Abstract
Background:
Chronic immune activation and CD4+ T cell depletion are significant pathogenic features of HIV infection. Expression of Fas ligand (FasL), a key mediator of activation-induced cell death in T cells, is elevated in people living with HIV-1 infection (PLWH). However, the epigenetic mechanisms underlying the enhanced induction of FasL expression in CD4+ T lymphocytes in PLWH are not completely elucidated. Hence, the current work examined the effect of HIV infection on FasL promoter-associated histone modifications and transcriptional regulation in CD4+ T lymphocytes in PLWH.
Method:
Flow cytometric analysis was performed to examine the Fas-FasL expression on total CD4+ T cells and naïve/memory CD4+ T cell subsets. Epigenetic FasL promoter histone modifications were investigated by chromatin immunoprecipitation-quantitative real-time polymerase chain reaction analysis using freshly isolated total CD4+ T lymphocytes from HIV-1 infected and noninfected individuals.
Results:
All naïve/memory CD4+ T cell subsets from PLWH showed markedly greater frequency of FasL expression. Notably, examination of functional outcome of FasL/Fas co-expression demonstrated the preferential susceptibility of Tcm and Tem subsets to activation-induced apoptosis. Importantly, these CD4+ T cells collectively demonstrated a distinct FasL promoter histone profile involving a coordinated cross-talk between histone H3 modifications leading to enhanced FasL gene expression. Specifically, levels of transcriptionally permissive histone H3K4-trimethylation (H3K4Me3) and histone H3K9-acetylation (H3K9Ac) were increased, with a concomitant decrease in the repressive H3K9-trimethylation (H3K9Me3).
Conclusion:
The present work demonstrates that epigenetic mechanisms involving promoter-histone modifications regulate transcriptional competence and FasL expression in CD4+ T cells from PLWH and render them susceptible to activation-induced cell death.
Keywords: epigenetic promoter modification, gene regulation, histone H3, CD4+ T cells, FasL, HIV
INTRODUCTION
The development of overt immune dysfunction is a significant feature of HIV pathogenesis that is intimately linked with CD4+ T-cell dynamics and reflects the complex interplay of direct viral cytopathogenicity and indirect effects due to chronic immune activation.1 Both antigen-specific and polyclonal activation driven by both viral and host products contribute to persistent, across-the-board activation, particularly in the chronic phase of the infection.2 Chronic activation sensitizes T cells to death ligands and activation-induced cell death (AICD), the resulting loss or failure to reconstitute CD4+ T cells is linked with impairment of cellular immune function and susceptibility to opportunistic functions.3 In this regard, aberrant apoptosis of both bystander and infected T cells during chronic HIV infection is a major contributing factor to on-going depletion of CD4+ T cells.4–7
Fas ligand (FasL) cross-links Fas causing a signaling cascade that can lead to apoptotic cell death or AICD of CD4+ T cells during HIV disease progression.8–10 Compared with uninfected individuals, elevated levels of both soluble and membrane-bound Fas and FasL are observed in people living with HIV-1 infection (PLWH) and correlate with disease progression.11–15 Both in vitro and in vivo studies have shown that the HIV/SIV-infected T cells undergo apoptosis using the Fas/FasL pathway.16–19 Significantly, HIV infection-mediated chronic immune activation and several viral proteins including gp120, Nef, and Tat, which contribute to accelerated apoptosis of CD4+ T cells, are shown to exert their pro-apoptotic effects by up-regulating activation-induced FasL expression.5,6,20,21 However, the molecular mechanisms mediated by HIV infection that underlie the development of susceptibility and sensitization of CD4+ T cells to enhanced activation-induced FasL expression and AICD remain largely undetermined.
In uninfected controls, FasL mRNA expression is marginal in resting primary CD4+ T cells and its induction is regulated at the level of transcriptional initiation in response to activation.22–25 The transcriptionally permissive or repressive state of the chromatin plays a major role in dictating the access of transcription factors to promoters of target genes and in turn activation of transcription.26 Chromatin remodeling occurs because of epigenetic modifications of histones, affecting the binding of specific transcription factors and gene transcription upon activation.27,28 Particularly, promoter histone methylation and acetylation have emerged as highly significant post-translational modifications (PTMs) in regulating the transition between transcriptionally repressive and permissive chromatin states.29,30 In this regard, we and others have identified the critical epigenetic mechanisms involving FasL promoter histone methylation and acetylation that regulate the binding of relevant transcription factors and FasL gene transcription in CD4+ T cells under physiologic and pathologic conditions.31–33 Several studies have demonstrated that cellular environmental inputs influence histone PTMs with resultant modulation of chromatin structure and gene expression.34,35 In view of this, histone PTMs are likely to serve as a clinically relevant interface through which the HIV infection associated cellular environment affects cellular phenotypes. Hence, the present study investigated FasL promoter-associated histone PTMs in CD4+ T cells from PLWH as potential epigenetic mechanism involved in abnormal FasL gene expression and susceptibility of CD4+ T cells to cell death.
Overall, the data show that in comparison to control CD4+ T cells without HIV infection, resting PLWH CD4+ T cells have a distinct FasL promoter histone PTM profile that is marked by a decrease in the transcriptionally repressive histone PTM, that is, histone H3 lysine9 trimethylation (H3K9Me3). Furthermore, T cell receptor (TCR) stimulation of PLWH CD4+ T cells results in significantly greater transcriptionally permissive promoter histone PTMs, including histone lysine4-trimethylation (H3K4Me3) and histone lysine9-acetylation (H3K9Ac), and FasL gene expression.
MATERIALS AND METHODS
Study Population
This was a cross-sectional study of PLWH managed at the University of Louisville. All procedures were in accordance with the ethical standards of the Helsinki Declaration (1964, 2008 amendment) of the World Medical Association, and were approved by the University of Louisville (UofL) Institutional Review Board (IRB# 08.0188). The HIV group included patients (n = 17) with an established diagnosis of HIV. All HIV patients were on ART treatment and had controlled viral load (HIV RNA < 400 copies/mL). The HIV-uninfected control group (n = 14) were age/gender/smoking status matched healthy subjects with no known diagnosis of HIV. The controls were also recruited at UofL. Trained personnel collected clinical data from patient medical records and entered these data into a secure, web-based data management system hosted by the University of Louisville. Blood samples were collected under UofL IRB-approved protocol (IRB # 08.0188) for peripheral blood mononuclear cells (PBMC) and CD4+ T lymphocyte isolation.
Human CD4+ T Lymphocyte Culture
PLWH and control CD4+ T lymphocytes from were isolated, cultured, and treated as described previously.36 A CD4+ T-cell purity of ≥90% CD4+ was determined by flow cytometry31
RNA Isolation and Real-Time Polymerase Chain Reaction Analysis
Total RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA), and quantitative real-time polymerase chain reaction (qPCR) was performed as described previously.37 The gene expression was analyzed by relative quantification using 2−ΔΔCt method by normalizing with 18S rRNA. Primers for mRNA analysis were as follows:
FasL- FP 5ʹ-TCTACCAGCCAGATGCACAC-3ʹ, RP 5ʹ-CAGAGGCATGGACCTTGAGT-3ʹ
18S rRNA- FP 5ʹ-CTCAACACGGGAAACCTCAC-3ʹ, RP 5ʹ-CGCTCCACCAACTAAGAACG-3ʹ
Flow Cytometric Analysis of PBMC
PBMC from normal controls or PLWH were thawed as described previously38 and cultured to assess Fas and FasL expression, active caspase 3/7, and live/dead in naïve and memory CD4 T-cell subsets. Live-or-Dye640/662 (Biotium, Fremont, CA) and CellEvent Caspase-3/7 Green flow cytometry assay kit (ThermoFisher) were used according to manufacturers’ instructions. Standard flow cytometric staining protocols were used to assess cell surface phenotypes using antibodies listed in Table 1, Supplemental Digital Content, http://links.lww.com/QAI/B554. After staining, cells were washed twice and fixed in 2% methanol-free formaldehyde (Polysciences, Warrington, PA) for 2–24 hours before acquisition on a LSRII flow cytometer (BD Biosciences, San Jose, CA). A total of 20,000 CD4+ lymphocytes were acquired for each sample. UltraComp eBeads (Thermo-Fisher) incubated with fluorochrome-conjugated mAb were used as compensation controls. Panel-specific fluorescence minus one controls were used to define negative events in gating strategies.39 Data were analyzed using FlowJo v10 software.
TABLE 1.
Demographic and Clinical Characteristics Between PLWH and Matched Controls
| Sample Variables | Control (n = 14), n (%)/Mean ± SD | PLWH (n = 17), n (%)/Mean ± SD | P |
|---|---|---|---|
| Demographics | |||
| Age in yr, mean (SD) | 46.86 ± 6.16 | 43.82 ± 12.48 | 0.5110 |
| Ethnicity/Racial distribution | |||
| Non-Hispanic- White | 3 (21.4%) | 7 (41.2%) | |
| Non-Hispanic- AA | 4 (28.6%) | 6 (35.3%) | |
| Non-Hispanic- other | 4 (28.6%) | 1 (5.9%) | |
| Hispanic- White | 1 (7.1%) | 2 (11.8%) | |
| Hispanic- AA | 2 (14.3%) | 1 (5.9%) | |
| Hispanic- other | 0 (0.0%) | 0 (0.0%) | |
| Sex | |||
| Male | 9 | 14 | |
| Female | 5 | 3 | |
| HIV infection history | |||
| HIV duration in yr | NA | 12.17 ± 10.59 | |
| No. of participants with HIV viral load (>20 copies/mL; VL: −207.5± 138.86) | NA | 6 | |
| No. of participants with HIV viral load (>20 copies/mL) | NA | 11 | |
| Antiretroviral treatment (n) | NA | 17 | |
| CD4+ T-cell counts (cells/µL) | NA | 639 ± 336.24 |
Chromatin Immunoprecipitation Assay
Histone modifications at the human FasL promoter were detected using a chromatin immunoprecipitation (ChIP) assay kit (EMDMillipore, MA) as per manufacturer’s protocol. ChIP antibodies to detect anti-acetyl-histone H3 Lys 9, anti-trimethyl-histone H3 Lys 9 and Lys 4, NF-κB (p65), NFAT, and RNA Polymerase II EMDMillipore, MA) were used for immunoprecipitation and control rabbit or mouse IgG (Cell Signaling Technology, Beverly, MA). ChIP-qPCR was performed as described previously.37 The following ChIP primers were used for analysis
-
ChIP- Primer for the region I of FasL promoter:
FP 5ʹ- TTCAGCTGCAAAGTGAGTGG −3ʹ
RP 5ʹ- CCTGTTGCTGACTGCTCAAG −3ʹ
-
ChIP- Primer for region II of FasL promoter:
FP 5ʹ- ACCTGTTTGGGTAGCACAGC −3ʹ
RP 5ʹ- TTGCAGCTGAAGCTGAGAAG −3ʹ
Quantikine sFasL ELISA Assay
The Quantikine Human Fas Ligand/TNFSF6 solid-phase ELISA (R&D Systems, MN) was used for the quantitative determination of human Fas Ligand concentrations in serum of study subjects as specified by the manufacturer.
Statistical Analyses
Data are presented as mean ± SD for the indicated number of independently performed experiments. The Student t-test was performed to examine differences between groups and significance was defined at P < 0.05. Correlations between parameters measured were calculated using the Spearman correlation coefficient. The significance level α was set at 0.05, and differences with P-value of less than 0.05 were considered significant. One-way analysis of variance (ANOVA) followed by post-hoc Bonferroni multiple comparison test was used where indicated. Graph Pad Prism version 8.3 software was used to analyze all data sets.
RESULTS
Previous studies have reported that FasL expression is up-regulated in HIV-infected CD4+ T lymphocytes and significantly contributes to their decline in the periphery.4,10,12,13 In the present study, we investigated the molecular epigenetic mechanisms regulating FasL promoter chromatin remodeling and transcriptional activation in CD4+ T lymphocytes occurring in PLWH.
Demographic and Clinical Characteristics of the Study Population
CD4+ T lymphocytes and serum were obtained from 14 uninfected healthy controls and 17 PLWH study population. The demographics of healthy control and PLWH study subjects are shown in Table 1. The average duration of HIV infection was 12.17± 10.59 years. There were no significant differences in age, sex, or racial distribution between control and PLWH. All PLWH were on ART therapy with controlled viral load (<400 copies/mL) and had a CD4+ T-cell count that ranged from 210 to 1318 cells/µL (average 639± 336.24) at the time of analysis.
HIV Infection Enhances Inducible FasL Expression in CD4+ T Cells
Initially, we confirmed the status of FasL in CD4+ T lymphocytes from PLWH by examining the FasL protein and mRNA levels. The serum concentrations of soluble FasL (sFasL) were significantly higher in PLWH in comparison to healthy controls (Fig. 1A). Freshly isolated total CD4+ T cells were stimulated via the T-cell receptor (TCR) using anti-CD3 and anti-CD28 antibodies for 24 hours and mRNA expression quantified by qPCR. Similar to earlier studies, there was significantly higher FasL mRNA expression after TCR stimulation in PLWH CD4+ T cells compared with control cells, (Fig. 1B).
FIGURE 1.
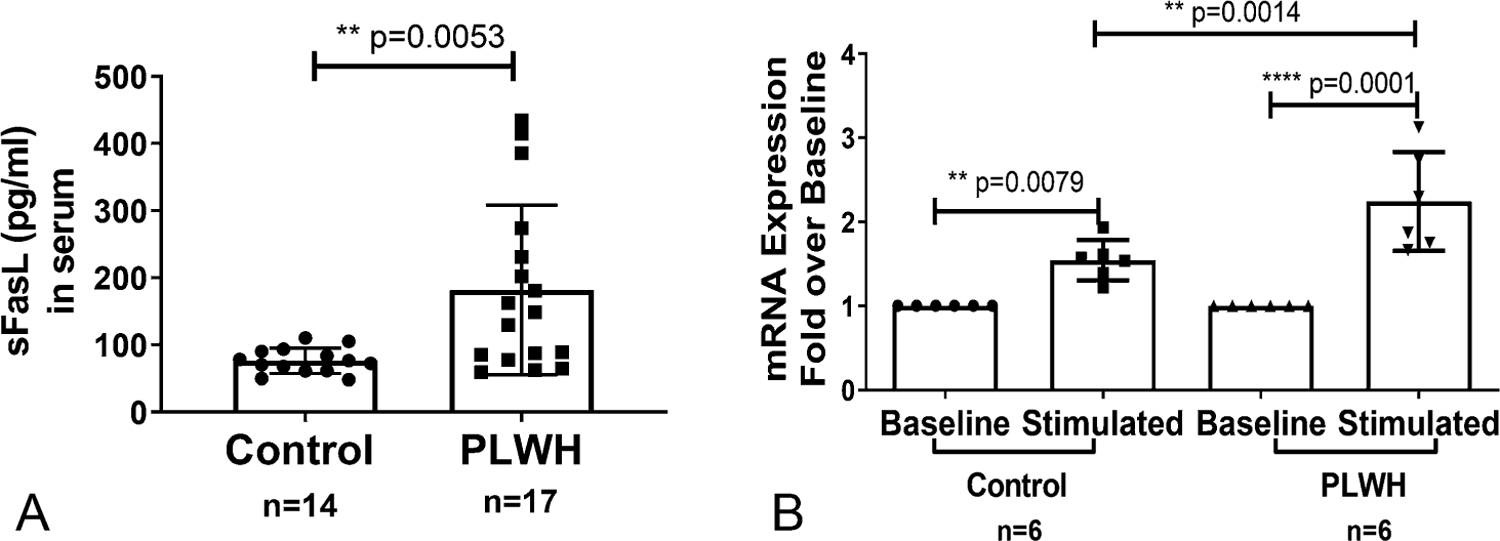
FasL expression in PLWH and noninfected control individuals: (A) sFASL levels were examined in the serum samples obtained from study subjects by ELISA. The Student t-test was performed between control (n = 14) and PLWH (n = 17) individuals. P < 0.01. B, Freshly isolated CD4+ T cells from a subset of control and PLWH individuals were either left untreated (baseline) or activated with anti-CD3/CD28 antibody (1 µg/mL each) for 24 hours (stimulated). Total RNA was isolated from and the mRNA levels were quantified by real-time qPCR. Data are expressed as fold-induction over baseline within each group. Results shown are mean ± SD (n = 6). Statistical analysis was performed with one-way ANOVA-Bonferroni correction for multiple comparisons as shown in the line bar with * = P < 0.05, ** = P < 0.01 and **** = P <0.0001.
FasL Gene Expression Negatively Correlates With Peripheral CD4+ T-Cell Counts
The functional relationship between FasL transcription and CD4+ T-cell count in PLWH was assessed by a nonparametric (Spearman coefficient) regression analysis (Fig. 2). Notably, a very strong negative correlation was observed between increased FasL expression (serum sFasL and mRNA) and decreased total CD4+ T-cell numbers (Figs. 2A, B).
FIGURE 2.
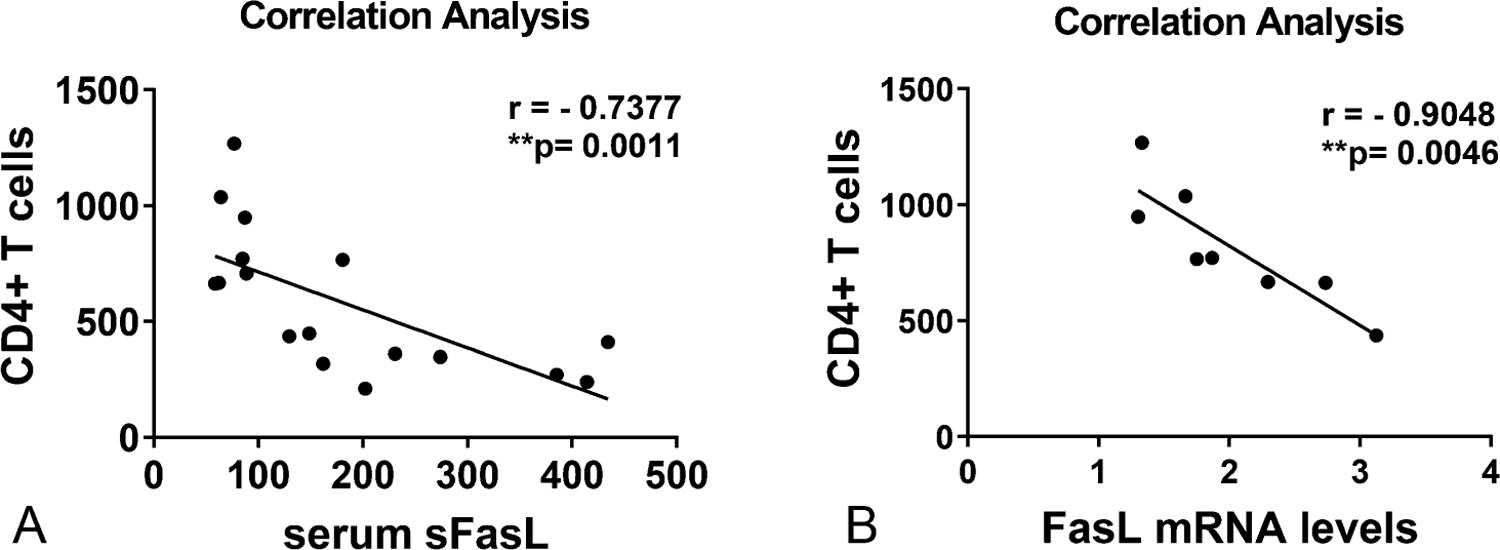
Correlation analysis of CD4+ T cells with FasL expression in PLWH individuals: Nonparametric regression analysis was performed between CD4+ T-cell numbers and FasL expression at both (A) sFasL protein (n = 17) and (B) mRNA levels (n = 8) in PLWH individuals. The Spearman correlation coefficient and significance were analyzed as Spearman r = −0.7377, P = **0.0011 and Spearman r = −0.9048, P = 0.0046, respectively.
HIV Infection Affects Naïve/Memory CD4+ T-Cell Distribution and FasL Expression
To understand whether the increase in the FasL expression may be associated with specific changes in T-cell subtypes, the FasL expression on 4 main subsets of T lymphocytes was evaluated by FACS analysis. PBMC-CD4+ T cells were gated as in Figure 1, Supplemental Digital Content, http://links.lww.com/QAI/B554. The CD4+ T-cell populations from PLWH had a significant decrease in naïve and Tcm (T central memory) population frequencies with a concomitant increase in Tem (T effector memory) cells compared with controls (Fig. 3A). The TEMRA (T effector memory cell re-expressing CD45RA) cells from PLWH showed an increasing trend but did not reach significance. The naïve/memory CD4+ T-cell subset distributions remained unaffected after TCR stimulation (Fig. 3A).
FIGURE 3.
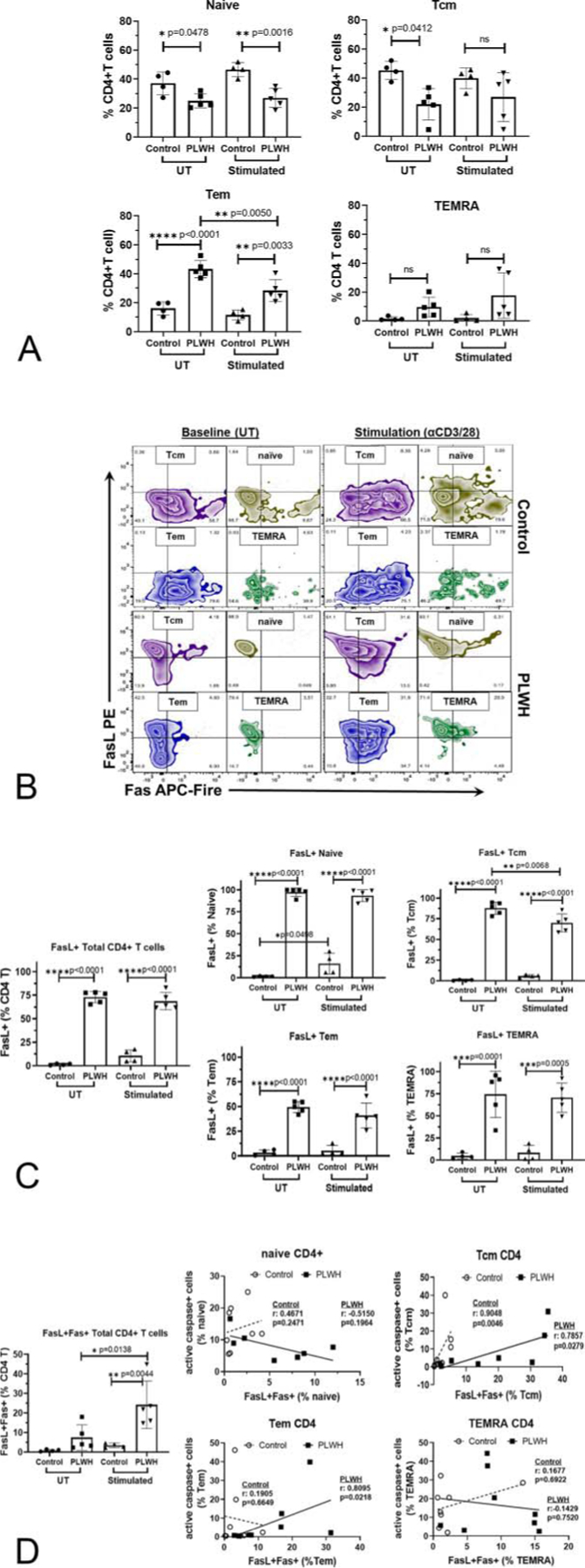
Naïve/memory T cell subset distribution and subset specific FasL expression in CD4+ T cells from controls and PLWH: PBMC cultures from healthy controls (control) or PLWH were cultured in medium alone (UT) or activated with anti-CD3/CD28 antibody (stimulated) for 48 hours and analyzed by flow cytometry. A, As gated in Figure 1, Supplemental Digital Content, http://links.lww.com/QAI/B554 total CD4+ T cells were assessed for naïve/memory T-cell subsets based on CD45RA and CCR7 expression and the percentage distribution of each subset is shown. B, Representative FACS plots were shown for Fas/FasL expression in naïve/memory CD4+ T cell subset in control and PLWH. C, The frequency of FasL expressing CD4+ T cells in total and each cell subsets was shown in controls (n = 4) and PLWH (n = 5). Results shown are mean ± SD. Statistical analysis was performed with one-way ANOVA-Bonferroni correction for multiple comparisons as shown in the line bar with * = P < 0.05, ** = P < 0.01 and **** = P <0.0001. D, Frequency distribution of FasL+Fas+ cells in total CD4+ T cells was evaluated (Left panel). Correlation analysis between FasL+Fas+ and active caspase 3/7+ apoptotic cells in naïve/memory CD4+ T-cell subsets was performed in healthy controls and PLWH with and without TCR activation for 72 hours. Nonparametric regression analysis for Spearman correlation coefficient and significance were analyzed and denoted on each graph for both controls and PLWH.
Representative surface expression of FasL and Fas in resting and TCR stimulated naïve/memory CD4+ T-cell subsets from PLWH and controls is shown in Figure 3B. The expression of FasL was present on >70% of total CD4+ T cells from PLWH compared with <2–10% from healthy controls. Furthermore, this striking phenotypic difference in expression of FasL was observed in all naïve/memory CD4+ T-cell subsets assessed from PLWH compared with controls (Fig. 3C). Overall, these data demonstrated that all naïve/memory CD4+ T-cell subsets from PLWH express FasL at a significantly greater frequency regardless of their activation status.
Co-expression of FasL-Fas Leads to Apoptosis in Tcm and Tem CD4+ T-Cell Subsets
In PLWH, the frequency of FasL+Fas+ cells was significantly increased upon activation in total and all naïve/memory CD4+ T-cell subsets (Fig. 3D and Fig. 2, Supplemental Digital Content, http://links.lww.com/QAI/B554). Co-expression of FasL and Fas on CD4+ T cells is often associated with activation-induced apoptosis. Hence, expression of active caspase 3/7 in naïve/memory CD4+ T-cell subsets was assessed and correlated with FasL+Fas+ expression (Fig. 3D). In PLWH, the frequency of FasL+Fas+ cells significantly and positively correlated with active caspase 3/7 only in Tcm and Tem subsets, but not in either naïve or TEMRA T cells (Fig. 3D). Notably, in control CD4+ T cells, FasL+Fas+ expression was very low in naïve/memory CD4+ T cell subsets, and only Tcm subset significantly correlated with active caspase 3/7. These data indicate that FasL+Fas+ memory (Tcm and Tem) but not naïve cells are likely the major contributors to the increased activation-induced apoptotic death in CD4+ T cells from PLWH.
HIV Infection Affects the Epigenetic Regulation of FasL Promoter in CD4+ T Cells
FasL expression is primarily regulated at the transcriptional level.31 Accordingly, FasL-promoter-associated histone modifications that affect chromatin modeling and transcriptional competence in a gene-specific manner were examined by ChIP analysis. Because all CD4+ T-cell subsets from PLWH demonstrated significant levels of FasL expression, promoter-associated epigenetic changes in the total CD4+ T-cell population were examined. Chromatin prepared from total CD4+ T lymphocytes from controls and PLWH was subjected to immunoprecipitation, using specific antibodies as described earlier.37 ChIP analysis evaluating promoter-histone modifications and transcription factor recruitment was performed focusing on 2 regions of the FasL promoter (denoted as region I and II—Fig. 4A) demonstrated to be relevant for the epigenetic regulation of FasL gene expression and associated with specific DNA bindingproteins.24,25,40
FIGURE 4.
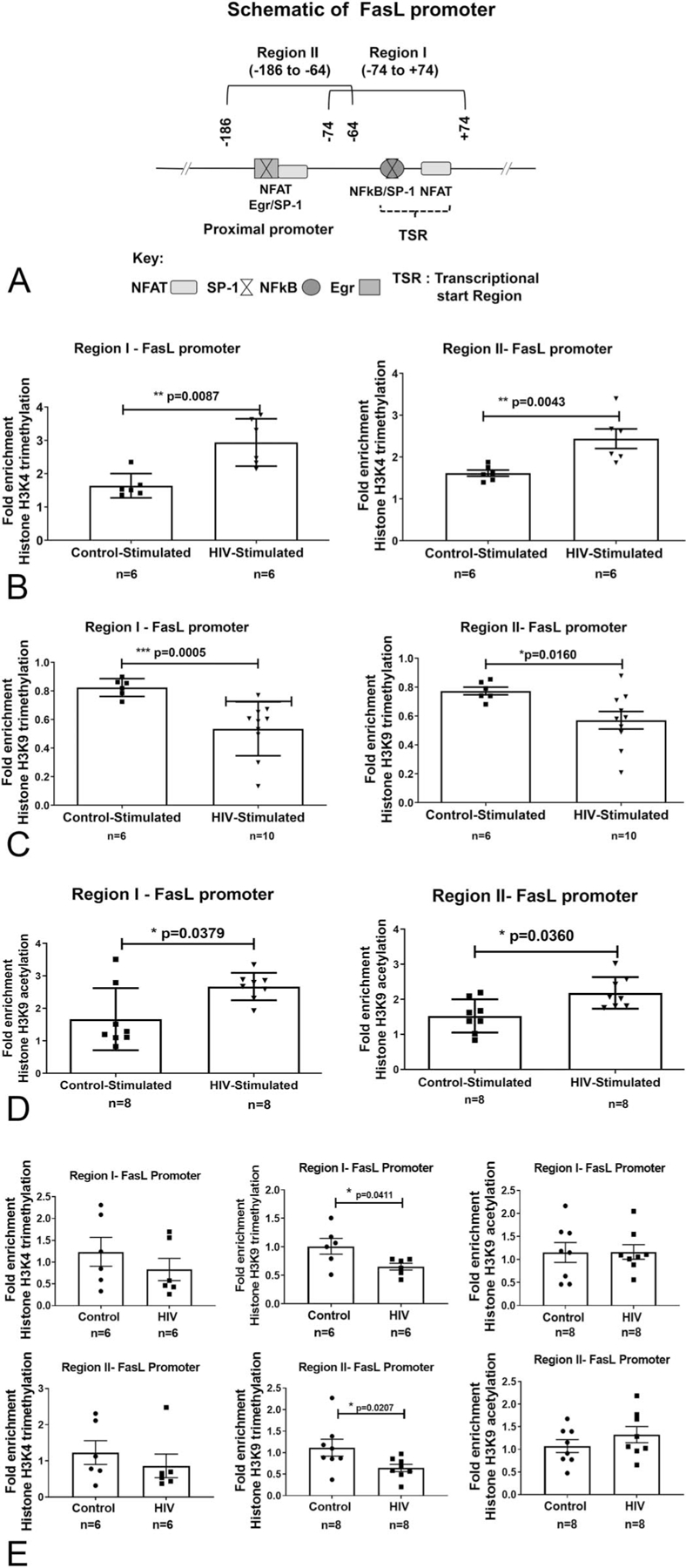
A, FasL promoter schematic and TCR stimulation-dependent histone H3 modifications at the FasL promoter in CD4+ T lymphocytes from control and PLWH individuals: (A) Locations of key transcription factor binding sites and ChIP-PCR primer pairs for analysis of epigenetic modifications are denoted as regions I and II. The coordinate locations shown are with respect to the transcription start site in REFSEQ NM_000639.1. B–D, Freshly isolated CD4+ T cells from PLWH and noninfected controls (n = 6–12) were examined by ChIP-qPCR analysis. The cells were either left untreated (baseline) or TCR stimulated for 6 hours (stimulated). FasL promoter-associated histone modifications were assessed by analyzing chromatin that was immunoprecipitated with (B) anti-trimethylated histone H3 lysine 4 (H3K4Me3), (C) anti-trimethylated histone H3 lysine 9 (H3K9Me3), or (D) anti-acetylated histone H3 lysine 9 (H3K9Ac) antibodies. E, ChIP analysis on chromatin from unstimulated cells was also performed to examine differences in the H3K4Me3, H3K9Me3, and H3K9Ac levels between CD4+ T cells from PLWH and HIV noninfected control individuals. Levels of histone modifications were measured using primers specific for regions I and II as shown in the schematic. Differences are expressed as fold over baseline after normalizing for input DNA. Results are represented as mean ± SD (n = 6–10). Statistical analysis was performed by one-way ANOVA with Bonferroni correction for multiple comparisons. * = P < 0.05, ** = P < 0.01 and **** = P <0.0001 compared with baseline.
TCR Activation Augments FasL Promoter H3K4-Trimethylation (H3K4Me3) in CD4+ T Cells from PLWH
Because the promoter status of H3K4Me3 PTM is critically linked to increased transcriptional activity,41–43 we initially examined the effect of HIV infection on the extent of H3K4Me3 at the FasL promoter. ChIP analysis was performed using anti-H3K4Me3-specific antibody on chromatin obtained from control and PLWH CD4+ T cells with and without TCR stimulation (6 hours). Levels of H3K4Me3 at the FasL promoter were interrogated by ChIP-qPCR assay using region-specific primers and the data was represented as fold-enrichment over baseline levels. In PLWH CD4+ T cells, the stimulation-induced FasL promoter H3K4Me3 levels were increased by 2.9-fold in Region I and 2.4-fold in Region II; whereas, the increase in control CD4+ T cells was ~1.6 fold in both regions I and II. This relative increase in transcriptionally permissive H3K4Me3 levels in PLWH CD4+ T cells also positively correlated with greater levels of FasL mRNA expression, compared with control CD4+ T cells (Fig. 4B).
TCR Activation Decreases FasL Promoter H3K9-Trimethylation (H3K9Me3) in CD4+ T Cells from PLWH
Because H3K4Me3 is known to impede transcriptionally repressive H3K9Me3, we next examined the H3K9-methylation status at the FasL promoter.42,44,45 Promoter ChIP analysis was done using an antihistone H3K9Me3 antibody as described earlier. The data showed that in contrast to H3K4Me3, activated CD4+ T cells from PLWH have significantly decreased levels of H3K9Me3 (Fig. 4C). Specifically, TCR activation led to a 50% reduction in H3K9Me3 present at both regions of the FasL promoter compared with nonactivated cells. Conversely, stimulated control cells exhibited significantly less of a reduction in H3K9Me3 levels at regions I and II, only 20% and 30% respectively.
In accordance with the high frequency of FasL+ CD4+ T cells in PLWH without activation, basal promoter H3K9Me3 levels in unstimulated CD4+ T cells showed those from PLWH had significantly lower levels of repressive H3K9Me3 than controls (Fig. 4E). These data indicate that CD4+ T cells from PLWH are primed or sensitized to an aberrant increase in FasL transcription.
TCR Activation Augments FasL Promoter H3K9-Acetylation in CD4+ T Cells from PLWH
Histone 3 lysine 9-acetylation (H3K9Ac) is among the key coordinated promoter histone modifications that culminate in the formation of transcriptionally permissive chromatin structures. Notably, studies have shown that H3K9-acetylation cooperates positively with H3K4Me3 and negatively with H3K9Me3 during transcriptional activation.46–49 Therefore, we also examined the alterations in FasL promoter H3K9Ac levels in control and PLWH CD4+ T lymphocytes. Similar to FasL promoter increased H3K4Me3 and decrease in H3K9Me3 levels, there was a significant increase in stimulation-induced H3K9Ac levels by 2.67-fold in Region I and 2.04-fold in region II compared with the increase of 1.78-fold and 1.52-fold in control CD4+ T cell regions I and II, respectively (Fig. 4D).
Together, these data indicate that a considerably greater percentage of CD4+ T lymphocytes in PLWH are committed to an activated transcriptional state leading to a significantly greater FasL expression.
Enhanced Transcriptional Activation Potential of FasL Promoter in CD4+ T Cells From PLWH
Following changes in histone modifications, transcriptional activation potential of FasL promoter was assessed by examining the recruitment of relevant transcription factors, and RNA polymerase-II (RNA Pol-II).
TCR Activation Increases Recruitment of Transcription Factors NFAT and NF-κB and RNA Pol-II to FasL Promoter in CD4+ T Cells From PLWH
The recruitment of NF-AT and NF-κB to regions I and II of the FasL promoter was assessed because these regions had accumulated significantly higher levels of transcriptionally permissive histone- H3-PTM in PLWH CD4+ T cells compared with controls. Specifically, the binding of NFAT to regions I and II (Fig. 5A) and, NF-κB (p65) (Fig. 5B) to the region I was evaluated by ChIP analysis. Both NFAT and NFκB showed increased promoter binding at their respective sites. Moreover, in accordance with the increase in transcription factor recruitment, establishment of enhanced transcriptional state of FasL promoter was further manifested by significantly increased RNA Pol–II binding at region I (TSS site) (Fig. 5C).
FIGURE 5.
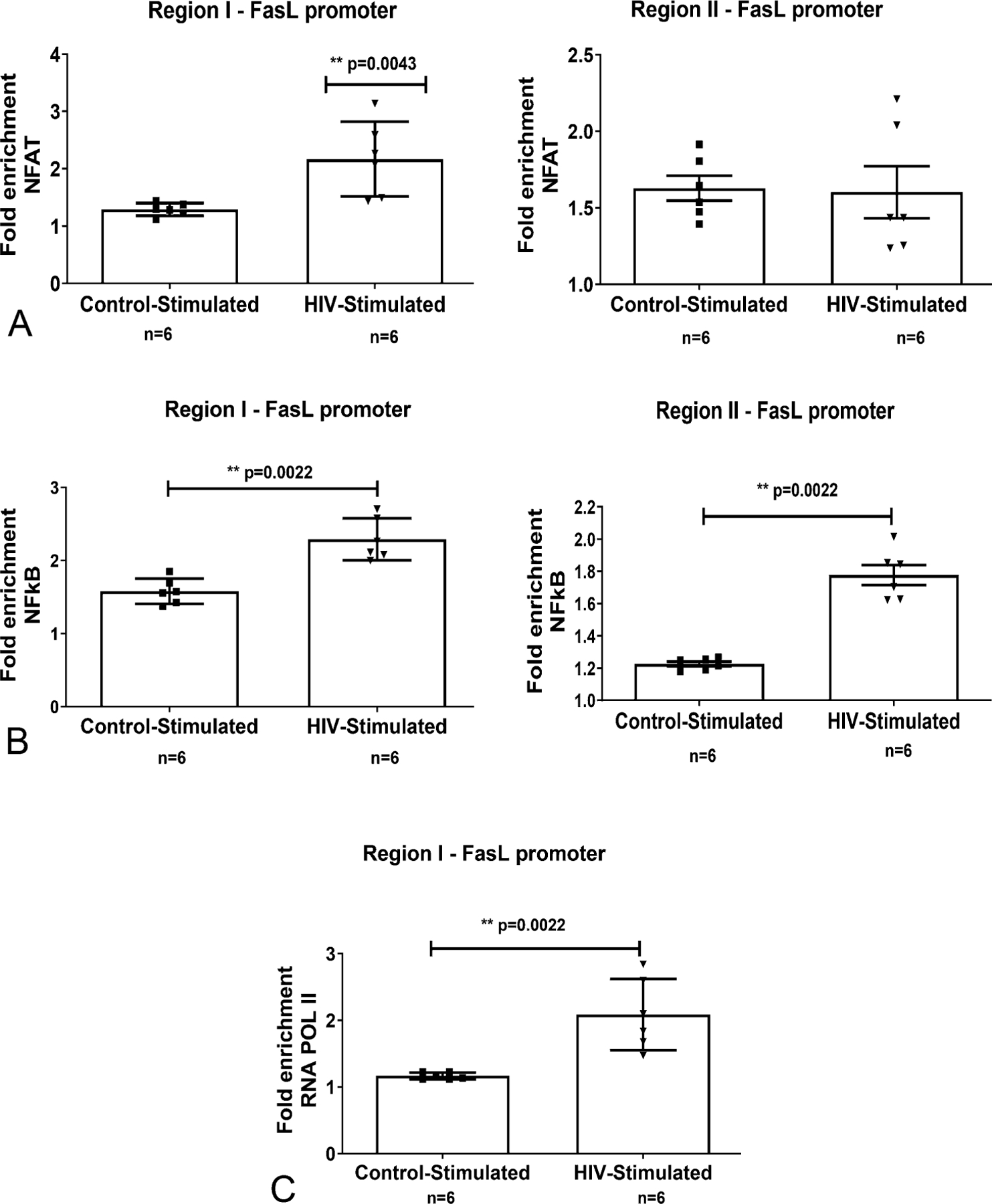
TCR activation-induced recruitment of transcription factors and RNA POL II at the FasL promoter in CD4+ T lymphocytes from control and HIV-infected individuals: Freshly isolated CD4+ T cells from control and PLWH individuals (n = 6–8) were untreated (baseline) or TCR activated with anti-CD3/CD28 antibodies (stimulated) for 12 hours. ChIP-qPCR quantification from (A) anti-NFAT (B) anti-NFκB (p65), (C) anti-RNA POL II immunoprecipitated chromatin was performed. Differences are expressed as fold over baseline after normalizing for input DNA. Results are represented as mean ± SD (n = 6–8). Statistical analysis was performed by one-way ANOVA with Bonferroni correction for multiple comparisons. * = P < 0.05, ** = P < 0.01 and **** = P <0.0001 compared with baseline.
Taken together, these epigenetic promoter histone modifications affecting transcriptional competence constitute a major mechanism regulating FasL expression and CD4+ T-cell survival in HIV infection.
DISCUSSION
A critical factor in immunodeficiency caused by HIV-infection is the loss of CD4+ T lymphocytes, leading to the impairment of multiple immune functions.17,18 Although data obtained from several experimental and clinical studies showed that HIV infection decreases CD4+ T lymphocytes, the associated molecular mechanism(s) have not been completely elucidated. Under physiologic conditions, the cellular homeostasis of CD4+ T lymphocytes is maintained by balancing the pro- and anti-apoptotic cell death mechanisms. In PLWH, studies have shown that this balance is shifted to be more pro-apoptotic.4,9,10 A predominant feature of this shift is the upregulation of FasL, a key apoptotic death-inducing molecule. In this regard, the examination of naïve/memory CD4+ T-cell subsets from virally controlled PLWH revealed that their distribution was not only skewed but they all expressed surface FasL at a markedly greater frequency compared with HIV noninfected controls. The observed alteration in naïve/memory T-cell subset distribution is in accordance with previous data from PLWH50,51; however, the presence of pronounced FasL expression in all the subsets is a significant finding. Hence, to address the molecular mechanisms underpinning aberrant FasL expression in PLWH, our work identifies epigenetic promoter-histone modifications that play a significant role in transcriptional regulation of FasL gene.
Epigenetic mechanisms involving histone PTMs affect chromatin structure and regulate the transcriptional status of promoters leading to either gene activation or repression.26,27,30 Examination of native FasL chromatin from unstimulated/resting CD4+ T cells from PLWH showed a significant decrease in the transcriptionally repressive H3K9Me3 modification (Fig. 4E). These data strongly suggest that this histone modification likely plays a role in priming the CD4+ T cells to increased transcriptional activation of FasL gene. This notion was further supported by a concomitant increase in the transcriptionally permissive (activating) H3K4Me3 and H3K9Ac modifications and FasL mRNA expression upon TCR stimulation in PLWH. The observed coordinated increase in promoter H3K4-methylation and H3K9-demethylation is further functionally correlated with an increase in H3K9Ac and constitutes a regulatory feature of initiated and actively transcribed genes.43,44,52–54 Moreover, these modifications are highly significant because they are mutually exclusive, functionally linked, and highly conserved.43,45,52,55 Our earlier work has demonstrated similar promoter histone modifications in CD4+ T cells exposed to alcohol leading to aberrant FasL expression.31 Taken together, these data demonstrate that coordinated regulation of H3 PTMs significantly contributes to HIV-infection associated development of susceptibility of CD4+ T cells to enhanced FasL expression in ART-treated virally suppressed PLWH.
Regarding increased FasL expression associated with HIV infection in PLWH, both viral and host factors play a contributory role. Particularly, HIV viral proteins including Tat, Nef, and gp120 are known to upregulate FasL expression in CD4+ T cells and are present in ART-treated and virally suppressed individuals.56–58 Furthermore, along with viral proteins, host factors such as inflammatory cytokines and oxidative stress components associated with HIV infection also increase FasL expression in CD4+ T cells.59–61 Accordingly, FasL promoter histone modifications observed in our PLWH study cohort could be instituted by HIV infection-associated viral and host factors. Furthermore, in relevance to activation-induced apoptosis in PLWH, our data demonstrate that marked increase in FasL expression also contributes to an increase in the frequency of activation-induced FasL and Fas double-positive cells in all naïve/memory CD4+ T-cell subsets. Notably, examination of functional outcome of FasL and Fas co-expression revealed a correlation with active caspase 3/7 only in Tcm and Tem subsets from PLWH, thus demonstrating their preferential susceptibility to activation-induced apoptosis. Similarly, CD4+ T-cell subset-dependent preferential susceptibility to Fas-induced apoptosis has also been reported in aged humans, albeit in naïve and Tcm cells.62 The development of this susceptibility of Tcm and Tem subsets likely plays an important role in the loss of CD4+ T cell homeostasis, characteristic of HIV pathogenesis. Overall, these data support the notion that even under conditions of controlled viremia, epigenetic histone modifications occurring in CD4+ T cells increase FasL expression and susceptibility to apoptosis, affecting CD4+ T-cell recovery in PLWH.
Supplementary Material
Acknowledgments
Supported by funding from RO1 AA024405, R01AA024405–02S1, P50 AA024337, U01AA026222.
Footnotes
The authors have no funding or conflicts of interest to disclose.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.jaids.com).
REFERENCES
- 1.Okoye AA, Picker LJ. CD4(+) T-cell depletion in HIV infection: mechanisms of immunological failure. Immunol Rev 2013;254:54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vidya Vijayan KK, Karthigeyan KP, Tripathi SP, et al. Pathophysiology of CD4+ T-cell depletion in HIV-1 and HIV-2 infections. Front Immunol 2017;8:580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saharia KK, Koup RA. T cell susceptibility to HIV influences outcome of opportunistic infections. Cell 2013;155:505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fevrier M, Dorgham K, Rebollo A. CD4+ T cell depletion in human immunodeficiency virus (HIV) infection: role of apoptosis. Viruses 2011;3:586–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cummins NW, Badley AD. Mechanisms of HIV-associated lymphocyte apoptosis: 2010. Cell Death Dis 2010;1:e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan ZT, Chen XL. Mechanisms of HIV envelope-induced T lymphocyte apoptosis. Virol Sin 2010;25:307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garg H, Joshi A. Host and viral factors in HIV-mediated bystander apoptosis. Viruses 2017;9:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alderson MR, Tough TW, Davis-Smith T, et al. Fas ligand mediates activation-induced cell death in human T lymphocytes. J Exp Med 1995; 181:71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poonia B, Pauza CD, Salvato MS. Role of the Fas/FasL pathway in HIV or SIV disease. Retrovirology 2009;6:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sloand EM, Young NS, Kumar P, et al. Role of Fas ligand and receptor in the mechanism of T-cell depletion in acquired immunodeficiency syndrome: effect on CD4+ lymphocyte depletion and human immunodeficiency virus replication. Blood 1997;89:1357–1363. [PubMed] [Google Scholar]
- 11.Debatin KM, Fahrig-Faissner A, Enenkel-Stoodt S, et al. High expression of APO-1 (CD95) on T lymphocytes from human immunodeficiency virus-1-infected children. Blood 1994;83:3101–3103. [PubMed] [Google Scholar]
- 12.Hosaka N, Oyaizu N, Than S, et al. Correlation of loss of CD4 T cells with plasma levels of both soluble form Fas (CD95) Fas ligand (FasL) in HIV-infected infants. Clin Immunol 2000;95:20–25. [DOI] [PubMed] [Google Scholar]
- 13.Badley AD, Dockrell DH, Algeciras A, et al. In vivo analysis of Fas/FasL interactions in HIV-infected patients. J Clin Invest 1998;102:79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fadeel B, Samuelsson A, Hachiya T, et al. Elevated serum levels of soluble Fas/APO-1 in human immunodeficiency virus-infected individuals. Blood 1996;88:4727–4730. [PubMed] [Google Scholar]
- 15.Hosaka N, Oyaizu N, Kaplan MH, et al. Membrane and soluble forms of Fas (CD95) and Fas ligand in peripheral blood mononuclear cells and in plasma from human immunodeficiency virus-infected persons. J Infect Dis 1998;178:1030–1039. [DOI] [PubMed] [Google Scholar]
- 16.Dianzani U, Bensi T, Savarino A, et al. Role of FAS in HIV infection. Curr HIV Res 2003;1:405–417. [DOI] [PubMed] [Google Scholar]
- 17.Badley AD, Pilon AA, Landay A, et al. Mechanisms of HIV-associated lymphocyte apoptosis. Blood 2000;96:2951–2964. [PubMed] [Google Scholar]
- 18.Alimonti JB, Ball TB, Fowke KR. Mechanisms of CD4+ T lymphocyte cell death in human immunodeficiency virus infection and AIDS. J Gen Virol 2003;84:1649–1661. [DOI] [PubMed] [Google Scholar]
- 19.Algeciras-Schimnich A, Vlahakis SR, Villasis-Keever A, et al. CCR5 mediates Fas- and caspase-8 dependent apoptosis of both uninfected and HIV infected primary human CD4 T cells. AIDS 2002;16:1467–1478. [DOI] [PubMed] [Google Scholar]
- 20.Muthumani K, Choo AY, Hwang DS, et al. HIV-1 Nef-induced FasL induction and bystander killing requires p38 MAPK activation. Blood 2005;106:2059–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uchiyama J, Kishi S, Yagita H, et al. Fas ligand-mediated depletion of CD4 and CD8 lymphocytes by monomeric HIV-1-gp120. Arch Virol 1997;142:1771–1785. [DOI] [PubMed] [Google Scholar]
- 22.Li-Weber M, Laur O, Hekele A, et al. A regulatory element in the CD95 (APO-1/Fas) ligand promoter is essential for responsiveness to TCR-mediated activation. Eur J Immunol 1998;28:2373–2383. [DOI] [PubMed] [Google Scholar]
- 23.Suda T, Takahashi T, Golstein P, et al. Molecular cloning and expression of the Fas ligand, a novel member of the tumor necrosis factor family. Cell 1993;75:1169–1178. [DOI] [PubMed] [Google Scholar]
- 24.Li-Weber M, Krammer PH. Function and regulation of the CD95 (APO-1/Fas) ligand in the immune system. Semin Immunol 2003;15:145–157. [DOI] [PubMed] [Google Scholar]
- 25.Kavurma MM, Khachigian LM. Signaling and transcriptional control of Fas ligand gene expression. Cell Death Differ 2003;10:36–44. [DOI] [PubMed] [Google Scholar]
- 26.Mellor J. The dynamics of chromatin remodeling at promoters. Mol Cell 2005;19:147–157. [DOI] [PubMed] [Google Scholar]
- 27.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011;21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strahl BD, Allis CD. The language of covalent histone modifications. Nature 2000;403:41–45. [DOI] [PubMed] [Google Scholar]
- 29.Verdone L, Caserta M, Di Mauro E. Role of histone acetylation in the control of gene expression. Biochem Cell Biol 2005;83:344–353. [DOI] [PubMed] [Google Scholar]
- 30.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell 2010;142:682–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghare SS, Joshi-Barve S, Moghe A, et al. Coordinated histone H3 methylation and acetylation regulate physiologic and pathologic fas ligand gene expression in human CD4+ T cells. J Immunol 2014;193: 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klebanoff CA, Scott CD, Leonardi AJ, et al. Memory T cell-driven differentiation of naive cells impairs adoptive immunotherapy. J Clin Invest 2016;126:318–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russ BE, Olshanksy M, Smallwood HS, et al. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8(+) T cell differentiation. Immunity 2014;41:853–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tammen SA, Friso S, Choi SW. Epigenetics: the link between nature and nurture. Mol Aspects Med 2013;34:753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benayoun BA, Pollina EA, Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat Rev Mol Cell Biol 2015;16:593–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghare S, Patil M, Hote P, et al. Ethanol inhibits lipid raft-mediated TCR signaling and IL-2 expression: potential mechanism of alcohol-induced immune suppression. Alcohol Clin Exp Res 2011;35:1435–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gobejishvili L, Avila DV, Barker DF, et al. S-adenosylmethionine decreases lipopolysaccharide-induced phosphodiesterase 4B2 and attenuates tumor necrosis factor expression via cAMP/protein kinase A pathway. J Pharmacol Exp Ther 2011;337:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez-Botran R, Joshi-Barve S, Ghare S, et al. Systemic cytokine and interferon responsiveness Patterns in HIV and HCV mono and co-infections. J Interferon Cytokine Res 2014;34:885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maecker HT, Trotter J. Flow cytometry controls, instrument setup, and the determination of positivity. Cytometry A 2006;69:1037–1042. [DOI] [PubMed] [Google Scholar]
- 40.Holtz-Heppelmann CJ, Algeciras A, Badley AD, et al. Transcriptional regulation of the human FasL promoter-enhancer region. J Biol Chem 1998;273:4416–4423. [DOI] [PubMed] [Google Scholar]
- 41.Vermeulen M, Mulder KW, Denissov S, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 2007;131:58–69. [DOI] [PubMed] [Google Scholar]
- 42.Binda O, LeRoy G, Bua DJ, et al. Trimethylation of histone H3 lysine 4 impairs methylation of histone H3 lysine 9: regulation of lysine methyltransferases by physical interaction with their substrates. Epigenetics 2010;5:767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol 2008;20:341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li F, Huarte M, Zaratiegui M, et al. Lid2 is required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin. Cell 2008;135:272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi L, Sun L, Li Q, et al. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc Natl Acad Sci U S A 2011;108:7541–7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications - writers that read. EMBO Rep 2015;16:1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murr R. Interplay between different epigenetic modifications and mechanisms. Adv Genet 2010;70:101–141. [DOI] [PubMed] [Google Scholar]
- 48.Nightingale KP, Gendreizig S, White DA, et al. Cross-talk between histone modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J Biol Chem 2007;282:4408–4416. [DOI] [PubMed] [Google Scholar]
- 49.Park JA, Kim AJ, Kang Y, et al. Deacetylation and methylation at histone H3 lysine 9 (H3K9) coordinate chromosome condensation during cell cycle progression. Mol Cell 2011;31:343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serrano-Villar S, Sainz T, Lee SA, et al. HIV-infected individuals with low CD4/CD8 ratio despite effective antiretroviral therapy exhibit altered T cell subsets, heightened CD8+ T cell activation, and increased risk of non-AIDS morbidity and mortality. PLoS Pathog 2014;10:e1004078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Breton G, Chomont N, Takata H, et al. Programmed death-1 is a marker for abnormal distribution of naive/memory T cell subsets in HIV-1 infection. J Immunol 2013;191:2194–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crump NT, Hazzalin CA, Bowers EM, et al. Dynamic acetylation of all lysine-4 trimethylated histone H3 is evolutionarily conserved and mediated by p300/CBP. Proc Natl Acad Sci U S A 2011;108:7814–7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roh TY, Cuddapah S, Cui K, et al. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci U S A 2006;103: 15782–15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature 1997;389:349–352. [DOI] [PubMed] [Google Scholar]
- 55.Turner BM. Reading signals on the nucleosome with a new nomenclature for modified histones. Nat Struct Mol Biol 2005;12:110–112. [DOI] [PubMed] [Google Scholar]
- 56.Mediouni S, Darque A, Baillat G, et al. Antiretroviral therapy does not block the secretion of the human immunodeficiency virus tat protein. Infect Disord Drug Targets 2012;12:81–86. [DOI] [PubMed] [Google Scholar]
- 57.Lee JH, Schierer S, Blume K, et al. HIV-nef and ADAM17-containing plasma extracellular vesicles induce and correlate with immune pathogenesis in chronic HIV infection. EBioMedicine 2016;6:103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferdin J, Goricar K, Dolzan V, et al. Viral protein Nef is detected in plasma of half of HIV-infected adults with undetectable plasma HIV RNA. PLoS One 2018;13:e0191613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iannello A, Samarani S, Debbeche O, et al. Potential role of interleukin-18 in the immunopathogenesis of AIDS: involvement in fratricidal killing of NK cells. J Virol 2009;83: 5999–6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gulow K, Kaminski M, Darvas K, et al. HIV-1 trans-activator of transcription substitutes for oxidative signaling in activation-induced T cell death. J Immunol 2005;174:5249–5260. [DOI] [PubMed] [Google Scholar]
- 61.Gasper-Smith N, Crossman DM, Whitesides JF, et al. Induction of plasma (TRAIL), TNFR-2, Fas ligand, and plasma microparticles after human immunodeficiency virus type 1 (HIV-1) transmission: implications for HIV-1 vaccine design. J Virol 2008;82:7700–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta S, Gollapudi S. CD95-mediated apoptosis in naive, central and effector memory subsets of CD4+ and CD8+ T cells in aged humans. Exp Gerontol 2008;43:266–274. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


