Key words: Covid-19, Cryptosporidium, New Zealand, nonpharmaceutical intervention, public health
Abstract
Coronavirus disease-2019 (Covid-19) nonpharmaceutical interventions have proven effective control measures for a range of respiratory illnesses throughout the world. These measures, which include isolation, stringent border controls, physical distancing and improved hygiene also have effects on other human pathogens, including parasitic enteric diseases such as cryptosporidiosis. Cryptosporidium infections in humans are almost entirely caused by two species: C. hominis, which is primarily transmitted from human to human, and Cryptosporidium parvum, which is mainly zoonotic. By monitoring Cryptosporidium species and subtype families in human cases of cryptosporidiosis before and after the introduction of Covid-19 control measures in New Zealand, we found C. hominis was completely absent after the first months of 2020 and has remained so until the beginning of 2021. Nevertheless, C. parvum has followed its typical transmission pattern and continues to be widely reported. We conclude that ~7 weeks of isolation during level 3 and 4 lockdown period interrupted the human to human transmission of C. hominis leaving only the primarily zoonotic transmission pathway used by C. parvum. Secondary anthroponotic transmission of C. parvum remains possible among close contacts of zoonotic cases. Ongoing 14-day quarantine measures for new arrivals to New Zealand have likely suppressed new incursions of C. hominis from overseas. Our findings suggest that C. hominis may be controlled or even eradicated through nonpharmaceutical interventions.
Introduction
Coronavirus disease-2019 (Covid-19) was declared a global pandemic by the World Health Organization (WHO) on 11 March 2020, leading to a variety of responses by different governments around the world. The nonpharmaceutical intervention strategy of New Zealand was particularly stringent and effectively eliminated a burgeoning Covid-19 outbreak through a nationwide lockdown, physical distancing, improved hand hygiene and ongoing 14-day quarantine for returning travellers (Robert, 2020; Baker et al., 2020a). A four-level alert system (New Zealand Government, 2020) was introduced on 21 March, beginning at level 2 and quickly moving to level 3 on 23 March and level 4 (i.e. the entire nation goes into self-isolation) on 25 March. The level 4 lockdown remained in place until 27 April, when it was reduced to level 3. By 11 May it was lowered to level 2 and finally decreased to level 1 on 8 June (Baker et al., 2020b). The primary intention of these measures in New Zealand was to reduce the transmission of Covid-19, but they also brought about a reduction in influenza and other respiratory viral infections in 2020 (Huang et al., 2021). Similar impacts have also been observed elsewhere (Fricke et al., 2021; Yang et al., 2021), but little attention to date has been given to parasitic enteric diseases.
Cryptosporidium species are obligatory intracellular intestinal parasites that infect a wide range of vertebrate hosts, causing a considerable burden of gastrointestinal disease (Xiao et al., 2004; Garcia and Hayman, 2016). In New Zealand, cryptosporidiosis is a notifiable disease with a higher incidence compared to other developed countries (Learmonth et al., 2004). Two species, C. hominis and C. parvum are responsible for the majority of human infections globally, and in New Zealand (Garcia-R and Hayman, 2017; Garcia-R et al., 2017; Garcia-R et al., 2020b). Of these, C. hominis is transmitted primarily through human-to-human contact whereas C. parvum is mainly zoonotic with ruminants as the primary reservoir host [(Learmonth et al., 2004; Feltus et al., 2006; Leitch and He, 2011) though see King et al. (2019)]. Clear seasonal peaks in cryptosporidiosis are present in New Zealand. An early autumnal peak for C. hominis associated with increased recreational water use and a spring peak for C. parvum associated with dairy cattle calving season is observed each year (Learmonth et al., 2004; Lake et al., 2008; Garcia-R et al., 2020b). Transmission of both species is through the faecal−oral route, following contact with infected hosts or water contaminated with the transmissive stages (oocysts) of the organism (Bouzid et al., 2013).
One of the difficulties with monitoring cryptosporidiosis is the lack of morphological characters to identify different Cryptosporidium species. Frequently, diagnosis is based on microscopy by the presence of oocysts in stool samples of infected patients or enzyme immunoassay and species-level identification is rarely attempted. Reliable species and subtype level identification is possible through molecular methods, though diagnostic laboratories moving to molecular diagnostics typically utilize qPCR and report presence/absence, without speciation. The most widely used marker for this purpose is the 60 kDa glycoprotein locus (gp60), upon which the subtyping of Cryptosporidium species is based (Feng and Xiao, 2017). This classification scheme has been used to study the epidemiology of human outbreaks of cryptosporidiosis in many previous studies (O'Brien et al., 2008; Chalmers et al., 2019). Here, we examine the disease dynamics of Cryptosporidium infections using gp60 for species identification and subtype family diversity patterns before, during and after lockdown measures in New Zealand.
Materials and methods
Anonymous Cryptosporidium-positive stool samples were collected between 2015 and 2021 through routine public health investigation activities from symptomatic humans visiting private and public general practitioner (GP) surgeries and hospitals throughout New Zealand. Samples were sent from accredited diagnostic laboratories to the Protozoa Research Unit at Hopkirk Research Institute (Massey University) for molecular analyses under a Ministry of Health contract. Stools were collected in 10-ml screw-cap tubes and stored at 4°C in the laboratory until DNA extraction.
DNA extraction was carried out 1–2 weeks after receiving samples at the Hopkirk Research Institute. Before DNA extraction, the oocysts of Cryptosporidium were physically disrupted using a beadbeater (Tissue Lyser II, Qiagen) at 30 Hz for 5 min. DNA extractions were performed using Zymo Quick-DNA Fecal/Soil Microbe kits following the manufacturer's instructions. A fragment of the gp60 gene was amplified using a nested PCR (Alves et al., 2003; Waldron et al., 2009) and amplification products were sequenced in both directions using Big Dye Terminator version 3.1 reagents on an ABI 3730XL automated DNA sequencer (Applied Biosystems, Foster City, California, USA).
Cryptosporidium gp60 sequences were classified into species and subtype families according to guidelines in Feng and Xiao (2017). While our sampling prior to 2020 included a wide range of regions throughout New Zealand, the 2020 samples were dominated by the Auckland region (118/130). For this reason, we filtered out results from elsewhere and limited our analyses to samples collected from the Auckland region. To check the representativeness of our dataset, we compared it with data from the national notifiable disease surveillance website (https://surv.esr.cri.nz/surveillance/annual_diseasetables.php).
Results
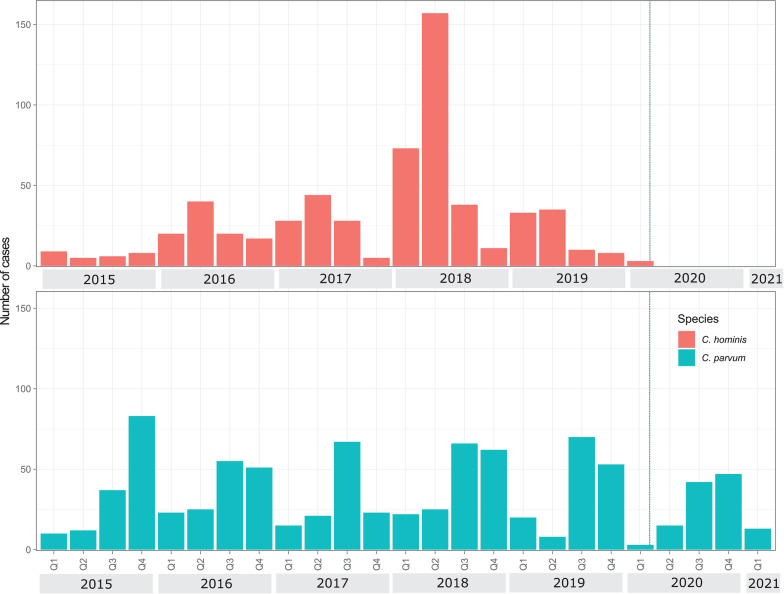
Our dataset included 1502 sequences received from 9 January 2015 to 3 March 2021 (Supplementary Table 1). Of these, C. parvum was the most common (n = 868), followed by C. hominis (598), Cryptosporidium cuniculus (28) and Cryptosporidium erinacei (8). Seasonal dynamics of C. hominis and C. parvum before 2020 and the impact of the Covid-19 lockdown are illustrated in Fig. 1. Cryptosporidium hominis was commonly found in the first (Q1) and second quarters (Q2) of the year between January and June while C. parvum was more common in the third (Q3) and fourth quarters (Q4) from July to December. The exception to this is in 2020, when no C. hominis was detected after 14 February 2020 (Fig. 1).
Fig. 1.
Cryptosporidium hominis and C. parvum quarterly cases from Auckland, New Zealand 2015–2021. Dotted line in Q1 2020 indicates the start of Covid-19 national lockdown.
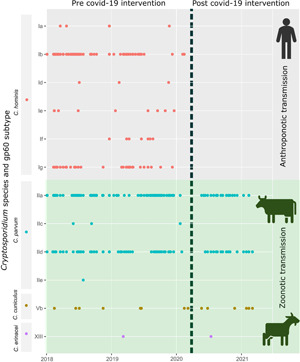
Further examination of gp60 subtype families shows the diversity and temporal dynamics of C. hominis and C. parvum (Fig. 2). Within C. hominis, subtype family Ib was detected consistently between 2015 and the beginning of 2020. Other gp60 subtype families in C. hominis appeared as sporadic outbreaks e.g. Id in 2016, Ig in 2017, 2018 and 2019 and/or single cases e.g. Ia in 2015–2019. Within C. parvum, subtype families IIa and IId were consistently detected whilst IIc and IIe appeared as sporadic single cases. Of the other zoonotic Cryptosporidium species, C. cuniculus and C. erinacei were only detected occasionally.
Fig. 2.
Timeline of Cryptosporidium gp60 subtype cases identified from Auckland, New Zealand 2015–2021. Number of cases per species are indicated on the y axis. Dotted line in 2020 indicates the start of Covid-19 national lockdown.
Our data are representative of the total number of cases reported nationally from the notifiable disease database from 2015 to 2019 (between 33% and 46%, Supplementary Table 1). However, in 2020 the number of cases reported to our lab dropped to a lower proportion (18%). This was probably caused by extra stress on diagnostic laboratories that did not send as many samples to our lab for molecular analyses during and following the Covid-19 outbreak. Data from outside Auckland are not included in our analyses due to low numbers in 2020, but in the few samples from outside Auckland which we do have, no post lockdown cases of C. hominis have been reported.
Discussion
In response to Covid-19, New Zealand underwent a nationwide lockdown beginning in late March 2020, with households spending over 7 weeks at home except for essential personal movements like supermarket and hospital visits. Since this time, our surveillance has revealed that C. hominis has not been detected in reported cases of cryptosporidiosis from the Auckland region. This absence has continued until the time of publication during the first quarter of 2021, which in previous years has been the beginning of peak season for C. hominis infections. In contrast, C. parvum and other less common zoonotic Cryptosporidium species have continued typical seasonal patterns. Our data suggest that the national measures for responding to the Covid-19 also interrupted C. hominis transmission. For instance, the closure of all child care centres and schools may have limited C. hominis transmission since these have been linked to previous outbreaks (Goñi et al., 2015). Ongoing managed isolation of visitors or citizens returning to New Zealand since then has also likely reduced new imported cases of cryptosporidiosis.
Prior to 2020, infections by C. hominis were reported seasonally (Garcia-R and Hayman, 2017; Garcia-R et al., 2020b). Furthermore, temporal dynamics of gp60 subtype families generally revealed sporadic infection patterns. Subtype families Ia, Id, Ie, If and Ig appeared at irregular intervals throughout the study period, which may indicate an importation by visitors or returning travellers from a global pool of C. hominis that fail to establish within New Zealand or an opportunistic behaviour of the parasite (Garcia-R and Hayman, 2017; Garcia-R et al., 2020a). The majority of C. hominis cases was subtype family Ib, which is also the most common subtype family globally (Feng et al., 2009; Putignani and Menichella, 2010; Segura et al., 2015). These findings imply that despite the hardiness of Cryptosporidium oocysts in the environment (Carey et al., 2004), historical C. hominis infections in New Zealand arise through importation and that the levels of sanitation and hand hygiene, as well as a low-density population, may be sufficient to prevent the establishment of endemic C. hominis infections. Similarly, improved hand hygiene on farms, especially during calving season may help control zoonotic C. parvum transmission in New Zealand (Thomas-Lopez et al., 2020).
C. parvum sequences were dominated by gp60 subtype families IIa and IId, which both occurred consistently throughout the study period, including during 2020. These C. parvum subtype families are primarily associated with ruminants and transmitted through them to humans (Mammeri et al., 2019; Nader et al., 2019). Both have been detected in cattle and sheep in New Zealand (Garcia-R et al., 2017) and evidence from overseas suggests that subtype family IId may be more prevalent in sheep and goat populations (Quílez et al., 2008), but further research is needed to identify if host-specific C. parvum subtype families occur in New Zealand's domestic animals. Overall, C. parvum transmission was not interrupted by Covid-19 control measures except perhaps by a small reduction in peak numbers during 2020 Q3 and Q4. Subtype family IIc is rare in New Zealand, with only six cases in our dataset. Previous research shows that unlike other C. parvum subtype families, IIc is an anthroponotic (human-adapted) and a potential emerging genotype (King et al., 2019; Nader et al., 2019). While no cases of subtype family IIc were detected after Covid-19 control measures, its rarity before Covid-19 prevents any strong conclusions from our data. Further investigation of gp60 diversity, i.e. trinucleotide repeat regions, may have revealed more details of the transmission patterns of cryptosporidiosis in New Zealand, but this would have been difficult to assess since we do not have descriptive epidemiology or exposure data for our cases. In C. parvum, previous research has demonstrated strong associations between animal contact and cryptosporidiosis, though human-to-human transmission is also possible (Chalmers et al., 2019). However, like C. hominis, we suggest that human to human C. parvum outbreaks in New Zealand are contained through basic sanitation measures.
Our study has limitations in the methodology of data collection stemming from significant changes to human behaviours during 2020 which may have influenced our results. The number of samples we have for 2020/2021 is lower than in previous years and adds uncertainty in the data. As well as potentially being due to a reduction in transmission and cases, since the symptoms of cryptosporidiosis do not closely match Covid-19, people with cryptosporidiosis may have been less likely to visit doctors immediately prior to, during and following Covid-19 restrictions compared with previous years. Furthermore, because symptoms usually resolve themselves without treatment, cryptosporidiosis is an under-reported disease throughout the developed world (Dreesman et al., 2007; Scallan et al., 2011). Despite being a notifiable disease in New Zealand it is likely that only a small proportion of cases are diagnosed, thus raising the potential for bias in reporting different Cryptosporidium species. The severity of cryptosporidiosis is impacted by many factors including host (concurrent infection, malnutrition, immunosuppression, age), environmental (dose of exposure) as well as species and genotype of Cryptosporidium (Weir, 2001). Of the species in our study, C. hominis is associated with more severe infections and higher shedding rates (Bushen et al., 2007), potentially making reporting and detection of C. hominis more likely than C. parvum. Once cryptosporidiosis cases are confirmed, the diagnostic laboratories send stool samples to our lab for genetic analyses, but this may not be nationally representative as evidenced by the lack of samples from outside the Auckland region during 2020. The reasons for this are unclear but may reflect a focus on essential services and over-stressed health systems not having time to send samples since the start of Covid-19.
In conclusion, our study revealed that nonpharmaceutical interventions greatly reduced C. hominis in Auckland, New Zealand. Since the March 2020 national lockdown, travel restrictions and mandatory visitor isolation have kept reported C. hominis numbers at zero. Ongoing monitoring of Cryptosporidium species and gp60 subtype families as international travel restrictions ease will provide further information on cryptosporidiosis in New Zealand. Finally, to further investigate the preliminary findings presented here we recommend future studies focus on descriptive epidemiology and exposure data for cases of human cryptosporidiosis in New Zealand.
Author contribution
DH conceived and designed the study. PO, AP and NV conducted data gathering. JG and MK performed analyses. MK wrote the original draft article and all coauthors contributed to editing.
Financial support
The authors thank the New Zealand Ministry of Health for funding. DH and MK acknowledge funding from the Royal Society Te Apārangi Rutherford Discovery Fellowship (MAU1701).
Conflicts of interest
The authors declare there are no conflicts of interest.
Ethical standards
Not applicable.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182021000974.
click here to view supplementary material
Data
Sequence data availability is outlined in Garcia-R et al. (2020b).
References
- Alves M, Xiao L, Sulaiman I, Lal AA, Matos O and Antunes F (2003) Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. Journal of Clinical Microbiology 41, 2744–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MG, Kvalsvig A and Verrall AJ (2020a) New Zealand's COVID-19 elimination strategy. The Medical Journal of Australia, 213, 198–200. e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MG, Wilson N and Anglemyer A (2020b) Successful elimination of COVID-19 transmission in New Zealand. New England Journal of Medicine 383, e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzid M, Hunter PR, Chalmers RM and Tyler KM (2013) Cryptosporidium pathogenicity and virulence. Clinical Microbiology Reviews 26, 115–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushen OY, Kohli A, Pinkerton RC, Dupnik K, Newman RD, Sears CL, Fayer R, Lima AAM and Guerrant RL (2007) Heavy cryptosporidial infections in children in northeast Brazil: comparison of Cryptosporidium hominis and Cryptosporidium parvum. Transactions of The Royal Society of Tropical Medicine and Hygiene 101, 378–384. [DOI] [PubMed] [Google Scholar]
- Carey CM, Lee H and Trevors JT (2004) Biology, persistence and detection of Cryptosporidium parvum and Cryptosporidium hominis oocyst. Water Research 38, 818–862. [DOI] [PubMed] [Google Scholar]
- Chalmers RM, Robinson G, Elwin K and Elson R (2019) Analysis of the Cryptosporidium spp. and gp60 subtypes linked to human outbreaks of cryptosporidiosis in England and Wales, 2009 to 2017. Parasites and Vectors 12, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreesman J, Villarroel-Conzales D, Cleves S, Reins H and Pulz M (2007) Regionally increased incidence of notified cryptosporidiosis cases due to different laboratory methods. Gesundheitswesen 69, 483–487. [DOI] [PubMed] [Google Scholar]
- Feltus DC, Giddings CW, Schneck BL, Monson T, Warshauer D and McEvoy JM (2006) Evidence supporting zoonotic transmission of Cryptosporidium spp. in Wisconsin. Journal of Clinical Microbiology 44, 4303–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y and Xiao L (2017) Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food and Waterborne Parasitology 8-9, 14–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Li N, Duan L and Xiao L (2009) Cryptosporidium genotype and subtype distribution in raw wastewater in Shanghai, China: evidence for possible unique Cryptosporidium hominis transmission. Journal of Clinical Microbiology 47, 153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke LM, Glöckner S, Dreier M and Lange B (2021) Impact of non-pharmaceutical interventions targeted at COVID-19 pandemic on influenza burden – a systematic review. Journal of Infection 82, 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-R JC and Hayman DT (2017) Evolutionary processes in populations of Cryptosporidium inferred from gp60 sequence data. Parasitology Research 116, 1855–1861. [DOI] [PubMed] [Google Scholar]
- Garcia-R JC, French N, Pita A, Velathanthiri N, Shrestha R and Hayman D (2017) Local and global genetic diversity of protozoan parasites: spatial distribution of Cryptosporidium and Giardia genotypes. PLoS Neglected Tropical Diseases 11, e0005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-R JC, Cox MP and Hayman DTS (2020a) Comparative genetic diversity of Cryptosporidium species causing human infections. Parasitology 147, 1532–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-R JC, Pita AB, Velathanthiri N, French NP and Hayman DTS (2020b) Species and genotypes causing human cryptosporidiosis in New Zealand. Parasitology Research 119, 2317–2326. [DOI] [PubMed] [Google Scholar]
- Garcia RJ and Hayman DT (2016) Origin of a major infectious disease in vertebrates: the timing of Cryptosporidium evolution and its hosts. Parasitology 143, 1683–1690. [DOI] [PubMed] [Google Scholar]
- Goñi P, Almagro-Nievas D, Cieloszyk J, Lóbez S, Navarro-Marí JM and Gutiérrez-Fernández J (2015) Cryptosporidiosis outbreak in a child day-care center caused by an unusual Cryptosporidium hominis subtype. Enfermedades Infecciosas y Microbiología Clínica, 33, 651–655. [DOI] [PubMed] [Google Scholar]
- Huang QS, Wood T, Jelley L, Jennings T, Jefferies S, Daniells K, Nesdale A, Dowell T, Turner N, Campbell-Stokes P, Balm M, Dobinson HC, Grant CC, James S, Aminisani N, Ralston J, Gunn W, Bocacao J, Danielewicz J, Moncrieff T, McNeill A, Lopez L, Waite B, Kiedrzynski T, Schrader H, Gray R, Cook K, Currin D, Engelbrecht C, Tapurau W, Emmerton L, Martin M, Baker MG, Taylor S, Trenholme A, Wong C, Lawrence S, McArthur C, Stanley A, Roberts S, Rahnama F, Bennett J, Mansell C, Dilcher M, Werno A, Grant J, van der Linden A, Youngblood B, Thomas PG, Webby RJ and Consortium NP (2021) Impact of the COVID-19 nonpharmaceutical interventions on influenza and other respiratory viral infections in New Zealand. Nature Communications 12, 1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King P, Tyler KM and Hunter PR (2019) Anthroponotic transmission of Cryptosporidium parvum predominates in countries with poorer sanitation: a systematic review and meta-analysis. Parasites and Vectors 12, 16–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake IR, Pearce J and Savill M (2008) The seasonality of human cryptosporidiosis in New Zealand. Epidemiology and infection 136, 1383–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learmonth JJ, Ionas G, Ebbett KA and Kwan ES (2004) Genetic characterization and transmission cycles of Cryptosporidium species isolated from humans in New Zealand. Applied and Environmental Microbiology 70, 3973–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch GJ and He Q (2011) Cryptosporidiosis − an overview. Journal of Biomedical Research, 25, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammeri M, Chevillot A, Chenafi I, Thomas M, Julien C, Vallée I, Polack B, Follet J and Adjou KT (2019) Molecular characterization of Cryptosporidium isolates from diarrheal dairy calves in France. Veterinary Parasitology: Regional Studies and Reports 18, 100323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader JL, Mathers TC, Ward BJ, Pachebat JA, Swain MT, Robinson G, Chalmers RM, Hunter PR, van Oosterhout C and Tyler KM (2019) Evolutionary genomics of anthroponosis in Cryptosporidium. Nature Microbiology 4, 826–836. [DOI] [PubMed] [Google Scholar]
- New Zealand Government (2020) New Zealand COVID-19 alert levels summary. https://covid19.govt.nz/assets/resources/tables/COVID-19-alert-levels-summary.pdf.
- O'Brien E, McInnes L and Ryan U (2008) Cryptosporidium GP60 genotypes from humans and domesticated animals in Australia, North America and Europe. Experimental Parasitology 118, 118–121. [DOI] [PubMed] [Google Scholar]
- Putignani L and Menichella D (2010) Global distribution, public health and clinical impact of the protozoan pathogen Cryptosporidium. Interdisciplinary Perspectives on Infectious Diseases 2010, 753512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quílez J, Torres E, Chalmers RM, Hadfield SJ, Del Cacho E and Sánchez-Acedo C (2008) Cryptosporidium genotypes and subtypes in lambs and goat kids in Spain. Applied and Environmental Microbiology 74, 6026–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert A (2020) Lessons from New Zealand's COVID-19 outbreak response. The Lancet Public Health 5, e569–e570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL and Griffin PM (2011) Foodborne illness acquired in the United States − major pathogens. Emerging Infectious Diseases 17, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura R, Prim N, Montemayor M, Valls ME and Muñoz C (2015) Predominant virulent IbA10G2 subtype of Cryptosporidium hominis in human isolates in Barcelona: a five-year study. PLoS ONE 10, e0121753–e0121753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Lopez D, Müller L, Vestergaard LS, Christoffersen M, Andersen A-M, Jokelainen P, Agerholm JS and Stensvold CR (2020) Veterinary students have a higher risk of contracting cryptosporidiosis when calves with high fecal Cryptosporidium loads are used for fetotomy exercises. Applied and Environmental Microbiology 86, e01250–e01220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron LS, Ferrari BC and Power ML (2009) Glycoprotein 60 diversity in C. hominis and C. parvum causing human cryptosporidiosis in NSW, Australia. Experimental Parasitology 122, 124–127. [DOI] [PubMed] [Google Scholar]
- Weir E (2001) The cryptic nature of cryptosporidiosis. Canadian Medical Association Journal 164, 1743–1743. [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Fayer R, Ryan U and Upton SJ (2004) Cryptosporidium taxonomy: recent advances and implications for public health. Clinical Microbiology Reviews 17, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Xiao X, Gu X, Liang D, Cao T, Mou J, Huang C, Chen L and Liu J (2021) Surveillance of common respiratory infections during COVID-19 pandemic demonstrates preventive effectiveness of non-pharmaceutical interventions. International Journal of Infectious Diseases. 105, 442–447. doi: 10.1016/j.ijid.2021.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0031182021000974.
click here to view supplementary material
Data Availability Statement
Sequence data availability is outlined in Garcia-R et al. (2020b).