Abstract
Skin development and patterning is dependent on factors that regulate the stepwise differentiation of dermal fibroblasts concomitant with dermal-epidermal reciprocal signaling, two processes that are poorly understood. Here we show that dermal EZH2, the methyltransferase enzyme of the epigenetic Polycomb Repressive Complex 2 (PRC2), is a new coordinator of both these processes. Dermal EZH2 activity is present during dermal fibroblast differentiation and is required for spatially restricting Wntβ-catenin signaling to reinforce dermal fibroblast cell fate. Later in development, dermal EZH2 regulates the expression of reticular dermal markers and initiation of secondary hair follicles. Embryos lacking dermal Ezh2 have elevated epidermal proliferation and differentiation that can be rescued by small molecule inhibition of retinoic acid (RA) signaling. Together, our study reveals that dermal EZH2 is acting like a rheostat to control the levels of Wnt/p-catenin and RA signaling to impact fibroblast differentiation cell autonomously and epidermal keratinocyte development non-cell autonomously, respectively.
Keywords: Embryonic Skin Development, Dermal fibroblast differentiation, epidermal-dermal cross talk, Polycomb Repressive Complex2, Non-cell autonomous
Graphical Abstract
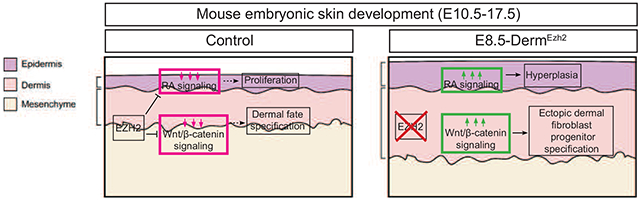
Ezh2 controls the differentiation of lower mesenchyme towards dermal fibroblast progenitors and proliferation of epidermal keratinocytes through regulation of Wnt/p-catenin and retinoic acid signaling, respectively.
Introduction
Differentiation of multipotent progenitors towards terminal fates is an essential component of development. Transcription factors aid in the execution of cell fate selection intrinsically by controlling the expression of fate-specific genes(Healy et al., 1999; Ito et al., 2000; Iwafuchi-Doi and Zaret, 2016; Zorick et al., 1996). Cell-cell communication can also affect differentiation of a target cell through extrinsic juxtracrine and paracrine factors(Chen et al., 2012; Daudet and Lewis, 2005; Dhouailly, 1975). The presence of both intrinsic transcription factors and cell-cell communication through extrinsic factors is evident and indispensable in cell fate selection during development of many organs including skin(Chen et al., 2012; Daudet et al., 2007; Daudet and Lewis, 2005). However, the mechanisms that control the temporal and spatial coordination of intrinsic and extrinsic factors during cell differentiation and cell-cell communication are not clearly understood. Although in vitro studies have identified that epigenetic complexes may regulate coordination of cell signaling of multiple pathways(Cieslik et al., 2013; Mohammad and Baylin, 2010), it remains unclear as to what extent and how epigenetic complexes orchestrate coordination of cellular heterogeneity and cell-cell communication in developing tissues. Here, we use the murine skin as a model to understand the role of developmental epigenetic regulators in generating dermal fibroblast heterogeneity and dermal-epidermal communication.
The murine dermis consists of dermal fibroblasts that undergo differentiation while reciprocally signaling overlying epidermal keratinocytes to induce appendages (Chen et al., 2012; Fu and Hsu, 2013; Ito et al., 2007). Murine dermal fibroblasts and their interaction with the epidermal keratinocytes serve as an excellent model system to address the question of whether Polycomb Repressive Complex 2 (PRC2) can coordinate cellular differentiation both cell autonomously through intrinsic factors and non-cell autonomously through inter-cellular communication. The differentiation of dermal fibroblasts and the reciprocal signaling to the epidermal keratinocytes are integral to skin patterning, development of appendages(Chen et al., 2012; Fu and Hsu, 2013) and healing(S. Werner and R. Grose, 2011; Werner et al., 2007). In mice, by embryonic day (E) 9.5, the dermal fibroblast (DF) precursors migrate from somites to sub-epidermal locations and differentiate into DF progenitors between E11.5 and E13.5(Atit et al., 2006; Chen et al., 2012; Ohtola et al., 2008; Thulabandu et al., 2018). The DF progenitors then differentiate into papillary fibroblast progenitors (PPs), reticular fibroblast progenitors (RPs), and hair-forming dermal papilla fibroblasts (DPs), with distinct gene expression profiles as early as E14.5(Driskell et al., 2013; Thulabandu et al., 2018) (Fig. 1A). PPs, differentiate into papillary fibroblasts, and DPs which are crucial for communicating to the epidermis and stimulating hair follicle morphogenesis and cycling(Driskell et al., 2013; Plikus et al., 2008). RPs differentiate into dermal white adipose tissue (DWAT), which aids in thermal insulation(Driskell et al., 2014, 2013), and reticular fibroblasts, which secrete dense collagen fibers to provide elasticity to the skin(Driskell and Watt, 2015). It is unknown whether PRC2 controls dermal fibroblast heterogeneity and impacts dermal signaling to the epidermis during skin development.
Figure 1: Ezh2 is efficiently deleted in the dermal mesenchyme by PDGFRαCreER:
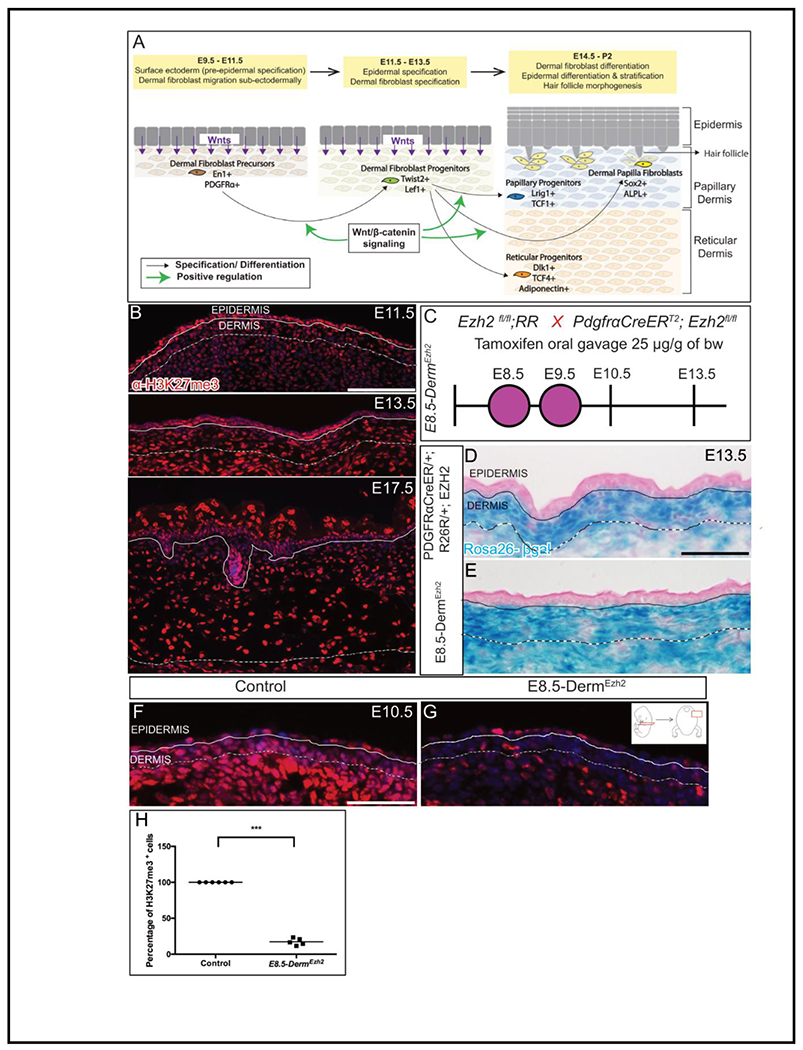
Schematic depicting the skin development program (A). Indirect immunofluorescence of H3K27me3 with DAPI counterstain at various stages of dermal development (B). Schematic illustration of tamoxifen gavage regimen (C) utilized in the study. βgalactosidase staining (D, E) depicting the region of Cre-ER recombinase of R26R expression. Indirect immunofluorescence of H3K27me3 with DAPI counterstain at E10.5 (F, G) and its quantification (H). Solid line demarcates the epidermis from dermis and dashed line demarcates the lower limit of the dermis. Scale bar=100 microns in B and 50 microns in D-G.
Polycomb group proteins (PcG) are one of the most well-studied epigenetic complexes that regulate cell differentiation through mediation of both intrinsic and extrinsic developmental signals(Laugesen and Helin, 2014; Mirzamohammadi et al., 2016; Wang et al., 2010). PcG regulates both intrinsic and extrinsic signals via genetic repression while also controlling the accessibility of the chromatin for various transcription factors by DNA methylation/hydroxymethylation, posttranslational histone modifications and higher-order chromatin folding(Aranda et al., 2015; Cheutin and Cavalli, 2014). Polycomb Repressive Complex2 (PRC2) is a PcG complex consisting of three core proteins: SUZ12, EED and EZH2. EZH2 is the catalytic component that mediates the trimethylation of Lysine27 on Histone 3 (H3K27me3) resulting in gene silencing. Deleting Ezh2 in epidermal basal keratinocytes leads to derepression of multiple late epidermal differentiation genes resulting in accelerated differentiation and precocious barrier acquisition(Ezhkova et al., 2009). Ablation of both Ezh1 and Ezh2 in epidermal basal keratinocytes leads to ectopic differentiation of keratinocytes into Merkel cells mediated by an increase in Sox2 expression(Bardot et al., 2013; Perdigoto et al., 2016). Deleting Ezh2 in cranial mesenchyme leads to upregulation of anti-osteogenic genes resulting in reduction of the size of skull bones(Dudakovic et al., 2015; Ferguson et al., 2018; Schwarz et al., 2014). These studies emphasize the ability of PcG to regulate differentiation cell autonomously. Besides, a previous study lends support to the non-cell autonomous regulation of PcG(Eun et al., 2014). Ablation of E(z), the drosophila homologue of EZH1/2, in somatic gonad cells leads to somatic cell marker expression in drosophila germ cells in a non-cell autonomous manner(Eun et al., 2014). These studies have demonstrated a requirement of PRC2 in regulating various intrinsic or extrinsic factors required for cell differentiation across different tissues. However, whether PRC2 coordinates various intrinsic and extrinsic factors in multipotential progenitor cells to simultaneously impact cell differentiation and cell-cell communication is not clear.
Multiple signaling pathways regulate the development of DF progenitors in vivo during cell fate selection and differentiation. Many of the key transcription factors and signaling factors involved in DF progenitor development are regulated by PRC2 in other cell types(Ferguson et al., 2018; Mirzamohammadi et al., 2016; Wang et al., 2010), and single gene deletions have not yielded strong dermal phenotypes. However, Wnt/Ezh2 controls the differentiation of lower mesenchyme towards dermal fibroblast progenitors and proliferation of epidermal-catenin signaling regulates several transcription factors expressed between E9.5 and E11.5, such as En1, Msx1, Msx2, Lef1, Twist1, Twist2 and is required for dermal fibroblast cell fate determination(Atit et al., 2006; Budnick et al., 2016; Chen et al., 2012; Driskell et al., 2013; Ohtola et al., 2008; Phan et al., 2020; Rognoni et al., 2016; Tran et al., 2010). Ablation or overexpression of β–catenin in DF precursors shows that canonical Wnt signaling is necessary and sufficient for specification of DF progenitors’ fate, respectively(Atit et al., 2006; Chen et al., 2012; Ohtola et al., 2008; Tran et al., 2010). Wnt signaling expressing DF progenitors give rise to dermal condensate cells and PPs that are precursors to DP cells and are required for hair follicle initiation in embryonic mouse skin(Gupta et al., 2019; Mok et al., 2019) . Wnt and BMP signaling are required to maintain DP’s ability to induce hair follicles(Rendl et al., 2008). RA signaling in DF progenitors is required to specify dermal condensate cells as shown by dermal fibroblasts restricted deletion of RA inhibitor, Cyp26b1 (Okano et al., 2012). Although, some of the signals governing the differentiation of DF precursors to DF progenitors have been identified, the mechanisms required to coordinate the spatio-temporal differentiation DF progenitors into PP, RP and DP lineages remain unidentified. However, whether PRC2 orchestrates these signals and factors in dermal fibroblasts is unknown.
Here, we tested the role of PRC2 in embryonic dermal development and dermal-epidermal signaling, via cell-type and temporal-specific deletion of Ezh2 in mouse DF precursors in vivo. Here, we found that Ezh2 is required to inhibit the differentiation of the lower mesenchyme to TWIST2+/LEF1+DF progenitors by spatially restricting Wnt/β-catenin signaling and not for generating the other lineages. Dermal PRC2 is required for timely induction of secondary hair follicles and dispensable for primary guard hair follicle initiation. We found that deletion of dermal Ezh2 aberrantly enhances RA signaling and causes epidermal hyperplasia by elevating proliferation. We show that RA signaling is a key driver and transient activation of RA signaling recapitulates the phenotype and inhibition of RA signaling rescues the epidermal hyperplasia in vivo. Thus, dermal PRC2 directed repression of the RA pathway is critical for the timely control of the epidermal proliferation and inhibition of accelerated differentiation. Altogether, these results demonstrate new and unexpected roles for dermal PRC2 to cell autonomously regulate the stepwise differentiation of dermal fibroblast cell types and non-cell autonomously regulate epidermal proliferation by maintaining critical levels of dermal Wnt/β-catenin and RA signaling.
Results
Ezh2 is active during early murine dermal development and can be efficiently ablated in dorsal dermal fibroblast precursors using PDGFRαCreER
To identify the spatio-temporal timing of PRC2 activity during dermal development, we queried PRC2-mediated H3K27me3 activity by immunostaining at various stages in mouse embryonic skin development (Fig. 1A). We observed persistent expression of H3K27me3 from as early as E11.5 through E17.5 in the differentiating DFs (Fig. 1B). Either Ezh1 or Ezh2 can catalyze the H3K27me3. However, Ezh1 is typically expressed in adult tissues, while Ezh2 is expressed in embryonic tissues suggesting that Ezh2 is the likely catalyzer in the developing dermis(Ezhkova et al., 2009; Laible et al., 1997; Margueron et al., 2008; Su et al., 2003). In line with this, we observed mesenchymal Ezh2 at E10.5 (Fig. S1A). This indicates that EZH2 is active during dermal development as the DF precursors begin differentiating into progenitors.
To address the role of Ezh2 in regulating DF differentiation, we used tamoxifen-inducible pan-mesenchymal PDGFRαCreER(Driskell et al., 2013; Rivers et al., 2008)to conditionally delete Ezh2 and activate Rosa26 β-galactosidase (R26R) in DF precursors and broadly in the dorsal mesenchyme at E8.5 and E9.5, (E8.5-DermEzh2) (Fig. 1A, 1C). As early as E10.5, we observed a 3.19-fold decrease in Ezh2 mRNA in DF precursors in E8.5-DermEzh2 as compared to Ezh2 heterozygous controls (Fig. S1A). The activity of PDGFRαCreER-mediated recombination of R26R is confined to the dorsal DF progenitors at E13.5 and absent in the epidermis as can be seen from β-gal staining (Fig. 1D, 1E). We visualized cells with Ezh2 methyltransferase activity, by determining the number of H3K27me3 expressing DF precursors and progenitors. We found an 85% decrease of H3K27me3+ DF precursor cells in E8.5-DermEzh2 mutants at E10.5 (Fig. 1F, G, H; n = 6 controls, 5 mutants) and 88% decrease in DF progenitors at E13.5 (Fig. S1B, C) while the keratinocyte H3K27me3 levels are unaffected at E10.5(Fig. S1D). This suggests that Ezh2 deletion in DF precursors is robust with our gavage regimen with PDGFRαCreER preceding DF differentiation.
Ezh2 deletion leads to upregulation of Wnt/β-catenin signaling and expansion of dermal fibroblast progenitors
Upon confirming the deletion of Ezh2 in DF precursors, we next sought to determine the effect of Ezh2 deletion on DF precursor differentiation into DF progenitors and the signals underlying dermal fibroblast fate selection (Fig. 1A, 2A’, 2A”). The expression domain of a pan-mesenchymal marker, PDGFRα, at E13.5 was comparable between control and E8.5-DermEzh2 suggesting that cells retained their mesenchymal identity (Fig. 2B, C; n= 4 controls, 4 mutants)(Driskell et al., 2013).We next sought to determine if DF precursor differentiation is altered in E8.5-DermEzh2 mutants. We examined the protein expression of DF progenitor markers, TWIST2 (DERMO1) and LEF1, at E12.5(Atit et al., 2006; Chen et al., 2012; Driskell et al., 2013; Ohtola et al., 2008; Phan et al., 2020; Rognoni et al., 2016; Tran et al., 2010). Compared to the control, the expression domain of TWIST2 and LEF1 proteins was expanded with an 84% increase in TWIST2+ cells (Fig. 2E–G; n = 4 controls, 8 mutants) and 40% increase in LEF1+ cells in the deeper layers of the E8.5-DermEzh2 dorsal mesenchyme (Fig. 2H–J; n = 3 controls, 5 mutants). DF progenitor cell density and cell proliferation at E12.5; and dermal thickness at E17.5 (Fig. S2A–E) were not significantly different between controls and the E8.5-DermEzh2 mutants. This indicates that dermal-Ezh2 deletion leads to an ectopic acquisition of deeper dorsal mesenchymal cells to DF progenitor identity.
Figure 2: Ezh2 restricts the specification of Wnt/ β-Catenin Signaling mediated DF progenitors to the dermis:
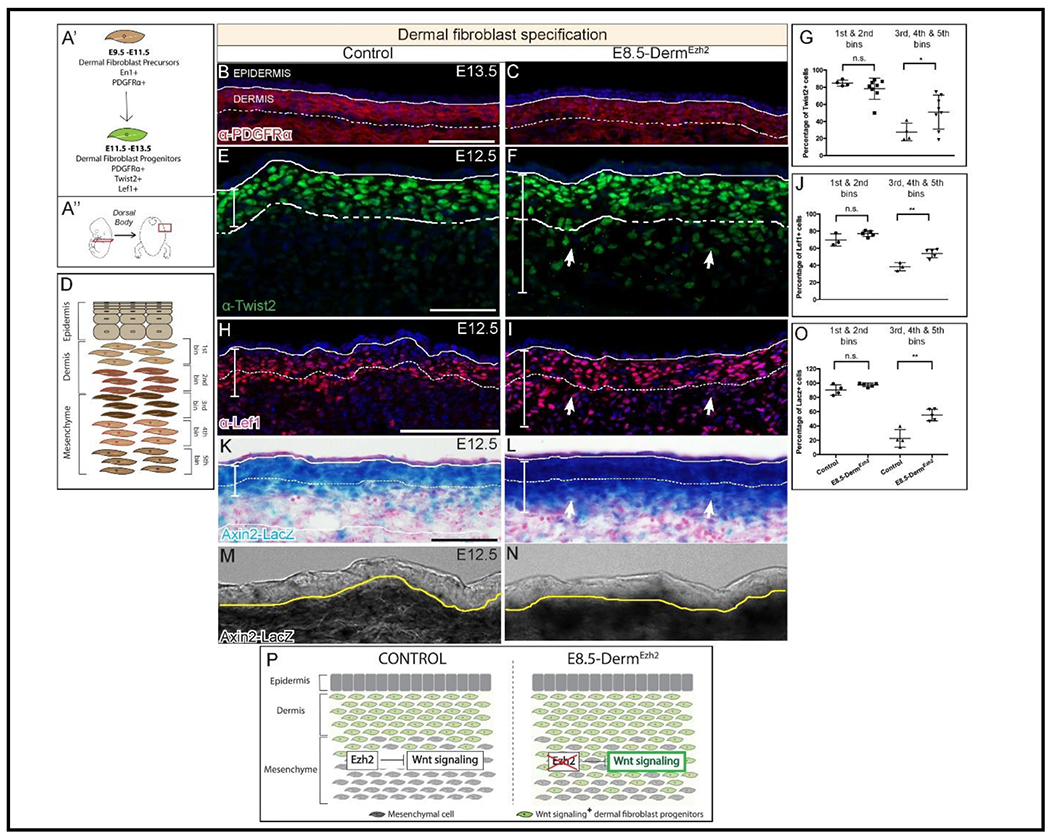
Schematic illustration of the temporal window (A’) and region of interest (A”). Indirect immunofluorescence with DAPI counterstain showing the expression of mesenchymal marker, PDGFRα (B, C), and dermal fibroblast progenitor markers TWIST2 (E, F), and LEF1 (H, I). Axin2-LacZ (β-galactosidase) staining (K, L) at E12.5 along with a digital zoom of a transmitted light channel using a confocal microscope (M, N). Schematic illustration of binning of dermis and dorsal mesenchyme employed to quantify cells (D). Quantification of TWIST2+ (G), LEF1+ (J), and β-galactosidase+ (O) cells in both controls and mutants. A summary schematic depicting the extent of Wnt/β-Catenin signaling domain in control and expansion in the E8.5-DermEzh2 mice (P). Solid line demarcates the epidermis from dermis and dashed line demarcates the lower limit of the dermis. Scale bar=100 microns in B, C, H, I, K, L and 50 microns in E and F.
Because Wnt/β-catenin signaling is known to be involved in cell fate decisions and both TWIST2 and LEF1 are among its targets, we further investigated whether there was an upregulation of Wnt/β-catenin_signaling(Chen et al., 2012; Ohtola et al., 2008; Phan et al., 2020; Rognoni et al., 2016; Tran et al., 2010). We utilized the Wnt/β-catenin signaling reporter, Axin2-LacZ, to look for changes in domains of Wnt signaling following loss of Ezh2(Jho et al., 2002; Yu et al., 2005). β-gal staining revealed an increase in the size of the domain of Axin2-LacZ expression in E8.5-DermEzh2 at E12.5. We identified a 144% increase in ectopic Axin2-LacZ+ cells in the deeper layers of the E8.5-DermEzh2 dorsal mesenchyme indicating an expansion of elevated canonical Wnt/β-catenin signaling (Figure 2K–P; n = 4 controls, 5 mutants). Thus, the increase in TWIST2+ and LEF1+ DF progenitors results from an expansion of Wnt/β-catenin signaling domain mediated differentiation of dorsal mesenchymal cells underlying the dermis. These data reveal a new role for dermal PRC2 in spatially restricting Wnt signaling activity and its target genes to DF progenitors in vivo during the dermal specification window.
Ablation of dermal Ezh2 promotes upregulation of some reticular progenitor markers and delays secondary dermal papilla formation
As early as E14.5, dermal fibroblast progenitors differentiate into its derivates such as PPs, RPs and DPs as seen by their distinct marker expression(Driskell et al., 2013; Thulabandu et al., 2018) (Fig. 1A). Many known signaling molecules and transcription factors that regulate dermal differentiation are targets of Wnt/β-catenin signaling(Driskell et al., 2013; Gupta et al., 2019; Philippeos et al., 2018; Tsai et al., 2014). From E12.5 to E14.5, the epidermis expresses canonical Wnt ligands which are required for Wnt/β-catenin signaling in DF progenitors and subsequently the Wnt-dependent differentiation of DF progenitors into PPs and DPs(Chen et al., 2012; Fu and Hsu, 2013).
Wnt/β-catenin signaling is upregulated in E8.5-DermEzh2 at E12.5 and persists until E13.5 as indicated by the relative quantity of Lef1 and Axin2 mRNA expression in whole skin (Fig. S2F, G, Table S1). The markers of PPs, RPs and DPs are known targets of Wnt/β-catenin signaling(Budnick et al., 2016; Chen et al., 2012) (Fig. 1A). So, we next examined the effect of increased Wnt/β-catenin signaling, due to Ezh2 ablation, on the differentiation of DF Progenitors (Fig. 1A). LRIG1 and TCF1 are markers of papillary progenitors, and DLK1 and TCF4 are markers of reticular progenitors(Driskell et al., 2013; Phan et al., 2020; Rognoni et al., 2016; Thulabandu et al., 2018). Histomorphometric analysis revealed that protein expression domain of LRIG1 and DLK1 was comparable between E16.5 controls and E8.5-DermEzh2 in the dermis (Fig S3A–G). At E17.5, though TCF1+ cells are comparable between controls and E8.5-DermEzh2 mutants (Fig 3A–C; n = 5 controls, 4 mutants), we found an increase of 35% in TCF4+ cells in the E8.5-DermEzh2 mutant dermis (Fig 3D–F; n = 3 controls, 4 mutants). In support of this result, we observed a 11-fold increase in the relative mRNA expression of another reticular progenitor marker, Adiponectin (Fig. 3H). However, there was no change in the relative quantities of other PP (Lrig1, Lef1, Prdm1) and RP markers (Dlk1, Colda1) between controls and E8.5-DermEzh2 mutants in whole skin at E16.5 (Fig. 3G, H). These embryos fail to survive birth for post-natal analysis (data not shown). Taken together, both histomorphometric and q-PCR analyses indicate that loss of Ezh2 promotes the expression of some RP lineage markers.
Figure 3: Dermal Ezh2 ablation promotes reticular dermal markers and delays secondary hair follicle initiation in E8.5-DermEzh2:
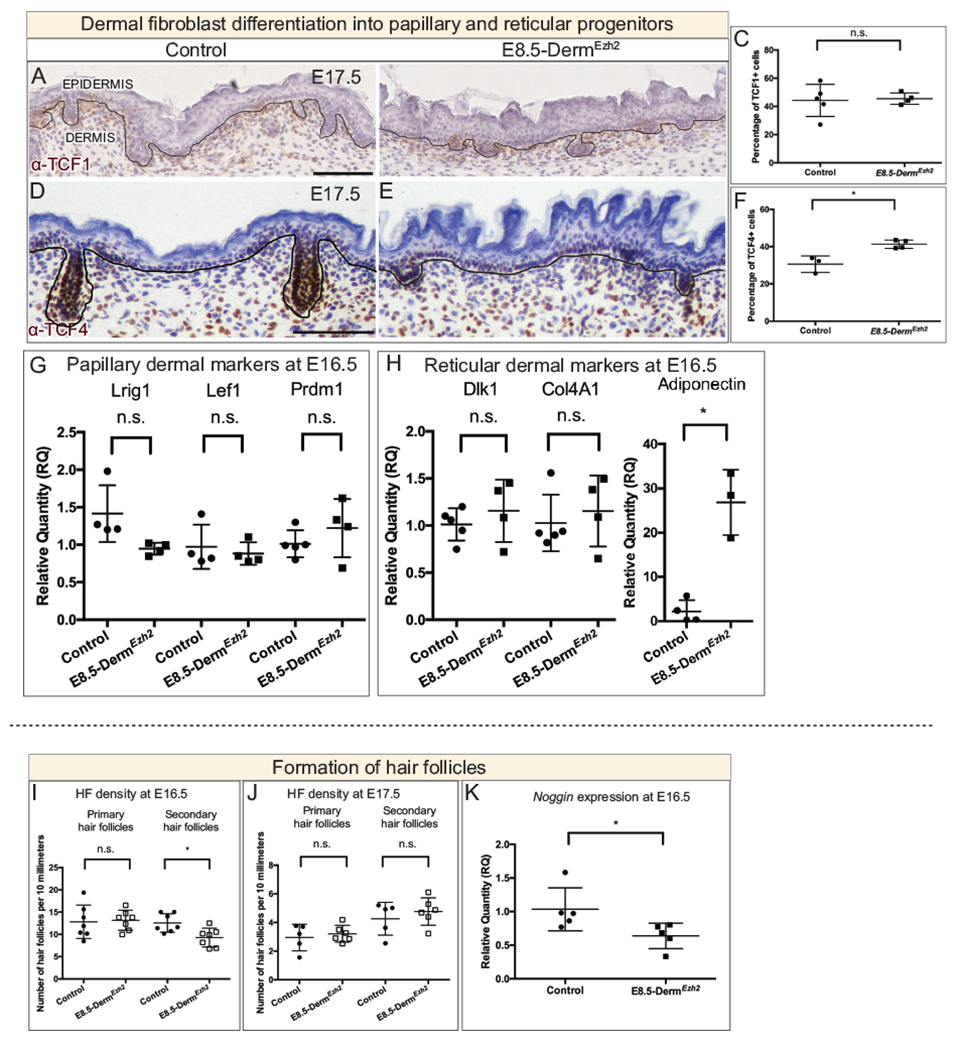
Immunohistochemistry (A, B, C, D) of a PP marker, TCF1 (A, B), and an RP marker, TCF4 (D, E), with hematoxylin counterstain. Quantification of TCF1+ (C) and TCF4+ (F) cells in the dermis. The relative quantity of papillary (G) and reticular (H) progenitor markers in E16.5 whole skin between controls and E8.5-DermEzh2. Hair follicle density at E16.5 (I) and E17.5 (J). The relative quantity of secondary HF initiation gene, Noggin (K). Solid line demarcates the epidermis from dermis. Scale bar=100 microns in A, B, D and E.
Next, we wanted to determine the effect on differentiation of DF progenitors towards hair follicle-forming dermal papilla fibroblasts. During development, the hair follicle-forming fibroblasts make dermal condensates in the dermis which initiate hair follicles (HF) by reciprocally signaling the overlying epidermal keratinocytes to differentiate into placodal cells(Chen et al., 2012; Dhouailly, 1975; Fu and Hsu, 2013; Kollar, 1970; Millar, 2005). HF initiation occurs in three waves: primary HF at E14.5, secondary HF at E16.5 and tertiary HF at E18.5(Schneider et al., 2009; Tsai et al., 2014). First, we assayed for DP differentiation by examining the expression domain of DP markers, Alkaline phosphatase(Chen et al., 2012), SOX2(Driskell et al., 2009), p75NTR(Rezza et al., 2016) and Tbx18(Rezza et al., 2016) which were comparable between controls and E8.5-DermEzh2 (Fig. S4A–E). Then, we assayed for placodal cell markers LEF1, SOX9 and Edar which were also comparable between controls and E8.5-DermEzh2 Fig. S4F–J). Additionally, we surveyed the stages and number of HF in transverse sections across the entire dorsum at the forelimb level. We observed that the primary HF density was comparable between controls and E8.5-DermEzh2 however, there was a 26% decrease in the secondary HF density at E16.5 (Fig. 3I). But by E17.5, the density of primary and secondary hair follicles is comparable between controls and E8.5-DermEzh2 (Fig. 3J). Noggin is required for secondary hair follicle initiation(Botchkarev et al., 2001; Botchkarev, 2003; Sharov et al., 2006). We found the relative quantities of Noggin mRNA was 38% lower in E8.5-DermEzh2 (Fig. 3K). These results suggest that dermal Ezh2 ablation delays secondary hair follicle initiation possibly by impacting NOGGIN-mediated gene network without affecting differentiation of DF progenitors. Thus, dermal PRC2-EZH2 activity is dispensable for primary hair follicle initiation and is required to regulate the gene network for secondary hair follicle initiation.
E8.5-DermEzh2 mutants exhibit epidermal hyperplasia
PRC2 activity is known to regulate various secreted factors crucial for epidermal differentiation(Mirzamohammadi et al., 2016; Wang et al., 2010). We next investigated the effect of dermal Ezh2 deletion on epidermal differentiation. We observed that E8.5-DermEzh2 mice exhibited hyperplastic epidermis throughout the dorsum skin at E16.5 (Fig 4 A, B). The epidermal hyperplasia is evident as early as E13.5 (Fig. S5A–C) and besides the dorsum, the hyperplasia exists at other locations of the skin such as head skin and vibrissae skin (Fig. S5D–G). We examined the expression of KERATIN14, KERATIN10, and FILLAGRIN that are respectively expressed in basal, spinous and granular layers of the E16.5 epidermis(Kulukian and Fuchs, 2013; Mack et al., 2005). We observed an expansion in the expression domain of all the three markers and it is more robust in the K10+ spinous layer (Fig. 4D, E, G–J). This hyperplasia is due to an increase in the number of cells: 37% increase in basal (K14+) and 41% increase in supra-basal (K10+ and FILAGGRIN+) layers (Fig. 4F, K; n = 4 controls, 4 mutants) and not due to periderm retention, which is known to delay epidermal barrier formation(M’Boneko and Merker, 1988; Okano et al., 2011). We found relative quantity of the periderm marker, Krtapl3, and barrier acquisition is comparable in control and E8.5-DermEzh2 skin at E17.5 by qPCR(Okano et al., 2011) (data not shown, Fig. S6A–D).
Figure 4: E8.5-DermEzh2 mice exhibit epidermal hyperplasia as a result of ectopic proliferation:
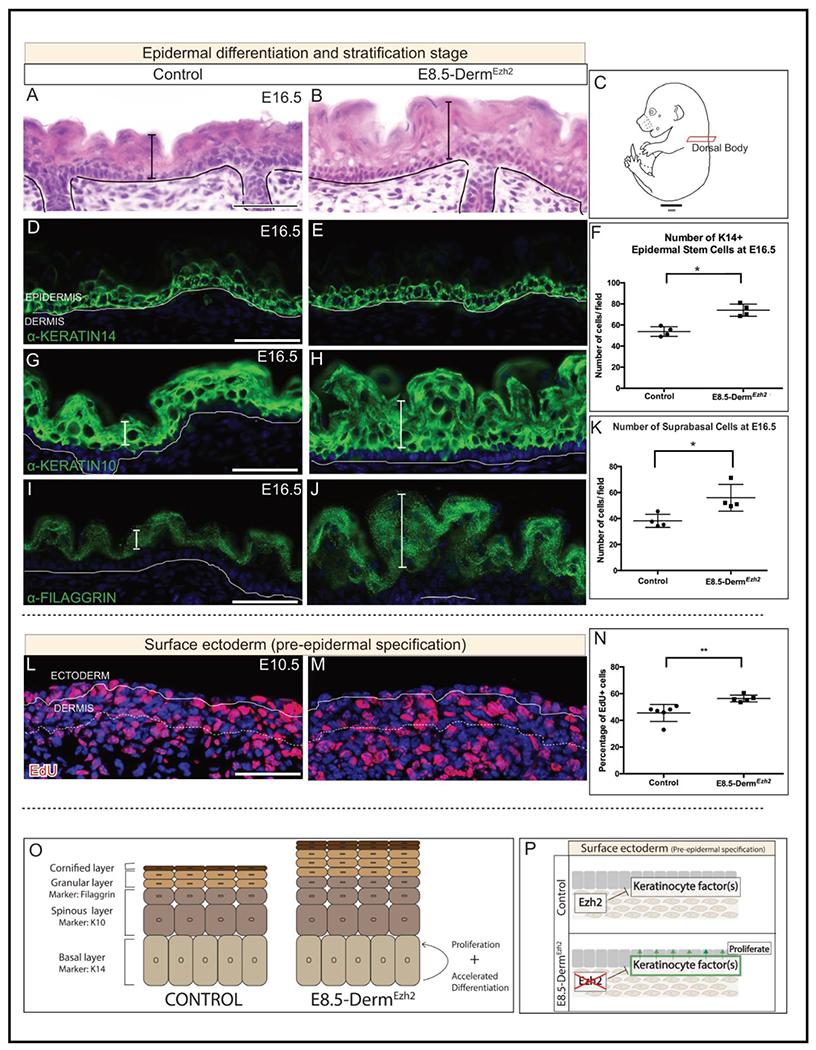
Hematoxylin and eosin staining of dorsal body (A, B) Schematic illustration of the regions of interest (C). Indirect immunofluorescence with DAPI counterstain at E16.5 (D, E, G-H, I, J) with markers of basal keratinocytes (D, E), spinous keratinocytes (G, H) and granular keratinocytes (I, J). Number of basal (F) and suprabasal cells per field (K) in both controls and mutants. Edu staining with DAPI counterstain at E10.5 (L, M). Proliferation index of EdU+ basal keratinocytes in controls and mutants (N). A summary schematic depicting the effect of dermal Ezh2 deletion on the epidermis (O). Schematic with the proposed model by which dermal Ezh2 ablation leads to epidermal hyperplasia (P). Solid line demarcates the epidermis from dermis and dashed line demarcates the lower limit of the dermis. Scale Bar= 50 microns.
We next sought to determine whether epidermal hyperplasia is a result of increased proliferation in E8.5-DermEzh2Epidermal hyperplasia is first observed at E13.5, therefore we analyzed the proliferation index of the surface ectoderm at E10.5 which are the precursors of epidermal basal keratinocytes. Compared to the average of 45.53% EdU+ proliferation index in controls, we found a significant increase at 56.37% in E8.5-DermEzh2 ectoderm (Fig. 4L, M, N; n = 6 controls, 5 mutants). Lastly, we also tested whether accelerated differentiation contributes to epidermal hyperplasia. In controls, FILLAGRIN protein is expressed in the granular layer at E16.5 in the controls (Fig. 4I). However, we found FILLAGRIN protein expression was expressed by E15.5 in E8.5-DermEzh2 mice (Fig. S6E, F). We identified that hyperplasia is a stage dependent phenotype where initiation of Ezh2 deletion at E9.5 doesn’t result in hyperplasic epidermis (Fig. S5H, I). These data together suggest that dermal Ezh2 ablation leads to elevated keratinocyte promoting factors in the skin that promote their proliferation and differentiation resulting in epidermal hyperplasia (Fig. 4O, P)
Dermal Ezh2 regulates cutaneous RA signaling which is sufficient to induce epidermal hyperplasia
Since dermal Ezh2 deletion leads to epidermal hyperplasia, we next took a candidate approach to investigate if known signaling factors involved in the regulation of epidermal proliferation and differentiation are dysregulated in E8.5-DermEzh2. Administration of RA results in epidermal proliferation and subsequent epidermal thickening in human skin(Kang et al., 1995) as seen in E8.5-DermEzh2. We queried whether RA signaling is dysregulated in E8.5-DermEzh2 skin. First, we utilized the transgenic RA signaling reporter allele, RARE-LacZ. β-gal staining revealed an increase in the intensity in E8.5-DermEzh2 dermis at E10.5 (Fig. 5A, B; n = 4 controls, 4 mutants). Second, we looked at the expression of an RA target, RAR-γ, and found that there was a clear increase in RAR-γ+ cells in both the epidermis and dermis at E12.5 (Fig. 5C, D; n = 3 controls, 3 mutants). Both these data suggest a possible elevation of RA signaling in E8.5-DermEzh2 skin.
Figure 5: Retinoic acid is regulated by Ezh2 and is sufficient to induce epidermal hyperplasia:
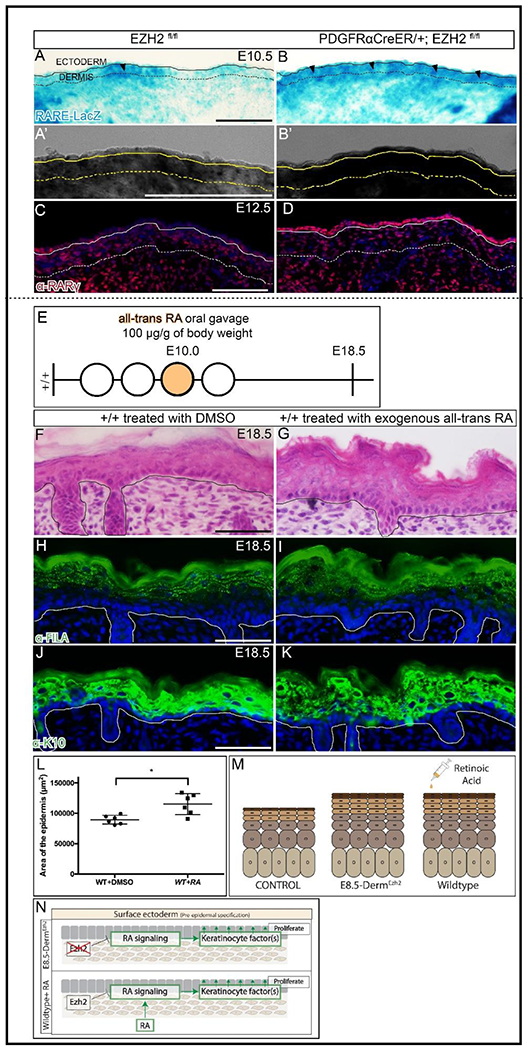
β-galactosidase staining of RARE-LacZ (A, B) along with a digital zoom of a transmitted light channel using a confocal microscope (A’, B’) at E10.5. Indirect immunofluorescence with DAPI counterstain showing the expression of RA signaling target gene, RARγ (C, D) at E12.5. Schematic illustration of Retinoic Acid (E) gavage regimen. Hematoxylin and eosin staining of dorsal skin (F, G) showing the histology of the epidermis. Indirect immunofluorescence of FILAGGRIN (H, I) and KERATIN10 (J, K) with DAPI counterstain at E18.5. Comparison of epidermal area between controls and mutants in Retinoic acid treated mice (L). A summary schematic depicting the epidermal hyperplasia phenotype (M) and the relationship between EZH2 and retinoic acid signaling (N). Solid line demarcates the epidermis from dermis and dashed line demarcates the lower limit of the dermis. Scale Bar= 100 microns in A-D and 50 microns in F-K.
Since retinoic acid signaling is possibly elevated in E8.5-DermEzh2 epidermis at E10.5, we first queried for the expression of key RA signaling components and found them to be comparable between controls and mutants. This is perhaps due to the transient action of RA signaling on the epidermis and is thus difficult to detect(Bok et al., 2011) (Table. S1, S2). Then, we tested the functional significance of this pathway, in causing epidermal hyperplasia, using a pharmacological approach. We administered a single dose of exogenous all-trans-RA (at-RA) orally to pregnant wild-type mice carrying E10.0 embryos, the time-point at which RA signaling is dysregulated in E8.5-DermEzh2 mutants (Fig. 5E). Compared to the vehicle-treated (DMSO) controls at E18.5, we found a 28.7% increase in epidermal area of at-RA-treated embryos (Fig. 5F, G, L; n = 6 controls, 6 mutants). Similar to E8.5-DermEzh2, at-RA-treated embryos demonstrated expansion of both K10+ spinous and FILAGGRIN+ granular layers of the epidermis at E18.5 (Fig. 5H–K). However in dermal fibroblast precursors, we found that RA signaling does not regulate the expression of PRC2 components as previously shown in osteoprogenitors(Ferguson et al., 2018) (Fig. S7A–D). These data demonstrate that a one-time administration of at-RA at E10.0 is sufficient to cause epidermal hyperplasia. In essence, dermal Ezh2 ablation leads to a possible elevation of RA signaling dependent keratinocyte factors which may mediate the epidermal hyperplasia seen in E8.5-DermEzh2 mutants.
Antagonizing RA signaling rescues epidermal hyperplasia in E8.5-DermEzh2 mice
To test that elevated RA signaling contributes to the epidermal hyperplasia observed in E8.5-DermEzh2 mice, we investigated whether antagonizing RA-signaling is sufficient to rescue epidermal hyperplasia. We orally administered BMS453, a small molecule inhibitor of RARα and RARγ16,58 that we validated in previous studies(Ferguson et al., 2018). At 3.5μg/g body weight dosing regimen of BMS453 (Fig. 6A), we rescued the epidermal area by 20.6% as compared to the vehicle-treated (DMSO) E8.5-DermEzh2 (Fig. 6B–F; n >= 4 for each genotype/treatment). Thus, the role of dermal Ezh2 is to ensure that RA signaling is carefully regulated to suppress keratinocyte targeted mitogenic factors and regulate normal epidermal proliferation.
Figure 6: Pharmacological inhibition of Retinoic acid signaling rescues the epidermal hyperplasia in E8.5-DermEzh2:
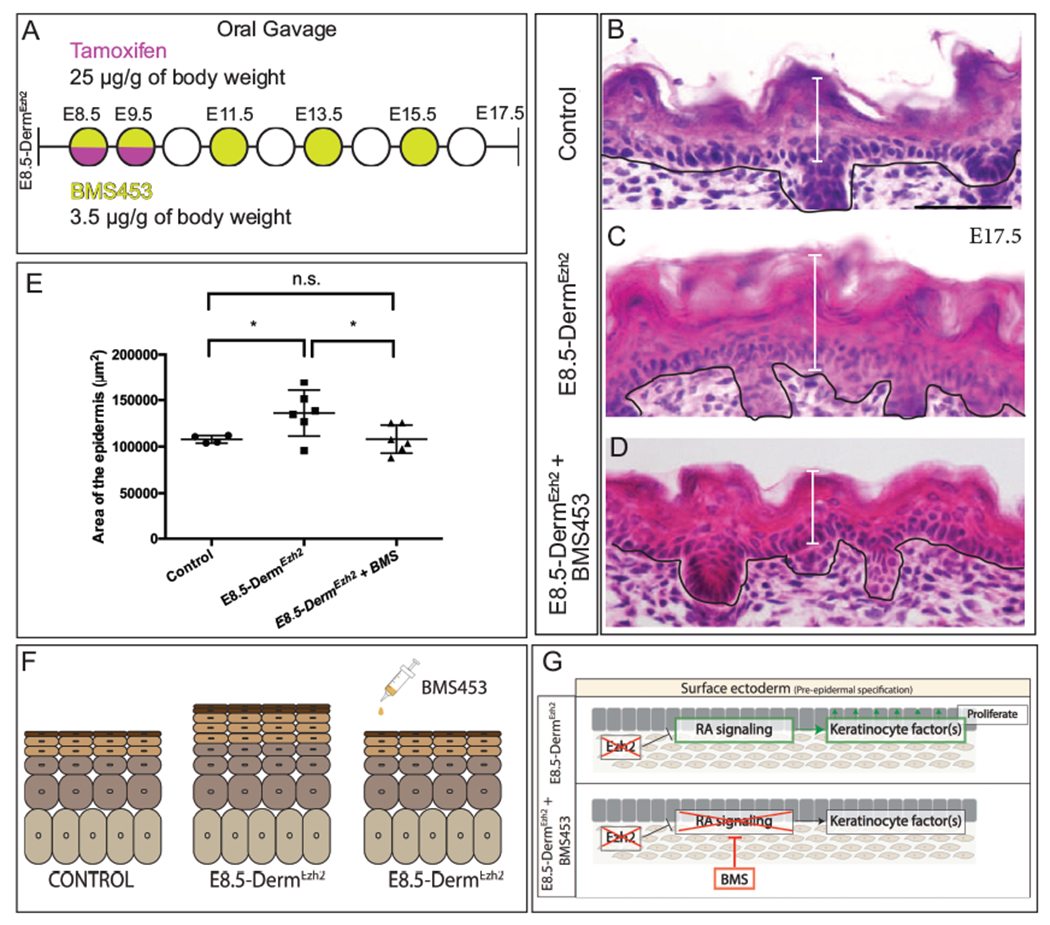
Schematic illustration BMS453 (A) gavage regimen. Hematoxylin and eosin staining of dorsal skin (B, C, D) showing the histology of the epidermis. Comparison of epidermal area between Controls, E8.5-DermEzh2 and BMS treated-E8.5-DermEzh2 mice (E). A summary schematic depicting the rescue of epidermal hyperplasia (F). The proposed model by which BMS453 is rescuing the hyperplasia in E8.5-DermEzh2 (G). Solid line demarcates the epidermal from the dermis. Scale Bar= 50 microns.
In summary, our data suggest a spatio-temporal regulation of RA and Wnt/β-catenin signaling by dermal EZH2 during dermal fibroblast differentiation and epidermal-dermal signaling. We show that ablation of Ezh2 (i) promotes Wnt/β-catenin signaling mediated differentiation of ectopic mesenchymal cells into DF progenitors (ii) promotes DF progenitor differentiation into RPs and (iii) delays secondary hair follicle formation (iv) promotes RA signaling which results in epidermal hyperplasia due to excess proliferation. Further, we rescued the epidermal hyperplasia in Ezh2 mutants by antagonizing RA signaling with BMS453.
Discussion
The role of PRC2 in directly regulating key developmental signaling factors and transcription factors is established in embryonic stem cells and tissue-specific stem cells(Boyer et al., 2006), but its role in coordinating spatio-temporal cell signaling events within and across tissue-specific progenitors is largely unexplored. Skin development and skin patterning with hair follicles is dependent on the coordination between the stepwise differentiation of dermal fibroblasts concomitant with dermal-epidermal reciprocal signaling. In this study, we examined the developmental role of dermal Ezh2 during dermal fibroblast cell fate selection and reciprocal signaling with the overlying epidermis. Here we report a new action of dermal EZH2 is acting like a rheostat of Wnt and RA cell signaling in the dermis. PRC2-EZH2 activity is required for spatially restricting Wnt/β-catenin signaling to reinforce DF specification and prevent lower dermal mesenchyme cells from erroneously adopting a dermal fibroblast fate. Additionally, we uncovered new roles for dermal Ezh2 to non-cell autonomously control epidermal proliferation and differentiation by maintaining critical RA signaling levels. We found that dermal PRC2 activity is a key component of the gene regulatory network of dermal fibroblasts differentiation and dermal-epidermal signaling.
Multiple studies have demonstrated the cell autonomous regulation of PRC2-EZH2 in various cell types. In the craniofacial mesenchyme, EZH2 directly represses anti-osteogenic factors such as Hox genes and Hand2 to allow for differentiation of osteoblasts into mineralized craniofacial bone(Dudakovic et al., 2015; Schwarz et al., 2014). Ezh2 deletion in limb mesenchyme causes an increase in cell death and alteration in axis specification factors(Wyngaarden et al., 2011). Eed ablation in committed cartilage progenitors, accelerates chondrocyte differentiation and premature ossification leading to a growth defect. Similarly, Ezh2 knockout in specified calvarial bone progenitors causes premature ossification of the suture mesenchyme causing craniosynostosis of the coronal suture(Dudakovic et al., 2015; Ferguson et al., 2018). In addition, Eun et al., (2015)(Eun et al., 2014) highlights the non-cell autonomous regulation of E(z), a homolog of EZH1/2 in drosophila, where ablation of E(z) in somatic gonad cells leads to somatic cell marker expression in germ cells. Although these studies demonstrated that EZH2 can exhibit cell and non-cell autonomous regulation in different contexts, they do not demonstrate whether and how EZH2 can simultaneously exhibit both during development. In this study, we found that dermal PRC2-EZH2 cell autonomously regulates differentiation of dermal fibroblast heterogeneity by inhibiting intrinsic effectors of Wnt/β-catenin signaling in the mesenchyme beneath the dermis. Simultaneously, PRC2-EZH2 non-cell autonomously regulates proliferation of epidermal basal keratinocytes by suppressing RA signaling mediated extrinsic factors.
Dermal EZH2 represses Wnt/β-catenin signaling activation in the mesenchymal cells, underlying dermis, during cell fate selection.
Wnt signaling plays a key role in mammalian embryonic dermal cell fate specification and later in differentiation of dermal condensate(Chen et al., 2012; Gupta et al., 2019; Tsai et al., 2014). Ectoderm Wnts are required for canonical Wnt signaling in mouse dermal fibroblasts precursors, however it is not known how the non-cell autonomous spatial restriction of Wnt signaling is achieved(Chen et al., 2012; Fu and Hsu, 2013). In this study we found, deletion of dermal Ezh2 causes expansion of Wnt signaling reporter and Wnt responsive-markers that are expressed by lineage-committed dermal fibroblast progenitor cells. However, the ectopic Wnt signaling is not sufficient to increase cell density and proliferation as seen in constitutive Wnt signaling mutants(Chen et al., 2012). Previous studies have demonstrated that Wnt/β-catenin signaling is required for dermal fibroblast cell fate(Atit et al., 2006; Chen et al., 2012; Ohtola et al., 2008; Tran et al., 2010). Wnt/β-catenin signaling at E11.5 is necessary and sufficient for Twist2 Dermo1 and Lef1 expression in DF progenitors, respectively(Atit et al., 2006; Chen et al., 2012; Ohtola et al., 2008; Tran et al., 2010). When Ezh2, a component of PRC2 is ablated in pre-adipocytes in vitro and chondrocytes in vivo, Wnts and its targets are ectopically de-repressed resulting in upregulation of Wnt/β-catenin signaling(Mirzamohammadi et al., 2016; Wang et al., 2010). Consistent with these studies, we show that dermal-Ehz2 ablation leads to-ectopic Wnt/β-catenin signaling at E12.5 which correlates with an expansion of TWIST2+ and LEF1+ domain of DF progenitors. Future studies will focus on rescuing the dermal Ezh2 mutants by restoring varying levels of Wnt signaling and its interaction with RA signaling using pharmacological and genetic approaches.
Both TCF1 and TCF4 are targets of Wnt signaling. It is important to note that these targets are differentially expressed depending on the level of Wnt signaling(Rudloff and Kemler, 2012). TCF1 is a Wnthi target gene and TCF4 is a Wntlo target gene. We found that E8.5-DermEzh2 mice have more TCF4+ cells and comparable TCF1+ cells with respect to controls. These data support the inference that a higher number of dermal cells are experiencing low levels of Wnt signaling ectopically. Previous studies have shown that upregulation of Wnt signaling by stabilizing β-catenin is sufficient to significantly expand the dermis. However, the dermal thickness of dermal Ezh2 mutants does not change, indicating that the level of upregulation of Wnt signaling seen in the dermal Ezh2 mutants is not sufficient to induce increase in dermal proliferation and thickening. This finding follows the “dimmer switch” model proposed by Nehila et al., (2020) where knocking out epigenetic repressors such as PRC2 does not completely eliminate the target activators/repressors, instead it serves as a rheostat to modulate expression level of activators/repressors(Nehila et al., 2020).
Loss of dermal Ezh2 promotes upregulation of reticular dermal markers and delays secondary hair follicle initiation.
Dermal fibroblast progenitors differentiate into PPs in the upper dermis, RPs in the lower dermis, and DC under hair follicle placodes(Driskell et al., 2013; Gupta et al., 2019; Philippeos et al., 2018). Driskell et al., (2013) (Driskell et al., 2013) showed that dermal fibroblast progenitors start to express distinct lineage markers in the dermis as early as E14.5. The markers of both PPs, RPs, and DC lineages are regulated by Wnt signaling as shown in published RNA sequencing dataset(Budnick et al., 2016; Philippeos et al., 2018). It is unclear from the literature on dermal Wnt signaling knockouts and constitutive Wnt signaling mutants when and how much Wnt signaling is required for differentiation of these dermal lineages and if epigenetic regulators control levels of dermal Wnt signaling(Chen et al., 2012; Fu and Hsu, 2013). The level of Wnt signaling elevation seen in Ezh2 knockout mice between E16.5 and E17.5 sufficiently induced changes in reticular dermal markers without altering papillary dermal markers. This can be explained in two possible ways: (i) that the threshold for modulating Wnt signaling is lower to induce changes in reticular dermal lineage markers than other lineages and/or (ii) Wnt signaling mediated PRC2 regulation of dermal markers at E16.5 is spatially restricted just to the reticular dermis while the papillary dermal marker regulation is independent of PRC2.
Ablation of β-catenin in DF precursors leads to a complete absence of DF progenitors and DP cells(Chen et al., 2012; Fu and Hsu, 2013) and stabilization of β-catenin leads to ectopic DP formation resulting in an increase in the number of hair follicles(Chen et al., 2012). In contrast to these studies, we found a delayed initiation of secondary hair follicles in dermal Ezh2 mutants. We show that the expression of DP markers, ALP and SOX2, are comparable between controls and E8.5-DermEzh2. So, two possible explanations of this effect are (i) that Ezh2 negatively impacts the gene regulatory network of secondary DP initiation, but not primary DP initiation or (ii) that dermal Ezh2 non-cell autonomously suppresses epidermal keratinocyte differentiation towards secondary follicular cell fate. These explanations are consistent with previous evidence suggesting that morphogenesis of the primary and secondary hair follicles is controlled by different mechanisms (Botchkarev et al., 2002; Headon and Overbeek, 1999; Zhang et al., 2009).
Loss of dermal Ezh2 induces ectopic proliferation and differentiation of epidermal keratinocytes in a non-cell autonomous manner
A unique phenotype of the dermal Ezh2 mutant is the epidermal hyperplasia due to an increase in proliferation and accelerated differentiation of basal keratinocytes in the epidermis. Our data strongly suggest that extrinsic factors from the dermis play a causal role in epidermal hyperplasia. Additionally, we found that exogenous at-RA exposure in WT embryos in vivo induced epidermal hyperplasia and RA signaling inhibitor treatment to E8.5-DermEzh2 mice rescued the hyperplasia. The association of embryonic murine epidermal hyperplasia and RA signaling was present in Cybp26b1−/− mutant and dermal Cyb26b1fl/del , a negative regulator of RA signaling(Okano et al., 2012).
There are several dermal signaling factors controlled by Ezh2 during skin development that can regulate epidermal proliferation and differentiation. Among these, we propose three plausible factors that may be involved in epidermal signaling thus causing RA-mediated hyperplasia, (i) Given that epidermal metabolites are solely transferred from the dermis, elevated transient RA signaling can result from the ectopic transfer of RA metabolite from dermis to the epidermis thus leading to excessive proliferation and accelerated differentiation(Al Tanoury et al., 2013; Levin, 1993; Noy and Blaner, 1991). (ii) PRC2 is known to epigenetically silence loci of Wnt ligands in mesenchymal derivatives, pre-adipocytes and chondrocytes(Mirzamohammadi et al., 2016; Wang et al., 2010). (iii) Wnt and RA signaling crosstalk in many contexts(Carpenter et al., 2015; Yasuhara et al., 2010). Thus, it is possible that Ezh2 ablation derepresses Wnts in the dermal mesenchyme and promotes RA-signaling effectors that further mediate keratinocyte proliferation and accelerated differentiation(Slavik et al., 2007). Future studies will need to combine genome-wide gene expression changes with chromatin confirmation changes in embryonic dermal fibroblasts to identify direct targets of dermal EZH2. Subsequent testing of candidate secreted factors with further contribute to our understanding of how dermal factors regulate epidermal development and differentiation. Additionally, future studies will focus on identifying dermal Ezh2 regulated RA-sensitive mediators and understanding the mechanism of BMS453 mediate rescue of epidermal hyperplasia.
In summary, our data supports that epigenetic regulators such as PRC2-EZH2 can simultaneously regulate intrinsic and extrinsic factors through different signaling pathways. Precisely, dermal Ezh2 regulates intrinsic factors such as TWIST2 and LEF1 via Wnt/β-catenin signaling to control DF progenitor differentiation. Aberrant activation of Wnt/β-catenin signaling occurs in dermal fibrosis and internal organ fibrosis(Bergmann and Distler, 2016; Hamburg and Atit, 2012). Thus, elevating PRC2 activity to repress Wnt signaling may be a new therapeutic approach to abrogate skin and organ fibrosis. Further, dermal Ezh2 directed down regulation of RA signaling mediated extrinsic factors is critical for the timely regulation of epidermal proliferation and differentiation. This approach can be employed to treat elevated RA signaling seen in psoriatic lesions(Didierjean et al., 1991; Siegenthaler et al., 1992, 1986). Our data are consistent that PRC2-EZH2 may serve as a new “dimmer switch” or rheostat. It modulates levels or spatial restriction of Wnt/β-catenin and RA signaling in vivo to impact cell differentiation and rate of proliferation in cell autonomous and non-autonomous ways.
Materials and Methods
Mice
The following mice were utilized for this project: PdgfrαCreER (JAX stock #018280)(Rivers et al., 2008), Rosa26-lacz reporter (JAX stock #003309)(Soriano, 1994), Ezh2 floxed (Ezh2fl/fl) (JAX stock #022616)(Shen et al., 2008), AxinLacZ reporter (JAX stock #009120)(Jho et al., 2002; Yu et al., 2005), and RARE-hsp68-LacZ reporter (JAX stock # 008477)(Rossant et al., 1991). Each of these genotypes was maintained in a mixed genetic background. The males and females were time mated and the vaginal plugs were checked daily. And a plug is assigned E0.5. Tamoxifen (Sigma Aldrich T5648) dissolved at 10mg/ml in com oil was gavaged to mice at 25μg/g of body weight on E8.5 and E9.5. For the rescue experiments, BMS453(Chen et al., 1995; Chung et al., 2011; Matt et al., 2003) (Cayman Chemical Company 19076) dissolved at 3.5μg/μl in DMSO and diluted in com oil was gavaged to mice at 3.5μg/g of body weight on E8.5, E9.5, E11.5, E13.5, and E15.5. To the wild type mice, at-retinoic acid (Sigma Aldrich R2625) dissolved at 100mg/ml in DMSO and diluted in peanut oil was gavaged to mice at 100μg/g of body weight on E9.5 and E10.5. For each experiment a minimum of 3 controls and 3 mutants were utilized. Case Western Reserve University Institutional Animal Care and Use Committee approved all animal procedures in accordance with AVMA guidelines (Protocol 2013–0156, Animal Welfare Assurance No. A3145-01).
Histology and stains
Cryosectioning: (i) For cryo embedding, we followed the protocol previously published(Atit et al., 2006; Ferguson et al., 2018).Once embedded, the embryos were cryo-sectioned at 10 μm and 20 μm thickness for LACZ staining in the transverse plane, (ii) For paraffin embedding, the embryos were fixed in 4% paraformaldehyde overnight, equilibrated with 70% ethanol overnight at 4°C, and processed for paraffin embedding. These embryos were then sectioned onto charged slides at 10 μm thickness.
Histological Stains: For β-Galactosidase detection, the cryo-sectioned slides were dried for 15mins, fixed with 4% paraformaldehyde for 5 mins and stained with X-gal compound (Amresco 0428) dissolved at 1mg/ml for 3-6hrs at 37°C as previously described(Atit et al., 2006). For Masson’s Trichrome staining, paraffin sectioned slides were stained with Weigert Iron Hematoxylin, Biebrich Scarlet-Acid Fuchsin Solution and Aniline Blue Solution using the standard protocol. For Alkaline phosphatase staining, cryosections were rinsed in PBS for 10 mins, fixed ice-cold Acetone on ice for 10 mins, then washed in NTMT (100mM Tris, pH9.4, 100mM NaCl, 60mM MgCl2, 0.02% Tween-20) for 10 mins at RT. Embryos were stained in 20μl/ml NBT/BCIP (Roche 11681451001) in the dark for 10 mins at RT. Slides were washed in PBS and mounted with aqueous mounting medium.
Immunostaining: For immunostaining, the slides were dried for 15 mins, washed in PBS, blocked with buffer (PBS + 1% BSA + 0.1% Tween20/Triton-X-100) with 10% goat or donkey serum for 1hr. Slides were incubated with primary antibodies overnight. Species-specific fluorescent or biotinylated secondary antibodies were incubated for 1hr to stain the slides. The primary antibodies utilized are listed in the resources table.
The EdU staining was implemented using the Click-iT Plus EdU kit (Thermo Fisher Scientific, C10639). The standard protocol suggested by the manufacturer was implemented.
Species-specific secondary antibodies utilized in this study are listed in the resources table. The TSA-Kit secondary antibody system (Perkin Elmer, NEL763E001KT) was utilized with Lef1 immunostaining.
Key resources table
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | Pdgfra-creER™ | JAX #018280 (Rivers et al., 2008) | Tg(Pdgfra-cre/ERT)467Dbe | https://www.jax.org/strain/018280 |
| Strain, strain background (Mus musculus) | R26R | JAX stock #003309 (Soriano, 1994) | Gt(ROSA)26Sortm1Sor | https://www.jax.org/strain/003309 |
| Strain, strain background (Mus musculus) | Ezh2fl | JAX stock #022616 (Shen et al., 2008) | Ezh2tm2Sho | https://www.jax.org/strain/022616 |
| Strain, strain background (Mus musculus) | ConductinlacZ | JAX stock #009120 (Jho et al., 2002; Yu et al., 2005) | Axin2tm1wbm | https://www.jax.org/strain/009120 |
| Strain, strain background (Mus musculus) | RARE-hsp68LacZ | JAX stock #008477 (Rossant et al., 1991) | Tg(RARE-Hspa1b/lacZ)12Jrt | https://www.jax.org/strain/008477 |
| Chemical agent | Tamoxifen | Sigma Aldrich | Cat: T5648 | 10mg/ml in corn oil |
| Chemical agent | BMS453 | Cayman chemical company | Cat: 19076 | 3.5μg/μl in DMSO |
| Chemical agent | all trans-retinoic acid | Sigma Aldrich | Cat: R2625 | 100mg/ml in DMSO |
| Antibody | rabbit anti-Twist2 | Abeam | Cat: ab66031 RRID:AB_2211862 | IF (1:200) |
| Antibody | goat anti-PDGFRα | R&D Systems | Cat: AF1062 RRID:AB_2236897 | IF (1:150) |
| Antibody | rabbit anti-H3K27me3 | Cell Signaling | Cat: 4395 RRID:AB_11220433 | IF (1:1000) |
| Antibody | rabbit anti-Lef1 | Cell Signaling | Cat: 2230, RRID:AB_823558 | IF (1:100) |
| Antibody | goat anti-Dlk1 | R&D Systems | Cat: AF1144 RRID:AB_2230586 | IF (1:50) |
| Antibody | goat anti-Lrig1 | R&D Systems | Cat: AF3688 RRID:AB_2138836 | IF (1:100) |
| Antibody | rabbit anti-TCF4 | Cell Signaling | Cat: 2569 RRID:AB_2199816 | IHC (1:100) |
| Antibody | rabbit anti-TCF1 | Cell Signaling | Cat: 2203 RRID:AB_2199302 | IHC (1:300) |
| Antibody | rabbit anti-K14 | Covance | Cat: PRB-155P RRID:AB_292096 | IF (1:2000) |
| Antibody | rabbit anti-K10 | Covance | Cat: PRB-159P RRID:AB_291580 | IF (1:500) |
| Antibody | rabbit anti-Filaggrin | Covance | Cat: PRB-417P RRID:AB_10064149 | IF (1:500) |
| Antibody | rabbit anti-Sox9 | Millipore | Cat: AB5535 RRID: AB_2239761_ | IF (1:1000) |
| Antibody | and rabbit anti-RARγ | Cell Signaling | Cat: D3A4 RRID:AB_10998934 | IF (1:1000) |
| Antibody | Goat Anti-Rabbit IgG (H+L) Antibody, Alexa Fluor 594 Conjugated | Molecular Probes | Cat: A-11012 RRID:AB_141359 | 1:800 (Secondary antibody) |
| Antibody | Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Molecular Probes | Cat: A-21207 RRID:AB_141637 | 1:500 (Secondary antibody) |
| Commercial assay or kit | TSA Cyanine 3 System - antibody amplification kit | Perkin Elmer | Cat: NEL704A001KT RRID:AB_2572409 | Kit |
| Commercial assay or kit | Click-iT Plus EdU kit | Thermo Fisher Scientific | Cat: C10639 | Kit |
| Sequence-based reagent | Ezh2 | Thermo Fisher Scientific | Taqman Assay Mm00468460_ml | qPCR primers Mus musculus |
| Sequence-based reagent | Cyp26b1 | Thermo Fisher Scientific | Taqman Assay Mm00558507_ml | qPCR primers Mus musculus |
| Sequence-based reagent | Twist2 | Thermo Fisher Scientific | Taqman Assay Mm00492147_ml | qPCR primers Mus musculus |
| Sequence-based reagent | Lef1 | Thermo Fisher Scientific | Taqman Assay Mm00550265_ml | qPCR primers Mus musculus |
| Sequence-based reagent | Twist1 | Thermo Fisher Scientific | Taqman Assay Mm00442036_ml | qPCR primers Mus musculus |
| Sequence-based reagent | Axin2 | Thermo Fisher Scientific | Taqman Assay Mm00443610_ml | qPCR primers Mus musculus |
| Sequence-based reagent | Suz12 | Thermo Fisher Scientific | Taqman Assay Mm01304152_ml | qPCR primers Mus musculus |
| Sequence-based reagent | Eed | Thermo Fisher Scientific | Taqman Assay Mm00469660_ml | qPCR primers Mus musculus |
| Sequence-based reagent | Tbx18 | Thermo Fisher Scientific | Taqman Assay Mm00470177_ml | qPCR primers Mus musculus |
| Sequence-based reagent | p75NTR | Thermo Fisher Scientific | Taqman Assay Mm00446296_ml | qPCR primers Mus musculus |
| Sequence-based reagent | Edar | Thermo Fisher Scientific | Taqman Assay Mm00839685_ml | qPCR primers Mus musculus |
| Sequence-based reagent | β-actin | Invitrogen | Cat:4352663 | qPCR primers Mus musculus |
| Software, algorithm | fiji-java6-20170530 | Schindelin et al., 2012 | RRID:SCR_002285 | https://fiji.sc/ |
| Software, algorithm | CellProfiler 3.1.5 | Carpenter et al., 2006 | RRID:SCR_007358 | http://cellnrofiler.org |
| Software, algorithm | Photoshop CC 19.1 | Adobe | RRID:SCR_014199 | https://www.adobe.com/products/photoshop.html |
| Software, algorithm | LAS X Core 3.5.7 | Leica Application Suite X | RRID:SCR_013673) | |
| Software, algorithm | Graphpad Prism v8.4.2 | Graphpad | SCR_002798 |
Images were taken on Olympus BX60 microscope with the Olympus DP70 digital camera using DC controller software. Confocal images were captured using Leica TCS SP8 (Leica Biosystems) using Application Suite X software (Leica Biosystems). Images were processed in Adobe Photoshop (www.adobe.com) , Fiji/ImageJ(Schindelin et al., 2012), and CellProfiler™ (Carpenter et al., 2006).
Barrier Assay: The embryos at E16 and E17.5 were equilibrated with a graded series of methanol and stained in 0.1% toluidine blue (Sigma-Aldrich T3260). The embryos were then washed in 1XPBS for 24hrs before recording the pictures.
Tissue harvesting and RT-qPCR
Ezh2fl/fl (control) and PdgfrαCreER; Ezh2fl/fl (mutant) were used to harvest skin for gene expression analyses. For the embryos staged between E10.5 and E12.5, the dorsal halves of the bodies devoid of internal organs (carcass) were harvested. And for the embryos staged between E13.5 and E17.5, the dorsal skin was collected.
Fluorescence Activated Cell Sorting (FACS): Just for flow sorting experiment, PdgfrαCreER; Rosa26-tdtomato; Ezh2fl/fl (heterozygous control) and PdgfrαCreER, Rosa26-tdtomato; Ezh2fl/fl (mutant) were used in place of aforementioned genotypes. The E10.5 tissue was collected directly into 0.25% Trypsin-EDTA (Thermo Fisher Scientific 25200056), minced, and homogenized for 4 mins at 37°C. The homogenized tissue was strained with 40μm filters (Fisher Scientific 22363547) and flow sorted into dermal (PdgfrαCreER; Rosa26-tdtomato positive) and epidermal (negative) fractions using FACS-ARIA-II (P65011000099) with an 85αm nozzle. Doublets and dead cells were excluded based on forward scatter, side scatter and DAPI (0.2 mg/ml) fluorescence.
Once the select tissue/cells were collected, RNA was isolated using RNeasy MinElute cleanup kit (Qiagen 74204) with DNase treatment (Qiagen 79254), and mRNA concentration was determined using Nanodrop (ND-8000-GL). Then the mRNA was converted to cDNA using the High Capacity RNA-to-cDNA Reverse Transcription Kit (Life Technologies, 4387406).
Relative quantity of mRNA expressed was determined by using 4ng of cDNA on a StepOnePlus Real-Time PCR System (Life Technologies). Commercially available Taqman probes from Invitrogen were utilized to implement q-PCR. CT values obtained were normalized to β-actin (Invitrogen 4352663) to obtain ΔCT values. Then, the ΔCT values were normalized to the average ΔCT values of the controls to obtain ΔΔCT values and relative mRNA levels(Schmittgen and Livak, 2008).
Histomorphometrics and automated quantification of protein expression in cells:
Demarcation of dermal boundary: (i) Between E10.5 and E13.5, we used the endogenous domain of TWIST2 expression in the controls to set the domains in the mutants and (ii) Between E16.5 and E17.5, we used the distance between the epidermis and the panniculus carnosus in the controls to set the domains in the mutants.
Dermal thickness: Photos of the Masson’s Trichrome stained tissue were loaded to Fiji(Schindelin et al., 2012). Using the line tool, a vertical line was drawn from the base of the epidermis (i) until the start of panniculus carnosus if present in the field (ii) until the end of the region with dense collagen staining if the panniculus carnosus was absent in the field. Lines were drawn at 5 fixed points in the field and measured for their length. The average of these 5 lengths represent the dermal thickness of the field. Each data point in the graph is an average obtained by analyzing between 3-6 sections per embryo.
Hair follicle staging: The hair follicles were staged as previously published(Hardy, 1992). Each data point in the graph is an average obtained by analyzing between 4-6 sections per embryo.
Epidermal area: The Hematoxylin and Eosin stained photos were loaded onto Fiji(Schindelin et al., 2012). Using a freeform line tool, a box was drawn around the epidermis and the area (μm2) of the box was obtained. Each data point in the graph is an average obtained by analyzing between 4-6 sections per embryo.
H3K27me3 immunostaining: Images were acquired on the Olympus BX-60 with DP72 camera. The first 4 layers of the dermis was manually cropped out of the photo using the Adobe Photoshop freeform pen tool. Then the resultant photo was manually counted for percent H3K27me3+ cells. Each data point in the graph is an average obtained by analyzing 2 sections per embryo.
TWIST2+ cells: Images were captured by Olympus BX-60 with DP72 camera at 20X. Using the freeform pen tool in Adobe Photoshop, three parallel lines were drawn to separate the epidermis, the first 6 layers of dermis, and the second 9 layers of dermis. Percent of cells that are TWIST2+ were manually counted in every region. Each data point in the graph is an average obtained by analyzing between 3-5 sections per embryo.
LEF1+ cells (at E12.5): Images were captured by Leica TCS SP8 gated STED 3X at 40X. Using the freeform pen tool in Adobe Photoshop, three parallel lines were drawn to crop the epidermis, the first 6 layers of dermis, and the underlying 9 layers of dermis into separate files. Using CellProfiler™ (Carpenter et al., 2006), we used these photos to count LEF1+ cells: (i) The red and blue channels were split (ii) Each channel was smoothened by Gaussian filter with automatic filter size (iii) the uneven illumination in each photo was calculated with a 60 block size and corrected (iii) The noise was reduced in blue channels (Size:7, Distance: 11, Cut-off distance: 0.065) and red channels (Size:7, Distance: 11, Cut-off distance: 0.09) using ‘ReduceNoise’ function (iv) The number of red and blue objects were identified with adaptive Otsu three-class thresholding with an object diameter between 5 and 40. Each data point in the graph is an average obtained by analyzing between 3-5 sections per embryo.
Axin-LacZ and Rare-LacZ staining: The DAPI fluorescence and LacZ staining in transmitted light were captured by Leica TCS SP8 gated STED 3X at 40X. The LacZ (transmitted light) channel was inverted and converted (pseudo-colored) to Red by default CellProfiler™ settings. The pseudo colored photo was then cropped using the freeform pen tool in Adobe Photoshop to separate the epidermis, the first 6 layers of dermis and the underlying 9 layers of dermis into separate files by drawing 3 parallel lines. Then these photos were used to count the DAPI and LacZ+ cells present in them. In order to do that, (i) the cropped photos were split into separate channels (ii) the DAPI and LacZ channels were both smoothened with a Gaussian diameter of 3 and rescaled the intensity to full (iii) Then DAPI objects were identified by adaptive Otsu three-class thresholding with an object diameter between 4 and 40 (iv) The LacZ objects were identified by manual thresholding of 0.35 and segmented by watershed method with default settings, (v) The DAPI objects that did not have 35% of their area overlapped with LacZ objects were masked (vi) The number of unmasked DAPI objects and the total DAPI objects were noted in a file. Each data point in the graph is an average obtained by analyzing between 3-6 sections per embryo.
DLK+ and LRIG1+ cells: The merged photos of DAPI and red channel were loaded to Fiji. The approximate size of a box that can enclose four layers of the dermis was determined by studying multiple photos. For each photo, 5 ‘region of interest’ files were created: 1 just with the epidermis and 4 non-overlapping boxes splitting the first 16 layers of the dermis. After creating these files, the fluorescence intensity in each region of interest was measured against the background. Then to count the number of cells in each region, the merged photo was manually loaded on to Fiji and (i) Splitted into RGB channels and the blue channel is selected (ii) Gaussian smoothed with a radius of 1 (iii) Thresholding of ‘Dark background’ is implemented with automatic settings (iv) segmented with watershed (v) The number of DAPI cells were identified by analyzing the particles with a size between 0.1 and infinity, and circularity between 0 and 1. Using the fluorescence intensity and number of DAPI cells in each region, the corrected fluorescence intensity was determined. Each data point in the graph is an average obtained by analyzing between 4-6 sections per embryo.
TCF1+ and TCF4+ cells: The brightfield photos were manually cropped to consist of just the dermis (devoid of hair follicles and other tissues). Color deconvolution plugin in Fiji was used to separate hematoxylin and DAB staining and each of the channels were saved in separate files. Then CellProfiler™ was used to count the number of hematoxylin and DAB cells by doing the following on both the files (i) The image was inverted to brighten the dark spots and vice versa (ii) The image was transformed to grayscale (iii) the noise was reduced in both DAB (Size: 7, Distance: 11, Cut-off distance: 0.075) and hematoxylin (Size:7, Distance: 11, Cut-off distance: 0.05) channels using ‘ReduceNoise’ function (iv) The number of DAB+ and Hematoxylin+ objects were identified with adaptive Otsu three class thresholding with an object diameter for DAB between 5 and 30 and for hematoxylin between 5 and 40. Each data point in the graph is an average obtained by analyzing between 4-6 sections per embryo.
LEF1 and SOX9 immunofluorescence (at E16.5): Images were captured by Olympus BX-60 with DP72 camera at 40X. Using the freeform pen tool in Adobe Photoshop, a line was drawn to separate the epidermis and the dermis. Each data point in the graph is an average obtained by analyzing between 3-5 sections per embryo.
Basal and suprabasal cell counts: The merged photos of DAPI and K14 staining were loaded to Adobe Photoshop. The cells that expressed K14 (basal cells) and the cells on top of K14+ cells (suprabasal cells) were manually counted. Each data point in the graph is an average obtained by analyzing between 4-6 sections per embryo.
EdU staining: At E10.5, images were taken on Olympus BX60 microscope with the Olympus DP70 digital camera. Using the freeform pen tool in Adobe Photoshop, 2 parallel lines were drawn to divide the epidermis and from the first 4 layers of dermis into separate files. And then the percent of Edu+ cells were manually counted. Each data point in the graph is an average obtained by analyzing between 4-6 sections per embryo.
Statistics:
All graphs (Mean ± SD) were generated using Prism 6 (GraphPad Software). All the statistical analyses were carried out in Microsoft excel using the Student’s t-test function using an unpaired, two-tail, with unequal variance. The p-values for statistical tests in all figures are represented as: * = P < 0.05, **= P < 0.01 and ***= P < 0.001.
Supplementary Material
FigureS1: PDGFRαCreER efficiently deletes mesenchymal Ezh2: The relative quantity of Ezh2 (A), in E10.5 flow sorted dorsal mesenchyme isolated from conditional heterozygous E8.5-DermEzh2 flox/+ controls and E8.5-DermEzh2. Indirect immunofluorescence of H3K27me3 with DAPI counterstain at E13.5 (B, C). Quantification of H3K27me3+ cells in the surface ectoderm at E10.5 (D). Solid line demarcates the epidermis from dermis and dashed line demarcates the lower limit of the dermis. Scale Bar= 50 microns in B and C.
FigureS2: Dermal density and thickness are comparable between controls and E8.5-DermEzh2 despite the persistence of Wnt signaling upregulation till E13.5. Dermal proliferation index at E10.5 (A) and cell density (B) at E12.5. Masson’s Trichrome staining (C, D) at E17.5 showing the thickness of the dermis and its quantification (E). The relative quantity of Wnt signaling target genes, Lef1 (F) and Axin2 (G) in E13.5 whole skin between controls and E8.5-DermEzh2. Scale Bar= 200 microns in C and D.
Figure S3: The PP marker, LRIG1, and the RP marker, DLK1, are comparable between controls and E8.5-DermEzh2 mutants: Indirect immunofluorescence with DAPI counterstain (A-D) showing papillary progenitor marker, LRIG1 (A, B), and reticular progenitor marker, DLK1 (C, D). Schematic illustration of binning of dermis employed to quantify LRIG1+/DLK1+ cells (E). Corrected fluorescence of LRIG1 (F) and DLK1 (G). Solid line demarcates the epidermis from dermis and dashed line demarcates the lower limit of the dermis. Scale Bar= 100 microns in A-D.
FigureS4: The dermal papilla marker expression is comparable between the controls and E8.5-DermEzh2 mice at E16.5. Alkaline phosphatase, a dermal papilla marker, staining sat E16.5 (A, B). Indirect immunofluorescence of dermal papilla marker, Sox2, with DAPI counterstain at E16.5 (C, D). The relative quantities of additional DP markers, Tbx18 and p75ntr (E). The relative quantity of a placode marker, Edar (F). Indirect immunofluorescence of additional placode markers, LEF1 (G, H) and SOX9 (I, J) with DAPI counterstain at E16.5. Solid line demarcates the epidermis from dermis. The arrows in G-J point towards the placode. Scale bar = 50 microns in A-D and G-J.
FigureS5: Epidermal hyperplasia in E8.5-DermEzh2 mice is evident at various locations and from E13.5 onwards: Hematoxylin and eosin staining of dorsal skin (A, B, D-I) showing the histology of the epidermis at E13.5 (A, B) and E16.5 (D-G). Quantification of the number of epidermal keratinocytes at E13.5 between controls and E8.5-DermEzh2 (C). Solid line demarcates the epidermis from dermis and dashed line demarcates the lower limit of the dermis. Scale Bar= 50 microns in A, B, D-G and 100 microns in H, I.
FigureS6: E8.5-DermEzh2 mice have intact barrier but exhibits accelerated differentiation: Whole mount toluidine blue O staining at E16 (A, B) and E17.5 (C, D). Indirect immunofluorescence of FILAGGRIN (E, F) with DAPI counterstain at E15.5. Solid line demarcates the epidermis from the dermis. Scale bar = 50 microns in A-D.
FigureS7: PRC2 components are not regulated by retinoic acid signaling. The relative quantity of Cyp26b1 (A), a target of RA signaling, and PRC2 components (B-D) in RA treated wild-type skin in comparison to vehicle control treated embryos at E10.5.
Dermal Ezh2 affects fibroblast heterogeneity
Dermal Ezh2 regulates epidermal proliferation leading to hyperplasia
Epidermal hyperplasia is rescued by RA signaling inhibitors
PRC2-EZH2 is acting as a rheostat to control Wnt/β-catenin and RA signaling levels
Acknowledgements:
Thanks to previous and current members of the Atit Lab for their input and feedback. We thank Gregg DiNuoscio and Vidhi Mendpara for genotyping animals. We thank Dr. Elena Ezhkhova, Dr. Peggy Myung, Dr. Martin Basch, and Dr. Claudia Mizutani for critical feedback. We thank Brian Zhang, Celine Opdycke and Charlotte Lo for help with manual and automated counting. We thank the SOM Light Microscopy Core Facility and the CWRU bio[box] shared instrumentation facility.
Funding:
This work was supported by NIH National Institute of Dental and Craniofacial Research: R01-01870 (RA) and NIH-National Institute of Arthritis, Musculoskeletal, and Skin Disease: R01-AR076938 (RA), NM-T32 AR-007505 (J.F.), and the NIH Grant S10-OD016164 CWRU SOM Light Microscopy Core Facility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest:
The authors declare that they have no competing interests.
References:
- Al Tanoury Z, Piskunov A, Rochette-Egly C, 2013. Vitamin a and retinoid signaling: Genomic and nongenomic effects. J. Lipid Res 54, 1761–1775. 10.1194/jlr.R030833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda S, Mas G, Croce L. Di, 2015. Regulation of gene transcription by Polycomb proteins 2, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA, 2006. B-Catenin Activation Is Necessary and Sufficient To Specify the Dorsal Dermal Fate in the Mouse. Dev. Biol 296, 164–176. 10.1016/j.ydbio.2006.04.449 [DOI] [PubMed] [Google Scholar]
- Bardot ES, Valdes VJ, Zhang J, Perdigoto CN, Nicolis S, Hearn SA, Silva JM, Ezhkova E, 2013. Polycomb subunits Ezh1 and Ezh2 regulate the Merkel cell differentiation program in skin stem cells. EMBO J. 32, 1990–2000. 10.1038/emboj.2013.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann C, Distler JHW, 2016. Canonical Wnt signaling in systemic sclerosis. Lab. Investig 96, 151–155. 10.1038/labinvest.2015.154 [DOI] [PubMed] [Google Scholar]
- Bok J, Raft S, Kong KA, Koo SK, Dräger UC, Wu DK, 2011. Transient retinoic acid signaling confers anterior-posterior polarity to the inner ear. Proc. Natl. Acad. Sci. U. S. A 108, 161–166. 10.1073/pnas.1010547108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev V. a., Botchkarev NV, Nakamura M, Huber O, Funa K, Lauster R, Paus R, Gilchrest B. a., 2001. Noggin is required for induction of the hair follicle. FASEB J. 15, 2205–2214. [DOI] [PubMed] [Google Scholar]
- Botchkarev VA, 2003. Bone morphogenetic proteins and their antagonists in skin and hair follicle biology. J. Invest. Dermatol 120, 36–47. 10.1046/j.1523-1747.2003.12002.x [DOI] [PubMed] [Google Scholar]
- Botchkarev VA, Botchkareva NV, Sharov AA, Gilchrest BA, Funa K, Huber O, 2002. Modulation of BMP signaling by noggin is required for induction of the secondary (Nontylotrich) hair follicles. J. Invest. Dermatol 118, 3–10. 10.1046/j.1523-1747.2002.01645.x [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R, 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353. 10.1038/nature04733 [DOI] [PubMed] [Google Scholar]
- Budnick I, Hamburg-Shields E, Chen D, Torre E, Jarrell A, Akhtar-Zaidi B, Cordovan O , Spitale RC, Scacheri P, Atit RP, 2016. Defining the identity of mouse embryonic dermal fibroblasts. Genesis 54, 415–430. 10.1002/dvg.22952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AC, Smith AN, Wagner H, Cohen-Taya Y, Rao S, Wallace V, Ashery-Padan R, Lang RA, 2015. Wnt ligands from the embryonic surface ectoderm regulate ‘bimetallic strip’ optic cup morphogenesis in mouse. Dev. 142, 972–982. 10.1242/dev.120022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Gotland P, Sabatini DM, 2006. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol, 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Jarrell a., Guo C, Lang R, Atit R, 2012. Dermal -catenin activity in response to epidermal Wnt ligands is required for fibroblast proliferation and hair follicle initiation. Development 139, 1522–1533. 10.1242/dev.076463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Penco S, Ostrowski J, Balaguer P, Pons M, Starrett JE, Reczek P, Chambon P, Gronemeyer H, 1995. RAR-specific agonist/antagonists which dissociate transactivation and API transrepression inhibit anchorage-independent cell proliferation. EMBO J. 14, 1187–1197. 10.1002/j.1460-2075.1995.tb07102.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin T, Cavalli G, 2014. Polycomb silencing: From linear chromatin domains to 3D chromosome folding. Curr. Opin. Genet. Dev 25, 30–37. 10.1016/j.gde.2013.ll.016 [DOI] [PubMed] [Google Scholar]
- Chung SSW, Wang X, Roberts SS, Griffey SM, Reczek PR, Wolgemuth DJ, 2011. Oral administration of a retinoic acid receptor antagonist reversibly inhibits spermatogenesis in mice. Endocrinology 152, 2492–2502. 10.1210/en.2010-0941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieślik M, Hoang SA, Baranova N, Chodaparambil S, Kumar M, Allison DF, Xu X, Wamsley JJ, Gray L, Jones DR, Mayo MW, Bekiranov S, 2013. Epigenetic coordination of signaling pathways during the epithelial-mesenchymal transition. Epigenetics and Chromatin 6, 1–22. 10.1186/1756-8935-6-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudet N, Ariza-McNaughton L, Lewis J, 2007. Notch signalling is needed to maintain, but not to initiate, the formation of prosensory patches in the chick inner ear. Development. 10.1242/dev.001842 [DOI] [PubMed] [Google Scholar]
- Daudet N, Lewis J, 2005. Two contrasting roles for Notch activity in chick inner ear development: Specification of prosensory patches and lateral inhibition of hair-cell differentiation. Development, 10.1242/dev.01589 [DOI] [PubMed] [Google Scholar]
- Dhouailly D, 1975. Formation of cutaneous appendages in dermo-epidermal recombinations between reptiles, birds and mammals. Wilhelm Roux’s Arch. Dev. Biol 10.1007/BF00848183 [DOI] [PubMed] [Google Scholar]
- Didierjean L, Durand B, Saurat J-H, 1991. Cellular retinoic acid-binding protein type 2 mRNA is overexpressed in human psoriatic skin as shown by in situ hybridization. Biochem. Biophys. Res. Commun 180, 204–208. 10.1016/S0006-291X(05)81277-X [DOI] [PubMed] [Google Scholar]
- Driskell RR, Giangreco A, Jensen KB, Mulder KW, Watt FM, 2009. Sox2-positive dermal papilla cells specify hair follicle type in mammalian epidermis. Development 136, 2815–2823. 10.1242/dev.038620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell RR, Jahoda CAB, Chuong CM, Watt FM, Horsley V, 2014. Defining dermal adipose tissue. Exp. Dermatol 23, 629–631. 10.1111/exd.12450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell RR, Lichtenberger BM, Hoste E, Kretzschmar K, Simons BD, Charalambous M, Ferron SR, Herault Y, Pavlovic G, Ferguson-Smith AC, Watt FM, 2013. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature 504, 277–81. 10.1038/naturel2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell RR, Watt FM, 2015. Understanding fibroblast heterogeneity in the skin. Trends Cell Biol. 25, 92–99. 10.1016/j.tcb.2014.10.001 [DOI] [PubMed] [Google Scholar]
- Dudakovic A, Camilleri ET, Xu F, Riester SM, McGee-Lawrence ME, Bradley EW, Paradise CR, Lewallen EA, Thaler R, Deyle DR, Larson AN, Lewallen DG, Dietz AB, Stein GS, Montecino MA, Westendorf JJ, Van Wijnen AJ, 2015. Epigenetic control of skeletal development by the histone methyltransferase Ezh2. J. Biol. Chem 290, 27604–27617. 10.1074/jbc.M115.672345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun SH, Shi Z, Cui K, Zhao K, Chen X, 2014. A Non-Cell Autonomous Role of E(z) to Prevent Germ Cells from Turning on a Somatic Cell Marker. Science (80-.). 343, 1513–1516. 10.1126/science.1246514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su I. hsin, Hannon G, Tarakhovsky A, Fuchs E, 2009. Ezh2 Orchestrates Gene Expression for the Stepwise Differentiation of Tissue-Specific Stem Cells. Cell 136, 1122–1135. 10.1016/j.cell.2008.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson JW, Devarajan M, Atit RP, 2018. Stage-specific roles of Ezh2 and Retinoic acid signaling ensure calvarial bone lineage commitment. Dev. Biol 443, 173–187. 10.1016/j.ydbio.2018.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Hsu W, 2013. Epidermal Wnt controls hair follicle induction by orchestrating dynamic signaling crosstalk between the epidermis and dermis. J. Invest. Dermatol 133, 890–898. 10.1038/jid.2012.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K, Levinsohn J, Linderman G, Chen D, Sun TY, Dong D, Taketo MM, Bosenberg M, Kluger Y, Choate K, Myung P, 2019. Single-Cell Analysis Reveals a Hair Follicle Dermal Niche Molecular Differentiation Trajectory that Begins Prior to Morphogenesis. Dev. Cell 48, 17–31.e6. 10.1016/j.devcel.2018.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburg EJ, Atit RP, 2012. Sustained B-catenin activity in dermal fibroblasts is sufficient for skin fibrosis. J. Invest. Dermatol 132, 2469–2472. 10.1038/jid.2012.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy MH, 1992. The secret life of the hair follicle. Trends Genet. 10.1016/0168-9525(92)90350-0 [DOI] [PubMed] [Google Scholar]
- Headon DJ, Overbeek PA, 1999. Involvement of a novel Tnf receptor homologue in hair follicle induction. Nat. Genet 22, 370–374. 10.1038/11943 [DOI] [PubMed] [Google Scholar]
- Healy C, Uwanogho D, Sharpe PT, 1999. Regulation and role of Sox9 in cartilage formation. Dev. Dyn [DOI] [PubMed] [Google Scholar]
- Ito M, Yang Z, Andl T, Cui C, Kim N, Millar SE, Cotsarelis G, 2007. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature 447, 316–320. 10.1038/nature05766 [DOI] [PubMed] [Google Scholar]
- Ito T, Cidaka N, Yazawa T, Okudela K, Hayashi H, Sudo T, Guillemot F, Kageyama R, Kitamura H, 2000. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development. [DOI] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Zaret KS, 2016. Cell fate control by pioneer transcription factors. Dev. 143, 1833–1837. 10.1242/dev.133900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jho E. -h., Zhang T, Domon C, Joo C-K, Freund J-N, Costantini F, 2002. Wnt/ -Catenin/Tcf Signaling Induces the Transcription of Axin2, a Negative Regulator of the Signaling Pathway. Mol. Cell. Biol 22, 1172–1183. 10.1128/mcb.22.4.1172-1183.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Duell EA, Fisher GJ, Datta SC, Wang ZQ, Reddy AP, Tavakkol A, Yi JY, Griffiths CEM, Elder JT, Voorhees JJ, 1995. Application of retinol to human skin in vivo induces epidermal hyperplasia and cellular retinoid binding proteins characteristic of retinoic acid but without measurable retinoic acid levels or irritation. J. Invest. Dermatol 105, 549–556. 10.1111/1523-1747.epl2323445 [DOI] [PubMed] [Google Scholar]
- Kollar EJ, 1970. The induction of hair follicles by embryonic dermal papillae. J. Invest. Dermatol 55, 374–378. 10.1111/1523-1747.ep12260492 [DOI] [PubMed] [Google Scholar]
- Kulukian A, Fuchs E, 2013. Spindle orientation and epidermal morphogenesis. Philos. Trans. R. Soc. Lond. B. Biol. Sci 368, 20130016. 10.1098/rstb.2013.0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T, 1997. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 16, 3219–3232. 10.1093/emboj/16.11.3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugesen A, Helin K, 2014. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell 14, 735–751. 10.1016/j.stem.2014.05.006 [DOI] [PubMed] [Google Scholar]
- Levin MS, 1993. Cellular retinol-binding proteins are determinants of retinol uptake and metabolism in stably transfected Caco-2 cells. J. Biol. Chem 268, 8267–8276. [PubMed] [Google Scholar]
- M’Boneko V, Merker HJ, 1988. Development and morphology of the periderm of mouse embryos (days 9-12 of gestation). Acta Anat. (Basel) [DOI] [PubMed] [Google Scholar]
- Mack JA, Anand S, Maytin EV, 2005. Proliferation and cornification during development of the mammalian epidermis. Birth Defects Res. Part C - Embryo Today Rev. 75, 314–329. 10.1002/bdrc.20055 [DOI] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D, 2008. Ezh1 and Ezh2 Maintain Repressive Chromatin through Different Mechanisms. Mol. Cell 32, 503–518. 10.1016/j.molcel.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matt N, Ghyselinck NB, Wendling O, Chambon P, Mark M, 2003. Retinoic acid-induced developmental defects are mediated by RARβ/RXR heterodimers in the pharyngeal endoderm. Development 130, 2083–2093. 10.1242/dev.00428 [DOI] [PubMed] [Google Scholar]
- Millar SE, 2005. An ideal society? Neighbors of diverse origins interact to create and maintain complex mini-organs in the skin. PLoS Biol. 3, 1873–1877. 10.1371/joumal.pbio.0030372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzamohammadi F, Papaioannou G, Inloes JB, Rankin EB, Xie H, Schipani E, Orkin SH, Kobayashi T, 2016. Polycomb repressive complex 2 regulates skeletal growth by suppressing Wnt and TGF-β signalling. Nat. Commun 7, 12047. 10.1038/ncomms12047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad HP, Baylin SB, 2010. Linking cell signaling and the epigenetic machinery. Nat. Biotechnol 10.1038/nbt1010-1033 [DOI] [PubMed] [Google Scholar]
- Mok KW, Saxena N, Heitman N, Grisanti L, Srivastava D, Muraro MJ, Jacob T, Sennett R, Wang Z, Su Y, Yang LM, Ma’ayan A, Omitz DM, Kasper M, Rendl M, 2019. Dermal Condensate Niche Fate Specification Occurs Prior to Formation and Is Placode Progenitor Dependent. Dev. Cell 48, 32–48.e5. 10.1016/j.devcel.2018.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehila T, Ferguson JW, Atit RP, 2020. Polycomb Repressive Complex 2: a Dimmer Switch of Gene Regulation in Calvarial Bone Development. Curr. Osteoporos. Rep 18, 378–387. 10.1007/s11914-020-00603-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy N, Blaner WS, 1991. Interactions of Retinol with Binding Proteins: Studies with Rat Cellular Retinol-Binding Protein and with Rat Retinol-Binding Protein. Biochemistry. 10.1021/bi00240a005 [DOI] [PubMed] [Google Scholar]
- Ohtola J, Myers J, Akhtar-Zaidi B, Zuzindlak D, Sandesara P, Yeh K, Mackem S, Atit R, 2008. -Catenin has sequential roles in the survival and specification of ventral dermis. Development 135, 2321–2329. 10.1242/dev.021170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano J, Levy C, Lichti U, Sun H-W, Yuspa SH, Sakai Y, Morasso MI, 2012. Cutaneous Retinoic Acid Levels Determine Hair Follicle Development and Downgrowth. J. Biol. Chem 287, 39304–39315. 10.1074/jbc.M112.397273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano J, Lichti U, Mamiya S, Aronova M, Zhang G, Yuspa SH, Hamada H, Sakai Y, Morasso MF, 2011. Increased retinoic acid levels through ablation of Cyp26b1 determine the processes of embryonic skin barrier formation and peridermal development 1. 10.1242/jcs.101550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdigoto CN, Dauber KL, Bar C, Tsai PC, Valdes VJ, Cohen I, Santoriello FJ, Zhao D, Zheng D, Hsu YC, Ezhkova E, 2016. Polycomb-Mediated Repression and Sonic Hedgehog Signaling Interact to Regulate Merkel Cell Specification during Skin Development. PLoS Genet. 12, 1–27. 10.1371/journal.pgen.1006151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan QM, Fine GM, Salz L, Herrera GG, Wildman B, Driskell IM, Driskell RR, 2020. Lef1 expression in fibroblasts maintains developmental potential in adult skin to regenerate wounds. Elife 9, 1–19. 10.7554/ELIFE.60066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippeos C, Telerman SB, Oulès B, Pisco AO, Shaw TJ, Elgueta R, Lombardi G, Driskell RR, Soldin M, Lynch MD, Watt FM, 2018. Spatial and Single-Cell Transcriptional Profiling Identifies Functionally Distinct Human Dermal Fibroblast Subpopulations. J. Invest. Dermatol 138, 811–825. 10.1016/j.jid.2018.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus MV, Mayer JA, de la Cruz D, Baker RE, Maini PK, Maxson R, Chuong C-M, 2008. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature 451, 340–4. 10.1038/nature06457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendl M, Polak L, Fuchs E, 2008. BMP signaling in dermal papilla cells is required for their hair follicle-inductive properties. Genes Dev. 22, 543–557. 10.1101/gad.1614408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezza A, Wang Z, Sennett R, Qiao W, Wang D, Heitman N, Mok KW, Clavel C, Yi R, Zandstra P, Ma’ayan A, Rendl M, 2016. Signaling Networks among Stem Cell Precursors, Transit-Amplifying Progenitors, and their Niche in Developing Hair Follicles. Cell Rep. 14, 3001–3018. 10.1016/j.celrep.2016.02.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers LE, Young KM, Rizzi M, Jamen F, Psachoulia K, Wade A, Kessaris N, Richardson WD, 2008. PDGFRA / NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice 11, 1392–1401. 10.1038/nn.2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognoni E, Gomez C, Pisco AO, Rawlins EL, Simons BD, Watt FM, Driskell RR, 2016. Inhibition of β-catenin signalling in dermal fibroblasts enhances hair follicle regeneration during wound healing. Development dev. 131797. 10.1242/dev.131797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, Zirngibl R, Cado D, Shago M, Giguere V, 1991. Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 5, 1333–1344. 10.1101/gad.5.8.1333 [DOI] [PubMed] [Google Scholar]
- Rudloff S, Kemler R, 2012. Differential requirements for β-catenin during mouse development. Dev. 139, 3711–3721. 10.1242/dev.085597 [DOI] [PubMed] [Google Scholar]
- Werner S and Grose R, 2011. Regulation of wound healing by growth factors and cytokines. Wound Heal. Process. Phases Promot 73–93. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ, 2008. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc 3, 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Schneider MR, Schmidt-Ullrich R, Paus R, 2009. The Hair Follicle as a Dynamic Miniorgan. Curr. Biol 19, R132–R142. 10.1016/j.cub.2008.12.005 [DOI] [PubMed] [Google Scholar]
- Schwarz D, Varum S, Zemke M, Schöler A, Baggiolini A, Draganova K, Koseki H, Schiibeler D, Sommer L, 2014. Ezh2 is required for neural crest-derived cartilage and bone formation. Dev. 141, 867–877. 10.1242/dev.094342 [DOI] [PubMed] [Google Scholar]
- Sharov AA, Sharova TY, Mardaryev AN, Di Vignano AT, Atoyan R, Weiner L, Yang S, Brissette JL, Dotto GP, Botchkarev VA, 2006. Bone morphogenetic protein signaling regulates the size of hair follicles and modulates the expression of cell cycle-associated genes. Proc. Natl. Acad. Sci. U. S. A 103, 18166–18171. 10.1073/pnas.0608899103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH, 2008. EZH1 Mediates Methylation on Histone H3 Lysine 27 and Complements EZH2 in Maintaining Stem Cell Identity and Executing Pluripotency. Mol. Cell 32, 491–502. 10.1016/j.molcel.2008.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegenthaler G, Saurat J-H, Hotz R, Camenzind M, Merot Y, 1986. Cellular Retinoic Acid- but Not Cellular Retinol-Binding Protein Is Elevated in Psoriatic Plaques. J. Invest. Dermatol 86, 42–45. 10.1111/1523-1747.ep12283788 [DOI] [PubMed] [Google Scholar]
- Siegenthaler G, Tomatis I, Didierjean L, Jaconi S, Saurat J-H, 1992. Overexpression of Cellular Retinoic Acid-Binding Protein Type II (CRABP-II) and Down-Regulation of CRABP-I in Psoriatic Skin. Dermatology 185, 251–256. 10.1159/000247462 [DOI] [PubMed] [Google Scholar]
- Slavik MA, Allen-Hoffmann BL, Liu BY, Alexander CM, 2007. Wnt signaling induces differentiation of progenitor cells in organotypic keratinocyte cultures. BMC Dev. Biol 7, 1–8. 10.1186/1471-213X-7-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P, 1994. Abnormal kidney development and hematological disorders in PDGF beta-receptor mutant mice. Genes Dev. 8, 1888–1896. 10.1101/gad.8.16.1888 [DOI] [PubMed] [Google Scholar]
- Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A, 2003. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol 4, 124–131. 10.1038/ni876 [DOI] [PubMed] [Google Scholar]
- Thulabandu V, Chen D, Atit RP, 2018. Dermal fibroblast in cutaneous development and healing. Wiley Interdiscip. Rev. Dev. Biol 7, e307. 10.1002/wdev.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TH, Jarrell A, Zentner GE, Welsh A, Brownell I, Scacheri PC, Atit R, 2010. Role of canonical Wnt signaling/ -catenin via Dermol in cranial dermal cell development. Development 137, 3973–3984. 10.1242/dev.056473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SY, Sennett R, Rezza A, Clavel C, Grisanti L, Zemla R, Najam S, Rendl M, 2014. Wnt/β-catenin signaling in dermal condensates is required for hair follicle formation. Dev. Biol 385, 179–188. 10.1016/j.ydbio.2013.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Jin Q, Lee J-E, Su I. -h., Ge K, 2010. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc. Natl. Acad. Sci 107, 7317–7322. 10.1073/pnas.1000031107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Krieg T, Smola H, 2007. Keratinocyte-fibroblast interactions in wound healing. J. Invest. Dermatol 127, 998–1008. 10.1038/sj.jid.5700786 [DOI] [PubMed] [Google Scholar]
- Wyngaarden LA, Delgado-Olguin P, Su I. hsin, Bruneau BG, Hopyan S, 2011. Ezh2 regulates anteroposterior axis specification and proximodistal axis elongation in the developing limb. Development 138, 3759–3767. 10.1242/dev.063180 [DOI] [PubMed] [Google Scholar]
- Yasuhara R, Yuasa T, Williams JA, Byers SW, Shah S, Pacifici M, Iwamoto M, Enomoto-Iwamoto M, 2010. Wnt/μ-Catenin and retinoic acid receptor signaling pathways interact to regulate chondrocyte function and matrix turnover. J. Biol. Chem 285, 317–327. 10.1074/jbc.M109.053926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HMI, Jerchow B, Sheu TJ, Liu B, Costantini F, Puzas JE, Birchmeier W, Hsu W, 2005. The role of Axin2 in calvarial morphogenesis and craniosynostosis. Development 132, 1995–2005. 10.1242/dev.01786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Tomann P, Andl T, Gallant NM, Huelsken J, Jerchow B, Birchmeier W, Paus R, Piccolo S, Mikkola ML, Morrisey EE, Overbeek PA, Scheidereit C, Millar SE, Schmidt-Ullrich R, 2009. Reciprocal Requirements for EDA/EDAR/NF-κB and Wnt/β-Catenin Signaling Pathways in Hair Follicle Induction. Dev. Cell 17, 49–61. 10.1016/j.devcel.2009.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorick TS, Syroid DE, Arroyo E, Scherer SS, Lemke G, 1996. The transcription factors SCIP and Krox-20 mark distinct stages and cell fates in Schwann cell differentiation. Mol. Cell. Neurosci 10.1006/mcne.1996.0052 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FigureS1: PDGFRαCreER efficiently deletes mesenchymal Ezh2: The relative quantity of Ezh2 (A), in E10.5 flow sorted dorsal mesenchyme isolated from conditional heterozygous E8.5-DermEzh2 flox/+ controls and E8.5-DermEzh2. Indirect immunofluorescence of H3K27me3 with DAPI counterstain at E13.5 (B, C). Quantification of H3K27me3+ cells in the surface ectoderm at E10.5 (D). Solid line demarcates the epidermis from dermis and dashed line demarcates the lower limit of the dermis. Scale Bar= 50 microns in B and C.
FigureS2: Dermal density and thickness are comparable between controls and E8.5-DermEzh2 despite the persistence of Wnt signaling upregulation till E13.5. Dermal proliferation index at E10.5 (A) and cell density (B) at E12.5. Masson’s Trichrome staining (C, D) at E17.5 showing the thickness of the dermis and its quantification (E). The relative quantity of Wnt signaling target genes, Lef1 (F) and Axin2 (G) in E13.5 whole skin between controls and E8.5-DermEzh2. Scale Bar= 200 microns in C and D.
Figure S3: The PP marker, LRIG1, and the RP marker, DLK1, are comparable between controls and E8.5-DermEzh2 mutants: Indirect immunofluorescence with DAPI counterstain (A-D) showing papillary progenitor marker, LRIG1 (A, B), and reticular progenitor marker, DLK1 (C, D). Schematic illustration of binning of dermis employed to quantify LRIG1+/DLK1+ cells (E). Corrected fluorescence of LRIG1 (F) and DLK1 (G). Solid line demarcates the epidermis from dermis and dashed line demarcates the lower limit of the dermis. Scale Bar= 100 microns in A-D.
FigureS4: The dermal papilla marker expression is comparable between the controls and E8.5-DermEzh2 mice at E16.5. Alkaline phosphatase, a dermal papilla marker, staining sat E16.5 (A, B). Indirect immunofluorescence of dermal papilla marker, Sox2, with DAPI counterstain at E16.5 (C, D). The relative quantities of additional DP markers, Tbx18 and p75ntr (E). The relative quantity of a placode marker, Edar (F). Indirect immunofluorescence of additional placode markers, LEF1 (G, H) and SOX9 (I, J) with DAPI counterstain at E16.5. Solid line demarcates the epidermis from dermis. The arrows in G-J point towards the placode. Scale bar = 50 microns in A-D and G-J.
FigureS5: Epidermal hyperplasia in E8.5-DermEzh2 mice is evident at various locations and from E13.5 onwards: Hematoxylin and eosin staining of dorsal skin (A, B, D-I) showing the histology of the epidermis at E13.5 (A, B) and E16.5 (D-G). Quantification of the number of epidermal keratinocytes at E13.5 between controls and E8.5-DermEzh2 (C). Solid line demarcates the epidermis from dermis and dashed line demarcates the lower limit of the dermis. Scale Bar= 50 microns in A, B, D-G and 100 microns in H, I.
FigureS6: E8.5-DermEzh2 mice have intact barrier but exhibits accelerated differentiation: Whole mount toluidine blue O staining at E16 (A, B) and E17.5 (C, D). Indirect immunofluorescence of FILAGGRIN (E, F) with DAPI counterstain at E15.5. Solid line demarcates the epidermis from the dermis. Scale bar = 50 microns in A-D.
FigureS7: PRC2 components are not regulated by retinoic acid signaling. The relative quantity of Cyp26b1 (A), a target of RA signaling, and PRC2 components (B-D) in RA treated wild-type skin in comparison to vehicle control treated embryos at E10.5.


