Abstract
The mechanisms by which transposable elements (TEs) can be horizontally transferred between animals are unknown, but viruses are possible candidate vectors. Here, we surveyed the presence of host-derived TEs in viral genomes in 35 deep sequencing data sets produced from 11 host–virus systems, encompassing nine arthropod host species (five lepidopterans, two dipterans, and two crustaceans) and six different double-stranded (ds) DNA viruses (four baculoviruses and two iridoviruses). We found evidence of viral-borne TEs in 14 data sets, with frequencies of viral genomes carrying a TE ranging from 0.01% to 26.33% for baculoviruses and from 0.45% to 7.36% for iridoviruses. The analysis of viral populations separated by a single replication cycle revealed that viral-borne TEs originating from an initial host species can be retrieved after viral replication in another host species, sometimes at higher frequencies. Furthermore, we detected a strong increase in the number of integrations in a viral population for a TE absent from the hosts’ genomes, indicating that this TE has undergone intense transposition within the viral population. Finally, we provide evidence that many TEs found integrated in viral genomes (15/41) have been horizontally transferred in insects. Altogether, our results indicate that multiple large dsDNA viruses have the capacity to shuttle TEs in insects and they underline the potential of viruses to act as vectors of horizontal transfer of TEs. Furthermore, the finding that TEs can transpose between viral genomes of a viral species sets viruses as possible new niches in which TEs can persist and evolve.
Keywords: horizontal transfer, virus, transposable element, insects, lepidopterans
Introduction
Like any other genome component, transposable elements (TEs) are vertically transmitted from one generation to the next through reproduction. But TEs can also bypass vertical transmission and cross species boundaries through a process not involving reproduction, called horizontal transfer (HT). The inference of HT of TEs (HTT) derives from the numerous observations that TE sequences from different host organisms show much lower genetic distance than expected given the divergence time of the hosts (Peccoud et al. 2018). Since the seminal report of the P element transfer between Drosophila willistoni and D. melanogaster (Daniels et al. 1990), dozens of studies have characterized HTTs involving many branches of the eukaryote tree (Schaack et al. 2010; Dotto et al. 2018). Large-scale HTT surveys in plants, insects, and vertebrates revealed recurrent transfers, which seeded a large fraction of the TE copies found today in these taxa (Bartolomé et al. 2009; El Baidouri et al. 2014; Ivancevic et al. 2018; Reiss et al. 2019; Zhang et al. 2020). Given the strong impact TEs have on genome structure and dynamics (Cordaux and Batzer 2009; Bourque et al. 2018), HTT is considered as an important process shaping the evolution of eukaryote genomes (Gilbert and Feschotte 2018).
Several important questions about HTT remain, perhaps first and foremost that of the factors facilitating these transfers. Large-scale studies have shown that HTT are more likely to occur between closely related species living in the same biogeographical realm than between more distantly related species living in different realms (Bartolomé et al. 2009; Peccoud et al. 2017). Interestingly, some host taxa such as teleost fish among vertebrates and moths and butterflies among arthropods seem to be more prone to within-phylum HTT than others, a trend that remains unexplained (Reiss et al. 2019; Zhang et al. 2020). Furthermore, many HTT events have been reported that involve parasites and their hosts, suggesting host–parasite relationships may facilitate HTT (Gilbert et al. 2010; Kuraku et al. 2012; Walsh et al. 2013; Guo et al. 2014; Suh et al. 2016). However, the molecular processes underlying HTT remain largely unknown.
Several scenarios have been proposed to explain how a TE can escape from a donor organism and enter the germline of a recipient host (Silva et al. 2004; Loreto et al. 2008; Schaack et al. 2010; Wallau et al. 2012). Experimental observations currently support two possible HTT routes. The first proposes that extracellular vesicles (EVs) could act as vectors of HTT between animals. These 50–500 nm membrane-derived vesicles are secreted by most cell types, may carry proteins, lipids and/or genetic material, and are naturally present in biological fluids (van Niel et al. 2018). EVs were shown to shuttle retrotransposons and to mediate their HT in laboratory conditions between different human cell lines or between cell culture media and mouse cell lines or embryos (Kawamura et al. 2019; Ono et al. 2019). However, the extent to which EVs may also shuttle TEs between species in natural conditions remains to be evaluated.
The second HTT route receiving some experimental support involves viruses (Gilbert and Cordaux 2017). Early studies using low-throughput targeted approaches identified TEs integrated in the genomes of several baculoviruses, which are double-stranded (ds) DNA viruses belonging to the Baculoviridae family, that were passaged in moth cell cultures (Miller and Miller 1982; Fraser et al. 1985, 1995) or whole larvae (Jehle et al. 1995, 1998). These pioneering works demonstrated that during infection, TEs can jump from the host genome to virus genomes, and that baculoviruses can receive and potentially carry a foreign genetic load from their host. More recent works using high-throughput sequencing showed that in addition to viral genomes, multiple host RNA sequences including TEs could be packaged in capsids of RNA viruses (Routh et al. 2012; Ghoshal et al. 2015; Eckwahl et al. 2016; Telesnitsky and Wolin 2016). These results further emphasized the potential role of some viruses as vectors of HTT and suggested that TEs may not have to be integrated into viral genomes to be shuttled by viruses. Using an ultra-deep sequencing approach, we revisited early works on baculoviruses and characterized the whole spectrum and frequency of host TEs integrated in genomes of the Autographa californica multiple nucleopolyhedrovirus (AcMNPV) purified from infected moth larvae (Gilbert et al. 2014, 2016). Our results revealed that a large diversity of TEs are able to jump from moth to virus genomes at each infection cycle, with an average of 4.8% of sequenced AcMNPV genomes carrying at least one host TE.
Studies of TEs segregating in baculovirus populations raised a number of outstanding questions. First, only a limited number of virus–host systems have been surveyed, such that it is still unclear whether the capacity of viral populations to carry host TEs is widespread among many viruses. Second, evidence exists showing that a virus-borne TE (TCl4.7) acquired by a baculovirus (CpGV) from a given host (the false codling moth Thaumatotibia (=Cryptophebia) leucotreta) can persist in the virus population during passages of the virus in another host (the codling moth Cydia pomonella) (Jehle et al. 1995). However, we were unable to detect any TE insertion shared between an initial AcMNPV population (called G0) replicated in the cabbage looper (Trichoplusia ni) and populations purified after ten successive infection cycles of the G0 on ten lines of the beet armyworm (Spodoptera exigua) (Gilbert et al. 2016). Thus, it is unclear whether persistence of virus-borne TEs is common among different viruses. In fact, we showed that each individual TE insertion segregates at low frequency in AcMNPV and was always purged out of the viral population within less than ten replication cycles (Gilbert et al. 2016). Third, the genome sequences of T. ni and S. exigua were not available at the time so we could not exclude that some of the AcMNPV-borne TEs originated from hosts other than T. ni or S. exigua in which the virus replicated before we conducted our study. Finally, resequencing an AcMNPV population using the long-read PacBio technology unveiled many full length TE copies integrated into viral genomes, suggesting that such copies have the capacity to encode the entire machinery necessary to transpose from viral genomes to other DNA molecules (Loiseau et al. 2020). Yet, more direct evidence supporting transposition of virus-borne TEs is still lacking.
Here, we monitored the presence, nature, and frequency of TEs in 35 deep-sequenced viral genomes obtained from 11 virus–host systems involving two iridovirus and four baculovirus species. The finding of moth TEs in non-AcMNPV baculoviruses and in the iridescent virus 6 suggests that the capacity to carry host TEs may be widespread among large dsDNA viruses. Importantly, our results also demonstrate that persistence of virus-borne TEs over more than one replication cycle occurs in different viruses (e.g., AcMNPV and the invertebrate iridescent virus 6 [IIV6] iridovirus) after two successive infection cycles in divergent species (e.g., a fly and a moth). Finally, we show that virus-borne TEs are able to transpose into other viral genomes during the course of an infection cycle.
Results
Overview of Sequenced Host–Virus Systems
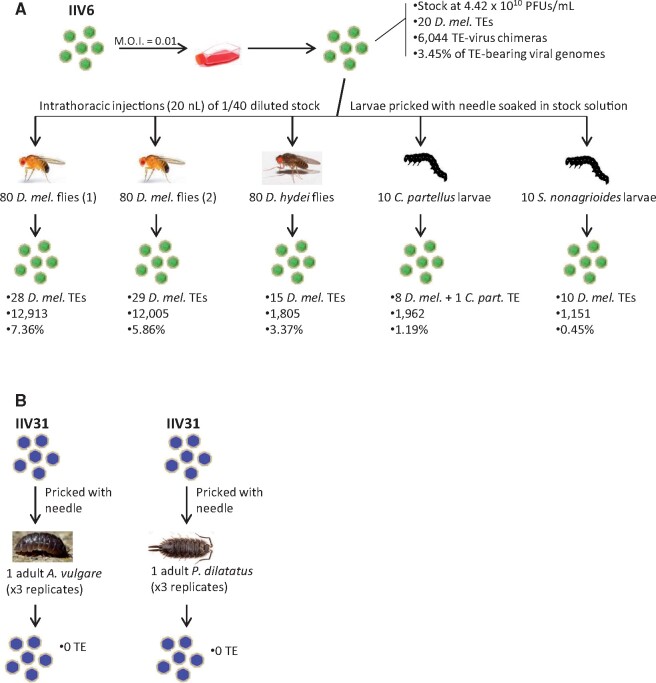
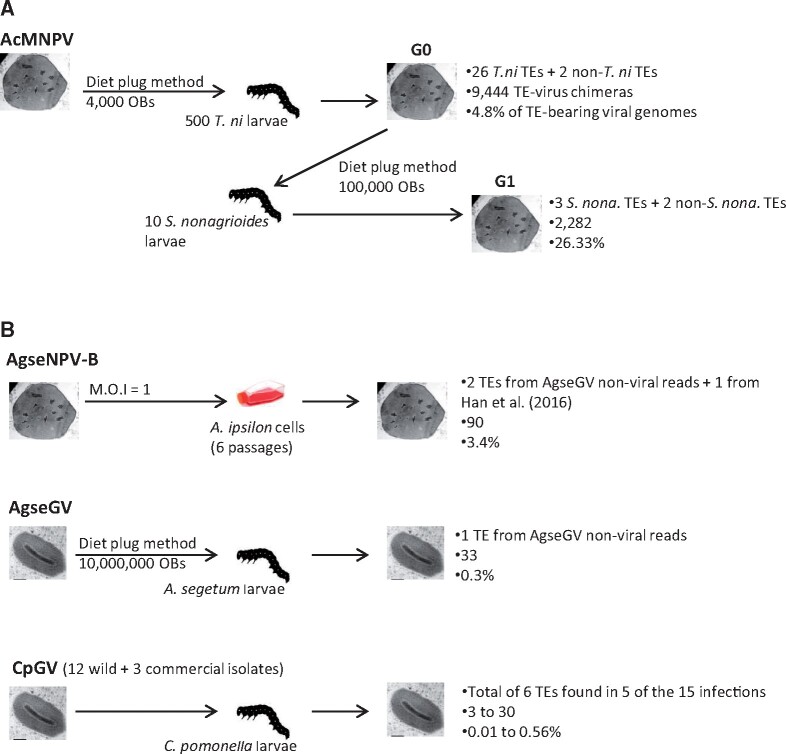
A total of 35 Illumina sequencing data sets were analyzed in this study (table 1). One data set was produced by sequencing genomes of the IIV6 purified from D. melanogaster S2 cells (fig. 1). IIV6 particles purified from S2 cells were then used to infect whole D. melanogaster flies, as well as D. hydei flies and two moth species, the spotted stalk borer (Chilo partellus) and the maize corn borer (Sesamia nonagrioides) (five data sets in total). In addition, six data sets were produced by sequencing genomes of the invertebrate iridescent virus 31 (IIV31) from two crustacean species (the terrestrial isopods Armadillidium vulgare and Porcellio dilatatus) (fig. 1). We also analyzed a population of the AcMNPV baculovirus initially replicated on T. ni (Chateigner et al. 2015) and here used to infect the maize corn borer (one data set) (fig. 2). Finally, we surveyed 22 baculovirus sequencing data sets produced as part of other studies (Gueli Alletti, Eigenbrod, et al. 2017; Gueli Alletti, Sauer, et al. 2017; Gueli Alletti et al. 2018; Fan, Jehle, et al. 2020; Fan, Wennmann, et al. 2020), including one data set of the Agrotis segetum granulovirus (AgseGV) purified from the turnip moth (Agrotis segetum), six data sets of the Agrotis segetum nucleopolyhedrovirus B (AgseNPV-B) purified from a cell line of the black cutworm (A. ipsilon), and 15 data sets of the Cydia pomonella granulovirus (CpGV) purified from the codling moth (Cydia pomonella) (fig. 2). All moth TEs found integrated into viral genomes in this study are listed in supplementary table S1, Supplementary Material online, and their sequence, as well as that of all TEs included in our library, is provided in supplementary file S1, Supplementary Material online.
Table 1.
Characteristics of the 35 Illumina Sequencing Data Sets Analyzed in This Study.
| Host | Virus | Average Read Depth over Virus Genome | Average Read Depth over Host Genome | Percentage of Viral Genomes Carrying a TE | Number of Chimeric Reads with PCR Duplicates | Number of Chimeric Reads without PCR Duplicates | Number of Insertion Points |
|---|---|---|---|---|---|---|---|
| Agrotis segetum larvae | AgseGV-DA | 7,997 | 0.0002 | 0.3 | 33 | 33 | 28 |
| Agrotis ipsilon cells | AgseNPV-pp0 | 839 | 0.15 | 0 | 0 | 0 | 0 |
| AgseNPV-pp1 | 1,939 | 0.0052 | 0 | 0 | 0 | 0 | |
| AgseNPV-pp3 | 2,348 | 0.043 | 0 | 0 | 0 | 0 | |
| AgseNPV-pp5 | 956 | 0.12 | 0 | 0 | 0 | 0 | |
| AgseNPV-pp7 | 2,930 | 0.0002 | 3.4 | 90 | 58 | 13 | |
| AgseNPV-pp10 | 1,245 | 0.018 | 0 | 0 | 0 | 0 | |
| Cydia pomonella larvae | CpGV-006 | 1,857 | 0.0002 | 0 | 0 | 0 | 0 |
| CpGV-ALE | 1,253 | 0.00005 | 0 | 0 | 0 | 0 | |
| CpGV-E2 | 3,790 | 0.007 | 0.01 | 6 | 6 | 6 | |
| CpGV-I07 | 3,342 | 0.016 | 0 | 0 | 0 | 0 | |
| CpGV-I12 | 3,441 | 0.003 | 0.009 | 5 | 5 | 5 | |
| CpGV-JQ | 1,092 | 0.00007 | 0 | 0 | 0 | 0 | |
| CpGV-KS1 | 1,446 | 0.00008 | 0 | 0 | 0 | 0 | |
| CpGV-KS2 | 942 | 0.00009 | 0.2 | 3 | 3 | 2 | |
| CpGV-M | 3,809 | 0.001 | 0.02 | 13 | 12 | 10 | |
| CpGV-R5 | 784 | 0.035 | 0 | 0 | 0 | 0 | |
| CpGV-S | 3,192 | 0.01 | 0.56 | 30 | 30 | 5 | |
| CpGV-V15 | 2,380 | 0.001 | 0 | 0 | 0 | 0 | |
| CpGV-WW | 523 | 0.0001 | 0 | 0 | 0 | 0 | |
| CpGV-ZY | 744 | 0.00004 | 0 | 0 | 0 | 0 | |
| CpGV-ZY2 | 1,152 | 0.00009 | 0 | 0 | 0 | 0 | |
| Adult Armadillidium vulgare individuals | IIV31 (1) | 133,086 | 0.19 | 0 | 0 | 0 | 0 |
| IIV31 (2) | 163,802 | 0.002 | 0 | 0 | 0 | 0 | |
| IIV31 (3) | 188,230 | 0.002 | 0 | 0 | 0 | 0 | |
| Adult Porcellio dilatatus individuals | IIV31 (1) | 112,750 | NA | 0 | 0 | 0 | 0 |
| IIV31 (2) | 220,537 | NA | 0 | 0 | 0 | 0 | |
| IIV31 (3) | 184,610 | NA | 0 | 0 | 0 | 0 | |
| Drosophila melanogaster S2 cells | IIV6 | 123,574 | 3.45 | 4.33 | 7,249 | 6,044 | 2,831 |
| Drosophila melanogaster flies (1) | IIV6 | 170,859 | 6.46 | 7.36 | 16,631 | 12,913 | 3,469 |
| Drosophila melanogaster flies (2) | 211,039 | 12.41 | 5.86 | 16,230 | 12,005 | 3,460 | |
| Adult Drosophila hydei individuals | IIV6 | 52,931 | 0.46 | 3.37 | 2,313 | 1,805 | 450 |
| Chilo partellus larvae | IIV6 | 325,206 | 0.24 | 1.19 | 5,041 | 1,962 | 22 |
| Sesamia nonagrioides larvae | IIV6 | 245,899 | 26.77 | 0.45 | 1,439 | 1,151 | 58 |
| Sesamia nonagrioides larvae | AcMNPV | 82,103 | 0.06 | 26.33 | 37,952 | 2,282 | 613 |
Note.—The percentage of viral genomes carrying a TE insertion was computed considering the number of viral genomes carrying a TE follows a Poisson distribution.
Fig. 1.
Overview of iridovirus infections and number and frequency of TEs found in viral genomes. (A)An IIV6 isolate was first replicated onto Drosophila melanogaster S2 cells and the resulting viral population was used to infect whole flies and moths. M.O.I., multiplicity of infection. (B) An IIV31 isolate was used to infect two pillbug species. The mode of infection, the number of TEs found integrated into viral genomes, the number of chimeric reads, and the percent of viral genomes carrying a TE are given for each experiment. All moth TEs found integrated into iridovirus genomes are provided in supplementary file S1, Supplementary Material online.
Fig. 2.
Overview of baculovirus infections and number and frequency of TEs found in viral genomes. (A) An AcMNPV isolate called “G0” purified from Trichoplusia ni larvae (Gilbert et al. 2016) was used to infect Sesamia nonagrioides larvae. The AcMNPV population purified from these larvae was called “G1.” (B) Illumina reads produced as part of other studies for three other baculoviruses were surveyed for the presence of TEs integrated into viral genomes (Gueli Alletti, Eigenbrod, et al. 2017; Gueli Alletti, Sauer, et al. 2017; Gueli Alletti et al. 2018; Fan, Jehle, et al. 2020; Fan, Wennmann, et al. 2020). The mode of infection (when appropriate), the number of TEs found integrated into viral genomes, the number of chimeric reads, and the percent of viral genomes carrying a TE are given for each experiment. M.O.I., multiplicity of infection. All moth TEs found integrated into baculovirus genomes are provided in supplementary data 1, Supplementary Material online.
Moth TEs in Baculoviruses Other Than AcMNPV
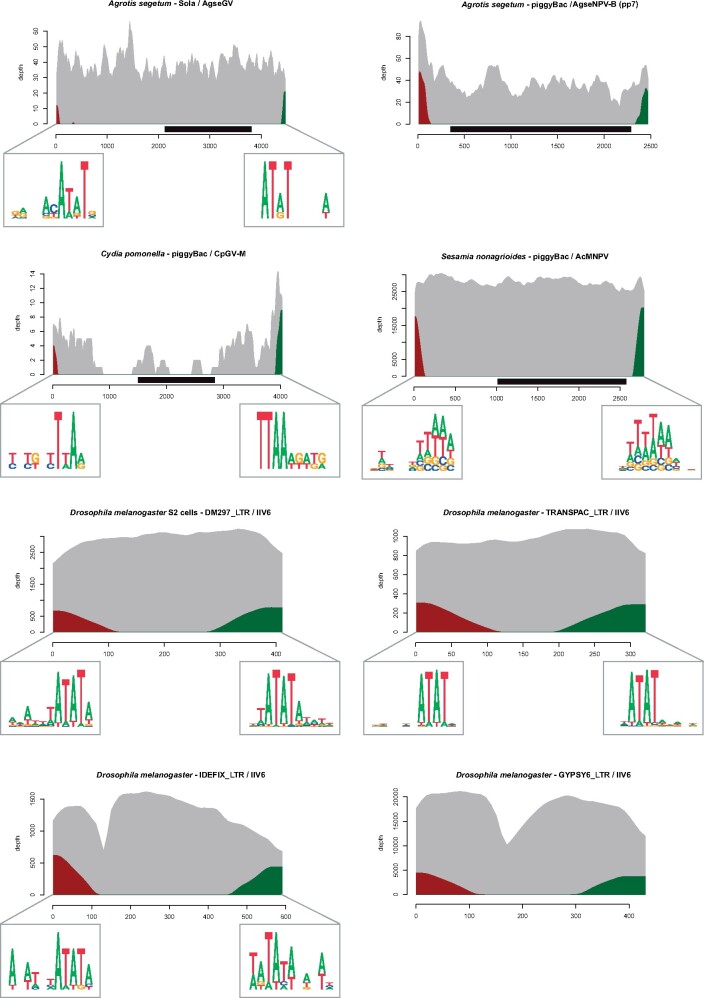
We first sought to assess whether TEs can transpose into genomes of baculoviruses other than AcMNPV using bulk deep sequencing, and to characterize the origin of such TEs as well as their frequency in viral populations. In brief, we found a total of six different TEs integrated in 7 out of 22 non-AcMNPV baculovirus data sets, which confirms that baculoviruses other than AcMNPV carry TEs integrated in their genomes (Jehle et al. 1995, 1998). More specifically, we began by searching for TEs integrated in genomes of six out of ten successive passages of AgseNPV-B replicated in cells of A. ipsilon. These genomes were sequenced at depths varying from 839× to 2,930×. We found TEs integrated in genomes of only one passage (pp7), with a total of 90 TE-virus chimeras covering 13 different positions in the viral genome and corresponding to a frequency of 3.4% of viral genomes carrying an insertion (table 1). The three involved TEs were class 2 DNA transposons from the piggybac and Sola superfamilies and an unclassified nonautonomous element (supplementary table S1, Supplementary Material online). Terminal inverted repeats were identified for all three TEs (supplementary table S2, Supplementary Material online) and all chimeric reads mapped to their extremities (supplementary table S1, Supplementary Material online; fig. 3; supplementary fig S1., Supplementary Material online). The three TEs were absent from the whole-genome sequence (WGS) of A. ipsilon (NCBI accession number: PNFC00000000.1), the species from which the cell line used to passage AgseNPV-B was established. The WGS of the A. ipsilon cell line is not available. Thus, the origin of the TEs found in pp7 cannot be easily explained. In any case, the piggybac and Sola TEs were identified in our assembly of nonviral reads obtained from the AgseGV/A. segetum larvae system and the nonautonomous element was previously identified in the plutellid moth Plutella xylostella (Han et al. 2016). The fact that we only retrieved these TEs in passage pp7 of the virus and not in earlier (pp0, pp1, pp3, pp5) or later (pp10) passages may be due to the stochasticity inherent to the detection of low frequency virus-borne TEs, which is likely pronounced given the relatively low depths at which these samples were sequenced.
Fig. 3.
Sequencing depth of some of the TEs found integrated into viral genomes. Sequencing depth by reads mapping entirely on the TEs is shown in gray. Sequencing depth by chimeric reads is shown in red on the 5′-end of the TEs and in green for the 3′-end of the TEs. Black rectangles represent TE genes annotated in autonomous TE sequences. TSDs are shown on each side of the elements using sequence logos for all TEs for which conserved TSDs were observed (sequencing depth of the other TEs uncovered in this study is shown in supplementary fig. S2, Supplementary Material online).
We then searched for TEs integrated in genomes of AgseGV replicated in A. segetum larvae and sequenced at 7,997×. A total of 33 TE-virus chimeras were detected, covering 28 different positions and corresponding to a frequency of 0.3% of viral genomes carrying an insertion (table 1). All chimeras mapped to the aforementioned Sola TE (supplementary table S1, Supplementary Material online).
We also detected TE-virus chimeras in 5 of the 15 CpGV isolates replicated in C. pomonella larvae and sequenced at depths varying from 523× to 3,809×. The number of chimeras varied from three (two different positions along the viral genome) to 30 (five different positions) and the frequency of viral genomes carrying a TE varied from 0.009% to 0.56% (table 1). The samples of the commercial isolates CpGV-006, -V15, and -R5 were free of TE insertions. In three of the five CpGV strains (CpGV-E2, CpGV-I12, CpGV-M), a single piggybac DNA TE, different from that detected into AgseNPV genomes, was integrated into viral genomes (supplementary table S1, Supplementary Material online). This piggybac was also found, together with a SHALINE-like element (non-LTR [long terminal repeat] retrotransposon) in the CpGV-S data set (supplementary table S1, Supplementary Material online). Finally, a Tc1/mariner DNA transposon was found integrated in the CpGV-KS2 strain. Terminal inverted repeats were identified in chimeric reads for the piggybac and Tc1/mariner elements (supplementary table S2, Supplementary Material online) and all chimeric reads mapped only to the extremities of the three TEs found integrated into CpGVs (fig. 3; supplementary fig. S2 and table S1, Supplementary Material online). All three TEs are present in the C. pomonella WGS, suggesting they transposed from the C. pomonella genome to that of the virus.
Retrotransposons from D. melanogaster S2 Cells Transpose into the IIV6 Iridovirus
To assess the potential of large dsDNA viruses other than baculoviruses to shuttle TEs between arthropods, we searched for TEs integrated in genomes of the IIV31 iridovirus purified from three P. dilatatus and three A. vulgare individuals (including one previously analyzed by Loiseau et al. [2020]) and in genomes of the IIV6 iridovirus after a passage on D. melanogaster S2 cells. Sequencing depths varied from 112,750× to 220,537× (table 1). Although all IIV31 data sets were found to be entirely devoid of TE-virus chimeras, we found thousands of low-frequency TE copies integrated in IIV6 genomes (table 1). This shows that the presence of unfixed TEs in viral populations is not a feature restricted to baculoviruses, but that it is also not systematic.
More specifically, we found 6,044 TE-virus chimeras in IIV6 genomes purified from D. melanogaster S2 cells, corresponding to 2,831 different positions along the viral genome (fig. 4) and 4.33% of sequenced IIV6 genomes carrying at least one TE (table 1). TE-virus junctions involved one D. melanogaster non-LTR retrotransposon and 19 LTR retrotransposons including five (MDG1_LTR, DM297_LTR, DM176, IDEFIX_LTR, and Drosophila_melanogaster: rnd-3_family-276#LTR/Gypsy) that accounted for 500 or more (up to 3,192) chimeras (supplementary table S1, Supplementary Material online).
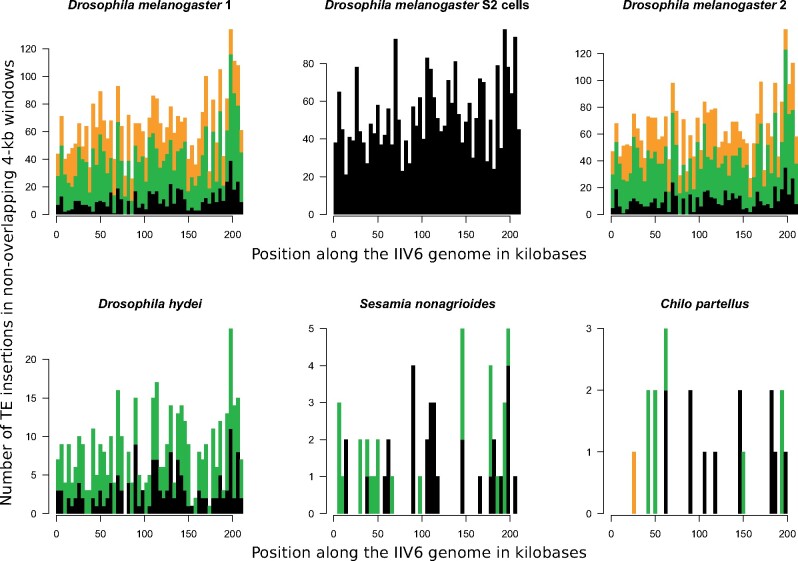
Fig. 4.
Stacked histograms showing the number of different insertions of TEs in 4-kb windows along the genome of the iridovirus IIV6 in a parental viral population (purified from Drosophila melanogaster S2 cells) and five IIV6 daughter populations purified from other hosts. The top central diagram illustrates TE insertions in the parental IIV6 purified from D. melanogaster S2 cells. The other five diagrams illustrate TE insertions in the daughter IIV6 populations purified from whole flies or moths (fig. 1). Black bars correspond to TE insertions shared by the parental IIV6 population and at least one daughter IIV6 population. These shared insertions correspond to the same TE at the exact same position in the parental IIV6 population purified from D. melanogaster S2 cells and other IIV6 populations purified from other hosts. Green bars correspond to TE insertions not shared with the parental IIV6 population. TEs involved in these unshared insertions were present in the parental IIV6 population but not at the position they were found in the daughter populations. These unshared insertions may be due to de novo virus-to-virus transposition during infection of Drosophila whole flies or moth larvae. Alternatively, they may correspond to TE insertions that were present in the parental population but were not detected by sequencing. Orange bars correspond to TEs that are absent from the parental IIV6 population. These are likely to result from de novo transposition from the host genome. Numbers of insertions are given in supplementary table S1, Supplementary Material online, for each TE.
As this is the first report of TEs segregating in an iridovirus population, we set out to independently validate the biological nature of TE-virus chimeras by comparing these chimeras with those involving human DNA added to the IIV6 DNA sample prior to constructing the sequencing library. We investigated the structure of chimeras occurring between human TEs and the IIV6 genome. These can only be technical as IIV6 has not been in contact with human cells. Given such chimeras are not generated by transposition, they are not expected to preferentially involve the extremities of TEs. Consistently, chimeras involving human TEs almost all (554 out of 560) mapped to internal parts of TE sequences (supplementary table S3, Supplementary Material online). By contrast, most chimeras involving D. melanogaster TEs (6,044 out of 6,105) mapped at the extremities of TE sequences (fig. 3; supplementary fig. S2 and table S1, Supplementary Material online). Together with the identification of TSDs for some TEs (examples are given in fig. 3), these results confirm the biological nature of the TE-virus chimeras we identified. Transposition of D. melanogaster LTR retrotransposons into IIV6 genomes is consistent with population genetics studies showing that most LTR elements are of recent origin and currently actively transposing in natural fly populations (Kofler et al. 2015).
LTR Retrotransposons from D. melanogaster Whole Flies Transpose into the IIV6 Iridovirus
The results presented above demonstrate that D. melanogaster TEs can transpose from a cell line (S2 cells) to IIV6 genomes. Here, we now show that TEs can also transpose from whole insects into IIV6 genomes. We infected two batches of 80 D. melanogaster adult flies with the IIV6 population purified from S2 cells and sequenced IIV6 viral genomes purified from these infections at 170,859× and 211,039× (fig. 1). We found more TE-virus chimeras in the two IIV6/adult flies’ batches (12,193 and 12,005 in batches 1 and 2, respectively) than in the parental IIV6/S2 cells data set (6,044) and a higher frequency of viral genomes carrying at least one TE (7.36% and 5.86% in batches 1 and 2, respectively) than in the parental IIV6/S2 cells population (4.33%) (table 1; supplementary table S1, Supplementary Material online). A total of 28 and 29 different TEs were identified in batches 1 and 2, respectively (supplementary tables S1 and S4, Supplementary Material online). All 20 TEs identified in the parental IIV6 purified from S2 cells were found to be integrated at 2,263 and 2,345 different positions in IIV6 from whole flies (batches 1 and 2, respectively) (black and green bars in D. melanogaster 1 and 2 graphs in fig. 4). In principle, these TE insertions could have two sources. First, they could have been present in the parental IIV6 purified from S2 cells and have persisted in the viral population during replication in whole flies, implying that IIV6 genomes bearing TE insertions were encapsidated again in whole flies. Second, the TE insertions could result from transposition of TE copies located in the genomes of whole flies. Given that the TE content of D. melanogaster S2 cells and whole flies genomes is likely very similar, it is difficult to assess whether and what portion of insertions of the shared TEs come from the parental IIV6 population or from whole flies.
Interestingly, the majority of TE-virus chimeras found in IIV6 purified from whole flies (6,154 and 5,706 in batches 1 and 2, respectively) map to GYPSY6_LTR and GYPS-7_Dsim, two LTR retrotransposons not detected in the parental IIV6 purified from S2 cells (orange bars in D. melanogaster 1 and 2 graphs in fig. 4; fig. 3; supplementary fig. S3 and tables S1 and S4, Supplementary Material online). Another six and seven TEs absent from IIV6 purified from S2 cells were found in batches 1 and 2, respectively, including one (DMLTR5) involved in more than 25 TE-virus chimeras in the two whole fly batches (supplementary fig. S3 and tables S1 and S4, Supplementary Material online). Given their absence in the parental IIV6/S2 cell data set, de novo integration of these TEs in IIV6 genomes is most likely due to transposition of TE copies located in the genomes of whole flies.
TEs from Other Hosts in IIV6
Here, we show that a TE from a moth host can transpose into IIV6 genomes, though at low frequency. We again used IIV6 particles purified from S2 cells to infect whole flies from another Drosophila species (D. hydei) and larvae from the C. partellus moth (fig. 1). We also reanalyzed another IIV6 sequencing data set from Loiseau et al. (2020) generated by infecting larvae from the S. nonagrioides moth with the IIV6 particles purified from S2 cells. In Loiseau et al. (2020), we only searched for S. nonagrioides TEs integrated into IIV6 genomes and found none. Here we extended our search to a much larger diversity of TEs, including D. melanogaster TEs. Sequencing depth varied from 52,931× to 325,206× (table 1). Across the three species, we found a single non-D. melanogaster TE integrated in IIV6, that is, a piggybac transposon involved in seven TE-virus chimeras in the C. partellus data set (supplementary table S1, Supplementary Material online). There is one copy identical to this TE over its full length in the C. suppressalis WGS and we independently validated its presence in the C. partellus genome by polymerase chain reaction (PCR) and Sanger sequencing (piggyBac_Chilo_partellus in supplementary file S1, Supplementary Material online). Thus, this TE likely transposed from C. partellus to IIV6 during replication of the virus in the moth larvae.
Persistence of Fly TEs during IIV6 Replication
Here, we demonstrate that many D. melanogaster TEs present in the IIV6 isolate extracted from D. melanogaster S2 cells can be retrieved in this virus after replication in another host. Contrasting with the absence or paucity of D. hydei, S. nonagrioides and C. partellus TEs integrated into IIV6 genomes, we found 1,805, 1,962 and 1,151 TE-virus chimeras involving D. melanogaster TEs in IIV6 genomes purified from the three respective hosts. All D. melanogaster TEs recovered in these IIV6 populations were present in the parental IIV6 purified from S2 cells and the eight most frequent TEs present in the IIV6/S2 cells data set (>0.1%) are also the most frequent in the daughter IIV6 populations (supplementary fig. S3 and table S4, Supplementary Material online). We verified that none of these D. melanogaster TEs is present in the genomes of D. hydei, S. nonagrioides, or C. partellus using BlastN similarity searches. Thus, we conclude that D. melanogaster TEs integrated into IIV6 genomes purified from these three hosts come from the parental IIV6 population purified from S2 cells. The various chimeric reads involving D. melanogaster TEs found in IIV6 purified from D. hydei, S. nonagrioides and C. partellus correspond to 450, 58 and 22 different positions along the IIV6 genome, respectively (table 1, fig. 4). Consistent with persistence of D. melanogaster TEs in IIV6 purified from the three other hosts, 149, 31, and 12 of these positions were shared with the parental IIV6 population purified from S2 cells, respectively. This implies that many TE-bearing IIV6 genomes persisted through one replication cycle and were again encapsidated in three different hosts. By contrast, 232, 27, and 10 insertions found in IIV6 purified from D. hydei, S. nonagrioides, and C. partellus were not found in the parental IIV6 purified from S2 cells. A majority (94% in D. hydei) of these insertions were supported by less than ten chimeric reads. Thus, insertions not shared with the parental IIV6 population purified from S2 cells may in fact have been present at very low frequency in this population but not sequenced. Interestingly, it is also possible that some of them were generated through virus-to-virus transposition during replication of the virus in D. hydei, S. nonagrioides, and C. partellus.
Persistence of TEs Integrated in AcMNPV Genomes
To assess whether TEs integrated into AcMNPV genomes can be recovered after one replication cycle, as observed for IIV6, we infected S. nonagrioides larvae with a viral population (termed “G0”) purified from T. ni larvae (Chateigner et al. 2015) and known to contain thousands of TE copies belonging to at least 29 TE families (Gilbert et al. 2016). Our search for TEs integrated into the AcMNPV population purified from S. nonagrioides (hereafter termed “G1”) unveiled a total of five TEs, two of which were also present in the G0 population: Tni_contig_27 (piggybac) and Tni_contig_21 (mariner) (supplementary table S1, Supplementary Material online). A BlastN similarity search of these two TEs in the S. nonagrioides genome did not reveal any significant hit and a PCR screening using three primer sets designed to amplify three regions of these elements supported their absence from S. nonagrioides. Thus, we conclude that Tni_contig_27 and Tni_contig_21 present in AcMNPV genomes purified from S. nonagrioides were most likely carried over from the G0 population. These results confirm that some TEs integrated into AcMNPV genomes can persist over at least one replication cycle, as observed for IIV6. Interestingly, aside from the shared presence of Tni_contig_27 and Tni_contig_21, the TE landscape of the G0 and G1 populations markedly differed, as none of the 27 other TEs present in the G0 population was recovered after replication on S. nonagrioides (fig. 5). This is in stark contrast with IIV6, for which TE contents and frequencies were relatively similar between populations purified from D. melanogaster S2 cells and D. hydei.
Fig. 5.
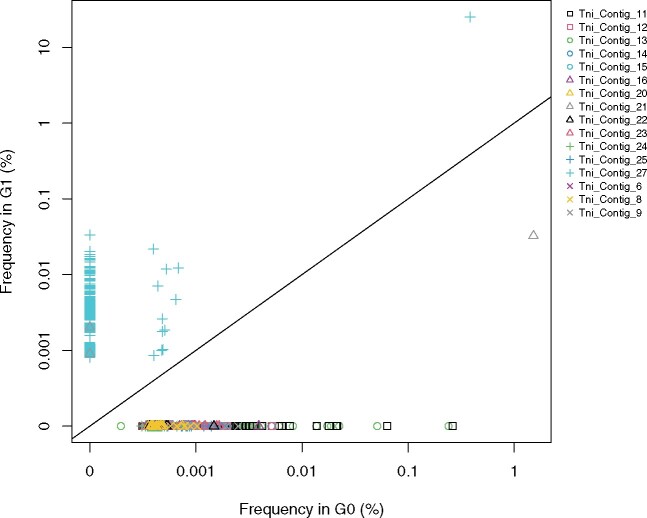
Frequency of specific TE insertions in an AcMNPV population purified from Trichoplusia ni (G0) plotted against the frequency of the same TE insertions in an AcMNPV population purified from Sesamia nonagrioides (G1). The AcMNPV baculovirus was first replicated and purified from T. ni larvae (Chateigner et al. 2015; Gilbert et al. 2016). This isolate was called generation 0 (G0). The G0 was here subsequently replicated on and purified from S. nonagrioides larvae. Frequency is expressed in logarithm. The zero on the Y and X axis are for illustrative purposes as zeros cannot be shown at the log scale. Only two TEs, Tni_Contig_27 (turquoise cross symbol) and Tni_Contig_21 (gray triangle symbol), have shared insertions (i.e., located at the same position in the viral genome) between both AcMNPV populations. Only TEs inserted at least at five different positions along the AcMNPV genome are shown.
AcMNPV-to-AcMNPV Transposition in Moths
Here, we compare the number of TE insertions in two AcMNPV populations separated by one replication cycle and found strong evidence supporting virus-to-virus transposition. A striking feature of the G1 population purified from S. nonagrioides is that 26.33% of AcMNPV genomes carried at least one TE insertion. This high frequency was mainly explained by Tni_contig_27, as the frequency of AcMNPV genomes bearing other TEs is only 0.04%. Compared with 0.38% in the G0 population purified from T. ni larvae (Gilbert et al. 2016), the frequency of genomes carrying a Tni_contig_27 insertion underwent a 69-fold increase during replication in S. nonagrioides. Similarly, the number of different Tni_contig_27 insertions was much higher in the G1 (n = 417) than in the G0 (n = 17) AcMNPV population (fig. 6). Though this difference may at first sight be explained by virus-to-virus transposition, two other hypotheses have to be considered. First, the G0 population was sequenced at a higher depth (124,221×) than the G1 population (82,103×), invalidating the hypothesis according to which many Tni_contig_27 insertions would have been missed in the G0 AcMNPV population. Second, fluctuations in insertion frequencies driven by genetic drift could explain the difference between the G0 and G1 populations. Under genetic drift, the frequency of an insertion has a probability of 0.5 to have increased during replication in S. nonagrioides larvae. In such a scenario, all 417 insertions found in the G1 population would have been present but missed because of their frequency being too low. Given that 12 out of the 17 G0 insertions are also found in the G1 population, this scenario would imply that a total of 417 + 17 – 12 = 422 Tni_contig_27 insertions were present in the G0 population. Assuming independence between insertions, the probability that as many as 417 insertions out of 422 increased in frequency due to genetic drift alone is almost null (P = 2.2 × 10−16, exact binomial test). Though inconsistent with drift alone, such a global increase could in principle be observed if all or most Tni_contig_27 insertions were beneficial to the virus. Such a scenario, whereby several hundreds of Tni_contig_27 insertions would be adaptive irrespective of their location along the AcMNPV genome, is highly unlikely. In fact, current evidence strongly suggests that the vast majority of TE insertions are deleterious to viruses as they very rarely reach high frequencies during viral replication in natural hosts (Gilbert et al. 2016; Gilbert and Cordaux 2017). This is best illustrated by the rare occurrence of TEs in consensus viral genomes (Sun et al. 2015; Gilbert and Cordaux 2017; Filée 2018). Thus, the nearly 25-fold increase in the number of Tni_contig_27 insertions observed in the G1 (from 17 to 417) is unlikely to be explained by fluctuation in insertion frequency. Instead, we contend that during replication in S. nonagrioides larvae, the Tni_contig_27 TE most likely transposed between AcMNPV genomes, and possibly within AcMNPV genomes as well.
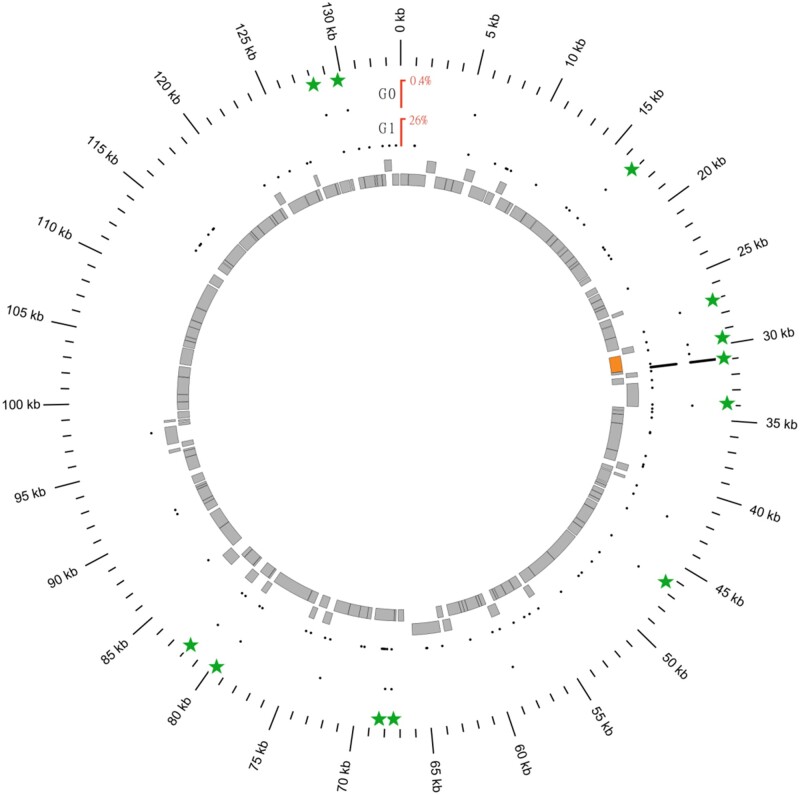
Fig. 6.
Insertion map of Tni_contig_27 in G0 and G1 populations along the AcMNPV genome. Green stars represent the 12 shared insertion points between both AcMNPV populations. Gray rectangles represent genes along the viral genome. The orange rectangle represents the Ac-GTA gene. Most Tni_Contig_27 insertions fall into the Ac-GTA gene for both AcMNPV populations although insertion points are scattered all along the viral genome.
High Frequency of a Single Tni_Contig_27 Insertion in the AcMNPV GTA Gene
In this section, we show that our ability to detect virus-to-virus transposition of the Tni_contig_27 TE is likely due to the fact that a single insertion of this TE underwent a sharp increase in frequency during replication of AcMNPV in S. nonagrioides. We noticed that although virus-to-virus transposition likely explains the increase in number of Tni_contig_27 insertions in the AcMNPV G1 population, it does not account for the overall increase in AcMNPV genomes bearing a Tni_contig_27 insertion (from 0.38% in the G0 to 26.29% in the G1 population). In fact, the vast majority of TE-virus chimeras involving Tni_contig_27 in the G1 population (1,069 out of 2,245 without PCR duplicates, or 35,189 out of 37,897 including PCR duplicates) map to a single insertion site, located at position 30,918 of the WP10 AcMNPV genome, in the AcMNPV global transactivator (Ac-GTA) gene (fig. 6). This insertion, together with 11 other Tni_contig_27 insertions (green stars in fig. 6), is present in the parental G0 population but at a much lower frequency. The frequency increase of this insertion during replication in S. nonagrioides larvae may be due to repeated integration of Tni_contig_27 at the exact same site and/or to preferential replication of viral genomes bearing this insertion. Our earlier analysis of ten AcMNPV populations obtained after infection of T. ni larvae with the G0 population did not reveal any frequency increase of this insertion (Gilbert et al. 2016). Repeated integration at position 30918 may have occurred but it is unlikely to solely explain the sharp increase in frequency of the Tni_contig_27 insertion during replication in S. nonagrioides larvae.
Instead, the frequency of this insertion may have mainly increased through replication of the viral genomes bearing it. It is noteworthy that Ac-GTA is involved in transcription regulation, DNA recombination and repair, chromatin unwinding, and other functions (Rohrmann 2019). Deletion of this gene from the Bombyx mori nucleopolyhedrovirus (BmNPV) only led to mild effects on the infection outcome (i.e., delayed killing time), suggesting that it may not be essential for viral replication in some contexts (Katsuma et al. 2008). We reasoned that the increase in frequency of the Tni_contig_27 insertion in this gene may be due to low functional constraints inducing relaxation of purifying selection. If true, other mutations inactivating this gene may also be expected to have increased in frequency during replication in S. nonagrioides. However, we found no evidence of other high-frequency TE insertions or high-frequency indels in Ac-GTA (fig. 6; supplementary fig. S4, Supplementary Material online). Thus, the strong increase in frequency of the Tni_contig_27 insertion in Ac-GTA is unlikely to be due to the absence of functional constraints acting on this gene in our experiment. The possibility that this insertion increased in frequency because of a positive effect on viral replication cannot be excluded at this stage. Another possibility worth testing in future experiments is that genomes bearing this insertion may have a higher propensity to be encapsidated compared with other genomes.
Origin of TEs Found in AcMNPV
Though our earlier study identified Tni_contig_27 and Tni_contig_21 into AcMNPV genomes purified from T. ni larvae, we were unable to trace the origin of these TEs because no WGS was available for T. ni at the time (Gilbert et al. 2016). A BlastN similarity search for these TEs against the two now available T. ni genomes (Chen et al. 2019; Talsania et al. 2019) revealed no hit. This result suggested the presence of Tni_contig_27 and Tni_contig_21 in the G0 AcMNPV population purified from T. ni larvae did not result from transposition of TEs located in the T. ni genome. Instead, the two TEs must have originated from an earlier replication cycle of AcMNPV on another host. In addition to Tni_contig_27 and Tni_contig_21, we found three S. nonagrioides DNA TEs (1 piggybac and 2 sola) involved in 5, 6, and 5 TE-AcMNPV chimeras, respectively (supplementary table S1, Supplementary Material online). Thus, TEs became integrated into AcMNPV genomes not only through virus-to-virus transposition but also through transposition of copies located in the S. nonagrioides genome. In turn, this result shows that TEs from yet another host can transpose into AcMNPV genomes during the course of an infection, though at lower rates than previously reported in T. ni or S. exigua (Gilbert et al. 2016).
HT of TEs Found Integrated into Viral Genomes
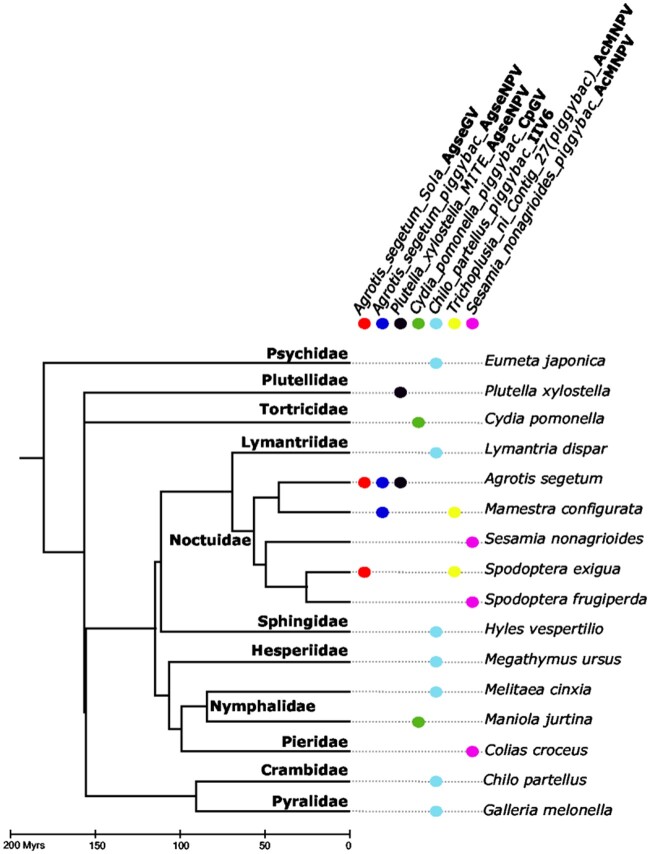
A previous survey showed that 8 of the 29 D. melanogaster TEs we found integrated into IIV6 genomes have undergone HT between D. melanogaster and its sister species D. simulans, which diverged less than 5 Ma (supplementary table S4, Supplementary Material online; Bartolomé et al. 2009). This further supports the recent origin and current activity of these D. melanogaster LTR retrotransposons. In addition, we found evidence that 7 out of the 12 non-Drosophila TEs integrated into viral genomes were involved in HTT (fig. 7). When BLAST searches were launched using these seven TEs as queries against all insect WGS available as of March 2020, we retrieved hits longer than 200 bp over at least 80% of the TE length and with >80% nucleotide identity. The synonymous distances (dS) calculated for each of these hits all fall below the 0.5% quantile of the distribution of orthologous gene dS (supplementary fig. S5, Supplementary Material online), strongly suggesting that these TEs were horizontally transferred between species, rather than vertically inherited from a common ancestor. Interestingly, all insect species in which we found these TEs are lepidopterans, perhaps reflecting the higher propensity of TEs to transfer horizontally between closely related species (Bartolomé et al. 2009; Peccoud et al. 2017). Furthermore, several lepidopteran species harboring these TEs are known to be susceptible to NPV (all noctuids included in fig. 7; Goulson 2003; Thézé et al. 2018) or IIV6 (Chilo moths; Fukaya and Nasu 1966), in agreement with the possibility that these viruses may have been involved in the transfers.
Fig. 7.
Timetree of lepidopteran species involved in HT of the TEs found integrated in viral genomes. TEs are shown as full colored circles. Names of TEs are on top of the figure, they correspond to the names used in supplementary table S1, Supplementary Material online, to which we added the name of the virus (in bold) in which the TEs were found. The sequence of each TE is provided in supplementary file S1, Supplementary Material online. TEs were considered present in a species when we recovered BlastN hits longer than 200 bp over at least 80% of the TE length and with >80% nucleotide identity. The synonymous distances (dS) calculated for each of these hits all fall below the 0.5% quantile of the distribution of orthologous gene dS (supplementary fig. S5, Supplementary Material online). Divergence times were taken from Kumar et al. (2017).
Discussion
The diversity and frequency of TEs integrated in baculovirus genomes previously assessed using ultra-deep sequencing identified 29 and 40 different TEs integrated in AcMNPV replicated in T. ni and S. exigua, respectively, with frequencies ranging from 1.1% to 14.3% of viral genomes carrying at least one host-borne TE (Gilbert et al. 2016; Loiseau et al. 2020). Here, we show that TEs from another host (S. nonagrioides) can transpose into AcMNPV, though at lower rates than in T. ni and S. exigua. We further report measurable frequencies of viral genomes carrying TEs in another NPV (AgseNPV) replicated in A. ipsilon cells as well as in two granuloviruses (AgseGV and CpGV) replicated in A. segetum and C. pomonella larvae, respectively. We also show that large numbers of D. melanogaster TEs, as well as a few C. partellus TEs, can integrate in genomes of the IIV6 iridovirus during replication in D. melanogaster cells or in whole flies and moths. Interestingly, earlier works (Bartolomé et al. 2009) and our own inferences of HTT based on comparisons of synonymous distances between TEs and host genes show that many TEs we found integrated in viral genomes have been transmitted through HT between insect species. Altogether, these results extend our knowledge of host–virus systems for which TEs have been demonstrated to jump into viral genomes. They are in line with previous studies that support a role for large dsDNA viruses as possible vectors of HTT between eukaryotes (Miller and Miller 1982; Carstens 1987; Fraser et al. 1995; Jehle et al. 1995, 1998; Sun et al. 2015; Gilbert and Cordaux 2017).
Highly Contrasted TE Contents in Populations of Different Viruses
Although this study demonstrates the capacity of some viruses to receive and carry TEs, it also reveals important variation in occurrence and frequency of TE integration into viral genomes across host–virus systems. No TE integration was detected in IIV31 iridoviruses purified from two isopod species (A. vulgare and P. dilatatus), in agreement with an earlier report (Loiseau et al. 2020). Although we retrieved many D. melanogaster TEs and a few from C. partellus in IIV6, no TE from D. hydei or S. nonagrioides transposed into genomes of this virus. We found moth TEs integrated in AgseNPVs and CpGVs purified from only 1 out of 6 and 4 out of 15 viral isolates, respectively. Thus, although baculoviruses and iridoviruses have the capacity to encapsidate and shuttle TE-bearing viral genomes, transposition into viral genomes does not systematically occur during infection. It may not even occur at all in some host–virus systems. We cannot exclude that some TE insertions may have been missed, especially in some relatively low sequencing-depth data sets (e.g., lower than 1,000× for six baculoviruses), or because our TE library may not include all host TEs due to unavailable WGS (e.g., C. partellus). Yet, we did not find any TE insertion in several ultra-deeply sequenced viral genomes that had been purified from hosts with available WGS (i.e., A. vulgare, S. nonagrioides, D. hydei; table 1). The absence of TEs in these data sets strongly suggests biological underpinnings.
Factors Possibly Influencing TE Content in Viral Populations
Multiple host and virus factors are likely involved in shaping the numbers of host TEs found integrated in viral genomes. On the host side, the level of TE activity may be involved. For example, the absence of A. vulgare, S. nonagrioides and D. hydei TEs in IIV31, AcMNPV and IIV6 genomes, respectively, could be due to the lack of currently active TEs in the genome of these three species. However, the TE landscape of A. vulgare and D. hydei (and TE expression patterns for A. vulgare) does not support this hypothesis, as a relatively large fraction of TE copies is nearly identical to their cognate family consensus sequence, which is highly suggestive of current activity (Becking et al. 2020; supplementary fig. S6, Supplementary Material online). In addition, the finding of three S. nonagrioides TEs integrated into AcMNPV genomes provides direct evidence that some TEs are indeed active in this moth. On the virus side, we are not aware of any major difference between baculovirus and iridovirus replication that could explain varying propensities of these viruses to receive and carry hosts TEs. In particular, replication involves transportation of viral DNA into the nucleus for both virus types. For baculoviruses, genome replication and assembly of nucleocapsids fully take place in the virogenic stroma within the nucleus (Rohrmann 2019). For iridoviruses, the first step of genome replication also occurs in the host nuclei, in which the viral polymerase synthesizes unit to twice unit sizes of the viral genome (Williams et al. 2005). The proximity of host and viral genomes during replication in host nuclei may facilitate host-to-virus transposition for the two virus types. However, among baculoviruses, the median frequency of viral genomes carrying a TE is significantly lower for GVs than for NPVs (Wilcoxon unpaired test, W = 0; P-value = 3.4 × 10−4). Complementation between genomes may be more frequent in multicapsid NPVs, such as AcMNPV and AgseNPV, which occlusion-derived virions (ODV) contain many nucleocapsids (and thus genomes), than in GVs which encapsidate only one genome per ODV and OB (Rohrmann 2019). Thus, defective TE-bearing NPV genomes may be more likely to be replicated than defective TE-bearing GVs, which may explain the trend we observe. In fact, many NPVs are known to be transmitted as multicapsid virions, whereas other large dsDNA viruses, including iridoviruses, are transmitted as monocapsid virions. This may in part explain why no A. vulgare and P. dilatatus TE and only few C. partellus TEs were found in iridovirus genomes. If true, NPVs would stand out as potentially being more efficient HTT vectors than other viruses. In turn, given that NPVs mainly infect lepidopterans (Goulson 2003; Thézé et al. 2018), their high propensity to shuttle TEs may explain why lepidopterans are seemingly more prone to HTT than other insects, as proposed by Reiss et al. (2019). However, the trend underlined above indicating a higher frequency of virus-borne TEs in NPVs than in GVs should be interpreted cautiously as it relies on a limited number of viruses. A more thorough assessment of the higher tolerance of multicapsid viruses to TEs should include single-capsid NPVs (such as the Helicoverpa zea nucleopolyhedrovirus for example). And in fact, the frequency of viral genomes carrying a TE is as high as or even higher in IIV6 purified from Drosophila S2 cells or whole flies (from 3.6% to 6%) than in NPVs, which contradicts the view that the mono-nucleocapsid nature of iridoviruses would dampen their ability to carry TEs. In this context, it is noteworthy that IIV6, which was originally sampled from the moth C. fumiferana (Fukaya and Nasu 1966), is not known to naturally infect flies. Thus, this artificial host–virus system involving a passage in cell culture and infection of whole flies by injection may have created favorable conditions to transposition and persistence of fly TEs in IIV6. In any case, our results call for future studies specifically dedicated to deciphering the relative contribution of host and virus factors involved in shaping transposition of host TEs into viral genomes, as well as TE persistence in viral populations over multiple replication rounds.
Persistence of Viral-Borne TEs over Two Successive Replication Cycles in Different Hosts
Importantly, this study provides direct evidence that TEs from a given insect species integrated into genomes of a viral population can be recovered after subsequent purification of the same virus from another species. Our previous study (Gilbert et al. 2016) showed that the number and frequency of TEs could be similar in a parental (G0) AcMNPV population replicated in one host (T. ni) and its daughter populations (G10) separated from G0 by ten successive infection cycles on another host (S. exigua). However, no TE was shared between the two populations (Gilbert et al. 2016). This result revealed a continuous dynamics of gain (via transposition) and loss (via drift and/or purifying selection) of TEs in AcMNPV populations and showed that the persistence of a given TE community in AcMNPV populations was less than ten infection cycles. Here, we found D. melanogaster TEs integrated in IIV6 genomes purified from two moths (C. partellus and S. nonagrioides) and another fly (D. hydei). We also recovered TEs present in the AcMNPV G0 data set in AcMNPV genomes purified from S. nonagrioides larvae. This demonstrates that TE-bearing viral genomes can be encapsidated, as shown earlier for the TCp3.2 and TCl4.7 TE in CpGV mutants MCp4 and MCp5, respectively (Jehle et al. 1995, 1998), and that persistence of virus-borne TEs over at least few infection cycles may be common among large dsDNA viruses. Interestingly, we observe large differences in TE diversity and number of viral genomes bearing TE insertions after one replication cycle of the same starting viral population in different hosts. For example, although the frequency of TE-bearing genomes in the IIV6 population purified from D. hydei (3.37%) is close to that of the parental IIV6 population purified from S2 cells (4.33%), it dropped down to 1.19% and 0.45% in IIV6 purified from C. partellus and S. nonagrioides, respectively. Furthermore, although many TEs originating from the genome of S2 cells were recovered in IIV6 purified from D. hydei, C. partellus and S. nonagrioides, only 2 out of 29 TEs initially found in AcMNPV purified from T. ni persisted during replication of this virus in S. nonagrioides. Such differences may be due to host–virus interactions other than host-to-virus transposition. Interstingly, the two transposon-carrying CpGV mutants identified in Jehle et al. (1995, 1998) showed similar virulence parameters, in terms of median lethal concentration and survival time, and virus progeny production as the wild-type (wt) CpGV, but they were efficiently out-competed by the wt-CpGV, when host larvae or cultured host cells were co-infected with one of the mutants and wt-CpGV (Arends et al. 2005). It would be interesting to further evaluate the role played by the host in shaping TE content and frequency in viral population in more controlled viral replication assays.
Virus-to-Virus Transposition
Interestingly, this study also showed that most of the TE insertions present in AcMNPV genomes purified from S. nonagrioides were generated by virus-to-virus transposition. This remarkable observation shows that virus-borne TEs can transpose in new genomes (here a virus), even if the recipient species is distantly related to the host from which the TEs originated (S. nonagrioides and T. ni diverged 60 Ma [Toussaint et al. 2012]). It confirms and extends earlier findings based on long-read sequencing showing that TEs can integrate as full-length copies into AcMNPV genomes and are thus likely able to transpose from the virus genome to another DNA molecule (Loiseau et al. 2020). Our results further indicate that only some of the TEs found in a given viral population may undergo measurable virus-to-virus transposition during subsequent infection cycles. Indeed, strong evidence of virus-to-virus transposition in AcMNPV purified from S. nonagrioides was found for only one TE (Tni_contig_27) out of the 29 present in the AcMNPV G0 population purified from T. ni. It is noteworthy that this TE is the second most frequent autonomous TE in the AcMNPV G0 population, being present in 0.38% of AcMNPV genomes (Gilbert et al. 2016). This suggests that the frequency of virus-to-virus transposition events involving a TE may depend on the TE frequency in the infecting viral population. Tni_contig_27 is the only TE present in the G0 population for which a single insertion underwent a sharp increase in frequency during replication in S. nonagrioides larvae. We speculate that the increase of the Tni_contig_27 insertion located in the Ac-GTA gene may not result from transposition. Instead, the frequency of this insertion may have increased through viral replication that may be driven by positive selection and/or by a higher propensity of viral genomes bearing this insertion to be encapsidated. In turn, the large number of Tni_contig_27 copies present in the viral population could have allowed this TE to reach a high enough rate of virus-to-virus transposition to be detectable by our approach. If so, many other TEs may have transposed between viral genomes at similar rates per TE copy, but these events would have been too rare to be detected as all these TEs were hundreds to thousands of times less frequent than Tni_contig_27 in the viral population. In any case, many TEs, host and virus factors are likely to shape the likelihood for a TE to undergo virus-to-virus transposition.
Altogether, our results suggest that viruses may not only serve as launching platforms for TEs to colonize naive cellular genomes (a hypothesis that still needs to be formally tested), but they may also be viewed as additional niches in which TEs could persist through time and evolve under different constraints than in cellular hosts. This is reminiscent of the molecular symbiosis hypothesis proposed by Filée (2018) to characterize the intricate interactions occurring between various types of mobile genetic elements and giant viruses infecting single-cell eukaryotes.
Materials and Methods
Infection of D. melanogaster S2 Cells with IIV6
The IIV6 viral strain used to infect D. melanogaster S2 cells is the one originally described in Fukaya and Nasu (1966). S2 cells were collected from two confluent T75 flasks and transferred into 50-ml tubes (fig. 1). The cells were counted and 2 × 108 cells were transferred to a fresh 50-ml tube. The cells were pelleted at 500 × g for 10 min at room temperature. Then, 5 ml of an IIV6 suspension was added to the cell pellet at a multiplicity of infection of 0.01. Cells were kept under the hood for 1 h with gentle inversions every 10 min so that the cells stayed in suspension. After 1 h, 1.25 ml (i.e., 0.5 × 108 cells) was added into each of the four T75 flasks containing 13.5 ml of complete medium. The cells were incubated at 25 °C for 5 days. Cells were detached by pipetting up and down with a 10-ml pipet and transferred into a 50-ml tube. The cells were broken down by three cycles of freezing at −20 °C and thawing at 37 °C. Cell debris were pelleted by centrifugation at 5,000 × g for 30 min at 4 °C. Supernatant was poured into an ULTRACLEAR tube (Beckman #361706) and underlaid with 1.5 ml of 30% (wt/wt) sucrose in 50 mM HEPES, pH 7, 0.1% BSA. Viral particles were pelleted at 35,000 × g for 90 min at 4 °C. The resulting virus pellet, showing the characteristic blue opalescence of insect iridoviruses, was resuspended in 300–400 µl 10 mM Tris-HCl buffer (pH 7.2), transferred to an Eppendorf tube and debris were pelleted by centrifugation for 5 min at 3,800 × g at 4 °C. The supernatant was aliquoted, the viral titer determined (4.42 × 1010 PFUs/ml) and this stock suspension was stored at −80 °C. Viral DNA was then extracted from an aliquot of this suspension using the QIAamp DNA Mini Kit (Qiagen).
Infection of D. melanogaster and D. hydei Flies with IIV6
Drosophila melanogaster and D. hydei flies were infected by intrathoracic injection of 20 nl of a 1/40 dilution of the IIV6 stock suspension purified from Drosophila S2 cells (see above and fig. 1). Injections were performed with a nanoject II nano injector. The flies were monitored for 2 weeks. The abdomen of flies in which the virus successfully replicated typically turned iridescent blue 10–15 days postinfection (supplementary fig. S1, Supplementary Material online). Infected flies were frozen in 1.5-ml Eppendorf tubes. For each fly species, viral particles were purified from a pool of 80 infected individuals. The 80 individuals were first grinded with a plastic pestle in Tris-HCl buffer (10 mM; pH 7.2) and two 5-min centrifugation steps at 500 × g were performed to eliminate most of host cells and tissues. Then, an ultracentrifugation step on sucrose cushion was performed at 35,000 × g for 90 min at 4 °C as described above. The pellet was resuspended in 100 µl of Tris-HCl buffer and viral DNA was extracted using the QIAamp DNA Mini Kit (Qiagen).
Infections of C. partellus with IIV6
Ten fourth instar C. partellus larvae were infected with the IIV6 stock suspension purified from Drosophila S2 cells (see above and fig. 1). Larvae were pricked using a thin needle soaked in the viral suspension. Fourteen days later, the larvae presented a purple iridescence and they finally died about 4 weeks after infection. Upon host death, viral particles were filtered through cheesecloth and two centrifugation steps were performed to eliminate most of host cells and tissues. Virus purification and DNA extraction were performed as for IIV6 in Drosophila flies (see above).
Infections of A. vulgare and P. dilatatus with IIV31
The IIV31 virus used to infect A. vulgare and P. dilatatus isopods was obtained through grinding a piece of cuticle from a naturally infected A. vulgare individual collected on the campus of the University of California Riverside (same viral isolate as in Loiseau et al. [2020]). Three P. dilatatus and two A. vulgare individuals were pricked with a thin needle soaked in the virus suspension (fig. 1). Four weeks postinfection, bluish dead isopods were individually placed and crushed in a 1.5-ml tube in a Tris-HCl buffer (10 mM; pH 7.2). Virus purification and DNA extraction were performed as for IIV6 in Drosophila flies (see above).
Infection of S. nonagrioides with AcMNPV
The AcMNPV-WP10 isolate (Chateigner et al. 2015) was used to infect ten fourth instar larvae of S. nonagrioides using the diet plug method (Sparks et al. 2008) (fig. 2). Each moth larva was fed ≈100,000 occlusion bodies (OBs) per 5-mm3 diet plug. Upon host death, which occurred 2–5 days postinfection, OBs were first filtered through cheesecloth, purified twice by centrifugation (10 min at 4,500 × g) with 0.1% sodium dodecyl sulfate, then distilled water, and finally resuspended in water. Approximately 1.5 × 1010 OBs were treated as described in Gilbert et al. (2014) to provide about 5 µg of high-quality dsDNA.
Infection of C. pomonella Larvae with CpGV
A total of 15 sequenced isolates from the CpGV collection of the Institute for Biological Control, Julius Kühn-Institut, Federal Research Centre for Cultivated Plants, Germany, were analyzed (fig. 2). The data sets included laboratory propagated CpGV strains originating from Mexico (CpGV-M) and Canada (CpGV-S) (Wennmann et al. 2020), England (CpGV-E2) and Iran (CpGV-I12 and -I0X) (Fan, Jehle, et al. 2020), as well as from China (CpGV-ALE, -JQ, -KS1, -KS2, -WW, -ZY and –ZY2) (Fan et al. 2019; Fan, Jehle, et al. 2020; Fan, Wennmann, et al. 2020). The isolates CpGV-0006, -V15, and -R5 were derived from commercial CpGV products used for codling moth control (Gueli Alletti, Sauer et al. 2017; Fan, Jehle, et al. 2020; Fan, Wennmann, et al. 2020). Stocks of CpGV OBs were obtained from propagation in third to fourth instar codling moth larvae or isolated from commercial products.
Infection of A. segetum Larvae with AgseGV
A previously sequenced AgseGV (isolate DA) was analyzed (Gueli Alletti, Eigenbrod, et al. 2017). Briefly, third to fourth instar A. segetum larvae were starved overnight and were fed the subsequent day with a small cube of artificial diet that contained 106 OBs of AgseGV (Gueli Alletti, Eigenbrod, et al. 2017). Larvae that consumed the entire piece within 12 h were transferred to virus-free diet and were checked daily for symptoms of viral infection. Larvae deceased by viral infection were collected and stored at −20 °C until virus purification. OB purification and DNA extraction were performed as described previously (Gueli Alletti, Eigenbrod, et al. 2017).
Serial Infections of AgseNPV-B in A. ipsilon Cell Line
The viral isolate AgseNPV-B PP2 was generated as a plaque purified clone (PP2) from an AgseNPV-B stock that was originally propagated in A. segetum (Gueli Alletti et al. 2018) (fig. 2). AgseNPV-B was then serially passaged six times (Gueli Alletti et al. 2018) in the A. ipsilon AiE1611T cell line (Harrison and Lynn 2008). For each serial infection, 4 × 104 cells/cm2 were infected with a multiplicity of infection of 1 pfu per cell starting with AgseNPV-B PP2 as the initial passage (PP2 #0). The virus-treated cells were incubated for 1 week at 26 °C. After the initial passage, the virus was used for another ten subsequent infections (PP2 #1 to #10). After each serial infection, the tissue culture infective dose (TCID50) was determined as described in O’Reilly et al. (1992). Viruses from passages #1, #3, #5, #7, and #10 OBs were harvested, purified, and had their DNA extracted (Gueli Alletti, Wennmann, Berner, Keilwagen, Jehle, unpublished data).
Sequencing of Viral Genomes
Viral genomes were sequenced in three batches. The first batch included the AcMNPV sample purified from S. nonagrioides and the IIV6 samples purified from D. hydei and D. melanogaster flies, as well as from D. melanogaster S2 cells. One microgram of human DNA was added to the 2 µg of IIV6 purified from S2 cells DNA before sequencing to characterize artificial chimeras involving human TEs and the IIV6 genome (see below). These samples were sequenced by Novogen. A library was constructed for each sample with the NEBnext kit (average insert size was 350 bp), which was sequenced on a HiSeqX instrument in 2 × 150 bp mode, generating 113.9–590.0 million paired-end reads. The second batch included the IIV6 samples purified from S. nonagrioides and from C. partellus, as well as the six IIV31 samples purified from three P. dilatatus and three A. vulgare individuals. These samples were sequenced by Génome Québec. A library was constructed for each sample with the NEB ultra II kit (average insert size was 260 bp), which was sequenced on a HiSeqX instrument in 2 × 150 bp paired-end mode, generating 246.1–330.9 million paired-end reads. The third batch included the 15 isolates of CpGV, the AgseGV-DA isolate, the AgseNPV-B PP2 isolate (= #0) and its five serial infections (#1, #3, #5, #7, and #10). Sequencing was performed on 50–100 ng purified viral DNA. Libraries were generated using a NexteraXT library preparation kit and sequencing was conducted with an Illumina NextSeq500 system (StarSEQ Ltd., Mainz, Germany) generating 0.3–10.9 million paired-end 150-bp-long reads (Gueli Alletti, Sauer, et al. 2017; Fan, Jehle, et al. 2020; Wennmann et al. 2020).
Detection of TEs Integrated in Viral Genomes
The aim of this study was to characterize as comprehensively as possible the diversity of host TEs that can become integrated into viral genomes. To this end, we searched for TE–virus junctions in sequencing reads using the approach developed by Gilbert et al. (2016). Reads were aligned separately on TE sequences and the viral genome using BlastN (-task megablast). Chimeric reads for which a portion aligned on a TE only and the other portion aligned on the viral genome only were then identified based on alignment coordinates. The R scripts used to detect and filter chimeric reads are provided in supplementary files S2 and S3, Supplementary Material online. Though artificial chimeras are certainly present in the various data sets analyzed here, several lines of evidence indicate that chimeras retained for counting and calculating the frequencies of TEs integrated into viral genomes are not artificial and result from bona fide integration that took place during replication of the virus. First, we considered that a TE was integrated into a viral genome only if we found at least three chimeric reads between a given TE and the virus (they were not required to map to the same position along the viral genome). Second, canonical transposition of a TE into a viral genome is expected to generate junction mapping in most cases at the very extremities of the TE sequences, that is, not anywhere along the TE sequence (Craig et al. 2015). Thus, if transposition into viral genomes occurred, chimeras covering the very extremities of the TE sequences should outnumber chimeras mapping internally to the TE sequences. This prediction was verified for all TEs (fig. 3, supplementary fig. S2 and table S1, Supplementary Material online). Third, the addition of human DNA to the IIV6 DNA extracted from S2 cells as a control to estimate the presence of artificial chimeras allowed us to validate our approach by showing that contrary to Drosophila TEs, artificial chimeras involving human TEs mapped almost exclusively internally to human TE sequences (see below). Fourth, many TEs are known to duplicate a small target sequence motif upon integration called target site duplication (TSD). As in our earlier study (Gilbert et al. 2016), we were able to identify TSD motifs for several TEs found integrated into viral genomes (fig. 3). Viral and TE sequences on which sequencing reads were aligned derive from WGSs and TE library that are fully described below. All moth TEs found integrated into viral genomes are provided in supplementary data 1, Supplementary Material online.
Viral Genomes Used for BLAST Searches
Genome sequences of four out of six viruses used in this study were retrieved from NCBI under accession number KR584663 (AgseGV-DA), KM102981 (AgseNPV-B), KM217575 (CpGV-M), and KM667940.1 (AcMNPV strain E2). For IIV31, we used the genome provided in the supplementary data of Loiseau et al. (2020). The IIV6 genome sequence available in NCBI was generated after replication on CF-124 cells of the C. fumiferana moth (Fukaya and Nasu 1966). As moths are distantly related to Drosophila flies, we reasoned that adaptation of IIV6 to S2 cells may have resulted in changes in its genome. We thus assembled the IIV6 genome de novo with the tadpole program (https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/tadpole-guide/, version of December 2018, options used: “k = 17”; “k = 31”; “k = 60”; “k = 90” with “mincov = 100”). The different assemblies obtained with 17, 31, 60, and 90 mers with 500× and 1,500× depth coverage were then fused with Geneious version 11.0.2 (https://www.geneious.com, last accessed on july 2020, options: de novo assembly, Geneious assembler, high sensitivity). The final assembly was annotated based on the available IIV6 (NC_003038.1) genome with the General Annotation Transfer Utility program (Tcherepanov et al. 2006).
TE Library
The TE library included all TE consensus sequences downloaded from Repbase in October 2017 (Bao et al. 2015), all those identified in 195 insect genomes by Peccoud et al. (2017) as well as all TEs identified integrated in the AcMNPV genome after replication of the virus onto T. ni and S. exigua moths (Gilbert et al. 2016). The library also included all D. melanogaster TE sequences downloaded from Flybase and all A. vulgare TE consensus sequences characterized in Chebbi et al. (2019). In addition, our TE library was complemented with the TE consensus sequences constructed using RepeatModeler2 (Flynn et al. 2020) from the WGS of A. ipsilon (NCBI accession number: PNFC00000000.1), C. suppressalis (PNFC00000000.1), C. pomonella (QFTL00000000.2), D. hydei (QMEQ00000000.2), and S. nonagrioides (Muller et al. 2021). No WGS was available for A. segetum and C. partellus moths and the P. dilatatus isopod. To increase our chances to identify TEs in viral genomes, the sequencing reads obtained from A. segetum and C. partellus infections were also aligned on all lepidopteran WGS available in LepBase (as of May 17, 2018) as well as on all Noctuidae WGS and transcriptomes available in NCBI (as of October 15, 2017). In addition, for A. segetum, C. partellus, and P. dilatatus, we aligned reads on contigs that we de novo assembled using all sequencing reads not mapping onto viral genomes, that is, reads resulting from sequencing residual host and nonviral/nonhost DNA that remained in the virus suspension after purification. Nonviral contig assembly was performed with the Tadpole assembler from the BBMap tools. Residual viral chunks were filtered out from the resulting contigs. All 248,482 TEs included in our library or characterized in partial assemblies of A. segetum, C. partellus, and P. dilatatus are provided in supplementary file S1, Supplementary Material online.
Inferring HTT between Insects
To assess whether moth TEs found integrated into viral genomes underwent HT during insect evolution, we used them as queries to perform BlastN searches (option megablast) on all 352 insect WGS available in GenBank as of March 2020. BlastN hits showing >90% nucleotide identity to the TE over >200 bp and >80% of its length were considered further. To assess whether such high levels of TE identity were due to vertical or horizontal transmission, we compared TE synonymous distances (dS) with the distribution of dS expected to occur under vertical transmission between each species pair of interest. The dS between TEs were calculated over the longest open reading frame found in the two copies involved in the best alignment for each species pair of interest. The distribution of dS expected under vertical transmission was generated for each species pair of interest by calculating dS between all single-copy and complete genes extracted from the WGS of each species using BUSCO version 4.0.2 (Simão et al. 2015; Waterhouse et al. 2018). All genes showing homology to the Insecta gene set (i.e., from 259 to 1,029 genes depending on species pairs) were used. MAFFT version 7.310 (Katoh and Standley 2013) was used to align nucleotide sequences of genes that are shared (orthologous) between species. Gene and TE alignments were analyzed in R version 3.6.1 (R Core Team 2019) with the “seqinr” package (Charif et al. 2005) to compute dS values.
Frequency of TEs in Viral Populations
The frequency of genomes carrying TE insertions was computed following the method described in Gilbert et al. (2016). Briefly, we considered the mean number of insertions per virus genome to be the frequency of chimeras among viral reads divided by two—as one insertion yields two chimeras—and multiplied by the ratio of the virus genome length over the length of a read. The latter was shortened to consider the minimum alignment length needed by BLAST to output a hit (see Gilbert et al. [2016] for details). The portion of viral genomes carrying insertions was computed as the probability that the number of inserted TEs into a viral genome exceeds zero, assuming that this number follows a Poisson distribution parameterized with the mean number of insertions per viral genome. To compute the frequencies of viral genomes carrying individual insertions in the AcMNPV-S. nonagrioides data set, another approach was used considering insertions distant by 5 bp or fewer as single insertion points and dividing the number of chimeric reads resulting from these insertions by the total number of reads covering the position (R script available in supplementary data 2, Supplementary Material online).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by Agence Nationale de la Recherche Grants ANR-15-CE32-0011-01, TransVir, and ANR-18-CE02-0021-01TranspHorizon (to C.G.).
Data Availability
The data underlying this article are available in the GenBank Nucleotide Database at https://www.ncbi.nlm.nih.gov/genbank/ and can be accessed with BioProject accession number PRJNA692820. The library of TEs used in this study to search for TE copies integrated into viral genomes is provided in supplementary file S1, Supplementary Material online, available at https://doi.org/10.6084/m9.figshare.14776041.v1. The R scripts used to identify chimeric reads, make figure 4 and supplementary figure S2 as well as supplementary table S1, are provided as supplementary files S2 and S3, Supplementary Material online.
References
- Arends HM, Winstanley D, Jehle JA.. 2005. Virulence and competitiveness of Cydia pomonella granulovirus mutants: parameters that do not match. J Gen Virol. 86(10):2731–2738. [DOI] [PubMed] [Google Scholar]
- Bao W, Kojima KK, Kohany O.. 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomé C, Bello X, Maside X.. 2009. Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol. 10(2):R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becking T, Gilbert C, Cordaux R.. 2020. Impact of transposable elements on genome size variation between two closely related crustacean species. Anal Biochem. 600:113770. [DOI] [PubMed] [Google Scholar]
- Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, Imbeault M, Izsvák Z, Levin HL, Macfarlan TS, et al. 2018. Ten things you should know about transposable elements. Genome Biol. 19(1):199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens EB. 1987. Identification and nucleotide sequence of the regions of Autographa californica nuclear polyhedrosis virus genome carrying insertion elements derived from Spodoptera frugiperda. Virology 161(1):8–17. [DOI] [PubMed] [Google Scholar]
- Charif D, Thioulouse J, Lobry JR, Perrière G.. 2005. Online synonymous codon usage analyses with the ade4 and seqinR packages. Bioinformatics 21(4):545–547. [DOI] [PubMed] [Google Scholar]
- Chateigner A, Bézier A, Labrousse C, Jiolle D, Barbe V, Herniou E.. 2015. Ultra deep sequencing of a baculovirus population reveals widespread genomic variations. Viruses 7(7):3625–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebbi MA, Becking T, Moumen B, Giraud I, Gilbert C, Peccoud J, Cordaux R.. 2019. The genome of Armadillidium vulgare (Crustacea, Isopoda) provides insights into sex chromosome evolution in the context of cytoplasmic sex determination. Mol Biol Evol. 36(4):727–741. [DOI] [PubMed] [Google Scholar]
- Chen W, Yang X, Tetreau G, Song X, Coutu C, Hegedus D, Blissard G, Fei Z, Wang P.. 2019. A high‐quality chromosome‐level genome assembly of a generalist herbivore. Mol Ecol Resour. 19(2):485–496. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA.. 2009. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 10(10):691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig NL, Chandler M, Gellert M, Lambowitz AM, Rice PA, Sandmeyer SB, editors. 2015. Mobile DNA III. Washington (DC: ): ASM Press. [Google Scholar]
- Daniels SB, Peterson KR, Strausbaugh LD, Kidwell MG, Chovnick A.. 1990. Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics 124(2):339–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto BR, Carvalho EL, da Silva AF, Dezordi FZ, Pinto PM, Campos TDL, Rezende AM, Wallau GDL. 2018. HTT-DB: new features and updates. Database 2018:bax102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckwahl MJ, Telesnitsky A, Wolin SL.. 2016. Host RNA packaging by retroviruses: a newly synthesized story. mBio. 7(1):e02025-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Baidouri M, Carpentier MC, Cooke R, Gao D, Lasserre E, Llauro C, Mirouze M, Picault N, Jackson SA, Panaud O.. 2014. Widespread and frequent horizontal transfers of transposable elements in plants. Genome Res. 24(5):831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Jehle JA, Wennmann JT.. 2020. Population structure of Cydia pomonellagranulovirus isolates revealed by quantitative analysis of genetic variation. Virus Evol. 6(2):veaa073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Wennmann JT, Wang D, Jehle JA.. 2019. Novel diversity and virulence patterns found in new isolates of Cydia pomonella granulovirus from China. Appl Environ Microbiol. 86:e02000-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Wennmann JT, Wang D, Jehle JA.. 2020. Single nucleotide polymorphism (SNP) frequencies and distribution reveal complex genetic composition of seven novel natural isolates of Cydia pomonellagranulovirus. Virology 541:32–40. [DOI] [PubMed] [Google Scholar]
- Filée J. 2018. Giant viruses and their mobile genetic elements: the molecular symbiosis hypothesis. Curr Opin Virol. 33:81–88. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF.. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 117(17):9451–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser MJ, Brusca JS, Smith GE, Summers MD.. 1985. Transposon-mediated mutagenesis of a baculovirus. Virology 145(2):356–361. [DOI] [PubMed] [Google Scholar]
- Fraser MJ, Cary L, Boonvisudhi K, Wang HG.. 1995. Assay for movement of Lepidopteran transposon IFP2 in insect cells using a baculovirus genome as a target DNA. Virology 211(2):397–407. [DOI] [PubMed] [Google Scholar]
- Fukaya M, Nasu S.. 1966. A Chilo iridescent virus (CIV) from the rice stem borer, Chilo suppressalis walker (Lepidoptera: Pyralidae). Appl Entomol Zool. 1(2):69–72. [Google Scholar]
- Ghoshal K, Theilmann J, Reade R, Maghodia A, Rochon D.. 2015. Encapsidation of host RNAs by cucumber necrosis virus coat protein during both agroinfiltration and infection. J Virol. 89(21):10748–10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Chateigner A, Ernenwein L, Barbe V, Bézier A, Herniou EA, Cordaux R.. 2014. Population genomics supports baculoviruses as vectors of horizontal transfer of insect transposons. Nat Commun. 5:3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Cordaux R.. 2017. Viruses as vectors of horizontal transfer of genetic material in eukaryotes. Curr Opin Virol. 25:16–22. [DOI] [PubMed] [Google Scholar]
- Gilbert C, Feschotte C.. 2018. Horizontal acquisition of transposable elements and viral sequences: patterns and consequences. Curr Opin Genet Dev. 49:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Peccoud J, Chateigner A, Moumen B, Cordaux R, Herniou EA.. 2016. Continuous influx of genetic material from host to virus populations. PLoS Genet. 12(2):e1005838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Schaack S, Pace JK, Brindley PJ, Feschotte C.. 2010. A role for host-parasite interactions in the horizontal transfer of transposons across phyla. Nature 464(7293):1347–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulson D. 2003. Can host susceptibility to baculovirus infection be predicted from host taxonomy or life history? Environ Entomol. 32(1):61–70. [Google Scholar]
- Gueli Alletti G, Carstens EB, Weihrauch B, Jehle JA.. 2018. Agrotis segetum nucleopolyhedrovirus but not Agrotis segetum granulovirus replicate in AiE1611T cell line of Agrotis ipsilon. J Invertebr Pathol. 151:7–13. [DOI] [PubMed] [Google Scholar]
- Gueli Alletti G, Eigenbrod M, Carstens EB, Kleespies RG, Jehle JA.. 2017. The genome sequence of Agrotis segetum granulovirus, isolate AgseGV-DA, reveals a new Betabaculovirus species of a slow killing granulovirus. J Invertebr Pathol. 146:58–68. [DOI] [PubMed] [Google Scholar]
- Gueli Alletti G, Sauer A, Weihrauch B, Fritsch E, Undorf-Spahn K, Wennmann J, Jehle J.. 2017. Using next generation sequencing to identify and quantify the genetic composition of resistance-breaking commercial isolates of Cydia pomonella granulovirus. Viruses 9(9):250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Gao J, Li F, Wang J.. 2014. Evidence of horizontal transfer of non-autonomous Lep1 Helitrons facilitated by host-parasite interactions. Sci Rep. 4:5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M-J, Zhou Q-Z, Zhang H-H, Tong X, Lu C, Zhang Z, Dai F.. 2016. iMITEdb: the genome-wide landscape of miniature inverted-repeat transposable elements in insects. Database 2016:baw148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RL, Lynn DE.. 2008. New cell lines derived from the black cutworm, Agrotis ipsilon, that support replication of the A. ipsilon multiple nucleopolyhedrovirus and several group I nucleopolyhedroviruses. J Invertebr Pathol. 99(1):28–34. [DOI] [PubMed] [Google Scholar]
- Ivancevic AM, Kortschak RD, Bertozzi T, Adelson DL.. 2018. Horizontal transfer of BovB and L1 retrotransposons in eukaryotes. Genome Biol. 19(1):85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehle JA, Fritsch E, Nickel A, Huber J, Backhaus H.. 1995. TCl4.7: a novel lepidopteran transposon found in Cydia pomonella granulosis virus. Virology 207(2):369–379. [DOI] [PubMed] [Google Scholar]
- Jehle JA, Nickel A, Vlak JM, Backhaus H.. 1998. Horizontal escape of the novel Tc1-like lepidopteran transposon TCp3.2 into Cydia pomonella granulovirus. J Mol Evol. 46(2):215–224. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuma S, Fujii T, Kawaoka S, Shimada T.. 2008. Bombyx mori nucleopolyhedrovirus SNF2 global transactivator homologue (Bm33) enhances viral pathogenicity in B. mori larvae. J Gen Virol. 89(Pt 12):3039–3046. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Sanchez Calle A, Yamamoto Y, Sato T-A, Ochiya T.. 2019. Extracellular vesicles mediate the horizontal transfer of an active LINE-1 retrotransposon. J Extracell Vesicles. 8(1):1643214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Nolte V, Schlötterer C.. 2015. Tempo and mode of transposable element activity in Drosophila. PLoS Genet. 11(7):e1005406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB.. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34(7):1812–1819. [DOI] [PubMed] [Google Scholar]
- Kuraku S, Qiu H, Meyer A.. 2012. Horizontal transfers of Tc1 elements between teleost fishes and their vertebrate parasites, lampreys. Genome Biol Evol. 4(9):929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau V, Herniou EA, Moreau Y, Lévêque N, Meignin C, Daeffler L, Federici B, Cordaux R, Gilbert C.. 2020. Wide spectrum and high frequency of genomic structural variation, including transposable elements, in large double-stranded DNA viruses. Virus Evol. 6:vez060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreto ELS, Carareto CMA, Capy P.. 2008. Revisiting horizontal transfer of transposable elements in Drosophila. Heredity (Edinb). 100(6):545–554. [DOI] [PubMed] [Google Scholar]
- Miller DW, Miller LK.. 1982. A virus mutant with an insertion of a copia-like transposable element. Nature 299(5883):562–564. [DOI] [PubMed] [Google Scholar]
- Muller H, Ogereau D, Da-Lage JL, Capdevielle C, Pollet N, Fortuna T, Kaiser JR, Gilbert L.. 2021. Draft nuclear genome and complete mitogenome of the Mediterranean corn borer, Sesamia nonagrioides, a major pest of maize. G3 (Bethesda) 7:jkab155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono R, Yasuhiko Y, Aisaki K-I, Kitajima S, Kanno J, Hirabayashi Y.. 2019. Exosome-mediated horizontal gene transfer occurs in double-strand break repair during genome editing. Commun Biol. 2:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly DR, Miller L, Luckow VA.. 1992. Baculovirus expression vectors: a laboratory manual. New York: W.H. Freeman. [Google Scholar]
- Peccoud J, Cordaux R, Gilbert C.. 2018. Analyzing horizontal transfer of transposable elements on a large scale: challenges and prospects. BioEssays 40(2):1700177. [DOI] [PubMed] [Google Scholar]
- Peccoud J, Loiseau V, Cordaux R, Gilbert C.. 2017. Massive horizontal transfer of transposable elements in insects. Proc Natl Acad Sci U S A. 114(18):4721–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. Vienna (Austria: ): R Foundation for Statistical Computing. Available from: https://www.R-project.org/. [Google Scholar]
- Reiss D, Mialdea G, Miele V, de Vienne DM, Peccoud J, Gilbert C, Duret L, Charlat S.. 2019. Global survey of mobile DNA horizontal transfer in arthropods reveals Lepidoptera as a prime hotspot. PLoS Genet. 15(2):e1007965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrmann GF. 2019. Baculovirus molecular biology. 4th ed. Bethesda (MD: ): National Center for Biotechnology Information (US; ). [PubMed] [Google Scholar]
- Routh A, Domitrovic T, Johnson JE.. 2012. Host RNAs, including transposons, are encapsidated by a eukaryotic single-stranded RNA virus. Proc Natl Acad Sci U S A. 109(6):1907–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaack S, Gilbert C, Feschotte C.. 2010. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. 25(9):537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JC, Loreto EL, Clark JB.. 2004. Factors that affect the horizontal transfer of transposable elements. Curr Issues Mol Biol. 6(1):57–71. [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Sparks W, Li H, Bonning B.. 2008. Protocols for oral infection of lepidopteran larvae with baculovirus. J Vis Exp. (19):888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh A, Witt CC, Menger J, Sadanandan KR, Podsiadlowski L, Gerth M, Weigert A, McGuire JA, Mudge J, Edwards SV, et al. 2016. Ancient horizontal transfers of retrotransposons between birds and ancestors of human pathogenic nematodes. Nat Commun. 7:11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Feschotte C, Wu Z, Mueller RL.. 2015. DNA transposons have colonized the genome of the giant virus Pandoravirus salinus. BMC Biol. 13:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talsania K, Mehta M, Raley C, Kriga Y, Gowda S, Grose C, Drew M, Roberts V, Cheng KT, Burkett S, et al. 2019. Genome assembly and annotation of the Trichoplusia ni Tni-FNL insect cell line enabled by long-read technologies. Genes 10(2):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherepanov V, Ehlers A, Upton C.. 2006. Genome Annotation Transfer Utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genomics 7:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesnitsky A, Wolin SL.. 2016. The host RNAs in retroviral particles. Viruses 8(8):235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thézé J, Lopez-Vaamonde C, Cory JS, Herniou EA.. 2018. Biodiversity, evolution and ecological specialization of baculoviruses: a treasure trove for future applied research. Viruses 10 (7):366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint EFA, Condamine FL, Kergoat GJ, Capdevielle-Dulac C, Barbut J, Silvain J-F, Le Ru BP.. 2012. Palaeoenvironmental shifts drove the adaptive radiation of a noctuid stemborer tribe (Lepidoptera, Noctuidae, Apameini) in the miocene. PLoS One 7(7):e41377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G, D'Angelo G, Raposo G.. 2018. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 19(4):213–228. [DOI] [PubMed] [Google Scholar]
- Wallau GL, Ortiz MF, Loreto ELS.. 2012. Horizontal transposon transfer in Eukarya: detection, bias, and perspectives. Genome Biol Evol. 4(8):801–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh AM, Kortschak RD, Gardner MG, Bertozzi T, Adelson DL.. 2013. Widespread horizontal transfer of retrotransposons. Proc Natl Acad Sci U S A. 110(3):1012–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM.. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 35(3):543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennmann JT, Fan J, Jehle JA.. 2020. Bacsnp: using single nucleotide polymorphism (SNP) specificities and frequencies to identify genotype composition in baculoviruses. Viruses 12(6):625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T, Barbosa‐Solomieu V, Chinchar VG.. 2005. A decade of advances in iridovirus research. Adv Virus Res. 65:173–248.. [DOI] [PubMed] [Google Scholar]
- Zhang H-H, Peccoud J, Xu M-R-X, Zhang X-G, Gilbert C.. 2020. Horizontal transfer and evolution of transposable elements in vertebrates. Nat Commun. 11:1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the GenBank Nucleotide Database at https://www.ncbi.nlm.nih.gov/genbank/ and can be accessed with BioProject accession number PRJNA692820. The library of TEs used in this study to search for TE copies integrated into viral genomes is provided in supplementary file S1, Supplementary Material online, available at https://doi.org/10.6084/m9.figshare.14776041.v1. The R scripts used to identify chimeric reads, make figure 4 and supplementary figure S2 as well as supplementary table S1, are provided as supplementary files S2 and S3, Supplementary Material online.