Abstract
Extreme halophilic Archaea thrive in high salt, where, through proteomic adaptation, they cope with the strong osmolarity and extreme ionic conditions of their environment. In spite of wide fundamental interest, however, studies providing insights into this adaptation are scarce, because of practical difficulties inherent to the purification and characterization of halophilic enzymes. In this work, we describe the evolutionary history of malate dehydrogenases (MalDH) within Halobacteria (a class of the Euryarchaeota phylum). We resurrected nine ancestors along the inferred halobacterial MalDH phylogeny, including the Last Common Ancestral MalDH of Halobacteria (LCAHa) and compared their biochemical properties with those of five modern halobacterial MalDHs. We monitored the stability of these various MalDHs, their oligomeric states and enzymatic properties, as a function of concentration for different salts in the solvent. We found that a variety of evolutionary processes, such as amino acid replacement, gene duplication, loss of MalDH gene and replacement owing to horizontal transfer resulted in significant differences in solubility, stability and catalytic properties between these enzymes in the three Halobacteriales, Haloferacales, and Natrialbales orders since the LCAHa MalDH. We also showed how a stability trade-off might favor the emergence of new properties during adaptation to diverse environmental conditions. Altogether, our results suggest a new view of halophilic protein adaptation in Archaea.
Keywords: ancestral protein resurrection, horizontal gene transfer, halophilic adaptation of proteins, malate dehydrogenase, Haloarchaea, Archaea
Introduction
The adaptation of organisms to fluctuating environments occurred through two evolutionary processes: amino acid replacements (replacement process, RP) that affect protein structure, function and dynamics (Tomatis et al. 2008; Liu and Bahar 2012), and, less frequently, horizontal gene transfer (HGT, the acquisition of a foreign functional coding gene), which can quickly modify the properties of an organism (Fournier et al. 2015). With respect to RP, a modern protein has evolved on a trajectory, through a series of now extinct ancestral states, via gradual primary sequence modifications. Sequence variability is shaped by the equilibrium between genetic drift and Darwinian selection, which will tend to clean out most of the mutations in order to maintain initial structure and function (Tokuriki and Tawfik 2009a). HGT has been detected in numerous organisms and is well documented as an important driving evolutionary force in prokaryotes and eukaryotes (Papke et al. 2004; Khomyakova et al. 2011; Andam et al. 2012; Naor et al. 2012; Kaminskiet al. 2013; Alexander et al. 2016). However, experimental studies that determine the properties of HGT genes are very rare (Savory et al. 2018). Here, we report the recovery and analysis of protein evolutionary trajectories, impacted both by gradual RP and HGT, which preserved structure and function while adapting to extreme physicochemical conditions.
A protein is a dynamical entity, which exists as a population of conformers (alternative substructures having slightly different free energies) represented statistically in specific proportions on various time scales (Frauenfelderet al. 2009; Lewandowski et al. 2015). Protein conformational landscapes are determined by hydrophobic, ionic, hydrogen bond and Van der Waals interactions between residues within the macromolecule as well as between the macromolecule and its solvent environment. Solvent interactions, in particular, influence folding pathways, stability, dynamics and solubility of the native state, so that proteins can be considered as dynamic protein–solvent complexes (Svergun et al. 1998; Bellissent-Funel et al. 2016). Mutational processes, therefore, impact protein–solvent interactions as well as intramolecular interactions. So far, however, we have not been aware of published work on the effects of amino acid replacements on protein–solvent interactions along an evolutionary trajectory. In order to address this fundamental question, we adopted halobacterial MalDH as a model enzyme and applied ancestral protein sequence resurrection (ASR) to access ancient proteins on the evolutionary pathway, in order to compare their properties with those from extant MalDH orthologs. ASR has been intensively used to investigate various functional changes in proteins and phenotypic adaptations to environmental constraints (Gaucher et al. 2003; Finnigan et al. 2012; Hobbs et al. 2012; Voordeckers et al. 2012; Mirceta et al. 2013; Hart et al. 2014; Kacar et al. 2017; Gonzales-Ordenes et al. 2018). MalDH is a ubiquitous enzyme involved in central metabolism, of which several forms have been characterized. In Archaea, MalDH is a constitutively expressed enzyme, which operates in the tri-carboxylic acid (Krebs) cycle to reversibly catalyze the conversion of malate into oxaloacetate linked to the oxidation/reduction of dinucleotide coenzymes (Madern 2002; Minárik et al. 2002).In order to maintain their efficiency within the metabolism, MalDHs are adapted to compete against the harsh conditions frequently encountered in extremophilic Archaea.
Here, we chose to investigate adaptation to high salinity. Recent phylogenetic studies have shown that the “salt-in” strategy used for thriving in hypersaline environments may have independently emerged on three occasions in Archaea (Aouad et al. 2018, 2019). Amongst these organisms, Halobacteria (a class belonging to Euryarchaeota), also frequently mentioned as Haloarchaea, are the most extensively studied (Oren 2002). Species from Halobacteria are adapted primarily to molar salt environments, with some species displaying secondary adaptation to other extreme physicochemical conditions, such as high pH (Falb et al. 2005), high (Kim et al. 2018), low temperature (DasSarma et al. 2013), or high pressure (Albuquerque et al. 2012). Early work suggested that halobacterial species obey the salt-in strategy, that is, the accumulation of high concentrations of mainly KCl in the cytoplasm to counterbalance the high osmotic pressure due to NaCl outside (Oren 2002). Analyses of ion transporter gene distribution have revealed, however, that the intracellular composition is better described as a complex, dynamic mixture of various salts and compatible solutes in order to accommodate salinity fluctuations encountered in the natural environment (Becker et al. 2014). The specific intracellular composition imposes further adaptation at the protein level to maintain solubility, stability and activity for a range of saline conditions (Madern, Ebel, Zaccai, et al. 2000).
Numerous studies have used tetrameric NADH-dependent MalDH from halophilic Archaea and Bacteria as a model to describe how solvent interactions govern protein properties (Costenaro et al. 2002; Irimia et al. 2003; Coquelle et al. 2010; Talon et al. 2014). Because HGT has been reported as a frequent phenomenon in Halobacteria (Papke et al. 2004; Khomyakova et al. 2011) and because some halobacterial MalDHs result clearly from duplication events, we performed ASR taking into account HGT events. In order to ensure maximum accuracy, we considered advanced phylogenetic models and large taxonomic samples; we estimated the halobacterial species phylogeny by analyzing simultaneously core genes, and inferred a joint MalDH phylogeny accounting for HGTs, duplications and losses. In addition, we applied indel-aware models to estimate ancestral MalDH sequences. Finally, we synthesized a number of key ancestral MalDHs.
Salt-dependent conformational stability and enzymatic properties of MalDHs from different modern lineages, together with their resurrected ancestors, were determined, by applying the same methodology as in our previous studies of the MalDH from Haloarcula marismortui. Furthermore, analytical ultracentrifugation (AUC) experiments demonstrated how changes of the oligomeric state equilibrium between active conformers could rescue the deleterious effect of low salt concentration in some lineages.
These are the main findings of our study: The last common ancestor MalDH of Halobacteria (LCAHa) does not necessitate multi-molar KCl in order to be stable and active, suggesting it was mildly halophilic; the paleo environment at the origin of Halobacteria in Archaea could not be inferred, because the KCl-concentration-dependent stabilities of MalDHs are not correlated with the respective environmental conditions of their hosts; the description of a tradeoff mechanism by which the haloalkaliphilic lineage of MalDH has emerged; and the unexpected experimental outcome that the modern MalDH from H. marismortui (the most studied halophilic enzyme to date) was the result of an ancestral HGT from an early haloalkaliphilic ancestor.
We concluded that solubility, stability and activity of MalDHs facing extreme salt conditions did not evolve concomitantly, and discuss, here, how these findings establish a new understanding of halophilic adaptation in MalDHs.
Results
Phylogenetic Inferences
Groussin et al. (2015) have shown the importance of taking into account the process of diversification within prokaryotes, in which both RP and HGT are accounted for when performing ASR. First, the species tree of Halobacteria was reconstructed from a large phylogenomic concatenation (see Materials and Methods). We examined several sources of reconstruction artifacts resulting from long branch attraction (LBA) artifacts and compositional biases to obtain a robust and strongly supported halobacteria species phylogeny (fig. 1 and supplementary fig. S1, Supplementary Material online). Halobacteria appeared to be subdivided into three main sub-lineages: Halobacteriales (group A, including H. marismortui: “H. mari” and Halomicrobuim mukohataei: “H. muko”), Haloferacales (group B, including Haloferax volcanii: “H. volc” and Halorubrum lacusprofundi: “H. lacu”) and Natrialbales(group C, a monophyletic group of alkaliphilic halobacterial species adapted to high pH environments and including Natrialba magadii: “N maga”), consistent with previous findings (Brochier-Armanet et al. 2011). Moreover, we inferred a strongly supported monophyly of species from groups B and C (see Supplementary Material section “Species tree reconstruction”).
Fig. 1.
Halobacteria species tree (pipes) and MalDH gene tree (dashed colored lines). The species tree is subdivided into three sublineages, groups A–C. Biochemical characterization of extant MalDHs were obtained for Haloarcula marismortui and Halomicrobuim mukohataei (group A), Haloferax volcanii and Halorubrum lacusprofundi (group B), and Natrialba magadii (group C). The halobacterial MalDH genes have encountered a complex history, with gene loss (crosses), gene transfers (dots), and gene duplication (squares). The ten ancestral MalDH sequences for which biochemical properties were studied are indicated on the MalDH tree nodes (Anc “N”). The examined ancestral MalDH existed in the last common ancestor of Halobacteria (LCAHa, Anc78), in LCAs of group A (Anc79), group B plus C (Anc80), group B (Anc81), and group C (Anc89). We also examined recent ancestors of H. volcanii (Anc82) and H. lacusprofundi (Anc84) MalDHs within group B, the MalDH ancestors of paralogous duplicates (Anc90) and of the copy inherited in N. magadii (Anc93) within group C, and finally the LCA (Anc88) of the transferred H. marismortui MalDH and of group C MalDHs.
In the second step of the analysis, a MalDH tree was reconstructed using the site heterogeneous Bayesian phylogenetic model CAT+GTR (Lartillot et al. 2009) (see Materials and Methods and supplementary material and fig. S2, Supplementary Material online). It reveals a weak phylogenetic signal and large incongruities with respect to the Halobacteria species phylogeny (supplementary fig. S1, Supplementary Material online).
Then, the reconstructed MalDH tree was reconciled with the species tree of Halobacteria (see Materials and Methods), yielding a final MalDH phylogeny correcting initial stochastic uncertainties and accounting for gene duplication, horizontal transfers and losses (DTL, Szöllosi, Rosikiewicz, et al. 2013; Szöllosi, Tannier, et al. 2013). This phylogeny, called the MalDH “joint-tree,” is presented in supplementary figure S3, Supplementary Material online and figure 1. Importantly, the MalDH joint-tree appears to be well supported (see posterior probabilities [PP] in supplementary fig. S3, Supplementary Material online). It reveals a highly supported HGT toward H. marismortui from an ancestral lineage related to the ancestor of group C species (PP = 0.99). This strong support, the short length of the H. mari MalDH branch (0.086 expected replacement per site, see supplementary fig. S3, Supplementary Material online) and biochemical observations (see below, enzymatic characterization) suggest that this clustering unlikely results from a LBA. The tree also describes a complex history of MalDH DTL events within the alkaliphilic Halobacteria (group C, see supplementary fig. S3, Supplementary Material online). No other DTL events are inferred during the evolution of MalDHs in groups A and B (fig. 1).
Subsequently, the MalDH protein sequences were aligned anew using PRANK (Löytynoja and Goldman 2008), considering indels as evolutionary events and the MalDH joint-tree as guide tree (see Materials and Methods).
Finally, the ancestral sequences were computed from the PRANK alignment, considering the MalDH joint-tree as a fixed topology, and using the CAT+GTR Bayesian model (Lartillot et al. 2009, see Materials and Methods). Control experiments were performed using other available methods, implemented in the Maximum Likelihood framework, in order to assess the accuracy of the ancestral sequence reconstructions (ASRs). Most noticeably, in each ten examined ancestral MalDHs, at least 90% of ancestral amino acid sites appear well identified (PP > 0.9), and only 2–6 sites are differently inferred depending on the applied method (see Supplementary Material, section “Ancestral MalDH reconstruction”). The MalDHs retained for biochemical characterizations span the oldest ancestors in Halobacteria and several recent ancestors close to the five extant MalDHs with known properties (fig. 1).
Evolution of Surface Residues and Protein Solubility
Acidic residue enrichment of proteomic composition in halophilic species, when compared with nonhalophiles is well established (Paul et al. 2008).The electrostatic field generated by charges and the solvent-accessible hydrophobic surface of a halophilic protein can be calculated using its crystal structure (Coquelle et al. 2010). These calculations were applied to a set of 15 structures of halophilic proteins, compared with their nonhalophilic counterparts. They showed that the negatively charged amino acid enrichment is a specific feature of the solvent-accessible surface that is compensated by a decrease in basic residues (Graziano and Merlino 2014).
Studies comparing MalDHs from extremely halophilic Bacteria and Archaea with nonhalophilic homologs have clearly demonstrated that this change in surface electrostatic properties is an efficient strategy to maintain protein solubility at high salt concentration (Solovyova et al. 2001; Coquelle et al. 2010; Talon et al. 2014). We calculated the ratio between negatively (D + E) and positively (R + K) charged amino acids for several modern and ancestral MalDHs. DE/KR ratios >2 indicate that the acidic amino acid surface enrichment is a common property of halobacterial MalDH shared since the root node Anc78 (five modern species, nine reconstructed MalDH ancestors characterized), whereas the nonhalophilic outgroup of modern MalDH displays nearly neutral surfaces (fig. 2A). A recent study of salt adaptation in Methanosarcinales has shown that the surface of ADP-dependent kinases changes its amino acid composition already in the moderately high environmental concentration of 0.5 M NaCl (Gonzales-Ordenes et al. 2018). The DE/KR ratio we calculated using Methanosarcinales MalDHs also indicates a moderate acidic enrichment (ratio of 1.5, supplementary table S4, Supplementary Material online). This suggests that, among other mechanisms, protein surface acidic enrichment could be a widespread strategy in moderate to extreme halophilic adaptation. According to most recent molecular phylogenies however, Halobacteria are closer to Methanomicrobiales than to Methanosarcinales (Aouad et al. 2018). Consequently, understanding if these phenotypes are convergent or diverged gradually from a common origin would deserve further scrutiny, which falls outside the scope of this study.
Fig. 2.
Acidic amino acid enrichment favors solubility in high KCl concentration. (A) The ratio between negatively charged residues over positively charged ones (DE/KR) for various modern and ancestral MalDH. High values of the ratio reflect acidic amino acid surface enrichment as it is illustrated by the electrostatic surface representation (small insets) of Haloarcula marismortui MalDH (pdb code 4J5K) and Picrophilus torridus MalDH (pdb code 4BVG). The positively and negatively charged surfaces are colored in blue and red, respectively. (B) Variations of the experimental sedimentation coefficient (s exp) at 3.8 M KCl, recorded at various protein concentrations, for Anc80 MalDH and H. volc MalDH (this work). Data for H. mari MalDH and for a nonhalophilic MalDH are from Coquelle et al. (2010).
In previous studies on H. mari MalDH, we have found that under given solvent conditions, constant or slightly decreasing sedimentation coefficient values over increasing protein concentration indicated interparticle repulsive effects favoring solubility of the tetrameric enzyme. In contrast, an increasing relation would indicate the general tendency of the protein to aggregate at high salt concentration (Solovyova et al. 2001; Coquelle et al. 2010). We applied the same methodology on the MalDH of H. volcanii and Anc80 in 3.8 M KCl. The negative slopes observed in figure 2B for ancestral and modern halobacterial MalDHs confirm that they are soluble as tetramers at high KCl concentration, whereas, under similar conditions, nonhalophilic proteins precipitate. To conclude, our data showed that acidic surface enrichment is an ancestral mechanism in Halobacteria to avoid protein precipitation in high salt conditions.
Salt Concentration Dependence of Conformational Stability in Modern and Ancestral MalDHs
Depending on their effects on the conformational stability of proteins, salts have been categorized into three mains groups: “salting-out,” “neutral,” and “salting-in” (Von Hippel and Schleich 1969). In a pioneering work, the effects of salts from these groups on of H. mari MalDH suggested that stability mechanisms accommodate to solvent conditions (Bonneté et al. 1994). The study revealed the additivity of stabilization forces in this halophilic protein, their subtle equilibria, as well as the rich palette of energy barriers competing against deleterious physico-chemical conditions. Since, numerous biophysical studies on the effects of physiological and nonphysiologically relevant salts, aimed at understanding halophily in H. mari MalDH, have revealed that stability strongly depends on the nature of solvent ions (Bonneté et al. 1994, Madern and Zaccai 1997; Ebel et al. 1999; Irimia et al. 2003; Madern and Ebel 2007).In particular, they have shown that ion charge density is the major determinant influencing the unfolding transition when the salt concentration decreases: higher ion charge densities induce stronger electrostatic interactions resulting in a stable protein fold in low salt concentration. Cation stabilizing efficiency of the folded form follows the series Ca2+ and Mg2+ stronger than K+, Na+, Li+, and stronger than Rb+, Cs+. The corresponding anion series is , OAc–, and F– stronger than Cl– stronger than Br– and I–.
Here, we applied the same comparative strategy using different salts to investigate in detail how stability evolved in various ancestral and modern MalDHs. The strategy offered the opportunity to understand fundamental processes enabling protein stability in various salts and to reveal the unsuspected effect of the various ions that, additionally to K+ and Cl–, populate the cytoplasm of halophilic cells. Hence, in this article, stability and stabilizing effects will refer to the protein propensity to remain folded in low salt concentrations, whereas instability and destabilizing effects will preclude protein folding in such conditions.
Supplementary figure S5, Supplementary Material online shows the salt concentration dependent stability of modern MalDHs, from H. marismortui, (H. mari) and four newly characterized species, Haloarcula mukohatei (H. muko), N. magadii (N. maga), H. volcanii (H. volc), and H. lacusprofundii (H. lacu), as well as from nine of their ancestors. In KCl, all the ancestral and modern enzymes display folded-unfolded transitions that are shifted toward lower concentration compared with H. mari MalDH (supplementary fig. S5B, Supplementary Material online). These stability curves help to determine the Mf½ parameter, which indicates the salt concentration at which 50% of a given halophilic MalDH is still folded and active.
Consequently, we show that multi-molar salt concentration is not mandatory for the stabilization of a halobacterial enzyme, in contrast to what has long been assumed. Our data, which present the first characterization of the salt requirements of an enzyme from various extant halophilic species, also clearly established that there is no correlation between Mf½ values in KCl and NaCl salt concentration in the environment (table 1).The shift of folded-unfolded transition curves toward lower salt concentration in KF (supplementary fig. S5A, Supplementary Material online) compared with KCl (supplementary fig. S5B, Supplementary Material online) indicates that the stabilizing effect induced by a high charge density anion acts on all ancestral and modern enzymes. Reciprocally, when Cs+, a weaker stabilizing cation, is used, all the modern and ancestral MalDH transition curves are shifted toward higher concentration, showing destabilization effects (supplementary fig. S5C, Supplementary Material online).
Table 1.
NaCl Concentration of the Considered Species Environment (Oren 2002) and Mf½ in KCl of the Corresponding MalDHs.
| Current Species | [NaCl] of the Medium | KCl Mf½ of MalDH |
|---|---|---|
| Haloferax volcanii | 2 M | 0.30 M |
| Haloarcula mukohatei | 4 M | 0.35 M |
| Halorubrum lacusprofundii | 4 M | 0.30 M |
| Natrialba magadii | 4 M | 0.80 M |
| Haloarcula marismortui | 4 M | 1.40 M |
Our results, moreover, show that the oldest considered ancestor of halobacterial MalDH (Anc78 MalDH) was a halophilic enzyme requiring salt to remain properly folded. This enzyme is stable between 0.8 M and 3.8 M KCl, with an Mf½ value of 0.5 M (supplementary fig. S5B, Supplementary Material online). Compared with the Mf½ value of 1.4 M for H. mari MalDH, considered as an obligate halophilic enzyme, Anc78 MalDH may be considered as moderately halophilic.
Distinct Evolutionary Trajectories on the Way to Modern Halobacterial MalDHs
Contrary to H. mari MalDH requiring high salt concentrations in order to be stable, five modern MalDH and their ancestors display various stability phenotypes in low salt. In this work, resurrected ancestral MalDHs provided insights into the evolution of these phenotypes. In previous work on paleo-environmental temperatures, (the temperature at which 50% of the resurrected protein is still folded) have been plotted as a function of ancestor age (Gaucher et al. 2003). We applied, here, an analogous approach, by plotting Mf½ values (determined using supplementary fig. S5, Supplementary Material online) versus the number of amino acid replacements accumulated from the oldest considered Anc78 MalDH. The number and nature of these replacements were determined from a sequence comparison between ancestral and modern enzymes (supplementary figs. S6–S8, Supplementary Material online). The results for physiological KCl (fig. 3, red triangles) show that MalDHs evolutionary trajectories differ over the distinct considered lineages.
Fig. 3.
Evolution of the conformational stability of ancestral and extant halobacterial MalDHs. The Mf½ values (y-axis) are plotted against the number of amino acid replacements accumulated since the LCAHa Anc78 MalDH (x-axis). Values were recorded in KCl (red), CsCl (blue), and KF (green). (A and B) Evolutionary path leading to two extant MalDH enzymes of group A species, H. muko and H. mari MalDHs, respectively. (C and D) Evolutionary path leading to two extant MalDH enzymes of group B species, H. lacu and H. volc. (E) Evolutionary path leading an extant MalDH enzyme of a group C species N. maga.
In group B, MalDHs encountered no DTL event and all reconstructed ancestral MalDHs are orthologs, transmitted vertically during species evolution. The Mf½ values of modern MalDH (H. lacu and H. volc) in this group are similar to the Mf½ value of theroot LCAHa Anc78 MalDH (fig. 3C and D). Varying stability of intermediate ancestral MalDHs (Anc80, Anc81, Anc82, and Anc84, see fig. 3) suggest genetic drift, where some destabilizing replacements are counterbalanced by other stabilizing replacements, leading to nearly neutral evolution with respect to protein stability. This genetic drift hypothesis might be further tested using Dn/Ds ratio measurement, providing that higher taxonomic sampling and DNA sequences could be analyzed. This however stands out of the scope of this study focusing on protein and ASR phenotypes characterization. Within group B, the unexpected case of Anc84 MalDH, a close ancestor of H. lacu MalDH, is of great interest: the protein shows non-obligate halophilic behavior and is stable at very low KCl concentration (0.064 M). This suggests a peculiar evolution pattern (see below).
In group C, the first part of the evolutionary trajectory toward N. maga MalDH is strongly destabilizing (fig. 3E), with KCl Mf½values increasing drastically between Anc78 MalDH and Anc90 MalDH, the ancestor of two paralogous MalDH duplicates in group C (fig. 1). However, in the second part of the trajectory, Mf½ indicate greater protein stability in relatively low KCl concentration for Anc93 MalDH (one of the paralogous copies deriving from Anc90) and N. maga MalDH (deriving from Anc93), with the latter modern MalDH being slightly less stable than Anc78 MalDH.
In group A, H. muko MalDH has followed the shortest evolutionary trajectory since the root Anc78 MalDH, diverging by 18 amino acid replacements (fig. 3A). Its KCl Mf½ values are similar to the ones recorded for Anc78 MalDH, and very distinct from the ones recorded for the H. mari MalDH, despite the fact that H. muko and H. mari are two related species in group A (fig. 1). It has been assumed that H. mari MalDH reflected typical behavior for a protein facing extreme environmental salt concentration (Madern, Ebel, Zaccai, et al. 2000). Our results now challenge this point of view, by showing that the low conformational stability of H. mari MalDH is not a phenotype shared by other modern MalDHs. Moreover, our phylogenetic inferences suggest that H. mari MalDH is highly divergent when compared with the LCAHa Anc78 MalDH, and that it most likely evolved by HGT from an extinct lineage related to the ancestor of the alkaliphilic group C (fig. 1). H. mari MalDH has diverged by 22 amino acid replacements from Anc88, its common ancestor with group C MalDHs, while retaining some of its properties, such as high instability (see also below).
Importantly, MalDH stability phenotypes were shown to depend, not only on the salt concentration, but also on the salt type. For example, in previous studies on H. mari MalDH, it has been found that the replacement of K+ by another monovalent cation is always destabilizing. We observed that this conclusion holds also for ancestral and modern MalDH. This suggests that the salt concentration dependence of halobacterial MalDH stability relies on a common ion-linked mechanism, shared since the root ancestor, Anc78.
Heterogeneous Stability Phenotypes of Modern and Ancestral MalDHs Are Modulated by Ions of High Charge Density
According to our previous studies, ions of high charge density enhance the stability of H. mari MalDH at low salt concentration (Madern and Zaccai 1997; Ebel et al. 1999; Irimia et al. 2003; Madern and Ebel 2007). We investigated how this observation applies to the various studied MalDHs, in different salts involving ions of higher charge density than Cl– and K+.
Anion Effects
We explored the effect of high charge density anions by comparing stability in KF and KCl. In contrast to CsCl, KF Mf½ trajectories for the different proteins do not vary consistently when compared with KCl. Mf½ values for the two salts are similar for Anc78, Anc88, Anc84 MalDHs, and H. volc MalDH. KF is slightly more stabilizing for Anc81 MalDH, H. muko, and H. lacu MalDHs. Most interestingly, KF is strongly stabilizing when compared with KCl not only for Anc80 MalDH and H. mari MalDH (fig. 3B), but also within group C, for Anc89, Anc90, Anc93 MalDHs, and N. maga MalDH (fig. 3C). This suggests particular phenotypes for some modern and ancestral MalDH related to group B and C species, where a higher charge density anion tends to favor stability in low salt concentration.
Previous AUC studies on wild type and mutants of H. mari MalDH have suggested an electrostatic interpretation for the KF effect. The method allows identifying the quaternary structure changes in a given solution: tetramers and/or dimers both being functional as it has been shown previously (Irimia et al. 2003; Madern and Ebel 2007). When analyzed with respect to the crystal structure of the tetrameric form (fig. 4), AUC sheds light on molecular mechanisms induced by the solvent. Below 2 M KCl (Mf½ ∼ 1.4 M), the H. mari MalDH tetramer dissociates and unfolds into inactive monomers (fig. 4). Because of its higher negative charge density compared with Cl–, F– strongly shifts the equilibrium toward lower salt concentration (KF Mf½ 0.5 M) suggesting stronger anion binding interactions between monomer to dimer and dimer to dimer interfaces in the tetramer (Irimia et al. 2003; Madern and Ebel 2007).
Fig. 4.
The oligomeric states of H. mari MalDH. The four monomers are shown in different colors. The crystal structure of the native tetrameric state is shown on the left. The central panel illustrates the crystallographic dimer associating monomers A and B. The right panel shows the crystallographic monomer. Because the dimer is a short-living form, and the monomer, an ensemble of non-homogeneous species, their structures have, so far, not been reported.
In order to assess whether or not the ion charge density hypothesis is valid for other MalDHs, we recorded sedimentation coefficient values (s20, w) on several ancestral and modern MalDH after 24 h incubation in various concentrations of KCl and KF. The results are reported in supplementary figure S10, Supplementary Material online. Importantly, previous studies on various tetrameric MalDH have shown that protein hydration, compactness and surface properties influence experimental sedimentation coefficients, which, nevertheless, oscillate around the theoretical shape values (Irimia et al. 2003; Madern and Ebel 2007; Coquelle et al. 2010). Thus, for the sake of clarity, when close to theoretical values for active folded forms, the experimental values were replaced by explicit abbreviations in table 2, that is, (T) for tetrameric MalDH of 130 kDa with s20, w =7.1S and (D) for a 65 kDa dimeric form with s20, w = 4.2S. When dissociated in solution, the compact monomer of 33 kDa with an expected s20, w = 2.8S is not stable, consequently, s20, w values below 4.2S should be seen only as indicative of the lack of quaternary structure, corresponding rather to a molten globule state labeled (M) (Madern, Ebel, Mevarech, et al. 2000). H. volc MalDH is the most stable modern MalDH yet tested (from 0.5 to 3.8 M KCl) with a sedimentation coefficient s20, w = 7.16 S for the active tetrameric form in 1.0 M KCl (supplementary fig. S10, Supplementary Material online). Recall that H. mari MalDH (KCl Mf½ of 1.4 M) dissociates below 1.0 M KCl as shown in table 2 (Irimia et al. 2003). This indicates that the shift of H. volc MalDH conformational stability toward lower KCl concentration, compared with H. mari MalDH, results from the preservation of tetramer integrity. The s20, w values at 0.2 M KCl (2.96 S) indicated that H. volc MalDH inactivation is concomitant with tetramer dissociation into inactive monomers (table 2, supplementary fig. S10, Supplementary Material online). Additionally, whereas H. volc MalDH is dissociated at 0.2 M KCl, it remains active in 0.2 M KF, displaying AUC data well fitted by a mixture of tetramers (7.2 S) and dimers (4.4 S, table 2, supplementary fig. S10, Supplementary Material online). The observation supports the electrostatic stabilization hypothesis proposed from previous measurements of H. mari MalDH stability in KF.H. volc MalDH stability in 0.2 M KF arises through the preservation of both tetrameric and dimeric active forms by F– binding. Furthermore, for Anc80 MalDH, we determined s20, w values of 2.2 S in 0.6 M KCl, and 4.3 and 7.4 S in 0.6 M KF. The data suggest that tetramer stabilization by the high charge density anion might therefore be an ancestral phenotype of group B and C MalDHs.
Table 2.
Effect of a High Charge Density Anion (F–) on the Quaternary Assembly of Three MalDHs, from AUC Experiments in Various Salt Concentrations.
|
H. volc MalDH |
H. mari MalDH |
Anc80 MalDH |
||||
|---|---|---|---|---|---|---|
| [Salt] | KCl | KF | KCl | KF | KCl | KF |
| 2 | — | — | T | T | T | — |
| 1 | T | — | M | — | — | — |
| 0.6 | — | — | M | D | M | T/D/M |
| 0.2 | M | T/D | — | — | — | — |
Note.—Active tetramer, dimer, and inactive monomer are abbreviated as indicated in the main text.
The amplitude of the KF stabilizing effect (compared with KCl) varies along evolutionary trajectories (fig. 3). To the exception of Anc84 MalDH in the H. lacusprofundii lineage (group B) and Anc88 MalDH in the N. magadii lineage (group C), this phenotype appears to be shared by all the examined ancestors in groups B and C (fig. 3) suggesting a similar mode of action as revealed by AUC.
Moreover, considering that, both the LCAHa Anc78 MalDH, and H. muko MalDH in group A display few differences in their respective stabilizing behaviors in KCl and in KF (see Mf½ in fig. 3), we propose that the stabilization by high charge density anions would have appeared in Anc80 MalDH. We cannot exclude that this stabilizing phenomenon pre-existed in Anc78 MalDH, and was further amplified during evolution. It was out of the scope of this study to analyze oligomeric state variation with sufficient accuracy in Anc78 MalDH, because of the small differences in salt concentration at which this effect may exist.
Cation Effects
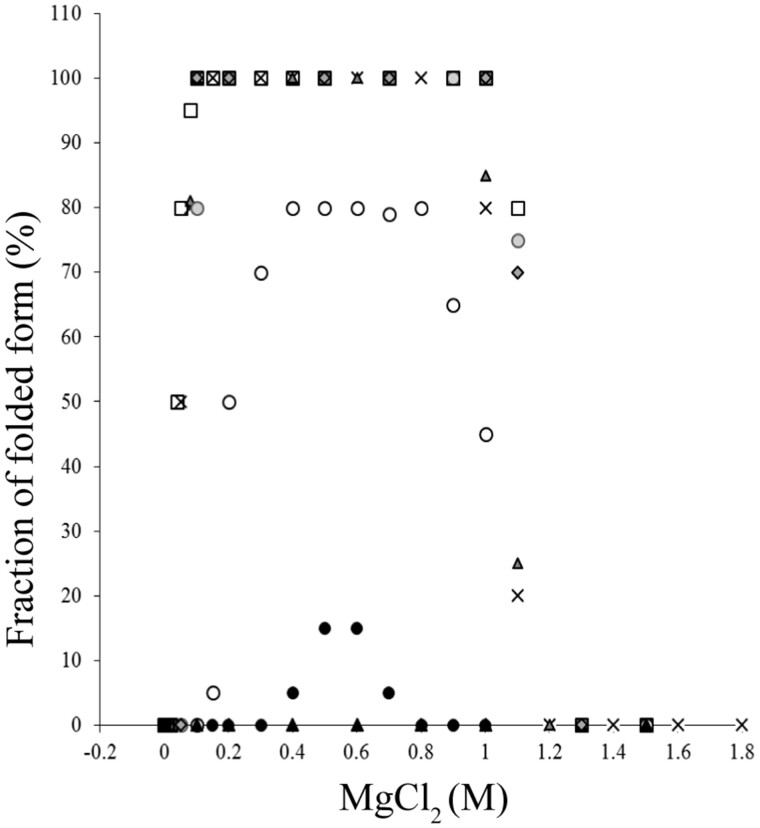
Magnesium chloride is another physiological salt that releases divalent cations of high charge density. Magnesium ions play an important role in numerous cellular mechanisms in all organisms. In Halobacteria, free Mg2+ intracellular concentration ranges within 0.05 M and 0.1 M (Oren 2002). Studies on the effects of MgCl2 on halophilic proteins are scarce, although it has been shown that divalent cations promote a favorable shift of conformational stability (i.e., Mf½ in MgCl2 < Mf½ in KCl) in H. mari MalDH, but also in H. volcanii isocitrate dehydrogenase toward lower salt concentration (Cendrin et al. 1993; Madern and Zaccai 1997; Ebel et al. 1999; Madern and Ebel 2007). The stability curves in MgCl2 displayed a bell shape (fig. 5), with deactivation occurring at low and high concentrations, with unfolding at the higher salt concentration due to the general effect of this chaotropic salt. We monitored whether or not such a phenomenon was detectable also in the modern and ancestral MalDHs examined in this study.
Fig. 5.
Salt-concentration dependent stability curves of various halobacterial MalDHs in MgCl2. Modern enzymes: Haloarcula mukohatei from group A (gray squares), Haloferax volcanii from group B (open squares), and Natrialba magadii from group C (black dots). Ancestral MalDHs: Anc78 (crosses), Anc80 (triangles), Anc88 (gray circles), Anc89 (open circles), and Anc90 (black triangles).
Here, we consider only the relevant change induced by MgCl2 on stability when its concentration decreases (i.e., below 0.6 M). Results established large differences between the studied enzymes (fig. 5). MgCl2 has a dramatic stabilizing effect on the ancestral Anc78, Anc80, and Anc88 MalDHs, and also on modern MalDH of H. muko (representative of group A) and H. volc (representative of group B). It was difficult to record a difference between Mf½ values recorded for these five enzymes, due to the very low MgCl2 concentration at which the transition toward the inactive state occurs. We nevertheless estimate that Mf½ lies below 0.1 M MgCl2. Our data show that the strong divalent cation stabilizing effect is an ancestral phenotype present at the root of halobacterial MalDHs Anc78. Interestingly, the curves for Anc89 and Anc90 MalDHs and N. maga MalDH (representative of alkaliphilic group C) in MgCl2 show that the divalent cation stabilizing effect is not efficient. The different values of folded fractions obtained at 0.6 M after 24 h incubation (fig. 5) suggest the dissociation and deactivation of these enzymes occurs at different rates.
To refine our understanding of MgCl2 stabilization effects on MalDHs, we recorded the s20, W at three low KCl concentrations, supplemented (or not) by MgCl2 (table 3).
Table 3.
Effect of a High Charge Density Cation (Mg2+) on the Quaternary Assembly of Several MalDHs, from AUC Experiments.
|
H. volc MalDH |
N. maga MalDH |
Anc80 MalDH |
Anc89 MalDH |
Anc90 MalDH |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| [KCl] | + MgCl2 (M) | + MgCl2 (M) | + MgCl2 (M) | + MgCl2 (M) | + MgCl2 (M) | |||||
| – | +0.05 | – | +0.50 | – | +0.05 | +0.05 | +0.5 | +0.50 | ||
| 0.6 | — | — | — | — | — | — | M | T | — | M |
| 0.2 | M | — | — | — | — | — | — | — | — | — |
| 0.1 | — | T | — | M | — | T | — | — | — | — |
Note.—Active tetramer, dimer and inactive monomer are abbreviated as indicated previously.
Recall that in molar concentration of KCl, Anc80, Anc89, Anc90 and H. volc MalDH, display s20, W values in agreement with the presence of the tetrameric state (supplementary fig. S10, Supplementary Material online). As discussed, in 0.1 M KCl (supplementary fig. S5, Supplementary Material online), all the enzymes are dissociated and inactive. On one hand, when the 0.1 M KCl solution is supplemented with 0.050 M MgCl2, Anc80 and H. volc MalDH behave as tetramers (table 3 and supplementary fig. S10, Supplementary Material online). On the other hand, within group C, Anc90 MalDH and N. maga MalDH sedimentation profiles at 0.6 and 0.1 M KCl, respectively, plus 0.5 M MgCl2, were well fitted by a population of inactive monomers, consistent with the stability profile measured in MgCl2 (fig. 5). When 0.5 M MgCl2 is added to the 0.6 M KCl solution, Anc89 MalDH is mainly tetrameric with s20, W = 7.4 S. At lower concentration of MgCl2 (0.05 M), the Anc89 MalDH s20, W value of 1.9 S shows that the enzyme is dissociated into monomers. The various oligomerization states induced by different concentration of MgCl2 likely reflect subtle changes in cation binding affinities between enzymes. It is out the scope of this work, however, to analyze this phenomenon in more detail. Our data demonstrate that, when it is considered as beneficial for MalDH stability, the effect of Mg2+ is due to its capacity to maintain the integrity of the active tetrameric state, even at very low KCl concentration. Importantly, as indicated by the absence of 4.2 S peaks in AUC (supplementary fig. S10, Supplementary Material online), dimeric active states are not stabilized by MgCl2, in contrast to anions of high charge density. This suggests that anion and cation binding sites exert different controls on monomer–monomer and dimer–dimer interfaces.
Altogether, our data suggest that the capacity to stay folded even at low KCl concentration in the presence of a secondary physiological salt, such as MgCl2 is an ancestral phenotype of halobacterial MalDH, possibly lost in group C MalDH. It should be considered as beneficial for halophilic lifestyle adaptation (see Discussion) because it has been conserved in lineages of group A and B. How this phenotype has been strongly weakened in group C, is addressed by a series of complementary investigations presented below.
The Halo-Alkaliphilic Phenotype of MalDH Is a Secondary Evolutionary Trait
Haloalkaliphilic Archaea are able to grow both in high salt concentration and high pH up to 11, with a matching cytoplasmic pH value (Falb et al. 2005). High pH is disruptive of electrostatic interactions and has a strong deleterious effect on protein stability (Dill 1990). We tested whether or not MalDH isolated from the haloalkaliphilic Archaea N. magadii and reconstructed ancestral enzymes in group C displayed specific features that may counterbalance this effect (Fig. 6). MalDH from H. marismortui and ancestors Anc80 and Anc88 MalDHs were considered for comparison. We incubated the enzymes for 24 h in 2.0 M KCl solutions buffered at different pH values ranging from pH 6 to pH 13, and monitored stability by using residual activity measurements. Anc80 and Anc88 and H. mari MalDH were unstable at pH 12, unlike Anc89, Anc90 and Anc93 and N. maga MalDH, which remained stable. Because of chemical incompatibility of high pH with sapphire optical cells, AUC data were not recorded. Consequently, we could not determine as previously to what extent the favorable stability changes results from equilibrium shifts between oligomers and higher. Nevertheless, data obtained with the Anc89, Anc90, Anc93 and N. maga MalDH at pH 12 suggest that some replacements were selected in Anc89 MalDH and conserved in the N. maga lineage, likely providing some beneficial effect in high pH environments. Intriguingly, the stability properties of group C MalDH, in high pH occurred concomitantly with the disappearance of the stabilizing effect of MgCl2, (see above). This observation is in agreement with the fundamental process of protein evolution, which has established that trade-offs between properties are responsible for the emergence of new phenotypes (Tokuriki et al. 2008).
Fig. 6.
pH-dependent stability of various modern and ancestral MalDHs. Measurements were done as indicated in Materials and Methods. The data are expressed in % with respect to the values recorded prior to incubation. (A) pH 7, (B) pH 11, and (C) pH 12. The number of amino acid replacements accumulated in the group C MalDH, with respect to Anc80 MalDH, is indicated.
We analyzed the amino acid replacement pattern of MalDH within group C (supplementary fig. S8, Supplementary Material online). From Anc88 to Anc89 MalDH, 13 replacements are observed. When their position is analyzed with respect to the crystal structure of H. mari MalDH (PDB code 2J5K), the replacement that occurred at position 239 (H–R) suggests its importance with respect to stability. Indeed, in contrast to other 12 positions, at position 239 the side chains of H239 in H. mari MalDH face each other across the interface in the dimer of active dimers (AB and CD) that makes up the tetrameric assembly. At this particular position, the replacement of an amino acid (H) with a pKa that is sensitive to pH variation, by R that is not, would have been a favorable replacement.
Evolution of Enzymatic Activity
Finally, we explored the enzymatic properties of MalDHs along the evolutionary pathways. We monitored the enzymatic activity at various concentration of their substrate (oxaloacetate, OAA) at KCl concentration of 3.8 M. Because exhaustive measurements of the steady state parameters of these enzymes were out of the scope of this study, we simply present the results in a comparative mode allowing to see the relative effect expressed as % of the maximal activity observed. The unexpected results indicate that MalDHs split into two groups, which differ by their capacity to be inhibited or not by increasing concentration of OAA (fig. 7). Activity profiles for both the deepest ancestors Anc78 and Anc80 MalDHs show a typical phenomenon of inhibition by an excess of substrate (fig. 7A). This effect can be, therefore considered as an ancestral feature in halobacterial MalDHs. In group B MalDHs, the maximal activity is reached with 0.4 mM of OAA; at higher concentration, the activity of all the enzymes is inhibited (fig. 7B), indicating that this property was conserved in this group. In contrast, in group C as well as in the ancestor of group C and H. mari MalDH (Anc88), the inhibition by high concentration of OAA is abolished (fig. 7C). In group A, the activity profiles of H. muko and H. mari MalDHs are completely different (fig. 7D). H. muko MalDH is representative of group A, its activity profile shows an inhibitory effect at OAA concentration higher than 0.5 mM. Because this is the ancestral behavior measured for the root MalDH Anc78, it could be seen as an inherited property shared by enzymes from group A. Our measurements showed that H. mari MalDH is not sensitive to inhibition by high concentration of OAA (fig. 7D), an observation in agreement with a previous work on this enzyme (Hecht et al. 1989). The peculiar behavior of H. mari MalDH is therefore consistent with an HGT from a lineage related to the group C ancestor. RP effects that explain the change in enzymatic activity of halobacterial MalDHs, is under investigation using site-directed mutagenesis.
Fig. 7.
Enzymatic activity profiles of ancestral and modern MalDHs from different groups. The results are expressed in % of maximal activity. (A) LCAHa Anc78 MalDH (closed triangles) and Anc80 MalDH (open triangles). (B) Anc81 MalDH (open circle), Anc84 MalDH (open square), H. volc MalDH (open triangle), and H. lacu MalDH (inverted triangle). (C) Anc82 MalDH (gray triangle), and Anc88 MalDHs (gray square) and N. maga MalDH (gray dots). (D) H. muko MalDH (open circle) and H. mari MalDH (black triangle).
An Intermediate Ancestral Halobacterial MalDH with Nonhalophilic-Like Properties
The unexpected stability down to 0.05 M KCl of Anc84 MalDH (from group B), in the lineage leading to H. lacu MalDH, was carefully analyzed. With an Mf½ value lower than 0.1 M KCl (see fig. 2), the enzyme is significantly more stable in low salt concentration than all other considered halobacterial MalDHs.
In 1.0 M KCl, the CD spectra of Anc84 MalDH and H. lacu MalDH display strong negative values of molar ellipticity at 210 nm and 222 nm, and positive values below 200 nm (fig. 8), typical of folded alpha + beta proteins, similar to those observed previously for H. mari MalDH at 4 M KCl (Madern, Ebel, Mevarech, et al. 2000). A similar spectrum indicates that Anc84 MalDH is still folded in 0.06 M KCl. In contrast, at the same low KCl concentration, H. lacu MalDH displays a CD spectrum typical of an unfolded protein. We tested that the Anc84 MalDH unfolds after dialysis in water as it is expected with a halophilic enzyme. The Anc84 MalDH behavior was further examined by monitoring its sedimentation coefficient (supplementary fig. S11, Supplementary Material online). As expected, at 1.0 M KCl, the s20, W value of 7.6 S indicates the presence of tetramer (table 4).In both 0.06 M and 0.1 M KCl, Anc84 MalDH forms dimers, displaying s20, W values of 4.8 S and 4.9 S. Surprisingly, the s20, W value in 0.1 M KF indicates that the Anc84 MalDH still forms a dimer, whereas with other MalDH, we always obtained stabilization of the tetrameric state (see previous section). In the presence of 0.05 M MgCl2, s20, W values are in agreement with the tetrameric state. Our data suggest, therefore, that the peculiar behavior of Anc84 MalDH results from beneficial replacements acting at the AD and BC interfaces (see fig. 4), which promote stronger binding between the active dimeric units AB and DC to make the tetramer, even at low salt concentration.
Fig. 8.
Circular dichroism signal of Anc84 MalDH (circles) and H. lacu MaDH (triangles), after incubation in 0.06 M (white) or 1.0 M (gray) KCl.
Table 4.
Oligomeric State Variation of Anc84 MalDH in Various Salt Conditions.
| Anc84 MalDH |
||||
|---|---|---|---|---|
| [Salt] | KCl | KF | KCl | |
| + 0.05 M MgCl2 | ||||
| 1.0 | T | — | — | |
| 0.1 | D | D | T | |
| 0.06 | D | — | — | |
| 0.00 | — | — | T | |
Note.—Abbreviations are as indicated previously. Data in the left columns help to see the putative effect of anions of high charge density. Columns on the right reflect the effect of divalent cation. The measurement done at low MgCl2 concentration in absence of KCl was done after dialysis of the sample.
Discussion
Salt-Dependent Stability of Halophilic MalDH Could Not Be Used as a Salinity Sensor
Understanding the evolution of organisms relies on the capacity to understand the processes that shaped cellular components under environmental selective pressures. Such questions may be appropriately addressed by using ASR approaches. Along these lines, several studies based on ASR support the hypothesis that, during early stages of prokaryotic evolution, high environmental temperature was a major determinant shaping the proteomes (Boussau et al. 2008; Groussin and Gouy 2011). In the framework of such a scenario, resurrection of several ancestral proteins from various bacterial and archaeal lineages revealed high resistance against thermal unfolding (Gaucher et al. 2003; Perez-Jimenez et al. 2011; Hobbs et al. 2012, Risso et al. 2013; Romero-Romero et al. 2016).These results are based on the reasonable assumption that the conformational stability of each protein is related directly to the environmental temperature of its host and could therefore be used as a molecular thermometer (Gaucher et al. 2003; Hobbs et al. 2012). Moreover, such a conclusion applied to the average paleo-environmental temperatures at the scale of the whole planet. Hence, in the case of temperature, application of ASR to the estimation of paleo-environmental conditions is possible because the conditions were global and effects on proteins, direct. Likewise, could the ASR and resurrection of ancestral halobacterial MalDHs achieved in our work help to infer the salt composition of ancient environments? Several limitations, in our opinion, unfortunately preclude direct interpretation of the estimated ancestral MalDH phenotypes as adaptations to paleo-environmental salt concentrations.
First, we found no correlation between the salt-concentration dependent stability of cytoplasmic MalDH from modern species (as indicated by Mf½ values, table 1) and the NaCl concentration of their natural habitats. Furthermore, recent studies have shown that the salt content of halobacterial cytoplasm cannot be described simply as containing a high KCl concentration. It corresponds rather to a complex ion mixture regulated by the nature, the number, and relative efficiency of membrane transporters for various ions and compatible solutes (Becker et al. 2014). In their natural environments, halobacterial species permanently face important variations in salinity due to rainfall and evaporation. In order to compensate for unfavorable variations in osmotic pressure, salt type and concentration within the cytoplasm can be transiently decorrelated from environmental fluctuations. Consequently, it is not possible to infer the salt composition of the paleo-environment that prevailed during the evolution of Halobacteria by using the ancestral MalDH conformational stability as a salinity sensor.
We assume that, in a near future, such estimates could rely on the examination of closely related halobacterial species and their genomic contents, including ion transporters and proteins regulating cytoplasmic ion content. Especially, since Halobacteria are thought to have diverged from methanogenic Archaea (Forterre et al. 2002). A recent publication has shown that the methanogen-to-halophile transition, in Archaea, has involved a new intermediate group of organisms called Hikarchaeia (Martijn et al. 2020). Experimental characterization of their enzymes would increase our understanding of halophilic adaptation.
Molecular Evolution in Halobacteria, a Case Study
We have, for the first time, experimentally discriminated at the molecular level how modern enzymes from extremophilic organisms have acquired their properties either through the modification of inherited genetic information (RP, genetic drift and Darwinian selection) or through non-genealogical acquisition of genetic material by HGT. We used halophilic Archaea as appropriate system models, in part, because they have a well-documented capacity for generating genetic variation through HGT (Ram-Mohan et al. 2014 refs therein). Our main results describing successive phenomena that shaped the adaptation of halobacterial MalDHs facing extreme salt concentrations are summarized in figure 9.
Fig. 9.
Summary of main results about halobacterial MalDH evolution. Left: schematic phylogenetic relationships among MalDH within Halobacteria. A–C refer to the various groups. Stars and circles correspond ancestral or modern MalDHs, respectively. The number of amino acid replacements accumulated since LCAHa Anc78 MalDH is indicated within each enzyme. Molecular properties of MalDHs: observed (+), absent (–), or not determined (ND). ND1 refers to refolding problems and ND2 to not done or failed experiments. Data on H. mari MalDH are obtained from Ebel et al. (1999) and Madern and Zaccai (1997). (*) Data obtained from halohandbook: https://haloarchaea.com/halohandbook/.
We examined the effects of RP on three different molecular properties of MalDHs. In particular, we tried to see whether solubility, conformational stability and enzymatic activity evolved concomitantly in order to fit the peculiar salt conditions encountered in the cytoplasm of modern cells.
Solubility
Acidic amino acid enrichment is a well-established structural feature of modern halophilic proteins that help them to stay soluble at high salt concentration (Coquelle et al. 2010, Talon et al. 2014). We showed that the strong acidic surface enrichment of halobacterial MalDH is a conserved ancestral phenotype, shared since the last common ancestor of the studied lineages. However, an acidic composition does not indicate a moderate or extreme halophilic lifestyle. Indeed, some studies using halophilic bacterial species have shown that a strict relationship between a highly acidic proteome and high salt in the environment does not hold (Deole et al. 2013; Oren 2013). Furthermore, even if acidic enrichment is a phenotype that favors solubility at high salt concentration, experimental studies have shown it is also beneficial at low salt concentration (Coquelle et al. 2010). Finally, previous attempts to classify pI variations (at the proteomic level) to different ecological classifications showed that extreme and moderate halophilic organisms could not be distinguished (Kiraga et al. 2007). These observations indicated that global acidic enrichment in a protein becomes favorable above a certain (low) threshold of salt concentration, and provides an important adaptive advantage in high environmental salt concentrations. Consequently, the use of protein acidity values as a proxy failed to predict with accuracy ancient environmental conditions in which the LCA of extant Halobacteria lived. One can only conclude that this shared ancestral phenotype was advantageous and remained conserved during the adaptation of modern species within Halobacteria to moderate or extreme halophilic lifestyles.
Conformational Stability
Theoretical and experimental studies indicated that evolutionary trajectories of enzyme oscillate between increasing and decreasing stability phenotype drifts (Hart et al. 2014; and reference therein). Along this line, monitored KCl Mf½ values revealed that MalDH conformational stabilities increase and decrease independently, with distinct amplitudes, during the evolution of the three analyzed lineages down to modern MalDH. The monitoring of oligomeric states revealed that this change in stability is mainly due to dissociation of the active tetrameric assembly into inactive monomers. We also found that anions of high charge density, such as fluoride, shift protein stability, in most of cases, toward lower salt concentration than chloride. In two alkaliphilic ancestral MalDHs (Anc89 and Anc90) and H. mari MalDH this ionic effect is strongly stabilizing with a Mf½ decrease of ∼1 M. This results in the capacity of these MalDHs to behave also as active dimers in the presence of strongly stabilizing anions. Numerous biophysical and structural investigations using the H. mari MalDHs strongly support the existence of weak and strong anion and cation binding sites that contribute to maintain the quaternary assembly (Madern and Zaccai 1997; Ebel et al. 1999; Madern and Ebel 2007). Our data demonstrate that when the salt concentration decreases to a given threshold, the tetrameric state of the various considered ancestral and modern MalDHs dissociates, triggering concomitant deactivation and unfolding. The KCl, CsCl and KF Mf½ variations along the evolutionary trajectories can be, therefore, well explained by affinity changes of ion-binding sites due to direct and indirect effects of amino acid replacements. Some of which are strongly beneficial, by lowering the rate of dissociation between oligomers when the salt concentration decreases. Although the various considered MalDHs behave differently according to salt type and concentration, the ion binding site mechanisms appear to be shared by all the halobacterial MalDH since the LCAHa. Additionally to the shared acidic surface enrichment, this ion-binding site phenotype might be interpreted as resulting from an adaptation of the LCAHa to a salted environment.
We observed that the stabilizing effect of cations of high charge density is effective on the two oldest considered LCA MalDHs (LCAHa: Anc78, and LCA of group B + C: Anc80) as well as on the modern MalDHs of H. volc and H. muko, belonging to species group A and B (fig. 8). These enzymes, at very low KCl concentration, in the presence of 100 mM MgCl2, can be stabilized only as tetramers. The data strongly suggest that 1) cation-binding sites at the interface between the dimer of active dimers (AB and CD) contribute to tetramer assembly, 2) Mg2+ may occupy these cation-binding sites with a higher efficiency than K+. Interestingly, we showed that Anc84 MalDH still forms a tetramer in pure solution of 0.05 M MgCl2. This suggests that amino acid replacements in its primary sequence have dramatically increased its binding affinity for Mg2+ through mechanisms that remain to be elucidated.
We noted that the stabilizing effect of Mg2+ is effective only on MalDHs from group A and group B. These are representative of halobacterial species living in salty environments at neutral pH, which do not show the loss of stabilization by Mg2+ occurring in the MalDH of the haloalkaliphilic group C LCA (Anc89), as well as in the haloalkaliphilic N. maga MalDH. Additionally, we showed that all modern and ancestral alkaliphilic MalDH acquired stabilization properties at high pH (fig. 8). This phenomenon likely relies on the acquisition of an amino acid (R239) in a structural key position, for which the ionization state and therefore the capacity to establish interactions is not abolished at high pH values. We noticed that this replacement occurred when the evolutionary trajectory is the most destabilizing with respect to the KCl requirement (KCl Mf½ of Anc89 MalDH = 1.4 M). This observation is in agreement with studies showing that, during an evolutionary process, protein stability changes do favor the acquisition of new properties (Tokuriki et al. 2008). Thus, even if the variation of MalDH conformational stability does not directly reflect variations of environmental salt concentrations, the identified drastic changes in stability phenotypes suggest an adaptation to haloalkaliphilic conditions in the LCA of Natrialbales order (Group C).
H. mari MalDH is unstable in MgCl2. It does not behave, however, as an alkaliphilic MalDH (this work and Madern and Zaccai 2007). Its primary sequence shows a histidine residue at position 239, implying that the decrease of Mg2+binding capacity of H. mari MalDH might be a convergent phenotype resulting from amino acid replacements occurring after it diverged from the haloalkaliphilic group C MalDHs (Anc88, see fig. 8).
Amino acid replacements do not only exert a local effect but also effects that propagate through the dynamical properties of the enzyme (Bigman and Levy 2018; Yu and Dalby 2018). Diverging amino acid replacements may have influenced the phenotypic properties of H. mari MalDH through long-range distance effects rather than specifically at the interface between dimers of active dimers. The ongoing description of a halophilic MalDH crystal structure with its associated solvation shell would provide the relevant information to describe such effects in detail.
Enzymatic Activity
According to the traditional comparison of properties of proteins isolated from extant organisms, it is commonly admitted that a stability–activity tradeoff drives adaptive evolution of enzymes (Somero 2004; Studer et al. 2014) even if this has been challenged by Nguyen et al. (2013). From previous comparative studies with its nonhalophilic homologs, it has been commonly admitted that the necessary flexibility of H. mari MalDH, required to function at high salt concentration, was the consequence of a compromise which has decreased its stability at low salt concentration. Latter, it was shown that a clear conclusion concerning the salt-concentration dependency of activity could not be established easily because most of the measurements have been done without taking into account the strong salt-concentration dependency of affinity for the substrate (Hecht et al. 1989; Madern et al. 2004). We therefore changed the strategy of analysis, by recording the substrate-concentration dependency of activity at a unique KCl concentration of 3.8 M for all the MalDHs under study. Thanks to this methodology, we discovered that ancestral and modern MalDHs display two kinds of phenotypes: some are sensitive to inhibition by high substrate concentration during their catalytic cycle; others are not (i.e., H. mari MalDH, alkaliphilic group C MalDH and their common MalDH ancestors). Our unexpected findings showed that oldest considered MalDH LCAs (LCAHa: Anc78, LCA of group B + C: Anc80), which are inhibited by high OAA substrate concentration, diverged into a non-inhibited enzyme in the ancestor of H. mari MalDH and of the alkaliphilic clade C MalDH (Anc88, see fig. 8). Interestingly, we analyzed that none of the amino acid replacement concerns those involved in substrate recognition and catalysis within the catalytic site (not shown), suggesting long range effects through change in protein dynamics, such as it has been observed in others enzymes (Zoi et al. 2016). Interestingly, this non-inhibited phenotype, appeared in a lineage related to the alkaliphilic halobacterial (group C) ancestor, is retained in the H. mari (group A) MalDH. This further demonstrates that H. mari acquired its MalDH through HGT and replacement of its inherited copy during evolution. In a recent work on the response of ADP-dependent kinases of Methanosarcinales facing moderate salt concentration, it has been shown that non-inhibited modern orthologs evolved from an inhibited ancestral state (Gonzales-Ordenes et al. 2018). We propose that the selection of enzymes less sensitive to inhibitory effects on their activity is one of the selective advantages that may contribute to adaptation to salty environments.
Conclusion: The Evolution of Halophilic Proteins
We are now able to propose a detailed scenario for the evolution of a halophilic enzyme, based on our biochemical, structural and evolutionary analyses.
Halobacteria are generally considered to be “salt-in” strategists—actively accumulating potassium and chloride ions to maintain an isosmotic cytoplasm in order to prevent loss of water to the hypersaline environment. Because it has been studied extensively, MalDH from H. marismortui has been considered for a long time to be the best model for a halophilic enzyme. In particular, a solvation shell of hydrated ions coordinated by carboxyl groups on the protein surface has been proposed to maintain H. mari MalDH stable and soluble at high salt (Madern et al. 2000) with a consequence that the structure would unfold already if the salt concentration fell to relatively high value (2 M KCl). By taking into account a large set of ancestral and modern orthologous halobacterial MalDHs, our study challenged the general validity of this model. We showed that H. mari MalDH instability at a relatively “high” salt concentration (2 M KCl) is a specific evolutionary fate, possibly inherited through an ancestral HGT, rather than the general behavior of halophilic enzymes. In contrast, most halobacterial MalDHs, as well as their oldest common ancestors, appear as moderate halophilic enzymes with a better tolerance for lower cytoplasmic salt concentration than observed with H. mari MalDH. In turn, within Halobacteria, some species, such as H. marismortui may have subsequently adapted to obligate extreme halophilic lifestyles, with possible loss of tolerance to cytoplasmic salt depletion. Moreover, our study did not show any evidence concerning a gradual shift in acidic amino acid composition. Because this surface modification is strongly beneficial to favor solubility at high salt concentration, the property was fixed over time. We found that most of ancestral and modern MalDHs could maintain their conformational stability at lower KCl concentration, especially in the presence of another physiological salt, such as MgCl2. This is in agreement with an intracellular measurement reporting an intracellular content of 100 mM MgCl2 in a halobacterial species (Matheson et al. 1976), and with genomics studies revealing the presence of several types of Mg2+ transporters such MgtE and CorA-like membrane proteins (Becker et al. 2014). We found that anions of high change density were also strongly efficient with respect to stability. Our data, which account for the complexity of the salt composition within the cytoplasm of Halobacteria, showed that the conformational stability of halophilic proteins varied over evolution. We propose that the capacity of a halophilic protein to stay folded even at low salt concentration may, therefore, be an efficient ancestral mechanism to survive osmotic down shock. Biophysical models of protein evolution explain that selection attempts to maintain the existing structure, function and dynamical properties when these remain beneficial in a new environment (DePristo et al. 2005; Tokuriki and Tawfik 2009b). In this framework, most of the amino acid replacements occurring during the evolution of MalDH in groups A and B would have nearly neutral effects with respect to stability.
The role of stabilizing versus destabilizing mutations, in the evolution of protein properties, is controversial (Bloom et al. 2006; Merski and Shoichet 2012, Bigman and Levy 2020). In the case of destabilizing mutations, their detrimental effects are buffered by the presence of molecular chaperones, so that they may promote evolvability (Geller et al. 2018). Here, we observed that an accumulation of destabilizing replacements, such as observed in group C MalDH ancestors, has rendered the enzyme prone to evolve new properties. Indeed, we found an unexpected molecular tradeoff in haloalkaliphilic group C showing that the selection of new enzymatic properties (i.e., nonsubstrate inhibited MalDH) and the loss of the capacity to enhance conformational stability at low salt by Mg2+ are concomitant with the adaptation to high pH environments. This evolutionary trait makes sense with respect to the specific chemical constraints encountered in alkaliphilic environments. Indeed, because of the high pH encountered in alkaliphilic environments divalent cations mainly exist as insoluble carbonates, consequently Ca2+ and Mg2+ availability is very low. In addition, MgCl2 inhibits cellular growth of species from the Natrialba genus (Tindall et al. 1980). When deprotonated malate and oxaloacetate exist, as reactive divalent anions, they may easily react with divalent cations, such as Ca2+ or Mg2+ (Butler 1998).
Because of this chemistry, one may reasonably consider that the concentration of oxaloacetate is higher in a haloalkaliphilic cytoplasm depleted in divalent cations than encountered in other halobacterial species; a situation which would have been less favorable for the functionality of an OAA-inhibited MalDH. We propose that the selection of MalDHs nonsensitive to substrate inhibition was advantageous during the expansion of species within Halobacteria to fit new environmental conditions, by preventing a decrease of metabolism efficiency.
Materials and Methods
Main Phylogenetic Reconstruction Steps
Several phylogenetic reconstruction steps are performed in order to obtain the ancestral MalDH sequences. First, a phylogenomics approach is used to estimate a species tree. Genes are gathered (section Reconstruction of gene families) and a species tree is reconstructed using ML methods (section Reconstruction of the species tree). Second, a collection of MalDH trees is computed using Bayesian methods (section Reconstruction of the MalDH sequence-only tree). Third the latter trees are reconciled with the species tree using a ML approach (section Species tree-gene tree reconciliation). Fourth, using the reconciled MalDH joint tree, MalDH sequence are aligned anew accounting for indel events and ancestral MalDH sequences are estimated using a Bayesian method (section ASR of MalDH). At each steps, control experiments are performed using ML or Bayesian models.
Reconstruction of Gene Families
From the collection of 51 proteomes, homologous gene families have been obtained with an all-against-all BLAST approach. The program Silix was then used to cluster protein sequences into homologous families (Miele et al. 2011). Default parameters were considered for the clustering: sequences having >30% similarity and >80% coverage were clustered together into a homologous gene family. Unicopy gene families having >80% of taxonomic coverage were conserved, these 240 gene families were further aligned with Prank (Löytynoja and Goldman 2005; Löytynoja and Goldman 2008), internally used in Guidance (Penn et al. 2010) to trim ambiguously aligned sites by taking into account the uncertainty of the guide tree during the alignment procedure. Ambiguously aligned sites were further trimmed with Gblocks (Castresana 2000), with standard options and with gaps allowed. Finally, the bppSeqMan program belonging to the bppSuite of programs (Duteil and Boussau 2008) was used to eliminate sites containing >20% of gaps.
Reconstruction of the Species Tree
The species tree has been inferred with a concatenation approach. After the concatenation of the 240 gene families, the final alignment contains 50,135 amino acid positions. Different strategies were employed to reconstruct the species tree. The LG replacement model (Le and Gascuel 2008) and a Gamma distribution with four categories were used in PhyML (Guindon et al. 2010) to reconstruct the tree with 100 bootstraps. The LG model assumes that the evolutionary process is constant between lineages and across sites. The CAT and CAT+GTR models (Lartillot and Philippe 2004) implemented in Phylobayes (version 3.3, Lartillot et al. 2009) were also employed. CAT and CAT+GTR assume that the process is heterogeneous among sites and constant between lineages. We computed 1,000,000 cycle long MCMC chains, saving a sample each 10 cycles. The 1,000 first samples were discarded as burnin. Two independent chains were executed for each experiment. The chain's convergence was assessed if the difference of posterior log likelihood across MCMC chains is less than three times the estimated standard error and the tree bipartition PP differ by <0.1. Finally, the COaLA replacement model was also used with bppML (Duteil and Boussau 2008) COaLA is site-homogeneous but implements a time-heterogeneous model that allows to model the variation of global compositions between lineages. As topology exploration is not feasible with time-heterogeneous models in bppML, COaLA was used to test alternative topologies regarding the position of the root of the Halobacteria clade. To discriminate between alternative topologies, AU tests were performed with Consel (Shimodaira and Hasegawa 2001).
To improve the resolution of the phylogeny of Halobacteria, and especially the position of its root, an elimination of the fast-evolving positions was realized to reduce systematic errors (Brinkmann et al. 2005; Philippe et al. 2005). We used the site-specific posterior rates computed by PhyML with the use of the Γ distribution to remove gradually the fast-evolving sites (by fractions of 10% of sites).
Reconstruction of the MalDH Sequence-Only Tree
The MalDH gene family was reconstructed with Silix (Miele et al. 2011). The 51 MalDH sequences were then aligned with Muscle (Edgar 2004) used in Guidance (Penn et al. 2010), which allows to trim ambiguously aligned sites owing to uncertainty in the guide tree topology. Remaining ambiguously aligned sites were removed with Gblocks (Castresana 2000). The Bayesian sequence-only tree was reconstructed using the CAT+GTR model implemented in PhyloBayes 3.3 (Lartillot et al. 2009). We ran 1,000,000 long independent MCMC chains, saving a sample each 10 cycles, and we discarded the 1,000 first samples as burnin. The chain's convergence was assessed if the tree bipartition PP differ by <0.05. The consensus sequence-only tree was then reconciled with the species tree.
Species Tree-Gene Tree Reconciliation
A prereleased version of the Amalgamated Likelihood Estimation (ALE) algorithm (ALE v0.1) (Szöllosi, Tannier, et al. 2013) was used to compute the ML MalDH joint tree. ALE uses the model described in (Szöllosi, Rosikiewicz, et al. 2013) to search for the best scenario of duplications, transfers and losses of genes and efficiently explores the space of joint trees that maximize the joint likelihood between the sequence and reconciliation information. ALE requires a time calibrated species tree to compute ML estimates of transfer rates. Divergence times were estimated using PhyloBayes 3.3 from the genomic concatenation having 50% of its fastest evolving sites filtered out (see above).
We used the CAT replacement model, the Log Normal relaxed molecular clock, no calibration point, and a flat root prior defined by a 1 billion years expectation and standard deviation. The species tree was rooted with the Thermoplasmatales. The MCMC chains were elongated for 1,000,000 cycles, a sample was saved every 10 cycles, and 1,000 first samples were discarded as burnin. Convergence is estimated to be reached when the difference of posterior log likelihood across MCMC chains is less than three times the estimated standard error. ALE also uses a sample of gene trees to compute conditional clade probabilities (Höhna and Drummond 2012), which can be used to approximately estimate the PP of a gene tree that can be amalgamated from clades present in the sample. The sample of posterior trees computed by PhyloBayes 3.3 on the MalDH alignment (see above) was provided to ALE.
ASR of MalDH
The MalDH joint tree was used as a final guide tree to Prank, which is very sensitive to the choice of the guide tree (Löytynoja and Goldman 2005). The Prank alignment and the joint tree were used to compute the ancestral sequences. Moreover, one of the key outputs of Prank is the inferred history of the insertion/deletion events leading to the extant gap pattern. This history is used to filter out from the computed ancestral sequences all ancestral sites that are inferred as gaps by Prank.
To reconstruct ancestral MalDH sequences in ML, the bppAncestor program (Dutheil and Boussau 2008) was used with the marginal reconstruction approach (Yang et al. 1995). For a given node at a given site, bppAncestor makes use of the ML estimates of branch lengths and model parameters obtained with bppML to compute the PP of each possible ancestral states. The state having the maximum PP is inferred as being the ML ancestral state. Site- and time-homogeneous models (LG and LG+Fopt), site-heterogeneous (EX2, EX3, EHO, UL2, UL3, EX_EHO [Le, Lartillot, et al. 2008], C10–C60 [Le, Gascuel, et al. 2008], and time-heterogeneous models COaLA with 1 or 2 parameters per branch [Groussin et al. 2013] were run with bppML to retain the best-fitting model in terms of AIC [Akaike 1973] and BIC [Schwarz 1978] values). Ancestral gaps inferred by Prank were then incorporated in the final ancestral sequences.
Several models available in PhyloBayes 3.3 were used to compute the ancestral sequences: LG, GTR, CAT, and CAT+GTR. As the latter model generally provides the best fit to the data, its estimation provided us the ancestral sequences to be synthesized. MCMC chains were run for 1,000,000 cycles, saving a sample each 10 cycles and discarding the first 1,000 sample as burnin. Two independent chains were checked for convergence using the tracecomp program provided in PhyloBayes 3.3. Convergence is estimated to be reached when the difference of posterior log likelihood across MCMC chains is less than three times the estimated standard error. The ancestral sequences were computed from the posterior distributions using the ancestral program of the PhyloBayes 3.3 suite. We used the midpoint rooting option (-midpointrooting) to provide an estimate at the root node. For each site and ancestor, the considered ancestral state has the maximal PP. Ancestral sequences obtained from two independent MCMC chains were compared and verified to be identical. As final step of the ancestral sequence inference, ancestral gaps inferred by prank are substituted to the inferred ancestral states, yielding the ancestral amino acid sequences to be synthesized.
Gene Synthesis, Protein Expression, and Purification
The complete gene synthesis of the various ancestral sequences was done by GeneCust (www.genecust.com). The resulting genes were cloned in pET 21a between the Nde1 and BamH1 sites. The resulting constructs were transformed in Escherichia coli BL21 DE3 pLysS strain. Cultivation in LB medium containing ampicillin, overexpression, refolding and purification were done accordingly to protocol used for H. mari MalDH (Irimia et al. 2003).
Analytical Ultracentrifugation
Sedimentation velocity experiments were performed using a Beckman XL-I analytical ultracentrifuge and an ANTI 50 rotor (Beckman Coulter). In order to analyze the change in oligomeric state as a function of salt concentration, the experiments were carried out at 20 °C for all MalDH at 0.1 mg/ml in 50 mM Tris pH 7.0, supplemented with the appropriate salt concentration. A volume of 400 µl was loaded into double-channel centerpiece of 1.2 cm optical path length and centrifuged at 130,000 × g (42,000 rpm). Scans were recorded at 280 nm. The partial specific volume of the polypeptide chain for all MalDHs was = 0.73 ml/g as calculated with the Sednterp software (free available at http://www.jphilo.mailway.com/). The solvent density, ρand the solvent viscosity ηat 20 °C for each tested conditions were calculated with the same software. Sedimentation profiles were analyzed by the size-distribution analysis of Sedfit (free available at http://www.analyticalultracentrifugation.com). In Sedfit, finite element solutions of the Lamm equation for a large number of discrete, independent species, for which a relationship between mass, sedimentation and diffusion coefficients, s and D, is assumed, are combined with a maximum entropy regularization to represent a continuous size-distribution (Schuck 2000). We used 200 generated sets of data on a grid of 300 radial points, calculated using fitted frictional ratio for sedimentation coefficients comprised between 1 and 10 S. For the regularization procedure, a confidence level of 0.68 was used.
Conformational Stability
Residual activity measurements using the standard assay (done in triplicates) and circular dichroism (CD) were used to determine conformational stability as in previous study (Ebel et al. 1999; Madern, Ebel, Mevarech, et al. 2000). For the determination of salt-dependent conformational stability, the proteins of interest were incubated at 20 °C for 24 h at 0.1 mg/ml in various salt concentrations buffered with 50 mM Tris–HCl pH7 prior to performing CD and activity assays. The data were expressed as % of the values recorded before incubation. The salt concentration at which 50% of the protein is still folded and active after incubation (Mf½) was graphically determined using the transitions curves. The confidence limits are ±0.1 M in KCl, CsCl, and KF. For the pH-dependent conformational stability, the protein were incubated at 2 M KCl with various buffers at 100 mM, that is, sodium citrate for pH5, MES for pH6, Tris–HCl for pH7 and pH8 and pH9, glycine for pH10, borate-NaOH for pH11, Na2HPO4 for pH12 and NAOH for pH13. Far-UV CD measurements were carried out on a JASCO J-810 thermostated spectropolarimeter. Far-UV spectra were recorded in 0.1-cm path length quartz cells. The spectra shown in this work represent the average of three accumulated consecutive scans.
Standard Enzymatic Assay
The activity was determined at 25 °C, by following the oxidation of NADH over 30 s by measuring the decrease in absorbance at 340 nm (Beckman DU 7500 Spectrophotometer) in 3.8 M KCl, 50 mM Tris–HCl buffered at pH7 supplemented with 1.0 mM OAA and 0.2 mM NADH. To test the substrate inhibitory effect, measurements were done at various concentration of OAA.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This manuscript is dedicated to the memory of Professor Dan Tawfik, who shaped the direction of research presented here through his groundbreaking work into the evolution of enzymes. We warmly acknowledge, Dr M. Facciotti (Davis University, USA) who gave us privileged access to genomic information prior publication and Drs Céline Brochier-Armanet, Christine Ebel, and Giuseppe Zaccai for fruitful discussion. IBS acknowledges integration into the Interdisciplinary Research Institute of Grenoble (IRIG, CEA). This study was supported by the Agence Nationale de la Recherche (ANR-10-BINF-01-01 and ANR-16-CE11-0011) and by the Centre National de la Recherche Scientifique (Project EOPV [Environnements Planétaires et Origines de la Vie]). We also used the platforms of the Grenoble Instruct-ERIC Center (ISBG: UMS 3518 CNRS-CEA-UGA-EMBL) with support from FRISBI (ANR-10-INBS-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB).
References
- Akaike H.1973. Information theory and an extension of the maximum likelihood principle. In: Petrov BN, Csaki F, editors. Second international symposium on information theory. Budapest (Hungary: ). New York (NY): Springer. p. 267–281. [Google Scholar]
- Albuquerque L, Taborda M, La Cono V, Yakimov M, da Costa MS.. 2012. Natrinema salaciae sp. nov, a halophilic archaeon isolated from the deep, hypersaline anoxic Lake Medee in the Eastern Mediterranean sea. Syst Appl Microbiol. 35(6):368–373. [DOI] [PubMed] [Google Scholar]
- Alexander WG, Wisecaver JH, Rokas A, Hittinger CT.. 2016. Horizontally acquired genes in early-diverging pathogenic fungi enable the use of host nucleosides and nucleotides. Proc Natl Acad Sci USA. 113(15):4116–4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andam CP, Harlow TJ, Papke RT, Gogarten JP.. 2012. Ancient origin of the divergent forms of leucyl-tRNA synthetases in the Halobacteriales. BMC Evol Biol. 12:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouad M, Borrel G, Brochier-Armanet C, Gribaldo S.. 2019. Evolutionary placement of Methanonatronarchaeia. Nat Microbiol. 4(4):558–559. [DOI] [PubMed] [Google Scholar]
- Aouad M, Taib N, Oudart A, lecocq M, Gouy M, Brochier-Armanet C.. 2018. Extreme halophilic archaea derive from two distinct methanogen Class II lineages. Mol Phylo Evol. 127:46–54. [DOI] [PubMed] [Google Scholar]
- Becker EA, Seitzer PM, Tritt A, Larsen D, Krusor M, Yao AI, Wu D, Madern D, Eisen JA, Darling AE, Facciotti MT.. 2014. Phylogenetically driven sequencing of extremely halophilic archaea reveals strategies for static and dynamic osmo-response. PLoS Genet. 10(11):e1004784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellissent-Funel M-C, Hassanali A, Havenith M, Henchman R, Pohl P, Sterpone F, van der Spoel D, Xu Y, Garcia AE.. 2016. Water determines the structure and dynamics of proteins. Chem Rev. 116(13):7673–7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigman LS, Levy Y.. 2018. Stability effects of protein mutations: the role of long-range contacts. J Phys Chem B. 122(49):11450–11459. [DOI] [PubMed] [Google Scholar]
- Bigman LS, Levy Y.. 2020. Proteins: molecules defined by their trade-offs. Curr Opin Struct Biol. 60:50–56. [DOI] [PubMed] [Google Scholar]
- Bloom JD, Labthavikul ST, Otey CR, Arnold FH.. 2006. Protein stability promotes evolvability. Proc Natl Acad Sci USA. 103(15):5869–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneté F, Madern D, Zaccai G.. 1994. Stability against denaturation mechanisms in halophilic malate dehydrogenase adapt to solvent conditions. J Mol Biol. 244(4):436–447. [DOI] [PubMed] [Google Scholar]
- Boussau B, Blanquart S, Necsulea A, Lartillot N, Gouy M.. 2008. Parallel adaptations to high temperatures in the Archaean eon. Nature 456(7224):942–945. [DOI] [PubMed] [Google Scholar]
- Brinkmann H, van der Giezen M, Zhou Y, de Raucourt GP, Philippe H.. 2005. An empirical assessment of long branch attraction artefacts in deep eukaryotic. Syst Biol. 54(5):743–757. [DOI] [PubMed] [Google Scholar]
- Brochier-Armanet C, Forterre P, Gribaldo S.. 2011. Phylogeny and evolution of the Archaea: one hundred genomes later. Curr Opin Microbiol. 14(3):274–281. [DOI] [PubMed] [Google Scholar]
- Butler JN.1998. Ionic equilibrium: solubility and pH calculations. New York: John Wiley & Sons, Inc. [Google Scholar]
- Castresana J.2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. [DOI] [PubMed] [Google Scholar]
- Cendrin F, Chroboczek J, Zaccai G, Eisenberg H, Mevarech M.. 1993. Cloning, sequencing, and expression in Escherichia coli of the gene coding for malate dehydrogenase of the extremely halophilic archaebacterium Haloarcula marismortui. Biochemistry 32(16):4308–4313. [DOI] [PubMed] [Google Scholar]
- Coquelle N, Talon R, Juers DH, Girard E, Kahn R, Madern D.. 2010. Gradual adaptive changes of a protein facing high salt concentrations. J Mol Biol. 404(3):493–505. [DOI] [PubMed] [Google Scholar]
- Costenaro L, Zaccai G, Ebel C.. 2002. Link between protein–solvent and weak protein–protein interactions gives insight into halophilic adaptation. Biochemistry 41(44):13245–13252. [DOI] [PubMed] [Google Scholar]
- DasSarma S, Capes MD, Karan R, DasSarma P.. 2013. Amino acid substitutions in cold-adapted proteins from Halorubrum lacusprofundii, an extremely halophilic microbe from Antarctica. PLoS One 8(3):e58587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deole R, Challacombe J, Raiford DW, Hoff WD.. 2013. An extremely halophilic proteobacterium combines a highly acidic proteome with a low cytoplasmic potassium content. J Biol Chem. 288(1):581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Weinreich DM, Hartl DL.. 2005. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet. 6(9):678–687. [DOI] [PubMed] [Google Scholar]
- Dill KA.1990. Dominant forces in protein folding. Biochemistry 29(31):7133–7155. [DOI] [PubMed] [Google Scholar]
- Dutheil J, Boussau B.. 2008. Non-homogeneous models of sequence evolution in the Bio++ suite of libraries and programs. BMC Evol Biol. 8:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel C, Faou P, Kernel B, Zaccai G.. 1999. Relative role of anions and cations in the stabilization of halophilic malate dehydrogenase. Biochemistry 38(28):9039–9047. [DOI] [PubMed] [Google Scholar]
- Edgar RC.2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falb M, Pfeiffer F, Palm P, Rodewald K, Hickmann V, Tittor J, Oesterhelt D.. 2005. Living with two extremes: conclusions from the genome sequence of Natronomas pharaonis. Genome Res. 15(10):1336–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnigan GC, Hanson-Smith V, Stevens TH, Thornton JW.. 2012. Evolution of increased complexity in a molecular machine. Nature 481(7381):360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P, Brochier C, Philippe H.. 2002. Evolution of the Archaea. Theor Popul Biol. 61(4):409–422. [DOI] [PubMed] [Google Scholar]
- Fournier GP, Andam CP, Gogarten JP.. 2015. Ancient horizontal gene transfer and the last common ancestors. BMC Evol Biol. 15:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfelder H, Chen G, Berendzen J, Fenimore PW, Jansson H, McMahon BH, Stroe IR, Swenson J, Young RD.. 2009. A unified model of protein dynamics. Proc Natl Acad Sci USA. 106(13):5129–5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher EA, Thomson JM, Burgan MF, Benner SA.. 2003. Inferring the paleoenvironment of ancient bacteria on the basis of resurrected proteins. Nature 425(6955):285–288. [DOI] [PubMed] [Google Scholar]
- Geller R, Pechmann S, Acevedo A, Andino R, Frydman J.. 2018. Hsp90 shapes protein and RNA evolution to balance trade-offs between protein stability and aggregation. Nat Commun. 9(1):1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales-Ordenes F, Cea PA, Fuentes-Ugarte N, Munoz SM, Zamora RA, Leonardo D, Garratt RC, Castro-Fernadez V, Guixé V.. 2018. ADP-dependent kinases from the archaeal order Methanosarcinales adapt to salt by a non-canonical evolutionarily conserved strategy. Front Microbiol. 9:1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano G, Merlino A.. 2014. Molecular bases of protein halotolerance. Biochim Biophys Acta. 1844(4):850–858. [DOI] [PubMed] [Google Scholar]
- Groussin M, Boussau B, Gouy M.. 2013. A branch-heterogeneous model of protein evolution for efficient inference of ancestral sequences. Syst Biol. 62(4):523–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groussin M, Gouy M.. 2011. Adaptation to environmental temperature is a major determinant of molecular evolutionary rates in Archaea. Mol Biol Evol. 28(9):2661–2674. [DOI] [PubMed] [Google Scholar]
- Groussin M, Hobbs JK, Szöllősi GJ, Gribaldo S, Arcus VL, Gouy M.. 2015. Toward more accurate ancestral protein genotype-phenotype reconstructions with the use of species tree-aware gene trees. Mol Biol Evol. 32(1):13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Hart KM, Harms MJ, Schmidt BH, Elya C, Thornton JW, Marqusee S.. 2014. Thermodynamic system drift in protein evolution. PLoS Biol. 12(11):e1001994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht K, Wrba A, Jaenicke R.. 1989. Catalytic properties of thermophilic lactate dehydrogenase and halophilic malate dehydrogenase at high temperature and low water activity. Eur J Biochem. 183(1):69–74. [DOI] [PubMed] [Google Scholar]
- Hobbs JK, Shepherd C, Saul DJ, Demetras NJ, Haaning S, Monk CR, Daniel RM, Arcus VL.. 2012. On the origin and evolution of thermophily: reconstruction of functional precambrian enzymes from ancestors of Bacillus. Mol Biol Evol. 29(2):825–835. [DOI] [PubMed] [Google Scholar]
- Höhna S, Drummond AJ.. 2012. Guided tree topology proposals for bayesian phylogenetic inference. Syst Biol. 61(1):1–11. [DOI] [PubMed] [Google Scholar]
- Irimia A, Ebel C, Madern D, Richard SB, Cosenza LW, Zaccai G, Vellieux FMD.. 2003. The oligomeric states of Haloarcula marismortui malate dehydrogenase are modulated by solvent components as shown by crystallographic and biochemical studies. J Mol Biol. 326(3):859–873. [DOI] [PubMed] [Google Scholar]
- Kacar B, Guy L, Smith E, Baross J.. 2017. Resurrecting ancestral genes in bacteria to interpret ancient biosignatures. Philos Trans A Math Phys Eng Sci. 375(2109):20160352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski L, Lurie-Weinberger MN, Allers T, Gophna U, Eichler J.. 2013. Phylogenetic- and genome-derived insight into the evolution of N-glycosylation in Archaea. Mol Phylogenet Evol. 68(2):327–339. [DOI] [PubMed] [Google Scholar]
- Khomyakova M, Bukmez O, Thomas LK, Erb TJ, Berg IA.. 2011. A methylaspartate cycle in Haloarchaea. Science 331(6015):334–337. [DOI] [PubMed] [Google Scholar]
- Kim YB, Kim JY, Song HS, Lee C, Ahn SW, Lee SH, Jung MY, Rhee J-K, Kim J, Hyun D-W, et al. 2018. Novel haloarchaeon Natrinema thermophila having the highest growth temperature among haloarchaea with a large genome size. Sci Rep. 8(1):7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiraga J, Mackiewicz P, Mackiewicz D, Kowalczuk M, Biecek P, Polak N, Smolarczyk K, Dudek MR, Cebrat S.. 2007. The relationships between the isoelectric point and length of proteins, taxonomy and ecology of organisms. BMC Genomics 8(1):163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S.. 2009. PhyloBayes 3. A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25(17):2286–2288. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H.. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 21(6):1095–2004. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O.. 2008. An improved general amino acid replacement matrix. Mol Biol Evol. 25(7):1307–1320. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O, Lartillot N.. 2008. Empirical profile mixture models for phylogenetic reconstruction. Bioinformatics 24(20):2317–2323. [DOI] [PubMed] [Google Scholar]
- Le SQ, Lartillot N, Gascuel O.. 2008. Phylogenetic mixture models for proteins. Philos Trans R Soc Lond B Biol Sci. 363(1512):3965–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski JR, Halse ME, Blackledge M, Emsley L.. 2015. Protein dynamics. Direct observation of hierarchical protein dynamics. Science 348(6234):578–581. [DOI] [PubMed] [Google Scholar]
- Liu Y, Bahar I.. 2012. Sequence evolution correlates with structural dynamics. Mol Biol Evol. 29(9):2253–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A, Goldman N.. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci USA. 102(30):10557–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A, Goldman N.. 2008. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science 320(5883):1632–1635. [DOI] [PubMed] [Google Scholar]
- Madern D.2002. Molecular evolution within the l-malate and l-lactate dehydrogenase super-family. J Mol Evol. 54(6):825–840. [DOI] [PubMed] [Google Scholar]
- Madern D, Camacho M, Rodríguez-Arnedo A, Bonete M-J, Zaccai G.. 2004. Salt-dependent studies of NADP-dependent isocitrate dehydrogenase from the halophilic archaeon Haloferax volcanii. Extremophiles 8(5):377–384. [DOI] [PubMed] [Google Scholar]
- Madern D, Ebel C.. 2007. Influence of an anion-binding site in the stabilization of halophilic malate dehydrogenase from Haloarcula marismortui. Biochimie 89(8):981–987. [DOI] [PubMed] [Google Scholar]
- Madern D, Ebel C, Mevarech M, Richard SB, Pfister C, Zaccai G.. 2000. Insights into the molecular relationships between malate and lactate dehydrogenases: structural and biochemical properties of monomeric and dimeric intermediates of a mutant of tetrameric L-[LDH-like] malate dehydrogenase from the halophilic archaeon Haloarcula marismortui. Biochemistry 39(5):1001–1010. [DOI] [PubMed] [Google Scholar]
- Madern D, Ebel C, Zaccai G.. 2000. Halophilic adaptation of enzymes. Extremophiles 4(2):91–98. [DOI] [PubMed] [Google Scholar]
- Madern D, Zaccai G.. 1997. Stabilisation of halophilic malate dehydrogenase from Haloarcula marismortui by divalent cations—effects of temperature, water isotope, cofactor and pH. Eur J Biochem. 249(2):607–611. [DOI] [PubMed] [Google Scholar]
- Martijn J, Schön ME, Lind AE, Vosseberg J, Williams TA, Spang A, Ettema TJG.. 2020. Hikarchaeia demonstrate an intermediate stage in the methanogen-to-halophile transition. Nat Commun. 11(1):5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson AT, Sprott GD, McDonald IJ, Tessier H.. 1976. Some properties of an unidentified halophile: growth characteristics, internal salt concentration, and morphology. Can J Microbiol. 22(6):780–786. [DOI] [PubMed] [Google Scholar]
- Merski M, Shoichet BK.. 2012. Engineering a model protein cavity to catalyze the Kemp elimination. Proc Natl Acad Sci USA. 109(40):16179–16183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele V, Penel S, Duret L.. 2011. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics 12:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minárik P, Tomásková N, Kollárová M, Antalík M.. 2002. Malate dehydrogenases–structure and function. Gen Physiol Biophys. 21(3):257–265. [PubMed] [Google Scholar]
- Mirceta S, Signore AV, Burns JM, Cossins AR, Campbell KL, Berenbrink M.. 2013. Evolution of mammalian diving capacity traced by myoglobin net surface charge. Science 340(6138):1234192. [DOI] [PubMed] [Google Scholar]
- Naor A, Lapierre P, Mevarech M, Papke RT, Gophna U.. 2012. Low barriers in halophilic archaea and the formation of recombinant hybrids. Curr Biol. 22(15):1444–1448. [DOI] [PubMed] [Google Scholar]
- Nguyen TH, Ranwez V, Pointet S, Chifolleau AMA, Doyon JP, Berry V.. 2013. Reconciliation and local gene tree rearrangement can be of mutual profit. Algorithms Mol Biol. 8(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V, Wilson C, Hoemberger M, Stiller JB, Agafonov RV, Kutter S, English J, Theobald DL, Kern D.. 2017. Evolutionary drivers of thermoadaptation in enzyme catalysis. Science 355(6322):289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A.2002. Halophilic microorganisms and their environments. The Netherlands: Kluwer Academic Publishers. [Google Scholar]
- Oren A.2013. Life at high salt concentrations, intracellular KCl concentrations and acidic proteomes. Front Microbiol. 4:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papke RT, Koenig JE, Rodriguez-Valera F, Doolittle WF.. 2004. Frequent recombination in a saltern population of Halorubrum. Science 306(5703):1928–1929. [DOI] [PubMed] [Google Scholar]
- Paul S, Bag SK, Das S, Harvill ET, Dutta C.. 2008. Molecular signature of hypersaline adaptation: insights from genome and proteome composition of halophilic prokaryotes. Genome Biol. 9(4):R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn O, Privman E, Landan G, Graur D, Pupko T.. 2010. An alignment confidence score capturing robustness to guide-tree uncertainty. Mol Biol Evol. 27(8):1759–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Jimenez R, Inglés-Prieto A, Zhao Z-M, Sanchez-Romero I, Alegre-Cebollada J, Kosuri P, Garcia-Manyes S, Kappock TJ, Tanokura M, Holmgren A, et al. 2011. Single-molecule paleoenzymology probes the chemistry of resurrected enzymes. Nat Struct Mol Biol. 18(5):592–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Delsuc F, Brinkmann H, Lartillot N.. 2005. Phylogenomics. Annu Rev Ecol Evol Syst. 36(1):541–562. [Google Scholar]
- Ram-Mohan N, Fullmer MS, Makkay AM, Wheeler RW, Ventosa A, Naor A, Gogarten JP, Papke RT.. 2014. Evidence from phylogenetic and genome-fingerprinting analyses suggests rapidly changing variation in Halorubrum and Haloarcula populations. Front Microbiol. 5:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso VA, Gavira JA, Mejia-Carmona DF, Gaucher EA, Sanchez-Ruiz JM.. 2013. Hyperstability and substrate promiscuity in laboratory resurrections of Precambrian β-lactamases. J Am Chem Soc. 135(8):2899–2902. [DOI] [PubMed] [Google Scholar]
- Romero-Romero ML, Risso VA, Martinez-Rodriguez S, Gaucher EA, Ibarra-Molero B, Sanchez-Ruiz JM.. 2016. Selection for protein kinetic stability connects denaturation temperatures to organismal temperatures and provides clues to archaean life. PLoS One 11(6):e0156657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savory FR, Milner DS, Miles DC, Richards TA.. 2018. Ancestral function and diversification of a horizontally acquired oomycete carboxylic acid transporter. Mol Biol Evol. 35(8):1887–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P.2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 78(3):1606–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G.1978. Estimating the dimension of a model. Ann Statist. 6:461–464. [Google Scholar]
- Shimodaira H, Hasegawa M.. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17(12):1246–1247. [DOI] [PubMed] [Google Scholar]
- Solovyova A, Schuck P, Costenaro L, Ebel C.. 2001. Non-ideality by sedimentation velocity of halophilic malate dehydrogenase in complex solvents. Biophys J. 81(4):1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somero GN.2004. Adaptation of enzymes to temperature: searching for basic “strategies”. Comp Biochem Physiol B Biochem Mol Biol. 139(3):321–333. [DOI] [PubMed] [Google Scholar]
- Studer RA, Christin PA, Williams MA, Orengo CA.. 2014. Stability–activity tradeoffs constrain the adaptive evolution of RubisCO. Proc Natl Acad Sci USA. 111(6):2223–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Richard S, Koch MHJ, Sayers Z, Kuprin S, Zaccai G.. 1998. Protein hydration in solution: experimental observation by X-ray and neutron scattering. Proc Natl Acad Sci USA. 95(5):2267–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szöllosi GJ, Rosikiewicz W, Boussau B, Tannier E, Daubin V.. 2013. Efficient exploration of the space of reconciled gene trees. Syst Biol. 62(6):901–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szöllosi GJ, Tannier E, Lartillot N, Daubin V.. 2013. Lateral gene transfer from the dead. Syst Biol. 62(3):386–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon R, Coquelle N, Madern D, Girard E.. 2014. An experimental point of view on hydration/solvation in halophilic proteins. Front Microbiol. 5:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindall BJ, Mills AA, Grant WD.. 1980. An alkaliphilic red halophilic bacterium with a low magnesium requirement from a Kenyan soda lake. Microbiology 116(1):257–260. [Google Scholar]
- Tokuriki N, Stricher F, Serrano L, Tawfik DS.. 2008. How protein stability and new functions trade off. PLoS Comput Biol. 4(2):e1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS.. 2009a. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol. 19(5):596–604. [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS.. 2009b. Protein dynamism and evolvability. Science 324(5924):203–207. [DOI] [PubMed] [Google Scholar]
- Tomatis PE, Fabiane SM, Simona F, Carloni P, Sutton BJ, Vila AJ.. 2008. Adaptive protein evolution grants organismal fitness by improving catalysis and flexibility. Proc Natl Acad Sci USA. 105(52):20605–20610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hippel PH, Schleich T.. 1969. The effects of neutral salts on the structure and conformational stability of macromolecules in solution. In: Timasheff SN, Fasman GD, editors. Structure and stability of biological macromolecules. New York: Marcel-Dekker. p. 417–574. [Google Scholar]
- Voordeckers K, Brown CA, Vanneste K, Van der Zande E, Voet A, Maere S, Verstrepen KJ.. 2012. Reconstruction of ancestral metabolic enzyme reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLoS Biol. 10(12):e1001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Kumar S, Nei M.. 1995. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141(4):1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Dalby PA.. 2018. Coupled molecular dynamics mediate long- and short-range epistasis between mutations that affect stability and aggregation kinetics. Proc Natl Acad Sci USA. 115(47):E11043–E11052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoi I, Suarez J, Antoniou D, Cameron SA, Schramm VL, Schwartz SD.. 2016. Modulating enzyme catalysis through mutations designed to alter rapid protein dynamics. J Am Chem Soc. 138(10):3403–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.