Summary
Inhibitors of cyclin-dependent kinases 4 and 6 (CDK4/6i) delay progression of metastatic breast cancer. However, complete responses are uncommon and tumors eventually relapse. Here we show that CDK4/6i can enhance efficacy of T cell-based therapies, such as adoptive T cell transfer or T cell-activating antibodies anti-OX40/anti-4-1BB, in murine breast cancer models. This effect is driven by the induction of chemokines CCL5, CXCL9 and CXCL10 in CDK4/6i-treated tumor cells facilitating recruitment of activated CD8+ T cells, but not Tregs, into the tumor. Mechanistically, chemokine induction is associated with metabolic stress that CDK4/6i treatment induces in breast cancer cells. Despite the cell cycle arrest, CDK4/6i-treated cells retain high metabolic activity driven by deregulated PI3K/mTOR pathway. This causes cell hypertrophy and increases mitochondrial content/activity associated with oxidative stress and inflammatory stress response. Our findings uncover a link between tumor metabolic vulnerabilities and anti-tumor immunity and support further development of CDK4/6i and immunotherapy combinations.
Introduction
CDK4 and CDK6 are cell cycle kinases that work in complex with cyclin D1 to phosphorylate tumor suppressor RB. Phosphorylation inhibits RB activity leading to its dissociation from E2F transcription factors. Free E2Fs, in turn, transactivate genes involved in DNA replication and cell cycle progression. Inhibitors of CDK4/6 (CDK4/6i), such as palbociclib, ribociclib, and abemaciclib, control tumor growth by blocking G1-to-S cell cycle transition in cancer cells. CDK4/6i are used for treatment of recurrent ER-positive, HER2-negative metastatic breast cancer (Cristofanilli et al., 2016; Turner et al., 2018).
Interestingly, several studies indicated that, in addition to direct effect on tumor cells, CDK4/6 inhibitors may influence immune cells in the tumor microenvironment (Petroni et al., 2020; Teh and Aplin, 2019). This includes suppression of regulatory T cell proliferation (Goel et al., 2017; Goel et al., 2016) and enhanced activation of tumor-infiltrating T cells (Deng et al., 2018; Schaer et al., 2018). Furthermore, CDK4/6i treatment may affect tumor cells in a way that may facilitate their recognition by the immune system (Goel et al., 2017). Accordingly, CDK4/6i was shown to augment immune checkpoint blockade therapy targeting PD-1/PD-L1 in mouse models (Deng et al., 2018; Goel et al., 2017; Schaer et al., 2018; Zhang et al., 2018).
These data suggest that CDK4/6i can promote activation of tumor-infiltrating T cells (TILs). However, breast cancer tumors often have low levels of TILs, which is referred to as “immunologically cold” tumor microenvironment. “Cold” tumors have tendency to respond poorly to anti-PD-1 therapy (Cristescu et al., 2018; Tumeh et al., 2014). Further, breast cancer patients with low TILs often have worse survival and have limited response to chemotherapy compared to patients with tumors that are highly infiltrated with T cells (Dushyanthen et al., 2015). Therefore, there is an unmet clinical need for therapeutic approaches that can improve recruitment of T cells into the tumor.
Trafficking, localization, and positioning of T cells is regulated by small proteins called chemokines (Chow and Luster, 2014). Chemokines secreted by tumor cells and by non-malignant cells within the tumor microenvironment (TME) play important role in shaping tumor immune landscape (Vilgelm and Richmond, 2019; Wellenstein and de Visser, 2018). A meta-analysis of 5,953 cancer specimens demonstrated positive correlation of CXCL9, CXCL10, and CXCL11 expression with density of tumor-infiltrating T and NK cells (Stoll et al., 2018). Similarly, CCR5 ligands, CCL4, and CCL5, were shown to facilitate recruitment of several anti-tumor immune cell types into the tumor including T cells and BATF3-dependent dendritic cells (Bottcher et al., 2018; Spranger et al., 2017; Vilgelm et al., 2016a). And vice versa, low expression of CCR5 ligands (CCL3, CCL4, CCL5) and CXCR3 ligands (CXCL9 and CXCL10) has been linked with limited T cells infiltration (Harlin et al., 2009). Importantly, chemokine expression is associated with effectiveness of immune checkpoint therapy (e.g., anti-PD1 and anti-PD-L1), as reviewed by Nagarsheth et al(Ayers et al., 2017; Nagarsheth et al., 2017). Here we investigated the role of chemokines in CDK4/6i-mediated modulation of tumor immune microenvironment.
Results
CDK4/6 inhibitor increases levels of T cells in mammary tumors
We used MMTV-PYMT murine mammary carcinoma model to study the effect of CDK4/6i palbociclib on tumor immune microenvironment. Transgenic MMTV-PYMT mice develop aggressive metastatic mammary tumors between 8–12 weeks of age (Guy et al., 1992) that recapitulate luminal BC subtype and express ERα and progesterone receptor (PR) similarly to human hormone receptor-positive luminal BC (Lin et al., 2003). Similar to human BC, PYMT tumors are not highly immunogenic based on virtually identical survival curves of PYMT mice on wild type and T cell-deficient (Rag−/−) backgrounds (DeNardo et al., 2009). Here we utilized a transplantable version of PYMT model where PYMT tumor cells are inoculated into 4th mammary fat pads of wild type female C57Bl/6 mice. Key experiments were repeated using transgenic MMTV-PYMT mice (FVB background) to determine whether our findings are consistent in distinct model types (transplanted vs spontaneous) and genetic backgrounds (C57Bl/6 vs FVB).
We first verified that CDK4/6i palbociclib was effective in PYMT model. Transplanted PYMT tumors grew slower in palbociclib-treated mice as compared to vehicle-treated animals (Fig. 1A). In agreement with cytostatic activity of CDK4/6i (Klein et al., 2018; Knudsen and Witkiewicz, 2017), palbociclib inhibited proliferation of tumor cells based on decreased Ki67 expression (Fig. 1B, C). Similarly, the growth of spontaneous tumors arising in mammary glands of PYMT-FVB transgenic mice was attenuated by palbociclib (Fig. S1A). Final tumor burden determined as combined weight of all tumor lesions per mouse was reduced in palbociclib group (Fig. S1B).
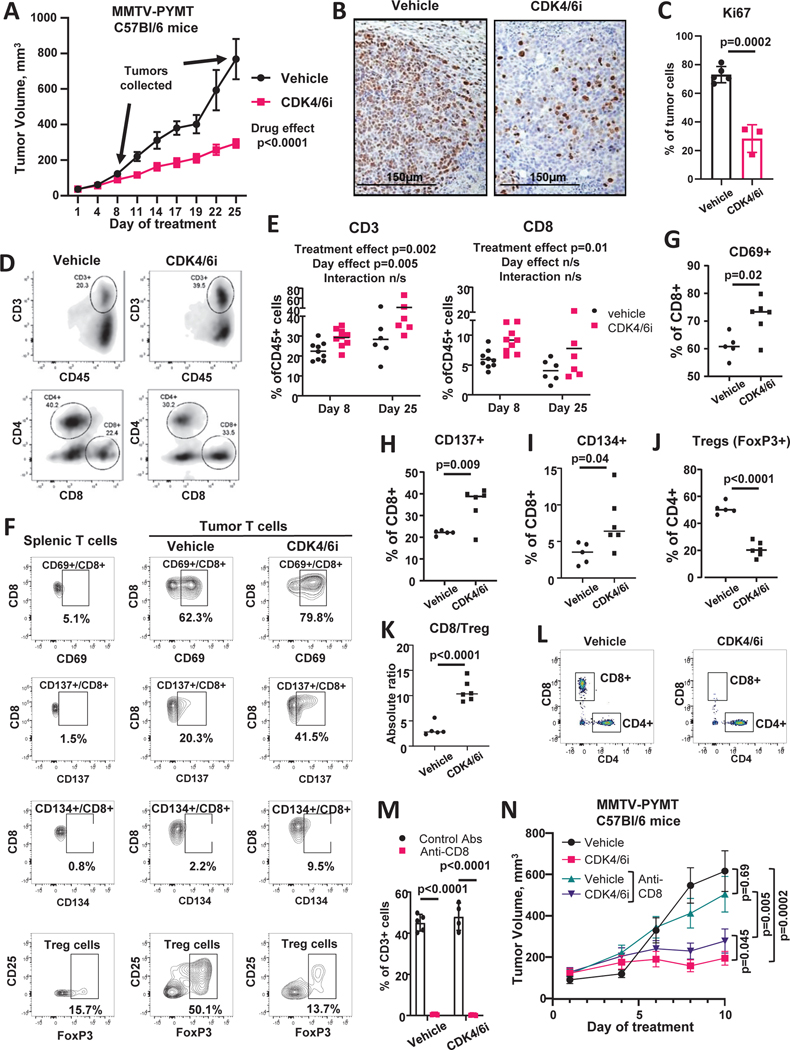
Figure 1. CDK4/6i treatment increases number and activity of T cells in mouse model of breast cancer.
A. Growth of PYMT tumors in female C57Bl/6 mice treated twice daily with 100 mg/kg CDK4/6i palbociclib or vehicle (n=20 mice/group, 10 mice/group sacrificed at day 8, mixed model statistics, data are represented as mean ± SEM). Arrows indicate days of tumor collection for immunophenotyping. B. Ki67 IHC staining in tumors from mice treated as in A. C. Quantified percentages on Ki67+ cells in tumors from mice treated as in A (n=3–5 individual tumors, t-test, data are presented as individual values and mean ± SD). D. Representative plots from flow cytometry analysis of tumor-infiltrating immune cells from mice shown in A. Tumors were collected at day 8 and 25. Cells were gated on live CD45+ cells. E. Percentages of CD3+ and CD8+ cells in tumors from mice shown in A (n=6–9 tumors/group, 2-way ANOVA). F. Representative plots from flow cytometry analysis of phenotypes of tumor-infiltrating T cells from mice treated twice daily with vehicle or 100 mg/kg palbociclib for 10 days. G-I. Percentages on CD69+ (G), CD137+ (H), and CD134+ (I) CD8 T cells from the experiment shown in F (n=5–6 tumors/group, t-test). Cells were gated on live/CD45+/CD3+/CD8+ population. J. Percentages of tumor-infiltrating Treg cells from the experiment in F (n=5–6 tumors/group, t-test). Cells were gated on live/CD45+/CD3+/CD4+ population. K. CD8/Treg cell ratios from the experiment shown in F (n=5–6 tumors/group, t-test). E-K. Data are presented as individual values and mean. L-N. Female C57Bl/6 mice bearing PYMT tumors were treated twice daily with vehicle or 100 mg/kg CDK4/6i palbociclib. Mice were injected IP with 100μg of CD8-depleting or isotype control antibodies on day 2 post palbociclib treatment initiation and every 2 days thereafter. Tumors were collected at day 10. There were 6 mice (12 tumors) in each treatment group. L. Representative plots of flow cytometry analysis of CD8+ and CD4+ T cells in blood. Cells were gated on live/CD45+/CD3+ population. M. Percentages of CD8+ cells of total CD3+ T cells in blood (n=4–5 mice/group, t-test with Holm-Sidak correction, data are presented as individual values and mean ± SD). N. Tumor growth over time (n=12 tumors/group, mixed model, data are represented as mean ± SEM). See also Fig. S1, Fig. S2, and Table S1.
Flow cytometry immunophenotyping was done to determine the effect of CDK4/6i treatment on the composition of tumor immune microenvironment. To assess the temporal variations of the CDK4/6i influence on immune profiles, as well as the effect of tumor size, tumors were analyzed at early (8 days, comparable size in vehicle and CDK4/6i groups) and late (25 days, reduced size in CDK4/6i group) time points (arrows on Fig. 1A indicate time points of tumor collection). Complete results are shown in Suppl. Table 1. The percentages of T cells were significantly increased (p=0.002) in tumors from CDK4/6i-treated mice as compared to vehicle group (Fig. 1D, E, Suppl. Table 1) across both time points. There was no significant interaction between treatment effect and day effect on T cell levels suggesting that CDK4/6i-mediated increase of T cells was consistent across tested time points and was not directly influenced by tumor size. Among T cells, CD8+ T cells accumulated in CDK4/6i-treated tumors (Fig. 1D, E, Suppl. Table 1). We also detected an increase of B cells after palbociclib treatment, however the overall levels of B cells were low. Several immune populations were differentially affected at early and late analysis time points, which may reflect the role of time and size in shaping of tumor immune landscape. For instance, the percentages of NK and NKT significantly decreased from early to late time point, however they were not significantly affected by palbociclib. There were also cell populations that were affected by palbociclib more prominently as the time went by (treatment and day interaction), such as tumor-associated macrophages, that were not dramatically different at day 8, but were reduced at day 25 (Table S1).
The increase of tumor-infiltrating T cells, and specifically, CD8+ T cells, by CDK4/6i treatment was also detected in spontaneous tumors from transgenic PYMT FVB mice (Fig. S1C), indicating that the effect of CDK4/6i on T cells in consistent in distinct models and genetic backgrounds. Notably, T cells were infiltrating throughout the tumor in both injected (Fig. S2A), and spontaneous (Fig. S1D) models. The density of T cells varied across tumor sections, with “hot” T cell-dense areas present in some of the tested sections of CDK4/6i-treated tumors (Fig. S2A, Fig. S1D).
We also characterized the effect of CDK4/6i on phenotype of tumor-infiltrating T cells using spectral cytometry. Higher fraction of tumor-infiltrating CD8+ T cells expressed activation marker CD69 in CDK4/6i-treated mice as compared to vehicle cohort (Fig. 1F, G). Furthermore, an increase in the percentages of CD8+ T cells expressing co-stimulatory receptors CD137 (4–1BB) and CD134 (OX40) was observed after CDK4/6i treatment (Fig. 1F, H, I). CDK4/6i decreased the percentages of CD4 T cells expressing markers of T regulatory cells (Treg), such as FoxP3+ and FoxP3+/CD25+ (Fig. 1F, J). Consequently, the CD8/Treg ratio was increased by CDK4/6i treatment (Fig. 1K). These data indicate that T cell infiltrate in CDK4/6i-treated tumors displays more prominent anti-tumor phenotype compare to vehicle cohort. Dimension reduction analysis using tSNE showed a decrease of the Treg cell cluster and an increase of active CD8+ T cell cluster characterized by high expression of CD69, CD137, and CD134 in CDK4/6i-treated tumors as compared to controls (Fig. S2B).
Of note, while PD-L1 expression on myeloid cells was not significantly affected by CDK4/6i treatment (Table S1), we observed an increase in the percentages of PD-L1 expressing CD45-negative tumor cells from 1.3% in vehicle group to 2.7% in CDK4/6i treatment group (Fig. S2C, p=0.005). Moreover, two out of six tumors in CDK4/6i treatment group had a proportion of CD45-negative tumor cells that lost MHCI expression (Fig. S2D). This effect may indicate compensatory feedback response to increased T cell infiltrate in CDK4/6i-treated tumors.
We next tested whether CD8+ T cells contributed to the overall anti-tumor activity of CDK4/6i. PYMT tumor bearing mice were treated with CDK4/6i or vehicle while also receiving injections of CD8-depleting or isotype control antibody. Levels of peripheral CD8 T cells were prominently reduced by anti-CD8 treatment (Fig. 1L, M). CDK4/6i inhibited tumor growth in both control and CD8+-depleted mice. However, tumor volumes were smaller in CDK4/6i-treated mice that received isotype control antibodies as compared to CD8-depleted mice (Fig. 1N). These data suggest that anti-tumor activity of CDK4/6i was partially dependent on CD8 T cells.
CDK4/6i treatment augments T cell-based therapies
We next tested if increased levels of activated T cells in CDK4/6i-treated tumors can be harnessed for cancer immunotherapy. Based on the increased expression of co-stimulatory receptors OX40 and 4–1BB on T cells from CDK4/6i-treated tumors, we hypothesized that CDK4/6i will sensitize tumors to antibodies that activate these receptors. Of note, agonistic OX-40 and 4–1BB antibodies are currently in clinical development (NCT03364348, NCT03217747). Mice inoculated with PYMT cells were treated with vehicle or palbociclib in the presence or absence of mouse-specific OX-40 and 4–1BB agonistic antibodies. No significant inhibition of tumor growth was evident after OX40/4–1BB stimulation in vehicle-treated mice. In contrast, combining palbociclib with OX40/4–1BB stimulation further reduced tumor growth as compared to palbociclib only or vehicle treatment cohorts (Fig. 2A). Mice treated with palbociclib and anti-OX40/4–1BB displayed lowest final tumor weight at sacrifice (Fig. 2B). We also tested whether combined palbociclib and anti-OX40/4–1BB therapy provided protection from tumor re-growth. PYMT-bearing mice were treated with palbociclib and OX-40/4–1BB antibodies for 25 days and monitored until their tumors reached the end point size (15 mm in diameter) or became perforated. The fraction of surviving mice was numerically higher in palbociclib and anti-OX40/4–1BB combination treatment cohort, however the differences were not statistically significant (p=0.098) (Fig. 2C). Based on this data we concluded that combining palbociclib and endogenous T cell stimulation provides a modest improvement of anti-tumor effect over single agent palbociclib.
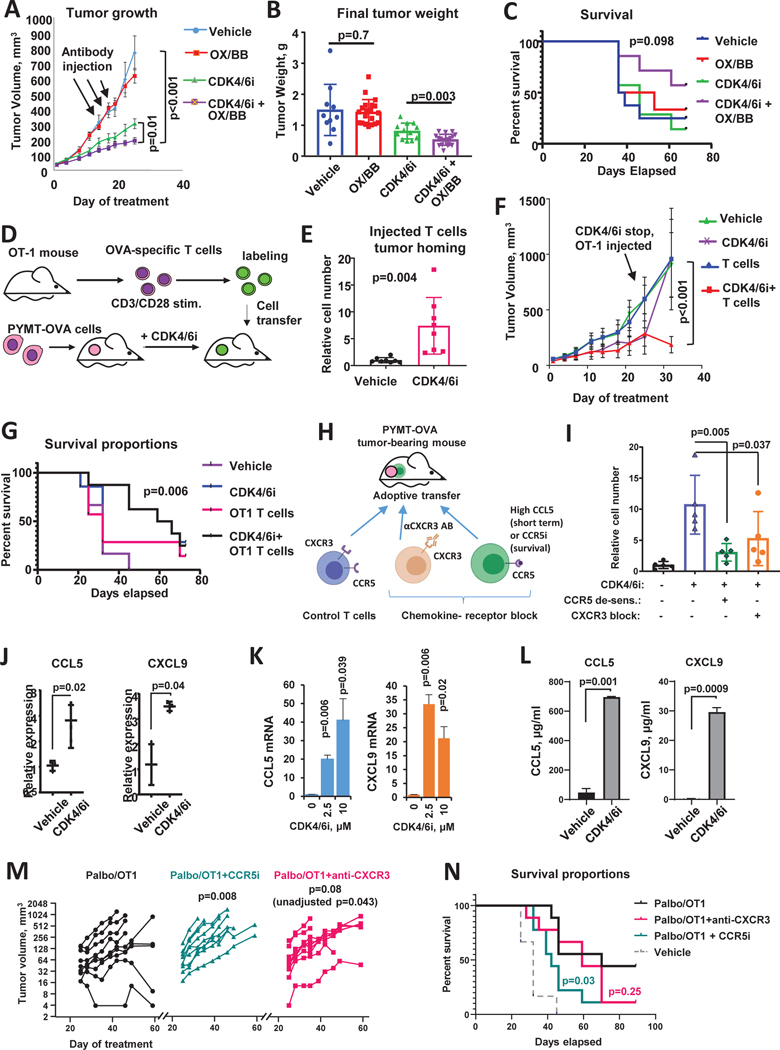
Figure 2. CDK4/6i improves immunotherapy response and promotes chemokine-mediated T cell tumor homing.
A. Tumor growth in C57Bl/6 mice implanted with OVA-expressing PYMT tumor cells and treated with CDK4/6i palbociclib (100mg/kg, wice a day), mix of agonistic antibodies for OX-40 and 4–1BB (OX/BB, 100μg each per mouse, once every 3 days), combination of both treatments, or vehicles for 25 days (n=8–10 mice, two tumors per mouse, mixed model statistics, data are represented as mean ± SEM). B. Final weight of tumors from experiment shown in A (n=10–20 tumors, one-way ANOVA, data are presented as individual values and mean ± SD). C. Survival analysis in C57Bl/6 mice implanted with OVA-expressing PYMT mammary tumor cells and treated as in A for 25 days and then followed till day 68 (n=10 mice/group, Gehan-Breslow-Wilcoxon test). D. Schematic of T cell recruitment experiment. OVA-expressing PYMT cells were implanted in 4th mammary fat pad on female C57Bl/6 mice. Mice were treated once a day with 100mg/kg palbociclib or vehicle for 2 weeks and then received IV injection of OVA-specific T cells. T cells derived from OT-1 mice were activated with anti-CD3/CD28 antibodies, and fluorescently labeled ex vivo. Tumors were collected 2 hours after cell transfer. E. Numbers of labeled T cells in tumors of C57Bl/6 mice from experiment described in D (n=8 tumors/group, t-test, data are presented as individual values and mean ± SD). F. Tumor growth in C57Bl/6 mice implanted with OVA-expressing PYMT mammary tumor cells and treated once a day with 100mg/kg CDK4/6i palbociclib or vehicle for 3 weeks followed by injection of OT-1 T cells or saline. Mice were followed until tumors reached end point size or became perforated (n=7 mice/group, mixed model, data are represented as mean ± SEM). G. Survival analysis of mice from in F. OT1 injection was given at day 25 (n=7, log-rank (Mantel-Cox) statistical comparison between Vehicle and CDK4/6i+OT1 groups is shown). Experiment was repeated with consistent results. H. Scheme of an in vivo T cell recruitment experiment. C57Bl/6 mice were inoculated with PYMT-OVA cells. Mice were treated with vehicle or CDK4/6i palbociclib at 100 mg/kg for 25 days followed by IV injection of activated OT-1 T cells. To block CCR5 and CXCR3-mediated migration, OT-1 T cells were treated with high dose of CCL5 or CXCR3-blocking antibody prior to transfer. T cells were labeled with cell tracker prior to injection into mice. I. Accumulation of fluorescent transferred T cells in PYMT tumors from experiment described in L (n=5 mice/group, one-way ANOVA, data are presented as individual values and mean ± SD). J. Real-time PCR analysis of CCL5 and CXCL10 mRNA in PYMT tumors from mice treated as in A (n=3 tumors, t-test after log transformation, data are presented as individual values and mean ± SD). K. CCL5 and CXCL9 mRNA expression in PYMT tumor cells treated with vehicle or CDK4/6i (palbociclib, 1μM) for 3 days in vitro (n=2, t-test, data are represented as mean ± SD). L. ELISA analysis of indicated chemokines in the conditioned medium of PYMT cells treated with vehicle or 1μm palbociclib for 5 days (n=2, t-test, data are represented as mean ± SD). M. Tumor volume change overtime with and without chemokine receptor blockade. Female C57Bl/6 mice were implanted with PYMT-OVA tumors and treated with palbociclib for 25 days and injected with activated OT-1 T cells (treatment scheduler was same as in F). A subgroup of mice received CCR5i maraviroc (10 mg/kg, once daily oral gavage) or CXCR3-blocking antibody (200 μM per mouse, IV injection every 3 days) on days 25–42. Mixed model with Dunnet’s post-test was used for statistical comparison of tumor growth between control CDK4/6i+OT-1 group and chemokine-receptor-inhibited groups (n=9 mice per group, individual data presented). p-value with and without adjustment for multiple comparison is sown for CXCR3-blockade group. N. Survival curves from experiment in M (n=9, log-rank (Mantel-Cox) statistical comparison between CDK4/6i+OT1 and chemokine-receptor inhibited groups).
We next evaluated if CDK4/6i can augment anti-tumor activity of adoptive cell therapy (ACT). OT-1-OVA model was used to recapitulate ACT conditions. PYMT tumor cell engineered to express model antigen OVA were injected into female C57Bl/6 mice. When tumors formed, mice were treated with vehicle or palbociclib for two weeks and then received injections of OVA-specific T cells derived from spleens and lymph nodes of OT-1 mice and activated ex vivo by CD3/CD28 stimulation (Fig. 2D). We first tested the recruitment of transferred T cells into vehicle and CDK4/6i-treated tumors. OT-1 T cells were fluorescently tagged prior to injection into mice to distinguish them from endogenous T cells. Tumors were analyzed 2 hours post injection (Fig. 2D). We found that tumors from CDK4/6i-treated mice recruited more T cells as compared to tumors in vehicle-treated mice (Fig. 2E). This suggests that CDK4/6i promotes recruitment of transferred T cells into the tumor.
We next investigated the impact of CDK4/6i on ACT efficacy. Palbociclib and vehicle pre-treated mice that received either mock injection or activated OT-1 T cells were monitored until their tumors reached end-point size or became perforated. Based on the first time point after T cell administration, palbociclib-treated mice that received mock injection had smaller tumors that accelerated immediately after treatment discontinuation. In contrast, tumors from palbociclib-treated mice that received OT-1 T cells remained slow growing (Fig. 2F). As a result, mice in combination treatment group exhibited improved survival as compared to control vehicle-treated mice (Fig. 2G, compare black and purple lines). No significant survival benefit was observed in OT-1 only or CDK4/6i only treatment groups (Fig. 2G). These data indicate that CDK4/6i pre-treatment improved recruitment of exogenous T cells into the tumor and augmented the outcome of ACT.
Chemokine receptors CCR5 and CXCR3 facilitate CDK4/6i-mediated T cell recruitment
Immune cell trafficking and localization are regulated by chemokine-chemokine receptor interactions. Activated T cells express chemokine receptors CCR5 and CXCR3 that recognize chemokines CCL5 and CXCL9, 10, and 11, respectively, secreted at the sites of inflammation (Griffith et al., 2014). Therefore, we investigated if CCR5 and CXCR3 facilitated recruitment of T cells into CDK4/6i-treated tumors by disabling these chemokine receptors on T cells prior to injection. Neutralizing antibody was used to block CXCR3. Incubation with high doses of CCL5 was used to inactivate CCR5. The latter causes internalization of CCR5 that persists for at least 4 hours after ligand withdrawal (Escola et al., 2010). T cells were fluorescently labeled and transferred into vehicle and CDK4/6i-treated C57Bl/6 mice bearing PYMT-OVA tumors (Fig. 2H). Both CCL5 de-sensitization and CXCR3 blockade abrogated homing of transferred T cells into CDK4/6i-treated tumors (Fig. 2I).
Based on these findings we next asked whether CDK4/6i treatment increased the levels of CCR5 and CXCR3 ligands at the tumor site. Indeed, we found that tumors in CDK4/6i-treated mice expressed higher levels of CCR5 ligand CCL5 and CXCR3 ligand CXCL9 as compared to controls (Fig. 2J). To determine whether malignant cells, rather than other cell types found within the TME, upregulated chemokines in response to CDK4/6i, we treated PYMT tumors cells with CDK4/6i palbociclib or vehicle in vitro. CDK4/6i treatment upregulated CCL5 and CXCL9 in PYMT cells on mRNA level (Fig. 2K) and increased their secretion into the culture medium. (Fig. 2L). These data suggest that CDK4/6i treatment promoted secretion of inflammatory chemokines by tumor cell resulting in improved CCR5 and CXCR3-mediated homing of T cells into the tumor.
To determine if chemokine-chemokine receptor interactions played role in CDK4/6i-mediated improvement of ACT outcome, we administered OT-1 T cells to PYMT-OVA tumor-bearing. Mice were pre-treated with CDK4/6i for 3 weeks (Fig. 2F, G) and received treatments blocking CCR5 and CXCR3-mediated trafficking. To block CCR5-CCL5 interaction, maraviroc was used, which is a small molecule antagonist of CCR5 (CCR5i). To inhibit CXCR3 function, CXCR3-neutralizing antibody was administered in vivo. Control mice received CDK4/6i and ACT only. Tumors in mice receiving chemokine receptor blockade exhibited upward growth patterns, while several tumors in control cohort decreased or stabilized after ACT administration. The overall growth was significantly accelerated in maraviroc-treated mice (p=0.008, Fig. 2M). CXCR3-inhibited mice exhibited similar trend, however after adjusting the p-value for multiple comparisons the effect was not significant (adj. p=0.08., raw p=0.043, Fig. 2M). Notably, CCL5i-co-treated mice exhibited worse survival in comparison to control (chemokine receptor-functional) mice (Fig. 2N). The effect of anti-CXCR3-treatement on survival was not significant. Of note, 4 out of 9 mice survived over 90 days after ACT administration in control group, as compared to 1 out 9 mice in chemokine receptor-inhibited groups (Fig. 2N). These data suggest that CDK4/6i-mediated improvement of ACT efficacy is, at least in part, dependent on chemokine receptors, especially CCR5.
CDK4/6i-treated human breast cancer cells produce T cell-recruiting chemokines
We next investigated whether our mouse model findings were relevant to human breast cancer. To test if CDK4/6i induced inflammatory chemokines in human model, ER+ breast cancer cells MCF7 were treated with CDK4/6i or vehicle control and the levels of secreted chemokines were measured in the conditioned media using chemokine array. Several chemokines were upregulated in CDK4/6i-treated cells, including CCR5 ligand CCL5 and CXCR3 ligand CXCL10 (Fig. 3A). Increased chemokine secretion was associated with increased expression of chemokine mRNA indicating upregulation on a transcriptional level (Fig. 3B). Similar to CXCL10, expression of two other CXCR3 ligands, CXCL9 and CXCL11, was increased after palbociclib treatment. We confirmed that 2 other structurally-distinct CDK4/6 inhibitors, ribociclib and abemaciclib, also upregulated chemokines in human breast cancer cells. In contrast, treatment with roscovitin that inhibits CDK1/2/5/7/9, but not CDK4 or CDK6, showed minimal effect on chemokine expression (Fig. 3B). This suggest that chemokine induction is an “on-target” effect of CDK4/6 inhibition. Chemokine induction was dose-dependent at the 3-day treatment time-point with the level of induction increasing progressively within the 0.1 μM to 10 μM range of palbociclib concentrations (Fig. 3C). Treatment was not toxic at these conditions based on propidium iodide cytotoxicity assay (Fig. S3A, B).
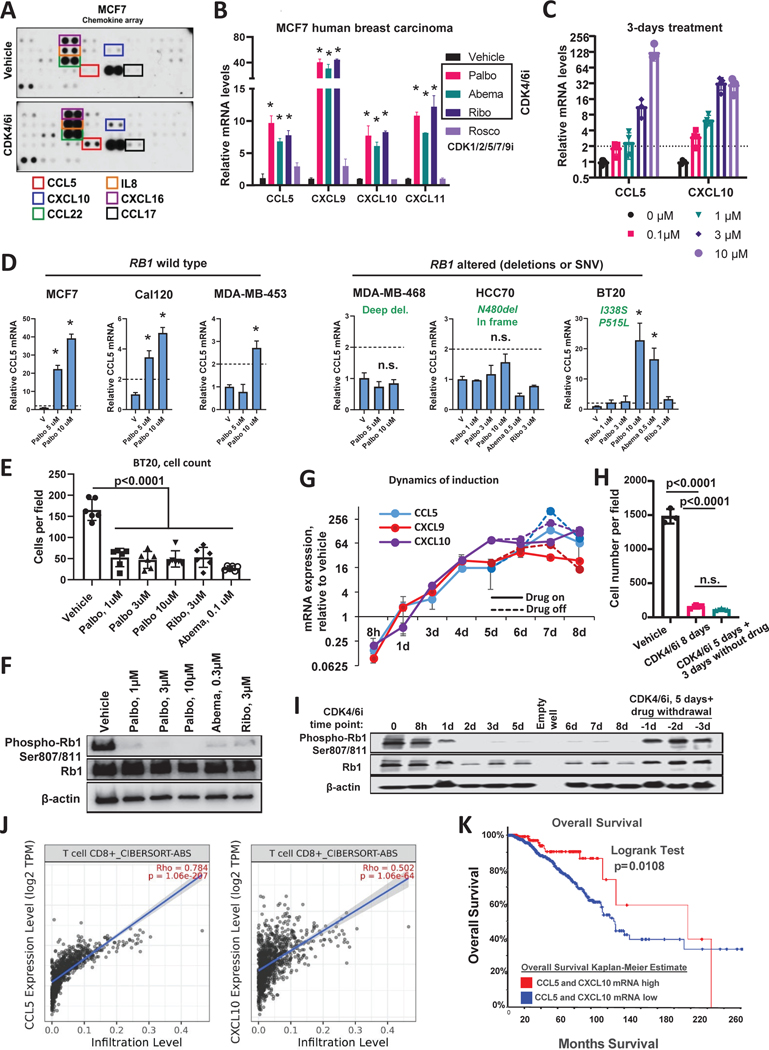
Figure 3. CDK4/6i-treated breast cancer cells secrete chemokines associated with T cell infiltrate and favorable prognosis in breast cancer patients.
A. Results of the cytokine array in conditioned media from MCF7 cells treated for 4 days with 1μM palbociclib or vehicle control. Proteins elevated after palbociclib (CDK4/6i) treatment are indicated with frames. B. Real-Time PCR detection of CCL5, CXCL9, 10, 11 mRNA in MCF7 cells treated with 1μM palbociclib, 3μM ribociclib, 0.2μM abemaciclib for 4 days. As a non-CDK4/6i control, cells were treated with 10μM roscovitin (n=3, t-test, data are represented as mean ± SD, *p<0.05). C. Results of the Real-Time PCR analysis of CCL5, CXCL9, and CXCL10 mRNA in MCF7 cells treated with palbociclib for three days at indicated concentrations (n=3, t-test, *p<0.05, data are presented as individual values, mean ± SD). D. Real-Time PCR analysis of CCL5 mRNA induction by palbociclib in six indicated cells lines with wild type or altered RB1 gene (n=2–3, one-way ANOVA, (n=3, t-test, data are represented as mean ± SD, *p<0.05). E. Numbers of BT20 cells after 5 days of treatment with indicated CDK4/6 inhibitors or vehicle. Cells were stained with crystal violet and imaged in transmitted light (n=6, one-way ANOVA, data are presented as individual values and mean ± SD). F. Western blot analysis of the inhibition of RB1 hyper-phosphorylation in BT20 cells treated as described in E. G. Real-time PCR detection of CCL5, CXCL9, and CXCL10 in MCF7 cells treated with 1μM palbociclib. Samples were collected at indicated time points ranging from 8 hours to 8 days. Solid lines represent samples that were continuously treated with palbociclib. Punctate lines indicate samples where palbociclib was removed from culture media after day 5 of treatment. H. Numbers of crystal violet stained MCF7 cells treated as described in G. Cells were treated with 1 μM palbociclib for 8 days continuously or treated for 5 days followed by drug wash out and additional 3 days of culture without drug. Control cells were treated with vehicle for 8 days (n=3, one-way ANOVA, data are presented as individual values and mean ± SD). I. Western blot analysis of RB phosphorylation in MCF7 cells treated as described in G. J. Correlation of CCL5 (right panel) or CXCL10 (left panel) mRNA with CD8+ T cell infiltrate computed by CIBERSORT in TCGA set of luminal A breast cancer tumors (N=1100). K. Comparison of overall survival in breast cancer patients with high expression of either CCL5, CXCL10 or both in their tumors (z score>+1 (median+1SD)) as compared to the rest of the cases in TCGA breast cancer dataset (n=1093, log-rank test). See also Fig. S3 and S4.
Inactivating mutations/deletions of RB1 gene are associated with CDK4/6i resistance (Alvarez-Fernandez and Malumbres, 2020). Therefore, we tested if chemokine induction by palbociclib was linked with RB1 status. We tested the induction of CCL5 by palbociclib in a panel of cell lines with wild type and deleted/mutated RB1. Information on RB1status was obtained from cBIO portal (Cancer Cell Line Encyclopedia (CCLE) dataset) (Cerami et al., 2012). Two or greater fold induction of CCL5 was detected in three tested RB1 wild type lines: MCF7, Cal120 and MDA-MB-453 (Fig. 3D). Two out of three RB1-altered lines showed no significant CCL5 induction after CDK4/6i treatment. Surprisingly, BT20 cells that have two amino acid substitutions in RB1 protein, I338S and P515L, exhibited strong induction of CCL5 mRNA after treatment with palbociclib. This induction was likely on target because another CDK4/6i abemaciclib also induced CCL5 in BT20 cells (Fig. 3D, right panel). To investigate this paradoxical activity further, we tested whether BT20 responded to CDK4/6i. Indeed, we found that cell numbers were reduced in cultures of BT20 cells treated with different CDK4/6 inhibitors as compared to vehicle treated cells (Fig. 3E). Furthermore, western blot analysis indicated reduction of the CDK4/6-mediated inhibitory RB1 phosphorylation by CDK4/6i treatment (Fig. 3F). Based on these data it is likely that the specific RB1 genetic alterations found in BT20 cells were not strongly detrimental to RB protein function, which is why these cells responded to CDK4/6i. This conclusion is consistent with data from other groups showing that BT20 cells are responsive to palbociclib (Finn et al., 2009). Collectively these data suggest that CDK4/6i-mediated chemokine induction is linked with RB inhibition.
We also showed that the induction of CCL5 and CXCL9/10 in response to CDK4/6i treatment was time-dependent. The levels of induction increased over time reaching a plateau around day 5 of continuous treatment (Fig. 3G). Notably, cells that were treated with palbociclib for 5 days continued to produce high levels of chemokines when drug was removed from the culture media (Fig. 3G, punctate lines). The numbers of cells were comparable after “5 days on + 3 days off” and “8 days on” treatments (Fig. 3H), suggesting that palbociclib-induced cell cycle arrest was not immediately reversible. In agreement with this conclusion, we detected an increased percentages of cells positive for senescence-associated β galactosidase (SA-βGal) after 5 days of CDK4/6i treatment (Fig. S4A, B), which is a marker of cellular senescence. Senescence is a type of persistent growth arrest that occurs in response to a variety of damaging conditions and is associated with increased secretion of many factors, including pro-inflammatory chemokines CCL5 and CXCL9,10,11 (Vilgelm et al., 2019). In contrast to cell cycle arrest, the levels of inhibitory RB1 phosphorylation showed full recovery to the baseline levels after palbociclib withdrawal, indicating reactivation of CDK4 and CDK6 (Fig. 3I). Altogether these data suggest that, although the effect of palbociclib on CDK4/6 activity was reversible, the damage done to the cell during the 5-day treatment was not. Since chemokine expression was also not reversible, we concluded that chemokine induction was not directly dependent in CDK4/6 activity, but rather was associated with cell damage induced by CDK4/6i treatment.
Chemokine expression is a favorable prognostic factor in breast cancer patients
Our results showed that CDK4/6i induced chemokines in breast cancer cells. Therefore, we sought to understand how elevated chemokine expression may impact patients with breast cancer. TIMER2.0 analysis of a TCGA dataset that included over a thousand of luminal A breast cancer samples (Li et al., 2020) demonstrated strong association of CCL5 and CXCL10 expression with CD8 T cell infiltrate estimated by CIBERSORT algorithm (Fig. 3J). Furthermore, breast cancer patients with elevated expression of CCL5 and CXCL10 in their tumors (defined by a z-score threshold of ± 1 SD) exhibited improved overall survival in comparison to patients with low-to-moderate chemokine expression (Fig. 3K). These data suggest that increased chemokine expression in CDK4/6i-treated tumors can have beneficial effect on patients’ outcome.
CDK4/6i inhibits expression of cell cycle mediators while promoting mTOR activity
We next investigated the mechanism of CDK4/6-mediated chemokine induction. Palbociclib and vehicle treated MCF7 cells were subjected to reverse phase protein array (RPPA) analysis that detects expression of 471 proteins and phosphoproteins. Proteins that were significantly up- or down-regulated after CDK4/6i treatment are presented on a heat map (Fig. 4A). Compete RPPA data are provided in Table S2. As expected, we observed down-regulation of proliferation-related proteins (phosphorylated RB, cyclin B1, Wee1, PLK1, etc.) along with up-regulation of cell cycle arrest inducer p21 in CDK4/6i-treated cells (Fig. 4A, blue frames). One pattern that caught our attention was the increased phosphorylation of mTOR kinase targets, such as S6, p70-S6K, and ULK1 in CDK4/6i-treated cells (Fig. 4A, red frames). Western blot analysis confirmed increased phosphorylation of S6 on residues S240–244, and S235–236, and 70-S6K on T389 after CDK4/6i treatment (Fig. 4B). These changes are indicative of the mammalian target of rapamycin complex 1 (mTORC1) activity (Liu and Sabatini, 2020).
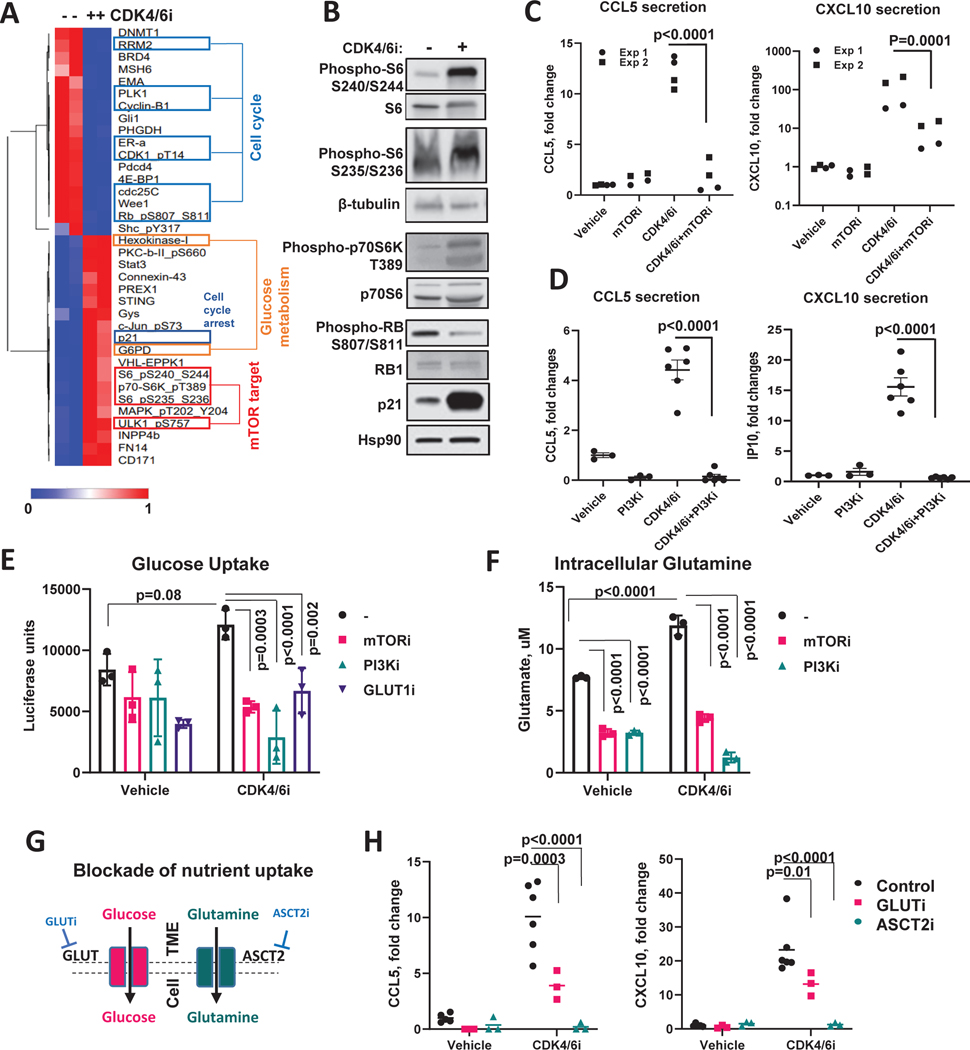
Figure 4. mTOR-regulated metabolic activity is required for CDK4/6i-mediated chemokine induction.
A. RPPA proteomic profiling of MCF7 cells treated with vehicle or 5μM CDK4/6i (Palbociclib) for 5 days. The heat map shows proteins that passed the 1.5-fold change and p<0.05 (multiple t-test) cutoff. B. Western blot validation of RPPA data. C. ELISA measurements of CL5 and CXCL10 in the conditioned medium of MCF7 cells treated with vehicle or 5 μM palbociclib ± 2 μM mTORi rapamycin for 5 days (n=4 biological replicates pooled from 2 independent experiments, 2-way ANOVA, individual data presented). D. ELISA measurements of CCL5 and CXCL10 in the conditioned medium of MCF7 cells treated with vehicle or 5 μM palbociclib ± 0.5 μM PI3Ki BKM120 (buparlisib) for 5 days (n=3–6 biological replicates, one-way ANOVA, data are presented as individual values, mean ± SEM). E. Results of the luciferase-based glucose uptake assay in MCF7 cells treated with vehicle or 1 μM CDK4/6i palbocilib ± 2 μM mTORi rapamycin, or 0.5 μM of PI3Ki BKM120 (buparlisib) for 5 days (n=3 biological replicates, one-way ANOVA, data are presented as individual values and mean ± SD). F. Same as E except intracellular glutamate was measured. G. Schematic of the experimental approach to block glucose and glutamine uptake for the experiment in H. H. ELISA measurements of CCL5 and CXCL10 in the conditioned media from MCF cells treated for 5 days with 5μM CDK4/6i palbociclib in the presence or absence of pharmacological inhibitors of glucose (1μM Bay876, GLUTi) or glutamine (10mM O-Benzyl-L-Serine, ASCTi) transporters (n=3–6 biological replicates, 2-way ANOVA, data are presented as individual values and mean). See also Fig. S5 and Table S2.
A number of prior studies showed the requirement of mTOR for chemokine induction in some immune cell subsets, such as monocytes and macrophages (Jin and Zhao, 2020; Lin et al., 2014; William et al., 2019). Therefore, we investigated whether mTOR was similarly involved in chemokine induction in breast cancer cells treated with CDK4/6i. MCF7 cells were treated with vehicle or palbociclib in the presence or absence of mTOR inhibitor rapamycin. We found that CDK4/6i-induced secretion of CCL5 and CXCL10 was abrogated in the presence of mTOR inhibitor, suggesting that mTOR activity was necessary for chemokine induction (Fig. 4C). Similarly, inhibition of upstream mTOR activator PI3K (Laplante and Sabatini, 2012) with a small molecule inhibitor BKM120 (buparlisib) abolished chemokine induction by CDK4/6i (Fig. 4D). In agreement with these experimental findings, cBIO analysis of the TCGA dataset of 960 breast carcinoma specimens showed higher content of mTOR activity marker phosphorylated S6 protein (residues S235/S236 and S240/S244) in tumors with elevated CCL5 and CXCL10 expression (Fig. S5A).
Prior studies indicated that targeting PI3K/mTOR pathway in the context of CDK4/6i treatment could improve tumor growth inhibition and prevent acquisition of resistance via tumor intrinsic mechanisms (O’Brien et al., 2020; Vilgelm et al., 2019; Yoshida et al., 2019). However, these preclinical studies used immunodeficient mice lacking T cells. Here we tested how combined CDK4/6 and mTOR inhibition affects tumor growth and immune infiltrate in immunocompetent model. Female C57Bl/6 mice were injected with PYMT tumor cells and treated with vehicle, CDK4/6i palbociclib, mTOR inhibitor everolimus (currently used for treatment of human breast cancer), or combination of both drugs (Fig. S5B). Tumor growth was most prominently inhibited in CDK4/6i and mTORi combination group. There was a significant benefit of combining CDK4/6i with mTORi over CDK4/6i single agent (p=0.048), but not over single agent mTORi (p=0.094). Flow cytometry analysis indicated a decrease of CD3+ T cells, including CD8+ T cells, in CDK4/6i and mTORi combination group as compared to CDK4/6i single agent (Fig. S5C). Considering the critical role of mTOR in immune system function (Weichhart et al., 2015) the mechanism of tumor immunome modulation by the mTORi is likely complex. Thus, while our results are in agreement with mTOR being a promising co-target in the context of CDK4/6i treatment, they also indicate that mTORi co-targeting may not be advisable in situations when T cell-enriched microenvironment is desired (e.i. if immunotherapy is planned). Dedicated future studies will be required to address the complex role of mTOR in anti-tumor immunity.
mTOR-regulated metabolic activity is required for CDK4/6i-mediated chemokine induction
We next sought to determine how mTOR regulated chemokine expression in CDK4/6i-treated cells. mTOR is a central regulator of cell metabolism (Sabatini, 2017; Saxton and Sabatini, 2017). Therefore, we investigated whether CDK4/6i-treated cells were metabolically active. RPPA indicated up-regulation of a number of metabolism-associate proteins in CDK4/6i-treated cells including hexokinase I (HK1), glycogen synthase (Gys), and glucose-6-phospate dehydrogenase (G6PD) (Fig. 4A orange frames). However, some metabolic proteins were differentially affected by CDK4/6i. For instance, a mediator of serine metabolism PHGDH was downregulated by palbociclib (Fig. 4A).
To further investigate the effect of CDK4/6i on cell metabolic activity, we measured glucose uptake and glutamine utilization in vehicle and palbociclib-treated MCF7 cells. Glucose and glutamine are critical nutrients required for cancer cell growth and their consumption reflects cell’s metabolic demands. Notably, CDK4/6i treatment did not inhibit nutrient consumption (Fig. 4D, E). In fact, the levels of intracellular glutamate, a product of glutamine conversion within the cells, was increased in CDK4/6i-treated cells as compared to vehicle-treated cells (Fig. 4E). Furthermore, inhibition of mTOR or its upstream regulator PI3K down-modulated glucose uptake and glutamine utilization in CDK4/6i-treated cells. These data suggest that mTOR is required to maintain metabolic activity in CDK4/6i-treated cells. To test if maintenance of high metabolic activity played role in CDK4/6-mediated chemokine induction we blocked nutrient uptake in breast cancer cells using pharmacological inhibitors of glucose transporter GLUT (GLUTi Bay876) or glutamine transporter ASCT2 (and O-Benzyl-L-serine, respectively) (Fig. 4F). Both inhibitors abrogated CCL5 and CXCL10 induction after CDK4/6i treatment (Fig. 4G).
CDK4/6i-mediated chemokine production is associated with cell hypertrophy, accumulation of mitochondria, and induction of ROS
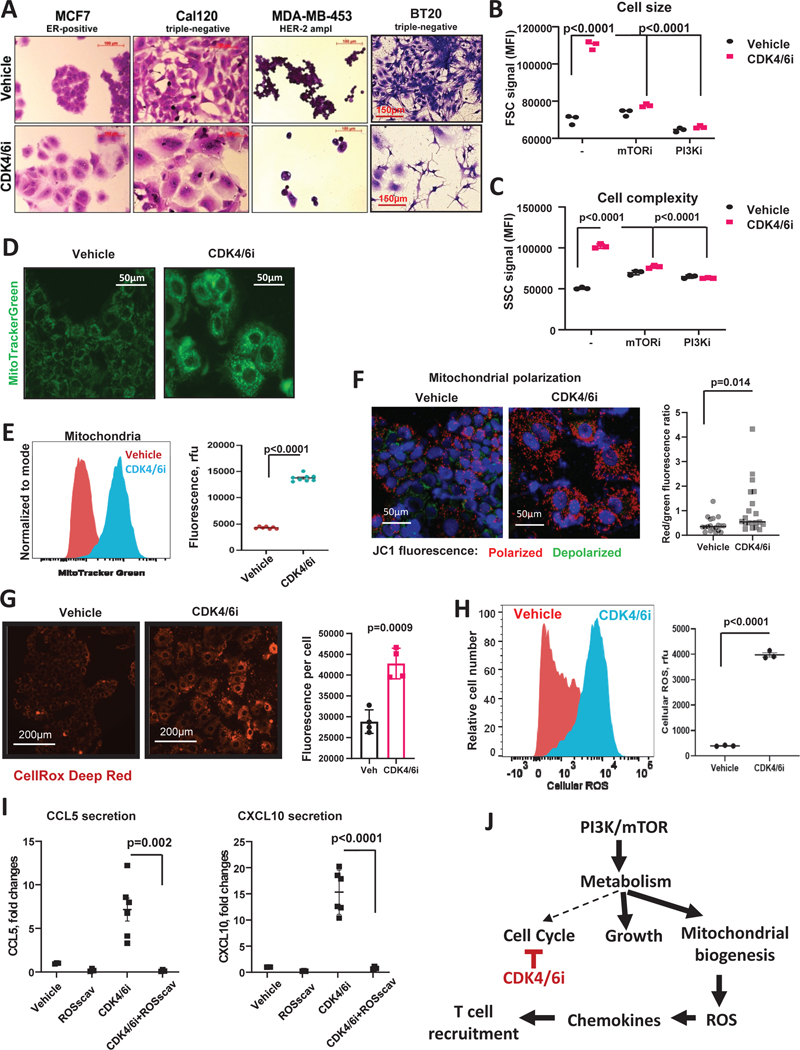
Generally, active metabolism and high mTOR activity are characteristics of fast proliferating cells. Indeed, mTOR promotes anabolic metabolism and biosynthesis to coordinate cell division with doubling of cell biomass and organelle biogenesis. Quiescence cells often inactivate the mTOR kinase due to the lack of need for cell growth (Valcourt et al., 2012). Therefore, we were surprised to detect activation of mTOR and metabolism in cells treated with CDK4/6i that inhibits proliferation. Hence we sought to understand how increased metabolic activity was utilized in CDK4/6i-treated cells. We found that palbociclib treatment caused cell hypertrophy in breast cancer lines (Fig. 5A). An increase in cell sizes after palbociclib was confirmed by flow cytometry (Fig. 5B). Cell growth phenotype was driven by PI3K/mTOR pathway as pharmacological inhibition of either mTOR or PI3K abrogated CDK4/6i-mediated cell size increase (Fig. 5B, Fig. S5F). CDK4/6i-treated cells also exhibited an increase of the cell complexity as per flow cytometry analysis, which indicates increased organelle content. Similar to the increased size, increased complexity was alleviated by mTOR and PI3K inhibition (Fig. 5C). We next investigated whether mitochondria were among the organelles accumulating in CDK4/6i-treated cells based on mTOR being a key driver of mitochondrial biogenesis (Morita et al., 2013). Indeed, mitochondrial content was increased in cells treated with CDK4/6i based on mitotracker staining detected by microscopy (Fig. 5D) and flow cytometry (Fig. 5E). To test whether mitochondria were functional in CDK4/6i-treated cells we used JC-1 mitochondrial potential probe. JC-1 forms aggregates in healthy polarized mitochondria that emit red fluorescence. Cells with depolarized mitochondria exhibit loss of a red punctate fluorescence and increase of diffuse green fluorescence from JC-1 monomers diffused in the cytoplasm. We detected strong red fluorescence in both vehicle and CDK4/6i-treated cells. Furthermore, red/green ratio was further increased after CDK4/6i treatment indicating high mitochondrial activity (Fig. 5F).
Figure 5. CDK4/6i-mediated chemokine production is associated with cell hypertrophy, accumulation of mitochondria, and induction of ROS.
A. Microscopic images of crystal violet stained cells that were treated for 5 days with 5μM of palbociclib, except for BT20, which were treated with 1 μM palbociclib. B. Flow cytometry measurement of forward-scattered light (FCS) parameter indicative of cell volume. MCF7 cells treated with vehicle or 1 μM palbociclib ± 2 μM mTORi rapamycin, or 0.5 μM of PI3Ki BKM120 (buparlisib) for 5 days (n=3, one-way ANOVA, data are presented as individual values and mean). C. Same as F, except side-scattered light (SSC) channel signals indicative of cell complexity (presence of granules and organelles) are shown. D. Fluorescent microscopy of mitochondria detected with MitoTracker Green in MCF7 cells treated with vehicle or 5μM CDK4/6i palbociclib for 5 days. E. Quantification of the MitoTracker staining shown in D using flow cytometry (n=6–8, one-way ANOVA, data are presented as individual values and mean). F. Mitochodrial potential analysis using JC-1 biosensor. Red punctate signal – aggregates of JC-1 in polarized mitochondria, green cytoplasmic staining – free JC-1 monomers. Random microscopic fields from 4 biological replicates quantified with ImageJ (n=19–20, t-test, data are presented as individual values and mean). G. Representative fluorescence microscopy images of cellular ROS detected with CellROX Deep Red in MCF7 cells treated with vehicle or 5μM palbociclib (CDK4/6i) for 5 days. Right panel shows average values of ROS fluorescence per cell from 4 biological replicates compared using t-test. Data are presented as individual values, mean ± SD. H. Representative histogram (left panel) from flow cytometry analysis of cellular ROS in MCF7 cells treated with vehicle or 5μM CDK4/6i palbociclib for 5 days. Right panel shows quantified data (n=3, t-test, data are presented as individual values, mean ± SEM). I. Levels of chemokines CCL5 and CXCL10 in the conditioned media from MCF7 cells treated with 5 mM of cellular ROS scavenger NAC, CDK4/6i (5μM), combination of both, or vehicle for 5 days (n=4–6, one-way ANOVA, data are presented as individual values, mean ± SEM). J. Proposed model CDK4/6i-mediated chemokine induction. Briefly, PI3K/mTOR pathway remains active in CDK4/6i-treated cells despite the decreased metabolic demands due to the cell cycle arrest. This leads to exacerbated cell growth and mitochondrial levels and activity associated with oxidative stress, which, in turn, promotes production of T cell-recruiting chemokines.
Mitochondria are a key source of intracellular ROS (Scialo et al., 2017). Furthermore, CDK4/6i has been shown to induce ROS in pancreatic cancer cells as a result of mTOR-mediated increase of glycolytic and oxidative metabolism and mitochondrial content (Franco et al., 2016), and we observed similar metabolic changes here in a model of breast cancer. Accordingly, we detected increased ROS levels after 5 day palbociclib treatment by fluorescence microscopy (Fig. 5G) and flow cytometry (Fig. 5H). ROS is a potent pro-inflammatory signal known to induce expression of chemokines and other inflammatory mediators (Sozzani et al., 2005). Therefore, we next evaluated whether ROS was involved in CDK4/6i-induced chemokine secretion. We measured cellular ROS production in MCF7 cells and found it to be increased after five days of palbociclib treatment. Removal of ROS using ROS scavenger N-acetylcysteine (NAC) completely abrogated CDK4/6i-mediated chemokine production (Fig. 5I). These data suggest that CDK4/6i-mediated chemokine induction in breast cancer cells requires ROS.
Collectively our data suggest that palbociclib inhibited cell cycle arrest without concurrently shutting down cell metabolism. Cells continued to uptake nutrients to supply energy/building blocks for mTOR-driven biosynthesis. The uncoupling of metabolic need and supply manifested in increased cell size and mitochondrial biogenesis and activity associated with accumulation of ROS, which, in turn, induced inflammatory chemokines that recruited T cells into the tumor (Fig. 5J).
Discussion
Here we show that CDK4/6i palbociclib promotes recruitment of T cells into mammary tumors by inducing expression of T cell-chemotactic chemokines in tumor cells. This finding has several clinical implications. First, the ability of CDK4/6i to enhance T cell tumor homing could make these agents useful for sensitizing patients to T cell-delivering therapies such as adoptive cell transfer of T cells and CAR T cells. Indeed, our pre-clinical model showed that CDK4/6i pre-treatment improved outcome and survival of mice that received transfer of activated tumor-specific T cells. Second, TCGA analysis showed that increased expression of chemokines CCL5 and CXCL10, which are induced by CDK4/6i, is an overall positive prognostic factor in breast cancer. Furthermore, recently published expression analysis of breast cancers tumors before and after treatment with CDK4/6i abemaciclib and aromatize inhibitor anastrosole (neoMONARCH study) showed CCL5 induction and overall enrichment of the chemokine/chemokine receptor expression signature after CDK4/6i treatment (Hurvitz et al., 2020), which is in agreement with our mouse model findings.
Present study uncovered the mechanism of CDK4/6i-mediated chemokine induction. We found it to be dependent on metabolic alterations that occurred in cells after CDK4/6i treatment. These changes included activation of mTOR, induction of glucose and glutamine metabolism accompanied by the increase in cell size and mitochondrial/lysosomal content. Metabolic activation after CDK4/6i treatment was reported previously (Warth et al., 2019; Warth et al., 2018; Zanuy et al., 2012). For example, CDK4/6i-treated pancreatic cancer cells exhibited accumulation of ATP, increased mitochondrial mass, activation of glycolytic and oxidative metabolism driven by the mTOR activation (Franco et al., 2016). Similarly, Cretella et al. showed that CDK4/6i treatment activated PI3K/AKT/mTOR pathway in TNBC cells, and co-inhibition of this pathway impaired glucose metabolism (Cretella et al., 2018). Inhibition of CDK4/6 with siRNA or specific pharmacologic inhibitor increased glutamine metabolism, mitochondrial content, and mTOR pathway activity in colon cancer cells (Tarrado-Castellarnau et al., 2017). Activation of mTOR was also reported in melanoma PDXs and in patients treated with CDK4/6i (Teh et al., 2018; Vilgelm et al., 2019). Here, we showed that mTOR-mediated metabolic reprogramming was critical for CDK4/6i-mediated chemokine induction. Blockade of metabolism by inhibiting the uptake of two key nutrients consumed by proliferating cancer cells, glucose and glutamine, inhibited CDK4/6i-mediated chemokine production.
Cell cycle machinery and metabolism are interconnected. Proliferating cells display high metabolic activity that ensures availability of metabolites for DNA synthesis and cell growth. This is in contrast to quiescent cells that have low metabolic demands (Valcourt et al., 2012). Surprisingly, despite being in a state of cell cycle arrest, CDK4/6i-treated breast cancer cells displayed phenotype of active metabolism. Thus, we postulated that excessive metabolic activity caused metabolic stress in CDK4/6i-treated cells due to nutrient excess. This could be analogous to chronic organismal over-nutrition (obesity) which can lead to various pathological conditions (Wellen and Thompson, 2010). One of the key markers of metabolic stress induced by nutrient excess is an excessive production of ROS and oxidative stress (Wellen and Thompson, 2010). Accordingly, we found increased ROS content in CDK4/6i-treated cells which was required for CDK4/6i-mediated chemokine production. Furthermore, CDK4/6i induced mitochondria content and activity, which are the major source intracellular ROS (Gogvadze et al., 2008).
Oxidative stress is a key link between metabolic stress and inflammatory stress response. For instance, ROS has been implicated as a key driver of chronic inflammation in many diseases associated with metabolic dysfunction, including atherosclerosis, diabetes mellitus, and stroke (Forrester et al., 2018). ROS play important role in regulation of inflammatory response associated with activation of pattern recognition receptors, NF-κB, and inflammasome (Forrester et al., 2018; Lugrin et al., 2014). Moreover, ROS are required for normal function of innate and adaptive immune cells, which further underlines its role as a key immune mediator (Chen et al., 2018; Kohchi et al., 2009; Yarosz and Chang, 2018). It is plausible that the biological significance of ROS-mediated induction of chemokines in context of CDK4/6i treatment is to alert the immune system of cell damage. Indeed, excessive ROS can damage cellular DNA leading to mutations that pose risk of oncogenic transformation; therefore, immune recognition of such cells can be beneficial.
Our data highlight an important role of inflammatory chemokines produced by malignant cells in regulation of the composition of tumor immune microenvironment, immunotherapy response and patient outcome. One can envision T-cell recruiting chemokines being useful as prognostic markers for patient stratification for immunotherapy treatment. Furthermore, with the advancement of the circulating tumor cell technology, longitudinal assessment of chemokine expression in tumor cell can be performed. This type of analysis can identify the perfect window for second hit immunotherapy administration in patients treated with CDK4/6i or other agents that induce chemokine expression in tumor cells, such as senescence-inducing drugs and epigenetic modulators (Dangaj et al., 2019; Vilgelm et al., 2016b; Vilgelm et al., 2015). The findings presented here raise a possibility of utilizing CDK4/6 inhibitors to enhance anti-tumor immunity and immunotherapy response in “cold” T-cell–excluded tumors.
STAR Methods
RESOURCE AVAILIABILITY
Lead Contact
Further information and requests for all original resources and reagents presented in this manuscript should be directed to and will be fulfilled by the Lead Contact, Anna Vilgelm (anna.vilgelm@osumc.edu)
Materials Availability
This study did not generate new unique reagents.
Data and Code availability
The published article includes all datasets generated during this study. Original data for Figure 4A is available in supplemental spreadsheet 1.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All animal experiments were approved by Vanderbilt University and the Ohio State University IACUC. Female mice were used in this study as breast cancer primarily affects female population. For transplanted model, mice were 7–14 weeks of age at the time of tumor inoculation. Transgenic PYMT mice were 8–13 weeks of age at the time of treatment initiation. To inoculate tumors in female C57Bl/6 mice, 100,000 PYMT or PYMT-Ova tumor cells were injected into 4th mammary fat pad as specified in figure legends. Palbociclib was administered by oral gavage at 100 mg/kg once or twice per day as indicated in figure legends. Palbociclib oral solution was prepared in 0.5% methylcellulose. OX-40 and 4–1BB agonistic antibodies and CD8-depleting antibody were diluted in PBS and administered by intraperitoneal injection at 100 μg/mouse. Injections were given every 3 days for a total of 4 doses. OT-1 cells were suspended in PBS and administered via retro-orbital injection at 8×106 cells per mouse. For in vivo chemokine receptor blockade studies mice were treated with 10 mg/kg CCR5i maraviroc (once a day by oral gavage) or CXCR3-blocking antibody (200 μg/mouse, intraperitoneal injection every 3 days). mTORi everolimus was administered via oral gavage once a day at 5mg/kg. Mouse body weight was assessed once a week, and tumor dimensions were measured twice a week with microcalipers (Fisherbrand Traceable). Tumor volume was calculated as 0.5 × length × width × width. Treatment began when tumors reached 50 to 150 mm3 volume on average. Mice were sacrificed when tumors exceeded 15 mm in diameter or became perforated.
Cell lines
MCF7 cells were purchased from the American Type Culture Collection. Cal120, MDA-MB-453, MDA-MB-468, HCC70, and BT20 cells were provided by Carlos Arteaga. PYMT murine mammary tumor cells syngeneic to C57Bl/6 mice were provided by Philip Owens (VA Eastern Colorado Health Care System, Denver, CO). PYMT-OVA cells syngeneic to C57Bl/6 mice were provided by David DeNardo (Washington University St. Louis, St. Louis MO). Cells were cultured in Dulbecco’s modified Eagle’s medium/F12 medium supplemented with 4.5g/L D-Glucose, L-Glutamine, 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin or 100x diluted Anti-Anti mixture (all from Gibco).
METHOD DETAILS
T cell activation and labeling
To activate T cells for ACT experiments, cells from spleens and lymph nodes of OT-1 mice were plated in 6-well plates coated with anti-CD3e Abs (5μg/mL, overnight at +4, clone 145–2C11, Biolegend) at 2×106 cells per well and incubated further in the presence of soluble anti-CD28 Abs (3 μg/mL, 37.51, Biolegend) for two days. Next, cells were collected and re-plated at 0.25×106 cells per well and incubated with 12.5 ng/ml of human recombinant IL-2 for another 3 days. Thirty minutes before injection into mice cells were labeled with CellTracker Green CMFDA Dye (ThermoFisher Scientific) as recommended by the manufacturer. To block chemokine-mediated trafficking, OT-1 cells were pre-treated with 100 ug/ml CXCR3-blocking antibody (clone CXCR3–173, Biolegend) or 500nM of murine recombinant CCL5 (Peprotech) for 1 hour before injection. Two hours post injections, mice were sacrificed, and tumors were analyzed for the presence of fluorescent cells using microscopy and flow cytometry.
Chemokine expression analysis
For mRNA expression analysis, RNA was extracted using RNA-easy mini kit (Quiagen). cDNA was prepared from 1 μg of RNA using iScript cDNA synthesis kit (Bio-rad). We used 2 μL of cDNA as a template for Real- Time PCR with SYBR-green master mix (Molecular Probes) and primers specific to CCL5, CXCL9, CXCL10, and β-actin purchased from Sigma. Primer sequences were obtained from PrimerBank70 (https://pga.mgh.harvard.edu/primerbank/). For analysis of secreted chemokines, ELISA kits for human CXCL10 (IP10) and CCL5 (RANTES), and mouse CXCL9 and CCL5 from Peprotech (Rocky Hills, NJ) or R&D (Minneapolis, MN) were utilized as instructed by the manufacturer. Human chemokine array was purchased from RayBiotech (Peachtree Corners, GA).
Proteomics and Western blot
Cells were pelleted by centrifugation at 15000g and sent to Reverse Phase Protein Array (RPPA) Core Facility at MD Anderson Cancer Center (Houston, TX) for lysis and RPPA analysis. RPPA protein expression was compared between CDK4/6i-treated and vehicle-treated cells using multiple t-test without adjusting for multiple comparisons. Threshold for proteins that were reported as up- or down-regulated was set at the level of ≥1.5-fold change and p<0.05. MORPHEUS web tool(https://software.broadinstitute.org/morpheus) was used for clustering of hits and heat map plotting.
Western blot was performed as described previously(Vilgelm et al., 2017). Antibodies were purchased from Cell Signaling and used as recommended by the vendor. Primary antibodies were applied overnight at 1:500 dilution and secondary antibody were hybridized for 2 hours at 1:5000 dilutions.
Flow cytometry
For detection of immune cells, tumors were excised, dissected into small pieces, and enzymatically digested using tumor tissue dissociation kit (Miltenyi, Germany) according to manufacturer’s recommendations. Cells were processed for viability staining using LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit or eBioscience™ Fixable Viability Dye eFluor™ 780 (Thermofisher) according to manufacturer’s instruction. Next, Fc receptor blocking was performed using Purified Rat Anti-Mouse CD16/CD32 Abs (BD biosciences) for 30 min followed by staining with fluorescently labeled antibodies to immune cell surface markers. Information on panels of antibodies, clones, fluorophores, and vendors is provided in Key Recourse Table. For experiments where Fox3 was evaluated, cells were processed using eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set (Thermofisher). After staining, cells were fixed in 0.5% buffered formaldehyde and analyzed on 4-laser Fortessa (BD Biosciences) for immunophenotyping panels, and on 5-laser Cytek Aurora (Cytek Biosciences) for T cell phenotype panel. Gating and t-SNE dimension reduction was done using FlowJo. In a separate series of in vitro experiments, flow cytometry was utilized to determine relative cell size using forward scatter (FSC) and cell complexity using side scatter (FSC). Amount of mitochondria were analyzed using MitoTracker Green (Invitrogen), respectively. Intracellular ROS levels were detected using CellRox Deep Red (Invitrogen) according to manufacturer’s recommendation.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD45 | BioLegend | 103116 |
| CD3 | Fisher | 460032-80 |
| CD4 | BioLegend | 100438 |
| CD8 | BioLegend | 100712 |
| NK1.1 | BioLegend | 108708 |
| CD19 | Miltenyi | 130-105-171 |
| CD44 | BioLegend | 103025 |
| CD62L | BioLegend | 104417 |
| TIM3 | BioLegend | 119721 |
| LAG3 | BioLegend | 125219 |
| CXCR3 | BioLegend | 126531 |
| CD8a | Thermofisher | 46-0081-82 |
| CD11C | BioLegend | 117330 |
| F4/80 | Thermofisher | 17-4801-80 |
| CD206 | BioLegend | 141705 |
| CD11b | BioLegend | 101217 |
| I-A/I-E | BioLegend | 107622 |
| Ly6G | BioLegend | 127617 |
| Ly-6C | BioLegend | 128035 |
| CD103 | BD | 564322 |
| PD-L1 | BD | 740614 |
| CD45 | BD | 564279 |
| CD69 | Invitrogen | 11-0691-85 |
| FOXP3 | BioLegend | 126404 |
| CD25 | Invitrogen | 56-0251-80 |
| MHC Class I | Invitrogen | 25-5958-80 |
| CD137 | Invitrogen | 46-1371-80 |
| CD134 | BD | 740945 |
| CD3e | BD | 563024 |
| CD8 | BioLegend | 100712 |
| CD11b | BioLegend | 101267 |
| CD8-depleting antibody (YTS 169.4) | BioXcell (West Lebanon, NH) | BPO 117 |
| Anti-OX-40 (OX-86) | BioXcell (West Lebanon, NH) | BE0031 |
| Anti-4-1BB (LOB12.3) | BioXcell (West Lebanon, NH) | BPO169 |
| Purified anti-mouse CD183 (CXCR3) | Biolegend | 126502 |
| Anti-CD3e | Biolegend | 100302 |
| Anti-CD28 | Biolegend | 102102 |
| Anti-mouse CD16/32 | Biolegend | 156604 |
| RB1 | Cell signaling | 9309S |
| Phospho- RB (S807/S811) rabbit antibody | Cell signaling | 8516S |
| S6 Ribosomal protein rabbit antibody | Cell signaling | 2217S |
| Phospho-S6 (S235/236) Ribosomal protein rabbit antibody | Cell signaling | 2211S |
| Phospho-S6 (S240/244) ribosomal protein rabbit antibody | Cell signaling | 5364T |
| Phospho-p70 S6 (T389) kinase rabbit antibody | Cell signaling | 9234T |
| p70 S6 kinase rabbit antibody | Cell signaling | 2708T |
| p21 Waf1/Cip1 rabbit antibody | Cell signaling | 2947S |
| β-actin rabbit antibody | Cell signaling | 4970S |
| β-tubulin rabbit antibody | Cell signaling | 2146S |
| HSP90 rabbit antibody | Cell signaling | 4874S |
| Bacterial and Virus Strains | ||
| NA | ||
| Biological Samples | ||
| NA | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dulbecco’s modified Eagle’s medium/F12 | Gibco | 11330032 |
| GlutaMAX | Gibco | 35050061 |
| Gibco™ Penicillin-Streptomycin | Gibco | 15-140-122 |
| Fetal Bovine Serum | Gibco | 16000036 |
| Palbociclib | Pfizer | NA |
| Palbociclib | LC laboratories | P-7766 |
| Rapamycin | Tocris | 1292 |
| N-acetylcysteine | Sigma | A9165 |
| benzylserine | Sigma | 13900 |
| Bay876 | Sigma | SML1774 |
| BKM120 | Sigma | 10296 |
| Maraviroc | Adooq | A10556 |
| CellTracker Green CMFDA Dye | Invitrogen | C2925 |
| Murine recombinant CCL5 | Peprotech | 25007 |
| MitoTracker Green | Invitrogen | T7514 |
| CellRox Deep Red | Invitrogen | C10422 |
| LysoTracker Deep Red | Invitrogen | L12492 |
| DAPI | Invitrogen | D1306 |
| Hoechst 33342 | Invitrogen | H1399 |
| JC-1 Mitochondrial Membrane Potential Dye | Thermo Fisher | 3168 |
| Fixable Viability Dye eFluor™ 780 | Invitrogen | 65-0865-14 |
| LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit | Thermofisher | L34957 |
| Everolimus | LC Laboratories | E-4040 |
| 10X Tris/Glycine/SDS Buffer | Biorad | 1610772 |
| Tris Buffer Saline | Fisher Bioreagents | BP2471-1 |
| Tween 20 | Fisher Bioreagents | BP 337-500 |
| Trans Blot Turbo RTA transfer kit | Biorad | 170-4270 |
| Bovine serum Albumin | Fisher Bioreagents | BP9706-100 |
| Protein assay dye reagent concentrate | Biorad | 5000006 |
| Critical Commercial Assays | ||
| Reverse Phase Protein Array (RPPA) | Core Facility at MD Anderson Cancer Center (Houston, TX) | NA |
| RNA-easy mini kit | Qiagen | 74104 |
| iScript cDNA synthesis kit | BioRad | 1708890 |
| Quantiline ELISA HumanCXCL10/IP-10 | R&D system | DIP100 |
| Quantiline ELISA Human CCL5/RANTES | R%D system | DRN00B |
| Tumor tissue dissociation kit | Miltenyi Biotech | 130-096-730 |
| FXP3/ Transcription Factor Staining Buffer Set | Invitrogen | 00-5523-00 |
| SYBR-green master mix | BioRad | 1725270 |
| Senescence β-Galactosidase Staining Kit | Sigma | CS0030 |
| Deposited Data | ||
| N/A | ||
| Experimental Models: Cell Lines | ||
| MCF7 Cell line | American Type Culture Collection | ATCC® HTB22™ |
| Cal120 cell line | Carlos L. Arteaga Vanderbilt-Ingram Cancer Center. | NA |
| MDA-MB-453 cell line | Carlos L. Arteaga Vanderbilt-Ingram Cancer Center. | NA |
| MDA-MB-468 cell line | Carlos L. Arteaga Vanderbilt-Ingram Cancer Center. | NA |
| HCC70 cell line | Carlos L. Arteaga Vanderbilt-Ingram Cancer Center. | NA |
| BT20 cell line | Carlos L. Arteaga Vanderbilt-Ingram Cancer Center. | NA |
| PYMT cell line | Philip Owens (VA Eastern Colorado Health Care System, Denver, CO) | NA |
| PYMT-OVA cell line | David DeNardo (Washington University St. Louis, St. Louis MO) | NA |
| Experimental Models: Organisms/Strains | ||
| C57BL/6 mice | The Jackson Laboratory | 000664 |
| C57BL/6 mice | Charles River/NCI | Strain code :027 |
| OT-1 mice | Marc Boothby, Richard O’Neil (Vanderbilt University, Nashville, TN) | NA |
| OT-1 mice | The Jackson Laboratory | 003831 |
| FVB-PYMT transgenic mice | Harold Moses (Vanderbilt University, Nashville, TN) | NA |
| Oligonucleotides | ||
| Primer: h CXCL10 F GTGGCATTCAAGGAGTACCTC | Sigma | NA |
| Primer: h CXCL10 R TGATGGCCTTCGATTCTGGATT | Sigma | NA |
| Primer: h CXCL9 F CCAGTAGTGAGAAAGGGTCGC | Sigma | NA |
| Primer: h CXCL9 R AGGGCTTGGGGCAAATTGTT | Sigma | NA |
| Primer: m CXCL9 F GGAGTTCGAGGAACCCTA | Sigma | NA |
| Primer: m CXCL9 R GGGATTTGTAGTAGTGGATCGTGC | Sigma | NA |
| Primer: m CCL5 F GCTGCTTTGCCTACCTCTCC | Sigma | NA |
| Primer: m CCL5 R TCGAGTGACAAACACGACTGC | Sigma | NA |
| Primer: h CCL5 F CCAGCAGTCGTCTTTGTCAC | Sigma | NA |
| Primer: h CCL5 R CTCTGGGTTGGCACACACTT | Sigma | NA |
| Recombinant DNA | ||
| NA | ||
| Software and Algorithms | ||
| ImageJ | NIH Image | https://imagej.nih.gov/ij/index.html |
| Graphpad’s Prism 7.03 software | GraphPad | https://www.graphpad.com/ |
| R (version3.3.0.) | Bioconductor | https://www.bioconductor.org/packages/devel/bioc/vignettes/limma/inst/doc/usersguide.pdf |
| Timer version 2.0 | Li et al, 2020 | http://timer.cistrome.org/ |
| Morpheus | Broad Institute | https://software.broadinstitute.org/morpheus/ |
| cBioPortal for Cancer Genomics version 3.6.3 | Cerami et al, 2012 | https://www.cbioportal.org/ |
| Other | ||
| N/A | ||
Fluorescence and bright-field microscopy
IHC staining was performed by Translational Pathology Shared Resources at Vanderbilt University (Nashville, TN). MitoTracker Green, LysoTracker Deep Red, DAPI, and CellRox Deep Red (all from Invitrogen) were used for fluorescent labeling of mitochondria, lysosomes, nuclei, and intracellular ROS, respectively. Senescence Cells Histochemical Staining Kit (Sigma-Aldrich) was used to detect SA-βGal-positive cells. To detect dead cells and cell DNA in live or dead cells Propidium Iodide (1μg/mL) and Hoechst 33342 (2μg/mL) were added directly into the cell media 20 minutes prior to imaging. JC-1 Mitochondrial Membrane Potential Dye was purchased from Thermofisher and used in accordance with manufacturer’s recommendations. To visualize stained tumor sections, cell populations, and intracellular content, EVOS M5000 digital inverted microscope (Thermo Fisher Scientific, Carlsbad, CA, USA) was utilized.
TCGA analysis
To study the association of CCL5 and CXCL10 expression with CD8+ T cell infiltration, Timer2.0 web resource for systematical analysis of immune infiltrates was used (http://timer.cistrome.org/). Expression of chemokines was correlated to CD8+ T cell infiltration estimated by CIBERSORT-Abs., which is a score of arbitrary units that reflects the absolute proportion of each cell type derived from computational analysis of the expression signatures of cell population of interest (Newman et al., 2015). TCGA breast cancer dataset (BRCA) was analyzed.
To query the association of CCL5 and CXCL10 chemokine expression with breast cancer patient survival or phosphorylation of mTOR target protein S6, cBioPortal was used (Cerami et al., 2012). TCGA Breast Invasive Carcinoma Firehose Legacy dataset was used for analysis. Samples with CCL5 and/or CXCL10 mRNA expression z-scores >1 were categorized as high expressing, and samples with z-cores <1 as low-intermediate expressing.
QUANTIFICATION AND STATISTICAL ANALYSIS
Standard t test and one-way ANOVA were used for analysis comparing two samples and multiple samples, respectively. To study the effect of two parameters (i.e. time and treatment) and their interaction, two-way ANOVA was applied. GraphPad’s Prism 7.03 software and R (version 3.3.0.) were used for statistical analysis. For in vivo experiments, we compared the progression of tumor volume (mm3) over time among groups of mice receiving different therapy with a linear mixed-effects regression model. To meet the normality assumptions for these parametric methods, a square root or a natural log transformation was implemented to ameliorate the heterogeneity evident in the data. Mixed models estimate corrected variance estimates in the presence of correlated measurements taken in the same mouse (such as left and right flank) and for repeated measures on the same tumor over time. The Akaike information criterion was used to select among competing correlation structures. Standard residual analysis and goodness-of-fit statistics were evaluated. Survival curves were plotted and compared using GraphPad’s Prism. All tests of statistical significance were two sided. The numbers of mice and biological relocates in experimental groups are indicated in figure legends. Animal experiments were performed once, except for experiments combining CDK4/6i with adoptive T cell transfer (Fig. 2G) and anti-OX-40/4–1BB (Fig. 2A) that were repeated twice with consistent results. In vitro experiments were repeated at least 3 times. Error bars represent standard deviation, except for graphs showing tumor volume change over time, where error bars represent standard error.
Supplementary Material
Table S2. Results of the RPPA analysis in MCF7 cells treated with vehicle or palbociclib. Related to Figure 4A. MCF7 cells treated with vehicle or 5μM CDK4/6i (Palbociclib) for 5 days using commercial RPPA assay service. Values are shown as provided by the vendor and represent data points normalized for protein loading and transformed to linear value.
Acknowledgements
This work was supported by grants from BCRF (IIDRP-16-001 to AEV), NCI SPORE in Breast Cancer (P50CA098131) pilot award (to AEV), NIH R37 CA233770-01 (to AEV), the Department of Veterans Affairs (5101BX000196-04 to AR), NIH (CA116021 and CA116021-S1 to AR), Senior Research Career Scientist Award (to AR). Support for core facilities used in this study was provided by Vanderbilt Ingram Cancer Center (P30 CA68485). The Reverse Phase Protein Array (RPPA) Core at MD Anderson Cancer Center (The University of Texas) is funded by NCI # CA16672.
Declaration of Interests
Carlos Arteaga receives or has received research grants from Pfizer, Lilly, and Takeda; holds minor stock options in Provista; serves or has served in an advisory role to Novartis, Merck, Lilly, Daiichi Sankyo, Taiho Oncology, OrigiMed, Puma Biotechnology, Immunomedics, AstraZeneca, Arvinas, and Sanofi; and reports scientific advisory board remuneration from the Susan G. Komen Foundation.
References:
- Alvarez-Fernandez M, and Malumbres M. (2020). Mechanisms of Sensitivity and Resistance to CDK4/6 Inhibition. Cancer Cell 37, 514–529. [DOI] [PubMed] [Google Scholar]
- Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V, et al. (2017). IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 127, 2930–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottcher JP, Bonavita E, Chakravarty P, Blees H, Cabeza-Cabrerizo M, Sammicheli S, Rogers NC, Sahai E, Zelenay S, and Sousa CRE (2018). NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell 172, 1022-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YX, Zhou ZY, and Min W. (2018). Mitochondria, Oxidative Stress and Innate Immunity. Front Physiol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow MT, and Luster AD (2014). Chemokines in cancer. Cancer Immunol Res 2, 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretella D, Ravelli A, Fumarola C, La Monica S, Digiacomo G, Cavazzoni A, Alfieri R, Biondi A, Generali D, Bonelli M, et al. (2018). The anti-tumor efficacy of CDK4/6 inhibition is enhanced by the combination with PI3K/AKT/mTOR inhibitors through impairment of glucose metabolism in TNBC cells . J Exp Clin Canc Res 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, Sher X, Liu XQ, Lu H, Nebozhyn M, et al. (2018). Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofanilli M, Turner NC, Bondarenko I, Ro J, Im SA, Masuda N, Colleoni M, DeMichele A, Loi S, Verma S, et al. (2016). Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. Lancet Oncol 17, 425–439. [DOI] [PubMed] [Google Scholar]
- Dangaj D, Bruand M, Grimm AJ, Ronet C, Barras D, Duttagupta PA, Lanitis E, Duraiswamy J, Tanyi JL, Benencia F, et al. (2019). Cooperation between Constitutive and Inducible Chemokines Enables T Cell Engraftment and Immune Attack in Solid Tumors. Cancer Cell 35, 885–900 e810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo GL, Mirick GR, Hok S, DeNardo SJ, Beckett LA, Adamson GN, and Balhorn RL (2009). Molecular specific and cell selective cytotoxicity induced by a novel synthetic HLA-DR antibody mimic for lymphoma and leukemia. Int J Oncol 34, 511–516. [PubMed] [Google Scholar]
- Deng JH, Wang ES, Jenkins RW, Li S, Dries R, Yates K, Chhabra S, Huang W, Liu HY, Aref AR, et al. (2018). CDK4/6 Inhibition Augments Antitumor Immunity by Enhancing T-cell Activation. Cancer Discov 8, 216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dushyanthen S, Beavis PA, Savas P, Teo ZL, Zhou C, Mansour M, Darcy PK, and Loi S. (2015). Relevance of tumor-infiltrating lymphocytes in breast cancer. BMC Med 13, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escola JM, Kuenzi G, Gaertner H, Foti M, and Hartley O. (2010). CC chemokine receptor 5 (CCR5) desensitization: cycling receptors accumulate in the trans-Golgi network. J Biol Chem 285, 41772–41780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, et al. (2009). PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res 11, R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester SJ, Kikuchi DS, Hernandes MS, Xu Q, and Griendling KK (2018). Reactive Oxygen Species in Metabolic and Inflammatory Signaling. Circ Res 122, 877-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco J, Balaji U, Freinkman E, Witkiewicz AK, and Knudsen ES (2016). Metabolic Reprogramming of Pancreatic Cancer Mediated by CDK4/6 Inhibition Elicits Unique Vulnerabilities. Cell Rep 14, 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel S, DeCristo MJ, Watt AC, BrinJones H, Sceneay J, Li BB, Khan N, Ubellacker JM, Xie SZ, Metzger-Filho O, et al. (2017). CDK4/6 inhibition triggers anti-tumour immunity. Nature 548, 471-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel S, Wang Q, Watt AC, Tolaney SM, Dillon DA, Li W, Ramm S, Palmer AC, Yuzugullu H, Varadan V, et al. (2016). Overcoming Therapeutic Resistance in HER2-Positive Breast Cancers with CDK4/6 Inhibitors. Cancer Cell 29, 255–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogvadze V, Orrenius S, and Zhivotovsky B. (2008). Mitochondria in cancer cells: what is so special about them? Trends Cell Biol 18, 165–173. [DOI] [PubMed] [Google Scholar]
- Griffith JW, Sokol CL, and Luster AD (2014). Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol 32, 659–702. [DOI] [PubMed] [Google Scholar]
- Guy CT, Cardiff RD, and Muller WJ (1992). Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol 12, 954–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlin H, Meng Y, Peterson AC, Zha YY, Tretiakova M, Slingluff C, McKee M, and Gajewski TF (2009). Chemokine Expression in Melanoma Metastases Associated with CD8(+) T-Cell Recruitment. Cancer Research 69, 3077–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurvitz SA, Martin M, Press MF, Chan D, Fernandez-Abad M, Petru E, Rostorfer R, Guarneri V, Huang CS, Barriga S, et al. (2020). Potent Cell-Cycle Inhibition and Upregulation of Immune Response with Abemaciclib and Anastrozole in neoMONARCH, Phase II Neoadjuvant Study in HR(+)/HER2(−) Breast Cancer. Clin Cancer Res 26, 566–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, and Zhao Q. (2020). Emerging role of mTOR in tumor immune contexture: Impact on chemokine-related immune cells migration. Theranostics 10, 6231–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein ME, Kovatcheva M, Davis LE, Tap WD, and Koff A. (2018). CDK4/6 Inhibitors: The Mechanism of Action May Not Be as Simple as Once Thought. Cancer Cell 34, 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen ES, and Witkiewicz AK (2017). The Strange Case of CDK4/6 Inhibitors: Mechanisms, Resistance, and Combination Strategies. Trends Cancer 3, 39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohchi C, Inagawa H, Nishizawa T, and Soma GI (2009). ROS and Innate Immunity. Anticancer Res 29, 817–821. [PubMed] [Google Scholar]
- Laplante M, and Sabatini DM (2012). mTOR Signaling. Cold Spring Harb Perspect Biol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, and Liu XS (2020). TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 48, W509–W514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, and Pollard JW (2003). Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol 163, 2113–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HY, Chang KT, Hung CC, Kuo CH, Hwang SJ, Chen HC, Hung CH, and Lin SF (2014). Effects of the mTOR inhibitor rapamycin on monocyte-secreted chemokines. BMC Immunol 15, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GY, and Sabatini DM (2020). mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol 21, 183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugrin J, Rosenblatt-Velin N, Parapanov R, and Liaudet L. (2014). The role of oxidative stress during inflammatory processes. Biol Chem 395, 203–230. [DOI] [PubMed] [Google Scholar]
- Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, et al. (2013). mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab 18, 698–711. [DOI] [PubMed] [Google Scholar]
- Nagarsheth N, Wicha MS, and Zou WP (2017). Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol 17, 559–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, and Alizadeh AA (2015). Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12, 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien NA, McDermott MSJ, Conklin D, Luo T, Ayala R, Salgar S, Chau K, DiTomaso E, Babbar N, Su F, et al. (2020). Targeting activated PI3K/mTOR signaling overcomes acquired resistance to CDK4/6-based therapies in preclinical models of hormone receptor-positive breast cancer. Breast Cancer Res 22, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni G, Formenti SC, Chen-Kiang S, and Galluzzi L. (2020). Immunomodulation by anticancer cell cycle inhibitors. Nat Rev Immunol 20, 669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini DM (2017). Twenty-five years of mTOR: Uncovering the link from nutrients to growth. Proc Natl Acad Sci U S A 114, 11818–11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, and Sabatini DM (2017). mTOR Signaling in Growth, Metabolism, and Disease. Cell 169, 361–371. [DOI] [PubMed] [Google Scholar]
- Schaer DA, Beckmann RP, Dempsey JA, Huber L, Forest A, Amaladas N, Li YX, Wang YC, Rasmussen ER, Chin D, et al. (2018). The CDK4/6 Inhibitor Abemaciclib Induces a T Cell Inflamed Tumor Microenvironment and Enhances the Efficacy of PD-L1 Checkpoint Blockade. Cell Rep 22, 2978–2994. [DOI] [PubMed] [Google Scholar]
- Scialo F, Fernandez-Ayala DJ, and Sanz A. (2017). Role of Mitochondrial Reverse Electron Transport in ROS Signaling: Potential Roles in Health and Disease. Front Physiol 8, 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani S, Bosisio D, Mantovani A, and Ghezzi P. (2005). Linking stress, oxidation and the chemokine system. Eur J Immunol 35, 3095–3098. [DOI] [PubMed] [Google Scholar]
- Spranger S, Dai D, Horton B, and Gajewski TF (2017). Tumor-Residing Batf3 Dendritic Cells Are Required for Effector T Cell Trafficking and Adoptive T Cell Therapy. Cancer Cell 31, 711-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll G, Pol J, Soumelis V, Zitvogel L, and Kroemer G. (2018). Impact of chemotactic factors and receptors on the cancer immune infiltrate: a bioinformatics study revealing homogeneity and heterogeneity among patient cohorts. Oncoimmunology 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrado-Castellarnau M, de Atauri P, Tarrago-Celada J, Perarnau J, Yuneva M, Thomson TM, and Cascante M. (2017). De novo MYC addiction as an adaptive response of cancer cells to CDK4/6 inhibition. Mol Syst Biol 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh JLF, and Aplin AE (2019). Arrested Developments: CDK4/6 Inhibitor Resistance and Alterations in the Tumor Immune Microenvironment. Clinical Cancer Research 25, 921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh JLF, Cheng PF, Purwin TJ, Nikbakht N, Patel P, Chervoneva I, Ertel A, Fortina PM, Kleiber I, HooKim K, et al. (2018). In Vivo E2F Reporting Reveals Efficacious Schedules of MEK1/2-CDK4/6 Targeting and mTOR-S6 Resistance Mechanisms (vol 8, pg 568, 2018). Cancer Discov 8, 1654–1654.30510016 [Google Scholar]
- Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. (2014). PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner NC, Slamon DJ, Ro J, Bondarenko I, Im SA, Masuda N, Colleoni M, DeMichele A, Loi S, Verma S, et al. (2018). Overall Survival with Palbociclib and Fulvestrant in Advanced Breast Cancer. New Engl J Med 379, 1926–1936. [DOI] [PubMed] [Google Scholar]
- Valcourt JR, Lemons JM, Haley EM, Kojima M, Demuren OO, and Coller HA (2012). Staying alive: metabolic adaptations to quiescence. Cell Cycle 11, 1680–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgelm AE, Cobb P, Malikayil K, Flaherty D, Andrew Johnson C, Raman D, Saleh N, Higgins B, Vara BA, Johnston JN, et al. (2017). MDM2 Antagonists Counteract Drug-Induced DNA Damage. EBioMedicine 24, 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgelm AE, Johnson CA, Prasad N, Yang JM, Chen SC, Ayers GD, Pawlikowski JS, Raman D, Sosman JA, Kelley M, et al. (2016a). Connecting the Dots: Therapy-Induced Senescence and a Tumor-Suppressive Immune Microenvironment. Jnci-J Natl Cancer I 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgelm AE, Johnson DB, and Richmond A. (2016b). Combinatorial approach to cancer immunotherapy: strength in numbers. J Leukoc Biol 100, 275–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgelm AE, Pawlikowski JS, Liu Y, Hawkins OE, Davis TA, Smith J, Weller KP, Horton LW, McClain CM, Ayers GD, et al. (2015). Mdm2 and aurora kinase a inhibitors synergize to block melanoma growth by driving apoptosis and immune clearance of tumor cells. Cancer Res 75, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgelm AE, and Richmond A. (2019). Chemokines Modulate Immune Surveillance in Tumorigenesis, Metastasis, and Response to Immunotherapy. Front Immunol 10, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgelm AE, Saleh N, Shattuck-Brandt R, Riemenschneider K, Slesur L, Chen SC, Johnson CA, Yang J, Blevins A, Yan C, et al. (2019). MDM2 antagonists overcome intrinsic resistance to CDK4/6 inhibition by inducing p21. Sci Transl Med 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth B, Palermo A, Rattray NJW, Lee NV, Zhu Z, Hoang LT, Cai Y, Mazurek A, Dann S, VanArsdale T, et al. (2019). Palbociclib and Fulvestrant Act in Synergy to Modulate Central Carbon Metabolism in Breast Cancer Cells. Metabolites 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth B, Raffeiner P, Granados A, Huan T, Fang M, Forsberg EM, Benton HP, Goetz L, Johnson CH, and Siuzdak G. (2018). Metabolomics Reveals that Dietary Xenoestrogens Alter Cellular Metabolism Induced by Palbociclib/Letrozole Combination Cancer Therapy. Cell Chem Biol 25, 291–300 e293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichhart T, Hengstschlager M, and Linke M. (2015). Regulation of innate immune cell function by mTOR. Nat Rev Immunol 15, 599–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, and Thompson CB (2010). Cellular Metabolic Stress: Considering How Cells Respond to Nutrient Excess. Mol Cell 40, 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellenstein MD, and de Visser KE (2018). Cancer-Cell-Intrinsic Mechanisms Shaping the Tumor Immune Landscape. Immunity 48, 399–416. [DOI] [PubMed] [Google Scholar]
- William M, Leroux LP, Chaparro V, Graber TE, Alain T, and Jaramillo M. (2019). Translational repression of Ccl5 and Cxcl10 by 4E-BP1 and 4E-BP2 restrains the ability of mouse macrophages to induce migration of activated T cells. Eur J Immunol 49, 1200–1212. [DOI] [PubMed] [Google Scholar]
- Yarosz EL, and Chang CH (2018). The Role of Reactive Oxygen Species in Regulating T Cell-mediated Immunity and Disease. Immune Netw 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Bu Y, Qie S, Wrangle J, Camp ER, Hazard ES, Hardiman G, de Leeuw R, Knudsen KE, and Diehl JA (2019). SLC36A1-mTORC1 signaling drives acquired resistance to CDK4/6 inhibitors. Sci Adv 5, eaax6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanuy M, Ramos-Montoya A, Villacanas O, Canela N, Miranda A, Aguilar E, Agell N, Bachs O, Rubio-Martinez J, Pujol MD, et al. (2012). Cyclin-dependent kinases 4 and 6 control tumor progression and direct glucose oxidation in the pentose cycle. Metabolomics 8, 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JF, Bu X, Wang HZ, Zhu YS, Geng Y, Nihira NT, Tan YY, Ci YP, Wu F, Dai XP, et al. (2018). Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 553, 91-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. Results of the RPPA analysis in MCF7 cells treated with vehicle or palbociclib. Related to Figure 4A. MCF7 cells treated with vehicle or 5μM CDK4/6i (Palbociclib) for 5 days using commercial RPPA assay service. Values are shown as provided by the vendor and represent data points normalized for protein loading and transformed to linear value.
Data Availability Statement
The published article includes all datasets generated during this study. Original data for Figure 4A is available in supplemental spreadsheet 1.