Summary
Ventilation is dependent upon pulmonary alveoli lined by two major epithelial cell types, alveolar type-1 (AT1) and 2 (AT2) cells. AT1 cells mediate gas exchange while AT2 cells synthesize and secrete pulmonary surfactants and serve as progenitor cells which repair the alveoli. We developed transgenic mice in which YAP was activated or deleted to determine its roles in alveolar epithelial cell differentiation. Postnatal YAP activation increased epithelial cell proliferation, increased AT1 cell numbers, and caused indeterminate differentiation of subsets of alveolar cells expressing atypical genes normally restricted to airway epithelial cells. YAP deletion increased expression of genes associated with mature AT2 cells. YAP activation enhanced DNA accessibility in promoters of transcription factors and motif enrichment analysis predicted target genes associated with alveolar cell differentiation. YAP participated with KLF5, NFIB, and NKX2-1 to regulate AGER. YAP plays a central role in a transcriptional network that regulates alveolar epithelial differentiation.
Subject areas: Genetics, Developmental genetics, Molecular biology
Graphical abstract
Highlights
-
•
YAP, TEAD, NKX2-1, KLF5 and NFIB interact to increase expression of AGER
-
•
Active YAP expression induces widespread increased accessibility of chromatin regions
-
•
YAP deletion enhances expression of mature AT2 cell markers
-
•
YAP activation increases expression of AT1 cell related genes and number of AT1 cells
Genetics; Developmental genetics; Molecular biology
Introduction
The respiratory epithelium is formed from embryonic endodermal progenitors that differentiate into a diversity of cell types that vary during development and along the anterior-posterior axis of the lung (Whitsett et al., 2019;Guo et al., 2019). Airways are lined by multiple epithelial cell types including club, goblet, ciliated, ionocytes, tuft, neuroendocrine, and basal cells (Rock et al., 2010;Perl et al., 2002;Rawlins et al., 2007;Cardoso, 2001;Tsao et al., 2009;Reynolds et al., 2002). The alveoli are lined by two major epithelial cell types, alveolar type 1 (AT1) and alveolar type 2 (AT2) cells. AT2 cells secrete surfactant proteins and lipids which serve important roles in innate host defense and in reduction of surface tension at the air/liquid interface. Subsets of AT2 cells also act as progenitor cells, which proliferate to self-renew or differentiate into AT1 cells that are required for efficient gas exchange with endothelial cells of the pulmonary microvasculature (Besnard et al., 2010;Lee et al., 2006;Cutz et al., 2000;Besnard et al., 2009;Bridges et al., 2003;Dahlin et al., 2004;Demling et al., 2006;Kasper et al., 1994;Matsuzaki et al., 2008;Lee et al., 2013;Park et al., 2004;Barkauskas et al., 2013). During lung morphogenesis, Sox9 expressing epithelial cells serve as alveolar progenitors. In the perinatal period, a subset of bi-potent (AT1/AT2) alveolar cells generate mature AT1 or AT2 cells (Rockich et al., 2013;Ustiyan et al., 2016;Guo et al., 2019;Treutlein et al., 2014). While cell turnover and proliferation are not highly active in the mature lung, the lung has a remarkable capacity to repair after injury (Warburton et al., 2001;Vaughan et al., 2015;Zuo et al., 2015;Lechner et al., 2017;Vaughan and Chapman, 2013;Hogan et al., 2014). During regeneration, a subset of Axin2+ AT2 cells act as facultative progenitors of both AT2 and AT1 populations (Zepp et al., 2017;Frank et al., 2016;Zacharias et al., 2018;Nabhan et al., 2018). YAP is dynamically regulated throughout development and regeneration (Isago et al., 2020;Lacanna et al., 2019;Liu et al., 2015;Mahoney et al., 2014;van Soldt et al., 2019), and recent studies demonstrated that Hippo/YAP signaling plays important roles in regulating proliferation and differentiation of both conducting airway and alveolar epithelial cells (Lange et al., 2015;Nantie et al., 2018).
Hippo/YAP signaling plays diverse and important roles in cell proliferation and differentiation to regulate organ size and regeneration. The Hippo/YAP pathway consists of MST1/2 (Stk4/Stk3) that direct phosphorylation of LATS. LATS then phosphorylates YAP to direct it to the cytoplasm, where it is sequestered or marked for 14-3-3 ubiquitination and degradation. In the absence of this phosphorylation, YAP is shuttled to the nucleus where it interacts with transcriptional co-factors, including TEADs 1-4, to regulate transcription of genes associated with cell proliferation, migration, and differentiation (Varelas, 2014;Yu et al., 2015;Zhao et al., 2011;Pfleger, 2017). YAP interacts with several developmental pathways to regulate cell behaviors (Piersma et al., 2015;van Soldt and Cardoso, 2020), including Wnt signaling to control cell proliferation and differentiation (Gokey et al., 2018). In the developing airway, YAP directs basal cell fate decisions, with both nuclear and cytoplasmic YAP playing independent roles (Mahoney et al., 2014;Lange et al., 2015;Zhao et al., 2014;van Soldt et al., 2019). Activation of YAP in primary human bronchial epithelial cells induced AT1 cell signature genes HOPX, PDPN, and AQP5 (Lange et al., 2015). In the embryonic lung, activation of YAP by deletion of LATS1/2 in AT2 cells increased AT1 cell numbers (Nantie et al., 2018). In idiopathic pulmonary fibrosis, a disease associated with failure of alveolar repair, activated YAP interacts with mTOR to direct cell migration and proliferation, contributing to aberrant gene expression in AT2 cells (Gokey et al., 2018). Although YAP is known to regulate AT1 cell differentiation, the underlying mechanisms remained unclear. Therefore, we sought to identify transcriptional mechanisms by which YAP regulates alveolar epithelial proliferation and differentiation in the postnatal lung.
Results
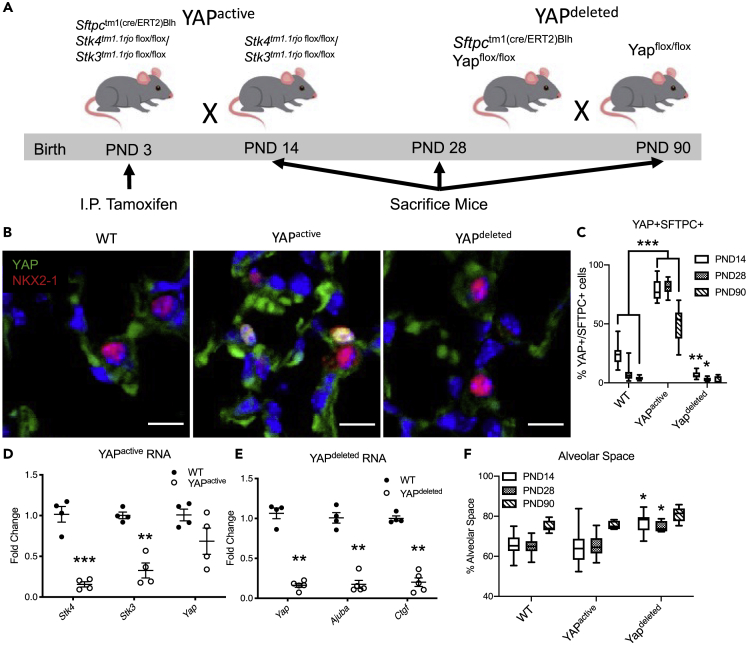
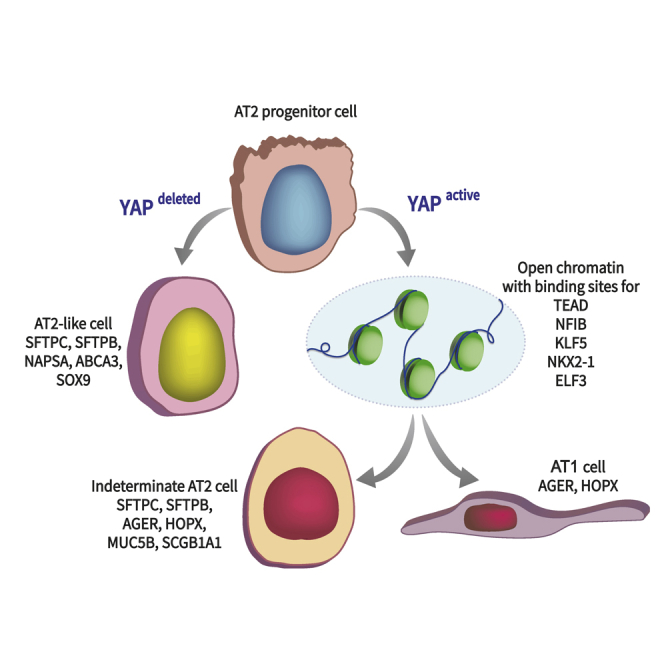
To determine the role of epithelial YAP activation or deletion during postnatal alveologenesis, tamoxifen inducible Sftpctm1(cre/ERT2)Blh driver was used to constitutively activate YAP by deleting Stk3flox/flox/Stk4flox/flox (hereafter referred to as YAPactive) or inhibiting YAP activity by deleting Yapflox/flox (hereafter referred to as YAPdeleted) (Figure 1A). Tamoxifen injected Cre-negative mice were used as wild type (WT) controls. To study the role of YAP during postnatal alveolar development, mice were exposed to tamoxifen at postnatal day 3 (PND3). Lung immunofluorescence staining, and gene expression were examined at PND14, 28, and 90 (Figure 1). Postnatal activation or inhibition of YAP did not affect survival, in contrast to previous findings, demonstrating embryonic lethality after deletion of Stk3/Stk4 in mice (Lange et al., 2015). Nuclear localization of YAP was increased in YAPactive mice. Nuclear YAP was detected in approximately 80% of AT2 cells (Figure 1C) and RNA analysis demonstrated an approximately 90% reduction in Stk3 and Stk4 at PND14 (Figures 1B and 1D). Nuclear YAP was detected in 20% of AT2 cells in PND14 WT mice and rarely detected in older WT or YAPdeleted mice (Figures 1B and 1C). Expression of known YAP transcriptional target genes (e.g., Ajuba and Ctgf) was decreased in CD45-, CD16/32-, Ter119-, CD90.2-, CD271-, CD31-, EPCAM+ cells (hereafter referred to as EPCAM+ or interchangeably as AT2) isolated from YAPdeleted mice, consistent with inhibition of YAP activity (Figure 1E). Alveolar spaces were significantly increased (P<0.05) in YAPdeleted lungs at PND14 and 28. Alveolar spaces in YAPactive lungs were not different from control lungs (Figure 1F).
Figure 1.
YAP activation and deletion in mouse AT2 cells
(A) Strategy and timeline of inducible deletion or activation of YAP. Mice were sacrificed at postnatal day (PND) 14, 28 and 90 for analysis.
(B) Immunofluorescence analysis of YAP (green) and NKX2-1 (red) and quantification of SFTPC+ AT2 cells with YAP+ nuclei at PND 14, 28, and 90 demonstrating increased nuclear YAP in YAPactive mouse lungs.
(D and E) qRT-PCR analysis of YAP-related genes in isolated EPCAM+ cells following YAP activation (D) or deletion (E).
(F) Quantification of alveolar space in WT, YAPactive and YAPdeleted mouse lungs at PND14, 28, and 90 demonstrating YAPdeleted mouse lungs have increased alveolar space. ∗ Indicates p < 0.05 as determined by two-way ANOVA followed by Sidak's multiple comparison test. Dot plot graph error bars represent S.E.M and whiskers of Box–Whisker plots represent min and max values. ∗∗p<0.001, ∗∗∗p<0.0001.
YAP activation and deletion alters the alveolar epithelial cell populations invivo
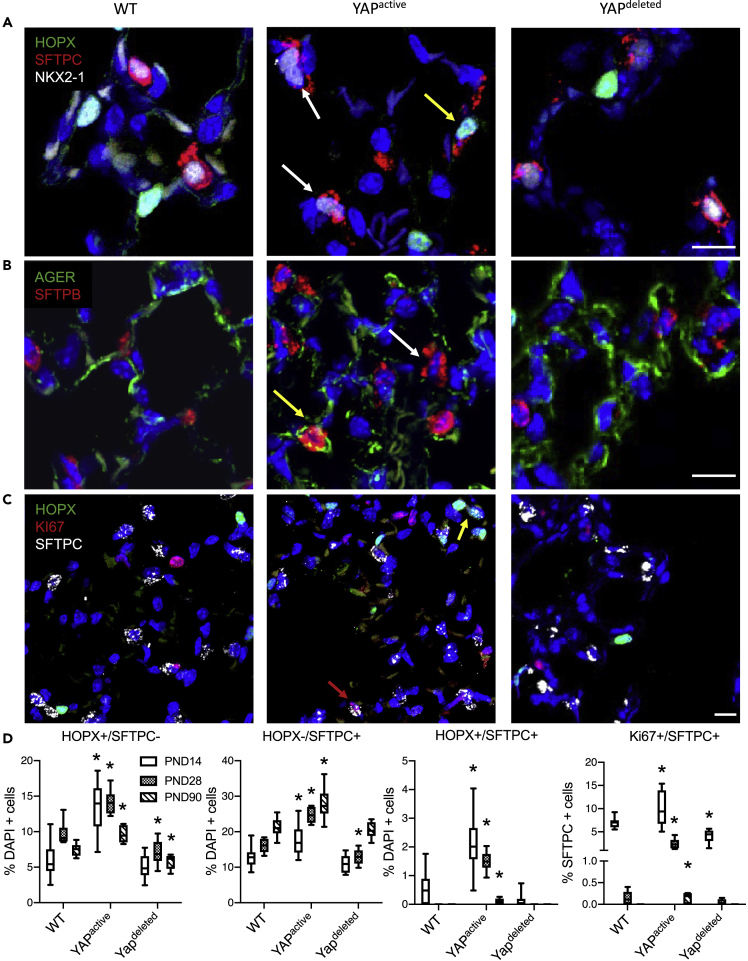
To identify the role of alveolar epithelial YAP activity during postnatal development, immunofluorescence staining of HOPX, AGER, SFTPC, SFTPB, and KI67 was assessed in mice at PND14, 28, and 90 (Figure 2). Proliferation of SFTPC+ cells, identified by KI67+ expression, was increased at PND14 and was sustained thereafter in YAPactive mice, times at which KI67+ cells were rarely detected in control mice. Activation of YAP increased numbers of NKX2-1+ SFTPC+SFTPB+ (AT2), and AGER+HOPX+ (AT1) cells (Figures 2A–2C). Along with AT2 and AT1 cells, abnormal alveolar epithelial cell subtypes were identified in YAPactive mice. Atypical HOPX+AGER+SFTPC+ AT1/AT2 “dual positive” cells were present in YAPactive lungs that persisted through PND90, perhaps indicating incomplete differentiation (Figure 2D). Atypical AT2 cell doublets of SFTPC+ cells were identified in YAPactive mice, of which one cell generally exhibited readily detectable KI67+ staining. These findings are consistent with the concept that YAP activates alveolar progenitor cell proliferation and that increased activity of YAP prevents normal AT2 and AT1 cell differentiation. In contrast, proliferation was significantly decreased at PND14 and the numbers of HOPX+ AT1 and SFTPC+ AT2 cells were significantly decreased (P<0.05) in YAPdeleted mice (Figure 2).
Figure 2.
YAP regulates alveolar epithelial cell differentiation
(A–C) Immunofluorescence analysis of proliferation (KI67), AT1 (HOPX, AGER), AT2 (SFTPC, SFTPB), and epithelial (NKX2.1) cell markers to assess alveolar epithelial cell proliferation and differentiation in YAPactive and YAPdeleted mouse lungs.
(D) Quantification of HOPX+/SFTPC- AT1, HOPX-/SFTPC+ AT2, HOPX+/SFTPC+ “dual positive” (yellow arrow) and KI67+/SFTPC+ proliferating AT2 cells (red arrows) in PND14 (N = 11 WT, 12 YAPactive, 10 YAPdeleted), 28 (N = 9 WT, 9 YAPactive, 8 YAPdeleted), and 90 (N = 8 WT, 8 YAPactive, 8 YAPdeleted). SFTPC+ “doublets” are indicated with white arrows. Whiskers of box-whisker plots represent min and max values. ∗Indicates p<0.05 as determined by Two-way ANOVA followed by Sidak's multiple comparisons test.
YAP activation and deletion alters the alveolar epithelial cell populations invitro
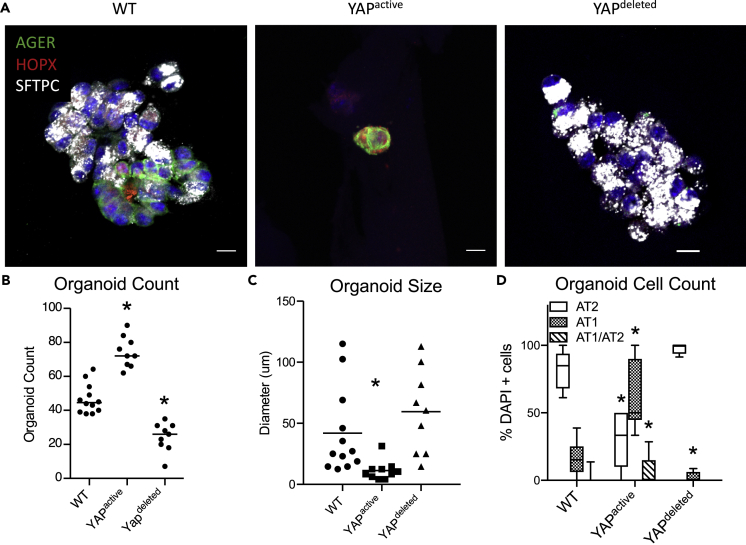
Alveolar organoids were produced from EPCAM+ cells isolated from YAPactive, YAPdeleted, and WT mice (cell population analyses available in Figure S1). EPCAM+ cells were isolated from mice of each genotype at PND14 and cultured for 21 days. Immunofluorescence analysis of SFTPC, AGER, and HOPX demonstrated that organoids produced from YAPactive EPCAM+ cells had increased numbers of AT1 cells, whereas a mixture of AT2 (SFTPC+) and AT1 (AGER + HOPX+) cells were produced from WT mice (Figure 3A). Organoid forming efficiency and size were assessed. YAPactive AT2 cells generated more organoids that were smaller than those produced from WT mice (Figures 3B and 3C). EPCAM+ cells isolated from YAPdeleted mice produced fewer organoids than those from WT cells. YAPdeleted EPCAM+ cells produced organoids that were enriched for SFTPC+ cells and lacked HOPX+ AT1 cells (Figure 3D).
Figure 3.
YAP regulates growth and differentiation of alveolar organoids
(A) Immunofluorescence analysis of AGER (green), HOPX (red), and SFTPC (white) of organoids generated with WT control, YAPactive, or YAPdeleted EPCAM+ cells.
(B) Quantification of organoids generated per well shows YAPactive cells (N = 9) generate more organoids, while YAPdeleted cells (N = 9) produced fewer than WT cells (N = 11).
(C) Analysis of average organoid size per well demonstrates YAPactive cells generated smaller organoids than those produced with YAPdeleted or WT EPCAM+ cells.
(D) Quantification of cell-type specific markers demonstrate a shift in alveolar epithelial cell differentiation with more AT1 cells present in YAPactive and less AT1 cells in YAPdeleted organoids. Whiskers of box-whisker plots represent min and max values. See also Figure S1 for AEC purity during cell preparation. ∗Indicates p<0.05 as determined by Two-way ANOVA followed by Sidak's multiple comparisons test.
Prediction of an alveolar epithelial cell transcriptional regulatory network
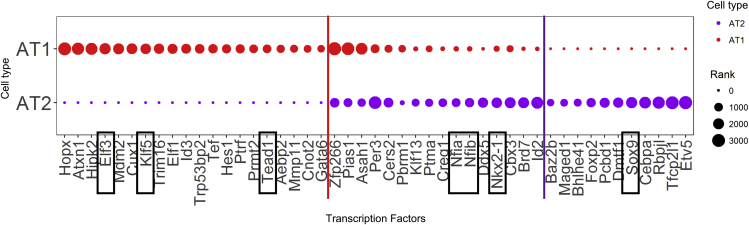
To further identify factors controlling AT1 and AT2 cell differentiation, we interrogated single-cell RNA sequencing data from E18.5 fetal mouse lung, a time of active AT1 and AT2 cell differentiation, to identify genes selectively expressed in AT1 and AT2 cells (Guo et al., 2019). We identified 401 AT1 associated genes and 403 AT2 associated genes (p <0.05 and fold change >1.5). Cell specific Transcription Regulatory Networks (TRN) were constructed using RNAs selectively expressed as potential targets and transcription factors expressed as potential regulators in AT1 or AT2 cells (Figure S2). The importance of a given transcription factor in the TRN was determined by measuring the combination of centrality and disruption using SINCERA (Guo et al., 2015). Transcription factors predicted to control AT2 cell differentiation included Etv5, Sox9, Rbpj1 and Tfcp2l1. Transcription factors including Nkx2-1 and chromatin modulators Nfia and Nfib, were predicted as key regulators in both AT1 and AT2 cells. Hopx, Elf3a, Klf5 and the YAP cofactor, Tead1, were predicted to regulate AT1 cell differentiation (Figure 4). The presence of Tead1 in the AT1 TRN supported the concept that YAP, via TEAD activity, may interact in a transcriptional network regulating AT1 cell gene expression and differentiation.
Figure 4.
Predicted alveolar epithelial cell transcriptional network
Prediction of alveolar epithelial cell transcriptional regulatory network (TRN) from previously published single cell RNA sequencing data. AT1 or AT2 specific differentially expressed genes were used as potential target genes and transcriptional factors commonly or selectively expressed in AT1 or AT2 were identified as potential transcription factor regulators. Transcription factors were ranked based on their nodes importance to the inferred AT1 or AT2 TRNs using a method combining the six node importance metrics as described in SINCERA, with a higher rank score corresponding to a transcription factor being more important to a given TRN. The transcription factors Elf3, Klf5 and Tead1 were predicted as AT1 selective, and Sox9 and Etv5 were AT2 selective. Nkx2-1, Nfia and Nfib were predicted as important in both AT1 and AT2 cells. See also Figure S2 for network information.
YAP deletion enhances expression of mature AT2 cell markers
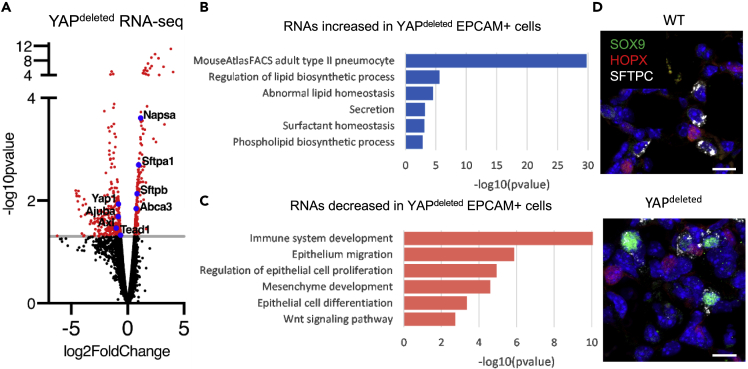
To assess the role of YAP in the regulation of AT2 cell proliferation and AT1/AT2 cell differentiation, RNA and Assay for Transposase Accessible Chromatin (ATAC) sequencing were performed using EPCAM+ cells isolated from YAPactive, WT, and YAPdeleted mouse lungs at PND14. ATAC-seq of isolated EPCAM+ cells demonstrated that deletion of YAP had little effect on chromatin accessibility (Figure S3A), consistent with the paucity of Yap expression in normal mature alveolar epithelial cells. RNAs selectively expressed in mature AT2 cells, including Abca3, Sftpb, and Napsa, were increased in YAPdeleted mice (Figure 5A). Deletion of YAP increased expression of genes involved in bioprocesses associated with mature AT2 cells, including “phospholipid biosynthetic process”, “surfactant homeostasis”, and “lipid metabolism” (Figure 5B). Consistent with deletion of Yap, expression of YAP target genes, including Axl, Ajuba, Tead1, and Tead4 was reduced. Genes involved in bioprocesses including “regulation of cell migration”, “positive regulation of cell population proliferation”, and “epithelial cell differentiation”, were suppressed consistent with reduction of YAP activity (Figure 5C). Since YAPdeleted cells expressed high levels of AT2 cell signature genes and SOX9 was predicted to regulate AT2 cell differentiation (Figure 4), lung tissues were stained for SOX9. While all AT1 and AT2 cells identified in WT mice were SOX9 negative, a readily detectable, abnormal population of SOX9+ SFTPC+ “AT2 like” cells was observed in YAPdeleted mice (Figure 5D).
Figure 5.
YAP deletion increases expression of AT2 cell signature genes
RNA-seq analyses were performed on EPCAM+ cells isolated from PND14 YAPdeleted (N = 5) and WT (N = 4) mice.
(A) Volcano plot of the differentially expressed genes (p < .05, FC > 1.5) shows increase in AT2 markers Abca3, Sftpb and Sftpa1, and decrease in known YAP targets Ajuba, Axl and Yap itself.
(B) Functional enrichment analyses of the induced genes associated with lipid and surfactant production.
(C) Functional enrichment analyses of the suppressed genes show decreased gene expression associated with Wnt signaling, Immune and Mesenchyme development, Epithelial cell migration and proliferation and Epithelial cell differentiation.
(D) Immunofluorescence analysis of SOX9 (green) and SFTPC (white) demonstrates the presence of SOX9+/SFTPC+ epithelial cells in YAPdeleted mice. See Figure S3 for further ATAC-seq quality control.
YAP activation increases expression of AT1 and proximal epithelial cell markers in AECs
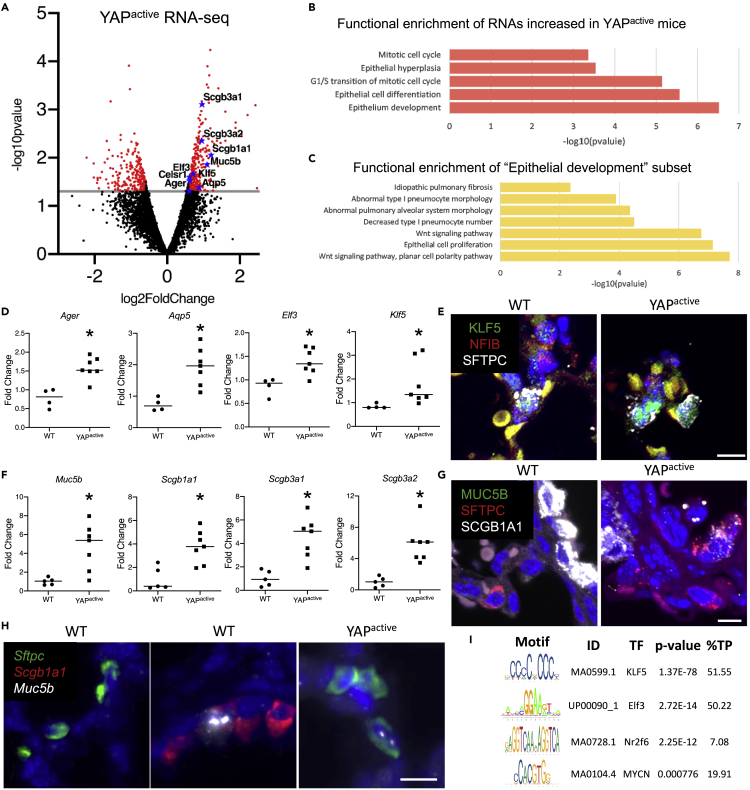
Expression of genes associated with epithelial cell proliferation, including Ccnd1 and Cndk1, were increased in EPCAM+ cells isolated from YAPactive mice. Ager and Aqp5, genes associated with AT1 cell differentiation, were increased (Figures 6A and 6D), consistent with an increased number of AT1 and AT1/AT2 cells in YAPactive mice. Genes induced in YAPactive cells were associated with “epithelial cell differentiation”, “mitotic cell cycle”, “epithelial hyperplasia”, and “epithelial development” (Figure 6B). Within genes involved in “epithelial development”, genes associated with “idiopathic pulmonary fibrosis” and “abnormal type 1 pneumocyte morphology” were increased in YAPactive EPCAM+ cells (Figure 6C). Genes associated with “mesenchymal development” and “extracellular matrix” were decreased in YAPactive cells (Table S1). Predicted AT1 regulators Klf5 and Elf3 RNAs were induced in YAPactive EPCAM+ cells as determined by qRT-PCR and RNAseq (Figures 6A and 6D). Immunofluorescence staining demonstrated that KLF5 was expressed in the “AT2 cell doublets” present in YAPactive mice and in normal AT2 cells (Figure 6E). Expression of genes associated with IPF, including proximal epithelial cell markers Muc5b, Scgb1a1, Scgb3a1, and Scgb3a2 were increased in YAPactive EPCAM+ cells by RNA-seq and qRT-PCR (Figures 6A and 6F). Immunofluorescence staining of SCGB1A1, MUC5B, and SFTPC identified SCGB1A1+/SFTPC+ cells in YAPactive mice, while none were detected in control mice with the antibodies used (Figures 6G and S4) consistent with abnormal epithelial cell differentiation. Analysis of MUC5B protein expression did not demonstrate co-expression in SFTPC+ AT2 cells (Figure S4), however, RNAScope analysis of Muc5b RNA expression showed the presence of Sftpc+/Muc5b+ cells in YAPactive mice (Figure 6H). Sftpc+/Scgb1a1+ cells were identified by RNAscope, perhaps representing BASCs cells, in both WT and YAPactive mice. Motif enrichment analyses of the promoters of genes induced by YAP were enriched for binding sites for transcription factors Klf5, Elf3, Nr2f6, and Mycn and these transcription factors were increased in YAPactive epithelial cells (Figure 6I).
Figure 6.
YAP activation increases expression of AT1 and proximal epithelial cell signature genes
RNA-seq analyses were performed on EPCAM+ cells isolated from D14 YAPactive (N = 6) and controls (N = 4) mice.
(A) Volcano plot of the differentially expressed genes (p < .05, FC > 1.5) shows increase in AT1 markers Ager and Aqp5 as well as proximal lung airway markers Muc5b, Scgb1a1, Scgb3a1 and Scgb3a2.
(B) Genes upregulated in the YAPactive epithelium are associated with mitotic cell cycle and epithelial cell differentiation and development.
(C) Functional enrichment analyses of the subset of genes associated with epithelium development revealed increases in the expression of genes associated with Wnt signaling, epithelium proliferation, IPF and abnormal AT1 differentiation.
(D) Realtime qRT-PCR analyses of isolated YAPactive epithelial cells demonstrate increased Ager, Aqp5, Elf3 and Klf5. ∗Indicates p<0.05 as determined by Welch's t-test.
(E) Immunofluorescence analysis of NFIB (red), KLF5 (green), and SFTPC (white) shows presence of KLF5 in SFTPC+ cells including the SFTPC+ AT2 cell doublets identified in YAPactive mice.
(F) qRT-PCR analysis of proximal cell signature genes demonstrate increased conducting airway epithelial cell markers in YAPactive epithelial cells. ∗Indicates p<0.05 as determined by Welch's t-test.
(G) Immunofluorescence analysis of MUC5B (green), SFTPC (red), and SCGB1A1 (white), shows the presence of SCGB1A1+/SFTPC+ cells in YAPactive mice.
(H) RNAscope Fluorescent in-situ hybridization of Muc5b, Scgb1a1, and Sftpc in WT and YAPactive mouse lungs demonstrating the presence of Sftpc+/Muc5b+ cells in YAPactive lungs.
(I) Promoter analyses of the genes increased in the YAPactive epithelium show that transcription factors Klf5, Elf3, Nr2f6, and Mycn RNAs are increased and have enriched binding sites in YAPactive induced genes. See Table S1 for pathways down regulated in YAPactive mice and Figure S4 for immunofluorescence images.
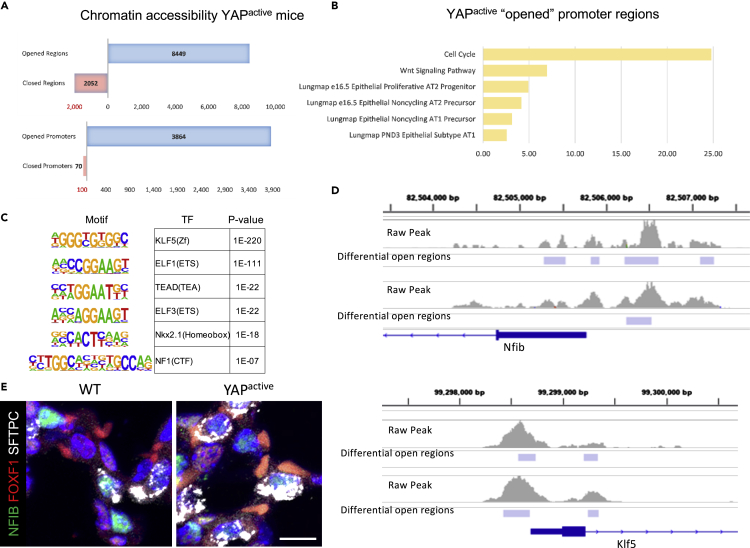
Active YAP expression induces widespread increased accessibility of chromatin regions
Since YAP influenced expression of many genes involved in pulmonary epithelial cell differentiation, we tested YAP altered chromatin accessibility in EPCAM+ cells using ATAC-seq. YAPactive epithelial cells had increased chromatin accessibility at more than 8400 genomic regions, of which 3864 were located in regions defined as gene promoters. Several genomic regions were less accessible in YAPactive mice, of which only 70 were in gene promoter regions (Figure 7A and S3B). Functional enrichment analyses of promoters opened in YAPactive epithelial cells were mapped to genes associated with “cell cycle”, “proliferative AT2 progenitor”, “AT1 cell precursor”, and “PND3 epithelial subtype AT1” (Figure 7B). Motif enrichment analyses of chromatin regions opened in YAPactive mice were enriched for binding sites for transcription factors KLF5, NKX2.1, ELF3 and NF1, and coincided with increased expression of Klf5 and Elf3 RNAs (Figure 7C). KLF5 and NFIB binding sites were identified in promoter regions of genes with YAP induced chromatin accessibility. Promoter regions of the Klf5 and Nfib genes were also opened (Figure 7D). Since the promoters of genes with altered expression in YAPactive epithelial cells were enriched for NFIB binding sites, immunofluorescence analysis was used to test whether NFIB was expressed in AT2 cells. Nuclear NFIB was detected in a subset of SFTPC+ cells in both YAPactive and WT mice (Figure 7E). Together, these data demonstrate that YAP regulates chromatin accessibility in putative Klf5 and Nfib regulatory regions, both being factors predicted to influence AT2/AT1 cell differentiation.
Figure 7.
YAP regulates chromatin accessibility of promoter regions in genes associated with alveolar differentiation
ATAC-seq was performed on EPCAM+ cells isolated from PND14 YAPactive (N = 3) and WT (N = 3) mice.
(A) YAP activation opened over 8400 regions of DNA and closed 2052 regions (p < .01). Over 3800 gene promoters (1.5kb of predicted transcriptional start site) were opened in YAPactive epithelial cells with only 70 promoters being closed.
(B) Functional enrichment analyses of genes with opened promoters opened in YAPactive mice show increased accessibility of signature genes for various AT1 and AT2 subtypes along with genes involved in Wnt signaling and cell cycle.
(C) Motif enrichment analyses of the regions opened in YAPactive mice show an enrichment for multiple transcription factors that were predicted regulators of the AT1 and/or AT2 cell TRN.
(D) IGV was used to visualize regions opened in Nfib and Klf5 promoter regions in YAPactive epithelial cells. ATAC-seq analyses were done using Mac2 on 2 litters, with YAPactive (N = 3) compared to Stk3flox/floxStk4flox/flox (N = 3) control littermates. Only regions altered in both litters were considered significant.
(E) Immunofluorescence analysis of NFIB (green), FOXF1 (red), and SFTPC (white) demonstrates NFIB in a subset of SFTPC+ cells in both WT and YAPactive mouse lungs. See Figure S3 for analysis of ATAC-seq quality.
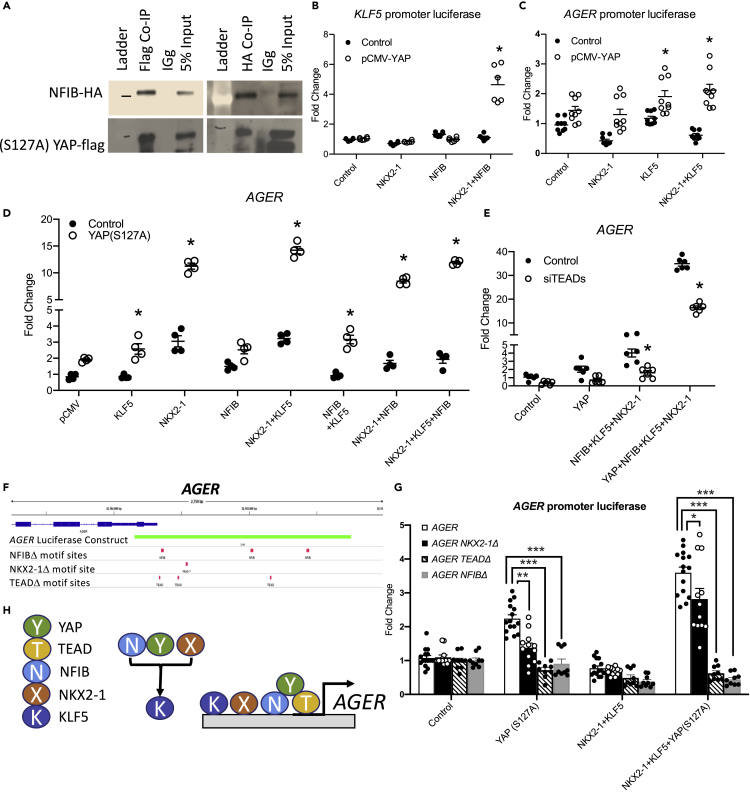
YAP and NFIB directly interact
NFI family members influence chromatin remodeling (Denny et al., 2016); thus, we hypothesized that NFIB may interact with the HIPPO/YAP pathway to open chromatin and influence alveolar epithelial cell gene expression. We co-expressed constitutively active, flag-tagged YAP(S127A) and HA-tagged NFIB in human bronchial epithelial (HBEC3) cells. Co-Immunoprecipitation assays of Flag-YAP(S127A) and HA-NFIB demonstrated that these proteins co-immunoprecipitated (Figures 8A and S6). We were unable to demonstrate that YAP directly interacts with NKX2-1 or KLF5 using available antibodies in HBEC3s; however, direct interactions between YAP and KLF5 (Zhi et al., 2012) and NKX2-1 (Otsubo et al., 2017) were previously shown in MCF10A breast cancer cells and C22 mouse airway progenitor cells, respectively.
Figure 8.
YAP interacts with NFIB, KLF5, and NKX2-1 to regulate gene expression
(A) Immunoprecipitation assay of HBEC3 cells co-expressing (S127A)YAP-FLAG and GFP-NFIB-HA constructs demonstrating YAP and NFIB co-precipitate.
(B) HBEC3 cells co-expressing (S127A)YAP, KLF5, and NKX2-1 activate KLF5 promoter luciferase activity.
(C) HBEC3 cells co-transfected with (S127A)YAP, NKX2-1, and KLF5 activate AGER promoter luciferase.
(D) HBEC3 cells co-expressing empty pCMV vector, KLF5, NKX2-1, NFIB, and (S127A)YAP increase AGER RNA assessed by qRT-PCR.
(E) AGER RNA was measured in HBEC3 cells co-transfected with pCMV empty vector (control), (S127A)YAP, NFIB, KLF5, and NKX2-1 with siRNAs targeting TEADs 1-4. AGER induction is partially inhibited by TEAD siRNAs. Graphs are representative of multiple (N > 3) experiments.
(F) A schematic of the AGER promoter and luciferase assay with locations of predicted TEAD, NKX2-1, and NFIB binding sites that were mutated to assess DNA binding of respective transcription factors to the AGER promoter.
(G) HBEC3 cells expressing NKX2-1, KLF5, and YAP in the presence of AGER luciferase constructs with five site mutations in predicted binding sites of TEAD (TEADΔ), NKX2-1 (NKX2-1Δ) or NFIB (NFIBΔ). Altering the predicted binding sites of NKX2-1 (p < 0.05), TEAD (p < 0.0001), or NFIB (p < 0.0001) significantly reduced AGER promoter activation by YAP, KLF5, and NKX2-1.
(H) A schematic of NFIB, KLF5, NKX2-1, YAP, and TEAD interacting on the AGER promoter to induce AGER transcription. See Figure S5 for further information and Figure S6 for full size co-immunoprecipitation blots. ∗Indicates p<0.05 as determined by Two-way ANOVA followed by Sidak's multiple comparisons test. ∗∗p<0.001, ∗∗∗p<0.0001.
YAP, NKX2-1, and NFIB induce KLF5 promoter activity
Since NFIB binding sites were identified in the promoter regions of the Klf5 gene that were opened in YAPactive mice, and Klf5 RNA was increased in YAPactive mice, we sought to identify whether YAP or NFIB regulated Klf5 promoter activity. A KLF5 luciferase construct containing 2kb of the KLF5 promoter region was used. Multiple TEAD binding sites were identified within this 2kb promoter region, along with predicted NKX2-1 and NFIB binding sites (Figure S5A and S5B). While NFIB and NKX2-1 expression was not sufficient to induce KLF5-luciferase activity, co-expression of NFIB, NKX2-1, and constitutively active (S127A)YAP markedly increased KLF5-luciferase activity demonstrating that YAP interacts with NKX2-1 and NFIB to regulate the KLF5 promoter in vitro (Figure 8B). Consistent with this finding, meta-analysis of publicly available YAP1 ChIP-seq data demonstrated that YAP binds to the KLF5 promoter at a site 3kb upstream of the KLF5 transcriptional start site in NCI H2052 and MDA MB 231 cells (Stein et al., 2015) (Figure S5B).
YAP, TEAD, NKX2-1, and NFIB interact to increase expression and promoter activity of the AT1 cell marker AGER
Since present findings supported a role for YAP and KLF5 in promoting AT1 cell-associated gene expression, we tested their effect on the expression of AGER, a known AT1 cell signature gene. Conserved KLF5, NFIB, NKX2-1, and TEAD binding sites were identified in the AGER promoter region located approximately 1.5kb from the transcriptional start site (Figure S5C). Co-expression of (S127A)YAP and KLF5 enhanced human AGER promoter luciferase activity (Figure 8C) and co-transfection of (S127A)YAP with KLF5 and/or NKX2-1 synergistically increased AGER RNA in HBEC3 cells, a cell line that normally does not express AGER (Figure 8D). To assess whether responses to YAP were mediated by TEAD, TEADs 1-4 were inhibited with siRNA in HBEC3 cells co-transfected with YAP, KLF5, and NKX2-1. The inhibition of TEADs reduced expression of AGER RNA (Figure 8E). To determine the role of the TEAD, NFIB, and NKX2-1 binding sites within the AGER promoter, site-directed mutagenesis was used to alter the binding sites of each transcription factor (Figure 8F). Mutagenesis of either NKX2-1, TEAD, or NFIB binding sites within the AGER promoter inhibited the YAP/NKX2-1/KLF5 induced AGER luciferase activity (Figures 8G and S5C). Taken together these data support a model in which YAP regulates chromatin remodeling and transcriptional activation in alveolar progenitor cells. YAP, in concert with NFIB, influences chromatin accessibility in promoter regions of genes regulating alveolar epithelial cell differentiation. YAP, KLF5, NFIB, and NKX2-1 interacted to activate the AT1 signature gene AGER, a process, at least in part, regulated by TEADs (Figure 8H). Taken together, YAP participates in a transcriptional and chromatin mediated regulatory network with KLF5, NFIB, and NKX2-1 to influence pulmonary epithelial cell proliferation and differentiation.
Discussion
YAP/TEAD interact with a complex of transcription factors
We used single cell RNA-seq data (Guo et al., 2019) to construct a gene regulatory network to predict the nodal importance of transcription factors regulating AT1 cell associated gene expression during late gestation, a time of alveolar epithelial cell proliferation and differentiation. KLF5, NFIB, NKX2-1, and TEAD1, the latter a mediator of YAP transcriptional activity, were strongly associated with AT1 gene expression. These findings are consistent with previous studies demonstrating critical roles for KLF5 (Wan et al., 2008), NFIB (Hsu et al., 2011), and NKX2-1 (Little et al., 2019;Wert et al., 2002) in alveolar formation, maturation, and differentiation. Together with YAP, these transcription factors and their target genes play critical roles in alveolar formation and repair. Present ATAC-seq data and gene expression studies further support the role of a YAP mediated regulatory network, wherein YAP promotes widespread chromatin changes in promoter regions of key transcription factors and predicted targets. Likewise, in vitro studies demonstrated that YAP was required for the coordinated transcriptional activities of NFIB, KLF5, and NKX2-1 to induce the AT1 signature gene AGER partially in a TEAD dependent manner. The transcription factor NKX2-1 plays a major role in regulating respiratory epithelial cell fate decisions. Recent findings demonstrate that NKX2-1 is present in AT1 cells, and deletion of NKX2-1 in AT2 cells leads to loss of respiratory epithelial cell identity, resembling gastrointestinal cells (Little et al., 2019). During the process of revising this work, a recent publication demonstrated findings consistent with the present work showing that NKX2-1 plays different roles in AT1 and AT2 cells, and that as AT1 or AT2 cells “mature”, NKX2-1 interacting partners change and this is in part regulated by YAP in AT1 cells (Little et al., 2021).The potential role of YAP/NKX2-1 interaction is unclear, however, in C22 mouse airway progenitor cells, YAP and NKX2-1 directly interact to negatively regulate the NKX2-1 target Col17a1 (Otsubo et al., 2017). Likewise, NKX2-1 directly binds NFIB in cultured lung epithelial cells (Hsu et al., 2011). Deletion of NFIB in the mouse lung results in alveolar simplification with loss of AT2 and AT1 cell differentiation (Hsu et al., 2011). YAP directly interacts with NFIB to regulate cancer cell proliferation (Pajtler et al., 2019); likewise, YAP and KLF5 directly interact to regulate cardiac progenitor cell proliferation and differentiation (Zhi et al., 2012). In the lung, deletion of KLF5 impairs perinatal lung sacculation and AT1 cell differentiation (Wan et al., 2008). KLF5 regulates epithelial cell differentiation in other organs including the gut (Bell et al., 2011, Bell et al., 2013). KLF5 regulates epithelial cell proliferation in several cancers and cancer cell lines, including A549 cells, and YAP/KLF5 interactions promote breast cancer proliferation (Zhi et al., 2012;Zhao et al., 2018). Collectively these findings strongly suggest that YAP, TEAD, KLF5, NFIB, and NKX2-1 participate in a gene network regulating lung epithelial proliferation and differentiation.
YAP/TEAD directly regulate AT1 cell differentiation
We have demonstrated that YAP regulates the promoter activity and transcription of the AT1 marker gene AGER, in a TEAD-dependent manner that is blocked by altering TEAD binding sites within the AGER promoter. We show that postnatal activation of YAP via deletion on Stk3/Stk4 is sufficient to increase AT1 cell differentiation. Present findings are consistent with previous reports demonstrating increased expression of AT1 signature genes in cultured airway epithelial cells (Lange et al., 2015), and previous reports that activation of YAP, via deletion of LATS1/2 at E17.5 or expression of YAP(S127A) at E15.5, increases numbers of AT1 cells at birth (Nantie et al., 2018). Recent lineage tracing studies and single cell analyses of the embryonic mouse lung demonstrated that during normal development, AT1 cell specification is completed by E17.5 (Frank et al., 2019). During revision, a recent manuscript has demonstrated that YAP knockout in AT1 cells leads to an “exaggerated” AT2 cell phenotype and activation of YAP leads to the presence of cells expressing AT1 and AT2 cell markers consistent with our findings (Penkala et al., 2021). Herein, we demonstrate that activation of YAP in the postnatal lung enhances AT1 cell numbers, indicating that AT1/AT2 specification and differentiation by bipotent progenitors can be activated after birth without injury in the perinatal lung.
YAP regulates progenitor cell fate decisions
We demonstrated that YAP enhanced proliferation and influenced differentiation of alveolar progenitor cells in the postnatal lung. Embryonic activation or deletion of YAP was lethal in transgenic mouse models (Dai et al., 2017; Lange et al., 2015; Mahoney et al., 2014; van Soldt and Cardoso, 2020). Deletion of the YAP homologue TAZ during embryonic development caused alveolar simplification and an emphysema-like phenotype, while deletion of YAP/TAZ impaired AT2 cell proliferation and AT1 cell differentiation (Lacanna et al., 2019; Mitani et al., 2009). Activation of nuclear YAP in cultured human airway cells and genetic models of YAP activation in mouse airway cells caused terminal differentiation and loss of basal progenitor cells (Lange et al., 2015; Mahoney et al., 2014; van Soldt et al., 2019). Nuclear YAP was dynamically regulated after naphthalene mediated depletion of airway club cells, with increased YAP observed during the proliferative phase of recovery followed by its rapid loss after epithelial differentiation and regeneration. YAP deletion during the proliferative phase of recovery caused failure of progenitor cell self-renewal (Lange et al., 2015), indicating the importance of the dynamic regulation of YAP during lung development and repair. Since recent studies demonstrated an important role for Wnt responsive Axin2+ alveolar progenitor cells in the repair of the alveoli following influenza infection (Frank et al., 2016; Zacharias et al., 2018), we assessed changes in chromatin accessibility in promoter regions of Wnt7b and Celsr1, genes mediating Wnt signaling (Wang et al., 2005). YAP enhanced transcription and opened chromatin in promoter regions of both genes, perhaps indicating crosstalk between YAP and Wnt signaling in alveolar epithelial progenitor cell differentiation (Piersma et al., 2015; Wang et al., 2014). YAP is known to regulate the β-catenin destruction complex and activate β-catenin in cancer cell lines (Deng et al., 2018; Azzolin et al., 2014). Conversely, Wnt5a and Wnt3a enhance YAP activity in cancer (Park et al., 2015). The mechanisms by which these developmental pathways intersect to regulate alveolar epithelial cell proliferation and cell fate decisions warrant further study.
Active epithelial YAP signaling induces a transcriptional network associated with IPF
The alveolar epithelium has a remarkable capacity to regenerate after acute injury, wherein subsets of AT2 cells self-renew and rapidly differentiate into mature AT2 and AT1 cells. Failures in normal repair processes contribute to pulmonary fibrosis and tissue remodeling associated with chronic interstitial lung disease including idiopathic pulmonary fibrosis (IPF), chronic obstructive pulmonary disease (COPD), and emphysema. Herein, we identify the Hippo/YAP pathway as an important regulator of alveolar epithelial cell proliferation and differentiation in the postnatal lung. Activation of YAP in in vitro organoids induced AT1 cell differentiation from isolated AT2 cells, while YAP deletion blocked AT1 cell and enhanced AT2 cell differentiation. Activation of YAP during the postnatal period of mouse lung development resulted in abnormal alveolar epithelial cell proliferation and “indeterminate” epithelial cell differentiation. Atypically differentiated cells co-express genes characteristic of AT1, AT2, and conducting airway epithelial cells, genes normally tightly restricted to distinct epithelial cell types, were identified in YAPactive mice. Previous findings demonstrate increased YAP activity and the presence of similar indeterminate, non-lineage restricted epithelial cells in lung tissues from patients with IPF, supporting a role for Hippo/YAP signaling in the pathogenic alveolar remodeling characteristic of interstitial lung diseases (Gokey et al., 2018; Xu et al., 2016). Activation of YAP in-vivo caused widespread chromatin accessibility changes in regulatory regions of transcription factors that regulate pulmonary epithelial cell gene expression. Functional enrichment analysis of YAP-mediated RNA expression and chromatin accessibility predicted a new role for YAP in a transcriptional network consisting of KLF5, NFIB, and NKX2-1 which interact to regulate alveolar epithelial cell differentiation.
Limitations of the study
Defining whether YAP directly or indirectly interacts with KLF5 or NKX2-1 to regulate transcription will be necessary to further elucidate the role of these interactions in AT1 cell differentiation. While YAP directly interacted with KLF5 and TTF1 (Zhi et al., 2012; Otsubo et al., 2017), we were not able to show this using the antibodies and methods we tested. Future experiments testing genetic interactions or ChIP experiments could shed light on how these transcription factors interact with YAP to regulate alveolar epithelial progenitor cell differentiation. Understanding how YAP differentially regulates Wnt-responsive AEPs compared to other AT2 cell subpopulations reveals a limitation within our study. Careful lineage tracing studies following YAP activation or deletion during development or repair models would provide further insight into the role of YAP in specific subsets of AT2 cell populations. The specific role of YAP in AT2 cell subtypes could also be tested utilizing our organoid model. Since the AT2 cells are YAPactive or YAPdeleted at the onset of organoid culture, future experiments are needed to define temporal changes in cell states regulated by YAP. Future organoid culture models could also address a limitation of our in-vitro analysis to assess how YAP, KLF5, NFIB, TEAD, and NKX2-1 interact. While using a more “alveolar” cell type may provide more direct analysis of AT2/AT1 cell differentiation, readily transfectable human alveolar epithelial cell lines are not currently available. We utilized immortalized proximal airway epithelial HBEC3 cells, which provide a readily transfectable cell line to assess the in-vitro interactions of these transcription factors in activating the AT1 cell marker AGER.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| YAP | Seven Hills Bioreagents | WRAB |
| SFTPC | Seven Hills Bioreagents | N/A |
| SFTPC | Seven Hills Bioreagents | 1231: RRID:AB_451721 |
| SFTPC | Santa Cruz | SC-7706:RRID:AB_2185507 |
| HOPX | Santa Cruz | SC-398703:RRID:AB_2687966 |
| AGER | R&D systems | AF1145:RRID:AB_354628 |
| NKX2-1 | Seven Hills Bioreagents | 1231:RRID:AB_2832953 |
| NKX2-1 | Seven Hills Bioreagents | N/A |
| SFTPB | Seven Hills Bioreagents | GP20 |
| KI67 | BD Biosciences | 556003:RRID:AB_396287 |
| SOX9 | Millipore | AB-5535: RRID:AB_2239761 |
| KLF5 | Seven Hills Bioreagents | N/A |
| NFIB | Novus Bio | NBP1-81000:RRID:AB_11027763 |
| MUC5B | Santa Cruz | SC-20119:RRID:AB_2282256 |
| SCGB1A1 | Seven Hills Bioreagents | N/A |
| FOXF1 | R&D systems | AF 4798:RRID:AB_2105588 |
| FLAG | Cell Signaling | 8146s:RRID:AB_10950495 |
| FLAG | Cell Signaling | 14793S:RRID:AB_2572291 |
| HA | Cell Signaling | 2362s:RRID:AB_2890916 |
| Deposited data | ||
| Yap_active_Atac1 | GSE154527 | GSM4672910 |
| Yap_active_Atac2 | GSE154527 | GSM4672911 |
| Yap_active_Atac3 | GSE154527 | GSM4672912 |
| Yap_active_control_Atac1 | GSE154527 | GSM4672913 |
| Yap_active_control_Atac2 | GSE154527 | GSM4672914 |
| Yap_active_control_Atac3 | GSE154527 | GSM4672915 |
| Yap_active_control_RNAseq1 | GSE154527 | GSM4672916 |
| Yap_active_control_RNAseq2 | GSE154527 | GSM4672917 |
| Yap_active_control_RNAseq3 | GSE154527 | GSM4672918 |
| Yap_active_control_RNAseq4 | GSE154527 | GSM4672919 |
| Yap_active_RNAseq1 | GSE154527 | GSM4672920 |
| Yap_active_RNAseq2 | GSE154527 | GSM4672921 |
| Yap_active_RNAseq3 | GSE154527 | GSM4672922 |
| Yap_active_RNAseq4 | GSE154527 | GSM4672923 |
| Yap_active_RNAseq5 | GSE154527 | GSM4672924 |
| Yap_active_RNAseq6 | GSE154527 | GSM4672925 |
| Yap_deleted_Atac1 | GSE154527 | GSM4672926 |
| Yap_deleted_Atac2 | GSE154527 | GSM4672927 |
| Yap_deleted_control_Atac1 | GSE154527 | GSM4672928 |
| Yap_deleted_control_RNAseq1 | GSE154527 | GSM4672929 |
| Yap_deleted_control_RNAseq2 | GSE154527 | GSM4672930 |
| Yap_deleted_control_RNAseq3 | GSE154527 | GSM4672931 |
| Yap_deleted_control_RNAseq4 | GSE154527 | GSM4672932 |
| Yap_deleted_RNAseq1 | GSE154527 | GSM4672933 |
| Yap_deleted_RNAseq2 | GSE154527 | GSM4672934 |
| Yap_deleted_RNAseq3 | GSE154527 | GSM4672935 |
| Yap_deleted_RNAseq4 | GSE154527 | GSM4672936 |
| Yap_deleted_RNAseq5 | GSE154527 | GSM4672937 |
| Experimental models: cell lines | ||
| HBEC3KT | Dr. John D. Minna | Multiple repositories:RRID:CVCL_X491 |
| Experimental models: organisms/strains | ||
| Sftpctm1(cre/ERT2)BlhStk3f/f/Stk4f/f | Generated through crossing below lines in house | Requestable |
| Sftpctm1(cre/ERT2)BlhYapf/f | Generated through crossing below lines in house | Requestable |
| Sftpctm1(cre/ERT2)Blh | Dr. Brigid Hogan, now available at The Jackson Laboratory | 028054:RRID:IMSR_JAX:028054 |
| Stk3f/f/Stk4f/f | Dr. Randy L. Johnson/The Jackson Laboratory | 017635:RRID:IMSR_JAX:017635 |
| Yapf/f | The Jackson Laboratory | 027929:RRID:IMSR_JAX:027929 |
| Oligonucleotides | ||
| YAP1 | ThermoFisher | Hs00902712_g1 |
| KLF5 | ThermoFisher | Hs00156145_m1 |
| AGER | ThermoFisher | Hs00542584_g1 |
| Yap1 | ThermoFisher | Mm01143263_m1 |
| Klf5 | ThermoFisher | Mm00456521_m1 |
| Nfib | ThermoFisher | Mm01257777_m1 |
| 18S | ThermoFisher | 4352930E |
| Nkx2-1 | ThermoFisher | Mm00447558_m1 |
| Ajuba | ThermoFisher | Mm00495049_m1 |
| Ctgf | ThermoFisher | Mm01192933_g1 |
| Stk4 | ThermoFisher | Mm00490480_m1 |
| Stk3 | ThermoFisher | Mm00451755_m1 |
| Ager | ThermoFisher | Mm01134790_g1 |
| Aqp5 | ThermoFisher | Mm00437578_m1 |
| Elf3 | ThermoFisher | Mm01295975_m1 |
| Muc5b | ThermoFisher | Mm00466391_m1 |
| Scgb1a1 | ThermoFisher | Mm00442046_m1 |
| Scgb3a1 | ThermoFisher | Mm00446493_m1 |
| Scgb3a2 | ThermoFisher | Mm00504412_m1 |
| Recombinant DNA | ||
| pCAGG:NFIB2:HA | Addgene | 112700:RRID:Addgene_112700 |
| pCDNA:KLF5 | Reference # 54 | Requestable |
| pCMV-flag (S127A)YAP | Addgene | 27370:RRID:Addgene_27370 |
| pCDNA:NKX2-1 | Reference # 79 | Requestable |
| pGL3:KLF5:Luciferase | Reference # 54 | Requestable |
| pLenti:AGER:Luciferase | ABM | C449 |
| pLenti:AGER NKX2-1Δ:Luciferase | ABM | C047 |
| pLenti:AGER NFIBΔ:Luciferase | ABM | C047 |
| pLenti:AGER TEADΔ:Luciferase | ABM | C047 |
Resource availability
Lead contact
Request for further information, resources and reagents should be directed to the lead contact Jason J. Gokey (Jason.j.gokey@vumc.org) by which request will be fulfilled.
Materials availability
All plasmids and mice within this study were acquired as per the following method section details. Request for further information should be directed to the lead contact.
Additional information
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Mice were housed and bred in accordance with protocols approved by the IACUC of Cincinnati Children's Research Foundation. To generate AT2 cell-specific activation or deletion of YAP, Sftpctm1(cre/ERT2)Blh heterozygous mice (a kind gift from Dr. Brigid Hogan) (Rock et al., 2011) were crossed with Stk3flox/floxStk4flox/flox (Lu et al., 2010) or Yapflox/flox mice (Zhang et al., 2010). SftpcCreert2 heterozygous and WT mouse pups were treated with tamoxifen at PND3. SftpcCreert2Stk3flox/floxStk4flox/flox or SftpcCreert2Yapflox/floxwere considered experimental mice and Stk3flox/floxStk4flox/flox or Yapflox/flox mice were used as tamoxifen injected WT littermate controls. All mice used for this study were not previously used for other procedures and were not used as breeding mice prior to study. Mice used throughout the project are listed as follows: Genotype, age sacrificed, and sex as N = male (female). Yapflox/flox PND14 N = 9(10), SftpcCreert2Yapflox/flox PND14 N = 15(14), Stk3flox/floxStk4flox/flox PND14 N = 10(11), SftpcCreert2Stk3flox/floxStk4flox/flox PND14 N = 16(15). Yapflox/flox PND28 N = 2(2), SftpcCreert2Yapflox/flox PND28 N = 4(4), Stk3flox/floxStk4flox/flox PND28 N = 3(2), SftpcCreert2Stk3flox/floxStk4flox/flox PND28 N = 4(5). Yapflox/flox PND90 N = 2(2), SftpcCreert2Yapflox/flox PND90 N = 4(4), Stk3flox/floxStk4flox/flox PND90 N = 2(2), SftpcCreert2Stk3flox/floxStk4flox/flox PND14 N = 4(4).
Method details
Immunofluorescence analysis
Mouse lungs were fixed in 4% PFA overnight and embedded in paraffin. Lung tissue sections (7μm) were deparaffinized and blocked in a 4% normal donkey serum in Phosphate Buffered Saline-0.1% Triton X-100 (PBST) (blocking agent) for 1 hr. Primary antibodies were diluted in blocking agent and incubated at 4°C overnight. Samples were washed three times in PBST and incubated in 1:200 dilution fluorescently conjugated secondary antibody for 1 hr at room temperature. Samples were washed three times in PBST, counterstained with DAPI for 15 min, washed in PBST three times and mounted in ProLong Gold (Thermofisher) antifade mounting media. Confocal imaging was performed on a Nikon A1R LUNV inverted confocal microscope at 20X and 60X magnification. Image analysis was performed using Nikon Elements software to count cell numbers and analyze alveolar space. RNAScope and YAP+ AT2 cell quantification imaging was acquired using a Keyence BZ-X710 with BZ-X Viewer software with a 40X objective. Image analysis was performed with automated HALO image analysis software. The total number of DAPI-positive and YAP-positive nuclei were quantified. To assess AT2 cell number, DAPI-positive nuclei having SFTPC were quantified using the HALO cell segmentation software feature, and AT2 cells having YAP-positive nuclei were quantified to identify the number of YAP active AT2 cells.
RNAScope
RNAScope technology (ACDBio) was used to perform fluorescent RNA in situ hybridization (RNA ISH) experiments according to manufacturer’s instructions. RNAScope probes against Sftpc (314101-C4), Scgb1a1 (420351-C3), and Muc5b (47991) were used.
Alveolar epithelial cell enrichment
Whole mouse lungs were incubated in dispase followed by addition of DNase and dissociation using a gentleMACS. Cells were centrifuged, resuspended in DMEM and passed through a 40μm filter to obtain single cell suspensions from the whole lung. AT2 cells were isolated using MACs magnetic bead (Miltenyi Biotec) with negative selection of CD45 (BioLegend), CD16/32 (Fisher), Ter119 (BioLegend), CD90.2 (BioLegend), CD271 (Miltenyi Biotec), and CD31 (BioLegend) (Corti et al., 1996), followed by positive selection of CD326 (Miltenyi Biotec). Due to subsequent identification of AT2 cells expressing markers associated with other epithelial cell types in YAPactive and YAPdeleted mice, isolated AT2 cells are interchangeably referred to as “EPCAM+ cells”.
Organoid culture
Organoids were produced by combining 5000 AT2 cells with 50,000 fibroblasts (isolated from PND14 wild type C57Bl6 mice and cultured for 3 passages). Cells were mixed in a 1:1 ratio with Matrigel and plated in 24 well plate transwell inserts. Cells were cultured for the first 24 hr with SAGM (Lonza) + Rock inhibitor (Y27632 Sigma), and subsequently cultured in SAGM without Rock inhibitor for 21 days. Organoids were then fixed in 4% PFA overnight, paraffin embedded, and processed for histological analysis. Brightfield images of organoid wells are available in Figure S1M.
Cell culture and transfections
HBEC3-KT (referred to as HBEC3) human bronchial epithelial cells were a kind gift from Dr John D. Minna (UT Southwestern) (Ramirez et al., 2004). HBEC3 cells were plated at 1.2x105 cells per well in 12-well culture plates and cultured in KSFM (Gibco, ThermoFisher). Cells were transfected at 50-60% confluence with plasmid DNA and Fugene HD (Promega) according to manufacturer’s instructions. Plasmids used were acquired as follows: pCAGG:NFIB2:HA (112700 Addgene), pCDNA:KLF5 (Wan et al., 2008), pCMV-flag (S127A)YAP (27370 Addgene) (Zhao et al., 2007), pCDNA:NKX2-1 (Maeda et al., 2006), pGL3:KLF5:Luciferase, pLenti:AGER:Luciferase (ABM). Site-mutagenesis of 5 base pairs within respective predicted transcription factor binding sites was performed by ABM Inc., to obtain Ager NKX2-1Δ, AGER TEADΔ, and AGER NFIBΔ from the parent pLenti:AGER:Luciferase construct. Cells were lysed for assays 48 hr after transfection.
Co-immunoprecipitation
HBEC3 cells were cultured at 2.4X105 in 6 well plates and transfected with pCMV-flag (S127A)YAP and/or pCAGG:NFIB-HA. Cells were lysed with 100μL of extraction buffer B as per Dynabeads protocol (Thermofisher). Samples were split into 3 equal groups and 5% input was saved. Samples were incubated overnight at 4°C with magnetic beads conjugated to HA, FLAG, or mouse IgG and the bound fraction was separated using magnetic μ-columns and eluted per manufacturer’s protocol (Miltenyi Biotec). Samples were separated on 10-20% Tris-glycine gradient gels (Thermofisher) and transferred onto PVDF membranes (Millipore Sigma) using an iBlot Gel Transfer Device (Thermofisher). Membranes were blocked with 5% milk/Tris Buffered Saline-0.1% Tween-20 (TBST) and incubated in primary antibody at 1:1000 overnight at 4°C. Blots were washed three times in TBST and incubated with TrueBlot HRP-conjugated secondary antibody at 1:1000 dilution (Rockland).
Gene promoter assays
Gene promoter-luciferase reporter constructs were transfected at 0.5μg/well with an empty vector and/or plasmid DNA expression constructs for transcription factor activators at ratios of 1:3, 1:10, or 1:24 activator:promoter DNA. Cells were harvested 48 hr after transfection by lysing with Luciferase Cell Culture Lysis 5X Reagent (Promega). Luciferase activity was assayed using the AutoLumat Plus (Berthold Technologies). Transfections were performed in triplicate and all experiments were repeated multiple times to generate N > 9 for all assays.
RNA analysis
RNA was extracted from HBEC3 cells using the RNeasy Micro kit (Qiagen) according to manufacturer’s instructions with an on-column DNase I digestion step. RNA was extracted from isolated mouse EPCAM+ cells using an RNAeasy Mini kit (Qiagen) with on-column DNAse I digestion as per manufacturer’s protocol. Sequencing was performed on RNA isolated from mouse EPCAM+ cells. RNA was reverse transcribed into cDNA (HBEC3 and isolated mouse EPCAM+ cells) using an iScript cDNA synthesis kit (Biorad). qRT-PCR assays were performed using either an Applied Biosystems Quantstudio 5 or a StepOne Plus Real-Time PCR System (ThermoFisher).
ATAC-seq analysis
For ATAC-seq analysis, 5X104 cells were used from purified EPCAM+ cells collected from WT, YAPactive or YAPdeleted mouse lungs. Remaining EPCAM+ cells from each mouse were used for RNA-seq. DNA fragments were isolated, barcoded, and sequenced following published protocols (Buenrostro et al., 2015). Sequencing was performed by Genewiz using an Illumina Hiseq 4000 with a 2X150bp sequencing strategy
Bioinformatics
Paired-end ATAC-seq and RNA-seq analyses were performed on EPCAM+ cells isolated from PND14 YAPactive, YAPdeleted and WT mouse lungs. ATAC-seq FASTQ files were processed in Galaxy (Afgan et al., 2018). Reads were trimmed and adaptors removed by TrimGalore prior alignment to 10mm with Bowtie2 (Langmead and Salzberg, 2012). Mitochondrial DNA was removed by using BAM filter (Barnett et al., 2011) and duplicate reads or reads larger than 100bp were filtered out by Picard MarkDuplicates (http://broadinstitute.github.io/picard). For initial ATAC-seq peak identification, individual mutants were compared to their age-matched controls using Homer (1v1, p < .01, FC > 2, YAPdeleted N = 2, Yapfloxlflox N = 1, YAPactive N = 2, Stk3flox/floxStk4flox/flox N = 1). Resulting replicate peak files were merged and annotated. YAPactive ATAC- BAM files were further analyzed with Macs2 using broad peak detections and a p < .01 cutoff. YAPactive (N = 3) were compared to WT (Stk3flox/floxStk4flox/flox N = 3) and litters were analyzed separately to create two differential peak files. Differential peak files were merged using Homer and only differential peaks present in both YAPactive litters were kept. Peaks were then annotated by Homer and separated into peaks within gene promoters or other regulatory regions. Functional enrichment analysis was performed on genes with differentially opened peaks in their promoter regions. Homer’s motif enrichment was performed on open promoter regions to determine enriched transcription factor binding sites. IGV viewer was used to visualize peaks and promoters. MACS2 was used to obtain total number of peaks, ATACseqQC (Ou et al., 2018) was used to calculate TSS enrichment score and bedtools intersect was used to calculate FRiP score. PCA analyses were done on ATAC-seq BAM files using DESeq and gene coding ATAC-seq. Differential peaks were visualized in a volcano plot.
RNA-seq FASTQ files generated from YAPactive (N = 6) and WT littermates (N = 4) or YAPdeleted (N = 5) and WT littermates (N = 4) were trimmed and adaptors were removed by TrimGalore. Trimmed FASTQ files were aligned and sorted by Bowtie2. Sorted BAM files were used to identify differentially expressed genes using DESeq and Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values were calculated by Cufflinks (Trapnell et al., 2010). Differentially expressed genes were identified using a p < .01, FC > 1.5 and FPKM>1 in over half of the replicates in at least one condition being compared. Subsets of differentially expressed genes were analyzed by Toppfun's functional enrichment analyses to predict altered biological functions (Chen et al., 2009). Promoter regions 1.5kb upstream of the predicted transcriptional start site of genes induced in YAPactive mice were analyzed using MEME suite’s AME coupled with Meme Suite’s Motif database. Promoter sequences were downloaded using the UCSC Table browser (Karolchik et al., 2004). Select motif locations in DNA sequences of interest were identified using Meme suites Fimo package with p < 0.0005 as cutoff (Grant et al., 2011). BRB was used for hierarchal clustering and dendrogram generation (Simon et al., 2007). RNA-seq analysis demonstrated background gene expression changes between Yapflox/flox“WT” AT2 cells and Stk3flox/flox/Stk4flox/flox“WT” AT2 cells which prevented direct comparisons between control mice.
AT1 or AT2 specific transcriptional regulatory networks (TRNs) were predicted by analyzing Fluidigm C1 based single-cell RNA sequencing of E18.5 mouse lungs previously sequenced and analyzed (Bridges et al., 2020). To infer the TRNs, AT1 or AT2 specific differentially expressed genes were used as potential target genes and transcriptional factors commonly or selectively expressed in AT1 or AT2 were identified as potential transcription factor regulators. AT1 or AT2 specific target genes were defined as genes with a Welch’s ttest p < 0.05 and fold change >1.5 in AT1 or AT2 when compared to all the other cells. For a given cell type, transcription factors with cell type frequency >70% were considered as commonly expressed and transcription factors with either their first or second top expression value occurring in a cell type coupled with a cell type frequency >40% were identified as selectively expressed. Transcription factors or transcription cofactors were defined based on Genomatix (MatBase 9.1 of Genomatix), IPA (Qiagen) and CIS-BP. Significance of interactions between transcription factors and target genes in a cell type were assessed based on a first-order conditional dependence driving force prediction method we previously developed (Du et al., 2015). The predicted AT1 TRN was constructed with 353 nodes (104 TF’s) and 2757 unique edges that passed the threshold (Sij<0.001). The predicted AT2 TRN was constructed with 363 nodes (53 TF’s) and 1353 unique edges that passed the threshold (Sij<0.001). Transcription factors were ranked based on the importance of the node to the inferred AT1 or AT2 TRNs using a method combining the six node importance metrics (Degree Centrality, Closeness Centrality, Betweenness Centrality, Disruptive Fragmentation Centrality, Disruptive Connection Centrality and Disruptive Distance Centrality) as described in SINCERA (Du et al., 2015).
Quantification and statistical analysis
Quantification and statistical analysis, outside the scope of RNA-seq and ATAC-seq bioinformatic analysis, was performed on GraphPad prism. For all N stated, N is equivalent to number of mice or experimental wells used for an experiment. For all multi-variate analysis, statistical significance was determined using Two-way ANOVA followed by Sidak’s or Tukey’s multiple comparison test. Single variant analysis was performed using Welch’s t test. P-values deemed significant can be found in the figure legend of each figure along with the definition of what error bar and whiskers represent. No samples were excluded from respective analyses of each figure.
Acknowledgments
The authors would like to thank JaymiSemona for assistance in mouse colony maintenance. We would like to thank Julia Bazzano and Chase Taylor for assistance in FACS analysis of isolated cells. This work was made possible by the Pulmonary Fibrosis Foundation, by an independent grant from Boehringer Ingelheim Pharmaceuticals, Inc. who provided the financial support. The authors meet criteria for authorship as recommended by the International Committee of Medical Journal Editors (ICMJE) and were fully responsible for all aspects of the trial and publication development This work was funded by the Pulmonary Fibrosis Foundation Scholars program (JJG), NIH T32 HL007752-24 training grant (JAW, JJG), U01 HL148856 (JAW, YX), U01HL134745 (JAW) and R01HL136722(JAW).
Author contributions
Conceptualization: JJG, JS, AS, JAW; Methodology: JJG, JS, AS; Validation: JJG, AS, JK; Formal Analysis: JJG, JS, AS, PS; Investigation: JJG, AS, JK; Data Curation: JS, PS; Writing Original Draft: JJG, JS, AS, JAW; Writing Review and Editing: JJG, JS, AS, YX, JAW; Visualization: JJG, JS, AS; Supervision: JJG, JAW.
Declaration of interests
The authors declare no competing interests.
Published: September 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102967.
Contributor Information
Jason J. Gokey, Email: jason.j.gokey@vumc.org.
Jeffrey A. Whitsett, Email: jeffrey.whitsett@cchmc.org.
Supplementalinformation
Data and code availability
RNA-seq and ATAC-seq data generated in this study are available through GEO, GSE 154527. See key resources table for additional accession information. This paper does not report original code. Requests of any data reported in this paper will be shared by the lead contact upon request.
References
- Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Gruning B.A. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46:W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzolin L., Panciera T., Soligo S., Enzo E., Bicciato S., Dupont S., Bresolin S., Frasson C., Basso G., Guzzardo V. YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158:157–170. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- Barkauskas C.E., Cronce M.J., Rackley C.R., Bowie E.J., Keene D.R., Stripp B.R., Randell S.H., Noble P.W., Hogan B.L. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett D.W., Garrison E.K., Quinlan A.R., Stromberg M.P., Marth G.T. BamTools: AC++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27:1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.M., Zhang L., Mendell A., Xu Y., Haitchi H.M., Lessard J.L., Whitsett J.A. Kruppel-like factor 5 is required for formation and differentiation of the bladder urothelium. Dev. Biol. 2011;358:79–90. doi: 10.1016/j.ydbio.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.M., Zhang L., Xu Y., Besnard V., Wert S.E., Shroyer N., Whitsett J.A. Kruppel-like factor 5 controls villus formation and initiation of cytodifferentiation in the embryonic intestinal epithelium. Dev. Biol. 2013;375:128–139. doi: 10.1016/j.ydbio.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard V., Matsuzaki Y., Clark J., Xu Y., Wert S.E., Ikegami M., Stahlman M.T., Weaver T.E., Hunt A.N., Postle A.D. Conditional deletion of Abca3 in alveolar type II cells alters surfactant homeostasis in newborn and adult mice. Am. J. Physiol. Lung Cell Mol. Physiol. 2010;298:L646–L659. doi: 10.1152/ajplung.00409.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard V., Wert S.E., Stahlman M.T., Postle A.D., Xu Y., Ikegami M., Whitsett J.A. Deletion of Scap in alveolar type II cells influences lung lipid homeostasis and identifies a compensatory role for pulmonary lipofibroblasts. J. Biol. Chem. 2009;284:4018–4030. doi: 10.1074/jbc.M805388200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges J.P., Sudha P., Lipps D., Wagner A., Guo M., DU Y., Brown K., Filuta A., Kitzmiller J., Stockman C. Glucocorticoid regulates mesenchymal cell differentiation required for perinatal lung morphogenesis and function. Am. J. Physiol. Lung Cell Mol. Physiol. 2020;319:L239–L255. doi: 10.1152/ajplung.00459.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges J.P., Wert S.E., Nogee L.M., Weaver T.E. Expression of a human surfactant protein C mutation associated with interstitial lung disease disrupts lung development in transgenic mice. J. Biol. Chem. 2003;278:52739–52746. doi: 10.1074/jbc.M309599200. [DOI] [PubMed] [Google Scholar]
- Buenrostro J.D., Wu B., Chang H.Y., greenleaf W.J. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr.Protoc. Mol. Biol. 2015;109:21 29 1–9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso W.V. Molecular regulation of lung development. Annu. Rev. Physiol. 2001;63:471–494. doi: 10.1146/annurev.physiol.63.1.471. [DOI] [PubMed] [Google Scholar]
- Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti M., Brody A.R., Harrison J.H. Isolation and primary culture of murine alveolar type II cells. Am. J. Respir. Cell Mol. Biol. 1996;14:309–315. doi: 10.1165/ajrcmb.14.4.8600933. [DOI] [PubMed] [Google Scholar]
- Cutz E., Wert S.E., Nogee L.M., Moore A.M. Deficiency of lamellar bodies in alveolar type II cells associated with fatal respiratory disease in a full-term infant. Am. J. Respir. Crit. Care Med. 2000;161:608–614. doi: 10.1164/ajrccm.161.2.9905062. [DOI] [PubMed] [Google Scholar]
- Dahlin K., Mager E.M., Allen L., Tigue Z., Goodglick L., Wadehra M., Dobbs L. Identification of genes differentially expressed in rat alveolar type I cells. Am. J. Respir.Cell Mol. Biol. 2004;31:309–316. doi: 10.1165/rcmb.2003-0423OC. [DOI] [PubMed] [Google Scholar]
- Dai Y., Jablons D., You L. Hippo pathway in lung development. J. Thorac. Dis. 2017;9:2246–2250. doi: 10.21037/jtd.2017.07.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demling N., Ehrhardt C., Kasper M., Laue M., Knels L., Rieber E.P. Promotion of cell adherence and spreading: A novel function of RAGE, the highly selective differentiation marker of human alveolar epithelial type I cells. Cell Tissue Res. 2006;323:475–488. doi: 10.1007/s00441-005-0069-0. [DOI] [PubMed] [Google Scholar]
- Deng F., Peng L., Li Z., Tan G., Liang E., Chen S., Zhao X., Zhi F. YAP triggers the Wnt/beta-catenin signalling pathway and promotes enterocyte self-renewal, regeneration and tumorigenesis after DSS-induced injury. Cell Death Dis. 2018;9:153. doi: 10.1038/s41419-017-0244-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny S.K., Yang D., Chuang C.H., Brady J.J., Lim J.S., Gruner B.M., Chiou S.H., Schep A.N., Baral J., Hamard C. Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell. 2016;166:328–342. doi: 10.1016/j.cell.2016.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Guo M., Whitsett J.A., Xu Y. 'LungGENS': A web-based tool for mapping single-cell gene expression in the developing lung. Thorax. 2015;70:1092–1094. doi: 10.1136/thoraxjnl-2015-207035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D.B., Peng T., Zepp J.A., Snitow M., Vincent T.L., Penkala I.J., Cui Z., Herriges M.J., Morley M.P., Zhou S. Emergence of a wave of Wnt signaling that regulates lung alveologenesis by controlling epithelial self-renewal and differentiation. Cell Rep. 2016;17:2312–2325. doi: 10.1016/j.celrep.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D.B., Penkala I.J., Zepp J.A., Sivakumar A., Linares-Saldana R., Zacharias W.J., Stolz K.G., Pankin J., Lu M., Wang Q. Early lineage specification defines alveolar epithelial ontogeny in the murine lung. Proc. Natl. Acad. Sci. U S A. 2019;116:4362–4371. doi: 10.1073/pnas.1813952116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokey J.J., Sridharan A., Xu Y., Green J., Carraro G., Stripp B.R., Perl A.T., Whitsett J.A. Active epithelial Hippo signaling in idiopathic pulmonary fibrosis. JCI Insight. 2018;3:e98738. doi: 10.1172/jci.insight.98738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C.E., Bailey T.L., Noble W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Du Y., Gokey J.J., Ray S., Bell S.M., Adam M., Sudha P., Perl A.K., Deshmukh H., Potter S.S. Single cell RNA analysis identifies cellular heterogeneity and adaptive responses of the lung at birth. Nat. Commun. 2019;10:37. doi: 10.1038/s41467-018-07770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Wang H., Potter S.S., Whitsett J.A., Xu Y. SINCERA: A pipeline for single-cell RNA-seq profiling analysis. Plos Comput. Biol. 2015;11 doi: 10.1371/journal.pcbi.1004575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B.L., Barkauskas C.E., Chapman H.A., Epstein J.A., Jain R., Hsia C.C., Niklason L., Calle E., Le A., Randell S.H. Repair and regeneration of the respiratory system: Complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15:123–138. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.C., Osinski J., Campbell C.E., Litwack E.D., Wang D., Liu S., Bachurski C.J., Gronostajski R.M. Mesenchymal nuclear factor I B regulates cell proliferation and epithelial differentiation during lung maturation. Dev. Biol. 2011;354:242–252. doi: 10.1016/j.ydbio.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isago H., Mitani A., Mikami Y., Horie M., Urushiyama H., Hamamoto R., Terasaki Y., Nagase T. Epithelial expression of YAP and TAZ is sequentially required in lung development. Am. J. Respir.Cell Mol. Biol. 2020;62:256–266. doi: 10.1165/rcmb.2019-0218OC. [DOI] [PubMed] [Google Scholar]
- Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper M., Haroske G., Muller M. Species differences in lectin binding to pulmonary cells: Soybean agglutinin (SBA) as a marker of type I alveolar epithelial cells and alveolar macrophages in mini pigs. Acta Histochem. 1994;96:63–73. doi: 10.1016/s0065-1281(11)80010-3. [DOI] [PubMed] [Google Scholar]
- Lacanna R., Liccardo D., Zhang P., Tragesser L., Wang Y., Cao T., Chapman H.A., Morrisey E.E., Shen H., Koch W.J. Yap/Taz regulate alveolar regeneration and resolution of lung inflammation. J. Clin.Invest. 2019;129:2107–2122. doi: 10.1172/JCI125014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A.W., Sridharan A., Xu Y., Stripp B.R., Perl A.K., Whitsett J.A. Hippo/Yap signaling controls epithelial progenitor cell proliferation and differentiation in the embryonic and adult lung. J. Mol.Cell Biol. 2015;7:35–47. doi: 10.1093/jmcb/mju046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- lechner A.J., Driver I.H., Lee J., Conroy C.M., Nagle A., Locksley R.M., Rock J.R. Recruited monocytes and type 2 Immunity promote lung regeneration following pneumonectomy. Cell Stem Cell. 2017;21:120–134 e7. doi: 10.1016/j.stem.2017.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Reddy R., Barsky L., Weinberg K., Driscoll B. Contribution of proliferation and DNA damage repair to alveolar epithelial type 2 cell recovery from hyperoxia. Am. J. Physiol. Lung Cell Mol. Physiol. 2006;290:L685–L694. doi: 10.1152/ajplung.00020.2005. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Kim J., Gludish D., Roach R.R., Saunders A.H., Barrios J., Woo A.J., Chen H., Conner D.A., Fujiwara Y. Surfactant protein-C chromatin-bound green fluorescence protein reporter mice reveal heterogeneity of surfactant protein C-expressing lung cells. Am. J. Respir.Cell Mol. Biol. 2013;48:288–298. doi: 10.1165/rcmb.2011-0403OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little D.R., Gerner-Mauro K.N., Flodby P., Crandall E.D., Borok Z., Akiyama H., Kimura S., Ostrin E.J., Chen J. Transcriptional control of lung alveolar type 1 cell development and maintenance by NK homeobox 2-1. Proc. Natl. Acad. Sci. U S A. 2019;116:20545–20555. doi: 10.1073/pnas.1906663116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little D.R., Lynch A.M., Yan Y., Akiyama H., Kimura S., Chen J. Differential chromatin binding of the lung lineage transcription factor NKX2-1 resolves opposing murine alveolar cell fates in vivo. Nat. Commun. 2021;12:2509. doi: 10.1038/s41467-021-22817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Lagares D., Choi K.M., Stopfer L., Marinkovic A., Vrbanac V., Probst C.K., Hiemer S.E., Sisson T.H., Horowitz J.C. Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 2015;308:L344–L357. doi: 10.1152/ajplung.00300.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Li Y., Kim S.M., Bossuyt W., Liu P., Qiu Q., Wang Y., Halder G., Finegold M.J., Lee J.S., Johnson R.L. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl. Acad. Sci. U S A. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y., Hunter T.C., Loudy D.E., Dave V., Schreiber V., Whitsett J.A. PARP-2 interacts with TTF-1 and regulates expression of surfactant protein-B. J. Biol. Chem. 2006;281:9600–9606. doi: 10.1074/jbc.M510435200. [DOI] [PubMed] [Google Scholar]
- Mahoney J.E., Mori M., Szymaniak A.D., Varelas X., Cardoso W.V. The hippo pathway effector Yap controls patterning and differentiation of airway epithelial progenitors. Dev.Cell. 2014;30:137–150. doi: 10.1016/j.devcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y., Besnard V., Clark J.C., Xu Y., Wert S.E., Ikegami M., Whitsett J.A. STAT3 regulates ABCA3 expression and influences lamellar body formation in alveolar type II cells. Am. J. Respir.Cell Mol. Biol. 2008;38:551–558. doi: 10.1165/rcmb.2007-0311OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani A., Nagase T., Fukuchi K., Aburatani H., Makita R., Kurihara H. Transcriptional coactivator with PDZ-binding motif is essential for normal alveolarization in mice. Am. J. Respir. Crit. Care Med. 2009;180:326–338. doi: 10.1164/rccm.200812-1827OC. [DOI] [PubMed] [Google Scholar]
- Nabhan A.N., Brownfield D.G., Harbury P.B., krasnow M.A., Desai T.J. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science. 2018;359:1118–1123. doi: 10.1126/science.aam6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantie L.B., Young R.E., Paltzer W.G., Zhang Y., Johnson R.L., Verheyden J.M., Sun X. Lats1/2 inactivation reveals Hippo function in alveolar type I cell differentiation during lung transition to air breathing. Development. 2018;145 doi: 10.1242/dev.163105. dev163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsubo K., Goto H., Nishio M., Kawamura K., Yanagi S., Nishie W., Sasaki T., Maehama T., Nishina H., Mimori K. MOB1-YAP1/TAZ-NKX2.1 axis controls bronchioalveolar cell differentiation, adhesion and tumour formation. Oncogene. 2017;36:4201–4211. doi: 10.1038/onc.2017.58. [DOI] [PubMed] [Google Scholar]
- Ou J., Liu H., Yu J., Kelliher M.A., Castilla L.H., Lawson N.D., Zhu L.J. ATACseqQC: A bioconductor package for post-alignment quality assessment of ATAC-seq data. BMC Genomics. 2018;19:169. doi: 10.1186/s12864-018-4559-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajtler K.W., Wei Y., Okonechnikov K., Silva P.B.G., Vouri M., Zhang L., Brabetz S., Sieber L., Gulley M., Mauermann M. YAP1 subgroup supratentorial ependymoma requires TEAD and nuclear factor I-mediated transcriptional programmes for tumorigenesis. Nat. Commun. 2019;10:3914. doi: 10.1038/s41467-019-11884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.W., Kim Y.C., Yu B., Moroishi T., Mo J.S., Plouffe S.W., Meng Z., Lin K.C., Yu F.X., Alexander C.M. Alternative Wnt signaling activates YAP/TAZ. Cell. 2015;162:780–794. doi: 10.1016/j.cell.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K.S., Whitsett J.A., diPalma T., Hong J.H., Yaffe M.B., Zannini M. TAZ interacts with TTF-1 and regulates expression of surfactant protein-C. J. Biol. Chem. 2004;279:17384–17390. doi: 10.1074/jbc.M312569200. [DOI] [PubMed] [Google Scholar]
- Penkala I.J., Liberti D.C., Pankin J., Sivakumar A., Kremp M.M., Jayachandran S., Katzen J., Leach J.P., Windmueller R., Stolz K. Age-dependent alveolar epithelial plasticity orchestrates lung homeostasis and regeneration. Cell Stem Cell. 2021 doi: 10.1016/j.stem.2021.04.026. S1934-5909(21)00184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl A.K., Wert S.E., Nagy A., Lobe C.G., Whitsett J.A. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc. Natl. Acad. Sci. U S A. 2002;99:10482–10487. doi: 10.1073/pnas.152238499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger C.M. The Hippo pathway: A master regulatory network important in development and dysregulated in disease. Curr.Top. Dev. Biol. 2017;123:181–228. doi: 10.1016/bs.ctdb.2016.12.001. [DOI] [PubMed] [Google Scholar]
- Piersma B., Bank R.A., Boersema M. Signaling in fibrosis: TGF-beta, WNT, and YAP/TAZ converge. Front.Med. (Lausanne) 2015;2:59. doi: 10.3389/fmed.2015.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez R.D., Sheridan S., Girard L., Sato M., Kim Y., Pollack J., Peyton M., Zou Y., Kurie J.M., Dimaio J.M. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- Rawlins E.L., Ostrowski L.E., Randell S.H., Hogan B.L. Lung development and repair: Contribution of the ciliated lineage. Proc. Natl. Acad. Sci. U S A. 2007;104:410–417. doi: 10.1073/pnas.0610770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds S.D., Reynolds P.R., Pryhuber G.S., Finder J.D., Stripp B.R. Secretoglobins SCGB3A1 and SCGB3A2 define secretory cell subsets in mouse and human airways. Am. J. Respir. Crit. Care Med. 2002;166:1498–1509. doi: 10.1164/rccm.200204-285OC. [DOI] [PubMed] [Google Scholar]
- Rock J.R., Barkauskas C.E., Cronce M.J., Xue Y., Harris J.R., Liang J., Noble P.W., Hogan B.L. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc. Natl. Acad. Sci. U S A. 2011;108:E1475–E1483. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock J.R., Randell S.H., Hogan B.L. Airway basal stem cells: A perspective on their roles in epithelial homeostasis and remodeling. Dis.Model Mech. 2010;3:545–556. doi: 10.1242/dmm.006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockich B.E., Hrycaj S.M., Shih H.P., Nagy M.S., Ferguson M.A., Kopp J.L., Sander M., Wellik D.M., Spence J.R. Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc. Natl. Acad. Sci. U S A. 2013;110:E4456–E4464. doi: 10.1073/pnas.1311847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., Lam A., Li M.C., Ngan M., Menenzes S., Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- Stein C., Bardet A.F., Roma G., Bergling S., Clay I., Ruchti A., Agarinis C., Schmelzle T., Bouwmeester T., Schubeler D., Bauer A. YAP1 exerts its transcriptional control via TEAD-mediated activation of enhancers. Plos Genet. 2015;11 doi: 10.1371/journal.pgen.1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B., Brownfield D.G., Wu A.R., Neff N.F., Mantalas G.L., Espinoza F.H., Desai T.J., Krasnow M.A., Quake S.R. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao P.N., Vasconcelos M., Izvolsky K.I., Qian J., Lu J., Cardoso W.V. Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development. 2009;136:2297–2307. doi: 10.1242/dev.034884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustiyan V., Zhang Y., Perl A.K., Whitsett J.A., Kalin T.V., Kalinichenko V.V. beta-catenin and Kras/Foxm1 signaling pathway are critical to restrict Sox9 in basal cells during pulmonary branching morphogenesis. Dev. Dyn. 2016;245:590–604. doi: 10.1002/dvdy.24393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Soldt B.J., Cardoso W.V. Hippo-Yap/Taz signaling: Complex network interactions and impact in epithelial cell behavior. Wiley Interdiscip. Rev. Dev. Biol. 2020;9:e371. doi: 10.1002/wdev.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Soldt B.J., Qian J., Li J., Tang N., LU J., Cardoso W.V. Yap and its subcellular localization have distinct compartment-specific roles in the developing lung. Development. 2019;146 doi: 10.1242/dev.175810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development. 2014;141:1614–1626. doi: 10.1242/dev.102376. [DOI] [PubMed] [Google Scholar]
- Vaughan A.E., Brumwell A.N., Xi Y., Gotts J.E., Brownfield D.G., Treutlein B., Tan K., Tan V., Liu F.C., Looney M.R. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2015;517:621–625. doi: 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan A.E., Chapman H.A. Regenerative activity of the lung after epithelial injury. Biochim.Biophys. Acta. 2013;1832:922–930. doi: 10.1016/j.bbadis.2012.11.020. [DOI] [PubMed] [Google Scholar]
- Wan H., Luo F., Wert S.E., Zhang L., Xu Y., Ikegami M., Maeda Y., Bell S.M., Whitsett J.A. Kruppel-like factor 5 is required for perinatal lung morphogenesis and function. Development. 2008;135:2563–2572. doi: 10.1242/dev.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Shu W., Lu M.M., Morrisey E.E. Wnt7b activates canonical signaling in epithelial and vascular smooth muscle cells through interactions with Fzd1, Fzd10, and LRP5. Mol.Cell Biol. 2005;25:5022–5030. doi: 10.1128/MCB.25.12.5022-5030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Ye J., Deng Y., Yan Z., Denduluri S., He T.C. Wnt versus Hippo: A balanced act or dynamic duo? Genes Dis. 2014;1:127–128. doi: 10.1016/j.gendis.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D., Tefft D., Mailleux A., Bellusci S., Thiery J.P., Zhao J., Buckley S., Shi W., driscoll B. Do lung remodeling, repair, and regeneration recapitulate respiratory ontogeny? Am. J. Respir.Crit. Care Med. 2001;164:S59–S62. doi: 10.1164/ajrccm.164.supplement_2.2106064. [DOI] [PubMed] [Google Scholar]
- Wert S.E., Dey C.R., Blair P.A., Kimura S., Whitsett J.A. Increased expression of thyroid transcription factor-1 (TTF-1) in respiratory epithelial cells inhibits alveolarization and causes pulmonary inflammation. Dev. Biol. 2002;242:75–87. doi: 10.1006/dbio.2001.0540. [DOI] [PubMed] [Google Scholar]
- Whitsett J.A., Kalin T.V., Xu Y., Kalinichenko V.V. Building and regenerating the lung cell by cell. Physiol. Rev. 2019;99:513–554. doi: 10.1152/physrev.00001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Mizuno T., Sridharan A., Du Y., Guo M., Tang J., Wikenheiser-Brokamp K.A., Perl A.T., Funari V.A., Gokey J.J. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight. 2016;1 doi: 10.1172/jci.insight.90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.X., Zhao B., Guan K.L. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias W.J., Frank D.B., Zepp J.A., Morley M.P., Alkhaleel F.A., Kong J., Zhou S., Cantu E., Morrisey E.E. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature. 2018;555:251–255. doi: 10.1038/nature25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepp J.A., Zacharias W.J., Frank D.B., Cavanaugh C.A., Zhou S., Morley M.P., Morrisey E.E. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell. 2017;170:1134–1148 e10. doi: 10.1016/j.cell.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Bai H., David K.K., Dong J., Zheng Y., Cai J., Giovannini M., Liu P., Anders R.A., PAN D. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Tumaneng K., Guan K.L. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat.Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Wei X., Li W., Udan R.S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., LI L. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Li Y., Qiu W., He F., Zhang W., Zhao D., Zhang Z., Zhang E., Ma P., Liu Y. C5a induces A549 cell proliferation of non-small cell lung cancer via GDF15 gene activation mediated by GCN5-dependent KLF5 acetylation. Oncogene. 2018;37:4821–4837. doi: 10.1038/s41388-018-0298-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Fallon T.R., Saladi S.V., Pardo-Saganta A., Villoria J., Mou H., Vinarsky V., Gonzalez-Celeiro M., Nunna N., Hariri L.P. Yap tunes airway epithelial size and architecture by regulating the identity, maintenance, and self-renewal of stem cells. Dev.Cell. 2014;30:151–165. doi: 10.1016/j.devcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi X., Zhao D., Zhou Z., Liu R., Chen C. YAP promotes breast cell proliferation and survival partially through stabilizing the KLF5 transcription factor. Am. J. Pathol. 2012;180:2452–2461. doi: 10.1016/j.ajpath.2012.02.025. [DOI] [PubMed] [Google Scholar]
- Zuo W., Zhang T., Wu D.Z., Guan S.P., Liew A.A., Yamamoto Y., Wang X., Lim S.J., Vincent M., Lessard M. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature. 2015;517:616–620. doi: 10.1038/nature13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and ATAC-seq data generated in this study are available through GEO, GSE 154527. See key resources table for additional accession information. This paper does not report original code. Requests of any data reported in this paper will be shared by the lead contact upon request.