Abstract
PURPOSE
Combined hepatocellular cholangiocarcinoma (cHCC-CCA) is a rare, aggressive primary liver carcinoma, with morphologic features of both hepatocellular carcinomas (HCC) and liver cholangiocarcinomas (CCA).
METHODS
The genomic profiles of 4,975 CCA, 1,470 HCC, and 73 cHCC-CCA cases arising from comprehensive genomic profiling in the course of clinical care were reviewed for genomic alterations (GA), tumor mutational burden, microsatellite instability status, genomic loss of heterozygosity, chromosomal aneuploidy, genomic ancestry, and hepatitis B virus status.
RESULTS
In cHCC-CCA, GA were most common in TP53 (65.8%), TERT (49.3%), and PTEN (9.6%), and 24.6% cHCC-CCA harbored potentially targetable GA. Other GA were predominantly associated with either HCC or CCA, including, but not limited to, TERT, FGFR2, IDH1, and presence of hepatitis B virus. On the basis of these features, a machine learning (ML) model was trained to classify a cHCC-CCA case as CCA-like or HCC-like. Of cHCC-CCA cases, 16% (12/73) were ML-classified as CCA-like and 58% (42/73) cHCC-CCA were ML-classified as HCC-like. The ML model classified more than 70% of cHCC-CCA as CCA-like or HCC-like on the basis of genomic profiles, without additional clinico-pathologic input.
CONCLUSION
These findings demonstrate the use of ML for classification as based on a targeted exome panel used during routine clinical care. Classification of cHCC-CCA by genomic features alone creates insights into the biology of the disease and warrants further investigation for relevance to clinical care.
INTRODUCTION
Combined hepatocellular carcinoma cholangiocarcinoma1 (cHCC-CCA) is a rare primary liver malignancy with morphologic features of both hepatocellular carcinomas (HCC) and liver cholangiocarcinomas (CCA). Estimates of the incidence of cHCC-CCA vary broadly from 0.4% to 5%,2-4 with reference to being a fraction of the incidence of primary liver tumors. For disseminated or recurrent cHCC-CCA, no consensus guidelines exist because of the rarity of this disease, which is prohibitive for conducting disease-specific clinical trials. By contrast, there are both numerous trials and consensus guidelines for the management of patients with HCC and CCA, and more recently the former often includes additional genomic inclusion criteria, for example, ongoing trials of FGFR2 inhibitors in FGFR2 fusion-positive CCA.5 Histologically, cHCC-CCA can be further subdivided into separate, combined, and mixed subtypes on the basis of morphology; however, these classifications have no impact on clinical care.6
CONTEXT
Key Objective
Combined hepatocellular cholangiocarcinoma (cHCC-CCA) is a rare cancer with morphologic features of both liver cholangiocarcinoma (CCA) and hepatocellular carcinoma (HCC). The rarity of cHCC-CCA poses challenges for evidence-guided management, and the origin of this disease remains enigmatic.
Knowledge Generated
This study analyzes 73 patients with cHCC-CCA who obtained comprehensive genomic profiling as part of routine clinical care and as compared to the comprehensive genomic profiling of 4,975 patients with CCA and patients with 1,470 HCC. We classify cHCC-CCA as HCC-like or CCA-like as based solely on the genomic profiles resulting from a targeted exome next generation sequencing assay used in the course of clinical care. Furthermore, we developed an artificial intelligence–driven machine learning classifier that leverages the well-known mutually exclusive genomics of CCA and HCC and demonstrate that genomic features alone can classify more than 70% of our cHCC-CCA cohort.
Relevance
This study builds on the seminal work of Xue et al (Cancer Cell 35:932-947.e8, 2019) who similarly classified cHCC-CCA using transcriptional profiling and whole-genome or whole-exome sequencing, all methodologies that are less commonly used in clinical care. Given the complexity and uncertain oncogenesis of this mixed tumor, this classifier can be used to further investigate the biology of cHCC-CCA and assessed for possible future clinical applications.
Numerous studies have defined the characteristic genomic landscapes of HCC and CCA.7-10 For cHCC-CCA, genomic studies are limited to small cohorts and have typically used whole-genome sequencing, whole-exome sequencing, and transcriptome sequencing, all retrospectively.11-13 The recent seminal study of Xue et al13 demonstrates the retrospective use of whole-genome sequencing and RNA sequencing on laser-capture microdissected samples, with additional specialized specimen handling for each cHCC-CCA subtype and additional immunohistochemistry studies. Importantly, this study suggests that the combined subtype of cHCC-CCA resembles CCA and the mixed subtype resembles HCC.
In this work, we used comprehensive genomic profiling (CGP) of formalin-fixed paraffin-embedded specimens of a large series of HCC, CCA, and cHCC-CCA cases, all sequenced as part of routine clinical care. We found genomic alterations (GA) enriched in HCC and CCA. On that basis, we built a machine learning (ML) model to classify cHCC-CCA as HCC-like or CCA-like by integrating genomic-derived data including alterations, aneuploidy, ancestry, complex biomarker signatures, and hepatitis B virus (HBV) status. We demonstrate that a genomic classification of cHCC-CCA in the course of clinical care is possible and may drive the utility of targeted therapy in this challenging disease.
METHODS
Comprehensive Genomic Profiling
This study was conducted in accordance with the Western Institutional Review Board–approved protocol no. 20152817. Clinical cHCC-CCA (N = 73), CCA (N = 4,975), and HCC (N = 1,470) cases, as diagnosed by the treating physician (and confirmed on hematoxylin and eosin–stained slides), underwent CGP in a Clinical Laboratory Improvement Amendments–certified, NY State–approved, College of American Pathologists-accredited laboratory (Foundation Medicine Inc, Cambridge, MA).14 Manual microdissection was performed if warranted on pathologist visual inspection.
CGP on 0.8-1.1 Mb of the coding genome was performed on hybridization-captured, adapter-ligation–based libraries, to identify GA (base substitutions, small insertions or deletions, copy-number alterations, and rearrangements) in exons and select introns in up to 404 genes, tumor mutational burden (TMB), microsatellite instability status (MSI), genomic loss of heterozygosity (gLOH), chromosomal aneuploidy, genomic ancestry, and HBV status.
TMB was calculated as the number of nondriver somatic coding mutations per megabase of genome sequenced.15 TMB high was defined as 20 mutations/Mb (mut/Mb) or higher. MSI status was determined by analyzing 114 intronic homopolymer repeat loci for length variability and MSI high was defined as described previously.16 gLOH high (gLOH-H) was classified as 16% gLOH or higher, as was used in the ARIEL3 poly (ADP-ribose) polymerase inhibitor trial in ovarian cancer.17,18
Genomic ancestry of patients was determined using a principal component analysis of genomic single-nucleotide polymorphisms trained on data from the 1,000 Genomes Project, and each patient was classified as belonging to one of the following super populations: AFR (African), AMR (Ad Mixed American), EAS (East Asian), EUR (European), and SAS (South Asian).19,20
All GA prevalence reported in this study only include alterations described as functional or pathogenic in literature and seen in the Catalogue of Somatic Mutations in Cancer21 repository or had a likely functional status (frameshift or truncation events in tumor suppressor genes). Variants of unknown significance were not studied.
Hepatitis B Virus Detection
Presence of HBV was determined by the identification of DNA sequences consistent with genomic HBV DNA. Sequencing reads left unmapped to the human reference genome (hg19) were de novo assembled by Velvet,22,23 and the assembled contigs were competitively aligned by BLASTn24 to the National Center for Biotechnology Information database of more than 3 million known viral nucleotide sequences. A positive viral status was determined by contigs at least 80 nucleotides in length and with at least 97% identity to the BLAST sequence.25
Statistical Analysis
All statistical analyses were performed using R software (R Foundation for Statistical Computing, Vienna, Austria, v3.6.0). Proportions of categorical variables were compared using the chi-square test or Fisher's exact test; chi-square test was used for contingency tables larger than 2 × 2.
The Wilcoxon-rank sum test and the Kruskal-Wallis test were used to test for differences between continuous variables. All P values are two-sided, and multiple hypothesis testing correction was performed using the Benjamini-Hochberg procedure to calculate the false discovery rate.
Machine Learning
A random forest26–based ML model was trained on a high-quality subset of CCA (n = 1,916) and HCC (n = 755), using genomic features (such as alterations, aneuploidy, signatures, ancestry, and HBV status) and clinicopathologic features (such as age at the time of CGP, sex of the patient, tissue biopsy site, and local or metastatic status of the tumor [Data Supplement]). Model performance was evaluated by 10-fold cross-validation of the training cohort and on an independent test cohort of 479 CCA and 189 HCC (Data Supplement). This model was then applied on each cHCC-CCA case to classify it as CCA-like, HCC-like, or ambiguous.
RESULTS
Patient and Specimen Characteristics
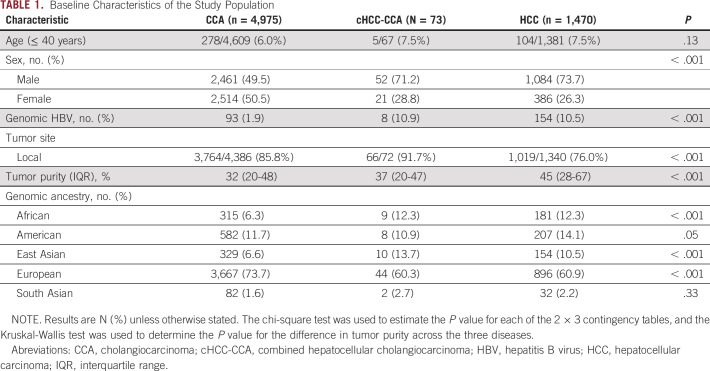
Liver biopsies were assayed for the majority of CCA, cHCC-CCA, and HCC cases. Patients with HCC were predominantly male, as described previously,27 and were enriched for younger patients and the African and East Asian genomic ancestry,28 whereas patients with CCA had a comparable sex prevalence, and were enriched for older patients and European genomic ancestry (Table 1). The cHCC-CCA cohort (N = 73) was 71.2% male and 60.3% of European ancestry, with a median age of 62 years (range: 22 years to 89+ years; Table 1).
TABLE 1.
Baseline Characteristics of the Study Population
Segregation of Genomic Alterations and Landscape of cHCC-CCA
When CCA was compared with HCC, genes were preferentially altered including ARID1A, BAP1, CDKN2A/B, FGFR2, IDH1, KRAS, and PBRM1 in CCA,9,10 and CTNNB1, MYC, and TERT in HCC7 (Figs 1A and 1B). Genomic HBV was also significantly associated with HCC compared to CCA (10.5% v 1.9%, P = 4.2e–42, odds ratio = 6.14).
FIG 1.
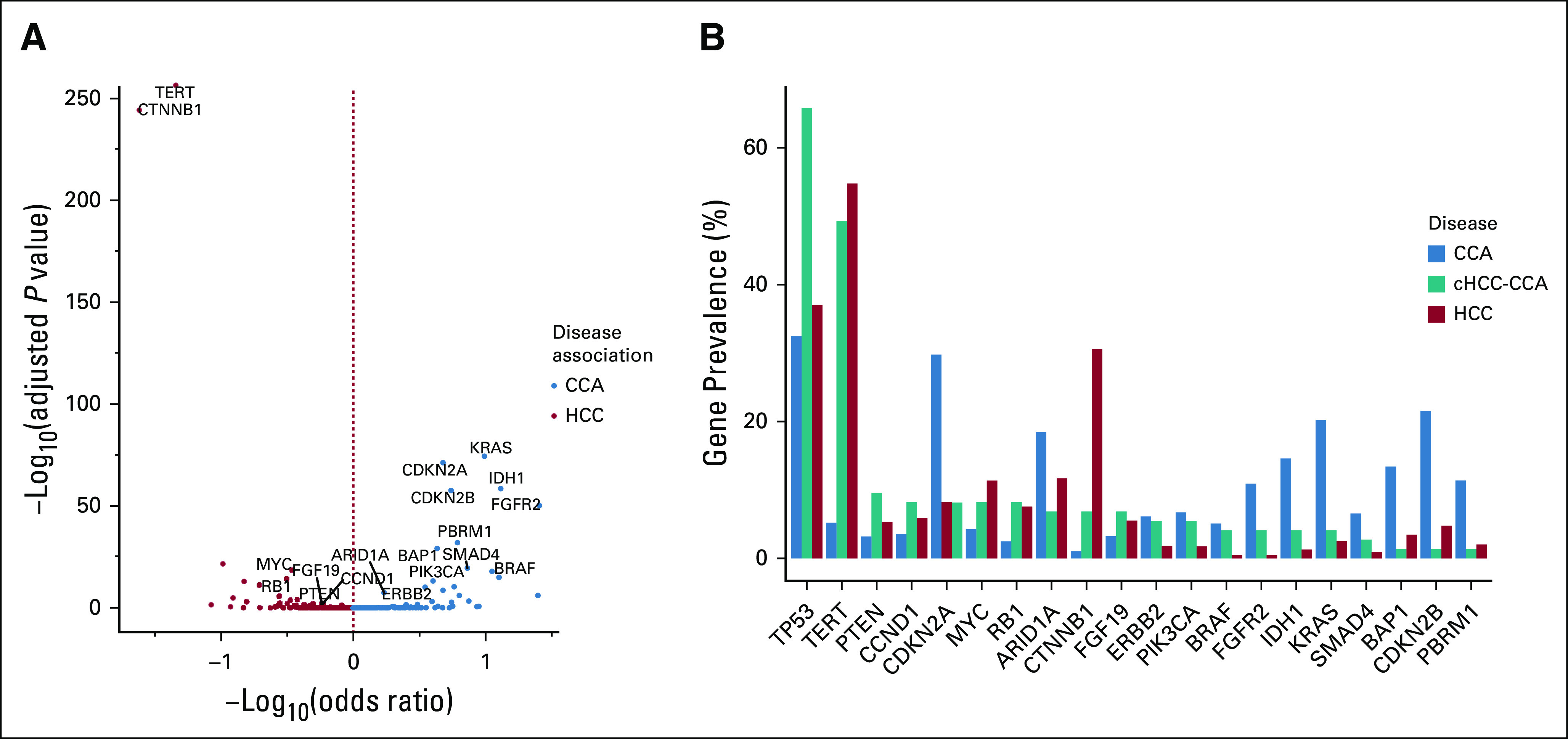
(A) Genes preferentially enriched in CCA or HCC. Volcano plot depicting the co-occurrence and mutual exclusivity of gene alterations between CCA and HCC. Only genes with an adjusted P value ≤ .05 and a prevalence ≥ 5% in either disease are labeled here. The two-tailed Fisher's exact test was used to evaluate the P values and odds ratios to determine associations between genes and disease. The Benjamini-Hochberg procedure was used to estimate the adjusted P values. (B) Prevalence of genes differentially enriched between CCA and HCC. The genes shown in (A), along with TP53, are depicted on the x-axis and are sorted in the decreasing order of their prevalence in cHCC-CCA. CCA, cholangiocarcinoma; cHCC-CCA, combined hepatocellular cholangiocarcinoma; HCC, hepatocellular carcinoma.
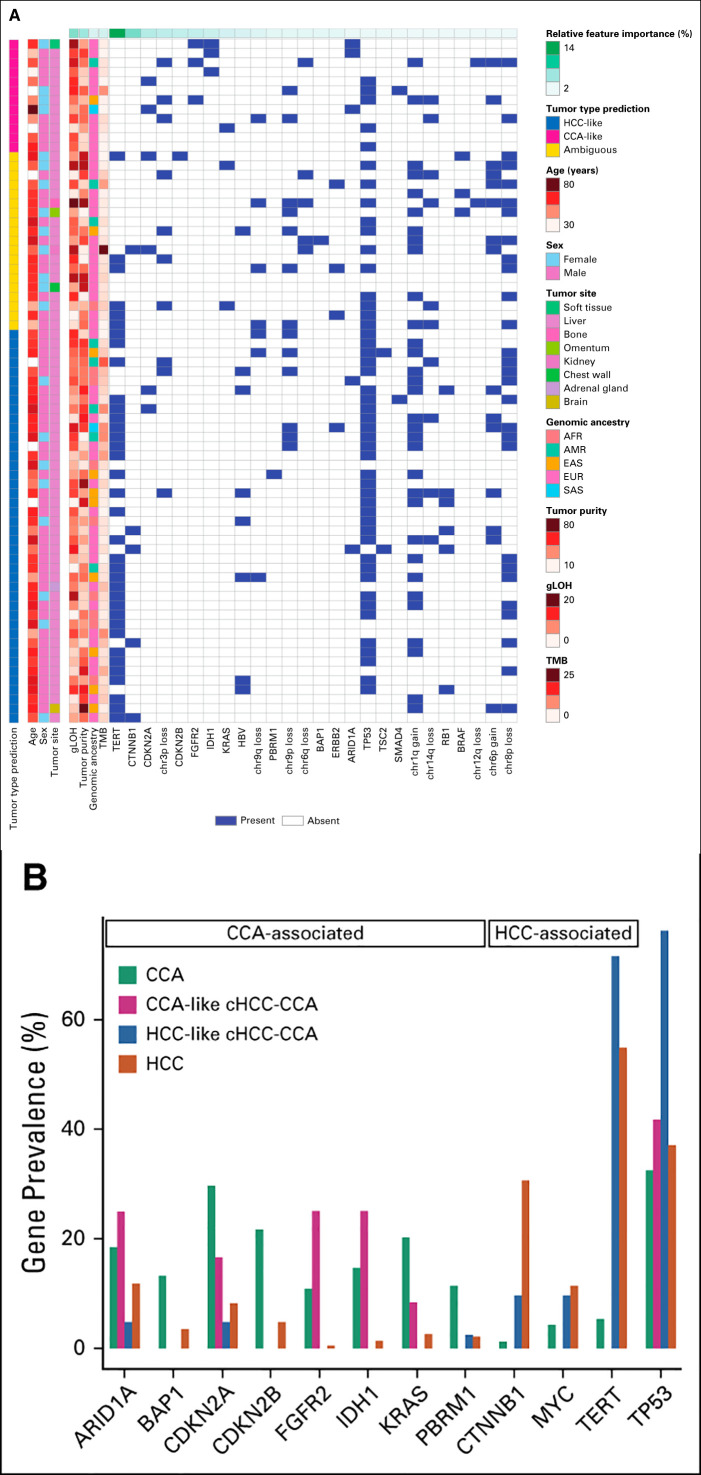
Among cHCC-CCA cases, we observed a median of 4 GA per tumor (range 0-14). Frequently altered genes in cHCC-CCA were TP53 (65.8%), TERT (49.3%), and PTEN (9.6%; Fig 2A). Within this cohort, the most commonly altered genes with GA that are linked to benefit from targeted therapies were BRCA2 (8.2%, 67% short variant, 25% were biallelic losses; 33% rearrangements), ERBB2 (5.5%, 75% amplifications), IDH1 (4.1%, 100% R132), BRAF (4.1%, 100% V600E), FGFR2 (4.1%, 67% fusions), and MET (2.7%, 100% amplifications), and accounted for 24.6% of cHCC-CCA.
FIG 2.
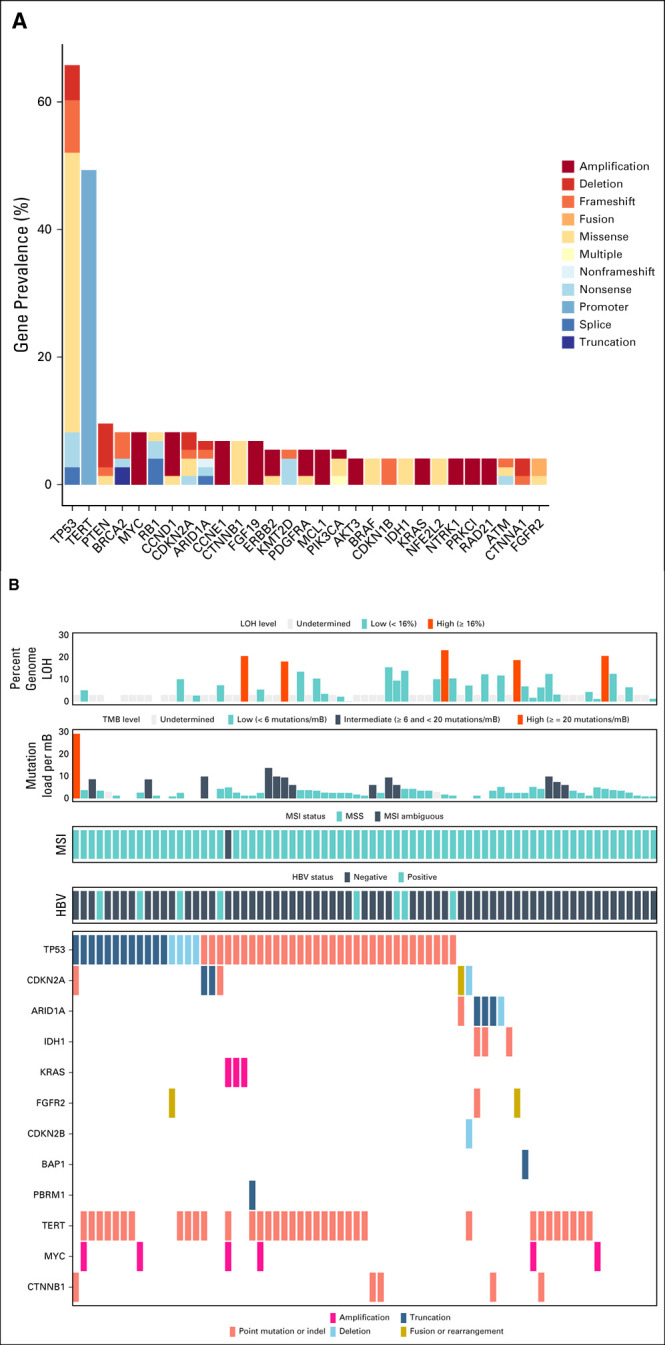
(A) Prevalence of genomic alterations in cHCC-CCA. Only gene alterations seen more than twice in the population (N = 73) are included in this plot. (B) Genomic characterization of cHCC-CCA, showing genes associated with CCA and HCC. The y-axis represents the genes associated with CCA and HCC (as described in Fig 1A and with a prevalence ≥ 10% in either disease; CCA-associated: ARID1A, BAP1, CDKN2A/B, FGFR2, IDH1, KRAS, and PBRM1; HCC-associated: CTNNB1, MYC, and TERT), and TP53, whereas the x-axis represents each of the individual 73 cHCC-CCA cases. CCA, cholangiocarcinoma; cHCC-CCA, combined hepatocellular cholangiocarcinoma; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; LOH, loss of heterozygosity; MSI, microsatellite instability; MSS, microsatellite instability status; TMB, tumor mutational burden.
Within the cHCC-CCA cohort, the prevalence of CCA- and HCC-enriched genes described above trended toward mutual exclusivity, possibly defining independent genomic subpopulations (Fig 2B). As such, the eight cases of cHCC-CCA positive for HBV were wild-type for genes often altered in CCA: ARID1A, BAP1, CDKN2B, FGFR2, IDH1, KRAS, and PBRM1.
Genomic Signatures
Biomarker signatures associated with response to immunotherapy like TMB high29 and MSI high30 were low in prevalence across the three diseases (Data Supplement). The median TMB was comparable across all three diseases: CCA (2.5 mut/Mb), HCC (3.5 mut/Mb), and cHCC-CCA (2.6 mut/Mb, Data Supplement). Alterations in the mismatch repair pathway were also seen in similar frequencies across all three diseases (data not shown).
However, for gLOH-H, which reflects genomic scarring associated with homologous recombination deficiency, 20.2% of CCA were gLOH-H, in contrast to 6.9% of HCC.17,18 Among cHCC-CCA, 13.8% were gLOH-H (Data Supplement). Median gLOH of CCA (10.5%) was higher than that of HCC (5.4%, P < .001) and comparable to cHCC-CCA (9.2%, P = .04; Data Supplement). However, GA in the homologous recombination repair pathway were seen in similar frequencies across all three diseases (data not shown).
Genomic Aneuploidy
Chromosome arm–level aneuploidy was highly prevalent and differentially enriched across the three forms of primary liver carcinomas. When CCA was compared with HCC, loss of 3p, 9p, 9q, and 6q and gain of 6p and 5q were significantly enriched in CCA and HCC, respectively (Fig 3A, Data Supplement). Within the cHCC-CCA population, the most frequent events were 8q gain (46.7%), 1q gain (40.0%), 8p loss (33.3%), and 17p loss (26.7%). In total, we identified 39 unique chromosome arm–level gains and 33 unique chromosome arm–level losses across the 60 cases (Fig 3B).
FIG 3.
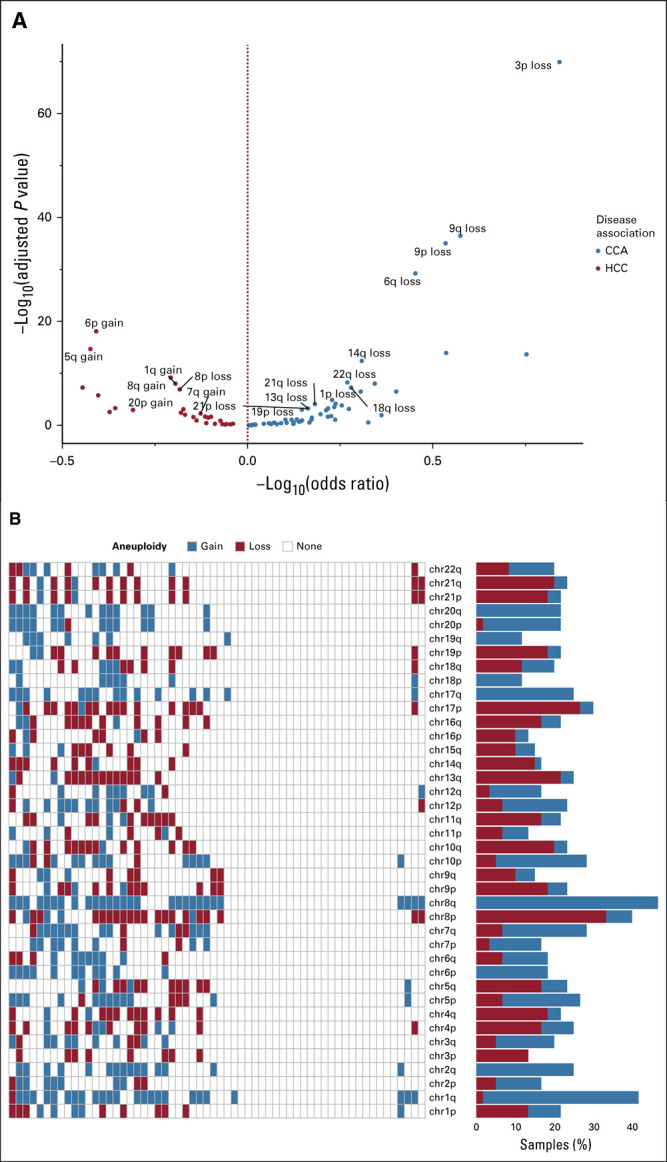
(A) Chromosomal arm–level aneuploidy preferentially enriched in CCA or HCC. Volcano plot depicting the co-occurrence and mutual exclusivity of aneuploidy events between CCA and HCC. Only aneuploidy events with an adjusted P value ≤ .01 and a prevalence ≥ 10% in at least one disease are labeled here. The two-tailed Fisher's exact test was used to evaluate the P values and odds ratios to determine associations between an event and disease. The Benjamini-Hochberg procedure was used to estimate the adjusted P values. (B) Landscape of chromosomal aneuploidy across cHCC-CCA. In the heatmap, the x-axis represents each cHCC-CCA case evaluated for aneuploidy and the y-axis represents the assessed aneuploidy events, sorted in the decreasing order of chromosome number and arm, from top to bottom. CCA, cholangiocarcinoma; cHCC-CCA, combined hepatocellular cholangiocarcinoma; HCC, hepatocellular carcinoma.
Machine Learning
The marked differences in the genomic features between CCA and HCC cases created an opportunity to build an ML model that could characterize a primary liver carcinoma as CCA-like or HCC-like.
We trained and tested a random forest–based classifier on a high-quality cohort of cases (training cohort: CCA n = 1,916 and HCC n = 755; independent test cohort: CCA n = 497 and HCC n = 189; Data Supplement). When the model was trained on genomic features alone, the model's sensitivity and specificity were 85.9% and 93.4%, respectively, and when the model was trained on genomic and clinicopathologic features, the model's sensitivity and specificity were 87.6% and 94.5%, respectively, based on 10-fold cross-validation of the training data (Data Supplement). Both the models performed similarly on the test cohort (Data Supplement), with a classification accuracy of 91% (95% CI, 88.8 to 93.2). When probed further, clinicopathologic features such as sex of the patient, biopsy site of the patient's tumor specimen, and age of the patient at the time of CGP testing were all found to be significantly associated with the presence or absence of genomic features including but not limited to TERT, CTNNB1, IDH1, and FGFR2 (Data Supplement), across the high-quality cohort of HCC and CCA. Subsequently, we chose the genomics-driven model for all further analyses.
The top genomic features that the classifier used to distinguish CCA from HCC were (ranked in their descending order of their classification power): TERT, CTNNB1, gLOH, tumor purity, CDKN2A, chromosome 3p loss, CDKN2B, FGFR2, IDH1, TMB, KRAS, genomic ancestry, genomic HBV, chromosome 9q loss, and PBRM1 (Data Supplement).
This classifier was then applied on each of the 73 cHCC-CCA cases, to place each individual case within the CCA-HCC spectrum (Fig 4A). A prediction call of CCA-like or HCC-like was made for 74% (54/73) of the cHCC-CCA cases. 0.61 was the probability cutoff used to separate HCC-like or CCA-like cHCC-CCA from ambiguous cHCC-CCA cases (Data Supplement); this value was derived from the probability threshold that maximized the Matthew's correlation coefficient31 in the HCC-CCA training cohort. In total, 16.4% (12/73) of the cHCC-CCA cases were classified as CCA-like, 57.5% (42/73) as HCC-like, and the remaining 26.3% (19/73) of the cHCC-CCA cases were classified as ambiguous.
FIG 4.
(A) RF-based classification of cHCC-CCA cases as CCA-like or HCC-like. Heatmap depicting the genomic and clinicopathologic features of the 73 cHCC-CCA cases, which were classified as CCA-like or HCC-like by a RF model. The top 30 features that the model uses to differentiate CCA from HCC are shown on the x-axis and the tumor type prediction of cHCC-CCA cases as CCA-like, HCC-like, or ambiguous is annotated along the y-axis. Clinicopathologic features such as age, sex, and tumor biopsy site are added for annotation purposes only. The relative feature importance of a feature, defined as the classification power of that feature with respect to those of the entire set of features used to train the model, is annotated along the x-axis. With the exception of gLOH, tumor purity, and genomic ancestry, features are sorted in the decreasing order of their importance. (B) Gene prevalence across CCA, CCA-like cHCC-CCA, HCC-like cHCC-CCA, and HCC. Prevalence of TP53-, CCA-, and HCC-associated genes, between CCA, HCC, and the machine learning–classified CCA-like cHCC-CCA and HCC-like cHCC-CCA cases. AFR, African; AMR, Ad Mixed American; EAS, East Asian; EUR, European; CCA, cholangiocarcinoma; cHCC-CCA, combined hepatocellular carcinoma cholangiocarcinoma; gLOH, genomic loss of heterozygosity; HCC, hepatocellular carcinoma; RF, random forest; SAS, South Asian; TMB, tumor mutational burden.
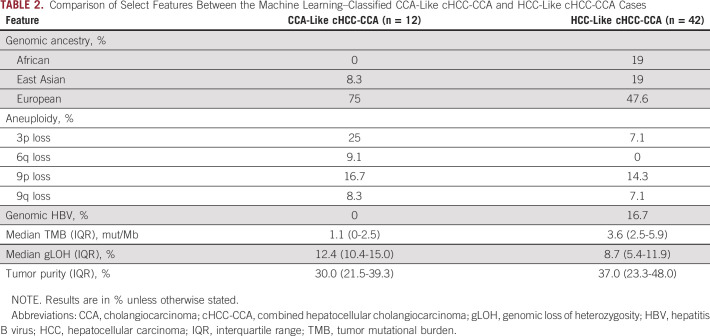
The CCA-like set of cHCC-CCA cohort had 25% each ARID1A, FGFR2, and IDH1, and were wild-type for TERT, CTNNB1, and MYC (Fig 4B) and lacked genomic HBV (Table 2). CCA-like cHCC-CCA cases had a median gLOH of 12.4% (Table 2). By contrast, the HCC-like cases harbored GA in TERT (71.4%), CTNNB1 (9.5%), and MYC (9.5%), and were wild-type for lDH1, FGFR2, and KRAS (Fig 4B). HCC-like cHCC-CCA cases had a median gLOH of 8.7% and 16.7% harbored genomic HBV (Table 2).
TABLE 2.
Comparison of Select Features Between the Machine Learning–Classified CCA-Like cHCC-CCA and HCC-Like cHCC-CCA Cases
DISCUSSION
The two most frequent forms of primary liver carcinomas (HCC and CCA) are ontologically, morphologically, and clinically distinct. However, as evoked by its name, cHCC-CCA is a poorly understood, aggressive rare primary liver cancer that exhibits morphologic characteristics of both HCC and CCA.32 As such, cHCC-CCA is an extremely challenging disease regarding both diagnosis and management.
We analyzed the genomic profiles of 73 cHCC-CCA, 4,975 CCA, and 1,470 HCC, all generated by targeted exome sequencing in the course of clinical care. As consistent with extensive previous studies, CCA and HCC differ strikingly in many characteristics, including the frequency of GA, genomic signatures, ancestry, and aneuploidy. Notable differentially enriched GA were CTNNB1, MYC, and TERT, and FGFR2 and IDH1, enriched in HCC and CCA, respectively.
We observed arm-level losses in 3p, 9p, 9q, and 6q, and arm-level gains in 5q and 6p as enriched in CCA and HCC, respectively, consistent with previous work. Loss of the 3p chromosomal arm (containing the BAP1, PBRM1 gene loci) has been described as a possible clonal event in CCA,10 whereas loss of 6q (containing ZNF292 and EEF1A1), 9p (containing CDKN2A, CDKN2B, and MTAP), and 9q have been described as recurrent aneuploidy events in CCA.33 Gain of the 6p chromosomal arm has been associated with late-stage HCC.34
We also identified multiple recurrent chromosome arm–level aneuploidy events in cHCC-CCA. A number of well-characterized tumor suppressors and oncogenes are known to be contained by these aberrant chromosomal segments, including MYC (8q gain)13,35; CHD1L, CKS1B, JTB, and SHC1 (1q gain)36; DLC1 (8p loss)37; SGCE, MET, and CDK6 (7q gain)13,35; TERT (5p gain)13; TP53 (17p loss)35; PTEN (10q loss)35; and RB1 (13q loss) as well other aberrations, notably, gain of the 2q, 5p, 17q, 20p, and 20q arms, and loss of the 21q arm whose functional implications remains to be evaluated.
Within individual cases, GA, signatures, and aneuploidy were strongly associated with either CCA or HCC, typically mutually exclusive for those associated with the other disease, suggesting that individual cases could be labeled as resembling either CCA or HCC. This led us to build a genomics-only–driven ML model to classify a cHCC-CCA case as CCA-like or HCC-like, which could then accurately classify more than 70% of the cases. Within our study cohort, the clinical characteristics examined were strongly associated with genomic features at a population level and as such did not add independent resolution to further strengthen classification in differentiating HCC from CCA.
This work complements the paradigm of Xue et al, but significantly differs from that seminal study by both methodology and the clinical validation status of the used assays. Xue et al used both whole-exome sequencing and transcriptional profiling, neither of which was annotated as validated for use in patient care. IHC, laser capture microdissection of areas with differing morphology, and a team of specialized pathologists for subtype classification were also used, and all this was performed on banked specimens dating from 2007 to 2018. By contrast, for the current study, the only assay used was a clinically validated targeted exome sequencing assay, at the request of a physician currently treating a patient with cHCC-CCA. If the results of this study are to become clinically impactful, then clincally validated targeted exome based sequencing assays would prove to be vital for future cHCC-CCA patients.
Furthermore, in this study, no histologic subclassification into the separate, combined, and mixed subtypes was performed, which is in contrast to Xue et al. However, the resolving power of the ML tool separated cHCC-CCA cases into HCC-like or CCA-like solely on the basis of genomic features, which indicates that subtype classification is not needed for classification.
The 19 ambiguous cHCC-CCA cases harbored genomic features associated with both CCA and HCC. Notable examples include a case with presence of genomic HBV, and wild-type FGFR2 and IDH1, all of which resemble HCC but also harbored a gLOH of 10.6% and 3p loss, which is more CCA-like. Another case harbors a GA in TERT, a TMB of 2.5 mut/Mb, gLOH of 4%, wild-type FGFR2 and IDH1, all consistent with HCC, but also an ERBB2 alteration, the latter being frequently associated with biliary tract cancer. Almost half of these ambiguous cases (8/19) had lower tumor purity, which is important as tumor purity is a strong feature differentiating CCA and HCC in this series. Low tumor purity reduces the sensitivity of the CGP test to detect low-allele-frequency GA, hence affecting classification accuracy.
One limitation is the inability to detect the RNA hepatitis C virus by the DNA-oriented targeted exome platform used here, or knowledge of infection with the virus from the clinical history of the patients. Association of HCC with hepatitis C virus varies by global geography. In the United States, CCA is associated with hepatitis C virus, whereas in Asia, the same disease is also associated with hepatitis B virus, perhaps secondary to endemic infection.38 Relatedly, the subdivision of cHCC-CCA into HCC-like and CCA-like cases on the basis of genomic findings alone mimics the findings of Xue et al, albeit with a clinically validated assay that can be used in the course of patient care. However, the clinical impact of these findings remains to be explored. In the terms of improving the performance of the classifier, further investigation can help benchmark the ML model's posterior probability threshold to delineate either HCC-like or CCA-like cHCC-CCA from ambiguous cHCC-CCA.
Another limitation of this study was the lack of characterization of clinicopathologic features such as serum alpha-fetoprotein levels, the presence of cirrhosis or ascites, and liver imaging–based features such as hepatic vein invasion, which are all typically observed in HCC but not in CCA. Inclusion of such features in the ML classifier could potentially enhance performance of the model and should be considered for further investigation.
Based on the findings in this study and Xue et al, the concept of cHCC-CCA itself should continue to be critically evaluated as to what the implications are for etiology of CCA-like and HCC-like cHCC-CCA. Unlike Xue et al, the genomic testing platform combined with the ML-classifier developed here allow for easy classification of cHCC-CCA in the course of clinical care by use of a clinically validated test. However, further investigation is needed to identify the clinical impact of these findings.
Karthikeyan Murugesan
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche Pharma AG
Patents, Royalties, Other Intellectual Property: Antibiotic resistance causation identification (US10629291B2) filed with Koninklijke Philips NV, Analytic prediction of antibiotic susceptibility (US20190279738A1) filed with Koninklijke Philips NV, Methods and devices for characterizing and treating combined hepatocellular cholangiocarcinoma, with Foundation Medicine Inc
Travel, Accommodations, Expenses: Foundation Medicine
Radwa Sharaf
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Meagan Montesion
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Jay A. Moore
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
James Pao
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Research Funding: Foundation Medicine
Patents, Royalties, Other Intellectual Property: Provisional patent submitted for Foundation Medicine, dealing with machine learning for gene predictions
Dean C. Pavlick
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Garrett M. Frampton
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Vivek A. Upadhyay
Employment: EQRx
Stock and Other Ownership Interests: EQRx
Consulting or Advisory Role: Foundation Medicine
Brian M. Alexander
Employment: Foundation Medicine
Leadership: Foundation Medicine
Stock and Other Ownership Interests: Roche
Research Funding: Lilly, Puma Biotechnology, Celgene
Open Payments Link: https://openpaymentsdata.cms.gov/physician/854258/summary
Vincent A. Miller
Employment: Foundation Medicine, EQRx
Leadership: Revolution Medicines
Stock and Other Ownership Interests: Foundation Medicine, Mirati Therapeutics, Revolution Medicines, EQRx
Patents, Royalties, Other Intellectual Property: Receives periodic royalties related to T790M patent awarded to Memorial Sloan Kettering Cancer Center
Milind M. Javle
Consulting or Advisory Role: QED Therapeutics, Oncosil, Incyte, Mundipharma EDO GmbH, AstraZeneca, Merck, EMD Serono
Other Relationship: Rafael Pharmaceuticals, Incyte, Pieris Pharmaceuticals, Merck, Merck Serono, Novartis, Seattle Genetics, BeiGene, QED Therapeutics, Bayer
Tanios S. Bekaii Saab
Consulting or Advisory Role: Amgen, Ipsen, Lilly, Bayer, Roche/Genentech, AbbVie, Incyte, Immuneering, Seattle Genetics, Pfizer, Boehringer Ingelheim, Janssen, Eisai, Daiichi Sankyo/UCB Japan, AstraZeneca, Exact Sciences, Natera, Treos Bio, Celularity, SOBI, BeiGene, Foundation Medicine
Patents, Royalties, Other Intellectual Property: Patent WO/2018/183488, Patent WO/2019/055687
Other Relationship: Exelixis, Merck, AstraZeneca, Lilly, Pancreatic Cancer Action Network
Lee A. Albacker
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Jeffrey S. Ross
Employment: Foundation Medicine
Leadership: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Consulting or Advisory Role: Celsius Therapeutics, Tango Therapeutics
Research Funding: Foundation Medicine
Siraj M. Ali
Employment: EQRx
Consulting or Advisory Role: Elevation Oncology, In8bio, Pillar Biosciences, Takeda, ArcherDx
Stock and Other Ownership Interests: In8bio, Pillar Biosciences, EQRx
Patents: Foundation Medicine
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented in part as a poster discussion (677PD) at the European Society of Medical Oncology (ESMO)'s Annual Congress, Barcelona, Spain, September 27-October 1, 2019.
AUTHOR CONTRIBUTIONS
Conception and design: Karthikeyan Murugesan, Garrett M. Frampton, Milind M. Javle, Tanios S. Bekaii Saab, Lee A. Albacker, Jeffrey S. Ross, Siraj M. Ali
Financial support: Brian M. Alexander
Administrative support: Brian M. Alexander
Collection and assembly of data: Karthikeyan Murugesan, Radwa Sharaf, Meagan Montesion, Jay A. Moore, Dean C. Pavlick, Garrett M. Frampton, Lee A. Albacker, Jeffrey S. Ross
Data analysis and interpretation: Karthikeyan Murugesan, Radwa Sharaf, James Pao, Dean C. Pavlick, Garrett M. Frampton, Vivek A. Upadhyay, Brian M. Alexander, Vincent A. Miller, Tanios S. Bekaii Saab, Lee A. Albacker, Jeffrey S. Ross, Siraj M. Ali
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by the authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Karthikeyan Murugesan
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche Pharma AG
Patents, Royalties, Other Intellectual Property: Antibiotic resistance causation identification (US10629291B2) filed with Koninklijke Philips NV, Analytic prediction of antibiotic susceptibility (US20190279738A1) filed with Koninklijke Philips NV, Methods and devices for characterizing and treating combined hepatocellular cholangiocarcinoma, with Foundation Medicine Inc
Travel, Accommodations, Expenses: Foundation Medicine
Radwa Sharaf
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Meagan Montesion
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Jay A. Moore
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
James Pao
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Research Funding: Foundation Medicine
Patents, Royalties, Other Intellectual Property: Provisional patent submitted for Foundation Medicine, dealing with machine learning for gene predictions
Dean C. Pavlick
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Garrett M. Frampton
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Vivek A. Upadhyay
Employment: EQRx
Stock and Other Ownership Interests: EQRx
Consulting or Advisory Role: Foundation Medicine
Brian M. Alexander
Employment: Foundation Medicine
Leadership: Foundation Medicine
Stock and Other Ownership Interests: Roche
Research Funding: Lilly, Puma Biotechnology, Celgene
Open Payments Link: https://openpaymentsdata.cms.gov/physician/854258/summary
Vincent A. Miller
Employment: Foundation Medicine, EQRx
Leadership: Revolution Medicines
Stock and Other Ownership Interests: Foundation Medicine, Mirati Therapeutics, Revolution Medicines, EQRx
Patents, Royalties, Other Intellectual Property: Receives periodic royalties related to T790M patent awarded to Memorial Sloan Kettering Cancer Center
Milind M. Javle
Consulting or Advisory Role: QED Therapeutics, Oncosil, Incyte, Mundipharma EDO GmbH, AstraZeneca, Merck, EMD Serono
Other Relationship: Rafael Pharmaceuticals, Incyte, Pieris Pharmaceuticals, Merck, Merck Serono, Novartis, Seattle Genetics, BeiGene, QED Therapeutics, Bayer
Tanios S. Bekaii Saab
Consulting or Advisory Role: Amgen, Ipsen, Lilly, Bayer, Roche/Genentech, AbbVie, Incyte, Immuneering, Seattle Genetics, Pfizer, Boehringer Ingelheim, Janssen, Eisai, Daiichi Sankyo/UCB Japan, AstraZeneca, Exact Sciences, Natera, Treos Bio, Celularity, SOBI, BeiGene, Foundation Medicine
Patents, Royalties, Other Intellectual Property: Patent WO/2018/183488, Patent WO/2019/055687
Other Relationship: Exelixis, Merck, AstraZeneca, Lilly, Pancreatic Cancer Action Network
Lee A. Albacker
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Jeffrey S. Ross
Employment: Foundation Medicine
Leadership: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Consulting or Advisory Role: Celsius Therapeutics, Tango Therapeutics
Research Funding: Foundation Medicine
Siraj M. Ali
Employment: EQRx
Consulting or Advisory Role: Elevation Oncology, In8bio, Pillar Biosciences, Takeda, ArcherDx
Stock and Other Ownership Interests: In8bio, Pillar Biosciences, EQRx
Patents: Foundation Medicine
No other potential conflicts of interest were reported.
REFERENCES
- 1.Nagtegaal ID, Odze RD, Klimstra D, et al. The 2019 WHO classification of tumours of the digestive system Histopathology 76:182–1882020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maximin S, Ganeshan DM, Shanbhogue AK, et al. Current update on combined hepatocellular-cholangiocarcinoma Eur J Radiol Open 1:40–482014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Connell LC, Harding JJ, Shia J, et al. Combined intrahepatic cholangiocarcinoma and hepatocellular carcinoma Chin Clin Oncol 5:66.2016 [DOI] [PubMed] [Google Scholar]
- 4.Stavraka C, Rush H, Ross P.Combined hepatocellular cholangiocarcinoma (cHCC-CC): An update of genetics, molecular biology, and therapeutic interventions J Hepatocell Carcinoma 6:11–212018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abou-Alfa GK, Sahai V, Hollebecque A, et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: A multicentre, open-label, phase 2 study Lancet Oncol 21:671–6842020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen RA, Lisa JR.Combined liver cell and bile duct carcinoma Am J Pathol 25:647–6551949 [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network Comprehensive and integrative genomic characterization of hepatocellular carcinoma Cell 169:1327–1341.e232017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujimoto A, Furuta M, Totoki Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer Nat Genet 48:500–5092016 [DOI] [PubMed] [Google Scholar]
- 9.Nakamura H, Arai Y, Totoki Y, et al. Genomic spectra of biliary tract cancer Nat Genet 47:1003–10102015 [DOI] [PubMed] [Google Scholar]
- 10.Farshidfar F, Zheng S, Gingras MC, et al. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-mutant molecular profiles Cell Rep 18:2780–27942017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moeini A, Sia D, Zhang Z, et al. Mixed hepatocellular cholangiocarcinoma tumors: Cholangiolocellular carcinoma is a distinct molecular entity J Hepatol 66:952–9612017 [DOI] [PubMed] [Google Scholar]
- 12.Wang A, Wu L, Lin J, et al. Whole-exome sequencing reveals the origin and evolution of hepato-cholangiocarcinoma Nat Commun 9:894.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue R, Chen L, Zhang C, et al. Genomic and transcriptomic profiling of combined hepatocellular and intrahepatic cholangiocarcinoma reveals distinct molecular subtypes Cancer Cell 35:932–947.e82019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing Nat Biotechnol 31:1023–10312013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden Genome Med 9:34.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trabucco SE, Gowen K, Maund SL, et al. A novel next-generation sequencing approach to detecting microsatellite instability and pan-tumor characterization of 1000 microsatellite instability–high cases in 67,000 patient samples J Mol Diagn 21:1053–10662019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coleman RL, Oza AM, Lorusso D, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): A randomised, double-blind, placebo-controlled, phase 3 trial Lancet 390:1949–19612017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swisher EM, Lin KK, Oza AM, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): An international, multicentre, open-label, phase 2 trial Lancet Oncol 18:75–872017 [DOI] [PubMed] [Google Scholar]
- 19.Newberg J, Connelly C, Frampton G. Determining patient ancestry based on targeted tumor comprehensive genomic profiling. Epidemiology. 2019;79 abstr 1599. [Google Scholar]
- 20.Carrot-Zhang J, Chambwe N, Damrauer JS, et al. Comprehensive analysis of genetic ancestry and its molecular correlates in cancer Cancer Cell 37639–654.e62020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forbes SA, Beare D, Boutselakis H, et al. COSMIC: Somatic cancer genetics at high-resolution Nucleic Acids Res 45:D777–D7832017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zerbino DR. Using the Velvet de novo assembler for short-read sequencing technologies. Curr Protoc Bioinformatics. 2010 doi: 10.1002/0471250953.bi1105s31. Chapter 11:Unit 11.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zerbino DR, Birney E.Velvet: Algorithms for de novo short read assembly using de Bruijn graphs Genome Res 18:821–8292008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool J Mol Biol 215:403–4101990 [DOI] [PubMed] [Google Scholar]
- 25.Knepper TC, Montesion M, Russell JS, et al. The genomic landscape of Merkel cell carcinoma and clinicogenomic biomarkers of response to immune checkpoint inhibitor therapy Clin Cancer Res 25:5961–59712019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breiman L.Random forests Mach Learn 455–322001 [Google Scholar]
- 27.El-Serag HB, Rudolph KL.Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis Gastroenterology 132:2557–25762007 [DOI] [PubMed] [Google Scholar]
- 28.Mittal S, El-Serag HB.Epidemiology of hepatocellular carcinoma: Consider the population J Clin Gastroenterol 47:S2–S62013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers Mol Cancer Ther 16:2598–26082017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carbone DP, Reck M, Paz-Ares L, et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer N Engl J Med 376:2415–24262017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chicco D, Jurman G.The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation BMC Genomics 21:6.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seehawer M, Heinzmann F, D'Artista L, et al. Necroptosis microenvironment directs lineage commitment in liver cancer Nature 562:69–752018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goeppert B, Toth R, Singer S, et al. Integrative analysis defines distinct prognostic subgroups of intrahepatic cholangiocarcinoma Hepatology 69:2091–21062019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwon SM, Kim DS, Won NH, et al. Genomic copy number alterations with transcriptional deregulation at 6p identify an aggressive HCC phenotype Carcinogenesis 34:1543–15502013 [DOI] [PubMed] [Google Scholar]
- 35.Niu ZS, Niu XJ, Wang WH.Genetic alterations in hepatocellular carcinoma: An update World J Gastroenterol 22:9069–90952016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L, Chan THM, Guan XY.Chromosome 1q21 amplification and oncogenes in hepatocellular carcinoma Acta Pharmacol Sin 31:1165–11712010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xue W, Krasnitz A, Lucito R, et al. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma Genes Dev 22:1439–14442008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan SA, Tavolari S, Brandi G.Cholangiocarcinoma: Epidemiology and risk factors Liver Int 39:19–312019suppl 1 [DOI] [PubMed] [Google Scholar]