ABSTRACT
Immune health requires innate and adaptive immune cells to engage precisely balanced pro- and anti-inflammatory forces. We employ the concept of chemical immunophenotypes to classify small molecules functionally or mechanistically according to their patterns of effects on primary innate and adaptive immune cells. The high-specificity, low-toxicity cyclin-dependent kinase 8 (CDK8) inhibitor 16-didehydro-cortistatin A (DCA) exerts a distinct tolerogenic profile in both innate and adaptive immune cells. DCA promotes regulatory T cells (Treg) and Th2 differentiation while inhibiting Th1 and Th17 differentiation in both murine and human cells. This unique chemical immunophenotype led to mechanistic studies showing that DCA promotes Treg differentiation in part by regulating a previously undescribed CDK8-GATA3-FOXP3 pathway that regulates early pathways of Foxp3 expression. These results highlight previously unappreciated links between Treg and Th2 differentiation and extend our understanding of the transcription factors that regulate Treg differentiation and their temporal sequencing. These findings have significant implications for future mechanistic and translational studies of CDK8 and CDK8 inhibitors.
KEYWORDS: CDK8, GATA3, T cell differentiation, T cells, cyclin-dependent kinases
INTRODUCTION
The immune system comprises innate and adaptive immune cells whose collaborative and coordinated responses maintain the healthy state. Each cell type can exert either pro- or anti-inflammatory forces. For example, innate immune cells can secrete either pro-inflammatory (e.g., gamma interferon [IFN-γ]) or anti-inflammatory (e.g., interleukin-10 [IL-10]) cytokines; similarly, CD4+ T cells can differentiate into either pro- (e.g., Th1 and Th17) or anti- (regulatory T cells [Treg]) inflammatory subsets (1–5). These pro- and anti- inflammatory forces must be precisely balanced; dysregulation of this balance can predispose to autoimmunity, infection, or cancer (3, 6).
We have previously demonstrated how small molecules can highlight novel pathways of immunoregulation in primary immune cells. For example, we showed that small-molecule inhibition of the dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A) promotes differentiation of murine and human CD4+ T cells into Tregs (7). We also showed that small-molecule inhibition of salt-induced kinases (SIKs) enhanced the production of IL-10 by murine and human myeloid cells (8). However, a comprehensive understanding of how both innate and adaptive immune cell function is modulated remains lacking for most small molecules.
Here, we investigate the effect of the natural-product-derived small molecule 16-didehydro-cortistatin A (DCA; Pubchem compound identification number [CID] 24898002) on murine and human CD4+ T cells. Cortistatin A was discovered as a marine sponge steroidal alkaloid and found to be a cyclin-dependent kinase 8 (CDK8) and CDK19 (CDK8/19) inhibitor (9–11). The analog DCA was subsequently found to be an equipotent and more easily synthesized compound (12). Recent studies pointing to DCA as the CDK8 inhibitor with the highest specificity and lowest toxicity highlight DCA as a CDK8 inhibitor of critical interest (13). CDK8 is an essential component of the CDK8 submodule of the Mediator coactivator complex, which regulates RNA polymerase II activity (14, 15). The CDK8 submodule facultatively binds the Mediator complex, phosphorylates transcription factors, and regulates specific pathways positively or negatively (15–18). CDK19 is a paralogous Mediator-associated kinase likely to be comparably inhibited by DCA, although CDK19 function is relatively less understood (18). CDK8 phosphorylates several immune-relevant transcription factors, including STAT1Ser727, STAT3Ser727, STAT5Ser730, c-JunSer243, and Notch (19–22). CDK8 regulates both innate and adaptive immune responses, and CDK8 inhibition typically exerts tolerogenic effects. We previously found that DCA promotes the production of IL-10 in myeloid cells by inhibiting CDK8 (20). Other innate immunity effects include improved tumor surveillance with deletion of CDK8 in NK cells (23, 24). In adaptive immune cells, CDK8/19 inhibitors promote the differentiation of anti-inflammatory Tregs (21, 25). Recent findings that CDK8 inhibition promotes Th17 differentiation suggest the first proinflammatory sequelae (26). How CDK8 regulates the differentiation to other T cell lineages (Th1 and Th2) remains less clear. Furthermore, much of the mechanistic work in T cells has focused on CDK8 phosphorylation of STAT5 and STAT3. The possibility of additional CDK8-regulated pathways in the context of T cell biology is suggested by our findings that CDK8 regulates myeloid cells by c-JunSer243 phosphorylation; however, the identity of these pathways remains incompletely elucidated (20). Understanding these pathways is essential to identify the patients who might benefit most from CDK8 inhibition therapy.
We demonstrate that DCA exerts a unique pattern of immunomodulation (i.e., chemical immunophenotype) compared to other known immunomodulatory small molecules. Using both small-molecule inhibitors and CRISPR-Cas9 (CRISPR-associated protein 9) knockout, we find that DCA inhibits CDK8 to promote the differentiation of both Treg and Th2 cells while suppressing the differentiation of proinflammatory Th1 and Th17 subsets. We show that DCA-driven Tregs are fully suppressive in the absence of concomitant tolerogenic effects on innate immune cells. Mechanistically, CDK8 inhibition by DCA regulates Treg/Th17/Th1 differentiation independent of effects on STAT1/STAT3 Ser727 phosphorylation. Notably, DCA’s unusual chemical immunophenotype directly leads us to find that DCA uniquely drives early temporal expression of FOXP3 at least in part via a CDK8-GATA3-FOXP3 pathway not previously described to regulate Treg differentiation. These findings further our mechanistic understanding of an emerging role for DCA as an immunomodulator that broadly drives tolerogenic programs in both innate and adaptive immune cells. These findings are discussed in the context of implications for future therapeutic use of CDK8 inhibitors.
RESULTS
DCA exerts tolerogenic effects on murine and human CD4+ T cell differentiation.
Given our previous observation that DCA promotes tolerogenic IL-10 production in innate immune cells (20), we determined whether DCA exerts tolerogenic effects on CD4+ T cell differentiation. We tested the effect of DCA on naive murine CD4+ T cells cultured under suboptimal pro-Treg or -Th2 conditions (Treglow and Th2low, respectively) as we described previously (7). DCA enhanced the differentiation of both Treg and Th2 cells (Fig. 1A). DCA increased (FOXP3+) Tregs specifically in cultures of fluorescence-activated cell sorting (FACS)-sorted naive CD4+ T cells but did not expand sorted naturally occurring thymic Tregs cultured under identical conditions, further demonstrating that the increase in Tregs is due to enhanced differentiation of Tregs (from naive CD4+ T cells) rather than expansion of existing Tregs (Fig. 1B). To examine whether these tolerogenic effects extended to inhibiting the differentiation of proinflammatory T cell lineages, we added DCA to murine T cells cultured under near-optimal pro-Th1 and -Th17 conditions (Th1hi and Th17hi, respectively). DCA significantly inhibited the differentiation of Th1 and Th17 cells (Fig. 1C). Notably, DCA promoted the differentiation of Treg and Th2 cells even under near-optimal Th17hi and Th1hi conditions, respectively (Fig. 1C, FACS plots). In the context of nonpolarizing Th0 conditions, DCA enhanced murine Treg and Th2 differentiation significantly, albeit modestly (Fig. 1D). Th1 differentiation was slightly reduced below the level of statistical significance, and Th17 cells were too infrequent to assess accurately (Fig. 1D). These results suggest that DCA can enhance Treg/Th2 differentiation in the absence of exogenous cytokines. Therefore, DCA exerts powerful and broad tolerogenic effects on murine T cell differentiation.
FIG 1.
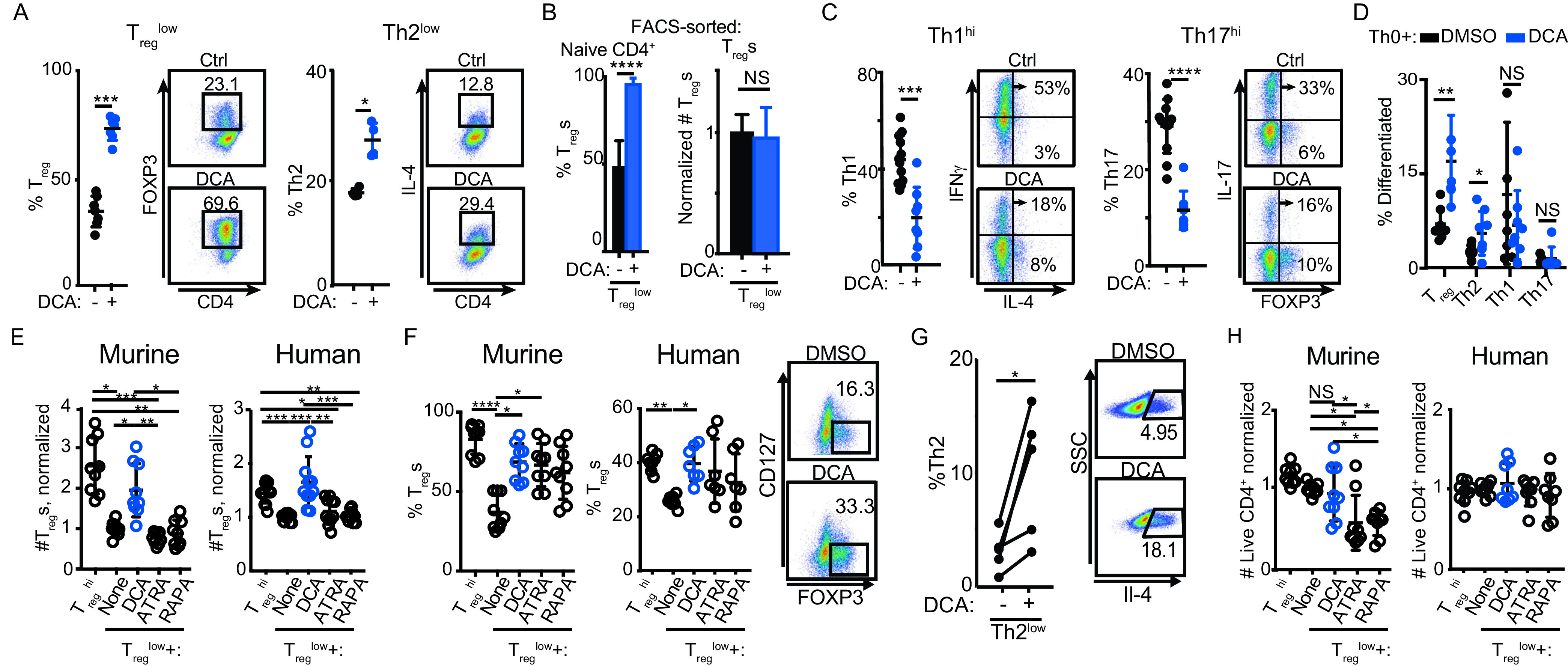
DCA broadly regulates differentiation of murine and human T cells. (A to D) Effect of DCA on murine naive CD4+ T cells cultured under suboptimal pro-Treg or -Th2 conditions (Treglow and Th2low, respectively) (A, B), near-optimal pro-Th1 or -Th17 conditions (Th1hi and Th17hi, respectively) (C), or neutral Th0 conditions (D) (n = 4 to 12 mice across 4 experiments). (B) Effect of DCA on FACS-sorted murine naive CD4+ T cells (left) versus FACS-sorted (FOXP3+) murine Tregs (right) cultured under Treglow conditions; FOXP3+ Tregs were quantitated at the end of the experiment (n = 4 to 6 mice across 4 experiments). (E, F) Effects of DCA, all-trans retinoic acid (ATRA), and rapamycin (RAPA) on numbers (E) and percentages (F) of Tregs generated from murine (n = 9 mice across 4 experiments) and human (n = 7 or 8 subjects across 3 experiments) naive CD4+ T cells cultured under Treglow conditions. (G) Effect of DCA on naive human CD4+ cells cultured under Th2low conditions (n = 5 subjects across 2 experiments). SSC, side scatter. (H) Comparison of how DCA, ATRA, and RAPA affect survival of murine (n = 9 mice across 4 experiments) and human (n = 7 or 8 subjects across 3 experiments) CD4+ T cells cultured under Treglow conditions. Mann-Whitney (A to D), Kruskal-Wallis (E, F, and H), and paired t test (G) were used. *, P < 0.05; **, P < 0.01; ***, P < 0.001 ****; P < 0.0001; NS, not significant.
We next investigated whether DCA similarly affects human Treg and Th2 differentiation by culturing human CD4+ T cells under (human-specific) suboptimal Treglow and Th2low conditions, respectively. DCA treatment enhanced both Treg and Th2 differentiation in human CD4+ T cells, pointing to concordant regulation in human and murine cells (Fig. 1E to G). We benchmarked the pro-Treg effect of DCA against the well-described Treg enhancers all-trans retinoic acid (ATRA) and rapamycin (RAPA) (27–33). In murine and human CD4+ T cells cultured under suboptimal Treglow conditions, DCA treatment enhanced the total number of Tregs significantly more than either ATRA or rapamycin (Fig. 1E). In addition, DCA enhanced the percentages of Tregs similarly to ATRA and rapamycin (Fig. 1F). These results highlight that DCA potently enhances Treg differentiation in both murine and human T cells and may reflect in part the lower cytotoxicity of DCA than of ATRA and rapamycin (Fig. 1H).
DCA identifies a novel CDK8 inhibition-driven chemical immunophenotype.
We compared DCA’s profile of tolerogenic effects against those of other tolerogenic compounds. We investigated the dose-response of murine CD4+ T cells to DCA and two other tolerogenic small molecules in the context of suboptimal pro-Treg, Th2, Th1, and Th17 conditions (Treglow, Th2low, Th1low, and Th17low, respectively) (7). These experiments showed that DCA enhanced the differentiation of both murine Treg and Th2 cells with similar dose-response relationships, supporting the involvement of a common mechanistic target (Fig. 2A). We wanted to understand whether DCA’s ability to enhance Treg and Th2 differentiation and myeloid IL-10 production represents a pattern common to many tolerogenic small molecules. We tested harmine, which we previously identified as a potent enhancer of Treg differentiation, and found that harmine enhanced the differentiation of Treg but not of Th2 cells (Fig. 2A) (7). We also tested HG-9-91-01, which we previously showed enhances myeloid cell production of IL-10 production by inhibiting salt-inducible kinases 1 to 3 (SIK1 to -3), and we found that HG-9-91-01 enhanced neither Treg nor Th2 differentiation (Fig. 2A) (8). Therefore, DCA, HG-9-91-01, and harmine exert distinct immune phenotypic profiles, which we term chemical immunophenotypes, reflecting the engagement of distinct pathways regulating tolerogenicity in innate and adaptive immune cells.
FIG 2.
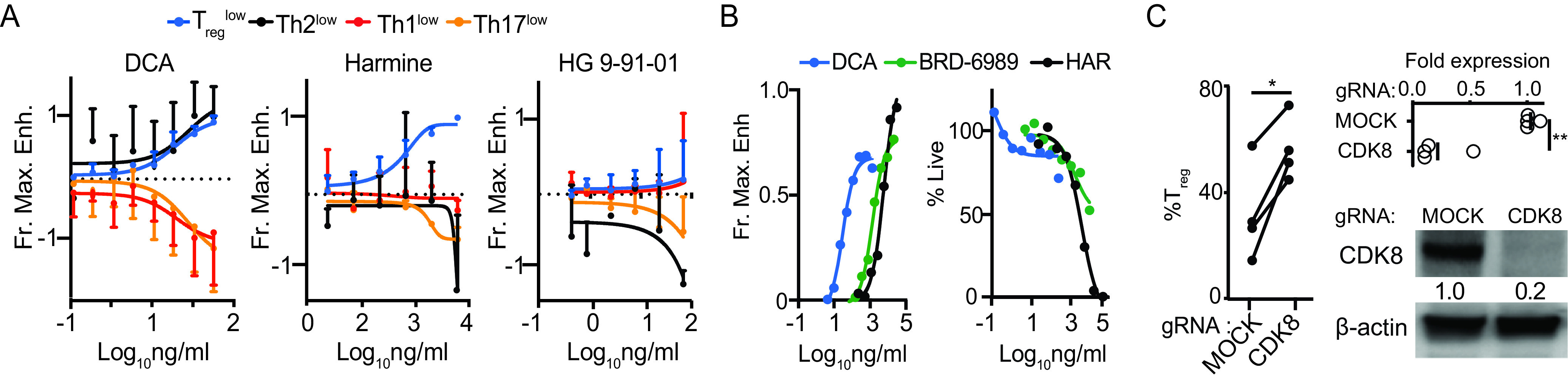
DCA describes a unique chemical immunophenotype. (A) Dose-response curves showing effects of DCA, harmine and HG-9-91-01 on murine CD4+ T cells cultured under suboptimal Treglow, Th2low, Th1low, and Th17low conditions (n = 3 to 5 mice across 3 to 5 experiments). Fractional maximal enhancement (Fr. Max. Enh.) was determined by increase in percentage of lineage-committed cells relative to maximal cytokine-driven enhancement, as previously reported (7). (B) Naive murine CD4+ T cell cultures showing dose-response for the CDK8 inhibitors DCA and BRD-6989 on Treg differentiation (left) and culture cellularity (right) (representative of 3 independent experiments). Harmine (HAR) is included for comparison. (C) Effect of CRISPR-Cas9 editing of CDK8, compared to no-guide control, on propensity of human CD4+ T cells to differentiate into Tregs (left, n = 4 subjects across 2 experiments). CDK8 expression was quantitated by qPCR (n = 4 subjects across 2 experiments) and Western blotting (representative of 3 independent subjects); paired t test was used. *, P < 0.05; **, P < 0.01.
In addition to our results with DCA, previous studies have shown that other CDK8/19 inhibitors, including CCT251921, Senexin A (25), and AS2863619 (21), also enhance Treg differentiation. We used two different approaches to further validate CDK8 as the Treg-relevant mechanistic target of DCA. First, we tested DCA alongside a structurally distinct small-molecule CDK8 inhibitor, BRD-6989 (20). Under Treglow conditions, both CDK8 inhibitors showed concentration-dependent enhancement of murine Treg differentiation, with a 50% effective concentration (EC50) for each compound similar to that observed for enhancing IL-10 production in bone-marrow-derived dendritic cells (BMDCs) (Fig. 2B) (20). These data are consistent with CDK8 50% inhibitory concentration (IC50) data for cortistatin A (11) and BRD-6989 (20). The EC50 of DCA was much lower than that of BRD-6989, driving its subsequent preferential use (Fig. 2B). Notably, DCA and BRD-6989 both exhibited modest impacts on the absolute numbers of T cells, even less than that observed with harmine, which we previously identified as one of the least cytotoxic Treg enhancers (Fig. 2B) (7). Second, we used CRISPR-Cas9 to knock out CDK8 in primary human CD4+ T cells. Efficient editing of CDK8 led to enhanced Treg differentiation comparable to the levels observed using DCA treatment (Fig. 1F and 2C). These results indicate that DCA enhances murine and human Treg differentiation at least in part by inhibiting CDK8.
DCA-driven Tregs are fully tolerogenic in the absence of DCA innate immune cell tolerogenic effects.
We next interrogated the suppressive capacity of DCA-driven Treg cells both in vitro and in vivo. Flow cytometric analysis of several proteins important for Treg function showed no significant difference between Tregs generated under Treghi or Treglow+DCA conditions (Fig. 3A). Using a standard in vitro suppression assay, we also observed no significant differences in the ability of Treghi- or Treglow+DCA-driven murine Treg cells to suppress the proliferation of cocultured responder CD4+ T cells (Student’s t test) (Fig. 3B, red and blue lines, respectively). We tested the capacity of DCA-driven Tregs to inhibit inflammation in vivo in two murine Treg transfer models in order to exclude confounding effects of systemically delivered DCA on endogenous innate immune cells. In an established model of type 1 diabetes, transfer of NOD-BDC2.5+ CD4+ T cells, specific for an epitope derived from the islet antigen chromogranin A, into NOD-scid recipients results in islet β-cell destruction and the onset of diabetes about 10 days later (Fig. 3C, black line) (34, 35). Coinjection of antigen-specific Treg cells, generated from naive NOD-BDC2.5.Foxp3IRES-GFP CD4+ T cells using either Treglow+DCA or Treghi conditions, significantly delayed the onset of diabetes to similar degrees (Fig. 3C, blue and red lines, respectively) (7). We observed similar results in a murine model of intestinal inflammation, where transfer of CD45RBhi CD4+ T cells into B10.RAG2−/− recipients resulted in the expansion of donor T cells and inflammation most prominently in the colon about 4 weeks later (Fig. 3D, black) (36, 37). Cotransfer of Treg cells generated from naive wild-type C57BL/6 CD4+ T cells using either Treglow+DCA or Treghi conditions resulted in significant and similar attenuation of intestinal inflammation (Fig. 3D, blue and red, respectively) (38). Together, these results demonstrate that DCA-driven Treg cells are fully functional and equivalent to Treghi-generated Treg cells (an adoptive cellular therapy-relevant gold standard comparison) both in vitro and in vivo, using model systems employing different genetic backgrounds and T cell specificities. Importantly, these experiments demonstrate that DCA exerts a strong Treg-intrinsic tolerogenic effect in the absence of concomitant effects on the innate immune compartment.
FIG 3.
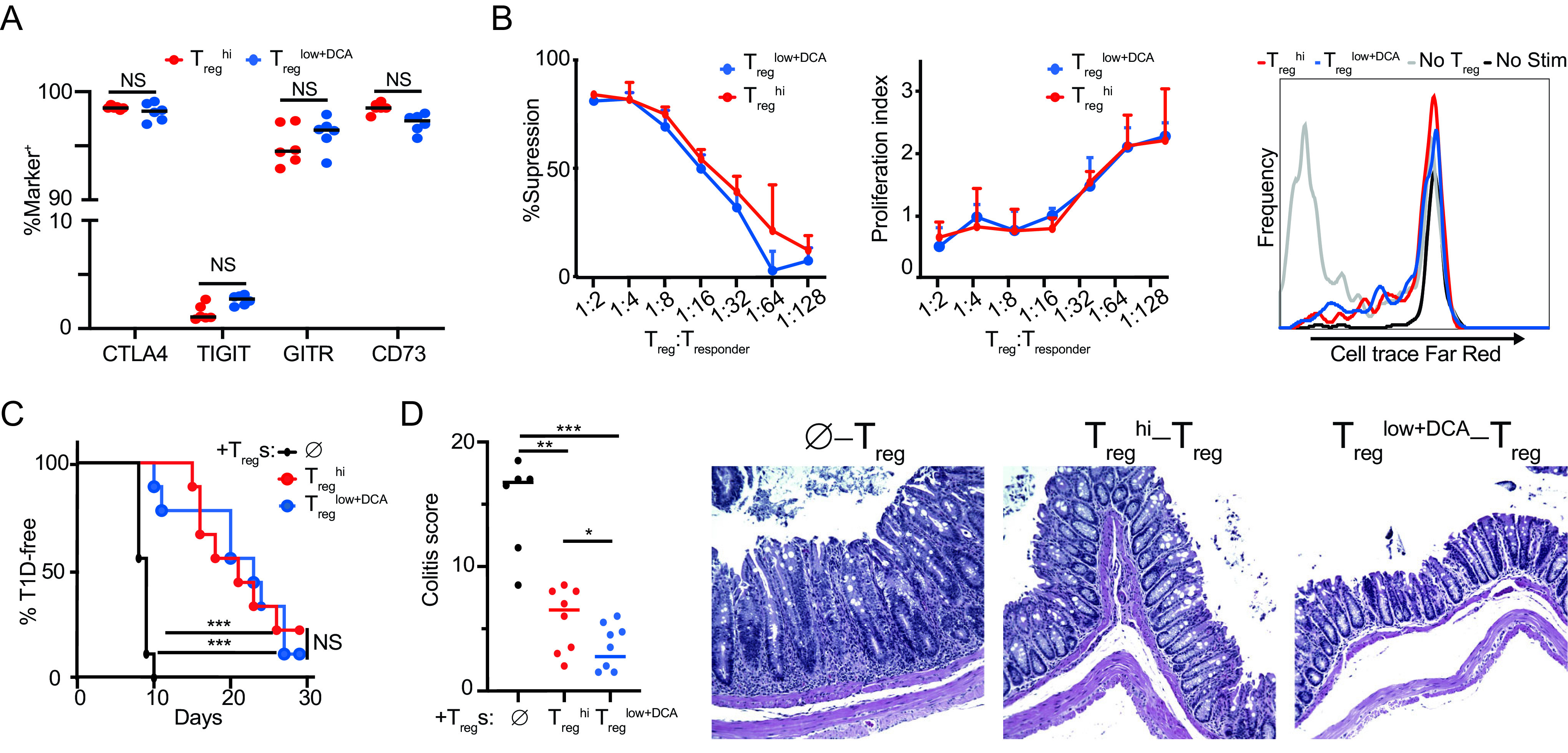
DCA enhances differentiation of functional Tregs. Phenotype and suppressive function of DCA-driven Tregs (Treglow+DCA) compared to Treghi-driven Tregs. (A) Expression of canonical Treg function-associated proteins. (B) Standard in vitro suppression assay (Tregs at 1:16) showing percentages of suppression (left), proliferation index (middle), and representative histograms (right) (n = 3 technical replicates, representative of 3 experiments; no difference at all points by Student’s t test). (C) NOD.BDC2.5 model of type 1 diabetes (n = 9 mice per condition across 2 experiments). (D) B10.RAG2−/− model of colitis (n = 7 or 8 mice per condition across 2 experiments). , no-Treg controls. Mann-Whitney (A, D) and Mantel-Cox (C) tests were used, *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant.
DCA exerts tolerogenic effects on T cell differentiation independent of regulating STAT1Ser727, STAT3Ser727, and c-JunSer243 phosphorylation.
We investigated key candidates that might account for DCA’s tolerogenic effects in CD4+ T cells. STAT1Ser727 and STAT3Ser727 are phosphorylated in several cell types by kinases, including CDK8 (39–42). Although the role of Ser727 phosphorylation in Th1/Th17/Treg differentiation is unclear, a potential contribution is suggested by the central role of STAT1Tyr701 and STAT3Tyr705 tyrosine phosphorylation in Th1 and Th17 differentiation, respectively (43–45). Recent studies argue that inhibition of CDK8 promotes Th17 differentiation by attenuating STAT3Ser727 phosphorylation, emphasizing the importance of investigating this pathway (26). DCA reduced IL-6-induced STAT3Ser727 phosphorylation in murine CD4+ T cells but did not reduce either STAT3Tyr705 phosphorylation or the expression of the hallmark Th17 transcription factor RORγt in cells cultured under Th17hi conditions; total STAT3 was slightly decreased (Fig. 4A to C). Primary CD4+ T cells from Stat3Ser727Ala mice, in which the Ser727Ala mutation abrogates STAT3Ser727 phosphorylation, showed reduced Th17 differentiation, highlighting a previously unappreciated role of STAT3Ser727 phosphorylation in this process (Fig. 4D) (40). DCA decreased Th17 differentiation in Stat3Ser727Ala cells, albeit to a lesser extent than in wild-type cells, possibly due in part to the lower baseline, showing that DCA can regulate Th17 differentiation via STAT3Ser727-independent mechanisms (Fig. 4D). DCA enhanced Treg differentiation comparably in both Stat3Ser727Ala and wild-type CD4+ T cells, demonstrating that DCA regulates Treg differentiation via mechanisms independent of STAT3Ser727 phosphorylation (Fig. 4E).
FIG 4.
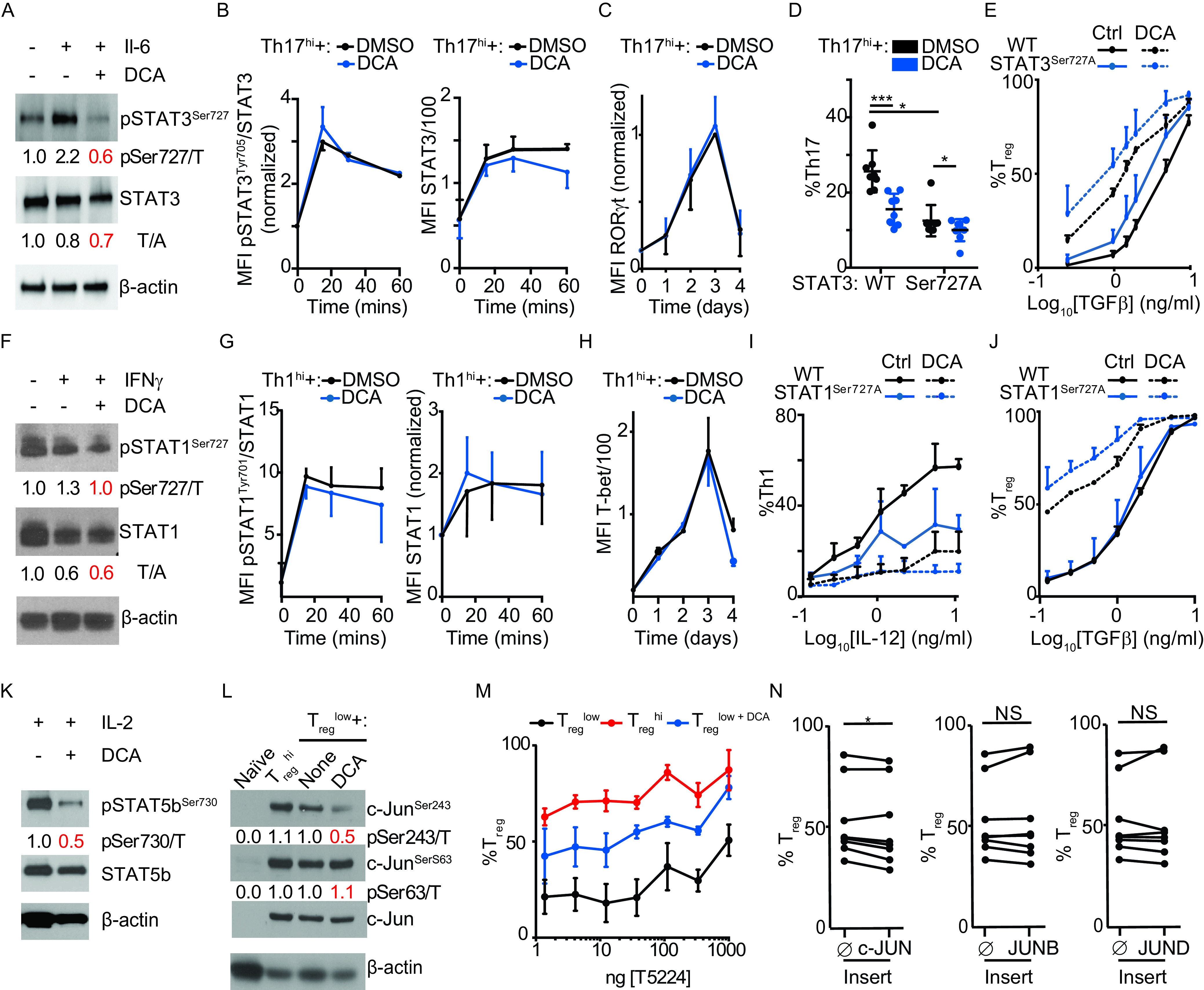
CDK8 inhibition does not impact canonical pathways of Treg differentiation. (A) Effect of DCA on IL-6-induced STAT3Ser727 phosphorylation in resting murine CD4+ T cells (representative of 3 independent experiments). (B, C) Effects of DCA on phospho-STAT3Tyr705 and total STAT3 (n = 2 mice across 2 experiments) (B) and RORγ t (n = 3 mice across 3 experiments) (C) in murine CD4+ T cells cultured under Th17hi conditions. MFI, mean fluorescence intensity. (D, E) Effects of DCA on Th17 (D) and Treg (E) differentiation in Stat3Ser727Ala naive murine CD4+ T cells (n = 8 mice across 3 experiments). WT, wild type. (F) Effect of DCA on IFN-γ-induced STAT1Ser727 phosphorylation in resting murine CD4+ T cells (representative of 2 independent experiments). (G, H) Effects of DCA on phospho-STAT1Tyr705 and total STAT1 (n = 2 mice across 2 experiments) (G) and T-bet (n = 3 mice across 3 experiments) (H) in murine CD4+ T cells cultured under Th1hi conditions. (I, J) Effects of DCA on Th1 (I) and Treg (J) differentiation in Stat1Ser727Ala naive murine CD4+ T cells (n = 4 mice across 2 experiments). (K) Effect of DCA on IL-2-induced STAT5bSer730 phosphorylation in resting murine CD4+ T cells (representative of 2 independent experiments). (L) Effects of DCA on c-Jun phosphorylation at Ser243 and Ser63 (representative of 3 independent experiments). (M) Effect of the c-Fos inhibitor T5224 on DCA’s ability to enhance Treg differentiation in murine CD4+ T cells cultured under Treglow conditions (n = 3 mice, representative of 2 experiments). The x axis shows nanograms per milliliter. (N) Effects of overexpressing AP-1 family members using NGFR-T2A-tagged lentivirus, compared to NGFR control (), on Treg differentiation in human CD4+ T cells cultured for 5 days under Treglow conditions (n = 8 subjects across 2 experiments). (A, F, K, and L) Ratio of phospho-STAT/c-Jun to total STAT/c-Jun (p/T) or of total STAT to β-actin (T/A) as indicated. Mann-Whitney test (D) or Wilcoxon matched-pair analysis (N) was used. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant.
We also showed that DCA reduced IFN-γ-induced phospho-STAT1Ser727 but not phospho-STAT1Tyr701 or total STAT1 in cells cultured under Th1hi conditions (Fig. 4F and G). The expression of the hallmark Th1 transcription factor T-bet in Th1hi cultures was also unaltered by DCA except at day 4, arguing against altered regulation by STAT1Ser727 (Fig. 4H). Primary CD4+ T cells from Stat1Ser727Ala mice, in which Ser727Ala mutation abrogates STAT1Ser727 phosphorylation, showed reduced Th1 differentiation, highlighting a previously unappreciated role of STAT1Ser727 phosphorylation in this process (Fig. 4I) (41). DCA decreased Th1 differentiation in Stat1Ser727Ala cells, albeit to a lesser extent than in wild-type cells, possibly due in part to the lower baseline, showing that DCA can regulate Th1 differentiation via STAT1Ser727-independent mechanisms (Fig. 4I). DCA enhanced Treg differentiation comparably in both Stat1Ser727Ala and wild-type CD4+ T cells, demonstrating that DCA regulates Treg and Th1 differentiation via mechanisms independent of STAT1Ser727 phosphorylation (Fig. 4J).
Consistent with previously published findings, we also found that treatment with DCA inhibited IL-2-induced phosphorylation of STAT5bSer730 (Fig. 4K) (21). We were unable to find Stat5bSer730Ala mice to perform similar studies as with STAT1 and STAT3 above.
We next investigated the overlap between characterized CDK8-regulated pathways in innate and adaptive immune cells. Similar to our findings in myeloid cells, T cells cultured under either Treglow or Treghi conditions showed increased phosphorylation of both c-JunSer243 and c-JunSer63; DCA specifically inhibited phosphorylation on the inhibitory site c-JunSer243 (Fig. 4L) (20). Unlike in myeloid cells, DCA’s tolerogenic pro-Treg effect in T cells was neither attenuated by the AP-1 inhibitor T-5224 nor enhanced by overexpression of multiple c-Jun family members (c-Jun, JunB, or JunD) (Fig. 4M and N). Together, these results indicate that DCA regulation of AP-1 transcription factors drives tolerogenicity in myeloid but not CD4+ T cells.
DCA enhances Foxp3 expression by engaging GATA3.
Temporal flow cytometric analysis of FOXP3 expression throughout the period of culture revealed indistinguishable kinetics between murine CD4+ T cells cultured under Treglow and Treghi conditions until day 2, with FOXP3+ cells increasing under Treghi conditions and decreasing under Treglow conditions thereafter (Fig. 5A) (7). Notably, DCA treatment increased FOXP3+ cells, as well as Foxp3 expression, significantly at early time points (days 1 and 2) compared to either Treglow or Treghi conditions (Fig. 5A and B). DCA treatment also drove concordant regulation of other FOXP3-regulated genes at day 2, including upregulation of Eos, Helios, and Cd25, as well as downregulation of Il2 expression (Fig. 5B) (46–49). These data suggest that DCA promotes murine Treg differentiation at least in part by enhancing early expression of FOXP3. This induction of key Treg transcription factors did not involve canonical Treg pathways; specifically, DCA neither enhanced phosphorylation of SMAD2/SMAD3 nor inhibited phosphorylation of the mTOR pathway members ribosomal S6/ribosomal S6 kinase (S6K), pointing to the involvement of a novel pathway(s) (Fig. 5C and D).
FIG 5.
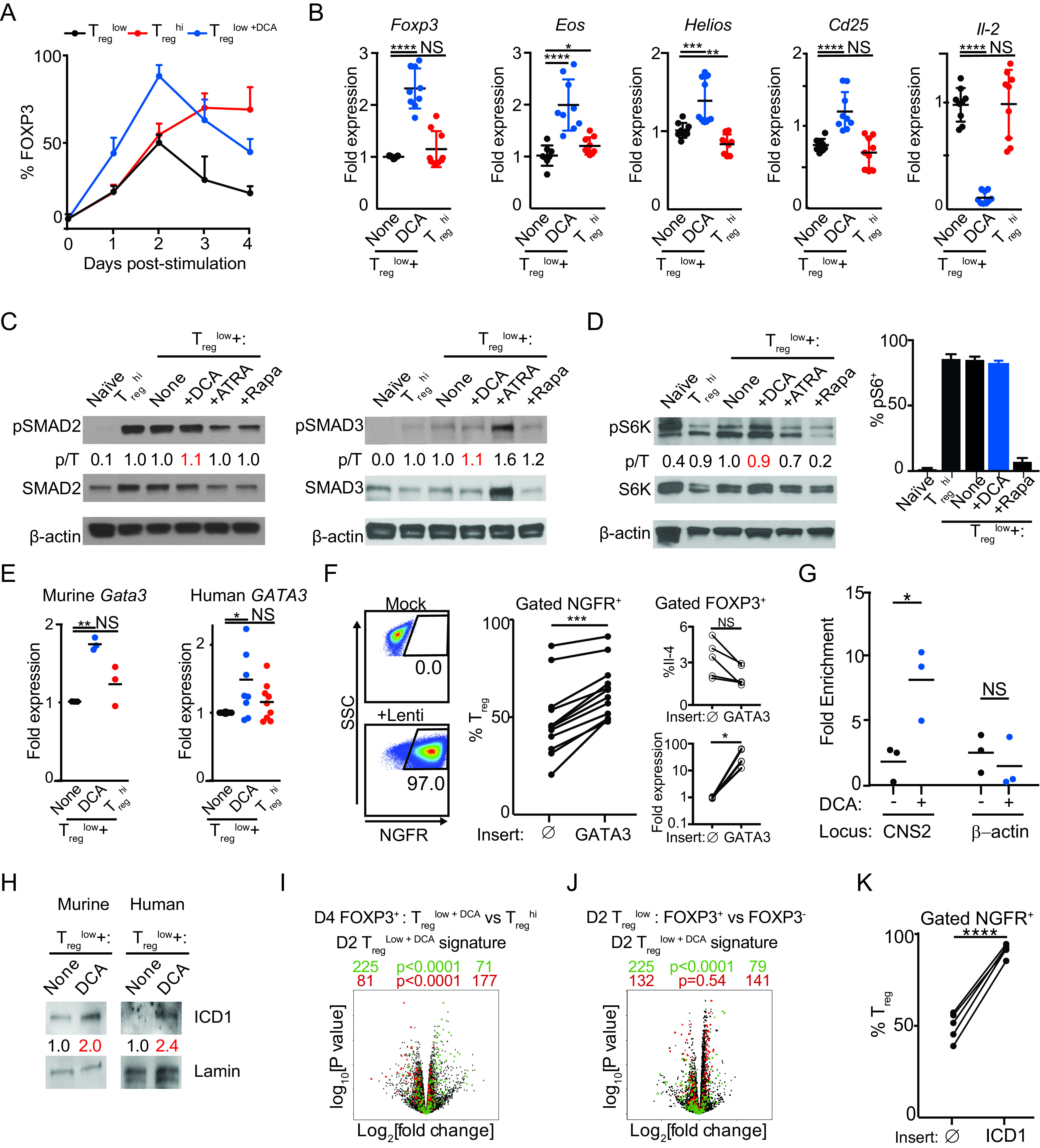
DCA drives novel early FOXP3 expression via a novel CDK8-Notch-GATA3 pathway. (A) Time course of FOXP3 expression in murine CD4+ T cells cultured under Treglow, Treglow+DCA, and Treghi conditions (n = 10 mice across 5 experiments). (B) Effect of DCA on expression of FOXP3-regulated genes in murine CD4+ T cells cultured for 2 days under Treglow conditions (n = 9 mice across 3 experiments). (C) Effects of DCA on phosphorylation of SMAD2 and SMAD3 in murine CD4+ T cells stimulated under indicated conditions (≥2 independent experiments). (D) Effect of DCA on mTOR activity, assessed by Western blotting of S6K phosphorylation (left) and flow cytometric analysis of S6 phosphorylation (right) in murine CD4+ T cells stimulated under the indicated conditions. (C, D) Ratios of phospho-SMAD/S6K to total SMAD/S6K (p/T) are indicated (representative of ≥2 experiments). (E) Effects of DCA on Gata3 expression in murine (n = 3 mice across 3 experiments) and human (n = 9 subjects across 3 experiments) CD4+ T cells, cultured for 2 days under Treglow conditions. (F) Effect of overexpressing GATA3 using NGFR-T2A-tagged lentivirus, compared to NGFR control (), on Treg differentiation in human CD4+ T cells cultured under Treglow conditions (n = 12 subjects across 3 experiments). Effects on Treg expression of IL-4 and GATA3 expression are also shown (right, n = 5 subjects across 2 experiments). (G) ChIP-qPCR quantitation of how DCA treatment impacts GATA3 binding to FOXP3 CNS2 in human CD4+ T cells cultured for 2 days under Treglow conditions (n = 3 subjects across 3 experiments). β-Actin is included as a control locus. (H) Effects of DCA on intranuclear levels of Notch1 intracellular domain (ICD1), normalized to nuclear Lamin B1 levels, in murine and human CD4+ T cells stimulated for 2 h under indicated conditions (representative of ≥2 independent experiments). (I, J) Volcano plots comparing gene expression in sorted mature murine Tregs generated under Treglow+DCA versus Treghi conditions (I) and murine CD4+ T cells cultured for 2 days under Treglow conditions (J), sorted FOXP3+ versus FOXP3− cells. DCA signature genes, obtained by comparing sorted FOXP3+ cells cultured for 2 days under Treglow+DCA versus Treglow conditions, are highlighted in green (downregulated) and red (upregulated). Numbers on the right and left reflect signature genes that are up- or downregulated, respectively, in the indicated comparison, with χ2 test P values in the middle. (K) Effect of overexpressing Notch ICD1 using NGFR-T2A-tagged lentivirus, compared to NGFR control (), on Treg differentiation in human CD4+ T cells cultured under Treglow conditions (n = 5 subjects across 3 experiments). Mann-Whitney test (B), Wilcoxon matched-pair analysis (E, F), and paired t test (G, K) were used. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; NS, not significant.
DCA’s unusual chemical immunophenotype led us to consider a mechanistic link between DCA-mediated enhancement of Treg and Th2 differentiation. GATA3, the hallmark Th2 transcription factor, is highly expressed beginning at the earliest time points of Th2 differentiation (50). Although not well studied in the context of Treg differentiation, previous findings that GATA3 binds the CNS2 enhancer element of the FOXP3 locus (about 4.5 kb downstream of the promoter) and regulates mature Treg physiology support the possibility that GATA3 could regulate FOXP3 expression earlier in Treg ontogeny (51–53). Consistent with this notion, DCA treatment of both murine and human CD4+ T cells cultured under Treglow conditions also enhanced early GATA3 expression at day 2 (Fig. 5E). The smaller increase in human than in mouse T cells could reflect effects of increased genetic diversity, although small changes in transcription factor levels can have large functional impacts. The similar levels of GATA3 expression in cells cultured under Treglow and Treghi conditions are consistent with the similar FOXP3 expression levels at this time point and emphasize the unusual features of DCA regulation. To validate the role of DCA-mediated upregulation of GATA3 in Treg differentiation, we generated lentiviral vectors to overexpress GATA3, including a truncated nerve growth factor receptor (NGFR) marker separated by a self-splicing T2A peptide to allow specific comparison of transduced cells. We performed these experiments using human T cells because these were more amenable to viral transduction. Overexpression of GATA3 consistently enhanced Treg differentiation in naive human CD4+ T cells cultured under Treglow conditions compared to the Treg differentiation in cells transduced with control (NGFR only) virus (Fig. 5F). Although lentiviral transduction drove GATA3 transcript levels higher than DCA, no increased expression of IL-4 was seen in GATA3-overexpressing Tregs (Fig. 5E and F). To further support the functional significance of DCA-enhanced GATA3, we performed chromatin immunoprecipitation (ChIP) experiments and found that DCA treatment of human CD4+ T cells cultured under Treglow conditions resulted in significantly increased binding of GATA3 specifically to FOXP3 CNS2 early in Treg differentiation (Fig. 5G). These results argue that GATA3 is an early regulator of FOXP3 expression and Treg differentiation that can be regulated by DCA.
We sought to better understand how DCA might regulate GATA3 expression. Previous studies showed that Notch can directly drive Gata3 expression and that enhanced Notch signaling promotes Treg differentiation (54, 55). Furthermore, CDK8 inhibits Notch signaling by phosphorylating the Notch1 signaling domain ICD1, leading to its degradation, leading us to hypothesize that DCA may drive Gata3 expression by enhancing Notch signaling in T cells (22). Consistent with this notion, we found that treatment with DCA led to increased intranuclear levels of ICD1 in both murine and human CD4+ T cells (Fig. 5H). Supporting the functional relevance of DCA-driven increased intracellular ICD1, we performed transcriptome sequencing (RNA-seq) analyses comparing FOXP3+ cells cultured for 2 days under Treglow versus Treglow+DCA conditions to define a 577-gene signature associated with DCA treatment. As previously described, we separately interrogated whether up- and downregulated genes in this signature were regulated in the same direction in the expression profiles of sorted mature FOXP3+ induced Tregs (iTregs) generated after 4 days of culture in Treglow+DCA versus Treghi conditions; we found statistically significant concordant regulation of both up- and downregulated signature genes, supporting Treg relevance of this signature (Fig. 5I) (7, 56). We also found that downregulated signature genes were concordantly regulated in FOXP3+ versus FOXP3− cells cultured for 2 days under Treglow conditions, supporting the FOXP3 relevance of this signature (Fig. 5J). Transcription factor target analysis of this DCA signature using the chromatin immunoprecipitation sequencing (ChIP-seq) result-based Gene Transcription Regulation Database (GTRD) in the Molecular Signatures database (MSigDB) revealed Mastermind-like (MAML) as the most enriched transcription factor (see Table S1 in the supplemental material) (57–59). MAML is recruited by DNA-bound ICD1, in complex with RBP-J, whereupon it recruits transcriptional coactivators (60). Therefore, enrichment of MAML binding sites is consistent with enrichment of ICD1 binding to DCA-regulated genes. Further supporting this notion, overexpression of ICD1 also enhances Treg differentiation (Fig. 5K). Together, these results demonstrate for the first time that inhibition of CDK8 by DCA drives FOXP3 expression and Treg differentiation at least in part by driving increased Notch and GATA3 signaling.
DISCUSSION
Here, we have demonstrated that DCA promotes Treg differentiation at least in part by engaging a previously unappreciated CDK8-GATA3-FOXP3 pathway. Our use of both novel small molecules (DCA and BRD-6989) and CRISPR-Cas9-mediated deletion point to CDK8 inhibition as a potential mechanism by which DCA exerts these effects. Genetic studies, in particular using conditional knockout mice, represent important future approaches to better define potential CDK8-independent roles of DCA, as well as the role of the CDK8 paralog CDK19, in Th differentiation. While previous studies have shown that GATA3 impacts mature Treg physiology, this is the first report, to our knowledge, that GATA3 can drive Treg differentiation (51, 61). Our findings extend prior studies showing that GATA3 protein levels are upregulated during Treg differentiation (62). This is further supported by our GATA3 overexpression experiments, although overexpression to levels more similar to those driven by DCA remains to be investigated. Whether GATA3 initiates, stabilizes, or amplifies FOXP3 expression and the precise cis-acting elements involved remain to be clearly dissected in future studies. Previous reports suggesting that GATA3 inhibits FOXP3 expression used significantly different experimental approaches, including secondary CD3 stimulation or removal of primary TCR stimulation, and did not exclude the contribution of IL-4 (63, 64). Given that GATA3 is a hallmark Th2 transcription factor that inhibits Th1 and, likely, Th17 differentiation, this CDK8-GATA3-FOXP3 pathway provides a parsimonious unifying mechanism to explain, at least in part, how DCA broadly regulates Treg, Th2, Th1, and Th17 differentiation (65–67). Given that Th2 cells can produce IL-10, this previously unappreciated link between Treg and Th2 differentiation may point to conserved (e.g., CDK8-related) anti-inflammatory signaling pathways that can engage distinct downstream effector pathways (68). We hypothesize that differences in the local cytokine milieu affect whether DCA enhances Treg or Th2 differentiation, for example, by modulating the epigenetic accessibility of FOXP3 and IL-4 loci. Additionally, where CDK8 effector pathways regulating tolerogenicity in innate (via phospho-c-JunSer243) versus adaptive immune cells diverge remains to be clearly defined (20).
Our discovery of DCA’s unusual temporal profile of enhancing early expression of FOXP3 and many FOXP3-regulated genes contrasts with the temporal profile of Treghi conditions and other Treg-enhancing compounds like harmine and reinforces a model of Treg differentiation that involves independently regulated early and late pathways (7). Whereas early pathways might involve transforming growth factor β (TGF-β) and other factors licensing cells to adopt the Treg fate and express FOXP3, late pathways might maintain and promote Treg lineage commitment. How these pathways overlap those regulating FOXP3’s stability in iTregs, which can be CNS2 dependent, remains to be clearly elucidated (69–72). Our data support a model where DCA largely enhances early pathways, including Notch-GATA3, to regulate FOXP3 expression. Whether DCA regulates Treg differentiation via Notch- and/or GATA3-independent pathways remains to be clearly elucidated. This suggests DCA may have particular therapeutic relevance to patients who have defects in early pathways of Treg differentiation and also raises the possibility of synergy with therapies that enhance late pathways of Treg differentiation.
Our findings exemplify how chemical immunotypes point to an important classification scheme that can inform both mechanistic and therapeutic hypotheses. DCA’s unique chemical immunophenotype (pro-Treg, pro-Th2, and pro-myeloid IL-10) is distinct from those of many other tolerogenic compounds, including SIK and DYRK1A inhibitors, which exert tolerogenic effects specifically in either innate or adaptive immune cells but not both (7, 20). Our novel finding that CDK8 inhibition promotes both Treg and Th2 differentiation directly informed our interrogation of GATA3 as a putative regulator of Treg differentiation. Additionally, our studies suggest value in monitoring tolerogenic effects, including impaired host-versus-tumor effects, in anticipated clinical use of CDK8 inhibitors as cancer therapeutics (73, 74).
The translational relevance of these data is reinforced by our finding that DCA promotes Treg differentiation in primary human CD4+ T cells. We note that Tregs generated using DCA are fully functional in vitro and in vivo. Importantly, our use of Treg transfer models specifically interrogates the functionality of DCA-driven Tregs without confounding anti-inflammatory effects on innate immune cells, which could confound the interpretation of models using systemic drug administration (21, 25). DCA and other CDK8 inhibitors may find utility as tolerogenic immunomodulators. Studies suggesting poor long-term tolerability of CDK8 inhibitors, together with our data showing that DCA affects early pathways in Treg differentiation, support this consideration (75). We recognize the utility of DCA in generating Tregs ex vivo, which would circumvent concerns regarding toxicity in vivo (75).
Our findings highlight the value of definitive interrogation of regulatory pathways. Prior knowledge drove the notion of CDK8-STAT interactions as key candidates to explain how CDK8 inhibition regulates T cell differentiation (19). Our experiments using Stat1Ser727Ala and Stat3Ser727Ala mice clearly demonstrate that DCA regulates Treg and, likely, also Th1 and Th17 differentiation independent of its effects on STAT1Ser727/STAT3Ser727 phosphorylation. Prior studies suggest that STAT1Ser727/STAT3Ser727 phosphorylation is required for full transcriptional activity (39–42). Consistent with this, we demonstrate a previously unappreciated role of STAT1Ser727 and STAT3Ser727 phosphorylation in positively regulating Th1 and Th17 differentiation, respectively. These findings differ from recent studies suggesting that CDK8 inhibition promotes human Th17 differentiation; possible explanations include differences in species, CDK8 inhibitor, and experimental approach (knock-in versus transduced allele) (26). This emphasizes the value of Ser-Ala STAT mutant mice in dissecting (CDK8-related) mechanistic hypotheses, including developing Stat5bSer730Ala mice to definitively define the role of CDK8-regulated STAT5bSer730 phosphorylation in Treg differentiation (21). These findings have important implications for disease pathobiology and precision therapy, for example, suggesting there may be synergy in therapeutically targeting STAT1Tyr701/STAT3Tyr705, STAT1Ser727/STAT3Ser727, and CDK8.
In summary, our studies highlight CDK8 as a regulator of innate and adaptive immune tolerogenicity that is targeted by the high-specificity, low-toxicity inhibitor DCA (13). We show for the first time that CDK8 regulates Th2 differentiation and human Treg differentiation. The unique chemical immunophenotype of DCA (pro-Treg/-Th2) directly informs the discovery of a novel CDK8-Notch-GATA3-FOXP3 axis that regulates early pathways of Treg differentiation and has further mechanistic and therapeutic implications. Our demonstration that DCA effectively enhances Treg differentiation compared to canonical Treg enhancers suggests utility in approaches to generate Tregs ex vivo for adoptive cellular therapy. In addition, the broadly tolerogenic effects of DCA suggest that it may be useful broadly in the setting of pathological inflammation or autoimmunity.
MATERIALS and METHODS
Mice.
BALB/c RRID:IMSR_JAX:000651, C57BL/6 000664RRID:IMSR_JAX:000664, Foxp3IRES-GFP RRID:IMSR_JAX:006772, CD45.1+/+002014RRID:IMSR_JAX:002014, NOD-scid, and NOD-BDC2.5 mice were purchased from Jackson Laboratory. NOD-BDC2.5.Foxp3IRES-GFP mice were from the JDRF Transgenic Core (Harvard Medical School, Boston, MA). C57BL/10-Rag2−/− mice were a kind gift from Brian Kelsall (37). Stat1Ser727Ala and Stat3Ser727Ala mice were previously described (40, 41). Mice were housed in the Benaroya Research Institute Vivarium in a specific-pathogen-free (SPF) animal room with unfettered access to food and water. All murine experiments were performed on male and female mice between 7 and 12 weeks of age, with the approval of the IACUC of Benaroya Research Institute (Seattle, WA).
Human samples.
Frozen PBMCs and fresh peripheral blood samples from healthy male and female subjects in similar ratios, ages 18 to 55, were obtained from the Benaroya Research Institute Immune Mediated Disease Registry and Repository. Human studies were approved by the Benaroya Research Institute’s Institutional Review Board, and all subjects signed written informed consent prior to inclusion in the study.
Cell lines.
293T cells used in lentiviral production were a generous gift from David Rawlings. 293T cells are of female provenance. They were cultured in Dulbecco modified Eagle medium (DMEM) medium (HyClone) supplemented with fetal bovine serum and GlutaMax (Thermo Fisher Scientific) at 37°C and 5% CO2. Cells were split every 3 days at a density of 7.5 × 104 cells per ml.
Small molecules and reagents.
16-Didehydro-cortistatin A (DCA) was a generous gift from P. Baran (The Scripps Research Institute) and was synthesized as previously reported (12, 76). Small-molecule reagents were confirmed to have ≥95% purity by high-performance liquid chromatography–mass spectrometry (HPLC-MS). Antibodies, chemical reagents, primers, and cytokines were sourced as listed in Table S2 in the supplemental material.
Cloning and plasmid preparation.
Coding sequences of GATA3, JUND, c-JUN, and JUNB were PCR amplified from pHAGE-GATA3, JunD-HA neo, pMIEG3-c-Jun, and pMIEG3-JunB (Addgene 116747, 58515, 40348, and 40349, gifts from Gordon Mills and Kenneth Scott, Kevin Janes, and Alexander Dent, respectively). PCR overhang extension was used to add (i) a self-splicing T2A sequence and (ii) 40-bp homology arms (HA) to permit cloning into EcoRV-digested pLKO.NGFR using a Gibson assembly ultrakit (Codex DNA, San Diego, CA). The primers used are listed in Table S2.
Murine T cell isolation and culture.
Unless otherwise noted, CD4+ CD62L+ naive T cells were isolated from spleens of 8- to 12-week-old mice, crushed through a 100-μm filter, processed using CD4 negative enrichment kits (Stemcell Technologies, Vancouver, Canada) and CD62L microbeads (Miltenyi Biotec, San Diego, CA) according to the manufacturer’s instructions, and confirmed to be >90% pure by flow cytometry. Cells were cultured on polystyrene 96-well plates (Corning, Corning, NY) precoated with anti-CD3 and anti-CD28 antibody using conditions outlined as follows. For all Treg, 1 μg/ml each anti-CD3 and anti-CD28 antibody and 2 μg/ml each anti-IL-12, anti-IFN-γ, and anti-IL-4 antibody. For Treglow, 2 ng/ml TGF-β, and for Treghi, 10 ng/ml TGF-β. For all Th17, 3 μg/ml each anti-CD3 and anti-CD28 antibody, 2 μg/ml each anti-IL-12, anti-IFN-γ, and anti-IL-4 antibody, and 0.25 ng/ml TGF-β. For Th17low, 5 ng/ml IL-6, and for Th17hi, 20 ng/ml each IL-6 and IL-1β. For all Th1, 1 μg/ml each anti-CD3 and anti-CD28 antibody. For Th1low, 0.25 ng/ml IL-12, and for Th1hi, 10 ng/ml IL-12. For all Th2, 1 μg/ml each anti-CD3 and anti-CD28 antibody and 2 μg/ml each anti-IL-12 and anti-IFN-γ antibody. For Th2low, 10 ng/ml IL-4, and for Th2hi, 100 ng/ml IL-4. DCA was supplemented at 100 nM as indicated. Amounts of 105 cells were cultured per well. The addition of DCA to Treglow conditions is abbreviated as Treglow+DCA. Treg and Th1 cultures were fed with equal volumes of IL-2-supplemented medium (20 ng/ml) and re-treated with compound at day 2, split 1:2 into IL-2-supplemented medium (10 ng/ml) at day 3, and analyzed at day 4. Th17 cultures were treated similarly except that no IL-2 was supplemented. Th2 cultures were treated similarly to Treg cultures except that they were additionally split 1:2 into IL-2-supplemented medium (10 ng/ml) at day 4 and day 5 and analyzed on day 6. To assess STAT1/STAT3/STAT5b Ser phosphorylation, cells were stimulated with 10 ng/ml IFN-γ plus 2 μg/ml anti-IL-4 antibody, 10 ng/ml IL-6 plus 2 μg/ml each anti-IL-4, anti-IL-12, and anti-IFN-γ antibody, and anti-CD3/CD28 antibody plus 100 ng/ml IL-2, respectively.
Human T cell isolation and culture.
Frozen PBMCs and fresh peripheral blood samples were obtained from the Benaroya Research Institute Immune Mediated Disease Registry and Repository. Human peripheral blood mononuclear cells were isolated from fresh whole blood by using Ficoll-Paque medium (GE Healthcare, Little Chalfont, United Kingdom). CD4+ CD45RA+ naive T cells were isolated using negative enrichment kits (Stemcell Technologies, Vancouver, Canada) according to the manufacturer’s instructions and confirmed to be >90% pure by flow cytometry. Cells were cultured on polystyrene 96-well plates (Corning, Corning, NY) precoated with anti-CD3 and anti-CD28 antibody using conditions as follows. For all Treg, 1 μg/ml each anti-CD3 and anti-CD28 antibody and 2 μg/ml each anti-IL-12, anti-IFN-γ, and anti-IL-4 antibody. For Treglow, 1 ng/ml TGF-β, and for Treghi, 10 ng/ml TGF-β. For all Th2, 1 μg/ml each anti-CD3 and anti-CD28 antibody and 2 μg/ml each anti-IL-12 and anti-IFN-γ antibody. For Th2low, 10 ng/ml IL-4, and for Th2hi, 100 ng/ml IL-4. DCA was supplemented at 100 nM as indicated. Amounts of 105 cells were cultured per well. Treg cultures were fed with equal volumes of IL-2-supplemented medium (20 ng/ml) and re-treated with compound at day 2, split 1:2 into IL-2-supplemented medium (10 ng/ml) at day 4, and analyzed at day 5. Th2 cultures were fed and split into medium supplemented with IL-2 and IL-4 (20 ng/ml each at day 2 and 10 ng/ml each thereafter) and compound as indicated to maintain ∼106 cells/ml, restimulated on days 7 and 14 on plates precoated with anti-CD3 and anti-CD28 antibody, and analyzed at day 21 as previously described (77).
Lentiviral production.
On day zero, 3.8 × 106 293T cells were plated in 10 ml DMEM plus 5% GlutaMax (Thermo Fisher) on a 10-cm plate. On day 1, cells were transfected with 1.5 μg pMD2G, 3 μg psPAX2 (kind gifts from David Rawlings), and 6 μg pLKO vector, mixed with 42 μg polyethyleneimine (PEI) transfection reagent (Polysciences, Inc.), and suspended in 0.5 ml diluent (10 mM HEPES, 150 mM NaCl, pH 7.05). Cells were washed with phosphate-buffered saline (PBS) and fed with fresh DMEM and GlutaMax on day 2. Viral supernatant was harvested on day 4, centrifuged (2,000 rpm for 5 min) to remove cellular debris, overlaid onto 5 ml of 10% sucrose in NTE (135 mM NaCl, 10 mM Tris-HCl [pH 7.50], 1 mM EDTA) in ultracentrifuge tubes (Beckman Coulter), and centrifuged at 25,000 rpm for 90 min at 4°C. Supernatant was removed, and the viral pellet resuspended in ice-cold NTE by shaking for 2 h at 4°C.
RNP complexing.
RNPs were generated by mixing 1.25 μg Cas9 protein (Aldevron, Fargo, ND) and 2.5 pmol each of 3 single guide RNAs (sgRNAs) (Synthego, Menlo Park, CA) with gentle swirling and incubating the mixture at 37°C for 15 min. The CDK8 guides used were CUCAUGCUGAUAGGAAG, UGUUUCUGUCUCAUGCUGAU, and UCUGUCUCAUGCUGAUAGGA.
CRISPR-Cas9 gene editing.
CRISPR-Cas9 gene editing was performed as previously described with modifications (78, 79). Briefly, human CD4+ CD45RA+ naive T cells were cultured on 96-well plates precoated with anti-CD3 and anti-CD28 antibody in DMEM supplemented with 5% fetal bovine serum, 20 ng/ml IL-2, and 2 μg/ml each of anti-IL-12, anti-IFN-γ, and anti-Il-4 antibody. Cells were harvested 2 days later, centrifuged (90 × g for 8 min), resuspended in buffer T, mixed with 20 μM each RNP complex, and electroporated (3 pulses of 1,600 V for 10 ms) using a Neon transfection system (Thermo Fisher, Waltham, MA). Cells were transferred into 90 μl TCM prewarmed to 37°C. After 24 h, cells were fed with medium supplemented with 100 ng/ml IL-2 and 1 ng/ml TGF-β. Cells were maintained for 5 additional days at a density of 1 × 106/ml and then analyzed by flow cytometry.
Flow cytometry.
Cells were stimulated with phorbol myristate acetate (PMA) and ionomycin (50 and 500 ng/ml, respectively) (Sigma-Aldrich, St. Louis, MO) in the presence of GolgiStop (BD Biosciences, San Jose, CA) 5 h prior to analysis as necessary. Cells were typically stained with LIVE/DEAD stain (Thermo Fisher, Waltham, MA) and anti-CD4 antibody prior to fixation and permeabilization, which was generally performed with Foxp3 fixation/permeabilization buffers (eBioscience, San Diego, CA). Phosflow cell lyse/fix buffer and perm buffer III (BD Biosciences, San Jose, CA) were used for phosphorylated-protein assessment. Intracellular staining was performed according to the manufacturer’s instructions. Counting beads (10 μm; Spherotech, Lake Forest, IL) were added at 5,000 per sample. Acquisition was performed on either a FACSCalibur or a FACSCanto (BD Biosciences, San Jose, CA). Cell sorting was performed using a FACSAria II (BD Biosciences, San Jose, CA). Data were analyzed using FlowJo software (Tree Star, Ashland, OR). Fractional maximal enhancement was determined by the increase in the percentage of lineage-committed cells relative to maximal cytokine-driven enhancement as previously reported (7). Fractional inhibition was calculated relative to the value for dimethyl sulfoxide (DMSO)-treated cells (7). STAT1/STAT3 phosphorylation was quantified as previously described (80).
In vitro proliferation and Treg suppression assays.
In vitro proliferation and Treg suppression assays were performed as previously described (81). Briefly, sorted CD45.1+ CD4+ CD62L+ responder T cells (Tresps) were labeled with CellTrace far red (Thermo Fisher, Waltham, MA) according to the manufacturer’s protocol and plated at 5 × 104 cells per well in 96-well U-bottom plates in the presence of anti-CD3 and anti-CD28 antibody-coupled beads (Dynabeads, Grand Island, NY) at 1:8 bead/cells. For Treg suppression assays, Tresps were cocultured with FACS-sorted and washed CD45.2+ Foxp3IRES-GFP+ Treg cells generated as indicated. Cells were analyzed by flow cytometry 3 days later. The proliferation index was calculated using FlowJo software (Tree Star, Ashland, OR) and represents the total number of divisions per cell that went into division.
Treg suppression. (i) Type 1 diabetes model.
Treg suppression in the type 1 diabetes model was assayed as previously described (7). Briefly, 5 × 104 sorted CD4+ CD62L+ naive T cells isolated from NOD-BDC2.5+ mice were injected intravenously into NOD-scid mice with or without 1 × 105 FACS-sorted and washed Treg cells generated from NOD-BDC2.5+FOXP3IRES-GFP mice as indicated (34, 35). Blood glucose levels were monitored with a handheld Contour glucometer (Bayer, Leverkusen, Germany) at days 3, 6, and 8 and every day following. Diabetes was diagnosed when blood sugar exceeded 250 mg/dl for 2 consecutive days.
(ii) CD45RBhi colitis model.
To assay Treg suppression in the CD45RBhi colitis model, 5 × 105 sorted CD4+ CD62L+ naive T cells isolated from CD45.1+ mice were injected intravenously into B10-Rag2−/− mice as previously described (37, 38). Five days later, the mice were injected with either PBS or 1.5 × 105 FACS-sorted and washed Treg cells generated from Foxp3IRES-GFP mice as indicated (38). Mice were monitored at least weekly for weight loss and morbidity per protocol. Mice were euthanized after 4 weeks, and the proximal, medial, and distal colon analyzed histologically by blinded observers for crypt depth, cellular infiltration, and crypt abscesses as previously described (82).
Histology.
Tissues were preserved in 10% formalin. Paraffin embedding and sectioning and staining with hematoxylin and eosin were performed by the Histology Core (Benaroya Research Institute, Seattle, WA).
Western blotting.
Cells were washed in PBS and lysed in either TNN lysis buffer, pH 8 (100 mM Tris-HCl, 100 mM NaCl, 1% NP-40, 1 mM dithiothreitol [DTT], 10 mM NaF), or radioimmunoprecipitation assay (RIPA) lysis buffer (150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl, pH 7.8) supplemented with DTT, protease inhibitors (Roche, Indianapolis, IN), and phosphatase inhibitors (Cell Signaling Technologies, Danvers, MA). Lysates were separated by SDS-PAGE using Tris-glycine gels loaded with about 1 × 106 cell equivalents per well and transferred onto polyvinylidene difluoride (PDVF) membrane (Millipore, Burlington, MA). Blots were blocked in either 5% milk (Nestle, Vervey, Switzerland) or bovine serum albumin (Sigma-Aldrich, St. Louis, MO) and visualized with Western Lightning plus-ECL (Perkin Elmer, Waltham, MA) and/or SuperSignal west femto substrate (Thermo Scientific, Waltham, MA) on CL-XPosure film (Thermo Scientific, Waltham, MA) according to the manufacturer’s instructions. Nuclear isolation was performed using the Nuclei EZ prep kit according to the manufacturer’s instructions (Sigma-Aldrich, St. Louis, MO). Fractions were subsequently lysed with Triton X-100 lysis buffer (1% Triton X-100, 150 mM NaCl, 50 mM Tris-HCl, pH 7.8). Band intensity was quantified by using ImageJ (83).
RNA isolation and reverse transcription-quantitative PCR (qRT-PCR).
RNA was isolated using RNeasy kits (Qiagen, Valencia, CA) and cDNA generated using iScript cDNA synthesis kits (Bio-Rad, Hercules, CA) according to the manufacturer’s directions. Real-time PCR was performed using an ABI 7500 fast real-time PCR system (Applied Biosystems, Foster City, CA). Cycling conditions were as follows: 1 cycle of 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Primers used are listed in Table S2. Quantitation was performed using the cycle threshold (ΔCT) method.
RNA-seq library preparation and sequencing.
RNA-seq libraries were generated from four Foxp3GFP littermate mice. On day zero, 1,000 naive CD4+ CD62L+ cells were sorted for RNA-seq. The remaining cells were cultured on plates precoated with anti-CD3 and anti-CD28 antibody under Treglow, Treghi, and Treglow+DCA conditions. On day 2, 250 FOXP3+ cells and 500 FOXP3− cells were sorted from cells cultured under Treglow, Treghi, and Treglow+DCA conditions. On day 4, 1,000 FOXP3+ cells were sorted from Treghi and Treglow+DCA cultures. Cells were sorted directly into lysis buffer from the SMART-Seq version 4 ultralow-input RNA kit for sequencing (TaKaRa) and frozen until all samples were ready for simultaneous processing. Reverse transcription was performed, followed by PCR amplification to generate full-length amplified cDNA. Sequencing libraries were constructed using the Nextera XT DNA sample preparation kit (Illumina) to generate Illumina-compatible barcoded libraries. Libraries were pooled and quantified using a Qubit fluorometer (Life Technologies). Dual-index, single-read sequencing of pooled libraries was carried out on a HiSeq 2500 sequencer (Illumina) with 58-base reads, using HiSeq version 4 cluster and SBS kits (Illumina) with a target depth of 5 million reads per sample.
Base-calling was performed automatically by Illumina real-time analysis software. Demultiplexing to generate FASTQ files was performed by bcl2fastq running on the Illumina BaseSpace platform. Subsequent processing was performed using the Galaxy platform. FASTQ reads were trimmed in two steps: (i) hard trimming to remove 1 3′-end base (FASTQ Trimmer tool, version 1.0.0), and (ii) quality trimming from both ends until the minimum base quality for each read was ≥30 (fastq-mcf version1.1.2). Reads were aligned to the GRCm38 mouse reference genome using STAR version 2.4.2a, with gene annotations from GRCm38 Ensembl release number 91 (84). Read counts per Ensembl gene ID were quantified using htseq-count version 0.4.1 (85). Sequencing, alignment, and quantitation metrics were obtained for FASTQ, BAM/SAM, and count files in Galaxy using FastQC version 0.11.3, Picard version 1.128, samtools version 1.2, and htseq-count version 0.4.1 (86). One library was excluded from downstream analysis for containing fewer than 1 million total reads.
Differential expression analysis.
The Limma R package was used to identify differentially expressed genes (87). RNA-seq data were processed using Tidyverse, Biomart, and EdgeR to generate relative expression values via TMM (trimmed mean of M values) normalization (genes with <5 counts in ≥10% of all libraries were excluded), followed by transformation using the limma-voom method (87–90). A linear model for gene expression based on culture condition, time point, or cell type was used. For each gene, a t statistic was computed using the empirical Bayes method within the Limma R package, which moderates the standard deviations between genes. A false discovery rate for each gene was calculated by applying the Benjamini-Hochberg multiple testing correction to P values calculated from t tests. Genes with a false discovery rate of less than 0.05 and a mean expression fold change greater than 1.5 were considered to be significantly differentially expressed. The raw RNA-seq data have been deposited to the Gene Expression Omnibus (GEO) with accession number GSE141933.
Pathway analysis.
Pathway analysis was performed using the Gene Set Enrichment Analysis Molecular Signature Database, or MSigDB, version 7.0 (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp), which uses the hypergeometric distribution on a background of all genes to calculate a P value (58, 91, 92).
Microscale ChIP assay.
The microscale ChIP assay was performed as described previously with few modifications (93). One hundred thousand naive CD4+ T cells were cultured under Treglow and Treglow+DCA conditions for 2 days and then harvested, washed with ice-cold PBS, fixed using 11% formaldehyde (diluted from 36% stock in 50 mM HEPES [pH 7.5], 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) at a concentration of 10% (vol/vol) for 10 min, quenched using 5% (vol/vol) 2.5 M glycine for 5 min, washed twice with 1 ml ice-cold PBS, and lysed in 50 μl lysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 1% SDS, 20 mg/ml sodium butyrate) supplemented with phenylmethylsulfonyl fluoride and protease inhibitor cocktail (Active Motif, Carlsbad, CA). DNA was sheared by sonication (Biorupter, Diagenode, Denville, NJ) into 200- to 500-bp fragments. Appropriate shearing (peak size, ∼250 bp) was verified by TapeStation (Agilent, Santa Clara, CA) analysis. Chromatin was precleared using 30 μl protein G-agarose beads (Active Motif) preblocked with bovine serum albumin according to the manufacturer’s instructions; beads were then removed by centrifugation. Chromatin was diluted with an equal volume of PBS, 4 μl of anti-GATA3 antibody or isotype control (Cell Signaling Technologies, Danvers, MA) was added, and the sample was incubated at 4°C with end-over-end rotation. Next, 30 μl of preblocked protein G-agarose beads was added and the sample incubated for 4 h at 4°C. The beads were then washed sequentially with 1 ml each low-SDS lysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 0.1% SDS, 20 mg/ml sodium butyrate), low-salt buffer (10 mM Tris-HCl n[pH 8], 1 mM EDTA, 50 mM NaCl), high-salt buffer (50 mM Tris-HCl [pH 8], 500 mM NaCl, 0.1% SDS, 0.5% Na-deoxycholate, 1% Nonidet-P40, 1 mM EDTA), LiCl buffer (50 mM Tris-HCl pH 8, 250 mM LiCl, 1 mM EDTA, 1% Nonidet-P40, 0.5% Na-deoxycholate), and 1 ml TE (10 mM Tris-HCl [pH 8], 1 mM EDTA). Beads were transferred to fresh tubes and centrifuged, and chromatin was eluted by incubating in 100 μl elution buffer (50 mM Tris-HCl [pH 8], 10 mM EDTA, 1% SDS) at 65°C with agitation. Chromatin was transferred to fresh tubes and incubated with 2 μl RNase A (Qiagen, 20 mg/ml) and 6 μl 5 M NaCl (Active Motif) for 30 min at 37°C followed by 2 μl proteinase K (0.2 mg/ml; Active Motif) at 65°C for 2 h. DNA was then purified by phenol-chloroform extraction and resuspended in nuclease-free water. Quantitative PCR was performed as described above. The primer sequences used were as follows: for FOXP3 CNS2, forward, 5′-GGACATCACCTACCACATCC-3′, and reverse, 5′-ACCACGGAGGAAGAGAAGAG-3′, and for β-actin, forward, 5′-TCCCCTCCTTTTGCGAAAA-3′, and reverse, 5′-CTCCCTCCTCCTCTTCCTCAA-3′.
Statistical analyses.
Statistical measures, including mean values, standard deviations, Student’s t test, Mantel-Cox test, Mann-Whitney test, one-way analysis of variance (ANOVA), and dose-response curve fitting (three-parameter dose-response curves), were performed using GraphPad Prism software and R. Where appropriate, unless otherwise stated, graphs display mean values ± standard deviations.
Data availability.
RNA-seq libraries generated in this study have been made available in the Gene Expression Omnibus (GEO) under accession number GSE141933.
ACKNOWLEDGMENTS
We express our deep appreciation to Anne Hocking, Karen Cerosaletti, Jessica Hamerman, and Daniel Campbell for helpful discussion. B10.Rag2−/− mice were a kind gift from Brian Kelsall. We acknowledge Tina Polintan for editorial assistance.
B.K. was supported by N.I.H. grant number K08 DK104021. Acquisition of key equipment was made possible by support from the Murdock Foundation.
B.K., A.A., R.J.X., P.S.L., V.H.G., and T.B.S. designed studies. A.A., K.G.M., K.J.F., T.B.S., L.J., A.F.S., and B.K. conducted experiments. A.A., K.J.F., K.G.M., Y.Z., and B.K. analyzed data. N.S.G., T.B.S., T.D., Y.Z., D.E.L., I.J.M., and Z.S.R. provided reagents. A.A. and B.K. wrote the manuscript.
We have no conflicts of interest to disclose.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Zigmond E, Varol C, Farache J, Elmaliah E, Satpathy AT, Friedlander G, Mack M, Shpigel N, Boneca IG, Murphy KM, Shakhar G, Halpern Z, Jung S. 2012. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity 37:1076–1090. 10.1016/j.immuni.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 2.Zigmond E, Jung S. 2013. Intestinal macrophages: well educated exceptions from the rule. Trends Immunol 34:162–168. 10.1016/j.it.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Josefowicz SZ, Lu L-F, Rudensky AY. 2012. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol 30:531–564. 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cretney E, Kallies A, Nutt SL. 2013. Differentiation and function of Foxp3(+) effector regulatory T cells. Trends Immunol 34:74–80. 10.1016/j.it.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 5.O’Shea JJ, Paul WE. 2010. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327:1098–1102. 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batlle E, Massagué J. 2019. Transforming growth factor-β signaling in immunity and cancer. Immunity 50:924–940. 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khor B, Gagnon JD, Goel G, Roche MI, Conway KL, Tran K, Aldrich LN, Sundberg TB, Paterson AM, Mordecai S, Dombkowski D, Schirmer M, Tan PH, Bhan AK, Roychoudhuri R, Restifo NP, O’Shea JJ, Medoff BD, Shamji AF, Schreiber SL, Sharpe AH, Shaw SY, Xavier RJ. 2015. The kinase DYRK1A reciprocally regulates the differentiation of Th17 and regulatory T cells. Elife 4:e05920. 10.7554/eLife.05920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundberg TB, Choi HG, Song J-H, Russell CN, Hussain MM, Graham DB, Khor B, Gagnon J, O’Connell DJ, Narayan K, Dancik V, Perez JR, Reinecker H-C, Gray NS, Schreiber SL, Xavier RJ, Shamji AF. 2014. Small-molecule screening identifies inhibition of salt-inducible kinases as a therapeutic strategy to enhance immunoregulatory functions of dendritic cells. Proc Natl Acad Sci U S A 111:12468–12473. 10.1073/pnas.1412308111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aoki S, Watanabe Y, Tanabe D, Arai M, Suna H, Miyamoto K, Tsujibo H, Tsujikawa K, Yamamoto H, Kobayashi M. 2007. Structure-activity relationship and biological property of cortistatins, anti-angiogenic spongean steroidal alkaloids. Bioorg Med Chem 15:6758–6762. 10.1016/j.bmc.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 10.Cee VJ, Chen DY-K, Lee MR, Nicolaou KC. 2009. Cortistatin A is a high-affinity ligand of protein kinases ROCK, CDK8, and CDK11. Angew Chem Int Ed Engl 48:8952–8957. 10.1002/anie.200904778. [DOI] [PubMed] [Google Scholar]
- 11.Pelish HE, Liau BB, Nitulescu II, Tangpeerachaikul A, Poss ZC, Da Silva DH, Caruso BT, Arefolov A, Fadeyi O, Christie AL, Du K, Banka D, Schneider EV, Jestel A, Zou G, Si C, Ebmeier CC, Bronson RT, Krivtsov AV, Myers AG, Kohl NE, Kung AL, Armstrong SA, Lemieux ME, Taatjes DJ, Shair MD. 2015. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature 526:273–276. 10.1038/nature14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi J, Manolikakes G, Yeh C-H, Guerrero CA, Shenvi RA, Shigehisa H, Baran PS. 2011. Scalable synthesis of cortistatin A and related structures. J Am Chem Soc 133:8014–8027. 10.1021/ja202103e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen M, Li J, Liang J, Thompson ZS, Kathrein K, Broude EV, Roninson IB. 2019. Systemic toxicity reported for CDK8/19 inhibitors CCT251921 and MSC2530818 is not due to target inhibition. Cells 8:1413. 10.3390/cells8111413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conaway RC, Conaway JW. 2011. Function and regulation of the Mediator complex. Curr Opin Genet Dev 21:225–230. 10.1016/j.gde.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CAS, Jin J, Cai Y, Washburn MP, Conaway JW, Conaway RC. 2004. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell 14:685–691. 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 16.Malumbres M. 2014. Cyclin-dependent kinases. Genome Biol 15:122. 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galbraith MD, Andrysik Z, Pandey A, Hoh M, Bonner EA, Hill AA, Sullivan KD, Espinosa JM. 2017. CDK8 kinase activity promotes glycolysis. Cell Rep 21:1495–1506. 10.1016/j.celrep.2017.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fant CB, Taatjes DJ. 2019. Regulatory functions of the Mediator kinases CDK8 and CDK19. Transcription 10:76–90. 10.1080/21541264.2018.1556915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bancerek J, Poss ZC, Steinparzer I, Sedlyarov V, Pfaffenwimmer T, Mikulic I, Dölken L, Strobl B, Müller M, Taatjes DJ, Kovarik P. 2013. CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity 38:250–262. 10.1016/j.immuni.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johannessen L, Sundberg TB, O’Connell DJ, Kolde R, Berstler J, Billings KJ, Khor B, Seashore-Ludlow B, Fassl A, Russell CN, Latorre IJ, Jiang B, Graham DB, Perez JR, Sicinski P, Phillips AJ, Schreiber SL, Gray NS, Shamji AF, Xavier RJ. 2017. Small-molecule studies identify CDK8 as a regulator of IL-10 in myeloid cells. Nat Chem Biol 13:1102–1108. 10.1038/nchembio.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akamatsu M, Mikami N, Ohkura N, Kawakami R, Kitagawa Y, Sugimoto A, Hirota K, Nakamura N, Ujihara S, Kurosaki T, Hamaguchi H, Harada H, Xia G, Morita Y, Aramori I, Narumiya S, Sakaguchi S. 2019. Conversion of antigen-specific effector/memory T cells into Foxp3-expressing Treg cells by inhibition of CDK8/19. Sci Immunol 4:eaaw2707. 10.1126/sciimmunol.aaw2707. [DOI] [PubMed] [Google Scholar]
- 22.Fryer CJ, White JB, Jones KA. 2004. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol Cell 16:509–520. 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Witalisz-Siepracka A, Gotthardt D, Prchal-Murphy M, Didara Z, Menzl I, Prinz D, Edlinger L, Putz EM, Sexl V. 2018. NK cell-specific CDK8 deletion enhances antitumor responses. Cancer Immunol Res 6:458–466. 10.1158/2326-6066.CIR-17-0183. [DOI] [PubMed] [Google Scholar]
- 24.Putz EM, Gotthardt D, Hoermann G, Csiszar A, Wirth S, Berger A, Straka E, Rigler D, Wallner B, Jamieson AM, Pickl WF, Zebedin-Brandl EM, Müller M, Decker T, Sexl V. 2013. CDK8-mediated STAT1-S727 phosphorylation restrains NK cell cytotoxicity and tumor surveillance. Cell Rep 4:437–444. 10.1016/j.celrep.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Z, Wang G, Lv Y, Wan YY, Zheng J. 2019. Inhibition of Cdk8/Cdk19 activity promotes Treg cell differentiation and suppresses autoimmune diseases. Front Immunol 10:01988. 10.3389/fimmu.2019.01988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Fabregas J, Wang L, Pohler E, Cozzani A, Wilmes S, Kazemian M, Mitra S, Moraga I. 2020. CDK8 fine-tunes IL-6 transcriptional activities by limiting STAT3 resident time at the gene loci. Cell Rep 33:108545. 10.1016/j.celrep.2020.108545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coombes JL, Siddiqui KRR, Arancibia-Cárcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. 2007. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med 204:1757–1764. 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, Cheroutre H. 2007. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science 317:256–260. 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 29.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. 2007. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med 204:1775–1785. 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haxhinasto S, Mathis D, Benoist C. 2008. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med 205:565–574. 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill JA, Hall JA, Sun CM, Cai Q, Ghyselinck N, Chambon P, Belkaid Y, Mathis D, Benoist C. 2008. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi Cells. Immunity 29:758–770. 10.1016/j.immuni.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sauer S, Bruno L, Hertweck A, Finlay D, Leleu M, Spivakov M, Knight ZA, Cobb BS, Cantrell D, O’Connor E, Shokat KM, Fisher AG, Merkenschlager M. 2008. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci U S A 105:7797–7802. 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall JA, Cannons JL, Grainger JR, Santos Dos LM, Hand TW, Naik S, Wohlfert EA, Chou DB, Oldenhove G, Robinson M, Grigg ME, Kastenmayer R, Schwartzberg PL, Belkaid Y. 2011. Essential role for retinoic acid in the promotion of CD4(+) T cell effector responses via retinoic acid receptor alpha. Immunity 34:435–447. 10.1016/j.immuni.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herman AE, Freeman GJ, Mathis D, Benoist C. 2004. CD4+CD25+ T regulatory cells dependent on ICOS promote regulation of effector cells in the prediabetic lesion. J Exp Med 199:1479–1489. 10.1084/jem.20040179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarbell KV, Yamazaki S, Olson K, Toy P, Steinman RM. 2004. CD25+ CD4+ T cells, expanded with dendritic cells presenting a single autoantigenic peptide, suppress autoimmune diabetes. J Exp Med 199:1467–1477. 10.1084/jem.20040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powrie F, Leach MW, Mauze S, Caddle LB, Coffman RL. 1993. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int Immunol 5:1461–1471. 10.1093/intimm/5.11.1461. [DOI] [PubMed] [Google Scholar]
- 37.Valatas V, He J, Rivollier A, Kolios G, Kitamura K, Kelsall BL. 2013. Host-dependent control of early regulatory and effector T-cell differentiation underlies the genetic susceptibility of RAG2-deficient mouse strains to transfer colitis. Mucosal Immunol 6:601–611. 10.1038/mi.2012.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341:569–573. 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kovarik P, Mangold M, Ramsauer K, Heidari H, Steinborn R, Zotter A, Levy DE, Müller M, Decker T. 2001. Specificity of signaling by STAT1 depends on SH2 and C-terminal domains that regulate Ser727 phosphorylation, differentially affecting specific target gene expression. EMBO J 20:91–100. 10.1093/emboj/20.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen Y, Schlessinger K, Zhu X, Meffre E, Quimby F, Levy DE, Darnell JE. 2004. Essential role of STAT3 in postnatal survival and growth revealed by mice lacking STAT3 serine 727 phosphorylation. Mol Cell Biol 24:407–419. 10.1128/MCB.24.1.407-419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varinou L, Ramsauer K, Karaghiosoff M, Kolbe T, Pfeffer K, Müller M, Decker T. 2003. Phosphorylation of the Stat1 transactivation domain is required for full-fledged IFN-gamma-dependent innate immunity. Immunity 19:793–802. 10.1016/S1074-7613(03)00322-4. [DOI] [PubMed] [Google Scholar]
- 42.Wen Z, Zhong Z, Darnell JE. 1995. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82:241–250. 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 43.Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. 2002. T-bet is a STAT1-induced regulator of IL-12R expression in naïve CD4+ T cells. Nat Immunol 3:549–557. 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 44.Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, Nguyen BV, Gadina M, Sher A, Paul WE, O’Shea JJ. 2001. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci U S A 98:15137–15142. 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, Dong C. 2007. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem 282:9358–9363. 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 46.Zheng Y, Josefowicz SZ, Kas A, Chu T-T, Gavin MA, Rudensky AY. 2007. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 445:936–940. 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 47.Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, Fraenkel E, Boehmer von H, Young RA. 2007. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature 445:931–935. 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu W, Ergun A, Lu T, Hill JA, Haxhinasto S, Fassett MS, Gazit R, Adoro S, Glimcher L, Chan S, Kastner P, Rossi D, Collins JJ, Mathis D, Benoist C. 2012. A multiply redundant genetic switch “locks in” the transcriptional signature of regulatory T cells. Nat Immunol 13:972–980. 10.1038/ni.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hori S, Nomura T, Sakaguchi S. 2003. Control of regulatory T cell development by the transcription factor Foxp3. Science 299:1057–1061. 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 50.Zheng W, Flavell RA. 1997. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89:587–596. 10.1016/S0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 51.Wohlfert EA, Grainger JR, Bouladoux N, Konkel JE, Oldenhove G, Ribeiro CH, Hall JA, Yagi R, Naik S, Bhairavabhotla R, Paul WE, Bosselut R, Wei G, Zhao K, Oukka M, Zhu J, Belkaid Y. 2011. GATA3 controls Foxp3+ regulatory T cell fate during inflammation in mice. J Clin Invest 121:4503–4515. 10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, Zhu J, Zhao K. 2011. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity 35:299–311. 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rudra D, deRoos P, Chaudhry A, Niec RE, Arvey A, Samstein RM, Leslie C, Shaffer SA, Goodlett DR, Rudensky AY. 2012. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat Immunol 13:1010–1019. 10.1038/ni.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. 2007. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity 27:100–110. 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mota C, Nunes-Silva V, Pires AR, Matoso P, Victorino RMM, Sousa AE, Caramalho I. 2014. Delta-like 1-mediated Notch signaling enhances the in vitro conversion of human memory CD4 T cells into FOXP3-expressing regulatory T cells. J Immunol 193:5854–5862. 10.4049/jimmunol.1400198. [DOI] [PubMed] [Google Scholar]
- 56.Joller N, Lozano E, Burkett PR, Patel B, Xiao S, Zhu C, Xia J, Tan TG, Sefik E, Yajnik V, Sharpe AH, Quintana FJ, Mathis D, Benoist C, Hafler DA, Kuchroo VK. 2014. Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses. Immunity 40:569–581. 10.1016/j.immuni.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yevshin I, Sharipov R, Kolmykov S, Kondrakhin Y, Kolpakov F. 2019. GTRD: a database on gene transcription regulation—2019 update. Nucleic Acids Res 47:D100–D105. 10.1093/nar/gky1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. 2003. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34:267–273. 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 60.Kitagawa M. 2016. Notch signalling in the nucleus: roles of Mastermind-like (MAML) transcriptional coactivators. J Biochem 159:287–294. 10.1093/jb/mvv123. [DOI] [PubMed] [Google Scholar]
- 61.Wang Y, Su MA, Wan YY. 2011. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity 35:337–348. 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mantel P-Y, Kuipers H, Boyman O, Rhyner C, Ouaked N, Rückert B, Karagiannidis C, Lambrecht BN, Hendriks RW, Crameri R, Akdis CA, Blaser K, Schmidt-Weber CB. 2007. GATA3-driven Th2 responses inhibit TGF-beta1-induced FOXP3 expression and the formation of regulatory T cells. PLoS Biol 5:e329. 10.1371/journal.pbio.0050329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei J, Duramad O, Perng OA, Reiner SL, Liu Y-J, Qin FX-F. 2007. Antagonistic nature of T helper 1/2 developmental programs in opposing peripheral induction of Foxp3+ regulatory T cells. Proc Natl Acad Sci U S A 104:18169–18174. 10.1073/pnas.0703642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hadjur S, Bruno L, Hertweck A, Cobb BS, Taylor B, Fisher AG, Merkenschlager M. 2009. IL4 blockade of inducible regulatory T cell differentiation: the role of Th2 cells, Gata3 and PU.1. Immunol Lett 122:37–43. 10.1016/j.imlet.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 65.Ouyang W, Ranganath SH, Weindel K, Bhattacharya D, Murphy TL, Sha WC, Murphy KM. 1998. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity 9:745–755. 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- 66.Yagi R, Junttila IS, Wei G, UrbanJF, Jr, Zhao K, Paul WE, Zhu J. 2010. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-γ. Immunity 32:507–517. 10.1016/j.immuni.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Hamburg JP, de Bruijn MJW, de Almeida CR, van Zwam M, van Meurs M, de Haas E, Boon L, Samsom JN, Hendriks RW. 2008. Enforced expression of GATA3 allows differentiation of IL-17-producing cells, but constrains Th17-mediated pathology. Eur J Immunol 38:2573–2586. 10.1002/eji.200737840. [DOI] [PubMed] [Google Scholar]
- 68.Fiorentino DF, Bond MW, Mosmann TR. 1989. Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J Exp Med 170:2081–2095. 10.1084/jem.170.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng Y, Arvey A, Chinen T, van der Veeken J, Gasteiger G, Rudensky AY. 2014. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell 158:749–763. 10.1016/j.cell.2014.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li X, Liang Y, LeBlanc M, Benner C, Zheng Y. 2014. Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell 158:734–748. 10.1016/j.cell.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kanamori M, Nakatsukasa H, Okada M, Lu Q, Yoshimura A. 2016. Induced regulatory T cells: their development, stability, and applications. Trends Immunol 37:803–811. 10.1016/j.it.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 72.Ohkura N, Hamaguchi M, Morikawa H, Sugimura K, Tanaka A, Ito Y, Osaki M, Tanaka Y, Yamashita R, Nakano N, Huehn J, Fehling HJ, Sparwasser T, Nakai K, Sakaguchi S. 2012. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity 37:785–799. 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 73.Menzl I, Witalisz-Siepracka A, Sexl V. 2019. CDK8—novel therapeutic opportunities. Pharmaceuticals 12:92–12. 10.3390/ph12020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, Chheda MG, Tamayo P, Finn S, Shrestha Y, Boehm JS, Jain S, Bojarski E, Mermel C, Barretina J, Chan JA, Baselga J, Tabernero J, Root DE, Fuchs CS, Loda M, Shivdasani RA, Meyerson M, Hahn WC. 2008. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature 455:547–551. 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Clarke PA, Ortiz-Ruiz M-J, TePoele R, Adeniji-Popoola O, Box G, Court W, Czasch S, Bawab El S, Esdar C, Ewan K, Gowan S, De Haven Brandon A, Hewitt P, Hobbs SM, Kaufmann W, Mallinger A, Raynaud F, Roe T, Rohdich F, Schiemann K, Simon S, Schneider R, Valenti M, Weigt S, Blagg J, Blaukat A, Dale TC, Eccles SA, Hecht S, Urbahns K, Workman P, Wienke D. 2016. Assessing the mechanism and therapeutic potential of modulators of the human Mediator complex-associated protein kinases. Elife 5:e20722. 10.7554/eLife.20722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi J, Shigehisa H, Guerrero CA, Shenvi RA, Li C-C, Baran PS. 2009. Stereodivergent synthesis of 17-alpha and 17-beta-alpharyl steroids: application and biological evaluation of D-ring cortistatin analogues. Angew Chem Int Ed Engl 48:4328–4331. 10.1002/anie.200901116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cousins DJ, Lee TH, Staynov DZ. 2002. Cytokine coexpression during human Th1/Th2 cell differentiation: direct evidence for coordinated expression of Th2 cytokines. J Immunol 169:2498–2506. 10.4049/jimmunol.169.5.2498. [DOI] [PubMed] [Google Scholar]
- 78.Roth TL, Puig-Saus C, Yu R, Shifrut E, Carnevale J, Li PJ, Hiatt J, Saco J, Krystofinski P, Li H, Tobin V, Nguyen DN, Lee MR, Putnam AL, Ferris AL, Chen JW, Schickel J-N, Pellerin L, Carmody D, Alkorta-Aranburu G, Del Gaudio D, Matsumoto H, Morell M, Mao Y, Cho M, Quadros RM, Gurumurthy CB, Smith B, Haugwitz M, Hughes SH, Weissman JS, Schumann K, Esensten JH, May AP, Ashworth A, Kupfer GM, Greeley SAW, Bacchetta R, Meffre E, Roncarolo M-G, Romberg N, Herold KC, Ribas A, Leonetti MD, Marson A. 2018. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 559:405–409. 10.1038/s41586-018-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aksoy P, Aksoy BA, Czech E, Hammerbacher J. 2019. Viable and efficient electroporation-based genetic manipulation of unstimulated human T cells. biorxiv 10.1101/466243. [DOI]
- 80.Chaudhry A, Samstein RM, Treuting P, Liang Y, Pils MC, Heinrich J-M, Jack RS, Wunderlich FT, Brüning JC, Muller W, Rudensky AY. 2011. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity 34:566–578. 10.1016/j.immuni.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Collison LW, Vignali DAA. 2011. In vitro Treg suppression assays. Methods Mol Biol 707:21–37. 10.1007/978-1-61737-979-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.De Jong YP, Comiskey M, Kalled SL, Mizoguchi E, Flavell RA, Bhan AK, Terhorst C. 2000. Chronic murine colitis is dependent on the CD154/CD40 pathway and can be attenuated by anti-CD154 administration. Gastroenterology 119:715–723. 10.1053/gast.2000.16485. [DOI] [PubMed] [Google Scholar]
- 83.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anders S, Pyl PT, Huber W. 2015. HTSeq–—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Pedersen T, Miller E, Bache S, Müller K, Ooms J, Robinson D, Seidel D, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H. 2019. Welcome to the Tidyverse. J Open Source Softw 4:1686. 10.21105/joss.01686. [DOI] [Google Scholar]
- 89.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. 2005. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21:3439–3440. 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 90.McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40:4288–4297. 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. 2015. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1:417–425. 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. 2005. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434:338–345. 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Seumois G, Chavez L, Gerasimova A, Lienhard M, Omran N, Kalinke L, Vedanayagam M, Ganesan APV, Chawla A, Djukanović R, Ansel KM, Peters B, Rao A, Vijayanand P. 2014. Epigenomic analysis of primary human T cells reveals enhancers associated with TH2 memory cell differentiation and asthma susceptibility. Nat Immunol 15:777–788. 10.1038/ni.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2. Download MCB.00085-21-s0001.pdf, PDF file, 0.2 MB (205.2KB, pdf)
Data Availability Statement
RNA-seq libraries generated in this study have been made available in the Gene Expression Omnibus (GEO) under accession number GSE141933.


