Abstract
Infectious diseases are considered as a pressing challenge to global public health. Accurate and rapid diagnostics tools for early recognition of the pathogen, as well as individualized precision therapy are essential for controlling the spread of infectious diseases. Aptamers, which were screened by systematic evolution of ligands by exponential enrichment (SELEX), can bind to targets with high affinity and specificity so that have exciting potential in both diagnosis and treatment of infectious diseases. In this review, we provide a comprehensive overview of the latest development of SELEX technology and focus on the applications of aptamer-based technologies in infectious diseases, such as targeted drug-delivery, treatments and biosensors for diagnosing. The challenges and the future development in this field of clinical application will also be discussed.
Keywords: aptamer, SELEX, infectious disease, precision medicine, biosensor
Introduction
Infectious diseases which result from pathogenic microorganisms become one of the most important illnesses in the world (Mosing et al., 2005; Giri et al., 2019). Because of the characteristics of contagion and epidemic, infectious diseases not only endanger public health but also pose serious threats and huge losses to social stability and economic development. Over the past decades, the sudden public health crises including Ebola hemorrhagic fevers, avian influenza, severe acute respiratory syndrome (SARS) or Middle East respiratory syndrome (MERS), as well as COVID-19, have swept out the world and caused a significant impact on society inevitably (Del Rio et al., 2014; Liu et al., 2017; Lycett et al., 2020; Wiersinga et al., 2020; Perra, 2021). Existing pathogen detection methods are difficult to achieve a balance between timeliness, accuracy and cost to meet the requirements of large-scale population screening. In addition, the existence of antibiotic-resistant microbes such as multidrug-resistant tuberculosis (MDR-TB), extensively drug-resistant tuberculosis (XDR-TB) and methicillin-and aminoglycoside-resistant Staphylococcus aureus (MARSA) brings greater challenges to the prevention and treatment of infectious diseases (Fauci et al., 2005; Tan Z. M. et al., 2020; Nang et al., 2021). Therefore, there is an urgent demand to develop rapid, economic, and sensitive early diagnostic assays for pathogens, and also adequate therapeutics of precision medicine for infectious diseases.
Aptamers, also known as “chemical antibodies”, are a class of single-stranded DNAs or RNAs which can target various ligands through non-covalent bonds. Those aptamers are synthetic screened in vitro by a selection procedure, commonly known as Systematic Evolution of Ligands by Exponential Enrichment (SELEX). DNA aptamers are more stable and widely used, while RNA aptamers are more likely to form complex structures such as stem, loop, hairpin, G-quadruplex and so on (Breaker, 1997; Lin and Patel, 1997; Wu et al., 2017). These folding 3D structures can increase sequence space coverage and improve space representation, which are beneficial for aptamer-target recognition, thus improving the specificity and affinity of the screened aptamers (Kinghorn et al., 2017). Aptamers have attracted considerable attention because of their exceptional merits such as low synthesis cost, easy chemical modification, high chemical stability and binding affinity, low immunogenicity, good repeatability and reusability (Hong and Sooter, 2015; Yu et al., 2021). To date, thousands of aptamers have been identified, which can be used to identify different targets with high affinity and specificity, such as small metal ions, organic molecules, amino acids, proteins, bacteria, viruses, whole cells and even animals (Cowperthwaite and Ellington, 2008; Zhou and Rossi, 2017). Based on the above advantages, nucleic acid aptamers have been explored as the most promising molecular recognition probes to widely applied in the field of the identification of infectious agents and the therapeutic of infectious diseases.
In this review, the recent advances of SELEX technologies for aptamer selecting of infectious pathogens will be overviewed. Then we will focus on a variety of aptamer-based biosensors for infection detecting and the state-of-the-art aptamer therapeutics and drug delivery systems in the precision treatment of infectious diseases. The current challenges and future prospects of aptamers will also be discussed to provide a direction for the research and development of aptamers.
Discovery of Specific Aptamers by Systematic Evolution of Ligands by Exponential Enrichment
SELEX was originally developed by Gold and Szostak in 1990 (Ellington and Szostak, 1990; Tuerk and Gold, 1990). Before selecting, an oligonucleotide library consisting of two constant regions at 5′ and 3′ ends and a random region in the middle should be synthesized. The primary library usually contains up to 1012–1015 different nucleic acid molecules, of which the random region is about 20–40 bp, and the constant regions that flanked are about 20 bp, including restriction endonuclease sites, primer binding sites and RNA promoter binding sites (Zimbres et al., 2013; Hong and Sooter, 2015). Currently, both DNA and RNA libraries are widely used for SELEX due to their distinct advantages.
General Systematic Evolution of Ligands by Exponential Enrichment Scheme
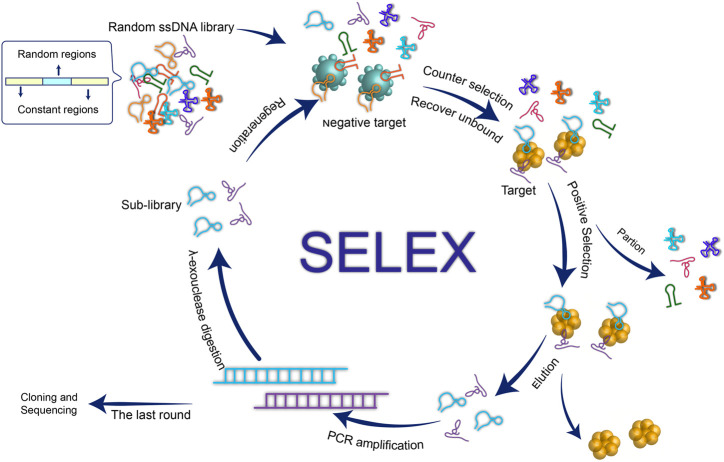
In brief, a typical SELEX comprises three critical stages: 1) incubate the target molecule with the combinatorial library of nucleic acids in vitro to form an aptamer-target complex; 2) partition the complex from the unbound nucleotides and separate the oligonucleotide chain that binds to the target molecule; 3) obtain a sub-library by employing PCR (DNA SELEX) or reverse transcription PCR (RNA SELEX) to amplifying the target-bound sequence (Figure 1). It is worth mentioning that the negative target is usually introduced to improve target specificity by recovering and amplifying the unbound oligonucleotide chain. In this way, the oligonucleotide chain obtained by iterative circles of selecting and PCR amplification is the nucleic acid aptamer of the target. After cloning and sequencing, the identification, binding ability and the secondary/tertiary structure of aptamer can be analyzed subsequently.
FIGURE 1.
schematic illustration of the aptamer generation by conventional SELEX process.
Novel Approaches for Aptamer Selection
Despite the conventional SELEX technology is well-established, the process is tedious and time-consuming, which typically takes up to 20 rounds. At present, various novel SELEX technologies have been developed to improve the shortcomings of the conventional one, further accelerate the speed and efficiency of high-affinity aptamer screening and shorten the selection period. In this respect, the rapid development of SELEX technology provides substantial potential for rapid response to public health emergencies.
The separation of aptamer-bound sequences from the unbound nucleotides is one of the most critical steps in the process of SELEX. Thus, the screening process can be accelerated by optimizing the binding and separation of the target molecule with libraries. Usually, immobilization is involved in binding the targets to a carrier and then incubated with the nucleic acid library (Van Dyke et al., 2016). Common carriers such as magnetic beads, affinity chromatography columns or microfluidic chips. Among them, considerable attention has been given to magnetic beads, as chemical modification of magnetic beads is easy and the magnetic separation method is convenient, fast and effective (McKeague et al., 2010; Hunniger et al., 2014; Xi et al., 2015; Duan et al., 2016; Ma et al., 2018). To date, a variety of aptamers have been successfully selected by magnetic SELEX (Oh et al., 2009; Lai et al., 2014; Wu J.-H. et al., 2018). For example, Hong et al. first proposed a magnetism-controlled chip for Ebola virus aptamers selection by integrating the magnetic bead-based SELEX (Mag-SELEX) with a microfluidic system and by this method they got an aptamer with low dissociation constants and reducing the selecting round to three (Hong et al., 2019). Another novel approach called Capillary Electrophoresis SELEX (CE-SELEX) is based on the difference of electrophoretic mobility between target bounded sequence with the unbound one (Mendonsa and Bowser, 2004a; Mendonsa and Bowser, 2004b). Mosing Renee K et al. successfully obtained aptamers candidate with high affinity to human immunodeficiency virus (HIV) reverse transcriptase by CE-SELEX only after four cycles (Mosing et al., 2005). Although in vitro SELEX technology for a single target has been quite mature, especially for proteins, the clinical applications of these aptamers are still limited. The reason is that the properties of a recombinant protein are not the same as those of natural proteins, including conformation, advanced structure and biological activity. Besides, aptamers screened for artificial recombinant proteins may be much less sensitive in identifying natural targets. To overcome this limitation, cell-SELEX, which employs the whole cell as a target, was first proposed in 2003 (Daniels et al., 2003). Recently, a series of aptamers against various pathogens were generated by cell-SELEX, such as Salmonella typhimurium, Escherichia coli, Vibrio parahaemolyticus, Trypanosoma cruzi, and so on (Duan et al., 2012; Nagarkatti et al., 2012; Dwivedi et al., 2013; Kim et al., 2013; Bitaraf et al., 2016; Saad et al., 2020). In addition to the methods mentioned above, many other innovative SELEX strategies have also been developed in the past decades, such as capture-SELEX, in vivo SELEX, high-throughput sequencing SELEX and so on. The principles and characteristics of those techniques are summarized in Table 1.
TABLE 1.
Summary of novel SELEX techniques for aptamer selecting.
| Techniques | Description | Advantages | Ref. |
|---|---|---|---|
| Magnetic bead-based SELEX | Immobilize the targets on the magnetic bead, accelerate the process by magnetic separation | Simple and fast operation; short cycle; low cost | Lou et al. (2009), Oh et al. (2009), Oh et al. (2011), Zhou et al. (2013b), Hunniger et al. (2014), Lai et al. (2014), Lai and Hong (2014), Lin et al. (2015), Ansari et al. (2017), Han et al. (2017), Paniel et al. (2017), Wu et al. (2018a), Hong et al. (2019), Leblebici et al. (2019) |
| Capillary Electrophoresis SELEX | According to the difference of electrophoretic mobility between target bounded sequence with the unbound one | Quick; economic; high efficiency; easy operation without washing procedures | Mendonsa and Bowser (2004b), Mosing et al. (2005), Mosing and Bowser (2009), Cella et al. (2010), Ruff et al. (2012), Jing and Bowser (2013), Yang and Bowser (2013), Dong et al. (2015), Zhu et al. (2019a), Zhu et al. (2019b) |
| Cell-SELEX | Employ the whole cell as targets | Remain the conformation and bioactivity of protein; obtain aptamers without knowing the molecular target on the cell surface; no need to purify protein before selection; explore new surface protein and biomarkers | Duan et al. (2012), Nagarkatti et al. (2012), Dwivedi et al. (2013), Kim et al. (2013), Bitaraf et al. (2016), He et al. (2019), Song et al. (2019), Gao et al. (2020), Saad et al. (2020), Lin et al. (2021) |
| Capture-SELEX | Immobilize the oligonucleotides on the solid substrate to further capture the targets | Simple operation of target immobilization; retain the natural structure of the target; especially suitable for small molecular targets | Stoltenburg et al. (2012), Duan et al. (2013), Spiga et al. (2015), Duan et al. (2017), Paniel et al. (2017), Lauridsen et al. (2018), Boussebayle et al. (2019a), Boussebayle et al. (2019b) |
| In vivo SELEX | Use living animal models as selection targets or conditions to generate aptamers | Aptamers were selected in a whole organism ideally; it holds promise for drug delivery and treatment in vivo | van Bel et al. (2014), Urak et al. (2016), Zhuo et al. (2017), Wang et al. (2018), Chen et al. (2019), Sola et al. (2020) |
| High-throughput sequencing SELEX | Conventional SELEX in conjunction with high-throughput sequencing system, sequencing across all the selection round instead of the last one | predominant efficiency and applicability | Roulet et al. (2002), Dittmar et al. (2012), Reiss et al. (2012), Ditzler et al. (2013), Dao et al. (2016), Ruan et al. (2017), Nitta et al. (2019), Asif and Orenstein (2020), Fan et al. (2020), Ishida et al. (2020) |
SELEX technology determines that the aptamer target molecules have a very wide range which covers a huge range from metal ions, compounds, peptides, nucleic acids, proteins to cells, and even includes some complex targets, such as viruses, bacteria and other pathogens. Based on SELEX technology, aptamers obtained from specific substances of the pathogens such as surface proteins or key enzymes in physiological processes, are expected to be used for pathogen diagnosis or infection treatment.
Application of Aptamers Biosensor in Infectious Diseases Diagnosis
Most infectious diseases progress rapidly, and a clear diagnosis is key for effective treatment. Also, efficient pathogen detection methods help control the spread of infectious diseases. Therefore, early, accurate and rapid pathogen diagnosis is of great significance. Currently, the routine diagnostics for infectious pathogen are bacterial culture, polymerase chain reaction (PCR) and immunological detection (Lampel et al., 2000; Byrne et al., 2009; Arvanitis et al., 2014; Rohr et al., 2016; Balmaseda et al., 2017). These approaches are relatively mature, but they inevitably have limitations such as time-consuming, high cost, and tedious operation. Alternatively, although antibody is indispensable in most routine tests, it inevitably meets many limitations, such as laborious and expensive production and identification, batch-to-batch variation, and its biological activity is susceptible to the environment such as pH and temperature (Jayasena, 1999; Nimjee et al., 2005; Rosenbaum et al., 2012; Yu et al., 2021). Aptamers have a very wide range of target molecules, which can be designed for early disease markers, creating conditions for the early detection of pathogens. At the same time, aptamers can be rapidly chemically synthesized in batches with a long shelf-life for storage at room temperature. The comparison of main characteristics between antibodies and aptamers is shown in Table 2.
TABLE 2.
The characteristics comparison of nucleic acid aptamers and antibodies (Jayasena, 1999; Zhou and Rossi, 2017; Zhuo et al., 2017; Yu et al., 2021).
| Features | Antibodies | Aptamers |
|---|---|---|
| Substance | polymer peptide | nucleic acid |
| Specificity | high | high |
| Affinity | high | high |
| Immunogenicity | high | no humoral immunity |
| Production cost | high | low |
| Stability | unstable | stable |
| Potential target | limited to immunogenic molecules | no limitation |
| Development time | 6–18 weeks | 2–6 weeks |
| Modification | limited | convenient |
| Batch-to-batch variation | high | low |
Due to those advantages, abundant research has integrated aptamers into biosensors as molecular recognition elements over the past decades. The biological signals received by aptamers can be transformed into the optical signal or electrical signal by the signal converter, then the output signal will be amplified by the electronic system and further be used for the detection of pathogenic microorganisms qualitatively or quantitatively. According to the pathogenic targets, we category these aptamer-based biosensors into three classes: bacteria, viruses and others.
Detection of Bacteria
The current diagnostic gold standard of bacterial identification is still bacterial culture, subsequent biochemical identification and serological typing (Rauch and Nauen, 2003; Andini et al., 2018; Takeuchi et al., 2018). However, they are time-consuming, usually several days, and have some limitations in the identification of some certain species. In addition, it is less convenient for point-of-care detection. To meet this need, aptamer-based biosensors were widely developed. As an example, an electrochemical sensor for the detection of Mycobacterium tuberculosis reference strain H37Rv ATCC 27294 using aptamer technology was introduced by Zhang X. et al. (2019). In this method, H37Rv aptamers layer modified on the bare Au interdigital electrode are recognition probe and oligonucleotides modified with gold nanoparticles (AuNPs-DNA) are signal probe. When H37Rv bacteria was present, it competitively bounds to aptamers, and the displacement of AuNPs-DNA dramatically changing the conductivity. The detection limit of this method is 100 CFU/ml, and it can be used for rapid detection of H37Rv only in 2 h. In one of recent studies, electrochemical aptasensor was devloped for the detection of Escherichia coli O157:H7 (E. coli) (Qaanei et al., 2021). In this system, the aptamer is employed to improve the selectivity while a reduced graphene oxide–poly (vinyl alcohol) and gold nanoparticles nanocomposite (AuNPs/rGO–PVA/GCE) is used to raise the sensor sensitivity. Consequently, this aptasensor is able to detect E. coli as low as 9.34 CFU/ml, with an excellent specificity.
Detection of Viruses
Many infectious diseases are caused by viral infection, such as acquired immunodeficient syndrome (AIDS), influenza and COVID-19, which has dealt a heavy blow to the world in 2020. These pathogens widely distribute in open systems and are endanger human health and the public environment. For example, the current COVID-19 pandemic in more than 100 countries around the world has infected an untold number of people and caused large numbers of deaths. Thus, it is urgently desirable for cost-effective, rapid and reliable diagnostic methods. Woo et al. designed an aptamer-based fluorescent sensor to detect SARS-CoV-2 RNA in human nasopharyngeal samples (Woo et al., 2020). They intelligently designed a one-pot, ligation-dependent isothermal reaction cascade that consists of a ligation reaction by SplintR ligase and subsequent transcription by T7 RNA polymerase. When target RNA existing, the RNA aptamers of the isothermal reaction products bind to fluorescent dyes and produce a significant fluorescence signal. The detection limit is 0.1 aM. Interestingly, only by redesigning the hybridization regions of the probes, a series of viruses including influenza viruses, MERS and SARS can be detected by this method. Another recent approach was proposed by Liu et al. (2020), which designed a qPCR amplification reaction triggered by two aptamers probes for ultrasensitive detection of serum COVID-19-associated antigens. This method exhibits excellent detection performance and can be conducted within 2 h. In another study by Babamiri et al. (2018), aptamer against HIV-1 was used for the development of an electrochemiluminescence (ECL) sensor. This aptamer-based biosensor showed excellent sensitivity and specificity, with a detection limit as low as 0.3 fM, and can be successfully applied to clinical serum samples analysis.
More importantly, some viruses have multiple subtypes and mutant quickly, which are highly infectious and transmissible pathogens (Shim, 2011; Li et al., 2019; Ma and Ma, 2020). Therefore, the detection of mutation is becoming a top priority in the field of methodological research. At present, molecular methods such as PCR and DNA sequencing are powerful tools to obtain information on mutant status, but these methods could not satisfy the expectation for extensive disease screening because of the need for special equipment and expensive consumables (Escadafal et al., 2014; Ma et al., 2015). Recently, Wang et al. established a highly sensitive platform for detecting SARS-CoV-2 and its mutated variants based on a CRISPR-Cas13 transcription amplification principle (Wang et al., 2021). They employed light-up RNA aptamers as the sensitive output of amplification signals, achieving sensing of as low as 82 copies of SARS-CoV-2. Moreover, this platform was applied to strictly identify the key mutation of the SARS-CoV-2 variant, D614G, which increases viral stability and flexibility and further enhances replication and transmission.
Detection of Other Pathogens
Aptamer-based biosensors are used to detect several other pathogens alternatively. Protozoan parasite infection remains one of the major public health problems in some underdeveloped and developing countries with poor sanitation and economic backwardness. Thus, the application of detections that require expensive equipment or complex laboratory sites is significantly limited in these areas. In this case, a deal of aptamer-based biosensors has been developed for the identification of parasites due to their low cost, simplicity, portability (Lee et al., 2012; Singh et al., 2019a; Singh et al., 2019b; Frezza et al., 2020; Minopoli et al., 2020). Take the diagnosis of malaria as an example, Singh et al. established instrument-based and instrument-free approaches for pan malaria and P. falciparum species based on aptamers specific to Plasmodium lactate dehydrogenase (PLDH) and Plasmodium falciparum glutamate dehydrogenase (PfGDH) respectively (Singh et al., 2019a). They successfully overcame the false-negative limitation of traditional microscopic examination of Giemsa-stained thick blood films and achieved an ultrasensitive detection with a low cost (∼0.10 $ per test) (Mikhail et al., 2011; Thongdee et al., 2014; Bin Dajem, 2015; Sumari et al., 2016; Habyarimana and Ramroop, 2020). In addition, Fu developed an indirect blocking enzyme linked aptamer assay (ib-ELAA) for the detection of Mycoplasma gallisepticum (M. gallisepticum), which was the major pathogen of chronic respiratory disease (Fu et al., 2021b; Fu et al., 2021a). In this method, they initially screened out the aptamer Apt-236 which can bind to PvpA protein of M. gallisepticum with high affinity, and further integrated Apt-236 into ib-ELAA and successfully applied in the detection of clinical chicken sera sample. Similarly, a great many of aptamer-based methods for the detection of pathogenic parasites (Homann et al., 2006; Bruno et al., 2014; Ospina-Villa et al., 2018; Ospina, 2020), mycoplasma (Fu et al., 2014; Liu Y. et al., 2019; Wan et al., 2020), several fungal species (McKeague et al., 2010; Barthelmebs et al., 2011; Ma et al., 2014; Wu S. et al., 2018; Liu M. et al., 2019; Han et al., 2021) have also been developed.
The comparison of representative aptasensor performance in the detection of the various pathogen is summarized in Table 3.
TABLE 3.
Comparison of aptasensor performance in the detection of various pathogen.
| Type of pathogen | Target | Aptamer | Detection method | Lod | Linear range | Detection time | Specificity | Ref. |
|---|---|---|---|---|---|---|---|---|
| Bacteria | Mycobacterium tuberculosis | H37Rv aptamers | Electrochemical | 100 CFU/ml | 1×102–1 × 107 CFU/ml | 2 h | 90% | Zhang et al. (2019a) |
| Staphylococcus aureus | S.aureus aptamer | Fluorescent | 39 CFU/ml | 80–8 × 106 CFU/ml | NR | high | Cai et al. (2019) | |
| L. monocytogenes | LM6-116 | Fluorescent | 10 CFU/ml | 10–1 × 106 CFU/ml | NR | high | Guo et al. (2020) | |
| Escherichia coli | E1 | Fluorescenct | 3.7 × 102 CFU/ml | 6×103–3.75 × 106 CFU/ml | 135 s | high | Zhang et al. (2019b) | |
| Viruses | SARS-CoV-2 | D614G variants aptamer | Fluorescent | 82 copies | 100–1,000 copies | 20 min | high | Wang et al. (2021) |
| HIV-1 | HIV aptamer | ECL | 0.3 fM | 3.3 fM–0.3 nM | NR | high | Babamiri et al. (2018) | |
| HBV | HBsAg aptamer | Chemiluminescent | 0.05 ng/ml | 1–225 ng/ml | NR | high | Xi et al. (2018) | |
| Influenza | Influenza nucleoprotein aptamer | lateral flow immunoassays | 0.26 pg/ml | 0.01–10 ng/ml | 10 min | high | Kang et al. (2019) | |
| Norovirus | Aptamer-6-FAM, Bt-Apt-Fc | Microfluidic | 100 pM | 100 pM - 3.5 nM | 35 min | high | Chand and Neethirajan, (2017) | |
| Other pathogens | P. falciparum | PLDH/PfGDH aptamers | Colorimetric | 0.55 pM/1.34 pM | 1 pM - 100 nM | 35 min | high | Singh et al. (2019a) |
| Trypanosoma cruzi | Apt68 | PCR | 0.33 parasites/ml | NR | NR | high | Dunning et al. (2014b) | |
| Leishmania | Leishmania aptamer | Fluorescent | ∼100 ng/2 ml sample | 0–1,000 ng | ∼1 h | NR | Bruno et al. (2014) |
NR, not report.
Application of Aptamers in Infectious Diseases Treatment
At present, the therapeutic of infectious diseases is mainly based on the principle of symptomatic treatment or specific anti-pathogen treatment. However, antimicrobial resistance, high viral genomes mutation variability and escaping the host immune response make most medications and vaccines inefficient (Finlay and McFadden, 2006; Labella and Merel, 2013; Dunning et al., 2014a; Fall-Malick et al., 2014; Ferir et al., 2014; Lazarevic, 2014; Marascio et al., 2014; Sahu, 2015; Wandtke et al., 2015). It is worth mentioning that the effect of antiviral therapy is not ideal for all patients and side effects caused by many existing antiviral drugs may lead to other diseases than primary affection. For instance, the most effective therapy for patients with hepatitis C (interferon alfa-2b plus ribavirin) benefits only about 50% of cases (Manns et al., 2001; Sarhan et al., 2020), whereas such therapeutic regimen usually be associated with numerous adverse effects (Fried, 2002; Nishimura et al., 2002; Fisher et al., 2004; Negro, 2010; Pazienza, 2011; Gull et al., 2019). Many studies confirmed that aptamer, as a promising candidate, can target the key molecules in bacterial physiological processes or viral surface proteins, and treat the infection effectively by inhibiting viruses penetrating the cells, disrupting the activity of enzymes related to viral replication or regulating immune response (Bellecave et al., 2008; Gopinath et al., 2012; Hwang et al., 2012; Torabi et al., 2020).
Aptamer-Based Therapeutics
Precision medicine holds great promise to harness the benefits of aptamers that can bound to targets with high specificity and affinity for targeted treatment of a variety of diseases. Such therapeutic aptamers function mainly in the following two ways: 1) aptamers function as antagonists to disrupt the function of a pathologic target protein and block the interaction of disease-associated targets by specifically binding to target; 2) aptamers function as agonists to increase the ability of the target receptors. For instance, Lee et al. reported an RNA aptamer against the Hepatitis C virus (HCV) nonstructural protein 5B can effectively inhibit HCV replication and suppressed HCV infectious virus particle formation (Lee et al., 2015). HIV integrase is considered necessary for retroviral replication, which is a primary target for the therapy of AIDS (Shoji et al., 2002). Thus, aptamers as potential anti-HIV integrase inhibitors have drawn much attention from researchers. Pang and his colleague designed an anti-HIV lentivirus vector consist of shRNA, ribozyme and RNA decoy (Pang et al., 2018). By screening aptamers against integrase and incorporating these aptamers in shRNA, they successfully observed interference and inhibition to transcription of HIV in cell cultures. Also, many other aptamers against Tat protein, gp120, reverse transcriptase, nucleocapsid protein were developed for further exploit research of antivirus therapy (Mufhandu et al., 2012; Aeksiri et al., 2014; Nguyen et al., 2020; Zhang et al., 2020). COVID-19 has wreaked havoc all over the world, but no specific treatment has been developed yet. Liu and his colleague developed an aptamer that specifically targets the spike protein of the coronavirus SARS-CoV-2, which is the critical role of viral infection (Liu et al., 2021). When the receptor-binding domain (RBD) of the spike protein of the coronavirus SARS-CoV-2 binds to the human angiotensin-converting enzyme 2 (ACE2), an infection cascade is triggered (Schmitz et al., 2021). They proved that this aptamer effectively protects host cells from infection by blocking the interaction between spike protein and ACE2 receptor. This exciting report is fueling hope in the field of COVID-19’s therapy, it also brings up new opportunities for aptamer-based treatment.
Despite aptamer-based therapy shows huge potential, their inherent physicochemical characteristics affect pharmacokinetic properties in some way, which may limit their widespread clinical application. The most critical problems are nuclease degradation and rapid renal filtration. Unmodified nucleic acid aptamers have an average half-life of fewer than 10 min for the susceptibility to nucleases which abundantly exist in biological fluids (Lakhin et al., 2013). To increase its biostability and prolong the in vivo half-life, chemical modifications are typically introduced such as replacing 2′-OH with fluoro (F), amino (NH2), or O-methyl (OCH3) groups at the 2′ position (Morita et al., 2018). Since the average diameter of 5–30 kDa aptamers is less than 5nm, which is smaller than the glomerular filtration threshold (i.e., 30–50 kDa), aptamers are inevitably rapidly excreted through renal filtration (Guo, 2010; Morita et al., 2018). To overcome this disadvantage, many macromolecular substances such as proteins, cholesterol, liposomes, high molecular mass PEG or nanomaterials are involved to modify aptamers, and there is indeed a significant improvement in some reports (Fisher et al., 1976; Drolet et al., 2000; Rusconi et al., 2004; Burmeister et al., 2005; Chen et al., 2015; Heo et al., 2016). However, chemical modification is a double-edged sword. Serious allergic responses caused by biomaterial, non-specific immune activation, tissue toxicity caused by drug metabolism and other undesirable side effects have been reported (Geary et al., 2003; Waring, 2010; Wong and Goldberg, 2014; Lincoff et al., 2016; Morita et al., 2016). Thus, it is necessary to cautiously improve and optimize the formulations or administration routines of aptamer therapy.
Aptamers as Intelligent Chemical Drug-Delivery Systems
The other ingenious anti-infective therapeutic strategy is to employ aptamers as intelligent messengers of therapeutic agents, such as small interfering RNA molecules and ribozymes (Dey et al., 2005; Romero-López et al., 2012; Zhu et al., 2012; Wandtke et al., 2015). In the broad area of drug delivery system, aptamers have also been extensively sought after due to the inherited merits: relatively small physical size, versatile structure, quick chemical production, flexible chemical modification, high stability, and lack of immunogenicity. Through chemical modification and bioconjugation, a variety of therapeutic agents increase their stability and bioactivity without change the primary characteristics (Fattal et al., 2018). The targeting of aptamer increases the local drug concentration, thus improving the therapeutic efficacy whilst reducing the systemic toxic and side effects of the drug. Therefore, many cell-specific aptamers are explored to conjugate with chemical entities including chemotherapeutic agents, siRNA, nanoparticles for targeted delivery of drugs.
For the development of aptamers as drug-delivery systems, one example is anti-gp120 aptamer for the treatment of AIDS (Zhou J. et al., 2013). A viral surface protein, gp120, is closely related to viral infection. HIV-1 infects target cells through binding gp120 to cellular receptor CD4 and chemokine receptors such as CCR5 or CXCR4. In this research, Zhou and his colleague employed an anti-gp120 aptamers as siRNA delivery vehicles, effective delivery viral inhibiting siRNA in vivo and potent inhibition of HIV-1 replication. Similarly, Pan et al. designed an ingenious system which employed bispecific circular aptamers (bc-apts) to specifically tether protein cargoes and cellular membrane proteins (Pan et al., 2020). This strategy achieved the specific delivery of functional therapeutic proteins, and the deactivation of functional proteins was also avoided. Furthermore, the bioactivity of the drug in the lesion was specifically increased. Yan et al. reviewed the design and application of aptamers as drug-delivery systems in the photodynamic platform of targeted therapy (Yan et al., 2021). In this review, they focused on the application of aptamers-targeted photodynamic therapies which achieve controlled and accurate delivery of therapeutic drugs to the lesion sites and obtained excellent photodynamic therapy efficiency. Another advantage of an aptamer-based targeted delivery system is that it can delay the evolution of resistance and improve the efficiency of the antimicrobials on already resistant pathogens. Ucak et al. used Staphylococcus aureus-specific aptamer to functionalize methicillin, which is an antibiotic for serious infectious caused by Gram-positive bacteria (Ucak et al., 2020). By limiting the amount and dosage during therapy, they proved that the novel delivery system was significantly effective in reducing minimum inhibitory concentration (MIC) values. Therefore, the aptamer-based targeted delivery system is a promising method for the treatment of infections caused by antibiotic-resistant bacteria. In addition to the examples above, multiple types of research have shown the extraordinary ability of aptamer in drug target-delivering (Nimjee et al., 2005; Shiang et al., 2013; Hahn, 2017; Chonco et al., 2018; Fattal et al., 2018; Tan X. et al., 2020).
Conclusion and Future Perspectives
Taking advantage of low cost, easy chemical modification, high specificity and binding affinity, low immunogenicity, aptamers have been used as an alternative to antibodies in the development of aptamer-based technologies in the past decades. The development of biomedical technology has enabled a comprehensive exploration of the screening technologies and practical applications of aptamers. In this review, we comprehensively discussed the recent progress in the development of SELEX technology and aptamer-based applied research in various types of infectious diseases.
Up to now, aptamers were intensively integrated into the biosensor strategies as molecular recognition elements. Compared with conventional diagnosis methods, aptamers-based biosensors strategies had obvious advancement in sensitivity and reliability which could improve diagnostic performance, thus lead to intervention at an earlier stage and avoid the spread of infectious diseases. Furthermore, as a class of single-stranded nucleic acid, aptamers showed outstanding advantages on cost and manufacturability, so that the development of aptamer-based biosensors could be conducive promote the popularization and improvement of infectious diseases diagnosis techniques in community hospitals. Last but not the least, the portability makes biosensors an alternative to traditional methods in point-of-care diagnostics and even more diverse medical settings such as epidemic areas. Aptamers could be also applied in biotherapy and drug-delivery systems. Due to low immunogenicity and high targeting ability, aptamer-based therapy could increase the drug concentration in local lesions, thus improve the therapeutic effect and reduce the toxic and side effects of drugs. Aptamers were also intelligent in solving problems of antimicrobial resistance and viral genomes mutation variability. Therefore, aptamers are expected to be promising in the therapeutic of infectious diseases.
However, there remain several challenges limiting the clinical application of aptamers. Firstly, aptamers for some complex pathogens are still limited because of the limitations of the current SELEX technology. Nevertheless, those problems can be ameliorated by optimizing the critical factors in the SELEX process in near future, such as the concentration of target molecules, the separation method of aptamers, PCR reaction conditions, the number of screening rounds and so on. Besides, the biostability and toxicity of aptamers, as well as the degradation of unmodified aptamers by nuclease in serum and rapid removal by renal filtration remain to be explored. The side effects caused by the metabolism of aptamer are also a problem to be reckoned with. Therefore, significant refinements of biochemical modification and rigorous administration routines of aptamer-based therapy are still needed in future research. Fortunately, in spite of all the challenges mentions above, the transition from aptamer-based basic research to clinical application is taking place, although slowly.
Overall, we foresee a promising prospect for aptamer-based technologies in precision medicine of infectious diseases. Shortly, with many research and development activities going on in this field, we envision that practical and commercial biosensors and novel drugs for clinical diagnosis and precise therapy are very close to realization, and consequently, that will significantly reduce the human diseases and economic burdens.
Author Contributions
YX and XJ wrote the manuscript. YZ and MM contributed to the literature research. MW and BY revised and approved the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant numbers 81672095); National Science and Technology Major Project of the Ministry of Science and Technology of China (Grant numbers 2018ZX10715,003-001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Aeksiri N., Songtawee N., Gleeson M. P., Hannongbua S., Choowongkomon K. (2014). Insight into HIV-1 Reverse Transcriptase-Aptamer Interaction from Molecular Dynamics Simulations. J. Mol. Model. 20 (8), 2380. 10.1007/s00894-014-2380-8 [DOI] [PubMed] [Google Scholar]
- Andini N., Hu A., Zhou L., Cogill S., Wang T.-H., Wittwer C. T., et al. (2018). A "Culture" Shift: Broad Bacterial Detection, Identification, and Antimicrobial Susceptibility Testing Directly from Whole Blood. Clin. Chem. 64 (10), 1453–1462. 10.1373/clinchem.2018.290189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari N., Ghazvini K., Ramezani M., Shahdordizadeh M., Yazdian-Robati R., Abnous K., et al. (2017). Selection of DNA Aptamers against Mycobacterium tuberculosis Ag85A, and its Application in a Graphene Oxide-Based Fluorometric Assay. Microchim Acta 185 (1), 21. 10.1007/s00604-017-2550-3 [DOI] [PubMed] [Google Scholar]
- Arvanitis M., Anagnostou T., Fuchs B. B., Caliendo A. M., Mylonakis E. (2014). Molecular and Nonmolecular Diagnostic Methods for Invasive Fungal Infections. Clin. Microbiol. Rev. 27 (3), 490–526. 10.1128/CMR.00091-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif M., Orenstein Y. (2020). DeepSELEX: Inferring DNA-Binding Preferences from HT-SELEX Data Using Multi-Class CNNs. Bioinformatics 36 (Suppl. l_2), i634–i642. 10.1093/bioinformatics/btaa789 [DOI] [PubMed] [Google Scholar]
- Babamiri B., Salimi A., Hallaj R. (2018). A Molecularly Imprinted Electrochemiluminescence Sensor for Ultrasensitive HIV-1 Gene Detection Using EuS Nanocrystals as Luminophore. Biosens. Bioelectron. 117, 332–339. 10.1016/j.bios.2018.06.003 [DOI] [PubMed] [Google Scholar]
- Balmaseda A., Stettler K., Medialdea-Carrera R., Collado D., Jin X., Zambrana J. V., et al. (2017). Antibody-based Assay Discriminates Zika Virus Infection from Other Flaviviruses. Proc. Natl. Acad. Sci. USA 114 (31), 8384–8389. 10.1073/pnas.1704984114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelmebs L., Jonca J., Hayat A., Prieto-Simon B., Marty J.-L. (2011). Enzyme-Linked Aptamer Assays (ELAAs), Based on a Competition Format for a Rapid and Sensitive Detection of Ochratoxin A in Wine. Food Control 22 (5), 737–743. 10.1016/j.foodcont.2010.11.005 [DOI] [Google Scholar]
- Bellecave P., Cazenave C., Rumi J., Staedel C., Cosnefroy O., Andreola M.-L., et al. (2008). Inhibition of Hepatitis C Virus (HCV) RNA Polymerase by DNA Aptamers: Mechanism of Inhibition of In Vitro RNA Synthesis and Effect on HCV-Infected Cells. Antimicrob. Agents Chemother. 52 (6), 2097–2110. 10.1128/AAC.01227-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bin Dajem S. M. (2015). Molecular Investigation of Mixed Malaria Infections in Southwest Saudi Arabia. Saudi Med. J. 36 (2), 248–251. 10.15537/smj.2015.2.10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitaraf F. S., Rasooli I., Mousavi Gargari S. L. (2016). DNA Aptamers for the Detection of Haemophilus Influenzae Type B by Cell SELEX. Eur. J. Clin. Microbiol. Infect. Dis. 35 (3), 503–510. 10.1007/s10096-015-2567-7 [DOI] [PubMed] [Google Scholar]
- Boussebayle A., Groher F., Suess B. (2019a). RNA-based Capture-SELEX for the Selection of Small Molecule-Binding Aptamers. Methods 161, 10–15. 10.1016/j.ymeth.2019.04.004 [DOI] [PubMed] [Google Scholar]
- Boussebayle A., Torka D., Ollivaud S., Braun J., Bofill-Bosch C., Dombrowski M., et al. (2019b). Next-level Riboswitch Development-Implementation of Capture-SELEX Facilitates Identification of a New Synthetic Riboswitch. Nucleic Acids Res. 47 (9), 4883–4895. 10.1093/nar/gkz216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker R. (1997). DNA Aptamers and DNA Enzymes. Curr. Opin. Chem. Biol. 1 (1), 26–31. 10.1016/s1367-5931(97)80105-6 [DOI] [PubMed] [Google Scholar]
- Bruno J. G., Richarte A. M., Phillips T., Savage A. A., Sivils J. C., Greis A., et al. (2014). Development of a Fluorescent Enzyme-Linked DNA Aptamer-Magnetic Bead sandwich Assay and Portable Fluorometer for Sensitive and Rapid Leishmania Detection in Sandflies. J. Fluoresc 24 (1), 267–277. 10.1007/s10895-013-1315-6 [DOI] [PubMed] [Google Scholar]
- Burmeister P. E., Lewis S. D., Silva R. F., Preiss J. R., Horwitz L. R., Pendergrast P. S., et al. (2005). Direct In Vitro Selection of a 2′-O-Methyl Aptamer to VEGF. Chem. Biol. 12 (1), 25–33. 10.1016/j.chembiol.2004.10.017 [DOI] [PubMed] [Google Scholar]
- Byrne B., Stack E., Gilmartin N., O’Kennedy R. (2009). Antibody-based Sensors: Principles, Problems and Potential for Detection of Pathogens and Associated Toxins. Sensors 9 (6), 4407–4445. 10.3390/s90604407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai R., Yin F., Zhang Z., Tian Y., Zhou N. (2019). Functional Chimera Aptamer and Molecular beacon Based Fluorescent Detection of Staphylococcus aureus with Strand Displacement-Target Recycling Amplification. Analytica Chim. Acta 1075, 128–136. 10.1016/j.aca.2019.05.014 [DOI] [PubMed] [Google Scholar]
- Cella L. N., Sanchez P., Zhong W., Myung N. V., Chen W., Mulchandani A. (2010). Nano Aptasensor for Protective Antigen Toxin of Anthrax. Anal. Chem. 82 (5), 2042–2047. 10.1021/ac902791q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chand R., Neethirajan S. (2017). Microfluidic Platform Integrated with Graphene-Gold Nano-Composite Aptasensor for One-step Detection of Norovirus. Biosens. Bioelectron. 98, 47–53. 10.1016/j.bios.2017.06.026 [DOI] [PubMed] [Google Scholar]
- Chen L., He W., Jiang H., Wu L., Xiong W., Li B., et al. (2019). In Vivo SELEX of Bone Targeting Aptamer in Prostate Cancer Bone Metastasis Model. Int. J. Nanomedicine 14, 149–159. 10.2147/IJN.S188003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Rashid F., Shah A., Awan H. M., Wu M., Liu A., et al. (2015). The Isolation of an RNA Aptamer Targeting to P53 Protein with Single Amino Acid Mutation. Proc. Natl. Acad. Sci. USA 112 (32), 10002–10007. 10.1073/pnas.1502159112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chonco L., Fernández G., Kalhapure R., Hernáiz M. J., García-Oliva C., Gonzalez V. M., et al. (2018). Novel DNA Aptamers against CCL21 Protein: Characterization and Biomedical Applications for Targeted Drug Delivery to T Cell-Rich Zones. Nucleic Acid Ther. 28 (4), 242–251. 10.1089/nat.2017.0689 [DOI] [PubMed] [Google Scholar]
- Cowperthwaite M. C., Ellington A. D. (2008). Bioinformatic Analysis of the Contribution of Primer Sequences to Aptamer Structures. J. Mol. Evol. 67 (1), 95–102. 10.1007/s00239-008-9130-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D. A., Chen H., Hicke B. J., Swiderek K. M., Gold L. (2003). A Tenascin-C Aptamer Identified by Tumor Cell SELEX: Systematic Evolution of Ligands by Exponential Enrichment. Proc. Natl. Acad. Sci. 100 (26), 15416–15421. 10.1073/pnas.2136683100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao P., Hoinka J., Takahashi M., Zhou J., Ho M., Wang Y., et al. (2016). AptaTRACE Elucidates RNA Sequence-Structure Motifs from Selection Trends in HT-SELEX Experiments. Cel Syst. 3 (1), 62–70. 10.1016/j.cels.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio C., Mehta A. K., Lyon G. M., 3rd, Guarner J. (2014). Ebola Hemorrhagic Fever in 2014: the Tale of an Evolving Epidemic. Ann. Intern. Med. 161 (10), 746–748. 10.7326/M14-1880 [DOI] [PubMed] [Google Scholar]
- Dey A. K., Griffiths C., Lea S. M., James W. (2005). Structural Characterization of an Anti-gp120 RNA Aptamer that Neutralizes R5 Strains of HIV-1. RNA 11 (6), 873–884. 10.1261/rna.7205405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar K. A., Jiang P., Park J. W., Amirikian K., Wan J., Shen S., et al. (2012). Genome-wide Determination of a Broad ESRP-Regulated Posttranscriptional Network by High-Throughput Sequencing. Mol. Cel Biol 32 (8), 1468–1482. 10.1128/MCB.06536-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzler M. A., Lange M. J., Bose D., Bottoms C. A., Virkler K. F., Sawyer A. W., et al. (2013). High-throughput Sequence Analysis Reveals Structural Diversity and Improved Potency Among RNA Inhibitors of HIV Reverse Transcriptase. Nucleic Acids Res. 41 (3), 1873–1884. 10.1093/nar/gks1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Tan Q., Ye W., Liu D., Chen H., Hu H., et al. (2015). Screening and Identifying a Novel ssDNA Aptamer against Alpha-Fetoprotein Using CE-SELEX. Sci. Rep. 5, 15552. 10.1038/srep15552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drolet D. W., Nelson J., Tucker C. E., Zack P. M., Nixon K., Bolin R., et al. (2000). Pharmacokinetics and Safety of an Anti-vascular Endothelial Growth Factor Aptamer (NX1838) Following Injection into the Vitreous Humor of Rhesus Monkeys. Pharm. Res. 17 (12), 1503–1510. 10.1023/a:1007657109012 [DOI] [PubMed] [Google Scholar]
- Duan N., Gong W., Wu S., Wang Z. (2017). An ssDNA Library Immobilized SELEX Technique for Selection of an Aptamer against Ractopamine. Analytica Chim. Acta 961, 100–105. 10.1016/j.aca.2017.01.008 [DOI] [PubMed] [Google Scholar]
- Duan N., Wu S., Chen X., Huang Y., Wang Z. (2012). Selection and Identification of a DNA Aptamer Targeted to Vibrio Parahemolyticus. J. Agric. Food Chem. 60 (16), 4034–4038. 10.1021/jf300395z [DOI] [PubMed] [Google Scholar]
- Duan N., Wu S., Chen X., Huang Y., Xia Y., Ma X., et al. (2013). Selection and Characterization of Aptamers against Salmonella typhimurium Using Whole-Bacterium Systemic Evolution of Ligands by Exponential Enrichment (SELEX). J. Agric. Food Chem. 61 (13), 3229–3234. 10.1021/jf400767d [DOI] [PubMed] [Google Scholar]
- Duan Y., Gao Z., Wang L., Wang H., Zhang H., Li H. (2016). Selection and Identification of Chloramphenicol-specific DNA Aptamers by Mag-SELEX. Appl. Biochem. Biotechnol. 180 (8), 1644–1656. 10.1007/s12010-016-2193-6 [DOI] [PubMed] [Google Scholar]
- Dunning J., Baillie J. K., Cao B., Hayden F. G., I I. S. A. R. E. (2014a). Antiviral Combinations for Severe Influenza. Lancet Infect. Dis. 14 (12), 1259–1270. 10.1016/S1473-3099(14)70821-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning J., Baillie J. K., Cao B., Hayden F. G., International Severe Acute R., Emerging Infection C. (2014b). Antiviral Combinations for Severe Influenza. Lancet Infect. Dis. 14 (12), 1259–1270. 10.1016/S1473-3099(14)70821-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi H. P., Smiley R. D., Jaykus L.-A. (2013). Selection of DNA Aptamers for Capture and Detection of Salmonella Typhimurium Using a Whole-Cell SELEX Approach in Conjunction with Cell Sorting. Appl. Microbiol. Biotechnol. 97 (8), 3677–3686. 10.1007/s00253-013-4766-4 [DOI] [PubMed] [Google Scholar]
- Ellington A. D., Szostak J. W. (1990). In Vitro selection of RNA Molecules that Bind Specific Ligands. Nature 346 (6287), 818–822. 10.1038/346818a0 [DOI] [PubMed] [Google Scholar]
- Escadafal C., Faye O., Sall A. A., Faye O., Weidmann M., Strohmeier O., et al. (2014). Rapid Molecular Assays for the Detection of Yellow Fever Virus in Low-Resource Settings. Plos Negl. Trop. Dis. 8 (3), e2730. 10.1371/journal.pntd.0002730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fall-Malick F.-Z., Tchiakpé E., Ould Soufiane S. A., Diop-Ndiaye H., Mouhamedoune Baye A., Ould Horma Babana A., et al. (2014). Drug Resistance Mutations and Genetic Diversity in Adults Treated for HIV Type 1 Infection in Mauritania. J. Med. Virol. 86 (3), 404–410. 10.1002/jmv.23860 [DOI] [PubMed] [Google Scholar]
- Fan L., Wang T., Hua C., Sun W., Li X., Grunwald L., et al. (2020). A Compendium of DNA-Binding Specificities of Transcription Factors in Pseudomonas syringae . Nat. Commun. 11 (1), 4947. 10.1038/s41467-020-18744-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattal E., Hillaireau H., Ismail S. I. (2018). Aptamers in Therapeutics and Drug Delivery. Adv. Drug Deliv. Rev. 134, 1–2. 10.1016/j.addr.2018.11.001 [DOI] [PubMed] [Google Scholar]
- Fauci A. S., Touchette N. A., Folkers G. K. (2005). Emerging Infectious Diseases: a 10-year Perspective from the National Institute of Allergy and Infectious Diseases. Emerg. Infect. Dis. 11 (4), 519–525. 10.3201/eid1104.041167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Férir G., Gordts S., Schols D. (2014). HIV-1 and its Resistance to Peptidic Carbohydrate-Binding Agents (CBAs): an Overview. Molecules 19 (12), 21085–21112. 10.3390/molecules191221085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay B. B., McFadden G. (2006). Anti-immunology: Evasion of the Host Immune System by Bacterial and Viral Pathogens. Cell 124 (4), 767–782. 10.1016/j.cell.2006.01.034 [DOI] [PubMed] [Google Scholar]
- Fisher M. E., Rossini M., Simmons E., Harris R. C., Moeckel G., Zent R. (2004). A Woman with Chronic Hepatitis C Infection and Nephrotic Syndrome Who Developed Multiple Renal Lesions after Interferon Alfa Therapy. Am. J. Kidney Dis. 44 (3), 567–573. 10.1016/s0272-6386(04)00828-5 [DOI] [PubMed] [Google Scholar]
- Fisher R. I., Jaffe E. S., Braylan R. C., Andersen J. C., Tan H. K. (1976). Immunoblastic Lymphadenopathy. Am. J. Med. 61 (4), 553–559. 10.1016/0002-9343(76)90337-5 [DOI] [PubMed] [Google Scholar]
- Frezza V., Pinto-Díez C., Fernández G., Soto M., Martín M. E., García-Sacristán A., et al. (2020). DNA Aptamers Targeting Leishmania Infantum H3 Protein as Potential Diagnostic Tools. Analytica Chim. Acta 1107, 155–163. 10.1016/j.aca.2020.02.012 [DOI] [PubMed] [Google Scholar]
- Fried M. W. (2002). Side Effects of Therapy of Hepatitis C and Their Management. Hepatology 36 (5 Suppl. 1), S237–s244. 10.1053/jhep.2002.3681010.1002/hep.1840360730 [DOI] [PubMed] [Google Scholar]
- Fu P., Sun Z., Yu Z., Zhang Y., Shen J., Zhang H., et al. (2014). Enzyme Linked Aptamer Assay: Based on a Competition Format for Sensitive Detection of Antibodies to Mycoplasma Bovis in Serum. Anal. Chem. 86 (3), 1701–1709. 10.1021/ac4042203 [DOI] [PubMed] [Google Scholar]
- Fu P., Wang F., Zhang Y., Qiao X., Zhang Y., Zhou W., et al. (2021a). The Application of Aptamer Apt-236 Targeting PvpA Protein in the Detection of Antibodies against Mycoplasma Gallisepticum. Anal. Methods 13 (27), 3068–3076. 10.1039/d1ay00515d [DOI] [PubMed] [Google Scholar]
- Fu P., Wang F., Zhang Y., Qiao X., Zhang Y., Zhou W., et al. (2021b). The Application of Aptamer Apt-236 Targeting PvpA Protein in the Detection of Antibodies against Mycoplasma Gallisepticum. Anal. Methods 13, 3068–3076. 10.1039/d1ay00515d [DOI] [PubMed] [Google Scholar]
- Gao T., Ding P., Li W., Wang Z., Lin Q., Pei R. (2020). Isolation of DNA Aptamers Targeting N-Cadherin and High-Efficiency Capture of Circulating Tumor Cells by Using Dual Aptamers. Nanoscale 12 (44), 22574–22585. 10.1039/d0nr06180h [DOI] [PubMed] [Google Scholar]
- Geary R. S., Yu R. Z., Watanabe T., Henry S. P., Hardee G. E., Chappell A., et al. (2003). Pharmacokinetics of a Tumor Necrosis Factor-Alpha Phosphorothioate 2’-O-(2-Methoxyethyl) Modified Antisense Oligonucleotide: Comparison across Species. Drug Metab. Dispos 31 (11), 1419–1428. 10.1124/dmd.31.11.1419 [DOI] [PubMed] [Google Scholar]
- Giri B. R., Mahato R. I., Cheng G. (2019). Roles of microRNAs in T Cell Immunity: Implications for Strategy Development against Infectious Diseases. Med. Res. Rev. 39 (2), 706–732. 10.1002/med.21539 [DOI] [PubMed] [Google Scholar]
- Gopinath S. C. B., Hayashi K., Kumar P. K. R. (2012). Aptamer that Binds to the gD Protein of Herpes Simplex Virus 1 and Efficiently Inhibits Viral Entry. J. Virol. 86 (12), 6732–6744. 10.1128/JVI.00377-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gull I., Aslam M. S., Tipu I., Mushtaq R., Ali T. Z., Athar M. A. (2019). Development of Latent Interferon Alpha 2b as a Safe Therapeutic for Treatment of Hepatitis C Virus Infection. Sci. Rep. 9 (1), 10867. 10.1038/s41598-019-47074-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P. (2010). The Emerging Field of RNA Nanotechnology. Nat. Nanotech 5 (12), 833–842. 10.1038/nnano.2010.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Zhao C., Liu Y., Nie H., Guo X., Song X., et al. (2020). A Novel Fluorescence Method for the Rapid and Effective Detection of Listeria Monocytogenes Using Aptamer-Conjugated Magnetic Nanoparticles and Aggregation-Induced Emission Dots. Analyst 145 (11), 3857–3863. 10.1039/d0an00397b [DOI] [PubMed] [Google Scholar]
- Habyarimana F., Ramroop S. (2020). Prevalence and Risk Factors Associated with Malaria Among Children Aged Six Months to 14 Years Old in Rwanda: Evidence from 2017 Rwanda Malaria Indicator Survey. Int. J. Environ. Res. Public Health 17 (21), 7975. 10.3390/ijerph17217975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn U. (2017). Charomers-Interleukin-6 Receptor Specific Aptamers for Cellular Internalization and Targeted Drug Delivery. Int. J. Mol. Sci. 18 (12), 2641. 10.3390/ijms18122641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B., Fang C., Sha L., Jalalah M., Al-Assiri M. S., Harraz F. A., et al. (2021). Cascade Strand Displacement Reaction-Assisted Aptamer-Based Highly Sensitive Detection of Ochratoxin A. Food Chem. 338, 127827. 10.1016/j.foodchem.2020.127827 [DOI] [PubMed] [Google Scholar]
- Han X., Zhang Y., Nie J., Zhao S., Tian Y., Zhou N. (2017). Gold Nanoparticle Based Photometric Determination of Tobramycin by Using New Specific DNA Aptamers. Microchim Acta 185 (1), 4. 10.1007/s00604-017-2568-6 [DOI] [PubMed] [Google Scholar]
- He J., Wang J., Zhang N., Shen L., Wang L., Xiao X., et al. (2019). In Vitro selection of DNA Aptamers Recognizing Drug-Resistant Ovarian Cancer by Cell-SELEX. Talanta 194, 437–445. 10.1016/j.talanta.2018.10.028 [DOI] [PubMed] [Google Scholar]
- Heo K., Min S.-W., Sung H. J., Kim H. G., Kim H. J., Kim Y. H., et al. (2016). An Aptamer-Antibody Complex (Oligobody) as a Novel Delivery Platform for Targeted Cancer Therapies. J. Controlled Release 229, 1–9. 10.1016/j.jconrel.2016.03.006 [DOI] [PubMed] [Google Scholar]
- Homann M., Lorger M., Engstler M., Zacharias M., Göringer H. U. (2006). Serum-stable RNA Aptamers to an Invariant Surface Domain of Live African Trypanosomes. Comb. Chem. High Throughput Screen. 9 (7), 491–499. 10.2174/138620706777935324 [DOI] [PubMed] [Google Scholar]
- Hong K. L., Sooter L. J. (2015). Single-Stranded DNA Aptamers against Pathogens and Toxins: Identification and Biosensing Applications. Biomed. Res. Int. 2015, 1–31. 10.1155/2015/419318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.-L., Xiang M.-Q., Tang M., Pang D.-W., Zhang Z.-L. (2019). Ebola Virus Aptamers: From Highly Efficient Selection to Application on Magnetism-Controlled Chips. Anal. Chem. 91 (5), 3367–3373. 10.1021/acs.analchem.8b04623 [DOI] [PubMed] [Google Scholar]
- Hünniger T., Wessels H., Fischer C., Paschke-Kratzin A., Fischer M. (2014). Just in Time-Selection: A Rapid Semiautomated SELEX of DNA Aptamers Using Magnetic Separation and BEAMing. Anal. Chem. 86 (21), 10940–10947. 10.1021/ac503261b [DOI] [PubMed] [Google Scholar]
- Hwang S.-Y., Sun H.-Y., Lee K.-H., Oh B.-H., Cha Y. J., Kim B. H., et al. (2012). 5′-Triphosphate-RNA-independent Activation of RIG-I via RNA Aptamer with Enhanced Antiviral Activity. Nucleic Acids Res. 40 (6), 2724–2733. 10.1093/nar/gkr1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida R., Adachi T., Yokota A., Yoshihara H., Aoki K., Nakamura Y., et al. (2020). RaptRanker: In Silico RNA Aptamer Selection from HT-SELEX experiment Based on Local Sequence and Structure Information. Nucleic Acids Res. 48 (14), e82. 10.1093/nar/gkaa484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasena S. D. (1999). Aptamers: an Emerging Class of Molecules that Rival Antibodies in Diagnostics. Clin. Chem. 45 (9), 1628–1650. 10.1093/clinchem/45.9.1628 [DOI] [PubMed] [Google Scholar]
- Jing M., Bowser M. T. (2013). Tracking the Emergence of High Affinity Aptamers for rhVEGF165 during Capillary Electrophoresis-Systematic Evolution of Ligands by Exponential Enrichment Using High Throughput Sequencing. Anal. Chem. 85 (22), 10761–10770. 10.1021/ac401875h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Yeom G., Jang H., Oh J., Park C.-J., Kim M.-G. (2019). Development of Replication Protein A-Conjugated Gold Nanoparticles for Highly Sensitive Detection of Disease Biomarkers. Anal. Chem. 91 (15), 10001–10007. 10.1021/acs.analchem.9b01827 [DOI] [PubMed] [Google Scholar]
- Kim Y. S., Song M. Y., Jurng J., Kim B. C. (2013). Isolation and Characterization of DNA Aptamers against Escherichia coli Using a Bacterial Cell-Systematic Evolution of Ligands by Exponential Enrichment Approach. Anal. Biochem. 436 (1), 22–28. 10.1016/j.ab.2013.01.014 [DOI] [PubMed] [Google Scholar]
- Kinghorn A., Fraser L., Liang S., Shiu S., Tanner J. (2017). Aptamer Bioinformatics. Int. J. Mol. Sci. 18 (12), 2516. 10.3390/ijms18122516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labella A. M., Merel S. E. (2013). Influenza. Med. Clin. North America 97 (4), 621–645, x. 10.1016/j.mcna.2013.03.001 [DOI] [PubMed] [Google Scholar]
- Lai H.-C., Wang C.-H., Liou T.-M., Lee G.-B. (2014). Influenza A Virus-specific Aptamers Screened by Using an Integrated Microfluidic System. Lab. Chip 14 (12), 2002–2013. 10.1039/c4lc00187g [DOI] [PubMed] [Google Scholar]
- Lai J.-C., Hong C.-Y. (2014). A Novel Protocol for Generating High-Affinity ssDNA Aptamers by Using Alternating Magnetic fields. J. Mater. Chem. B 2 (26), 4114–4121. 10.1039/c3tb21729a [DOI] [PubMed] [Google Scholar]
- Lakhin A. V., Tarantul V. Z., Gening L. V. (2013). Aptamers: Problems, Solutions and Prospects. Acta Naturae 5 (4), 34–43. 10.32607/20758251-2013-5-4-34-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampel K. A., Orlandi P. A., Kornegay L. (2000). Improved Template Preparation for PCR-Based Assays for Detection of Food-Borne Bacterial Pathogens. Appl. Environ. Microbiol. 66 (10), 4539–4542. 10.1128/AEM.66.10.4539-4542.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauridsen L. H., Doessing H. B., Long K. S., Nielsen A. T. (2018). A Capture-SELEX Strategy for Multiplexed Selection of RNA Aptamers against Small Molecules. Methods Mol. Biol. 1671, 291–306. 10.1007/978-1-4939-7295-1_18 [DOI] [PubMed] [Google Scholar]
- Lazarevic I. (2014). Clinical Implications of Hepatitis B Virus Mutations: Recent Advances. World J. Gastroenterol. 20 (24), 7653–7664. 10.3748/wjg.v20.i24.7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblebici P., Leirs K., Spasic D., Lammertyn J. (2019). Encoded Particle Microfluidic Platform for Rapid Multiplexed Screening and Characterization of Aptamers against Influenza A Nucleoprotein. Analytica Chim. Acta 1053, 70–80. 10.1016/j.aca.2018.11.055 [DOI] [PubMed] [Google Scholar]
- Lee C. H., Lee S. H., Kim J. H., Noh Y. H., Noh G. J., Lee S. W. (2015). Pharmacokinetics of a Cholesterol-Conjugated Aptamer against the Hepatitis C Virus (HCV) NS5B Protein. Mol. Therapy-Nucleic Acids 4, e254. 10.1038/mtna.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Song K.-M., Jeon W., Jo H., Shim Y.-B., Ban C. (2012). A Highly Sensitive Aptasensor towards Plasmodium Lactate Dehydrogenase for the Diagnosis of Malaria. Biosens. Bioelectron. 35 (1), 291–296. 10.1016/j.bios.2012.03.003 [DOI] [PubMed] [Google Scholar]
- Li G., Zhou L., Zhang C., Shi Y., Dong D., Bai M., et al. (2019). Insulin-Like Growth Factor 1 Regulates Acute Inflammatory Lung Injury Mediated by Influenza Virus Infection. Front. Microbiol. 10, 2541. 10.3389/fmicb.2019.02541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. H., Patei D. J. (1997). Structural Basis of DNA Folding and Recognition in an AMP-DNA Aptamer Complex: Distinct Architectures but Common Recognition Motifs for DNA and RNA Aptamers Complexed to AMP. Chem. Biol. 4 (11), 817–832. 10.1016/s1074-5521(97)90115-0 [DOI] [PubMed] [Google Scholar]
- Lin H.-I., Wu C.-C., Yang C.-H., Chang K.-W., Lee G.-B., Shiesh S.-C. (2015). Selection of Aptamers Specific for Glycated Hemoglobin and Total Hemoglobin Using On-Chip SELEX. Lab. Chip 15 (2), 486–494. 10.1039/c4lc01124d [DOI] [PubMed] [Google Scholar]
- Lin N., Wu L., Xu X., Wu Q., Wang Y., Shen H., et al. (2021). Aptamer Generated by Cell-SELEX for Specific Targeting of Human Glioma Cells. ACS Appl. Mater. Inter. 13 (8), 9306–9315. 10.1021/acsami.0c11878 [DOI] [PubMed] [Google Scholar]
- Lincoff A. M., Mehran R., Povsic T. J., Zelenkofske S. L., Huang Z., Armstrong P. W., et al. (2016). Effect of the REG1 Anticoagulation System versus Bivalirudin on Outcomes after Percutaneous Coronary Intervention (REGULATE-PCI): a Randomised Clinical Trial. The Lancet 387 (10016), 349–356. 10.1016/S0140-6736(15)00515-2 [DOI] [PubMed] [Google Scholar]
- Liu M., Li X., Li B., Du J., Yang Z. (2019a). A Fluorometric Aptamer-Based Assay for Ochratoxin A by Using Exonuclease III-Assisted Recycling Amplification. Microchim Acta 187 (1), 46. 10.1007/s00604-019-3992-6 [DOI] [PubMed] [Google Scholar]
- Liu R., He L., Hu Y., Luo Z., Zhang J. (2020). A Serological Aptamer-Assisted Proximity Ligation Assay for COVID-19 Diagnosis and Seeking Neutralizing Aptamers. Chem. Sci. 11 (44), 12157–12164. 10.1039/d0sc03920a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. J., Zhao M., Liu K., Xu K., Wong G., Tan W., et al. (2017). T-cell Immunity of SARS-CoV: Implications for Vaccine Development against MERS-CoV. Antiviral Res. 137, 82–92. 10.1016/j.antiviral.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wang Y. l., Wu J., Qi J., Zeng Z., Wan Q., et al. (2021). Neutralizing Aptamers Block S/RBD‐ACE2 Interactions and Prevent Host Cell Infection. Angew. Chem. 133 (18), 10361–10366. 10.1002/ange.202100345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Jiang W., Yang S., Hu J., Lu H., Han W., et al. (2019b). Rapid Detection of Mycoplasma-Infected Cells by an ssDNA Aptamer Probe. ACS Sens. 4 (8), 2028–2038. 10.1021/acssensors.9b00582 [DOI] [PubMed] [Google Scholar]
- Lou X., Qian J., Xiao Y., Viel L., Gerdon A. E., Lagally E. T., et al. (2009). Micromagnetic Selection of Aptamers in Microfluidic Channels. Proc. Natl. Acad. Sci. 106 (9), 2989–2994. 10.1073/pnas.0813135106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycett S. J., Pohlmann A., Staubach C., Caliendo V., Woolhouse M., Beer M., et al. (2020). Genesis and Spread of Multiple Reassortants during the 2016/2017 H5 Avian Influenza Epidemic in Eurasia. Proc. Natl. Acad. Sci. USA 117 (34), 20814–20825. 10.1073/pnas.2001813117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Wang W., Chen X., Xia Y., Wu S., Duan N., et al. (2014). Selection, Identification, and Application of Aflatoxin B1 Aptamer. Eur. Food Res. Technol. 238 (6), 919–925. 10.1007/s00217-014-2176-1 [DOI] [Google Scholar]
- Ma X., Xu H., Shi L., Yang P., Zhang L., Sun X., et al. (2015). A Multiplex PCR Assay for the Detection of Five Influenza Viruses Using a Dual Priming Oligonucleotide System. BMC Infect. Dis. 15, 93. 10.1186/s12879-015-0818-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Li X., Li W., Liu Z. (2018). Glycan-Imprinted Magnetic Nanoparticle-Based SELEX for Efficient Screening of Glycoprotein-Binding Aptamers. ACS Appl. Mater. Inter. 10 (47), 40918–40926. 10.1021/acsami.8b14441 [DOI] [PubMed] [Google Scholar]
- Ma Y., Ma J. (2020). Immunotherapy against Prion Disease. Pathogens 9 (3), 216. 10.3390/pathogens9030216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manns M. P., McHutchison J. G., Gordon S. C., Rustgi V. K., Shiffman M., Reindollar R., et al. (2001). Peginterferon Alfa-2b Plus Ribavirin Compared with Interferon Alfa-2b Plus Ribavirin for Initial Treatment of Chronic Hepatitis C: a Randomised Trial. The Lancet 358 (9286), 958–965. 10.1016/s0140-6736(01)06102-5 [DOI] [PubMed] [Google Scholar]
- Marascio N., Torti C., Liberto M., Focà A. (2014). Update on Different Aspects of HCV Variability: Focus on NS5B Polymerase. BMC Infect. Dis. 14 (Suppl. 5), S1. 10.1186/1471-2334-14-S5-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeague M., Bradley C. R., Girolamo A. D., Visconti A., Miller J. D., Derosa M. C. (2010). Screening and Initial Binding Assessment of Fumonisin B1 Aptamers. Int. J. Mol. Sci. 11 (12), 4864–4881. 10.3390/ijms11124864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonsa S. D., Bowser M. T. (2004a). In Vitro evolution of Functional DNA Using Capillary Electrophoresis. J. Am. Chem. Soc. 126 (1), 20–21. 10.1021/ja037832s [DOI] [PubMed] [Google Scholar]
- Mendonsa S. D., Bowser M. T. (2004b). In Vitro selection of High-Affinity DNA Ligands for Human IgE Using Capillary Electrophoresis. Anal. Chem. 76 (18), 5387–5392. 10.1021/ac049857v [DOI] [PubMed] [Google Scholar]
- Mikhail A. F., Leslie T. J., Mayan M. I., Zekria R., Mohammad N., Hasanzai M. A., et al. (2011). Field Trial of Three Different Plasmodium Vivax- Detecting Rapid Diagnostic Tests with and without Evaporative Cool Box Storage in Afghanistan. Malar. J. 10, 169. 10.1186/1475-2875-10-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minopoli A., Della Ventura B., Lenyk B., Gentile F., Tanner J. A., Offenhäusser A., et al. (2020). Ultrasensitive Antibody-Aptamer Plasmonic Biosensor for Malaria Biomarker Detection in Whole Blood. Nat. Commun. 11 (1), 6134. 10.1038/s41467-020-19755-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y., Kamal M., Kang S.-A., Zhang R., Lokesh G. L., Thiviyanathan V., et al. (2016). E-selectin Targeting PEGylated-Thioaptamer Prevents Breast Cancer Metastases. Mol. Ther. - Nucleic Acids 5 (12), e399. 10.1038/mtna.2016.103 [DOI] [PubMed] [Google Scholar]
- Morita Y., Leslie M., Kameyama H., Volk D., Tanaka T. (2018). Aptamer Therapeutics in Cancer: Current and Future. Cancers 10 (3), 80. 10.3390/cancers10030080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosing R. K., Bowser M. T. (2009). Isolating Aptamers Using Capillary Electrophoresis-SELEX (CE-SELEX). Methods Mol. Biol. 535, 33–43. 10.1007/978-1-59745-557-2_3 [DOI] [PubMed] [Google Scholar]
- Mosing R. K., Mendonsa S. D., Bowser M. T. (2005). Capillary Electrophoresis-SELEX Selection of Aptamers with Affinity for HIV-1 Reverse Transcriptase. Anal. Chem. 77 (19), 6107–6112. 10.1021/ac050836q [DOI] [PubMed] [Google Scholar]
- Mufhandu H. T., Gray E. S., Madiga M. C., Tumba N., Alexandre K. B., Khoza T., et al. (2012). UCLA1, a Synthetic Derivative of a Gp120 RNA Aptamer, Inhibits Entry of Human Immunodeficiency Virus Type 1 Subtype C. J. Virol. 86 (9), 4989–4999. 10.1128/Jvi.06893-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkatti R., Bist V., Sun S., Fortes de Araujo F., Nakhasi H. L., Debrabant A. (2012). Development of an Aptamer-Based Concentration Method for the Detection of Trypanosoma Cruzi in Blood. PLoS One 7 (8), e43533. 10.1371/journal.pone.0043533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nang S. C., Azad M. A. K., Velkov T., Zhou Q., Li J. (2021). Rescuing the Last-Line Polymyxins: Achievements and Challenges. Pharmacol. Rev. 73 (2), 679–728. 10.1124/pharmrev.120.000020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negro F. (2010). Adverse Effects of Drugs in the Treatment of Viral Hepatitis. Best Pract. Res. Clin. Gastroenterol. 24 (2), 183–192. 10.1016/j.bpg.2009.10.012 [DOI] [PubMed] [Google Scholar]
- Nguyen P. D. M., Zheng J., Gremminger T. J., Qiu L., Zhang D., Tuske S., et al. (2020). Binding Interface and Impact on Protease Cleavage for an RNA Aptamer to HIV-1 Reverse Transcriptase. Nucleic Acids Res. 48 (5), 2709–2722. 10.1093/nar/gkz1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimjee S. M., Rusconi C. P., Sullenger B. A. (2005). Aptamers: an Emerging Class of Therapeutics. Annu. Rev. Med. 56, 555–583. 10.1146/annurev.med.56.062904.144915 [DOI] [PubMed] [Google Scholar]
- Nishimura S., Miura H., Yamada H., Shinoda T., Kitamura S., Miura Y. (2002). Acute Onset of Nephrotic Syndrome during Interferon-α Retreatment for Chronic Active Hepatitis C. J. Gastroenterol. 37 (10), 854–858. 10.1007/s005350200141 [DOI] [PubMed] [Google Scholar]
- Nitta K. R., Vincentelli R., Jacox E., Cimino A., Ohtsuka Y., Sobral D., et al. (2019). High-Throughput Protein Production Combined with High- Throughput SELEX Identifies an Extensive Atlas of Ciona Robusta Transcription Factor DNA-Binding Specificities. Methods Mol. Biol. 2025, 487–517. 10.1007/978-1-4939-9624-7_23 [DOI] [PubMed] [Google Scholar]
- Oh S. S., Qian J., Lou X., Zhang Y., Xiao Y., Soh H. T. (2009). Generation of Highly Specific Aptamers via Micromagnetic Selection. Anal. Chem. 81 (13), 5490–5495. 10.1021/ac900759k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. S., Ahmad K. M., Cho M., Kim S., Xiao Y., Soh H. T. (2011). Improving Aptamer Selection Efficiency through Volume Dilution, Magnetic Concentration, and Continuous Washing in Microfluidic Channels. Anal. Chem. 83 (17), 6883–6889. 10.1021/ac201269f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ospina J. D. (2020). Los aptámeros como novedosa herramienta diagnóstica y terapéutica y su potencial uso en parasitología. Biomedica 40 (Suppl. 1), 148–165. 10.7705/biomedica.4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ospina-Villa J., López-Camarillo C., Castañón-Sánchez C., Soto-Sánchez J., Ramírez-Moreno E., Marchat L. (2018). Advances on Aptamers against Protozoan Parasites. Genes 9 (12), 584. 10.3390/genes9120584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Yang Y., Li L., Li X., Li Q., Cui C., et al. (2020). A Bispecific Circular Aptamer Tethering a Built-In Universal Molecular Tag for Functional Protein Delivery. Chem. Sci. 11 (35), 9648–9654. 10.1039/d0sc02279a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang K. M., Castanotto D., Li H., Scherer L., Rossi J. J. (2018). Incorporation of Aptamers in the Terminal Loop of shRNAs Yields an Effective and Novel Combinatorial Targeting Strategy. Nucleic Acids Res. 46 (1), e6. 10.1093/nar/gkx980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniel N., Istamboulié G., Triki A., Lozano C., Barthelmebs L., Noguer T. (2017). Selection of DNA Aptamers against Penicillin G Using Capture-SELEX for the Development of an Impedimetric Sensor. Talanta 162, 232–240. 10.1016/j.talanta.2016.09.058 [DOI] [PubMed] [Google Scholar]
- Pazienza V. (2011). Ophthalmological Complications in Hepatitis C Virus Infection: Side Effect of Interferon Therapy or a Direct Role of HCV?. Biomed. Pharmacother. 65 (4), 317–318. 10.1016/j.biopha.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Perra N. (2021). Non-pharmaceutical Interventions during the COVID-19 Pandemic: A Review. Phys. Rep. 913, 1–52. 10.1016/j.physrep.2021.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qaanei M., Taheri R. A., Eskandari K. (2021). Electrochemical Aptasensor for Escherichia coli O157:H7 Bacteria Detection Using a Nanocomposite of Reduced Graphene Oxide, Gold Nanoparticles and Polyvinyl Alcohol. Anal. Methods 13 (27), 3101–3109. 10.1039/d1ay00563d [DOI] [PubMed] [Google Scholar]
- Rauch N., Nauen R. (2003). Identification of Biochemical Markers Linked to Neonicotinoid Cross Resistance inBemisia Tabaci (Hemiptera: Aleyrodidae). Arch. Insect Biochem. Physiol. 54 (4), 165–176. 10.1002/arch.10114 [DOI] [PubMed] [Google Scholar]
- Reiss D. J., Howard F. M., Mobley H. L. T. (2012). A Novel Approach for Transcription Factor Analysis Using SELEX with High-Throughput Sequencing (TFAST). PLoS One 7 (8), e42761. 10.1371/journal.pone.0042761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr U.-P., Binder C., Dieterle T., Giusti F., Messina C. G. M., Toerien E., et al. (2016). The Value of In Vitro Diagnostic Testing in Medical Practice: A Status Report. PLoS One 11 (3), e0149856. 10.1371/journal.pone.0149856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-López C., Berzal-Herranz B., Gómez J., Berzal-Herranz A. (2012). An Engineered Inhibitor RNA that Efficiently Interferes with Hepatitis C Virus Translation and Replication. Antiviral Res. 94 (2), 131–138. 10.1016/j.antiviral.2012.02.015 [DOI] [PubMed] [Google Scholar]
- Rosenbaum C. D., Carreiro S. P., Babu K. M. (2012). Here Today, Gone Tomorrow…and Back Again? A Review of Herbal Marijuana Alternatives (K2, Spice), Synthetic Cathinones (Bath Salts), Kratom, Salvia Divinorum, Methoxetamine, and Piperazines. J. Med. Toxicol. 8 (1), 15–32. 10.1007/s13181-011-0202-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulet E., Busso S., Camargo A. A., Simpson A. J. G., Mermod N., Bucher P. (2002). High-throughput SELEX-SAGE Method for Quantitative Modeling of Transcription-Factor Binding Sites. Nat. Biotechnol. 20 (8), 831–835. 10.1038/nbt718 [DOI] [PubMed] [Google Scholar]
- Ruan S., Swamidass S. J., Stormo G. D. (2017). BEESEM: Estimation of Binding Energy Models Using HT-SELEX Data. Bioinformatics 33 (15), 2288–2295. 10.1093/bioinformatics/btx191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff P., Pai R. B., Storici F. (2012). Real-Time PCR-Coupled CE-SELEX for DNA Aptamer Selection. ISRN Mol. Biol. 2012, 1–9. 10.5402/2012/939083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi C. P., Roberts J. D., Pitoc G. A., Nimjee S. M., White R. R., Quick G., Jr., et al. (2004). Antidote-mediated Control of an Anticoagulant Aptamer In Vivo . Nat. Biotechnol. 22 (11), 1423–1428. 10.1038/nbt1023 [DOI] [PubMed] [Google Scholar]
- Saad M., Chinerman D., Tabrizian M., Faucher S. P. (2020). Identification of Two Aptamers Binding to Legionella pneumophila with High Affinity and Specificity. Sci. Rep. 10 (1), 9145. 10.1038/s41598-020-65973-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu G. K. (2015). Potential Implication of Residual Viremia in Patients on Effective Antiretroviral Therapy. AIDS Res. Hum. Retroviruses 31 (1), 25–35. 10.1089/AID.2014.0194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarhan M., El-Bitar A. M. H., Hotta H. (2020). Potent Virucidal Activity of Honeybee "Apis mellifera" Venom against Hepatitis C Virus. Toxicon 188, 55–64. 10.1016/j.toxicon.2020.10.014 [DOI] [PubMed] [Google Scholar]
- Schmitz A., Weber A., Bayin M., Breuers S., Fieberg V., Famulok M., et al. (2021). A SARS‐CoV‐2 Spike Binding DNA Aptamer that Inhibits Pseudovirus Infection by an RBD‐Independent Mechanism**. Angew. Chem. Int. Ed. 60 (18), 10279–10285. 10.1002/anie.202100316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiang Y.-C., Ou C.-M., Chen S.-J., Ou T.-Y., Lin H.-J., Huang C.-C., et al. (2013). Highly Efficient Inhibition of Human Immunodeficiency Virus Type 1 Reverse Transcriptase by Aptamers Functionalized Gold Nanoparticles. Nanoscale 5 (7), 2756–2764. 10.1039/c3nr33403a [DOI] [PubMed] [Google Scholar]
- Shim B. S. (2011). Current Concepts in Bacterial Sexually Transmitted Diseases. Korean J. Urol. 52 (9), 589–597. 10.4111/kju.2011.52.9.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji Y., Shimada J., Mizushima Y. (2002). Drug Delivery System to Control Infectious Diseases. Curr. Pharm. Des. 8 (6), 455–465. 10.2174/1381612023395934 [DOI] [PubMed] [Google Scholar]
- Singh N. K., Jain P., Das S., Goswami P. (2019a). Dye Coupled Aptamer-Captured Enzyme Catalyzed Reaction for Detection of Pan Malaria andP. falciparumSpecies in Laboratory Settings and Instrument-free Paper-Based Platform. Anal. Chem. 91 (6), 4213–4221. 10.1021/acs.analchem.9b00670 [DOI] [PubMed] [Google Scholar]
- Singh N. K., Thungon P. D., Estrela P., Goswami P. (2019b). Development of an Aptamer-Based Field Effect Transistor Biosensor for Quantitative Detection of Plasmodium Falciparum Glutamate Dehydrogenase in Serum Samples. Biosens. Bioelectron. 123, 30–35. 10.1016/j.bios.2018.09.085 [DOI] [PubMed] [Google Scholar]
- Sola M., Menon A. P., Moreno B., Meraviglia-Crivelli D., Soldevilla M. M., Cartón-García F., et al. (2020). Aptamers against Live Targets: Is In Vivo SELEX Finally Coming to the Edge?. Mol. Ther. - Nucleic Acids 21, 192–204. 10.1016/j.omtn.2020.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Wang X., Xu K., Li Q., Ning L., Yang X. (2019). Selection of Highly Specific Aptamers to Vibrio Parahaemolyticus Using Cell-SELEX Powered by Functionalized Graphene Oxide and Rolling circle Amplification. Analytica Chim. Acta 1052, 153–162. 10.1016/j.aca.2018.11.047 [DOI] [PubMed] [Google Scholar]
- Spiga F. M., Maietta P., Guiducci C. (2015). More DNA-Aptamers for Small Drugs: A Capture-SELEX Coupled with Surface Plasmon Resonance and High-Throughput Sequencing. ACS Comb. Sci. 17 (5), 326–333. 10.1021/acscombsci.5b00023 [DOI] [PubMed] [Google Scholar]
- Stoltenburg R., Nikolaus N., Strehlitz B. (2012). Capture-SELEX: Selection of DNA Aptamers for Aminoglycoside Antibiotics. J. Anal. Methods Chem. 2012, 1–14. 10.1155/2012/415697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumari D., Grimberg B. T., Blankenship D. A., Mugasa J., Mugittu K., Moore L., et al. (2016). Application of Magnetic Cytosmear for the Estimation of Plasmodium Falciparum Gametocyte Density and Detection of Asexual Stages in Asymptomatic Children. Malar. J. 15, 113. 10.1186/s12936-016-1170-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi N., Segawa S., Ishiwada N., Ohkusu M., Tsuchida S., Satoh M., et al. (2018). Capsular Serotyping of Haemophilus Influenzae by Using Matrix-Associated Laser Desorption Ionization-Time of Flight Mass Spectrometry. J. Infect. Chemother. 24 (7), 510–514. 10.1016/j.jiac.2018.02.007 [DOI] [PubMed] [Google Scholar]
- Tan X., Jia F., Wang P., Zhang K. (2020a). Nucleic Acid-Based Drug Delivery Strategies. J. Controlled Release 323, 240–252. 10.1016/j.jconrel.2020.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z. M., Lai G. P., Pandey M., Srichana T., Pichika M. R., Gorain B., et al. (2020b). Novel Approaches for the Treatment of Pulmonary Tuberculosis. Pharmaceutics 12 (12), 1196. 10.3390/pharmaceutics12121196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongdee P., Chaijaroenkul W., Kuesap J., Na-Bangchang K. (2014). Nested-PCR and a New ELISA-Based NovaLisa Test Kit for Malaria Diagnosis in an Endemic Area of Thailand. Korean J. Parasitol. 52 (4), 377–381. 10.3347/kjp.2014.52.4.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabi R., Ranjbar R., Halaji M., Heiat M. (2020). Aptamers, the Bivalent Agents as Probes and Therapies for Coronavirus Infections: A Systematic Review. Mol. Cell Probes 53, 101636. 10.1016/j.mcp.2020.101636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gold L. (1990). Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 249 (4968), 505–510. 10.1126/science.2200121 [DOI] [PubMed] [Google Scholar]
- Ucak S., Sudagidan M., Borsa B. A., Mansuroglu B., Ozalp V. C. (2020). Inhibitory Effects of Aptamer Targeted Teicoplanin Encapsulated PLGA Nanoparticles for Staphylococcus aureus Strains. World J. Microbiol. Biotechnol. 36 (5), 69. 10.1007/s11274-020-02845-y [DOI] [PubMed] [Google Scholar]
- Urak K. T., Shore S., Rockey W. M., Chen S.-J., McCaffrey A. P., Giangrande P. H. (2016). In Vitro RNA SELEX for the Generation of Chemically-Optimized Therapeutic RNA Drugs. Methods 103, 167–174. 10.1016/j.ymeth.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bel N., Das A. T., Berkhout B. (2014). In Vivo SELEX of Single-Stranded Domains in the HIV-1 Leader RNA. J. Virol. 88 (4), 1870–1880. 10.1128/JVI.02942-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. W., Beyer M. D., Clay E., Hiam K. J., McMurry J. L., Xie Y. (2016). Identification of Preferred DNA-Binding Sites for the Thermus Thermophilus Transcriptional Regulator SbtR by the Combinatorial Approach REPSA. PLoS One 11 (7), e0159408. 10.1371/journal.pone.0159408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q., Liu X., Zeng Z., Chen Z., Liu Y., Zu Y. (2020). Aptamer Cocktail to Detect Multiple Species of Mycoplasma in Cell Culture. Int. J. Mol. Sci. 21 (11), 3784. 10.3390/ijms21113784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandtke T., Woźniak J., Kopiński P. (2015). Aptamers in Diagnostics and Treatment of Viral Infections. Viruses 7 (2), 751–780. 10.3390/v7020751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang Y., Yang H., Qin M., Ding X., Liu R., et al. (2018). In Vivo SELEX of an Inhibitory NSCLC-specific RNA Aptamer from PEGylated RNA Library. Mol. Ther. - Nucleic Acids 10, 187–198. 10.1016/j.omtn.2017.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang Y., Chen J., Wang M., Zhang T., Luo W., et al. (2021). Detection of SARS-CoV-2 and its Mutated Variants via CRISPR-Cas13-Based Transcription Amplification. Anal. Chem. 93 (7), 3393–3402. 10.1021/acs.analchem.0c04303 [DOI] [PubMed] [Google Scholar]
- Waring M. J. (2010). Lipophilicity in Drug Discovery. Expert Opin. Drug Discov. 5 (3), 235–248. 10.1517/17460441003605098 [DOI] [PubMed] [Google Scholar]
- Wiersinga W. J., Rhodes A., Cheng A. C., Peacock S. J., Prescott H. C. (2020). Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19). JAMA 324 (8), 782–793. 10.1001/jama.2020.12839 [DOI] [PubMed] [Google Scholar]
- Wong E., Goldberg T. (2014). Mipomersen (Kynamro): a Novel Antisense Oligonucleotide Inhibitor for the Management of Homozygous Familial Hypercholesterolemia. P T 39 (2), 119–122. [PMC free article] [PubMed] [Google Scholar]
- Woo C. H., Jang S., Shin G., Jung G. Y., Lee J. W. (2020). Sensitive Fluorescence Detection of SARS-CoV-2 RNA in Clinical Samples via One-Pot Isothermal Ligation and Transcription. Nat. Biomed. Eng. 4 (12), 1168–1179. 10.1038/s41551-020-00617-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-H., Wang C.-H., Ma Y.-D., Lee G.-B. (2018a). A Nitrocellulose Membrane-Based Integrated Microfluidic System for Bacterial Detection Utilizing Magnetic-Composite Membrane Microdevices and Bacteria-specific Aptamers. Lab. Chip 18 (11), 1633–1640. 10.1039/c8lc00251g [DOI] [PubMed] [Google Scholar]
- Wu S., Liu L., Duan N., Wang W., Yu Q., Wang Z. (2018b). A Test Strip for Ochratoxin A Based on the Use of Aptamer-Modified Fluorescence Upconversion Nanoparticles. Microchim Acta 185 (11), 497. 10.1007/s00604-018-3022-0 [DOI] [PubMed] [Google Scholar]
- Wu X., Shaikh A., Yu Y., Li Y., Ni S., Lu A., et al. (2017). Potential Diagnostic and Therapeutic Applications of Oligonucleotide Aptamers in Breast Cancer. Int. J. Mol. Sci. 18 (9), 1851. 10.3390/ijms18091851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z., Gong Q., Wang C., Zheng B. (2018). Highly Sensitive Chemiluminescent Aptasensor for Detecting HBV Infection Based on Rapid Magnetic Separation and Double-Functionalized Gold Nanoparticles. Sci. Rep. 8 (1), 9444. 10.1038/s41598-018-27792-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z., Huang R., Li Z., He N., Wang T., Su E., et al. (2015). Selection of HBsAg-specific DNA Aptamers Based on Carboxylated Magnetic Nanoparticles and Their Application in the Rapid and Simple Detection of Hepatitis B Virus Infection. ACS Appl. Mater. Inter. 7 (21), 11215–11223. 10.1021/acsami.5b01180 [DOI] [PubMed] [Google Scholar]
- Yan J., Gao T., Lu Z., Yin J., Zhang Y., Pei R. (2021). Aptamer-Targeted Photodynamic Platforms for Tumor Therapy. ACS Appl. Mater. Inter. 13 (24), 27749–27773. 10.1021/acsami.1c06818 [DOI] [PubMed] [Google Scholar]
- Yang J., Bowser M. T. (2013). Capillary Electrophoresis-SELEX Selection of Catalytic DNA Aptamers for a Small-Molecule Porphyrin Target. Anal. Chem. 85 (3), 1525–1530. 10.1021/ac302721j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Alkhamis O., Canoura J., Liu Y., Xiao Y. (2021). Advances and Challenges in Small‐Molecule DNA Aptamer Isolation, Characterization, and Sensor Development. Angew. Chem. Int. Ed. 60 (31), 16800–16823. 10.1002/anie.202008663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Fang X., Liu X., Ou H., Zhang H., Wang J., et al. (2020). Discovery of sandwich Type COVID-19 Nucleocapsid Protein DNA Aptamers. Chem. Commun. 56 (70), 10235–10238. 10.1039/d0cc03993d [DOI] [PubMed] [Google Scholar]
- Zhang X., Feng Y., Duan S., Su L., Zhang J., He F. (2019a). Mycobacterium Tuberculosisstrain H37Rv Electrochemical Sensor Mediated by Aptamer and AuNPs-DNA. ACS Sens. 4 (4), 849–855. 10.1021/acssensors.8b01230 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhu L., He P., Zi F., Hu X., Wang Q. (2019b). Sensitive Assay of Escherichia coli in Food Samples by Microchip Capillary Electrophoresis Based on Specific Aptamer Binding Strategy. Talanta 197, 284–290. 10.1016/j.talanta.2019.01.040 [DOI] [PubMed] [Google Scholar]
- Zhou J., Neff C. P., Swiderski P., Li H., Smith D. D., Aboellail T., et al. (2013a). Functional In Vivo Delivery of Multiplexed Anti-HIV-1 siRNAs via a Chemically Synthesized Aptamer with a Sticky Bridge. Mol. Ther. 21 (1), 192–200. 10.1038/mt.2012.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Rossi J. (2017). Aptamers as Targeted Therapeutics: Current Potential and Challenges. Nat. Rev. Drug Discov. 16 (3), 181–202. 10.1038/nrd.2016.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Wang J., Zhang J., Li C., Tian Y., Wang J. (2013b). Selection and Identification of Streptomycin-specific Single-Stranded DNA Aptamers and the Application in the Detection of Streptomycin in Honey. Talanta 108, 109–116. 10.1016/j.talanta.2013.01.064 [DOI] [PubMed] [Google Scholar]
- Zhu C., Li L., Yang G., Fang S., Liu M., Ghulam M., et al. (2019a). Online Reaction Based Single-step Capillary Electrophoresis-Systematic Evolution of Ligands by Exponential Enrichment for ssDNA Aptamers Selection. Analytica Chim. Acta 1070, 112–122. 10.1016/j.aca.2019.04.034 [DOI] [PubMed] [Google Scholar]
- Zhu C., Yang G., Ghulam M., Li L., Qu F. (2019b). Evolution of Multi-Functional Capillary Electrophoresis for High-Efficiency Selection of Aptamers. Biotechnol. Adv. 37 (8), 107432. 10.1016/j.biotechadv.2019.107432 [DOI] [PubMed] [Google Scholar]
- Zhu Q., Shibata T., Kabashima T., Kai M. (2012). Inhibition of HIV-1 Protease Expression in T Cells Owing to DNA Aptamer-Mediated Specific Delivery of siRNA. Eur. J. Med. Chem. 56, 396–399. 10.1016/j.ejmech.2012.07.045 [DOI] [PubMed] [Google Scholar]
- Zhuo Z., Yu Y., Wang M., Li J., Zhang Z., Liu J., et al. (2017). Recent Advances in SELEX Technology and Aptamer Applications in Biomedicine. Int. J. Mol. Sci. 18 (10), 2142. 10.3390/ijms18102142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimbres F. M., Tárnok A., Ulrich H., Wrenger C. (2013). Aptamers: Novel Molecules as Diagnostic Markers in Bacterial and Viral Infections?. Biomed. Res. Int. 2013, 1–7. 10.1155/2013/731516 [DOI] [PMC free article] [PubMed] [Google Scholar]