Abstract
PURPOSE
Tissue-based comprehensive genomic profiling (CGP) is increasingly used for treatment selection in patients with advanced cancer; however, tissue availability may limit widespread implementation. Here, we established real-world CGP tissue availability and assessed CGP performance on consecutively received samples.
MATERIALS AND METHODS
We conducted a post hoc, nonprespecified analysis of 32,048 consecutive tumor tissue samples received for StrataNGS, a multiplex polymerase chain reaction (PCR)–based comprehensive genomic profiling (PCR-CGP) test, as part of an ongoing observational trial (NCT03061305). Sample characteristics and PCR-CGP performance were assessed across all tested samples, including exception samples not meeting minimum input quality control (QC) requirements (< 20% tumor content [TC], < 2 mm2 tumor surface area [TSA], DNA or RNA yield < 1 ng/µL, or specimen age > 5 years). Tests reporting ≥ 1 prioritized alteration or meeting TC and sequencing QC were considered successful. For prostate carcinoma and lung adenocarcinoma, tests reporting ≥ 1 actionable or informative alteration or meeting TC and sequencing QC were considered actionable.
RESULTS
Among 31,165 (97.2%) samples where PCR-CGP was attempted, 10.7% had < 20% TC and 59.2% were small (< 25 mm2 tumor surface area). Of 31,101 samples evaluable for input requirements, 8,089 (26.0%) were exceptions not meeting requirements. However, 94.2% of the 31,101 tested samples were successfully reported, including 80.5% of exception samples. Positive predictive value of PCR-CGP for ERBB2 amplification in exceptions and/or sequencing QC-failure breast cancer samples was 96.7%. Importantly, 84.0% of tested prostate carcinomas and 87.9% of lung adenocarcinomas yielded results informing treatment selection.
CONCLUSION
Most real-world tissue samples from patients with advanced cancer desiring CGP are limited, requiring optimized CGP approaches to produce meaningful results. An optimized PCR-CGP test, coupled with an inclusive exception testing policy, delivered reportable results for > 94% of samples, potentially expanding the proportion of CGP-testable patients and impact of biomarker-guided therapies.
INTRODUCTION
Molecular profiling of patient tumor specimens is increasingly important as more therapies are indicated in biomarker-defined patient populations.1 Next-generation sequencing (NGS) is the diagnostic method of choice to assess relevant biomarkers simultaneously from formalin-fixed paraffin-embedded (FFPE) tumor tissue2-10 or circulating cell-free DNA (cfDNA) liquid biopsy sample.11-14 The US Center for Medicare and Medicaid Services has deemed tissue-based comprehensive genomic profiling (CGP) by NGS—which includes evaluation of single-nucleotide variants, short insertions and deletions (indels), copy number amplifications and deep deletions, gene fusions, microsatellite instability (MSI), and tumor mutation burden (TMB)—medically necessary for patients with advanced solid tumors (NCD CAG-00450N and LCD L38045).
CONTEXT
Key Objective
Comprehensive genomic profiling (CGP) on tumor tissue can guide treatment for patients with advanced solid tumors. Whether tissue requirements for leading tests (eg, ≥ 25 mm2 tumor surface area and ≥ 20% tumor content) limit adoption and affect real-world performance is unclear. Here, we determined tissue characteristics and CGP performance in > 30,000 consecutively received tissue samples tested by polymerase chain reaction (PCR)–based comprehensive genomic profiling.
Knowledge Generated
Among > 30,000 consecutively tested tissue samples, 59% had < 25 mm2 tumor surface area and 11% had < 20% tumor content. PCR-based CGP and a broad exception testing policy (performing testing on samples not meeting minimum input requirements) successfully reported 94% of samples, including 81% of such exception samples.
Relevance
The majority of samples received for tissue-based CGP testing in a real-world cohort are small. An optimized PCR-CGP test and testing of exception samples may increase the proportion of patients who can undergo tissue CGP to guide biomarker-based therapies.
Successful FFPE tissue CGP requires nucleic acid isolation of adequate quantity and quality from tumor cells. Challenges affecting real-world CGP applicability include minute specimens, samples with low tumor content (TC), and low-quality nucleic acid (affected by sample age and fixation).15 To maximize reportability, most CGP tests have tumor size (generally in mm2 of tumor surface area [TSA]), TC, and nucleic acid yield or input requirements.3,5,15 However, tissue CGP test failure rates of 30%-50% in clinical trial cohorts, where testing is attempted on all received samples (or those received below input requirements are considered failures), suggest that current CGP approaches, which are largely based on hybrid capture (HC) library preparation, may only be applicable for a subset of specimens.16-20
Herein, we characterized sample attributes of > 30,000 consecutive real-world samples submitted for CGP and the performance of a multiplex polymerase chain reaction (PCR)–based CGP test, StrataNGS, applied to consecutively received and tested samples, including those below sample input requirements.
MATERIALS AND METHODS
Patient Cohort
All samples received for testing from February 13, 2017, to June 25, 2020, from the Strata Trial (NCT03061305), a 100,000-patient observational study for patients with advanced solid tumors, were included. Sample, sequencing quality control (QC) metrics, and clinically reported biomarker results were retrieved from StrataPOINT, a de-identified Strata Trial Results Database (Data Supplement).
CGP Testing
Samples were tested with StrataNGS, the current version of which is a 429-gene polymerase chain reaction–based comprehensive genomic profiling (PCR-CGP) laboratory-developed test for FFPE tumor tissue samples performed on coisolated DNA and RNA, which has been validated on more than 1,900 FFPE tumor samples, and is covered for Medicare beneficiaries (21,22 and Tomlins et al, manuscript submitted). Earlier StrataNGS versions used during the described study period (Data Supplement) were essentially the same, but only report prioritized mutations from 57 genes (v3) or did not include TMB (v2)22; as specimen requirements have not changed, all received samples during the described study period were included.
StrataNGS requires one FFPE block or 10 × 5 µm-thick unstained slides. Minimum sample input QC requirements are TSA ≥ 2 mm2, TC ≥ 20%, time from sample acquisition < 5 years, and ≥ 1 ng/µL for both DNA and RNA; however, samples not meeting these requirements but with identifiable and isolatable tumor are deemed exceptions and testing is attempted. PCR-CGP data are processed using in-house–developed bioinformatics pipelines and sequencing QC assessments are performed per variant type and a final molecularly informed TC is determined. For samples failing ≥ 1 sequencing QC assessments or with a final TC < 20% (the StrataNGS limits of detection [LOD] for most alteration types), positive alterations may still be called via an expert molecular pathology review process; however, other alterations cannot be definitively ruled out, thus yielding a partial test result. Additional test details and sample QC metric definitions are provided in the Data Supplement; the formal analytical and clinical validation of the current test is described separately (Tomlins et al manuscript submitted).
Reportability, Actionability, and Positive Predictive Value
Tests with ≥ 1 reported prioritized alteration or passing all sequencing QC assessments and having TC ≥ 20% were considered successfully reported. Pan-cancer actionability is described in the Data Supplement. For prostate cancer and non–small-cell lung cancer (NSCLC) adenocarcinoma specific analyses, only reports that could rule in (by being positive for an actionable or exclusionary biomarker) or rule out (by passing all sequencing QC metrics and TC > 20%) biomarker-directed therapy were considered informative, as described in the Data Supplement.
RESULTS
Characteristics of Samples Received for CGP
CGP testing was performed by a single Clinical Laboratory Improvement Amendments–certified, College of American Pathologists–accredited laboratory (Strata Oncology, Ann Arbor, MI) as part of an observational clinical trial evaluating the impact of solid tumor sequencing in the advanced-cancer setting using a previously validated PCR-CGP test (StrataNGS). Across 28 diverse US health systems, 31,165 consecutive unique solid-tumor samples (from 30,565 unique patients) were received for CGP testing between February 13, 2017, and June 25, 2020; an additional 883 samples (Fig 1A) were rejected for various reasons, most commonly scant or no identifiable tissue or tumor (Data Supplement). Rejected samples were excluded from further analysis.
FIG 1.
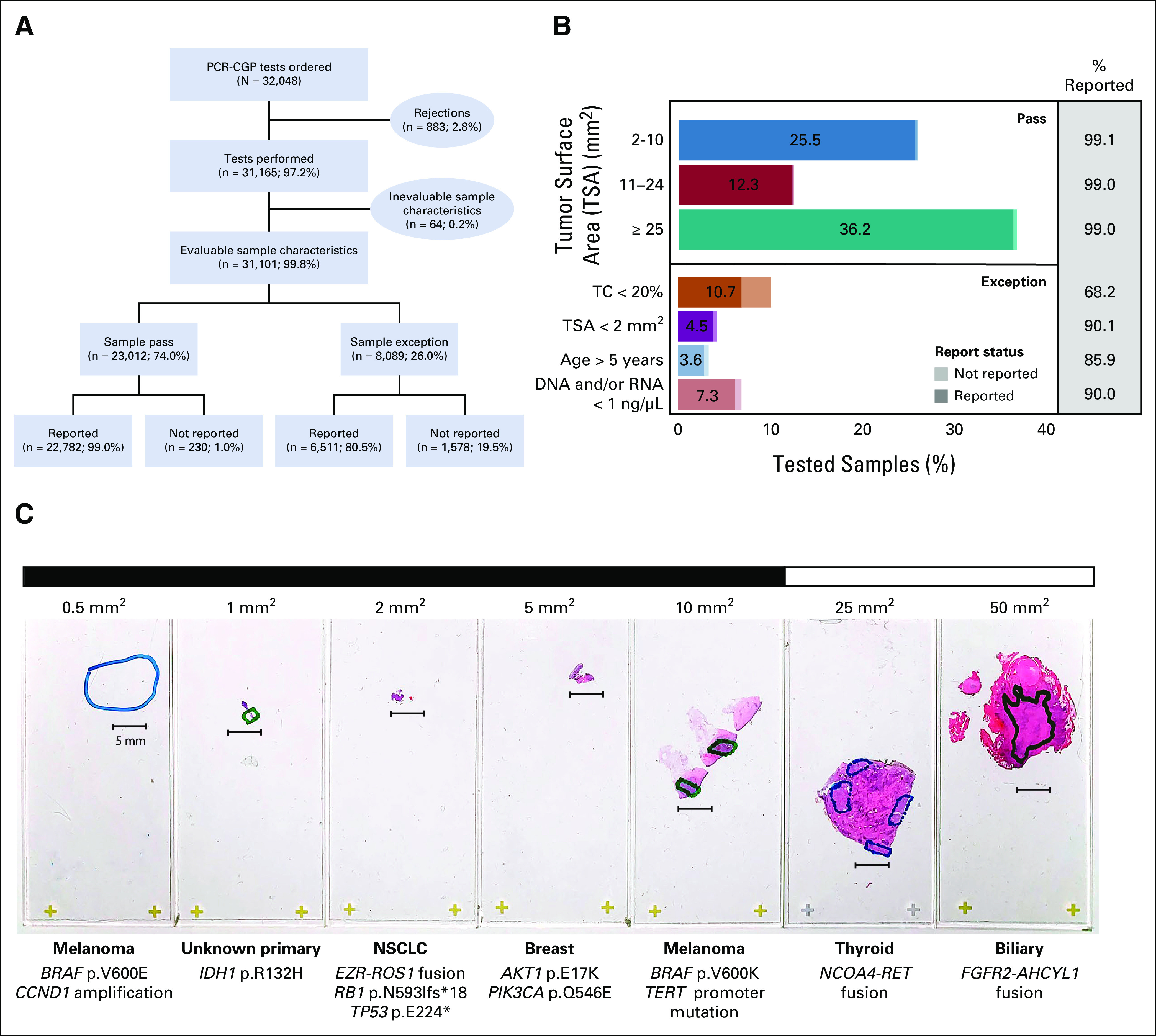
(A) Breakdown of consecutive PCR-CGP tests ordered from a single commercial clinical sequencing provider between February 13, 2017, and June 25, 2020, including the number of samples rejected before testing, the number of tests performed, the number of samples with evaluable input characteristics, and the number of PCR-CGP tests successfully reported. Samples were grouped into those meeting (pass) or not meeting (exception) PCR-CGP input requirements. (B) For all samples with evaluable input characteristics (n = 31,101), the distribution of samples per characteristic is shown. For samples passing all input requirements (pass), samples are stratified by TSA; exception samples were stratified by indicated sample attribute (TC < 20%; TSA < 2 mm2; age > 5 years: specimen collected > 5 years before PCR-CGP; and DNA and/or RNA concentration < 1 ng/µL). For each sample characteristic category, the proportion of the total number of samples with evaluable input characteristics is shown within the bar; the percentage of successfully reported samples is indicated by darker shading in the stacked bar chart and displayed numerically in the gray box at right. (C) Representative successfully reported samples received for PCR-CGP across a TSA range (small [< 25 mm2] and large [≥ 25 mm2] samples indicated). Cancer types and selected prioritized alterations are shown. CGP, comprehensive genomic profiling; NSCLC, non–small-cell lung cancer; PCR-CGP, multiplex polymerase chain reaction–based comprehensive genomic profiling; QC, quality control; TC, tumor content; TSA, tumor surface area.
Sample characteristics from all 31,165 consecutively tested samples are shown in Table 1, with demographics in the Data Supplement. Notably, 10.7% of tested samples had a final TC < 20% (Table 1), a common minimum TC requirement for CGP tests (including the PCR-CGP test), because corresponding LOD can preclude exclusion of certain variants or variant types below that TC.3,5 Only 40.8% of samples had TSA ≥ 25 mm2, with 44.7% of samples having ≤ 10 mm2 TSA (Table 1). Importantly, TSA ≥ 25 mm2 is the minimum TSA requirement for several leading commercial HC-CGP tests, including the only US Food and Drug Administration (FDA)–approved tissue CGP companion diagnostic device (FoundationOne CDx).3,5,23,24
TABLE 1.
Characteristics of 31,165 Specimens Received for CGP
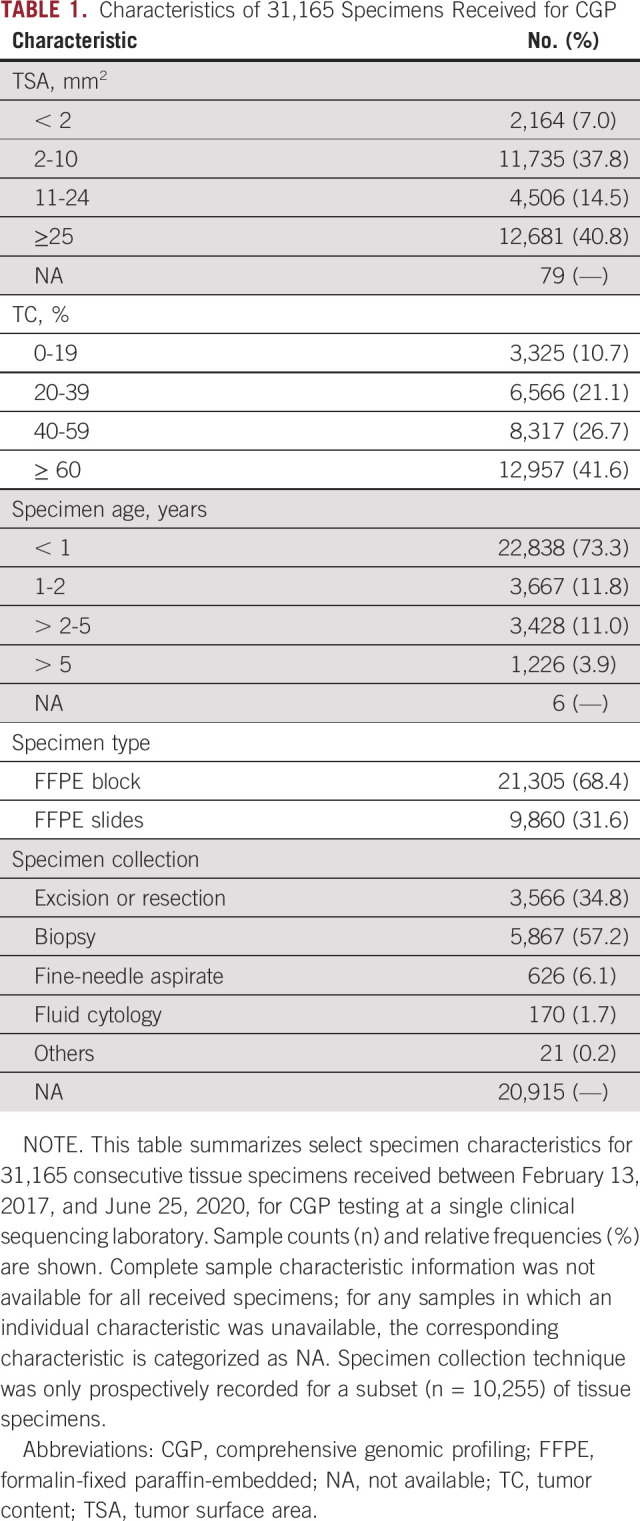
As expected, the majority of samples were from biopsies (57.2%); however, cytology cell blocks from fine-needle aspirations and fluid cytology comprised 7.8% of samples (Table 1). As expected, nucleic acid yield was associated with TSA; as among samples with < 2 mm2 TSA, only 44.5% and 51.7% yielded > 1 ng/µL DNA and RNA, respectively (Data Supplement).
Pan-Tumor CGP Experience
Given our previous experience that PCR-CGP could often deliver partial results even in very poor quality samples, CGP was attempted on all 31,165 tumor samples using the PCR-CGP test, including exception samples not passing input requirements (those with TC < 20%, TSA < 2 mm2, specimen age > 5 years, or DNA and/or RNA concentration < 1 ng/µL); median turnaround time from sample receipt to report release was 7 business days (interquartile range 6-9). Of these 31,165 samples, 31,101 (99.8%) were evaluable for passing sample input requirements and were further considered for assessing sample characteristic impact on PCR-CGP reportability (Fig 1A and Data Supplement).
As shown in Figure 1 and the Data Supplement, 29,293 of 31,101 (94.2%) samples were successfully reported, defined as having at least one reported prioritized alteration or passing all sequencing QC assessments and ≥ 20% final TC. Among the 23,012 (74.0%) samples passing all input requirements, 22,782 (99.0%) were successfully reported. Reportability did not vary by sample size (Fig 1B and Data Supplement), demonstrating that this PCR-CGP test is suitable for minute samples with ≥ 2 mm2 TSA when other input requirements are met. Notably, among 8,089 (26.0%) exception samples, 6,511 (80.5%) were still successfully reported (Figs 1A and 1B). Samples with TC < 20% comprised the largest (10.7%) and poorest performing exception category (68.2% successfully reported), as expected given that < 20% TC samples automatically fail QC because of the overall LOD, and thus all such samples without reported prioritized alterations are deemed test failures (not reported) as the presence of variants cannot be excluded. Samples not meeting other input requirements had decreased reportability (85.9%-90.1%) relative to QC-passing samples, but again, reportable results were still provided for most (Fig 1B). The impact of sample characteristics on individual variant class performance is described in the Data Supplement. Representative successfully tested samples across the TSA range are shown in Figure 1C, and results stratified by cancer type and potential biomarker–based actionability are shown in Figure 2. Clinicopathologic and biomarker findings from all samples are provided (Data Supplement).
FIG 2.
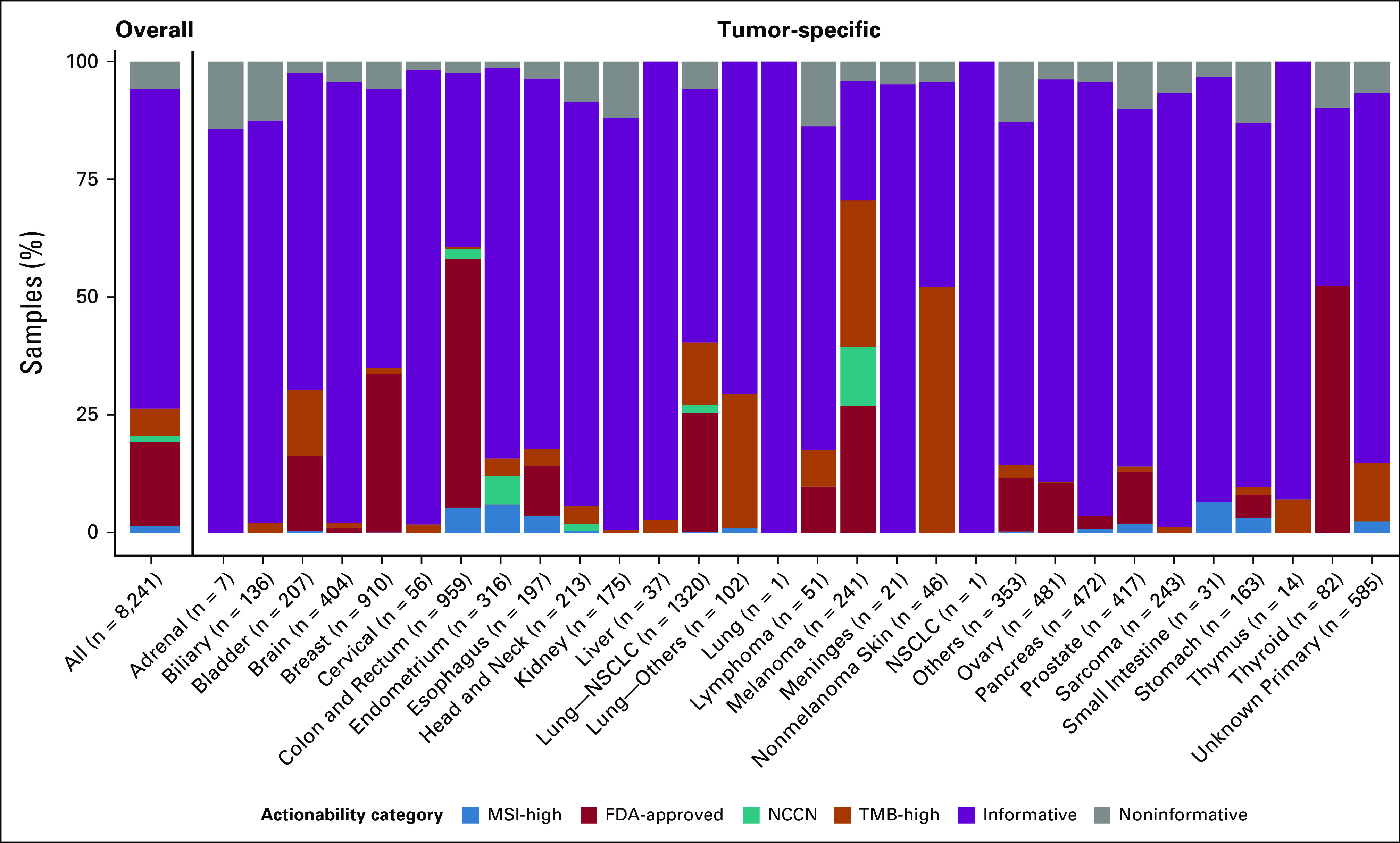
Pan-cancer assessment of potential actionability from PCR-CGP testing. All sample QC input-evaluable samples profiled between January 1 and June 25, 2020 (n = 8,241), were stratified by tumor type and assigned to one actionability class on the basis of MSI-H status, presence of an FDA-approved (within cancer type) biomarker, presence of an NCCN guideline–recommended (within cancer type) biomarker, and other TMB-H (≥ 10 mutations/megabase as TMB-H) using the associated therapy logic used in current StrataNGS reporting. Samples without one of these biomarkers were considered informative if at least one prioritized biomarker was reported or the sample passed all sequencing QC metrics with ≥ 20% TC. All other samples were considered test failures. CGP, comprehensive genomic profiling; FDA, US Food and Drug Administration; MSI-H, microsatellite instability–high; NCCN, National Comprehensive Cancer Network; NSCLC, non–small-cell lung cancer; PCR-CGP, multiplex polymerase chain reaction–based comprehensive genomic profiling; QC, quality control; TC, tumor content; TMB, tumor mutation burden; TSA, tumor surface area.
To address the positive predictive value (PPV) of prioritized biomarkers reported from exception and/or sequencing QC samples, we determined the PPV of PCR-CGP for ERBB2 amplification in all exception and/or sequencing QC-failure breast cancer samples with orthogonal clinical ERBB2 amplification status. As shown in the Data Supplement, the PPV in these 60 samples was 96.7%, similar to 98.5% PPV for ERBB2 amplifications determined in the PCR-CGP test clinical validation (only including sample and sequencing QC-passing samples; Tomlins et al, manuscript in review); genomic data from a true-positive 1.5-mm2, TC-exception sample are shown in Figure 3. Likewise, an example report from a successfully reported TC-exception NSCLC sample harboring expert-reviewed prioritized TP53 mutation and EML4-ALK fusion is shown in the Data Supplement. Additionally, as shown in the Data Supplement, biomarker frequencies were highly correlated (overall Pearson r = 0.990; per tumor type r = 0.897-0.999) to those from MSK-IMPACT, an independent, large, single-institution, advanced solid-tumor profiling experience using an FDA-cleared HC-CGP test.7 Lastly, no significant changes in the percentage of samples meeting sample QC metrics or reportability were observed across the study period, consistent with continued desire for CGP testing of challenging tissue samples (Data Supplement).
FIG 3.
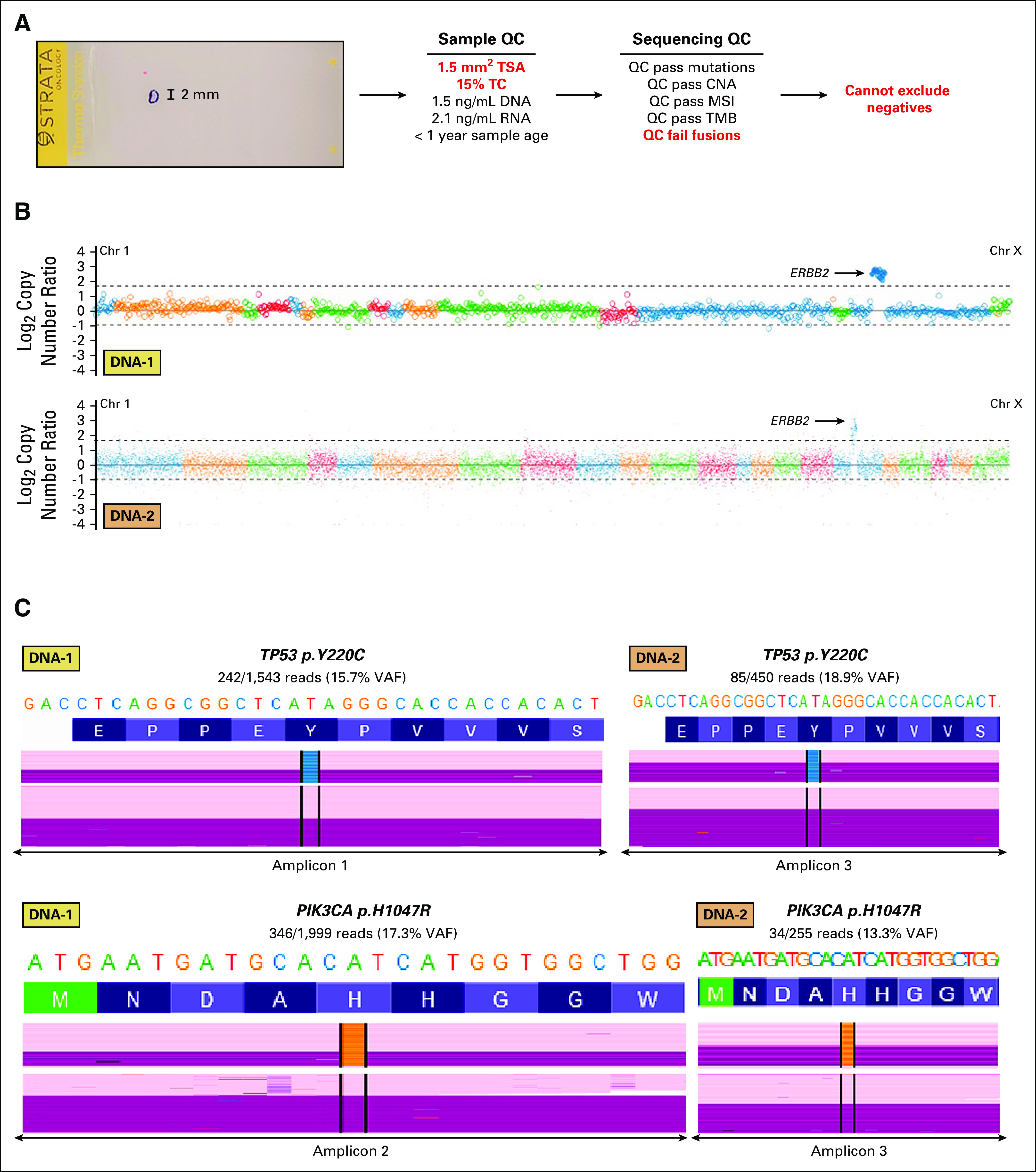
Underlying genomic data supporting a reported ERBB2 amplification in a breast cancer TC and tumor size exception sample. (A) Hematoxlyin and eosin slide from a 1.5-mm2 TSA lung biopsy from a patient with metastatic breast cancer submitted for PCR-CGP. The inked region indicates the region for microdissection, and a scale bar is shown. Sample QC and sequencing QC metrics are shown, with sample exception and failing QC metrics in red. As the overall molecular profile supports a TC of 15%, below the PCR-CGP's overall limit of detection of TC ≥ 20%, positive results can be reported by expert review; however, negative results cannot be asserted and hence the test is partially reported. (B) Genome-wide copy number profiles from DNA panel 1 (top) and DNA panel 2 (bottom) are shown. Individual amplicon-level log2 copy-number ratio (v a pseudomatched normal profile) is plotted for each targeted gene, with data colored by chromosome (chromosome 1 to X from left to right). TC correction has not been applied. Thresholds for calling amplifications and deep deletions are shown by gray dashed lines. The ERBB2 amplification is indicated. (C) Read-level support for reported prioritized TP53 and PIK3CA mutations is shown from DNA panel 1 (left) and DNA panel 2 (right). Reference nucleotides and amino acids are shown on top, with coverage and nonreference allele distributions below. Forward and reverse strand reads are shown in pink and purple, respectively (randomly downsampled reads are shown; nonvariant-containing reads are compressed). Nonreference bases are colored (black = deletion, light purple = insertion, A = green, C = blue, G = orange, and T = red). Variant and total reads are shown, along with the VAF. CGP, comprehensive genomic profiling; chr, chromosome; CNA, copy number alteration; MSI, microsatellite instability; PCR-CGP, multiplex polymerase chain reaction–based comprehensive genomic profiling; QC, quality control; TC, tumor content; TMB, tumor mutation burden; TSA, tumor surface area; VAF, variant allele frequency.
Prostate Carcinoma Experience
The FDA approval of pembrolizumab for all advanced microsatellite instability–high (MSI-H) solid tumors and the approval of rucaparib (for BRCA1/2) and olaparib (for BRCA1/2, ATM, and 11 other potential homologous recombination deficiency [HRD] genes) for metastatic castration-resistant prostate cancer led the National Comprehensive Cancer Network to recommend MSI-H and HRD gene testing for all men with metastatic castration-resistant prostate cancer given relatively high frequency of these alterations.17,25-30
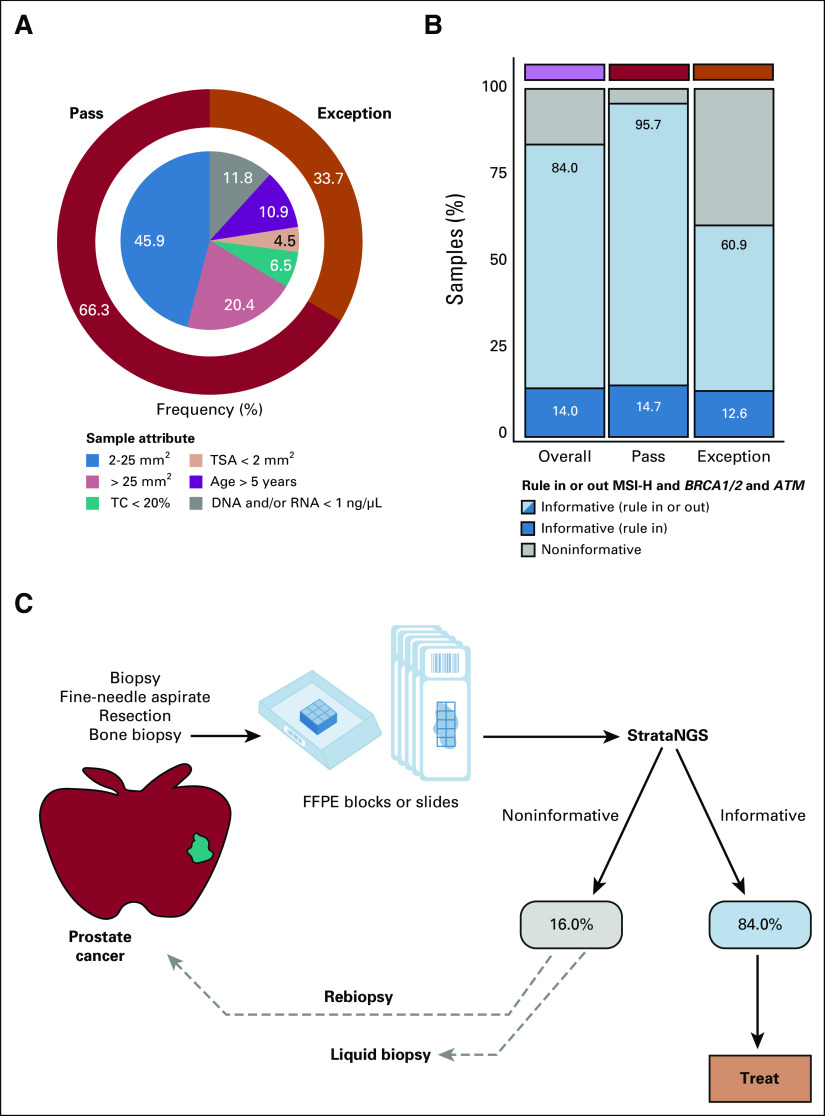
As shown in Figure 4A and the Data Supplement, among 1,344 prostate cancer samples, although the overall exception proportion was similar to the pan-tumor input-evaluable cohort (33.7% v 26.0%), prostate cancer had the highest frequency of age exception samples (10.9%) and overall frequency of samples age > 5 years (14.9%; Data Supplement), consistent with the frequent delay between diagnosis and recurrence after definitive therapy and/or androgen deprivation therapy. To assess therapy selection performance, we separated reports into those yielding informative therapy selection results (able to rule in or out biomarker-guided therapy) for MSI status and HRD gene (BRCA1, BRCA2, and ATM) alterations and those yielding noninformative results where additional testing (eg, either by liquid biopsy or obtaining and testing another sample) would be required. Overall, 84.0% of prostate cancer samples yielded informative results (including 60.9% of exception samples; Fig 4B and Data Supplement). Importantly, the positive MSI-H and HRD biomarker rate was similar between samples meeting input requirements versus exceptions (14.7% v 12.6%; Fig 2B and Data Supplement). Together, this suggests that approximately 84% of patients with advanced prostate cancer desiring CGP have sufficient tissue samples for informative PCR-CGP, minimizing the potential need to obtain and test a new sample or pursue liquid biopsy testing (Fig 4C).
FIG 4.
(A) Donut plot characterizing the composition of consecutively tested, sample input characteristic-evaluable prostate cancer samples (n = 1,344) from the overall PCR-CGP test cohort. The outer ring indicates the percentage of samples meeting (pass: dark red or not meeting (exception: orange) PCR-CGP input requirements. In the inner pie chart, samples passing all input requirements are stratified by TSA; exception samples are stratified by indicated sample attribute (TC < 20%; TSA < 2 mm2; age > 5 years: specimen collected > 5 years before PCR-CGP; and DNA and/or RNA concentration < 1 ng/µL). (B) The proportion of tested samples (overall and by sample input requirement category) for which an informative result (able to rule in or out actionable alterations) was reported. To be considered informative (total of light and dark blue), the test must have reported (1) either MSI-H or a deleterious mutation and/or copy number deep deletion in MSH2/6, BRCA1/2, or ATM (dark blue) or (2) tested definitively negative for these biomarkers by meeting all sequencing QC metrics and having TC ≥ 20%, the PCR-CGP test's overall limit of detection (light blue). The percent of total informative and informative rule in are indicated. (C) Potential real-world prostate cancer testing paradigm on the basis of sample characteristics and PCR-CGP performance characteristics observed in this cohort. Patients with noninformative test results in this cohort were not followed to determine whether rebiopsy or liquid biopsy testing was pursued. CGP, comprehensive genomic profiling; FFPE, formalin-fixed paraffin-embedded; MSI-H, microsatellite instability–high; PCR-CGP, multiplex polymerase chain reaction–based comprehensive genomic profiling; QC, quality control; TC, tumor content; TSA, tumor surface area.
NSCLC Adenocarcinoma Experience
CGP is especially relevant in NSCLC given the large number of recommended biomarkers required to guide therapy (Data Supplement). Among 1,144 NSCLC adenocarcinoma samples, the exception proportion was greater than the overall pan-tumor cohort (40.6% v 26.0%), with 21.6% having TC < 20% and 12.1% having TSA < 2 mm2 (Fig 5A and Data Supplement). Yet, 87.9% of NSCLC adenocarcinoma samples yielded informative (able to rule in or out biomarker-guided therapy) results (Fig 5B and Data Supplement), including 98.4% of samples meeting input requirements and 72.6% of exceptions. Importantly, overall informative biomarker frequencies in this NSCLC adenocarcinoma cohort were similar to those observed in NSCLC adenocarcinoma from the MSK-IMPACT cohort (Pearson correlation coefficient r2 = 0.96, P < .001; Figs 5C and Data Supplement).
FIG 5.
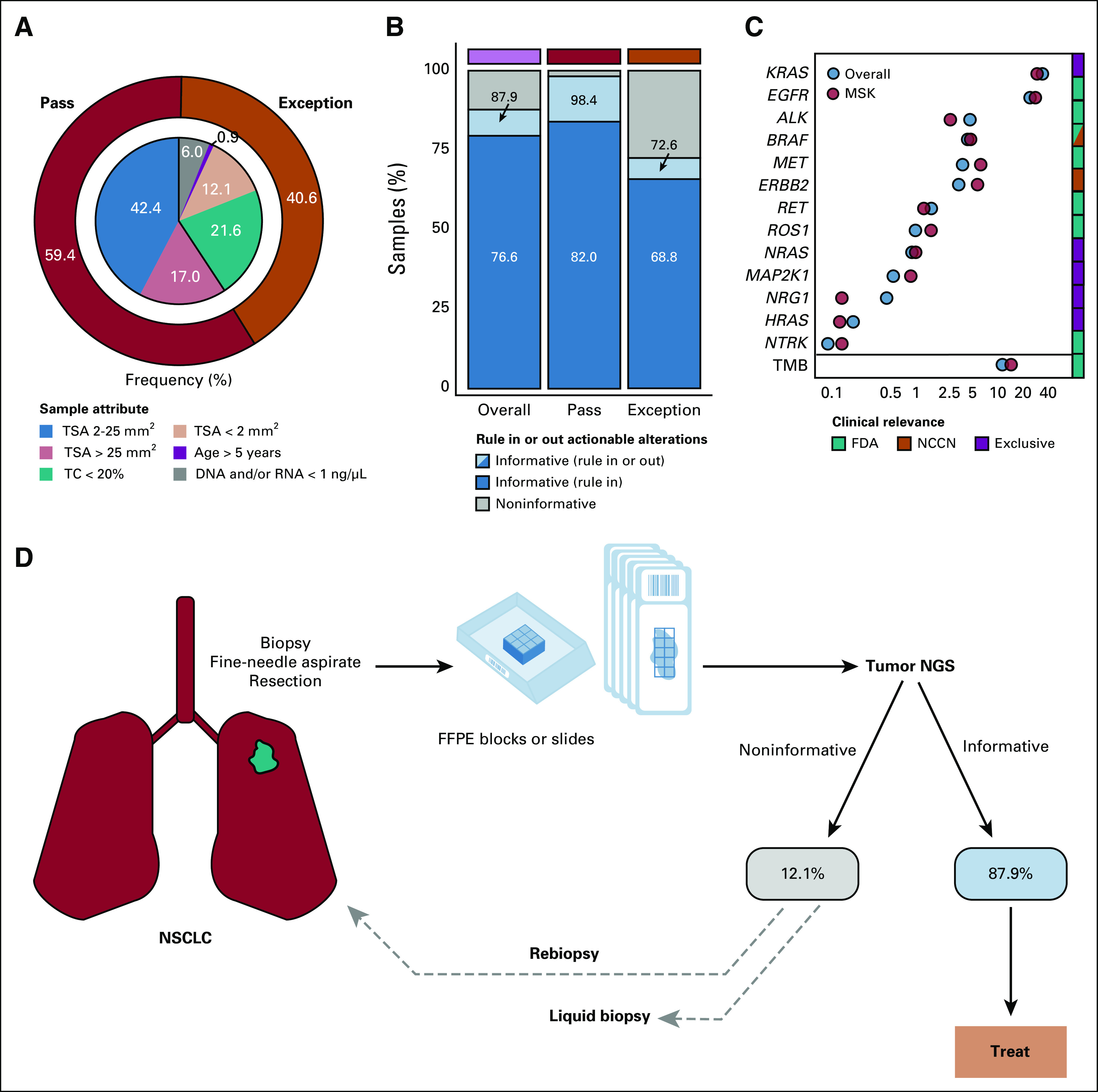
(A) Donut plot characterizing the composition of consecutively tested, sample input characteristic-evaluable NSCLC adenocarcinoma samples (n = 1,142) from the overall PCR-CGP test cohort. The outer ring indicates the percentage of samples meeting (pass: dark red) or not meeting (exception: orange) PCR-CGP input requirements. In the inner pie chart, samples passing all input requirements are stratified by TSA; exception samples are stratified by indicated sample attribute (TC < 20%; TSA < 2 mm2; age > 5 years: specimen collected > 5 years before PCR-CGP; and DNA and/or RNA concentration < 1 ng/µL). (B) The proportion of tested samples (overall and by sample input requirement category) for which an informative result (able to rule in or out actionable alterations) was reported. To be considered informative, the test must have either reported a therapy selection and/or mutually exclusive biomarker (as in C and the Data Supplement) or tested definitively negative for all such biomarkers by meeting all sequencing QC metrics and having TC ≥ 20% (the PCR-CGP test's overall limit of detection). (C) Reported actionable biomarker frequencies from this PCR-CGP NSCLC adenocarcinoma cohort (overall) are shown along with those from an external single-institution cohort (MSK-IMPACT; MSK: light red). The color bar at right indicates whether testing positive for each corresponding biomarker is associated with an FDA-approved (green) or NCCN-recommended (orange) targeted therapy or thought to be mutually exclusive (purple) with known LUAD therapy selection biomarkers. TMB frequencies are presented separately from gene-specific biomarkers given the expected overlap between TMB-high and some therapy selection or actionable biomarkers (for this analysis, samples with more than one biomarker were counted in each group). (D) Potential real-world NSCLC adenocarcinoma testing paradigm on the basis of sample characteristics and PCR-CGP performance characteristics observed in this cohort. Patients with noninformative test results in this cohort were not followed to determine whether rebiopsy or liquid biopsy testing was pursued. CGP, comprehensive genomic profiling; FDA, US Food and Drug Administration; FFPE, formalin-fixed paraffin-embedded; LUAD, lung adenocarcinoma; MSK, Memorial Sloan Kettering; NCCN, National Comprehensive Cancer Network; NGS, next-generation sequencing; NSCLC, non–small-cell lung cancer; PCR-CGP, multiplex polymerase chain reaction–based comprehensive genomic profiling; QC, quality control; TC, tumor content; TMB, tumor mutation burden; TSA, tumor surface area.
Like prostate cancer, TC < 20% NSCLC adenocarcinoma samples had the lowest informative rate (Data Supplement), as negative results cannot be definitively asserted in this sub-LOD setting, which can particularly affect detection of nonmutation biomarkers given the frequent difficulty of knowing the true TC in the absence of TC-defining mutations. However, in contrast to the low actionable biomarker frequency in prostate cancer, actionable or informative biomarkers are frequent in NSCLC adenocarcinoma. Hence, the positive informative biomarker detection rate in NSCLC adenocarcinoma TC-exception samples (58.7%) is only marginally less than that in samples meeting input requirements (82.0%), and all other sample exception groups had positive detection rates of 79.7%-81.2% (Data Supplement). These results suggest that approximately 88% of patients with advanced NSCLC adenocarcinoma desiring CGP have sufficient tissue samples for informative PCR-CGP, minimizing the potential need for rebiopsy or liquid biopsy (Fig 5C).
DISCUSSION
Herein, we present the tissue characteristics and PCR-CGP test performance from more than 30,000 consecutively tested solid-tumor samples from patients with advanced cancer submitted from 28 diverse US health systems through a multi-institutional observational clinical trial. Importantly, testing during the study period was not restricted by tumor type and was provided at no cost to patients. Sites were provided with minimal sample submission requirements and PCR-CGP testing was attempted for essentially all samples with identifiable tumor, providing a unique view on real-world tumor tissue availability and CGP test performance.
Unexpectedly, we found that most submitted samples were limited, with 10.7% having < 20% TC and 44.7% with TSA ≤ 10 mm2. Despite these challenges, PCR-CGP reported results for 94.2% of all tested tumor samples, including 80.5% of exception samples not meeting input criteria. Specifically, among NSCLC adenocarcinoma, we found that limited tissue was even more pronounced with 21.6% of samples having < 20% TC and an additional 12.1% having < 2 mm2 TSA; however, PCR-CGP testing successfully reported treatment selection informative results in 87.9% of samples. Similar results were observed in prostate carcinoma, where despite a substantially lower positive informative biomarker rate, PCR-CGP produced treatment selection informative results in 84.0% of samples.
We attribute PCR-CGP's high reportability rates to two main factors. First, multiplex PCR-based CGP library preparation method enabled minimal input (eg, TSA 2 mm2) versus leading commercially available HC-CGP tests requiring ≥ 25 mm.2,3,5,23,24 Notably, only 40.8% of samples in our total received cohort met this requirement and the proportion was even smaller in input-evaluable NSCLC samples (23.6%, lung—NSCLC; Data Supplement). Consistent with these findings, in clinical trials testing available FFPE tissue samples from patients with advanced NSCLC or CRPC, HC-CGP failure rates of approximately 30%-40% have been reported.16-20 Thus, the PCR-CGP test, with its lower input requirements, has the potential to expand the proportion of testable tumor samples.
Second, as PCR-CGP can generate some data on nearly all samples and because of our belief that even a single biomarker may be highly actionable (regardless of the ability to assess all CGP variant classes), we used a liberal exception testing policy, where we attempted testing on nearly all samples with identifiable tumor, even if not meeting all input requirements. To maximize actionable insights from the available tissue, this necessitated PCR-CGP bioinformatic pipeline development and QC metrics optimized for minute, low-quality samples, and reporting included expert-level variant review. As expected, low-TC samples were the most challenging (68.2% reportability), given the inability to exclude the presence of alterations in such samples on the basis of test LOD (data and an example report from two such samples are shown in Fig 3 and the Data Supplement2). Likewise, samples failing more than one sample QC metric were less frequently reportable (Data Supplement), which may guide decisions on rebiopsy or liquid biopsy testing. However, the high PPV (96.7%) for ERBB2 amplification in exception and/or sequencing QC-failure breast cancer samples supports the potential clinical utility of our approach.
Liquid biopsy represents an alternative CGP methodology when no tissue is available or procurement is difficult. Although highly specific for treatment selection,16 as evidenced by the recent FDA approval of both the FoundationOne Liquid CDx and Guardant360 CDx cfDNA CGP tests, cfDNA sensitivity is challenged by the lack of circulating tumor DNA in some patients and differentiating an informative negative test from a lack of detectable cfDNA (eg, does an NSCLC cfDNA test identifying a TP53 mutation at 0.5% variant allele frequency [VAF] exclude the possibility of an EGFR exon 19 deletion?) given the prevalence of clonal hematopoiesis (CHIP) and the poor PPV of de novo alterations at VAFs < 1%.31-36 For example, in NSCLC, the Guardant360 CDx test showed only 67.4%-77.7% positive predictive agreement versus tissue-based EGFR testing (exon 19 deletions/p.L858R/p.T790M).33 In prostate cancer, liquid biopsy may be particularly appealing in patients with very old diagnostic tissue or bone-only disease.37 Encouragingly, sensitivity for actionable BRCA1/2 mutations was 93% in a comparison of cfDNA FoundationACT/One Liquid versus FoundationOne tissue testing in CRPC rucaparib screening studies; however, cfDNA-exclusive mutations had low VAF relative to cfDNA-based TC,38 suggesting that they may be subclonal and thus of unclear therapeutic relevance. Likewise, BRCA2 deep deletion detection is critical but challenging as detection (via tissue or cfDNA) requires 30%-40% TC; however, < 25% of patients in the cfDNA-based rucaparib studies had ≥ 35% cfDNA TC38 versus 79.5% of the 2,045 prostate cancer samples having ≥ 35% tissue TC herein. Last, CHIP is particularly relevant in prostate cancer as a recent cfDNA-based laboratory-developed test found 10% of men harbored CHIP variants in olaparib-associated HRD genes, most frequently ATM.39 These results complicate interpreting efficacy of poly (ADP-ribose) polymerase inhibitor trials enrolling men with cfDNA-based ATM variants40 and olaparib treatment selection. Hence, these issues highlight the difficult decisions clinicians face when deciding between tissue versus liquid testing.36,37
A limitation of our study is the lack of head-to-head testing with HC-based tissue tests and/or liquid biopsy testing, necessitating carefully designed future studies to directly compare real-world performance. Likewise, patients with noninformative PCR-CGP testing in this cohort were not followed to determine whether rebiopsy or liquid biopsy was pursued and impact on clinical management. Additionally, the PCR-CGP test used herein does not report a global HRD assessment.41 Last, PCR-CGP testing treatment response has not been determined, and thus, our approach's clinical impact is unclear.
The growing compendium of biomarker-guided targeted therapies and immunotherapies makes clear the importance of CGP for treatment selection in patients with advanced cancer. Our study demonstrates that although most patients desiring CGP in a real-world cohort have challenging tissue specimens, optimized approaches including PCR-CGP and broadly testing sample exceptions can maximize actionable information.
Scott A. Tomlins
Employment: Strata Oncology
Leadership: Strata Oncology
Stock and Other Ownership Interests: Strata Oncology, Javelin Oncology
Consulting or Advisory Role: Janssen, Astellas Medivation, Strata Oncology
Research Funding: Astellas Medivation
Patents, Royalties, Other Intellectual Property: I am a coinventor on a patent issued to the University of Michigan on ETS gene fusions in prostate and am included in the royalty stream. The diagnostic field of use was licensed to Hologic/Gen-Probe (who sublicensed some rights to Ventana/Roche) and is licensed to LynxDx. I am a coinventor on a patent issued to Strata Oncology related to MSI determination and checkpoint inhibitor benefit
Travel, Accommodations, Expenses: Strata Oncology, Genzyme
Daniel H. Hovelson
Employment: Strata Oncology
Elizabeth C. Dees
Consulting or Advisory Role: Novartis, Strata Oncology, G1 Therapeutics
Research Funding: Novartis, Genentech/Roche, Pfizer, Merck, H3 Biomedicine, Meryx Pharmaceuticals
Travel, Accommodations, Expenses: G1 Therapeutics
Mark E. Burkard
Consulting or Advisory Role: Pointcare genomics, Strata Oncology, Novartis
Research Funding: AbbVie, Strata Oncology, Puma Biotechnology, Loxo, Merck, Arcus Ventures, Apollomics, Elevation Oncology, Genentech
Patents, Royalties, Other Intellectual Property: I have a patent for implantable/localized drug delivery device that can sample the tumor microenvironment and deliver drug, I have a patent for a method to detect recombination events with CRISPR-mediated editing, and I have a patent for conducting expansion microscopy without specialized equipment
Michael Guarino
Stock and Other Ownership Interests: Johnson and Johnson, Johnson and Johnson
Travel, Accommodations, Expenses: McKesson, ARMO BioSciences, AstraZeneca, BMS, De Novo Pharmaceuticals
Marc R. Matrana
Consulting or Advisory Role: Strata Oncology
Speakers' Bureau: Bristol Myers Squibb, AstraZeneca, Merck, Eisai, Genentech, Janssen, Exelixis
Eddy S. Yang
Consulting or Advisory Role: Strata Oncology, AstraZeneca, Bayer, Clovis Oncology, Lilly
Research Funding: Lilly, Novartis, Clovis Oncology, Puma Biotechnology
Benjamin M. Parsons
Consulting or Advisory Role: Celgene, Amgen, AstraZeneca
Speakers' Bureau: Amgen, Celgene, AstraZeneca
Open Payments Link: https://openpaymentsdata.cms.gov/physician/795031
Michael A. Thompson
Stock and Other Ownership Interests: Doximity
Consulting or Advisory Role: Celgene, VIA Oncology, Takeda, GlaxoSmithKline, Strata Oncology, Syapse, Adaptive Biotechnologies, AbbVie, GRAIL, Epizyme, Janssen Oncology
Research Funding: Takeda, Bristol Myers Squibb, TG Therapeutics, Cancer Research and Biostatistics, AbbVie, PrECOG, Strata Oncology, Lynx Biosciences, Denovo Biopharma, ARMO BioSciences, GlaxoSmithKline, Amgen
Patents, Royalties, Other Intellectual Property: UpToDate, Peer Review for Plasma Cell Dyscrasias (Editor: Robert Kyle)
Travel, Accommodations, Expenses: Takeda, GlaxoSmithKline, Syapse
Other Relationship: Doximity
Open Payments Link: https://openpaymentsdata.cms.gov/physician/192826/summary
William J. Edenfield
Consulting or Advisory Role: Chimerix
Suresh Nair
Stock and Other Ownership Interests: Moderna Therapeutics, Novavax, Biontech, Gilead Sciences
Research Funding: Bristol Myers Squibb, Merck, Nektar
Adedayo Onitilo
Consulting or Advisory Role: Kite, a Gilead company, Envision Communications
Speakers' Bureau: GlaxoSmithKline, Puma Biotechnology, Kite/Gilead, AbbVie
Robert Siegel
Research Funding: Merck, Mirati Therapeutics, GRAIL, Altor BioScience, Galera Therapeutics, Apollomics, Strata Oncology, Arcus Biosciences, Bristol Myers Squibb, Cancer Insight, Puma Biotechnology, Conjupro Biotherapeutics, Razor Genomics, Sanofi, Seattle Genetics
Other Relationship: American Board of Internal Medicine (ABIM)
William J. Irvin
Research Funding: Merck, Altor BioScience, Odonate Therapeutics, Boston Biomedical, Novartis, Pfizer, Seattle Genetics, Altor BioScience, AstraZeneca
Arvinda Padmanabhan
Speakers' Bureau: Clovis Oncology, Roche
Anneliese Gonzalez
Research Funding: Novartis, Radius Health, Astellas Pharma
Paul Harms
Research Funding: Q32 Bio
Jennifer Hipp
Employment: Strata Oncology
Stock and Other Ownership Interests: Strata Oncology
Consulting or Advisory Role: PathAI
Kat Kwiatkowski
Employment: Strata Oncology
Stock and Other Ownership Interests: Strata Oncology, Epizyme, Loxo, Editas Medicine, Intuitive Surgical
Khalis Mitchell
Employment: Strata Oncology
Stock and Other Ownership Interests: Mirati Therapeutics
Javed Siddiqui
Consulting or Advisory Role: Strata Oncology, LynxDx
Hana Vakil
Employment: Strata Oncology
Stock and Other Ownership Interests: Strata Oncology
D. Bryan Johnson
Employment: Strata Oncology
Stock and Other Ownership Interests: Strata Oncology
Daniel R. Rhodes
Employment: Strata Oncology, Javelin Oncology
Leadership: Strata Oncology, Javelin Oncology
Stock and Other Ownership Interests: Strata Oncology, Javelin Oncology
Patents, Royalties, Other Intellectual Property: I am paid royalties from the University of Michigan on license revenues related to a patent on prostate cancer gene fusions
No other potential conflicts of interest were reported.
PREPRINT VERSIONPreprint version available on https://www.medrxiv.org/content/10.1101/2020.11.19.20233866v1.
DISCLAIMER
Authors who are employed by the study sponsor Strata Oncology were involved in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation of the manuscript; and the decision to submit the manuscript for publication.
PRIOR PRESENTATION
Presented at the American Society of Clinical Oncology 2020 Annual Meeting, Chicago, IL.
SUPPORT
The Strata Trial and analyses presented in this article were sponsored by Strata Oncology (Ann Arbor, MI).
DATA SHARING STATEMENT
De-identified participant and molecular data (including raw data) are made available to all Strata Trial partnered institutions. De-identified participant and biomarker data are provided in the Data Supplement.
AUTHOR CONTRIBUTIONS
Conception and design: Scott A. Tomlins, Kat Kwiatkowski, D. Bryan Johnson, Daniel R. Rhodes
Administrative support: Khalis Mitchell
Provision of study materials or patients: Jennifer M. Suga, Elizabeth C. Dees, Mark E. Burkard, Michael Guarino, Jamil Khatri, Malek M. Safa, Eddy S. Yang, Benjamin M. Parsons, Jennifer N. Slim, Michael A. Thompson, William J. Edenfield, Adedayo Onitilo, Robert Siegel, Alan Miller, William J. Irvin, Abdul Hai Mansoor, Andrew Kellum, Paul Harms
Collection and assembly of data: Scott A. Tomlins, Jennifer M. Suga, Daniel M. Anderson, Han A. Koh, Elizabeth C. Dees, Brendan McNulty, Mark E. Burkard, Michael Guarino, Jamil Khatri, Malek M. Safa, Marc R. Matrana, Eddy S. Yang, Alex R. Menter, Jennifer N. Slim, Michael A. Thompson, Leon Hwang, William J. Edenfield, Suresh Nair, Adedayo Onitilo, Alan Miller, Timothy Wassenaar, William Schulz, Arvinda Padmanabhan, Anneliese Gonzalez, Andrew Kellum, Paul Harms, Jayson Falkner, Andrew Fischer, Jennifer Hipp, Kat Kwiatkowski, Lorena Lazo de la Vega
Data analysis and interpretation: Scott A. Tomlins, Daniel H. Hovelson, Elizabeth C. Dees, Eddy S. Yang, Benjamin M. Parsons, Michael A. Thompson, William J. Edenfield, Robert Siegel, William J. Irvin, Vallathucherry Harish, Stephanie Drewery, Travis Reeder, Javed Siddiqui, Hana Vakil, D. Bryan Johnson
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by the authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Scott A. Tomlins
Employment: Strata Oncology
Leadership: Strata Oncology
Stock and Other Ownership Interests: Strata Oncology, Javelin Oncology
Consulting or Advisory Role: Janssen, Astellas Medivation, Strata Oncology
Research Funding: Astellas Medivation
Patents, Royalties, Other Intellectual Property: I am a coinventor on a patent issued to the University of Michigan on ETS gene fusions in prostate and am included in the royalty stream. The diagnostic field of use was licensed to Hologic/Gen-Probe (who sublicensed some rights to Ventana/Roche) and is licensed to LynxDx. I am a coinventor on a patent issued to Strata Oncology related to MSI determination and checkpoint inhibitor benefit
Travel, Accommodations, Expenses: Strata Oncology, Genzyme
Daniel H. Hovelson
Employment: Strata Oncology
Elizabeth C. Dees
Consulting or Advisory Role: Novartis, Strata Oncology, G1 Therapeutics
Research Funding: Novartis, Genentech/Roche, Pfizer, Merck, H3 Biomedicine, Meryx Pharmaceuticals
Travel, Accommodations, Expenses: G1 Therapeutics
Mark E. Burkard
Consulting or Advisory Role: Pointcare genomics, Strata Oncology, Novartis
Research Funding: AbbVie, Strata Oncology, Puma Biotechnology, Loxo, Merck, Arcus Ventures, Apollomics, Elevation Oncology, Genentech
Patents, Royalties, Other Intellectual Property: I have a patent for implantable/localized drug delivery device that can sample the tumor microenvironment and deliver drug, I have a patent for a method to detect recombination events with CRISPR-mediated editing, and I have a patent for conducting expansion microscopy without specialized equipment
Michael Guarino
Stock and Other Ownership Interests: Johnson and Johnson, Johnson and Johnson
Travel, Accommodations, Expenses: McKesson, ARMO BioSciences, AstraZeneca, BMS, De Novo Pharmaceuticals
Marc R. Matrana
Consulting or Advisory Role: Strata Oncology
Speakers' Bureau: Bristol Myers Squibb, AstraZeneca, Merck, Eisai, Genentech, Janssen, Exelixis
Eddy S. Yang
Consulting or Advisory Role: Strata Oncology, AstraZeneca, Bayer, Clovis Oncology, Lilly
Research Funding: Lilly, Novartis, Clovis Oncology, Puma Biotechnology
Benjamin M. Parsons
Consulting or Advisory Role: Celgene, Amgen, AstraZeneca
Speakers' Bureau: Amgen, Celgene, AstraZeneca
Open Payments Link: https://openpaymentsdata.cms.gov/physician/795031
Michael A. Thompson
Stock and Other Ownership Interests: Doximity
Consulting or Advisory Role: Celgene, VIA Oncology, Takeda, GlaxoSmithKline, Strata Oncology, Syapse, Adaptive Biotechnologies, AbbVie, GRAIL, Epizyme, Janssen Oncology
Research Funding: Takeda, Bristol Myers Squibb, TG Therapeutics, Cancer Research and Biostatistics, AbbVie, PrECOG, Strata Oncology, Lynx Biosciences, Denovo Biopharma, ARMO BioSciences, GlaxoSmithKline, Amgen
Patents, Royalties, Other Intellectual Property: UpToDate, Peer Review for Plasma Cell Dyscrasias (Editor: Robert Kyle)
Travel, Accommodations, Expenses: Takeda, GlaxoSmithKline, Syapse
Other Relationship: Doximity
Open Payments Link: https://openpaymentsdata.cms.gov/physician/192826/summary
William J. Edenfield
Consulting or Advisory Role: Chimerix
Suresh Nair
Stock and Other Ownership Interests: Moderna Therapeutics, Novavax, Biontech, Gilead Sciences
Research Funding: Bristol Myers Squibb, Merck, Nektar
Adedayo Onitilo
Consulting or Advisory Role: Kite, a Gilead company, Envision Communications
Speakers' Bureau: GlaxoSmithKline, Puma Biotechnology, Kite/Gilead, AbbVie
Robert Siegel
Research Funding: Merck, Mirati Therapeutics, GRAIL, Altor BioScience, Galera Therapeutics, Apollomics, Strata Oncology, Arcus Biosciences, Bristol Myers Squibb, Cancer Insight, Puma Biotechnology, Conjupro Biotherapeutics, Razor Genomics, Sanofi, Seattle Genetics
Other Relationship: American Board of Internal Medicine (ABIM)
William J. Irvin
Research Funding: Merck, Altor BioScience, Odonate Therapeutics, Boston Biomedical, Novartis, Pfizer, Seattle Genetics, Altor BioScience, AstraZeneca
Arvinda Padmanabhan
Speakers' Bureau: Clovis Oncology, Roche
Anneliese Gonzalez
Research Funding: Novartis, Radius Health, Astellas Pharma
Paul Harms
Research Funding: Q32 Bio
Jennifer Hipp
Employment: Strata Oncology
Stock and Other Ownership Interests: Strata Oncology
Consulting or Advisory Role: PathAI
Kat Kwiatkowski
Employment: Strata Oncology
Stock and Other Ownership Interests: Strata Oncology, Epizyme, Loxo, Editas Medicine, Intuitive Surgical
Khalis Mitchell
Employment: Strata Oncology
Stock and Other Ownership Interests: Mirati Therapeutics
Javed Siddiqui
Consulting or Advisory Role: Strata Oncology, LynxDx
Hana Vakil
Employment: Strata Oncology
Stock and Other Ownership Interests: Strata Oncology
D. Bryan Johnson
Employment: Strata Oncology
Stock and Other Ownership Interests: Strata Oncology
Daniel R. Rhodes
Employment: Strata Oncology, Javelin Oncology
Leadership: Strata Oncology, Javelin Oncology
Stock and Other Ownership Interests: Strata Oncology, Javelin Oncology
Patents, Royalties, Other Intellectual Property: I am paid royalties from the University of Michigan on license revenues related to a patent on prostate cancer gene fusions
No other potential conflicts of interest were reported.
REFERENCES
- 1.Cheng ML, Berger MF, Hyman DM, et al. Clinical tumour sequencing for precision oncology: Time for a universal strategy Nat Rev Cancer 18:527–5282018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaubier N, Bontrager M, Huether R, et al. Integrated genomic profiling expands clinical options for patients with cancer Nat Biotechnol 37:1351–13602019 [DOI] [PubMed] [Google Scholar]
- 3.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing Nat Biotechnol 31:1023–10312013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson DR, Wu YM, Lonigro RJ, et al. Integrative clinical genomics of metastatic cancer Nature 548:297–3032017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vanderwalde A, Spetzler D, Xiao N, et al. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients Cancer Med 7:746–7562018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood DE, White JR, Georgiadis A, et al. A machine learning approach for somatic mutation discovery Sci Transl Med 10:eaar7939.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients Nat Med 23:703–7132017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dy GK, Nesline MK, Papanicolau-Sengos A, et al. Treatment recommendations to cancer patients in the context of FDA guidance for next generation sequencing BMC Med Inform Decis Mak 19:14.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris SM, Subramanian J, Gel ES, et al. Performance of next-generation sequencing on small tumor specimens and/or low tumor content samples using a commercially available platform PLoS One 13:e0196556.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sholl LM, Do K, Shivdasani P, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population JCI Insight 1:e87062.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodhouse R, Li M, Hughes J, et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin PLoS One 15:e0237802.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab Nat Med 24:1441–14482018 [DOI] [PubMed] [Google Scholar]
- 13.Odegaard JI, Vincent JJ, Mortimer S, et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies Clin Cancer Res 24:3539–35492018 [DOI] [PubMed] [Google Scholar]
- 14.Georgiadis A, Durham JN, Keefer LA, et al. Noninvasive detection of microsatellite instability and high tumor mutation burden in cancer patients treated with PD-1 blockade Clin Cancer Res 25:7024–70342019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown NA, Elenitoba-Johnson KSJ.Enabling precision oncology through precision diagnostics Annu Rev Pathol 15:97–1212020 [DOI] [PubMed] [Google Scholar]
- 16.Aggarwal C, Thompson JC, Black TA, et al. Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer JAMA Oncol 5:173–1802019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Bono J, Mateo J, Fizazi K, et al. Olaparib for metastatic castration-resistant prostate cancer N Engl J Med 382:2091–21022020 [DOI] [PubMed] [Google Scholar]
- 18.Gilson C, Ingleby F, Gilbert DC, et al. Targeted next-generation sequencing (tNGS) of metastatic castrate-sensitive prostate cancer (M1 CSPC): A pilot molecular analysis in the STAMPEDE multi-center clinical trial. J Clin Oncol. 2019;37 suppl; abstr 5019. [Google Scholar]
- 19.Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden N Engl J Med 378:2093–21042018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Si H, Kuziora M, Quinn KJ, et al. A blood-based assay for assessment of tumor mutational burden in first-line metastatic NSCLC treatment: Results from the MYSTIC study Clin Cancer Res 27:1631–16402021 [DOI] [PubMed] [Google Scholar]
- 21.File DM, Morgan KP, Khagi S.Durable near-complete response to olaparib plus temozolomide and radiation in a patient with ATM-mutated glioblastoma and MSH6-deficient lynch syndrome JCO Precis Oncol 4:841–8472020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller TI, Zoumberos NA, Johnson B, et al. A genomic survey of sarcomas on sun-exposed skin reveals distinctive candidate drivers and potentially targetable mutations Hum Pathol 102:60–692020 [DOI] [PubMed] [Google Scholar]
- 23.Foundation Medicine . FoundationOne CDx Specimen Instructions. 2020. https://assets.ctfassets.net/w98cd481qyp0/6qYLg8jUuEYEvUytoBz8p6/d9dec1344d2af0b697ee819e04a73bbc/F1CDx_Specimen_Instructions_062020.pdf [Google Scholar]
- 24.Caris Molecular Intelligence . Molecular Intelligence Specimen Preparation Instructions. 2020. https://www.carismolecularintelligence.com/wp-content/uploads/2017/12/TN0252-v9_Specimen_Prep_Instructions.pdf [Google Scholar]
- 25.Abida W, Patnaik A, Campbell D, et al. Rucaparib in men with metastatic castration-resistant prostate cancer harboring a BRCA1 or BRCA2 gene alteration J Clin Oncol 38:3763–37722020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade Science 357:409–4132017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Comprehensive Cancer Network . Prostate Cancer (Version 2.2020) 2020. https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1459 [Google Scholar]
- 28.Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer Nature 487:239–2432012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A, White TA, MacKenzie AP, et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers Proc Natl Acad Sci USA 108:17087–170922011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer Cell 162:454.2015 [DOI] [PubMed] [Google Scholar]
- 31.Stetson D, Ahmed A, Xu X, et al. Orthogonal comparison of four plasma NGS tests with tumor suggests technical factors are a major source of assay discordance JCO Precis Oncol 3:1–92019 [DOI] [PubMed] [Google Scholar]
- 32.Razavi P, Li BT, Brown DN, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants Nat Med 25:1928–19372019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.US Food and Drug Administration . PMA P200010: FDA Summary of Safety and Effectiveness Data. Silver Spring, MD, US Food: Drug Administration; 2020. [Google Scholar]
- 34.US Food and Drug Administration . PMA P190032: FDA Summary of Safety and Effectiveness Data. Silver Spring, MD, US Food: Drug Administration; 2020. [Google Scholar]
- 35.Deveson IW, Gong B, Lai K, et al. Evaluating the analytical validity of circulating tumor DNA sequencing assays for precision oncology Nat Biotechnol 10.1038/s41587-021-00857-zepub ahead of print on 2021 April 12, 2021] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aggarwal C, Rolfo CD, Oxnard GR, et al. Strategies for the successful implementation of plasma-based NSCLC genotyping in clinical practice Nat Rev Clin Oncol 18:56–622021 [DOI] [PubMed] [Google Scholar]
- 37.Ng SWS, Wyatt AW.Building confidence in circulating tumour DNA assays for metastatic castration-resistant prostate cancer Nat Rev Urol 18:255–2562021 [DOI] [PubMed] [Google Scholar]
- 38.Tukachinsky H, Madison RW, Chung JH, et al. Genomic analysis of circulating tumor DNA in 3,334 patients with advanced prostate cancer identifies targetable BRCA alterations and AR resistance mechanisms Clin Cancer Res 27:3094–31052021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jensen K, Konnick EQ, Schweizer MT, et al. Association of clonal hematopoiesis in DNA repair genes with prostate cancer plasma cell-free DNA testing interference JAMA Oncol 7:107–1102021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abida W, Campbell D, Patnaik A, et al. Non-BRCA DNA damage repair gene alterations and response to the PARP inhibitor rucaparib in metastatic castration-resistant prostate cancer: Analysis from the phase II TRITON2 study Clin Cancer Res 26:2487–24962020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stover EH, Fuh K, Konstantinopoulos PA, et al. Clinical assays for assessment of homologous recombination DNA repair deficiency Gynecol Oncol 159:887–8982020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
De-identified participant and molecular data (including raw data) are made available to all Strata Trial partnered institutions. De-identified participant and biomarker data are provided in the Data Supplement.