Abstract
Disease susceptibility and resistance are important factors for the conservation of endangered species, including elephants. We analyzed pathology data from 26 zoos and report that Asian elephants have increased neoplasia and malignancy prevalence compared with African bush elephants. This is consistent with observed higher susceptibility to tuberculosis and elephant endotheliotropic herpesvirus (EEHV) in Asian elephants. To investigate genetic mechanisms underlying disease resistance, including differential responses between species, among other elephant traits, we sequenced multiple elephant genomes. We report a draft assembly for an Asian elephant, and defined 862 and 1,017 conserved potential regulatory elements in Asian and African bush elephants, respectively. In the genomes of both elephant species, conserved elements were significantly enriched with genes differentially expressed between the species. In Asian elephants, these putative regulatory regions were involved in immunity pathways including tumor-necrosis factor, which plays an important role in EEHV response. Genomic sequences of African bush, forest, and Asian elephant genomes revealed extensive sequence conservation at TP53 retrogene loci across three species, which may be related to TP53 functionality in elephant cancer resistance. Positive selection scans revealed outlier genes related to additional elephant traits. Our study suggests that gene regulation plays an important role in the differential inflammatory response of Asian and African elephants, leading to increased infectious disease and cancer susceptibility in Asian elephants. These genomic discoveries can inform future functional and translational studies aimed at identifying effective treatment approaches for ill elephants, which may improve conservation.
Keywords: elephants; EEHV, tuberculosis, genomes; cancer; conservation; disease
Introduction
Elephants (family Elephantidae) first appear in the fossil record ∼5 − 10 million years ago (Ma), and three species roam today: the African bush elephant (Loxodonta africana), the African forest elephant (L. cyclotis), and the Asian elephant (Elephas maximus). These species are the only surviving members of the once diverse proboscidean clade of afrotherian mammals (Shoshani 1998). Straight-tusked elephants (genus Paleoloxodon) and mammoths (Mammuthus) went extinct around 34,000 and 4,300 years ago, respectively (Vartanyan et al. 1993; Stuart 2005). All remaining elephant species are considered endangered and threatened with extinction (Williams et al. 2020; Gobush, Edwards, Balfour, et al. 2021; Gobush, Edwards, Maisels, et al. 2021). A recent assessment of forest elephants concluded that they are critically endangered (Gobush, Edwards, Balfour, et al. 2021; Gobush, Edwards, Maisels, et al. 2021). Pressures faced in the wild by elephants include poaching and habitat loss, and elephants under human care are threatened by infectious disease (Greenwald et al. 2009; Long et al. 2016). The broad scale decline of these charismatic megafauna requires a combination of sustained conservation efforts and improved veterinary care in order to maintain healthy breeding populations.
The uniqueness of elephants stem from their many identifiable traits: their prehensile trunks, ivory tusks, intelligence with long-term memory, and the fact that they are among the largest of terrestrial mammals both living and extinct (Shoshani 1998; Larramendi 2015). Given their long lifespans of ∼80 years for Asian (Chapman et al. 2019) and ∼65 years for African elephants (Moss 2001), and their lengthy gestation periods of 22 months, disease defense is also an important trait for elephants, with urgent ramifications for elephant conservation (Greenwald et al. 2009; Long et al. 2016). For instance, Asian elephants are uniquely threatened by an acute hemorrhagic disease resulting from infection with elephant endotheliotropic herpesvirus (EEHV) (Long et al. 2016; Srivorakul et al. 2019). Although fatalities in African elephant calves from EEHV have been reported, mortality rates are higher for Asian elephants, suggesting a genetic component for increased EEHV lethality. Asian elephants also are more susceptible to tuberculosis (TB) infection (Mycobacterium tuberculosis and M. bovis) (Greenwald et al. 2009). Understanding the functional immunological and molecular basis of elephant disease response may improve their conservation through improved therapeutic interventions and veterinary care. However, the genomic mechanisms underlying disease defenses in elephants have not yet been studied in detail, even though genomics has been identified as a key tool for elephant conservation (Roca et al. 2015).
Elephants under human care and in the wild are also sometimes diagnosed with cancer, although elephant cancer mortality rates are low compared with humans (Abegglen et al. 2015; Boddy et al. 2020). Cancer defenses in elephants are thought to be mediated by an enhanced apoptotic response of elephant cells to DNA damage, associated with extensive amplification of retrogene copies of the tumor suppressor gene TP53 (Abegglen et al. 2015; Caulin et al. 2015; Sulak et al. 2016). Among extant elephant species, TP53 retrogenes have been studied in African bush and Asian elephants, but not forest elephants. It is therefore unclear if interspecific variation in TP53 retrogene copy numbers contributes to differences in cancer defenses between elephant species. Also, it is not known whether the observed differential responses to EEHV and TB between Asian and African elephants also hold with regard to their respective cancer susceptibilities. Detailed analyses of cancer prevalence and mortality in elephants may provide insights into how elephants evolved to handle disease.
Here, we analyze data from three living and two extinct elephant species in comparative and population genomic frameworks. We sequenced the genomes of two African bush and one Asian elephant and report new and/or improved genome assemblies for each species. Our goals were 1) to estimate changes in putative regulatory regions contributing to functional differences between Asian and African bush elephants; 2) to analyze TP53 retrogene copy numbers in both living and extinct elephant populations to understand their evolution and function; and 3) to determine regions of elephant genomes under selection and their associated genetic pathways. We identified genomic regions under selection in elephants related to many iconic elephant traits including cancer defenses, and annotated functional genetic differences between Asian and African elephants enriched in immunity pathways, particularly those related to elephant EEHV response. We also assess pathology reports from 26 zoos accredited by the Association of Zoos and Aquariums (AZA), including better documentation of cancer rates in elephants. The differential disease defense response inferred by our genomic and pathology analyses has implications for both the health and conservation of elephants.
Results and Discussion
Asian Elephants Suffer From Higher Rates of EEHV, TB, and Malignant Cancers Than African Elephants
We reanalyzed data from Greenwald et al. (2009) and found that in addition to increased susceptibility to EEHV compared with African elephants, Asian elephants were also significantly more vulnerable to TB infection (Fisher’s Exact Test, P = 2.52e−04; χ2 test, P = 6.84e−04; supplementary fig. 1, Supplementary Material online). To compare rates of neoplasia and malignancy between Asian and African elephants, we collected and analyzed pathology data from 26 AZA-accredited zoos in the USA, which included diagnoses from 76 different elephants (n = 35 African and n = 41 Asian). We found that 5.71% of the African elephants and 41.46% of the Asian elephants were diagnosed with neoplasia (which included benign and malignant tumors) (Fisher’s Exact Test, P = 3.78e−04; χ2 test, P = 8.95e−04) (table 1; supplementary table 1, Supplementary Material online).
Table 1.
Cancer Diagnoses and Prevalence in African and Asian Elephants.
| Neoplasia and Malignant Prevalence | |||||
|---|---|---|---|---|---|
| Species | Total Individuals | Neoplasia |
Malignant |
||
| Cases | % | Cases | % | ||
| Asian elephant | 41 | 17 | 41.46 | 6 | 14.63 |
| African elephant | 35 | 2 | 5.71 | 0 | 0 |
| Total elephants | 76 | 19 | 25 | 6 | 7.89 |
| Neoplasia Diagnoses in African/Asian Elephants | |||||
| Species | Sex | Age | Lesion Type | Lesion Site | Malignant |
| African elephant | F | 28 | Polyp | Vagina | N |
| African elephant | NA | NA | Adenoma | Parathyroid | N |
| Asian elephant | F | 40 | Polyp | Vagina | N |
| Asian elephant | F | 45 | Polyp | Vulva | N |
| Asian elephanta | F | 30 | Polyp | Vagina | N |
| 40 | Spindle cell tumor | Leg | N | ||
| Asian elephanta | F | 50 | Polyp | Uterus | N |
| 50 | Leiomyoma | Uterus | N | ||
| Asian elephant | F | 39 | Leiomyoma | Uterus | N |
| Asian elephant | F | 39 | Mast cell tumor | Abdomen | N |
| Asian elephant | M | 35 | Papilloma | Trunk | N |
| Asian elephant | F | 36 | Papilloma | Skin | N |
| Asian elephant | F | 50 | Adenocarcinoma | Breast | Y |
| Asian elephanta | F | 59 | Adenocarcinoma | Uterus | Y |
| 59 | Leiomyoma | Uterus | N | ||
| Asian elephant (7)b | NA | NA | Adenocarcinoma (2) | Uterus (2) | Y |
| Undifferentiated malignant neoplasm (1) | Uterus (1) | Y | |||
| Leiomyosarcoma (1) | Lung (1) | Y | |||
| Sarcoma (1) | Liver (1) | Y | |||
| Leiomyoma (4) | Uterus (4) | N | |||
| Leiomyoma (1) | Stomach (1) | N | |||
| Microadenoma (1) | Pituatary gland (1) | N | |||
Individual elephants diagnosed with multiple tumors.
These 7 elephants were diagnosed with 11 tumors. Information to link multiple tumors to individual elephants was not available for these elephants.
Sixty-nine percent of elephant tumors were benign, and 14.63% of Asian elephants were diagnosed with malignant tumors compared with zero in African elephants (Fisher’s Exact Test, P = 0.028; χ2 test, P = 0.053). In contrast, the lifetime risk of developing malignant cancer is 39.5% for humans (Howlander et al. 2020) and the lifetime risk of developing benign tumors is even higher, with 70 − 80% of women developing uterine fibroids (leiomyomas) alone (Zimmermann et al. 2012). Asian elephants are also reported to have a high prevalence of uterine leiomyomas (Montali et al. 1997), including 17% in our data set. Our results confirm that 1) malignant cancer rates in elephants are lower than in humans and 2) Asian elephants are diagnosed with both neoplasia and malignancies more often than African elephants in zoos.
Elephant-Specific Accelerated Genomic Regions Are Enriched for Immune Pathways and Correlate with Species-Specific Gene Expression Patterns
To explore the genomic mechanisms governing disease response and other traits across elephant species, we sequenced at 94× coverage and assembled the genome of an Asian elephant (“Icky,” born in Myanmar and currently under human care), with a final scaffold N50 of 2.77 Mb (GCA_014332765.1) (table 2; supplementary information, supplementary tables 2 and 3, Supplementary Material online). We also improved the African bush elephant genome assembly with Hi-C libraries (supplementary information, supplementary fig. 2 and table 4, Supplementary Material online). These genomic resources for elephants have been made publicly available.
Table 2.
Summary Statistics for the Asian Elephant Genome Assembly.
| Feature | Contigs | Scaffolds |
|---|---|---|
| Assembly length | 2.98 Gb | 3.13 Gb |
| Longest | 731 kb | 14.6 Mb |
| Number | 90,662 | 6,954 |
| N50 | 79.8 kb | 2.77 Mb |
| L50 | 10,736 | 336 |
| Percent genome in gaps | 0.09 | 4.88 |
| BUSCO results | C: 91.5% [D: 0.4%], F: 5.7%, M: 2.8%, n: 4,104 | |
BUSCO, Benchmarking Universal Single Copy Orthologs; C, complete; D, duplicated; F, fragmented; M, missing.
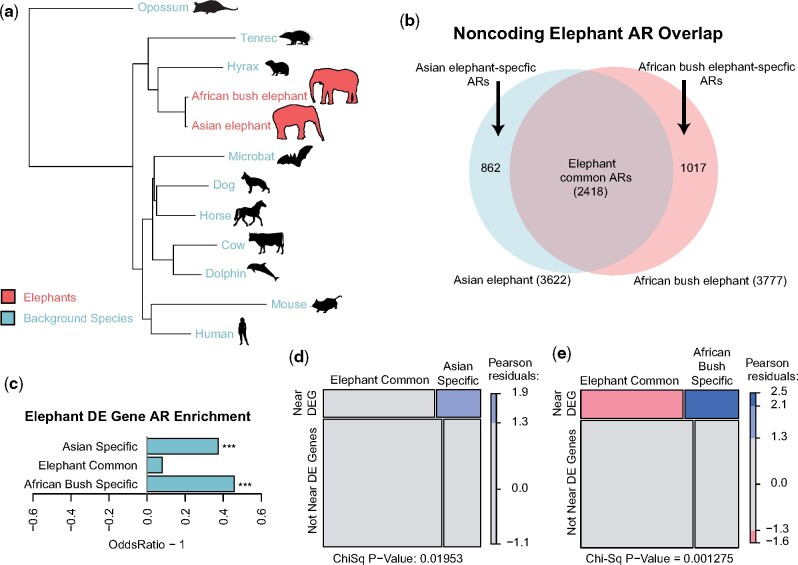
We used the Asian and African elephant genome assemblies to generate a whole genome alignment (WGA) with 10 other mammals, which we used to define accelerated genomic regions unique to Asian and African elephants. We first defined 676,509 regions of 60 bp in length present in Asian and African elephants and conserved in the 10 background species with phastCons (Siepel et al. 2005; Hubisz et al. 2011) (conserved regions, fig. 1a). Asian and African elephants likely diverged ∼5 Ma (Palkopoulou et al. 2018), and since differences between closely related mammals are primarily due to changes in noncoding regulatory genomic regions (Pollard et al. 2010; Hubisz et al. 2011; Booker et al. 2016), we focused on the 376,899 noncoding conserved regions. We tested these for accelerated substitution rates in elephants with phyloP (Pollard et al. 2010; Hubisz et al. 2011) and found 3,622 regions with significantly increased nucleotide substitution rates in the Asian elephant, whereas 3,777 regions were accelerated in the African bush elephant (q-value < 0.10). We found 2,418 accelerated regions (ARs) were shared between both species, with 862 being Asian elephant-specific and 1,017 being African bush elephant-specific (fig. 1b).
Fig. 1.
Accelerated regions in elephant genomes. (a) Using a whole genome alignment of 12 mammals we defined genomic regions that were accelerated (ARs) in two elephant species (red), yet conserved in the set of background species (blue). Branch lengths are given in terms of mean substitutions per site at 4-fold degenerate sites (neutral model). (b) Among the ARs detected in elephants, we found ARs common to both elephant species as well as ARs specific to either Asian or African bush elephants. (c) Differentially expressed (DE) genes were much more likely to be found in Asian elephant-specific (Fisher’s Exact Test, P = 2.05e−4) and African elephant-specific (P = 8.30e−7) ARs than in common ARs. (d, e) Species-specific ARs disproportionately overlap DE gene regulatory regions relative to the common ARs (χ2 test, P = 0.019 and P = 0.001, respectively).
Accelerated regions common to Asian and African bush elephants were likely driven by changes predating the evolutionary divergence of the two elephants, whereas ARs specific to each species may point to regulatory regions driving gene expression level changes that impact distinguishing phenotypes. Using available African bush elephant and Asian elephant peripheral blood mononuclear cell (PBMC) RNA-Seq data (Reddy et al. 2015; Ferris et al. 2018), we defined 5,034 differentially expressed (DE) elephant genes (false discovery rate or FDR < 0.01). Both Asian elephant- and African bush elephant-specific ARs were significantly enriched near DE genes relative to conserved regions (Fisher’s Exact Test, P = 2.05e−4, P = 8.30e−7, respectively). Meanwhile, the 2,418 ARs common to both elephants were not significantly enriched near DE genes (fig. 1c). This pattern remained robust with subsets of increasingly significantly DE genes based on adjusted P-values (supplementary fig. 3, Supplementary Material online). Asian elephant- and African bush elephant-specific ARs disproportionately overlapped DE gene regulatory regions relative to the common ARs (χ2 test, P = 0.019 and P = 0.001, respectively; fig. 1d and e), suggesting that some ARs reflect changes in regulatory regions that alter elephant PBMC gene expression patterns.
We functionally annotated the elephant species-specific and common ARs by testing for biological process (BP) gene ontology (GO) term enrichments (supplementary data 1, Supplementary Material online). Based on a likelihood ratio test that compared general linearized models (see Materials and Methods), 605 out of 607 (99.6%) GO terms were uniquely enriched in the elephant ARs in contrast to ARs found in other mammalian lineages (Ferris et al. 2018). Therefore, the enrichment of these GO terms in the elephant ARs are significant in elephants compared with other mammals.
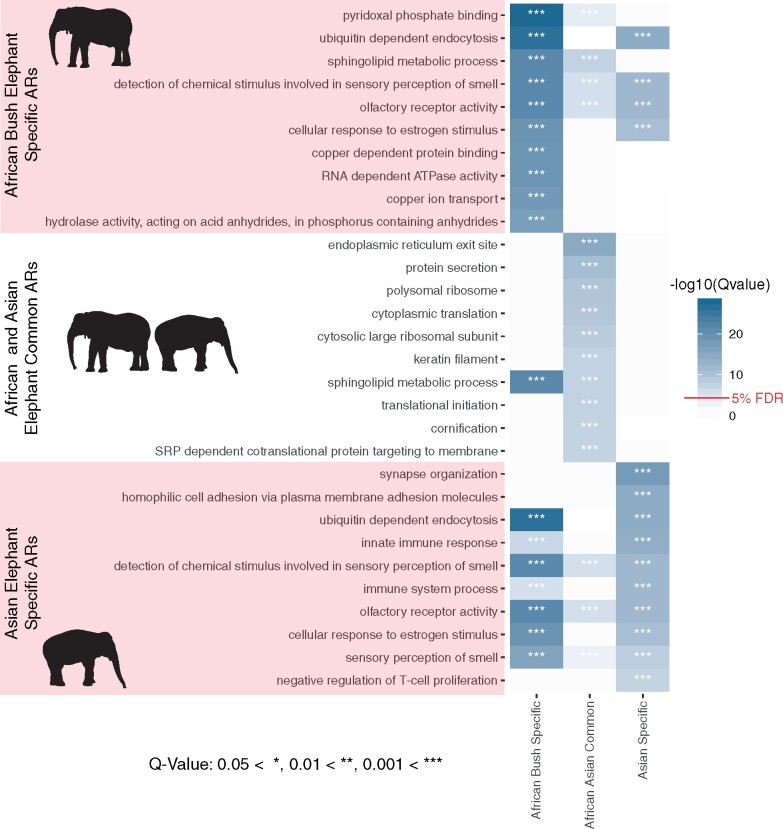
Of 18,056 BP GO terms, 252 were significantly enriched in Asian elephant-specific ARs and 275 were enriched in African elephant-specific ARs (q-value < 0.05). Of note, “olfactory receptor activity” and “detection of chemical stimulus involved in sensory perception of smell” were 22- and 28-fold enriched in Asian (q-value = 5.56e−25) and African (q-value = 1.6e−15) elephant ARs, respectively (fig. 2; supplementary figs. 4 and 5, supplementary data, Supplementary Material online). This is perhaps related to uniquely acute elephant olfaction abilities compared with other mammals (Plotnik et al. 2019). Many of the significantly enriched GO terms were also related to the immune system in both elephant species (fig. 2; supplementary figs. 4 and 5; supplementary data, Supplementary Material online). The broad term “immune system process” was 4.5- and 2.8-fold enriched with Asian and African elephant-specific ARs (q-value = 4.87e−12 and 7.84e−06, respectively), but not significantly enriched with elephant common ARs. Our results suggest 1) many of the species-specific ARs alter gene expression patterns and transcription factor binding networks that lead to differences in immune function, and 2) Asian elephant ARs are more enriched in immune pathways than African elephant-specific ARs.
Fig. 2.
Top 10 Biological Process gene ontology (GO) terms most significantly enriched with elephant accelerated genomic regions (ARs). The 10 most significantly enriched GO terms in terms of −log10(q-value) for each set of ARs (African elephant-specific, African and Asian elephant common, Asian elephant-specific), and their overlap. “Innate immune response” and “immune system process” are in the top 10 most significantly enriched GO terms for Asian elephant-specific ARs, are significantly enriched in African elephant-specific ARs but not in the top 10, and are not significantly enriched in the common ARs. “Negative regulation of T-cell proliferation” was only in the top 10 significantly enriched GO terms for the Asian elephant-specific ARs.
We found 109 GO terms significantly enriched with elephant common ARs (q-value < 0.05, supplementary fig. 6, supplementary data, Supplementary Material online), many of which were related to cancer, including “sphingolipid metabolic process” which was in the 10 most significantly enriched GO terms for both elephant common (5.7-fold enrichment, q-value = 4.69e−08) and African elephant-specific (17.3-fold enrichment, q-value = 4.18e−22) ARs (fig. 2). Sphingolipid metabolites mediate the signaling cascades involved in apoptosis (Hetz et al. 2002), necrosis (Hetz et al. 2002), senescence (Venable et al. 1995), and inflammation (Snider et al. 2010). We found 2.9- and 3.6-fold enrichments for “tumor-necrosis factor (TNF)-mediated signaling pathway” (q-value = 4.75e−04) and “positive regulation of TNF production” (q-value = 1.75e−03) in the common ARs, and a 21.5-fold enrichment of “negative regulation of TNF secretion” in African elephant-specific ARs (q-value = 5.01e−04). TNF is a cytokine involved in cell differentiation and death that can induce the necrosis of cancer cells (Balkwill 2009). The upregulation of TNF-alpha has been associated with increased apoptosis in EEHV-infected Asian elephant PBMCs as well (Srivorakul et al. 2019).
In a check for reproducibility, we found that the number of African elephant-specific ARs assigned to each gene was correlated with previous studies (Ferris et al. 2018) (R = 0.82). The gene most enriched with previously defined noncoding African elephant ARs was the DNA repair gene FANCL (7.3-fold enrichment; q-value = 2.16e−56), which mediates the E3 ligase activity of the Fanconi anemia core complex, a master regulator of DNA repair (Moldovan and D’Andrea 2009). We found that FANCL was the third most significantly enriched gene in both African and Asian elephant ARs relative to conserved regions with 4.6- and 4.9-fold enrichments (q-value = 1.27e−14 and 4.46e−16). Of 50 African elephant ARs and 51 Asian elephant ARs assigned to FANCL, 43 are common to both elephant species suggesting their acceleration predates African-Asian elephant divergence. These results suggest noncoding cis-regulatory elements have regulated cancer resistance adaptations throughout elephant evolution, particularly in the ancestor of modern elephants and the lineage leading to the African bush elephant.
Evolution of TP53 and Its Retrogenes in Elephant Genomes
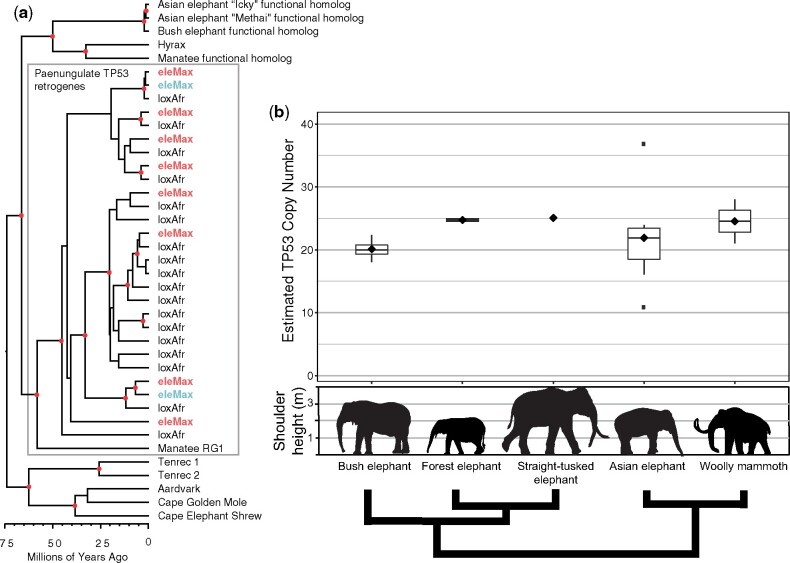
To understand the origins and evolution of elephant TP53 retrogenes, we incorporated TP53 homologs from 44 mammalian genomes, including Icky the Asian elephant, an additional genome assembly of an Asian elephant (Clavijo et al. 2017; Dudchenko et al. 2017, 2018) (“Methai,” born in Thailand and living at the Houston Zoo, assembly available at www.dnazoo.org, last accessed September 2020), and the African bush elephant assembly presented in this study (supplementary table 5, Supplementary Material online) in a molecular clock analysis. We estimated that TP53 retrogene copies originated in the paneungulate ancestor of manatees and elephants ∼55 − 60 Ma (95% highest posterior density or HPD 41.3–75.2 Ma) (fig. 3a, supplementary fig. 7, Supplementary Material online). A subsequent TP53 expansion began ∼45 Ma (95% HPD 30.7–60.1 Ma) in a common ancestor of Asian and African elephants, and continued throughout elephantid evolution.
Fig. 3.
Evolution of TP53 in elephants and other afrotherians. (a) Phylogeny of TP53 sequences extracted from afrotherian genomes. TP53 retrogenes (Manatee RG1, eleMax, and loxAfr) appeared early in the evolution of paenungulates ∼55–60 million years ago (Ma), followed by subsequent amplification in the elephant lineage ∼45 Ma. Red dots indicate estimated nodes with posterior probability ≥90%. Red eleMax indicates TP53 retrogene sequences extracted from the “Methai” Asian elephant assembly, and blue eleMax indicates TP53 retrogene sequences extracted from the Asian elephant assembly “Icky” presented in this study. (b) TP53 copy number estimates based on read counts from three living (Asian elephant, bush, and forest elephants) and two extinct (straight-tusked elephant and woolly mammoth) elephant species. Shoulder height estimates from Larramendi (2015). Phylogeny is schematic only and represents relationships from Palkopoulou et al. (2018).
In addition to the TP53 functional homolog in each elephant species, we annotated 18 retrogene copies of TP53 in the African bush elephant genome assembly, and 2 in the genome of Icky the Asian elephant that phylogenetically clustered with 2 of the 9 retrogenes validated in Methai’s genome assembly (fig. 3a). Taken overall, we estimated between nine and 11 TP53 retrogenes in the Asian elephant genome based on the available assemblies.
We also mapped whole genome shotgun data from the three living elephant species as well as that for two extinct elephant species (table 3), and used normalized read counts to estimate TP53 copy numbers in elephant genomes (fig. 3b;supplementary table 6, Supplementary Material online). Based on read depth, African bush elephants (n = 4) have ∼19–23 TP53 copies in their genomes, and Asian elephant genomes (n = 7) contain from 10 to 37, similar to previous estimates (Abegglen et al. 2015; Caulin et al. 2015; Sulak et al. 2016). We estimated ∼21–24 TP53 copy number variants in forest elephant genomes (n = 2). The woolly mammoths (n = 2) contained between 19 and 28 TP53 copies in their genomes, which was slightly higher than previous estimates (Sulak et al. 2016). Meanwhile, the straight-tusked elephant genome contained ∼22 − 25 TP53 copies.
Table 3.
Summary of Elephant Whole-Genome Shotgun Resequencing Data Utilized in This Study, Mapped to the African Bush Elephant Reference Assembly (loxAfr3.0).
| Species | Name | Geographic Origin | Source | No. Mapped Reads | Prop. Reads Properly Paired | Peak Read Depth |
|---|---|---|---|---|---|---|
| Loxodonta africana | Watoto | Kenya | ERR2260496 | 874,537,386 | 0.99 | 26× |
| Swazi | South Africa | ERR2260497 | 1,014,067,450 | 0.93 | 30× | |
| Hi-Dari | Kenya | SRR11869865, SRR11869866,(Current study) | 1,072,817,612 | 0.97 | 38× | |
| Christie | Zimbabwe | SRR12799664, SRR12799663 (Current study; Abegglen et al. 2015) |
1,031,044,341 | 0.98 | 36× | |
| Loxodonta cyclotis | DS1546 | Central African Republic | ERR2260495 | 852,948,500 | 0.96 | 24× |
| Coco | Sierra Leone | ERR2260500 | 981,145,080 | 0.99 | 30× | |
| Elephas maximus | Moola | Myanmar | ERR2260498 | 1,188,021,033 | 0.99 | 36× |
| Chendra | Borneo | ERR2260499 | 981,228,188 | 0.99 | 30× | |
| Icky | Myanmar | SRR11577048 (Current study) |
898,020,572 | 0.97 | 32× | |
| Parvathy | India | SRR2008170 | 872,535,345 | 0.96 | 26× | |
| Asha | India | SRR2009586 | 977,136,495 | 0.94 | 29× | |
| Uno | Assam, India | SRR2012205, SRR2012206, SRR2012207 | 912,606,191 | 0.96 | 26× | |
| Jayaprakash | Karnataka, India | SRR2912975 | 475,023,505 | 0.94 | 13× | |
| Palaeoloxodon antiquus | NA | Germany | ERR2260504 | 916,662,984 | NA | 7× |
| Mammuthus primigenius | NA | Oimyakon, Russia | ERR852028 | 617,446,606 | NA | 10× |
| NA | Wrangel Island, Russia | ERR855944 | 760,223,385 | NA | 16× |
The lower estimates of Asian elephant TP53 copy numbers based on the genome assemblies compared with read counting may be due to poorly resolved repetitive regions typical of mammalian genomes, which hamper graph-based de novo genome assemblers (Clavijo et al. 2017). Subsequent refinement of Asian elephant genome assemblies using long read sequencing may better resolve these regions. In the meantime, our results suggest that copy number estimates based on read depth are useful approximations for approaches validated from genomic DNA (Abegglen et al. 2015).
Genetic Variation in Elephant TP53 and Its Retrogenes
We found a high degree of sequence conservation relative to the rest of the genome in the TP53 paralogs both within and between the three living elephants (supplementary table 7, Supplementary Material online). For instance, the proportion of polymorphic sites in putatively neutrally evolving ancestral repeats was 0.013, but across 12 annotated TP53 paralogs was 0.004. Despite the high genome-wide heterozygosity found in forest elephants (supplementary materials, Supplementary Material online), we found very little genetic variation in TP53 paralogs for the species, with just a single segregating site in three of the retrogenes. Across all species, we detected zero nonsynonymous SNPs for the functional homolog (Ensembl Gene ID ENSLAFG00000007483) and ENSLAFG00000028299, or “retrogene 9,” whose protein expression is stabilized by DNA damage in human cells (Abegglen et al. 2015). We annotated variants in the 12 TP53 paralogs based on the Ensembl bush elephant genome annotation (loxAr3) and found few variants affecting gene function (supplementary table 8, Supplementary Material online), consisting of mostly missense mutations. There were no variants of high or moderate impact on gene function annotated in the functional homolog (ENSLAFG00000007483), with the majority of variants occurring downstream, in introns, or upstream of the gene.
Positive Selection Scans of Elephant Genomes
We used the aligned whole genome shotgun sequences from the three extant elephant species (Loxodonta africana, n = 4; L. cyclotis, n = 2; Elephas maximus, n = 7; table 3) to call variants with freebayes v1.3.1-12 (Garrison and Marth 2012), genotyping 41,296,555 biallelic single nucleotide polymorphisms (SNPs), averaging one SNP every 77 bases and with a genome-wide transition-transversion ratio of 2.46. Altogether, we annotated 290,965 exonic, 11,245,343 intronic, and 32,512,650 intergenic SNPs across the 13 elephant genomes. We corroborated our assessment of the demographic history of each living elephant species with summary population genomics statistics with results largely consistent with Palkopoulou et al. (2018) (supplementary figs. 8 and 9 and supplementary materials, Supplementary Material online).
To determine the importance of selection in modern elephants, we scanned the genomes of the three extant species for selective sweeps using SweeD v3.3.1 (Nielsen et al. 2005; Pavlidis et al. 2013). For Asian elephants, this yielded 24,394 selectively swept outlier regions meeting our statistical thresholds based on neutral expectations (see Materials and Methods; supplementary fig. 10 and supplementary materials, Supplementary Material online), comprising ∼0.07% of the genome and overlapping 1,611 gene annotations. Out of the 41,204 regions experiencing putative selective sweeps in bush elephants (∼1.3% of the genome), we detected 2,882 protein-coding genes. We estimated 4,099 protein-coding genes involved in the 51,249 regions involved in putative selective sweeps in forest elephants (∼1.6% of the genome). We found 242 protein-coding genes that overlapped regions with evidence of positive selection and were shared in all 3 of the living elephant species, which were enriched for pathways related to many iconic elephant traits such as memory, tusk development, and somatic maintenance including cancer defenses (supplementary fig. 11 and supplementary materials, Supplementary Material online).
Significant GO terms from shared outlier regions across all three elephant species were related to cancer, including cell adhesion (9-fold enrichment, FDR = 0.007), cell−cell signaling (3-fold enrichment, FDR = 0.01), and cell communication (2-fold enrichment, FDR = 0.0001). Significant protein-protein interactions were found associated with EGF-like domain (UniProt keyword enrichment, 13 out of 209 genes, FDR = 4.2e−04; and INTERPRO protein domain enrichment, 13 out of 191 genes, FDR = 2.6e−04). The EGF-like domain contains repeats which bind to apoptotic cells and play a key role in their clearance (Park et al. 2008). Our selective sweep results are consistent with those from the AR analysis and suggest ongoing selection in pathways involved with somatic maintenance and in particular apoptosis, a possible key mechanism for cancer suppression in elephants.
Elephant Health, Conservation, Evolution, and Genomics
Our study expands the knowledge of elephant evolution and disease defense, highlighting differences and similarities between living and extinct species. We found that elephant tumors tend to be benign, indicating strong genetic defenses to prevent malignant transformation in Asian and African elephants. Asian elephants reported in our study developed benign and malignant neoplasms at higher rates than African elephants, and therefore may benefit from increased monitoring for tumors. Even though our data originates from zoo elephants, these differences most likely reflect true genetic differences as the AZA Species Survival Plan (SSP) (https://www.aza.org/species-survival-plan-programs) for elephants maximizes and maintains genetic diversity similar to wild populations via the careful selection of mate pairs and studbook documentation (Lei et al. 2008, 2012). Together with the fact that many elephants in zoos are wild born, it is likely that wild Asian elephants share increased susceptibility to neoplasia.
Although collecting cancer prevalence data in wild elephants is challenging due to decomposition and predator consumption, future data from wild elephants and genomic analysis of benign and malignant tumors will be crucial to further understand the evolutionary basis of differences in cancer risk between elephant species. This information could benefit the survival of individual elephants and assist with selecting the best treatment interventions when the rare elephant tumor is diagnosed under human care or in the wild. More than half of the elephant tumors reported here were found in reproductive organs (table 1). Even benign reproductive tumors can affect reproduction and conservation; therefore, future studies to understand their impact and to develop preventative and treatment measures are needed.
We found some of the highest TP53 copy numbers in the smallest species of elephants, suggesting that TP53 copy number has not increased linearly with body mass during elephant evolution as a response to increased cancer risk, as has been suggested (Sulak et al. 2016). We also found that genetic variation at some TP53 retrogenes is highly conserved relative to other parts of the genome in living elephant populations. This is despite an estimated ∼50 million year divergence of some elephant TP53 paralogs, which predates the Asian-African elephant divergence of ∼5 Ma. A lack of polymorphisms in an ancient gene family with high intraspecific sequence similarity (92% mean pairwise similarity in bush elephant and 88% in Asian elephant) may be the result of gene conversion (Chen, Cooper, et al. 2007). We suggest that while there may be a core set of TP53 retrogenes conferring the bulk of cancer suppression in elephants, particularly “retrogene 9” (Abegglen et al. 2015), more research is needed to determine the relative roles of concerted evolution and selection in the maintenance of elephant TP53 retrogenes.
The enrichment of immune pathways in Asian elephant ARs relative to African elephants may reflect exposure to novel pathogens during the migration of Asian elephant ancestors out of Africa, which had occurred by ∼2.7 − 3.6 Ma (Vidya et al. 2009). Regulatory elements may play a role in the increased infectious susceptibility and inflammatory response in Asian elephants, particularly in the mediation of the TNF cytokine. Asian elephant calves are much more susceptible than African elephant calves to cytokine storm triggered by EEHV infection (Hayward 2012). Compared with African elephants, we found that Asian elephant ARs are enriched for BP GO terms “interleukin-1 beta (IL-1β) production” (q-value = 0.036), “interleukin-18 (IL-18) production” (q-value = 0.00073), and “neutrophil activation involved in immune response” (q-value = 2.44e−05) (supplementary data, Supplementary Material online). IL-1β, IL-18, and neutrophils, combined with TNF-alpha, are potent mediators of innate immunity. Uncontrolled activation of these factors leads to immune-induced disease pathogenesis through excessive inflammation. In humans and other mammals, neutrophil activation directly contributes to tissue damage through the release of neutrophil extracellular traps (NETs) (Jorch and Kubes 2017; Goggs et al. 2020). Functional studies comparing cytokine secretion and NET release in response to infectious agents may confirm that genetic differences in innate immunity contribute to differences in disease susceptibility and outcomes between Asian and African elephant species.
In addition to maintaining the health of elephants under human care through improved breeding and species survival plans, conservation efforts can benefit from genomic studies that identify genetic variants associated with traits such as disease defense (Supple and Shapiro 2018). Our study provides an example of how genomics can inform functional immunological and molecular studies, which may lead to improved conservation and medical care for elephants. This type of genetic information could provide important evolutionary insights that may one day be translated into human patients with infections or cancer.
Materials and Methods
Cancer Data Collection
Pathology and necropsy records were collected with consent from 26 zoological institutions across the United States over the span of 26 years. All participating institutions were deidentified and anonymized. Data were collected with permissions from Buffalo Zoo, Dallas Zoo, El Paso Zoo, Fort Worth Zoo, Gladys Porter Zoo, Greenville Zoo, Jacksonville Zoo and Gardens, Louisville Zoological Garden, Oakland Zoo, Oklahoma City Zoo and Botanical Garden, Omaha’s Henry Doorly Zoo and Aquarium, The Phoenix Zoo, Point Defiance Zoo and Aquarium, San Antonio Zoological Society, Santa Barbara Zoological Gardens, Sedgwick County Zoo, Seneca Park Zoo, Toledo Zoological Gardens, Utah’s Hogle Zoo, Woodland Park Zoo, Zoo Atlanta, Zoo Miami and three other anonymous zoos. Neoplasia was diagnosed by board certified veterinary pathologists and cancers were identified from pathological services at Northwest ZooPath, Monroe, WA. Published pathology data from San Diego Zoo were also included (Boddy et al. 2020). Neoplasia prevalence was estimated by the number of elephants that were diagnosed with tumors (benign or malignant) in respect to all elephants documented within our database.
De Novo Assembly and Annotation of the Asian Elephant Genome
A whole blood sample was drawn in an EDTA tube from the Asian elephant (“Icky,” North American studbook number 199) from the Ringling Bros. Center for Elephant Conservation, and DNA libraries were constructed and sequenced at the University of Utah Genomics Core Facility. Paired-end DNA libraries were constructed with the TruSeq Library Prep kit for a target insert size of 200 bp, and mate-paired libraries were constructed for target sizes of 3, 5, 8, and 10 kb using the Nextera Mate Pair Library kit. Genomic DNA was sequenced on an Illumina HiSeq2500. Raw reads were trimmed for base quality score =>30 and the removal of with Trimmomatic v0.33 (Bolger et al. 2014) and SeqClean (Chen, Lin, et al. 2007). Genome assembly was carried out using ALLPATHS-LG (MacCallum et al. 2009; Gnerre et al. 2011). The expected gene content was assessed by searching for 4,104 mammalian single-copy orthologs (mammalia_odb9) using BUSCO v3.0.2 (Simão et al. 2015).
We annotated and masked repeats in the resulting assembly using both the de novo method implemented in RepeatModeler v1.0.11 (Smit et al. 2015a) and a database method using RepeatMasker v4.07 (Smit et al. 2015b) with a library of known mammalian repeats from RepBase (Jurka et al. 2005). Modeled repeats were used in a Blast search against Swiss-Prot (UniProt Consortium 2015) to identify and remove false positives. We then generated gene models for the Asian elephant assembly using MAKER2 (Holt and Yandell 2011), which incorporated 1) homology to publicly available proteomes of cow, human, mouse, and all mammalian entries in UniProtKB/Swiss-Prot, and 2) ab initio gene predictions based on species-specific gene models in SNAP (11/29/2013 release) (Korf 2004), species-specific and human gene models in Augustus v3.0.2 (Stanke et al. 2008), and EvidenceModeler (Haas et al. 2008). Final gene calls were functionally annotated by using InterProScan 5 (Jones et al. 2014) to identify protein domains and a Blastp search of all annotated proteins to UniProt proteins to assign putative orthologs with an e-value cutoff of 1e−6.
Tissue Collection, DNA Extraction, and Genome Sequencing of African Bush Elephants
The African bush elephant assembly was improved with the addition of Hi-C sequencing libraries. First, a whole blood sample was drawn (in an EDTA tube) from a wild-born female named Swazi (animal ID: KB13542, North American studbook number 532) at the San Diego Zoo Safari Park in Escondido, CA. We selected this individual because her genome was originally sequenced by the Broad Institute (Palkopoulou et al. 2018). Three Hi-C libraries were constructed and sequenced to ∼38× genome coverage and used for scaffolding with HiRise (Putnam et al. 2016) at Dovetail Genomics in Santa Cruz, CA, with the most recent version of the African bush elephant assembly (loxAfr4.0) as an input. DNA was extracted from fresh frozen subcutaneous skin necropsy tissue samples from an African bush elephant named Hi-Dari (animal ID 00003, North American studbook number 33) at the Hogle Zoo in Salt Lake City, UT using a ThermoFisher PureLink Genomic Mini DNA Kit at the University of Utah. Two pieces of tissue were digested and extracted separately and pooled followed extraction. DNA concentration was measured by PicoGreen (8.66 ng/μl) with a total volume of 200 μl in 10 mM pH8.0. DNA sequencing libraries were generated using the Illumina TruSeq Library Prep Kit for a 350 bp mean insert size, and sequenced on two lanes the Illumina HiSeq2500 platform at Huntsman Cancer Institute’s High-Throughput Genomics Core (Salt Lake City, UT).
TP53 Evolution in African and Asian Elephants
To determine TP53 copy numbers and evolutionary patterns across placental mammals, we exported the TP53 human peptide from Ensembl (July 2019), and used it as a query in reciprocal BLAT searches (Kent 2002) of 44 mammalian genome assemblies (supplementary table 6, Supplementary Material online), validated with a BlastX query of the human peptide database on NCBI in order to ensure the top hit was human TP53 with ≥70% protein identity, following Tollis et al. (2020). Accepted nucleotide sequences were aligned with MAFFT (Katoh and Standley 2013), and we weighted and filtered out unreliable columns in the alignment with GUIDANCE2 (Sela et al. 2015) using 100 bootstrap replicates.
We reconstructed the phylogeny of all aligned mammalian TP53 homologs and estimated their divergence times in a Bayesian framework with BEAST 2.5 (Bouckaert et al. 2014) using the HKY substitution model, a relaxed lognormal molecular clock model, and a Yule Model tree prior. We used a normal prior distribution for the age of Eutheria (offset to 105 million years or My with the 2.5% quantile at 101 Ma, and the 97.5% quantile at 109 Ma) and lognormal prior distributions for the following node calibrations from the fossil record (Benton et al. 2015): Boreoeutheria (offset the minimum age to 61.6–164 Ma and the 97.5% quantile to 170 Ma), Euarchontoglires (56–164 Ma), Primates (56–66 Ma), Laurasiatheria (61.6–164 Ma), and Afrotheria (56–164 Ma). We monitored proper MCMC mixing with Tracer v1.7.1 (Rambaut et al. 2018) to ensure an effective sampling size of at least 200 from the posterior distributions of each parameter and ran 2 independent chains. The final MCMC chain was run for 100,000,000 generations, and we logged parameter samples every 10,000 generations to collect a total of 10,000 samples from the posterior distribution. We then collected 10,000 of the resulting trees, ignored the first 10% as burn-in, and calculated the maximum clade credibility tree using TreeAnnotator.
Detection of ARs in African and Asian Elephant Genomes
We generated a multiple alignment (whole genome alignment or WGA) of 12 mammalian genome assemblies. First, we downloaded publicly available pairwise syntenic alignments of opossum (monDom5), mouse (mm10), dolphin (turTru1), cow (bosTau7), dog (canFam3), horse (equCab2), microbat (myoLuc1), tenrec (echTel2), and hyrax (proCap1) to the human reference (hg19) from the UCSC Genome Browser (Kent et al. 2002). We also computed two additional de novo pairwise syntenic alignments with the human genome as a target and the two elephant genome assemblies reported here as queries using local alignments from LASTZ v1.02 (Harris 2007) using the following options from the UCSC Genome Browser for mammalian genome alignments: –hspthresh 2200 –inner 2000 –ydrop 3400 –gappedthresh 10000 –scores HOXD70, followed by chaining to form gapless blocks and netting to rank the highest scoring chains (Kent et al. 2003). We then constructed a multiple sequence alignment with MULTIZ v11.2 (Blanchette et al. 2004) with human as the reference species.
To define elephant ARs, we used functions from the R package rphast v1.6 (Hubisz et al. 2011). We used phyloFit with the substitution model “REV” to estimate a neutral model based on autosomal 4-fold degenerate sites from the WGA. We then used phastCons to define 60 bp autosomal regions conserved in the 10 nonelephant species in the WGA with the following options: expected.length = 45, target.coverage = 0.3, rho = 0.31. We further selected regions with aligned sequence for both African and Asian elephants that have aligned sequence present for at least 9 of the 10 nonelephant species. We tested the resulting 676,509 regions for acceleration in each elephant species relative to the 10 nonelephant species with phyloP using the following options: mode = “ACC.” We used the Q-Value method (Storey 2003) to adjust for multiple testing. Statistically significant ARs were defined with an FDR threshold of 10%. We defined regions significantly accelerated in the Asian elephant, but not the African bush elephant as Asian elephant-specific ARs and conversely defined African bush elephant-specific ARs. Our previous studies of ARs suggest no significant relationship between genome quality and number of ARs discovered (Ferris et al. 2018; Ferris and Gregg 2019).
To define genes DE between Asian and African elephants we took advantage of the closeness between the two species. The Asian elephant is more closely related to the African elephant than humans are to chimpanzees (0.01186 substitutions/100 bp vs. 0.01277 substitutions/100 bp based on 4-fold degenerate sites from our WGA). For the purpose of defining DE genes, chimpanzee RNA-Seq reads have been aligned to human transcriptome indices (Marchetto et al. 2019). We aligned African bush elephant PBMC reads (four technical replicates) from a previous study (Ferris et al. 2018) and publicly available Asian elephant PBMC data from a single individual (Reddy et al. 2015) (one replicate) to an African elephant (loxAfr3) transcriptome index with the STAR aligner (Dobin et al. 2013). After counting reads for each elephant gene with featureCounts (Liao et al. 2014), we normalized counts with the TMM method and defined significant DE genes with edgeR (Robinson et al. 2010) correcting for multiple testing with the Benjamini-Hochberg method (FDR < 0.01). The DE gene list was minimally affected by modest FDR cutoff changes. We note differences in the cell preps, RNA purification methods and sex of the Asian and African elephants as potential confounding factors in defining DE genes. The African elephant PBMC RNA was purified with a Ribo-Zero depletion step, whereas the Asian elephant RNA was purified by Poly-A selection. A study comparing the 2 RNA purification methods shows a high gene expression correlation (0.931) between the 2 methods and detects 410 genes as DE when contrasting these purification methods (Zhao et al. 2014).
Potential regulatory regions for elephant DE genes were defined with custom R scripts implementing logic detailed by McLean et al. (2010) based on transcription start site (TSS) locations obtained for protein-coding genes with the R package biomaRt (Durinck et al. 2005) for the African bush elephant genome (loxAfr3) with basal distances of 5 kb upstream and 1 kb downstream an extension distance of 100 kb. We chose this extension distance because the majority of conserved enhancers are located within 100 kb of a TSS (Villar et al. 2015). We used the R package LOLA (Sheffield and Bock 2016) to test for enrichment of ARs relative to CRs in the potential regulatory regions of DE genes in the loxAfr3 genome. BPs and associated elephant orthologs of human genes were obtained with biomaRt. The resulting P-values were q-value corrected for multiple testing. We used the same potential regulatory regions and LOLA to test for GO enrichments.
We compared elephant AR set GO enrichments with GO enrichments from previously published AR sets for 5 mammalian species (13-lined ground squirrel, naked mole rat, orca, bottlenose dolphin, and little brown bat) (Ferris et al. 2018). These AR sets were lifted over from hg19 coordinates to loxAfr3 coordinates. Numbers of ARs and background CRs overlapping potential regulatory regions of genes included in and excluded from each GO term were calculated with LOLA. We used generalized linear models with binomial distributions to compare elephant AR enrichments in each GO term to AR enrichments for the five other mammals. We contrasted models without and with an interaction term distinguishing the elephant AR set from the others. The two models are
where gt is a binary value {0, 1} indicating gene regions excluded from or included in a given GO term set; ce is a binary value {0, 1} indicating nonelephant or elephant ARs study; ar is the number of ARs in a given category. For each GO term with significant AR enrichments for at least one of the three elephant AR sets in the earlier analysis, we determined the significance of the enrichment in each elephant AR set relative to the other mammal AR sets by comparing the two models by likelihood ratio test. The likelihood ratio test P-values are reported in the supplementary data, Supplementary Material online.
Whole Genome Sequence Analysis of Living Elephants
We obtained ∼15–40× whole-genome sequencing data from multiple individuals from across the modern range of living elephants from public databases (Abegglen et al. 2015; Lynch et al. 2015; Reddy et al. 2015; Palkopoulou et al. 2018), and the WGS libraries for “Hi-Dari” and “Icky” as well as a third African elephant named “Christie” (table 3). We also obtained sequence data from a straight-tusked elephant (Palkopoulou et al. 2018) and two woolly mammoths (Palkopoulou et al. 2015). Sequences were quality checked using FastQC and trimmed for base quality score and adapter sequences were removed with Trimmomatic where necessary. Reads from each individual were mapped to the L. africana genome (loxAfr3.0, Ensembl version) using bwa-mem v077 (Li and Durbin 2009). Alignments were filtered to include only mapped reads and sorted by position using Samtools v0.0.19 (Li et al. 2009), and we removed PCR duplicates using MarkDuplicates in picard v1.125 (DePristo et al. 2011). Single-end reads from the ancient samples were mapped to loxAfr3.0 with bwa-aln following Palkolpoulou et al. (2018).
We estimated the number of TP53 paralogs present in the genome of each elephant by calculating the average read depth of annotated sites in Ensembl TP53 exons and whole genes with Samtools, dividing the total average genome coverage, multiplied by the number of TP53 annotations (n = 12). We called variants in the living elephant species (n = 13) by incorporating the .bam files using freebayes v1.3.1-12 (Garrison and Marth 2012), with extensive filtering to avoid false positives as follows with vcffilter from vcflib (https://github.com/vcflib/vcflib, last accessed July 2019): Phred-scale probability that a REF/ALT polymorphism exists at a given site (QUAL) > 20, the additional contribution of each observation should be 10 log units (QUAL/AO > 10), read depth (DP > 5), reads must exist on both strands (SAF > 0 and SAR > 0), and at least two reads must be balanced to each side of the site (RPR > 1 and RPL > 1). We then removed indels from the .vcf file and filtered to only include biallelic SNPs that were genotyped in every individual using VCFtools v0.1.17 (Danecek et al. 2011) (–remove-indels –min-alleles 2 –max-alleles 2 –max-missing-count 0) and bcftools v1.9 (Danecek and McCarthy 2017) (–v snps –m 1). We annotated the biallelic SNPs using SnpEff v4.3 (Cingolani et al. 2012) based on loxAfr3 (Ensembl), and calculated diversity statistics including per-individual heterozygosity, nucleotide diversity and Tajima’s D in 10 kb windows with VCFtools, and the fixation index FST with PopGenome v2.7.1 (Pfeifer et al. 2014). A detailed description of population genomics and selective sweep analyses is included in the supplementary materials, Supplementary Material online.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We acknowledge Leigh Duke for data coordination and Trent Fowler and Rosann Robinson for assistance with sample collection. We acknowledge the collections and veterinary staff at the San Diego Zoo Safari Park and Utah’s Hogle Zoo for sample collection. We acknowledge the following institutions for sharing data and/or resources: Buffalo Zoo, Dallas Zoo, El Paso Zoo, Fort Worth Zoo, Gladys Porter Zoo, Greenville Zoo, Jacksonville Zoo and Gardens, Louisville Zoological Garden, Oakland Zoo, Oklahoma City Zoo and Botanical Garden, Omaha’s Henry Doorly Zoo and Aquarium, The Phoenix Zoo, Point Defiance Zoo and Aquarium, San Antonio Zoological Society, Santa Barbara Zoological Gardens, Sedgwick County Zoo, Seneca Park Zoo, Toledo Zoological Gardens, Utah’s Hogle Zoo, Woodland Park Zoo, Zoo Atlanta, Zoo Miami and three other anonymous zoos. We acknowledge Huntsman Cancer Institute’s High-Throughput Genomics Core and the Monsoon computing cluster at Northern Arizona University (https://nau.edu/high-performance-computing/). We acknowledge help from Dr Kenneth Boucher of the Huntsman Cancer Institute in designing generalized linear models. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number U54CA217376. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. J.D.S., L.M.A., and this research are supported through the EP53 Research Program and its generous funding provided to Huntsman Cancer Institute by the State of Utah. J.D.S. is supported by Hyundai Hope on Wheels, Soccer for Hope Foundation, Li-Fraumeni Syndrome Association, and Kneaders Hope Fights Childhood Cancer. J.D.S. also was previously supported by the Edward B. Clark, MD Endowed Chair in Pediatric Research and currently by the Helen Clise Presidential Endowed Chair in Li-Fraumeni Syndrome Research. L.M.A. and this research is supported by the Department of Pediatrics Research Enterprise (University of Utah).
Data Availability
Short-read sequence data generated for this study has been shared under NCBI Bioproject PRJNA622303, and the genome assembly for Icky the Asian elephant is available on NCBI (GCA_014332765.1). Other data sets including the updated African elephant genome assembly, annotation files for Asian elephant, multiple genome alignments, TP53 alignments and phylogeny, .vcf files, and selective sweep results have been deposited to Zenodo (https://zenodo.org/record/4033444#.X5cISFNKhGp).
Conflict of Interest
Dr J.D.S. is co-founder, shareholder, and employed by PEEL Therapeutics, Inc., a company developing evolution-inspired medicines based on cancer resistance in elephants. Dr L.M.A. is share-holder and consultant to PEEL Therapeutics, Inc.
References
- Abegglen LM, Caulin AF, Chan A, Lee K, Robinson R, Campbell MS, Kiso WK, Schmitt DL, Waddell PJ, Bhaskara S, et al. 2015. Potential mechanisms for cancer resistance in elephants and comparative cellular response to DNA damage in humans. JAMA. 314(17):1850–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill F.2009. Tumour necrosis factor and cancer. Nat Rev Cancer. 9(5):361–371. [DOI] [PubMed] [Google Scholar]
- Benton MJ, Donoghue PCJ, Asher RJ, Friedman M, Near TJ, Vinther J.. 2015. Constraints on the timescale of animal evolutionary history. Palaeontol. Electron. 18:1–106. [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AFA, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14(4):708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy AM, Abegglen LM, Pessier AP, Aktipis A, Schiffman JD, Maley CC, Witte C.. 2020. Lifetime cancer prevalence and life history traits in mammals. Evol Med Public Health. 2020(1):187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker BM, Friedrich T, Mason MK, VanderMeer JE, Zhao J, Eckalbar WL, Logan M, Illing N, Pollard KS, Ahituv N.. 2016. Bat accelerated regions identify a bat forelimb specific enhancer in the HoxD locus. PLOS Genet. 12(3):e1005738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ.. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLOS Comput Biol. 10(4):e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulin AF, Graham TA, Wang L-S, Maley CC.. 2015. Solutions to Peto’s paradox revealed by mathematical modelling and cross-species cancer gene analysis. Phil Trans R Soc B. 370(1673):20140222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman SN, Jackson J, Htut W, Lummaa V, Lahdenperä M.. 2019. Asian elephants exhibit post-reproductive lifespans. BMC Evol Biol. 19:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-M, Cooper DN, Chuzhanova N, Férec C, Patrinos GP.. 2007. Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet. 8(10):762–775. [DOI] [PubMed] [Google Scholar]
- Chen Y-A, Lin C-C, Wang C-D, Wu H-B, Hwang P-I.. 2007. An optimized procedure greatly improves EST vector contamination removal. BMC Genomics. 8:416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM.. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavijo BJ, Accinelli GG Wright J, Heavens D, Barr K, Yanes L, Di-Palma F.. 2017. W2RAP: a pipeline for high quality, robust assemblies of large complex genomes from short read data. bioRxiv. 110999. [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST. et al. ; 1000 Genomes Project Analysis Group. 2011. The variant call format and VCFtools. Bioinformatics. 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, McCarthy SA.. 2017. BCFtools/csq: haplotype-aware variant consequences. Bioinformatics. 33(13):2037–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, et al. 2017. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 356(6333):92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, Shamim MS, Batra SS Durand NC Musial NT Mostofa R, Pham M, Hilaire BGS, Yao W, Stamenova E, et al. 2018. The Juicebox Assembly Tools module facilitates de novo assembly of mammalian genomes with chromosome-length scaffolds for under $1000. bioRxiv. 254797. [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W.. 2005. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 21(16):3439–3440. [DOI] [PubMed] [Google Scholar]
- Ferris E, Abegglen LM, Schiffman JD, Gregg C.. 2018. Accelerated evolution in distinctive species reveals candidate elements for clinically relevant traits, including mutation and cancer resistance. Cell Rep. 22(10):2742–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris E, Gregg C.. 2019. Parallel accelerated evolution in distant hibernators reveals candidate cis elements and genetic circuits regulating mammalian obesity. Cell Rep. 29(9):2608–2620.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G.. 2012. Haplotype-based variant detection from short-read sequencing. http://arxiv.org/abs/1207.3907. Accessed September 2019.
- Gnerre S, MacCallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, et al. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 108(4):1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobush KS, Edwards CTT, Balfour D, Wittemyer G, Maisels F, Taylor RD.. 2021. Loxodonta africana. IUCN Red List Threat. Species e.T181008073A181022663.
- Gobush KS, Edwards CTT, Maisels F, Wittemyer G, Balfour D, Taylor RD.. 2021. Loxodonta cyclotis. IUCN Red List Threat. Species e.T181007989A181019888.
- Goggs R, Jeffery U, LeVine DN, Li RHL.. 2020. Neutrophil-extracellular traps, cell-free DNA, and immunothrombosis in companion animals: a review. Vet Pathol. 57(1):6–23. [DOI] [PubMed] [Google Scholar]
- Greenwald R, Lyashchenko O, Esfandiari J, Miller M, Mikota S, Olsen JH, Ball R, Dumonceaux G, Schmitt D, Moller T, et al. 2009. Highly accurate antibody assays for early and rapid detection of tuberculosis in African and Asian elephants. Clin Vaccine Immunol. 16(5):605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR.. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9(1):R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS.2007. Improved pairwise alignment of genomic DNA. PhD Thesis.
- Hayward GS.2012. Conservation: clarifying the risk from herpesvirus to captive Asian elephants. Vet Rec. 170(8):202–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz CA, Hunn M, Rojas P, Torres V, Leyton L, Quest AFG.. 2002. Caspase-dependent initiation of apoptosis and necrosis by the Fas receptor in lymphoid cells: onset of necrosis is associated with delayed ceramide increase. J Cell Sci. 115(Pt 23):4671–4683. [DOI] [PubMed] [Google Scholar]
- Holt C, Yandell M.. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 12:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlander N, Noone A, Kraphco M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis D, et al. 2020. SEER Cancer Statistics Review, 1975-2017. Bethesda (MD: ): National Cancer Institute. Available from: https://seer.cancer.gov/csr/1975_2017/. [Google Scholar]
- Hubisz MJ, Pollard KS, Siepel A.. 2011. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief Bioinform. 12(1):41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics. 30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorch SK, Kubes P.. 2017. An emerging role for neutrophil extracellular traps in noninfectious disease. Nat Med. 23(3):279–287. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J.. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110(1-4):462–467. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ.2002. BLAT–the BLAST-like alignment tool. Genome Res. 12(4):656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D.. 2003. Evolution’s cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci U S A. 100(20):11484–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D.. 2002. The human genome browser at UCSC. Genome Res. 12(6):996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I.2004. Gene finding in novel genomes. BMC Bioinformatics. 5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larramendi A.2015. Shoulder height, body mass, and shape of proboscideans. Acta Palaeontol. Pol. 61:537–574. [Google Scholar]
- Lei R, Brenneman RA, Louis EE.. 2008. Genetic diversity in the North American captive African elephant collection. J. Zool. 275(3):252–267. [DOI] [PubMed] [Google Scholar]
- Lei R, Brenneman RA, Schmitt DL, Louis EE.. 2012. Genetic diversity in North American captive Asian elephants. J Zool. 286(1):38–47. [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup 2009. 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30(7):923–930. [DOI] [PubMed] [Google Scholar]
- Long SY, Latimer EM, Hayward GS.. 2016. Review of elephant endotheliotropic herpesviruses and acute hemorrhagic disease. Ilar J. 56(3):283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch VJ, Bedoya-Reina OC, Ratan A, Sulak M, Drautz-Moses DI, Perry GH, Miller W, Schuster SC.. 2015. Elephantid genomes reveal the molecular bases of woolly mammoth adaptations to the Arctic. Cell Rep. 12(2):217–228. [DOI] [PubMed] [Google Scholar]
- MacCallum I, Przybylski D, Gnerre S, Burton J, Shlyakhter I, Gnirke A, Malek J, McKernan K, Ranade S, Shea TP, et al. 2009. ALLPATHS 2: small genomes assembled accurately and with high continuity from short paired reads. Genome Biol. 10(10):R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Hrvoj-Mihic B, Kerman BE, Yu DX, Vadodaria KC, Linker SB, Narvaiza I, Santos R, Denli AM, Mendes AP, et al. 2019. Species-specific maturation profiles of human, chimpanzee and bonobo neural cells. eLife. 8:e37527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G.. 2010. GREAT improves functional interpretation of cis -regulatory regions. Nat Biotechnol. 28(5):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan G-L, D’Andrea AD.. 2009. How the Fanconi anemia pathway guards the genome. Annu Rev Genet. 43:223–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montali RJ, Hildebrandt TB Goritz F, Hermes R, Ippen R, Ramsay E.. 1997. Ultrasonography and pathology of genital tract leiomymoas in captive Asian elephants: implications for reproductive soundness. Erkrangungen Zootiere. 38. Available from: http://repository.si.edu/xmlui/handle/10088/6159. [Google Scholar]
- Moss CJ.2001. The demography of an African elephant (Loxodonta africana) population in Amboseli. J Zool. 255(2):145–156. [Google Scholar]
- Nielsen R, Williamson S, Kim Y, Hubisz MJ, Clark AG, Bustamante C.. 2005. Genomic scans for selective sweeps using SNP data. Genome Res. 15(11):1566–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palkopoulou E, Lipson M, Mallick S, Nielsen S, Rohland N, Baleka S, Karpinski E, Ivancevic AM, To T-H, Kortschak RD, et al. 2018. A comprehensive genomic history of extinct and living elephants. Proc Natl Acad Sci U S A. 115(11):E2566–E2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palkopoulou E, Mallick S, Skoglund P, Enk J, Rohland N, Li H, Omrak A, Vartanyan S, Poinar H, Götherström A, et al. 2015. Complete genomes reveal signatures of demographic and genetic declines in the woolly mammoth. Curr Biol. 25(10):1395–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Živkovic D, Stamatakis A, Alachiotis N.. 2013. SweeD: likelihood-based detection of selective sweeps in thousands of genomes. Mol Biol Evol. 30(9):2224–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Kim S-Y, Jung M-Y, Bae D-J, Kim I-S.. 2008. Epidermal growth factor-like domain repeat of stabilin-2 recognizes phosphatidylserine during cell corpse clearance. Mol Cell Biol. 28(17):5288–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer B, Wittelsbürger U, Ramos-Onsins SE, Lercher MJ.. 2014. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol Biol Evol. 31(7):1929–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnik JM, Brubaker DL, Dale R, Tiller LN, Mumby HS, Clayton NS.. 2019. Elephants have a nose for quantity. Proc Natl Acad Sci U S A. 116(25):12566–12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A.. 2010. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20(1):110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, O'Connell BL, Stites JC, Rice BJ, Blanchette M, Calef R, Troll CJ, Fields A, Hartley PD, Sugnet CW, et al. 2016. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 26(3):342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA.. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 67(5):901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PC, Sinha I, Kelkar A, Habib F, Pradhan SJ, Sukumar R, Galande S.. 2015. Comparative sequence analyses of genome and transcriptome reveal novel transcripts and variants in the Asian elephant Elephas maximus. J Biosci. 40(5):891–907. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca AL, Ishida Y, Brandt AL, Benjamin NR, Zhao K, Georgiadis NJ.. 2015. Elephant natural history: a genomic perspective. Annu Rev Anim Biosci. 3:139–167. [DOI] [PubMed] [Google Scholar]
- Sela I, Ashkenazy H, Katoh K, Pupko T.. 2015. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43(W1):W7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield NC, Bock C.. 2016. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics. 32(4):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoshani J.1998. Understanding proboscidean evolution: a formidable task. Trends Ecol Evol. 13(12):480–487. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15(8):1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley RM, Green P.. 2015a. RepeatModeler Open-1.0 2008-2015. Available from: http://www.repeatmasker.org.
- Smit AFA, Hubley RM, Green P.. 2015b. RepeatMasker Open-4.0 2013-2015. Available from: http://www.repeatmasker.org.
- Snider AJ, Orr Gandy KA, Obeid LM.. 2010. Sphingosine kinase: role in regulation of bioactive sphingolipid mediators in inflammation. Biochimie. 92(6):707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivorakul S, Guntawang T, Kochagul V, Photichai K, Sittisak T, Janyamethakul T, Boonprasert K, Khammesri S, Langkaphin W, Punyapornwithaya V, et al. 2019. Possible roles of monocytes/macrophages in response to elephant endotheliotropic herpesvirus (EEHV) infections in Asian elephants (Elephas maximus). PLOS One. 14(9):e0222158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D.. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 24(5):637–644. [DOI] [PubMed] [Google Scholar]
- Storey JD.2003. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Stat. 31:2013–2035. [Google Scholar]
- Stuart AJ.2005. The extinction of woolly mammoth (Mammuthus primigenius) and straight-tusked elephant (Palaeoloxodon antiquus) in Europe. Quat Int. 126-128:171–177. [Google Scholar]
- Sulak M, Fong L, Mika K, Chigurupati S, Yon L, Mongan NP, Emes RD, Lynch VJ.. 2016. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. eLife. 5:1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supple MA, Shapiro B.. 2018. Conservation of biodiversity in the genomics era. Genome Biol. 19(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, Schneider-Utaka AK, Maley CC.. 2020. The evolution of human cancer gene duplications across mammals. Mol Biol Evol. 37(10):2875–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. 2015. UniProt: a hub for protein information. Nucleic Acids Res. 43:D204–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanyan SL, Garutt VE, Sher AV.. 1993. Holocene dwarf mammoths from Wrangel Island in the Siberian Arctic. Nature. 362(6418):337–340. [DOI] [PubMed] [Google Scholar]
- Venable ME, Lee JY, Smyth MJ, Bielawska A, Obeid LM.. 1995. Role of ceramide in cellular senescence. J Biol Chem. 270(51):30701–30708. [DOI] [PubMed] [Google Scholar]
- Vidya TNC, Sukumar R, Melnick DJ.. 2009. Range-wide mtDNA phylogeography yields insights into the origins of Asian elephants. Proc Biol Sci. 276(1658):893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M, Park TJ, Deaville R, Erichsen JT, Jasinska AJ, et al. 2015. Enhancer evolution across 20 mammalian species. Cell. 160(3):554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, Tiwari SK, Goswami VR, de Silva S, Kumar A, Baskaran N, Yoganand K, Menon V.. 2020. Elephas maximus. IUCN Red List Threat. Species e.T7140A45818198.
- Zhao W, He X, Hoadley KA, Parker JS, Hayes DN, Perou CM.. 2014. Comparison of RNA-Seq by poly (A) capture, ribosomal RNA depletion, and DNA microarray for expression profiling. BMC Genomics. 15:419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann A, Bernuit D, Gerlinger C, Schaefers M, Geppert K.. 2012. Prevalence, symptoms and management of uterine fibroids: an international internet-based survey of 21,746 women. BMC Womens Health. 12:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Short-read sequence data generated for this study has been shared under NCBI Bioproject PRJNA622303, and the genome assembly for Icky the Asian elephant is available on NCBI (GCA_014332765.1). Other data sets including the updated African elephant genome assembly, annotation files for Asian elephant, multiple genome alignments, TP53 alignments and phylogeny, .vcf files, and selective sweep results have been deposited to Zenodo (https://zenodo.org/record/4033444#.X5cISFNKhGp).