Abstract
The Hui people are unique among Chinese ethnic minorities in that they speak the same language as Han Chinese (HAN) but practice Islam. However, as the second-largest minority group in China numbering well over 10 million, the Huis are under-represented in both global and regional genomic studies. Here, we present the first whole-genome sequencing effort of 234 Hui individuals (NXH) aged over 60 who have been living in Ningxia, where the Huis are mostly concentrated. NXH are genetically more similar to East Asian than to any other global populations. In particular, the genetic differentiation between NXH and HAN (FST = 0.0015) is only slightly larger than that between northern and southern HAN (FST = 0.0010), largely attributed to the western ancestry in NXH (∼10%). Highly differentiated functional variants between NXH and HAN were identified in genes associated with skin pigmentation (e.g., SLC24A5), facial morphology (e.g., EDAR), and lipid metabolism (e.g., ABCG8). The Huis are also distinct from other Muslim groups such as the Uyghurs (FST = 0.0187), especially, NXH derived much less western ancestry (∼10%) compared with the Uyghurs (∼50%). Modeling admixture history indicated that NXH experienced an episode of two-wave admixture. An ancient admixture occurred ∼1,025 years ago, reflecting the intensive west–east contacts during the late Tang Dynasty, and the Five Dynasties and Ten Kingdoms period. A recent admixture occurred ∼500 years ago, corresponding to the Ming Dynasty. Notably, we identified considerable sex-biased admixture, that is, excess of western males and eastern females contributing to the NXH gene pool. The origins and the genomic diversity of the Hui people imply the complex history of contacts between western and eastern Eurasians.
Keywords: Hui, genetic admixture, population structure, Muslim, natural selection, whole-genome sequencing
Introduction
With a population size of 10.5 million, the Huis are the largest of the ten Muslim minorities and the second largest of the 55 ethnic minorities in China. There was a long-lasting debate about the origins of the Hui people. According to historical records, Islam has been first introduced into China during the Tang Dynasty about 1,400 years ago. Merchants and political emissaries migrated along the Silk Road from Arab, Persia, and Central Asia to China, and later some of them became permanent residents. Moreover, during the Yuan Dynasty about 600 years ago, group after group of Islamic-oriented people from Arab, Persia, and Central Asia either were forced to move to or voluntarily migrated to China (Gladney 1997; Chen 1999). Historians believed that the present Hui people were descendants of those immigrants (Gladney 1997; Chen 1999). However, physical measurements indicate that the Hui people have the common physical features of East Asians (Dai et al. 1996). Historical studies failed to estimate the exact contribution of western Eurasian ancestry to the Hui people. It remains unclear whether the origins of the Hui people involved massive population migration or not.
Previous genetic studies of the Hui people were predominantly based on autosomal InDel (Zhou et al. 2020), autosomal STR (Deng et al. 2011; Yao et al. 2016), Y chromosome (Xie et al. 2019), X chromosome (Meng et al. 2014), and mtDNA (Yao et al. 2004). Moreover, most of these studies mainly aimed to evaluate the power of those markers in forensic applications. A previous study suggested there was an affinity between the Hui and East Asian populations (Yao et al. 2016). However, because of the limited sample size and number of genetic markers, previous studies failed to reveal the admixture history of the Hui population. Moreover, the maternal mtDNA profile showed that 6.7% of the lineages in Hui people living in Xinjiang originated from western populations (Yao et al. 2004). The paternal Y chromosome profile showed that nearly 30% of the lineages in the Hui were of western origin (Wang et al. 2019). The estimated genetic contribution of western ancestry varied based on different genetic markers, failing to comprehensively understand the extent to which western ancestry contributed to the Hui population. Moreover, to our best knowledge, the admixture history of the Hui population has not been modeled, in particular, admixture time was not estimated by any published genomic studies.
Recent studies have demonstrated that leveraging shared haplotype has the power to uncover previously unrecognized fine-scale population structure within populations, yielding novel insight into the demographic history of British (Leslie et al. 2015), Japanese (Takeuchi et al. 2017), and Han Chinese populations (Xu et al. 2009; Chiang et al. 2018; Cao et al. 2020). However, the genetic structure within the Hui population has not yet been studied.
In this study, we made the very first effort in whole-genome sequencing of 234 Hui individuals aged over 60 who have been living in the Ningxia Hui Autonomous Region (NXH), Northwestern China, where the Hui people are mostly concentrated in. With this unprecedented data set, we comprehensively assessed the genomic diversity, fine-scale population structure, and further inferred the genetic origins and admixture history of the Hui people. Our results are expected to advance the understanding of the complex admixture history of the Hui peoples, intensive contacts between western and eastern Eurasians. In addition, the whole-genome data of 234 Huis of age over 60 serve as a useful control data set for future genetic association studies of late-onset diseases such as hypertension, Type-2 diabetes, cardiovascular heart diseases, and so on.
Results
Genetic Affinity of NXH in the Context of Global Populations
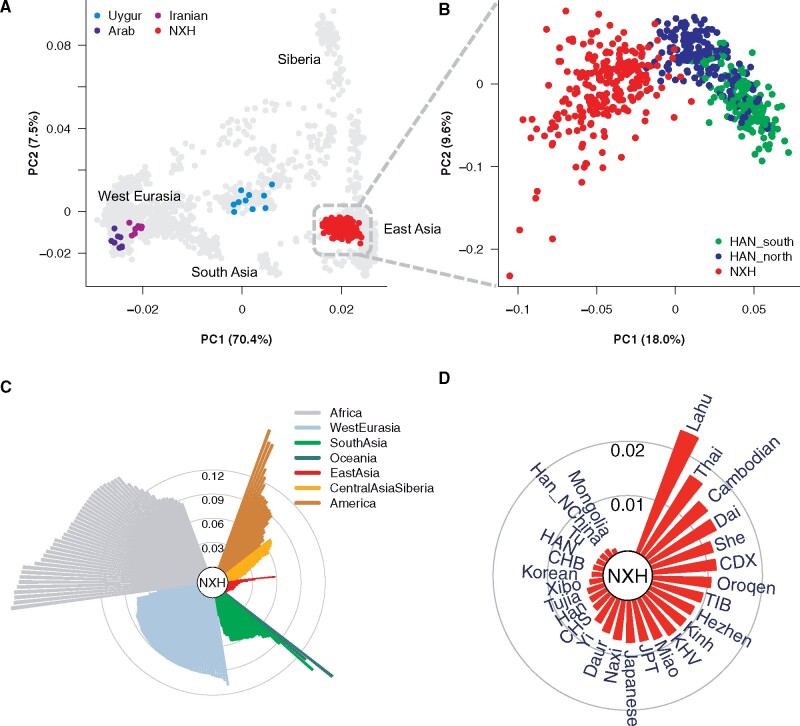
We analyzed the whole-genome data of NXH together with the available data of contemporary populations to understand the genomic diversity of NXH and its relationship with worldwide populations. Principal component analysis (PCA) showed that the genetic coordinates of Eurasian individuals on the PC plot were highly correlated with their geographical locations (fig. 1A). NXH sits between East Asian and West Eurasian clusters on the PC plot but much closer to the East Asians, indicating genetic admixture of both eastern and western Eurasian ancestry, with greater ancestry contribution from East Asian populations. Notably, unlike many other Chinese Muslim populations such as the Uyghurs, NXH did not cluster together with the Central Asian populations. Interestingly, though NXH clustered together with East Asian populations, NXH was genetically distinguishable from the Han Chinese population (FST = 0.0015) (fig. 1B and supplementary fig. S5, Supplementary Material online), suggesting they were two distinct groups in terms of genetic makeup. Besides, NXH had a closer relationship with the northern Han Chinese people represented by CHB (FST = 0.0017) than the southern Han Chinese people represented by CHS (FST = 0.0037).
Fig. 1.
Genetic affinity of NXH in the context of worldwide populations. (A) PCA of 230 NXH samples with samples from other Eurasian populations. Geographical regions from where the samples were collected are labeled on the plot. The number in the bracket represents the variance explained by each PC accounting for the top 10 PCs. (B) PCA of 230 NXH and 341 Han Chinese samples. (C) A fan-like chart showing genetic differences (FST) between NXH and other global populations. Each branch represents one pairwise comparison between NXH and 1 of 205 worldwide populations. Lengths are proportional to FST value, which is indicated by gray circles. The populations are classified by geographical regions and indicated with colors showing in the legend. (D) A fan-like chart showing FST between NXH and East Asian populations.
Genetic differences measured using unbiased FST between NXH and other global populations also showed that NXH had the closest relationship with East Asian populations, followed by Central Asian and Siberian, South Asian, and West Eurasian populations (fig. 1C). Particularly, NXH was most closely related to populations living in northern East Asia, such as Mongolian (FST = 0.0007), northern Han Chinese (FST = 0.0013), Tu in Qinghai of China (FST = 0.0014), and so on (fig. 1D). Besides, NXH did not show a very close relationship with Arab (FST = 0.089), Iranian (FST = 0.080), and Central Asian populations. NXH was also distinct from other Chinese Muslim groups such as the Uyghurs (FST = 0.0187). This genetic difference between NXH and other populations was further confirmed by PCA (fig. 1A and supplementary fig. S6, Supplementary Material online) and outgroup f3 analysis (supplementary fig. S7, Supplementary Material online).
Ancestral Makeup of NXH
PCA suggested that there was some gene flow from west Eurasian populations into NXH. The result of the f3 test in the form of f3 (source1, source2; NXH) (supplementary table S4, Supplementary Material online) confirmed that NXH was an admixed population and East Asian populations and West Eurasian populations, which had the lowest f3 values, could be used as representative of ancestral populations for further analysis. To reveal the genetic makeup of NXH, we carried out ADMIXTURE analysis for NXH with other Eurasian populations, assuming the number of ancestral groups (K) from 2 to 20.
As K increased, a more complex population structure was revealed. However, we did not observe any dominant ancestral component specific to NXH (supplementary fig. S8, Supplementary Material online). We found the results assuming four ancestral populations for Eurasian populations best explain the genetic ancestry composition of NXH, that is, East Asian (EA), Siberian (SIB), West Eurasian (WE), and South Asian (SA).
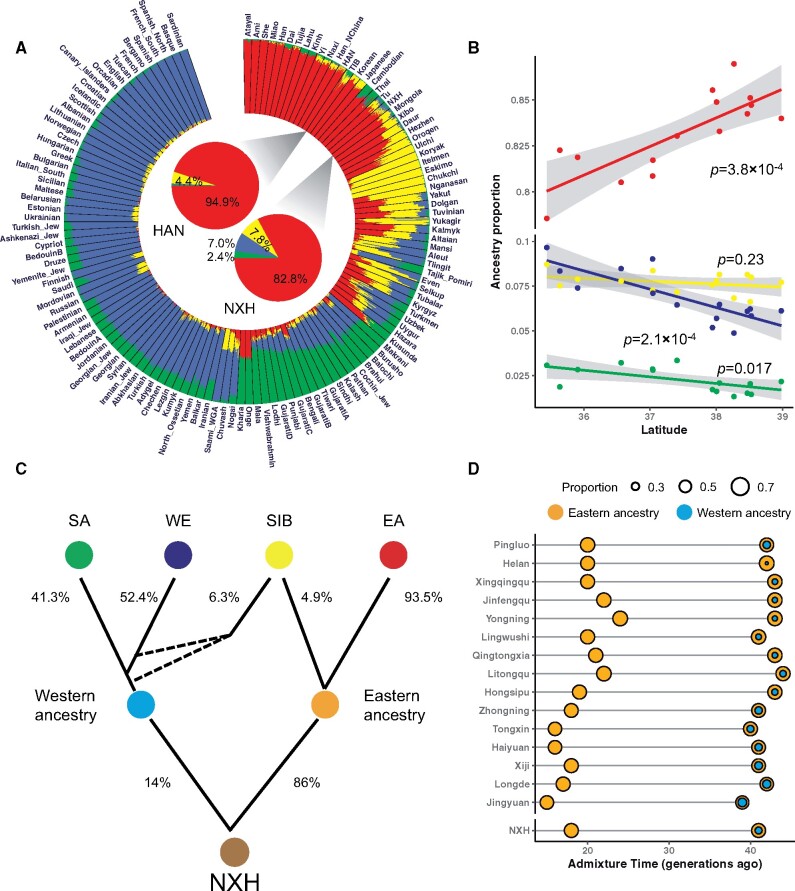
We performed replicate ADMIXTURE analyses for NXH with other Eurasian populations to evaluate the reliability of our results (supplementary table S5, Supplementary Material online). When two ancestral Eurasian populations (K = 2) were assumed, the genetic contribution from the eastern ancestry and the western ancestry to NXH was 0.915 and 0.085, respectively, which was consistent with that estimated by f4 statistics (supplementary table S6, Supplementary Material online). At K = 4, each Eurasian individual was estimated as a descendent of four ancestries: EA, SIB, WE, and SA, which returned an admixture pattern consistent with geography. NXH shared the majority of ancestry with EA (∼82.8%), SIB (∼7.8%), WE (∼7.0%), and SA (∼2.4%) (fig. 2A). HAN shared ancestry with EA (∼94.9%) and SIB (∼4.4%), little with WE and SA (<1%). The components of WE and SA, which account for nearly 10% of the NXH genome, were nearly absent (<1%) in HAN. This was the main difference in the genetic makeup of NXH and HAN.
Fig. 2.
Ancestral makeup and admixture history of NXH. (A) ADMIXTURE result of NXH with other Eurasian populations at K = 4. The results of the population-level admixture of NXH and HAN are highlighted and displayed in the two pie charts in the center of the circle plot. (B) Correlations between the proportions of four ancestries and the latitude of NXH across regions. Each point represents one region in Ningxia. The gray region indicates the 95% CI for each regression fit. (C) The most likely admixture topology of four ancestral components is inferred based on the AHG and GLOEBOTROTTER results. The dashed lines indicate the accurate topology remains unknown. The admixture proportion is inferred by GLOBETROTTER. SA: South Asian, WE: West Eurasian, EA: East Asian, SIB Siberian. (D) The admixture events of western and eastern ancestries in NXH are estimated by MultiWaver 2.0. HAN and CEU were used as representatives for eastern and western ancestries, respectively. Each line represents one target population. Each circle on the line represents one admixture event, where the color indicates the source ancestry and the size is proportional to the admixture proportion of the corresponding ancestry.
We further inferred the sequence of admixture events for those four ancestries. Interestingly, the EA ancestral component positively correlated with latitude (COR = 0.80, P = 0.0004). WE and SA ancestral components negatively correlated with latitude (COR = −0.82, P = 0.002; COR = −0.60, P = 0.017, respectively). The SIB ancestral component showed no correlation with latitude (COR = −0.33, P = 0.23) (fig. 2B). These results suggested that WE and SA ancestral components jointly formed the western ancestry before it contributed to the NXH gene pool in a south-to-north direction. In contrast, the scenario of eastern ancestry was more complex, of which EA ancestral component contributed to the NXH gene pool in a north-to-south direction, whereas SIB ancestral component contributed to NXH gene pool via both western and eastern ancestries (fig. 2C). Moreover, both the admixture history graph (AHG) and GLOBETROTTER results suggested that SIB ancestry might be introduced into the gene pool of NXH via both western and eastern ancestries (fig. 2C and supplementary fig. S9 and table S7, Supplementary Material online).
Assuming the above model, we would expect some present-day populations showing admixture signals of EA-SIB and WE-SA-SIB. First, we searched for populations showing EA-SIB admixture. The Han Chinese population (HAN) was the best-guess representative of ancestral origins of eastern ancestry inferred with GLOBETROTTER (supplementary table S7, Supplementary Material online). Additionally, some HAN samples had the closest ratio of EA/SIB ancestry with NXH (supplementary fig. S10, Supplementary Material online). Next, we looked for populations showing WE-SA-SIB admixture. Lezgin and Kumyk populations were the best-guess representatives of ancestral origins of western ancestry inferred with GLOBETROTTER (supplementary table S7, Supplementary Material online). The proportion of WE, SA, SIB ancestral components in these two populations highly correlated with that in NXH (COR = 0.999, P = 0.02; COR = 0.997, P = 0.05 for Lezgin and Kumyk populations, respectively) (supplementary fig. S11, Supplementary Material online).
Based on the above analysis, we proposed an “admixture of admixture” model for NXH (fig. 2C). In brief, admixture first occurred among the ancestral populations of WE, SA, and SIB ancestry in the West, whereas admixture also occurred between the ancestral populations of EA and SIB ancestry in the East; followed by further contacts of the mixed Western ancestries (WE-SA-SIB) and the mixed Eastern ancestries (EA-SIB). Eventually, these ancestral populations of mixed ancestries experienced more complex admixture and formed the gene pool of present-day NXH. We also employed qpGraph to explore the admixture graph of NXH. The best admixture graph (maximum |Z| = 2.1) inferred by qpGraph confirmed four ancestral components in NXH and the “admixture of admixture” model for NXH (supplementary fig. S12, Supplementary Material online). The detailed admixture history is modeled and described in the following sections.
Admixture Time
We further reconstructed the history of the admixture between eastern ancestry and western ancestry in NXH. We applied MultiWaver 2.0, which infers the admixture history based on the length distribution of ancestral tracks, to investigate the potential of multiple admixture waves from multiple source populations in the history of NXH. The ancestral tracks were inferred using HAPMIX, with HAN and CEU as representatives of the eastern ancestry and western ancestry, respectively. The results showed that the “multi 1-2 model” fitted the data well, indicating there were two discrete admixture events (fig. 2D). The first wave of admixture was estimated to have occurred 1,025 years (41 generations) ago and the second wave of admixture was estimated to have occurred 500 years (20 generations) ago. The time of the second admixture event was consistent with the results estimated by other methods, such as ALDER and GLOBETROTTER (supplementary fig. S13 and tables S7 and S8, Supplementary Material online).
Sex-Biased Admixture
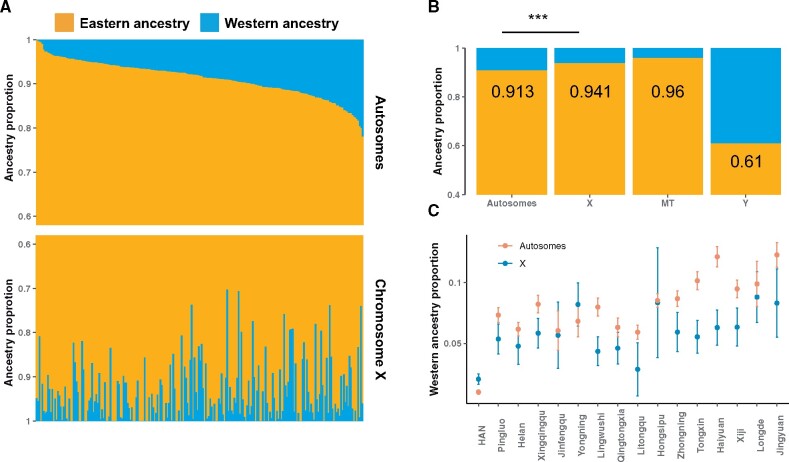
To investigate whether there was a sex-biased admixture in the history of NXH, we compared the admixture results obtained from autosomes, X chromosome, mtDNA, and Y chromosome. We estimated the admixture proportion assuming two major ancestral components, that is, western and eastern (fig. 3 and supplementary tables S2 and S3, Supplementary Material online). The estimated genetic contribution of the western ancestry into NXH was 8.6% for autosomes, 5.9% for X chromosome, 3.6% for mtDNA, and 39.3% for Y chromosome, respectively. The results of Y chromosome and mtDNA were consistent with the previous studies (Yao et al. 2004; Wang et al. 2019; Xie et al. 2019). Additionally, though the difference in genetic contribution was small, there was a significant difference in admixture proportions between autosomes and X chromosome (Student’s t-test, P < ). This pattern was consistent across different regions in Ningxia (fig. 3C). These results indicated that the admixture of NXH was sex biased to the combination of Eastern females and Western males.
Fig. 3.
Sex-biased admixture in NXH people. (A) The genetic makeup of NXH individuals is estimated based on markers on Autosomes and X chromosome. The samples were ordered from highest to lowest by the eastern ancestry proportion based on Autosomes. (B) The ancestral makeup of the NXH people is estimated based on markers on different chromosomes assuming two ancestries. The “***” indicates P < 0.001. (C) The genetic contribution of western ancestry in NXH people in different regions. The error bar indicates the one standard error.
Fine-Scale Genetic Structure of NXH
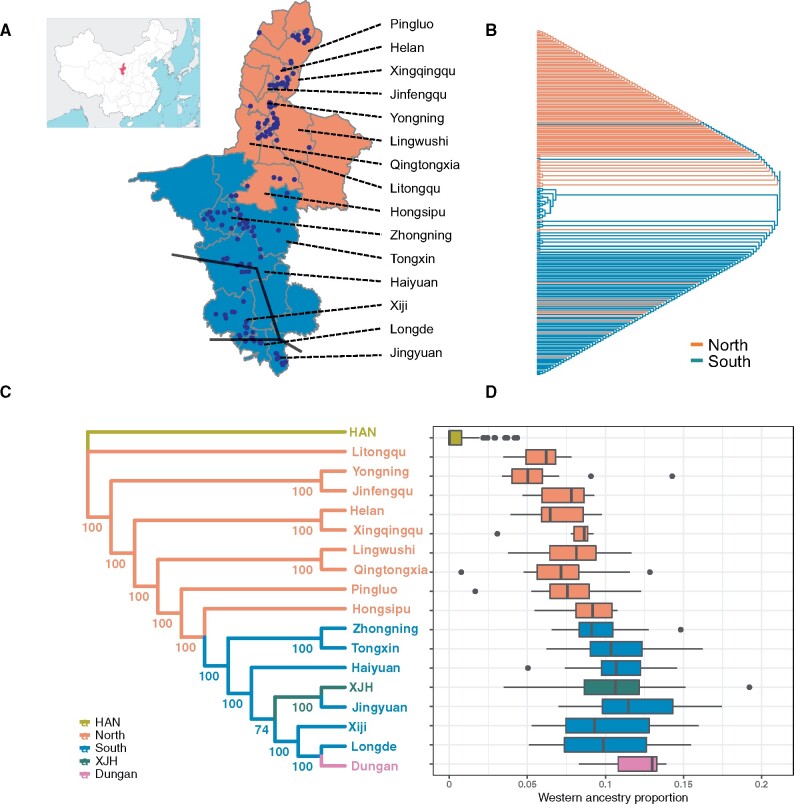
We identified two major genetic clusters using ChromoPainter and fineSTRUCTURE, which are largely corresponding to the geographical distribution of NXH individuals (fig. 4A and B and supplementary table S9, Supplementary Material online). One cluster mainly includes samples from northern Ningxia, and the other mainly includes samples from southern Ningxia. The result indicated that geography might play an important role in the genetic differentiation of NXH.
Fig. 4.
Fine-scale genetic structure exists in NXH people. (A) The geographical distribution of NXH samples. The small panel in the top left indicates the location of Ningxia in China. Ningxia could be divided geographically into the southern and northern regions, which is indicated with different colors. The regions from which samples were collected were labels on the right side. Each point represents one sample. The dark lines indicate the Silk Road across Ningxia. Map boundary data from National Catalogue Service for Geographic Information (http://www.webmap.cn). (B) The phylogenetic tree of all the 230 NXH samples inferred by fineSTRUCTURE. The samples were colored based on the geographical region from which they were collected. (C) The neighbor-joining phylogenetic tree of Hui samples from different regions in Ningxia, Xinjiang (XJH), and Dungan based on pairwise FST. The bootstrap values based on 100 replicates are shown. The samples in Ningxia are grouped based on the regions from which they were collected. The regions were colored as in the legend. (D) The genetic proportion of western ancestry in Hui samples in different regions and Han samples based on ADMIXTURE for Eurasian populations assuming two ancestries.
The average pairwise FST between regional NXH populations was 0.0007, with the maximum 0.002 between Yongning in northern Ningxia and Zhongning in southern Ningxia and the minimum close to 0 between some regions (supplementary fig. S14, Supplementary Material online). According to the Neighbor-Joining phylogeny tree based on the pairwise FST, the regional populations located in southern and northern Ningxia, respectively, clustered together (fig. 4C). Moreover, fine-scale geographical regions clustered tightly and negatively correlated with the geographical distance. These results were also confirmed by allele-sharing distance (ASD) and identity by descent (IBD) (supplementary fig. S14, Supplementary Material online). ADMIXTURE analysis showed that there was a significant correlation between admixture proportions and the latitude of the NXH in different regions (fig. 2B and supplementary fig. S15, Supplementary Material online). In particular, from north to south, EA ancestral component decreased from 87.0% (Yongning) to 78.5% (Jingyuan), whereas WE and SA ancestral components increased from 4.9% (Yongning) to 9.6% (Jingyuan), from 1.3% (Yongning) to 3.4% (Hongsipu), respectively. These results clearly showed that there was a south-to-north cline in the genetic makeup of the NXH in different regions of Ningxia. Although there was no significant difference within southern or northern NXH, a significant difference in the genetic makeup between southern NXH and northern NXH was observed (supplementary fig. S16, Supplementary Material online). These results were also consistent with that of PCA (supplementary figs. S17 and S18, Supplementary Material online).
Despite the genetic difference as observed in the above analyses, the length distribution of the ancestral segments was similar between northern and southern NXH (supplementary fig. S19, Supplementary Material online). These results indicated northern and southern NXH-shared admixture history; the difference in the genetic makeup of southern and northern NXH was likely due to a south-to-north migration in history.
In addition to NXH, we also collected Hui samples from Xinjiang (XJH) in China and samples from Uzbekistan (Dungan). Genetic differentiation measured by FST indicated that the Hui people from different geographical regions are closely related, suggesting a common origin of the Hui people (supplementary fig. S20, Supplementary Material online). Notably, the genetic difference between some regions in southern and northern NXH (FST = 0.0020) was even larger than that between XJH and NXH (FST = 0.0003) (fig. 4C).
We further investigated the genetic makeup of the Hui people in different regions. We used the ADMIXTURE result of all the Hui samples together with other Eurasian populations at K = 2 (fig. 4D). Dungan and Jingyuan Hui in southern Ningxia had the highest western ancestry proportions (∼12%), followed by XJH (∼10%), Yongning Hui in northern Ningxia had the lowest western ancestry proportion (∼5.9%). There was no significant difference between the Dungan and the southern NXH (supplementary fig. S16, Supplementary Material online). The pattern was also observed in the PCA result (supplementary figs. S17 and S18, Supplementary Material online).
Additionally, the western ancestry proportion in the Dungan samples ranged from 0.083 to 0.139, which was within the range of 0.007 ∼ 0.174 for NXH. The western ancestry proportion in the Uzbekistan individuals surrounding the Dungan people ranged from 0.461 to 0.710 (supplementary fig. S21, Supplementary Material online). Moreover, it seems that there was no significant difference in the admixture time among the Huis in different regions (supplementary fig. S22, Supplementary Material online). These results suggested that there was no considerable gene flow between Dungan and surrounding populations. Additionally, we did not find some SNPs extremely highly differentiated (FST > 0.1) between southern NXH and northern NXH (supplementary fig. S23, Supplementary Material online).
Differentiation between NXH and HAN Induced by Admixture
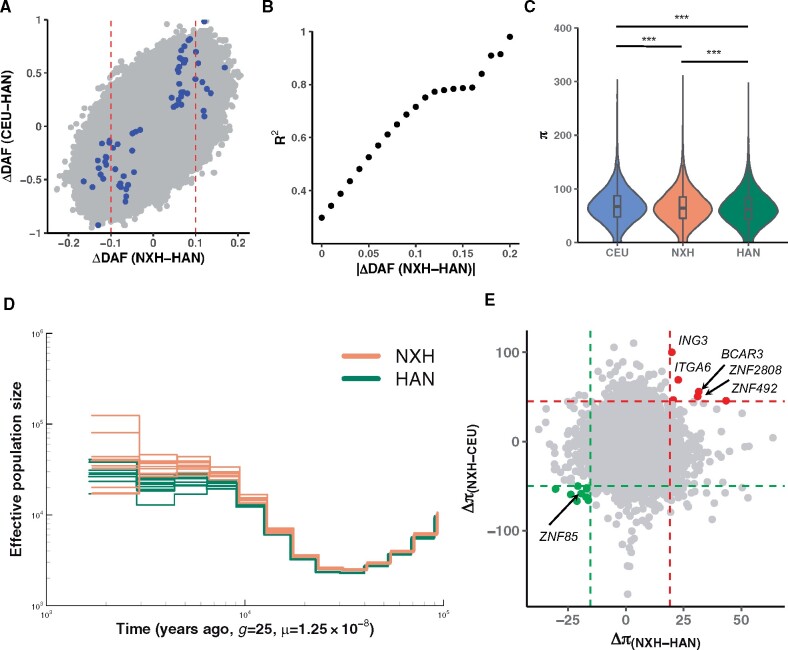
The allele frequency difference between NXH and HAN is highly correlated with that between the western ancestry (represented by CEU) and the eastern ancestry (represented by HAN) (fig. 5A), indicating the differentiation between NXH and HAN attributed to the western ancestry. On the whole genome level, 29.8% of the total difference between NXH and HAN could be explained by the difference between CEU and HAN. The larger differentiation between NXH and HAN, the greater the contribution of western ancestry to the differentiated SNPs. For example, the contribution was about 71.6% and 98.0% for SNPs with allele frequency differences between NXH and HAN larger than 0.1 and 0.2, respectively (fig. 5B). Besides, SNPs highly differentiated between NXH and HAN tended to show larger differentiation between CEU and HAN (supplementary fig. S24, Supplementary Material online). These results suggested that the differentiation between NXH and HAN was largely induced by genetic admixture.
Fig. 5.
The impact of admixture on the genome diversity of NXH. (A) The correlation between allele frequency difference between NXH and HAN and that between western ancestry (CEU) and eastern ancestry (HAN). The red line indicates the top 1% threshold of the empirical distribution of the absolute difference between NXH and HAN. The colored points indicate they are highly differentiated functional variants between NXH and HAN. (B) The x axis indicates the threshold which is used to prune the data set. SNPs with absolute allele frequency larger than the threshold were used to estimate the correlation. The y axis indicates the correlation (measured by R2) between allele frequency difference between NXH and HAN and that between CEU and HAN for the pruned data set. (C) The nucleotide diversity (π) of CEU, NXH, and HAN. An equal number of samples were selected from each population. The “***” indicates P < 0.001. (D) Estimated changes in historical effective population size. Four genomes (eight haploid genomes) were randomly selected from one of the two populations with deeply sequenced genomes in this study. This analysis was repeated 10 times. We showed the results based on an absolute estimation of time under the assumption of a slow mutation rate of 0.5 × 10−9 per site per year. (E) The difference in nucleotide diversity (π) between NXH and HAN and between NXH and CEU populations. Each point represents one nonoverlap 100 kb window on the genome. The dashed line indicates the difference of this window is significant with P < 0.001 of the empirical distribution. Gene names from the significant windows are labeled on the plot.
NXH showed greater genetic differentiation with HAN (FST = 0.0015) than that between northern and southern Han Chinese populations (FST = 0.001). We further searched for genomic loci that highly differentiated between NXH and HAN. In sum, we identified a total of 65 potentially functional SNPs, of which the genetic consequence was high or moderate predicted by variant effect predictor (VEP) (supplementary fig. S25 and table S10, Supplementary Material online). For these 65 potentially functional SNPs, 74.2% of the difference between NXH and HAN could be attributed to the difference between the western ancestry and the eastern ancestry. Moreover, most of the signals are well-known differences between west Eurasian and East Asian populations. Among these 65 SNPs, the rs1264454 ranked at the top (FST = 0.071) and located in the SLC24A5 gene, which was associated with pigmentation and was subjected to natural selection in European populations (Lamason et al. 2005). Another SNP rs3827760 ranked the fourth (FST = 0.055) and located in the EDAR gene, which was a famous gene underlying selection in East Asian populations and associated with hair morphology and facial morphology (Sabeti et al. 2007; Kamberov et al. 2013). Interestingly, the SNPs rs4148211 located in ABCG5/ABCG8 gene region showed high genetic differentiation between NXH and HAN (FST = 0.033). ABCG5/ABCG8 were well-known genes that regulate lipid metabolism by suppressing intestinal absorption and promoting bile excretion of sterols (Calandra et al. 2011). Adaptation to different diets is a well-known selective pressure for human evolution. The Hui and Han people in China have different dietary habits. For example, the Hui people don’t eat pork, which was the most common meat for the Han people. This might lead to the genetic differentiation of certain associated genes between the Hui and Han populations.
The Impact of Admixture on the Genome Diversity of NXH
The distribution of joint allele frequency spectra of NXH and others also confirmed Hui was an admixed population with both western and eastern ancestries (supplementary fig. S26, Supplementary Material online). Besides, we did not find any variants showing low frequency in both HAN and CEU but common in NXH, as expected as NXH are an admixed population. Admixture might introduce some alleles that were absent in HAN or other East Asian populations, hereafter refers to non-HAN allele. There were a total of 2,564 SNPs with non-HAN alleles. The frequencies of most non-HAN alleles were low (<0.1) in NXH (supplementary fig. S27, Supplementary Material online). This might indicate that the effect of western ancestry on the genome of NXH was relatively limited, as only 10% of the NXH gene pool was derived from populations of western ancestry.
The admixture was expected to influence the population diversity. We calculated the nucleotide diversity π for the NXH, HAN, and CEU populations (fig. 5C). The genetic diversity of NXH was significantly larger than that of HAN (Wilcoxon test, p < 1.9 × ) and significantly lower (Wilcoxon test, p < 1.12 × ) than that of CEU. One possible explanation was that the genetic contribution of western ancestry increased the population diversity of NXH, but to a limited extent. We applied MSMC2 to estimate the effective population size (Ne) of NXH and HAN. HAN showed an overall smaller Ne than NXH populations, which was confirmed by an estimation based on linkage disequilibrium (LD) (fig. 5D and supplementary fig. S28, Supplementary Material online). It is not clear to what extent the estimation was affected by admixture, as MSMC2 assumes the target population to be a nonadmixed population. These results may only suggest that the diversity of NXH was larger than that of HAN. We also identified some regions in which the genetic diversity was significantly higher in NXH than its ancestral populations represented by HAN and CEU populations (fig. 5E, supplementary tables S11 and S12, Supplementary Material online). One region contained the ITGA6 gene, which was associated with the integrin pathway. Interestingly, the genetic contribution from archaic groups in NXH (∼1.49%) was significantly lower than that in HAN (∼1.52%) (Student’s t-test, P < 2 × ), the total length of the whole genome covered by archaic introgression was larger in NXH than in HAN (supplementary fig. S29 and S30, Supplementary Material online), which could be due to that some archaic sequences in NXH genome were introduced via the western ancestry. These results indicated the influence of genetic admixture on the genome diversity of NXH.
We also calculated the runs of homozygosity (ROH) to measure the inbreeding of NXH and HAN. The segments of ROH were clustered into different categories by length: short (500 ∼ 1,500 kb) and long (>1,500 kb). For short ROH, NXH had a lower total length of ROH than HAN did. For the long segments, the number and the total length in NXH were significantly larger than that of HAN (supplementary fig. S31, Supplementary Material online). The admixed population was expected to have fewer ROH, and the consanguineous population carries long ROH (Ceballos et al. 2018). NXH people practice Islam and are thought to prefer consanguinity (Jia 2006).
Discussion
We have presented a comprehensive characterization of genetic variation in 234 NXH samples of age over 60, which is the first whole-genome sequencing study of this ethnic group. Considering the unique history of the Hui population in China, the whole-genome data generated in this study is of great significance for the genomic studies of East Asian populations and Muslim populations and serves as a useful control data set for genetic association studies of late-onset diseases.
With this unprecedented data, we comprehensively revealed the genetic origins, admixture history, and population structure of NXH. Our results showed that NXH was most closely related to East Asian populations compared with other global populations. Moreover, NXH shared the majority of genetic makeup with the Han Chinese population. Interestingly, although the Hui and Han Chinese peoples were very similar in appearance (Zheng et al. 1997), they were genetically distinguishable from each other, which could attribute to the western ancestry in NXH. Remarkable differences in the genetic makeup between NXH and HAN were observed. Specifically, four major ancestral components were identified in the NXH, which potentially were derived from ancestral populations in East Asia, Siberia, West Eurasia, and South Asia. In contrast, two major ancestral components were identified in HAN. Modeling admixture history indicated that these four ancestral components were derived from two earlier admixed populations. The eastern ancestry consisted of East Asian and Siberian ancestral components. The western ancestry consisted of West Eurasian, South Asian, and Siberian ancestral components. The population movement and gene flow between Siberia and West Eurasia across the Eurasian steppe has been reported by several studies (Pugach et al. 2016; Sikora et al. 2019), which suggested the possibility that the Siberian ancestral component through western ancestry. Moreover, our simplified modeling of isolation by distance showed that it is unlikely to explain the history of NXH (supplementary fig. S32, Supplementary Material online), thus supporting the admixture model we proposed for NXH (supplementary fig. S12, Supplementary Material online). Besides, the admixture between eastern and western ancestries was sex-biased, with more Eastern females and Western males. Merchants, emissaries, and soldiers migrated from Arab, Persia, and Central Asia into China and those people were mainly males. Moreover, the Hui people practice endogamy and the marriage occurred mainly within the Huis. Intermarriage generally involves a Han Chinese converting to Islam, especially, marriages between Hui male and Han female were more frequent (Jia 2006).
The distribution of the Huis is a result of the genetic origin, migration, and admixture history of this group. We observed a south-to-north cline within NXH. Specifically, the samples in southern Ningxia regions had a higher western ancestry proportion, which can be attributed to a few factors. First, the old Silk Road went through the southern Ningxia, which was the main route of the gene flow between East Asian and West Eurasian (Gladney 1997) (fig. 4A), which might have resulted in differentiated admixture between southern and northern NXH. Second, the Hui account for a higher total population percentage in southern Ningxia than in northern Ningxia, resulting in relatively fewer intermarriages between the Huis and Han Chinese in the south compared with that in the north. Indeed, intermarriage between Huis and other ethnic groups is much less frequent in southern than in northern Ningxia (Yang 2002).
Interestingly, among the Hui people in Northwestern China, the genetic differentiation between southern and northern NXH was even larger than that between NXH and XJH, though Ningxia and Xinjiang were further apart geographically. Moreover, the western ancestry contribution was lower in XJH (∼10%) than that in some regions southern NXH (∼12%), although slightly higher than that in northern NXH (<9%). These results seemly unexpected, because compared with Ningxia, Xinjiang was geographically closer to Central Asia and West Eurasia and was the gateway for western people from Central Asia to enter the interior. However, it would be reasonable if historical documents were referred to, which recorded that the XJH were mainly immigrants from Northwestern China. For example, it is believed that the history of the Huis settlement in Xinjiang began after the suppression of the Junggar rebellion in the twentieth year of the Qing Emperor Qianlong (1755) (Li 2010).
Also, our results suggested that Dungan was genetically more closely related to southern NXH. According to historical documents, Dungan were the Hui people who migrated from China into Central Asia in the year 1867. ADMIXTURE analysis suggested that there was no considerable gene flow between Dungan and surrounding populations after Dungan people migrated from China into Central Asia, which could due to that they speak the Sino-Tibetan language, whereas most of the surrounding populations speak Turkic language.
We evaluated the effect of some confounding factors on the inference of fine-scale population structure. ASD was commonly used to measure the genetic differentiation at the individual level. Compared with FST, it is not necessary to separate individuals into groups with different genetic backgrounds to estimate the allele frequency. Pairwise distance within one population was expected to be less than that between populations. However, we found that this was not true for the admixed population (supplementary fig. S33, Supplementary Material online). We performed a simulation to investigate whether this was due to the effect of admixture. Genetic distance within groups having higher western ancestry proportion was larger than that between groups having lower western ancestry proportion (supplementary figs. S33 and S34, Supplementary Material online). This was consistent with the larger ASD among samples from southern Ningxia (supplementary fig. S35, Supplementary Material online). The genetic distance between regions in southern Ningxia was larger than that between regions in northern Ningxia. This could partially explain the pattern observed in the estimated effective migration surfaces (EEMS) result that the lowest effective migration rate was among the southern regions in Ningxia (supplementary fig. S35, Supplementary Material online) and that genetic distance between NXH and HAN was less than genetic distance within NXH (supplementary fig. S33, Supplementary Material online).
We also found that enough markers were needed to detect fine-scale population structure (supplementary fig. S36, Supplementary Material online). Additionally, estimating admixture time is important to understand the history of the admixed population. We observed the admixture times inferred by ALDER and GLOBETROTTER are inconsistent with that inferred by MultiWaver 2.0. Both ALDER and GLOBETROTTER estimated that the admixture of NXH occurred 20 generations ago, whereas MultiWaver 2.0 identified an additional ancient admixture event that occurred 41 generations ago. Indeed, according to the recorded history, the admixture of the Hui population may have started as early as 1,400 years ago during the Tang and Song dynasties (Chen 1999). Previous studies also suggested that MultiWaver 2.0 was more powerful than other methods in modeling ancient and complex admixture. For example, an ancient admixture of the Uyghurs that occurred in the Bronze Age was identified by MultiWaver 2.0, and has been confirmed by some recent ancient DNA studies (Feng et al. 2017; Ning et al. 2019). MultiWaver 2.0 leveraged the information of length distribution of ancestral tracks and required the ancestral segments of local ancestry inference as input. However, many local ancestry inference methods need a priori admixture time. Our results showed that the marker density would affect the process to choose the optimized parameter, which would indirectly affect the result inferred based on the length distribution of ancestral tracks (supplementary fig. S36, Supplementary Material online). These findings valued the whole genome sequencing data in exploring the history of admixed populations such as the Huis.
Our results suggested a complex scenario of genetic origin, admixture history, and population structure in the Hui population. The two-wave model we proposed here does not necessarily mean there were only two admixture events in history. Rather, it suggested population admixture occurred at least more than once. Moreover, the first wave of admixture was estimated to occur 1,025 years (41 generations) ago, which might suggest the admixture event begins to occur 1,025 years ago at the latest. Furthermore, the admixture time could be underestimated (Leslie et al. 2015). The ancient admixture we identified, that is, occurred over 1,025 years ago, is roughly corresponding to the late Tang Dynasty, and the Five Dynasties and Ten Kingdoms period. However, the intensive contact between western and eastern peoples might be common in the early Tang Dynasty according to historical records. Similarly, the time of a recent admixture that occurred nearly 500 years ago as we inferred in this study, corresponding to the Ming Dynasty, might be also underestimated. Again, according to the recorded history, west–east contacts were more frequent during the Mongolian Conquests in the 13th and 14th centuries. The concordance would be improved if we adopted a longer generation time, 29–30 years, as a previous study suggested (Fenner 2005). Nonetheless, we believe history could be more complex than the simplified models as we presented in this study. We should point out that the Hui is one of the most distributed populations in China, the samples studied here were mainly descendants of the Hui people in Northwestern China, in which more than 51% of the Hui people are concentrated. Whether the Hui genomes in this study fully represent the diversity of the Hui China has not been evaluated. Further efforts in developing more sophisticated methods and recruiting more diverse population samples are expected to uncover a more comprehensive picture of the diversity, origins, and history of the Hui people.
Materials and Methods
Populations and Samples
Peripheral blood samples of 234 Hui individuals aged over 60 were collected from 15 (Jingyuan, Longde, Xiji, Haiyuan, Tongxin, Zhongning, Hongsipu, Litongqu, Qingtongxia, Lingwushi, Yongning, Jinfengqu, Xingqingqu, Helan, and Pingluo) of the 22 county administrative regions in the Ningxia Hui Autonomous Region (NX) of China (supplementary fig. S1 and table S1, Supplementary Material online). The samples enrolled in this study were chosen almost uniformly from each geographical region, ranging from 6 to 24 with a median of 12. Peripheral blood samples were also collected from 39 individuals from the Xinjiang Uygur Autonomous Region (XJ) of China. Each individual was the offspring of a nonconsanguineous marriage of members of the same nationality within three generations. Informed consent was obtained from all participants. This study was approved by the Biomedical Research Ethics Committee of the Shanghai Institutes for Biological Sciences. All the procedures were in accordance with the ethical standards of the Responsible Committee on Human Experimentation (approved by the Biomedical Research Ethics Committee of the Shanghai Institutes for Biological Sciences, No. ER-SIBS-261408) and the Helsinki Declaration of 1975 (revised in 2000).
Genome Sequencing and Data Processing
Whole-genome sequencing, with 36 high target coverage (30×) and 198 medium target coverage (10×) for 150 bp paired-end reads was carried out on an Illumina HiSeq X Ten according to Illumina-provided protocols with standard library preparation at WuXi NextCODE (Shanghai). Each sample of high coverage was run on a unique lane with at least 90 GB of data that had passed filtering and each sample of medium coverage was run on a unique lane with at least 30 GB of data that had passed filtering. For high coverage data, reads data were quality controlled so that 80% of the bases achieved at least a base quality score of 30. For medium coverage data, reads data were quality controlled so that 80% of the bases achieved at least a base quality of 10 (supplementary fig. S2, Supplementary Material online). Reads were merged, adaptor trimmed, and mapped to the human reference genome (GRCh37) with the Burrows-Wheeler Aligner (Li and Durbin 2010). Variant calling was carried out with the HaplotypeCaller module in the Genome Analysis Toolkit (GATK) version 3.8 (McKenna et al. 2010; DePristo et al. 2011).
Among these sequenced samples, we observed 18,112,647 single-nucleotide variants (SNV), consisting 17,503,234 for autosomes, 592,432 for X chromosome, and 16,981 for Y chromosome. Particularly, we discovered 1,573,001 (8.7%) novel SNVs compared with the dbSNP153 version. Unsurprisingly, most of the novel variants were extremely rare. Singleton accounted for 87.2% of all the novel variants. 9.7% of the novel variants reached MAF < 0.01. 1.6% had 0.01 <=MAF < 0.05. And 1.5% of the novel variants were common (supplementary fig. S3, Supplementary Material online).
As an additional quality control, the transition versus transversion ratio for the Hui genome was 2.103. The variants in the call set were annotated using Ensembl VEP (McLaren et al. 2016) with the corresponding VEP-compiled annotation database (v86_GRCh37).
We used Beagle v4.1 software, which could take genotype likelihoods as input (Browning and Yu 2009), to perform the LD-based genotype refinement. Low-quality variants with Dosage R2 < 0.3 estimated by Beagle were removed, including 416,802 SNVs on the autosomes and X chromosome. The genotypes from Beagle were used for further analysis. We did not observe any subtle batch effect between high-coverage (30×) sequencing and medium-coverage (10×) sequencing data (supplementary fig. S4, Supplementary Material online).
Genotyping
Thirty-nine XJH samples were genotyped using Illumina HumanOmniZhongHua-8 Beadchip. To study the genetic variation of the Hui population in a broader context, we also obtained genotype data of 13 Dungan from a previous study (Jeong et al. 2019), typed on Affymetrix Axiom Human Origins 1 Array.
Quality Control
Genetic relatedness for all pairs of Hui samples was estimated using PLINK v1.90 (Chang et al. 2015). We preferentially removed one individual with a higher genotype missing rate from each pair with a coefficient of relationship larger than 0.125. This step removed five individuals, including four whole-genome sequencing samples and one Dungan sample. After quality control, there were 281 individuals left for further analysis.
Public and Published Data
The Affymetrix Human Origins Array data set (Lazaridis et al. 2014) contains 600,841 SNPs for 2,345 individuals from 203 present-day populations. This data set was obtained with a signed letter permitting full data access and was used for comparison with Hui under a global context. The 1000 Genomes Project Phase 3 data set (Auton et al. 2015) was also included for the larger sample size of some worldwide populations. We also included some published high coverage whole-genome data for East Asian populations, like TIB and HAN (Lu et al. 2016).
We generated several data panels that were used for analytical purposes as described later. We merged the whole-genome sequencing data including NXH and HAN with data from 1000 Genome Project Phase 3 by extracting biallelic autosome SNPs called in both data sets (Panel 1). This data set was used for analysis that needs higher marker density, for example, analysis of local ancestry inference and fine-scale population structure within NXH. The microarray data of Hui samples were imputed using Beagle version 4.1 (Browning and Browning 2016) with a reference panel, which contained 1,025 East Asian samples, including 234 whole-genome sequencing Hui samples in this study. We merged the whole genome sequencing data, the imputation data, the 1000 Genomes Project Phase 3, and Human Origins data set, which resulted in 433,214 SNPs. This data set contained all the Hui samples and was used for most of the analysis (Panel 2).
The final combined data were phased using Beagle version 4.1 (Browning and Browning 2007) with 1000 Genomes Project Phase 3 data as a reference panel. HapMap phase II genetic map was used to calculate genetic distance between markers (Frazer et al. 2007).
Determining Y Chromosomal and mtDNA Lineages
The mtDNA haplogroups were classified using HaploGrep2 (Weissensteiner et al. 2016) based on PhyloTree17. The NRY haplogroups were classified based on known Y-SNP markers from ISOGG Y-DNA phylogenetic tree 2019–2020. To compare the frequency distribution of worldwide populations, we combined sublineages under each major paternal or maternal haplogroup (supplementary tables S2 and S3, Supplementary Material online). To compare maternal and paternal lineages of the Hui population, we collected haplogroup data of different populations from the literature based on the division of ancestral composition. Then we applied ADMIX2.0 (Dupanloup and Bertorelle 2001) to calculate ancestry proportion based on haplogroup results for both mtDNA and NRY genetic markers.
Principal Component Analysis
PCA was performed at the individual level using EIGENSOFT version 6.1 (Patterson et al. 2006). To investigate the fine-scale population structure, we carried out a series of PCA by gradually removing “outliers” on the plot of the first two principal components and re-analyzing the remaining samples based on the same set of SNV markers. We removed LD by thinning the SNPs to be at least 150 kb apart, resulting in 16,217 SNPs. We relaxed the criteria to 10 kb to get a higher marker density data set including 250,409 SNPs, which was used to analyze fine-scale population structure within NXH.
Admixture
We applied ADMIXTURE version 1.3.0 (Alexander et al. 2009) on the merged data set of Human Origins, Hui, and HAN data, which consist of individuals from 205 populations on the same SNPs as PCA. We run ADMIXTURE by assuming the number of ancestries (K) from 2 to 20. For each K, we repeated the analysis 30 times with different random seeds and picked the run with the highest log-likelihood score to avoid the local minimum. To reveal the genetic makeup of the Hui population, for each K, we identified the ancestral component (>1%) in the Hui population. For each ancestral component, we identified the representative reference individuals and populations, which were used in some further analysis, like GLOBETROTTER. We also estimated the genetic makeup of the Eurasian population based on 2,514 markers on the X chromosome. To compare the results based on Autosomes and X chromosome, we rerun ADMIXTURE based on the same number of markers randomly chosen from the SNPs on the Autosomes. The data visualization was carried out using AncestryPainter (Feng et al. 2018).
Admixture History Graph
We have identified four ancestral components in NXH. We used a method called AHG (Pugach et al. 2016) to detect the sequence of admixture events. In this study, we examined all possible trios (a combination of three ancestries) from these four ancestries, and for each trio, we chose the one that produces the least absolute value of Pearson correlation coefficient instead of covariance used in the original study. Then the full graph was reconstructed based on the ordering of likely configurations.
ADMIXTURE result for NXH with other Eurasian populations at K = 4 was used to estimate the admixture proportion of each ancestral component in NXH samples. For each trio, we did 1,000 replicates with 230 individuals randomly sampled with replacement for each replicate. We examined the model that best fits the data.
FST Analysis
Genetic distance within Hui and between Hui and other populations was measured with FST according to Weir and Cockerham (1984), which accounts for the difference in the sample size of each population. We randomly chose ten samples from each population to calculate pairwise FST. For a population with a sample size of less than 10, all the samples were used. We repeated 100 times to calculate the pairwise FST and confidence interval. SNPs with a missing rate larger than 0.05 in any population were removed. There were 407,368 SNPs remained for the analysis. When calculating pairwise FST between sample collection regions within Ningxia, we chose six samples from each collection region, which was the minimum sample size of the sample regions. We used a similar strategy to calculate the confidence interval except that we chose six samples from each population.
The neighbor-joining phylogeny tree (Saitou and Nei 1987) was constructed using MEGA 6.0 (Tamura et al. 2013) based on an unbiased FST matrix. The visualization of the phylogenetic tree was done by R package “ggtree” (Yu et al. 2017).
Allele Sharing Distance
ASD (Gao and Martin 2009) was used to measure the genetic difference at the individual level and to explore the genetic relationship between pairwise sampling regions in Ningxia. The genetic difference between regions was defined as the average distance of pairwise individuals from different regions. The genetic difference within one region was defined as the average of the pairwise distance of individuals of that region. A neighbor-joining phylogeny tree was constructed using ASD between regions.
We also performed systematic simulations to confirm the pattern observed in the analysis. The simulation data were generated using the forward-time simulator Admixsim (Yang et al. 2020). We applied the two-way admixture model with western ancestry proportion ranging from 0.05 to 0.9 and assuming that the admixture event occurred 20 generations ago, which was a simplified admixture model for NXH.
Identity by Descendent Analysis
The pairwise IBD sharing was inferred using refinedIBD implemented in Beagle 4.1 (Browning and Browning 2013) using default parameters except that the trim was adjusted as the author suggested. The IBD sharing between regions was calculated using a similar strategy to ASD.
EEMS Analysis
We also used a method called EEMS (Petkova et al. 2016) to quantify the relative migration rate within NXH. We approximated the outline of Ningxia with a polygon of near coordinates with the “polyline” function in Google Maps. We then ran EEMS with several demes 100, 200, and 500 to investigated whether this change affects the result. We independently ran five repeats for each deme. For each repeat, we ran the model for 2,000,000 iterations, with 1,000,000 burn-in steps to allow the model to converge to a maximum log-likelihood configuration. R package “rEEMSplots” was used to plot the result of 500 demes.
f Statistics
All the f statistics were calculated using ADMIXTOOLS 7.0 (Patterson et al. 2012). We used the f3 test in the form of f3 (source1, source2; NXH) to formally check whether NXH was an admixed population. All the possible combinations of worldwide populations were used as references. More negative values denoted reference populations were closer to true mixing ancestral populations. We also computed outgroup f3 (NXH, X; Ju hoan North), according to Raghavan (Raghavan et al. 2014), to examine for relatedness between NXH and non-African populations. We applied f4 ratio estimation in the form f4 (Papuan, Ju hoan North; NXH, Y)/f4 (Papuan, Ju hoan North; X, Y) to estimate the contribution of eastern ancestry to NXH, where X was one population from East Asia and Y was one population from West Eurasia. We modeled population relationship and admixture using qpGraph in ADMIXTOOLS. We calculated f2, f3, f4 statistics measuring allele sharing of two, three, and four populations. Models with maximum |Z| for f4 statistics < 3 were considered as acceptable. Mubti, English, Mala, Chukchi, Han, and NXH populations were chosen in this analysis.
Chromosome Painting, Population Clustering, and Admixture
We employed a set of haplotype-based methods—ChromoPainter (Lawson et al. 2012), fineSTRUCTRE (Lawson et al. 2012), GLOBETROTTER (Hellenthal et al. 2014)—to explore fine-scale population structure within NXH and to describe admixture events in NXH. First, we applied ChromoPainter (v2) to get the haplotype painting for all target individuals and copying vectors for all individuals, which involves two steps. We run ChromoPainter to estimate switch rate Ne and the global mutation rate μ using ten expectation–maximization (EM) iterations on all chromosomes for all individuals. We obtained the final parameter estimates Ne and μ by averaging the estimates for all chromosomes. Next, we run using these parameter estimates and without EM iterations to paint each haplotype in the target individual with haplotypes from surrogate individuals. We sampled 10 paintings per haplotype for target individuals and obtained copying vectors for all individuals. We used GLOBETROTTER to describe the admixture events.
For diverse purposes, we performed GLOBETROTTER analysis with a different strategy to set the donor populations and the target population: 1) The donor populations were ethnic groups. We used all the 132 Eurasian populations containing 1,844 individuals as donor populations. 2) The donor groups were the clusters inferred using fineSTRUCTURE analysis. FineSTRUCTURE assigns individual samples into natural genetic groups based on haplotype similarity. Clusters with a sample size of less than five were excluded. There are 83 clusters with 1397 samples. 3) According to ADMIXTURE analysis for Eurasia populations at K = 4, we chose 30 individuals with the highest proportion as representatives for each ancestral component. These chosen individuals were used as donor samples. This analysis could help to understand how the four ancestries contributed to NXH.
We also used different strategies to group the target samples: 1) all the Hui individuals were regarded as a whole population. 2) The Hui individuals were grouped according to their geography. We inferred the admixture event for each region to see whether there were significant differences among regions. 3) We also randomly chose 10, 30, and 100 samples from the total Hui samples to investigate whether the sample size affects the result. We did repeats for each chosen number. For all the analyses, the target population was not used as donor groups. To obtain a confidence interval, 100 bootstrap re-sample procedures were used for the analysis.
Local Ancestry Inference
HAPMIX (Price et al. 2009) was employed to infer ancestral tracks with CEU and CHB as a proxy of western and eastern ancestries, respectively. We also used HAN as a proxy of eastern ancestry. To improve the accuracy of local ancestry inference, we only used those 2,035,121 SNPs that were highly differentiated between CEU and HAN (FST> 0.1). Before we did the analysis, we chose the best parameter including admixture proportion and admixture time for the Hui population. The admixture proportion parameter “theta” was set according to the results of the ADMIXTURE and f4 ratio. We optimized the admixture time parameter (T) at a granularity of 5 from 10 to 100. The result showed that T = 40 provided the best fit of the data. We chose the parameter T that produced the largest log-likelihoods, indicating the best fit of the data. Next, we used the estimated parameter to infer ancestral tracks for all Hui samples.
We re-run the HAMPIX analysis using different numbers of SNPs to evaluate the effect of marker density on the inference of the best parameter. To reduce bias, a data set with a smaller number of SNPs was a subset of the data set with a larger number of SNPs.
Estimation of Admixture Time
We additionally used ALDER (v1.03) (Loh et al. 2013), which measures the decay of admixture LD, to estimate the time since admixture for all the Hui samples. One advantage of ALDER is that it could be used to identify the source of gene flow by comparing weighted LD curves for different reference populations.
We also used MultiWaver 2.0 (Ni et al. 2019), which leveraged the length distribution of ancestral tracks to infer admixture parameters, like admixture model and admixture time. MultiWaver 2.0 could describe the admixture scenario under the model with multiple waves that occurred from multiple source populations. We carried out this analysis with all the default parameters except that all segments with a length of less than 0.01 Morgan were not used for the analysis.
Runs of Homozygosity
SNVs with a missing rate >0.05 or minor allele frequency (MAF) < 0.05 in any population (NXH, HAN) were removed. We used the filtered data set to estimate runs of homozygosity (ROH). ROH was identified with PLINK v.1.90 (Purcell et al. 2007) using a window of 2,000 kb and sliding it across the genome allowing for one heterozygous per window. The minimum length of ROH was set to 500 kb.
Genetic Diversity
We calculated nucleotide diversity π (Nei and Li 1979) for CEU, NXH, and HAN. An equal number of samples were chosen from each population to avoid the bias caused by the sample size. We divided the whole genome into nonoverlap 100 kb windows. For each window, we calculated the average nucleotide diversity for each population,
where n is the number of haplotypes, kij is the total difference between haplotype i and haplotype j. We removed the major histocompatibility complex region (chr6:28477797–33448356), which has unusually high genetic diversity.
Estimation of Effective Population Size (Ne)
We further explored the demographic history of NXH using LD between 700,000 SNPs (McEvoy et al. 2011). R2 was calculated using PLINK v1.90 to measured LD between SNP pairs. The observed pairwise LD was binned into one of 50 recombination distance categories up to 0.25 cm with incremental upper boundaries of 0.005 cm. We did not include the first category since these may have been particularly affected by gene conversion, for which the methods do not account.
We also applied multiple sequentially Markovian coalescent (MSMC2) (Malaspinas et al. 2016) to infer the long-term effective population size. We used only those SNVs with allele state identical to those of genotypes called by bamCaller.py from the VCF result from GATK analysis. Beagle 4.1 was used to phase the data. We calculated the absolute time estimation by assuming a slow mutation rate of 0.5 × per site per year (1.25 × per base per human generation for a generation time of 25 years).
Scan for Selection
We computed FST using biallelic SNVs that were polymorphism in NXH and HAN. The 1% and 0.1% threshold for the whole genome FST between NXH and HAN was 0.026 and 0.04, respectively. We applied the following criteria to identify these candidates SNVs:
FST between NXH and HAN > 0.026 and FST between NXH and CHB > 0.04 or FST between NXH and HAN > 0.04 and FST between NXH and CHB > 0.026.
The consequence of this SNV predicted by VEP should be moderate or high.
Archaic Introgression
We used both SPrime (Browning et al. 2018) and ArchaicSeeker (Lu et al. 2016) to detect archaic introgression in the present-day populations. Archaic genomes used in this analysis included Altai Neanderthal (Prufer et al. 2014) and Denisovan (Meyer et al. 2012) populations.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors thank all of the participants who contributed to this research. This study was supported by the National Key Research and Development Program (2016YFC0906403), the National Natural Science Foundation of China (NSFC) (32030020, 31771388, 31525014, 91731303, 31961130380, 81760525, and 32041008), the Strategic Priority Research Program (XDB38000000) of the Chinese Academy of Sciences (CAS), the UK Royal Society-Newton Advanced Fellowship (NAF\R1\191094), and the Shanghai Municipal Science and Technology Major Project (2017SHZDZX01). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
S.X. conceived and supervised the project. W.Y. collected the samples. Y.L. performed the extraction of the genomic DNA and coordinated the genome sequencing. Y.G., Y.P., and D.L. did variant calling. X.M. performed population genetic analysis. Y.L. and H.C analyzed Y haplotype and mtDNA. X.M. drafted the manuscript. S.X. revised the manuscript.
Data Availability
The data underlying this article are available in the National Omics Data Encyclopedia (NODE) at https://www.biosino.org/node/ and can be accessed with accession number OEP001722. Requests for access to data may be directed to xushua@picb.ac.cn.
References
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR, et al. 2015. A global reference for human genetic variation. Nature 526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR.. 2013. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194(2):459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR.. 2016. Genotype imputation with millions of reference samples. Am J Hum Genet. 98(1):116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Yu ZX.. 2009. Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies. Am J Hum Genet. 85(6):847–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL.. 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 81(5):1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL, Zhou Y, Tucci S, Akey JM.. 2018. Analysis of human sequence data reveals two pulses of archaic Denisovan admixture. Cell 173(1):53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calandra S, Tarugi P, Speedy HE, Dean AF, Bertolini S, Shoulders CC.. 2011. Mechanisms and genetic determinants regulating sterol absorption, circulating LDL levels, and sterol elimination: implications for classification and disease risk. J Lipid Res. 52(11):1885–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Li L, Xu M, Feng Z, Sun X, Lu J, Xu Y, Du P, Wang T, Hu R, et al. 2020. The ChinaMAP analytics of deep whole genome sequences in 10,588 individuals. Cell Res. 30(9):717–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos FC, Joshi PK, Clark DW, Ramsay M, Wilson JF.. 2018. Runs of homozygosity: windows into population history and trait architecture. Nat Rev Genet. 19(4):220–234. [DOI] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell S, Lee JJ.. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LK.1999. Zhongguo minzushi gangyao (The compendium of Chinese nationality histories). Beijing (China: ): China Financial & Economic Press. [Google Scholar]
- Chiang CWK, Mangul S, Robles C, Sankararaman S.. 2018. A comprehensive map of genetic variation in the world’s largest ethnic group-Han Chinese. Mol Biol Evol. 35(11):2736–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Xi R, Zhao J.. 1996. Preliminary study of the physical features of Hui nationality in the town of Linxia. Acta Anthropol Sin. 233–240. [Google Scholar]
- Deng YJ, Zhu BF, Shen CM, Wang HD, Huang JF, Li YZ, Qin HX, Mu HF, Su J, Wu J, et al. 2011. Genetic polymorphism analysis of 15 STR loci in Chinese Hui ethnic group residing in Qinghai province of China. Mol Biol Rep. 38(4):2315–2322. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupanloup I, Bertorelle G.. 2001. Inferring admixture proportions from molecular data: extension to any number of parental populations. Mol Biol Evol. 18(4):672–675. [DOI] [PubMed] [Google Scholar]
- Feng Q, Lu D, Xu S.. 2018. Ancestry painter: a graphic program for displaying ancestry composition of populations and individuals. Genomics Proteomics Bioinformatics. 16(5):382–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Lu Y, Ni X, Yuan K, Yang Y, Yang X, Liu C, Lou H, Ning Z, Wang Y, et al. 2017. Genetic history of Xinjiang’s Uyghurs suggests Bronze Age multiple-way contacts in Eurasia. Mol Biol Evol. 34(10):2572–2582. [DOI] [PubMed] [Google Scholar]
- Fenner JN.2005. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol. 128(2):415–423. [DOI] [PubMed] [Google Scholar]
- Gao XY, Martin ER.. 2009. Using allele sharing distance for detecting human population stratification. Hum Hered. 68(3):182–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladney DC.1997. Ethnic identity in China: the making of a Muslim minority nationality. San Diego (CA: ): Harcourt Brace College. [Google Scholar]
- Hellenthal G, Busby GBJ, Band G, Wilson JF, Capelli C, Falush D, Myers S.. 2014. A genetic atlas of human admixture history. Science 343(6172):747–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449(7164):U851–U853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C, Balanovsky O, Lukianova E, Kahbatkyzy N, Flegontov P, Zaporozhchenko V, Immel A, Wang C-C, Ixan O, Khussainova E, et al. 2019. The genetic history of admixture across inner Eurasia. Nat Ecol Evol. 3(6):966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z.2006. The marriage customs among China’s ethnic minority groups. Beijing, China: China Intercontinental Press. [Google Scholar]
- Kamberov YG, Wang SJ, Tan JZ, Gerbault P, Wark A, Tan LZ, Yang YJ, Li SL, Tang K, Chen H, et al. 2013. Modeling recent human evolution in mice by expression of a selected EDAR variant. Cell 152(4):691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason RL, Mohideen M, Mest JR, Wong AC, Norton HL, Aros MC, Jurynec MJ, Mao XY, Humphreville VR, Humbert JE, et al. 2005. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310(5755):1782–1786. [DOI] [PubMed] [Google Scholar]
- Lawson DJ, Hellenthal G, Myers S, Falush D.. 2012. Inference of population structure using dense haplotype data. PLoS Genet. 8(1):e1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I, Patterson N, Mittnik A, Renaud G, Mallick S, Kirsanow K, Sudmant PH, Schraiber JG, Castellano S, Lipson M, et al. 2014. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513(7518):409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie S, Winney B, Hellenthal G, Davison D, Boumertit A, Day T, Hutnik K, Royrvik EC, Cunliffe B, Lawson DJ, et al. 2015. The fine-scale genetic structure of the British population. Nature 519(7543):309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.2010. A study of the Hui nationality’s history in Xinjiang province. J Beifang Ethnic Univ. 2010(06):41–46. [Google Scholar]
- Loh PR, Lipson M, Patterson N, Moorjani P, Pickrell JK, Reich D, Berger B.. 2013. Inferring admixture histories of human populations using linkage disequilibrium. Genetics 193(4):1233–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu DS, Lou HY, Yuan K, Wang XJ, Wang YC, Zhang C, Lu Y, Yang X, Deng LA, Zhou Y, et al. 2016. Ancestral origins and genetic history of Tibetan highlanders. Am J Hum Genet. 99(3):580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaspinas AS, Westaway MC, Muller C, Sousa VC, Lao O, Alves I, Bergstrom A, Athanasiadis G, Cheng JY, Crawford JE, et al. 2016. A genomic history of Aboriginal Australia. Nature 538(7624):207–214. [DOI] [PubMed] [Google Scholar]
- McEvoy BP, Powell JE, Goddard ME, Visscher PM.. 2011. Human population dispersal “Out of Africa” estimated from linkage disequilibrium and allele frequencies of SNPs. Genome Res. 21(6):821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F.. 2016. The Ensembl variant effect predictor. Genome Biol. 17(1):Article number 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng HT, Han JT, Zhang YD, Liu WJ, Wang TJ, Yan JW, Huang JF, Du WA, Guo JX, Wang HD, et al. 2014. Diversity study of 12 X-chromosomal STR loci in Hui ethnic from China. Electrophoresis 35(14):2001–2007. [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prufer K, de Filippo C, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 338(6104):222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Li WH.. 1979. Mathematical-model for studying genetic-variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 76(10):5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni XM, Yuan K, Liu C, Feng QD, Tian L, Ma ZM, Xu SH.. 2019. MultiWaver 2.0: modeling discrete and continuous gene flow to reconstruct complex population admixtures. Eur J Hum Genet. 27(1):133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning C, Wang CC, Gao SZ, Yang Y, Zhang X, Wu XY, Zhang F, Nie ZZ, Tang YP, Robbeets M, et al. 2019. Ancient genomes reveal Yamnaya-related ancestry and a potential source of Indo-European speakers in Iron Age Tianshan. Curr Biol. 29(15):2526–2532. [DOI] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo YT, Mallick S, Rohland N, Zhan YP, Genschoreck T, Webster T, Reich D.. 2012. Ancient admixture in human history. Genetics 192(3):1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D.. 2006. Population structure and eigenanalysis. PLoS Genet. 2(12):e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova D, Novembre J, Stephens M.. 2016. Visualizing spatial population structure with estimated effective migration surfaces. Nat Genet. 48(1):94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Tandon A, Patterson N, Barnes KC, Rafaels N, Ruczinski I, Beaty TH, Mathias R, Reich D, Myers S.. 2009. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 5(6):e1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505(7481):43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugach I, Matveev R, Spitsyn V, Makarov S, Novgorodov I, Osakovsky V, Stoneking M, Pakendorf B.. 2016. The complex admixture history and recent southern origins of Siberian populations. Mol Biol Evol. 33(7):1777–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, Moltke I, Rasmussen S, Stafford TW, Orlando L, Metspalu E, et al. 2014. Upper palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 505(7481):87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie XH, Byrne EH, McCarroll SA, Gaudet R, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449(7164):913–U912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M.. 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425. [DOI] [PubMed] [Google Scholar]
- Sikora M, Pitulko VV, Sousa VC, Allentoft ME, Vinner L, Rasmussen S, Margaryan A, Damgaard PD, de la Fuente C, Renaud G, et al. 2019. The population history of northeastern Siberia since the Pleistocene. Nature 570(7760):182–188. [DOI] [PubMed] [Google Scholar]
- Takeuchi F, Katsuya T, Kimura R, Nabika T, Isomura M, Ohkubo T, Tabara Y, Yamamoto K, Yokota M, Liu XY, et al. 2017. The fine-scale genetic structure and evolution of the Japanese population. PLoS One. 12(11):e0185487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CC, Lu Y, Kang LL, Ding HQ, Yan S, Guo JX, Zhang Q, Wen SQ, Wang LX, Zhang MF, et al. 2019. The massive assimilation of indigenous East Asian populations in the origin of Muslim Hui people inferred from paternal Y chromosome. Am J Phys Anthropol. 169(2):341–347. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC.. 1984. Estimating F-STATISTICS for the analysis of population structure. Evolution 38(6):1358–1370. [DOI] [PubMed] [Google Scholar]
- Weissensteiner H, Pacher D, Kloss-Brandstatter A, Forer L, Specht G, Bandelt HJ, Kronenberg F, Salas A, Schonherr S.. 2016. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 44(W1):W58–W63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie MK, Song F, Li JN, Lang M, Luo HB, Wang Z, Wu J, Li CZ, Tian CC, Wang WZ, et al. 2019. Genetic substructure and forensic characteristics of Chinese Hui populations using 157 Y-SNPs and 27 Y-STRs. Forensic Sci Int Genet. 41:11–18. [DOI] [PubMed] [Google Scholar]
- Xu SH, Yin XY, Li SL, Jin WF, Lou HY, Yang L, Gong XH, Wang HY, Shen YP, Pan XD, et al. 2009. Genomic dissection of population substructure of Han Chinese and its implication in association studies. Am J Hum Genet. 85(6):762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yuan K, Ni X, Zhou Y, Guo W, Xu S.. 2020. AdmixSim: a forward-time simulator for various complex scenarios of population admixture. Front Genet. 11:601439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.2002. Investigation and study on marital status of the Hui nationality in cities of Ningxia—taking Yinchuan Wuzhong and Lingwu as examples. Res Hui. 2002(01):39–47. [Google Scholar]
- Yao HB, Wang CC, Tao XL, Shang L, Wen SQ, Zhu BF, Kang LL, Jin L, Li H.. 2016. Genetic evidence for an East Asian origin of Chinese Muslim populations Dongxiang and Hui. Sci Rep. 6(1):38656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao YG, Kong QP, Wang CY, Zhu CL, Zhang YP.. 2004. Different matrilineal contributions to genetic structure of ethnic groups in the Silk Road region in China. Mol Biol Evol. 21(12):2265–2280. [DOI] [PubMed] [Google Scholar]
- Yu GC, Smith DK, Zhu HC, Guan Y, Lam TTY.. 2017. GGTREE: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 8(1):28–36. [Google Scholar]
- Zheng L, Zhu Q, Wang Q, Gao Q, Li C, Li W, Chen Z, Yang Z, Li S.. 1997. The physical characters of Hui nationality in Ningxia. Acta Anthropol Sin. 16:12–22. [Google Scholar]
- Zhou BY, Wen SQ, Sun HL, Zhang H, Shi RM.. 2020. Genetic affinity between Ningxia Hui and eastern Asian populations revealed by a set of InDel loci. R Soc Open Sci. 7(1):190358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the National Omics Data Encyclopedia (NODE) at https://www.biosino.org/node/ and can be accessed with accession number OEP001722. Requests for access to data may be directed to xushua@picb.ac.cn.