Abstract
The kinetochore plays an essential role in facilitating chromosome segregation during cell division. This massive protein complex assembles onto the centromere of chromosomes and enables their attachment to spindle microtubules during mitosis. The kinetochore also functions as a signaling hub to regulate cell cycle progression, and is crucial to ensuring the fidelity of chromosome segregation. Despite the fact that kinetochores are large and robust molecular assemblies, they are also highly dynamic structures that undergo structural and organizational changes throughout the cell cycle. This review will highlight our current understanding of kinetochore structure and function, focusing on the dynamic processes that underlie kinetochore assembly.
Introduction
Cell division is the fundamental process by which one cell gives rise to two genetically identical daughter cells. In eukaryotes, each cell division cycle requires that the genetic material be properly duplicated and segregated across the dividing cell. It is critical that the DNA is equally distributed between the two cells, as chromosome gains or losses can be highly detrimental to an organism. Accurate chromosome segregation depends upon the macromolecular structure called the kinetochore [1], which is assembled upon a specific region of each chromosome known as the centromere. The kinetochore is comprised of multiple individual proteins that assemble together to form a network of interdependent sub-complexes. This large protein assembly can be divided into two distinct regions: the inner kinetochore, which associates with centromere DNA and is assembled throughout the cell cycle; and the outer kinetochore, which binds directly to spindle microtubules and is assembled only in mitosis.
The kinetochore is a massive protein assembly with more than 110 different components and around 400 molecules of each protein at a single human kinetochore, that together form a structure that is roughly 5-10-fold larger than a ribosome [2, 3]. However, despite its large size, the kinetochore is a remarkably dynamic structure, exhibiting changes in its assembly, dynamics, organization, and function throughout the cell cycle. Our understanding of kinetochore assembly often focuses on the recruitment of the outer kinetochore in mitosis. However, a subset of kinetochore components are present at centromeres constitutively, and the manner in which these protein complexes adapt to the events of the cell cycle is also important to fully understand the principles of kinetochore assembly.
During interphase, three critical dynamic events must occur that result in substantial changes to the centromere. First, during G1, new molecules of the centromere-specific histone CENP-A are deposited into centromeric chromatin, resulting in a substantial increase of CENP-A molecules at centromeres (Figure 1A). During S phase, centromeric DNA undergoes a dramatic structural change, similar to other chromosomal regions, as chromosomes are unwound and replicated, resulting in at least the temporary eviction of chromatin-associated factors (Figure 1B). Finally, during the G2/M transition, chromosomal DNA undergoes further structural changes as chromosomes become condensed with sister chromatids remaining bound together at centromeres (Figure 1C,D). In addition to navigating DNA compaction during the G2/M transition, the kinetochore must assemble its outer structure to prepare for the formation of microtubule attachments in mitosis. The constitutive centromere components that comprise the inner kinetochore must remain assembled despite these vast changes and disruptions that occur to centromeric chromatin over the course of the cell cycle. Once in mitosis, the kinetochore recruits additional dynamically associated proteins that are required to enable the diverse mitotic kinetochore functions. Here, we review kinetochore assembly in the context of the constantly changing centromere chromatin status and discuss how kinetochore organization and function are altered to facilitate these changes and confer the diverse functions of the kinetochore throughout the cell cycle.
Figure 1. Dynamic events throughout the cell cycle affect centromere structure.
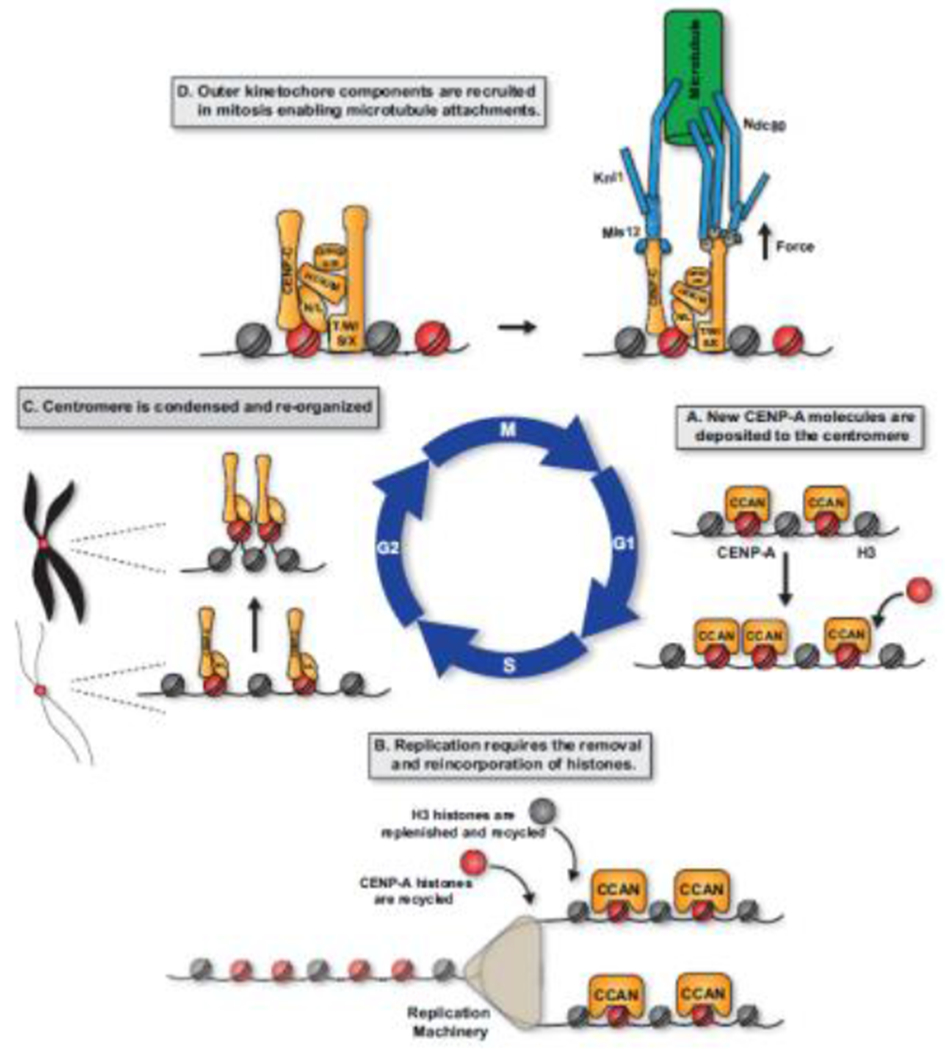
A. In G1, new CENP-A molecules are deposited at centromeres. This process is tightly regulated to ensure this only happens once each cell division cycle. B. During S phase, centromere DNA must be replicated. Because no new molecules of CENP-A are deposited, this involves the distribution of previously deposited CENP-A molecules such that CENP-A molecules are diluted and H3.3 histones are incorporated as placeholders to fill in gaps. Throughout S-phase, the CCAN remains assembled upon centromeric CENP-A and helps to retain CENP-A centromere following passage of the replication fork. C. In late G2, chromatin is condensed to form the distinct chromosome structures observed in mitosis. In addition, centromere DNA must reorganize to orient the centromere proteins to recruit of outer kinetochore components and binding to spindle microtubules. D. In mitosis, following nuclear envelope breakdown and phosphorylation by CDK1, outer kinetochore proteins are recruited to the centromere completing the assembly of the kinetochore and enable kinetochores to interact directly with spindle microtubules and facilitate chromosome segregation.
The centromere locus
An understanding of kinetochore assembly and organization first requires a consideration of the centromere, the chromosomal locus upon which the kinetochore is built. Despite providing the functional platform for kinetochore assembly, centromere structure, position, and sequence vary widely across organisms [4], For example, in the budding yeast S. cerevisiae, centromeric DNA is 125 base pairs, with a defined sequence that is both necessary and sufficient for centromere function and kinetochore assembly, referred to as a point centromere. Conversely, other commonly studied organisms, including mammals, Drosophila, and fission yeast, have centromeres that consist of long arrays of repetitive DNA sequences that can span ten’s of thousands to millions of base pairs in length, termed regional centromeres. Finally, the centromeres found in some plants, nematodes, and insect species are not restricted to a single position, but instead span the complete chromosome, a centromere type called holocentric. Holocentricity makes it such that kinetochores assemble along the entire length of the chromosome [5].
Regional centromeres are often composed of repetitive DNA elements including retro-transposon elements and small units of repetitive sequences called satellite DNA that can span kilobases to megabases of DNA [4, 6]. The organization of these centromeres can be divided into two regions: 1) the core centromere region, which is comprised of homogenous ordered repeats (largely satellite DNA) upon which the kinetochore is assembled, and 2) the pericentromere, which flanks the core centromere and is made of less ordered repetitive sequences that are heterochromatic [7]. In humans, the repeating unit of the core centromere region is known as α-satellite DNA. The α-satellite sequence is 171-bp in length, where each unit is arranged in a head-to-tail manner forming higher-order repeats throughout the core centromere [7–11]. Subsets of α-satellite repeats additionally contain a 17 bp motif known as the CENP-B box [12, 13], which serves to recruit the centromere protein (CENP), CENP-B. Regional centromeres promote the assembly of larger kinetochore structures that bind to multiple microtubules during mitosis [2]. In fact, kinetochore size has been shown to scale with centromere length, which can ultimately impact kinetochore function as chromosomes with larger kinetochores are prone to mis-segregate in mitosis [14].
Although regional centromeres can be highly structured, the underlying DNA sequence is not sufficient to generate a functional centromere [15]. Indeed, although the presence of repetitive sequences at regional centromeres is common, there are exceptions to this, such as the non-repetitive centromeres found in horse, orangutan, and chicken chromosomes [16–20]. Instead, regional centromeres are considered to be epigenetically defined based on the presence of the centromere-specific histone CENP-A, a variant of the canonical histone protein H3 [21]. The presence of CENP-A at centromeres is not unique to organisms with regional centromeres. CENP-A can be found at the centromeres of many organisms, including those with holocentric and point centromeres. In fact, CENP-A is present at the centromere of most model organisms including S. cerevisiae, S. pombe, D. melanogaster, C. elegans, and humans [22–25]. However, there are notable exceptions, including kinetoplastids and some insects such as Lepidoptera, which lack centromere-specific histones [26–28]. The precise mechanism by which centromeres are specified in these organisms is an active area of research. Recent work in Lepidoptera suggests that kinetochores are specifically assembled at regions of the genome with low transcriptional activity and have adapted to rely on the function of other centromere proteins [29, 30].
In addition to CENP-A, multiple factors act to collectively specify the centromere region. Among these, are the proteins that localize to centromeres throughout the cell cycle and form the inner kinetochore, collectively referred to as the Constitutive Centromere Associated Network (CCAN) [31, 32]. The CCAN is composed of 16 proteins that can be further organized into 5 distinct groups based on their physical associations and dependencies for centromere localization: CENP-C, CENPL/N, CENP-H/I/K/M, CENP-O/P/Q/U/R, and CENP-T/W/S/X [33]. Together, these sub-complexes function to bind specifically to CENP-A and recruit the components of the outer kinetochore. Importantly, although the features that define and characterize the centromere can vary dramatically between species, the requirement for constitutively bound centromere proteins to maintain centromere identity and function throughout the cell cycle remains constant.
Kinetochore assembly during G1 – refreshing centromere chromatin
The first dynamic changes to centromere chromatin and kinetochore assembly state occur in Gl, when new CENP-A histones are deposited at the centromere. CENP-A is both necessary and sufficient for the recruitment of all known kinetochore proteins [34–42], and thus this deposition process is a critical event for the specification and maintenance of centromere identity. The deposition of new CENP-A molecules involves the coordination of multiple factors. Importantly, the cell must distinguish between CENP-A nucleosomes and canonical H3 nucleosomes, which share ~62% sequence identity in their histone fold domain [21]. To achieve this, CENP-A deposition depends on a dedicated chaperone called HJURP (Holliday Junction Recognizing Protein) in vertebrates [43–45]. Functional orthologs of HJURP also exist in S. cerevisiae and S. pombe, called Scm3, and in D. melanogaster, called Call [46–56]. The HJURP chaperone binds specifically to the CENP-A targeting domain (CATD), a region that distinguishes CENP-A from canonical histone H3 [57–60]. In humans, this domain of CENP-A is both necessary and sufficient for targeting CENP-A to centromeres although not sufficient to promote kinetochore assembly [34, 61–64]. HJURP binds to the CATD of soluble CENP-A through its N-terminal CENP-A binding domain [43, 44, 57]. Once bound to CENP-A, HJURP is targeted to centromere regions through its interaction with the Misl8 complex (Misl8α, Mis 18β, and Misl8BPl) [65–69], with Misl8BPl binding directly to HJURP via its central domain [39, 70, 71]. In humans, Misl8BPl also binds to the inner kinetochore protein CENP-C and through this interaction is targeted to the centromere [72–74]. Given that CENP-C localizes to centromeres through its direct interaction with CENP-A, this is one mechanism to ensure that CENP-A is exclusively deposited at centromeres in a self-propagating manner. Although the general paradigms of CENP-A propagation are conserved across vertebrates, the precise interactions vary between organisms. For example, in chicken and Xenopus, Misl8BPl centromere localization does not depend on CENP-C, and instead involves a unique motif that is similar to the CENP-A binding domain in CENP-C to directly interact with CENP-A [75, 76]. This Misl8BPl-CENP-A interaction provides an alternative strategy for CENP-A to promote centromere specification and its self-propagation.
The timing for deposition of new CENP-A molecules is tightly coordinated with cell cycle progression in a way that is distinct from canonical H3 nucleosomes. Whereas H3 nucleosomes are deposited during S phase, the deposition of new CENP-A in vertebrates occurs in early G1 [61, 77–79]. To enact this temporal restriction, two kinases function as the gatekeepers of CENP-A deposition: Cyclin Dependent Kinase (CDK) and Polo-like Kinase 1 (PLK1). In human cells, CDK phosphorylation of Misl8BPl results in decreased centromere localization and prevents its interaction with Misl8α and Mis18β [68, 74, 80, 81]. CDK phosphorylation of HJURP has also been shown to negatively impact its ability to localize to centromeres [81, 82]. Therefore, deposition of new CENP-A molecules can only occur following chromosome segregation, when CDK activity is at its lowest. Conversely, the kinase activity of PLK1 acts to promote CENP-A deposition through phosphorylation of upstream targets, including the Misl8 complex [74]. Disrupting this temporal control of CENP-A deposition has negative effects on mitotic progression, highlighting the importance of this regulatory network on centromere function [74].
Once incorporated into centromere chromatin, CENP-A remains remarkably stable in rapidly dividing cells [83–85], with individual CENP-A molecules persisting over multiple divisions [78, 86]. It is currently not well understood how centromeric chromatin adapts to the changes in CENP-A occupancy throughout the cell cycle, although the presence of chromatin remodeling factors and transcription at the centromere may play a role [87]. Additionally, CENP-A has been shown to acquire post-translational modifications throughout the cell cycle that help coordinate different aspects of CENP-A function [88].
Given that maintenance of centromere CENP-A relies on passage through the cell cycle, it is interesting to consider how it is that the centromere is maintained in cells that are not actively dividing. Non-transformed cells and many cells in an organism can enter a state of proliferative hibernation termed quiescence or GO [89]. Quiescent cells can persist in a non-dividing state for weeks, months, even decades and maintain their proliferative potential. Therefore, it is critical to retain the presence of centromere components to mark this chromosomal locus. In particular, as the centromere is specified primarily by the presence of CENP-A, if CENP-A is evicted from chromosomes, it would eliminate centromere identity and prevent future chromosome segregation. Thus, quiescent cells must maintain the protein components at centromeres over extended periods. One possibility to explain this is that CENP-A and other CCAN components are exceptionally stable. Indeed, studies in mouse oocytes blocking new CENP-A synthesis have indicated that CENP-A protein remains stable over extended periods [84]. Alternatively, quiescent cells may alter CCAN dynamics to promote the deposition of CENP-A and refresh centromere proteins. By studying this in quiescent cultured cells and starfish oocytes, our lab has found that CENP-A undergoes a gradual, but consistent turnover at centromeres [90]. This dynamic behavior requires the canonical CENP-A deposition machinery and may be promoted by ongoing transcription at centromeres to evict CENP-A molecules providing “holes” to enable new CENP-A deposition. The failure to deposit new CENP-A molecules in quiescent cells results in defective chromosome segregation when quiescent cells return to proliferation. In contrast to CENP-A, some other CCAN components are lost from centromeres in quiescent cells [90] suggesting the potential for additional changes in centromere organization in non-dividing cells. Interestingly, CENP-A is differentially maintained in diverse cell types in a manner that correlates with replicative potential [91]. For example, in work by Swartz et. al. [90], terminally differentiated cells, such as adult mouse cardiomyocytes, lose CENP-A at their centromeres. In contrast, adult mouse hepatocytes, which maintain the capacity to divide and regenerate, maintain centromeric CENP-A. Together, this supports the importance of CENP-A maintenance at the centromere to facilitate cell division.
Together, the dynamic localization and maintenance of CENP-A molecules in the early cell cycle stages and in non-dividing cells provides the foundation onto which the remaining kinetochore architecture is assembled in the subsequent cell cycle phases. Importantly, the large number of CENP-A nucleosomes deposited during G1 represents a substantial change to centromere chromatin, and likely also requires the subsequent recruitment of CENP-A binding factors.
Retaining centromere assembly during Replication and Transcription
As the cell cycle proceeds, the next challenge that centromeres face is the need to replicate the underlying DNA during S phase (Figure 1B). DNA replication requires the displacement of existing nucleosomes to allow for the passage of the replication fork and DNA synthesis [92]. Therefore, the centromere must accommodate the passage of the replication fork while maintaining its identity and function. CENP-A histones are stably inherited across multiple divisions, suggesting that centromere nucleosomes are retained through DNA replication. Given that no new CENP-A molecules are deposited during S phase, CENP-A nucleosomes must be recycled and conservatively distributed between the newly replicated sister centromeres [78]. It is thought that the gaps left due to the 2-fold reduction in the number of CENP-A nucleosomes are instead filled with histone H3.3 [93]. Interestingly, the position and distribution of CENP-A nucleosomes within the centromere are maintained, implicating a positional memory of CENP-A nucleosomes within the centromere [93, 94], although the CENP-A domain can migrate slightly across divisions [95]. This raises the question as to how CENP-A nucleosomes are distributed during DNA replication. Recent work has suggested that the distribution of CENP-A nucleosomes involves HJURP, which transiently associates with CENP-A during S phase, in conjunction with Mcm2, a helicase involved in DNA replication [96]. This parallels the mechanism by which canonical H3 nucleosomes are deposited in non-centromere chromatin, which also involves Mcm2 and distinct H3/H4 nucleosome chaperones [92].
The passage of a replication fork through centromere DNA has the potential to be highly destabilizing. In fact, DNA replication has been shown to evict CENP-A nucleosomes erroneously incorporated at chromosome arms [94]. So how is it that CENP-A nucleosomes remain enriched at the centromere following DNA replication? A key distinction of CENP-A at centromeres is the presence of the associated CCAN components, which remain assembled on CENP-A nucleosomes during S phase/G2 [94]. Recent work has suggested that the CCAN contributes to centromere specific recycling/retention of CENP-A nucleosomes specifically at centromeres. Specifically, the rapid depletion of CENP-C in early S phase results in a significant loss of CENP-A nucleosomes at centromeres following a round of DNA replication [94]. Together, this supports the idea that the CCAN plays a critical role in supporting the reincorporation of CENP-A nucleosomes at centromeres during DNA replication. How the CCAN remains assembled on CENP-A nucleosomes and interacts with the replication machinery remains an important open question.
Importantly, DNA polymerase is not the only enzyme that acts at centromeres and has the ability to disrupt the underlying chromatin. Despite the fact that centromeres lack coding genes, centromeres are actively transcribed. RNA polymerase II has been found at centromeres in mitosis [97] and centromeric chromatin contains histone modifications, such as H3K4me2, that are permissive for transcription [98, 99]. Importantly, centromere transcription occurs at low levels throughout the cell cycle, including during mitosis [97, 100]. The presence of ongoing centromere transcription during mitosis may play an active role in centromere function [87, 101]. For example, centromere transcription by RNA polymerase could destabilize nucleosomes and CCAN components. In fact, centromere transcription has been implicated in promoting the turnover of CENP-A [90]. Alternatively, the presence of RNA polymerase II at mitotic centromeres may be due to the persistence of cohesin at centromere regions [102], but may not reflect an active role [103].
Together, the CCAN and CENP-A nucleosomes maintain a reciprocal relationship to ensure centromere identity is maintained despite drastic changes to the DNA structure in S phase. Whereas the CCAN components require the presence of CENP-A within the centromere for their localization, these proteins also help maintain CENP-A localization, including during DNA replication. Additionally, although replication and transcription represent a challenge to centromere identity, it also performs a beneficial role in centromere maintenance by preventing the accumulation of CENP-A at ectopic chromosome regions.
The Inner Kinetochore
The inner kinetochore, or CCAN, is assembled specifically at the centromere due to its interactions with CENP-A. A distinguishing characteristic of the CCAN is that it remains present at the centromere throughout the cell cycle. Therefore, once assembled the interactions between CCAN components must be robust enough so that the entire assembly is maintained at every cell cycle state. Indeed, each of the CCAN sub-complexes (CENP-C, CENP-L/N, CENP-H/I/K/M, CENP-T/W/S/X, and CENP-O/P/Q/U/R) [104–106] has a distinct role in CCAN organization, and together form a hierarchy of interactions that promote the stability of this protein complex (Figure 2A).
Figure 2. Organization of the Inner Kinetochore.
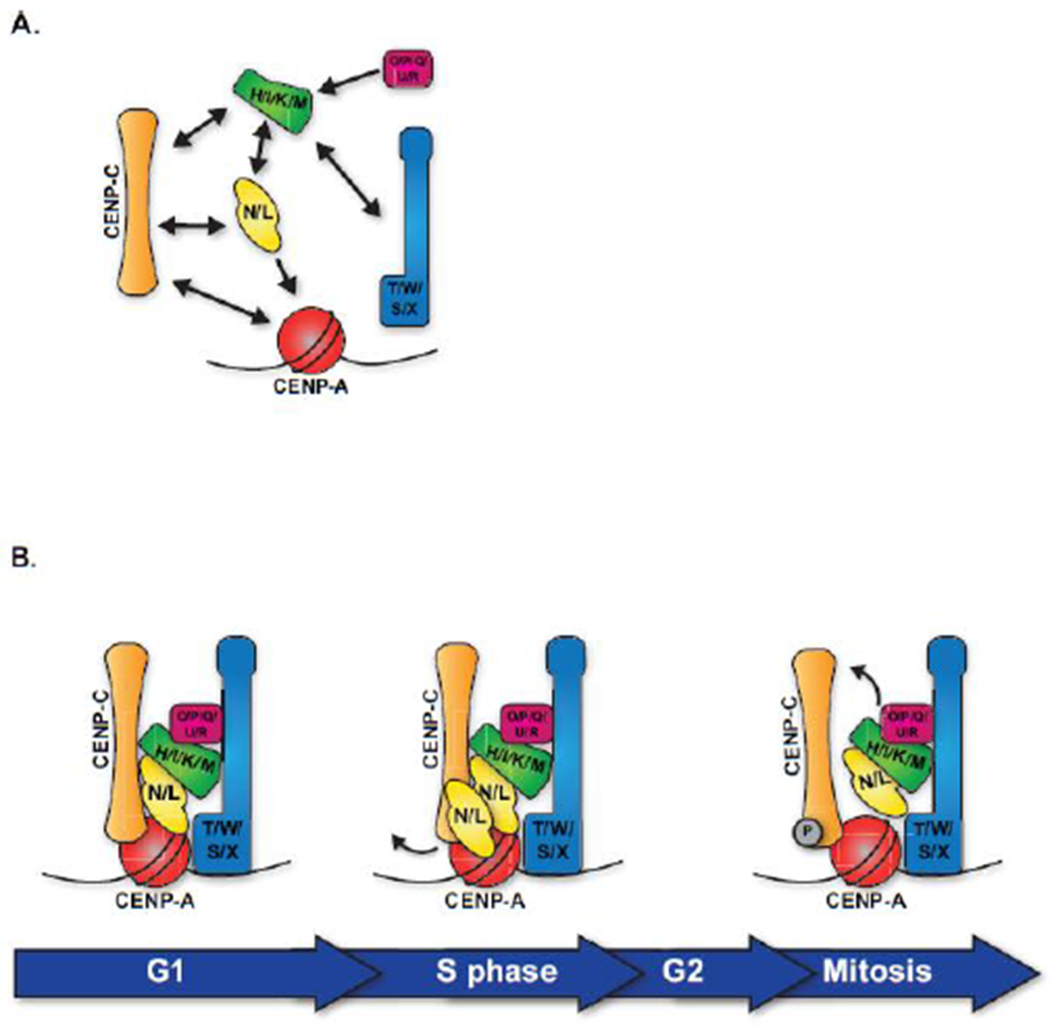
A. The Inner kinetochore is made up of 5 distinct groups: CENP-C, CENP-N/L, CENP-H/I/K/M, CENP-T/W/S/X, and CENP-O/P/Q/U/R. These sub complexes are largely interdependent for their localization to the centromere. Direct interactions between inner kinetochore sub-complexes, as defined by biochemical reconstitutions and functional dependencies, are represented by arrows. B. Although all components of the CCAN are present at centromeres constitutively, these proteins undergo reorganization and changes throughout the cell cycle. This model represents snapshots of inner kinetochore organization at distinct cell cycle phases.
CENP-C plays a central role in kinetochore organization as it interacts with multiple components of the inner kinetochore, including CENP-A, and can recruit components of the outer kinetochore directly [107–111]. In fact, for some organisms, such as D. melanogaster and C. elegans, CENP-C is the only identified CCAN component [26, 31, 112]. Based on its network of interactions, CENP-C is considered to be an organizing scaffold within the CCAN [107, 108, 113–115]. In addition to CENP-C, CENP-N is the only other protein within the kinetochore that can interact directly with CENP-A [114, 116], CENP-C and CENP-N interact with CENP-A in a distinct manner [114], providing the potential for both proteins to interact with CENP-A simultaneously. Within the CCAN, CENP-N forms an obligate complex with CENP-L where together they interact directly with CENP-HIKM and CENP-C [108, 115, 117], The CENP-HIKM complex also sits at the core of the CCAN as it has been shown to directly interact with CENP-C, CENP-LN, and CENP-TWSX [104, 115, 118], The CENP-TWSX complex contains histone fold domains that promote centromere recruitment by conferring DNA binding abilities to this complex in addition to facilitating protein interactions [32, 107, 108, 119, 120], Like CENP-C, the CENP-TWSX complex interacts directly with proteins of the outer kinetochore and plays an important role in their regulated recruitment in mitosis [32, 121–123], Finally, the CENP-OPQUR complex is recruited to the kinetochore through its interaction with CENP-C/HIKM/LN [118], The CENP-OPQUR complex is the most peripheral CCAN component, as depletion of this complex does not affect the localization of the other CCAN components [106, 108, 118], The CENP-OPQUR complex facilitates functions that are important for mitotic progression, including recruitment of CENP-E and PLK1, and has been suggested to interact directly with microtubules [118, 124–130], The interdependencies that exist between the CCAN components enable this complex of proteins to adapt to changes that occur in the centromere so that they remain assembled throughout the cell cycle.
The Dynamic Nature of the Inner Kinetochore
The naming of the CCAN as “constitutive” centromere factors can imply that they are unchanged throughout the cell cycle. However, despite their constitutive localization, the association of individual components of the CCAN to the centromere is instead quite dynamic. This includes cell cycle changes in the abundance of individual CCAN components [131–133], the timing by which new proteins are incorporated at centromeres [85, 132, 134, 135], and changes in the physical associations and functional relationships between the CCAN sub-complexes (Figure 2B).
Of the CCAN proteins, CENP-C and CENP-N provide important examples of these distinct cell cycle behaviors. Although CENP-C is always present at centromeres, it displays dynamic and changing properties. First, CENP-C levels at kinetochores remain relatively constant throughout the cell cycle but increase upon entry into mitosis [133], The molecular requirements for CENP-C localization to centromeres also differ between interphase and mitosis. In interphase, CENP-C localization depends on its interaction with CENP-HIKM and CENP-L/N as depletion of either complex result in the loss of CENP-C at the centromere [108, 136]. In contrast, during mitosis CENP-C can localize to kinetochores in the absence of CENP-LN and CENP-HIKM. Given the important role that CENP-C plays in CCAN organization, this change in its localization requirements suggests that there is a cell cycle-dependent reorganization of the CCAN. Recent work in chicken cells found that CDK-dependent phosphorylation of CENP-C in mitosis promotes its interaction with CENP-A [137, 138]. Thus, the requirements for CENP-C localization change throughout the cell cycle, such that its interaction with other CCAN subunits is important in interphase, but its interaction with CENP-A becomes more critical in mitosis. This change may be important in adapting the CCAN for the recruitment of outer kinetochore components, or rewiring CCAN organization to accommodate interphase events at centromeres.
The CENP-L/N complex also plays a critical role in the organization of the CCAN and displays distinct cell cycle dependent behaviors. CENP-N interacts directly with the CATD of CENP-A [116, 139]. This interaction is mediated by the N-terminus of CENP-N, which is both necessary and sufficient for CENP-N localization to the kinetochore during interphase, but not mitosis [108, 116]. The C-terminus of CENP-N, which meditates its direct interaction with CENP-L and indirectly with the CCAN, is critical for the mitotic localization of CENP-N [108, 140]. Therefore, the localization of CENP-N relies more on its interactions with the CCAN in mitosis, a behavior that directly contrasts with that of CENP-C. Additionally, CENP-N displays distinct localization behaviors throughout the cell cycle. Unlike CENP-C, CENP-N is enriched at kinetochores specifically during S-phase and then dissociates during the G2/M transition such that its levels are low in mitosis [141, 142]. Finally, CENP-N has been shown to associate dynamically with centromeres during G1 and early S-phase [141]. However, the basis and the significance of this change in CENP-N kinetochore levels remain unclear. One aspect of CENP-N behavior that may contribute to this difference in CENP-N association is the fact that CENP-N loading appears to be dependent on chromatin state. In vitro assays suggest that CENP-N more readily interacts with CENP-A nucleosomes present in open chromatin where the RG loop is exposed, as compared to a more compact chromatin state where the RG loop is not accessible [143]. Therefore, the underlying changes in chromatin state that occur throughout the cell cycle may affect the ability of CENP-N to remain stably bound at kinetochores.
The fact that CENP-N and CENP-C, two components essential for CCAN recruitment and CENP-A recognition, have distinct cell cycle dependent behaviors suggests the CCAN complex as a whole undergoes substantial changes. However, it remains unclear how these behaviors and differences in centromere associations affect CCAN organization and function. The underlying mechanisms that contribute to the varying association behaviors within the CCAN have not been elucidated, but post-translational modifications, such as phosphorylation, may play a role [137, 138, 144].
Role of CENP-B in Centromere Function
Given the critical requirement to maintain the assembly of inner kinetochore components at centromeres, it is important to highlight the potential contributions that CENP-B plays in maintaining centromere function. CENP-B is the only protein within the inner kinetochore that binds to a specific DNA sequence - the 17 bp CENP-B box present in the alpha-satellite sequences [12]. The functional importance of CENP-B at centromeres has been debated given that it is not essential for viability in either cell culture or mouse models and that it is absent from both neocentromeres and the Y chromosome [145]. However, chromosomes that lack CENP-B have been shown to mis-segregate more frequently [146] suggesting that CENP-B plays important roles in centromere function. Within the centromere, CENP-B has been shown to interact directly with CENP-A, and through this interaction increases the stability of CENP-A in reconstituted nucleosomes [147–149]. CENP-B and CENP-C also interact directly. This interaction has been proposed to serve as an additional recruitment mechanism for CENP-C independently from CENP-A, as depletion of CENP-B results in a decrease of CENP-C at kinetochores [147]. In fact, in the absence of CENP-A, CENP-B is sufficient to maintain CENP-C at the kinetochore and facilitate proper chromosome segregation [150], at least for a short period of time. Recent work has supported a model in which CENP-B becomes more important at centromeres in the absence of CENP-A [146]. Therefore, the presence of CENP-B at the centromere may help to ensure centromere function in conjunction with CENP-A by providing an additional mechanism to tether the inner kinetochore to the centromere.
Higher order centromere structure and changes to chromatin state
A major consideration for understanding the assembly and dynamics of the kinetochore is the higher order organization of the underlying centromere DNA and chromatin (Figure 1C). The centromere does not exist as a linear segment of DNA. Instead, centromere DNA displays a complex three-dimensional organization, similar to other chromosomal loci. Within the centromere, CENP-A nucleosomes are interspersed with H3 nucleosomes [93, 151, 152]. At human centromeres, quantification of CENP-A molecules indicate that ~100 CENP-A nucleosomes are dispersed throughout the centromere (likely over ~100 kb) at a ratio of ~1:25 relative to H3 [153]. Therefore, to enable kinetochore assembly and proper microtubule attachment in mitosis, as chromosomes are compacted and condensed at mitotic entry, the centromere must be organized to position CENP-A nucleosomes together facing away from the chromosome to facilitate kinetochore assembly. How this occurs remains unclear, but multiple models have been proposed [15, 154, 155]. Although the mechanism and precise structure of centromere chromatin upon entry into mitosis is not known, CENP-A and the CCAN confer properties to centromere chromatin that are important for enacting its mitotic function. For example, in C. elegans, loss of CENP-A causes defects in chromosome structure and affects the dynamics of chromosome condensation [156]. A similar behavior has also been shown in human cells, where CENP-A chromatin withstands unfolding to a higher degree than the surrounding heterochromatin and mitotic CENP-A enriched chromatin can withstand unfolding to a higher degree than interphase chromatin [151]. This suggests that the presence of CENP-A distinctly impacts chromatin architecture at the centromere, and that this may confer an added rigidity that allows the centromere to withstand mitotic forces.
The presence and association of the CCAN at the centromere also contributes to centromere rigidity [151]. Reciprocally, the changes that happen to centromere chromatin upon mitotic entry can affect the organization of the CCAN as demonstrated by changes in CENP-C interdependencies and the effect that CENP-A chromatin state has on CENP-N binding in vitro [143]. Efforts to reconstitute the kinetochore suggest that two CCAN complexes (containing CENP-C/HIKM/LN) are capable of binding a single CENP-A nucleosome, forming a symmetric structure [115]. Recent structural work reconstituting the core centromere nucleosome complex (CCNC) consisting of CENP-A, CENP-N, and CENP-C from human and chickens has provided insights into the potential changes in CCAN associations at centromeres between interphase and mitosis [138, 142]. In work reconstituting the human CCNC, Allu and colleagues [142] observed two CCNC populations - one in which two CENP-C and CENP-N molecules bind to CENP-A, and one in which only a single molecule of CENP-N and two molecules of CENP-C are bound to the CENP-A nucleosome. Given that this corresponds to the differing ratios of CENP-C and CENP-N present at centromeres in interphase and mitosis, it is possible that these two structures represent the cell cycle differences in CCAN centromere associations. Alternatively, studies analyzing centromere proteins from chicken cells support an alternative model for the changes in CCNC assembly throughout the cell cycle [138]. In this work, the authors argue that CENP-C and CENP-N do not bind CENP-A simultaneously and instead do so in a mutually exclusive and cell cycle-dependent manner. In this model, CENP-N binds directly to CENP-A nucleosomes during interphase, whereas CENP-C does so in mitosis. A key distinction between these two studies is the CENP-C fragments used to reconstitute the CCNC. Ariyoshi and colleagues utilized a longer CENP-C C-terminal fragment, which resulted in structures where CENP-C obscured the RG loop in CENP-A preventing CENP-N binding. Importantly, CENP-C was phosphorylated in these experiments, modeling mitotic CENP-C [137]. Together, these studies provide molecular insights into the mechanisms that may underlie CCAN reorganization during mitosis. However, future studies will be required to determine the physiological relevance of these two proposed mechanisms, including structural studies with full length CENP-C and CENP-N. Overall, progression into mitosis requires that the centromere adapt a structure that will withstand the forces transmitted during chromosome segregation and provide a platform for outer kinetochore assembly.
The Outer Kinetochore
The next critical phase in kinetochore assembly occurs during entry into mitosis when the CCAN directs recruitment of the outer kinetochore, which in turn interacts directly with spindle microtubules. Outer kinetochore assembly is a highly controlled process that occurs through multiple regulatory layers (Figure 3). First, the core components of the outer kinetochore are recruited upon mitotic entry in a manner that depends upon the phospho-regulation of inner kinetochore components to create a platform for outer kinetochore assembly. Second, kinetochores alter their functional properties at different stages within mitosis by altering outer kinetochore composition, adding or removing specific components to enable distinct activities. Finally, under circumstances where kinetochores fail to interact with microtubules, the outer kinetochore seeds formation of an extended fibrous corona structure.
Figure 3. Organization and assembly of the outer kinetochore.
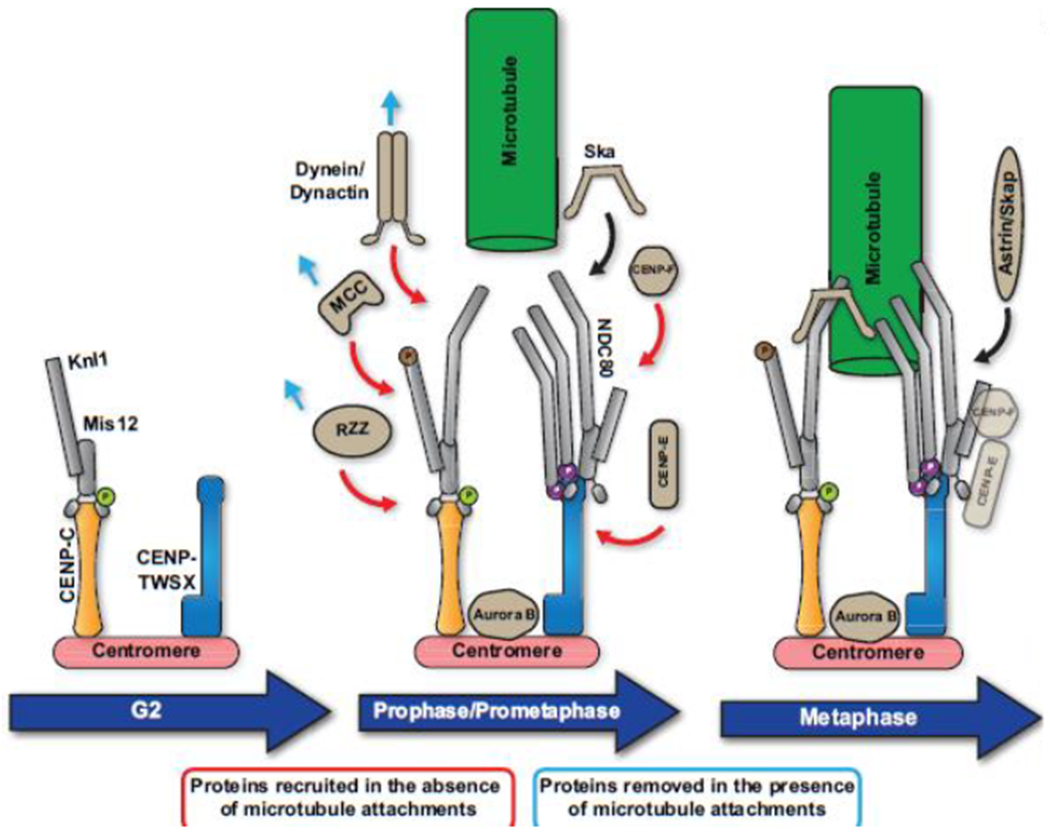
Recruitment of outer kinetochore components occurs in a step-wise manner. In late G2, Mis12/Knl1 are recruited through their interaction with CENP-C. This interaction is enabled by Aurora B phosphorylation of the Mis12 complex. Ndc80 is then recruited to the kinetochore following entry into mitosis. Once in mitosis, additional proteins are transiently recruited to the kinetochore to enable the multiple kinetochore functions in mitosis. In the absence of a microtubule interaction, proteins involved in the spindle assembly checkpoint (MCC) and the fibrous corona (RZZ, spindly, CENP-E, CENP-F, dynein/dynactin) are recruited to kinetochore’s As kinetochore-microtubule interactions are established, the Ska1 complex and Astrin/SKAP are recruited promote Ndc80 binding to microtubules. Once stable kinetochore-microtubule interactions are established and are properly bi-oriented, the spindle assembly checkpoint is satisfied and chromosome segregation occurs. CENP-E and CENP-F perform additional roles in chromosome segregation and remain at the kinetochore until anaphase.
The core of the outer kinetochore is made up of 10 proteins that assemble into the three sub-complexes Knl1, Mis12, and Ndc80 - also referred to as the KMN network [157–167]. In particular, the four-subunit Ndc80 complex acts as the primary kinetochore receptor for microtubule binding [157, 162, 168]. The KMN network is recruited in a stepwise manner throughout the cell cycle via two distinct pathways mediated by CENP-C and CENP-T. Each of these pathways is sufficient to recruit the outer kinetochore in vertebrates [41, 133, 169]. The coordinated assembly of the outer kinetochore relies on the activity of cell cycle-associated kinases and cell cycle-specific cellular events [170]. The Mis12 complex and Knl1 are the first to associate to the kinetochore starting in late S phase [133, 171] (Figure 3). This recruitment is mediated through the direct interaction between Mis12 and CENP-C [109–111, 121, 172]. Following their initial association, Mis12 and Knl1 levels gradually increase and are maximally enriched at metaphase [133]. The recruitment of Mis12 is facilitated by phosphorylation of Mis12 by the mitotic kinase Aurora B, which results in a conformational change that promotes the interaction between Mis12 and CENP-C [109, 173]. Upon entry into mitosis and onset of nuclear envelope breakdown, Ndc80 is recruited to the kinetochore by two distinct mechanisms [133, 171] (Figure 3). First, the Mis12 complex, via its interaction with CENP-C, recruits a single molecule of Ndc80 upon mitotic entry [174, 175]. Second, CENP-T can directly bind to two molecules of Ndc80 [123, 176, 177], following phosphorylation of two distinct sites of CENP-T by mitotic CDK1 [121, 123, 169, 176]. Additionally, CENP-T has been shown to independently recruit Mis12 following CDK phosphorylation, therefore indirectly recruiting an additional molecule of Ndc80 [169, 176]. Although both CENP-C and CENP-T recruit outer kinetochore proteins, work in chicken cells has suggested that CENP-T plays a more critical role in the recruitment of the KMN network. In this work, researchers found that expressing a CENP-C mutant that lacks its Mis12 complex binding domain did not affect mitotic progression in chicken cells. In contrast, CENP-T mutants that lack the ability to bind the Ndc80 or Mis12 complexes in the absence of endogenous CENP-T resulted in cell death [172]. Further work is needed to determine whether this is true in mammalian cells.
Although Ndc80 is essential for mediating kinetochore/microtubule interactions, additional microtubule-binding proteins are recruited to the outer kinetochore throughout mitosis to strengthen and stabilize this interaction. The first of these proteins is the Skal complex, which functions to enhance the microtubule binding activity of Ndc80 [178–180]. Recruitment of the Ska1 to kinetochores requires Ndc80 and the presence of kinetochore/microtubule interactions [181, 182]. The Ska1 complex is first recruited during prometaphase and accumulates at kinetochores throughout chromosome congression [133, 183, 184]. Once kinetochores have established stable attachments to microtubules and are properly bi-oriented, the Astrin-SKAP protein complex is recruited to kinetochores [185–188]. The Astrin-SKAP localizes to bi-oriented kinetochores at metaphase and persists into anaphase, such that this complex is thought to play an important role in stabilizing proper kinetochore/microtubule attachments [186, 188–190]. Additional proteins also associate with kinetochores to regulate spindle microtubule dynamics and facilitate microtubule organization (see [191, 192]).
Finally, under conditions where kinetochores fail to generate microtubule interactions, kinetochores undergo a dramatic additional assembly step and physical transformation where they can expand to form a crescent-like structure on its surface called the fibrous corona [193–196]. This fibrous structure forms at kinetochores that lack attached microtubules and compacts once kinetochore-microtubule interactions are established [193, 197]. This process involves a complex network of proteins, including the motor proteins CENP-E [198], the dynein/dynactin motor complex [199], the microtubule binding protein CENP-F [200], and the RZZ (Rod-ZW10-Zwilch) complex [201, 202]. This structure is thought to function to increase the likelihood of microtubule capture and promote Spindle Assembly Checkpoint (SAC) signaling in the presence of unattached kinetochores [195, 197]. The formation of the fibrous corona also precedes the formation of end-on attachments in unperturbed cells [195]. The molecular mechanisms underlying kinetochore expansion have implicated the RZZ complex and Spindly protein, which together have been shown to oligomerize in vitro [203–205]. Depletion of Spindly or RZZ complex components prevents kinetochore expansion in the absence of microtubules [203–205]. It remains unclear how the RZZ complex is tethered to kinetochores, although Knl1 and Bub1 have been proposed to contribute to this process [203, 206]. Once end-on attachments are established, the majority of the fibrous corona components are stripped from the kinetochore in a dynein-dependent manner, resulting in compaction of the kinetochore structure [207–211]. However, CENP-E and CENP-F perform additional roles in chromosome segregation and remain at the kinetochore until anaphase [212, 213]. This general process of kinetochore compaction is important for proper mitotic progression as disruption of this process results in erroneous microtubule attachments as well as chromosome mis-segregation [14, 195, 205, 211]. Therefore, this regulated morphological change in the outer kinetochore is critical to ensure faithful chromosome segregation.
Ensuring the fidelity of mitosis
In mitosis, the kinetochore functions as a scaffold to recruit signaling proteins and monitor kinetochore-microtubule attachments. For faithful chromosome segregation to occur, sister kinetochores must attach to microtubules from opposing spindle poles to establish bi-orientation. Through a process called error correction, kinetochore-microtubule attachments are monitored by the activity of kinetochore-associated kinases and phosphatases [214]. Together, the dynamic association of these factors modulates kinetochore function in mitosis to not only form interactions with microtubules but also to ensure that kinetochores undergo proper chromosome segregation.
Although changes in kinetochore composition can dynamically alter kinetochore function, some kinetochore activities are modulated without changes to kinetochore assembly state. For example, Aurora B kinase regulates kinetochore-microtubule interactions by destabilizing the interaction between components of the KMN and microtubules. Ndc80 is a critical substrate of Aurora B within the KMN, where Aurora B phosphorylates the regulatory N-terminal tail of Ndc80, decreasing its affinity for microtubules and therefore disrupting erroneous interactions with microtubules [157, 168, 215]. Similarly, Aurora B phosphorylates the Ska1 complex reducing its binding affinity for microtubules [179, 216]. Once proper kinetochore-microtubule attachments have been achieved, dephosphorylation of these substrates is mediated by the activity of two mitotic phosphatases: PP1 and PP2A-B56 [217–219]. Once dephosphorylated, Ndc80 and Ska1 are able to establish stable interactions with microtubules [157, 215, 220–222].
Accurate chromosome segregation is also monitored by an additional set of proteins that collectively make up the Spindle Assembly Checkpoint (SAC), which associate preferentially with unattached kinetochores. The key function of the SAC is to prevent anaphase onset until all chromosomes are stably attached to spindle microtubules [223]. This checkpoint is regulated by the activity of two complexes called the mitotic checkpoint complex (MCC), which prevents anaphase onset, and the Anaphase Promoting Complex/Cyclosome (APC/C), an E3 ubiquitin ligase that induces anaphase onset and mitotic exit through the targeted degradation of important mitotic proteins [224–226], In addition to its role in mediating kinetochore-microtubule interactions, the KMN acts as a scaffold for the recruitment of spindle assembly checkpoint proteins. For example, the MCC is recruited to the kinetochore by phosphorylation of Knl1 by Mps1, a kinase that is critical for initiating the SAC response, as Mps1 phosphorylation activity is required for the recruitment of downstream SAC components [227, 228], Once stable microtubule attachments have been achieved and sister kinetochores are bi-oriented, the SAC is inactivated to allow for mitotic progression [229], The inactivation of the SAC involves the disassembly of the MCC, removal of SAC proteins by dynein, and dephosphorylation of kinetochore and SAC components by the PP1, localized to the kinetochore through its interaction with Knl1 and PP2A-B56 [223, 230].
In summary, the kinetochore continues to be a dynamic structure even once it is fully assembled in mitosis. Through the coordinated recruitment of regulatory proteins, the kinetochore is able to perform the many functions required to facilitate proper chromosome segregation.
Conclusions
The kinetochore is a dynamic molecular protein complex that changes in composition and structure throughout the cell cycle to enable the kinetochore to perform distinct functions at each cell cycle stage. During interphase, the minimal kinetochore structure present at the centromere, the CCAN, performs the important function of maintaining centromere identity through its role in maintaining CENP-A at the centromere. As the cell progresses into mitosis, the CCAN reorganizes to enable the recruitment of the outer kinetochore. Once in mitosis, the kinetochore functions to recruit proteins required to facilitate the formation of kinetochore-microtubule interactions, promote checkpoint signaling to ensure proper attachments are made, and expand to enable kinetochore-microtubule interactions to be formed. Given the importance of the regulation of these protein-protein interactions, future work in understanding the proteins and regulatory mechanisms that enable kinetochore function and disassembly will provide a more comprehensive understanding of the kinetochore’s role in chromosome segregation and centromere maintenance.
Acknowledgements
The authors thank Océane Marescal and Ally Nguyen for critical reading of the manuscript, and B. Knowles and members of the Cheeseman lab for their support. The work in the Cheeseman lab is supported by grants from the NIH/National Institute of General Medical Sciences (R35GM126930), the Moore Foundation, the National Science Foundation (2029868), and a Pilot award from the Global Consortium for Reproductive Longevity and Equity (GCRLE), and an NSF graduate research fellowship to APN.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none
References
- [1].Cheeseman IM, The kinetochore, Cold Spring Harb Perspect Biol 6(7) (2014) a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Musacchio A, Desai A, A Molecular View of Kinetochore Assembly and Function, Biology (Basel) 6(1) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].de la Cruz J, Karbstein K, Woolford JL Jr., Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo, Annu Rev Biochem 84 (2015) 93–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].McKinley KL, Cheeseman IM, The molecular basis for centromere identity and function, Nat Rev Mol Cell Biol 17(1) (2016) 16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Maddox PS, Oegema K, Desai A, Cheeseman IM, “Holo”er than thou: Chromosome segregation and kinetochore function in C. elegans, Chromosome Res 12(6) (2004) 641–53. [DOI] [PubMed] [Google Scholar]
- [6].Kit S, Equilibrium sedimentation in density gradients of DNA preparations from animal tissues, J Mol Biol 3 (1961) 711–6. [DOI] [PubMed] [Google Scholar]
- [7].Aldrup-Macdonald ME, Sullivan BA, The past, present, and future of human centromere genomics, Genes (Basel) 5(1) (2014) 33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Vissel B, Choo KH, Human alpha satellite DNA--consensus sequence and conserved regions, Nucleic Acids Res 15(16) (1987) 6751–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Manuelidis L, Chromosomal localization of complex and simple repeated human DNAs, Chromosoma 66(1) (1978) 23–32. [DOI] [PubMed] [Google Scholar]
- [10].Manuelidis L, Complex and simple sequences in human repeated DNAs, Chromosoma 66(1) (1978) 1–21. [DOI] [PubMed] [Google Scholar]
- [11].Waye JS, Willard HF, Human beta satellite DNA: genomic organization and sequence definition of a class of highly repetitive tandem DNA, Proc Natl Acad Sci U S A 86(16) (1989) 6250–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T, A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite, J Cell Biol 109(5) (1989) 1963–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Muro Y, Masumoto H, Yoda K, Nozaki N, Ohashi M, Okazaki T, Centromere protein B assembles human centromeric alpha-satellite DNA at the 17-bp sequence, CENP-B box, J Cell Biol 116(3) (1992) 585–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Drpic D, Almeida AC, Aguiar P, Renda F, Damas J, Lewin HA, Larkin DM, Khodjakov A, Maiato H, Chromosome Segregation Is Biased by Kinetochore Size, Curr Biol 28(9) (2018) 1344–1356 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fukagawa T, Earnshaw WC, The centromere: chromatin foundation for the kinetochore machinery, Dev Cell 30(5) (2014) 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shang WH, Hori T, Toyoda A, Kato J, Popendorf K, Sakakibara Y, Fujiyama A, Fukagawa T, Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences, Genome Res 20(9) (2010) 1219–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang SP, Wang Z, Chinwalla AT, Minx P, Mitreva M, Cook L, Delehaunty KD, Fronick C, Schmidt H, Fulton LA, Fulton RS, Nelson JO, Magrini V, Pohl C, Graves TA, Markovic C, Cree A, Dinh HH, Hume J, Kovar CL, Fowler GR, Lunter G, Meader S, Heger A, Ponting CP, Marques-Bonet T, Alkan C, Chen L, Cheng Z, Kidd JM, Eichler EE, White S, Searle S, Vilella AJ, Chen Y, Flicek P, Ma J, Raney B, Suh B, Burhans R, Herrero J, Haussler D, Faria R, Fernando O, Darre F, Farre D, Gazave E, Oliva M, Navarro A, Roberto R, Capozzi O, Archidiacono N, Della Valle G, Purgato S, Rocchi M, Konkel MK, Walker JA, Ullmer B, Batzer MA, Smit AF, Hubley R, Casola C, Schrider DR, Hahn MW, Quesada V, Puente XS, Ordonez GR, Lopez-Otin C, Vinar T, Brejova B, Ratan A, Harris RS, Miller W, Kosiol C, Lawson HA, Taliwal V, Martins AL, Siepel A, Roychoudhury A, Ma X, Degenhardt J, Bustamante CD, Gutenkunst RN, Mailund T, Dutheil JY, Hobolth A, Schierup MH, Ryder OA, Yoshinaga Y, de Jong PJ, Weinstock GM, Rogers J, Mardis ER, Gibbs RA, Wilson RK, Comparative and demographic analysis of orang-utan genomes, Nature 469(7331) (2011) 529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Piras FM, Nergadze SG, Magnani E, Bertoni L, Attolini C, Khoriauli L, Raimondi E, Giulotto E, Uncoupling of satellite DNA and centromeric function in the genus Equus, PLoS Genet 6(2) (2010) e1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, Blocker H, Distl O, Edgar RC, Garber M, Leeb T, Mauceli E, MacLeod JN, Penedo MC, Raison JM, Sharpe T, Vogel J, Andersson L, Antczak DF, Biagi T, Binns MM, Chowdhary BP, Coleman SJ, Della Valle G, Fryc S, Guerin G, Hasegawa T, Hill EW, Jurka J, Kiialainen A, Lindgren G, Liu J, Magnani E, Mickelson JR, Murray J, Nergadze SG, Onofrio R, Pedroni S, Piras MF, Raudsepp T, Rocchi M, Roed KH, Ryder OA, Searle S, Skow L, Swinburne JE, Syvanen AC, Tozaki T, Valberg SJ, Vaudin M, White JR, Zody MC, P. Broad Institute Genome Sequencing, T. Broad Institute Whole Genome Assembly, Lander ES, Lindblad-Toh K, Genome sequence, comparative analysis, and population genetics of the domestic horse, Science 326(5954) (2009) 865–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sanyal K, Baum M, Carbon J, Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique, Proc Natl Acad Sci U S A 101(31) (2004) 11374–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sullivan KF, Hechenberger M, Masri K, Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere, J Cell Biol 127(3) (1994) 581–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S, A histone-H3-like protein in C. elegans, Nature 401(6753) (1999) 547–8. [DOI] [PubMed] [Google Scholar]
- [23].Takahashi K, Chen ES, Yanagida M, Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast, Science 288(5474) (2000) 2215–9. [DOI] [PubMed] [Google Scholar]
- [24].Henikoff S, Ahmad K, Platero JS, van Steensel B, Heterochromatic deposition of centromeric histone H3-like proteins, Proc Natl Acad Sci U S A 97(2) (2000) 716–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M, A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis, Genes Dev 9(5) (1995) 573–86. [DOI] [PubMed] [Google Scholar]
- [26].Drinnenberg IA, deYoung D, Henikoff S, Malik HS, Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects, eLife 3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Navarro AP, Cheeseman IM, Chromosome Segregation: Evolving a Plastic Chromosome-Microtubule Interface, Curr Biol 30(4) (2020) R174–R177. [DOI] [PubMed] [Google Scholar]
- [28].Akiyoshi B, Gull K, Discovery of unconventional kinetochores in kinetoplastids, Cell 156(6) (2014) 1247–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cortes-Silva N, Ulmer J, Kiuchi T, Hsieh E, Cornilleau G, Ladid I, Dingli F, Loew D, Katsuma S, Drinnenberg IA, CenH3-Independent Kinetochore Assembly in Lepidoptera Requires CCAN, Including CENP-T, Curr Biol 30(4) (2020) 561–572 e10. [DOI] [PubMed] [Google Scholar]
- [30].Senaratne AP, Muller H, Fryer KA, Kawamoto M, Katsuma S, Drinnenberg IA, Formation of the CenH3-Deficient Holocentromere in Lepidoptera Avoids Active Chromatin, Curr Biol 31(1) (2021) 173–181 e7. [DOI] [PubMed] [Google Scholar]
- [31].Cheeseman IM, Desai A, Molecular architecture of the kinetochore-microtubule interface, Nat Rev Mol Cell Biol 9(1) (2008) 33–46. [DOI] [PubMed] [Google Scholar]
- [32].Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, Cheeseman IM, Fukagawa T, CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore, Cell 135(6) (2008) 1039–52. [DOI] [PubMed] [Google Scholar]
- [33].Hara M, Fukagawa T, Critical Foundation of the Kinetochore: The Constitutive Centromere-Associated Network (CCAN), Prog Mol Subcell Biol 56 (2017) 29–57. [DOI] [PubMed] [Google Scholar]
- [34].Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LE, Cleveland DW, A two-step mechanism for epigenetic specification of centromere identity and function, Nat Cell Biol 15(9) (2013) 1056–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Liu ST, Rattner JB, Jablonski SA, Yen TJ, Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells, J Cell Biol 175(1) (2006) 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Logsdon GA, Barrey EJ, Bassett EA, DeNizio JE, Guo LY, Panchenko T, Dawicki-McKenna JM, Heun P, Black BE, Both tails and the centromere targeting domain of CENP-A are required for centromere establishment, J Cell Biol 208(5) (2015) 521–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH, Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores, Dev Cell 10(3) (2006) 303–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mendiburo MJ, Padeken J, Fulop S, Schepers A, Heun P, Drosophila CENH3 is sufficient for centromere formation, Science 334(6056) (2011) 686–90. [DOI] [PubMed] [Google Scholar]
- [39].Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR, HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore, J Cell Biol 194(2) (2011) 229–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Van Hooser AA, Ouspenski HC II, Gregson DA, Starr TJ, Yen ML, Goldberg K, Yokomori WC, Earnshaw KF, Sullivan BR Brinkley, Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A, J Cell Sci 114(Pt 19) (2001) 3529–42. [DOI] [PubMed] [Google Scholar]
- [41].Hori T, Shang WH, Takeuchi K, Fukagawa T, The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly, J Cell Biol 200(1) (2013) 45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF, In vitro centromere and kinetochore assembly on defined chromatin templates, Nature 477(7364) (2011) 354–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G, HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres, Cell 137(3) (2009) 485–97. [DOI] [PubMed] [Google Scholar]
- [44].Foltz DR, Jansen LE, Bailey AO, Yates JR 3rd, Bassett EA, Wood S, Black BE, Cleveland DW, Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP, Cell 137(3) (2009) 472–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bernad R, Sanchez P, Rivera T, Rodriguez-Corsino M, Boyarchuk E, Vassias I, Ray-Gallet D, Arnaoutov A, Dasso M, Almouzni G, Losada A, Xenopus HJURP and condensin II are required for CENP-A assembly, J Cell Biol 192(4) (2011) 569–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Aravind L, Iyer LM, Wu C, Domain architectures of the Scm3p protein provide insights into centromere function and evolution, Cell Cycle 6(20) (2007) 2511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE, Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization, Proc Natl Acad Sci U S A 104(25) (2007) 10571–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL, Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore, Mol Cell 26(6) (2007) 853–65. [DOI] [PubMed] [Google Scholar]
- [49].Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, Allshire RC, Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin, Mol Cell 33(3) (2009) 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C, Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes, Cell 129(6) (2007) 1153–64. [DOI] [PubMed] [Google Scholar]
- [51].Chen CC, Dechassa ML, Bettini E, Ledoux MB, Belisario C, Heun P, Luger K, Mellone BG, CAL1 is the Drosophila CENP-A assembly factor, J Cell Biol 204(3) (2014) 313–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Williams JS, Hayashi T, Yanagida M, Russell P, Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin, Mol Cell 33(3) (2009) 287–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC, Common ancestry of the CENP-A chaperones Scm3 and HJURP, Cell 137(7) (2009) 1173–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF, Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation, J Cell Biol 183(5) (2008) 805–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Schittenhelm RB, Althoff F, Heidmann S, Lehner CF, Detrimental incorporation of excess Cenp-A/Cid and Cenp-C into Drosophila centromeres is prevented by limiting amounts of the bridging factor Cal1, J Cell Sci 123(Pt 21) (2010) 3768–79. [DOI] [PubMed] [Google Scholar]
- [56].Rosin L, Mellone BG, Co-evolving CENP-A and CAL1 Domains Mediate Centromeric CENP-A Deposition across Drosophila Species, Dev Cell 37(2) (2016) 136–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Shuaib M, Ouararhni K, Dimitrov S, Hamiche A, HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres, Proc Natl Acad Sci U S A 107(4) (2010) 1349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, Li G, Xu RM, Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP, Genes Dev 25(9) (2011) 901–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bassett EA, DeNizio J, Barnhart-Dailey MC, Panchenko T, Sekulic N, Rogers DJ, Foltz DR, Black BE, HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly, Dev Cell 22(4) (2012) 749–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zhou Z, Feng H, Zhou BR, Ghirlando R, Hu K, Zwolak A, Miller Jenkins LM, Xiao H, Tjandra N, Wu C, Bai Y, Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3, Nature 472(7342) (2011) 234–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Black BE, Brock MA, Bedard S, Woods VL Jr., Cleveland DW, An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes, Proc Natl Acad Sci U S A 104(12) (2007) 5008–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL Jr., Cleveland DW, Structural determinants for generating centromeric chromatin, Nature 430(6999) (2004) 578–82. [DOI] [PubMed] [Google Scholar]
- [63].Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW, Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain, Mol Cell 25(2) (2007) 309–22. [DOI] [PubMed] [Google Scholar]
- [64].Goutte-Gattat D, Shuaib M, Ouararhni K, Gautier T, Skoufias DA, Hamiche A, Dimitrov S, Phosphorylation of the CENP-A amino-terminus in mitotic centromeric chromatin is required for kinetochore function, Proc Natl Acad Sci U S A 110(21) (2013) 8579–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M, Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1, Dev Cell 12(1) (2007) 17–30. [DOI] [PubMed] [Google Scholar]
- [66].Maddox PS, Hyndman F, Monen J, Oegema K, Desai A, Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin, J Cell Biol 176(6) (2007) 757–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Spiller F, Medina-Pritchard B, Abad MA, Wear MA, Molina O, Earnshaw WC, Jeyaprakash AA, Molecular basis for Cdk1-regulated timing of Mis18 complex assembly and CENP-A deposition, EMBO Rep 18(6) (2017) 894–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Pan D, Klare K, Petrovic A, Take A, Walstein K, Singh P, Rondelet A, Bird AW, Musacchio A, CDK-regulated dimerization of M18BP1 on a Mis18 hexamer is necessary for CENP-A loading, Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Korntner-Vetter M, Lefevre S, Hu XW, George R, Singleton MR, Subunit interactions and arrangements in the fission yeast Mis16-Mis18-Mis19 complex, Life Sci Alliance 2(4) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Perpelescu M, Hori T, Toyoda A, Misu S, Monma N, Ikeo K, Obuse C, Fujiyama A, Fukagawa T, HJURP is involved in the expansion of centromeric chromatin, Mol Biol Cell 26(15) (2015) 2742–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Wang J, Liu X, Dou Z, Chen L, Jiang H, Fu C, Fu G, Liu D, Zhang J, Zhu T, Fang J, Zang J, Cheng J, Teng M, Ding X, Yao X, Mitotic regulator Mis18beta interacts with and specifies the centromeric assembly of molecular chaperone holliday junction recognition protein (HJURP), J Biol Chem 289(12) (2014) 8326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Dambacher S, Deng W, Hahn M, Sadic D, Frohlich J, Nuber A, Hoischen C, Diekmann S, Leonhardt H, Schotta G, CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin, Nucleus 3(1) (2012) 101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Moree B, Meyer CB, Fuller CJ, Straight AF, CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly, J Cell Biol 194(6) (2011) 855–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].McKinley KL, Cheeseman IM, Polo-like kinase 1 licenses CENP-A deposition at centromeres, Cell 158(2) (2014) 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].French BT, Westhorpe FG, Limouse C, Straight AF, Xenopus laevis M18BP1 Directly Binds Existing CENP-A Nucleosomes to Promote Centromeric Chromatin Assembly, Dev Cell 42(2) (2017) 190–199 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Hori T, Shang WH, Hara M, Ariyoshi M, Arimura Y, Fujita R, Kurumizaka H, Fukagawa T, Association of M18BP1/KNL2 with CENP-A Nucleosome Is Essential for Centromere Formation in Non-mammalian Vertebrates, Dev Cell 42(2) (2017) 181–189 e3. [DOI] [PubMed] [Google Scholar]
- [77].Shelby RD, Vafa O, Sullivan KF, Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites, J Cell Biol 136(3) (1997) 501–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Jansen LE, Black BE, Foltz DR, Cleveland DW, Propagation of centromeric chromatin requires exit from mitosis, J Cell Biol 176(6) (2007) 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Schuh M, Lehner CF, Heidmann S, Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase, Curr Biol 17(3) (2007) 237–43. [DOI] [PubMed] [Google Scholar]
- [80].Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE, Cdk activity couples epigenetic centromere inheritance to cell cycle progression, Dev Cell 22(1) (2012) 52–63. [DOI] [PubMed] [Google Scholar]
- [81].Stankovic A, Guo LY, Mata JF, Bodor DL, Cao XJ, Bailey AO, Shabanowitz J, Hunt DF, Garcia BA, Black BE, Jansen LET, A Dual Inhibitory Mechanism Sufficient to Maintain Cell-Cycle-Restricted CENP-A Assembly, Mol Cell 65(2) (2017) 231–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Muller S, Montes de Oca R, Lacoste N, Dingli F, Loew D, Almouzni G, Phosphorylation and DNA binding of HJURP determine its centromeric recruitment and function in CenH3(CENP-A) loading, Cell Rep 8(1) (2014) 190–203. [DOI] [PubMed] [Google Scholar]
- [83].Falk SJ, Guo LY, Sekulic N, Smoak EM, Mani T, Logsdon GA, Gupta K, Jansen LE, Van Duyne GD, Vinogradov SA, Lampson MA, Black BE, Chromosomes. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere, Science 348(6235) (2015) 699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Smoak EM, Stein P, Schultz RM, Lampson MA, Black BE, Long-Term Retention of CENP-A Nucleosomes in Mammalian Oocytes Underpins Transgenerational Inheritance of Centromere Identity, Curr Biol 26(8) (2016) 1110–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S, Dynamics of inner kinetochore assembly and maintenance in living cells, J Cell Biol 180(6) (2008) 1101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Bodor DL, Valente LP, Mata JF, Black BE, Jansen LE, Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes, Mol Biol Cell 24(7) (2013) 923–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Mitra S, Srinivasan B, Jansen LET, Stable inheritance of CENP-A chromatin: Inner strength versus dynamic control, J Cell Biol 219(10) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Srivastava S, Foltz DR, Posttranslational modifications of CENP-A: marks of distinction, Chromosoma 127(3) (2018) 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Marescal O, Cheeseman IM, Cellular Mechanisms and Regulation of Quiescence, Dev Cell 55(3) (2020) 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Swartz SZ, McKay LS, Su KC, Bury L, Padeganeh A, Maddox PS, Knouse KA, Cheeseman IM, Quiescent Cells Actively Replenish CENP-A Nucleosomes to Maintain Centromere Identity and Proliferative Potential, Dev Cell 51(1) (2019) 35–48 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Lee SH, Itkin-Ansari P, Levine F, CENP-A, a protein required for chromosome segregation in mitosis, declines with age in islet but not exocrine cells, Aging (Albany NY) 2(11) (2010) 785–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Zhang W, Feng J, Li Q, The replisome guides nucleosome assembly during DNA replication, Cell Biosci 10 (2020) 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Dunleavy EM, Almouzni G, Karpen GH, H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G(1) phase, Nucleus 2(2) (2011) 146–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Nechemia-Arbely Y, Miga KH, Shoshani O, Aslanian A, McMahon MA, Lee AY, Fachinetti D, Yates JR 3rd, Ren B, Cleveland DW, DNA replication acts as an error correction mechanism to maintain centromere identity by restricting CENP-A to centromeres, Nat Cell Biol 21(6) (2019) 743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Hori T, Kagawa N, Toyoda A, Fujiyama A, Misu S, Monma N, Makino F, Ikeo K, Fukagawa T, Constitutive centromere-associated network controls centromere drift in vertebrate cells, J Cell Biol 216(1) (2017) 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Zasadzinska E, Huang J, Bailey AO, Guo LY, Lee NS, Srivastava S, Wong KA, French BT, Black BE, Foltz DR, Inheritance of CENP-A Nucleosomes during DNA Replication Requires HJURP, Dev Cell 47(3) (2018) 348–362 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Chan FL, Marshall OJ, Saffery R, Kim BW, Earle E, Choo KH, Wong LH, Active transcription and essential role of RNA polymerase II at the centromere during mitosis, Proc Natl Acad Sci U S A 109(6) (2012) 1979–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Bergmann JH, Jakubsche JN, Martins NM, Kagansky A, Nakano M, Kimura H, Kelly DA, Turner BM, Masumoto H, Larionov V, Earnshaw WC, Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function, J Cell Sci 125(Pt 2)(2012) 411–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC, Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore, EMBO J 30(2) (2011) 328–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Bury L, Moodie B, Ly J, McKay LS, Miga KH, Cheeseman IM, Alpha-satellite RNA transcripts are repressed by centromere-nucleolus associations, Elife 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Perea-Resa C, Blower MD, Centromere Biology: Transcription Goes on Stage, Mol Cell Biol 38(18) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Perea-Resa C, Bury L, Cheeseman IM, Blower MD, Cohesin Removal Reprograms Gene Expression upon Mitotic Entry, Mol Cell 78(1) (2020) 127–140 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Novais-Cruz M, Alba Abad M, van IWF, Galjart N, Jeyaprakash AA, Maiato H, Ferras C, Mitotic progression, arrest, exit or death relies on centromere structural integrity, rather than de novo transcription, Elife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR 3rd, Desai A, Fukagawa T, The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres, Nat Cell Biol 8(5) (2006) 446–57. [DOI] [PubMed] [Google Scholar]
- [105].Hori T, Okada M, Maenaka K, Fukagawa T, CENP-O class proteins form a stable complex and are required for proper kinetochore function, Mol Biol Cell 19(3) (2008) 843–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR 3rd, Cleveland DW, The human CENP-A centromeric nucleosome-associated complex, Nat Cell Biol 8(5) (2006) 458–69. [DOI] [PubMed] [Google Scholar]
- [107].Klare K, Weir JR, Basilico F, Zimniak T, Massimiliano L, Ludwigs N, Herzog F, Musacchio A, CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores, J Cell Biol 210(1) (2015) 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].McKinley KL, Sekulic N, Guo LY, Tsinman T, Black BE, Cheeseman IM, The CENP-L-N Complex Forms a Critical Node in an Integrated Meshwork of Interactions at the Centromere-Kinetochore Interface, Mol Cell 60(6) (2015) 886–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Petrovic A, Keller J, Liu Y, Overlack K, John J, Dimitrova YN, Jenni S, van Gerwen S, Stege P, Wohlgemuth S, Rombaut P, Herzog F, Harrison SC, Vetter IR, Musacchio A, Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores, Cell 167(4) (2016) 1028–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Screpanti E, De Antoni A, Alushin GM, Petrovic A, Melis T, Nogales E, Musacchio A, Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore, Curr Biol 21(5) (2011) 391–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM, CENP-C is a structural platform for kinetochore assembly, Curr Biol 21(5) (2011) 399–405. [DOI] [PubMed] [Google Scholar]
- [112].van Hooff JJ, Tromer E, van Wijk LM, Snel B, Kops GJ, Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics, EMBO Rep 18(9)(2017) 1559–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Tanaka K, Chang HL, Kagami A, Watanabe Y, CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis, Dev Cell 17(3) (2009) 334–43. [DOI] [PubMed] [Google Scholar]
- [114].Carroll CW, Milks KJ, Straight AF, Dual recognition of CENP-A nucleosomes is required for centromere assembly, J Cell Biol 189(7) (2010) 1143–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Weir JR, Faesen AC, Klare K, Petrovic A, Basilico F, Fischbock J, Pentakota S, Keller J, Pesenti ME, Pan D, Vogt D, Wohlgemuth S, Herzog F, Musacchio A, Insights from biochemical reconstitution into the architecture of human kinetochores, Nature 537(7619) (2016) 249–253. [DOI] [PubMed] [Google Scholar]
- [116].Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF, Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N, Nat Cell Biol 11(7) (2009) 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Basilico F, Maffini S, Weir JR, Prumbaum D, Rojas AM, Zimniak T, De Antoni A, Jeganathan S, Voss B, van Gerwen S, Krenn V, Massimiliano L, Valencia A, Vetter IR, Herzog F, Raunser S, Pasqualato S, Musacchio A, The pseudo GTPase CENP-M drives human kinetochore assembly, Elife 3 (2014) e02978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Pesenti ME, Prumbaum D, Auckland P, Smith CM, Faesen AC, Petrovic A, Erent M, Maffini S, Pentakota S, Weir JR, Lin YC, Raunser S, McAinsh AD, Musacchio A, Reconstitution of a 26-Subunit Human Kinetochore Reveals Cooperative Microtubule Binding by CENP-OPQUR and NDC80, Mol Cell 71(6) (2018) 923–939 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Nishino T, Takeuchi K, Gascoigne KE, Suzuki A, Hori T, Oyama T, Morikawa K, Cheeseman IM, Fukagawa T, CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold, Cell 148(3) (2012) 487–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Takeuchi K, Nishino T, Mayanagi K, Horikoshi N, Osakabe A, Tachiwana H, Hori T, Kurumizaka H, Fukagawa T, The centromeric nucleosome-like CENP-T-W-S-X complex induces positive supercoils into DNA, Nucleic Acids Res 42(3) (2014) 1644–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM, Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes, Cell 145(3) (2011) 410–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Schleiffer A, Maier M, Litos G, Lampert F, Hornung P, Mechtler K, Westermann S, CENP-T proteins are conserved centromere receptors of the Ndc80 complex, Nat Cell Biol 14(6) (2012) 604–13. [DOI] [PubMed] [Google Scholar]
- [123].Nishino T, Rago F, Hori T, Tomii K, Cheeseman IM, Fukagawa T, CENP-T provides a structural platform for outer kinetochore assembly, EMBO J 32(3) (2013) 424–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Bancroft J, Auckland P, Samora CP, McAinsh AD, Chromosome congression is promoted by CENP-Q-and CENP-E-dependent pathways, J Cell Sci 128(1) (2015) 171–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Hua S, Wang Z, Jiang K, Huang Y, Ward T, Zhao L, Dou Z, Yao X, CENP-U cooperates with Hec1 to orchestrate kinetochore-microtubule attachment, J Biol Chem 286(2) (2011) 1627–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Amaro AC, Samora CP, Holtackers R, Wang E, Kingston IJ, Alonso M, Lampson M, McAinsh AD, Meraldi P, Molecular control of kinetochore-microtubule dynamics and chromosome oscillations, Nat Cell Biol 12(4) (2010) 319–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Kang YH, Park CH, Kim TS, Soung NK, Bang JK, Kim BY, Park JE, Lee KS, Mammalian polo-like kinase 1-dependent regulation of the PBIP1-CENP-Q complex at kinetochores, J Biol Chem 286(22) (2011) 19744–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Kang YH, Park JE, Yu LR, Soung NK, Yun SM, Bang JK, Seong YS, Yu H, Garfield S, Veenstra TD, Lee KS, Self-regulated Plk1 recruitment to kinetochores by the Plk1-PBIP1 interaction is critical for proper chromosome segregation, Mol Cell 24(3) (2006) 409–22. [DOI] [PubMed] [Google Scholar]
- [129].Singh P, Pesenti ME, Maffini S, Carmignani S, Hedtfeld M, Petrovic A, Srinivasamani A, Bange T, Musacchio A, BUB1 and CENP-U, Primed by CDK1, Are the Main PLK1 Kinetochore Receptors in Mitosis, Mol Cell 81(1) (2021) 67–87 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Nguyen AL, Fadel MD, Cheeseman IM, Differential Requirements for the CENP-O Complex Reveal Parallel PLK1 Kinetochore Recruitment Pathways, Mol Biol Cell (2021) mbcE20110751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].McClelland SE, Borusu S, Amaro AC, Winter JR, Belwal M, McAinsh AD, Meraldi P, The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity, EMBO J 26(24) (2007) 5033–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Eskat A, Deng W, Hofmeister A, Rudolphi S, Emmerth S, Hellwig D, Ulbricht T, Doring V, Bancroft JM, McAinsh AD, Cardoso MC, Meraldi P, Hoischen C, Leonhardt H, Diekmann S, Step-wise assembly, maturation and dynamic behavior of the human CENP-P/O/R/Q/U kinetochore sub-complex, PLoS One 7(9) (2012) e44717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Gascoigne KE, Cheeseman IM, CDK-dependent phosphorylation and nuclear exclusion coordinately control kinetochore assembly state, J Cell Biol 201(1) (2013) 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Hellwig D, Munch S, Orthaus S, Hoischen C, Hemmerich P, Diekmann S, Live-cell imaging reveals sustained centromere binding of CENP-T via CENP-A and CENP-B, J Biophotonics 1(3) (2008) 245–54. [DOI] [PubMed] [Google Scholar]
- [135].Prendergast L, van Vuuren C, Kaczmarczyk A, Doering V, Hellwig D, Quinn N, Hoischen C, Diekmann S, Sullivan KF, Premitotic assembly of human CENPs -T and -W switches centromeric chromatin to a mitotic state, PLoS Biol 9(6) (2011) e1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Fukagawa T, Mikami Y, Nishihashi A, Regnier V, Haraguchi T, Hiraoka Y, Sugata N, Todokoro K, Brown W, Ikemura T, CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells, EMBO J 20(16) (2001) 4603–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Watanabe R, Hara M, Okumura EI, Herve S, Fachinetti D, Ariyoshi M, Fukagawa T, CDK1-mediated CENP-C phosphorylation modulates CENP-A binding and mitotic kinetochore localization, J Cell Biol 218(12) (2019) 4042–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Ariyoshi M, Makino F, Watanabe R, Nakagawa R, Kato T, Namba K, Arimura Y, Fujita R, Kurumizaka H, Okumura EI, Hara M, Fukagawa T, Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C, EMBO J (2021) e105671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Guo LY, Allu PK, Zandarashvili L, McKinley KL, Sekulic N, Dawicki-McKenna JM, Fachinetti D, Logsdon GA, Jamiolkowski RM, Cleveland DW, Cheeseman IM, Black BE, Centromeres are maintained by fastening CENP-A to DNA and directing an arginine anchor-dependent nucleosome transition, Nat Commun 8 (2017) 15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Pentakota S, Zhou K, Smith C, Maffini S, Petrovic A, Morgan GP, Weir JR, Vetter IR, Musacchio A, Luger K, Decoding the centromeric nucleosome through CENP-N, Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Hellwig D, Emmerth S, Ulbricht T, Doring V, Hoischen C, Martin R, Samora CP, McAinsh AD, Carroll CW, Straight AF, Meraldi P, Diekmann S, Dynamics of CENP-N kinetochore binding during the cell cycle, J Cell Sci 124(Pt 22) (2011) 3871–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Allu PK, Dawicki-McKenna JM, Van Eeuwen T, Slavin M, Braitbard M, Xu C, Kalisman N, Murakami K, Black BE, Structure of the Human Core Centromeric Nucleosome Complex, Curr Biol 29(16) (2019) 2625–2639 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Fang J, Liu Y, Wei Y, Deng W, Yu Z, Huang L, Teng Y, Yao T, You Q, Ruan H, Chen P, Xu RM, Li G, Structural transitions of centromeric chromatin regulate the cell cycle-dependent recruitment of CENP-N, Genes Dev 29(10) (2015) 1058–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Ohta S, Kimura M, Takagi S, Toramoto I, Ishihama Y, Identification of Mitosis-Specific Phosphorylation in Mitotic Chromosome-Associated Proteins, J Proteome Res 15(9) (2016) 3331–41. [DOI] [PubMed] [Google Scholar]
- [145].Gamba R, Fachinetti D, From evolution to function: Two sides of the same CENP-B coin?, Exp Cell Res 390(2) (2020) 111959. [DOI] [PubMed] [Google Scholar]
- [146].Dumont M, Gamba R, Gestraud P, Klaasen S, Worrall JT, De Vries SG, Boudreau V, Salinas-Luypaert C, Maddox PS, Lens SM, Kops GJ, McClelland SE, Miga KH, Fachinetti D, Human chromosome-specific aneuploidy is influenced by DNA-dependent centromeric features, EMBO J 39(2) (2020) e102924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW, DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function, Dev Cell 33(3) (2015) 314–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Fujita R, Otake K, Arimura Y, Horikoshi N, Miya Y, Shiga T, Osakabe A, Tachiwana H, Ohzeki J, Larionov V, Masumoto H, Kurumizaka H, Stable complex formation of CENP-B with the CENP-A nucleosome, Nucleic Acids Res 43(10) (2015) 4909–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Hasson D, Panchenko T, Salimian KJ, Salman MU, Sekulic N, Alonso A, Warburton PE, Black BE, The octamer is the major form of CENP-A nucleosomes at human centromeres, Nat Struct Mol Biol 20(6) (2013) 687–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Hoffmann S, Dumont M, Barra V, Ly P, Nechemia-Arbely Y, McMahon MA, Herve S, Cleveland DW, Fachinetti D, CENP-A Is Dispensable for Mitotic Centromere Function after Initial Centromere/Kinetochore Assembly, Cell Rep 17(9) (2016) 2394–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC, A super-resolution map of the vertebrate kinetochore, Proc Natl Acad Sci U S A 107(23) (2010) 10484–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Sullivan LL, Boivin CD, Mravinac B, Song IY, Sullivan BA, Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells, Chromosome Res 19(4) (2011) 457–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, Jansen LE, The quantitative architecture of centromeric chromatin, Elife 3 (2014) e02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Vargiu G, Makarov AA, Allan J, Fukagawa T, Booth DG, Earnshaw WC, Stepwise unfolding supports a subunit model for vertebrate kinetochores, Proc Natl Acad Sci U S A 114(12)(2017) 3133–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Schalch T, Steiner FA, Structure of centromere chromatin: from nucleosome to chromosomal architecture, Chromosoma 126(4) (2017) 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Maddox PS, Portier N, Desai A, Oegema K, Molecular analysis of mitotic chromosome condensation using a quantitative time-resolved fluorescence microscopy assay, Proc Natl Acad Sci U S A 103(41) (2006) 15097–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A, The conserved KMN network constitutes the core microtubule-binding site of the kinetochore, Cell 127(5) (2006) 983–97. [DOI] [PubMed] [Google Scholar]
- [158].Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR 3rd, Oegema K, Desai A, A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension, Genes Dev 18(18) (2004) 2255–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Shevchenko A, Hyman A, Oegema K, KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans, Genes Dev 17(19) (2003) 2421–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Kline SL, Cheeseman IM, Hori T, Fukagawa T, Desai A, The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation, J Cell Biol 173(1) (2006) 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Wigge PA, Kilmartin JV, The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation, J Cell Biol 152(2) (2001) 349–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Bharadwaj R, Qi W, Yu H, Identification of two novel components of the human NDC80 kinetochore complex, J Biol Chem 279(13) (2004) 13076–85. [DOI] [PubMed] [Google Scholar]
- [163].McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, Stukenberg PT, The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity, Genes Dev 17(1) (2003) 101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Obuse C, Iwasaki O, Kiyomitsu T, Goshima G, Toyoda Y, Yanagida M, A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1, Nat Cell Biol 6(11) (2004) 1135–41. [DOI] [PubMed] [Google Scholar]
- [165].Westermann S, Cheeseman IM, Anderson S, Yates JR 3rd, Drubin DG, Barnes G, Architecture of the budding yeast kinetochore reveals a conserved molecular core, J Cell Biol 163(2) (2003) 215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Nekrasov VS, Smith MA, Peak-Chew S, Kilmartin JV, Interactions between centromere complexes in Saccharomyces cerevisiae, Mol Biol Cell 14(12) (2003) 4931–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Pinsky BA, Tatsutani SY, Collins KA, Biggins S, An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase, Dev Cell 5(5) (2003) 735–45. [DOI] [PubMed] [Google Scholar]
- [168].DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED, Kinetochore microtubule dynamics and attachment stability are regulated by Hec1, Cell 127(5) (2006) 969–82. [DOI] [PubMed] [Google Scholar]
- [169].Rago F, Gascoigne KE, Cheeseman IM, Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T, Curr Biol 25(5) (2015) 671–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Gascoigne KE, Cheeseman IM, Kinetochore assembly: if you build it, they will come, Curr Opin Cell Biol 23(1) (2011) 102–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Cheeseman IM, Hori T, Fukagawa T, Desai A, KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates, Mol Biol Cell 19(2) (2008) 587–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Hara M, Ariyoshi M, Okumura EI, Hori T, Fukagawa T, Multiple phosphorylations control recruitment of the KMN network onto kinetochores, Nat Cell Biol 20(12) (2018) 1378–1388. [DOI] [PubMed] [Google Scholar]
- [173].Dimitrova YN, Jenni S, Valverde R, Khin Y, Harrison SC, Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly, Cell 167(4) (2016) 1014–1027 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Malvezzi F, Litos G, Schleiffer A, Heuck A, Mechtler K, Clausen T, Westermann S, A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors, EMBO J 32(3) (2013) 409–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Petrovic A, Pasqualato S, Dube P, Krenn V, Santaguida S, Cittaro D, Monzani S, Massimiliano L, Keller J, Tarricone A, Maiolica A, Stark H, Musacchio A, The MIS12 complex is a protein interaction hub for outer kinetochore assembly, J Cell Biol 190(5) (2010) 835–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Huis In ’t Veld PJ, Jeganathan S, Petrovic A, Singh P, John J, Krenn V, Weissmann F, Bange T, Musacchio A, Molecular basis of outer kinetochore assembly on CENP-T, Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Pekgoz Altunkaya G, Malvezzi F, Demianova Z, Zimniak T, Litos G, Weissmann F, Mechtler K, Herzog F, Westermann S, CCAN Assembly Configures Composite Binding Interfaces to Promote Cross-Linking of Ndc80 Complexes at the Kinetochore, Curr Biol 26(17) (2016) 2370–8. [DOI] [PubMed] [Google Scholar]
- [178].Gaitanos TN, Santamaria A, Jeyaprakash AA, Wang B, Conti E, Nigg EA, Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3, EMBO J 28(10) (2009) 1442–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Schmidt JC, Arthanari H, Boeszoermenyi A, Dashkevich NM, Wilson-Kubalek EM, Monnier N, Markus M, Oberer M, Milligan RA, Bathe M, Wagner G, Grishchuk EL, Cheeseman IM, The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments, Dev Cell 23(5) (2012) 968–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Jeyaprakash AA, Santamaria A, Jayachandran U, Chan YW, Benda C, Nigg EA, Conti E, Structural and functional organization of the Ska complex, a key component of the kinetochore-microtubule interface, Mol Cell 46(3) (2012) 274–86. [DOI] [PubMed] [Google Scholar]
- [181].Cheerambathur DK, Prevo B, Hattersley N, Lewellyn L, Corbett KD, Oegema K, Desai A, Dephosphorylation of the Ndc80 Tail Stabilizes Kinetochore-Microtubule Attachments via the Ska Complex, Dev Cell 41(4) (2017) 424–437 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Janczyk PL, Skorupka KA, Tooley JG, Matson DR, Kestner CA, West T, Pornillos O, Stukenberg PT, Mechanism of Ska Recruitment by Ndc80 Complexes to Kinetochores, Dev Cell 41(4) (2017) 438–449 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Welburn JP, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR 3rd, Cheeseman IM, The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility, Dev Cell 16(3) (2009) 374–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Auckland P, Clarke NI, Royle SJ, McAinsh AD, Congressing kinetochores progressively load Ska complexes to prevent force-dependent detachment, J Cell Biol 216(6) (2017)1623–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Gruber J, Harborth J, Schnabel J, Weber K, Hatzfeld M, The mitotic-spindle-associated protein astrin is essential for progression through mitosis, J Cell Sci 115(Pt 21) (2002) 4053–9. [DOI] [PubMed] [Google Scholar]
- [186].Manning AL, Bakhoum SF, Maffini S, Correia-Melo C, Maiato H, Compton DA, CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity, EMBO J 29(20) (2010) 3531–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Thein KH, Kleylein-Sohn J, Nigg EA, Gruneberg U, Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity, J Cell Biol 178(3) (2007) 345–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Schmidt JC, Kiyomitsu T, Hori T, Backer CB, Fukagawa T, Cheeseman IM, Aurora B kinase controls the targeting of the Astrin-SKAP complex to bioriented kinetochores, J Cell Biol 191(2) (2010) 269–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Kern DM, Monda JK, Su KC, Wilson-Kubalek EM, Cheeseman IM, Astrin-SKAP complex reconstitution reveals its kinetochore interaction with microtubule-bound Ndc80, Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Mack GJ, Compton DA, Analysis of mitotic microtubule-associated proteins using mass spectrometry identifies astrin, a spindle-associated protein, Proc Natl Acad Sci U S A 98(25) (2001) 14434–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Monda JK, Cheeseman IM, The kinetochore-microtubule interface at a glance, J Cell Sci 131(16) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Lawrence EJ, Zanic M, Rice LM, CLASPs at a glance, J Cell Sci 133(8) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Rieder CL, The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber, Int Rev Cytol 79 (1982) 1–58. [DOI] [PubMed] [Google Scholar]
- [194].McEwen BF, Arena JT, Frank J, Rieder CL, Structure of the colcemid-treated PtK1 kinetochore outer plate as determined by high voltage electron microscopic tomography, J Cell Biol 120(2) (1993) 301–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Magidson V, Paul R, Yang N, Ault JG, O’Connell CB, Tikhonenko I, McEwen BF, Mogilner A, Khodjakov A, Adaptive changes in the kinetochore architecture facilitate proper spindle assembly, Nat Cell Biol 17(9) (2015) 1134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Jokelainen PT, The ultrastructure and spatial organization of the metaphase kinetochore in mitotic rat cells, J Ultrastruct Res 19(1) (1967) 19–44. [DOI] [PubMed] [Google Scholar]
- [197].Magidson V, He J, Ault JG, O’Connell CB, Yang N, Tikhonenko I, McEwen BF, Sui H, Khodjakov A, Unattached kinetochores rather than intrakinetochore tension arrest mitosis in taxol-treated cells, J Cell Biol 212(3) (2016) 307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Cooke CA, Schaar B, Yen TJ, Earnshaw WC, Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase, Chromosoma 106(7) (1997) 446–55. [DOI] [PubMed] [Google Scholar]
- [199].Wordeman L, Steuer ER, Sheetz MP, Mitchison T, Chemical subdomains within the kinetochore domain of isolated CHO mitotic chromosomes, J Cell Biol 114(2) (1991) 285–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Rattner JB, Rao A, Fritzler MJ, Valencia DW, Yen TJ, CENP-F is a .ca 400 kDa kinetochore protein that exhibits a cell-cycle dependent localization, Cell Motil Cytoskeleton 26(3) (1993) 214–26. [DOI] [PubMed] [Google Scholar]
- [201].Basto R, Scaerou F, Mische S, Wojcik E, Lefebvre C, Gomes R, Hays T, Karess R, In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis, Curr Biol 14(1) (2004) 56–61. [DOI] [PubMed] [Google Scholar]
- [202].Starr DA, Williams BC, Hays TS, Goldberg ML, ZW10 helps recruit dynactin and dynein to the kinetochore, J Cell Biol 142(3) (1998) 763–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Rodriguez-Rodriguez JA, Lewis C, McKinley KL, Sikirzhytski V, Corona J, Maciejowski J, Khodjakov A, Cheeseman IM, Jallepalli PV, Distinct Roles of RZZ and Bub1-KNL1 in Mitotic Checkpoint Signaling and Kinetochore Expansion, Curr Biol 28(21) (2018) 3422–3429 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Pereira C, Reis RM, Gama JB, Celestino R, Cheerambathur DK, Carvalho AX, Gassmann R, Self-Assembly of the RZZ Complex into Filaments Drives Kinetochore Expansion in the Absence of Microtubule Attachment, Curr Biol 28(21) (2018) 3408–3421 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Sacristan C, Ahmad MUD, Keller J, Fermie J, Groenewold V, Tromer E, Fish A, Melero R, Carazo JM, Klumperman J, Musacchio A, Perrakis A, Kops GJ, Dynamic kinetochore size regulation promotes microtubule capture and chromosome biorientation in mitosis, Nat Cell Biol 20(7) (2018) 800–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Silio V, McAinsh AD, Millar JB, KNL1-Bubs and RZZ Provide Two Separable Pathways for Checkpoint Activation at Human Kinetochores, Dev Cell 35(5) (2015) 600–613. [DOI] [PubMed] [Google Scholar]
- [207].Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED, Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation, J Cell Biol 155(7) (2001) 1159–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Wojcik E, Basto R, Serr M, Scaerou F, Karess R, Hays T, Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein, Nat Cell Biol 3(11) (2001) 1001–7. [DOI] [PubMed] [Google Scholar]
- [209].Mische S, He Y, Ma L, Li M, Serr M, Hays TS, Dynein light intermediate chain: an essential subunit that contributes to spindle checkpoint inactivation, Mol Biol Cell 19(11) (2008) 4918–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Sivaram MV, Wadzinski TL, Redick SD, Manna T, Doxsey SJ, Dynein light intermediate chain 1 is required for progress through the spindle assembly checkpoint, EMBO J 28(7) (2009) 902–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Gassmann R, Holland AJ, Varma D, Wan X, Civril F, Cleveland DW, Oegema K, Salmon ED, Desai A, Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells, Genes Dev 24(9) (2010) 957–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Brown KD, Wood KW, Cleveland DW, The kinesin-like protein CENP-E is kinetochore-associated throughout poleward chromosome segregation during anaphase-A, J Cell Sci 109 (Pt 5) (1996) 961–9. [DOI] [PubMed] [Google Scholar]
- [213].Liao H, Winkfein RJ, Mack G, Rattner JB, Yen TJ, CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis, J Cell Biol 130(3) (1995) 507–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Lampson MA, Grishchuk EL, Mechanisms to Avoid and Correct Erroneous Kinetochore-Microtubule Attachments, Biology (Basel) 6(1) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Welburn JP, Vleugel M, Liu D, Yates JR 3rd, Lampson MA, Fukagawa T, Cheeseman IM, Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface, Mol Cell 38(3) (2010) 383–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Maciejowski J, Drechsler H, Grundner-Culemann K, Ballister ER, Rodriguez-Rodriguez JA, Rodriguez-Bravo V, Jones MJK, Foley E, Lampson MA, Daub H, McAinsh AD, Jallepalli PV, Mps1 Regulates Kinetochore-Microtubule Attachment Stability via the Ska Complex to Ensure Error-Free Chromosome Segregation, Dev Cell 41(2) (2017) 143–156 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Liu D, Vleugel M, Backer CB, Hori T, Fukagawa T, Cheeseman IM, Lampson MA, Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase, J Cell Biol 188(6) (2010) 809–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Posch M, Khoudoli GA, Swift S, King EM, Deluca JG, Swedlow JR, Sds22 regulates aurora B activity and microtubule-kinetochore interactions at mitosis, J Cell Biol 191(1)(2010) 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Schleicher K, Porter M, Ten Have S, Sundaramoorthy R, Porter IM, Swedlow JR, The Ndc80 complex targets Bod1 to human mitotic kinetochores, Open Biol 7(11) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR 3rd, Chan CS, Drubin DG, Barnes G, Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p, Cell 111(2) (2002) 163–72. [DOI] [PubMed] [Google Scholar]
- [221].DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF, Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites, Mol Biol Cell 16(2) (2005) 519–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].DeLuca KF, Lens SM, DeLuca JG, Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis, J Cell Sci 124(Pt 4) (2011) 622–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Musacchio A, The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics, Curr Biol 25(20) (2015) R1002–18. [DOI] [PubMed] [Google Scholar]
- [224].London N, Biggins S, Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint, Genes Dev 28(2) (2014) 140–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Musacchio A, Salmon ED, The spindle-assembly checkpoint in space and time, Nat Rev Mol Cell Biol 8(5) (2007) 379–93. [DOI] [PubMed] [Google Scholar]
- [226].London N, Biggins S, Signalling dynamics in the spindle checkpoint response, Nat Rev Mol Cell Biol 15(11) (2014) 736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Pachis ST, Kops G, Leader of the SAC: molecular mechanisms of Mps1/TTK regulation in mitosis, Open Biol 8(8) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Caldas GV, DeLuca JG, KNL1: bringing order to the kinetochore, Chromosoma 123(3) (2014) 169–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Dou Z, Prifti DK, Gui P, Liu X, Elowe S, Yao X, Recent Progress on the Localization of the Spindle Assembly Checkpoint Machinery to Kinetochores, Cells 8(3) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Wurzenberger C, Gerlich DW, Phosphatases: providing safe passage through mitotic exit, Nat Rev Mol Cell Biol 12(8) (2011) 469–82. [DOI] [PubMed] [Google Scholar]


