Abstract
Xupu goose, a breed from Hunan province, produces high quality and quantity of meat and liver. However, its egg production rate is low, with poor reproductive traits but strong broody performance. These characteristics decrease the economic value of Xupu goose significantly. Here, RNA-seq was used to analyze the transcriptome changes of ovaries of Xupu goose at different stages to explore the molecular mechanism of reproduction from the pre-laying period to the broody period. A total of 258 genes were differentially expressed in the 3 stages. These genes are associated with inflammation, reproduction, mutual recognition and adhesion between cells, and cytoskeleton formation, and so on. In particular, we report, for the first time, the expression patterns of MRP126, serglycin, TXNIP, and FZD2 during the pre-laying, egg-laying, and broody periods of goose ovaries. Functional analysis by GO annotation revealed that GO terms were mainly involved in actin, cell signal transduction and regulation, and cellular components. Three pathways, including focal adhesion (gga04510), ECM-receptor interaction (gga04512), and N-Glycan biosynthesis (gga00510), were significantly enriched in the three groups. These findings provide a basis for further exploration of profiles of goose ovaries to improve egg production of Xupu goose.
Key words: Xupu goose, ovarian transcriptome, pre-laying period, laying period, broody period
INTRODUCTION
Goose grows rapidly with low input requirements to yield highly valuable products. China is an important goose production exporter in the world's goose consumer market to meet the high annual demand for goose meat. However, most goose species have long broody periods which are associated with low egg production. As a result, the economic value of the geese industry declines. China has a number of domesticated geese species, with various unique characteristics. Xupu goose, an excellent breed in Hunan province, has excellent performance on fattening and liver production, therefore, is of high economic value (Dai et al., 2016). However, the annual egg production by Xupu goose is only 30 eggs as compared with Huoyan goose (120 eggs/year) and Sichuan goose (70 eggs/year) (Yao et al., 2019). The high broodiness of the Xupu goose significantly decreases the economic benefits of its breeding.
Broodiness, a unique characteristic of most domestic geese, is associated with atrophy of ovaries and fallopian tubes, which consequently terminate egg production (Romanov et al., 2002). Mounting evidence has shown that various environmental factors, such as length of photoperiod (Marsden et al., 1966; Geng et al., 2014) and environmental temperature (Thomason et al., 1976) induce the nesting behavior of female poultry. Estrogen (E2), progesterone (P4), prolactin (PRL), vasoactive intestinal peptide (VIP), dopamine (AD), and follicle-stimulating hormone (FSH) secreted by the hypothalamic-pituitary-gonadal axis (Sharp et al., 1984; Onagbesan et al., 2009; Zhou et al., 2010; Yu et al., 2016b) have direct effects on nesting behavior. Heredity is also a fundamental factor that modulates broodiness. Emerging reports demonstrate that broodiness is a polygenic trait regulated by at least 2 dominant autosomal genes, implicated in multiple signal transduction pathways, including GnRH and Wnt pathways (Yu et al., 2016a; Gumułka et al., 2020). Heritability of nesting sex for waterfowl is only 0.116. Conventional genetic breeding methods have some shortcomings, for example, the lower efficiency of genetic breeding in the later period and an increasing in the brooding ability of offspring. Therefore, it would be imperative to explore the molecular mechanisms underlying reproductive biology of geese. The molecular mechanisms of reproductive biology can be adopted to improve the non-broody goose breeds; this approach would improve the economic value of goose breeding.
RNA-seq is a highly sensitive, high throughput, and genome-wide analysis method compared to conventional DNA microarray analysis. The whole-goose genome map was published in 2015, and it provided a genetic basis for RNA-seq analysis studies on geese breeding. Previous reports indicate that multiple organs such as the pituitary (Ye et al., 2019), uterus, and ovary (Yu et al., 2016b; Liu et al., 2018) regulate the broody behavior. Changes in the ovarian transcriptome during the goose reproductive cycle can be explored with RNA-seq analysis. Although differentially expressed genes, including ND1, heat shock protein 70 (HSP70) and MAPK have been reported in previous studies (Gao et al., 2015), the studies only compared the transcriptional changes of the ovaries at 2 stages of the reproductive cycle, such as egg-laying vs. nesting (Xu et al., 2013), egg-laying vs. pre-egg laying (Kang et al., 2009; Ding et al., 2015), and egg-laying vs. confinement (laying period and ceased period) (Luan et al., 2014).
In the present study, we used next-generation sequencing technology to sequence the transcriptome of ovaries and compare changes in pre-laying, laying, and broody stages. RNA-seq approach was employed to explore the dynamic changes of broody-related genes and pathways in the 3 stages of the reproductive cycle. Candidate regulatory genes on the broody behavior were screened out, and related pathways were evaluated through KEGG enrichment and GO enrichment analyses. The findings provide a basis for breeding low nesting geese.
MATERIALS AND METHODS
Ethics Statement
All animal experimental protocols in this study complied with guidelines for animal welfare and were approved by Jiangsu Administrative Committee for Laboratory Animals (Permission number: SYXK-SU-2007-0005).
Animals, Feeding, and Ovarian Collection
Xupu geese were purchased from the National Gene Bank of Waterfowl Resources (Jiangsu, China). Geese were raised on the ground under the same environmental condition. The goose family breeding dwellings are including outdoor water and land sports field and indoor house. The indoor house is equipped with an automatic all-flock feed and water line device. The geese were fed twice daily by an automatic all-flock feed line (8:00, 16:00) with water and all-flock feed based on their age. The indoor temperature, humidity, and lighting time can be adjusted automatically. The geese were subjected to a standard light regimen of 17 h light (17 L:7D) throughout the experimental period. All geese were fasted and deprived of water 12 h before anesthetization with CO2 following guidelines by Jiangsu laboratory animal welfare. The ovarian samples were obtained from 3 stages of the reproductive cycle (5 geese per group), including the pre-laying period (90 days old, named T1), the egg-laying period (180 days old, named T2), and the broody period (300 days old, named T3) in the afternoon. The whole ovary including the small and large yellow follicles was swiftly sampled and immediately frozen in liquid nitrogen for further analyses.
Total RNA Isolation, cDNA Library Construction, and RNA-seq
Total RNA of follicles was extracted with Triozl reagent (15596-026, Invitrogen, CA) following the manufacturer's protocol. RNA purity and concentration were determined using NanoPhotometer spectrophotometer (NP80, IMPLEN, CA), and Qubit RNA Assay Kit (Q10211, Invitrogen) in Qubit2.0 fluorometer (Q32866, Life Technologies, CA). RNA Nano 6000 Assay Kit (5067-1511, Agilent Technologies, CA) of the Bioanalyzer 2100 system (G2939BA, Agilent Technologies) and 1% agarose gel were used to assess RNA integrity and whether it was degraded.
Total RNA (3 μg) from each sample was purified using poly-T oligo-attached magnetic beads. Pure RNA was fragmented using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X) (E7525S, NEB, MA). AMPure XP system (A63881, Beckman Coulter, Indianapolis, Indiana) was employed to determine cDNA fragment size, and approximately 250 to 300 bp was selected for library construction. Agilent Bioanalyzer 2100 system was used to assess the quality of libraries. Index-coded samples were clustered on a cBot Cluster Generation System (SY-312-2001, Illumina, CA) using TruSeq PE Cluster Kit v3-cBot-HS (PE-401-3001, Illumina). Following cluster generation, library preparations were sequenced on an Illumina Hiseq platform (HiSeq 2500, Illumina) and 125 bp/150 bp paired-end reads were generated.
RNA-Seq Reads Quality Control and Mapping
Raw data in fastq format were processed using in-house Perl scripts to generate clean data for downstream analyses. The content of Q20, Q30, and GC in the clean data was established. We applied Hisat2v2.0.5 to construct the reference genome index and align the clean data to the reference genome. The number of reads mapped to each gene was calculated with the featureCounts v1.5.0-p3 program.
Screening of Differentially Expressed Genes and Enrichment Analysis
Differential expression analysis of the genes in the three groups (5 biological replicates per group) was performed with the DESeq2 R package (1.16.1). The resulting P-values were adjusted using Benjamini and Hochberg's approach to control the false discovery rate. Differential expression of genes was evaluated using the following thresholds: |log2-fold change| of ≥ 1 and an adjusted P-value of ≤ 0.05. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed to determine the key pathways of the differentially expressed genes (DEGs). ClusterProfiler R package was used to test the statistical enrichment of DEGs in KEGG pathways and GO terms. P-values less than 0.05 denoted statistical significance.
Quantitative Real-Time PCR
Seven DEGs were selected randomly to validate the result of high-throughput RNA-seq analysis by quantitative real-time PCR (qRT-PCR) as previously described (Everaert et al., 2017). qRT-PCR analysis was conducted on the ABI 7500 Real-Time PCR System (4351105, Applied Biosystems, Foster City, Cafeteria) with ChamQ SYBR qPCR Master Mix (2 ×) (Q331-02, Vazyme, Nanjing, China). Thermocycling parameters used for qRT-PCR were as follows: 95°C for 10 min, 40 cycles at 95°C for 10 s, 60°C for 40 s, and 95°C for 15 s, followed by a melting curve from 60°C for 60 s, 95°C for 30 s, and 60°C for 15 s. Gene expression values were estimated using the 2−ΔΔCt method and normalized using GADPH.
RESULTS
Transcriptome Analysis of Ovary From the Geese During the Pre-laying period, Egg-laying Period, or Broody Period
RNA-seq method was employed to explore the transcriptional difference of the 3 periods of the reproductive cycle. Total RNA was extracted from ovary of Xupu geese at different ages. Fifteen cDNA sequencing libraries (5 pre-laying period samples, 5 egg-laying period samples, and 5 broody period samples) were prepared and sequenced on the Illumina Hiseq platform. More than 4.1 × 107 clean reads per sample were generated after filtering. The clean reads were characterized by more than 92% of Q30 with a GC content of approximately 50% (Table 1). In addition, more than 82% of clean reads were perfectly mapped to the reference goose genomes (ftp://ftp.ensembl.org/pub/release-101/fasta/anser_cygnoides/) to generate a read count value. All subsequent analyses were based on mapped reads and raw data were submitted to the NCBI database (SUB8312645).
Table 1.
Summary of Illumina RNA-seq data.
| Group | Sample | Total reads | Clean reads | Total mapped Clean data* (Gb) |
Q30 (%) |
GC (%) |
Total map Rate (%) |
|---|---|---|---|---|---|---|---|
| Pre-laying Period |
T1_1 | 53724448 | 52649734 | 7.9 | 92.88 | 50.38 | 85.64 |
| T1_2 | 48537944 | 47388374 | 7.11 | 93.07 | 50.78 | 83.74 | |
| T1_3 | 45525954 | 44780912 | 6.72 | 92.87 | 50.13 | 82.56 | |
| T1_4 | 46500408 | 45719558 | 6.86 | 92.73 | 50.58 | 84.26 | |
| T1_5 | 53724448 | 52649734 | 7.9 | 92.88 | 50.38 | 86.38 | |
| Egg-laying Period |
T2_1 | 45357328 | 44660502 | 6.7 | 92.68 | 50.47 | 83.08 |
| T2_2 | 46630778 | 45276860 | 6.79 | 94.77 | 53.11 | 82.2 | |
| T2_3 | 46595642 | 45170742 | 6.78 | 94.43 | 52.62 | 83.18 | |
| T2_4 | 47682596 | 46355658 | 6.95 | 94.7 | 52.4 | 81.34 | |
| T2_5 | 46931022 | 45638700 | 6.85 | 94.63 | 52.31 | 82.83 | |
| Broody Period |
T3_1 | 43081800 | 41777300 | 6.27 | 93.62 | 51.94 | 85.06 |
| T3_2 | 43133282 | 41865106 | 6.28 | 93.55 | 54.13 | 84.84 | |
| T3_3 | 45730386 | 44495388 | 6.67 | 93.37 | 51.73 | 84.13 | |
| T3_4 | 42388562 | 41154010 | 6.17 | 93.56 | 51.72 | 84.84 | |
| T3_5 | 46141102 | 44868082 | 6.73 | 93.28 | 51.71 | 84.51 |
Clean data were obtained from raw data by removing reads containing adapter, ploy-N and low-quality reads.
Validation of RNA-Seq Data Using qPCR
Following RNA-seq analysis, 7 highly expressed DEGs were selected randomly for further validation with qRT-PCR (Table 2). qRT-PCR analysis data showed that most trends of upregulation or downregulation for the selected 7 genes corroborated with results from RNA-seq analysis (Figure 1). qRT-PCR findings validated the accuracy and reliability of RNA-seq analysis results.
Table 2.
Validation of DGEs by qRT-PCR.
| Gene name | Primers | Description | Product size (bp) | |
|---|---|---|---|---|
| GCK | FP | CGGCACGCTCTACAAGC | glucokinase (hexokinase 4) | 188 |
| RP | GCAAACCTCCCTCCTCCT | |||
| ITGB2 | FP | GGGCTCCTCCACATTTC | integrin 2C beta 2 (complement component 3 receptor 3 and 4 subunit) 2C transcript variant X1 | 102 |
| RP | TTCAGATTGCTGCTCCTTT | |||
| LCP1 | FP | CACAGAGGATGGCAGGA | lymphocyte cytosolic protein 1 (L-plastin) | 129 |
| RP | ATCCCACCAATAGCACAGA | |||
| MPEG1 | FP | CAGAGGCCCCAAGGTTT | macrophage expressed 1 | 140 |
| RP | CATGTCGTGGTGGGTCA | |||
| PTAFR | FP | GCACTGGGGCTTTGTCT | platelet activating factor receptor | 144 |
| RP | GCTGACTTTGACCTGCCT | |||
| STRA6 | FP | GCAGGACAACACATTTCCC | stimulated by retinoic acid 6 | 128 |
| RP | GGCGTTTCACCAGCAAG | |||
| SLA | FP | AGAGTGCCCTTCACTTGC | Src-like-adaptor | 64 |
| RP | TTACCCCTCTGGTTGTCCT | |||
| GAPDH | FP | TGGCATCCAAGGAGTAAGC | house-keeping gene for qRT-PCR | 72 |
| RP | GGGCTCCAACAAAGGGT | |||
Abbreviation: DEGs, differentially expressed genes; qRT-PCR, quantitative real-time PCR.
Figure 1.
Validation of sequencing data by qPRC. Validation of the results for pre-laying period (T1), egg-laying period (T2) and broody period (T3). (A) T1 vs. T2; (B) T1 vs. T3; (C) T2 vs. T3. All data are presented as means ± SEM, x-axis individual genes whereas y-axis represents the fold change in expression determined by RNA-seq (gray bars) or qPCR (black bars). The representative genes are GCK (glucokinase), ITGB2 (integrin subunit beta 2), LCP1 (lymphocyte cytosolic protein 1), MPEG1 (macrophage expressed 1), PTAFR (platelet-activating factor receptor), SLA (Src-like-adaptor), STRA6 (stimulated by retinoic acid 6) and GAPDH (glyceraldehyde-3-phosphate dehydrogenase, house-keeping gene for qPCR).
DEGs in the Pre-laying Period, Egg-laying Period, or Broody Period
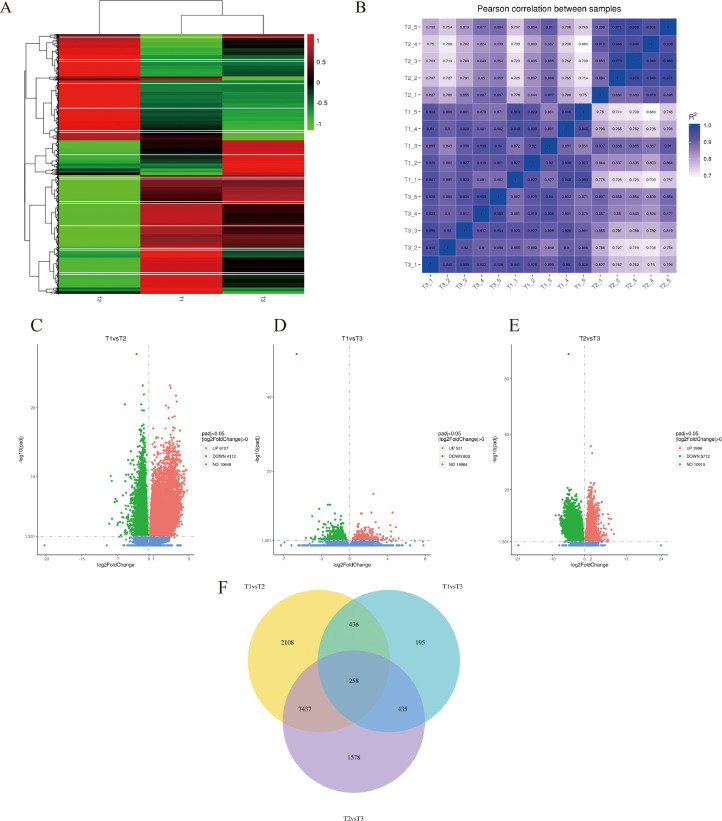
Total RNA was extracted from ovarian tissue of 3 important reproduction periods to give an overview of goose ovarian changes and allow for the exploration of the related gene and pathways of broodiness. A heatmap of DEGs (Figure 2A) and Pearson correlation coefficient of the samples (Figure 2B) demonstrated good sample repeatability in each group. Analysis using DESeq2 R package revealed 6,127 upregulated and 4,112 downregulated DEGs in T1 vs. T2 (Figure 2C), 521 upregulated and 803 downregulated DEGs in T1 vs. T3 (Figure 2D), and 3998 upregulated and 5712 downregulated DEGs inT2 vs. T3 (Figure 2E). A Venn diagram (Figure 2F) depicted common DEGs among T1, T2, and T3 periods. Of the 12,447 DEGs, 258 DEGs showed significant differences between the 3 groups (Supplementary material 1). These 258 DEGs are implicated in multiple physiological pathways, including inflammation, reproduction, mutual recognition and adhesion between cells, and formation of the cytoskeleton. Notably, expression patterns of MRP126, serglycin, TXNIP, and FZD2 genes have, for the first time, been reported in the avian ovary from the pre-laying period to the broody period.
Figure 2.
Differentially expressed genes from the three reproduction periods. (A) Heat maps of DEGs from ovaries during pre-egg period (T1), laying-egg period (T2) and broody period (T3). The read counts of each cellular mRNA were normalized by the sum of the total reads. Colors from white to red represent upregulated cellular genes; colors from white to green represent downregulated cellular genes. (B) Pearson correlation analysis of T1, T2, and T3 groups. Volcano plot of corrected P values as a function of weighted fold change for mRNAs in T1 vs. T2 (C); T1 vs. T3 (D); (C) T2 vs. T3 (E). The vertical dotted line delimits up- and downregulation. Red plots represent significant upregulated genes and green plots represent significant downregulated genes (|log2-fold change|≥0, corrected P < 0.05). (F) A Venn diagram showed the relationships among T1, T2, and T3 groups of DEGs. A total of 258 DEGs were identified in all 3 groups. Abbreviation: DEGs, differentially expressed genes.
GO Enrichment Analysis of DEGs
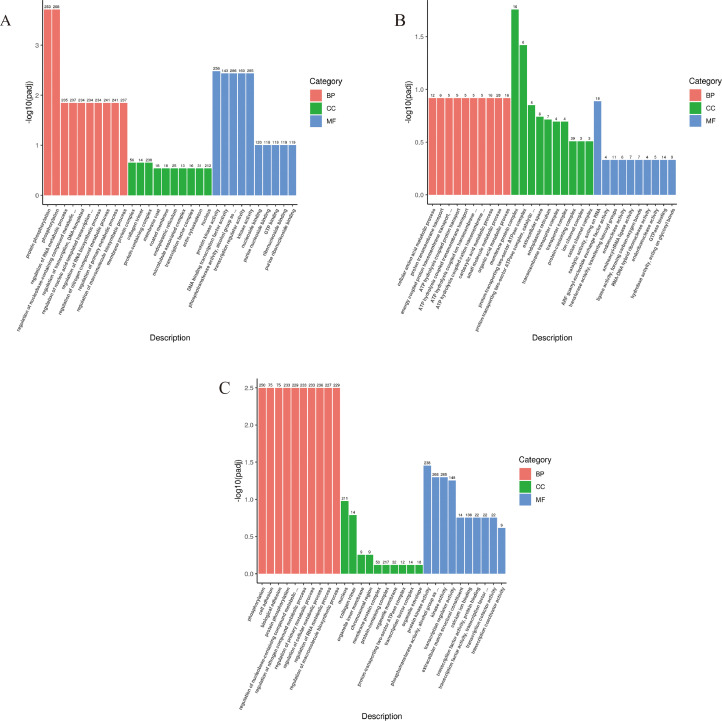
We employed the GOseq R package for GO enrichment analysis of DEGs (Figure 3). In total, 69 GO terms were enriched in T1 vs. T2, including 25 molecular functions (MF) terms, 11 cell components (CC), and 33 biological processes (BP) terms (Supplementary material 2, P-value < 0.05). The top 10 significantly enriched GO terms in T1 vs. T2 were primarily associated with the regulation of synthesis and metabolism (8/10) and protein phosphorylation (2/10) (Table 3). Moreover, 59 GO terms were enriched between T2 vs. T3, including 36 BP terms, 5 CC terms, and 18 MF terms (Supplementary material 2, P-value < 0.05). The top 10 significantly enriched GO terms in T2 vs. T3 were implicated in protein phosphorylation (2/10), cell or biological adhesion (2/10), and regulation of multiple compound metabolic processes (6/10) (Table 3). Analysis of DEGs in T1 with T3 showed 79 enriched GO terms, including 48 BP terms, 13 CC terms, and 18 MF terms (Supplementary material 2, P-value < 0.05). The top 10 significantly enriched GO terms were implicated in ATP hydrolysis coupled-related compounds transport (4/10), transmembrane transport (3/10), and others (3/10) (Table 3). Of note, 39 GO terms were significantly enriched in both T1 vs. T2 and T2 vs. T3 and were mainly involved in the regulation of signaling or biosynthetic process (18/39), cellular components (4/39), protein polymerization (5/39), nucleic acid synthesis, and transcription (8/39), and others (4/39). These data are in support of the view that these GO terms may play critical roles in the entire ovulation cycle.
Figure 3.
GO terms enrichment analysis of DEGs in ovaries. Histogram charts show the top 30 significantly enriched GO terms of T1 vs. T2 (A), T1 vs. T3 (B), T2 vs. T3 (C), which are classified as biological process (BP, red bar), cellular component (CC, green bar) and molecular function (MF, blue bar). Abbreviations: DEGs, differentially expressed genes; GO, gene ontology.
Table 3.
Top 10 enriched GO terms in T1 vs. T2, T1 vs. T3 and T2 vs. T3.
| Comparison between two groups | Category | GO ID | Description | GeneRatio | BgRatio | P value | Count | Up | Down | |
|---|---|---|---|---|---|---|---|---|---|---|
| T1 vs. T2 |
BP | GO:0006468 | protein phosphorylation | 253/2434 | 407/4798 | 8.19E-07 | 253 | 105 | 148 | |
| BP | GO:0016310 | phosphorylation | 268/2434 | 434/4798 | 8.48E-07 | 268 | 113 | 155 | ||
| MF | GO:0004672 | protein kinase activity | 256/4089 | 414/7920 | 1.12E-05 | 256 | 106 | 150 | ||
| MF | GO:0003700 | DNA binding transcription factor activity | 143/4089 | 221/7920 | 4.66E-05 | 143 | 54 | 89 | ||
| MF | GO:0016773 | phosphotransferase activity, alcohol group as acceptor | 286/4089 | 474/7920 | 5.21E-05 | 286 | 114 | 172 | ||
| MF | GO:0140110 | transcription regulator activity | 160/4089 | 251/7920 | 5.54E-05 | 160 | 60 | 100 | ||
| MF | GO:0016301 | kinase activity | 285/4089 | 473/7920 | 6.20E-05 | 285 | 116 | 169 | ||
| BP | GO:0051252 | regulation of RNA metabolic process | 235/2434 | 398/4798 | 0.000311 | 235 | 102 | 133 | ||
| BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 237/2434 | 402/4798 | 0.000333 | 237 | 104 | 133 | ||
| BP | GO:0006355 | regulation of transcription, DNA-templated | 234/2434 | 397/4798 | 0.000371 | 234 | 101 | 133 | ||
| T1 vs. T3 |
CC | GO:0098796 | membrane protein complex | 16/186 | 88/2755 | 0.000188 | 16 | 5 | 11 | |
| MF | GO:0140098 | catalytic activity, acting on RNA | 18/585 | 104/7917 | 0.000544 | 18 | 2 | 16 | ||
| CC | GO:0016469 | proton-transporting two-sector ATPase complex | 6/186 | 18/2755 | 0.000818 | 6 | 0 | 6 | ||
| BP | GO:0006520 | cellular amino acid metabolic process | 12/340 | 63/4790 | 0.001322 | 12 | 1 | 11 | ||
| BP | GO:1902600 | proton transmembrane transport | 6/340 | 20/4790 | 0.002019 | 6 | 0 | 6 | ||
| BP | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient | 5/340 | 15/4790 | 0.002899 | 5 | 0 | 5 | ||
| BP | GO:0015991 | ATP hydrolysis coupled proton transport | 5/340 | 15/4790 | 0.002899 | 5 | 0 | 5 | ||
| BP | GO:0090662 | ATP hydrolysis coupled transmembrane transport | 5/340 | 15/4790 | 0.002899 | 5 | 0 | 5 | ||
| BP | GO:0099131 | ATP hydrolysis coupled ion transmembrane transport | 5/340 | 15/4790 | 0.002899 | 5 | 0 | 5 | ||
| BP | GO:0099132 | ATP hydrolysis coupled cation transmembrane transport | 5/340 | 15/4790 | 0.002899 | 5 | 0 | 5 | ||
| T2 vs. T3 |
BP | GO:0016310 | phosphorylation | 250/2298 | 433/4775 | 1.67E-05 | 250 | 142 | 108 | |
| BP | GO:0007155 | cell adhesion | 75/2298 | 112/4775 | 3.63E-05 | 75 | 44 | 31 | ||
| BP | GO:0022610 | biological adhesion | 75/2298 | 112/4775 | 3.63E-05 | 75 | 44 | 31 | ||
| BP | GO:0006468 | protein phosphorylation | 233/2298 | 406/4775 | 5.78E-05 | 233 | 134 | 99 | ||
| BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 229/2298 | 400/4775 | 8.33E-05 | 229 | 113 | 116 | ||
| BP | GO:0051171 | regulation of nitrogen compound metabolic process | 233/2298 | 408/4775 | 8.96E-05 | 233 | 114 | 119 | ||
| BP | GO:0080090 | regulation of primary metabolic process | 233/2298 | 408/4775 | 8.96E-05 | 233 | 114 | 119 | ||
| BP | GO:0031323 | regulation of cellular metabolic process | 236/2298 | 414/4775 | 9.45E-05 | 236 | 115 | 121 | ||
| BP | GO:0051252 | regulation of RNA metabolic process | 227/2298 | 397/4775 | 9.99E-05 | 227 | 113 | 114 | ||
| BP | GO:0010556 | regulation of macromolecule biosynthetic process | 229/2298 | 401/4775 | 0.000104 | 229 | 114 | 115 | ||
Abbreviation: GO, gene ontology.
KEGG Pathway Enrichment Analysis of DEGs
Functional classification of the DEGs using KEGG pathway enrichment analysis (Figure 4) demonstrated that the DEGs were associated with 18 pathways in T1 vs. T2, 11 pathways in T1 vs. T3, and 5 pathways in T2 vs. T3 (Supplementary material 3). The top 5 KEGG pathways in T1 vs. T2 were focal adhesion (gga04510), regulation of actin cytoskeleton (gga04810), salmonella infection (gga05132), protein processing in the endoplasmic reticulum (gga04141), and influenza A (gga05164) (Table 4). The top 5 pathways in T1 vs. T3 were lysosome (gga04142), N-glycan biosynthesis (gga00510), spliceosome (gga03040), types of N-glycan biosynthesis (gga00513), and sphingolipid metabolism (gga00600) (Table 4). Moreover, the top 5 pathways in T2 relative to T3 were focal adhesion (gga04510), ECM-receptor interaction (gga04512), N-glycan biosynthesis (gga00510), TGF-beta signaling pathway (gga04350), and endocytosis (gga04144) (Table 4).
Figure 4.
KEGG pathway enrichment analysis of DEGs in ovaries. Bubble charts represent the top 20 significantly enriched KEGG pathways of T1 vs. T2 (A), T1 vs. T3 (B), T2 vs. T3 (C). Size of each circle represents the number of DEGs in each pathway (larger circles represent more DEGs) and the color represents the P value of each pathway. Abbreviations: DEGs, differentially expressed genes; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table 4.
Top 5 enriched KEGG pathways in T1 vs. T2, T1 vs. T3 and T2 vs. T3.
| Comparison between two groups | KEGG ID | Description | P value | P adj | Count | Up | Down |
|---|---|---|---|---|---|---|---|
| T1 vs. T2 |
gga04510 | Focal adhesion | 0.000662 | 0.099952 | 125 | 22 | 103 |
| gga04810 | Regulation of actin cytoskeleton | 0.001571 | 0.118588 | 121 | 29 | 92 | |
| gga05132 | Salmonella infection | 0.002563 | 0.129018 | 135 | 41 | 94 | |
| gga04141 | Protein processing in endoplasmic reticulum | 0.005158 | 0.150095 | 91 | 26 | 65 | |
| gga05164 | Influenza A | 0.005641 | 0.150095 | 75 | 14 | 61 | |
| T1 vs. T3 |
gga04142 | Lysosome | 9.53E-10 | 1.34E-07 | 32 | 1 | 31 |
| gga00510 | N-Glycan biosynthesis | 0.001436 | 0.101227 | 11 | 2 | 9 | |
| gga03040 | Spliceosome | 0.002174 | 0.102192 | 18 | 4 | 14 | |
| gga00513 | Various types of N-glycan biosynthesis | 0.004959 | 0.174818 | 9 | 2 | 7 | |
| gga00600 | Sphingolipid metabolism | 0.007867 | 0.221839 | 10 | 2 | 8 | |
| T2 vs. T3 |
gga04510 | Focal adhesion | 0.000213 | 0.032159 | 120 | 93 | 27 |
| gga04512 | ECM-receptor interaction | 0.020819 | 0.757549 | 51 | 40 | 11 | |
| gga00510 | N-Glycan biosynthesis | 0.02695 | 0.757549 | 29 | 14 | 15 | |
| gga04350 | TGF-beta signaling pathway | 0.036209 | 0.757549 | 52 | 37 | 15 | |
| gga04144 | Endocytosis | 0.03976 | 0.757549 | 121 | 63 | 58 |
Abbreviation: KEGG, Kyoto Encyclopedia of Genes and Genomes.
Comparative analysis of enriched KEGG pathways in the three periods revealed that three common pathways, including focal adhesion (gga04510), ECM-receptor interaction (gga04512), and N-glycan biosynthesis (gga00510) were enriched between the groups. Taken together, these findings help define the potential central role of these enriched pathways in the entire ovulation cycle.
DISCUSSION
Unlike in mammals, goose follicles do not undergo atresia. This is the reason why avian species have long-term continuous egg production, which is influenced by ovarian follicle development and ovulation. Although Xupu goose (Anser cygnoides domesticus) is a nationally and commercially important farm animal in Hunan Province, its strong broody and poor egg-laying performance are limiting its economic value in the farm industry. In this study, ovary tissues were collected from Xupu gooses in the pre-laying period, laying period, and broody period. The total RNA was sequenced on the Illumina MiSeq platform to reveal differentially expressed gene transcripts in the ovary of Xupu geese from the pre-laying period to the broody period. We reported transcriptome changes of ovarian tissue in the entire reproduction period of Xupu geese. KEGG and GO analyses further revealed the key genes and pathways implicated in the laying cycle and brooding of Xupu geese. Analysis showed, 258 genes are significantly differentially expressed in T1 vs. T2, T1 vs. T3, and T2 vs. T3, and are implicated in inflammation, reproduction, mutual recognition and adhesion between cells, and the establishment of the cytoskeleton.
The broody behavior of goose is a significant aspect of their reproduction. It influences egg production with the degeneration of follicles. Mounting evidence has shown that the degeneration of follicles is associated with autophagy, apoptosis, and homeostasis imbalance (Yu et al., 2016b). Autophagy is implicated in both cell survival and cell death. Herein, we found that the expression of several autophagy-related genes, including AMBRA1, ATGs, DRAM1, MAP1LC3A, SOGA1, UVRAG, VPS13A, VPS13C, and WDFY3 were altered in the ovary at different stages. In support of our findings, Yu et al. (2016c) also revealed that the expression of some autophagy relative genes, including BECN1, TP63, and ATGs, were alerted in the broody follicles. Autophagy is a multistep process highly regulated by a number of the conserved autophagy-related genes (ATGs) (Liu et al., 2010). Emerging evidence indicates that ATG genes are crucial in autophagosome formation and autophagy regulation, and are also associated with several key pathological and physiological processes (Levine and Kroemer, 2008). In particular, ATG4B, a mammalian homologue of yeast Atg4, has been implicated in the processing of LC3. Its protein and mRNA expressions were ubiquitous in rat tissues (Yoshimura et al., 2006). Our results showed lower mRNA relative expression of Atg4B significantly in the laying period as compared with the pre-laying period and broody period. Moreover, a few DEGs and GO terms were associated with apoptosis, a form of programmed cell death, or “cellular suicide.” All peptidases of the C14 family have a strict requirement for the amino acid in P1. The apoptosis cascade, predominant in animal cells, is primarily regulated by the caspases. In this study, we revealed significant alteration of the relative expression of some genes belonging to the Peptidase C14 family, in these 3 periods. They included CASP10, CASP6, CASP7, CFLAR, CASP9, and CASP8. Of note, the mRNA relative expression of CASP10, CASP6 and CASP7 significantly increased in the laying period as compared with the pre-laying period and broody period. CASP6 is traditionally recognized as an crucial molecule in programmed cell apoptosis, it cleaves the nuclear structural protein NuMA (nuclear mitotic apparatus protein) and the lamin A/C proteins and induces nuclear shrinkage and fragmentation (Li and Yuan, 2008). Indeed, CASP6 exerts crucial regulatory effects on non-apoptotic cellular events, such as modification of cell cycle entry (Richards et al., 2008). Caspase-10, a close homolog of caspase-8, is a highly conserved caspase throughout evolution (Eckhart et al., 2008). It is currently assumed that caspase-8 and caspase-10 have redundant functions in cell death signaling, though the potential function of caspase-10 as a substitute for caspase-8 is controversial (Fischer et al., 2006). Caspase-3 is the most important executioner caspase, activated by both intrinsic and extrinsic pathways (Seervi and Xue, 2015).
In the recent past, differences in the mRNA levels of CASP3 among primordial, primary, and secondary follicles, and the magnitude were revealed, though they varied according to species (duck and goose) and the stages of development (Hu et al., 2021). Contrary to previous works, CASP3 expression differed in the 3 stages of our experiment, but the difference was not statistically significant. These results suggest a role for apoptosis and autophagy in different reproductive stages, but warrant further exploration of the specific role and underlying mechanism.
Of note, this is the first report on the expression pattern of MRP126, serglycin, TXNIP, and FZD2 genes in the avian ovary. Bukovsky and Presl (1979) reported the association between the immune system and the regulation of ovulation. Mounting reports show that several mediators of LH-induced signaling cascades are associated with inflammation, and the process leading to ovulation and that of the inflammatory response are similar (Duffy et al., 2019; Ernst et al., 2020). In the present study, MRP126 was significantly upregulated in the pre-laying period vs. egg-laying period and significantly downregulated in the egg-laying vs. broody period. We also reported a higher relative expression of MRP126 in the broody period as compared with that in the pre-laying period. MRP126 protein is a co-orthologue of calgranulin, expressed solely in birds and reptiles (Loes et al., 2018). High expression levels of MRP126 protein have previously been reported in chicken tissues, including heterophils (avian counterparts of mammalian neutrophils), caecum, and macrophages, following bacteria exposure (Matulova et al., 2013; Rychlik et al., 2014). A similar pattern of expression of MRP126 and calgranulin is observed when birds or mammals are infected by bacteria. Although there is no previous data on MRP126 expression in the avian ovary, studies reported expression profiles of calgranulin protein in the mammalian ovary. Mammals express 3 distinct members of the calgranulins family, including S100A8 (calgranulin A), S100A9 (calgranulin B), and S100A12 (calgranulin C) (Bozzi and Nolan, 2020). S100A8 is primarily expressed in oocytes within cysts/plasmodia where it induces oocytes or ovarian somatic cells to form primordial follicles (Teng et al., 2015). S100A9 forms a heterodimer with S100A8 under calcium (Teigelkamp et al., 1991) and the resultant complex contributes to inflammatory processes (Hessian et al., 1993). mRNA expression of S100A9 is upregulated in both granulosa cells and residual ovarian cells 6 h after hCG injection and then sharply declines by 12 h post-hCG injection. In situ hybridization analysis shows that S100A9 mRNA is expressed predominantly in cells located in the interstitial and stroma layer of preovulatory ovaries. However, data on the specific expression pattern of S100A9 in different parts of the ovary is scanty. Besides, S100A9 up-regulation demonstrates its significant role in leukocyte trafficking during inflammatory responses of ovulation (Jo et al., 2004). This is the first study to report MRP126 expression profile in avian ovary, and implicate MRP126 in ovulation induced inflammatory response.
Serglycin was first reported as a secretory product of a rat yolk sac tumor (Oldberg et al., 1981). Herein, we the first group to report the serglycin mRNA profile in ovarian tissue of goose during the whole ovulation cycle. Serglycin expression during the pre-laying period was significantly lower, as compared with its expression in the other 2 periods. Besides, serglycin was significantly upregulated during the egg-laying period and then significantly downregulated during the broody period. Several lines of evidence had revealed that serglycin is primarily expressed in cells with hematopoietic lineage, including neutrophils, lymphocytes, monocytes, macrophages, and mast cells (Elliott et al., 1993; Niemann et al., 2004). It also interacts with various inflammatory mediators (Kolset and Tveit, 2008). Serglycin has further been implicated in uterine decidual function (Keith Ho et al., 2001), extravasation of peripheral blood natural killer cells into the endometrium (Santoni et al., 2008), and signaling within the decidua, or between the placenta and the decidua (Schick, 2010). Ferrazza (Ferrazza et al., 2017) explored the protein expression profile of bovine follicular fluid at different development stages and reported serglycin upregulation in early follicular development and a negative correlation of serglycin expression level with progesterone concentration in follicular fluid (Ferrazza et al., 2017). The expression pattern of serglycin supports the view that it plays a role in the ovulation-induced inflammatory response.
In addition, the functions of the ovary are influenced by metabolism. There is previous evidence that obesity, insulin resistance, oxidative stress, and reproductive hormone imbalance induce ovarian dysfunction (Robker, 2008; Gu et al., 2015). Our analysis showed that TXNIP was highly expressed in the 3 periods, in particular, TXNIP was significantly upregulated in the egg-laying and broody period as compared with the pre-laying period. TXNIP (thioredoxin-interacting protein) is a redox-sensitive signaling protein implicated in glucose metabolism (Patwari et al., 2006), and it is associated with insulin resistance and insulin secretion (Wu et al., 2014). Studies have reported high expression TXNIP in cumulus cells, oocytes, and granulosa cells of several species (Lee et al., 2013; Salhab et al., 2013). Obesity leads to downregulation of TXNIP expression in metaphase II of oocytes compared with expression levels of the normal group, which may reduce development and quality of oocyte (Ruebel et al., 2017). In addition, Chutkow (Chutkow et al., 2008) revealed that TXNIP modulated hepatic glucose production and global glucose homeostasis (Chutkow et al., 2008), whereas Parikh (Parikh et al., 2007) found that low TXNIP levels improved glucose uptake in skeletal muscle (Parikh et al., 2007). These data suggest that TNXIP plays multiple roles in the process of oocyte maturation. Our study is the first to explore the TNXIP mRNA profile in the ovary of a goose during the reproductive cycle, and future research direction will be to explore the specific function of TNXIP in the ovary.
The Wnt pathway is a conserved signaling pathway implicated in the regulation of ovarian development and function. Wnt signaling components are expressed in the human ovary from early to mid-gestation. Of note, canonical Wnt signaling is only observed in oocytes of primordial follicles (Bothun and Woods, 2019). FZD2 is linked to the Wnt pathway. In this study, the relative expression of FZD2 was at its peak during the egg-laying period and then declined during the broody period. FZD2 was first reported by (Zhao et al., 1995) in developing ovaries. In the same study, high expression levels of the frizzled receptor were reported throughout ovary development, an implication that Wnt-signaling was mediated via the non-canonical pathway. The profiles of Wnt signaling components have previously been explored in the human ovary from development to adulthood. As depicted by the germ cell nests, FZD2 was expressed at all time-points of ovarian development but strictly in low cytoplasmic levels in adult tissue (Bothun and Woods, 2019). In the past decade, Wang (Wang et al., 2010) explored the expression pattern of FZD2 in mouse ovaries during the oestrous cycle and reported the highest FZD2 mRNA and protein levels in murine oocytes and granulosa cells during the proestrus stage which significantly decreased from oestrus to diestrus stages. Additionally, the expression profiles of FZD2 in both human and mouse ovary tissues demonstrated that FZD2 was involved in the regulation of follicular growth, oocyte maturation, and luteinization in mammals. This is the first study to report the expression pattern of FZD2 in the avian ovary from the pre-laying period to the broody period. However, the role of FZD2 in the development and regulation of ovulation should be explored further.
In summary, this study reports on the entire profile of transcriptome changes in ovaries from the pre-laying period to the broody period. Candidate genes implicated in the whole cycle have also been identified. These findings demonstrate a role for inflammation, reproduction, mutual recognition and adhesion between cells, and the establishment of the cytoskeleton, in the ovarian cycle changes. These data will provide valuable information to improve Xupu goose breeding with low nestability in the future.
ACKNOWLEDGMENTS
This work was supported by Protection of livestock and poultry genetic resources ([2020-SJ-011]); Jiangsu Provincial Agricultural Science and Technology Independent Innovation Fund (CX (18)1004); and the earmarked fund for Jiangsu Agricultural Industry Technology System
The funding bodies did not play direct roles in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript. The funding bodies did not play direct roles in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
DISCLOSURES
The authors declare no conflict of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2021.101403.
Appendix. Supplementary materials
REFERENCES
- Bothun A.M., Woods D.C. Dynamics of WNT signaling components in the human ovary from development to adulthood. Histochem. Cell. Biol. 2019;151:115–123. doi: 10.1007/s00418-018-1729-y. [DOI] [PubMed] [Google Scholar]
- Bozzi A.T., Nolan E.M. Avian MRP126 restricts microbial growth through Ca(II)-Dependent Zn(II) sequestration. Biochemistry. 2020;59:802–817. doi: 10.1021/acs.biochem.9b01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukovsky A., Presl J. Ovarian function and the immune system. Med. Hypotheses. 1979;5:415–436. doi: 10.1016/0306-9877(79)90108-7. [DOI] [PubMed] [Google Scholar]
- Chutkow W.A., Patwari P., Yoshioka J., Lee R.T. Thioredoxin-interacting protein (Txnip) is a critical regulator of hepatic glucose production. J. Biol. Chem. 2008;283:2397–2406. doi: 10.1074/jbc.M708169200. [DOI] [PubMed] [Google Scholar]
- Dai Q.Z., Lin Q., Jiang G.T. Phylogenetic studies of four Anser cygnoides (Anserini: Anserinae) in Hunan province of China based on complete mitochondrial DNA sequences. Mitochondrial. DNA. A. DNA. Mapp. Seq. Anal. 2016;27:2464–2465. doi: 10.3109/19401736.2015.1033699. [DOI] [PubMed] [Google Scholar]
- Ding N., Han Q., Zhao X.Z., Li Q., Li J., Zhang H.F., Gao G.L., Luo Y., Xie Y.H., Su J., Wang Q.G. Differential gene expression in pre-laying and laying period ovaries of Sichuan White geese (Anser cygnoides) Genet. Mol. Res. 2015;14:6773–6785. doi: 10.4238/2015.June.18.20. [DOI] [PubMed] [Google Scholar]
- Duffy D.M., Ko C., Jo M., Brannstrom M., Curry T.E. Ovulation: parallels with inflammatory processes. Endocr. Rev. 2019;40:369–416. doi: 10.1210/er.2018-00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhart L., Ballaun C., Hermann M., VandeBerg J.L., Sipos W., Uthman A., Fischer H., Tschachler E. Identification of novel mammalian caspases reveals an important role of gene loss in shaping the human caspase repertoire. Mol. Biol. Evol. 2008;25:831–841. doi: 10.1093/molbev/msn012. [DOI] [PubMed] [Google Scholar]
- Elliott J.F., Miller C.L., Pohajdak B., Talbot D., Helgason C.D., Bleackley R.C., Paetkau V. Induction of a proteoglycan core protein mRNA in mouse T lymphocytes. Mol. Immunol. 1993;30:749–754. doi: 10.1016/0161-5890(93)90146-3. [DOI] [PubMed] [Google Scholar]
- Ernst E.H., Amoushahi M., Sørensen A.S., Kragstrup T.W., Ernst E., Lykke-Hartmann K. Distinct expression patterns of TLR transcripts in human oocytes and granulosa cells from primordial and primary follicles. J. Reprod. Immunol. 2020;140 doi: 10.1016/j.jri.2020.103125. [DOI] [PubMed] [Google Scholar]
- Everaert C., Luypaert M., Maag J.L.V., Cheng Q.X., Mestdagh P. Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci. Rep. 2017;7:1588. doi: 10.1038/s41598-017-01617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrazza R.A., Garcia H.D.M., Schmidt E., Carmichael M.Mihm, Souza F.F., Burchmore R., Sartori R., Eckersall P.D., Ferreira J.C.P. Quantitative proteomic profiling of bovine follicular fluid during follicle development. Biol. Reprod. 2017;97:835–849. doi: 10.1093/biolre/iox148. [DOI] [PubMed] [Google Scholar]
- Fischer U., Stroh C., Schulze-Osthoff K. Unique and overlapping substrate specificities of caspase-8 and caspase-10. Oncogene. 2006;25:152–159. doi: 10.1038/sj.onc.1209015. [DOI] [PubMed] [Google Scholar]
- Gao G., Li Q., Zhao X., Ding N., Han Q., Su J., Wang Q. Transcriptome profiling of the hypothalamus during prelaying and laying periods in Sichuan white geese (Anser cygnoides) Anim. Sci. J. 2015;86:800–805. doi: 10.1111/asj.12356. [DOI] [PubMed] [Google Scholar]
- Geng A.L., Xu S.F., Zhang Y., Zhang J., Chu Q., Liu H.G. Effects of photoperiod on broodiness, egg-laying and endocrine responses in native laying hens. Br. Poult. Sci. 2014;55:264–269. doi: 10.1080/00071668.2013.878782. [DOI] [PubMed] [Google Scholar]
- Gu L., Liu H., Gu X., Boots C., Moley K.H., Wang Q. Metabolic control of oocyte development: linking maternal nutrition and reproductive outcomes. Cell. Mol. Life Sci. 2015;72:251–271. doi: 10.1007/s00018-014-1739-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumułka M., Hrabia A., Avital-Cohen N., Andres K., Rozenboim I. The effect of parachlorophenylalanine treatment on the activity of gonadal and lactotrophic axes in native Polish crested chickens stimulated to broodiness. Poult. Sci. 2020;99:2708–2717. doi: 10.1016/j.psj.2019.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessian P.A., Edgeworth J., Hogg N. MRP-8 and MRP-14, two abundant Ca(2+)-binding proteins of neutrophils and monocytes. J. Leukoc. Biol. Suppl. 1993;53:197–204. [PubMed] [Google Scholar]
- Hu S., Zhu M., Wang J., Li L., He H., Hu B., Hu J., Xia L. Histomorphology and gene expression profiles during early ovarian folliculogenesis in duck and goose. Poult. Sci. 2021;100:1098–1108. doi: 10.1016/j.psj.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo M., Gieske M.C., Payne C.E., Wheeler-Price S.E., Gieske J.B., Ignatius I.V., Curry T.E., Jr., Ko C. Development and application of a rat ovarian gene expression database. Endocrinology. 2004;145:5384–5396. doi: 10.1210/en.2004-0407. [DOI] [PubMed] [Google Scholar]
- Kang B., Guo J.R., Yang H.M., Zhou R.J., Liu J.X., Li S.Z., Dong C.Y. Differential expression profiling of ovarian genes in prelaying and laying geese. Poult. Sci. 2009;88:1975–1983. doi: 10.3382/ps.2008-00519. [DOI] [PubMed] [Google Scholar]
- Ho Keith, C. H., McGrath K.E., Brodbeck K.C., Palis J., Schick B.P. Serglycin proteoglycan synthesis in the murine uterine decidua and early embryo. Biol. Reprod. 2001;64:1667–1676. doi: 10.1095/biolreprod64.6.1667. [DOI] [PubMed] [Google Scholar]
- Kolset S.O., Tveit H. Serglycin–structure and biology. Cell. Mol. Life Sci. 2008;65:1073–1085. doi: 10.1007/s00018-007-7455-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.Y., Lee H.S., Kim E.Y., Ko J.J., Yoon T.K., Lee W.S., Lee K.A. Thioredoxin-interacting protein regulates glucose metabolism and affects cytoplasmic streaming in mouse oocytes. Plos One. 2013;8:e70708. doi: 10.1371/journal.pone.0070708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yuan J. Caspases in apoptosis and beyond. Oncogene. 2008;27:6194–6206. doi: 10.1038/onc.2008.297. [DOI] [PubMed] [Google Scholar]
- Liu B., Cheng Y., Liu Q., Bao J.K., Yang J.M. Autophagic pathways as new targets for cancer drug development. Acta Pharmacol. Sin. 2010;31:1154–1164. doi: 10.1038/aps.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang J., Li L., Han C., He H., Xu H. Transcriptome analysis revealed the possible regulatory pathways initiating female geese broodiness within the hypothalamic-pituitary-gonadal axis. Plos One. 2018;13 doi: 10.1371/journal.pone.0191213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loes A.N., Bridgham J.T., Harms M.J. Coevolution of the toll-like receptor 4 complex with calgranulins and lipopolysaccharide. Front. Immunol. 2018;9:304. doi: 10.3389/fimmu.2018.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan X., Liu D., Cao Z., Luo L., Liu M., Gao M., Zhang X. Transcriptome profiling identifies differentially expressed genes in Huoyan goose ovaries between the laying period and ceased period. Plos One. 2014;9 doi: 10.1371/journal.pone.0113211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden S.J., Lucas L.M., Wilson S.P. The influence of daylength and environment on reproduction, broodiness, and mortality of turkeys. Poult. Sci. 1966;45:668–675. doi: 10.3382/ps.0450668. [DOI] [PubMed] [Google Scholar]
- Matulova M., Varmuzova K., Sisak F., Havlickova H., Babak V., Stejskal K., Zdrahal Z., Rychlik I. Chicken innate immune response to oral infection with Salmonella enterica serovar Enteritidis. Vet. Res. 2013;44:37. doi: 10.1186/1297-9716-44-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann C.U., Cowland J.B., Klausen P., Askaa J., Calafat J., Borregaard N. Localization of serglycin in human neutrophil granulocytes and their precursors. J. Leukoc. Biol. 2004;76:406–415. doi: 10.1189/jlb.1003502. [DOI] [PubMed] [Google Scholar]
- Oldberg A., Hayman E.G., Ruoslahti E. Isolation of a chondroitin sulfate proteoglycan from a rat yolk sac tumor and immunochemical demonstration of its cell surface localization. J. Biol. Chem. 1981;256:10847–10852. [PubMed] [Google Scholar]
- Onagbesan O., Bruggeman V., Decuypere E. Intra-ovarian growth factors regulating ovarian function in avian species: a review. Anim. Reprod. Sci. 2009;111:121–140. doi: 10.1016/j.anireprosci.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Parikh H., Carlsson E., Chutkow W.A., Johansson L.E., Storgaard H., Poulsen P., Saxena R., Ladd C., Schulze P.C., Mazzini M.J., Jensen C.B., Krook A., Björnholm M., Tornqvist H., Zierath J.R., Ridderstråle M., Altshuler D., Lee R.T., Vaag A., Groop L.C., Mootha V.K. TXNIP regulates peripheral glucose metabolism in humans. PLoS Med. 2007;4:e158. doi: 10.1371/journal.pmed.0040158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwari P., Higgins L.J., Chutkow W.A., Yoshioka J., Lee R.T. The interaction of thioredoxin with Txnip. Evidence for formation of a mixed disulfide by disulfide exchange. J. Biol. Chem. 2006;281:21884–21891. doi: 10.1074/jbc.M600427200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S., Watanabe C., Santos L., Craxton A., Clark E.A. Regulation of B-cell entry into the cell cycle. Immunol. Rev. 2008;224:183–200. doi: 10.1111/j.1600-065X.2008.00652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robker R.L. Evidence that obesity alters the quality of oocytes and embryos. Pathophysiology. 2008;15:115–121. doi: 10.1016/j.pathophys.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Romanov M.N., Talbot R.T., Wilson P.W., Sharp P.J. Genetic control of incubation behavior in the domestic hen. Poult. Sci. 2002;81:928–931. doi: 10.1093/ps/81.7.928. [DOI] [PubMed] [Google Scholar]
- Ruebel M.L., Cotter M., Sims C.R., Moutos D.M., Badger T.M., Cleves M.A., Shankar K., Andres A. Obesity modulates inflammation and lipid metabolism oocyte gene expression: a single-cell transcriptome perspective. J. Clin. Endocrinol. Metab. 2017;102:2029–2038. doi: 10.1210/jc.2016-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychlik I., Elsheimer-Matulova M., Kyrova K. Gene expression in the chicken caecum in response to infections with non-typhoid Salmonella. Vet. Res. 2014;45:119. doi: 10.1186/s13567-014-0119-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salhab M., Dhorne-Pollet S., Auclair S., Guyader-Joly C., Brisard D., Dalbies-Tran R., Dupont J., Ponsart C., Mermillod P., Uzbekova S. In vitro maturation of oocytes alters gene expression and signaling pathways in bovine cumulus cells. Mol. Reprod. Dev. 2013;80:166–182. doi: 10.1002/mrd.22148. [DOI] [PubMed] [Google Scholar]
- Santoni A., Carlino C., Stabile H., Gismondi A. Mechanisms underlying recruitment and accumulation of decidual NK cells in uterus during pregnancy. Am. J. Reprod. Immunol. 2008;59:417–424. doi: 10.1111/j.1600-0897.2008.00598.x. [DOI] [PubMed] [Google Scholar]
- Schick B.P. Serglycin proteoglycan deletion in mouse platelets: physiological effects and their implications for platelet contributions to thrombosis, inflammation, atherosclerosis, and metastasis. Prog. Mol. Biol. Transl. Sci. 2010;93:235–287. doi: 10.1016/S1877-1173(10)93011-1. [DOI] [PubMed] [Google Scholar]
- Seervi M., Xue D. Mitochondrial cell death pathways in caenorhabiditis elegans. Curr. Top. Dev. Biol. 2015;114:43–65. doi: 10.1016/bs.ctdb.2015.07.019. [DOI] [PubMed] [Google Scholar]
- Sharp P.J., MacNamee M.C., Talbot R.T., Sterling R.J., Hall T.R. Aspects of the neuroendocrine control of ovulation and broodiness in the domestic hen. J. Exp. Zool. 1984;232:475–483. doi: 10.1002/jez.1402320314. [DOI] [PubMed] [Google Scholar]
- Teigelkamp S., Bhardwaj R.S., Roth J., Meinardus-Hager G., Karas M., Sorg C. Calcium-dependent complex assembly of the myeloic differentiation proteins MRP-8 and MRP-14. J. Biol. Chem. 1991;266:13462–13467. [PubMed] [Google Scholar]
- Teng Z., Wang C., Wang Y., Huang K., Xiang X., Niu W., Feng L., Zhao L., Yan H., Zhang H., Xia G. S100A8, an oocyte-specific chemokine, directs the migration of ovarian somatic cells during mouse primordial follicle assembly. J. Cell. Physiol. 2015;230:2998–3008. doi: 10.1002/jcp.25032. [DOI] [PubMed] [Google Scholar]
- Thomason D.M., Leighton A.T., Jr., Mason J.P., Jr. A study of certain environmental factors and mineral chelation on the reproductive performance of young and yearling turkey hens. Poult. Sci. 1976;55:1343–1355. doi: 10.3382/ps.0551343. [DOI] [PubMed] [Google Scholar]
- Wang S.B., Xing B.S., Yi L., Wang W., Xu Y.X. Expression of Frizzled 2 in the mouse ovary during oestrous cycle. J. Anim. Physiol. Anim. Nutr. 2010;94:437–445. doi: 10.1111/j.1439-0396.2009.00927.x. [DOI] [PubMed] [Google Scholar]
- Wu J., Wu Y., Zhang X., Li S., Lu D., Li S., Yang G., Liu D. Elevated serum thioredoxin-interacting protein in women with polycystic ovary syndrome is associated with insulin resistance. Clin. Endocrinol. (Oxf.). 2014;80:538–544. doi: 10.1111/cen.12192. [DOI] [PubMed] [Google Scholar]
- Xu Q., Zhao W., Chen Y., Tong Y., Rong G., Huang Z., Zhang Y., Chang G., Wu X., Chen G. Transcriptome profiling of the goose (Anser cygnoides) ovaries identify laying and broodiness phenotypes. Plos One. 2013;8:e55496. doi: 10.1371/journal.pone.0055496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Yang Y.Z., Gu T.T., Cao Z.F., Zhao W.M., Qin H.R., Xu Q., Chen G.H. Comparison of the broody behavior characteristics of different breeds of geese. Poult. Sci. 2019;98:5226–5233. doi: 10.3382/ps/pez366. [DOI] [PubMed] [Google Scholar]
- Ye P., Ge K., Li M., Yang L., Jin S., Zhang C., Chen X., Geng Z. Egg-laying and brooding stage-specific hormonal response and transcriptional regulation in pituitary of Muscovy duck (Cairina moschata) Poult. Sci. 2019;98:5287–5296. doi: 10.3382/ps/pez433. [DOI] [PubMed] [Google Scholar]
- Yoshimura K., Shibata M., Koike M., Gotoh K., Fukaya M., Watanabe M., Uchiyama Y. Effects of RNA interference of Atg4B on the limited proteolysis of LC3 in PC12 cells and expression of Atg4B in various rat tissues. Autophagy. 2006;2:200–208. doi: 10.4161/auto.2744. [DOI] [PubMed] [Google Scholar]
- Yu J., He K., Ren T., Lou Y., Zhao A. High-throughput sequencing reveals differential expression of miRNAs in prehierarchal follicles of laying and brooding geese. Physiol. Genomics. 2016;48:455–463. doi: 10.1152/physiolgenomics.00011.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Lou Y., He K., Yang S., Yu W., Han L., Zhao A. Goose broodiness is involved in granulosa cell autophagy and homeostatic imbalance of follicular hormones. Poult. Sci. 2016;95:1156–1164. doi: 10.3382/ps/pew006. [DOI] [PubMed] [Google Scholar]
- Yu J., Lou Y., Zhao A. Transcriptome analysis of follicles reveals the importance of autophagy and hormones in regulating broodiness of Zhedong white goose. Sci. Rep. 2016;6:36877. doi: 10.1038/srep36877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Lee C.C., Baldini A., Caskey C.T. A human homologue of the Drosophila polarity gene frizzled has been identified and mapped to 17q21.1. Genomics. 1995;27:370–373. doi: 10.1006/geno.1995.1060. [DOI] [PubMed] [Google Scholar]
- Zhou M., Du Y., Nie Q., Liang Y., Luo C., Zeng H., Zhang X. Associations between polymorphisms in the chicken VIP gene, egg production and broody traits. Br. Poult. Sci. 2010;51:195–203. doi: 10.1080/00071661003745786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.