt EJ, Harper JW & Kopito RR (2011) Defining human ERAD networks through an integrative mapping strategy, Nat Cell Biol. 14, 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bircham PW, Maass DR, Roberts CA, Kiew PY, Low YS, Yegambaram M, Matthews J, Jack CA & Atkinson PH (2011) Secretory pathway genes assessed by high-throughput microscopy and synthetic genetic array analysis, Mol Biosyst. 7, 2589–98. [DOI] [PubMed] [Google Scholar]
- 24.Tang X, Snowball JM, Xu Y, Na CL, Weaver TE, Clair G, Kyle JE, Zink EM, Ansong C, Wei W, Huang M, Lin X & Whitsett JA (2017) EMC3 coordinates surfactant protein and lipid homeostasis required for respiration, J Clin Invest. 127, 4314–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian S, Wu Q, Zhou B, Choi MY, Ding B, Yang W & Dong M (2019) Proteomic Analysis Identifies Membrane Proteins Dependent on the ER Membrane Protein Complex, Cell Rep. 28, 2517–2526 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wideman JG (2015) The ubiquitous and ancient ER membrane protein complex (EMC): tether or not?, F1000Res. 4, 624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anghel SA, McGilvray PT, Hegde RS & Keenan RJ (2017) Identification of Oxa1 Homologs Operating in the Eukaryotic Endoplasmic Reticulum, Cell Rep. 21, 3708–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Donnell JP, Phillips BP, Yagita Y, Juszkiewicz S, Wagner A, Malinverni D, Keenan RJ, Miller EA & Hegde RS (2020) The architecture of EMC reveals a path for membrane protein insertion, Elife. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai L, You Q, Feng X, Kovach A & Li H (2020) Structure of the ER membrane complex, a transmembrane-domain insertase, Nature. 584, 475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pleiner T, Tomaleri GP, Januszyk K, Inglis AJ, Hazu M & Voorhees RM (2020) Structural basis for membrane insertion by the human ER membrane protein complex, Science. 369, 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller-Vedam LE, Brauning B, Popova KD, Schirle Oakdale NT, Bonnar JL, Prabu JR, Boydston EA, Sevillano N, Shurtleff MJ, Stroud RM, Craik CS, Schulman BA, Frost A & Weissman JS (2020) Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients, Elife. 9, e62611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moller S, Croning MD & Apweiler R (2001) Evaluation of methods for the prediction of membrane spanning regions, Bioinformatics. 17, 646–53. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Anishchenko I, Park H, Peng Z, Ovchinnikov S & Baker D (2020) Improved protein structure prediction using predicted interresidue orientations, Proc Natl Acad Sci U S A. 117, 1496–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wickles S, Singharoy A, Andreani J, Seemayer S, Bischoff L, Berninghausen O, Soeding J, Schulten K, van der Sluis EO & Beckmann R (2014) A structural model of the active ribosome-bound membrane protein insertase YidC, Elife. 3, e03035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohler R, Boehringer D, Greber B, Bingel-Erlenmeyer R, Collinson I, Schaffitzel C & Ban N (2009) YidC and Oxa1 form dimeric insertion pores on the translating ribosome, Mol Cell. 34, 344–53. [DOI] [PubMed] [Google Scholar]
- 36.Kumazaki K, Chiba S, Takemoto M, Furukawa A, Nishiyama K, Sugano Y, Mori T, Dohmae N, Hirata K, Nakada-Nakura Y, Maturana AD, Tanaka Y, Mori H, Sugita Y, Arisaka F, Ito K, Ishitani R, Tsukazaki T & Nureki O (2014) Structural basis of Sec-independent membrane protein insertion by YidC, Nature. 509, 516–20. [DOI] [PubMed] [Google Scholar]
- 37.Wu X, Siggel M, Ovchinnikov S, Mi W, Svetlov V, Nudler E, Liao M, Hummer G & Rapoport TA (2020) Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex, Science. 368, eaaz2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voorhees RM, Fernandez IS, Scheres SH & Hegde RS (2014) Structure of the mammalian ribosome-Sec61 complex to 3.4 A resolution, Cell. 157, 1632–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xin Y, Zhao Y, Zheng J, Zhou H, Zhang XC, Tian C & Huang Y (2018) Structure of YidC from Thermotoga maritima and its implications for YidC-mediated membrane protein insertion, FASEB J. 32, 2411–2421. [DOI] [PubMed] [Google Scholar]
- 40.Kumazaki K, Kishimoto T, Furukawa A, Mori H, Tanaka Y, Dohmae N, Ishitani R, Tsukazaki T & Nureki O (2014) Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase, Sci Rep. 4, 7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKenna MJ, Sim SI, Ordureau A, Wei L, Harper JW, Shao S & Park E (2020) The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase, Science. 369, eabc5809. [DOI] [PMC free article] [PubMed] [Google Scholar]


