Abstract
Colon capsule endoscopy (CCE) is a minimally invasive alternative to conventional colonoscopy. Most studies on CCE focus on colorectal neoplasia detection. The development of automated tools may address some of the limitations of this diagnostic tool and widen its indications for different clinical settings. We developed an artificial intelligence model based on a convolutional neural network (CNN) for the automatic detection of blood content in CCE images. Training and validation datasets were constructed for the development and testing of the CNN. The CNN detected blood with a sensitivity, specificity, and positive and negative predictive values of 99.8 %, 93.2 %, 93.8 %, and 99.8 %, respectively. The area under the receiver operating characteristic curve for blood detection was 1.00. We developed a deep learning algorithm capable of accurately detecting blood or hematic residues within the lumen of the colon based on colon CCE images.
Introduction
Capsule endoscopy has become the main instrument for evaluation of patients with suspected small-bowel bleeding. Colonoscopy is routinely performed for the investigation of suspected lower gastrointestinal bleeding; however, it is invasive, potentially painful, frequently requires sedation, and is associated with a risk of perforation 1 .
Colon capsule endoscopy (CCE) has been recently introduced as a minimally invasive alternative to conventional colonoscopy when the latter is contraindicated, unfeasible, or unwanted by the patient. The application of CCE has been most extensively studied in the setting of colorectal cancer screening 2 . However, a single CCE examination may produce 50 000 images, the review of which is time-consuming, requiring approximately 50 minutes for completion 3 . Additionally, abnormal findings may be restricted to a small number of frames, thus contributing to the risk of overlooking important lesions.
Convolutional neural networks (CNN) are a type of deep learning algorithm tailored for image analysis. This artificial intelligence (AI) architecture has demonstrated high performance levels in diverse medical fields 4 5 . Recent studies have reported a high diagnostic yield of CNN-based tools for the detection of luminal blood in small-bowel capsule endoscopy 6 . The use of CCE for investigation of conditions other than colorectal neoplasia has not been evaluated. Detection of blood content is important when reviewing CCE images and, to date, no AI algorithm has been developed for the detection of colonic luminal blood or hematic residues in CCE images. The aim of this pilot study was to develop and validate a CNN-based algorithm for the automatic detection of colonic luminal blood or hematic vestiges in CCE examinations.
Methods
Study design
We retrospectively reviewed CCE images obtained between 2010 and 2020 at São João University Hospital, Porto, Portugal. The full-length video of all participants (n = 24) was reviewed (total number of frames 3 387 259). A total of 5825 images of the colonic mucosa were ultimately extracted. Inclusion and labeling of frames were performed by two experienced gastroenterologists who had each read more than 1000 capsule endoscopies (M.M.S. and H.C.) prior to the study. Significant frames were included regardless of image quality and bowel cleansing quality. A final decision on the frame labeling required undisputed consensus between the two gastroenterologists. The study was approved by the ethics committee of São João University Hospital/Faculty of Medicine of the University of Porto (No. CE 407/2020). A team with Data Protection Officer certification (Maastricht University) confirmed the nontraceability of data and conformity with general data protection regulations.
CCE procedure
In all patients, the procedures were conducted using the PillCam Colon 2 system (Medtronic, Minneapolis, Minnesota, USA). This system was launched in 2009 and no hardware modifications were introduced during the study period. Therefore, image quality remained unaltered between 2010 and 2020, with no difference in quality between the image frames used to train the CNN and those used to test the model. The images were reviewed using PillCam software v9 (Medtronic). Bowel preparation was performed according to previously published recommendations 7 . In brief, a solution consisting of 4 L of polyethylene glycol solution was used in a split-dosage regimen (2 L in the evening before and 2 L on the morning of capsule ingestion). Two boosters consisting of a sodium phosphate solution were applied after the capsule had entered the small bowel and with a 3-hour interval.
Development of the CNN
A CNN was developed for automatic identification of blood or hematic residues within the lumen of the colon. From the collected pool of images (n = 5825), 2975 had evidence of luminal blood or hematic residues and 2850 showed normal mucosa or mucosal lesions. This pool of images was split to form training and validation image datasets. The training dataset comprised 80 % of the consecutively extracted images (n = 4660); the remaining 20 % were used as the validation dataset (n = 1165). The validation dataset was used for assessing the performance of the CNN ( Fig. 1 ).
Fig. 1.
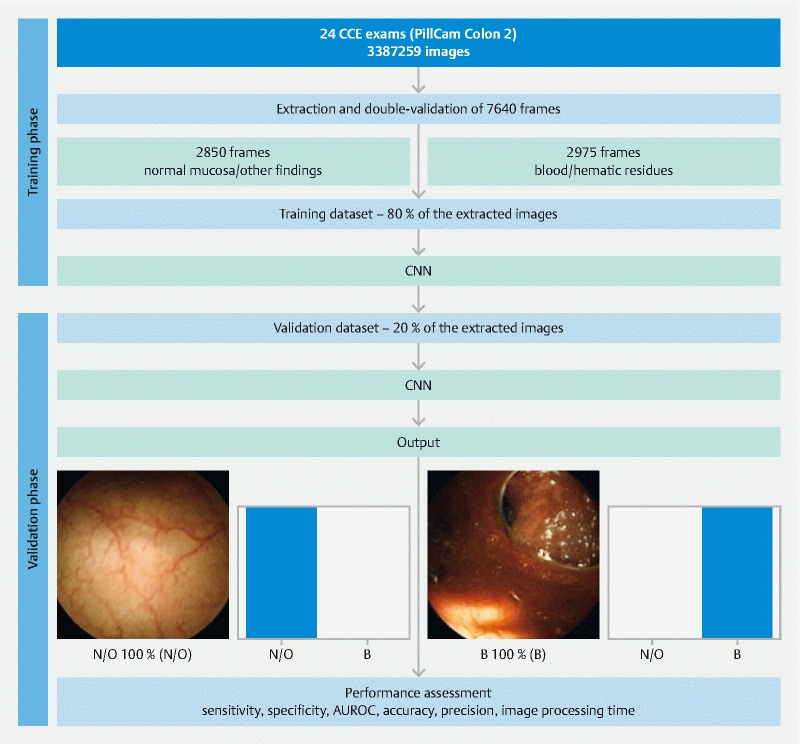
Study flow chart for the training and validation phases. N/O, normal mucosa/other findings; B, blood or hematic residues; AUROC, area under the receiver operating characteristic curve. PillCam Colon 2 (Medtronic, Minneapolis, Minnesota, USA).
To create the CNN, we used the Xception model with its weights trained on ImageNet. To transfer this learning to our data, we kept the convolutional layers of the model. We used Tensorflow 2.3 and Keras libraries to prepare the data and run the model. Each inputted frame had a resolution of 512 × 512 pixels. For each image, the CNN calculated the probability for each category (normal colonic mucosa/other findings vs. blood/hematic residues). The category with the highest probability score was outputted as the CNN’s predicted classification ( Fig. 2 ).
Fig. 2.
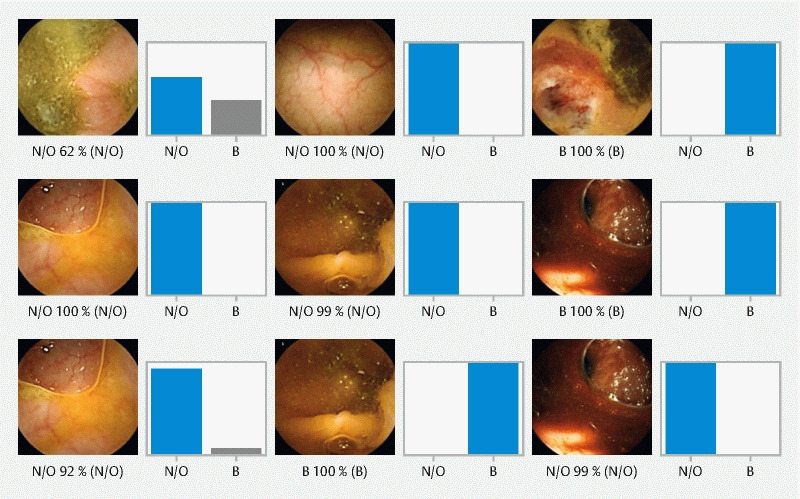
Output obtained from the application of the convolutional neural network. The bars represent the probability estimated by the network. The finding with the highest probability was outputted as the predicted classification. Blue bars represents a correct prediction; gray bars represent an incorrect prediction. N/O, normal mucosa/other findings; B, blood or hematic residues.
Model performance and statistical analysis
The primary outcome measures included sensitivity, specificity, positive and negative predictive values, and accuracy. Moreover, we used receiver operating characteristic (ROC) curve analysis and area under the ROC curve (AUROC) to measure the performance of our model for distinction between the categories. The network’s classification was compared with that recorded by the gastroenterologists (gold standard). Sensitivities, specificities, and precisions are presented as means and standard deviations (SDs). ROC curves are graphically represented and AUROC was calculated as mean and 95 % confidence interval (CI), assuming normal distribution of these variables.
The computational performance of the network was also determined by calculating the time required for the CNN to provide output for all images in the validation dataset. Statistical analysis was performed using SciKit learn v. 0.22.2.
Results
Construction of the CNN
A total of 24 patients who underwent CCE were enrolled in the study. A total of 5825 frames were extracted, 2975 containing blood and 2850 showing normal mucosa/other findings. The training dataset comprised 80 % of the total image pool; the remaining 20 % (n = 1165) were used for testing the model. The latter subset of images comprised 595 images (51.1 %) with evidence of blood or hematic residues and 570 images (48.9 %) with normal colonic mucosa/other findings. The CNN evaluated each image and predicted a classification (normal mucosa/other findings or blood/hematic residues), which was compared with the classification provided by the gastroenterologists. The network demonstrated its learning ability, with accuracy increasing as data were repeatedly inputted to the multi-layer CNN ( Fig. 3 ).
Fig. 3.
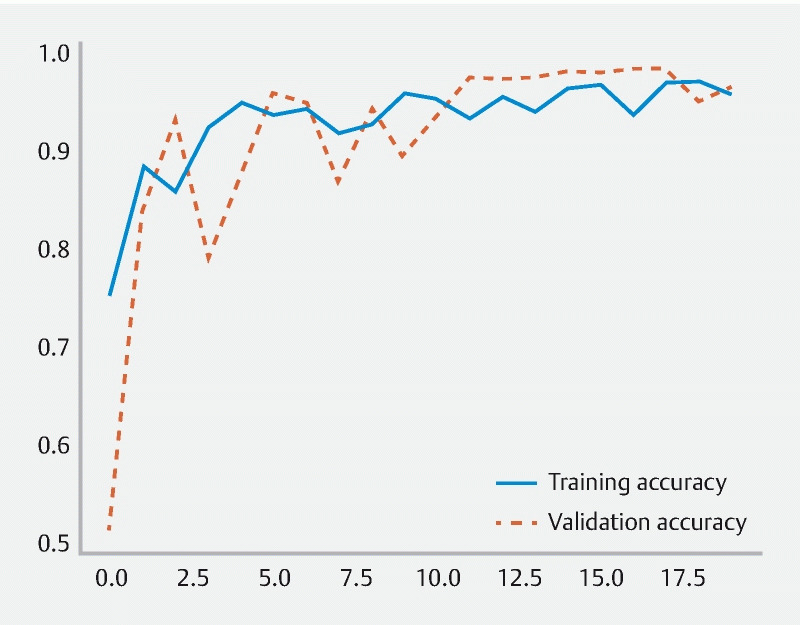
Evolution of the accuracy of the convolutional neural network during training and validation phases as the training and validation datasets were repeatedly inputted into the neural network.
Performance of the CNN
The performance of the CNN is shown in Table 1 . Overall, the mean (SD) sensitivity and specificity were 99.8 % (4.7 %) and 93.2 % (4.7 %), respectively. The positive predictive value and negative predictive value were 93.8 % (4.2 %) and 99.8 % (4.2 %), respectively ( Table 1 ). The overall accuracy of the CNN was 96.6 %. The AUROC for detection of blood was 1.00 (95 %CI 0.99–1.00) ( Fig. 4 ).
Table 1. Confusion matrix of the automatic detection vs. expert classification.
| Expert | ||||
| Blood/hematic residues | Normal/other findings | |||
| CNN | Blood/hematic residues | 594 | 39 | |
| Normal/other findings | 1 | 531 | ||
|
Sensitivity
1
99.8 % (4.7 %) |
Specificity
1
93.2 % (4.7 %) |
PPV
1
93.8 % (4.2 %) |
NPV
1
99.8 % (4.2 %) |
|
CNN, convolutional neural network; PPV, positive predictive value; NPV, negative predictive value.
Expressed as mean (standard deviation).
Fig. 4.

Receiver operating characteristic analyses of the network’s performance in the detection of blood vs. normal colonic mucosa/other findings. B, blood or hematic residues; ROC, receiver operating characteristic.
Computational performance of the CNN
The CNN completed the reading of the dataset in 9 seconds. This translates into an approximated reading rate of 129 frames per second. At this rate, review of a full-length CCE video containing an estimated 50 000 frames would require approximately 6 minutes.
Discussion
We developed a CNN for automatic detection of blood in the lumen of the colon in CCE images. Our AI model was highly sensitive, specific, and accurate for the detection of blood in CCE images.
The application of AI tools in the field of capsule endoscopy has been generating increasing interest. The development of AI tools for automatic detection of a wide array of lesions has provided promising results 8 . Recently, Aoki et al. reported high performance of a CNN for the detection of blood content in images of small-bowel capsule endoscopy, outperforming currently existing software tools for screening the presence of blood in capsule endoscopy images 6 . The development of AI tools for automatic detection of lesions in CCE images is in its early stages, and mainly focuses on automatic detection of colorectal neoplasia. To date, two studies have reported the development of CNN-based models for detection of colorectal neoplasia, with high performance levels 9 10 .
The role of CCE in everyday clinical practice has not been fully established. Most studies focus on detection of polyps for colorectal cancer screening. Besides its application in the screening setting, CCE may be a noninvasive alternative to conventional colonoscopy for other common indications, including the investigation of lower gastrointestinal bleeding and colonic lesions other than polyps. To the best of our knowledge, this is the first study to develop a CNN for automatic detection of blood content in CCE images. Our network detected the presence of blood in CCE images with high sensitivity, specificity, and accuracy (99.8 %, 93.2 %, and 96.6 %, respectively). The development of automated AI tools for CCE has the potential to improve its diagnostic yield and time efficiency, thus contributing to CCE acceptance. Moreover, increased diagnostic capacity may widen the indications for CCE. These results suggest that AI-enhanced CCE may be a useful examination for evaluation of patients with lower gastrointestinal bleeding, particularly when conventional colonoscopy is contraindicated or unwanted by the patient.
This study has several limitations. First, it was a retrospective proof-of-concept study involving images collected at a single center. Second, the tool was only tested in still frames; assessment of performance using full-length videos is required before clinical application of these tools. Third, although a large pool of images was reviewed, the number of patients included in the study was small. Thus, subsequent prospective multicenter studies with larger numbers of CCE examinations are desirable before this model can be applied to clinical practice. Furthermore, these tools should be regarded as supportive rather than substitutive in a real-life clinical setting.
In conclusion, we developed a CNN-based model capable of detecting blood content in CCE images with high sensitivity and specificity. We believe that the implementation of AI tools to clinical practice will address some of the limitations of CCE, mainly the time required for reading, thus lessening the burden on gastroenterologists and boosting the acceptance of CCE to routine clinical practice.
Footnotes
Competing interests The authors declare that they have no conflict of interest.
References
- 1.Niikura R, Yasunaga H, Yamada A et al. Factors predicting adverse events associated with therapeutic colonoscopy for colorectal neoplasia: a retrospective nationwide study in Japan. Gastrointest Endosc. 2016;84:971–982. doi: 10.1016/j.gie.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 2.Spada C, Pasha S F, Gross S A et al. Accuracy of first- and second-generation colon capsules in endoscopic detection of colorectal polyps: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2016;14:1533–1543. doi: 10.1016/j.cgh.2016.04.038. [DOI] [PubMed] [Google Scholar]
- 3.Eliakim R, Yassin K, Niv Y et al. Prospective multicenter performance evaluation of the second-generation colon capsule compared with colonoscopy. Endoscopy. 2009;41:1026–1031. doi: 10.1055/s-0029-1215360. [DOI] [PubMed] [Google Scholar]
- 4.Esteva A, Kuprel B, Novoa R A et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gargeya R, Leng T. Automated identification of diabetic retinopathy using deep learning. Ophthalmology. 2017;124:962–969. doi: 10.1016/j.ophtha.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Aoki T, Yamada A, Kato Y et al. Automatic detection of blood content in capsule endoscopy images based on a deep convolutional neural network. J Gastroenterol Hepatol. 2020;35:1196–1200. doi: 10.1111/jgh.14941. [DOI] [PubMed] [Google Scholar]
- 7.Spada C, Hassan C, Galmiche J P et al. Colon capsule endoscopy: European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy. 2012;44:527–536. doi: 10.1055/s-0031-1291717. [DOI] [PubMed] [Google Scholar]
- 8.Iakovidis D K, Koulaouzidis A. Software for enhanced video capsule endoscopy: challenges for essential progress. Nat Rev Gastroenterol Hepatol. 2015;12:172–186. doi: 10.1038/nrgastro.2015.13. [DOI] [PubMed] [Google Scholar]
- 9.Blanes-Vidal V, Baatrup G, Nadimi E S. Addressing priority challenges in the detection and assessment of colorectal polyps from capsule endoscopy and colonoscopy in colorectal cancer screening using machine learning. Acta Oncol. 2019;58:S29–S36. doi: 10.1080/0284186X.2019.1584404. [DOI] [PubMed] [Google Scholar]
- 10.Yamada A, Niikura R, Otani K et al. Automatic detection of colorectal neoplasia in wireless colon capsule endoscopic images using a deep convolutional neural network. Endoscopy. 2020 doi: 10.1055/a-1266-1066. [DOI] [PubMed] [Google Scholar]


