Abstract
Mounting evidence has indicated that circular RNAs (circRNAs) serve essential roles in the tumorigenesis and development of various types of cancer. However, the biological functions and the underlying mechanisms of circRNAs in breast cancer (BC) remain largely elusive. In the present study, the expression pattern of circRNAs in three pairs of BC tissues and adjacent normal tissues was determined using a circRNA microarray. The expression and prognostic value of circOMA1 were evaluated by reverse transcription-quantitative PCR in 64 pairs of BC tissues and adjacent normal tissues. Survival curves were generated by the Kaplan-Meier method, and statistical significance was estimated using the log-rank test. A series of in vitro functional experiments were then performed to investigate the role of circOMA1 in the tumorigenesis of BC. The results revealed that the expression levels of circOMA1 were upregulated in BC tissues, and its expression was markedly associated with tumor size and lymph node metastasis. Receiver operating characteristic analysis demonstrated that the expression of circOMA1 could be used to discriminate between BC tissues and adjacent normal tissues. Functionally, overexpression of circOMA1 promoted the viability, migration and invasion of BC cells, whereas circOMA1 knockdown had the opposite effect. Mechanistic investigations showed that circOMA1 promoted the progression of BC by sponging microRNA (miR)-1276 and upregulating sirtuin 4 (SIRT4) expression. In conclusion, circOMA1 may act as an oncogenic circRNA in BC via regulation of the miR-1276/SIRT4 axis.
Keywords: circular RNA OMA1, breast cancer, microRNA-1276, sirtuin 4
Introduction
Breast cancer (BC) is a common malignancy with the highest incidence and the second highest mortality rate worldwide (1). Numerous factors contribute to the occurrence of BC, including age, obesity, alcoholism and exposure to radiation (2). Although substantial progress has been made in the treatment of BC in recent decades, the overall prognosis of patients with BC remains poor. Therefore, there is an urgent need to discover additional mechanisms underlying the progression of BC and identify more effective therapeutic targets.
Circular RNAs (circRNAs) are a novel type of noncoding RNA (ncRNA) with covalently closed loop characteristics (3). It has been verified that circRNAs are derived from pre-mRNA transcripts (4). In addition, it has been suggested that circRNAs serve essential roles in gene regulation and may thereby be involved in the progression of various human diseases, including BC (5). For example, Wu et al (6) reported that circIRAK3 could sponge microRNA (miRNA/miR)-3607 to enhance the metastasis of BC. Furthermore, circMYO9B has been shown to increase the proliferation and invasiveness of BC by sponging miR-4316 and upregulating forkhead box P4 (7). However, the functions and underlying mechanisms of circRNAs are still not fully understood in BC.
miRNAs are another group of ncRNAs with a length of 20–25 nucleotides. miRNAs can bind to the 3′ untranslated region (UTR) of mRNAs and thereby inhibit the expression of genes (8). Accumulating evidence has indicated that dysregulation of miRNAs is involved in the progression of various types of cancer, including BC. For example, miR-93 has been reported to act as a tumor suppressor via inhibition of WASP family member 3 and metastasis of BC (9). miR-6744 has also been reported to target the N-acetyltransferase 1 enzyme and promote anoikis in breast cancer (10). However, little is currently known about the upstream regulatory mechanisms of miRNAs.
Sirtuin 4 (SIRT4) is a mitochondrial matrix protein that belongs to the SIRT family (11). SIRT4 has been shown to participate in various biological activities, such as cellular proliferation, migration, metabolism and inflammation (12). According to previous studies, SIRT4 is strongly associated with the tumorigenesis of various tumors, including prostate cancer, liver cancer, thyroid cancer and neuroblastoma (13–15). However, the role of SIRT4 in BC is still unclear.
The aim of this study was to identify circRNAs associated with the tumorigenesis of BC. circOMA1 was identified as a novel circRNA that acted as an oncogene in BC. The diagnostic and therapeutic value of circOMA1 and its molecular mechanism were also examined.
Materials and methods
Clinical samples
This prospective study involved 64 patients who underwent tumor resection at The Zhenghai Longsai Hospital for primary BC between December 2018 and October 2019. All patients were newly diagnosed with primary BC. Patients who were <18 years old or >75 years old, presented with initial distant metastasis, complicated with other malignancies or received neoadjuvant therapy were excluded from the study. Tumor and matched, adjacent normal tissue were collected and stored in liquid nitrogen. The present study was approved by the Ethics Committee of Zhenghai Longsai Hospital (Ningbo, China) and was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all participants prior to the study.
Cell culture
Human normal breast epithelial cells (MCF-10A), BC cells (MDA-MB-468, MDA-MB-231, BT-549 and MCF-7) and 293T cells were obtained from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. All cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin (all Gibco; Thermo Fisher Scientific, Inc.). The cells were cultured in a humidified atmosphere containing 5% CO2 at 37°C.
Cell transfection
Three different short hairpin RNA (shRNA) molecules against circOMA1 (shRNA#1, shRNA#2, shRNA#3), SIRT4 (sh-SIRT4) and negative control (sh-NC) were subcloned into the GV248 (hU6-MCS-Ubiquitin-EGFP-IRES-puromycin) vector (Shanghai GeneChem Co., Ltd.). The following sequences were used: i) sh-circOMA1#1, 5′-ACCUCAGGUAGUCAAGUGATT-3′; ii) sh-circOMA1#2, 5′-AUGAUUCUUAAACCAGUACAG-3′; iii) sh-circOMA1#3 5′-CUAACAAGAAGGAAGUAGU-3′ iv) sh-SIRT4, 5′-TCCTATACAGCTACGGCTC-3′; and v) sh-NC′ 5′-TTCTCCGAACGTGTCACGT-3′. In order to overexpress SIRT4 or circOMA1, full-length SIRT4 and circOMA1 were synthesized by Shanghai GenePharma Co., Ltd. and were subcloned into the pcDNA3.1 vector (Shanghai GenePharma Co., Ltd.). The empty vector was used as a negative control. The plasmids were used to transfect 293 cells using the ViraPower kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's guidelines to generate the lentivirus. miR-1276 mimics/inhibitors and negative control (NC) mimics/inhibitors were purchased from Shanghai GenePharma Co., Ltd., and transfection was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The miR sequences were: i) miR-NC mimics, 5′-UUGUACUACACAAAAGUACUG-3′; ii) miR-1276 mimics, 5′-UAAAGAGCCCUGUGGAGACA-3′; iii) miR-NC inhibitor, 5′-UCACAACCUCCUAGAAAGAGUAGA-3′; and iv) miR-1276 inhibitor, 5′-UGUAUCCACAGGGCUCUUUA-3′; For the plasmids or miRNA mimics/inhibitors transfection, MCF-7 and MDA-MB-478 cells were seeded into 6-well plates at a density of 1×106 cells/well and cultured to 80% confluence. For each well, 1 µg of plasmids or 20 nM miRNA mimics/inhibitor were mixed with Lipfectamine® 2000 at a ratio of 1:4. The cells were then cultured for 24 h and transfection efficiency was assessed. For lentivirus infection, MCF-7 and MDA-MB-478 cells were seeded into 6-well plates at a density of 1×106 cells/well and cultured to 80% confluence. The lentiviral vector solution was mixed with 2 ml culture medium to 5×106 transduction units (TU), then added to each well and incubated overnight before the medium was changed. Transduction efficiency was assessed 24 h later.
For co-transfection of sh-circOMA1 and pcDNA3.1 SIRT4, MCF-7 and MDA-MB-478 cells were seeded into 6-well plates at a density of 1×106 cells/well and cultured to 80% confluence. For each well, 1 µg plasmid was mixed with Lipfectamine® 2000 at a ratio of 1:4 and 1 ml total volume of lentiviral vector solution (5×106 TU) was added to MCF-7 and MDA-MB-478 cells at the same time,. The culture medium was changed with fresh medium 4 I mh later. The cells were then kept in the incubator and subjected to different assays at indicated time point. The cells were screened using puromycin (2 µg/ml) for 3 days.
RNA purification and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from breast cancer and adjacent normal tissue samples or cells (MCF-10A, MDA-MB-468, MDA-MB-231, BT-549 and MCF-7) using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The quantity and quality of RNA were measured using a NanoDrop ND 2000 Spectrophotometer (NanoDrop; Thermo Fisher Scientific Inc.). Total RNA was reverse transcribed into cDNA using PrimeScript RT Reagent (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol.
The expression levels of circOMA1 and miRNAs were evaluated using SYBR Premix Ex Taq (Takara Biotechnology). GAPDH was used as internal control for circOMA1/SIRT4 and U6 was used as internal control for and miR-1276, respectively. The following primers were used: i) circOMA1 forward, 5′-AACCCAAGATGCCAGAATGGT-3′ and reverse, 5′-TTGATGACAGCCCCGTGAG-3′; ii) SIRT4 forward, 5′-TGTGGTGAACTGACTCCTCGTGCTGAGC-3′ and reverse, 5′-CGGAAGTTTTCTTTCACTAGCAGCGAGG-3′; iii) GAPDH forward, 5′-CTGGGCTACACTGAGCACC-3′ and reverse, 5′-AAGTGGTCGTTGAGGGCAATG-3′; and iv) U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′ The reaction conditions were as follows: 95°C for 10 min for 1 cycle, followed by denaturation at 95°C for 30 sec, annealing at 56°C for 1 min and extension at 72°C for 30 sec for a total of 40 cycles. The relative gene expression was calculated using the 2−ΔΔCq method (16), and the samples were run in triplicate.
Actinomycin D and RNase R treatment
To test the stability of circOMA1 expression, actinomycin D (2 mg/ml; Sigma-Aldrich; Merck KGaA) or DMSO was added to inhibit transcription. Total RNA (2 µg) was treated with or without 3U/µg of RNase R (Sigma-Aldrich; Merck KGaA). The resulting RNA was purified with the RNeasy MinElute Cleanup kit (Qiagen, Inc.)
circRNA microarray
Total RNA purified from three pairs of BC and adjacent normal tissues was subjected to circRNA array analysis. Purification of RNA and microarray hybridization were conducted according to the standard protocols from Arraystar, Inc. Briefly, total RNA was treated with RNase R at 37°C for 1 h (Sigma-Aldrich; Merck KGaA) to remove linear RNA and to enrich circRNAs. Subsequently, antisense RNA was amplified and labeled using the Arraystar Super RNA labeling kit (cat. no. AS-LE-005; Arraystar, Inc.) according to the manufacturer's protocol. Briefly, first-strand cDNA synthesis was conducted using an oligo(dT)24 primer containing a T7 promoter sequence. After RNA template degradation and second-strand cDNA synthesis, antisense RNA was transcribed using biotinylated ribonulceotides and T7 RNA polymerase provided in the kit. The labeled RNAs were then hybridized using Arraystar circRNA Array (v2.0; Arraystar, Inc.), and hybridization was scanned using the G2505C scanner (Agilent Technologies, Inc.).
RNA pull-down assay
RNA pull-down assays were performed using the protocol from GeneSeed. Briefly, 1×106 MCF-7 or MDA-MB-478 cells were fixed in 2% formaldehyde at room temperature for 20 min, then sonicated at room temperature for 15 min. Subsequently, the supernatant was incubated with biotinylated circOMA1 or miR-1276 probes or control probe (Guangzhou RiboBio Co., Ltd.) and magnetic streptavidin Dynabeads (Sigma-Aldrich; Merk KGaA) at 4°C for 1 h in an orbital shaker. The lysates were then discarded and total RNA was extracted for RT-qPCR analysis using as aforementioned
RNA-binding protein immunoprecipitation (RIP) assay
RIP experiments were conducted using the Magna RIP RNA-binding protein immunoprecipitation kit (cat. no. 17-10523; EMD Millipore) according to the manufacturer's guide. Briefly, 1×107 MCF-7 or MDA-MB-478 cells were transfected with Flag-Ago2 or Flag-GFP plasmids, then cultured for 24 h. The cells were then collected and re-suspended in 0.1 ml RIPA lysis buffer (Beyotime Institute of Biotechnology). The cell lysates were incubated with 5 antibody against Flag (cat. no. F7425) or IgG-coated beads (cat. no. I8600) (both from Sigma-Aldrich; Merck KGaA) with rotation at 4°C overnight. After treating with proteinase K, the immunoprecipitated RNA was extracted with the RNeasy MinElute Cleanup kit (Qiagen, Inc.) and reverse transcribed PrimeScript RT Reagent (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. The abundance of circOMA1 was measured by RT-PCR as aforementioned.
Cell viability assay
Cell viability was assessed using the Cell Counting Kit (CCK)-8 assay (Beyotime Institute of Biotechnology) at 24, 48 and 72 h. MCF-7 and MDA-MB-478 cells were seeded into 96-well plates at a density of 1×104 cells/well. Subsequently 10 µl CCK-8 solution was added to each well and incubated at 37°C for a further 2 h. The absorbance was measured at 450 nm using a microplate reader (Biotek Corporation).
Cellular apoptosis assay
Cell apoptosis was assessed using the Annexin V-fluorescein isothiocyanate/propidium iodide (PI) detection kit (Invitrogen), according to the manufacturer's instructions. Briefly, 1×105 MCF-7 and MDA-MB-478 cells were stained with Annexin V/PI at 4°C for 15 min in the dark. Cells were then washed with PBS and resuspended in binding buffer provided in the kit. The status of early and late apoptotic cells (Annexin V+) was determined by flow cytometry (FACSCalibur; BD Biosciences). The data were analyzed using the FlowJo 10.0.7 (FlowJo LLC)
Migration and invasion assays
For migration and invasion assays, similar procedures were performed. Briefly, 5×105 cells were seeded into 8-µm pore Transwell chambers (Corning, Inc.) For the invasion assay, the Transwell chamber was precoated with Matrigel (Thermo Fisher Scientific, Inc.) before cell seeding. The chambers were then inserted into a 24-well plate; the upper chambers were filled with 200 µl serum-free medium, whereas the bottom chambers were filled with 500 µl complete medium. After incubation for 24 h at room temperature, cells were fixed in 4% paraformaldehyde (Beyotime Institute of Biotechnology) for 10 min and stained with 0.1% crystal violet (Beyotime Institute of Biotechnology) for 10 min at room temperature. Non-migrating cells in the upper chambers were removed; migratory and invasive cells were counted in three randomly selected fields and images were captured using an IX70 inverted optical microscope (magnification, ×100; Olympus Corporation).
Subcellular fractionation assay
The location of RNAs was evaluated using the PARIS™ kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Briefly, cells were suspended in cytoplasm lysis buffer (cat. no. 78840; Thermo Fisher Scientific, Inc.) and centrifuged at 500 × g for 5 min at room temperature. The cytoplasmic supernatant was then collected, and the pellet was resuspended in nucleus lysis buffer at 4°C for 1 h, followed by centrifugation at 500 × g for 10 min at room temperature. The RNAs derived from cytoplasmic and nuclear extracts were extracted using TRIzol, according to the manufacturer's instructions. The expression levels of GAPDH (cytoplasmic control), U6 (nuclear control) and circOMA1 in the nucleus and cytoplasm were assessed by RT-qPCR as aforementioned.
Dual-luciferase reporter assay
Dual-luciferase reporter assays were performed by cotransfection of cells with recombinant luciferase reporter vectors and the indicated transfection plasmids. Partial fragments of circOMA1 and SIRT4 3′-UTRs containing wild-type (wt) or mutant (mut) miR-1276 interacting sites were obtained. Subsequently, these fragments were subcloned into psiCHECK luciferase reporter vectors (Promega Corporation). Co-transfection was performed using Lipofectamine 2000. Briefly, MCF-7 and MDA-MB-468 were seeded into 6-well plates at a density of 1×106 cells/well., Cells were co-transfected 24 h later with 1 µg circOMA-WT/MUT or SIRT4-WT/MUT plasmids and 100 pmol miR-1276 mimics or miR-NC mimics using Lipofectamine 2000. After transfection, cells were cultured at 37°C for 24 h. Luciferase activity was monitored after using the Dual Luciferase Reporter Assay system (Promega Corporation). The data were normalized to Renilla luciferase activity. The mut 3′-UTRs were constructed using the QuickChange™ II Site-Directed Mutagenesis kit (Stratagene; Agilent Technologies, Inc.) according to the manufacturer's protocol. All experiments were conducted in triplicate and repeated at least three times.
Western blotting
Total cellular proteins were obtained using RIPA lysis buffer (Beyotime Institute of Biotechnology) supplemented with a proteinase and phosphatase inhibitor cocktail (Sigma-Aldrich). Total proteins (20 µg) were separated by SDS-PAGE on 12% gels and were transferred onto PVDF membranes (EMD Millipore). The membranes were blocked with skimmed milk for 1 h at room temperature and were then incubated with primary antibodies overnight at 4°C. Subsequently, the membranes were washed three times with PBS and incubated with the corresponding HRP-conjugated secondary antibody at room temperature for 1 h. The membranes were visualized using an ECL Prime Western Blotting kit (Beyotime Institute of Biotechnology). The following primary and secondary antibodies were used: i) Anti-SIRT4 (cat. no. ab10140; 1:1,000; Abcam); ii) anti-tubulin (cat. no. 2146; 1:5,000; Cell Signaling Technology); iii) anti-mouse, HRP-linked antibody (cat. no. 7076; 1:5,000; Cell Signaling Technology, Inc.).
Bioinformatical analysis
Prediction of the circOMA1-miRNA-target gene was performed using the StarBase 3.0 (http://starbase.sysu.edu.cn) and CircInteractome (https://circinteractome.nia.nih.gov/).
Statistical analysis
Statistical analyses were performed with SPSS 12.0 (IBM Corp.). Data are presented as the mean ± SD. Differences between two groups were assessed using a paired Student's t-test. Differences between multiple groups were analysed using one-way ANOVA followed by Tukey's post hoc test. The diagnostic value of circOMA1 was evaluated using receiver operating characteristic (ROC) curves as described previously (17). Survival curves for patients with BC were generated using the Kaplan-Meier method, and the difference was analyzed by log-rank test. The χ2 test was used to compare the clinicopathological characteristics of patients. Pearson's correlation was performed to assess correlation. P<0.05 (two-tailed) was considered to indicate a statistically significant difference. All statistical analyses were performed using the Prism 7.0 (GraphPad Software, Inc.).
Results
circOMA1 is upregulated in BC tissues and is associated with poor prognosis in patients with BC
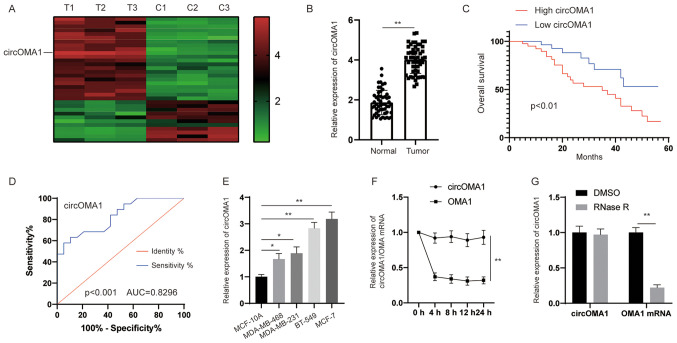
To identify the potential dysregulated circRNAs in BC, circRNA expression profiling was performed in BC tissues and adjacent normal tissues. The circRNA microarray assay revealed that hsa_circRNA_0000073 was the most upregulated circRNA in BC tissues (Fig. 1A). According to the human reference genome (https://www.ncbi.nlm.nih.gov/grc/human), hsa_circRNA_0000073, which was derived from the gene OMA1, was termed circOMA1. Subsequently, the expression levels of circOMA1 were measured in 64 pairs of BC tissues and adjacent normal tissues. The results revealed that the expression levels of circOMA1 were significantly higher in BC tissues compared with those in normal tissues (Fig. 1B). According to the median ratio of circOMA1 expression, patients with BC were divided into two groups (high and low expression of circOMA1). Kaplan-Meier analysis and log-rank test revealed that patients with BC and high expression of circOMA1 had poorer overall survival compared with in patients with BC and low expression of circOMA1 (Fig. 1C). In addition, to assess whether the expression of circOMA1 may be used discriminate between BC tissues and adjacent normal tissues, receiver operating characteristic (ROC) analysis was performed. An area under the ROC curve of 0.8296 indicated that the expression of circOMA1 could discriminate between BC tissues and adjacent normal tissues (Fig. 1D). Furthermore, the expression levels of circOMA1 were higher in BC cells than those in normal breast epithelial cells (Fig. 1E). RNA stability analysis demonstrated that the circular transcript circOMA1 was much more stable than the linear mRNA transcript in BC cells incubated with a transcription inhibitor (actinomycin D) (Fig. 1F). After exposure to RNase R or control (DMSO), RT-qPCR results demonstrated that the expression levels of the linear form of OMA1 were significantly decreased under RNase R treatment, whereas there was little change in circOMA1 expression levels. Resistance to digestion by RNase R exonuclease further confirmed that OMA1 RNA species were present in circular form (Fig. 1G). circOMA1 expression was also revealed to be closely associated with tumor size and lymph node metastasis in patients (Table I). These data suggested that circOMA1 expression was increased in BC tissues and cell lines, and may be associated with poor prognosis of BC. Among the BC cell lines assessed, circOMA1 expression was the highest in MCF-7, whereas it was lowest in MDA-MB-468 cells. Therefore, circOMA1 overexpression was induced in MDA-MB-468 cells and circOMA1 expression was knocked down in MCF-7 cells, in order to elucidate the function of circOMA1 in BC cells.
Figure 1.
circOMA1 is upregulated in BC tissues and cells. (A) A heat map indicated the upregulated and downregulated circRNAs. (B) Expression levels of circOMA1 in BC tissues and adjacent normal tissues. (C) Prognosis analysis of patients with BC and high or low circOMA1 expression, as determined by Kaplan-Meier and log-rank test. (D) ROC analysis was used to discriminate between BC tissues and adjacent normal tissues. (E) Expression levels of circOMA1 in BC cells and normal breast cells. *P<0.05, **P<0.01. (F) RT-qPCR analysis of circOMA1 and OMA1 RNA after treatment with Actinomycin D at the indicated time points. (G) Expression levels of circOMA1 and OMA1 in MCF-7 cells administrated RNase R or control (DMSO). Data are presented as the mean ± SD. Experiments were performed at least three times. **P<0.01. BC, breast cancer; circ/circRNA, circular RNA; AUC, area under the ROC curve; ROC, receiver operating characteristic.
Table I.
Association between the circOMA1 expression and the clinicopathological features of patients of breast cancer.
| Expression of circOMA1 | |||||
|---|---|---|---|---|---|
|
|
|||||
| Parameters | Numbers (n=64) | High (n=32) | Low (n=32) | χ2 | P-value |
| Age, years | 0.068 | 0.794 | |||
| ≤50 | 23 | 12 | 11 | ||
| >50 | 41 | 20 | 21 | ||
| Tumor size, cm | 6.478 | 0.011 | |||
| ≤2 | 26 | 8 | 18 | ||
| >2 | 38 | 24 | 14 | ||
| TNM stage | 1.697 | 0.193 | |||
| I–II | 41 | 18 | 23 | ||
| III- IV | 23 | 14 | 9 | ||
| Lymphatic metastasis | 6.667 | 0.01 | |||
| Yes | 24 | 17 | 7 | ||
| No | 40 | 15 | 25 | ||
| T stage | 1.564 | 0.211 | |||
| T1 | 31 | 13 | 18 | ||
| T2-T3 | 33 | 19 | 14 | ||
| N stage | 1.036 | 0.309 | |||
| N0 | 38 | 21 | 17 | ||
| N1-N3 | 26 | 11 | 15 | ||
circOMA1, circular RNA OMA1.
circOMA1 promotes the tumorigenesis of BC cells
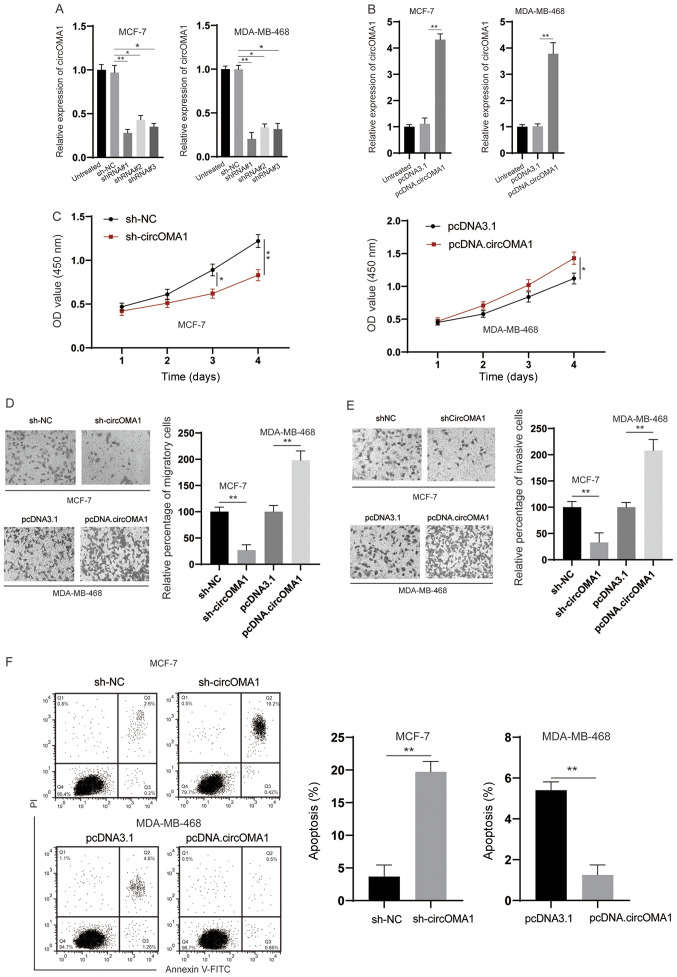
To examine the biological functions of circOMA1, shRNA was used to knock down circOMA1 in BC cells (Fig. 2A). Among the three shRNAs tested, shRNA#1 was the most efficient and was therefore chosen for subsequent experiments. In addition, the expression levels of circOMA1 were increased by transfection of BC cells with the circOMA1 expression vector (Fig. 2B). The CCK-8 assay revealed that silencing circOMA1 inhibited proliferation, whereas overexpression of circOMA1 promoted the proliferation of BC cells (Fig. 2C). Subsequently, Transwell assays indicated that knockdown of circOMA1 suppressed migration/invasion, whereas overexpression of circOMA1 increased the migration/invasion of BC cells (Fig. 2D and E). Flow cytometric analysis of early and late apoptotic cells demonstrated that knockdown of circOMA1 increased apoptosis, whereas overexpression of circOMA1 reduced apoptosis of BC cells (Fig. 2F). Taken together, these data indicated that circOMA1 promoted the progression of BC.
Figure 2.
circOMA1 promotes the tumorigenesis of BC. (A) MCF-7 cells were transfected as indicated and the expression of circOMA1 was evaluated by RT-qPCR. (B) MDA-MB-468 cells were transfected with pcDNA3.1 or pcDNA.circOMA1, and the expression of circOMA1 was measured by RT-qPCR. (C) BC cells were transfected as indicated and the proliferation of cells was measured by Cell Counting Kit-8 assay at different time points. (D) BC cells were transfected as indicated and cellular migration was assessed (magnification, ×100). (E) BC cells were transfected as indicated and cellular invasion was assessed (magnification, ×100). (F) BC cells were transfected as indicated and cellular apoptosis was assayed. Data are presented as the mean ± SD. Experiments were performed at least three times. *P<0.05, **P<0.01. BC, breast cancer; circ, circular RNA; RT-qPCR, reverse transcription-quantitative PCR; sh, short hairpin RNA; NC, negative control; OD, optical density; PI, propidium iodide.
circOMA1 acts as a sponge of miR-1276
To further analyze the role of circOMA1, a subcellular fractionation assay was conducted, and it was revealed that circOMA1 was mainly distributed in the cytoplasm rather than the nucleus (Fig. 3A). The cytoplasmic location of circOMA1 strongly indicated that circOMA1 may be involved in post-transcriptional regulation. A previous study has demonstrated that circRNA molecules act as competing endogenous RNAs (ceRNAs) by blocking the Ago2-mediated silencing complex (18). Therefore, an RNA-protein immunoprecipitation assay was conducted; the results revealed that circOMA1 could bind Ago2 protein in BC cells (Fig. 3B). Based on these findings, it was hypothesized that circOMA1 might regulate gene expression at the post-transcriptional level. To verify this, bioinformatics tools (StarBase 3.0 and CircInteractome) were used, and several candidate miRNAs were identified. To test this hypothesis, an RNA pull-down assay was performed and revealed that a specific probe against circOMA1 could enrich circOMA1 over 30-fold compared with the control (Fig. 3C). Furthermore, miR-1276 was significantly increased in the circOMA1 probe group, whereas there was little change in the expression levels of other miRNAs in BC cells (Fig. 3D). Thus, miR-1276 was chosen for subsequent experiments. Dual-luciferase reporter assays revealed that the luciferase activities of the circOMA1 wt reporter were significantly decreased when cells were transfected with miR-1276 mimics compared with those transfected with the miR-NC or circOMA1 mut luciferase reporter (Fig. 3E). In addition, it was revealed that silencing circOMA1 led to the upregulation of miR-1276 (Fig. 3F), whereas overexpression of circOMA1 induced downregulation of miR-1276 in BC cells (Fig. 3G). The expression levels of miR-1276 were also downregulated in BC tissues compared with those in normal tissues (Fig. 3H), and there was a negative correlation between miR-1276 and circOMA1 expression in BC tissues (Fig. 3I). Moreover, to further analyze the role of miR-1276, miR-1276 mimics and miR-1276 inhibitor were used to increase or decrease miR-1276 levels, respectively (Fig. 3J). It was demonstrated that miR-1276 mimics inhibited the tumorigenesis of BC cells, whereas the miR-1276 inhibitor had the opposite effects of silencing circOMA1 on the proliferation, migration, invasion and apoptosis of BC cells (Fig. 3K-N). Taken together, these data suggested that circOMA1 may act as a sponge of miR-1276.
Figure 3.
circOMA1 acts as sponge of miR-1276. (A) Subcellular fraction analysis of the location of circOMA1 in BC cells. (B) Ago2 RNA-protein immunoprecipitation assay was used for the detection of circOMA1 in BC cells expressing Flag-Ago2 or Flag-tag. (C) RNA pull-down assay was used for the detection of circOMA1 using a circOMA1-specific probe in BC cells. (D) Relative expression of circOMA1 putative binding miRNAs was examined by reverse transcription-quantitative PCR analysis using a circOMA1-specific probe. (E) Predictive binding sites between circOMA1 and miR-1276; relative luciferase activity was assessed in BC cells co-transfected with wt or mut luciferase reporters and miR-1276 mimics or corresponding NC. (F) BC cells were transfected with sh-NC or sh-circOMA1, and the expression of miR-1276 was assessed. (G) BC cells were transfected with pcDNA3.1 or pcDNA.circOMA1, and the expression of miR-1276 was assessed. (H) Expression of miR-1276 was assessed in BC tissues and adjacent normal tissues. (I) Correlation between the expression of miR-1276 and circOMA1 was analyzed in BC tissues. (J) BC cells were transfected as indicated and the expression levels of miR-1276 were measured. (K) BC cells were transfected as indicated and cell proliferation was measured by Cell Counting Kit-8 assay at different time points. *P<0.05, **P<0.01 vs. NC group. (L) BC cells were transfected as indicated and cellular migration was assessed (magnification, ×100). (M) BC cells were transfected as indicated and cellular invasion was assessed (magnification, ×100). (N) BC cells were transfected as indicated and cellular apoptosis was assessed. Data are presented as the mean ± SD. Experiments were performed at least three times. *P<0.05, **P<0.01. BC, breast cancer; circ/circRNA, circular RNA; sh, short hairpin RNA; NC, negative control; miR, microRNA; mut, mutant; wt, wild-type; OD, optical density; PI, propidium iodide; FITC, fluorescein isothiocyanate.
miR-1276 targets SIRT4 in BC cells
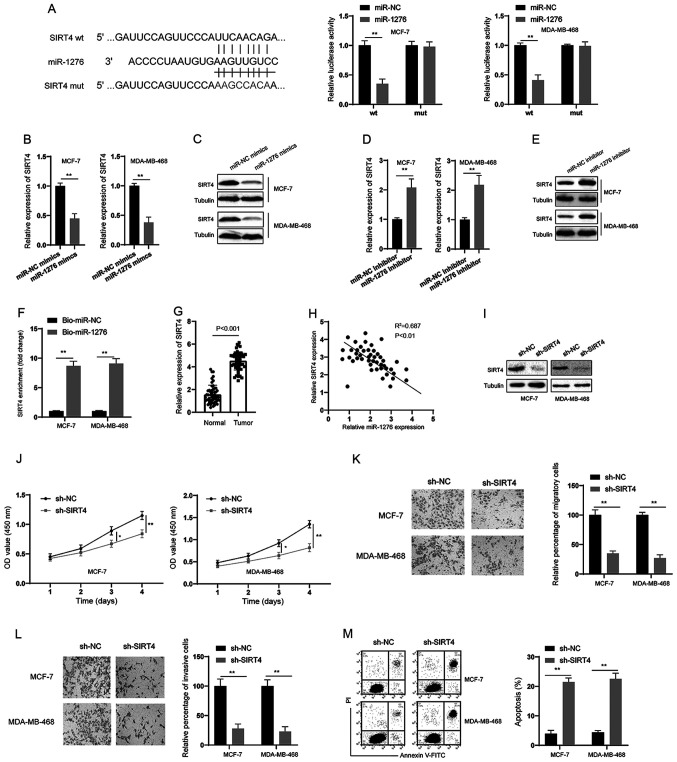
Through bioinformatics analysis, SIRT4 was predicted to be a direct target of miR-1276 (Fig. 4A). Subsequently, a dual-luciferase reporter assay was conducted, and it was revealed that miR-1276 could inhibit the luciferase activity of the wt reporter but had no effect on the mut reporter system in BC cells (Fig. 4A). In addition, miR-1276 mimics decreased the expression of SIRT4 at both the mRNA and protein levels in BC cells (Fig. 4B and C). Conversely, the miR-1276 inhibitor increased the expression of SIRT4 at both the mRNA and protein levels in BC cells (Fig. 4D and E). RNA pull-down assays also demonstrated that miR-1276 could markedly enrich SIRT4 in BC cells (Fig. 4F). Furthermore, SIRT4 was significantly upregulated in BC tissues compared with in normal tissues (Fig. 4G), and there was a negative correlation between miR-1276 and SIRT4 expression in BC tissues (Fig. 4H). Subsequently, shRNA was used to knock down SIRT4 in BC cells (Fig. 4I). Similar to the observations made following knockdown of circOMA1, silencing SIRT4 suppressed the proliferation, migration and invasion, and promoted the apoptosis of BC cells (Fig. 4J-M). These data suggested that miR-1276/SIRT4 acted downstream of circOMA1.
Figure 4.
SIRT4 is a target of miR-1276. (A) Predictive binding sites between 3’ untranslated region of SIRT4 and miR-1276; relative luciferase activity was assessed in BC cells co-transfected with wt or mut luciferase reporters and miR-1276 mimics or corresponding NC. (B and D) BC cells were transfected as indicated and the mRNA expression levels of SIRT4 were measured. (C and E) BC cells were transfected as indicated and the protein expression levels of SIRT4 were measured. (F) RNA pull-down assay was used for the detection of SIRT4 using miR-1276 and miR-NC probes in BC cells. (G) mRNA expression levels of SIRT4 were measured in BC tissues and adjacent normal tissues. (H) Correlation analysis of the expression of SIRT4 and miR-1276 in BC tissues. (I) BC cells were transfected as indicated and the protein expression levels of SIRT4 were measured by western blotting. (J) BC cells were transfected as indicated and proliferation of BC cells was measured by Cell Counting Kit-8 assay at different time points. (K) BC cells were transfected as indicated and cellular migration was assessed (magnification, ×100). (L) BC cells were transfected as indicated and cellular invasion was assessed (magnification, ×100). (M) BC cells were transfected as indicated and cellular apoptosis was assessed. Data are presented as the mean ± SD. Experiments were performed at least three times. *P<0.05, **P<0.01. BC, breast cancer; sh, short hairpin RNA; NC, negative control; miR, microRNA; mut, mutant; wt, wild-type; SIRT4, sirtuin 4; OD, optical density; PI, propidium iodide; FITC, fluorescein isothiocyanate.
circOMA1 exerted its oncogenic effects via the miR-1276/SIRT4 axis
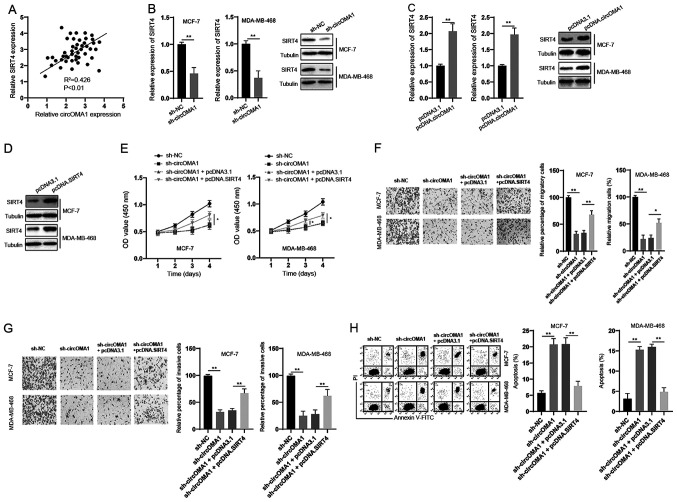
The correlation between circOMA1 and SIRT4 expression was assessed and a positive correlation was detected between them in BC tissues (Fig. 5A). In vitro experiments demonstrated that silencing circOMA1 led to the downregulation of SIRT4 at both the mRNA and protein levels in BC cells (Fig. 5B). Moreover, overexpression of circOMA1 induced an upregulation of SIRT4 at both the mRNA and protein levels in BC cells (Fig. 5C). To further analyze the role of SIRT4 in the function of circOMA1, SIRT4 was overexpressed in BC cells via transfection with the pcDNA3.1 vector containing full-length SIRT4 (pcDNA.SIRT4) (Fig. 5D). The CCK-8 assay revealed that overexpression of SIRT4 reversed the inhibitory effects of circOMA1 knockdown on the proliferation of BC cells (Fig. 5E). In addition, overexpression of SIRT4 inhibited the effects of circOMA1 knockdown on the migration, invasion and apoptosis of BC cells (Fig. 5F-H). Taken together, these data suggested that circOMA1 exerted its oncogenic effects via regulation of the miR-1276/SIRT4 axis in BC cells.
Figure 5.
Overexpression of SIRT4 abrogates the effects of circOMA1 silencing in BC cells. (A) Correlation between circOMA1 and SIRT4 expression was analyzed in BC tissues. (B) BC cells were transfected as indicated and the mRNA expression levels of SIRT4 were measured by reverse transcription-quantitative PCR. (C) BC cells were transfected as indicated and the protein expression levels of SIRT4 were measured by western blotting. (D) BC cells were transfected with pcDNA3.1 or pcDNA.SIRT4, and protein expression levels of SIRT4 were measured by western blotting. (E) BC cells were transfected as indicated and cell viability was measured by Cell Counting Kit-8 assay. *P<0.05 vs. sh-circOMA1 + pcDNA3.1. (F) BC cells were transfected as indicated and cellular migration were assessed (magnification, ×100). (G) BC cells were transfected as indicated and cellular invasion were assessed (magnification, ×100). (H) BC cells were transfected as indicated and cellular apoptosis was measured by flow cytometry. Data are presented as the mean ± SD. Experiments were performed at least three times. *P<0.05, **P<0.01. BC, breast cancer; circ, circular RNA; sh, short hairpin RNA; NC, negative control; SIRT4, sirtuin 4; OD, optical density; PI, propidium iodide; FITC, fluorescein isothiocyanate.
Discussion
BC is one of the most prevalent types of cancer with a high incidence and poor prognosis. In recent years, accumulating evidence has indicated that circRNAs, a new type of ncRNA, may have essential roles in the initiation and progression of various types of cancer, including BC. For example, circSKA3 was demonstrated to bind integrin β1 to induce invadopodium formation and thereby enhance invasion of BC (19). Another recent study reported that circCDYL was able to promote autophagy and increase the progression of BC (20).
The present study examined the expression pattern and prognostic value of circOMA1 in BC. The results demonstrated that the expression levels of circOMA1 were significantly higher in BC tissues compared with those in normal tissues. Higher expression of circOMA1 was also associated with poorer survival of patients with BC. In vitro functional studies revealed that circOMA1 acted as an oncogene in BC cells. The present findings are in accordance with a previous study, which reported that circOMA1 acted as an oncogene and promoted the progression of nonfunctioning pituitary adenomas (21). It is well documented that circRNAs can function as ceRNAs to interact with miRNAs and thus modulate the expression of target genes. In the present study, the interaction between circOMA1 and miR-1276 was verified. It was observed that silencing circOMA1 increased miR-1276 expression, whereas overexpression of circOMA1 inhibited the expression of miR-1276. Furthermore, a dual-luciferase reporter assay confirmed the binding between circOMA1 and miR-1276. To date, knowledge about the role of miR-1276 in tumors is limited. A previous study reported that miR-1276 could bind to lncRNA HCG1, and inhibit the proliferation and migration of gastric cancer cells (22). However, another study reported that miR-1276 exhibited high expression tendencies in liver cancer tissues compared with in normal liver tissues (23). The present study revealed that miR-1276 was downregulated in BC tissues and acted as a tumor suppressor in BC cells. These discrepancies may be due to the different cancer types, and further investigation into the role of miR-1276 in different types of cancer is required.
It is well documented that miRNAs exert their functions via inhibition or deregulation of targeted mRNAs (24). The present study aimed to investigate the target genes of miR-1276, and SIRT4 was identified as a potential candidate. SIRT4 belongs to the SIRT family and has been shown to serve controversial roles in tumors. It has been demonstrated that overexpression of SIRT4 may inhibit the proliferation of HeLa and Burkitt lymphoma cells by inhibiting glutamine metabolism in vitro (25,26). In addition, SIRT4 has been shown to inhibit the tumorigenesis of colorectal and lung cancer cells (27,28). However, SIRT4 may act as an oncogene in cancer. For example, in a previous study, SIRT4 protein levels were higher in esophageal cancer tissue compared with those in adjacent nontumor esophageal tissue (29). The present study indicated that SIRT4 acted as an oncogene in BC, and these findings are in line with a previous study that also demonstrated that SIRT4 was upregulated in BC tissues and promoted the migration, proliferation and invasion of BC cells (30). Moreover, the present study revealed that overexpression of SIRT4 abrogated the effects of circOMA1 silencing on the proliferation, migration, invasion and apoptosis of BC cells. Collectively, it was concluded that circOMA1 was involved in BC progression via sponging miR-176 and upregulating SIRT4.
In conclusion, the present study revealed that the expression levels of circOMA1 were increased in BC and that circOMA1 was associated with poor prognosis of BC. Functional investigations demonstrated that circOMA1 may be involved in the proliferation, migration, invasion and apoptosis of BC cells. Furthermore, the pivotal role of the circOMA1/miR-1276/SIRT4 axis in BC was revealed in the current study. Therefore, circOMA1 may have potential as a therapeutic target and novel prognostic biomarker for BC.
Acknowledgements
The authors would like to thank Dr Renxi Zhang (Zhejiang University, School of Medicine) for his helpful suggestions.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LXu was responsible for the acquisition of data, KX and LXiang analysed and interpreted the data, JY conceived and designed this study, drafted the manuscript and revised it. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Zhenghai Longsai Hospital. Written informed consent was obtained from all participants prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. Erratum in: CA Cancer J Clin 70: 313, 2020. [DOI] [PubMed] [Google Scholar]
- 2.Nagini S. Breast Cancer: Current Molecular Therapeutic Targets and New Players. Anticancer Agents Med Chem. 2017;17:152–163. doi: 10.2174/1871520616666160502122724. [DOI] [PubMed] [Google Scholar]
- 3.Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440–441. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 5.Jahani S, Nazeri E, Majidzadeh-A K, Jahani M, Esmaeili R. Circular RNA; a new biomarker for breast cancer: A systematic review. J Cell Physiol. 2020;235:5501–5510. doi: 10.1002/jcp.29558. [DOI] [PubMed] [Google Scholar]
- 6.Wu J, Jiang Z, Chen C, Hu Q, Fu Z, Chen J, Wang Z, Wang Q, Li A, Marks JR, et al. CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer Lett. 2018;430:179–192. doi: 10.1016/j.canlet.2018.05.033. [DOI] [PubMed] [Google Scholar]
- 7.Wang N, Gu Y, Li L, Wang F, Lv P, Xiong Y, Qiu X. Circular RNA circMYO9B facilitates breast cancer cell proliferation and invasiveness via upregulating FOXP4 expression by sponging miR-4316. Arch Biochem Biophys. 2018;653:63–70. doi: 10.1016/j.abb.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Shibuya N, Kakeji Y, Shimono Y. MicroRNA-93 targets WASF3 and functions as a metastasis suppressor in breast cancer. Cancer Sci. 2020;111:2093–2103. doi: 10.1111/cas.14423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malagobadan S, Ho CS, Nagoor NH. MicroRNA-6744-5p promotes anoikis in breast cancer and directly targets NAT1 enzyme. Cancer Biol Med. 2020;17:101–111. doi: 10.20892/j.issn.2095-3941.2019.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, Maechler P, Verdin E. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282:33583–33592. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- 12.Sebastián C, Satterstrom FK, Haigis MC, Mostoslavsky R. From sirtuin biology to human diseases: An update. J Biol Chem. 2012;287:42444–42452. doi: 10.1074/jbc.R112.402768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li T, Li Y, Liu T, Hu B, Li J, Liu C, Liu T, Li F. Mitochondrial PAK6 inhibits prostate cancer cell apoptosis via the PAK6-SIRT4-ANT2 complex. Theranostics. 2020;10:2571–2586. doi: 10.7150/thno.42874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Li H, Zhao ZB, Zhu W, Feng PP, Zhu XW, Gong JP. SIRT4 silencing in tumor-associated macrophages promotes HCC development via PPARδ signalling-mediated alternative activation of macrophages. J Exp Clin Cancer Res. 2019;38:469. doi: 10.1186/s13046-019-1456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Guo Y, Gao J, Yuan X. Tumor-suppressive function of SIRT4 in neuroblastoma through mitochondrial damage. Cancer Manag Res. 2018;10:5591–5603. doi: 10.2147/CMAR.S172509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Δ Δ C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Obuchowski NA. ROC analysis. AJR Am J Roentgenol. 2005;184:364–372. doi: 10.2214/ajr.184.2.01840364. [DOI] [PubMed] [Google Scholar]
- 18.Thomson DW, Dinger ME. Endogenous microRNA sponges: Evidence and controversy. Nat Rev Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 19.Du WW, Yang W, Li X, Fang L, Wu N, Li F, Chen Y, He Q, Liu E, Yang Z, et al. The Circular RNA circSKA3 binds integrin β1 to induce invadopodium formation enhancing breast cancer invasion. Mol Ther. 2020;28:1287–1298. doi: 10.1016/j.ymthe.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang G, Ling Y, Mehrpour M, Saw PE, Liu Z, Tan W, Tian Z, Zhong W, Lin W, Luo Q, et al. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol Cancer. 2020;19:65. doi: 10.1186/s12943-020-01152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du Q, Hu B, Feng Y, Wang Z, Wang X, Zhu D, Zhu Y, Jiang X, Wang H. circOMA1-Mediated miR-145-5p Suppresses Tumor Growth of Nonfunctioning Pituitary Adenomas by Targeting TPT1. J Clin Endocrinol Metab. 2019;104:2419–2434. doi: 10.1210/jc.2018-01851. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Huang H, Xu X, Wang H, Wang J, Yao Z, Xu X, Wu Q, Xu F. LncRNA HCG11 promotes proliferation and migration in gastric cancer via targeting miR-1276/CTNNB1 and activating Wnt signaling pathway. Cancer Cell Int. 2019;19:350. doi: 10.1186/s12935-019-1046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong DD, Dang YW, Lin P, Wen DY, He RQ, Luo DZ, Feng ZB, Chen G. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 2018;16:220. doi: 10.1186/s12967-018-1593-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Afonso-Grunz F, Müller S. Principles of miRNA-mRNA interactions: Beyond sequence complementarity. Cell Mol Life Sci. 2015;72:3127–3141. doi: 10.1007/s00018-015-1922-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeong SM, Lee A, Lee J, Haigis MC. SIRT4 protein suppresses tumor formation in genetic models of Myc-induced B cell lymphoma. J Biol Chem. 2014;289:4135–4144. doi: 10.1074/jbc.M113.525949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K, Li YH, Wang X, Laurent G, German NJ, et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. 2013;23:450–463. doi: 10.1016/j.ccr.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu L, Dong Q, He J, Wang X, Xing J, Wang E, Qiu X, Li Q. SIRT4 inhibits malignancy progression of NSCLCs, through mitochondrial dynamics mediated by the ERK-Drp1 pathway. Oncogene. 2017;36:2724–2736. doi: 10.1038/onc.2016.425. [DOI] [PubMed] [Google Scholar]
- 28.Miyo M, Yamamoto H, Konno M, Colvin H, Nishida N, Koseki J, Kawamoto K, Ogawa H, Hamabe A, Uemura M, et al. Tumour-suppressive function of SIRT4 in human colorectal cancer. Br J Cancer. 2015;113:492–499. doi: 10.1038/bjc.2015.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakahara Y, Yamasaki M, Sawada G, Miyazaki Y, Makino T, Takahashi T, Kurokawa Y, Nakajima K, Takiguchi S, Mimori K, et al. Downregulation of SIRT4 Expression Is Associated with Poor Prognosis in Esophageal Squamous Cell Carcinoma. Oncology. 2016;90:347–355. doi: 10.1159/000445323. [DOI] [PubMed] [Google Scholar]
- 30.Huang G, Lin Y, Zhu G. SIRT4 is upregulated in breast cancer and promotes the proliferation, migration and invasion of breast cancer cells. Int J Clin Exp Pathol. 2017;10:11849–11856. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.