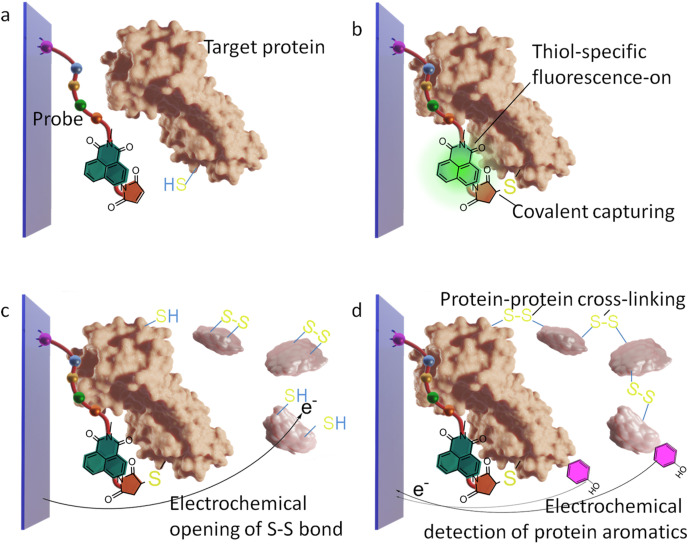
Abstract
The poor situational awareness about the spreading of the virus especially in the underdeveloped regions calls for novel virus assays of low cost and simple operation. Currently, such assays are exclusively restricted to nucleic acid detection. In this investigation, a virus protein serum assay has been proposed in a one-step and reagent-less route. Specifically, in this assay, the main protease of the virus is targeted by a short probe mimicking its substrate. While the probe-protein interaction brings them together, a fluorescent thiol targeting molecule reacts with the free thiol groups on the target protein near the probe, generating a fluorescence signal proportional to the concentration of the target. This induces an electroactive 2D peptide nano-network on the sensing surface only in the presence of the target protein. The sensitivity of the method is enhanced through potential electrochemical scanning during incubation with serum samples. The successful detection of the virus marker protein in the serum of the infected patients encourages further development of incorporation of this method into clinical practice.
Keywords: Thiol probe, Main protease, SARS-Cov-2, Peptide nano-network, COVID-19
1. Introduction
COVID-19 vaccination is successfully an ongoing process in many countries(Krammer, 2020); still, some impoverished regions are in dire need of this vaccine(Sbarra et al., 2021). So, large scale surveillance becomes the first line of defense in combating the virus, providing all the important situation awareness(Hozé et al.). In this data network of global awareness, the poorly developed regions usually have a weak link. The simple, fast, one-step, reagent-less and low-cost SARS-Cov-2 assays are urgently required to provide effective coverage for these regions. The current gold standard, the nucleic acid test, needs several amplification steps, introducing human errors since the medical staff works under stress(Basile et al., 2020). On the other hand, the traditional antibody-based assays seem to have fewer chances of replacing the DNA test. This is due to the relatively high cost of biologically produced antibodies and enzymes(Weinstein et al., 2020). Hence, researchers have developed artificial targeting molecules such as aptamers and small molecular drug leads, but these have rarely been applied in clinical practice. The less strong binding affinity (Kalra et al., 2018) of these targeting molecules than antibodies is the main obstacle to their clinical application. This necessitates the requirement of a simple and effective bio-assay to check the viral spreading.
This study attempts to design a SARS-Cov-2 assay using synthetic molecules only. A strong target-binding and effective signal amplification were realized in the detection using clinical samples. The detailed design of this assay strategy has been shown in Scheme 1 . The surface-immobilized probe is a short peptide sequence mimicking the substrate of a virus marker protein, the main protease of SARS-Cov-2. This protease is highly conservative and responsible for building the replication machine of the virus(Zhang et al., 2020). It thus does not tend to mutate rapidly like the Spike protein. Virus protease from the clinical samples can recognize the surface substrate probes and bind with them. However, this molecular recognition event does not inherently generate any readable signal output. This might be why traditional immuno-assays always follow the conventional sandwich design where an additional reagent is required to mark the surface captured target protein. But we aim to achieve a reagent-less detection route. For this, the reactivity of amino acid side chains is considered. These are two cysteine residues with free thiol groups(Zhang et al., 2020) located near the substrate-binding pocket of the target protein. The substrate-mimicking probe is extended by a recently developed fluorescent “signal on” detector molecule(Liu et al., 2016). This is a recently developed “signal-on” probe, which has a maleimide group that can react with thiol to recover the quenched fluorescence of the conjugated 1,8-naphthalimide. This fluorophore can be activated by visible light, avoiding UV radiation that can lead to large background interference. This molecule forms covalent bonding with the free thiol groups, leading to “lighting”. In this way, using one probe alone, not only the target protein is irreversibly attached to the sensing surface, but the covalent attachment also generates a signal in proportion to the amount of surface captured protein molecules. This covalent attachment is indispensable in clinical application, as it allows thorough denaturing on the sensing layer to remove the non-specific interfering species. High robustness can therefore be achieved at the same time when complicated assaying steps involving additional reagents are also not needed.
Scheme 1.
The proposed method to detect the viral protease, not drawn to scale. The protease model is adopted from the crystal structure from the reference (Zhang et al., 2020).
But the sensitivity may not be sufficient in this design, while additional biosensing reagents are avoided to simplify the assay maximally. Hence, the well-established electrochemical pre-treatment in mass spectrometry/chromatography protein analysis can be employed. In this, a simple potentiostat module can electrochemically reduce S–S bonds in proteins, and the formed free thiol can crosslink the protein molecules together(Kraj et al., 2013; Stocks and Melanson, 2018). The molecules in the clinical serum sample can then be used to enhance the signal since the surface probe does not have such a function. The probe molecules are immobilized on a conductive glass slide. Previous reports have successfully used a serum disease marker protein to crosslink the most abundant serum species, the albumin, together(Jiang et al., 2021). Therefore, using similar methods, the virus protease covalently captured on the sensing surface can serve as the covalent anchoring point of the crosslinked serum proteins (mainly albumin). The protease contains Zinc finger-like domains with free thiol groups(Maiti, 2020). The extensive 2D nano-network of the electrochemically crosslinked peptide chains can then be interrogated electrochemically, leading to cathodic stripping or oxidation of reactive groups such as the tyrosine moieties(Zhang et al., 2019). In both the above methods of direct fluorescent detection and amplified electrochemical detection, the equipment is not expensive, and the fluorescent detection can be done with ELISA fluorescent plate reader, while the handheld electrochemical module has already become commercially available. The electrochemical scanning is applied during incubation with the clinical serum sample, so that no extra step is required before collecting signal readout. Thus, a low-cost, one-step, reagent-less bio-assay using only synthesizable simple molecules are developed, enabling not only antibody-like strong targeting of the virus marker protein, but also reasonable signal amplification, using the interfering species in the serum as signal amplifier.
2. Experimental
2.1. Chemicals and reagents
The peptide probe (sequence shown in Scheme S1) and the ferrocene tagged control probe were manufactured by Jinan V.R. Biotechnology, purity> 95%. The main protease of SARS-Cov-2, also known as SARS-Cov-3CL protease was received in the form of HEPES buffer solution from the R&D system. 11-Mercaptoundecanoic acid (MUA), 9-Mercapto-1-nonanol (MN), ferrocene and cystine were from Sigma-Aldrich. All other chemicals were of analytical grade. The peptide probe solution was prepared by dissolving the powder to 10 μM with 10 mM phosphate buffer solution (PBS) (pH 7.4). The HEPES buffer solution of the target protease was serially diluted with “blank” serum samples from a healthy volunteer to prepare standard samples for establishing working curves. All solutions are prepared with double distilled water (dd water), which has been purified by Milli-Q purification system (Branstead, USA) with a resistivity of 18MΩ•cm. Approved by the local ethics committee, clinical serum samples of patients infected with SARS-Cov-2 were retrieved from the sample database of the P2.5 clinical laboratory of the Nanjing Second Hospital. The patients were randomly picked from the sample bank, and since only the marker protein in their blood samples were under concern, the past medical history of the patients were not disclosed. These 9 patients were incidentally all male, aged from 21 to 56. And the sample bank collected these samples from a wide area not restricted to Nanjing, during the outbreak in the first half of 2020. The blood samples were stored in heparin vial, before testing, the samples were centrifugated for 15 min at 2500 RPM, and then the supernatant was collected. Strictly adhering to the safety regulations, all experiments were carried out in the enclosed area of the laboratory.
2.2. Fabrication of the sensing surface
The conductive indium tin oxide (ITO) coated glass (from Hangzhou DCT hardware) was cut into small pieces to fit the small cuvette for fluorescence measurement. In order to fix the probe, the ITO glass slide was continuously immersed in dd water, ethanol and methanol, and subjected to bath ultrasonic treatment to remove any organic impurities absorbed on the surface. The end of the probe (Scheme S1) contained a 16-phosphohexadecanoic acid residue (Sigma-Aldrich), which could self-assemble on the ITO surface. To start this process, drop the probe stock solution onto the newly cleaned glass slide, and then mark the area covered by the stock solution drop with a marker pen. Then the self-assembly process was carried out for 16 h in a refrigerator at 4°C in the dark. Finally, the glass slide was rinsed with a large amount of dd water, dried in a high-purity nitrogen stream, and then sent to the test sample.
2.3. Detection of the virus protease
Serum spiked samples or clinical serum samples were incubated with the slide presenting the sensing surface at 37 °C for proper time as detailed in the results and discussion section. Thorough detergent rinsing was then applied to the slide, and after this rinsing step, the slide was ready for subsequent fluorescent measurements. For cross-linking method, during incubation, the sensing slide was kept at an open circuit potential of −0.5 V.
2.4. Experimental measurements
The MicroCal ITC200 system (GE Healthcare Life Sciences) was used for isothermal titration calorimetry (ITC) measurement. The titration was performed at 25°C. The titration schedule consisted of 38 consecutive 1 μL injections with at least 120 s between each two injections. The heat of dilution measured by titrating over saturation from each data set was subtracted. Before titration, all solutions were degassed. Origin 7.0 software was used to analyze the data. The electrochemical measurement was carried out on a CHI660D potentiostat (CH Instruments) with a conventional three-electrode system: the glass slide was used as the working electrode, the saturated calomel electrode (SCE) was used as the reference electrode, and the platinum wire was used as the counter electrode. Cyclic voltammetric scanning was applied under a rate of potential scanning of 1 mV/s, the range of scanning is as shown in Figure S1. Square wave voltammetric scanning rounds were conducted under the following parameter setting: scan range, as shown in Figure S3, step potential, 5 mV, frequency, 15 Hz, amplitude, 25 mV. The electrolyte vial is a self-made type of approximately 100 μL. A QM-4/2005 fluorescence spectrometer (Photon Technology International, Inc., Birmingham, NJ) equipped with a xenon lamp was used to measure the fluorescence emission spectrum of the surface. The light source and the detector were located on the same plane and were at right angles to each other. The glass slide was kept in a water-filled test tube at 60° from the bottom of the test tube for fluorescence measurement. The surface with the gold film should be away from the light source and towards the detector. The activating wavelength of 1,8-naphthalimide is 345 nm. The SPR measurement was performed by an Autolab ESPRIT system (Echo Chemie B.V., The Netherlands) equipped with a 670 nm monochromatic p-polarized light source. Atomic force microscopy (AFM) imagery results were obtained on an ex situ Agilent 5500 AFM system. The samples were interrogated at a set scanning rate of 0.5–1 Hz, and the setting was in a tapping mode. AFM tips used were of a resonant frequency in the range from 160 to 260 kHz.
3. Results and discussion
3.1. Validation of the sensing mechanism
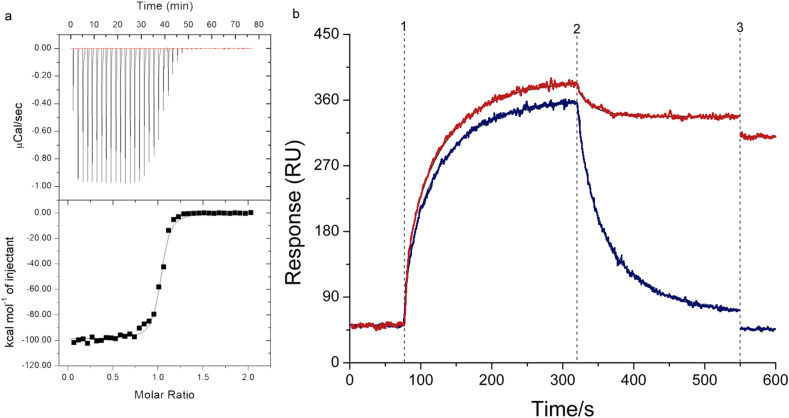
The proposed synthetic probe is first tested in targeting the virus protease in the bulk solution phase. Since the fluorescent thiol-targeting molecule may readily react with the protease if being placed near to the free thiol on the protease molecule, the thiol reactive maleimide group in this fluorescent-targeting molecule is removed, resulting in a control probe that always give out fluorescent signal. As the maleimide, before reacting with thiol group, can quench the fluorophore. Using this control probe that can only non-covalently target the virus protease, isothermal titration calorimetry (ITC) measurement is conducted. And as shown in Fig. 1 a, the substrate-mimicking sequence seems to work properly in docking with the target virus, showing a typical saturated binding curve (the calculated affinity is 6.82 (±0.43) × 10−6 M). This step is the pre-requisite for subsequent covalent thiol-targeting, as only molecular recognition of the probe can bring the thiol-targeting molecule to the vicinity of the free thiol groups around the substrate binding pocket of the protease. To examine this covalent step, surface plasma resonance (SPR) spectra are recorded for the designed probe as well as for the control probe. As shown in Fig. 1b, in the blue line corresponding to the control probe, a saturated binding curve can be observed. While in the red line representing the designed probe, the binding is generally similar to the control, but the signal response is higher, showing the possible existence of a second source of SPR signal, the response of which is superimposed on the first. This might be due to the covalent coupling. As the non-covalent molecular recognition is actually fast and real-time association and dissociation, so the signal recorded is smaller than the covalent curve. To verify this, mild surfactant is first added to wash the sensing surface. And dramatic reduction of SPR signal can be observed in the case of the control probe, indicating that many non-covalently surface-captured protease molecules are disrupted by the surfactant when interacting with the probe, and is washed away. In contrast, the red line shows a signal drop much smaller, suggesting that some of the surface-captured protein molecules have already formed covalent linkage with the probe. But this process is not as fast as the non-covalent recognition, since there are still considerable protease molecules being washed-off. And more non-covalently attached protease molecules can still hold fast, since the surfactant is mild. So in a second proving step, de-naturing detergent is added. This can destroy the conformation of the non-covalently surface-captured proteins, resulting in total loss of protein SPR response in the blue line, as expected, but again the signal drop in the red line is limited, and the remaining signal can be postulated as from the covalently bound protease molecules on the surface. The kinetics of the SPR for the red curve (kon=1.03 (±0.21) × 108 M−1s−1, koff=1.82(±0.13) × 10−1 s−1) results an apparent affinity of 1.77(±0.02) × 109 M.
Fig. 1.
Validation of the covalent detection mechanism. (a) Isothermal titration calorimetry (ITC) results obtained by titrating 0.1 mM control probe into 0.01 mM viral protease. (b) Surface Plasma Resonance (SPR) time course recorded during detecting the target protease using the designed probe (red), and the control probe (blue). The points 1–3 represents the start of adding the protease, the mild surfactant and the de-naturing. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Fluorescent detection as the intermediate step
Then the fluorescent signal of covalent bonding between the probe and the target protein can be directly studied. But before that step, the surface density of the probe molecules should be studied. To this end, the fluorescent thiol-targeting molecules is swapped out, and replaced by an electrochemical reporter molecule, ferrocene. The result is shown in Figure S1, which is the cyclic voltammetric I–V curve recorded on the ferrocene-tagged probe modified sensing surface. While knowing the potential scanning rate, using the electrochemical response of ferrocene, the area below the curves can be calculated to give the surface density of the probe. This is based on the Faraday law of electrolysis. And estimation of the order of magnitude is enough to guarantee that the average distance between each two probes are far larger than the size of the target protein. This, combined with the high ion strength of the serum, can prevent undesirable mutual stereo-hindrance during the non-covalent molecular recognition between the probe and the target protein. Because the assay only involves one step of incubation, there are not many experimental conditions to be optimized.
3.3. Induced cross-linking for improved sensitivity
The fluorescent signal of probe-protein covalent binding is then first employed to trace the time line of probe-protein incubation. As shown in Fig. 2 a and b, this covalent process is indeed slower than the non-covalent interaction represented by the blue curve in Fig. 1b. It takes over 1 h for most non-covalently surface-captured protease molecules to form the fluorescent covalent bond. Under this optimized condition, the direct fluorescent detection of the virus protease in serum diluted sample is studied. As shown in Figure S2, quantitative detection can be realized in serum samples, and a working curve can be established between fluorescent peak repsonse and the logarithm of protease concentration. In the absence of the target protein, the main types of serum proteins cannot generate evident interfering noise (Fig. 2c), since only the probe-protease molecular recognition can bring the fluorescent thiol-targeting molecule within range of free thiol groups on protein. To study this point in depth, control experiments using blank serum samples are conducted. As shown in Figure S3a, it needs far longer time for the non-specific proteins in the serum to form covalent-link with the maleimide group of the probe. In Figure S3b, cysteine molecules are added directly into the serum, and what looks similar to the original reporting constructing the fluorescent thiol-targeting molecules is achieved. These results can be viewed in light of the fact that the large protein molecule itself effectively enlarges the distance between the thiol group and the probe. Since only correctly oriented protein can put its free thiol groups in contact with the probe. And only the probe-target molecular recognition can induce the correct orientation effectively.
Fig. 2.
Detection of the protease using the thiol sensitive probe. (a) Time course of probe and protease incubation recordede as the peak fluorescent response of the probe. Inset is the fluorescent peaks plotted as a function of incubation time. Error bars represents the standard deviation (n=3). (b) Quantitative fluorescent detection of the protease using the probe. Inset is the calibration curve, the linear working curve about the logarithm of protease concentration has been included. Error bars are of the same meaning as (a). (c) Specificity with respect to various control targets. The concentration of all control targets is 33 nM.
To improve the sensitivity of detection, electrochemical potential scanning is applied to the sensing surface during incubation, to induce cross-linking of the serum proteins for signal amplification. The effect of this cross-linking is first noted morphologically using atomic force microscopy (AFM). It is evident that after cross-linking (Fig. 3 a), an extensive web-like morphology is developed, if compared with the probe-modified surface before incubation (Fig. 3b). But this cross-linking is non-specific, only that the probe does not contain reactive amino acids that can cross-link with the interfering serum proteins. So, in the absence of the target protease, the blank buffer is incubated with the sensing surface. The time course is recorded in Figure S4a, the background signal is negligible. This is because the probe is peptidomimetic but not the amino acids that can react with the serum protein, and the Thiol group is also absent in the probe. As a proof, the solution for thorough rinsing is collected after the slide has been incubated for long time with the blank serum. The solution is centrifugated; and the parameter is set so that normally, proteins won't be fractionated. But this time there is sediment, which, if collected and dried on a working electrode, can generate the characteristic electrochemical response of aromatic amino acid oxidation (Figure S4b). This result confirms that the electrochemical scanning is able to cross-link proteins to form larger oligomers, but they cannot anchor to the surface slide. In Figure S4c, the time course of detecting the target protease at relative high and extremely diluted concentrations are studied, and it can be concluded that 50 min incubation under electrochemical agitation is the optimal for maximizing the signal difference between different concentrations. As shown in Figure S4, the peptide probe can work specifically for SARS-Cov-3CL protease.
Fig. 3.
Atomic Force Microscopy (AFM) results before (a)/after (b) electrochemical cross-linking among the proteins of the serum sample on the sensing slide.
3.4. Analytical performance
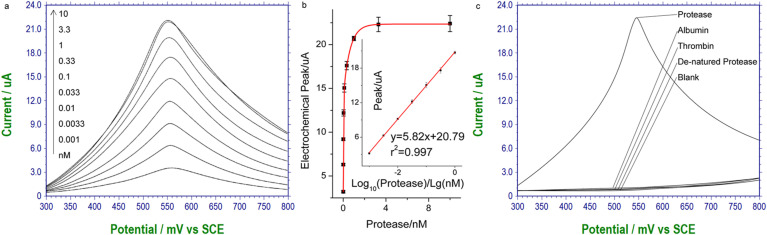
Under this optimized condition, the detection of the target using the amplified electrochemical response of aromatic amino acid oxidation is carried out in serum diluted signal. As shown in Fig. 4 a and b, quantitative detection is possible, while Fig. 4c shows negligible noise in the absence of the target protease and the presence of various serum proteins. The clinically required sensitivity and limit of detection (LOD) of SARS-Cov-3CL protease is equivalent to around 182 pM, while the LOD of the proposed method is 1 pM. The performance of this method also compares favorably with commercially available methods (Table S1). This amplified electrochemical signal ensures higher sensitivity compared with the above fluorescent method as shown in Fig. 2, so it is chosen for the final clinical test. Using this method, the clinical serum samples are detected, serum from healthy volunteers serves as the control (Fig. 5 ), and the detected serum level of the viral protease in the patients are clustered in the range of 0.16 nM–1.6 nM. It can be seen that the method is well adapted to detect the presence of this unique protease in the serum environment.
Fig. 4.
Analytical performance in detecting the viral protease in serum sample using the electrochemical cross-linking. (a) Quantitative electrochemical results of tyrosine oxidation after cross-linking among the target protease and serum proteins. The peak currents in (a) is plotted as a function of protease concentration in (b), and the linear range has also been included. (c) Specificity of similar meaning to Fig. 3b, control targets are all of 3.3 nM.
Fig. 5.
Detection of clinical serum samples with the serum of healthy volunteers as the control. The method adopted is that described in Fig. 4. Briefly, the serum samples of 9 infected patients, as well as similar samples collected via vein extract from 9 male healthy volunteers are dropped onto probe modified sensing slides to induce the covalent capturing of the target viral protease. Then electrochemical potential scanning at 1 mV/s between 0 and 0.8 V is applied to the slide to induce cross-linking with the serum proteins. Finally, using the electrochemical signal of aromatic amino acid of the serum proteins, the protease contained in the sample is detected as signal readout.
4. Conclusion
In conclusion, we have developed a maximally simplified detection method that requires no antibodies and enzymes. Using this method we have successfully detected a marker protein of the SARS-Cov-2 virus in serum clinical samples in a one-step and reagent-less fashion. Specifically, a mimic of the substrate of the target protease is used to form the main sequence of the probe, while a fluorescent thiol-targeting molecule is responsible for signal generation. The interaction between the probe and the target protein brings the two molecules to the vicinity of each other, so that the thiol-sensitive molecule can specifically target the free thiol group on the target protein, forming new covalent bond. In consequence, the fluorophore previously quenched by the thiol-targeting group can now gives out fluorescent signal in proportion to concentration of the target protein. This method has been further improved by amplifying the signal readout using the interfering proteins in the serum. Through electrochemical potential scanning, an electroactive 2D nanonetwork can be formed on the sensing surface, only in the presence of the captured target protein molecules as the surface anchoring points. This method is capable of detecting the virus protease in clinical serum samples of the SARS-Cov-2 infected patients. These results point to clinical application of the proposed method in combat against COVID-19 in the near future.
5. Supplementary data
Supplementary data can be found in the online version.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (21705061, 81602737), the Jiangsu Provincial Key Medical Discipline (Laboratory) (ZDXKA2016017), and the Innovation Capacity Development Plan of Jiangsu Province (BM2018023).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113579.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Basile K., Maddocks S., Kok J., Dwyer D.E. Accuracy amidst ambiguity: false positive SARS-CoV-2 nucleic acid tests when COVID-19 prevalence is low. Pathology. 2020;52(7):809–811. doi: 10.1016/j.pathol.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozé, N., Paireau, J., Lapidus, N., Tran Kiem, C., Salje, H., Severi, G., Touvier, M., Zins, M., de Lamballerie, X., Lévy-Bruhl, D., Carrat, F., Cauchemez, S., Monitoring the Proportion of the Population Infected by SARS-CoV-2 Using Age-Stratified Hospitalisation and Serological Data: a Modelling Study. The Lancet Public Health. [DOI] [PMC free article] [PubMed]
- Jiang S., Hägglund P., Carroll L., Rasmussen L.M., Davies M.J. Crosslinking of human plasma C-reactive protein to human serum albumin via disulfide bond oxidation. Redox Biology. 2021;41:101925. doi: 10.1016/j.redox.2021.101925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra P., Dhiman A., Cho W.C., Bruno J.G., Sharma T.K. Simple methods and rational design for enhancing aptamer sensitivity and specificity. Frontiers in Molecular Biosciences. 2018;5(41) doi: 10.3389/fmolb.2018.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraj A., Brouwer H.-J., Reinhoud N., Chervet J.-P. A novel electrochemical method for efficient reduction of disulfide bonds in peptides and proteins prior to MS detection. Anal. Bioanal. Chem. 2013;405(29):9311–9320. doi: 10.1007/s00216-013-7374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586(7830):516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Liu T., Huo F., Li J., Chao J., Zhang Y., Yin C. A fast response and high sensitivity thiol fluorescent probe in living cells. Sensor. Actuator. B Chem. 2016;232:619–624. [Google Scholar]
- Maiti B.K. Can papain-like protease inhibitors halt SARS-CoV-2 replication? ACS Pharmacology & Translational Science. 2020;3(5):1017–1019. doi: 10.1021/acsptsci.0c00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbarra A.N., Rolfe S., Nguyen J.Q., Earl L., Galles N.C., Marks A., Abbas K.M., Abbasi-Kangevari M., Abbastabar H., Abd-Allah F., Abdelalim A., Abdollahi M., Abegaz K.H., Abiy H.A.A., Abolhassani H., Abreu L.G., Abrigo M.R.M., Abushouk A.I., Accrombessi M.M.K., Adabi M., Adebayo O.M., Adekanmbi V., Adetokunboh O.O., Adham D., Afarideh M., Aghaali M., Ahmad T., Ahmadi R., Ahmadi K., Ahmed M.B., Alanezi F.M., Alanzi T.M., Alcalde-Rabanal J.E., Alemnew B.T., Ali B.A., Ali M., Alijanzadeh M., Alinia C., Alipoor R., Alipour V., Alizade H., Aljunid S.M., Almasi A., Almasi-Hashiani A., Al-Mekhlafi H.M., Altirkawi K.A., Amare B., Amini S., Amini-Rarani M., Amiri F., Amit A.M.L., Amugsi D.A., Ancuceanu R., Andrei C.L., Anjomshoa M., Ansari F., Ansari-Moghaddam A., Ansha M.G., Antonio C.A.T., Antriyandarti E., Anvari D., Arabloo J., Arab-Zozani M., Aremu O., Armoon B., Aryal K.K., Arzani A., Asadi-Aliabadi M., Asgari S., Atafar Z., Ausloos M., Awoke N., Quintanilla B.P.A., Ayanore M.A., Aynalem Y.A., Azadmehr A., Azari S., Babaee E., Badawi A., Badiye A.D., Bahrami M.A., Baig A.A., Bakhtiari A., Balakrishnan S., Banach M., Banik P.C., Barac A., Baradaran-Seyed Z., Baraki A.G., Basu S., Bayati M., Bayou Y.T., Bedi N., Behzadifar M., Bell M.L., Berbada D.A., Berhe K., Bhattarai S., Bhutta Z.A., Bijani A., Birhanu M., Bisanzio D., Biswas A., Bohlouli S., Bolla S.R., Borzouei S., Brady O.J., Bragazzi N.L., Briko A.N., Briko N.I., Nagaraja S.B., Butt Z.A., Cámera L.A., Campos-Nonato I.R., Car J., Cárdenas R., Carvalho F., Castaldelli-Maia J.M., Castro F., Chattu V.K., Chehrazi M., Chin K.L., Chu D.-T., Cook A.J., Cormier N.M., Cunningham B., Dahlawi S.M.A., Damiani G., Dandona R., Dandona L., Danovaro M.C., Dansereau E., Daoud F., Darwesh A.M., Darwish A.H., Das J.K., Weaver N.D., De Neve J.-W., Demeke F.M., Demis A.B., Denova-Gutiérrez E., Desalew A., Deshpande A., Desta D.M., Dharmaratne S.D., Dhungana G.P., Dianatinasab M., Diaz D., Dipeolu I.O., Djalalinia S., Do H.T., Dorostkar F., Doshmangir L., Doyle K.E., Dunachie S.J., Duraes A.R., Kalan M.E., Leylabadlo H.E., Edinur H.A., Effiong A., Eftekhari A., El Sayed I., El Sayed Zaki M., Elema T.B., Elhabashy H.R., El-Jaafary S.I., Elsharkawy A., Emamian M.H., Enany S., Eshrati B., Eskandari K., Eskandarieh S., Esmaeilnejad S., Esmaeilzadeh F., Esteghamati A., Etisso A.E., Farahmand M., Faraon E.J.A., Fareed M., Faridnia R., Farioli A., Farzadfar F., Fattahi N., Fazlzadeh M., Fereshtehnejad S.-M., Fernandes E., Filip I., Fischer F., Foigt N.A., Folayan M.O., Foroutan M., Fukumoto T., Fullman N., Gad M.M., Geberemariyam B.S., Gebrehiwot T.T., Gebrehiwot A.M., Gebremariam K.T., Gebremedhin K.B., Gebremeskel G.G., Gebreslassie A.A., Gedefaw G.A., Gezae K.E., Ghadiri K., Ghaffari R., Ghaffarifar F., Ghajarzadeh M., Gheshlagh R.G., Ghashghaee A., Ghiasvand H., Gholamian A., Gilani S.A., Gill P.S., Girmay A., Gomes N.G.M., Gopalani S.V., Goulart B.N.G., Grada A., Guimarães R.A., Guo Y., Gupta R., Hafezi-Nejad N., Haj-Mirzaian A., Haj-Mirzaian A., Handiso D.W., Hanif A., Haririan H., Hasaballah A.I., Hasan M.M., Hasanpoor E., Hasanzadeh A., Hassanipour S., Hassankhani H., Heidari-Soureshjani R., Henry N.J., Herteliu C., Heydarpour F., Hollerich G.I., Rad E.H., Hoogar P., Hossain N., Hosseini M., Hosseinzadeh M., Househ M., Hu G., Huda T.M., Humayun A., Ibitoye S.E., Ikilezi G., Ilesanmi O.S., Ilic I.M., Ilic M.D., Imani-Nasab M.H., Inbaraj L.R., Iqbal U., Irvani S.S.N., Islam S.M.S., Islam M.M., Iwu C.J., Iwu C.C.D., Jadidi-Niaragh F., Jafarinia M., Jahanmehr N., Jakovljevic M., Jalali A., Jalilian F., Javidnia J., Jenabi E., Jha V., Ji J.S., John O., Johnson K.B., Joukar F., Jozwiak J.J., Kabir Z., Kabir A., Kalani H., Kalankesh L.R., Kalhor R., Kamal Z., Kanchan T., Kapoor N., Karami M., Matin B.K., Karch A., Karimi S.E., Kayode G.A., Karyani A.K., Keiyoro P.N., Khader Y.S., Khafaie M.A., Khammarnia M., Khan M.S., Khan E.A., Khan J., Khan M.N., Khatab K., Khater M.M., Khatib M.N., Local Burden of Disease Vaccine Coverage, C Mapping routine measles vaccination in low- and middle-income countries. Nature. 2021;589(7842):415–419. doi: 10.1038/s41586-020-03043-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocks B.B., Melanson J.E. In-source reduction of disulfide-bonded peptides monitored by ion mobility mass spectrometry. J. Am. Soc. Mass Spectrom. 2018;29(4):742–751. doi: 10.1007/s13361-018-1894-1. [DOI] [PubMed] [Google Scholar]
- Weinstein M.C., Freedberg K.A., Hyle E.P., Paltiel A.D. Waiting for certainty on covid-19 antibody tests — at what cost? N. Engl. J. Med. 2020;383(6):e37. doi: 10.1056/NEJMp2017739. [DOI] [PubMed] [Google Scholar]
- Zhang K., Yang Q., Fan Z., Zhao J., Li H. Platelet-driven formation of interface peptide nano-network biosensor enabling a non-invasive means for early detection of Alzheimer's disease. Biosens. Bioelectron. 2019;145:111701. doi: 10.1016/j.bios.2019.111701. [DOI] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.