Abstract
The ectopic proliferation, migration and invasion of vascular smooth muscle cells (VSMCs) contributes to the progression of various human vascular diseases. Accumulating evidence has demonstrated that microRNAs (miRs) exert vital functions in the proliferation and invasion of VSMCs. The current study aimed to elucidate the functions of miR-125a-5p and miR-7 in VSMCs and investigate the associated molecular mechanisms. The results of EdU and reverse transcription-quantitative PCR assays revealed that platelet-derived growth factor (PDGF)-BB enhanced the proliferation of VSMCs and significantly reduced the expression of miR-125a-5p and miR-7. miR-125a-5p or miR-7 overexpression significantly ameliorated PDGF-BB-induced proliferation, migration and invasion of VSMCs. Furthermore, the results demonstrated that epidermal growth factor receptor (EGFR) may be a target mRNA of miR-125a-5p and miR-7 in VSMCs. The results of western blot analysis indicated that co-transfection of miR-125a-5p mimics or miR-7 mimics distinctly decreased the protein expression of EGFR in EGFR-overexpressed VSMCs. Moreover, rescue experiments indicated that EGFR overexpression alleviated the suppressive impact of the miR-125a-5p and miR-7 s on the growth, migration and invasion of VSMCs. In conclusion, the current study identified that miR-125a-5p and miR-7 repressed the growth, migration and invasion of PDGF-BB-stimulated VSMCs by, at least partially, targeting EGFR. The current study verified that miR-125a-5p and miR-7 may be used as feasible therapeutic targets for cardiovascular diseases.
Keywords: micro RNA-125a-5p, micro RNA-7, epidermal growth factor receptor, growth, migration, invasion vascular smooth muscle cells
Introduction
As a major constituent of the vascular system, vascular smooth muscle cells (VSMCs) crucially influence the processes of sustaining vascular structure and blood pressure (1,2). The ectopic proliferation and migration of VSMCs have been demonstrated to be implicated in the development of various cardiovascular diseases, including pulmonary artery hypertension, vein graft failure and atherosclerosis (3,4). Under normal circumstances, VSMCs remain in a non-proliferative and non-migrating states (5). However, the proliferation and migration of VSMCs can be markedly elevated in the presence of extracellular factors, including basic fibroblast growth factor, tumor necrosis factor-α and platelet-derived growth factor (PDGF)-BB (6,7). Of these, PDGF-BB, a well-known potent mitogen, contributes effectively to increase VSMC proliferation and migration (8). Investigating the associated regulatory mechanisms underlying VSMC proliferation may be useful for the prevention of cardiovascular diseases. Inhibition of HSF1-FAM3A-ATP signaling pathway in VSMC can alleviate Ang II (angiotensin II)-induced constriction and vascular remodeling, hypertension, and cardiac hypertrophy in mice, thus HSF1 inhibitors may be used to treat hypertension (9). Dysfunction of VSMCs is crucial in the pathogenesis of proliferative cardiovascular diseases, and knockdown of miR-146a could significantly inhibit the proliferative and migratory properties of VSMCs in vitro via regulating NF-κB expression (10). VSMCs lose differentiation markers and gain uncontrolled proliferative activity during the early stages of atherosclerosis, and in response to vascular injury or alterations in local environmental cues, differentiated/contractile VSMCs are capable of switching to a dedifferentiated phenotype characterized by increased proliferation, migration and extracellular matrix synthesis together with decreased expression of contractile markers (11).
MicroRNAs (miRNAs or miRs) are small non-coding RNAs that regulate genes by binding to the 3′-untranslated regions (3′-UTRs) of target mRNA (9). Accumulating research has indicated that miRNAs exert pivotal effects in the development and occurrence of vascular diseases (12). For instance, miR-24 was reported to attenuate vascular remodeling in high-glucose (HG)-induced VSMCs by inhibiting their proliferation and migration (13). miR-19 was identified to serve protective effects against myocardial infarction (14). Additionally, miR-130a was demonstrated to modulate the proliferation and migration of VSMCs in various vascular tissues (15). The results of these previous studies revealed that miRNAs are crucial regulators of vascular function maintenance. Recently, miR-125a-5p has been demonstrated to be implicated in the progression of multiple regulatory processes, including VSMCs proliferation (16). miR-125b is involved in vascular calcification in vitro and in vivo by, at least partly, targeting SP7 (17). miR-125a-5p is highly expressed in VSMCs and inhibits the PDGF-BB pathway by targeting E26 transformation-specific (ETS)-1 and is, therefore, a potential regulator of the phenotypic switch of VSMCs (16). It has been reported that miR-7 is closely correlated with biological processes (18). miR-7 was reported to regulate autophagy and the ubiquitin-proteasome system in human muscle cells. Low levels of miR-7 promoted both processes and high levels of miR-7 repressed them. Furthermore, miR-7 was demonstrated to inhibit tumor metastasis and reverse epithelial-mesenchymal transition through protein kinase/extracellular signal-regulated kinase (ERK)1/2 inactivation by targeting epidermal growth factor receptor (EGFR) in epithelial ovarian cancer (18). However, the possible roles of miR-125a-5p and miR-7 in VSMCs growth and migration remain to be elucidated.
EGFR, a member of ErbB receptor family, is extensively distributed in human tissues (15). EGFR overexpression is associated with diverse solid tumors, including ovarian cancer and colorectal cancer (19). EGFR has been demonstrated to activate the ERK pathway, which manages cellular processes, such as cell growth and differentiation (20). More importantly, EGFR has been indicated to influence the proliferation and migration of VSMCs (21). However, it is unclear whether EGFR is affected by miR-125a-5p and miR-7 in VSMCs.
The current study investigated the functional role of miR-125a-5p or miR-7 on cell growth, migration and invasion in VSMCs treated by PDGF-BB. The present study hypothesized that miR-125a-5p and miR-7 were involved in the proliferation and migration of VSMCs by targeting EGFR. Therefore, the current study aimed to establish the functional roles of miR-125a-5p and miR-7 on the proliferation of VSMCs through EGFR.
Materials and methods
Cell culture and treatment
VSMCs, which were derived from the thoracic aortas of rats, were obtained from American Type Culture Collection. VSMCs were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) at 5% CO2 and 37°C. VSMCs were then incubated at 37°C with PDGF-BB (Sigma-Aldrich; Merck KGaA) at various concentrations (0, 5, 10, 20 and 40 ng/ml) for 24 h prior to subsequent experiments.
miR-125a-5p mimics (5′-UCCCUGAGACCCUUUAACCUGUGA-3′), miR-125a-5p inhibitors (5′-UGCCAGUCUCUAGGUCCCUGAGAC-3′), miR-7 mimics (5′-UGGAAGACUAGUGAUUUUGUUGU-3′), miR-7 inhibitors (5′-UUGGAUGUUGGCCUAGUUCUGUGU-3′), NC mimics (5′-UGAACAGUGUUACGUACGAUACC-3′), NC inhibitor (5′-GGUUCGUACGUACACUGUUCA-3′), pcDNA-EGFR and pcDNA3.1 empty vectors were commercially synthesized by Shanghai GenePharma Co., Ltd. Cell transfections were conducted using Lipofectamine® 2000 reagent (Beyotime Institute of Biotechnology), according to the manufacturer's protocol. The concentration of miR-125a-5p mimics was 50 nM, and the concentration of miR-125a-5p inhibitor was 100 nM. At 48 h post-transfection, cells were incubated at 37°C with 20 ng/ml PDGF-BB for another 24 h prior to subsequent experiments (22,23).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA from VSMCs was collected using TRIzol® reagent (Thermo Fisher Scientific, Inc.) and subjected to TaqMan one-step reverse transcription (Applied Biosystems; Thermo Fisher Scientific, Inc.). RT-qPCR was conducted on an ABI Prism 7500 (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacture's protocol. The relative expressions of miRNAs were quantified using 2−ΔΔCq method (24). U6 was used as the control. The sequences for the primers used are as follows: miRNA-125a-5p forward, 5′-GCTCCCTGAGACCCT-3′ and reverse, 5′-GAGCAGGCTGGAGAA-3′; miR-7 forward, 5′-CTGTTACTATGGTAGCGACACTG-3′ and reverse, 5′-CACACTGGAGGATTACATTCCC-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The reaction conditions were 94 °C for 3 min, 94°C for 45 sec, 57°C for 45 sec and 72°C for 45 sec for 30 cycles, and final extension at 72°C for 10 min.
Cell counting kit-8 (CCK-8) and EdU assays
Cell growth was analyzed using CCK-8 and EdU assays. VSMCs (seeding density, 2×103) were maintained in 96-well plates for 24 and 48 h at 37°C in a CO2 incubator. A total of 10 µl CCK-8 (Beyotime Institute of Biotechnology) was added and VSMCs were incubated at 37°C for 4 h. Absorbance was analyzed at 450 nm using a microplate reader (Bio-Rad Laboratories, Inc.).
For the EdU assays, VSMCs (seeding density, 2×103) were grown in 96-well plates at 37°C for 24 h. Cells were then incubated with 50 µM EdU at 37°C for 2 h and fixed with 4% formaldehyde at room temperature for 30 min. The nuclei were counterstained for 15 min at room temperature with 100 ng/ml DAPI. EdU-positive cells were observed using a fluorescence microscope (magnification, ×200; Olympus Corporation). EdU-positive cells were quantified by ImageJ software version 4.3 (National Institutes of Health).
Wound healing assay
VSMCs (seeding density, 5×104) were grown in 6-well plates up to 100% confluence and scratched by a sterile 200 µl pipette tip. Cells were then washed with serum-free medium three times and cultured for 24 h at 37°C. Images were obtained using an optical microscope (magnification, ×100; BX-51; Olympus Corporation).
Transwell assay
Transwell assays were used to determine the migratory and invasive capacity of VSMCs. For the migration assays, cells (seeding density, 5×104) were suspended in 200 µl serum-free DMEM and placed into the upper chamber of the Transwell inserts (8 µm pore size; Corning, Inc.). A total of 600 µl DMEM supplemented with 20% FBS was added to the lower chamber of the Transwell. The cells were incubated for 24 h at 37°C.
For the invasion assays, the upper chamber of the Transwell was pre-coated with Matrigel (BD Biosciences) for 60 min at 37°C prior to the assay. Cells (seeding density, 5×104) were suspended in 200 µl serum-free DMEM and placed into the upper chamber of the Transwell inserts (8 µm pore size; Corning, Inc.). At 24 h post-incubation, non-migrating cells were removed using a cotton swab. Cells that migrated into the lower chamber of the Transwell were fixed in 5% glutaraldehyde for 30 min at 4°C and stained with 0.1% crystal violet for 10 min at room temperature. A total of six nonoverlapping visual fields were randomly selected for cell counting, and images were captured using a light microscope (magnification, ×100).
Luciferase reporter assay
Luciferase reporter assay were performed as previously described (19). TargetScan (www.targetscan.org/vert_72/) was used to investigate the putative target genes of miR-125a-5p and miR-7. Luciferase reporter assays were used to research the association between EGFR and miR-125a-5p or miR-7 in VSMCs. Wild- and mutant-type EGFR vectors (EGFR-WT or EGFR-MUT) were synthesized by Shanghai GenePharma Co., Ltd. and cloned into luciferase genes (Shanghai GenePharma, Co., Ltd.). VSMCs (5×104) were plated in 24-well plates for 24 h at 37°C, followed by transfection with EGFR-WT or EGFR-MUT vectors and miR-125a-5p mimics, miR-125a-5p inhibitors, miR-7 mimics, miR-7 inhibitors or NCs using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h post-transfection, luciferase activity was measured using a Dual Luciferase reporter assay (Promega Corporation) and normalized to Renilla luciferase.
Western blotting
Total protein was extracted from VSMCs using RIPA lysis buffer (Bio-Rad Laboratories, Inc.) and protein concentration was measured with a BCA kit (Beyotime Institute of Biotechnology). Protein (20 µg) was isolated using 10% SDS-PAGE gels and transferred to PVDF membranes. The membranes were blocked for 1 h at room temperature in Tris Buffer Saline Tween supplemented with 0.5% Tween and 5% skimmed milk. Subsequently, the membranes were treated with primary antibodies overnight at 4°C and then incubated with horseradish peroxidase (HRP)-conjugated rabbit anti-mouse Immunoglobulin G (IgG) H&L antibodies (1:2,000; cat. no. ab6728) or HRP-conjugated goat anti-rabbit IgG H&L antibodies (1:2,000; cat. no. ab6721) at room temperature for 2 h. The primary antibodies used were as follows: Anti-p38 mitogen-activated protein kinase (38 kDa, p38; 1:1,000; cat. no. ab227426), anti-phosphorylated-p38 (38 kDa, p-p38; 1:1,000; cat. no. ab45381), anti-matrix metalloproteinase-2 (74 kDa, MMP-2; 1:1,000; cat. no. ab97779), anti-MMP-9 (78 kDa, 1:1,000; cat. no. ab73734), anti-EGFR (134 kDa, 1:1,000; cat. no. ab131498) and anti-GAPDH (37 kDa, 1:2,000; cat. no. ab9485) all antibodies from Abcam. Protein bands were visualized using an ECL kit (Beyotime Institute of Biotechnology) and quantified using ImageJ software version 4.3 (National Institutes of Health).
Statistical analysis
Data was analyzed using GraphPad Prism software (version 5.0; GraphPad Software, Inc.) and presented as the mean ± standard deviation. Each experiment was performed in triplicate. Student's t-test was used to compare the difference between two groups and one-way ANOVA analysis followed by Tukey's post-hoc test was used to compare the differences between multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-125a-5p and miR-7 are downregulated in PDGF-BB-treated VSMCs
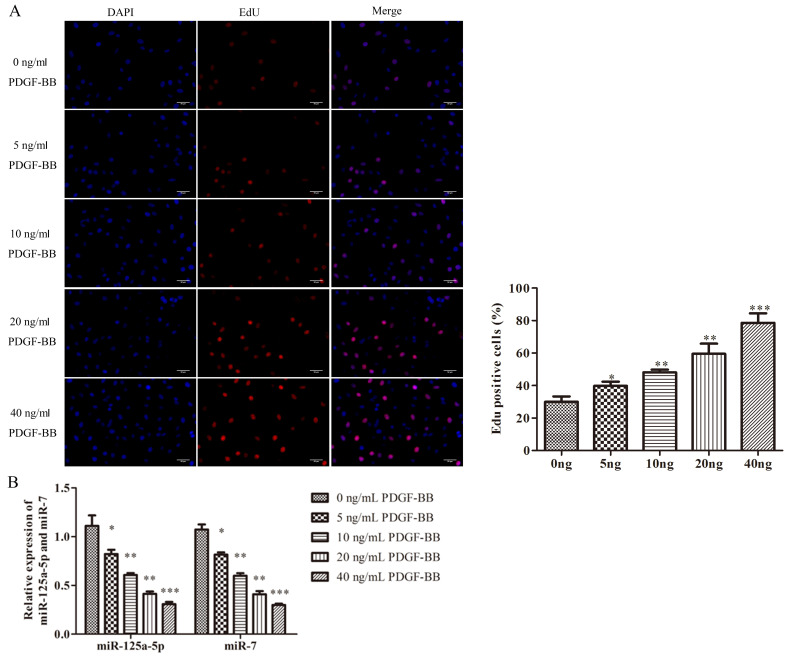
It has been well-documented that PDGF-BB is the most potent stimuli that triggers the proliferation of VSMCs (7). EdU assays were performed to investigate the effects of PDGF-BB on VSMCs. The viability of VSMCs was promoted by PDGF-BB in a concentration-dependent manner, indicating that PDGF-BB increased the proliferation of VSMCs successfully (Fig. 1A). Furthermore, the expression of miR-125a-5p and miR-7 was examined by RT-qPCR to determine the possible role of miR-125a-5p or miR-7 in PDGF-BB-treated VSMCs. PDGF-BB significantly decreased the expression of miR-125a-5p and miR-7 in a concentration-dependent manner, indicating that miR-125a-5p and miR-7 were downregulated in PDGF-BB-treated VSMCs (Fig. 1B). The results revealed that dysregulated miR-125a-5p and miR-7 may be associated with the proliferative state of VSMCs.
Figure 1.
miR-125a-5p and miR-7 were downregulated in PDGF-BB-treated VSMCs. (A) VSMCs were incubated with PDGF-BB (0, 5, 10, 20 and 40 ng/ml) for 24 h. Cell growth was detected using EdU assays (magnification, ×100). (B) VSMCs were incubated at various concentrations of PDGF-BB and then cultured in an additional 20 ng/ml PDGF-BB. miR-125a-5p and miR-7 levels were measured via reverse transcription-quantitative PCR. *P<0.05 vs. 0 ng/ml PDGF-BB; **P<0.01 vs. 0 ng/ml PDGF-BB; ***P<0.001 vs. 0 ng/ml PDGF-BB. miR, microRNA; PDGF-BB, platelet-derived growth factor BB; VSMCs, vascular smooth muscle cells.
miR-125a-5p and miR-7 restrain the growth of PDGF-BB-treated VSMCs
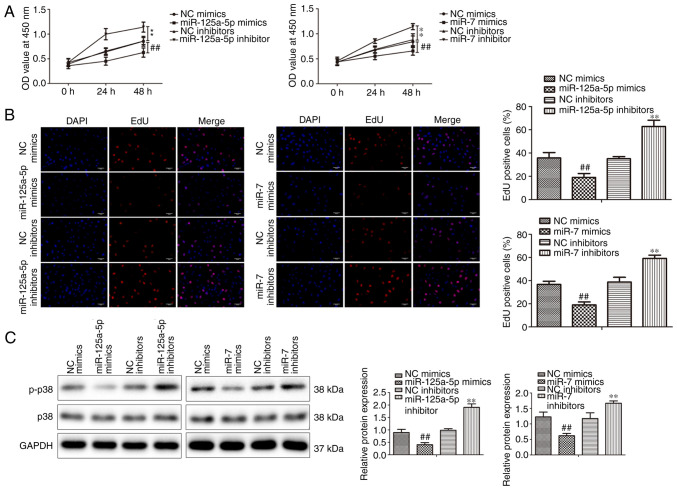
VSMCs were transfected with miR-125a-5p and miR-7 mimics or inhibitors or NCs and incubated with PDGF-BB to investigate the biological impacts of miR-125a-5p and miR-7 on the growth of PDGF-BB-treated VSMCs. CCK-8 assays indicated that miR-125a-5p and miR-7 mimics significantly decreased the proliferation of VSMCs compared with mimic NC groups in a time-dependent manner (Fig. 2A). Additionally, the inhibition of miR-125a-5p and miR-7 significantly promoted the proliferation of VSMCs compared with inhibitor NC groups. The results of the EdU assays revealed that the miR-125a-5p and miR-7 mimics significantly suppressed the proliferation of VSMCs, while inhibitors demonstrated the opposite effect (Fig. 2B). Furthermore, the protein expression of cell proliferation marker p38 (21) was assessed via western blotting. miR-125a-5p and miR-7 mimics significantly inhibited p38 phosphorylation, while inhibitors promoted p38 phosphorylation (Fig. 2C). p38 expression was not markedly different between groups. In summary, the results indicated that miR-125a-5p and miR-7 inhibited the proliferation of PDGF-BB-treated VSMCs.
Figure 2.
miR-125a-5p and miR-7 inhibited the PDGF-BB-stimulated proliferation of VSMCs. PDGF-BB-treated VSMCs were transfected with miR-125a-5p mimics or inhibitors, miR-7 mimics or inhibitors or negative controls. Cell growth was detected by Cell Counting kit-8 (A) and (B) EdU assays (magnification, ×100). (C) The expressions of proliferative genes were examined using western blotting. ##P<0.01 vs. NC mimics; **P<0.01 vs. NC inhibitors. miR, microRNA; PDGF-BB, platelet-derived growth factor BB; VSMCs, vascular smooth muscle cells; NC, negative control; OD, optical density; p-p38, anti-phosphorylated-p38.
miR-125a-5p and miR-7 repress the migration and invasion of PDGF-BB-treated VSMCs
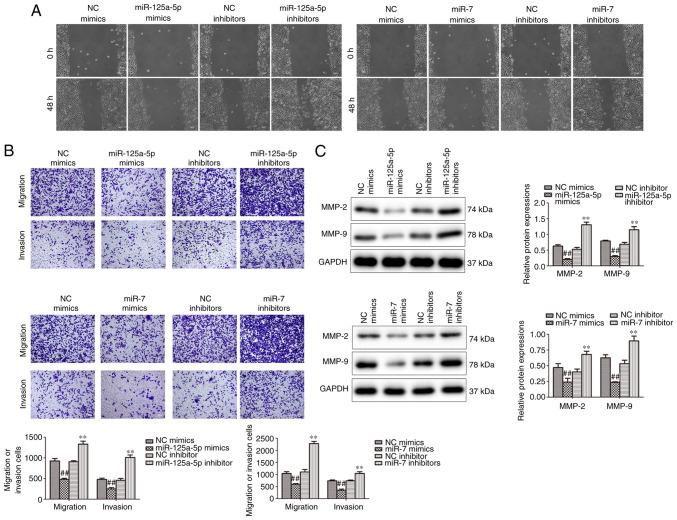
Wound healing and Transwell assays were performed to analyze the effect of miR-125a-5p and miR-7 on the migration and invasion of PDGF-BB-treated VSMCs. The results demonstrated that miR-125a-5p and miR-7 mimics markedly attenuated migration, while inhibition promoted the migration of PDGF-BB-treated VSMCs (Fig. 3A). Furthermore, Transwell assays verified that miR-125a-5p and miR-7 mimics markedly decreased the migration and invasion in PDGF-BB-treated VSMCs, while inhibitors promoted migration and invasion (Fig. 3B). Furthermore, the expression of MMP-2 and MMP-9, which modulate the migration of VSMCs, were determined by western blotting (25). The results demonstrated that MMP-2 and MMP-9 protein levels were significantly reduced in the miR-125a-5p and miR-7 mimic groups and significantly increased in the inhibitor groups compared with their corresponding NCs (Fig. 3C). The results indicated that miR-125a-5p and miR-7 decreased the migration and invasion of VSMCs stimulated by PDGF-BB.
Figure 3.
miR-125a-5p and miR-7 inhibited the migration and invasion of PDGF-BB-treated VSMCs. The influence of miR-125a-5p and miR-7 on cell migration and invasion of PDGF-BB-treated VSMCs was analyzed by (A) wound healing and (B) Transwell assays (magnification, ×100). (C) The expression of migratory protein was determined by western blotting. ##P<0.01 vs. NC mimics; **P<0.01 vs. NC inhibitors. miR, microRNA; PDGF-BB, platelet-derived growth factor BB; VSMCs, vascular smooth muscle cells; NC, negative control; MMP, matrix metalloproteinase.
EGFR is a direct target of miR-125a-5p and miR-7 in VSMCs
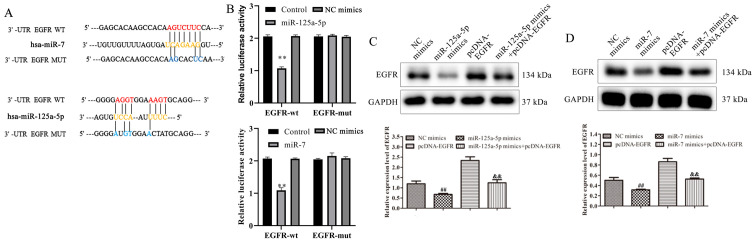
Bioinformatics analysis was used to predict potential target genes to further investigate the underlying mechanisms of miR-125a-5p and miR-7 in the proliferation and migration of VSMCs. According to the results, EGFR was reported to contain binding sites for miR-125a-5p and miR-7 (Fig. 4A). To confirm the binding sites, luciferase reporter assays were conducted. Co-transfection with miR-125a-5p or miR-7 mimics and EGFR-WT decreased the relative luciferase activity of VSMCs (Fig. 4B). The results of the mutated EGFR target sequences did not demonstrate significant differences in luciferase activity. To investigate the association between miR-125a-5p or miR-7 and EGFR, miR-125a-5p or miR-7 mimics and pcDNA-EGFR were co-transfected into PDGF-BB-treated VSMCs. The results of western blotting indicated that EGFR expression was significantly decreased in miR-125a-5p and miR-7 mimics group compared with NC mimics, while overexpression of EGFR reversed the miR-125a-5p or miR-7 mimic-mediated EGFR inhibition (Fig 4C and D). The results demonstrated that EGFR was identified as a target gene of miR-125a-5p and miR-7 and that these miRNAs negatively regulated EGFR expression by binding to its 3′-UTR.
Figure 4.
EGFR was a direct target of miR-125a-5p and miR-7 in VSMCs. (A) Potential binding sites of miR-125a-5p and miR-7 in the EGFR 3′-UTR. (B) The association between EGFR and miR-125a-5p or miR-7 was evaluated using luciferase reporter assays. Effect of (C) miR-125a-5p and (D) miR-7 on EGFR expression in VSMCs was determined by western blotting. **P<0.01 vs. controls; ##P<0.01 vs. NC mimics; &&P<0.01 vs. pcDNA-EGFR. EGFR, epidermal growth factor receptor; miR, microRNA; VSMCs, vascular smooth muscle cells; UTR, untranslated region; NC, negative control; WT, wild type; MUT, mutant.
miR-125a-5p and miR-7 inhibits cell proliferation by targeting EGFR
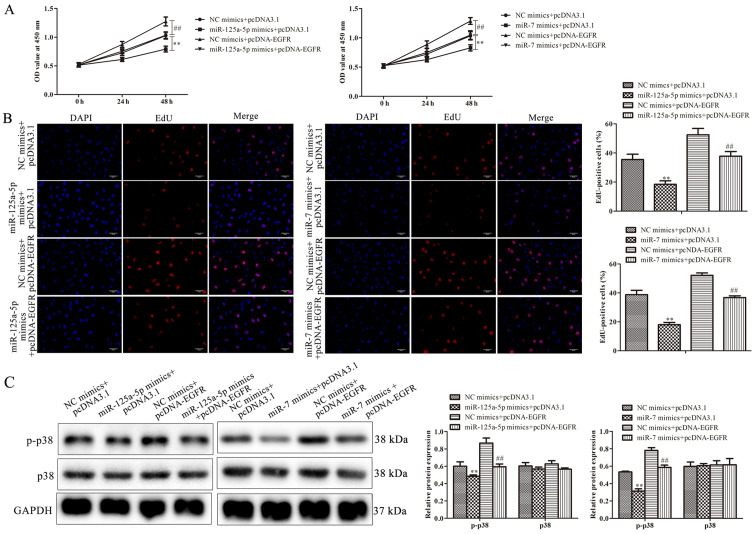
To further determine whether EGFR was involved in miR-125a-5p- and miR-7-mediated cell growth of PDGF-BB-treated VSMCs, VSMCs were co-transfected with pcDNA-EGFR and miR-125a-5p or miR-7 mimics, followed by PDGF-BB treatment. The results of the CCK-8 assay revealed that EGFR overexpression significantly increased the proliferation of PDGF-BB-treated VSMCs compared with the NC group (Fig. 5A). Similar to these results, EdU assays also indicated that miR-125a-5p and miR-7 mimics reduced the proliferation of PDGF-BB-treated VSMCs, while EGFR overexpression increased proliferation (Fig. 5B). Cells co-treated with pcDNA-EGFR and miR-125a-5p or miR-7 mimics reversed EGFR-mediated proliferation (Fig. 5B). miR-125a-5p or miR-7 mimics reduced the level of p-p38, while pcDNA-EGFR had the opposite effect; however, cells co-treated with pcDNA-EGFR eliminated the decrease in p-p38 expression induced by miR-125a-5p or miR-7 mimics (Fig. 5C). The results indicated that EGFR was associated with the inhibition of PDGF-BB-treated VSMC proliferation mediated by miR-125a-5p or miR-7.
Figure 5.
miR-125a-5p and miR-7 inhibited PDGF-BB-treated VSMCs proliferation by targeting EGFR. PDGF-BB-treated VSMCs were transfected with miR-125a-5p mimics, miR-7 mimics or the combination of mimics and pcDNA-EGFR. Cell growth was measured using (A) cell counting kit-8 and (B) EdU assays. (C) The expression of proliferative genes as assessed by western blotting. **P<0.01 vs. NC mimics + pcDNA3.1 empty vector; ##P<0.01 vs. NC mimics + pcDNA-EGFR. miR, microRNA; PDGF-BB, platelet-derived growth factor BB; VSMCs, vascular smooth muscle cells; EGFR, epidermal growth factor receptor; NC, negative control; OD, optical density; p-p38, anti-phosphorylated-p38.
miR-125a-5p and miR-7 suppress migration and invasion by targeting EGFR
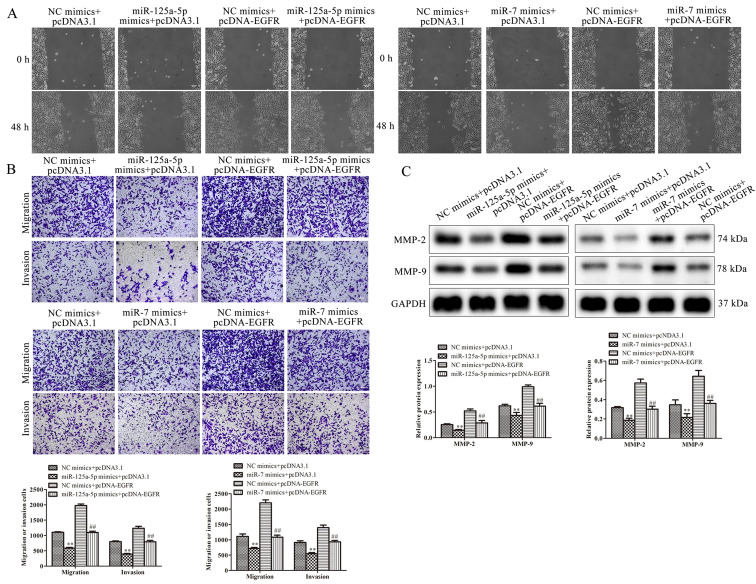
The effect of EGFR and miR-125a-5p or miR-7 on the migration and invasion of VSMCs was investigated. The results demonstrated that miR-125a-5p and miR-7 mimics decreased migration, while EGFR overexpression increased the migration of PDGF-BB-treated VSMCs. Cells co-treated with EGFR markedly ameliorated the suppressive function of miR-125a-5p and miR-7 mimics on migration (Fig. 6A). Furthermore, the results of Transwell assays revealed that miR-125a-5p and miR-7 mimics decreased migration and invasion, while EGFR upregulation reversed this result (Fig. 6B). These results were consistent with those of wound healing. The expression of migratory-associated proteins, including MMP-2 and MMP-9, were examined using western blotting. The expression of MMP-2 and MMP-9 was significantly reduced in VSMCs transfected with miR-125a-5p and miR-7 mimics compared with the NC group, while cells co-transfected with EGFR exhibited attenuated MMP-2 and MMP-9 levels (Fig. 6C). In summary, the results demonstrated that miR-125a-5p and miR-7 repressed the migration and invasion of PDGF-BB-induced VSMCs by regulating EGFR.
Figure 6.
miR-125a-5p and miR-7 inhibited the migration and invasion of PDGF-BB-treated VSMCs by targeting EGFR. Migration and invasion were evaluated by (A) wound healing and (B) Transwell assays in PDGF-BB-treated VSMCs. (C) The expression of migratory proteins was determined by western blotting. **P<0.01 vs. NC mimics + pcDNA3.1 empty vector; ##P<0.01 vs. NC mimics + pcDNA-EGFR. miR, microRNA; PDGF-BB, platelet-derived growth factor BB; VSMCs, vascular smooth muscle cells; EGFR, epidermal growth factor receptor; NC, negative control; MMP, matrix metalloproteinase.
Discussion
The results of the current study revealed that miR-125a-5p and miR-7 were downregulated in PDGF-BB-treated VSMCs, while upregulation of miR-125a-5p and miR-7 significantly suppressed cell growth and migration in vitro. The transfected miRNAs impeded the growth and migration of PDGF-BB-treated VSMCs by targeting EGFR. Therefore, miR-125a-5p and miR-7 may be novel regulators implicated in the proliferation and migration of VSMCs. However, a limitation of the current study was that other cells lines or types of cell were not investigated.
Accumulating evidence has indicated that miR-125a-5p modulates the progression of cancer, the differentiation of skeletal muscle cells and the inhibition of angiogenesis (26–31). miR-125a-5p has been established as a tumor suppressor that prevents the development of human diseases including colorectal and bladder cancer (32–34). Similarly, miR-7 acts as a tumor inhibitor in glioblastoma, gastric, liver, breast, head and neck, non-small cell lung carcinoma, melanoma, cervical, prostate, colorectal, thyroid, ovarian and schwannoma carcinoma (35). However, the roles of miR-125a-5p and miR-7 in VSMCs are yet to be elucidated. The current study demonstrated that miR-125a-5p and miR-7 were associated with the proliferation and migration of PDGF-BB-treated VSMCs. Overexpression of miR-125a-5p and miR-7 suppressed the cell growth and migration of PDGF-BB-treated VSMCs, while downregulation of miR-125a-5p and miR-7 exerted the opposite effect. Additionally, the expression of the miRNAs were negatively associated with expression of p-p38, MMP-2 and MMP-9, which are proliferation and migration marker genes of VSMCs. It has been demonstrated that MMP-9 degrades collagen IV, collagen V and gelatin in the extracellular matrix and promotes tumor invasion and metastasis (36–39). Li et al (40) examined the effect of MMP-9 on the angiogenesis of gastric cancer cells using gelatin zymography, the results of which indicated that MMP-9 was positively associated with angiogenesis progression in patients with gastric cancer (40). These results were consistent with that of the current study. The results verified that miR-125a-5p and miR-7 regulated the proliferation and migration of VSMCs.
It is well documented that miRNAs modulate the biological functions of cells by downregulating mRNA targets post-transcriptionally (41). Due to the suppression of miR-125a-5p and miR-7 on the growth and migration of VSMCs, the current study hypothesized that target gene(s) should be positively associated with cell proliferation and migration. To explore the mechanism underlying the inhibition of the proliferation and migration of VMSCs mediated by miR-125a-5p and miR-7, the putative target gene for both miRNAs was identified using bioinformatics tools. EGFR was considered a candidate target for miR-125a-5p and miR-7 in VMSCs. As a member of ErbB family, EGFR serves an important role in various pathophysiological processes, including cell tumorigenesis and transformation (42,43). The association between EGFR and miR-125a-5p or miR-7 was further validated by luciferase reporter assays and western blotting. EGFR had a negative association with the expression of miR-125a-5p and miR-7 in VSMCs. Previous studies have demonstrated that miR-125 and miR-7 target numerous genes, including EGFR (44,45). The results of the current study are therefore consistent with these. In VSMCs, EGFR is involved with cell growth, migration, inflammation and vascular remodeling, consequently triggering vascular diseases, including atherosclerosis and hypertension (46). In the current study, overexpression of EGFR accelerated cell growth and migration of PDGF-BB-treated VSMCs. Furthermore, the results demonstrated that EGFR overexpression abrogated the miR-125a-5p and miR-7-mediated inhibition of VSMC proliferation and migration. In summary, the results verified that miR-125a-5p and miR-7 decreased the migration and invasion of PDGF-BB-treated VSMCs by targeting EGFR. However, a limitation of the current study is that the effect of miR-125a-5p and miR-7 on EGFR luciferase activity and protein level were not investigated. These should be researched in future studies.
In conclusion, the results demonstrated that the expression of miR-125a-5p and miR-7 was decreased in PDGF-BB-stimulated VSMCs. Following this, EGFR was reported to be a target of miR-125a-5p and miR-7, and overexpression of EGFR reversed the inhibition of VSMC proliferation, migration and invasion mediated by miR-125a-5p and miR-7. The results indicated that miR-125a-5p and miR-7 may serve as novel molecular targets in the proliferation and migration of VSMCs.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author's contributions
HZ and XL conceived and designed the study. SL and YH performed the literature search and acquisition of data. DG and YW contributed to analysis and interpretation of data. HZ and XL drafted and revised the manuscript. HZ and XL confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lacolley P, Regnault V, Nicoletti A, Li Z, Michel JB. The vascular smooth muscle cell in arterial pathology: A cell that can take on multiple roles. Cardiovasc Res. 2012;95:194–204. doi: 10.1093/cvr/cvs135. [DOI] [PubMed] [Google Scholar]
- 2.Chen Q, Zhang H, Liu Y, Adams S, Eilken H, Stehling M, Corada M, Dejana E, Zhou B, Adams RH. Endothelial cells are progenitors of cardiac pericytes and vascular smooth muscle cells. Nat Commun. 2016;7:12422. doi: 10.1038/ncomms12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Régent A, Ly KH, Lofek S, Clary G, Tamby M, Tamas N, Federici C, Broussard C, Chafey P, Liaudet-Coopman E, et al. Proteomic analysis of vascular smooth muscle cells in physiological condition and in pulmonary arterial hypertension: Toward contractile versus synthetic phenotypes. Proteomics. 2016;16:2637–2649. doi: 10.1002/pmic.201500006. [DOI] [PubMed] [Google Scholar]
- 4.Bennett MR, Sinha S, Owens GK. Vascular smooth muscle cells in atherosclerosis. Circ Res. 2016;118:692–702. doi: 10.1161/CIRCRESAHA.115.306361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.In This Issue. Proc Natl Acad Sci. 2007;104:7731–7732. doi: 10.1073/iti1907104. doi: 10.1073/iti1907104. [DOI] [Google Scholar]
- 6.Kato Y, Yokoyama U, Fujita T, Umemura M, Kubota T, Ishikawa Y. Epac1 deficiency inhibits basic fibroblast growth factor-mediated vascular smooth muscle cell migration. J Physiol Sci. 2019;69:175–184. doi: 10.1007/s12576-018-0631-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim SH, Yun SJ, Kim YH, Ha JM, Jin SY, Lee HS, Kim SJ, Shin HK, Chung SW, Bae SS. Essential role of krüppel-like factor 5 during tumor necrosis factor α-induced phenotypic conversion of vascular smooth muscle cells. Biochem Biophys Res Commun. 2015;463:1323–1327. doi: 10.1016/j.bbrc.2015.06.123. [DOI] [PubMed] [Google Scholar]
- 8.Shawky NM, Segar L. Sulforaphane inhibits platelet-derived growth factor-induced vascular smooth muscle cell proliferation by targeting mTOR/p70S6kinase signaling independent of Nrf2 activation. Pharmacol Res. 2017;119:251–264. doi: 10.1016/j.phrs.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiang R, Chen J, Li S, Yan H, Meng Y, Cai J, Cui Q, Yang Y, Xu M, Geng B, Yang J. VSMC-specific deletion of FAM3A attenuated ang ii-promoted hypertension and cardiovascular hypertrophy. Circ Res. 2020;126:1746–1759. doi: 10.1161/CIRCRESAHA.119.315558. [DOI] [PubMed] [Google Scholar]
- 10.Dong S, Xiong W, Yuan J, Li J, Liu J, Xu X. MiRNA-146a regulates the maturation and differentiation of vascular smooth muscle cells by targeting NF-κB expression. Mol Med Rep. 2013;8:407–412. doi: 10.3892/mmr.2013.1538. [DOI] [PubMed] [Google Scholar]
- 11.Frismantiene A, Philippova M, Erne P, Resink TJ. Smooth muscle cell-driven vascular diseases and molecular mechanisms of VSMC plasticity. Cell Signal. 2018;52:48–64. doi: 10.1016/j.cellsig.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Zhang L, Wang Y, Ding H, Xue S, Qi H, Li P. MicroRNAs or long noncoding RNAs in diagnosis and prognosis of coronary artery disease. Aging Dis. 2019;10:353–366. doi: 10.14336/AD.2018.0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai W, Zhang J, Yang J, Fan Z, Liu X, Gao W, Zeng P, Xiong M, Ma C, Yang J. MicroRNA-24 attenuates vascular remodeling in diabetic rats through PI3K/Akt signaling pathway. Nutr Metab Cardiovasc Dis. 2019;29:621–632. doi: 10.1016/j.numecd.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Gao F, Kataoka M, Liu N, Liang T, Huang ZP, Gu F, Ding J, Liu J, Zhang F, Ma Q, et al. Therapeutic role of miR-19a/19b in cardiac regeneration and protection from myocardial infarction. Nat Commun. 2019;10:1802. doi: 10.1038/s41467-019-09530-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu WH, Hu CP, Chen XP, Zhang WF, Li XW, Xiong XM, Li YJ. MicroRNA-130a mediates proliferation of vascular smooth muscle cells in hypertension. Am J Hypertens. 2011;24:1087–1093. doi: 10.1038/ajh.2011.116. [DOI] [PubMed] [Google Scholar]
- 16.Gareri C, Iaconetti C, Sorrentino S, Covello C, De Rosa S, Indolfi C. miR-125a-5p modulates phenotypic switch of vascular smooth muscle cells by targeting ETS-1. J Mol Biol. 2017;429:1817–1828. doi: 10.1016/j.jmb.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Goettsch C. miRNA-125b and their targets in cardiovascular calcification. Proc Physiol Soc. 2013;13:1234–1246. [Google Scholar]
- 18.Fan X, Liu M, Tang H, Leng D, Hu S, Lu R, Wan W, Yuan S. MicroRNA-7 exerts antiangiogenic effect on colorectal cancer via ERK signaling. J Surg Res. 2019;240:48–59. doi: 10.1016/j.jss.2019.02.035. [DOI] [PubMed] [Google Scholar]
- 19.Yano S, Kondo K, Yamaguchi M, Richmond G, Hutchison M, Wakeling A, Averbuch S, Wadsworth P. Distribution and function of EGFR in human tissue and the effect of EGFR tyrosine kinase inhibition. Anticancer Res. 2003;23:3639–3650. [PubMed] [Google Scholar]
- 20.Clauditz TS, Gontarewicz A, Lebok P, Tsourlakis MC, Grob TJ, Münscher A, Sauter G, Bokemeyer C, Knecht R, Wilczak W. Epidermal growth factor receptor (EGFR) in salivary gland carcinomas: Potentials as therapeutic target. Oral Oncol. 2012;48:991–996. doi: 10.1016/j.oraloncology.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Lee JS, Kim SY, Kwon CH, Kim YK. EGFR-dependent ERK activation triggers hydrogen peroxide-induced apoptosis in OK renal epithelial cells. Arch Toxicol. 2006;80:337–346. doi: 10.1007/s00204-005-0052-2. [DOI] [PubMed] [Google Scholar]
- 22.Schreier B, Döhler M, Rabe S, Schneider B, Schwerdt G, Ruhs S, Sibilia M, Gotthardt M, Gekle M, Grossmann C. Consequences of epidermal growth factor receptor (ErbB1) loss for vascular smooth muscle cells from mice with targeted deletion of ErbB1. Arterioscler Thromb Vasc Biol. 2011;31:1643–1652. doi: 10.1161/ATVBAHA.111.223537. [DOI] [PubMed] [Google Scholar]
- 23.Dong X, Hu H, Fang Z, Cui J, Liu F. CTRP6 inhibits PDGF-BB-induced vascular smooth muscle cell proliferation and migration. Biomed Pharmacother. 2018;103:844–850. doi: 10.1016/j.biopha.2018.04.112. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-(Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Ye G, Huang K, Yu J, Zhao L, Zhu X, Yang Q, Li W, Jiang Y, Zhuang B, Liu H, et al. MicroRNA-647 Targets SRF-MYH9 axis to suppress invasion and metastasis of gastric cancer. Theranostics. 2017;7:3338–3353. doi: 10.7150/thno.20512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun L, Lian JX, Meng S. MiR-125a-5p promotes osteoclastogenesis by targeting TNFRSF1B. Cell Mol Biol Lett. 2019;24:23. doi: 10.1186/s11658-019-0146-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, Ma Y, Liu C, Li P, Yu T. Reduced miR-125a-5p level in non-small-cell lung cancer is associated with tumour progression. Open Biol. 2018;8:180118. doi: 10.1098/rsob.180118. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Vo DT, Karanam NK, Ding L, Saha D, Yordy JS, Giri U, Heymach JV, Story MD. miR-125a-5p functions as tumor suppressor microRNA and is a marker of locoregional recurrence and poor prognosis in head and neck cancer. Neoplasia. 2019;21:849–862. doi: 10.1016/j.neo.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, An X, Bao L, Li Y, Pan Y, He J, Liu L, Zhu X, Zhang J, Cheng J, Chu W. MiR-125a-3p-KLF15-BCAA regulates the skeletal muscle branched-chain amino acid metabolism in nile tilapia (Oreochromis niloticus) during starvation. Front Genet. 2020;11:852. doi: 10.3389/fgene.2020.00852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Che P, Liu J, Shan Z, Wu R, Yao C, Cui J, Zhu X, Wang J, Burnett MS, Wang S, Wang J. miR-125a-5p impairs endothelial cell angiogenesis in aging mice via RTEF-1 downregulation. Aging Cell. 2014;13:926–934. doi: 10.1111/acel.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng X, Wu Z, Xu K, Qiu Y, Su X, Zhang Z, Zhou M. Interfering histone deacetylase 4 inhibits the proliferation of vascular smooth muscle cells via regulating MEG3/miR-125a-5p/IRF1. Cell Adh Migr. 2019;13:41–49. doi: 10.1080/19336918.2018.1506653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang X, Qiu J, Kang H, Wang Y, Qian J. miR-125a-5p suppresses colorectal cancer progression by targeting VEGFA. Cancer Manag Res. 2018;10:5839–5853. doi: 10.2147/CMAR.S161990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Zhang D, Lv J, Wang S, Zhang Q. MiR-125a-5p suppresses bladder cancer progression through targeting FUT4. Biomed Pharmacother. 2018;108:1039–1047. doi: 10.1016/j.biopha.2018.09.100. [DOI] [PubMed] [Google Scholar]
- 34.Xu L, Li Y, Yin L, Qi Y, Sun H, Sun P, Xu M, Tang Z, Peng J. miR-125a-5p ameliorates hepatic glycolipid metabolism disorder in type 2 diabetes mellitus through targeting of STAT3. Theranostics. 2018;8:5593–5609. doi: 10.7150/thno.27425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horsham JL, Ganda C, Kalinowski FC, Brown RA, Epis MR, Leedman PJ. MicroRNA-7: A miRNA with expanding roles in development and disease. Int J Biochem Cell Biol. 2015;69:215–224. doi: 10.1016/j.biocel.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Ha YM, Nam JO, Kang YJ. Pitavastatin regulates ang II induced proliferation and migration via IGFBP-5 in VSMC. Korean J Physiol Pharmacol. 2015;19:499–506. doi: 10.4196/kjpp.2015.19.6.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang BF, Jiang H, Chen J, Guo X, Hu Q, Yang S. KDM3A inhibition attenuates high concentration insulin-induced vascular smooth muscle cell injury by suppressing MAPK/NF-κB pathways. Int J Mol Med. 2018;41:1265–1274. doi: 10.3892/ijmm.2017.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park HJ, Kim MK, Kim Y, Bae SS, Kim HJ, Bae SK, Bae MK. Gastrin-releasing peptide promotes the migration of vascular smooth muscle cells through upregulation of matrix metalloproteinase-2 and −9. BMB Rep. 2017;50:628–633. doi: 10.5483/BMBRep.2017.50.12.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi S, Perrino S, Miao X, Lamarche-Vane N, Brodt P. The chemokine CCL7 regulates invadopodia maturation and MMP-9 mediated collagen degradation in liver-metastatic carcinoma cells. Cancer Lett. 2020;483:98–113. doi: 10.1016/j.canlet.2020.03.018. [DOI] [PubMed] [Google Scholar]
- 40.Li TJ, Jiang YM, Hu YF, Huang L, Yu J, Zhao LY, Deng HJ, Mou TY, Liu H, Yang Y, et al. Interleukin-17-producing neutrophils link inflammatory stimuli to disease progression by promoting angiogenesis in gastric cancer. Clin Cancer Res. 2017;23:1575–1585. doi: 10.1158/1078-0432.CCR-16-0617. [DOI] [PubMed] [Google Scholar]
- 41.Wang P, Guan Q, Zhou D, Yu Z, Song Y, Qiu W. miR-21 inhibitors modulate biological functions of gastric cancer cells via PTEN/PI3K/mTOR pathway. DNA Cell Biol. 2018;37:38–45. doi: 10.1089/dna.2017.3922. [DOI] [PubMed] [Google Scholar]
- 42.Schreier B, Schwerdt G, Heise C, Bethmann D, Rabe S, Mildenberger S, Gekle M. Substance-specific importance of EGFR for vascular smooth muscle cells motility in primary culture. Biochim Biophys Acta. 2016;1863:1519–1533. doi: 10.1016/j.bbamcr.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 43.Spano JP, Lagorce C, Atlan D, Milano G, Domont J, Benamouzig R, Attar A, Benichou J, Martin A, Morere JF, et al. Impact of EGFR expression on colorectal cancer patient prognosis and survival. Ann Oncol. 2005;16:102–108. doi: 10.1093/annonc/mdi006. [DOI] [PubMed] [Google Scholar]
- 44.Zhou X, Hu Y, Dai L, Wang Y, Zhou J, Wang W, Di W, Qiu L. MicroRNA-7 inhibits tumor metastasis and reverses epithelial-mesenchymal transition through AKT/ERK1/2 inactivation by targeting EGFR in epithelial ovarian cancer. PLoS One. 2014;9:e96718. doi: 10.1371/journal.pone.0096718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin T, Ren Q, Zuo W, Jia R, Xie L, Lin R, Zhao H, Chen J, Lei Y, Wang P, et al. Valproic acid exhibits anti-tumor activity selectively against EGFR/ErbB2/ErbB3-coexpressing pancreatic cancer via induction of ErbB family members-targeting microRNAs. J Exp Clin Cancer Res. 2019;38:150. doi: 10.1186/s13046-019-1160-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schreier B, Gekle M, Grossmann C. Role of epidermal growth factor receptor in vascular structure and function. Curr Opin Nephrol Hypertens. 2014;23:113–121. doi: 10.1097/01.mnh.0000441152.62943.29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.