Summary
Cellulose is one of the most abundant organic polymers in nature. It contains multiple β‐1,4‐glucan chains synthesized by cellulose synthases (CesAs) on the plasma membrane of higher plants. CesA subunits assemble into a pseudo‐sixfold symmetric cellulose synthase complex (CSC), known as a ‘rosette complex’. The structure of CesA remains enigmatic. Here, we report the cryo‐EM structure of the homotrimeric CesA7 from Gossypium hirsutum at 3.5‐angstrom resolution. The GhCesA7 homotrimer shows a C3 symmetrical assembly. Each protomer contains seven transmembrane helices (TMs) which form a channel potentially facilitating the release of newly synthesized glucans. The cytoplasmic glycosyltransferase domain (GT domain) of GhCesA7 protrudes from the membrane, and its catalytic pocket is directed towards the TM pore. The homotrimer GhCesA7 is stabilized by the transmembrane helix 7 (TM7) and the plant‐conserved region (PCR) domains. It represents the building block of CSCs and facilitates microfibril formation. This structure provides insight into how eukaryotic cellulose synthase assembles and provides a mechanistic basis for the improvement of cotton fibre quality in the future.
Keywords: cellulose synthase, GhCesA, homotrimer, cotton fibres
Introduction
Cellulose is a highly abundant natural biopolymer composed of hydrogen‐bonded β‐1,4 glucans. It is predominantly synthesized by algae and plants, and is also found in some bacteria (Matthysse and Thomas, 1995; Grimson et al., 1996; Keegstra, 2010; Saxena and Brown, 2005). Cellulose is an essential component of plant cell walls, which provide mechanical support to plants and protect them from biotic and abiotic stresses (Cantu et al., 2008; Wang et al., 2016). It is widely used for various industrial products, including textiles, paper and biofuel (McFarlane et al., 2014).
Cellulose microfibrils are synthesized at the plasma membrane by the cellulose synthase complex (CSC) (Somerville et al., 2004). The CSC forms a rosette shape and is assembled symmetrically as six rosette subunits; each of the rosette subunits is proposed to be formed by the multimeric complexes of cellulose synthases (CesAs) (Doblin et al., 2002; Lampugnani et al., 2019; Somerville, 2006). CesAs catalyse the polymerization of β‐1,4 glucans in each rosette subunit and align them into protofibrils, which are subsequently bundled into microfibrils (Cosgrove, 2005; Saxena and Brown, 2005). Initial models proposed that each rosette subunit contains six CesAs (Doblin et al., 2002; Herth, 1983). Recently, Nixon and colleagues suggested that three CesAs constitute one subunit in moss using improved transmission electron microscopy images coupled with computer modelling (Nixon et al., 2016). This model was supported by the recently reported cryo‐electron microscopy (cryo‐EM) structure of the CesA homotrimer from Populus tremula x tremuloides (Purushotham et al., 2020). However, structures of CesAs from other plants have not been reported and whether they consistently form a trimer in the CSC subunit remains elusive.
Plant CesA is a membrane‐embedded Glycosyltransferase Family 2 (GT‐2) enzyme (Cantarel et al., 2009). It consists of an N‐terminal domain (NTD) with two zinc finger motifs, a transmembrane domain (TM), a glycosyltransferase (GT) domain, a plant‐conserved region (PCR) and a class‐specific region (CSR) (Pear et al., 1996). The zinc finger motifs are probably responsible for the oligomerization of CesA (Kurek et al., 2002). The GT domain is inserted between TM2 and TM3. It possesses conserved D, D, D, and QXXRW motifs and catalyses the polymerization of glucan using UDP‐activated glucose (UDP‐Glc) as the substrate. The PCR and CSR of plant CesA are inserted into the GT domain (Somerville, 2006; Vergara and Carpita, 2001). The PCR contains two antiparallel α‐helices and is speculated to interact with other domains in CesA (Rushton et al., 2017). The cysteine‐rich CSR was believed to facilitate the membrane localization of CesA via S‐acylation of cysteines (Kumar et al., 2016; Somerville, 2006; Vergara and Carpita, 2001). However, the mechanism underlying the synthesis and translocation of the glucan chain directed by plant CesA remains largely unknown.
Cotton fibre (Gossypium hirsutum) has a secondary cell wall of almost pure cellulose and is a primary material used for textile manufacturing (Brett, 2000). Thirty‐four CesA homologs identified in cotton are divided into six sub‐classes (Figure S1), among which GhCesA4, GhCesA7 and GhCesA8 predominantly participate in secondary cell wall development (Wang et al., 2019; Yuan et al., 2015). In this study, we report the cryo‐EM structure of oligosaccharide‐bound trimeric GhCesA7 at 3.5 Å resolution. The transmembrane region of GhCesA has seven transmembrane helices (TMs), of which TMs 1‐6 form a channel containing an oligosaccharide. In this structure, the catalytic core of the GT domain faces the entrance of the transmembrane channel and the PCR domain locates at the periphery of the GT domain. The PCR in the GhCesA7 protomer is close to the other PCRs of two neighbouring protomers and is associated with trimer formation. This structure offers insights into how CesAs organize into a trimer to generate the protofibril and provides a framework for future improvement of cotton fibre quality and yield.
Results
Structure determination of trimeric GhCesA7
The 34 CesA homologs from cotton were fused with different tags and recombinantly expressed in Expi293 cells. After initial screening, the GhCesA7 fused with an N‐terminal 3 × flag showed a high level of expression compared to the other constructs and was chosen for further purification. This recombinant GhCesA7 protein showed good homogeneity in size exclusion chromatography (SEC) in the presence of glyco‐diosgenin (GDN) (Figure 1a and Figure S2a). The protein was shown to possess cellulose synthase activity in vitro (described below). The details of recombinant GhCesA7 protein expression, purification and cryo‐EM image acquisition are described in the Materials and Methods. The cryo‐EM structure of the trimeric GhCesA7 (residues 24‐1,042) was determined at a resolution of 3.5 Å (Table S1, Figure S2b and S3). In the GhCesA7 trimer, the threefold symmetry axis of protomers is perpendicular to the plane of the membrane. The overall structure of the trimeric GhCesA7 is approximately 90 Å perpendicular and 120 Å parallel to the membrane (Figure 1b). Each protomer possesses seven α‐helical TMs (TM1 to TM7) and a bulky intracellular region (Figure 1c and Figure S4). The two regions of one protomer bend outward and form a large cavity with that of the other two protomers (Figure 1b and 1c). The bulky cytoplasmic region is composed of the GT, PCR and CSR domains and located between TM2 and TM3. Three helices (α11, α17, α21) lay on the interface between the TM region and the intracellular region (designated IF1, IF2 and IF3, respectively) (Figure 1c). The NTD (residues 24‐234), the residues 629 to 674 in the CSR region, and the residues 912 to 930 after IF3 lack well‐defined densities, which is indicative of their flexible nature.
Figure 1.
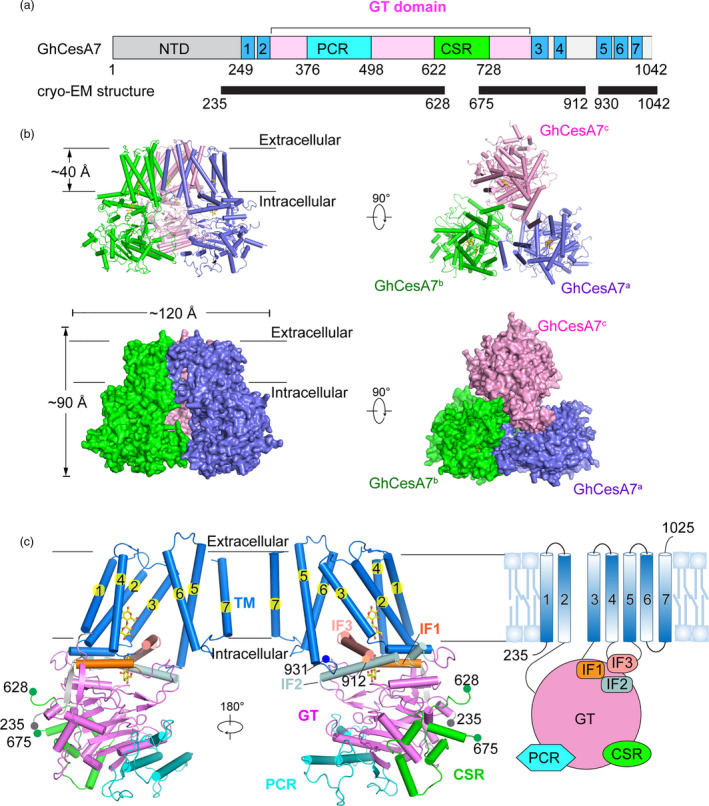
Cryo‐EM structure of GhCesA7. (a) Schematic of the domains of GhCesA7 (Uniprot: L7NUA2). Seven transmembrane helices are coloured blue. PCR and CSR within the GT domain (pink) are in cyan and green. NTD of GhCesA7 is coloured grey. Black bar labelled with exact residue number indicates the region with a model built by cryo‐EM experimentation. (b) The overall structure of GhCesA7 in side view and top view. Structures are shown as cartoon and surface coloured slate, green, and pink for each protomer. The glucan in each protomer is shown as a stick and coloured yellow. (c) Structure of a GhCesA7 protomer. The transmembrane region is coloured blue, and each helix is numbered. The NTD, PCR, CSR and GT domains are coloured as in (a). The IF1‐3 are coloured orange, pale cyan and salmon, respectively
The catalytic GT domain of GhCesA7
The cytoplasmic region of GhCesA7 contains the catalytic GT domain and two extra plant‐specific domains (PCR and CSR). The GT domain of GhCesA7 exhibits a typical GT‐A fold (Figure S5a). The seven β‐strands (β3, β4, β6, β7, β9, β11, β12) of the GT domain are organized into a β‐sheet and surrounded by seven helices (α4, α8, α9, IF1, α12, α16, η1). The conserved D, D, D and Q/RXXRW motifs in the catalytic core are located at β4, β7, α16 of the GT domain and IF2, respectively (Figure 2a, right panel, Figure S4, and S5a). To establish a structure‐function correlation, we employed a scintillation counting assay to detect the synthesis of high molecular weight glucose polymer. Compared to the wild‐type GhCesA7, a significant reduction in the signal, which is indicative of severely disrupted GhCesA7 cellulose synthase activity, was observed in three mutants (D540N, D742N, and W784A; Figures S5b and S5c). This suggests that D540, D742 and W784 play key roles in the synthesis of glucan. The analysis of the D374 site was not successful as the GhCesA protein with this mutation was not obtained.
Figure 2.
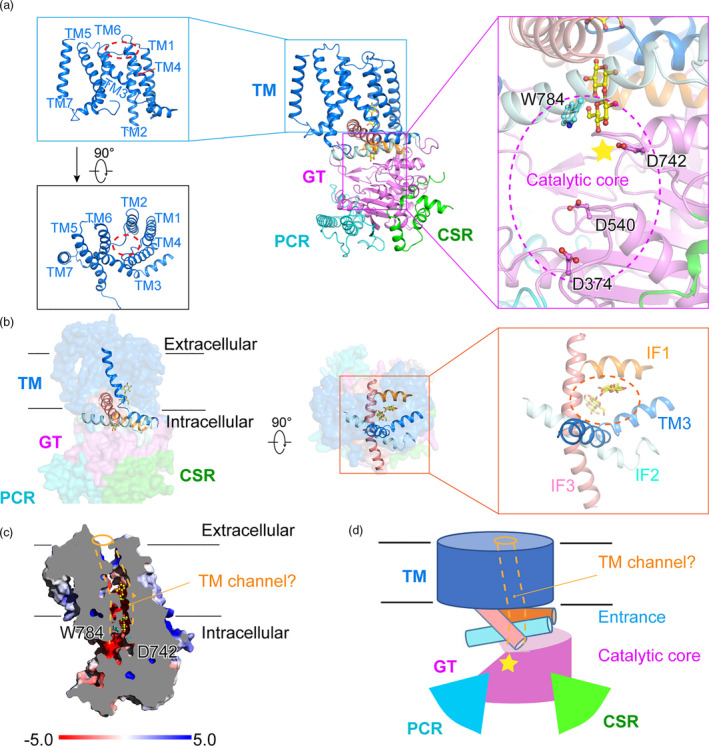
TM channel, IF1‐3 and the cytoplasmic region of GhCesA7. (a) Organization of the transmembrane helices (TMs) and the catalytic core of a protomer. The side view and top view of TMs are shown in left panels. The channel formed by TM1‐6 is indicated by a red circle. The catalytic core is circled and highlighted with a star in the right panel with key residues shown as sticks. (b) Surface structure of GhCesA7 with IF1‐3 and TM3 shown as cartoon in side and top view. The TM channel entrance formed by IF1‐3 and TM3 is highlighted with a circle. (c) A cut‐open view of GhCesA7. The electrostatic surface potential was calculated by PyMOL. The glucans occupying the channel are shown as yellow sticks. (d) The model of TM channel. The majority of the channel is formed by TMs. The IF1‐3 is located at the entrance and attached to the GT domain. The TMs, GT, PCR, CSR and IF1‐3 are coloured blue, violet, cyan, green, orange, pale cyan and salmon, respectively
Glucan translocating channel of GhCesA7
Two discontinuous disaccharide‐like densities, which are separated by one glucose‐sized space and surrounded by TM1‐TM6, are found near the catalytic core (Figure 2a and Figure S6). These densities may be a synthesized pentad glucan (Purushotham et al., 2020). We modelled the densities as two cellobioses, for cellobiose was incorporated during protein purification. The nonreducing end of the cellobiose lies near W784 of the Q/RXXRW motif in IF2 and the catalytic residue D742 in the GT domain (Figure 2a, right panel). Therefore, we consider this inner space formed by TM1‐TM6 to be the glucan translocating channel (Figure 2a, left panel). The N‐terminal short helix of TM3 bends at P823 and runs parallel to the membrane (Figure S8). This helix together with IF1‐IF3 forms the entrance of the glucan translocating channel, towards which the catalytic core of the GT domain faces (Figure 2b, 2c, Figure S7a and S7b).
Each IF contains three characteristic sides. The first side interacts with the TMs mainly via hydrophobic residues. The second one faces the cytoplasm and is enriched in hydrophilic residues. The third one is the inner surface of the channel entrance. The Phe and Trp residues on this surface, such as F592 and F593 of IF1, W784 of IF2, and W883 and W890 of IF3, guard the access of the newly added glucose moiety of the glucan chain (Figure S9). IF3 also possesses a ‘GGISA’ sequence, which allows it to bend towards TM5/6 and confers flexibility to the entrance (Figure S7c and S7d). This structure may be associated with the elongation of glucan.
The glucan translocating channel formed by TM1‐TM6 is 40 Å in length. It is enriched in aromatic residues, including F267 of TM1, F293 and W297 of TM2, Y832 of TM3, F862 and F866 of TM4, F978, F979 and W982 of TM6. This feature suggests that the glucan product is mainly associated with these amino acids via stacking interactions (Figure S9). The existence of P823 and P828 of TM3, P943 and P944 of TM5, and P989 of TM6 allows the transmembrane helices to bend outwards and facilitates the anchoring of IF3 between TM3 and TM5/TM6 (Figure S7a).
The plant unique PCR and CSR domains of GhCesA7
The plant‐specific PCR and CSR domains, which connect to the GT domain via long flexible loops, protrude from both sides of the GT domain at a distance from the membrane (Figure 1c). The PCR domain (residues 376 to 498) is located between β4 and β6 of the GT domain (Figure S4). Two antiparallel α‐helices (designated PCR‐ α1 and PCR‐ α2), which are linked by a short α‐helix (PCR‐ αs), form a coiled‐coil structure followed by a β strand (Figure S10). PCR‐ α1 and PCR‐ α2 interact with each other via stacking interactions between aromatic residues, including F390, W394, F397 and F401 in PCR‐ α1, and F427, Y438 and F441 in PCR‐ α2. PCR‐ αs turns back towards the α4 of the GT domain (Figure S10). The CSR domain (residues 622 to 728) locates between α12 and α16 of the GT domain (Figure 1c and Figure S4). The density of residues 629 to 674 is missing and only four helices (α13, α14, α15 and η2) located at the periphery of the GT domain are observed, at a distance from the trimer’s threefold symmetry axis. This domain may be related to the interactions between rosette units or the interactions with other proteins.
Trimer formation of GhCesA7
Two regions, TM7 and PCR, participate in the trimerization of GhCesA7 (Figure 3a). PCR‐ α1 and PCR‐ α2 from three protomers form a triangular plane. The side chains of R431, R432 and K435 in PCR‐ α2 point to the threefold symmetry axis and form a positively charged core. A conical density was found in the core, which appears to buttress the triangular plane (Figure S11). We speculate that this density might be a negatively charged molecule (e.g. nucleotide or inositol phosphate). Consistent with this observation, a R362K mutation of AtCesA8 in Arabidopsis (corresponding to R432 of GhCesA7) led to reduced cellulose content (Taylor et al., 2003). TM7 is not involved in the formation of the glucan translocating channel, but it is close to TM4 and TM6 of the neighbouring protomer (Figure 1 and 3a). Besides, an unidentified density was found at the centre of the trimer near the cytoplasmic face of the membrane, which is coordinated by the positively charged residue R1000 located at the loop between TM6 and TM7 (Figure S12). This molecule is predicted to be negatively charged and may play a role in stabilizing the conformation between TM6 and TM7. We speculate that these two unidentified molecules may glue the trimers of CesA together or play regulatory roles in cellulose synthesis. This is reminiscent of the bacterial BcsA‐BcsB complex, which was activated by cyclic di‐GMP (Morgan et al., 2014). Identification of these molecules in the future study will provide new insights into plant cellulose synthesis.
Figure 3.
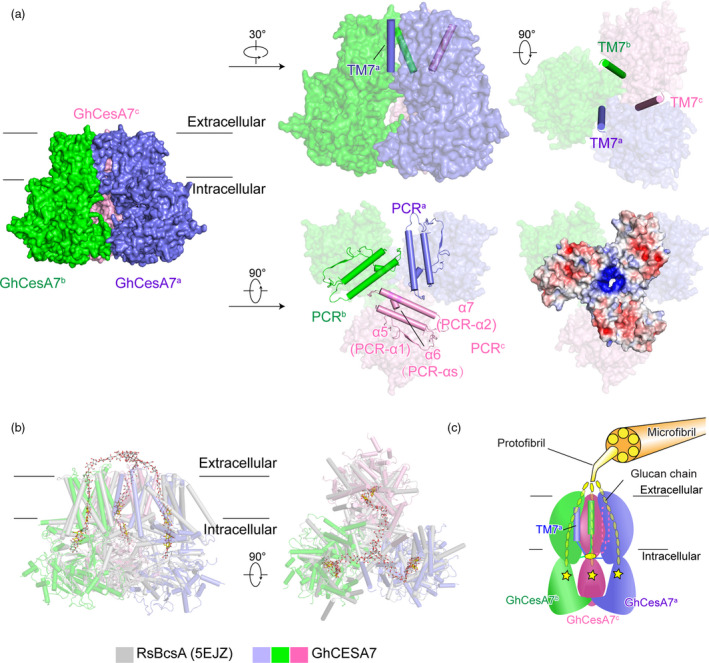
The trimerization of GhCesA7. (a) The TM7 and PCR of GhCesA7. Side view and top view of TM7 are shown as surface with TM7 in cartoon. The PCR are shown as cartoon and surface. The electrostatic surface potential of PCR was calculated by PyMOL. (b) Structure superimposition of GhCesA7 with RsBcsA (5EJZ). (c) Proposed model of trimeric GhCesA7 assembled for protofibril synthesis. Trimerization of GhCesA produces a protofibril with one glucan chain synthesized by one protomer. The catalytic core is highlighted with a star
Comparison between GhCesA7 and bacterial BcsA
In plants, the cellulose synthase complex produces multiple glucan chains which are subsequently bundled into protofibrils and microfibrils. In bacteria, amorphous cellulose is mainly synthesized by the BcsA‐BcsB complex (McNamara et al., 2015; Römling, 2002). The membrane protein BcsA is the catalytically active subunit containing a GT domain and eight TMs. BcsA facilitates the translocation of the polysaccharide through a narrow channel formed by TM3‐TM8 (Morgan et al., 2013). The four characteristic conserved motifs D, D, D and Q/RXXRW align well with those of BcsA (Figure S13), indicating that the eukaryotic and prokaryotic cellulose synthases share a conserved catalytic mechanism. TM1‐TM6 of GhCesA7 form a channel fold similar to TM3‐TM8 of BcsA (Figure S13). Moreover, glucan‐like density in the GhCesA7 channel is well aligned with the glucan near the catalytic core in BcsA (Figure 3b). To gain insights into the elongation of glucan chain, we modelled the trimeric GhCesA7 with three glucan‐bounded BcsA molecules. Three glucan chains point to and gather in the trimer’s threefold symmetry axis. These structural features raise the possibility that the glucan chains from eukaryotic trimeric CesA are bundled into the protofibrils at the periphery of the TM (Figure 3b). Six protofibrils may be bundled into microfibrils according to the ‘hexamer of trimers’ model (Figure 3c).
Discussion
Knowledge of how CesAs assemble in the CSC will facilitate the future genetic manipulation of plant cell wall synthesis and help improve feedstocks for the production of sustainable fuels and chemicals. The precise number and class of CesAs in a rosette CSC remained unclear for years. Previous studies proposed that CSC contains 18, 24 or 36 CesAs (Jarvis, 2013; Kieber and Polko, 2019), which suggested that one rosette subunit comprised the trimer, tetramer or hexamer of CesAs in a homo‐ or hetero‐ pattern. In this study, we report a trimeric structure of GhCesA7 and find that the TM7 and PCR domains of GhCesA7 are associated with trimer formation (Figure 3). This observation is consistent with the latest reported PttCesA8 trimer (Purushotham et al., 2020) (Figure S14), and this and GhCesA7 adopt almost identical conformations, with a root‐mean‐square deviation (RMSD) of 1.24 Å over 712 Cα atoms. TM7 and PCR are highly conserved among plant CesAs from different species (Figure S4, S11b and S12b), which suggests similar mechanisms underly the assembly of heterotrimers and homo‐trimers. Nevertheless, we cannot rule out the existence of the tetramer or hexamer CesA complexes in one rosette subunit since both of the solved CesAs structures were obtained using proteins surrounded by a belt of detergent. The natural components which affect the oligomerization of CSC might be lost during purification.
Cellulose synthase interactive (CSI) proteins, as a link between CSCs and cortical microtubules, have been shown to be involved in primary cell wall formation (Lei et al., 2015). PttCesA8 and GhCesA7 both participate in secondary cell wall development (Zhang et al., 2018). Interestingly, the NTD (residues 1‐156) of trimeric PttCesA8, representing a cytosolic stalk formation, interacts with CSI (Purushotham et al., 2020). However, we were unable to model the NTD of GhCesA7 due to poor density. Moreover, the sequences of CSCs show a high variability in the NTD (Figure S4, S14c). Whether GhCesA7 interacts with CSI via the NTD requires further investigation.
Cotton fibre is a renewable resource and the mature fibre comprises more than 90% cellulose (Lee et al., 2007). Fibre development facilitates the expansion of the primary cell wall and the formation of the secondary cell wall (Haigler et al., 2012). Six classes of cotton CesAs (CesA1, CesA3, CesA4, CesA6, CesA7 and CesA8) involve in cellulose synthesis (Figure S1). Previously, we found that GhCesA4, GhCesA7 and GhCesA8 predominantly contribute to the cellulose production of the cotton secondary cell wall, especially in the 20 d post‐anthesis fibre (Wang et al., 2019; Yuan et al., 2015). Taylor et al., (2003) have reported that CesA4, CesA7 and CesA8 of Arabidopsis are coexpressed in the same cells, and all three proteins interact in detergent‐solubilized extracts, which suggests CesA4, CesA7 and CesA8 can form a heterotrimer. At the secondary cell wall synthesis stage of cotton fibre development, the fragments per kilobase per million (FPKM) ratio of GhCesA4, GhCesA7 and GhCesA8 was approximately at 1.0:1.1:0.8 based on our RNA‐seq data (https://cottonfgd.org/), which indicates that secondary cell wall CSC perhaps undertakes a heterotrimeric conformation in cotton. We are trying heterologous coexpression of GhCesA4, GhCesA7 and GhCesA8 in a mammalian cell system to determine whether they can conform a heterotrimer.
Crispr/Cas9 cotton mutants of each CesA isoform are at an early stage of evaluation and will be reported on in the future. GhCesA7 mutant cotton fibres are finer and longer than wild type, indicating improved fibre quality (unpublished data). We have also overexpressed the CesA isoforms in cotton. CesAs fused with GFP tags are being overexpressed in transgenic cotton, and endogenous natural CSC will be isolated to verify whether the CSC exists as a heterotrimer or a homotrimer. With the results in vitro and in vivo, we hope to elucidate further the structure and activity of CSC and determine the most appropriate CSC structure for optimum cotton fibre development and quality. Therefore, the structure of GhCesA7 will not only provide a milestone for further studies on the molecular mechanisms of fibre cellulose synthesis, but also pave a path for cotton fibre improvement.
Experimental Procedures
Expression of GhCesAs
The coding sequences of 34 full‐length Gossypium hirsutum CesAs (GhCesAs) were codon‐optimized, synthesized (General Biosystems Inc.) and subcloned into pMLink vector with an N‐terminal 3 × Flag tag, respectively. Expi293FTM cells (Invitrogen) were cultured in SMM 293TI medium (Sino Biological Inc.) at 37°C under 5% CO2 and 80% humidity in a ZCZY‐CS8 shaker (Shanghai Zhichu Instrument co., Ltd). The cells were diluted into 2.0 × 106 cells per mL after reaching a density of 3.5 × 106 ~ 4.0 × 106 cells per mL. The diluted cells were transfected by the pMLink plasmid containing GhCesA and 25‐kDa linear polyethylenimines (PEIs) (Polysciences). For one litre of cell culture, approximately 4 mg plasmid was pre‐mixed with 12 mg PEIs in 50 mL of fresh medium. The mixture was incubated for 15‐30 min before its addition into the diluted cell culture. Transfected cells were harvested after 48 hours.
Preparation of transfected cells and initial expression tests of GhCesAs
Transfected cells were centrifuged at 2,000 × g for 20 min and washed with cold PBS. The cells were then resuspended in the lysis buffer (20 mM Tris‐HCl, pH 7.4 and 150 mM NaCl) supplemented with 5 mM cellobiose (Sigma) and 1 mM phenylmethylsulphonyl fluoride (PMSF). To solubilize membrane, 1% (w/v) n‐Dodecyl β‐D‐maltoside (DDM, Bluepus) was added to the cell lysate and incubated at 4°C for 2 hours. The cell debris was removed by centrifugation at 100,000 x g for 1 h, and the supernatant was applied to anti‐Flag G1 affinity resin (Genescript). After 1 h incubation by gentle rotation at 4°C, the resin was washed with 20 column volumes of lysis buffer with the addition of 0.01% DDM and 1 mM PMSF. The protein was eluted with lysis buffer plus 0.01% DDM and 250 μg/mL Flag peptide (Genescript). The eluent was subjected to 10% SDS‐polyacrylamide gel electrophoresis (SDS‐PAGE) and stained with Coomassie brilliant blue (Bio‐Rad).
Purification of GhCesA7
For cryo‐EM sample preparation, the full‐length GhCesA7 was subjected to boundary screening. One of the N‐terminus truncated GhCesA7 boundary (residues 24‐1042) exhibited an improved protein yield. The membrane fraction was solubilized at 4°C for 2 hours with 1% (w/v) DDM, 0.25% (w/v) Asolectin (Sigma) and 0.1% (w/v) cholesteryl hemisuccinate (CHS, Anatrace). The supernatant was incubated with anti‐Flag G1 affinity resin at 4°C for 1 h. The resin was rinsed with the lysis buffer supplemented with 0.02% (w/v) glycol‐diosgenin (GDN, Anatrace) and 1 mM PMSF. The protein was eluted with the lysis buffer plus 0.02% (w/v) GDN and 250 μg/mL Flag peptide. The eluate was concentrated and further purified by Superose‐6 increase 10/300 column (GE Healthcare) in the buffer containing 20 mM Tris pH 8.0, 150 mM NaCl, 5 mM cellobiose, and 0.02% (w/v) GDN. The peak fractions were pooled, concentrated to 3 mg/mL using a 100‐kDa MWCO centrifugal device (Milipore), and immediately used for cryo‐EM grid preparation.
For the activity assay, wild‐type GhCesA7 and its variants were purified through anti‐Flag affinity resin and Superose‐6 increase 10/300 column without the addition of cellobiose in the process of cell resuspension, membrane solubilization and purification.
Grid preparation and data acquisition
Cryo grids were prepared using Vitrobot Mark Ⅳ (Thermo Fisher Scientific). Aliquots of 3.5 μL of GhCesA were dropped onto plasma cleaned holey carbon grids (Quantifoil Cu R1.2/1.3, 300 mesh). After blotting for 3.5 s at 100% humidity at 8°C, grids were plunged into liquid ethane cooled with liquid nitrogen.
Micrograph stacks were automatically collected with AutoEmation (Lei and Frank, 2005) on a Titan Krios at 300 kV equipped with K3 Summit direct electron detector (Gatan) at a nominal magnification of 81,000 × with defocus values from −1.8 μm to −2.5 μm. Each stack was exposed in the counting mode for 2.56 s with an exposing time of 0.08 s per frame, resulting in a total of 32 frames per stack. The total dose rate was about 50 e‐/Å2 for each stack. The stacks were motion corrected with MotionCor2 (Zheng et al., 2017) with a binning factor of 2, resulting in a pixel size of 1.087 Å, while dose weighting was performed simultaneously. The defocus values were estimated using CTFFIND4 (Rohou and Grigorieff, 2015).
Cryo‐EM data processing
After motion correction, 2,715,441 particles were automatically picked from 5,116 micrographs using ‘Blob Picker’. The best class averages of trimeric particles were selected for heterogeneous refinement. An initial model was generated using good particles from the best class following another round of heterogeneous refinement with a binning factor of 1. Subsequently, non‐uniform refinement was conducted and C3 symmetry restraints were applied. 2D classification and 3D classification were performed using cryoSPARC (Punjani et al., 2017). Local resolution variations were estimated by ResMap (Kucukelbir et al., 2014).
Model building and refinement
The atomic model for GhCesA7 trimer lacking NTD was built in COOT (Emsley et al., 2010) and refined with PHENIX (Adams et al., 2010). The structure of GhCesA7 was validated through the examination of Molprobity (Davis et al., 2007) scores. Resolution was estimated with the gold standard Fourier shell correlation 0.143 criterion. High‐resolution images were prepared using PyMOL.
Activity assays
About 4 mg of GhCesA7 was incubated in the presence of 5 mM UDP‐glucose (Sigma), and 0.25 µCi UDP‐[3H]‐Glc (American Radiolabeled Chemicals, Inc) in a 200 μL reaction buffer containing 20 mM Tris pH 7.4, 100 mM NaCl and 20 mM MgCl2. After incubation overnight at 30°C, the water‐insoluble polymer was pelleted by centrifugation at 14,000 g at room temperature for 20 min and washed at least twice in 60% ethanol. The pellet was then gently resuspended in 60% ethanol and subsequently transferred into a 96‐well plate. The amount of synthesized cellulose in the pellet was quantified by scintillation counting (Beckman) as previously described (Purushotham et al., 2016).
Accession Numbers
The structure factors and atomic coordinates of trimeric GhCesA7 have been deposited at the Protein Data Bank (www.rcsb.org) under accession code PDB 7D5K. The cryo‐EM density map of this trimer has been deposited at the Electron Microscopy Data Bank (www.ebi.ac.uk/pdbe/emdb/) with accession code EMD‐30583.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
X.L Zhang., P. Yin., and L.L Tu., conceived the project. P. Yin., and L.L Tu., designed the experiments. X.N Zhang., Y. Xue., C. Zhou., Y.F Nie., S. Men., C.C Shen., and D.L Zhang., performed protein expression and purification. Y. Xue., and Q. Wang., prepared the cryo‐EM grids and collected the EM data. X.N Zhang carried out the enzyme assays. Z.Y Guan determined the structure. All authors analysed the data and contributed to the manuscript preparation. C.C Shen., P. Yin., L.L Tu., and X.L Zhang., wrote the manuscript.
Supporting information
Figure S1. Phylogenetic tree of GhCesAs. Phylogenetic tree constructed using MEGA7 with 500 bootstrap replicates based on amino acid sequences of the 34 protein‐encoding Gossypium hirsutum genes from six classes (CesA1, CesA3, CesA4, CesA6, CesA7 and CesA8) from the CottonGen database (https://www.cottongen.org/). GhCesA7 D7 in the red box is the gene used in this study. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to each branch.
Figure S2. Purification and data processing. (a) Gel filtration chromatography (Superose‐6 increase 10/300 column) of GDN‐solubilized GhCesA7. The fraction indicated by dashed lines was pooled and concentrated for sample preparation immediately. (b) Outline of cryo‐EM data processing. Based on the FSC value of 0.143, the final reconstruction has an average resolution of 3.5 Å. Details are presented in Materials and Methods.
Figure S3. Cryo‐EM analysis. (a) FSC curves before (blue) and after (orange) the application of soft mask. The overall resolution at the 0.5 criterion is marked. (b) The EM density map of the color‐coded GhCesA7 showing the local resolution estimated by ResMap. (c) The electron microscopy maps for the transmembrane helices of the GhCesA7.
Figure S4. Sequence alignment of CesAs. The sequences (Uniprot code: GhCesA7, L7NUA2; AtCesA7, Q9SWW6; OsCesA8, Q84ZN6; PpCesA7, Q3Y4F5; RsBcsA, Q3J125 and PttCesA8 (PDB accession code: 6WLB)) are aligned using MAFFT (https://www.ebi.ac.uk/Tools/msa/mafft/). The conserved residues are colored yellow, and the secondary structural elements are indicated above the sequences by ENDscript program. Residues involved in the catalytic reaction are highlighted with blue dots. The transmembrane helices are colored slate. And the NTD, PCR, CSR, IF1, IF2 and IF3 are in grey, cyan, green, orange, sky‐blue, and salmon, respectively.
Figure S5. Topology diagrams and mutagenesis analysis of the GT domain. (a) Secondary elements are labeled. D, D, D, W motif is highlighted by green triangles. Coloring scheme for β‐strand: blue, α‐helix: red. (b) Reaction catalyzed by cellulose synthase and the structure of cellulose building block. Coloring scheme for carbon: yellow, oxygen: red. (c) Catalytic activity of each GhCesA7 variant. CPM, count per minute. Each dot represents a biological replicate; bars indicate ± SEM. Image is generated by GraphPad Prism7.
Figure S6. Density in the TM channel. Density mesh in the TM channel can accommodate two cellobiose molecules. GhCesA7 is in the same color scheme as in Figure 1c. Cellobiose are shown as sticks in yellow.
Figure S7. The bent TM2‐3, TM5‐6 and IF3. (a)(c) The TMs and IF3 are bent due to the existence of Prolines which are showed as sticks and sequential flexible residues between α21 and α22, respectively. (b)(d) Sequence alignments of TMs and IF3 from different organisms (GhCesA7, Gossypium hirsutum; PttCesA8, Populus tremula × tremuloides; AtCesA7, Arabidopsis thaliana; OsCesA8, Oryza sativa; PpCesA7, Physcomitrella patens; RsBcsA, Rhodobacter sphaeroides). The key conserved Prolines are highlighted in yellow and the flexible residues of the bent region of IF3 are highlighted in the purple boxed. Similar residues and identical residues are written with blue and red characters.
Figure S8. Hydropathy of IF1‐3. (a) Entrance formed by IF1‐3. The face interacted with TMs is presented in right‐up panel with hydrophobic residues shown in sticks. The bottom panel shows the face towards cytosol with hydrophilic residues shown in sticks. (b) Sequence alignment of IF1‐3 of CesAs from different organisms as Figure S7. Color legend indicates the hydropathy of residues. Similar residues are written with blue characters. Identical residues are written with black characters and highlighted in yellow.
Figure S9. Residues involved in the TM channel and catalytic activity. Residues surrounding glucans or related to the catalytic reaction are shown as sticks and labeled based on their location. TMs, blue; IF1, orange; IF2, cyan; IF3, magenta; GT domain, deep pink. The cellobiose molecules occupying the channel are colored in yellow.
Figure S10. Helices in PCR. The α6 (PCR‐αs) is bent back to GT domain and residues that face towards each helix in PCR are labeled and shown as sticks.
Figure S11. Density in the center of PCRs in trimeric GhCesA7. (a) Side and bottom views of the uncharacterized density located in the center of trimeric PCR. Coordinating residues are labeled and shown as sticks. (b) Sequence alignment of helix 7 (PCR‐α2). Residues interacting with the uncharacterized density are written with black characters and highlighted in yellow.
Figure S12. Density in the center of loops between TM6 and TM7. (a) Top view of the density in the axis of trimeric loop between TM6 and TM7. R1000 in each protomer interacting with the uncharacterized density is shown in the right panel. (b) Sequence alignment of the same region with the conserved Arginine highlighted in yellow.
Figure S13. Alignment of GhCesA7 and RsBcsA. RsBcsA (PDB: 5EJZ) is colored gray and GhCesA7 is colored as in Figure 1c. TMs are aligned and labeled in the top panels (with green outline) and the channel is indicated with an orange circle. GT domains are aligned and the catalytic core is presented in the bottom panels in which UDP (5EJZ) and residues involved are labeled and shown as sticks.
Figure S14. Alignment with PttCesA8. (a) Overall structure alignment of trimeric GhCesA7 and PttCesA8 (PDB: 6WLB). PttCesA8 is colored gray and GhCesA7 is colored as in Figure 1b. (b) Alignment of the protomers from GhCesA7 and PttCesA8. The glucans within the protomer are located at the same region. (c) Sequence alignment of NTD from PttCesA8 and GhCesA. Similar residues and identical residues are indicated with blue and red characters.
Table S1. Statistics of Cryo‐EM data collection and refinement.
Acknowledgements
We are grateful to the Cryo‐EM Facility Center of Westlake University for providing technical support during cryo‐EM data acquisition. We thank the research associates at the Center for Protein Research and Public Laboratory of Electron Microscopy, Huazhong Agricultural University, for technical support. This work was supported by funds from the Ministry of Science and Technology of China (2018YFA0507700), the National Natural Science Foundation of China (31830062 and 31671739) and the Fundamental Research Funds for the Central Universities (2662017PY031).
Zhang, X. , Xue, Y. , Guan, Z. , Zhou, C. , Nie, Y. , Men, S. , Wang, Q. , Shen, C. , Zhang, D. , Jin, S. , Tu, L. , Yin, P. and Zhang, X. (2021) Structural insights into homotrimeric assembly of cellulose synthase CesA7 from Gossypium hirsutum . Plant Biotechnol J, 10.1111/pbi.13571
Contributor Information
Lili Tu, Email: lilitu@mail.hzau.edu.cn.
Ping Yin, Email: yinping@mail.hzau.edu.cn.
Xianlong Zhang, Email: xlzhang@mail.hzau.edu.cn.
References
- Adams, P.D. , Afonine, P.V. , Bunkoczi, G. , Chen, V.B. , Davis, I.W. , Echols, N. , Headd, J.J. et al.(2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett, C.T. (2000) Cellulose microfibrils in plants: biosynthesis, deposition, and integration into the cell wall. Int. Rev. Cytol. 199, 161–199. [DOI] [PubMed] [Google Scholar]
- Cantarel, B.L. , Coutinho, P.M. , Rancurel, C. , Bernard, T. , Lombard, V. and Henrissat, B. (2009) The Carbohydrate‐Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu, D. , Vicente, A.R. , Labavitch, J.M. , Bennett, A.B. and Powell, A.L. (2008) Strangers in the matrix: plant cell walls and pathogen susceptibility. Trends Plant Sci. 13, 610–617. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2005) Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861. [DOI] [PubMed] [Google Scholar]
- Davis, I.W. , Leaver‐Fay, A. , Chen, V.B. , Block, J.N. , Kapral, G.J. , Wang, X. , Murray, L.W. et al.(2007) MolProbity: all‐atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin, M.S. , Kurek, I. , Jacob‐Wilk, D. and Delmer, D.P. (2002) Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiology. 43, 1407–1420. [DOI] [PubMed] [Google Scholar]
- Emsley, P. , Lohkamp, B. , Scott, W.G. and Cowtan, K. (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson, M.J. , Haigler, C.H. and Blanton, R.L. (1996) Cellulose microfibrils, cell motility, and plasma membrane protein organization change in parallel during culmination in Dictyostelium discoideum. J. Cell Sci. 109(Pt 13), 3079–3087. [DOI] [PubMed] [Google Scholar]
- Haigler, C.H. , Betancur, L. , Stiff, M.R. and Tuttle, J.R. (2012) Cotton fiber: a powerful single‐cell model for cell wall and cellulose research. Frontiers In Plant Science. 3, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herth, W. (1983) Arrays of plasma‐membrane "rosettes" involved in cellulose microfibril formation of Spirogyra. Planta 159, 347–356. [DOI] [PubMed] [Google Scholar]
- Jarvis, M.C. (2013) Cellulose biosynthesis: counting the chains. Plant Physiol. 163, 1485–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra, K. (2010) Plant cell walls. Plant Physiol. 154, 483–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber, J.J. and Polko, J. (2019) The Regulation of Cellulose Biosynthesis in Plants. Plant Cell. 31(2), 282–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir, A. , Sigworth, F.J. and Tagare, H.D. (2014) Quantifying the local resolution of cryo‐EM density maps. Nat. Methods 11, 63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, M. , Wightman, R. , Atanassov, I. , Gupta, A. , Hurst, C.H. , Hemsley, P.A. and Turner, S. (2016) S‐Acylation of the cellulose synthase complex is essential for its plasma membrane localization. Science 353, 166–169. [DOI] [PubMed] [Google Scholar]
- Kurek, I. , Kawagoe, Y. , Jacob‐Wilk, D. , Doblin, M. and Delmer, D. (2002) Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc‐binding domains. Proc Natl Acad Sci. USA 99, 11109–11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani, E.R. , Flores‐Sandoval, E. , Tan, Q.W. , Mutwil, M. , Bowman, J.L. and Persson, S. (2019) Cellulose Synthesis ‐ Central Components and Their Evolutionary Relationships. Trends Plant Sci. 24, 402–412. [DOI] [PubMed] [Google Scholar]
- Lee, J.J. , Woodward, A.W. and Chen, Z.J. (2007) Gene expression changes and early events in cotton fibre development. Ann Bot. 100, 1391–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, J. and Frank, J. (2005) Automated acquisition of cryo‐electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J. Struct. Biol. 150, 69–80. [DOI] [PubMed] [Google Scholar]
- Lei, L. , Singh, A. , Bashline, L. , Li, S. , Yingling, Y.G. and Gu, Y. (2015) CELLULOSE SYNTHASE INTERACTIVE1 Is Required for Fast Recycling of Cellulose Synthase Complexes to the Plasma Membrane in Arabidopsis. Plant Cell. 27, 2926–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse, A.g. , Thomas, D.l. (1995) Mechanism of Cellulose Synthesis in Agrobacterium tumefaciens. J. Bacteriol. 177, 1076–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane, H.E. , Doring, A. and Persson, S. (2014) The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 65, 69–94. [DOI] [PubMed] [Google Scholar]
- McNamara, J.T. , Morgan, J.L. and Zimmer, J. (2015) A molecular description of cellulose biosynthesis. Annu. Rev. Biochem. 84, 895–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, J.L. , McNamara, J.T. and Zimmer, J. (2014) Mechanism of activation of bacterial cellulose synthase by cyclic di‐GMP. Nat. Struct. Mol. Biol. 21, 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, J.L. , Strumillo, J. and Zimmer, J. (2013) Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon, B.T. , Mansouri, K. , Singh, A. , Du, J. , Davis, J.K. , Lee, J.G. , Slabaugh, E. et al.(2016) Comparative Structural and Computational Analysis Supports Eighteen Cellulose Synthases in the Plant Cellulose Synthesis Complex. Sci. Rep. 6, 28696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear, J.R. , Kawagoe, Y. , Schreckengost, W.E. , Delmer, D.P. and Stalker, D.M. (1996) Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci. USA 93, 12637–12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani, A. , Rubinstein, J.L. , Fleet, D.J. and Brubaker, M.A. (2017) cryoSPARC: algorithms for rapid unsupervised cryo‐EM structure determination. Nat. Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Purushotham, P. , Cho, S.H. , Diaz‐Moreno, S.M. , Kumar, M. , Nixon, B.T. , Bulone, V. and Zimmer, J. (2016) A single heterologously expressed plant cellulose synthase isoform is sufficient for cellulose microfibril formation in vitro. Proc Natl Acad Sci. USA 113, 11360–11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushotham, P. , Ho, R. and Zimmer, J. (2020) Architecture of a catalytically active homotrimeric plant cellulose synthase complex. Science 369(6507), 1089–1094. [DOI] [PubMed] [Google Scholar]
- Rohou, A. and Grigorieff, N. (2015) CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling, U. (2002) Molecular biology of cellulose production in bacteria. Res. Microbiol. 153(4), 205–212. [DOI] [PubMed] [Google Scholar]
- Rushton, P.S. , Olek, A.T. , Makowski, L. , Badger, J. , Steussy, C.N. , Carpita, N.C. and Stauffacher, C.V. (2017) Rice Cellulose SynthaseA8 Plant‐Conserved Region Is a Coiled‐Coil at the Catalytic Core Entrance. Plant Physiol. 173, 482–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, I.M. and Brown, R.M. Jr .(2005) Cellulose biosynthesis: current views and evolving concepts. Ann Bot. 96, 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville, C. (2006) Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol. 22, 53–78. [DOI] [PubMed] [Google Scholar]
- Somerville, C. , Bauer, S. , Brininstool, G. , Facette, M. , Hamann, T. , Milne, J. , Osborne, E. et al.(2004) Toward a Systems Approach to Understanding Plant Cell Walls. Science 306, 2206–2211. [DOI] [PubMed] [Google Scholar]
- Taylor, N.G. , Howells, R.M. , Huttly, A.K. , Vickers, K. and Turner, S.R. (2003) Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci. USA 100, 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara, C.E. and Carpita, N.C. (2001) β‐D‐Glycan synthases and the CesA gene family: lessons to be learned from the mixed‐linkage (1→3), (1→4)β‐D‐glucan synthase. Plant Mol. Biol. 47, 145–160. [PubMed] [Google Scholar]
- Wang, M. , Tu, L. , Yuan, D. , Zhu, D. , Shen, C. , Li, J. , Liu, F. et al.(2019) Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 51, 224–229. [DOI] [PubMed] [Google Scholar]
- Wang, T. , McFarlane, H.E. and Persson, S. (2016) The impact of abiotic factors on cellulose synthesis. J. Exp. Bot. 67, 543–552. [DOI] [PubMed] [Google Scholar]
- Yuan, D. , Tang, Z. , Wang, M. , Gao, W. , Tu, L. , Jin, X. , Chen, L. et al.(2015) The genome sequence of Sea‐Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 5, 17662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Dominguez, P.G. , Kumar, M. , Bygdell, J. , Miroshnichenko, S. , Sundberg, B. , Wingsle, G. and et al.(2018) Cellulose Synthase Stoichiometry in Aspen Differs from Arabidopsis and Norway Spruce. Plant Physiol. 177, 1096–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, S.Q. , Palovcak, E. , Armache, J.P. , Verba, K.A. , Cheng, Y. and Agard, D.A. (2017) MotionCor2: anisotropic correction of beam‐induced motion for improved cryo‐electron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phylogenetic tree of GhCesAs. Phylogenetic tree constructed using MEGA7 with 500 bootstrap replicates based on amino acid sequences of the 34 protein‐encoding Gossypium hirsutum genes from six classes (CesA1, CesA3, CesA4, CesA6, CesA7 and CesA8) from the CottonGen database (https://www.cottongen.org/). GhCesA7 D7 in the red box is the gene used in this study. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to each branch.
Figure S2. Purification and data processing. (a) Gel filtration chromatography (Superose‐6 increase 10/300 column) of GDN‐solubilized GhCesA7. The fraction indicated by dashed lines was pooled and concentrated for sample preparation immediately. (b) Outline of cryo‐EM data processing. Based on the FSC value of 0.143, the final reconstruction has an average resolution of 3.5 Å. Details are presented in Materials and Methods.
Figure S3. Cryo‐EM analysis. (a) FSC curves before (blue) and after (orange) the application of soft mask. The overall resolution at the 0.5 criterion is marked. (b) The EM density map of the color‐coded GhCesA7 showing the local resolution estimated by ResMap. (c) The electron microscopy maps for the transmembrane helices of the GhCesA7.
Figure S4. Sequence alignment of CesAs. The sequences (Uniprot code: GhCesA7, L7NUA2; AtCesA7, Q9SWW6; OsCesA8, Q84ZN6; PpCesA7, Q3Y4F5; RsBcsA, Q3J125 and PttCesA8 (PDB accession code: 6WLB)) are aligned using MAFFT (https://www.ebi.ac.uk/Tools/msa/mafft/). The conserved residues are colored yellow, and the secondary structural elements are indicated above the sequences by ENDscript program. Residues involved in the catalytic reaction are highlighted with blue dots. The transmembrane helices are colored slate. And the NTD, PCR, CSR, IF1, IF2 and IF3 are in grey, cyan, green, orange, sky‐blue, and salmon, respectively.
Figure S5. Topology diagrams and mutagenesis analysis of the GT domain. (a) Secondary elements are labeled. D, D, D, W motif is highlighted by green triangles. Coloring scheme for β‐strand: blue, α‐helix: red. (b) Reaction catalyzed by cellulose synthase and the structure of cellulose building block. Coloring scheme for carbon: yellow, oxygen: red. (c) Catalytic activity of each GhCesA7 variant. CPM, count per minute. Each dot represents a biological replicate; bars indicate ± SEM. Image is generated by GraphPad Prism7.
Figure S6. Density in the TM channel. Density mesh in the TM channel can accommodate two cellobiose molecules. GhCesA7 is in the same color scheme as in Figure 1c. Cellobiose are shown as sticks in yellow.
Figure S7. The bent TM2‐3, TM5‐6 and IF3. (a)(c) The TMs and IF3 are bent due to the existence of Prolines which are showed as sticks and sequential flexible residues between α21 and α22, respectively. (b)(d) Sequence alignments of TMs and IF3 from different organisms (GhCesA7, Gossypium hirsutum; PttCesA8, Populus tremula × tremuloides; AtCesA7, Arabidopsis thaliana; OsCesA8, Oryza sativa; PpCesA7, Physcomitrella patens; RsBcsA, Rhodobacter sphaeroides). The key conserved Prolines are highlighted in yellow and the flexible residues of the bent region of IF3 are highlighted in the purple boxed. Similar residues and identical residues are written with blue and red characters.
Figure S8. Hydropathy of IF1‐3. (a) Entrance formed by IF1‐3. The face interacted with TMs is presented in right‐up panel with hydrophobic residues shown in sticks. The bottom panel shows the face towards cytosol with hydrophilic residues shown in sticks. (b) Sequence alignment of IF1‐3 of CesAs from different organisms as Figure S7. Color legend indicates the hydropathy of residues. Similar residues are written with blue characters. Identical residues are written with black characters and highlighted in yellow.
Figure S9. Residues involved in the TM channel and catalytic activity. Residues surrounding glucans or related to the catalytic reaction are shown as sticks and labeled based on their location. TMs, blue; IF1, orange; IF2, cyan; IF3, magenta; GT domain, deep pink. The cellobiose molecules occupying the channel are colored in yellow.
Figure S10. Helices in PCR. The α6 (PCR‐αs) is bent back to GT domain and residues that face towards each helix in PCR are labeled and shown as sticks.
Figure S11. Density in the center of PCRs in trimeric GhCesA7. (a) Side and bottom views of the uncharacterized density located in the center of trimeric PCR. Coordinating residues are labeled and shown as sticks. (b) Sequence alignment of helix 7 (PCR‐α2). Residues interacting with the uncharacterized density are written with black characters and highlighted in yellow.
Figure S12. Density in the center of loops between TM6 and TM7. (a) Top view of the density in the axis of trimeric loop between TM6 and TM7. R1000 in each protomer interacting with the uncharacterized density is shown in the right panel. (b) Sequence alignment of the same region with the conserved Arginine highlighted in yellow.
Figure S13. Alignment of GhCesA7 and RsBcsA. RsBcsA (PDB: 5EJZ) is colored gray and GhCesA7 is colored as in Figure 1c. TMs are aligned and labeled in the top panels (with green outline) and the channel is indicated with an orange circle. GT domains are aligned and the catalytic core is presented in the bottom panels in which UDP (5EJZ) and residues involved are labeled and shown as sticks.
Figure S14. Alignment with PttCesA8. (a) Overall structure alignment of trimeric GhCesA7 and PttCesA8 (PDB: 6WLB). PttCesA8 is colored gray and GhCesA7 is colored as in Figure 1b. (b) Alignment of the protomers from GhCesA7 and PttCesA8. The glucans within the protomer are located at the same region. (c) Sequence alignment of NTD from PttCesA8 and GhCesA. Similar residues and identical residues are indicated with blue and red characters.
Table S1. Statistics of Cryo‐EM data collection and refinement.


