We identify a peptide derived from Aspergillus nidulans recognized by CD4+ T cells derived from the lungs of Löfgren’s syndrome (LS) patients and provide evidence that A. nidulans plays a role in the pathogenesis of LS.
Abstract
Löfgren’s syndrome (LS) is an acute form of sarcoidosis characterized by a genetic association with HLA-DRB1*03 (HLA-DR3) and an accumulation of CD4+ T cells of unknown specificity in the bronchoalveolar lavage (BAL). Here, we screened related LS-specific TCRs for antigen specificity and identified a peptide derived from NAD-dependent histone deacetylase hst4 (NDPD) of Aspergillus nidulans that stimulated these CD4+ T cells in an HLA-DR3–restricted manner. Using ELISPOT analysis, a greater number of IFN-γ– and IL-2–secreting T cells in the BAL of DR3+ LS subjects compared with DR3+ control subjects was observed in response to the NDPD peptide. Finally, increased IgG antibody responses to A. nidulans NDPD were detected in the serum of DR3+ LS subjects. Thus, our findings identify a ligand for CD4+ T cells derived from the lungs of LS patients and suggest a role of A. nidulans in the etiology of LS.
Introduction
Granulomatous lung diseases encompass an array of disorders caused by both infectious and noninfectious agents in combination with genetic susceptibility. Sarcoidosis is an inflammatory granulomatous disease that primarily affects the lungs in ∼95% of cases (Baughman et al., 2001). Although sarcoidosis is thought to be a noninfectious disorder, the etiology remains unknown. The lack of a known inciting antigen in sarcoidosis represents a huge gap in our understanding of the disease and our ability to prevent and treat it. There has been speculation regarding causative agents, including self-antigens such as vimentin (Kinloch et al., 2018; Wahlström et al., 2007), bacterial-derived antigens from species such as Mycobacterium (Chen et al., 2008; Drake et al., 2007; Gupta et al., 2007; Oswald-Richter et al., 2009; Song et al., 2005) or Cutibacterium (Eishi et al., 2002), and multiple environmental antigens (Demirkok et al., 2006; Newman et al., 2004). However, the majority of the evidence linking these factors to sarcoidosis is correlative, and no CD4+ T cells specific for the above-mentioned antigens have been identified. Thus, studies to identify etiologic antigens in sarcoidosis are needed.
Löfgren’s syndrome (LS) is an acute form of sarcoidosis that presents with bilateral hilar lymphadenopathy, fever, erythema nodosum, and arthritis (Grunewald and Eklund, 2009). In Sweden, the majority of LS patients express HLA-DRB1*03 (HLA-DR3; Grunewald, 2012) and have expansions of CD4+ T cells in the bronchoalveolar lavage (BAL) expressing TCRα variable region (TRAV)12-1 (Grunewald et al., 2000, 1992, 1994). We previously demonstrated preferential pairing of TRAV12-1 with TCRβ variable region (TRBV)2 in BAL CD4+ T cells from HLA-DR3+ LS patients and that TRAV12-1 and TRBV2 were the most expanded variable regions relative to control subjects (Mitchell et al., 2017). Due to the homogeneity of disease presentation, LS represents an ideal subset of sarcoidosis to delineate T cell epitopes and interrogate the origins of disease. Here, we identified LS-specific αβTCRs that recognized a peptide derived from the NAD-dependent protein deacetylase hst4 (NDPD) of an airborne mold species, Aspergillus nidulans. A. nidulans NDPD stimulated CD4+ T cells from the BAL of the majority of DR3+ LS patients, and increased IgG antibody responses to A. nidulans NDPD were detected in the serum of DR3+ LS subjects. Thus, we have identified and validated a T cell epitope and the corresponding infectious organism potentially involved in the pathogenesis of LS.
Results
CDR3β motifs expressed on lung CD4+ T cells of HLA-DR3+ LS subjects
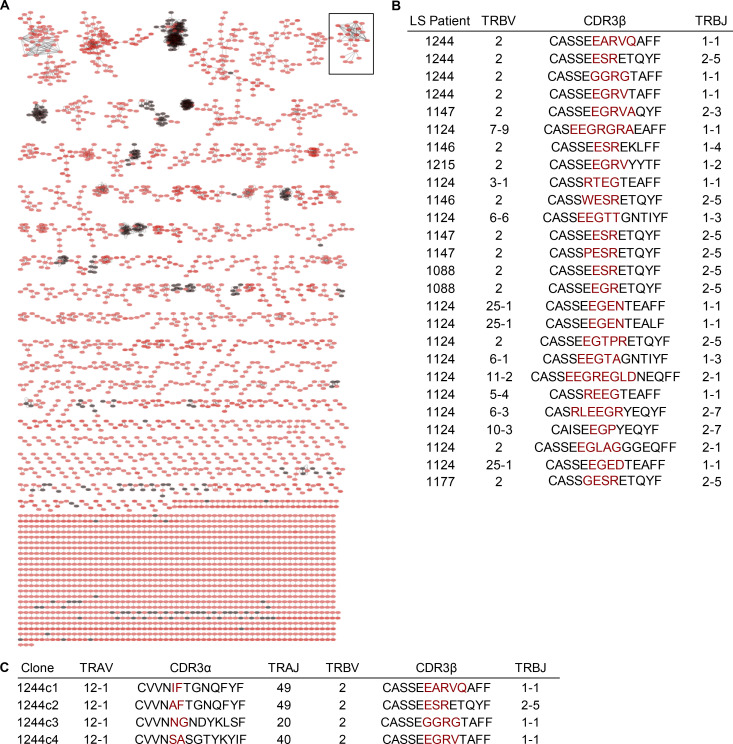
To identify disease-relevant TCR sequences present in the lungs of LS subjects, we previously performed bulk TCR sequencing of TRB genes from BAL CD4+ T cells obtained from eight HLA-DR3+ LS subjects and six control subjects (Mitchell et al., 2017). We also performed single-cell PCR amplification of TRB and TRA genes from TRAV12-1–expressing BAL CD4+ T cells from HLA-DR3+ LS patient 1244 to interrogate disease-specific αβTCR pairs (Mitchell et al., 2017). To identify related TCRs, we clustered TRB sequences from all subjects based on their predicted binding to shared epitopes using the Grouping of Lymphocyte Interactions by Paratope Hotspots (GLIPH) algorithm (Glanville et al., 2017). GLIPH identified several TCR clusters that included TRB sequences from both HLA-DR3+ LS (red circles) and control subjects (black circles; Fig. 1 A). However, the majority of TCR clusters identified by GLIPH were found in HLA-DR3+ LS patients and not in HLA-DR3+ non-LS control subjects.
Figure 1.
Clusters of CDR3β sequences derived from BAL CD4+ T cells of LS patients. (A) CDR3β sequences were clustered based on GLIPH analysis. Red circles represent CDR3β sequences derived from BAL CD4+ T cells of nine HLA-DR3+ LS patients, while black circles are CDR3β sequences derived from BAL CD4+ T cells of six control subjects. CDR3β sequences were obtained from BAL CD4+ T cells by iRepertoire bulk PCR and single-cell PCR. (B) CDR3β amino acid sequences and gene usage contained within the expanded cluster enclosed within the black square in A, highlighting TCR sequences only found in LS patients. (C) TCR gene segment usage and junctional region amino acid sequences of the LS-specific CDR3β motifs present in LS patient 1244 contained in the cluster shown in B along with the paired CDR3α amino acid sequences. Red denotes nongermline-encoded amino acids.
We focused on the sixth largest LS-specific TCR cluster (outlined in Fig. 1 A) because it contained sequences from the majority of LS patients (seven of nine), no controls, and four of the TRB sequences originated from the single-cell TCR analysis of patient 1244, giving complete αβTCR pairs (Fig. 1, B and C). These related 1244 TRB sequences all expressed TRBV2, which we have previously shown to be enriched in BAL CD4+ T cells from LS subjects (Mitchell et al., 2017). While using multiple TRB genes, half of the other TCR sequences present in this LS-specific cluster also expressed TRBV2 (Fig. 1 B). The TRA genes paired with the TRBV2 genes from patient 1244 were also highly related, and all expressed the LS-associated TRAV12-1 gene (Fig. 1 C; Grunewald et al., 2000, 1992, 1994). These data provide compelling evidence that these αβTCR pairs (1244c1–c4; Fig. 1 C) share a conserved specificity and are, therefore, ideal candidates for T cell epitope discovery.
Specific peptide requirements for LS-specific TCRs
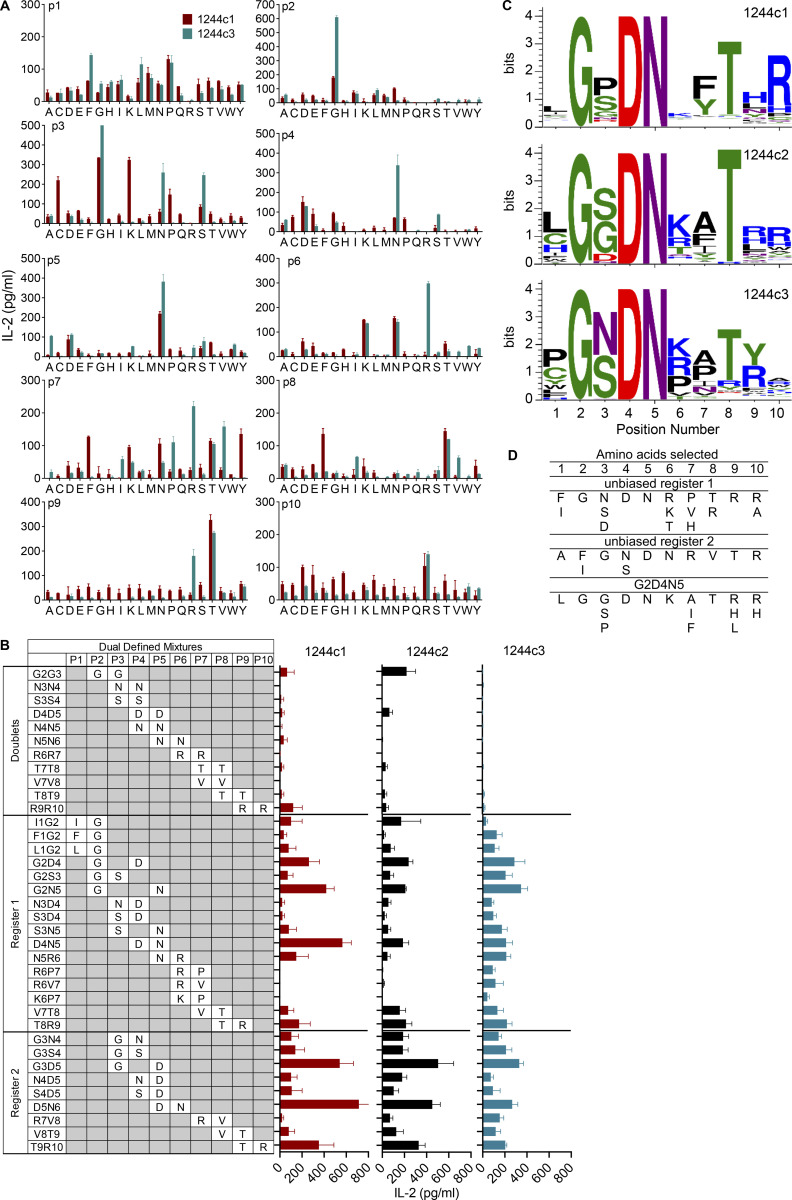
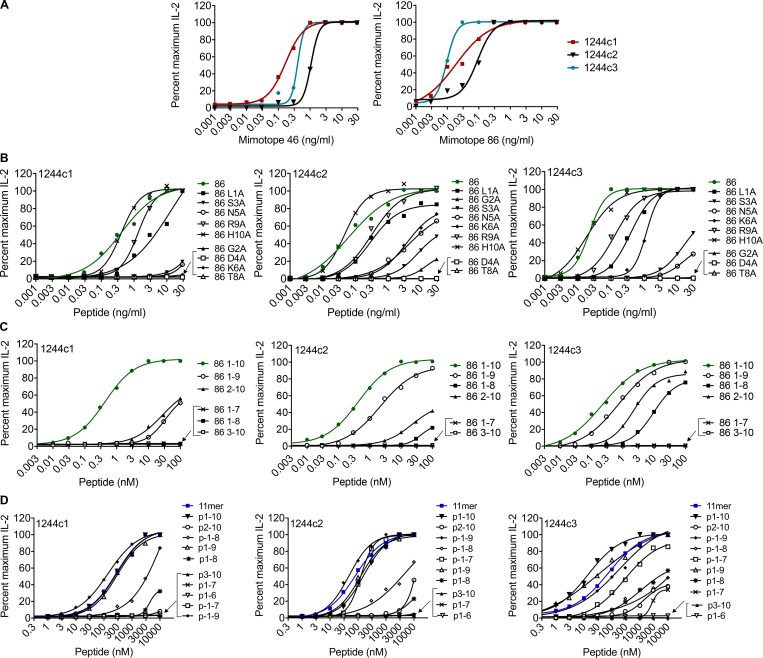
We generated immortalized T cell hybridomas expressing the four LS-specific 1244c1–c4 αβTCRs and confirmed that these TCRs were not specific to several previously identified candidate antigens. Thus, none of these 1244 hybridomas demonstrated reactivity to 74 overlapping 15-mers derived from mKatG, 17 overlapping 15-mers from ESAT-6, or 4 HLA-DR3–binding vimentin peptides (all tested at 10 μg/ml) using HLA-DR3–transfected murine kidney fibroblasts (Falta et al., 2013) as APCs (data not shown). Next, we used an unbiased decapeptide positional scanning library (PSL; Hemmer et al., 1999; Pinilla et al., 1994) to investigate T cell epitopes. The unbiased PSL allowed for a systematic evaluation of amino acid preferences at all positions of a decapeptide in a standard T cell activation assay. IL-2 secretion from hybridomas 1244c1 and 1244c3 in response to the decapeptide PSL is shown in Fig. 2 A. The initial PSL screen yielded high IL-2 responses to the same amino acid in adjacent positions, and this pattern was repeated along the decapeptide. For example, a glycine (G) fixed at positions 2 and 3 showed high activity for both hybridomas, as did an asparagine (N) at positions 4–6, arginine (R) at positions 6 and 7, and threonine (T) at positions 8 and 9 (Fig. 2 A). To resolve whether these repetitive amino acid preferences were due to register shifting of the peptides in the HLA-DR3–binding groove, we synthesized dual-defined mixtures composed of varying combinations of duplicate amino acids (Fig. 2 B). The lack of response of the hybridomas to the doublet mixtures confirmed that register shifting was occurring (Fig. 2 B). Additional dual-defined mixtures were synthesized to determine the correct peptide register, denoted as register 1 or register 2 in Fig. 2 B. Mixtures from both register 1 (G2D4, G2N5, and D4N5) and register 2 (G3D5 and D5N6) were highly stimulatory (Fig. 2 B), demonstrating that peptides in either register could be recognized by the 1244 TCRs.
Figure 2.
Deconvolution strategy to define peptides that stimulate LS-specific TCRs. (A) IL-2 responses of LS-specific 1244c1 (red bars) and 1244c3 (blue bars) CD4+ T cell hybridomas after stimulation with a decapeptide PSL presented by a fibroblast cell line (B6DK10) expressing HLA-DR3 after a 24-h incubation. In each mixture, one position is fixed with each of the 20 amino acids, which is denoted on the x axis with the single-letter code. Unfixed positions in each decapeptide mixture consist of an equimolar mixture of 19 amino acids (cysteine is excluded). Peptide mixtures were tested at 200 μg/ml, and IL-2 responses were measure by ELISA. Data are representative of two independent experiments. (B) LS-specific 1244c1, 1244c2, and 1244c3 CD4+ T cell hybridomas were stimulated with 37 dual-defined PSL mixtures at 100 μg/ml. Fixed amino acids and their positions in each decapeptide mixture are labeled. Unfixed positions in each decapeptide mixture consist of an equimolar mixture of 19 amino acids (cysteine is excluded). Dual-defined doublet mixtures are included to test register shifting in the unbiased PSL results. In the remaining dual-defined mixtures, two different TCR peptide recognition registers are apparent, denoted as register 1 and register 2. IL-2 responses of the LS-specific 1244c1 (red), 1244c2 (black), and 1244c3 (blue) CD4+ hybridomas stimulated with a dual-defined PSL are shown. Data are representative of three independent experiments. (C) Sequence logos summarizing LS-specific 1244c1 (top), 1244c2 (middle), and 1244c3 (bottom) CD4+ hybridoma responses to the G2D4N5 PSL are shown. The x axis denotes the position of the decapeptide, and the y axis denotes the relative IL-2 activity of each TCR when stimulated with peptide mixtures at 200 μg/ml. The color of each amino acid represents its chemical property: green, polar; purple, neutral; blue, basic; red, acidic; and black, hydrophobic. (D) Selection of amino acids for each peptide position for the 1244 LS-specific TCRs based on the most active mixtures from the unbiased PSL and G2D4N5 PSL.
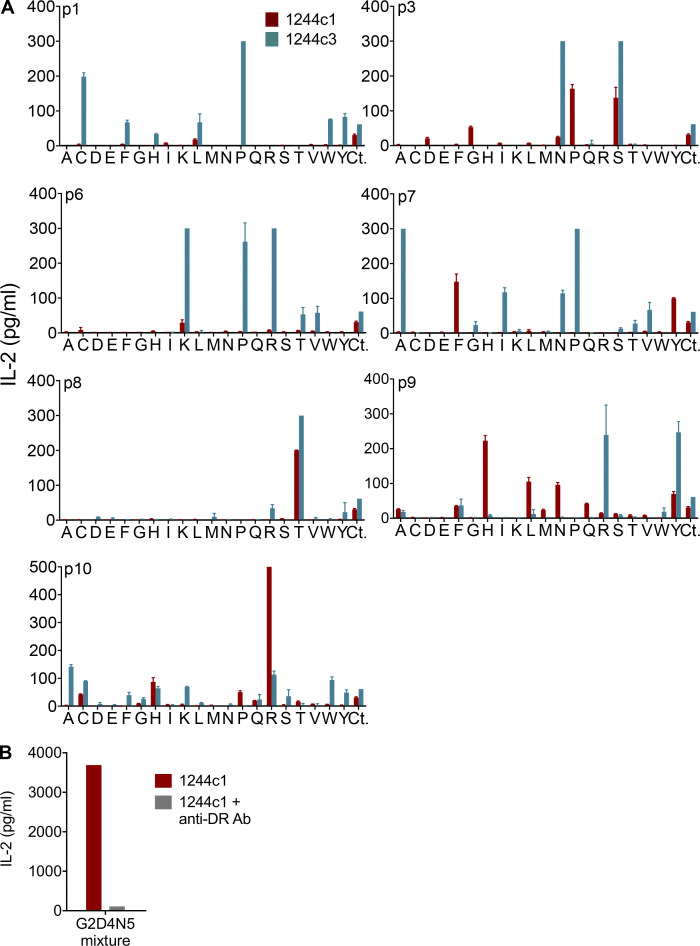
Since register 1 peptides aligned with amino acids known to bind to HLA-DR3 anchor positions (e.g., isoleucine/leucine [I/L]1, D4, R/lysine [K]6; Rammensee et al., 1995; Sidney et al., 1992), we synthesized a G2D4N5-biased library (G2D4N5 PSL) to perform a positional scan of the remaining seven peptide positions (140 mixtures) in the context of fixed amino acids at p2 (G), p4 (aspartic acid [D]), and p5 (N). The 1244 hybridomas shared similar amino acid profiles for most positions. For example, preferences for a serine (S) at the p3 position and a T at p8 were observed (Fig. S1 A). In addition, positively charged R and K were the preferred amino acids at p6, indicative of predicted anchor residues at the P6 position of HLA-DR3 (Rammensee et al., 1995; Sidney et al., 1992). Sequence logos quantitatively summarize amino acid preferences at all peptide positions for 1244c1, c2, and c3 (Fig. 2 C). Importantly, IL-2 activity induced by the G2D4N5 mixture was abolished by an anti–HLA-DR antibody, confirming that TCR recognition was HLA-DR restricted (Fig. S1 B).
Figure S1.
Biased G2D4N5 PSL results. (A) IL-2 responses of the LS-specific 1244c1 and 1244c3 54ζ hybridoma cells stimulated with a biased G2D4N5 decapeptide PSL are shown. Decapeptide mixtures were fixed at G2, D4, and N5, and one additional peptide position (p1, p3, p6, p7, p8, p9, and p10) was scanned with each of the 20 amino acids (140 mixtures). Unfixed positions in each decapeptide mixture consisted of an equimolar mixture of 19 amino acids (excluding cysteine). Each panel shows IL-2 responses (pg/ml) of T cell hybridomas 1244c1 (red bars) and 1244c3 (blue bars) to the decapeptide mixtures compared with the G2D4N5 mixture (labeled Ct) at 200 μg/ml with no additional fixed positions. Data are representative of two independent experiments. (B) IL-2 responses for 1244c1 5415 hybridoma cells after stimulation with 200 μg/ml G2D4N5 mixture with or without 20 μg/ml anti–HLA-DR blocking antibody (L243). Data are representative of two independent experiments.
Mimotopes completing the ligand for LS-specific TCRs
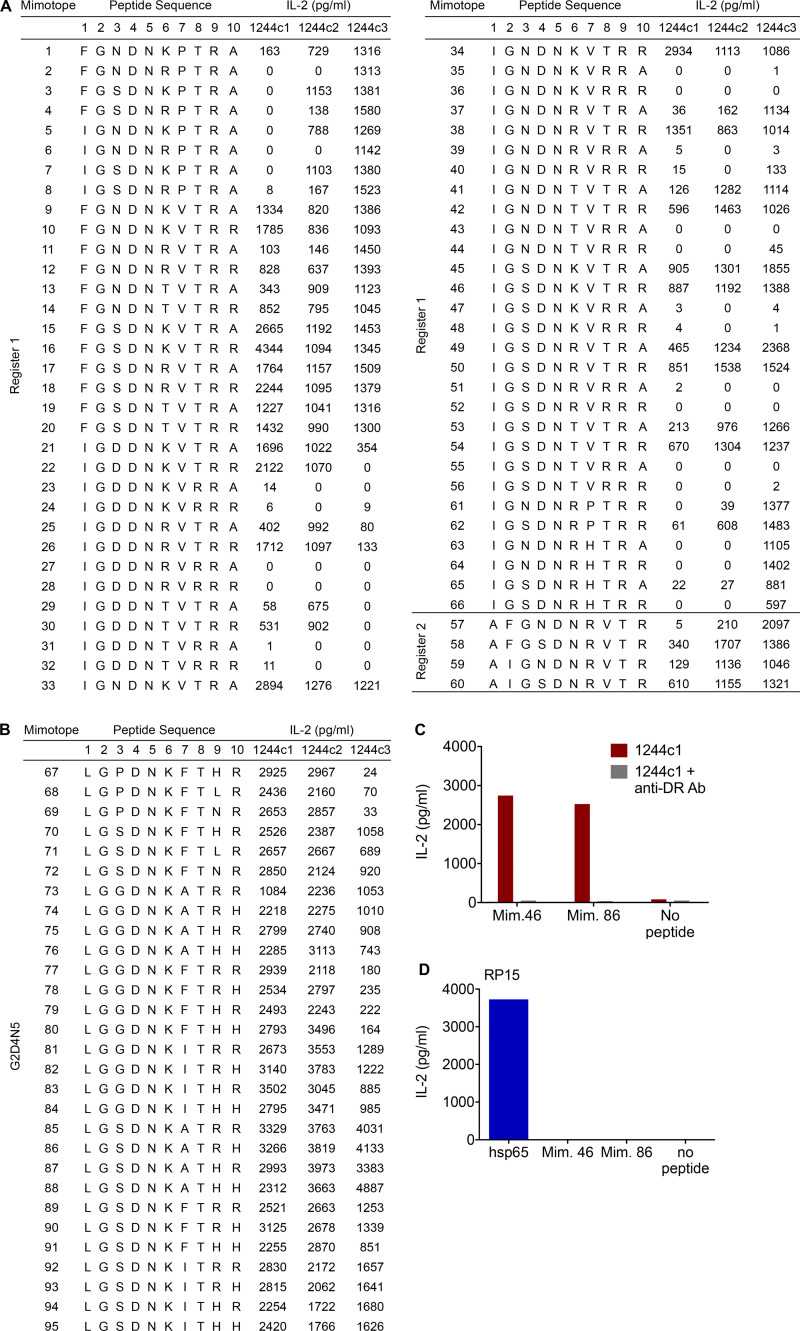
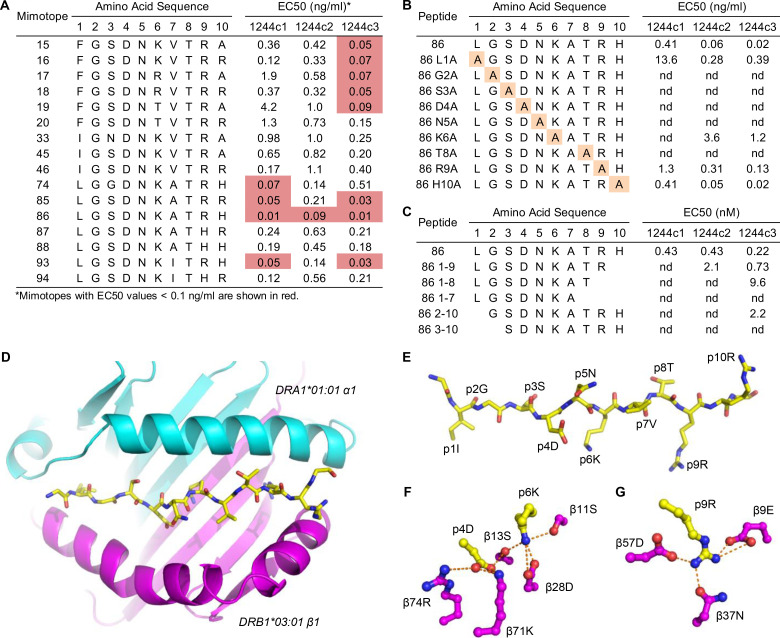
To identify peptides that stimulate the 1244 TCRs, we selected amino acids for each position based on both the unbiased and the G2D4N5 PSL scans and synthesized two sets of peptides (66 from unbiased registers 1 and 2 and 29 from the G2D4N5 PSL data). Amino acids chosen at each position are shown in Fig. 2 D, and peptide sequences and 1244c1, 1244c2, and 1244c3 hybridoma responses to each peptide (2 ng/ml) are listed in Fig. S2. Overall, these three TCRs recognized the majority of the peptides (Fig. S2, A and B). Dose–response curves were performed for the 16 most stimulatory mimotopes (peptides that mimic the naturally occurring epitope and stimulate TCRs), and peptide concentration inducing half-maximal IL-2 response (EC50) values for each TCR are shown in Fig. 3 A, with representative curves shown in Fig. S3 A. EC50 values of these 16 mimotopes fell within a narrow range (0.01–4.21 ng/ml; Fig. 3 A). We focused on mimotopes 46 and 86 due to the presence of the correct HLA-DR3 anchor residues (I/L at p1, D at p4, and K at p6) and highly stimulatory responses with all of the 1244 TCRs. Similar to the G2D4N5 PSL, addition of an anti–HLA-DR mAb abolished IL-2 responses to both mimotope 46 and 86, confirming HLA-DR restriction of these TCRs (Fig. S2 C). Additionally, a hybridoma expressing an HLA-DR3–restricted TCR (RP15) specific for a peptide derived from heat shock protein 65 (hsp65) of Mycobacterium leprae (hsp[3–13]; Struyk et al., 1995; Van Schooten et al., 1989) did not respond to mimotope 46 or 86 (Fig. S2 D). These data confirmed that mimotopes with cross-reactivity to the related set of LS-specific TCRs could be readily identified.
Figure S2.
1244 mimotope responses. (A) IL-2 responses (pg/ml) for all mimotopes synthesized based on the unbiased PSL results and dual-defined mixtures, noted as register 1 and register 2. LS-specific 1244c1, 1244c2, and 1244c3 5415 hybridoma cells were stimulated with mimotopes at 2 ng/ml. Data are representative of three independent experiments. (B) Additional mimotopes were synthesized based on the G2D4N5-biased PSL results, and corresponding IL-2 values for each peptide tested at 2 ng/ml with the LS-specific 1244c1, 1244c2, and 1244c3 5415 hybridoma cells are shown. Data are representative of two independent experiments. (C) IL-2 responses for 1244c1 5415 hybridoma cells after stimulation with 1 μg/ml mimotope 46 and 86 with or without 20 μg/ml anti–HLA-DR blocking antibody (L243). Data are representative of two independent experiments. (D) IL-2 responses for the hsp65–specific hybridoma RP15 after stimulation with media or 5 μg/ml of either hsp65 (3–13) of M. leprae, mimotope 46, or mimotope 86. Data are representative of two independent experiments. In A–D, HLA-DR3–expressing B6DK10 fibroblasts were used as APCs, and IL-2 responses were measured by ELISA after a 20–24-h incubation. Mim., mimotope.
Figure 3.
Delineation of highly stimulatory LS-specific mimotopes. (A) EC50 values are shown for the LS-specific 1244c1, 1244c2, and 1244c3 5415 hybridoma cells for the 16 mimotopes with the overall highest activities. (B) 1244c1, 1244c2, and 1244c3 hybridoma cells were incubated with peptides with a single A substitution of each position of mimotope 86, and EC50 values were calculated. (C) 1244c1, 1244c2, and 1244c3 5415 hybridoma cells were incubated with minimal epitope peptides of mimotope 86, and EC50 values were calculated. IL-2 responses were measured by ELISA. EC50 values were calculated by generating dose–response curves of IL-2 responses after stimulation with peptides ranging from 0.001 to 30 ng/ml. Data for A–C are representative of three independent experiments. (D) Ribbon representation of the full HLA-DR3α (cyan) and β (magenta) extracellular domains and a wireframe representation of the mimotope 46 peptide (CPK coloring) viewed from the top down are shown. (E) Wireframe representation of mimotope 46 (CPK coloring) with individual amino acids in the peptide labeled. Downward-facing anchor residues at amino acid positions p1, p4, p6, and p9 are shown, with upward-facing TCR interacting residues at p2, p3, p5, p7, p8, and p10. (F and G) Wireframe representations of the P4 and P6 (F) and P9 (G) pockets are shown (CPK coloring). Dashed orange lines connect oxygens in the P4, P6, and P9 pockets that form hydrogen bonds or salt bridges to each other or to the nitrogens of the R and K guanidinium groups. nd, nonstimulatory.
Figure S3.
Mimotope dose–response curves. (A–D) IL-2 dose–response curves for 1244c1, 1244c2, and 1244c3 5415 hybridomas stimulated with mimotope 46 and mimotope 86 (A), mimotope 86 peptides with separate A substitutions at each amino acid position (B), and mimotope 86 (C) and NDPD 11-mer (D) peptides of varying length to determine the minimal epitope capable of TCR activation. HLA-DR3–expressing B6DK10 fibroblast cells were used as APCs, and IL-2 responses were measured by ELISA after incubating 20–24 h. All peptides were tested at a range of 0.001–30 ng/ml. Data are representative of three independent experiments.
Peptide length variants and alanine (A) substitutions of mimotope 86 were performed to determine a minimal epitope and critical amino acids for TCR activation. Either an L to A substitution at the p1 position of the peptide or deletion of the p1 amino acid shifted the dose–response curve to the right (Fig. S3, B and C), resulting in significant increases in the EC50 values (Fig. 3, B and C). Substitutions at p9 and p10 had minimal effects on EC50 values (Fig. 3 B and Fig. S3 B), while deletion of these amino acid residues more significantly reduced peptide activity (Fig. 3 C and Fig. S3 C). Both the A scan and the minimal epitope peptides demonstrated that the core of the peptide (positions 2–8: GSDNKAT) was crucial for peptide recognition. Replacing any of these positions with an A or deleting G2 or T8 abolished peptide activity (Fig. 3, B and C; and Fig. S3, B and C). Thus, for optimal TCR activity, the full decapeptide is required (LGSDNKATRH); however, the minimal epitope needed for TCR stimulation is p2–p8 (GSDNKAT). Additionally, peptide length variants and A substitutions at each position of mimotope 46 were tested, and the critical amino acid requirements were analogous to those of mimotope 86 (data not shown).
Structural analysis of HLA-DR3–mimotope 46
To validate our A scan and minimal epitope findings, we produced a soluble version of HLA-DR3 (DRA1*01:01, DRB1*03:01) with a covalently tethered mimotope 46. This peptide only differed from mimotope 86 at three positions, with conservative substitutions at p1 (L to I) and p7 (A to valine [V]). We crystallized this complex and solved its structure to a resolution of 2.45 Å. The statistics for the data collection and refined model are shown in Table S1. The overall structure of the HLA-DR3–mimotope 46 complex was similar to other MHCII/peptide complexes (Fig. 3 D), with a well-defined electron density for the bound peptide. The four amino acids at p1, p4, p6, and p9 are the major anchor residues for the binding of mimotope 46 to HLA-DR3 (Fig. 3 E). Isoleucine at p1 (p1I) occupied the top portion of a large pocket lined with hydrophobic amino acids and was stabilized by van der Waals contacts. The space below p1I could accommodate the phenylalanine (F) present in some of mimotopes shown in Fig. 3 A and indicated that p1I was not significantly contributing binding energy between the mimotope and HLA-DR3. p4D and p6K were the major determinants of peptide binding. The carboxylate of p4D formed salt bridges with the α-amino group of the K at position 71 of the HLA-DR3 β-chain (Kβ71) with a distance of 2.7 Å (Fig. 3 F). In addition, p4D formed hydrogen bonds with Rβ74 and Sβ13. p6K sits in the P6 pocket with a usual conformation and moves toward p4D with a distance of 4.3 Å. Compared with HLA-DR3–CLIP structure (Ghosh et al., 1995), p6K inserted into the bottom of the P6 pocket (Fig. 3 F). The effects of the intricate interactions between p4D and p6K (Fig. 3 F) resulted in bulging of the p5N side chain out of the peptide binding groove. Furthermore, p5N formed hydrogen bonds with p4D and Rβ74 and is predicted to be a central hot spot for T cell activation, as suggested by the PSL screening (Fig. 2 C). The α-amino group of p6K formed salt bridges with Dβ28. The distance between Nζ of p6K and OD1 atom of Dβ28 was only 2.5 Å. In addition, p6K formed short hydrogen bonds with Sβ11 and Sβ13 of 3.1 and 2.8 Å, respectively. We predict that the p6R of mimotope 86 would occupy the P6 pocket in a similar manner. The α-amino group of p9R formed salt bridges with Dβ57 and Eβ9 (Fig. 3 G). The unique properties of the HLA-DR3 molecule contribute to the selection of charged amino acids in the mimotopes with high binding energy in the P4, P6, and P9 pockets. The highly conserved amino acids from the PSL screening (Fig. 2 C) are consistent with the structure, with G2, N5, and T8 all being solvent exposed and potential ligands for LS-specific TCRs (Fig. 3 E).
LS-specific TCRs recognize a peptide derived from A. nidulans
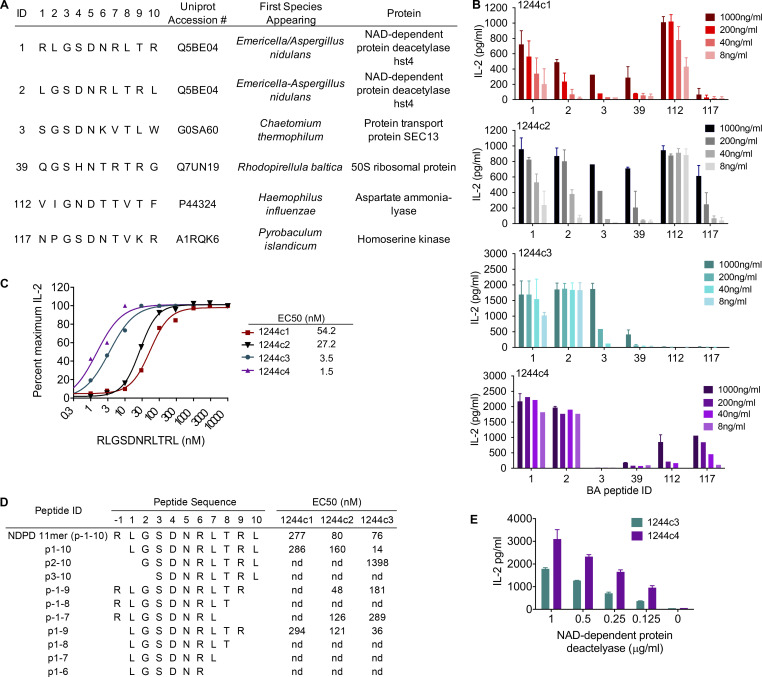
We performed a biometrical analysis to identify candidate naturally occurring peptides capable of stimulating the LS-specific T cell hybridomas. Scoring matrices were created based on the stimulatory potential of each fixed amino acid at each position of the PSL, as previously described (Falta et al., 2021; Zhao et al., 2001). To account for different peptide registers in the unbiased matrix, the stimulatory responses of the dual-defined mixtures in either register 1 or register 2 were integrated into the scoring matrices using the harmonic mean model (Santos et al., 2011) to create “harmonic boost” register 1 and register 2 (Planas et al., 2018). A total of 16 matrices were created, and an independent biometrical analysis was performed for each and applied to all possible overlapping decapeptides within the SwissProt database containing curated proteins from all organisms to rank and score naturally occurring peptides. The resulting lists of potential antigens were consolidated by choosing the top 50 overall peptides in both registers and the remaining top 10 in each individual matrix that were not included in the previous top 50.
Table S2 lists 125 peptides according to their ranking in the overall analysis, amino acid sequence, accession number, first species of origin, protein source, and IL-2 secretion by the three 1244 LS-specific T cell hybridomas in response to peptide. We focused on peptides that induced >100 pg/ml of IL-2 in at least one of the 1244 TCRs (Table S2). Only six naturally occurring peptides fit this criterion (Table S2 and Fig. 4 A). We also expanded our investigation to include the fourth related 1244 TCR, 1244c4 (TCR sequence shown in Fig. 1 C). Of the six peptides, the only ones that activated all four of the 1244 LS-specific T cell hybridomas were overlapping versions of a peptide derived from NDPD (209–218 and 210–219) of A. nidulans (Fig. 4 B). Importantly, these were the highest ranked peptides (1 and 2) in the biometrical analysis (Table S2). We combined the two peptide sequences to create an 11-mer (209–219) derived from A. nidulans NDPD and generated dose–response curves for this peptide with all four TCRs (Fig. 4 C). Hybridomas 1244c3 and c4 had substantially lower EC50 values (10–50-fold) compared with c1 and c2. Assays to define the minimal epitope demonstrated that all positions of the peptide except for p1 and p10 were required for T cell activation, with the minimal A. nidulans epitope being p1–p9 (LGSDNRLTR; Fig. 4 D). Compared with mimotope 86, the NDPD 11-mer was more dependent on L at p1, likely to provide additional anchoring of the peptide to the HLA-DR3 binding groove.
Figure 4.
LS-specific TCRs recognize NDPD derived from A. nidulans. (A) Amino acid sequences, UniProt accession numbers, and associated species and proteins of the naturally occurring peptides that stimulated the LS-specific 1244 TCRs. (B) IL-2 secretion by 1244c1 (top panel), 1244c2 (second panel), 1244c3 (third panel), and 1244c4 (bottom panel) 5415 hybridomas stimulated with peptides shown in A, tested at 1,000, 200, 40, and 8 ng/ml. (C) IL-2 dose–response curves and calculated EC50 values for 1244c1, 1244c2, 1244c3, and 1244c4 5415 hybridomas in response to the NDPD 11-mer peptide (RLGSDNRLTRL) tested at a range of 0.3–10,000 nM. (D) 1244c1, 1244c2, and 1244c3 5415 hybridoma cells were incubated with minimal epitope peptides of NDPD 11-mer, and EC50 values were calculated. (E) IL-2 secretion by 1224c3 and 1244c4 in response to the NDPD recombinant protein from 0.125 to 1.0 μg/ml. IL-2 responses were measured by ELISA after a 20–24-h incubation. Data for B–E are representative of at least two independent experiments. nd, nonstimulatory.
Validation of NDPD derived from A. nidulans as an antigen in LS
To prove that NDPD derived from A. nidulans is a target of the adaptive immune response in LS, we ascertained whether NDPD protein could be processed to yield the stimulatory HLA-DR3–binding epitopes. Recombinant purified NDPD protein (amino acids 117–410) was tested at varying concentrations using HLA-DR3–transfected fibroblasts as APCs. IL-2 secretion was observed from hybridomas 1244c3 and 1244c4 at concentrations of protein ranging from 0.125 to 1 μg/ml (Fig. 4 E), indicating that NDPD can be processed and presented to activate the 1244 TCRs.
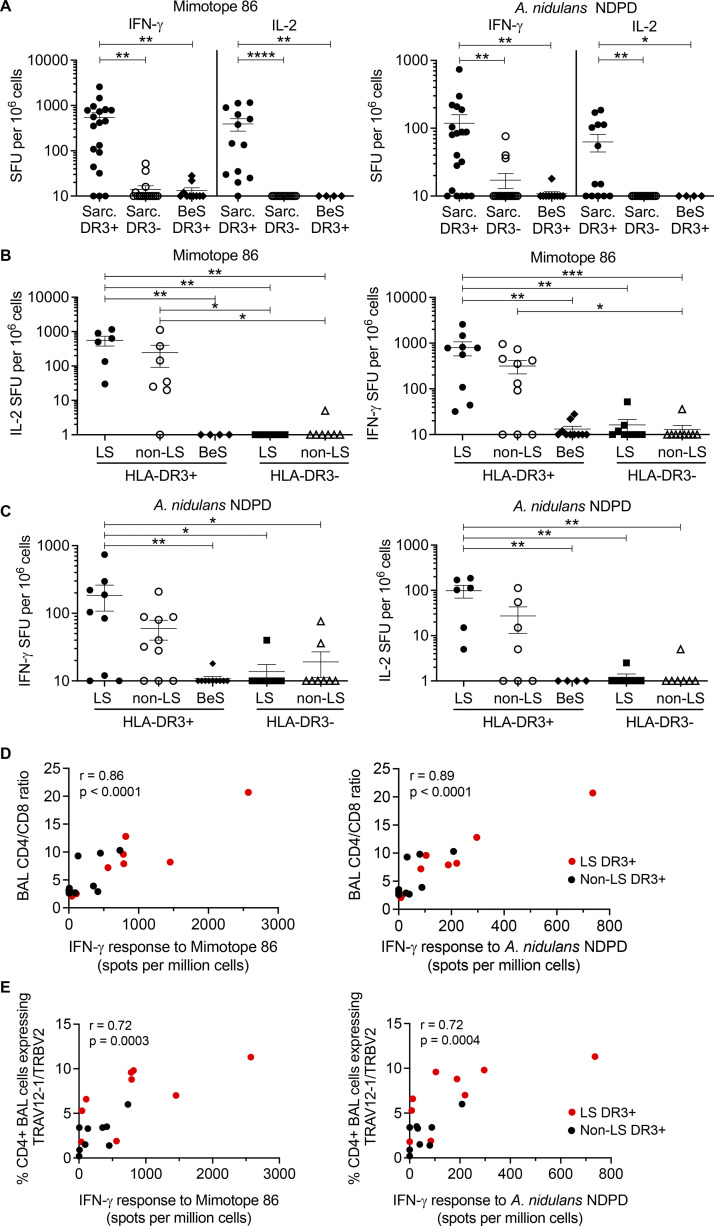
To validate the presence of T cells reactive against both mimotope 86 and A. nidulans NDPD in LS patients, we performed IFN-γ and IL-2 ELISPOT assays on BAL cells from 9 DR3+ LS, 10 DR3+ non-LS sarcoidosis, 10 DR3+ beryllium-sensitized (BeS), 8 DR3− LS, and 9 DR3− non-LS sarcoidosis subjects. Demographics of the validation cohort are shown in Table 1. First, we grouped subjects based on the diagnosis of sarcoidosis (LS and non-LS) and DR3 expression (Fig. 5 A). In response to both mimotope 86 and the A. nidulans NDPD peptide, the number of IFN-γ– and IL-2–secreting cells in BAL was significantly higher in the DR3+ sarcoidosis patients compared with either DR3− subjects or DR3+ BeS subjects (Fig. 5 A). IFN-γ and IL-2 responses for mimotope 86 were significantly higher than for NDPD, consistent with the 1244 hybridoma responses (Fig. 5 A).
Table 1. Demographics of the validation cohort.
| LS HLA-DR3+ (n = 10) | Non-LS HLA-DR3+ (n = 11) | LS HLA-DR3− (n = 9) | Non-LS HLA-DR3− (n = 12) | BeS HLA-DR3+ (n = 10) | |
|---|---|---|---|---|---|
| Sex (M/F) | 8/2 | 9/3 | 7/2 | 7/5 | 7/3 |
| Age (yr) | 37 (33–44) | 46 (39–55) | 45 (36–55) | 40 (34–46) | 57 (51–64) |
| Chest radiographical stage a | N/A | ||||
| 0 | 0 | 0 | 1 | 0 | |
| I | 5 | 5 | 5 | 6 | |
| II | 5 | 3 | 2 | 3 | |
| III | 0 | 2 | 0 | 2 | |
| IV | 0 | 0 | 0 | 0 | |
| Smoking status | |||||
| Non | 6 | 9 | 6 | 8 | 3 |
| Former | 3 | 1 | 1 | 1 | 6 |
| Current | 1 | 1 | 2 | 3 | 1 |
| BALF cell concentration (106 cells/liter) | 177 (144–329) | 201 (157–341) | 128 (97–282) | 153 (108–208) | 22 (19–29) |
| % BALF macrophages | 68 (42–87) | 75 (60–78) | 71 (55–83) | 77 (60–82) | 93 (87–96) |
| % BALF lymphocytes | 30 (12–50) | 21 (18–38) | 20 (14–36) | 19 (13–37) | 5.6 (2.8–11) |
| % BALF neutrophils | 1.0 (0.8–1.6) | 1.3 (0.3–1.6) | 1.6 (0.8–3.3) | 1.4 (0.6–2.6) | 0.9 (0.2–1.5) |
| BAL CD4/CD8 ratio | 8 (2–14) | 3.5 (2.9–9.3) | 6.3 (3.3–15.3) | 7.8 (3–12.5) | N/A |
| % TRAV12-1/TRBV2–expressing BAL CD4+ T cells | 6.8 (4.5–10) | 3.4 (1.4–4.7) | 0.1 (0.1–0.1) | 0.1 (0.1–0.2) | N/A |
Data are counts or median (25th–75th percentile). BALF, BAL fluid; N/A, not available.
Chest radiography staging as follows: stage 0 = normal chest radiography, stage I = enlarged lymph nodes, stage II = enlarged lymph nodes with parenchymal infiltrates, stage III = parenchymal infiltrates without enlarged lymph nodes, and stage IV = signs of pulmonary fibrosis.
Figure 5.
Mimotope 86 and A. nidulans NDPD peptides stimulate sarcoidosis BAL cells in an HLA-DR3–restricted manner. (A) IFN-γ and IL-2 ELISPOT responses (shown as SFUs per million cells) to mimotope 86 (left) and A. nidulans NDPD (right) at 5 μg/ml are shown. The sarcoidosis (Sarc) group includes LS and non-LS subjects. For IFN-γ ELISPOTs, subject numbers are as follows: Sarc DR3+ (n = 19), Sarc DR3− (n = 17), and BeS DR3+ (n = 10). For the IL-2 ELISPOTS, subject numbers are as follows: Sarc DR3+ (n = 13), Sarc DR3− (n = 14), and BeS DR3+ (n = 4). (B) IFN-γ (left) and IL-2 (right) ELISPOT responses for mimotope 86 at 5 μg/ml. (C) IFN-γ (left) and IL-2 (right) ELISPOT responses for A. nidulans NDPD at 5 μg/ml. In B and C, subjects are divided into DR3+ LS (IFN-γ, n = 9; IL-2, n = 6), DR3+ non-LS sarcoidosis (IFN-γ, n = 10; IL-2, n = 7), DR3− LS (IFN-γ, n = 9; IL-2, n = 7), DR3− non-LS sarcoidosis (IFN-γ, n = 9; IL-2, n = 7), and DR3+ BeS (IFN-γ, n = 10; IL-2, n = 4). Data are expressed in A-C as mean SFUs of duplicate wells, with bars representing mean ± SEM. (D and E) Correlation of IFN-γ responses to mimotope 86 (left) and A. nidulans NDPD (right) with BAL CD4/CD8 ratios (D) and percentage of TRAV12-1/TRBV-2–expressing CD4+ T cells (E) in the BAL of DR3+ LS (red) or non-LS sarcoidosis (black) subjects. The correlation coefficient (r) and P value are shown. Statistical significance was determined by one-way ANOVA with multiple comparisons in A–C and Spearman rank correlation in D and E. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Next, we divided the sarcoidosis cohort based on an LS diagnosis and DR3 expression. Although not significantly different, there was a threefold increase in the number of NDPD- and mimotope 86–responsive BAL cells among DR3+ LS patients compared with DR3+ non-LS sarcoidosis subjects for both IFN-γ and IL-2 secretion (Fig. 5, B and C). Compared to DR3+ BeS, DR3− LS, and DR3− non-LS sarcoidosis subjects, the number of IFN-γ– and IL-2–secreting BAL cells in DR3+ LS subjects in response to mimotope 86 was significantly increased (Fig. 5 B). In addition, a significantly increased number of NDPD-induced IFN-γ– and IL-2–secreting cells was seen in DR3+ LS subjects compared with DR3+ BeS, DR3− LS, and DR3− non-LS sarcoidosis subjects (Fig. 5 C). Taken together, these data confirm a mimotope 86 and an A. nidulans NDPD-specific immune response in the lungs of DR3+ LS patients.
To determine the relationship between immunological markers of sarcoidosis and BAL T cell responses to mimotope 86 and A. nidulans NDPD peptides, we correlated the frequency of IFN-γ–secreting T cells in BAL of DR3+ sarcoidosis patients with immunological markers of disease activity (Fig. 5, D and E). High numbers of antigen-specific cells in the BAL of sarcoidosis patients (non-LS, black dots; LS, red dots) were associated with the BAL CD4/CD8 ratio (mimotope 86: r = 0.86, P < 0.0001; A. nidulans NDPD: r = 0.89, P < 0.0001; Fig. 5 D) and percentage of BAL CD4+ T cells expressing TRAV12-1/TRBV2 (mimotope 86: r = 0.72, P < 0.0003; A. nidulans NDPD: r = 0.72, P < 0.0005; Fig. 5 E). These data suggest that A. nidulans NDPD-specific T cell responses are directly related to lung infiltration of disease-specific CD4+ T cells in sarcoidosis.
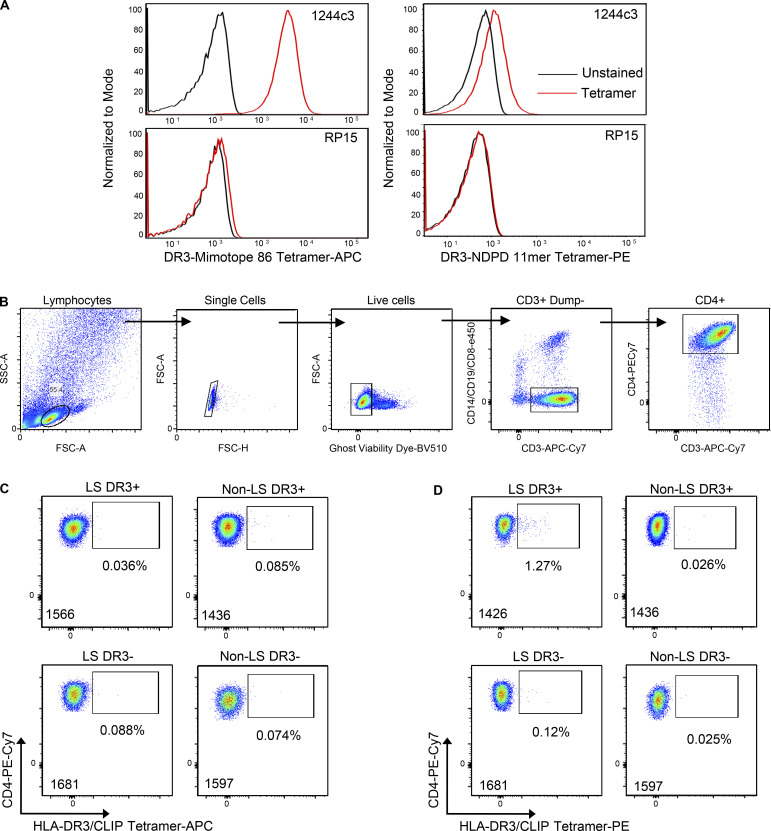
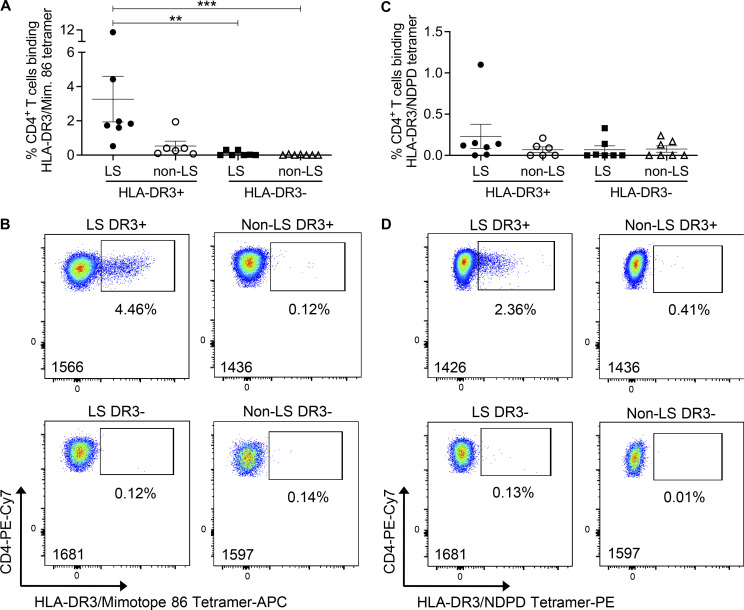
To assess the breadth of this T cell specificity in the BAL of sarcoidosis patients, HLA-DR3 tetramers were constructed with mimotope 86 and NDPD (209–218). These tetramers stained hybridomas expressing the 1244 TCRs (Fig. S4 A) but did not stain the HLA-DR3–restricted, hsp65-specific RP15 hybridoma (Fig. S4 A), confirming their specificity. Next, we investigated the frequency of tetramer binding ex vivo BAL CD4+ T cells from DR3+ and DR3− LS and non-LS sarcoidosis patients. Using a gating strategy shown in Fig. S4 B, the frequency of mimotope 86–specific CD4+ T cells in the BAL of HLA-DR3+ LS patients was significantly higher than DR3− LS and non-LS sarcoidosis subjects and was increased, although not significantly, compared with DR3+ non-LS patients, with frequencies ranging from 0.52% to 10.8% (mean, 3.2%) after subtraction of HLA-DR3–CLIP background staining (Fig. 6, A and B; and Fig. S4 C). Despite elevated IFN-γ and IL-2 ELISPOT responses in the BAL of DR3+ non-LS sarcoidosis subjects, we did not identify significantly increased HLA-DR3–mimotope 86 tetramer-binding CD4+ T cells compared with DR3− LS and non-LS sarcoidosis subjects (Fig. 6, A and B). Although HLA-DR3–A. nidulans NDPD tetramer staining was detectable in LS patient 1426, no significantly increased HLA-DR3–A. nidulans NDPD tetramer staining in DR3+ LS patients was detected compared with the other groups (Fig. 6, C and D).
Figure S4.
HLA-DR3–peptide tetramer staining of LS-specific 1244 TCR-expressing hybridomas and ex vivo BAL cells. (A) Representative histograms of LS-specific 1244c3 and RP15 (specific for HLA-DR3 p3–p13 of hsp65 of M. leprae) T cell hybridoma staining with HLA-DR3–peptide tetramers (A. nidulans NDPD 11-mer, 1 μg/ml; mimotope 86, 10 μg/ml). (B) Representative density plots showing the gating strategy to isolate live CD4+ T cells from the BAL to analyze tetramer staining on CD4+ T cells. (C and D) Representative density plots of HLA-DR3–CLIP–APC (10 μg/ml; C) and HLA-DR3–CLIP–PE (1 μg/ml; D) tetramer staining of CD4+ T cells used to determine background frequencies of HLA-DR3–peptide tetramer–binding cells. The patient number is listed in the lower left corner of each density plot. FSC, forward scatter; SSC, side scatter.
Figure 6.
Increased frequency of HLA-DR3–mimotope 86–binding CD4+ T cells in the BAL of HLA-DR3+ LS patients. (A and C) Summary of the frequency of tetramer staining of ex vivo BAL CD4+ T cells from DR3+ LS (n = 7), DR3+ non-LS (n = 6), DR3− LS (n = 7), DR3− non-LS (n = 7) sarcoidosis subjects stained with HLA-DR3–mimotope 86 (A) and HLA-DR3–A. nidulans NDPD (C) tetramers. Values recorded are after subtraction of background HLA-DR3–CLIP staining. Bars represent mean ± SEM. Statistical significance was determined by one-way ANOVA with multiple comparisons. **, P < 0.01, ***, P < 0.001. (B and D) Representative density plots of HLA-DR3–mimotope 86 (B) and HLA-DR3–A. nidulans NDPD (D) tetramer staining of ex vivo BAL cells from DR3+ LS, DR3+ non-LS, DR3− LS, and DR3− non-LS sarcoidosis subjects are shown. The patient number is listed in the lower left corner of each density plot. Mim. mimotope.
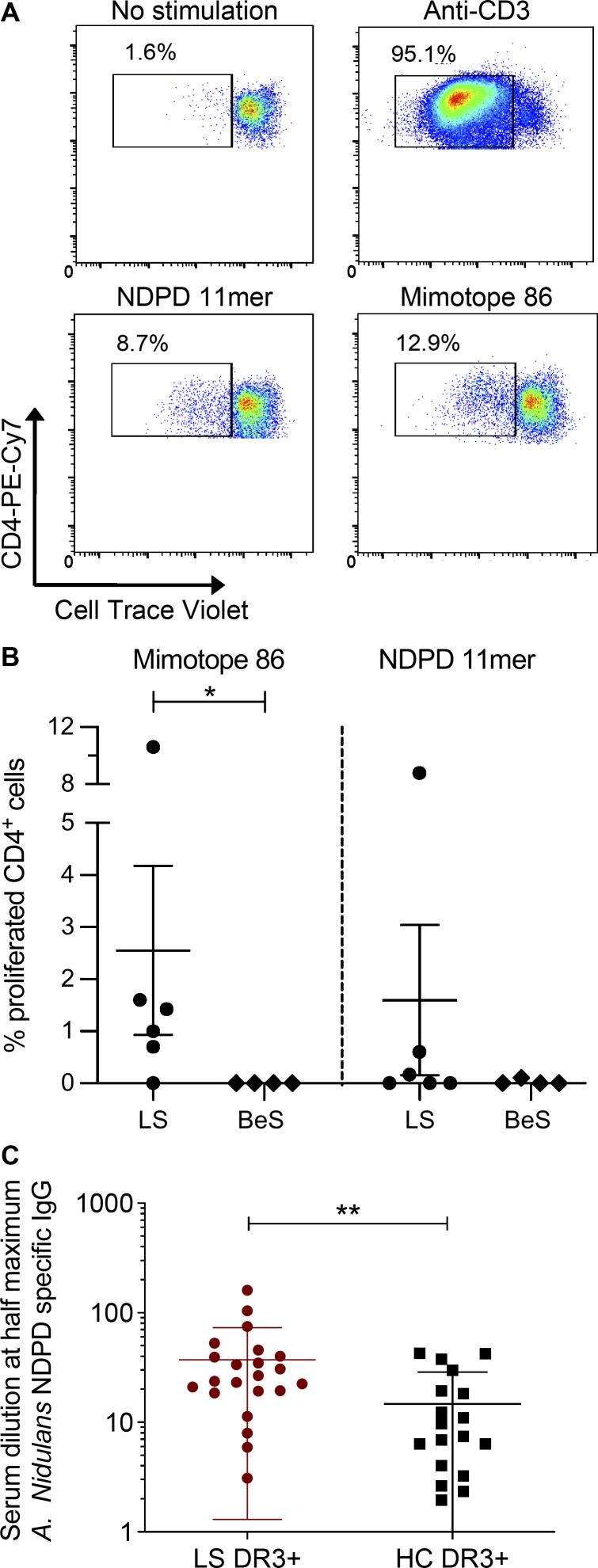
To determine whether DR3+ LS subjects had A. nidulans–derived NDPD peptide and mimotope 86–responsive CD4+ T cells in blood, we measured the proliferative response after stimulation with the A. nidulans NDPD and mimotope 86 peptides. Representative density plots from a DR3+ LS subject show proliferation in response to no stimulation, plate-bound anti-CD3, NDPD peptide, or mimotope 86 (all at 10 μg/ml) as seen in Fig. 7 A. Compared with CD4+ T cells from BeS subjects, a significantly greater percentage of CD4+ T cells from DR3+ LS subjects underwent cell division after exposure to mimotope 86 (P = 0.04; Fig. 7 B). Conversely, although NDPD peptide–specific proliferation of CD4+ T cells was seen in one of the six DR3+ LS subjects, no statistically significant differences were noted (Fig. 7 B).
Figure 7.
T cell proliferation and IgG specific for A. nidulans NDPD in the blood of DR3+ LS patients. (A) Representative density plots showing the frequency of proliferated CD4+ T cells (measured by loss of Cell Trace Violet) derived from blood of a DR3+ LS subject after stimulation with media alone, plate-bound anti-CD3 (10 μg/ml), 5 μg/ml A. nidulans NDPD peptide, or 5 μg/ml mimotope 86 peptide. (B) Summary of the frequency of CD4+ T cell proliferation in response to A. nidulans NDPD peptide or mimotope 86 peptide, after subtraction of background proliferation with no stimulation, in DR3+ LS (n = 6) and DR3+ BeS (n = 4) subjects. (C) Levels of IgG specific for A. nidulans NDPD were measured by ELISA in threefold serum dilutions (1–2,187) of HLA-DR3+ LS (n = 22) and HLA-DR3+ healthy control (HC) subjects (n = 18), and the serum dilution that elicited 50% of the maximum antibody response for each subject was determined using nonlinear regression curves. Statistical significance in B and C was determined by Mann-Whitney test. *, P < 0.05; **, P < 0.01.
In addition to characterizing T cell responses to A. nidulans antigens, we investigated other facets of adaptive immunity directed against A. nidulans. Serum levels of IgG specific for A. nidulans NDPD were significantly elevated in HLA-DR3+ LS subjects compared with healthy controls (Fig. 7 C). These findings suggest that the response to A. nidulans in HLA-DR3+ LS patients extends beyond T cell activation in the lungs, suggesting a systemic immune response directed against NDPD of A. nidulans.
Immunization with A. nidulans NDPD or mimotope 86 peptide induces CD4+ T cell–specific immune responses in HLA-DR3 transgenic mice
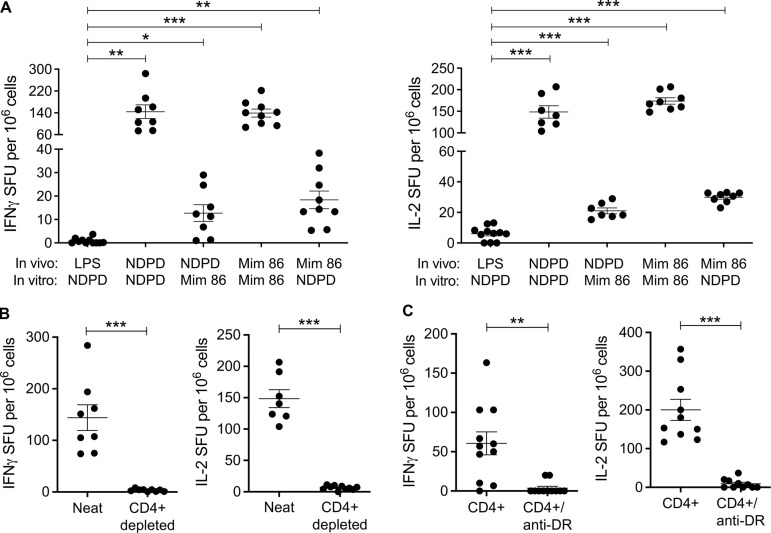
To test the immunogenicity of the A. nidulans NDPD peptide, MHCII-deficient HLA-DR3 transgenic (Tg) mice were immunized intraperitoneally with 10 μg NDPD peptide in the presence of LPS (10 μg) as an adjuvant. Additional mice were similarly immunized with mimotope 86. At day 10, splenocytes were harvested and stimulated ex vivo with either the NDPD or the mimotope 86 peptide. Ex vivo stimulation with the same peptide as used in the immunization resulted in a significant increase in the number of IFN-γ– (Fig. 8 A, left) and IL-2– (Fig. 8 A, right) secreting cells compared with the LPS control group. Importantly, HLA-DR3 Tg mice immunized with either the NDPD or the mimotope 86 peptide showed T cell cross-reactivity to mimotope 86 or NDPD peptide, respectively, as demonstrated by significantly increased IFN-γ and IL-2 secretion compared with LPS controls (Fig. 8 A, left and right).
Figure 8.
CD4+ T cell–specific immune response to A. nidulans NDPD or mimotope 86 peptide in HLA-DR3 Tg mice. HLA-DR3 Tg mice were immunized with NDPD or mimotope 86 peptide in the presence of LPS, and splenocytes were harvested at day 10. (A) Splenocytes were stimulated with 5 μg/ml NDPD or mimotope 86 peptide, and IFN-γ and IL-2 responses were measured by ELISPOT. (B) Total and CD4-depleted splenocytes were stimulated with 5 μg/ml NDPD peptide. (C) CD4+ T cells purified from splenocytes were cultured with irradiated naive splenocytes from HLA-DR3 Tg mice and stimulated with 5 μg/ml NDPD peptide in the presence or absence of 10 μg/ml anti–HLA-DR mAb. For B and C, IFN-γ– (left) and IL-2– (right) secreting cells were examined by ELISPOT. Data from A–C are representative of two independent experiments. Statistical significance was determined by one-way ANOVA with multiple comparisons in A and Mann-Whitney test in B. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Mim, mimotope.
To prove that the NDPD responses were CD4+ T cell specific, we immunized HLA-DR3 Tg mice with 10 μg NDPD peptide and 10 μg LPS, harvested splenocytes, and depleted CD4+ T cells (Fig. 8 B). Compared with neat splenocytes, CD4-depleted splenocytes did not secrete IFN-γ or IL-2 in response to NDPD peptide (5 μg/ml; Fig. 8 B). In a complementary experiment, purified CD4+ T cells from similarly immunized HLA-DR3 Tg mice were combined with irradiated naive splenocytes and stimulated with 5 μg/ml NDPD peptide in the presence or absence of 10 μg/ml anti–HLA-DR mAb. As shown in Fig. 8 C, CD4+ T cells secreted IFN-γ and IL-2 in response to NDPD peptide stimulation, and the cytokine response was HLA-DR3 restricted. Overall, these murine studies confirmed that CD4+ T cells recognize the A. nidulans NDPD peptide in an HLA-DR3–restricted manner and that a subset of naturally occurring NDPD-specific CD4+ T cells can cross-react with the mimotopes 86 peptide discovered using PSLs.
Discussion
In this study, we identified a related set of LS-specific TCRs that recognized a peptide derived from NDPD expressed in A. nidulans. Using the webtool Immune Epitope Database for reported epitopes and their corresponding antigens (Vita et al., 2019), neither this epitope nor protein has been previously reported. We validated the presence of A. nidulans NDPD-specific T cells in ex vivo BAL cells of HLA-DR3+ LS and non-LS patients and found IgG antibody responses to A. nidulans NDPD in the serum of DR3+ LS subjects. Taken together, our findings suggest that A. nidulans may be involved in the pathogenesis of LS.
Aspergillus is a ubiquitous airborne organism that most humans are exposed to in everyday environments, yet the majority of individuals remain unaffected. Although DR3+ LS patients show no signs of an active pulmonary Aspergillus infection, our data indicate that there may be a hypersensitivity response directed against the A. nidulans NDPD epitope that causes excessive inflammation in the lungs. There is also a seasonal clustering of LS, particularly in the spring (Bardinas et al., 1989; Glennås et al., 1995; Grunewald and Eklund, 2007), which may coincide with higher environmental Aspergillus exposure. Recently, Clarke et al. (2018) investigated microbial lineages in sarcoidosis and identified limited enrichment for Aspergillus in the BAL. Although healthy individuals have T cells in blood that are reactive to common Aspergillus species, including A. nidulans (Bacher et al., 2019), our ELISPOT data from blood of DR3+ and DR3− control subjects do not show the presence of IFN-γ– or IL-2–secreting T cells in response to stimulation with the A. nidulans NDPD epitope. Several previous studies have linked exposure to fungal byproducts to more severe cases of sarcoidosis. Higher levels of fungal biomass were detected in residences of sarcoidosis patients compared with controls (Terčelj et al., 2014, 2011b), and the amount of fungal biomass detected was directly related to fungal cell wall component β-glucan in the BAL of sarcoidosis patients (Terčelj et al., 2014). Additionally, a significant association was found among β-glucan levels in lymph nodes, granuloma formation in the lungs, and the size of the mediastinal lymph nodes in sarcoidosis patients (Terčelj et al., 2017). Finally, when compared with corticosteroids in the treatment of sarcoidosis, antifungal medication was found to be equally effective, as determined by decreased granuloma mass and inflammatory markers (Terčelj et al., 2007, 2011a). These studies, along with our current study, support a role for fungal exposure, particularly Aspergillus, in the pathogenesis of sarcoidosis and LS.
NAD-dependent protein deacetylase is an intranuclear histone-interacting protein in the SIR2 family of proteins involved in transcriptional silencing (Buck et al., 2004). Interestingly, the protein homology between A. nidulans NDPD and the corresponding protein in Aspergillus fumigatus (the most common Aspergillus species found in humans) is only ∼60%, and the particular epitope corresponding to the NDPD 11-mer differs at amino acid positions p2–p4 (RLAQENRLTRL), which are crucial for peptide recognition by the LS-specific TCRs. Additionally, the corresponding human protein (NAD-dependent protein deacetylase sirtuin-2) is only ∼20% homologous to A. nidulans NDPD, and there is no similar peptide within this protein corresponding to the NDPD 11-mer. As an illustration of this point, the harmonic mean boost matrix for A. nidulans NDPD was run against all NDPD proteins in the UniProt database, and the A. nidulans NDPD peptide identified scored 10% higher than the next highest peptide. We cannot completely exclude the possibility that sarcoidosis patients express an immunogenic fusion peptide, similar to the hybrid insulin peptides found in type 1 diabetes patients (Arribas-Layton et al., 2020; Baker et al., 2019; Delong et al., 2016). The SwissProt database screened for the biometrical analysis does not include fusion peptides. However, because the LS-specific TCR recognition requirements for the mimotopes and the A. nidulans NDPD peptide are so specific (i.e., removing or replacing any of the core amino acids ablates peptide recognition), it is unlikely that these TCRs would recognize an alternate fusion or posttranslationally modified peptide.
Our results show a stronger TCR response to the mimotope 86 peptide compared with the naturally occurring A. nidulans NDPD epitope, evident by the lower EC50 values and the ∼10-fold higher number of IFN-γ and IL-2 spot-forming units (SFUs) elicited by mimotope 86 in the BAL of LS patients. These results are not surprising given that mimotope 86 was created by unbiased determination of the optimal amino acids at every position of the decapeptide for recognition by the LS-specific TCRs. This disparity in responses between the peptides can help to elucidate the lack of HLA-DR3–A. nidulans NDPD tetramer staining in the majority of LS patients despite their significantly higher IFN-γ and IL-2 ELISPOT responses compared with control subjects. ELISPOT assays have the ability to detect precursor frequencies in the range of ∼1 in 250,000 cells and are far more sensitive than flow-based assays, such as MHCII tetramer staining, which usually requires a precursor frequency in the target population of ∼0.04% for detection. We identified ∼800 IFN-γ SFUs per million cells (0.08%) in the BAL from HLA-DR3+ LS patients after stimulation with 5 μg/ml of the mimotope 86 peptide, which hovers around the threshold of detection for tetramer staining. Thus, we are able to detect significant HLA-DR3–mimotope 86–binding CD4+ T cells in the BAL of HLA-DR3+ LS patients. Conversely, we identified ∼200 IFN-γ SFUs per million cells (0.02%) in the BAL from HLA-DR3+ LS patients after stimulation with 5 μg/ml of the NDPD 11-mer, which is below the level of detection for tetramer staining, explaining our ability to only detect HLA-DR3–NDPD 11-mer–binding CD4+ T cells in the BAL in one HLA-DR3+ LS patient.
While sarcoidosis has a broad array of clinical manifestations, LS is diagnosed based on the presence of bilateral hilar lymphadenopathy, fever, erythema nodosum, and ankle arthritis as well as an expansion of TRAV12-1/TRBV2–expressing CD4+ T cells in the BAL of HLA-DR3+ subjects. Non-LS sarcoidosis patients expressing HLA-DR3 do not share this exact set of symptoms, but they do have increased frequencies of TRAV12-1/TRBV2–expressing CD4+ T cells in the lung compared with HLA-DR3− sarcoidosis population. The expansions of these oligoclonal CD4+ T cells suggest that both LS and non-LS sarcoidosis in HLA-DR3–expressing patients could be driven by the same antigenic stimuli. Our data indicate that this is a definite possibility, with HLA-DR3+ sarcoidosis patients (LS and non-LS) showing significantly higher IL-2 and IFN-γ production in the BAL after stimulation with mimotope 86 and A. nidulans NDPD peptides compared with control subjects. However, a threefold increase in NDPD- and mimotope 86-responsive cells was seen in HLA-DR3+ LS subjects. Although the ratio of CD4/CD8 T cells and the frequency of TRAV12-1/TRBV2–expressing CD4+ T cells in the BAL were higher in LS versus non-LS HLA-DR3+ patients, these characteristics significantly correlate with mimotope 86 and A. nidulans NDPD peptide responses in both HLA-DR3+ patient groups. Taken together, these data suggest that LS and non-LS sarcoidosis patients expressing HLA-DR3 may represent a spectrum of the same disease rather than two distinct clinical entities.
The unknown etiology of sarcoidosis has hindered progress in our understanding of disease pathogenesis and development of effective diagnostic tools and treatments. This study provides evidence that A. nidulans may play a role in the pathogenesis of LS and possibly a greater subset of HLA-DR3+ sarcoidosis patients. Sarcoidosis is mainly diagnosed by exclusion of similar conditions and lacks a specific test; thus, our findings may lead to the development of a specific diagnostic tool and treatments for both HLA-DR3+ LS and non-LS sarcoidosis patients.
Materials and methods
Study population
Demographics of the 13 sarcoidosis patients (9 LS and 4 non-LS) and 3 control subjects included in the discovery cohort were previously described (Mitchell et al., 2017). Table 1 shows the demographic data of the validation cohort, which consisted of 10 DR3+ LS, 11 DR3+ non-LS sarcoidosis, 10 DR3+ BeS, 9 DR3− LS, and 12 DR3− non-LS sarcoidosis subjects. All patients were HLA typed, and sarcoidosis was diagnosed according to criteria established by the World Association of Sarcoidosis and Other Granulomatous Disorders (Crouser et al., 2020). The diagnosis of BeS was established based on a history of beryllium exposure, an abnormal blood beryllium lymphocyte proliferation test on two occasions, and the absence of granulomatous inflammation or other abnormalities on lung biopsy (Mroz et al., 1991; Newman, 1996). Informed consent was obtained from all subjects, and ethical approval was granted from the Stockholm County Regional Ethical Committee and the human subject institutional review boards at the University of Colorado Anschutz Medical Campus and National Jewish Health.
TCR clustering
iRepertoire sequencing analysis and single-cell PCR of TCR genes for DR3+ LS patient 1244 using the Illumina MiSeq platform have been previously described (Mitchell et al., 2017). TCR sequences presented in this article are listed according to International ImMunoGeneTics Information System nomenclature (http://www.imgt.org) and are available in Gene Expression Omnibus under accession no. GSE100378. CDR3β amino acid sequences were used for a clone network analysis using the GLIPH program (Glanville et al., 2017). GLIPH uses an algorithm to cluster CDR3β amino acid sequences into convergence groups, defined as a set of TCRs from one or more individuals predicted to bind to the same antigen in a similar manner through similar TCR contacts. After inputting all available CDR3β amino acid sequences from LS DR3+ patients and control subjects, the network analysis outputted CDR3βs as nodes and global, local, or singleton interactions as edges. Network clusters were generated using Cytoscape 3.7.2.
HLA-DR3–transfected fibroblast lines
Gene constructs encoding HLA-DRA*01:01 and HLA-DRB1*03:01 were cotransfected into a mouse fibroblast line, designated B6DK10, derived from C57BL/6 mice that were deficient for MHCII and invariant chain genes (Huseby et al., 2003). Cells were stained with an anti–HLA-DR antibody (clone L243) and sorted for high HLA-DR expression using a BD FACSAria. Fibroblasts expressing HLA-DR3 were adapted to grow in 0.5% serum in OptiPRO SFM (Gibco BRL Life Technologies) supplemented with 4 mM GlutaMAX (Gibco BRL Life Technologies) and 50 μg/ml hygromycin B (Millipore Sigma) to maintain HLA-DR expression.
Generation of mouse hybridomas expressing TCRs from human T cell clones
TCRs derived from BAL CD4+ T cells of LS DR3+ patient 1244 were introduced into two different TCR-negative mouse recipient hybridoma cell lines. The primary hybridoma used was a 5KC cell line (White et al., 1993) that expresses a mutated human CD4 molecule having substitutions at positions p40 and p45 (Gln40Tyr and Thr45Trp; Wang et al., 2011) to enhance HLA interactions. It has been previously shown (Michels et al., 2017) and confirmed in our laboratory (data not shown) that these mutations enhance TCR responses compared with hybridomas with native human CD4. The second hybridoma line, 54ζ, expresses wild-type human CD4 (Falta et al., 2021, 2013). To generate TCR vectors, the extracellular variable domains of the expressed TRA and TRB genes of each T cell clone were synthesized as gBlock gene fragments (Integrated DNA Technologies). These TCR gene fragments contained homologous overlapping nucleotide sequences that allowed cloning into a murine stem cell virus retroviral plasmid separated by a linker encoding the mouse Cα domain connected to the porcine teschovirus-1 2A (p2A) peptide using a Gibson Assembly Cloning Kit (New England Biolabs). Phoenix 293T cells were transfected with the murine stem cell virus plasmids along with the pCL-Eco packaging vector to produce replication-incompetent retroviral particles. TCR-negative 5415 and 54ζ hybridoma cells were transduced with filtered retroviral supernatant using a spinfection protocol as previously described (Bowerman et al., 2011; Falta et al., 2021).
PSLs, mixtures, and peptides
A decapeptide, N-terminal–acetylated, C-terminal–amidated, L–amino acid PSL was synthesized as previously described (Falta et al., 2013). Briefly, this library consists of 200 mixtures prepared in an OX9 format, where O represents a specific amino acid at a defined position and X represents an equal molar mixture of 19 natural amino acids (excluding cysteine) in each of the remaining nine positions. Each OX9 mixture consists of 3.2 × 1011 different decapeptides, and the total number of peptides in the library is 6.4 × 1012. Mixtures containing multiple fixed positions, including dual-defined mixtures and a biased G2D4N5 PSL, were synthesized as previously described (Hemmer et al., 1999; Pinilla et al., 1994).
Individual peptides were synthesized using GenScript peptide library services. All peptides were dissolved in DMSO at high concentration before making a working stock in PBS. Recombinant A. nidulans NDPD (UniProt under accession no. AN1126; amino acids 117–410) was produced using BacPower Guaranteed Bacterial Protein Expression (90% purity; GenScript). Overlapping 15-mers derived from mKatG (n = 74) and ESAT-6 (n = 17) and four HLA-DR3–binding vimentin peptides were generously provided by Drs. David Moller (Johns Hopkins University, Baltimore, MD), Wonder Drake (Vanderbilt University, Nashville, TN), and J. Grunewald, respectively.
T cell hybridomas and activation assays
For activation assays, T cell hybridomas (1 × 105 cells per well) were cultured in serum-free medium (OptiPRO SFM; Gibco BRL Life Technologies) supplemented with 4 mM GlutaMAX (Gibco BRL Life Technologies) in 96-well flat-bottomed tissue culture plates (Falcon) in the presence of B6DK10 fibroblasts expressing HLA-DR3 (5 × 104 cells per well). Initial experiments involving the unbiased decapeptide PSL and dual-defined mixtures were performed using mixtures at 200 μg/ml in duplicate wells with the 5415 hybridomas. G2D4N5 PSL mixtures were tested at 200 μg/ml with the 54ζ hybridomas. G2D4N5 mixtures were also tested in 5415 (data not shown), and positive amino acids were nearly identical to those shown for 54ζ. Mimotopes were initially tested at 2 ng/ml with 5415 hybridomas. For dose–response curves to mimotopes and A scan peptides, peptides were added at concentrations ranging from 0.001 to 30 ng/ml with 5415 hybridomas. The full set of biometrical analysis peptides was initially tested at 10 μg/ml with 54ζ hybridomas, and positive peptides were screened with 5415 hybridomas at indicated concentrations. Recombinant NAD-dependent protein deacetylase was tested at indicated concentrations. For all experiments, supernatants were harvested after 20–24 h, and mouse IL-2 was measured by ELISA (eBioscience). In some experiments, the concentration of peptide that provided EC50 for each hybridoma was determined using nonlinear regression (sigmoidal-fit, GraphPad Prism; GraphPad Software).
Crystal structure determination of HLA-DR3–mimotope 46
HLA-DR3 α and β chains with covalently attached mimotope 46 were cloned into a two-promoter Escherichia coli baculovirus transfer vector as previously described (Crawford et al., 1998; Dai et al., 2010). The construct was incorporated into Sapphire Baculovirus DNA (Orbigen) by standard homologous recombination techniques to produce a stock of baculovirus encoding HLA-DR3–mimotope 46. To produce HLA-DR3–mimotope 46 protein, High Five insect cells at 5–10 × 105/ml were infected with 10 ml of the baculovirus stock in 1 liter TMN-FH medium with 5% FBS and cultured in spinner flasks at 27°C. After 18–24 h of culture, spinner flasks were transferred to 19°C and incubated for 6 d. HLA-DR3–mimotope 46 protein was purified from the supernatant by immunoaffinity chromatography using the anti-zipper mAb 2H11. Eluted protein was further purified by Superdex 200 Increase 10/300 GL column (GE Healthcare) in PBS buffer. HLA-DR3–mimotope 46 protein was further processed with thrombin protein and purified by Mono Q ion exchange chromatography. HLA-DR3–mimotope 46 was crystallized at 4°C by sitting drop vapor diffusion against 1 ml of mother liquor containing 18% wt/vol PEG20k and 0.1 M citrate (pH 5.0). X-ray diffraction data were collected under liquid nitrogen cryogenic conditions at 100°K. HLA-DR3–mimotope 46 crystals were flash cooled in liquid nitrogen after a short soak in a cryoprotection solution consisting of the reservoir solution with an addition of 20% (wt/vol) glycerol. X-ray data were measured at synchrotron beamline ID-24C at the Advanced Photon Source of Argonne National Laboratory using the PILATUS detector. The data were indexed, integrated, scaled, and merged using HKL2000 (Otwinowski and Minor, 1997). The structure was solved by molecular replacement method using Phaser (McCoy et al., 2007) software and further refined by refmac5 (Murshudov et al., 1997). Building of the structure was performed with Coot (Emsley et al., 2010). The Research Collaboratory for Structural Bioinformatics Protein Database accession no. for the coordinate and structure factor files for the structure is 7N19.
Biometrical analysis (scoring matrix and database searches)
Scoring matrices were created based on the stimulatory potential of each fixed amino acid at each decapeptide position of the unbiased PSL as previously described (Zhao et al., 2001). To account for the different peptide registers in the unbiased matrix, the stimulatory responses of the dual-defined mixtures in either register 1 or register 2 were integrated into the scoring matrices using the harmonic mean model (Santos et al., 2011) to create “harmonic boost” register 1 and register 2 (Planas et al., 2018). For each hybridoma in each register as well as a combined matrix for all the hybridomas, two harmonic mean matrices were generated using the boosted value of IL-2 (pg/ml) and the logarithm of that value. A total of 16 matrices were created using different combinations of different hybridoma screening data. An independent biometrical analysis was performed for each matrix and applied to all possible overlapping decapeptides in the protein sequences within the SwissProt database containing curated proteins from all organisms to rank and score naturally occurring peptides. The predicted stimulatory potential of a peptide, or score, was calculated by summing the matrix values associated with each amino acid in each position of the peptide. The sum of the maximum values at each position was defined as the maximum matrix score. The scoring matrix was applied to rank, according to their stimulatory score, all of the overlapping peptides within each protein sequence of a SwissProt protein database (downloaded July 2, 2018) as previously described (Hemmer et al., 1999; Zhao et al., 2001). Subsequent analysis showed that there was significant redundancy among the various matrices and that a more streamlined approach focused on the activities of the dual-defined mixtures was sufficient to identify antigens of interest. The resulting lists of potential antigens were consolidated by choosing the top 50 overall peptides in both registers and the remaining top 10 in each individual matrix that were not included in the previous top 50.
ELISAs and ELISPOTs
IFN-γ and IL-2 production from BAL cells was measured using the human IFN-γ ELISPOT kit (3420-4AST-2; Mabtech) or human IL-2 ELISPOT kit (551282; BD Biosciences). 2.5 × 105 BAL cells from LS, non-LS sarcoidosis, or control patients were incubated in duplicate with either 95% purified mimotope 86 peptide or A. nidulans NDPD peptide at 5 μg/ml each, anti-CD3 (1 μg/ml) or PHA (2.5 μg/ml) as a positive control, or medium alone in 200 μl in 96-well ELISPOT plates precoated with IFN-γ mAb or coated with IL-2 mAb overnight at 37°C. Cells were incubated in RPMI 1640 medium supplemented with 10% FBS (Seradigm), 100 U/ml penicillin, 100 μg/ml streptomycin, 290 g/ml L-glutamine, and 10% Hepes. After 42–48 h, cells were removed, IFN-γ and IL-2 were detected, and spots were visualized as previously described (Martin et al., 2011). ELISPOT plates were analyzed with a CTL Immunospot Analyzer (Cellular Technology Ltd), and results were reported as SFUs per well minus SFUs in unstimulated wells.
To measure IgG specific for A. nidulans NDPD in serum, 96-well ELISA plates were coated with 10 μg/ml recombinant NDPD (GenScript) and incubated overnight at 4°C. Plates were washed, and threefold serial dilutions of serum (1:1–1:2,187) were added and incubated for 1 h at 25°C. Plates were washed, and anti-human IgG conjugated to HRP was added for 1 h at room temperature. Plates were developed, and the OD at 405 nm was measured. The serum dilution that elicited 50% of the maximum OD 405-nm value for each subject was determined using nonlinear regression (sigmoidal-fit, GraphPad Prism; GraphPad Software).
Tetramer production and staining
The HLA-DR3–mimotope 86, HLA-DR3–A. nidulans NDPD, and HLA-DR3–CLIP tetramers were produced at the National Institutes of Health Tetramer Core Facility (Emory University, Atlanta, GA). Tetramers were purified using an AKTA fast protein liquid chromatography size-exclusion column. For staining, hybridoma cells were incubated in 25 μl of culture medium containing the HLA-DR3–mimotope 86–APC, HLA-DR3–A. nidulans NDPD–PE, and HLA-DR3–CLIP tetramer (1–10 μg/ml) for 1 h at 37°C in a humidified 10% CO2 incubator, with gentle mixing every 20 min. To assess HLA-DR3–tetramer binding in BAL samples, 1–3 × 106 BAL cells from LS or control patients were incubated in 25 μl of C-IMDM with either 1 μg/ml PE-labeled HLA-DR3–A. nidulans NDPD or HLA-DR3–CLIP tetramer or 10 μg/ml APC-labeled HLA-DR3–mimotope 86 or DR3–CLIP for 1 h at 37°C in a humidified 10% CO2 incubator, with gentle mixing every 20 min. After 1 h, cells were stained for 30 min at 4°C with the following mAbs: anti–CD3-APC-Cy7 (OKT3), anti–CD4-PECy7 (OKT4), anti–CD8-eF450 (SK1), anti–CD14-eF450 (61D3), and anti–CD19-eF450 (all from Invitrogen) and Ghost Dye Violet 510 (Tonbo Biosciences). The viable mononuclear cell population was evaluated for fluorescence intensity on a FACSCanto II flow cytometer (BD Biosciences), and data were analyzed with FlowJo software (Tree Star). Cells expressing CD8, CD14, and CD19 were excluded via a dump gate, and T cells positively staining for CD3 and CD4 were analyzed for tetramer binding.
Proliferation assays
Cell Trace Violet Proliferation Dye (Thermo Fisher Scientific) was added to peripheral blood mononuclear cells according to the manufacturer’s instructions. After labeling, cells were added to a 48-well tissue culture plate with one of the following stimuli: media alone, plate-bound anti-CD3 (10 μg/ml), mimotope 86 (5 μg/ml), and NDPD peptide (5 μg/ml) in complete RPMI media. Cells were incubated for 5 d at 37°C and stained for 30 min at 4°C with anti–CD3-APC-Cy7 (OKT3; Invitrogen), anti–CD4-PE-Cy7 (OKT4; Invitrogen), and Ghost Dye Violet 510 (Tonbo Biosciences). The viable mononuclear cell population was evaluated for fluorescence intensity on a FACSCanto II flow cytometer (BD Biosciences), and data were analyzed with FlowJo software (Tree Star). Ghost dye–negative CD3+CD4+ T cells were analyzed for the loss of Cell Trace Violet to determine proliferation.
Immunizations of HLA-DR3 Tg murine
HLA-DR3 Tg mice were kindly provided by Dr. Govindarajan Rajagopalan (Yale University, New Haven, CT; Mangalam et al., 2007). Animals were housed and bred at the University of Colorado Biological Resource Center. 6–8-week-old male and female HLA-DR3 Tg mice were immunized intraperitoneally with either 10 μg mimotope 86 peptide or NDPD peptide and 10 μg LPS (Serotype O55:B5 S-form; ENZO Life Sciences) in 200 μl PBS. Mice immunized with LPS alone were used as negative controls. At day 10, splenocytes were collected. Splenocytes from mice immunized with mimotope 86 peptide or NDPD peptide were incubated for 24 h with either mimotope 86 peptide or NDPD peptide at 5 μg/ml. In some experiments, CD4+ T cells (5 × 104 cells/well) were purified using CD4+ T cell selection microbeads (STEMCELL Technologies) and cocultured with 5 × 105 irradiated naive splenocytes and 5 μg/ml NDPD peptide in the presence or absence of an anti–HLA-DR mAb (10 μg/ml, clone L243, BioLegend). A purity check demonstrated that >90% of the purified cells expressed CD4. Splenocytes depleted of CD4+ T cells (5 × 105 cells/well) were stimulated in the presence of NDPD peptide (5 μg/ml). IFN-γ and IL-2 ELISPOTs were performed as previously described (Mack et al., 2014). All experiments were approved by the institutional animal care and use committee of the University of Colorado Anschutz Medical Campus in accordance with National Institutes of Health guidelines.
Statistical analysis
Statistical tests were performed using GraphPad Prism 6.0 software. Data are presented as mean ± SEM as stated in the figure legends. Statistical significance for parametric or nonparametric tests were determined as indicated. P < 0.05 was considered statistically significant.
Online supplemental material
Fig. S1 shows IL-2 responses (pg/ml) of T cell hybridomas 1244c1 and 1244c3 to the biased G2D4N5 PSL. Fig. S2 shows the IL-2 responses (pg/ml) of T cell hybridomas 1244c1, 1244c2, and 1244c3 to the mimotopes. Fig. S3 shows the dose–responses curves for T cell hybridomas 1244c1, 1244c2, and 1244c3 stimulated with mimotopes 46 and 86, mimotope 86 peptides with separate A substitutions at each amino acid position, and mimotope 86 and NDPD 11-mer peptides of varying length. Fig. S4 shows representative HLA-DR3–peptide tetramer staining of LS-specific 1244 TCR-expressing hybridomas and ex vivo BAL cells. Table S1 provides data collection and refinement statistics for the HLA-DR3–mimotope 46 structure. Table S2 lists the naturally occurring peptides that were screened for their ability to stimulate T cell hybridomas 1244c1, 1244c2, and 1244c3.
Supplementary Material
provides data collection and refinement statistics for the HLA-DR3–mimotope 46 structure.
lists the naturally occurring peptides that were screened for their ability to stimulate T cell hybridomas 1244c1, 1244c2, and 1244c3.
Acknowledgments
This work was supported by grants from the National Institutes of Health (HL136137, HL102245, HL62410, ES02534) to A.P. Fontenot, Swedish Heart-Lung Foundation (20190478) to J. Grunewald, Swedish Research Council (2019-01034) to J. Grunewald, King Gustaf V and Queen Victoria’s Foundation of Freemasons to J. Grunewald, and Regional Agreement on Medical Training and Clinical Research between Stockholm County Council and the Karolinska Institutet (20180120) to J. Grunewald.
Author contributions: S.A. Greaves, J. Grunewald, and A.P. Fontenot conceptualized the study and wrote the manuscript. L.A. Maier and J. Grunewald provided patient samples and data analysis. S.A. Greaves and A. Ravindran performed the experiments with contributions from R.G. Santos, L. Chen, M.T. Falta, A.N. Tinega, and A.M. Mitchell. R.G. Santos and C. Pinilla provided peptide libraries and data analysis. L. Chen, Y. Wang, and S. Dai generated the HLA-DR3 structure. S.M. Atif, D.G. Mack, and A.P. Fontenot performed the murine experiments. All authors reviewed and edited the manuscript.
References
- Arribas-Layton, D., Guyer P., Delong T., Dang M., Chow I.T., Speake C., Greenbaum C.J., Kwok W.W., Baker R.L., Haskins K., and James E.A.. 2020. Hybrid insulin peptides are recognized by human T cells in the context of DRB1*04:01. Diabetes. 69:1492–1502. 10.2337/db19-0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher, P., Hohnstein T., Beerbaum E., Röcker M., Blango M.G., Kaufmann S., Röhmel J., Eschenhagen P., Grehn C., Seidel K., et al. 2019. Human anti-fungal Th17 immunity and pathology rely on cross-reactivity against Candida albicans. Cell. 176:1340–1355.e15. 10.1016/j.cell.2019.01.041 [DOI] [PubMed] [Google Scholar]
- Baker, R.L., Rihanek M., Hohenstein A.C., Nakayama M., Michels A., Gottlieb P.A., Haskins K., and Delong T.. 2019. Hybrid insulin eptides are autoantigens in type 1 diabetes. Diabetes. 68:1830–1840. 10.2337/db19-0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardinas, F., Morera J., Fité E., and Plasencia A.. 1989. Seasonal clustering of sarcoidosis. Lancet. 2:455–456. [DOI] [PubMed] [Google Scholar]
- Baughman, R.P., Teirstein A.S., Judson M.A., Rossman M.D., Yeager H. Jr., Bresnitz E.A., DePalo L., Hunninghake G., Iannuzzi M.C., Johns C.J., et al. Case Control Etiologic Study of Sarcoidosis (ACCESS) research group . 2001. Clinical characteristics of patients in a case control study of sarcoidosis. Am. J. Respir. Crit. Care Med. 164:1885–1889. 10.1164/ajrccm.164.10.2104046 [DOI] [PubMed] [Google Scholar]
- Bowerman, N.A., Falta M.T., Mack D.G., Kappler J.W., and Fontenot A.P.. 2011. Mutagenesis of beryllium-specific TCRs suggests an unusual binding topology for antigen recognition. J. Immunol. 187:3694–3703. 10.4049/jimmunol.1101872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, S.W., Gallo C.M., and Smith J.S.. 2004. Diversity in the Sir2 family of protein deacetylases. J. Leukoc. Biol. 75:939–950. 10.1189/jlb.0903424 [DOI] [PubMed] [Google Scholar]
- Chen, E.S., Wahlström J., Song Z., Willett M.H., Wikén M., Yung R.C., West E.E., McDyer J.F., Zhang Y., Eklund A., et al. 2008. T cell responses to mycobacterial catalase-peroxidase profile a pathogenic antigen in systemic sarcoidosis. J. Immunol. 181:8784–8796. 10.4049/jimmunol.181.12.8784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, E.L., Lauder A.P., Hofstaedter C.E., Hwang Y., Fitzgerald A.S., Imai I., Biernat W., Rękawiecki B., Majewska H., Dubaniewicz A., et al. 2018. Microbial lineages in sarcoidosis. A metagenomic analysis tailored for low-microbial content samples. Am. J. Respir. Crit. Care Med. 197:225–234. 10.1164/rccm.201705-0891OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, F., Kozono H., White J., Marrack P., and Kappler J.. 1998. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity. 8:675–682. 10.1016/S1074-7613(00)80572-5 [DOI] [PubMed] [Google Scholar]
- Crouser, E.D., Maier L.A., Wilson K.C., Bonham C.A., Morgenthau A.S., Patterson K.C., Abston E., Bernstein R.C., Blankstein R., Chen E.S., et al. 2020. Diagnosis and detection of sarcoidosis. An official American Thoracic Society clinical practice guideline. Am. J. Respir. Crit. Care Med. 201:e26–e51. 10.1164/rccm.202002-0251ST [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, S., Murphy G.A., Crawford F., Mack D.G., Falta M.T., Marrack P., Kappler J.W., and Fontenot A.P.. 2010. Crystal structure of HLA-DP2 and implications for chronic beryllium disease. Proc. Natl. Acad. Sci. USA. 107:7425–7430. 10.1073/pnas.1001772107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delong, T., Wiles T.A., Baker R.L., Bradley B., Barbour G., Reisdorph R., Armstrong M., Powell R.L., Reisdorph N., Kumar N., et al. 2016. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. 351:711–714. 10.1126/science.aad2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirkok, S.S., Basaranoglu M., and Akbilgic O.. 2006. Seasonal variation of the onset of presentations in stage 1 sarcoidosis. Int. J. Clin. Pract. 60:1443–1450. 10.1111/j.1742-1241.2006.00773.x [DOI] [PubMed] [Google Scholar]
- Drake, W.P., Dhason M.S., Nadaf M., Shepherd B.E., Vadivelu S., Hajizadeh R., Newman L.S., and Kalams S.A.. 2007. Cellular recognition of Mycobacterium tuberculosis ESAT-6 and KatG peptides in systemic sarcoidosis. Infect. Immun. 75:527–530. 10.1128/IAI.00732-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eishi, Y., Suga M., Ishige I., Kobayashi D., Yamada T., Takemura T., Takizawa T., Koike M., Kudoh S., Costabel U., et al. 2002. Quantitative analysis of mycobacterial and propionibacterial DNA in lymph nodes of Japanese and European patients with sarcoidosis. J. Clin. Microbiol. 40:198–204. 10.1128/JCM.40.1.198-204.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley, P., Lohkamp B., Scott W.G., and Cowtan K.. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falta, M.T., Pinilla C., Mack D.G., Tinega A.N., Crawford F., Giulianotti M., Santos R., Clayton G.M., Wang Y., Zhang X., et al. 2013. Identification of beryllium-dependent peptides recognized by CD4+ T cells in chronic beryllium disease. J. Exp. Med. 210:1403–1418. 10.1084/jem.20122426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falta, M.T., Crawford J.C., Tinega A.N., Landry L.G., Crawford F., Mack D.G., Martin A.K., Atif S.M., Li L., Santos R.G., et al. 2021. Beryllium-specific CD4+ T cells induced by chemokine neoantigens perpetuate inflammation. J. Clin. Invest. 131:e144864. 10.1172/JCI144864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, P., Amaya M., Mellins E., and Wiley D.C.. 1995. The structure of an intermediate in class II MHC maturation: CLIP bound to HLA-DR3. Nature. 378:457–462. 10.1038/378457a0 [DOI] [PubMed] [Google Scholar]
- Glanville, J., Huang H., Nau A., Hatton O., Wagar L.E., Rubelt F., Ji X., Han A., Krams S.M., Pettus C., et al. 2017. Identifying specificity groups in the T cell receptor repertoire. Nature. 547:94–98. 10.1038/nature22976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glennås, A., Kvien T.K., Melby K., Refvem O.K., Andrup O., Karstensen B., and Thoen J.E.. 1995. Acute sarcoid arthritis: occurrence, seasonal onset, clinical features and outcome. Br. J. Rheumatol. 34:45–50. 10.1093/rheumatology/34.1.45 [DOI] [PubMed] [Google Scholar]
- Grunewald, J. 2012. HLA associations and Löfgren’s syndrome. Expert Rev. Clin. Immunol. 8:55–62. 10.1586/eci.11.76 [DOI] [PubMed] [Google Scholar]
- Grunewald, J., and Eklund A.. 2007. Sex-specific manifestations of Löfgren’s syndrome. Am. J. Respir. Crit. Care Med. 175:40–44. 10.1164/rccm.200608-1197OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald, J., and Eklund A.. 2009. Löfgren’s syndrome: human leukocyte antigen strongly influences the disease course. Am. J. Respir. Crit. Care Med. 179:307–312. 10.1164/rccm.200807-1082OC [DOI] [PubMed] [Google Scholar]
- Grunewald, J., Janson C.H., Eklund A., Ohrn M., Olerup O., Persson U., and Wigzell H.. 1992. Restricted V alpha 2.3 gene usage by CD4+ T lymphocytes in bronchoalveolar lavage fluid from sarcoidosis patients correlates with HLA-DR3. Eur. J. Immunol. 22:129–135. 10.1002/eji.1830220120 [DOI] [PubMed] [Google Scholar]
- Grunewald, J., Olerup O., Persson U., Ohrn M.B., Wigzell H., and Eklund A.. 1994. T-cell receptor variable region gene usage by CD4+ and CD8+ T cells in bronchoalveolar lavage fluid and peripheral blood of sarcoidosis patients. Proc. Natl. Acad. Sci. USA. 91:4965–4969. 10.1073/pnas.91.11.4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald, J., Berlin M., Olerup O., and Eklund A.. 2000. Lung T-helper cells expressing T-cell receptor AV2S3 associate with clinical features of pulmonary sarcoidosis. Am. J. Respir. Crit. Care Med. 161:814–818. 10.1164/ajrccm.161.3.9906001 [DOI] [PubMed] [Google Scholar]
- Gupta, D., Agarwal R., Aggarwal A.N., and Jindal S.K.. 2007. Molecular evidence for the role of mycobacteria in sarcoidosis: a meta-analysis. Eur. Respir. J. 30:508–516. 10.1183/09031936.00002607 [DOI] [PubMed] [Google Scholar]
- Hemmer, B., Gran B., Zhao Y., Marques A., Pascal J., Tzou A., Kondo T., Cortese I., Bielekova B., Straus S.E., et al. 1999. Identification of candidate T-cell epitopes and molecular mimics in chronic Lyme disease. Nat. Med. 5:1375–1382. 10.1038/70946 [DOI] [PubMed] [Google Scholar]
- Huseby, E.S., Crawford F., White J., Kappler J., and Marrack P.. 2003. Negative selection imparts peptide specificity to the mature T cell repertoire. Proc. Natl. Acad. Sci. USA. 100:11565–11570. 10.1073/pnas.1934636100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinloch, A.J., Kaiser Y., Wolfgeher D., Ai J., Eklund A., Clark M.R., and Grunewald J.. 2018. In situ humoral immunity to vimentin in HLA-DRB1*03+ patients with pulmonary sarcoidosis. Front. Immunol. 9:1516. 10.3389/fimmu.2018.01516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack, D.G., Falta M.T., McKee A.S., Martin A.K., Simonian P.L., Crawford F., Gordon T., Mercer R.R., Hoover M.D., Marrack P., et al. 2014. Regulatory T cells modulate granulomatous inflammation in an HLA-DP2 transgenic murine model of beryllium-induced disease. Proc. Natl. Acad. Sci. USA. 111:8553–8558. 10.1073/pnas.1408048111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangalam, A., Rodriguez M., and David C.. 2007. A new humanized HLA transgenic mouse model of multiple sclerosis expressing class II on mouse CD4 T cells. Ann. N. Y. Acad. Sci. 1103:112–117. 10.1196/annals.1394.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, A.K., Mack D.G., Falta M.T., Mroz M.M., Newman L.S., Maier L.A., and Fontenot A.P.. 2011. Beryllium-specific CD4+ T cells in blood as a biomarker of disease progression. J. Allergy Clin. Immunol. 128:1100–6.e5. 10.1016/j.jaci.2011.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy, A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., and Read R.J.. 2007. Phaser crystallographic software. J. Appl. Cryst. 40:658–674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels, A.W., Landry L.G., McDaniel K.A., Yu L., Campbell-Thompson M., Kwok W.W., Jones K.L., Gottlieb P.A., Kappler J.W., Tang Q., et al. 2017. Islet-derived CD4 T cells targeting proinsulin in human autoimmune diabetes. Diabetes. 66:722–734. 10.2337/db16-1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, A.M., Kaiser Y., Falta M.T., Munson D.J., Landry L.G., Eklund A., Nakayama M., Slansky J.E., Grunewald J., and Fontenot A.P.. 2017. Shared αβ TCR usage in lungs of sarcoidosis patients with Löfgren’s syndrome. J. Immunol. 199:2279–2290. 10.4049/jimmunol.1700570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroz, M.M., Kreiss K., Lezotte D.C., Campbell P.A., and Newman L.S.. 1991. Reexamination of the blood lymphocyte transformation test in the diagnosis of chronic beryllium disease. J. Allergy Clin. Immunol. 88:54–60. 10.1016/0091-6749(91)90300-D [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin A.A., and Dodson E.J.. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255. 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- Newman, L.S. 1996. Significance of the blood beryllium lymphocyte proliferation test. Environ. Health Perspect. 104(Suppl 5):953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, L.S., Rose C.S., Bresnitz E.A., Rossman M.D., Barnard J., Frederick M., Terrin M.L., Weinberger S.E., Moller D.R., McLennan G., et al. ACCESS Research Group . 2004. A case control etiologic study of sarcoidosis: environmental and occupational risk factors. Am. J. Respir. Crit. Care Med. 170:1324–1330. 10.1164/rccm.200402-249OC [DOI] [PubMed] [Google Scholar]
- Oswald-Richter, K.A., Culver D.A., Hawkins C., Hajizadeh R., Abraham S., Shepherd B.E., Jenkins C.A., Judson M.A., and Drake W.P.. 2009. Cellular responses to mycobacterial antigens are present in bronchoalveolar lavage fluid used in the diagnosis of sarcoidosis. Infect. Immun. 77:3740–3748. 10.1128/IAI.00142-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z., and Minor W.. 1997. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326. 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- Pinilla, C., Appel J.R., and Houghten R.A.. 1994. Investigation of antigen-antibody interactions using a soluble, non-support-bound synthetic decapeptide library composed of four trillion (4 x 10(12) sequences. Biochem. J. 301:847–853. 10.1042/bj3010847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas, R., Santos R., Tomas-Ojer P., Cruciani C., Lutterotti A., Faigle W., Schaeren-Wiemers N., Espejo C., Eixarch H., Pinilla C., et al. 2018. GDP-l-fucose synthase is a CD4+ T cell-specific autoantigen in DRB3*02:02 patients with multiple sclerosis. Sci. Transl. Med. 10:eaat4301. 10.1126/scitranslmed.aat4301 [DOI] [PubMed] [Google Scholar]
- Rammensee, H.G., Friede T., and Stevanoviíc S.. 1995. MHC ligands and peptide motifs: first listing. Immunogenetics. 41:178–228. 10.1007/BF00172063 [DOI] [PubMed] [Google Scholar]
- Santos, R.G., Giulianotti M.A., Dooley C.T., Pinilla C., Appel J.R., and Houghten R.A.. 2011. Use and implications of the harmonic mean model on mixtures for basic research and drug discovery. ACS Comb. Sci. 13:337–344. 10.1021/co100065a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney, J., Oseroff C., Southwood S., Wall M., Ishioka G., Koning F., and Sette A.. 1992. DRB1*0301 molecules recognize a structural motif distinct from the one recognized by most DR beta 1 alleles. J. Immunol. 149:2634–2640. [PubMed] [Google Scholar]
- Song, Z., Marzilli L., Greenlee B.M., Chen E.S., Silver R.F., Askin F.B., Teirstein A.S., Zhang Y., Cotter R.J., and Moller D.R.. 2005. Mycobacterial catalase-peroxidase is a tissue antigen and target of the adaptive immune response in systemic sarcoidosis. J. Exp. Med. 201:755–767. 10.1084/jem.20040429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struyk, L., Hawes G.E., Haanen J.B., de Vries R.R., and van den Elsen P.J.. 1995. Clonal dominance and selection for similar complementarity determining region 3 motifs among T lymphocytes responding to the HLA-DR3-associated Mycobacterium leprae heat shock protein 65-kd peptide 3-13. Hum. Immunol. 44:220–227. 10.1016/0198-8859(95)00110-7 [DOI] [PubMed] [Google Scholar]
- Terčelj, M., Rott T., and Rylander R.. 2007. Antifungal treatment in sarcoidosis--a pilot intervention trial. Respir. Med. 101:774–778. 10.1016/j.rmed.2006.08.005 [DOI] [PubMed] [Google Scholar]
- Terčelj, M., Salobir B., Zupancic M., and Rylander R.. 2011a. Antifungal medication is efficient in the treatment of sarcoidosis. Ther. Adv. Respir. Dis. 5:157–162. 10.1177/1753465811401648 [DOI] [PubMed] [Google Scholar]
- Terčelj, M., Stopinšek S., Ihan A., Salobir B., Simčič S., Wraber B., and Rylander R.. 2011b. In vitro and in vivo reactivity to fungal cell wall agents in sarcoidosis. Clin. Exp. Immunol. 166:87–93. 10.1111/j.1365-2249.2011.04456.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terčelj, M., Stopinšek S., Ihan A., Salobir B., Simčič S., and Rylander R.. 2014. Fungal exposure and low levels of IL-10 in patients with sarcoidosis. Pulm. Med. 2014:164565. 10.1155/2014/164565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terčelj, M., Salobir B., and Rylander R.. 2017. β-glucan in the lymph nodes in sarcoidosis and in Kveim-Siltzbach test reagent. Sarcoidosis Vasc. Diffuse Lung Dis. 34:130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schooten, W.C., Elferink D.G., Van Embden J., Anderson D.C., and De Vries R.R.. 1989. DR3-restricted T cells from different HLA-DR3-positive individuals recognize the same peptide (amino acids 2-12) of the mycobacterial 65-kDa heat-shock protein. Eur. J. Immunol. 19:2075–2079. 10.1002/eji.1830191116 [DOI] [PubMed] [Google Scholar]
- Vita, R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., and Peters B.. 2019. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 47(D1):D339–D343. 10.1093/nar/gky1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlström, J., Dengjel J., Persson B., Duyar H., Rammensee H.G., Stevanović S., Eklund A., Weissert R., and Grunewald J.. 2007. Identification of HLA-DR-bound peptides presented by human bronchoalveolar lavage cells in sarcoidosis. J. Clin. Invest. 117:3576–3582. 10.1172/JCI32401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.X., Li Y., Yin Y., Mo M., Wang Q., Gao W., Wang L., and Mariuzza R.A.. 2011. Affinity maturation of human CD4 by yeast surface display and crystal structure of a CD4-HLA-DR1 complex. Proc. Natl. Acad. Sci. USA. 108:15960–15965. 10.1073/pnas.1109438108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J., Pullen A., Choi K., Marrack P., and Kappler J.W.. 1993. Antigen recognition properties of mutant V beta 3+ T cell receptors are consistent with an immunoglobulin-like structure for the receptor. J. Exp. Med. 177:119–125. 10.1084/jem.177.1.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Gran B., Pinilla C., Markovic-Plese S., Hemmer B., Tzou A., Whitney L.W., Biddison W.E., Martin R., and Simon R.. 2001. Combinatorial peptide libraries and biometric score matrices permit the quantitative analysis of specific and degenerate interactions between clonotypic TCR and MHC peptide ligands. J. Immunol. 167:2130–2141. 10.4049/jimmunol.167.4.2130 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
provides data collection and refinement statistics for the HLA-DR3–mimotope 46 structure.
lists the naturally occurring peptides that were screened for their ability to stimulate T cell hybridomas 1244c1, 1244c2, and 1244c3.