Abstract
The intestinal microbiota of an organism can significantly alter outcome data in otherwise identical experiments. Occasionally, animals may require sedation or anesthesia for scientific or health-related purposes, and certain anesthetics, such as ketamine, can profoundly affect the gastrointestinal system. While many factors can alter the gut microbiome (GM), the effects of anesthetics on the composition or diversity of the GM have not been established. The goal of the current study was to determine whether daily administration of ketamine would significantly alter the microbiome of CD1 mice. To achieve this goal, female CD1 mice received daily injections of ketamine HCl (100 mg/kg) or the equivalent volume of 0.9% saline for 10 consecutive days. Fecal samples were collected before the first administration and 24 h after the final dose of either ketamine or saline. Samples were analyzed by 16S rRNA sequencing to identify changes between groups in diversity or composition of GM. The study found no significant changes to the GM after serial ketamine administration when treated mice were housed with controls. Therefore, ketamine administration is unlikely to alter the GM of a CD1 mouse and should not serve be a confounding factor in reproducibility of research.
Abbreviations: GM, gut microbiota; IP, intraperitoneal; OTU, operational taxonomic unit
Reproducibility in science has been under increasing scrutiny in the past decade, with one recent report suggesting more than 70% of researchers have failed to reproduce another’s experimental results.2 Another survey conducted by the American Society for Cell Biology reported similar results and that the most common reason (55%) for not resolving reproducibility issues was that the issue was deemed ‘not important enough to pursue’.1 Underlying issues with reproducibility may be poor experimental design, inadequate technical training, and pressure to publish in high-impact journals.2 To combat this lack of reproducibility, in 2014 the NIH released plans to enhance reproducibility efforts.7 This plan implemented a series of web-based training modules and webinars along with a variety of resources that can be accessed by anyone performing research.
Although the NIH has made tremendous efforts to enhance reproducibility, differences in the gut microbiome of genetically identical animals are often overlooked as a factor. The intestinal microbiota consists of the microorganisms present in a particular environment in the gut.34,35 These microorganisms have a symbiotic relationship with the host and are important in maintaining homeostasis, aiding digestion, regulating metabolism, and influencing immunity.5,38 Differences in the microbiome can significantly alter outcome data, leading to the proposition that every publication should include microbiome data.3,34 Although different vendors may offer genetically similar mice, differences in the microbiome can cause otherwise identical experiments to produce different results.11,14 In addition, changes in the microbiome can drastically alter disease expression in models of obesity, type 2 diabetes, and inflammatory bowel disease and can alter cognition and behavior in mice.8,28 Husbandry conditions such as the pH or treatment of the water, bedding, and feed can all affect the microbiome.32,36 Other factors that can change the microbiome include antibiotic treatment, diet changes, aging, and pregnancy.6,23
Because stress can also affect the microbiome, any manipulation of an animal can introduce a confounding factor in research, resulting in potential inadvertent influences on the data.22,25,26 Anesthesia is a common stressor that research animals experience frequently. Animals are often sedated or anesthetized to facilitate experimental manipulations and health examinations, including treatment administration and sample collection. The process of anesthesia encompasses more than simply the act of being anesthetized; for research animals, anesthesia involves handling, either an injection or exposure to anesthetic gas before the event, and recovery and can include additional manipulation and housing in areas outside of the home cage. These are potentially stressful events for animals and therefore could alter the GM. Few studies explicitly investigate the effect of anesthesia or sedation on the gut microbiota.
Ketamine, a short-acting dissociative anesthetic that works as an NMDA receptor antagonist, is commonly used in conjunction with other sedatives or as the sole agent in various species.8 For this project, we assessed ketamine alone to determine whether it might contribute to microbiota changes when used in an anesthetic cocktail and how it might affect the microbiota in sedation protocols. Ketamine can cause intestinal cramping in people, hypersalivation, weight loss, and tolerance over time.8,10,15 The influence of ketamine on the gastrointestinal system, along with known vascular effects, raises the concern that it could alter the gut microbiome, ultimately causing dysbiosis.8,9 One study showed that small daily doses of ketamine, similar to those used to treat human depression, altered the GM in rats.16 In addition, a study using (R)-ketamine,31 a less potent ketamine analogue that is used in people for its antidepressant effects,29 showed that a single dose of ketamine altered the GM in mice. However, that study31 assessed only a single antidepressant dose of ketamine and had additional confounding factors that are known to alter the GM. Our current study uses a larger sample size of unmanipulated mice. Ketamine administration is repeated across days to simulate conditions for animals that may be repeatedly anesthetized for research purposes, such as sample collection.
The objective of our study was to determine whether serial ketamine administration, mimicking regular sedation or anesthetic events that may occur during an experiment, would change the composition or diversity of the GM in CD1 mice. To test this, we used 20 female CD1 mice and injected them intraperitoneally (IP) with 100 mg/kg ketamine HCl or the same volume of 0.9% saline daily for 10 consecutive days. Fecal samples were collected immediately before the first injection and 24 h after the final injection. We hypothesized that daily ketamine administration would significantly alter the microbiome.
Materials and Methods
Animals.
This study was conducted at an AAALAC-accredited facility under an IACUC-approved protocol in compliance with the Guide for the Care and Use of Laboratory Animals, 8th ed.21 Twenty female 6 to 8-wk-old CD1 mice (weight, 26.4 × 5.8 g) were obtained from the Mutant Mouse Resource and Research Center (Columbia, MO). The GM of these mice is considered to have high richness (GMHSD,17 a GM originated from Envigo Corporation, Indianapolis, IN facilities) relative to other commercial species. Richness refers to the total number of amplicon sequence variants, which represent unique bacterial taxonomies. The housing environment was maintained at 22 ± 2 °C, with a relative humidity of 30% to 70% on a 14:10-h light:dark cycle (lights on, 0600 CST). Mice were group-housed with 4 animals per standard polypropylene shoebox cages (7.70 × 12.17 × 5.25 in., Thoren, Hazleton, PA) on PAPERCHIP bedding (Shepherd Specialty Papers, Watertown, TN) and had unrestricted access to a commercial rodent diet (Formulab Diet 5008, Purina) and autoclaved water that was treated with sulfuric acid with a pH range of 2.3 to 2.7. Cotton squares were supplied for each cage for enrichment. Colony health was evaluated every 3 mo through sentinel exposure to dirty bedding. All sentinels were seronegative for Mouse hepatitis virus, Minute virus of mice, Mouse parvovirus, Parvovirus NS-1, Theiler murine encephalomyelitis virus, Murine rotavirus, Mycoplasma pulmonis, Sendai virus. PCR testing was negative for fur mites and pinworms.
Mice were divided into 2 treatment groups (n = 10 per group); one group received daily ketamine injections; the other group received equivalent daily injections of saline. Three mice from the ketamine treatment group died due to inadvertent ketamine injection into the liver and were not included in the analysis because paired data (before and after) could not be obtained from these mice.
Drugs.
Pharmaceutical grade ketamine HCl (100 mg/mL) and 0.9% sterile saline were purchased from MWI Animal Health (Boise, ID). Drugs were drawn directly from the undiluted pharmaceutical stock using a 25-gauge needle and 1 mL syringe at a dose of 100 mg/kg or volume of 1 mL/kg.
Weights.
Individual mice within a cage were identified by placing a mark on their tail using a permanent marker. All mice were weighed immediately before any treatments to obtain baseline measurements using a digital gram scale. Mice were weighed before starting the experiment, every other day during the treatment period, and immediately after euthanasia. To weigh mice, a weigh basket was placed on the gram scale, and the scale was tared. The mouse was then picked up by the base of the tail and placed in the weigh basket, and the weight was recorded. Immediately after weighing, mice were given their daily injection and either returned to their cage (saline control group) or monitored (ketamine treatment group).
Pre- and post-treatment fecal collection.
Fecal samples were collected from all mice by being placed individually in a clean polypropylene shoebox cage (7.70 × 12.17 × 5.25 in., Thoren, Hazleton, PA) without bedding. Each mouse had unrestricted access to the cage; once an adequate fecal sample was produced, 1 to 2 fecal pellets were collected with an autoclaved wooden toothpick. Samples were placed in an autoclaved 1 mL microfuge tube (Thermo Fisher Scientific, Waltham, MA) labeled with each mouse number and stored in a −20 °C freezer until DNA extraction. The cage was cleaned with 10% bleach between mice and allowed to dry completely. A new toothpick and microfuge tube were used for each sample.
Serial drug administration.
Mice were divided into 2 groups (n = 10 per group): serial ketamine treatment group (100 mg/kg) and a 0.9% saline control group. 100 mg/kg dose of ketamine was chosen because it is a dose used for sedation in laboratory mice.20 Mice were housed 4 per cage, with 2 of the mice in the ketamine treatment group and the other 2 in the saline control group to act as in cage controls and simulate conditions in which not all mice undergo sedation in an experimental group. Mice were manually restrained and injected IP daily with their respective treatments at 1600 CST. Mice in the ketamine treatment group were given a dose of 100 mg/kg or volume of 1 mL/kg for 10 consecutive days. Mice in the saline control group were given an identical volume (1 mL/kg) of saline during the same time period. IP injections were given by 3 of the authors (SG, RD and RG) using a 25-gauge needle and 1 mL syringe. Injections were alternated between the lower right and lower left abdominal quadrant of the mouse to minimize discomfort. After ketamine injection, mice were placed in a clean cage without bedding on a circulating hot water blanket (Midwest Veterinary Supply, Des Moines, IA). Eyes were lubricated with sterile veterinary eye lubricant (Covetrus, Des Moines, IA), and mice were monitored continuously until recovery of consciousness. Respiratory rate and pedal withdrawal reflex were assessed on each mouse every 15 min as part of our standard recovery procedure until consciousness was regained. Once mice were able to ambulate, they were returned to their home cage. Final fecal samples were collected 24 h after final ketamine or saline injections. After fecal sample collection on day 11, animals were euthanized via CO2 overdose and cervical dislocation.
Fecal DNA extraction.
Fecal DNA extraction was performed as previously described using a PowerFecal kit (Qiagen, Germantown, MD).9 The manufacturer’s instructions were followed, with the exception that samples were homogenized in the provided bead tubes using a TissueLyser II (Qiagen, Venlo, Netherlands) for 3 min at 30 s rather than by using the vortex adapter as described in the protocol. The protocol was then resumed by eluting in 100 µL of elution buffer (Qiagen). Fluorometry (Qubit 2.0, Life Technologies, Carlsbad CA) and quant-iT BR dsDNA reagent kits (Invitrogen, Carlsbad, CA) was used to quantify DNA.
16S rRNA library preparation, sequencing and informatics.
The MU Informatics Research Core Facility performed all assembly, binning, and annotation of DNA sequences as previously described.11,18,26 Briefly, the V4 hypervariable region of the 16S rRNA gene was amplified using previously developed universal primers to create bacterial 16S rRNA amplicons. Amplicons were purified and evaluated using an automated electrophoresis system (Fragment Analyzer, Advanced Analytical, Ankeny, IA). Quantification was performed using a fluorometer (Qubit 2.0, Life Technologies, Carlsbad, CA). Finally, all samples were sequenced by using a desktop sequencer (MiSeq, Illumina, San Diego, CA). Operational taxonomic units (OTU) were identified and given taxonomic assignments using BLAST against the SILVA database of 16S rRNA sequences and taxonomy.11,12,18,26
Data availability.
All 16S rRNA amplicon sequencing data presented here have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject ID PRJNA705656.
Statistics.
All samples were included in the analysis. All indices used in analyses were calculated and analyzed using either Past Software 3.2419 or SigmaPlot 13.0 (SYSTAT, San Jose, CA).37 To determine whether daily ketamine administration would alter weight, mouse weights were measured throughout the study, and end weights were compared with the starting weight of each individual mouse. A 2-factor mixed-design ANOVA was performed on the pre- and posttreatment weights for both the ketamine and control treatment groups. To test for differences in GM richness and diversity (α diversity) associated with ketamine administration, Shannon and Simpson indices were calculated, and a paired student’s t test was used to compare respective indices. For all statistical analysis P values less than 0.05 were considered significant.
To assess richness (that is, total number of distinct taxonomies detected) and diversity (that is, a combination of richness and evenness of distribution), the Shannon H and Simpson 1-D indices were used. Differences within groups and between time points were compared using a paired-Student t test, while differences between groups were compared using an unpaired-Student t test. A 2-factor mixed-design ANOVA was used to assess for changes in weight between groups. Differences in community composition were measured by one- or 2-way permutational multivariate ANOVA (PERMANOVA) using Jaccard and Bray–Curtis distances, which are unweighted and weighted similarities, respectively. Weighted and unweighted UniFrac distances were also calculated for each pre- and post-sample and compared. Unweighted similarities are based on the proportions of shared features (for example, taxa) between samples, while weighted similarities also factor in the similarity between samples in the relative abundance of shared features. UniFrac distances build upon Bray–Curtis and Jaccard similarities by incorporating phylogenetic relationships of detected taxa. For example, the greater the distance between samples, the more dissimilar the composition.
Results
Body weights.
The analysis detected no significant change in weight over the time of the experiment (P = 0.2404), within experimental groups (P = 0.6144), or over time and between the groups (P = 0.3002).
To compare sequencing data from each animal, high-quality amplicon sequence reads (that is, 249 bp sequences of variable 4 (V4) region of the 16S rRNA gene, amplified via PCR) from each sample were counted. All samples yielded excellent community coverage with a mean (± SD) of 151,807 (± 20,077) sequence reads per sample. No significant differences were found in the number of sequence reads between treatment groups or time-points.
Bacterial richness and diversity.
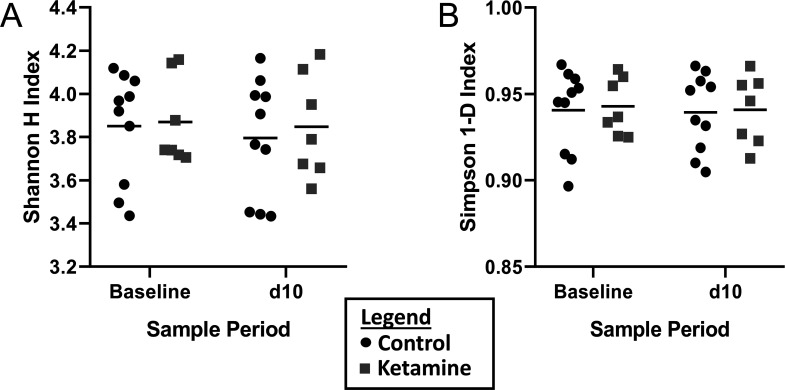
To test for differences in GM richness and diversity (α diversity) associated with ketamine administration, Shannon and Simpson indices were calculated, and a paired student’s t test was used to compare respective indices. No significant differences were detected as measured by the Shannon or Simpson indices between baseline and posttreatment control groups (P = 0.5244 and 0.8634 respectively), or ketamine groups (P = 0.8585 and 0.8714 respectively).
To assess changes in the GM after either daily ketamine or daily saline injections, the pre- to post-injection GM composition ratios were compared for the saline and the ketamine groups using a 2-tailed Student t test. The ratios were derived from the Shannon and Simpson indices and were not significantly different (P = 0.8262 and 0.9371 respectively). These data indicate no significant difference in the richness and diversity of either group after daily administration of either ketamine or saline, indicating no significant differences in the composition of the GM after treatment.
Bacterial composition heterogeneity (β diversity).
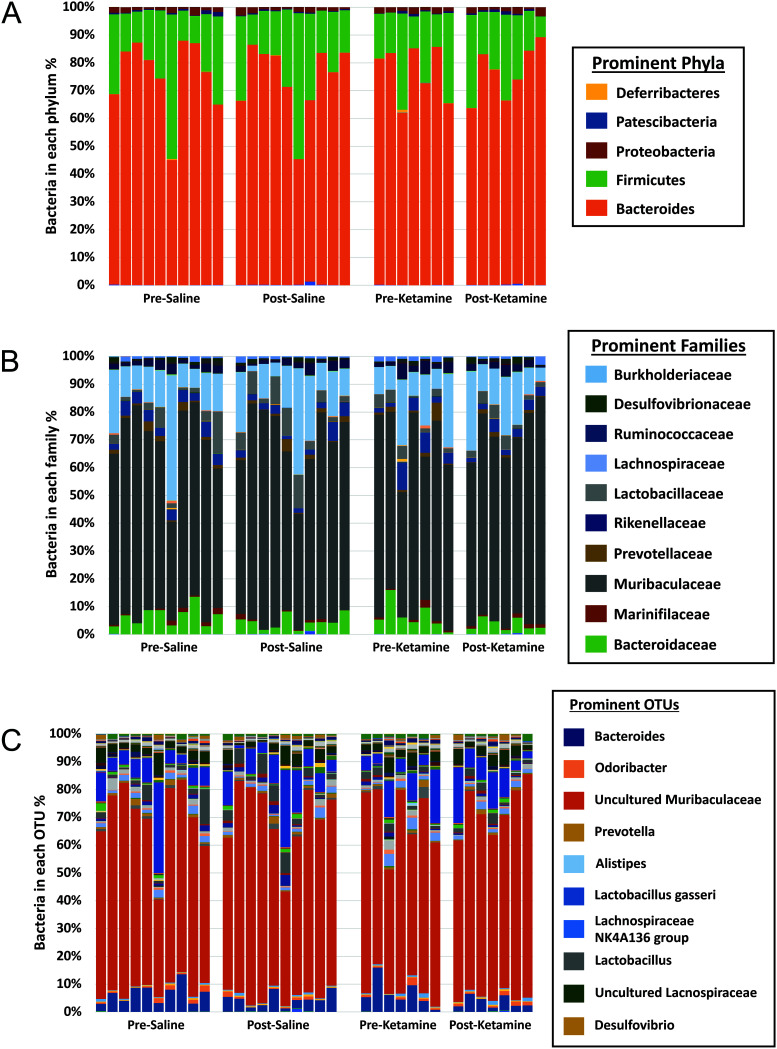
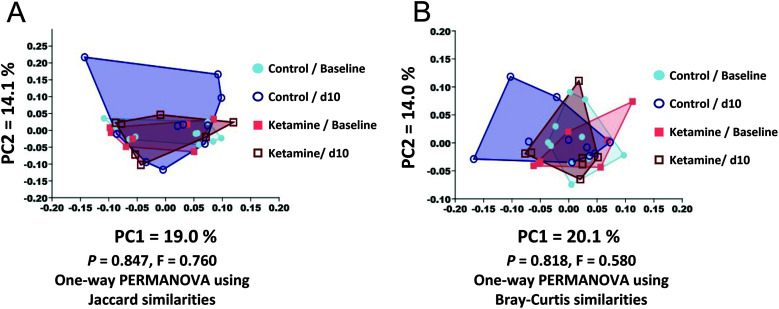
Bar charts that compared the GM composition and relative abundance of bacteria at the phylum, family, and OTU level (Figure 1) revealed the typical subtle variability seen in laboratory mouse cohorts, but did not yield any obvious differences between or within experimental groups over time. To further assess differences in composition, we performed principal coordinate analysis (PCoA) (Figure 2). Subjectively, no differences were observed between groups. A one-way PERMANOVA analysis was used to test for significant differences between groups using Bray–Curtis and Jaccard similarities, respectively. No differences were detected between groups using weighted Bray–Curtis similarities (P = 0.818) or unweighted Jaccard similarities (P = 0.847) (Figure 3). To further determine whether composition of the GM changed between groups, the average UniFrac distance between the pre- and posttreatment samples for the experimental and control groups were compared using a 2-tailed Student t test. Neither the weighted nor unweighted UniFrac distances were statistically different between the groups receiving saline and ketamine (P = 0.2164 and 0.6067 respectively), thus indicating that any differences between groups in overall community structure were masked by the variability within groups.
Figure 1.
Stacked bar charts showing relative abundance of bacterial phyla (A), families (B), and operational taxonomic units (C) in presaline, postsaline, preketamine, and postketamine samples. Note that bars are in numerical order by sample, such that the first bar within each treatment group represents mouse 1 in pre- and post- samples.
Figure 2.
Richness and evenness of taxonomic distribution (represented here by the α-diversity) is calculated as the Shannon H (A) and the Simpson 1-D (B) indices. Group means are represented as horizontal black lines. There was no significant difference between baseline and d10 control groups as measured by the Shannon H or Simpson 1-D indices (P = 0.5244 and 0.8634 respectively), nor in the ketamine group (P = 0.8585 and 0.8714 respectively).
Figure 3.
Compositional similarity between and within groups, expressed as β-diversity, is represented using principal coordinate analyses based on Jaccard (A) or Bray-Curtis (B) similarities of feces. Using one-way PERMANOVA analyses, there were no differences between or within groups using weighted Bray-Curtis similarities (P = 0.818) or unweighted Jaccard similarities (P = 0.847).
Discussion
Our study shows that repeated ketamine or saline administration does not significantly alter the GM in CD1 mice, causing us to reject our original hypothesis and accept our null hypothesis. Although no significant differences in the microbiome were observed, researchers can use these results when designing an experiment. For example, a researcher who is concerned that anesthesia may alter results could choose to use ketamine as an anesthetic or sedative. In addition, if a subset of mice in a study requires sedation for health purposes, ketamine administration is unlikely to alter the microbiome.
Visual inspection of bacterial composition using stacked bar charts revealed subtle differences among mice of both cohorts. These findings highlight the biologic variability that is common to murine GM even within cohorts.17 Such studies often reveal mice (for example, mouse #6 in this study) whose GM appears to have distinct compositional features as compared with other mice, yet larger sample sizes reveal that these ‘outlier’ samples truly fall within the spectrum of ‘normal’. This highlights the need for adequate sample sizes to permit interpretation of the overall impact of an experimental intervention. In this study, no explanation could be identified to account for the unusual characteristics of mouse #6, but an overall interpretation of the effects of ketamine was nonetheless possible.
Although we did not specifically measure the duration of anesthesia/sedation, the duration subjectively decreased with successive timepoints of the experiment. Mice remained unconscious for approximately 15 to 30 min and were considered conscious once their righting reflex returned and they were able to ambulate. Each mouse achieved a plane of presurgical anesthesia with the duration of anesthesia decreasing across days, suggesting that mice became tolerant to ketamine with repeated administration.13 Based on multiple studies showing changes in pharmacokinetics of ketamine after repeated administration in several species, we speculated that mice became tolerant to serial ketamine administration4,15,24,30,33
Three mice died immediately after ketamine injection. All injections in these mice were completed by the same individual, who had the least experience with injection techniques. Upon necropsy, intrahepatic hemorrhage was seen in 2 of the 3 mice. Collectively, this suggests that ketamine was inadvertently injected into the liver. While an unfortunate outcome, these deaths highlight the value of experience with necessary techniques.
For the current study, we cohoused the experimental and control mice to mimic a situation in which only a subset of mice may undergo anesthesia or sedation as part of the experimental design or for clinical purposes. Theoretically, because mice are coprophagic, any shifts in GM of the mice that received ketamine could have been diluted by and/or spread to mice that received saline. This could be an important determinant of the results of our study, which indicate that ketamine administration does not significantly alter the GM in mice cohoused with mice not undergoing anesthetic treatments. Previous studies that examined the effects of ketamine on the microbiome did observe a change with ketamine administration.29,36 However, these studies differ from ours in multiple ways. Our primary focus was on the GM and adequate sample size. A previous study that found negligible differences in the GM of mice also subjected mice to behavioral tests; these tests themselves could have caused a shift in the GM.29 Our study focused on the GM as the main variable and attempted to remove other potential confounding factors that might influence the composition of the GM. Furthermore, we used an outbred stock with more genetic variation and repeated ketamine administration as opposed to a single dose. These differences highlight the myriad factors that can modulate the microbiome, and why the GM is such an important consideration for the reproducibility of research. Ultimately, our study showed that when exposed to ketamine administration daily for 10 d, injected and cohoused do not have altered GM compositions. Thus, ketamine has no significant effect on the GM of mice that receive ketamine for experimental or clinical purposes, and thus likely is not a confounding factor in research involving the GM.
The purpose of this study was to determine whether repeated ketamine administration affects the GM after anesthetic treatments. Ketamine is a commonly used anesthetic in various animal species, whether used as a sole agent for sedation or for induction followed by inhalant anesthesia for a deeper anesthetic plane. While ketamine may frequently be used in a cocktail with other anesthetic agents such as xylazine, ketamine was used alone in our study to fully assess its effect on the GM without additional confounding factors. While acute changes in the microbiome might be anticipated, studies assessing the long-term effects of repeated ketamine administration may provide more insight. A limitation of the current study was the inclusion of only female mice. Sex is a known contributing factor in GM composition; we chose to eliminate this additional variable and perform this study using only female mice.27 Female mice were selected to minimize the potential for intracage aggression. Our study is also limited in that only one form of anesthesia was tested. We chose ketamine due to its profound gastrointestinal effects and its prevalent use in research animals. However, other methods of anesthesia, such as inhalation anesthesia and anesthetic cocktails, should also be considered.10 Moreover, we used mice with a high richness microbiome representative of many mice in contemporary research colonies. However, a GM of lower richness could be more sensitive to pharmacological pressures; the response of sparse GM is unknown. Future studies assessing both sexes, other anesthetic agents, and other microbiota compositions may be useful.
When designing experiments and analyzing research data, attention should be taken to ensure reproducibility. Researchers can control many independent factors in their experiments, including sedation and anesthesia. These procedures may concurrently create other independent factors, such as sedation-associated changes in the GM. Minimizing alterations in the GM during experiments is important because these changes can alter results. Because many environmental and experimental factors have been associated with alterations in the GM, we sought to determine whether ketamine administration would also cause changes. Our study indicates that repeated administration of ketamine to female CD1 mice has little impact on the GM and is an ideal drug choice for experiments in which the GM is important and ketamine use is required.
Acknowledgments
Support for SG was provided in part by the Joseph E Wagner Fellowship in Laboratory Animal Medicine, support for RD was provided in part by NIH grant T32OD011126, support for both RD and SG was also provided by the Enhanced Infrastructure for Biomedical Researchers Using Animal Models— a grant from the University of Missouri Life Sciences Mission Enhancement Program. Funding for the project was provided by the MU College of Veterinary Medicine’s Phi Zeta Research Grant funding program.
Technical support was provided by Rebecca Dorfmeyer and Giedre Turner as well as the MU DNA Core and Informatics Research Core facilities. Additional supplies were provided by the MMRRC supported by the NIH grant U42OD010918
References
- 1.ASCB Organization. [Internet]. 2014. Results of ASCB member survey on reproducibility. [Cited 18 July 2021]. Available at: https://www.ascb.org/wp-content/uploads/2015/11/final-survey-results-without-Q11.pdf
- 2.Baker M.2016.1,500 scientists lift the lid on reproducibility.Nature 533:452–454. 10.1038/533452a. [DOI] [PubMed] [Google Scholar]
- 3.Bleich A, Kornerup Hansen A.2012.Time to include the gut microbiota in the hygienic standardization of laboratory rodents.Comp Immunol Microbiol Infect Dis 35:81–92. 10.1016/j.cimid.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Bree MM, Feller I, Corssen G.1967.Safety and tolerance of repeated anesthesia with CI 581 (ketamine) in monkeys.Anesth Analg 46:596–600. 10.1213/00000539-196709000-00021. [DOI] [PubMed] [Google Scholar]
- 5.Bull MJ, Plummer NT.2014.Part 1: The human gut microbiome in health and disease.Integr Med (Encinitas) 13:17–22. [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng C, Wei H, Yu H, Xu C, Jiang S, Peng J.2018.Metabolic syndrome during perinatal period in sows and the link with gut microbiota and metabolites.Front Microbiol 9:1–13. 10.3389/fmicb.2018.01989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins FS, Tabak LA.2014.Policy: NIH plans to enhance reproducibility.Nature 505:612–613. 10.1038/505612a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craven R.2007.Ketamine.Anaesthesia 62 Suppl 1:48–53. 10.1111/j.1365-2044.2007.05298.x. [DOI] [PubMed] [Google Scholar]
- 9.Cryan J, Dinan T.2012.Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour.Nat Rev Neurosci 13:701–712. 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 10.Dholakia U, Clark-Price SC, Keating SCJ, Stem AW.2017.Anesthetic effects and body weight changes associated with ketamine-xylazine-lidocaine administered to CD-1 mice.PLoS One 12:1–11. 10.1371/journal.pone.0184911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ericsson AC, Davis JW, Spollen W, Bivens N, Givan S, Hagan CE, McIntosh M, Franklin CL.2015.Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice.PLoS One 10:1–19. 10.1371/journal.pone.0116704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ericsson AC, Personett AR, Turner G, Dorfmeyer RA, Franklin CL.2017.Variable colonization after reciprocal fecal microbiota transfer between mice with low and high richness microbiota.Front Microbiol 8:1–13. 10.3389/fmicb.2017.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flecknell PA.1996.Laboratory animal anaesthesia: a practical introduction for research workers and technicians. San Diego (CA):Elsevier. [Google Scholar]
- 14.Franklin CL, Ericsson AC.2017.Microbiota and reproducibility of rodent models.Lab Anim (NY)46:114–122. 10.1038/laban.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerb SA, Cook JE, Gochenauer AE, Young CS, Fulton LK, Grady AW, Freeman KB.2019.Ketamine tolerance in Sprague–Dawley rats after chronic administration of ketamine, morphine, or cocaine.Comp Med 69:29–34. 10.30802/AALAS-CM-18-000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Getachew B, Aubee J, Schottenfed R, Csoka A, Thompson K, Tizabi Y.2018.Ketamine interactions with gut-microbiota in rats: relevance to its antidepressant and anti-inflammatory properties.BMC Microbiol 18:1–10. 10.1186/s12866-018-1373-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hart ML, Ericsson AC, Lloyd KC, Grimsrud KN, Rogala AR, Godfrey VL, Nielsen JN, Franklin CL.2018.Development of outbred CD1 mouse colonies with distinct standardized gut microbiota profiles for use in complex microbiota targeted studies.Sci Rep 8:1–11. 10.1038/s41598-018-28448-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart ML, Meyer A, Johnson PJ, Ericsson AC.2015.Comparative evaluation of DNA extraction methods from feces of multiple host species for downstream next-generation sequencing.PLoS One 10:1–16. 10.1371/journal.pone.0143334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammer O, Harper DA, Ryan PD. [Internet].2001. Past: Paleontological statics software package for education and data analysis. Palaeontological Association. [Cited 19 March 2020]. https://palaeo-electronica.org/2001_1/past/issue1_01.htm.
- 20.Hawk TA, Leary SL, Morris TH.2005.Formulary for laboratory animals. Ames, (IA):Blackwell Publishing. [Google Scholar]
- 21.Institute for Laboratory Animal Research.2011.Guide for the care and use of laboratory animals 8th ed.Washington (DC):National Academies Press. [Google Scholar]
- 22.Justice MJ, Dhillon P.2016.Using the mouse to model human disease: increasing validity and reproducibility.Dis Model Mech 9:101–103. 10.1242/dmm.024547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SM, Kim N, Yoon H, Nam RH, Lee SH.2018.Microbial changes and host response in F344 rat colon depending on sex and age following a high-fat diet.Front Microbiol 9:1–17. 10.3389/fmicb.2018.02236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livingston A, Waterman AE.1978.The development of tolerance to ketamine in rats and the significance of hepatic metabolism.Br J Pharmacol 64:63–69. 10.1111/j.1476-5381.1978.tb08641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma BW, Bokulich NA, Castillo PA, Kananurak A, Underwood MA.2012.Routine habitat change: a source of unrecognized transient alteration of intestinal microbiota in laboratory mice.PLoS One 7:1–11. 10.1371/journal.pone.0047416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montonye DR, Ericsson AC, Busi SB, Lutz C, Wardwell K, Franklin CL.2018.Acclimation and institutionalization of the mouse microbiota following transportation.Front Microbiol 9:1–13. 10.3389/fmicb.2018.01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris A.2018.Microbiota drives sex-specific differences.Nat Rev Endocrinol 15:4. 10.1038/s41574-018-0127-9. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen TL, Vieira-Silva A, Liston A, Raes J.2015.How informative is the mouse for human gut microbiota research? Dis Model Mech 8:1–16. 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu Y, Yang C, Ren Q, Ma M, Dong C, Hashimoto K.2017.Comparison of (R)-ketamine and lanicemine on depression-like phenotype and abnormal composition of gut microbiota in a social defeat stress model.Sci Rep 7:1–10. 10.1038/s41598-017-16060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pouget P, Wattiez N, Rivaud-Péchoux S, Gaymard B.2010.Rapid development of tolerance to sub-anesthetic dose of ketamine; an oculomotor study in macaque monkeys.Psychopharmacology (Berl)209:313–318. 10.1007/s00213-010-1797-8. [DOI] [PubMed] [Google Scholar]
- 31.Réus GZ, de Moura AB, Borba LA, Abelaira HM, Quevedo J.2019.Strategies for treatment-resistant depression: lessons learned from animal models.Mol Neuropsychiatry 5:178–189. 10.1159/000500324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasada T, Hinoi T, Saito Y, Adachi T, Takakura Y, Kawaguchi Y, Sotomaru Y, Sentani K, Oue N, Yasui W, Ohdan H.2015.Chlorinated water modulates the development of colorectal tumors with chromosomal instability and gut microbiota in apc-deficient mice.PLoS One 10:1–15. 10.1371/journal.pone.0132435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selvaggi G, Spagnolo AG, Elander A.2017.A review of illicit psychoactive drug use in elective surgery patients: detection, effects, and policy.Int J Surg 48:160–165. 10.1016/j.ijsu.2017.10.074. [DOI] [PubMed] [Google Scholar]
- 34.Servick K.2016.Of mice and microbes.Science 353:741–743. 10.1126/science.353.6301.741. [DOI] [PubMed] [Google Scholar]
- 35.Shreiner A, Kao JY, Young VB.2015.The gut microbiome in health and in disease.Curr Opin Gastroenterol 31:69–75. 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sofi HM, Gudi R, Karumuthil-Melethil S, Perez N, Johnson B, Vasu C.2013.pH of drinking water influences the composition of gut microbiome and type 1 diabetes incidence.Diabetes 63:632–644. 10.2337/db13-0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Systat Software. [Internet]. 2021. Download SigmaPlot Version 13. [Cited 18 July 2021]. https://systatsoftware.com/products/sigmaplot/sigmaplot-version-13/
- 38.Thaiss CA, Zmora N, Levy M, Elinav E.2016.The microbiome and innate immunity.Nature 535:65–74. 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All 16S rRNA amplicon sequencing data presented here have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject ID PRJNA705656.