Abstract
A few reports have indicated that a single gene confers resistance to bacterial blight, sheath blight and rice blast. In this study, we identified a novel disease resistance mutant gene, methyl esterase‐like (osmesl) in rice. Mutant rice with T‐DNA insertion displayed significant resistance to bacterial blight caused by Xanthomonas oryzae pv. oryzae (Xoo), sheath blight caused by Rhizoctonia solani and rice blast caused by Magnaporthe oryzae. Additionally, CRISPR‐Cas9 knockout mutants and RNAi lines displayed resistance to these pathogens. Complementary T‐DNA mutants demonstrated a phenotype similar to the wild type (WT), thereby indicating that osmesl confers resistance to pathogens. Protein interaction experiments revealed that OsMESL affects reactive oxygen species (ROS) accumulation by interacting with thioredoxin OsTrxm in rice. Moreover, qRT‐PCR results showed significantly reduced mRNA levels of multiple ROS scavenging‐related genes in osmesl mutants. Nitroblue tetrazolium staining showed that the pathogens cause ROS accumulation, and quantitative detection revealed significantly increased levels of H2O2 in the leaves of osmesl mutants and RNAi lines after infection. The abundance of JA, a hormone associated with disease resistance, was significantly more in osmesl mutants than in WT plants. Overall, these results suggested that osmesl enhances disease resistance to Xoo, R. solani and M. oryzae by modulating the ROS balance.
Keywords: JA, OsTrxm, ROS, rice blast, sheath blight, Xoo
Introduction
Plants experience a wide variety of biotic and abiotic stresses during a life cycle, and diseases greatly affect crop yield and quality (Zhang and Wang, 2013). An early sign of successful cell infection in response to pathogens is a burst of reactive oxygen species (ROS) (Torres et al., 2006). Previous studies have shown that elevated ROS in plants can lead to local production of programmed cell death (PCD) in leaves, thereby preventing further infection (Muhlenbock et al., 2008; Ruan et al., 2019). rrsRLK encodes a receptor‐like kinase required for ROS scavenging, and mutation of this gene results in reduced activity of enzymes involved in ROS scavenging in vivo, leading to H2O2 accumulation, which enhances defence resistance to bacterial blight (Yoo et al., 2018). microRNA528 (miR528) negatively regulates rice virus defence resistance by cleaving L‐ascorbate oxidase (AO) mRNA and reduces AO‐mediated ROS accumulation (Wu et al., 2017). One function of thioredoxin is to maintain ROS homeostasis in vivo. Tomato LeCITRX (Cf‐9‐interacting thioredoxin) negatively regulates plant disease defence resistance through cell death and ROS accumulation mediated by resistance protein Cf‐9 (Rivas et al., 2004). Therefore, ROS is an effector that is directly or indirectly regulated by different types of genes to improve plant defence resistance when the plant is infected by pathogens.
The homologs of OsMESL in Arabidopsis belong to AtMES (methyl esterase) family. Previous studies have shown that AtMES is involved in the systemic acquired resistance (SAR) process, while its subfamily members have different functions. For example, AtMES‐1, AtMES‐2, AtMES‐7 and AtMES‐9 can hydrolyse MeSA to generate SA, and free SA acts as a long‐range SAR signal to generate a defence resistance response. Knockdown or RNA interference (RNAi)‐mediated silencing of these several genes exhibits an accumulation of MeSA in response to SAR (Vlot et al., 2008), and StMES1 of potato has a similar function (Manosalva et al., 2010). AtMES17 is unable to use MeSA as a substrate, but it exhibits strong hydrolysis to MeIAA in vitro (Yang et al., 2008). AtMES16, which belongs to the same subfamily as AtMES17, is functionally different. AtMES16 is involved in the degradation process of chloroplasts in senescing leaves of arabidopsis and specifically catalyses the O134‐demethylation of primary fluorescent chlorophyll catabolites (PFCCs) (Banala et al., 2010; Christ et al., 2012). However, AtMES11 which has the most similar homology with OsMESL and its subfamily members are yet to be reported, and they have no hydrolytic activity.
In this study, a rice OsMESL gene was identified. The mutants and RNAi plants showed significant defence resistance to Xoo and sheath blight. By contrast, the complementary plants lost defence resistance to the pathogen, and the overexpression plants showed the same phenotype with wild‐type (WT) ZH11. Protein interaction experiments showed that OsMESL can interact with OsTrxm in vivo and in vitro, thereby indicating that OsMESL may be involved in the ROS pathway response to pathogens. These results suggest that OsMESL is a key regulator of pathogen stress.
Results
Identification and characterization of osmesl
In this study, a T‐DNA insertion mutant was identified that demonstrated significant resistance to Xoo (Figure 1a) and Rhizoctonia solani (Figure 1b). We named this mutant osmesl after performing the alignment analysis with the homologous protein sequence in Arabidopsis. Measurement of the lesion length and areas of detached infected leaves indicated that the disease resistance conferred by osmesl is significantly higher than that by WT. Additionally, in vivo inoculation experiments of sheath blight conducted through the toothpick embedding method indicated enhanced resistance in osmesl mutants compared with WT, and the percentage disease index (PDI) of sheath blight was significantly lower in osmesl mutants than in WT (Figure 1c).
Figure 1.
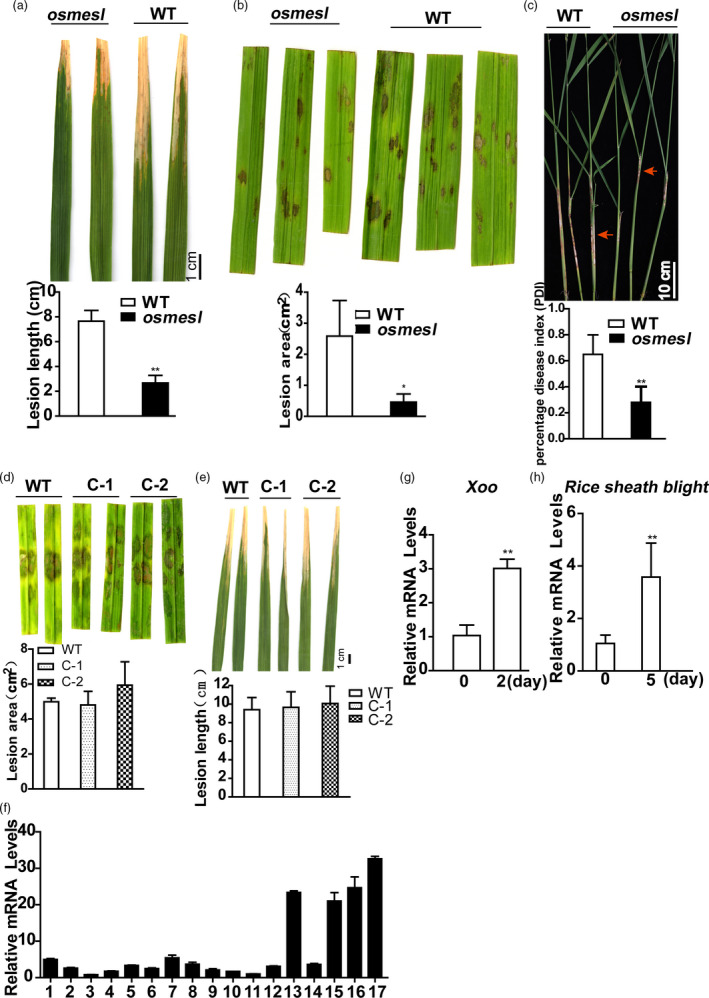
Phenotypes of mutant osmesl and its complementation lines, and expression pattern of OsMESL. (a) Phenotypes of osmesl and WT after inoculation with Xoo, and lesion length (cm) measurements at 10 days post‐inoculation. Values are means ± SD (n = 16). (b) Detached leaf phenotypes of osmesl and WT after inoculation with R. solani, lesion areas of sheath blight measured by photoshop. Values are means ± SD (n = 3). (c) Phenotypes of osmesl and WT by toothpick method after inoculation with R. solani. The percentage disease index. Values are means ± SD (n = 13). (d) Detached leaf phenotypes of WT, C‐1 and C‐2 after inoculation with R. solani, lesion areas of sheath blight measured by photoshop. Values are means ± SD (n = 3). (e) Phenotypes of WT, C‐1 and C‐2 after inoculation with Xoo, and lesion length (cm) measurements at 10 days post‐inoculation. Values are means ± SD (n = 10). (f) Detection of OsMESL expression in different tissues at varying developmental stages in WT. 1, Callus at 15 days after induction; 2, callus at 15 days after passage; 3, plumule at 24 h after germinating; 4, radicle at 48 h after germination; 5, seedling at three‐leaf stage; 6, aboveground part at 2 tillering stage; 7, stem at 5 days before heading; 8, stem at heading stage; 9, young panicle which length <1 mm; 10, young panicle which length was 3–5 mm; 11, young panicle which length was 10–15 mm; 12, panicled spike; 13, stamen at 1 day before flowering; 14, spikelet at 3 days after flowering; 15, flag leaf of booting stage which the panicle length was 40–50 mm; 16, flag leaf at 5 days before heading; 17, flag leaf at 14 days after flowering. (g–h) Expression of OsMESL can be induced by Xoo and R. solani in WT. Values are means ± SD (n = 3) (*P ≤ 0.05, **P ≤ 0.01, Student’s t‐test).
Functional complementation of the osmesl mutant with OsMESL genomic DNA
We obtained the entire OsMESL coding region from BAC genome to construct a complementation vector for verifying the phenotype of osmesl. Furthermore, we genetically transformed the callus induced by osmesl‐homozygous positive seeds to create transgenic plants. The expression of complementary lines was detected in transgenic positive plants (Figure S1). The lines with restored expression levels, complement‐1 (C‐1) and complement‐2 (C‐2) were selected for the inoculation experiment. Results indicated that complementary lines could recover the phenotypes infected with R. solani (Figure 1d) and Xoo (Figure 1e).
Expression pattern of OsMESL
Real‐time quantitative PCR results showed that OsMESL is constitutively expressed in all rice tissues. OsMESL exhibited low expression in callus, plumule, radicle, seedling, stem, young panicle and inflorescence; high expression in leaf and stamen; and the highest expression in flag leaf 14 days after heading (Figure 1f).
We analysed the expression pattern of response of OsMESL to pathogens in WT ZH11. The WT plants were inoculated with both pathogens, and the control was treated with water. The results indicated that OsMESL can be strongly induced by Xoo (Figure 1g) and R. solani (Figure 1h).
Subcellular localization of OsMESL protein
The localization of OsMESL in chloroplasts was predicted using protein subcellular localization prediction software WoLF PSORT (https://wolfpsort.hgc.jp/). We constructed a subcellular localization vector of OsMESL to determine the OsMESL subcellular localization. We used the cauliflower mosaic virus 35S promotor (CaMV35S) to drive OsMESL‐mCherry and transformed rice protoplasts. Chloroplast gave white fluorescence, mCherry gave red signals and CFP gave cyan fluorescence. SCAMP was a marker located in the cell membrane. The results indicated overlapping of red fluorescence with cyan and white fluorescences. These results suggested that OsMESL is located in the chloroplasts and cell membrane (Figure S2).
RNAi and CRISPR‐Cas9 of OsMESL in rice enhanced the resistance to pathogen infection
We induced RNAi interference, CRISPR–Cas9 knockout and overexpression of OsMESL in rice to further validate the phenotypes of the T‐DNA insertion mutants. The overexpression lines, namely OE‐1 and OE‐2, RNAi lines, namely RNAi‐1 (R‐1) and RNAi‐2 (R‐2), which were single‐copy lines (Figure S3), and OsMESL–Cas9 KO plants were selected for further study. mRNA levels were assessed, and the mRNA level of the overexpression lines was found to be significantly higher than that of WT, whereas mRNA level of the RNAi lines was notably lower (Figure 2a). The pathogen infection test showed that the RNAi lines exhibit enhanced resistance to Xoo; however, no difference was observed between the OE lines and WT (Figure 2b). We obtained homozygous CRISPR–Cas9 mutants of OsMESL by sequencing in T0 transgenetic plants (Figure 2c). Inoculation with two pathogens to OsMESL‐cas9 KO T1 transgenic plants with two pathogens, the OsMESL–Cas9 KO mutants were also found to acquire resistance to Xoo (Figure 2d) and R. solani that not only in detached leaves (Figure 2e) but also in the living plant (Figure 2f).
Figure 2.
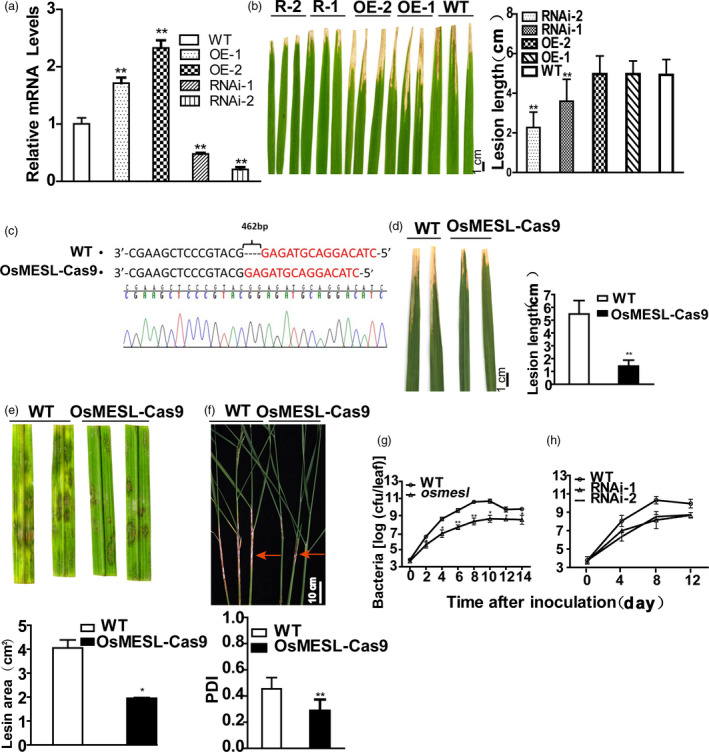
OsMESL‐RNAi and KO plants show resistance to Xoo and sheath blight, and Xoo growth was inhibited in OsMESL‐RNAi and KO plants. (a) mRNA expression levels of RNAi and OE lines. Values are means ± SD (n = 3). (b) Phenotypes and lesion length of RNAi‐1, RNAi‐2, OE‐1, OE‐2 and WT after inoculating with Xoo. Values are means ± SD (n = 10). (c) Sequecning of OsMESL KO plants for homozygous positive. (d) Phenotype and lesion length of OsMESL KO and WT inoculated with Xoo. Values are means ± SD (n = 13). (e) Detached leaves of OsMESL‐Cas9 plants inoculated with R. solani. Values are means ± SD (n = 3). (f) Phenotypes of OsMESL‐Cas9 plants and WT by toothpick method after inoculation with R. solani and the percentage disease index. Values are means ± SD (n = 12). (g–h) Xoo growth curve in WT, osmesl and RNAi lines. Values are means ± SD (n = 3) (*P ≤ 0.05, **P ≤ 0.01, Student’s t‐test).
To understand the growth rate of the pathogen in rice leaves, growth curves were plotted by inoculating leaves with Xoo. The bacterial growth rate in WT was found to be 7.95‐ to 104.27‐fold higher than in osmesl mutants between day 2 and day 8 (Figure 2g). However, the bacterial growth rates in WT were found to be 77.82‐ and 68.74‐fold higher than in RNAi‐1 and RNAi‐2 plants, respectively, on day 8 (Figure 2h). These results suggested that osmesl and RNAi plants may enhance the resistance to Xoo in rice by affecting the growth of the pathogen.
H2O2 content increased and defence‐related gene expression activated in osmesl mutants
Chloroplasts are the key components of the early immune response and ROS production (de Torres Zabala et al., 2015). Nitroblue tetrazolium (NBT) can react with superoxide anion to form blue formazan. Hence, we performed NBT staining on leaves of osmesl and WT plants to verify whether chloroplast‐localized OsMESL is involved in the ROS pathway. The result indicated higher content in leaves of osmesl plants than in those of WT plants before inoculation (Figure 3a). We further determined the mRNA expression levels of ROS scavenging genes in osmesl plants. The results showed that the mRNA expression levels of genes involved in ROS scavenging, namely, AOX1a, AOX1b, SODA1, SODB, CatA and POD1, in osmesl plants than in WT plants (Figure 3b).
Figure 3.
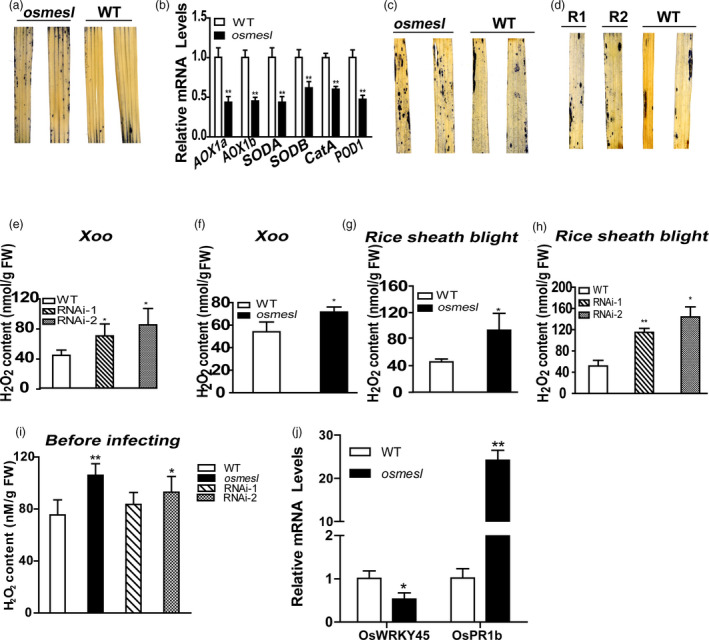
osmesl showed reduced ROS scavenging capacity and increased expression of genes involved in disease defence resistance. (a) NBT staining of osmesl and WT before inoculation with Xoo. (b) mRNA levels of genes involved in ROS scavenging. Values are means ± SD (n = 3). (c–d) NBT staining leaves of WT, osmesl, RNAi‐1 and RNAi‐2. (e–h) Quantitative determination of H2O2 in WT, osmesl, RNAi‐1 and RNAi‐2 after inoculating with Xoo and R. solani, respectively. Values are means ± SD (n = 3). (i) Quantitative determination of H2O2 in WT, osmesl, RNAi‐1 and RNAi‐2 before inoculating (n = 5). (j) Expression levels of genes involved in disease defense resistance. Values are means ± SD (n = 3) (*P ≤ 0.05, **P ≤ 0.01, Student’s t‐test).
Subsequently, NBT staining after inoculation with Xoo revealed increased ROS accumulation in osmesl plants and RNAi lines (Figure 3c,d). Further, we determined H2O2 content in osmesl, RNAi lines and WT plants after 48 h inoculation with Xoo or R. solani, respectively. The results indicated higher H2O2 content in osmesl plants than in WT plants (71.6 nM/g vs 54.13 nM/g FW) after Xoo infection, and the H2O2 content in RNAi‐1 and RNAi‐2 was found to be 70.23nM/g FW and 85.11nM/g FW, respectively, which was significantly higher than that found in WT plants (44.07 nM/g FW) (Figure 3e,f). The H2O2 content in osmesl plants was found to be higher than in WT plants after inoculation with R. solani (98.41 vs. 48.06 nM/g FW). Similarly, the H2O2 content in RNAi‐1 and RNAi‐2 was 115.05 and 144.14 nM/g FW, respectively, which was significantly higher than that found in WT plants after inoculation with R. solani (51.67 nM/g FW) (Figures 3g,h). We assessed whether H2O2 accumulates in the osmesl and RNAi plants before the infection. The H2O2 content in osmesl, RNAi‐1 and RNAi‐2 plants was 106, 83.56 and 92.99 nM/g FW, respectively, which was higher than that observed in WT plants (75.43 nM/g FW) (Figure 3i).
Further, we studied whether the expression of genes involved in disease resistance is altered in osmesl mutant plants. The mRNA levels of disease resistance‐related genes were determined in osmesl plants. The results indicated significantly increased OsPR1b expression in osmesl plants (Figure 3j). Previous studies have reported that the suppression of OsWRKY45‐1 expression in japonica rice can enhance resistance to bacterial leaf blight caused by Xoo and bacterial leaf streak caused by Xoc (Tao et al., 2009). Therefore, we detected the expression of Os WRKY45‐1 in osmesl and WT plants. The results indicated that the expression of Os WRKY45‐1 is significantly decreased in osmesl plants compared with WT plants, after inoculation with Xoo (Figure 3i). These results suggested that the resistance to pathogens in osmesl plants may involve the ROS pathway and affect the expression of several disease resistance‐related genes.
OsMESL interacts with OsTrxm
We selected a membrane‐based yeast two‐hybrid system for yeast two‐hybrid screening because of the localization of OsMESL in the cell membrane and chloroplast. The results indicated that OsMESL could interact with OsTrxm in yeast cells (Figure 4a). To further validate the interaction between OsMESL and OsTrxm, we performed a luciferase activity assay in rice protoplasts. The results indicated significantly higher luciferase activity in the experimental group was than in the control group, which further indicated that the two proteins could interact in vivo (Figure 4b). Additionally, we observed an interaction between OsMESL and OsTrxm in rice protoplasts through the bimolecular fluorescence complementation (BiFC) experiment by using splited mCherry (Figure 4c); the control group showed negative results. Moreover, the Co‐IP assay verified the interaction between OsMESL and OsTrxm in Nicotiana benthamiana (Figure 4d).
Figure 4.
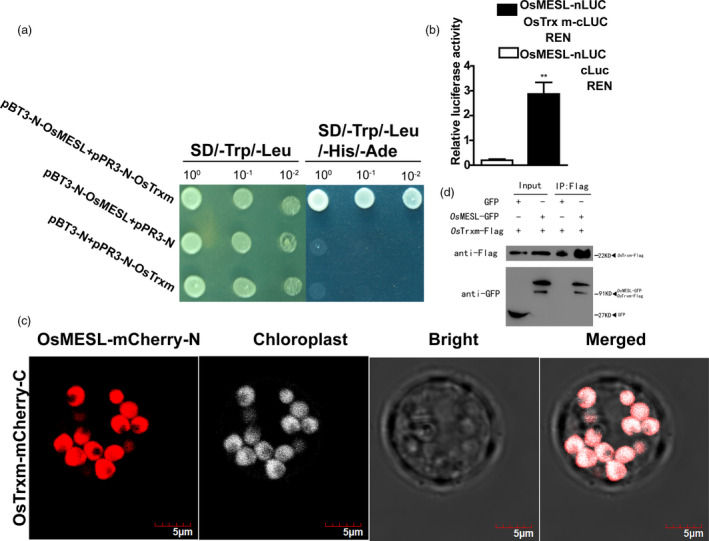
OsMESL interacts with OsTrxm in vivo and in vitro. (a) OsMESL interacts with OsTrxm in yeast NMY51. pBT3‐N‐OsMESL and vector pPR3‐N or vector pBT3‐N and pPR3‐N‐OsTrxm as the controls cotransformed into yeast, respectively. (b) OsMESL interacts with OsTrxm in bimolecular luciferase enzyme activity assay. Values are means ± SD (n = 3). (c–d) OsMESL interacts with OsTrxm in a BiFC assay. mCherry, red fluorescence. Chloroplast, white autofluorescence. Scale bars, 5 μm. (d) FLAG tagged OsTrxm was immunopreciptated using an anti‐FLAG antibody and co‐immunopreciptated OsMESL‐GFP was detected by anti‐GFP antibody. (**P ≤ 0.01, Student’s t‐test).
OsTrxm knockout plants enhance defence to pathogens
A study reported that OsTrxm plays an important role in the redox regulation of chloroplast and that RNAi of OsTrxm increases the H2O2 content and developmental defects, such as semid‐warfism, pale‐green leaves and abnormal chloroplast structure, in rice plants (Chi et al., 2008).
We first examined the mRNA expression level of OsTrxm in osmesl mutants to understand whether the mRNA expression level of OsTrxm is altered. The results showed a significantly lower expression of OsTrxm in osmesl mutants than in WT plants (Figure 5a). Furthermore, we measured the protein content of OsTrxm in osmesl mutants, OE and RNAi lines by using the Western blot assay. We observed that the protein content of OsTrxm in osmesl and RNAi‐2 plants is lower than in WT. Therefore, we constructed OsTrxm‐Cas9 knockout and 35S::Trxm plants to determine the phenotypes. A DNA sequence of 139bp length, which included parts of the sequence of the first exon and first intron, was knocked out (Figure 5c). Inoculation with pathogens Xoo and R. solani revealed increased resistance to Xoo (Figure 5d) and sheath blight (Figure 5e) in OsTrxm–Cas9 homozygous mutant plants, compared with the control. However, the OsTrxm‐OE lines displayed no difference in resistance to both pathogens (Figure S4). These results illustrated that OsTrxm may be crucial for the involvement of osmesl in ROS‐mediated disease resistance pathways.
Figure 5.
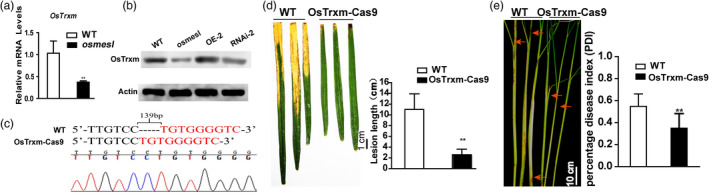
OsTrxm‐Cas9 plants show a defence resistance to Xoo and sheath blight. (a) OsTrxm expression level in osmesl. Values are means ± SD (n = 3). (b) Dection of OsTrxm protein level of osmesl, OE‐2, RNAi‐2 by Western blot. (c) Sequencing of OsTrxm KO plants for homozygous positive. (d) Phenotype of OsTrxm‐Cas9 plants after inoculating with Xoo, and lesion length (cm) measurements at 10 days post‐inoculation. Values are means ± SD (n = 13). (e) Phenotype of OsTrxm‐Cas9 plants after inoculating with R. solani. Values are means ± SD (n = 13) (**P ≤ 0.01, Student’s t‐test).
JA signalling pathway activation in osmesl mutant
The phytohormone JA has an important role in regulating immune responses (Galis et al., 2009; Yan and Xie, 2015; Yu et al., 2020). Studies have shown that the overexpression of a OsTrxm interacting protein BAS1, a 2‐Cys peroxiredoxins, increases the plant tolerance to alkyl hydroperoxides. BAS1 could control the synthesis of JA by reducing 13‐hydroperoxy linolenic acid (Baier and Dietz, 1997; Zhang et al., 2011). Thus, we assessed whether the decreased OsTrxm expression affects the JA pathway in osmesl mutants. First, we assessed the expression of certain genes in the JA synthesis and signalling pathway in osmesl mutants. The expression of AOC and LOX2, the key genes involved in the JA synthesis pathway, was found to be significantly higher, whereas that of JMT1, an MeJA synthesis gene, was found to be significantly lower in osmesl mutants than in WT plants (Figure 6a). These results indicated that the JA content may be higher in osmesl mutants than in WT plants. Subsequently, we quantitatively determined the JA content of osmesl mutants and WT plants at the seedling stage. The JA content in osmesl mutants was found to be higher than in WT plants (11.63 vs 5.01 ng/g FW) (Figure 6b). Thus, the results indicate that the resistance to pathogens conferred by osmesl may involve the JA signalling pathway.
Figure 6.
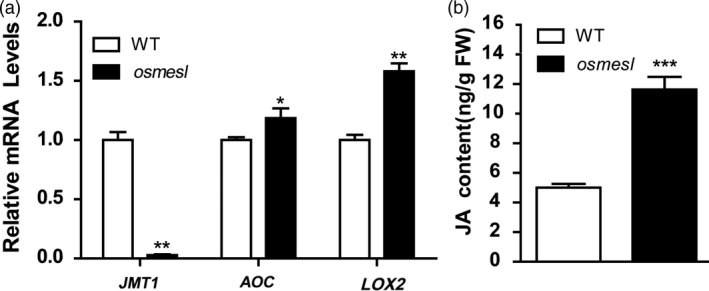
JA quantitative determination and related gene expression level. (a) JA signaling pathway‐related gene expression level. Values are means ± SD (n = 3). (b) JA content in osmesl and WT. Values are means ± SD (n = 3) (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, Studment’s t‐test).
Activation of defence response and inhibition of ROS scavenging‐related gene expression revealed through RNA‐Seq
To further unravel the function of OsMESL in rice, heading stage leaves in WT and osmesl plants were used for the transcriptome analysis. A total of 3602 differentially expressed genes (DEGs) were identified (P value < 0.05). Of these genes, 2052 were up‐regulated and 1550 were down‐regulated in osmesl plants. GO terms were significantly involved in metabolic process, response to stress, catalytic activity, response to biotic stimulus and response to abiotic stimulus (Figure S5). A set of DEGs associated with defence response [namely WRKY24 (Yokotani et al., 2018), WRKY77 (Lan et al., 2013), Pi9 (Qu et al., 2006), WRKY76 (Liang et al., 2017), MADS26 (Khong et al., 2015), PR1a (Agrawala, 2001), JIPR10 (Jwa et al., 2001), RSPR10 (Hashimoto et al., 2004) and DR8 (Wang et al., 2006)] and ROS scavenging [namely AOX1a (alternative oxidase), POD1 (peroxidase), CATB (catalase), MT2b (metallothionein) (Wong et al., 2004), APX1 (ascorbate peroxidases), APX3, APX8 (Singh and Shah, 2014)] was found to be significantly down‐regulated in osmesl plants (Table S2). Genes involved in the JA synthesis pathway, namely GH3.5 (jasmonic acid amino acid synthase), OPR1 and OPR7 (12‐oxo‐phytodienoate reductase gene) (Sobajima et al., 2007; Tani et al., 2008), LOX1, LOX2, LOX5, LOX6, LOX8 and LOX9 (lipoxygenase gene) (Zhang et al., 2018), were found to be up‐regulated in the transcriptome data. Meanwhile, the mRNA level of the OsTrxm interaction protein, BAS1, was found to be significantly lower in osmesl mutants than in WT in the transcriptome data, which indicated that the expression of BAS1 is repressed in mutants. The expression of the lipid hydroperoxide lyase gene (HPL3), which negatively regulates JA synthesis, was found to be decreased in the transcriptome data. HPL3 negatively regulates JA synthesis mainly by hydrolysing hydroperoxide linolenic acid to produce green leaf volatiles (Liu et al., 2012b; Tong et al., 2012). Thus, the results suggest that the mutation of OsMESL might affect the expression of defence response‐ and ROS scavenging‐related genes to some extent.
Discussion
In this study, resistance to Xoo and sheath blight was observed in the disease‐resistant mutant osmesl, RNAi and OsMESL knockout plants. Complementation of osmesl can restore the phenotype sensitive to pathogens, which suggests that the phenotype of osmesl plants is indeed because of OsMESL mutation.
Accumulation of ROS in osmesl mutants confers broad‐spectrum disease resistance
Plants can adopt various strategies to resist pathogen infection. In maize, ZmFBL41 gene knockout confers resistance to banded leaf and sheath blight by accumulating lignin (Li et al., 2019a). OsXa13 protein interacts with two copper transporters, COPT1 and COPT5, and co‐ordinates the distribution of copper, which is toxic to Xoo in rice (Yuan et al., 2010). Similarly, OsXA3/XA26 interacts with a triosephosphate isomerase (TPI), OsTPI1.1, and modulates ROS production through the glycolysis pathway (Liu et al., 2018). Chloroplasts, one of the major sources of ROS, are the main sites that are involved in production. is unstable and its subsequent catalys is by dismutases generates H2O2, which in turn is hydrolysed by peroxidases or catalases (CAT) to produce H2O and O2 (Krieger‐Liszkay, 2005). Recent studies have revealed that ROS can also act as a signalling molecule that regulates various stresses, such as plant growth and development, hypersensitive response and pathogen‐induced SAR (Baxter et al., 2014; Zhang et al., 2014).
ROS plays an important role in plant–pathogen interactions (Heller and Tudzynski, 2011). ROS‐related plant defence responses include direct killing of pathogens that activates PCD and provides a suitable redox environment for the initiation of immune responses (Vellosillo et al., 2010). Moreover, ROS offer wide possibilities for broad‐spectrum disease resistance (Li et al., 2019b; Li et al., 2017). Protein interaction experiments have demonstrated that OsMESL can interact with OsTrxm, which is localized in chloroplasts. Previous studies have shown that thioredoxin (TRX), an antioxidant modulator, can improve the tolerance of TRX‐deficient yeast to oxidative stress (Issakidis‐Bourguet et al., 2001). TRX localized in chloroplast regulates the activity and functions of various enzymes present in chloroplasts. A commonly studied TRX target protein is NADPH–malate dehydrogenase (NADPH–MDH), which is involved in exporting the reducing energy from chloroplasts to the cytosol in the form of malate with complete inactivation of the oxidized state of NADP–MDH, and its activity is strictly dependent on TRX (Arner and Holmgren, 2000; Lemaire et al., 2007; Motohashi et al., 2001). XopI, an F‐box effector of a Xoo strain BAI3, acts as an adapter to form a ternary complex, which regulates the immune response in rice through proteasomal degradation of OsTrxh2 (Ji et al., 2020). OsTrxm RNAi plants exhibit certain phenotypes, such as increased H2O2 levels, abnormal chloroplast structure and disrupted ROS scavenging (Chi et al., 2008). In the present study, OsTrxm expression was significantly reduced in osmesl mutants, and the mutants exhibited ROS accumulation before and after infection by pathogens. Given that previous studies did not address the relationship between OsTrxm and disease resistance, we assessed whether the resistance to pathogens is because of increased ROS levels in osmesl mutants and the decreased expression level of OsTrxm. H2O2, JA and defence‐related genes PR1b and LOX2 were increased in the OsTrxm knockout plants (Figures S6A–D). Further, OsTrxm knockout plants were inoculated with Xoo or R. solani. In OsTrxm knockout plants, resistance to pathogens was found to be enhanced. Furthermore, we explored whether osmesl mutants are resistant to rice blast. We induced rice blast in osmesl mutants and found that osmesl mutants are significantly resistant to rice blast (Figure S7A,B). Similarly, RNAi‐1 and RNAi‐2 also displayed resistance to rice blast (Figures S7C,D). These results suggest that OsMESL is involved in broad‐spectrum resistance to pathogens in rice.
Resistance to pathogens by osmesl involves the JA signalling pathway
JA, as a hormone for plant defence responses, plays an important role in disease defence resistance. The relationship between JA and ROS has not been well studied. Previous studies have reported that rice WRKY13 activates the glutathione/glutaredoxin system, which maintains the ROS balance, but inhibits JA biosynthesis in pathogen‐induced defence responses (Qiu et al., 2007). The exogenous application of methyl jasmonate to castor bean leaves has been found to induce ROS accumulation and decrease the activities of ROS scavenging enzymes, namely superoxide dismutase, CAT and guaiacol peroxidase. Recent studies have found that JA production can be induced by H2O2 treatment in Arabidopsis (Camejo et al., 2016; Hieno et al., 2019; Singh et al., 2016). In tomato leaves, wounding or JA treatment has been found to significant increase the activity of plasma membrane NADPH oxidase and ROS accumulation; however, the accumulation of ROS was found to be blocked by pre‐treatment with plasma membrane NADPH oxidase inhibitor or H2O2 scavengers (Li et al., 2002; Wasternack et al., 2006). These studies illustrated a certain relationship between JA and ROS. In the present study, the key genes, AOC and LOX2, of the JA synthesis pathway were significantly up‐regulated in osmesl mutants. However, the expression of JMT1, a MeJA synthase gene, was significantly decreased. These results indicated that the JA signalling pathway is activated in osmesl mutants. Quantitative determination of JA revealed that the JA content in osmesl mutants is twice of that in WT plants at the seedling stage. We speculated that OsMESL regulates ROS scavenging and involves activation of the JA signalling pathway by inducing ROS imbalance.
Previous studies have reported that BAS1, an interaction protein of OsTrxm, has a reductase effect on H2O2 and alkyl hydroperoxides. Additionally, JA synthesis can be regulated by reducing 13‐hydroperoxide linolenic acid. JA is synthesized from linolenic acid, which undergoes a series of oxidation reactions, followed by reduction reactions, to finally produce JA. Our transcriptome data indicated a decreased BAS1 expression in the mutant. We speculate that the decreased BAS1 expression could decrease the reducing potential of linolenic acid, further facilitating the oxidative response to linolenic acid and promoting the initiation of JA synthesis. Meanwhile, the key genes of OsAOC and OsLOX2 in JA synthesis pathway up‐regulated in osmesl mutants. In osmesl mutants, ROS scavenging is impaired and the reduction force in vivo is low. Previous studies have reported that the mRNA level of BAS1 decreases with increasing concentrations of glutathione and ascorbic acid; however, two of the most important antioxidants in vivo and oxidative stressors, such as methyl viologen and light, slightly induce its expression (Dietz, 1994). However, in osmesl mutants, the expression of BAS1 was instead significantly reduced in the environment with low reducing power, which seems to be contradictory to the function of BAS1. A possible reason for decreased BAS1 expression could be the impaired OsTrxm. In fact, the expression of BAS1 was down‐regulated in OsTrxm–Cas9 plants (Figure S8). Similarly, the RNA‐Seq data indicated that HPL3, which could hydrolyse the hydroperoxy polyunsaturated fatty acid, down‐regulated in osmesl mutants; hydroperoxy polyunsaturated fatty acid is the substrate of JA synthesis. OsTrxm was down‐regulated after MeJA induction in Zea mays (Zhang et al., 2015). Considering that OsMESL has an N‐myristoylation site at the N‐terminal, we speculated that OsMESL plays an important role in the stability of OsTrxm. The western blot analysis showed that the reduced OsMESL gene expression decreases the protein content of OsTrxm. These results suggest that osmesl confers pathogen resistance through the ROS and JA pathways by affecting OsTrxm protein stability.
In summary, we proposed a working model for the role of OsMESL (Figure 7). OsMESL somehow assists OsTrxm in ROS scavenging. When OsMESL expression is reduced, the ROS scavenging capacity of OsTrxm is attenuated, which results in ROS accumulation. In addition, the increase in JA synthesis could be attributed to the decreased expression of BAS1, and the expression of some resistance gene is induced in osmes mutantsl. Finally, repressed OsMESL confers resistance to different pathogens.
Figure 7.
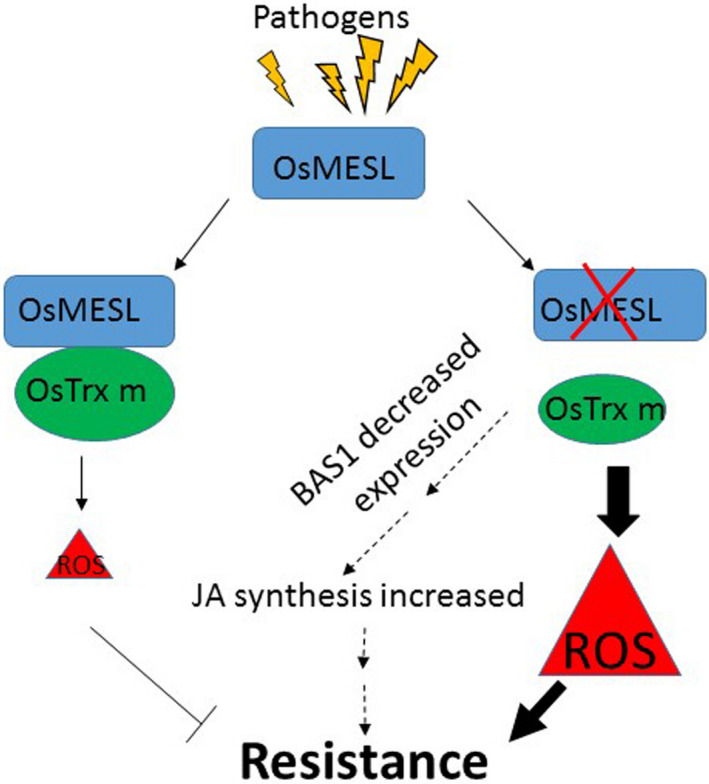
Model for the role of OsMESL in the resistance to Xoo and R. solani. OsMESL assists OsTrxm in ROS scavenging. When OsMESL expression is supressed, the OsTrxm ROS scavenging capacity is attenuated, and resulting in ROS accumulation. In addition, JA synthesis is increased due to BAS1 decreased expression, and some resistance gene induced expression in osmesl. Finally, these results lead to the resistance to the pathogens.
Materials and methods
Plant material and growth condition
In this study, the japonica rice (Oryza sativa) cultivar (Zhonghua 11, ZH11) was used as the WT control, and transformation was performed on this cultivar. The osmesl T‐DNA insertion mutant was procured from Kyung Hee University, Republic of Korea. The mutant family number was 3A‐03499. The primers used for the genotype detection were MESLT‐F, MESLT‐R and 2715L2. The field was sown and transplantation was performed after approximately 1 month, with 10 plants per row for a total of 3 rows. Agronomic character investigation and sample collection were conducted in the field under natural conditions during summer in Wuhan.
Samples from different parts of various tissues were obtained for analysing the expression pattern of OsMESL in WT rice. The samples were as follows: (1) callus 15 days after induction, (2) callus 15 days after passage, (3) plumule 24 h after germination, (4) radicle 48 h after germination, (5) seedling at the three‐leaf stage, (6) part above the ground at 2 tillering stage, (7) stem 5 days before heading, (8); stem at the heading stage, (9) young panicle with length of <1 mm, (10) young panicle with length of 3–5 mm, (11) young panicle with length of 10–15 mm, (12) panicled spike, (13) stamen 1 day before flowering, (14) spikelet 3 days after flowering, (15) flag leaf of booting stage with panicle length 40–50 mm, (16) flag leaf 5 days before heading and (17) flag leaf 14 days after flowering.
Vector construction and genetic transformation
The total RNA from WT ZH11 leaves was extracted and reverse transcribed to form the cDNA. The OsMESL (LOC_Os07g41230) gene was amplified from full‐length cDNA and constructed onto the pU1301 vector with cauliflower mosaic virus (CaMV) 35S RNA as the promoter. The RNAi vector amplified a 227‐bp fragment from the cDNA sequence of OsMESL and constructed in the pDS1301 vector. The BAC clone containing the OsMESL gene was selected from the Nipponbare BAC library in our laboratory, and the clone number was A0053G01. The target fragment was ligated into the final vector pC2301 by using a restriction enzyme to construct the complementation vector. OsMESL–Cas9 and OsTrxm–Cas9 were used to drive the expression of gRNA by using U3 and U6a promoters through dual‐target knockdown. WT ZH11 was transformed using Agrobacterium‐mediated genetic transformation (Lin and Zhang, 2005). Table S1 lists all the primers used in the experiment.
RNA extraction and qRT‐PCR analysis
The tissue samples, such as leaves, were immediately frozen in liquid nitrogen after sampling from the field. The total RNA was extracted with TRIzol reagent (TransGen Biotech, Beijing, China) as per the manufacturer’s instructions. Approximately 2 µg of total RNA was reverse transcribed, and the first strand of cDNA was obtained by reverse transcription by using Superscript III reverse transcriptase kit (Invitrogen, California, USA) as per manufacturer’s instructions. qRT‐PCR experiments were performed using FastStar Universal SYBR Green Master (ROX) (Roche, Basel, Switzerland) kits on an ABI 7500 system (Thermo Fisher Scientific, California, USA). The relative expression was calculated. The rice actin gene was used as an internal reference (Livak and Schmittgen, 2001). The primers used in the experiment are listed in Table S1.
Subcellular localization assays
The full‐length cDNA of OsMESL was constructed into the pM999‐mCherry vector through homologous recombination. The OsMES11–pM999‐mCherry plasmid was transformed into rice protoplasts for transient expression. Images were captured using a confocal fluorescence microscope (Olympus FV1200). Cyan fluorescence from the CFP channel, red fluorescence from the mCherry channel and chloroplast fluorescence were observed at the excitation wavelengths of 405, 559 and 488 nm, respectively, and the fluorescence acquisition wavelengths of 460–500, 570–618 and 655–755 nm, respectively.
Protein interaction analysis
The membrane system yeast two‐hybrid library constructed in our laboratory was used to screen the library. The entire rice plant at the seedling stage was used for constructing the cDNA library. The ligation vector was pPR3‐N, and OsMESL constructed on the pBT3‐N vector was used to screen the library. OsTrxm was constructed in pBT3‐N for interaction validation in yeast. All the restriction sites used in the study were SfiI. PEG4000 was used to transform the yeast strain NMY51; the yeast culture temperature was 30 °C, and the 3‐AT screening concentration was 25 mm.
The BiFC assay was performed in rice protoplast. Rice protoplast isolation and transformation were performed according to a previously described method (Tang et al., 2012; Xie et al., 2015). The rice OsMESL gene was cloned into the 1300s‐mCherry‐N vector, and OsTrxm was cloned into the 1300s‐mCherry‐C vector (Fan et al., 2008). Rice protoplasts were isolated from 12‐day‐old seedlings of ZH11 (Oryza sativa spp. japonica). First, the rice protoplasts were isolated by digesting the rice sheath strips in digestion solution (10 mm MES, pH 5.7; 0.6 m mannitol; 1 mm CaCl2; 5 mm beta‐mercaptoethanol; 0.1% BSA; 0.3% cellulase RS; and 0.75% macerozume R10) for 4 h with gentle agitation. Further, the protoplasts were incubated in W5 solution for 30 min, precipitated through centrifugation at 100 ×g for 8 min and finally resuspended in MMG solution. For transformation, 7 μg of each plasmid was joined together and gently mixed with 100 μL of protoplasts and 110 μL of PEG–CaCl2 solution, and the mixture was further incubated at room temperature for 15 min in dark. W5 solution (twice the volume of reaction mixture) was added to stop the transformation. The transformed protoplasts were collected through centrifugation and further resuspended in WI solution. The transformed protoplasts were incubated for 12 h in dark. Finally, the protoplasts were collected through centrifugation at 100 ×g for 8 min and immediately subjected to the fluorescence analysis (Olympus FV1200).
The protein interaction of luciferase activity recovery method was performed in rice protoplasts. Plasmids with N‐ and C‐terminal LUC fusion genes were cotransformed with 35S:REN in the ratio 10:10:1, with the latter as the internal control. The REN and LUC activities were detected using REN and LUC assay substrates (Promega, Madison, WI, USA), respectively, after 20‐h incubation. Relative activity of the LUC reporter was expressed as the ratio of LUC to GUS (Wang et al., 2015).
Co‐immunoprecipitation assay (Co‐IP) was performed in the Nicotiana benthamiana system. The OsMESL coding region was constructed in fusion with a C‐terminal GFP tag in pC1300S‐GFP, and OsTrxm was constructed in fusion with a C‐terminal FLAG tag in PU2301‐Flag. The total protein was extracted from the leaves of N. benthamiana for the Co‐IP assay. The Co‐IP samples were subjected to western blot by using anti‐GFP (1:10,000 dilution; ab290, Abcam, Cambridge, UK) and anti‐FLAG (1:10,000 dilution; F3165, Sigma‐Aldrich, St. Louis, MO, USA) antibodies.
Overall, 50 μg of each sample was used for western blot. Actin (1:2000 dilution, #A01050, Abbkine) was used as the control. Antibodies were prepared using 73 C‐terminal amino acids of OsTrxm as the antigen. Anti‐OsTrxm was used in 1:1000 dilution for western blot.
Pathogen inoculation
The blight species was Philippine species PXO99 (race 6). Flag leaves at the booting stage of rice were selected for the inoculation experiment by using the leaf‐clipping method (Zhou et al., 2002). Disease was scored by measuring the lesion length 10 days after inoculation. Bacterial growth in rice leaves was measured by counting the colony‐forming units (Sun et al., 2004).
The pathogen of sheath blight was RH‐9 virulent strain. Two inoculation methods were used in this study. Flag leaves with the same growth status at the booting stage in field were selected in vitro. Sheath blight fungi were placed in the middle of the detached leaves, moisturized, cultured at 28 °C for 48 h, and further photographed. The relative lesion area was measured using Photoshop. In the fields, toothpick embedding method was used to inoculate R. solani. Disease severity was measured as the relative lesion height and expressed as the percentage disease index (Molla et al., 2013).
NBT staining and quantitative determination of H2O2
NBT (Sigma‐Aldrich, St. Louis, MO, USA) powder was dissolved in sterile ddH2O. The leaves, which were grafted with the disease for approximately 7 days, were stained in dark at 28 °C. Subsequently, the leaves were destained with 95% ethanol. Finally, 40% glycerol was used for rehydration for photography.
Flag leaves after 48 h of inoculation with Xoo or R. solani were subjected to quantitative determination of H2O2 in the tillering stage. The part of leaves approximately 10 cm below the incision was obtained and immediately placed into liquid nitrogen for H2O2 determination. Overall, 3 biological replicates per sample were used. The whole flag leaf before infection was subjected to quantitative determination of H2O2 in the late tillering stage. Overall, 5 biological replicates were used per sample. The procedures were performed using Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Thermo Fisher Scientific, CAS: A22188) according to the manufacturer's instructions.
Determination of endogenous JA in rice
The parts of osmesl and WT ZH11 plant seedlings above the ground at the three‐leaf stage were immediately frozen in liquid nitrogen (four biological replicates per sample). The samples from OsTrxm‐Cas9 plants were collected in the late stage of rice maturation. Hormone extraction assays were performed using a method described previously (Liu et al., 2012a).
Transcriptome sequencing and data analysis
RNA samples from WT and osmesl plants at the heading stage were used for transcriptome sequencing, and each sample was pooled for total RNA isolation (6 biological replicates). Transcriptome sequencing was performed in CapitalBio Technology Corporation by using the Illumina HiSeq X ten system according to the standard procedure. Raw reads were subjected to quality check by using ‘FastP’ (Chen et al., 2018). Sample‐wise high‐quality reads were aligned to the rice reference genome (http://rice.plantbiology.msu.edu/) by using ‘HISAT2’ (Kim et al., 2015). Aligned reads were subsequently used to create a reference annotation‐based transcript assembly to identify uniquely aligned reads in each sample. The results were subjected to ‘featureCounts v1.5.0’ (Liao et al., 2014) to obtain the read count of all samples. Differences in the gene expression were identified using ‘DESeq2 1.2.5R’ (Love et al., 2014). The thresholds for P value and FDR (adjusted P value or Q‐value) were set to 0.05 for identifying significant differences in the expression. Gene ontology was derived from the msu7 rice genome database.
Accession numbers
The sequence data from this article can be found in the RGAP database (http://rice.plantbiology.msu.edu) under the following accession numbers: OsMESL, LOC_Os07g41230; OsTrxm, LOC_Os12g08730; SODB, LOC_Os06g05110; SODA1, LOC_Os05g25850; CatA, LOC_Os02g02400; POD1, LOC_Os07g02440; AOX1a, LOC_Os04g51150; AOX1b, LOC_Os04g51160; PR1a, LOC_Os07g03710; PR1b, LOC_Os01g28450; PR10, WRKY45‐1, LOC_Os05g25770; JMT1, LOC_Os06g20920; AOC, LOC_Os03g32314; LOX2, LOC_Os03g08220; BAS1, LOC_Os02g33450 and Actin, LOC_Os03g50885.
CONFLICT OF INTERESTS
The authors have no conflicts of interest to declare.
Author contributions
Y.L. and B.H. conceived and designed the experiments. B.H. performed the experiments and wrote the manuscript. Y.L. edited and revised the manuscript. Y.Z. provided assistance with the writing of the manuscript, and Z.Z. provided assistance with the luciferase activity assay. All other authors participated in the discussion of the results and commented on the manuscript.
Supporting information
Figure S1 The expression level of complementary lines.
Figure S2 Subcellular localization of OsMESL protein.
Figure S3 Detectiion of copy numbers in OsMESL‐RNAi lines, OsMESL‐OE lines and osmeslcomplementation lines
Figure S4 Phenotypes of OsTrxm OE lines inoculated with Xoo and R. solani.
Figure S5 GO analysis of DEGs in osmesl and WT.
Figure S6 Detection of disease resistance‐related indicators.
Figure S7 osmesl and RNAi lines showed resistance to rice blast.
Figure S8 Expression level of OsBAS1 in OsTrxm‐Cas9 mutant.
Table S1 Primers used in this study.
Table S2 Differentially expressed of defence response‐ and ROS scavenging‐related genes in osmesl plants.
Acknowledgements
We thank Dr. Hongbo Liu (Huazhong Agricultural University, China) for technical assistance in JA level quantification analyses, and Dr. Meng Yuan (Huazhong Agricultural University, China) and Qianhua Shen (Institute of Genetice and Developmental Biology, Chinese Academy of Sciences, China) for some suggestions in writing, and Dr. Kabin Xie (Huazhong Agricultural University, China) for rice blast inoculation. This work was supported by the The National Key Research and Development Program of China (2016YFD0100600), the National Program of Transgenic Variety Development of China (2016ZX08001001) and the Fundamental Research Funds for the Central Universities (Grant no. 2662015PY115).
Hu, B. , Zhou, Y. , Zhou, Z. , Sun, B. , Zhou, F. , Yin, C. , Ma, W. , Chen, H. and Lin, Y. (2021) Repressed OsMESL expression triggers reactive oxygen species mediated broad‐spectrum disease resistance in rice. Plant Biotechnol. J., 10.1111/pbi.13566
References
- Agrawala, G.K. , Rakwal, R. , Jwac, N.‐S. and Agrawala, V.P. (2001) Signalling molecules and blast pathogen attack activates rice OsPR1a and OsPR1b genes: A model illustrating components participating during defence/stress response. Plant Physiol. Biochem. 39, 1095–1103. [Google Scholar]
- Arner, E.S. and Holmgren, A. (2000) Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267, 6102–6109. [DOI] [PubMed] [Google Scholar]
- Baier, M. and Dietz, K.J. (1997) The plant 2‐Cys peroxiredoxin BAS1 is a nuclear‐encoded chloroplast protein: its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J. 12, 179–190. [DOI] [PubMed] [Google Scholar]
- Banala, S. , Moser, S. , Muller, T. , Kreutz, C. , Holzinger, A. , Lutz, C. and Krautler, B. (2010) Hypermodified fluorescent chlorophyll catabolites: source of blue luminescence in senescent leaves. Angew. Chem. Int. Ed. Engl. 49, 5174–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter, A. , Mittler, R. and Suzuki, N. (2014) ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. [DOI] [PubMed] [Google Scholar]
- Camejo, D. , Guzman‐Cedeno, A. and Moreno, A. (2016) Reactive oxygen species, essential molecules, during plant‐pathogen interactions. Plant Physiol. Biochem. 103, 10–23. [DOI] [PubMed] [Google Scholar]
- Chen, S. , Zhou, Y. , Chen, Y. and Gu, J. (2018) fastp: an ultra‐fast all‐in‐one FASTQ preprocessor. Bioinformatics, 34, i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, Y.H. , Moon, J.C. , Park, J.H. , Kim, H.S. , Zulfugarov, I.S. , Fanata, W.I. , Jang, H.H. et al. (2008) Abnormal chloroplast development and growth inhibition in rice thioredoxin m knock‐down plants. Plant Physiol. 148, 808–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ, B. , Schelbert, S. , Aubry, S. , Sussenbacher, I. , Muller, T. , Krautler, B. and Hortensteiner, S. (2012) MES16, a member of the methylesterase protein family, specifically demethylates fluorescent chlorophyll catabolites during chlorophyll breakdown in Arabidopsis. Plant Physiol. 158, 628–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz, M.‐B.‐A.‐K.‐J. (1994) The plant 2‐Cys peroxiredoxin BAS1 is a nuclear‐encoded chloroplast protein: its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J. 12, 179–190. [DOI] [PubMed] [Google Scholar]
- Fan, J.Y. , Cui, Z.Q. , Wei, H.P. , Zhang, Z.P. , Zhou, Y.F. , Wang, Y.P. and Zhang, X.E. (2008) Split mCherry as a new red bimolecular fluorescence complementation system for visualizing protein‐protein interactions in living cells. Biochem. Bioph. Res. Co. 367, 47–53. [DOI] [PubMed] [Google Scholar]
- Galis, I. , Gaquerel, E. , Pandey, S.P. and Baldwin, I.T. (2009) Molecular mechanisms underlying plant memory in JA‐mediated defence responses. Plant Cell Environ. 32, 617–627. [DOI] [PubMed] [Google Scholar]
- Hashimoto, M. , Kisseleva, L. , Sawa, S. , Furukawa, T. , Komatsu, S. and Koshiba, T. (2004) A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol. 45, 550–559. [DOI] [PubMed] [Google Scholar]
- Heller, J. and Tudzynski, P. (2011) Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu. Rev. Phytopathol. 49, 369–390. [DOI] [PubMed] [Google Scholar]
- Hieno, A. , Naznin, H.A. , Inaba‐Hasegawa, K. , Yokogawa, T. , Hayami, N. , Nomoto, M. , Tada, Y. et al. (2019) Transcriptome analysis and identification of a transcriptional regulatory network in the response to H2O2. Plant Physiol. 180, 1629–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issakidis‐Bourguet, E. , Mouaheb, N. , Meyer, Y. and Miginiac‐Maslow, M. (2001) Heterologous complementation of yeast reveals a new putative function for chloroplast m‐type thioredoxin. Plant J. 25, 127–135. [DOI] [PubMed] [Google Scholar]
- Ji, H. , Liu, D. , Zhang, Z. , Sun, J. , Han, B. and Li, Z. (2020) A bacterial F‐box effector suppresses SAR immunity through mediating the proteasomal degradation of OsTrxh2 in rice. Plant J. 104, 1054–1072. [DOI] [PubMed] [Google Scholar]
- Jwa, N.S. , Kumar Agrawal, G. , Rakwal, R. , Park, C.H. and Prasad Agrawal, V. (2001) Molecular cloning and characterization of a novel Jasmonate inducible pathogenesis‐related class 10 protein gene, JIOsPR10, from rice (Oryza sativa L.) seedling leaves. Biochem Bioph Res Co 286, 973–983. [DOI] [PubMed] [Google Scholar]
- Khong, G.N. , Pati, P.K. , Richaud, F. , Parizot, B. , Bidzinski, P. , Mai, C.D. , Bes, M. et al. (2015) OsMADS26 negatively regulates resistance to pathogens and drought tolerance in rice. Plant Physiol. 169, 2935–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Langmead, B. and Salzberg, S.L. (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger‐Liszkay, A. (2005) Singlet oxygen production in photosynthesis. J Exp Bot 56, 337–346. [DOI] [PubMed] [Google Scholar]
- Lan, A. , Huang, J. , Zhao, W. , Peng, Y. , Chen, Z. and Kang, D. (2013) A salicylic acid‐induced rice (Oryza sativa L.) transcription factor OsWRKY77 is involved in disease resistance of Arabidopsis thaliana . Plant Biol. (Stuttg.) 15, 452–461. [DOI] [PubMed] [Google Scholar]
- Lemaire, S.D. , Michelet, L. , Zaffagnini, M. , Massot, V. and Issakidis‐Bourguet, E. (2007) Thioredoxins in chloroplasts. Curr. Genet. 51, 343–365. [DOI] [PubMed] [Google Scholar]
- Li, L. , Li, C. , Lee, G.I. and Howe, G.A. (2002) Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc. Natl Acad. Sci. USA, 99, 6416–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N. , Lin, B. , Wang, H. , Li, X. , Yang, F. , Ding, X. , Yan, J. et al. (2019a) Natural variation in ZmFBL41 confers banded leaf and sheath blight resistance in maize. Nat. Genet. 51, 1540–1548. [DOI] [PubMed] [Google Scholar]
- Li, W. , Chern, M. , Yin, J. , Wang, J. and Chen, X. (2019b) Recent advances in broad‐spectrum resistance to the rice blast disease. Curr. Opin. Plant Biol. 50, 114–120. [DOI] [PubMed] [Google Scholar]
- Li, W. , Zhu, Z. , Chern, M. , Yin, J. , Yang, C. , Ran, L. , Cheng, M. et al. (2017) A natural allele of a transcription factor in rice confers broad‐spectrum blast resistance. Cell, 170,114–126.e15. [DOI] [PubMed] [Google Scholar]
- Liang, X. , Chen, X. , Li, C. , Fan, J. and Guo, Z. (2017) Metabolic and transcriptional alternations for defense by interfering OsWRKY62 and OsWRKY76 transcriptions in rice. Sci. Rep. 7, 2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y. , Smyth, G.K. and Shi, W. (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lin, Y.J. and Zhang, Q. (2005) Optimising the tissue culture conditions for high efficiency transformation of Indica rice. Plant Cell Rep. 23, 540–547. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Li, X. , Xiao, J. and Wang, S. (2012a) A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice‐bacterium interaction. Plant Methods, 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Li, F. , Tang, J. , Wang, W. , Zhang, F. , Wang, G. , Chu, J. et al. (2012b) Activation of the jasmonic acid pathway by depletion of the hydroperoxide lyase OsHPL3 reveals crosstalk between the HPL and AOS branches of the oxylipin pathway in rice. PLoS One, 7, e50089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Cao, Y. , Zhang, Q. , Li, X. and Wang, S. (2018) A Cytosolic triosephosphate isomerase is a key component in XA3/XA26‐mediated resistance. Plant Physiol. 178, 923–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. and Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosalva, P. , Park, S. , Forouhar, F. , Tong, L. , Fry, W. and Klessig, D. (2010) Methyl esterase 1 (StMES1) is required for systemic acquired resistance against Phytophthora infestans in potato. Phytopathology, 100, S77. [DOI] [PubMed] [Google Scholar]
- Molla, K.A. , Karmakar, S. , Chanda, P.K. , Ghosh, S. , Sarkar, S.N. , Datta, S.K. and Datta, K. (2013) Rice oxalate oxidase gene driven by green tissue‐specific promoter increases tolerance to sheath blight pathogen (Rhizoctonia solani) in transgenic rice. Mol. Plant Pathol. 14, 910–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi, K. , Kondoh, A. , Stumpp, M.T. and Hisabori, T. (2001) Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc. Natl Acad. Sci. USA, 98, 11224–11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenbock, P. , Szechynska‐Hebda, M. , Plaszczyca, M. , Baudo, M. , Mateo, A. , Mullineaux, P.M. , Parker, J.E. et al. (2008) Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. (vol 20, pg 2339, 2008). Plant Cell, 20, 3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, D. , Xiao, J. , Ding, X. , Xiong, M. , Cai, M. , Cao, Y. , Li, X. et al. (2007) OsWRKY13 mediates rice disease resistance by regulating defense‐related genes in salicylate‐ and jasmonate‐dependent signaling. Mol. Plant Microbe Int. 20, 492–499. [DOI] [PubMed] [Google Scholar]
- Qu, S. , Liu, G. , Zhou, B. , Bellizzi, M. , Zeng, L. , Dai, L. , Han, B. et al. (2006) The broad‐spectrum blast resistance gene Pi9 encodes a nucleotide‐binding site‐leucine‐rich repeat protein and is a member of a multigene family in rice. Genetics, 172, 1901–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas, S. , Rougon‐Cardoso, A. , Smoker, M. , Schauser, L. , Yoshioka, H. and Jones, J.D. (2004) CITRX thioredoxin interacts with the tomato Cf‐9 resistance protein and negatively regulates defence. EMBO J. 23, 2156–2165. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ruan, B. , Hua, Z. , Zhao, J. , Zhang, B. , Ren, D. , Liu, C. , Yang, S. et al. (2019) OsACL‐A2 negatively regulates cell death and disease resistance in rice. Plant Biotechnol. J. 17, 1344–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, I. and Shah, K. .(2014) Evidences for structural basis of altered ascorbate peroxidase activity in cadmium‐stressed rice plants exposed to jasmonate. Biometals, 27, 247–263. [DOI] [PubMed] [Google Scholar]
- Singh, R. , Singh, S. , Parihar, P. , Mishra, R.K. , Tripathi, D.K. , Singh, V.P. , Chauhan, D.K. et al. (2016) Reactive oxygen species (ROS): beneficial companions of plants' developmental processes. Front. Plant Sci. 7, 1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobajima, H. , Tani, T. , Chujo, T. , Okada, K. , Suzuki, K. , Mori, S. , Minami, E. et al.(2007) Identification of a jasmonic acid‐responsive region in the promoter of the rice 12‐oxophytodienoic acid reductase 1 gene OsOPR1. Biosci Biotech. Bioch. 71, 3110–3115. [DOI] [PubMed] [Google Scholar]
- Sun, X. , Cao, Y. , Yang, Z. , Xu, C. , Li, X. , Wang, S. and Zhang, Q. .(2004) Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase‐like protein. Plant J. 37, 517–527. [DOI] [PubMed] [Google Scholar]
- Tang, N. , Zhang, H. , Li, X. , Xiao, J. and Xiong, L. (2012) Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 158, 1755–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani, T. , Sobajima, H. , Okada, K. , Chujo, T. , Arimura, S. , Tsutsumi, N. , Nishimura, M. et al. (2008) Identification of the OsOPR7 gene encoding 12‐oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta 227, 517–526. [DOI] [PubMed] [Google Scholar]
- Tao, Z. , Liu, H. , Qiu, D. , Zhou, Y. , Li, X. , Xu, C. and Wang, S. .(2009) A pair of allelic WRKY genes play opposite roles in rice‐bacteria interactions. Plant Physiol. 151, 936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, X. , Qi, J. , Zhu, X. , Mao, B. , Zeng, L. , Wang, B. , Li, Q. et al. (2012) The rice hydroperoxide lyase OsHPL3 functions in defense responses by modulating the oxylipin pathway. Plant J. 71, 763–775. [DOI] [PubMed] [Google Scholar]
- Torres, M.A. , Jones, J.D. and Dangl, J.L. (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres Zabala, M. , Littlejohn, G. , Jayaraman, S. , Studholme, D. , Bailey, T. , Lawson, T. , Tillich, M. et al. (2015) Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nat Plants, 1, 15074. [DOI] [PubMed] [Google Scholar]
- Vellosillo, T. , Vicente, J. , Kulasekaran, S. , Hamberg, M. and Castresana, C. (2010) Emerging complexity in reactive oxygen species production and signaling during the response of plants to pathogens. Plant Physiol. 154, 444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot, A.C. , Liu, P.P. , Cameron, R.K. , Park, S.W. , Yang, Y. , Kumar, D. , Zhou, F. et al. (2008) Identification of likely orthologs of tobacco salicylic acid‐binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana . Plant J. 56, 445–456. [DOI] [PubMed] [Google Scholar]
- Wang, G. , Ding, X. , Yuan, M. , Qiu, D. , Li, X. , Xu, C. and Wang, S. .(2006) Dual function of rice OsDR8 gene in disease resistance and thiamine accumulation. Plant Mol. Biol. 60, 437–449. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Sun, S. , Jin, J. , Fu, D. , Yang, X. , Weng, X. , Xu, C. et al. (2015) Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl Acad. Sci. USA, 112, 15504–15509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. , Stenzel, I. , Hause, B. , Hause, G. , Kutter, C. , Maucher, H. , Neumerkel, J. et al. (2006) The wound response in tomato–role of jasmonic acid. J. Plant Physiol. 163, 297–306. [DOI] [PubMed] [Google Scholar]
- Wong, H.L. , Sakamoto, T. , Kawasaki, T. , Umemura, K. and Shimamoto, K. .(2004) Down‐regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol. 135, 1447–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Yang, R. , Yang, Z. , Yao, S. , Zhao, S. , Wang, Y. , Li, P. et al.(2017) ROS accumulation and antiviral defence control by microRNA528 in rice. Nat Plants, 3, 16203. [DOI] [PubMed] [Google Scholar]
- Xie, K. , Minkenberg, B. and Yang, Y. .(2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA‐processing system. Proc. Natl Acad. Sci. USA, 112, 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, C. and Xie, D. .(2015) Jasmonate in plant defence: sentinel or double agent? Plant Biotechnol. J. 13, 1233–1240. [DOI] [PubMed] [Google Scholar]
- Yang, Y. , Xu, R. , Ma, C.J. , Vlot, A.C. , Klessig, D.F. and Pichersky, E. .(2008) Inactive methyl indole‐3‐acetic acid ester can be hydrolyzed and activated by several esterases belonging to the AtMES esterase family of Arabidopsis. Plant Physiol. 147, 1034–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani, N. , Shikata, M. , Ichikawa, H. , Mitsuda, N. , Ohme‐Takagi, M. , Minami, E. and Nishizawa, Y. (2018) OsWRKY24, a blast‐disease responsive transcription factor, positively regulates rice disease resistance. J. Gen. Plant Pathol. 84, 85–91. [Google Scholar]
- Yoo, Y. , Park, J.C. , Cho, M.H. , Yang, J. , Kim, C.Y. , Jung, K.H. , Jeon, J.S. et al. (2018) Lack of a cytoplasmic RLK, required for ROS homeostasis, induces strong resistance to bacterial leaf blight in rice. Front. Plant Sci. 9, 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Zhou, Y.F. , Feng, Y.Z. , He, H. , Lian, J.P. , Yang, Y.W. , Lei, M.Q. et al. (2020) Transcriptional landscape of pathogen‐responsive lncRNAs in rice unveils the role of ALEX1 in jasmonate pathway and disease resistance. Plant Biotechnol. J. 18, 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, M. , Chu, Z. , Li, X. , Xu, C. and Wang, S. .(2010) The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell, 22, 3164–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. and Wang, S. (2013) Rice versus Xanthomonas oryzae pv. oryzae: a unique pathosystem. Curr. Opin. Plant Biol. 16, 188–195. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Du, L. and Poovaiah, B.W. .(2014) Calcium signaling and biotic defense responses in plants. Plant Signal. Behav. 9, e973818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Bao, Y. , Shan, D. , Wang, Z. , Song, X. , Wang, Z. , Wang, J. et al. (2018) Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiol. 177, 352–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Su, J. , Duan, S. , Ao, Y. , Dai, J. , Liu, J. , Wang, P. et al. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast‐related processes. Plant Methods, 7, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y.T. , Zhang, Y.L. , Chen, S.X. , Yin, G.H. , Yang, Z.Z. , Lee, S. , Liu, C.G. et al. (2015) Proteomics of methyl jasmonate induced defense response in maize leaves against Asian corn borer. BMC Genom. 16, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B. , Peng, K. , Zhaohui, C. , Wang, S. and Zhang, Q. (2002) The defense‐responsive genes showing enhanced and repressed expression after pathogen infection in rice (Oryza sativa L.). Sci. China C Life Sci. 45, 449–467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The expression level of complementary lines.
Figure S2 Subcellular localization of OsMESL protein.
Figure S3 Detectiion of copy numbers in OsMESL‐RNAi lines, OsMESL‐OE lines and osmeslcomplementation lines
Figure S4 Phenotypes of OsTrxm OE lines inoculated with Xoo and R. solani.
Figure S5 GO analysis of DEGs in osmesl and WT.
Figure S6 Detection of disease resistance‐related indicators.
Figure S7 osmesl and RNAi lines showed resistance to rice blast.
Figure S8 Expression level of OsBAS1 in OsTrxm‐Cas9 mutant.
Table S1 Primers used in this study.
Table S2 Differentially expressed of defence response‐ and ROS scavenging‐related genes in osmesl plants.


