Abstract
A highly acute disease characterized as visceral gout broke out in Muscovy ducklings in Henan province (China) in June 2020, with a mortality rate of up to 61%. In this study, common pathogenic agents were screened using reverse-transcription polymerase chain reaction or polymerase chain reaction. The results found the novel goose astrovirus (GoAstV) to be the pathogenic agent. We isolated the GoAstV, which has been designated as HNNY0620, using the Leghorn male chicken hepatocellular carcinoma (LMH) cell line and sequenced the complete genome. The phylogenetic tree showed that the amino acid (aa) sequences of ORF1a and ORF2 and the completed nucleotide sequences of the HNNY0620 strain were clustered in the GoAstV-I clade. ORF1a aa and whole-genome sequences were genetically close to TAstV-2 and DHV-3, whereas the ORF2 aa sequences were clustered with TAstV-2 and DHV2. Both the duck-origin GoAstVs and HNNY0620 harbored some special mutations, but ORF1a in 700 (I/T), ORF1b in 288 (F/L), and ORF2 in 306 (A/T) were only found in HNNY0620. These results suggest that the host range of GoAstV is diffusing, which can potentially affect other waterfowl.
Key words: Muscovy-ducklings, genomic sequence analysis, mutation analysis, GoAstV, gout
INTRODUCTION
Astroviruses (AstVs) are single-stranded, positive-sense, and non-enveloped RNA viruses that belong to the Astroviridae family. The length of their genomes range from 6.8 kb to 7.9 kb, comprising of a 5′-untranslated region (UTR), 3 open reading frames (ORF1a, ORF1b, and ORF2), a 3′-UTR, and a poly-A tail (Pantin-Jackwood et al., 2011). ORF1a encodes nonstructural proteins, ORF1b encodes an RNA-dependent RNA polymerase (RdRp), and ORF2 encodes the viral capsid protein that encapsulates the viral genome (Jiang B. et al., 1993; Arias and Rebecca, 2017).
AstVs are classified into the MamAstV (infecting mammals) and the AvAstV genuses (infecting avian and wild birds). The MamAstV genus viruses, including human, cattle, sheep, pig, boar, cat, and deer AstVs, primarily causes mild gastroenteritis and encephalitis (Smits et al., 2010; Reuter et al., 2011; Xiao et al., 2012; Céline L Boujon et al., 2017; Roach and Ryan, 2021), whereas the AvAstV viruses, including duck, chicken, turkey, and goose AstVs, lead to duck hepatitis and self-limited diarrhea (Todd D et al., 2009; Espinoza et al., 2016; Victoria, 2017; Jin et al., 2018).
Goose gout due to a novel AstV was first reported in 2017 in Shandong province, China and is characterized by urate deposition in the viscera, joint cavity, and other interstitial tissues (Yang et al., 2018; Zhang et al., 2018a). Duck gout resulting from novel AstVs was reported in the Shandong province in 2019 and is characterized by visceral gout, with a 30% mortality rate (Chen et al., 2020a).
In this study, with similar gout symptoms in Muscovy-ducklings in the Henan province, we performed the isolation and whole-genome sequencing of the AstVs caused gout.
MATERIALS AND METHODS
Case Review and Sample Processing
The commercial Muscovy-duck farm (feeding approximately 6,000 birds), from where the ducklings were sourced, is located in the southwest Henan province, China. Five-day-old Muscovy ducklings showed the symptoms of joint swelling, claudication, growth retardation, and recumbency. The anatomical characteristics included urate deposits in the pericardium, liver capsule, ureter, serosal surface, muscle, and hypodermis, with the pericardium being the most conspicuous. Moreover, swelling and necrosis were observed on in the liver surface. The morbidity and mortality rates of the ducklings infected with GoAstV decreases as their age increases. Tissues from the liver, spleen, and kidney were collected from the diseased Muscovy-ducklings from the Henan province with their clinical characteristics and autopsy results revealing gout.
Virus Screening and Isolation
The viral DNA and RNA were extracted from pooled tissue samples from the same single bird using a commercial kit (EasyPure Viral DNA/RNA Kit; TransGen Biotechnology, Inc., Beijing, China) as per the manufacturer's instructions for the detection of common duck-origin pathogenic agents, including fowl adenovirus (FAdV); goose parvovirus (GPV), hemorrhagic polyomavirus (GHPV), and AstV (GoAstV); and duck AstV (DAstV), circovirus (DCV), reovirus (DRV), and Tembusu virus (DTMV), using reverse-transcription polymerase chain reaction (RT-PCR) and polymerase chain reaction (PCR). The primers previously reported were used (Chen et al., 2020b; Wei et al., 2020).
Viruses from each sample were isolated on the male Leghorn chicken hepatocellular carcinoma (LMH) cell line as described previously and further tested for other viruses as previously mentioned (Zhang et al., 2018b). The whole-genome sequencing of the isolated GoAstV was performed using RNAs obtained from positive cell cultures using previously reported primer sets. PCR and sequencing were performed at least 3 times (Jin et al., 2018).
Sequence Analysis
Sequence identity searches were completed performed via BLASTx in GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi), pairwise comparison was carried out using ClustalW (http://www.genome.jp/tools/clustalw/), and transmembrane domains in ORF1a were predicted by TMHMM (http://www.cbs.dtu.dk/services/TMHMM). Phylogenetic analysis was performed using the maximum likelihood method with the Tarnura-Nei model of complete nucleotide sequence substitution and the neighbor-joining method with the Jones-Taylor-Thornton matrix-based model of the amino acid (aa) substitution of ORF1a and ORF2 and 1,000 replicates with MEGA 6.06 (Tamura et al., 2013). The comparison of aa sequence variants and same gene segments from all GoAstV were implemented using MEGA 6.06 (Tamura et al., 2013). Pairwise nucleotide sequence identities of the whole genome and pairwise aa sequence identities of ORF1a and ORF2 were implemented and visualized using SDT v1.2 (Muhire et al., 2014). The stem ring structures of nucleic acids were analyzed using Rfam (http://rfam.xfam.org/).
RESULTS
Sample Screening
We collected a total of 19 visceral samples with gout characteristics from the Muscovy-duck farm. We found that the common duck-origin pathogenic agents FAdV, GPV, DRV, GHPV, DAstV, DCV, DRV, and DTMUV were negative except for GoAstV in all samples; cell culture was performed to isolate the virus. The GoAstV strain isolated in this study was designated as HNNY0620.
Characterization of the Full Genome and Genes
The full-length genome segments of the HNNY0620 strain were obtained and deposited in GenBank (Accession no. MZ367612). The complete genome of the HNNY0620 strain is 7,166 nt in length, with a typical genomic characteristic of AstVs, including a 5′-UTR (18 nt), 3 ORFs [ORF1a (3,255 nt), ORF1b (1,551 nt), and ORF2 (2,115 nt)], a 3′-UTR (206 nt), and a poly-A tail (18 nt).
As reported previously (Sajewicz-Krukowska and Katarzyna, 2016; Chen et al., 2020b), the sequence construction was the same as that of the HN01 strain and contained the same characteristics and conserved aa motifs as those in other AstVs: a serine protease motif located at position 672 (GNSG); a nuclear localization signal motif at positon 773 (KKKGKTK) of ORF1a; and 4 RdRp motifs at positions 265 (DWTR), 327 (GNPSG), 377 (YGDD), and 405 (FGMWVK) in ORF1b. ORF1a contained 4 predicted TM domains as well as a predicted zinc finger protein model. Similar to other known classical avian AstVs, the HNNY0620 strain also had an overlapping region between ORF1a and ORF1b (3,259–3,265 nt), which was a highly conserved heptameric−ribosome frameshift sequence (5′-AAAAAAC-3′) and had a downstream hairpin structure (3,273–3,298 nt) as predicted via RNA folding analysis. One 18-nt spacer was observed between the ORF1b stop codon and the ORF2 start codon. A stem-loop-II-like motif including of 43 nt (6,953−6,995 nt) was found adjacent to 10 nt of ORF2 in the 3′-UTR of the genome based on the Rfam analysis.
Analysis of the Phylogenetic Tree and Identity
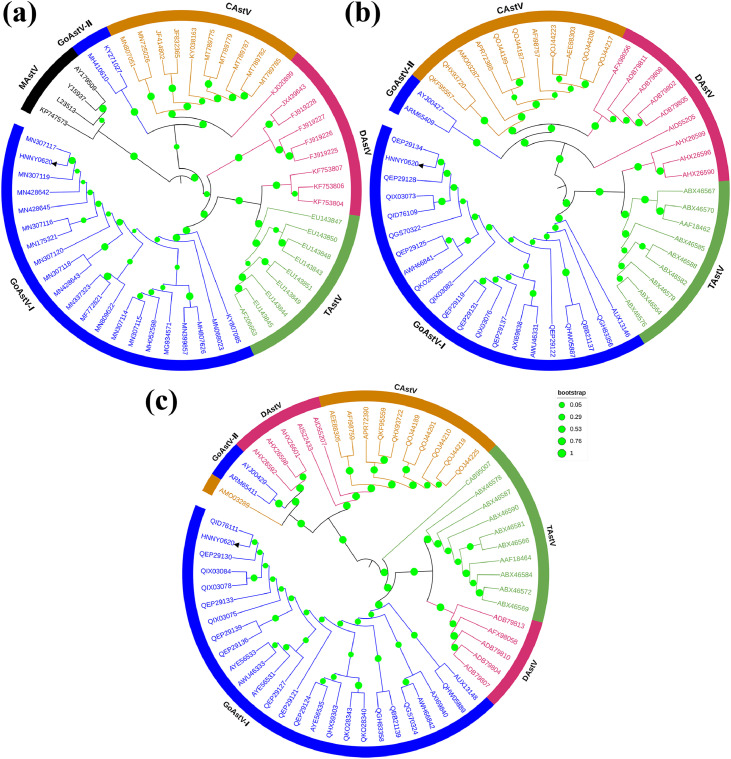
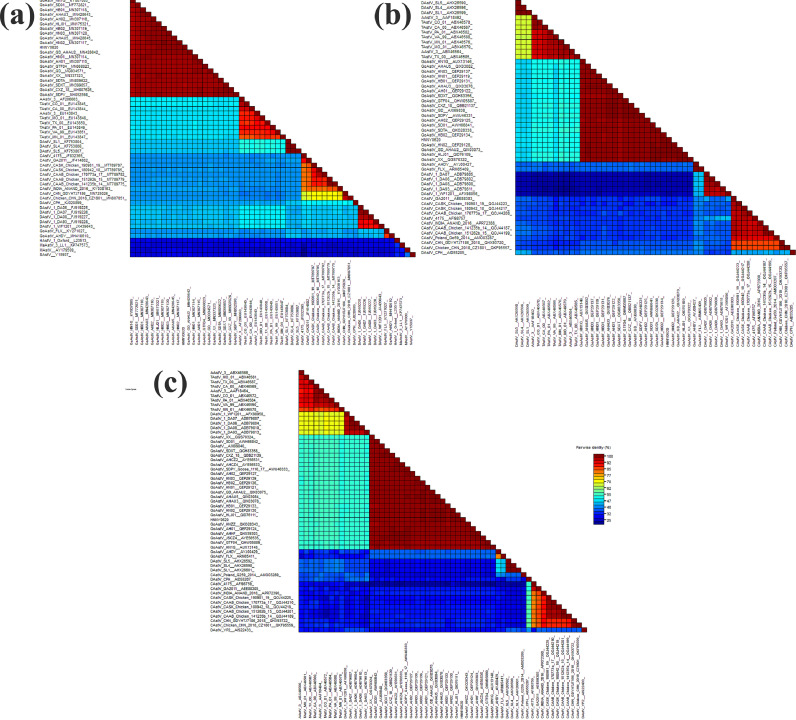
As shown in Figures 1A–1C, a phylogenetic tree was created based on the whole-genome and aa sequences of ORF1a and ORF2 that were deduced from the nucleotide sequences, representative of avian and non-avian AstV species, obtained from GenBank. The whole-genome and aa sequences from the ORF1a phylogenetic tree showed that the HNNY0620 strain was clustered with the TAstV-2 and DHV-3 AstV strains. However, the ORF2 phylogenetic tree showed that the HNNY0620 strain was adjacent to the TAstV-2 and DHV-2 AstV strains. As described in Figures 2A–2C, the HNNY0620 strain was highly similar to goose-origin AstVs with respect to the genome (96.3−99.2%) and aa sequences (ORF1a: 96.0−99.8%; ORF2: 98.1−99.7%) and to the duck-origin GoAstV (SDXT) (96.9, 96.0, and 99.0% for the whole-genome, ORF1a, and ORF2 sequences, respectively). However, the HNNY0620 strain shared lower identity with FLX and AHDY with respect to the whole-genome (55.7−55.9%) and aa sequences (ORF1a: 65.1%; ORF2: 39.2−39.6%) than the other waterfowl avian-derived AstVs.
Figure 1.
Phylogenetic tree of the whole sequence (A), ORF1a (B), and ORF2 (C) of the HNNY0620 strain (▲) and other AstVs.
Figure 2.
Identity of the whole sequence (A), ORF1a (B), and ORF2 (C) of all goose-origin AstV and some avian-origin (non–goose-origin) AstVs and MamAstV.
Compared with the duck AstVs causing duck hepatitis, the identity of the whole genome of the nucleotide were 54.6 to 60.5%, and the ORF1a and ORF2 aa were 58.0 to 71.9% and 33.1 to 56.9%, respectively. The strains of SL5, SL4, and SL1 of DHV-3 showed the highest identity with the HNNY0620 strain in the whole sequence. And HNNY0620 strain shared the highest identity with DA06 in ORF1a, and DA07 and DA06 in ORF2. Compared with turkey AstVs, the identity of the whole sequence, ORF1a, and ORF2 were 53.1 to 63.4%, 57.6 to 72.1%, and 57.1 to 58.1%, respectively. The HNNY0620 strain had low similarity to chicken AstVs in the nucleotide (51.1−55.5%) and aa sequences (ORF1a: 63.0−63.5%; ORF2: 27.9−36.0%).
Mutation Analysis
We analyzed the mutation sites of ORF1a, ORF1b, and ORF2 compared with SDXT (MN399857) and SDTA (MN809622), which are duck-origin GoAstVs, and to HN1G (KY807085), HN01 (MN307114), and HLJ01 (MN175321), which are goose-origin GoAstVs. As Tables 1−3 show, some special mutations were observed in both duck-origin GoAstV and HNNY0620; however, ORF1a in 5 (G/C), 700 (I/T); ORF1b in 288 (F/L); and ORF2 in 306 (A/T) were found only in the HNNY0620 strain.
Table 2.
ORF1b mutation sites, original amino acid residues, and corresponding amino acid residues after mutation.
| Position | Strain name |
|||||
|---|---|---|---|---|---|---|
| HN01 | SDXT | SDTA | HNNY0620 | HN1G | HLJ01 | |
| 28 | D | D→H | D | D | D | D |
| 114 | S | S→R | S | S | S | S |
| 167 | K | K | K→R | K | K | K |
| 225 | K | K | K→R | K | K | K |
| 288 | F | F | F | F→L | F | F |
| 319 | G | G→R | G | G | G | G |
| 423 | M | M→I | M | M | M | M |
Note: “→” represents the mutation that changed from the preceding amino acid residue to the subsequent amino acid residue.
Table 1.
ORF1a mutation sites, original amino acid residues, and corresponding amino acid residues after mutation.
| Position | Strain name |
|||||
|---|---|---|---|---|---|---|
| HN01 | SDXT | SDTA | HNNY0620 | HN1G | HLJ01 | |
| 5 | G | - | - | G→C | G | - |
| 183 | S | S→T | S | S | S | S |
| 200 | W | W→S | W | W | W | W |
| 309 | I | I→T | I | I | I | I |
| 426 | I | I→L | I | I | I | I |
| 520 | A | A | A→I | A | A | A |
| 640 | T | T→K | T | T | T | T |
| 700 | I | I | I | I→T | I | I |
| 723 | G | G→R | G | G | G | G |
| 724 | F | F→V | F | F | F | F |
| 985 | S | S→T | S | S | S | S |
| 993 | N | N→K | N | N | N | N |
| 997 | K | K→E | K | K | K | K |
| 1,015 | R | R→G | R | R | R | R |
| 1,028 | F | F→L | F | F | F | F |
“-” Represents without any amino acid residues.
“→” Represents the mutation that changed from the preceding amino acid residue to the subsequent amino acid residue.
Table 3.
ORF2 mutation sites, original amino acid residues, and corresponding amino acid residues after mutation.
| Position | Strain name |
|||||
|---|---|---|---|---|---|---|
| HN01 | SDXT | SDTA | HNNY0620 | HN1G | HLJ01 | |
| 249 | S | S→W | S | S | S | S |
| 295 | D | D→A | D | D | D | D |
| 306 | A | A | A | A→T | A | A |
| 573 | N | N | N→D | N | N | N |
Note: “→” represents the mutation that changed from the preceding amino acid residue to the subsequent amino acid residue.
Analysis of the Gene Segments of Different GoAstVs
The GoAstV-II cluster can result in clinical symptoms that are highly similar to those produced by the GoAstV-I cluster, although their identities were low. Based on the whole-genome analysis of strains grouped from GoAstV-I and GoAstV-II, some segments, including 1,324–1,333, 1,341–1,351, 1,468–1,477, 1,876–1,889, 2,504–2,519, 2,561–2,578, 3,426–3,436, 3,693–3,706, 4,374–4,384, 4,551–4,561, 4,641–4,654, 4,753–4,766, 7,158–7,168, 7,123–7,132, and 7,205–7,251, were highly conserved (≥10 nt in length) among the strains isolated from the ducks in this study compared with the HN01 and FLX strains isolated from geese.
DISCUSSION
Since 2017, when gosling gout induced by novel GoAstV was discovered in the Shandong province, gout symptoms related to GoAstV infection have persisted in mainland China; and caused enormous financial losses to the Chinese goose industry. Several gout-causing GoAstV strains have been recently observed in geese (Yang et al., 2018; Zhang et al., 2018b; Miaomiao Liu et al., 2019; Chen et al., 2020a,b; Wang et al., 2020), and most of the GoAstV strains in GenBank are goose-origin strains. The AstVs detected in ducks were once associated only with hepatitis prior to the reports of ducks with gout (Chen et al., 2020a; Wei et al., 2020). To the best of our knowledge, this is the first study to report on the Muscovy duck-origin GoAstV, expanding the variety of hosts of GoAstV.
The whole-genome sequence length of the HNNY0620 strain is 7,166 nt, with a classical AstVs construction: 5′-UTR-ORF1a-ORF1b-ORF2-3′UTR-poly(A). The phylogenetic analysis of the deduced aa sequences of ORFs and the whole-genome sequences revealed that the HNNY0620 strain can be classified into the novel GoAstV-I clade. A similar phylogenetic relation has been found in the strains of SDTA from duck-origin and SDXT from Cherry Valley duckling-origin GoAstV (Chen et al., 2020a; Wei et al., 2020). Of note, all strains from the cluster of FLX (representing GoAstV-II) and HN01 (representing GoAstV-I) can lead to the clinical symptoms of urate deposition in the viscera and joint cavity, although their identity was low. In the present study, all sequences of the GoAstV (from GoAstV-I and GoAstV-II) strains were compared together, and several highly conserved gene segments (≥10-nt) were found. They were speculated to get relation to the special similarly clinical characteristic. Furthermore, the reverse genetic and virus rescue tests of more strains originating from different hosts need to be completed to verify whether these gene segments play an important role in pathogenic mechanisms.
Recently, another study on duck-origin GoAstV causing spontaneous gout disease in Cherry Valley ducklings has indicated that cross-host transmission from goose to duck may have occurred (Chen et al., 2020a). In this study, the identity of duck-origin and goose-origin GoAstVs from gout causing GoAstV-I was high (nt ≥96.9%, aa of ORF1a ≥96.0%, and ORF2 ≥98.1%) (Chen et al., 2020b). Thus far, although only 2 reports on ducklings infected with GoAstV have been published (Chen et al., 2020a), the high mortality of ducklings was the same as that for goslings infected with GoAstVs, suggesting that the virulence of GoAstV was unchanged during cross-species spread. Therefore, GoAstV may also infect other species of aquatic birds.
To explore the mutations of duck-origin GoAstV, the SDXT, SDTA, and HNNY0620 strains were compared to HN01 (goose-origin GoAstV) and analyzed. The results found several special mutations in ORF2 in 249 (S/W) and 295 (D/A) of SDXT, 573 (N/D) of SDTA, and 306 (A/T) of HNNY0620 of only duck GoAstVs. However, whether these mutation sites are related to the recognition site of the host as well as with pathogenicity and virulence must be explored further.
CONCLUSIONS
To our knowledge, this is the first study to report the novel GoAstV causing gout in Muscovy ducks. Based on the genome and phylogenetic relationship, the HNNY0620 strain should be classified into the GoAstV-I group. The present study also suggests that GoAstV may be easily transmissible across hosts in waterfowl.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (Grant nos. 31802185 and 31870917), the Scientific and Technological Project of Henan province (Grant no. 182102310077), the Key Scientific and Technological Project of the Education Department of Henan province (Grant no. 18A230011), the program for Innovative Research Team of Science and Technology in University of Henan Province (Grant no. 20IRTSTHN024), and the Technological Project of Nanyang Normal University (Grant nos. 18046, 2019QN009, and 2020CX011), Guangdong Basic and Applied Basic Research Foundation (2019A1515012006).
Ethical statement: The autopsy and sampling protocols for dead birds were approved by the South China Agricultural University Committee for Animal Experiments (approval ID: SYXK-2014-0136).
Data availability statement: All data generated or analyzed during this study are included in this article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
DISCLOSURES
The authors have no competing interests to declare.
REFERENCES
- Arias C.F., Rebecca M.D. The astrovirus capsid: a review. Viruses. 2017;9:15. doi: 10.3390/v9010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zhang B., Yan M., Diaoand Y., Yi T. First report of a novel goose astrovirus outbreak in Cherry Valley ducklings in China. Transbound. Emerg. Dis. 2020;67:1019–1024. doi: 10.1111/tbed.13418. [DOI] [PubMed] [Google Scholar]
- Chen Q., Xu X., Yu Z., Sui C., Zuo K., Zhi G., Ji J., Yao L., Kan Y., Biand Y., Qingmei X. Characterization and genomic analysis of emerging astroviruses causing fatal gout in goslings. Transbound. Emerg. Dis. 2020;67:865–876. doi: 10.1111/tbed.13410. [DOI] [PubMed] [Google Scholar]
- Céline L Boujon M.C.K., Daniel Wüthrich S.W., Dennis Jakupovicand R.B., Torsten S. Indication of cross-species transmission of astrovirus associated with encephalitis in sheep and cattle. Emerg. Infect. Dis. 2017;23:1604–1608. doi: 10.3201/eid2309.170168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza L.L., Beserra L.A.R., Soaresand R.M., Fabio G. Turkey astrovirus type 1 (TAstV-1) and chicken astrovirus (CAstV) detection in Brazilian chicken flocks. Avian. Dis. 2016;60:681–687. doi: 10.1637/11403-030816-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Jiang B., Monroe S.S., Koonin E.V., Stine S.E., Glass R.I. RNA sequence of astrovirus: distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc. Natl. Acad. Sci. U.S.A. 1993;90:10539–10543. doi: 10.1073/pnas.90.22.10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M., Wang X., Ning K., Liuand N., Dabing Z. Genetic characterization of a new astrovirus in goslings suffering from gout. Arch. Virol. 2018;163:2865–2869. doi: 10.1007/s00705-018-3932-5. [DOI] [PubMed] [Google Scholar]
- Miaomiao Liu Y.Z., Dongmei Hu X.H., Haifeng Xiongand K.Q., Hongmei L. Clinical and histologic characterization of co-infection with Astrovirus and goose parvovirus in goslings. Avian. Dis. 2019;63:731–736. doi: 10.1637/aviandiseases-D-19-00110. [DOI] [PubMed] [Google Scholar]
- Muhire B.M., Varsaniand A., Darren P.M. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One. 2014;9 doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantin-Jackwood M.J., Strother K.O., Mundt E., Zsak L., Day J.M., Spackman S.E. Molecular characterization of avian astroviruses. Arch. Virol. 2011;156:235–244. doi: 10.1007/s00705-010-0849-z. [DOI] [PubMed] [Google Scholar]
- Reuter G., Pankovicsand P., Akos B. Identification of a novel astrovirus in a domestic pig in Hungary. Arch. Virol. 2011;156:125–128. doi: 10.1007/s00705-010-0827-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach S.N., Ryan A.L. Intra- and cross-species transmission of astroviruses. Viruses. 2021;13:1127. doi: 10.3390/v13061127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajewicz-Krukowska J, Katarzyna D. Nearly full-length genome sequence of a novel astrovirus isolated from chickens with 'white chicks' condition. Arch. Virol. 2016;161:2581–2587. doi: 10.1007/s00705-016-2940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits S.L., van Leeuwen M., Kuiken T., Hammer A.S., Simonand J.H., Albert D.M.E.O. Identification and characterization of deer astroviruses. J. Gen. Virol. 2010;91:2719–2722. doi: 10.1099/vir.0.024067-0. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipskiand A., Sudhir K. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd D., Smyth V.J., Ball N.W., Donnelly B.M., Wylie M., Knowles N.J., Adair B.M. Identification of chicken enterovirus-like viruses, duck hepatitis virus type 2 and duck hepatitis virus type 3 as astroviruses. Avian. Pathol. 2009;38:21–30. doi: 10.1080/03079450802632056. [DOI] [PubMed] [Google Scholar]
- Victoria J.S. A review of the strain diversity and pathogenesis of chicken astrovirus. Viruses. 2017;9:29. doi: 10.3390/v9020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Bai C., Zhang D., Yang K., Yu Z., Jiang S., Geand K., Yongdong L. Genomic and phylogenetic characteristics of a novel goose astrovirus in Anhui Province, Central-Eastern China. Gene. 2020;756 doi: 10.1016/j.gene.2020.144898. [DOI] [PubMed] [Google Scholar]
- Wei F., Yang J., Wang Y., Chen H., Diaoand Y., Yi T. Isolation and characterization of a duck-origin goose astrovirus in China. Emerg. Microbes. Infect. 2020;9:1046–1054. doi: 10.1080/22221751.2020.1765704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Halburand P.G., Tanja O. Complete genome sequence of a newly identified porcine astrovirus genotype 3 strain US-MO123. J. Virol. 2012;86:13126. doi: 10.1128/JVI.02426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Tian J., Tangand Y., Youxiang D. Isolation and genomic characterization of gosling gout caused by a novel goose astrovirus. Transbound. Emerg. Dis. 2018;65:1689–1696. doi: 10.1111/tbed.12928. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Cao Y., Wang J., Fu G., Sun M., Zhang L., Meng L., Cui G., Huang Y., Huand X., Jingliang S. Isolation and characterization of an astrovirus causing fatal visceral gout in domestic goslings. Emerg. Microbes. Infect. 2018;7:71. doi: 10.1038/s41426-018-0074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ren D., Li T., Zhou H., Liu X., Wang X., Lu H., Gao W., Wang Y., Zou X., Sunand H., Jianqiang Y. An emerging novel goose astrovirus associated with gosling gout disease, China. Emerg. Microbes. Infect. 2018;7:152. doi: 10.1038/s41426-018-0153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]