Abstract
Purpose
Studies have shown that multiple genes influence antibiotic susceptibility, but the relationship between genotypic and phenotypic antibiotic susceptibility is unclear. We sought to analyze the concordance between the presence of antibiotic resistance (ABR) genes and antibiotic susceptibility results in urine samples collected from patients with symptomatic urinary tract infection (UTI).
Patients and Methods
Urine samples were collected from patients presenting to 37 geographically disparate urology clinics across the United States from July 2018 to February 2019. Multiplex polymerase chain reaction was used to detect 27 ABR genes. In samples containing at least one culturable organism at a concentration of ≥ 104 cells per mL, pooled antibiotic susceptibility testing (P-AST), which involves simultaneous growing all detected bacteria together in the presence of antibiotic and then measure susceptibility, was performed against 14 antibiotics. The concordance rate between the ABR genes and the P-AST results was generated for the overall group. The concordance rates for each antibiotic between monomicrobial and polymicrobial infection were compared using chi-square test.
Results
Results from ABR gene detection and P-AST of urine samples from 1155 patients were included in the concordance analysis. Overall, there was a 60% concordance between the presence or absence of ABR genes and corresponding antimicrobial susceptibility with a range of 49–78% across antibiotic classes. Vancomycin, meropenem, and piperacillin/tazobactam showed significantly lower concordance rates in polymicrobial infections than in monomicrobial infections.
Conclusion
Given the 40% discordance rate, the detection of ABR genes alone may not provide reliable data to make informed clinical decisions in UTI management. However, when used in conjunction with susceptibility testing, ABR gene data can offer valuable clinical information for antibiotic stewardship.
Keywords: urinary tract infection, antibiotic resistance, antibiotic resistance genes, bacteria, culture, antibiotic susceptibility test, polymerase chain reaction
Introduction
Urinary tract infection (UTI) is common among geriatric populations1 and contributes to high healthcare costs in the United States.2 Current clinical urine tests, such as standard urine culture (SUC) are limited in their scope and parameters. Additionally, turnaround time can reach up to 3–5 days for SUC and antibiotic susceptibility testing (AST),3–5 which leads to a reliance on empiric treatment.5 To effectively treat geriatric patients, developing novel tests to accurately and rapidly identify uropathogens and assess antibiotic susceptibility is necessary to determine who will require what therapy.3–5 Amidst the rising rate of multidrug resistant organisms, a variety of technologies used for rapid phenotypic growth-based antibiotic susceptibility testing, such as digital time-lapse microscopy, morphokinetic cellular analysis, and MALDI-TOF direct-on-target microdroplet growth assays, have been investigated. However, most of these technologies are still far from being used in routine clinical diagnostics.6–9 During the past decade, advances in polymerase chain reaction (PCR) technology and other DNA target amplification techniques have resulted in molecular diagnostics becoming a revolutionary method in the field of infectious disease.3–5 Clinicians and researchers have started using these more sensitive and specific tests to detect microorganisms10,11 and antibiotic resistance (ABR) genes12–14 to understand urobiome and more effectively manage UTIs. Whereas the role of nucleic acid-based methods for detecting pathogens is well established, its role in the determination of antibiotic susceptibility is less clear.
Innate or acquired antibiotic resistance is a significant obstacle in UTI management. Innate antibiotic resistance is typically chromosome-encoded and includes mechanisms such as nonspecific efflux pumps, antibiotic-inactivating enzymes, and permeability barriers.15,16 Acquired resistance results from horizontal transfer of plasmid encoded ABR genes, including those that encode specific efflux pumps and enzymes that modify targeted antibiotics.17,18 While an increasing number of ABR genes are known, the detection of an ABR gene does not guarantee that gene’s activity since the regulation of many of the ABR genes has not been thoroughly investigated.19–21 Further research is needed to elucidate how frequently the presence of an ABR gene is correlated with its activity. Here, we analyze the concordance between the ABR gene and antibiotic susceptibility in urine samples collected from symptomatic patients with UTI.
Materials and Methods
Study Design and Participants
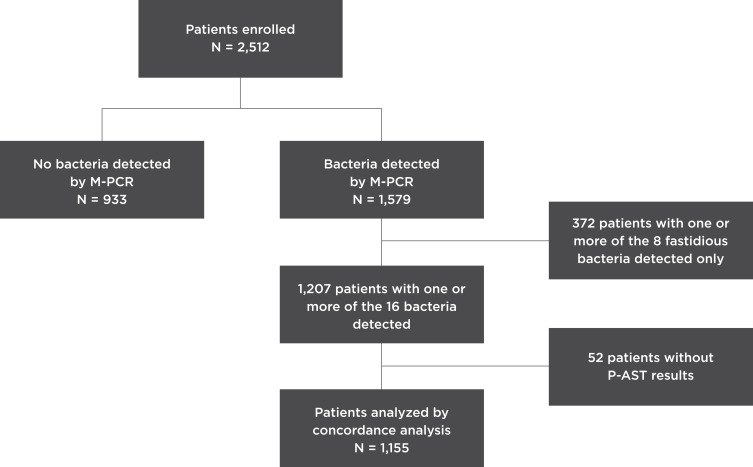
This concordance study investigated a subset of a prospective UTI study cohort comprising 2512 consecutive patients presenting with UTI symptoms enrolled by 75 physicians from 37 urology clinics from July 26, 2018 to February 27, 2019 (Figure 1). All patients provided written informed consent, and the study was approved by the Western Institutional Review Board (20181661). The study was conducted in accordance with the Declaration of Helsinki. The patient selection criteria were patients 60 years or older who presented to the urology clinics with a suspicion of acute cystitis, complicated UTI, persistent UTI, recurrent UTIs, prostatitis, or pyelonephritis. Additionally, we included patients of any age who presented with a history of interstitial cystitis. Patients were excluded if they did not have UTI symptoms, took antibiotics for any reason other than a UTI at the time of enrollment, had urinary diversion, performed self-catheterization, or had a chronic (≥10 days) indwelling catheter at the time of consultation. A 3-mL urine sample from each participant was obtained for bacterial detection via multiplex PCR (M-PCR, see Bacterial Detection by M-PCR). Patients were excluded from the study if the method used to collect their urine sample was not properly documented, they did not meet the collection criteria (see Urine Collection, Storage and DNA Extraction) for testing, or the collected urine sample was less than 3 mL. Patients were also excluded if they did not have results from the M-PCR test.
Figure 1.
Study design and participant selection.
Abbreviation: P-AST, pooled antibiotic susceptibility testing.
Among the 2512 patients, M-PCR detected bacteria in the urine samples of 1579 patients. Among them, 372 patient samples contained exclusively fastidious bacteria that were deemed unculturable based on laboratory standards; as a result, susceptibility testing could not be performed. These fastidious bacteria include Actinotignum schaalii, Aerococcus urinae, Alloscardovia omnicolens, Corynebacterium riegelii, Mycoplasma genitalium, Mycoplasma hominis, Pantoea agglomerans, Ureaplasma urealyticum, and Viridans group streptococci (VGS).
Pooled antibiotic susceptibility testing (P-AST, see Detection of Antibiotic Susceptibility by P-AST) results could not be generated for 52 patients because the bacteria failed to thrive during testing. Therefore, the concordance analysis was conducted using a subset (N = 1155) of the total study cohort. This subset only includes patients with positive bacterial identifications by M-PCR, antimicrobial susceptibility results from P-AST, and results from the ABR detection by M-PCR (Figure 1).
Urine Collection, Storage and DNA Extraction
Midstream clean catch or catheterized urine were collected with gray top tubes (Cat. # BD364953, VWR, Radnor, PA) and stored at room temperature.
DNA was extracted from urine samples using a KingFisher/MagMAX automated DNA extraction instrument and MagMAX DNA Multi-Sample Ultra Kit (Thermo Fisher, Carlsbad, CA). Briefly, 400 µL aliquots of urine were transferred to 96-deep-well plates, sealed, and centrifuged to concentrate the samples by removing the supernatant. Enzyme Lysis Mix (220 µL/well) was added, followed by incubation for 20 min at 65°C. Proteinase K Mix was added (50 µL/well), and the samples were incubated for 30 min at 65°C. Lysis buffer (125 µL/well) and DNA Binding Bead Mix (40 µL/well) were added and shaken for at least 5 min. Finally, the 96-deep-well plate was loaded into the KingFisher/MagMAX instrument for DNA extraction, following standard operating procedures.
Bacterial Detection by M-PCR
Bacteria in the urine samples were detected using the Pathnostics Guidance® UTI Test, an M-PCR assay described previously.22,23 Briefly, the DNA extracted from patient samples was mixed with a universal PCR master mix and amplified using TaqMan technology in a Life Technologies 12K Flex Open Array System. DNA samples were spotted in duplicate on 112-format OpenArray chips. Positive controls were included in the form of plasmids containing bacterial target DNA. Candida tropicalis was used as a control for PCR inhibition. The Pathnostics data analysis tool was used to sort data, assess data quality, summarize control sample data, identify positive assays, calculate concentrations, and generate results.
The following bacteria and bacterial groups were detected using the Pathnostics Guidance® UTI Test. Not all organisms detected by this test were readily cultivatable using standard culture protocols; therefore, among the following detected bacteria, only the 16 listed in bold were included in the analysis: Acinetobacter baumannii,Actinotignum schaalii, Aerococcus urinae, Alloscardovia omnicolens, Citrobacter freundii, Citrobacter koseri, Corynebacterium riegelii, Klebsiella aerogenes, Enterococcus faecalis, Escherichia coli, Klebsiella oxytoca, Klebsiella pneumoniae, Morganella morganii, Mycoplasma genitalium, Mycoplasma hominis, Pantoea agglomerans, Proteus mirabilis, Providencia stuartii, Pseudomonas aeruginosa, Serratia marcescens, Staphylococcus aureus, Streptococcus agalactiae,Ureaplasma urealyticum, Coagulase-negative staphylococci group (CoNS), and VGS. Patient samples containing any of the 16 cultivatable bacteria were analyzed using pooled sensitivity analysis testing (P-AST) and concordance analyses.
The quantities of each of the bacterial species were determined using the standard curve method, as described previously.22 Briefly, standard curves of each of the bacteria were generated from testing replicates of a bacterial culture dilution series (Supplementary Table 1) of known concentrations; constants necessary for the quantitation of each bacterial species in unknown samples were established from the standard curves. The PCR cycle values of a target bacterium from a patient sample were compared to the standard curve, and the concentration (cells/mL) of the target bacterial species in the sample was extrapolated. A bacterium with a quantity of ≥10,000 cells/mL was defined as “positive” or “detected,” and those with < 10,000 cells/mL were defined as “negative” or “not detected”; this cut-off was validated for reliable bacteria detection and corresponding antibiotic susceptibility testing for the Pathnostics Guidance® UTI Test.
ABR Gene Detection by M-PCR
We tested 27 ABR genes associated with resistance to six classes of antibiotics (Table 1) including 6 carbapenem resistance genes (VIM, KPC, IMP-1 group, OXA-23, OXA-40, and OXA-48), 6 ampicillin resistance genes (DHA, MOX/CMY, BIL/LAT/CMY, AmpC, FOX, and ACC), 2 fluoroquinolone resistance genes (QnrA, QnrB), 3 vancomycin resistance genes (vanA1, vanA2, and vanB), 9 extended-spectrum beta-lactamases resistance genes (CTX-M group 1, CTX-M group 2, CTX-M group 8/25, CTX-M group 9, OXA-1, GES, SHV, TEM, and VEB), and 1 methicillin resistance gene (mecA). Each position of a 112-format OpenArray chip was coated with a probe targeting one ABR gene, with the exception of DHA, MOX/CMY, BIL/LAT/CMY, AmpC, FOX, and ACC, where the probes targeting DHA, MOX/CMY, and BIL/LAT/CMY share one position, and the probes targeting AmpC, FOX, and ACC share a position). Two additional ABR genes, ErmA and ErmB, associated with resistance to the macrolide antibiotic class were also included on the OpenArray chip as part of the ABR gene testing; however, results from these two genes were not included in the concordance analysis as no macrolide antibiotics were part of the P-AST testing.
Table 1.
Antibiotic Resistance (ABR) Genes Tested in the Study, Their Groups and Antibiotic Classes
| Antibiotic Names and Classes | Resistance Gene Group | ABR Genes |
|---|---|---|
| Aminopenicillin (Ampicillin) Beta-lactamase Inhibitor Combination (Ampicillin/Sulbactam, Amoxicillin/Clavulanate, Piperacillin/Tazobactam) Cephalosporins (Cefepime, Ceftazidime, Ceftriaxone, Cefoxitin, Cefazolin, Cefaclor) |
Ampicillin resistance genes | AmpC, FOX, ACC |
| DHA, MOX/CMY, BIL/LAT/CMY | ||
| Extended-spectrum beta-lactamases resistance genes | CTX-M group 1 | |
| CTX-M group 2 | ||
| CTX-M group 8/25 | ||
| CTX-M group 9 | ||
| OXA-1 | ||
| SHV | ||
| TEM | ||
| VEB | ||
| GES * | ||
| Methicillin resistance genes | mecA | |
| Fluoroquinolones (Levofloxacin, Ciprofloxacin) | Quinolone and fluoroquinolone resistance genes | QnrA |
| QnrB | ||
| Glycopeptides (Vancomycin) | Vancomycin resistance genes | vanA1 |
| vanA2 | ||
| vanB | ||
| Carbapenems (Meropenem) | Carbapenems resistance genes | IMP-1 group * |
| KPC | ||
| OXA-23 * | ||
| OXA-40 | ||
| OXA-48 | ||
| VIM |
Notes: ABR genes marked with * are the genes that were tested but not detected.
A bacterium was considered “positive” for an ABR gene if the cycle number (Ct) of that gene was above a particular threshold. We determined the thresholds by first comparing a series of negative samples, extraction control samples, and specificity samples (genomic DNA of a nontarget organism/gene) and selecting the lowest Ct from these assays. Second, we tested a plasmid dilution series (ThermoFisher provided plasmids for each target ABR gene). The lower limit of detection (LLoD) for each ABR gene assay was set as the lowest plasmid concentration in which 50% or more of the replicates were detected with Ct values below the cycle number determined in the first step. We then set the threshold Ct for each target ABR gene assay as the cycle equivalent of the established LLoD for that gene. ABR genes with Ct values no higher than the Ct threshold were defined as “positive” or “detected,” and those with Ct values higher than the threshold were defined as “negative” or “not detected.” For the mecA gene, the “positive” status was limited to patients with S. aureus detection.
Detection of Antibiotic Susceptibility by P-AST
A total of 18 antibiotics were evaluated in the P-AST assay. Four antibiotics (gentamycin, nitrofurantoin, tetracycline, and trimethoprim/sulfamethoxazole) were not associated with any of the ABR genes on the Pathnostics Guidance® UTI Test detection panel. Therefore, 14 antibiotics representing six antibiotic classes (aminopenicillins, beta-lactamase inhibitor combinations, cephalosporins, fluoroquinolones, carbapenems, and glycopeptides) were evaluated in the concordance analysis (Table 1). The antibiotics were purchased from Sigma Aldrich: amoxicillin/clavulanate (Cat. # A8523-1G/33454-100MG), ampicillin (Cat. # A5354-10ML), ampicillin/sulbactam (Cat. # A5354-10ML/1623670-250MG), cefaclor (Cat. # PHR1283-1G), cefazolin (Cat. # C5020-1G), cefepime (Cat. # PHR1763-1G), cefoxitin (Cat. # C4786-1G), ceftazidime (Cat. # A6987-5G), ceftriaxone (Cat. # C5793-1G), ciprofloxacin (Cat. # 17850–5G-F), levofloxacin (Cat. # 28266–10G-F), meropenem (Cat. # M2574-10MG), piperacillin/tazobactam (Cat. # P8396-1G/1643383-200MG), and vancomycin (Cat. # SBR00001-10ML).
The proprietary P-AST was performed, as described previously.14 Briefly, 1 mL aliquot of patient urine sample was added to a 1.7 mL microcentrifuge tube. After centrifugation, the supernatant was aspirated and discarded. Next, 1 mL of Mueller Hinton Growth Media was added to the microcentrifuge tube, combined with the pellet of the sample at the bottom, and incubated at 35°C in a non-CO2 incubator for 6 h. Turbidity of the pre-culture mixture was measured with the Densi-Chek plate reader (Biomerieux, France).
When the turbidity read reached ≥ 0.5 McF, a 500 µL aliquot of the pre-culture mixture was added to a 50-mL conical tube containing 29.5 mL of Mueller Hinton liquid broth media. This diluted sample was then distributed across wells in 96-well plates containing various antibiotics at varying concentrations. This spec plate was then sealed with a breathable membrane and incubated for 12 to 16 h at 35°C in a non-CO2 incubator, in a single layer without plate-stacking. At the end of the incubation, OD600 was measured with the Infinite M Nano absorbance plate reader (Tecan, Switzerland). The measurements were compared to the established threshold value and a proprietary algorithm to determine resistance to a given antibiotic.
If the turbidity of the pre-culture mixture was less than 0.5 McF after the 6h of incubation, it would be incubated for another maximum of 10 hrs. If it failed to reach the required OD600 at the end of the extended incubation, it did not proceed to the step of culturing with antibiotics and was reported as “failure to thrive.”
Statistics
We collected and summarized the demographic and clinical patient information, bacterial species detection frequency, ABR gene presence or absence, and antibiotic resistance rates for the entire study cohort (Table 2). For each antibiotic, we coded the ABR gene status as “present” if one or more of the associated ABR genes were detected. Bacterium–antibiotic combinations in which the bacterium possesses innate resistance to the antibiotic were not included in the concordance analysis, as indicated by an “X” in Supplementary Table 2.
Table 2.
Patient Demographic and Clinical Information
| Demographic & Clinical Information | All |
|---|---|
| N = 1155 (%) | |
| Sex | |
| Female | 784 (67.9%) |
| Male | 371 (32.1%) |
| Age | |
| ≤60, n (%) | 8 (0.7%) |
| >60, n (%) | 1147 (99.3%) |
| Mean ± standard deviation | 74.3 ± 8.5 |
| Min, Max age | 31, 100 |
| UTI Symptoms, n (%) | |
| Dysuria | 395 (34.2%) |
| Urine cloudy or strong smell | 201 (17.4%) |
| Pain/Pelvic discomfort | 331 (28.7%) |
| Fever | 23 (2.0%) |
| LUTS | 803 (69.5%) |
| Urinary incontinence | 415 (35.9%) |
| Gross hematuria | 232 (20.1%) |
| Antibiotic Usage in the Last 3 Weeks, n (%) | 162 (14.3%) |
| Positive Urine Analysis or Dipsticks Results, n (%) | 1028 (89.0%) |
Abbreviations: UTI, urinary tract infection; LUTS, lower urinary tract symptoms.
We determined the concordance rates between the presence of ABR gene susceptibility test results for each antibiotic, each antibiotic class, monomicrobial infections, polymicrobial infections, and the overall group. We compared the concordance rates for each antibiotic between monomicrobial and polymicrobial samples using the chi-square test. The analyses were performed using SAS version 9.4.
Results
Patient Demographics and Clinical Information
A total of 1155 patients were included in the concordance analyses: 784 (67.9%) females and 371 (32.1%) males. The median age was 74.3 years, and 99.3% of patients were over the age of 60 years. All patients presented with UTI symptoms, including dysuria, cloudy or strong-smelling urine, pain or pelvic discomfort, fever, and/or other non-specific lower urinary tract symptoms (LUTS). Blood, leukocytes, and/or nitrites were detected in the urine of 89.0% of the study population. Only 14.3% of patients were taking antibiotics in the three-week period before enrollment into the study (Table 2).
Bacterial Infection, ABR Genes, and Antibiotic Resistance Among 1155 Symptomatic Patients with UTI
M-PCR detected monomicrobial infections in 886 (76.7%) patients and polymicrobial infections (two or more bacteria) in 269 (23.3%) patients. The most commonly detected bacterial species in the urine samples of patients were E. coli (564, 48.8%), CoNS (282, 24.4%), and E. faecalis (242, 21.0%). Detection frequencies for each bacterium are presented in Supplementary Table 3.
M-PCR detected 24 ABR genes 470 times in 419 (36.2%) patient samples; 379 (90.5%) samples contained only one ABR gene, 29 (6.92%) contained two ABR genes, and 11 (2.63%) contained three ABR genes. The most frequently identified ABR genes were TEM, SHV, and CTX-M group 1 gene, which were detected in 205 (17.7%), 100 (8.7%), and 47 (4.1%) patients, respectively (Supplementary Table 4). Nine ABR genes, GES, IMP-1 group, PER-1, PER-2, IMP-7, OXA-72, OXA-58, NDM-1, and OXA-23, were not detected in any patient samples.
Antibiotic resistance rates ranged from 6.2% (for piperacillin/tazobactam) to 50.9% (for ceftriaxone) (Table 3). ABR gene detection ranged from 0.3% (for meropenem, levofloxacin, and ciprofloxacin-associated ABR genes) to 36.8% (for amoxicillin/clavulanate-associated ABR genes) (Table 3).
Table 3.
Antibiotic Resistance by Pooled Antibiotic Susceptibility Testing (P-AST) and Antibiotic Resistance (ABR) Gene Presence for All 14 Antibiotics Analyzed
| Antibiotics (Total N = 1155) | Sample size | Antibiotic Resistance by P-AST | Detection of Associated ABR Genes |
|---|---|---|---|
| Amoxicillin/Clavulanate | 1084 | 29.1% | 36.8% |
| Ampicillin | 916 | 48.9% | 30.1% |
| Ampicillin/Sulbactam | 1067 | 31.8% | 36.6% |
| Cefaclor | 1153 | 49.3% | 35.7% |
| Cefazolin | 1109 | 38.2% | 35.8% |
| Cefepime | 1154 | 40.9% | 35.7% |
| Cefoxitin | 1132 | 44.9% | 36.0% |
| Ceftazidime | 1155 | 46.3% | 35.7% |
| Ceftriaxone | 1110 | 50.9% | 36.5% |
| Ciprofloxacin | 1154 | 35.4% | 0.3% |
| Levofloxacin | 1153 | 29.7% | 0.3% |
| Meropenem | 1155 | 24.1% | 0.3% |
| Piperacillin/Tazobactam | 1155 | 6.2% | 35.7% |
| Vancomycin | 354 | 44.1% | 0.8% |
Among the 372 patients that contained exclusively eight fastidious bacteria, 15 ABR genes were detected in 26 patients (7.0%). Twenty-four samples contained one ABR gene, one sample contained two ABR genes, and one contained three ABR genes. These patients were excluded from the concordance analyses because their P-AST results were not available. The ABR genes detected in the 372 samples are associated with resistance to the following classes of antibiotics: aminopenicillins, beta-lactamase inhibitor/antibiotic combinations, glycopeptides, fluoroquinolones, carbapenems, and cephalosporins.24–32 Supplementary Table 5 lists the prevalence of each ABR gene among the 372 patients.
Overall Concordance Between the Presence of ABR Genes and Antibiotic Susceptibility
The results show an overall concordance rate of 60% between ABR gene presence M-PCR and antibiotic susceptibility by P-AST. Two concordance circumstances were observed: ABR gene absent/antibiotic-susceptible (48%) and ABR gene present/antibiotic-resistant (12%). The 40% discordance included two circumstances: ABR gene absent/antibiotic-resistant (15%) and ABR gene present/antibiotic-susceptible (25%) (Table 4).
Table 4.
Overall Concordance Between the Presence of Antibiotic Resistance (ABR) Genes Detected by Multiplex Polymerase Chain Reaction and Antibiotic Susceptibility Detected Using Pooled Antibiotic Susceptibility Testing (P-AST) of Urine Samples from Symptomatic Patients with Urinary Tract Infection (UTI)
| Status | ABR Gene and Phenotype Agree | ABR Gene and Phenotype Disagree | ||
|---|---|---|---|---|
| Detail | Sensitive, ABR Genes Absent |
Resistant, ABR Genes Present |
Sensitive, ABR Genes Present |
Resistant, ABR Genes Absent |
| % | 48% | 12% | 25% | 15% |
| Overall % | 60% | 40% | ||
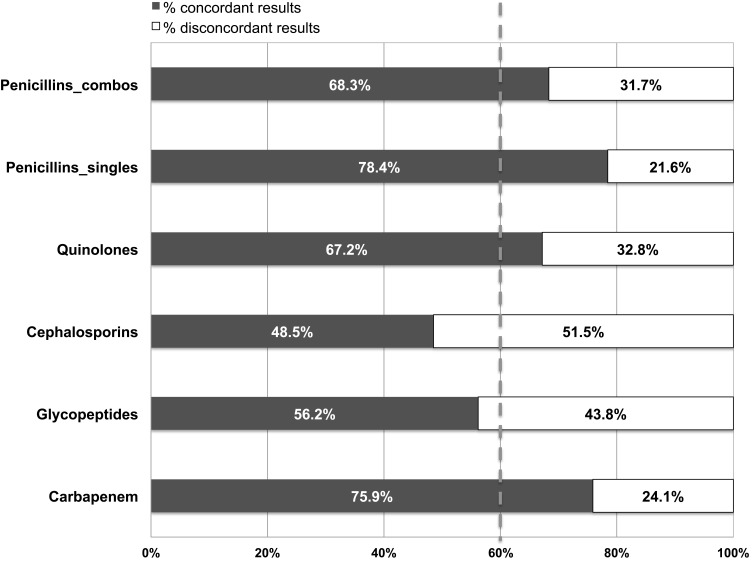
Some antibiotic categories showed higher concordance than the overall concordance rate. For example, aminopenicillins, beta-lactamase inhibitor/antibiotic combinations, fluoroquinolones, and carbapenems had concordance rates of ≥67.2%; whereas, cephalosporins only exhibited a concordance rate of 48.5% (Figure 2).
Figure 2.
Concordance between the presence of antibiotic resistance genes (ABR) detected by multiplex polymerase chain reaction (M-PCR) and antibiotic susceptibility detected by pooled antibiotic susceptibility testing (P-AST) of urine samples from symptomatic patients with urinary tract infection (UTI). The dashed line represents the weighted average concordance across all samples (60%).
Abbreviations: Combo, combination antibiotics, including Ampicillin/Sulbactam, Amoxicillin/Clavulanate, and Piperacillin/Tazobactam.
Concordance by Antibiotic and Monomicrobial versus Polymicrobial Infectious Status
A total of 14 antibiotics were included in the concordance analysis. The overall concordance rates ranged from 44.7% (ceftriaxone) to 78.4% (ampicillin). Similar concordance rates were observed among most antibiotics for monomicrobial and polymicrobial infections. However, the concordance rates of three antibiotics, namely, vancomycin, meropenem, and piperacillin/tazobactam, were significantly lower for polymicrobial infections than for monomicrobial infections with absolute differences of 9.3% (p = 0.002), 13.1% (p < 0.0001), and 19.0% (p = 0.02), respectively (Table 5). In the polymicrobial dataset, the higher discordance rates for vancomycin and meropenem were due to an increased phenotypic resistance rate in the absence of ABR gene detection in polymicrobial infections than in monomicrobial infections. The higher discordance for piperacillin/tazobactam was due to an increased level of ABR gene detection in the presence of the sensitive phenotype (Supplementary Table 6).
Table 5.
Concordance Between the Presence of Antibiotic Resistance (ABR) Genes and Antibiotic Susceptibility of Urine Samples from Symptomatic Patients with Urinary Tract Infection (UTI) by Antibiotic and Infection Status (Monomicrobial vs. Polymicrobial)
| Antibiotics | Number of Associated ABR Genes Tested | Concordance Rate in All (N = 1155) | Concordance Rate in Monomicrobial Specimens (n = 886) | Concordance Rate in Polymicrobial Specimens (n = 269) | p value (Monomicrobial vs. Polymicrobial) |
|---|---|---|---|---|---|
| Amoxicillin /Clavulanate | 16 | 66.8% | 67.9% | 62.7% | 0.13 |
| Ampicillin | 16 | 78.4% | 79.0% | 75.7% | 0.34 |
| Ampicillin /Sulbactam | 16 | 74.0% | 74.9% | 71.0% | 0.24 |
| Cefaclor | 16 | 48.3% | 48.9% | 46.3% | 0.45 |
| Cefazolin | 16 | 55.7% | 56.6% | 52.7% | 0.29 |
| Cefepime | 16 | 50.3% | 51.8% | 45.4% | 0.07 |
| Cefoxitin | 16 | 46.9% | 47.1% | 46.3% | 0.82 |
| Ceftazidime | 16 | 45.4% | 46.0% | 43.1% | 0.40 |
| Ceftriaxone | 16 | 44.7% | 43.4% | 49.0% | 0.12 |
| Ciprofloxacin | 2 | 64.4% | 65.3% | 61.3% | 0.23 |
| Levofloxacin | 2 | 70.1% | 71.3% | 66.2% | 0.11 |
| Meropenem | 6 | 75.9% | 78.1% | 68.8% | 0.002 |
| Piperacillin /Tazobactam | 16 | 64.3% | 67.4% | 54.3% | <0.0001 |
| Vancomycin | 3 | 56.2% | 58.5% | 39.5% | 0.02 |
Note: Statistically significant p values (p < 0.05) were shown in bold fonts.
Discussion
Antibiotic resistance, particularly among uropathogens, is an increasingly important clinical problem. Increasing antibiotic resistance recently has been observed to trimethoprim/sulfamethoxazole, a widely used first-line treatment of uncomplicated UTIs. The widespread use of fluoroquinolones, especially ciprofloxacin, in outpatients is the cause of a continuous increase in resistance to these drugs.33 Consequently, the development of resistance has led to escalating costs in patient care, increasing number of hospital stays, and a demonstrable higher mortality rate.34,35 Many common UTI pathogens in clinical practice have been reported to demonstrate significant levels of resistance to first-line antibiotics. Some are even reported to be multidrug resistant.34,35 Reliable and rapid microbial identification along with resistance information are essential for the management of UTIs and antibiotic stewardship.2 Rapid testing may decrease the frequency of empirical therapies, which have been suggested to be inappropriate in more than 20% of community-acquired bacteremic UTIs.36 Therefore, rapid testing may reduce the use of antibiotics, which may lead to improved antibiotic stewardship. M-PCR-based tests have been developed for clinical use to detect ABR genes in UTI cases. However, the detection of an ABR gene may not translate into phenotypic resistance, nor can ABR gene absence guarantee susceptibility.
We acquired urine samples from 1155 patients with UTI symptoms from 37 different urology clinics in the United States. We used an M-PCR-based test to detect ABR genes and a P-AST test to obtain antibiotic susceptibility results. We then determined the concordance rate between the ABR gene status (present or absent) and antibiotic susceptibility results. The overall concordance rate was 60%: 48% lacked ABR genes and producing sensitive susceptibility results and 12% contained ABR genes and produced resistant susceptibility results. The other 40% were discordant: 15% of the cases demonstrated resistance but lacked ABR genes and 25% of the samples contained ABR genes but produced sensitive susceptibility results.
Although the 60% concordance rate validates the involvement of ABR genes in antibiotic resistance, the 40% discordance rate clearly demonstrates the limitations of making clinical decisions based on the presence or absence of ABR genes. Here, we used an M-PCR OpenArray chip to identify 27 ABR genes commonly used in clinical assays. This represents less than 0.62% of the more than 4336 identified ABR genes.37 Additionally, ABR genes are continuously being discovered through ongoing research.38,39 Thus, incorporating all ABR genes into a single assay is extraordinarily difficult if not impossible. Furthermore, antibiotic resistance may be conferred via ABR genes not included in the testing panel.
Alternatively, the presence of ABR genes did not confer phenotypic resistance in 15% of samples. Multiple factors could have contributed to the observed discordance. First, PCR assays detect ABR genes at the DNA level. For an ABR gene to generate resistance, the bacterium must first transcribe the gene into messenger RNA. The ribosomes then must translate the messenger RNA into protein; then, in some cases, the protein must be activated.40 If mutations occur, for example, in the gene promoter region, the protein would not be produced, thus yielding no antibiotic resistance. In other instances, mutational changes in the coding region of an ABR gene are susceptible to mutations, such as frameshifts, which would result in failure to produce the protein product, preventing the bacteria from generating the antibiotic-resistant phenotype.41 From these observations, one can conclude that the detection of ABR genes alone is not entirely reliable in predicting bacterial antibiotic response.
The concordance rate differed among antibiotic classes. For example, the concordance rates were as high as 78.4% for single-agent penicillin’s and as low as 48.5% for cephalosporins. At the individual antibiotic level, there was a significant mismatch between ABR genes and P-AST results for five antibiotics, namely, piperacillin/tazobactam, meropenem, ciprofloxacin, levofloxacin, and vancomycin. Fewer ABR genes were targeted for ciprofloxacin, levofloxacin, meropenem, and vancomycin than for cephalosporins, beta-lactamase inhibitor combinations, and penicillin. Thus, it is possible that additional ABR genes associated with resistance to ciprofloxacin, levofloxacin, meropenem, and vancomycin, were not included in the testing panel. In the case of piperacillin/tazobactam, the rate of ABR gene detection was higher than the rate of resistance from P-AST results. Resistance was detected in only 6.2% of cases, whereas 35.7% contained ABR resistance genes. Cabot et al41 demonstrated that resistance to piperacillin/tazobactam involves AmpC overexpression, as well as two additional ABR genes, mexB and mexY, which were not targeted in this study. Therefore, it is likely that the samples that tested positive for ABR genes were negative for mexB or mexY, or did not overexpress AmpC, failing to produce a piperacillin/tazobactam-resistant phenotype. Interestingly, the clinical findings of Patterson et al42 and Lee et al43 show that, unlike other antibiotics, the increased use of piperacillin/tazobactam did not produce increased resistance. This phenomenon could be ascribed to the fact that several events are necessary to convey resistance against piperacillin/tazobactam.
We observed similar concordance between monomicrobial and polymicrobial specimens for most antibiotics (11/14). However, three antibiotics, vancomycin, meropenem, and piperacillin/tazobactam, exhibited significantly lower concordance rates in polymicrobial infections than in monomicrobial infections. For vancomycin and meropenem, the high discordance rates involved higher bacterial resistance relative to ABR gene detection. Vollstedt et al23 reported that the odds of resistance increased relative to the number of species detected for these two antibiotics. The discordance may result from interactions among organisms in a polymicrobial sample. Conversely, piperacillin/tazobactam discordance in polymicrobial samples may have resulted from increased detection of ABR genes relative to the rate of resistance. Vollstedt et al23 also reported that the odds of resistance to piperacillin/tazobactam decreased with the presence of additional organisms. As discussed earlier, increased discordance may result from the need to overexpress AmpC, and perhaps, the overexpression of AmpC is reduced with the introduction of additional species. Regardless, these three antibiotics are relatively strong and are often reserved for highly resistant bacterial infections.41
We were unable to generate susceptibility results using P-AST for 372 samples that contained exclusively fastidious bacteria that do not grow in the culture conditions used. Therefore, these samples were excluded from the concordance rate analysis. However, we used M-PCR to detect ABR genes in the samples and detected 15 ABR genes (Supplementary Table 3). The fastidious growth of these bacteria also renders traditional urine culture and isolate-based antimicrobial susceptibility tests unfeasible. Therefore, ABR gene detection may provide clinically valuable information for patients with an exclusively fastidious bacterial infection.
While we tested for a relatively large number of ABR genes, we could only detect the genes for which we had primers. Additionally, we detected the presence of ABR genes but did not quantify them. Therefore, we could not evaluate the concentration of the ABR genes relative to the bacterial bio-load or examine potential impacts on antimicrobial susceptibility. To address these limitations in future studies, we plan to include updated ABR gene testing panels and employ quantitative approaches.
Conclusions
We observed a 60% concordance rate between the presence or absence of ABR genes and the P-AST test results in our multi-institutional study with a large sample size of 1155 patients with symptomatic UTIs. In the remaining 40% of cases where discordance was observed, reliance on the ABR gene detection without phenotypic data can potentially lead to inappropriate antimicrobial therapy. In order to improve antimicrobial stewardship, physicians should utilize ABR gene detection and antibiotic susceptibility test results in conjunction to enhance clinical treatment outcomes, particularly with P-AST results, which takes into consideration of bacterial interactions.
Acknowledgments
All funding was provided by Pathnostics and Thermo Fisher. The authors would like to thank Drs. Kirk J. Wojno, Kevin Cline, Laurence Belkoff, Aaron Milbank, Neil Sherman, Rashel Haverkorn, Natalie Gaines, Neal Shore, Howard Korman, Mohammad Jafri, Patrick Keating, Bridget Makhlouf, Dylan Hazelton, Stephany Hindo, David Wenzler, Mansour Sabry, and Meghan Campbell for participating in the overall prospective UTI study. The authors would like to thank Dr. Alan Wolfe for the scientific review. The authors would also like to thank Enago (www.enago.com) for the English language review.
Abbreviations
ABR, antibiotic resistance; AST, antimicrobial susceptibility testing; ESBL, extended-spectrum beta-lactamases; M-PCR, multiplex polymerase chain reaction; P-AST, pooled antibiotic susceptibility testing; UTI, urinary tract infection; CoNS, Coagulase negative staphylococci group; VGS, Viridans group streptococci; LLoD, lower limit of detection; LUTS, lower urinary tract symptoms; Ct, cycle number.
Disclosure
David Baunoch, Patrick Cacdac, and Natalie Luke are employees of Pathnostics Inc. David Baunoch reports grants from Thermo Fisher, during the conduct of the study; In addition, David Baunoch has a patent 12-216,751 licensed to Pathnostics. Natalie Luke reports grants from Thermo Fisher, outside the submitted work; In addition, Natalie Luke has patents: Assay for the Comprehensive Identification of Antibiotic Sensitivity (US Pat. No. 10,160,991; issued), Methods for Treating Polymicrobial Infections (16/848,651 & US 2020/0347433; pending), and Resistance Genes and P-AST (63/047,846; pending) to Pathnostics. Dicken SC Ko and Larry T Sirls are consultant for Pathnostics, outside the submitted work. The authors report no other conflicts of interest in this work.
References
- 1.Alpay Y, Aykin N, Korkmaz P, Gulduren HM, Caglan FC. Urinary tract infections in the geriatric patients. Pak J Med Sci. 2018;34(1):67–72. doi: 10.12669/pjms.341.14013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simmering JE, Tang F, Cavanaugh JE, Polgreen LA, Polgreen PM. The increase in hospitalizations for urinary tract infections and the associated costs in the United States, 1998–2011. Open Forum Infect Dis. 2017;4(1):ofw281. doi: 10.1093/ofid/ofw281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daly A, Baunoch D, Rehling K, et al. Utilization of M-PCR and P-AST for diagnosis and management of urinary tract infections in home-based primary care. JOJ Uro Nephron. 2020;7(2):555707. [Google Scholar]
- 4.Davenport M, Mach KE, Shortliffe LMD, Banaei N, Wang TH, Liao JC. New and developing diagnostic technologies for urinary tract infections. Nat Rev Urol. 2017;14(5):296–310. doi: 10.1038/nrurol.2017.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price TK, Hilt EE, Dune TJ, Mueller ER, Wolfe AJ, Brubaker L. Urine trouble: should we think differently about UTI? Int Urogynecol J. 2018;29(2):205–210. doi: 10.1007/s00192-017-3528-8 [DOI] [PubMed] [Google Scholar]
- 6.Idelevich EA, Becker K. How to accelerate antimicrobial susceptibility testing. Clin Microbiol Infect. 2019;25(11):1347–1355. doi: 10.1016/j.cmi.2019.04.025 [DOI] [PubMed] [Google Scholar]
- 7.Fredborg M, Andersen KR, Jorgensen E, et al. Real-time optical antimicrobial susceptibility testing. J Clin Microbiol. 2013;51(7):2047e53. doi: 10.1128/JCM.00440-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Descours G, Desmurs L, Hoang TLT, et al. Evaluation of the accelerate pheno™ system for rapid identification and antimicrobial susceptibility testing of gram-negative bacteria in bloodstream infections. Eur J Clin Microbiol Infect Dis. 2018;37(8):1573e83. doi: 10.1007/s10096-018-3287-6 [DOI] [PubMed] [Google Scholar]
- 9.Idelevich EA, Sparbier K, Kostrzewa M, Becker K. Rapid detection of antibiotic resistance by MALDI-TOF mass spectrometry using a novel direct-on-target microdroplet growth assay. Clin Microbiol Infect. 2018;24(7):738e43. doi: 10.1016/j.cmi.2017.10.016 [DOI] [PubMed] [Google Scholar]
- 10.Wojno KJ, Baunoch D, Luke N, et al. Multiplex PCR based Urinary Tract Infection (UTI) analysis compared to traditional urine culture in identifying significant pathogens in symptomatic patients. Urology. 2020;136:119–126. doi: 10.1016/j.urology.2019.10.018 [DOI] [PubMed] [Google Scholar]
- 11.Mouraviev V, McDonald M. An implementation of next generation sequencing for prevention and diagnosis of urinary tract infection in urology. Can J Urol. 2018;25(3):9349–9356. [PubMed] [Google Scholar]
- 12.Anjum MF, Zankari E, Hasman H. Molecular methods for detection of antimicrobial resistance. Microbiol Spectr. 2017;5(6). doi: 10.1128/microbiolspec.ARBA-0011-2017 [DOI] [PubMed] [Google Scholar]
- 13.Do TT, Tamames J, Stedtfeld RD, et al. Antibiotic resistance gene detection in the microbiome context. Microb Drug Resist. 2018;24(5):542–546. doi: 10.1089/mdr.2017.0199 [DOI] [PubMed] [Google Scholar]
- 14.Boolchandani M, D’Souza AW, Dantas G. Sequencing-based methods and resources to study antimicrobial resistance. Nat Rev Genet. 2019;20(6):356–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fajardo A, Martínez-Martín N, Mercadillo M, et al. The neglected intrinsic resistome of bacterial pathogens. PLoS One. 2008;3(2):e1619. doi: 10.1371/journal.pone.0001619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox G, Wright GD. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int J Med Microbiol. 2013;303(6–7):287–292. doi: 10.1016/j.ijmm.2013.02.009 [DOI] [PubMed] [Google Scholar]
- 17.Bismuth R, Zilhao R, Sakamoto H, Guesdon JL, Courvalin P. Gene heterogeneity for tetracycline resistance in Staphylococcus spp. Antimicrob Agents Chemother. 1990;34(8):1611–1614. doi: 10.1128/AAC.34.8.1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Hoek AH, Mevius D, Guerra B, Mullany P, Roberts AP, Aarts HJ. Acquired antibiotic resistance genes: an overview. Front Microbiol. 2011;2:203. doi: 10.3389/fmicb.2011.00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Girgis HS, Hottes AK, Tavazoie S. Genetic architecture of intrinsic antibiotic susceptibility. PLoS One. 2009;4(5):e5629. doi: 10.1371/journal.pone.0005629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki S, Horinouchi T, Furusawa C. Prediction of antibiotic resistance by gene expression profiles. Nat Commun. 2014;5(1):5792. doi: 10.1038/ncomms6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duran N, Ozer B, Duran GG, Onlen Y, Demir C. Antibiotic resistance genes & susceptibility patterns in staphylococci. Indian J Med Res. 2012;135(3):389–396. [PMC free article] [PubMed] [Google Scholar]
- 22.Vollstedt A, Baunoch D, Wojno KJ, et al. Multisite prospective comparison of multiplex polymerase chain reaction testing with urine culture for diagnosis of urinary tract infections in symptomatic patients. J Sur Urol. 2020:JSU–102. doi: 10.29011/JSU-102.100002 [DOI] [Google Scholar]
- 23.Vollstedt A, Baunoch D, Wolfe A, et al. Bacterial interactions as detected by Pooled Antibiotic Susceptibility Testing (P-AST) in polymicrobial urine specimens. J Sur Urol. 2020:JSU–101. doi: 10.29011/JSU-101.100001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rivoarilala OL, Garin B, Andriamahery F, Collard JM. Rapid in vitro detection of CTX-M groups 1, 2, 8, 9 resistance genes by LAMP assays. PLoS One. 2018;13(7):e0200421. doi: 10.1371/journal.pone.0200421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chérif T, Saidani M, Decré D, et al. Cooccurrence of multiple ampC β-lactamases in Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis in Tunisia. Antimicrob Agents Chemother. 2015;60(1):44–51. doi: 10.1128/AAC.00828-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Queenan AM, Bush K. Carbapenemases: the versatile β-Lactamases. Clin Microbiol Rev. 2007;20(3):440–458. doi: 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bauernfeind A, Schneider I, Jungwirth R, Sahly H, Ullmann U. A novel type of ampC beta-lactamase, ACC-1, produced by a Klebsiella pneumoniae strain causing nosocomial pneumonia. Antimicrob Agents Chemother. 1999;43(8):1924–1931. doi: 10.1128/AAC.43.8.1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto T, Tanaka M, Baba R, et al. Physical and functional mapping of Tn2603, a transposon encoding ampicillin, streptomycin, sulfonamide, and mercury resistance. Molec. Gen. Genet. 1981;181(4):464–469. doi: 10.1007/BF00428737 [DOI] [PubMed] [Google Scholar]
- 29.Tran JH, Jacoby GA. Mechanism of plasmid-mediated quinolone resistance. PNAS. 2002;99(8):5638–5642. doi: 10.1073/pnas.082092899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthew M, Hedges RW, Smith JT. Types of β-lactamase determined by plasmids in gram-negative bacteria. J. Bacteriol. 1979;138(3):657–662. doi: 10.1128/jb.138.3.657-662.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Negri MC, Lipsitch M, Blázquez J, Levin BR, Baquero F. Concentration-dependent selection of small phenotypic differences in TEM beta-lactamase-mediated antibiotic resistance. Antimicrob Agents Chemother. 2000;44(9):2485–2491. doi: 10.1128/AAC.44.9.2485-2491.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis. 2006;42(Suppl 1):S25–34. doi: 10.1086/491711 [DOI] [PubMed] [Google Scholar]
- 33.Kot B. Antibiotic resistance among uropathogenic Escherichia coli. Pol J Microbiol. 2019;68(4):403–415. doi: 10.33073/pjm-2019-048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waller TA, Pantin SAL, Yenior AL, Pujalte GGA. Urinary tract infection antibiotic resistance in the United States. Prim Care. 2018;45(3):455–466. doi: 10.1016/j.pop.2018.05.005 [DOI] [PubMed] [Google Scholar]
- 35.Paul R. State of the globe: rising antimicrobial resistance of pathogens in urinary tract infection. J Glob Infect Dis. 2018;10(3):117–118. doi: 10.4103/jgid.jgid_104_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baerheim A. Empirical treatment of uncomplicated cystitis. BMJ. 2001;323(7323):1197–1198. doi: 10.1136/bmj.323.7323.1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alcock BP, Raphenya AR, Lau TTY, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D517–D525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi SM, Kim SH, Kim HJ, et al. Multiplex PCR for the detection of genes encoding aminoglycoside modifying enzymes and methicillin resistance among Staphylococcus species. J Korean Med Sci. 2003;18(5):631–636. doi: 10.3346/jkms.2003.18.5.631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martineau F, Picard FJ, Lansac N, et al. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000;44(2):231–238. doi: 10.1128/AAC.44.2.231-238.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kime L, Randall CP, Banda FI, et al. Transient silencing of antibiotic resistance by mutation represents a significant potential source of unanticipated therapeutic failure. mBio. 2019;10(5):e01755–19. doi: 10.1128/mBio.01755-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cabot G, Ocampo-Sosa A, Tubau F, et al. Overexpression of ampC and efflux pumps in pseudomonas aeruginosa isolates from bloodstream infections: prevalence and impact on resistance in a spanish multicenter study. Antimicrob Agents Chemother. 2011;55(5):1906–1911. doi: 10.1128/AAC.01645-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patterson J, Hardin T, Kelly C, Garcia R, Jorgensen J. Association of antibiotic utilization measures and control of multiple-drug resistance in Klebsiella pneumonia. Infect Control Hosp Epidemiol. 2000;21(7):455–458. doi: 10.1086/501787 [DOI] [PubMed] [Google Scholar]
- 43.Lee J, Oh C, Choi E, Lee H. The impact of the increased use of piperacillin/tazobactam on the selection of antibiotic resistance among invasive Escherichia coli and Klebsiella pneumoniae isolates. Int. J. Infect. Dis. 2013;17(8):e638–e643. doi: 10.1016/j.ijid.2013.01.030 [DOI] [PubMed] [Google Scholar]