Abstract
Centromeres are functionally conserved chromosomal loci essential for proper chromosome segregation during cell division, yet they show high sequence diversity across species. Despite their variation, a near universal feature of centromeres is the presence of repetitive sequences, such as DNA satellites and transposable elements (TEs). Because of their rapidly evolving karyotypes, gibbons represent a compelling model to investigate divergence of functional centromere sequences across short evolutionary timescales. In this study, we use ChIP-seq, RNA-seq, and fluorescence in situ hybridization to comprehensively investigate the centromeric repeat content of the four extant gibbon genera (Hoolock, Hylobates, Nomascus, and Siamang). In all gibbon genera, we find that CENP-A nucleosomes and the DNA-proteins that interface with the inner kinetochore preferentially bind retroelements of broad classes rather than satellite DNA. A previously identified gibbon-specific composite retrotransposon, LAVA, known to be expanded within the centromere regions of one gibbon genus (Hoolock), displays centromere- and species-specific sequence differences, potentially as a result of its co-option to a centromeric function. When dissecting centromere satellite composition, we discovered the presence of the retroelement-derived macrosatellite SST1 in multiple centromeres of Hoolock, whereas alpha-satellites represent the predominate satellite in the other genera, further suggesting an independent evolutionary trajectory for Hoolock centromeres. Finally, using de novo assembly of centromere sequences, we determined that transcripts originating from gibbon centromeres recapitulate the species-specific TE composition. Combined, our data reveal dynamic shifts in the repeat content that define gibbon centromeres and coincide with the extensive karyotypic diversity within this lineage.
Keywords: chromosome evolution, centromeres, gibbon, primate genomics, transposable elements, satellite DNA
Introduction
Centromeres are essential genomic loci that support the assembly of the kinetochore, thereby facilitating spindle attachment and faithful segregation of chromosomes to daughter cells during mitosis and meiosis. Comparative studies across deep branches of eukaryotic lineages have shown a stark contrast between the functional conservation of the components at the interface of the inner kinetochore and the rapid evolution of centromeric DNA sequences (Henikoff et al. 2001). The centromeric DNA of humans and other great apes largely consists of ∼171 bp long AT-rich alpha-satellite monomers that are tandemly repeated in blocks and further arranged into larger repetitive units, known as higher order repeat arrays (Willard 1985; Waye and Willard 1987; Alexandrov et al. 1993; Rudd et al. 2003). Although the presence of alpha-satellites is a predominantly conserved characteristic of primate centromeres, the identification of large-scale, variable centromere haplotypes in humans suggests the genomic landscape of such satellites is mutable (Langley et al. 2019). In addition to centromeric satellites, recent work in phylogenetically divergent species, such as marsupials (Longo et al. 2009; Ferreri et al. 2011; Renfree et al. 2011; Johnson et al. 2018), Drosophila (Chang et al. 2019), plants (Zhong et al. 2002), and fungi (Yadav et al. 2018) show that centromeric retroelements may be a defining element of functional centromeric chromatin. Retroelements, mobile elements that propagate via an RNA intermediate, have been proposed as major players in centromere evolution because they can provide the material for generation of new satellite families. Such phenomena have been observed in plants (Kapitonov and Jurka 1999; Cheng and Murata 2003), Drosophila (Heikkinen et al. 1995), and cetaceans (Kapitonov et al. 1998). Furthermore, centromeric retroelements have been implicated in facilitating chromosome evolution through the introduction of large-scale genomic rearrangements as specific classes of centromeric retroelement have been found to be enriched at evolutionary breakpoints (Longo et al. 2009).
Gibbons, the endangered small apes in the Hominoidea superfamily, a taxonomic group co-occupied by both small and great apes (Cunningham and Mootnick 2009), present a unique opportunity to study the evolution of functional centromere sequences in the context of rapid karyotypic change. Gibbons diverged from the common Hominoidea ancestor only ∼17 million years ago (Mya), and a more recent (∼5 Mya) radiation event gave rise to the four extant genera. Despite their recent divergence and species radiation, gibbons have accumulated an unusually high number of chromosomal rearrangements compared with other lineages, suggesting a rapid rate of karyotype evolution (Carbone, Vessere et al. 2006; Roberto et al. 2007; Carbone et al. 2009, 2012, 2014; Girirajan et al. 2009). Moreover, drastically different karyotypes are found in each of the four extant gibbon genera (Hoolock [2n = 38], Nomascus [2n = 52], Hylobates [2n = 44], and Symphalangus [2n = 50]), derived by translocations, fusions, fissions, and inversions, often concomitant with centromere inactivation and formation of evolutionary new centromeres (Capozzi et al. 2012). These rapid karyotypic changes, which have been extensively mapped in gibbons, offer a unique opportunity to study centromeres that are variable within the same species and across different genera.
In our previous investigations of the evolutionary history of gibbon chromosomes, we discovered a nonautonomous, gibbon-specific composite retrotransposon, named LAVA (after its three component elements: LINE-Alu-VNTR-Alulike), that depends on the L1 machinery to retrotranspose in the host genome. Based on structural characterizations of LAVA subfamilies (Lupan et al. 2015), the average length of LAVA elements ranges from 1,661 to 2,557 bp. The Variable Number Tandem Repeat (VNTR) region of LAVA is the most variable portion of the LAVA, with an average length that can range from 546 to 1,377bp across LAVA sub families and is generally inversely correlated to the age of the element (Lupan et al. 2015). Although the LAVA element can be found in the genomes of all gibbon species examined to date, it has expanded in almost all centromeres of only two gibbon species belonging to the Hoolock genus (Carbone et al. 2012; Hara et al. 2012). Thus, expansion of the LAVA retroelement in gibbon centromeres likely occurred recently and after divergence of the four gibbon genera. In fact, the LAVA centromeric expansion in Hoolock has occurred recently enough not to have yet equally impacted all chromosomes, as four Hoolock chromosomes lack LAVA expansions (Carbone et al. 2012).
To determine if recently expanded repetitive sequences are linked to centromeric function, we completed an in-depth, cross-genera characterization of the sequences binding three DNA-interacting centromeric proteins: CENP-A, the histone that demarcates centromeric chromatin (Yoda et al. 2000; Van Hooser et al. 2001); as well as CENP-B and CENP-C, two inner kinetochore proteins that lie at the interface between centromeric chromatin and kinetochore−spindle complex (Okada et al. 2007; Klare et al. 2015). Combining next generation sequencing, de novo sequence assembly, repeat annotation methods and fluorescence in situ hybridization (FISH), the putatively functional centromeric repeat content was characterized and compared across the four gibbon genera. Instead of the homogeneous satellite arrays that typify primate centromeres, all four gibbon genera showed diverse transposable element (TE) content in CENP-A chromatin and the functional part of the centromere that forms the inner kinetochore (CENP-B and CENP-C). Across the Hoolock karyotype, LAVA and SST1 (a retroelement-derived macrosatellite found in primates) define highly divergent functional centromere forms compared with the other genera. Finally, a subset of transcriptionally active retroelements, including LAVA, define functional elements within centromeres and recapitulate the largest variation among centromeres of gibbons.
Results
Centromeres of All Four Gibbon Genera Are Enriched in SINEs and Depleted in LINEs
We investigated the repeat composition of gibbon centromeres using chromatin immunoprecipitation sequencing (ChIP-seq) targeting centromere proteins (CENPs) known to demarcate active centromeric chromatin, including the centromeric histone variant CENP-A, and two additional proteins, CENP-B and CENP-C (Yoda et al. 2000; Van Hooser et al. 2001; Okada et al. 2007; Klare et al. 2015). CENP-A is the centromere-specific H3 variant (Earnshaw and Rothfield 1985) that replaces conventional H3 in a subset of nucleosomes (Blower et al. 2002) and is required for kinetochore assembly. Via its histone chaperone, HJURP, CENP-A is assembled into centromeric chromatin during late-telophase, early G1 in humans every cell cycle (Jansen et al. 2007; Foltz et al. 2009), demarcating the chromosomal site for kinetochore formation and spindle attachment. CENP-B is known to bind CENP-B box containing sequences within a subset of functional centromeric sequences (Amor et al. 2004; Henikoff et al. 2015; Aldrup-MacDonald et al. 2016) and is found at centromeres throughout the cell cycle (Earnshaw et al. 1989). However, it is unknown when in the cell cycle loading of CENP-B actively occurs (Gamba and Fachinetti 2020). CENP-C is proposed to serve as a centromere marker that may target CENP-A as part of the constitutive centromere-associated network (CCAN), however, it is distinguished from CENP-A in the timing and mode of deposition. Although CENP-A deposition is limited to late-telophase/early-G1 (Jansen et al., 2007), CENP-C targets centromeres throughout the cell cycle (Earnshaw et al. 1989; Du et al. 2010). Thus, each of these three CENPs can independently serve as a marker for centromere function, yet their predicted binding profiles are unknown.
ChIP-seq was carried out on gibbon lymphoblastoid cell lines (LCLs) established previously (Carbone et al. 2014; Lazar et al. 2018; Okhovat et al. 2020) and for this study, from four different gibbons species: Nomascus leucogenys (NLE), Hoolock leuconedys (HLE), Hylobates moloch (HMO), and Symphalangus syndactylus (SSY), each representing one of the four extant gibbon genera (supplementary table S1, Supplementary Material online). Alignments of CENP-A, CENP-B, and CENP-C protein sequences between human and gibbons indicates high likelihood of cross-reactivity of human-specific antibodies with gibbon (95% identity across CENP-A with only 1 amino acid difference in the peptide used to generate the antibody, 98% identity across CENP-B, and 100% across CENP-C proteins). To independently validate successful immunoprecipitation of DNA bound to each CENP in each species, FISH was used to visualize each ChIP-seq library on mitotic chromosome spreads from corresponding LCLs. Each ChIP-seq library probe localized to centromeres in the four species examined, indicative of successful CENP immunoprecipitation for all three antibodies in each species (fig. 1A; supplementary fig. 1, Supplementary Material online). In addition to centromeric enrichment, pericentromeric and telomeric probe localization of variable intensity was observed in some species and chromosomes, similar to the hybridization patterns previously observed using a centromeric alpha-satellite probe (Cellamare et al. 2009). Cross-hybridization of ChIP-seq DNA to noncentromeric loci, which was most easily visible in Symphalangus (SSY) (fig. 1A; supplementary fig. 1, Supplementary Material online), likely reflects occasional sequence similarities between centromeric and noncentromeric repeats that cannot be distinguished by FISH.
Fig. 1.
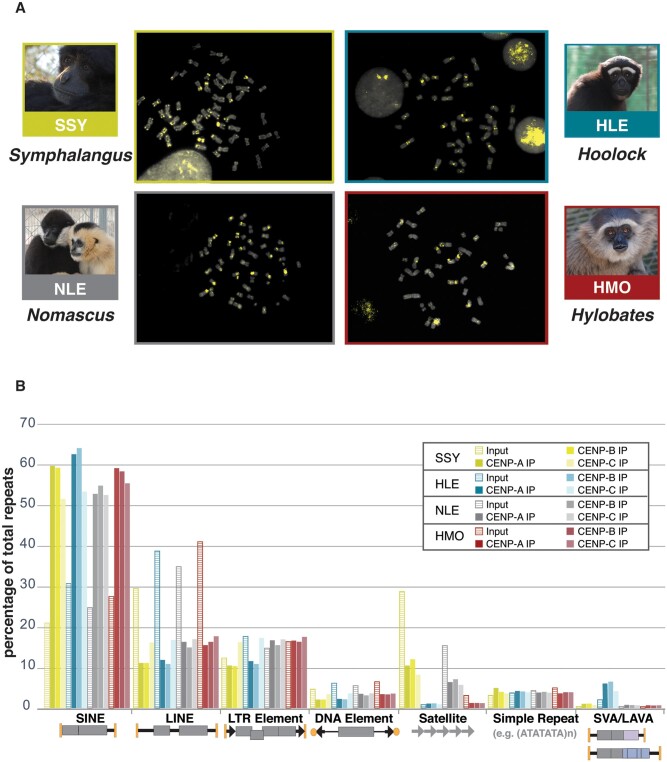
Validation of CENP-A ChIP, and characterization of repeat composition of CENP-A, CENP-B, and CENP-C bound DNA. (A) DNA FISH using CENP-A ChIP library as a probe shows signals (yellow) prevalently localized on centromeric regions on DAPI stained metaphase chromosome preparations for each gibbon genus. Pictures of gibbon species used in this study are shown. FISH validation of CENP-B and CENP-C ChIP libraries can be found in supplementary figure S1, Supplementary Material online. (B) The percentage of all repeats composed of each repeat family (as classified by RepeatMasker) is shown in CENP-A, CENP-B, and CENP-C ChIP-seq libraries for each genus. Schematic structures of repeat classes are shown below the x-axis.
To characterize the repeat content of CENP-A, CENP-B, and CENP-C bound DNA, the repeat composition of 2.5 million randomly subsampled read pairs from each ChIP-seq library were annotated using RepeatMasker (Tempel 2012). The repeat content of corresponding control libraries constructed from chromatin that did not undergo immunoprecipitation (i.e., input libraries) was similarly annotated and used to estimate genome-wide repeat composition per species. The overall abundance and repeat composition of CENP-A, CENP-B, and CENP-C libraries were broadly similar across gibbon genera, but largely different from their corresponding input libraries (fig. 1B; supplementary table S2, Supplementary Material online). Consistent with the known genetic structure of complex, regional centromeres [reviewed in Hartley and O’Neill (2019)], gibbon ChIP-seq libraries had significantly higher repeat content compared with corresponding inputs. On average, 43% of trimmed read sequences were annotated as repeats across all ChIP libraries versus 36% in input reads, indicating gibbon centromeres are enriched for, but not solely characterized by, repeats (paired t-test P < 0.0001; fig. 1B; supplementary table S2, Supplementary Material online).
Most repeat classes, including SINEs, LINEs, LTRs, and DNA elements, displayed relatively similar abundance in each ChIP-seq data set across the four gibbon genera. Representing 52-64% of annotated repeats, SINEs were the most prevalent repeat class in CENP-A, CENP-B, and CENP-C bound DNA pools. The percentage of SINE elements was approximately double those found in the corresponding input samples, indicating enrichment of SINEs in the functional compartments of gibbon centromeres. LINEs, the most abundant repeat class genome-wide, were depleted in CENP-bound DNA and constituted only 11-17% of the annotated repeats in CENP ChIP-seq libraries across all four genera (fig. 1B; supplementary table S2, Supplementary Material online).
The Gibbon-Specific LAVA Retrotransposon Is Enriched in Hoolock Centromeres
The prevalence of the repeat class defined as “retrotransposon” by RepeatMasker (representing non-LINE/LTR/SINE retrotransposon elements) varied among gibbon genera, with HLE distinctly showing higher prevalence (fig. 1B; supplementary table S2, Supplementary Material online). Indeed, the HLE input had ∼10 times the relative abundance of retrotransposon repeats compared with the inputs of SSY, HMO, and NLE (1.9% of HLE input repeats, vs. 0.19 ± 0.04% [mean ± standard deviation (SD)] of input repeats in SSY, HMO, NLE). HLE CENP-bound DNA showed an almost 3-fold enrichment in the proportion of repeats annotated as retrotransposons (5.41 ± 1.33%) relative to input and ∼11-fold enrichment relative to CENP-bound DNA from SSY, HMO, and NLE (0.51 ± 0.17% of repeats; mean ± SD).
Earlier cross-species FISH experiments showed centromeric expansion of the gibbon-specific retrotransposon LAVA exclusively in HLE (Carbone et al. 2012), hence we wanted to determine if our CENP-bound DNA annotated as retrotransposons might include this element and if this element is enriched in gibbon centromeres. Using in silico simulations, we first verified that RepeatMasker could identify LAVA repeats from short-read sequences reliably and with low false negative and false positive rates (supplementary table S3, Supplementary Material online). These analyses revealed that ∼50% of LAVA repeats may be misannotated as “SVA_A,” likely due to large regions of homology between LAVA and SVA, particularly at the VNTR region (fig. 2A; supplementary table S3, Supplementary Material online). Given that gibbon genomes contain a negligible number of SVA element insertions (Wang et al. 2005; Ianc et al. 2014), gibbon repeats classified as “SVA_A” by RepeatMasker are likely LAVA elements. Thus, reads annotated as either LAVA or SVA_A were collectively classified as LAVA in this study.
Fig. 2.
Retrotransposons drive centromere-protein binding DNA variation across gibbon genera. (A) The SVA and the gibbon-specific composite LAVA element are depicted. Full-length LAVA (mean length 1.6-2.6 kb) is comprised of portions of other repeats (CT-rich, Alu-like, and VNTR [mean length 0.5-1.4 kb] all derived from SVA, as well as fractions of AluSz and L1ME5) and flanked by target site duplications. Region of high identity between SVA and LAVA is indicated by a gray box. (B) Sankey diagram demonstrates the amount of retrotransposons annotated as SVA (SVA_A) and LAVA in each ChIP-seq library. (C) LAVA frequency in CENP-A ChIP-seq data sets is plotted against LAVA enrichment relative to input. HLE individuals (circled) cluster separately from the other species indicating LAVA is more abundant genome-wide and at the centromeres in this species.
Inter-genera comparison of retrotransposon annotations revealed higher abundance and enrichment of LAVA elements (relative to input) in CENP-A and CENP-B, but not CENP-C, ChIP-seq libraries from HLE compared with the three other genera (figs. 1B, 2B , and 2C; supplementary fig. 2A and table S2, Supplementary Material online). To confirm these findings, CENP-A ChIP-seq and analyses were repeated as above on three additional HLE gibbons and revealed the same levels of LAVA enrichment, indicating that LAVA enrichment in CENP-A and CENP-B bound centromeric DNA is HLE-specific (fig. 2C). These observations, together with our previous findings using whole-genome shotgun sequences (Okhovat et al. 2020), indicate that the LAVA element is overall more prevalent in the HLE genome compared with other genera, and that LAVA is distinctly enriched in HLE centromeric regions demarcated by CENP-A and CENP-B DNA binding.
LAVA Elements in the Hoolock Genome Are Structurally Distinct from Other Genera
Motivated by the higher abundance of LAVA elements in HLE, we sought to determine if HLE LAVA elements are also structurally different from those found in NLE, HMO, and SSY. In lieu of centromeric LAVA sequences, we took advantage of LAVA repeat annotations in our short read sequences originating from centromeres (i.e., CENP ChIP-seq) versus reads from the rest of the genome (i.e., input library) across all four gibbon species. We used Repstat (Johnson et al. 2018), which is an analysis tool that can compare short read data from a specific repeat to their consensus sequence and test for statistically significant deviations from this consensus, among different data sets. Compared with LAVA reads identified in the libraries of the three non-HLE genera, portions of reads annotated as LAVA in the CENP and input libraries from HLE were significantly longer and had more substitutions (i.e., higher percent divergence) relative to the consensus LAVA sequence (adjusted P < 0.01) (fig. 3A; supplementary table S4, Supplementary Material online). Pairwise comparisons among HMO, NLE, and SSY libraries revealed no difference in the inferred length distribution and sequence of portions of reads annotated as LAVA repeats (adjusted P > 0.05), with only a few exceptions (fig. 3A; supplementary table S4, Supplementary Material online). Overall, these findings indicate that the sequence and structure of LAVA elements in the HLE genome and centromeres are distinct from those found in other gibbon species.
Fig. 3.
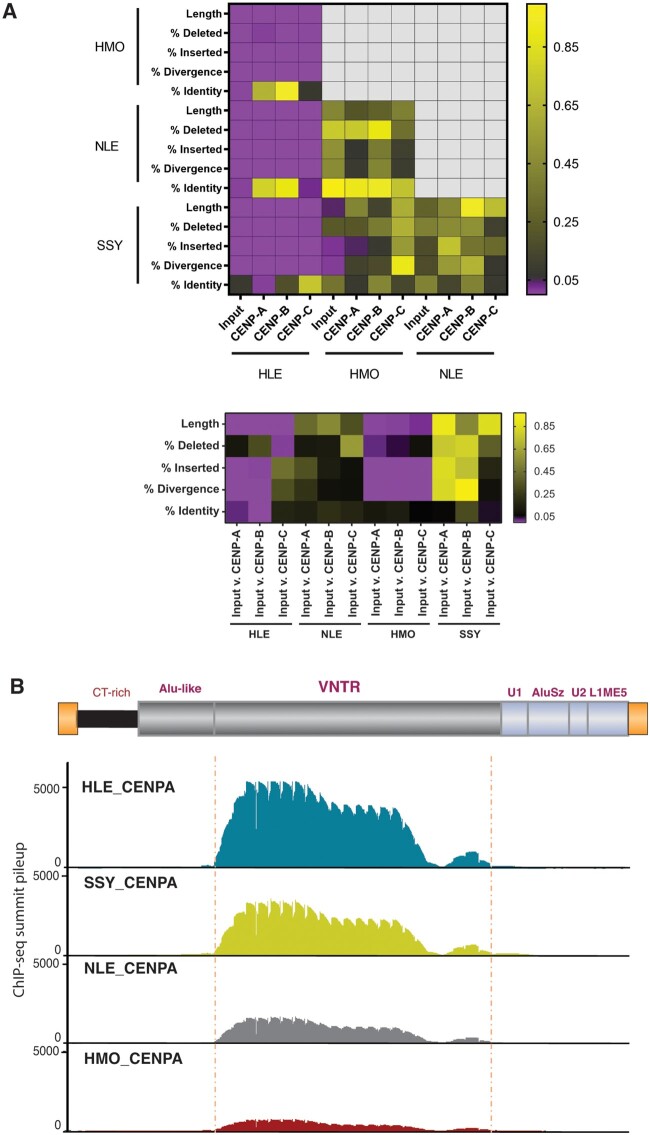
CENP-bound LAVAs in HLE differ from the rest of the genome and from other genera. (A) Heatmap of structural variation in LAVA among different ChIP-seq libraries performed in pairwise comparisons among species (top) and between input and ChIP-IP libraries (bottom). As indicated, color corresponds to P value reported by Repstat. Comparisons in purple are significant under a P value of 0.05. (B) CENP-A ChIP-seq summit pileups against the consensus LAVA sequence are shown for each species. The LAVA element scheme on top represents annotations of the consensuses sequence. The position of the significant meta-summits across libraries is marked with dashed vertical lines. CENP-B and CENP-C ChIP-seq summit pileups are in supplementary figure S2, Supplementary Material online.
Repstat analyses of LAVA-annotated sequencing reads across HLE libraries revealed that portions of reads annotated as LAVA repeats in all three CENP protein bound DNA libraries were longer compared with those identified in the HLE input library (adjusted P < 0.01) (fig. 3A; supplementary table S4, Supplementary Material online). Moreover, compared with input, reads annotated as LAVA in the CENP-A and CENP-B, but not CENP-C, ChIP-seq data sets were significantly less diverged relative to the consensus LAVA sequence (adjusted P < 0.05). Among the three HLE CENP ChIP-seq libraries, LAVA annotated reads identified in the CENP-C library were shorter (adjusted P < 0.01), and had significantly less sequence identity/more insertions relative to the LAVA consensus sequence compared with LAVA repeats annotated in either HLE CENP-B or CENP-A libraries (adjusted P < 0.01) (fig. 3A; supplementary table S4, Supplementary Material online). There were no significant differences in the length or sequence of reads annotated as LAVA between HLE CENP-A and CENP-B libraries (adjusted P > 0.05) (fig. 3A; supplementary table S4, Supplementary Material online). Together, these results suggest that the structure and sequence composition of centromeric LAVA elements in HLE, particularly those putatively bound by CENP-A and CENP-B, differ significantly from LAVA sequences found elsewhere in the HLE genome. These patterns are likely due to differences in insertion preference of specific LAVA elements into centromeres, differences in the evolutionary trajectories of LAVA following centromeric insertion and CENP interaction, or both.
The structure and sequence of centromeric and noncentromeric LAVA repeats in NLE, SSY, and HMO were compared to test if structurally distinct centromeric LAVA are present only in HLE, and as demonstrated in earlier FISH assays (Carbone et al. 2012) and in our ChIP-seq analyses herein, are thus associated with the centromeric LAVA expansion exclusively found in HLE. Compared with input libraries, no difference in the length or sequence divergence from consensus was detected in LAVA reads found in any CENP library from NLE and SSY gibbons (adjusted P > 0.05). In contrast, within the HMO species, LAVA-annotated sequences in all three CENP-bound DNA pools were significantly shorter and more diverged from the LAVA consensus compared with sequences annotated as LAVA in the input library (adjusted P < 0.05) (fig. 3A; supplementary table S4, Supplementary Material online). Altogether, whereas we observed distinct structures in centromeric LAVA across genera, the strongest differences were observed in HLE.
Gibbon Centromere Proteins Putatively Bind LAVA Elements at CENP-B Box Enriched VNTR Sequences
Since LAVA is a composite retrotransposon (fig. 2A), we examined whether centromere proteins preferentially bind specific repeat components within the LAVA sequence. Meta-summit analyses, previously optimized for LAVA (Fernandes et al. 2020; Okhovat et al. 2020), were completed for CENP ChIP-seq data sets across all four gibbon genera, since even non-HLE libraries showed >2-fold enrichment of LAVA elements in CENP libraries relative to input (fig. 3B; supplementary fig. 2B, Supplementary Material online). Briefly, CENP-A, CENP-B, and CENP-C ChIP-seq reads were aligned to the gibbon reference genome (Nleu3.0) (Carbone et al. 2014) and both unique and multimapping reads were used to identify significant narrow peak summits that overlapped LAVA elements annotated in the reference genome. For each sample, the overlapping summits were mapped to the consensus LAVA sequence to generate a pileup of summit positions along the consensus element. Summits across this summit pileup, i.e., “meta-summits,” represent putative CENP binding sites within the composite LAVA element. In total, the number of significant CENP summits per gibbon CENP library ranged from 6,380 to 158,933, and 1-3% of all summits overlapped LAVA elements across samples (supplementary table S5, Supplementary Material online). The high cross-species and cross-library variation in the total number of CENP peaks overlapping LAVA was reflected in the summit pileup height differences. Across all genera, the meta-summits (i.e., putative binding sites) of CENP-A, CENP-B, and CENP-C were located within the VNTR region of LAVA (fig. 3B; supplementary fig. 2A, Supplementary Material online), indicating this repeat component may be putatively bound by CENPs.
We then mined ChIP-seq data for the presence of detectable CENP-B box motifs in gibbon centromeres. Human CENP-A ChIP-seq data (Hasson et al. 2013) were used as a positive control to validate reliability of our pipeline. Briefly, broad human CENP-A ChIP-seq peaks were identified within the human genome and sequences corresponding to these broad peaks showed overall enrichment of the CENP-B box motif (JASPAR database, MA0637.1) (Masumoto et al. 1989; Jolma et al. 2013) relative to shuffled sequences. Next, CENP peaks were sorted based on their fold-enrichment relative to input (i.e., peak height) and we compared enrichment of CENP-B box motifs between high- and low-height peaks. Since ChIP-seq height may be a proxy for binding strength/frequency, the expectation was that higher peaks would be associated with higher prevalence of the CENP-B box motifs. As expected, a significant association was found between CENP-B box motifs and CENP-A peak height in human CENP-A bound DNA sequences (adjusted P < 0.05; supplementary table S6, Supplementary Material online).
Using a similar approach to investigate the presence of the CENP-B box motif in gibbon CENP-bound DNA, we first identified 139,315-383,872 broad peaks across all gibbon CENP ChIP-seq libraries. Overall, sequences of these broad peaks were not significantly enriched in the CENP-B box motif in any of the CENP libraries when compared with shuffled sequences (adjusted P > 0.05). However, when peaks were sorted by height, we found higher peaks had significantly more CENP-B box motifs compared with weaker peaks in at least one CENP pull-down library in each genus (adjusted P < 0.05; supplementary table S6, Supplementary Material online). In order to test our hypothesis that LAVA was co-opted in gibbons to become a functional centromeric element, we aimed to also investigate CENP-B box enrichment in this element. Due to unavailability of centromere sequences in the reference genome, we used LAVA elements present elsewhere in the genome and found that among putative CENP binding sites overlapping LAVA elements (i.e., CENP ChIP-seq summits overlapping LAVA in meta-summit analysis above), 7-16% contained predicted CENP-B box motifs (supplementary table S6, Supplementary Material online), and the majority of these motifs (∼88% on average) were located in the VNTR region of LAVA (supplementary fig. 2C, Supplementary Material online). In addition, the DNA sequences of CENP peak summits within LAVA repeats were collectively strongly enriched in the CENP-B box motif (Benferroni adjusted P < 0.01; supplementary table S6, Supplementary Material online), linking CENP-DNA binding to a specific DNA recognition motif within the LAVA retroelement. Although further studies and contiguous centromere sequence assemblies are required to fully characterize the presence, enrichment and functionality of the CENP-B box motif in gibbon centromeres, our analyses suggest that some gibbon centromeres may contain this motif, albeit likely at a much lower prevalence compared with human centromeres.
Centromeric Alpha-Satellite and SST1 Repeats Vary across Gibbon Genera, Particularly in Hoolock
We observed high variation in satellite class abundance among ChIP-seq libraries across genera, with HLE libraries (which have the highest LAVA abundance) showing the lowest satellite prevalence (fig. 1B). In general, satellite repeats were less abundant among HLE and HMO CENP ChIP-seq and input repeats (0.6-3% of repeats), compared with SSY and NLE [14.8 ± 9.4% and 8.4 ± 4.6% of repeats, respectively; (mean ± SD); supplementary table S2, Supplementary Material online]. Except for HLE, which had equally low satellite DNA abundance in both input and CENP ChIP-seq libraries, all other gibbon genera showed around 3-fold lower satellite repeat content in their CENP ChIP repeat composition compared with input, suggesting that despite being present in gibbon centromeres, satellites are not highly enriched in these loci compared with the rest of the genome. These data support previous FISH analyses showing a lack of satellite enrichment in HLE centromeres (Carbone et al. 2012), but are in contrast to observations that centromeres of great apes are highly enriched in satellites, particularly alpha-satellite repeats (Alkan et al. 2007).
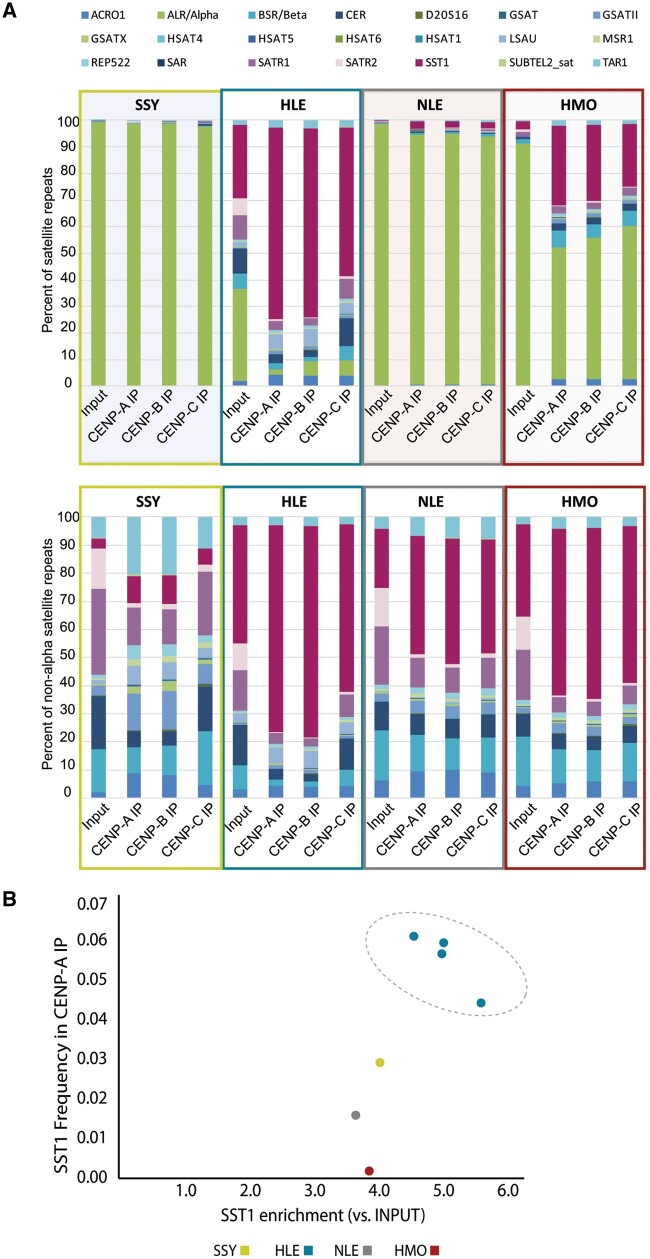
Within the satellite class of repeats, the alpha-satellite family was responsible for most of the variation observed across gibbon genera and libraries (fig. 4A) and was the most prevalent family of satellites in the genomes and centromeres of SSY and NLE gibbons, representing 94-99% of all satellite repeats per library of these two genera. However, this repeat family was less abundant in the centromeres of HMO (constituting <58% of satellites in CENP ChIP-seq data sets), and the least abundant in the HLE both genome-wide and in the centromeres (representing <6% of satellites in CENP libraries; fig. 4A; supplementary table S2, Supplementary Material online).
Fig. 4.
Centromere protein bound satellite distribution varies across gibbon genera. (A). Overview of annotated satellite repeats in each species-specific CENP-A, CENP-B, and CENP-C ChIP library. Each satellite classification (shown in color-coded key at top) is shown as a percentage of total satellites annotated via RepeatMasker with (top) and without (bottom) alpha-satellite content. SSY, NLE, and HMO samples show high alpha-satellite content, whereas SST1 is the most abundant centromeric satellite in HLE. (B) The higher absolute and relative abundance of SST1 repeats in HLE CENP-A ChIP-seq data sets were validated in three biological replicates (circled).
Based on sequence library annotations, the lack of alpha-satellites in HLE libraries is partially offset by the presence of a retroelement-derived macro-satellite family known as SST1 (also called MER22; fig. 4A) (Fatyol et al. 2000). The SST1 satellite family accounted for 56-72% of satellite repeats across HLE CENP libraries, whereas making up <30% of satellites in HMO CENP libraries and <3% of CENP satellites in SSY and NLE (fig. 4A). The distinctly higher absolute and relative abundance of SST1 repeats in HLE CENP ChIP-seq data sets, particularly in CENP-A and CENP-B libraries, was validated in all three biological HLE CENP-A replicates (fig. 4B; supplementary fig. 3A, Supplementary Material online). Although non-HLE gibbon genera have a smaller number of SST1 repeats genome-wide than HLE, each genera showed relative enrichment of SST1 repeats in CENP ChIP-seq data sets compared with their input (fig. 4B; supplementary table S2, Supplementary Material online). Thus, while SST1 elements are likely present in some centromeres across all gibbon genera, they are more prevalent in HLE centromeres.
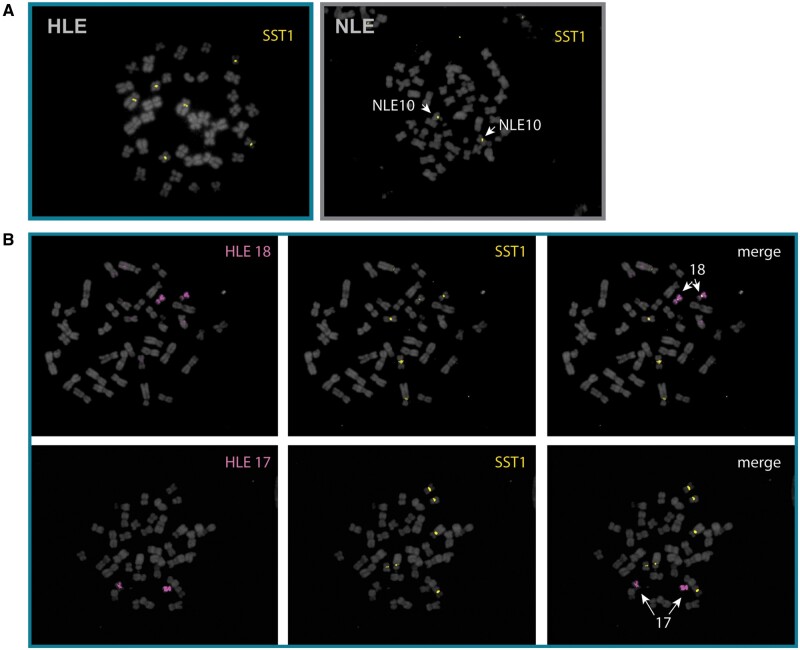
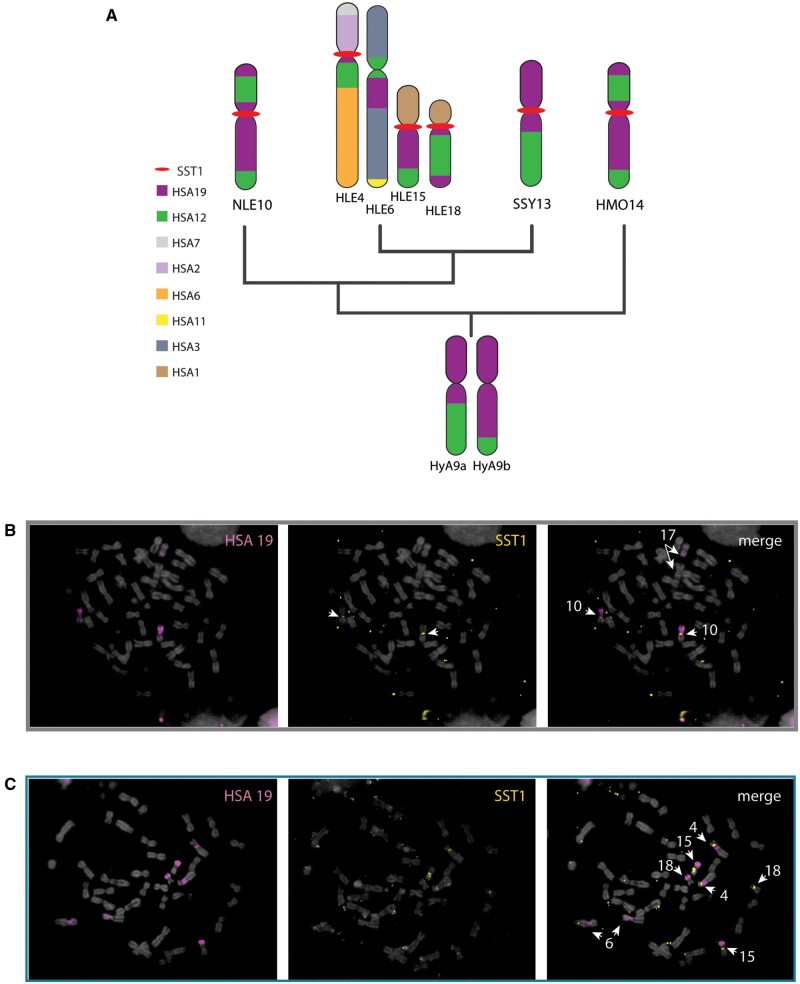
FISH to metaphase preparations using genus-specific SST1 probes (supplementary table S7, Supplementary Material online) confirmed the centromeric localization of SST1 on three chromosome pairs in HLE, but on only one chromosome pair in each of the other three species (fig. 5A; supplementary fig. 3B, Supplementary Material online). The three HLE chromosomes that contained SST1-rich centromeres were identified by inverse-DAPI banding and chromosome painting with HLE-specific painting probes (Nie et al. 2001) as HLE4, HLE15 and HLE18 (fig. 5B). SST1, found on the q arms of human chromosomes 19 and 4 (Tremblay et al. 2010), is consistent with the predicted synteny between gibbons and human based on previous FISH (Roberto et al. 2007; Capozzi et al. 2012) (fig. 6A). We used FISH with an HSA19 chromosome paint to verify SST1 hybridization patterns and found overlap between the gibbon SST1 centromeric signal and HSA19 on gibbon chromosomes NLE10 (fig. 6B), and HLE15 and HLE18 (fig. 6C). The gibbon SST1 centromeric signal and the HSA19 paint also revealed an unknown synteny with HLE4 (fig. 6A and C). The SST1 signal is absent in HLE6, despite HLE6 having a noncentromeric syntenic block within the proximal long arm of HSA19 (fig. 6A and C). Thus, SST1 centromeric signal seems to be linked to centromere-specific rearrangements distinctive to each gibbon lineage. Of note, the three chromosomes containing SST1 repeats in HLE were previously shown to have weak or no centromeric LAVA expansion (Carbone et al. 2012) confirming that an inverse relationship may exist between centromeric LAVA and satellite abundance.
Fig. 5.
Characterization of SST1 localization on gibbon chromosomes by FISH. (A) DNA FISH with SST1 as a probe on metaphase chromosomes from HLE (left) and NLE (right) and counterstained with DAPI. SST1 (yellow) signal is found on three chromosome pairs in HLE and one chromosome pair in NLE. (B) FISH with chromosome paints HLE18 and HLE17 (pink) identified the small chromosome pair with centromeric SST1 signals (yellow) as HLE18 whereas HLE17 appears depleted of SST1.
Fig. 6.
SST1 associates with centromere-specific chromosome rearrangements homologous to human chromosome 19. (A) Predicted synteny of human chromosome 19 (purple) to HLE, NLE, SSY and HMO chromosomes is depicted. Phylogenetic relationship are as per (Shi and Yang 2018). Chromosome homologies to human chromosomes and Hylobates ancestral synteny are shown based on Capozzi et al. (2012). Both inferred ancestral chromosome heteromorphs are shown (HyA9a and HyA9b). SST1 localization is indicated with red bar. Whole-chromosome paints of human chromosome 19 (pink) on NLE (B) and HLE (C) metaphase chromosomes cohybridized with SST1 (yellow); relevant chromosomes identified by inverted DAPI are indicated (white) in merged image (right).
Similar to our analysis of LAVA repeats, we used Repstat (Johnson et al. 2018) to compare sequences and structural characteristics of reads annotated as SST1 repeats within and across species. Overall, very few differences were found in SST1 structure across libraries within each species, indicating that SST1 satellites have similar structures in both centromeric and noncentromeric regions within each genus. Of the few significant structural differences found across libraries, most were found in CENP-C libraries in HLE and NLE (fig. 7; supplementary table S4, Supplementary Material online), whereas no significant differences in SST1 sequences were identified in HMO and SSY (adjusted P > 0.05) (fig. 7; supplementary table S4, Supplementary Material online), albeit low abundance of SST1 in the SSY genome may have affected significance of comparisons. When the structure of SST1 repeats was compared across gibbon genera, SST1-annotated regions in HLE were identified as the most distinct. For example, SST1 read annotations across all HLE ChIP-seq data sets were significantly longer than those found in the respective NLE, HMO, and SSY ChIP-seq data sets (adjusted P < 0.01). Very few differences were identified in SST1 structure and sequence among the non-HLE species (fig. 7; supplementary table S4, Supplementary Material online).
Fig. 7.
Centromere proteins in gibbons bind similar SST1 elements across genera. Heatmap of structural variation analysis of SST1 among different ChIP-seq libraries are shown for pairwise comparisons among species (top) and between input and ChIP-IP (bottom). Color corresponds to P values reported by Repstat as indicated. Comparisons in purple are significant under a P value of 0.05.
Patterns of Expression of Centromeric LAVA and SST1 Differ across Gibbon Genera
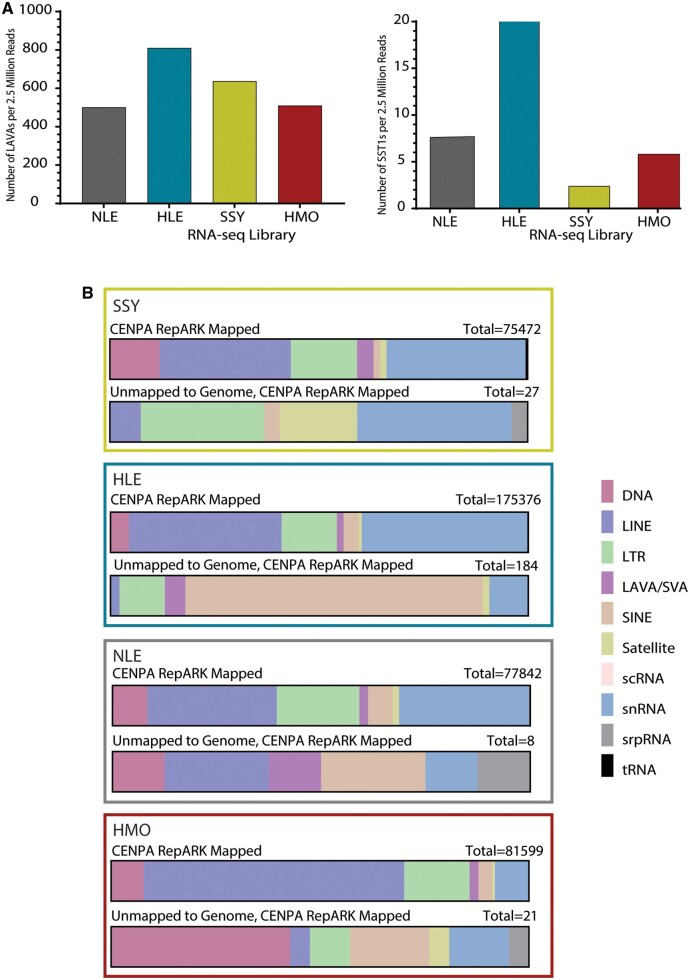
Our analyses indicated that both LAVA and SST1 repeats have expanded, and may be functional in gibbon centromeres, particularly in HLE. To test if putatively functional retrotransposons in centromeres are transcriptionally active, we characterized the repeat composition of the transcriptome across gibbon genera. Total RNA sequencing (RNA-seq) data were generated from LCLs of the same four gibbons that were used for our ChIP-seq assays (supplementary table S1, Supplementary Material online). Consistent with findings in input and CENP ChIP-seq data sets, a significantly higher number of reads annotated as SVA_A/LAVA (one-sided z test, P < 0.00001), with 1.3-1.6 times more LAVAs annotated per million reads in HLE than any other library (fig. 8A; supplementary table S8, Supplementary Material online) was observed. Similarly, more SST1 elements were identified in the HLE RNA-seq library compared with the other genera (one-sided z test, P < 0.00001), with 6.0-18.9 times more SST1 repeats annotated per million reads in HLE than any other library (fig. 8A; supplementary table S8, Supplementary Material online). Thus, LAVA and SST1 transcript abundance varies across gibbon genera in a pattern consistent with centromeric repeat composition observed in the CENP ChIP-seq and FISH results.
Fig. 8.
Genome-wide and centromere-derived expression profiles for repeats vary across genera. (A) Combined number of LAVA (left) and SST1 (right) repeats annotated per one million reads across the RNA-seq library of each species. (B) Repeat annotation of centromere-derived RNA-seq reads for each genus. Each repeat classification is shown as a percentage of total annotated repeats mapped to CENP-A RepARK contigs (top of each pair) and as percentage of total annotated repeats mapped to RepARK contigs that did not map to the NLE3.0 assembly (bottom of each pair). Key for repeat annotations shown to the right.
To further delineate centromere-specific transcripts in the absence of centromere-spanning gibbon genome assemblies, ab initio contig assemblies were generated from each CENP-A, CENP-B, and CENP-C ChIP-seq data set using RepARK, a program designed to generate de novo repeat libraries from whole-genome next generation sequencing reads (Koch et al. 2014). Assemblies generated from CENP libraries had an average of 2,926 (±1,256; SD) contigs and an average contig length of 163 bp (± 471; SD) (supplementary table S9, Supplementary Material online). We then identified RNA-seq reads unable to map to Nleu3.0 (4-5% of reads in each species), reasoning that, since the reference genome lacks centromere sequences, these unmapped reads would be enriched for centromere-specific transcripts. Unmapped reads were then aligned to the CENP ChIP-seq derived contigs, resulting in transcripts that can be ascribed to centromeres in each gibbon species (supplementary fig. 4 and table S9, Supplementary Material online). Across RNA-seq libraries from all four species, 0.27 ± 0.17% of the total RNA-seq reads did not align to the reference genome but did align to at least one of the assembled CENP ChIP-seq contigs, representing putative centromere-specific transcripts (supplementary table S9, Supplementary Material online). The most common repeat families identified among the putative centromere-specific transcripts across species included: Simple repeats, low complexity repeats, Alus, SVAs, LINE elements, and ERV-Ks (fig. 8B; supplementary table S10 and fig. 5, Supplementary Material online). Notably, Alu, SVA, and LINE elements are all TEs that constitute the composite LAVA element. Consistent with our CENP ChIP-seq results (figs. 1B and 4), SST1-annotated transcripts mapped to HLE CENP-A, CENP-B, and CENP-C ChIP-seq contigs, as well as HMO CENP-A and CENP-C ChIP-seq contigs, suggesting that this satellite element is actively transcribed in centromeres. Although our approach does not provide a comprehensive annotation of the transcriptional landscape of gibbon centromeres, these results clearly indicate that centromere repeats are transcribed in species-specific patterns that reflect the underlying sequence and repeat composition.
Discussion
The scarcity of centromere sequences in genome assemblies has long hindered comparative studies on the function and evolution of centromere DNA. Although advanced long-read sequencing technologies are reducing centromeric sequence deficiencies in model organism reference genome assemblies (Jain et al. 2018; Johnson et al. 2018; Chang et al. 2019; Miga et al. 2020), alternative strategies are still needed for cross-species centromere studies. To compare centromere repeats across the four extant genera of gibbons, a group of endangered small apes that has experienced rapid chromosome and centromere evolution (Carbone, Vessere et al. 2006; Roberto et al. 2007; Carbone et al. 2009, 2012, 2014; Girirajan et al. 2009), we integrated centromere-specific ChIP-seq, traditional cytogenetic, and centromere-focused RNA-seq analyses. Overall, these combined analyses highlighted high variability in abundance, sequences, and transcription of satellite and retroelement DNA among the gibbon genera.
Targeting DNA at the inner kinetochore of the centromere using ChIP-seq for CENP-A, -B, and -C, we found that centromeres of the Hoolock genus, represented by the Eastern Hoolock (HLE), are unique among gibbons with respect to both retroelements and satellites. In addition to a higher prevalence of the gibbon-specific LAVA retroelement within HLE centromeres, we found significant differences in the length and sequence of centromeric LAVA read annotations compared with those found in chromosome arms. In particular, within the HLE genome, LAVA read annotations originating from regions putatively bound by CENPs were significantly longer than noncentromeric LAVA reads (fig. 3A). As the VNTR portion of LAVA (fig. 2A) is found to be variable across insertions (Lupan et al. 2015), it is likely that its expansions within HLE centromeres may have resulted in longer centromeric LAVA elements. Of note, our metasummit analysis indicated that CENPs preferentially bind LAVA elements at the VNTR and at sequences enriched in the predicted CENP-B box motif (fig. 3B).
To date, CENP-B box motifs have been found in satellite sequences, linking this motif to centromere function. However, the evolution of the evolution of putative CENP-B box motifs in LAVA does not necessarily reflect the selective order of events by which sequences become involved in centromere identity. For example, a recent study in New World Monkeys indicates that the acquisition of CENP-B box binding capacity in centromeric sequences occurred independently in different lineages after the gain of both the satellite elements themselves and centromere function (Thongchum et al. 2020). Thus, CENP-B is proposed to be a stabilizing factor for previously established centromeres (Gamba and Fachinetti 2020), increasing stability under stress in mitotic cells (Dumont et al. 2020) or segregation efficiencies during assymetrical segregation in meiosis (Chmatal et al. 2017; Iwata-Otsubo et al. 2017). In this context, it is likely that the acquisition of retroelements to support centromere function precedes the emergence of functional CENP-B boxes within the retroelement sequences themselves. The prediction of CENP-B boxes within LAVA elements and the capacity to bind CENP-B, as indicated based on our ChIP-seq data, support the observation that LAVA elements are centromeric and have acquired centromere function (as also evident by CENP-A and CENP-C binding). Although our predictions do not indicate any mechanism by which LAVA elements initially acquired centromere function, the evolution of CENP-B box motifs in some centromeric LAVA sequences indicates selective pressure for stabilization of these elements within centromeres via CENP-B binding activity, analogous to the role alpha-satellites play in centromere stabilization in humans.
It should be noted that in lieu of centromeric LAVA sequences, our analysis of the length and sequence of centromeric LAVA elements relied solely on short-read sequences. Moreover, our meta-summit analyses were carried out by aligning ChIP-seq reads to the current gibbon reference genome (Nleu3.0) and are thus based on the assumption that centromeric LAVA repeats bear high sequence similarity to LAVA repeats present in the reference genome. Deriving centromere-spanning genome assemblies in the future will allow for better characterization of centromeric LAVA sequences and identification centromere-specific variants, and thus may reveal additional CENP-B-binding sites. Moreover, whereas the enrichment of the CENP-B box motif in LAVA sequences and enrichment of LAVA in CENP-B ChIP-seq data is intriguing, these findings are inconsistent with a previous study reporting that gibbon genomes lack CENP-B box motifs (Suntronpong et al. 2016). Unlike centromeres characterized by highly homogenized satellite arrays, gibbon centromeres seem to be characterized by a diverse group of repeat types and this heterogeneity makes the detection of a statistically significant enrichment more challenging. Thus, further studies are required to confirm if the predicted CENP-B box motifs within centromeric LAVA elements are a site of CENP-B-DNA interaction.
In contrast to the alpha-satellite-rich centromere structures described for many other primates (Mahtani and Willard 1990; Warburton et al. 1996; Alkan et al. 2007; Sujiwattanarat et al. 2015; Miga et al. 2020), we observed that satellites, including alpha-satellites, comprise only a small percentage (0.7-12.0%) of CENP-bound repeats in all gibbon genera. Previous cytogenetic data gathered for HLE had shown that presence of centromeric LAVA expansions on a chromosome is mutually exclusive to centromeric alpha-satellites (Carbone et al. 2012), suggesting a potentially conflictual relationship between the two repeats within gibbon centromeres. The exclusive relationship between satellites and retroelements may be linked to rapid chromosome change and centromere turnover as our observations in gibbon are similar to the finding of satellite-free, retroelement-enriched centromeres identified in Equus species (Nergadze et al. 2018), another species group characterized by rapid karyotype and centromere evolution (Carbone, Nergadze et al. 2006).
We show that while the most prevalent satellite repeat family in SSY, NLE and HMO gibbons are alpha-satellites, the vast majority of HLE satellites bound by centromeric proteins are annotated as the retroelement-derived macrosatellite SST1 (Fatyol et al. 2000; Tremblay et al. 2010), underscoring, once again, differences in the prevalence and composition of centromeric satellite repeats between Hoolock and the other gibbon genera. Although the sequence and structure of SST1 repeats appear relatively unchanged across SSY, NLE, and HMO, significant variation exists for SST1 repeats between these three species and HLE. Further, SST1 maps to three chromosomes in HLE (HLE 4, 15, and 18) but only one chromosome in the other gibbon species (NLE 10, HMO 14, and SSY 13); each of these gibbon chromosomes carries syntenic blocks homologous to segments of human chromosome 19 (figs. 5 and 6), which span the centromeres (i.e., both p and q arm). We therefore infer that the SST1 sequence within the portion syntenic to human chromosome 19 was likely centromeric on the ancestral Hylobatidae chromosome (HyA9) (Capozzi et al. 2012). The SST1-containing segment of HSA19 then remained as a stable centromere on a single chromosome pair except in HLE, where it was involved in complex interchromosomal rearrangements, resulting in utilization of the repeat as both putative functional centromeric material and as evolutionary breakpoints on three chromosomes lacking LAVA expansions. In humans, SST1 forms a large, noncentromeric array on the q arm of HSA19 (Tremblay et al. 2010), yet it is centromeric in more distantly related primate lineages (Fatyol et al. 2000). It is therefore possible, given SST1 mediated rearrangements with centromeric locations in related species, that SST1 might represent an ancestral centromeric retroelement.
In addition to analyzing the DNA sequences binding to the inner kinetochore, we inquired about possible transcription of centromeric DNA. Active transcription of centromeric repeats has been detected in several organisms (reviewed in Smurnova and De Wulf 2018), with transcription of centromeric DNAs playing a pivotal role in the maintenance of proper centromere function by promoting CENP-A deposition (Bergmann et al. 2011; Chan et al. 2012). Implicated in the processes required for key centromere protein localization, interruption of centromeric satellite DNA transcription leads to chromosomal mis-segregation (Chueh et al. 2009; Rošić et al. 2014). Using de novo ChIP-seq assemblies as centromere-proxies, we determined that among the centromere-derived transcripts in all gibbon genera there is a spectrum of transcribed repeats comprised predominately of retroelements. Given the lack of species-specific, contiguous genomic sequence, our estimates are likely an under-representation of the centromeric transcription present in these species; however, the proportions of these centromere-transcript annotations are broadly reflecting those we defined by ChIP-seq. Furthermore, using Nleu3.0 as a filter to remove repeats shared among genera and noncentromeric repeats from our analyses, we were able to define a small number of species-specific centromere transcripts. For example, HLE has a high level of HLE-specific SINE-derived transcripts (predominantly Alu) within the RNA-seq data. Given their overall high abundance in ChIP-seq (fig. 1B) and RNA-seq (fig. 8B) data, we expect these SINEs to be found across several HLE centromeres. However, the location of these elements has not yet been assigned to centromeres of specific chromosomes. Ongoing long-read-based genome assembly work in this species will afford discrete, chromosome-specific centromeric maps to further refine chromosome-specific content and transcriptional activity. Although a specific role for centromere transcription remains to be established in gibbons, herein we show that repeats found in functional centromeric domains of gibbons across the karyotype are transcriptionally active, supporting the model that centromeric transcription is integral to centromere protein specification and function.
The historical belief that satellite DNAs are the predominant functional component of eukaryotic centromeres, with large tandem arrays present in animals, plants, and fungi (reviewed in McKinley and Cheeseman 2016), is challenged by the finding that neocentromeres devoid of satellites can successfully recruit CENP-A (Voullaire et al. 1993; du Sart et al. 1997; Barry et al. 1999; Amor et al. 2004). In fact, many species are now known to have some centromeres that lack satellite arrays altogether, including Equus species of horse, zebra, and donkey (Wade et al. 2009; Nergadze et al. 2018), orangutan (Locke et al. 2011), chicken (Shang et al. 2010), and potato (Gong et al. 2012). Moreover, long-read genome assemblies spanning centromeric contigs in Drosophila (Chang et al. 2019) and koala (Johnson et al. 2018) has shown that retroelements, not satellites, are the predominate CENP-A binding sequences. A model has been proposed to explain these shifts in repeat content (Klein and O’Neill 2018), wherein satellite arrays, such as alpha-satellite, can become stabilized and homogenize across a karyotype (as observed for great apes). Destabilization events lead to the seeding or invasion of centromeres by TEs, which can be co-opted to become functional centromere sequences. Recent work has shown that retroelement insertions are common following neocentromere formation in maize (Schneider et al. 2016), and evolutionary new centromeres in Equus asinus (Nergadze et al. 2018) and wallabies (Longo et al. 2009) are found in retroelement-rich regions. Combined, these observations suggest that retroelement insertion might favor CENP-A deposition or, vice versa, CENP-A chromatin might provide a “safe heaven” for retroelements.
In line with the observations made by others, our data suggest that satellites are neither the dominant nor exclusive functional centromeric sequences. Rather, our identification of retroelement-enriched gibbon centromeres in Hoolock and low abundance of satellite DNA in all four gibbon genera further supports the hypothesis that retroelements are powerful determinants of centromeric function and this might be particularly true in the context of rapid chromosome evolution. In the future, access to affordable ultralong read sequencing technologies, as well as availability of high-quality genus- or species-specific genome assemblies, will allow for a more comprehensive view into the extraordinary DNA diversity that shapes functionally stable centromeres in gibbons and other organisms.
Materials and Methods
Establishment of LCLs
We used previously established Epstein Barr Virus (EBV) transformed LCLs from a NLE (Lazar et al. 2018), and established new LCL for HLE, HMO, and SSY gibbons according to previous protocols (Lazar et al. 2018). Briefly, opportunistic whole blood samples were collected in sodium heparin tubes from each gibbon during routine check-ups at the Gibbon Conservation Center (Santa Clarita, CA). We used Ficoll-Paque PLUS (GE Healthcare) to isolate lymphocytes from the blood. Next, we transformed 3-9 × 106 lymphocytes with EBV from the marmoset cell line B95-8 (ATCC CRL-1612), using a standard protocol. To do so, we incubated the cells with EBV for 2 h at 37 °C, then diluted them with RPMI-1640 (Corning cellgro) supplemented with 10% FBS (Hyclone), 1× MEM Nonessential Amino Acids Solution (Corning cellgro), 1 mM Sodium pyruvate (Corning cellgro) 1% Pen-Strep (Corning cellgro), and 2 mM l-glutamine (Hyclone). Finally, we grew cells undisturbed for 10-12 days and started feeding the cells with the same supplemented RPMI-1640 as soon as signs of transformation were observed under the microscope.
Chromatin Immunoprecipitation Sequencing
We used LCLs (described above) from four gibbons each belonging to one of the four extant gibbon genera (Hoolock, Nomascus, Hylobates, and Symphalangus) and performed CENP-A, CENP-B, and CENP-C ChIP-seq using the Magnify ChIP kit (ThermoFisher Scientific) according to the manufacturer’s instructions, with minor modifications. Briefly, per ChIP assay we fixed 3 × 105 cells with 1% formaldehyde at room temperature for 10 min. Fixation was quenched with glycine, and cells were washed three times with cold 1× PBS. Cells were lysed for 10 min on ice using the Magnify Lysis buffer, in presence of Proteinase inhibitor cocktail. Lysed cells were then sonicated using the Bioruptor Pico sonicator (Diagenode) for 12 cycles (30 s on/off) and spun down to remove cell debris. A 1% aliquot was taken from the chromatin as input, and the rest was incubated with the appropriate antibody [2 µg of CENP-A (ab13939), 4 µg of CENP-B (ab25734), or 2 µg of CENP-C (ab50974)] at 4 °C, with overnight rotation. The next day we incubated samples with Dynabead protein A/G rotating at 4 °C for 2 h, followed by bead washes, reverse-crosslink and DNA purification according to Magnify ChIP kit protocol. Concentrations of all ChIP and input samples were measured using the Qubit dsDNA High Sensitivity kit (ThermoFisher Scientific). ChIP replicates were pooled to 2 ng and used to construct sequencing libraries using the NEBNext Ultra II library construction kit (New England BioLabs) without size selection. Libraries were quantified and QC’d using the Bioanalyzer High Sensitivity DNA kit (Agilent) and paired-end sequenced on the Illumina NextSeq and HiSeq platforms.
In order to test reproducibility of LAVA’s distribution in HLE centromeres we repeated CENP-A ChIP on LCL from three additional unrelated HLE gibbons (supplementary table S1, Supplementary Material online).
In Silico Validation of RepeatMasker LAVA Annotations
Since our study focused heavily on the complex composite LAVA retrotransposon, we decided to first validate that the RepeatMasker tool (Tempel 2012) is able to reliably annotate this element from short-read sequences. We used in silico simulated short-read data sets to estimate RepeatMasker’s false-negative and false-positive rates in identifying LAVA repeats. Briefly, to estimate false negative rates of our analysis we used wgsim (Li 2011) to simulate three data sets, each containing 2.5 million 75 bp paired-end reads (indel rate = 0.02, single nucleotide mutation rate = 0.05), from the sequences of 1,204 LAVA elements annotated during the construction of the gibbon reference genome (Nleu3.0) (Carbone et al. 2014). These reads, which are expected to only include LAVA repeats, were annotated using RepeatMasker. The %counts of repeat not annotated as the SVA family (either LAVA or SVA_A) is an estimate of the false-negative rate of our analysis. To estimate the false positive rate of our analysis, we first used custom bash scripts and BEDtools (Quinlan and Hall 2010) to mask and remove all LAVA sequences present in the gibbon reference genome. We then used wgsim (Li 2011) to simulate three short-read data sets, each containing 2.5 million random 75 bp paired-end reads (indel rate = 0.02, single nucleotide mutation rate = 0.05) from the modified genome sequence. We annotated these reads, which are expected to lack LAVA repeats, using RepeatMasker and calculated percent of reads annotated as LAVA repeats. The false-positive rate is estimated by the mean %count of repeats annotated as SVA family (including SVA_A or LAVA annotations) across simulated data sets.
ChIP-Seq Data Processing, Repeat Annotation and Repeat Structure Comparison
The quality of raw ChIP-seq data sets were assessed using FastQC (Andrews 2010) and reads were trimmed using Trimmomatic (Bolger et al. 2014) to remove adaptor sequences, as well as low quality and short reads (2:30:10:26:true SLIDINGWINDOW:4:20 TRAILING:20 LEADING:20 MINLEN:50). Next, we used Seqtk (Shen et al. 2016) to randomly select 2.5 million trimmed read pairs from each ChIP sample. The random subsets of reads were converted into fasta format using manual bash scripts and their repeats were annotated using RepeatMasker version 4.0.3 and the gibbon (Hylobates sp.) 20150807 Repbase library (Jurka et al. 2005). We used a custom script to group repeats in each library based on repeat class and family and to calculate the %count and %length (of total repeats) they constituted. Since overall results from %count and %length calculations were highly similar within data sets, we only report the %count in the main text of this study, but both numbers are reported in supplementary table S2, Supplementary Material online. Considering the high homology between SVA_A and LAVA and the fact that only few copies of SVA elements exist in the gibbon genome, repeats annotated as SVA_A using RepeatMasker are likely LAVA repeats and were annotated as such in this study.
To compare the sequence and structure of repeats across libraries, 50,000 reads were subsampled from HLE ChIP-seq libraries and 100,000 reads were subsampled from HMO, NLE, and SSY ChIP-seq libraries using Seqtk (Shen et al. 2016). Fewer reads were required to be sampled from HLE due to the enrichment of SST1 and LAVA. Subsampled libraries were annotated using RepeatMasker using the previously described annotation pipeline. We then used Repstat (Johnson et al. 2018) with two-sided Wilcoxon signed-rank test, to identify significant differences in repeat structures annotated by RepeatMasker across libraries. Since the subsampled SSY input library only contained one repeat annotated as SST1, this library was excluded from downstream Repstat analysis of SST1 repeats.
Identifying Putative CENP Binding Sites within the LAVA Element and Examining CENP-B Motif Enrichment
We used an approach originally described by Fernandes et al. (2020) and adapted for the LAVA element in Okhovat et al. (2020) to identify putative CENP bindings sites in the consensus LAVA sequence. Briefly, we used Bowtie2 (Langmead and Salzberg 2012) with the –very-sensitive settings to align the trimmed reads from each input and CENP ChIP-seq library to the gibbon reference genome (Nleu3.0). We used these alignments, which consisted of both unique and multimapping reads, to identify significant ChIP-seq peaks using MACS2 (Zhang et al. 2008) (-f BAMPE –keep-dup = 2 –nomodel -q 0.01 -g 2.8e9). For each CENP library, we used BEDtools (Quinlan and Hall 2010) to identify all CENP peak summits overlapping LAVA elements annotated in the reference genome (Carbone et al. 2014). We extended each of these summits 50 bp in each direction and mapping them to the consensus LAVA sequence (from the Repase library) using BLAT (Kent 2002) to generated a pileup of summits. The summit pileups generated were then used by MACS2 (Zhang et al. 2008) (–keep- dup all –nomodel –call-summits –extsize 50) to identify summits of the summit pileups (“meta-summits”), which represent putative binding sites of CENP inside the LAVA element.
To validate our approach in detecting CENP-B box motifs, we first analyzed public human CENP-A ChIP-seq data as positive control. Human CENP-A ChIP-seq reads were aligned to Hg38 using Bowtie2 with the “–very-sensitive” paired-end settings (Langmead and Salzberg 2012). We identified significant (q < 0.01) broad peaks using Model-based Analysis of ChIP-seq 2 (MACS2) (Zhang et al. 2008), whereas allowing a maximum of two duplicate reads per data set. We then used AME from the MEME suite (Machanick and Bailey 2011) to examine enrichment of CENP-B box motifs (JASPAR database, MA0637.1) (Jolma et al. 2013) in these broad peaks using two slightly different approaches. In the first approach, we examined overall enrichment of the CENP-B box motif in sequences corresponding to all broad peak sequences, relative to shuffled versions of the same sequences. In the second approach, we sorted CENP peaks based on their fold-enrichment and compared enrichment of CENP-B box motifs between high- and low-enrichment peaks. A similar approach was then used to investigate presence of CENP-B box motifs in gibbon centromeres by aligning all libraries to the Nleu3.0 genome. P values calculated for enrichment analyses were adjusted for multiple testing using the Bonferroni method.
To characterize prevalence of the CENP-B-box motif at putative CENP binding sites within the LAVA element, we extracted 100 bp sequences centered at CENP ChIP-seq summits overlapping LAVA (identified in the meta-summit analysis above) and used the Find Individual Motif Occurrences (FIMO) tool of the MEME suite (Machanick and Bailey 2011) to annotate all significant (P < 0.05) CENPB-box motifs. We used the predicted motifs to calculate the percentage of summits containing the CENPB-box motif in each CENP library. Next, we selected 20 of the LAVA elements with highest number of predicted CENPB-box motifs and determined the percentage of total CENP-B-box motifs that where located in the VNTR. Lastly, to examine statistical significance of enrichment of the CENP-B box motif in the putative binding sites of CENP within the LAVA element, we used the Motif Enrichment Analysis pipeline (AME) of the MEME suite (Machanick and Bailey 2011), and compared prevalence of the CENP-B box motif (MA0637.1) in the 100 bp summit sequences (described above) relative to shuffled versions of the same sequences (scrambled while preserving the 2-mer frequencies).
SST1 Probe Design and FISH
We used our CENP-A ChIP-seq data to generate genus-specific repeat consensus sequences for SST1. First, we extracted CENP-A reads annotated as SST1 repeats by RepeatMasker, in each genus. R1 and R2 reads were combined within each genus and aligned to the reference SST1 consensus sequence (obtained from the RepeatMasker repeat library), using default single-end bwa (Li and Durbin 2009) settings. Next, we used a combination of SAMtools mpileup and bcftools (Li et al. 2009) to characterize sequence variations in each genus. Lastly, we used bcftools (Li et al. 2009) to modify the reference SST1 sequence based on genus-specific sequence variations and construct a genus-specific centromere SST1 consensus sequence. PCR primers used for probe development were designed based on genus-specific HLE SST1 consensus sequences. The consensus SST1 and primer sequences can be found in supplementary table S7.
Each 50 µl PCR reaction contained 0.2 µM forward and reverse primers, 0.125 mM deoxyribonucleotide triphosphate (dNTPs), 2× PCR buffer, Taq polymerase, and 100 ng genomic DNA isolated from each gibbon LCL. PCR settings consisted of an initial 3 min denaturation at 94 °C, followed by 35 cycles of 30 s at 94 °C, 30 at 58 °C and 1 min at 72 °C with a final 5 min extension at 72 °C. PCR products were purified using the QIAquick PCR purification kit (Qiagen) and PCR labeled in 25 µL reactions containing 1× GoTaq buffer (Promega), 0.2 mM dGTTPs/dATTPs/dCTTPs, 0.15 mM dTTPs, 0.2 nM forward and reverse primers, 0.625 U of GoTaq (Promega), 0.04 mM digoxigenin-deoxyuridine triphosphatase (dig-dUTPs, Enzo) and 100 ng PCR product. PCR labeling consisted of a 30 s initial denaturation at 98 °C, followed by 25 cycles of 10 s at 98 °C, 30 s at 60 °C, and 30 s at 72 °C with a final extension of 5 min at 72 °C.
FISH was carried out on metaphase spreads prepared per standard protocols (Rooney and Czepulkowski 1992). Briefly, 200-500 ng of SST1 probe was rehydrated in Hybrisol VII (MP Biomedicals) and hybridized on slides overnight in a humid chamber at 37 °C following denaturation at 80 °C for 5 min. Prior to hybridization, slides were treated with RNase A (0.1 mg/mL in 2× SSC) for 15 min at 37 °C, 0.192 M HCl for 10 min, and denatured in 70% formamide/2× SSC at 72 °C for 2 min. Post hybridization washes were performed once in 2× SSC room temperature to remove the coverglass, 0.4× SSC/0.3% NP40 for 2 min at 72 °C and once in 2× SSC/0.1% NP40 at room temperature. The reaction was blocked in 4× SSC/0.2% Tween 20/5% BSA for 30 min at 37 °C and detection was using 1:500 antidigoxigenin fluorescein following the manufacturer’s instructions for 30 min at 37 °C. Excess reagents were removed by rinsing slides three times in 4× SSC/0.2% Tween 20 for 5 min at 45 °C, rinsing in distilled water, and dehydrating in an ethanol row. Slides were counterstained with a 1:5 dilution of DAPI in Vectashield (Vector Laboratories, Inc.). Images were captured on an Olympus AX70 microscope using CytoVision software (Leica Biosystems Richmond, Inc.).
Chromosome painting was carried out on metaphase spreads using Aquarius Whole Chromosome Painting probes (Cytocell Ltd). Probes were hybridized to RNase A treated slides overnight in a humid chamber at 37 °C following slide and probe codenaturation at 72 °C for 5 min on a Hybaid in-situ block. Post hybridization washes were performed once in 2× SSC RT to remove the coverslip, 0.4× SSC at 60 °C for 2 min and once in 2× SSC/0.05% Tween 20 for 1 min at room temperature. Slides were rinsed in distilled water, dehydrated in ethanol and counterstained with a 1:5 dilution of DAPI in Vectashield (Vector Laboratories, Inc.). Images were captured using an Olympus AX70 microscope and CytoVision software (Leica Biosystems Richmond, Inc.).
Generating Ab Initio CENP Contigs From ChIP-Seq Reads
To generate longer reads that may improve contig assembly, we used PANDAseq (Masella et al. 2012) with default settings to merge read pairs that had >25 bp overlap with each other (quality threshold = 0.6). The combination of merged reads and unmerged read-pairs were then used by RepARK (Koch et al. 2014) to generate de novo assemblies according to developer’s instructions. A k-mer size of 37 bp was used for RepARK assemblies, after the optimal k-mer size was identified using the K-mer Analysis Toolkit (Mapleson et al. 2017).
RNA-Sequencing and Identification of Centromere-Specific Reads
Total RNA was extracted from fresh frozen cell pellets of the same four gibbons used for CENP ChIP assay, using the mirVana™ Total RNA Isolation kit (ThermoFisher Scientific). High-quality RNA was subjected to lllumina Tru-seq stranded total RNA library preparation, including Ribo-depletion. Libraries were sequenced on the NextSeq 500 v2, paired-end 75 bp (High Output kit), with a target depth of ∼65 M total paired-end reads/sample and an average insert length of 260 bp. RNA-seq reads were preprocessed similar to ChIP-seq reads described above, without subsequence subsampling. In order to broadly identify centromere RNA-seq reads, RNA-seq libraries was aligned to each respective RepARK assembly using Burrows−Wheeler Aligner’s default bwa-mem algorithm (Li and Durbin 2009). Aligned reads were separated and converted into FASTA format using Bedtools (Quinlan and Hall 2010) and Seqtk (Shen et al. 2016). Repeats were annotated using RepeatMasker (described previously). In order to identify unique, centromere-derived reads from RNA-seq libraries, each RNA-seq library was aligned to Nleu3.0 using Burrows−Wheeler Aligner’s default bwa-mem algorithm (Li and Durbin 2009). Because this genome assembly does not contain reference centromere sequences, unaligned reads were assumed to contain reads corresponding to centromere-specific transcripts. Unaligned reads were separated using SAMtools (Li et al. 2009) and converted into FASTA format using Bedtools (Quinlan and Hall 2010) and Seqtk (Shen et al. 2016).
To identify centromeric reads, unaligned reads were then mapped to CENP RepARK contig assemblies described above using the bwa-mem algorithm. Unaligned reads were removed from the remaining pool using SAMtools and the remaining aligned reads processed to FASTA format using Bedtools (Quinlan and Hall 2010) and Seqtk (Shen et al. 2016). Duplicate reads produced during alignment were removed using the FASTX-Toolkit (Hannon 2010). Repeats in the remaining centromere-derived reads were annotated using RepeatMasker, as previously described.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
Authors would like to thank Kimberly Nevonen, Christine McCann, Judy Brown and Nicole Pauloski for assistance with experiements, and the staff at the Gibbon Conservation Center (Santa Clarita, CA), especially the director Gabriella Skollar, who provided the opportunistic gibbon blood samples used for the establishment of gibbon LCLs. We also acknowledge the KCVI Epigenetics Consortium and the Exacloud cluster at OHSU, and the Center for Genome Innovation and Computational Biology Core at UConn. This work was supported by the National Science Foundation (grant number 1613856 to L.C. and R.O.) and the National Institutes of Health (grant 5R01GM123312-02 to R.O. and R01HG010333 to L.C.).
Data Availability
The ChIP-seq and RNA-seq data generated in this work are available at the Gene Expression Omnibus (GEO) database (accession number GSE161217). Scripts are available at https://github.com/gabriellehartley/CENP-Gibbon-Analysis/.
References
- Aldrup-MacDonald ME, Kuo ME, Sullivan LL, Chew K, Sullivan BA.. 2016. Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. Genome Res. 26(10):1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov IA, Medvedev LI, Mashkova TD, Kisselev LL, Romanova LY, Yurov YB.. 1993. Definition of a new alpha satellite suprachromosomal family characterized by monomeric organization. Nucleic Acids Res. 21(9):2209–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C, Ventura M, Archidiacono N, Rocchi M, Sahinalp SC, Eichler EE.. 2007. Organization and evolution of primate centromeric DNA from whole-genome shotgun sequence data. PLoS Comput Biol. 3(9):e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor DJ, Bentley K, Ryan J, Perry J, Wong L, Slater H, Choo KH.. 2004. Human centromere repositioning “in progress”. Proc Natl Acad Sci U S A. 101(17):6542–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S.2010. FastQC: a Quality Control Tool for High Throughput Sequence Data. Available from: http://www.bioinformatics.babraham.ac.uk. Accessed May 21, 2021.
- Barry AE, Howman EV, Cancilla MR, Saffery R, Choo KH.. 1999. Sequence analysis of an 80 kb human neocentromere. Hum Mol Genet. 8(2):217–227. [DOI] [PubMed] [Google Scholar]
- Bergmann JH, Rodríguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC.. 2011. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 30(2):328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH.. 2002. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2(3):319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capozzi O, Carbone L, Stanyon RR, Marra A, Yang F, Whelan CW, de Jong PJ, Rocchi M, Archidiacono N.. 2012. A comprehensive molecular cytogenetic analysis of chromosome rearrangements in gibbons. Genome Res. 22(12):2520–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Harris RA, Gnerre S, Veeramah KR, Lorente-Galdos B, Huddleston J, Meyer TJ, Herrero J, Roos C, Aken B et al. 2014. Gibbon genome and the fast karyotype evolution of small apes. Nature. 513(7517):195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Harris RA, Mootnick AR, Milosavljevic A, Martin DI, Rocchi M, Capozzi O, Archidiacono N, Konkel MK, Walker JA et al. 2012. Centromere remodeling in Hoolock leuconedys (Hylobatidae) by a new transposable element unique to the gibbons. Genome Biol Evol. 4(7):648–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Harris RA, Vessere GM, Mootnick AR, Humphray S, Rogers J, Kim SK, Wall JD, Martin D, Jurka J et al. 2009. Evolutionary breakpoints in the gibbon suggest association between cytosine methylation and karyotype evolution. PLoS Genet. 5(6):e1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Nergadze SG, Magnani E, Misceo D, Cardone MF, Roberto R, Bertoni L, Attolini C, Piras MF, de Jong P et al. 2006. Evolutionary movement of centromeres in horse, donkey, and zebra. Genomics 87(6):777–782. [DOI] [PubMed] [Google Scholar]
- Carbone L, Vessere GM, ten Hallers BFH, Zhu B, Osoegawa K, Mootnick A, Kofler A, Wienberg J, Rogers J, Humphray S et al. 2006. A high-resolution map of synteny disruptions in gibbon and human genomes. PLoS Genet. 2(12):e223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellamare A, Catacchio CR, Alkan C, Giannuzzi G, Antonacci F, Cardone MF, Della Valle G, Malig M, Rocchi M, Eichler EE et al. 2009. New insights into centromere organization and evolution from the white-cheeked gibbon and marmoset. Mol Biol Evol. 26(8):1889–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan FL, Marshall OJ, Saffery R, Kim BW, Earle E, Choo KHA, Wong LH.. 2012. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc Natl Acad Sci U S A. 109(6):1979–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-H, Chavan A, Palladino J, Wei X, Martins NMC, Santinello B, Chen C-C, Erceg J, Beliveau BJ, Wu C-T et al. 2019. Islands of retroelements are major components of Drosophila centromeres. PLOS Biol. 17(5):e3000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZJ, Murata M.. 2003. A centromeric tandem repeat family originating from a part of Ty3/gypsy-retroelement in wheat and its relatives. Genetics 164(2):665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmatal L, Schultz RM, Black BE, Lampson MA.. 2017. Cell biology of cheating-transmission of centromeres and other selfish elements through asymmetric meiosis. Prog Mol Subcell Biol. 56:377–396. [DOI] [PubMed] [Google Scholar]
- Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KHA, Wong LH.. 2009. LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet. 5(2):e1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C, Mootnick A.. 2009. Gibbons. Curr Biol. 19(14):R543–544. [DOI] [PubMed] [Google Scholar]
- du Sart D, Cancilla MR, Earle E, Mao JI, Saffery R, Tainton KM, Kalitsis P, Martyn J, Barry AE, Choo KH.. 1997. A functional neo-centromere formed through activation of a latent human centromere and consisting of non alpha satellite DNA. Nat Genet. 16(2):144–153. [DOI] [PubMed] [Google Scholar]
- Du Y, Topp CN, Dawe RK.. 2010. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 6(2):e1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont M, Gamba R, Gestraud P, Klaasen S, Worrall JT, De Vries SG, Boudreau V, Salinas-Luypaert C, Maddox PS, Lens SM et al. 2020. Human chromosome-specific aneuploidy is influenced by DNA-dependent centromeric features. EMBO J. 39(2):e102924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Ratrie IIH, Stetten G.. 1989. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma 98(1):1–12. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N.. 1985. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91(3 − 4):313–321. [DOI] [PubMed] [Google Scholar]
- Fatyol K, Illes K, Diamond DC, Janish C, Szalay AA.. 2000. Mer22-related sequence elements form pericentric repetitive DNA families in primates. Mol Gen Genet. 262(6):931–939. [DOI] [PubMed] [Google Scholar]
- Fernandes JD, Zamudio-Hurtado A, Clawson H, Kent WJ, Haussler D, Salama SR, Haeussler M.. 2020. The UCSC repeat browser allows discovery and visualization of evolutionary conflict across repeat families. Mob DNA. 11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreri GC, Brown JD, Obergfell C, Jue N, Finn CE, O'Neill MJ, O'Neill RJ.. 2011. Recent amplification of the kangaroo endogenous retrovirus, KERV, limited to the centromere. J Virol. 85(10):4761–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR III, Bassett EA, Wood S, Black BE, Cleveland DW.. 2009. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137(3):472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamba R, Fachinetti D.. 2020. From evolution to function: two sides of the same CENP-B coin? Exp Cell Res. 390(2):111959. [DOI] [PubMed] [Google Scholar]
- Girirajan S, Chen L, Graves T, Marques-Bonet T, Ventura M, Fronick C, Fulton L, Rocchi M, Fulton RS, Wilson RK et al. 2009. Sequencing human–gibbon breakpoints of synteny reveals mosaic new insertions at rearrangement sites. Genome Res. 19(2):178–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Wu Y, Koblizkova A, Torres GA, Wang K, Iovene M, Neumann P, Zhang W, Novak P, Buell CR et al. 2012. Repeatless and repeat-based centromeres in potato: implications for centromere evolution. Plant Cell 24(9):3559–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon. 2010. FASTX-Toolkit [Internet]. Available from: http://hannonlab.cshl.edu/fastx_toolkit/. Accessed May 21, 2021.
- Hara T, Hirai Y, Jahan I, Hirai H, Koga A.. 2012. Tandem repeat sequences evolutionarily related to SVA-type retrotransposons are expanded in the centromere region of the western hoolock gibbon, a small ape. J Hum Genet. 57(12):760–765. [DOI] [PubMed] [Google Scholar]
- Hartley G, O’Neill R.. 2019. Centromere repeats: hidden gems of the genome. Genes 10(3):223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson D, Panchenko T, Salimian KJ, Salman MU, Sekulic N, Alonso A, Warburton PE, Black BE.. 2013. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat Struct Mol Biol. 20(6):687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen E, Launonen V, Muller E, Bachmann L.. 1995. The pvB370 BamHI satellite DNA family of the Drosophila virilis group and its evolutionary relation to mobile dispersed genetic pDv elements. J Mol Evol. 41(5):604–614. [DOI] [PubMed] [Google Scholar]
- Henikoff JG, Thakur J, Kasinathan S, Henikoff S.. 2015. A unique chromatin complex occupies young alpha-satellite arrays of human centromeres. Sci Adv. 1(1):e1400234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS.. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293(5532):1098–1102. [DOI] [PubMed] [Google Scholar]
- Ianc B, Ochis C, Persch R, Popescu O, Damert A.. 2014. Hominoid composite non-LTR retrotransposons-variety, assembly, evolution, and structural determinants of mobilization. Mol Biol Evol. 31(11):2847–2864. [DOI] [PubMed] [Google Scholar]
- Iwata-Otsubo A, Dawicki-McKenna JM, Akera T, Falk SJ, Chmatal L, Yang K, Sullivan BA, Schultz RM, Lampson MA, Black BE.. 2017. Expanded satellite repeats amplify a discrete CENP-A nucleosome assembly site on chromosomes that drive in female meiosis. Curr Biol. 27(15):2365–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Olsen HE, Turner DJ, Stoddart D, Bulazel KV, Paten B, Haussler D, Willard HF, Akeson M, Miga KH.. 2018. Linear assembly of a human centromere on the Y chromosome. Nat Biotechnol. 36(4):321–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW.. 2007. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 176(6):795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RN, O'Meally D, Chen Z, Etherington GJ, Ho SYW, Nash WJ, Grueber CE, Cheng Y, Whittington CM, Dennison S et al. 2018. Adaptation and conservation insights from the koala genome. Nat Genet. 50(8):1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G et al. 2013. DNA-binding specificities of human transcription factors. Cell 152(1-2):327–339. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J.. 2005. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110(1-4):462–467. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Holmquist GP, Jurka J.. 1998. L1 repeat is a basic unit of heterochromatin satellites in cetaceans. Mol Biol Evol. 15(5):611–612. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J.. 1999. Molecular paleontology of transposable elements from Arabidopsis thaliana. Genetica 107(1-3):27–37. [PubMed] [Google Scholar]
- Kent WJ.2002. BLAT - the BLAST-like alignment tool. Genome Res. 12(4):656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klare K, Weir JR, Basilico F, Zimniak T, Massimiliano L, Ludwigs N, Herzog F, Musacchio A.. 2015. CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J Cell Biol. 210(1):11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein SJ, O’Neill RJ.. 2018. Transposable elements: genome innovation, chromosome diversity, and centromere conflict. Chromosome Res. 26:5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch P, Platzer M, Downie BR.. 2014. RepARK—de novo creation of repeat libraries from whole-genome NGS reads. Nucleic Acids Res. 42(9):e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley SA, Miga KH, Karpen GH, Langley CH.. 2019. Haplotypes spanning centromeric regions reveal persistence of large blocks of archaic DNA. eLife 8:e42989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar NH, Nevonen KA, O'Connell B, McCann C, O'Neill RJ, Green RE, Meyer TJ, Okhovat M, Carbone L.. 2018. Epigenetic maintenance of topological domains in the highly rearranged gibbon genome. Genome Res. 28(7):983–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li. 2011. wgsim - read simulator for next generation sequencing [Internet].
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang S-P, Wang Z, Chinwalla AT, Minx P et al. 2011. Comparative and demographic analysis of orangutan genomes. Nature 469(7331):529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo MS, Carone DM, Green ED, O'Neill MJ, O’Neill RJ, NISC Comparative Sequencing Program. 2009. Distinct retroelement classes define evolutionary breakpoints demarcating sites of evolutionary novelty. BMC Genomics. 10: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupan I, Bulzu P, Popescu O, Damert A.. 2015. Lineage specific evolution of the VNTR composite retrotransposon central domain and its role in retrotransposition of gibbon LAVA elements. BMC Genomics 16(1): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL.. 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27(12):1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahtani MM, Willard HF.. 1990. Pulsed-field gel analysis of alpha-satellite DNA at the human X chromosome centromere: high frequency polymorphisms and array size estimate. Genomics 7(4):607–613. [DOI] [PubMed] [Google Scholar]
- Mapleson D, Garcia Accinelli G, Kettleborough G, Wright J, Clavijo BJ.. 2017. KAT: a K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics 33(4):574–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD.. 2012. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 13(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T.. 1989. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol. 109(5):1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM.. 2016. The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol. 17(1):16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga KH, Koren S, Rhie A, Vollger MR, Gershman A, Bzikadze A, Brooks S, Howe E, Porubsky D, Logsdon GA et al. 2020. Telomere-to-telomere assembly of a complete human X chromosome. Nature 585(7823):79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nergadze SG, Piras FM, Gamba R, Corbo M, Cerutti F, McCarter JGW, Cappelletti E, Gozzo F, Harman RM, Antczak DF et al. 2018. Birth, evolution, and transmission of satellite-free mammalian centromeric domains. Genome Res. 28(6):789–799.,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie W, Rens W, Wang J, Yang F.. 2001. Conserved chromosome segments in Hylobates hoolock revealed by human and H. leucogenys paint probes. Cytogenet Cell Genet. 92(3-4):248–253. [DOI] [PubMed] [Google Scholar]
- Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H.. 2007. CENP-B controls centromere formation depending on the chromatin context. Cell 131(7):1287–1300. [DOI] [PubMed] [Google Scholar]
- Okhovat M, Nevonen KA, Davis BA, Michener P, Ward S, Milhaven M, Harshman L, Sohota A, Fernandes JD, Salama SR et al. 2020. Co-option of the lineage-specific LAVA retrotransposon in the gibbon genome. Proc Natl Acad Sci U S A. 117(32):19328–19338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfree MB, Papenfuss AT, Deakin JE, Lindsay J, Heider T, Belov K, Rens W, Waters PD, Pharo EA, Shaw G et al. 2011. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 12(8):R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberto R, Capozzi O, Wilson RK, Mardis ER, Lomiento M, Tuzun E, Cheng Z, Mootnick AR, Archidiacono N, Rocchi M et al. 2007. Molecular refinement of gibbon genome rearrangements. Genome Res. 17(2):249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney DE, Czepulkowski BH.. 1992. Human Cytogenetics: A Practical Approach. Oxford: IRL Press. [Google Scholar]
- Rošić S, Kohler F, Erhardt S.. 2014. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J Cell Biol. 207(3):335–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd MK, Schueler MG, Willard HF.. 2003. Sequence organization and functional annotation of human centromeres. Cold Spring Harb Symp Quant Biol. 68:141–150. [DOI] [PubMed] [Google Scholar]
- Schneider KL, Xie Z, Wolfgruber TK, Presting GG.. 2016. Inbreeding drives maize centromere evolution. Proc Natl Acad Sci U S A. 113(8):E987–E996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang W-H, Hori T, Toyoda A, Kato J, Popendorf K, Sakakibara Y, Fujiyama A, Fukagawa T.. 2010. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 20(9):1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Le S, Li Y, Hu F.. 2016. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One. 11(10):e0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C-M, Yang Z.. 2018. Coalescent-based analyses of genomic sequence data provide a robust resolution of phylogenetic relationships among major groups of gibbons. Mol Biol Evol. 35(1):159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smurova K, De Wulf P.. 2018. Centromere and pericentromere transcription: roles and regulation… in sickness and in health. Front Genet. 9:674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujiwattanarat P, Thapana W, Srikulnath K, Hirai Y, Hirai H, Koga A.. 2015. Higher-order repeat structure in alpha satellite DNA occurs in New World monkeys and is not confined to hominoids. Sci Rep. 5:10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntronpong A, Kugou K, Masumoto H, Srikulnath K, Ohshima K, Hirai H, Koga A.. 2016. CENP-B box, a nucleotide motif involved in centromere formation, occurs in a new world monkey. Biol Let. 12:20150817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel S.2012. Using and understanding RepeatMasker. Methods Mol Biol. 859:29–51. [DOI] [PubMed] [Google Scholar]
- Thongchum R, Nishihara H, Srikulnath K, Hirai H, Koga A.. 2020. The CENP-B box, a nucleotide motif involved in centromere formation, has multiple origins in New World monkeys. Genes Genet Syst. 94(6):301–306. [DOI] [PubMed] [Google Scholar]
- Tremblay DC, Alexander Moseley, Jr.G S, Chadwick BP.. 2010. Expression, tandem repeat copy number variation and stability of four macrosatellite arrays in the human genome. BMC Genomics 11:632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR.. 2001. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 114(Pt 19):3529–3542. [DOI] [PubMed] [Google Scholar]
- Voullaire LE, Slater HR, Petrovic V, Choo KH.. 1993. A functional marker centromere with no detectable alpha satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am J Hum Genet. 52(6):1153–1163. [PMC free article] [PubMed] [Google Scholar]
- Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR et al.; Broad Institute Whole Genome Assembly Team. 2009. Genome sequence, comparative analysis and population genetics of the domestic horse (Equus caballus). Science 326(5954):865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA.. 2005. SVA elements: a hominid-specific retroposon family. J Mol Biol. 354(4):994–1007. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Haaf T, Gosden J, Lawson D, Willard HF.. 1996. Characterization of a chromosome-specific chimpanzee alpha satellite subset: evolutionary relationship to subsets on human chromosomes. Genomics 33(2):220–228. [DOI] [PubMed] [Google Scholar]
- Waye JS, Willard HF.. 1987. Nucleotide sequence heterogeneity of alpha satellite repetitive DNA: a survey of alphoid sequences from different human chromosomes. Nucleic Acids Res. 15(18):7549–7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard HF.1985. Chromosome-specific organization of human alpha satellite DNA. Am J Hum Genet. 37(3):524–532. [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Sun S, Billmyre S, Thimmappa RB, Shea BC, Lintner T, Bakkeren R, Cuomo G, Heitman CA, Sanyal J K.. 2018. RNAi is a critical determinant of centromere evolution in closely related fungi. Proc Natl Acad Sci U S A. 115(12):3108–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T.. 2000. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc Natl Acad Sci U S A. 97(13):7266–7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9(9):R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong CX, Marshall JB, Topp C, Mroczek R, Kato A, Nagaki K, Birchler JA, Jiang J, Dawe RK.. 2002. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14(11):2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ChIP-seq and RNA-seq data generated in this work are available at the Gene Expression Omnibus (GEO) database (accession number GSE161217). Scripts are available at https://github.com/gabriellehartley/CENP-Gibbon-Analysis/.