Abstract
The post-translational methylation of the α-N-terminal amino group of protein was first documented over 40 years ago, but the functional significance of this modification has been underexplored relative to lysine and arginine methylation. The increase in reports implicates α-N-terminal methylation as a widespread and critical regulator of mitosis, chromatin interactions, DNA repair, and translation fidelity. Here, we summarize advances in the current understanding of protein α-N-terminal methylation biological functions and mechanisms across eukaryotic organisms. Also, we describe the recent literature on substrate recognition and the discovery of potent and selective inhibitors for protein N-terminal methyltransferases. Lastly, we summarized the emergent crosstalk between α-N-terminal methylation and other N-terminal modifications.
Keywords: N-terminal methylation, post-translational modification, bisubstrate inhibitor, peptidomimetic, epigenetic modification
1. Introduction
Protein α-N-terminal (Nα) methylation is a post-translational modification catalyzed by N-terminal methyltransferases (NTMTs) using S-adenosyl-L-methionine (SAM) as a co-factor. Following cleavage of initiating methionine, NTMTs install 1–3 methyl group(s) on the newly exposed Nα amino group with concomitant generation of S-adenosine-L-homocysteine (SAH). While the occurrence of Nα methylation was identified over 40 years ago in ribosome subunits, histone H2B, and myosin light chain proteins in evolutionarily distant species, this modification was largely neglected until the recent discovery of Nα methylation on the regulator of chromosome condensation 1 (RCC1) [1,2]. Subsequent identification of NTMT1/NRMT1 in human, YBR261C/Tae1 in yeast, NTMT2, and human methyltransferase like 13 (METTL13) prompted detection of Nα methylation on the tumor suppressor retinoblastoma, oncoprotein SET (also known as TAF-Iα, I2PP2A), centromere protein A and B (CENP-A/B), damaged DNA-binding protein 2 (DDB2), poly(ADP-ribose) polymerase 3 (PARP3), Obg-like ATPase 1 (OLA1), mortality factor 4-like protein 1 (MRG15), and eukaryotic elongation factor 1 alpha (eEF1A) [2–13]. The increasing occurrences of Nα methylation across various eukaryotic organisms have brought to light the important role of this post-translational modification in the regulation of mitosis, chromatin interactions, DNA repair, tRNA transport, and maintenance of genome stability [9–14]. Dysregulation of the enzymes responsible for catalyzing Nα methylation has been implicated in the pathogenesis of various diseases including breast, colorectal, pancreatic, and lung cancers [15]. However, a comprehensive understanding of the molecular mechanisms and pathways of Nα methylation remains unclear. Here, we focus on the current knowledge of Nα methylation’s biological functions and mechanisms in eukaryotes. We also describe recent findings in substrate recognition and inhibitors of NTMTs. Furthermore, we comment on emerging findings on the crosstalk between Nα methylation and other post-translational modifications.
2. Function of α-N-terminal methylation
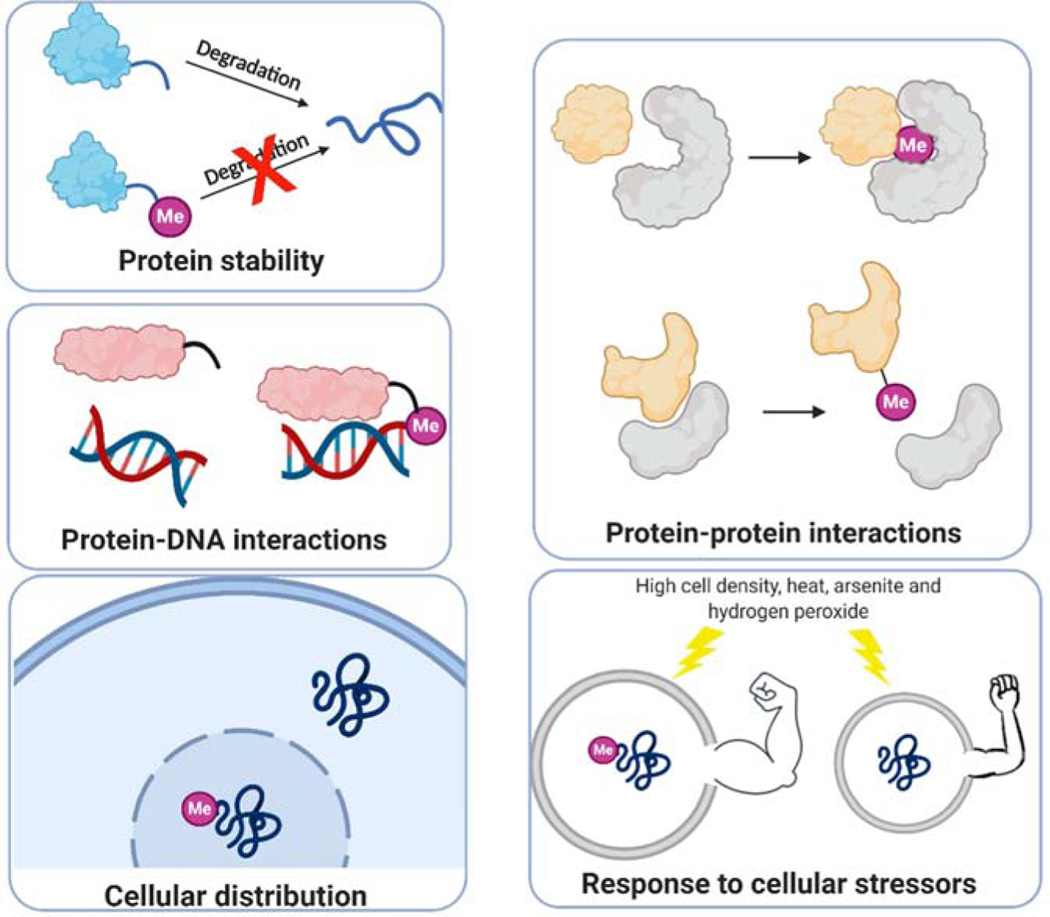
Early reports of Nα methylation implied its effect on protein stability, as the Nα methyl group was inferred as a blocking group to prevent methylated proteins from degradation via the N-degron pathway [16]. For example, Nα methylation was found to increase the stability of cytochrome c-557 to protect it from aminopeptidases [17]. While this well-accepted role of Nα methylation in protein stability remains rational, recent discoveries of Nα methylation have unveiled additional cellular functions across new dimensions.
Nα methylation has been implicated in a variety of biological processes through the regulation of protein-protein and protein-DNA interactions. Consequently, such interactions lead to changes in downstream effects, cellular distribution, and tolerance to the environment. The chemical basis of Nα methylation function lies in the lower basicity of the Nα amine (pKa = 6–8) compared to side-chain aliphatic amines (pKa ~ 10.5) [18]. This confers a unique opportunity for NTMTs to modulate not only the hydrophobicity of the α-amino group, but also the charge state. Different methylation states (mono-, di, and trimethylation) have been observed in biochemical assays and on RCC1 in cells, but full methylation at the Nα amine is commonly observed for most endogenous substrates. Monomethylation has marginal effects on the pKa of the Nα amino group, with an increase in pKa by up to 0.5 units [19]. Undoubtedly, full methylation on the Nα amino group generates a permanent positive charge. This fixed positive charge at the Nα site enhances its ability to bind to proteins or nucleic acids through electrostatic interactions with negatively charged functional groups or cation-π interactions with aromatic rings. These strengthened interactions may result in distinct downstream effects, depending on specific contexts.
2.1. Protein-protein interactions
Proteins that are known to be Nα methylated are often components of large macromolecular complexes, such as subunits of the macromolecular complexes comprising the ribosome, histone H2B of Tetrahymena, and Drosophila, vertebrate striated muscle myosin light chains, and cytochrome c-557 of the protozoan Crithidia oncopelti [19]. The installation of Nα-trimethylalanine (Me3Ala) broadly occurs on both myosin alkali and regulatory light chains in vertebrate striated skeletal and cardiac muscles [20]. Compared to unmethylated myosin, the presence of Me3Ala on the extended N-terminal tail (1–13 amino acids) of myosin alkali light chain A1 resulted in a two-fold increased binding affinity of A1-actin, but slightly enhanced inhibition on the catalytic efficiency of actin-activated myosin ATPase activity [21]. Thus, Nα methylation may work with other basic residues within the N-terminal tail of A1 to fine-tune the mobility between actin and myosin filaments. Likewise, the first Gly residue of centromere protein A (CENP-A) is primarily trimethylated by NTMT1. The proportion of Me3Gly increased with cell cycle progression and reached 90% during mitosis. Nα methylation of CENP-A is essential for the formation of the constitutive centromere associated protein network (CCAN), specifically for the recruitment of CENP-T and CENP-I at the centromere [22]. Moreover, Nα methylation of CENP-A is critical for the maintenance of faithful chromosome segregation to stabilize the dipolar microtubule spindle through CENP-T [22]. In contrast, Nα methylation on Myosin Regulatory Light Chain 9 (MYL9) weakened the interaction between MYL9 and Cofilin-1, an actin-modulating protein, according to pulldown studies with N-terminal unmodified and methylated 14-mer peptides [23]. However, it remains obscure whether the decreased interaction of methylated MYL9 with Cofilin-1 is direct or indirect.
2.2. Protein–DNA interactions
Nα methylation of RCC1 (also known as Ran guanine nucleotide exchange factor) was shown to regulate the RCC1-chromatin interaction through a bimodal attachment mechanism. Specifically, the N-terminal tail of RCC1 imposes an inhibitory role on the association of the core portion of RCC1 with histones H2A or H2B [2]. Once Ran binds to RCC1, a conformational change of Nα-methylated RCC1 exposes the histone-binding surface of RCC1 and promotes the Nα-methylated RCC1 tail to interact with negatively charged DNA [2,6,24]. This electrostatic interaction occurs in a Nα methylation-dependent manner. Loss of Nα methylation reduced the binding of RCC1 to DNA, causing mitotic defects [6]. Similarly, centromere protein B (CENP-B) was predominantly trimethylated at the first Gly residue. This Nα-trimethylation strengthened the interaction of CENP-B to the CENP-B box (a 17-bp DNA motif) within human α-satellite DNA and mouse centromeric minor satellite DNA, playing an important role in the organization and maintenance of centromere activity. Notably, methylation-defective CENP-B mutants were able to bind to the CENP-B box but exhibited a ~30% loss in binding [11]. MYL9 functions as a transcriptional activator of ICAM1 (intercellular adhesion molecule 1) in the nucleus. Nα methylation on MYL9 promotes ICAM1 transcription through enriched interaction between MYL9 and ICAM1 promoter gene, while NTMT1 knockout cells showed decreased ICAM1 transcription [23]. Although MYL9 can directly interact with the ICAM1 promoter gene, it remains elusive whether the Nα methylation-promoted interaction is direct or indirect. The determination of a co-crystal structure between any aforementioned Nα-methylated proteins and DNA would elucidate the molecular details of this putative interaction.
2.3. Cellular distribution
Nα methylation can also modulate the cellular distribution of proteins through the modulation of protein-protein and/or protein-DNA interactions. One case is DDB2, an early UV damage recognition factor, which harbors an N-terminal APK motif methylated by NTMT1. DDB2 recognizes UV-induced DNA damage in chromatin via recruitment to UV light-induced cyclobutane pyrimidine dimer (CPD) foci after exposure to UV irradiation [25]. Nα methylation promoted the nuclear localization of DDB2 to CPD foci and activation of ataxia telangiectasia mutated (ATM) to stimulate CPD repair in HEK293T cells, suggesting a protective role of Nα methylation against UV damage [10]. The differential cellular distribution of Nα-methylated proteins urges us to speculate that Nα-methylated proteins may interact with trafficking proteins to govern their subcellular localization. Conversely, Nα methylation on MYL9 does not affect the subcellular distribution of nuclear MYL9 but promotes its nuclear transcriptional regulator activity [23]. These results suggest that the effect of Nα methylation on protein cellular distribution is context-dependent and produces variable effects for different NTMT substrates.
2.4. Cellular Stress Response
The Nα methylation levels of NTMT1 substrates increase under stressful environments including increased cell density and heat shock. For example, Nα methylation levels of CENP-B were increased in HEK293T cells under cellular stresses including high cell density, heat, and arsenite treatment [11]. A similar phenomenon was also observed for the Nα methylation on Drosophila melanogaster histone H2B [26]. On the other hand, loss of Nα methylation on Rpt-1 in yeast slowed growth rate and displayed an increased sensitivity to hydrogen peroxide or canavanine stress [27]. However, the detailed mechanism of how Nα methylation regulates the aforementioned responses requires additional examination. Nevertheless, such responses to extracellular environment alterations are an important feature of epigenetics, supporting Nα methylation as an epigenetic modification.
3. Recognition and Mechanism
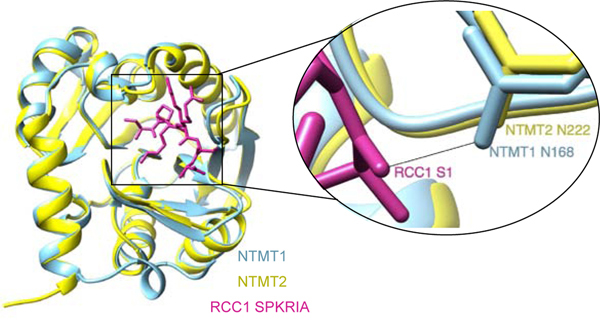
NTMT1 and NTMT2 share over 50% sequence similarity as two functional writers for Nα methylation in humans. Co-crystal structures of NTMT1/2 in complex with its SAH cofactor and substrate peptides provided a clear view of their similar substrate-binding regions (Figure 3), supporting their preference for the canonical X-P-K/R motif. First, a conserved Asn residue (N168 in NTMT1 and N222 in NTMT2) forms a critical backbone hydrogen bond with the X1 carbonyl oxygen. A negatively-charged substrate-binding pocket is capable of accommodating various X1 residues except for the negatively charged D or E [8,28,29]. Second, a conserved Trp residue (W136 in NTMT1 and W191 in NTMT2) and another aromatic residue (Y34 in NTMT1 and F90 in NTMT2) form a stacking interaction with P2 [10,29,30]. Third, two Asp residues (D177/180 in NTMT1 and D232/235 in NTMT2) form electrostatic interactions with K3/R3 [28–30]. Despite its high similarity to NTMT1, NTMT2 primarily monomethylates X-PKRIA peptides but can fully methylate G/P-PKRIA peptides [31]. Co-crystal structures of both NTMT1/2 in complex with their substrate peptides indicate that the fourth residue R4 does not directly interact with the enzymes. Thus, substrate residues downstream of the X-P-K/R motif have been suggested to play a minor contribution to substrate recognition. Notably, recent substrate profiling of NTMT1 inferred the importance of the downstream residue [7]. The N89 residue of NTMT2 acts as a gatekeeper for its catalytic activities and product preference [31]. However, little is known regarding the physiological substrates of NTMT2, which precludes the effort to elucidate its biological roles.
Figure 3.
NTMT1/2 structural similarity. NTMT1 (blue) is overlayed with NTMT2 (yellow) in complex with SPKRIA peptide with a close-up image of the conserved NTMT1 N168 and NTMT2 N222 hydrogen bonding with S1 of RCC1.
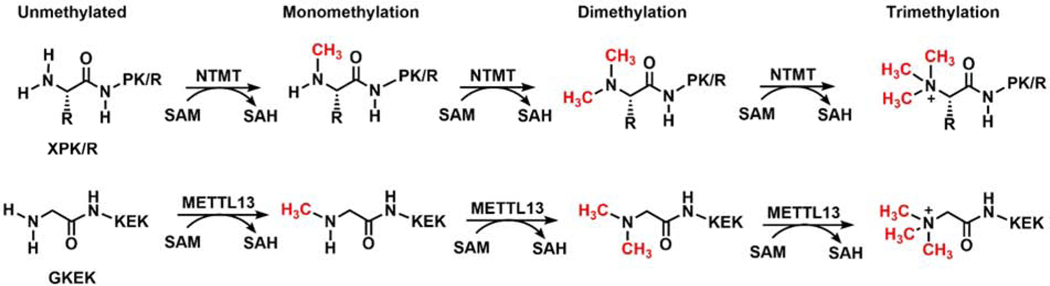
After Nα methylation on RCC1 was first identified through mass spectrometry (MS) and subsequent discovery of the writer NTMT1, most biologically validated NTMT1 substrates contain the N-terminal consensus sequence X-P-K/R (X = S/P/A/G) (Figure 1) [28–30], except MYL9 starting with SSK [10,30]. Recent activity-based substrate profiling of NTMT1 with an artificial cofactor Hey-SAM reported 72 potential targets. A subsequent MS methylation assay confirmed that a 10-mer peptide (GSKRRRATSP) derived from protein polybromo 1 (PB1) can be methylated, but only OLA1 containing a canonical APK sequence was validated in a cellular context [7]. These exceptions to the X-P-K/R consensus sequence imply that additional unidentified proteins may undergo Nα methylation. The recent discovery of Nα methylation on the novel GKEK motif of yeast and human eEF1A by METTL13 offers additional evidence for noncanonical Nα methylation (Figure 1) [9]. METTL13 is a dual specific methyltransferase that contains two distinct domains: a C-terminal methyltransferase domain (MT13-C) for Nα methylation on Gly1 and an N-terminal methyltransferase domain (MT13-N) for Lys55 methylation on eEF1A [9]. Although the mechanisms and interactions involved in METTL13 substrate recognition remain to be studied, the discovery of Nα methylation on noncanonical motifs ratifies Nα methylation as a widespread modification.
Figure 1.
General scheme for protein Nα methylation.
Kinetic studies have indicated that NTMT1 methylation follows a Bi-Bi catalytic mechanism, whereby either SAM or protein substrate can first bind to NTMT1 [32]. Moreover, the co-crystal structure of NTMT1-SAH-SPKRIA illustrated the formation of a ternary complex of NTMT1 with both substrate and cofactor to exemplify the Bi-Bi mechanism [29]. The existence of a ternary complex structure of NTMT2-SAH-SPKRIA (PDB: 6DUB), along with its high structural similarity to the active site of NTMT1, makes it reasonable to predict that NTMT2 also follows a Bi-Bi mechanism [31]. Since NTMTs catalyze the transfer of 1–3 methyl groups from SAM to Nα amines to yield methylated protein with different methylation states, the progression of this multi-step methylation may follow either a distributive or processive mechanism. There have been contradictory observations to support either mechanism. Biochemical studies using NTMT1 with peptides derived from RCC1 and SET protein demonstrated a distributive mechanism [32,33]. Additional support for a distributive mechanism comes from the different distribution of all methylation states of endogenous RCC1 throughout mitotic progression in HeLa cells [2,30]. Immunoprecipitation studies showed that CENP-A starting with a GPR sequence is predominantly trimethylated during mitosis, suggestive of a processive mechanism [12]. Notably, we observed that methylation of a GPKRIA peptide substrate yields predominant trimethylation with marginal mono- and dimethylation levels [34]. Thus, NTMT1 methylation progression may go through either processive or distributive mechanisms, dependent on the first amino acid and/or the influence of residues C-terminal to the recognition motif. The mass-spectrometry analysis of eEF1A methylation status in HeLa cells revealed that MT13-C acts as a processive enzyme because Nα-trimethylated eEF1A is the predominant form with trace amounts of mono- and dimethylated states observed [9]. However, it is too early to draw a definite conclusion since the distribution of methylation states may vary as in the case of RCC1 methylation.
4. Regulation and Crosstalk
The occurrence of Nα methylation currently appears to be a static process, as the existence of an “eraser” for this modification remains mysterious. However, assumptions of irreversibility were also held for lysine methylation until the identification of the first histone demethylase in 2004 [35]. An investigation into the possibility of Nα methylation erasers would be an interesting avenue to explore to understand the dynamic regulation of Nα methylation. A recent study demonstrated that NTMT1 protein expression may be regulated by readers, writers, and erasers involved in the N6-methyladenosine (m6A) modification of mRNA, providing a critical first piece of evidence for the regulation of Nα methylation [5]. The reversibility of Nα methylation would explain the differential distribution of Nα methylation states, even if NTMTs are processive methyltransferases, as well as introduce additional dimensions that govern the interplay among different modifications at the Nα position.
Similar to histone tails, emerging reports suggest the crosstalk between Nα methylation and other modifications including Nα acetylation, methylation on lysine or arginine side chains, phosphorylation, and m6A modifications in RNA. The differential impact of Nα-methylation and Nα-acetylation on the subcellular localization of MYL9 is the first report on the interplay between methylation and acetylation at the same site [23,30]. Besides Nα methylation, the side chain of K3 is commonly trimethylated in ribosomal subunits L11 and L12, and cytochrome c-557, but the relationship between Me3K3 and Nα methylation remains enigmatic [36–38]. Nα methylation on H2B has been inferred to be negatively regulated by H3R2 methylation in D. melanogaster [39]. Both Nα methylation and Ser phosphorylation on the N-terminal tail of RCC1 were concurrent during mitosis [2,40]. In asynchronous HeLa cells, S1 phosphorylation decreased about 25% on the RCC1 Nα methylation-defective mutant compared to wild type RCC1, suggesting that Nα-methylation has a positive effect on phosphorylation of S1 [2]. While in mitotic cells, no significant change was observed on the total phosphorylation levels on two Ser residues (S1 and S10) regardless of Nα methylation, though the phosphorylation level on S2 increased 10% in the absence of Nα methylation [2]. The Nα methylation of MRG15 was recently found to be modulated by modifiers of m6A, unveiling a novel regulation of Nα methylation by the m6A-based epitranscriptome [5].
5. Inhibitors
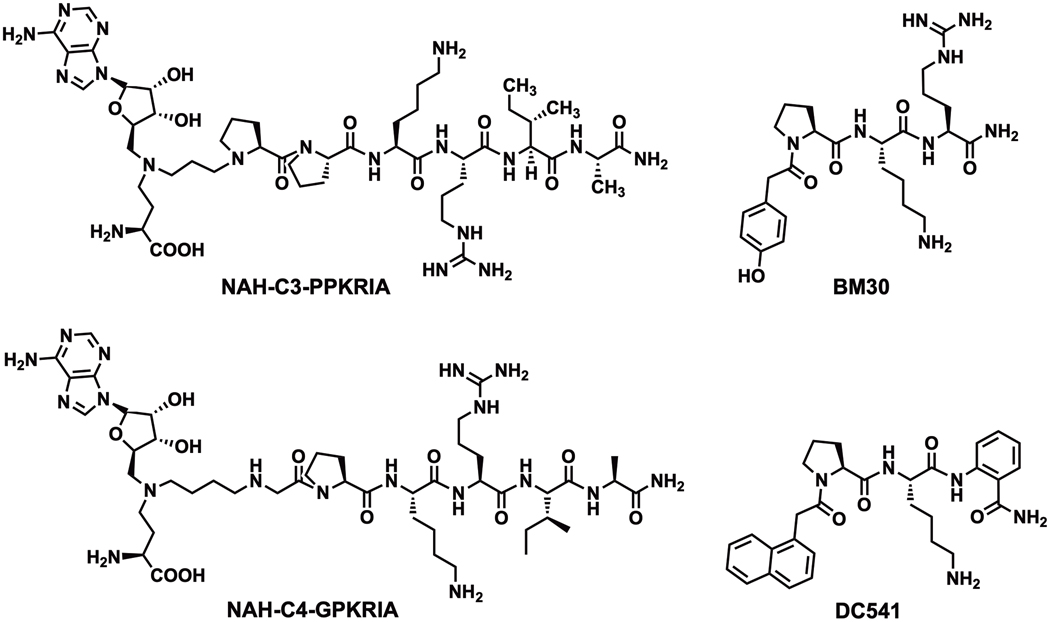
Besides the important biological roles of Nα methylation discussed in section Function of α-N-terminal methylation, dysregulation of NTMT1 and METTL13 has been implicated across various cancers and aging processes. Thus, the development of specific and potent NTMT inhibitors is important to probe the biological functions of Nα methylation. Currently, two different inhibitor types have been reported for NTMTs. The first type is the bisubstrate inhibitor that is designed based on the NTMT1 kinetic mechanism [32]. A series of bisubstrate analogs that covalently link a SAM analog (NAH) with a peptide substrate moiety through variable linkers have been developed. A 3-C atom linker covalently connecting NAH to a PPKRIA peptide derived from mouse RCC1 yielded the inhibitor NAH-C3-PPKRIA (Ki =39 ± 9.5 nM) (Figure 4), which displayed a 600-fold selectivity over methyltransferases including G9a, SETD7, and NNMT [41]. The binary complex of NTMT1-NAH-C3-PPKRIA, the first NTMT1-inhibitor co-crystal structure, clearly illustrated its engagement with both substrate and cofactor binding sites. Recent investigations using a series of NAH-GPKRIA bisubstrate analogs revealed the plasticity of the NTMT1 transition state, as it can accommodate linkers ranging from a 2-C to 4-C atom linker. Among them, NAH-C4-GPKRIA is the most and selective potent inhibitor (Ki,app = 130 ± 40 pM) to date (Figure 4), displaying more than 3,000-fold selectivity for other methyltransferases and even for its homolog NTMT2 [42]. Although bisubstrate analogs provide valuable insights to understand the mechanism of NTMT1, their poor cell permeability limits their applications for cellular studies that probe the physiological roles of NTMT1 [34]. Recently, the tetra-peptidomimetic BM30 has been reported to selectively inhibit NTMT1/2 at sub-micromolar activity by targeting the unique substrate-binding site, exhibiting over 100-fold selectivity among a panel of 41 methyltransferases [43]. Guided by the co-crystal structure of BM30 in complex with NTMT1 (PDB ID: 6WH8), a cell-potent and selective inhibitor DC541 (IC50 of 0.34 ± 0.02 μM) was recently obtained by introduction of a naphthyl group and an ortho-aminobenzoic amide [44]. Furthermore, DC541 displayed a cellular inhibition on the Nα methylation level of RCC1 and SET protein at 30 μM in human colorectal cancer cells without any significant cytotoxicity up to 1 mM [44], providing the first cell-potent probe to study biological functions of NTMT1/2. Besides the aforementioned inhibitors developed through rational design, ongoing efforts on high-throughput screening of small molecule inhibitors are expected to be reported in the future.
Figure 4.
Bisubstrate and peptidomimetic inhibitors of NTMT1.
6. Conclusions
Protein methylation is an important epigenetic modification that plays a crucial role in regulating diverse biological processes ranging from gene expression to signal transduction. Dysregulation of protein methylation has been implicated in numerous human disorders including cancer, inflammation, neurodegenerative, and cardiovascular diseases. Thus, targeting methylation-modifying enzymes has attracted burgeoning interest. Notably, inhibitors that modulate protein lysine and arginine methylation have begun to demonstrate therapeutic promise in cancers. In stark contrast, Nα methylation remains underexplored despite the increasing implications of Nα methylation in vital physiological and pathological processes across diverse species. Thus, it is imperative to comprehensively elucidate Nα methylation-mediated pathways. Specifically, the number of validated physiological substrates for NTMTs remains low considering the prediction of ~ 300 substrates. The detection of Nα methylation remains challenging due to the conserved Lys and Arg on the 3rd and 4th position since the commonly used trypsin would generate a tripeptide or tetrapeptide upon cleavage. To speed up the substrate identification process, there is a need for high quality and commercially available antibodies to specifically recognize Nα methylation. As the reported anti-Me2-PPK antibody was also able to recognize Nα methylation on A/S-PK motifs, it is rational to propose that there will be no major obstacles in the development of a pan-Nα methylation antibody despite the variability of the X1 residue. Despite recent progress on deciphering the role of Nα methylation, there remains a lack of systematic approaches to examine its functions. We speculate that Nα methylation may play a fine-tuning regulatory role through interaction with the protein, DNA, or both. Moreover, this fine-tuning feature may also be affected by neighboring modifications since Nα methylation normally occurs on tails enriched with basic residues. Furthermore, cell-potent and specific inhibitors for NTMT1/2 and METTL13 are required to probe their physiological functions. Currently, only the peptidomimetic inhibitor DC541 has been shown to exhibit inhibition on cellular Nα methylation levels. However, its modest inhibition suggests that further optimization of DC541 is needed to improve its cell-potency. Meanwhile, there is no inhibitor available for METTL13. Specific inhibitors targeting either MT13-C or MT13-N would be valuable to differentiate the function of Nα methylation from Lys55 methylation.
Figure 2.
Functions of Nα methylation across different biological processes.
Acknowledgments
Funding: This work was supported by the National Institutes of Health R01GM117275 (R.H.), U01CA214649 (R.H.).
Abbreviations
- Nα
α-N-terminal
- NTMT1
N-terminal methyltransferase 1
- SAM
S-adenosyl-L-methionine
- SAH
S-adenosine-L-homocysteine
- RCC1
regulator of chromosome condensation 1
- METTL13
methyltransferase like 13
- DDB2
DNA-binding protein 2
- eEF1A
eukaryotic elongation factor 1 alpha
- MYL9
myosin regulatory light chain 9
- CENP-A
centromere protein A
- CENP-B
centromere protein B
- OLA1
Obg-like ATPase 1
- PARP3
poly(ADP-ribose) polymerase 3
- MRG15
mortality factor 4-like protein 1
- ICAM1
intercellular adhesion molecule 1
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen R, Brosius J, Wittmann-Liebold B, Schäfer W: Occurrence of methylated amino acids as N-termini of proteins from Escherichia coli ribosomes. J Mol Biol 1977, 111:173–181. doi: 10.1016/S0022-2836(77)80121-6 [DOI] [PubMed] [Google Scholar]
- 2.Chen T, Muratore TL, Schaner-Tooley CE, Shabanowitz J, Hunt DF, Macara IF: N-terminal α-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat Cell Biol 2007, 9:596–603. doi: 10.1038/ncb1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petkowski JJ, Bonsignore LA, Tooley JG, Wilkey DW, Merchant ML, Macara IG, Schaner Tooley CE: NRMT2 is an N-terminal monomethylase that primes for its homologue NRMT1. Biochem J 2013, 456:453–462. doi: 10.1042/BJ20131163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang R: Chemical Biology of Protein N-Terminal Methyltransferases. ChemBioChem 2019, doi: 10.1002/cbic.201800615. doi: 10.1002/cbic.201800615 *This review provides an overview of N-terminal methylation, a post-translational modification identified in prokaryotes over 40 years ago. Here, the senior corresponding author outlines the history of N-terminal methylation, with a comparison of this PTM across prokaroyic and eukaryotic organisms.
- 5. Bade D, Cai Q, Li L, Yu K, Dai X, Miao W, Wang Y: Modulation of N-terminal methyltransferase 1 by an N6-methyladenosine-based epitranscriptomic mechanism. Biochem Biophys Res Commun 2021, 546:54–58. doi: 10.1016/j.bbrc.2021.01.088 **This seminal study provides the first evidence for regulation of protein Nα methylation through the modulation of NTMT1 expression and methylation activity by m6A reader, writer, and eraser proteins. Additionally, the results validated that MRG15 is N-terminally methylated by NTMT1 and is an NTMT1 substrate.
- 6.Hitakomate E, Hood FE, Sanderson HS, Clarke PR: The methylated N-terminal tail of RCC1 is required for stabilisation of its interaction with chromatin by Ran in live cells. BMC Cell Biol 2010, 11. doi: 10.1186/1471-2121-11-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jia K, Huang G, Wu W, Shrestha R, Wu B, Xiong Y, Li P: In vivo methylation of OLA1 revealed by activity-based target profiling of NTMT1. Chem Sci 2019, 10:8094–8099. doi: 10.1039/c9sc02550b *Using activity-based substrate profiling, the authors identified Obg-like ATPase (OLA1) as a novel NTMT1 substrate. OLA1 was found to be fully methylated in normal 293FT cells. This study provided the first evidence of post-translational modification on OLA1.
- 8.Webb KJ, Lipson RS, Al-Hadid Q, Whitelegge JP, Clarke SG: Identification of protein n-terminal methyltransferases in yeast and humans. Biochemistry 2010, 49:5225–5235. doi: 10.1021/bi100428x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jakobsson ME, Małecki JM, Halabelian L, Nilges BS, Pinto R, Kudithipudi S, Munk S, Davydova E, Zuhairi FR, Arrowsmith CH, et al. : The dual methyltransferase METTL13 targets N terminus and Lys55 of eEF1A and modulates codon-specific translation rates. Nat Commun 2018, 9:3411. doi: 10.1038/s41467-018-05646-y **Using quantitative mass spectrometry proteomics screens, METTL13 gene knockout studies, and in vitro studies, this study identified METTL13 as the methyltransferase responsible for the Nα methylation of eEF1A. The authors demonstrated that Nα methylation and Lys55 methylation of eEF1A modulates codon-specific translation rates. The eEF1A protein substrate bears a unique GKEK methylation motif, highlighting the need to expand the canonical Nα methylation motif beyond XPK.
- 10.Dai X, Wang Y, Fu L, Wang Z, Gan N, Cai Q: α- N -Methylation of Damaged DNA-binding Protein 2 (DDB2) and Its Function in Nucleotide Excision Repair. J Biol Chem 2014, 289:16046–16056. doi: 10.1074/jbc.m114.558510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai X, Otake K, You C, Cai Q, Wang Z, Masumoto H, Wang Y: Identification of novel α-N-methylation of CENP-B that regulates its binding to the centromeric DNA. J Proteome Res 2013, 12:4167–4175. doi: 10.1021/pr400498y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey AO, Panchenko T, Sathyan KM, Petkowski JJ, Pai PJ, Bai DL, Russell DH, Macara IG, Shabanowitz J, Hunt DF, et al. : Posttranslational modification of CENP-A influences the conformation of centromeric chromatin. Proc Natl Acad Sci U S A 2013, 110:11827–11832. doi: 10.1073/pnas.1300325110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai X, Rulten SL, You C, Caldecott KW, Wang Y: Identification and functional characterizations of N-terminal α- N -methylation and phosphorylation of serine 461 in human poly(ADP-ribose) polymerase 3. J Proteome Res 2015, 14:2575–2582. doi: 10.1021/acs.jproteome.5b00126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.England JR, Huang J, Jennings MJ, Makde RD, Tan S: RCC1 Uses a Conformationally Diverse Loop Region to Interact with the Nucleosome: A Model for the RCC1-Nucleosome Complex. J Mol Biol 2010, 398:518–529. doi: 10.1016/j.jmb.2010.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tooley JG, Schaner Tooley CE: New roles for old modifications: Emerging roles of N-terminal post-translational modifications in development and disease. Protein Sci 2014, 23:1641–1649. doi: 10.1002/pro.2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Varshavsky A: N-degron and C-degron pathways of protein degradation. Proc Natl Acad Sci U S A 2019, 116:358–366. doi: 10.1073/pnas.1816596116 *This perspective offers a comprehensive overview of proteolytic pathways that target the N- and C- termini of proteins for degradation. The substrate recognition, function, and interaction partners involved in these pathways are described in detail. The author proposes that these multifunctional systems be denoted “N-degron” and “C-degron” pathways for protein degradation.
- 17.Smith GM, Pettigrew GW: Identification of N,N-Dimethylproline as the N-Terminal Blocking Group of Crithidia oncopelti Cytochrome c557. Eur J Biochem 1980, 110:123–130. doi: 10.1111/j.1432-1033.1980.tb04847.x [DOI] [PubMed] [Google Scholar]
- 18.Grimsley GR, Scholtz JM, Pace CN: A summary of the measured pK values of the ionizable groups in folded proteins. Protein Sci 2009, 18:247–251. doi: 10.1002/pro.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stock A, Clarke S, Clarke C, Stock J: N-terminal methylation of proteins: Structure, function and specificity. FEBS Lett 1987, 220:8–14. doi: 10.1016/0014-5793(87)80866-9 [DOI] [PubMed] [Google Scholar]
- 20.Henry GD, Trayer IP, Brewer S, Levine BA: The widespread distribution of α-N-trimethylalanine as the N-terminal amino acid of light chains from vertebrate striated muscle myosins. Eur J Biochem 1985, 148:75–82. doi: 10.1111/j.1432-1033.1985.tb08809.x [DOI] [PubMed] [Google Scholar]
- 21.Hayashibara T, Miyanishi T: Binding of the Amino-Terminal Region of Myosin Alkali 1 Light Chain to Actin and Its Effect on Actin-Myosin Interaction. Biochemistry 1994, 33:12821–12827. doi: 10.1021/bi00209a013 [DOI] [PubMed] [Google Scholar]
- 22.Sathyan KM, Fachinetti D, Foltz DR: α-amino trimethylation of CENP-A by NRMT is required for full recruitment of the centromere. Nat Commun 2017, 8:1–15. doi: 10.1038/ncomms14678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nevitt C, Tooley JG, Schaner Tooley CE: N-terminal acetylation and methylation differentially affect the function of MYL9. Biochem J 2018, 475:3201–3219. doi: 10.1042/BCJ20180638 **This study identified MYL9 as the first confirmed protein to undergo both Nα methylation and Nα acetylation in vivo. The N-terminal acetylation and methylation of MYL9 were found to have differential functions, with Nα methylation promoting the nuclear role of MYL9 as a transcription factor, and Nα acetylation promoting the cytoplasmic roles of MYL9. The MYL9 N-terminal methylation motif of SSK deviates from the canonical XPK motif. Identification of Nα methylation on this noncanonical motif expands the functional implications of this modification and supports Nα methylation as a widespread modification in cellular processes.
- 24.Hao Y, Macara IG: Regulation of chromatin binding by a conformational switch in the tail of the Ran exchange factor RCC1. J Cell Biol 2008, 182:827–836. doi: 10.1083/jcb.200803110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moser J, Volker M, Kool H, Alekseev S, Vrieling H, Yasui A, Van Zeeland AA, Mullenders LHF: The UV-damaged DNA binding protein mediates efficient targeting of the nucleotide excision repair complex to UV-induced photo lesions. DNA Repair (Amst) 2005, 4:571–582. doi: 10.1016/j.dnarep.2005.01.001 [DOI] [PubMed] [Google Scholar]
- 26.Desrosiers R, Tanguay RM: Methylation of Drosophila histones at proline, lysine, and arginine residues during heat shock. J Biol Chem 1988, 263:4686–4692. [PubMed] [Google Scholar]
- 27.Kimura Y, Kurata Y, Ishikawa A, Okayama A, Kamita M, Hirano H: N-Terminal methylation of proteasome subunit Rpt1 in yeast. Proteomics 2013, 13:3167–3174. doi: 10.1002/pmic.201300207 [DOI] [PubMed] [Google Scholar]
- 28.Schaner Tooley CE, Petkowski JJ, Muratore-Schroeder TL, Balsbaugh JL, Shabanowitz J, Sabat M, Minor W, Hunt DF, MacAra IG: NRMT is an α-N-methyltransferase that methylates RCC1 and retinoblastoma protein. Nature 2010, 466:1125–1128. doi: 10.1038/nature09343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong C, Mao Y, Tempel W, Qin S, Li L, Loppnau P, Huang R, Min J: Structural basis for substrate recognition by the human N-terminal methyltransferase 1. Genes Dev 2015, 29:2343–2348. doi: 10.1101/gad.270611.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petkowski JJ, Schaner Tooley CE, Anderson LC, Shumilin IA, Balsbaugh JL, Shabanowitz J, Hunt DF, Minor W, MacAra IG: Substrate specificity of mammalian N-terminal α-amino methyltransferase NRMT. Biochemistry 2012, 51:5942–5950. doi: 10.1021/bi300278f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong C, Dong G, Li L, Zhu L, Tempel W, Liu Y, Huang R, Min J: An asparagine/glycine switch governs product specificity of human N-terminal methyltransferase NTMT2. Commun Biol 2018, 1:1–9. doi: 10.1038/s42003-018-0196-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richardson SL, Mao Y, Zhang G, Hanjra P, Peterson DL, Huang R: Kinetic mechanism of protein N-terminal methyltransferase 1. J Biol Chem 2015, 290:11601–11610. doi: 10.1074/jbc.M114.626846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richardson SL, Hanjra P, Zhang G, Mackie BD, Peterson DL, Huang R: A direct, ratiometric, and quantitative MALDI-MS assay for protein methyltransferases and acetyltransferases. Anal Biochem 2015, 478:59–64. doi: 10.1016/j.ab.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen D, Dong G, Noinaj N, Huang R: Discovery of Bisubstrate Inhibitors for Protein N-Terminal Methyltransferase 1. J Med Chem 2019, 62:3773–3779. doi: 10.1021/acs.jmedchem.9b00206 **This study is fundamental in that it reported the first crystal structure of NTMT1 in complex with an inhibitor. The NTMT1-NAH-C3-PPKRIA co-crystal structure verified that it occupies both the substrate and cofactor binding sites. This bisubstrate inhibitor displayed over a 600-fold selectivity over a panel of methyltransferases, rendering the development of bisubstrate analogs as an attractive strategy to produce potent and selective NTMT1 inhibitors.
- 35.Shi YY, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi YY: Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119:941–953. doi: 10.1016/j.cell.2004.12.012 [DOI] [PubMed] [Google Scholar]
- 36.Clarke SG: The ribosome: A hot spot for the identification of new types of protein methyltransferases. J Biol Chem 2018, 293:10438–10446. doi: 10.1074/jbc.AW118.003235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Webb KJ, Laganowsky A, Whitelegge JP, Clarke SG: Identification of two SET domain proteins required for methylation of lysine residues in yeast ribosomal protein Rpl42ab. J Biol Chem 2008, 283:35561–35568. doi: 10.1074/jbc.M806006200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polevoda B, Martzen MR, Das B, Phizicky EM, Sherman F: Cytochrome c methyltransferase, Ctm1p, of yeast. J Biol Chem 2000, 275:20508–20513. doi: 10.1074/jbc.M001891200 [DOI] [PubMed] [Google Scholar]
- 39.Villar-Garea A, Forne I, Vetter I, Kremmer E, Thomae A, Imhof A: Developmental regulation of N-terminal H2B methylation in Drosophila melanogaster. Nucleic Acids Res 2012, 40:1536–1549. doi: 10.1093/nar/gkr935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hutchins JRA, Moore WJ, Hood FE, Wilson JSJ, Andrews PD, Swedlow JR, Clarke PR: Phosphorylation Regulates the Dynamic Interaction of RCC1 with Chromosomes during Mitosis. Curr Biol 2004, 14:1099–1104. doi: 10.1016/j.cub.2004.05.021 [DOI] [PubMed] [Google Scholar]
- 41.Zhang G, Richardson SL, Mao Y, Huang R: Design, synthesis, and kinetic analysis of potent protein N-terminal methyltransferase 1 inhibitors. Org Biomol Chem 2015, 13:4149–4154. doi: 10.1039/c5ob00120j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen D, Dong C, Dong G, Srinivasan K, Min J, Noinaj N, Huang R: Probing the Plasticity in the Active Site of Protein N-terminal Methyltransferase 1 Using Bisubstrate Analogs. J Med Chem 2020, 63:8419–8431. doi: 10.1101/2020.04.13.039990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mackie BD, Chen D, Dong G, Dong C, Parker H, Schaner Tooley CE, Noinaj N, Min J, Huang R: Selective Peptidomimetic Inhibitors of NTMT1/2: Rational Design, Synthesis, Characterization, and Crystallographic Studies. J Med Chem 2020, 63:9512–9522. doi: 10.1021/acs.jmedchem.0c00689 *The rational design strategy used in this study yielded BM30, the first potent peptidomimetic inhibitor for NTMT1 and 2. The high specificity of BM30 to the NTMT1/2 active site is exemplified in its over 100-fold selectivity to NTMT1/2 across a panel of 41 methyltransferases. Further optimization of BM30 produced a cell-permeable analog DC432 that decreased the N-terminal methylation level of RCC1 and SET proteins in HCT116 cells. The design strategy used in this study thus has the capability to produce cell-potent inhibitors useful for probing NTMT function.
- 44. Chen D, Dong G, Deng Y, Noinaj N, Huang R: Structure-based Discovery of Cell-Potent Peptidomimetic Inhibitors for Protein N-terminal Methyltransferase 1. ACS Med Chem Lett 12, 2021, 485–495. doi: 10.1021/acsmedchemlett.1c00012 *The first cell-potent peptidomimetic inhibitor DC541 was reported, providing a valuable tool for the research community to study NTMT1 functions.