Abstract
Wildlife is known to be a source of high-impact pathogens affecting people. However, the distribution, genetic diversity, and zoonotic potential of Cryptosporidium, Enterocytozoon bieneusi, and Giardia duodenalis in wildlife are poorly understood. Here, we conducted the first molecular epidemiological investigation of these three pathogens in wildlife in Zhejiang and Shanghai, China. Genomic DNAs were derived from 182 individual fecal samples from wildlife and then subjected to a nested polymerase chain reaction–based sequencing approach for detection and characterization. Altogether, 3 (1.6%), 21 (11.5%), and 48 (26.4%) specimens tested positive for Cryptosporidium species, E. bieneusi, and G. duodenalis, respectively. Sequence analyses revealed five known (BEB6, D, MJ13, SC02, and type IV) and two novel (designated SH_ch1 and SH_deer1) genotypes of E. bieneusi. Phylogenetically, novel E. bieneusi genotype SH_deer1 fell into group 6, and the other genotypes were assigned to group 1 with zoonotic potential. Three novel Cryptosporidium genotypes (Cryptosporidium avian genotype V-like and C. galli-like 1 and 2) were identified, C. galli-like 1 and 2 formed a clade that was distinct from Cryptosporidium species. The genetic distinctiveness of these two novel genotypes suggests that they represent a new species of Cryptosporidium. Zoonotic assemblage A (n = 36) and host-adapted assemblages C (n = 1) and E (n = 7) of G. duodenalis were characterized. The overall results suggest that wildlife act as host reservoirs carrying zoonotic E. bieneusi and G. duodenalis, potentially enabling transmission from wildlife to humans and other animals.
Keywords: Cryptosporidium, Enterocytozoon bieneusi, Giardia duodenalis, genotypes, wildlife, prevalence, zoonotic potential
Introduction
Wildlife has been an important source of various high-impact pathogens affecting people, and zoonoses originated in wildlife remain a major public health issue around the world (Kruse et al., 2005). Novel diseases continue to emerge, and the responsible pathogens are often from unexpected wildlife, such as Ebola and Marburg virus (Bats; Amman et al., 2015; Jones et al., 2015); HIV-1 and HIV-2 (Primates; Gao et al., 1999); Nipah, Hendra, and Menangle virus (Bats; Field et al., 2007); West Nile virus (Mosquitoes; CDC, 1999, 2000); SARS (severe acute respiratory syndrome)–like virus (Bats; Li et al., 2005); and 2019 novel coronavirus (COVID-19) (Wildlife; Liu et al., 2020; Xiao et al., 2020), highlighting the important role of wildlife in the transmission of zoonotic pathogens. Currently, the ongoing 2019 novel coronavirus pandemic has resounded the alarm on pathogens in wildlife. Thus, it is crucial to screen and identify potentially zoonotic pathogens in wildlife from different geographical regions for prediction, prevention, and control of zoonotic diseases outbreaks in humans (Cunningham et al., 2017).
Infectious diarrhea remains a major public health concern worldwide (Walker et al., 2012; Liu et al., 2015; GBD, 2016). It kills more than 2,000 children every day, more than AIDS, malaria, and measles (Liu et al., 2015; Chingwaru and Vidmar, 2018; Lemos et al., 2018; Maniga et al., 2018). Multiple pathogens including viruses, bacteria (Blander et al., 2017), fungi (Liguori et al., 2015; Hallen-Adams and Suhr, 2017), and protists (Lanata et al., 2013; Blander et al., 2017) are responsible for diarrhea. Among protists, Cryptosporidium species, Giardia duodenalis, and Enterocytozoon bieneusi are the most common etiological pathogens of the intestinal disease and are known to cause large disease outbreaks in humans, especially for Cryptosporidium and G. duodenalis (Karanis et al., 2007; Baldursson and Karanis, 2011; Decraene et al., 2012; Ryan and Cacciò, 2013; Checkley et al., 2015).
Currently, ∼40 named Cryptosporidium species and close to 50 genotypes have been reported (Feng et al., 2018; Haghi et al., 2020). There are ∼20 species of Cryptosporidium identified in humans (Xiao, 2010), of which Cryptosporidium parvum and Cryptosporidium hominis are the most common species infecting humans (Feng et al., 2018). C. parvum has a broad host range that includes humans and various animal species. By contrast, C. hominis is mainly restricted to humans, non-human primates, and equine animals (Feng et al., 2018). G. duodenalis is recognized as a species complex consisting of eight assemblages (A–H). Assemblages A and B can infect humans and other mammals, assemblages C and D are frequently found in dogs and other canids, assemblage E in hoofed animals, assemblage F in cats, assemblage G in rodents, and assemblage H in pinnipeds (Ryan and Zahedi, 2019). Until recently, assemblages C to H were considered host-specific, except that assemblages C, D, E, and F are occasionally found in humans (i.e., assemblage E has been found in human samples more frequently than F) (Gelanew et al., 2007; Broglia et al., 2013; Liu et al., 2014; Štrkolcová et al., 2015; Scalia et al., 2016; Zahedi et al., 2017). Among 14 species of microsporidia infecting humans, E. bieneusi is the most common microbe causing diarrhea. E. bieneusi can infect a broad host range, including mammals, birds, reptiles (Squamata), and insects (Diptera). Currently, there are more than 600 genotypes, and most genotypes can be found in both humans and animals, showing zoonotic potential (Li et al., 2019a, b; Zhang, 2019; Zhang et al., 2021). The three enteric eukaryotic agents can infect humans through the fecal–oral route, via direct contact with infected individuals or ingestion of contaminated water or food (Yu et al., 2020).
The three microbes can be identified or characterized at species, subspecies, and/or genotypic level using molecular techniques. Currently, small subunit ribosomal DNA (SSU rDNA) has been wildly used for Cryptosporidium species identification, whereas a genetic marker in the 60-kDa glycoprotein (gp60) gene has been commonly used for differentiating Cryptosporidium at the genotypic and subgenotypic levels (Abeywardena et al., 2015). For G. duodenalis, triose-phosphate isomerase (tpi), β-giardin (bg), glutamate dehydrogenase (gdh), elongation factor 1-alfa (ef1-α), and SSU rDNA are commonly used for genotypic identification (Ryan and Cacciò, 2013). As ef1-α and SSU rDNA are relatively problematic, and they cannot discriminate G. duodenalis subtypes within assemblages accurately and are thus not useful for transmission analyses (Traub et al., 2004). Internal transcribed spacer (ITS) of nuclear ribosomal DNA is sufficiently variable for the identification and genotypic characterization of E. bieneusi (Santín et al., 2009).
Using the approach above, we have explored the microbes from various animals including wild deer (Zhang et al., 2018b; Koehler et al., 2020), marsupials (Zhang et al., 2018c), domestic alpacas (Koehler et al., 2018), cattle (Zhang et al., 2018a), goats and sheep (Zhang et al., 2020), companion cats and dogs (Zhang et al., 2019), and humans (Zhang et al., 2018e). We also established a new phylogenetic classification system of overall 600 E. bieneusi genotypes (Zhang et al., 2021). The present study aims to identify three pathogens (Cryptosporidium species, E. bieneusi, and G. duodenalis) in wildlife in Zhejiang and Shanghai, characterize their genotypes and analyze their zoonotic potential. The findings in this study would help to understand the genetic diversity of the three agents and provide critical information for future global strategies to prevent outbreaks of their zoonoses.
Materials and Methods
Samples and DNA Isolation
In total, 182 fecal samples were collected from 48 species of zoo animals from Zhejiang zoo (n = 52) and Shanghai Wild Animal Park (n = 130) from May 2018 to August 2020 (Supplementary Table 1). Some fecal samples were collected from wildlife rectum directly, whereas others were fresh deposited fecal samples. Genomic DNA was extracted directly from 0.1 to 0.4 g of each of the 182 fecal samples using the FastDNA SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA, United States) according to the manufacturer’s recommendations. The extracted DNA was stored at −20°C for further polymerase chain reaction (PCR) assay.
Detection of Cryptosporidium Species, E. bieneusi and G. duodenalis
Nested PCR-Based Sequencing of Cryptosporidium Species SSU rDNA
The small subunit of ribosomal nuclear DNA locus (target length 830 bp) of each sample was screened for identification of Cryptosporidium species (i.e., primers are listed in Table 1). In the first run, PCR contained 25 μL of 2 × PCR buffer for KOD FX (Mg2+ plus) (Toyobo, Japan), 2 mM dNTPs, 100 nM (each) primer, 1.0 U KOD FX, and 1 μL of DNA template in a total 50 μL reaction mixture. A total of 35 cycles were carried out, each consisting of 94°C for 45 s, 55°C for 45 s, and 72°C for 1 min, with an initial hot start at 94°C for 3 min, and a final extension at 72°C for 7 min. A secondary PCR product was then amplified from 2 μL of the primary PCR products with the same cycling conditions as the first run, except for 60°C annealing temperature (Xiao et al., 1999, 2001; Jiang et al., 2005).
TABLE 1.
PCR primers (forward and reserve) used for the amplification of Cryptosporidium, Enterocytozoon bieneusi, and Giardia duodenalis in this study.
| Species (genetic marker) | Primers (5′-3′) | Length (∼bp) | References |
| Cryptosporidium | TTC TAG AGC TAA TAC ATG CG | 1,325 | Xiao et al., 1999 |
| (SSU rDNA) | CCC ATT TCC TTC GAA ACA GGA | Xiao et al., 2001 | |
| GGA AGG GTT GTA TTT ATT AGA TAA AG | 830 | Jiang et al., 2005 | |
| CTC ATA AGG TGC TGA AGG AGT A | |||
| E. bieneusi | MSP-1 (TGA ATG KGT CCC TGT) | 590 | Katzwinkel-Wladarsch et al., 1996 |
| (ITS) | MSP-2B (GTT CAT TCG CAC TAC T) | ||
| MSP-3 (GGA ATT CAC ACC GCC CGT CRY TAT) | 508 | ||
| MSP-4B (CCA AGC TTA TGC TTA AGT CCA GGG AG) | |||
| G. duodenalis | AL3543 (AAA TTA TGC CTG CTC GTC G) | 605 | Sulaiman et al., 2003 |
| (tpi) | AL3546 (CAA ACC TTT TCC GCA AAC C) | ||
| AL3544 (CCC TTC ATC GGT GGT AAC TT) | 532 | ||
| AL3545 (GTG GCC ACC ACT CCC GTG CC) | |||
Nested PCR-Based Sequencing of E. bieneusi ITS
Individual genomic DNA samples were subjected to nested PCR-coupled sequencing of the ITS (243–245 bp) region (i.e., only 243–245-bp fragment of the ITS was used for further phylogenetic analyses) using an established technique (Katzwinkel-Wladarsch et al., 1996). Nested PCR (in 50 μL) was conducted in a standard buffer containing 3.0 μM MgCl2, 0.4 mM dNTPs, 50 pmol of each primer, 1.25 U of Ex Taq DNA (TaKaRa Bio Inc., Beijing, China), and DNA template—except for the negative (no-template) control. The cycling conditions for both primary and secondary (nested) PCRs were as follows: 94°C for 5 min (initial denaturation), followed by 35 cycles of 94°C for 45 s (denaturation), 54°C for 45 s (annealing), and 72°C for 1 min (extension), followed by 72°C for 10 min (final extension).
Nested PCR-Based Sequencing of G. duodenalis TPI
G. duodenalis assemblages were identified and characterized by nested PCR-based sequencing of the tpi gene (∼530 bp) using the established methods (Sulaiman et al., 2003). PCR was carried out in a volume of 50 μL containing 3.0 μM MgCl2, 0.4 mM dNTPs, 50 pmol of each primer, 1.25 U of Ex Taq DNA (TaKaRa Bio Inc., Beijing, China), and DNA template. A cycling protocol of 94°C for 5 min (initial denaturation), followed by 35 cycles of 94°C for 45 s (denaturation), 50°C for 45 s (annealing), 72°C for 1 min (extension), and a final extension of 72°C for 10 min. The secondary amplification was achieved using the same cycling conditions, except for the annealing temperature of 55°C for 30 s.
Known test-positive, test-negative, and no-template controls were included in each PCR run. The secondary PCR products were examined by gel electrophoresis on a 1.5% agarose gel containing 4S Green Plus Nucleic Acid Stain (Sangon Biotech, Shanghai, China) and directly sequenced using second-round PCR primers in both directions. All sequences obtained (GenBank accession nos. Cryptosporidium: MW168840-MW168842; E. bieneusi: MT895455-MT895461 and G. duodenalis: MW048593-MW048601) were inspected for quality and compared with reference sequences acquired from the GenBank database.
Phylogenetic Analysis
Obtained sequences from this and previous studies were aligned over a consensus length of 735 (Cryptosporidium), 459 (G. duodenalis), and 270 (E. bieneusi; after trimming, approximately 243-bp fragment of the ITS was analyzed) positions using previously established methods (Zhang et al., 2018d) and then subjected to phylogenetic analyses using the Bayesian inference (BI) and Monte Carlo Markov Chain methods in MrBayes v.3.2.3 (Huelsenbeck and Ronquist, 2001). The Akaike Information Criteria test in jModeltest v.2.1.7 (Darriba et al., 2012) was used to evaluate the likelihood parameters set for BI analysis. Posterior probability (pp) values were calculated by running 2,000,000 generations with four simultaneous tree-building chains, with trees saved every one-hundredth generation. A 50% majority-rule consensus tree for each analysis was constructed based on the final 75% of trees generated by BI. The clades and subclades were assigned and named using an established classification system (Santín and Fayer, 2009, 2011; Feng and Xiao, 2011; Karim et al., 2015; Li W. et al., 2015; Koehler et al., 2016; Li et al., 2019a, b; Ryan and Zahedi, 2019).
Results
Molecular Detection of Cryptosporidium Species Based on SSU rDNA Gene
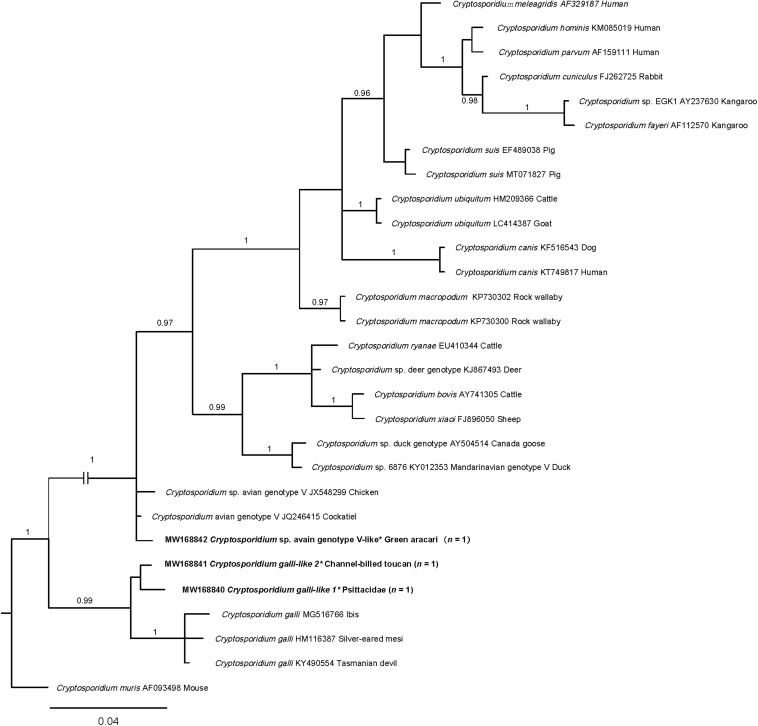
In total, three fecal DNA samples were identified Cryptosporidium species with the prevalence of 1.6% (3/182) (Table 2). They were all novel SSU rDNA sequences (i.e., < 100% identity with a sequence on GenBank) uniquely form the zoo in Zhejiang (Table 3). The three novel SSU rDNA sequences were assigned to the most closely related species or genotypes of Cryptosporidium based on sequence identity, representing Cryptosporidium galli-like 1 (from a species of Psittacidae) and C. galli-like 2 (channel-billed toucan) and Cryptosporidium avian genotype V-like (green aracari). C. galli-like 1 and 2 differed by 18 bp (763/781; 97.7%) and 17 bp (765/778; 98.3%) from the sequences representing C. galli (GenBank accession no. MG516766), and Cryptosporidium avian genotype V-like differed by 7 bp (780/787; 99.1%) from a sequence with GenBank accession no. JX548292 (Cryptosporidium avian genotype V).
TABLE 2.
Prevalence of Cryptosporidium species, Enterocytozoon bieneusi, and Giardia duodenalis in Shanghai Wild Animal Park and Zhejiang zoo of China.
| Species | Prevalence of each species (%) | Total no. of positive/total no. of samples | Prevalence in each region (%) (no. of positive/no. of samples) |
|
|
|||
| Shanghai | Zhejiang | ||
| Cryptosporidium | 1.6 3/182 | 0 | 1.6 (3/52) |
| E. bieneusi | 11.5 21/182 | 14.6 (19/130) | 3.8 (2/52) |
| G. duodenalis | 26.4 48/182 | 30.8 (40/130) | 15.4 (8/52) |
TABLE 3.
Summary of all pathogen species, genotypes, and/or assemblages identified in wildlife in Zhenjiang and Shanghai, China, using PCR-based sequencing of particular genetic markers.
| Species/genotype/assemblage identified by PCR based on sequencing (positivity no.) | Genetic marker used | GenBank accession no. | Host (Latin name) | Positivity no. for each wild animal species |
| Cryptosporidium species V-like | (1) SSU | MW168842* | Green aracari (Pteroglossus viridis) | (1) |
| C. galli-like 1 | (2) SSU | MW168841* | Psittacidae (species unknown) | (1) |
| C. galli-like 2 | MW168840* | Channel-billed toucan (Ramphastos vitellinus) | (1) | |
| Giardia duodenalis A | (40) tpi | MW048593 | Alpaca (Vicugna pacos) | (2) |
| MW048598* | Siberian tiger (Panthera tigris altaica) | (2) | ||
| MW048599* | Black-necked Crane (Grus nigricollis) | (2) | ||
| MW048600* | Blue-headed macaw (Propyrrhura couloni) | (3) | ||
| MW048601* | Cheetah (Acinonyx jubatus) | (3) | ||
| MW048594a | Fennec fox (Vulpes zerda) | (2) | ||
| MW048595a | Giant Eland (Tragelaphus derbianus) | (1) | ||
| MW048596a | Giraffe (Giraffa camelopardalis) | (3) | ||
| MW048597a | Golden takin (Budorcas taxicolor bedfordi) | (1) | ||
| Great pied hornbill (Buceros bicomis) | (1) | |||
| Hippopotamus (Hippopotamus amphibious) | (1) | |||
| Lion (Panthera leo) | (2) | |||
| Malabar pied hornbill (Anthracoceros coronatus) | (1) | |||
| Snub-nosed monkey (Rhinopithecus roxellana) | (8) | |||
| Ostrich (Struthio camelus) | (2) | |||
| Peafowl (Pavo cristatus) | (2) | |||
| Scarlet macaw (Ara macao) | (1) | |||
| Sika deer (Cervus Nippon) | (1) | |||
| Sun parakeet (Aratinga solstitialis) | (1) | |||
| Tiger (Panthera tigris tigris) | (1) | |||
| G. duodenalis C | (1) tpi | MW048604* | Spotted hyena (Crocuta crocuta) | (1) |
| G. duodenalis E | (5) tpi | MW048602 | Giraffe (Giraffa camelopardalis) | (4) |
| Kangaroo (Macropus species) | (1) | |||
| (2) tpi | MW048603* | Giraffe (Giraffa camelopardalis) | (2) | |
| Enterocytozoon bieneusi BEB6 | (3) ITS | MT895455 | Alpaca (Vicugna pacos) | (1) |
| Fallow deer (Dama dama) | (1) | |||
| Red deer (Cervus elaphus) | (1) | |||
| E. bieneusi D | (8) ITS | MT895457 | Siberian tiger (Panthera tigris altaica) | (2) |
| Lion (Panthera leo) | (2) | |||
| Snub-nosed monkey (Rhinopithecus roxellana) | (2) | |||
| Tiger (Panthera tigris tigris) | (2) | |||
| E. bieneusi MJ13 | (1) ITS | MT895460 | Red-and-green macaw (Ara chloropterus) | |
| E. bieneusi SC02 | (3) ITS | MT895459 | Great pied hornbill (Buceros bicomis) | (2) |
| Red-and-green macaw (Ara chloropterus) | (1) | |||
| E. bieneusi SH_ch1 | (2) ITS | MT895458* | Cheetah (Acinonyx jubatus) | (2) |
| E. bieneusi SH_deer1 | (1) ITS | MT895456* | Sika deer (Cervus Nippon) | (1) |
| E. bieneusi type IV | (1) ITS | MT895461 | Chestnut-fronted macaw (Ara severa) | (1) |
| E. bieneusi BEB6-like | (1) ITS | MT895462a | Red deer (Cervus elaphus) | (1) |
| E. bieneusi MJ17-like | (1) ITS | MT895463a | Brown bear (Ursus arctos pruinosus) | (1) |
*Novel genotypes. aMixed (indeterminate) genotypes.
The three SSU rDNA sequences were aligned with selected representative sequences in particular clades and subjected to the phylogenetic analysis (Figure 1). Genotypes Cryptosporidium avian genotype V-like clustered with genotype V with strong statistical support (pp = 1). Cryptosporidium galli-like 1 and 2 fell in one group and clustered with a clade of C. galli (pp = 0.99).
FIGURE 1.
Relationships among Cryptosporidium taxa inferred from the phylogenetic analysis of partial small subunit ribosomal rDNA gene (SSU rDNA) sequence data by Bayesian inference (BI). Posterior probabilities are indicated at all major nodes. Bold font indicates Cryptosporidium species or genotypes characterized from fecal DNA samples in this study. In parentheses are the numbers of samples representing a particular species, genotype, and sequence (GenBank accession numbers indicated). Novel genotypes (∗). Scale bar represents the number of substitutions per site. Most clades were strongly supported (pp = 0.96–1.00). pp < 0.95 was not shown.
E. bieneusi Genotype Characterizations Based on ITS Region
Enterocytozoon DNA was specifically detected by nested PCR of ITS in 21 of 182 (11.5%) fecal samples from zoo animals in Zhejiang (3.8%; 2/52) and Shanghai (14.6%; 19/130) (Table 2), including 10 mammal species: Alpaca (Vicugna pacos), amur tiger (Panthera tigris altaica), brown bear (Ursus arctos pruinosus), cheetah (Acinonyx jubatus), fallow deer (Dama dama), lion (Panthera leo), red deer (Cervus elaphus), sika deer (Cervus Nippon), snub-nosed monkey (Rhinopithecus species), tiger (Panthera tigris tigris), and three species of birds: Chestnut-fronted macaw (Ara severa), great pied hornbill (Buceros bicomis), and red-and-green macaw (Ara chloropterus) (Table 3).
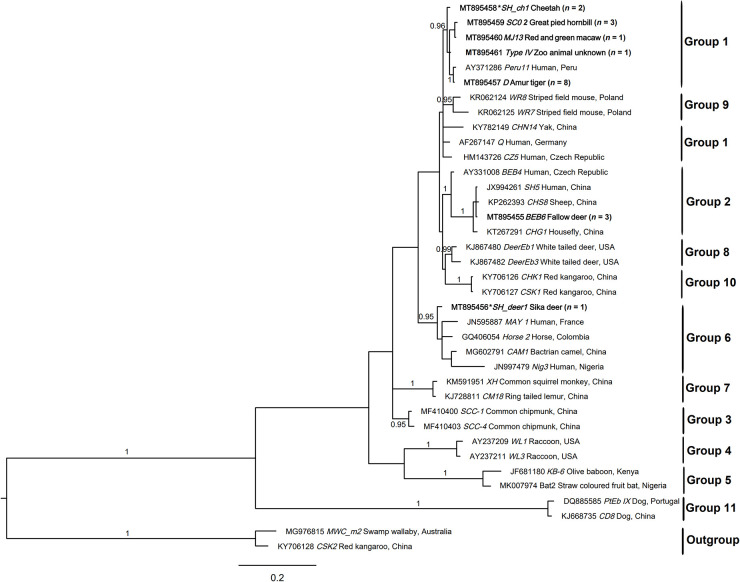
The 21 ITS amplicons (243 bp) were aligned to reference sequences in the GenBank database, and seven distinct genotypes were identified, including five known (BEB6, D, MJ13, SC02, and type IV) and two novel genotypes (designated SH_ch1 and SH_deer1) (Table 3). Novel genotype SH_ch1 (n = 2; from cheetahs) differed by 1 bp (242/243; 99.6%) from the sequence representing genotypes and KIN-1 (GenBank number MT231508). Novel genotype SH_deer1 (n = 1; from a sika deer) showed 8-bp (234/242; 99.7%) differences from the sequence with GenBank accession number KF261802. Two ambiguous sequences were derived from two amplicons, each containing multiple genotypes.
The eight ITS sequences representing seven distinct genotypes were aligned with sequences representing 10 groups of E. bieneusi and subjected to phylogenetic analysis (Figure 2). Genotypes BEB6, D, MJ13, SC02, SH_ch1, and type IV could be assigned to group 1 (pp = 0.96). Novel genotype SH_deer1 clustered with genotypes in group 6 with strong statistical support (pp = 0.95).
FIGURE 2.
Relationships among the genotypes of Enterocytozoon bieneusi recorded in the wildlife in this study inferred from the phylogenetic analysis of sequence data for the internal transcribed spacer (ITS) of nuclear ribosomal DNA by Bayesian inference (BI). Statistically significant posterior probabilities (pps) are indicated on branches. Individual GenBank accession numbers precede genotype designation (in italics) followed by sample and locality descriptions. The Enterocytozoon bieneusi genotypes identified and characterized from fecal DNA samples in the present study are indicated in bold type. Clades were assigned group names based on the classification system established by Karim et al. (2015) and Li et al. (2019a). The scale bar represents the number of substitutions per site. The E. bieneusi genotypes PtEbIX (DQ885585) and CD8 (KJ668735) from dogs were used as outgroups. All the groups were strongly supported (pp = 0.96–1). pp < 0.95 were not shown.
G. duodenalis Assemblages Identification Based on tpi Gene
Sequencing of all tpi amplicons identified 48 of 182 (26.4%) individual fecal samples to contain Giardia based on direct sequence comparisons, including 8 (15.4%; 8/52) in Zhejiang zoo and 40 (30.8%; 40/130) in Shanghai Wild Animal Park (Table 2). Genetic assemblages A (n = 36), C (n = 1), and E (n = 7) of G. duodenalis were characterized, and four amplicons contained mixed indeterminate genotypes. In total, eight distinct sequence types for tpi were defined (Table 3), including four representing Giardia sub-assemblage A (i.e., one known type from 16 species of wildlife and three novel sequence types from cheetah, fennec fox, lion, and snub-nosed monkey), one novel sequence type from a spotted hyena defined as assemblage C, and two novel distinct sequence types all representing assemblage E from giraffes.
The eight distinct tpi sequences representing four distinct assemblages or sub-assemblages were aligned with sequences representing Giardia assemblages A–G and subjected to the phylogenetic analysis (Figure 3). A novel sequence type (GenBank accession no. MW048604) clustered with assemblage C with strong statistical support (pp = 1.00).
FIGURE 3.
Relationships among Giardia taxa inferred from the phylogenetic analysis of partial triose-phosphate isomerase gene (tpi) sequence data by Bayesian inference (BI). Posterior probabilities are indicated at all major nodes. Bold font indicates Giardia species or genotypes characterized from fecal DNA samples in this study. In parentheses are the numbers of samples representing a particular species, genotype, and sequence (GenBank accession numbers indicated). Novel genotypes (*). Scale bar represents the number of substitutions per site. All groups were strongly supported (pp = 0.96–1.00). pp < 0.95 were not shown.
Discussion
The zoonotic enteric pathogens Cryptosporidium, Enterocytozoon, and Giardia have been reported in captive, wild, and zoo animals around the world (Leśniańska et al., 2016; Li N. et al., 2018; Amer et al., 2019). Their ability to spread via contaminated food, water, or direct contact with humans (e.g., zookeeper) poses a risk to public health.
Cryptosporidium
PCR-based sequencing of all three amplicons from 182 fecal DNA samples (1.6%; 3/182) revealed three operational taxonomic units (OTUs) of Cryptosporidium from three birds (channel-billed toucan, green aracari, and an unknown species of Psittacidae). Their SSU rDNA sequences were aligned (over a consensus length of 735 positions) with publicly available sequences, representing 14 species and an outgroup C. muris (Figure 1). Phylogenetic analyses of SSU rDNA data revealed that Cryptosporidium avian genotype V-like clustered with the genotypes C. galli and Cryptosporidium avian genotype V, which are typically found in birds (Xiao et al., 2004), and novel OTUs (genotypes C. galli-like 1 and 2 grouped, with strong nodal support (pp = 0.99). This analysis clearly showed that C. galli-like 1 and 2 represent a new and distinct clade. As the sequence variation (0–1.2%) within novel C. galli-like group was substantially less than differences (2.7–3.7%) between C. galli group and C. galli-like 1 and 2 upon pairwise comparison (Figure 1 and Supplementary Table 2), we propose that the latter two genotypes may represent a novel species of Cryptosporidium. However, it should be cautious to draw this conclusion. Definitely, further histological and morphological studies are needed. Sequencing SSU rDNA from many more representatives of Cryptosporidium to conduct a comprehensive phylogenetic analysis is also required.
E. bieneusi
E. bieneusi was identified in three wildlife fecal DNA samples in Zhejiang zoo (3.8%; 2/52) and 19 in Shanghai Wild Animal Park (14.6%; 19/130), with a total prevalence of 11.5% (21/182). Similarly, Li J. et al. (2015) and Yu et al. (2017) studied the prevalence of E. bieneusi in Shanghai wildlife animal park and reported 44.8% (30/67) and 69.1% (38/55), respectively. These cited prevalences are all higher than that in our study; however, they uniquely focused on the populations of non-human primates. By contrast, Li et al. (2016) studied 70 different wildlife species (272 fecal samples) in Chengdu zoo and Bifengxia zoo with prevalences of 10.6% (21/198) and 29.7% (22/74), respectively, both of which are higher than that in Zhejiang zoo, but E. bieneusi positivity in Chengdu zoo was lower than that in Shanghai, indicating that E. bieneusi might be widespread in Shanghai wild animal park. Internationally, the overall prevalences of E. bieneusi in farmed and/or captive wildlife and zoo animals globally ranged from 1.4% in Australia (Zhang et al., 2018c) to 53.3% in China (Yu et al., 2020). The variety of E. bieneusi prevalences might be due to host species, health status, and immunity of animals; management; locations; sample size; and environmental factors—season, temperature, sunlight, and humidity.
In total, five known (BEB6, D, MJ13, SC02, and type IV) and two novel genotypes (designated SH_ch1 and SH_deer1) were identified in this study. The predominant genotype here was genotype D (38.1%; 8/21), followed by BEB6 and SC02 (each 14.3%; 3/21), SH_ch1 (9.5%; 2/21), and four other genotypes (each 4.8%; 1/21). Genotype D is frequently identified in humans and nearly 70 species of animals, including birds (Anseriformes, Columbiformes, Falconiformes, Galliformes, Gruiformes, and Passeriformes) and mammals (Artiodactyla, Carnivora, Lagomorpha, Perissodactyla, Primates, and Rodentia) (Zhang, 2019; Zhang et al., 2021), indicating that genotype D has the capability of intra-species transmission. Similarly, genotype BEB6 has also been found in humans and 23 animal species (Zhang, 2019; Zhang et al., 2021), and fallow deer (reported here) is the first record of this genotype. Genotype SC02 was found in human and bear (Wu et al., 2018), giant panda (Li W. et al., 2018), horse (Deng et al., 2016b), Pallas’s squirrel, raccoon (Li et al., 2016), red-bellied tree squirrel (Deng et al., 2016a), rhesus macaque (Zhong et al., 2017), and wild boar (Li et al., 2017); great pied hornbill (Buceros bicomis) identified in this study is the first such published record. Similarly, red-and-green macaw (Ara chloropterus) is the first host record of genotype MJ13. Predominant genotypes BEB6, D, and SC02 were also found in water samples (Ayed et al., 2012; Li et al., 2012; Huang et al., 2017; Li W. et al., 2018), indicating that they might spread via E. bieneusi spores–contaminated water.
Phylogenetic analyses revealed that novel genotype SH_deer1 clustered with genotypes CAM1 (camel), horse 2 (horse), MAY 1 (human), and Nig3 (human), falling into group 6. Previously, genotypes in this group were predominantly found in animals. Thus, group 6 was typically considered as the host-adapted group. However, with more genotypes from this group identified in humans (Akinbo et al., 2012; Qi et al., 2018), demonstrating that group 6 revealed zoonotic potential. Additionally, we have also created a phylogeny using all nearly 600 unique genotypes from all published studies employing complete ITS sequences, with the aim of assessing the relationships of the genotypes and the validity of groups (Zhang et al., 2021), proving the zoonotic potential of group 6. The overall results indicate that wildlife carrying zoonotic genotypes have the capacity to transmit from them to humans.
G. duodenalis
In the present study, 48 wildlife tested positive for G. duodenalis with a total prevalence of 26.37% (48/182), which was higher than that of Cryptosporidium (1.6%; 3/182) and E. bieneusi (11.5%; 21/182), indicating that G. duodenalis is more widely spread than the other two microbes. The prevalences of G. duodenalis in Shanghai Wild Animal Park and in Zhejiang were 30.8% (40/130) and 15.4% (8/52), respectively, both of which were higher than that in a number of studies of G. duodenalis globally (Matsubayashi et al., 2005; Lallo et al., 2009; Beck et al., 2011b; Majewska et al., 2012; Oates et al., 2012; Aghazadeh et al., 2015; Reboredo-Fernández et al., 2015; Adriana et al., 2016; Mynarova et al., 2016; Mateo et al., 2017; Helmy et al., 2018). Additionally, the prevalence of G. duodenalis in wild animals worldwide ranged from 1.1% in zoo in Japan (Matsubayashi et al., 2005) to 29.0% in Zagreb zoo in Croatia (Beck et al., 2011a); 30.8% (40/130) here in wildlife in Shanghai is the highest prevalence around the world. The overall results indicate relatively high G. duodenalis infections in zoo animals in this study. However, it cannot be entirely excluded that G. duodenalis cysts might only pass through the gastrointestinal tract (pseudoparasitism), as identification of G. duodenalis DNA from fecal samples is not a direct evidence of infection.
In total, three assemblages A, C, and E of G. duodenalis were characterized. Zoonotic assemblage A is predominant (75%; 36/48) in this study, followed by genotype E (14.58%; 7/48) and C (2.08%; 1/48). Genotype A has been reported in humans and a large number of animal species with the capacity of cross-species transmission (Ryan and Zahedi, 2019). In this study, assemblage E was mostly identified in giraffe, except for one positivity in kangaroo. This is the first time that kangaroo was recorded in the G. duodenalis assemblage E. This assemblage has been mainly reported in hoofed animals, but it was also detected in human specimens in Brazil (Fantinatti et al., 2016), Egypt (Foronda et al., 2008), and Australia (Zahedi et al., 2017), posing less risk to public health. Phylogenetically, the novel tpi sequence found in spotted hyena (Crocuta crocuta) clustered with assemblage C (Figure 3), which has been frequently reported in canids and occasionally reported in humans (Hopkins et al., 1997; Monis et al., 1998). The overall results indicate that zoo animals can harbor zoonotic G. duodenalis and potentially act as a host reservoir for human infections of giardiasis.
Conclusion
Exploring the genetic composition of Cryptosporidium species E. bieneusi and G. duodenalis populations in animals and humans is important for understanding transmission patterns of enteric disease and for its prevention and control. By conducting the present molecular-phylogenetic investigation of three pathogens target sequences derived from fecal samples (n = 182) from zoo animals in China, we found (phylogenetically) a novel species of Cryptosporidium. We also identified genotypes or assemblages (E. bieneusi: BEB6, D, MJ13, SC02, SH_ch1, SH_deer1, and type IV; G. duodenalis: A, C, and E), all of which have zoonotic potential. The overall results indicate that wildlife carrying zoonotic E. bieneusi and G. duodenalis can potentially transmit the pathogens to humans, thus posting a public health risk.
Data Availability Statement
The datasets generated for this study can be found in GenBank under the accession numbers, Cryptosporidium: MW168840-MW168842; E. bieneusi: MT895455-MT895461 and G. duodenalis: MW048593-MW048601.
Ethics Statement
Sample collections were carried out by colleges from the Shanghai Wild Animal Park and Zhejiang Zoo. Animals were handled in accordance with the Animal Ethics Procedures and Guidelines of the People’s Republic of China.
Author Contributions
YZ, RM, LY, ZC, YF, XC, YH, and HG: sample collection. YZ and LY: designed the study and performed the experiments. YZ: analysis and interpretation and wrote the manuscript. ZC: review the draft and supervision of project. All authors read and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank colleagues from zoos in Zhejiang and Shanghai for donating fecal samples.
Footnotes
Funding. This study was supported in part by the National Key Research and Development Program of China (Grant No. 2017YFD0500401), Shanghai Science and Technology Commission Scientific Research Project (Grant No. 20140900400), National Risk Assessment Project for Quality and Safety of Agricultural Products (Grant No. GJFP2019027), International Postdoctoral Exchange Fellowship Program (Talent-Introduction Program), Scientific Research Project of Special Training Program for Scientific and Technological Talents of Ethnic Minorities in Xinjiang (Grant No. 2020D03030), China Postdoctoral Science Foundation (2021M693455), and Shanghai Agriculture Applied Technology Development Program, China (Grant No. G20050304).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.692837/full#supplementary-material
The information regarding fecal samples (sample codes are given) collected from various species of wildlife (host and Latin name are given) located in Zhejiang and Shanghai from May 2018 to August 2020.
Pairwise comparison of sequence differences in the small subunit of nuclear ribosomal RNA gene (SSU rDNA; 735 bp) among Cryptosporidium species from samples tested in the present study (cf. Figure 1).
References
- Abeywardena H., Jex A. R., Gasser R. B. (2015). A perspective on Cryptosporidium and Giardia, with an emphasis on bovines and recent epidemiological findings. Adv. Parasitol. 88 243–301. 10.1016/bs.apar.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adriana G., Zsuzsa K., Oana D. M., Gherman C. M., Viorica M. (2016). Giardia duodenalis genotypes in domestic and wild animals from Romania identified by PCR-RFLP targeting the gdh gene. Vet. Parasitol. 217 71–75. 10.1016/j.vetpar.2015.10.017 [DOI] [PubMed] [Google Scholar]
- Aghazadeh M., Elson-Riggins J., Reljic S., De Ambrogi M., Huber D., Majnaric D., et al. (2015). Gastrointestinal parasites and the first report of Giardia spp. in a wild population of European brown bears (Ursus arctos) in Croatia. Vet. Arhiv. 85 201–210. [Google Scholar]
- Akinbo F. O., Okaka C. E., Omoregie R., Dearen T., Leon E. T., Xiao L. H. (2012). Molecular epidemiologic characterization of Enterocytozoon bieneusi in HIV-infected persons in Benin city, Nigeria. Am. J. Trop. Med. Hyg. 86 441–445. 10.4269/ajtmh.2012.11-0548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amer S., Kim S., Han J. I., Na K. J. (2019). Prevalence and genotypes of Enterocytozoon bieneusi in wildlife in Korea: a public health concern. Parasit. Vectors 12:160. 10.1186/s13071-019-3427-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amman B. R., Jones M. E., Sealy T. K., Uebelhoer L. S., Schuh A. J., Bird B. H., et al. (2015). Oral shedding of Marburg virus in experimentally infected Egyptian fruit bats (Rousettus aegyptiacus). J. Wildl. Dis. 51 113–124. 10.7589/2014-08-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayed L. B., Yang W., Widmer G., Cama V., Ortega Y., Xiao L. H. (2012). Survey and genetic characterization of wastewater in Tunisia for Cryptosporidium spp., Giardia duodenalis, Enterocytozoon bieneusi, Cyclospora cayetanensis and Eimeria spp. J. Water Health 10 431–444. 10.2166/wh.2012.204 [DOI] [PubMed] [Google Scholar]
- Baldursson S., Karanis P. (2011). Waterborne transmission of protozoan parasites: review of worldwide outbreaks-an update 2004-2010. Water Res. 45 6603–6614. 10.1016/j.watres.2011.10.013 [DOI] [PubMed] [Google Scholar]
- Beck R., Sprong H., Bata I., Lucinger S., Pozio E., Cacciò S. M. (2011a). Prevalence and molecular typing of Giardia spp. in captive mammals at the zoo of Zagreb, Croatia. Vet. Parasitol. 175 40–46. 10.1016/j.vetpar.2010.09.026 [DOI] [PubMed] [Google Scholar]
- Beck R., Sprong H., Lucinger S., Pozio E., Caccio S. M. (2011b). A large survey of Croatian wild mammals for Giardia duodenalis reveals a low prevalence and limited zoonotic potential. Vector Borne Zoonot. 11 1049–1055. 10.1089/vbz.2010.0113 [DOI] [PubMed] [Google Scholar]
- Blander J. M., Longman R. S., Iliev I. D., Sonnenberg G. F., Artis D. (2017). Regulation of inflammation by microbiota interactions with the host. Nat. Immunol. 18:851. 10.1038/ni.3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broglia A., Weitzel T., Harms G., Cacció S. M., Nöckler K. (2013). Molecular typing of Giardia duodenalis isolates from German travellers. Parasitol. Res. 112 3449–3456. 10.1007/s00436-013-3524-y [DOI] [PubMed] [Google Scholar]
- CDC (1999). Outbreak of West Nile-like viral encephalitis-New York, 1999. MMWR Morb. Mortal. Wkly. Rep. 48 845–849. [PubMed] [Google Scholar]
- CDC (2000). Update: West Nile Virus activity-Eastern United States, 2000. MMWR Morb. Mortal. Wkly. Rep. 49 1044–1047. [PubMed] [Google Scholar]
- Checkley W., White A. C. J., Jaganath D., Arrowood M. J., Chalmers R. M., Chen X. M., et al. (2015). A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect. Dis. 15 85–94. 10.1016/S1473-3099(14)70772-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chingwaru W., Vidmar J. (2018). Culture, myths and panic: three decades and beyond with an HIV/AIDS epidemic in Zimbabwe. Glob. Public Health 13 249–264. 10.1080/17441692.2016.1215485 [DOI] [PubMed] [Google Scholar]
- Cunningham A. A., Daszak P., Wood J. L. N. (2017). One health, emerging infectious diseases and wildlife: two decades of progress? Philos. Trans. R. Soc. Lond. B Biol. Sci. 372:20160167. 10.1098/rstb.2016.0167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Taboada G. L., Doallo R., Posada D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decraene V., Lebbad M., Botero-Kleiven S., Gustavsson A. M., Löfdahl M. (2012). First reported foodborne outbreak associated with microsporidia, Sweden, October 2009. Epidemiol. Infect. 140 519–527. 10.1017/S095026881100077X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Li W., Yu X., Gong C., Liu X., Zhong Z., et al. (2016a). First report of the human-pathogenic Enterocytozoon bieneusi from red-bellied tree squirrels (Callosciurus erythraeus) in Sichuan, China. PLoS One 11:e0163605. 10.1371/journal.pone.0163605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Li W., Zhong Z., Gong C., Liu X., Huang X., et al. (2016b). Molecular characterization and multilocus genotypes of Enterocytozoon bieneusi among horses in southwestern China. Parasit. Vectors 9:561. 10.1186/s13071-016-1844-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantinatti M., Bello A. R., Fernandes O., Da-Cruz A. M. (2016). Identification of Giardia lamblia assemblage E in humans points to a new anthropozoonotic cycle. J. Infect. Dis. 214 1256–1259. 10.1093/infdis/jiw361 [DOI] [PubMed] [Google Scholar]
- Feng Y. Y., Ryan U. M., Xiao L. H. (2018). Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 34 997–1011. 10.1016/j.pt.2018.07.009 [DOI] [PubMed] [Google Scholar]
- Feng Y., Xiao L. (2011). Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin. Microbiol. Rev. 24 110–140. 10.1128/CMR.00033-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field H. E., Mackenzie J. S., Daszak P. (2007). Henipaviruses: emerging paramyxoviruses associated with fruit bats. Curr. Top. Microbiol. Immunol. 315 133–159. 10.1007/978-3-540-70962-6_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foronda P., Bargues M. D., Abreu-Acosta N., Periago M. V., Valero M. A., Valladares B., et al. (2008). Identification of genotypes of Giardia intestinalis of human isolates in Egypt. Parasitol. Res. 103 1177–1181. 10.1007/s00436-008-1113-2 [DOI] [PubMed] [Google Scholar]
- Gao F., Bailes E., Robertson D. L., Chen Y., Rodenburg C. M., Michael S. F., et al. (1999). Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397 436–441. 10.1038/17130 [DOI] [PubMed] [Google Scholar]
- GBD (2016). Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the global burden of disease study 2015. Lancet 388 1545–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelanew T., Lalle M., Hailu A., Pozio E., Cacciò S. M. (2007). Molecular characterization of human isolates of Giardia duodenalis from Ethiopia. Acta Trop. 102 92–99. 10.1016/j.actatropica.2007.04.003 [DOI] [PubMed] [Google Scholar]
- Haghi M. M., Khorshidvand Z., Khazaei S., Foroughi-Parvar F., Ghasemikhah R. (2020). Cryptosporidium animal species in Iran: a systematic review and meta-analysis. Trop. Med. Health 48:97. 10.1186/s41182-020-00278-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallen-Adams H. E., Suhr M. J. (2017). Fungi in the healthy human gastrointestinal tract. Virulence 8 352–358. 10.1080/21505594.2016.1247140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmy Y. A., Spierling N. G., Schmidt S., Rosenfeld U. M., Reil D., Imholt C., et al. (2018). Occurrence and distribution of Giardia species in wild rodents in Germany. Parasit. Vectors 11:213. 10.1186/s13071-018-2802-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins R. M., Meloni B. P., Groth D. M., Wetherall J. D., Reynoldson J. A., Thompson R. C. (1997). Ribosomal RNA sequencing reveals differences between the genotypes of Giardia isolates recovered from humans and dogs living in the same locality. J. Parasitol. 83 44–51. 10.2307/3284315 [DOI] [PubMed] [Google Scholar]
- Huang C., Hu Y., Wang L., Wang Y., Li N., Guo Y., et al. (2017). Environmental transport of emerging human-pathogenic Cryptosporidium species and subtypes through combined sewer overflow and wastewater. Appl. Environ. Microbiol. 83:e00682-17. 10.1128/AEM.00682-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J. P., Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Jiang J., Alderisio K. A., Xiao L. (2005). Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl. Environ. Microbiol. 71 4446–4454. 10.1128/AEM.71.8.4446-4454.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. E. B., Schuh A. J., Amman B. R., Sealy T. K., Zaki S. R., Nichol S. T., et al. (2015). Experimental inoculation of Egyptian rousette bats (Rousettus aegyptiacus) with viruses of the Ebolavirus and Marburgvirus genera. Viruses 7 3420–3442. 10.3390/v7072779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanis P., Kourenti C., Smith H. (2007). Waterborne transmission of protozoan parasites: a worldwide review of outbreaks and lessons learnt. J. Water Health 5 1–38. 10.2166/wh.2006.002 [DOI] [PubMed] [Google Scholar]
- Karim M. R., Dong H., Li T., Yu F., Li D., Zhang L., et al. (2015). Predomination and new genotypes of Enterocytozoon bieneusi in captive nonhuman primates in zoos in China: high genetic diversity and zoonotic significance. PLoS One 10:e0117991. 10.1371/journal.pone.0117991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzwinkel-Wladarsch S., Lieb M., Helse W., Löscher T., Rinder H. (1996). Direct amplification and species determination of microsporidian DNA from stool specimens. Trop. Med. Int. Health 1 373–378. 10.1046/j.1365-3156.1996.d01-51.x [DOI] [PubMed] [Google Scholar]
- Koehler A. V., Haydon S. R., Jex A. R., Gasser R. B. (2016). Cryptosporidium and Giardia taxa in faecal samples from animals in catchments supplying the city of Melbourne with drinking water (2011 to 2015). Parasit. Vectors 9:315. 10.1186/s13071-016-1607-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler A. V., Rashid M. H., Zhang Y., Vaughan J. L., Gasser R. B., Jabbar A. (2018). First cross-sectional, molecular epidemiological survey of Cryptosporidium, Giardia and Enterocytozoon in alpaca (Vicugna pacos) in Australia. Parasit. Vectors 11:498. 10.1186/s13071-018-3055-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler A. V., Zhang Y., Wang T., Haydon S. R., Gasser R. B. (2020). Multiplex PCRs for the specific identification of marsupial and deer species from scats as a basis for non-invasive epidemiological studies of parasites. Parasit. Vectors 13:144. 10.1186/s13071-020-04009-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse H., Kirkemo A. M., Handeland K. (2005). Wildlife as source of zoonotic infections. Emerg. Infect. Dis. 10 2067–2072. 10.3201/eid1012.040707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallo M. A., Pereira A., Araujo R., Favorito S. E., Bertolla P., Bondan E. F. (2009). Occurrence of Giardia, Cryptosporidium and microsporidia in wild animals from a deforestation area in the state of Sao Paulo, Brazil. Cienc. Rural 39 1465–1470. 10.1590/S0103-84782009005000085 [DOI] [Google Scholar]
- Lanata C. F., Fischer-Walker C. L., Olascoaga A. C., Torres C. X., Aryee M. J., Black R. E. (2013). Global causes of diarrheal disease mortality in children < 5 years of age: a systematic review. PLoS One 8:e72788. 10.1371/journal.pone.0072788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos D. R. Q., Franco A. R., de Oliveir Garcia M. H., Pastor D., Bravo-Alcântara P., de Moraes J. C., et al. (2018). Risk analysis for the reintroduction and transmission of measles in the post-elimination period in the Americas. Rev. Panam. Salud Pública 41:e157. 10.26633/RPSP.2017.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leśniańska K., Perec-Matysiak A., Hildebrand J., Bunkowska-Gawlik K., Pirog A., Popiolek M. (2016). Cryptosporidium spp. and Enterocytozoon bieneusi in introduced raccoons (Procyon lotor)-first evidence from Poland and Germany. Parasitol. Res. 115 4535–4541. 10.1007/s00436-016-5245-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Qi M., Chang Y., Wang R., Li T., Dong H., et al. (2015). Molecular characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in captive wildlife at Zhengzhou Zoo, China. J. Eukaryot. Microbiol. 62 833–839. 10.1111/jeu.12269 [DOI] [PubMed] [Google Scholar]
- Li N., Ayinmode A. B., Zhang H. W., Feng Y. Y., Xiao L. H. (2018). Host-adapted Cryptosporidium and Enterocytozoon bieneusi genotypes in straw-colored fruit bats in Nigeria. Int. J. Parasitol. Parasites Wildl. 8 19–24. 10.1016/j.ijppaw.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Xiao L. H., Wang L., Zhao S., Zhao X., Duan L., et al. (2012). Molecular surveillance of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi by genotyping and subtyping parasites in wastewater. PLoS Negl. Trop. Dis. 6:e1809. 10.1371/journal.pntd.0001809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Deng L., Wu K., Huang X., Song Y., Su H., et al. (2017). Presence of zoonotic Cryptosporidium scrofarum, Giardia duodenalis assemblage A and Enterocytozoon bieneusi genotypes in captive Eurasian wild boars (Sus scrofa) in China: potential for zoonotic transmission. Parasit. Vectors 10:10. 10.1186/s13071-016-1942-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Deng L., Yu X., Zhong Z., Wang Q., Liu X., et al. (2016). Multilocus genotypes and broad host-range of Enterocytozoon bieneusi in captive wildlife at zoological gardens in China. Parasit. Vectors 9:395. 10.1186/s13071-016-1668-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Feng Y. Y., Santín M. (2019a). Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol. 35 436–451. 10.1016/j.pt.2019.04.004 [DOI] [PubMed] [Google Scholar]
- Li W., Feng Y. Y., Zhang L. X., Xiao L. H. (2019b). Potential impacts of host specificity on zoonotic or interspecies transmission of Enterocytozoon bieneusi. Infect. Genet. Evol. 75:104033. 10.1016/j.meegid.2019.104033 [DOI] [PubMed] [Google Scholar]
- Li W., Li Y., Song M., Lu Y., Yang J., Tao W., et al. (2015). Prevalence and genetic characteristics of Cryptosporidium, Enterocytozoon bieneusi and Giardia duodenalis in cats and dogs in Heilongjiang province, China. Vet. Parasitol. 208 125–134. 10.1016/j.vetpar.2015.01.014 [DOI] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J. H., et al. (2005). Bats are natural reservoirs of SARS-like coronaviruses. Science 310 676–679. 10.1126/science.1118391 [DOI] [PubMed] [Google Scholar]
- Li W., Zhong Z., Song Y., Gong C., Deng L., Cao Y., et al. (2018). Human-pathogenic Enterocytozoon bieneusi in captive giant pandas (Ailuropoda melanoleuca) in China. Sci. Rep. 8:6590. 10.1038/s41598-018-25096-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguori G., Lamas B., Richard M. L., Brandi G., da Costa G., Hoffmann T. W., et al. (2015). Fungal dysbiosis in mucosa-associated microbiota of Crohn’s disease patients. J. Crohns Colitis 10 296–305. 10.1093/ecco-jcc/jjv209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A., Yang F., Shen Y., Zhang W., Wang R., Zhao W., et al. (2014). Genetic analysis of the gdh and bg genes of animal-derived Giardia duodenalis isolates in northeastern China and evaluation of zoonotic transmission potential. PLoS One 9:e95291. 10.1371/journal.pone.0095291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Oza S., Hogan D., Perin J., Rudan I., Lawn J. E., et al. (2015). Global, regional, and national causes of child mortality in 2000–13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet 385 430–440. 10.1016/S0140-6736(14)61698-6 [DOI] [PubMed] [Google Scholar]
- Liu P., Jiang J. Z., Wan X. F., Hua Y., Li L., Zhou J., et al. (2020). Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. 16:e1008421. 10.1371/journal.ppat.1008421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska A. C., Solarczyk P., Moskwa B., Cabaj W., Jankowska W., Nowosad P. (2012). Giardia prevalence in wild cervids in Poland. Ann. Parasitol. 58 207–209. [PubMed] [Google Scholar]
- Maniga J. N., Aliero A. A., Ibrahim N., Okech M. A., Mack M. C. (2018). Plasmodium falciparum malaria clinical and parasitological outcomes after in-vivo artemether-lumefantrine (AL) treatment at Bushenyi district Uganda. Int. J. Trop. Dis. Health 29 1–17. 10.9734/IJTDH/2018/39642 [DOI] [Google Scholar]
- Mateo M., de Mingo M. H., de Lucio A., Morales L., Balseiro A., Espí A., et al. (2017). Occurrence and molecular genotyping of Giardia duodenalis and Cryptosporidium spp. in wild mesocarnivores in Spain. Vet. Parasitol. 235 86–93. 10.1016/j.vetpar.2017.01.016 [DOI] [PubMed] [Google Scholar]
- Matsubayashi M., Takami K., Kimata I., Nakanishi T., Tani H., Sasai K., et al. (2005). Survey of Cryptosporidium spp. and Giardia spp. infections in various animals at a zoo in Japan. J. Zoo Wildl. Med. 36 331–335. 10.1638/04-032.1 [DOI] [PubMed] [Google Scholar]
- Monis P. T., Andrews R. H., Mayrhofer G., Mackrill J., Kulda J., Isaac-Renton J. L., et al. (1998). Novel lineages of Giardia intestinalis identified by genetic analysis of organisms isolated from dogs in Australia. Parasitology 116 7–19. 10.1017/S0031182097002011 [DOI] [PubMed] [Google Scholar]
- Mynarova A., Foitova I., Kvac M., Kvetonova D., Rost M., Morrogh-Bernard H., et al. (2016). Prevalence of Cryptosporidium spp., Enterocytozoon bieneusi, Encephalitozoon spp. and Giardia intestinalis in wild, semi-wild and captive orangutans (Pongo abelii and Pongo pygmaeus) on Sumatra and Borneo, Indonesia. PLos One 11:e0152771. 10.1371/journal.pone.0152771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates S. C., Miller M. A., Hardin D., Conrad P. A., Melli A., Jessup D. A., et al. (2012). Prevalence, environmental loading, and molecular characterization of Cryptosporidium and Giardia isolates from domestic and wild animals along the central California coast. Appl. Environ. Microbiol. 78 8762–8772. 10.1128/AEM.02422-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M., Li J., Zhao A., Cui Z., Wei Z., Jing B., et al. (2018). Host specificity of Enterocytozoon bieneusi genotypes in bactrian camels (Camelus bactrianus) in China. Parasit. Vectors 11:219. 10.1186/s13071-018-2793-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboredo-Fernández A., Ares-Mazás E., Cacciò S. M., Gómez-Couso H. (2015). Occurrence of Giardia and Cryptosporidium in wild birds in Galicia (northwest Spain). Parasitology 142 917–925. 10.1017/S0031182015000049 [DOI] [PubMed] [Google Scholar]
- Ryan U. M., Cacciò S. M. (2013). Zoonotic potential of Giardia. Int. J. Parasitol. 43 943–956. 10.1016/j.ijpara.2013.06.001 [DOI] [PubMed] [Google Scholar]
- Ryan U., Zahedi A. (2019). Molecular epidemiology of giardiasis from a veterinary perspective. Adv. Parasitol. 106 209–254. 10.1016/bs.apar.2019.07.002 [DOI] [PubMed] [Google Scholar]
- Santín M., Fayer R. (2009). Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J. Eukaryot. Microbiol. 56 34–38. 10.1111/j.1550-7408.2008.00380.x [DOI] [PubMed] [Google Scholar]
- Santín M., Fayer R. (2011). Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res. Vet. Sci. 90 363–371. 10.1016/j.rvsc.2010.07.014 [DOI] [PubMed] [Google Scholar]
- Santín M., Trout J. M., Fayer R. (2009). A longitudinal study of Giardia duodenalis genotypes in dairy cows from birth to 2 years of age. Vet. Parasitol. 162 40–45. 10.1016/j.vetpar.2009.02.008 [DOI] [PubMed] [Google Scholar]
- Scalia L. A. M., Fava N. M. N., Soares R. M., Limongi J. E., da Cunha M. J. R., Pena I. F., et al. (2016). Multilocus genotyping of Giardia duodenalis in Brazilian children. Trans. R. Soc. Trop. Med. Hyg. 110 343–349. 10.1093/trstmh/trw036 [DOI] [PubMed] [Google Scholar]
- Štrkolcová G., Mad’án M., Hinney B., Goldová M., Mojžišová J., Halánová M. (2015). Dog’s genotype of Giardia duodenalis in human: first evidence in Europe. Acta Parasitol. 60 769–799. 10.1515/ap-2015-0113 [DOI] [PubMed] [Google Scholar]
- Sulaiman I. M., Fayer R., Bern C., Gilman R. H., Xiao L. (2003). Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg. Infect. Dis. 9 1444–1452. 10.3201/eid0911.030084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub R. J., Monis P. T., Robertson I., Irwin P., Mencke N., Thompson R. C. (2004). Epidemiological and molecular evidence supports the zoonotic transmission of Giardia among humans and dogs living in the same community. Parasitology 128 253–262. 10.1017/S0031182003004505 [DOI] [PubMed] [Google Scholar]
- Walker C. L. F., Aryee M. J., Boschi-Pinto C., Black R. E. (2012). Estimating diarrhea mortality among young children in low and middle income countries. PLoS One 7:e29151. 10.1371/journal.pone.0029151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Han J. Q., Shi L. Q., Zou Y., Li Z., Yang J. F., et al. (2018). Prevalence, genotypes, and risk factors of Enterocytozoon bieneusi in Asiatic black bear (Ursus thibetanus) in Yunnan province, southwestern China. Parasitol. Res. 117 1139–1145. 10.1007/s00436-018-5791-0 [DOI] [PubMed] [Google Scholar]
- Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.-J., et al. (2020). Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 583 286–289. 10.1038/s41586-020-2313-x [DOI] [PubMed] [Google Scholar]
- Xiao L. H. (2010). Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124 80–89. 10.1016/j.exppara.2009.03.018 [DOI] [PubMed] [Google Scholar]
- Xiao L. H., Bern C., Limor J., Sulaiman I., Roberts J., Checkley W., et al. (2001). Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 183 492–497. 10.1086/318090 [DOI] [PubMed] [Google Scholar]
- Xiao L. H., Fayer R., Ryan U., Upton S. J. (2004). Cryptosporidium taxonomy: recent advances and implications for public health. Clin. Microbiol. Rev. 17 72–97. 10.1128/CMR.17.1.72-97.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L. H., Morgan U. M., Limor J., Escalante A., Arrowood M., Shulaw W., et al. (1999). Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65 3386–3391. 10.1128/AEM.65.8.3386-3391.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Wu Y., Li T., Cao J., Wang J., Hu S., et al. (2017). High prevalence of Enterocytozoon bieneusi zoonotic genotype D in captive golden snub-nosed monkey (Rhinopithecus roxellanae) in zoos in China. BMC Vet. Res. 13:158. 10.1186/s12917-017-1084-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Wen X., Huang X., Yang R., Guo Y., Feng Y. Y., et al. (2020). Molecular characterization and zoonotic potential of Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium sp. in farmed masked palm civets (Paguma larvata) in southern China. Parasit. Vectors 13:403. 10.1186/s13071-020-04274-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahedi A., Field D., Ryan U. (2017). Molecular typing of Giardia duodenalis in humans in Queensland – first report of assemblage E. Parasitology 144 1154–1161. 10.1017/S0031182017000439 [DOI] [PubMed] [Google Scholar]
- Zhang Y. (2019). Molecular Epidemiological Investigations of Enterocytozoon Bieneusi. Ph.D. thesis. Melbourne, VIC: The University of Melbourne. [Google Scholar]
- Zhang Y., Koehler A. V., Wang T., Gasser R. B. (2021). Enterocytozoon bieneusi of animals—with an ‘Australian twist’. Adv. Parasitol. 111 1–73. 10.1016/bs.apar.2020.10.001 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Koehler A. V., Wang T., Cunliffe D., Gasser R. B. (2019). Enterocytozoon bieneusi genotypes in cats and dogs in Victoria, Australia. BMC Microbiol. 19:183. 10.1186/s12866-019-1563-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Koehler A. V., Wang T., Haydon S. R., Gasser R. B. (2018a). Enterocytozoon bieneusi genotypes in cattle on farms located within a water catchment area. J. Eukaryot. Microbiol. 66 553–559. 10.1111/jeu.12696 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Koehler A. V., Wang T., Haydon S. R., Gasser R. B. (2018b). First detection and genetic characterisation of Enterocytozoon bieneusi in wild deer in Melbourne’s water catchments in Australia. Parasit. Vectors 11:2. 10.1186/s13071-017-2577-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Koehler A. V., Wang T., Haydon S. R., Gasser R. B. (2018c). New operational taxonomic units of Enterocytozoon in three marsupial species. Parasit. Vectors 11:371. 10.1186/s13071-018-2954-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Koehler A. V., Wang T., Robertson G. J., Bradbury R. S., Gasser R. B. (2018d). Enterocytozoon bieneusi genotypes in people with gastrointestinal disorders in Queensland and Western Australia. Infect. Genet. Evol. 65 293–299. 10.1016/j.meegid.2018.08.006 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Koehler A. V., Wang T., Robertson G. J., Bradbury R. S., Gasser R. B. (2018e). Enterocytozoon bieneusi genotypes in people with gastrointestinal disorders in Queensland and Western Australia. Infect. Genet. Evol. 65 293–299. 10.1016/j.meegid.2018.08.006 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Mi R. S., Yang J. B., Wang J. X., Chen Z. G. (2020). Enterocytozoon bieneusi genotypes in farmed goats and sheep in Ningxia, China. Infect. Genet. Evol. 89:104559. 10.1016/j.meegid.2020.104559 [DOI] [PubMed] [Google Scholar]
- Zhong Z., Li W., Deng L., Song Y., Wu K., Tian Y., et al. (2017). Multilocus genotyping of Enterocytozoon bieneusi derived from nonhuman primates in southwest China. PLoS One 12:e0176926. 10.1371/journal.pone.0176926 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The information regarding fecal samples (sample codes are given) collected from various species of wildlife (host and Latin name are given) located in Zhejiang and Shanghai from May 2018 to August 2020.
Pairwise comparison of sequence differences in the small subunit of nuclear ribosomal RNA gene (SSU rDNA; 735 bp) among Cryptosporidium species from samples tested in the present study (cf. Figure 1).
Data Availability Statement
The datasets generated for this study can be found in GenBank under the accession numbers, Cryptosporidium: MW168840-MW168842; E. bieneusi: MT895455-MT895461 and G. duodenalis: MW048593-MW048601.