Abstract
The COVID-19 pandemic brought attention to studies about viral infections and their impact on the cell machinery. SARS-CoV-2, for example, invades the host cells by ACE2 interaction and possibly hijacks the mitochondria. To better understand the disease and to propose novel treatments, crucial aspects of SARS-CoV-2 enrolment with host mitochondria must be studied. The replicative process of the virus leads to consequences in mitochondrial function, and cell metabolism. The hijacking of mitochondria, on the other hand, can drive the extrusion of mitochondrial DNA (mtDNA) to the cytosol. Extracellular mtDNA evoke robust proinflammatory responses once detected, that may act in different pathways, eliciting important immune responses. However, few receptors are validated and are able to detect and respond to mtDNA. In this review, we propose that the mtDNA and its detection might be important in the immune process generated by SARS-CoV-2 and that this mechanism might be important in the lung pathogenesis seen in clinical symptoms. Therefore, investigating the mtDNA receptors and their signaling pathways might provide important clues for therapeutic interventions.
Keywords: SARS-CoV-2, Mitochondria, Innate receptors, Cytokine storm
Background
At the end of 2019, a new severe acute respiratory syndrome coronavirus (SARS-CoV-2) emerged, and by the middle of 2020 it was already a worldwide pandemic [1,2]. SARS-CoV-2 is a RNA virus and its viral proteins interact with angiotensin-converting enzyme carboxypeptidase 2 (ACE2) and TMPRSS2 proteins of the host to enter the cells and begin the viral replication causing the COVID-19 disease [3]. The virus is capable of causing a severe acute respiratory syndrome lead by an exacerbated immune response triggering a cytokine storm profile [4].
Several recent data have directly linked SARS-CoV-2 infection with the metabolic status and the mitochondrial function of the host cells, and there is commending evidence about an intrinsic relationship between the SARS-CoV-2 viral cycle and the mitochondrial compartment. Risk groups for COVID-19 comprehend people with metabolic diseases like diabetes, obesity and elder people, that have in common decreased mitochondrial function and metabolic alterations [5,6]. Although the main immune considerations established so far regarding mitochondria are mainly related to the maintenance of metabolism in leukocytes [7,8] and the signaling pathways related to the detection of viral RNA (by RIG-I/MAVS for example), other possible immunomodulatory mechanisms, such as the release of mitochondrial damage associated with molecular patterns (DAMPs) into the intracellular or extracellular environment [9,10], were recently discovered. The main DAMP related to this organelle is mitochondrial DNA (mtDNA), the result of the endosymbiont origin that occurred more than 1 billion years ago [11]. Disruption of mtDNA signaling trough diverse pattern recognition receptors (PRRs) is an evolved strategy for a variety of RNA viruses, like dengue and influenza, among others.
As discussed throughout this article, mitochondria have different effects on immune responses [12,13] as mtDNA can be directly recognized as a DAMP, and can be linked to systemic inflammation and acute lung injury (Figure 1) [14]. Herein, we highlight a possible novel role for mtDNA in the COVID-19 pathogenesis and give a glimpse about the receptors responsible for its detection and how this can be connected with a worse prognosis. In this review, we hypothesize that mtDNA can affect inflammatory processes and influence SARS-CoV-2 treatment approaches.
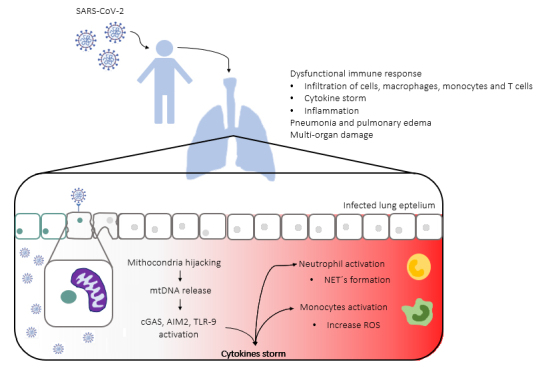
Figure 1. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lung pathogenesis. SARS-CoV-2 infected cells express the angiotensin-converting enzyme 2 (ACE2) receptor and TMPRSS2, seen in cells of the respiratory tract, among other tissues. The SARS-CoV-2 virus during the replication process hijacks the mitochondria, which leads to the release of mitochondrial DNA (mtDNA) into the cytosol of the cell. The mtDNA presented in the cytosol can be recognized by several receptors as damage-associated molecular patterns (DAMP), such as cGAS, AIM2, TLR9, activating a signal cascade. The signals activated by the DAMP-recognition will be identified by other cells, such as neighbor epithelial cells, endothelial cells, macrophages, among others, triggering a pro-inflammatory response. Meanwhile, the release of mtDNA is also inducing the NETs formation in neutrophils and increasing the production of reactive oxygen species (ROS) in infected monocytes. The activated pro-inflammatory response in different immune cells attracts other cells to the infection site - macrophages, monocytes, T cells - boosting the inflammation and enhancing the cytokine production, which leads to the cytokine storm and ultimately damaging the site. The high concentration of immune cells and the cytokine reach other organs, which eventually causes multi-organ damage.
The pathological roles of mtDNA
mtDNA, unlike genomic DNA, is present in several circular copies of approximately 16.4kbp and it is considered rich in unmethylated CpG sequences, the target of receptors such as TLR9. mtDNA is compartmentalized within the inner membrane of the mitochondria in healthy cells. However, in some pathologies, cell death events, or mitochondrial stress, mtDNA escapes from mitochondrial boundaries and becomes available in the cytosol or extracellular environment [15,16]. An overview of the mechanisms that lead to the presence of mtDNA outside the mitochondria has been extensively recently reviewed [17].
mtDNA can activate immune responses after being extruded in classically inflammatory cell death pathways such as necrosis, pyroptosis or necroptosis.
Necroptotic cell death can decrease pathogen replication, but it may also release damaged mitochondria in this process [18] which possibly increases the level of mtDNA and inflammation. Likewise, necroptosis is capable of induce lung pathogenesis. It perturbs the bronchial epithelial integrity, as seen in mice infected with influenza [19], and, in humans, necrotic cell death in response to H1N1 infection is related to Acute Respiratory Distress Syndrome (ARDS) [20]. Curiously, necroptosis can be induced by accessory protein open reading frame 3a (Orf3a), presented in SARS-CoV [21], an protein also present in the SARS-CoV-2 genome. Although it has been shown that SARS-CoV-2 Orf3a can induce apoptosis, it is not clear whether it can also stimulate necroptosis [22]. Besides, mtDNA has been shown to be important in several other cellular events such as inducing apoptosis [23], neutrophil extracellular traps formation (NETs) [24], renal fibrosis and chronic and acute renal diseases [25,26] and in inflammatory events in the lung and hepatocytes (Figure 1) [27-29].
In 2014, two distinct groups showed that, during apoptosis in a caspase 3/7 knockout model, through the permeabilization of mitochondrial membrane by pores formed by BAK and BAX, mtDNA can be released in the cytosol [30,31]. The cytosolic mtDNA activates mainly the cGAS-STING pathway (discussed below), and this leads to the induction of type I interferons (IFNs) production (Figure 2) [30,31]. It was discovered that during apoptosis, the effector caspases are responsible for suppressing the pro-inflammatory response as they can directly cleave IRF3 and cGAS, therefore inhibiting its function as a PRR, consequently decreasing the production of type I IFNs [32]. Also, it was observed that even in the presence of effector caspases, the permeabilization of the mitochondrial outer membrane by BAX-BAK gradually increases, allowing the release of mtDNA [33]. mtDNA can also be released by the action of the N-terminal portion of the gasdermin-D protein after stimulation of cells with LPS [34]. Cleaved gasdermin-D can be formed as part of the action of the inflammasome complexes [35]. Interestingly, cleaved gasdermin-D can also direct to pyroptotic cell death, thus additionally corroborating mtDNA release. This indicates that the activation of inflammasome complexes can be directly linked to the release of mtDNA in the cytosol, which may suggest a new impact of these receptors directly with the mitochondrial homeostasis, and consequently with cell metabolism.
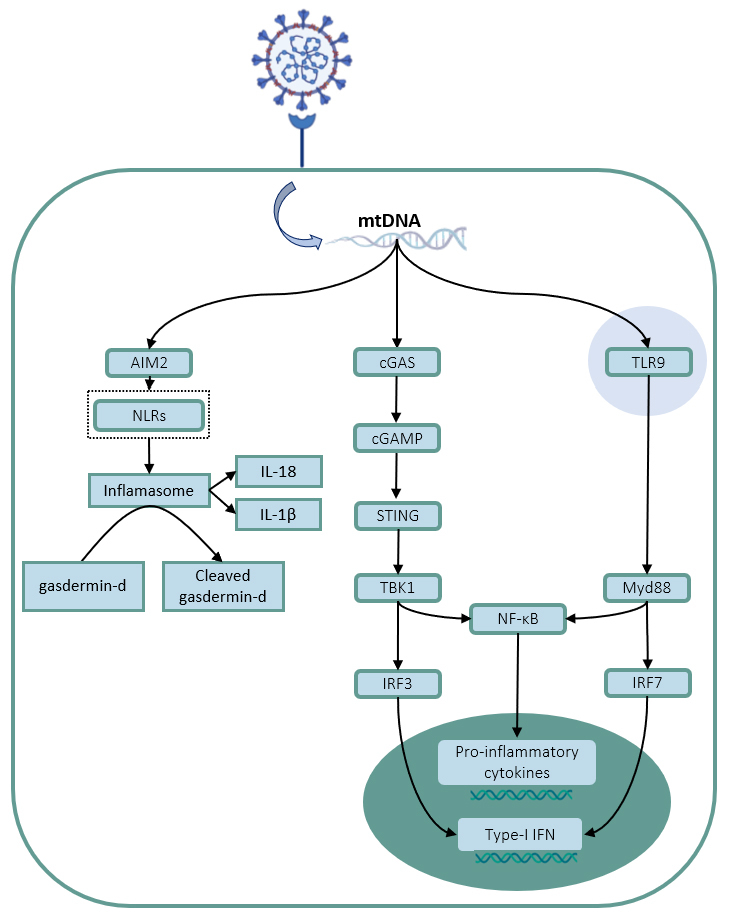
Figure 2. Molecular mechanisms involved in mtDNA recognition on the cytosol and endosome. SARS-CoV-2 infection leads to mtDNA releases. The TLR9, inside the endosome, recognizes mtDNA and triggers the Myd88 pathway, consequently inducing the release of pro-inflammatory factors and type-I IFNs, seen as well in cGAS pathway, but as a response of STING activation. The mtDNA is recognized by AIM2 leading to inflammasome activation, IL-18 and IL-1β production and cleave gasdermin-D.
On mitochondria, mtDNA and SARS-CoV-2
The SARS-CoV-2 infection might be directly related with the mitochondrial status on the cells that are being infected, what could help explain why people with chronic diseases are on higher risk groups. Glycolysis and HIF-1α stabilizations have been shown to be fundamental to the replicative process of the virus in monocytes, with their inhibition abruptly decreasing the viral load in these cells [36]. Importantly, SARS-CoV-2 infected monocytes had an increased level of mitochondrial reactive oxygen species (mtROS) [36]. mtROS can directly oxidize the mitochondrial DNA, and therefore reduce mitochondrial bioenergetics and ATP supply for the cell [37]. Oxidized mtDNA and mtROS are both strong inductors of the NLRP3 inflammasome, a multiproteic complex that leads to the active forms of the cytokines IL-1β and IL-18 by cleaving pro-caspase1 into active caspase1 [38,39]. In COVID-19 patients, NLRP3 has been shown to be strictly associated with disease severity. NLRP3 is activated in patients with COVID-19 and both IL-18 and active caspase 1 can be associated with distinct levels of the disease [40]. Additionally, mtDNA when oxidized fails to undergo clearance by enzymatic repair mechanism, thus becoming more persistent in the cell and increasing their chances of activating an immune response.
SARS-CoV-2 is also proposed to have direct contact with mitochondria. SARS-CoV infection alter the functionality of the mitochondria and their accessory protein open reading frame 9b (Orf9b) is localized in the mitochondria [41]. SARS-CoV Orf3b, Orf7a and Orf8a are also in direct contact with mitochondria [42]. These proteins are mainly related to inducing apoptotic pathways in the infected cells [43,44]. Curiously, SARS-CoV-2 also presents amino acid sequences for the proteins Orf7a, Orf8a, and Orf9b, analogous to SARS-CoV [42]. In fact, a relationship of SARS-CoV-2 with mitochondria may be even more intricate, given that the predicted localization of the viral RNA is both the nucleus an the mitochondria, suggesting that this relationship is important to viral replication, with the virus being able to hijack the mitochondria and replicate inside this organelle [45]. It is very possible that if this really happens mtDNA can be easily released by exosomes, one of the main mechanism that the SARS-CoV-2 uses to exit the cell [46,47]. These circulating exosomes containing mtDNA can be a trigger to a more pronounced systemic inflammation.
There is an association of a higher mtDNA plasma levels with ARDS in patients during critical conditions like sepsis or trauma [48]. Interestingly, circulating free mtDNA has also been shown as a predictive marker for COVID-19 pathogenesis [49]. It presented itself as a very accurate predictor of the principal severe outcomes of COVID-19, mortality, intensive care unit admission and intubation. In the article that proposed this marker, the levels of mtDNA were a more precise biomarker than the current clinically utilized, like D-dimer and C reactive Protein [49]. This data indicates that this DAMP can be, in fact, pivotal for the observed lung pathogenesis.
Regarding the interactome of SARS-CoV-2 proteins with the host cell, Gordon et al. [50] showed that the proteins Orf9c, NSP4 and NSP8 can also directly interact with mitochondria. Among the proteins found to interact with SARS-CoV-2 proteome there are mitochondrial ribosomal proteins, assembled proteins of the NADH dehydrogenase complex and proteins with intrinsic relationship with variety of mitochondrial metabolic pathways [42,50]. The article also presented results showing that a very considerable amount of the interactome was directed to pathways regarding endomembrane compartments and vesicle trafficking [50]. In addition, the SARS-CoV protein NSP3 was found to interact with the structural mitochondrial protein prohibitin (PHB) and modulate the cell survival signaling [51]. Although the result of most of the interactions presented above are still unknown, there is a clear presence of the SARS-CoV-2 in the mitochondria, inferring an important role of this organelle for the viral replication.
The mtDNA receptors and their role in inflammatory processes - do they constitute a possible link with SARS-CoV-2?
In viral diseases, it is thought that the activation of mtDNA receptors are not restricted only to DNA viruses, since important pathological events such as mitochondrial stress [52] and cell death in the inflammatory microenvironment are triggered with reasonable frequency in these situations. The extravasation of mtDNA into the cytosol or into the extracellular environment is observed in infections involving RNA viruses, such as dengue [53], Zika (ZIKV) [54], influenza virus [55], and even possibly in the infection caused by SARS-CoV-2 [49,56].
As mtDNA presence in the cytosol can develop several different immune responses. The study of different receptors involved in mtDNA sensing can affect directly in our knowledge of the inflammatory processes. Here, we briefly describe the mechanisms involved in the receptors that already have been shown to elicit an immune response to mtDNA (Figure 3).
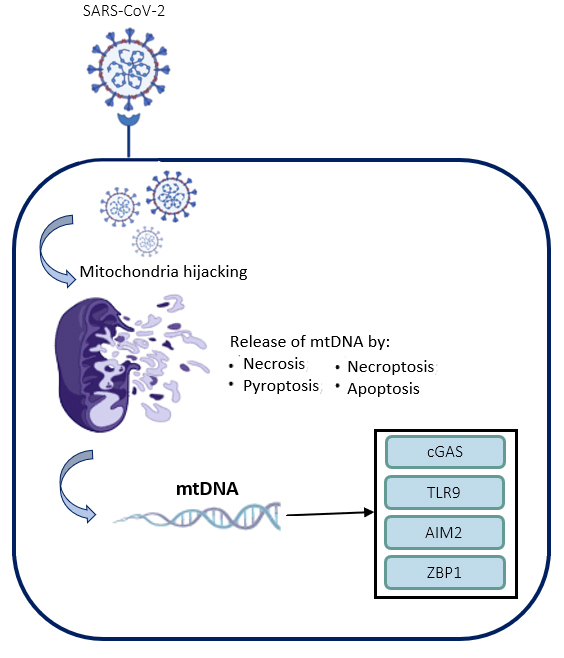
Figure 3. DNA receptors and SARS-CoV-2 mechanisms upon infection. Scheme showing the hijacking of mitochondria due SARS-CoV-2 entry. The mitochondria release mtDNA as consequence of different process, such as necrosis, pyroptosis, necroptosis and apoptosis. The mtDNA can be recognized by TLR9, cGAS, AIM, ZBP1.
Cyclic GMP-AMP synthase - stimulator of interferon genes (cGAS)
One of the main mechanisms that evolved to respond to the detection of cytosolic DNA is the protein cGAS and its downstream pathway. cGAS is currently regarded as the cytosolic DNA sensor that most corroborates with the detection and signaling of DNA - both endogenous and exogenous - as well as with the production of type I IFNs in the most diverse conditions and cell [57-60]. Upon recognizing DNA, cGAS catalyzes the formation of cyclic GMP-AMP (cGAMP) from cytosolic ATP and GTP [61]. The cGAMP produced by cGAS is a peculiar cyclic dinucleotide containing mixed phosphodiester linkages, connecting the 2’ hydroxyl and 5’ phosphate regions of GMP, with the 5 ‘phosphate and 3’ hydroxyl regions of AMP, respectively [58, 60-62]. Once recognized it leads to the activation of STING, an endoplasmic reticulum protein. After being activated STING leads to the activation of the transcription factors Interferon Responsible Factor 3 (IRF3) and NF-kB and to the induction of type I IFNs and inflammatory cytokines, respectively [60,63-65].
The conformational mechanisms that activate STING, as well as those that lead to the recruitment of transcription factors in its pathway, are not yet fully established. It was shown that STING needs to be phosphorylated in its Ser366 residue by the Tank Binding Kinase 1 (TBK1), and that TBK1 is also responsible for the subsequent phosphorylation and activation of the IRF3 transcription factor [57,63,66,67]. Additionally, STING can recruit IKK kinase complex and lead to phosphorylation and translocation of NF-kB to the nucleus (Figure 2) [57].
The extravasation of mtDNA into the extracellular environment, or into the cytosol is a process seen in different RNA viruses. Therefore several viral strategies have emerged to try to circumvent the cGAS-STING pathway [68]. The Dengue Virus for example is capable of expressing the NS2B protease, responsible for leading to lysosomal degradation of cGAS [69], ZIKV is also capable of degrading cGAS by the action of NS1 and caspase-1 [54], and SARS-CoV and HCoV-NL63, two coronaviruses, express papain-like proteases capable of inhibiting STING-mediated IRF3 activation [70]. Interestingly, the SARS-CoV-2 virus also expresses a similar papain-like protein [71].
Still, in a more general context, it was recently discovered that the cytokine IL-1β, present in a plethora of inflammatory processes and derived from inflammasome cleavage of pro-IL-1β, is capable of inducing the release of mtDNA and consequent activation of the cGAS-STING pathway in pulmonary epithelial cells A549 and the myeloid lineage THP-1 [72]. Additionally, Gkirtzimanaki et al. [73] also showed that INF-α leads the release of mtDNA in monocytes derived from patients with lupus. In this case, cytosolic mtDNA is derived from changes in mitochondrial metabolism, inducing an increase in ROS and lysosomal pH, culminating in an inefficient mitophagy in the monocytes analyzed. mtDNA in this case also induces the cGAS-STING pathway and leads to an increase in the cellular inflammatory profile [73]. Although in this case patients with untreated lupus were compared with healthy controls, representing in this a condition of chronic inflammation vs homeostasis, it would be important for a better understanding of the immune system to delimit the temporal variable and what is the necessary basal level of chronic inflammation for this phenotype to be observed. These responses may indicate fundamental insights into diseases where type I IFNs are important, as apparently is the case with SARS-CoV-2 disease.
Toll-like 9 (TLR9)
TLR’s comprehend a family of several membrane associated PRRs capable of sensing a variety of different stimuli, ranging from pathogen associated molecular patterns like LPS to damage associated molecular patterns like extracellular ATP. TLR9 is normally present in endosomes and is activated in the presence of unmethylated CpG of both double and single strand DNA structures [74]. DNA rich in unmethylated CpG structures is a common characteristic of several different pathogens as well as of mtDNA, and it is present in viruses such as Epstein-Barr virus [75], Herpes Simplex viruses (HSV-1 and HSV-2)[76,77] and Cytomegalovirus (CMV) [78]. After the recognition and binding of DNA by TLR9, the receptor dimerizes and allow the binding of the adaptor protein MyD88 [74,79,80]. This complex activates a signaling cascade that stimulates the translocation of NF-κB and IRF3 to the nucleus, leading to the expression of genes responsible for the production of cytokines and chemokines, such as type I IFNs (Figure 2) [81,82]. The detection of mtDNA by the TLR9 receptor, similar to cGAS, also leads to a plethora of different responses. The recognition of mtDNA by TLR9 in the lung induces inflammation via NF-kB pathway [83,84]. The mtDNA/TLR9 axis was also shown to be able to be important in the induction of cardiomyopathy [85], promote ischemia-reperfusion injury [86], induce apoptosis [87], muscle inflammation [88], and vascular disfunction [89,90].
Absent in melanoma 2 (AIM2)
The AIM2 sensor was initially described in 2008 [91] after observations that macrophages deficient in the ASC protein, an essential protein for the function and activity of the inflammasome complexes, failed to induce the cytokine IL-1β and cell death after the cells were transfected with double stranded DNA (dsDNA) [91,92]. It was observed that this event did not happen to macrophages lacking the NLRP3, NLRP6 or NLRP12 inflammasome proteins, therefore giving rise to the AIM2 function [92].
AIM2 is a multimeric cytoplasmic sensor, belonging to the AIM2 like receptors protein family [93] and is able to recognize double-stranded DNA both from self, viruses and bacteria, and thus form the inflammasome complex. AIM2 is able to induce the active forms of caspase-1 and the cytokines IL-1β and IL-18 and also lead to cell death by pyroptosis via cleavage of the gasdermin-D protein [94].
AIM2 can be activated independent of the DNA sequence or GC content [95,96]. Interestingly, AIM2 binds directly to the two DNA strands, both between the larger and the smaller grooves, which explains, in theory, why their activation does not occur with single-stranded DNA [96]. Interestingly, in patients with type 2 diabetes the level of circulating mtDNA is significantly higher compared to healthy patients [97]. It was observed that this extracellular mtDNA is the driver for the activation of macrophages, and the subsequent induction of the cytokine IL-1β, contributing to the establishment of chronic inflammation [97]. AIM2 has also been observed to sense mtDNA from events of mitochondrial stress, such as increased cell levels of cholesterol, which lead to the extravasation of mtDNA [98], and in non-alcoholic fatty liver disease, driving inflammation and hepatocyte pyroptosis [99].
ZBP1
Z-DNA binding protein 1 (ZBP1), also referred as DAI (DNA-dependent activator of IFN-regulatory factors), is a cytosolic DNA sensor related to the initiation of the innate immune response. It activation can lead to programmed cell death and inflammation [100,101]. ZBP1 when associated with DNA recruits TBK1 that regulates the activation of IRF3 and induces the expression of type I IFNs [101]. ZBP1 needs oligomerization/multimerization to initiate its signaling transduction [102].
Murine L929 fibroblasts lacking ZBP1 express less NF-kB after infection with HSV-1, which reveals a possible importance of this sensor for activating a proinflammatory response [101]. However, the role of ZBP1 as an important DNA sensor has been questioned, as mice lacking ZBP1 (Zbp1 -/-) were able to generate an innate and adaptive immune response after DNA vaccination and after exposition to double-stranded B-form of DNA [103,104].
Recently, the ZBP1 DNA sensor was included in the select group of direct mtDNA receptors. Low levels of chronic oxidative stress in smoking-derived tissue damage models, have also been shown to be able to not only cause damage to mtDNA but also lead to its presence in the cytosol [52]. In this case, ZBP1 is capable of binding to mtDNA in pulmonary epithelial cells and induce inflammation via TBK1 and by stabilizing the IRF3 transcription factor [52].
It is of paramount importance for immunology to unveil how and when each receptor is preferentially activated to seek possible new therapeutic approaches, as well as to unravel the processes of why and how mtDNA is released from cells.
Finally, recent data is linking each one of the mtDNA receptors presented above with the SARS-CoV-2 infection.
In array analysis, the level of TLR9 was upregulated in response to SARS-CoV infection, even more than other TLRs such as TLR2 and TLR4 [105]. TLR9 can in fact explain a huge variety of the symptoms observed in COVID-19, and in specific cases be hypothesized as the leading cause of the hyperinflammatory process [106]. AIM2 can be activated in monocytes infected with SARS-CoV-2 in an antibody mediated manner, causing pyroptotic cell death [107] and in pangolins, carriers of coronaviruses and together with bats, focus of studies of the SARS-CoV-2 zoonotic transmission to humans, lack the ZBP1 protein [108]. The authors of this study hypothesize that this difference in the innate immune system is probably a selection factor for pangolins to carry out coronaviruses, switching from a mechanism of immune virus combat and resistance, to a more tolerogenic state.
Regarding cGAS, SARS-CoV-2 proteins Orf3a and 3CL are able to interact and inhibit cGAS-STING activity. Orf3a bind to STING independently of it C or N terminal regions, and inhibit NF-kB activity and downstream gene expression [109]. 3CL protein inhibit K63-ubiquitin-mediated modification of STING and also decreases the function of NF-kB. Interestingly in the study that reported this results, both Orf3a and 3CL did not interfered with IRF3 activity [109].
Conclusion
The relationship of mitochondria with the immune system is complex as different mitochondrial dynamics and metabolic pathways result in very different phenotypes of immune cells [7]. In addition, mitochondria can serve as inner DAMP generator, releasing ATP, mtDNA, and other molecules that can activate the immune system [10]. We have focused in the present study in the mtDNA, which have been shown to elicit a powerful proinflammatory response upon its detection. A large number of processes can be responsible for its extrusion of the mitochondrial compartment, including a variety of RNA viruses that have even evolved mechanisms to circumvent the mtDNA detection.
As novel functions and interactions are discovered almost on a daily basis, the receptors here reviewed can possibly contribute to the understanding of the inflammatory process in the SARS-CoV-2 lung pathogenesis. The presence of mtDNA in cytosol or in the extracellular environment is highly associated with an exacerbated inflammation in the lungs, one of the principal clinical manifestations of SARS-CoV-2 [28,110-113]. Furthermore, a vast number of the receptors reviewed here are directly associated with lung inflammatory diseases [111,112]. Additionally, each of the receptors reviewed here have presented significant evidence of relationship with COVID-19. The release and presence of mtDNA outside mitochondria due to SARS-CoV-2 infection might reflect one important pathogenic mechanism and exploring the role of mtDNA in clinical patients together with its receptors can be beneficial to unravel a new mechanism of the disease and to open new treatment possibilities.
Abbreviations
ACE2: angiotensin-converting enzyme 2; AIM2: absent in melanoma 2; AMP: adenosine monophosphate; ARDS: acute respiratory distress syndrome; ASC: caspase recruitment domain; ATP: adenosine triphosphate; BAK: pro-apoptotic Bcl-2 family member; BAX: pro-apoptotic Bcl-2 family member; cGAMP: cyclic GMP-AMP; cGAS: cyclic GMP-AMP (cGAMP) synthase; CgP: 5'-C-phosphate-G-3’; CMV: cytomegalovirus; COVID-19: corona virus disease 19; CpG: cytosine-phosphate-guanine; DAI: DNA-dependent activator of IFN-regulatory factors; DAMP: damage-associated molecular pattern molecule; DNA: deoxyribonucleic acid; dsDNA: double-stranded DNA; GC: guanine-cytosine; GMP: guanosine monophosphate; GTP: guanosine triphosphate; H1N1: human influenza A strain; HCoV-NL63: Human coronavirus NL63; HIF-1α: hypoxia-inducible factor-1 alpha; HSV-1: herpes simplex virus type 1; HSV-2: herpes simplex virus type 2; IFN: interferon; IFN-α: interferon alpha; IKK: inhibitor of nuclear factor‐κB (IκB) kinase; IL-18: interleukin 18; IL-1β: interleukin 1 beta; IRF3: interferon responsible factor 3; LPS: lipopolysaccharides; MAVS: mitochondrial antiviral-signaling protein; mtDNA: mitochondrial DNA; mtROS: mitochondrial reactive oxygen species; MyD88: myeloid differentiation factor 88; NADH: nicotinamide adenine dinucleotide reduced; NET: neutrophil extracellular traps; NF-kB: nuclear factor kappa B; NLRP12: NOD-like receptor family pyrin domain containing 12; NLRP3: NOD-like receptor family pyrin domain containing 3; NLRP6: NOD-like receptor family pyrin domain containing 6; NS1: nonstructural protein 1; NS2B: nonstructural protein 2B; NSP3: nonstructural protein 3; NSP4: nonstructural protein 4; NSP8: nonstructural protein 8; Orf3a: open reading frame 3a; Orf7b: open reading frame 7b; Orf8b: open reading frame 8b; Orf9b: open reading frame 9b; Orf9c: open reading frame 9c; PAMP: pathogen associated molecular pattern; pH: potential hydrogen; PHB: prohibitin; PRR: pattern recognition receptors; RIG-I: retinoic acid-inducible gene I; RNA: ribonucleic acid; ROS: reactive oxygen species; SARS-CoV: severe acute respiratory syndrome coronavirus; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; Ser366: serine residue 366; STING: stimulator of interferon genes; TBK1: TANK-binding kinase 1; TLR: toll-like receptor; TLR2: toll-like receptor 2; TLR4: toll-like receptor 4; TLR9: toll-like receptor 9; TMPRSS2: transmembrane serine protease 2; ZBP1: Z-DNA binding protein 1; Zikv: Zika virus.
Footnotes
Availability of data and materials: Not applicable.
Funding: This work was supported by the National Council for Scientific and Technological Development (CNPq), São Paulo Research Foundation (FAPESP - grants n. 2017/05264-7, 2015/21644-9, 2020/04408-8, 2021/00946-8), and the Coordination for the Improvement of Higher Education Personnel (CAPES) - Financial Code 001.
Ethics approval : Not applicable.
Consent for publication: Not applicable.
References
- Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman M, Chambers RC, Singer M, Brown JS. SARS-CoV-2 pandemic: clinical picture of COVID-19 and implications for research. Thorax. 2020;75(8):614–616. doi: 10.1136/thoraxjnl-2020-215024. [DOI] [PubMed] [Google Scholar]
- Goyal P, Ringel JB, Rajan M, Choi JJ, Pinheiro LC, Li HA, Wehmeyer GT, Alshak MN, Jabri A, Schenck EJ, Chen R, Satlin MJ, Campion TR, Jr, Nahid M, Plataki M, Hoffman KL, Reshetnyak E, Hupert N, Horn EM, Martinez FJ, Gulick RM, Safford MM. Obesity and COVID-19 in New York city: a retrospective cohort study. Ann Intern Med. 2020;173(10):855–858. doi: 10.7326/M20-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korakas E, Ikonomidis I, Kousathana F, Balampanis K, Kountouri A, Raptis A, Palaiodimou L, Kokkinos A, Lambadiari V. Obesity and COVID-19: immune and metabolic derangement as a possible link to adverse clinical outcomes. Am J Physiol Endocrinol Metab. 2020;319(1):E105–E109. doi: 10.1152/ajpendo.00198.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski L, Chaib M, Rathmell JC. Immunometabolism: from basic mechanisms to translation. Immunol Rev. 2020;295(1):5–14. doi: 10.1111/imr.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Luan HH, Medzhitov R. An evolutionary perspective on immunometabolism. Science. 2019;363(6423):eaar3932. doi: 10.1126/science.aar3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K, Hisata S, Choi AMK. The roles of mitochondrial damage-associated molecular patterns in diseases. Antioxid Redox Signal. 2015;23(17):1329–1350. doi: 10.1089/ars.2015.6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grazioli S, Pugin J. Mitochondrial damage-associated molecular patterns: from inflammatory signaling to human diseases. Front Immunol. 2018;9:832. doi: 10.3389/fimmu.2018.00832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C, Wei X, Wei Y. Mitochondrial DNA in the regulation of innate immune responses. Protein Cell. 2016;7(1):11–16. doi: 10.1007/s13238-015-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biacchesi S, LeBerre M, Lamoureux A, Louise Y, Lauret E, Boudinot P, Brémont M. Mitochondrial antiviral signaling protein plays a major role in induction of the fish innate immune response against RNA and DNA viruses. J Virol. 2009;83(16):7815–7827. doi: 10.1128/JVI.00404-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Nuevo A, Zorzano A. The sensing of mitochondrial DAMPs by non-immune cells. Cell Stress. 2019;3(6):195–207. doi: 10.15698/cst2019.06.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464(7285):104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LS, Hong Z, Wu W, Xiong S, Zhong M, Gao X, Rehman J, Malik AB. mtDNA activates cGAS signaling and suppresses the YAP-mediated endothelial cell proliferation program to promote inflammatory injury. Immunity. 2020;52(3):475–486.e5. doi: 10.1016/j.immuni.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vringer E, Tait SWG. Mitochondria and inflammation: cell death heats up. Front Cell Dev Biol. 2019;7:100. doi: 10.3389/fcell.2019.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley JS, Tait SW. Mitochondrial DNA in inflammation and immunity. EMBO Rep. 2020;21(4):e49799. doi: 10.15252/embr.201949799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A, Fadeel B. Mitochondria released by cells undergoing TNF-α-induced necroptosis act as danger signals. Cell Death Dis. 2014;5(7):e1312. doi: 10.1038/cddis.2014.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue-Gervais IG, Labbé K, Dagenais M, Dupaul-Chicoine J, Champagne C, Morizot A, Skeldon A, Brincks EL, Vidal SM, Griffith TS, Saleh M. Cellular inhibitor of apoptosis protein cIAP2 protects against pulmonary tissue necrosis during influenza virus infection to promote host survival. Cell Host Microbe. 2014;15(1):23–35. doi: 10.1016/j.chom.2013.12.003. [DOI] [PubMed] [Google Scholar]
- Mauad T, Hajjar LA, Callegari GD, da Silva LFF, Schout D, Galas FRBG, Alves VAF, Malheiros DMAC, Auler JOC, Ferreira AF, Borsato MRL, Bezerra SM, Gutierrez PS, Caldini ETEG, Pasqualucci CA, Dolhnikoff M, Saldiva PHN. Lung pathology in fatal novel human influenza A (H1N1) infection. Am J Respir Crit Care Med. 2010;181(1):72–79. doi: 10.1164/rccm.200909-1420OC. [DOI] [PubMed] [Google Scholar]
- Yue Y, Nabar NR, Shi CS, Kamenyeva O, Xiao X, Hwang IY, Wang M, Kehrl JH. SARS-Coronavirus open reading frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9(9):904. doi: 10.1038/s41419-018-0917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Shu T, Wu D, Mu J, Wang C, Huang M, Han Y, Zhang XY, Zhou W, Qiu Y, Zhou X. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell Mol Immunol. 2020;17(8):881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Xia H, Liang Y, Ye Y, Lu Y, Xu X, Duan A, He J, Chen Z, Wu Y, Wang X, Zheng C, Liu Z, Shi S. Toll-like receptor 9 can be activated by endogenous mitochondrial DNA to induce podocyte apoptosis. Sci Rep. 2016;6(1):22579. doi: 10.1038/srep22579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Mao Y, Xu B, Zhang X, Fang C, Ma Y, Men K, Qi X, Yi T, Wei Y, Wei X. Induction of neutrophil extracellular traps during tissue injury: involvement of STING and Toll-like receptor 9 pathways. Cell Prolif. 2019;52(3):e12579. doi: 10.1111/cpr.12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H, Inoue T, Ouchi H, Jao TM, Inoue R, Nishi H, Fujii R, Ishidate F, Tanaka T, Tanaka Y, Hirokawa N, Nangaku M, Inagi R. Mitochondrial damage causes inflammation via cGAS-STING signaling in acute kidney injury. Cell Rep. 2019;29(5):1261–1273.e6. doi: 10.1016/j.celrep.2019.09.050. [DOI] [PubMed] [Google Scholar]
- Chung KW, Dhillon P, Huang S, Sheng X, Shrestha R, Qiu C, Kaufman BA, Park J, Pei L, Baur J, Palmer M, Susztak K. Mitochondrial damage and activation of the STING pathway lead to renal inflammation and fibrosis. Cell Metab. 2019;30(4):784–799.e5. doi: 10.1016/j.cmet.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez I, Santoro N, Chen Y, Hoque R, Ouyang X, Caprio S, Shlomchik MJ, Coffman RL, Candia A, Mehal WZ. Hepatocyte mitochondrial DNA drives nonalcoholic steatohepatitis by activation of TLR9. J Clin Invest. 2016;126(3):859–864. doi: 10.1172/JCI83885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JZ, Liu Z, Liu J, Ren JX, Sun TS. Mitochondrial DNA induces inflammation and increases TLR9/NF-κB expression in lung tissue. Int J Mol Med. 2014;33(4):817–824. doi: 10.3892/ijmm.2014.1650. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bueno M, Zank D, Buendia-Roldán I, Fiedler K, Mays BG, Alvarez D, Sembrat J, Kimball B, Bullock JK, Martin JL, Nouraie M, Kaufman BA, Rojas M, Pardo A, Selman M, Mora AL. PINK1 attenuates mtDNA release in alveolar epithelial cells and TLR9 mediated profibrotic responses. Mukhopadhyay P, editor. PLoS One. 2019;14(6):e0218003. doi: 10.1371/journal.pone.0218003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A, Jackson R, Harman CCD, Li T, West AP, de Zoete MR, Wu Y, Yordy B, Lakhani SA, Kuan CY, Taniguchi T, Shadel GS, Chen ZJ, Iwasaki A, Flavell RA. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159(7):1563–1577. doi: 10.1016/j.cell.2014.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ, McArthur K, Metcalf D, Lane RM, Cambier JC, Herold MJ, van Delft MF, Bedoui S, Lessene G, Ritchie ME, Huang DCS, Kile BT. Apoptotic caspases suppress mtDNA induced STING-mediated type I IFN production. Cell. 2014;159(7):1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning X, Wang Y, Jing M, Sha M, Lv M, Gao P, Zhang R, Huang X, Feng JM, Jiang Z. Apoptotic caspases suppress type I interferon production via the cleavage of cGAS, MAVS, and IRF3. Mol Cell. 2019;74(1):19–31.e7. doi: 10.1016/j.molcel.2019.02.013. [DOI] [PubMed] [Google Scholar]
- Riley JS, Quarato G, Cloix C, Lopez J, O’Prey J, Pearson M, Chapman J, Sesaki H, Carlin LM, Passos JF, Wheeler AP, Oberst A, Ryan KM, Tait SW. Mitochondrial inner membrane permeabilisation enables mt DNA release during apoptosis. EMBO J. 2018;37(17):e99238. doi: 10.15252/embj.201899238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, Lieberman J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535(7610):153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The Inflammasomes. Cell. 2010;140(6):821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Codo AC, Davanzo GG, Monteiro LB, de Souza GF, Muraro SP, Virgilio-da-Silva JV, Prodonoff JS, Carregari VC, de CAO, Biagi Junior, Crunfli F, Jimenez Restrepo JL, Vendramini PH, Reis-de-Oliveira G, Bispo dos Santos K, Toledo-Teixeira DA, Parise PL, Martini MC, Marques RE, Carmo HR, Borin A, Coimbra LD, Boldrini VO, Brunetti NS, Vieira AS, Mansour E, Ulaf RG, Bernardes AF, Nunes TA, Ribeiro LC, Palma AC, Agrela MV, Moretti ML, Sposito AC, Pereira FB, Velloso LA, Vinolo MAR, Damasio A, Proença-Módena JL, Carvalho RF, Mori MA, Martins-de-Souza D, Nakaya HI, Farias AS, Moraes-Vieira PM. Elevated glucose levels favor SARS-CoV-2 infection and monocyte response through a HIF-1α/glycolysis-dependent axis. Cell Metab. 2020;32(3):437–446.e5. doi: 10.1016/j.cmet.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan Y, Xin Y, Tian G, Zhou J, Liu X. Mitochondrial ROS-modulated mtDNA: a potential target for cardiac aging. Oxid Med Cell Longev. 2020:9423593. doi: 10.1155/2020/9423593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, Ramanujan VK, Wolf AJ, Vergnes L, Ojcius DM, Rentsendorj A, Vargas M, Guerrero C, Wang Y, Fitzgerald KA, Underhill DM, Town T, Arditi M. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36(3):401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward GA, McGraw KL, McLemore AF, Abbas-Aghababazadeh F, Hou HA, Le Trinh T, Vincelette ND, Meyer BS, Onimus AH, Calescibetta A, Eksioglu EA, Wei S, List AF. Oxidized mitochondrial DNA is a catalyst and biomarker of pyroptotic cell death in myelodysplastic syndromes. Blood. 2018;132(1):3076 [Google Scholar]
- Rodrigues TS, de Sá KSG, Ishimoto AY, Becerra A, Oliveira S, Almeida L, Gonçalves AV, Perucello DB, Andrade WA, Castro R, Veras FP, Toller-Kawahisa JE, Nascimento DC, de Lima MHF, Silva CMS, Caetite DB, Martins RB, Castro IA, Pontelli MC, de Barros FC, do Amaral NB, Giannini MC, Bonjorno LP, Lopes MIF, Santana RC, Vilar FC, Auxiliadora-Martins M, Luppino-Assad R, de Almeida SCL, de Oliveira FR, Batah SS, Siyuan L, Benatti MN, Cunha TM, Alves-Filho JC, Cunha FQ, Cunha LD, Frantz FG, Kohlsdorf T, Fabro AT, Arruda E, de Oliveira RDR, Louzada-Junior P, Zamboni DS. Inflammasomes are activated in response to SARS-cov-2 infection and are associated with COVID-19 severity in patients. J Exp Med. 2021;218(3):e20201707. doi: 10.1084/jem.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Qi HY, Boularan C, Huang NN, Abu-Asab M, Shelhamer JH, Kehrl JH. SARS-Coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J Immunol. 2014;193(6):3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Chaubey G, Chen JY, Suravajhala P. Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am J Physiol. 2020;319(2):C258–C267. doi: 10.1152/ajpcell.00224.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Ping YH, Lee HC, Chen KH, Lee YM, Chan YJ, Lien TC, Jap TS, Lin CH, Kao LS, Chen YMA. Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J Infect Dis. 2007;196(3):405–415. doi: 10.1086/519166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YX, Tan THP, Lee MJR, Tham PY, Gunalan V, Druce J, Birch C, Catton M, Fu NY, Yu VC, Tan YJ. Induction of apoptosis by the severe acute respiratory syndrome Coronavirus 7a protein is dependent on its interaction with the Bcl-XL protein. J Virol. 2007;81(12):6346–6355. doi: 10.1128/JVI.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KE, Fazal FM, Parker KR, Zou J, Chang HY. RNA-GPS predicts SARS-CoV-2 RNA residency to host mitochondria and nucleolus. Cell Syst. 2020;11(1):102–108.e3. doi: 10.1016/j.cels.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberis E, Vanella VV, Falasca M, Caneapero V, Cappellano G, Raineri D, Ghirimoldi M, De Giorgis V, Puricelli C, Vaschetto R, Sainaghi PP, Bruno S, Sica A, Dianzani U, Rolla R, Chiocchetti A, Cantaluppi V, Baldanzi G, Marengo E, Manfredi M. Circulating exosomes are strongly involved in SARS-CoV-2 infection. Front Mol Biosci. 2021;8:632290. doi: 10.3389/fmolb.2021.632290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanpour M, Rezaie J, Nouri M, Panahi Y. The role of extracellular vesicles in COVID-19 virus infection. Infect Genet Evol. 2020;85:104422. doi: 10.1016/j.meegid.2020.104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangalmurti N, Hunter CA. Cytokine storms: understanding COVID-19. Immunity. 2020;53(1):19–25. doi: 10.1016/j.immuni.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scozzi D, Cano M, Ma L, Zhou D, Zhu JH, O’Halloran JA, Goss C, Rauseo AM, Liu Z, Sahu SK, Peritore V, Rocco M, Ricci A, Amodeo R, Aimati L, Ibrahim M, Hachem R, Kreisel D, Mudd PA, Kulkarni HS, Gelman AE. Circulating mitochondrial DNA is an early indicator of severe illness and mortality from COVID-19. JCI Insight. 2021;6(4):e143299. doi: 10.1172/jci.insight.143299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O’Meara MJ, Rezelj VV, Guo JZ, Swaney DL, Tummino TA, Hüttenhain R, Kaake RM, Richards AL, Tutuncuoglu B, Foussard H, Batra J, Haas K, Modak M, Kim M, Haas P, Polacco BJ, Braberg H, Fabius JM, Eckhardt M, Soucheray M, Bennett MJ, Cakir M, McGregor MJ, Li Q, Meyer B, Roesch F, Vallet T, Mac Kain A, Miorin L, Moreno E, Naing ZZC, Zhou Y, Peng S, Shi Y, Zhang Z, Shen W, Kirby IT, Melnyk JE, Chorba JS, Lou K, Dai SA, Barrio-Hernandez I, Memon D, Hernandez-Armenta C, Lyu J, Mathy CJP, Perica T, Pilla KB, Ganesan SJ, Saltzberg DJ, Rakesh R, Liu X, Rosenthal SB, Calviello L, Venkataramanan S, Liboy-Lugo J, Lin Y, Huang XP, Liu YF, Wankowicz SA, Bohn M, Safari M, Ugur FS, Koh C, Savar NS, Tran QD, Shengjuler D, Fletcher SJ, O’Neal MC, Cai Y, Chang JCJ, Broadhurst DJ, Klippsten S, Sharp PP, Wenzell NA, Kuzuoglu-Ozturk D, Wang HY, Trenker R, Young JM, Cavero DA, Hiatt J, Roth TL, Rathore U, Subramanian A, Noack J, Hubert M, Stroud RM, Frankel AD, Rosenberg OS, Verba KA, Agard DA, Ott M, Emerman M, Jura N, von Zastrow M, Verdin E, Ashworth A, Schwartz O, d’Enfert C, Mukherjee S, Jacobson M, Malik HS, Fujimori DG, Ideker T, Craik CS, Floor SN, Fraser JS, Gross JD, Sali A, Roth BL, Ruggero D, Taunton J, Kortemme T, Beltrao P, Vignuzzi M, García-Sastre A, Shokat KM, Shoichet BK, Krogan NJ. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornillez-Ty CT, Liao L, Yates 3rd JR, Kuhn P, Buchmeier MJ. Severe acute respiratory syndrome Coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J Virol. 2009;83(19):10314–10318. doi: 10.1128/JVI.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny B, Marcatti M, Ahmad A, Montalbano M, Brunyánszki A, Bibli SI, Papapetropoulos A, Szabo C. Mitochondrial DNA damage and subsequent activation of Z-DNA binding protein 1 links oxidative stress to inflammation in epithelial cells. Sci Rep. 2018;8(1):914. doi: 10.1038/s41598-018-19216-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Sundström KB, Chew JJ, Bist P, Gan ES, Tan HC, Goh KC, Chawla T, Tang CK, Ooi EE. Dengue virus activates cGAS through the release of mitochondrial DNA. Sci Rep. 2017;7(1):3594. doi: 10.1038/s41598-017-03932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Liu Q, Wu Y, Ma L, Zhang Z, Liu T, Jin S, She Y, Li YP, Cui J. Zika virus elicits inflammation to evade antiviral response by cleaving cGAS via NS 1‐caspase‐1 axis. EMBO J. 2018;37(18):e99347. doi: 10.15252/embj.201899347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama M, Koshiba T, Ichinohe T. Influenza A virus M2 protein triggers mitochondrial DNA-mediated antiviral immune responses. Nat Commun. 2019;10(1):4624. doi: 10.1038/s41467-019-12632-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtscher J, Cappellano G, Omori A, Koshiba T, Millet GP. Mitochondria: in the crossfire of SARS-CoV-2 and immunity. iScience. 2020;23(10):101631. doi: 10.1016/j.isci.2020.101631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MM, Shu HB. Annual review of immunology innate immune response to cytoplasmic DNA: mechanisms and diseases. 2020;38:79–98. doi: 10.1146/annurev-immunol-070119-115052. [DOI] [PubMed] [Google Scholar]
- Motwani M, Pesiridis S, Fitzgerald KA. DNA sensing by the cGAS-STING pathway in health and disease. Nat Rev Genet. 2019;20(11):657–674. doi: 10.1038/s41576-019-0151-1. [DOI] [PubMed] [Google Scholar]
- Li T, Chen ZJ. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med. 2018;215(5):1287–1299. doi: 10.1084/jem.20180139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Omura H, Ishitani R, Nureki O. Cyclic GMP-AMP as an endogenous second messenger in innate immune signaling by cytosolic DNA. Annu Rev Biochem. 2017;86(1):541–566. doi: 10.1146/annurev-biochem-061516-044813. [DOI] [PubMed] [Google Scholar]
- Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3(5):1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Röhl I, Hopfner KP, Ludwig J, Hornung V. CGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498(7454):380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, Chen ZJ. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347(6227):aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341(6152):1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Shang G, Gui X, Zhang X, Bai XC, Chen ZJ. Structural basis of STING binding with and phosphorylation by TBK1. Nature. 2019;567(7748):394–398. doi: 10.1038/s41586-019-1000-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5(214):ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglesham JB, Kranzusch PJ. Conserved strategies for pathogen evasion of cGAS-STING immunity. Curr Opin in Immunol. 2020;66(27-34) doi: 10.1016/j.coi.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre S, Luthra P, Sanchez-Aparicio MT, Maestre AM, Patel J, Lamothe F, Fredericks AC, Tripathi S, Zhu T, Pintado-Silva J, Webb LG, Bernal-Rubio D, Solovyov A, Greenbaum B, Simon V, Basler CF, Mulder LCF, García-Sastre A, Fernandez-Sesma A. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat Microbiol. 2017;2:17037. doi: 10.1038/nmicrobiol.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Xing Y, Chen X, Zheng Y, Yang Y, Nichols DB, Clementz MA, Banach BS, Li K, Baker SC, Chen Z. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS One. 2012;7(2):e30802. doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Mukherjee R, Grewe D, Bojkova D, Baek K, Bhattacharya A, Schulz L, Widera M, Mehdipour AR, Tascher G, Geurink PP, Wilhelm A, van der Heden van Noort GJ, Ovaa H, Müller S, Knobeloch KP, Rajalingam K, Schulman BA, Cinatl J, Hummer G, Ciesek S, Dikic I. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587(7835):657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarreberg LD, Esser-Nobis K, Driscoll C, Shuvarikov A, Roby JA, Gale M., Jr Interleukin-1β induces mtDNA release to activate innate immune signaling via cGAS-STING. Mol Cell. 2019;74(4):801–815.e6. doi: 10.1016/j.molcel.2019.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkirtzimanaki K, Kabrani E, Nikoleri D, Polyzos A, Blanas A, Sidiropoulos P, Makrigiannakis A, Bertsias G, Boumpas DT, Verginis P. IFNα impairs autophagic degradation of mtDNA promoting autoreactivity of SLE monocytes in a STING-dependent fashion. Cell Rep. 2018;25(4):921–933.e5. doi: 10.1016/j.celrep.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, Lien E, Nilsen NJ, Espevik T, Golenbock DT. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5(2):190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- Fiola S, Gosselin D, Takada K, Gosselin J. TLR9 contributes to the recognition of EBV by primary monocytes and plasmacytoid dendritic cells. J Immunol. 2010;185(6):3620–3631. doi: 10.4049/jimmunol.0903736. [DOI] [PubMed] [Google Scholar]
- Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J Exp Med. 2003;198(3):513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug A, Luker GD, Barchet W, Leib DA, Akira S, Colonna M. Herpes simplex virus type 1 activates murine natural interferon-producing cells through Toll-like receptor 9. Blood. 2004;103(4):1433–1437. doi: 10.1182/blood-2003-08-2674. [DOI] [PubMed] [Google Scholar]
- Varani S, Cederarv M, Feld S, Tammik C, Frascaroli G, Landini MP, Söderberg-Nauclér C. Human cytomegalovirus differentially controls B cell and T cell responses through effects on plasmacytoid dendritic cells. J Immunol. 2007;179(11):7767–7776. doi: 10.4049/jimmunol.179.11.7767. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Kaisho T, Takeda K, Akira S. The roles of Toll-like receptor 9, MyD88, and DNA-dependent protein kinase catalytic subunit in the effects of two distinct CpG DNAs on dendritic cell subsets. J Immunol. 2003;170(6):3059–3064. doi: 10.4049/jimmunol.170.6.3059. [DOI] [PubMed] [Google Scholar]
- Latz E, Verma A, Visintin A, Gong M, Sirois CM, Klein DCG, Monks BG, McKnight JC, Lamphier MS, Duprex WP, Espevik T, Golenbock DT. Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat Immunol. 2007;8(7):772–779. doi: 10.1038/ni1479. [DOI] [PubMed] [Google Scholar]
- Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4(7):499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Zhang JZ, Liu Z, Liu J, Ren JX, Sun TS. Mitochondrial DNA induces inflammation and increases TLR9/NF-κB expression in lung tissue. Int J Mol Med. 2014;33(4):817–824. doi: 10.3892/ijmm.2014.1650. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jing R, Hu ZK, Lin F, He S, Zhang SS, Ge WY, Dai HJ, Du XK, Lin JY, Pan LH. Mitophagy-mediated mtDNA release aggravates stretching-induced inflammation and lung epithelial cell injury via the TLR9/MyD88/NF-κB pathway. Front Cell Dev Biol. 2020;8:819. doi: 10.3389/fcell.2020.00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, Oyabu J, Murakawa T, Nakayama H, Nishida K, Akira S, Yamamoto A, Komuro I, Otsu K. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485(7397):251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Liu S, Cheng J, Wang L, Liu J, Gong J. Exogenous administration of mitochondrial DNA promotes ischemia reperfusion injury via TLR9-p38 MAPK pathway. Regul Toxicol Pharmacol. 2017;89:148–154. doi: 10.1016/j.yrtph.2017.07.028. [DOI] [PubMed] [Google Scholar]
- Bao W, Xia H, Liang Y, Ye Y, Lu Y, Xu X, Duan A, He J, Chen Z, Wu Y, Wang X, Zheng C, Liu Z, Shi S. Toll-like Receptor 9 Can be activated by endogenous mitochondrial DNA to induce podocyte apoptosis. Sci Rep. 2016;6(1):22579. doi: 10.1038/srep22579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Nuevo A, Díaz‐Ramos A, Noguera E, Díaz‐Sáez F, Duran X, Muñoz JP, Romero M, Plana N, Sebastián D, Tezze C, Romanello V, Ribas F, Seco J, Planet E, Doctrow SR, González J, Borràs M, Liesa M, Palacín M, Vendrell J, Villarroya F, Sandri M, Shirihai O, Zorzano A. Mitochondrial DNA and TLR9 drive muscle inflammation upon Opa1 deficiency. EMBO J. 2018;37(10):e96553. doi: 10.15252/embj.201796553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulopoulou S, Matsumoto T, Bomfim GF, Webb RC. Toll-like receptor 9 activation: a novel mechanism linking placenta-derived mitochondrial DNA and vascular dysfunction in pre-eclampsia. Clin Sci. 2012;123(7):429–435. doi: 10.1042/CS20120130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy CG, Wenceslau CF, Goulopoulou S, Ogbi S, Baban B, Sullivan JC, Matsumoto T, Webb RC. Circulating mitochondrial DNA and Toll-like receptor 9 are associated with vascular dysfunction in spontaneously hypertensive rats. Cardiovasc Res. 2015;107(1):119–130. doi: 10.1093/cvr/cvv137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muruve DA, Pétrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, Parks RJ, Tschopp J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452(7183):103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458(7237):514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma BR, Karki R, Kanneganti TD. Role of AIM2 inflammasome in inflammatory diseases, cancer and infection. Eur J Immunol. 2019;49(11):1998–2011. doi: 10.1002/eji.201848070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458(7237):509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, Hornung V, Latz E, Bowie AG, Fitzgerald KA, Xiao TS. Structures of the HIN domain: DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36(4):561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae JH, Jo SI, Kim SJ, Lee JM, Jeong JH, Kang JS, Cho NJ, Kim SS, Lee EY, Moon JS. Circulating cell-free mtDNA contributes to AIM2 inflammasome-mediated chronic inflammation in patients with type 2 diabetes. Cells. 2019;8(4):328. doi: 10.3390/cells8040328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang EV, Mcdonald JG, Russell DW, Cyster JG. Oxysterol restraint of cholesterol synthesis prevents AIM2 inflammasome activation. Cell. 2017;171(5):1057–1065.e11. doi: 10.1016/j.cell.2017.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zhou J, Che J, Wang H, Yang W, Zhou W, Zhao H. Mitochondrial DNA enables AIM2 inflammasome activation and hepatocyte pyroptosis in non-alcoholic fatty liver disease. Am J Physiol Liver Physiol. 2021;320(6):G1034–G1044. doi: 10.1152/ajpgi.00431.2020. [DOI] [PubMed] [Google Scholar]
- Kuriakose T, Man SM, Subbarao Malireddi RK, Karki R, Kesavardhana S, Place DE, Neale G, Vogel P, Kanneganti TD. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci Immunol. 2016;1(2):aag2045. doi: 10.1126/sciimmunol.aag2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448(7152):501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Wang Z, Choi MK, Ban T, Yanai H, Negishi H, Lu Y, Tamura T, Takaoka A, Nishikura K, Taniguchi T. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci U S A. 2008;105(14):5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, Akira S. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451(7179):725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- Kuriakose T, Kanneganti TD. ZBP1: innate sensor regulating cell death and inflammation. Trends Immunol. 2018;39(2):123–134. doi: 10.1016/j.it.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng LFP, Hibberd ML, Ooi EE, Tang KF, Neo SY, Tan J, Murthy KRK, Vega VB, Chia JM, Liu ET, Ren EC. A human in vitro model system for investigating genome-wide host responses to SARS coronavirus infection. BMC Infect Dis. 2004;4(1):34. doi: 10.1186/1471-2334-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezemer GFG, Garssen J. TLR9 and COVID-19: a multidisciplinary theory of a multifaceted therapeutic target. Front Pharmacol. 2021;11:601685. doi: 10.3389/fphar.2020.601685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junqueira C, Crespo Â, Ranjbar S, Ingber J, Parry B, Ravid S, de Lacerda LB, Lewandrowski M, Clark S, Ho F, Vora SM, Leger V, Beakes C, Margolin J, Russell N, Gehrke L, Adhikari UD, Henderson L, Janssen E, Kwon D, Sander C, Abraham J, Filbin M, Goldberg MB, Wu H, Mehta G, Bell S, Goldfeld AE, Lieberman J. SARS-CoV-2 infects blood monocytes to activate NLRP3 and AIM2 inflammasomes, pyroptosis and cytokine release. medRxiv. 2021 [Google Scholar]
- Fischer H, Tschachler E, Eckhart L. Pangolins lack IFIH1/MDA5, a cytoplasmic RNA sensor that initiates innate immune defense upon Coronavirus infection. Front Immunol. 2020;11:939. doi: 10.3389/fimmu.2020.00939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui Y, Su J, Shen S, Hu Y, Huang D, Zheng W, Lou M, Shi Y, Wang M, Chen S, Zhao N, Dong Q, Cai Y, Xu R, Zheng S, Yu XF. Unique and complementary suppression of cGAS-STING and RNA sensing- triggered innate immune responses by SARS-CoV-2 proteins. Signal Transduct Target Ther. 2021;6(1):123. doi: 10.1038/s41392-021-00515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotz MJ, Qing D, Shashaty MGS, Zhang P, Faust H, Sondheimer N, Rivella S, Worthen GS, Mangalmurti NS. Red blood cells homeostatically bind mitochondrial DNA through TLR9 to maintain quiescence and to prevent lung injury. Am J Respir Crit Care Med. 2018;197(4):470–480. doi: 10.1164/rccm.201706-1161OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuck JL, Obiako BO, Gorodnya OM, Pastukh VM, Kua J, Simmons JD, Gillespie MN. Mitochondrial DNA damage-associated molecular patterns mediate a feed-forward cycle of bacteria-induced vascular injury in perfused rat lungs. Am J Physiol Cell Mol Physiol. 2015;308(10):L1078–L1085. doi: 10.1152/ajplung.00015.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmerzoug S, Ryffel B, Togbe D, Quesniaux VFJ. Self-DNA Sensing in lung inflammatory diseases. Trends Immunol. 2019;40(8):719–734. doi: 10.1016/j.it.2019.06.001. [DOI] [PubMed] [Google Scholar]
- Ma R, Ortiz Serrano TP, Davis J, Prigge AD, Ridge KM. The cGAS-STING pathway: the role of self-DNA sensing in inflammatory lung disease. FASEB J. 2020;34(10):13156–13170. doi: 10.1096/fj.202001607R. [DOI] [PMC free article] [PubMed] [Google Scholar]


