Abstract
Magnetic particle-based immunoassays are widely used in microbiology-related assays for both microbial capture, separation, analysis, and detection. Besides facilitating sample operation, the implementation of micro-to-nanometer scale magnetic beads as a solid support potentially shortens the incubation time (for magnetic immuno capture) from several hours to less than an hour. Analytical technologies based on magnetic beads offer a rapid, effective and inexpensive way to separate and concentrate the target analytes prior to detection. Magneto-immuno separation uses magnetic particles coated with specific antibodies to capture target microorganisms, bear the corresponding antigens, and subsequently separate them from the sample matrix in a magnetic field. The method has been proven effective in separating various types of pathogenic bacteria from environmental water samples and in eliminating background interferences. Magnetic particles are often used to capture target cells (pathogenic bacteria) from samples. In most commercially available assays, the actual identification and quantitation of the captured cells is then performed by classical microbiological assays. This review highlights the most sensitive analytic methods (i.e., long-range surface plasmon resonance and electrochemical impedance spectroscopy) to detect magnetically tagged bacteria in conjunction with magnetic actuation.
Keywords: magneto-immuno separation, long-range surface plasmon resonance, electrochemical impedance spectroscopy, periodic magnetic actuation
Introduction
Bacteria are prokaryotic microorganisms generally of a few micrometers in length with various shapes such as rod, spiral and sphere. As a main cause of infectious diseases, pathogenic bacteria are the world's biggest life threat of children and young adults, causing 45% of death in all and 63% in early childhood in non-industrialized countries[1]. Therefore, pathogenic bacteria are essential detection and identification targets in medicine, food and environment safety and security. Swift detection of bacteria is also becoming increasingly important for anti-bioterrorism measures.
Bacterial infection is a common cause of morbidity and mortality worldwide. Escherichia coli O157:H7, salmonellae, Campylobacter jejuni, and Listeria monocytogenes are the leading cause of bacterial foodborne and waterborne illnesses, while other bacteria such as Cronobacter sakazakii can also lead to life-threatening infections[2]. Despite the availability of antibiotics, these infections are frequently misdiagnosed or there is an unacceptable delay in diagnosis. Currently, bacterial detection relies upon laboratory-based techniques such as cell culture, microscopic analysis, and biochemical assays.
In case of infection, the appropriate antibiotic should be prescribed as soon as the infection symptoms occur, or microbial contamination of food or water should be rapidly assessed. Yet the current methods for pathogenic bacteria detection are culture-based, which usually takes longer than 24 hours and requires laboratory analysis because the target bacteria coexist with a mixed population of dominating background cells (as in the case of blood or food matrices) or/and are in a low concentration (e.g. 1 bacterium/mL). Molecular detection technologies have been advanced for rapid detection of microbial pathogens in clinical specimens with PCR as the most sensitive method. Especially, when specific pathogens that are difficult to culture in vitro or require a long cultivation period are expected to be present in specimens, the diagnostic value of PCR has been proven significant[3]. However, the application of PCR in clinical specimens has many potential pitfalls due to the susceptibility of PCR to inhibitors, contamination and experimental conditions. For instance, the sensitivity and specificity of a PCR assay are dependent on target genes, primer sequences, PCR techniques, DNA extraction procedures, and PCR product detection methods[3–4]. Alternative to molecular methods, magneto-immunoassays involving immunocapture, magnetic separation and concentration have been advanced sequentially to support and speed up the detection of pathogenic bacteria either in bio-samples, food or water-bodies[5–6]. Immunoassays (the largest in vitro diagnostic technology worldwide) have an analytical error rate of approximately 11%[7], which is considerably higher than the rate of the molecular (e.g. PCR) assays, the latter usually taken as method of reference. This paper aims to review the state-of-the-art magneto-immunoassays in reducing the detection time with the emphasis on the most sensitive ones. Immunoassays refer to antibody (Ab)-based approaches based on the specific binding affinity of the antibody-antigen pair. Once produced and tested for specificity, Abs are typically mounted onto a support system (magnetic beads, nylon supports, cantilevers, plastic, etc.). However, other magneto-affinity capturing assays use aptamers[8], or generic ligands[9], instead of Abs. Notably, Abs, especially monoclonal antibodies, assure high affinity, while some aptamers may present a rather limited affinity, because it is based on systematic evolution of ligands by exponential enrichment.
Immunomagnetic separation is used in diagnostic microbiology to capture and detect pathogenic cells out of body fluids or cultured cells by using superparamagnetic particles (MPs) coated with binding molecules specific to the target cell. The magnetic core of MP is usually coated with a shell/matrix not only to protect against degradation or non-specific binding, but also to functionalize the bead surface with specific molecules. Proteins may bind/adsorb to hydrophobic surfaces, such as those of polymer-coated beads, and form a monolayer resistant to washing. A strong covalent binding between the particle surface and the protein is achieved through specific groups (–COOH-NH2, –CONH2, –OH) at the particle surface, which bind to –NH2 or –SH groups on the proteins via an activating reagent[10]. Also, streptavidin, biotin, histidine, protein A, and protein G, can be grafted onto the bead surface for specific biorecognition reactions[10]. Notably, the protocols of MP functionalization for affinity binding to the target cells (including the concentration of the Abs) are provided by the MP manufacturer. However, the ratio specific/non-specific binding depends on the quality of affinity molecules (Abs) functionalized on MPs, on the physicochemical parameters (e.g., pH, ionic strength) and last but not the least, on sample complexity. Therefore, to reduce the false negative and false positive results affecting magneto-immunoassays in complex bio-samples, MP coatings robust to non-specific binding as well as adjustment of sample parameters by tailored buffers must be considered.
Magnetic manipulation of MPs and aggregates of magnetically labelled target cells is suitable for integration in lab-on-a-chip applications[11]. The MP/aggregate's traveling velocity in a liquid environment depends on the size and magnetic susceptibility, as well as on the magnetic field gradient and the viscosity of the medium[8,10]. Consequently, the magnetic force acting on MPs (of volume V) in the linear susceptibility regime can be expressed as a function of the derivative of the magnetic induction (B):
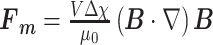 |
1 |
with Δ as the difference in magnetic susceptibility between the MPs and the surrounding liquid medium and µ0 as the permeability of free space. When the MPs eventually move at a constant velocity, the magnetic and viscous forces become equal and the value for the velocity difference between the MP and the liquid (Δv) becomes[8,10]:
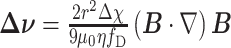 |
2 |
with fD as the drag coefficient of the particle (when no solid wall is in the MP vicinity fD ≈ 3).
The magnetic induction of an MP acting as a magnetic dipole depends on the strength, and direction of an MP magnetic moment m is given by[10]:
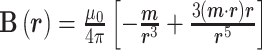 |
3 |
While the magnetic moment of an MP is smaller than that of a larger ferromagnetic microparticle, the advantages of superparamagnetic particles include: (1) the "switch off" magnetic effects by removing the magnetic induction field and (2) the possibility to be used for detection as labels assessed by a magnetic field sensor. Moreover, when applying the magnetic field, the MPs will interact via mutual magnetic dipole interaction to form magnetic supraparticle with chain-like structures. Following bacteria capture and separation from sample, detection could be performed by exploiting the properties of the magnetically labelled cells, using either the magnetic, or the optical/electrochemical methods[12].
Several analytical methods for magnetic field analysis based on MPs have been advanced for biosensing (immuno-detection)[13–14]. Superconducting quantum interference device (SQUID) magnetometer using magnetic markers can be used in a liquid phase without separating bound/free MPs[15]. Giant magnetoresistive (GMR) biosensing, used in both basic science research and clinical diagnostics, is based on the principle that stray field from MPs bound on sensor surface alters the magnetization in free layer, thus changing the resistance of GMR sensors[16–17]. These sensors, which can transduce changes in the local magnetic field into electrical signals, have been used as the read head in hard disk drives, current sensors, magnetic memory and biosensors[18]. Magnetic relaxation switching (MRS) is another emerging tool for immunoassays. In an external uniform magnetic field, the existence of the target in the sample will result in the aggregation of the dispersed antibody-conjugated MPs, thus leading to a local heterogeneous magnetic field that alters the transverse relaxation time of the surrounding water molecules dependent on the amount of target in the sample. Coupled with magnetic separation, MRS could reach a sensitivity of 102 cfu/mL for bacteria (Salmonella enterica) detection[19].
Complementary analytic methods to appraise aggregates of magnetically labelled target cells involve surface plasmon resonance (SPR), electrochemical impedance spectroscopy (EIS), enzyme-linked immuno-sorbent assay (ELISA), and lateral flow tests (e.g. immune-chromatographic assays)[20] with detection limits ranging around 105 bacteria/mL by ELISA[20–21], 107 bacteria/mL by lateral flow assays[9], about 102 bacteria/mL by EIS combined with periodic magnetophoresis[8], about 100 bacteria/mL by SPR, and about 50 bacteria/mL by long-range SPR (LRSPR), respectively[20–29]. Similarly, the time of analysis ranges from 10 minutes (lateral flow tests), and to several hours (EIS and ELISA)[20–26]. In the following part, the label-free methods SPR and EIS, which present the lowest detection limits, will be shortly discussed.
Detection of pathogenic bacteria using magneto-immunoassay in conjunction with SPR
SPR is a label-free optical sensing method for detecting analytes. Surface plasmon waves (SPWs) are excited at the interface between a thin, highly reflecting metal layer and a sample by coupling through a substrate with a high refractive index[27]. SPWs are transverse waves with an oscillating electric field normal to the surface. The electric field component of the p-polarized light is required to satisfy the boundary conditions to excite SPR. At the SPR angle, the photon energy is transferred to SPW, which results in energy loss of the reflected intensity. This phenomenon can be observed as a sharp dip in the reflected light intensity. Outside the metal, an evanescent electric wave, part of the internally reflected light beam, penetrates into the lower refractive index medium. The evanescent wave, as the "sensing" component, can interact optically with compounds close to or at the surface. The conventional SPR sensor is based on the Kretschmann structure, in which a thin film of gold (Au) is coated on the prism to stimulate surface plasmons. The SPR phenomenon can be used to measure the change in optical reflectivity of a metallic film, which arises due to changes in refractive index in the region close to the metal surface, within the penetration range of the evanescent wave[27,29]. The SPR systems comprise a source of plane-polarized light which then passes through a glass prism, the bottom of which contacts the transducer surface (typically a thin film of gold). Magnetic attraction of the aggregates of bacteria bound to magnetic nanoparticles to the transducer surface changes its refractive index, which in turn alters the angle corresponding to the dip of the reflectivity curve (the SPR angle), as sketched in Fig. 1[23].
Figure 1.
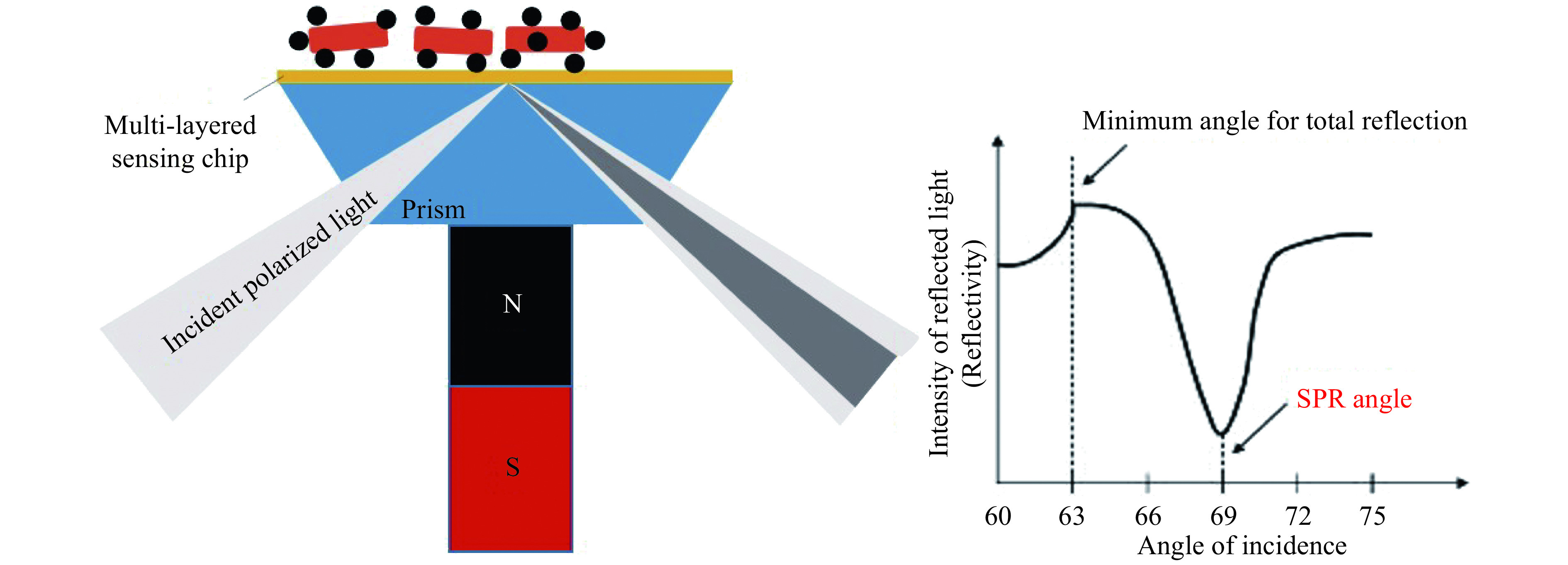
Setup to assess bacteria affinity bound to magnetic nanoparticles, magnetically attracted by an SPR module (with a long-range SPR sensing chip) and the corresponding angle resolved reflectivity curve (qualitative representation).
The figure was adapted from ref.[23] with permission from Copyright (2012) American Chemical Society.
The sensitive dependence of the SPR angular shift on the refractive index at the sensor sample interface supports the exquisite detection limits of the SPR assays.
Due to the narrow penetration depth of the evanescent field (usually limited to approximately 200 nm in standard SPR assays[26]), multi-layered SPR chips may allow for a highly increased penetration depth beyond 1000 nm, hence it is named LRSPR[29].
Due to the highly increased penetration depth as well as the steeper and sharper angle resolved reflectivity curve (SPR dip profile), LRSPR can assess large analytes with increased sensitivity, as the case of bacteria affinity bound to magnetic nanoparticles allowing for a detection limit of 50 to 100 bacteria/mL[21,23].
Detection of pathogenic bacteria using EIS in conjunction with magnetic actuation
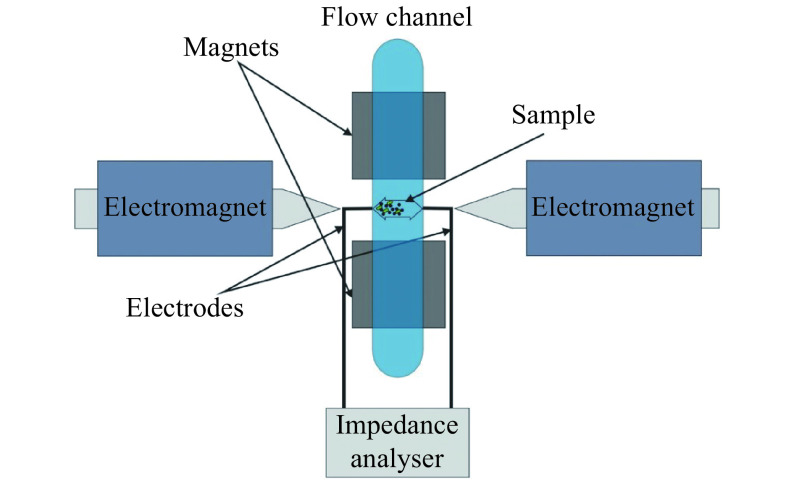
This section presents an effective method based on EIS in conjunction with magneto-immuno separation and periodic magnetic actuation to identify and quantitate target cells (pathogenic bacteria)[8]. The electrochemical impedance spectroscopy can reveal the (dynamics of) sample's electrical structure, and therefore is suitable for quantitative analysis of cells and bio-interfaces. By applying a small alternating current (AC) voltage at a given frequency and measuring the resulting current, the voltage to current ratio gives the complex impedance Z*. Notably, EIS assays are amenable to label-free analysis. EIS provides the impedance of biological systems arranged between two or more electrodes at different AC frequencies. It has been successfully used to assess: (i) aggregates of MPs and bacteria, (ii) living cells, either in suspension or adhered at an electrode, and tissues[8,30–37], as well as (iii) cellular biosensors genetically engineered to feature highly increased sensitivity[38–39].
After magneto-immuno separation and aggregates formation, the suspension of MP-aggregates is injected in the measurement chamber. Appropriate actuation of the magnetic field using a quadrupolar arrangement is applied to determine periodic displacement of MP-aggregates (driven by magnetic and viscous forces, as described by equation 1 and 2) and coverage of the working electrodes monitored by EIS at alternating current frequencies within 400 kHz to 6 MHz. This frequency range is chosen to avoid electrode polarization and achieve an optimal signal to noise ratio[8]. The setup of the system is sketched in Fig. 2.
Figure 2.
Setup to assess bacteria affinity bound to magnetic nanoparticles using EIS in conjunction and periodic magnetic actuation.
The figure was reproduced from ref.[8] with permission from the Royal Society of Chemistry. EIS: electrochemical impedance spectroscopy.
The method relates the concentration of bacterial cells to the amplitude of the oscillating electrical impedance in conjunction with the displacement of magnetic particle-bacteria aggregates (MP-aggregates) within a sample when applying a periodic magnetic actuation field. The oscillations of MP-aggregates between the two micro-electrodes are measured via electrical impedance assays at several AC frequencies using a highly sensitive custom-made impedance analyzer (see ref.[8] and references therein). As in the case of LRSPR, the bacteria quantitation is related to the size of the aggregates magnetically attracted on/covering the sensing chip (working-electrode).
However, besides the information on bacterial load, EIS can provide an additional quantitative measure on the bacterial viability status, which is complementary to SPR. This feature is based on EIS label-free assessment of the cell membrane integrity as revealed by the related variation of sample electrical impedance components and can be addressed using either an EIS setup, as already reported[8], or a setup deploying EIS in conjunction with optical microscopy and especially effective for low bacterial load[40].
The assay presents a detection limit of about 100 bacteria/mL (E. coli O157:H7 spiked in PBS)[8].
Assay's selectivity is tested by injecting a sample of MP incubated with Salmonella typhimurium (106 cells/mL). The lack of MP-aggregates following MP incubation with S. typhimurium bacteria is demonstrated by obtaining a similar EIS response when analyzing the sample (unbound) MP[8].
Table 1 summarizes the key metrics of the technologies chosen as examples and provides related references.
Table 1. Magnetic, SPR and EIS sensors based on magnetic nanoparticles.
| Sensor | MP composition | NP size | Bacterial target | Detection limit
(cell/mL) |
Detection range
(cell/mL) |
Ref. |
| MP: superparamagnetic particle; NP: nanoparticle; ND: no data; GMR: giant magnetoresistive; SQUID: superconducting quantum interference device; MRS: magnetic relaxation switching; SPR: surface plasmon resonance; LRSPR: long-range SPR; EIS: electrochemical impedance spectroscopy; L. monocytogenes: Listeria monocytogenes; P. aeruginosa: Pseudomonas aeruginosa; S. enterica: Salmonella enterica; S. enteritidis: Salmonella enteritidis; E. coli: Escherichia coli. | ||||||
| GMR | Streptavidin conjugated MPs | 1 µm | ND | 104 | ND | [41] |
| SQUID | Biotin conjugated MPs | 50 nm | L. monocytogenes | ~108 | ND | [42] |
| MRS | Carboxyl group modified Fe3O4 MPs | ~10 nm | P. aeruginosa | 50 | 102−106 | [43] |
| MRS | Carboxyl group modified Fe2O3/Fe3O4 MPs | 30 nm and 250 nm | S. enterica | 102 | 104−108 | [19] |
| SPR | Carboxyl group modified Fe3O4 MPs | 6 nm | S. enteritidis | 14 | 14–1.4×109 | [44] |
| LRSPR | Polysaccharide conjugated MPs | 200 nm | E. coli | 50 | 102−107 | [23] |
| EIS | Carboxyl group modified Fe3O4 MPs | 2.7 µm | E. coli | 102 | 102−104 | [8] |
Conclusions
Detection of specific pathogenic bacteria is based on the magneto-immuno capture/separation rather than on the specificity of the analytical methods, except for the molecular-based ones (e.g. PCR). Successful detection of pathogenic bacteria is supported by the vivid evolution of bio affinity-based sensors to replace the currently used diagnostic techniques. However, large scale applications are limited by the cost of the sensors and antibodies as well as the assay reproducibility.
The enhanced sensitivity of the lab-on-a-chip platforms based on SPR and EIS in conjunction with affinity functionalized MPs for both capture and detection simplifies sample preparation by saving steps like sample concentration and/or enrichment. It also has advantages such as the simple and portable design, a low detection limit and fast analysis. Such platforms can be used to detect other target cells (e.g. fungi) with minor modifications, e.g., changing the capturing step via a different affinity material (either antibodies or aptamers) bound to the magnetic beads. Though both EIS- and SPR-based magneto-immune assays feature high sensitivity, their (linear) detection range is inherently limited. EIS assays allow fine detection limit by deploying micrometer-size working electrodes. High bacterial load would lead to MP-aggregates with a coverage susceptible to exceed electrode area, hence limiting its applicability to quantitate a large bacterial concentration (beyond 107 cell/mL). In case of SPR, a similar restriction occurs due to the height of the aggregate layer on top of the sensing chip, for high bacterial loads may overpass the limited depth extension of the evanescent field.
Immunomagnetically isolated target organisms from sample matrices allow for the use of any endpoint detection method. The use of magnetic actuation processes for integration purposes is proceeding steadily. However, advancement of actuation principles is expected to further enhance system integration and overall analytical performance. Several magnetic actuation processes supporting both improved bacteria capture and detection assays have been qualitatively demonstrated but are not yet practically implemented.
While SPR- and EIS-based detection methods are highlighted in this review, we see many avenues for further innovation allowing for rapid and effective capturing, separation and detection using affinity (immune-) coated magnetic particles. MPs are fundamentally suited for developing miniaturized biosensing systems and allow a range of novel concepts supporting portable, point-of-care detection system. Such systems will allow rapid quantitative decentralized in vitro diagnostic testing with desktop-sized and hand-held instruments and a user-friendly "sample-in result-out" type of performance. Benefitting of these properties, magnetic actuation-based biosensing systems can improve diagnostic workflows, patient monitoring and disease management, with positive impacts on the quality, accessibility and cost-effectiveness of future healthcare[45].
Footnotes
CLC number:R318.08, Document code: A
The authors reported no conflict of interests.
References
- 1.Zang P, Gong AJ, Zhang PR, et al Targeting druggable enzymome by exploiting natural medicines: an in silico–in vitro integrated approach to combating multidrug resistance in bacterial infection . Pharm Biol. 2016;54(4):604–618. doi: 10.3109/13880209.2015.1068338. [DOI] [PubMed] [Google Scholar]
- 2.Aly MA, Domig KJ, Kneifel W, et al Immunogold nanoparticles for rapid plasmonic detection of C. sakazakii . Sensors. 2018;18(7):2028. doi: 10.3390/s18072028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamamoto Y PCR in diagnosis of infection: detection of bacteria in cerebrospinal fluids. Clin Diagn Lab Immunol. 2002;9(3):508–514. doi: 10.1128/cdli.9.3.508-514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abram TJ, Cherukury H, Ou CY, et al Rapid bacterial detection and antibiotic susceptibility testing in whole blood using one-step, high throughput blood digital PCR. Lab Chip. 2020;20(3):477–489. doi: 10.1039/C9LC01212E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reimhult E Nanoparticle-triggered release from lipid membrane vesicles. New Biotechnol. 2015;32(6):665–672. doi: 10.1016/j.nbt.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Cudjoe KS. Immunomagnetic particle-based techniques: overview[M]//Batt CA, Tortorello ML. Encyclopedia of Food Microbiology. 2nd ed. London: Academic Press, 2014: 351–357.
- 7.Yu HW, Halonen MJ, Pepper IL. Chapter 12 - immunological methods[M]//Pepper IL, Gerba CP, Gentry TJ. Environmental Microbiology. 3rd ed. San Diego: Academic Press, 2015: 245–269.
- 8.David S, Polonschii C, Gheorghiu M, et al Assessment of pathogenic bacteria using periodic actuation. Lab Chip. 2013;13(16):3192–3198. doi: 10.1039/c3lc50411e. [DOI] [PubMed] [Google Scholar]
- 9.Lee JJ, Jeong KJ, Hashimoto M, et al Synthetic ligand-coated magnetic nanoparticles for microfluidic bacterial separation from blood. Nano Lett. 2014;14(1):1–5. doi: 10.1021/nl3047305. [DOI] [PubMed] [Google Scholar]
- 10.Gijs MAM, Lacharme F, Lehmann U Microfluidic applications of magnetic particles for biological analysis and catalysis. Chem Rev. 2010;110(3):1518–1563. doi: 10.1021/cr9001929. [DOI] [PubMed] [Google Scholar]
- 11.Rapoport E, Montana D, Beach GSD Integrated capture, transport, and magneto-mechanical resonant sensing of superparamagnetic microbeads using magnetic domain walls. Lab Chip. 2012;12(21):4433–4440. doi: 10.1039/c2lc40715a. [DOI] [PubMed] [Google Scholar]
- 12.Ha Y, Ko S, Kim I, et al Recent advances incorporating superparamagnetic nanoparticles into immunoassays. ACS Appl Nano Mater. 2018;1(2):512–521. doi: 10.1021/acsanm.7b00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galkin OY, Besarab OB, Pysmenna MO, et al Modern magnetic immunoassay: biophysical and biochemical aspects. https://doi.org/10.15421/021806 Regul Mech Biosyst. 2018;9(1):47–55. [Google Scholar]
- 14.Huang HT, Garu P, Li CH, et al Magnetoresistive biosensors for direct detection of magnetic nanoparticle conjugated biomarkers on a chip. SPIN. 2019;9(2):1940002. doi: 10.1142/S2010324719400022. [DOI] [Google Scholar]
- 15.Kuma H, Oyamada H, Tsukamoto A, et al Liquid phase immunoassays utilizing magnetic markers and SQUID magnetometer. Clin Chem Lab Med. 2010;48(9):1263–1269. doi: 10.1515/CCLM.2010.259. [DOI] [PubMed] [Google Scholar]
- 16.Gaster RS, Xu L, Han SJ, et al Quantification of protein interactions and solution transport using high-density GMR sensor arrays. Nat Nanotechnol. 2011;6(5):314–320. doi: 10.1038/nnano.2011.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishna VD, Wu K, Perez AM, et al Giant magnetoresistance-based biosensor for detection of influenza a virus. Front Microbiol. 2016;7:400. doi: 10.3389/fmicb.2016.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adem S, Jain S, Sveiven M, et al Giant magnetoresistive biosensors for real-time quantitative detection of protease activity. Sci Rep. 2020;10(1):7941. doi: 10.1038/s41598-020-62910-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YP, Xianyu YL, Wang Y, et al One-step detection of pathogens and viruses: combining magnetic relaxation switching and magnetic separation. ACS Nano. 2015;9(3):3184–3191. doi: 10.1021/acsnano.5b00240. [DOI] [PubMed] [Google Scholar]
- 20.Tanchou V. Review of methods for the rapid identification of pathogens in water samples, ERNCIP thematic area Chemical & Biological Risks in the Water Sector, Task7, deliverable1[R]. Luxembourg: Publications Office of the European Union, 2014: 1–38.
- 21.Silva NFD, Magalhães JMCS, Freire C, et al Electrochemical biosensors for Salmonella: state of the art and challenges in food safety assessment . Biosens Bioelectron. 2018;99:667–682. doi: 10.1016/j.bios.2017.08.019. [DOI] [PubMed] [Google Scholar]
- 22.Ahmed A, Rushworth JV, Hirst NA, et al Biosensors for whole-cell bacterial detection. Clin Microbiol Rev. 2014;27(3):631–646. doi: 10.1128/CMR.00120-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Knoll W, Dostalek J Bacterial pathogen surface Plasmon resonance Biosensor advanced by long-range surface Plasmons and magnetic nanoparticle assays. Anal Chem. 2012;84(19):8345–8350. doi: 10.1021/ac301904x. [DOI] [PubMed] [Google Scholar]
- 24.Leva-Bueno J, Peyman SA, Millner PA A review on impedimetric immunosensors for pathogen and biomarker detection. Med Microbiol Immunol. 2020;209(3):343–362. doi: 10.1007/s00430-020-00668-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi JH, Lee JH, Son J, et al Noble metal-assisted surface Plasmon resonance immunosensors. Sensors. 2020;20(4):1003. doi: 10.3390/s20041003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou F, Wang XX, Qi FJ, et al Magneto-plamonic nanoparticles enhanced surface Plasmon resonance TB sensor based on recombinant gold binding antibody. Sens Actuators B: Chem. 2017;250:356–363. doi: 10.1016/j.snb.2017.04.162. [DOI] [Google Scholar]
- 27.Kim SH, Koh K. Functional dyes for surface Plasmon resonance-based sensing system[M]//Kim SH. Functional Dyes. Amsterdam: Elsevier, 2006: 185–213.
- 28.Vlček J, Pištora J, Lesňák M Design of plasmonic-waveguiding structures for sensor applications. Nanomaterials. 2019;9(9):1227. doi: 10.3390/nano9091227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isaacs S, Abdulhalim I long-range surface Plasmon resonance with ultra-high penetration depth for self-referenced sensing and ultra-low detection limit using diverging beam approach. Appl Phys Lett. 2015;106(19):193701. doi: 10.1063/1.4921200. [DOI] [Google Scholar]
- 30.Gheorghiu E Characterizing cellular systems by means of dielectric spectroscopy. BioElectroMagnetics. 1996;17(6):475–482. doi: 10.1002/(SICI)1521-186X(1996)17:6<475::AID-BEM7>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 31.Gheorghiu E, Asami K Monitoring cell cycle by impedance spectroscopy: experimental and theoretical aspects. Bioelectrochem Bioenerg. 1998;45(2):139–143. doi: 10.1016/S0302-4598(98)00084-1. [DOI] [Google Scholar]
- 32.Gheorghiu M, Gersing E, Gheorghiu E Quantitative analysis of impedance spectra of organs during ischemia. Ann N Y Acad Sci. 1999;873(1):65–71. doi: 10.1111/j.1749-6632.1999.tb09450.x. [DOI] [PubMed] [Google Scholar]
- 33.Vrinceanu D, Gheorghiu E Shape effects on the dielectric behaviour of arbitrarily shaped particles with particular reference to biological cells. Bioelectrochem Bioenerg. 1996;40(2):167–170. doi: 10.1016/0302-4598(96)05068-4. [DOI] [Google Scholar]
- 34.Gheorghiu E On the limits of ellipsoidal models when analyzing dielectric behavior of living cells emphasis on red blood cells. Ann N Y Acad Sci. 1999;873(1):262–268. doi: 10.1111/j.1749-6632.1999.tb09474.x. [DOI] [PubMed] [Google Scholar]
- 35.Gheorghiu E, Balut C, Gheorghiu M Dielectric behaviour of gap junction connected cells: a microscopic approach. Phys Med Biol. 2002;47(2):341–348. doi: 10.1088/0031-9155/47/2/312. [DOI] [PubMed] [Google Scholar]
- 36.Sandu T, Vrinceanu D, Gheorghiu E Linear dielectric response of clustered living cells. Phys Rev E. 2010;81(2):021913. doi: 10.1103/PhysRevE.81.021913. [DOI] [PubMed] [Google Scholar]
- 37.Vosika ZB, Lazovic GM, Misevic GN, et al Fractional calculus model of electrical impedance applied to human skin. PLoS One. 2013;8(4):e59483. doi: 10.1371/journal.pone.0059483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gheorghiu M, Stănică L, Tegla MGG, et al Cellular sensing platform with enhanced sensitivity based on optogenetic modulation of cell homeostasis. Biosens Bioelectron. 2020;154:112003. doi: 10.1016/j.bios.2019.112003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gheorghiu M, Stanica L, Polonschii C, et al Modulation of cellular reactivity for enhanced cell-based biosensing. Anal Chem. 2020;92(1):806–814. doi: 10.1021/acs.analchem.9b03217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang FN, Wang SP, Yang YZ, et al Imaging single bacterial cells with electro-optical impedance microscopy. ACS Sens. 2020 doi: 10.1021/acssensors.0c00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giraud M, Delapierre FD, Wijkhuisen A, et al Evaluation of in-flow magnetoresistive chip cell-counter as a diagnostic tool. Biosensors. 2019;9(3):105. doi: 10.3390/bios9030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grossman HL, Myers WR, Vreeland VJ, et al Detection of bacteria in suspension by using a superconducting quantum interference device. Proc Natl Acad Sci USA. 2004;101(1):129–134. doi: 10.1073/pnas.0307128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jia F, Xu L, Yan WJ, et al A magnetic relaxation switch aptasensor for the rapid detection of Pseudomonas aeruginosa using superparamagnetic nanoparticles . Microchim Acta. 2017;184(5):1539–1545. doi: 10.1007/s00604-017-2142-2. [DOI] [Google Scholar]
- 44.Liu X, Hu YX, Zheng S, et al Surface Plasmon resonance immunosensor for fast, highly sensitive, and in situ detection of the magnetic nanoparticles-enriched Salmonella enteritidis . Sens Actuators B: Chem. 2016;230:191–198. doi: 10.1016/j.snb.2016.02.043. [DOI] [Google Scholar]
- 45.van Reenen A, de Jong AM, den Toonder JMJ, et al Integrated lab-on-chip biosensing systems based on magnetic particle actuation–a comprehensive review. Lab Chip. 2014;14(12):1966–1986. doi: 10.1039/C3LC51454D. [DOI] [PubMed] [Google Scholar]