Abstract
The relationship between the three domains of life—Archaea, Bacteria, and Eukarya—is one of Biology’s greatest mysteries. Current favored models imply two ancestral domains, Bacteria and Archaea, with eukaryotes originating within Archaea. This type of models has been supported by the recent description of the Asgardarchaeota, the closest prokaryotic relatives of eukaryotes. However, there are many problems associated with any scenarios implying that eukaryotes originated from within the Archaea, including genome mosaicism, phylogenies, the cellular organization of the Archaea, and their ancestral character. By contrast, all models of eukaryogenesis fail to consider two relevant discoveries: the detection of membrane coat proteins, and of phagocytosis-related processes in Planctomycetes, which are among the bacteria with the most developed endomembrane system.
Consideration of these often overlooked features and others found in Planctomycetes and related bacteria suggest an evolutionary model based on a single ancestral domain. In this model, the proximity of Asgard and eukaryotes is not rejected but instead, Asgard are considered as diverging away from a common ancestor instead of on the way toward the eukaryotic ancestor. This model based on a single ancestral domain solves most of the ambiguities associated with the ones based on two ancestral domains. The single-domain model is better suited to explain the origin and evolution of all three domains of life, blurring the distinctions between them. Support for this model as well as the opportunities that it presents not only for reinterpreting previous results, but also for planning future experiments, are explored.
Keywords: evolution, eukaryogenesis, Asgard, Planctomycetes, PVC superphylum, tree of life
Introduction
Deciphering the Relationship between the Three Domains of Life: Difficult and Still Unsolved
Life on earth is classified into three domains: Archaea, Bacteria, and Eukarya. The first two domains are prokaryotes, so named to differentiate them from the eukaryotes, which are characterized by a developed cellular organization (the name derives from the Greek “eu” for “true” and “karyon” for “nucleus”). The eukaryotic cell has many apparently unique features, and there is a tremendous gap between prokaryotic and eukaryotic cells in terms of their complexity and development. Despite this apparent gap, few features are truly eukaryotic only, which suggests a mixed contribution from the other two domains. It is now well accepted that the eukaryotic cell is an evolutionary mosaic composed of bacterial and archaeal characteristics, as well as eukaryotic innovations. The role of bacterial “capture” by the ancestor of the eukaryotic cell, that is, endosymbiosis, leading to the development of the mitochondria, has been recognized as a result of the work of Lynn Margulis (Cornish-Bowden 2017), and this process is commonly accepted as one of the main steps in eukaryogenesis. However, the nature of both the host and guest in this inaugural event is not clear (Martijn et al. 2018). Thus, how the three domains are related and how they emerged via eukaryo-, archaeo-, and bacterio-genesis, are still unknown.
There is considerable agreement that life began some 4–3.5 billion years ago but, the very nature of the last universal common ancestor (LUCA) is still a mystery, although it appears likely that it was a prokaryote-like organism of considerable genomic and organizational complexity. Indeed, it is well accepted now that LUCA was a prokaryotic cell using nucleic acids as genetic material, ribosomes for template-directed protein synthesis based on a genetic code for 20 amino acids, and membranes that allowed for energy generation by chemiosmotic coupling (Krupovic et al. 2020). However, if it was a bacterium or an archaeon, or a hybrid between both, is still unknown, with important implications, owing to the major differences between central systems in the two prokaryotic domains, for example, the replication and membrane machineries.
Deciphering the origin of the three domains of life is difficult as it is one of the most ancient events. Most recent analyses provide estimates for the origin of the first eukaryotic cell in the range of 1–2 billion years (Dacks et al. 2016). However, the last eukaryotic common ancestor (LECA) could have been the result of an abrupt change brought about by the introduction of the mitochondrial precursor in a prokaryotic host, or, on the opposite, of a gradual increase in complexity from a prokaryote developing eukaryotic features and slowly accumulating them over time (Martin et al. 2015; Dacks et al. 2016).
This conundrum is mostly due to the difficulties inherent with reconstructing ancestral relationships and states based on extant organisms which are derived as compared with the ancestral states. For example, if eukaryotes and archaea have shared a common ancestor, this internal node is not accessible to us and can only be guessed at based on the features (e.g., sequences) of extant organisms and their inferred relationship. There are however many difficulties associated with ancient phylogenies and the reconstruction of ancestral characters, that is, associated to internal nodes. One important tool in the reconstruction of phylogenies and ancestral nodes is the concept of clade-specific genes or proteins, for example, eukaryotic signature proteins (ESPs) are those proteins that are found in their majority in eukaryotic proteomes, although some such proteins can sometimes be found in the other two domains, for various reasons. This is different from the origin of the genes which refers to its most likely source based on phylogenetic inferences. For example, archaeal genes of bacterial origin refer to genes found in their majority in archaeal genomes but whose phylogeny indicates an origin from bacteria.
Two-Domain Scenarios Uplifted by the Discovery of the Asgardarchaeota Superphylum
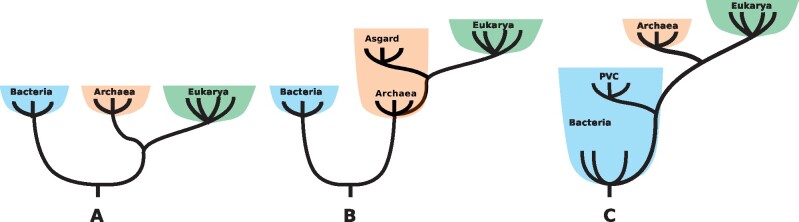
The discovery of the third domain of life, the Archaea, by Carl Woese and colleagues initially led to versions of the tree of life based on three ancestral domains with independent origins, with Archaea more closely related to Eukarya (Fox et al. 1980; fig. 1A). In these models, Eukarya is the sister group to Archaea and cellularization events are independent, the Archaea, the Bacteria, and the Eukarya, each arising from a “progenote” phase of evolution. Improved phylogenetic tools combined with increased taxon sampling have led to different scenarios proposing only two ancestral prokaryotic domains (2D models; Lake et al. 1984). In these, Eukarya is sister to one of the archaeal lineages and branches within the Archaea, and so the LECA is an archaeon (fig. 1B). Variations of 2D models that are mostly derived from the genome mosaicism of eukaryotes are syntrophy models, which are based on interactions between various prokaryotes (López-García and Moreira 2020). In most currently accepted 2D models, there are only two cellularization events, one at the base of Bacteria and another one before the inception of the common ancestor of archaea. These are the main models, variations, and outsiders exist; for review, see Martin et al. (2015) and Martin (2017a).
Fig. 1.
The evolving view of the tree of life. (A) The 3D tree of life, (B) 2D tree of life, and (C) the PVC-based 1D tree of life. Archaea, Bacteria, and Eukarya are colored red, blue, and green, respectively.
The discovery of the Asgardarchaeota, an archaeal superphylum, has been pivotal for addressing many unanswered questions in eukaryogenesis (Spang et al. 2015). In addition to the Euryarchaeota, there are currently three archaeal superphyla: Asgard, DPANN, and TACK. Asgard archaea currently consist of Lokiarchaeota, Thorarchaeota, Odinarchaeota, and Heimdallarchaeota (Baker et al. 2020). In addition to encoding more ESPs than other archaea, phylogenetic analyses using various genes placed Eukarya within the Asgard archaea, with Heimdallarchaeota as the closest archaeal relative of the Eukarya (MacLeod et al. 2019).
The genomic data derived from the Asgard lineages, combined with phylogenomic analyses, support the conclusion that this superphylum comprises the closest prokaryotic relatives of eukaryotes. This close phylogenetic relationship between Asgard and Eukarya is strengthened by the presence of a number of ESPs that are unique to Asgard proteomes. Although some ESPs had previously been discovered in other archaeal lineages, only with a patchy distribution and not as complete as for Asgard, the repertoire of these proteins in the Asgard dwarfed all previous estimates and some of them are the most closely related orthologs to eukaryotes so far. These ESPs include the information processing machineries, as well as some of the proteins that are typically associated with a developed cell organization and endomembrane system, such as tubulin; actin and actin-binding proteins, such as profilins and gelsolin/cofilin; proteins related to vesicle coats, such as COPII’s Sec23/Sec24; the endosomal sorting complex required for transport (ESCRT; although homologs for ESCRT complexes III have been known in non-Asgard archaea for some time [Samson et al. 2017]); and their regulatory systems, including small GTPases, SNAREs, BAR, and longin domain proteins. The discovery of these proteins has led to much speculation concerning the complexity of the intracellular organization of the Asgard (Dey et al. 2016; Klinger et al. 2016). However, most of the eukaryotic systems encoded in Asgard archaea are incomplete (raising the question of the utility of incomplete systems) and are yet to be functionally characterized (with few exceptions [Akıl and Robinson 2018; Akıl et al. 2020]).
Any doubt about the existence, or sequence purity, of these organisms, initially identified from metagenomic surveys, has been eliminated by the isolation and visualization of the first Asgardarchaea. It took a decade to isolate Candidatus Prometheoarchaeum syntrophicum strain MK-D1, due to its extreme slow growth (Imachi et al. 2020). Its genome confirmed previous observations; it contains many ESPs, phylogenetic analyses supported a closer relationship between Asgard and Eukarya, and evidence for expression of the many ESPs has been found. In addition, although not unique in prokaryotes, the cells of MK-D1 present numerous evaginations, or external extensions, of the cell membrane, which could promote syntrophic relationships with other prokaryotes. Metabolic reconstructions of various Asgard members have suggested that they have limited biosynthetic capacities (Spang et al. 2019). The combination of deficient metabolism with these membrane evaginations has been interpreted as supporting symbiogenetic models based on metabolic symbioses or syntrophies (Bulzu et al. 2019; Spang et al. 2019; López-García and Moreira 2020). Even more, these observations are supportive of a creative variant of 2D models, the “inside-out” hypothesis (Baum and Baum 2014), in which it is proposed that the eukaryotic endomembrane system developed by evagination of the external membrane of an archaeal host that would surround the future symbiont and fuse to form the eukaryotic endomembrane system. Importantly, despite high expectations, visualization of the Asgard cell did not reveal any internal membrane or any type of eukaryote-like cellular organization.
Hence, the discovery of the Asgard has been important. It confirmed the close relationship between Eukarya and Archaea, and gave support to 2D scenarios with Eukarya branching within Asgard (for more information, see recent research and review articles, e.g., López-García and Moreira 2020; Stairs and Ettema 2020).
The Limitations of 2D Scenarios
There are, however, issues with the proposed origin of eukaryotes from within the Asgard which includes the following: the only Asgard cell visualized so far is very small in size, 0.5 µm, they lack any sign of cellular complexity, that is, membrane organization, and they are metabolically deficient, requiring symbiotic associations. Clearly, this does not look like the complex LECA. More generally, there are many additional problems associated with a speculated origin of the Eukarya within the Archaea, that is, with any 2D model. These are related to genome mosaicism, phylogenies, and the many biochemical, metabolic, and cellular differences between Archaea and Eukarya, including lipid biocompatibility. Ultimately, the very question of the Archaeal ancestry is one of the main issues.
Genome Mosaicism
The genomes of eukaryotes, unlike those of bacteria, are mosaic when it comes to the origin of the genes comprising them. Eukaryotic genomes are composed of three unequal parts. The first part is formed by genes that only have eukaryotic homologues, which code for the ESPs. The second part is formed by genes that have an archaeal origin, reflecting the closest relationship between these two domains. The third part is composed of genes that have a bacterial origin. This constitutes the well-recognized mixed heritage of eukaryotic genomes: genes involved in the eukaryotic genetic apparatus and information processing, such as ribosomes, polymerases, and topoisomerases, tend to reflect an archaeal origin, whereas genes involved in eukaryotic biochemical and metabolic processes tend to reflect a bacterial origin (Pisani et al. 2007). Lateral gene transfer (LGT) is often invoked to explain most of this mosaicism, often without clear evidence, and without considering the fact that the amount of LGT required seems unbearable (Ku and Martin 2016; Martin 2017b).
Recognizing this mosaicism, most 2D models invoke only two partners: an archaeal host and the alphaproteobacteria-related ancestor of mitochondria as the guest (López-García and Moreira 2020). Consequently, three predictions follow from any 2D scenario, ignoring the unlikely possibility of an unsustainable amount of LGT: 1) host genes (of archaeal origin) must predominate the guest (bacterial) genes in eukaryotic genomes; 2) the majority of the genes of bacterial origin must be related to the symbiont and thus to alpha-proteobacteria; and 3) the bacterial contribution to archaeal genomes, if any, should be minimal, as in these scenarios, Bacteria and Archaea are domains with independent origins.
These predictions, however, do not stand up to scrutiny. First, eukaryotic genomes harbor more bacterial genes (56% of the prokaryote-like ones) than archaeal genes (44%) overall, which holds true across all eukaryotes (Martijn et al. 2018). As Brueckner and Martin wrote, “if eukaryotes were to be classified by genome-based democratic principle, they would […] have to be grouped with bacteria, not archaea (Brueckner and Martin 2020).” Second, the main sources of this fraction of genes with bacterial origin are not related to mitochondria. Instead, these genes are of mixed origin from various bacterial sources, including but not restricted to alphaproteobacteria. This has been argued to support symbiogenetic models of eukaryogenesis (López-García and Moreira 2020). However, even elaborate 2D models involve only a few additional bacteria and do not address the mixed origin and multiple sources of most of these genes. And third, similarly to what is observed for eukaryotes, an important part of most archaeal genomes is related to bacteria (Nelson-Sathi et al. 2012; Deschamps et al. 2014; Nelson-Sathi et al. 2015). Even the genomes of the Asgard members are mosaics composed primarily of bacterial and archaeal components, followed by eukaryotic components (Spang et al. 2015).
Despite the fact that the extent of these gene lists has been contested, the chimeric nature of archaeal genomes is a reality (Groussin et al. 2016; Kapust et al. 2018). From the perspective of 2D models, this important bacterial contribution to archaeal genomes has been interpreted as the result of massive independent gene acquisitions from bacteria at the origins of major archaeal clades (Nelson-Sathi et al. 2012; 2015). Thus, a recurrent theme in 2D models is the convenient but unconvincing invocation of massive LGT events. Such events, however, directly implies that Archaea, or at least the major clades, postdates Bacteria posing a serious problem to 2D scenarios. Importantly, the opposite is not true. A pattern corresponding to the acquisition of a significant amount of archaeal genes at the origin of bacterial groups is not detected, indicating that gene transfers from Archaea to Bacteria, although not denied here, do not correspond to the origin of the major bacterial groups.
Phylogenies and the Tree of Life
Although the closest relationship between Archaea and Eukarya versus Bacteria is not disputed, the proposed relationship with the eukaryotes branching within Asgard is still under debate (Cunha et al. 2017; Fournier and Poole 2018; Spang et al. 2018; Cavalier-Smith and Chao 2020). The phylogenetic placement of Eukarya within the Archaea and closely related to the Asgard indeed appears only in a few phylogenies. However, most of these are unrooted or consider other archaea as the outgroup, providing a 2D-centric framework of experimentation and reflecting a profound bias toward 2D models. In addition, interpreting a small number of phylogenetic trees too literally is problematic as it ignores the fact that a majority of genes in eukaryotic genomes have a bacterial origin. Of course, there are phylogenies in which eukaryotic sequences branch inside Archaea, and even within the Asgard clade. However, there are still more genes showing a bacterial origin, even if their phylogenies are inconclusive (Pittis and Gabaldón 2016). This is related to the difficulty of phylogenetic reconstructions across such a vast timespan. Eventually, it is important to note that those genes showing Asgard proximity to the eukaryotes are not the ones usually taken as evolutionary markers and, as such, their reliability and accuracy are still to be determined.
Although Asgard affiliate strongly with Eukarya in some phylogenetic analyses, the exact evolutionary relationship between the two groups has yet to be determined. For example, a recent comparative analysis of 162 nearly complete Asgard genomes concluded that phylogenetic analysis does not strongly support an origin of eukaryotes within Asgard (Liu et al. 2021). Once again, extensive LGT was invoked. Hence, the exact nature of the relationship between Eukarya and Archaea cannot be considered as solved by a limited number of phylogenies extracted from the minority gene set. The pattern of inconclusive phylogenies is pointing toward wrong premises of 2D-centric analyses. This is supported by the anomalous phylogenetic behavior of ribosomal proteins in Asgard metagenome-assembled genomes (Garg et al. 2021).
Biochemical and Metabolic Differences
It is well-known that Eukarya and Archaea are more closely related in their information manipulation systems, which constitutes one of the bases of 2D scenarios. By contrast, a direct consequence of the “silent bacterial majority” in eukaryotic genomes is that their metabolism and cell biology are closer to those of Bacteria than to those of Archaea (Pittis and Gabaldón 2016; López-García and Moreira 2020). In the pool of metabolic genes in eukaryotic genomes, 68% have a bacterial origin (Brueckner and Martin 2020). Thus, the majority of the eukaryotic metabolism is of bacterial origin. These observations raise an interesting question rarely addressed in-depth by 2D scenarios: How is an archaeal metabolism changed into a bacteria-like one? If the host was an archaeon, presumably with an archaeal metabolism, and the end result was the first eukaryotic cell with a bacteria-derived metabolism, which mechanism explains how the metabolism of the host was modified so profoundly from archaea-like to bacteria-like?
Similarly, early consideration of the major differences between Archaea lipids and the ones of the other two domains led to suggest a difficult evolutionary transition (Wächtershäuser 2003). Membrane components consist of a hydrophilic head group (glycerol) and long hydrophobic tails (fatty acid chain). In archaea, the chemical composition of the head group and tail, and the connection between them, are different in archaea than in bacteria and eukaryotes. Although all other living organisms use glycerol-3-phosphate esters of long-chain fatty acids, archaea use glycerol-1-phosphate ethers of branched isoprenoids. The recent report of bacteria with a putative mixed membrane suggests that the evolutionary transition might not have been so difficult (Villanueva et al. 2021). In addition, in 2D scenarios, bacterial and archaeal lipids have independent origins. But, both eukaryotic glycerol-3-phosphate and isoprenoid biosynthesis genes seem to be of bacterial origin (Villanueva et al. 2017; Hoshino and Gaucher 2018). This apparent contradiction can only be explained by invoking again large LGT events.
Cellular Differences
There has been much expectation concerning the first observation of Asgard cells, due to the presence of specific cellular organization and endomembrane system-associated ESPs in their genomes. Surprisingly, MK-D1, the only Asgard isolate that has been visualized thus far, has turned out to be relatively basic in its cellular organization. It is very similar to prokaryotes, showing a complete lack of internal membrane organization. In addition, no genes have been found in the Asgard genomes to code for proteins with the 2-fold types typical of membrane coat proteins (MCPs) (Spang et al. 2015; Zaremba-Niedzwiedzka et al. 2017). This is highly relevant, as the eukaryotic endomembrane system is based on MCPs, which fuse an N-terminal beta-propeller combined to a C-terminal stacked pairs of alpha-helices fold in the same protein (Devos et al. 2004). This architecture is found exclusively in membrane-interacting proteins, almost always in eukaryotes (with few exceptions) and these proteins were necessary for the origin of the eukaryotic endomembrane system. Detecting these two folds in two different proteins, even if in close genomic proximity, is not comparable to detecting both in the same protein (Zaremba-Niedzwiedzka et al. 2017). Thus, the lack of observation of even a minimal endomembrane system in MK-D1 is congruent with the absence of MCP-like proteins in Asgard proteomes. In addition, Archaea, and Asgard in particular, are small (Imachi et al. 2020), which is not compatible with the enlarged size required to engulf the ancestor of the mitochondria (Hampl et al. 2019). Lastly, it is well-established that the LECA was complex (Koonin 2015). However, the Asgard ancestor had limited biosynthetic capacities and partner dependencies, that is, it was not complex, was most likely metabolically deficient and required associations of the symbiotic type (Bulzu et al. 2019; Spang et al. 2019; López-García and Moreira 2020). Thus, the Asgard ancestor appears unsuited to be the ancestor of a complex LECA. It is, however, likely that the last archaeal common ancestor, LACA, was complex and that most archaeal supergroups evolved by gene losses (Csurös and Miklós 2009; Koonin and Yutin 2014; Kellner et al. 2018). Note that defining “complexity” is difficult, but genome size or the number of metabolic processes or pathways can be used as a proxy.
The Archaeal Ancestry Misconception
Finally, a fundamental issue linked to all 2D scenarios is the initial mistake of claiming Archaea as ancestral (Woese et al. 1990). There is currently no evidence that Archaea is ancestral, and this proposal sounds now like an educated guess at best. It is in fact equally, if not more, likely that Archaea are derived. First of all, shared fundamental features of the three domains (such as the use of DNA/RNA for conservation and expression of information, the genetic code, the transcription machinery, ribosomal translation, ATP as an energy conservation molecule, amongst many others) point strongly toward a unique origin of life. Second, Woese declared the Archaea to be ancestral based on his belief that the 16S rRNA molecule was a regular molecular clock (Fox et al. 1980). We know now that this is not the case. Clocks are not constant (Williams et al. 2020). Finally, a growing list of archaeal fundamental features appear to be derived from bacteria, such as lipids (Coleman et al. 2019), C1 metabolism as methanogenesis and methylotrophy (Adam et al. 2019), the kynurenine pathway for NAD+ biosynthesis (Bulzu et al. 2019), and many others (Nelson-Sathi et al. 2015).
1D Scenarios
Thus, despite the undisputed proximity between Asgard and Eukarya, there are many problems associated with any 2D scenarios. Here, I suggest that most of these issues could be solved by looking at the situation from a different angle. I argue that an alternative scenario, based on a single bacterial ancestral domain, is more likely and explains most of the issues associated with 2D models. A bacterial rooting of the tree of life is seldom considered in current hypothesis, despite evidence supporting it, for example, based on paralogue duplication studies (Lake et al. 2009; Fournier and Gogarten 2010; Williams et al. 2015). In 3D scenarios, Archaea, Bacteria, and Eukarya are sister lineages, and all cells originated from an ancestor that preceded diversification of any domains, with different possible cellularization events (fig. 1A). In 2D scenarios, Eukarya emerged from within the archaeal radiation, but Archaea and Bacteria are sister lineages that originated from an ancestor that preceded their diversification (fig. 1B). In single-domain (1D) scenarios, both Archaea and Eukarya originated from within the bacterial radiation. Bacteria is the sole ancestral domain, and the other two are derived (fig. 1C;table 1). In this scenario, there is only one cellularization event, at the base of Bacteria.
Different 1D scenarios have been presented previously (Cavalier-Smith 2002; de Duve 2007; Devos and Reynaud 2010; Forterre 2011; Reynaud and Devos 2011; Forterre 2013). The most developed such scenario is arguably the neomura hypothesis, which is based on the major difference in cell wall between Bacteria and both Eukarya and Archaea, and proposing that the clade composed of Archaea and Eukarya evolved from Bacteria where one of the most obvious changes was the modification of the peptidoglycan cell wall (Cavalier-Smith 1987). The initial version of this hypothesis nested the Neomura clade (Archaea and Eukarya) as a sister group to Actinobacteria, principally due to the numerous features shared between eukaryotes and this group, including cholesterols and proteasomes (Cavalier-Smith 2002). However, the bacterial clade of origin of the Neomura is still unclear and various proposals have been made.
1D Scenarios Are Supported by Features of the PVC Bacteria
In addition, two of the discoveries most relevant for key steps of eukaryogenesis are supportive of 1D scenario and consistently ignored in any 2D models. Those steps are the development of the endomembrane system and of phagocytosis and both phenomena are related to Planctomycetes bacteria.
Based on the homology between many components of the eukaryotic endomembrane system and those of the bacterial periplasm, Günter Blobel and Christian de Duve separately suggested that the eukaryotic endomembrane system is the result of the internalization of the bacterial periplasm (Blobel 1980; de Duve 2007). In addition, the emergence and development of the eukaryotic endomembrane system has been based on MCPs (Devos et al. 2004). Planctomycetes have always been of interest for scenarios of eukaryogenesis because of their developed endomembrane system (DEMS) (Fuerst and Nisbet 2004; Forterre 2011), which is arguably one of the most developed among the prokaryotes (Santarella-Mellwig et al. 2013; Acehan et al. 2014; Devos 2014; Boedeker et al. 2017). In these bacteria, the cytoplasmic membrane sends invaginations toward the inside of the cytoplasm, forming a complex organization and representing “true” internalization of the periplasm, as hypothesized by Blobel and de Duve (Santarella-Mellwig et al. 2013; Devos 2014). However, without a molecular link between the eukaryotic and bacterial endomembrane systems, the Planctomycetes remained a curiosity of the prokaryotic world. The detection of proteins showing the structural signature of MCPs in the proteomes of various Planctomycetes (and related species), and the demonstration that these proteins sustain their endomembranes, changed this situation considerably (Santarella-Mellwig et al. 2010). Hence, like eukaryotes, some Planctomycetes have a DEMS based on MCPs (fig. 2). Although there is no sequence signal between the Planctomycetes and eukaryotic MCPs, the presence of these proteins in prokaryotes is unique and represents the first molecular link between the eukaryotic and prokaryotic endomembrane systems. Whether their structural and functional similarities argue in favor of an evolutionary relationship, the exact nature of this relationship, convergent or divergent, remains to be determined (Devos 2012).
Fig. 2.
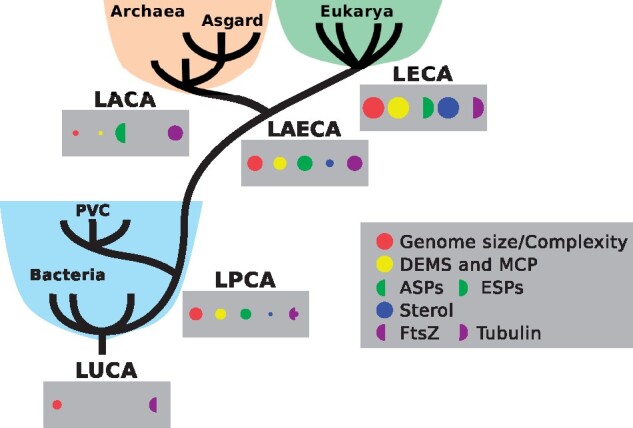
PVC-based one-domain (1D) tree of life and its implications. Last common ancestors in the PVC-based 1D scenarios. The LUCA and LPCA are the last universal and PVC common ancestors, respectively, whereas the LAECA, LACA, and LECA are the last archaeal and eukaryotic, archaeal only, and eukaryotic only common ancestors, respectively. Domains are colored as in figure 1. Traits are represented by filled disks or half-disks. Half-disks represent the transition from one form, left, to the next, right, as in the case of FtsZ to tubulin. Size of the disks or half-disks is related to the development of those features, the bigger the more developed. DEMS, developed endomembrane system; ASP, Archaea signature proteins.
The second discovery relevant to eukaryogenesis is the development of phagocytosis. Again, Planctomycetes display related phenotypes. Initially, it was shown that the planctomycete Gemmata obscuriglobus was able to internalize whole proteins before degrading them internally in a process reminiscent of eukaryotic endocytosis (Lonhienne et al. 2010). This observation was then extended to other molecules, such as dextran, and to another Planctomycetes species, Planctopirus limnophila, suggesting that this capability is more global (Boedeker et al. 2017). Recently, it was shown that a divergent Planctomycetes, Candidatus Uab amorphum, is able to phagocytose other bacteria (Shiratori et al. 2019). Despite the authors’ carefulness in their use of the term “phagocytosis-like” and in their interpretation, the phenotype appears similar to phagocytosis at least in function. Related to their phagocytic capabilities, Planctomycetes are bigger than typical bacteria; the spherical G. obscuriglobus measures a few microns (Santarella-Mellwig et al. 2013), and Ca. Uab amorphum is even bigger, reaching ∼4–5 microns in diameter (Shiratori et al. 2019).
Thus, Planctomycetes display two features of the prokaryotic world that are among the most relevant to the process of eukaryogenesis, and there is a possible, although not demonstrated, evolutionary connection between the bacterial and eukaryotic phenomena through their MCPs. Although these phenomena represent a unique case in microbiology, they appear to have had limited impact—thus far—on the scientific community. Even if the bacterial and eukaryotic MCPs are not homologous, the Planctomycetes at least illustrate the possibility of intermediate steps in the development of the eukaryotic endomembrane system from the bacterial periplasm, including phagocytosis, which supports any bacteria-based 1D scenario.
In addition, these bacteria and their relatives in the Planctomycetes-Verrucomicrobia-Chlamydiae (PVC) superphylum display additional features usually not found in bacteria and more often associated with eukaryotes or archaea, or both (Devos and Reynaud 2010; Reynaud and Devos 2011; Rivas-Marín and Devos 2018; Wiegand et al. 2018table 1 ) suggesting a PVC-based 1D scenario where the ancestor of the PVC bacteria diverged by developing some features that would later become known as archaeal, eukaryotic, or shared between them (fig. 2 and table 1). These features include the ones described hereafter. Sterol synthesis, previously thought to have developed during eukaryogenesis, has now been convincingly shown to be of bacterial origin (Santana-Molina et al. 2020). The planctomycete G. obscuriglobus is one of the few bacteria that produces sterol (although this list is constantly growing [Wei et al. 2016]) and is the only bacteria thus far for which sterol essentiality has been described (Rivas-Marin et al. 2019). Tubulin-related proteins have been described in Verrucomicrobia (Pilhofer et al. 2007; 2011) and proteins containing a tubulin-like domain have been detected in Planctomycetes too (Makarova and Koonin 2010). Similarly, the genome of Cand. U. amorphum, the phagocyte-like planctomycete, encodes an actin-related protein that is phylogenetically related to its Asgard homolog (Shiratori et al. 2019). However, the phylogenetic location of these proteins, tubulin- and actin-like, relative to their eukaryotic and archaeal homologs is still undefined. The anammox Planctomycetes, a divergent group, has a wide range of hydrocarbon chains that are either ether- or ester-linked to the glycerol backbone, the so-called ladderane membrane lipids. Few reports have followed up on this discovery, but it provides a possible solution to the issue of lipid transition (Villanueva et al. 2021). In addition, C1 transfer metabolism genes have been detected in Planctomycetes, and it has been suggested that they could be related to the origin of methanogenesis (Chistoserdova et al. 2004; but see also Adam et al. [2019] for an alternative interpretation). Similarly, members of the Verrucomicrobia and Lentisphaerae phyla have some of the enzymes from the mevalonate pathway of isoprenoid biosynthesis, which is usually associated with archaea and eukaryotes (Hoshino and Gaucher 2018). Homologs to eukaryotic serine/threonine kinases and E2 ubiquitin-conjugating enzymes were detected in Planctomycetes species (Arcas et al. 2013). Planctomycetes also contain more intrinsically disordered protein fragments, intermediary in terms of numbers between prokaryotes and eukaryotes (Bordin et al. 2018). In addition, members of the Verrucomicrobia divide by binary fission using FtsZ, demonstrating that the last PVC common ancestor (LPCA) had the ftsZ gene and divided by binary fission, as most bacteria and the last bacterial common ancestor (LBCA). However, all members of the Chlamydiae and Planctomycetes have lost the ftsZ gene, as well as others from the division and cell wall (dcw) cluster, and they divide by asymmetric division (Rivas-Marín et al. 2016, 2020). Their mechanisms of division are currently unknown. It is known that loss of ftsZ and development of a new mode of cell division occurred during eukaryogenesis. Hence, the LPCA is a bacteria that started accumulating features that will later be recognized as specific to one, or both, of the Archaea and Eukarya domains (Reynaud and Devos 2011, fig. 2).
Table 1.
Differences between Three-Domain (3D), Two-Domain (2D), and One-Domain (1D) Scenarios of the Tree of Life and Their Implications.
| Feature | 3D Scenarios | 2D Scenarios | 1D Scenarios |
|---|---|---|---|
| Ancestral domain(s) | A, B, and E | B and A | B only, A and E are derived from B |
| Origin | All independent origin | E = A + internalized B | E = derived B + internalized B, A = derived B |
| Lipids | Two ancestral lipids (B/E and A) | Two ancestral lipids (B/E and A) plus complete lipid overhaul in LECA, from A to B lipids | Only one ancestral type of lipids, B. A lipids are derived from B |
| Ancestral genome manipulation machinery | Three kinds: A and E are different from B | Two kinds: A and B | One kind B, A and E are derived |
| Metabolism | A metabolism different from B, E | A metabolism changed to B in E | A metabolism derived from divergent B |
| Ancestor complexity | A and B appeared not complex, E appeared complex | A and B appeared not complex | B not complex |
| Complexity development | Few changes | Increased in E from A, few changed in A and B | Increase of complexity from B to LAECA, then increased in E, erosion in A |
| Phylogeny | All monophyletic | A paraphyletic, E and B monophyletic | B paraphyletic, A and E monophyletic |
| LGT | Many | Many | Few |
Note.—A, Archaea; B, Bacteria; E, Eukarya; LAECA, last archaeal and eukaryotic common ancestor.
Although the relevance of some of these features for eukaryogenesis has been disputed (McInerney et al. 2011), the objectivity and accuracy of these arguments have been criticized (Forterre 2011; Reynaud and Devos 2011; Devos 2012; Cavalier-Smith and Chao 2020), and the implications of these features for eukaryogenesis have not been adequately addressed. Unfortunately, in most cases, the proteins associated with these features have not been identified, which impedes any evaluation of their evolutionary relationship. Despite the presence of some of these features in other bacteria (McInerney et al. 2011), the members of the PVC superphylum are the only ones to combine so many in a single group of related organisms (Reynaud and Devos 2011; fig. 2).
Reconciling Both Prokaryotic Superphyla with Eukaryotes in the 1D Scenario
Thus, the LECA seems to be surrounded by two prokaryotic superphyla. On the one hand, Asgard have currently the highest number of ESPs and the phylogenies of some of these ESPs place the Asgard as the closest archaea to the Eukarya. On the other hand, the PVC superphylum is the bacterial group that so far contains the most eukaryotic but also archaea-related features. Both superphyla have led to scenarios of relationships between the three domains that have different but equally valid support. How can these two apparently opposed type of scenarios be reconciled? Could the archaeal proximity to the eukaryotes be reconciled with a bacterial origin of both phyla?
In the PVC-based 1D scenario, bacteria derived from the LPCA diverged by developing some features that would later become known as archaeal, eukaryotic, or shared between them (fig. 2 and table 1). In this scenario, there is a line, or community, of organisms (O’Malley et al. 2019)—related to PVC bacteria and intermediate between the ancestor of Archaea and Eukarya, on its way to the last archaeal and eukaryotic common ancestor (LAECA)—that had already developed some of their signature characteristics (those shared between the ancestor of eukaryotes and archaea; fig. 2), that is, on its way to complexity.
In PVC-based 1D model, the LPCA was more complex than classical bacteria with features not usually observed in them (table 1). However, the LPCA is not more eukaryotic than archaeal. In this scenario, the LPCA is, in fact, the first archaeal and eukaryotic common ancestor, the FAECA. The “relative complexity” of the LPCA is in agreement with a gain of complexity toward the LAECA. Of course, complexity does not have the same meaning in different domains, but their “relative complexities” (as compared with that of other members of their domain) probably meet somewhere in the middle in some intermediary, bacteria-derived organism, en route toward Eukarya and Archaea (fig. 2). It is these intermediary organisms, between the LPCA and the LAECA, that should be the focus of future research.
Thus, in this scenario, the PVC bacteria are the sister taxa to the LAECA lineage and the root of the Archaea and eukaryotes is possibly, although not necessarily, located between the Archaea and Eukaryotic lineages. Notice that deviation from this prediction would not necessarily falsify the hypothesis. The discovery of an archaeon with a DEMS, possibly sustained by MCP(s) would also be in agreement with this hypothesis (fig. 2).
Importantly, a phylogenetic signal has recently been detected in ribosomal proteins, which supports the PVC-based 1D scenario (Cavalier-Smith and Chao 2020). It has been argued that ribosomal proteins, when treated appropriately, return more informative, accurate, and correct phylogenies than any other molecular markers. Comprehensive and thorough phylogenies based on these proteins have now robustly placed the PVC superphylum (and Sphingobacteria, including Bacteroidetes) at the base of the Archaea-Eukarya domain. Although still limited, this result could have a significant impact as it is one of the first phylogenetic analyses based on a coherent data set that supports a PVC-based 1D scenario. In agreement with this analysis, an intriguing short stem signal was detected for the Verrucomicrobia/Chlamydiae group of eukaryotic family genes in another phylogenomics analysis, possibly indicating a closest proximity between these groups (Pittis and Gabaldón 2016).
1D Scenario Addresses the Deficiencies of 2D Scenarios
As most 1D scenarios, the PVC-based 1D model proposes that the host that engulfed the proto-mitochondrion was an organism derived from a bacterium but with divergent features that are now recognized as strictly eukaryotic, archaeal, or both (fig. 2). These features kept diverging until the lineages split; one line became the Archaea, whereas the other eventually ended up with the internalization of a symbiotic partner, the mitochondria. This proposal immediately explains why Eukarya is closer to Archaea than to Bacteria: they are derived from a common ancestor, the LAECA, which was bacteria derived and not yet an archaeon or a eukaryote. This also immediately explains the mixed bacterial contribution to eukaryotic and archaeal genomes, as this common ancestor is bacteria derived (fig. 2).
This proximity would be particularly noticeable today in genes related to information processing. Possibly due to their central role in the cell, these genes that have evolved together have diverged less after the split of the two lines. This shared evolution would have led to a seemingly mixed signal, which is now interpreted as indicating a mosaic origin. The difficulties in identifying the group of origin, expressed in the apparently mixed origin of bacteria-like genes, are proposed to be the result of signal blurring after accelerated divergence, which clearly happened in the line diverging from bacteria and is a well-known issue in phylogenetics, combined with long diverging time as these events happened between 3 and 1 billion years.
The mixed origin of archaeal genomes is currently explained by massive LGT from bacteria that occurred at the origin of the Archaea (Martin et al. 2015). In the PVC-based 1D scenario, the majority of the inferred LGT events are in fact the result of continuous evolution in a bacteria-derived line, as in eukaryotes. This is supported by the strikingly similar bacterial mosaicism of archaeal and eukaryotic genomes, underlying the similarity between the evolutionary processes that has been noted before and has been suggested to reflect similar evolutionary processes, despite the differences between their evolutionary endpoints (Akanni et al. 2015).
The issue of the complexity of the ancestor of each domain is also solved in a 1D scenario where the LAECA diverged from bacteria headed toward more complexity (fig. 2; see note about complexity before). There is thus a continuous increase in complexity from Bacteria to Eukarya, only interrupted in the line branching out toward Archaea. Complexity then increased in Eukarya whereas it decreased in Archaea.
However, the metabolism of this intermediary organism, or population thereof, would still be mostly bacteria like, with a minimally DEMS that was possibly not yet differentiated but already sustained by MCPs (González-Sánchez et al. 2015, fig. 2).
The PVC-Based 1D Scenarios Support the Proximity of Asgard to Eukaryotes
The fact that the genomes of all members of the Asgard superphylum are enriched in genes encoding ESPs is often taken as directly supporting the claim that the archaeal ancestor of eukaryotes already contained several building blocks for the subsequent evolution of eukaryotic complexity—such as tubulin, actin and its regulators, longin, and histones (Grau-Bové et al. 2015; Stairs and Ettema 2020). This allegedly supports 2D scenarios, as despite the presence of some of these genes in bacteria, the Asgard ones are closer to the eukaryotic ones and claimed as ancestral. But what if it were the other way around? It is also possible that these ESPs are in fact remnants of what was present in their common “complex” ancestor, the LAECA. This would suggest that the archaea are in fact becoming less, rather than more, complex. This is supported by the fact that archaeal evolution is likely to have implied massive losses from a “complex” ancestor (Koonin 2015). In 1D scenarios, archaea are not ancestral and the so-called archaeal ancestry of many eukaryotic features (e.g., histone, actin, and tubulin) is interpreted as conservation from the LAECA in a reductive context. In 1D scenarios, archaea have lost most of the eukaryotic features, with Asgard having lost the least and diverged less, immediately explaining the two facts related with Asgard, their sheer number of ESPs and the phylogenetic proximity of these ESPs to eukaryotes (fig. 2). Therefore, 1D scenarios do not contradict the proximity of Asgard to Eukarya, but rather interpret it differently. Phylogenomic analysis of the structure of the archaeal population in terms of genes’ presence and absence, gains and losses, could resolve these issues. The presence of incomplete systems, pathways or complexes in Asgard also raises the question of the utility of incomplete sets of molecular players.
In this scenario, Asgard members are still the closest prokaryotes to eukaryotes, but this does not imply that the Asgard are the closest to the root of the Archaea. It just means that Asgard have diverged less, both in terms of sequence and of gene loss.
Interpretation
It is important to consider the possibility of PVC-based 1D scenarios, because it has various profound implications—for ancestral state and associated conditions, the age of the domains, LGT inference, assumptions of ancestrality, conserved genes and their context, among other things (table 1).
First, eukaryotes are not the result of a symbiotic merger between an archaeon and a bacterium, but are rather the result of diverging bacteria until the split with archaea. Eukaryotes are still the result of a symbiosis between the premitochondria and their ancestor, but this ancestor was not an archaeon, it was a profoundly modified bacterium. Second, understanding the origin and evolution of archaea is highly relevant to eukaryogenesis, but archaea are not ancestral to eukaryotes. Instead, archaea and eukaryotes have shared a common ancestor, the LAECA. Archaea is not an ancestral domain. One of the consequences is that methanogenesis is probably an ancestral feature of archaea, but not to life, that is, it was not in the LUCA. The connection with methylotrophy is obvious, intriguing and worthy of further consideration (Chistoserdova et al. 2004; Sorokin et al. 2017; Adam et al. 2019). Similarly, the eukaryotic genome is not a genome with contributions from both prokaryotic domains. Instead, it is a genome with contributions from the ancestral bacteria, but also shared with archaea that developed on the lineage of their common ancestor. The many features conserved between archaea and eukaryotes are not ancestral in archaea but are derived from a common ancestor and thus ancestral in the LAECA. Another important implication of 1D scenarios is that the extensive amount of LGT that has been inferred from bacteria to archaea (Nelson-Sathi et al. 2015) might possibly represent mostly remnants of a continuous line of divergence from a bacterial ancestor that was lost in some archaea and kept divergent in others.
I suggest that the branching of eukaryotes within archaea in some phylogenetic reconstructions is the product of phylogenetic artefacts associated to strong divergence and ancient events, including, but not limited to long branch attraction.
Syntrophic interactions are widespread in nature, and it is possible that such interactions between an archaeon and various bacteria are at the origin of the eukaryotes (López-García and Moreira 2020). However, a bacterial origin of Archaea (and Eukarya) argues against this possibility and resolves most of the issues associated with syntrophy scenarios.
Importantly, the PVC-based 1D scenario does not reject the phylogenetic signal between Asgard and Eukarya but suggests that new ways of analyzing the relationship between the three domains are needed. The relationship between Archaea and Eukarya is undeniable. But it is equally important to look at the relationship between these two domains and Bacteria. 2D-centric analyses will only search for more support for 2D scenarios. Even the creative proposal of considering Asgard as early diverging members of a broader group including eukaryotes and named “Eukaryomorpha” is still 2D centric (Fournier and Poole 2018). Significantly, the authors concluded that “resolving [the debate over 2 vs. 3 domains of life] depends upon identifying the root of Archaea with respect to Bacteria.” The consideration of bacterial sequences is often avoided in order to remedy the requirements of phylogeny reconstruction methods. Although technically understandable, this de facto eliminates the contemplation of 1D models. In fact, one of the few analyses considering bacterial sequences recovers significant signal consistent with the PVC-based 1D hypothesis proposed here (Cavalier-Smith and Chao 2020). The signal is weak, but this is expected as dealing with one of the most ancient relationships in the tree of life. In fact, it is expected that the signal between PVC bacteria and eukaryotes would be much less clear than between archaea and eukaryotes as the former separated much earlier than the latter. Indeed, the length of the time lag between the LAECA and the LECA is unknown, but could be anything between 1 and 2 billion years (Dacks et al. 2016). The signal between the LECA or the LACA and the LAECA is less eroded than the one between them and the LPCA, as the time elapsed is shorter, that is, the former split is younger than the latter. The signal with the archaeal domain is easier to observe, because it is younger than the one with bacteria, but the signal with the PVC bacteria is much older and thus mostly erased, requiring different, more powerful methodologies to be revealed (Santarella-Mellwig et al. 2010; Cavalier-Smith and Chao 2020).
It should be noted that 1D and 3D scenarios as presented here are similar in an unrooted perspective. 3D models are derived from the progenote that Woese imagined as ancestral to all life forms, including bacteria. A bacterial rooting was not envisioned by Woese due to his bias toward ancestrality of the archaea. Passing beyond this bias, and accepting the possibility of a bacterial root, leads to 1D scenario in agreement with 3D scenario.
The proposal might appear to be Planctomycetes centric. This is however not the case and most likely a reflection of the bias in the exploration of the PVC superphylum. The Planctomycetes phylum is so far the free-living bacteria most explored in this superphylum (Wiegand et al. 2020). It is important to explore more the whole PVC, including noncultivable organisms. There is a bias toward chlamydia due to their human pathogenicity, but they are intracellular pathogens with highly reduced and divergent genomes. The exploration of environmental chlamydia is highly relevant (Collingro et al. 2020). Although the last decade witnessed a substantial expansion of PVC bacteria diversity, a broader genomic exploration in nature is still needed.
In conclusion, the Archaea remains central to understanding the history of life, and they are linked to the origin of the eukaryotes. Asgardarchaeota is also relevant, due to its closer proximity to Eukarya. I believe that Asgard is the nearest prokaryote to, but not the prokaryotic ancestor of, the eukaryotic lineage. 1D scenarios do not contradict the proximity of Asgard to Eukarya, but they interpret this proximity differently. The PVC-based 1D scenario of the origin of life is gradual, as opposed to a big jump. It is minimalist regarding LGT; rather than denying them, it attempts to avoid excessive recourse to them. In addition, the PVC-based 1D scenario predicts that archaea are as derived as eukaryotes. Finally, this scenario is mitochondria late, in agreement with the shorter branches of alphaproteobacterial genes with respect to other bacterial genes found in eukaryotic genomes (Pittis and Gabaldón 2016).
The PVC-based 1D scenario provides a coherent path of evolution of the three domains of life, addressing most of the inconsistencies of current scenarios and taking into account most recent discoveries in the three domains. Future exploration of prokaryotic diversity should focus on the line between the LPCA and the LAECA, which is currently the main gap in the history of life on earth (fig. 2).
Acknowledgments
Thanks to many members of the CABD, UPO, Seville, SP, including members and visitors of the microPlatypus group and in particular JR Martinez, F Casares and N Maeso, for support, comments, and criticisms. Very special thanks to Carlos Santana-Molina for providing data, endless discussions, and critical comments on the concept and the manuscript, as well as for initial suggestions on the artwork. Special thanks to Elena Rivas-Marin. D.P.D. was supported by the Spanish Ministry of Economy and Competitiveness (Grant No. BFU2016-78326-P) and is currently supported by the “Moore-Simons Project on the Origin of the Eukaryotic Cell” (Grant No. 9733). We thank all reviewers as well as the MBE editorial team for critical comments, suggestions, and corrections of the manuscript. This manuscript is dedicated to Thomas Cavalier-Smith (October 1942–March 2021), University of Oxford, who inspired most of our evolutionary thinking and whose recent version of the neomura hypothesis merged with the PVC superphylum complementary to this proposal.
Data Availability
No new data were generated or analyzed in support of this research.
References
- Acehan D, Santarella-Mellwig R, Devos DP.. 2014. A bacterial tubulovesicular network. J Cell Sci. 127(Pt 2):277–280. [DOI] [PubMed] [Google Scholar]
- Adam PS, Borrel G, Gribaldo S.. 2019. An archaeal origin of the Wood–Ljungdahl H 4 MPT branch and the emergence of bacterial methylotrophy. Nat Microbiol. 4(12):2155–2163. [DOI] [PubMed] [Google Scholar]
- Akanni WA, Siu-Ting K, Creevey CJ, McInerney JO, Wilkinson M, Foster PG, Pisani D.. 2015. Horizontal gene flow from Eubacteria to Archaebacteria and what it means for our understanding of eukaryogenesis. Philos Trans R Soc Lond B Biol Sci. 370(1678):20140337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akıl C, Robinson RC.. 2018. Genomes of Asgard archaea encode profilins that regulate actin. Nature 562(7727):439–443. [DOI] [PubMed] [Google Scholar]
- Akıl C, Tran LT, Orhant-Prioux M, Baskaran Y, Manser E, Blanchoin L, Robinson RC.. 2020. Insights into the evolution of regulated actin dynamics via characterization of primitive gelsolin/cofilin proteins from Asgard archaea. Proc Natl Acad Sci U S A. 117(33):19904–19913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcas A, Cases I, Rojas AM.. 2013. Serine/threonine kinases and E2-ubiquitin conjugating enzymes in Planctomycetes: unexpected findings. Antonie Van Leeuwenhoek. 104(4):509–520. [DOI] [PubMed] [Google Scholar]
- Baker BJ, De Anda V, Seitz KW, Dombrowski N, Santoro AE, Lloyd KG.. 2020. Diversity, ecology and evolution of Archaea. Nat Microbiol. 5(7):887–900. [DOI] [PubMed] [Google Scholar]
- Baum DA, Baum B.. 2014. An inside-out origin for the eukaryotic cell. BMC Biol. 12:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G.1980. Intracellular protein topogenesis. Proc Natl Acad Sci U S A. 77(3):1496–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedeker C, Schüler M, Reintjes G, Jeske O, van Teeseling MCF, Jogler M, Rast P, Borchert D, Devos DP, Kucklick M, et al. 2017. Determining the bacterial cell biology of Planctomycetes. Nat Commun. 8:14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordin N, González-Sánchez JC, Devos DP.. 2018. PVCbase: an integrated web resource for the PVC bacterial proteomes. Database 2018:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueckner J, Martin WF.. 2020. Bacterial genes outnumber archaeal genes in eukaryotic genomes. Genome Biol Evol. 12(4):282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulzu P-A, Andrei A, Salcher MM, Mehrshad M, Inoue K, Kandori H, Beja O, Ghai R, Banciu HL.. 2019. Casting light on Asgardarchaeota metabolism in a sunlit microoxic niche. Nat Microbiol. 4(7):1129–1137. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T.1987. The origin of eukaryotic and archaebacterial cells. Ann N Y Acad Sci. 503:17–54. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T.2002. The neomuran origin of Archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int J Syst Evol Microbiol. 52(Pt 1):7–76. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE-Y.. 2020. Multidomain ribosomal protein trees and the planctobacterial origin of neomura (eukaryotes, Archaebacteria). Protoplasma 257(3):621–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L, Jenkins C, Kalyuzhnaya MG, Marx CJ, Lapidus A, Vorholt JA, Staley JT, Lidstrom ME.. 2004. The enigmatic planctomycetes may hold a key to the origins of methanogenesis and methylotrophy. Mol Biol Evol. 21(7):1234–1241. [DOI] [PubMed] [Google Scholar]
- Coleman GA, Pancost RD, Williams TA.. 2019. Investigating the origins of membrane phospholipid biosynthesis genes using outgroup-free rooting. Genome Biol Evol. 11(3):883–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingro A, Köstlbacher S, Horn M.. 2020. Chlamydiae in the environment. Trends Microbiol. 28(11):877–888. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A.2017. Lynn Margulis and the origin of the eukaryotes. J Theor Biol. 434:1. [DOI] [PubMed] [Google Scholar]
- Csurös M, Miklós I.. 2009. Streamlining and large ancestral genomes in Archaea inferred with a phylogenetic birth-and-death model. Mol Biol Evol. 26(9):2087–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha VD, Gaia M, Gadelle D, Nasir A, Forterre P.. 2017. Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes. PLoS Genet. 13(6):e1006810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacks JB, Field MC, Buick R, Eme L, Gribaldo S, Roger AJ, Brochier-Armanet C, Devos DP.. 2016. The changing view of eukaryogenesis - fossils, cells, lineages and how they all come together. J Cell Sci. 129(20):3695–3703. [DOI] [PubMed] [Google Scholar]
- Deschamps P, Zivanovic Y, Moreira D, Rodriguez-Valera F, López-García P.. 2014. Pangenome evidence for extensive interdomain horizontal transfer affecting lineage core and shell genes in uncultured planktonic thaumarchaeota and euryarchaeota. Genome Biol Evol. 6(7):1549–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos DP.2012. Regarding the presence of membrane coat proteins in bacteria: confusion? What confusion? Bioessays 34(1):38–39. [DOI] [PubMed] [Google Scholar]
- Devos DP.2014. PVC bacteria: variation of, but not exception to, the Gram-negative cell plan. Trends Microbiol. 22(1):14–20. [DOI] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP.. 2004. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2(12):e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos DP, Reynaud EG.. 2010. Evolution. Intermediate steps. Science 330(6008):1187–1188. [DOI] [PubMed] [Google Scholar]
- Dey G, Thattai M, Baum B.. 2016. On the archaeal origins of eukaryotes and the challenges of inferring phenotype from genotype. Trends Cell Biol. 26(7):476–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Duve C.2007. The origin of eukaryotes: a reappraisal. Nat Rev Genet. 8(5):395–403. [DOI] [PubMed] [Google Scholar]
- Forterre P.2011. A new fusion hypothesis for the origin of Eukarya: better than previous ones, but probably also wrong. Res Microbiol. 162(1):77–91. [DOI] [PubMed] [Google Scholar]
- Forterre P.2013. The common ancestor of archaea and Eukarya was not an archaeon. Archaea 2013:372396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier GP, Gogarten JP.. 2010. Rooting the ribosomal tree of life. Mol Biol Evol. 27(8):1792–1801. [DOI] [PubMed] [Google Scholar]
- Fournier GP, Poole AM.. 2018. A briefly argued case that Asgard archaea are part of the Eukaryote tree. Front Microbiol. 9:1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox GE, Stackebrandt E, Hespell RB, Gibson J, Maniloff J, Dyer TA, Wolfe RS, Balch WE, Tanner RS, Magrum LJ, et al. 1980. The phylogeny of prokaryotes. Science 209(4455):457–463. [DOI] [PubMed] [Google Scholar]
- Fuerst JA, Nisbet EG.. 2004. Buds from the tree of life: linking compartmentalized Prokaryotes and Eukaryotes by a non-hyperthermophile common ancestor and implications for understanding archaean microbial communities. Int J Astrobiol. 3(3):183–187. [Google Scholar]
- Garg SG, Kapust N, Lin W, Knopp M, Tria FDK, Nelson-Sathi S, Gould SB, Fan L, Zhu R, Zhang C, et al. 2021. Anomalous phylogenetic behavior of ribosomal proteins in metagenome-assembled Asgard archaea. Genome Biol Evol. 13(1):evaa238. doi:10.1093/gbe/evaa238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Sánchez JC, Costa R, Devos DP.. 2015. A multi-functional tubulovesicular network as the ancestral eukaryotic endomembrane system. Biology 4(2):264–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau-Bové X, Sebé-Pedrós A, Ruiz-Trillo I.. 2015. The eukaryotic ancestor had a complex ubiquitin signaling system of archaeal origin. Mol Biol Evol. 32(3):726–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groussin M, Boussau B, Szöllõsi G, Eme L, Gouy M, Brochier-Armanet C, Daubin V.. 2016. Gene acquisitions from bacteria at the origins of major archaeal clades are vastly overestimated. Mol Biol Evol. 33(2):305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampl V, Čepička I, Eliáš M.. 2019. Was the mitochondrion necessary to start Eukaryogenesis? Trends Microbiol. 27(2):96–104. [DOI] [PubMed] [Google Scholar]
- Hoshino Y, Gaucher EA.. 2018. On the origin of isoprenoid biosynthesis. Mol Biol Evol. 35(9):2185–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, Takano Y, Uematsu K, Ikuta T, Ito M, et al. 2020. Isolation of an archaeon at the prokaryote–eukaryote interface. Nature 577(7791):519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust N, Nelson-Sathi S, Schönfeld B, Hazkani-Covo E, Bryant D, Lockhart PJ, Röttger M, Xavier JC, Martin WF.. 2018. Failure to recover major events of gene flux in real biological data due to method misapplication. Genome Biol Evol. 10(5):1198–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner S, Spang A, Offre P, Szöllősi GJ, Petitjean C, Williams TA.. 2018. Genome size evolution in the Archaea. Emerg Top Life Sci. 2(4):595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger CM, Spang A, Dacks JB, Ettema TJG.. 2016. Tracing the archaeal origins of eukaryotic membrane-trafficking system building blocks. Mol Biol Evol. 33(6):1528–1541. [DOI] [PubMed] [Google Scholar]
- Koonin EV.2015. Origin of eukaryotes from within archaea, archaeal eukaryome and bursts of gene gain: eukaryogenesis just made easier? Philos Trans R Soc Lond B Biol Sci. 370(1678):20140333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Yutin N.. 2014. The dispersed archaeal eukaryome and the complex archaeal ancestor of eukaryotes. Cold Spring Harb Perspect Biol. 6(4):a016188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Dolja VV, Koonin EV.. 2020. The LUCA and its complex virome. Nat Rev Microbiol. 18(11):661–670. [DOI] [PubMed] [Google Scholar]
- Ku C, Martin WF.. 2016. A natural barrier to lateral gene transfer from prokaryotes to eukaryotes revealed from genomes: the 70% rule. BMC Biol. 14(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake JA, Henderson E, Oakes M, Clark MW.. 1984. Eocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc Natl Acad Sci U S A. 81(12):3786–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake JA, Skophammer RG, Herbold CW, Servin JA.. 2009. Genome beginnings: rooting the tree of life. Philos Trans R Soc Lond B Biol Sci. 364(1527):2177–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Makarova KS, Huang W-C, Wolf YI, Nikolskaya AN, Zhang X, Cai M, Zhang C-J, Xu W, Luo Z, et al. 2021. Expanding diversity of Asgard archaea and the elusive ancestry of eukaryotes. Nature 593(7860):553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonhienne TGA, Sagulenko E, Webb RI, Lee K-C, Franke J, Devos DP, Nouwens A, Carroll BJ, Fuerst JA.. 2010. Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci U S A. 107(29):12883–12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-García P, Moreira D.. 2020. The syntrophy hypothesis for the origin of eukaryotes revisited. Nat Microbiol. 5(5):655–667. [DOI] [PubMed] [Google Scholar]
- MacLeod F, Kindler GS, Wong HL, Chen R, Burns BP.. 2019. Asgard archaea: diversity, function, and evolutionary implications in a range of microbiomes. AIMS Microbiol. 5(1):48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV.. 2010. Two new families of the FtsZ-tubulin protein superfamily implicated in membrane remodeling in diverse bacteria and archaea. Biol Direct. 5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martijn J, Vosseberg J, Guy L, Offre P, Ettema TJG.. 2018. Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 557(7703):101–105. [DOI] [PubMed] [Google Scholar]
- Martin WF.2017a. Symbiogenesis, gradualism, and mitochondrial energy in eukaryote origin. Period Biol. 119(3):141–158. [Google Scholar]
- Martin WF.2017b. Too much Eukaryote LGT. BioEssays 39(12):1700115. [DOI] [PubMed] [Google Scholar]
- Martin WF, Garg S, Zimorski V.. 2015. Endosymbiotic theories for eukaryote origin. Philos Trans R Soc Lond B Biol Sci. 370(1678):20140330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney JO, Martin WF, Koonin EV, Allen JF, Galperin MY, Lane N, Archibald JM, Embley TM.. 2011. Planctomycetes and eukaryotes: a case of analogy not homology. Bioessays 33(11):810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson-Sathi S, Dagan T, Landan G, Janssen A, Steel M, McInerney JO, Deppenmeier U, Martin WF.. 2012. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc Natl Acad Sci U S A. 109(50):20537–20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson-Sathi S, Sousa FL, Roettger M, Lozada-Chávez N, Thiergart T, Janssen A, Bryant D, Landan G, Schönheit P, Siebers B, et al. 2015. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517(7532):77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley MA, Leger MM, Wideman JG, Ruiz-Trillo I.. 2019. Concepts of the last eukaryotic common ancestor. Nat Ecol Evol. 3(3):338–344. [DOI] [PubMed] [Google Scholar]
- Pilhofer M, Ladinsky MS, McDowall AW, Petroni G, Jensen GJ.. 2011. Microtubules in bacteria: ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton. PLoS Biol. 9(12):e1001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilhofer M, Rosati G, Ludwig W, Schleifer K-H, Petroni G.. 2007. Coexistence of tubulins and ftsZ in different Prosthecobacter species. Mol Biol Evol. 24(7):1439–1442. [DOI] [PubMed] [Google Scholar]
- Pisani D, Cotton JA, McInerney JO.. 2007. Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol Biol Evol. 24(8):1752–1760. [DOI] [PubMed] [Google Scholar]
- Pittis AA, Gabaldón T.. 2016. Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature 531(7592):101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud EG, Devos DP.. 2011. Transitional forms between the three domains of life and evolutionary implications. Proc Biol Sci. 278(1723):3321–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-Marín E, Canosa I, Devos DP.. 2016. Evolutionary cell biology of division mode in the bacterial planctomycetes-verrucomicrobia-chlamydiae superphylum. Front Microbiol. 7:1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-Marín E, Devos DP.. 2018. The paradigms they are a-Changin’: past, present and future of PVC bacteria research. Antonie Van Leeuwenhoek. 111(6):785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-Marin E, Peeters SH, Claret Fernández L, Jogler C, van Niftrik L, Wiegand S, Devos DP.. 2020. Non-essentiality of canonical cell division genes in the planctomycete Planctopirus limnophila. Sci Rep. 10(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-Marin E, Stettner S, Gottshall EY, Santana-Molina C, Helling M, Basile F, Ward NL, Devos DP.. 2019. Essentiality of sterol synthesis genes in the planctomycete bacterium Gemmata obscuriglobus. Nat Commun. 10(1):2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Dobro MJ, Jensen GJ, Bell SD.. 2017. The structure, function and roles of the archaeal ESCRT apparatus. In: Löwe J, Amos LA, editors. Prokaryotic cytoskeletons: filamentous protein polymers active in the cytoplasm of bacterial and archaeal cells. Subcellular biochemistry Cham: Springer International Publishing. p. 357–377. [Google Scholar]
- Santana-Molina C, Rivas-Marin E, Rojas AM, Devos DP.. 2020. Origin and evolution of polycyclic triterpene synthesis. Mol Biol Evol. 37(7):1925–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarella-Mellwig R, Franke J, Jaedicke A, Gorjanacz M, Bauer U, Budd A, Mattaj IW, Devos DP.. 2010. The compartmentalized bacteria of the planctomycetes-verrucomicrobia-chlamydiae superphylum have membrane coat-like proteins. PLoS Biol. 8(1):e1000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarella-Mellwig R, Pruggnaller S, Roos N, Mattaj IW, Devos DP.. 2013. Three-dimensional reconstruction of bacteria with a complex endomembrane system. PLoS Biol. 11(5):e1001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratori T, Suzuki S, Kakizawa Y, Ishida K.. 2019. Phagocytosis-like cell engulfment by a planctomycete bacterium. Nat Commun. 10(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin DY, Makarova KS, Abbas B, Ferrer M, Golyshin PN, Galinski EA, Ciordia S, Mena MC, Merkel AY, Wolf YI, et al. 2017. Discovery of extremely halophilic, methyl-reducing euryarchaea provides insights into the evolutionary origin of methanogenesis. Nat Microbiol. 2:17081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Eme L, Saw JH, Caceres EF, Zaremba-Niedzwiedzka K, Lombard J, Guy L, Ettema TJG.. 2018. Asgard archaea are the closest prokaryotic relatives of eukaryotes. PLoS Genet. 14(3):e1007080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJG.. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521(7551):173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Stairs CW, Dombrowski N, Eme L, Lombard J, Caceres EF, Greening C, Baker BJ, Ettema TJG.. 2019. Proposal of the reverse flow model for the origin of the eukaryotic cell based on comparative analyses of Asgard archaeal metabolism. Nat Microbiol. 4(7):1138–1148. [DOI] [PubMed] [Google Scholar]
- Stairs CW, Ettema TJG.. 2020. The archaeal roots of the eukaryotic dynamic actin cytoskeleton. Curr Biol. 30(10):R521–R526. [DOI] [PubMed] [Google Scholar]
- Villanueva L, von Meijenfeldt FAB, Westbye AB, Yadav S, Hopmans EC, Dutilh BE, Damsté JSS.. 2021. Bridging the membrane lipid divide: bacteria of the FCB group superphylum have the potential to synthesize archaeal ether lipids. Isme J. 15(1):168–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva L, Schouten S, Sinninghe Damsté JS.. 2017. Phylogenomic analysis of lipid biosynthetic genes of Archaea shed light on the “lipid divide”. Environ Microbiol. 19(1):54–69. [DOI] [PubMed] [Google Scholar]
- Wächtershäuser G.2003. From pre-cells to Eukarya–a tale of two lipids. Mol Microbiol. 47(1):13–22. [DOI] [PubMed] [Google Scholar]
- Wei JH, Yin X, Welander PV.. 2016. Sterol synthesis in diverse bacteria. Front Microbiol. 7:990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand S, Jogler M, Boedeker C, Pinto D, Vollmers J, Rivas-Marín E, Kohn T, Peeters SH, Heuer A, Rast P, et al. 2020. Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology. Nat Microbiol. 5(1):126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand S, Jogler M, Jogler C.. 2018. On the maverick planctomycetes. FEMS Microbiol Rev. 42(6):739–760. [DOI] [PubMed] [Google Scholar]
- Williams TA, Cox CJ, Foster PG, Szöllősi GJ, Embley TM.. 2020. Phylogenomics provides robust support for a two-domains tree of life. Nat Ecol Evol. 4(1):138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Heaps SE, Cherlin S, Nye TMW, Boys RJ, Embley TM.. 2015. New substitution models for rooting phylogenetic trees. Philos Trans R Soc Lond B Biol Sci. 370: PMC4571574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML.. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 87(12):4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, et al. 2017. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541(7637):353–358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analyzed in support of this research.